Abstract
Here we present a detailed protocol for in situ multiple fluorescence monitoring of adenosine-5′-triphosphate (ATP) and guanosine-5′-triphosphate (GTP) in MCF-7 breast cancer cells by using graphene oxide nanosheet (GO-nS) and DNA/RNA aptamers. FAM-labeled ATP aptamer and Cy5-modified GTP aptamer are used to construct the multiple aptamer/GO-nS sensing platform through 'π-π stacking' between aptamers and GO-nS. Binding of aptamers to GO-nS guarantees the fluorescence resonance energy transfer between fluorophores and GO-nS, resulting in 'fluorescence off'. When the aptamer/GO-nS are transported inside the cells via endocytosis, the conformation of the aptamers will change on interaction with cellular ATP and GTP. On the basis of the fluorescence 'off/on' switching, simultaneous sensing and imaging of ATP and GTP in vitro and in situ have been realized through fluorescence and confocal microscopy techniques. In this protocol, we describe the synthesis of GO and GO-nS, preparation of aptamer/GO-nS platform, in vitro detection of ATP and GTP, and how to use this platform to realize intracellular ATP and GTP imaging in cultured MCF-7 cells. The preparation of GO-nS is anticipated to take 7–14 d, and assays involving microscopy imaging and MCF-7 cells culturing can be performed in 2–3 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Herschman, H.R. Molecular imaging: looking at problems, seeing solutions. Science 302, 605–608 (2003).
Baker, M. Cellular imaging: taking a long, hard look. Nature 466, 1137–1140 (2010).
Massoud, T.F. & Gambhir, S.S. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 17, 545–580 (2003).
Imamura, H. et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. USA 106, 15651–15656 (2009).
Tsuboi, T., Lippiat, J.D., Ashcroft, F.M. & Rutter, G.A. ATP-dependent interaction of the cytosolic domains of the inwardly rectifying K+ channel Kir6.2 revealed by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 101, 76–81 (2004).
Berg, J., Hung, Y.P. & Yellen, G. A genetically encoded fluorescent reporter of ATP: ADP ratio. Nat. Methods 6, 161–166 (2009).
Wang, W.U., Chen, C., Lin, K.H., Fang, Y. & Lieber, C.M. Label-free detection of small-molecule-protein interactions by using nanowire nanosensors. Proc. Natl. Acad. Sci. USA 102, 3208–3212 (2005).
Bell, C.J., Manfredi, G., Griffiths, E.J. & Rutter, G.A. Luciferase expression for ATP imaging: application to cardiac myocytes. Methods Cell Biol. 80, 341–352 (2007).
Jose, D.A. et al. Colorimetric sensor for ATP in aqueous solution. Org. Lett. 9, 1979–1982 (2007).
Lee, D.H., Kim, S.Y. & Hong, J.I. A fluorescent pyrophosphate sensor with high selectivity over ATP in water. Angew. Chem. Int. Ed. 43, 4777–4780 (2004).
Sancenón, F., Descalzo, A.B., Martínez-Mánñz, R., Miranda, M.A. & Soto, J. A colorimetric ATP sensor based on 1,3,5-triarylpent-2-en-1,5-diones. Angew. Chem. Int. Ed. 40, 2640–2643 (2001).
Mizukami, S., Nagano, T., Urano, Y., Odani, A. & Kikuchi, K. A fluorescent anion sensor that works in neutral aqueous solution for bioanalytical application. J. Am. Chem. Soc. 124, 3920–3925 (2002).
Schneider, S.E., O'Neil, S.N. & Anslyn, E.V. Coupling rational design with libraries leads to the production of an ATP selective chemosensor. J. Am. Chem. Soc. 122, 542–543 (2000).
Wang, S.L. & Chang, Y.T. Combinatorial synthesis of benzimidazolium dyes and its diversity directed application toward GTP-selective fluorescent chemosensors. J. Am. Chem. Soc. 128, 10380–10381 (2006).
McCleskey, S.C., Griffin, M.J., Schneider, S.E., McDevitt, J.T. & Anslyn, E.V. Differential receptors create patterns diagnostic for ATP and GTP. J. Am. Chem. Soc. 125, 1114–1115 (2003).
Kwon, J.Y. et al. Fluorescent GTP-sensing in aqueous solution of physiological pH. J. Am. Chem. Soc. 126, 8892–8893 (2004).
Neelakandan, P.P., Hariharan, M. & Ramaiah, D. A supramolecular ON-OFF-ON fluorescence assay for selective recognition of GTP. J. Am. Chem. Soc. 128, 11334–11335 (2006).
Xu, Z. et al. Unique sandwich stacking of pyrene-adenine-pyrene for selective and ratiometric fluorescent sensing of ATP at physiological pH. J. Am. Chem. Soc. 131, 15528–15533 (2009).
Ojida, A. et al. Bis(Dpa-Zn-II) appended xanthone: excitation ratiometric chemosensor for phosphate anions. Angew. Chem. Int. Ed. 45, 5518–5521 (2006).
Li, C., Numata, M., Takeuchi, M. & Shinkai, S. A sensitive colorimetric and fluorescent probe based on a polythiophene derivative for the detection of ATP. Angew. Chem. Int. Ed. 44, 6371–6374 (2005).
Nutiu, R. & Li, Y.F. Structure-switching signalling aptamers. J. Am. Chem. Soc. 125, 4771–4778 (2003).
Nutiu, R. & Li, Y.F. In vitro selection of structure-switching signaling aptamers. Angew. Chem. Int. Ed. 117, 1085–1089 (2005).
Li, N. & Ho, C.-M. Aptamer-based optical probes with separated molecular recognition and signal transduction modules. J. Am. Chem. Soc. 130, 2380–2381 (2008).
Liu, J. & Lu, Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. Int. Ed. 45, 90–94 (2006).
Zayats, M., Huang, Y., Gill, R., Ma, C. & Willner, I. Label-free and reagentless aptamer-based sensors for small molecules. J. Am. Chem. Soc. 128, 13666–13667 (2006).
Geim, A.K. Graphene: status and prospects. Science 324, 1530–1534 (2009).
Liu, X.Q., Aizen, R., Freeman, R., Yehezkeli, O. & Willner, I. Multiplexed aptasensors and amplified DNA sensors using functionalized graphene oxide: application for logic gate operations. ACS Nano 6, 3553–3563 (2012).
Liu, B.W., Sun, Z.Y., Zhang, X. & Liu, J.W. Mechanisms of DNA sensing on graphene oxide. Anal. Chem. 85, 7987–7993 (2013).
Huang, P.J. & Liu, J.W. Molecular beacon lighting up on graphene oxide. Anal. Chem. 84, 4192–4198 (2012).
Chen, D., Feng, H. & Li, J. Graphene oxide: preparation, functionalization, and electrochemical applications. Chem. Rev. 112, 6027–6053 (2012).
Wang, Y., Li, Z.H., Wang, J., Li, J.H. & Lin, Y.H. Graphene and graphene oxide: biofunctionalization and applications in biotechnology. Trends Biotechnol. 29, 205–212 (2011).
Wang, Y., Lu, J., Tang, L.H., Chang, H.X. & Li, J.H. Graphene oxide amplified electrogenerated chemiluminescence of quantum dots and its selective sensing for glutathione from thiol-containing compounds. Anal. Chem. 81, 9710–9715 (2009).
Liu, Z., Robinson, J.T., Sun, X.M. & Dai, H.J. PEGylated nanographene oxide for delivery of water-insoluble cancer drugs. J. Am. Chem. Soc. 130, 10876–10877 (2008).
Lu, C.H. et al. Using graphene to protect DNA from cleavage during cellular delivery. Chem. Commun. 46, 3116–3118 (2010).
Chang, H.X., Tang, L.H., Wang, Y., Jiang, J.H. & Li, J.H. Graphene fluorescence resonance energy transfer aptasensor for the thrombin detection. Anal. Chem. 82, 2341–2346 (2010).
Wang, Y. et al. Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells,. J. Am. Chem. Soc. 132, 9274–9276 (2010).
Nel, A.E. et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mat. 8, 543–557 (2009).
Wang, Y. et al. In situ live cell sensing of multiple nucleotides exploiting DNA/RNA aptamers and graphene oxide nanosheets. Anal. Chem. 85, 6775–6782 (2013).
Liu, J., Cao, Z. & Lu, Y. Functional nucleic acid sensors. Chem. Rev. 109, 1948–1998 (2009).
Fang, X. & Tan, W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc. Chem. Res. 43, 48–57 (2010).
Huizenga, D.E. & Szostak, J.W. A DNA aptamer that binds adenosine and ATP. Biochemistry 34, 656–665 (1995).
Carothers, J.M., Oestreich, S.C. & Szostak, J.W. Aptamers selected for higher-affinity binding are not more specific for the target ligand. J. Am. Chem. Soc. 128, 7929–7937 (2006).
Davis, J.H. & Szostak, J.W. Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc. Natl. Acad. Sci. USA 99, 11616–11621 (2002).
Carothers, J.M., Oestreich, S.C., Davis, J.H. & Szostak, J.W. Informational complexity and functional activity of RNA structures. J. Am. Chem. Soc. 126, 5130–5137 (2004).
Carothers, J.M., Davis, J.H., Chou, J.J. & Szostak, J.W. Solution structure of an informationally complex high-affinity RNA aptamer to GTP. RNA 12, 567–579 (2006).
Li, Q. & Chang, Y.T. A protocol for preparing, characterizing and using three RNA-specific, live cell imaging probes: E36, E144 and F22. Nat. Protoc. 1, 2922–2932 (2006).
Pittet, M.J., Swirski, F.K., Reynolds, F., Josephson, L. & Weisslede, R. Labeling of immune cells for in vivo imaging using magnetofluorescent nanoparticles. Nat. Protoc. 1, 74–79 (2006).
Zrazhevskiy, P., True, L.D. & Gao, X.H. Multicolor multicycle molecular profiling with quantum dots for single-cell analysis. Nat. Protoc. 8, 1852–1869 (2013).
Robinson, K.M., Janes, M.S. & Beckman, J.S. The selective detection of mitochondrial superoxide by live-cell imaging. Nat. Protoc. 3, 941–947 (2008).
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (no. 21235004, no. 21327806, no. 21305046), the National Basic Research Program of China (No. 2011CB935704) and the Tsinghua University Initiative Scientific Research Program. This work was also partially supported by a laboratory-directed research and development program at Pacific Northwest National Laboratory (PNNL). We are very grateful to T.J. Weber, D. Hu, C.-T. Lin and A.S. Lea (PNNL) for their help in experimental work and for helpful discussion.
Author information
Authors and Affiliations
Contributions
Y.W., Z.L., J.L. and Y.L. designed the protocol and carried out the experiments. Y.W., L.T. and J.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Characterization of GO-nS.
(a) AFM images of GO-nS. (b) Cross-sectional plot indicated with the blue line, lateral average over 400 nm. (c) TEM image of GO-nS dispersed on a carbon film. (d) Size distribution histogram of GO-nS. This figure is reproduced with permission from our previously published work38, copyright 2013 American Chemical Society.
Supplementary Figure 2 XRD pattern of GO-nS.
Powder X-ray diffraction (XRD) measurements were performed on Bruker D8-Advance X-ray powder diffract meter using a graphite monochrometer with Cu Ka radiation (k = 1.5406 Å). This figure is reproduced with permission from our previously published work38, copyright 2013 American Chemical Society.
Supplementary Figure 3 Raman spectra of GO-nS.
Raman spectra were obtained using a confocal microprobe Raman system (Renishaw, RM2000). This figure is reproduced with permission from our previously published work38, copyright 2013 American Chemical Society.
Supplementary Figure 4 FTIR spectrum of GO-nS.
The FTIR spectrum (800-4,000 cm-1) was measured using a Perkin Elmer Fourier transform infrared spectroscopy spectrometer with pure KBr as the background. This figure is reproduced with permission from our previously published work38, copyright 2013 American Chemical Society.
Supplementary Figure 5 Single nucleotide detection based on single-aptamer/GO-nS complex.
(a) Fluorescence emission spectra of 100 nM ATP aptamer-FAM quenched with 3 μg/mL GO-nS (black bottom line) and fluorescence recovery by addition of ATP with concentration ranging from 10 μM to 2.5 mM (from bottom to top). Inset: linear relationship between (F-F0)/F0 (where F0 and F are the fluorescence intensity without and with the presence of ATP, respectively) and ATP concentration. (b) Fluorescence emission spectra of 100 nM GTP aptamer-Cy5 quenched with 3μg/mL GO-nS (red bottom line) and fluorescence recovery by addition of GTP with concentration ranging from 0.01 to 2 mM (from bottom to top) in reaction buffer for 1 h at 25 °C. Inset: linear relationship between (F-F0)/F0 (relative fluorescence intensity, where F0 and F are the fluorescence intensity without and with the presence of GTP) and GTP concentration. Error bars were obtained from three parallel experiments. This figure is reproduced from published work37,38. Copyright 2010 and 2013, American Chemical Society.
Supplementary Figure 6 High-resolution cell image.
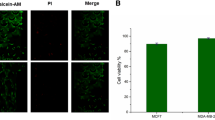
MCF-7 cells were cultured with 100 nM aptamer-FAM/GO-nS for 6 h with recommended media at 37 °C. Picture A and B were captured from two parallel experiments under the same operation conditions, respectively.
(Fluorescence imaging was carried out on an intensified charge-coupled device(ICCD)-based real-time fluorescence microscopy system, which was composed mainly of a TE2000 inverted microscope (Nikon, Japan) with a Nikon 100× N.A. 1.3 oil objective, an I-PentaMAX Gen IV ICCD camera (Roper Scientific, USA), and an xenon arc lamp in the Lambda DG-4 Wavelength Switcher (Sutter Instrument, USA). Selective excitation was produced through a 484 ± 7.5 nm band-pass filter. Fluorescence was monitored and quantitatively analyzed with the MetaMorph software (Universal Imaging Co., USA). Excitation light was adequately attenuated. Photo bleaching and cell damage were minimized by auto-synchronizing the exciting shutter and the exposure shutter, i.e. cells were exposed to light only during exposure but not at the interval. This ICCD fluorescence micro-imaging system possessed the advantages of high-performance thermoelectric cooling function to reduce noise, and a gain high enough to detect extremely low light.)
Supplementary Figure 7 Cleavage protection assay of GO-nS.
Agarose gel electrophoresis image for enzymatic cleavage protection assay: lane 1, DNA size ladder for 100 bp; lane 2, aptamer-FAM; lane 3, aptamer-FAM reacted with DNase I for 15 min; lane 4, aptamer-FAM reacted with DNase I for 40 min; lane 5, aptamer-FAM/GO-nS; lane 6, aptamer-FAM/GO-nS incubated with DNase I for 15 min; lane 7, aptamer-FAM/GO-nS incubated with DNase I for 40 min. Aptamer concentration is 100 nM, GO-nS is 3 μg/mL, and DNase I is 0.2 units/μL. Excitation wavelength for SYBR Green I: 494 nm.
Supplementary Figure 8 Cell viability assay.
Cell viability determined using a trypan blue assay after treatment of MCF-7 cells with 1 – 9 μg/ml GO-nS for 12 h (red), 24 h (green) or 72 h (blue). Values represent the mean ± se, n=3. *Significantly different from control, p < 0.05. This figure is adapted with permission from our previously published work38, copyright 2013 American Chemical Society.
Supplementary information
Supplementary Figure 1
Characterization of GO-nS. (PDF 144 kb)
Supplementary Figure 2
XRD pattern of GO-nS. (PDF 66 kb)
Supplementary Figure 3
Raman spectra of GO-nS. (PDF 81 kb)
Supplementary Figure 4
FTIR spectrum of GO-nS.. (PDF 93 kb)
Supplementary Figure 5
Single nucleotide detection based on single-aptamer/GO-nS complex. (PDF 115 kb)
Supplementary Figure 6
High-resolution cell image. (PDF 499 kb)
Supplementary Figure 7
Cleavage protection assay of GO-nS. (PDF 112 kb)
Supplementary Figure 8
Cell viability assay. (PDF 110 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Tang, L., Li, Z. et al. In situ simultaneous monitoring of ATP and GTP using a graphene oxide nanosheet–based sensing platform in living cells. Nat Protoc 9, 1944–1955 (2014). https://doi.org/10.1038/nprot.2014.126
Published:
Issue date:
DOI: https://doi.org/10.1038/nprot.2014.126
This article is cited by
-
Combination of multiple nucleic acid aptamers for precision detection of tumors based on optical methods
Journal of Cancer Research and Clinical Oncology (2023)
-
Homogeneous luminescent quantitation of cellular guanosine and adenosine triphosphates (GTP and ATP) using QT-LucGTP&ATP assay
Analytical and Bioanalytical Chemistry (2023)
-
Catalytic nanozyme Zn/Cl-doped carbon quantum dots as ratiometric fluorescent probe for sequential on-off-on detection of riboflavin, Cu2+ and thiamine
Scientific Reports (2022)
-
Simultaneous probing of dual intracellular metabolites (ATP and paramylon) in live microalgae using graphene oxide/aptamer nanocomplex
Microchimica Acta (2022)
-
A fluorescence nanoplatform for the determination of hydrogen peroxide and adenosine triphosphate via tuning of the peroxidase-like activity of CuO nanoparticle decorated UiO-66
Microchimica Acta (2022)