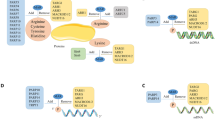
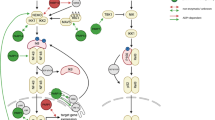
Abstract
ADP-ribosylation of proteins was first described in the early 1960's, and today the function and regulation of poly(ADP-ribosyl)ation (PARylation) is partially understood. By contrast, little is known about intracellular mono(ADP-ribosyl)ation (MARylation) by ADP-ribosyl transferase (ART) enzymes, such as ARTD10. Recent findings indicate that MARylation regulates signalling and transcription by modifying key components in these processes. Emerging evidence also suggests that specific macrodomain-containing proteins, including ARTD8, macroD1, macroD2 and C6orf130, which are distinct from those affecting PARylation, interact with MARylation on target proteins to 'read' and 'erase' this modification. Thus, studying macrodomain-containing proteins is key to understanding the function and regulation of MARylation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
28 June 2013
On page 443 of this article, the incorrect references were cited at the end of the following sentence: "Four years ago, macrodomains were shown to be able to bind PAR that has been synthesized in response to DNA damage18,19,20." The authors meant to refer to references 20, 25 and 26 at the end of this sentence. This has been corrected online. The authors apologize to the authors of references 25 and 26 for this mistake and for any confusion caused to readers.
References
Holbourn, K. P., Shone, C. C. & Acharya, K. R. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 273, 4579–4593 (2006).
Corda, D. & Di Girolamo, M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 22, 1953–1958 (2003).
Yates, S. P., Jorgensen, R., Andersen, G. R. & Merrill, A. R. Stealth and mimicry by deadly bacterial toxins. Trends Biochem. Sci. 31, 123–133 (2006).
Otto, H. et al. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics 6, 139 (2005).
Hottiger, M. O., Hassa, P. O., Luscher, B., Schuler, H. & Koch-Nolte, F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 35, 208–219 (2010).
Schreiber, V., Dantzer, F., Ame, J. C. & de Murcia, G. Poly(ADP-ribose): novel functions for an old molecule. Nature Rev. Mol. Cell Biol. 7, 517–528 (2006).
Gibson, B. A. & Kraus, W. L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nature Rev. Mol. Cell Biol. 13, 411–424 (2012).
Kleine, H. et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell 32, 57–69 (2008).
Altmeyer, M., Messner, S., Hassa, P. O., Fey, M. & Hottiger, M. O. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 37, 3723–3738 (2009).
Haenni, S. S. et al. Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int. J. Biochem. Cell Biol. 40, 2274–2283 (2008).
Messner, S. et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 38, 6350–6362 (2010).
Chapman, J. D., Gagne, J. P., Poirier, G. G. & Goodlett, D. R. Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 12, 1868–1880 (2013).
Sharifi, R. et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 32, 1225–1237 (2013).
Tao, Z., Gao, P. & Liu, H. W. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J. Am. Chem. Soc. 131, 14258–14260 (2009).
Kalisch, T., Ame, J. C., Dantzer, F. & Schreiber, V. New readers and interpretations of poly(ADP-ribosyl)ation. Trends Biochem. Sci. 37, 381–390 (2012).
Zaja, R., Mikoc, A., Barkauskaite, E. & Ahel, I. Molecular insights into poly(ADP-ribose) recognition and processing. Biomolecules 3, 1–17 (2012).
Pehrson, J. R. & Fried, V. A. MacroH2A, a core histone containing a large nonhistone region. Science 257, 1398–1400 (1992).
Li, G. Y. et al. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl Acad. Sci. USA 107, 9129–9134 (2010).
Ahel, I. et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451, 81–85 (2008).
Ahel, D. et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325, 1240–1243 (2009).
Slade, D. et al. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 477, 616–620 (2011).
Kim, I. K. et al. Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element. Nature Struct. Mol. Biol. 19, 653–656 (2012).
Dunstan, M. S. et al. Structure and mechanism of a canonical poly(ADP-ribose) glycohydrolase. Nature Commun. 3, 878 (2012).
Satoh, M. S. & Lindahl, T. Role of poly(ADP-ribose) formation in DNA repair. Nature 356, 356–358 (1992).
Gottschalk, A. J. et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl Acad. Sci. USA 106, 13770–13774 (2009).
Timinszky, G. et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nature Struct. Mol. Biol. 16, 923–929 (2009).
Kleine, H. & Luscher, B. Learning how to read ADP-ribosylation. Cell 139, 17–19 (2009).
Huambachano, O., Herrera, F., Rancourt, A. & Satoh, M. S. Double-stranded DNA binding domain of poly(ADP-ribose) polymerase-1 and molecular insight into the regulation of its activity. J. Biol. Chem. 286, 7149–7160 (2011).
Hsiao, S. J. & Smith, S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 90, 83–92 (2008).
Aravind, L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 26, 273–275 (2001).
Callow, M. G. et al. Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS ONE 6, e22595 (2011).
Zhang, Y. et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nature Cell Biol. 13, 623–629 (2011).
MacDonald, B. T., Tamai, K. & He, X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 (2009).
Pleschke, J. M., Kleczkowska, H. E., Strohm, M. & Althaus, F. R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275, 40974–40980 (2000).
Gagne, J. P. et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 36, 6959–6976 (2008).
Krietsch, J. et al. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Aspects Med. 23 Dec 2012 (10.1016/j.mam.2012.12.005).
Tucker, J. A. et al. Structures of the human poly (ADP-ribose) glycohydrolase catalytic domain confirm catalytic mechanism and explain inhibition by ADP-HPD derivatives. PLoS ONE 7, e50889 (2012).
Oka, S., Kato, J. & Moss, J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 281, 705–713 (2006).
Mueller-Dieckmann, C. et al. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl Acad. Sci. USA 103, 15026–15031 (2006).
Rosenthal, F. et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nature Struct. Mol. Biol. 20, 502–507 (2013).
Hottiger, M. O. et al. Progress in the function and regulation of ADP-ribosylation. Sci. Signal. 4, mr5 (2011).
Yu, M. et al. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene 24, 1982–1993 (2005).
Kleine, H. et al. Dynamic subcellular localization of the mono-ADP-ribosyltransferase ARTD10 and interaction with the ubiquitin receptor p62. Cell Commun. Signal. 10, 28 (2012).
Verheugd, P. et al. Regulation of NF-κB signaling by the mono-ADP-ribosyltransferase ARTD10. Nature Commun. 4, 1683 (2013).
Chen, Z. J. Ubiquitination in signaling to and activation of IKK. Immunol. Rev. 246, 95–106 (2012).
Herzog, N. et al. Caspase-dependent cleavage of the mono-ADP-ribosyltransferase ARTD10 interferes with its pro-apoptotic function. FEBS J. 280, 13 (2013).
Feijs, K. L. et al. ARTD10 substrate identification on protein microarrays: regulation of GSK3β by mono-ADP-ribosylation. Cell Commun. Signal. 11, 5 (2013).
Wu, D. & Pan, W. GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem. Sci. 35, 161–168 (2010).
Goenka, S. & Boothby, M. Selective potentiation of Stat-dependent gene expression by collaborator of Stat6 (CoaSt6), a transcriptional cofactor. Proc. Natl Acad. Sci. USA 103, 4210–4215 (2006).
Mehrotra, P. et al. PARP-14 functions as a transcriptional switch for Stat6-dependent gene activation. J. Biol. Chem. 286, 1767–1776 (2011).
Goenka, S., Cho, S. H. & Boothby, M. Collaborator of Stat6 (CoaSt6)-associated poly(ADP-ribose) polymerase activity modulates Stat6-dependent gene transcription. J. Biol. Chem. 282, 18732–18739 (2007).
Yang, J. et al. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 21, 4950–4958 (2002).
Mehrotra, P. et al. Poly (ADP-ribose) polymerase 14 and its enzyme activity regulates TH2 differentiation and allergic airway disease. J. Allergy Clin. Immunol. 131, 521–531.e12 (2013).
Macpherson, L. et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 41, 1604–1621 (2012).
Jwa, M. & Chang, P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1α-mediated unfolded protein response. Nature Cell Biol. 14, 1223–1230 (2012).
Di Paola, S., Micaroni, M., Di Tullio, G., Buccione, R. & Di Girolamo, M. PARP16/ARTD15 is a novel endoplasmic-reticulum-associated mono-ADP-ribosyltransferase that interacts with, and modifies karyopherin-ss1. PLoS ONE 7, e37352 (2012).
Hetz, C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nature Rev. Mol. Cell Biol. 13, 89–102 (2012).
Karlberg, T., Thorsell, A. G., Kallas, A. & Schuler, H. Crystal structure of human ADP-ribose transferase ARTD15/PARP16 reveals a novel putative regulatory domain. J. Biol. Chem. 287, 24077–24081 (2012).
Anantharaman, V., Koonin, E. V. & Aravind, L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 30, 1427–1464 (2002).
Till, S. & Ladurner, A. G. Sensing NAD metabolites through macro domains. Front. Biosci. 14, 3246–3258 (2009).
Han, W., Li, X. & Fu, X. The macro domain protein family: structure, functions, and their potential therapeutic implications. Mutat. Res. 727, 86–103 (2011).
Karras, G. I. et al. The macro domain is an ADP-ribose binding module. EMBO J. 24, 1911–1920 (2005).
Chen, D. et al. Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J. Biol. Chem. 286, 13261–13271 (2011).
Peterson, F. C. et al. Orphan macrodomain protein (human C6orf130) is an O-acyl-ADP-ribose deacylase: solution structure and catalytic properties. J. Biol. Chem. 286, 35955–35965 (2011).
Tong, L. & Denu, J. M. Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose. Biochim. Biophys. Acta 1804, 1617–1625 (2010).
Neuvonen, M. & Ahola, T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 385, 212–225 (2009).
Egloff, M. P. et al. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 80, 8493–8502 (2006).
Saikatendu, K. S. et al. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1′′-phosphate dephosphorylation by a conserved domain of nsP3. Structure 13, 1665–1675 (2005).
Shull, N. P., Spinelli, S. L. & Phizicky, E. M. A highly specific phosphatase that acts on ADP-ribose 1′′-phosphate, a metabolite of tRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res. 33, 650–660 (2005).
Putics, A., Filipowicz, W., Hall, J., Gorbalenya, A. E. & Ziebuhr, J. ADP-ribose-1”-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 79, 12721–12731 (2005).
Pines, A. et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J. Cell Biol. 199, 235–249 (2012).
Aguiar, R. C. et al. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood 96, 4328–4334 (2000).
Takeyama, K. et al. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J. Biol. Chem. 278, 21930–21937 (2003).
Yan, Q. et al. BAL1 and its partner E3 ligase, BBAP, link PARP activation, ubiquitylation and double-strand DNA repair independent of ATM, MDC1 and RNF8. Mol Cell. Biol. 33, 845–857 (2013).
Aguiar, R. C., Takeyama, K., He, C., Kreinbrink, K. & Shipp, M. A. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 280, 33756–33765 (2005).
Jankevicius, G. et al. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nature Struct. Mol. Biol. 20, 508–514 (2013).
Gagne, J. P. et al. Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res. 40, 7788–7805 (2012).
Dani, N. et al. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc. Natl Acad. Sci. USA 106, 4243–4248 (2009).
Forst, A. H. et al. Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure 21, 462–475 (2013).
Filippakopoulos, P. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012).
Ono, T., Kasamatsu, A., Oka, S. & Moss, J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc. Natl Acad. Sci. USA 103, 16687–16691 (2006).
Moss, J., Oppenheimer, N. J., West, R. E. Jr & Stanley, S. J. Amino acid specific ADP-ribosylation: substrate specificity of an ADP-ribosylarginine hydrolase from turkey erythrocytes. Biochemistry 25, 5408–5414 (1986).
Wang, Y. et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci. Signal. 4, ra20 (2011).
Williams, J. C., Chambers, J. P. & Liehr, J. G. Glutamyl ribose 5-phosphate storage disease. A hereditary defect in the degradation of poly(ADP-ribosylated) proteins. J. Biol. Chem. 259, 1037–1042 (1984).
Williams, J. C. et al. Progressive neurologic deterioration and renal failure due to storage of glutamyl ribose-5-phosphate. N. Engl. J. Med. 311, 152–155 (1984).
Allen, M. D., Buckle, A. M., Cordell, S. C., Lowe, J. & Bycroft, M. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330, 503–511 (2003).
Kustatscher, G., Hothorn, M., Pugieux, C., Scheffzek, K. & Ladurner, A. G. Splicing regulates NAD metabolite binding to histone macroH2A. Nature Struct. Mol. Biol. 12, 624–625 (2005).
Patel, C. N., Koh, D. W., Jacobson, M. K. & Oliveira, M. A. Identification of three critical acidic residues of poly(ADP-ribose) glycohydrolase involved in catalysis: determining the PARG catalytic domain. Biochem. J. 388, 493–500 (2005).
Cho, S. H. et al. PARP-14, a member of the B aggressive lymphoma family, transduces survival signals in primary B cells. Blood 113, 2416–2425 (2009).
Ma, N. F. et al. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology 47, 503–510 (2008).
Brockschmidt, A. et al. CHD1L: a new candidate gene for congenital anomalies of the kidneys and urinary tract (CAKUT). Nephrol. Dial. Transplant. 27, 2355–2364 (2012).
Kapoor, A. et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 468, 1105–1109 (2010).
Sporn, J. C. et al. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene 28, 3423–3428 (2009).
Park, E. & Griffin, D. E. The nsP3 macro domain is important for Sindbis virus replication in neurons and neurovirulence in mice. Virology 388, 305–314 (2009).
Chaurushiya, M. S. & Weitzman, M. D. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amst.) 8, 1166–1176 (2009).
Haince, J. F., Ouellet, M. E., McDonald, D., Hendzel, M. J. & Poirier, G. G. Dynamic relocation of poly(ADP-ribose) glycohydrolase isoforms during radiation-induced DNA damage. Biochim. Biophys. Acta 1763, 226–237 (2006).
Acknowledgements
The authors thank their colleagues for insightful discussions and A. Ladurner and I. Ahel for providing manuscripts prior to publication. They apologize to researchers whose work could not be included owing to space restrictions. The work in the author's laboratory was supported by the START program of the Medical School of the Rheinisch-Westfaelische Technische Hochschule (RWTH) Aachen University and by the Deutsche Forschungsgemeinschaft DFG (LU466/15-1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Rights and permissions
About this article
Cite this article
Feijs, K., Forst, A., Verheugd, P. et al. Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat Rev Mol Cell Biol 14, 443–451 (2013). https://doi.org/10.1038/nrm3601
Published:
Issue date:
DOI: https://doi.org/10.1038/nrm3601
This article is cited by
-
Mono-ADP-ribosylation by PARP10 inhibits Chikungunya virus nsP2 proteolytic activity and viral replication
Cellular and Molecular Life Sciences (2023)
-
Fundamental mechanisms of the stem cell regulation in land plants: lesson from shoot apical cells in bryophytes
Plant Molecular Biology (2021)
-
A mycorrhizae-like gene regulates stem cell and gametophore development in mosses
Nature Communications (2020)
-
Engineering Af1521 improves ADP-ribose binding and identification of ADP-ribosylated proteins
Nature Communications (2020)
-
Comparative analysis of MACROD1, MACROD2 and TARG1 expression, localisation and interactome
Scientific Reports (2020)