Abstract
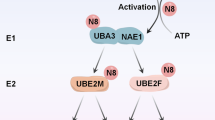
Neddylation is a post-translational modification that controls the cell cycle and proliferation by conjugating the ubiquitin-like protein NEDD8 to specific targets. Here we report that glycyl-tRNA synthetase (GlyRS), an essential enzyme in protein synthesis, also plays a critical role in neddylation. In human cells, knockdown of GlyRS, but not knockdown of a different tRNA synthetase, decreased the global level of neddylation and caused cell-cycle abnormality. This function of GlyRS is achieved through direct interactions with multiple components of the neddylation pathway, including NEDD8, E1, and E2 (Ubc12). Using various structural and functional approaches, we show that GlyRS binds the APPBP1 subunit of E1 and captures and protects activated E2 (NEDD8-conjugated Ubc12) before the activated E2 reaches a downstream target. Therefore, GlyRS functions as a chaperone that critically supports neddylation. This function is probably conserved in all eukaryotic GlyRS enzymes and may contribute to the strong association of GlyRS with cancer progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kamitani, T., Kito, K., Nguyen, H.P. & Yeh, E.T. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 272, 28557–28562 (1997).
Osaka, F. et al. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19, 3475–3484 (2000).
Tateishi, K., Omata, M., Tanaka, K. & Chiba, T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 155, 571–579 (2001).
Kerscher, O., Felberbaum, R. & Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 (2006).
Liakopoulos, D., Doenges, G., Matuschewski, K. & Jentsch, S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 17, 2208–2214 (1998).
Osaka, F. et al. A new NEDD8-ligating system for cullin-4A. Genes Dev. 12, 2263–2268 (1998).
Furukawa, M., Zhang, Y., McCarville, J., Ohta, T. & Xiong, Y. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20, 8185–8197 (2000).
Huang, D.T. et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell 33, 483–495 (2009).
Enchev, R.I., Schulman, B.A. & Peter, M. Protein neddylation: beyond cullin–RING ligases. Nat. Rev. Mol. Cell Biol. 16, 30–44 (2015).
Soucy, T.A., Dick, L.R., Smith, P.G., Milhollen, M.A. & Brownell, J.E. The NEDD8 conjugation pathway and its relevance in cancer biology and therapy. Genes Cancer 1, 708–716 (2010).
Welchman, R.L., Gordon, C. & Mayer, R.J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6, 599–609 (2005).
Vijay-Kumar, S., Bugg, C.E. & Cook, W.J. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 194, 531–544 (1987).
Bublitz, C. The properties of a glycyl-soluble-RNA synthetase from chick embryo. Biochim. Biophys. Acta 113, 158–166 (1966).
Zhou, Q. et al. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat. Struct. Mol. Biol. 17, 57–61 (2010).
Huang, D.T. et al. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat. Struct. Mol. Biol. 11, 927–935 (2004).
Eletr, Z.M., Huang, D.T., Duda, D.M., Schulman, B.A. & Kuhlman, B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat. Struct. Mol. Biol. 12, 933–934 (2005).
Xie, W., Nangle, L.A., Zhang, W., Schimmel, P. & Yang, X.L. Long-range structural effects of a Charcot-Marie-Tooth disease-causing mutation in human glycyl-tRNA synthetase. Proc. Natl. Acad. Sci. USA 104, 9976–9981 (2007).
Scott, D.C. et al. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell 157, 1671–1684 (2014).
Duhovny, D., Nussinov, R. & Wolfson, H.J. Efficient unbound docking of rigid molecules. Algorithms Bioinform 2452, 185–200 (2002).
Schneidman-Duhovny, D., Inbar, Y., Nussinov, R. & Wolfson, H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–W367 (2005).
Guo, R.T., Chong, Y.E., Guo, M. & Yang, X.L. Crystal structures and biochemical analyses suggest a unique mechanism and role for human glycyl-tRNA synthetase in Ap4A homeostasis. J. Biol. Chem. 284, 28968–28976 (2009).
Pagano, M. et al. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269, 682–685 (1995).
Toyoshima, H. & Hunter, T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 78, 67–74 (1994).
Soucy, T.A. et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009).
Hengst, L. & Reed, S.I. Translational control of p27Kip1 accumulation during the cell cycle. Science 271, 1861–1864 (1996).
Petroski, M.D. & Deshaies, R.J. Function and regulation of cullin–RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005).
Cardozo, T. & Pagano, M. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 (2004).
Stanley, D.J. et al. Inhibition of a NEDD8 cascade restores restriction of HIV by APOBEC3G. PLoS Pathog. 8, e1003085 (2012).
Mazauric, M.H. et al. Glycyl-tRNA synthetase from Thermus thermophilus: wide structural divergence with other prokaryotic glycyl-tRNA synthetases and functional inter-relation with prokaryotic and eukaryotic glycylation systems. Eur. J. Biochem. 251, 744–757 (1998).
Dil Kuazi, A. et al. NEDD8 protein is involved in ubiquitinated inclusion bodies. J. Pathol. 199, 259–266 (2003).
Mori, F. et al. Accumulation of NEDD8 in neuronal and glial inclusions of neurodegenerative disorders. Neuropathol. Appl. Neurobiol. 31, 53–61 (2005).
Antonellis, A. et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet. 72, 1293–1299 (2003).
Yao, P. & Fox, P.L. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol. Med. 5, 332–343 (2013).
Dubourg, O. et al. The G526R glycyl-tRNA synthetase gene mutation in distal hereditary motor neuropathy type V. Neurology 66, 1721–1726 (2006).
Motley, W.W. et al. Charcot-Marie-Tooth-linked mutant GARS is toxic to peripheral neurons independent of wild-type GARS levels. PLoS Genet. 7, e1002399 (2011).
He, W. et al. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature 526, 710–714 (2015).
Stum, M. et al. An assessment of mechanisms underlying peripheral axonal degeneration caused by aminoacyl-tRNA synthetase mutations. Mol. Cell. Neurosci. 46, 432–443 (2011).
Chairatvit, K. & Ngamkitidechakul, C. Control of cell proliferation via elevated NEDD8 conjugation in oral squamous cell carcinoma. Mol. Cell. Biochem. 306, 163–169 (2007).
Salon, C. et al. Altered pattern of Cul-1 protein expression and neddylation in human lung tumours: relationships with CAND1 and cyclin E protein levels. J. Pathol. 213, 303–310 (2007).
Podust, V.N. et al. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc. Natl. Acad. Sci. USA 97, 4579–4584 (2000).
Nakayama, K.I. & Nakayama, K. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 (2006).
Kobayashi, A. et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24, 7130–7139 (2004).
Kamura, T. et al. Activation of HIF1α ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. USA 97, 10430–10435 (2000).
Winston, J.T. et al. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 13, 270–283 (1999).
Nishitani, H. et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 25, 1126–1136 (2006).
Sarantopoulos, J. et al. Phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with advanced solid tumors. Clin. Cancer Res. 22, 847–857 (2016).
Swords, R.T. et al. Pevonedistat (MLN4924), a first-in-class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: a phase 1 study. Br. J. Haematol. 169, 534–543 (2015).
Györffy, B. et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 (2010).
Williams, T.F., Mirando, A.C., Wilkinson, B., Francklyn, C.S. & Lounsbury, K.M. Secreted threonyl-tRNA synthetase stimulates endothelial cell migration and angiogenesis. Sci. Rep. 3, 1317 (2013).
Kim, D.G. et al. Interaction of two translational components, lysyl-tRNA synthetase and p40/37LRP, in plasma membrane promotes laminin-dependent cell migration. FASEB J. 26, 4142–4159 (2012).
Kim, D.G. et al. Chemical inhibition of prometastatic lysyl-tRNA synthetase-laminin receptor interaction. Nat. Chem. Biol. 10, 29–34 (2014).
Huang, D.T. & Schulman, B.A. Expression, purification, and characterization of the E1 for human NEDD8, the heterodimeric APPBP1-UBA3 complex. Methods Enzymol. 398, 9–20 (2005).
He, W. et al. Dispersed disease-causing neomorphic mutations on a single protein promote the same localized conformational opening. Proc. Natl. Acad. Sci. USA 108, 12307–12312 (2011).
Sajish, M. et al. Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-γ and p53 signaling. Nat. Chem. Biol. 8, 547–554 (2012).
Xu, X. et al. Unique domain appended to vertebrate tRNA synthetase is essential for vascular development. Nat. Commun. 3, 681 (2012).
Jin, J., Li, X., Gygi, S.P. & Harper, J.W. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447, 1135–1138 (2007).
Chalmers, M.J. et al. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 78, 1005–1014 (2006).
Pascal, B.D. et al. HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 23, 1512–1521 (2012).
Acknowledgements
We thank B. Schulman (St. Jude Children's Research Hospital) and W. Harper (Harvard Medical School) for providing plasmids and advice on the project. We thank W. He and H. Zhou (The Scripps Laboratories for tRNA Synthetase Research) for providing plasmids of GlyRS. We thank P. Schimmel, M.H. Nawaz, and other members of The Scripps Laboratories for tRNA Synthetase Research for advice, discussion, and technical support. This research was supported by US National Institutes of Health grant R01GM088278 (X.-L.Y.).
Author information
Authors and Affiliations
Contributions
X.-L.Y. and Z.M. designed experiments, analyzed data, and wrote the manuscript. Z.M. performed the molecular cloning, binding analysis, structural docking, bioinformatic analysis, and additional biochemical analysis. Z.M. and Q.Z. carried out protein purification. Z.M. and Z.L. performed the cell-cycle analysis. J.L. and P.R.G. performed the HDX analysis. Y.S. and L.S. contributed to biochemical analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Characterization of the GlyRS-NEDD8 interaction.
(a) Hydrogen-deuterium exchange (HDX) analysis of GlyRS in the presence of NEDD8. Changes in deuterium incorporation resulting from the NEDD8 interaction are mapped to the sequence and the crystal structure of GlyRS (PDB 2PME). (b,c) Di-glycine motif of NEDD8 is not essential for GlyRS. Biolayer interferometry analysis comparing GlyRS binding to the full-length NEDD8 (b) and the truncated NEDD8 lacking the C-terminal di-glycine motif (NEDD8ΔGG) (c). His-tagged NEDD8 proteins were immobilized to the Ni-NTA sensor tips. Tag-free GlyRS (yellow green, 250 nM; orange,500 nM) were used to detect the interaction with NEDD8.
Supplementary Figure 2 GlyRS does not affect Ube2F conjugation.
(a) Western blot analysis to detect N8-conjugation of Ube2F in HeLa cells transfected with plasmids expressing shRNAs against GlyRS (shGlyRS) or SerRS (shSerRS) or a control vector. (b) Biolayer interferometry analysis to compare the binding of Ubc12 and Ube2F (both at 1.0 µM) to immobilized GST-GlyRS.
Supplementary Figure 3 The catalytic domain of GlyRS is the major binding site for Ubc12.
(a) In vitro neddylation assay with γ-32P-ATP labeled NEDD8. (b) In vitro neddylation assay using fluorescein modified NEDD8. (c) GST pull-down analysis to detect the interaction of GST-Ubc12 with GlyRS and its truncated fragments. (d) Hydrogen-deuterium exchange (HDX) analysis of GlyRS in the presence of Ubc12. Changes in deuterium incorporation resulting from the Ubc12 interaction are mapped to the sequence and the crystal structure of GlyRS (PDB 2PME).
Supplementary Figure 4 GlyRS stabilizes Ubc12N8.
Time courses of Ubc12N8 degradation in the absence and presence of WT or Δ232-238 GlyRS. Gels were stained by coomassie blue and quantified with ImageJ.
Supplementary Figure 6 GlyRS facilitates cullin neddylation.
(a) Modeling analysis suggesting that GlyRS is unlikely to interfere with NEDD8 transferring from Ubc12 to cullin. The complex structure of cullin1-Rbx1-Ubc12N8 is adapted from PDB 4P5O. (b) Biolayer interferometry analysis to detect the binding of cullin1-Rbx1 (5 μg/mL) or GlyRS (5 μg/mL) or both to the immobilized Ubc12N8. Black dotted lines indicate the calculated sum of the binding curves for cullin1-Rbx1 and for GlyRS, respectively. (c) In vitro neddylation assay to detect the activity of WT and Δ232-238 GlyRS in promoting cullin1 neddylation.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 986 kb)
Supplementary Data Set 1
Combined raw gels and blots (PDF 6568 kb)
Supplementary Data Set 2
Model of the GlyRS–Ubc12N8 complex (PDB 651 kb)
Supplementary Data Set 3
Model of the GlyRS–E1 complex (PDB 950 kb)
Rights and permissions
About this article
Cite this article
Mo, Z., Zhang, Q., Liu, Z. et al. Neddylation requires glycyl-tRNA synthetase to protect activated E2. Nat Struct Mol Biol 23, 730–737 (2016). https://doi.org/10.1038/nsmb.3250
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/nsmb.3250