Abstract
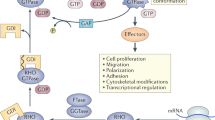
NOTCH1 is frequently mutated in T-cell acute lymphoblastic leukaemia (T-ALL), and can stimulate T-ALL cell survival and proliferation. Here we explore the hypothesis that Notch1 also alters T-ALL cell migration. Rho GTPases are well known to regulate cell adhesion and migration. We have analysed the expression levels of Rho GTPases in primary T-ALL samples compared with normal T cells by quantitative PCR. We found that 5 of the 20 human Rho genes are highly and consistently upregulated in T-ALL, and 3 further Rho genes are expressed in T-ALL but not detectable in normal T cells. Of these, RHOU expression is highly correlated with the expression of the Notch1 target DELTEX-1. Inhibition of Notch1 signalling with a γ-secretase inhibitor (GSI) or Notch1 RNA interference reduced RhoU expression in T-ALL cells, whereas constitutively active Notch1 increased RhoU expression. In addition, Notch1 or RhoU depletion, or GSI treatment, inhibits T-ALL cell adhesion, migration and chemotaxis. These results indicate that NOTCH1 mutation stimulates T-ALL cell migration through RhoU upregulation that could contribute to the leukaemia cell dissemination.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Pieters R, Carroll WL . Biology and treatment of acute lymphoblastic leukemia. Pediatr Clin North Am 2008; 55: 1–20.
Aifantis I, Raetz E, Buonamici S . Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol 2008; 8: 380–390.
Crazzolara R, Kreczy A, Mann G, Heitger A, Eibl G, Fink FM et al. High expression of the chemokine receptor CXCR4 predicts extramedullary organ infiltration in childhood acute lymphoblastic leukaemia. Br J Haematol 2001; 115: 545–553.
Bhojwani D, Howard SC, Pui CH . High-risk childhood acute lymphoblastic leukemia. Clin Lymphoma Myeloma 2009; 9 (Suppl 3): S222–S230.
Weng AP, Ferrando AA, Lee W, Morris 4th JP, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
van Grotel M, Meijerink JP, van Wering ER, Langerak AW, Beverloo HB, Buijs-Gladdines JG et al. Prognostic significance of molecular-cytogenetic abnormalities in pediatric T-ALL is not explained by immunophenotypic differences. Leukemia 2008; 22: 124–131.
Hayday AC, Pennington DJ . Key factors in the organized chaos of early T cell development. Nat Immunol 2007; 8: 137–144.
Vicente R, Swainson L, Marty-Gres S, De Barros SC, Kinet S, Zimmermann VS et al. Molecular and cellular basis of T cell lineage commitment. Semin Immunol 2010; 22: 270–275.
Aster JC, Blacklow SC, Pear WS . Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol 2011; 223: 262–273.
Ferrando AA . The role of NOTCH1 signaling in T-ALL. Hematology Am Soc Hematol Educ Program 2009; 353–361.
Buonamici S, Trimarchi T, Ruocco MG, Reavie L, Cathelin S, Mar BG et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature 2009; 459: 1000–1004.
Van Hennik PB, Hordijk PL . Rho GTPases in hematopoietic cells. Antioxid Redox Signal 2005; 7: 1440–1455.
Tybulewicz VL, Henderson RB . Rho family GTPases and their regulators in lymphocytes. Nat Rev Immunol 2009; 9: 630–644.
Boureux A, Vignal E, Faure S, Fort P . Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol 2007; 24: 203–216.
Chang FK, Sato N, Kobayashi-Simorowski N, Yoshihara T, Meth JL, Hamaguchi M . DBC2 is essential for transporting vesicular stomatitis virus glycoprotein. J Mol Biol 2006; 364: 302–308.
Aspenstrom P, Ruusala A, Pacholsky D . Taking Rho GTPases to the next level: the cellular functions of atypical Rho GTPases. Exp Cell Res 2007; 313: 3673–3679.
Vega FM, Ridley AJ . Rho GTPases in cancer cell biology. FEBS Lett 2008; 582: 2093–2101.
Ellenbroek SI, Collard JG . Rho GTPases: functions and association with cancer. Clin Exp Metastasis 2007; 24: 657–672.
Symons M, Segall JE . Rac and Rho driving tumor invasion: who's at the wheel? Genome Biol 2009; 10: 213.
Parri M, Chiarugi P . Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal 2010; 8: 23.
Heasman SJ, Carlin LM, Cox S, Ng T, Ridley AJ . Coordinated RhoA signaling at the leading edge and uropod is required for T cell transendothelial migration. J Cell Biol 2010; 190: 553–563.
O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med 2007; 204: 1813–1824.
Palomero T, Odom DT, O’Neil J, Ferrando AA, Margolin A, Neuberg DS et al. Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia. Blood 2006; 108: 986–992.
Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 2007; 13: 1203–1210.
Wolfe MS . Secretase in biology and medicine. Semin Cell Dev Biol 2009; 20: 219–224.
Heasman SJ, Ridley AJ . Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol 2008; 9: 690–701.
Takesono A, Heasman SJ, Wojciak-Stothard B, Garg R, Ridley AJ . Microtubules regulate migratory polarity through Rho/ROCK signaling in T cells. PLoS One 2010; 5: e8774.
Infante E, Heasman SJ, Ridley AJ . Statins inhibit T-acute lymphoblastic leukemia cell adhesion and migration through Rap1b. J Leukoc Biol 2011; 89: 577–586.
Aspenstrom P, Fransson A, Saras J . Rho GTPases have diverse effects on the organization of the actin filament system. Biochem J 2004; 377: 327–337.
Chuang YY, Valster A, Coniglio SJ, Backer JM, Symons M . The atypical Rho family GTPase Wrch-1 regulates focal adhesion formation and cell migration. J Cell Sci 2007; 120: 1927–1934.
Aronheim A, Broder YC, Cohen A, Fritsch A, Belisle B, Abo A . Chp, a homologue of the GTPase Cdc42Hs, activates the JNK pathway and is implicated in reorganizing the actin cytoskeleton. Curr Biol 1998; 8: 1125–1128.
Brazier H, Pawlak G, Vives V, Blangy A . The Rho GTPase Wrch1 regulates osteoclast precursor adhesion and migration. Int J Biochem Cell Biol 2009; 41: 1391–1401.
Fort P, Guemar L, Vignal E, Morin N, Notarnicola C, de Santa Barbara P et al. Activity of the RhoU/Wrch1 GTPase is critical for cranial neural crest cell migration. Dev Biol 2011; 350: 451–463.
Weisz Hubsman M, Volinsky N, Manser E, Yablonski D, Aronheim A . Autophosphorylation-dependent degradation of Pak1, triggered by the Rho-family GTPase, Chp. Biochem J 2007; 404: 487–497.
Arias-Romero LE, Chernoff J . A tale of two Paks. Biol Cell 2008; 100: 97–108.
Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 2002; 1: 75–87.
Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC et al. Direct inhibition of the NOTCH transcription factor complex. Nature 2009; 462: 182–188.
Tao W, Pennica D, Xu L, Kalejta RF, Levine AJ . Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev 2001; 15: 1796–1807.
Weerkamp F, van Dongen JJ, Staal FJ . Notch and Wnt signaling in T-lymphocyte development and acute lymphoblastic leukemia. Leukemia 2006; 20: 1197–1205.
Fueller F, Kubatzky KF . The small GTPase RhoH is an atypical regulator of haematopoietic cells. Cell Commun Signal 2008; 6: 6.
Wang H, Zeng X, Fan Z, Lim B . RhoH plays distinct roles in T-cell migrations induced by different doses of SDF1α. Cell Signal 2010; 22: 1022–1032.
Berthold J, Schenkova K, Rivero F . Rho GTPases of the RhoBTB subfamily and tumorigenesis. Acta Pharmacol Sin 2008; 29: 285–295.
Berthold J, Schenkova K, Ramos S, Miura Y, Furukawa M, Aspenstrom P et al. Characterization of RhoBTB-dependent Cul3 ubiquitin ligase complexes-evidence for an autoregulatory mechanism. Exp Cell Res 2008; 314: 3453–3465.
Riou P, Villalonga P, Ridley AJ . Rnd proteins: multifunctional regulators of the cytoskeleton and cell cycle progression. Bioessays 2010; 32: 986–992.
Gomez del Pulgar T, Benitah SA, Valeron PF, Espina C, Lacal JC . Rho GTPase expression in tumourigenesis: evidence for a significant link. Bioessays 2005; 27: 602–613.
Ridley AJ . Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol 2006; 16: 522–529.
Huang M, Prendergast GC . RhoB in cancer suppression. Histol Histopathol 2006; 21: 213–218.
Ono Y, Fukuhara N, Yoshie O . Transcriptional activity of TAL1 in T cell acute lymphoblastic leukemia (T-ALL) requires RBTN1 or -2 and induces TALLA1, a highly specific tumor marker of T-ALL. J Biol Chem 1997; 272: 4576–4581.
Graux C, Cools J, Michaux L, Vandenberghe P, Hagemeijer A . Cytogenetics and molecular genetics of T-cell acute lymphoblastic leukemia: from thymocyte to lymphoblast. Leukemia 2006; 20: 1496–1510.
Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, Sanchez-Madrid F . Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol 2009; 19: 434–446.
Schmittgen TD, Livak KJ . Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3: 1101–1108.
de Hoon MJ, Imoto S, Nolan J, Miyano S . Open source clustering software. Bioinformatics 2004; 20: 1453–1454.
Saldanha AJ . Java Treeview--extensible visualization of microarray data. Bioinformatics 2004; 20: 3246–3248.
Acknowledgements
This work was supported by Leukaemia and Lymphoma Research UK, Cancer Research UK and King's College London British Heart Foundation Centre of Excellence. EI was supported by the Department of Health via National Institute for Health Research (NIHR) Comprehensive Biomedical Research Centre Award to Guy's and St Thomas’ NHS Foundation Trust in partnership with King's College London and King's College Hospital NHS Foundation Trust. We are grateful to Sarah Heasman for scientific discussion and guidance; Ritu Garg and Katrina Soderquest for technical assistance; Katherine Lawler for advice on statistical analysis; and Matthew Arno and Estibaliz Aldecoa-otalora Astarloa for assistance and advice on qPCR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Bhavsar, P., Infante, E., Khwaja, A. et al. Analysis of Rho GTPase expression in T-ALL identifies RhoU as a target for Notch involved in T-ALL cell migration. Oncogene 32, 198–208 (2013). https://doi.org/10.1038/onc.2012.42
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/onc.2012.42
Keywords
This article is cited by
-
OVOL2 impairs RHO GTPase signaling to restrain mitosis and aggressiveness of Anaplastic Thyroid Cancer
Journal of Experimental & Clinical Cancer Research (2022)
-
Transcriptional and post-transcriptional regulation of the genes encoding the small GTPases RhoA, RhoB, and RhoC: implications for the pathogenesis of human diseases
Cellular and Molecular Life Sciences (2018)
-
JMJD3 and NF-κB-dependent activation of Notch1 gene is required for keratinocyte migration during skin wound healing
Scientific Reports (2017)
-
RNA-sequencing analysis reveals new alterations in cardiomyocyte cytoskeletal genes in patients with heart failure
Laboratory Investigation (2014)
-
The CD46-Jagged1 interaction is critical for human TH1 immunity
Nature Immunology (2012)