Abstract
Polymorphous adenocarcinoma (PAC) and cribriform adenocarcinoma of (minor) salivary gland (CASG) are salivary gland tumors with overlapping spectrum of morphology. Whether these represent distinct entities or a histologic spectrum of the same tumor remains contentious. PACs harbor recurrent PRKD1 E710D hotspot mutations in >70% of cases, whereas 80% of CASGs display rearrangements involving PRKD1, PRKD2, or PRKD3 (PRKD1/2/3). We studied the molecular and morphologic features of 37 PACs/CASGs, seeking to identify the associations among genotype, histologic phenotype, and classification. DNA was subjected to Sanger sequencing analysis of the PRKD1 hotspot locus. Fluorescence in situ hybridization (FISH) analysis for PRKD1/2/3 was performed using dual-color break-apart probes. Tumors were classified into four categories as described previously: PAC, CASG, tumor with indeterminate features (TIF), and tumor with a predominant papillary pattern (TPPP). PRKD1 E710D hotspot mutations were identified in 56%, 20%, 43% and 0% of PACs, CASGs, TIFs, and TPPPs, respectively. FISH demonstrated PRKD1/2/3 rearrangements in 13%, 78%, 36%, and 75% of PACs, CASGs, TIFs, and TPPPs, respectively. Histologically, fusion-positive tumors were associated with a high percentage of papillary growth, low percentage of single filing arrangement, a propensity of base of tongue location, and frequent (50%) lymph node metastasis, compared with the mutation-related tumors which had negligible nodal metastasis risk. Our results demonstrated that (1) PACs/CASGs are underpinned by genetic alterations affecting PRKD genes; (2) despite the associations between PAC and PRKD1 hotspot mutations and CASG and PRKD1/2/3 fusion, such distinction is not absolute; and (3) there is of a novel genotypic-phenotypic association whereby fusion-positive tumors are usually located in the base of the tongue, show papillary architecture and have a high risk of nodal metastasis. Genetic analysis of PRKD genes appears to be useful characterizing this spectrum of tumors, not only histologically but also clinically identifying those tumors with high risk of nodal metastasis.
Similar content being viewed by others
Introduction
Polymorphous adenocarcinoma is the second most common malignancy arising in the minor salivary glands [1]. Polymorphous adenocarcinoma is infiltrative, displaying cytologic uniformity, and an array of architectural patterns, including tubular, fascicular, cribriform, papillary, and solid [2]. Although most polymorphous adenocarcinomas follow an indolent course, some may have an aggressive behavior with local and even distant recurrence [3,4,5,6].
In 1999, Michal et al. has described cribriform adenocarcinoma of (minor) salivary glands [7]. In the original and subsequent series, Cribriform adenocarcinoma displayed a high risk (>70%) of lymph node metastasis and a propensity to base of tongue location. Histologically, it was described as a tumor with optically clear nuclei, lobulated growth, and a predominantly cribriform and solid architecture with peripheral palisading, clefts, and glomeruloid formation [7,8,9,10,11,12].
We have shown previously that classic polymorphous adenocarcinoma is underpinned by recurrent PRKD1 E710D hotspot mutations [13], and that a subset of polymorphous adenocarcinoma lacking PRKD1 mutations harbors rearrangements involving genes of the PRKD family, including PRKD1, PRKD2, and PRKD3 (PRKD1/2/3) [5]. Moreover, PRKD1/2/3 rearrangements have also been described in the majority of CASGs [5]. The fact that polymorphous adenocarcinoma and cribriform adenocarcinoma harbor alterations of the same gene family but through different mechanisms and that polymorphous adenocarcinoma may contain either PRKD1 mutation or PRKD1/2/3 fusions suggests that these entities likely share an overlapping molecular pathogenesis.
In the current study, we conducted a detailed genotype–phenotype correlation of 37 polymorphous adenocarcinoma/cribriform`adenocarcinoma spectrum of tumors. The aims of the study were twofold. First, we sought to investigate whether polymorphous adenocarcinoma/cribriform adenocarcinoma are distinct or related at the genetic level. Second, we aimed to investigate if the underlying molecular alterations may predict the tumor histologic phenotype and/or clinical behavior.
Materials and methods
Study cohort, classification, and clinicopathologic review
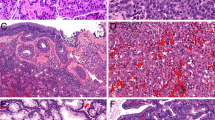
The clinicopathologic characteristics and classification of all 37 cases included in this series have been previously reported in the study by Xu et al. [14]. In brief, following institutional review board approval, tumors of polymorphous adenocarcinoma spectrum were retrieved from the archives of the Department of Pathology at Memorial Sloan Kettering Cancer Center. Samples were anonymized prior to analysis. All cases were previously reviewed by three head and neck pathologists (NK, BX, and RG) to reach a consensus diagnosis [14] using criteria put forward by the World Health Organization fourth edition (2017) [15] and prior publications by Skalova et al. [11] and Michal et al. [7]. The cases were classified as (i) polymorphous adenocarcinoma, (ii) cribriform adenocarcinoma of salivary gland, (iii) tumor with a predominant papillary architecture, and (iv) tumors with indeterminate features as previously described in our prior publication [14]. Histologically, all tumors demonstrated cytologic uniformity characterized by tumor cells with open chromatin and a variable degree of nuclear clearing. Classic polymorphous adenocarcinoma contained diverse architectural patterns, in particular single filing and trabecular growth, arranging in a targetoid fashion (Fig. 1a). Cribriform adenocarcinoma of salivary gland demonstrated lobulated growth with a predominant solid or cribriform architecture (Fig. 1b). Tumors with at least 50% of papillary growth were defined as tumor with a predominant papillary architecture (Fig. 1d). Tumors with indeterminate features were tumors within the spectrum of polymorphous adenocarcinoma/cribriform adenocarcinoma but difficult to be subclassified into any of the other three categories (Fig. 1c). Most of the tumors with indeterminate features exhibited mixed features of cribriform adenocarcinoma and polymorphous adenocarcinoma.
Histologic spectrum of polymorphous adenocarcinoma of the salivary glands harbors genetic alterations affecting PRKD Genes. Representative hematoxylin and eosin (H&E) photomicrographs of a a polymorphous adenocarcinoma, b a cribriform adenocarcinoma of the salivary gland, c a tumor with indeterminate features between polymorphous adenocarcinoma and cribriform adenocarcinoma of the salivary gland and d a tumor with predominant papillary pattern. e Heatmap depicting PRKD1 E710D hotspot mutations and PRKD1/2/3 rearrangements identified in the polymorphous adenocarcinoma, cribriform adenocarcinoma, tumor with indeterminate features, and tumor with predominant papillary pattern included in this study. Clinicopathologic characteristics are depicted in phenobars (top). Scale bars, 300 μm. PAC polymorphous adenocarcinoma, CASG cribriform adenocarcinoma of the salivary gland, TIF, tumor with indeterminate features between PACs and CASGs
A detailed clinicopathologic review was conducted previously [14] to collect the following clinical and pathologic information: age, sex, site of the primary tumor, architectural patterns, mitotic index, tumor size, tumor necrosis, percentage of tumor cells exhibiting nuclear clearing, nodal metastasis at the time of primary resection, and clinical outcome including recurrence and mortality.
Microdissection and DNA extraction
Tumor and matching normal tissues were microdissected from consecutive 8-μm-thick sections of representative formalin-fixed paraffin-embedded blocks under a stereomicroscope (Olympus SZ61) to ensure a tumor cell content >80%, as previously described [16]. DNA was extracted using the DNAeasy Blood and Tissue Kit (Qiagen) according to the manufacturers’ instructions and quantified using the Qubit Fluorometer assay (Life Technologies).
Sanger sequencing
The assessment of the PRKD1 E710D hotspot mutation was conducted by Sanger sequencing using previously described primers pairs [13]. PCR amplification was performed using the AmpliTaq Gold 360 Master Mix (Life Technologies), as previously described [13]. PCR fragments were purified using ExoSAP-IT (Thermo Fisher Scientific) and submitted to Sanger sequencing. Sequence electropherograms of the forward and reverse strands were analyzed using Mutation Surveyor (SoftGenetics) and the mutations identified were manually curated. All analyses were performed in duplicate.
Fluorescence in situ hybridization (FISH)
All cases except for PMC01 and PMC11, for which insufficient material was available, were reviewed, representative areas were selected and a tissue microarray (TMA) was constructed at the MSKCC Pathology Core by sampling two tumor cores from each case, as previously described [17]. The TMA was subjected to FISH analysis for PRKD1, PRKD2, and PRKD3 using dual-color break-apart probes following validated protocols at the MSKCC Molecular Cytogenetics Core, as previously described [18]. The probe mix consisted of bacterial artificial chromosome clones mapping to 5′ PRKD1 (RP11-269C4, RP11-777L23; red), 3′ PRKD1 (RP11-684G15, RP11-942P15; green), 5′ PRKD2 (RP11-846M4, RP11-611I8; red), 3′ PRKD2 (RP11-194H9, RP11-210G11; green), 5′ PRKD3 (RP11-695L15, RP11-278G12; red), and 3′ PRKD3 (RP11-1130K21, RP11-142K18; green). A minimum of 50 interphase nuclei was analyzed for PRKD1/2/3 rearrangements. Cases were considered positive for rearrangement if a separation between the 5′ (red) and 3′ (green) signals (>2 signal width apart) was identified in >15% tumor cells.
Statistical analysis
All statistical analyses were performed using the SPSS software 24.0 (IBM Corporation, New York, NY, U.S.). The clinicopathologic features and recurrence-free survival of mutation-positive tumors and fusion-positive tumors were compared using appropriate statistical tests, i.e., Fisher’s exact test for categorical variables, two-tailed Student’s t-test for continuous variables, and log-rank test for disease-free survival. P values <0.05 were considered as statistically significant.
Results
Cases
Our study included 37 salivary gland tumors, which upon central histopathologic review based on previously described criteria [14], were classified as polymorphous adenocarcinoma (n = 9, Fig. 1a), cribriform adenocarcinoma (n = 10, Fig. 1b), tumor with indeterminate features (n = 14, Fig. 1c) and tumor with predominant papillary pattern (n = 4, Fig. 1d). The tumors affected 12 males (32%) and 25 (68%) females (Table 1). The median age at diagnosis was 61 years old (range 22–83) (Table 1). The majority (95%; 35/37) of tumors originated in minor salivary glands, with the palate being the most frequent site of origin for all tumor types [polymorphous adenocarcinoma (7/9; 78%), cribriform adenocarcinoma (4/10; 40%), tumor with indeterminate features (6/14; 42.9%), and tumor with predominant papillary pattern (2/4; 50%); Fig. 1e and Table 1].
Polymorphous adenocarcinoma/cribriform adenocarcinoma spectrum of tumors harbors recurrent genetic alterations in PRKD genes
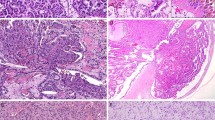
DNA from all 37 cases was subjected to Sanger sequencing of the PRKD1 E710 hotspot locus. Our analysis revealed that polymorphous adenocarcinoma, cribriform adenocarcinoma, and tumor with indeterminate features harbored PRKD1 E710D hotspot mutations in 56% (5/9), 20% (2/10), and 43% (6/14) of cases, respectively, whereas the four tumors with predominant papillary pattern tested lacked PRKD1 mutations (Figs. 1e, 2a and Table 1).
PRKD1 E710D hotspot mutations and PRKD1/2/3 rearrangements in the histologic spectrum of polymorphous adenocarcinoma of the salivary gland. a Representative Sanger sequencing electropherograms of the PRKD1 E710D hotspot locus in polymorphous adenocarcinomas (PACs), cribriform adenocarcinomas of the salivary gland (CASGs), tumors with indeterminate features between PACs and CASGs (TIF) and tumors with predominant papillary pattern (TPPP). Arrows point to the altered base. b Representative micrographs of the fluorescence in situ hybridization (FISH) analysis of PRKD3 and PRKD1 in a PAC and a CASG using dual-color break-apart probes (red, 5′ probe; green, 3′ probe). c Frequency of PRKD1 E710D hotspot mutations and PRKD1/2/3 rearrangements in polymorphous adenocarcinoma (n = 9), cribriform adenocarcinoma (n = 10), tumor with indeterminate features (n = 14), and tumor with predominant papillary pattern (n = 4)
Next, all cases were subjected to FISH analysis for rearrangements involving PRKD1/2/3 using dual-color break-apart probes, except for PMC01 and PMC11, for which no additional material was available. This FISH analysis revealed the presence of PRKD1/2/3 rearrangements in 13% (1/8), 78% (7/9), 36% (5/14), and 75% (3/4) of polymorphous adenocarcinoma, cribriform adenocarcinoma, tumors with indeterminate features, and tumors with predominant papillary patterns interrogated, respectively (Figs. 1e, 2b), which corresponded to 25% (1/4) of polymorphous adenocarcinomas, 88% (7/8) of cribriform adenocarcinomas, 63% (5/8) tumors with indeterminate features, and 75% (3/4) of tumors with predominant papillary pattern lacking PRKD1 E710D hotspot mutations. In agreement with previous reports, PRKD1 E710D hotspot mutations were mutually exclusive with rearrangements in PRKD1/2/3 (Figs. 1e, 2c and Table 1). Notably, we identified seven cases, including three polymorphous adenocarcinomas, one cribriform adenocarcinoma, three tumors with indeterminate features, and one tumor with predominant papillary pattern, lacking PRKD1 E710D hotspot mutations and PRKD1/2/3 rearrangements (Figs. 1e, 2c and Table 1). The determination of the genetic underpinning of these cases warrants further study.
Fusion-positive tumors had distinct histologic features and a propensity for lymph node spread
The clinicopathologic features and clinical outcomes were compared between fusion-positive and mutation-positive tumors regardless of their consensus histologic classification (Table 2). Fusion-positive tumors commonly involved the palate (7/16, 44%) and base of tongue (5/16, 31%), whereas mutation-positive tumors most frequently originated from the palate (10/14, 71%) and rarely occurred in base of tongue (1/14, 7%). Two tumors tested originated from the parotid gland. Interestingly, both tumors harbored a fusion involving PRKD gene; one was classified as tumor with indeterminate features and showed a PRKD2 rearrangement, whereas the other was a tumor with predominant papillary pattern and harbored a PRKD3 rearrangement (Fig. 1e).
Compared with tumors harboring a PRKD1 hotspot mutation, fusion-positive tumors were associated with statistically significantly higher frequency of tumor necrosis (31% vs. 0%, p = 0.045), higher percentage of tumor cells with nuclear chromatin clearing (mean ± standard errors of mean, 81% ± 4% vs. 51% ± 6%, p < 0.001), higher rate of tumors with at least 10% papillae (75% vs. 7%, p < 0.001), higher percentage of papillary growth within the tumor (28% ± 6% vs. 2% ± 1%, p < 0.001) and lower percentage of cells arranged in single files (2% ± 1% vs. 7% ± 1%, p = 0.001). Other characteristics, e.g., sex, age, tumor size, cribriform, solid, reticular, tubular, and tubuloreticular patterns did not differ between mutation-positive and fusion-positive tumors (p > 0.05).
Among the 37 tested tumors, 9 (24%) were associated with nodal metastasis at the time of primary resection, including 8 of 16 (50%) fusion-positive tumors, 0 of 14 (0%) mutation-positive tumors, and 1 of 7 (14%) wild-type tumors. Notably, PRKD1/2/3 fusion tumors were associated with a propensity for nodal metastasis, whereas tumors harboring the PRKD1 hotspot mutation rarely involved the lymph nodes (p = 0.003). The primary tumor sites of the eight fusion-positive tumors with nodal metastasis were base of tongue (n = 3, 37.5%), palate (n = 3, 37.5%), and parotid (n = 2, 25%).
Three patients recurred, including two patients with fusion-positive tumors (one with distant metastasis to lung and one with local recurrence) and one patient with a mutation-positive tumor developed regional recurrence. No death of disease was found during the follow-up period (median follow-up period 70 months, range 1–295 months). The recurrence-free survival did not differ between these two groups (log-rank test, p = 0.630).
Discussion
There is an ongoing controversy regarding polymorphous adenocarcinoma and cribriform adenocarcinoma of salivary gland and whether they constitute the same tumor or two distinct entities. Indeed, the last edition of the World Health Organization Classification of Head and Neck Tumors [15] has considered cribriform adenocarcinoma of salivary gland as a variant of polymorphous adenocarcinoma, despite that some authors may regard cribriform adenocarcinoma as a different entity [5, 10,11,12, 19]. Moreover, these tumors seem to display an overlapping morphologic spectrum which may pose challenges to their histologic separation. Here, we demonstrate that akin to polymorphous adenocarcinoma [13], salivary gland tumors in the histologic spectrum of polymorphous adenocarcinoma, such as cribriform adenocarcinoma, tumor with indeterminate features, and tumor with predominant papillary pattern harbor recurrent genetic alterations targeting PRKD genes, including PRKD1 E710D hotspot mutations and PRKD1/2/3 rearrangements. These findings show that polymorphous adenocarcinoma, cribriform adenocarcinoma, tumor with indeterminate features, and tumor with predominant papillary pattern display marked genetic overlap, and suggest that they may represent a spectrum of lesions driven by PRKD gene alterations, rather than separate entities.
We identified genetic alterations targeting PRKD genes in the majority (78.4%) of tumors in the histologic spectrum of polymorphous adenocarcinoma included in this study. We observed the frequency of PRDK1 E710D hotspot mutations to be numerically higher in classic polymorphous adenocarcinoma and PRKD1/2/3 rearrangements to be more frequent in cribriform adenocarcinoma and in tumors with predominant papillary pattern. These findings suggest that, in the context of salivary gland tumors, the presence of rearrangements involving PRKD1/2/3 genes associated with a diagnosis of cribriform adenocarcinoma, a tumor that has been previously linked to base of tongue location and high frequency of nodal metastasis [7, 11].
Importantly, we found PRKD1 E710D somatic hotspot mutations and rearrangements in PRKD1/2/3 to be mutually exclusive. These findings support the notion that the histologic spectrum of polymorphous adenocarcinomas might constitute a convergent phenotype [20] driven by genetic alterations affecting PRKD genes.
We did not identify genetic alterations in PRKD genes in 21.6% (n = 7) of the cases studied. Of note, whether tumors lacking PRKD1 E710D hotspot mutations or PRKD1/2/3 rearrangements harbor mutations in the kinase domain of PRKD2 or PRKD3 was not interrogated. It is worth mentioning, nevertheless, that we did not identify somatic mutations affecting the kinase domain of PRKD2 or PRKD3 in PRKD1-wild-type polymorphous adenocarcinoma in a previous study [21]. It is plausible that tumors lacking PRKD1 E710D hotspot mutations and PRKD1/2/3 rearrangements might harbor mutations in other regions of PRKD1 or genetic or epigenetic alterations targeting other genes in the diacylglycerol and protein kinase C signaling cascade. Further analyses with whole-genome sequencing polymorphous adenocarcinoma/cribriform adenocarcinoma and RNA sequencing are warranted; regrettably, representative frozen samples from these PRKD1/2/3 wild-type tumors were unavailable.
Our study was the first to demonstrate a strong genotype–phenotype correlation in polymorphous adenocarcinoma/cribriform adenocarcinoma. Fusion-positive tumors as a group, regardless of the rendered consensus diagnosis, was associated with a higher rate of base of tongue involvement, higher percentage of chromatin clearing and papillary growth, lower rate of single cell file architecture (i.e., cells arranged in single files), and a significantly higher rate (50%) of nodal metastasis, compared with the mutation-positive tumors, in which none of the tumors had nodal metastasis at the time of primary resection. Such genotype–phenotype correlation coincided with the original observation by Skalova et al. [7, 11] that cribriform adenocarcinoma of salivary gland is characterized with base of tongue location, prominent chromatin clearing, and significant risk (over 60%) of nodal metastasis. Interestingly, among the eight fusion-positive tumors with N1 disease at primary resection, only three (37.5%) originated from base of tongue, while the remaining five (63%) were located in the palate and parotid gland. In addition, we reported two parotid fusion-positive tumors with nodal metastasis at presentation. These findings highlight the fact that fusion-positive tumors with N1 disease may occur outside of base of tongue, and even in major salivary glands. Given the high frequency of nodal metastasis in fusion-positive tumors, molecular testing for PRKD1/2/3 fusion may be of clinical relevance pre-operatively to risk-stratify patients with cribriform adenocarcinoma/polymorphous adenocarcinoma spectrum of tumors to determine the necessity of regional lymph node dissection. Nevertheless, the classification of a given tumor as a polymorphous adenocarcinoma or a cribriform adenocarcinoma is possible based on the morphology alone and without molecular testing when the hallmark histologic features of these tumors which were previously detailed are identified. Such distinction may be helpful to provide clinical guidance in particular when lymph node dissection is considered such as in case of cribriform adenocarcinoma or tumors with predominant papillary pattern.
We have previously reported that tumors with ≥10% papillae were associated with worse clinical outcome [14]. In this study, 10% or more papillae was significantly more commonly seen in fusion-positive tumors. Such evidence suggests that the percentage of papillae may be predictive of the underlying genetic alteration in PRKD genes as well as the clinical outcome. Therefore, it is prudent to include the percentage of papillae in the pathology report of these tumors. Interestingly, despite the designated name of cribriform adenocarcinoma of salivary gland, the actual percentage of tumor area displaying a cribriform pattern did not differ significantly between fusion-positive and mutation-positive tumors. We have previously demonstrated that 30% cribriform pattern was a significant independent prognostic factor in polymorphous adenocarcinoma/cribriform adenocarcinoma spectrum of tumors [14]. It appears that cribriform architecture may be prognostically relevant but does not seem to predict the underlying molecular alterations.
Our study has several limitations. Due to insufficient material, we did not determine the partner gene in cases harboring PRKD1/2/3 rearrangements. Moreover, due to the relatively small size of our cohort and rarity of recurrence events, our study may have a limited statistical power to assess the prognostic significance of PRKD1 E710D hotspot mutations or PRKD1/2/3 rearrangements in predicting clinical outcomes in the polymorphous adenocarcinoma/CASG spectrum of tumor. Because we did not perform whole exome and RNA sequencing, further studies are required to define the molecular basis of the wild-type PRKD1 cases that do not harbor rearrangements in PRKD1/2/3.
Taken together, our study shows that polymorphous adenocarcinoma/cribriform adenocarcinoma spectrum of tumors shares histologic and genetic features, and that PRKD1/2/3 fusion is associated with high percentage of papillary growth and a high risk of nodal metastasis. These findings support the notion that these tumors represent a spectrum of related lesions driven by genetic alterations in PRKD genes, rather than separate entities. Fusion-positive tumors, however, appear to be more aggressive clinically and may require additional treatments (e.g., neck dissection). Larger, multicentric studies to define the clinical behavior of these tumors stratified according to their driving genetic alteration are warranted.
References
Waldron CA, el-Mofty SK, Gnepp DR. Tumors of the intraoral minor salivary glands: a demographic and histologic study of 426 cases. Oral Surg Oral Med Oral Pathol. 1988;66:323–33.
Evans HL, Luna MA. Polymorphous low-grade adenocarcinoma: a study of 40 cases with long-term follow up and an evaluation of the importance of papillary areas. Am J Surg Pathol. 2000;24:1319–28.
Mills SE, Garland TA, Allen MS Jr. Low-grade papillary adenocarcinoma of palatal salivary gland origin. Am J Surg Pathol. 1984;8:367–74.
Castle JT, Thompson LD, Frommelt RA, Wenig BM, Kessler HP. Polymorphous low grade adenocarcinoma: a clinicopathologic study of 164 cases. Cancer. 1999;86:207–19.
Weinreb I, Zhang L, Tirunagari LM, Sung YS, Chen CL, Perez-Ordonez B, et al. Novel PRKD gene rearrangements and variant fusions in cribriform adenocarcinoma of salivary gland origin. Genes Chromosomes Cancer. 2014;53:845–56.
Kimple AJ, Austin GK, Shah RN, Welch CM, Funkhouser WK, Zanation AM, et al. Polymorphous low-grade adenocarcinoma: a case series and determination of recurrence. Laryngoscope. 2014;124:2714–9.
Michal M, Skalova A, Simpson RH, Raslan WF, Curik R, Leivo I, et al. Cribriform adenocarcinoma of the tongue: a hitherto unrecognized type of adenocarcinoma characteristically occurring in the tongue. Histopathology. 1999;35:495–501.
Gnepp DR. Salivary gland tumor “wishes” to add to the next WHO tumor classification: sclerosing polycystic adenosis, mammary analogue secretory carcinoma, cribriform adenocarcinoma of the tongue and other sites, and mucinous variant of myoepithelioma. Head Neck Pathol. 2014;8:42–9.
Michal M, Kacerovska D, Kazakov DV. Cribriform adenocarcinoma of the tongue and minor salivary glands: a review. Head Neck Pathol. 2013;7:S3–11.
Laco J, Kamaradova K, Vitkova P, Sehnalkova E, Dvorakova S, Vaclavikova E, et al. Cribriform adenocarcinoma of minor salivary glands may express galectin-3, cytokeratin 19, and HBME-1 and contains polymorphisms of RET and H-RAS proto-oncogenes. Virchows Arch. 2012;461:531–40.
Skalova A, Sima R, Kaspirkova-Nemcova J, Simpson RH, Elmberger G, Leivo I, et al. Cribriform adenocarcinoma of minor salivary gland origin principally affecting the tongue: characterization of new entity. Am J Surg Pathol. 2011;35:1168–76.
Cocek A, Hronkova K, Voldanova J, Sach J, Skalova A, Ambrus M, et al. Cribriform adenocarcinoma of the base of the tongue and low-grade, polymorphic adenocarcinomas of the salivary glands. Oncol Lett. 2011;2:135–8.
Weinreb I, Piscuoglio S, Martelotto LG, Waggott D, Ng CK, Perez-Ordonez B, et al. Hotspot activating PRKD1 somatic mutations in polymorphous low-grade adenocarcinomas of the salivary glands. Nat Genet. 2014;46:1166–9.
Xu B, Aneja A, Ghossein R, Katabi N. Predictors of outcome in the phenotypic spectrum of polymorphous low-grade adenocarcinoma (PLGA) and cribriform adenocarcinoma of salivary gland (CASG): a retrospective study of 69 patients. Am J Surg Pathol. 2016;40:1526–37.
Fonseca I, Assaad A, Katabi N, Seethala RR, Weinreb I, Wenig BM. Polymorphous adenocarcinoma. In: El-naggar AK, Chan JKC, Grandis JR, Takata T, Slootweg PJ, (eds.) WHO Classification of Head and Neck Tumours. 4th ed. Lyon: International Agency for Research on Cancer; 2017.
Geyer FC, Li A, Papanastasiou AD, Smith A, Selenica P, Burke KA, et al. Recurrent hotspot mutations in HRAS Q61 and PI3K-AKT pathway genes as drivers of breast adenomyoepitheliomas. Nat Commun. 2018;9:1816.
Wilkerson PM, Dedes KJ, Samartzis EP, Dedes I, Lambros MB, Natrajan R, et al. Preclinical evaluation of the PARP inhibitor BMN-673 for the treatment of ovarian clear cell cancer. Oncotarget. 2017;8:6057–66.
Piscuoglio S, Burke KA, Ng CK, Papanastasiou AD, Geyer FC, Macedo GS, et al. Uterine adenosarcomas are mesenchymal neoplasms. J Pathol. 2016;238:381–8.
Majewska H, Skalova A, Weinreb I, Stodulski D, Hyrcza M, Stankiewicz C, et al. Giant cribriform adenocarcinoma of the tongue showing PRKD3 rearrangement. Pol J Pathol. 2016;67:84–90.
Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–8.
Piscuoglio S, Fusco N, Ng CK, Martelotto LG, da Cruz Paula A, Katabi N, et al. Lack of PRKD2 and PRKD3 kinase domain somatic mutations in PRKD1 wild-type classic polymorphous low-grade adenocarcinomas of the salivary gland. Histopathology. 2016;68:1055–62.
Acknowledgements
Research reported in this publication was partly funded by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (grant no. P30CA008748). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. JSR-F is partly funded by the Breast Cancer Research Foundation and BW by Cycle for Survival.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
JSR-F reports personal/consultancy fees from VolitionRx, Page.AI, Goldman Sachs, Grail, Ventana Medical Systems, Invicro, Roche Diagnostics, and Genetech, outside the scope of the submitted work. The remaining authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sebastiao, A.P.M., Xu, B., Lozada, J.R. et al. Histologic spectrum of polymorphous adenocarcinoma of the salivary gland harbor genetic alterations affecting PRKD genes. Mod Pathol 33, 65–73 (2020). https://doi.org/10.1038/s41379-019-0351-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41379-019-0351-4
This article is cited by
-
Nodal Metastasis in Low Grade Salivary Gland Carcinoma: A Retrospective Analysis of Incidence, Histologic Spectrum and Outcome
Head and Neck Pathology (2025)
-
Unraveling complexity and leveraging opportunities in uncommon breast cancer subtypes
npj Breast Cancer (2025)
-
Clinicopathological Features of 26 Intraoral Polymorphous Adenocarcinomas from a Single Brazilian Institution
Head and Neck Pathology (2024)
-
Molecular pathology in diagnosis and prognostication of head and neck tumors
Virchows Archiv (2024)
-
Speicheldrüsenkarzinome – ein aktueller Überblick
Die Pathologie (2023)