Abstract
The aim of this study was to assess the performance of fluorescence in situ hybridization (FISH) in identifying the copy number profiles of the three key peritoneal mesothelioma tumor suppressor genes BAP1, CDKN2A, and NF2, with particular emphasis on minute homozygous deletions, a copy number abnormality recently unveiled at the 3p21 (BAP1) chromosomal region using high-throughput methods. FISH was performed on 75 formalin-fixed-paraffin-embedded peritoneal mesotheliomas and recognized two types of monoallelic loss (monosomy, and hemizygous deletion) and two types of biallelic loss (canonical homozygous deletion with a complete loss of FISH signal and homozygous deletion with diminished signal). Diminished FISH signals revealed deletions occurring within the genomic region covered by the gene-specific probe and affected all three tumor suppressors. BAP1 homozygous deletions with diminished signal outnumbered canonical homozygous deletions (13 vs 3): conversely, canonical homozygous deletions were prevalent for CDKN2A (2 vs 14). Diminished signal homozygous deletion was the only pattern of biallelic loss observed for NF2 (2 cases). Hemizygous deletion mainly affected BAP1 (21 vs 6), while monosomy was prevalent for CDKN2A (14 vs 7) and particularly for NF2 where it accounts for all monoallelic losses. FISH/immunohistochemistry (BAP1, CDKN2A, and MTAP) correlation showed that all homozygous deletions, including those with diminished signals, resulted in a null BAP1 and CDKN2A immunophenotype but only canonical CDKN2A homozygous deletions resulted in MTAP loss of expression. BAP1 hemizygous deletion, but not monosomy, was also invariably associated with loss of protein expression whereas neither type of CDKN2A monoallelic loss correlated with p16 or MTAP immunohistochemistry. Array comparative genomic hybridization performed on a spontaneously emerging peritoneal mesothelioma cell line provided support for the interpretation of the FISH patterns and allowed us to extend the number of chromatin remodeling factors involved in mesothelioma to SETD7 and PCGF5, two previously unreported genes.
Similar content being viewed by others
Introduction
Malignant mesothelioma is a rare and aggressive neoplasia arising from mesothelium-covered body cavities, mostly pleura (80%) and peritoneum (10–20%) [1]. Over the last few years, the panorama of genomic alterations in mesothelioma has been broadened by the introduction of high-throughput technologies such as next generation sequencing [2], including targeted next generation sequencing [3], whole exon sequencing [4], array comparative genomic hybridization [5], and integrations of different approaches [6, 7].
In addition to the canonical cyclin-depended kinase inhibitor 2A (CDKN2A) and Neurofibromin 2 (NF2) genes, the most frequent alterations in mesothelioma involve chromatin regulatory genes such as DDX3X, ARID1A, SETD2, PBRM1, SMARCC1, and particularly, the BRCA1 associated protein 1 (BAP1), which is the most frequently deregulated [8,9,10]. Interestingly, four of these genes (BAP1, SETD2, PBRM1, and SMARCC1) map to the 3p21 genomic region. A recent combined custom array comparative genomic hybridization and target next generation sequencing study of 3p21 [8] has revealed a complex copy number abnormality profile characterized by multiple noncontiguous minute deletions in BAP1 and the flanking genes mentioned above. BAP1 small intragenic deletions have been also frequently detected by multiplex ligation-dependent probe amplification [11]. Moreover, BAP1 can be inactivated by disruptive chromosome translocation/gene fusion [12], a mechanism that cannot be detected by mutational or copy number analysis, including multiplex ligation-dependent probe amplification. Given the variety of possible mechanisms involved in BAP1 inactivation and the difficulty in detecting all possible inactivation events by using a single molecular biology approach, immunohistochemistry is still the most widely used and most reproducible means for assessing the gene status [13].
Although fluorescence in situ hybridization (FISH) is the technique of choice for assessing CDKN2A [14] and, to a lesser extent NF2 [15, 16], it has been rarely used to evaluate BAP1 [17], probably due to the fact that BAP1 is mostly inactivated by a mutation in mesothelioma. For CDKN2A alterations, the most frequently reported cause of biallelic loss is a large 9p21 homozygous deletion that can be detected by FISH at different rates in pleural versus peritoneal mesothelioma (more than 80% versus only 20–30%, respectively) [15], while mutation seems to be infrequent at both tumor occurrence sites [7, 16]. The studies addressing the usefulness of the immunohistochemistry detection of the p16 protein encoded by CDKN2A gene in mesothelioma have exhibited variable results and low specificity [18, 19]. However, immunohistochemistry evaluation of the protein product of the methylthioadenosine phosphorylase (MTAP), a 9p21.3 gene often codeleted with CDKN2A, have exhibited both high sensitivity and specificity suggesting MTAP immunohistochemistry as a reliable surrogate for CDKN2A deletion [20].
The NF2 locus, less extensively studied, is known to be targeted by various mechanisms of biallelic inactivation: typically, one allele is frequently lost as a result of monosomy 22 or a large chromosome 22 deletions detectable by FISH [16], whereas the other is targeted by more subtle inactivation events such as mutations, small internal gene deletions, and disruptive translocations [7, 12, 21].
The majority molecular studies have been performed on pleural mesothelioma, the most common site of the disease, whereas only few studies have been carried out on peritoneal mesothelioma which accounts for only 10–20% of all mesotheliomas. Studies have reported a lower mutational burden in peritoneal versus pleural mesothelioma with the possible exception of BAP1 [9, 16, 22,23,24].
Here we gained insights into the molecular landscape of peritoneal mesothelioma and tested the efficacy and feasibility of FISH in detecting genomic losses at the three chosen tumor suppressor genes loci. Taking advantage of the availability of a spontaneous cell line obtained in our laboratory from one patient with peritoneal epithelioid mesothelioma we first profiled it by means of array comparative genomic hybridization. The comparative genomic hybridization data were then integrated with the classical cytogenetic and FISH analysis performed on the established cell line, the first in vitro passage of tumor tissue culture (primary culture) and the formalin-fixed tumor tissue sample. FISH analysis was then extended to all series (further 74 formalin-fixed consecutive cases) of peritoneal mesotheliomas. FISH results were complemented by immunohistochemistry analyses of BAP1, P16, and MTAP. Array comparative genomic hybridization was shown to be essential for the correct interpretation of the FISH patterns observed in the index case and, consequently, in the full series of surgical specimens, and it also revealed further deletions targeting chromatin remodeling genes not described so far.
Materials and methods
Case material
The case material consisted of 75 consecutive formalin-fixed and paraffin-embedded tissue sections obtained from surgical resections from patients with peritoneal mesothelioma. Fifty-two of the 75 specimens were arranged in tissue micro-arrays bearing 4–12 spots for each case. The sex ratio was 1.27 (42 males, 33 females) with a median age of 58 (from 22 to 77) and a mean age of 54.8.
All cases presented mesothelioma involving the entire peritoneal cavity. All of the cases were reviewed by SP: the diagnoses were made on the basis of morphological and immunophenotypical criteria including calretinin, WI1, cytocheratin 5/6 [25], and podoplanin as proposed in the World Health Organization classification [26] and vascular endothelial growth factor receptor 2 as proposed by Miettinen [27]. Seventy-one tumors were epithelioid, two were sarcomatoid, and two were biphasic.
A fresh tumor sample from case #1, an epithelioid mesothelioma hereinafter referred as the index case, was processed as described elsewhere [28], to obtain a primary cell culture. After the 50th passage, the cell culture was considered as established cell line.
Array comparative genomic hybridization
High-resolution oligo array comparative genomic hybridization analysis was carried out on the established cell line. Genomic DNA was extracted using a DNeasy Tissue Kit (Qiagen, Valencia, CA, USA) including RNase treatment, and its quality was assessed by using NanoVue (VWR) and agarose gel electrophoresis. The molecular karyotype of the samples was assessed by using the Agilent platform (Agilent Technologies, Santa Clara, CA) with SurePrint G3 Human CGH arrays at the resolution of 180 K and the Agilent Genomic DNA ULS labeling kit in accordance with the manufacturer’s protocol. The image was acquired using the Agilent G2565CA Microarray Scanner System and the raw data were processed using Feature Extraction Software. The data were analyzed using Agilent’s Genomic Workbench Standard Edition 6.5.0.58. Copy number abnormalities (CNAs) were defined either as gain or loss based on the log2 ratio distribution in Genomic Workbench software. Copy number variations found in the Database of Genomic Variants (http://dgv.tcag.ca) and the regions containing no genes were excluded.
Chromosome preparation and G banding
Chromosomes were prepared from the established mesothelioma cell line and from the primary cell culture using standard procedures, and G banding was obtained by means of Wright staining. Ten metaphase spreads from both the established cell line and primary culture were analyzed using an Olympus BX41 microscope (Tokyo, Japan). The metaphases were captured and karyotypes analyzed using McType software.
Immunohistochemistry
The immunohistochemical analyses were carried out using an automated immunostaining device (BenchMark Ultra, Ventana Medical Systems, Tucson, AZ, USA) in accordance with the manufacturer’s instructions. BAP1 protein expression was investigated using the sc-28383 antibody (Santa Cruz, Dallas, TX) at 1/100 dilution. MTAP immonohistochemistry was performed with Abnova (clone 2G4) (Tebu-bio, Le-Perray-en-Yvelines, France) at 1/200 dilution. P16 expression was determined using the CINtec p16INK4a histology kit (Roche, Basel, Switzerland).
FISH
The CDKN2A Spectrum Orange/CEP9 Spectrum Green (Abbott Molecular, Des Plains, IL) was used to assess CDKN2A copy number. The FISH probes used for BAP1 evaluation were spectrum green-labeled bacterial artificial chromosome clone RP11–630O10 (3p21.1) and spectrum orange-labeled clone RP11-584A6 covering the MITF gene (3p13) as the control. Those used for NF2 were spectrum green-labeled clone RP11-506F7 (22q12.1) and spectrum orange-labeled clone RP11- 551L12 covering the PDGFB gene (22q13.1) as the control.
Bacterial artificial chromosome clones were obtained from C.H.O.R.I. (bac-pac resources. Children’s Hospital Oakland Research Institute, CA) and labeled by means of nick translation (Nick Translation Reagent Kit, Abbott Molecular) in accordance with the manufacturer’s instructions and were validated on normal metaphase spreads.
The FISH experiments were performed on chromosome preparations from both the primary mesothelioma culture and established cell line as well as on the formalin-fixed samples using standard protocols. Formalin-fixed paraffin-embedded samples included the 75 peritoneal mesothelioma samples and four of normal mesothelium (Omenta) samples as controls.
Ten metaphase samples from both cell cultures, a minimum of 50 tumor nuclei from the formalin-fixed tissue samples and 200 nuclei from each of the controls were scored using a Leica DM 6000B (Wetzlar, Germany) microscope at ×100 magnification and the appropriate fluorescence filters. The images were captured using Cytovision software (v. 7.0, Leica). Cut-off values for formalin-fixed FISH assessment were based on the proportion of cells carrying the abnormal pattern: 30% for Homozygous and Hemizygous deletions, and 40% for monosomy.
Morphometric evaluation of FISH signals
A tentative morphometric evaluation of FISH signal size was carried out for three hybridization experiments, one for each tumor suppressor gene: BAP1 and CDKN2A from the case index, and NF2 from case #55. A representative image (TIFF format) for each tissue FISH was analyzed using software tools available through ImageJ (http://rsb.info.nih.gov/ij/). After spitting the color channels, the tumor suppressor gene-specific fluorochrome was selected (Spectrum Green for BAP1 and NF2, Spectrum Orange for CDKN2A), image thresholds were determined in order to isolate the FISH signals, the “particle analysis” ImageJ software tool was then applied to measure each individual FISH signal. The FISH signals from nontumor cells and from tumor cells were compared using a t-test. Data were processed using R environment for statistical computing and graphics (R Foundation, Vienna, Austria - http://www.R-project.org/). P values < 0.05 will be consider statistically significant.
Statistical analysis
We used Fisher’s exact bilateral test to evaluate the association between FISH/Immunohistochemistry alterations (BAP1 homozygous deletion, BAP1 hemyzigous deletion, CDKN2A homozygous deletion, NF2 Monosomy, NF2 homozygous deletion, BAP1 immunohistochemistry, and P16 immunohistochemistry) and sex/age (<58 versus ≥58). P values < 0.05 were consider statistically significant.
Results
Index case
Established cell line. Karyotype and comparative genomic hybridization
The mesothelioma cell line exhibited a pseudo-diploid karyotype (45, XY, +Y, −3, −3, +der(6)add(6)(q14)−10, −13, −18, −20, add(22)(q12.1), +3mar) (Fig. 1b). Array comparative genomic hybridization showed 17 CNAs, consisting of three gains and 14 losses (two of which were homozygous).
Index case BAP1: Established cell line, primary culture and formalin-fixed paraffin-embedded tumor. a Established cell line array comparative genomic hybridization profile for 3p21.1 locus showing the 145 kb Homozygous deletion (log2 ratio = −4) involving BAP1 gene. SEMA3G gene is also biallelically deleted while PBRM1 gene appear to be within the single copy (log2 ratio = −1) loss region. The vertical green line on the right indicates the position of the 180 kb RP11-630O10 clone employed for BAP1 FISH analysis. b Karyogram of the established cell line. The karyotype is described in the text. Red arrow indicates Marker 1 (mar1), green arrow indicates der (22) add (22)(q12.2). c Partial metaphase spread from the established cell line hybridized with BAP1 (Spectrum Green) and MITF (Spectrum Orange) probe mixture (counterstained with DAPI). BAP1 single signal on the der (22). MITF single signal on the mar1. d G banded partial metaphase from index case primary culture: Green arrow indicates der (22), Red arrow indicates mar1 and double Red arrow indicates the ring chromosome. e Partial metaphase spread from the index case primary culture hybridized with BAP1 (Spectrum green) and MITF (Spectrum Orange) probe mixture (counterstained with DAPI). BAP1 single signal on the der (22) indicated with green arrow. MITF single signal on the mar1, indicated with red arrow and two further MITF signals on the ring chromosome indicated with double red arrow. f Tumor tissue section from the index case hybridized with BAP1 (Spectrum Green) and MITF (Spectrum Orange) probe mixture counterstained with DAPI. Tumor cells display a variable number of MITF signals and a single BAP1 signal. A nontumor cell and a tumor cell are highlighted in white and yellow boxes, respectively, and shown at higher magnification on the right. Notice loss of association between BAP1 and MITF signals in the tumor cells coherent with cytogenetic profile. BAP1 FISH signal is diminished in the tumor cell compared with nontumor cells. Figure 1F inset: Box plot representing the distribution of FISH signal size in nontumor (WT) and tumor (dim) cells. p = 0.001419. g Tumor tissue section BAP1 immunohistochemistry indicate complete loss of expression. Positive controls represented by lymphocytes and stromal cells
All of the gains were single copy (log2 ratio = 0.5) and involved large chromosomal areas: chromosome 3 (from 3q13.31 to 3q 28, Fig. 1a); chromosome 6 (from the short arm telomere to q14.1); and the whole of chromosome Y.
Single-copy losses (log2 ratio = −1) were divided into large and focal losses, the latter defined as losses of ≤2 Megabases (Mb).
There were five large regions of single-copy loss: chromosome 3 from p14.3 to q13.31; chromosome 10 from the short arm telomere to 10q21.1; the whole chromosome 13; chromosome 18q from q12.3 to the long arm telomere; and chromosome 22q from q12.1 to the telomere.
Focal single-copy losses were identified in seven regions: 3p21.31 (2 Mb), 3p21.1 (1.5 Mb.) (Fig. 1a), two small noncontiguous regions in 4q13.3 (280 and 600 Kb), 4q31.1 (1 Mb); 10q23.31 (2 Mb), and 20 p13 (670 kb).
Four chromatin remodeling/oncosuppressor genes presented monoallelic losses: SMARCC1 in 3p21.31, PBRM1 in 3p21.1 (Fig. 1a), SETD7 in 4q31, and PCGF5 in 10q23.31.
Finally, there were two homozygous deletions, one of 180 kb within the monosomic 3p21.1 region, involved BAP1 and SEMA3G (Fig. 1a) and the other, focused on CDKN2A in 9p21.3(143 kb) (Fig. 2a), consisted of a sharp switch from a disomy (log2 ratio = 0) down to a homozygous loss (log2 ratio < −2). This particular deletion profile, previously described in mesothelioma cell lines [6], suggests a mechanism involving monoallelic deletion followed by duplication of the deleted chromosome and loss of the normal one. Of note, CDKN2A neighbor gene MTAP, was not involved in the focal loss (Fig. 2a).
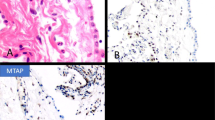
Index case CDKN2A: Established cell line array comparative genomic hybridization and formalin-fixed paraffin-embedded tumor FISH. a Established cell line detailed array comparative genomic hybridization profile for 9p21.3 locus showing the 143 kb homozygous deletion (log2 ratio 2 = − 4) involving CDKN2A gene. Notice the two break points ‘sharp’ profile of the homozygous deletion characterized by sudden shift from a normal copy number status to a homozygous deletion (see text). The vertical red line on the right indicates the position of the 222 kb Vysis locus specific CDKN2A FISH probe employed. Notice that MTAP gene in not included in the region of homozygous deletion. b Tumor tissue section from the index case hybridized with the Vysis CDKN2A Spectrum Orange/CEP9 Spectrum Green probe (counterstained with DAPI). Tumor cells display two signals of both CDKN2A and CEP9. A nontumor cell and a tumor cell are highlighted in white and yellow boxes, respectively, and shown at higher magnification. CDKN2A FISH signals are diminished in the tumor cells compared with nontumor cells. Figure 1B inset: Box plot representing the distribution of FISH signal size in nontumor(WT) and tumor(dim) cells (p = 0.000126). c Tumor tissue section from the index case. Immunohistochemistry indicating P16 complete loss of expression (upper panel) and preservation of MTAP expression (lower panel)
Cytogenetic and FISH profiles of primary culture, established cell line and original tumor. Focus on tumor suppressor genes
Cytogenetic
Classical cytogenetics showed that the primary cell culture was characterized by multiple related clones that all share chromosomal markers with the established cell line: in particularly, a der (22) add 22q(12.1), and a large sub-metacentric marker chromosome (mar1) (Fig. 1b, d). The majority of metaphases from the primary culture contained marker ring chromosomes not present in the established cell line (Fig. 1d) and appeared to vary in size between clones. Chromosome 9 homologs appeared to be normal in both the established cell line, and the primary culture.
FISH: BAP1/MITF. Immunohistochemistry: BAP1
The der 22 and mar1 respectively carried single-copy BAP1 and MITF signals in both the established cell line and the primary culture (Fig. 1c, e). The primary cell line often carried from two to six copies of the MITF gene on the ring chromosome marker (Fig. 1e) which were presumably lost in long term culture.
The FISH profile of the index tissue sample (Fig. 1f) closely mirrored that of the primary cell culture; different clones showed 1–10 MITF signals in addition to the single BAP1. However, the single BAP1 signal observed in the tumor cells appeared to be diminished in comparison with the full-size BAP1 signals present in the surrounding nontumor cells (1F). Morphometric comparison of FISH signals of tumor (13 signals) versus nontumor cells (34 signals) by dedicated ImageJ software tool confirmed a significant (p = 0.001419) decrease of size in tumor cells (inset Fig. 1f). This finding is in line with the comparative genomic hybridization profile which shows a 3p21.1 homozygously deleted region spanning only part of the probe used for the FISH analysis (Fig. 1a). BAP1 immohistochemistry was negative throughout the tissue sample (Fig. 1g).
FISH: CDKN2A/CEP9. Immunohistochemistry: P16 and MTAP
In both the established cell line and the primary culture, chromosome 9 seemed to have a normal FISH profile, with two centromeric signals and two identical size CDKN2A signals (not shown). This observation is in line with its comparative genomic hybridization profile which showed a two-break point homozygous deletion spanning only part of the genomic region covered by probe (Fig. 2a), which indicates identical deletions on both chromosome 9 homologs. As was seen for BAP1, only comparison of the tissue section of CDKN2A FISH signals of tumor cells with those of the surrounding nontumor cells revealed the diminished pattern (Fig. 2b). This observation was supported by ImageJ software analysis (17 tumor versus 9 nontumor FISH signals, p = 0.000126) (inset Fig. 2b). The immunohistochemical profile, indicating loss of p16 and maintenance of MTAP expression (Fig. 2c), was consistent with the comparative genomic hybridization results and further supported the notion of focal CDKN2A deletion.
FISH: NF2/PDGFB
NF2/PDGFB FISH showed single-copy signals of both probes carried by the normal chromosome 22 in all of the metaphases of the primary culture and established cell line (not shown), whereas no NF2/PDGFB signals were observed on der 22. This pattern is in line with the chromosome 22 comparative genomic hybridization profile. The tumor tissue sample had a monosomic FISH pattern.
It is worth noting that all of the critical gene losses (i.e., BAP1 and CDKN2A diminished signals, and the monosomy-like loss of NF2) were present in all of the tumor cells from the original tumor and were maintained in the established cell line.
Formalin-fixed paraffin-embedded series
FISH pattern classification
On the basis of the comparative genomic hybridization profiles and the FISH strategy applied, four patterns have been detected: canonical homozygous deletion, homozygous deletion with diminished signals, hemizygous deletion (corresponding to a chromosomal loss involving one copy of BAP1, CDKN2A or NF2, but no loss of the respective control probes MITF, CEP9, and PDGFB), and monosomy (corresponding to the loss of one copy of both the tumor suppressor gene and control probe). Figure 3 illustrates FISH, immunohistochemistry, and clinical data for the series.
Formalin-fixed paraffin-embedded series: Histological, Clinical, Immunohistochemical, and FISH features. Each column represents a patient. Rows indicate (from top to bottom): Histological type, Age at diagnosis, Sex, BAP1/MITF FISH, BAP1 immunohistochemistry, CDKN2A/ CEP9 FISH, P16 immunohistochemistry, MTAP immunohistochemistry, and NF2/PDGFB FISH. Patient cohort is presented according with the number of FISH ‘hits’ or abnormalities reported: BAP1, CDKN2A, NF2 triple hits: cases 1–14. BAP1 and CDKN2A double hits: cases 15–26. BAP1 and NF2 double hits cases 27–29. CDKN2A and NF2 double hits cases 30–35. BAP1 sole hits cases 36–49. CDKN2A sole hits cases 50–54. NF2 sole hits 55–57. No hits for all the remaining. Case #1 is the index case
BAP1
FISH pattern
Evidence of BAP1 loss was found in 43 cases (57%). The pattern was consistent with BAP1 homozygous deletion in 16 cases (21%), 13 of which showed one (8 cases) (Fig. 4a) or more (5 cases) diminished signals. No full-strength BAP1 signals were observed in the cases carrying diminished signals. This finding underlines the frequent occurrence of deletions within the 180 kb chromosomal region coved by the BAP1-specific probe (Fig. 1a). Twenty-one cases (28%) showed hemizygous deletion, a pattern that can also be ascribed to a 3p chromosome deletion. The remaining six cases (8%) showed a monosomic pattern of BAP1 loss. MITF copy number gains were observed in six of the BAP1 homozygous deletion cases (including the index case), and four of the hemizygous deletion cases. A balanced gain of BAP1 and MITF (chromosome 3 polysomy) was seen in 13 cases. Finally, one case (#65) showed amplification of the MITF gene with more than 10 copies together with two full-size copies of BAP1.
Formalin-fixed paraffin-embedded series. FISH pattern and Immunohistochemistry: BAP1. a Case #6. Tumor tissue section hybridized with BAP1 (Spectrum Green) and MITF (Spectrum Orange) probe mixture (counterstained with DAPI). Tumor cells display three copies of MITF and a single diminished copy of BAP1. A nontumor cell and a tumor cell are highlighted in white and yellow boxes, respectively, and shown at higher magnification. Notice the association between BAP1 and MITF is preserved in the tumor cells. b Case #6. Tumor tissue section. BAP1 immunohistochemistry: loss of BAP1 expression
No BAP1/MITF alterations (including diminished signals) were observed in the normal mesothelium controls.
Correlations between FISH and immunohistochemistry findings
Fifty-five of the 75 cases (73%) had a null BAP1 immunophenotype. All of the 37 cases with BAP1 homozygouos deletions (including those with diminished signals) or hemizygous deletions were negative for BAP1 immunohistochemistry, versus only three of the six (50%) monosomic cases (Fig. 3). Finally, 15 out of 32 cases showing no BAP1 loss by FISH, were negative by immunohistochemistry (46.8%).
CDKN2A/p16
FISH patterns
Evidence of CDKN2A loss was found in 37 out of the 72 (51%) cases evaluated (three cases failed FISH). CDKN2A homozygous deletions were observed in 16 cases (22%), 14 of which showed the canonical pattern with the complete loss of FISH signal (Fig. 5d). Only two cases (including the index case) had a diminished FISH pattern, both with two diminished CDKN2A signals of similar size (Fig. 5a).Of note, the correct interpretation of FISH findings in such cases required a comparison between tumor cells and the surrounding nontumor cells. Seven cases (9%) showed hemizygous deletions and 14 (19%) monosomy. A balanced gain of both CDKN2A and centromere 9 (chromosome 9 polysomy) was observed in five cases.
Formalin-fixed paraffin-embedded series. FISH patterns and immunohistochemistry: CDKN2A/MTAP. a Case #14. Tumor tissue section hybridized with the Vysis CDKN2A Spectrum Orange/CEP9 Spectrum Green probe (counterstained with DAPI). Tumor cells display two CDKN2A signals and three CEP9 signals. A nontumor cell and a tumor cells are highlighted in white and yellow boxes, respectively, and shown at higher magnification (top panels). Notice the size of both CDKN2A FISH signals are greatly diminished in the tumor cells compared with nontumor cells. b Case #14. Tumor tissue section. P16 Complete loss of expression. c Case #14. Tumor tissue section MTAP immunohistochemistry. Positivity. d Case #51. Tumor tissue section hybridized with the Vysis CDKN2A Spectrum Orange/CEP9 Spectrum Green probe (counterstained with DAPI). Canonical CDKN2A homozygous deletion: Tumor cells display multiple CEP9 and no CDKN2A signals. Internal control present. e Case #51. Tumor tissue section P16 immunohistochemistry. Complete loss of expression. f Case #51. Tumor tissue section MTAP immunohistochemistry. Complete loss of expression
No CDKN2A/CEP9 alterations (including diminished signals) were observed in the normal mesothelium controls.
Correlations between FISH and immunohistochemistry (CDKN2A and MTAP)
P16 immonohistochemistry was evaluable in 70 cases. The loss of p16 expression was observed in 37 out of the 68 cases (54%) evaluated by both FISH and immonohistochemistry. All of the 16 homozygous deletion cases (including the two diminished signals cases) were negative by immunohistochemistry (Fig. 5b, e) but, differently from BAP1, there was no correlation between CDKN2A hemizygous deletion and immunohistochemistry as three out of six cases (50%) were negative; only 7 of the 14 (50%) monosomic cases showed loss of p16 expression. Finally, 11 out of the 31 (35%) cases which showed no loss of CDKN2A upon FISH were p16 immunohistochemistry-negative.
MTAP immonohistochemistry was evaluated in 70 cases. Loss of MTAP expression was exclusively observed in the cases with canonical homozygous deletion (13 cases) (Fig. 5d, f) whereas both cases with diminished signals were positive (Fig. 2c, Fig. 5c). All nondeleted cases, monosomies and hemizygous deletions were MTAP positive by immonohistochemistry. Considering CDKN2A FISH as the gold standard method, p16 immonohistochemistry appears to have high sensitivity (100%) but very low specificity (42%) whereas MTAP immonohistochemistry detection appears to be highly specific (100%) but less sensitive (86.6%).
NF2 FISH pattern
The most frequently detected abnormality was NF2/PDGFB monosomy, which was observed in 24 of the 71 cases tested (34%). No hemizygous deletion was observed, however, two cases (3%) presented a pattern consistent with NF2 homozygous loss with a single diminished NF2 signal and a single full-strength control probe signal (not shown). ImageJ software analysis of FISH images from case #55 supported this observation (p = 0.0003171). As in the case of BAP1 and CDKN2A, this pattern can be ascribed to a small deletion within the genomic area covered by the probe used. Copy gains involving both NF2 and PDGFB (chromosome 22 polysomy) were observed in seven cases (9%). No NF2/PDGFB alterations (including diminished signals) were observed in the normal mesothelium controls.
Formalin-fixed, paraffin-embedded series FISH data overview: hit groups
Based on the number of FISH losses (referred to as hits), the patient samples were divided into four classes: three hits in 14/71(19.7%), two hits in 21/71 (29.5%), one hit in 22/71 (30.9%), and no hits in 16/71 (22,5%) (Fig. 3).
Figure 3 shows the FISH patterns and the corresponding immunophenotypes by hit groups. BAP1 seems to be the most affected gene (43/75, 57.3%) and was also the most frequent sole loss (14, 63.9% of the 22 samples in the single-hit group); remarkably, 32% of the BAP1-deleted cases (14/43) showed no other alteration. On the contrary, only 13% of the CDKN2A (5/37) and 11.5% of the NF2 deletions (3/26) were sole abnormalities.
The most frequent double hit was BAP1/CDKN2A (12 cases) followed by CDKN2A/NF2 (six cases) and BAP1/NF2 (three cases)
Statistical analysis showed no correlation between sex or age (<58 versus ≥58) with any of the FISH/immunohistochemistry features evaluated with the exception of BAP1 hemizygous deletion which appeared to be more frequent (p < 0.03) in the ≥58 age group
Discussion
To the best of our knowledge, this is the first study describing the performance of FISH to detect the presence of minute deletions involving three key tumor suppressor genes in routinely processed specimens of peritoneal mesothelioma. To do this, we took advantage of the spontaneous emergence of a cell line from one of our cases, whose array comparative genomic hybridization profile not only supported the interpretation of the FISH patterns observed in the case material (75 formalin-fixed, paraffin-embedded specimens) included in this study, but also allowed us to extend the number of genes involved in chromatin remodeling in peritoneal mesothelioma.
The established cell line comparative genomic hybridization profile showed a prevalence of losses (14, two of which were homozygous) and just three gains, in line with previously published findings [5]. In particular, BAP1 and CDKN2A were targeted by homozygous deletion while NF2 by monoallelic loss. Chromosome 3 exhibited a complex profile, particularly at the 3p21 band with a focal homozygous loss involving BAP1 and SEMA3G, a class 3 semaphorin frequently deleted in epithelioid mesothelioma, and single-copy loss of SMARCC1 and PBRM1, both members of the SWI/SNF complex, known to be biallelically inactivated in a subset of mesotheliomas [8, 29].
The involvement of the 3p21 chromosomal region in mesothelioma has been reported since the time of the early cytogenetic studies [30]. More recently, a study using high-density array comparative genomic hybridization and target next generation sequencing specifically concentrated on this region, revealed a complex pattern of multiple noncontiguous minute deletions targeting a cluster of oncosuppressors involved in chromatin regulation including: SETD2, SMARCC1, BAP1, and PBRM1 [8]. Our comparative genomic hybridization findings support the role of these tumor suppressor genes in peritoneal mesothelioma and, interestingly, have revealed focal deletions of two previously unreported genes encoding chromatin remodeling factors: SETD7 at 4q31.1 and PCGF5 at 10q23.31.
PCGF5 is a member of a polycomb group multiprotein PRC1-like complex [31]. Interestingly, the deregulation of EZH2, a member of the PRC2 complex, is involved in peritoneal mesothelioma cell stemness and plasticity [32].
SETD7 is a hystone methyltransferase [33] closely related to SETD2, one of the 3p21 frequently inactivated genes in mesothelioma. These remodeling factors, by compacting and de-compacting chromatin fibers, control the proper execution of critical processes such as gene transcription DNA replication and repair. They have recently emerged as potential gatekeepers for the maintenance of genome integrity [34] and are likely to be important players in the mesothelioma genetic scenario [35].
The comparative genomic hybridization profile of our cell line was crucial in order to assess the performance of FISH in detecting losses at 3 key tumor suppressor genes (BAP1, CDKN2A, and NF2). FISH analysis was performed in the primary culture, established cell line and the original formalin-fixed paraffin-embedded tumor tissue samples. However, only the tumor tissue samples allowed us to recognize the decrease in BAP1 and CDNK2A FISH signals (diminished) thanks to the presence of the internal control provided by nontumor cells (not represented in the cell cultures). NF2 comparative genomic hybridization and FISH profiles were also coherent as they showed large chromosome 22 deletion and monosomy NF2/PDGFB, respectively. The peculiar pattern of diminished BAP1 and CDKN2A exhibited by tumor cells, in addition to suggesting possible biological contribution to malignant phenotype, warned us to be cautious in interpreting the FISH results. In fact, when we extended the FISH analysis to the entire tissue section series of 75 cases, we were able to recognize the pattern of diminished signals for all of the tested tumor suppressor genes. Diminished FISH signals indicate deletions occurring within the probe covered genomic area and leading to the loss of a consistent portion of it [36].
In the case of CDKN2A homozygous deletions are often large as they involve the neighbor MTAP gene, hence the use of MTAP immunohistochemistry as a valid surrogate of 9p21 homozygous deletion [20]. However, about 10% of CDKN2A homozygous deletions have been observed to be smaller and not involving MTAP gene [37, 38]; this is in line with our FISH/Iimmunohistochemistry results showing the diminished pattern in 2 out of 16 homozygously deleted cases (12.5%) both with a coherent, p16 negative/MTAP positive, immunohistochemisty profile (Figs. 2 and 5a–c).
In BAP1, we can hypothesize that the multiple noncontiguous minute deletions already reported for this genomic area [8], singularly are likely to be under the FISH sensitivity threshold, but, cumulatively, may result in diminished FISH signals.
Our FISH analysis revealed a similar occurrence of biallelic losses in BAP1 (21.3%) and CDKN2A (21.9%) but dramatic differences in term of FISH pattern: diminished signal homozygous deletion was prevalent for BAP1 (81.2%) and rare for CDKN2A (12.5%). We also found that the impact of these gene deletions on the corresponding protein expression was the same as shown by the correlations between the FISH and immonohistochemistry findings (Fig. 4).
We also found that the pattern of monoallelic deletion, due to hemizygous deletion or monosomy, was very different in BAP1 and CDKN2A: 77.7% of all the monoallelic losses in BAP1 were hemizygous deletions, whereas 66% of all the monoallelic losses in CDKN2A were monosomies.
The impact of the tumor suppressor genes monoallelic deletion on protein expression was again profoundly different: whereas monosomies seemed equally associated with a positive or null immunophenotype for both BAP1 and CDKN2A, hemyzigous deletions appeared highly predictive of loss of expression of BAP1 but not of p16 or MTAP. BAP1 hemyzigous deletions were the second most frequent abnormality observed in our case material and most frequently the sole alteration, suggesting that a recurrent break point in the chromosomal region between BAP1 to MITF genes may be a frequent event in peritoneal mesothelioma. Furthermore, BAP1 hemyzigous deletions was the only alteration significantly associated with a clinical feature (age ≥ 58) in our series. It is worth mentioning that the fragile histidine triad (FHIT) gene site, which is known to be a target of environmental carcinogenesis, maps to 3p14.2 chromosome band between BAP1 (3p21.1) and MITF (3p13) and may play a role in chromosome 3 instability. Interestingly, the reduced expression of FHIT has been described in mesothelioma tumors and derived cell lines [39].
Our results indicate a high frequency of monosomic losses (33.8%) of NF2, and are in line with previously published observations [16]. To the best of our knowledge, no FISH-based studies have reported NF2 homozygous deletion. It seems that the biallelic NF2 loss, frequently observed in mesothelioma, is achieved by monosomy chromosome 22 coupled with mutation, intragenic deletions or disruptive translocations focused on the remaining allele [7]. Here we identified two cases (2.8%) with NF2 homozygous deletion, both of which had a single diminished FISH signal that, as in the case of BAP1 and CDKN2A, could be interpreted as a deletion occurring within the genomic region covered by the FISH probe.
In addition to underscoring the relevance of genes involved in epigenetic processes, this paper described the first use of FISH to investigate in detail the status of the three critical onco-suppressors in a consecutive series of routinely processed peritoneal mesothelioma tissue specimens. Our findings indicate that: (i) interpreting the FISH pattern of BAP1 is challenging because most of the cases with BAP1 homozygous losses exhibited a pattern characterized by the presence of diminished FISH signals; (ii) diminished signal homozygous deletion is not exclusive for BAP1 as CDKN2A and NF2 show the same FISH pattern (albeit at low frequency); and (iii) diminished FISH signal, is invariably associated with the loss of protein expression, at least in the case of BAP1 and CDKN2A genes. Our approach to assessing BAP1 copy numbers reveals frequent chromosome 3 deletions (28% of cases), leading to hemizygous type losses that seem to be biologically significant insofar as they are invariably associated with the loss of protein expression. Cumulatively homozygous deletions (including those with diminished signal) and hemyzigous deletions account for 69.8% of BAP1 immunohistochemistry negative cases, thus indicating that FISH is highly specific but not very sensitive. Such limitation is shared by all of the molecular techniques used to investigate BAP1 status as no single approach is likely to reveal all of the possible events targeting the gene, thus, immunohistochemistry remains the best methods for assessing BAP1 alteration in mesothelioma. By contrast, FISH is generally considered the gold standard to detect CDKN2A alterations as p16 immunohistochemistry performs poorly in term of specificity. However, MTAP immunohistochemistry appears to be a valid surrogate of 9p21 homozygous deletion, although restrict to the canonical FISH pattern.
Taken together, the results of this study indicate that the routine application of a selected FISH strategy can improve our knowledge and refine the FISH/immunohistochemistry correlations of the three key tumor suppressor genes of peritoneal mesothelioma. They also support the epigenetic relevance of the genes involved, a promising area of study that should be further pursued using dedicated methodologies.
References
Comertpay S, Pastorino S, Tanji M, et al. Evaluation of clonal origin of malignant mesothelioma. J Transl Med. 2014;12:301.
Bueno R, De Rienzo A, Dong L, et al. Second generation sequencing of the mesothelioma tumor genome. PLoS ONE. 2010;5:e10612.
Lo Iacono M, Monica V, Righi L, et al. Targeted next-generation sequencing of cancer genes in advanced stage malignant pleural mesothelioma: a retrospective study. J Thorac Oncol. 2015;10:492–9.
Guo G, Chmielecki J, Goparaju C, et al. Whole-exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer Res. 2015;75:264–9.
Chirac P, Maillet D, Leprêtre F, et al. Genomic copy number alterations in 33 malignant peritoneal mesothelioma analyzed by comparative genomic hybridization array. Hum Pathol. 2016;55:72–82.
Klorin Geula, Rozenblum Ester, Glebov Oleg, et al. Integrated high-resolution array CGH and SKY analysis of homozygous deletions and other genomic alterations present in malignant mesothelioma cell lines. Cancer Genet. 2013;206:191–205.
Hmeljak J, Sanchez-Vega F, Hoadley KA, et al. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov. 2018;8:1548–65.
Yoshikawa Y, Emi M, Hashimoto-Tamaoki T, et al. High-density array-CGH with targeted NGS unmask multiple noncontiguous minute deletions on chromosome 3p21 in mesothelioma. Proc Natl Acad Sci USA. 2016;113:13432–7.
Joseph NM, Chen YY, Nasr A, et al. Genomic profiling of malignant peritoneal mesothelioma reveals recurrent alterations in epigenetic regulatory genes BAP1, SETD2, and DDX3X. Mod Pathol. 2017;30:246–54.
Kato S, Tomson BN, Buys TP, et al. Genomic landscape of malignant mesotheliomas. Mol Cancer Ther. 2016;15:2498–507.
Emi M, Yoshikawa Y, Sato C, et al. Frequent genomic rearrangements of BRCA1 associated protein-1 (BAP1) gene in Japanese malignant mesothelioma-characterization of deletions at exon level. J Hum Genet. 2015;60:647–9.
Bueno R, Stawiski EW, Goldstein LD, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48:407–16.
Carbone M, Gaudino G. Yang recent insights emerging from malignant mesothelioma genome sequencing. Jh Thorac Oncol. 2015;10:409–11.
Hwang HC, Sheffield BS, Rodriguez S, et al. Utility of BAP1 Immunohistochemistry andp16 (CDKN2A) FISH in the diagnosis of malignant mesothelioma in effusion cytology specimens. Am J Surg Pathol. 2016;40:120–6.
Takeda M, Kasai T, Enomoto Y, Takano M, et al. Comparison of genomic abnormality in malignant mesothelioma by the site of origin. J Clin Pathol. 2014;67:1038–43.
Singhi AD, Krasinskas AM, Choudry HA, et al. The prognostic significance of BAP1, NF2, and CDKN2A in malignant peritoneal mesothelioma. Mod Pathol. 2016;29:14–24.
Cigognetti M, Lonardi S, Fisogni S, et al. BAP1 (BRCA1-associated protein 1) is a highly specific marker for differentiating mesothelioma from reactive mesothelial proliferations. Mod Pathol. 2015;28:1043–57.
Chiosea S, Krasinskas A, Cagle PT, Mitchell KA, Zander DS, Dacic S. Diagnostic importance of 9p21 homozygous deletion in malignant mesotheliomas. Mod Pathol. 2008;21:742–7.
Kratzke RA, Otterson GA, Lincoln CE, et al. Immunohistochemical analysis of the p16INK4 cyclin-dependent kinase inhibitor in malignant mesothelioma. J Natl Cancer Inst. 1995;87:1870–5.
Chapel DB, Schulte JJ, Berg K, Churg A, Dacic S, Fitzpatrick C, et al. MTAP immunohistochemistry is an accurate and reproducible surrogate for CDKN2A fluorescence in situ hybridization in diagnosis of malignant pleural mesothelioma. Mod Pathol. 2019. https://doi.org/10.1038/s41379-019-0310-0.
Thurneysen C, Opitz I, Kurtz S, et al. Functional inactivation of NF2/merlin in human mesothelioma. Lung Cancer. 2009;64:140–7.
Ugurluer G, Chang K, Gamez ME, et al. Genome-based mutational analysis by next generation sequencing in patients with malignant pleural and peritoneal mesothelioma. Anticancer Res. 2016;36:2331–8.
Leblay N, Leprêtre F, Le Stang N, et al. BAP1 is altered by copy number loss, mutation, and/or loss of protein expression in more than 70% of malignant peritoneal mesotheliomas. J Thorac Oncol. 2017;12:724–33.
Alakus Hakan, Yost ShawnE, Woo Brian, et al. BAP1 mutation is a frequent somatic event in peritoneal malignant mesothelioma. J Transl Med. 2015;13:122.
Gibbs AR. Tumours of the serosal membranes: armed forces institute of pathology atlas of tumor pathology. fourth series, Fascsicle 3. Edited by Churg A, Cagle PT, Roggli VL. DC: Washington, 2006, p. 147.
Travis WD. Brambilla E, Burke AP, Marx A, Nicholson AG. WHO classification of tumors of the lung, pleura, thymus and heart. 4th ed. Lyon: International Agency on Cancer Research; 2015. p. 412.
Miettinen M, Rikala MS, Rysz J, Lasota J, Wang ZF. Vascular endothelial growth factor receptor 2 (VEGFR2) as a marker for malignant vascular tumors and mesothelioma—Immunohistochemical study of 262 vasular endothelial and 1640 non vascular tumors. Am J Surg Pathol. 2012;4:629–39.
Bosotti R, Magnaghi P, Di Bella S, Cozzi L, Cusi C, Bozzi F, et al. Establishment and genomic characterization of the new chordoma cell line Chor-IN-1. Sci Rep. 2017;7:9226.
Yoshikawa Y, Sato A, Tsujimura T, et al. Frequent deletion of 3p21.1 region carrying semaphorin 3G and aberrant expression of the genes participating in semaphorin signaling in the epithelioid type of malignant mesothelioma cells. Int J Oncol. 2011;39:1365–74.
Lu YY, Jhanwar SC, Cheng JQ, et al. Deletion mapping of the short arm of chromosome 3 in human malignant mesothelioma. Genes Chromosomes Cancer. 1994;9:76–80.
Si S, Nakajima-Takagi Y, Aoyama K, et al. Loss of Pcgf5 affects global H2A monoubiquitination but not the function of hematopoietic stem and progenitor cells. PLoS ONE. 2016;11:e0154561.
Bozzi F, Brich S, Dagrada GP, et al. Epithelioid peritoneal mesothelioma: a hybrid phenotype within a mesenchymal-epithelial/epithelial-mesenchymal transition framework. Oncotarget. 2016;7:75503–17.
Batista IAA, Helguero LA. Biological processes and signal transduction pathways regulated by the protein methyltransferase SETD7 and their significance in cancer. Signal Transduct Target Ther. 2018;3:19.
Papamichos-Chronakis M, Peterson CL. Chromatin and the genome integrity network. Nat Rev Genet. 2013;14:62–75.
McLoughlin KC, Kaufman AS, Schrump DS. Targeting the epigenome in malignant pleural mesothelioma. Transl Lung Cancer Res. 2017;6:350–65.
Gardiner A, Parker H, Glide S, et al. A new minimal deleted region at 11q22.3 reveals the importance of interpretation of diminished FISH signals and the choice of probe for ATM deletion screening in chronic lymphocytic leukemia. Leuk Res. 2012;36:307–10.
Illei PB, Rusch VW, Zakowski MF, Ladanyi M. Homozygous deletion of CDKN2A and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas. Clin Cancer Res. 2003;9:2108–13.
Hamasaki M, Matsumoto S, Abe S, et al. Low homozygous/high heterozygous deletion status by p16 FISH correlates with a better prognostic group than high homozygous deletion status in malignant pleural mesothelioma. Lung Cancer. 2016;99:155–61.
Pylkkänen L, Wolff H, Stjernvall T, et al. Reduced Fhit protein expression in human malignant mesothelioma. Virchows Arch. 2004;444:43–8.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Brich, S., Bozzi, F., Perrone, F. et al. Fluorescence in situ hybridization (FISH) provides estimates of minute and interstitial BAP1, CDKN2A, and NF2 gene deletions in peritoneal mesothelioma. Mod Pathol 33, 217–227 (2020). https://doi.org/10.1038/s41379-019-0371-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41379-019-0371-0
This article is cited by
-
Clinical and molecular validation of BAP1, MTAP, P53, and Merlin immunohistochemistry in diagnosis of pleural mesothelioma
Modern Pathology (2022)
-
Hierarchal Bayes model with AlexNet for characterization of M-FISH chromosome images
Medical & Biological Engineering & Computing (2021)
-
Molecular characterization of diffuse malignant peritoneal mesothelioma
Modern Pathology (2020)