Abstract
The opioid overdose epidemic has rapidly expanded in North America, with rates accelerating during the COVID-19 pandemic. No existing study has demonstrated prospective opioid overdose at a population level. This study aimed to develop and validate a population-level individualized prospective prediction model of opioid overdose (OpOD) using machine learning (ML) and de-identified provincial administrative health data. The OpOD prediction model was based on a cohort of approximately 4 million people in 2017 to predict OpOD cases in 2018 and was subsequently tested on cohort data from 2018, 2019, and 2020 to predict OpOD cases in 2019, 2020, and 2021, respectively. The model’s predictive performance, including balanced accuracy, sensitivity, specificity, and area under the Receiver Operating Characteristics Curve (AUC), was evaluated, achieving a balanced accuracy of 83.7, 81.6, and 85.0% in each respective year. The leading predictors for OpOD, which were derived from health care utilization variables documented by the Canadian Institute for Health Information (CIHI) and physician billing claims, were treatment encounters for drug or alcohol use, depression, neurotic/anxiety/obsessive-compulsive disorder, and superficial skin injury. The main contribution of our study is to demonstrate that ML-based individualized OpOD prediction using existing population-level data can provide accurate prediction of future OpOD cases in the whole population and may have the potential to inform targeted interventions and policy planning.
Similar content being viewed by others
Introduction
Opioid overdose (OpOD) is a rapidly growing epidemic in North America. In Canada, 47 162 people died from apparent opioid-related overdoses between January 2016 and March 2024 [1]. During the COVID-19 pandemic, overdose rates accelerated, with a 24% increase in opioid-related poisonings in the first half of 2022 alone [2]. Similarly, the United States is facing a crisis, reporting 212 892 opioid-related deaths between 2017 and 2020 [3]. Recently, from 2023 to 2024, opioid-related deaths showed signs of reduction overall, yet opioids continue to account for most drug overdoses, and rates continue to increase in some regions [1, 4]. Identifying actionable intervention strategies would be an important step in supporting people at risk of OpOD and their communities.
One potential solution is the early identification of OpOD cases and associated risk factors through individualized prediction and analysis based on large-scale health data by leveraging advanced computational methods like machine learning (ML) so that potential timely intervention and support can be delivered to a population in need of help. Rising research interests in the clinical applications of OpOD prediction and other adverse outcomes have emerged due to the increased availability of cross-linked administrative health data and Electronic Health Records (EHR) [5]. The use of ML further enables individual-level predictions of OpOD, opioid use disorder and other adverse outcomes related to opioids [6,7,8,9] and associated risk factors for OpOD, including pain-related symptoms, mental disorders, and demographic, community and environmental factors [10, 11]. However, the lack of population-level representative data usually limits such predictions to a small portion of the population, a problem exacerbated by non-universal or non-stratified access to health insurance [12], potentially introducing bias and affecting prediction generalizability and reliability.
In the current study, we developed a series of longitudinal population cohorts based on provincial administrative health data from the Government of Alberta, Canada, which have been routinely collected and maintained. We aimed to develop an OpOD prediction model by applying ML to the baseline cohort in 2017 to predict OpOD cases in 2018 and to longitudinally test this model in the subsequent three years, demonstrating a framework for individual-level prospective OpOD prediction in a representative population in North America.
Methods
Study cohort
Four cohorts of all residents in Alberta, Canada (approximately 4 million) were developed based on data retrieved from fiscal years 2017, 2018, 2019, 2020, and 2021 (April 1st to March 31st of the following year). For each cohort, we included all individuals with an active Alberta Health Care Insurance Plan (AHCIP) status who had used the system in the two years before the corresponding cohort year.
Data source
Alberta has a health care system that offers universal access to medically necessary hospital and health care services. All new and existing Alberta residents are provided a unique identifier under the AHCIP for access to insured health care services, allowing deterministic linkage of different administrative data sources and robust databases for this study. Data in the cohorts included de-identified individual-level information of different types (e.g., demographic, socio-economic, health utilization) that were collected by the Alberta Ministry of Health and cross-linked. The linked administrative health data were prepared based on AHCIP Practitioner Claims (e.g., physician office’s claim codes, patient demographics), the National Ambulatory Care Reporting System (NACRS) (e.g., hospital and emergency department visits data), the Canadian Institute of Health Information Discharge Abstract Database (DAD) (e.g., administrative, clinical and demographic information on hospital discharges), the AHCIP Population Registry Database (e.g., demographics of patients), Alberta Pharmaceutical Information Network (PIN) database (e.g., drug prescription and use history), data developed from the Canadian Institute of Health Information population grouping methodology (CIHI) [13] (e.g., individual-level risk scores of patient’s health conditions), and Alberta Health Services Drug Supplement Plan database (AHSDSP) (to identify prescription opioids and patient consumption). All databases accessed include information on all Alberta residents covered under the AHCIP, ensuring comprehensive coverage of the province’s population.
Outcome definition
The prospective OpOD outcome was derived from the fiscal year following the cohort (e.g., OpOD events in 2018 for the 2017 cohort), from AHCIP Practitioner Claims, NACRS and DAD, based on the International Classification of Diseases, Ninth Revision (ICD-9) code 965.0 (poisoning by opiates and related narcotics) in practitioner claims data, and International Statistical Classification of Diseases and Related Health Problems, Tenth Revision (ICD-10), code T40.0 (poisoning by, and adverse effect of opium), T40.1 (poisoning by, and adverse effect of heroin), T40.2 (poisoning by, adverse effect of and underdosing of opioids), T40.3 (poisoning by, adverse effect of and underdosing of methadone), T40.4 (poisoning by, adverse effect of and underdosing of other synthetic narcotics) and T40.6 (poisoning by, adverse effect of and underdosing of other and unspecified narcotics), in ambulatory, inpatient and outpatient data [8, 14]. The OpOD status was binary coded and labeled as 1 if a patient had at least 1 incident of OpOD in the fiscal year following the cohort and 0 if no incident was found in the administrative health records. Note that the OpOD outcome might include both fatal and non-fatal cases and did not capture subjects that did not access health care.
Predicting variables
Candidate predicting variables or “features” for ML, were developed based on the cohort’s fiscal year, with a total of 368 features, including health system utilization indicators (e.g., number of family physician visits), demographics (e.g., age, sex), opioid specific indicators (e.g., opioid use disorder), substance use and related disorders (e.g., alcohol, nicotine), and CIHI groupers [15] that identified other physical and mental health indicators (e.g., chronic pain, hepatitis, depression). In our model, we used the ‘relative importance’ of these features to measure the strength of each predictor’s impact on the outcome, quantifying how much each feature contributes to the overall prediction accuracy of the model [16]. Higher values indicate a stronger influence on the model’s predictions (maximum 1), with these metrics calculated through an ensemble model to determine which features are most critical in predicting opioid overdose.
To address potential overlap in predictor data across years, we developed the predictors independently for each cohort year. This approach minimizes the risk of bias from overlapping data points while maintaining the utility of the model. In a real-world setting, annual data updates allow for continuous retraining of the model, ensuring it remains responsive to population trends and emerging patterns.
Data preparation and modeling pipeline
SAS 9.4 and SAS Viya Data Studio software were used for data preparation [17]. Features representing the frequency of occurrence and binary risk indicator had no missing data. Zero was interpreted as zero occurrence or lack of evidence.
The prepared data were then processed through a modeling pipeline (Fig. 1) developed using SAS Viya Model Studio software, version V.03.05. Because OpOD is a rare event in population-level data (e.g., 0.10% of the population in 2017), there is a severe class imbalance that would impact model building [18]. Class imbalance refers to a situation in ML where the number of observations in one class significantly outweighs the observations in the other class; in this case, the instances of OpOD compared to no OpOD. This imbalance can lead to biases in the model as a model can simply predict all cases as the majority class to produce high accuracy. In our exploration of the imbalanced data, we ran several single ML models (e.g., Gradient Boosting) on the data, all achieving a roughly 0% sensitivity.
To address the class imbalance in our ML pipeline, we used an under-sampling technique and devised 50 subsamples that each included all OpOD subjects within the cohort, joined with a stratified random sample drawn from subjects with no OpOD records, matched by age, sex, and sample size. The number of 50 subsamples was chosen to achieve a stable estimation of model performance with the limited computational resources that we had. In our pilot analysis, model performance started to stabilize after 10 subsamples, and our computational resources would not allow more subsamples than 50 at the time of model training. Other features were not used for sample matching. Each of the subsamples had a 1:1 ratio of OpOD and NoOpOD subjects (e.g., 4 002 OpOD and 4 002 No OpOD) and included the same predictive features, allowing the ML model to learn characteristics from OpOD subjects. The 50 subsamples were further split into training and validation sets, where 70% of the data were randomly used for model training and 30% of the data were used for validation. These splits ran through Gradient boosting nodes in SAS Viya to learn base models for classifying OpOD, optimizing for logistic loss, a measure of how well the model could make a correct classification.
Each of the 50 models was set to perform auto-tuning, which performed adjustments to the following parameters: number of trees, number of inputs to consider for split, learning rates, subsample rates, and L1 & L2 regularization [17]. A Gradient boosting model is a machine learning algorithm that iteratively combines multiple weak decision trees to create a stronger, more accurate model by adjusting the weights of the trees and reducing errors in the predictions. The parameters optimization method used for our model was a grid search algorithm, with the initial values used for the baseline model set to default values provided by the SAS model [17].
The 50 models trained and validated using the subsamples were then put through an ensemble node. The ensemble node took the average of the estimated prediction probabilities to combine the models and determine the top contributing features. By averaging the probabilities, the ensemble model reduces the impact of individual model biases or errors to improve the prediction accuracy. A predicted probability of 0.5 was used as a threshold to classify OpOD (>= 0.5) and No OpOD (<0.5). The entire 2018, 2019, and 2020 cohorts were used as testing data to evaluate the ensemble model. The top five features of the models were evaluated based on the ranking of feature importance in the ensemble model. In SAS, relative feature importance is a metric used to quantify the contribution of each feature to the predictive performance of the model. It helps to identify the more important features driving the model predictions and is valuable for interpreting model outputs [16, 17]. Relative feature importance is calculated by dividing the RSS-based importance, the reduction in residual sum of squares due to a variable, of each variable by the maximum RSS-based importance among all variables [16]. The selection of the Gradient boosting algorithm, the number of resamples, and the ensemble method to solve class-imbalance issues in the pipeline were based on the availability of algorithms and computation power in a highly resource-restricted software analysis environment.
Results
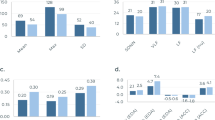
For each cohort (2017–2020), the prevalence rate of OpOD in the population remained under 0.2% (Table 1). Despite the large imbalance in our data, the final ensemble model obtained an Area Under the Receiver Operating Characteristics Curve (AUC) of 89.2%, a balanced accuracy of 82.7%, an average sensitivity of 75.8%, and a specificity of 89.7%, from the reserved 2017 validation data. The trained model was then applied to the full 2018–2020 cohorts (N = 4 095 364 – 4 203 233). In each of these cohorts, the OpOD outcomes were extracted from the year after the cohort year to verify the prospective prediction of our model. The sensitivity in the subsequent years 2018–2020 achieved 78.1, 68.4, and 77.9%, respectively, while specificity was 89.3, 94.8, and 92.1%, respectively, corresponding to balanced accuracy of prospective prediction at 83.7, 81.6, and 85.0% (Table 1).
Top predictive features
From the 368 features, the top five predictors include drug/alcohol use/dependence (CIHI Q07), depression (CIHI Q04), neurotic/anxiety/obsessive-compulsive disorder (CIHI Q11), Superficial skin injury/contusion/non-serious burns (CIHI I43), and Depression from billing claims data. Note that depression from billing claims data was developed by the authors, independent of CIHI’s method of developing the depression indicator [15]. These features had relative importance of 1.00, 0.60, 0.46, 0.40, and 0.36, respectively (Table 2). A score of 1.00 for drug/alcohol dependence indicates it was the strongest predictor, with other features ranked relative to this benchmark. Other top-ranked features are consistent with risk factors reported in the literature, including substance use and substance use related health utilization, mood and anxiety disorder related claims, and physician health indicators such as back pain and skin wounds.
Discussion
In this study, we developed an ML model based on the cohort of 2017 to prospectively predict OpOD in 2018 in the general population. We report a model performance achieving a balanced accuracy of 83.7, 81.6, and 85.0% and AUC of 89.2, 86.9, and 89.8%, when testing the model in 2018, 2019, and 2020 for predicting OpOD cases in 2019, 2020 and 2021, respectively. To our best knowledge, this is the first study using population-level data to make a prospective prediction of individual-level opioid overdose and verify it longitudinally within a fixed time window. These results demonstrate that ML-based individualized OpOD prediction using existing population data can provide accurate prediction of future OpOD cases in the whole population.
The performance of our model is comparable to that of other large-scale prospective OpOD models using smaller and less representative samples. For example, Lo-Ciganic et al. [9] built a three-month prospective prediction OpOD model using the US Medicaid data and showed a gradient boosting model performed at 84.1% balanced accuracy for internal validation and 82.8% for external validation within the same state. Note that our study population included the general population, and thus modeling performance is not directly comparable to studies focused on predicting OpOD in patient population with a prior episode [19]. Comparing modeling performance requires consideration of factors such as the definition of the future OpOD outcome, the predictors, and the scope of the study sample.
It is also worth noting that Canada has a publicly funded, universal health care system that routinely collects data in all provinces, and the Canadian Institute for Health Information synthesized the data and made nationally representative data accessible [13]. Compared to the US, where up to 9% of the population is uninsured [20], universal health coverage in Canada provides broader coverage of the population and consistently collected objective indicators.
Our model identified several stable predictors across training models at a population level. Notably, predictors such as CIHI Drug/alcohol use/dependence (Q07) and CIHI Depression (Q04) surfaced as consistent indicators across the training models, reinforcing their relevance as significant risk factors for OpOD, aligning with findings from Ellis et al. [10], who also identified similar risk factors in opioid-related outcomes. Despite the known linkage between these factors and OpOD, none of them alone could accurately predict future OpOD cases at the individual level. Our study demonstrates that with a data-driven approach, we can extract sufficient information between all potential contributing factors and make such OpOD predictions for the future and for the whole population.
The model’s consistent predictors and accuracy offer an opportunity to develop targeted education, prevention or intervention strategies for people at high risk of OpOD at the population level and promote proper utilization of known risk factors. Pending more converging evidence and support from future studies as well as overcoming challenges in practical applications, the models could be integrated into clinical workflows as a decision-support tool, flagging patients at risk and prompting clinicians to review their history in health records. This use case could enhance decision-making and prevent adverse outcomes.
Our successful demonstration of OpOD prediction using a representative, longitudinal data set also suggests the feasibility of developing a population-level OpOD risk screening tool when appropriate. Such a tool is especially valuable given that many OpOD cases may be associated with non-prescription opioids but interact with the health system previously with other needs (e.g., mental health, pain management, and other substance dependence). The model developed using population-level data has the opportunity to understand the profile of high-risk cases, which further leads to the potential for generalizations.
For applications in public sectors, our model could potentially help inform policy on which communities should be engaged in policy planning, predict changes in OpOD rates, prioritize interventions for individuals at most significant risk, and help model costs associated with OpOD rates. With the option to adjust the weights of predictors and prediction threshold according to policy needs, our model may facilitate more efficient resource allocation and better substance use support programs and community outreach initiatives. All these potential applications of such a prediction model require further engagement of stakeholders and community members in future studies.
Limitations
The current study presents several limitations to be considered when interpreting the results and drawing conclusions. Firstly, as the study relies on available administrative health records and ICD-code-based algorithms to identify OpOD, there might be missing or incorrect information that could affect the quality and validity of the results, including missing OpOD events not interfacing with health care, missing broader public health data such as vital statistics and laboratory tests. However, the ICD-code-base algorithm has been shown to have good specificity but moderate sensitivity [21, 22]. Secondly, the data used in this study were drawn from one province in Canada. Thus, caution may be needed to generalize and interpret data from other regions of North America and the world. However, population-based health administration data were routinely collected and synthesized in all Canadian provinces, and the methodology of this study applies to other Canadian provinces. Thus, the results could be potentially generalizable across Canada and verified by future studies. Thirdly, the study employed an under-sampling technique to address the class imbalance, which might have resulted in the exclusion of some relevant data, even though we tried to mitigate this by using 50 different sampling models. Lastly, the practical application of predictive models in clinical settings is challenging due to the inherent false-positive rates caused by the low prevalence of opioid overdose. However, the primary value of the model may not only be in its direct prediction accuracy but also in its ability to identify high-risk groups. These groups can be prioritized for preventive interventions, such as education, early treatment of contributing conditions, and community outreach, where the consequences of false positives are more controllable. This population-level stratification approach may offer a feasible path to leveraging predictive models for public health impact. Future studies should explore alternative techniques for handling class imbalance to mitigate the potential loss of information. Our model demonstrates decent predictive accuracy compared to existing studies, with balanced accuracy in the 80% range, yet it is important to note that a significant number of overdoses remain undetected, which could represent substantial numbers in a large population. Additionally, even though the model achieved a high specificity, due to the low prevalence of OpOD, between 5 to 11% false positive rate (corresponding to between 89.3 to 94.8% specificity in our three testing cohorts) are still substantial in the whole population. Thus, it is not yet practical to use these predictive tools in clinical settings; for example, physicians might be hesitant to provide opioid medications when indicated for fear of further increasing the risk of subsequent opioid overdose [23]. In our data exploration, we could select a higher probability threshold for classification at the expense of missing more subjects that will experience OpOD in the following year. Adjusting cutoffs would likely provide more clinical utility, with various use cases benefiting from different cutoff values. For example, in primary care settings, a higher sensitivity may be preferred to ensure that few cases are missed, while in population-level risk screening settings, a higher specificity may be more important to avoid unnecessary interventions (see Fig. 2 to visualize sensitivity and specificity trade-offs). Future research and applications may need to specify such trade-offs depending on different clinical and policy needs.
Model prediction performance tested on cohort data from 2018, 2019, and 2020 to predict opioid overdose cases in 2019, 2020, and 2021, respectively, as shown with the Receiver Operating Characteristics (ROC) Curves. The x-axis represents 1-specificity in a range of 0-1 and y-axis represents sensitivity in a range of 0-1, where 1 equals 100%.
Conclusion
Our study demonstrates that ML-based individualized opioid overdose prediction using existing population-level data can accurately predict future opioid overdose cases in the whole population. Such prediction may have the potential to inform targeted interventions and policy planning. Our findings emphasize the need for a multidisciplinary approach to address the complex interplay of factors contributing to opioid overdose risk and show the promise of leveraging population-level data for more effective prevention and intervention strategies for opioid overdose.
Data availability
Due to privacy policy restrictions, individualized data cannot be shared. Data can be accessed with permission from the Ministry of Health in Alberta, Canada through www.alberta.ca/health-research or health.inforequest@gov.ab.ca.
References
Public Health Agency of Canada. Federal, provincial, and territorial Special Advisory Committee on Toxic Drug Poisonings. Opioid- and stimulant-related harms in Canada. 2024. https://health-infobase.canada.ca/substance-related-harms/opioids-stimulants/. Accessed 19 December 2024.
Public Health Agency of Canada. Opioid-related harms in Canada. 2023. https://health-infobase.canada.ca/substance-related-harms/opioids/. Accessed 27 August 2023.
National Institute on Drug Abuse. Number of national drug overdose deaths involving select prescription and illicit drugs. 2022. https://nida.nih.gov/research-topics/trends-statistics/overdose-death-rates. Accessed 26 January 2023.
National Center for Health Statistics. Provisional drug overdose death counts. 2024. https://www.cdc.gov/nchs/nvss/vsrr/drug-overdose-data.htm#xd_co_f=ODllMzNkNjktYjMwYi00ODI1LTg5MzYtMzMyNWI5NDc3M2U2~. Accessed 19 December 2024.
Tseregounis IE, Henry SG. Assessing opioid overdose risk: a review of clinical prediction models utilizing patient-level data. Transl Res. 2021;234:74–87.
Liu YS, Kiyang L, Hayward J, Zhang Y, Metes D, Wang M, et al. Individualized prospective prediction of opioid use disorder. Can J Psychiatry. 2022. https://doi.org/10.1177/07067437221114094/ASSET/IMAGES/LARGE/10.1177_07067437221114094-FIG2.JPEG.
Sharma V, Kulkarni V, Jess E, Gilani F, Eurich D, Simpson SH, et al. Development and validation of a machine learning model to estimate risk of adverse outcomes within 30 days of opioid dispensation. JAMA Netw Open. 2022;5:e2248559.
Lo-Ciganic W-H, Huang JL, Zhang HH, Weiss JC, Wu Y, Kwoh CK, et al. Evaluation of machine-learning algorithms for predicting opioid overdose risk among medicare beneficiaries with opioid prescriptions. JAMA Netw Open. 2019;2:e190968.
Lo-Ciganic W-H, Donohue JM, Yang Q, Huang JL, Chang C-Y, Weiss JC, et al. Developing and validating a machine-learning algorithm to predict opioid overdose in Medicaid beneficiaries in two US states: a prognostic modelling study. Lancet Digit Heal. 2022;4:e455–65.
Ellis RJ, Wang Z, Genes N, Ma’Ayan A. Predicting opioid dependence from electronic health records with machine learning. BioData Min. 2019;12:3. https://doi.org/10.1186/s13040-019-0193-0.
Schell RC, Allen B, Goedel WC, Hallowell BD, Scagos R, Li Y, et al. Identifying predictors of opioid overdose death at a neighborhood level with machine learning. Am J Epidemiol. 2022;191:526–33.
Berchick ER, Barnett JC, Upton RD. Health insurance coverage in the United States, 2018. Washington, DC: US Department of Commerce, US Census Bureau; 2019. https://www.census.gov/content/dam/Census/library/publications/2019/demo/p60-267.pdf.
Weir S, Steffler M, Li Y, Shaikh S, Wright JG, Kantarevic J. Use of the population grouping methodology of the canadian institute for health information to predict high-cost health system users in Ontario. Can Med Assoc J. 2020;192:E907–12.
Dunn KM. Opioid prescriptions for chronic pain and overdose. Ann Intern Med. 2010;152:85.
Canadian Institute for Health Information. Codes and classifications. 2023. https://www.cihi.ca/en/submit-data-and-view-standards/codes-and-classifications. Accessed 27 August 2023.
SAS Institute Inc. The HPSPLIT procedure. SAS/STAT® 151 User’s Guide, Cary, NC: SAS Institute Inc; 2018. https://support.sas.com/documentation/onlinedoc/stat/151/hpsplit.pdf. Accessed 19 December, 2024
SAS Institute Inc. Exploring SAS® Viya®: Data Mining and Machine Learning. Cary, NC, USA: SAS Institute Inc.; 2019 https://support.sas.com/content/dam/SAS/support/en/books/free-books/exploring-sas-viya-data-mining-machine-learning.pdf. Accessed 19 December, 2024
Cartus AR, Samuels EA, Cerdá M, Marshall BDL. Outcome class imbalance and rare events: An underappreciated complication for overdose risk prediction modeling. Addiction. 2023;118:1167–76.
Dong X, Rashidian S, Wang Y, Hajagos J, Zhao X, Rosenthal RN, et al. Machine learning based opioid overdose prediction using electronic health records. AMIA Annu Symp Proc AMIA Symp. 2019;2019:389–98.
Berchick ER, Hood E, Barnett JC. Health insurance coverage in the United States: 2017. Curr Popul Reports Washingt DC US Gov Print Off. 2018. https://www.census.gov/content/dam/Census/library/publications/2018/demo/p60-264.pdf.
Green CA, Perrin NA, Hazlehurst B, Janoff SL, DeVeaugh‐Geiss A, Carrell DS, et al. Identifying and classifying opioid‐related overdoses: a validation study. Pharmacoepidemiol Drug Saf. 2019;28:1127–37.
Mbutiwi FIN, Yapo APJ, Toirambe SE, Rees E, Plouffe R, Carabin H. Sensitivity and specificity of international classification of diseases algorithms (ICD-9 and ICD-10) used to identify opioid-related overdose cases: a systematic review and an example of estimation using Bayesian latent class models in the absence of gold standards. Can J Public Heal. 2024;115:770–83.
Desveaux L, Saragosa M, Kithulegoda N, Ivers NM. Understanding the behavioural determinants of opioid prescribing among family physicians: a qualitative study. BMC Fam Pract. 2019;20:59.
Acknowledgements
The interpretation and conclusions contained herein are those of the researchers and do not necessarily represent the views of the Government of Alberta. Neither the Government nor Alberta Health expresses any opinion in relation to this study.
Funding
This research was undertaken, in part, thanks to funding from the Canada Research Chairs program, BBRF Young Investigator Grant from Brain & Behavior Research Foundation, Canadian Institute for Advanced Research (CIFAR), Alberta Innovates, the Institute for Advancements in Mental Health, Mental Health Foundation, Mental Health Research Canada, MITACS Accelerate program, Simon & Martina Sochatsky Fund for Mental Health, Howard Berger Memorial Schizophrenia Research Fund, the Abraham & Freda Berger Memorial Endowment Fund, the Alberta Synergies in Alzheimer’s and Related Disorders (SynAD) program, University Hospital Foundation and University of Alberta. The funding sources had no impact on the design and conduct of the study, the collection, management, analysis, and interpretation of the data, the preparation, review, or approval of the manuscript, or the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
YSL and DVP participated in the formal analysis and writing the original draft. LK, YS, DM, MW, JH, YZ, and BC participated in conceptualization, results interpretation, draft review & editing. KD, DTE, SP, RG, and AG participated in editing the draft. BC supervised the study and provided financial support for the trainees.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics committee approval
The present study was approved by the University of Alberta Health Ethics Board (Pro00072946). Informed consent was waived due to the minimal risks of secondary analysis, as the Ministry of Health had already anonymized the cross-linked data before analysis. All methods were performed in accordance with the relevant guidelines and regulations.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Y.S., Pierce, D.V., Metes, D. et al. Population-level individualized prospective prediction of opioid overdose using machine learning. Mol Psychiatry 30, 4122–4127 (2025). https://doi.org/10.1038/s41380-025-02992-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41380-025-02992-4