Abstract
Natural Killer (NK) cells can target and destroy cancer cells, yet tumor microenvironments typically suppress NK cell recruitment and cytotoxicity. The epidermal growth factor receptor (EGFR) is a potent oncogene that can activate survival, migration, and proliferation pathways, and clinical data suggests it may also play an immunomodulating role in cancers. Recent work has demonstrated a novel role for nuclear EGFR (nEGFR) in regulating transcriptional events unique from the kinase domain. Using a novel peptide therapeutic (cSNX1.3) that inhibits retrograde trafficking of EGFR and an EGFR nuclear localization mutant, we discovered that nEGFR suppresses NK cell recruitment and cytotoxicity. RNA-Seq analysis of breast cancer cells treated with cSNX1.3 or modified to lack a nuclear localization sequence (EGFRΔNLS) revealed the EGF-dependent induction of NK activating receptor ligands, while kinase inhibition by erlotinib did not impact these genes. NanoString analysis of tumor-bearing WAP-TGFα transgenic mice treated with cSNX1.3 demonstrated an increase in immune cell populations and activating genes. Additionally, immunohistochemistry confirmed an increase in NK cells upon cSNX1.3 treatment. Finally, cSNX1.3 treatment was found to enhance NK cell recruitment and cytotoxicity in vitro. Together, the data demonstrate a unique immunomodulatory role for nEGFR.
Similar content being viewed by others
Introduction
Natural killer (NK) cells are cytotoxic lymphocytes known to have an anti-tumor and anti-metastatic effect [1,2,3]. Their functions are regulated by activating (NKp46, NKp44, NKp30, NKp80, NKG2D CD16, 2B4, DNAM1) [3] and inhibitory (CD161, KLRG1, PD1, TIM3, LAG3, CD96, TIGIT) [2] receptors that regulate tumor recognition and cytotoxic activity. Current NK cell-based therapies focus on enhancing NK cell potency and NK cell specificity against the tumor cells [1, 4, 5]. However, cancer cells are efficient at evading the immune system and altering the tumor immune microenvironment in a way that can impair the recruitment and function of NK cells [6, 7]. This includes the suppression of cytokine expression (IL-2, IL-12, IL-15, IL-18) [7] and shedding of ligands (MICA, MICB, ULBPs, B7-H6) [2], a process by which enzymes known as “sheddases” cut and release the extracellular domains of integral membrane proteins [8]. These alterations in cytokine expression and shedding of ligands contribute to diminished NK cell anti-tumor activity. This type of immune suppression can also be driven by oncogenes such as the epidermal growth factor receptor (EGFR) [9].
In breast cancer, EGFR is overexpressed [10, 11] and can retro-translocate to the nucleus [12,13,14,15,16,17], potentially contributing to the underwhelming success of conventional EGFR tyrosine kinase inhibitor (TKI) and antibody-based therapies [18,19,20,21,22] in breast cancer. EGFR nuclear translocation is ligand-dependent and requires retrograde endosomal trafficking from the membrane to the nucleus via the endoplasmic reticulum (ER). Once trafficked to the ER, EGFR interacts with the sec61 translocon, HSP70 and importin 1β, after which it is shuttled through the nuclear pore [23]. In the nucleus, EGFR acts as a co-transcription factor and can regulate the expression of genes such as cyclin D1 and iNOS [15, 24]. Previous studies have shown that when EGFR retro-translocation is targeted, it is more effective at inhibiting cell survival than targeting EGFR’s kinase activity with the use of tyrosine kinase inhibitors [25]. Although inhibiting nEGFR reduces cell survival and migration [24,25,26], little is known about how nEGFR impacts the tumor immune microenvironment in breast cancer. There are several RTKs (insulin receptor, FGFR, ErbB2, ErbB3, ErbB4) [27] that also retro-translocate [28,29,30,31] to the nucleus and are associated with immune regulation [32, 33], but it is unknown how nEGFR directly impacts immune regulation.
In the current study, we have discovered that by inhibiting the retro-translocation of EGFR, we can enhance NK cell activation via the regulation of activating NK receptor ligands. Furthermore, blocking nEGFR promotes NK cell infiltration into the tumors of an EGFR-dependent breast cancer mouse model. Importantly, this type of activated immune response is elicited when the function of EGFR in the nucleus is blocked, but not when kinase activity is blocked, indicating this to be driven by nEGFR regulation. This study is the first to demonstrate the role of nEGFR in the regulation of NK cells in the tumor microenvironment, contributing to our understanding of how nEGFR promotes the progression of breast cancer.
Results
Nuclear EGFR drives a unique transcriptional profile
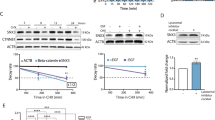
We recently reported that a peptide-based therapeutic, cSNX1.3, can block the interaction between EGFR and Sorting Nexin 1, thereby reducing the nuclear localization of EGFR [25]. Using single cell tracing of Crispr/Cas9 modified EGFR to express an EGFR-mVenus fusion protein, we validated the strong reduction of nEGFR in T47D (T47D-mVenus) breast cancer cells treated with cSNX1.3 compared to control cPTD4 treated cells (Fig. 1a). To compare the mechanism of action of cSNX1.3 with tyrosine kinase inhibition, we performed a cell survival assay on MDA-MB-468 (Fig. 1b) and T47D-mVenus (Supplementary Fig. s1a) breast cancer cells. While both cSNX1.3 and erlotinib significantly reduce cell survival, cSNX1.3 has a stronger impact – notably, combining cSNX1.3 and erlotinib had an additive effect (as determined by the Bliss independence model). We next set out to evaluate the EGF-dependent transcription profiles induced by nEGFR versus EGFR kinase-driven transcription through the inhibition of nEGFR (via cSNX1.3 treatment) or inhibition of the EGFR tyrosine kinase (via erlotinib treatment). We identified all genes that were significantly differentially expressed (Padj < 0.05 and log2FC > 1) in the cells treated with cSNX1.3 + EGF (referred to as cSNX1.3) relative to EGF-only controls (referred to as EGF) and visualized their expression across all three conditions (Fig. 1c). Together with the Principal Component Analysis (PCA) plot of the data (Supplementary Fig. s1b), this analysis showed that cSNX1.3 and erlotinib have different effects on gene expression. There was a total of 1130 differentially expressed genes with Padj < 0.05 in the cSNX1.3 vs. EGF comparison and 6688 differentially expressed genes in the erlotinib vs. EGF comparison, with the 832 genes in common showing largely opposite effects (Fig. 1c).
a nEGFR tracking in T47D EGFR-YFP cells. T47D cells expressing the EGFR-YFP fusion protein were serum starved overnight, then treated with 100 ng/ml of EGF, 10 μM of cSNX1.3, or 10 μM of the control peptide cPTD4 and imaged every 10 min for 24 h. Images were quantified via MATLAB. Data represents the mean ± SEM of the integrated intensity of EGFR-YFP in the nucleus of T47D cells, ±SEM. Dash line represents the time of drug added at 30 min. cSNX1.3 treatment is statistically different from cPTD4 and EGF treatments; p < 0.0001. Statistics were analyzed using one-way ANOVA, Dunnett’s post hoc. b Cell viability after cSNX1.3 and erlotinib treatment. MDA-MB-468 cells were treated with the drug treatments shown above for 72 h. Cell viability was determined with an MTT assay and normalized to vehicle control. Data represents the percentage of cell viability ± SEM of 3 biological replicates. Statistics were analyzed via one-way ANOVA, Dunnett’s post hoc; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, compared to cSNX1.3 alone. The Bliss independence model was used to test for synergy, resulting in a score of 3.499. c Heatmap; MDA-MB-468 cells were serum starved for 12 h, stimulated with 10 ng/mL EGF, and treated with cSNX1.3 (10 μM) or erlotinib (15 μM) for 12 h. Samples were collected and next generation sequencing was performed by GENEWIZ from Azenta Life Sciences. Significantly differentially expressed (Padj < 0.05 and abs(log2FC)>1) genes in the cells treated with cSNX1.3 + EGF (referred to as cSNX1.3) are visualized using their z-score across three different conditions (cSNX1.3, erlotinib, and EGF control). Venn diagram; number of significant (Padj < 0.05) differentially expressed genes shared by both cSNX1.3 and erlotinib treatments. d Functional enrichment in the significant (Padj < 0.05) differentially expressed genes between cSNX1.3 and erlotinib treatments. GO term enrichment analysis of cSNX1.3 with log2FC > 1 (top, left). GO term enrichment analysis of erlotinib with log2FC > 1 (top, right) and log2FC < -1 (bottom). e Natural killer cell-mediated cytotoxicity genes regulated by cSNX1.3 or erlotinib. Log2FC of significant (Padj < 0.05) genes associated with natural killer cell-mediated cytotoxicity in cSXN1.3 and erlotinib. The human gene set used for identifying natural killer cell-mediated cytotoxicity genes can be found under the standard name KEGG_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY.
To help determine the biological functions perturbed by each treatment, we performed GO term enrichment analysis. Treatment with erlotinib resulted in changes in cell growth, migration, and MAP kinase activity, as expected (log2FC < -1). Alternatively, treatment with cSNX1.3 resulted in changes in the regulation of lymphocyte-mediated immunity and leukocyte-mediated immunity, specifically NK cell-mediated immunity, among others (log2FC > 1; Fig. 1d, arrows). GO term analysis did not uncover any statistically significant functional associations among the genes with log2FC < -1 in the cSNX1.3 treatment. While 832 genes are common between cSNX1.3 and erlotinib, those that are involved in NK cell-mediated cytotoxicity and function were downregulated by erlotinib as opposed to upregulated by cSNX1.3 (Fig. 1e). Additionally, we verified that cSNX1.3 increased expression of NKG2D ligands RAET1H, RAET1G, and RAET1L by RT-PCR and qPCR (RAET1L) (Supplementary Fig. s1c, Fig. s1d). The upregulation of these genes upon blocking EGFR nuclear translocation indicates the potential role of nEGFR in regulating the immune system, specifically in inhibiting the function of NK cells.
Directly targeting the NLS of EGFR impacts the expression of NK cell activation genes
To further validate that the effects of cSNX1.3 treatment were specific to blocking nEGFR accumulation, we generated EGFR-GFP constructs that were either wildtype (EGFR.GFPWT) or harbored mutations in its NLS (EGFR.GFPΔNLS). The tripartite NLS is found within the juxtamembrane domain of EGFR and is composed of three basic amino acid clusters. To generate the mutant cell line, two arginines and one lysine were mutated to aspartic acids, as was done previously [34, 35] (Fig. 2a). Endogenous EGFR expression was knocked down using a lentiviral shRNA construct targeting the 3’-UTR of EGFR (Supplementary Fig. 2), followed by transfection of either EGFR.GFPWT or EGFR.GFPΔNLS into MDA-MB-468 cells. EGF treatment and bulk RNA-Seq analysis revealed a total of 6118 genes that were differentially expressed with Padj < 0.05 in the ΔNLS sample vs. WT. When compared to the cSNX1.3 vs. EGF differentially expressed genes, 686 genes were found to be in common (Fig. 2b).
a Diagram depicting the NLS in wildtype EGFR (WT; top) and EGFR mutant (ΔNLS; bottom). Three amino acids (two arginines and one lysine) in the tripartite sequence (645–657) were changed to aspartic acids (646 R > D; 653 K > D; 657 R > D). b Venn diagram; significantly differentially expressed (Padj < 0.05) genes shared by both cSNX1.3 and ΔNLS (ΔNLS relative to its WT control). c Functional enrichment in the significantly (Padj < 0.05) differentially expressed genes of the ΔNLS group. GO term enrichment analysis of ΔNLS with log2FC > 1 (left) and log2FC < -1 (right). d Natural killer cell-mediated cytotoxicity genes regulated from directly targeting the NLS of EGFR. Log2FC of significantly (Padj < 0.05) differentially expressed genes associated with natural killer cell-mediated cytotoxicity in both ΔNLS and cSNX1.3. The human gene set used for identifying natural killer cell-mediated cytotoxicity genes can be found under the standard name KEGG_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY. e Schematic of chromosome 6, which encodes for major histocompatibility complex genes (Class I, Class II, Class III, and Class-I related).
To determine the biological functions associated with nEGFR accumulation, we performed GO term enrichment analysis on the differentially expressed genes. We found that directly targeting the NLS of EGFR impacts biological processes involved in cell cycle signaling and the immune response (Fig. 2c). Considering that NK cell-mediated cytotoxicity is a biological process that was impacted by treatment with cSNX1.3, we examined the change in expression of genes known to be regulators of NK cell-mediated cytotoxicity in the ΔNLS and cSNX1.3 treatment conditions. Blocking nEGFR by directly targeting its NLS resulted in the downregulation of HLA genes (HLA-A; HLA-B; HLA-C; HLA-E; HLA-G) known to be important inhibitory ligands for NK cells, and the upregulation of ULBP (ULBP1; ULBP3) and MIC (MICA; MICB) genes, ligands for the NKG2D receptor known to be involved in the detection and elimination of damaged, transformed, and pathogen-infected cells [36]. Similarly, treatment with cSNX1.3 impacts the regulation of MICA and the ULBP (ULBP4; ULBP6) genes (Fig. 2d). These MHC complex and related genes are all part of chromosome 6 (Fig. 2e). Overall, the results suggest that nEGFR impacts recognition of tumor cells by NK cells through the modulation of both activating and inhibitory NK receptor ligands.
Targeting nEGFR results in modulation of the tumor immune microenvironment
To evaluate differential gene expression in vivo, we utilized mammary tumors generated in the WAP-TGFα transgenic mouse model [37]. Tumor-bearing females were established and then treated with cSNX1.3 or cPTD4 peptide once a tumor reached approximately 100 mm3 or 250 mm3. The mice remained in the study for 4 weeks or until they reached maximal tumor burden. Tumors grew in 1 or more of the 10 mammary fat pads. Results from the study showed an overall regression in tumor volume of mice treated with cSNX1.3 (ΔV = –1.8 mm3 and –3.6 mm3, respectively) compared to cPTD4 peptide control treatment (ΔV = 38.9 mm3 and 7.6 mm3, respectively) (Fig. 3a). The tumors that started at ≥ 100 mm3 had a significant change in tumor volume between cSNX1.3 and cPTD4 (P < 0.01). At the end of the study, mice were sacrificed and tumors were collected and paraffin embedded for gene expression profiling (NanoString). Using the nCounter mouse PanCancer IO (immune-oncology) 360 panel, the differentially expressed genes were used as marker genes to specify and quantify the different cell populations present in our samples. We used ROSALIND to perform a filtering of Cell Type Profiling, retaining all results that had scores with P < 0.05. The results showed a greater abundance of multiple immune cell types (NK cells, cytotoxic cells, macrophages, B cells, T cells, Th1 cells) in the tumor tissue from the mice treated with cSNX1.3 as compared to cPTD4 (Fig. 3b). Note that cytotoxic cells include both cytotoxic T cells and cytotoxic NK cells with marker genes – Prf1, Gzma, Gzmb, Nkg7, Klrd1, Klrk1, Klrb1, Gzma, NanoString Codeset: NS_Gzmb, Nkg7, Klrd1, Klrk1, Klrb1, Ctsw – as annotated in the NanoString Codeset: NS_Mm_IO_360_v1.0.
ϯTumors from the WAP-TGFα female mice treated with cSNX1.3 (n = 6) or cPTD4 (n = 5) were fixed and paraffin embedded. The embedded tissue was used for NanoString analysis. a Change in tumor volume per day of mice treated with cSNX1.3 or cPTD4, ±SEM. Statistics were analyzed using an unpaired t test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. b Heatmap demonstrating cell type score for each cSNX1.3 and cPTD4 sample. Red represents upregulation and blue represents downregulation of gene expression. c Volcano plot showing the differentially expressed genes in cSNX1.3 vs. cPTD4. Blue dots represent downregulated genes (4) with a log2FC < 0.5 and red dots represent upregulated genes (n = 41) with a log2FC > 0.5. Scatter plots showing Log2 normalized expression of genes found to be ϯϯsignificant (Padj ≤ 0.05) between cSNX1.3 and cPTD4. ϯ11 tumors that started from the ≥100 mm3 arm of the study (left) were used for NanoString analysis. ϯϯUnpaired t test; correction for multiple testing using Benjamini-Hochberg method in R.
Of the 45 genes that were discovered to be differentially expressed, 16 genes were statistically significant (Padj. < 0.05) when cSNX1.3 and cPTD4 treatments were compared. Specifically, one gene was found to be downregulated in response to cSNX1.3 treatment that is involved in DNA repair (Exo1) and 15 genes were upregulated in response to cSNX1.3 treatment that are involved in immunity (Fig. 3c). The gene ontology attributes corresponding to these genes are associated with immune response-activating signal transduction, leukocyte activation, and regulation of cytokine production (Supplementary Table 1).
Blocking nEGFR enhances NK cell recruitment and cytotoxicity
Considering the alteration of NK cell activation and recruitment genes by RNA-Seq in vitro and NanoString analysis in WAP-TGFα tumors in vivo, we next evaluated the impact of cSNX1.3 treatment on NK cell recruitment and activation directly. We first evaluated how cSNX1.3 treatment altered the recruitment of NK cells in the WAP-TGFα model by immunohistochemistry (IHC). To address the presence of NK cells in the tumor microenvironment, we analyzed tumors derived from cSNX1.3 (n = 9) and cPTD4 (n = 9) treated mice with for NK cell infiltration and EGFR expression (Fig. 4a). While NK cells were observed in almost all cSNX1.3-treated samples, they were rarely observed in cPTD4 controls. To quantify NK cell infiltration, total nuclei and NKp46 positive cells were counted using image segmentation of 50 stitched images per slide and quantified with MatLab (Fig. 4b and Supplementary Fig. 4). Percent of NK cells per sample was determined by calculating the number of NKp46 cells/(NKp46 cells + nuclei). Next, we evaluated all individual tumors to compare their growth rate to the infiltration of NK cells observed (Fig. 4c). We found a striking concordance between NK cell infiltration and low to negative growth rates compared to no NK cell infiltration and high growth rates. Overall, our results show that cSNX1.3 treatment results in increased NK cell recruitment associated with low to negative tumor growth in WAP-TGFα transgenic tumors.
a Tumors from WAP-TGFα female mice treated with cSNX1.3 (n = 9) or cPTD4 (n = 9) were fixed in 10% buffered formalin and paraffin embedded (tumors from both ≥100 mm3 and ≥250 mm3 groups were used for IHC). Slides were incubated with NKp46 primary antibody for NK cell detection, an anti-EGFR antibody, and counterstained with methyl green (nuclei). Black arrow heads indicate examples of positive staining for each. ×40 magnification for all images. b Quantification; identification of methyl green and positive NKp46 stain. Bar graph showing NK cells/(NK cells + nuclei) for each experimental group. c Table including the mice used for IHC, alongside their average tumor growth rate and the number of cells positive for NKp46 stain and methyl green (nuclei). d MDA-MB-468 cells were labeled with DiO, stimulated with EGF (10 ng/ml), treated with cSNX1.3 (10 μM) or cPTD4 (10 μM), and co-cultured with NK-92 cells labeled with DiD. Cells were tracked for 24 h and total intensity of both DiO and DiD staining is shown. e Target cell count after drug treatment and primary NK cell co-culture. MDA-MB-468 target cells were seeded at a density of 50,000 cells/well (Donors 1–3) or 400,000 cells/well (Donor 4). Target cells were stimulated with EGF (10 ng/ml) and treated with either cSNX1.3 (10 μM), erlotinib (15 μM), or cPTD4 (10 μM) for 24 h, and pre-activated primary NK cells were co-cultured with the target cells at an E:T ratio of 4:1 for 4 h.
To determine how cSNX1.3 impacts the recruitment of human NK cells in vitro, MDA-MB-468 cells were labeled with DiO (Fig. 4d, green), stimulated with EGF, treated with cSNX1.3 or cPTD4, and co-cultured with NK-92 cells labeled with DiD (Fig. 4d, red). Live-cell imaging was performed and total intensity of DiO and DiD was tracked over a period of 24 h. In cells treated with cSNX1.3, we observed the NK-92 cells that were originally in suspension being recruited to the adhered tumor cells over the 24-h period, a phenomenon we did not observe with the cells treated with cPTD4 (3-fold increase in NK-92 cell recruitment compared to cPTD4) (Fig. 4d).
To determine if cSNX1.3 also results in the induction of NK cell-mediated cytotoxicity, we next evaluated cSNX1.3-dependent NK cell killing against target cells in vitro. MDA-MB-468 target cells were stimulated with EGF (10 ng/ml) and treated with cSNX1.3 (10 μM), cPTD4 (10 μM), or erlotinib (15 μM) for 24 h and then co-cultured with primary human donor NK effector cells at a 4:1 E:T ratio for an additional 4 h. Compared to cSNX1.3 alone, cells treated with cSNX1.3 and exposed to NK cells had at least a 50% decrease in viable cells compared to either cSNX1.3 alone, cPTD4, or erlotinib (Fig. 4e; note cells were limited from donor 3 and the erlotinib control was not performed). While there is significant sample-to-sample variability due to the use of primary human donor NK cells, each sample did respond to cSNX1.3 treatment with at least a 50% tumor cell killing.
Discussion
In the present report, we have identified a novel function of nEGFR in regulating NK cell recruitment and cytotoxicity. By utilizing a targeted therapeutic, cSNX1.3, and a nEGFR mutant cell line, we found a significant difference in nEGFR and EGFR tyrosine kinase-driven gene transcription. Importantly, we found that several genes encoding NK cell activating ligands were specifically altered by nEGFR. We show that NK cells are preferentially recruited to tumors and cells treated with cSNX1.3 and that cSNX1.3 treatment also results in enhanced NK cell killing of EGFR-expressing breast cancer cells. Together, these data demonstrate a novel role for nEGFR in regulating NK cells.
While we find that erlotinib treatment impacts gene pathways long associated with EGFR function (i.e. MAP kinase pathways), blocking nEGFR transcription – either via cSNX1.3 treatment or genetically ablating the nuclear localization sequence—does not impact these canonical signaling pathways. In fact, we observe a strikingly different set of transcriptional targets from nEGFR, specifically those genes that regulate NK cells. Despite other studies showing that cancers with mutated EGFR [38, 39] treated with erlotinib leads to increased tumor killing by NK cells [9], our studies do not show a reliable increase in NK cell killing by erlotinib, especially when compared to the NK cell killing induced by cSNX1.3.
The cSNX1.3 peptide interacts with a sequence on EGFR that has significant homology with the other erbB receptors (including erbB2, erbB3 and erbB4), and erbB3 and erbB4 are expressed and functional in these cells (Supplementary Fig. 5) [40, 41]. Furthermore, the sequence of cSNX1.3 has significant homology between Snx1 and Snx2 (Supplementary Fig. 5). Therefore it is probable that cSNX1.3 impacts additional protein-protein interactions and represents a strong rationale for comparing transcriptional effects to a genetic mutation such as the ΔNLS. The ΔNLS cell line was created through the knockdown of endogenous EGFR and re-expression of the EGFR-GFP plasmids (both wildtype and mutant; Supplementary Fig. 2). Since the erbB receptors work through homo- and hetero-dimerization (and as noted above erbB3 and erbB4 are expressed in these cells), this approach also potentially represents non-EGFR-dependent transcriptional effects. Together, these factors allow for non-overlapping impacts of the two approaches. Yet, both approaches found alterations in NK cell gene expression, specifically the ULBP genes. This striking overlap (and difference from erlotinib treatment) led us to focus on the potential impact of nEGFR in NK cell recruitment and activation. Evaluation of the NanoString gene expression profiling and RNA-seq demonstrate additional cell type involvement in the response to cSNX1.3 loss of nuclear EGFR (Fig.1 and Fig. 3). Specifically, B and T lymphocytes and macrophages are found to be increased by gene expression profiling (Fig. 3). As the innate and adaptive cells of the immune system work in synergy, it is probable that there is crosstalk between the arms. For example, alterations to NK cell recruitment and activation can alter the overall inflammatory nature and induce a recruitment of T lymphocytes [42]. Alternatively, it is possible that cSNX1.3 is directly impacting macrophage activity, as EGFR is expressed and functional in these cells in regulating macrophage polarity [43]. While it will be interesting to investigate the role of nuclear EGFR in these additional immune cell types, the current in vitro data demonstrates that nuclear EGFR can directly impact the expression of NK cell activation antigens in tumor cells. The in vitro killing assays demonstrate that NK cells can induce tumor cell killing of cSNX1.3-treated tumor cells in the absence of additional immune mediators. Together, these data show a direct role for tumor cell nuclear EGFR in modulating NK cell activity, although other impacts on the immune system are likely.
Overall, NK cells are an important component of the anti-tumor immune response, capable of controlling progression and metastasis [4, 12, 44, 45]. However, breast cancer cells have developed mechanisms of escape from NK cell-mediated immunity by impairing their recruitment and cytotoxic functions. Although attempts at activating the tumor immune microenvironment have been made using antibodies and TKIs, the impact of nEGFR on immunosuppression has yet to be explored. We now show that nEGFR can regulate NK cell recruitment and cytotoxicity in EGFR-dependent breast cancer. Our findings suggest a suppressive role of nEGFR in the immune system process, specifically in NK cell function. Several avenues remain to be explored, including opportunities to better understand the mechanism by which nEGFR impacts NK cell recruitment. Insights gained from our study may help direct the development of combinatorial therapies, utilizing nEGFR inhibitors with immunotherapies.
Materials and methods
Cell lines
MDA-MB-468, NK-92, and primary NK cell lines
MDA-MB-468, T47D and NK-92 cell lines are from ATCC (and maintained according to ATCC guidelines). Leukoreduction chambers were obtained from anonymous healthy platelet donors and NK cells were purified (>95% CD56 + CD3-) using RosetteSep (Stem Cell Technologies) negative selection and Ficoll centrifugation, as described [46]. Primary NK cells were grown in RPMI-1640 medium supplemented with 10% heat inactivated human AB serum, 2 mM glutamine, 1% penicillin-streptomycin, 100 mM HEPES, 1x nonessential amino acids, and maintained in 1 ng/mL of IL-15 (Miltenyi Biotech). Cell lines are tested for mycoplasma every 6 months.
EGFRWT and EGFRΔNLS cell lines in MDA-MB-468 cells
EGFR-GFP plasmid (a gift from Graham Carpenter) was subjected to mutagenesis using the Q5 Site-Directed Mutagenesis Kit (New England BioLabs). EGFR knockdown was created using lentiviral particles containing isopropyl-β-D-1-thiogalactopyranoside (IPTG)-inducible short hairpin RNA (shRNA) sequences and puromycin resistance (MilliporeSigma). EGFR knockdown: Construct A (5ʹ-CCGGAGAATGTGGAATACCTAAGGCTCGAGCCTTAGGTATTCCACATTCTCTTTTTG 3ʹ) Construct B (5ʹ- CCGGGCTGCTCTGAAATCTCCTTTACTCGAGTAAAGGAGATTTCAGA GCAGCTTTTTG-3ʹ). Nonspecific shRNA: Construct C (5ʹ-GCGCGATAGCGCTAATAATTT 3ʹ. Cells were incubated with hexadimethrine bromide (8 μg/mL) to increase transduction efficiency before viral particles were added at an MOI of 1. Transduced cells were then incubated with 1 mM IPTG for 2 days to establish knockdown and then remained in IPTG for the duration of the experiment.
Mouse studies
All animal experiments were performed with approval by the University of Arizona Institutional Animal Care and Use Committee. WAP-TGFα transgenic mice (The Jackson Laboratory; Tg(WapTgfa)215Bri) were backcrossed to C57BL/6 J background. WAP-TGFα heterozygous males were bred with C57BL/6 J and offspring were genotyped using primers against the WAP-TGFα transgene. WAP-TGFα heterozygous females were entered into either the cPTD4 (control) or cSNX1.3 (treatment) groups of the study when tumors reached either 150 mm3 or 250 mm3, as indicated. Randomization was used to determine how mice were allocated to experimental groups*. Mice were added to alternating control or treatment arms as they developed tumors. Injections were administered intravenously at 10 μg/g body weight 3x/week for 4 weeks. Mice were sacrificed when tumors reached a total burden of 2000 mm3, when a single tumor reached a diameter of 2 cm, or after 4 weeks of drug treatment. Mice were monitored 2x/week.
*Researchers were not blinded to the experimental arm allocation.
Immunohistochemistry (IHC) of mouse tumor tissues
Tumors were fixed in 10% neutral buffered formalin and paraffin embedded. Tissues were sectioned by the Tissue Acquisition and Cellular/Molecular Analysis Core at the University of Arizona Cancer Center. Anti-NK cell antibody NKp46 (cat. #PA5-102860; ThermoFisher Scientific) and anti-EGFR antibody EP38Y (ab52894; Abcam) were used to assess NK cells and total EGFR. Deparaffinization, rehydration, antigen retrieval and primary and secondary antibody reactions were performed according to the antibody’s manufacture specifications.
Immunohistochemistry image segmentation and quantification
A custom MATLAB script was used to *segment IHC images and quantify methyl green and NKp46 counterstained cells. Briefly, RGB images were split into individual channels and Cellpose [47] was used to identify methyl green (channel 1) positive cells. The “nuclei” Cellpose model was used. Parameters were optimized to limit false positives for the identification of methyl green counterstained objects. Key parameters are as follows. Methyl green: (ImageCellDiameter = 40, CellThreshold = -0.5, and FlowErrorThreshold = 0.75).
For NKp46 counterstained cells, channel 2 of the original RGB images was used. A custom MATLAB script was used for image processing. Cellpose was not used for the identification of NKp46 counterstained objects as many false positives were present. Instead, background subtraction followed by Gaussian smoothing was performed. Otsu’s method was then used to automate image thresholding and create a binary image for segmentation. Finally, segmented objects were filtered based on circularity and size. Examples of segmented images are in Supplementary Fig. 4. All custom written scripts described are available upon request.
*Researchers were blinded to treatment groups to avoid bias.
NK cell killing assay
MDA-MB-468 target cells were seeded at a density of 50,000 cells/well in a 24-well plate or 400,000 cells/well in a 6-well plate and allowed to adhere for 24 h. The target cells were then stimulated with EGF (10 ng/ml) and treated with either cSNX1.3 (10 μM) or cPTD4 (10 μM) for an additional 24 h. Prior to being co-cultured with the target cells, primary NK effector cells were pre-activated with IL-15 (50 ng/ml) for 2 h, washed, and resuspended in 1 ng/ml of IL-15 for the remainder of the assay. After 24 h of drug treatment, the pre-activated primary NK cells were co-cultured with the target cells at an effector:target (E:T) ratio of 4:1 for 4 h. After 4 h, NK cells were washed off with PBS and the remaining target cells were trypsinized and counted.
Cell viability
MDA-MB-468 cells were treated with the indicated drugs and incubated for 72 h. The drug-containing media was decanted from the plate and replaced with media containing 250 μg/mL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) for 80 min. The resulting formazan crystals were dissolved in 100% DMSO and the absorbance at 540 nm was detected using a Biotech Synergy LX plate reader. To test for synergy, the Bliss independence model was used. A score of < –10 indicates the interaction between two drugs is likely to be antagonistic, a score between -10 and 10 indicates the interaction is likely to be additive, and a score >10 indicates the interaction is likely to be synergistic.
Live-cell nuclear EGFR tracking
T47D cells were modified with the eFlut plasmid system [48] to stably express mVenus [49] fused to the endogenous EGFR gene using recombinant Cas9 (Alt-R™ S.p. Cas9 Nuclease V3, IDT). T47D EGFR-mVenus cells were serum starved overnight and incubated with SPY650-DNA Probe (Spirochrome), treated with the indicated drugs and imaged every 10 min for 24 h using a Nikon Eclipse Ti microscope. Cells were imaged with the ET-EYFP and Filter Set for 800 ms (Chroma). The SPY650 DNA dye was imaged using the ET-Cy5 Filter Set for 20 ms (Chroma). Images were acquired with the Hamamatsu ORCA-Flash 4.0 camera and quantified via MATLAB.
CELLCYTE real-time live imaging
MDA-MB-468 target cells were seeded at a density of 300,000 cells/well in a 6-well plate and labeled with VybrantTM DiO NK-92 and effector cells were labeled with VybrantTM DiD and co-cultured with the target cells along with drug treatment at an E:T ratio of 2:1. EGF (10 ng/ml) and cSNX1.3 (10 μM) or cPTD4 (10 μM) were added to the media and all cells were tracked for 24 h using the CELLCYTE XTM Live Cell Imager and Analyzer.
Bioinformatics analysis
Quality of raw reads was assessed using FastQC, and reads were trimmed using TrimGalore 0.6.7 with a Phred score quality threshold of 20 (the default) and removing Illumina adapters. Trimmed reads were then aligned to the GRCh38 reference human genome using STAR 2.7.9a with default parameters. Aligned reads were then quantified using featureCounts from the Rsubread package, along with the GRCh38 gtf file, and specifying that the sequencing was paired end but not strand specific.
The R package DESeq2 was used to test for differential expression between the different group comparisons and to select all genes with a Padj value < 0.05. The R package clusterProfiler was used to evaluate functional enrichment in the differentially expressed genes between the different groups. We ran overrepresentation analysis on Gene Ontology using the inputs: differentially expressed genes, the universe, the species-specific annotation database (org.Hs.eg.db), the BP ontology, a p value adjustment method (Benjamini-Hochberg), and p value and q-value cutoffs (0.05). The R package ComplexHeatmap was used to create a heatmap visualizing the vst-normalized data created using the DESeq2 package.
NanoString analysis
As described in Jordan et al. [50]. Formalin-fixed paraffin-embedded (FFPE) murine tumor specimens were sectioned and for each tumor block there were two, 20 μm sections. RNA was extracted with High Pure FFPET RNA Isolation Kit (Roche) and the RNA quantity and quality were determined with an RNA ScreenTape on a TapeStation 4150 (Agilent Technologies). RNA concentration was calculated by comparing the RNA ladder and the percentage of RNA fragments greater than 200 bp. RNA (150 ng) was combined with hybridization buffer and the reporter CodeSet for the Murine PanCancer IO 360 Panel (NanoString Technologies) and incubated for 20 h at 65 °C. The hybridized reaction was analyzed on an nCounter SPRINT Profiler (NanoString Technologies).
mRNA expression, normalization, fold changes and p values were calculated using ROSALIND. Normalization. Normalization involves dividing counts within a lane by the geometric mean of the normalizer probes from the same lane. Housekeeping probes used for normalization were selected based on the geNorm algorithm as implemented in the NormqPCR Bioconductor package in R. Differential expression. Differential expression is calculated based on user specified groups. Fold changes and p values are calculated using the method described in the nCounter Advanced Analysis 2.0 User Manual. P value adjustment was performed using the Benjamini-Hochberg of estimating false discovery rates. Cell type profiler. The method to quantify abundance of cell populations uses marker genes expressed stably and specifically in given cell types. ROSALIND filters results to include those that have scores with p values ≤ 0.05. Algorithm details of all processes can be found in the nCounter Advanced Analysis 2.0 User Manual.
Data availability
RNA-Seq data is available in the Gene Expression Omnibus under accession number GSE273890.
References
Chiossone L, Dumas P-Y, Vienne M, Vivier E. Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol. 2018;18:671–88.
Wu S-Y, Fu T, Jiang Y-Z, Shao Z-M. Natural killer cells in cancer biology and therapy. Mol Cancer. 2020;19:120.
Thomas M, Boname JM, Field S, Nejentsev S, Salio M, Cerundolo V, et al. Down-regulation of NKG2D and NKp80 ligands by Kaposi’s sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc Natl Acad Sci. 2008;105:1656–61.
Baysal H, De Pauw I, Zaryouh H, Peeters M, Vermorken JB, Lardon F, et al. The right partner in crime: unlocking the potential of the anti-EGFR antibody cetuximab via combination with natural killer cell chartering immunotherapeutic strategies. Front Immunol. 2021;12:737311.
Berrien-Elliott MM, Jacobs MT, Fehniger TA. Allogeneic natural killer cell therapy. Blood. 2023;141:856–68.
Riggan L, Shah S, O’Sullivan TE. Arrested development: suppression of NK cell function in the tumor microenvironment. Clin Transl Immunol. 2021;10:e1238.
Laskowski TJ, Biederstädt A, Rezvani K. Natural killer cells in antitumour adoptive cell immunotherapy. Nat Rev Cancer. 2022;22:557–75.
Endoh H, Ishibashi Y, Yamaki E, Yoshida T, Yajima T, Kimura H, et al. Immunohistochemical analysis of phosphorylated epidermal growth factor receptor might provide a surrogate marker of EGFR mutation. Lung Cancer. 2009;63:241–6.
Dominguez C, Tsang K-Y, Palena C. Short-term EGFR blockade enhances immune-mediated cytotoxicity of EGFR mutant lung cancer cells: rationale for combination therapies. Cell Death Dis. 2016;7:e2380–e.
Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–45.
Hashmi AA, Naz S, Hashmi SK, Irfan M, Hussain ZF, Khan EY, et al. Epidermal growth factor receptor (EGFR) overexpression in triple-negative breast cancer: association with clinicopathologic features and prognostic parameters. Surg Exp Pathol. 2019;2:81–90.
Lin S-Y, Makino K, Xia W, Matin A, Wen Y, Kwong KY, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–8.
Kamio T, Shigematsu K, Sou H, Kawai K, Tsuchiyama H. Immunohistochemical expression of epidermal growth factor receptors in human adrenocortical carcinoma. Hum Pathol. 1990;21:277–82.
Marti U, Ruchti C, Kämpf J, Thomas GA, Williams ED, Peter HJ, et al. Nuclear localization of epidermal growth factor and epidermal growth factor receptors in human thyroid tissues. Thyroid. 2001;11:137–45.
Lo H-W, Hsu S-C, Ali-Seyed M, Gunduz M, Xia W, Wei Y, et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–89.
Lo H-W, Xia W, Wei Y, Ali-Seyed M, Huang S-F, Hung M-C. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–48.
Psyrri A, Yu Z, Weinberger PM, Sasaki C, Haffty B, Camp R, et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res. 2005;11:5856–62.
Baselga J, Albanell J, Ruiz A, Lluch A, Gascón P, Guillém V, et al. Phase II and tumor pharmacodynamic study of gefitinib in patients with advanced breast cancer. J Clin Oncol. 2005;23:5323–33.
Dickler MN, Cobleigh MA, Miller KD, Klein PM, Winer EP. Efficacy and safety of erlotinib in patients with locally advanced or metastatic breast cancer. Breast Cancer Res Treat. 2009;115:115–21.
Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, et al. TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol. 2012;30:2615–23.
Douillard J-Y, Ostoros G, Cobo M, Ciuleanu T, Mccormack R, Webster A, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Cancer. 2014;110:55–62.
Kim C, Liu SV. First-line EGFR TKI therapy in non-small-cell lung cancer: looking back before leaping forward. Ann Oncol. 2019;30:1852–5.
Wang S-C, Hung M-C. Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin Cancer Res. 2009;15:6484–9.
Bitler BG, Goverdhan A, Schroeder JA. MUC1 regulates nuclear localization and function of the epidermal growth factor receptor. J Cell Sci. 2010;123:1716–23.
Atwell B, Chen C-Y, Christofferson M, Montfort WR, Schroeder J. Sorting nexin-dependent therapeutic targeting of oncogenic epidermal growth factor receptor. Cancer Gene Ther. 2023;30:267–76.
Bitler BG, Menzl I, Huerta CL, Sands B, Knowlton W, Chang A, et al. Intracellular MUC1 peptides inhibit cancer progression. Clin Cancer Res. 2009;15:100–9.
Maisel SA, Schroeder J. Wrong place at the wrong time: how retrograde trafficking drives cancer metastasis through receptor mislocalization. J Cancer Metastasis Treatment. 2019;5:1–14.
Batista TM, Cederquist CT, Kahn CR. The insulin receptor goes nuclear. Cell Res. 2019;29:509–11.
Giri DK, Ali-Seyed M, Li L-Y, Lee D-F, Ling P, Bartholomeusz G, et al. Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor. Mol Cell Biol. 2005;25:11005–18.
Maher PA. Nuclear Translocation of fibroblast growth factor (FGF) receptors in response to FGF-2. J cell Biol. 1996;134:529–36.
Reif R, Adawy A, Vartak N, Schröder J, Günther G, Ghallab A, et al. Activated ErbB3 translocates to the nucleus via clathrin-independent endocytosis, which is associated with proliferating cells. J Biol Chem. 2016;291:3837–47.
Liu S, Geng R, Lin E, Zhao P, Chen Y. ERBB1/2/3 expression, prognosis, and immune infiltration in cutaneous melanoma. Front Genet. 2021;12:602160.
Makhijani P, Basso PJ, Chan YT, Chen N, Baechle J, Khan S, et al. Regulation of the immune system by the insulin receptor in health and disease. Front Endocrinol. 2023;14:1128622.
Hart MR, Su H-Y, Broka D, Goverdhan A, Schroeder JA. Inactive ERBB receptors cooperate with reactive oxygen species to suppress cancer progression. Mol Ther. 2013;21:1996–2007.
Hsu S-C, Hung M-C. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–40.
Zingoni A, Molfetta R, Fionda C, Soriani A, Paolini R, Cippitelli M, et al. NKG2D and its ligands: “One for All, All for One”. Front Immunol. 2018;9:476.
Sandgren EP, Schroeder JA, Qui TH, Palmiter RD, Brinster RL, Lee DC. Inhibition of mammary gland involution is associated with transforming growth factor alpha but not c-myc-induced tumorigenesis in transgenic mice. Cancer Res. 1995;55:3915–27.
Mallmann-Gottschalk N, Sax Y, Kimmig R, Lang S, Brandau S. EGFR-specific tyrosine kinase inhibitor modifies nk cell-mediated antitumoral activity against ovarian cancer cells. Int J Mol Sci. 2019;20:4693.
Ayeni D, Miller B, Kuhlmann A, Ho P-C, Robles-Oteiza C, Gaefele M, et al. Tumor regression mediated by oncogene withdrawal or erlotinib stimulates infiltration of inflammatory immune cells in EGFR mutant lung tumors. J Immunother Cancer. 2019;7:172.
Rau A, Lieb WS, Seifert O, Honer J, Birnstock D, Richter F, et al. Inhibition of tumor cell growth and cancer stem cell expansion by a bispecific antibody targeting EGFR and HER3. Mol Cancer Therapeutics. 2020;19:1474–85.
Sundvall M, Korhonen A, Vaparanta K, Anckar J, Halkilahti K, Salah Z, et al. Protein inhibitor of activated STAT3 (PIAS3) protein promotes SUMOylation and nuclear sequestration of the intracellular domain of ErbB4 protein. J Biol Chem. 2012;287:23216–26.
Roda JM, Parihar R, Magro C, Nuovo GJ, Tridandapani S, Carson WE. Natural killer cells produce T cell–recruiting chemokines in response to antibody-coated tumor cells. Cancer Res. 2006;66:517–26.
Zhao G, Liu L, Peek RM, Hao X, Polk DB, Li H, et al. Activation of epidermal growth factor receptor in macrophages mediates feedback inhibition of M2 polarization and gastrointestinal tumor cell growth. J Biol Chem. 2016;291:20462–72.
Wang X, Semba T, Manyam GC, Wang J, Shao S, Bertucci F, et al. EGFR is a master switch between immunosuppressive and immunoactive tumor microenvironment in inflammatory breast cancer. Sci Adv. 2022;8:eabn7983.
Selenz C, Compes A, Nill M, Borchmann S, Odenthal M, Florin A, et al. EGFR inhibition strongly modulates the tumour immune microenvironment in EGFR-driven non-small-cell lung cancer. Cancers. 2022;14:3943.
Romee R, Schneider SE, Leong JW, Chase JM, Keppel CR, Sullivan RP, et al. Cytokine activation induces human memory-like NK cells. Blood. 2012;120:4751–60.
Stringer C, Wang T, Michaelos M, Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nat Methods. 2021;18:100–6.
Stewart-Ornstein J, Lahav G. Dynamics of CDKN1A in single cells defined by an endogenous fluorescent tagging toolkit. Cell Rep. 2016;14:1800–11.
Kremers GJ, Goedhart J, van Munster EB, Gadella TW. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry. 2006;45:6570–80.
Jordan KR, Sikora MJ, Slansky JE, Minic A, Richer JK, Moroney MR, et al. The capacity of the ovarian cancer tumor microenvironment to integrate inflammation signaling conveys a shorter disease-free interval. Clin Cancer Res. 2020;26:6362–73.
Acknowledgements
AE was supported by the IMSD program (NIH T32GM139779), by the Marshall Foundation, and by the Hispanic Serving Institution Initiatives at the University of Arizona (for funding support). RH was supported by the Biochemistry, Molecular and Cellular Biology Training Grant (T32GM136536). CP was supported by the Biochemistry, Molecular and Cellular Biology Training Grant (T32GM008659). DOC and SK were supported by the MARC Biomedical Research and Training Grant (T34GM008718). BGB was supported by NIH/NCI R37CA261987. TAF was supported by P50CA171963, P30CA91842, Lymphoma Research Foundation, Leukemia and Lymphoma Society, Rising Tide Foundation for Cancer Research, and Melanoma Research Alliance. ALP was supported by RO1GM130864. MP was supported by NIH R01CA251729. Work performed by the Tissue Acquisition core at the University of Arizona Cancer Center was supported by NIH P30 CA023074 and the University of Arizona Imaging Cores - Optical Core Facility (RRID:SCR_023355). We would like to acknowledge Mireya L. Herrera-Herrera for her support with qPCR.
Author information
Authors and Affiliations
Contributions
AE and JS conceived the project, led the data interpretation, and wrote the manuscript. MP provided guidance for the writing of the manuscript and bioinformatics expertise. AE and MP performed RNA-Seq analysis and MP revised the analysis codes. AE performed immunohistochemistry, NK cell experiments, qPCR, and NanoString analysis. BGB performed NanoString. RH performed cell viability experiments. CP performed IHC segmentation analysis and ALP revised the analysis codes. SC performed RT-PCR experiments. BA maintained and treated the WAP-TGFα mice with the assistance of BS. AJ, SK, and DOC created the EGFR-GFP constructs. TAF donated primary NK cells and provided NK cell expertise. All authors reviewed the manuscript. JS provided the funding for the project.
Corresponding authors
Ethics declarations
Competing interests
TAF has licensed patents, equity, consulting, and potential royalty interests in Wugen; equity interests in Indapta Therapeutics and Orca Bio; and serves as a consultant for Affimed, AI Proteins, and Smart Immune; and has research funding from HCW biologics, Wugen, and Affimed.
Ethics approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations. No human subjects were involved in this research. All animal studies were approved by the University of Arizona Institutional Animal Care and Use Committee, protocol approval #18-427.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Escoto, A., Hecksel, R., Parkinson, C. et al. Nuclear EGFR in breast cancer suppresses NK cell recruitment and cytotoxicity. Oncogene 44, 288–295 (2025). https://doi.org/10.1038/s41388-024-03211-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-024-03211-0