Abstract
Nasopharyngeal carcinoma (NPC) is a special histological and ethical type of head and neck cancer with unsatisfactory clinical outcome. Thus, exploring effective molecular targets is critical for NPC treatment. We observed increased expression levels of synaptotagmin-7 (SYT7) in NPC tissues, which correlated with unfavorable prognoses. Furthermore, knockdown of SYT7 in NPC cells suppressed proliferation and migration rates, and enhanced apoptosis. In contrast, overexpression of SYT7 accelerated NPC tumor growth. Using whole-genome gene arrays and immunoprecipitation-mass spectrometry assays, aldehyde dehydrogenase 1 family member A3 (ALDH1A3), a regulator of glycolytic metabolism, was identified as a critical downstream target of SYT7. Mechanistically, SYT7 binds and promotes ALDH1A3 deubiquitination, resulting in decreased ALDH1A3 degradation. Notably, we also observed an increased expression of ALDH1A3 in NPC. More importantly, the knockdown of ALDH1A3 resulted in suppressed proliferation, migration, glycolysis, and promoted apoptosis, all of which could be restored by the overexpression of SYT7 in NPC cells. Taken together, we found that SYT7 increases ALDH1A3-mediated STAT3 activation and glycolysis, contributing to NPC progression, which provides a possible molecular mechanism for the development of targeted therapeutics interventions.
Similar content being viewed by others
Introduction
Nasopharyngeal carcinoma (NPC) is a distinctive form of head and neck cancer that highly prevalents in Southeast Asia and Southern China [1, 2]. Locoregionally metastasis occurs in more than 70% newly diagnosed NPC patients [3, 4]. Owing to significant advancements in early diagnosis techniques, chemotherapy strategies and surgery interventions, the prognosis of NPC has significantly improved [2,3,4,5,6,7,8,9]. Despite these advancements, distant metastases and recurrences continue to pose the most substantial challenges in NPC treatment and remain the primary causes of mortality [1, 5, 8]. Therefore, a deeper comprehension understanding of the pathogenesis and metastatic mechanism of NPC on the molecular basis is crucial for the development of targeted therapeutics.
Synaptotagmin-7 (SYT7) is a calcium sensor in the process of cell exocytosis and synaptic transmission [10,11,12,13]. SYT7 also plays a critical role in serials biological processes, such as cell apoptosis, proliferation, cytokine production, and transcriptional regulation. Accumulating evidence has revealed the significance of SYT7 in the initiation and progression of cancer, indicating that targeting SYT7 may be a potential strategy for cancer treatment [14,15,16,17]. Nevertheless, whether SYT7 takes part in the progression of NPC remains unraveled.
In this study, we demonstrate the upregulation of SYT7 expression in NPC. We further observed that knockdown of SYT7 resulted in a decreased proliferation rate and enhanced apoptosis in NPC cells. Additionally, we identify that aldehyde dehydrogenase 1 family member A3 (ALDH1A3) as a key downstream target of SYT7, which promotes NPC progression by modulating signal transducer and activator of transcription 3 (STAT3) activation and aerobic glycolysis. Our findings suggest that SYT7 promotes NPC progression by modulating STAT3 activation and aerobic glycolysis, thereby highlighting a potential molecular pathway for targeted intervention.
Results
The expression level of SYT7 is closely associated with cell proliferation and migration, and unfavorable clinical outcome in NPC
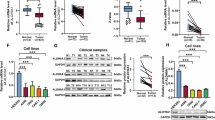
To investigate the role of SYT7 in nasopharyngeal carcinogenesis, immunohistochemistry (IHC) staining for SYT7 was conducted on both NPC and non-cancerous nasopharyngeal tissue microarray (TMA). As shown in Fig. 1A and Table S1, SYT7 expression levels were significantly higher in NPC tissue samples than those in non-cancerous nasopharyngeal tissue samples (P < 0.001). Moreover, SYT7 expression levels demonstrated significant associations with several patient characteristics, such as clinical stage (P < 0.001), tumor infiltration (P < 0.001), lymph node status (P < 0.001), distant metastasis (P < 0.01), and recurrence status (P < 0.001) in the NPC TMA (Tables S2 and S3). Elevated SYT7 expression was inversely correlated with the overall survival rate of NPC patients (P < 0.001) (Fig. 1B), indicating that SYT7 may be considered a prognostic marker for patients with NPC. SYT7 expression was appraised in normal nasopharyngeal cell line NP-69 and various NPC cell lines. NPC cells showed significant higher SYT7 mRNA levels than that in NP-69 cells (Fig. S1A).
A Representative images of immunohistochemistry staining for SYT7 in NPC tissue samples and adjacent non-cancerous tissue samples. B Kaplan–Meier curves depicting the overall survival rate of NPC patients with high or low levels of SYT7 expression (N = 105). C The mRNA level of SYT7 in C666-1 cells treated with all three shSYT7 constructs. D The protein level of SYT7 was detected by immunoblot in C666-1 and HONE-1 cell lines after infection with shCtrl and shSYT7 lentivirus. E The MTT assay was used for indicated stable cell lines to generate cell growth curves. F Images of colony formation in stable shCtrl and shSYT7 C666-1 or HONE-1 cell lines. G Wound-healing assays revealed the migration rates of stable shCtrl and shSYT7 C666-1 or HONE-1 cells at indicated time points after infection. H Transwell assays assessing the migration rates of shCtrl and shSYT stable C666-1 or HONE-1 cells. Magnification: ×200. I Flow cytometry assay showed the effect of SYT7 knockdown on cell apoptosis. The experiments were repeated at least thrice. Data are expressed as mean ± SD, *P < 0.05, **P < 0.01 and ***P < 0.001.
To further explore the biological function of SYT7 in NPC cells, short hairpin (sh)RNA lentiviral particles targeting negative control (shCtrl) and SYT7 (shSYT7) were constructed and added to C666-1 or HONE-1 cells. As shown in Figs. 1C and S1B, C, all three shSYT7 lentiviruses effectively depleted SYT7 mRNA in C666-1 and HONE-1 cells. Based on its efficacy, shSYT7-1 was selected for subsequent experiments. Moreover, SYT7 protein levels were significantly decreased in both C666-1 and HONE-1 stable cell lines (Fig. 1D). It was shown that SYT7 knockdown led to a significantly decrease in cell proliferation rates and colonies numbers in NPC cells, as demonstrated by in 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT) and colony formation assays (P < 0.001, Fig. 1E, F). Using wound-healing and transwell assays, we also observed that the migration ability was significantly inhibited after SYT7 knockdown in NPC cells (P < 0.01, Fig. 1G, H). SYT7 depletion also resulted in increased cell apoptosis in NPC cells (P < 0.001, Fig. 1I). These findings collectively show that SYT7 functions as an oncogenic molecule, promoting NPC cell proliferation and migration while suppressing apoptosis.
ALDH1A3 is a critical downstream target of SYT7-mediated NPC carcinogenesis
To further investigate the downstream targets of SYT7 in NPC, whole-genome gene expression assays were performed on stable shCtrl and shSYT7 C666-1 cells to identify the differential expressed genes (DEGs). Knockdown of SYT7 resulted in 503 upregulated DEGs and 608 downregulated DEGs (Fig. 2A, B). Using ingenuity pathway analysis (IPA) assay, the DEGs showed significant enrichment in “GNRH signaling” and “IL-1 signaling” (Figs. 2C and S2). IPA further revealed that genes including ADCY7, AK2, CAB39, CCND1, ALDH1A3, DUSP4, NRIP2, GNAI2, GNAI3, GNB5, SPDEF, IL6ST, INSR, JUN, and HHIP, are potential downstream interactors of SYT7 in NPC (Fig. 2D and Table S4). Among these genes, SPDEF, NRIP2, ALDH1A3, and HHIP were identified as the genes most likely to be co-expressed with SYT7 (Fig. 2E). Consistent with the DEGs results, the shSYT7 C666-1 cells exhibited significantly reduced protein expression of all four genes (SPDEF, NRIP2, ALDH1A3, and HHIP) (Fig. 2F). Given its established association with cancer progression and metastasis [18,19,20], ALDH1A3 was selected as the primary candidate downstream target of SYT7 for further investigation.
A Heatmap of DEGs in stable shCtrl and shSYT7 C666-1 cells generated using RNA-sequencing (n = 3). B Volcano plot of DEGs between shCtrl and shSYT7 C666-1 cells. C Classic pathway enrichment analysis based on IPA. D An IPA-based interaction network for SYT7 and the DEGs. E Co-expression of SYT7 with various genes. F The protein expression levels of SPDEF, NRIP2, ALDH1A3 and HHIP were detected by immunoblot in shCtrl and shSYT7 C666-1 cells. G Schematic diagram illustration of the IP-MS procedure. Created in BioRender. H KEGG pathway enrichment analysis of Top 20 interacting proteins revealed that significant enrichment of interacting proteins in the “Proteasome pathway”.
To further investigate the interacting proteins associated with SYT7, we conducted immunoprecipitation-mass spectrometry assays (IP-MS) (Fig. 2G). The IP-MS results identified 311 proteins as interacting proteins. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of Top 20 high-confidence SYT7-interacting proteins revealed significant enrichment in the “Proteasome pathway” (Fig. 2H). Notably, among the top 20 most likely interacting proteins with SYT7, ALDH1A3 was also identified (Table S5). These findings prompted us to further explore the role of ALDH1A3 and the proteasome pathway in SYT7-mediated NPC carcinogenesis.
SYT7 knockdown promotes ALDH1A3 ubiquitination in NPC
The protein expression of ALDH1A3 was significantly reduced following SYT7 knockdown in NPC cells (Figs. 3B and S3A). However, ALDH1A3 mRNA levels remained relatively unaffected (Fig. 3A), indicating that ALDH1A3 is regulated by SYT7 mainly through post-translational modifications. Thus, we treated stable shCtrl and shSYT7 NPC cells along with a protein synthesis inhibitor, cycloheximide (CHX). SYT7 knockdown increased the degradation rate of ALDH1A3 (Figs. 3C and S3C) without affecting ALDH1A3 mRNA levels (Fig. S3B). Furthermore, MG-132, a proteasome inhibitor, increased the ALDH1A3 protein levels in both shCtrl and shSYT7 NPC cells. More importantly, the downregulation of ALDH1A3 protein after SYT7 depletion was partially rescued by MG-132 (Figs. 3D and S3D), indicating that ALDH1A3 is regulated via proteasome-mediated degradation. The lysosomal protease inhibitor, Chloroquine (CQ), did not change the expression level of ALDH1A3 both in shCtrl and shSYT7 NPC cells, indicating the lysosomal-dependent proteolysis is not involved in SYT7 mediated proteolysis in NPC (Figs. 3E and S3E). Previous studies have reported that ALDH1A3 can be deubiquitinated in cancer cells [20, 21]. Therefore, we examined the ubiquitination status of ALDH1A3 following SYT7 knockdown. Co-IP assays showed that SYT7 binds to ALDH1A3 (Fig. 3F), and SYT7 knockdown promoted ALDH1A3 ubiquitination (Fig. 3G). Taken together, these data suggest that SYT7 functions by deubiquitinating ALDH1A3 in NPC cells.
A, B The expression levels of ALDH1A3 associated with SYT7 were determined using A RT-qPCR, and B WB in C666-1 and HONE-1 cells. C At 0, 6, 12, and 24 h after CHX treatment, immunoblot was conducted to detect ALDH1A3 protein levels in C666-1 cells and HONE-1 cells subjected to SYT7 knockdown, with DMSO used as a negative control. D Indicated NPC cells with or without SYT7 knockdown were treated with 20 μM MG-132 for 6 h, and then indicated protein levels were determined via immunoblot. E shCtrl and shSYT7 stable C666-1 and HONE-1 cells were treated with DMSO or 30 μM Chloroquine for 2 h. F endogenous immunoprecipitation was performed by using ALDH1A3 antibody or IgG in C666-1 and HONE-1 cells, and then immunoblotted with SYT7 antibody. G C666-1 cells infected with shSYT7 or shCtrl lentivirus were treated with MG-132, and the ubiquitination of ALDH1A3 was detected via immunoblotted with an anti-Ubiquitin (Ub) antibody. The data are expressed as the mean ± SD, *P < 0.05 and **P <0.01; the experiments were repeated at least thrice.
ALDH1A3 is a bona fide downstream target of SYT7 in NPC
To further determine the functional role of ALDH1A3 on SYT7-mediated regulation of NPC, we established four groups of stable C666-1 or HONE-1 cell lines were constructed: a negative control group (Vector+shCtrl), a SYT7 overexpression group (SYT7+shCtrl), an ALDH1A3 knockdown group (Vector+shALDH1A3), and a SYT7 overexpression plus ALDH1A3 knockdown group (SYT7+shALDH1A3). As illustrated in Figs. 4A, B and S4, effective infection and knockdown were reached in four groups in NPC cells. SYT7 overexpression promoted cell proliferation in both the SYT7 overexpression (SYT7+shCtrl) and the simultaneous SYT7 overexpression and ALDH1A3 knockdown (SYT7+shALDH1A3) groups compared to their respective controls (Fig. 4C, D). Moreover, the loss of ALDH1A3 dramatically attenuated and reversed the upregulated proliferation induced by SYT7 overexpression, indicating a downstream role of ALDH1A3 in the SYT7-mediated regulation of NPC. Similar trends were also observed in the wound-healing assay, where ALDH1A3 depletion restored cell migration in both the control and SYT7 overexpression groups (Fig. 4E, F). These results collectively highlight the critical involvement of ALDH1A3 in mediating the oncogenic effects of SYT7 in NPC.
A, B Relative protein levels of SYT7 and ALDH1A3, along with their quantifications in stable cell lines of C666-1 or HONE-1, as detected by immunoblot. C, D Effects of SYT7 overexpression, ALDH1A3 knockdown, and simultaneous SYT7 overexpression and ALDH1A3 knockdown on proliferation in C666-1 C and HONE-1 cells D, as analyzed by a Celigo-based cell counting assay. E, F Effects of SYT7 overexpression, ALDH1A3 knockdown, and simultaneous SYT7 overexpression and ALDH1A3 knockdown on the migration of C666-1 cells E and HONE-1 cells F, as analyzed via wound-healing assays 24 h or 48 h after scratching. The data are expressed as the mean ± SD, from three independent biological replicates. P values determined by two-tailed t test, *P < 0.05 and **P < 0.01.
SYT7 promotes NPC cell glycolysis and xenograft growth through ALDH1A3
ALDH1A3 is a critical enzyme involved in glucose metabolism [22,23,24]. Thus, we examined glycolytic activity in stable shCtrl and shSYT7 C666-1 cells. As shown in Fig. 5A–C, SYT7 knockdown resulted in a significant reduction in glucose consumption, lactic acid production and ATP levels. As expected, SYT7 overexpression increased glucose consumption, lactic acid and ATP levels compared to the negative control cells (Fig. 5D–F). In addition, ALDH1A3 knockdown resulted in a significant decrease in glucose consumption, lactic acid and ATP production compared with negative control cells. More importantly, ALDH1A3 knockdown reversed the increased glycolytic activities in SYT7 overexpression groups, indicating that SYT7 promotes glycolysis via ALDH1A3 in NPC cells.
A–C Glucose consumption, lactic acid production and ATP content were measured in C666-1 cells before and after SYT7 knockdown. D–F The production of glucose, lactic acid and ATP were assessed in four stable cell lines before and after overexpressing SYT7 or/and knockdown ALDH1A3. G Photographs and H volumes of tumors from the SYT7+shALDH1A3, SYT7+shCtrl, Vector+shALDH1A3, and Vector+shCtrl groups. Tumor volumes were calculated on the indicated days after post-inoculation. I Tumor weights from the SYT7+shALDH1A3, SYT7+shCtrl, Vector+shALDH1A3, and Vector+shCtrl groups. J H&E staining and K a proliferation marker, Ki-67, IHC staining of xenograft tumors from all four groups. Magnification: ×200 or ×400. Error bars indicate Mean ± SD. P values determined by two-tailed t test, *P < 0.05, **P < 0.01 and ***P < 0.001.
To explore the impact of SYT7 to ALDH1A3 in vivo, we used a subcutaneous xenograft tumor mouse model (n = 5/group). As shown in Fig. 5G, H SYT7 overexpression significantly promoted NPC growth in both the SYT7 overexpression (SYT7+shCtrl) group and the simultaneous SYT7 overexpression and ALDH1A3 knockdown (SYT7+shALDH1A3) group compared to their respective counterparts. Moreover, loss of ALDH1A3 dramatically reduced NPC growth and rescued the tumor-promoting effects induced by SYT7 overexpression (Fig. 5G, H). Consistently, the results of tumor weight measurements (Fig. 5I), hematoxylin and eosin (H&E) staining (Fig. 5J), and Ki-67 (a proliferation marker) IHC staining (Fig. 5K) further validated the results of the tumor growth analysis. These results are consistent with in vitro experimental results and support the hypothesis that SYT7 promotes NPC growth by upregulating ALDH1A3 expression in vivo.
STAT3 signaling is involved in the SYT7-ALDH1A3-mediated NPC tumorigenesis
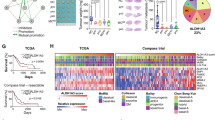
SYT7 mRNA levels from GEO datasets showed a positive correlation with ALDH1A3 expression, IL-6 and STAT3 mRNA levels, respectively (Fig. 6A). Considering the importance of IL-6/JAK/STAT3 signaling in NPC [25,26,27], we examined whether STAT3 signaling is involved in SYT7-ALDH1A3 axis-mediated regulation of NPC. We observed that both SYT7 and ALDH1A3 depletion led to significant suppressed STAT3 activity in NPC cells (Figs. 6B, C and S5). Furthermore, SYT7 overexpression led to a significant upregulation of p-STAT3, indicating enhanced STAT3 activity (Fig. 6D). Moreover, SYT7 overexpression led to an increased cell proliferation rate (Fig. 6E) and decreased apoptosis percents (Fig. 6F, G) in NPC cells, all of which could be reversed by a STAT3 activation inhibitor, S31-201. Since S31-201 did not change the protein levels of either SYT7 or ALDH1A3, we can conclude that STAT3 signaling acts as a downstream regulator in the SYT7-ALDH1A3 axis-mediated progression of NPC.
A SYT7, ALDH1A3, IL-6 and STAT3 mRNA levels in NPC were extracted from GEO datasets (GSE13597, GSE34573 and GSE61218). Correlations between SYT7 and each gene’s mRNA levels were analyzed by Spearman’s rank correlation analysis and visualized by using the ggplot2 package in R. B Immunoblot showing STAT3 and p-STAT3 in C666-1 (left) and HONE-1 (right) cells with or without SYT7 knockdown. C STAT3 and p-STAT3 were detected via western blotting in C666-1 (left) and HONE-1 (right) cells with or without ALDH1A3 knockdown. D Protein levels or E Cell proliferation of C666-1 (left) and HONE-1 (right) cells with stable SYT7 overexpression treated with a STAT3 inhibitor, S31-201 (100 μM), for 24 h were observed by immunoblot or MTT assays. F, G Flow cytometry was employed to determine the apoptotic percentages of C666-1 (left) and HONE-1 (right) cells that stably overexpressed SYT7 and were treated with S31-201. The data are expressed as the mean ± SD, from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001.
Discussion
Accumulating evidence has shown that SYT7 is overexpressed in various malignancies, including glioblastoma, gastric cancer, and lung cancer [14, 16, 28]. Previous studies also mentioned the involvement of SYT7 in head and neck squamous cell carcinoma [17] and thyroid cancer [17, 29]. However, the functional role and clinical significance of SYT7 in NPC, another independent type of head and neck cancer, remains unexplored. To the best of our knowledge, this is the first study to define the role of SYT7 in NPC.
Our current study demonstrates that SYT7 promotes NPC cell proliferation and migration while suppresses apoptosis in vitro, and facilitates the growth of NPC xenograft tumors in vivo. In addition, increased expression of SYT7 was significantly associated with unfavorable overall survival rates in NPC patients. These results indicate that SYT7 is a potential prognostic biomarker for NPC.
Though the function of SYT7 has been documented in the aforementioned malignancies, the downstream signal regulation of SYT7 remain relatively unexplored. Dong et al. reported that SYT7 is account for increased proliferation and decreased apoptosis in thyroid cancer cells, which are attributed to the inhibition of HMGB3 ubiquitination and degradation [29]. To clarify the specific mechanisms underlying SYT7’s function in NPC, a comprehensive whole-genome analysis and IP-MS were performed, both indicating ALDH1A3 is a potential downstream target of SYT7 in NPC cells. In addition, endogenous Co-IP analysis revealed that SYT7 interacts with ALDH1A3 in NPC cells. Mechanistically, SYT7 knockdown promoted the ubiquitination and degradation of ALDH1A3, an effect that was mitigated by the proteasome inhibitor MG132. Previous studies have shown that the deubiquitinating of ALDH1A3 by deubiquitinating enzymes (DUBs), such as ubiquitin-specific protease 22 (USP22) or USP9X, plays a critical role in promoting tumorigenicity and maintaining chemoresistance in various malignancies [20, 21]. The present study indicates that SYT7 functions as a DUB of ALDH1A3 in NPC, thereby stabilizing ALDH1A3 and contributing to tumor progression.
ALDH1A3, a member of the aldehyde dehydrogenase superfamily, is a functional marker of pancreas β-cell dedifferentiation and cancer stem cells [20, 30,31,32,33,34,35,36]. Its upregulation has been reported in many malignancies, including colorectal cancer, pancreatic cancer, and glioblastoma [20, 22, 33,34,35, 37, 38]. However, the role of ALDH1A3 in NPC has not been explored. Here, we demonstrated that ALDH1A3 knockdown significantly inhibited cell proliferation and migration. Furthermore, the tumor-promoting effects of SYT7 overexpression, including amplified cell growth, enhanced migration ability, and resistance to cell apoptosis, were effectively reversed by ALDH1A3 depletion. In summary, these data collectively suggest that ALDH1A3 is a downstream target of SYT7 in NPC.
ALDH1A3 is also recognized as a key enzyme involved in glucose metabolism in cancer cells [22, 23]. Thus, we examined the effects of SYT7 and ALDH1A3 on the glycolytic activity in NPC cells. Consistent with previous studies, ALDH1A3 knockdown suppressed glucose uptake and decreased lactic acid and ATP levels in NPC cells. Similarly, SYT7 knockdown inhibited glycolytic activity, whereas SYT7 overexpression had the opposite effect. Moreover, increased glycolytic activity in SYT7 overexpression cells was rescued by ALDH1A3 knockdown, indicating that ALDH1A3-mediated glycolysis is a critical downstream mechanism underlying the oncogenic effects of SYT7 in NPC.
As a cytoplasmic transcription factor, STAT3 transmit signals from the plasma membrane into nucleus [39]. STAT3 is involved in various oncogenic cellular processes, including promoting of tumor cell proliferation, angiogenesis, and suppressing of antitumor immunity [40]. STAT3 phosphorylation induces its dimerization, nuclear translocation and activation [41]. Elevated STAT3 activity is common in most malignancies [41]. Moreover, STAT3 targeting as a cancer treatment strategy has been widely investigated in preclinical and clinical trials [39,40,41,42]. Studies have also confirmed the activation of STAT3 signaling in NPC [25, 43,44,45,46]. Using STAT3 inhibitors in cancers, including NPC, have shown promising results in preclinical trials [40, 47, 48]. Our results showed that the STAT3 inhibitor S3I-201 did not change the expression levels of either SYT7 or ALDH1A3. However, S3I-201 treatment reversed the oncogenic effects of SYT7 upregulation, indicating a new interpretation of STAT3 targeting strategy in NPC.
Emerging evidence has demonstrated the critical involvement of both ALDH1A3 and STAT3 in tumorigenesis across various cancer types. Studies demonstrated that regulation of ALDH1A3 activity and expression by STAT3 pathway in breast cancer and ovarian cancer [49, 50]. Our data demonstrate that ALDH1A3 determines the phosphorylation of STAT3 in NPC. These collective findings indicate the existence of a reciprocal regulatory relationship between ALDH1A3 and STAT3 in a cancer-type dependent manner. Moreover, accumulating studies also indicated synergistic effects between glycolysis and STAT3 activation in cancer cells [51]. Thus, both of STAT3 signaling and glycolysis are considered as downstream pathways underlying SYT7-ALDH1A3 axis-mediated carcinogenesis in NPC (Fig. 7).
SYT7 stabilizes ALDH1A3 by inhibiting its ubiquitin-mediated proteolysis. ALDH1A3 enhances STAT3 phosphorylation, indicating activation of the IL-6/STAT3 signaling pathway. Additionally, ALDH1A3 promotes glycolysis in NPC. The synergistic effects of STAT3 activation and enhanced glycolysis drive NPC progression. Created in BioRender.
In summary, this study demonstrates that SYT7 functions as an oncoprotein in NPC, driving tumor progression through its regulation of ALDH1A3-mediated glycolysis and STAT3 signaling pathways. Therefore, SYT7 may serve as a potential biomarker for the prediction and treatment of NPC.
Materials and methods
Tissue microarray (TMA)
TMA of formalin-fixed NPC tissues (HNasN110Su01) were provided by Outdo Biotech Company Co., Ltd (Shanghai, China). All tissue samples in TMA were collected from 2010 to 2011. Related clinical characteristics of NPC patients in TMA were listed in Table S2. The non-cancerous nasopharyngeal tissues were collected from patients pathologically diagnosed in Zibo Central Hospital (Zibo, China). This study was conducted in accordance with the Declaration of Helsinki, and all participants signed informed consent before enrollments. All procedures in this study were approved by Zibo Central Hospital Medical Ethics Committee.
Immunohistochemical (IHC) staining
TMA and tissue sections were deparaffinized, antigen-retrieved, and blocked following standard protocols. The sections were subjected to incubation with anti-SYT7 antibody (Creative Biolabs; SRP09489) in a cold room overnight, then with secondary antibody at 37 °C for 1 h. As mentioned previously [52], TMA and tissue sections were stained with diaminobenzidine (Sigma-Aldrich; D12384), then evaluated based on the H-score under a Leica microscope.
Cell culture and stable cell lines establishment and selection
The human NPC cell lines C666-1, 5-8 F, HONE-1 and CNE-2Z, along with the normal nasopharygeal cell line NP-69, were acquired from the American Type Culture Collection (Manassas, VA, USA) and iCell Bioscience Inc (iCell Bioscience, Shanghai, China). All NPC cells were cultured and maintained in RPMI-1640 medium containing 10% fetal bovine serum (FBS).
shRNA lentiviral vectors targeting indicated sequences in shSYT7 and shALDH1A3 were listed in Supplementary Table S6, which were constructed into pLKO.1-puro vector system and confirmed by sequencing (Genechem, Shanghai, China). Vectors stably overexpressing of SYT7 were cloned into pCDH-MSCV vector system (Bioscience, Shanghai, China). Cells were infected with the aforementioned lentiviral vector particles and subjected to puromycin selection for 7 days.
MTT assay
As previously described [53], NPC cells were seeded and treated with 20 μM MTT solution (Genview, El Monte, CA, USA) for 4 h daily over 5 days. Cell viability was assessed by detecting the optical density at 490 nm. For MTT assays involving the STAT3 inhibitor, S3I-201 (Selleck Chemicals, Houston TX, USA), indicated stable cell lines of NPC were cultured with 100 μM S3I-201 for 24 h before analysis.
Cell colony formation assay
Stable shCrtl and shSYT7 C666-1 or HONE-1 cells (1 × 103 cells/well) were seeded and cultured in six-well plates for 5 days. After fixation with 4% paraformaldehyde and staining with Giemsa, the colonies were counted and photographed with a digital camera.
Cell apoptosis assay
Stable NPC cell lines were seeded in triplicate in six-well dishes (2 × 105 cells/well). Using the Annexin V Apoptosis Detection Kit APC (eBioscience, San Diego, CA, USA), cells were resuspended and treated with 5 μL Annexin V-APC. Up to 1 × 104 cells were calculated and analyzed by a FACSCalibur flow cytometer (BD, San Jose, CA, USA).
Wound-healing assay
C666-1 or HONE-1 cells were seeded in triplicate in 6-well dishes (3 × 104 cells/well) and cultured until logarithmic growth phase. Scratches were carried out using a 96-pin replicator. Cells were rinsed gently with medium 2–3 times and then cultured with 0.5% FBS RPMI-1640 for 24 h or 48 h. Finally, migration rates were measured under a fluorescence microscope.
Transwell assay
Following the manufacturer’s protocol (Corning Inc., NY, USA), the upper chamber was used to culture C666-1 or HONE-1 cells (5 × 104 cells/well) in 100 μL of serum-free RPMI-1640 medium. The lower chamber received an aliquot of 600 μL RPMI-1640 containing 30% FBS. After incubated for 24 h, the cells on the upper membrane were immobilized, treated with 400 μL of Giemsa buffer for 20 min, and photographed.
Quantitative real-time RT-PCR
Total cellular RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and the quality was determined by a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, MA, USA). Using PrimeScript RT reagent (Takara, Beijing, China), cDNA was synthesized accordingly. qPCRs were carried out based on the related primers in Supplementary Table S7 and qPCR SYBR Green Master Mix Kit (Vazyme, Nanjing, China). GAPDH was used as an internal control. The relative SYT7 and ALDH1A3 mRNA levels were calculated using the 2‑ΔΔCT method.
Microarray analysis and Ingenuity Pathway Analysis (IPA)
As described previously [53], mRNA samples from shCtrl and shSYT7 C666-1 cells were extracted and qualified. Following the manufacturer’s instructions, the whole-genome gene expression data of each sample were collected using a PrimeView Human Gene Expression Array (Thermo Fisher Scientific). Differentially expressed genes (DEGs) that showed |Fold change | ≥1.3 and a less than 0.05 of false discovery rate were classified as statistically significant. Function and pathway enrichment studies of DEGs were classified and analyzed using IPA (QIAGEN), with |Z score | ≥2 considered as statistically significant.
Celigo cell counting assay
NPC cells (1 × 103 cells/well) were seeded into RPMI-1640 medium with 10% FBS and counted daily for 5 days, as described previously [53]. Using a Celigo image cytometer (Nexcelom Bioscience, MA, USA), cell numbers were calculated and proliferation curves were generated.
Glucose content assay, lactic acid assay and ATP assay
Using a Glucose Content Assay Kit (Solarbio, Beijing, China), total glucose content levels were measured according to the protocol provided by the manufacturer. Briefly, C666-1 cells were digested with trypsin and homogenized in 200 μL of dH2O. After sonication, the samples were boiled at 100 °C for 10 min and centrifuged at 8000 rpm for 10 min, the supernatant was collected for further measurement. The optical density at 505 nm was utilized to quantify the glucose levels.
As mentioned previously [54], lactic acid content and ATP levels in C666-1 cells were measured respectively by a Lactic Acid Content Assay Kit (Solarbio) and an ATP assay kit (Solarbio) following the instructions from the manufacturer.
Western blotting
As previously described [53], proteins in the indicated stable NPC cells were harvested, lysed with RIPA buffer, separated and transferred to PVDF membranes. For protein degradation and stability assays, NPC cells were cultured in medium containing 20 μM of MG-132 (Sigma-Aldrich, St. Louis, MO, USA) for 6 h, 20 μg/mL of cycloheximide (CHX, Sigma-Aldrich) for 3 h or 30 μM of Chloroquine (CQ, Sigma-Aldrich) for 3 h, respectively, and cell lysates were collected at indicated time points. Same amount of Dimethyl sulfoxide (DMSO) was added to medium as negative control. Immunoblotting was conducted after cell harvesting. All primary and secondary antibodies employed are listed in Supplementary Table S8.
Co-immunoprecipitation (IP) assay
For co-IP assays, 1.0 mg of protein lysates were incubated with either an anti-ALDH1A3 or IgG antibody at 4 °C for 12 h. Subsequently, 20 μL of protein A sepharose beads (Millipore, CA, USA) were added into the lysates and incubated at cold room for 2 h. The resolved immunocomplexes were immunoblotted with either anti-Ubiquitin antibody or anti-SYT7 antibody.
Immunoprecipitation-mass spectrometry assays (IP-MS)
As mentioned previously, 1.0 mg protein lysates from C666-1 cells were incubated with pretreated anti-SYT7 or IgG beads at cold room for 2 h. Then, the resolved immunocomplexes were separated in 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) gel. After dyeing and washing, the gel was sent to Beijing Biotech-pack Co., LTD for further IP-MS analysis by using Easy-nLC 1200/QExactive Liquid Chromatograph (Thermo Fisher Scientific, USA).
The raw spectrometric data were under bioinformatics and statistical analysis by using MaxQuant v2.6.7.0. Interacting proteins were defined as |Fold change | ≥5. KEGG and GO pathway enrichment were performed by “clusterprofiler” package in R language about proteins interacted with SYT7.
In vivo xenograft animal model
Six-week-old female nude BALB/c mice were provided by Gem Pharmatech (Nanjing, China). The mice were maintained with the consent of the Institutional Animal Care and Use Committee of Zibo Central Hospital. Mice were randomly assigned to four groups: SYT7+shALDH1A3 group (n = 5), SYT7+shCtrl group (n = 5), Vector+shALDH1A3 group (n = 5), and Vector+shCtrl group (n = 5). stable C666-1 (4 × 106/mouse) cells from each group was prepared and resuspended in 200 μL PBS, and subcutaneously injected into dorsal flanks of nude mice. Tumor size was monitored every 7 days using calipers. Mice were euthanized after 32 days. The tumor tissues were collected, weighed and then photographed. H&E and Ki-67 staining were performed according to the previously mentioned IHC procedure. All animal experimental protocols were approved by the IACUC and Zibo Central Hospital Medical Ethics Committee (Prot. n. 202211009).
Statistical analysis
All experiments were conducted at least for three independent times, and data were quantified as the mean ± standard deviation (SD). Means of two groups were evaluated by GraphPad Prism v10.1.2 (GraphPad Software, San Diego, CA, USA) using the two-tailed Student’s t test, while comparisons among multiple groups were compared by one-way ANOVA. Difference in SYT7 protein levels between para-carcinoma tissue and NPC tissue in TMAs were compared by chi-square test. The relationship between SYT7 protein levels and patients’ characteristics were analyzed by Mann−Whitney U analysis and Spearman’s rank correlation analysis, while Kaplan−Meier survival analysis was used to generate survival curves using SPSS 26.0 (IBM Corp., Armonk, NY, USA). P < 0.05 was defined as statistically significant.
Data availability
The related data are available from the corresponding author upon request.
References
Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80.
Wong KCW, Hui EP, Lo KW, Lam WKJ, Johnson D, Li L, et al. Nasopharyngeal carcinoma: an evolving paradigm. Nat Rev Clin Oncol. 2021;18:679–95.
Zhang Y, Chen L, Hu GQ, Zhang N, Zhu XD, Yang KY, et al. Gemcitabine and cisplatin induction chemotherapy in nasopharyngeal carcinoma. New Engl J Med. 2019;381:1124–35.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Au KH, Ngan RKC, Ng AWY, Poon DMC, Ng WT, Yuen KT, et al. Treatment outcomes of nasopharyngeal carcinoma in modern era after intensity modulated radiotherapy (IMRT) in Hong Kong: a report of 3328 patients (HKNPCSG 1301 study). Oral Oncol. 2018;77:16–21.
Liu YP, Wen YH, Tang J, Wei Y, You R, Zhu XL, et al. Endoscopic surgery compared with intensity-modulated radiotherapy in resectable locally recurrent nasopharyngeal carcinoma: a multicentre, open-label, randomised, controlled, phase 3 trial. Lancet Oncol. 2021;22:381–90.
Petit C, Lee A, Ma J, Lacas B, Ng WT, Chan ATC, et al. Role of chemotherapy in patients with nasopharynx carcinoma treated with radiotherapy (MAC-NPC): an updated individual patient data network meta-analysis. Lancet Oncol. 2023;24:611–23.
Tang XR, Li YQ, Liang SB, Jiang W, Liu F, Ge WX, et al. Development and validation of a gene expression-based signature to predict distant metastasis in locoregionally advanced nasopharyngeal carcinoma: a retrospective, multicentre, cohort study. Lancet Oncol. 2018;19:382–93.
Lin DC, Meng X, Hazawa M, Nagata Y, Varela AM, Xu L, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46:866–71.
Jackman SL, Turecek J, Belinsky JE, Regehr WG. The calcium sensor synaptotagmin 7 is required for synaptic facilitation. Nature. 2016;529:88–91.
Wu D, Bacaj T, Morishita W, Goswami D, Arendt KL, Xu W, et al. Postsynaptic synaptotagmins mediate AMPA receptor exocytosis during LTP. Nature. 2017;544:316–21.
Chen C, Satterfield R, Young SM Jr., Jonas P. Triple function of synaptotagmin 7 ensures efficiency of high-frequency transmission at central GABAergic synapses. Cell Rep. 2017;21:2082–9.
Zhang X, Li N, Zhang J, Zhang Y, Yang X, Luo Y, et al. 5-IP(7) is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis. Nat Metab. 2021;3:1400–14.
Liu X, Li C, Yang Y, Liu X, Li R, Zhang M, et al. Synaptotagmin 7 in twist-related protein 1-mediated epithelial–mesenchymal transition of non-small cell lung cancer. EBioMedicine. 2019;46:42–53.
Li L, Zhao S, Liu Z, Zhang N, Pang S, Liu J, et al. Sunitinib treatment promotes metastasis of drug-resistant renal cell carcinoma via TFE3 signaling pathway. Cell Death Dis. 2021;12:220.
Kanda M, Tanaka H, Shimizu D, Miwa T, Umeda S, Tanaka C, et al. SYT7 acts as a driver of hepatic metastasis formation of gastric cancer cells. Oncogene. 2018;37:5355–66.
Fu Y, Tian G, Zhang Z, Yang X. SYT7 acts as an oncogene and a potential therapeutic target and was regulated by DeltaNp63alpha in HNSCC. Cancer Cell Int. 2021;21:696.
Poturnajova M, Kozovska Z, Matuskova M. Aldehyde dehydrogenase 1A1 and 1A3 isoforms—mechanism of activation and regulation in cancer. Cell Signal. 2021;87:110120.
Yamashita D, Minata M, Ibrahim AN, Yamaguchi S, Coviello V, Bernstock JD, et al. Identification of ALDH1A3 as a viable therapeutic target in breast cancer metastasis-initiating cells. Mol Cancer Ther. 2020;19:1134–47.
Chen Z, Wang HW, Wang S, Fan L, Feng S, Cai X, et al. USP9X deubiquitinates ALDH1A3 and maintains mesenchymal identity in glioblastoma stem cells. J Clin Investig. 2019;129:2043–55.
Yun X, Zhang K, Wang J, Pangeni RP, Yang L, Bonner M, et al. Targeting USP22 suppresses tumorigenicity and enhances cisplatin sensitivity through ALDH1A3 downregulation in cancer-initiating cells from lung adenocarcinoma. Mol Cancer Res. 2018;16:1161–71.
Nie S, Qian X, Shi M, Li H, Peng C, Ding X, et al. ALDH1A3 accelerates pancreatic cancer metastasis by promoting glucose metabolism. Front Oncol. 2020;10:915.
Huang X, Hou Y, Weng X, Pang W, Hou L, Liang Y, et al. Diethyldithiocarbamate-copper complex (CuET) inhibits colorectal cancer progression via miR-16-5p and 15b-5p/ALDH1A3/PKM2 axis-mediated aerobic glycolysis pathway. Oncogenesis. 2021;10:4.
Duan JJ, Cai J, Guo YF, Bian XW, Yu SC. ALDH1A3, a metabolic target for cancer diagnosis and therapy. Int J Cancer. 2016;139:965–75.
Ho Y, Tsao SW, Zeng M, Lui VW. STAT3 as a therapeutic target for Epstein-Barr virus (EBV): associated nasopharyngeal carcinoma. Cancer Lett. 2013;330:141–9.
Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–46.
Cheng JZ, Chen JJ, Xue K, Wang ZG, Yu D. Clinicopathologic and prognostic significance of VEGF, JAK2 and STAT3 in patients with nasopharyngeal carcinoma. Cancer Cell Int. 2018;18:110.
Xiao B, Li J, Fan Y, Ye M, Lv S, Xu B, et al. Downregulation of SYT7 inhibits glioblastoma growth by promoting cellular apoptosis. Mol Med Rep. 2017;16:9017–22.
Dong S, Pan J, Shen YB, Zhu LX, Chen L, Zhu F, et al. SYT7 plays a role in promoting thyroid cancer by mediating HMGB3 ubiquitination. Endocr Relat Cancer. 2022;29:175–89.
Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA. 2013;110:8644–9.
Son J, Du W, Esposito M, Shariati K, Ding H, Kang Y, et al. Genetic and pharmacologic inhibition of ALDH1A3 as a treatment of beta-cell failure. Nat Commun. 2023;14:558.
Wang Z, Mohan R, Chen X, Matson K, Waugh J, Mao Y, et al. microRNA-483 protects pancreatic beta-cells by targeting ALDH1A3. Endocrinology. 2021;162:bqab031.
Lang T, Xu J, Zhou L, Zhang Z, Ma X, Gu J, et al. Disruption of KDM4C-ALDH1A3 feed-forward loop inhibits stemness, tumorigenesis and chemoresistance of gastric cancer stem cells. Signal Transduct Target Ther. 2021;6:336.
Wakimoto H. Deubiquitinating ALDH1A3 key to maintaining the culprit of aggressive brain cancer. J Clin Investig. 2019;129:1833–5.
Vidovic D, Huynh TT, Konda P, Dean C, Cruickshank BM, Sultan M, et al. ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ. 2020;27:363–78.
Pan M, Li M, You C, Zhao F, Guo M, Xu H, et al. Inhibition of breast cancer growth via miR-7 suppressing ALDH1A3 activity concomitant with decreasing breast cancer stem cell subpopulation. J Cell Physiol. 2020;235:1405–16.
Gan C, Pierscianek D, El Hindy N, Ahmadipour Y, Keyvani K, Sure U, et al. The predominant expression of cancer stem cell marker ALDH1A3 in tumor infiltrative area is associated with shorter overall survival of human glioblastoma. BMC Cancer. 2020;20:672.
Duan JJ, Wang D, Cai J, Chen JJ, Zheng XX, Chen TQ, et al. An aldehyde dehydrogenase 1A3 inhibitor attenuates the metastasis of human colorectal cancer. Cancer Lett. 2022;536:215662.
Zou S, Tong Q, Liu B, Huang W, Tian Y, Fu X. Targeting STAT3 in cancer immunotherapy. Mol Cancer. 2020;19:145.
Thilakasiri PS, Dmello RS, Nero TL, Parker MW, Ernst M, Chand AL. Repurposing of drugs as STAT3 inhibitors for cancer therapy. Semin Cancer Biol. 2021;68:31–46.
Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15:234–48.
Zhao L, Hu H, Gustafsson JA, Zhou S. Nuclear receptors in cancer inflammation and immunity. Trends Immunol. 2020;41:172–85.
Li JY, Xiao T, Yi HM, Yi H, Feng J, Zhu JF, et al. S897 phosphorylation of EphA2 is indispensable for EphA2-dependent nasopharyngeal carcinoma cell invasion, metastasis and stem properties. Cancer Lett. 2019;444:162–74.
Zhang CX, Ye SB, Ni JJ, Cai TT, Liu YN, Huang DJ, et al. STING signaling remodels the tumor microenvironment by antagonizing myeloid-derived suppressor cell expansion. Cell Death Differ. 2019;26:2314–28.
Si Y, Xu J, Meng L, Wu Y, Qi J. Role of STAT3 in the pathogenesis of nasopharyngeal carcinoma and its significance in anticancer therapy. Front Oncol. 2022;12:1021179.
Lin Y, Zhou X, Yang K, Chen Y, Wang L, Luo W, et al. Protein tyrosine phosphatase receptor type D gene promotes radiosensitivity via STAT3 dephosphorylation in nasopharyngeal carcinoma. Oncogene. 2021;40:3101–17.
Pan Y, Zhou F, Zhang R, Claret FX. Stat3 inhibitor Stattic exhibits potent antitumor activity and induces chemo- and radio-sensitivity in nasopharyngeal carcinoma. PLoS ONE. 2013;8:e54565.
Luo F, Lu FT, Qiu MZ, Zhou T, Ma WJ, Luo M, et al. Gemcitabine and APG-1252, a novel small molecule inhibitor of BCL-2/BCL-XL, display a synergistic antitumor effect in nasopharyngeal carcinoma through the JAK-2/STAT3/MCL-1 signaling pathway. Cell Death Dis. 2021;12:772.
Arnold KM, Opdenaker LM, Flynn NJ, Appeah DK, Sims-Mourtada J. Radiation induces an inflammatory response that results in STAT3-dependent changes in cellular plasticity and radioresistance of breast cancer stem-like cells. Int J Radiat Biol. 2020;96:434–47.
Lu T, Bankhead A 3rd, Ljungman M, Neamati N. Multi-omics profiling reveals key signaling pathways in ovarian cancer controlled by STAT3. Theranostics. 2019;9:5478–96.
Li YJ, Zhang C, Martincuks A, Herrmann A, Yu H. STAT proteins in cancer: orchestration of metabolism. Nat Rev Cancer. 2023;23:115–34.
Wang Z, Li B, Li S, Lin W, Wang Z, Wang S, et al. Metabolic control of CD47 expression through LAT2-mediated amino acid uptake promotes tumor immune evasion. Nat Commun. 2022;13:6308.
Xiang XX, Liu YL, Kang YF, Lu X, Xu K. MEX3A promotes nasopharyngeal carcinoma progression via the miR-3163/SCIN axis by regulating NF-kappaB signaling pathway. Cell Death Dis. 2022;13:420.
Peng M, Yang D, Hou Y, Liu S, Zhao M, Qin Y, et al. Intracellular citrate accumulation by oxidized ATM-mediated metabolism reprogramming via PFKP and CS enhances hypoxic breast cancer cell invasion and metastasis. Cell Death Dis. 2019;10:228.
Acknowledgements
We thank Editage (www.ediatge.cn) for language editing.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81600695 and 81702687), China Postdoctoral Science Foundation (2023M741859), Natural Science Foundation of Shandong Province (ZR2024MH344 and ZR2020MC062) and Postdoctoral Innovation Project of Shandong Province (SDCXZG-202400030).
Author information
Authors and Affiliations
Contributions
Kai X: writing and funding acquisition. YK, JW and YH: project administration. WZ, WT and CL: validation, conceptualization, and formal analysis. YL: funding acquisition and validation. XX: supervision, writing, and funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, K., Kang, Y., Wang, J. et al. SYT7 accelerates nasopharyngeal carcinoma progression via ALDH1A3-mediated STAT3 signaling activation. Oncogenesis 14, 16 (2025). https://doi.org/10.1038/s41389-025-00558-1
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41389-025-00558-1