Abstract
Squamous cell carcinoma (SCC) poses a significant global health challenge due to the lack of effective treatments. Boron neutron capture therapy (BNCT), a targeted particle therapy, has shown promising results in various cancers. SLC7A5, a transporter of essential amino acids and boronophenylalanine (BPA) used in BNCT, emerges as a potential therapeutic target. However, its expression across different histological subtypes and the role of SLC7A5 inhibition in developing drug resistance to BPA-BNCT remain poorly understood. Our study reveals elevated SLC7A5 expression in most SCCs, particularly in lung squamous cell carcinoma (LUSC), where it is significantly higher compared to other lung cancer subtypes. Increased SLC7A5 expression and a higher tumor-to-normal (T/N) ratio in LUSC are associated with poor patient prognosis. SLC7A5 knockdown in LUSC cells reduces colony formation and induces apoptosis. RNA-seq analysis of SLC7A5 knockout LUSC cells shows downregulated mTORC1 signaling, reduced expression of other amino acid transporters, and upregulated autophagy genes, indicating a potential cancer metabolic shift. Furthermore, SLC7A5 knockout LUSC cells demonstrate resistance to BPA-BNCT but sensitivity to the autophagy inhibitor chloroquine. Post-BPA-BNCT treatment, surviving wild-type LUSC cells exhibit reduced SLC7A5 levels and increased sensitivity to chloroquine, highlighting a vulnerability in BPA-BNCT-resistant cells. Our findings elucidate the interplay between SLC7A5, mTOR signaling, and autophagy pathways, providing insights into potential strategies to overcome drug resistance in BPA-BNCT therapy.

Similar content being viewed by others
Introduction
Solute carrier family 7 member 5 (SLC7A5), known as LAT1 (L-type amino acid transporter), is the catalytic subunit responsible for amino acid transport [1]. It functions with SLC3A2, also called CD98hc or 4F2hc, which acts as the heavy chain subunit necessary for the complex’s proper functioning and membrane localization. SLC7A5 predominantly transports large neutral amino acids, including phenylalanine, tyrosine, tryptophan, and leucine [2]. Through its transport function, SLC7A5 ensures amino acids’ intracellular availability, facilitating crucial cellular processes such as protein synthesis, energy production, and maintenance of cellular homeostasis [3]. Notably, SLC7A5 is frequently overexpressed in various cancers, suggesting its potential as a promising therapeutic target in cancer treatment [4]. Despite the development of several inhibitors targeting SLC7A5 for cancer treatment, their efficacy remains a subject of ongoing validation [5,6,7,8].
Boron neutron capture therapy (BNCT) is a radiation treatment modality that exploits cancer cells’ preferential uptake of boron-10 conjugated drugs over normal tissues. After uptake, nonradioactive boron-10 atoms accumulate within tumor cells and disintegrate upon exposure to epithermal neutrons, releasing energy over a short range, typically less than 10 μm [9]. Boronophenylalanine (BPA), a phenylalanine analog primarily transported into cells by the SLC7A5 transporter, is the most commonly used drug in BNCT [10]. Given that tumors exhibit higher expression of SLC7A5 compared to non-tumorous tissues, BPA accumulates preferentially in cancer cells, where the boron neutron capture reaction occurs [11, 12]. Despite the efficacy of BPA-BNCT in targeting cancer cells, the mechanisms underlying cancer cell resistance to this therapy remain poorly understood.
mTOR (mechanistic target of rapamycin), a critical regulator of cell growth, proliferation, and survival, responds to nutritional status, growth factors, and stress signals [13]. The small GTPase RHEB is crucial in activating mTORC1, a complex pivotal in cellular processes [14]. Furthermore, mTORC1 has emerged as a critical player in regulating autophagy, a cellular catabolic process essential for responding to environmental and physiological stresses [15]. Autophagy involves the sequestration of portions of cytoplasm within double-membrane vesicles called autophagosomes, followed by cargo degradation and resulting amino acid recycling into the cytosol [16]. Rapamycin, an mTOR inhibitor, binds to FKBP12 to form a complex that inhibits mTORC1, reducing protein synthesis and cell growth while enhancing autophagy [17, 18]. It has well-known anticancer and immunosuppressive properties [19]. Additionally, rapamycin has been found to influence the radiosensitivity of cultured tumor cells in BPA-BNCT therapy [20]. Although autophagy is one of the earliest cellular responses to radiation [21], the interactions between mTORC1, autophagy pathways, and their modulation by SLC7A5 deficiency or BPA-BNCT treatment in cancers remain unclear.
This study investigated the expression of SLC7A5 in different cancers, revealing high expression levels in lung squamous cell carcinoma (LUSC). Subsequently, SLC7A5 knockout LUSC cells were generated to explore the impact of SLC7A5 loss on mTORC1 and autophagy signaling and its association with resistance to BPA-BNCT treatment.
Materials and methods
Chemicals and reagents
RPMI-1640 (Gibco™, 31800-014, USA) and fetal bovine serum (FBS, Gibco™, 10437-028, USA) were procured from GIBCO-BRL. Chloroquine was obtained from Cell Signaling Technology Inc. (CST, 14774, USA), while rapamycin was purchased from Selleck Chemicals LLC (Selleckchem, S1039, USA). BPA-fructose was synthesized by Taiwan Biotech Inc. and provided by the Nuclear Science and Technology Development Center at National Tsing Hua University.
Cell culture
LUSC cell lines H2170 (ATCC, CRL-5928), H520 (ATCC, HTB-182), and the lung carcinoma cell line A549 (ATCC, CCL-185) were obtained from the American Type Culture Collection (https://www.atcc.org/) [22]. H2170-RAPA cells were derived from H2170 cells treated with the mTORC1 inhibitor rapamycin (20 μM) for 1 month, and surviving cells were pooled. All cells were cultured in RPMI-1640 medium supplemented with L-glutamine (4 mM), sodium pyruvate (1 mM), HEPES (10 mM), sodium bicarbonate (23.8 mM), penicillin/streptomycin (1%), and FBS (10%). The cells were maintained at 37 °C with 5% CO2 in a humidified atmosphere. Short tandem repeat (STR) profiling was used to authenticate all cell lines within the last three years.
Plasmids
The shRNA clone of pLKO.1-shSLC7A5 (TRC N0000043009) was obtained from the National RNAi Core Facility, Academia Sinica (Taiwan). The pLKO.1-Scrambled shRNA (#136035) was sourced from Addgene (USA).
Reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cultured cells using the TRIzol reagent (Invitrogen™, 15596026, USA) following the manufacturer’s protocol. Subsequently, cDNA synthesis was performed using 1 µg of total RNA and the PrimeScript™ RT Reagent Kit (Takara Bio USA, RR037B, USA) according to the manufacturer’s instructions. For quantitative analysis, 1000 ng of cDNA was mixed with Taqman or UPL probes and corresponding primers in each reaction. The reactions were performed in triplicate using the StepOnePlus system from Applied Biosystems. The expression levels of the genes were normalized to the housekeeping gene (18S rRNA). Relative gene expression was analyzed using the 2-ΔΔCT method. The primer and probe sequences are provided in Table S1.
Immunoblotting
Cells were collected using a lysis buffer containing a cOmplete™, EDTA-free Protease Inhibitor Cocktail (Roche, 04693132001, Switzerland). Immunoblotting was conducted using the following primary antibodies: anti-SLC7A5 (Cell Signaling, CST#5347, USA), anti-SLC3A2 (Bioss, bs-6659R, USA), anti-SQSTM1 (GeneTex Inc., GTX100685, USA), anti-LC3B (GeneTex Inc., GTX127375, USA), anti-alpha Tubulin (GeneTex Inc., GTX112141, USA), and anti-GAPDH (GeneTex Inc., GTX100118, USA). Horseradish peroxidase (HRP)-conjugated secondary antibodies were employed, and then protein bands were detected using chemiluminescence and imaging with an Image Quant LAS 4000 mini chemiluminator.
Clonogenic assay
Cells were seeded in 6-well plates at a density of 500 to 2000 cells per well, with three independent replicates for each condition. The plates were then incubated for 2 weeks at 37 °C with 5% CO2 to allow colony formation. Following incubation, the colonies were fixed and stained using a mixture of 3.7% formaldehyde, 80% methanol, and 0.25% crystal violet (Fisher Chemical, C58125, USA) for 20 min at room temperature. The area of the stained colonies was subsequently quantified using Image J software.
Cell-cycle analysis
Cells were seeded at a density of 3 × 10⁵ cells in 60 mm dishes, with three independent replicates, and incubated overnight. Subsequently, the cells were fixed by incubating them overnight with 70% ethanol at −20 °C. Following fixation, the cells were stained with 4’,6-diamidino-2-phenylindole (DAPI) for 30 min at room temperature. The stained cells were then analyzed using flow cytometry according to the manufacturer’s instructions.
Apoptosis analysis
Cells were plated at a density of 1×105 cells in a 60-mm dish, with three independent replicates. After 48 h of incubation, the cells were harvested and fixed in 500 μL of Annexin V binding buffer, supplemented with 1 μL of Annexin V-FITC (Elabscience®, E-CK-A111, USA) and 5 μL of 7-AAD-PC5.5 (Elabscience®, CK-A162, USA). Following a 20-min incubation at room temperature, flow cytometry analysis was performed on 10,000 cells, following the manufacturer’s instructions.
Generation of SLC7A5 knockout and knockdown cell lines
The CRISPR/Cas9 system was used to generate SLC7A5 knockout H2170 cells. A guide RNA (gRNA) targeting the first exon of SLC7A5 (sequence: GGCCGGTGCGCAGAGCATGGCGG) was designed by Dharmacon Edit-R™ CRISPR-Cas9 Gene Engineering System (https://horizondiscovery.com/en). Before Cas9/gRNA delivery, H2170 cells were synchronized with 200 ng/mL nocodazole (Cayman Chemical Inc., 13857, USA) for 17 h to enhance transfection efficiency. On the day of electroporation, crRNA and tracrRNA (150 pmol each) were mixed to form the gRNA complex. Cas9 protein (30 pmol) and gRNA (150 pmol) were then combined in Opti-MEM™ I Reduced Serum Medium at room temperature for 15 min to assemble Cas9/gRNA ribonucleoprotein (RNP) complexes. Synchronized cells (1×105) were collected, washed, and resuspended in Opti-MEM (Gibco, 31985070, USA), followed by adding the RNP complexes. Electroporation was performed using the Super Electroporator NEPA21 Type II (Nepa Gene Inc., Japan) under optimized voltage conditions. Post-electroporation, cells were transferred to 24-well plates with culture medium and incubated at 37 °C with 5% CO2 for 48 h. High-resolution melting quantitative PCR (HRM-qPCR) was employed alongside the T7 Endonuclease I (T7E1) assay to assess the efficiency of CRISPR/Cas9 gene editing. The T7E1 assay detects mismatches caused by indels at the target site, visualized via gel electrophoresis. HRM-qPCR distinguishes wild-type and indel-containing DNA, enabling screening for SLC7A5 knockout cells. Gene editing was confirmed, and single-cell clones were isolated, cultured, and analyzed by HRM-qPCR and sequencing. Protein expression was assessed by immunoblotting, and positive clones were selected for further experiments. We assessed the impact of SLC7A5 knockdown on cell growth and apoptosis in pooled H2170 cells transduced with lentiviral particles encoding SLC7A5-targeting shRNA (shSLC7A5). Knockdown of SLC7A5 in pooled populations induced apoptosis, thereby limiting the feasibility of downstream analyses. To overcome this, stable SLC7A5 knockdown H2170 cells were generated through clonal selection following lentiviral transduction. A clone with confirmed SLC7A5 downregulation was selected for further experiments.
Immunohistochemistry
The immunohistochemical analysis of SLC7A5 expression in lung cancer samples was conducted using a lung cancer tissue array slide (Superbiochips, CC5, Korea). After deparaffinization, endogenous peroxidase activity was blocked by incubating the slide in methanol with 3% hydrogen peroxide (H₂O₂). Antigen retrieval was performed using sodium citrate buffer and autoclave heating. To prevent non-specific binding, the slide was incubated with 2.5% FBS. Primary antibodies against SLC7A5/LAT (Abcam, ab208776, UK) were then applied, followed by incubation with secondary antibodies. The antigen-antibody complexes were visualized using 3,3’-diaminobenzidine (DAB), and the tissue sections were counterstained with hematoxylin. Immunohistochemical staining intensities and percentages were evaluated by Dr. Shien-Tung Pan at the Department of Pathology, China Medical University Hospital. The intensity of staining was scored on a scale from 0 (no staining) to 3 (strong staining), while the percentage of stained cells was scored as 0 (<10%), 1 (11-50%), 2 (51-80%), and 4 (>80%). The tissue staining patterns were quantified using the Immunoreactive Score (IRS) assessment.
Confocal immunofluorescence microscopy
1 × 105 cells/well were seeded in 8-well chambered coverslips (Ibidi Inc., 80806, Germany) and incubated at 37 °C in a 5% CO2 incubator for 24 h. After incubation, the cells were treated with chloroquine (30 μM) for 24 h. Subsequently, the cells were stained with an anti-LC3B polyclonal antibody (1/100 dilution; GeneTex Inc., GTX127375, USA), followed by incubation with Goat anti-Rabbit IgG (H + L) secondary antibodies, DyLight™ 488 (1/200 dilution; Thermo Scientific™, 35552, USA). The nucleus was stained with Hoechst 33342 (Thermo Scientific™, 62249, USA). The slides were then analyzed under a confocal laser-scanning microscope (FluoView FV10i; Olympus, Japan), and the images were processed using FV31s-SW viewer (Version 2.4.1.198; Olympus, Japan). The number of autophagosome puncta was quantified using ImageJ software.
Boron neutron capture therapy (BNCT)
Cells were seeded at a density of 2.0 × 105 cells per well, with three independent replicates, in a 6-well polystyrene plate. After incubation for one day, cells were treated with or without a BPA-fructose solution (25 µg/mL) for 4 h, followed by three washes with cold PBS. Subsequently, the cells were irradiated with an epithermal neutron flux of 1.28 × 109 nth/cm2s at an ambient temperature of 25 ± 3 °C. This irradiation was conducted at the Tsing Hua Open Pool Reactor at the Nuclear Science and Technology Development Center, National Tsing Hua University, Taiwan.
Public domain data and statistical analysis
The public gene expression profiling and RNA-seq datasets used in this study were analyzed as described [23]. The sources of these datasets are listed in Table S2. Statistical analyses were conducted using the Student’s t-test for comparing two samples or one-way analysis of variance (ANOVA) for comparing multiple samples. The overall survival of lung cancer patients was analyzed using the log-rank test. A linear regression test examined the correlation between genes in the scatter plot results. A p-value of less than 0.05 was considered statistically significant. For cell experiments, statistical analyses were carried out using Prism software (GraphPad Inc., USA). Statistical significance was defined as a p-value less than 0.05 (p < 0.05).
Ethics statement
The CC5 lung cancer tissue array used in this study was obtained from SuperBiochips and acquired in accordance with the supplier’s ethical sourcing standards. This commercially available array contains no identifiable patient information and does not require additional consent for research use. All experimental procedures, including the acquisition and analysis of publicly available datasets and commercial biospecimens, were conducted in accordance with relevant institutional, national, and international ethical guidelines and regulations.
Results
Elevated SLC7A5 expression in lung squamous cell carcinoma
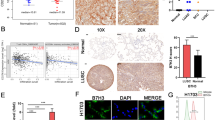
Using RNA-seq analysis from the TCGA database, we investigated SLC7A5 expression across various cancers. Our results revealed significantly higher expression levels in squamous cell type cancers, such as lung squamous cell carcinoma (LUSC), head-and-neck squamous cell carcinoma, esophageal carcinoma, and cervical squamous cell carcinoma, compared to other tumor types (Fig. 1A). Additionally, RNA-seq analysis demonstrated increased SLC7A5 expression in most LUSC or lung adenocarcinoma (LUAD) tumors compared to normal tissues (Fig. 1B and Fig. S1A, S1B). Given the histologic diversity of lung cancer, we utilized gene expression profiling analysis and RNA-seq assays from Gene Expression Omnibus Series (GSE) and Cancer Cell Line Encyclopedia (CCLE) datasets to assess SLC7A5 expression in different histologic subtypes. Our findings revealed that LUSC exhibited the highest SLC7A5 expression among all subtypes of lung cancer (Fig. 1C and Fig. S1C). To compare the expression of SLC7A5 protein in LUSC and LUAD, immunohistochemical staining for SLC7A5 expression was performed in lung tumors from a tissue array. The result showed that SLC7A5 expression was significantly higher in LUSC tumors than in LUAD tumors, indicating a specific association between SLC7A5 and LUSC (Fig. 1D). A high tumor-to-normal (T/N) ratio of targeted gene expression is required to develop targeted therapy. The T/N ratio of SLC7A5 expression in LUSC and LUAD tumors was examined. In paired tumorous versus non-tumorous tissues, tumors harbored higher SLC7A5 expression than the corresponding non-tumorous tissues from LUSC and LUAD (Fig. 1E and Fig. S1D). Additionally, the T/N ratio of SLC7A5 was more elevated in lung squamous cell carcinoma than in lung adenocarcinoma (Fig. 1F). The data demonstrate that LUSC displays high SLC7A5 expression and contains a high T/N ratio for SLC7A5, which could serve as a therapeutic target in LUSC. Furthermore, we investigated the prognostic significance of SLC7A5 in non-small cell lung cancer (NSCLC), correlating SLC7A5 expression with overall survival in patients with either LUSC or LUAD. Kaplan–Meier analysis demonstrated that high SLC7A5 expression was associated with poor overall survival outcomes in LUSC and LUAD patients (Fig. 1G and Fig. S1E). These findings suggest the potential of SLC7A5 as a prognostic factor in NSCLC.
A RNA-seq analysis of SLC7A5 expression in the specified cancers from the TCGA database. HNSCC head and neck squamous cell carcinoma, ESCA esophageal carcinoma, LUSC lung squamous cell carcinoma, CESC cervical squamous cell carcinoma, COAD colorectal adenocarcinoma, READ rectum adenocarcinoma, BUC bladder urothelial carcinoma, LUAD lung adenocarcinoma, UCS uterine carcinosarcoma, PAAD pancreatic adenocarcinoma, OV ovarian serous cystadenocarcinoma, KIRC kidney renal clear cell carcinoma, HCC liver hepatocellular carcinoma, PRAD prostate adenocarcinoma, THCA thyroid carcinoma. B Percentage of high SLC7A5 expression in tumors versus normal tissues in LUSC from the cBioPortal database. C Gene expression profiling analysis showing SLC7A5 expression in different histologic subtypes of lung cancer tumors from the GSE32036 database. LUSC Lung squamous cell carcinoma, LUAD lung adenocarcinoma, SCLC small cell lung cancer, LCC large cell carcinoma. *p < 0.05; ***, p < 0.001. D Representative images (upper) and quantitative analysis (lower) of SLC7A5 expression by immunohistochemistry assay of LUSC and LUAD tissues from the lung cancer tissue array (Superbiochips, CC5). ***p < 0.001. E RNA-seq analysis revealing SLC7A5 expression in paired LUSC primary tumor and solid normal tissue samples from the TCGA-LUSC database. ***p < 0.001. F Tumor-to-normal (T/N) ratio of SLC7A5 expression in LUSC and LUAD from the TCGA database. **p < 0.01. G Kaplan–Meier analysis demonstrating the correlation of SLC7A5 expression with the overall survival of LUSC from the TCGA-LUSC database. *p < 0.05.
Knockdown of SLC7A5 induces cell death in lung squamous cell carcinoma cells
To investigate the role of SLC7A5 in LUSC cells, SLC7A5 was knocked down by specific shRNA against SLC7A5 in H2170 LUSC cells (Fig. 2A). Clonogenic assays showed that SLC7A5-silenced cells displayed lower colony-forming ability than control H2170 cells (Fig. 2B). SLC7A5 was also knocked down in LUSC H520 and lung carcinoma A549 cells (Fig. S2A). Similarly, the knockdown of SLC7A5 decreased colony-forming ability in H520 and A549 cells (Fig. S2B). SLC7A5 interacts with SLC3A2 (also known as 4F2hc or CD98) to form a heteromeric amino acid transporter complex, which promotes the stability and translocation of SLC7A5 to the plasma membrane. RT-qPCR and immunoblotting were used to investigate whether silencing SLC7A5 affects SLC3A2 expression. The results revealed that SLC7A5 knockdown reduced SLC3A2 expression in H2170 cells (Fig. 2C). To examine the impact of SLC7A5 expression on the survival of LUSC cells, SLC7A5-silenced H2170 cells were subjected to an annexin V apoptosis assay. The results showed that the knockdown of SLC7A5 increased both early and late-stage apoptosis ratios in H2170 cells (Fig. 2D). To assess the influence of SLC7A5 deprivation on the proliferation of LUSC cells, SLC7A5-silenced H2170 cells were subjected to cell-cycle analysis. The result showed no significant effect comparing the S phase ratio between H2170 WT and SLC7A5-silenced cells (Fig. 2E). These findings suggest that SLC7A5 expression regulates cellular survival in lung cancer cells.
A RT-qPCR (upper) and immunoblotting (lower) analyses of SLC7A5 expression in H2170 cells transduced with lentiviral particles encoding shRNA against SLC7A5 (shSLC7A5) or scrambled control (SC). ***p < 0.001. B Clonogenic assay assessing colony growth of H2170 cells transduced with shSLC7A5 or scrambled control (SC). Colonies were stained with crystal violet (bottom) and quantified by ImageJ software (top). *p < 0.05. C RT-qPCR (upper) and immunoblotting (lower) analyses of SLC3A2 expression in H2170 cells transduced with shSLC7A5 or scrambled control (SC). ***p < 0.001. D Annexin V flow cytometric analysis assessing the apoptotic rate of H2170 cells transduced with shSLC7A5 or scrambled control (SC). **p < 0.01. E Flow cytometric analysis of cell cycle distribution in H2170 cells transduced with shSLC7A5 or scrambled control (SC). ns, p > 0.05.
Decreased amino acid transporter expression in SLC7A5-deficient lung squamous cell carcinoma cells
To investigate the effect of SLC7A5 loss of function on LUSC cells, SLC7A5 was knocked out using the CRISPR-Cas9 system in H2170 cells, followed by a selection of single-cell clones harboring SLC7A5 mutations or deletions (Fig. S3A). Immunoblotting assays confirmed the loss of SLC7A5 expression in the SLC7A5 knockout (KO) clones (Fig. 3A). Phase-contrast imaging analysis showed that SLC7A5 knockout and wild-type H2170 cells shared similar cellular morphology (Fig. S3B). Clonogenic assays displayed no significant difference in colony-forming ability between SLC7A5 knockout and wild-type H2170 cells (Fig. 3B). Annexin V apoptosis assays showed no significant difference in apoptotic rates between SLC7A5 knockout and wild-type H2170 cells (Fig. 3C and Fig. S3C). Moreover, cell-cycle analysis showed no difference in S-phase cell-cycle entry between SLC7A5 knockout and wild-type H2170 cells (Fig. 3D). To identify pathways affected by SLC7A5 knockout in LUSC cells, RNA-seq analysis was applied to SLC7A5 knockout versus wild-type H2170 cells. The results revealed that the knockout of SLC7A5 in H2170 cells decreased the expression of amino acid transporter genes SLC3A2, SLC1A5, SLC1A4, and SLC38A5 (Fig. 3E). RT-qPCR confirmed that SLC7A5 knockout cells harbored less SLC3A2, SLC1A5, SLC1A4, and SLC38A5 expression than wild-type H2170 cells (Fig. 3F). Correlation analysis showed positive associations of SLC7A5 with SLC3A2, SLC1A5, and SLC1A4 expression in LUSC tumors (Fig. 3G). These data suggest that the expression of a group of amino acid transporters related to SLC7A5 is interconnected, and LUSC cells lacking SLC7A5 exhibit reduced expression of these amino acid transporters.
A Immunoblotting analysis of SLC7A5 expression in wild-type H2170 (WT) and its SLC7A5 knockout cells (KO1 and KO2), which harbor differential deletion/mutation regions in the SLC7A5 coding region. B Clonogenic assay assessing colony growth of H2170 and its SLC7A5 knockout cells (KO1 and KO2). Colonies were stained with crystal violet (bottom) and quantified by ImageJ software (top). ns, p > 0.05. C Annexin V flow cytometric analysis assessing the apoptotic rate in H2170 and its SLC7A5 knockout cells (KO1 and KO2). ns, p > 0.05. D Flow cytometric analysis of the cell-cycle distribution in H2170 and its SLC7A5 knockout cells (KO1 and KO2). ns, p > 0.05. E RNA-seq analysis assessing the indicated amino acid transporter gene (SLC3A2, SLC1A5, SLC1A4, and SLC38A5) expression in H2170 and its SLC7A5 knockout cells (KO1 and KO2). F RT-qPCR analysis assessing amino acid transporter gene (SLC3A2, SLC1A5, SLC1A4, and SLC38A5) expression in H2170 and its SLC7A5 knockout cells (KO1 and KO2). **p < 0.01; ***p < 0.001. G Scatter plot analysis assessing the correlation between indicated amino acid transporter genes SLC7A5, SLC3A2, SLC1A5, and SLC1A4 expression in lung squamous cell carcinoma from the TCGA database. ***p < 0.001.
Crosstalk of SLC7A5 expression with mTORC1 signaling in lung squamous cell carcinoma
As SLC7A5 knockout H2170 cells display low amino acid transporter gene expression, RNA-seq data from SLC7A5 knockout versus wild-type 2170 cells were subjected to gene set enrichment analysis (GSEA) to identify potential genes and pathways affected by low amino acid transporter expression in SLC7A5 knockout cells. GSEA showed that the mTORC1 signaling gene set was downregulated in SLC7A5 knockout compared with wild-type H2170 cells (Fig. 4A). mTORC1 active effector genes, such as AKT3, RHEB, mTOR, and RPTOR, were expressed lower in SLC7A5 knockout cells than in wild-type H2170 cells (Fig. 4B). On the other hand, mTORC1 suppressive effector genes, such as TSC1 and TSC2, were more highly expressed in SLC7A5 knockout H2170 cells (Fig. 4B). RT-qPCR analysis confirmed that AKT3 and RHEB were expressed lower in SLC7A5-deficient cells than in wild-type H2170 cells (Fig. 4C). Scatter plot analysis revealed positive associations of SLC7A5 expression with RHEB transcript and phospho-S6 protein levels in LUSC tumors (Fig. 4D, E). These data suggest that SLC7A5 expression may be coupled with mTORC1 signaling to regulate cellular anabolism in LUSC cells. To test this hypothesis, wild-type H2170 LUSC cells were treated with the mTORC1 inhibitor rapamycin (20 μM) for one month, and surviving cells were pooled and named H2170-RAPA. H2170-RAPA cells displayed a higher tolerance ability than parental H2170 cells (Fig. S4A). Given that mTORC1 signaling suppresses autophagy, H2170-RAPA underwent RT-qPCR analysis to assess autophagy marker genes LC3B and SQSTM1 expression levels. The results showed that LC3B and SQSTM1 were expressed more highly in H2170-RAPA than in parental H2170 cells (Fig. 4F, upper). To compare the autophagy flux between H2170-RAPA and parental H2170 cells, both cell types were treated with chloroquine to block autophagy flux. Subsequently, they were subjected to immunoblotting for LC3B-I, primarily located in the cytoplasm, and LC3B-II, which is membrane-bound and is formed from LC3-I during autophagosome formation and elongation. The results revealed that under chloroquine treatment, H2170-RAPA cells displayed higher LC3B-II expression than parental H2170 cells, supporting that blocking mTORC1 signaling activates autophagy in LUSC cells (Fig. 4F, lower). RT-qPCR and immunoblotting analysis showed that SLC7A5 was expressed lower level in H2170-RAPA compared to parental H2170 cells, indicating that blocking mTORC1 signaling inhibits SLC7A5 expression in LUSC cells (Fig. 4G). These data suggest the presence of crosstalk between SLC7A5 expression and mTORC1 signaling. These findings indicate that SLC7A5 interacts with mTORC1 signaling to regulate cellular anabolism in LUSC cells.
A Gene set enrichment analysis (GSEA) of RNA-seq data assessing mTORC1 signaling-associated gene expression in wild-type H2170 (WT) versus its SLC7A5 knockout cells (SLC7A5-KO). B Gene set of the mTORC1 signaling pathway in wild-type H2170 (WT) versus its SLC7A5 knockout cells (KO) from RNA-seq data. C RT-qPCR analysis assessing mTOR pathway-related gene (AKT3 and RHEB) expression in wild-type H2170 and SLC7A5 knockout cells (KO1 and KO2). ***p < 0.001. D Scatter plot analysis assessing the correlation between SLC7A5 and RHEB expression in LUSC tumors from the TCGA-LUSC database. **p < 0.01. E Scatter plot analysis assessing the correlation of SLC7A5 with phospho-S6 (pS240S244 and pS235S236) protein expression in LUSC tumors from the TCGA-LUSC database. *p < 0.05. F RT-qPCR analysis (upper) assessing LC3B and SQSTM1 expression in H2170 in the presence or absence of rapamycin (RAPA, 20 μM) for one month. Immunoblotting analysis (lower) assessing LC3B-I and LC3B-II expression in wild-type (WT) H2170 and H2170-RAPA cells treated with or without chloroquine (CQ). **p < 0.01. G RT-qPCR (upper) and immunoblotting (lower) analysis assessing SLC7A5 expression in H2170 and H2170-RAPA cells. ***p < 0.001.
SLC7A5 expression regulates autophagy in lung squamous cell carcinoma cells
As SLC7A5 knockout LUSC cells display low mTORC1 signaling, which regulates autophagy, we examined whether SLC7A5 expression regulates autophagy. Autophagy marker gene expression was examined in SLC7A5 knockout LUSC cells using RNA-seq analysis, revealing that SLC7A5 knockout cells exhibited higher expression levels of SQSTM and LC3B compared to wild-type H2170 cells (Table S3). This result was further confirmed by RT-qPCR assays (Fig. 5A). LC3B-II-mediated autophagy puncta formation was monitored by immunofluorescence staining with antibodies against LC3B in SLC7A5 knockout and wild-type H2170 cells treated with chloroquine to block autophagy flux. Immunofluorescence staining assays showed that under chloroquine treatment, more LC3B-II-mediated puncta accumulated in SLC7A5 knockout than in wild-type H2170 cells (Fig. 5B). Immunoblotting analysis confirmed that under chloroquine treatment, more LC3B-II and SQSTM1 expression was present in SLC7A5 knockout than in wild-type H2170 cells (Fig. 5C). To validate these observations, we analyzed stable SLC7A5 knockdown H2170 cells. RT-qPCR confirmed reduced SLC7A5 expression in these cells (Fig. S5A). Similar to the knockout model, stable SLC7A5 knockdown H2170 cells displayed increased LC3B-II puncta formation and protein levels following chloroquine treatment compared to control H2170 cells (Fig. S5B, S5C). Collectively, these results suggest that SLC7A5 expression regulates autophagy in LUSC cells.
A RT-qPCR analysis assessing autophagy pathway-related gene (LC3B and SQSTM1) expression in H2170 and SLC7A5 knockout cells (KO1 and KO2). *p < 0.05; **p < 0.01. B Representative images (left) of confocal analysis for assessing LC3B-II mediated fluorescent puncta formation in H2170 and its SLC7A5 knockout cells (KO1 and KO2) treated with or without chloroquine (CQ). LC3B-II mediated fluorescent puncta levels (right) in chloroquine-treated groups were normalized to the corresponding untreated group. C Immunoblotting analysis of LC3B expression (upper) and SQSTM1 expression (lower) in H2170 and its SLC7A5 knockout cells (KO1 and KO2) treated with or without chloroquine (CQ).
SLC7A5 expression affects the effects of BNCT and autophagy inhibitors in lung squamous cell carcinoma
Considering that SLC7A5 knockout LUSC cells demonstrate elevated autophagy activity, we explored whether SLC7A5 expression influences the efficacy of the autophagy inhibitor chloroquine. Clonogenic assays unveiled an increased number of chloroquine-resistant colonies in wild-type H2170 cells compared to SLC7A5 knockout H2170 cells, suggesting that SLC7A5-deficient LUSC cells depend more on the autophagy catabolic pathway than their wild-type counterparts (Fig. 6A). Since BPA is an analog of phenylalanine primarily transported into cells by the SLC7A5 coded amino acid transporter, we tested whether SLC7A5 deficiency affects the efficacy of BPA-BNCT in LUSC cells. Clonogenic assays showed that the treatment of SLC7A5 wild-type H2170 cells with BPA rendered the cells sensitive to BNCT, whereas SLC7A5 knockout H2170 cells were resistant to BPA-BNCT (Fig. 6B). Similarly, stable SLC7A5 knockdown H2170 cells showed greater resistance to BPA-BNCT compared to control cells (Fig. S5D). To study BPA-BNCT resistance, surviving cells from BPA-BNCT-treated wild-type H2170 cells were pooled, propagated, and named H2170-BNCT. RT-qPCR and immunoblotting assays revealed that H2170-BNCT cells displayed lower SLC7A5 expression than wild-type H2170 cells (Fig. 6C). Moreover, RT-qPCR and immunoblotting assays showed that H2170-BNCT cells displayed higher LC3B expression and elevated autophagy flux than wild-type H2170 cells (Fig. 6D). Long-term rapamycin treatment induced autophagy flux in H2170 cells, making them more susceptible to the autophagy inhibitor chloroquine than untreated H2170 cells (Fig. S4B). Clonogenic assays further demonstrated heightened sensitivity of H2170-BNCT cells to chloroquine compared to parental H2170 cells (Fig. 6E). Moreover, cotreatment with chloroquine enhanced the efficacy of BPA-BNCT in suppressing the growth of H2170 cells (Fig. 6F). These findings strongly suggest that the loss of SLC7A5 and the concurrent increase in autophagic activity play pivotal roles in conferring resistance to BPA-BNCT in LUSC cells.
A Clonogenic assay assessing colony growth of H2170 and its SLC7A5 knockout cells (KO1 and KO2) treated with chloroquine (CQ, 30 μM). Colonies were stained with crystal violet (bottom) and quantified by ImageJ software (top). *p < 0.05; **p < 0.01. B Clonogenic assay assessing colony growth of H2170 and its SLC7A5 knockout cells (KO1 and KO2) treated with or without BPA-BNCT. Colonies were stained with crystal violet (bottom) and quantified by ImageJ software (top). ns, p > 0.5; *p < 0.05. C RT-qPCR (upper) and immunoblotting (lower) analysis assessing SLC7A5 expression in wild-type (WT) H2170 and its BPA-BNCT-treated surviving cells (BPA-BNCT). **p < 0.01. D RT-qPCR analysis (upper) assessing LC3B expression in wild-type (WT) H2170 and its BPA-BNCT-treated surviving cells (BPA-BNCT). Immunoblotting analysis (lower) assessing LC3B-I and LC3B-II expression in wild-type (WT) H2170 and its BPA-BNCT-treated surviving cells (BPA-BNCT) in the presence or absence of chloroquine (CQ). ***p < 0.001. E Clonogenic assay (lower) assessing colony growth of wild-type (WT) H2170 and BPA-BNCT-treated surviving cells (BPA-BNCT) in the presence or absence of chloroquine (CQ, 20 μM). Colonies were stained with crystal violet (bottom) and quantified by Image. *p < 0.05. F Clonogenic assay assessing colony growth of wild-type (WT) H2170 treated with or without BPA-BNCT in the presence or absence of chloroquine (CQ, 20 μM). *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
SLC7A5 has been proposed as both a biomarker and a therapeutic target in numerous cancers; nevertheless, its expression across different cancer histological subtypes remains inadequately understood. Furthermore, key questions persist regarding the impact of targeting SLC7A5 on drug resistance, particularly in therapies such as BPA-BNCT, and the strategies needed to overcome this resistance. Here, we demonstrate that SLC7A5 is highly expressed in squamous cell carcinoma, including LUSC, and its elevated expression correlates with poor prognosis in LUSC patients. Notably, knocking down SLC7A5 in LUSC cells induced cellular apoptosis, an effect that was not replicated under SLC7A5 knockout conditions. RNA-seq analysis further unveiled a downregulation in mTORC1 signaling and an upregulation in autophagy pathway genes in SLC7A5 knockout LUSC cells. Intriguingly, the knockout of SLC7A5 conferred resistance to BPA-BNCT in LUSC cells, characterized by heightened autophagy flux, yet retained sensitivity to autophagy inhibitors. These findings suggest a complex interplay between SLC7A5-mediated amino acid metabolism, mTORC1 signaling, and the autophagy pathway, offering potential insights into biomarkers and therapeutic targets for effective LUSC treatment strategies.
SLC7A5 is known to be overexpressed in solid tumors, including lung cancer [24, 25], head-and-neck carcinoma [26], esophagus cancer [27], cervical cancer [28], bladder cancer [29], colorectal cancer [30, 31], prostate cancer [32], breast cancer [33], gastric cancer [34], and pancreatic cancer [35]. Despite this recognition, the expression patterns of SLC7A5 in distinct cancer histological subtypes remain unclear. In our investigation, we observed pronounced expression of SLC7A5 in cancers predominantly characterized by squamous cell carcinoma histological types, such as those found in head-and-neck, lung, esophagus, and cervical cancers. Studies focusing on non-small cell lung cancer (NSCLC) have indicated a higher prevalence of SLC7A5-positive tumors in squamous cell carcinoma and large cell carcinoma as opposed to adenocarcinoma [36,37,38]. Moreover, SLC7A5 expression in NSCLC strongly correlates with Ki-67 labeling index levels, and its high expression predicts a poor overall survival outcome in patients [38]. Upon closer examination, our immunohistochemical staining data revealed significantly higher SLC7A5 expression levels in squamous cell carcinoma compared to adenocarcinoma within the context of lung cancer. This observation was further supported by RNA-seq analysis, which confirmed elevated SLC7A5 expression, specifically in squamous cell carcinoma of the lung, compared to other lung cancer subtypes. Furthermore, this heightened expression of SLC7A5 in LUSC was associated with a poorer prognosis in patients. Our findings and those from other studies highlight the potential of SLC7A5 as a biomarker in LUSC, delineating highly proliferative tumors with an adverse prognosis.
Due to its high expression in various cancers, SLC7A5 has been proposed as a promising therapeutic target [29]. In this study, we observed that silencing SLC7A5 reduced colony-forming ability and induced apoptosis in lung cancer cells. Additionally, LUSC tumors exhibit a high tumor-to-normal (T/N) ratio of SLC7A5 expression. Our findings and those of other studies underscore the potential of SLC7A5 as a therapeutic target in LUSC and other cancer types. Although several SLC7A5 inhibitors have been developed, their efficacy in preclinical studies and clinical trials, particularly in squamous cell carcinomas, requires further investigation [8, 39]. In addition, SLC7A5 has been suggested as the BPA-based drug transporter for BNCT in the clinical treatment of head-and-neck cancer with promising results [10, 40,41,42]. BPA-BNCT applied to lung adenocarcinoma treatment showed significant tumor regression, demonstrating the feasibility of BPA-BNCT for lung tumor treatment [43, 44]. Here, we showed that LUSC has a higher T/N ratio of SLC7A5 expression than lung adenocarcinoma. Moreover, BPA-BNCT application primarily decreased colony-forming ability in LUSC cells, while knockout of SLC7A5 blocked the BPA-BNCT-mediated colony inhibitory effect. These results suggest that SLC7A5 expression plays a crucial role in mediating the effects of BPA-BNCT, further indicating the potential of BPA-BNCT as an effective therapeutic strategy in LUSC treatment.
In this work, we observed distinct effects between the knockdown and knockout of SLC7A5 expression in LUSC cells. Specifically, the knockdown of SLC7A5 resulted in decreased colony-forming ability and increased apoptosis, whereas these phenotypes were not observed in the SLC7A5 knockout LUSC cells. Such differential phenotypes between specific gene knockout and knockdown models are commonly attributed to gene expression changes and subsequent compensatory mechanisms in knockout models compared to knockdown models [45]. This phenomenon is often associated with genetic or epigenetic-mediated pathway switching, leading to compensation, which is a hallmark of the development of drug resistance in cancer-targeted therapies that pharmacologically block driver genes in cancers [46,47,48]. Our data revealed significant downregulation of amino acid transporters and mTORC1 pathway genes, alongside upregulation of autophagy genes in the SLC7A5 knockout LUSC cells, indicating a pathway switch in these cells. Previous reports have highlighted the critical role of the glutamine pool and the expression of SLC7A5/SLC3A2, SLC1A5, and SLC38A2 in maintaining high mTOR activity and a cancer-like metabolic profile [49, 50]. Similarly, SLC1A4 and SLC38A5 contribute to sustaining the cellular glutamine pool, crucial for the influx of large neutral amino acids facilitated by the SLC7A5/SLC3A2 antiport [46, 49, 51, 52]. Our observation of SLC7A5 knockout cells displaying reduced levels of SLC3A2, SLC1A5, SLC1A4, and SLC38A5 suggests a potential coupling between SLC7A5 expression and the levels of these transporters in maintaining the prerequisites for mTOR activation and cancer metabolism in LUSC.
mTOR is crucial in regulating autophagy, mainly by controlling mTORC1 signaling. Under sufficient amino acid supply conditions, mTORC1 signaling strongly inhibits autophagy [53]. Autophagy is a fundamental cellular process responsible for digesting and recycling damaged macromolecules and organelles, providing energy and molecular building blocks in response to nutrient and environmental stress [54]. In our investigation, we found that long-term treatment of LUSC cells with the mTORC1 inhibitor rapamycin led to an enrichment of LUSC cells displaying high expression levels of autophagy genes such as LC3B and SQSTM1, accompanied by reduced SLC7A5 expression. This observation supports the idea of crosstalk between the amino acid anabolism mediated by SLC7A5/mTORC1 and the catabolic signaling of autophagy in LUSC cells. This intricate interplay between amino acid metabolism and autophagy pathways underscores the complexity of cellular processes governing nutrient sensing and recycling in cancer cells, highlighting potential avenues for targeted interventions in LUSC and other malignancies. Tatebe et al. reported that rapamycin treatment diminished the efficacy of BPA-BNCT in the SAS head and neck cancer cell line, likely due to rapamycin’s suppression of BPA uptake into cancer cells [20]. However, the precise mechanism remained unclear. Our findings that rapamycin decreases SLC7A5 expression while enhancing autophagy signaling, thus inhibiting BPA uptake, may offer a mechanistic explanation for the rapamycin-induced resistance to BPA-BNCT.
Despite its high response rate in head-and-neck cancer, BPA-BNCT often encounters resistance, with around 75% of patients experiencing tumor progression within a year [10]. However, the underlying mechanism behind this resistance remains unclear. Our study sheds light on this aspect by revealing that SLC7A5 knockout LUSC cells exhibit resistance to BPA-BNCT treatment, while wild-type LUSC cells that survive BPA-BNCT show reduced SLC7A5 expression. These findings suggest that the loss of SLC7A5 expression could contribute to the development of drug resistance against BPA-BNCT therapy. Further investigation revealed that SLC7A5 knockout LUSC cells, although resistant to BPA-BNCT, were sensitive to treatment with the autophagy inhibitor chloroquine. Interestingly, wild-type LUSC cells surviving after BPA-BNCT treatment exhibited heightened autophagy flux and were more susceptible to chloroquine treatment than parental LUSC cells. These results imply that BPA-BNCT drug-resistant cells may be vulnerable to autophagy inhibitors. Combinational treatments with the autophagy inhibitor enhanced BPA-BNCT efficacy, significantly reducing the colony-forming ability of LUSC cells. These findings have significant implications for improving the effectiveness of BPA-BNCT therapy and overcoming drug resistance in LUSC and potentially other cancers. Further research and clinical studies are warranted to validate these findings and optimize treatment protocols for better patient outcomes.
Our data indicate that SLC7A5 couples with the mTOR pathway to mediate LUSC tumor progression, suggesting its potential as both a prognosis marker and therapeutic target. However, targeting SLC7A5 in LUSC may trigger a pathway switch, increasing reliance on autophagy. These results highlight the importance of understanding the interplay between SLC7A5/mTOR amino acid anabolism and autophagy catabolism. Moreover, our findings suggest that autophagy pathways could serve as potential drug targets for reducing BPA-BNCT resistance.
Data availability
The datasets generated and/or analyzed during the current study are available in the Supplementary Information. Additional data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
Kahlhofer J, Teis D. The human LAT1-4F2hc (SLC7A5-SLC3A2) transporter complex: Physiological and pathophysiological implications. Basic Clin Pharm Toxicol. 2023;133:459–72.
Kanai Y, Segawa H, Miyamoto K, Uchino H, Takeda E, Endou H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J Biol Chem. 1998;273:23629–32.
Park G, Fukasawa K, Horie T, Masuo Y, Inaba Y, Tatsuno T, et al. l-Type amino acid transporter 1 in hypothalamic neurons in mice maintains energy and bone homeostasis. JCI Insight. 2023;8:e154925.
Kanai Y. Amino acid transporter LAT1 (SLC7A5) as a molecular target for cancer diagnosis and therapeutics. Pharm Ther. 2022;230:107964.
Kim CS, Cho SH, Chun HS, Lee SY, Endou H, Kanai Y, et al. BCH, an inhibitor of system L amino acid transporters, induces apoptosis in cancer cells. Biol Pharm Bull. 2008;31:1096–100.
Oda K, Hosoda N, Endo H, Saito K, Tsujihara K, Yamamura M, et al. L-type amino acid transporter 1 inhibitors inhibit tumor cell growth. Cancer Sci. 2010;101:173–9.
Wempe MF, Rice PJ, Lightner JW, Jutabha P, Hayashi M, Anzai N, et al. Metabolism and pharmacokinetic studies of JPH203, an L-amino acid transporter 1 (LAT1) selective compound. Drug Metab Pharmacokinet. 2012;27:155–61.
Yun DW, Lee SA, Park MG, Kim JS, Yu SK, Park MR, et al. JPH203, an L-type amino acid transporter 1-selective compound, induces apoptosis of YD-38 human oral cancer cells. J Pharm Sci. 2014;124:208–17.
Monti Hughes A, Hu N. Optimizing boron neutron capture therapy (BNCT) to treat cancer: an updated review on the latest developments on boron compounds and strategies. Cancers. 2023;15:4091.
Wang LW, Liu YH, Chu PY, Liu HM, Peir JJ, Lin KH, et al. Boron neutron capture therapy followed by image-guided intensity-modulated radiotherapy for locally recurrent head and neck cancer: a prospective phase I/II trial. Cancers. 2023;15:2762.
Detta A, Cruickshank GS. L-amino acid transporter-1 and boronophenylalanine-based boron neutron capture therapy of human brain tumors. Cancer Res. 2009;69:2126–32.
Suzuki M. Boron neutron capture therapy (BNCT): a unique role in radiotherapy with a view to entering the accelerator-based BNCT era. Int J Clin Oncol. 2020;25:43–50.
Kennedy BK, Lamming DW. The mechanistic target of rapamycin: the grand conducTOR of metabolism and aging. Cell Metab. 2016;23:990–1003.
Yang H, Jiang X, Li B, Yang HJ, Miller M, Yang A, et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552:368–73.
Paquette M, El-Houjeiri L, Pause A. mTOR pathways in cancer and autophagy. Cancers. 2018;10:18.
Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40.
Alfarsi LH, El-Ansari R, Craze ML, Masisi BK, Mohammed OJ, Ellis IO, et al. Co-expression effect of SLC7A5/SLC3A2 to predict response to endocrine therapy in oestrogen-receptor-positive breast cancer. Int J Mol Sci. 2020;21:1407.
Guduru SKR, Arya P. Synthesis and biological evaluation of rapamycin-derived, next generation small molecules. Medchemcomm. 2018;9:27–43.
Ballou LM, Lin RZ. Rapamycin and mTOR kinase inhibitors. J Chem Biol. 2008;1:27–36.
Tatebe H, Masunaga SI, Nishimura Y. Effect of rapamycin on the radio-sensitivity of cultured tumor cells following boron neutron capture reaction. World J Oncol. 2020;11:158–64.
Roy A, Bera S, Saso L, Dwarakanath BS. Role of autophagy in tumor response to radiation: Implications for improving radiotherapy. Front Oncol. 2022;12:957373.
Chou YT, Lee CC, Hsiao SH, Lin SE, Lin SC, Chung CH, et al. The emerging role of SOX2 in cell proliferation and survival and its crosstalk with oncogenic signaling in lung cancer. Stem Cells. 2013;31:2607–19.
Lee CJ, Sung PL, Kuo MH, Tsai MH, Wang CK, Pan ST, et al. Crosstalk between SOX2 and cytokine signaling in endometrial carcinoma. Sci Rep. 2018;8:17550.
Imai H, Kaira K, Oriuchi N, Yanagitani N, Sunaga N, Ishizuka T, et al. L-type amino acid transporter 1 expression is a prognostic marker in patients with surgically resected stage I non-small cell lung cancer. Histopathology. 2009;54:804–13.
Takeuchi K, Ogata S, Nakanishi K, Ozeki Y, Hiroi S, Tominaga S, et al. LAT1 expression in non-small-cell lung carcinomas: analyses by semiquantitative reverse transcription-PCR (237 cases) and immunohistochemistry (295 cases). Lung Cancer. 2010;68:58–65.
Toyoda M, Kaira K, Ohshima Y, Ishioka NS, Shino M, Sakakura K, et al. Prognostic significance of amino-acid transporter expression (LAT1, ASCT2, and xCT) in surgically resected tongue cancer. Br J Cancer. 2014;110:2506–13.
Kobayashi H, Ishii Y, Takayama T. Expression of L-type amino acid transporter 1 (LAT1) in esophageal carcinoma. J Surg Oncol. 2005;90:233–8.
Uno K, Kuwabara H, Terado Y, Kojima K, Kawakami T, Kamma H, et al. Divergent expression of L-type amino acid transporter 1 during uterine cervical carcinogenesis. Hum Pathol. 2011;42:1660–6.
Maimaiti M, Sakamoto S, Yamada Y, Sugiura M, Rii J, Takeuchi N, et al. Expression of L-type amino acid transporter 1 as a molecular target for prognostic and therapeutic indicators in bladder carcinoma. Sci Rep. 2020;10:1292.
Ogawa H, Kaira K, Motegi Y, Yokobori T, Takada T, Katoh R, et al. Role of amino acid transporter expression as a prognostic marker in patients with surgically resected colorectal cancer. Anticancer Res. 2019;39:2535–43.
Zhang L, Hu DL, Tang B, Cheng Y, Jiao C, Cheng L, et al. NFE2L3 Inhibition induces cell cycle arrest at the G0/G1 phase in colorectal cancer cells through downregulating CCND1 and pRb1-ser807/811. Dis Markers. 2019;2019:2829798.
Yanagisawa N, Satoh T, Hana K, Ichinoe M, Nakada N, Endou H, et al. L-amino acid transporter 1 may be a prognostic marker for local progression of prostatic cancer under expectant management. Cancer Biomark. 2015;15:365–74.
Bartlett JM, Thomas J, Ross DT, Seitz RS, Ring BZ, Beck RA, et al. Mammostrat as a tool to stratify breast cancer patients at risk of recurrence during endocrine therapy. Breast Cancer Res. 2010;12:R47.
Ichinoe M, Mikami T, Yoshida T, Igawa I, Tsuruta T, Nakada N, et al. High expression of L-type amino-acid transporter 1 (LAT1) in gastric carcinomas: comparison with non-cancerous lesions. Pathol Int. 2011;61:281–9.
Kaira K, Sunose Y, Arakawa K, Ogawa T, Sunaga N, Shimizu K, et al. Prognostic significance of L-type amino-acid transporter 1 expression in surgically resected pancreatic cancer. Br J Cancer. 2012;107:632–8.
Kaira K, Oriuchi N, Otani Y, Shimizu K, Tanaka S, Imai H, et al. Fluorine-18-alpha-methyltyrosine positron emission tomography for diagnosis and staging of lung cancer: a clinicopathologic study. Clin Cancer Res. 2007;13:6369–78.
Kaira K, Oriuchi N, Imai H, Shimizu K, Yanagitani N, Sunaga N, et al. Prognostic significance of L-type amino acid transporter 1 expression in resectable stage I-III nonsmall cell lung cancer. Br J Cancer. 2008;98:742–8.
Kaira K, Oriuchi N, Imai H, Shimizu K, Yanagitani N, Sunaga N, et al. Prognostic significance of L-type amino acid transporter 1 (LAT1) and 4F2 heavy chain (CD98) expression in surgically resectable stage III non-small cell lung cancer. Exp Ther Med. 2010;1:799–808.
Okano N, Naruge D, Kawai K, Kobayashi T, Nagashima F, Endou H, et al. First-in-human phase I study of JPH203, an L-type amino acid transporter 1 inhibitor, in patients with advanced solid tumors. Invest N Drugs. 2020;38:1495–506.
Koivunoro H, Kankaanranta L, Seppälä T, Haapaniemi A, Mäkitie A, Joensuu H. Boron neutron capture therapy for locally recurrent head and neck squamous cell carcinoma: an analysis of dose response and survival. Radiother Oncol. 2019;137:153–8.
Malouff TD, Seneviratne DS, Ebner DK, Stross WC, Waddle MR, Trifiletti DM, et al. Boron neutron capture therapy: a review of clinical applications. Front Oncol. 2021;11:601820.
Suzuki M, Kato I, Aihara T, Hiratsuka J, Yoshimura K, Niimi M, et al. Boron neutron capture therapy outcomes for advanced or recurrent head and neck cancer. J Radiat Res. 2014;55:146–53.
Farías RO, Bortolussi S, Menéndez PR, González SJ. Exploring boron neutron capture therapy for non-small cell lung cancer. Phys Med. 2014;30:888–97.
Suzuki M, Suzuki O, Sakurai Y, Tanaka H, Kondo N, Kinashi Y, et al. Reirradiation for locally recurrent lung cancer in the chest wall with boron neutron capture therapy (BNCT). Int Cancer Conf J. 2012;1:235–8.
El-Brolosy MA, Stainier DYR. Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet. 2017;13:e1006780.
Hwang W, Chiu YF, Kuo MH, Lee KL, Lee AC, Yu CC, et al. Expression of neuroendocrine factor VGF in lung cancer cells confers resistance to EGFR kinase inhibitors and triggers epithelial-to-mesenchymal transition. Cancer Res. 2017;77:3013–26.
Kuo MH, Lee AC, Hsiao SH, Lin SE, Chiu YF, Yang LH, et al. Cross-talk between SOX2 and TGFβ signaling regulates EGFR-TKI tolerance and lung cancer dissemination. Cancer Res. 2020;80:4426–38.
Sabnis AJ, Bivona TG. Principles of resistance to targeted cancer therapy: lessons from basic and translational cancer biology. Trends Mol Med. 2019;25:185–97.
Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34.
Taylor PM. Role of amino acid transporters in amino acid sensing. Am J Clin Nutr. 2014;99:223S–230S.
Scalise M, Console L, Cosco J, Pochini L, Galluccio M, Indiveri C. ASCT1 and ASCT2: brother and sister?. SLAS Discov. 2021;26:1148–63.
Wang Z, Yemanyi F, Blomfield AK, Bora K, Huang S, Liu CH, et al. Amino acid transporter SLC38A5 regulates developmental and pathological retinal angiogenesis. Elife. 2022;11:73105.
Rabanal-Ruiz Y, Otten EG, Korolchuk VI. mTORC1 as the main gateway to autophagy. Essays Biochem. 2017;61:565–84.
Gómez-Virgilio L, Silva-Lucero MD, Flores-Morelos DS, Gallardo-Nieto J, Lopez-Toledo G, Abarca-Fernandez AM, et al. Autophagy: a key regulator of homeostasis and disease: an overview of molecular mechanisms and modulators. Cells. 2022;11:2262.
Acknowledgements
We are grateful for the support of epithermal neutron irradiation from the Nuclear Science and Technology Development Center, National Tsing Hua University, Hsinchu, Taiwan.
Funding
We acknowledge support from National Tsing Hua University, National Science and Technology Council (109-2320-B-007-003-MY3; 112-2320-B-007-005), Executive Yuan, Taiwan.
Author information
Authors and Affiliations
Contributions
Project design: Yu-Ting Chou, Min-Yuan Chou. Data collection: Yen-Ju Lin, Yu-Cheng Wu, Yu-Jui Liu, Wei-Chen Yeh, Yi-Chen Huang, Shen-Ao Zhang, Alvin Chen, Ming-Yi Zheng. Data analysis: Hong-Ming Liu, Fong-In Chou, Shien-Tung Pan. Paper writing: Yu-Ting Chou, Yen-Ju Lin, Yu-Cheng Wu.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lin, YJ., Wu, YC., Liu, YJ. et al. Targeting SLC7A5 in lung squamous cell carcinoma: implications for cancer metabolism shift and boron neutron capture therapy resistance. Oncogenesis 14, 26 (2025). https://doi.org/10.1038/s41389-025-00568-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41389-025-00568-z