Abstract
The dynamic regulation of chromatin accessibility is one of the prominent characteristics of eukaryotic genome. The inaccessible regions are mainly located in heterochromatin, which is multilevel compressed and access restricted. The remaining accessible loci are generally located in the euchromatin, which have less nucleosome occupancy and higher regulatory activity. The opening of chromatin is the most important prerequisite for DNA transcription, replication, and damage repair, which is regulated by genetic, epigenetic, environmental, and other factors, playing a vital role in multiple biological progresses. Currently, based on the susceptibility difference of occupied or free DNA to enzymatic cleavage, solubility, methylation, and transposition, there are many methods to detect chromatin accessibility both in bulk and single-cell level. Through combining with high-throughput sequencing, the genome-wide chromatin accessibility landscape of many tissues and cells types also have been constructed. The chromatin accessibility feature is distinct in different tissues and biological states. Research on the regulation network of chromatin accessibility is crucial for uncovering the secret of various biological processes. In this review, we comprehensively introduced the major functions and mechanisms of chromatin accessibility variation in different physiological and pathological processes, meanwhile, the targeted therapies based on chromatin dynamics regulation are also summarized.
Similar content being viewed by others
Introduction
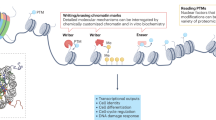
Chromatin, a linear complex containing the genetic material, is composed of DNA, histone, non-histone protein, and a small amount of RNA, encapsulated in the interphase nucleus of eukaryote. It can be divided into euchromatin and heterochromatin according to the compression degree. Nucleosome is the basic structural element of chromatin, containing an octamer histone core (two molecules of H2A, H2B, H3, and H4, encircled by ~147 bp of DNA) and a linker histone (H1).1 The nucleosomes are densely arranged in facultative and constitutive heterochromatin, while depleted at active regions, such as enhancers, promoters, transcribed gene bodies, DNA replication loci and damage repair sites.2,3 Chromatin accessibility refers to the physical contact permissibility of nuclear macromolecules with chromatinized DNA, which is mainly determined by distribution and occupancy of nucleosomes, as well as other DNA-binding factors.4,5,6 The accessible regions only comprise ~2–3% of the whole genome and more than 90% of these regions are yet captured by transcription factors (TFs). The accessibility of specific locus usually reflects its regulatory capacity3 (Fig. 1).
Dynamics of chromatin accessibility in eukaryote. The chromatin of eukaryote is multistage compressed and encapsulated in mitotic interphase nuclei. Majority of the chromatin is highly condensed, which is usually inaccessible. The remaining region is dynamically bound by histones, transcription factors, and other chromatin interaction molecules, characterizing by dynamic accessible, which is crucial for DNA transcription, replication, damage repair, and so on. This picture was drawn by Freescience
The orderly gene expression, DNA replication and damage repair play an important role in maintaining organism homeostasis, and aberrant of which usually results in a variety of diseases, such as cardiovascular diseases, liver diseases, nervous system disorders, diabetes, and neoplasms.7 At the different stages of physiological and pathological processes, the gene expression profiles are altered accordingly, which are strictly regulated by TFs.8,9 Generally, the TF-mediated gene expression, the orderly DNA replication and damage repair require an appropriate openness state of chromatin.3,10,11 Hence, chromatin accessibility plays a critical role in multiple biological processes. The chromatin accessibility is determined by multiple regulatory factors and targeting the chromatin dynamics has obtained many research achievements and improved the treatment of some diseases.
Acquainting the history and actuality of chromatin accessibility research can provide great guidance and help for further investigation. Therefore, in this review, we will introduce the history of chromatin accessibility investigation, common research methods, major mechanisms involved in chromatin accessibility regulation, their functions in different physiological and pathological processes, and targeting therapy strategies in human.
History of chromatin accessibility research
The history of chromatin accessibility investigation is more than five decades. (Fig. 2) The occupied DNA is insusceptibility to enzymatic cleavage, methylation, transposition, and has distinct solubility, which can be applied to investigate the chromatin accessibility. In 1973, Hewish and colleagues found that digesting the nuclear DNA in situ by endonucleases formed conservative periodic stripes, while the free DNA couldn’t. The following work revealed that the DNase hypersensitivity sites (DHSs) within genome were mainly determined by nucleosome distribution.12,13,14,15 With the invention of polymerase chain reaction (PCR) technology, a series of quantitative methods are applied to measure site-specific chromatin accessibility.16,17 In 2005, the nucleosome positions in the promoter of human p16 gene were clarified based on the CpG DNA methylation footprinting.18 In 2006, two research groups first measured the genome-wide DHSs using tiled DNA microarrays.19,20 In 2007, Giresi et al. invented FAIRE (formaldehyde-assisted isolation of regulatory elements) to detect the chromatin accessibility.21 Through combining with next-generation sequencing, many high-throughput methods are applied to research the genome-wide chromatin accessibility, such as MNase-seq,22 DNase-seq,23 FAIRE-seq,24 and NOMe-seq.25 Tn5 is a hyperactive transposase that can simultaneously fragment and tag the open genome with designated sequencing adapters. Based on this, Buenrostro et al. invented ATAC-seq (transposase-accessible chromatin using sequencing) in 2013 which simplified and flourished the investigation of chromatin accessibility.26 (Fig. 3) Importantly, ATAC-seq has significant advantage to detect the chromatin accessibility of few cells or single cell.27
The key discovery in chromatin accessibility research. The research on the accessibility of chromatin can trace back to 1973. Following, many achievements have been obtained in this field. From detecting general accessibility alteration, specific loci changes, to the genome-wide accessibility state. Recently, we are able to detect spatial chromatin accessibility, organelle chromatin accessibility, as well as different epigenetic modifications, transcriptome, and chromatin state in the same specimen at the single-cell resolution. This picture was drawn by Freescience
The principal methods to measure chromatin accessibility. The free and occupied DNA are distinct in hypersensitive to MNase- and DNase-mediated cleavage, solubility in different solvents, DNA methylation, as well as transposase-mediated transposition. Combining with high-throughput sequencing, MNase-seq (a), DNase-seq (b), FAIRE-seq (c), NOMe-seq (d), and ATAC-seq (e) are widely used in genome-wide accessibility studies. This picture was drawn by Freescience
At present, researchers can detect the chromatin accessibility of single-cell,28 intractable tissue samples,29 single-cell in intractable tissue samples,30 and organelle genome.31 Particularly, the emergence of multimodal detection techniques makes it possible to directly detect the epigenetic modifications, chromatin states, and gene expression in the same sample at single-cell resolution.32,33,34 By changing the sequencing strategy, it’s also possible to detect the chromatin states and interactions in the relatively long genomic regions.35,36 What’s more, uncovering the property of the spatial heterogeneity is very meaningful for the understanding of life and the treatment of diseases. In 2016, Chen et al. detected the chromatin accessibility in situ.37 In 2021, Thornton et al. detected the spatial chromatin accessibility at the single-cell resolution.38 Recently, many novel strategies have been reported to detect the spatial chromatin accessibility.39,40 In summary, new techniques are emerging which make it possible to investigate chromatin accessibility in a faster, more precise, larger scale, higher throughput, and lower cost manner. Meanwhile, the standardization of operation and data analysis processing has brought much convenience to the research.41,42 Here, we presented a general process for multimodal detection based on 10× Genomics single-cell sequencing platform (Fig. 4).
A general process for multimodal single-cell detection of chromatin dynamics. Currently, 10× Genomics sequencing platform is widely applied to obtain the gene expression and epigenetic information in single cell. The process is divided into single-cell nucleus preparation (a), library construction and sequencing (b), and data analysis (c). The sample sources include various tissues or cultured cells. After obtaining single cell suspension, the intact nuclei were obtained by gentle lysis and centrifugation. By adjusting the combination of enzymes, we can obtain gene expression, DNA methylation, histone modification, and chromatin accessibility profiles in the same single cell. Based on the significantly different genes, we can divide cells into different clusters and construct the pseudotemporal differentiation paths, as well as analyze the correlation and difference between gene expression, epigenetic modifications, and chromatin accessibility. This picture was drawn by Freescience
Regulation of chromatin accessibility
The accessibility of chromatin is determined by nucleosome distribution, histone modification, DNA methylation, non-histone protein occupancy, non-coding RNAs, chromatin 3D structure, and so on. In this section, we will introduce the main mechanisms that regulate the chromatin accessibility.
Nucleosome remodeling regulates chromatin accessibility
As the basic chromatin structure, nucleosome positioning is closely associated with chromatin accessibility. ATP-dependent chromatin remodeling complexes are master regulators of nucleosome mobilization. They can switch the “close” or “‘open” state of chromatin by hydrolyzing ATP as energy. According to the distinctive core domains, they are divided into four families, including switch/sucrose-non-fermenting (SWI/SNF), nucleosome remodeling and deacetylation (NuRD), imitation switch (ISWI), and INO80 family.11 In this part, we will introduce their mechanisms involved in chromatin accessibility regulation (Fig. 5a).
The primary remodellers of chromatin accessibility. The chromatin accessibility is determined by multiple regulators, mainly including chromatin remodellers (a), DNA methylation (b), histone modifications (c, d), pioneer transcription factors (e), non-coding RNAs (f), DNA sequence (g), and chromatin 3D structure (h). This picture was drawn by Freescience
SWI/SNF family
The SWI/SNF complex is the most intensively investigated nucleosome remodeller. This family contains at least 15 subunits, which are divided into BRG1/BRM-associated factor complex (BAF), polybromo-associated BAF complex (PBAF), and non-canonical BAF (ncBAF), depending on different assembly. They can promote or suppress gene transcription by binding with different cofactors.43 In 1997, researchers purified partial SWI/SNF complex from rat live tissue and HeLa cells. They found that glucocorticoid receptor (GR) activated SWI/SNF complex to regulate nucleosome disruption in a glucocorticoid response element (GRE) dependent manner. The remolded nucleosome occupancy facilitated the entrance of nuclear factor 1 (NF1) to its target sites, but had no influence on the binding of GR to GREs.44 BRM and BRG1 (also called SMARCA2 and SMARCA4, respectively) are the core subunits of SWI/SNF complexes that contain ATPase activity. SMARCA2 and SMARCA4 have distinct effect on the promoter accessibility. In the liver of cholic acid feeding mouse, when treated with farnesoid X receptor (FXR) agonists, SMARCA4 interacted with FXR which subsequently increased the promoter accessibility and transcription of small heterodimer partner (SHP), meanwhile SMARCA2 interacted with SHP to reduce the promoter accessibility and transcription of cytochrome P450 family 7 subfamily A member 1 (CYP7A1) and SHP.45 Hemogen can promote the entrance of GATA1/LDB1 complex to its binding motifs by recruiting SMARCA4 while expelling NuRD complex.46 Garry et al. revealed that the histone reader PHF7 recruited SWI/SNF complex to the cardiac super enhancers by directly binding with SMARCD3 to facilitate the chromatin opening and expression of target genes in fibroblasts.47 AT-rich interaction domain 1A (ARID1A) is another highly conserved subunit of SWI/SNF complex, containing DNA binding ability.48 In regenerating liver, lack of ARID1A remolded the histone modification (H3k4me2 and H3k27ac) and decreased chromatin accessibility, which blocked the entrance of hepatocyte nuclear factor 4α (HNF4α), CCAAT enhancer binding protein α (C/EBPα), forkhead box A2 (FoxA2), and E2F transcription factor 4 (E2F4), to their target genes.49 Its deficiency also promoted the expression of many cancer stem cell (CSC)-like markers.50 In human liver cancer, most of the SWI/SNF members are upregulated. SMARCD1, which increased most significantly, can activate the mechanistic target of rapamycin kinase (mTOR) signaling pathway.51 Meanwhile, the mTOR complex 1 (mTORC1) can remodel the chromatin accessibility by promoting ubiquitination-dependent degradation of ARID1A.52
NuRD family
The NuRD family, also called Mi-2/CHD (Chromodomain, Helicase, DNA binding) complex, contains both histone deacetylase and ATP-dependent chromatin remodeling activities. They can promote transcription by remodeling nucleosomes or suppress that by driving histone deacetylation.53 Matsui et al. indicated that FoxAs and PRDM1 recruited NuRD to maintain an accessible nucleosome state during human endoderm differentiation.54 During somatic reprogramming, NuRD interacted with Sall4 to reduce the chromatin accessibility of anti-reprogramming genes.55 The core ATPase-containing subunits of NuRD complexes include CHD3, CHD4, and CHD5.56,57 During B lymphopoiesis, CHD4 reduced the accessibility and expression of many non-B cell lineage genes to convoy the B lymphopoiesis.58 In rhabdomyosarcoma, CHD4-containing NuRD complex located to the super-enhancers, establishing a permissive chromatin architecture for the entrance of the tumor driving fusion protein PAX3-FoxO1.59 Besides, Shi et al. revealed that CHD4, interacting with SMYD1, suppressed the chromatin accessibility and expression of glycolysis-, hypoxia-, and angiogenesis-related genes during heart development.60 Depletion of CHD4 resulted in a globally increased DNA accessibility and induced spontaneous DNA damage in Ewing sarcoma.61 RBBP4/7, HDAC1/2, and MTA3 also are important subunits of NuRD complexes. Zhang et al. demonstrated that inhibiting HDAC2 reduced the chromatin accessibility of HDAC2/NuRD binding motifs in HDAC1-deficient neuroblastoma.62 Price et al. revealed that DLX1 recruited NuRD by interacting with RBBP4/7 to reduce the chromatin accessibility and expression of Olig2 during subpallium development.63 Chanda et al. indicated that NO synthase-induced NO promoted the S-nitrosylation of MTA3 to decrease NuRD activity. Inhibiting NO synthase reduced DNA accessibility in induced pluripotent stem cells (iPSCs).64
ISWI family
There are at least 16 different ISWI remodellers assembling with diverse subunits. Generally, the ISWI remodellers relocate the nucleosomes, instead of evicting them.65,66 In human, SNF2L and SNF2H (also called SMARCA1 and SMARCA5, respectively) are the core subunits of the ISWI family that contain ATPase activity. Goodwin et al. indicated that inactivation of SMARCA1 increased the accessibility of Fos/Jun binding motifs at the promoter regions to activate the ERK signaling pathway in cerebellar granule neuron precursor.67 Jiang et al. revealed that the phosphorylated 40S ribosomal protein SA (RPSA) recruited SMARCA5 to increase the chromatin accessibility and expression of NF-κB-targeted genes.68 Additionally, the ISWI complex also regulates the DNA repair progress by remodeling chromatin accessibility.69
INO80 family
The INO80 family contain INO80 (Inositol requiring 80) and SWR1 (SWI2/SNF2-related 1) complexes. Cai et al. found that INO80, interacting with Yin-Yang 1 (YY1), increased the accessibility of YY1 binding motifs and facilitated the YY1-induced transcription.70 Ren et al. demonstrated that overexpression of INO80 remodeled the nucleosome landscape and TF binding sites accessibility of the cardiac genes in cardiomyocyte.71 In non-small-cell lung cancer (NSCLC), upregulated INO80 increased the genome accessibility and expression of lung cancer-associated genes.72 Additionally, HELLS (also called SMARCA6, a member of the SWI2/SNF2 family) increased the nucleosome occupancy of multiple tumor suppressors in hepatocellular carcinoma (HCC) cells.73 Besides, INO80 and SWR1 remodeled H2A-H2B assembly in the transcriptional start site (TSS) to regulate gene transcription.74,75 The INO80 complexes also ensured the DNA damage repair by maintaining the chromatin accessibility in the damaged loci.76
DNA methylation regulates chromatin accessibility
DNA methylation also participates in the regulation of chromatin accessibility, which always acts as an obstruction for gene transcription. In this part, we will introduce the DNA methylation-mediated chromatin accessibility variation (Fig. 5b).
The DNA methylation is dynamically regulated by methyltransferase and demethylase. DNA (cytosine-5)-methyltransferase (DNMT) and enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) are methyltransferases. Inhibition of DNMT and EZH2 widely reduced DNA methylation and increased the chromatin accessibility in HCC cells.77 During liver-to-pancreas transdifferentiation, the accessibility of the pancreatic TFs binding motifs was increased, corresponding with reduced DNA methylation. Knockdown of DNMT1 promoted liver-to-pancreas transdifferentiation by increasing the expression of pancreatic-specific genes.78 Huang et al. identified a round of DNA demethylation and increasing of chromatin accessibility at meiosis initiation during human spermatogenesis. They confirmed that the reduced expression of DNMT chaperone ubiquitin-like, containing PHD and RING finger domains 1 (UHRF1) contributed to the variation.79 Guo et al. also indicated that UHRF1 recruited to the promoter of miR-26b to increase its DNA methylation, leading to reduced chromatin accessibility and miR-26b expression in abdominal aortic aneurysm.80
Tet methylcytosine dioxygenase 1 (TET1) is a demethylase. It interacted with TEA domain TF 1/4 (TEAD1/4) to enhance the regional DNA demethylation and H3K27ac of Yes1-associated transcriptional regulator (YAP)-derived genes.81 Deng et al. found that RNA m6A modification were inversely correlated with DNA 5mC modification both in normal and cancer cells. When binding with m6A-modified RNA, reader protein FXR1 recruited TET1 to the transcriptional loci to demethylate DNA, resulting in increased chromatin accessibility and gene transcription.82 5-hydroxymethylcytosine (5hmC) is an active DNA modification that modified by TETs. Li et al. indicated that the 5hmC level is dynamically regulated during pancreatic differentiation of human embryonic stem cells (hESCs) which corresponded with chromatin accessibility and gene transcription activity.83
The plasma cell-free DNA (cfDNA) exists in human peripheral blood. Studies indicated that low level of DNA methylation also increased nucleosome accessibility of cfDNA which was in line with nuclease sensibility, cutting site, and size distribution.84,85 Perinatal expression of deiodinase 2 (D2) also remolded the gene expression and DNA methylation pattern in adult mouse hepatocytes. D2 insufficiently reduced the overall chromatin accessibility which was in line with increased DNA methylation, but the modification enzyme is unclear.86
Histone modification regulates chromatin accessibility
Histones are the basic structural proteins of chromosome, rich in alkaline amino acid (arginine and lysine), which can facilitate the binding of acid DNA. Up to now, a great number of histone post-translation modifications (PTMs) are confirmed, such as phosphorylation,87 methylation,88 ubiquitylation,89 acetylation,90 lactylation,91 and crotonylation,92 which make up the histone code. The PTM landscapes of histones are closely related to the accessible state of chromatin. In this part, we will introduce the chromatin accessibility variant determined by histone modification. (Fig. 5c, d)
Histone phosphorylation
Phosphorylation is one of the most common and important PTMs in histones. The histones are phosphorylated and dephosphorylated periodically corresponding with the condensation and unwrapping of chromatin during mitosis, which indicates that histone phosphorylation is a crucial regulator of chromatin dynamics.
It’s well confirmed that phosphorylation of H3 at different serine or threonine residues can increase the accessibility of chromatin to facilitate gene transcription and DNA repair.93,94 Covalently closed circular DNA (cccDNA) is the hereditary material of the hepatitis B virus (HBV), which forms a microchromosomes in the hepatocyte nuclei. Ming et al. indicated that high mobility group nucleosome binding domain 1 (HMGN1) promoted the accessibility and transcription of cccDNA by competitively combining with H1 and promoting H3 phosphorylation.95 The H3 phosphorylation also orchestrates other PTMs, such as H3K9me3 and H3K36me3.96
Phosphorylation of H2AX is an important indicator for DNA damage repair. Researches indicated that the phosphorylated H2AX, called γH2AX, specifically recruited to the DNA double strand break sites, maintain the accessible of damaged DNA to facilitate the following repair pathway.97 H2A.Z is another variant of H2A. Fuglerud et al. indicated SRY-box transcription factor 9 (Sox9)-induced open chromatin was main located in H2A.Z enriched regions, and the H3S28 phosphorylation prevent the binding of Sox9 with chromatin.98
Histone H1 is the linker histone, regulating the higher-ordered chromatin structure. Researches indicate that the expression level of H1 is closely related to the chromatin structure.99 Meanwhile, the phosphorylation of H1 regulates its affinity to chromatin.100 The ex vivo research indicated that partial phosphorylation of H1 increased chromatin accessibility.101 However, in vivo evidence for the direct relationship between H1 phosphorylation and chromatin accessibility is still lacking.
Histone methylation
Histone methylation, including monomethylation, dimethylation, and trimethylation, usually occur on the lysine or arginine residues at the N-terminus of H3 and H4. Their role on chromatin structure and gene expression are site- and quantity-dependent.102
Kochat et al. indicated that downregulation of the methyltransferase EZH2 reduced H3K27me3 modification of many hepatic development-related genes, promoting the reprogramming of bone marrow progenitor cells to hepatocytes.103 Boonsanay et al. confirmed that loss of the methyltransferase SUV420H2 reduced H4K20me3 modification and increased chromatin accessibility predominantly in colorectal cancer organoids.104 Disruptor of telomeric silencing 1-like (DOT1L) is the only known methyltransferase catalyzing the methylation of H3K79.105 Researches indicated that DOT1L-induced H3K79me2/3 modification was important to maintain the accessible state of chromatin both in MLL-AF4 and mouse embryonic stem cells.106,107 Yang et al. indicated that the H3K4-specific methyltransferase MLL2 promoted the H3K4me modification to increase the accessibility of GR-targeted genes in ARPE-19 cells.108 PRMT5 is an arginine methyltransferase. Dacwag et al. revealed that PRMT5 catalyzed H3R8me2 modification, facilitating the binding of SWI/SNF complex with the promoter of target genes and enhancing their accessibility.109 Recent research indicated that SMARCA4 facilitated the binding of PRMT5 with the promoter of FoxO1 to maintain its H3R2 methylation and accessible state.110 These researches indicated that PRMT5 and SWI/SNF complex may cooperate to maintain the chromatin accessibility.
The JmjC domain-containing family (JMJDs) is a histone demethylase superfamily. In the same research, Kochat et al. they demonstrated that the upregulated JMJD3 was accountable for the reduction of H3K27me3 modification.103 Synergistically utilization of donafenib (multi-kinase inhibitor) and GSK-J4 (JMJD3 inhibitor) increased the promoter accessibility and expression of heme oxygenase 1 (HMOX1) by reducing H3K27me3 modification and simultaneously enhancing H3K4me1 and H3K27ac modifications.111 Additionally, JMJD1C enhanced the promoter accessibility of lipogenic genes by reducing the H3K9me3 modification.112 Lysine demethylase 1A (KDM1A) and KDM5B are histone demethylases. KDM1A exacerbated metabolic dysfunction-associated steatotic liver disease (MASLD) by increasing chromatin accessibility.113 KDM5B inhibited Nfkbia expression by reducing its promoter H3K4me3 modification and accessibility.114 Zhang et al. demonstrated that KDM4 (JMJD2) reduced the H3K9me2/3 and H3K36me2/3 modification to maintain the accessibility and expression of aging-related genes in prostate cancer cells.115
Histone ubiquitination
Ubiquitination is an universal modification to determine the protein stability, location, activation, and interaction. The ubiquitination of histone can remodel the chromatin structure, transcript elongation, and DNA damage repair.
Segala et al. revealed that the E3 ligase complex RNF20/RNF40 catalyzed the monoubiquitylation of histone H2B (H2Bub1) to block the INO80-mediated eviction of H2A.Z in the inducible enhancer regions and reduce their accessibility.116 Hooda et al. indicated that RNF20 insufficiency-induced H2Bub1 loss promoted the chromatin accessibility and expression of immune signaling pathways.117 Lin et al. also indicated that RNF20 increased the H2Bub1 to enhance the promoter accessibility and expression of many genes.118 Yin et al. indicated that the H2AK121ub modification was located in the less accessible but still permissive chromatin regions at transcriptional regulation locus.119 The loss of deubiquitinating enzyme BAP1 promoted H2AK119ub modification, remodeling the chromatin accessibility to perturb the transcriptomic pattern in human ductal liver organoids.120 Zhang et al. revealed that the H2BK120ub modification was important for the maintenance of accessible chromatin fiber to facilitate DNA repair.121 Additionally, the unbiquitination of histones always synergy with other modifications. Huang et al. demonstrated that the H2BK123ub inhibited Jhd2-induced H3K4 demethylation to maintain chromatin accessibility.122 Similarly, Worden et al. indicated that H2BK123ub facilitated DOT1L-induced histone H3K79me to increase the chromatin accessibility.123
At present, the research on histone ubiquitination and chromatin accessibility mainly focuses on H2B and H2A. The ubiquitination modification on other histones and their functions on chromatin accessibility still need to be investigated.
Histone acylation
Lysine acetylation is the most common acylation modification types on histones. Acetylation reduces the positive charge of histones to weaken their DNA binding capacity and inhibit the formation of higher chromatin structures, usually corresponding with higher accessibility and transcription activity.
Multiple acetylation modification sites have been clarified in different histones, which are dynamically regulated by acetyltransferase and deacetylases.124 In transforming growth factor β (TGFβ) treated cholangiocytes, SMAD family members recruited the histone acetyltransferase KAT2A to increase the H3K9ac modification and chromatin accessibility in the promoters of hematopoietic stem cell (HSC)-activating genes.125 Mutation of KAT6A reduced the H3K23ac modification and chromatin accessibility of HOXC gene cluster in dermal fibroblasts.126 Muthukrishnan, et al. confirmed that the histone acetyltransferase P300 increased the H3K27ac modification and chromatin accessibility of specific genes in glioma CSCs.127 The TFs c-Jun and Klf5 can recruit CBP/P300 to the promoter of special genes, which increases their H3K27ac modification, accessibility, and expression.128,129 Samata et al. indicated that H4K16ac was essential to maintain chromatin accessibility for the zygotic genome activation.130
The acetylation on histones can be eliminated by histone deacetylases. In the progression of metabolic steatohepatitis (MASH), methyltransferase-like 3 (METTL3) interacted with HDAC1/2 to remove the H3K9ac and H3K27ac modification in the promoters of CD36 and C-C motif chemokine ligand 2 (CCL2).131 HDAC8 reduced H3K27ac modification at the enhancers of CCL4 in HCC cells.132 SPI1 recruits HDAC1 to the active enhancers which globally reduces the promoter acetylation, chromatin accessibility, and RNA pol II occupancy in leukemic cells.133 Nucleosome assembly protein 1-like 2 (NAP1L2) recruits the deacetylase SIRT1 to evict H3K14ac modification on promoters of osteogenic genes and reduce their accessibility and expression in bone marrow mesenchymal stem cells (BMSCs).134 P53 recruits SIRT1 to reduce the H3K27ac modification, promoter accessibility, and transcription of Neat1.135 Yuan et al. indicated that ZCWPW1 inhibited HDAC-induced H3K9 deacetylation to maintain the openness of chromatin during meiotic double-strand break repair.136
The upregulation of acetyl-CoA metabolism can provide more substrate for acetylation. Liu et al. revealed that vitamin B1 increased the histone acetylation and chromatin accessibility by facilitating acetyl-CoA metabolism.137 Pyruvate dehydrogenase 1α (PDHE1α) drives acetyl-CoA production by catalyzing pyruvate. It increases the acetyl-CoA level in the DNA damage sites to facilitate histone acetylation and chromatin accessibility.138
Recently, many novel acylation modifications on histone are constantly identified which participate into the regulation of chromatin dynamics. In 2019, Zhao group identified lactylation on histones which can remodel the expression atlas in macrophage.139 Recently, Merkuri et al. demonstrated that glycolysis induced the histone lactylation at neural crest related genes, which increased their chromatin accessibility and expression.140 Trujillo et al. also indicated that lactylation of histone increased the chromatin accessibility to promote inflammatory signaling in macrophages.141 Jing et al. demonstrated that, succinylation of H4K77 (H4K77succ) promoted DNA unwrapping from the histone surface, facilitating the binding of proteins with the nucleosome DNA.142 Additionally, serotonylation has been confirmed to exclude from constitutive heterochromatic regions which hints that serotonylation is a potential active marker for chromatin.143
In addition, other factors also regulate chromatin accessibility by remolding the histone modification profiles. For example, the HBV X protein (HBx) increases the H3K27ac modification and super enhancer accessibility of ETS variant TF 4 (ETV4), thereby promotes its expression.144 Interleukin 6 (IL-6) and tumor necrosis factor α (TNF-α) treatment increase the H3K4me3 modification and promoter accessibility of microtubule-associated serine/threonine kinase like (MASTL).145 Mitochondrial stress increases the H3K4me1 and H3K27ac modifications as well as the chromatin accessibility in amphiregulin (AREG) enhancer regions.146 Knockdown of nuclear autoantigenic sperm protein (NASP) decreases H3K9me1 modification and enhances chromatin accessibility globally.147 Sox4 modifies the landscapes of H3K27ac, H3K4me1, and H3K4me3 modifications during the hepatocytes to biliary transdifferentiation.148 ARID1A deletion decreases H3K4me3 and chromatin accessibility on the promoters of fatty acid oxidation (FAO)-related genes.149 The co-transcription factor (co-TF) VGLL1, binds with TEAD4 to increase the accessibility and histone acetylation modification at target gene loci.150 Mineral dust-induced gene (MDIG) can promote the H3K9me3-to-H3K9me1 transformation of OTX2 promoter which facilitates the entrance of Myc.151 However, these regulators cannot modify histone directly, the direct participant involved in these processes needs further elucidation.
Pioneer transcription factors regulate chromatin accessibility
Early studies have shown that TFs can enhance chromatin openness.152 Now, we know that majority of these TFs are pioneer transcription factors (PTFs). Different from conventional TFs, PTFs can bind with closed chromatin regions and enhance the accessibility of local chromatin which facilitate the entrance of other tissue-specific TFs to control cell fate and function (Fig. 5e).
The members of FoxA family are the most important PTFs in liver. They remold chromatin accessibility to regulate the development and function of liver.153 FoxAs can expel linker histone H1, thereby maintaining the accessibility of liver-specific enhancers and promoters to permit the entrance of other liver-specific TFs,154 such as HNF4α, FXR, and liver X receptor α (LXRα).155,156 Research indicated that FoxA2 binding sites are located in nucleosome-free regions which showed hypersensitive to MNase.157 In the adult liver, HNF4α, but not FoxA2, is required for maintaining the chromatin structure.158 HNF4α enriched H3K4me1 and H3K27ac modification to maintain opening chromatin at active transcriptional regions.159 HNF4α also enhanced the accessibility of basic helix-loop-helix ARNT-like 1 (BMAL1) binding motifs to regulate liver circadian rhythms.160 Exogenous expression of Yamanaka factors (Sox2, Oct-3/4, Klf4, and c-Myc) in adult somatic cells can facilitate them dedifferentiating into iPSCs, which involves many epigenetic remodeling.161,162 Research indicated that inducible expression of Yamanaka factors promoted the expression of DNA topoisomerase II alpha (TOP2A) to modify the chromatin accessibility.163 Fuglerud et al. indicated that Sox9 can unwrap chromatin at special sites in closed chromatin. It promoted the expression of endothelial-to-mesenchymal transition genes by increasing their chromatin accessibility.98
Research also revealed that FoxA2 binding alone did not increase the permissive of chromatin. Only synergistically binding with other cofactors can FoxA2 enhance chromatin accessibility.164 These results indicate that some PTFs are more likely to play a positioning function. When bind to specific chromatin site, they recruit other chromatin regulators to synergistically regulate the local chromatin accessibility.
Non-coding RNAs regulate chromatin accessibility
The non-coding RNAs, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), circular non-coding RNAs (circRNAs), and so on, perform many important biological functions. Researches have confirmed that they are master regulators of chromatin accessibility (Fig. 5f).
MiR-137 is a neuropsychiatric-disorder-associated miRNA that located in the microglial nucleus. Li et al. indicated that miR-137 decreased the chromatin accessibility by competitively binding with microglial master transcription factor Pu.1 to suppress the binding of Pu.1 with chromatin.165 Boos et al. indicated that the hypoxia-induced lncRNA LINC00607 was essential for normal endothelial function and angiogenic sprouting. It regulated the chromatin accessibility around the binding motifs of ETS transcription factors ERG, by directly interacting with the chromatin remodeller SMARCA4.166 In the earlier research of the same group, they found that SMARCA4 can interact with lncRNA MANTIS, to facilitate the binding of RNA Polymerase II to DNA.167 Ma et al. indicated that the lncRNA HOTAIR, increased the chromatin accessibility and expression of metastasis-related genes in breast cancer cells, but the detail mechanism requires to reveal.168 Additionally, circRNA circTmem241 recruited methyltransferase Ash1l to the promoter of Elk3 and enhanced its chromatin accessibility and transcription in innate lymphoid cells.169 Besides, synthetic RNA increased the chromatin accessibility of albumin gene in vitro but its function in vivo is also unclear.170
Dueva et al. indicated that single-stranded RNA in the nucleus maintain an open chromatin structure by interacting with histone tails to neutralize the positive charge on histones.171 In 2020, He et al. proposed a concept, chromosome-associated regulatory RNAs (carRNAs), including promoter-associated RNAs, enhancer RNAs, repeat RNAs, and so on, which can regulate the chromatin structure in mammalian cells.172 Members of long interspersed element-1 (LINE1) family are representative carRNAs, that have been confirmed to regulate chromatin structure.173 He et al. indicated that m6A modification decreased the carRNA level, especially LINE1 to reduce the chromatin accessibility and nascent RNA transcription.172 Li et al. indicated that the m6A modification stabilized the super-enhancer RNAs, which consequently recruited H3K4 methyltransferase MLL1 to promote H3K4me3 modification and accessibility accessibility in specific genes.174
Cis-regulatory element regulates chromatin accessibility
The cis-regulatory elements, which specifically bound by DNA binding proteins (TF, co-TF, transcription inhibitor, transposase, etc) are nonnegligible regulators of chromatin accessibility. In vivo researches have showed that nucleosomes exhibit significant DNA sequence preferences.1 Li et al. indicated that mutation of human enhancers altered their accessibility.175 The single nucleotide polymorphisms (SNPs) also determine the chromatin accessibility and TF affinity.176,177 Spisak et al. edited the SNPs in prostate cancer cells significantly remodeling the chromatin accessibility, histone modification, and TF affinity.178 Recently, Mononen et al. demonstrated that the genetically driven differences in the expression pattern, H3K27ac modification, and chromatin accessibility were more pronounced than those induced by diet in mouse liver.176 (Fig. 5g)
The three-dimensional (3D) structure of chromatin regulates its accessibility
The chromatin is orderly and dynamically encapsulated in the nucleus, and the 3D structure plays an important role in regulating its accessibility. (Fig. 5h) The lamin A/C variation-induced morphology change of nucleus is closely related to chromatin accessibility, epigenetic modification, and gene expression.179 Besides, the nuclear matrix protein, heterogeneous nuclear ribonucleoprotein U (hnRNPU) has been reported to regulate chromatin accessibility.180 Mitochondrial TF A (TFAM) remolded the chromatin accessibility by inducing the polymerization of nuclear actin.181
At present, the chromatin conformation capture (3C) technologies allow us to measure the 3D structure of chromatin directly. These 3D structures mainly include compartments,182 loop domains,183 topologically associated domains (TADs),184 and so on. Cohesin and CCCTC-binding factor (CTCF) are the key architectural proteins that regulate the formation of TAD and loop domain.185 Xie et al. indicated that Cohesin, CTCF together with BRD2 protected the architectural boundaries of accessible chromatin regions.186 Chen et al. demonstrated that the TAD boundaries were more accessible, contained higher transcriptional capacity and more DNA double-strand breaks (DSBs).187 Li et al. indicated that the chromatin 3D structure and accessibility determined the pluripotent state of embryonic stem cells (ESCs).188 The variation of the chromatin accessibility and higher structure also coincided with the neuron development. Wahl et al. revealed that SATB2 remodeled the chromatin 3D structure and accessibility both independent and in cooperation with CTCF in cortical neurons.189 Additionally, many researches have indicated that the feature of chromatin 3D structure and accessibility were distinct in different diseases, such as breast cancer,190 glioma,191 and Alzheimer’s disease (AD).192 It indicates that the chromatin 3D structure could be an potential disease biomarker and therapeutic target.
It’s sure that the higher structure and distribution of chromatin are closely related with chromatin accessibility and gene expression. However, whether the variation of chromatin 3D structure is a cause or consequence of chromatin accessibility change is still controversial,193 hence, further investigation on this field is required urgently.
Environmental factors regulate chromatin accessibility
The surrounding environment is an important regulator of biological process. Many environmental factors can directly regulate gene expression. In this section, we summarized the function of environmental factors on chromatin accessibility.
Chemical exposure-induced epigenetic alteration is attracting more and more attention in human health. The environmental chemicals can widely remold the chromatin accessibility and TF binding patterns. Israel et al. indicated that exposure to genotoxic carcinogen 1,3-butadiene widely remolded the histone acetylation, chromatin accessibility, and gene expression patterns in lung, kidney, and liver tissues of C57BL/6J and CAST/EiJ mice.194 Hexavalent chromium (Cr(VI)) is well-clarified respiratory carcinogens by forming protein-Cr-DNA adducts. VonHandorf et al. indicated that Cr(VI) dysregulated the nucleosome occupancy at specific genome locus, blocking the activator protein-1 (AP-1) and CTCF-targeting motifs in mouse liver.195 Acrolein is abundant in cigarette smoke. Chen et al. indicated that acrolein exposure increased the chromatin accessibility through compromising the delivery of H3 into chromatin.196 Bisphenol F also remodeled the hepatic transcriptome, metabolome, and chromatin accessibility to trigger MASLD.197 Ionizing radiation is an ubiquitous environmental pathogenic factor. Dahl et al. found that radiation induced globally chromatin accessibility alteration in mouse liver.198 In mouse hepatoma cells, dioxin-induced promoter accessibility of CYP1A1 facilitated the formation of AhR/Arnt heteromer.199,200 These studies have suggested that the harmful environmental factor-induced chromatin accessibility alteration played an important role in disease progression. Besides, environmental factor is irreplaceable for the regulation of our daily routine. The related role and mechanism in these progresses are remained to be elucidated.
In addition to mentioned above, many other regulators may also participate in the remolding of chromatin accessibility. For example, folic acid increased the accessibility of IGF2 promoter during embryonic development of broiler.201 The mechanical signals of the extracellular matrix also regulates the chromatin accessibility.202 However, their variation, manifestation, function, and mechanism in the physiological and pathological processes are required further investigation.
Chromatin accessibility in physiological processes
In the previous section, we introduced the major molecular mechanisms of chromatin accessibility regulation. The variation of chromatin accessibility participates in many physiological processes, such as early embryogenesis, organ development, tissue regeneration, aging, circadian rhythms, and so on. Researchers have constructed the chromatin accessibility atlas of different human organs and identified the specific regulatory networks in different cells. In this section, we will introduce the detail functions and mechanisms of chromatin accessibility variant in some physiological processes (Fig. 6).
Chromatin accessibility variation is involved in multiple physiological processes. The chromatin accessibility is dynamically varied in many physiological processes, such as embryonic and organ development, tissue regeneration, organ function execution, circadian rhythm, gender difference, senescence, and so on. This picture was drawn by Freescience
Chromatin accessibility in early embryonic development
All mammals are originated from the fusion of male and female gametes, called zygote. After fertilization, the embryo is constantly dividing, orderly forming morula, blastocyst, gastrula, and finally differentiated into various tissues and organs. Following fertilization, the highly specialized epigenetic modification of sperm and oocytes are reprogrammed to facilitate the establishment of a totipotent state which is required for the embryo development. The division of human embryo in the first 3 days is mainly sustained by maternally inherited factors, and a major wave of embryonic genome activation (EGA) arises at the 4-cell (4C) to 8-cell (8C) stage.203 The epigenetic modification of embryo chromatin changes significantly which is in line with its accessibility variation during embryogenesis. It’s consistent in many researches that the accessibility regions of human embryonic genome are increased progressively from the 2-cell (2C) to blastocyst stage.204,205 Most of the accessible regions are located in the promoters, CpG islands, and enhancers. Although some genes containing accessible promoter in the 2C stage do not transcribed until at the 8C stage, their are more highly expressed in the subsequent stages than those gain accessibility later. It indicated that accessibility state poised in the early stage is critical for the high expression of some critical genes.206
It is of great significance to clarify the contribution of the remodellers on chromatin accessibility variation during the early embryonic development. Samata et al. demonstrated that the H4K16ac modification was intergenerationally chromatin modification from oocytes to fertilized embryos. It’s indispensable to maintain the chromatin accessibility for zygotic genome activation. Maternal depletion of the acetyltransferase MOF resulted in H4K16ac loss and downregulation of post-zygotically expressed genes.130 Treating the 8C embryo with the transcription inhibitor α-amanitin resulted in a similar distal regulatory element accessibility pattern with pre-EGA stage, indicating that transcription is indispensable for chromatin accessibility variation, at least in distal elements at EGA stage.205 In human embryos, the direct evidence about the relationship between chromatin accessibility and its remodeller is yet to be revealed, but some leads are promising. The inverse relationship between accessibility and DNA methylation occurs at all stages of human early embryogenesis, and the expression pattern of many gene families with repetitive elements are coincident with their methylation modification and chromatin accessibility during preimplantation development.204 Besides, the subunits of SNF2-family regulate Oct-4 in naive human pluripotent stem cells to orchestrate the expression of blastocyst lineage genes.207 The H3K9me3 modification landscape is also varied during preimplantation embryo development.208 Given that SNF2 complex, DNA hypermethylation, and H3K9me3 are regulators or indicators of chromatin accessibility, they could contribute to the chromatin variation in human early embryogenesis.
Chromatin accessibility in organ development
The human body consists of many tissues and organs, all of which are derived from the epiblast of the blastocyst. Meanwhile, the maintenance and differentiation of somatic stem cells are also crucial for maintaining organ homeostasis. The determination of cell fate is crucial for the orderly development of different tissues and organs. Multiple researches have indicated that the chromatin accessibility is an important regulator of cell fate. In undifferentiated pluripotent cells, the tissue-specific cis-regulatory elements usually reside in the closed, silent chromatin regions that are hard for TFs entrance. Besides, the chromatin accessibility is distinct in different tissues and organs of mammals. These differences are mainly determined by tissue-specific PTFs, which can bind with heterochromatin and permanently change the epigenetic chromatin modification and stably maintain the accessibility of tissue-specific genes.209 In this part, we will discuss the variation and function of chromatin accessibility during different organ development.
Heart development
Heart is the first functional organ during embryonic development. The epigenetic modification, which can determine the chromatin accessibility, is a master regulator of cardiac development.
In 2022, Ameen et al. constructed a single-cell resolution chromatin accessibility atlas of human fetal heart tissues. They defined a series of cell types in the heart by different TFs and identified the developmental trajectories of human fetal heart. Meanwhile, by comparing the chromatin accessibility profiles of congenital heart disease (CHD) cases and normal controls, they identified many potential CHD-inducing factors, among which they confirmed that loss of JARID2 impaired heart development.210 Researches have indicated that H2Bub modification was critical for cardiac development, and the mutations of E3 ligases RNF20, which regulates H2Bub modification, commonly occur in CHD patients.211,212 Recently, Lin et al. indicated that RNF20 increased the promoter accessibility and expression of cell-cell connections and actin organization related genes by monoubiquitylating histone H2B to promote postnatal cardiomyocyte polarization.118 The TF FoxK1 is specifically expressed in developing cardiac and skeletal muscles.213 It can promote the proliferation of myogenic stem cell following injury.214 Sierra-Pagan et al. indicated that FoxK1 also promoted the development of mesodermal progenitor cells by orchestrating the chromatin accessibility of cardiac developmental genes, especially inhibiting the Wnt/β-catenin signal pathway.215 GATA4/5/6 are essential and conserved TFs that regulate heart development.216 Song et al. indicated that GATA5/6 determined the balance of cardiac and pharyngeal development. GATA5/6 regulated globally chromatin accessibility to orchestrate the expression of cardiac and pharyngeal regulatory genes.217 Arrieta, et al. indicated that knockdown the circadian protein BMAL1 in ventricular myocytes impaired the postnatal development of rat heart. The loss of BMAL1 decreased the accessibility of Per2 and Sik1 promoter.218 Zhong et al. indicated that knockout c-Jun facilitated the differentiation of hESCs into cardiomyocytes in vitro. C-Jun deficiency increased the chromatin accessibility of hESCs. Mechanically, loss of c-Jun increased the expression of RBBP5 and SETD1B, increasing H3K4me3 deposition on cardiogenesis-related genes.219 Krup et al. indicated that knockout Mesp1 impaired the differentiation of cardiac mesoderm cells to cardiomyocytes. ScATAC-seq analysis revealed that Mesp1-KO cells showed strikingly divergent regulatory landscapes compared with controls. Mesp1 insufficiency reduces the promoter accessibility and expression of cardiac differentiation-driving genes.220 The TFs Wt1a and Wt1b blocked cardiomyocyte differentiation by reducing the chromatin accessibility of cardiomyocyte-specific genes.221 Fang et al. revealed that deletion of TET2/3 decreased the global 5hmC modification and chromatin accessibility, perturbing YY1 binding to disrupt cardiac-specific transcription.222 Meier et al. constructed the single-cell transcriptome and chromatin accessibility profiles of human epicardioids. Through combining lineage tracing, they found that epicardioids were derived from first heart and juxtacardiac field progenitors. Additionally, the in vitro treated epicardioids can mimic left ventricular hypertrophy and fibrosis, offering an unique testing ground for heart epicardial development, disease, and regeneration.223
Chromatin accessibility variation is also associated with the pacemaker development in the sinoatrial node (SAN).224 Expression of GATA4, Tbx5, and Mef2c combining with or without Hand2 can reprogram fibroblasts into induced cardiomyocyte-like myocytes (iCLMs) or pacemaker-like myocytes (iPM), respectively.225,226 Fernandez-Perez et al. indicated that Hand2 promoted the expression of pacemaker-specific genes by increasing their promoter accessibility.227 Galang et al. compared the accessible chromatin atlas of cardiac pacemaker cells with that of right atrial cardiomyocytes. They found that the accessibility of an Isl1 enhancer was increased in the SAN, which promoted the expression of Isl1, thus facilitating SAN development. By analyzing the ATAC-seq peak around Isl1 enhancer, they speculated that this Isl1 enhancer also regulated SAN development in human.228 van Eif et al. analyzed the human accessible chromatin both in pluripotent stem cell-derived SAN-like pacemaker cells (SANLPCs) and ventricle-like cells. They confirmed that the Isl1 locus was more accessible in SANLPCs. Besides, they identified that the Tbx3 enhancer is also enriched. Specific-deletion of the homologous region in mouse model impaired the SAN development.229
Liver development
The orderly development plays a vital role in maintaining liver homeostasis. During liver development, the chromatin structure, epigenetic modifications, and gene expression profile are changing correspondingly (Fig. 7).
Chromatin accessibility regulates the development, regeneration, and transdifferentiation of liver. During the development of liver, the chromatin accessibility of pluripotent genes and liver-specific genes are reduced and increased, respectively. During the repair of damaged liver, the chromatin accessibility of pluripotency and proliferation-related genes are increased. During the transdifferentiation of liver, the specific gene accessibility of origin tissue is always reduced and that of the aim tissue is correspondingly increased. This picture was drawn by Freescience
FoxA and GATA families can remold the chromatin structure of liver-specific genes, facilitating the access of hepatic TFs to their target genes.230,231 Reizel et al. demonstrated that FoxAs increased the enhancer accessibility of HNF4α-targeting genes to maintain liver homeostasis.232 Besides, the DNA methylation profiles are altering during development, manifested as de novo methylated of pluripotency genes and demethylation of tissue-specific genes, which are remarkably consistent with chromatin accessibility and gene expression.233 In broiler, folic acid injection decreased DNMT1-induced methylation and loosened the promoter of IGF2 to enhance its expression, subsequently facilitating embryonic growth and liver development.201 During zebrafish liver development, the nuclear morphology of hepatocytes was changing. Meanwhile, the chromatin accessibility of development-related genes in the larval liver was enriched. UHRF1 and DNMT1 are required for maintaining appropriate nuclear morphology, and their mutation lead to DNA hypomethylation, loss of lamin B2, and large dysmorphic nuclei in hepatocytes.234 Hepatocyte and cholangiocyte are the main cell types that originated from bipotential hepatoblast in liver.235 Yang et al. constructed the histone modification and chromatin accessibility profiles during hepatoblast differentiation. They found that the differentiation pathways of hepatoblasts can be determined by chromatin accessibility patterns which were mostly synchronous with H3K27ac on enhancers, and H3K27me3 on promoters. EZH2 and JMJD3, which methylate or demethylate H3K27, respectively, had distinct functions on hepatoblast-to-hepatocyte or hepatoblast-to-cholangiocyte differentiation, while the histone acetyltransferase P300 promoted both progresses.236
Brain development
In 2021, under the BRAIN Initiative Cell Census Network (BICCN) of National Institutes of Health (NIH), a series of outstanding achievements in brain research have been obtained, providing a spatially resolved cell-type atlas of the motor cortex in different mammals depending on the single-cell transcriptomes, epigennetic modification, and chromatin accessibility.237,238,239,240 The revelation of neurocyte definition and spatial distribution, transcriptomes and epigenetic markers, as well as the differences among mammals, will construct a solid foundation for the investigation of nervous system evolution, development, and functional execution. In 2023, the Bing Ren group comprehensively analyzed the chromatin accessibility in human brain by single-nucleus ATAC-seq (snATAC-seq). They defined many cell types in brain depending on the single-cell chromatin accessibility atlas. They also identified the specific expressed genes in different cell types and their regulatory networks. What’s more, they predicted the disease-relevant cell types for many neuropschiatric disorders.241 Similarly, the accessible regions in the neurons of drosophila brain preferentially drive the expression of genes in neuronal subsets which are distinct in different neuronal sub-types.242 Herring et al. constructed the single-cell resolution chromatin accessibility atlas of human brain from gestation to adulthood, which revealed the cell types and chromatin dynamics during the mature of human prefrontal cortex. They also defined some regulatory drivers of neurological and psychiatric diseases.243
The nervous system is composed of a variety of cell types. Pavlou et al. indicated that the differentiated astrocytes had distinct expression pattern and chromatin accessibility compared with multipotent neural stem cells (NSCs). They found that the inflammatory condition increased the chromatin accessibility and facilitated the expression of inflammatory response genes. The enriched accessible regions were recognized by Rarg and Dlx1, while the reduced regions were recognized by Tcf21.244 LHX2 is a well-confirmed regulator both in early development of hippocampal primordium (Hcp) and neocortical primordium (Ncp).245 Suresh et al. demonstrated that the chromatin of Hcp was more accessible than Ncp, and consistent with increased active histone marks, H3K27ac, H3K4me1, and H3K4me3, in the enriched loci. Loss of LHX2 didn’t effect the global chromatin accessibility in Ncp but striking reduced the accessibility in Hcp.246 Berg et al. identified that the Hopx-CreERT2 line was an embryonic origin of adult dentate neural progenitors. They found that these dentate neural progenitors contained a distinct chromatin accessibility signature compared with the mature dentate gyrus. The genes located in the ATAC-seq peaks are mainly enriched in signal transduction and nervous system development, and the top four TF binding motifs are Zfp354c, Bcl6, Zbtb18, and YY1, all of which have been reported to regulate somatic stem cells.247 Cerebellar is one of the main parts of human central nervous system and its development is subtly orchestrated. Zhong et al. established the integrative spatiotemporal development landscape of human fetal cerebellar by systematically using spatial transcriptomics, single-cell transcriptomics, and single-cell chromatin accessibility. They found that not only progenitor cells at different locations displayed differential gene expression and chromatin accessibility profiles, but also differentiated neurons showed distinctive spatial-temporal molecular signatures.248 Liu et al. indicated that ARID1A orchestrated the chromatin accessibility and expression of neurogenic and cardiogenic genes in hESCs to facilitate neurogenesis and block cardiogenesis.249
Lung development
In 2022, He et al. established a single-cell atlas of human fetal lung based on multi-omics analysis, including scATAC-seq, scRNA-seq. They identified lung cells in different differentiation states and mapped human lung development accordingly.250 Sox9 is one of the important biomarkers of pluripotent cell in the respiratory buds, which can differentiated into both airway and alveolar epithelium.251 Based on the chromatin accessibility and expression pattern variation during respiratory bud development, Khattar et al. revealed that PI3K signaling was essential for the epithelial differentiation of Sox9+ progenitors.252 Little et al. indicated that NKX2-1 had an opposite impact on the cell fate of lung alveolar type 1 (AT1) and AT2 cells. It interacted with different co-TFs to remodel the chromatin accessibility in there cell types.253 FoxF1 is a key factor regulating alveolar capillary development. Guo et al. detected the chromatin accessibility atlas in alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV). They confirmed the FoxF1 regulatory network in ACDMPV and provided some potential therapy targets for this disease.254
Development of other organs or cells
Miao et al. established the chromatin accessibility atlas in mouse kidney, and analyzed the differentiation trajectory of nephron progenitor. They found that FoxL1 was sustainedly expressed during nephron progenitor differentiation, and the expression of Hfn4a and Tfap2b was associated with proximal and distal fates, respectively. Additionally, they indicated that the chromatin accessibility feature can reflect the development mechanism of human kidney, meanwhile the H3K27ac and H3Kme1 modifications may be the main regulators.255 Erythropoietin (EPO) is an important peptide hormone regulating erythropoiesis. Riou et al. found that erythropoiesis was enhanced in APC and ARID1A co-deletion mice liver. Mechanically, APC deletion activated β-catenin signaling and ARID1A deletion increased the promoter accessibility of EPO, which synergistically boosted its transcription.256 Prepro-B is the first stage of common lymphoid progenitor cells differentiation into B-lineage cells. Recent research indicated that PTEN promoted the B lineage differentiation mainly by suppressing PU.1. PTEN loss blocked the prepro-B to B cell differentiation while promoted it differentiation into T and myeloid lineages depending on chromatin accessibility remolding.257 Atoh1 is a master TF to determine the fate specification of cochlear hair cells. Through ATAC-seq analysis, Luo et al. identified two novel enhancers of Atoh1 to regulate its expression in cochlear hair cells.258 What’s more, the chromatin accessibility plays an important role in the adaptation of people to the local environment.259
It is difficult and restricted to investigate human development in vivo, and the organoid technology can solve part of the problems. Wahle et al. established a human retinal organoid to mimic the retinal development. According to the chromatin accessibility profile, they inferred the gene regulatory network contributing to retinal organoid development.260 Kanton et al. established stem cell-derived cerebral organoids of human, chimpanzee, and macaque to reveal the substantial changes of human brain during evolution. They analyzed the cell composition and reconstructed the entire course of cerebral differentiation trajectories in organoids by scRNA-seq. They found that the neuronal development of human was slower than that of the other two primates. The pseudotemporal alignment of differentiation paths indicated that the expression of human-specific genes resolved to distinct cell states are along progenitor-to-neuron lineages in the cortex. Further, using scATAC-seq, they confirmed that the variation of chromatin accessibility contribute to these differences.261 Similarly, Trevino et al. developed cortical and subpallial spheroids derived from human iPSCs. By comparing the expression and chromatin accessibility profiles with primary human tissues, both of these spheroids can mimic the early development of human forebrain.262 However, the research of Herring et al. revealed that there are few postnatal maturity neurons in the long-term brain organoids,243 which indicates that the culture system of organoids requires improvement to better mimic the in vivo situation.
Chromatin accessibility in tissue regeneration
Most of the somatic cells possess a restricted regenerative capacity, and there are significant distinct in each organ. When injured, the tightly regulated regeneration process will be stimulated. In this part, we will discuss the functions and mechanisms of chromatin accessibility in the regeneration processes in different organs.
Heart regeneration
The matured cardiomyocytes are almost nonproliferative, blocking cardiac regeneration after injury. The zebrafish heart has a robust regenerative capacity.263 Cao et al. constructed the chromatin accessibility, H3K27ac modification, and expression landscapes of zebrafish heart during regeneration. They identified multiple enhancers with varied accessibility and the corresponding TFs.264 Wang et al. indicated that Keratin5 (Krt5) altered genome accessibility at the loci of Pax3a, Acta2, and Bmp4 to promote their expression and zebrafish heart regeneration.265 Beisaw et al. also found that the AP-1 binding motifs were enriched most significantly in the gain accessibility regions during zebrafish heart regeneration. Inhibiting AP-1 leaded to defects in cardiomyocyte proliferation and decreased chromatin accessibility at cardiac regeneration genes.266 Quaife-Ryan et al. constructed the transcriptome and chromatin landscape of infarcted and noninfarcted neonatal and adult mouse hearts. The chromatin accessibility largely mirrored the transcriptional state. Besides, they confirmed that the neonatal hearts had a higher regenerative capacity than that of adult hearts.267 Boogerd et al. revealed that ARID1A suppressed YAP-induced proliferation of cardiomyocytes by remodeling the H3K27ac landscape in mice. Inhibiting ARID1A promoted the proliferation of border zone cardiomyocytes after ischemic injury.268
Cardiomyocytes reprogrammed from other cell types is a potential strategy for cardiac regeneration.269 Zhang et al. reprogrammed human urine cells into cardiomyocyte-like cells by heterogenous expressing MEF2C, MESP1, Tbx5, and MYOCD, but without GATA4. They found that these TFs, especial MYOCD, remodeled the chromatin accessibility landscape to facilitate the expression of multiple cardiac-specifc genes.270
Liver regeneration
As the metabolic center, the liver is more likely to be exposed to harmful substances, hence the liver maintains an extraordinary regeneration ability to restore its structure and function effectively after damaged. Regeneration failure will lead to severe liver diseases and complications. (Fig. 7)
During liver regeneration, the chromatin structure is dynamically regulated by many epigenetic events. When injured, the hepatocytes dedifferentiate into liver progenitor-like cells (LPLCs) to participate in liver regeneration. Sox2, Oct-3/4, Klf4, and c-Myc (also called Yamanaka factors or 4F) are the key factor that promote the reprogramming of differentiated cells.161 Hishida et al. demonstrated that inducible expression of 4F in hepatocytes specifically promoted partial reprogramming of differentiated hepatocytes into a progenitor state which showed a stronger proliferation ability. They found that 4F can promoted the expression of TOP2A to modify the chromatin accessibility of adult hepatocytes.163 Li et al. revealed that ARID1A increased the chromatin accessibility of LPLC-enriched genes and consistently facilitated the binding of YAP to boost the regeneration progress. ARID1A deletion hindered the regeneration of injured liver.271 However, an earlier research by Sun et al. found that the expression of ARID1A was reduced in regenerating tissues. Hepatocyte-specific deletion of ARID1A increased the tissue repair of injured liver. Compared with WT mouse, the proliferation ability, tissue damage, fibrosis, and organ function following chemical injuries or surgical resection were improved in ARID1A deficiency ones. Mechanically, the absented ARID1A was replaced by ARID1B in the altered SWI/SNF complex which has distinct function on targeting genes. Lack of ARID1A remolded the histone modification and chromatin structure. These variation block the entrance of the differentiation-related TFs, HNF4a and C/EBPα, as well as the cell cycle and mitosis repressive factor, E2F4, to the their target genes. Reduced H3K4me2 marks in the promoters of HNF4a and C/EBPα-targeting genes, while increased H3k4me2 and H3k27ac marks in that of E2F4-targeting genes were observed, which in line with the transcriptional activity of these genes. They also found that global ARID1A knockout potentiated the healing of injured ear soft tissue.49 These contradictory results may be due to their different liver injury molds or other unknown reasons, which confirmed the complex functions of ARID1A from another perspective. Additionally, in the Fah null mouse repopulation model, the chromatin structure of hepatocytes are altered significantly during liver regeneration. The chromatin accessibility of proliferation related genes, such as CTCF-targeting genes, were increased while that of hepatocyte differentiation and metabolism-related genes, such as HNF4α-driving genes, were decreased significantly in repopulating hepatocytes.272 DINO is a damage-induced lncRNA which amplifies p53 signaling by directly interacting with p53 and increases its stability.273 Khanal et al. demonstrated that knockout of nuclear receptor subfamily 2 group E member 3 (NR2E3) decreased the expression of DINO by reducing DINO chromatin accessibility and consequently lowered the p53 protein level. Compared with the WT mouse, the NR2E3 KO mouse showed more severe liver injuries and reduced recovery ability under hepatotoxicity.274 MDIG is a mineral dust exposure-induced gene that first identified in alveolar macrophages.275 The subsequent study confirmed that it is a JmjC domain containing protein and mediates the demethylation of H3K9me3 to H3K9me1.276 Recently, Du et al. demonstrated that liver-specific MDIG-deletion significantly prolonged the recovery process of both partial hepatectomy and carbon tetrachloride (CCl4)-treated mouse liver. Mechanically, MDIG promoted the OTX2-induced expression of Myc. The MDIG-induced H3K9me3 demethylation of OTX2 promoter enhanced its chromatin accessibility and subsequently facilitated the entrance of Myc to form a positive regulatory loop.151
Intercellular transdifferentiation plays an important role in maintaining organism homeostasis. Fibroblasts, with strong plasticity and proliferation ability, are the ideal doner cell for transdifferentiation. HNF4α and FoxAs play important roles in the development and homeostasis of the liver. Horisawa et al. indicated that FoxAs and HNF4α sequentially and cooperatively bind to the loci of liver-specific gene to induce the mouse embryo fibroblast (MEF) to hepatocyte reprogramming procession. The FoxAs, act as PTFs, increase the chromatin accessibility and recruit HNF4α to its target genes.156 Hepatic stellate cells (hPSCs) are intrahepatic resident fibroblasts. Ma et al. indicated that the chromatin accessibility, H3K27ac modification and gene expression pattern were different in human primary hepatocytes and hPSC-derived hepatocytes. The expression of thyroid receptor THRB had no significant differences in these two type cells, but THRB binding motifs were significantly enriched in primary hepatocytes compared with that in hPSC-derived hepatocytes. Thyroid hormone T3 treatment increased the binding of THRB with CYP3A4 proximal enhancer to promote its expression and subsequently facilitated hepatocytes maturation. Even so, they found that engrafting the hPSC-derived hepatocytes into undamaged liver of immunocompromised mice didn’t disrupt the normal histology of the liver.277 Other research also indicated that the fibroblast-derived hepatocytes had similar morphology and function with that of hepatocytes, which can apply for the reconstitution of damaged hepatic tissues.278,279 These research indicated that the fibroblast-derived hepatocytes are important tissue source for liver repair. Except transdifferentiation into hepatocytes, great many of researches have confirmed that forced expression of cell fate-determining TFs can convert fibroblasts into multiple cell types, such as neuronal cells and cardiomyocytes.280,281
Other cell types also can transdifferentiation into hepatocytes. Mukhopadhyay et al. confirmed that adult bone marrow progenitor cells, Lin-, participated in liver regeneration of hemophilia A mouse model through engraftment and lineage conversion.282,283 In the following work, they found that Lin− cells can be partially reprogrammed to hepatocyte-like cells. The Lin−-derived hepatocyte-like cell showed a distinct transcriptional pattern compared with the original Lin− cell. Active histone marks (H3K9Ac and H3K4me3) or repressive marks (H3K9me3 and H3K27me3) were enhanced or reduced, respectively, in the promoters of hepatic TFs, such as C/EBPα/β, HNF1α/3α/3β/4α/6, and GATA4, which were crucial for hepatic development. Further, they confirmed that the upregulated demethylase JMJD3, and down-regulated methyltransferase EZH2, were account for the reduced H3K27me3 modification and reprogramming of bone marrow progenitor cells to hepatocytes.103
Besides, biliary is an important part of liver. Multiple researches show that hepatocytes also can transdifferentiate into biliary epithelial cells (BECs). Seirup et al. revealed that the primary hepatocytes cultured in vitro undergo dedifferentiation gradually. They demonstrated that the epigenetic modification and chromatin accessibility of primary hepatocytes changed rapidly when cultured in vitro. Additionally, based on the areas of open chromatin, they identified the TFs involved in the different stages of dedifferentiation, including RXR, Fox, STAT, Fos, Jun, and MAF-related factor families.284 Katsuda et al. demonstrated that Sox4 promoted the reprogramming of adult hepatocytes to biliary both in vitro and vivo through altering the histone modifications, including H3K27ac, H3K4me1, and H3K4me3. Sox4 attenuated the activity of hepatocyte enhancers and evicted the hepatic TFs, HNF4a and RXRα, meanwhile opened chromatin in biliary-driving regions. They also found that Sox4 and Sox9 were targets of YAP, the key biliary-reprogramming driving factor.148 Merrell et al. indicated that the chromatin accessibility was changing during BECs reprogramming. Re-expression of BEC-specific genes and silence of hepatocyte-specific genes were distinctive characteristics during hepatocyte-to-BEC reprogramming. The expression panel of these genes are in accord with the open state of chromatin. The binding sites of hepatocyte-specific TFs, such as HNF4α, FoxA, and C/EBP, were more accessible in hepatocytes. Conversely, in BECs or reprogrammed cells, accessible sites were enriched for biliary-specific TFs, such as TEAD and HNF1β. Additionally, they revealed that AP-1 and NFκB played an important role in hepatocyte-to-BEC reprogramming.285 Besides, Har-Zahav et al. demonstrated that the global DNA methylation was reduced during liver-to-pancreas transdifferentiation, which increases the accessible of the pancreatic TF binding motifs. DNMT1 knockdown improved the efficiency of liver-to-pancreas transdifferentiation by increasing the expression of pancreatic-specific genes.78
Nervous system regeneration
Many diseases and traumatism may damage the nervous system. The regeneration of axons is essential for the functional recovery of the nervous system from injury. However, the axon growth activity is gradually sealed during neurons mature which is partially correlated with reduced chromatin accessibility of axon pro-regenerative genes.286 Some animals, such as zebrafish, possess a robust axon regeneration ability induced by injury. The regeneration progress is strictly regulated by the regeneration-associated signaling.287 Dhara et al. indicated that the chromatin accessibility landscape was varied during axon regeneration in zebrafish. The binding motifs of Jun were opened to facilitate the expression of target genes and regeneration progress.288 Although the regenerative capacity of the central nervous system (CNS) is restricted, the peripheral nerve has a robust regeneration and repair ability in many mammals. The dorsal root ganglia sensory neurons, containing a regeneration-competent peripheral axonal branch and a regeneration-incompetent central axonal branch, are the ideal material to investigate the mechanism of nerve regeneration.289 Palmisano et al. established the dorsal root ganglia injury model at the peripheral or central axonal branch to clarify the mechanism leading to the regeneration ability difference between the central and peripheral nervous system. The comprehensive analysis of RNA-seq, ATAC-seq, and ChIP-seq indicated that the regeneration-associated pathway was activated corresponding with chromatin accessibility, H3K9ac, H3K27ac, and H3K27me3 modification variation in the peripheral axonal injury, but there was no significant change in the central axonal injury.290
In addition to the organs mentioned above, chromatin accessibility also regulates the regeneration of other organs, such as lung,291 pancreas,78 kidney,292 and bone.293 Differentiation, dedifferentiation, transdifferentiation, and proliferation of specific cells play an important role in the homeostasis and regeneration of organs. As summarized above, chromatin accessibility variation has been shown to participate in these processes. Uncovering the function and mechanism of these progresses are of great significance for the treatment of dysplastic disease, as well as for the rapid and effective recovery or replacement of damaged organs.
Chromatin accessibility regulates function execution
Each organ has its own unique function, and the expression of many tissue-specific genes plays an important role in the maintenance of their functional homeostasis. Multiple researches have indicated that chromatin accessibility is a master regulator of tissue-specific expressed genes.
Liver function
Liver is one of the most vital organs in vertebrate, performing multiple functions, including detoxification, lipid metabolism, glycometabolism, bile secretion, hematopoiesis, immunity homeostasis, and so on.
The function of liver is precisely determined by multiple factors, such as mental state, hormonal readiness, positive and negative feedbacks of metabolic materials, and so on. Many metabolic regulatory signals have been shown to control the expression of downstream genes by regulating chromatin accessibility. The nuclear receptor superfamily, including LXRs, FXRs, and peroxisome proliferator-activated receptors (PPARs), are master regulators of metabolic progresses. Bideyan et al. indicated that knock-out of LXRα/β widely remolded the chromatin accessibility and transcription patterns in mice liver. In addition to regulating the expression of their target genes, loss of LXRα/β can also remold the binding motif accessibility of other TFs, such as reducing the accessibility of CTCF/CTCFL, HNF6/CUX2, HNF1/HNF1β, ATF4/CHOP, nuclear receptor family, and Fox family, while enhancing the accessible of DR1/4 motif and nuclear receptor half-site motif (AGGTCA).294 Kain et al. indicated that ligand-activated FXR and LXRα enhanced the expression of target genes by increasing their chromatin accessibility. Mechanically, FoxA2 bound to highly condensed chromatin and interacted with ligand-activated FXR and LXRα to remold the chromatin structure. Additionally, FoxA2 reduced the competing binding of PPARα to ensure the activation of FXR and LXRα signals.155
Glucocorticoids (GCs) are useful therapeutic for many diseases by activating their receptor GR. NCoR1 and NCoR2 are two nuclear receptor corepressors. Hauck et al. indicated that liver-specific dual loss of NCoR1 and NCoR2 leaded to hypoglycemia in mice. They found NCoR1 and NCoR2 maintained the accessibility of GR binding motif to regulate glucoregulatory genes.295 Grøntved et al. demonstrated that occupied by C/EBPβ maintained the permissive of most GR binding sites, required for the GC-induced response.296 In mouse liver, the HNF4α binding motifs and GRE lie adjacent within open chromatin regions. Loss of HNF4α decreases the recruitment of GR to GRE by reducing chromatin accessibility which consequently remodels the expression of GCs-responsive genes.297 ANGPTL4 increases the transportation of triglycerides from white adipose tissue to liver, participating in hypertriglyceridemia and hepatic steatosis.298 Koliwad et al. indicated that ANGPTL4 was a direct target of GR. Dexamethasone treatment increased the expression of ANGPTL4, coinciding with increased DNase I susceptibility and histone H4 acetylation of GRE in ANGPTL4 promoter.299 Besides, earlier research has confirmed that estrogen increase the promoter accessibility of vitellogenin gene B1 in male xenopus laevis hepatocytes.300
Goldstein et al. confirmed that fasting massively remolded the hepatic chromatin accessibility. Thousands of fasting-induced or -repressed DHSs following 24 h of food deprivation were clarified which were strongly overlapped with enhanced or reduced H3K27ac modification. C/EBPβ, GR, PPARα, and cAMP responsive element binding protein 1 (CREB1) are key TFs to regulate the expression of fasting-induced genes.301 Korenfeld et al. also found that fasting promoted the expression of many amino acid catabolism genes, most of which were in line with open chromatin.302 Caloric restriction improves metabolic adaptation and reduces the occur of many age-associated diseases.303 Fan et al. indicated that short-term caloric restriction remolded the chromatin accessibility profile and HNF4α entrance locus to reprogram fatty acid and bile acid metabolism.304 SMARCDs, the SWI/SNF factors, also regulate nutrient and metabolic signaling in liver.305
Fetal liver is one of the most important early hematopoietic organs. Farlik et al. constructed the hematopoietic lineage based on DNA methylation maps. They found that hematopoietic cells derived from different tissues, and in different differentiation stages exhibited distinct DNA methylation profiles. Compared the chromatin accessibility dataset which published by another research,306 they found that in all cell types, the HSC-specific accessible regions had low DNA methylation level. The reduced DNA methylation of differentiation-related regions, which showed higher accessibility, were only detected in differentiated cells but progenitors. Meanwhile, the differentiated cells derived from different progenitors had different hypomethylation patterns.307 Zhu et al. determined the differentiation trajectory from hemogenic endothelial cells to HSC precursors by single-cell RNA-seq and ATAC-seq. They found that Runx1 contains two promoters which have distinct accessibility in different differentiation stages. It regulated the transition efficiency of hemogenic endothelial precursors to hemogenic endothelial cells.308 Applying similar method, Anna Maria Ranzoni and colleagues identified the differentiation trajectory from HSCs to multipotent progenitors and confirmed three highly proliferative oligopotent progenitor populations. They also found that the changes of chromatin accessibility preceded transcriptional priming during HSCs differentiation.309 Hemogen, also called erythroid differentiation-associated gene (EDAG), plays an important role in promoting proliferation and differentiation of hematopoietic cells through activating GATA1, the critical TF for erythropoiesis. Guo et al. indicated that in fetal liver cells, hemogen enhanced the accessibility of GATA1/LDB1 complex-binding motifs by recruiting coactivator SMARCA4 while expelling corepressor NuRD complex.46
The liver microenvironment is involved in regulating immune cell differentiation. The epithelial cells can’t prime circulating naïve CD8+ T cells for the hindering of endothelial barrier limiting antigen, but the liver epithelial cell is an exception for its slow blood flow, fenestrations, and basement membrane loss, allowing the direct interaction of CD8+ T cells with hepatocytes, which usually leads to T cell unresponsiveness and dysfunction.310 Bénéchet et al. indicated that hepatocyte-activated CD8+ T cells exhibited distinct chromatin accessibility and transcriptome compared with normally activated CD8+ T cells, triggering a defective differentiation program.311 Besides, hemolysis releases toxic cell-free hemoglobin and heme, while macrophages phagocytize damaged erythrocytes to reverse the toxic effects.312 In mouse and human liver, heme exposed macrophages transformed into erythrophagocytes to suppress inflammatory response. Based on the alterations of chromatin accessibility and transcriptome, they found that heme/NRF2 signaling drove the transformation progress.313
Brain function
Memory is one of the important functions of the brain. Feierman et al. indicated that the H2B variant H2BE is enriched at the promoter regions of synaptic genes, increasing their chromatin accessibility and expression to regulate long-term memory.314 The immune homeostasis in the brain is determined by many immune cells, such as microglia and CD4+ T cells. Li et al. indicated that the neuropsychiatric-disorder-associated miRNA miR-137 regulated adult microglial homeostasis and brain function by remodeling chromatin accessibility. MiR-137 competitively bound with Pu.1 to block its binding with chromatin, which consequently inhibited the expression of Jun dimerization protein 2 (Jdp2) to impair phagocytosis and pro-inflammatory response in microglia.165 Recently, Elizaldi et al. identified a distinct subset of CCR7+ CD4+ T cells that was similar with the lymph node central memory cells. The chromatin accessibility features at the Bcl6, CD28, and CCR7 loci were exceeding resembled in both types of cells. Reducing the CCR7+ CD4+ T cell frequencies in the brain significantly activated microglial and neurodegenerative pathways.315 Besides, Chang et al. demonstrated that long light exposure remodeled the chromatin accessibility in the hypothalamus of male quail to regulate their reproductive axis activation.316
The chromatin accessibility also regulated the function of heart,317 ovary,318 lung,319 and pancreas.317 Currently, investigation focusing on the chromatin accessibility and physiological function execution of different organs is still lacking. However, the chromatin accessibility variation under pathological condition can give us many clues to speculate the truth during the physiological function execution.
Chromatin accessibility regulates the circadian rhythm
The expression of many genes exhibits circadian rhythmicity. The heterodimer BMAL1::CLOCK is one of the core mammalian circadian clock TFs, driving the first circadian rhythms feedback loop and REV-ERBs, which drives the second feedback loop by inhibiting BMAL1.320 Zhu et al. demonstrated that during the circadian cycle, ROR and/or BMAL1 promoted decondensation of global chromatin, facilitating REV-ERB entrance to repress circadian gene expression.321 Recently, Qu et al. demonstrated that HNF4α remolded the liver-specific circadian transcriptome. Mechanically, HNF4α facilitated the binding of BMAL1 to its target genes by enhancing their H3K4me1 and H3K27ac modification, as well as chromatin accessibility. Besides, they also found that HNF4α has distinct function on the rhythm regulation in other tissues, which hinted that HNF4α was a novel rhythm regulator.160 Takeda et al. indicated that RORγ increased the H3K9ac modification and accessibility of Npas2 promoter, cooperating with REV-ERBα to regulate the circadian expression of Npas2.322 Hassan et al. demonstrated that FXR deletion reduced the accessibility of some TF binding motifs, including NF-Y/CBF, Sp1, and Klf, which consequently leaded to dysregulation of the bile acid metabolism and circadian rhythm pathways in hepatocyte.323 Besides, air pollution (PM2.5) exposure disrupted the chromatin accessibility and expression profiles of circadian genes in liver and brown adipose tissue through remolding histone acetylation.324 There researches demonstrated that circadian rhythms and chromatin accessibility mutually regulated each other, but more mechanisms require to be revealed.
Chromatin accessibility regulates aging
Aging is a physiological process that regulated by genetic, epigenetic, environmental, and other factors. The transcriptome, histone modification, and chromatin accessibility are changing with aging.
Early research has indicated that the liver chromatin of young rats is more sensitive to different DNA endonucleases than that of old rats, indicating globally decreased chromatin accessibility with aging.325 Chen, et al. found that the overall variant of histone H3 occupancy was limiting, however, the decreased H3 occupancy regions were enriched in pro-inflammatory genes.326 Ding et al. also indicated that the increased chromatin accessibility was due to the upregulation of inflammation and immune response genes in the senescent liver progenitor-like cells (HepLPCs). Meanwhile, they found that the depletion of FOS like 2 (FOSL2) delayed HepLPCs aging, indicating that FOSL2 may be a key TF which regulated by aging-induced chromatin accessibility.327 Bozukova et al. found that the chromatin accessibility of promoter regions was increased in aging murine liver, however the transcriptional output was not increased correspondingly. They found that promoter-proximal pausing of RNA polymerase II was significantly reduced in aging liver.328 Additionally, Hu et al. revealed that the expression of histone chaperone, NAP1L2 was in creased in the senescent BMSCs both in vitro and in vivo. Mechanically, NAP1L2 recruited SIRT1 to the promoter of osteogenic genes, such as Sp7, Runx2, and Bglap, reducing their H3K14ac modification, chromatin accessibility, and expression to suppression the differentiation of BMSCs into osteogenic.134 Hu et al. indicated that ZKSCAN3 inhibited senescence of human mesenchymal stem cells (hMSCs) by maintaining the stability of heterochromatin. However, it is downregulated in the senescent cells which increased the chromatin accessibility and gene transcription leading to repetition-induced sequences.329
Sex-related chromatin accessibility variation
Multiple researches have indicated that there are many sex-determined differences in chromatin accessibility and gene expression patterns. The distinct pituitary growth hormone (GH) secretion and plasma GH profiles, which are pulsatile in males and persistent in females, are the important reason for sex-based differences of gene expression pattern in liver.330 Signal transducer and activator of transcription 5 (STAT5) is the critical TF that responds to the periodic stimulation of GH.331 Rampersaud et al. indicated that pulsatile GH regulated the male-biased chromatin accessibility to permit the periodic binding of STAT5. They also found that the binding of Bcl6, CUX2, FoxA1, and FoxA2 to their target locus was sex-dependent.332 Besides, the chromatin accessibility, TF binding, and expression of long intergenic noncoding RNAs (lincRNAs) were distinct in males and females, which were regulated by GH.333 Additionally, Hao et al. demonstrated that the expression pattern of miRNAs in liver are characterized by sex-biased chromatin accessibility. For example, the expression of miR-1948 (locating in male-biased DHSs) and miR-802 (locating in female-biased DHSs) were higher in males and females, respectively.334 These studies indicate that the differences in chromatin accessibility are one of the secrets between males and females.
As mentioned above, chromatin accessibility plays an important role in a variety of physiological processes. However, there still are many aspect required to reveal, such as the variation of chromatin accessibility changes during cognitive formation, emotion changes, environmental perception, and so on. Besides, at present, more and more attention has been paid to the role of lifestyles on our health, hence it’s important to investigate the different lifestyles, such as exercise, routine, diet, and stress induced chromatin accessibility variation. Elucidating their detail functions and mechanisms are of great significance for the maintenance of human health and the treatment of different diseases.
Chromatin accessibility in pathological processes
Epigenetic abnormality is one of the major causes for many diseases, which often leads to anomalous change of chromatin accessibility. The chromatin accessibility is significantly altered in various diseases, indicating that it is an important regulator of disease occurrence and development. In this section, we will discuss the chromatin accessibility variation in different diseases (Fig. 8).
Chromatin accessibility variation is involved in multiple pathological processes. The chromatin accessibility state is dysregulated in multiple diseases. The aberrant chromatin accessibility widely aggravates the initiation and progression of numerous diseases, such as cardiovascular disease, cancer, digestive system disease, nervous system disease, endocrine disease, respiratory disease, and infectious disease. This picture was drawn by Freescience
Chromatin accessibility variation in cardiovascular disease
Cardiovascular diseases are the major cause of mortality across the world. Heart failure is the most common symptom. Dilated cardiomyopathy is one of the major causes of severe heart failure and heart transplantation, which is closely associated with LMNA mutation.335 Chromatin accessibility analysis revealed that the TF TEAD1 was critical for cardiomyocyte mature. However, the mutated LMNA gene, coding Q353R-Lamin A/C, hijacked TEAD1 at the nuclear membrane, causing downregualtion of TEAD1 target genes to dysregulate cardiac development.336 Shi et al. indicated that the expression of WTAP was decreased both in human and mice heart failure tissues. Specific deletion of WTAP in mice cardiomyocyte (WTAP-CKO) caused dilated cardiomyopathy and even neonatal death. They found that WTAP loss remodeled the chromatin accessibility in the heart. The accessibility of Mef2a/b/c/d binding motifs was decreased while that of TEAD4 was increased in the WTAP-CKO mice. Additionally, WTAP can promote the expression of Mef2c by binding to its promoter directly.337 Myocardial infarction is another important inducer of heart failure. Kuppe et al. constructed a spatial multi-omic map of human myocardial infarction. Through snRNA-seq and snATAC-seq analysis, they found that MYH7 and NPPB were upregulated in the pre-stressed and stressed ventricular cardiomyocytes, respectively. Meanwhile, ANKRD1 was increased in both states.338 MYH7, NPPB, and ANKRD1 are the confirmed cardiomyocyte-associated stress genes. Hence, their investigation provided an important reference for the research on myocardial infarction. Besides, Kirkland et al. indicated that the loss of Lamin C with age reduced the global chromatin accessibility, downregulated the cytoskeletal regulators and myogenic transcription factors, resulting in age-dependent cardiac decline.179 Mitral valve prolapse is a fatal heart disease. Kyryachenko et al. established the chromatin accessibility atlas in human pathogenic and nonpathogenic mitral valves. They identified many mitral valve prolapse risk loci based on the chromatin accessibility variation which provided an indicator for mitral valve prolapse research.339 Ventricular arrhythmia is one of the symptoms of long QT syndrome type 7, which is mainly caused by the mutation of KCNJ2 gene.340 Recently, Chen et al. repaired the mutant in hiPSCs derived from human patients. Through comparing the chromatin accessibility between mutant or repaired cardiomyocytes during differentiation progress, they found that the accessibility of the TF ZNF528 was significantly increased in the repaired cardiomyocytes. Inhibiting ZNF528 reduced the expression of the pathogenicity associated genes, KCNJ2, CTTN, and ATP1B1.341
Vascular dysfunction is another main type of cardiovascular disease. Cheng et al. indicated that specific knockout of ZEB2 in smooth muscle cells accelerated the formation of atherosclerotic plaques in human coronary arteries. ZEB2 insufficiency disrupts both NOTCH and TGFβ signal pathway by remodeling the chromatin accessibility of related genes.342 Besides, hypertension widely remodels the chromatin accessibility and expression profiles of vascular smooth muscle and endothelial cells.167,343
Cardiovascular diseases always lead to systemic aberrant that involve multiple organs. Uncovering the function and mechanism of chromatin accessibility alteration in cardiovascular diseases will improve the treatment of these diseases.
Chromatin accessibility variation in cancer
Cancer is another leading threaten to human health and life. During the initiation and progression of cancer, the expression profiles of tumor cells are altered significantly which are always in line with the chromatin accessibility state. In 2018, Chang group established the chromatin accessibility landscape of 23 primary human cancers, and screened out lots of cancer risk locus.344 In this part, we will discuss the functions and mechanisms of chromatin accessibility alteration in different cancer malignant phenotypes (Fig. 9).
Chromatin accessibility participates in the malignant progression of different cancers. The chromatin accessibility state is dysregulated in multiple cancers which promotes many malignant progression, including tumorigenesis, proliferation, metastasis, chemoresistance, angiogenesis, stemness, immune escape, pro-tumor inflammation, pro-tumor senescence, metabolic reprogramming, heterogeneity, and so on. This picture was drawn by Freescience
Tumorigenesis
Tumorigenesis is a slow and complex process, which is mainly caused by the gradual activation of oncogenic genes and the inactivation of tumor suppressor genes. These progresses are determined by genetic, epigenetic, environmental, and other factors, which always remold the chromatin accessibility in cancer cells.
In liver, MASH is the leading tumorigenesis inducer. Dechassa et al. identified that the accessible chromatin atlas was altered in the MASH-derived HCC. ANXA2 and PDLIM7 were upregulated most significantly which corresponded with enhanced active chromatin marks, H3K4me1 and H3K27ac.345 Wu et al. indicated that YAP-TEAD complex promoted the expression of TET1. Consequently TET1 enhanced the DNA demethylation, H3K27ac modification, and chromatin opening of YAP-targeting genes by directly interacting with TEAD. The loss of TET1 reversed the YAP-derived tumorigenesis.81 TCPOBOP (1, 4-bis [2-(3, 5-dichloropyridyloxy)] benzene) is a highly specific agonist ligand of the nuclear receptor constitutive androstane receptor (CAR).346 TCPOBOP-induced activation of CAR triggers diversity of pathogenic responses in liver. Lodato et al. demonstrated that TCPOBOP induced widely variation in chromatin accessibility and gene expression. The changes in gene expression were in line with the accessibility of chromatin, although the transcription priming of some genes lag behind the chromatin variation.347 BAP1 is H2AK119 mono-deubiquitinase. Artegiani et al. indicated that BAP1 loss resulted in aberrant morphology of human ductal liver organoids which showed reduced cell polarity, disrupted epithelium organization, and enhanced motility. Mechanically, BAP1 loss promoted H2AK119ub modification and perturbed the transcriptome pattern of junctional and cytoskeleton components, through regulating the chromatin accessibility of related genes. Restoration of BAP1 activity can rescue its deficiency-induced disorders.120
The SWI/SNF complexes can promote or suppress cancer development by binding with different cofactors.348 ARID1A is originally hypothesized to be an anti-cancer gene, for it’s the most frequently mutated SWI/SNF component in cancer.349 Sun et al. demonstrated that ARID1A has dual function on initiation and development of liver cancer. It promoted tumor initiation in multiple mouse models through enhancing cytochrome P450 induced reactive oxygen species.48 In contrast, in the AKT/NRAS-driven hepatocarcinogenesis model, mTORC1 promoted YAP-induced transcriptome by remodeling chromatin accessibility in an ARID1A-dependent manner. Mechanically, mTORC1 directly interacted with ARID1A and enhanced its ubiquitination-mediated degradation. Ectopic expression of ARID1A can antagonize AKT/NRAS-driven hepatocarcinogenesis.52 These research indicates the complex function of ARID1A in tumor initiation.
Additionally, chromatin accessibility also regulate the initiation of other tumors, such as lung cancer,350 breast cancer,350 pancreatic cancer,351 colorectal cancer,352 and prostatic cancer.353 These researches indicate that chromatin remodeling is a significant feature of tumorigenesis and facilitates tumor initiation through various mechanisms.
Tumor proliferation
Continuous proliferation is the most significant malignant phenotype of tumor cells. Sun et al. have demonstrated that ARID1A promoted liver tumorigenesis. In the same work, they found that ARID1A inhibited tumor growth and metastasis through decreasing the chromatin accessibility and expression of metastasis-related genes.48 MASTL has been report to regulate mitosis in many cancer cells. Liye Cao and colleagues demonstrated that MASTL level was significantly increased in HCC tissues compared with non-tumor liver tissues. IL-6 and TNF-α were able to induce MASTL expression and promoted the proliferation of HCC cells. Mechanistically, upon IL-6 and TNF-α stimulation, the H3K4me3 modification and chromatin accessibility of MASTL promoter were increased remarkably.145 NASP is a regulator of cell cycle and proliferation. Kang et al. demonstrated that the expression of NASP was increased in cancer tissues compared with normal tissues and required for the cell proliferation and tumor formation. NASP knockdown decreased H3K9me1 modification and enhanced chromatin accessibility globally which induced the expression of tumor suppressor BACH2 and RunX1T1 to induce replication defect and enhance apoptosis of HCC cells.147 The nuclear matrix protein hnRNPU participates in three-dimensional architecture maintenance of hepatocyte genome.354 Xu et al. demonstrated that lncRNA RP11-386G11.10 enhanced lipid accumulation by upregulating hnRNPU, promoting the proliferation and metastasis of HCC cells. Additionally, ZBTB7A, the downstream of hnRNPU, promoted RP11-386G11.10 transcription to form a positive feedback loop.355 The extracellular matrix (ECM), providing structural and biochemical support to tumor cells, is also closely related with tumor progression. MDIG has been confirmed to promote the regeneration of damaged liver by remolding the H3K9me3 modification and chromatin accessibility of OTX2.151 Additionally, researches indicated that the expression of MDIG was usually higher in HCC tissues than normal ones and promoted the growth, metastasis, and drug resistance of HCC cells by promoting the expression of p21 and cell division cycle 6 (CDC6) in a H3K9me3 demethylation-dependent manner.356,357
HCC and iCCA are the two main histological subtypes of primary liver cancer derived from hepatocytes or cholangiocytes, respectively. Although they share several common risk factors, there still are distinct characteristics. Recently, Craig et al. compared the chromatin accessibility differences of HCC and iCCA through scATAC-seq. The POU and ETS factors binding motifs were enriched in iCCA while that of nuclear receptors were enriched in HCC.358 Besides, Li et al. found that the expression of microfibrillar-associated protein 5 (MFAP5), a ECM glycoprotein, was increasesed in intrahepatic cholangiocarcinoma (iCCA) tissues compared with para-carcinoma tissues. MFAP5 promoted the proliferation of iCCA cells by activating NOTCH1 pathway both in vitro and in vivo. Additionally, they indicated that MFAP5 can remold the chromatin accessibility. The accessible peaks were in line with the upregulated genes, which can be reversed by the NOTCH inhibitor FLI-06.359 Recently, Peng et al. indicated that the expression of ΔNp63α was also increased in iCCA tissues and promoted the proliferation, migration, and invasion of iCCA cells. The existence of ΔNp63α increased the accessibility of its binding sites to change the transcriptome. Additionally, ΔNp63α was able to bind with the chromatin structural protein YY1 in iCCA cells.360
Additionally, research indicated that loss of H4K20me3 modification correlated with poor prognosis of colorectal cancer (CRC).361 Boonsanay et al. demonstrated that the expression of lysine methyltransferase SUV420H2 was decreased in right-sided colon cancer and correlated with poor outcomes. SUV420H2 insufficiency reduced the H4K20me3 modification and increased chromatin accessibility of the Wnt-related genes to promote the growth of cancer organoids.104 Beside, chromatin accessibility also regulates the proliferative signaling in lung cancer,362 pancreatic cancer,363 prostate cancer,364 breast cancer,365 and so on.
Tumor metastasis
Metastasis is the leading cause of cancer-related death. The epigenetics and epitranscriptomics of metastatic tumor cells are altered remarkably. As the most frequently integrated viral gene of HBV, HBx plays an important role in remodeling epigenetic modification during HCC pathogenesis. Zheng et al. indicated that HBx/ETV4/DVL2/β-catenin axis promoted the migration and invasion of HCC cells. Mechanically, HBx promoted the transcription of ETV4 by increasing the H3K27ac modification and chromatin accessibility of super-enhancers upper ETV4 promoter. As a TF, ETV4 promoted the transcription of DVL2.144 Wu et al. found that the expression of lncRNA lncMER52A was increased in HCC tissues. The increased expression was due to the enriched H3K4me3 and H3K27ac modification which facilitated YY1 binding to its promoter. Subsequently, sufficient lncMER52A promoted the migration, invasion, and metastasis of HCC cells by suppressing the ubiquitin-mediated degradation of p120-catenin.366 Huang et al. indicated that the deficiency of TFAM increased the metastasis of liver cancer. TFAM loss can remold the chromatin accessibility by inducing the polymerization of nuclear actin. The accessible chromatin peaks were enriched in the locus of IL‐6, fibronectin 1 (FN1), TGFβ, integrin β3 (ITGB3), and so on.181 Besides, in contrast to promoting tumor initiation, presence of ARID1A inhibited HCC metastasis.48
Additionally, the upregulated HDAC8 promoted the brain metastasis of melanoma by enhancing the accessibility of c-Jun binding motifs. It’s interesting that, as a histone deacetylase, HDAC8 increased the H3K27ac modification at these regions by inactivating the acetyltransferase P300. Hence, the deeper mechanisms need to be elucidated.367 Through CRISPR-Cas9 screening, Pierce et al. confirmed that LKB1 was a master regulator of chromatin accessibility. Its mutation globally remodeled the chromatin accessibility landscape to drive metastatic phenotype in lung cancer.368 What’s more, in breast cancer,369 pancreatic cancer,370 and colorectal cancer,371 the chromatin accessibility variation also is a master indicator and regulator.
Tumor cell stemness
In tumor tissue, cancer stem cells (CSCs) are a small group of cells which can promote the treatment resistance, recurrence, growth, and metastasis of many tumors. Many investigations have confirmed that chromatin accessibility variation plays an important role in regulating the stemness of tumor cells.
Wang et al. found that loss of ARID1A increased the proliferation, self-renewal and stemness of hepatocytes. ARID1A deficiency promoted the expression of many CSC-like markers through modifying the chromatin accessibility. Among the verified genes, Jag2 was a key gene that accountable for ARID1A deficiency induced stemness of HCC cells.50 HBV infection can hijack the gene expression system of host to ensure viral gene expression and replication, such as inhibiting the expression of RNA helicase DDX5.372 The HBV-induced expression of miR106b~25 and miR17~92 directly targeted the 3′UTR of DDX5 to suppress its expression. DDX5 knockdown enhanced viral biosynthesis and CSC-like properties of hepatocytes. Mechanically, DDX5 loss increased the expression and chromatin accessibility of Wnt pathway-associated genes. Inhibition of miR106b~25 and miR17~92 restored DDX5 expression in HBV-infected hepatocytes.373 Song et al. reported that in human liver CSCs, circHULC increased the expression of CARM1 by enhancing the interaction of RNA polymerase II with the CARM1 promoter and promoter-enhancer binding loop. The increased CARM1 methylated PKM2 which consequently increased the transcription of SIRT1 to promote the growth of liver CSCs by enhancing autophagy.374 The increased promoter accessibility of CBX2 and CEP55 facilitated their expression which consequently triggered stem cell-like phenotype of HCC cells.375 Besides, the hypoxia condition and PLD2 overexpression increased the accessibility of stemness genes, promoting the stemness and chemoresistance of ovarian cancer cells.376 Hagiwara et al. indicated that MUC1-C, interacting with NRF2 and PBRM1, increased the chromatin accessibility and expression of SLC7A11, G6PD, and PGD to maintain the stemness of prostate cancer cells.377
The existence of CSCs is a major challenge in cancer therapy. Now, we can confirm that the chromatin accessibility of CSCs is significantly different with that of common tumor cells, which brings us some promising directions for targeting CSCs.
Tumor immune escape
Cancer cells can remold the immune microenvironment to promote cancer progression. The chromatin accessibility is involved in the formation of immunosuppressive microenvironment, resulting in tumor tolerance.
Infiltration of regulatory T cells (Tregs, expressing FoxP3) is the main trigger of cancer immune escape. The CC chemokine receptor 4 (CCR4) is a crucial chemokine receptor that mediates Tregs infiltrating into the tumor microenvironment through binding to its ligands CCL17 or CCL22.378 Gao et al. demonstrated that CCR4+ Tregs was the predominant Tregs in HBV+ HCC, and displayed enhanced immunosuppressive ability. The chromatin accessibility of CCR4- and CCR4+ Tregs were distinct, and the expression and promoter accessibility of FoxP3, CCR4, HNF1α, and programmed cell death 1 (PD-1) were increased in CCR4+ Tregs. CCR4 N-terminal extracellular recombinant protein (N-CCR4-Fc) or C-021 (CCR4 antagonist) can attenuate CCR4+ Tregs-mediated immunosuppression both in vitro and vivo.379 Besides, the H3K27ac and H3K4me1 modification landscapes as well as chromatin accessibility were significantly distinct in tumor-infiltrating Treg cells (TITRs) and peripheral blood Treg cells (PBTRs). The expression of genes involved in immune response, cell migration, and leukocyte differentiation were positively concordant with the chromatin accessibility. The accessible motifs that recognized by AP-1 family at the enhancer regions were enriched in TITRs.380 Yang et al. revealed that the H3K27ac and H3K4me1 modification landscapes were different in HDAC8 high and low HCC cells. HDAC8 inhibited the infiltration of CD8+ T cells by reducing the expression of CCL4, one of the T cell-trafficking chemokines. It reduced the H3K27ac modification of CCL4 enhancer. Knockdown or pharmacological inhibition of HDAC8 increased the infiltration of CD8+ T cells and enhanced the therapeutic effect of PD-L1 blockade for HCC.132 Zhang et al. found that combined treatment of DNMT and EZH2 inhibitors inhibited proliferation of human HCC cells and upregulated antitumor immune response. The DNMT and EZH2 inhibitors significantly decreased the global promoter DNA methylation and increased the chromatin accessibility, which strongly promoted the expression of immune response and interferon-stimulated genes.77 Zhang et al. reported that WD40 repeat-containing protein 6 (WDR6) reprogrammed the tumor immune microenvironment by increasing the expression of TNF-α to promote the growth and metastasis of HCC. It suppressed UVRAG-initiated autophagic degradation of p65 to promote TNF-α expression. Knockdown of WDR6 decreased the chromatin accessibility of the TNF-α locus in Hepa1-6 cells. TNF-α supplementation can revert the WDR6 knockdown reduced infiltration of myeloid-derived suppressor cells.381 Chromatin assembly factor 1 (CAF-1), which governs histone H3.1 incorporation into chromatin to facilitate heterochromatin establishment, is crucial for DNA replication during mitosis. Recently, Chan et al. demonstrated that the expression of CAF-1 was increased in HCC. CAF-1 deficiency enhanced antitumor immune response and the treatment effect of immune checkpoint inhibitor. Mechanically, knockout of CAF-1 increased the chromatin accessibility of endogenous retrovirus elements (ERVs), also known as viral mimicry, which activated dsRNA viral sensing and STING pathways in HCC cells.382
The innate myeloid cells, including monocytes and macrophages, can be treated to strengthen their immunocompetence.383 Wang et al. found that influenza A virus (IAV) infection-treated macrophages gained tissue-specific and long-lasting antitumor immunity and resistant to cancer-induced immunosuppression. They revealed that IAV treatment remodeled the chromatin accessibility atlas of macrophages, among which the accessibility of immune activation genes were increased significantly.384
Tumor chemoresistance
Acquired resistance to chemotherapy is another challenge in cancer treatment. Tumor cells in the senescent state tend to have higher chemotherapy resistance ability. Zhang et al. indicated that the senescent prostate cancer cells displayed reduced H3K9me2/3 and H3K36me2/3 modification, which were caused by the upregulated demethylases, KDM4. They confirmed that KDM4 promoted the expression of aging-related genes by increasing their promoter accessibility. Inhibiting KDM4 minimized chemoresistance and obtained better prognosis.115 Quiescent CSCs are highly resistant to chemotherapy. Yang et al. indicated that the nuclear protein DEK increased the global chromatin accessibility of breast CSCs. Thus, it activated and suppressed the Myc and P53 signaling, respectively, to trigger the quiescence exit of breast CDCs.385 Zhang et al. found that many chromatin regions in the anlotinib-resistant lung cancer cells were more accessible than that in the wild-type control, among which, the TFAP2A binding motifs were enriched most significantly. Knockdown TFAP2A sensitized anlotinib-resistant cells to anlotinib through weakening the PDGFR, VEGFR, and TGF-β signaling pathways.386 Altered metabolic pattern also contributes to chemoresistance. Ku et al. confirmed that the expression of PRMT1 was increased in pancreatic cancer, remodeling chromatin accessibility and promoting glycolysis. Inhibiting PRMT1 increased the susceptibility to gemcitabine both in vitro and vivo.387
Tumor heterogeneity
The prominent heterogeneity of cancer cells are also challenging the treatment. Distinct chromatin accessibility status were confirmed in different cancer cell lines, animal models, and patients, which correspond to different malignancy and treatment sensitivity.388 B6C3F1 and C57BL/6J mouse have distinct susceptible in phenobarbital-mediated liver tumor promotion.389,390 To clarify the mechanism leading to this discrepancy, researchers detected the chromatin accessibility variation of B6C3F1 and C57BL/6J mouse liver after treated by phenobarbital. It showed that these mouse strains maintained differential chromatin accessibility maps in livers after phenobarbital treatment. The B6C3F1-specific DHSs were mainly enriched in Klf4, Wnt/β-catenin, and Src binding motifs, while that enriched in C57BL/6J mainly included Ctnna1, as well as cell cycle, differentiation, and metabolism regulators. The expression pattern of mRNA and protein under phenobarbital treatment was consistent with the chromatin states.391 The human hepatoblastoma can divided into well-differentiated pediatric hepatoblastoma Class 1 (C1) and undifferentiated highly proliferative Class 2 (C2) which have favorable or poor prognosis, respectively. The C2 tumors have higher YAP1 expression, proliferation ability, and stem markers than that of C1 tumors.392 Smith et al. indicated that conditional expression of YAP1 cooperating with constitutive expression of β-catenin remolded the chromatin structure and drove the formation of hepatoblastoma in vivo, which can mimic the subtypes of human pediatric hepatoblastoma.393 In mouse hepatoblastoma model, Rodríguez et al. found that the accessibility of multiple tumor-related cis-regulatory elements had changed in a H3K27ac dependent manner. The YAP1-targeting enhancers were significantly enriched among these open chromatin.394 Wang et al. detected the single-cell transcriptomic, proteomic, and epigenetic landscapes of many HCC cell lines. They found that the gene expression patterns and chromatin accessibility were distinct in different cell lines. Based on the chromatin accessibility of the epidermal mesenchymal transformation (EMT) markers, the metastatic potential of HCC cell lines was highly correlated with their EMT scores.395 The cancer somatic mutations are tightly associated with DNA replication, and the replication timing are regulated by chromatin structure. The early replicating regions (ERR) are enriched within accessible chromatin and have higher transcription ability, while late replicating regions (LRR) are arranged in the inaccessible portions of the genome.396 Yaacov et al. indicated that the mutational profiles of tumors were different between ERR and LRR in multiple cancers including liver cancer. Both replication timing and chromatin accessibility can contribute to mutagenesis.397 Chirag Nepal, et al. indicated that many genes in HCC had alternative promoters which possess distinct DNA sequences, DNA methylation, chromatin architecture, H3K4me3, H3K27ac, H3K4me1, and H3K27me3 modifications. The comprehensive function of these variation leaded to the changes in the expression of numerous genes.398 Lymphoid neoplasms also are heterogeneous diseases.399 Corces et al. identified thirteen human primary blood cell types depending on the chromatin accessibility and transcriptional landscapes. They found significant heterogeneity in these cell types which reflected the hematopoietic differentiation and AML progression.306 Rendeiro et al. detected the chromatin accessibility of 88 chronic lymphocytic leukemia (CLL) samples from fifty-five patients which also presented remarkable heterogeneity. In particular, they found that the mutant status of IGHV genes can distinguish the aggressive of CLL. The chromatin accessibility and transcriptional atlas were distinct between the IGHV mutated and unmutanted samples.400 Similarly, Wang et al. identified the chromatin accessibility heterogeneity in sixty-one relapsed B-lineage ALL (B-ALL) patients.401 In small-cell lung cancer (SCLC), Yang et al. indicated that cancer cells derived from different metastatic model presented distinct chromatin accessibility landscapes which triggered by different transcription factors.402 Recently, Yang et al. measured the chromatin accessibility of twenty-three breast cancer cell lines which shown significant heterogeneity and each cell type expressed different key transcription factors to determine their unique chromatin accessibility atlas.403
At present, the rising of single-cell omics make us much more easier to understand the heterogeneity of tumor cells.404 The various differentiation status of tumor cells is one of the important reasons for tumor heterogeneity. Through applying single-cell multi-omics including ATAC-seq, Roehrig et al. identified a continuum of cell differentiation states in hepatoblastomas (HB).405 Desmoplastic small round cell tumor (DSRCT) is definitely mainly triggered by the chimeric transcription factor EWSR1::WT1 but still present heterogeneity. Henon et al. indicated that the accessibility of EWSR1::WT1 binding sites were variational in different DSRCT patients which drove the heterogeneity.406 By using similar investigation strategy, many researchers have identified the heterogeneity in other cancers, such as colorectal cancer,407 gastric cancer,408 and glioblastoma.409 Furthermore, the emergence of spatial single-cell omics allows us to reveal the heterogeneity of tumors meanwhile also to define their spatial distribution.38 For instance, Mathur et al. constructed a 3D spatial atlas of GBM cells and revealed the spatial feature of genomic, epigenomic, and microenvironmental heterogeneity.410 The emergence of various novel technologies make us keep unraveling the mysteries of tumor heterogeneity, however, more efforts are still required to clarify the mechanism and significance, thus proposing novel therapeutic strategies.
In addition to regulating tumor malignant phenotypes mentioned above, chromatin accessibility is also involved in the regulation of tumor metabolic reprogramming,411 angiogenesis,386 inflammatory response,412 and pro-tumor senescence.413 In summary, owing to spatial confined, we mainly focused on the functions and mechanisms of chromatin accessibility in HCC, but this does not mean that its role in other cancers can be ignored. Instead, we should devote more efforts to uncover the specific mechanisms of chromatin remodeling in tumorigenesis and development, so as to provide potential targets for more precise and effective cancer therapy.
Chromatin accessibility variation in liver diseases
Liver diseases are the considerable burden of human health. In the preceding parts, we have summarized the functions and mechanisms of chromatin accessibility in cancer initiation and progression, especially in liver cancer. In this part, we will describe the function and mechanism of chromatin accessibility variant in some common non-neoplastic liver diseases.
Lipid deposition in liver
Lipid accumulation is the initiation of many liver diseases. High-fat diet (HFD) can remold the chromatin accessibility in the liver of many vertebrates. Arginine ureahydrolase, arginase 2 (ARG2) is a glucose withdrawal-induced factor, which can partially alleviate the therapeutic metabolic sequelae of caloric restriction. ArcA, a bacterial virulence factor, has higher arginine binding affinity than that of ARG2. Zhang et al. found that both pharmacological activating ARG2 and overexpressing arcA in hepatocyte-induced thermogenesis and insulin sensitivity, attenuated steatosis and inflammation in obesity mice model. Mechanistically, the enhanced arginine catabolism activated systemic autophagic flux and FGF21 secretion. Additionally, scATAC-seq revealed that systemic arginine deprivation altered the chromatin accessibility landscape of hepatocyte and reduced the infiltration of inflammatory macrophagocytes in WT but not in autophagy deficient mice.414 The thyroid hormone triiodothyronine (T3) is an important regulator of multiple metabolic progression. The type 2 deiodinase (D2) which expressed in fetal and neonatal mouse liver, activates thyroxine by removing one iodine atom from thyroxine (also called deiodination) to product the active thyroid hormone triiodothyronine (T3).415 Fonseca et al. reported that perinatal expression of D2 remolded the gene expression panel and DNA methylation pattern in adult mouse hepatocytes. The mice that hepatocyte-specific knockout of D2 were resistant to HFD-induced obesity, hypertriglyceridemia, and liver steatosis.86 Recently, they found that the overall chromatin accessibility was reduced in D2 insufficient mouse which was in line with increased DNA methylation.416 Alharthi et al. reported that membrane-bound O-acyltransferase domain containing 7 (MBOAT7) reduced the accessible of inflammatory-related chromatin in COVID-19 and metabolic-associated fatty liver disease. However, MBOAT7 deficiency in metabolic steatohepatitis patients and SARS-CoV-2-induced downregulation of MBOAT7 make it insufficient to inhibit inflammatory responses in these diseases. They also found that MBOAT7 SNP rs8736 could be a disease risk of inflammation-induced liver injure.417 Besides, JMJD1C promoted liver lipogenesis both in vitro and vivo under insulin treatment. Mechanically, insulin-induced α-ketoglutarate accumulation and mTOR-mediated T505 phosphorylation of JMJD1C increased its demethylase activity, meanwhile USF-1 facilitated the binding of JMJD1C with the promoters of lipogenic genes. Consequently, JMJD1C enhanced the promoter accessibility of these genes by reducing H3K9me3 modification.112
Metabolic dysfunction-associate steatotic liver disease (MASLD)
According to the latest consensus, NAFLD and NASH are replaced by MASLD and MASH, respectively. Hepatic steatosis is one of the leading characteristics of many liver diseases. Research has indicated that the initiation and development of fatty-induced hepatopathy are closely accompanied by chromatin accessibility variation. In this part, we will discuss the detailed mechanisms and functions in MASLD.
MASLD is an important subtype of fatty liver diseases, which can develop into MASH, hepatic fibrosis, cirrhosis, and even liver cancer. The mitochondrial signaling is a master epigenetic regulator, dysfunction of which promotes MASLD.418 In hepatocytes, mitochondrial stress promoted the expression of AREG, a EGFR ligand. Mechanically, mitochondrial stress increased transcription activity of c-Jun and YAP1 to increase H3K4me1 and H3K27ac modification, as well as accessibility of AREG enhancer region.146 Yin et al. found that RPA1 was down-regulated in the livers of fatty liver patients. The loss of RPA1 promoted occurrence of hepatic steatosis, fatty liver, and even spontaneous HCC. Hepatic deficiency of RPA1 suppressed the transcription of FAO-related genes through modulating the chromatin accessibility landscape, and subsequently inhibited FAO.419 Bone morphogenetic protein 9 (Bmp9), is a member of the BMP superfamily. It is reported that Bmp9 was down-regulated in HFD fed mice. Ablation of Bmp9 induced lipid accumulation in HCC cells and promoted liver steatosis through reducing the expression of PPARα.420 Sun et al. demonstrated that Bmp9 attenuated HFD-induced obesity, hepatic steatosis, and macrophage infiltration by reducing the expression of Cers6, Fabp4, and Cidea, which is involved in glucose and lipid metabolism, as well as Fos, Tlr1, and CCL2, which involved in inflammatory response. Mechanically, Bmp9 treatment reduces the promoter accessibility of Cers6, CCL2, Fabp4, and Fos, meanwhile inhibits TNF and Toll-like receptor signaling pathway.421 The aberrant glyoxylate metabolism also promotes the progress of steatosis. Alanine-glyoxylate aminotransferase (Agxt) is a key enzyme in glyoxylate detoxification. Gianmoena et al. indicated that Agxt was downregulated in mouse and human steatotic hepatocytes which was insufficient to detoxificate the mitochondrial glyoxylate. They found that the hypermethylation of Agxt locus reduced its chromatin accessibility in steatotic hepatocytes, and rescue expression of Agxt reduced the production of oxalate in the fatty liver.422 Recently, Yang et al. indicated that KDM1A was upregulated in MASLD tissues and promoted the expression of lipid metabolism, glucose metabolism, and inflammation-related genes by increasing their loci accessibility.113
Increasing researches demonstrate that Th17 cells promote the pathogenesis of multiple inflammatory disease, such as Crohn’s disease,423 cardiovascular disease,424 and experimental encephalomyelitis.425 Recently, researchers characterized a CXCR3+ population of inflammatory hepatic Th17 cells (ihTh17) which were sufficient to exacerbate the pathogenesis of MASLD in mouse. The pathogenic potential of ihTh17 correlated with increased chromatin accessibility, glycolysis, IFNγ and TNFα production. In ihTh17, the open chromatin was enriched in the regulatory regions of metabolic and immune activation-related genes, including CXCR3, STAT1, GAPDH, and PKM2. Intercepting glycolysis by 2-DG or cell-specific deletion of PKM2 reversed ihTh17-induced inflammatory and MASLD severity. Moreover, the ihTh17-like cells are also enriched in human MASLD liver, and positively correlate with MASLD severity. However, the source of ihTh17 cells and the mechanisms of their temporal emergence and maintenance are still unclear.426
Metabolic dysfunction-associated steatohepatitis (MASH)
Many factors can lead to hepatitis, such as severe hepatic steatosis, excessive drinking, metabolic dysfunction, hepatitis virus infection, autoimmune, and drug damage. According to the latest consensus, the metabolic disturbance-induced steatohepatitis is collectively named as MASH instead of NASH.
MASH is becoming the most important subtype of hepatitis. The variation of chromatin accessibility is one marked feature of MASH. Li et al. found that the protein level of METTL3 was increased in the cytosol but significantly decreased in the nuclei of MASH hepatocytes. They indicated that reduced nuclear METTL3 protein contributed to the MASLD-to-MASH transition in vivo. Specific deletion of METTL3 in liver increased the expression of CD36 and CCL2 which enhanced free fatty acid uptake or inflammation response, respectively. Mechanically, METTL3, interacting with HDAC1/2, removed the H3K9ac and H3K27ac modification of CD36 and CCL2 promoters to suppress their transcription. Hence, insufficient of nuclear METTL3 increased the chromatin accessibility of CD36 and CCL2 promoter. Overexpression of METTL3 or inhibition of CD36 and CCL2 can ameliorate MASH progression.131 Pérez-Schindler et al. revealed that the molecular signatures were similar in MASH and hepatocyte lipotoxicity, but they had distinct impact on chromatin accessibility in liver. The global chromatin accessibility of MASH patients was increased slightly, but decreased strongly in hepatocyte lipotoxicity. They indicated that MAF bZIP transcription factor K (MAFK) and transcription factor 4 (TCF4) can contribute to the expression of lipotoxicity-related genes in hepatocyte.427 Patatin-like phospholipase domain-containing 3 (PNPLA3), which resides in the membrane of lipid droplets, remodels the lipid droplets after eating through its triglyceride hydrolase activity. The I148M mutant PNPLA3 (PNPLA3 I148M) has less than 20% catalytic activity and higher protein stability, but still accumulated to the surface of lipid droplets to block triglyceride hydrolysis.428 Schwartz et al. revealed that momelotinib reduced the expression and promoter accessibility of PNPLA3 in human and mouse hepatocytes, as well as human hPSCs. Momelotinib treatment reduced BMP signaling-induced expression of PNPLA3 and alleviated the PNPLA3 I148M-mediated triglyceride accumulation in MASH model.429
Choline-deficient, L-amino acid-defined, high-fat diet (CDAHFD) has been proven to rapidly replicate the full pathologies of MASH, including steatosis, inflammation, and fibrosis of liver. Xiong et al. demonstrated that liver-specific hnRNPU inactivation exacerbated CDAHFD-induced MASH, including enhanced inflammation response and hepatocyte injury. HnRNPU inactivation remolded the chromatin accessibility and transcriptome in hepatocyte, such as the CYP4A10 locus was significantly reduced and Ntrk2 was enhanced in hnRNPU LKO livers. Ntrk2 encodes the TrkB receptor tyrosine kinase. They found that hnRNPU deficiency promoted the expression of a truncated isoform of TrkB, named TrkB-T1. TrkB-T1 can aggravate stress-induced hepatocyte injury. Besides, increased TrkB-T1 expression was observed in liver biopsies of MASH patients. Brain-derived neurotrophic factor (BDNF), the ligand of TrkB can inhibit the expression of TrkB-T1 to relieve liver injury. They speculated that BDNF could trigger endocytosis and degradation of TrkB-T1.180 Besides, treating zebrafish larvae with thioacetamide (a hepatotoxin) can be applied to establish steatohepatitis model.430 Research indicated that thioacetamide treatment remolded the chromatin accessibility landscape of zebrafish liver cells, especial endothelial cells.431
Additionally, liver lacking ARID1A is more susceptible to fatty hepatopathy. Moore et al. indicated that ARID1A deficiency increased lipogenesis and inhibited FAO by disorganizing the expression homeostasis of related genes. Suppressing de novo lipogenesis by SREBP inhibitor delayed ARID1A and PTEN loss-triggered fatty liver. ChIP-seq indicated that the ARID1A binding peaks were significant associated with C/EBPα binding peaks, H3K27ac and H3K4me3 modification in the promoters of most upregulated genes.432 Additionally, Qu et al. reported that hepatocyte-specific deletion of ARID1A significantly increased the susceptibility of hepatic steatosis, insulin resistance, and inflammation response in HFD-feeding mice. Mechanically, ARID1A deletion decreased H3K4me3 modification and chromatin accessibility on the promoters of FAO-related genes.149 Besides, the SWI/SNF complexes SMARCA2 and SMARCA4 have also been reported to promote the proinflammatory cytokine production in MASH mouse by remolding epigenetic modifications.433
In addition to mentioned above, other factors also regulate the chromatin accessibility in MASH liver. Vertical sleeve gastrectomy (VSG) is one of bariatric surgeries which is effectively to improve MASLD and MASH by promoting weight loss. Du et al. demonstrated that VSG reduced the expression of genes in inflammatory pathways and induced the expression of lipid metabolic genes. VSG remolded the chromatin accessibility which was in line with the expression pattern of related genes.434 Kupffer cells, the liver resident macrophages, regulate the progression liver diseases, including MASLD and MSAH. Seidman et al. indicated that MASH-induced Kupffer cell death recruited blood monocytes, leading to increased myeloid cells diversity in liver. MASH remolded the chromatin accessibility of Kupffer cells and selectively impaired LXR signaling pathway.435
Alcoholic liver disease
Excessive alcohol consumption always leads to alcohol-related liver disease, including alcoholic fatty liver, hepatitis, and so on. The alcohol induced epigenetic modification variation is one of the main mechanism. Massey et al. indicated that the hepatic metabolome, especially carbohydrate metabolism of alcoholic hepatitis patients was dysregulated which partially promoted the expression of hexokinase domain containing 1 (HKDC1). The H3K4me1, H3K4me3, and H3K27ac modification of HKDC1 promoter was enriched significantly in alcoholic hepatitis patients.436 Increased infection susceptibility is a remarkable characteristic of severe alcohol-related liver disease. Weichselbaum et al. found that the function of monocytes and dendritic cells was impaired in patients with severe alcoholic hepatitis and the pro-inflammatory cytokines were correspondingly reduced, leading to a higher infection and mortality risk. Further, they indicated that compared with healthy control, monocytes derived from patients with severe alcoholic hepatitis displayed a distinct chromatin accessibility landscape which was in line with an immunosuppressive transcriptome pattern.437 However, at present, the chromatin accessibility landscapes in alcohol-related liver diseases are still unclear. As alcohol consumption is one of the leading causes of liver diseases, the research about this respect is of great significance.
Viral hepatitis
Chronic HBV infection is one of the leading etiologic inducement for HCC. The hereditary material of HBV, called cccDNA, forms microchromosomes in the hepatocyte nuclei. HMGN1 promotes HBV replication and expression of virulence proteins. It increased the accessibility and transcription of cccDNA by competitively combining with H1 and promoting H3 phosphorylation, respectively.95 As the most frequently integrated viral gene, HBx plays an important role in HCC pathogenesis which partial through remolding the epigenetic modification.438 Zheng et al. indicated that HBx/ETV4/DVL2/β-catenin axis promoted the migration and invasion of HCC cells. Mechanically, HBx promoted the transcription of ETV4 by increasing the H3K27ac modification and chromatin accessibility of super-enhancers upper ETV4 promoter. As a TF, ETV4 promoted the transcription of DVL2.144 Additionally, HBV infection reduces the expression of DDX5 to increase the chromatin accessibility and expression of multiple genes involved in Wnt signaling pathway in HCC cells.373 Besides, the HBV-induced differentiation of CD8+ T cells can be primed by Kupffer cells or hepatocytes, but the latter possesses negligible immune activity. Bénéchet et al. indicated that they have different chromatin landscapes, contributing to the difference in immune activity.311 Yates et al. demonstrated that chronic HCV- and HIV-infection exhausted CD8+ T cells. Their infection induced an irreversible chromatin accessibility variation in CD8+ cells, disrupting TOX and HIF1α signaling.439 However, whether HBV infection has a similar effect requires further investigation.
At present, the association between chromatin accessibility and other hepatitis viruses, including HAV, HCV, HDV, and HEV, is rarely reported. Considering the important role of these hepatitis viruses in a variety of liver diseases, it’s of great significance to further investigate the chromatin dynamics regulated by these viruses.
Hepatic fibrosis
Uncontrolled hepatitis often progresses to liver fibrosis, even cirrhosis and liver cancer. Research has confirmed that the chromatin accessibility is gradually changing from healthy, steatosis, to fibrotic liver.440
The hPSCs only comprise ~10% of the liver, but great numbers of studies have confirmed that their abnormal activation and proliferation are one of the primary triggers for liver fibrosis. Countering with our initial intuitive, Brougham-Cook et al. indicated that the expression of intracellular collagen I and lysyl oxidase (LOX) in activated hPSCs were higher in soft microenvironments (1 kPa, mimicking healthy tissue) than stiff microenvironments (25 kPa, mimicking fibrotic tissue).441 Their following work demonstrated that the global chromatin accessibility of hPSCs cultured in soft substrates was higher that in stiff substrates. Among which, the C/EBPβ binding motifs were significantly enriched and the HSD11b1 promoter was more accessible in soft substrate-cultured cells. Correspondingly, knockdown of C/EBPβ or HSD11B1 alleviated fibrogenic phenotype under soft condition.442 However, more investigations have indicated that the activation of hPSCs requires a stiff environment.443,444,445 Hence, the detailed ECM condition that regulates the chromatin accessibility in liver requires further research. Liver sinusoidal endothelial cells also play an important role in maintaining the normal function of liver. Winkler et al. found that sinusoidal capillarization was increased significantly in mice that endothelial-specific knockout GATA4, meanwhile these mice showed increased profibrotic angiocrine, hPSC overactivation, perisinusoidal liver fibrosis, and reduced liver regeneration. Further, they revealed that Myc and PDGFB were direct targets of GATA4. Endothelial GATA4 deficiency increased the accessibility of Myc and PDGFB gene loci which subsequently increased their expression. PDGFB is a target of Myc, they also found that GATA4 deficiency increases the binding of Myc to PDGFB promoter.446
Transforming growth factor-beta (TGFβ) is a master regulator of fibrosis in many organs. Aseem et al. indicated that TGFβ treatment remolded the landscapes of H3K27ac and H3K9ac modification, and promoted the expression of hPSC-activating genes, such as SERPINE1 and FN1 in cholangiocytes. By comparing the differentially expressed genes enriched by H3K27ac and H3K9ac modification, they found that the H3K9ac enriched genes are more specific. The H3K9ac modification enhanced the chromatin accessibility of TGFβ-stimulating genes. Mechanically, SAMDs, downstream of TGFβ signaling, recruited the acetyltransferase KAT2A to increase the H3K9ac modification in the promoters of target genes. Pharmacologic inhibition of H3K9ac or knockout of KAT2A alleviated biliary fibrosis in vivo.125 Besides, Tang et al. demonstrated that the expression of YBX1 was increased in several liver fibrosis models, and aggravated CCl4-induced liver fibrosis through activating hPSCs and extracellular matrix deposition. It promoted the expression of multiple fibrosis-related genes by increasing their chromatin accessibility.447
Other liver diseases
Primary sclerosing cholangitis (PSC) is an immune-mediated liver disease and the ingredient of intrahepatic T cells is aberrant in PSC patients. Poch et al. identified a novel T cell cluster named tissue-resident naive-like CD4+ T cells which were expanded in the liver of PSC patients. According to the transcriptome and T cell receptor, this population is similar to circulating naive T cells, but expressed a set of tissue residency genes. Through comparing the accessible chromatin regions of naive CD4+ T cells from PSC patients and healthy donors, they revealed that the PSC-derived naive CD4+ T cells were predisposed to polarize toward Th17 cells. Several genes that regulate T cell activation and Th17 polarization were identified, such as STAT3, MAF, TLE1, and RUNX1.448 In polycystic liver disease, Ji et al. found that the overall chromatin accessibility was increased significantly which activated cyst-associated genes in male and female cholangiocytes. Interestingly, there were significant different manifestations between females and males. The increased chromatin accessibility in male was induced by H3K9me3 elimination and H3K9ac enrichment, and in female is due to H3K27ac enrichment.449 In channel catfish, aeromonas hydrophila infection increased the serum cortisol levels which regulated the expression of infection-induced genes in liver by affecting chromatin accessibility.450 Iron homeostasis is critical for many physiological processes, dysregulation of which leads to many diseases. A recent study indicated that excess iron in liver cells increased the chromatin accessibility of cell cycle genes, most of which are tightly linked to HCC.451 Graft-versus-host disease (GVHD), caused by alloreactive T cells infiltration, is the one of the main adverse reactions of organ transplantation.452 Inhibitor of DNA binding 3 (ID3) is a DNA-binding inhibitor that regulates T cell fate determination. Wang et al. indicated that ID3 promoted GVHD in the mouse model. ID3 inhibited the expression of PD-1 by reducing its promoter accessibility to facilitate the infiltration of alloreactive T cells. ID3 knockout led to increased liver infiltration of PD-1+ T cells. It’s of great meaningful that targeting ID3 reduced the GVHD of CAR-T cells transplantation but don’t compromise the anti-tumor activity.453
Liver is one of the most important organs of human being, playing many vital roles, and its lesions often cause serious outcomes. The studies discussed above have partially revealed the functions and mechanisms of chromatin accessibility variation in some liver diseases. However, in numerous other liver diseases, such as bacterial infection, parasitic disease, drug-induced liver damage, autoimmune liver disease, and so on, the chromatin accessibility-related function and mechanism remain to be elucidated.
Chromatin accessibility variation in nervous system disease
The nervous system disorders are affecting more than one third of the world’s population. These disorders are complex and multiinducible, making it is hard to clarify the specific regions matching disorders. Fortunately, the correlation between the accessibility of specific region and neurological disease is revealing constantly.
Alzheimer’s disease (AD) is a degenerative disease of the central nervous system (CNS), which is the most common form of dementia, mainly occurring in old or pre-old age. Many researches have identified the chromatin accessibility feature in AD patients, screened out many AD risk genes.192 The differences in cis-regulatory elements (CREs) are perceived as the main cause of phenotypic differentiation among primate relatives.454 Recently, Hu et al. identified many highly conserved rapidly evolving conserved elements (RECEs) in primates which regulated the development of adult human brain. Additionally, they found that these RECEs were generally more accessible in the oligodendrocytes from AD patients compared with control group, indicating that these RECEs may drive brain aging and AD.455 Zhang et al. indicated that the postmortem brain of AD patients contained much more double-stranded DNA breaks (DSBs) compared with that in non-demented individuals. These DSBs leaded in unique SNPs, chromatin accessibility, and gene expression profiles in AD brains.456 Through analyzing the different open chromatin regions in neurons and non-neurons brains, Bendl et al. found that the binding motifs for USF2 were perturbed most significantly. Downregulation of USF2 disturbed the lysosomal function which was corresponded with the accumulation of abnormal proteins in AD.192
Autism spectrum disorder (ASD) is a common neurodevelopmental disorder, and the dysregulation of CHD8 is the most important causal factor.457 Shi et al. indicated that CHD8+/− neurons showed a globally increased chromatin accessibility. The genes with varied chromatin accessibility and expression are similar with the genes mutated in probands for ASD, schizophrenia, and intellectual disability. Further, they found that insufficient of CHD8 reduced functional synapses and leaded to reduced spontaneous firing rates in neurons.458 NASP is a histone-binding protein occurring in two major forms, sNASP and tNASP which is expressed in somatic cells or testis and embryonic tissues, respectively.459 It has reported that NASP regulates chromatin accessibility in HCC cells.147 Recently, Zhang et al. identified a nonsense mutant of NASP (tNASP-Q289X) in a Chinese nuclear family with ASD. They found that both tNASP KO and tNASP-Q289X increased the chromatin accessibility and affected the gene expression of neural and immune response signal in the brain.460
Additionally, addiction is an obstacle for heroin to treat human disease. FYN, a member of the protein-tyrosine kinase family, regulates the drug-seeking behavior and relapse.461 Egervari et al. found that heroin remodeled the chromatin accessibility in striatum. Especially, the accessibility of FYN promoter is increased in the heroin users compared to controls.462 Chronic cerebral hypoperfusion (CCH) can activate microglial to promote chronic inflammation-induced neuronal impair.463 Zhang et al. indicated that CCH remodeled the chromatin accessibility profile in the cortex, and the PU.1 binding motifs were enriched most significantly in the ATAC peaks. The regulator network of PU.1 may provide potential targets for the early intervention of CCH-induced neuroimmune response.464
Besides, other CNS disorder are also involved in chromatin accessibility variation, such as Parkinson’s disease,465 stroke,466 infection,315 and so on. However, the detail roles and mechanisms are required further investigation.
Chromatin accessibility variation in diabetes
Diabetes is a chronic endocrine disease, characterized by persistent hyperglycemia. The mainly cause of diabetes including insulin secretion defect and cells respond poorly to insulin which leading to type 1 diabetes (T1D) and type 2 diabetes (T2D), respectively. The chromatin accessibility is a master regulator of diabetes.
Autoimmune-induced β cells cytolysis is the main pathogenesis of T1D, and the β cell-specific CD8+ T cells maintained stem cell memory phenotype which preserved for long periods, challenging the pancreatic islet transplant.467 Abdelsamed et al. confirmed that the β cell-specific CD8+ T cells possessed a similar DNA methylation and chromatin accessibility landscapes compared with naive CD8+ T cells to maintain a stem-like state.468 The haploinsufficiency of BACH2 is main cause of congenital autoimmunity and immunodeficiency.469 Robertson et al. screened out many novel T1D risk variants by genome-wide association studies, and by mapping the chromatin accessibility variation, they confirmed that SNP rs72928038 at the promoter of BACH2, leading to reduced promoter accessibility in CD4+ T cells.470 Similarly, Chiou et al. confirmed that the SNP rs7795896 in the enhancer of CFTR reduced its accessibility and expression in pancreatic ductal cells, indicating a high-risk of T1D.471
Currently, there are more studies focused on T2D. Wang et al. indicated that T2D suppressed the insulin production and secretion signaling pathways in pancreatic β cells.472 Xue et al. knocked out twenty T2D-associated genes in β cells, and they found that the loss of many genes can remodel the chromatin accessibility and expression profiles significantly. Among which, HNF4α was a key TF for the maintenance of β cells physiological functions. Besides, CP, PCSK1N, RNASE1, and GSTA2 regulated insulin production. TAGLN3 and DHRS2 regulated the lipotoxicity sensitivity of β cells.473 Qiao et al. indicated that knockout STING suppressed Pax6 expression and nuclear localization, while the accessibility of Pax6 targets was correspondingly reduced, leading to disturbed glucose-stimulated insulin secretion.474 Reduced IGF2BP2 expression and accessibility of its binding sites may contribute to T2D.475 Glucocorticoid also increases the chromatin accessibility at glucocorticoid receptor binding sites to regulate the expression of T2D-associated genes.476 Besides, Qadir et al. indicated that the sex-based difference in gene accessibility and expression were predominantly enriched in sex chromosomes of cultured islets derived from nondiabetic donors, meanwhile in islets from T2D, there was much more sex-based difference in autosomes, and the females displayed more variation compared with male donors. The differentially expressed genes in males and females were enriched in insulin secretion pathways, oxidative phosphorylation and electron transport chain pathways, respectively, which corresponded with the sex-based pathogenesis of T2D.477 Additionally, Wang et al. indicated that HFD-feeding remodeled the chromatin accessibility and transcriptome through suppressing CTCF expression, which was corresponded with decreased CTCF expression in β cells from obese and diabetic mice and humans. Further, they confirmed that both early dietary intervention and restoration of CTCF expression reversed β cell dysfunction.478 Besides, Wei. et al. indicated that Vitamin D remodeled chromatin accessibility landscape by orchestrating the interaction of BRD7 and BRD9 binding with BAF chromatin remodeling complexes to protect β cells.479 Through genome-wide accessibility studies, many T2D-associated variation loci have been screened out in human,480 but their role in T2D is required investigation.
Diabetes usually leads to multiple organ abnormalities in patients. Bansal et al. indicated that T2D dysregulated the fibrotic and transport-associated genes in kidney proximal tubule epithelial cells by remodeling their DNA methylation and chromatin accessibility. The most disturbed TF binding sites included HNF4α, SMAD3, and CTCF.481 Cavalli et al. detected the difference of chromatin accessibility in normal and T2D livers. Based on the differential accessible regions, they identified a novel enhancer of acyl-CoA thioesterase 1 (ACOT1) gene, which catalyzed the hydrolysis of acyl-CoA to free fatty acid and coenzyme A (CoA). The increased expression of ACOT1 in T2D liver was in line with the more accessible chromatin structure. Meanwhile, multi-omics study indicated that the ACOT1 level was an indicator of T2D.482
Chromatin accessibility variation in other diseases
In addition to mentioned above, the chromatin accessibility variation is also involved in many other diseases. Such as, in T-cell acute lymphoblastic leukemia (T-ALL), the chromatin is more accessible than that in normal human immature T cells.483 In idiopathic pulmonary fibrosis myofibroblast, the increased accessibility and expression of TWIST1 promotes fibrosis progress.484 Raphael et al. demonstrated that the accessibility of IL-33 promoter was increased in airway cells of chronic obstructive pulmonary disease patients, facilitating IL-33 expression and aggravating the disease.485 Besides, chromatin accessibility variation is also involved in the progression of chronic kidney disease,486 AIDS,487 COVID-19,488 osteoporosis,489 and so on.
It’s difficult to list all the diseases that occur in our organs and systems, but based on the available research, we can confirm that the initiation and development of numerous diseases are accompanied by chromatin accessibility variation, which are both disease-induced outcomes and disease triggers. Therefore, chromatin accessibility is an ideal indicator of morbidity, target for treatment, and biomarker for prognosis. What’s more, research strategies targeting single organ or single disease are already abundant, however, most diseases are often caused by systemic abnormalities and many diseases also cause systemic lesions. At present, the single-approach treatment often causes severe side effects which seriously reduce the curative effect of disease and the life quality of patients. Therefore, it’s of great significance to ensure the life quality of patients while ensuring the treatment effect of disease by every means. In recent years, the importance of integrative medicine in maintaining human health and disease treatment has obtained more and more attention. Hence, comprehensive research on systemic variation of chromatin accessibility in various disease processes may help to reveal the pathogenesis and development mechanisms of these diseases as well as propose better treatment strategies. Besides, the traditional Chinese medicine (TCM) has an innate advantage in disease treatment through a systemic manner, therefore, it is of great significance to investigate the alteration of chromatin accessibility during the treatment of diseases by TCM. Comprehensively revealing the function and mechanism of chromatin remodeling in different diseases allows us to treat these diseases more timely and effectively, however, there still are many challenges to be solved.
Targeting chromatin accessibility for disease therapy
As we known, the chromatin accessibility varies significantly during various pathological process, and given its powerful function in the initiation and progression of different diseases, the chromatin remodellers are potential targets for disease treatment. In this section, we will discuss the disease treatment strategies based on targeting different chromatin regulators (Table 1).
Therapeutic opportunity by targeting nucleosome remodeller
Nucleosome remodeller can wildly regulate the nucleosome distribution and chromatin accessibility, participating in many pathological processes. Thus, targeting these nucleosome remodellers is an opportunity to treat some diseases.
Targeting SWI/SNF complexes
There are many inhibitors have been found and characterized to target different subunits or domains of SWI/SNF complexes. Hydrolyzing ATP as energy is the common characteristic of different nucleosome remodellers. Hence, targeting ATPase activity is the primary strategy to block their function for disease treatment. Researches indicated that insufficient SMARCA2 and/or SMARCA4, the ATPase activity subunits of SWI/SNF complexes, sensitized the cancer cells to chemotherapy or radiotherapy. Active DNA-dependent ATPase A Domain inhibitor (ADAADi) is the first identified inhibitor directly targeting SWI/SNF catalytic activity.490 It has been reported to suppress the progression of many cancers.491,492 BRM014, an allosteric ATPase inhibitor of SMARCA2/4 varying the chromatin accessibility very rapidly.493 FHD-286, another allosteric inhibitor of SMARCA2/4, has been showed to inhibit AML.494
Up to date, at least five SWI/SNF subunits bear bromodomains, including Family IV and VIII. The bromodomains are important reader of epigenetic code on chromatin, indicating that targeting the bromodomains can effectively inhibit SWI/SNF complex activity. SMARCA2/4 and PBRM1 bear Family VIII bromodomains. PFI-3 is the first identified inhibitor selectively binding to the bromodomains of SMARCA2/4 and PBRM1. Although applying PFI-3 alone has limited effect on some cancer cells, it significantly sensitized of these cancer cells to chemotherapy.495,496 PB16 and GNE-235 are two promising PBRM1 selective inhibitors, but their biological function remains to be elucidated.497,498 BRD7 and BRD9, assembled into different SWI/SNF complexes, contain Family IV bromodomains.499 BRD9 or BRD7 is considered as oncogene or tumor suppressive, respectively,500,501 thus more studies are focused on targeting BRD9. LP99 is the first identified drug binding to the bromodomains of BRD7 and BRD9 to evict both proteins from acetylated histones.502 The BRD9 selective bromodomains inhibitors, such as BI-7271, BI-7273, BI-9564,503 and I-BRD9,504 significantly inhibit the progression of many cancers. Recently, compounds 1-78 and 2-77 were identified as the first BRD7-selective inhibitors which can suppress the proliferation of prostate cancer cells.505
PROTAC is an effective method for specific protein degradation in vivo. Panditharatna et al. constructed an ATPase degrader by linking a SMARCA4 ATPase inhibitor to a phthalimide, called JQ-dS-4, facilitating ubiquitin E3 ligase CRBN-mediated degradation, which suppressed the progression of glioma.506 ACBI1 and AU-15330 are two PROTACs that link the Family VIII bromodomains with E3 ligase VHL to promote the degradation of SMARCA2/4, and PBRM1.507,508 A947 and PRT3789 selectively degrade SMARCA2 to suppress SMARCA4 mutant solid tumors.509,510 GSK39,511 dBRD9,512 C6,513 CFT8634,514 and FHD-609515 are CRBN-based PROTACs for BRD9, all of which showed anti-cancer activity. Besides, VZ-185, a VHL-based PROTAC, degrades both BRD7 and BRD9.516
Targeting NuRD complex
The chromodomain helicase DNA-binding proteins (CHDs), containing ATPase activity, are the core subunits of NuRD complexes. Oyama et al. indicated that the SMARCA5/CHD4 dual inhibitor, ED2-AD101, sensitized ovarian cancer cells to cisplatin by reducing the expression of multi-drug resistance 1 (MDR1).517 MBD2 is a methylated DNA-binding protein within the NuRD complex. Wyhs et al. screened out four inhibitors, mitoxantrone, idarubicin, NF449, and aurintricarboxylic acid, to inhibit the binding of MBD2 with methylated DNA.518 HDAC1 and HDAC2 are the important subunits of different NuRD complexes. Trichostatin A (TSA) and SAHA (also called as Vorinostat), two broad spectrum HDAC inhibitors, are applied in many clinical trials. Both TSA and SAHA have been reported to remodel the chromatin accessibility.519,520 Up to date, many HDAC1/2 selective inhibitors have been found, such as Cmpd60,521 mocetinostat (MGCD0103),522 romidepsin (FK228),523 tacedinaline (CI-994), santacruzamate A (CAY10683),524 and so on, which have the potential to treat cancer and hypertension, as well as alleviate aging.
Targeting ISW1 complex
SNF2H (also called SMARCA5) is one of the core subunits of ISW1 complex, containing ATPase activity. Kishtagari et al. identified a SMARCA5/CHD4 dual inhibitor, ED2-AD101, suppressed the proliferation of AML cells and promoted the epithelial differentiation of many solid tumor cells.525 BPTF is the largest subunit of ISW1 complex, containing bromodomains. BI-2536 and bromosporine are the first two discovered inhibitors of BPTF, but their specificity are limited.526,527 Zhang et al. screened out a selective BPTF bromodomain inhibitor, called DCB29, but the biological function is unknown.528 Xu et al. identified another BPTF bromodomain inhibitor, named C620-0696, which suppressed the progression of NSCLC mainly by inhibiting c-Myc transcription.529
Targeting INO80 complex
RUVBL1 and RUVBL2, containing ATPase activity, are parts of INO80 and SWR1 complexes. Assimon et al. identified an allosteric inhibitor of the ATPase domain of RUVBL1/2, named CB-6644, which can suppress the progression of many cancers.530 Besides, Nano et al. indicated that sorafenib inhibited the ATPase activity of RUVBL1/2 complex by directly interacting with RUVBL2.531
Currently, there are many chromatin remodeller inhibitors, but satisfactory results are rarely achieved in clinical application. What’s more, the functions of most inhibitors on chromatin accessibility, in particular at specific sites, are still require further investigation.
Therapeutic opportunity by targeting DNA methylation modifier
The methylation of DNA is dynamically regulated by DNA methyltransferases and demethylases which have been identified as inhibition targets for various diseases. DNA methyltransferase enzymes (DNMTs) catalyzed the CpG islands methylation. 5-azacytidine (azacytidine) and 5-aza-2’-deoxycytidine (decitabine) are two cytidine analogs that specifically inhibit DNA methyltransferases.532 Bowler et al. indicated that azacitidine promoted the degradation of DNMTs, in particular DNMT1.533 Kapuganti et al. indicated that the promoter of CLU was hypomethylated, leading to reduced expression of CLU in the blood and lens capsules of pseudoexfoliation patients. Treating human lens epithelial cells with decitabine promoted CLU expression by facilitating its promoter accessible to the TF Sp1.534 Nanaomycin A can sensitize HCC cells to sorafenib by inhibiting DNMT3B.535 Rodems et al. indicated that DNMT inhibitors decitabine or guadecitabine (SGI-110) enhanced the antitumor immunity by increasing the accessibility and expression of HLA-I in prostate cancer cells, while combination with the HDAC inhibitor LBH-589 (LBH) getting a better effect.536 Additionally, combined treating by DNMT inhibitor decitabine and HDAC inhibitor SAHA increased the chromatin accessibility and sensitized solid tumor cells to cisplatin, doxorubicin, and irradiation.537 Combined applying decitabine and EZH2 inhibitor GSK126, suppressed the proliferation and upregulated anti-tumor immune response in human HCC cells.77 Besides, many natural bioactive compounds have been reported to inhibit DNMTs in many cancers.538
The removement of 5mC modification dependents on the ten-eleven translocation protein family (TETs) and thymine DNA glycosylase (TDG).539 Researches have indicated that the accumulation of oncometabolite, fumarate, succinate, and 2-hydroxyglutarate (2-HG) induced by hydratase (FH), succinate dehydrogenase (SDH), and isocitrate dehydrogenase 1/2 (IDH1/2) mutations, respectively, significantly inhibited both α-ketoglutarate (α-KG)-dependent histone and DNA demethylases,540,541 which linking metabolism and epigenetics.542 Verdura et al. indicated that decarboxymethyl oleuropein aglycone (DOA), a naturally phenolic compound in olive oil, can inhibit the mutant IDH1, IDH1-R132H, to reverse hypermethylation of DNA and H3K9 through restoring the 2-HG-suppressed demethylases.543 Multiple researches have indicated that targeting IDH-mutant to restore DNA and histone demethylation is a promising anti-cancer strategy.544 Xu et al. indicated that MAGI2-AS3 suppressed the proliferation and migration of breast cancer cells by increasing the demethylation of MAGI2 promoter and facilitating its expression. But, the TET1 inhibitor, bobcat339 hydrochloride, reversed the inhibitory effect of MAGI2-AS3.545
Therapeutic opportunity by targeting histone modifier
The epigenetic modifications on histones and their functions are complex, which orchestrated by multiple modifiers. Lots of histone modification-targeting molecules have been characterized and even applied in clinic. Some of which have been reported to remodel the chromatin accessibility.
CBP/P300 is the best-studied acyltransferase, mediating many types of acylation on histones and non-histone proteins. Recently, Welsh et al. indicated that the P300 inhibitor GNE-781 sensitized multiple myeloma (MM) to immunomodulatory imide drugs (IMiD) through blocking the super-enhancers accessibility and suppressing the expression of Myc, IRF4 and POU2AF1.546 Lindqvist et al. indicated that inhibiting P300 by GNE-049 reversed HIV latency by reducing the H3K27ac modification on integrated viral DNA.547 Ebrahimi et al. indicated that the CBP/P300 bromodomain inhibitors, CBP30 and I-CBP112, facilitated cellular reprogramming by decreasing the H3K27ac modification, chromatin accessibility, and expression of somatic-specific genes.548 A-485 inhibits the acetyltransferase activity of P300 to targeting enhancer accessibility in leukemia stem cells.549 Besides, Hu et al. revealed that the histone acetyltransferase (HAT) inhibitor, anacardic acid, alleviated paraquat-induced pulmonary fibrosis by reducing the expression of IL-6 in macrophages.550 MG149 inhibits MOF to attenuate MKL1-induced cardiac ischemia-reperfusion injury in mice.551 MG149 also decreases the total lysine 2-hydroxyisobutyrylation (Khib) modification by inhibiting TIP60 in pancreatic cancer.552
Currently, there are more researches on histone deacetylase inhibitors. Through high-throughput small molecule screening, Pattenden et al. identified a series of HDAC inhibitors, such as SAHA, TSA, panobinostat (LBH-589), givinostat (ITF2357), abexinostat (PCI-24781), tubastatin A (HDAC6 selective), PCI-34051 (HDAC8 selective), and so on, which can remodel the chromatin accessibility.553 Yang et al. indicated that the HDAC8 inhibitor, PCI-34051, increased the infiltration of CD8+ T cells and enhanced the therapeutic effect of PD-L1 blockade in HCC mice model.132 Similarly, the HDAC6 inhibitor, tubastatin A, also inhibits the expression of PD-L1 in melanoma cells. Besides, Tiwari et al. indicated that epigallocatechin gallate (EGCG) increased the chromatin accessibility by inhibiting HDAC which alleviated radiation-induced hemopoietic system injury in mice.
Targeting histone methylation has also obtained many achievement. Treating with DOT1L inhibitor, EPZ-5676, reduced H3K79me2/3 modification globally in leukemia cells.106 EPZ-5676 also reduced the H3K79me2 modification and accessibility of Sox2-binding enhancers to impair neuronal differentiation.107 Combined utilization of donafenib and the JMJD3 inhibitor, GSK-J4, suppressed the proliferation of liver cancer by enhancing ferroptosis both in vitro and in vivo.111 Ku et al. revealed that furamidine (FM) inhibited PRMT1, reducing the H4R3me2 modification, chromatin accessibility, and expression of glycolysis-related genes to reverse the chemoresistance of pancreatic cancer.387 Treated the CRC organoids by the histone demethylase inhibitor, methylstat, significantly restore the H4K20me3 and H3K27me3 levels in a dose-dependent manner, meanwhile reduced organoids growth.104
Therapeutic opportunity by targeting TF or co-TF
The transcription and cotranscription factors (TFs and co-TFs) can occupy the cis-acting element specifically to determine the local accessibility of chromatin. Thus, targeting TF and co-TF is promising strategies to remodel chromatin accessibility.
All intracellular receptors are trans-acting factors, such as the androgen receptor (AR). AR inhibitors are applied for the treatment of prostate cancer, but drug resistance is a major clinical problem. Leppänen et al. revealed that the AR inhibitor, enzalutamide (ENZ), increased the chromatin accessibility and expression of SIX2, facilitating stem-like reprogramming, neuroendocrine differentiation, and AR inhibitors resistance.554 ONECUT2 was a prostate cancer trigger through remodel the chromatin accessibility landscape independent of AR. The activation of ONECUT2 was an novel mechanisms of drug resistance, and inhibiting ONECUT2 by CSRM-617 reshaped chromatin accessibility, expression profile, and cell lineage of prostate cancer cells.555 MYCi975 is a Myc inhibitor that disrupts Myc/Max dimerization.556 Holmes et al. demonstrated that MYCi975 reduced H3K27ac and chromatin accessibility at the binding motifs of CTCF and FOX family transcription factors.557 AZD4573, an CDK9 inhibitor, remodeled the global promoter and enhancer accessibility as well as inhibited the proliferation and induced the apoptosis of lymphoma cells.558 Seredinski et al. indicated that the acriflavine, inhibited DNA topoisomerase, increasing the locus accessibility and expression of many lncRNAs to suppress the growth of endothelial cells.559 Additionally, other selective inhibitor also attenuated the recruitment of TFs to their binding motifs, such as HLM006474 (E2F inhibitor), S3I-201 (STAT3 inhibitor),560 Am80 (KLF5 inhibitor),561 gliotoxin (NOTCH2 inhibitor),562 and so on.
Besides, targeting the specific kinases of the TFs also is a viable strategy for chromatin remodeling, such as XAV-939 (TNKS1/2 inhibitor) and VT02956 (LATS inhibitor) to determine WNT/β-catenin- and Hippo/YAP-mediated chromatin dynamics, respectively.563,564
As mentioned above, there are many targeting compounds to remodel chromatin structure, however, most of them have not yet entered clinical or preclinical trials. Even if some inhibitors have got clinical trials, the therapeutic effect are still not satisfactory. The main reasons include the low specificity and bioavailability of these inhibitors. In addition, most of the targets are multifunctional and existed in different complexes, therefore, simply incapacitating these targets may cause a variety of functional disturbances leading to serious side effects. Hence, in parallel with searching for more selective inhibitors, screening out inhibitors to specifically target the assembly of specific complexes may be a more promising strategy to reverse the dysregulation chromatin accessibility.
Conclusion and perspective
The conformity of structure and function is one of the basic characteristics of living organisms. The hereditary information of eukaryotes is contained in chromosomes, which contain DNA, histones, non-histone proteins, and a little RNA, forming a multilevel structure encapsulated in the nucleus. Nucleosomes, the basic structure of chromatin, are highly enriched in inaccessible regions, but relatively reduced in open regions, which correspond with their transcription potential, DNA replication, and repair capacity.
The chromatin accessibility is dynamically regulated. Nucleosome localization and occupancy are the most dominated parameters of chromatin accessibility. The open portion of chromatin is not completely free DNA, but dynamically occupied by histones and other DNA-binding factors which regulate the opening state of chromatin. Additionally, the RNA polymerase can relocate nucleosomes during transcription elongation, rather than completely expel them, hinting that dynamically occupied by nucleosomes, but free DNA is required for gene transcription.565 Besides, chromatin openness and local transcriptional activity are not completely consistent. For example, nucleosomes, located at the TSS, usually inhibit transcription, but can also promote RNA polymerase entry when the nucleosomes are located appropriately.566 Recent research indicated that the nucleosomes located in TSS are essential for transcription by recruiting the transcription preinitiation complex.567 Some enhancer and promoter regions are accessible but showed low transcription activity for the lack of active modification markers, suggesting that the advanced topological structure and modification of local chromatin are also required for transcription.568 In addition, many epigenetic modifications are believed to indicate chromatin accessibility, but it’s also not absolute corresponded.328 Hence, the code of chromatin accessibility, nucleosome occupancy, epigenetic modification, and transcription activity requires further research to decode.
Chromatin accessibility participates in many cellular progresses, such as alternative splicing, DNA repair, immune and inflammatory responses, which are widely involved in embryogenesis, neurodegenerative disease, cardiovascular disease, diabetes, COVID-19 infection, and so on. Changes in chromatin accessibility and gene transcription are sequential, and the variation of chromatin accessibility always precede that of transcription.347 Hence, chromatin accessibility based information can raise an earlier prediction for the initiation, progression, and prognosis of many diseases. Up to now, several chromatin accessibility-based strategies have been applied in the clinical or preclinical diagnosis and treatment of different diseases. Many disease susceptibility risk loci are screened out by genome-wide association study, combined with high-throughput chromatin accessibility detection, we can comprehensively analyze the correlation between diseases and the accessibility variation at specific sites. CRISPR has great advantages in specific site editing, and CRISPR-based therapeutics have been used for the treatment of some diseases.569 The existing CRISPR-based treatment is mainly focused on DNA editing. Currently, scientists have achieved CRISPR-based epigenetic editing to control gene expression, having a much lower genomic mutation risk, which has a greater advantage in therapeutic application.570 Additionally, given the specificity of TFs to their binding motifs, they also are ideal candidate targets for site-specific epigenetic editing. Similar to PROTAC, a chemical inducer assembled Bcl6 with CDK9, phosphorylating RNA polymerases to promote the transcription of specific genes.571 It suggests that we can apply chemical inducer of proximity that can assemble the protein complexes with transcription factor to regulate the local chromatin accessibility. This strategy allow us to utilize the “enemy” (disease inducer) to defeat the “enemy” (disease) through precision striking. Besides, non-coding RNA drugs have become an important research direction for targeted therapy.572 Considering the role of non-coding RNAs in chromatin accessibility regulation, targeting non-coding RNA-mediated chromatin remodeling could be a promising field.
The laboratory results are always encouraging, however, the outcomes of clinical application are more about unsatisfactory, and the main reasons at least include the limitations in specificity and bioavailability. Fortunately, the high-throughput drug screening strategies allow us to obtain potential drugs more effectively. Meanwhile, the experimental model that can better imitate the real situation of patients, such as organoids, could further enhance the clinical application prospects of various drugs. Besides, the organism-wide chromatin accessibility profiles can pinpoint the regulatory variants during the development and disease progress, helping to propose more precise treatment strategies. In summary, yet there are many challenges, it’s sure that the investigation on chromatin accessibility will bring more and more help and surprise to our life science research and disease treatment.
Data availability
All the data of this study were available from the corresponding authors on reasonable request.
References
Kaplan, N. et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458, 362–366 (2009).
Lee, C. K. et al. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36, 900–905 (2004).
Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
McBryant, S. J., Adams, V. H. & Hansen, J. C. Chromatin architectural proteins. Chromosome Res. 14, 39–51 (2006).
Routh, A., Sandin, S. & Rhodes, D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA 105, 8872–8877 (2008).
Fyodorov, D. V. et al. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 19, 192–206 (2018).
Forouzanfar, M. H. et al. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 386, 2287–2323 (2015).
Macchi, F. & Sadler, K. C. Unraveling the epigenetic basis of liver development, regeneration and disease. Trends Genet. 36, 587–597 (2020).
Suarez, R. et al. Epigenetics in obesity and diabetes mellitus: new insights. Nutrients 15, 811 (2023).
Struhl, K. Non-canonical functions of enhancers: regulation of RNA polymerase III transcription, DNA replication, and V(D)J recombination. Trends Genet. 40, 471–479 (2024).
Eustermann, S. et al. Energy-driven genome regulation by ATP-dependent chromatin remodellers. Nat. Rev. Mol. Cell Biol. 25, 309–332 (2024).
Hewish, D. R. & Burgoyne, L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem. Biophys. Res. Commun. 52, 504–510 (1973).
Wu, C. et al. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell 16, 797–806 (1979).
Kornberg, R. D. Chromatin structure: a repeating unit of histones and DNA. Science 184, 868–871 (1974).
Horz, W. et al. Specific cleavage of chromatin by restriction nucleases. Nucleic Acids Res. 3, 3213–3226 (1976).
Mueller, P. R. & Wold, B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246, 780–786 (1989).
Rao, S., Procko, E. & Shannon, M. F. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J. Immunol. 167, 4494–4503 (2001).
Fatemi, M. et al. Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res. 33, e176 (2005).
Crawford, G. E. et al. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods 3, 503–509 (2006).
Sabo, P. J. et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods 3, 511–518 (2006).
Giresi, P. G. et al. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome. Res. 17, 877–885 (2007).
Albert, I. et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446, 572–576 (2007).
Boyle, A. P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008).
Gaulton, K. J. et al. A map of open chromatin in human pancreatic islets. Nat. Genet. 42, 255–259 (2010).
Kelly, T. K. et al. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome. Res. 22, 2497–2506 (2012).
Buenrostro, J. D. et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Sun, Y., Miao, N. & Sun, T. Detect accessible chromatin using ATAC-sequencing, from principle to applications. Hereditas 156, 29 (2019).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Peng, S. et al. Successful ATAC-seq from snap-frozen equine tissues. Front. Genet. 12, 641788 (2021).
Strzelecki M., et al. Isolation of nuclei from flash-frozen liver tissue for single-cell multiomics. J Vis Exp. 190, e64792 (2022).
Lareau, C. A. et al. Mitochondrial single-cell ATAC-seq for high-throughput multi-omic detection of mitochondrial genotypes and chromatin accessibility. Nat. Protoc. 18, 1416–1440 (2023).
Barcenas-Walls, J. R. et al. Nano-CUT&Tag for multimodal chromatin profiling at single-cell resolution. Nat. Protoc. 19, 791–830 (2024).
Guo, F. et al. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 27, 967–988 (2017).
Xu, W. et al. ISSAAC-seq enables sensitive and flexible multimodal profiling of chromatin accessibility and gene expression in single cells. Nat. Methods 19, 1243–1249 (2022).
Lin, J. et al. scNanoCOOL-seq: a long-read single-cell sequencing method for multi-omics profiling within individual cells. Cell Res. 33, 879–882 (2023).
Ye, F. et al. Fast and flexible profiling of chromatin accessibility and total RNA expression in single nuclei using Microwell-seq3. Cell Discov. 10, 33 (2024).
Chen, X. et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods 13, 1013–1020 (2016).
Thornton, C. A. et al. Spatially mapped single-cell chromatin accessibility. Nat. Commun. 12, 1274 (2021).
Mangiameli, S. M. et al. Photoselective sequencing: microscopically guided genomic measurements with subcellular resolution. Nat. Methods 20, 686–694 (2023).
Deng, Y. et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature 609, 375–383 (2022).
Minnoye, L. et al. Chromatin accessibility profiling methods. Nat. Rev. Methods Prim. 1, 10 (2021).
Grandi, F. C. et al. Chromatin accessibility profiling by ATAC-seq. Nat. Protoc. 17, 1518–1552 (2022).
Trotter, K. W. & Archer, T. K. The BRG1 transcriptional coregulator. Nucl. Recept. Signal. 6, e4 (2008).
Ostlund, F. A. et al. Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol. Cell Biol. 17, 895–905 (1997).
Miao, J. et al. Functional specificities of Brm and Brg-1 Swi/Snf ATPases in the feedback regulation of hepatic bile acid biosynthesis. Mol. Cell Biol. 29, 6170–6181 (2009).
Guo, X. et al. Hemogen/BRG1 cooperativity modulates promoter and enhancer activation during erythropoiesis. Blood 139, 3532–3545 (2022).
Garry, G. A. et al. The histone reader PHF7 cooperates with the SWI/SNF complex at cardiac super enhancers to promote direct reprogramming. Nat. Cell Biol. 23, 467–475 (2021).
Sun, X. et al. Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell 32, 574–589 (2017).
Sun, X. et al. Suppression of the SWI/SNF component arid1a promotes mammalian regeneration. Cell Stem Cell 18, 456–466 (2016).
Wang, L. et al. Inhibition of Arid1a increases stem/progenitor cell-like properties of liver cancer. Cancer Lett. 546, 215869 (2022).
Zhou, Y. et al. Enhanced SMARCD1, a subunit of the SWI/SNF complex, promotes liver cancer growth through the mTOR pathway. Clin. Sci. 134, 1457–1472 (2020).
Zhang, S. et al. mTORC1 promotes ARID1A degradation and oncogenic chromatin remodeling in hepatocellular carcinoma. Cancer Res. 81, 5652–5665 (2021).
Clapier, C. R. & Cairns, B. R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 (2009).
Matsui, S. et al. Pioneer and PRDM transcription factors coordinate bivalent epigenetic states to safeguard cell fate. Mol. Cell 84, 476–489 (2024).
Wang, B. et al. The NuRD complex cooperates with SALL4 to orchestrate reprogramming. Nat. Commun. 14, 2846 (2023).
Hoffmeister, H. et al. CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Nucleic Acids Res. 45, 10534–10554 (2017).
Kolla, V. et al. The tumour suppressor CHD5 forms a NuRD-type chromatin remodelling complex. Biochem. J. 468, 345–352 (2015).
Arends, T. et al. CHD4 is essential for transcriptional repression and lineage progression in B lymphopoiesis. Proc. Natl. Acad. Sci. USA 116, 10927–10936 (2019).
Marques, J. G. et al. NuRD subunit CHD4 regulates super-enhancer accessibility in rhabdomyosarcoma and represents a general tumor dependency. Elife 9, e54993 (2020).
Shi, W. et al. CHD4 and SMYD1 repress common transcriptional programs in the developing heart. Development 151, dev202505 (2024).
Graca, M. J. et al. The cromatin remodeler CHD4 sustains ewing sarcoma cell survival by controlling global chromatin architecture. Cancer Res. 84, 241–257 (2024).
Zhang, Y. et al. Collateral lethality between HDAC1 and HDAC2 exploits cancer-specific NuRD complex vulnerabilities. Nat. Struct. Mol. Biol. 30, 1160–1171 (2023).
Price, J. D. et al. DLX1 and the NuRD complex cooperate in enhancer decommissioning and transcriptional repression. Development 149, dev199508 (2022).
Chanda, P. K. et al. Nuclear S-nitrosylation defines an optimal zone for inducing pluripotency. Circulation 140, 1081–1099 (2019).
Langst, G. et al. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97, 843–852 (1999).
Yan, L. et al. Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat. Struct. Mol. Biol. 26, 258–266 (2019).
Goodwin, L. R. et al. Impaired SNF2L chromatin remodeling prolongs accessibility at promoters enriched for Fos/Jun binding sites and delays granule neuron differentiation. Front. Mol. Neurosci. 14, 680280 (2021).
Jiang, Y. et al. Nuclear RPSA senses viral nucleic acids to promote the innate inflammatory response. Nat. Commun. 14, 8455 (2023).
Aydin, O. Z., Vermeulen, W. & Lans, H. ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle 13, 3016–3025 (2014).
Cai, Y. et al. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 14, 872–874 (2007).
Ren, Z. et al. INO80-dependent remodeling of transcriptional regulatory network underlies the progression of heart failure. Circulation 149, 1121–1138 (2024).
Zhang, S. et al. INO80 is required for oncogenic transcription and tumor growth in non-small cell lung cancer. Oncogene 36, 1430–1439 (2017).
Law, C. T. et al. HELLS regulates chromatin remodeling and epigenetic silencing of multiple tumor suppressor genes in human hepatocellular carcinoma. Hepatology 69, 2013–2030 (2019).
Mizuguchi, G. et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 (2004).
Zhang, M. et al. Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling. Science 381, 313–319 (2023).
Jiang, Y. et al. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc. Natl. Acad. Sci. USA 107, 17274–17279 (2010).
Zhang, L. et al. DNMT and EZH2 inhibitors synergize to activate therapeutic targets in hepatocellular carcinoma. Cancer Lett. 548, 215899 (2022).
Har-Zahav, A. et al. The role of DNA demethylation in liver to pancreas transdifferentiation. Stem Cell Res. Ther. 13, 476 (2022).
Huang, Y. et al. Single-cell multi-omics sequencing of human spermatogenesis reveals a DNA demethylation event associated with male meiotic recombination. Nat. Cell Biol. 25, 1520–1534 (2023).
Guo, X. et al. LXRalpha promotes abdominal aortic aneurysm formation through UHRF1 epigenetic modification of miR-26b-3p. Circulation 150, 30–46 (2024).
Wu, B. K. et al. YAP induces an oncogenic transcriptional program through TET1-mediated epigenetic remodeling in liver growth and tumorigenesis. Nat. Genet. 54, 1202–1213 (2022).
Deng, S. et al. RNA m(6)A regulates transcription via DNA demethylation and chromatin accessibility. Nat. Genet. 54, 1427–1437 (2022).
Li, J. et al. Decoding the dynamic DNA methylation and hydroxymethylation landscapes in endodermal lineage intermediates during pancreatic differentiation of hESC. Nucleic Acids Res. 46, 2883–2900 (2018).
An, Y. et al. DNA methylation analysis explores the molecular basis of plasma cell-free DNA fragmentation. Nat. Commun. 14, 287 (2023).
Sun, K. et al. Size-tagged preferred ends in maternal plasma DNA shed light on the production mechanism and show utility in noninvasive prenatal testing. Proc. Natl. Acad. Sci. USA 115, E5106–E5114 (2018).
Fonseca, T. L. et al. Perinatal deiodinase 2 expression in hepatocytes defines epigenetic susceptibility to liver steatosis and obesity. Proc. Natl. Acad. Sci. USA 112, 14018–14023 (2015).
Oki, M., Aihara, H. & Ito, T. Role of histone phosphorylation in chromatin dynamics and its implications in diseases. Subcell. Biochem. 41, 319–336 (2007).
Dimitrova, E., Turberfield, A. H. & Klose, R. J. Histone demethylases in chromatin biology and beyond. EMBO Rep. 16, 1620–1639 (2015).
Wang, H. et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 (2004).
Xia, J. K. et al. Roles and regulation of histone acetylation in hepatocellular carcinoma. Front. Genet. 13, 982222 (2022).
Lv, X., Lv, Y. & Dai, X. Lactate, histone lactylation and cancer hallmarks. Expert Rev. Mol. Med. 25, e7 (2023).
Tan, M. et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).
Brehove, M. et al. Histone core phosphorylation regulates DNA accessibility. J. Biol. Chem. 290, 22612–22621 (2015).
Armache, A. et al. Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature 583, 852–857 (2020).
Ming, T. et al. Chromatin binding protein HMGN1 promotes HBV cccDNA transcription and replication by regulating the phosphorylation of histone 3. Antivir. Res. 221, 105796 (2024).
Udugama, M. et al. Histone H3.3 phosphorylation promotes heterochromatin formation by inhibiting H3K9/K36 histone demethylase. Nucleic Acids Res. 50, 4500–4514 (2022).
Natale, F. et al. Identification of the elementary structural units of the DNA damage response. Nat. Commun. 8, 15760 (2017).
Fuglerud, B. M. et al. SOX9 reprograms endothelial cells by altering the chromatin landscape. Nucleic Acids Res. 50, 8547–8565 (2022).
Fan, Y. et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123, 1199–1212 (2005).
Dou, Y. et al. Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin. J. Cell Biol. 158, 1161–1170 (2002).
Lopez, R. et al. Linker histone partial phosphorylation: effects on secondary structure and chromatin condensation. Nucleic Acids Res. 43, 4463–4476 (2015).
Michalak, E. M. et al. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 20, 573–589 (2019).
Kochat, V. et al. JMJD3 aids in reprogramming of bone marrow progenitor cells to hepatic phenotype through epigenetic activation of hepatic transcription factors. PLoS ONE 12, e173977 (2017).
Boonsanay, V. et al. Loss of SUV420H2-dependent chromatin compaction drives right-sided colon cancer progression. Gastroenterology 164, 214–227 (2023).
Feng, Q. et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058 (2002).
Godfrey, L. et al. DOT1L inhibition reveals a distinct subset of enhancers dependent on H3K79 methylation. Nat. Commun. 10, 2803 (2019).
Ferrari, F. et al. DOT1L-mediated murine neuronal differentiation associates with H3K79me2 accumulation and preserves SOX2-enhancer accessibility. Nat. Commun. 11, 5200 (2020).
Yang, L. et al. MLL2 regulates glucocorticoid receptor-mediated transcription of ENACalpha in human retinal pigment epithelial cells. Biochem. Biophys. Res. Commun. 525, 675–680 (2020).
Dacwag, C. S. et al. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol. Cell Biol. 27, 384–394 (2007).
Jbeli, A. H. et al. Brg1/PRMT5 nuclear complex epigenetically regulates FOXO1 in IPF mesenchymal progenitor cells. Am. J. Physiol. Lung Cell Mol. Physiol. 326, L344–L352 (2024).
Zheng, C. et al. Donafenib and GSK-J4 synergistically induce ferroptosis in liver cancer by upregulating HMOX1 expression. Adv. Sci. 10, e2206798 (2023).
Viscarra, J. A. et al. Histone demethylase JMJD1C is phosphorylated by mTOR to activate de novo lipogenesis. Nat. Commun. 11, 796 (2020).
Yang, Z. et al. Histone demethylase KDM1A promotes hepatic steatosis and inflammation by increasing chromatin accessibility in NAFLD. J. Lipid Res. 65, 100513 (2024).
Zhang, Y. et al. Histone demethylase KDM5B licenses macrophage-mediated inflammatory responses by repressing Nfkbia transcription. Cell Death Differ. 30, 1279–1292 (2023).
Zhang, B. et al. KDM4 orchestrates epigenomic remodeling of senescent cells and potentiates the senescence-associated secretory phenotype. Nat. Aging 1, 454–472 (2021).
Segala, G. et al. Monoubiquitination of histone H2B blocks eviction of histone variant H2A.Z from inducible enhancers. Mol. Cell 64, 334–346 (2016).
Hooda, J. et al. Early loss of histone H2B monoubiquitylation alters chromatin accessibility and activates key immune pathways that facilitate progression of ovarian cancer. Cancer Res. 79, 760–772 (2019).
Lin, C. Y. et al. Epigenetic regulator RNF20 underlies temporal hierarchy of gene expression to regulate postnatal cardiomyocyte polarization. Cell Rep. 42, 113416 (2023).
Yin, X. et al. H2AK121ub in Arabidopsis associates with a less accessible chromatin state at transcriptional regulation hotspots. Nat. Commun. 12, 315 (2021).
Artegiani, B. et al. Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell Stem Cell 24, 927–943 (2019).
Zhang, F. et al. Arsenite binds to the RING finger domains of RNF20-RNF40 histone E3 ubiquitin ligase and inhibits DNA double-strand break repair. J. Am. Chem. Soc. 136, 12884–12887 (2014).
Huang, F. et al. Interaction of the Jhd2 histone H3 Lys-4 demethylase with chromatin is controlled by histone H2A surfaces and restricted by H2B ubiquitination. J. Biol. Chem. 290, 28760–28777 (2015).
Worden, E. J. et al. Mechanism of cross-talk between H2B ubiquitination and H3 methylation by Dot1L. Cell 176, 1490–1501 (2019).
Barnes, C. E., English, D. M. & Cowley, S. M. Acetylation & Co: an expanding repertoire of histone acylations regulates chromatin and transcription. Essays Biochem. 63, 97–107 (2019).
Aseem, S. O. et al. Epigenomic evaluation of cholangiocyte transforming growth factor-beta signaling identifies a selective role for histone 3 lysine 9 acetylation in biliary fibrosis. Gastroenterology 160, 889–905 (2021).
Singh, M. et al. KAT6A mutations in Arboleda-Tham syndrome drive epigenetic regulation of posterior HOXC cluster. Hum. Genet. 142, 1705–1720 (2023).
Muthukrishnan, S. D. et al. P300 promotes tumor recurrence by regulating radiation-induced conversion of glioma stem cells to vascular-like cells. Nat. Commun. 13, 6202 (2022).
Jiang, C. et al. Targeting c-Jun inhibits fatty acid oxidation to overcome tamoxifen resistance in estrogen receptor-positive breast cancer. Cell Death Dis. 14, 653 (2023).
Dou, C. et al. The transcriptional activator Klf5 recruits p300-mediated H3K27ac for maintaining trophoblast stem cell pluripotency. J. Mol. Cell Biol. 15, mjad045 (2024).
Samata, M. et al. Intergenerationally maintained histone H4 lysine 16 acetylation is instructive for future gene activation. Cell 182, 127–144 (2020).
Li, X. et al. The methyltransferase METTL3 negatively regulates nonalcoholic steatohepatitis (NASH) progression. Nat. Commun. 12, 7213 (2021).
Yang, W. et al. A selective HDAC8 inhibitor potentiates antitumor immunity and efficacy of immune checkpoint blockade in hepatocellular carcinoma. Sci. Transl. Med. 13, eaaz6804 (2021).
Gregoricchio, S. et al. HDAC1 and PRC2 mediate combinatorial control in SPI1/PU.1-dependent gene repression in murine erythroleukaemia. Nucleic Acids Res. 50, 7938–7958 (2022).
Hu, M. et al. NAP1L2 drives mesenchymal stem cell senescence and suppresses osteogenic differentiation. Aging Cell 21, e13551 (2022).
Yuan, J. et al. Ezh2 competes with p53 to license lncRNA Neat1 transcription for inflammasome activation. Cell Death Differ. 29, 2009–2023 (2022).
Yuan, S. et al. The histone modification reader ZCWPW1 promotes double-strand break repair by regulating cross-talk of histone modifications and chromatin accessibility at meiotic hotspots. Genome Biol. 23, 187 (2022).
Liu, W. X. et al. Maternal vitamin B1 is a determinant for the fate of primordial follicle formation in offspring. Nat. Commun. 14, 7403 (2023).
Zhang, J. et al. PARylated PDHE1alpha generates acetyl-CoA for local chromatin acetylation and DNA damage repair. Nat. Struct. Mol. Biol. 30, 1719–1734 (2023).
Zhang, D. et al. Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019).
Merkuri, F., Rothstein, M. & Simoes-Costa, M. Histone lactylation couples cellular metabolism with developmental gene regulatory networks. Nat. Commun. 15, 90 (2024).
Trujillo, M. N. et al. Lactoylglutathione promotes inflammatory signaling in macrophages through histone lactoylation. Mol. Metab. 81, 101888 (2024).
Jing, Y. et al. Semisynthesis of site-specifically succinylated histone reveals that succinylation regulates nucleosome unwrapping rate and DNA accessibility. Nucleic Acids Res. 48, 9538–9549 (2020).
Lukasak, B. J. et al. TGM2-mediated histone transglutamination is dictated by steric accessibility. Proc. Natl. Acad. Sci. USA 119, e2086295177 (2022).
Zheng, C. et al. HBx increases chromatin accessibility and ETV4 expression to regulate dishevelled-2 and promote HCC progression. Cell Death Dis. 13, 116 (2022).
Cao, L. et al. Inflammatory cytokine-induced expression of MASTL is involved in hepatocarcinogenesis by regulating cell cycle progression. Oncol. Lett. 17, 3163–3172 (2019).
Hino, Y. et al. Mitochondrial stress induces AREG expression and epigenomic remodeling through c-JUN and YAP-mediated enhancer activation. Nucleic Acids Res. 50, 9765–9779 (2022).
Kang, X. et al. NASP antagonize chromatin accessibility through maintaining histone H3K9me1 in hepatocellular carcinoma. Biochim. Biophys. Acta. Mol. Basis Dis. 1864, 3438–3448 (2018).
Katsuda, T. et al. Cellular reprogramming in vivo initiated by SOX4 pioneer factor activity. Nat. Commun. 15, 1761 (2024).
Qu, Y. L. et al. Arid1a regulates insulin sensitivity and lipid metabolism. EBioMedicine 42, 481–493 (2019).
Yang, Y. et al. VGLL1 cooperates with TEAD4 to control human trophectoderm lineage specification. Nat. Commun. 15, 583 (2024).
Du, J. et al. MDIG-mediated H3K9me3 demethylation upregulates MYC by activating OTX2 and facilitates liver regeneration. Signal Transduct. Target Ther. 8, 351 (2023).
Lomvardas, S. & Thanos, D. Opening chromatin. Mol. Cell 9, 209–211 (2002).
Heslop, J. A. & Duncan, S. A. FoxA factors: the chromatin key and doorstop essential for liver development and function. Genes Dev. 34, 1003–1004 (2020).
Iwafuchi-Doi, M. et al. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell 62, 79–91 (2016).
Kain, J. et al. Pioneer factor Foxa2 enables ligand-dependent activation of type II nuclear receptors FXR and LXRalpha. Mol. Metab. 53, 101291 (2021).
Horisawa, K. et al. The dynamics of transcriptional activation by hepatic reprogramming factors. Mol. Cell 79, 660–676 (2020).
Li, Z. et al. The nucleosome map of the mammalian liver. Nat. Struct. Mol. Biol. 18, 742–746 (2011).
Thakur, A. et al. Hepatocyte nuclear factor 4-Alpha Is essential for the active epigenetic state at enhancers in mouse liver. Hepatology 70, 1360–1376 (2019).
Thakur, A. et al. HNF4A guides the MLL4 complex to establish and maintain H3K4me1 at gene regulatory elements. Commun. Biol. 7, 144 (2024).
Qu, M. et al. HNF4A defines tissue-specific circadian rhythms by beaconing BMAL1::CLOCK chromatin binding and shaping the rhythmic chromatin landscape. Nat. Commun. 12, 6350 (2021).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Hishida, T. et al. In vivo partial cellular reprogramming enhances liver plasticity and regeneration. Cell Rep. 39, 110730 (2022).
Cernilogar, F. M. et al. Pre-marked chromatin and transcription factor co-binding shape the pioneering activity of Foxa2. Nucleic Acids Res. 47, 9069–9086 (2019).
Li, Z. et al. Nuclear microRNA-mediated transcriptional control determines adult microglial homeostasis and brain function. Cell Rep. 43, 113964 (2024).
Boos, F. et al. The endothelial-enriched lncRNA LINC00607 mediates angiogenic function. Basic Res. Cardiol. 118, 5 (2023).
Leisegang, M. S. et al. Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation 136, 65–79 (2017).
Ma, Q. et al. Inducible lncRNA transgenic mice reveal continual role of HOTAIR in promoting breast cancer metastasis. Elife 11, e79126 (2022).
Liu, N. et al. Circular RNA circTmem241 drives group III innate lymphoid cell differentiation via initiation of Elk3 transcription. Nat. Commun. 13, 4711 (2022).
Wang, X. et al. Effects of RNAs on chromatin accessibility and gene expression suggest RNA-mediated activation. Int. J. Biochem. Cell Biol. 79, 24–32 (2016).
Dueva, R. et al. Neutralization of the positive charges on histone tails by RNA promotes an open chromatin structure. Cell Chem. Biol. 26, 1436–1449 (2019).
Liu, J. et al. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586 (2020).
Jachowicz, J. W. et al. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 49, 1502–1510 (2017).
Li, R. et al. Super-enhancer RNA m(6)A promotes local chromatin accessibility and oncogene transcription in pancreatic ductal adenocarcinoma. Nat. Genet. 55, 2224–2234 (2023).
Li, S., Ovcharenko, I. & Ovcharenko, I. Human enhancers are fragile and prone to deactivating mutations. Mol. Biol. Evol. 32, 2161–2180 (2015).
Mononen, J. et al. Genetic variation is a key determinant of chromatin accessibility and drives differences in the regulatory landscape of C57BL/6J and 129S1/SvImJ mice. Nucleic Acids Res. 52, 2904–2923 (2023).
Pandey, G. K. et al. Liver regulatory mechanisms of non-coding variants at lipid and metabolic trait loci. HGG Adv. 5, 100275 (2024).
Spisak, S. et al. A biallelic multiple nucleotide length polymorphism explains functional causality at 5p15.33 prostate cancer risk locus. Nat. Commun. 14, 5118 (2023).
Kirkland, N. J. et al. Age-dependent Lamin changes induce cardiac dysfunction via dysregulation of cardiac transcriptional programs. Nat. Aging 3, 17–33 (2023).
Xiong, J. et al. hnRNPU/TrkB defines a chromatin accessibility checkpoint for liver injury and nonalcoholic steatohepatitis pathogenesis. Hepatology 71, 1228–1246 (2020).
Huang, Q. et al. TFAM loss induces nuclear actin assembly upon mDia2 malonylation to promote liver cancer metastasis. EMBO J. 41, e110324 (2022).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Sanborn, A. L. et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA 112, E6456–E6465 (2015).
Xie, L. et al. BRD2 compartmentalizes the accessible genome. Nat. Genet. 54, 481–491 (2022).
Chen, B. et al. Stratifying TAD boundaries pinpoints focal genomic regions of regulation, damage, and repair. Brief. Bioinform 25, bbae306 (2024).
Li, N. et al. Single-cell 3D genome structure reveals distinct human pluripotent states. Genome. Biol. 25, 122 (2024).
Wahl, N. et al. SATB2 organizes the 3D genome architecture of cognition in cortical neurons. Mol. Cell 84, 621–639 (2024).
Kim, T. et al. Comparative characterization of 3D chromatin organization in triple-negative breast cancers. Exp. Mol. Med. 54, 585–600 (2022).
Wang, J. et al. Epigenomic landscape and 3D genome structure in pediatric high-grade glioma. Sci. Adv. 7, eabg4126 (2021).
Bendl, J. et al. The three-dimensional landscape of cortical chromatin accessibility in Alzheimer’s disease. Nat. Neurosci. 25, 1366–1378 (2022).
Moindrot, B., Imaizumi, Y. & Feil, R. Differential 3D genome architecture and imprinted gene expression: cause or consequence? Biochem. Soc. Trans. 52, 973–986 (2024).
Israel, J. W. et al. Tissue- and strain-specific effects of a genotoxic carcinogen 1,3-butadiene on chromatin and transcription. Mamm. Genome. 29, 153–167 (2018).
VonHandorf, A. et al. Chromium disrupts chromatin organization and CTCF access to its cognate sites in promoters of differentially expressed genes. Epigenetics 13, 363–375 (2018).
Chen, D. et al. Cigarette smoke component acrolein modulates chromatin assembly by inhibiting histone acetylation. J. Biol. Chem. 288, 21678–21687 (2013).
Fan, Y. et al. Multi-omics approach characterizes the role of Bisphenol F in disrupting hepatic lipid metabolism. Environ. Int. 187, 108690 (2024).
Dahl, H. et al. Dose rate dependent reduction in chromatin accessibility at transcriptional start sites long time after exposure to gamma radiation. Epigenetics 18, 2193936 (2023).
Wu, L. & Whitlock, J. J. Mechanism of dioxin action: ah receptor-mediated increase in promoter accessibility in vivo. Proc. Natl. Acad. Sci. USA 89, 4811–4815 (1992).
Okino, S. T. & Whitlock, J. J. Dioxin induces localized, graded changes in chromatin structure: implications for Cyp1A1 gene transcription. Mol. Cell Biol. 15, 3714–3721 (1995).
Liu, Y. et al. Effect of in ovo folic acid injection on hepatic IGF2 expression and embryo growth of broilers. J. Anim. Sci. Biotechnol. 7, 40 (2016).
Koester, J. et al. Niche stiffening compromises hair follicle stem cell potential during ageing by reducing bivalent promoter accessibility. Nat. Cell Biol. 23, 771–781 (2021).
Braude, P., Bolton, V. & Moore, S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332, 459–461 (1988).
Gao, L. et al. Chromatin accessibility landscape in human early embryos and its association with evolution. Cell 173, 248–259 (2018).
Wu, J. et al. Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 557, 256–260 (2018).
Li, L. et al. Single-cell multi-omics sequencing of human early embryos. Nat. Cell Biol. 20, 847–858 (2018).
Huang, X. et al. OCT4 cooperates with distinct ATP-dependent chromatin remodelers in naive and primed pluripotent states in human. Nat. Commun. 12, 5123 (2021).
Yu, H. et al. Dynamic reprogramming of H3K9me3 at hominoid-specific retrotransposons during human preimplantation development. Cell Stem Cell 29, 1031–1050 (2022).
Zaret, K. S. & Mango, S. E. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 37, 76–81 (2016).
Ameen, M. et al. Integrative single-cell analysis of cardiogenesis identifies developmental trajectories and non-coding mutations in congenital heart disease. Cell 185, 4937–4953 (2022).
Robson, A. et al. Histone H2B monoubiquitination regulates heart development via epigenetic control of cilia motility. Proc. Natl. Acad. Sci. USA 116, 14049–14054 (2019).
Zaidi, S. et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–223 (2013).
Garry, D. J. et al. Persistent expression of MNF identifies myogenic stem cells in postnatal muscles. Dev. Biol. 188, 280–294 (1997).
Shi, X. & Garry, D. J. Sin3 interacts with Foxk1 and regulates myogenic progenitors. Mol. Cell Biochem. 366, 251–258 (2012).
Sierra-Pagan, J. E. et al. FOXK1 regulates Wnt signalling to promote cardiogenesis. Cardiovasc. Res. 119, 1728–1739 (2023).
Sam, J. et al. Specificity, redundancy and dosage thresholds among gata4/5/6 genes during zebrafish cardiogenesis. Biol. Open 9, bio053611 (2020).
Song, M. et al. GATA4/5/6 family transcription factors are conserved determinants of cardiac versus pharyngeal mesoderm fate. Sci. Adv. 8, eabg834 (2022).
Arrieta, A. et al. Circadian control of histone turnover during cardiac development and growth. J. Biol. Chem. 300, 107434 (2024).
Zhong, H. et al. c-JUN is a barrier in hESC to cardiomyocyte transition. Life Sci. Alliance 6, e202302121 (2023).
Krup, A. L. et al. A Mesp1-dependent developmental breakpoint in transcriptional and epigenomic specification of early cardiac precursors. Development 150, dev201229 (2023).
Marques, I. J. et al. Wt1 transcription factor impairs cardiomyocyte specification and drives a phenotypic switch from myocardium to epicardium. Development 149, dev200375 (2022).
Fang, S. et al. Tet inactivation disrupts YY1 binding and long-range chromatin interactions during embryonic heart development. Nat. Commun. 10, 4297 (2019).
Meier, A. B. et al. Epicardioid single-cell genomics uncovers principles of human epicardium biology in heart development and disease. Nat. Biotechnol. 41, 1787–1800 (2023).
Mandla, R., Jung, C. & Vedantham, V. Transcriptional and epigenetic landscape of cardiac pacemaker cells: insights into cellular specialization in the sinoatrial node. Front. Physiol. 12, 712666 (2021).
Ieda, M. et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 (2010).
Nam, Y. J. et al. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development 141, 4267–4278 (2014).
Fernandez-Perez, A. et al. Hand2 selectively reorganizes chromatin accessibility to induce pacemaker-like transcriptional reprogramming. Cell Rep. 27, 2354–2369 (2019).
Galang, G. et al. ATAC-Seq reveals an Isl1 enhancer that regulates sinoatrial node development and function. Circ. Res. 127, 1502–1518 (2020).
van Eif, V. et al. Genome-wide analysis identifies an essential human TBX3 pacemaker enhancer. Circ. Res. 127, 1522–1535 (2020).
Iwafuchi-Doi, M. & Zaret, K. S. Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679–2692 (2014).
Mayran, A. & Drouin, J. Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem. 293, 13795–13804 (2018).
Reizel, Y. et al. Collapse of the hepatic gene regulatory network in the absence of FoxA factors. Genes Dev. 34, 1039–1050 (2020).
Reizel, Y. et al. Postnatal DNA demethylation and its role in tissue maturation. Nat. Commun. 9, 2040 (2018).
Madakashira, B. P. et al. Nuclear organization during hepatogenesis in zebrafish requires Uhrf1. Genes 12, 1081 (2021).
El, S. G. et al. Single-cell murine genetic fate mapping reveals bipotential hepatoblasts and novel multi-organ endoderm progenitors. Development 145, dev168658 (2018).
Yang, L. et al. The default and directed pathways of hepatoblast differentiation involve distinct epigenomic mechanisms. Dev. Cell 58, 1688–1700 (2023).
BRAIN Initiative Cell Census Network (BICCN). A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021).
Bakken, T. E. et al. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119 (2021).
Berg, J. et al. Human neocortical expansion involves glutamatergic neuron diversification. Nature 598, 151–158 (2021).
Ziffra, R. S. et al. Single-cell epigenomics reveals mechanisms of human cortical development. Nature 598, 205–213 (2021).
Li, Y. E. et al. A comparative atlas of single-cell chromatin accessibility in the human brain. Science 382, eadf7044 (2023).
Merrill, C. B. et al. Iterative assay for transposase-accessible chromatin by sequencing to isolate functionally relevant neuronal subtypes. Sci. Adv. 10, eadi4393 (2024).
Herring, C. A. et al. Human prefrontal cortex gene regulatory dynamics from gestation to adulthood at single-cell resolution. Cell 185, 4428–4447 (2022).
Pavlou, M. et al. Transcriptional and chromatin accessibility profiling of neural stem cells differentiating into astrocytes reveal dynamic signatures affected under inflammatory conditions. Cells 12, 948 (2023).
Chou, S. J. & Tole, S. Lhx2, an evolutionarily conserved, multifunctional regulator of forebrain development. Brain Res. 1705, 1–14 (2019).
Suresh, V. et al. Regulation of chromatin accessibility and gene expression in the developing hippocampal primordium by LIM-HD transcription factor LHX2. PLoS Genet. 19, e1010874 (2023).
Berg, D. A. et al. A common embryonic origin of stem cells drives developmental and adult neurogenesis. Cell 177, 654–668 (2019).
Zhong, S. et al. Single-cell epigenomics and spatiotemporal transcriptomics reveal human cerebellar development. Nat. Commun. 14, 7613 (2023).
Liu, J. et al. Genome-wide studies reveal the essential and opposite roles of ARID1A in controlling human cardiogenesis and neurogenesis from pluripotent stem cells. Genome Biol. 21, 169 (2020).
He, P. et al. A human fetal lung cell atlas uncovers proximal-distal gradients of differentiation and key regulators of epithelial fates. Cell 185, 4841–4860 (2022).
Morrisey, E. E. & Hogan, B. L. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev. Cell 18, 8–23 (2010).
Khattar, D. et al. PI3K signaling specifies proximal-distal fate by driving a developmental gene regulatory network in SOX9+ mouse lung progenitors. Elife 11, e67954 (2022).
Little, D. R. et al. Differential chromatin binding of the lung lineage transcription factor NKX2-1 resolves opposing murine alveolar cell fates in vivo. Nat. Commun. 12, 2509 (2021).
Guo, M. et al. Single cell multiomics identifies cells and genetic networks underlying alveolar capillary dysplasia. Am. J. Respir. Crit. Care Med. 208, 709–725 (2023).
Miao, Z. et al. Single cell regulatory landscape of the mouse kidney highlights cellular differentiation programs and disease targets. Nat. Commun. 12, 2277 (2021).
Riou, R. et al. ARID1A loss in adult hepatocytes activates beta-catenin-mediated erythropoietin transcription. Elife 9, e53550 (2020).
Xu, Z. et al. PTEN regulates hematopoietic lineage plasticity via PU.1-dependent chromatin accessibility. Cell Rep. 42, 112967 (2023).
Luo, Z. et al. Three distinct Atoh1 enhancers cooperate for sound receptor hair cell development. Proc. Natl. Acad. Sci. USA 119, e2119850119 (2022).
Xin, J. et al. Chromatin accessibility landscape and regulatory network of high-altitude hypoxia adaptation. Nat. Commun. 11, 4928 (2020).
Wahle, P. et al. Multimodal spatiotemporal phenotyping of human retinal organoid development. Nat. Biotechnol. 41, 1765–1775 (2023).
Kanton, S. et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422 (2019).
Trevino, A. E. et al. Chromatin accessibility dynamics in a model of human forebrain development. Science 367, eaay1645 (2020).
Lepilina, A. et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127, 607–619 (2006).
Cao, Y. et al. Identification of enhancer regulatory elements that direct epicardial gene expression during zebrafish heart regeneration. Development 149, dev200133 (2022).
Wang, X. et al. Keratin5-cytoskeleton-BMP4 network regulates cell phenotype conversions during cardiac regeneration. Exp. Cell Res. 418, 113272 (2022).
Beisaw, A. et al. AP-1 contributes to chromatin accessibility to promote sarcomere disassembly and cardiomyocyte protrusion during zebrafish heart regeneration. Circ. Res. 126, 1760–1778 (2020).
Quaife-Ryan, G. A. et al. Multicellular transcriptional analysis of mammalian heart regeneration. Circulation 136, 1123–1139 (2017).
Boogerd, C. J. et al. Cardiomyocyte proliferation is suppressed by ARID1A-mediated YAP inhibition during cardiac maturation. Nat. Commun. 14, 4716 (2023).
Sadahiro, T., Yamanaka, S. & Ieda, M. Direct cardiac reprogramming: progress and challenges in basic biology and clinical applications. Circ. Res. 116, 1378–1391 (2015).
Zhang, X. et al. MYOCD is required for cardiomyocyte-like cells induction from human urine cells and fibroblasts through remodeling chromatin. Stem Cell Rev. Rep. 18, 2414–2430 (2022).
Li, W. et al. A homeostatic arid1a-dependent permissive chromatin state licenses hepatocyte responsiveness to liver-injury-associated yap signaling. Cell Stem Cell 25, 54–68 (2019).
Wang, A. W. et al. The dynamic chromatin architecture of the regenerating liver. Cell Mol. Gastroenterol. Hepatol. 9, 121–143 (2020).
Schmitt, A. M. et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 48, 1370–1376 (2016).
Khanal, T. et al. NR2E3 is a key component in p53 activation by regulating a long noncoding RNA DINO in acute liver injuries. FASEB J. 33, 8335–8348 (2019).
Zhang, Y. et al. The Human mineral dust-induced gene, mdig, is a cell growth regulating gene associated with lung cancer. Oncogene 24, 4873–4882 (2005).
Lu, Y. et al. Lung cancer-associated JmjC domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3. Cell Cycle 8, 2101–2109 (2009).
Ma, H. et al. The nuclear receptor THRB facilitates differentiation of human PSCs into more mature hepatocytes. Cell Stem Cell 29, 795–809 (2022).
Sanal, M. G. Cell therapy from bench to bedside: hepatocytes from fibroblasts—the truth and myth of transdifferentiation. World J. Gastroenterol. 21, 6427–6433 (2015).
Pfisterer, U. et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. USA 108, 10343–10348 (2011).
Pang, Z. P. et al. Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223 (2011).
Wada, R. et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA 110, 12667–12672 (2013).
Yadav, N. et al. The therapeutic effect of bone marrow-derived liver cells in the phenotypic correction of murine hemophilia A. Blood 114, 4552–4561 (2009).
Kochat, V. et al. Donor antigen-primed regulatory T cells permit liver regeneration and phenotype correction in hemophilia A mouse by allogeneic bone marrow stem cells. Stem Cell Res. Ther. 6, 129 (2015).
Seirup, M. et al. Rapid changes in chromatin structure during dedifferentiation of primary hepatocytes in vitro. Genomics 114, 110330 (2022).
Merrell, A. J. et al. Dynamic transcriptional and epigenetic changes drive cellular plasticity in the liver. Hepatology 74, 444–457 (2021).
Venkatesh, I. et al. Developmental chromatin restriction of pro-growth gene networks acts as an epigenetic barrier to axon regeneration in cortical neurons. Dev. Neurobiol. 78, 960–977 (2018).
Patodia, S. & Raivich, G. Role of transcription factors in peripheral nerve regeneration. Front. Mol. Neurosci. 5, 8 (2012).
Dhara, S. P. et al. Cellular reprogramming for successful CNS axon regeneration is driven by a temporally changing cast of transcription factors. Sci. Rep. 9, 14198 (2019).
Neumann, S. & Woolf, C. J. Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron 23, 83–91 (1999).
Palmisano, I. et al. Epigenomic signatures underpin the axonal regenerative ability of dorsal root ganglia sensory neurons. Nat. Neurosci. 22, 1913–1924 (2019).
Paris, A. J. et al. STAT3-BDNF-TrkB signalling promotes alveolar epithelial regeneration after lung injury. Nat. Cell Biol. 22, 1197–1210 (2020).
Suzuki, N. et al. Adrenergic receptor signaling induced by Klf15, a regulator of regeneration enhancer, promotes kidney reconstruction. Proc. Natl. Acad. Sci. USA 119, e2090629177 (2022).
Ma, L. et al. A Janus-ROS healing system promoting infectious bone regeneration via sono-epigenetic modulation. Adv. Mater. 36, e2307846 (2024).
Bideyan, L. et al. Integrative analysis reveals multiple modes of LXR transcriptional regulation in liver. Proc. Natl. Acad. Sci. USA 119, e2122683119 (2022).
Hauck, A. K. et al. Nuclear receptor corepressors non-canonically drive glucocorticoid receptor-dependent activation of hepatic gluconeogenesis. Nat. Metab. 6, 825–836 (2024).
Grontved, L. et al. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J. 32, 1568–1583 (2013).
Hunter, A. L. et al. HNF4A modulates glucocorticoid action in the liver. Cell Rep. 39, 110697 (2022).
Xu, A. et al. Angiopoietin-like protein 4 decreases blood glucose and improves glucose tolerance but induces hyperlipidemia and hepatic steatosis in mice. Proc. Natl. Acad. Sci. USA 102, 6086–6091 (2005).
Koliwad, S. K. et al. Angiopoietin-like 4 (ANGPTL4, fasting-induced adipose factor) is a direct glucocorticoid receptor target and participates in glucocorticoid-regulated triglyceride metabolism. J. Biol. Chem. 284, 25593–25601 (2009).
Marilley, D. et al. Regulation of the vitellogenin gene B1 promoter after transfer into hepatocytes in primary cultures. Mol. Cell Endocrinol. 141, 79–93 (1998).
Goldstein, I. et al. Transcription factor assisted loading and enhancer dynamics dictate the hepatic fasting response. Genome Res. 27, 427–439 (2017).
Korenfeld, N. et al. Fasting hormones synergistically induce amino acid catabolism genes to promote gluconeogenesis. Cell Mol. Gastroenterol. Hepatol. 12, 1021–1036 (2021).
Madeo, F. et al. Caloric restriction mimetics against age-associated disease: targets, mechanisms, and therapeutic potential. Cell Metab. 29, 592–610 (2019).
Fan, Y. et al. Caloric restriction remodels the hepatic chromatin landscape and bile acid metabolism by modulating the gut microbiota. Genome Biol. 24, 98 (2023).
Wang, R. R. et al. The SWI/SNF chromatin-remodeling factors BAF60a, b, and c in nutrient signaling and metabolic control. Protein Cell 9, 207–215 (2018).
Corces, M. R. et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016).
Farlik, M. et al. DNA methylation dynamics of human hematopoietic stem cell differentiation. Cell Stem Cell 19, 808–822 (2016).
Zhu, Q. et al. Developmental trajectory of prehematopoietic stem cell formation from endothelium. Blood 136, 845–856 (2020).
Ranzoni, A. M. et al. Integrative single-cell RNA-seq and atac-seq analysis of human developmental hematopoiesis. Cell Stem Cell 28, 472–487 (2021).
Wong, Y. C. et al. Immune outcomes in the liver: Is CD8 T cell fate determined by the environment? J. Hepatol. 63, 1005–1014 (2015).
Benechet, A. P. et al. Dynamics and genomic landscape of CD8(+) T cells undergoing hepatic priming. Nature 574, 200–205 (2019).
Buehler, P. W., Humar, R. & Schaer, D. J. Haptoglobin therapeutics and compartmentalization of cell-free hemoglobin toxicity. Trends Mol. Med. 26, 683–697 (2020).
Pfefferle, M. et al. Hemolysis transforms liver macrophages into antiinflammatory erythrophagocytes. J. Clin. Investig. 130, 5576–5590 (2020).
Feierman, E. R. et al. Histone variant H2BE enhances chromatin accessibility in neurons to promote synaptic gene expression and long-term memory. Mol. Cell 10, S1097–S2765 (2024).
Elizaldi, S. R. et al. Chronic SIV-induced neuroinflammation disrupts CCR7+CD4+ T cell immunosurveillance in the rhesus macaque brain. J. Clin. Investig. 134, e175332 (2024).
Chang, J. et al. The dynamic landscape of chromatin accessibility and active regulatory elements in the mediobasal hypothalamus influences the seasonal activation of the reproductive axis in the male quail under long light exposure. BMC Genom. 25, 197 (2024).
Moonen, J. R. et al. KLF4 recruits SWI/SNF to increase chromatin accessibility and reprogram the endothelial enhancer landscape under laminar shear stress. Nat. Commun. 13, 4941 (2022).
Bianco, S. et al. The ovulatory signal precipitates LRH-1 transcriptional switching mediated by differential chromatin accessibility. Cell Rep. 28, 2443–2454 (2019).
An, X. et al. Genomic structural variation is associated with hypoxia adaptation in high-altitude zokors. Nat. Ecol. Evol. 8, 339–351 (2024).
Takahashi, J. S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017).
Zhu, B. et al. Coactivator-dependent oscillation of chromatin accessibility dictates circadian gene amplitude via REV-ERB loading. Mol. Cell 60, 769–783 (2015).
Takeda, Y. et al. Retinoic acid-related orphan receptor gamma directly regulates neuronal PAS domain protein 2 transcription in vivo. Nucleic Acids Res. 39, 4769–4782 (2011).
Hassan, H. M. et al. Regulation of chromatin accessibility by the farnesoid X Receptor Is essential for circadian and bile acid homeostasis in vivo. Cancers 14, 6191 (2022).
Palanivel, R. et al. Exposure to air pollution disrupts circadian rhythm through alterations in chromatin dynamics. iScience 23, 101728 (2020).
Chaurasia, P. & Thakur, M. K. Nucleosomal organization of the rat liver satellite DNA-containing chromatin during aging. Mech. Ageing Dev. 95, 63–70 (1997).
Chen, Y. et al. Remodeling of the H3 nucleosomal landscape during mouse aging. Transl. Med. Aging 4, 22–31 (2020).
Ding, M. et al. Integration of ATAC-Seq and RNA-Seq reveals FOSL2 drives human liver progenitor-like cell aging by regulating inflammatory factors. BMC Genom. 24, 260 (2023).
Bozukova, M. et al. Aging is associated with increased chromatin accessibility and reduced polymerase pausing in liver. Mol. Syst. Biol. 18, e11002 (2022).
Hu, H. et al. ZKSCAN3 counteracts cellular senescence by stabilizing heterochromatin. Nucleic Acids Res. 48, 6001–6018 (2020).
Waxman, D. J. & O’Connor, C. Growth hormone regulation of sex-dependent liver gene expression. Mol. Endocrinol. 20, 2613–2629 (2006).
Waxman, D. J. & Holloway, M. G. Sex differences in the expression of hepatic drug metabolizing enzymes. Mol. Pharm. 76, 215–228 (2009).
Rampersaud, A., Connerney, J. & Waxman, D. J. Plasma growth hormone pulses induce male-biased pulsatile chromatin opening and epigenetic regulation in adult mouse liver. Elife 12, RP91367 (2023).
Melia, T. et al. Hepatic long intergenic noncoding RNAs: high promoter conservation and dynamic, sex-dependent transcriptional regulation by growth hormone. Mol. Cell Biol. 36, 50–69 (2016).
Hao, P. & Waxman, D. J. Functional roles of sex-biased, growth hormone-regulated microRNAs miR-1948 and miR-802 in young adult mouse liver. Endocrinology 159, 1377–1392 (2018).
Fatkin, D. et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 341, 1715–1724 (1999).
Yamada, S. et al. TEAD1 trapping by the Q353R-Lamin A/C causes dilated cardiomyopathy. Sci. Adv. 9, eade7047 (2023).
Shi, L. et al. Downregulation of Wtap causes dilated cardiomyopathy and heart failure. J. Mol. Cell Cardiol. 188, 38–51 (2024).
Kuppe, C. et al. Spatial multi-omic map of human myocardial infarction. Nature 608, 766–777 (2022).
Kyryachenko, S. et al. Chromatin accessibility of human mitral valves and functional assessment of MVP risk loci. Circ. Res. 128, e84–e101 (2021).
Kimura, H. et al. Phenotype variability in patients carrying KCNJ2 mutations. Circ. Cardiovasc. Genet. 5, 344–353 (2012).
Chen, P. et al. Transcriptome and open chromatin analysis reveals the process of myocardial cell development and key pathogenic target proteins in Long QT syndrome type 7. J. Transl. Med. 22, 307 (2024).
Cheng, P. et al. ZEB2 shapes the epigenetic landscape of atherosclerosis. Circulation 145, 469–485 (2022).
Ranasinghe, A. et al. Altered smooth muscle cell histone acetylome by the SPHK2/S1P axis promotes pulmonary hypertension. Circ. Res. 133, 704–719 (2023).
Corces, M. R. et al. The chromatin accessibility landscape of primary human cancers. Science 362, eaav1898 (2018).
Dechassa, M. L. et al. Identification of chromatin-accessible domains in non-alcoholic steatohepatitis-derived hepatocellular carcinoma. Mol. Carcinog. 57, 978–987 (2018).
Tzameli, I. et al. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol. Cell Biol. 20, 2951–2958 (2000).
Lodato, N. J., Rampersaud, A. & Waxman, D. J. Impact of CAR agonist ligand TCPOBOP on mouse liver chromatin accessibility. Toxicol. Sci. 164, 115–128 (2018).
Mittal, P. & Roberts, C. The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat. Rev. Clin. Oncol. 17, 435–448 (2020).
Wu, J. N. & Roberts, C. W. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 3, 35–43 (2013).
Abatti, L. E. et al. Epigenetic reprogramming of a distal developmental enhancer cluster drives SOX2 overexpression in breast and lung adenocarcinoma. Nucleic Acids Res. 51, 10109–10131 (2023).
Wang, T. et al. Lactate-induced protein lactylation: a bridge between epigenetics and metabolic reprogramming in cancer. Cell Prolif. 56, e13478 (2023).
Zhang, Q. et al. Lactobacillus plantarum-derived indole-3-lactic acid ameliorates colorectal tumorigenesis via epigenetic regulation of CD8(+) T cell immunity. Cell Metab. 35, 943–960 (2023).
Grbesa, I. et al. Reshaping of the androgen-driven chromatin landscape in normal prostate cells by early cancer drivers and effect on therapeutic sensitivity. Cell Rep. 36, 109625 (2021).
Fan, H. et al. The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome. Res. 28, 192–202 (2018).
Xu, K. et al. A novel lncRNA RP11-386G11.10 reprograms lipid metabolism to promote hepatocellular carcinoma progression. Mol. Metab. 63, 101540 (2022).
Huo, Q. et al. Dysfunction of IKZF1/MYC/MDIG axis contributes to liver cancer progression through regulating H3K9me3/p21 activity. Cell Death Dis. 8, e2766 (2017).
Zhang, L. et al. ZNF143-Mediated H3K9 trimethylation upregulates CDC6 by activating MDIG in hepatocellular carcinoma. Cancer Res. 80, 2599–2611 (2020).
Craig, A. J. et al. Genome-wide profiling of transcription factor activity in primary liver cancer using single-cell ATAC sequencing. Cell Rep. 42, 113446 (2023).
Li, J. H. et al. MFAP5 facilitates the aggressiveness of intrahepatic Cholangiocarcinoma by activating the Notch1 signaling pathway. J. Exp. Clin. Cancer Res. 38, 476 (2019).
Peng, A. et al. DeltaNp63alpha facilitates proliferation and migration, and modulates the chromatin landscape in intrahepatic cholangiocarcinoma cells. Cell Death Dis. 14, 777 (2023).
Ozgur, E. et al. Plasma histone H4 and H4K20 trimethylation levels differ between colon cancer and precancerous polyps. Vivo 33, 1653–1658 (2019).
Zhao, Z. et al. The landscape of cryptic antisense transcription in human cancers reveals an oncogenic noncoding RNA in lung cancer. Sci. Adv. 9, eadf3264 (2023).
Mello, S. S. et al. Multifaceted role for p53 in pancreatic cancer suppression. Proc. Natl. Acad. Sci. USA 120, e2083030176 (2023).
Lv, S. et al. Integrated analysis reveals FOXA1 and Ku70/Ku80 as targets of ivermectin in prostate cancer. Cell Death Dis. 13, 754 (2022).
Schauwecker, S. M. et al. Histone H1 and chromosomal protein HMGN2 regulate prolactin-induced STAT5 transcription factor recruitment and function in breast cancer cells. J. Biol. Chem. 292, 2237–2254 (2017).
Wu, Y. et al. An LTR retrotransposon-derived long noncoding RNA lncMER52A promotes hepatocellular carcinoma progression by binding p120-catenin. Cancer Res. 80, 976–987 (2020).
Emmons, M. F. et al. HDAC8-mediated inhibition of EP300 drives a transcriptional state that increases melanoma brain metastasis. Nat. Commun. 14, 7759 (2023).
Pierce, S. E. et al. LKB1 inactivation modulates chromatin accessibility to drive metastatic progression. Nat. Cell Biol. 23, 915–924 (2021).
Llorente, A. et al. MAF amplification licenses ERalpha through epigenetic remodelling to drive breast cancer metastasis. Nat. Cell Biol. 25, 1833–1847 (2023).
Liu, Y. et al. The CTCF/LncRNA-PACERR complex recruits E1A binding protein p300 to induce pro-tumour macrophages in pancreatic ductal adenocarcinoma via directly regulating PTGS2 expression. Clin. Transl. Med. 12, e654 (2022).
Li, S. et al. Chromatin accessibility dynamics in colorectal cancer liver metastasis: uncovering the liver tropism at single cell resolution. Pharm. Res. 195, 106896 (2023).
Zhang, H. et al. RNA helicase DEAD box protein 5 regulates Polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology 64, 1033–1048 (2016).
Mani, S. et al. Restoration of RNA helicase DDX5 suppresses hepatitis B virus (HBV) biosynthesis and Wnt signaling in HBV-related hepatocellular carcinoma. Theranostics 10, 10957–10972 (2020).
Song, S. et al. CircHULC accelerates the growth of human liver cancer stem cells by enhancing chromatin reprogramming and chromosomal instability via autophagy. Cell Signal. 109, 110772 (2023).
Meng, Q. et al. Prognostic hub gene CBX2 drives a cancer stem cell-like phenotype in HCC revealed by multi-omics and multi-cohorts. Aging 15, 12817–12851 (2023).
Munoz-Galvan, S. et al. Essential role of PLD2 in hypoxia-induced stemness and therapy resistance in ovarian tumors. J. Exp. Clin. Cancer Res. 43, 57 (2024).
Hagiwara, M. et al. MUC1-C activates the PBAF chromatin remodeling complex in integrating redox balance with progression of human prostate cancer stem cells. Oncogene 40, 4930–4940 (2021).
Curiel, T. J. et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 10, 942–949 (2004).
Gao, Y. et al. Intratumoral stem-like CCR4+ regulatory T cells orchestrate the immunosuppressive microenvironment in HCC associated with hepatitis B. J. Hepatol. 76, 148–159 (2022).
Zhuo, B. et al. Integrative epigenetic analysis reveals AP-1 promotes activation of tumor-infiltrating regulatory T cells in HCC. Cell Mol. Life Sci. 80, 103 (2023).
Zhang, H. et al. Targeting WDxR motif reprograms immune microenvironment and inhibits hepatocellular carcinoma progression. EMBO Mol. Med. 15, e15924 (2023).
Chan, F. F. et al. Inhibition of CAF-1 histone chaperone complex triggers cytosolic DNA and dsRNA sensing pathways and induces intrinsic immunity of hepatocellular carcinoma. Hepatology 80, 295–311 (2024).
Divangahi, M. et al. Trained immunity, tolerance, priming and differentiation: distinct immunological processes. Nat. Immunol. 22, 2–6 (2021).
Wang, T. et al. Influenza-trained mucosal-resident alveolar macrophages confer long-term antitumor immunity in the lungs. Nat. Immunol. 24, 423–438 (2023).
Yang, Y. S. et al. Exosomal DEK removes chemoradiotherapy resistance by triggering quiescence exit of breast cancer stem cells. Oncogene 41, 2624–2637 (2022).
Zhang, L. L. et al. Chromatin accessibility analysis reveals that TFAP2A promotes angiogenesis in acquired resistance to anlotinib in lung cancer cells. Acta. Pharm. Sin. 41, 1357–1365 (2020).
Ku, B. et al. PRMT1 promotes pancreatic cancer development and resistance to chemotherapy. Cell Rep. Med. 5, 101461 (2024).
Carter, B. & Zhao, K. The epigenetic basis of cellular heterogeneity. Nat. Rev. Genet. 22, 235–250 (2021).
Goldsworthy, T. L. & Fransson-Steen, R. Quantitation of the cancer process in C57BL/6J, B6C3F1 and C3H/HeJ mice. Toxicol. Pathol. 30, 97–105 (2002).
Aydinlik, H. et al. Selective pressure during tumor promotion by phenobarbital leads to clonal outgrowth of beta-catenin-mutated mouse liver tumors. Oncogene 20, 7812–7816 (2001).
Vitobello, A. et al. Drug-induced chromatin accessibility changes associate with sensitivity to liver tumor promotion. Life Sci. Alliance 2, e201900461 (2019).
Sumazin, P. et al. Genomic analysis of hepatoblastoma identifies distinct molecular and prognostic subgroups. Hepatology 65, 104–121 (2017).
Smith, J. L. et al. YAP1 withdrawal in hepatoblastoma drives therapeutic differentiation of tumor cells to functional hepatocyte-like cells. Hepatology 73, 1011–1027 (2021).
Rodriguez, T. C. et al. Multiomics characterization of mouse hepatoblastoma identifies yes-associated protein 1 target genes. Hepatology 78, 58–71 (2023).
Wang, S. et al. Single-cell multiomics reveals heterogeneous cell states linked to metastatic potential in liver cancer cell lines. iScience 25, 103857 (2022).
Farkash-Amar, S. & Simon, I. Genome-wide analysis of the replication program in mammals. Chromosome Res. 18, 115–125 (2010).
Yaacov, A. et al. Cancer mutational processes vary in their association with replication timing and chromatin accessibility. Cancer Res. 81, 6106–6116 (2021).
Nepal, C. & Andersen, J. B. Alternative promoters in CpG depleted regions are prevalently associated with epigenetic misregulation of liver cancer transcriptomes. Nat. Commun. 14, 2712 (2023).
Duran-Ferrer, M. & Martin-Subero, J. I. Epigenomic characterization of lymphoid neoplasms. Annu. Rev. Pathol. 19, 371–396 (2024).
Rendeiro, A. F. et al. Chromatin accessibility maps of chronic lymphocytic leukaemia identify subtype-specific epigenome signatures and transcription regulatory networks. Nat. Commun. 7, 11938 (2016).
Wang, H. et al. Chromatin accessibility landscape of relapsed pediatric B-lineage acute lymphoblastic leukemia. Nat. Commun. 14, 6792 (2023).
Yang, D. et al. Intertumoral heterogeneity in SCLC is influenced by the cell type of origin. Cancer Discov. 8, 1316–1331 (2018).
Yang, L. et al. Identification of lineage-specific epigenetic regulators FOXA1 and GRHL2 through chromatin accessibility profiling in breast cancer cell lines. Cancer Gene Ther. 31, 736–745 (2024).
Hu, Y. et al. Single-cell sequencing technology applied to epigenetics for the study of tumor heterogeneity. Clin. Epigenetics 15, 161 (2023).
Roehrig, A. et al. Single-cell multiomics reveals the interplay of clonal evolution and cellular plasticity in hepatoblastoma. Nat. Commun. 15, 3031 (2024).
Henon, C. et al. Single-cell multiomics profiling reveals heterogeneous transcriptional programs and microenvironment in DSRCTs. Cell Rep. Med. 5, 101582 (2024).
Liu, Z. et al. Single-cell chromatin accessibility analysis reveals the epigenetic basis and signature transcription factors for the molecular subtypes of colorectal cancers. Cancer Discov. 14, 1082–1105 (2024).
Bian, S. et al. Integrative single-cell multiomics analyses dissect molecular signatures of intratumoral heterogeneities and differentiation states of human gastric cancer. Natl. Sci. Rev. 10, nwad94 (2023).
Raviram, R. et al. Integrated analysis of single-cell chromatin state and transcriptome identified common vulnerability despite glioblastoma heterogeneity. Proc. Natl. Acad. Sci. USA 120, e2083976176 (2023).
Mathur, R. et al. Glioblastoma evolution and heterogeneity from a 3D whole-tumor perspective. Cell 187, 446–463 (2024).
Nguyen, T. et al. Aurora kinase A inhibition reverses the Warburg effect and elicits unique metabolic vulnerabilities in glioblastoma. Nat. Commun. 12, 5203 (2021).
Bhattacharya, A. et al. MUC1-C intersects chronic inflammation with epigenetic reprogramming by regulating the set1a compass complex in cancer progression. Commun. Biol. 6, 1030 (2023).
Miyata, K. et al. Chromatin conformational changes at human satellite II contribute to the senescence phenotype in the tumor microenvironment. Proc. Natl. Acad. Sci. USA 120, e1989921176 (2023).
Zhang, Y. et al. Pegylated arginine deiminase drives arginine turnover and systemic autophagy to dictate energy metabolism. Cell Rep. Med. 3, 100498 (2022).
Bianco, A. C. & Silva, J. E. Intracellular conversion of thyroxine to triiodothyronine is required for the optimal thermogenic function of brown adipose tissue. J. Clin. Investig. 79, 295–300 (1987).
Fonseca, T. L. et al. Neonatal thyroxine activation modifies epigenetic programming of the liver. Nat. Commun. 12, 4446 (2021).
Alharthi, J. et al. A metabolic associated fatty liver disease risk variant in MBOAT7 regulates toll like receptor induced outcomes. Nat. Commun. 13, 7430 (2022).
Mansouri, A., Gattolliat, C. H. & Asselah, T. Mitochondrial dysfunction and signaling in chronic liver diseases. Gastroenterology 155, 629–647 (2018).
Yin, Q. et al. RPA1 controls chromatin architecture and maintains lipid metabolic homeostasis. Cell Rep. 40, 111071 (2022).
Yang, Z. et al. CRISPR-mediated BMP9 ablation promotes liver steatosis via the down-regulation of PPARalpha expression. Sci. Adv. 6, eabc5022 (2020).
Sun, Q. J. et al. The role of bone morphogenetic protein 9 in nonalcoholic fatty liver disease in mice. Front. Pharm. 11, 605967 (2020).
Gianmoena, K. et al. Epigenomic and transcriptional profiling identifies impaired glyoxylate detoxification in NAFLD as a risk factor for hyperoxaluria. Cell Rep. 36, 109526 (2021).
Ramesh, R. et al. Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J. Exp. Med. 211, 89–104 (2014).
Wang, Z. et al. Increased Th17 cells in coronary artery disease are associated with neutrophilic inflammation. Scand. Cardiovasc. J. 45, 54–61 (2011).
Yang, Y. et al. T-bet is essential for encephalitogenicity of both Th1 and Th17 cells. J. Exp. Med. 206, 1549–1564 (2009).
Moreno-Fernandez, M. E. et al. PKM2-dependent metabolic skewing of hepatic Th17 cells regulates pathogenesis of non-alcoholic fatty liver disease. Cell Metab. 33, 1187–1204 (2021).
Perez-Schindler, J. et al. Characterization of regulatory transcriptional mechanisms in hepatocyte lipotoxicity. Sci. Rep. 12, 11477 (2022).
BasuRay, S. et al. Accumulation of PNPLA3 on lipid droplets is the basis of associated hepatic steatosis. Proc. Natl. Acad. Sci. USA 116, 9521–9526 (2019).
Schwartz, B. E. et al. Discovery and targeting of the signaling controls of pnpla3 to effectively reduce transcription, expression, and function in pre-clinical NAFLD/NASH settings. Cells 9, 2247 (2020).
Amali, A. A. et al. Thioacetamide induced liver damage in zebrafish embryo as a disease model for steatohepatitis. J. Biomed. Sci. 13, 225–232 (2006).
Migdal, M. et al. Multi-omics analyses of early liver injury reveals cell-type-specific transcriptional and epigenomic shift. BMC Genom 22, 904 (2021).
Moore, A. et al. Arid1a loss drives nonalcoholic steatohepatitis in mice through epigenetic dysregulation of hepatic lipogenesis and fatty acid oxidation. Hepatology 69, 1931–1945 (2019).
Tian, W. et al. Brahma-related gene 1 bridges epigenetic regulation of proinflammatory cytokine production to steatohepatitis in mice. Hepatology 58, 576–588 (2013).
Du, J. et al. Vertical sleeve gastrectomy reverses diet-induced gene-regulatory changes impacting lipid metabolism. Sci. Rep. 7, 5274 (2017).
Seidman, J. S. et al. Niche-specific reprogramming of epigenetic landscapes drives myeloid cell diversity in nonalcoholic steatohepatitis. Immunity 52, 1057–1074 (2020).
Massey, V. et al. Integrated multiomics reveals glucose use reprogramming and identifies a novel hexokinase in alcoholic hepatitis. Gastroenterology 160, 1725–1740 (2021).
Weichselbaum, L. et al. Epigenetic basis for monocyte dysfunction in patients with severe alcoholic hepatitis. J. Hepatol. 73, 303–314 (2020).
Park, I. Y. et al. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology 132, 1476–1494 (2007).
Yates, K. B. et al. Epigenetic scars of CD8(+) T cell exhaustion persist after cure of chronic infection in humans. Nat. Immunol. 22, 1020–1029 (2021).
Kang, B. et al. The chromatin accessibility landscape of nonalcoholic fatty liver disease progression. Mol. Cells 45, 343–352 (2022).
Brougham-Cook, A. et al. High throughput interrogation of human liver stellate cells reveals microenvironmental regulation of phenotype. Acta. Biomater. 38, 240–253 (2022).
Jain, I., Brougham-Cook, A. & Underhill, G. H. Effect of distinct ECM microenvironments on the genome-wide chromatin accessibility and gene expression responses of hepatic stellate cells. Acta. Biomater. 167, 278–292 (2023).
Wells, R. G. The role of matrix stiffness in regulating cell behavior. Hepatology 47, 1394–1400 (2008).
Olsen, A. L. et al. Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 301, G110–G118 (2011).
Zhao, Y. Q. et al. Mechanical homeostasis imbalance in hepatic stellate cells activation and hepatic fibrosis. Front. Mol. Biosci. 10, 1183808 (2023).
Winkler, M. et al. Endothelial GATA4 controls liver fibrosis and regeneration by preventing a pathogenic switch in angiocrine signaling. J. Hepatol. 74, 380–393 (2021).
Tang, Z. et al. Y-box binding protein 1 promotes chromatin accessibility to aggravate liver fibrosis. Cell Signal. 109, 110750 (2023).
Poch, T. et al. Single-cell atlas of hepatic T cells reveals expansion of liver-resident naive-like CD4(+) T cells in primary sclerosing cholangitis. J. Hepatol. 75, 414–423 (2021).
Ji, R. et al. Multi-omics profiling of cholangiocytes reveals sex-specific chromatin state dynamics during hepatic cystogenesis in polycystic liver disease. J. Hepatol. 78, 754–769 (2023).
Jiang, H. et al. The early function of cortisol in liver during Aeromonas hydrophila infection: dynamics of the transcriptome and accessible chromatin landscapes. Front. Immunol. 13, 989075 (2022).
Radushkevitz-Frishman, T., Charni-Natan, M. & Goldstein, I. Dynamic chromatin accessibility during nutritional iron overload reveals a BMP6-independent induction of cell cycle genes. J. Nutr. Biochem. 119, 109407 (2023).
Sacirbegovic, F. et al. Graft-versus-host disease is locally maintained in target tissues by resident progenitor-like T cells. Immunity 56, 369–385 (2023).
Wang, Y. et al. Tissue-infiltrating alloreactive T cells require Id3 to deflect PD-1-mediated immune suppression during GVHD. Blood 143, 166–177 (2023).
King, M. C. & Wilson, A. C. Evolution at two levels in humans and chimpanzees. Science 188, 107–116 (1975).
Hu, B. et al. Deciphering the role of rapidly evolving conserved elements in primate brain development and exploring their potential involvement in Alzheimer’s Disease. Mol. Biol. Evol. 41, msae001 (2024).
Zhang, X. et al. Landscape of double-stranded DNA breaks in postmortem brains from Alzheimer’s disease and non-demented individuals. J. Alzheimers Dis. 94, 519–535 (2023).
Turner, T. N. et al. Genomic patterns of de novo mutation in simplex autism. Cell 171, 710–722 (2017).
Shi, X. et al. Heterozygous deletion of the autism-associated gene CHD8 impairs synaptic function through widespread changes in gene expression and chromatin compaction. Am. J. Hum. Genet. 110, 1750–1768 (2023).
Richardson, R. T. et al. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J. Biol. Chem. 275, 30378–30386 (2000).
Zhang, S. et al. NASP gene contributes to autism by epigenetic dysregulation of neural and immune pathways. J. Med. Genet. 61, 677–688 (2024).
Gibb, S. L. et al. Ethanol-induced increase in Fyn kinase activity in the dorsomedial striatum is associated with subcellular redistribution of protein tyrosine phosphatase alpha. J. Neurochem. 119, 879–889 (2011).
Egervari, G. et al. Chromatin accessibility mapping of the striatum identifies tyrosine kinase FYN as a therapeutic target for heroin use disorder. Nat. Commun. 11, 4634 (2020).
Zhang, Y. et al. Microglia: the hub of intercellular communication in ischemic stroke. Front. Cell Neurosci. 16, 889442 (2022).
Zhang, Z. et al. Integrated analysis of chromatin and transcriptomic profiling identifies PU.1 as a core regulatory factor in microglial activation induced by chronic cerebral hypoperfusion. Mol. Neurobiol. 61, 2569–2589 (2024).
Adams, L. et al. A single-nuclei paired multiomic analysis of the human midbrain reveals age- and Parkinson’s disease-associated glial changes. Nat. Aging 4, 364–378 (2024).
Mohan, K. et al. Age-associated changes in endothelial transcriptome and epigenetic landscapes correlate with elevated risk of cerebral microbleeds. J. Am. Heart Assoc. 12, e31044 (2023).
Yeo, L. et al. Autoreactive T effector memory differentiation mirrors beta cell function in type 1 diabetes. J. Clin. Investig. 128, 3460–3474 (2018).
Abdelsamed, H. A. et al. Beta cell-specific CD8(+) T cells maintain stem cell memory-associated epigenetic programs during type 1 diabetes. Nat. Immunol. 21, 578–587 (2020).
Afzali, B. et al. BACH2 immunodeficiency illustrates an association between super-enhancers and haploinsufficiency. Nat. Immunol. 18, 813–823 (2017).
Robertson, C. C. et al. Fine-mapping, trans-ancestral and genomic analyses identify causal variants, cells, genes and drug targets for type 1 diabetes. Nat. Genet. 53, 962–971 (2021).
Chiou, J. et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 594, 398–402 (2021).
Wang, G. et al. Integrating genetics with single-cell multiomic measurements across disease states identifies mechanisms of beta cell dysfunction in type 2 diabetes. Nat. Genet. 55, 984–994 (2023).
Xue, D. et al. Functional interrogation of twenty type 2 diabetes-associated genes using isogenic human embryonic stem cell-derived beta-like cells. Cell Metab. 35, 1897–1914 (2023).
Qiao, J. et al. A distinct role of STING in regulating glucose homeostasis through insulin sensitivity and insulin secretion. Proc. Natl. Acad. Sci. USA 119, e2101848119 (2022).
Greenwald, W. W. et al. Pancreatic islet chromatin accessibility and conformation reveals distal enhancer networks of type 2 diabetes risk. Nat. Commun. 10, 2078 (2019).
Aylward, A. et al. Glucocorticoid signaling in pancreatic islets modulates gene regulatory programs and genetic risk of type 2 diabetes. PLoS Genet. 17, e1009531 (2021).
Qadir, M. M. F. et al. Single cell regulatory architecture of human pancreatic islets suggests sex differences in β cell function and the pathogenesis of type 2 diabetes. Res Sq. Preprint (2024).
Wang, R. R. et al. Dietary intervention preserves beta cell function in mice through CTCF-mediated transcriptional reprogramming. J. Exp. Med. 219, e20211779 (2022).
Wei, Z. et al. Vitamin D switches BAF complexes to protect beta cells. Cell 173, 1135–1149 (2018).
Chiou, J. et al. Single-cell chromatin accessibility identifies pancreatic islet cell type- and state-specific regulatory programs of diabetes risk. Nat. Genet. 53, 455–466 (2021).
Bansal, A. et al. Integrative omics analyses reveal epigenetic memory in diabetic renal cells regulating genes associated with kidney dysfunction. Diabetes 69, 2490–2502 (2020).
Cavalli, M. et al. The thioesterase ACOT1 as a regulator of lipid metabolism in type 2 diabetes detected in a multi-omics study of human liver. OMICS 25, 652–659 (2021).
Erarslan-Uysal, B. et al. Chromatin accessibility landscape of pediatric T-lymphoblastic leukemia and human T-cell precursors. EMBO Mol. Med. 12, e12104 (2020).
Valenzi, E. et al. Single-nucleus chromatin accessibility identifies a critical role for TWIST1 in idiopathic pulmonary fibrosis myofibroblast activity. Eur. Respir. J. 62, 2200474 (2023).
Raphael, H. E. et al. Activator protein transcription factors coordinate human IL-33 expression from noncanonical promoters in chronic airway disease. JCI Insight 9, e174786 (2024).
Sullivan, K. M. & Susztak, K. Unravelling the complex genetics of common kidney diseases: from variants to mechanisms. Nat. Rev. Nephrol. 16, 628–640 (2020).
Le-Bury, G. et al. HIV-1 active and latent infections induce disparate chromatin reorganization and transcriptional regulation of mRNAs and lncRNAs in SupT1 cells. mBio 14, e261923 (2023).
Zhang, B. et al. Multimodal single-cell datasets characterize antigen-specific CD8(+) T cells across SARS-CoV-2 vaccination and infection. Nat. Immunol. 24, 1725–1734 (2023).
Morris, J. A. et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 51, 258–266 (2019).
Dutta, P. et al. Global epigenetic changes induced by SWI2/SNF2 inhibitors characterize neomycin-resistant mammalian cells. PLoS ONE 7, e49822 (2012).
Rakesh, R. et al. Altering mammalian transcription networking with ADAADi: an inhibitor of ATP-dependent chromatin remodeling. PLoS ONE 16, e251354 (2021).
Muthuswami, R. et al. BRG1 is a prognostic indicator and a potential therapeutic target for prostate cancer. J. Cell Physiol. 234, 15194–15205 (2019).
Schick, S. et al. Acute BAF perturbation causes immediate changes in chromatin accessibility. Nat. Genet. 53, 269–278 (2021).
Fiskus, W. et al. BRG1/BRM inhibitor targets AML stem cells and exerts superior preclinical efficacy combined with BET or Menin inhibitor. Blood 143, 2059–2072 (2024).
Lee, D. et al. The Bromodomain Inhibitor PFI-3 Sensitizes Cancer Cells to DNA Damage by Targeting SWI/SNF. Mol. Cancer Res 19, 900–912 (2021).
Wu, Q. et al. Targeting the chromatin remodeling enzyme BRG1 increases the efficacy of chemotherapy drugs in breast cancer cells. Oncotarget 7, 27158–27175 (2016).
Shishodia, S. et al. Selective and cell-active PBRM1 bromodomain inhibitors discovered through NMR fragment screening. J. Med. Chem. 65, 13714–13735 (2022).
Cochran, A. G. & Flynn, M. GNE-235: a lead compound selective for the second bromodomain of PBRM1. J. Med. Chem. 66, 13116–13134 (2023).
Filippakopoulos, P. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012).
Brien, G. L. et al. Targeted degradation of BRD9 reverses oncogenic gene expression in synovial sarcoma. Elife 7, e41305 (2018).
Li, M. et al. BRD7 inhibits enhancer activity and expression of BIRC2 to suppress tumor growth and metastasis in nasopharyngeal carcinoma. Cell Death Dis. 14, 121 (2023).
Clark, P. G. et al. LP99: discovery and synthesis of the first selective BRD7/9 bromodomain inhibitor. Angew. Chem. Int. Ed. Engl. 54, 6217–6221 (2015).
Martin, L. J. et al. Structure-based design of an in vivo active selective BRD9 inhibitor. J. Med. Chem. 59, 4462–4475 (2016).
Zhou, L. et al. Targeting BRD9 by I-BRD9 efficiently inhibits growth of acute myeloid leukemia cells. Transl. Cancer Res. 10, 3364–3372 (2021).
Ordonez-Rubiano, S. C. et al. Rational design and development of selective BRD7 bromodomain inhibitors and their activity in prostate cancer. J. Med. Chem. 66, 11250–11270 (2023).
Panditharatna, E. et al. BAF complex maintains glioma stem cells in pediatric H3K27M glioma. Cancer Discov. 12, 2880–2905 (2022).
Farnaby, W. et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 15, 672–680 (2019).
Xiao, L. et al. Targeting SWI/SNF ATPases in enhancer-addicted prostate cancer. Nature 601, 434–439 (2022).
Cantley, J. et al. Selective PROTAC-mediated degradation of SMARCA2 is efficacious in SMARCA4 mutant cancers. Nat. Commun. 13, 6814 (2022).
Michael Hulse, et al. Preclinical characterization of PRT3789, a potent and selective SMARCA2 targeted degrader. Cancer Res. 3263, abstract. 82, (12_Supplement) (2022).
Theodoulou, N. H. et al. Discovery of I-BRD9, a selective cell active chemical probe for bromodomain containing protein 9 inhibition. J. Med. Chem. 59, 1425–1439 (2016).
Kurata, K. et al. BRD9 degradation disrupts ribosome biogenesis in multiple myeloma. Clin. Cancer Res. 29, 1807–1821 (2023).
Zhang, J. et al. Structure-based identification of new orally bioavailable BRD9-PROTACs for treating acute myelocytic leukemia. Eur. J. Med. Chem. 26, 115872 (2023).
Katrina L. Jackson, et al. The discovery and characterization of CFT8634: a potent and selective degrader of BRD9 for the treatment of SMARCB1-perturbed cancers. Cancer Res. 82, (12_Supplement), abstract. ND9 (2022).
Claudia Dominici, et al. Investigation of FHD-609, a potent degrader of BRD9, in preclinical models of acute myeloid leukemia (AML). Mol. Cancer Ther. 22, (12_Supplement), abstract. A49 (2023).
Zoppi, V. et al. Iterative design and optimization of initially inactive proteolysis targeting chimeras (PROTACs) identify VZ185 as a potent, fast, and selective von hippel-lindau (VHL) based dual degrader probe of BRD9 and BRD7. J. Med. Chem. 62, 699–726 (2019).
Oyama, Y. et al. CHD4 regulates platinum sensitivity through MDR1 expression in ovarian cancer: a potential role of CHD4 inhibition as a combination therapy with platinum agents. PLoS ONE 16, e251079 (2021).
Wyhs, N. et al. Time-resolved fluorescence resonance energy transfer assay for discovery of small-molecule inhibitors of methyl-CpG binding domain protein 2. J. Biomol. Screen 19, 1060–1069 (2014).
Choi, S. H. et al. TSA promotes CRISPR/Cas9 editing efficiency and expression of cell division-related genes from plant protoplasts. Int. J. Mol. Sci. 22, 7817 (2021).
He, S. et al. Histone deacetylase inhibitor SAHA improves high salinity tolerance associated with hyperacetylation-enhancing expression of ion homeostasis-related genes in cotton. Int. J. Mol. Sci. 21, 7105 (2020).
Tammaro, A. et al. HDAC1/2 inhibitor therapy improves multiple organ systems in aged mice. iScience 27, 108681 (2024).
Fournel, M. et al. MGCD0103, a novel isotype-selective histone deacetylase inhibitor, has broad spectrum antitumor activity in vitro and in vivo. Mol. Cancer Ther. 7, 759–768 (2008).
Nakajima, H. et al. FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp. Cell Res. 241, 126–330 (1998).
Beckers, T. et al. Distinct pharmacological properties of second generation HDAC inhibitors with the benzamide or hydroxamate head group. Int. J. Cancer 121, 1138–1148 (2007).
Kishtagari, Ashwin. et al. A first-in-class inhibitor of ISWI-mediated (ATP-dependent) transcription repression releases terminal-differentiation in AML cells while sparing normal hematopoiesis. Blood 132, 216 (2018).
Mishra, N. K. et al. Fluorinated aromatic amino acids are sensitive 19F NMR probes for bromodomain-ligand interactions. ACS Chem. Biol. 9, 2755–2760 (2014).
Picaud, S. et al. Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia. Sci. Adv. 2, e1600760 (2016).
Zhang, D. et al. Discovery of alkoxy benzamide derivatives as novel BPTF bromodomain inhibitors via structure-based virtual screening. Bioorg. Chem. 86, 494–500 (2019).
Xu, J. et al. Compound C620-0696, a new potent inhibitor targeting BPTF, the chromatin-remodeling factor in non-small-cell lung cancer. Front. Med. 14, 60–67 (2020).
Assimon, V. A. et al. CB-6644 Is a selective inhibitor of the RUVBL1/2 complex with anticancer activity. ACS Chem. Biol. 14, 236–244 (2019).
Nano, N. et al. Sorafenib as an Inhibitor of RUVBL2. Biomolecules 10, 605 (2020).
Christman, J. K. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21, 5483–5495 (2002).
Bowler, E. H. et al. Proteomic analysis of azacitidine-induced degradation profiles identifies multiple chromatin and epigenetic regulators including Uhrf1 and Dnmt1 as sensitive to azacitidine. J. Proteome Res. 18, 1032–1042 (2019).
Kapuganti, R. S. et al. Role of clusterin gene 3′-UTR polymorphisms and promoter hypomethylation in the pathogenesis of pseudoexfoliation syndrome and pseudoexfoliation glaucoma. Biochim. Biophys. Acta. Gene Regul. Mech. 1866, 194980 (2023).
Lai, S. C. et al. DNMT3b/OCT4 expression confers sorafenib resistance and poor prognosis of hepatocellular carcinoma through IL-6/STAT3 regulation. J. Exp. Clin. Cancer Res. 38, 474 (2019).
Rodems, T. S. et al. Reversible epigenetic alterations regulate class I HLA loss in prostate cancer. Commun. Biol. 5, 897 (2022).
Li, J. et al. Epigenetic targeting drugs potentiate chemotherapeutic effects in solid tumor therapy. Sci. Rep. 7, 4035 (2017).
Aanniz, T. et al. Natural bioactive compounds targeting DNA methyltransferase enzymes in cancer: mechanisms insights and efficiencies. Chem. Biol. Interact. 392, 110907 (2024).
DeNizio, J. E. et al. TET-TDG Active DNA Demethylation at CpG and Non-CpG Sites. J. Mol. Biol. 433, 166877 (2021).
Xu, W. et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011).
Xiao, M. et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 26, 1326–1338 (2012).
Raineri, S. & Mellor, J. IDH1: linking metabolism and epigenetics. Front. Genet. 9, 493 (2018).
Verdura, S. et al. An olive oil phenolic is a new chemotype of mutant isocitrate dehydrogenase 1 (IDH1) inhibitors. Carcinogenesis 40, 27–40 (2019).
Park, J. W. & Turcan, S. Epigenetic reprogramming for targeting IDH-mutant malignant gliomas. Cancers 11, 1616 (2019).
Xu, X. et al. MAGI2-AS3 inhibits breast cancer by downregulating DNA methylation of MAGI2. J. Cell Physiol. 236, 1116–1130 (2021).
Welsh, S. J. et al. Transcriptional heterogeneity overcomes super-enhancer disrupting drug combinations in multiple myeloma. Blood Cancer Discov. 5, 34–55 (2024).
Lindqvist, B. et al. Chromatin maturation of the HIV-1 provirus in primary resting CD4+ T cells. PLoS Pathog. 16, e1008264 (2020).
Ebrahimi, A. et al. Bromodomain inhibition of the coactivators CBP/EP300 facilitate cellular reprogramming. Nat. Chem. Biol. 15, 519–528 (2019).
Pan, F. et al. Enhancer remodeling drives MLL oncogene-dependent transcriptional dysregulation in leukemia stem cells. Blood Adv. 7, 2504–2519 (2023).
Hu, L. et al. Epigenetic regulation of interleukin 6 by histone acetylation in macrophages and its role in paraquat-induced pulmonary fibrosis. Front. Immunol. 7, 696 (2016).
Yu, L. et al. Megakaryocytic leukemia 1 bridges epigenetic activation of NADPH oxidase in macrophages to cardiac ischemia-reperfusion injury. Circulation 138, 2820–2836 (2018).
Lu, Y. et al. Global landscape of 2-hydroxyisobutyrylation in human pancreatic cancer. Front. Oncol. 12, 1001807 (2022).
Pattenden, S. G. et al. High-throughput small molecule screen identifies inhibitors of aberrant chromatin accessibility. Proc. Natl. Acad. Sci. USA 113, 3018–3023 (2016).
Leppanen, N. et al. SIX2 promotes cell plasticity via Wnt/beta-catenin signalling in androgen receptor independent prostate cancer. Nucleic Acids Res. 52, 5610–5623 (2024).
Qian, C. et al. ONECUT2 acts as a lineage plasticity driver in adenocarcinoma as well as neuroendocrine variants of prostate cancer. Nucleic Acids Res. 52, 7740–7760 (2024).
Han, H. et al. Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell 36, 483–497 (2019).
Holmes, A. G. et al. A MYC inhibitor selectively alters the MYC and MAX cistromes and modulates the epigenomic landscape to regulate target gene expression. Sci. Adv. 8, eabh3635 (2022).
Thieme, E. et al. CDK9 inhibition induces epigenetic reprogramming revealing strategies to circumvent resistance in lymphoma. Mol. Cancer 22, 64 (2023).
Seredinski, S. et al. DNA topoisomerase inhibition with the HIF inhibitor acriflavine promotes transcription of lncRNAs in endothelial cells. Mol. Ther. Nucleic Acids 27, 1023–1035 (2022).
Yoon, J. et al. E2F and STAT3 provide transcriptional synergy for histone variant H2AZ activation to sustain glioblastoma chromatin accessibility and tumorigenicity. Cell Death Differ. 29, 1379–1394 (2022).
Liu, L. et al. Identification of a KLF5-dependent program and drug development for skeletal muscle atrophy. Proc. Natl Acad. Sci. USA 118, e2102895118 (2021).
Hubmann, R. et al. Targeting nuclear NOTCH2 by gliotoxin recovers a tumor-suppressor NOTCH3 activity in CLL. Cells 9, 1484 (2020).
Xu, T. et al. OCT4 regulates WNT/beta-catenin signaling and prevents mesoendoderm differentiation by repressing EOMES in porcine pluripotent stem cells. J. Cell Physiol. 238, 2855–2866 (2023).
Ma, S. et al. Transcriptional repression of estrogen receptor alpha by YAP reveals the Hippo pathway as therapeutic target for ER(+) breast cancer. Nat. Commun. 13, 1061 (2022).
Clark, D. J. & Felsenfeld, G. A nucleosome core is transferred out of the path of a transcribing polymerase. Cell 71, 11–22 (1992).
Lorch, Y., LaPointe, J. W. & Kornberg, R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49, 203–210 (1987).
Chen, X. et al. Structures of +1 nucleosome-bound PIC-Mediator complex. Science 378, 62–68 (2022).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Chavez, M. et al. Advances in CRISPR therapeutics. Nat. Rev. Nephrol. 19, 9–22 (2023).
Nunez, J. K. et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell 184, 2503–2519 (2021).
Sarott, R. C. et al. Relocalizing transcriptional kinases to activate apoptosis. Science 386, eadl5361 (2024).
Niu, D., Wu, Y. & Lian, J. Circular RNA vaccine in disease prevention and treatment. Signal Transduct. Target Ther. 8, 341 (2023).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82203598), and Natural Science Foundation of Chongqing (No. cstc2021jcyj-msxmX0536).
Author information
Authors and Affiliations
Contributions
Guanbin Song conceived and designed this work. Yang Chen drafted this manuscript. Rui Liang and Yong Li are the figure and table makers. Lingli Jiang and Di Ma helped to collect the material. Qing Luo revised the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interest.
Consent for publication
All authors have read the manuscript and consent to publication
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, Y., Liang, R., Li, Y. et al. Chromatin accessibility: biological functions, molecular mechanisms and therapeutic application. Sig Transduct Target Ther 9, 340 (2024). https://doi.org/10.1038/s41392-024-02030-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41392-024-02030-9