Abstract
Major depressive disorder (MDD) is a common disease affecting 300 million people worldwide. The existing drugs are ineffective for approximately 30% of patients, so it is urgent to develop new antidepressant drugs with novel mechanisms. Here, we found that norisoboldine (NOR) showed an antidepressant efficacy in the chronic social defeat stress (CSDS) depression model in the tail suspension, forced swimming, and sucrose consumption tests. We then utilized the drug-treated CSDS mice paradigm to segregate and gain differential protein groups of CSDS versus CON (CSDSCON), imipramine (IMI)-treated versus CSDS (IMICSDS), and NOR-treated versus CSDS (NORCSDS) from the prefrontal cortex. These protein expression alterations were first analyzed by ANOVA with p < 0.05. The protein cluster 1 and cluster 3, in which the pattern of protein levels similar to the mood pattern, showed enrichment in functions and localizations related to mitochondrion, ribosome and synapses. Further GO analysis of the common proteins for NORCSDS groups and NORIMI groups supported the findings from ANOVA analysis. We employed Protein-Protein interaction (PPI) analysis to examine the proteins of NORCSDS and NORIMI, revealing an enrichment of the proteins associated with the mitochondrial ribosomal and synaptic functions. Further independent analysis using parallel reaction monitoring (PRM) revealed that Cox7c, Mrp142, Naa30, Ighm, Apoa4, Ssu72, Mrps30, Apoh, Acbd5, and Cdv3, exhibited regulation in the NOR-treated group to support the homeostasis of mitochondrial functions. Additionally, Dcx, Arid1b, Rnf112, and Fam3c, were also observed to undergo modulation in the NOR-treated groups to support the synaptic formation and functions. These findings suggest that the proteins involved in depression treatment exert effects in strengthen the mitochondrial and synaptic functions in the mice PFC. Western blot analysis supported the data that the levels of Mrpl42, Cox7c, Naa30, Rnf112, Dcx Apoa4, Apoh and Fam3c were altered in the CSDS mice, and rescued by NOR treatment, supporting the PRM data. NOR treatment also rescued the NLRP3 inflammasome activation in CSDS mice. In summary, the current proteomic research conducted on the prefrontal cortex has provided valuable insights into the specific and shared molecular mechanisms underlying pathophysiology and treatment to CSDS-induced depression, shedding light on the therapeutic effects of Norisoboldine.
Similar content being viewed by others
Introduction
Major depressive disorder (MDD) is a recurrent and disabling mental disorder. About 30% of patients will become treatment-resistant depression (TRD), although a variety of antidepressants are used currently [1]. It is partially because its pathophysiological and pharmacological mechanisms remain poorly elucidated [2]. Therefore, there is a significant clinical demand for drugs that can effectively treat refractory depression [1]. In recent decades, proteomics technique has become a powerful tool to uncover the pathophysiology of neuropsychiatric disorders [3]. Proteomic method, due to their open-ended nature, can make valuable contributions to the investigation of the molecular neurobiology of MDD and mechanisms of treatment.
Current research on the pathophysiology and treatment of MDD highlights several key mechanisms, including mitochondrial mechanism, neuroimmune mechanism, and synaptic plasticity mechanism. 1) Mitochondria mechanism is that because both MDD and stress are associated with changes in mitochondrial biogenesis, redox imbalance, increased oxidative damages of cellular macromolecules, and apoptosis revealed by literature Meta-analysis in both humans and animal studies [4]. 2) Neuroimmune mechanism is based on that patients with the refractory depression show all the main characteristics of chronic inflammatory response, including increased expression of proinflammatory cytokines and their receptors in peripheral blood and cerebrospinal fluid, and increased levels of reactants, chemokines and soluble adhesion molecules in the acute phase [5]. Elevated level of inflammatory factors, such as C-reactive protein and proinflammatory cytokines, have been detected in the serum of patients with refractory depression [6]. 3) Synaptic plasticity mechanism is particularly significant in understanding the impact of chronic stress damaged brain function. It has been discovered that chronic stress can result in damage to excitatory synapses, leading to adverse effects on cognition and mood. The critical mechanism(s) of antidepressant efficacy is restoration of the function of stress-harmed synapses within these same circuits [6]. These three mechanisms, namely mitochondrial functions, inflammation, and synaptic plasticity, are closely interconnected. Healthy mitochondria is essential for regulating inflammation, while inflammation can increase the vulnerability of synapses and neurons to the detrimental effects of stress. This interplay highlights the importance of maintaining mitochondrial health in modulating inflammation and protecting synapses and neurons from stress-relevant damage [7].
Among various animal models, chronic social defeat stress (CSDS) is one of the most commonly employed models for investigating possible causes and treatments for depression [8]. The prefrontal cortex (PFC) plays a crucial role in integrating cognitive evaluations, such as assessing the controllability of a stressor [9] or the valence of sensory stimuli [10], particularly supported by the data from using functional magnetic resonance imaging (fMRI) [6]. In animal models, chronic stress impairs attentional set-shifting tasks mediated by the PFC and decision making in reward-based tasks [11], mimicking the human deficits seen in MDD patients [12]. There is compelling evidence indicating that treatments capable of alleviating depression symptoms also have the potential to reverse deficits observed in the PFC [13]. While the prefrontal cortical plasticity in the CSDS depression animal model is known to be potentially abnormal, and the antidepressant treatment has been shown to correct the abnormality, the underlying molecular basis of these effects may be intrinsically different and not yet fully understood.
Norisoboldine (NOR) is the primary isoquinoline alkaloid constituent of the root of Lindera aggregata [14]. It is well established that NOR has anti-inflammatory and renal protective efficacy [15]. In particular, recent studies have found that the binding of NOR and Aryl Hydrocarbon Receptor (AHR) has the activity of inhibiting inflammasome [16]. However, its antidepressant effect remains to be illustrated. To evaluate the improvement effect of NOR on the depressive-like behavior of mice, this study used imipramine as a positive control drug. Imipramine is a tricyclic antidepressant that blocks the reuptake of norepinephrine and serotonin in the central nervous system, increasing the concentration of these two neurotransmitters in the synaptic cleft, thereby exerting an antidepressant effect. Imipramine is a classic antidepressant that has been clinically used and researched for many years. Imipramine is also a commonly used positive control drug that can be used to evaluate the antidepressant effect of other drugs or substances.
In this study, we have identified the chemicals with antidepressant properties through the behavioral tests. To probe into depression- and anti-depressant-related proteins, we utilized the prefrontal cortex tissues from the normal control (CON), CSDS, imipramine (IMI)-treated, and NOR-treated mouse groups. The differentially regulated genes of NOR-treated versus CSDS (NORCSDS) and CSDS versus CON (CSDSCON); IMI-treated versus CSDS (IMICSDS) were segregated as the three different gene groups related to the pathophysiology and treatment of depression. The resulting protein data were comparatively analyzed, offering the significant molecular basis associated with depression and antidepressant behavioral phenotypes.
Materials and methods
Experimental animals and drug treatment
All procedures were performed in accordance with the National Institutes of Health protocols for the use and care of laboratory animals (ISBN:0-309-05377-3). The animal protocols were approved by the Animal Ethics Review Board of Beijing Huada Protein R & D Center Co., Ltd. The ethics approval number is BPI20221011-1. Male CD1 (9-10 weeks old) or C57BL/6J mice (7-8 weeks old) were purchased from Vital River Co. Ltd. (Beijing, China). Animal treatments were performed as described in the supplemental materials. In the experiment with normal animals in Fig. 1, the control (CON) groups were injected with saline to compare with imipramine group, whereas the control group was injected with 1%DMSO in saline compared with the rest.
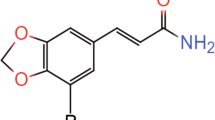
A1 The chemical structure of Norisoboldine (NOR). A2 Schematic timeline of the experimental procedures. A3 After the i.p. injection (5 mg/kg NOR and 10.0 mg/kg NOR) for sixty minutes, NOR significantly reduced the immobility time compared with the C57 mice in the tail suspension test (TST) (N = 9–10 per group). A4 After 5 days of injection, NOR significantly decreased the immobility time in the FST compared with the C57 mice (N = 9–10 per group). B1 Schematic timeline of the experimental procedures. B2 After CSDS procedure, sucrose preference index were determined to select the depression model mice (N = 6–14 per group). B3 After 60 minutes of NOR treatment, NOR significantly reduced the immobility time in the TST compared with the untreated CSDS mice (N = 7–14 per group). B4 After 4 days of NOR treatment, NOR significantly enhanced the sucrose preference index in the SPT compared with the untreated CSDS mice (N = 4–6 per group). B5 After 5 days of NOR treatment, NOR significantly reduced the immobility time in the FST compared with the untreated CSDS mice (N = 7–18 per group). Data were analyzed by one-way ANOVA followed by post hoc Tukey’s test and are presented as the mean ± SEM (*p < 0.05, **p < 0.01, ***p < 0.001).
Chronic social defeat stress (CSDS) procedures
CSDS procedure was followed as previously described [17] and detailed in the supplemental materials. In our study, the susceptibility versus the resistance in the CSDS mice were 80% and 20%, respectively.
Behavioral testing
As previously described [18], we conducted the tail suspension test (TST), forced swimming test (FST) and sucrose preference test (SPT), to investigate animal behaviors as detailed in the supplemental materials.
Protein extraction, trypsin digestion and LC-MS analysis
After the behavioral tests were completed, the prefrontal cortex tissues were separated from the brain. The service of proteomics analysis was provided by PTM Biolabs, Inc. as described in the supplemental material in details.
ANOVA ananlysis
Single factor analysis of variance (ANOVA) was used to test the significance of the difference between the means of two or more samples as described in the supplemental materials.
Bioinformatics analysis
For bioinformatics analysis, gene ontology (GO) enrichment and functional classification were constituted with the DAVID Bioinformatics Resources (https://david.ncifcrf.gov). GO categorization of the proteins was conducted, including biological process (GO-BP), cellular compartments (GO-CC), and molecular function (GO-MF).
Protein-protein interaction (PPI) network analysis
The functional PPI network was explored in Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) and visualized with Cytoscape as described in the supplemental materials.
Parallel reaction monitoring (PRM) MS assay
The PFC proteins were extracted and digested. The resulting peptides were collected and analyzed with the Q-Exacutive mass spectrometer coupled online to Easy-nLC 1000 UPLC system as described in the supplemental materials.
Western blot (WB) analysis
For WB analysis, mice were euthanized and brain sampled were acquired. The Western Blot analysis were performed using standard method as described in the supplemental materials in details.
Statistical analysis
The statistical analysis was conducted with SPSS 24.0. All the results were analyzed using ANOVA test or Student’s t test and expressed as means ± standard error. Data were considered to be statistically significant at p < 0.05.
Results
NOR demonstrated antidepressant effects on depression-like animal behaviors
To investigate whether NOR (Fig. 1A1) produces antidepressant effects, mice were intraperitonially (i.p.) injected with different concentrations of NOR (2.5 mg/kg, 5 mg/kg, and 10 mg/kg) for 60 min, 3 days or 5 days (Fig. 1A2). Sixty minutes after NOR treatment, the mice were subjected to TST. The duration of immobility in the NOR-treated groups was significantly lower than that of the controls in the TST (p < 0.05, Fig. 1A3). The positive control imipramine also showed an antidepressant effect (p < 0.05, Fig. 1A3. To further examine the antidepressant effect of NOR after long-term treatment, we treated mice with NOR (2.5 mg/kg, 5 mg/kg, and 10 mg/ kg) for 5 consecutive days, and FST were performed after the treatments. We found that the immobility times for FST in the NOR-treated groups were significantly reduced compared to the controls in a dose-dependent manner (p < 0.05, Fig. 1A4). Imipramine also demonstrated an antidepressant effect (p < 0.001, Fig. 1A4). With the antidepressant effects of NOR, we further conducted the open field test (OFT) to determine whether NOR causes locomotor hyperactivity in mice. We performed OFT after 3 consecutive days of NOR treatment with 5 mg/kg and 10 mg/kg doses. NOR treatments were 60 min before testing. The total distance traveled were 20.04 ± 2.11 cm, 21.35 ± 4.63 cm, 25.19 ± 4.36 cm, for CON, NOR-L (low dose) and NOR-H (high dose), respectively (ANOVA analysis p > 0.05, N = 8 per group). The time traveled in the center area showed no significant differences (ANOVA analysis p > 0.05, N = 8 per group) in the CON (26.96 ± 8.01 sec.), NOR-L (24.58 + 16.75 sec.), and NOR-H (24.52 + 7.03 sec.), which suggested that NOR does not cause locomotor hyperactivity in mice.
NOR reverses chronic social defeat stress (CSDS)-induced depression-like behaviors
CSDS is a well-established animal model of depression-like behaviors [19]. According to our findings, the dose of 10 mg/kg NOR demonstrated the best antidepressant efficacy in the FST and TST tests. Therefore, we used a dose of 10 mg/kg NOR to study the antidepressant effect of NOR in CSDS depression model mice (Fig. 1B1). After repeated social defeat for 10 days by aggressive CD1 mice, the mice showed reduced sucrose preference index than the average of control were used as depression model mice in the SPT (Fig. 1B2). In the TST, we examined the effects of NOR (10 mg/kg) administration for 60 min in the CSDS model mice after treatment in the TST. After 60 min of NOR treatment, the immobility time for the untreated CSDS groups was significantly higher than that of the controls in the TST (p < 0.01, Fig. 1B3). NOR treatment for 60 min significantly reduced the immobility time (p < 0.01) in CSDS mice in the TST (Fig. 1B3). The SPT were performed to determine the hedonic-like behavior of the NOR-treated CSDS mice after 4 days of treatment. The sucrose preference index were significantly increased in both of the high and low doses of the NOR-treated groups (p < 0.01, Fig. 1B4). After 5 days of treatment, in the CSDS-induced groups, the immobility time was significantly higher (p < 0.05) than that of the controls in the FST (Fig. 1B5), and NOR-treatment for 5 days significantly reduced the immobility time (p < 0.01) induced by CSDS (Fig. 1B5). These results showed that NOR exerts an antidepressant effect on CSDS-induced depression-like behaviors.
Prefrontal proteomics analysis of the NOR-treated CSDS mice
In the present work, conducting the assessments of behavioral tests, we gained prefrontal cortical protein samples from the mice of all CON, CSDS, NOR, and IMI groups. The protein extracts from three animals per group were used for the LC-MS analysis. In total, 4938 nonredundant proteins were relatively quantified across all 4 sample pools based upon the FDR less than 0.01 (Fig. 2B). The protein quantitative principal component analysis results of all samples are presented in Fig. 2C, where the degree of aggregation between samples suggested that the samples had significant differences. As described in Fig. 2A, a label free quantification experiment was designed and performed on the three animals from the CON, CSDS, NOR + CSDS, and IMI + CSDS groups. The prefrontal cortical proteins from the four groups (three animal sample for each group) were trypsin digested to be applied to the relative quantitation. The peptides were characterized by LC-MS analysis. The relative protein abundances were analyzed using the signature ion ratio. We comparatively characterized the global expression changes in the mouse PFC proteomes of the three groups CSDSCON, NORCSDS, IMICSDS following the Volcano plot with the fold change of >1.2 or <0.833, and p < 0.05 (Fig. 2D–F).
A Flow diagram of quantitative proteomics. The prefrontal cortex tissue samples from the four groups [including CON, CSDS, NOR-treated CSDS, IMI-treated CSDS)] were used. B Overview of Protein Identification. C Principal Component Analysis(PCA). D–F Volcano plots of differentially expressed proteins in comparison between the CSDS and CON(CSDSCON), NOR-treated CSDS(NORCSDS), IMI-treated CSDS(IMICSDS) groups. Green spots represent downregulated proteins, and orange spots represent upregulated proteins. Gray spots represent nondifferentially expressed proteins.
ANOVA differential protein expression clustering analysis
The ANOVA analysis (p < 0.05) was conducted for all CON, CSDS, NOR and IMI four groups. We conducted GO classification analysis on all of ANOVA identified differential proteins and the top twenty categories were presented (Fig. 3A–D). The Gene Ontology Biological Process (GO-BP), Gene Ontology Cellular Component (GO-CC), and the Gene Ontology Molecular Functions (GO-MF) category analysis showed that the all differentiated proteins from ANOVA were significantly enriched in mitochondrial, ribosomal and synaptic location and functions. GO-BP enriched categories included regulation of protein metabolic process, negative regulation of neuronal death, ribose phosphate metabolic process, ribose phosphate biosynthetic process, regulation of vesicle mediated transport, and protein containing complex assembly (Fig. 3B). The GO-CC category analysis revealed that the proteins were enriched in mitochondrion, inner mitochondrial membrane protein complex, ribosome, riboneucleoprotein complex, synapse, neuron projection and so on (Fig. 3C). The GO-MF category analysis showed that the proteins were enriched in CoA ligase activity, disulfide oxidoreductase activity, RNA binding, structural constituent of ribosome, cytoskeletal protein binding, tubulin binding and so on (Fig. 3D).
A Heatmap diagrams displaying the expression pattern and the levels of differentially expressed proteins after ANOVA analysis. Hierarchical clustering of the differentially expressed proteins representing log2 of their fold changes in the IMI, NOR groups in relative to the CSDS group, and CSDS group in relative to the CON group. Red represents upregulation, and blue represents downregulation with darker shades indicating greater changes in expression. B–D GO-BP, GO-CC, and GO-MF of all ANOVA identified differential proteins; E–G GO-BP, GO-CC, and GO-MF of the cluster 1; H–J GO-BP, GO-CC, and GO-MF of the cluster 2; K–M GO-BP, GO-CC, and GO-MF of the cluster 3. The vertical axis is GO function description information, and the horizontal axis is the proportion of differential protein in this function type compared with Fold enrichment level of identified protein after Log2 conversion. The color of dots represents P value of enrichment significance, blue represents strong enrichment significance, and the size of dots represents the number of different proteins in GO functional classes.
Based on the expression trend, differential proteins are divided into multiple clusters, with each cluster represented by a different color. Among the three clusters, the cluster 1 and cluster 3 protein expression levels agreed with the mood pattern, that the protein levels in the CSDS depression model group was altered than the CON, and in the antidepressant IMI-treatment and the novel antidepressant NOR-treatment groups, the protein levels were rescued (Fig. 3A). Because of the importance of cluster 1 and cluster 3, we conducted GO classification analysis on cluster 1 and cluster 3 proteins based on the clustering results of ANOVA analysis in each comparative group, and the results in the form of a bubble chart (Fig. 3). The GO-BP category analysis showed that the proteins in cluster 1 were significantly enriched in mitochondria-related biological process, including tricarboxylic acid metabolic process, oxidative phosphorylation, respiratory electron transport chain, cellular respiration, mitochondrial transmembrane transport, aerobic respiration, carboxylic acid metabolic process. Moreover, they were also associated with synaptic functions, such as receptor localization to synapse, postsynapse organization, and protein localization to membrane (Fig. 3E). The GO-CC category analysis revealed that the proteins belonging to cluster1 were significantly enriched in cellular compartments related to mitochondria, such as, respiratory chain complex, mitochondrial respirasome, respirasome, oxidoreductase complex, mitochondrial inner membrane, mitochondrial matrix, mitochondrial membrane, mitochondrial envelope, and mitochondrion. Additionally, these proteins in cluster 1 were also found to be associated with synaptic functions, which exhibited representation in categories such as presynaptic membrane, presynapse, synapse, and myelin sheath (Fig. 3F). The GO-MF category analysis showed that the proteins in cluster 1 were also enriched in mitochondria-related functions, including NADH dehydrogenase (ubiquinone) activity, NADH dehydrogenase (quinone) activity, NADH dehydrogenase activity, oxidoreductase activity acting on NAD(P)H, electron transfer activity, and oxidoreductase activity (Fig. 3G).
As to the GO analysis for cluster 3, the GO-BP analysis showed that the proteins in cluster 3 were enriched in ribosomeand endoplasmic reticulum-related biological process, including peptide transport, ribose phosphate metabolic process, ribosome biogenesis, ribosomal large subunit biogenesis, protein localization to endoplasmic reticulum (Fig. 3K). The GO-CC analysis revealed that the proteins belonging to cluster 3 were significantly enriched in cellular compartments related to ribosome, such as ribonucleoprotein complex, ribosome, ribosomal subunit, cytosolic small ribosomal subunit, large ribosomal subunit, cytosolic ribosome, polysomal ribosome and so on (Fig. 3L). The GO-MF analysis showed that the proteins in cluster 3 were also enriched in ribosome-related functions, including RNA binding, rRNA binding, mRNA 5’UTR binding, poly(U) RNA binding, structural constituent of ribosome, polyA binding, large ribosomal large RNA binding, etc. (Fig. 3M).
Taken together, proteins within the cluster 1 and cluster 3 exhibited significantly enrichment in mitochondrial, ribosomal and synaptic protein categories (Fig. 3). However, proteins within the cluster 2 did not exhibit enrichment in mitochondria-, ribosome- and synapse-related protein category (Fig. 3E, F, G). These suggests the antidepressant behavior specifically involved in crucial biological processes related to mitochondrial, ribosomal, and synaptic functions.
GO analysis of NORCSDS and NORIMI with the fold change greater than 1.2 fold or less than 0.833 fold and p < 0.05
The differentially expressed proteins in prefrontal cortex were identified among the NORCSDS, NORIMI, and a total of 153, 103 proteins were gained based upon the fold change (greater than 1.2 fold or less than 0.833 fold) and p < 0.05, respectively. Among the identified proteins, we found that there were 107 upregulated and 46 downregulated proteins in the NORCSDS group, 72 upregulated and 31 downregulated proteins in the NORIMI group.
To systematically identify the potential biological functions and processes of these differentially expressed proteins by NOR treatment, they were subjected to analysis using the GO pathway databases, and the top ten enriched items were presented in the GO-BP, GO-CC, GO-MF categories as shown in Fig. 4. The GO-BP, GO-CC and GO-MF category analysis revealed that many proteins are associated with mitochondrial, ribosomal and synaptic functions such as mitochondrion, regulation of reactive oxygen species metabolic process, synapse, mRNA processing, RNA splicing, mRNA transport, endoplasmic reticulum, structural constituent of ribosome, and RNA binding, and others, supporting the ANOVA data.
A The GO analysis for differential proteins of NORCSDS. B The GO analysis for differential proteins of NORIMI. The top 10 most significant enriched entries in the three categories, including biological process (GO-BP), cellular component (GO-CC), and molecular function (GO-MF). The terms in the same category are sorted using the p value.
The GO enrichment analysis was also performed on the 103 differential proteins for NORIMI group for the comparison of NOR to the traditional medicine IMI. The top ten enriched items in the GO-BP, GO-CC, GO-MF categories are presented in Fig. 4B, and revealed that most of these proteins were involved in synaptic, mitochondrial and ribosomal regulation. The categories included synapse, negative regulation of axon regeneration, dendrites, glutamatergic synapse, cellular response to oxidative stress, neuron apoptotic process, endoplasmic reticulum, mRNA processing, mRNA transport, RNA binding, mRNA binding and so on (Fig. 4B). These data suggested that both NOR and IMI modulated synaptic, mitochondrial and ribosomal functions, however, may mediate though different proteins (Fig. 4).
Analysis of protein and functional interaction networks
The GO analysis in Figs. 3 and 4 showed that the terms such as “mitochondrion”, “synapse”, “dendrite”, “extracellular space”, and “ribosome” were frequently associated with the differential proteins in NORCSDS (153) and NORIMI (103) groups. After performing a comparison with the STRING protein network interaction database, we extracted the protein interactions with a confidence score greater than 0.4. The high confidence protein interaction network was visualized using Cytoscape software, allowing us to explore the relationship among the proteins. Furthermore, key biological processes that proteins may participate in were annotated using the STRING database. Among the 153 differential proteins in NORCSDS, we found that 9 proteins were annotated to “synapse”, 15 proteins to “mitochondrion”, 11 proteins to “extracellular space”, 10 proteins to “dendrite”, 2 proteins to cellular response to oxidative stress” and 4 proteins to ribosome (Fig. 5A). Similarly, among the 103 differential proteins in NORIMI, 10 proteins were annotated to “synapse”,10 proteins to “mitochondrion”, 1 proteins to ribosome, 6 proteins to “extracellular space”, 7 proteins to “dendrite”, 2 proteins to “cellular response to oxidative stress” (Fig. 5B).
The differentially expressed proteins from NORCSDS (A) and NORIMI (B) were analyzed by PPI. A network diagram displays a network of specific functions interacting with proteins. Blue dots represent proteins, red dots represent specific functions, and solid lines represent interactions between proteins or protein annotations to corresponding functions.
PRM-based and western blot analysis of NORCSDS and IMICSDS common proteins
In this study, 50 common differentially expressed proteins of interest from the mitochondrial regulation or synaptic functions were selected to be further independently validated using the PRM assay. Regarding the proteins related to the mitochondrial function, we observed significant downregulation of Cox7c expression levels in the CSDS group compared to the Ctrl group. Interestingly, in the NOR-treated group, Cox7c, Mrp142, Naa30 were significantly upregulated compared to the CSDS group. Additionally, the expression level of Ighm and Apoa4 was significantly upregulated in the CSDS group compared to the control. In contrast, Ighm, Apoa4, Ssu72, Mrps30, Apoh, Acbd5, and Cdv3 were down-regulated in the NOR-treated group compared to the CSDS (Fig. 6). As to the synapse-related genes, the expression levels of Dcx, Arid1b, and Rnf112 were significantly upregulated in the NOR-treated group compared to the CSDS, whereas Fam3c were down-regulated in the NOR-treated group (Fig. 6). In addition, the expressions of Fam3c were reduced in the IMI-treated group as compared with the CSDS group. These findings suggest that NOR treatment may have differential effects on the expression of proteins related to mitochondrial function compared to the control and CSDS groups. We further confirmed these data to detect the levels of the important proteins Mrpl42, Dcx, Cox7c, Rnf112, Fam3c, Apoh, Naa30 and Apoa4 with Western blot analysis using the protein samples isolated from prefrontal cortex after NOR and imipramine treatment (Fig. 7A). In the prefrontal cortex, the levels of Mrpl42, Cox7c, Naa30, Rnf112 and Dcx were down-regulated and Apoa4, Apoh and Fam3c levels were up-regulated in the CSDS mice, however, NOR treatment reversed the changes of CSDS by significantly increasing the levels of Mrpl42, Cox7c, Naa30, Rnf112 and Dcx, and down-regulating the level of Apoa4, Apoh and Fam3c, supporting the PRM data (Fig. 7A). We also examined the Mrpl42, Dcx, and Apoa4 levels in the hippocampus, and obtained similar results as in prefrontal cortex for Mrpl42 and Dcx (Fig. 7B). But there was no significant difference in Apoa4 expression after NOR treatment, suggesting the inflammatory regulation may be brain region specific. Intriguingly, NOR treatment also rescued the up regulation of NLRP3 inflammasome activity in CSDS indicated by Western Blot of NLRP3, matured form of Caspase 1 and IL-1β (Fig. 7C).
Protein extracts of prefrontal cortex from the mice of the four groups, CON, CSDS, NOR-treated CSDS, and IMI-treated CSDS were examined by PRM analysis. Mitochondrial proteins Cox7c, Mrp142, Naa30, Ighm, Apoa4, Ssu72, Mrps30, Apoh, Acbd5, and Cdv3 and synaptic proteins, including Dcx, Fam3c, Arid1b, and Rnf112 showed significant difference (N = 4–6 per group, *p < 0.05, **p < 0.01; #p < 0.05).
Protein extracts of prefrontal cortex (A) or hippocampus (B) from the mice of the four groups, CON, CSDS, NOR-treated CSDS, and IMI-treated CSDS were examined by western blot analysis. C The NLRP3 inflammasome activation was determined by western blot with anti-NLRP3, anti-IL-1β, anti-caspase1 antibodies. α-tubulin was used as a loading control. *P < 0.05, **P < 0.01 (one-way ANOVA, post hoc LSD test); #p < 0.05 (Student’s T test).
Discussion
Given the limited efficacy and various adverse effects associated with current SSRI antidepressants, there is an urgent need for new therapeutic approaches that target specific underlying disease mechanisms, which are not adequately addressed by serotonergic- and/or noradrenergic-targeting antidepressants [1]. In recent years, there has been growing interest in phytochemical compounds, such as norisoboldine (NOR), an isoquinoline alkaloidt found in the root of Lindera aggregata, due to their strong anti-inflammatory and tissue-protective activities [20]. NOR, known as a natural aryl hydrocarbon receptor (AHR) agonist and an anti-arthritis alkaloid isolated from Radix Linderae, has been shown to attenuate osteoclast differentiation and inflammatory bone erosion in an AHR-dependent manner, and to inhibit the activation of NLRP3 inflammasome [21]. In our study, we investigated the antidepressant effect of NOR (Fig. 1), which is consistent with previous findings indicating that the activation of AHR by the metabolites of tryptophan pathway, such as KYN (kynurenine) and KYNA (kynurenic acid), exhibits antidepressant effects [22]. This suggests that norisoboldine may exert its antidepressant properties through similar mechanisms involving AHR activation.
Chronic social defeat stress is recognized as an important risk factor in the onset of depression and anxiety disorders [9], and imipramine is a positive SSRI control in this study. Of note, proteins regulated uniquely in the four phenotypes may underlie the observed behavioral differences. To unravel the possible protein dysregulations related to the four behavioral phenotypes, we carried out an ANOVA analysis of the proteomes of prefrontal cortex proteins. Intriguingly, the distinct protein dysfunctional profiles in all CON, CSDS, NOR and IMI four groups might represent differences of the three behavioral phenotypes. Based on the expression trend, differential proteins are divided into multiple clusters, with each cluster represented by a different color. The protein expression levels of Cluster 1 and Cluster 3 aligned with the observed mood pattern. In the CSDS depression model group, the protein levels were down-regulated or up-regulated, reflecting a state depressed mood. Conversely, in the IMI-treatment and the novel NOR-treatment groups, the protein levels were up-regulated or down-regulated, indicating an improvement in mood (Fig. 3). Consistent with the GO analysis of the all ANOVA differential proteins (Fig. 3A–D), GO-CC, GO-BP, and GO-MF category analysis in Cluster 1 (Fig. 3E–G) and Cluster 3 (Fig. 3H–J) all showed that the proteins enriched in the categories related to mitochondrial, ribosomal and the synaptic functions (Fig. 3). Previous transcriptomic study from Dr. Nestler’s group showed that in the limbic brain region, particularly PFC, treatment with imipramine or ketamine both reversed susceptibility and induced resilience-associated molecular adaptations [23]. We found that many differential proteins strengthened mitochondrial functions, which may enhance the resilience of the cells. Conducing molecular investigations on these four sub-group protein profiles would enhance our current the underlying mechanisms of pathophysiology and treatment of depression. Moreover, it would provide us with valuable new insights into this field.
Upon analyzing the 153 common differential proteins in the NORCSDS group for the differential proteins using GO pathway databases, we observed significant enrichment of proteins in the top ten items in GO-CC category. These enriched proteins included the mitochondrion, synapse and endoplasmic reticulum (Fig. 4). Likewise, the GO-CC analysis of the 103 common differential proteins in the NORIMI comparison revealed significant enrichment in various cellular components, including synapse, endoplasmic reticulum, dendrite, intracellular membrane bound organelle, glutamatergic synapse, and other related categories (Fig. 4B). Our findings align with recent evidence that has expanded our understanding the pathogenesis of depression, particularly highlighting the role of mitochondrial dysfunction, suggesting that the combined effects of social stress on mitochondria and inflammation may synergistically contribute to the development of stress-related depression [24, 25]. Moreover, the exploration excitatory synapses in pathogenesis and pharmacological treatment of depression has gained significant importance. Understanding the regulation of excitatory synapses can uncover novel depression mechanisms underlying depression and identified potential targets for the pharmacotherapy [26, 27]. It is worth noting that mitochondrial function plays a critical role in regulating to the excitatory synapses within the brain. By influencing excitatory synaptic activity, mitochondrial function contributes to the overall synaptic plasticity and neuronal communication associated with depression. In addition, recently, ribosomal changes were highlighted as the focus in the development of depression [28]. The PCA analysis appears to only separate the NOR group from the rest groups (Fig. 2C), suggesting NOR has distinct proteomic alterations. NOR shows distinct capacity in the heatmap as compared to that by IMI. These data raise an intriguing possibility that NOR and IMI may have distinct mechanisms to achieve antidepressant effects (Fig. 3). This is consistent with the fold of changes in the heat map of Cluster I, II, and III (Fig. 3A) and the fact only a small fraction of proteomic hits overlapping with CSDSCON and IMICSDS (Fig. 4).
During the GO analysis, several categories such as “mitochondrion, ribosome, synapse, dentrite, extracellular space, cellular response to oxidative stress” were consistently enriched. To further investigate the proteins involved in these categories, we performed PPI analysis. Among the 153 differential proteins in NORCSDS, we found that 9 proteins were annotated to “synapse”, 15 proteins to “mitochondrion”, 11 proteins to “extracellular space”, 10 proteins to “dendrite”, 2 proteins to cellular response to oxidative stress” and 4 proteins to ribosome(Fig. 5A). These PPI data provide additional confirmation that the differential proteins were enriched in the proteins related to mitochondrial, ribosomal and synaptic functions.
The PRM was performed to confirm the proteomic findings. COX7C is a mitochondrial long-lived protein that forms a stable contact site between complexes I and IV, and is required for complex IV and supercomplex assembly [29]. It was significantly decreased in the PFC of CSDS and increased after NOR treatment, suggesting the mitochondrial function in ATP production was enhanced after NOR treatment. In addition, knockdown of Naa30 induced the loss of mitochondrial membrane potential and fragmentation of mitochondria [30]. Naa30 was significantly decreased in the PFC of CSDS and increased after NOR treatment, suggesting the mitochondrial integrity was well maintained after NOR treatment. MRPL42 mediates the proliferation of the glial cells [31]. MRPL42 was significantly decreased in the PFC of CSDS and increased after NOR treatment, suggesting a role in glial homeostasis. Moreover, Ighm (immunoglobulin heavy constant mu) is involved in response to oxidative stress [32]. Ighm was increased in the PFC of CSDS and down-regulated by NOR treatment suggesting the recovery from oxidative stress after NOR treatment. Apoa4 is an inflammation stimulated protein, which is upregulated in CSDS mice and down-regulated by NOR treatment, suggesting an inhibition in inflammation response [33]. Other proteins, Ssu72 [34], Mrps30 [35], Apoh [36], Acbd5 [37], and Cdv3 [38] were identified to be involved in a variety of important mitochondrion-related functions, including autophagy [39], regulation of mitochondrial functions [34], mitoribosomal proteins [35], mitochondrial fat metabolism [36], ER-mitochondria tethering [37], and oxygen tension [38], respectively. The findings highlight the potential significance of these proteins in the pathophysiology of depression and provide insights into their specific roles or pathways in mitochondrial-related functional processes.
Regarding the synapse-related proteins Dcx, Arid1b, Fam3c, and Rnf112, which exhibited unique regulation in the CSDS or NOR-treated groups (Fig. 6B). Knockdown of Dcx (doublecortin, a crucial effector of migration) resulted in misplaced neurons failing to properly form dendrites, spines, and functional glutamatergic and GABAergic synapses [40]. NOR treatment enhanced Dcx expression, suggesting an increase in synaptic formation. RNF112 (neurolastin) is a dynamin family GTPase that affects spine density [41]. Increased RNF112 expression after NOR treatment implied an enrichment in spine density. Arid1b (AT-rich interactive domain 1B) demonstrated a critical role for interneuron development [42]. Upregulated Arid1b expression after NOR treatment suggested a support for interneuron development.
This study aimed to give a holistic investigation of the pathways involved in the antidepressant action. The data showed that the NOR treatment affected a pharmacological protein network involving in multiple signaling pathways to strengthen mitochondrial, ribosomal, and synaptic functions. This interplay highlights the importance of maintaining mitochondrial and ribosomal health in modulating inflammation and protecting synapses and neurons from stress-relevant damage. These phenotype-specific dysregulated proteins, analyzed independently using PRM, primarily involve the functions of enhancing mitochondrial function, ribosomal function and synaptic formation, which have been implicated in the pathophysiology and treatment of mood disorders in previous studies [24, 25, 27, 43]. Various pathogenic processes, including various infections, autophagy, metabolism and brain-gut axis, are associated with ER stress and mitochondrial dysfunction [26]. Indeed, the inflammasome activation, which is directly associated with dysregulation of mitophagy and related to the pathogenesis of depression, was enhanced in CSDS and rescued by NOR treatment (Fig. 7B), which is consistent with previous findings [17]. As to the limitation of the study, the proteomic method we use can only detect about 5000 proteins in top abundance, and the total protein amount of the human body is more than 20,000 proteins. This is may be one of the reason that in the present study, we did not catch the proteins, such as inflammasome, which could be affected by NOR according to previous study [17].
Our unbiased profile highlighted the importance of mitochondria and synapses as key subgroups for the protein candidates in the prefrontal cortex, indicating their relevance to the pathophysiology and treatment of depression. These findings provide valuable insights into the underlying mechanisms of depression and offer potential targets for drug development in the field of major depressive disorder.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD047064.
References
Stachowicz K, Sowa-Kućma M. The treatment of depression - searching for new ideas. Front Pharmacol. 2022;13:988648.
Sabatini S, Martyr A, Gamble LD, Jones IR, Collins R, Matthews FE, et al. Profiles of social, cultural, and economic capital as longitudinal predictors of stress, positive experiences of caring, and depression among spousal carers of people with dementia. Aging Ment Health. 2023;27:1335–43.
Fernandes BS, Dai Y, Jia P, Zhao Z. Charting the proteome landscape in major psychiatric disorders: From biomarkers to biological pathways towards drug discovery. Eur Neuropsychopharmacol. 2022;61:43–59.
Rappeneau V, Wilmes L, Touma C. Molecular correlates of mitochondrial dysfunctions in major depression: Evidence from clinical and rodent studies. Mol Cell Neurosci. 2020;109:103555.
Foley ÉM, Parkinson JT, Mitchell RE, Turner L, Khandaker GM. Peripheral blood cellular immunophenotype in depression: a systematic review and meta-analysis. Mol Psychiatry. 2023;28:1004–19.
Thompson SM. Plasticity of synapses and reward circuit function in the genesis and treatment of depression. Neuropsychopharmacology. 2023;48:90–103.
Duman RS, Aghajanian GK, Sanacora G, Krystal JH. Synaptic plasticity and depression: new insights from stress and rapid-acting antidepressants. Nat Med. 2016;22:238–49.
Radley JJ, Herman JP. Preclinical Models of Chronic Stress: Adaptation or Pathology? Biol Psychiatry. 2023;94:194–202.
Wang W, Liu W, Duan D, Bai H, Wang Z, Xing Y. Chronic social defeat stress mouse model: Current view on its behavioral deficits and modifications. Behav Neurosci. 2021;135:326–35.
Amat J, Baratta MV, Paul E, Bland ST, Watkins LR, Maier SF. Medial prefrontal cortex determines how stressor controllability affects behavior and dorsal raphe nucleus. Nat Neurosci. 2005;8:365–71.
Berridge KC, Kringelbach ML. Pleasure systems in the brain. Neuron. 2015;86:646–64.
Dias-Ferreira E, Sousa JC, Melo I, Morgado P, Mesquita AR, Cerqueira JJ, et al. Chronic stress causes frontostriatal reorganization and affects decision-making. Science. 2009;325:621–5.
Liston C, McEwen BS, Casey BJ. Psychosocial stress reversibly disrupts prefrontal processing and attentional control. Proc Natl Acad Sci USA. 2009;106:912–7.
Soares JM, Sampaio A, Ferreira LM, Santos NC, Marques F, Palha JA, et al. Stress-induced changes in human decision-making are reversible. Transl Psychiatry. 2012;2:e131.
Wei ZF, Lv Q, Xia Y, Yue MF, Shi C, Xia YF, et al. Norisoboldine, an Anti-Arthritis Alkaloid Isolated from Radix Linderae, Attenuates Osteoclast Differentiation and Inflammatory Bone Erosion in an Aryl Hydrocarbon Receptor-Dependent Manner. Int J Biol Sci. 2015;11:1113–26.
Tong B, Yuan X, Dou Y, Wu X, Chou G, Wang Z, et al. Norisoboldine, an isoquinoline alkaloid, acts as an aryl hydrocarbon receptor ligand to induce intestinal Treg cells and thereby attenuate arthritis. Int J Biochem Cell Biol. 2016;75:63–73.
Lv Q, Wang K, Qiao SM, Dai Y, Wei ZF. Norisoboldine, a natural aryl hydrocarbon receptor agonist, alleviates TNBS-induced colitis in mice, by inhibiting the activation of NLRP3 inflammasome. Chin J Nat Med. 2018;16:161–74.
Golden SA, Covington HE 3rd, Berton O, Russo SJ. A standardized protocol for repeated social defeat stress in mice. Nat Protoc. 2011;6:1183–91.
Kim HD, Call T, Carotenuto S, Johnson R, Ferguson D. Testing Depression in Mice: a Chronic Social Defeat Stress Model. Bio Protoc. 2017;7:e2203.
Kraeuter AK, Guest PC, Sarnyai Z. The Forced Swim Test for Depression-Like Behavior in Rodents. Methods Mol Biol. 2019;1916:75–80.
Bao H, Sun L, Zhu Y, Ran P, Hu W, Zhu K, et al. Lentinan produces a robust antidepressant-like effect via enhancing the prefrontal Dectin-1/AMPA receptor signaling pathway. Behav Brain Res. 2017;317:263–71.
Zang X, Zheng X, Hou Y, Hu M, Wang H, Bao X, et al. Regulation of proinflammatory monocyte activation by the kynurenine-AhR axis underlies immunometabolic control of depressive behavior in mice. FASEB J. 2018;32:1944–56.
Bagot RC, Cates HM, Purushothaman I, Vialou V, Heller EA, Yieh L, et al. Ketamine and Imipramine Reverse Transcriptional Signatures of Susceptibility and Induce Resilience-Specific Gene Expression Profiles. Biol Psychiatry. 2017;81:285–95.
Hollis F, Pope BS, Gorman-Sandler E, Wood SK. Neuroinflammation and Mitochondrial Dysfunction Link Social Stress to Depression. Curr Top Behav Neurosci. 2022;54:59–93.
Du J, Zhu M, Bao H, Li B, Dong Y, Xiao C, et al. The Role of Nutrients in Protecting Mitochondrial Function and Neurotransmitter Signaling: Implications for the Treatment of Depression, PTSD, and Suicidal Behaviors. Crit Rev food Sci Nutr. 2016;56:2560–78.
Fries GR, Saldana VA, Finnstein J, Rein T. Molecular pathways of major depressive disorder converge on the synapse. Mol Psychiatry. 2023;28:284–97.
Samojedny S, Czechowska E, Pańczyszyn-Trzewik P, Sowa-Kućma M. Postsynaptic Proteins at Excitatory Synapses in the Brain-Relationship with Depressive Disorders. Int J Mol Sci. 2022;23:11423.
Zhang J, Xie S, Chen Y, Zhou X, Zheng Z, Yang L, et al. Comprehensive analysis of endoplasmic reticulum stress and immune infiltration in major depressive disorder. Front Psychiatry. 2022;13:1008124.
Krishna S, Arrojo E, Drigo R, Capitanio JS, Ramachandra R, Ellisman M, et al. Identification of long-lived proteins in the mitochondria reveals increased stability of the electron transport chain. Dev Cell. 2021;56:2952–2965.e2959.
Van Damme P, Kalvik TV, Starheim KK, Jonckheere V, Myklebust LM, Menschaert G, et al. A Role for Human N-alpha Acetyltransferase 30 (Naa30) in Maintaining Mitochondrial Integrity. Mol Cell Proteom MCP. 2016;15:3361–72.
Hao C, Duan H, Li H, Wang H, Liu Y, Fan Y, et al. Knockdown of MRPL42 suppresses glioma cell proliferation by inducing cell cycle arrest and apoptosis. Biosci Rep. 2018;38:BSR20171456.
Martina JA, Puertollano R. Protein phosphatase 2A stimulates activation of TFEB and TFE3 transcription factors in response to oxidative stress. J Biol Chem. 2018;293:12525–34.
Lee HH, Cho YI, Kim SY, Yoon YE, Kim KS, Hong SJ, et al. TNF-α-induced Inflammation Stimulates Apolipoprotein-A4 via Activation of TNFR2 and NF-κB Signaling in Kidney Tubular Cells. Sci Rep. 2017;7:8856.
Woo YD, Koh J, Ko JS, Kim S, Jung KC, Jeon YK, et al. Ssu72 regulates alveolar macrophage development and allergic airway inflammation by fine-tuning of GM-CSF receptor signaling. J Allergy Clin Immunol. 2021;147:1242–60.
Cheong A, Lingutla R, Mager J. Expression analysis of mammalian mitochondrial ribosomal protein genes. Gene Expr Patterns GEP. 2020;38:119147.
Fujiwara K, Takeuchi S, Okamura-Ikeda K, Motokawa Y. Purification, characterization, and cDNA cloning of lipoate-activating enzyme from bovine liver. J Biol Chem. 2001;276:28819–23.
Schrader M, Kamoshita M, Islinger M. Organelle interplay-peroxisome interactions in health and disease. J Inherit Metab Dis. 2020;43:71–89.
Piltti J, Bygdell J, Qu C, Lammi MJ. Effects of long-term low oxygen tension in human chondrosarcoma cells. J Cell Biochem. 2018;119:2320–32.
Tong M, Saito T, Zhai P, Oka SI, Mizushima W, Nakamura M, et al. Alternative Mitophagy Protects the Heart Against Obesity-Associated Cardiomyopathy. Circ Res. 2021;129:1105–21.
Martineau FS, Sahu S, Plantier V, Buhler E, Schaller F, Fournier L, et al. Correct Laminar Positioning in the Neocortex Influences Proper Dendritic and Synaptic Development. Cereb Cortex. 2018;28:2976–90.
Lomash RM, Gu X, Youle RJ, Lu W, Roche KW. Neurolastin, a Dynamin Family GTPase, Regulates Excitatory Synapses and Spine Density. Cell Rep. 2015;12:743–51.
Jung EM, Moffat JJ, Liu J, Dravid SM, Gurumurthy CB, Kim WY. Arid1b haploinsufficiency disrupts cortical interneuron development and mouse behavior. Nat Neurosci. 2017;20:1694–707.
Lai H, Guo Y, Tian L, Wu L, Li X, Yang Z, et al. Protein Panel of Serum-Derived Small Extracellular Vesicles for the Screening and Diagnosis of Epithelial Ovarian Cancer. Cancers. 2022;14:3719.
Acknowledgements
This study was supported by the Science and Technology Innovation Fund of China “Brain science and brain-like research” (No. 2021ZD0200600, 2021ZD0202003); CAMS Innovation Fund for Medical Sciences (CIFMS, no. 2021-I2M-1-028); International Cooperation Project of Qinghai Province (2020-HZ-803). We thank the PTM Biolabs, Inc. for the technical support of the proteomics.
Author information
Authors and Affiliations
Contributions
LL performed the experiments, analyzed the data and wrote and revised part of the manuscript; WK performed the experiments and analyzed the data; YZ performed the experiments for the revision and analyzed the data; TW performed the experiments for the revision and analyzed the data; FY helped revising the manuscript critically for important intellectual content; TJ contributed to conception and design of the study, and helped revising the manuscript critically for important intellectual content; GW contributed to conception and design of the study, and helped revising the manuscript critically for important intellectual content; JD contributed to conception and design of the study, analyzed the data and wrote and revised the manuscript. All the authors listed have reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, L., Kan, W., Zhang, Y. et al. Quantitative proteomics combined independent PRM analysis reveals the mitochondrial and synaptic mechanism underlying norisoboldine’s antidepressant effects. Transl Psychiatry 14, 400 (2024). https://doi.org/10.1038/s41398-024-03127-z
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41398-024-03127-z