Abstract
The death of fungal cells has been studied in a variety of contexts including responses to antifungal drugs, during fungal developmental processes, in response to bacterial or mycoviral fungal pathogens, and during non-self-recognition between distinct strains of the same species (allorecognition). Some of the genetic determinants and molecular mechanisms of fungal cell death processes are now beginning to be understood in detail. Recent advances have uncovered fungal cell death machinery that shares ancestry with key actors of immune cell death in other eukaryotic and prokaryotic taxa. Transkingdom evolutionary links include fungal molecular sensors such as NOD-like receptors and signaling domains related to the TIR (Toll/interleukin-1 receptor) family, which are a staple of immunity throughout the tree of life. Moreover, cell death executioner proteins homologous to the pore-forming proteins that mediate mammalian necroptosis and pyroptosis are also abundant and widespread in fungi, particularly in Ascomycota. These findings prompt us to speculate on the possible origins of fungal cell death and to reconsider fungal innate immunity beyond allorecognition. This review discusses historical landmarks and major recent discoveries regarding the regulation of cell death processes in fungi through the lens of immunity.
Similar content being viewed by others
Facts
-
Fungal regulated cell death involves molecular machinery that is distantly related to cell death pathways in other eukaryotic and prokaryotic taxa.
-
Many fungal cell death determinants have been identified through studies of heterokaryon incompatibility (HI), a prophylactic cell death process occurring during conspecific non-self-recognition.
-
Fungal genomes encode a diverse repertoire of NOD-like receptors, some of which are involved in cell death and non-self-recognition.
-
Fungal amyloid-signaling and HeLo/HELL membrane-targeting domains establish an evolutionary link with necroptosis and RCD in plants.
-
Fungal gasdermins (fGSDMs) and their cognate proteolytic regulators draw evolutionary parallels with pyroptosis.
Open questions
-
What molecular cues are recognized by each of the numerous fungal NOD-like receptors?
-
What is the full range of diversity of defense-related cell death and immunity mechanisms that exist across fungal species?
-
What is the level of interconnectedness of the cell death pathways constituting fungal immunity systems?
-
Are distinct fungal cell death pathways associated with specific morphological hallmarks?
Regulated cell death (RCD) refers to cellular suicide in both multicellular and unicellular organisms. RCD relies on genetic determinants and signaling pathways, providing points of genetic and pharmacological control and with stimulus- or context-dependent specificity to the process [1, 2]. In animals, a variety of RCD pathways participate in the maintenance of homeostasis by eliminating damaged, malfunctioning and potentially harmful cells from the body [3, 4]. Here we refer to fungal cell death as RCD, as defined by the functional and sequential interactions between the involved molecular players and associated with specific morphological and biochemical hallmarks of cell disintegration [4].
Regulated cell death is a shared defense strategy between eukaryotic and prokaryotic taxa
Comparisons between immune systems of distant taxa provide an opportunity to understand the underlying, universal principles of immunity and identify major taxa-specific distinctions. Regulated cell death is a shared defense strategy between eukaryotic [5] and prokaryotic taxa [6]. Simply put, the strategy consists of eliminating infected cells to limit or prevent the multiplication and spread of a pathogen. In the case of single-cell prokaryotes including many bacteria and archaea, the benefits of cell death occur at the population level, notably preventing the propagation of phages [7, 8]. In plants, the hypersensitive cell death response (HR) activates at the point of penetration by a plant pathogen (e.g. bacteria, virus, fungi) and helps contain the infection providing resistance to the pathogen invasion, playing a key role in the plant immune system [9,10,11].
Multicellular animals (Metazoa) are a sister clade of Fungi, and together form the eukaryotic supergroup of Opisthokonta [12, 13]. In animals and particularly in mammals, many distinct RCD pathways have been extensively characterized, since the introduction of the term apoptosis in 1972 [14]. Apoptotic cell death plays an essential role in animal development, homeostasis and immunity [15,16,17]. Cells undergoing apoptosis are dismantled in an orderly manner, with minimal dispersal of cellular content [18], while other mammalian RCD pathways, like necroptosis [19,20,21] and pyroptosis [22,23,24], are lytic and highly inflammatory. Necroptosis, pyroptosis and ferroptosis are three of the best understood pathways and also core cell death programs of mammalian immunity [4, 25,26,27]. Remarkably, exploration of fungal immune defense-related RCD has yielded thus far, several findings establishing evolutionary ties between some fungal RCD-controlling proteins and crucial players in mammalian pyroptosis and necroptosis.
This review describes current fungal RCD research with an emphasis on recent discoveries that tie fungal cell death-controlling proteins to immune systems across the tree of life. A particular focus is placed on comparisons within Opisthokonta, where the metazoan pathways are characterized in great detail. Comparisons are further extended to bacterial and plant immunity genes, underscoring the ancient evolutionary origins of some cell death programs. By considering the historical context, I discuss our evolving understanding of fungal RCD and propose that a comparative immunology framework offers an ambitious, productive, and systematizing approach for future research on fungal regulated cell death.
Heterokaryon Incompatibility is a fungal prophylactic cell death response that restricts mycoparasitism
Cell death in fungi has been investigated in the context of fungal pathogenesis by examining fungal responses to antifungal drugs and other antimicrobial compounds [28, 29]. Conversely, fungal RCD has also been investigated in fungi infected by fungal viruses (mycoviruses) [30] and during fungal–bacterial interactions [31,32,33]. In addition, RCD plays an important role in various physiological processes in fungi. These include the differentiation of specialized cellular structures for reproduction (protoperithecia) [34], for host invasion purposes (appressoria) [35,36,37], and during sexual sporulation in both filamentous fungi [38, 39] and yeast [40].
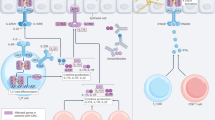
A major part of our current understanding of fungal RCD stems from historical studies of conspecific non-self-discrimination (also known as allorecognition), which occurs during somatic cell fusion between genetically distinct fungal colonies [41,42,43]. Notably, this phenomenon has been compared to graft rejection in mammals, where the immune system recognizes and rejects non-self tissues [44, 45]. Allorecognition has also been studied in basal metazoans (sponges and corals) [46, 47], social amoebae [48], and bacteria [49]. In fungal species like Neurospora crassa, non-self recognition and discrimination between distinct strains can occur at a distance (pre-contact) [50], at the point of contact before cellular fusion [51], or after somatic fusion of two strains. Incompatibility between strains may arise independently at any of these three stages due to genetic factors. In the first two cases, which occur prior to cellular fusion, the strains remain viable. However, if two genetically incompatible strains undergo somatic cell fusion (anastomosis) to form a heterokaryon, a lytic cell death process is triggered within the heterokaryon [42, 52, 53]. This abortive anastomosis event is known as heterokaryon incompatibility (HI) and is characterized by extensive vacuolization, plasma membrane shrinkage, elevated reactive oxygen species (ROS) levels, and lipid droplet production [52, 54] (Fig. 1). The heterokaryotic fusion cells undergoing HI are sequestered from the rest of the colony by plugging the large channels through which cytoplasm and organelles normally flow [55]. New septa (cell membrane partitions) form de novo in the dying cells before lysis completes and the cells disintegrate, severing the connection between incompatible strains [54].
Regulated cell death (RCD) occurs post-anastomosis – a cellular fusion process. The illustration shows a blue hyphal cell from one colony fusing with a red hyphal cell from a genetically distinct, incompatible individual. Generally, the newly formed heterokaryon is rapidly isolated from both colonies – symbolized by keylocks – by plugging the pores that connect adjacent cells in the hyphae. The HI cell death process is characterized by massive vacuolization, production of reactive oxygen species (ROS), and the formation of lipid droplets. Additionally, cell wall thickening and de novo septation occur—although these features are not depicted in the cartoon. Subsequently, the HI cell death culminates in plasma membrane shrinkage and rupture, releasing the cytoplasm of the dead fusion cells.
het genes control regulated cell death during heterokaryon incompatibility
Exploration of the cell death mechanisms occurring during HI has led to the discovery of the genetic bases of fungal RCD. Early classical genetic studies identified several genetic loci responsible for cell death during somatic cell fusion between incompatible fungal strains [41]. In most species – including Neurospora crassa, Podospora anserina, and Aspergillus fumigatus – the genes are named het (short for heterokaryon incompatibility), whereas in Cryphonectria parasitica, they are called vic genes (for vegetative incompatibility). The genetic interactions between het genes define heterokaryon incompatibility (HI) systems. Each HI system is independent of the others in its ability to trigger incompatibility, and their total number per species typically does not exceed a dozen – often fewer. Such HI systems can arise from interactions between allelic variants of a single gene or between unrelated genes, which may be located at distant loci or, more commonly, represent two tightly linked genes [41, 56] (Fig. 2).
A HI is triggered by genes termed het or vic (vegetative incompatibility), which define independent ‘incompatibility systems’ when sharing the cytoplasm of a heterokaryotic cell originating from the fusion of genetically incompatible strains, shown as blue and red colonies. HI systems can be formed by the antagonistic alleles of the same gene or by different genes. In the latter case, the RCD-inducing genes may be at the same locus in the genomes of the incompatible strains (HI system between idiomorphs) or consist of two antagonistic genes situated at different loci. Some HI systems rely on tightly linked genes, in which the RCD-inducing interactions among these het gene products can be symmetrical or asymmetrical (indicated by arrows). An HI system (V/V1) has also been described in P. anserina, in which the absence of a specific locus in one strain can lead to RCD in heterokaryons formed with another strain that possesses this locus. B Molecularly characterized HI systems in three model ascomycete species used for investigating fungal allorecognition and cell death. The characterization of almost all HI systems in these species reveals evolutionary parallels with genes and protein domains that regulate immunity and cell death throughout the tree of life, as indicated by the fuchsia highlighting each HI system representation. Genes establishing the evolutionary link are underscored. Genetic loci containing two het genes (het-V, het-Z, and het-B in P. anserina) are highlighted in purple. The molecular characterization of het/vic genes is presently exhaustive for both P. anserina and C. parasitica; however, several genetically mapped het loci in N. crassa remain to be explored and characterized molecularly (not shown in the figure).
Heterokaryon incompatibility limits the spread of fungal viruses (mycoviruses)
HI limits cytoplasmic exchanges between incompatible fungal strains, thereby preventing the horizontal transmission of mycoviruses and other genetic elements [57,58,59,60,61,62]. Mycoviruses can impair infected strains by reducing their fitness and virulence, resulting in hypovirulent fungal strains [63]. Therefore, some researchers are harnessing the features of HI systems, and mycoviruses, as biocontrol strategies to combat plant-pathogenic fungi [63, 64]. HI has been found to limit the spread of viral and other selfish cytoplasmic elements (such as senescence plasmids) in Aspergillus niger [65], Neurospora crassa [66], Podospora anserina [67], and Cryphonectria parasitica [57].
In effort to develop anti-fungal strategies against chestnut blight disease, the relationship between HI systems and mycoviral spread has been most thoroughly explored in the pathogenic fungus Cryphonectria parasitica. Studies of C. parasitica HI systems have led to the molecular characterization of six distinct incompatibility systems [68, 69]. The efficiency of Cryphonectria HI systems in restricting mycoviral transmission – defined as the infection of a virus-free recipient strain through somatic cellular fusion with a virus-infected donor – varied among the six vic loci [70]. Heteroallelism at each of five Cryphonectria HI systems individually restricted mycoviral transmission between strains, with some vic loci reducing transmission rates by 80%. In contrast, the remaining one of the six HI systems had no impact on viral transmission under the tested conditions [70]. When tested independently, gene disruptions that inactivate any one of these five HI-controlling systems – and thereby abolish prophylactic HI cell death – result in increased viral transmission between strains in both laboratory [57, 68, 69] and natural environments [58]. By simultaneously shutting down four of the five virus-restricting HI systems, Zhang and Nuss successfully generated super mycovirus donor strains capable of transmitting the virulence-attenuating CHV1/EP713 mycovirus (Cryphonectria hypovirus 1, isolated from strain EP713) to a broad range of wild isolates [57].
For some Cryphonectria HI systems composed of two tightly linked genes (Fig. 2), disruption of a single gene can increase mycoviral transmission without completely abolishing the cell death event between the incompatible strains [57, 69]. These findings, along with the discovery that mycoviral transmission frequencies can depend on the mycoviral donor-recipient status of a strain [70], suggest that some HI systems may be under selective pressure to specifically block mycoviral infection. As expected for virus-host interactions, some mycoviruses have also evolved mechanisms to suppress HI systems, facilitating their horizontal intraspecies transmission [71]. Although the precise molecular details of viral counter-defense mechanisms remain unclear, some viruses appear to downregulate the transcription of fungal allorecognition genes [71]. An important recent study shows that plant hosts can also suppress HI systems in the pathogenic fungus Sclerotinia sclerotiorum, thereby facilitating mycoviral transmission and reducing the virulence of this fungal pathogen [72].
Heterokaryon incompatibility as a defense against resource plundering and cheater exploitation
HI cell death can also protect fungal colonies from other hazards, such as resource plundering [73] and cheater genotypes that exploit the genomes of conspecific non-self-strains [74, 75]. These detrimental situations arise because anastomosis is a key aspect of fungal lifestyles, and many fungal species are characterized by a multinucleate, syncytial organization. An example of resource plundering has been described between strains of N. crassa, which develop specialized ‘paternal’ cells (conidia) and ‘maternal’ reproductive structures (protoperithecia) to undergo the sexual cycle. Debets et al. reported that some paternal nuclei gain access to the maternal colony when conidia from the paternal line vegetatively fuse with the maternal protoperithecia-developing strain, thereby exploiting the resources that the maternal colony dedicates to sexual differentiation and sporulation [73]. The success of this ‘resource plundering’ strategy is significantly reduced by allorecognition-mediated cell death between the two strains [73]. Notably, the mating type locus defines one of the somatic HI systems in N. crassa [76, 77]. A similar context and logic apply to N. crassa nuclei carrying mutations that reduce investment in somatic functions such as conidiation, while providing reproductive advantages (‘cheaters’) compared to non-cheaters [75]. HI reduces heterokaryon formation with such cheater genotypes, further exemplifying the benefits of this prophylactic cell death as a potent defense strategy against parasitism.
Evolutionary signatures of het genes highlight the adaptive role of heterokaryon incompatibility in nature
The importance of HI in nature is reflected in the evolutionary hallmarks of het genes. Notably, het genes are generally highly polymorphic, and the alleles of many appear to be under balancing selection [78]. As a result, het alleles within a given HI system tend to be maintained at nearly equal frequencies in wild populations [79,80,81,82]. This pattern is thought to arise from the interdependency between the adaptive value of each het allele and its frequency. That is, strains harboring rarer alleles can recognize and reject a wider range of incompatible partners, conferring a selective advantage over more common variants. Over time, these initially rare alleles increase in frequency, displacing previously common alleles, which in turn become rarer [78]. Some heterokaryon incompatibility (HI) systems even transcend species boundaries, leading to trans-species polymorphism – where the same alleles are shared by closely related species rather than strictly sorted along species lines [79, 83]. Both balancing selection and trans-species polymorphism have been associated with immunity-related genes in animals and plants [84,85,86,87]. These evolutionary hallmarks underscore the significant benefits het genes confer in nature, enabling fungi to maintain robust defenses against conspecific non-self and its associated hazards.
Heterokaryon Incompatibility reveals a treasure trove of genes inducing fungal RCD
To date, molecular characterization of het genes has led to two striking observations: (i) het genes are evolutionarily related to innate immune genes that regulate RCD in both prokaryotic and eukaryotic organisms, including mammals; and (ii) they belong to large and widespread gene families. The gene families containing the most het genes often comprise dozens or even hundreds of genes per genome and their numbers can vary greatly between species. One large gene family, to which many het genes belong, encodes fungal NOD-like receptors (NLRs) [88] (Fig. 3). NOD-like receptors are ancient intracellular sensors that couple immune detection to regulated cell death across animals, plants, and prokaryotes. Frequently, het genes also encode proteins containing evolutionary distant TIR (Toll/interleukin-1 receptor) domains [83], which are found on immunity proteins in animals, plants, and bacteria [89]. In fungi, such TIR-like domains are also known as HET domains – a name derived from their frequent association with identified het genes [83, 90]. Other het genes are members of the fungal gasdermin (fGSDM) family [91, 92], distant evolutionary relatives of mammalian gasdermins, executioner proteins of pyroptosis [93]. A smaller group of characterized het genes belongs to a large gene family encoding proteins with membrane-targeting coiled-coil domains [94, 95], which are related to the necroptosis executioner protein MLKL [96] (Fig. 2).
NLRs are defined by their central nucleotide-binding (NB) and oligomerization (NOD) domains. NODs generally belong to either the NB-ARC family or the NACHT family. A Founding members of the NB-ARC family include mammalian APAF-1 (Apoptotic protease activating factor 1), its homolog CED-4 (Cell death protein 4) from the roundworm Caenorhabditis elegans, and plant R (resistance) proteins. B Founding members of the NACHT family include three animal proteins – NAIP (Neuronal apoptosis inhibitory protein), CIITA (class II, major histocompatibility complex transactivator), and TEP1 (Telomerase associated protein 1) – and HET-E (heterokaryon gene), which is a protein encoded in the genome of the mold Podospora anserina. C NOD-like receptors encoded in the genome of the ascomycete Podospora anserina, a fungal model organism for the study of regulated cell death and NLR biology. The hnwd (HET-NACHT-WD40) gene family comprises five paralogs, three of which (het-r, het-d, and het-e) control heterokaryon incompatibility (HI). The family is characterized by the presence of a HET domain—likely related to the TIR (Toll/interleukin-1 receptor/resistance protein) domain—at the N-termini of its five members. The WD40 pseudo-repeats of the hnwd genes exhibit strong internal conservation and are proposed to evolve in a concerted manner. Repeats can be exchanged between alleles of the same gene or between different members of the gene family. D Interallelic genetic interactions define the het-e/het-c HI system in P. anserina. Different alleles of het-e (e.g., het-e1, het-e2, het-e3), which differ almost exclusively in the number and composition of their WD40 repeats, trigger HI in the presence of distinct alleles of the het-c gene. The het-c gene encodes a glycolipid transfer protein that is broadly conserved across fungi. Some functionally characterized het-e alleles (e.g., het-e4) appear inactive in heterokaryon incompatibility (HI) and typically carry a low number of WD40 repeats, while others (e.g., het-e2) can induce RCD in the presence of different het-c alleles. Experimental data have demonstrated that the specificity of recognition between het-e and het-c allelic variants depends on the WD40 domains of the NLRs, and the cell-death reaction relies on a functional HET/TIR domain. The het-D paralog is involved in a very similar HI system with het-c. E Cartoon representation of 45 P. anserina NOD-like receptors. Genes directly involved in heterokaryon incompatibility (HI) are highlighted in red, and those indirectly involved are shown in orange. Abbreviations: HEL (HeLo-like in fungal proteins), PAT (Patatin-like phospholipase), SBL (SesB-like lipase), GBL (Goodbye-like domain), UDP (UDP-glucose phosphorylase domain), HET (Heterokaryon determinants domain), ASM (Amyloid Signaling Motif), CARD (Caspase recruitment domain), BIR (Baculovirus IAP repeat), TR (Telomerase, Ro and Vault or TROVE domain), vWA (von Willebrand A domain), TPR (Tetratricopeptide repeat), ANK (Ankyrin repeat), and Unk (unknown).
Fungal het genes are part of broad gene families with proposed roles in RCD and immunity
Importantly, the vast majority of fungal NLRs, fGSDMs, and TIR/HET domain-encoding genes are not involved in allorecognition, indicating broader roles beyond incompatibility responses. Consistent with this, genetic mapping in the few model species used to study HI has consistently identified only six to a dozen het loci per species [41, 43]. To clarify the relationship between het genes and other members of their gene families, it has been proposed that HI-defining loci were co-opted from large pre-existing gene pools primarily involved in xenorecognition (discrimination between species) and innate immunity [97,98,99]. This hypothesis is further supported by uncovered functional similarities between gene members across kingdoms – such as fGSDMs, NLRs, and TIR-containing receptors from other taxa – which include known regulators of immune cell death in the context of xenorecognition [97]. Aligned with this perspective, although het genes govern prophylactic cell death in heterokaryotic chimeras, most members in these gene families (NLRs, fGSDMs, TIR/HET domain-encoding genes) are likely involved in the regulation of RCD in other contexts, including direct defense against pathogens and antagonists that target fungi. Together, the het genes and the (non-het) members of the gene families would form the allorecognition and xenorecognition molecular mediators, respectively, of the fungal immune system [97,98,99].
NOD-like receptors and the STAND superfamily: conserved intracellular mediators of immunity and cell death
Signal-transducing ATPases with numerous domains (STANDs) are a superfamily of intracellular receptors conserved in prokaryotes and eukaryotes, associated with non-self discrimination, immunity, and regulated cell death [100]. The NTPase module binds and hydrolyzes nucleotides, playing an important role in the oligomerization of the sensors, which can be activated by endogenous or exogenous signals [101]. Two of the major families of STAND NTPases are the NB-ARC family (nucleotide-binding domain found in human APAF-1, certain plant R proteins, and in C. elegans CED-4) [102], and the NACHT family NTPases, named after the four founding STAND members (NAIP, CIITA, HET-E, and TP1) [103]. The NB-ARC and NACHT domains are collectively referred to as the nucleotide-binding and oligomerization domain (or NOD), and proteins that contain these domains are commonly termed NOD-like receptors (NLRs) [104,105,106] (Fig. 3).
In the classical tripartite STAND architecture, the NOD module is spanned by an N-terminal signaling (or accessory) domain and superstructure-forming pseudo-repeats (20-40-amino acid length) in the C-terminal region [100]. These repeats are proposed to regulate activation of the STAND sensors in response to interactions with diverse types of molecules [107]. Leucine-rich repeats (LRRs) are among the most frequently identified sequences in such architectures and are characteristic of the NBs-LRR family (nucleotide-binding site with leucine-rich repeats), controlling immune cell death in animals and plants [108,109,110,111,112,113]. NBs-LRRs are also frequently referred to as NLRs (the same acronym as for NOD-like receptors) [104,105,106]. However, NOD-like receptors more broadly designate immunity-related STAND proteins that exhibit more diverse architectures than the classical NBs-LRR receptors [114, 115]. In prokaryotes, for example, such NLR proteins play immune roles by protecting against phages [115, 116]. These microbial NLRs are proposed to induce RCD in infected bacterial cells to hinder phage propagation – a process termed abortive infection [115]. Gao et al. have demonstrated that some bacterial and archaeal STANDs – named Avs for anti-viral STANDs – can directly recognize hallmark phage proteins and share signaling mechanisms similar to those of their homologs in metazoans and plants [116].
NOD-like receptors in fungi: from molecular diversity to roles in heterokaryon incompatibility
Fungal genomes encode an abundant and highly diverse repertoire of NOD-like receptors, many of which carry WD40 repeats similar to those of APAF-1 [94, 114, 117]. Ankyrin (ANK) and tetratricopeptide repeats (TPRs) frequently substitute for WD40 repeats in a variety of fungal NLR-like proteins [114]. However, no leucine-rich repeats were identified, and the classical NOD-like architecture of NBs-LRR – prevalent in plants and animals – appeared absent from the analyzed fungal genomes [88]. The fungal NLR-like proteins (hereafter, simply referred to as NLRs) exhibit greater architectural diversity than homologs from other eukaryotic taxa, with more than 14 distinct types of N-terminal signaling modules [94]. Among this diversity, approximately 20% of the fungal NLRs possess a central NB-ARC NOD domain, while most of the remaining receptors carry a central NACHT domain [114]. Based on the type of NOD domain, fungal NLRs therefore resemble those of animals—where NACHT domains predominate—more than those of plants, in which NB-ARC domains are more common [114]. Beyond their structural diversity, the distribution of NLR genes varies across fungal lineages: they are generally abundant in ascomycete and basidiomycete species but absent from yeast genomes, including Saccharomyces cerevisiae [114].
Several fungal NLRs have been experimentally characterized in the context of heterokaryon incompatibility, and the family is proposed to play a broader role in fungal immunity and immune cell death [88, 118] (Table 1). NLR genes have been found to determine the outcome of somatic cell fusions between conspecific strains in N. crassa [119], A. fumigatus [80], C. parasitica [69], and P. anserina [119,120,121,122]. In the latter, two paralogs of the HNWD family (HET-NACHT-WD40 domain organization), reminiscent of the APAF-1 architecture, trigger RCD when co-expressed with incompatible allelic variants of an endogenous glycolipid transfer protein (GLTP) known as HET-C [123, 124] (Fig. 3). The cell death process initiated by the incompatibility between certain HNWD proteins (HET-E and HET-D) and HET-C (GLTP) depends on polymorphisms in the WD40 repeats of the fungal NLRs [121, 125, 126] (Fig. 3). This has led to a model proposing direct binding between the WD40 sensory domain and the HET-C GLTP [127]. The interaction appears analogous to the one described between the WD40 repeats of APAF-1 and cytochrome c [128]. Nevertheless, HI-related RCD – and specifically NLR-controlled cell death in this context – is generally lytic and thus fundamentally distinct from the apoptotic programs described in animals.
Two of the most comprehensively characterized fungal NLRs are the HI-controlling gene plp-1 from N. crassa [119] and the HI-associated gene nwd2 from P. anserina [129, 130]. PLP-1 (patatin-like phospholipase-1) contains an NB-ARC domain with TPR repeats and an N-terminal patatin phospholipase domain. The PLP-1 NLR triggers cell death in germinating asexual spores and hyphae of N. crassa in the presence of an incompatible variant of the SEC-9 protein, which is involved in vesicle transport (t-SNARE). Heller et al. have shown that the two proteins interact physically, and that RCD depends on the activity of the patatin domain and the ATPase-binding motif of the NB-ARC domain [119]. PLP-1 also interacts with itself during the cell death process, suggesting that the protein oligomerizes in a manner similar to characterized NLRs from metazoans, plants, and bacteria. A striking observation about the PLP-1/SEC-9 HI system is its apparent independent emergence in multiple fungal lineages, including P. anserina [119], C. parasitica [69], and B. cinerea [131]. An explanation for the apparent convergent evolution has been proposed, suggesting that PLP-1 and SEC-9 function as partners outside the context of allorecognition, and that the NLR might be surveilling the state (or presence) of the t-SNARE protein. This model is inspired by the ‘guard’ strategy employed by some plant NLRs to provide immune defense against pathogens [132, 133]. However, in fungi, these genes accumulate polymorphisms in strains that evolve independently, potentially generating incompatible PLP-1/SEC-9 combinations when such strains undergo somatic cell fusion. The ‘guard hypothesis’ has also been invoked to explain other NLR-based HI systems in P. anserina [88].
NWD2 (NACHT-WD40s) is a member of the nwd family of NLRs in P. anserina [125], whose reference genome encodes 77 different NLR genes (Fig. 3) [114]. Three different nwd genes encode allorecognition determinants (het-e, het-d, and het-r), while nwd2 is involved only indirectly in an HI system (reviewed below) [134]. The discovery and characterization of nwd2 have established that some fungal RCD pathways are related to necroptosis [20] and to plant cell death programs like the hypersensitive response [11, 135](Fig. 4). These findings have greatly contributed to the integration of HI and fungal RCD into the field of comparative immunology [97, 98].
In mammals, necroptosis depends upon the formation of a protein complex termed the necrosome, formed by the RIP1 and RIP3 kinases. The RIP homotypic interaction motif (RHIM) is a short amyloidogenic sequence that mediates necrosome formation. Activated RIPK3 in the necrosome phosphorylates the pseudokinase domain of MLKL, the executioner of necroptosis. Phosphorylated MLKL aggregates and punctures the plasma membrane, forming a transmembrane cation channel, ultimately inducing cell death. The four-helical bundle (4HB) domain plays an essential role in MLKL membrane targeting. An octameric MLKL complex, composed of two tetramers, opens a cation channel to induce necroptotic cell death. The membrane-targeting module has been characterized in immunity-related cell death processes in plants and fungi. In plants, 4HB homologs can be found in some NLRs and in recently characterized plant MLKL-like proteins. These plant and fungal homologs carry distinct names – RPW8 (Resistant to Powdery Mildew) coiled-coil domain (CCRPW8) and HeLo (found in HET-s/LopB) domain. Plant NLRs carrying the CCRPW8 play an important role in the plant immune system, controlling localized necrotic cell death in response to pathogens, termed the hypersensitive response (HR). Some such plant NLRs have been shown to form a pentameric complex named the resistosome to induce HR. The HeLo domain has also been experimentally characterized in fungi. Some fungal NLRs control HeLo-domain cell death effectors through signal transduction involving amyloid-forming domains, some of which appear homologous to mammalian RHIM. HeLo (and CCRPW8) rely on an extreme N-terminal α-helix to attack the plasma membrane. While fungal HeLo domains are shown to act as pore-forming proteins in vitro, it is currently unknown if they form cation channels.
Evolutionary links between fungal cell death, mammalian necroptosis, and the hypersensitive response in plants
Necroptosis is involved in the pathophysiology of a range of human diseases [136] and is considered an important anti-viral immune cell death [137]. Necroptotic cell death can be triggered by a variety of cell surface receptors – such as TNFR (tumor necrosis factor receptor) or certain Toll-like receptors (e.g., TLR4) [138] – as well as by intracellular sensors of nucleic acids [139,140,141]. The necroptotic pathway is typically engaged when apoptosis and the pro-apoptotic caspase-8 are inhibited, in some cases directly by the invading pathogen [142]. The initiation of necroptosis requires the formation of a large multimeric heterocomplex of the kinases RIPK1 (receptor-interacting protein kinase 1) and RIPK3, known as the necrosome [143, 144]. Oligomerization of the necrosome is mediated by an 18–22 amino acid-long RIP homotypic interaction motif (RHIM), conserved in both RIPK1 and RIPK3 [145,146,147]. Once recruited into the necrosome, RIPK3 phosphorylates the mixed-lineage kinase-like (MLKL) protein, which then oligomerizes to form a cation channel in the plasma membrane, culminating in necroptotic cell death [96, 148, 149] (Fig. 4). MLKL functions as a cell death executioner, and its involvement in RCD is a hallmark of necroptosis.
In the following paragraphs, we review the evolutionary links between necroptosis and recently described fungal RCD processes.
A subset of fungal NLRs use amyloid signaling to activate cell death effectors
The RHIM domain, or motif, which is essential for the formation of the necrosome, forms a functional amyloid [145]. Amyloids are highly ordered, strictly cooperative, β-sheet-rich protein structures often associated with neurodegenerative diseases [150, 151]. Many amyloid aggregates are the result of protein misfolding. Functional amyloids, however, are integral to various biological processes and are shaped by evolutionary forces [152,153,154].
In fungi, amyloid domains have been integrated into RCD pathways as signal-transducing modules between a NOD-like receptor and a downstream signaling partner [155]. In general, the two or three genes that constitute the functional unit are genomically clustered [129]. The first such gene cluster described, identified in P. anserina, comprises the nwd2 NLR-encoding gene, and het-S, which encodes a pore-forming protein [129, 130]. The short amyloid signal-transducing sequence, consisting of 21 amino acids and termed R0 (repeat zero), is situated at the extreme N-terminus of NWD2 [129, 130]. HET-S carries two C-terminal amyloid pseudo-repeats – R1 and R2 – and an N-terminal HeLo domain, which targets the plasma membrane when the protein is activated by the NWD2 NLR [156,157,158]. The three amyloid-forming repeats (R0, R1, and R2) exhibit strong sequence similarity and appear to adopt the same amyloid fold [130]. A model for the signaling mechanism proposes that activated NWD2 oligomerizes, bringing the R0 motifs into proximity within the NLR protein complex, which in turn promotes the cooperative folding and the emergence of the amyloid fold. The newly formed amyloid then serves as a structural template for the R1-R2 pseudo-repeats of the HET-S cell death effector [129, 130, 159]. The transconformation of HET-S C-terminal amyloid domain triggers the activity of the cytotoxic HeLo domain [159]. The discovery of NLR-based amyloid signaling, along with insights into its molecular mechanism, has been advanced through studies of a naturally occurring cytotoxic-dead HET-S variant, known as HET-s (small s) [134]. HET-s is a model prion protein extensively used to investigate the fundamental properties of amyloid folds [160].
Natural diversity of fungal signaling amyloids includes RHIM-like motifs
A high-resolution SSNMR structure of the HET-S/s amyloid domain has been reported, revealing a left-handed β-solenoid (β helix) fold [161]. The triangular, highly hydrophobic amyloid core of HET-S/s bears slight similarity to the N-shaped amyloid core of RIPK3 RHIM homotypic oligomers [162]. An evolutionary relationship between RHIMs and the HET-S R1-R2 amyloid core-forming repeats has previously been proposed based on molecular modeling [163]. NLR-dependent signaling amyloids in fungi have been undergone extensive diversification [155]. Among the diverse amyloidogenic sequences, one widespread signaling amyloid – named pseudo-palindromic, or ‘PP’ – appears to share significant sequence similarity with RHIM [129]. The PP (fungal RHIM) domain was first characterized as a functional signaling amyloid using a three-gene cluster from Chaetomium globosum [95], and more recently, a two-gene cluster from P. anserina [164]. In the latter case, Bardin et al. explored HELLP (HeLo-like PP) and PNT1 (named after the NLR architecture PP-NB-ARC-TPR), encoded by a two-gene cluster analogous to het-S/nwd2. The authors show that P. anserina PP/RHIM domain can ‘cross-seed’ – that is, signal-transduce between two distinct amyloids – in vivo with human RIPK1 and RIPK3 RHIMs [164]. PP/RHIM signaling has also been reported in bacteria, where, similarly to fungi, a much greater diversity of signaling amyloids has been uncovered compared to metazoans [165]. The increased diversity of the signaling amyloids likely contributes to higher specificity in the signaling process, as suggested by experimental data from the HET-S-related amyloid motifs (HRAM) subfamily [166, 167].
Evolutionary links between fungal HeLo domains and the membrane-targeting mammalian protein MLKL
The necrosome is activated by phosphorylation of the MLKL protein, the cell death executioner of necroptosis [168,169,170]. Phosphorylation by RIPK3 of a regulatory loop in the pseudokinase domain of MLKL unleashes its N-terminal, membrane-targeting four-helix bundle (4HB) domain [171, 172]. Phosphorylated MLKL oligomerizes and localizes to the cell periphery, where the 4HB domain perforates the plasma membrane to form ion channels [173,174,175]. Remarkably, an evolutionary link has been established between the 4HB domain of MLKL and the HeLo and HeLo-like (HELL) domains of HET-S and HELLP, respectively [95]. Several other fungal domain annotations have been grouped with the membrane-targeting HeLo and HELL in the superfamily of 4HB MLKL homologs [94]. Using AlphaFold molecular modeling, Wojciechowski et al. have established that the helical folds of the HeLo and HeLo-like domains present the highest similarity with MLKL 4HB [94]. The main difference between the generated models and the structure of 4HB domain lies in the variable lengths of the helix located at the extreme N-terminus (helix 1) in some molecular models [94]. The N-terminal helix of the HET-S HeLo domain has been shown to play a key role in membrane targeting [158] and it is hypothesized that a similar mechanism of membrane disruption and pore formation is conserved for the other members of the protein family [94].
The fungal HeLo and HeLo-like domains have also been conserved in plants [94, 95]. In plants, HeLo homologs occur as N-terminal domains on a subset of NLRs [176]. Recently, the structure of one such NLR – ZAR1 (HOPZ-ACTIVATED RESISTANCE 1) – has been solved with cryo-EM, revealing the formation of a pentameric signaling hub, termed a ‘resistosome’ [177, 178]. The HeLo/4HB homologous domains of ZAR1 molecules cluster atop of the doughnut-shaped resistosome, forming a calcium-permeable channel [179]. Noteworthy, a large number of fungal NLRs present similar protein architectures, where HeLo, HeLo-like, SesA or Goodbye domains are directly integrated at the N-terminus of the molecular receptors [88, 94, 114]. Thus, in fungi, some NLRs use signaling amyloids like RHIM to activate downstream effector or cell death-inducing domains, while in other cases, these domains are integrated to the receptor in an alternate ‘all-in-one’ architecture. Intriguingly, plants also possess in their immune arsenal bona fide MLKL homologs [180]. Future work should be focused on exploring more broadly the HeLo/4HB proteins in fungi and clarify the extent of conservation between necroptosis and fungal necroptotic-like cell death.
Microbial gasdermins draw evolutionary parallels with mammalian pyroptosis
Recent findings indicate that the fungal RCD machinery can rely on cell death programs resembling mammalian pyroptosis [181, 182]. Like necroptosis, pyroptosis is a highly inflammatory and lytic cell death in mammals, with diverse immune and physiological roles [22, 183, 184]. The pyroptotic cell death pathway relies on a family of pore-forming proteins, known as gasdermins, which act as executioners of cell death [185, 186]. The gasdermins are regulated by proteolytic cleavage, exerted by different caspases and some serine proteases, depending on the specific gasdermin [187]. In humans, the gasdermin family comprises six members (GSDMA-GSDME and Pejvakin) with GSDMD being one of the first and best-characterized gasdermin proteins [188, 189]. GSDMD is specifically cleaved by pro-inflammatory caspases (CASP1, CASP4, and CASP5), with CASP1 acting notably downstream of various NLR-based inflammasomes [189]. The caspase removes the inhibitory C-terminal domain of GSDMD and liberates the N-terminal pore-forming domain, which then binds to the plasma membrane and oligomerizes into highly ordered pores [190,191,192] (Fig. 5). Gasdermin homologs have been identified in species across the metazoan branch, including in some basal clades, and in both fungi and bacteria [92, 193]. Remarkably, these distant microbial gasdermins have retained the immune-related role – as determinants of allorecognition in fungi, and as components of anti-phage defense systems in bacteria [92, 193].
A Shown are cartoon representations of gasdermin pores with known structures. The two allelic variants RCD-1-1 and RCD-1-2 from N. crassa are currently the only known gasdermins to form heteromeric pores, composed of eleven alternating RCD-1-1/RCD-1-2 dimers. The RCD-1 fGSDM is not controlled by proteolytic cleavage, like most other gasdermins, but by the coexistance in the same cell of the two allelic variants. This coexistance occurs only when N. crassa strains from the rcd-1-1 and rcd-1-2 antagonistic genotypes undergo cellular fusion. B The majority of gasdermins in microorganisms (bacteria and fungi) are genomically clustered with protease-encoding genes. These multidomain proteases are putative molecular sensors that control the adjecently encoded gasdermin. PDB IDs: GSDMD (6VFE), GSDMA3 (6CB8), TrichoGSDM (8JYW), bGSDM (8SL0), fGSDM (8JYZ).
Fungal gasdermins (fGSDMs) in allorecognition: cell death mechanisms and structural insights
Gasdermin-like genes are abundant in fungal genomes and have been uncovered through studies of heterokaryon incompatibility (HI) and allorecognition systems [92, 193]. Fungal gasdermin (fGSDM)-based allorecognition systems have been described in at least two different ascomycete species, Podospora anserina and Neurospora crassa [181, 182, 194]. These two fGSDM-based HI systems – like HI systems in general – are proposed to have emerged from broader signaling pathways and to have been evolutionary selected for their prophylactic cell death function [97, 98]. Under such a model, the het-Q1/het-Q2 system in P. anserina and the rcd-1-1/rcd-1-2 in N. crassa likely derive from pre-existing fGSDM-mediated cell death pathways (Fig. 6). This would explain the striking differences between the two HI systems; notably, in N. crassa, the allorecognition cell death is triggered by the co-expression of two divergent gasdermin alleles (rcd-1-1 and rcd-1-2) within the same cell, whereas the HI system in P. anserina involves a gasdermin-encoding gene (het-Q1) and a serine protease-encoding gene (het-Q2) [182, 194]. The het-Q1 and het-Q2 genes are idiomorphs (different genes located at the same locus), and genomic rearrangements have also been reported for the rcd-1 locus [181, 194]. The two fGSDM-based incompatibility systems have recently been reviewed in greater depth elsewhere [193].
Fungal genomes encode a variety of putative molecular sensors controlling pore-forming proteins, such as fGSDMs. Some of these sensors belong to the NOD-like receptor (NLR) family. A subset of NLRs use signaling amyloids – some of which appear related to the metazoan necroptosis-controlling RHIM (RIP homotypic interaction motif) – to activate downstream membrane-targeting proteins. The latter are characterized by a helical bundle domain, named HeLo-like (HELL) or HeLo, evolutionary related to the four-helix bundle (4HB) of the necroptosis executioner protein MLKL (Mixed lineage kinase domain-like pseudokinase). The signaling mechanism is based on ‘amyloid templating’ or the transmission of structural information from the NLR-based signalosome to the RCD executioner protein. Certain NLR proteins carry a HeLo/HELL domain at their N-terminal end. This ‘all-in-one’ architecture is analogous to some plant NLRs, which carry a coiled-coil (CC) domain as an N-terminal effector/signaling module. Remarkably, some HELL domains are integral to proteins with kinase/pseudokinase domain, reminiscent of the original MLKL architecture. Similar diversity of molecular receptors has been uncovered in the fungal pyroptotic-like pathways. A subset of the fGSDM-controlling proteins are NLR sensors, carrying an S8 serine protease or a CHAT caspase-like domain. The protease domains can frequently be directly fused to superstructure-forming repeats (WD40, TPR or ANK) or other predicted kinase-like domains. Notably, the fungal necroptotic-like and pyroptotic-like RCD pathways are often encoded in two- or three-gene clusters, sharing thus some characteristics with prokaryotes. The number of such clusters can vary widely between species. A major open question regarding these RCD pathways in fungi is related to the nature of stimuli that activate the identified diverse putative sensors.
A notable difference between fungal and metazoan gasdermins lies in the length of their inhibitory C-terminal domains, where no sequence or structural similarity has been identified between the two clades [182, 194]. In metazoans, the inhibitory domain is typically of similar length to the pore-forming domain, which itself shares some sequence homology and structural similarity with the cytotoxic N-terminal domain of fGSDMs. In contrast, microbial gasdermins possess a much shorter inhibitory domain relative to their membrane-targeting cytotoxic domain [194]. For example, the proteolytically processed HET-Q1 gasdermin loses a ~ 5-kDa fragment from its C-terminus, likely cleaved at residue F238 by the subtilisin-like serine protease HET-Q2 [194]. Such short inhibitory domains have been described in bacterial gasdermins, whose inhibition mechanisms differ significantly from those of mammalian gasdermins [195].
Several functional similarities have been identified between fungal and metazoan gasdermins, including oligomer formation, lipid recognition profiles, and regulation by proteolysis [182, 194]. In vivo, RCD-1-1 (257 aa) and RCD-1-2 (244 aa) show plasma membrane affinity, which depends on a patch of positively charged residues situated on a pair of amphipathic α-helices [182]. Similar structural features of functional importance are conserved in human GSDMD and murine GSDMA3 [190, 196]. When co-expressed heterologously, the two RCD-1 allelic variants physically interact to trigger pyroptotic-like cell death in human 293 T cells, underscoring the autonomy of the allorecognition system [182]. Li et al. have recently reported the crystal structures of both monomeric RCD-1 variants, which share nearly identical protein folds [197]. The structures the two gasdermins most closely resemble those of mammalian gasdermins. Remarkably, the authors have also solved a Cryo-EM structure of heteromeric transmembrane pore formed by alternating RCD-1-1 and RCD-1-2 subunits (Fig. 5). The uncovered mechanism of pore-formation relies on the heterodimerization of the two allelic variants in a lipid environment, before shaping the observed 11-fold pore complex [197, 198]. This mechanism provides a molecular explanation for the allorecognition cell death process, as the two allelic variants coexist only after somatic fusion of a strain from the rcd-1-1 genotype with a strain from the rcd-1-2 genotype.
Protease-clustered fGSDM genes and the ancient origins of pyroptotic-like cell death
Most fGSDMs appear to be regulated by proteolytic cleavage, and it has been reported that more than 80% of fGSDM genes are genomically clustered with genes encoding a protease domain [194]. The encoded proteases represent multidomain molecular sensors, with a large fraction of the proteins carrying super-structure-forming motifs (WD40, ANK, and TPR) next to the protease domain. A small subset of the molecular sensors consists of NLR proteins with the protease domain as the N-terminal signaling module [194]. The majority of protease domains have been classified as serine proteases and approximately 17% as cysteine proteases, among which is the caspase-like CHAT domain (Caspase HetF Associated with TPRs) [199]. CHAT proteases belong to the same clade as mammalian caspases and appear to adopt folds showing a high structural similarity to the separase family of cysteine proteases [199, 200]. The functional relation between the fGSDMs and the protease-carrying receptors has been explored within a two-gene cluster from P. anserina, demonstrating the activation of the gasdermin by a TPR-bearing CHAT protease [194]. Similar genomic clustering of gasdermin genes and protease-encoding genes has been reported in bacterial and archaeal genomes [195, 201]. These findings suggest that pyroptotic-like cell death has an extremely ancient evolutionary origin and that fGSDMs, considering their abundance and distribution, are important actors in the fungal cell death machinery. Future research will be needed to uncover the regulatory mechanisms controlling the protease-bearing proteins that activate fGSDMs, and to explore the roles of these two-gene clusters in other biological processes such as secretion.
Comparative immunology in Fungi: the end of the beginning
In summary, it is now clear that fungal genomes harbor a variety of genes with evolutionary ties to molecular players integral to all three core mammalian immune-related RCD pathways: apoptosis, necroptosis and pyroptosis. Most of these evolutionary parallels extend from bacteria to metazoans, highlighting the trans-kingdom history of the cell death machinery. This conservation encompasses all aspects of regulated cell death, including execution, signal transduction and signal perception. Considering the latter, the NOD-like receptor family emerges as a significant thread in the evolutionary history of immune sensing. Fungal NLR proteins participate in both necroptotic- and pyroptotic-like regulated cell death, and in some cases, they display protein architectures similar to those of the pro-apoptotic mammalian NLR-like protein APAF-1. In addition, several signaling N-terminal domains of fungal NLRs share ancestry with immune signaling modules in other taxa, including plants and bacteria. Two notable examples are the HET domain, proposed as a distant relative to the Toll-like receptor (TIR) domain [83, 114], and members of the purine nucleoside phosphorylases and uridine phosphorylases (PNP_UDP) family [94]. Both of these signaling modules exhibit hydrolase activity (to NAD+ and ATP, respectively) and have been shown to function in immune-related cell death in different eukaryotic kingdoms as well as bacteria [202,203,204,205,206]. These findings further extend the evolutionary ties between eukaryotic and prokaryotic RCD genes, while also expanding the fungal immune arsenal.
At present, as the outlines of the fungal immune-related gene families become clearer, many questions remain regarding the description and conceptualization of the fungal immune system. In the first instance, while some evolutionary links have been unveiled between fungal RCD and the core innate immune pathways in metazoans (necroptosis and pyroptosis), the cellular characterization of the fungal cell death pathways remains incomplete, complicating comparisons, analogies, and the adoption of a common vocabulary within the Opisthokonts. A better understanding of the RCD manifestations at the cellular level, triggered by different molecular receptors, would be needed to eventually delineate distinct cell death pathways. Most of the presently characterized cell death processes, especially in the context of heterokaryon incompatibility, appear to be lytic (leakage of cytoplasm), drawing parallels with necroptosis, pyroptosis, and PANoptosis. However, it is unclear if specific hallmarks can be associated with different allorecognition systems and inputs from distinct molecular sensors or following the activation of specific cell death–inducing proteins, allowing us to identify separate RCD programs.
In the perspective of comparing the innate immune mechanisms of distant organisms, we would need to decipher the ones operating in fungi in much greater details, including their regulation, roles, and integration with other fundamental physiological processes. Novel experimental model systems, recently developed to study fungal-bacterial interactions [207], may help uncover more specifics about fungal RCD in response to heterospecific non-self. Despite our limited understanding of the fungal immune system, it appears to display features of both eukaryotic and prokaryotic lineages. The observed genomic clustering in many fungal genomes of sensors and cell death executioner genes resembles the numerous bacterial anti-phage operons clustered together in defense islands [208]. It is conceivable that microorganisms, independently of their eukaryotic or prokaryotic classification, share certain immunity-related adaptations setting them apart from differentiated higher eukaryotes (i.e. plants, animals), due to morphological similarities (limited differentiation of somatic cells), common ecological niches, and similar lifestyles.
As evidence accumulate for the prokaryotic origin of the molecular RCD machinery [209, 210], and its role in the emergence of multicellularity appears highly plausible [211, 212], fungi occupy a particular space on the multicellularity spectra with their syncytial – sharing cytoplasm and nuclei between cells – filamentous networks as organismal body structure. Besides the syncytial aspect of fungi, their ability to undergo regular vegetative anastomosis, or cellular fusions between distinct fungal colonies, adds some taxon-level idiosyncrasy to the notion of ‘individuality’, which is of paramount importance in immunology [213]. The extent to which cellular organization and different lifestyles shape the evolution and mechanisms of immune cell death and broadly of immunity [214], are particularly intriguing questions to explore in the framework of trans-kingdom comparative immunology. Finally, elucidating the molecular mechanisms of fungal RCD could open the door to designing new strategies and exploiting these endogenous cell death programs to thwart some pathogenic fungal species, of major importance for human health, agriculture and food security [215, 216].
References
Galluzzi L, Bravo-San Pedro JM, Vitale I, Aaronson SA, Abrams JM, Adam D, et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ. 2015;22:58–73.
Tang D, Kang R, Berghe TV, Vandenabeele P, Kroemer G. The molecular machinery of regulated cell death. Cell Res. 2019;29:347–64.
Nagata S, Tanaka M. Programmed cell death and the immune system. Nat Rev Immunol. 2017;17:333–40.
Bertheloot D, Latz E, Franklin BS. Necroptosis, pyroptosis and apoptosis: an intricate game of cell death. Cell Mol Immunol. 2021;18:1106–21.
Salguero-Linares J, Coll NS. Cell death as a defense strategy against pathogens in plants and animals. PLoS Pathog. 2023;19:e1011253.
Lopatina A, Tal N, Sorek R. Abortive infection: bacterial suicide as an antiviral immune strategy. Annu Rev Virol. 2020;7:371–84.
Millman A, Melamed S, Leavitt A, Doron S, Bernheim A, Hör J, et al. An expanded arsenal of immune systems that protect bacteria from phages. Cell Host Microbe. 2022;30:1556–1569.e5.
Rousset F, Sorek R. The evolutionary success of regulated cell death in bacterial immunity. Curr Opin Microbiol. 2023;74:102312.
Jones JDG, Staskawicz BJ, Dangl JL. The plant immune system: From discovery to deployment. Cell. 2024;187:2095–116.
Coll NS, Epple P, Dangl JL. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18:1247–56.
Balint-Kurti P. The plant hypersensitive response: concepts, control and consequences. Mol Plant Pathol. 2019;20:1163–78.
Wainright PO, Hinkle G, Sogin ML, Stickel SK. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science. 1993;260:340–2.
Ocaña-Pallarès E, Williams TA, López-Escardó D, Arroyo AS, Pathmanathan JS, Bapteste E, et al. Divergent genomic trajectories predate the origin of animals and fungi. Nature. 2022;609:747–53.
Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–57.
Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516.
Formigli L, Conti A, Lippi D. Falling leaves”: a survey of the history of apoptosis. Minerva Med. 2004;95:159–64.
Ekert PG, Vaux DL. Apoptosis and the immune system. Br Med Bull. 1997;53:591–603.
Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–41.
Seo J, Nam YW, Kim S, Oh D-B, Song J. Necroptosis molecular mechanisms: Recent findings regarding novel necroptosis regulators. Exp Mol Med. 2021;53:1007–17.
Dhuriya YK, Sharma D. Necroptosis: a regulated inflammatory mode of cell death. J Neuroinflammation. 2018;15:199.
Chen D, Yu J, Zhang L. Necroptosis: an alternative cell death program defending against cancer. Biochim Biophys Acta. 2016;1865:228–36.
Yu P, Zhang X, Liu N, Tang L, Peng C, Chen X. Pyroptosis: mechanisms and diseases. Signal Transduct Target Ther. 2021;6:128.
Chen X, He W-T, Hu L, Li J, Fang Y, Wang X, et al. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res. 2016;26:1007–20.
Liu X, Lieberman J. Inflammasome-independent pyroptosis. Curr Opin Immunol. 2024;88:102432.
Jorgensen I, Rayamajhi M, Miao EA. Programmed cell death as a defence against infection. Nat Rev Immunol. 2017;17:151–64.
Yuan J, Ofengeim D. A guide to cell death pathways. Nat Rev Mol Cell Biol. 2024;25:379–95.
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72.
Schuster M, Kilaru S, Steinberg G. Azoles activate type I and type II programmed cell death pathways in crop pathogenic fungi. Nat Commun. 2024;15:4357.
Struyfs C, Cammue BPA, Thevissen K. Membrane-interacting antifungal peptides. Front Cell Dev Biol. 2021;9:649875.
Chau S, Gao J, Diao AJ, Cao SB, Azhieh A, Davidson AR, et al. Diverse yeast antiviral systems prevent lethal pathogenesis caused by the L-A mycovirus. Proc Natl Acad Sci USA. 2023;120:e2208695120.
Götze S, Vij R, Burow K, Thome N, Urbat L, Schlosser N, et al. Ecological niche-inspired genome mining leads to the discovery of crop-protecting nonribosomal lipopeptides featuring a transient amino acid building block. J Am Chem Soc. 2023;145:2342–53.
Trunk K, Peltier J, Liu Y-C, Dill BD, Walker L, Gow NAR, et al. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol. 2018;3:920–31.
Deveau A, Bonito G, Uehling J, Paoletti M, Becker M, Bindschedler S, et al. Bacterial-fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev. 2018;42:335–52.
Pinan-Lucarré B, Paoletti M, Dementhon K, Coulary-Salin B, Clavé C. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol Microbiol. 2003;47:321–33.
Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580–3.
Liu X-H, Xu F, Snyder JH, Shi H-B, Lu J-P, Lin F-C. Autophagy in plant pathogenic fungi. Semin Cell Dev Biol. 2016;57:128–37.
Ryder LS, Talbot NJ. Regulation of appressorium development in pathogenic fungi. Curr Opin Plant Biol. 2015;26:8–13.
Raju NB, Perkins DD. Programmed ascospore death in the homothallic ascomycete Coniochaeta tetraspora. Fungal Genet Biol. 2000;30:213–21.
Saupe SJ, Johannesson H. On the mechanistic basis of killer meiotic drive in fungi. Annu Rev Microbiol. 2022;76:305–23.
Eastwood MD, Meneghini MD. Developmental coordination of gamete differentiation with programmed cell death in sporulating yeast. Eukaryotic Cell. 2015;14:858–67.
Paoletti M. Vegetative incompatibility in fungi: From recognition to cell death, whatever does the trick. Fungal Biol Rev. 2016;30:152–62.
Saupe SJ. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Mol Biol Rev. 2000;64:489–502.
Daskalov A, Heller J, Herzog S, Fleiβner A, Glass NL. Molecular mechanisms regulating cell fusion and heterokaryon formation in filamentous fungi. In: Heitman J, Howlett BJ, Crous PW, Stukenbrock EH, James TY, Gow NAR, editors. The Fungal Kingdom. Washington, DC, USA: ASM Press; 2017. p. 215–29.
Callemeyn J, Lamarthée B, Koenig A, Koshy P, Thaunat O, Naesens M. Allorecognition and the spectrum of kidney transplant rejection. Kidney Int. 2022;101:692–710.
Marino J, Paster J, Benichou G. Allorecognition by T lymphocytes and allograft rejection. Front Immunol. 2016;7:582.
Taketa DA, De Tomaso AW. Botryllus schlosseri allorecognition: tackling the enigma. Dev Comp Immunol. 2015;48:254–65.
Nydam ML. Evolution of allorecognition in the tunicata. Biology. 2020;9:129.
Kundert P, Shaulsky G. Cellular allorecognition and its roles in Dictyostelium development and social evolution. Int J Dev Biol. 2019;63:383–93.
Tipping MJ, Gibbs KA. Peer pressure from a Proteus mirabilis self-recognition system controls participation in cooperative swarm motility. PLoS Pathog. 2019;15:e1007885.
Heller J, Zhao J, Rosenfield G, Kowbel DJ, Gladieux P, Glass NL. Characterization of greenbeard genes involved in long-distance kind discrimination in a microbial eukaryote. PLoS Biol. 2016;14:e1002431.
Gonçalves AP, Heller J, Span EA, Rosenfield G, Do HP, Palma-Guerrero J, et al. Allorecognition upon fungal cell-cell contact determines social cooperation and impacts the acquisition of multicellularity. Curr Biol. 2019;29:3006–3017.e3.
Glass NL, Kaneko I. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryotic Cell. 2003;2:1–8.
Glass NL, Jacobson DJ, Shiu PK. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu Rev Genet. 2000;34:165–86.
Pinan-Lucarré B, Paoletti M, Clavé C. Cell death by incompatibility in the fungus Podospora. Semin Cancer Biol. 2007;17:101–11.
Jedd G, Pieuchot L. Multiple modes for gatekeeping at fungal cell-to-cell channels. Mol Microbiol. 2012;86:1291–4.
Kaneko I, Dementhon K, Xiang Q, Glass NL. Nonallelic interactions between het-c and a polymorphic locus, pin-c, are essential for nonself recognition and programmed cell death in Neurospora crassa. Genetics. 2006;172:1545–55.
Zhang D-X, Nuss DL. Engineering super mycovirus donor strains of chestnut blight fungus by systematic disruption of multilocus vic genes. Proc Natl Acad Sci USA. 2016;113:2062–7.
Stauder CM, Nuss DL, Zhang D-X, Double ML, MacDonald WL, Metheny AM, et al. Enhanced hypovirus transmission by engineered super donor strains of the chestnut blight fungus, Cryphonectria parasitica, into a natural population of strains exhibiting diverse vegetative compatibility genotypes. Virology. 2019;528:1–6.
Caten CE. Vegetative incompatibility and cytoplasmic infection in fungi. J Gen Microbiol. 1972;72:221–9.
Myers JM, James TY. Mycoviruses. Curr Biol. 2022;32:R150–5.
Hough B, Steenkamp E, Wingfield B, Read D. Fungal viruses unveiled: a comprehensive review of mycoviruses. Viruses. 2023;15:1202.
Biella S, Smith ML, Aist JR, Cortesi P, Milgroom MG. Programmed cell death correlates with virus transmission in a filamentous fungus. Proc Biol Sci. 2002;269:2269–76.
Nuss DL. Hypovirulence: mycoviruses at the fungal-plant interface. Nat Rev Microbiol. 2005;3:632–42.
García-Pedrajas MD, Cañizares MC, Sarmiento-Villamil JL, Jacquat AG, Dambolena JS. Mycoviruses in biological control: from basic research to field implementation. Phytopathology. 2019;109:1828–39.
van Diepeningen AD, Debets AJ, Hoekstra RF. Heterokaryon incompatibility blocks virus transfer among natural isolates of black Aspergilli. Curr Genet. 1997;32:209–17.
Debets F, Yang X, Griffiths AJ. Vegetative incompatibility in Neurospora: its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr Genet. 1994;26:113–9.
Debets AJM, Dalstra HJP, Slakhorst M, Koopmanschap B, Hoekstra RF, Saupe SJ. High natural prevalence of a fungal prion. Proc Natl Acad Sci USA. 2012;109:10432–7.
Zhang D-X, Spiering MJ, Dawe AL, Nuss DL. Vegetative incompatibility loci with dedicated roles in allorecognition restrict mycovirus transmission in chestnut blight fungus. Genetics. 2014;197:701–14.
Choi GH, Dawe AL, Churbanov A, Smith ML, Milgroom MG, Nuss DL. Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics. 2012;190:113–27.
Cortesi P, McCulloch CE, Song H, Lin H, Milgroom MG. Genetic control of horizontal virus transmission in the chestnut blight fungus, Cryphonectria parasitica. Genetics. 2001;159:107–18.
Wu S, Cheng J, Fu Y, Chen T, Jiang D, Ghabrial SA, et al. Virus-mediated suppression of host non-self recognition facilitates horizontal transmission of heterologous viruses. PLoS Pathog. 2017;13:e1006234.
Hai D, Li J, Jiang D, Cheng J, Fu Y, Xiao X, et al. Plants interfere with non-self recognition of a phytopathogenic fungus via proline accumulation to facilitate mycovirus transmission. Nat Commun. 2024;15:4748.
Debets AJM, Griffiths AJF. Polymorphism of het-genes prevents resource plundering in Neurospora crassa. Mycol Res. 1998;102:1343–9.
Gonçalves AP, Heller J, Rico-Ramírez AM, Daskalov A, Rosenfield G, Glass NL. Conflict, competition, and cooperation regulate social interactions in filamentous fungi. Annu Rev Microbiol. 2020;74:693–712.
Bastiaans E, Debets AJM, Aanen DK. Experimental evolution reveals that high relatedness protects multicellular cooperation from cheaters. Nat Commun. 2016;7:11435.
Shiu PK, Glass NL. Molecular characterization of tol, a mediator of mating-type-associated vegetative incompatibility in Neurospora crassa. Genetics. 1999;151:545–55.
Jacobson DJ. Control of mating type heterokaryon incompatibility by the tol gene in Neurospora crassa and N. tetrasperma. Genome. 1992;35:347–53.
Hedrick PW. Balancing selection. Curr Biol. 2007;17:R230–1.
Milgroom MG, Smith ML, Drott MT, Nuss DL. Balancing selection at nonself recognition loci in the chestnut blight fungus, Cryphonectria parasitica, demonstrated by trans-species polymorphisms, positive selection, and even allele frequencies. Heredity. 2018;121:511–23.
Auxier B, Zhang J, Marquez FR, Senden K, van den Heuvel J, Aanen DK, et al. The Narrow Footprint of Ancient Balancing Selection Revealed by Heterokaryon Incompatibility Genes in Aspergillus fumigatus. Mol Biol Evol. 2024;41:msae079.
Wu J, Saupe SJ, Glass NL. Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proc Natl Acad Sci USA. 1998;95:12398–403.
Zhao J, Gladieux P, Hutchison E, Bueche J, Hall C, Perraudeau F, et al. Identification of allorecognition loci in Neurospora crassa by genomics and evolutionary approaches. Mol Biol Evol. 2015;32:2417–32.
Clavé C, Dheur S, Ament-Velásquez SL, Granger-Farbos A, Saupe SJ. het-B allorecognition in Podospora anserina is determined by pseudo-allelic interaction of genes encoding a HET and lectin fold domain protein and a PII-like protein. PLoS Genet. 2024;20:e1011114.
Koenig D, Hagmann J, Li R, Bemm F, Slotte T, Neuffer B, et al. Long-term balancing selection drives evolution of immunity genes in Capsella. eLife. 2019;8.
Nydam ML, Stephenson EE, Waldman CE, De Tomaso AW. Balancing selection on allorecognition genes in the colonial ascidian Botryllus schlosseri. Dev Comp Immunol. 2017;69:60–74.
Cornetti L, Fields PD, Du Pasquier L, Ebert D. Long-term balancing selection for pathogen resistance maintains trans-species polymorphisms in a planktonic crustacean. Nat Commun. 2024;15:5333.
Kloch A, Wenzel MA, Laetsch DR, Michalski O, Bajer A, Behnke JM, et al. Signatures of balancing selection in toll-like receptor (TLRs) genes - novel insights from a free-living rodent. Sci Rep. 2018;8:8361.
Daskalov A, Dyrka W, Saupe SJ. 6 NLR Function in Fungi as Revealed by the Study of Self/Non-self Recognition Systems. In: Benz JP, Schipper K, editors. Genetics and Biotechnology. Cham: Springer International Publishing; 2020. p. 123–41.
Essuman K, Milbrandt J, Dangl JL, Nishimura MT. Shared TIR enzymatic functions regulate cell death and immunity across the tree of life. Science. 2022;377:eabo0001.
Paoletti M, Clavé C. The fungus-specific HET domain mediates programmed cell death in Podospora anserina. Eukaryotic Cell. 2007;6:2001–8.
Broz P, Pelegrín P, Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol. 2020;20:143–57. Mar
Daskalov A, Louise Glass N. Gasdermin and gasdermin-like pore-forming proteins in invertebrates, fungi and bacteria. J Mol Biol. 2021;434:167273.
Kovacs SB, Miao EA. Gasdermins: effectors of pyroptosis. Trends Cell Biol. 2017;27:673–84.
Wojciechowski JW, Tekoglu E, Gąsior-Głogowska M, Coustou V, Szulc N, Szefczyk M, et al. Exploring a diverse world of effector domains and amyloid signaling motifs in fungal NLR proteins. PLoS Comput Biol. 2022;18:e1010787.
Daskalov A, Habenstein B, Sabaté R, Berbon M, Martinez D, Chaignepain S, et al. Identification of a novel cell death-inducing domain reveals that fungal amyloid-controlled programmed cell death is related to necroptosis. Proc Natl Acad Sci USA. 2016;113:2720–5.
Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang J-G, Alvarez-Diaz S, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–53.
Paoletti M, Saupe SJ. Fungal incompatibility: evolutionary origin in pathogen defense? Bioessays. 2009;31:1201–10.
Daskalov A. Emergence of the fungal immune system. iScience. 2023;26:106793.
Gaspar ML, Pawlowska TE. Innate immunity in fungi: Is regulated cell death involved? PLoS Pathog. 2022;18:e1010460.
Leipe DD, Koonin EV, Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol. 2004;343:1–28.
Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. Wheel of life, wheel of death: a mechanistic insight into signaling by STAND Proteins. Structure. 2009;17:172–82.
van der Biezen EA, Jones JD. The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol. 1998;8:R226–7.
Koonin EV, Aravind L. The NACHT family - a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem Sci. 2000;25:223–4.
Kim YK, Shin JS, Nahm MH. NOD-Like receptors in infection, immunity, and diseases. Yonsei Med J. 2016;57:5–14.
Platnich JM, Muruve DA. NOD-like receptors and inflammasomes: a review of their canonical and non-canonical signaling pathways. Arch Biochem Biophys. 2019;670:4–14.
Proell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS ONE. 2008;3:e2119.
Jernigan KK, Bordenstein SR. Tandem-repeat protein domains across the tree of life. PeerJ. 2015;3:e732.
Eitas TK, Dangl JL. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol. 2010;13:472–7.
Jones JDG, Vance RE, Dangl JL. Intracellular innate immune surveillance devices in plants and animals. Science. 2016; 2;354.
Bentham A, Burdett H, Anderson PA, Williams SJ, Kobe B. Animal NLRs provide structural insights into plant NLR function. Ann Bot. 2017;119:827–702.
Ting JP-Y, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8:372–9.
Xi Y, Cesari S, Kroj T. Insight into the structure and molecular mode of action of plant paired NLR immune receptors. Essays Biochem. 2022;66:513–26.
Contreras MP, Lüdke D, Pai H, Toghani A, Kamoun S. NLR receptors in plant immunity: making sense of the alphabet soup. EMBO Rep. 2023;24:e57495.
Dyrka W, Lamacchia M, Durrens P, Kobe B, Daskalov A, Paoletti M, et al. Diversity and variability of NOD-like receptors in fungi. Genome Biol Evol. 2014;6:3137–58.
Kibby EM, Conte AN, Burroughs AM, Nagy TA, Vargas JA, Whalen LA, et al. Bacterial NLR-related proteins protect against phage. Cell. 2023;186:2410–2424.e18.
Gao LA, Wilkinson ME, Strecker J, Makarova KS, Macrae RK, Koonin EV, et al. Prokaryotic innate immunity through pattern recognition of conserved viral proteins. Science. 2022;377:eabm4096.
Bonometti L, Charriat F, Hensen N, Miñana-Posada S, Johannesson H, Gladieux P. Genomic organization, domain assortments, and nucleotide-binding domain diversity of NLR proteins in Sordariales fungi. PLoS Genet. 2025;21:e1011739.
Uehling J, Deveau A, Paoletti M. Do fungi have an innate immune response? An NLR-based comparison to plant and animal immune systems. PLoS Pathog. 2017;13:e1006578.
Heller J, Clavé C, Gladieux P, Saupe SJ, Glass NL. NLR surveillance of essential SEC-9 SNARE proteins induces programmed cell death upon allorecognition in filamentous fungi. Proc Natl Acad Sci USA. 2018;115:E2292–301.
Espagne E, Balhadère P, Penin M-L, Barreau C, Turcq B. HET-E and HET-D belong to a new subfamily of WD40 proteins involved in vegetative incompatibility specificity in the fungus Podospora anserina. Genetics. 2002;161:71–81.
Chevanne D, Bastiaans E, Debets A, Saupe SJ, Clavé C, Paoletti M. Identification of the het-r vegetative incompatibility gene of Podospora anserina as a member of the fast evolving HNWD gene family. Curr Genet. 2009;55:93–102.
Ament-Velásquez SL, Vogan AA, Granger-Farbos A, Bastiaans E, Martinossi-Allibert I, Saupe SJ, et al. Allorecognition genes drive reproductive isolation in Podospora anserina. Nat Ecol Evol. 2022;6:910–23.
Saupe S, Descamps C, Turcq B, Bégueret J. Inactivation of the Podospora anserina vegetative incompatibility locus het-c, whose product resembles a glycolipid transfer protein, drastically impairs ascospore production. Proc Natl Acad Sci USA. 1994;91:5927–31.
Saupe S, Turcq B, Bégueret J. A gene responsible for vegetative incompatibility in the fungus Podospora anserina encodes a protein with a GTP-binding motif and G beta homologous domain. Gene. 1995;162:135–9.
Paoletti M, Saupe SJ, Clavé C. Genesis of a fungal non-self recognition repertoire. PLoS ONE. 2007;2:e283.
Ament-Velásquez SL, Furneaux B, Dheur S, Granger-Farbos A, Stelkens R, Johannesson H, et al. Reconstructing NOD-like receptor alleles with high internal conservation in Podospora anserina using long-read sequencing. Microb Genom. 2025;11:001442.
Espagne E, Balhadère P, Bégueret J, Turcq B. Reactivity in vegetative incompatibility of the HET-E protein of the fungus Podospora anserina is dependent on GTP-binding activity and a WD40 repeated domain. Mol Gen Genet. 1997;256:620–7.
Yuan S, Akey CW. Apoptosome structure, assembly, and procaspase activation. Structure. 2013;21:501–15.
Daskalov A, Paoletti M, Ness F, Saupe SJ. Genomic clustering and homology between HET-S and the NWD2 STAND protein in various fungal genomes. PLoS ONE. 2012;7:e34854.
Daskalov A, Habenstein B, Martinez D, Debets AJM, Sabaté R, Loquet A, et al. Signal transduction by a fungal NOD-like receptor based on propagation of a prion amyloid fold. PLoS Biol. 2015;13:e1002059.
Arshed S, Cox MP, Beever RE, Parkes SL, Pearson MN, Bowen JK, et al. The Bcvic1 and Bcvic2 vegetative incompatibility genes in Botrytis cinerea encode proteins with domain architectures involved in allorecognition in other filamentous fungi. Fungal Genet Biol. 2023;169:103827.
Cesari S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2018;219:17–24.
Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–33.
Daskalov A, Saupe SJ. As a toxin dies a prion comes to life: A tentative natural history of the [Het-s] prion. Prion. 2015;9:184–9.
Morel JB, Dangl JL. The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 1997;4:671–83.
Khoury MK, Gupta K, Franco SR, Liu B. Necroptosis in the pathophysiology of disease. Am J Pathol. 2020;190:272–85.
Nailwal H, Chan FK-M. Necroptosis in anti-viral inflammation. Cell Death Differ. 2019;26:4–13.
Liu T, Zong H, Chen X, Li S, Liu Z, Cui X, et al. Toll-like receptor 4-mediated necroptosis in the development of necrotizing enterocolitis. Pediatr Res. 2022;91:73–82.
Brault M, Olsen TM, Martinez J, Stetson DB, Oberst A. Intracellular nucleic acid sensing triggers necroptosis through Synergistic Type I IFN and TNF Signaling. J Immunol. 2018;200:2748–56.
Maelfait J, Liverpool L, Bridgeman A, Ragan KB, Upton JW, Rehwinkel J. Sensing of viral and endogenous RNA by ZBP1/DAI induces necroptosis. EMBO J. 2017;36:2529–43.
Schock SN, Chandra NV, Sun Y, Irie T, Kitagawa Y, Gotoh B, et al. Induction of necroptotic cell death by viral activation of the RIG-I or STING pathway. Cell Death Differ. 2017;24:615–25.
Mocarski ES, Upton JW, Kaiser WJ. Viral infection and the evolution of caspase 8-regulated apoptotic and necrotic death pathways. Nat Rev Immunol. 2011;12:79–88.
Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–23.
Qi W, Yuan J. RIPK1 and RIPK3 form mosaic necrosomes. Nat Cell Biol. 2022;24:406–7.
Riebeling T, Kunzendorf U, Krautwald S. The role of RHIM in necroptosis. Biochem Soc Trans. 2022;50:1197–205.
Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao Y-S, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–50.
Sun X, Yin J, Starovasnik MA, Fairbrother WJ, Dixit VM. Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J Biol Chem. 2002;277:9505–11.
Wang H, Sun L, Su L, Rizo J, Liu L, Wang L-F, et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell. 2014;54:133–46.
Xia B, Fang S, Chen X, Hu H, Chen P, Wang H, et al. MLKL forms cation channels. Cell Res. 2016;26:517–28.
Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–66.
Greenwald J, Riek R. Biology of amyloid: structure, function, and regulation. Structure. 2010;18:1244–60.
Badtke MP, Hammer ND, Chapman MR. Functional amyloids signal their arrival. Sci Signal. 2009;2:pe43.
Pham CLL, Kwan AH, Sunde M. Functional amyloid: widespread in Nature, diverse in purpose. Essays Biochem. 2014;56:207–19.
Deshmukh M, Evans ML, Chapman MR. Amyloid by design: intrinsic regulation of microbial amyloid assembly. J Mol Biol. 2018;430:3631–41.
Loquet A, Saupe SJ. Diversity of amyloid motifs in NLR signaling in fungi. Biomolecules. 2017;7:38.
Balguerie A, Dos Reis S, Ritter C, Chaignepain S, Coulary-Salin B, Forge V, et al. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 2003;22:2071–81.
Greenwald J, Buhtz C, Ritter C, Kwiatkowski W, Choe S, Maddelein M-L, et al. The mechanism of prion inhibition by HET-S. Mol Cell. 2010;38:889–99.
Seuring C, Greenwald J, Wasmer C, Wepf R, Saupe SJ, Meier BH, et al. The mechanism of toxicity in HET-S/HET-s prion incompatibility. PLoS Biol. 2012;10:e1001451.
Riek R, Saupe SJ. The HET-S/s Prion Motif in the Control of Programmed Cell Death. Cold Spring Harb Perspect Biol. 2016;8:a023515.
Wan W, Stubbs G. Fungal prion HET-s as a model for structural complexity and self-propagation in prions. Proc Natl Acad Sci USA. 2014;111:5201–6.
Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–6.
Wu X, Ma Y, Zhao K, Zhang J, Sun Y, Li Y, et al. The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3. Proc Natl Acad Sci USA. 2021;118:e2022933118.
Kajava AV, Klopffleisch K, Chen S, Hofmann K. Evolutionary link between metazoan RHIM motif and prion-forming domain of fungal heterokaryon incompatibility factor HET-s/HET-s. Sci Rep. 2014;4:7436.
Bardin T, Daskalov A, Barrouilhet S, Granger-Farbos A, Salin B, Blancard C, et al. Partial Prion Cross-Seeding between Fungal and Mammalian Amyloid Signaling Motifs. MBio. 2021;12.
Dyrka W, Coustou V, Daskalov A, Lends A, Bardin T, Berbon M, et al. Identification of NLR-associated Amyloid Signaling Motifs in Bacterial Genomes. J Mol Biol. 2020;432:6005–27.
Daskalov A, Dyrka W, Saupe SJ. Theme and variations: evolutionary diversification of the HET-s functional amyloid motif. Sci Rep. 2015;5:12494.
Daskalov A, Martinez D, Coustou V, El Mammeri N, Berbon M, Andreas LB, et al. Structural and molecular basis of cross-seeding barriers in amyloids. Proc Natl Acad Sci USA. 2021;118:e2014085118.
Linkermann A, Kunzendorf U, Krautwald S. Phosphorylated MLKL causes plasma membrane rupture. Mol Cell Oncol. 2014;1:e29915.
Sun L, Wang H, Wang Z, He S, Chen S, Liao D, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–27.
Murphy JM. The Killer Pseudokinase Mixed Lineage Kinase Domain-Like Protein (MLKL). Cold Spring Harb Perspect Biol. 2020;12:a036376.
Hildebrand JM, Tanzer MC, Lucet IS, Young SN, Spall SK, Sharma P, et al. Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc Natl Acad Sci USA. 2014;111:15072–7.
Rodriguez DA, Weinlich R, Brown S, Guy C, Fitzgerald P, Dillon CP, et al. Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ. 2016;23:76–88.
Huang D, Zheng X, Wang ZA, Chen X, He WT, Zhang Y, et al. The MLKL channel in necroptosis is an octamer formed by tetramers in a dyadic process. Mol Cell Biol. 2017;37:e00497-16.
Samson AL, Zhang Y, Geoghegan ND, Gavin XJ, Davies KA, Mlodzianoski MJ, et al. MLKL trafficking and accumulation at the plasma membrane control the kinetics and threshold for necroptosis. Nat Commun. 2020;11:3151.
Zhang Y, Liu J, Yu D, Zhu X, Liu X, Liao J, et al. The MLKL kinase-like domain dimerization is an indispensable step of mammalian MLKL activation in necroptosis signaling. Cell Death Dis. 2021;12:638.
Tamborski J, Krasileva KV. Evolution of plant nlrs: from natural history to precise modifications. Annu Rev Plant Biol. 2020;71:355–78.
Burdett H, Bentham AR, Williams SJ, Dodds PN, Anderson PA, Banfield MJ, et al. The Plant “Resistosome”: Structural Insights into Immune Signaling. Cell Host Microbe. 2019;26:193–201.
Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, et al. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science. 2019;364:eaav5870.
Bi G, Su M, Li N, Liang Y, Dang S, Xu J, et al. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell. 2021;184:3528–3541.e12.
Mahdi LK, Huang M, Zhang X, Nakano RT, Kopp LB, Saur IML, et al. Discovery of a family of mixed lineage kinase domain-like proteins in plants and their role in innate immune signaling. Cell Host Microbe. 2020;28:813–824.e6.
Daskalov A, Gladieux P, Heller J, Glass NL. Programmed cell death in Neurospora crassa is controlled by the allorecognition Determinant rcd-1. Genetics. 2019;213:1387–400. Oct 21
Daskalov A, Mitchell PS, Sandstrom A, Vance RE, Glass NL. Molecular characterization of a fungal gasdermin-like protein. Proc Natl Acad Sci. 2020;117:18600–07.
Liu Z, Wang C, Lin C. Pyroptosis as a double-edged sword: The pathogenic and therapeutic roles in inflammatory diseases and cancers. Life Sci. 2023;318:121498.
Shi J, Gao W, Shao F. Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends Biochem Sci. 2017;42:245–54.
Wang C, Ruan J. Mechanistic Insights into Gasdermin Pore Formation and Regulation in Pyroptosis. J Mol Biol. 2022;434:167297.
Ramos-Junior ES, Morandini AC. Gasdermin: A new player to the inflammasome game. Biomed J. 2017;40:313–6.
Zahid A, Ismail H, Jin T. Molecular and structural aspects of gasdermin family pores and insights into gasdermin-elicited programmed cell death. Biochem Soc Trans. 2021;49:2697–710.
Zou J, Zheng Y, Huang Y, Tang D, Kang R, Chen R. The versatile gasdermin family: their function and roles in diseases. Front Immunol. 2021;12:751533.
Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–5.
Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–8.
Sborgi L, Rühl S, Mulvihill E, Pipercevic J, Heilig R, Stahlberg H, et al. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 2016;35:1766–78.
Wang K, Sun Q, Zhong X, Zeng M, Zeng H, Shi X, et al. Structural mechanism for GSDMD targeting by autoprocessed caspases in pyroptosis. Cell. 2020;180:941–955.e20.
Zheng Q, Daskalov A. Microbial gasdermins: More than a billion years of pyroptotic-like cell death. Seminars Immunol. 2023;69:101813.
Clavé C, Dyrka W, Turcotte EA, Granger-Farbos A, Ibarlosa L, Pinson B, et al. Fungal gasdermin-like proteins are controlled by proteolytic cleavage. Proc Natl Acad Sci USA. 2022;119:e2109418119.
Johnson AG, Wein T, Mayer ML, Duncan-Lowey B, Yirmiya E, Oppenheimer-Shaanan Y, et al. Bacterial gasdermins reveal an ancient mechanism of cell death. Science. 2022;375:221–5.
Ruan J, Xia S, Liu X, Lieberman J, Wu H. Cryo-EM structure of the gasdermin A3 membrane pore. Nature. 2018;557:62–7.
Li Y, Hou Y, Sun Q, Zeng H, Meng F, Tian X, et al. Cleavage-independent activation of ancient eukaryotic gasdermins and structural mechanisms. Science. 2024;eadm9190.
Ren K, Farrell JD, Li Y, Guo X, Xie R, Liu X, et al. Mechanisms of RCD-1 pore formation and membrane bending. Nat Commun. 2025;16:1011.
Aravind L, Koonin EV. Classification of the caspase-hemoglobinase fold: detection of new families and implications for the origin of the eukaryotic separins. Proteins. 2002;46:355–67.
Cui N, Zhang J-T, Li Z, Liu X-Y, Wang C, Huang H, et al. Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase. Nat Commun. 2022;13:7549.
Wein T, Millman A, Lange K, Yirmiya E, Hadary R, Garb J, et al. CARD domains mediate anti-phage defence in bacterial gasdermin systems. Nature. 2025;639:727–34.
Jia A, Huang S, Song W, Wang J, Meng Y, Sun Y, et al. TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity. Science. 2022;377:eabq8180.
Ofir G, Herbst E, Baroz M, Cohen D, Millman A, Doron S, et al. Antiviral activity of bacterial TIR domains via immune signalling molecules. Nature. 2021;600:116–20.
Essuman K, Summers DW, Sasaki Y, Mao X, Yim AKY, DiAntonio A, et al. TIR Domain Proteins Are an Ancient Family of NAD + -Consuming Enzymes. Curr Biol. 2018;28:421–430.e4.
Horsefield S, Burdett H, Zhang X, Manik MK, Shi Y, Chen J, et al. NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science. 2019;365:793–9.
Rousset F, Yirmiya E, Nesher S, Brandis A, Mehlman T, Itkin M, et al. A conserved family of immune effectors cleaves cellular ATP upon viral infection. Cell. 2023;186:3619–3631.e13.
Lastovetsky OA, Krasnovsky LD, Qin X, Gaspar ML, Gryganskyi AP, Huntemann M, et al. Molecular dialogues between early divergent fungi and bacteria in an antagonism versus a mutualism. MBio. 2020;11:e02088-20.
Makarova KS, Wolf YI, Snir S, Koonin EV. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J Bacteriol. 2011;193:6039–56.
La SR, Ndhlovu A, Durand PM. The ancient origins of death domains support the “original sin” hypothesis for the evolution of programmed cell death. J Mol Evol. 2022;90:95–113.
Rousset F. Innate immunity: the bacterial connection. Trends Immunol. 2023;44:945–53.
Iranzo J, Lobkovsky AE, Wolf YI, Koonin EV. Virus-host arms race at the joint origin of multicellularity and programmed cell death. Cell Cycle. 2014;13:3083–8.
Durand PM, Barreto Filho MM, Michod RE. Cell death in evolutionary transitions in individuality. Yale J Biol Med. 2019;92:651–62. Dec 20
Pradeu T. Immunology and individuality. eLife. 2019;8:e47384.
Eberl G, Pradeu T. Towards a general theory of immunity? Trends Immunol. 2018;39:261–3. Apr
Kulkarni M, Stolp ZD, Hardwick JM. Targeting intrinsic cell death pathways to control fungal pathogens. Biochem Pharmacol. 2019;162:71–8.
Case NT, Berman J, Blehert DS, Cramer RA, Cuomo C, Currie CR, et al. The future of fungi: threats and opportunities. G3. 2022;12:jkac224.
Acknowledgements
The author wishes to thank everyone who provided critical feedback during the writing of this article.
Funding
The author wishes to acknowledge funding from CNRS ATIP-Avenir (France).
Author information
Authors and Affiliations
Contributions
AD conceptualized and wrote the manuscript, designed and created figures.
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Daskalov, A. Regulated cell death in fungi from a comparative immunology perspective. Cell Death Differ (2025). https://doi.org/10.1038/s41418-025-01570-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41418-025-01570-z