Abstract
It is still challenging to predict the efficacy of cisplatin-based therapy, particularly in relation to the activation of macroautophagy/autophagy in oral squamous cell carcinoma (OSCC). We studied the effect of selected chromatin remodeling genes on the cisplatin resistance and their interplay with autophagy in 3-dimensional tumor model and xenografts. We analyzed gene expression patterns in the cisplatin-sensitive UMSCC1, and a paired cisplatin-resistant UM-Cis cells. Many histone protein gene clusters involved in nucleosome assembly showed significant difference of expression. Gain- and loss-of-function analyses revealed an inverse correlation between cisplatin resistance and HIST1H3D expression, while a positive correlation was observed with HIST3H2A or HIST3H2B expression. In UM-Cis, HIST3H2A- and HIST3H2B-mediated chromatin remodeling upregulates autophagy status, which results in cisplatin resistance. Additionally, knockdown of HIST3H2A or HIST3H2B downregulated autophagy-activating genes via chromatin compaction of their promoter regions. MiTF, one of the key autophagy regulators upregulated in UM-Cis, negatively regulated transcription of HIST1H3D, suggesting an interplay between chromatin remodeling-dependent cisplatin resistance and autophagy. On comparing the staining intensity between cisplatin-sensitive and –insensitive tissues from OSCC patients, protein expression pattern of the selected histone protein genes were matched with the in vitro data. By examining the relationship between autophagy and chromatin remodeling genes, we identified a set of candidate genes with potential use as markers predicting chemoresistance in OSCC biopsy samples.
Similar content being viewed by others
Introduction
Oral squamous cell carcinoma (OSCC) is the most common type of head and neck squamous cell carcinoma (HNSCC). Despite recent advances in OSCC treatments, the 5-year survival rate remains between 50–60% without substantial improvement [1]. The demand for chemoradiation has increased due to the aging population and longer life expectancy. However, its application is constrained by the presence of complications or toxicity [2, 3]. One of the main reasons for the limited effectiveness of chemotherapy is drug resistance, which contributes significantly to cancer recurrence and is a leading cause of death in cancer patients [4]. Platinum-based chemotherapy has been used to treat many solid tumors [5,6,7,8]. Cisplatin, one of the most common platinum compounds, interacts with DNA to form crosslinked DNA adducts, which trigger a series of intracellular events culminating in cell death [9, 10]. Cisplatin alone or in combination with other compounds is used in chemotherapy regimens, including first-line treatments of several cancer types, in approximately 50% of cancer patients [11]. However, the effectiveness of cisplatin is often hindered by the development of drug resistance in treating tumors, including OSCC [11]. To understand the mechanisms behind this resistance, researchers often generate cisplatin-resistant cell lines from parental cancer cells and compare their gene expression profiles.
One of the mechanisms by which tumor cells develop cisplatin resistance is through cytoprotective macroautophagy/autophagy. Autophagy can be upregulated in response to a range of stresses, including chemotherapy. Although autophagy is considered a protective mechanism for cisplatin-resistant phenotypes, the effect of autophagy on chemotherapy is context-dependent based on the tumor microenvironment [12,13,14]. The regulation of autophagy by specific histone proteins in tumor cells’ cisplatin resistances remains poorly understood. Together with transcription factors, chromatin remodeling due to fluctuations of histone proteins fine-tunes the transcriptional regulation of gene expression by making gene loci accessible to or hidden from the transcription machinery ultimately determining cell fate [15, 16]. Therefore, it is necessary to evaluate whether the perturbation of histone proteins leads to changes in biological pathways involved in cisplatin resistance in cancer cells.
We established a cisplatin-resistant cell line from the UMSCC1, one of the representative oral squamous cancer cells, followed by DNA microarray analysis. Significant changes in expression were observed in numerous genes related to nucleosome assembly. While extensive studies have explored drug responses in vitro, translating those findings to a clinical context is challenging [17]. We validated cisplatin efficacy in OSCC patients, categorizing them into two groups: cisplatin-sensitive and -insensitive. We then compared the expression profiles of candidate genes in tissue samples from these two groups. We specifically examined how changes in core histone proteins influenced chromatin assembly and induced autophagy in OSCC cells, in the context of cisplatin resistance. Our overall goal was to provide additional input that can be utilized in strategies to improve cisplatin efficacy in patients with OSCC.
Materials and methods
Two-dimensional (2D) and three-dimensional (3D) cell culture
OSCC cells were cultured in DMEM containing 10% FBS and 1% penicillin-streptomycin solution, at 37 °C in a 5% carbon dioxide (CO2) humidified atmosphere. The cell lines were tested for contamination every two months using the BioMycoX® Mycoplasma PCR detection kit (CellSafe, D-100). For 3D spheroid formation, cells were seeded into a 96-well U-bottom ultra-low attachment plate (Corning Incorporated, 7007; 4000 cells/well) and cultured for 2–3 days to form spheroids with uniform sizes in each well ( > 300 μm in diameter). The spheroid size was determined by measuring the surface area of each group containing 6–8 spheroids using a Cell3iMager scanner CC-5000 (SCREEN Holdings Co., Ltd. Kyoto, Japan). Each spheroid’s surface area was the same at the beginning of the experiments, within the 5% error range.
Organoid culture
Organoid culture was performed using mouse xenografts derived from UMSCC1, UM-Cis, or FaDu cells. Tongue tissue from patients with OSCC was also used for organoid culture. Tissue processing and organoid culture were performed as described by Driehuis et al. [18, 19]. More detailed information is provided in the Supplementary methods.
Mouse xenograft model
We compared cisplatin efficacy in xenograft mice (6-week-old male BALB/c; Hyochang Science, Daegu, Korea). Cancer cells (2 × 106/100 µL DMEM) from each cell line were injected subcutaneously into the right and left dorsal sides of mice. After 27 days, tumor formation was observed, and cisplatin was injected for 4 weeks. Detailed information is presented in the Supplementary methods.
Chromatin accessibility assay
To evaluate nucleosome remodeling between UMSCC1 and UM-Cis cells, chromatin compaction was compared using the EpiQuik™ Chromatin Accessibility Assay Kit (Epigentek, P-1047-48). Changes in chromatin structure were identified by the degree of the Ct shift between nuclease-treated and untreated samples, following qPCR [20]. Heterochromatin-like compact DNA is not accessible to nucleases, resulting in only a slight shift of Ct between the digested sample and the undigested control, whereas decondensed DNA is accessible to nucleases, which is typically reflected by a larger Ct shift between experimental and control samples. Detailed information is presented in the Supplementary methods.
Ethics approval and consent to participate
Human tissue specimens were used after receiving written informed consent from the patients, with approval from the Institutional Research Ethics Committee of Kyungpook National University Hospital (KNUH201704011) and adherence to the principles of the Declaration of Helsinki. All experimental protocols with mice followed the ARRIVE guidelines (Animal Research: Reporting of In Vivo Experiments) and were approved by the Animal Ethics Committee of Kyungpook National University (2017-94-2).
Results
Characterization of cisplatin-resistant UM-Cis cells
The proliferation rate of UM-Cis was decreased compared to that of UMSCC1 (Fig. S1A). Incubation with cisplatin resulted in significantly increased viability or decreased apoptosis of UM-Cis cells relative to UMSCC1 (Fig. S1B). In 3D spheroids, the size was significantly decreased with cisplatin treatment relative to a vehicle control in UMSCC1. However, no similarly significant decrease in spheroid size was observed in UM-Cis (Fig. S1C). Mouse xenografts derived from UM-Cis displayed significant resistance to cisplatin compared to UMSCC1-derived tumors (Fig. S1D). Representative hematoxylin and eosin (H&E) staining images of tumor tissues are shown in Fig. S1E. Both tumor tissues showed strong reactivity with the KRT13 antibody, confirming their squamous epithelial origin.
DNA microarray analysis and gene ontology enrichment
Following quantile normalization of raw DNA microarray data, the expression profile of UMSCC1 and UM-Cis cells was obtained and submitted to the Gene Expression Omnibus repository (GSE197561). A total of 797 differentially expressed genes (526 upregulated and 271 downregulated) were obtained between the two groups, using an average fold change of ≥ 2-fold and a p-value < 0.05 as our selection criteria. Shown in Fig. S2A is the heatmap using the z-score normalized data for genes with significant difference in expression > 4-fold. The 797 gene list was uploaded to the online software DAVID for biological process and cellular compartment analyses. According to biological process enrichment, nucleosome assembly was the most significant functional annotation term (p < 0.0001; gene count, 16; Fig. S2B). Telomere organization, which is closely related to nucleosome assembly, was the third most important pathway in biological processes (p < 0.0001; gene count, 7; Fig. S2B). In addition, GO cellular compartment enrichment revealed terms related to the nucleosome to be the most significantly different (Fig. S2C). As the role of nucleosome assembly in the mechanism of cisplatin resistance is not well understood, we focused on examining this biological process further within the context of OSCC.
Differential expression of nucleosome assembly-related genes between UMSCC1 and UM-Cis cells
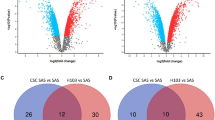
We identified several nucleosome assembly-related genes with >2-fold significant difference in expression (Fig. 1A). Of these, 14 genes were downregulated, while 28 genes were upregulated in UM-Cis. The mRNA and protein expression patterns of four genes: H3 clustered histone-4 (H3C4, also known as HIST1H3D), H2A clustered histone-25 (H2AC25, also known as HIST3H2A), H2B clustered histone-26 (H2BC26, also known as HIST3H2B and HIST3H2BB), and nucleosome assembly protein 1-like-2 (NAP1L2) were consistent with the microarray data (red and blue asterisks represent increased or decreased expression in UM-Cis, respectively; Fig. 1B, C).
A Heatmap for nucleosome assembly-related genes in the two groups with row z-score normalization (Fold change > 2, p < 0.05). The asterisks indicate representative genes consistent with the qPCR analysis in UMSCC1 and UM-Cis cells (Red asterisks denote increased expression in UM-Cis cells; blue asterisks denote decreased expression in UM-Cis cells). B The mRNA expression level of the nucleosome assembly-related genes. C Protein expression at the same samples. Results represent the mean ± standard deviation of three independent experiments (*p < 0.05, **p < 0.01, ***p < 0.001).
Effect of candidate gene knockdown on cisplatin efficacy in OSCC spheroids
siHIST1H3D pretreatment in UMSCC1 and FaDu spheroids led to an observable increase in cisplatin tolerance (Fig. S3A), and reduced HIST1H3D mRNA levels in both spheroids for 14 days (Fig. S3B). On the contrary, the knockdown of HIST3H2A or HIST3H2B was associated with a concomitant reduction in cisplatin resistance for UM-Cis spheroids (Fig. S3C). The knockdown effect of the siRNA was sustained for 14 days in both cisplatin or vehicle control-treated groups (Fig. S3D). The effect of these siRNAs on cisplatin efficacy were more pronounced in spheroids than in 2D cell cultures.
Inverse correlation of cisplatin resistance and HIST1H3D expression level in organoids and xenografts
We confirmed the organoids’ tumor identity using immunostaining against KRT13 (Fig. 2A). The siHIST1H3D-transfected UMSCC1 organoids displayed increased tolerance to cisplatin compared to the control siRNA (Fig. 2B). siHIST1H3D-mediated gene knockdown effectively persisted for up to seven days in all cislpatin- or vehicle-treated organoids (Fig. 2C). Mouse xenografts derived from siHIST1H3D-transfected UMSCC1 spheroids showed signifcant resistance to cisplatin (Fig. 2D). Anti-KRT13 immunostaining in xenografts is shown in Fig. 2E. The knockdown effect of HIST1H3D in xenografts was maintained in the extracted tumor tissues (Fig. 2F). We repeated this experiment with organoids cultured from tongue OSCC tissues of patients and FaDu cell-derived xenografts. These organoids showed strong signal with anti-KRT13 antibody (Fig. S4A). Pretreatment with siHIST1H3D and cisplatin for seven days led to a significant increase in drug tolerance in both organoids (Fig. S4B), similar to UMSCC1 observations. The knockdown effect of the siRNA was sustained for seven days in both cisplatin or vehicle control-treated groups (Fig. S4C, D).
A IF staining in fixed organoids using an anti-KTR13 antibody, a representative squamous epithelial marker. B Organoids pretreated with siHIST1H3D or control siRNA, followed by cisplatin treatment for seven days. C siHIST1H3D efficiency in organoids, evaluated by IF staining and qPCR. Results represent the mean ± standard deviation of three independent experiments (*p < 0.05, **p < 0.01). D Cisplatin efficacy in mice xenografts derived from UMSCC1 spheroids transfected with siHIST1H3D or control siRNA. Tumor volume was measured till sacrifice (***p < 0.0005). E IF staining of mice tumor tissues with anti-KRT13 antibody. F Protein and mRNA expression of HIST1H3D in xenografts showing the efficiency of siHIST1H3D in xenografts (**p < 0.005).
In UM-Cis spheroids, HIST1H3D-overexpression vector efficiency was maintained for 14 days (Fig. 3B), during which, cisplatin treatment resulted in a significant decrease of spheroid size, in comparison to the control vector group (Fig. 3A). Similarly, mouse xenografts developed from HIST1H3D-overexpressing UM-Cis spheroids markedly decreased in size compared to control vector group with cisplatin treatment (Fig. 3C). Anti-KRT13 immunostaining in xenografts is shown in Fig. 3D. The overexpression effect of HIST1H3D in xenografts was maintained in the extracted tumor tissues in both cisplatin or vehicle control-treated groups (Fig. 3E). These results suggest that the mechanism of cisplatin resistance is sensitive to fluctuations in HIST1H3D activity.
A UM-Cis spheroids transfected with HIST1H3D-overexpression vector or control vector, followed by cisplatin treatment for 14 days. B mRNA expression level in spheroids transfected with HIST1H3D-overexpression vector for 14 days. Results represent the mean ± standard deviation of three experiments (*p < 0.05, **p < 0.005). C Cisplatin efficacy in mice xenografts derived from UM-Cis spheroids transfected with HIST1H3D-overexpression vector or control vector. Tumor volume was measured till sacrifice. D IF staining of mice tumor tissues with anti-KRT13 antibody. E Protein and mRNA expression of HIST1H3D in xenografts showing the efficiency of HIST1H3D-overexpression vector (*p < 0.05).
Correlation between protein expression of candidate genes and cisplatin sensitivity in clinical OSCC tissues
We investigated the relationship between HIST1H3D, HIST3H2A, and HIST3H2B protein expression and cisplatin sensitivity in patients with OSCC. Patients were classified as either sensitive or insensitive to cisplatin monotherapy based on changes in tumor size and survival. Figure 4A displays staining results from six tissue samples in each group. On comparing the staining intensity, tissues from cisplatin-sensitive patients exhibited a robust signal for HIST1H3D, whereas tissues from cisplatin-resistant patients displayed a comparatively weaker response (Fig. 4B). Conversely, the expression patterns of HIST3H2A and HIST3H2B were opposite, with higher levels in the cisplatin-resistant group (Fig. 4C, D). These results affirm the positive correlation between HIST1H3D expression, and the negative correlation between HIST3H2A and HIST3H2B expression and cisplatin sensitivity in OSCC tissues, supporting our findings.
A, C Tissues from patients with OSCC showing cisplatin-sensitive and cisplatin-insensitive tissues were stained with anti-HIST1H3D, anti-HIST3H2A, and anti-HIST3H2B antibodies. B, D The level of protein expression on each specimen was scored as 0, 1, 2, and 3 (0 = negative, 1 = weak, 2 = intermediate, and 3 = strong) according to its staining intensity. **p < 0.01.
Kaplan–Meier analysis revealed that high HIST1H3D expression was strongly associated with longer patient survival, while higher levels of HIST3H2A or HIST3H2B were linked to shorter survival (Fig. S5A). Our results regarding the mRNA expression of the three candidate genes in patient-derived tissues might be indirectly associated with post-treatment survival in OSCC patients. Disease-free survival analysis revealed no significant difference in the survival rates between the groups with high and low expression of the candidate genes (Fig. S5B).
HIST1H3D is a target of transcriptional downregulation by MiTF
Our search for candidate transcription factors regulating HIST1H3D identified microphthalmia-associated transcription factor (MiTF) as a key regulator. We induced siRNA-mediated knockdown or vector-assisted overexpression of MiTF in UM-Cis, and UMSCC1/FaDu cells, respectively, followed by HIST1H3D protein expression analysis. In both cases, MiTF protein expression was consistently inversely related to HIST1H3D level, suggesting that MiTF might negatively regulate HIST1H3D (Figs. 5A, S6A). We investigated chromatin immunoprecipitation (ChIP) using UM-Cis cells expressing high levels of enodgenous MiTF. A schematic of the promoter region containing the consensus MiTF-binding sites is shown in Fig. 5B. The MiTF-bound HIST1H3D promoter sequences increased significantly compared to IgG control (Fig. 5C). Following luciferase reporter assay in UMSCC1 cells transfected with an MiTF-overexpression vector, luciferase activity from the HIST1H3D promoter region significantly decreased under MiTF overexpression (Fig. 5D). We conducted an analysis of the TCGA dataset using cBioPortal to validate the relationship between HIST1H3D and MiTF expression in tissues from HNSCC patients. As depicted in Fig. 5E, there was an inverse correlation between MiTF mRNA levels and HIST1H3D expression, suggesting that MiTF would function as a negative regulator of HIST1H3D. Together our findings suggest that MiTF could bind directly to the HIST1H3D promoter and negatively regulate the expression of HIST1H3D.
A Protein expression in cells following exogenous overexpression or knockdown of MiTF in UMSCC1 or UM-Cis cells, respectively. B Schematic representation of the promoter regions of the HIST1H3D gene showing the consensus MiTF binding sites for ChIP analysis (black arrowheads in 1 and 2). C Gel electrophoresis image of PCR products from Chromatin IP using a MiTF antibody. Fold enrichment of DNA sequences that are associated with the target protein by qPCR. D Dual luciferase activity shows the direct binding of the 3′ regions of HIST1H3D gene and MiTF protein in UMSCC1 cells. E MiTF mRNA level in relation to HIST1H3D expression levels in HNSCC patient tissues. The data were obtained using the TCGA dataset through cBioportal. F UMSCC1 spheroids transfected with MiTF-overexpression vector, followed by cisplatin treatment. G MiTF-overexpression vector efficiency in spheroids after 14 days. H UMSCC1 organoids transfected with MiTF-overexpression vector, followed by cisplatin treatment. I mRNA and protein expression of MiTF in organoids showing the efficiency of MiTF-overexpression vector. J UM-Cis spheroids pretreated with siMiTF, followed by cisplatin treatment. K MiTF overexpression efficiency in spheroids after 14 days. Results represent the mean ± standard deviation of three independent experiments. (*p < 0.05, **p < 0.01, ***p < 0.005).
Regarding cisplatin resistance, MiTF overexpression in spheroids and organoids derived from UMSCC1 (Fig. 5F, H) and FaDu (Fig. S6B, D) significantly increasd the tolerance of cisplatin. Conversely, siMiTF pretreatment in UM-Cis spheroids reduced cisplatin resistance (Fig. 5J). Knockdown or overexpression efficiency of MiTF is shown in Fig. 5G, I and K; Fig. S6C, E.
Interplay between histone protein expression and autophagy
As MiTF is a key regulator of autophagy induction, we explored the potential interplay between autophagy and histone proteins in the context of cisplatin resistance. A heatmap of autophagy-associated genes with > 2-fold significant difference is shown in Fig. 6A. Several genes involved in autophagy initiation, such as autophagy-related (ATG) were upregulated in UM-Cis. Protein levels of autophagy markers were higher in UM-Cis (Fig. 6B). UM-Cis displayed a pronounced distribution of cytoplasmic microtubule-associated proteins 1 A/1B light chain 3B (MAP1LC3B; abbreviated to LC3B) in punctate formations (Fig. 6C). We treated chloroquine (CQ), an autophagic flux inhibitor in both cells. Lipidated LC3B (LC3B-II) was increased in both CQ-treated cells. However, LC3B-II levels in UM-Cis were consistently higher than UMSCC1 in both baseline and CQ-treated conditions (Fig. 6D). Furthermore, LC3B-lysosome co-localization, a key marker of autophagy activation, was significantly higher in UM-Cis cells compared to UMSCC1 (Fig. 6E). Knockdown of HIST3H2A or HIST3H2B in UM-Cis resulted in downregulated MiTF mRNA with a consequent increase in HIST1H3D mRNA (Fig. 6F). This knockdown also led to lower levels of ATG13, LC3B, and MiTF proteins, while increasing HIST1H3D expression (Fig. 6G). The significant decrease of LC3B puncta levels under these conditions was also recorded in UM-Cis (Fig. 6H). These findings suggest that alteration of histone levels regulated autophagy-associated gene expression and subsequent autophagic activity in UM-Cis cells.
A Heatmap of genes involved in autophagy from DNA microarray dataset. B Autophagy-related proteins expression in UMSCC1 and UM-Cis cells. C LC3B puncta in both cells. D Comparison of autophagy flux under CQ treatment by LC3B protein expression level. E Representative images of co-localization of LC3B with lysosome using LysoTrackerTM Deep Red. F, G mRNA and protein expression in UM-Cis cells transfected with siHIST3H2A or siHIST3H2B. H LC3B puncta in UM-Cis cells transfected with siHIST3H2A or siHIST3H2B. Results represent the mean ± standard deviation of three independent experiments. (*p < 0.05, **p < 0.01).
In UM-Cis, pretreatment with both CQ or a lysosomal inhibitor, bafilomycin A1 (BF-A1), resulted in comparable decreases in cisplatin resistance (Fig. 7A), supporting evidence for a link between autophagy activation and cisplatin resistance. LIVE/DEAD cell viability assay also showed that pretreatment with autophagy inhibitors reduced cisplatin resistance and increased cell death (Fig. 7B).
A UM-Cis cell viability pretreated with autophagy inhibitors such as CQ or BF-A1, followed by cisplatin treatment. B LIVE/DEAD staining of UM-Cis cells pretreated with autophagy inhibitors. Dead cells were counted in each condition. Results represent the mean ± standard deviation of three experiments (**p < 0.01).
Effects of histone proteins and autophagy on cisplatin resistance in YD-38/CIS cell line
Additional experiments were conducted with YD-38/CIS, a cisplatin-resistant cells derived from the parental OSCC cell line YD-38 (Fig. S7A). The expression patterns of HIST1H3D, HIST3H2A, and HIST3H2B in the two cell lines were consistent with those observed in UMSCC1 and UM-Cis (Fig. S7B). Moreover, pretreatment with either the HIST1H3D-overexpression vector or siHIST3H2A + siHIST3H2B in YD-38/CIS led to a reduction in cisplatin resistance (Fig. S7C). Additionally, autophagy was more activated in YD-38/CIS compared to YD-38 (Fig. S7D), and pretreatment with autophagy inhibitors resulted in reduced cisplatin resistance and increased cell death (Fig. S7E, F). These findings collectively suggest that HIST1H3D, HIST3H2A, and HIST3H2B genes might be involved in cisplatin resistance of YD-38/CIS along with autophagy.
Chromatin structure modification
To further evaluate how protein level fluctuations of specific histones affect autophagy status, we analyzed the accessibility of chromatin due to histone-dependent chromatin remodeling in the UMSCC1 and UM-Cis. As shown in Fig. 8A, UM-Cis exhibited a lower Ct shift with internal control primers between nuclease-digested and undigested samples, suggesting more condensed chromatin structures. Next, the promoter region of HIST1H3D was significantly more condensed in UM-Cis than in UMSCC1 (Fig. 8B). In contrast, the promoters of HIST3H2A and HIST3H2B were less compact in UM-Cis, supporting the differential expression of these genes between the two cell lines. We examined the accessibility of the selected promoter regions of autophagy-facilitating genes for transcription. All tested gene promoter regions were less condensed in UM-Cis than in UMSCC1 (Fig. 8C), which may partially explain the increased expression of autophagy-promoting genes in UM-Cis. We repeated chromatin accessibility assay following treatment of UM-Cis with siHIST3H2A or siHIST3H2B. In the siRNA-transfected cells, the promoter regions of autophagy-promoting genes became significantly more condensed compared to control cells (Fig. 8D), indicating a possible downregulation of these genes following the knockdown of either HIST3H2A or HIST3H2B. Altogether, these results demonstrate that histone proteins may regulate autophagy-modulating genes at the transcriptional level, in the context of cisplatin resistance (Fig. 8E).
A, B qPCR data showing the chromatin accessibility of specific genes’ region in UMSCC1 and UM-Cis cells. The lower Ct shift suggests more condensed chromatin conformations. C qPCR data showing the chromatin accessibility of autophagy-associated promoter regions. D qPCR data showing the chromatin accessibility of autophagy-associated promoter regions after transfection of UM-Cis cells with siHIST3H2A or siHIST3H2B. Results represent the mean ± standard deviation of three independent experiments (*p < 0.05, **p < 0.01, ***p < 0.005). E Schematic representation of the molecular mechanism exerting the cisplatin resistance in UM-Cis cells via the interplay between histone proteins and autophagy.
Discussion
Numerous attempts are aimed at providing more effective chemotherapeutic regimens for cancer patients [21]. Previous data showed that several genes are associated with cisplatin resistance in OSCC cell-based experiments [22,23,24,25,26]. Nevertheless, there exists a substantial gap in correlating these gene expression patterns with drug response to predict chemotherapeutic efficacy in patients. Moreover, autophagy activation, a recognized mechanism contributing to cisplatin resistance in cancer, lacks comprehensive understanding within the context of OSCC. Our aim is to identify the mechanism of cisplatin resistance in OSCC and subsequently decipher the expression patterns of genes that can predict cisplatin efficacy in biopsy samples. This approach holds the potential to reduce unnecessary patient discomfort associated with drug side effects.
Chromatin conformation, in which histones play an important role, is essential for mediating various transcriptional responses that affect disease status, including cancer [27,28,29,30,31,32,33,34]. Kamur et al. suggest that understanding eukaryotic genomes requires looking beyond the 3D structure of chromatin fibers. They highlight the importance of examining 4D genomics, which considers how gene expression is regulated in response to environmental factors such as developmental stages, growth conditions, and diseases [35]. Alterations in chromatin assembly lead to perturbations in the expression of genes involved in intracellular functions such as autophagy, a primary pathway for cisplatin resistance [36,37,38]. Furthermore, the nucleus is a major regulator of autophagy through transcriptional and epigenetic modifications [39, 40]. Therefore, the nucleosome assembly status is likely involved in regulating drug resistance in cancer. In this study, many genes involved in nucleosome assembly exhibited significantly different expression patterns between cisplatin-sensitive and –insensitive cells. As the mechanisms of cisplatin resistance in the context of nucleosome assembly remain uncharacterized, we focused on further investigating this biological process.
Chromatin can exist in more compact (inaccessible) or relaxed (accessible) conformations, depending on the function being supported in the genome, either locally or globally. The H2A/H2B dimers contribute to nucleosome compaction, which results in decreased gene activity [41, 42]. Cisplatin-resistant cancer cells show significant changes in their transcription profile, as a result of epigenetic alterations including histone remodeling [37]. However, the link between changes in histone protein transcription levels and drug resistance in cancer is not well understood. In UM-Cis, HIST3H2A and HIST3H2B were upregulated, leading to a greater degree of global chromatin compaction compared to UMSCC1. However, the promoter regions of several autophagy-activating genes showed more relaxed chromatin structures, likely resulting in higher expression. Furthermore, knockdown of either HIST3H2A or HIST3H2B resulted in the promoter regions becoming more condensed. Collectively, the chromatin state in UM-Cis was altered by the changes in histone protein expression, and the promoter regions of autophagy genes were locally decondensed. Consequently, autophagy was upregulated and contributed to the development of cisplatin resistance in OSCC.
Unlike most histone protein-coding genes, HIST1H3D expression was significantly reduced in UM-Cis due to negative transcriptional regulation by MiTF. Vyas et al. suggested that MiTF activation plays a significant role in developing cisplatin resistance [43]. Our data support a strong relationship between specific histone proteins expression level and autophagy activation. The ultimate goal was to verify the results by tracking chemotherapy outcomes in patients who donated tissues for the trial. Specifically, we examined HIST1H3D, HIST3H2A, and HIST3H2B protein expression in cisplatin-sensitive and insensitive OSCC tissues. However, due to the small number of patients receiving cisplatin monotherapy, each group included only six tissue samples. Despite this limitation, the expression patterns of these histone protein genes in patient tissues strongly supported our findings. Moving forward, it is crucial to expand our research through retrospective studies, incorporating a larger number of patient tissues. Future studies will delve into the molecular mechanisms responsible for autophagy activation by these histone proteins, and explore potential associations with other cisplatin resistance-related genes. These investigations are crucial for understanding the mechanisms underlying cisplatin resistance that depends on histone proteins.
In conclusion, we demonstrated that chromatin assembly changes may be related to cisplatin resistance via autophagy regulation in OSCC. We identified that the mRNA or protein expression of HIST3H2A, HIST3H2B, and HIST1H3D genes may be used as indicators to predict, to some degree, the cisplatin efficacy in patients with OSCC. This may in turn lead to the design of chemotherapeutic regimens with improved efficiency, based on a more personalized treatment plan according to the unique genetic landscape of each patient.
Data availability
The expression profile of UMSCC1 and UM-Cis was submitted to the Gene Expression Omnibus repository (GSE197561). The raw/processed data required to reproduce these findings are available with the corresponding author and can be given upon reasonable request.
References
Nicolatou-Galitis O, Psyrri A. Trismus and reduced quality of life in patients with oral squamous cell carcinoma who received postoperative radiotherapy alone or combined with chemotherapy. J Clin Oncol. 2017;35:29–36.
Fasano M, D’Onofrio I, Belfiore MP, Angrisani A, Caliendo V, Della Corte CM, et al. Head and neck squamous cell carcinoma in elderly patients: role of radiotherapy and chemotherapy. Cancers. 2022;14:472.
Teymoortash A, Ferlito A, Halmos GB. Treatment in elderly patients with head and neck cancer: a challenging dilemma. HNO. 2016;64:217–20.
Ramos A, Sadeghi S, Tabatabaeian H. Battling chemoresistance in cancer: root causes and strategies to uproot them. Int J Mol Sci. 2021;22:9451.
Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9–23.
Lokich J, Anderson N. Carboplatin versus cisplatin in solid tumors: an analysis of the literature. Ann Oncol. 1998;9:13–21.
Hack J, Crabb SJ. Platinum-based chemotherapy ‘rechallenge’ in advanced non-ovarian solid malignancies. Clin Oncol. 2022;34:e329–e344.
Zhang C, Xu C, Gao X, Yao Q. Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics. 2022;12:2115–32.
Cepeda V, Fuertes MA, Castilla J, Alonso C, Quevedo C, Perez JM. Biochemical mechanisms of cisplatin cytotoxicity. Anticancer Agents Med Chem. 2007;7:3–18.
Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–79.
Chen SH, Chang JY. New insights into mechanisms of cisplatin resistance: from tumor cell to microenvironment. Int J Mol Sci. 2019;20:4136.
Xu J, Gewirtz DA. Is autophagy always a barrier to cisplatin therapy? Biomolecules. 2022;12:463.
O’Donovan TR, O’Sullivan GC, McKenna SL. Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy. 2011;7:509–24.
Magnano S, Hannon Barroeta P, Duffy R, O’Sullivan J, Zisterer DM. Cisplatin induces autophagy-associated apoptosis in human oral squamous cell carcinoma (OSCC) mediated in part through reactive oxygen species. Toxicol Appl Pharmacol. 2021;427:115646.
Sproul D, Gilbert N, Bickmore WA. The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet. 2005;6:775–81.
Davis PK, Brackmann RK. Chromatin remodeling and cancer. Cancer Biol Ther. 2003;2:22–29.
Robert BM, Dakshinamoorthy M, Ganapathyagraharam Ramamoorthy B, Dhandapani M, Thangaiyan R, Muthusamy G, et al. Predicting tumor sensitivity to chemotherapeutic drugs in oral squamous cell carcinoma patients. Sci Rep. 2018;8:15545.
Driehuis E, Kolders S, Spelier S, Lohmussaar K, Willems SM, Devriese LA, et al. Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov. 2019;9:852–71.
Driehuis E, Kretzschmar K, Clevers H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat Protoc. 2020;15:3380–409.
Vincent ZL, Mitchell MD, Ponnampalam AP. Regulation of MT1-MMP/MMP-2/TIMP-2 axis in human placenta. J Inflamm Res. 2015;8:193–200.
Elshimali YI, Wu Y, Khaddour H, Wu Y, Gradinaru D, Sukhija H, et al. Optimization of cancer treatment through overcoming drug resistance. J Cancer Res Oncobiol. 2018;1:107.
Sun L, Wang K, Peng L, Zhang J, Yang J, Zhao J, et al. Naa10p enhances chemosensitivity to cisplatin in oral squamous cell carcinoma cells. Cancer Manag Res. 2021;13:1843–51.
Ito Y, Narita N, Nomi N, Sugimoto C, Takabayashi T, Yamada T, et al. Suppression of poly(rC)-binding protein 4 (PCBP4) reduced cisplatin resistance in human maxillary cancer cells. Sci Rep. 2015;5:12360.
Yamano Y, Uzawa K, Saito K, Nakashima D, Kasamatsu A, Koike H, et al. Identification of cisplatin-resistance related genes in head and neck squamous cell carcinoma. Int J Cancer. 2010;126:437–49.
Xiong P, Li YX, Tang YT, Chen HG. Proteomic analyses of Sirt1-mediated cisplatin resistance in OSCC cell line. Protein J. 2011;30:499–508.
Wu HT, Chen WT, Li GW, Shen JX, Ye QQ, Zhang ML, et al. Analysis of the differentially expressed genes induced by cisplatin resistance in oral squamous cell carcinomas and their interaction. Front Genet. 2019;10:1328.
Hu Z, Chen K, Xia Z, Chavez M, Pal S, Seol JH, et al. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014;28:396–408.
Larson K, Yan SJ, Tsurumi A, Liu J, Zhou J, Gaur K, et al. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012;8:e1002473.
Sen P, Shah PP, Nativio R, Berger SL. Epigenetic mechanisms of longevity and aging. Cell. 2016;166:822–39.
Yimit A, Adebali O, Sancar A, Jiang Y. Differential damage and repair of DNA-adducts induced by anti-cancer drug cisplatin across mouse organs. Nat Commun. 2019;10:309.
Cruft HJ, Mauritzen CM, Stedman E. Abnormal properties of histones from malignant cells. Nature. 1954;174:580–5.
Piscopo M, Campisi G, Colella G, Bilancione M, Caccamo S, Di Liberto C, et al. H3 and H3.3 histone mRNA amounts and ratio in oral squamous cell carcinoma and leukoplakia. Oral Dis. 2006;12:130–6.
Rui Y, Peng WJ, Wang M, Wang Q, Liu ZL, Chen YQ, et al. HIST1H3D: a promising therapeutic target for lung cancer. Int J Oncol. 2017;50:815–22.
Sakamoto R, Nitta T, Kamikawa Y, Sugihara K, Hasui K, Tsuyama S, et al. The assessment of cell proliferation during 9,10-dimethyl-1,2-benzanthracene-induced hamster tongue carcinogenesis by means of histone H3 mRNA in situ hybridization. Med Electron Microsc. 2004;37:52–61.
Kumar S, Kaur S, Seem K, Kumar S, Mohapatra T. Understanding 3D genome organization and its effect on transcriptional gene regulation under environmental stress in plant: a chromatin perspective. Front Cell Dev Biol. 2021;9:774719.
Lu YX, Regan JC, Esser J, Drews LF, Weinseis T, Stinn J, et al. A TORC1-histone axis regulates chromatin organisation and non-canonical induction of autophagy to ameliorate ageing. Elife. 2021;10:e62233.
Gasiorkiewicz BM, Koczurkiewicz-Adamczyk P, Piska K, Pekala E. Autophagy modulating agents as chemosensitizers for cisplatin therapy in cancer. Invest New Drugs. 2021;39:538–63.
Wu T, Wang MC, Jing L, Liu ZY, Guo H, Liu Y, et al. Autophagy facilitates lung adenocarcinoma resistance to cisplatin treatment by activation of AMPK/mTOR signaling pathway. Drug Des Devel Ther. 2015;9:6421–31.
Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15:65–74.
Baek SH, Kim KI. Epigenetic control of autophagy: nuclear events gain more attention. Mol Cell. 2017;65:781–5.
Hansen JC, Wolffe AP. A role for histones H2A/H2B in chromatin folding and transcriptional repression. Proc Natl Acad Sci USA. 1994;91:2339–43.
Kniazeva AS, Armeev GA, Shaytan AK. H2A-H2B histone dimer plasticity and its functional implications. Cells. 2022;11:2837.
Vyas A, Gomez-Casal R, Cruz-Rangel S, Villanueva H, Sikora AG, Rajagopalan P, et al. Lysosomal inhibition sensitizes TMEM16A-expressing cancer cells to chemotherapy. Proc Natl Acad Sci USA. 2022;119:e2100670119.
Acknowledgements
The authors thank Professor Soeun Jeon (Department of Anesthesia and Pain Medicine, School of Dentistry, Kyungpook National University) and Mr. Dong-Yeon Kim (Department of Microbiology and Immunology, School of Dentistry, Kyungpook National University) for their assistance with the Kaplan-Meier survival analysis used in this report. YD-38/CIS cell line was kindly provided by professor Pil-Young Yun (Seoul National University Bundang Hospital, Department of Oral and Maxillofacial Surgery).
Funding
This work was supported by the Basic Science Research Program from the National Research Foundation (NRF) of Korea, funded by the Korean Government (MIST 2017R1A6A3A11031132, 2022R1A2C2006728) and in part by the ICT & Future Planning (2021R1A2C4002660).
Author information
Authors and Affiliations
Contributions
S-HH, S-YC, KYL, and SYO designed the study and experiments. S-H H, SYO, T-GK, and JK, performed experiments and analyzed data. S-YC, KYL, T-GK, JWK, STL, and TGK provided guidance on the organoid-related experiments. S-HH, SYO, KYL, H-JL, T-GK, and TGK performed statistical analyses. S-HH, S-YC, SYO, JK, H-JL, JWK, and STL wrote the manuscript. All authors gave final approval of the version to be published.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Boris Zhivotovsky
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Oh, S.Y., Kim, J., Lee, K.Y. et al. Chromatin remodeling-driven autophagy activation induces cisplatin resistance in oral squamous cell carcinoma. Cell Death Dis 15, 589 (2024). https://doi.org/10.1038/s41419-024-06975-1
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41419-024-06975-1
This article is cited by
-
Streptococcus mutans-derived extracellular vesicles promote skin wound healing via tRNA cargo
Journal of Nanobiotechnology (2025)
-
MTMR6 downregulation contributes to cisplatin resistance in oral squamous cell carcinoma
Cancer Cell International (2025)
-
Silencing of SCEL promotes progression of oral squamous cell carcinoma via activating TGF-β/Smad pathway
Discover Oncology (2025)
-
A two-decade bibliometric analysis of drug resistance in oral cancer research: patterns, trends, and future directions
Discover Oncology (2025)