Abstract
Post-translational addition of O-linked N-acetylglucosamine (O-GlcNAc) to proteins is commonly associated with a variety of stress responses and cellular processes in eukaryotes, but its potential roles in bacteria are unclear. Here, we show that protein HmwC acts as an O-GlcNAc transferase (OGT) responsible for O-GlcNAcylation of multiple proteins in Yersinia pestis, a flea-borne pathogen responsible for plague. We identify 64 O-GlcNAcylated proteins (comprising 65 sites) with differential abundance under conditions mimicking the mammalian host (Mh) and flea vector (Fv) environments. Deletion of hmwC, encoding a putative OGT, structurally distinct from any existing member of the GT41 family, results in reduced O-GlcNAcylation, reduced growth, and alterations in virulence properties and survival under stress. Purified HmwC can modify target proteins in vitro using UDP-GlcNAc as sugar donor. One of the target proteins, OsdY, promotes Y. pestis survival under oxidative stress conditions. Thus, our results support that regulation of antioxidative responses through O-GlcNAcylation may be a conserved process shared by prokaryotes and eukaryotes.
Similar content being viewed by others
Introduction
Glycosylation modifications play a critical role in regulating various biological processes. These modifications can be categorized into two types: N-glycosylation, which occurs on aspartate residues, and O-glycosylation, which typically occurs on serine/threonine residues but can also involve other target residues1. N-glycosylation is relatively conserved in prokaryotes and eukaryotes and follows a well-defined N-linked glycosylation pathway. In contrast, O-glycosylation uses different sugar donors in eukaryotes and prokaryotes, resulting in structural and functional differences in glycosylated proteins2,3,4. O-linked N-acetylglucosamine (O-GlcNAc) is a simple monosaccharide modification on the side chains of serine and threonine residues, which is commonly associated with a variety of stress responses and cellular processes5,6,7,8,9. This modification is a dynamic process that cycles rapidly during cellular activity. The reversible addition or removal of GlcNAc is mediated by two conserved enzymes O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA)10,11,12,13,14, by using uridine diphosphate-GlcNAc (UDP-GlcNAc) as the direct donor sugar nucleotide15. O-GlcNAcylation occurs on a range of nucleus and cytoplasm proteins and dysregulation of this modification is implicated in various human diseases such as cancer, diabetes and Alzheimer’s disease5,16,17,18. Similar enzyme systems regulating O-GlcNAcylation have been identified in various bacterial species, including Listeria Monocytogenes, Streptococcus pneumoniae, Xanthomonas campestris, Thermobaculum terrenum, etc19,20,21,22. A noteworthy difference between prokaryotic and eukaryotic O-GlcNAcylation system lies in that there is only one OGT in eukaryotes and knockout of OGT has been shown to be embryonic lethality9. However, it is common to possess multiple types and functional OGT enzymes in prokaryotes, and OGT-knockout bacteria are still able to survive19,23,24. Despite of these progresses, related research in bacteria is currently very limited.
The Carbohydrate Active Enzymes (CAZy; http://www.cazy.org) database classifies OGT and HMW1C-like protein as members of the glycosyl transferase family 41(GT41)22,25,26. In bacterial enzymes, TtOGT in T. terrenum, XcOGT in X. campestris and HMW1C in Haemophilus influenzae are the representative enzymes belonging to GT41 family22,27. The N-terminus of TtOGT and XcOGT contains a series of tetratricopeptide repeat (TPR) domains, while the C-terminus is a glycosyltransferase domain with GT-B topology. HMW1C is a recently discovered glycosyltransferase belonging to the GT41 family. It contains an N-terminal all α-domain (AAD) fold and a C-terminal GT-B fold with two Rossmann-like domains. Notably, what sets HMW1C apart from other members of the GT41 family is the absence of the typical TPR fold that characterizes this enzyme family. HMW1C exhibits remarkable versatility in its enzymatic activities. It possesses both N-glycosyltransferase activity, which catalyzes the formation of an N-glycosidic bond with glucose and galactose on asparagine residues, and O-glycosyltransferase activity, which produces a disaccharide structure on the substrate proteins28,29,30. The dual functionality of HMW1C allows for the modification of proteins through diverse glycosylation patterns, thereby expanding its potential biological roles and emphasizing its significance in cellular processes involving protein glycosylation. Yersinia pestis is the causative agent of plague, a disease that has been responsible for millions of deaths throughout history31,32. Nowadays, this deadly pathogen continues to pose a significant threat by causing sporadic outbreaks of plague every year and potentially being misused as a bioterrorism agent33. Y. pestis is a flea-borne pathogen and its survival and replication in macrophages during the early stages of infection is a crucial event for the successful establishment of a systemic infection after entering the host via fleas34,35,36,37,38. Therefore, the transition from the flea vector to the mammalian host requires Y. pestis to regulate physiological processes rapidly to adapt to the hostile host environment39,40,41,42. In our study, we have revealed the significance of HmwC as a noteworthy OGT in Y. pestis. Notably, HmwC displays a unique function and structure that differs significantly from other members of the GT41 family.
Quantitative proteomics analysis for protein O-GlcNAcylation has been performed for Y. pestis grown under Fv (26 °C in calcium-containing TMH) and Mh (37 °C in calcium-free TMH), which simulate the environment of fleas and mammals32,43, respectively. We have successfully identified 64 O-GlcNAcylated sites in 65 proteins, exhibiting varying levels of abundance. We have also discovered that HmwC functions as an OGT in Y. pestis and play crucial roles in regulating various biological process. By conducting a comparative O-GlcNAcome analysis of ΔhmwC and the wild-type Y. pestis, we have identified several potential substrates of HmwC. We further provide evidence supporting the functional O-GlcNAcylation of two proteins, Yp_3614 and Yp_0610, which are referred to as oxidative stress defense protein of Yersinia (OsdY) and Alginate lyase (AlgL) in this study, respectively. Specifically, our study has revealed HmwC-mediated O-GlcNAcylation plays a critical role in regulating antioxidative response of Y. pestis.
Results
O-GlcNAcylation Proteome Analysis of Y. pestis Grown Under the Mh or Fv Conditions
When Y. pestis were grown under 7 different conditions (Fig. 1a), the O-GlcNAcylated protein profiles were found to undergo great alterations, especially under the conditions of Mh, 10 °C, hyperosmotic stress, or Fe2+ deprivation, compared to that of bacteria grown at 26 °C (Fig. 1b and Supplementary Fig. 1a). These results suggest that O-GlcNAcylation may play some important regulatory roles in Y. pestis to response to external stimuli. It is well known that the switch from flea vector to mammalian hosts leads to the activation of numerous virulence mechanisms in Y. pestis. Thus, we conducted quantitative proteomics for O-GlcNAcylation of Y. pestis cultured under Fv and Mh conditions, respectively (Fig. 1c). The secondary MS data was analyzed using Maxquant (v1.6.15.0), and the Y. pestis 91001 database was used as the reference (downloaded from NCBI on Oct 27, 2020, GenBank assembly accession: GCA_000007885.1). A total of 127 proteins (comprising 178 sites) with different abundances were identified, of which 64 proteins (comprising 65 sites) were quantifiable (Supplementary Fig. 1b and Supplementary Data 1). Applying a cutoff value of 1.2 for the fold change of a protein, there were a total of 23 sites of 22 proteins with increased abundances while 20 sites of 20 proteins with decreased abundances under the Mh condition in comparison to the Fv condition (Fig. 1d and Supplementary Data 2). Among the proteins with known functions, more than 70% were cytoplasmic proteins, and relatively few periplasmic and outer membrane proteins (Fig. 1e). GO term functional classification analysis showed that most bacterial proteins with altered O-GlcNAcylation at 37 °C in Y. pestis were catalytic proteins, binding proteins and transporters (Fig. 1f ).
a Workflow for obtaining bacterial protein samples of Y. pestis grown under various conditions. b The O-GlcNAcylations of the wild-type Y. pestis strain were found to be significantly different under seven different growth conditions. The experiment was independently repeated at least three times, yielding consistent results. Protein samples were obtained using the method described in a, separated on a 12% SDS-PAGE gel, and immunoblotted with a mouse monoclonal anti-O-GlcNAc antibody. Long and short exposures were used to provide comprehensive information. Lanes 1-7 correspond to Fv, Mh, 10 °C in TMH, 42 °C in TMH, 37 °C in pH 6 TMH without calcium, 26 °C in TMH with 2.5% NaCl, and 37 °C in TMH without Ca2+ and Fe2+. c Technical roadmap for quantitative proteomic analysis of O-GlcNAcylation of protein in Y. pestis grown under the Fv and Mh conditions. d Statistics plot of the proteins and sites with different abundances under the Fv and Mh conditions in triplicates. Proteins with fold change (FC) ≥ 1.2 were considered to be up-regulated. e Statistical plot predicting the subcellular localization of the differentially modified proteins. f Statistical plot showing the distribution of differentially modified proteins in secondary Gene Ontology (GO) annotations. BP biological process, CC cellular component and MF stands for molecular function.
HmwC Affects the O-GlcNAcylation in Y. pestis
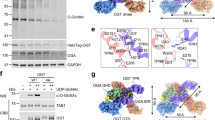
The different O-GlcNAcylated protein profiles in Y. pestis grown under various conditions suggest that certain proteins are functioning as OGTs to regulate the protein O-GlcNAcylation in response to various stimuli. hmwC in Y. pestis was predicted to be O-linked N-acetylglucosamine transferase according to the annotation of CO92 genome. To investigate whether this gene was responsible for the observed protein O-GlcNAcylation in Y. pestis, we generated a hmwC mutant (denoted as ΔhmwC). In comparison to the wild-type Y. pestis, global protein O-GlcNAcylation was substantially reduced in ΔhmwC under all the tested conditions, including Fv, Mh, hyperosmotic stress and Fe2+ deprivation (Fig. 2a and Supplementary Fig. 2a). These findings indicates that HmwC indeed contributes significantly to the protein O-GlcNAcylation probably by functioning as an OGT in Y. pestis. However, O-GlcNAcylation of protein was not entirely eliminated in ΔhmwC, implying that OGTs other than HmwC might exist in Y. pestis.
a O-GlcNAcylation of proteins in ΔhmwC was significantly lower than that in WT. The experiment was independently repeated at least three times, yielding consistent results. Equal protein samples obtained from the wild-type Y. pestis and ΔhmwC grown under different conditions were separated by 12% SDS-PAGE and immunoblotted with a mouse anti-O-GlcNAc antibody to detect O-GlcNAcylated proteins. b Protein structure prediction and domain division of HmwC by AlphaFold2. Structural domains were divided according to protein annotations and represented with different colors. c The secondary structure of HmwC based on PDB:3Q3E (https://doi.org/10.2210/pdb3Q3E/pdb) template alignment using Espript 3.0 online tool. Identical and conserved residues were highlighted in red background and light red font. The helices and folds observed in the structures were indicated above the sequences. d Phylogenetic tree of HmwC homologous sequences in bacteria. The homologous sequences of HmwC in different species were retrieved using BLAST at NCBI. Sequences with more than 40% identity were selected for phylogenetic tree analysis. HmwC is represented by Y. pestis in group I and highlighted in red font. ApHMW1C by A. pleuropneumoniae and HMW1C by H. influenzae in group III are highlighted in purple font. The evolutionary history was inferred using the Neighbor-Joining method. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analyzed. The evolutionary distances were computed using the p-distance method and are in the units of the number of amino acid differences per site. Evolutionary analyses were conducted in MEGA11. The phylogenetic tree was beautified using the iTOL online tool. e Proteins in Group I and HMW1C in H. influenzae and ApHMW1C in A. pleuropneumoniae (Group III) were aligned with Uniprot, and the key sites were highlighted with a black box. Thr-438 of ApHMW1C and Ser-435 of HmwC were labeled in the tertiary structure of the protein.
The diagram depicting the tertiary structure modeling of HmwC illustrates the division of its domains (Fig. 2b). By searching the PDB database, we found that the sequence of HmwC was closest to the ApHMW1C (PDB: 3Q3E), which has a resolved crystal. The amino acid sequence identity between HmwC and ApHMW1C is approximately 41% (Fig. 2c). ApHMW1C and HMW1C are representatives of GT41 family HMW1C-like proteins. However, the phylogenetic tree analysis of HmwC homologous proteins revealed that HmwC (in Group I) and ApHMW1C, HMW1C (in Group III) were located in separate branches (Fig. 2d). Sequence alignment revealed a significant distinction in the UDP-binding pocket between ApHMW1C/HMW1C and HmwC. Thr-438 of ApHMW1C was the key residue for the UDP binding pocket27 in proteins of Group III, corresponding to the residue Ser-435 of HmwC (conserved in proteins of Group I), which indicates the different usage of their sugar donors (Fig. 2e).
Growth of the hmwC mutant in the nutrient-limited TMH medium was significantly delayed
The growth of ΔhmwC in LB medium at 26 °C was similar to that of the wild-type Y. pestis strain (Fig. 3a). When grown in the nutrient-limited TMH medium, hmwC mutant exhibited significantly reduced reproduction rate, delayed logarithmic phase, and a lower density of cells at the plateau phase. Normal growth was restored upon complementation of HmwC in ΔhmwC (Fig. 3b).
Analysis of the influence of hmwC deletion on bacterial proliferation and growth in LB medium (a), TMH medium (b), and LB medium with 2.5% NaCl (d). Y. pestis strains were grown at 26 °C and the absorbance at OD620nm were continuously monitored every 2 h of for a total of 32 h. c Y. pestis strains were grown in 24-well plates. First measure the OD620nm value of the bacterial solution. Then the biofilm was stained with 0.1% crystal violet solution and decolorized with dimethyl sulfoxide, and the OD570nm value of the decolorized solution was measured. The amount of biofilm was expressed as OD570nm/OD620nm ratio. e Equal bacterial solutions of WT, ΔhmwC, and ΔhmwC/HmwC strain were incubated parallelly at 50 °C for 10, 20, and 40 min, then the viable count of bacteria was determined by plating the bacterial dilutions onto a Hottinger’s agar plate in triplicate. f Equal bacterial solutions of WT, ΔhmwC, and ΔhmwC/HmwC strain were placed parallelly at −20 °C or 4 °C for 24 h, then the viable count of bacteria was determined as described above. g Circular filter paper with 100 mM or 200 mM hydrogen peroxide solution was placed in the center of semi-solid petri dish, and the diameter of the inhibition circle was measured to reflect the hydrogen peroxide resistance. h WT, ΔhmwC, and ΔhmwC/HmwC strain cultures diluted parallelly with pH 3.5 20 mM glucose minimal medium were incubated at room temperature for 30 min and 60 min, then the viable count of bacteria was determined as described above. i RAW264.7 were infected with WT, ΔhmwC, and ΔhmwC/HmwC strain at a MOI of 10. At 0.5 hpi, gentamicin was added to kill the extracellular bacteria. At 0.5, 2, 4, 8, and 24 hpi., the viable count of intracellular bacteria was determined as described above. j Each 6–8-week-old female mice was subcutaneously challenged with 100 μl of the wild-type Y. pestis strain or ΔhmwC suspension (~100 CFU) (n = 10/group), and the survival of challenged mice was observed for 14 consecutive days. A log-rank test was used to calculate the statistical significance. k Livers, spleens and lungs of the challenged mice were harvested when the s.c. challenged Bal b/c mice were moribund, i.e. 4 dpi., and the collected tissues were homogenized in sterile PBS to measure live bacteria, as described above. One-way ANOVA was performed to analyze the significance of difference in bacterial loads in the different tissues. c n = 10 biological replicates in each group, (j, k) n = 10 mice in each group. a, b, d–i n = 3 biological replicates in each group. Data are presented as mean values ±SEM. p values were calculated by two-tailed Student’s t test. Note that some error bars are too small to be visualized.
The hmwC mutant exhibits significantly enhanced biofilm formation ability
Biofilm formation can provide protection to bacteria against various external stressors44,45. In the case of Y. pestis, biofilm formation is crucial for its transmission via fleas. The crystal violet assay results demonstrated that the amount of biofilm produced by the ΔhmwC strain was almost 12 times higher than that of the wild-type strain (Fig. 3c).
Deletion of hmwC leads to increased intracellular survival of Y. pestis
Bacterial cells of Y. pestis strains were resuspended in a 20 mM glucose minimal medium and then subjected to different stimuli for a period of time to assess their tolerance. Surprisingly, ΔhmwC exhibited significantly increased tolerance to hyperosmotic (Fig. 3d), heat and cold (Fig. 3e, f ), hydrogen peroxide (Fig. 3g), and acidic environments (Fig. 3h) compared to the wild-type strain. To investigate the role of HmwC in the intracellular survival of Y. pestis, RAW264.7 cells were infected with either the wild-type or ΔhmwC strain, and the survival percentage of bacteria was analyzed by plating and counting the live bacteria from the lysed RAW264.7 cells. Consistent with the results of the in vitro phenotypic analysis, a significant increase in replication was observed for the ΔhmwC strain compared to the wild-type strain at 2 h post-infection (hpi) (p < 0.01). In contrast, the wild-type strain was cleared much more rapidly than the ΔhmwC strain at 4 hpi (p < 0.001) and 8 hpi (p < 0.01) (Fig. 3i). Consistently, the ΔhmwC strain also showed increased resistance to hydrogen peroxide (Fig. 3g), one of the representative stresses normally present in macrophages. These findings suggest that the deletion of hmwC significantly enhances the resistance of Y. pestis to various adverse stimuli, as well as its replication and survival in macrophages.
Deletion of hmwC leads to slightly improved virulence of Y. pestis
We next want to investigate the role of HmwC in the development of bubonic plague. To this end, mice were subcutaneously challenged with 100 CFU of either the wild-type Y. pestis or ΔhmwC strain. Mice infected with ΔhmwC showed slightly earlier onset of symptoms and mortality compared to those infected with the wild-type strain (Fig. 3j). However, the difference was not statistically significant (Log-rank test, ns). The bacterial counts in the liver, lung, and spleen of mice infected with ΔhmwC were significantly higher by one to two orders of magnitude than those of the wild-type strain in the respective organs (Fig. 3k). These findings suggest that the deletion of hmwC results in increased virulence in mice. Taken together, our phenotypic assay results indicate the importance of HmwC in regulation of the pathogenesis of Y. pestis.
Comparative proteomics analysis for O-GlcNAcylation between the wild type and the hmwC mutant of Y. pestis
Protein O-GlcNAcylation was analyzed by MS-based proteomics methods for the bacterial total proteins of the wild-type Y. pestis and ΔhmwC grown under the Fv and Mh conditions, respectively. The comparative proteomics analysis revealed that the abundances of 103 proteins were significantly increased and 77 were significantly decreased in ΔhmwC compared to the wild-type Y. pestis under the Fv condition, while 123 proteins were significantly increased and 61 were significantly decreased under the Mh condition (p < 0.05) (Fig. 4a, b, Supplementary Fig. 2b, c, d, and Supplementary Data 3). Proteins with |Log2 FC| ≥ 0.585 (~1.5 × FC) were considered to be different in abundance. Of these, 65 proteins showed different abundances under both Fv and Mh conditions, while 115 and 119 proteins (about 2/3 of the total) exclusively showed different abundances under the Fv or Mh conditions, respectively (Fig. 4c, d). The KEGG pathways of sulfur metabolism, biofilm formation, bacterial secretion system, and ABC transporters were significantly enriched under both Fv and Mh conditions. However, the carbon fixation pathways and pyruvate metabolism were only enriched under the Fv condition, while the C5-branched dibasic acid metabolism, protein export, and oxidative phosphorylation pathways were only enriched under the Mh condition (Fig. 4e).
Heatmap (a) and statistics plot (b) of the proteins with different abundances between the different samples under Fv and Mh conditions (n = 3 biologically replicates). Proteins with a p value ≤ 0.05 and fold change (FC) ≥ 1.5 were considered to be up-regulated. p values were calculated by two-tailed Student’s t test. c Venn diagram displaying the proteins with differential abundance between ΔhmwC and WT under Fv or Mh conditions. d Volcano plot displaying the regulation of proteins with different abundances between ΔhmwC and WT under Fv and Mh conditions. Red and blue dots, proteins with significantly up or down differences. Several important global regulators or virulence factors were labeled with protein names. e KEGG pathway enrichment analysis of proteins with different abundances between ΔhmwC and WT under Fv and Mh. The barplot shows the significance of the p values (color) and the number of proteins (length) in the enriched KEGG pathways. p values calculated by cumulative hypergeometric test.
On the other hand, O-GlcNAcome results showed a significant difference in the abundance of O-GlcNAcylated proteins or sites between WT and ΔhmwC, under both Fv and Mh conditions (Fig. 4b). Protein quantification from three biological samples was reproducible (Fig. 5a and Supplementary Fig. 3a). All the O-GlcNAcylated proteins or sites reported had a |Log2 FC| ≥ 0.263 (~1.2 × FC) at least. Specifically, there were a total of 26 O-GlcNAcylated sites of 26 proteins with increased abundance and 6 sites of 6 proteins with decreased abundance under Fv condition respectively. Under the Mh condition, there were a total of 11 O-GlcNAcylated sites of 11 proteins with increased abundance while 9 sites in 9 proteins with decreased abundance respectively (Fig. 5b and Supplementary Data 4). Additionally, 10 O-GlcNAcylated proteins or sites with different abundances were shared by Y. pestis grown under Fv and Mh conditions (Fig. 5c). The O-GlcNAcylated proteins with different abundances between ΔhmwC and WT under Fv and Mh were significantly enriched in GO terms related to the cell, membrane, organelle, and protein-containing complex (Fig. 5d, e and Supplementary Fig. 3b, c).
a Principal component analysis of O-GlcNAc proteomics data and statistical plot (b) showing differentially O-GlcNAcylated peptides and proteins between ΔhmwC and WT strains grown under Fv or Mh conditions. c Venn diagram analysis of the O-GlcNAcylated proteins in b. Proteins were illustrated in red for up-regulation, blue for down-regulation, and gray for those with opposite direction in alterations under the two conditions. d Statistical plot for the prediction and classification of subcellular localization of differentially O-GlcNAcylated proteins. e Statistical plot showing the GO term enrichment analysis of the O-GlcNAcylated proteins in b. f LC-MS/MS analyses of the corresponding peptide sequences of the substrate proteins OsdY (left) and AlgL (right). The letter “S” with “he” above in the figure represents the O-GlcNAcylated site. g Glycosylation of OsdY by HmwC. To define the specificity of sugar donor utilization by HmwC, glycosylation assays were carried out in the reaction buffer with or without HmwC in the presence of different UDP activated sugars. After the glycosylation reactions, samples were separated by SDS-PAGE, and the gel was stained with Coomassie Blue (up) or subjected to detection using the Pierce™ Glycoprotein Staining Kit (down). Glycosylated OsdY protein was indicated by arrow: ‘a’ is glycosylated OsdY reacted with UDP-GlcNAc and ‘b’ was glycosylated positive control. The experiment was independently repeated at least three times, yielding consistent results. h The interactions enable the stable presence of UDP-GlcNAc in the protein HmwC bind-pocket. Pink represents UDP, green represents GlcNAc, and blue represents active amino acid sites. Ser-435, Leu-492, Asn-517, Ser-277, Agr-280, His-276, and Gly-367 of HmwC form 10 hydrogen bonds (blue solid line) with UDP-GlcNAc. Pro-369 exhibits strong hydrophobic interactions (gray dotted line) with UDP-GlcNAc, while the active group amino in Lys-438 forms a salt bridge (yellow dotted line) with UDP in the molecular ligand.
We hypothesized that the proteins exhibiting a significant decrease in the abundance of O-GlcNAcylated peptides in ΔhmwC, compared to the WT strain, may represent potential substrates of HmwC. Among the potential substrates, Yp_3614 was annotated as a putative oxidative stress defense protein, which we designated as OsdY, representing the Oxidative Stress Defense protein of Yersinia. However, its functional activity has not been supported by any experimental evidence so far. The annotated MS2 spectra from LC-MS/MS analysis revealed that the Ser-83 residue of OsdY and Ser-5 residue of AlgL underwent O-GlcNAcylation (Fig. 5f). The theoretical fragment ion types of target peptides predicted by Prosit, as shown in Supplementary Table 1 (filtered for Relative Intensity >0.2, https://www.proteomicsdb.org/prosit/), closely matched the detected spectra in actual experiments. To assess whether HmwC was able to glycosylate OsdY, we performed an in vitro glycosylation assay using purified HmwC and OsdY by providing UDP-glucose, UDP-galactose, or UDP-GlcNAc as sugar donor. In the presence of UDP-GlcNAc, but not UDP-glucose and UDP-galactose, a retarded mobility for OsdY was observed (Fig. 5g), suggesting that HmwC utilizes UDP-GlcNAc as a donor to glycosylate substrates. According to our molecular docking analysis, the binding energy between the HmwC and UDP-GlcNAC was determined to be -10.4 kcal/mol, indicating a stable and high-affinity binding between UDP-GlcNAC and HmwC (Fig. 5h).
HmwC of Y. pestis is an OGT capable of O-GlcNAcylating of substrates
We further employ Yp_3614, designated as OsdY, and Yp_0610, designated as AlgL (alginate lyase family protein), as substrates to validate the O-GlcNAc transferase activity of HmwC. The results of in vitro O-GlcNAcylation experiments clearly demonstrate that HmwC is capable of O-GlcNAcylating OsdY in the presence of UDP-GlcNAc (Fig. 6a). By sharp contrast, when the Ser-83 residue was mutated to Ala, the band of GlcNAcylated OsdY completely disappeared (Fig. 6a). Similar results were observed for the modification of the AlgL Ser-5 residue mediated by HmwC (Fig. 6b). After adding of varying amounts of purified recombinant HmwC to the reaction system, we observed that the modification levels of OsdY and AlgL were directly proportional to the concentration of HmwC (Fig. 6c, d). These results suggest that HmwC is a novel O-GlcNAc transferase in Y. pestis.
a, b HmwC O-GlcNAcylates OsdY at Ser-83 and AlgL at Ser-5. Purified recombinant GST-HmwC was incubated with His-OsdY, His-OsdYSer83A, His-AlgL or His-AlgLSer5A in the mixture containing UDP-GlcNAc, OGT assay buffer, and ddH2O at 37 °C for 2 h, then 2xSDS loading buffer was added to stop the reaction. All the samples were separated in 12% SDS-PAGE and immunoblotted with mouse monoclonal anti-GST, anti-His, or anti-O-GlcNAc antibodies. c, d The O-GlcNAcylation of OsdY and AlgL showed dose-dependent on concentration of HmwC. e D217 and H276 of HmwC were predicted to be the key residues for substrate binding and enzyme activity, respectively. Protein structure prediction was accomplished using Alphafold2 software (Supplementary Data 6). f GST pull-down assay of binding affinity between OsdY and HmwC and its mutants. Purified recombinant His-tagged OsdY was incubated at 4 °C overnight with glutathione-sepharose 4B beads conjugated with GST-HmwC, GST-HmwC-D217A and GST-HmwC-H276A, respectively. Then the beads were intensively washed and analyzed by SDS-PAGE and immunoblotting. g GST pull-down assay of binding affinity between AlgL and HmwC and its mutants, using the method the same as described in f. h, i Analysis of the O-GlcNAcylation of OsdY and AlgL by HmwC, HmwC-D217 or HmwC-H276A was carried out according to the method described in a. All above experiments were performed at least in triplicate with similar results, and a representative result was shown here.
The crystal structure of ApHMW1C has previously been reported, in which Asp-215 has been identified as key binding site and His-277 as catalytic site27. Sequence alignment analysis of HmwC and ApHMW1C revealed that Asp-217 and His-276 of HmwC, mapping to Asp-215 and His-277 of ApHMW1C, respectively, indicating that they might play crucial roles in substrate recognition and catalytic activity (Fig. 6e, Supplementary Fig. 4). As expected, when the Asp-217 residue was mutated, the interaction affinity between HmwC and OsdY was significantly reduced by approximately 60% (Fig. 6f), resulting in a decrease in the O-GlcNAcylation level of OsdY (Fig. 6h). In contrast, mutation of the His-276 residue of HmwC did not affect the binding affinity between HmwC and OsdY (Fig. 6f), but led to the disappearance of O-GlcNAcylation of OsdY (Fig. 6h), consisting with the cognition that this residue is a crucial catalytic site for glycosyltransferase activity of HmwC. We conducted similar experiments on another modified substrate of HmwC, AlgL, and obtained similar results (Fig. 6g, i).
O-GlcNAcylation of OsdY plays critical roles in oxidative stress response of Y. pestis
OsdY, encoded by yp_3614, is an uncharacterized conserved protein in Y. pestis, whereas its homologous protein YggE in E. coli has been reported to be involved in oxidative stress defense46. The proteomics data suggested that the O-GlcNAcylation of OsdY occurred on the Ser-83 residue. To investigate the functional role of this site-specific O-GlcNAcylation, we generated an osdY null mutant (ΔosdY), a mutant with a point mutation at 83 serine to alanine (ΔosdYS83A), and their respective complemented strains. The substitution of serine with alanine at position 83 in OsdY protein does not seem to influence its overall structure, as predicated by Alphafold2 (Supplementary Fig. 5). Subsequently, by quantifying the relative mRNA expression level of osdY in both the WT and ΔosdYS83A strains, we observed no significant difference (Fig. 7a), indicating that the mutation does not lead to a decrease of osdY expression. Compared to WT, ΔosdYS83A exhibited a similar growth rate in LB (Fig. 7b), but significantly decreased acid resistance (Fig. 7c). RAW264.7 cells were infected with WT, ΔosdYS83A and ΔosdYS83A/OsdY, and bacterial survival percentage was analyzed by plating and counting of living bacteria released from RAW264.7. A significant replication had been found for WT at 2 hpi. in comparison to ΔosdYS83A. Besides, ΔosdYS83A was cleared much more rapidly than WT in RAW264.7 at 4 and 8 hpi. (Fig. 7d), indicating that Ser-83 of OsdY significantly promoted the replication and survival of Y. pestis in macrophages. Additionally, both ΔosdY and ΔosdYS83A exhibited an enhanced biofilm formation capability compared to WT (Fig. 7e). When exposed to H2O2 with a concentration ranging from 200 to 400 nM, the bacteriostasis circle radius of the ΔosdYS83A strain exhibited a significant increase (p < 0.001) compared to the WT strain (Fig. 7f ). However, the phenotype was restored to normal when the ΔosdYS83A strain was complemented with plasmid overexpressing OsdY (p < 0.05). We further complement the ΔosdY strain by introducing OsdY or OsdYS83A expressing plasmids. We found that the antioxidant capacity of ΔosdY/OsdY was even a little higher than that of the WT strain, probably due to the overexpression of OsdY, while that of ΔosdY/OsdYS83A remained to be lower. No significant difference was observed between ΔosdY and ΔosdYS83A strains (Fig. 7f). We further conducted measurements of H2O2 content in various Y. pestis strains, including WT, ΔosdYS83A, ΔosdY, ΔosdYS83A /OsdY, ΔosdY/OsdY, and ΔosdY/OsdYS83A using a H2O2 Assay kit (Fig. 7g). The results revealed the WT strain exhibited an approximate H2O2 content of 5.4 pmol/103 cells, while the ΔosdY and ΔosdYS83A mutants displayed more than five-fold higher than that of the WT strain, with values of 26.53 and 21.44 pmol/103 cells, respectively. Moreover, complementary expression of OsdY in both the ΔosdY and ΔosdYS83A restored the H2O2 content similar to that in WT, while overexpressing the OsdYS83A in ΔosdY has no significant impacts on H2O2 content compared with ΔosdY (18.46 pmol/103 cell). These findings suggest that OsdY is involved in antioxidant stress function in Y. pestis and Ala substitution of Ser-83 crucial for O-GlcNAcylation abolished its antioxidant activity.
a No significant difference in relative mRNA expression level of osdY between WT and ΔosdYS83A. b The mutation of osdY-Ser83A had no significant effect on the proliferation rate of Y. pestis in TMH medium. c The mutation of osdY-Ser83A led to decreased acid resistance of Y. pestis. d The mutation of osdY-Ser83A led to decreased survival of Y. pestis in macrophages. a osdY or osdY-Ser83A mutations resulted in increased biofilm formation of Y. pestis, which could be compensated by overexpression of OsdY. f The mutation of osdY or osdY-Ser83A resulted in decreased antioxidant capacity of Y. pestis, which could be compensated by overexpression of OsdY but not by overexpression of the OsdYS83A. g Determination of the hydrogen peroxide content in various Y. pestis strains as indicated. Ser-83 of OsdY is a critical residue for its antioxidant activity in Y. pestis. a–d, f, g n = 3 biological replicates in each group. e n = 10 biological replicates in each group. Data are presented as mean values ±SEM. p values were calculated by two-tailed Student’s t test. Note that some error bars are too small to be visualized.
Discussion
Our study reports that HmwC, the first OGT discovered in Y. pestis, can O-GlcNAcylate a group of proteins. Notably, HmwC exhibits distinct characteristics compared to all known GT41 family members, such as TtOGT, XcOGT, and HMW1C. The amino acid sequence of HmwC shares 41.88% identity with HMW1C, while exhibiting only 21.81% and 22.31 similarity with XcOGT and TtOGT, respectively. However, similar to XcOGT and TtOGT, HmwC utilizes UDP-GlcNAc as sugar donor, instead of UDP-hexose used by HMW1C. The UDP binding pockets of XcOGT and TtOGT show substantial differences compared to that of HMW1C. Specifically, Asn-385 in XcOGT (mapping to Gln-839 of hOGT) forms a hydrogen bond with the α-phosphate group of UDP, while the corresponding residue Thr-464 in HMW1C does not directly contact with UDP27. The phylogenetic analysis shows that HmwC and HMW1C are located in distinct branches (Fig. 2d). Furthermore, the critical UDP-binding residues in the proteins within each branch exhibit significant difference (Fig. 2e), well explaining their distinct specificity in utilizing donor sugars for O-GlcNAcylation of substrates. The residues that are highly conserved and critical for the binding of sugar donor and catalytic activity for GT41 family members have been identified in HmwC by sequence alignment. Our GST pull-down assay results proved that the Asp-217 in HmwC, corresponding to Asp-242 of HMW1C and Asp-215 of ApHMW1C, is crucial for substrate binding (Supplementary Fig. 4a). In vitro GlcNAcylation analysis results verified that His-276 is crucial for glycotransferase activity of HmwC, corresponding to His-303 of HMW1C, His-277 of ApHMW1C, His-218 of XcOGT and His-558 of hOGT (Supplementary Fig. 4b).
Through O-GlcNAcome analysis, we have identified an oxidative defense protein, OsdY, to be the substrate protein of HmwC and found that its anti-oxidant activity is likely to be regulated by O-GlcNAcylation at Ser-83 by HmwC. This is the first confirmation of the oxidative defense activity of protein encoded by yp_3614, namely OsdY in Y. pestis. YggE, a homologous protein of OsdY in E. coli has been demonstrated to participate in oxidative stress response. These findings suggest that the conserved oxidative stress defense proteins present in a range of bacteria may also undergo O-GlcNAcylation when exerting their function. The previous findings in eukaryotes have also indicated that hydrogen peroxide serves as a classic stress signal capable of inducing alterations in intracellular O-GlcNAcylation33,34,35,36,38. Our results hint that the regulation of anti-oxidant activity through O-GlcNAcylation can be a conserved cellular process shared by both prokaryotes and eukaryotes.
As an OGT, HmwC has the ability to O-GlcNAcylate not only OsdY but also a variety of other substrates, including AlgL as identified in this study, as well as potential substrates yet to be discovered. Consequently, mutation of hmwC may have a significant impact on various physiological phenotypes of Y. pestis. Deletion of hmwC in Y. pestis results in several notable effects, such as a reduced growth rate in nutritionally limited media, increased resistance to adverse stimuli like acid, cold and heat shock, enhanced biofilm formation, as well as higher intracellular viability in macrophages and increased virulence in mice. Notably, similar phenotypic alterations have been observed in the TtOGT mutant of T. terrenum19. These findings suggest that the hmwC gene may not confer beneficial effects to Y. pestis in terms of virulence and resistance to stresses, as its deletion actually increases these capabilities in phenotype analysis. However, the fact that hmwC is highly conserved in Y. pestis, as well as in its ancestor Y. pseudotuberculosis, suggests that this gene has undergone a positive selection and may play an important role in the different stage of its lifecycle during evolution. We speculate that although the hmwC mutant strain exhibited increased virulence, the role of HmwC in regulating bacterial rapid response to stress stimuli might confer advantages in facing of detrimental environments during evolution.
OGT is discovered originally in the animal kingdom and its knockout is embryotic lethal9. Two OGT homologs, SPINDLY and SECRET AGENT, are subsequently discovered in plants47,48, which exhibits some degree of functional redundancy but their simultaneous knockout is lethal. By contrast, knockout of OGT in prokaryotes is not lethal. The contrast between the effects of OGT knockout in animals and prokaryotes indicates a difference in the function of O-GlaNAcylation between these two domains. Furthermore, this comparison suggests that there may be more than one OGT in Y. pestis because protein O-GlcNAcylation in the ΔhmwC strain, although reduced compared to the wild-type strain, is not completely eliminated. In addition, we did not find any consensus sequence associated with O-GlcNAcylation in proteins examined (Supplementary Fig. 6a, b), suggesting that substrate specificity may not be solely dependent on primary sequences surrounding the O-GlcNAcylation sites. Alternatively, it is possible that the O-GlcNAc transferases may recognize other characteristics.
In conclusion, our study represents the first discovery of an O-GlcNAc enzyme system in Y. pestis, with the successful identification of the pivotal OGT enzyme, HmwC. Notably, HmwC exhibits characteristics distinct from all known members of the GT41 family. We have not only validated the enzymatic activity of HmwC but also identified the key residues responsible for substrate recognition and catalytic activity. Moreover, we demonstrated that HmwC is capable of catalyzing O-GlcNAcylation on multiple proteins, including OsdY that has been shown to be involved in oxidative response of Y. pestis. This discovery is particularly significant as OsdY is a widely conserved protein in bacteria, and our findings provide novel insights and evidence into the mechanisms underlying bacterial oxidative stress response and regulation.
Methods
Culture conditions of bacteria strains
Y. pestis strain biovar Microtus 20149 isolated from the rodent Brandt’s vole (Microtus brandti) was used in this study. And its related mutants and complementary strains were cultured in Luria-Bertani (LB) broth at 26 °C or chemically defined TMH medium with or without 2.5 mM calcium at 26 °C or 37 °C, depending on the requirements of specific experiments. E. coli DH5 α and BL21 (DE3) were grown in LB broth containing antibiotics at the following concentrations: 100 mg/ml ampicillin, 20 mg/ml chloramphenicol, and 50 mg/ml kanamycin.
Antibodies and reagents
O-GlcNAc (PTM-952) antibody was obtained from PTM BioLab (Hangzhou, China). Antibodies for His-Tag (#2365) and GST-Tag (#2622) were obtained from Cell Signaling Technology (Danvers, MA, USA). IRDye 800CW-conjugated goat anti-rabbit antibody (C90529-19) and IRDye 800CW-conjugated goat anti-mouse antibody (C81106-03) were from LI-COR Biosciences (Lincoln, NE, USA). Goat anti-mouse IgG (H + L) secondary antibody (31430) was from Thermo Fisher Scientific (Waltham, MA, USA).
Pierce BCA Protein Assay Kit (#23227), Pierce Top 12 Abundant Protein Depletion Spin Columns (85164), TMT labeling reagent (A44521), acetonitrile (400060) and Ubl-specific protease 1 (12588018), Pierce™ Glycoprotein Staining Kit (24562) were Thermo Fisher Scientific products. Glutathione affinity gel (17-0756-01) and PD-10 desalting columns (17085101) was obtained from GE Healthcare (Pittsburgh, PA. USA). Ni-NTA agarose (30210) was obtained from Qiagen (Valencia, CA, USA). The Immobilon-P transfer membrane (IPVH00010) and Immobilon-NC transfer membrane (HATF00010) were purchased from Millipore (Bedford, MA, USA). Protease Inhibitor Cocktail III (ab201117), V (ab287909) and IV (ab201118) were purchased from Abcam (Cambridge, UK). Tetraethylammonium bromide (140023), Seppro ® Rat Spin Columns (SEP130), DL-dithiothreitol (D0632), UDP-GlcNAc (U4375) and trichloroacetic acid (T6399) were from Sigma-Aldrich (St Louis, MO, USA). ProteoMiner™ Protein Enrichment Small-Capacity Kit (#1633006) was Bio-Rad (Hercules, CA, USA) product. UDPG (U303319) and UDP-Galactose (U303352) were from Aladdin (Shanghai, China). Trypsin (VA9000) was from Promega (Beijing, China) and Formic acid (09676-500 ML) was from Fluka (Seelze, Germany).
Construction of the Y. pestis mutants and the complemented strains
Y. pestis strains and plasmids used in this study were listed in Supplementary Data 5. The coding sequence of hmwC was replaced with a kanamycin resistance cassette by the λRed recombination system. The coding sequences of yp_3614 and yp_3614-Ser83 were eliminated with suicide vector pDS132 by homologous recombination. The mutant cassettes were consisted of approximately 300 bp homologous sequences at both sides of mutant genes and cloned into pDS132. The recombinant pDS132 plasmids were transformed into E. coli S17λpir. Then, yp_3614 and yp_3614-Ser83 deletions were introduced by conjugation and allelic exchange. The plasmid pBAD24 was used to construct complemented strains. DNA fragments generated by PCR containing the coding sequences of hmwC and yp_3614 were cloned into pBAD24. The recombinant plasmids were introduced into the corresponding mutants separately to obtain c complemented mutant strains and the expression of the cloned genes were induced by 0.2% arabinose.
Quantitative polymerase chain reaction
The mRNA expression level of the osdY gene was analyzed by quantitative polymerase chain reaction. Total RNA was extracted from bacteria by TRIzol extraction method and subsequently digested by DNAase I. The mRNA samples ware then reverse transcribed to cDNA using Super-Script III reverse transcriptase (Invitrogen, USA). Real-time PCR amplification was performed using a 7500 Real-Time PCR system (Applied Biosystems, USA). The mRNA expression level was normalized to the expression level of osdY in WT. Each experiment was performed in triplicate.
Expression and purification of recombinant proteins
Plasmids used in this study were listed in Supplementary Data 5. The plasmids pSUMO-HmwC, pSUMO-HmwCD217A, pSUMO-HmwCH276A, pSUMO-OsdY, pSUMO-OsdYS83A, pSUMO-AlgL, pSUMO-AlgLS5A, pGEX-4T-2-HmwC, pGEX-4T-2-HmwCD217A and pGEX-4T-2-HmwCH276A were constructed and introduced into E. coli BL21 (DE3) strains for protein purification. E. coli BL21 (DE3) strains were grown in LB medium with appropriate antibiotics. When the OD600nm of the cultures reached 0.6, protein expression was induced overnight with 1 mM IPTG at 100 rpm in an incubator at 20 °C. His-tagged proteins were purified by affinity chromatography using Ni-NTA beads and eluted with 250 mM imidazole solution. GST-tagged proteins were purified by glutathione sepharose beads 4B and eluted with 10 mM reduced glutathione (pH 8.0). Finally, the eluted solutions containing the purified proteins were exchanged to phosphate-buffered saline (PBS) using PD-10 desalting columns and partial His-tagged proteins were excised by ULP1 (Ubl-specific protease 1).
Protein extraction
Wide type Y. pestis and ΔhmwC were grown at 26 °C in TMH medium to OD620nm of 1.0, then shifted to seven different conditions for 4 h’s incubation, respectively, which included: 26 °C in TMH, 37 °C in TMH without calcium, 10 °C in TMH, 42 °C in TMH, 37 °C in pH 6 TMH without calcium, 26 °C in TMH with 2.5% NaCl, and 37 °C TMH without Ca2+ and Fe2+. The bacterial pellets were washed twice with PBS then resuspended with 4 times the volume of lysis buffer (1% SDS, 1% protease inhibitor) and lysed by sonication. Centrifuge at 12,000 g for 10 min at 4 °C then the supernatants were transferred to new centrifuge tubes and the protein concentrations were determined using the BCA kit.
Detection of O-GlcNAcylation in Y. pestis Under Different Conditions
Equal amounts of proteins in pellets prepared under different conditions were boiled in 5×SDS loading buffer, 20 μg protein per lane was separated by 12% SDS-PAGE. Then the proteins on the gel were transferred onto an immobilon-NC membrane and subjected to immunoblot analysis with primary antibody anti-O-GlcNAc antibody and peroxidase conjugated goat anti-Mouse IgG, (H + L). The short-exposure and long-exposure images of the immunoblotting results were obtained using the automatic chemiluminescence imaging system (Tanon 5200).
Mass spectrometry analysis of O-GlcNAcylation proteomics
Trypsin digestion
The protein solution was reduced with 5 mM dithiothreitol for 30 min at 56 °C and alkylated with 11 mM iodoacetamide for 15 min at room temperature in darkness. The protein samples were diluted by 100 mM TEAB to urea concentration less than 2 M. Trypsin was added at 1:50 trypsin-to-protein mass ratio for the first digestion and 1:100 trypsin-to-protein mass ratio for a second 4 h-digestion.
TMT labeling
Peptide was desalted by Strata X C18 SPE column and vacuum-dried. Peptide was reconstituted in 0.5 M TEAB and processed for TMT kit. One unit of TMT reagent were thawed and reconstituted in acetonitrile. The peptide mixtures were then incubated for 2 h at room temperature and pooled, desalted and dried by vacuum centrifugation.
O-GlcNAcylated peptides enrichment
Peptides were dissolved in IP buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl, 0.5% NP-40, pH 8.0), and the supernatant was transferred to the pre-washed O-GlcNAc pan antibody-conjugated beads, placed on a rotary shaker at 4 °C overnight. After incubation, the beads were washed four times with IP buffer and twice with deionized water. Finally, the peptides bound to the beads were eluted off using 0.1% trifluoroacetic acid eluent, and the collected eluate was dried by vacuum freezing and desalted.
HPLC fractionation
The tryptic peptides were fractionated into fractions by high pH reverse-phase HPLC using Thermo Betasil C18 column (5 μm particles, 10 mm ID, 250 mm length). Briefly, peptides were first separated with a gradient of 8% to 32% acetonitrile (pH 9.0) over 60 min into 60 fractions. Then, the peptides were combined into 6 fractions and dried by vacuum centrifuging.
LC-MS/MS analysis
The tryptic peptides were dissolved in 0.1% formic acid (solvent A), directly loaded onto a home-made reversed-phase analytical column (15 cm length, 75 μm i.d.). The gradient was comprised of an increase from 6% to 23% solvent B (0.1% formic acid in 98% acetonitrile) over 26 min, 23% to 35% in 8 min, and climbing to 80% in 3 min then holding at 80% for the last 3 min, all at a constant flow rate of 400 nL/min on an EASY-nLC 1000 UPLC system. The peptides were subjected to NSI source followed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo) coupled online to the UPLC. The electrospray voltage applied was 2.0 kV. The m/z scan range was 350 to 1800 for full scan, and intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were then selected for MS/MS using NCE setting as 28 and the fragments were detected in the Orbitrap at a resolution of 17,500. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans with 15.0 s dynamic exclusion. Automatic gain control (AGC) was set at 5E4. Fixed first mass was set as 100 m/z.
Mass Spectrometry for Comparative Proteomics contained the sections of Trypsin Digestion, TMT Labeling, HPLC Fractionation, and LC-MS/MS Analysis as described above.
Protein identification and quantification
The secondary mass spectrometry data were retrieved using Maxquant 1.6.15.0. Search parameter settings: the database was Y. pestis 91001 database (downloaded from NCBI on OCT 27, 2020, GenBank assembly accession: GCA_000007885.1), inverse libraries were added to calculate the false positive rate (FDR), and common contamination libraries were added to the database to eliminate the effect of contaminating proteins in the identification results, the enzyme digestion mode was set to Trypsin/P, the number of missed cut sites was set to 2, the minimum length of peptide was set to 7 amino acid residues, the maximum number of peptide modifications was set to 5, the mass error tolerance of the primary parent ion was set to 20.0 ppm and 5 ppm for first search and main search, respectively, and the mass error tolerance of the secondary fragment ion was set to 20.0 ppm. Carbamidomethyl(C) was set as fixed modification and variable modifications were set as [‘Acetyl (Protein Nterm)’, ‘Oxidation (M)’, ‘Deamidation (NQ)’, ‘O-HexNAc’]. The quantification method was set to TMT-6plex, and the FDR for protein identification and PSM identification were set to 1%. The identification data were filtered using localization probability >0.75. The quantitative values of each sample in multiple replicates were obtained separately by multiple experiments. The relative quantification of the differential modification for the modification sites between two samples, and the significant p-value for that differential modification were calculated separately. For the biological replicate samples, statistical analyses such as principal component analysis (PCA), relative standard deviation (RSD) and pearson correlation coefficient were used to assess the quantitative protein reproducibility, respectively.
Western blot
The supernatant was boiled in 5×SDS loading buffer and the bacterial pellet was washed with PBS and boiled in 2×SDS loading buffer. Then, equal proteins were separated on a 12% SDS-PAGE gel, transferred onto an immobilon-P transfer membrane and subjected to immunoblot analysis with appropriate primary antibody and secondary antibody. The images of the immunoblotting results were obtained using the Odyssey SA imaging system (LI-COR).
Detection of HmwC expression
Wide type Y. pestis, ΔhmwC, and ΔhmwC/HmwC were grown at 26 °C in TMH medium with or without calcium to OD620nm = 1.0, then shifted respectively to 26 °C (Fv) and 37 °C (Mh) for another 4 h’s incubation. The protein HmwC in bacterial pellets were detected by WB with anti-SPINDLY mouse monoclonal antibody and IRDye 800CW-conjugated goat anti-mouse IgG.
In vitro O-GlcNAcylation assays
For the in vitro O-GlcNAcylation assay, a reaction mixture containing 5 μL purified proteins (OsdY, AlgL, OsdYS83A or AlgLS5A), 0-20 μL OGTs (HmwC, HmwCD217A or HmwCH276A), 24-44 μL ddH2O, 6 μL UDP-GlcNAc, 5 μL 10×OGT assay buffer was incubated in a 37 °C shaker for 2 h. Reactions were quenched with 5×SDS loading buffer and boiled for 10 mins. Then the proteins were detected by WB with anti-His, anti-SPINDLY, and anti-O-GlcNAc mouse monoclonal primary antibodies, and 2nd antibody IRDye 800CW-conjugated goat anti-mouse IgG.
GST pull-down assay
The GST-tagged recombinant proteins GST-HmwC, GST-HmwCD217A and GST-HmwCH276A were incubated with glutathione-Sepharose 4B beads for 4 h at 4 °C, respectively, and then the beads were washed thoroughly with PBS, followed by the addition of His-OsdY and His-AlgL, respectively, and then incubated overnight at 4 °C. After washing beads, the bound proteins were analyzed by WB with anti-GST and anti-His monoclonal antibodies.
Glycoprotein staining
After electrophoresis, the gel was completely immersed in 100 mL of 50% methanol, soaked for 30 min, and then washed twice with 3% acetic acid. The gel was then transferred to 25 mL of oxidation solution for 15 min and washed twice with 3% acetic acid. Subsequently, the gel was transferred to 25 mL of glycoprotein dye and shaken for 15 minutes. The gel was then transferred to 25 mL of reducing solution for 5 minutes. Finally, the gel was thoroughly washed with 3% acetic acid followed by ultrapure water. Glycoproteins were visualized as magenta bands. It is recommended that these steps be performed in a fume hood.
Characterization of the phenotypes of the hmwC mutant
Growth rate determination
Wild-type Y. pestis, mutants, and complemented strains were cultured in Luria-Bertani (LB) broth at 26 °C or chemically defined TMH medium. Then, bacterial cultures were diluted 1:20 in fresh LB, LB containing 2.5% NaCl and TMH medium, respectively, and incubated continuously in a 220 rpm 26 °C shaker. Bacterial growth was monitored every 2 h by measuring the absorbance of OD620nm for a total of 32 h.
Acid resistance assay
Bacteria cells of Y. pestis strains cultured in LB to OD620nm = 1.0 were resuspended with 20 mM glucose minimal medium. Bacterial resuspensions diluted 1:100 with pH 3.5 minimal medium were regarded as experimental groups, while those diluted with pH 6.5 minimal medium were regarded as the control. After incubation at room temperature for 30 min (and 60 min in a parallel group), the number of living bacteria was determined by plating the bacterial dilutions onto a Hottinger’s agar plate in triplicate.
Biofilm formation determination
The bacterial solutions of Y. pestis strains cultured to OD620nm = 1.0 as described previously were left at 4 °C overnight, then diluted 1:20 in fresh LB medium, added to 24-well plate at 1 ml per well, and incubated continuously in a 220 rpm 26 °C shaker for 24 h. The bacteriosphere formed at the junction of the liquid and gas phases was biofilm. The OD620nm value of bacterial solutions in 24-well plate was measured. After gently washing the biofilm with deionized water and fixing it at 80 °C for 15 min, 3 ml of 0.1% crystalline violet staining solution was added to each well for 15 min, and after washing away the free crystalline violet, the stained biofilm was dissolved with 3 ml of dimethyl sulfoxide and the OD570nm value was determined. The amount of biofilm formation was expressed as OD570nm/OD620nm value.
Heat-shock and cold-shock tolerance assay
The bacterial solutions of Y. pestis strains cultured to OD620nm = 1.0 were resuspended and diluted 1:100 with 20 mM glucose minimal medium described previously50. 100 μl of bacterial solution was added in parallel to each EP tube and those placed at 26 °C were the control groups. For heat-shock experiment, bacterial solutions placed in a 50 °C water bath for 10 min, 20 min and 40 min, respectively, were the experimental groups. For cold-shock experiment, the bacterial solution placed at −20 °C and 4 °C for 24 h were the experimental groups. Finally, the number of living bacteria was determined by plating the bacterial dilutions onto a Hottinger’s agar plate in triplicate.
Hydrogen peroxide resistance assay
The bacterial solutions of Y. pestis strains cultured to OD620nm = 1.0 were adjusted to OD620nm = 0.8 in parallel with PBS. 200 μl of the above bacterial solution was mixed thoroughly with 8 ml of 0.7% semi-solid medium respectively, and the mixture was quickly spread flat on the base medium until solidified. Circular filter paper with a diameter of 6 mm was placed in the center of the petri dish and 5 μl of hydrogen peroxide solution of appropriate concentration was added to each paper piece. After that the petri dishes were incubated at 26 °C for 16 h and the diameter of the inhibition circle was measured to reflect the hydrogen peroxide resistance of Y. pestis strains.
Detection of hydrogen peroxide (H2O2) content in Y. pestis
The quantification of hydrogen peroxide content was performed using a Hydrogen Peroxide Assay Kit (Beyotime Biotechnology, China). Y. pestis was cultured in LB to OD620nm = 1.0, followed by centrifugation to collect bacteria cells. Aliquot 1 mL of the reagent I to resuspend 5×106 bacteria, and the bacteria were then sonicated for lysis, followed by centrifugation at 8000 g for 10 minutes at 4 °C. The supernatant was collected and the hydrogen peroxide concentration were determined analyzed according to the manufacturer’s instructions.
Determination of survival capability in macrophages
The wild-type Y. pestis, ΔhmwC and ΔhmwC/HmwC were cultured to OD620nm = 1.0, then shifted to 37 °C for an additional 2 h. RAW264.7 were seeded into 24-well plates at a density of 5 × 105 cells/well before infection. RAW264.7 were infected with Y. pestis strains at a multiplicity of infection (MOI) of 10. At 0.5 hpi, gentamicin at 30 μg/ml was added to each well to kill the extracellular bacteria. After 0.5 h of continued incubation, the culture medium was replaced with fresh DMEM containing 10% FBS and 2 μg/ml gentamicin until the end of the experiment. At 0.5, 2, 4, 8 and 24 hpi., the RAW264.7 were thoroughly washed with PBS and lysed in sterile H2O containing 0.1% Triton X-100. The number of living bacteria was determined as described previously. This experiment was independently performed three times in biological triplicates.
Animal infection and the survival curves
Bal b/c mice were purchased from Vitalriver Company (Beijing, China). Mice were maintained in an animal facility with an ambient temperature of 20-25 °C and a relative humidity of 30-60%. The bacterial solutions of wild-type Y. pestis and ΔhmwC cultured until log phase were adjusted to OD620nm = 1.0 in parallel with PBS. 6 to 8-week-old female mice (n = 10/group) were used in this experiment, and each was subcutaneously challenged with 100 μl of bacterial suspension (about 100 CFU), and the survival of challenged mice was observed for 14 consecutive days. Survival curves for each group of mice were plotted using GraphPad Prism 8.0. To determine the bacterial burden in organs, livers, spleens and lungs of the challenged mice were harvested when the mice were moribund, i.e. 4 dpi., and the collected tissues were homogenized in sterile PBS to measure live bacteria, as described previously. Statistics of bacterial burden measurements were performed using the two-tailed unpaired Student’s t-test and p values ≤ 0.05 were considered significant. All the animal experiments were conducted in strict accordance with the Guidelines for the Welfare and Ethics of Laboratory Animals of China (license number IACUC-IME-2022-025) and approved by the Institutional Animal Care Committee of Beijing Institute of Epidemiology and Microbiology.
Prediction of protein structure
The alignment of protein amino acid sequences was completed using Espript 3.0 online tool The tertiary structures of protein were predicted using AlphaFold/2.3.1 software and visualized by PyMOL2.3.0 software.
Molecular docking methods
Auto Dock Tools 1.5.6 was used to hydrogenate the target protein molecule and the optimal conformation UDP-GlcNAC molecule. Auto Dock Vina v.1.2.0 software was used to perform molecular docking simulation of the target protein and ligand molecule using a Lamarckian genetic algorithm as the docking algorithm, semi-flexible docking as the docking method, exhaustiveness was set to 8, and maximum number of output conformations was set to 9. PyMOL2.3.0 software was used for visualization of the docking results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the data set identifier PXD042927 and PXD042928. All data generated or analyzed during this study are included in this published article and supplementary information files. Sequences of bacteria strains used were download from NCBI database (https://www.ncbi.nlm.nih.gov/assembly). Source data are provided with this paper.
References
Torres, C. R. & Hart, G. W. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 (1984).
Olivier, N. B., Chen, M. M., Behr, J. R. & Imperiali, B. In vitro biosynthesis of UDP-N,N’-diacetylbacillosamine by enzymes of the Campylobacter jejuni general protein glycosylation system. Biochemistry 45, 13659–13669 (2006).
Sharon, N. Celebrating the golden anniversary of the discovery of bacillosamine, the diamino sugar of a Bacillus. Glycobiology 17, 1150–1155 (2007).
Azadi, P. & Heiss, C. Mass spectrometry of N-linked glycans. Methods Mol. Biol. 534, 37–51 (2009).
Hart, G. W., Housley, M. P. & Slawson, C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 (2007).
Wells, L., Vosseller, K. & Hart, G. W. Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science 291, 2376–2378 (2001).
Wang, Z., Gucek, M. & Hart, G. W. Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl Acad. Sci. USA 105, 13793–13798 (2008).
Zeidan, Q. & Hart, G. W. The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J. Cell Sci. 123, 13–22 (2010).
Hart, G. W., Slawson, C., Ramirez-Correa, G. & Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 (2011).
Macauley, M. S. & Vocadlo, D. J. Increasing O-GlcNAc levels: an overview of small-molecule inhibitors of O-GlcNAcase. Biochim. Biophys. Acta 1800, 107–121 (2010).
Wang, Y., Zhu, J. & Zhang, L. Discovery of cell-permeable O-GlcNAc transferase inhibitors via tethering in situ click chemistry. J. Med. Chem. 60, 263–272 (2017).
Nagel, A. K. & Ball, L. E. O-GlcNAc transferase and O-GlcNAcase: achieving target substrate specificity. Amino Acids 46, 2305–2316 (2014).
Haltiwanger, R. S., Holt, G. D. & Hart, G. W. Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins. Identification of a uridine diphospho-N-acetylglucosamine:peptide beta-N-acetylglucosaminyltransferase. J. Biol. Chem. 265, 2563–2568 (1990).
Dong, D. L. & Hart, G. W. Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 269, 19321–19330 (1994).
Hanover, J. A., Krause, M. W. & Love, D. C. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta 1800, 80–95 (2010).
Copeland, R. J., Bullen, J. W. & Hart, G. W. Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am. J. Physiol. Endocrinol. Metab. 295, E17–E28 (2008).
Singh, J. P., Zhang, K., Wu, J. & Yang, X. O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 356, 244–250 (2015).
Banerjee, P. S., Lagerlof, O. & Hart, G. W. Roles of O-GlcNAc in chronic diseases of aging. Mol. Asp. Med. 51, 1–15 (2016).
Ostrowski, A., Gundogdu, M., Ferenbach, A. T., Lebedev, A. A. & van Aalten, D. M. Evidence for a functional O-linked N-acetylglucosamine (O-GlcNAc) system in the thermophilic bacterium thermobaculum terrenum. J. Biol. Chem. 290, 30291–30305 (2015).
Thomasen, R. S. S. et al. Absence of N-acetylglucosamine glycosylation on listeria monocytogenes wall teichoic acids promotes fatty acid tolerance by repulsion from the bacterial surface. Front. Microbiol. 13, 897682 (2022).
Shi, W. W. et al. Structure of a novel O-linked N-acetyl-D-glucosamine (O-GlcNAc) transferase, GtfA, reveals insights into the glycosylation of pneumococcal serine-rich repeat adhesins. J. Biol. Chem. 289, 20898–20907 (2014).
Martinez-Fleites, C. et al. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat. Struct. Mol. Biol. 15, 764–765 (2008).
Khayatan, B. et al. A putative O-linked beta-N-acetylglucosamine transferase is essential for hormogonium development and motility in the filamentous cyanobacterium nostoc punctiforme. J. Bacteriol. 199, e00075–17 (2017).
Shen, A., Kamp, H. D., Gründling, A. & Higgins, D. E. A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Dev. 20, 3283–3295 (2006).
Jinek, M. et al. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat. Struct. Mol. Biol. 11, 1001–1007 (2004).
Lazarus, M. B., Nam, Y., Jiang, J., Sliz, P. & Walker, S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567 (2011).
Kawai, F. et al. Structural insights into the glycosyltransferase activity of the Actinobacillus pleuropneumoniae HMW1C-like protein. J. Biol. Chem. 286, 38546–38557 (2011).
Gross, J. et al. 3rd: The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J. Biol. Chem. 283, 26010–26015 (2008).
Grass, S. et al. 3rd: The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog. 6, e1000919 (2010).
Choi, K. J., Grass, S., Paek, S., Geme, J. W. St 3rd & Yeo, H. J. The Actinobacillus pleuropneumoniae HMW1C-like glycosyltransferase mediates N-linked glycosylation of the Haemophilus influenzae HMW1 adhesin. PLoS ONE 5, e15888 (2010).
Perry, R. D. & Fetherston, J. D. Yersinia pestis-etiologic agent of plague. Clin. Microbiol. Rev. 10, 35–66 (1997).
Yang, R. et al. Yersinia pestis and Plague: some Knowns and Unknowns. Zoonoses 3, 5 (2023).
Yang, R. & Butler, T. Discovery of the plague pathogen: lessons learned. Adv. Exp. Med. Biol. 918, 27–33 (2016).
Cornelis, G. R. et al. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62, 1315–1352 (1998).
Hinnebusch, B. J. et al. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296, 733–735 (2002).
Viboud, G. I. & Bliska, J. B. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 59, 69–89 (2005).
Galan, J. E. & Wolf-Watz, H. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573 (2006).
Zhao, Y. et al. Single-cell transcriptomics of immune cells in lymph nodes reveals their composition and alterations in functional dynamics during the early stages of bubonic plague. Sci. China Life Sci. 66, 110–126 (2023).
Han, Y. et al. Microarray analysis of temperature-induced transcriptome of Yersinia pestis. Microbiol. Immunol. 48, 791–805 (2004).
Bolin, I., Portnoy, D. A. & Wolf-Watz, H. Expression of the temperature-inducible outer membrane proteins of yersiniae. Infect. Immun. 48, 234–240 (1985).
Suomalainen, M. et al. Temperature-induced changes in the lipopolysaccharide of Yersinia pestis affect plasminogen activation by the pla surface protease. Infect. Immun. 78, 2644–2652 (2010).
Hinnebusch, B. J., Fischer, E. R. & Schwan, T. G. Evaluation of the role of the Yersinia pestis plasminogen activator and other plasmid-encoded factors in temperature-dependent blockage of the flea. J. Infect. Dis. 178, 1406–1415 (1998).
Mehigh, R. J., Sample, A. K. & Brubaker, R. R. Expression of the low calcium response in Yersinia pestis. Microb. Pathog. 6, 203–217 (1989).
Van Acker, H. et al. Biofilm-grown Burkholderia cepacia complex cells survive antibiotic treatment by avoiding production of reactive oxygen species. PLoS ONE 8, e58943 (2013).
Guo, L. et al. Reactive oxygen species inhibit biofilm formation of Listeria monocytogenes. Microb. Pathog. 127, 183–189 (2019).
Kim, S. Y. et al. The gene yggE functions in restoring physiological defects of Escherichia coli cultivated under oxidative stress conditions. Appl. Environ. Microbiol. 71, 2762–2765 (2005).
Thornton, T. M., Swain, S. M. & Olszewski, N. E. Gibberellin signal transduction presents ellipsisthe SPY who O-GlcNAc’d me. Trends Plant Sci. 4, 424–428 (1999).
Hartweck, L. M., Scott, C. L. & Olszewski, N. E. Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161, 1279–1291 (2002).
Zhou, D. et al. Defining the genome content of live plague vaccines by use of whole-genome DNA microarray. Vaccine 22, 3367–3374 (2004).
Liu, W. et al. ProteiN ACETYLATION MEDiated by YfiQ and CobB is involved in the virulence and stress response of yersinia pestis. Infect. Immun. 86, e00224–18 (2018).
Acknowledgements
We thank Hui Li at the National Center for Protein Sciences at Peking University in Beijing, China, for assistance with providing experimental instrument support. This work was supported by grants from the National Key Research and Development Program of China (2022YFC2303503) and the National Natural Science Foundation of China (No. 32270189).
Author information
Authors and Affiliations
Contributions
Z.D. designed the study. S.C. performed the experiments unless otherwise noted. Y.R., T.W., and Y. Zhang analyzed the proteomics data. Y.T., Y. Zhou, H.C., G.W., and Yu Zhang performed the mouse experiments. R.Y. and Y.S. reviewed the manuscript and raised constructive suggestions. Z.D., S.C., and R.Y. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cao, S., Wang, T., Ren, Y. et al. A protein O-GlcNAc glycosyltransferase regulates the antioxidative response in Yersinia pestis. Nat Commun 15, 7062 (2024). https://doi.org/10.1038/s41467-024-50959-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-50959-w