Abstract
Enoblituzumab, an immunotherapeutic agent targeting CD276, shows both safety and efficacy in activating T cells and oligodendrocyte-like cells against various cancers. Preclinical studies and mouse models suggest that therapies targeting CD276 may outperform PD1/PD-L1 blockade. However, data from mouse models indicate a significant non-responsive population to anti-CD276 treatment, with the mechanisms of resistance still unclear. In this study, we evaluate the activity of anti-CD276 antibodies in a chemically-induced murine model of head and neck squamous cell carcinoma. Using models of induced and orthotopic carcinogenesis, we identify ITGB6 as a key gene mediating differential responses to anti-CD276 treatment. Through single-cell RNA sequencing and gene-knockout mouse models, we find that ITGB6 regulates the expression of the tumor-associated chemokine CX3CL1, which recruits and activates PF4+ macrophages that express high levels of CX3CR1. Inhibition of the CX3CL1-CX3CR1 axis suppresses the infiltration and secretion of CXCL16 by PF4+ macrophages, thereby reinvigorating cytotoxic CXCR6+ CD8+ T cells and enhancing sensitivity to anti-CD276 treatment. Further investigations demonstrate that inhibiting ITGB6 restores sensitivity to PD1 antibodies in mice resistant to anti-PD1 treatment. In summary, our research reveals a resistance mechanism associated with immune checkpoint inhibitor therapy and identifies potential targets to overcome resistance in cancer treatment.
Similar content being viewed by others
Introduction
Immune checkpoint inhibitors (ICIs), such as those targeting programmed cell death 1 (PD1) and its ligand (PD-L1) or cytotoxic T lymphocyte-associated protein 4 (CTLA4), have revolutionized solid tumor therapy, significantly improving patient survival rates1,2. Compared to PD-L1, CD276 (B7 homolog 3 protein) is more robustly and specifically expressed in various epithelial-origin cancers, making it a promising target for anti-cancer therapy3. For instance, we have previously demonstrated that CD276 is enriched in the cancer stem cells (CSCs) of head and neck squamous cell carcinoma (HNSCC) and can serve as a therapeutic target for HNSCC4,5. Moreover, multiple clinical trials have confirmed the antitumor effectiveness and safety of enoblituzumab, a humanized Fc-engineered CD276-targeting antibody, in patients with localized prostate cancer (PCa), advanced HNSCC, and non-small cell lung cancer (NSCLC)6,7. Despite the advancements in α-CD276-based immunotherapy, the response rates remain suboptimal, and the molecular mechanisms underlying this resistance are not yet fully understood8.
The failure of the immune response is largely governed by the interaction between the immune system and solid tumors. Previous studies have suggested that the negative regulatory function of CD276 is achieved through its binding to receptors expressed on activated T cells, thereby inhibiting their proliferation and production of immunosuppressive cytokines9. Additionally, various studies have highlighted the critical role of myeloid innate immune cells, such as granulocytes, monocytes, macrophages, and dendritic cells (DCs), in shaping the immune responses against cancer cells10. For example, CD276 influences the migration and differentiation of M2 macrophages via the CCL2-CCR2 axis, promoting ovarian cancer progression11. This underscores the significant role of innate immunity in the efficacy of α-CD276 cancer immunotherapy. In recent years, PF4+ macrophages—a subset of tumor-associated macrophages (TAMs) characterized by their expression of Platelet Factor 4—have garnered significant attention for their crucial functions within the tumor microenvironment and their potential effects on immunoregulation12,13,14. As central regulators, these macrophages modulate inflammation and immune responses by secreting a wide range of cytokines and chemokines15,16. This secretion profoundly influences the recruitment and activation of various immune cells, playing a critical role in tumor immunosurveillance. However, despite their recognized importance, the precise roles and mechanisms of PF4+ macrophages in cancer immunotherapy remain incompletely understood.
CD8+ T lymphocytes, also known as cytotoxic T cells, are pivotal in antitumor immunity due to their ability to identify and destroy cancer cells. Their critical role in tumor suppression underpins various cancer immunotherapy strategies, such as checkpoint inhibition and adoptive cell transfer. Specifically, GZMB+ CD8+ T cells, characterized by their expression of Granzyme B, represent a highly activated cytotoxic state and exhibit an enhanced capacity to recognize and eradicate tumor cells17,18. Recent research has also highlighted a unique subset of CXCR6+ CD8+ T cells within the tumor immune landscape, emphasizing their vital contribution to targeting and eliminating malignant cells19,20. However, the efficacy of these cells is significantly influenced by their complex interactions with myeloid-derived innate immune cells within the tumor microenvironment21,22. Despite their crucial role, the precise regulatory mechanisms controlling the effectiveness of CXCR6+ CD8+ T cells remain largely unclear, highlighting an urgent need for further research to unravel the complexities of tumor immunology23,24.
Integrins are multifunctional αβ-heterodimers that mediate interactions between tumor cells and their microenvironment, including immune cells, stromal cells, and the extracellular matrix (ECM). Dysregulation of these heterodimeric transmembrane receptors has been linked to tumor growth, invasion, angiogenesis, and therapy resistance25. Recent studies have demonstrated that inhibition of αvβ3 integrin can suppress PD-L1 expression, thereby facilitating tumor immune evasion26. Furthermore, ITGB6 has been identified as a critical player in tumor immune evasion, particularly in colorectal cancer. Its expression activates latent TGF-β, which leads to the suppression of antitumor immune responses and resistance to immune checkpoint blockade therapies. Previous studies have demonstrated that antibody-mediated inhibition of integrin αvβ6 can trigger a robust cytotoxic T cell response. This approach effectively overcomes the resistance of ITGB6-expressing tumors to PD1 blockade therapy, significantly enhancing the efficacy of anti-PD1 treatment27.
In this work, our study aims to further elucidate the role of ITGB6 in mediating resistance to CD276 antibody therapy. We identify ITGB6 as a key gene driving this resistance and characterize a specific subset of PF4+ macrophages recruited by ITGB6 through the CX3CL1-CX3CR1 axis. These macrophages are significantly correlated with poorer patient survival and promote the exhaustion of tumor-specific CXCR6+CD8+ T cells via the CXCL16-CXCR6 axis. This mechanism facilitates tumor immune evasion and contributes to the progression of resistance to immunotherapy. In summary, our findings underscore the importance of ITGB6 in the immune evasion strategies employed by tumors. They suggest that combining immune checkpoint inhibitors with anti-ITGB6 therapy could be a promising and safe approach to overcoming immune evasion and enhancing the therapeutic success of current immunotherapies for HNSCC.
RESULTS
Murine HNSCC displays different responses upon α-CD276 treatment
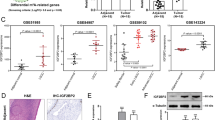
To study the therapeutic effect of a CD276 blocking antibody in an immunocompetent mouse model of head and neck squamous cell carcinoma (HNSCC), we first treated wild-type (WT) C57BL/6 mice with 4-nitroquinoline 1-oxide (4NQO) for 16 weeks, followed by an additional 8 weeks of normal drinking water. Mice bearing similarly sized HNSCC lesions were then selected for α-CD276 antibody or control IgG treatment (Fig. 1A). The α-CD276 antibody was administered intraperitoneally every other day at a dose of 10 mg/kg for five doses, as previously described5. Mice were monitored every four days from the initial α-CD276 injection using magnetic resonance imaging (MRI) to track tumor growth (Fig. 1B). Resistance was defined according to the response evaluation criteria in solid tumors (RECIST) as tumors demonstrating an initial therapeutic response by MRI, followed by growth to >120% of the original tumor size (Fig. 1B). We found that 38.9% of animals (7 out of 18) were responsive to the α-CD276 therapy, showing partial reduction in tumor progression or near-complete tumor eradication. In contrast, 61.1% of animals (11 out of 18) were not sensitive to α-CD276 treatment, with tumors growing rapidly (Fig. 1C). Animals that became symptomatic or lost more than 20% of their original body weight were sacrificed (Fig. 1C). Forty-one days after the initial antibody injection, tongues from HNSCC-bearing mice were excised (Fig. 1D). Formalin-fixed and paraffin-embedded sections of the tongue tumor lesions were examined histologically (Fig. 1D). Compared to the control and resistant groups, the sensitive group demonstrated a significant decrease in tumor incidence, area, and grading (Supplementary Fig. 1A–C)4,5. Considering that IHC analysis conducted after 41 days of treatment may more accurately represent the state of residual disease compared to initial antitumor mechanisms, we followed the previously established methods for mouse tumor induction and treatment. Subsequently, we selected six mice from each group at both 13 and 27 days following α-CD276 treatment for additional analysis. We hypothesized that disparities in treatment outcomes with the α-CD276 antibody could be attributed to variations in CD276 expression. To test this, we conducted immunohistochemical (IHC) staining of CD276, finding no significant differences between the three groups at the three distinct time points (Supplementary Fig. 1D). Ki67, a marker indicative of cell proliferation, showed no significant differences in proliferative activity among the three tumor cell groups at all observed time points (Supplementary Fig. 1E). However, IHC staining for cleaved-caspase-3 indicated elevated levels of apoptosis in the sensitive samples compared to the resistant ones, with all three time points showing higher cellular apoptosis levels in the sensitive group in tumor cells. There were no significant differences in apoptosis levels within the stromal cells (Supplementary Fig. 1F, G). Immunofluorescence co-staining for CD8 and GZMB at these time points demonstrated a significant increase in the infiltration of both CD8+ T cells and CD8+ GZMB+ T cells in the sensitive group compared to the resistant and IgG control groups (Supplementary Fig. 1H, I).
A The experimental design of the HNSCC tumorigenesis model and treatment strategy. B Representative examples of head and neck magnetic resonance imaging (MRI) for different groups (IgG = 8, Resistant = 11, Sensitive = 7 mice). The dashed area is the boundary of the tumor. D, days. C Tumor growth curves of each mouse receiving Anti-CD276 antibody or control IgG treatment. Measurements were taken every 4 days (IgG = 8, Resistant = 11, Sensitive = 7 mice). D Representative images of 4NQO-induced HNSCC (upper) and corresponding HE staining(lower) (IgG = 8, Resistant = 11, Sensitive = 7 mice). Scale bar, 1 mm (upper), 100 μm (lower). E Workflow strategy diagram for the isolation of HNSCC cells and single-cell RNA sequencing (IgG = 6, Resistant = 3, Sensitive = 3 mice). F UMAP plot showing the results after unbiased clustering. Subpopulations of epithelial cells, endothelial cells, fibroblast cells, and immune cells were identified, with each cell type colored. G Copy number of epithelial cells was annotated using CopyKAT. Cells are colored by ploidy status. H UMAP plots showing the subclusters of epithelial cells. I Heatmap of characteristic genes in the subclusters of epithelial cells. Each cell cluster is represented by three specifically expressed genes. J The UMAP plots showing the Scissor-selected epithelial cells. The red and gray dots are Scissor+ (cluster 1, worse survival) and Scissor− (cluster 0, good survival) cells. K The proportions of subclusters of epithelial cells in Scissor 0 and Scissor 1. L Volcano plot displaying the -Log10 FDR vs Log2 fold-change of genes differentially expressed between DEG1 (Scissor 0 and Scissor 1, left) and DEG2 (CD276R and CD276S, right) in Epithelial cells. R Resistant, S Sensitive, M Venn diagram displaying the overlap of DEGs in Epithelial cells. The P values in Supplementary Data 1 and 2 were calculated by the Wilcoxon rank sum test. P values are two-sided and exact. Adjustments for multiple comparisons were made using the Bonferroni correction method. Source data and exact p values are provided as a Source Data file.
To further investigate whether the reduction in tumor growth following α-CD276 treatment in the induced tumor model was mediated by CD8 or CD4 T cells, the mice from the sensitive group were systematically divided into four groups and treated with anti-CD4 and anti-CD8 depleting antibodies, respectively (Supplementary Fig. 2A). Histological analyses indicated that depleting CD8+ T cells significantly reversed the growth inhibition mediated by α-CD276, whereas depletion of CD4+ T cells showed no difference (Supplementary Fig. 2B). Analysis of the overall lesion quantity and area further confirmed that depletion of CD8+ T cells notably attenuated the tumor growth inhibition mediated by CD276 blockade, whereas depletion of CD4+ T cells did not have an impact (Supplementary Fig. 2C).
To search for systematic biomarkers responsive to α-CD276 treatment, we performed single-cell RNA sequencing (scRNA-seq) on samples pooled from three fresh tissue collections after microdissection, including one resistant, one sensitive, and two control pooled samples (Fig. 1E). ScRNA-seq yielded 21,079 high-quality transcriptomes after quality control and filtering (Fig. 1F and Supplementary Fig. 2D). Using characteristic canonical cell markers, four major cell types were identified: epithelial cells (Krt17, Krt13, Krt6a, Krt14, and Krt5), immune cells (Cd74, Lyz2, H2-Aa, H2-Ab1, and C1qb), fibroblasts (Col1a2, Col3a1, Col1a1, Igfbp5, and Col5a2), and endothelial cells (Vim, Pecam1, Lrg1, Ackr1, and Ly6c1) (Fig. 1F and Supplementary Fig. 2E). The proportions of each cell type were similar across different groups (Supplementary Fig. 2F).
To understand the heterogeneity and molecular characteristics of epithelial cells responsible for the efficacy of α-CD276 treatment, we first used copyKAT28 to distinguish malignant cells from non-malignant cells. We successfully identified 16,674 aneuploid tumor cell subpopulations with distinct gene expression profiles (Fig. 1G). Subsequently, we extracted the malignant cells and divided them into four different subclusters: cell cycle, differentiated, partial epithelial-to-mesenchymal transition (p-EMT), and stress subclusters, consistent with our previous study (Fig. 1H and S2G)29. Figure 1I demonstrates the top three genes preferentially expressed in each subcluster. We then performed Scissor analysis30, which utilizes phenotype data collected from bulk assays to identify the most phenotype-associated cell subpopulations from single-cell data. We identified 286 Scissor+ tumor cells related to poor prognosis in HNSCC patients, most of which displayed a p-EMT phenotype (Fig. 1J, K). We compared the differentially expressed genes (DEGs) between Scissor+ tumor cells and other background cells, identifying 150 upregulated genes and 92 downregulated genes (Fig. 1L). Next, we compared the DEGs between CD276 resistant (CD276R) and CD276 sensitive (CD276-S) Scissor+ tumor cells, identifying 26 upregulated genes and 78 downregulated genes (Fig. 1L). By combining these two sets of DEGs, we found three commonly upregulated genes: ITGB6, PRNP, and CAV1, suggesting that these genes might be involved in the resistance to α-CD276 treatment in HNSCC (Fig. 1M, Supplementary Data 1, Supplementary Data 2).
ITGB6 is a crucial gene that mediates divergent responses to α-CD276 treatment
To address that, we used the mouse oral cancer cell line Moc1 to generate stable knockdown cells for CAV1, ITGB6, and PRNP (Supplementary Fig. 3A). Knockdown and control Moc1 cells were subcutaneously injected into the base of the tongues of C57BL/6 mice, which were then treated with either IgG control or α-CD276 antibody (Supplementary Fig. 3B, C). We observed that knockdown of all three genes slightly affected tumor growth, while treatment with the α-CD276 antibody led to a significant inhibition of tumor growth overall (Fig. 2A, B). Unexpectedly, we detected a slightly additive effect on tumor growth after PRNP and CAV1 depletion upon α-CD276 treatment (Fig. 2A, B). However, the tumor size in mice with ITGB6 depletion and α-CD276 treatment decreased by 53.24% (Fig. 2A, B), supporting the notion that loss of ITGB6 is critical for increasing the sensitivity of HNSCC to α-CD276 treatment.
A, B Growth curves (A) and representative images (B) of HNSCC in mice transplanted in situ in different treatment groups. Data are expressed as mean ± standard deviation (SD) (n = 6 mice). C, D Representative images of ITGB6 IHC staining (C) and percentage of ITGB6+ cells (D) in different treatment groups. Scale bar, 50 μm. Data are presented as mean ± SD (n = 8 mice). P values are presented by two-tailed unpaired Student’s t test. E Representative image of 4NQO-induced HNSCC (upper) and display of histological characteristics of HNSCC in different groups by H&E (down) (n = 8 mice). Scale bar, 1 mm (upper) and 100 μm (lower). F Representative immunofluorescence (IF) staining images of CD8 (green) and GZMB (red) (upper). Statistical analysis of the ratio of CD8+ cells and CD8+ GZMB+ cells to CD8+ cells (CD8T killing capacity) in different treatment groups (down). Data are expressed as mean ± SD (n = 8 mice). Scale bar, 25 μm. P values were calculated by two-tailed unpaired Student’s t test. G, H Representative images of KI67 (G) and Caspase-3 (H) IHC staining (left) and quantitation of the percentage of KI67+ (G) and Caspase-3+ (H) cells (right) in different treatment groups within tumor cells. Scale bar, 100 μm. Data are shown as mean ± SD (n = 8 mice). P values were calculated by two-tailed unpaired Student’s t-test. I Quantitation of the percentage of Caspase-3+ cells in different treatment groups within stromal cells. Data are shown as mean ± SD (n = 8 mice). P values were calculated by two-tailed unpaired Student’s t-test. J Display of histological characteristics of HNSCC in different groups by H&E (down) and the representative image of 4NQO-induced HNSCC (upper) (n = 8 mice). Scale bar, 100 μm (down) and 1 mm (upper). K, L Quantification of tongue SCC number (J) and lesion area (K) in different treatment groups. Data are presented as mean ± SD (n = 8 mice). P values were calculated by two-tailed unpaired Student’s t-test. Source data and exact p values are provided as a Source Data file.
To investigate the relationship between ITGB6 and CD276 expression, we performed statistical analyses on the IHC scores of ITGB6 and CD276 from samples collected at three different time points across control, resistant, and sensitive groups. Results showed significantly lower ITGB6 IHC scores in the sensitive group compared to the other groups. However, no significant correlation was found between ITGB6 and CD276 IHC scores among all groups (Supplementary Fig. 3D–F).
To assess how CD276 blockade affects CD8+ or CD4+ T cell-dependent antitumor responses in a transplanted mouse tumor model, mice injected with ITGB6-knockout Moc1 cell lines were allocated into four groups, receiving treatments with anti-CD4 and anti-CD8 depleting antibodies, respectively (Supplementary Fig. 3G). Histological examination revealed that CD8+ T cell depletion significantly mitigated the aggressive tumor growth inhibition induced by anti-CD276 treatment, whereas CD4+ T cell depletion had no notable effect on this inhibition (Supplementary Fig. 3H). Tumor volume analyses further confirmed that CD8+ T cell depletion substantially reduced the antitumor effect of CD276 blockade, while CD4+ T cell depletion had no discernible impact (Supplementary Fig. 3I).
To further explore the impact of ITGB6 on the progression of HNSCC, we employed the CRISPR-Cas9 system to knock out ITGB6 in HNSCC cells with high ITGB6 expression (SCC15 cell line: sgGFP, sgITGB6-1, sgITGB6-2), as depicted in Supplementary Fig 4A. Following the deletion of ITGB6, we assessed the expression of CD276 protein and found that the knockout of ITGB6 did not alter the expression of CD276 protein (Supplementary Fig. 4A). Experiments examining cell proliferation and clonogenicity suggested that the knockout of ITGB6 had no significant effect on the proliferation or colony-forming capabilities of HNSCC cells (Supplementary Fig. 4B, C). Additionally, when compared to the sgGFP control cells, the migration and invasion capabilities of ITGB6 gene knockout cells showed no significant changes (Supplementary Fig. 4D, E). Concurrently, flow cytometry was utilized to examine the apoptosis levels of ITGB6 gene knockout and control HNSCC cells, revealing that ITGB6 gene knockout did not impact the apoptosis levels of HNSCC cells (Supplementary Fig. 4F). In summary, these data support the conclusion that ITGB6 does not have a direct impact on the progression of HNSCC.
ITGB6 protein expression was then assessed at different stages of spontaneous mouse HNSCC. Overall, we observed an increasing trend of ITGB6 protein levels along with HNSCC development and progression (Supplementary Fig. 5A, B). Importantly, we noticed a dramatic elevation of ITGB6 protein level in resistant tumors compared with control or sensitive tumors (Fig. 2C, D and S5C). To further characterize the role of ITGB6 in HNSCC, we generated Itgb6 conditional knockout (Itgb6flox) mice, in which the exon2 and 3 of Itgb6 gene is floxed (Supplementary Fig. 5D). Itgb6flox/flox mice were crossed with K14creER mice to generate K14creER; Itgb6flox/flox conditional knockout (ITGB6-cKO) mice, allowing us to knockout Itgb6 gene in the oral epithelium upon Tamoxifen administration. K14creER; Itgb6wt/wt were used as control (ITGB6-ctl). To examine the effect of Itgb6 gene during oral epithelium homeostasis, we harvested tongue tissues after Tamoxifen administration from 1- and 12-month-old ITGB6-ctl and ITGB6-cKO mice. Of note, epithelium-specific deletion of ITGB6 in ITGB6-cKO had no noticeable consequences on normal oral epithelium tissue (Supplementary Fig. 5E, F). In ITGB6-ctl mice, a low level of epithelial ITGB6 expression was seen at 1-month and 12-month of age; conversely, ITGB6 expression was almost absent in ITGB6-cKO mice, confirming the knockout efficiency (Supplementary Fig. 5G).
Four-weeks old ITGB6-ctl and ITGB6-cKO mice were injected with Tamoxifen and then treated with 4NQO to initiation HNSCC formation. Analogous to the Moc1 transplantation experiments, the tongues of 4NQO-treated ITGB6-ctl mice exhibited a more malignant phenotype in terms of tumors and lesions at the conclusion of the 24th week, as compared to their ITGB6-cKO counterparts (Fig. 2E). The visible tumor incidence, tumor area and tumor grade showed significant differences between ITGB6-ctl and ITGB6-cKO groups (Supplementary Fig. 5H–J). The immunofluorescence co-staining results of CD8 and GZMB demonstrated that, compared to the ITGB6-ctl group, the ITGB6-cKO group exhibited a significant increase in the infiltration of CD8-positive T cells, although there was no significant difference in the infiltration of GZMB+ CD8+ T cells between the two groups (Fig. 2F). Additionally, compared to ITGB6-ctl tumors, ITGB6-cKO tumors showed no significant difference in Ki67 levels but exhibited a notable upregulation in active-caspase-3 levels within the tumor cells (Fig. 2G, H). However, there was no significant difference in apoptosis levels of stromal cells (Supplementary Fig. 2I). Thus, in the chemically induced HNSCC model, cell proliferation and malignant evolution appeared affected by the loss of ITGB6. To assess the degree to which the ablation of ITGB6 relies on CD8+ T or CD4+ T cells for its antitumor response, mice from the ITGB6-cKO group were randomly divided into four groups and treated with anti-CD4 and anti-CD8 depleting antibodies. Histological analyses revealed that depleting CD8+ T cells significantly reversed the aggressive growth inhibition mediated by anti-CD276 while depleting CD4+ T cells did not exhibit any impact on the aggressive growth inhibition mediated by anti-CD276 (Fig. 2J). Analysis of the overall lesion quantity and area showed that the depletion of CD8+ T cells significantly weakened the tumor growth inhibition mediated by CD276 blockade, whereas the depletion of CD4+ T cells demonstrated no effect (Fig. 2K, L). Regulatory T cells (Tregs) are pivotal immunosuppressive elements within tumors, with their activity modulated by TGFβ, which is subsequently regulated by ITGB6. To further investigate the impact of ITGB6 deficiency on tumor progression through the modulation of Treg infiltration, flow cytometry analyses were conducted on mice from the ITGB6-ctl and ITGB6-cKO groups. These analyses revealed no significant differences in Treg proportions between the groups, indicating that ITGB6 loss does not influence Treg levels (Supplementary Fig. 5K, L).
ITGB6 is essential for resistance to anti-CD276 treatment in mouse HNSCC
Next, we examined whether depletion of ITGB6 can overcome the resistance of α-CD276 treatment. We employed 4NQO treatment on both ITGB6-ctl and ITGB6-cKO mice to induce the formation of HNSCC, and at the 26-week mark, selected mice with comparable lesion areas for α-CD276 intervention. Before α-CD276 treatment, mice are selected in the wt and ko group for similar lesion size. Thirteen days post-treatment, we obtained eight resistant mice from each group. We then injected Tamoxifen or corn oil into resistant ITGB6-ctl and ITGB6-cKO mice for 5 days and harvested tongue tissues 41 days after the initial α-CD276 treatment (Supplementary Fig. 5M). We monitored the mice every four days from the day of the first α-CD276 injection. The results demonstrated that while Itgb6 ablation partially mitigates tumor progression, the combination of Itgb6 knockout and α-CD276 treatment synergistically enhances therapeutic efficacy, as evidenced by a notable reduction in tumor size (Fig. 3A). Compared to the control group (ITGB6-ctl) treated with IgG, the ITGB6-ctl combined with α-CD276 did not significantly impede the tumor’s relentless growth. In contrast, the ITGB6-cKO cohort treated with IgG exhibited moderate tumor growth attenuation, and this retardation was substantially accentuated in the ITGB6-cKO group receiving α-CD276 treatment (Fig. 3B–E). No significant variations in Ki67 expression were observed across the treatment cohorts (Fig. 3F). Enhanced expression of caspase-3 in tumor cells was noted in the ITGB6-cKO group treated with IgG, and this elevation was further amplified in the ITGB6-cKO cohort receiving α-CD276 therapy (Fig. 3G). However, there was no significant difference in apoptosis levels of stromal cells (Fig. 3H). Immunofluorescence staining was performed to assess CD8+ T cell infiltration and activation in tumor tissues from control (CTL) and ITGB6 conditional knockout (cKO) mice treated with either IgG or α-CD276 antibody. Quantitative analysis indicated that continued treatment with the CD276 antibody in resistant mice did not affect CD8+ T cell infiltration. In contrast, ITGB6 knockout significantly enhanced CD8+ T cell infiltration. Furthermore, compared to the cKO+IgG treatment group, the cKO+CD276 treatment group exhibited a substantial increase in the proportion of GZMB+CD8+ T cells (Fig. 3I).
A The tumor growth curves of different groups of mice were plotted. Measurements were taken every 4 days. Data are shown as mean ± SD (n = 8 mice). P values were calculated by one-way ANOVA and Tukey HSD test. B Display of histological characteristics of HNSCC in different groups by H&E (left) and the representative image of 4NQO-induced HNSCC (right) (n = 8 mice). Scale bar, 100 μm (left) and 1 mm (right). C Quantification of primary tumor incidence in different treatment groups (n = 8 mice). Data are presented as percentages. Statistical significance was assessed using the Pearson chi-square test. P value is exact and two-sided. D, E Quantification of tongue lesion area (D) and SCC number (E) in different treatment groups. Data are shown as mean ± SD (n = 8 mice). P values were calculated by one-way ANOVA with Tukey’s multiple comparison test. F Representative images of KI67 IHC staining (left) and quantitation of the percentage of KI67+cells (right) in different treatment groups. Scale bar, 50 μm. Data are shown as mean ± SD (n = 8 mice). P values were calculated one-way ANOVA with Tukey’s multiple comparison test. G Representative images of Caspase-3 IHC staining (left) and quantitation of the percentage of Caspase-3+cells (right) in different treatment groups. Scale bar, 50 μm. Data are shown as mean ± SD (n = 8 mice). P values were calculated by one-way ANOVA with Tukey’s multiple comparison test. H Quantitation of the percentage of Caspase-3+ cells in different treatment groups within stromal cells. Data are shown as mean ± SD (n = 8 mice). P values were calculated by one-way ANOVA with Tukey’s multiple comparison test. I Representative immunofluorescence (IF) staining images of CD8 (green) and GZMB (red) (left). Statistical analysis of the ratio of CD8+ cells (mid) and CD8+ GZMB+ cells to CD8+ cells (CD8T killing capacity) in different treatment groups (right). Scale bar, 25 μm. Data are expressed as mean ± SD (n = 8 mice). P values were calculated by one-way ANOVA with Tukey’s multiple comparison test. Source data and exact p values are provided as a Source Data file.
ITGB6 mediates the interaction between tumor cells and PF4+ macrophages
To explore the mechanism of ITGB6-mediated α-CD276 resistance in HNSCC, tumors from ITGB6-ctl and ITGB6-cKO mice that displayed treatment resistance were harvested thirteen days after Tamoxifen injection for scRNA-seq analysis (Fig. 4A). We obtained a total of 22,376 cells (5939 cells from CTL-1, 5567 cells from CTL-2, 6186 cells from cKO-1, and 5195 cells from cKO-2) (Fig. 4A). The average UMI and gene counts were 5931 and 2125, respectively (Supplementary Fig. 6A). After data integration and dimensionality reduction, the visualization results demonstrated a significant overlap in the distribution of ITGB6-ctl and ITGB6-cKO samples (Fig. 4B). We identified four major cell types: epithelial cells (Krt14, Krt5, and Lgals7), immune cells (Ptprc, Cd3d, and Cd52), fibroblasts (Col1a1, Col1a2, and Dcn), and endothelial cells (Vim, Pecam1, and Chd5) (Fig. 4B, C). CopyKAT analysis revealed 932 aneuploid cells (Fig. 4D, E). Since CD276 is one of the most important immune checkpoints31, to understand how ITGB6 is involved in the α-CD276-mediated immune response of HNSCC, we isolated the immune cell population and annotated them into six major immune cell types: CD4 Th cells, Tregs, CD8 T cells, γδT cells, Monocytes/Macrophages/Dendritic cells (MoMΦDC), and neutrophils, based on their marker gene expression (Fig. 4F and S6B). We found that the percentages of MoMΦDC and γδT cells were decreased in ITGB6-cKO mice, while the percentages of neutrophils, CD8 T cells, and CD4 T cells were increased in ITGB6-cKO mice compared to ITGB6-ctl mice (Fig. 4F). Additionally, we characterized the detailed alterations in interactions between tumor cells and immune cells using CellPhoneDB analysis32. Apart from intercommunication among tumor cells, the number of ligand-receptor pairs identified by CellPhoneDB analysis was highest between tumor cells and the MoMΦDC population (Fig. 4G).
A Strategy for the treatment of HNSCC mice and single-cell RNA sequencing (n = 2 mice for each group). B UMAP plot showing four cell clusters from mice HNSCC tissues, colored by cell cluster. D, days. C Heatmap of signature genes for four cell clusters. Each cell cluster is represented by three specifically expressed genes. D Copy number of epithelial cells was estimated using CopyKAT. E UMAP embedding of the inferred CopyKAT diploid and aneuploid copy number profiles (all epithelial cells). D, days. F The UMAP diagram of the immune cell subpopulation (left) and the cell ratio between the two groups (right). G Heatmap showing the ligand-receptor interaction between tumor cells and immune cell subpopulations based on CellPhoneDB. H UMAP plot of macrophage subpopulation, colored by clustering. I Circle plot showing the differences in the number and strength of intercellular communications between the two groups by CellChat. Both the strength and number of intercellular communications between PF4+ macro and tumor cells were significantly reduced. (The blue line represents intercellular communication downregulation, red represents up-regulation, and the width of the line represents the number of up- or downregulation). J, K Dot plot showing the expression of receptor-ligand pairs between PF4+ macrophages and tumor cells in WT and cKO HNSCC tissues by CellChat (J) and CellPhoneDB (K) analysis. Data are presented as communication probabilities. The size of the dots represents the communication probability, and the color scale represents the communication strength. P values were calculated using a permutation test, assessing the significance of cell–cell communication by comparing observed mean expression with a null distribution generated by random permutations. P values are two-sided and exact, with significance indicated (0.01 < P < 0.05 and P < 0.01) (J). Data are presented as mean expression levels. The size of the dots represents the -log10(P value), and the color scale represents the mean expression levels. P values were calculated using a permutation test, which generates a null distribution by randomly shuffling cell labels to determine the specificity of interactions. P values are two-sided and exact, with significance indicated (0.01 < P < 0.05 and P < 0.01) (K). Source data and exact p values are provided as a Source Data file.
To further investigate, we divided and annotated 1,001 MoMΦDC cells into subtypes: Thbs1+ monocytes, PF4+ macrophages, Prdx1+ macrophages, Lars2+ macrophages, Ccl17+ DCs, and Clec9a DCs, with PF4+ macrophages being the most abundant (Fig. 4H and S6C). The cell-specific markers were shown in a heatmap (Supplementary Fig. 6D). We then performed CellChat analysis, which quantitatively infers intercellular communication networks using mass action models and enables the visualization of cellular interactions33. Both the number and strength of interactions between tumor cells and PF4+ macrophages were reduced in ITGB6-cKO tumors compared to ITGB6-ctl tumors (Fig. 4I). Interestingly, quantitative calculation of ligand-receptor networks by CellChat revealed that the CX3CL1-CX3CR1 interaction, which is essential for the recruitment of macrophages in cancer, was dramatically decreased between tumor cells and PF4+ macrophages in ITGB6-cKO tumors compared to ITGB6-ctl tumors (Fig. 4J). This ligand-receptor interaction result was validated using CellPhoneDB analysis (Fig. 4K).
Knockout of ITGB6 greatly reduced the mRNA expression of CX3CL1 in tumor cells (Supplementary Fig. 6E). In addition, IHC staining of CX3CL1 similarly showed that knockdown of Itgb6 significantly downregulated CX3CL1 protein expression (Supplementary Fig. 6F). Furthermore, our western blot experiments confirmed that the knockout of ITGB6 results in the downregulation of the expression level of CX3CL1 (Supplementary Fig. 6G). To probe the possible mechanism of how ITGB6 mediates CX3CL1 expression, we first performed SCENIC analysis and found 22 downregulated inferred transcription factors (Supplementary Fig. 6H, Supplementary Data 3). In addition, we used tools from online UCSC Genome Bioinformatics Site (http://genome.ucsc.edu/) and found 25 predicted transcription factors at the promoter of Cx3cl1 gene (score >10 and relative score >0.95) (Supplementary Data 4). Of note, Stat3 was the common transcription factor between SCENIC analysis and online analysis (Supplementary Fig. 6I). Subsequently, to investigate whether STAT3 can bind to the promoter of Cx3cl1 gene, we used Moc1 cells and performed chromatin Immunoprecipitation quantitative real-time PCR (ChIP-qPCR) assay using antibodies against STAT3 or IgG (control). We observed enriched occupancy of STAT3 at the promoter regions of CX3CL1 gene compared to the IgG control, whereas STAT3 knockdown significantly reduced the binding of STAT3 to the promoter of Cx3cl1 (Supplementary Fig. 6J). To clarify how ITGB6 regulates STAT3, we further focused on all downstream signaling pathways related to the integrin family, including FAK/SRC, ERK1/2, NFKB1, SAPK/JNK, PI3K/AKT, and P38 MAPK34,35,36,37. Among these pathways, we found that ITGB6 knockout can reduce the phosphorylation of FAK/SRC (Supplementary Fig. 6K). Our scRNA-seq data revealed that CX3CR1 is specifically expressed in PF4+ macrophages (Supplementary Fig. 6L). To validate that, we conducted immunofluorescence (IF) staining and detected co-expression of CX3CR1 and PF4 (Supplementary Fig. 6M). To further investigate whether the ITGB6-CX3CL1 axis could recruit other suppressive cell types, such as Tregs, via STAT3 activation, flow cytometric analyses were undertaken. The results showed that, in comparison with the control group, the Treg proportion in the ITGB6-cKO group receiving α-CD276 treatment exhibited no significant differences (Supplementary Fig. 6N).
Signature of PF4+ macrophages and its association with clinicopathological parameters in HNSCC
To delineate the functional implications of PF4+ macrophages in tumorigenesis, we derived a distinct gene signature for these cells using single-cell RNA sequencing (Supplementary Fig. 7A, Supplementary Data 5). Leveraging the TCGA HNSCC dataset, we formulated a multivariate regression model based on this unique gene signature. Subsequent analyses disclosed a marked upregulation of individual genes within the high-risk PF4_high cohort as opposed to the low-risk PF4_low cohort (Supplementary Fig. 7B). Kaplan–Meier log-rank analysis further substantiated this divergence between the two groups (Fig. 5A). To validate these inferences, we conducted a parallel investigation using the TCGA Pan-Cancer dataset, which similarly revealed congruent gene expression profiles and survival disparities between the PF4_high and PF4_low groups (Fig. 5B and S7C). Additional validation was sought through analysis of a renal carcinoma dataset subjected to immunotherapy, yielding analogous results (Fig. 5C and S7D). Subsequent verification was executed in a patient cohort from the First Affiliated Hospital of Sun Yat-sen University (FAH-SYSU), encompassing 81 HNSCC cases, for which comprehensive demographic, pathological, and clinical metadata were compiled (Supplementary Data 6). Immunohistochemical assays were conducted on these tissue specimens to determine PF4 protein expression levels and their associations with diverse clinicopathological variables (Fig. 5D). Based on the immunohistochemical quantification of PF4, patients were categorized into PF4_high and PF4_low subsets. While no discernible association was noted between PF4 expression and either T-stage or nodal metastasis (Supplementary Fig. 7E, G), PF4 expression demonstrated a significant correlation with tumor stage and histological grade (Supplementary Fig. 7F, H). Kaplan–Meier log-rank analysis within this cohort revealed an inverse relationship between elevated PF4 levels and overall patient survival (Fig. 5D).
A Kaplan–Meier curve depicting the overall survival (OS) of patients with HNSCC (TCGA-Cohort) in the model group based on PF4 signature (n = 259 in PF4_low and n = 260 in PF4_high). P values were calculated by log-rank test. B Kaplan–Meier curve depicting the overall survival (OS) of patients with Pan-Cancer (TCGA-Cohort) in the model group based on PF4 signature. P values were calculated by log-rank test (n = 4892 in PF4_low and n = 4892 in PF4_high). C Kaplan–Meier curve depicting the overall survival (OS) of patients with Immunotherapy-treated renal cancer (GEO-Cohort) in the model group based on PF4 signature (n = 155 in PF4_low and n = 156 in PF4_high)55. P values were calculated by log-rank test. D Representative images of PF4 IHC staining in PF4_low (upper) and PF4_high (lower) (left) groups with HNSCC (FAH-SYSU-Cohort) (n = 40 in PF4_low and n = 41 in PF4_high). Scale bar, 100 μm. Kaplan–Meier curve depicting the overall survival (OS) of patients with HNSCC (FAH-SYSU-Cohort) in the model group based on PF4 expression (right). P values were calculated by log-rank test. E The experimental design of the HNSCC tumorigenesis model and treatment strategy (left) and representative image of PF4 IHC staining in CTL (upper) and DTR (down) groups (right). F Representative image of 4NQO-induced HNSCC in different groups (n = 6 mice). Scale bar, 1 mm. G Display of histological characteristics of HNSCC in different groups by H&E (n = 6 mice). Scale bar, 100 μm. H, I Quantification of tongue lesion area (H) and SCC number (I) in different treatment groups. Data are shown as mean ± SD (n = 6 mice). P values were calculated by one-way ANOVA with Tukey’s multiple comparison test. J Representative images of ITGB6 IHC staining in ITGB6_low (left) and ITGB6_high (right) groups with HNSCC (FAH-SYSU-Cohort) (n = 81 biological replicates). Scale bar, 100 μm. K Representative images of CD276 IHC staining in CD276_low (upper) and CD276_high (down) groups with HNSCC (FAH-SYSU-Cohort) (n = 81 biological replicates). Scale bar, 100 μm. L–N Correlation of PF4 IHC score with ITGB6 IHC score (L) and CD276 IHC score (M) (n = 81 for each group). And the correlation of ITGB6 IHC score with CD276 IHC score (N) (n = 81 for each group). Data are presented as scatter plots with a linear regression line. The shaded area represents the 95% confidence interval. Pearson correlation coefficients (R values) were calculated to assess the linear relationship between the variables. P values are exact and two-sided. Source data and exact p values are provided as a Source Data file.
Flow cytometric analyses have shown a significant decrease in PF4+ macrophage proportions after ITGB6 deletion in drug-resistant mouse models (Supplementary Fig. 7I, J). Concurrently, IF colocalization studies of CD8 and GZMB revealed an increased presence of CD8+ T cells and CD8+ GZMB+ T cells following ITGB6 ablation in these models (Supplementary Fig. 7K, L). To examine the impact of PF4+ cell depletion on cellular response to α-CD276, Pf4CreERT; DTA mice were generated (Fig. 5E). We employed a consistent methodology by initially inducing resistance to α-CD276 antibody treatment in Pf4CreERT; DTA mice, subsequently randomizing them into four distinct groups, each consisting of six mice (Fig. 5E). IHC results show that tamoxifen treatment markedly reduced PF4+ macrophage presence (Fig. 5E). Comparative analyses of lesion number, area, and histology indicated that, unlike the control group treated with IgG (Pf4-ctl), combined treatment with Pf4-ctl and α-CD276 did not significantly alter tumor growth dynamics (Fig. 5F–I). However, the Pf4-cKO group treated with IgG demonstrated moderate tumor growth reduction, which was further enhanced by α-CD276 treatment (Fig. 5F–I). To further explore the relationships between ITGB6, CD276, and PF4 in human samples, ITGB6 and CD276 IHC staining were conducted on patient tissues. Samples were classified into high and low expression categories based on their IHC scores (Fig. 5J, K). Correlation analyses revealed a significant association between PF4 and ITGB6 scores in these samples, aligning with single-cell analysis findings (Fig. 5L). However, no significant correlation was found between PF4 and CD276 scores (Fig. 5M), while the relationship between CD276 and ITGB6 scores mirrored that observed in SCC15 cells, indicating no significant correlation between ITGB6 and CD276 scores in patient samples (Fig. 5N)
Resistance of α-CD276 treatment by ITGB6 is mediated by CX3CL1-CX3CR1 axis
To test whether CX3CL1-CX3CR1 axis is functionally relevant to ITGB6-mediated resistance, we treated resistant animals with α-ITGB6 (264RAD), α-CX3CL1, α-CX3CR1, and α-ITGB6 plus CX3CL1 recombinant protein (Supplementary Fig. 6A). All antibodies were tested for their efficacy based on previously published literature. To confirm that PF4+ macrophages are derived from monocytes and are locally recruited during tumor development, they were characterized using an anti-CCL2 antibody. Flow cytometry results indicated that, compared to the control group, the α-CCR2 treatment group exhibited a significantly reduced proportion of PF4+ macrophages, as well as a significant reduction in overall macrophages (Supplementary Fig. 6B). Mice Mice treated with vehicle were used as controls. Our results demonstrated that α-ITGB6, α-CX3CL1, and α-CX3CR1 were able to sensitize mouse HNSCC to α-CD276 treatment (Fig. 6A). In the presence of recombinant CX3CL1 protein, the effects of α-ITGB6 on HNSCC resistance were significantly compromised (Fig. 6A). Additionally, treatments with α-ITGB6, α-CX3CL1, and α-CX3CR1 reduced the number and area of 4NQO-induced lesions compared with the control group, while recombinant CX3CL1 protein abolished the effect of α-ITGB6 (Fig. 6B, C). Compared with the control group, the α-CX3CL1 group showed significantly inhibited cell proliferation, but there was no significant difference between the other groups (Fig. 6D, E). Conversely, treatments with α-ITGB6, α-CX3CL1, and α-CX3CR1 induced a significant increase in apoptotic events in tumor cells compared to the control group, while CX3CL1 could reverse this phenotype (Fig. 6F, G). Immunofluorescence co-staining of CD8 and GZMB revealed that treatments with α-ITGB6, α-CX3CL1, and α-CX3CR1 significantly elevated the infiltration ratios of both CD8+ T cells and GZMB+ CD8+ T cells compared to the control group, while CX3CL1 reversed this phenotype (Fig. 6H–J). Flow cytometry results showed that treatments with α-ITGB6, α-CX3CL1, and α-CX3CR1 suppressed the infiltration of PF4+ macrophages in tumors, whereas CX3CL1 treatment increased the proportion of PF4+ macrophages (Fig. 6K, L). Additionally, analysis of all macrophage subpopulations indicated that inhibition of ITGB6, CX3CL1, or CX3CR1 reduces all subpopulations of tumor-associated monocyte-derived macrophages. Interestingly, CX3CL1 could reverse this phenotype (Fig. 6M).
A Representative images of 4NQO-induced HNSCC (upper) and corresponding HE staining (lower) in different treatment groups (n = 8 mice). Scale bar, 1 mm (upper), 100 μm (lower). B, C Quantification of SCC number (B) and tongue lesion area (C) in different treatment groups. Data are presented as mean ± SD (n = 8 mice). P values are presented by one-way ANOVA with Tukey’s multiple comparison test. D–G. Representative images of KI67 (D) and Caspase-3 (F) IHC staining and quantitation of the percentage of KI67+ (E) and Caspase-3+ (G) cells in different treatment groups. Scale bar, 50 μm. Data are shown as mean ± SD (n = 8 mice). P values are presented by one-way ANOVA with Tukey’s multiple comparison test. H–J Representative immunofluorescence (IF) staining images of CD8 (green) and GZMB (red) (H). Statistical analysis of the ratio of CD8+ cells (I) and CD8+ GZMB+ cells to CD8+ cells (CD8T killing capacity) (J) in different treatment groups. Scale bar, 25 μm. Data are expressed as mean ± SD (n = 8 mice). P values are presented by one-way ANOVA with Tukey’s multiple comparison test. K, L Representative flow plots show the frequency of the F4/80+PF4+ macrophages (K) and quantification of the proportions of F4/80+PF4+cells (L) in different treatment groups. Data are shown as mean ± SD (n = 8 mice). P values are presented by one-way ANOVA with Tukey’s multiple comparison test. M Representative flow plots show the frequency of the F4/80+ macrophages in different treatment groups. Data are shown as mean ± SD (n = 8 mice). P values are presented by one-way ANOVA with Tukey’s multiple comparison test. Source data and exact p values are provided as a Source Data file.
To exclude the non-specific effect of the chemical inhibitor, we also generated CX3CL1 conditional knockout mice (Supplementary Fig. 8C). Similarly, we crossed Cx3cl1flox/flox mice with K14creER mice to obtain K14creER; Cx3cl1wt/wt control (CX3CL1-ctl) and K14creER; Cx3cl1flox/flox conditional knockout (CX3CL1-cKO) mice. Loss of Cx3cl1 in the oral epithelium did not show discernible oral epithelium defect (Supplementary Fig. 8D, E). The knockout efficiency of Cx3cl1 gene was validated via IHC staining (Supplementary Fig. 8F).
To examine whether genetic ablation of CX3CL1 can lead to reverse of drug resistance, we treated control, ITGB6-cKO and CX3CL1-cKO HNSCC-bearing mice with α-CD276 (Supplementary Fig. 8G). Compared with control group, ITGB6-cKO and CX3CL1-cKO group showed reduced level of malignancies, while ITGB6-cKO plus CX3CL1 protein group demonstrated a resistant phenotype (Supplementary Fig. 8H–J). Although the proliferation ratios were comparable between all groups, the apoptotic cells were significantly increased in the ITGB6-cKO and CX3CL1-cKO groups in tumor cells. And there were no significant differences in apoptosis levels within the stromal cells (Supplementary Fig. 8K–N). Altogether, our data clearly demonstrated that CX3CL1-CX3CR1 is involved in ITGB6-mediated α-CD276 treatment resistance of HNSCC. To assess whether 264RAD can mimic the in vivo knockdown effects of ITGB6 by suppressing the expression of CX3CL1 and CXCL16, Western blot analyses were performed. The findings demonstrated that 264RAD significantly reduced the expression levels of CX3CL1 and CXCL16 in mice (Supplementary Fig. 8O).
Induction of exhaustion in CXCR6+ CD8+ T cells by PF4+ macrophages
To investigate the influence of PF4+ macrophages on the tumor immune microenvironment, we conducted CellChat analysis. Consistent with our prior observations regarding the alterations in CD8 T-cell infiltration and cytotoxic functions in α-CD276 treated, drug-resistant mice, the interaction frequency and intensity between PF4+ macrophages and CD8+ T cells were attenuated in ITGB6-cKO tumors (Fig. 7A). Intriguingly, quantitative assessment of ligand-receptor networks via CellChat revealed a significant reduction in the CXCL16-CXCR6 interactions between PF4+ macrophages and CD8+ T cells in ITGB6-cKO tumors compared to ITGB6-ctl (Fig. 7B). Upon classification of CD8+ T cells from our single-cell data, we identified three distinct subpopulations: CXCR6_low, cytotoxic CXCR6_high, and suppressive CXCR6_high (Fig. 7C). Monocle3 pseudotime trajectory analysis of these subpopulations suggested a possible linear CD8+ T cell developmental trajectory (Fig. 7D). Scoring of these subgroups for cytotoxic (GZMB, GNLY, IFNG, and PRF1), suppressive (CTLA4, PDCD1, LAG3, TIGIT, and HAVCR2), and early developmental (TCF7, CCR7, SELL, and LEF1) markers showed significant differences between them (Supplementary Fig. 9A–F). Compared to the ITGB6-ctl mice, ITGB6-cO mice exhibited a significantly elevated proportion of cytotoxic CXCR6_high cells and a concomitantly reduced proportion of suppressive CXCR6_high cells (Fig. 7E).
A Circle plot showing the differences in the number and strength of intercellular communications between the two groups by CellChat (n = 2 mice for each group). Both the strength and number of intercellular communications between PF4+ macrophages and CD8+ T cells were significantly reduced. (The blue line represents intercellular communication downregulation, red represents up-regulation, and the width of the line represents the number of up- or downregulation). B Dot plot showing the expression of receptor-ligand pairs between PF4+ macrophages and CD8+ T cells in CTL and cKO HNSCC tissues by CellChat analysis. C Data are presented as communication probabilities. The size of the dots represents the communication probability, and the color scale represents the communication strength. P values were calculated using a permutation test, assessing the significance of cell–cell communication by comparing observed mean expression with a null distribution generated by random permutations. P values are two-sided and exact, with significance indicated (0.01 < P < 0.05 and P < 0.01) (J). D UMAP plots showing the subclusters of epithelial cells. E Pseudotime evolution trajectories inferred by Monocle3 after reclustering of CD8+ T cells. F Proportion of three CD8+ T-cell subclusters. G UMAP (left) and pseudotime evolution trajectories inferred by Monocle3 (right) of three CD8+ T-cell subclusters with HNSCC (GEO-Cohort1)56 (n = 18 biological replicates). H UMAP (left) and pseudotime evolution trajectories inferred by Monocle3 (right) of three CD8+ T-cell subclusters with HNSCC (GEO-Cohort2)57 (n = 63 biological replicates). I Representative immunofluorescence (IF) staining images of CD8 (red) and GZMB (green) (left). Statistical analysis of the ratio of CD8+ cells (mid) and CD8+ GZMB+ cells to CD8+ cells (CD8T killing capacity) (right) in different groups with HNSCC (FAH-SYSU-Cohort) (n = 40 in PF4_low and n = 41 in PF4_high). Scale bar, 25 μm. Data are expressed as mean ± SD. P values were calculated by two-tailed unpaired Student’s t-test. J Representative immunofluorescence (IF) staining images of CD8 (red) and PDCD1 (green) (left). Statistical analysis of the ratio of CD8+ cells (mid) and CD8+ PDCD1+ cells to CD8+ cells (CD8T exhaustion) (right) in different groups with HNSCC (FAH-SYSU-Cohort) (n = 40 in PF4_low and n = 41 in PF4_high). Scale bar, 25 μm. Data are expressed as mean ± SD. P values were calculated by two-tailed unpaired Student’s t-test. J, K. Correlation of PF4 IHC score with the ratio of CD8+GZMB+ cells (J) and CD8+PDCD1+ cells (K) (n = 81 biological replicates). Data are presented as scatter plots with a linear regression line. The shaded area represents the 95% confidence interval. Pearson correlation coefficients (R values) were calculated to assess the linear relationship between the variables. P values are exact and two-sided. Source data and exact p values are provided as a Source Data file.
To validate the reliability of this classification, we conducted confirmatory analyses in two independent single-cell cohorts of HNSCC. In summary, these three subtypes were consistently present in the CD8+ T cells across both cohorts, each exhibiting significant differences in their respective scores (Fig. 7F, G, S9G-L). To further probe the association of CXCR6 with these cytotoxic and suppressive profiles, we performed CXCR6 correlation analyses on both human HNSCC cohorts as well as the TCGA HNSCC dataset. Our findings indicated that CXCR6 was positively correlated with both cytotoxic and suppressive markers, namely GZMB, PRF1, CTLA4, PDCD1, LAG3, TIGIT, and HAVCR2, suggesting the coexistence of both cytotoxic and suppressive subtypes within CXCR6+ CD8+ T cells (Supplementary Fig. 10A–F, Supplementary Data 7–9).
Additional validation was conducted in the HNSCC cohort from SYSU FAH. Immunofluorescent co-staining of GZMB and CD8 revealed that compared to the PF4_high group, the proportion of CD8+ and GZMB+ CD8+ T cells was significantly elevated in the PF4_low group, while PDCD1+ CD8+ T cells were markedly reduced (Fig. 7H, I). Correlational analyses between PF4 IHC scores and the proportions of GZMB+ CD8+ and PDCD1+ CD8+ T cells indicated a significant inverse relationship with GZMB+ CD8+ T cells and a positive correlation with PDCD1+ CD8+ T cells (Fig. 7J, K). These results suggest that PF4+ macrophages may be facilitating the transition of cytotoxic CXCR6+ CD8+ T cells toward their suppressive CXCR6+ CD8+ counterparts.
CXCL16 serves as a critical factor for cytotoxic functionality of CXCR6+ CD8+ T cells while excessive stimulation promotes their exhaustion
To investigate the influence of PF4+ macrophages on the functional capacities of CXCR6+ CD8+ T cells, we employed a co-culture system (Supplementary Fig. 11A). Initially, tumor tissues were dissociated into single-cell suspensions, followed by the separate Fluorescence Activated Cell Sorting (FACS) of PF4+ macrophages and CD8+ T cells. Subsequently, the CD8+ T cells were stimulated with CD3/CD28 agonists and cultured in a medium enriched with M-CSF/IL4 for PF4+ macrophages (Supplementary Fig. 11A). Following established methodologies, we first edited the Moc1 cell line to generate an anti-CD3-expressing variant which enabled the Moc1 cells to effectively mediate CD8+ T cell-specific cytotoxicity in vitro. Quantitative Moc1 cells were incorporated into the co-culture system, and the ratio of PF4+ macrophages to CD8+ T cells was meticulously adjusted. Data demonstrated that at a 1:1 cell ratio, the proportion of GZMB+ CXCR6+ CD8+ T cells peaked, while PDCD1+ CXCR6+ CD8+ T cells were minimally represented (Fig. 8A, B). Fluctuations in cell ratios led to varied declines in GZMB+ CXCR6+ CD8+ T cells, whereas the PDCD1+ CXCR6+ CD8+ T cells increased (Fig. 8A, B). Notably, in the absence of PF4+ macrophages, the proportions of either GZMB+ or PDCD1+ CXCR6+ CD8+ T cells were significantly different than when PF4+ macrophages were present in suboptimal quantities. To validate whether alterations in the state of CXCR6+ CD8+ T cells were induced by CXCL16, recombinant CXCL16 at different concentrations was added to the Moc1 and CD8+ T cell co-culture system. The findings revealed that at a concentration of 100 ng/ml, the proportion of GZMB+ CXCR6+ CD8+ T cells was at its zenith, and PDCD1+ CXCR6+ CD8+ T cells were at their nadir (Fig. 8C, D). When the concentration of CXCL16 reached 800 ng/ml, the peak of PDCD1+ CXCR6+ CD8+ T cells was observed, and the proportion of GZMB+ CXCR6+ CD8+ T cells substantially declined (Fig. 8C, D). These observations suggest that optimal levels of CXCL16 are indispensable for the cytotoxic role of CXCR6+ CD8+ T cells, whereas excessive CXCL16 drives them toward exhaustion.
A Representative flow plots show the frequency of the CXCR6+ CD8+ GZMB+ T cells (left) and quantification of the proportions of CXCR6+ CD8+ GZMB+ T cells (right) in groups with different PF4+ Macrophage and CD8+ T-cell proportions. Data are shown as mean ± SD (n = 3 biological replicates). P values are presented by one-way ANOVA with Tukey’s multiple comparison test. B Representative flow plots show the frequency of the CXCR6+ CD8+ PDCD1+ T cells (left) and quantification of the proportions of CXCR6+ CD8+ PDCD1+ T cells (right) in groups with different PF4+ Macrophage and CD8+ T-cell proportions. Data are shown as mean ± SD (n = 3 biological replicates). P values are presented by one-way ANOVA with Tukey’s multiple comparison test. C, D Representative flow plots show frequency of the CXCR6+ CD8+ GZMB+ T cells (C) and quantification of the proportions of CXCR6+ CD8+ GZMB+ T cells (D) in groups with different concentrations of CXCL16 recombinant protein. Data are shown as mean ± SD (n = 3 biological replicates). P values are presented by one-way ANOVA with Tukey’s multiple comparison test. E, F Representative flow plots show the frequency of the CXCR6+ CD8+ PDCD1+ T cells (E) and quantification of the proportions of CXCR6+ CD8+ PDCD1+ T cells (F) in groups with different concentrations of CXCL16 recombinant protein. Data are shown as mean ± SD (n = 3 biological replicates). P values are presented by one-way ANOVA with Tukey’s multiple comparison test. G Representative images of 4NQO-induced HNSCC (upper) and corresponding HE staining(lower) in groups with different concentrations of CXCL16 recombinant protein. Scale bar, 1 mm (upper), 100 μm (lower). H–K Representative images of KI67 (H) and Caspase-3 (J) IHC staining and quantitation of the percentage of KI67+ (I) and Caspase-3+ (K) cells in groups with different concentrations of CXCL16 recombinant protein. Scale bar, 50 μm. Data are shown as mean ± SD (n = 8 mice). P values are presented by one-way ANOVA with Tukey’s multiple comparison test. Source data and exact p values are provided as a Source Data file.
To investigate the regulatory mechanisms underlying Cxcl16 expression in PF4+ macrophages, we initially executed a differential gene expression analysis. The results demonstrated that the ITGB6-ctl group was predominantly enriched in pathways such as NFKB1 and TNF, while the ITGB6-cKO group displayed a heightened enrichment in oxidative phosphorylation signaling pathways (Supplementary Fig. 11B, C). We then conducted a SCENIC analysis and identified 18 putative transcription factors that were downregulated (Supplementary Fig. 11D, Supplementary Data 10). In addition, we used tools from the online UCSC Genome Bioinformatics Site (http://genome.ucsc.edu/) and found 29 predicted transcription factors at the promoter of Cx3cl1 gene (score >10 and relative score >0.95) (Supplementary Data 11). Of note, Nfkb1 and Klf4 were the common transcription factors between SCENIC analysis and online analysis (Supplementary Fig. 11E). We further focused on all downstream signaling pathways related to the integrin family, including ERK1/2, NFKB1, RELA, and P38 MAPK34,35,36,37. Among these pathways, we found that ITGB6 knockout can reduce the phosphorylation of NFKB1 in PF4+ Macrophages (Supplementary Fig. 11F). Subsequently, to investigate whether Nfkb1 can bind to the promoter of Cxcl16 gene, we used Moc1 cells and performed chromatin Immunoprecipitation quantitative real-time PCR (ChIP-qPCR) assay using antibodies against Nfkb1 or IgG (control). We observed enriched occupancy of Nfkb1 at the promoter regions of the Cxcl16 gene compared to the IgG control, whereas Nfkb1 knockdown significantly reduced the binding of Nfkb1 to the promoter of Cx3cl1 (Supplementary Fig. 11G).
To further corroborate these findings in a murine model, mice treated with cKO and α-CD276 related to Fig. 2 were randomly allocated into three groups: control, 4 µg/ml CXCL16, and 8 µg/ml CXCL16 (Supplementary Fig. 11H). Succinctly, in comparison to the control group, two other treatments exacerbated the tumor’s progressive development, significantly enlarging the lesion area (Fig. 8G and S11I). Moreover, the severity and malignancy of tumors were higher in the 8 µg/ml CXCL16 and CXCL16 inhibitor groups compared to the 4 µg/ml CXCL16 group (Fig. 8G and S11I). Furthermore, the expression of Ki67 remained unchanged across all treatment groups, whereas the expression of active-caspase-3 was notably downregulated in the 4 µg/ml CXCL16 group and even more so in the 8 µg/ml CXCL16 and CXCL16 inhibitor groups (Fig. 8H–K).
To further assess the influence of the CXCR6 receptor on CD8+ T cell functionality and state, a differential gene expression analysis was carried out. The results revealed that the GZMB+ CXCR6+ CD8+ T cell subgroup was predominantly enriched in pathways associated with the immune response and antigen processing and presentation, indicative of immune activation. Conversely, the PDCD1+ CXCR6+ CD8+ T cell group displayed greater enrichment in lectin C and glycolysis pathways (Supplementary Fig. 12A, B). A subsequent SCENIC analysis showed that CREM gene expression was significantly elevated in the PDCD1+ CXCR6+ CD8+ T cell group (Supplementary Fig. 12C). To further determine CREM’s role in CD8+ T cell depletion, SCENIC analysis was applied to CD8+ T cells data from Robert L. Ferris et al., confirming high CREM activity in the PDCD1+ CXCR6+ CD8+ T cell subgroup (Supplementary Fig. 12D). To explore the possibility that CREM activity in this group is regulated by the CXCL16-CXCR6 axis, CD8+ T cells were isolated from tumor tissues and subjected to activation with 100 ng/ml and 800 ng/ml concentrations of CXCL16 recombinant protein (Supplementary Fig. 12E). Findings indicated that the low CXCL16 concentration group showed a significant decrease in the PDCD1+ CXCR6+ CD8+ T cell fraction and an increase in the GZMB+ CXCR6+ CD8+ T cell fraction compared to the high concentration group (Supplementary Fig. 12F, G). Western blot analysis of sorted PDCD1+ CXCR6+ CD8+ T cells and GZMB+ CXCR6+ CD8+ T cells indicated that the levels of CREM were significantly increased in the PDCD1+ CXCR6+ CD8+ T cell subset, suggesting that CREM might mediate the exhaustion process of CXCR6+ CD8+ T cells (Supplementary Fig. 12H). To further investigate whether this change is mediated through the CXCL16-CXCR6 axis, we added 100 ng/ml and 800 ng/ml of recombinant CXCL16 protein to the cultures of sorted CD8+ T cells. The results showed that with increasing concentrations of CXCL16, the expression levels of CREM were significantly upregulated in GZMB+ CXCR6+ CD8+ T cells. Additionally, CREM was already highly expressed in PDCD1+ CXCR6+ CD8+ T cells without the addition of CXCL16, and its expression further increased with higher concentrations of CXCL16 (Supplementary Fig. 12I).
ITGB6 facilitates tolerance to α-PD1 treatment in mice
To explore the role of ITGB6 in mediating resistance to α-PD1 therapy, we employed a comparable approach and administered α-PD1 or IgG to 4NQO-induced carcinogenic mice for 13 days. The α-PD1 was delivered intraperitoneally at a dose of 10 mg/kg every four days, consistent with previous protocols, amounting to four total doses (Fig. 9A). Tumor progression was monitored biweekly from the initial α-PD1 administration via MRI (Fig. 9B). Resistance was characterized by an early response in MRI imaging followed by an increase to more than 120% of the initial size, according to RECIST criteria (Fig. 9B). Thirteen days after the first injection, tongues of the HNSCC-bearing mice were removed (Fig. 9C), revealing reduced malignancy, invasion area, and tumor count in the αPD1-sensitive group compared to the control and resistant groups (Fig. 9C). IHC analysis showed a marked decrease in ITGB6 expression in the sensitive group (Fig. 9D). Furthermore, increased infiltration of PF4+ macrophages was observed in the resistant group (Fig. 9E). To determine ITGB6’s contribution to α-PD1 treatment efficacy, 12 αPD1-resistant mice were divided into two groups: one continued with the PD1 antibody while the other was treated with a combination of PD1 and the ITGB6 antibody 264RAD. This method notably restored α-PD1 treatment responsiveness in the mice (Fig. 9F, G). The overall analysis of lesion quantity and area demonstrated that ITGB6 inhibition reinstates α-PD1 treatment sensitivity in mice (Fig. 9H, I).
A The experimental design of the HNSCC tumorigenesis model and treatment strategy. w, weeks. B Representative examples of head and neck magnetic resonance imaging (MRI) for different groups (n = 8 mice). The dashed area is the boundary of the tumor. D, days. C Representative images of 4NQO-induced HNSCC (upper) and corresponding HE staining (lower) (left). Quantification of SCC number (upper) and tongue lesion area (lower) (right) in different treatment groups. Scale bar, 1 mm (upper), 100 μm (lower). Data are shown as mean ± SD (n = 8 mice). P values are presented by one-way ANOVA with Tukey’s multiple comparison test. D, E Representative images of ITGB6 (D), PF4 (E) IHC staining (left) and quantification of the percentage of ITGB6+ (D), PF4+ (E) (right) in different treatment groups. Scale bar, 50 μm. Data are presented as mean ± SD (n = 8 mice). P values are presented by one-way ANOVA with Tukey’s multiple comparison test. F Tumor growth curves for each mouse treated with either the combination of α-PD1 and α-ITGB6 or the control α-PD1 treatment, with measurements recorded every four days (n = 8 mice). G Representative images of 4NQO-induced HNSCC (upper) and corresponding HE staining(lower). Scale bar, 1 mm (upper), 100 μm (lower). H, I Quantification of tongue lesion area (left) and SCC number (right) in different groups. Data are shown as mean ± SD (n = 8 mice). P values were calculated by two-tailed unpaired Student’s t-test. Source data and exact p values are provided as a Source Data file.
Discussion
Results from multiple Phase I/II trials have demonstrated the safety and encouraging tumor shrinkage in patients with NSCLC, HNSCC, PCa, melanoma, and bladder cancer following enoblituzumab treatment. However, a proportion of patients do not benefit from the treatment due to unknown resistance mechanisms. Currently, a variety of factors have been associated with the efficacy of ICI therapy, such as tumor immunogenicity, T cell function, IFNγ production, immune checkpoint expression, and the tumor microenvironment38,39. In an effort to better understand the mechanisms underlying resistance to CD276 blockade, we used a 4NQO-induced spontaneous HNSCC mouse model, which shares great mutational similarity with tobacco-related human cancers, including HNSCC, esophageal cancer, and lung cancer40. By leveraging scRNA-seq data and HNSCC patient cohorts, we identified a subpopulation of malignant tumor cells expressing high levels of ITGB6, which was associated with resistance to CD276 blockade. This finding suggests the utility of ITGB6 as a predictive biomarker for patient stratification to improve α-CD276 therapeutic outcomes.
The cell surface receptor integrin αvβ6 is poorly expressed or completely absent in healthy adult tissue epithelia but is upregulated during the carcinogenesis of many epithelial cancers41. In our study, the loss of ITGB6 in mouse oral epithelium showed minimal effect on the development and progression of HNSCC in vivo. However, ITGB6 loss led to increased sensitivity to α-CD276 treatment. Recently, αvβ6 has been identified as an effective factor for cancer cells to escape the T-cell α-tumor immune response. For example, Bagati et al. demonstrated that integrin αvβ6 stimulates TGF-β from a latent precursor to induce SOX4 expression, and targeting integrin αvβ6 promotes T cell-mediated immunity in ICI-resistant triple-negative breast cancer25. Similarly, another study showed that the inhibition of αvβ6 integrin prevented the pro-tumor effects of TGF-β on T cells and enhanced immune checkpoint blockade therapy in colorectal cancers27. However, unlike in other cancers, our study showed that αvβ6 integrin is involved in the regulation of tumor-associated macrophages (TAMs), suggesting an alternative mechanism for αvβ6 in regulating the immune response in HNSCC.
In this study, we dissected the cellular and molecular crosstalk between tumor cells and TAMs regulated by αvβ6 integrin. We found that αvβ6 integrin is responsible for the secretion of CX3CL1 by tumor cells and the subsequent infiltration and activation of PF4+ macrophages, which express CX3CR1, in HNSCC. TAMs play an essential role in extracellular matrix remodeling, tumor growth, metastasis, and immunosuppression, and act as the main contributors to ICI resistance by blocking effective antitumor immune responses42,43. Functionally, TAMs can contribute to T cell dysfunction and depletion through the secretion of cytokines and metabolites4,44. To date, many therapeutic agents targeting TAMs to enhance the efficacy of ICI are in preclinical and clinical trials8,45,46.
Previous studies have shown that the CX3CL1/CX3CR1 axis is involved in the recruitment of monocytes/macrophages in cancer47. Indeed, we found that both genetic and chemical inhibition of the CX3CL1/CX3CR1 axis suppressed the infiltration of PF4+ macrophages in HNSCC. In human colon carcinoma, the expression of CX3CR1 in TAMs has been shown to be essential for cell survival and tumor metastasis48. During mouse cutaneous SCC formation, CX3CR1+ TAMs displayed M2 phenotypes with the expression of angiogenic factors and Wnt3a33. Additionally, other studies have proven a critical role of the CX3CL1-CX3CR1 axis in ICI response in various types of cancers49,50. In our study, we found that the CX3CL1/CX3CR1 axis in PF4+ macrophages also participates in chemokine secretion, which affects the cytotoxic function of T cells. Concurrently, multiple studies have indicated that CXCR6+ CD8+ T cells play a pivotal role in tumor immunity19,22. Our findings reveal that within the tumor microenvironment, CXCR6+ CD8+ T cells exist in both cytotoxic and exhausted states, with PF4+ macrophages serving as a critical factor promoting their transition toward exhaustion. Previous research has shown that CXCL16 secreted by DC3 enables cytotoxic T cells to survive within the tumor milieu51, a phenomenon corroborated in our study. However, excessive secretion of CXCL16 by PF4+ macrophages encourages the CXCR6+ CD8+ T cells to progress toward an exhausted state.
Mechanistically, our murine and human HNSCC data consistently demonstrate that the CREM gene is highly expressed in exhausted CXCR6+ CD8+ T cells. CREM is a crucial transcription factor in the immune system, significantly influencing T cell function, autoimmunity, and cancer immune evasion. CREM plays a vital role in T cell activation, differentiation, and cytokine production. Recent studies highlight CREM’s involvement in regulatory T cell (Treg) function, modulating FOXP3 expression, which is critical for maintaining immune tolerance51,52. In cancer, CREM contributes to tumor immune evasion by fostering an immunosuppressive microenvironment. Targeting CREM pathways may enhance the effectiveness of cancer immunotherapies by preventing tumor-induced immune suppression53. Notably, a recent study has also highlighted the close association between CREM and the function of CD8+ T cells20. Consequently, our findings highlight the potential of targeting CREM to restore the functionality of exhausted CXCR6+ CD8+ T cells and improve therapeutic outcomes in HNSCC. By inhibiting CREM, it may be possible to reprogram exhausted T cells, enhancing their cytotoxic capabilities and thereby improving the efficacy of α-CD276 treatment. This approach could potentially overcome resistance mechanisms and lead to better clinical outcomes for patients with HNSCC.
In summary, our findings show that ITGB6 promoted cancer resistance to α-CD276 immunotherapy through the recruitment of PF4+ macrophages via CX3CL1-CX3CR1 axis which consequently resulted in the progression of CXCR6+ CD8+ T cells toward an exhausted state and provide fundamental insight into how integrin components regulate innate immunity. Hence, our work provides a predictive biomarker for α-CD276 immunotherapy response, and suggests a therapeutic modality that may sensitize HNSCC.
Methods
Ethics statement
All animal experiments described in this study were reviewed and approved by the Animal Care and Use Committee of Sun Yat-sen University (SYSU-IACUC-2023-000861). The Committee limits tumor growth to no more than 10% of the animal’s original body weight and the average tumor diameter to no more than 20 mm.
A cohort containing 81 HNSCC specimens was obtained from the Department of Oral and Maxillofacial Surgery, The First Affiliated Hospital of Sun Yat-sen University. Tumor samples were collected with the patients’ written informed consent and the study was approved by the Medical Ethics Committee, The First Affiliated Hospital of Sun Yat-sen University. Informed consent was obtained from all participants, including consent to publish potentially identifying clinical information. All data in this study were anonymized and processed in compliance with relevant privacy protection regulations.
Animals
WT C57BL/6, Itgb6fl/fl, Cx3cl1fl/fl, and PF4CreERT2 mice were purchased from The GemPharmatech (Nanjing, China). K14CreER (005107) mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). All animal subjects were accommodated under specific pathogen-free conditions, maintained on a 12-h light/dark circadian cycle, and granted ad libitum access to sustenance and hydration. All animal protocols received ethical clearance from the Institutional Animal Care and Use Committee (IACUC) of the First Affiliated Hospital of Sun Yat-sen University, under the protocol number SYSU-IACUC-2023-000861. For the induction of oral carcinogenesis, mice aged beyond four weeks were administered 4-nitroquinoline 1-oxide (4NQO) via drinking water at a concentration of 10 mg/mL.
MRI acquisition and interpretation
All mice were examined using a 3.0T MR scanner (MAGNETOM Vida, Siemens Healthcare, Erlangen, Germany) and a 16-channel mouse coil (CG-MUC48-H300-AS, Shanghai Chenguang Medical Technology Co., Ltd., Shanghai, China). The scan protocol includes axial T2WI TSE, coronal T2WI TSE, and axial T1WI StarVIBE. The sequence parameters are detailed in Supplementary Data 13.
Tumor models and in vivo treatments
In our in vivo experiments, we ensured no bias in the selection of the sex of the mice. Each experimental group was randomly assigned with an approximately equal distribution of male and female mice to maintain gender balance. To induce oral squamous cell carcinoma in mice, a carcinogenic solution containing 1,2-propanediol and 4NQO (10 mg/mL, Sigma, N8141) was prepared. Subsequently, 8-week-old mice underwent a 16-week feeding regimen followed by an 8-week period of water consumption. A 2-week treatment phase involving α-CD276 antibody and IgG isotype control was then administered in a drug treatment experiment. Throughout the treatment period, meticulous monitoring was conducted to assess the development and progression of oral cancer. Following the treatment phase, a precise collection of head and neck squamous cell carcinoma samples was performed for thorough analysis. Subsequent validation experiments involved the meticulous collection and comprehensive analysis of squamous cell carcinoma samples from both wild-type and ITGB6ko mice. Furthermore, in the Krt14CreER, Itgb6flox transgenic mouse model, a subgroup of mice exhibiting resistance to CD276 antibody treatment was randomly divided into four groups for further investigation. These groups received specific interventions: (1) administration of tamoxifen followed by a 6-week CD276 antibody treatment, (2) administration of tamoxifen followed by a 6-week IgG isotype control treatment, (3) no intervention followed by a 6-week CD276 antibody treatment, and (4) no intervention followed by a 6-week IgG isotype control treatment. A similar experimental design and grouping were implemented in the Krt14CreER, CX3CL1flox transgenic mouse model to assess the response of mice resistant to CD276 antibody treatment. In the in vivo studies involving CXCL16, concentrations of CXCL16 solutions were administered at 4 and 8 micrograms per milliliter, respectively. Following the completion of the treatment regimens, a meticulous collection of head and neck squamous cell carcinoma samples was performed for comprehensive analysis.
In our CD276 blockade therapy experiments, we treated mice through a regimen of intraperitoneal injections. More precisely, mice received either the carrier IgG control (InVivoMAb rat IgG1 isotype control, BioXcell, Catalog #BE0088, at a dosage of 10 mg/kg body weight) or the anti-CD276 antibody (enoblituzumab, MCE, Catalog #HY-P9966, also at 10 mg/kg body weight), following a four-day cycle for six weeks. For the specific depletion of CD8+ T lymphocytes, we performed intraperitoneal injections of anti-CD8 (InVivoPlus anti-mouse CD8α, BioXcell, Catalog #BP0061, at 100 μg per mouse). Similarly, CD4+T cells were depleted using anti-CD4 (InVivoPlus anti-mouse CD4, BioXcell, Catalog #BE0119, at 100 μg per mouse) every 4 days, extending over six weeks in the induced model and 20 days in the transplanted model. In the experiment described in Fig. 6, mice were treated via intraperitoneal injection every four days for a total of 28 days. Specifically, the treatment protocol included the administration of anti-CD276 antibody (enoblituzumab, MCE, catalog number: HY-P9966, 10 mg/kg), anti-mouse CX3CL1 (Fractalkine) antibody (BioLegend, Cat# 938903, 1 mg/kg), anti-mouse CX3CR1 antibody (GeneTex, Cat. No. GTX27200, 1 mg/kg), and anti-mouse ITGB6 antibody (Anti-Integrin αvβ6 ADCC Enhanced Antibody, Creative Biolabs, Cat# AFC-431CL, 10 mg/kg). Tumor volume was measured using calipers or MRI, with maximum diameters recorded in three orthogonal directions (length, width, height). The tumor volume was then calculated using the cuboid formula V = (π / 6) × (L × W × H)/1000.
Cell lines and cell culture
The mouse Moc1 cell line was purchased from KERAFAST, INC. (Boston, MA, USA, Cat#EWL001-FP) and HNSCC cell lines SCC15 were provided by Cheng Wang from the Department of Maxillofacial Surgery, Guanghua Stomatological Hospital, Sun Yat‐sen University, with the cell line STR authentication report. All cell lines were cultured in DMEM/F12 (GIBCO, C11330500BT) media supplemented with 10% fetal bovine serum (FBS), 1% penicillin/ streptomycin (GIBCO, A3160801). Lentiviral particles with shRNA targeting CAV1, ITGB6, and PRNP were transfected into 293T cells along with packaging plasmids using the Lipofectamine 2000 reagent (GIBCO, 25200072) following the standard lentivirus production protocol as instructed by the manufacturer. In the co-culture system, concentrations of CXCL16 solutions were administered at 0, 50, 100, 200, 400, and 800 ng per milliliter.
Mouse transplantation experiment
For in vivo experiments, 5 × 105 cells were suspended in 100 µL of a 1:1 mixture of PBS and matrigel (Corning, 354234) and administered directly into the base of the tongue in situ. of C57 mice obtained from The Jackson Laboratory. Tumor size was measured biweekly using a digital caliper. Starting from day 8 after tumor injection, mice were intraperitoneally treated with α-CD276 antibody (10 mg/kg) every 4 days or the IgG isotype control. On day 41 after tumor injection, mice were euthanized by CO2 suffocation followed by cervical dislocation, and the tumors were subsequently harvested.
H&E, immunohistochemistry, and immunofluorescence staining
For hematoxylin and eosin (H&E) staining, tumor tissues were obtained from mice and immediately fixed in formalin after dissection. The fixed tissues were dehydrated in ethanol and embedded in paraffin. The processed tissues were sectioned and stained H&E with solution. The sections were scanned using a digital slide scanner (KF-PRO-005-EX) and analyzed with K-viewer software.
For IHC staining, mice tissues were fixed in paraformaldehyde, embedded in paraffin, and dewaxed/rehydrated. Blocking was performed using 5% BSA (Sango Biotech, Shanghai, China). The tissue slides were incubated with primary antibodies against CD276 (14058, CST, Guangzhou, China, 1:200), MKi67 (NB500-170, Novus, Guangzhou, China, 1:200), CD8 (ab52625, Abcam, Guangzhou, China, 1:200), PF4 (ab303494, Abcam, Guangzhou, China, 1:200), GZMB (A2557, abclonal, Guangzhou, China, 1:200) and Cleaved-Caspase-3 (9661 S, CST, Guangzhou, China, 1:200), CX3CL1(ab25093, Abcam, Guangzhou, China, 1:200), CX3CR1(13885-1-AP, Proteintech, Wuhan, China, 1:200), ITGB6 (ab229552, Abcam, Guangzhou, China, 1:200) overnight at 4 °C. Subsequently, the slides were incubated with a secondary antibody for 1 h at room temperature. Signal visualization was achieved using a DAB solution, resulting in brown color development. The slides underwent counterstaining with hematoxylin and were subsequently scanned using a digital slide scanner (KF-PRO-20 MAGSCANCNER KFBIO, KFBIO, Zhejiang, China).
For IF staining, tissue samples were meticulously fixed in paraformaldehyde, followed by permeabilization with 0.1% Triton X-100 in PBS, blocked with 5% BSA in PBS. Incubation with primary antibodies against specific targets was carried out overnight at 4 °C. Subsequently, fluorophore-conjugated secondary antibodies were employed to facilitate signal detection. Nuclei were counterstained using DAPI, enabling precise localization within the tissue samples. High-resolution imaging was conducted using a state-of-the-art fluorescence microscope, employing meticulous adjustments of exposure time, gain, and resolution settings. Rigorous image analysis encompassed the quantification of fluorescence intensity and the implementation of colocalization analysis techniques. The primary antibodies employed in the experiment included.
Western blotting
Mice tissue and cell lysates were prepared by the radioimmunoprecipitation assay (RIPA) lysis buffer supplemented with protease and phosphatase inhibitors. The proteins were concentrated, denatured, and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were then transferred onto a membrane, followed by blocking to prevent non-specific binding. Incubation with primary antibodies against target proteins was conducted overnight at 4 °C. After washing, the membrane was probed with HRP-conjugated secondary antibodies. Protein bands were visualized using enhanced chemiluminescence (ECL, Merck Millipore, IPVH00010) reagents, and the signal was detected and captured. Band intensities were quantified using image analysis software, with normalization to a loading control protein. Blots were probed with antibodies against CAV1 (ab192869, Abcam, 1:1000), PRNP (ab52604, Abcam, 1:1000), ITGB6 (ab187155, Abcam, 1:1000), FAK (3285S, CST, 1:1000), p-FAK (3284S, CST, 1:1000), SRC (AF6161, Affinity Biosciences, 1:1000), p-STAT3 (AP0474, Abclonal, 1:1000), AKT (10176-2-AP, Proteintech, 1:1000), p-AKT (66444-1-Ig, Proteintech, 1:1000), SAPK/JNK (9252, CST, 1:1000), p-SAPK/JNK (9251, CST, 1:1000), ERK1/2 (9102, CST, 1:1000), p-ERK1/2 (9101, CST, 1:1000), p38 MAPK (9212, CST, 1:1000), p-p38 MAPK (9211, CST, 1:1000), PI3K p85 alpha (AF6241, Affinity Biosciences, 1:1000), p-PI3K p85 alpha (AF3241, Affinity Biosciences, 1:1000), NF-κB p65 (8242, CST, 1:1000), p-NF-κB p65 (3033, CST, 1:1000), NF-κB p50 (3035, CST, 1:1000), p-NF-κB p50 (3035S, CST, 1:1000), KLF4 (ab215036, Abcam, 1:1000), CD276 (A17216, Abclonal, 1:1000), and GAPDH (2118S, CST, 1:2000) after washing re-probed with an anti-rabbit horseradish peroxidase-conjugated secondary antibody (SA00001-2, Proteintech). The bands were visualized using ECL (180-5001, Tanon).
Single-cell RNA sequencing
Fresh tumor tissue was finely minced into small pieces and dissociated at 37 °C using the Tumor Dissociation Kit, mouse (130-096-730, Miltenyi) on a gentleMACS Octo Dissociator. Tissue was filtered after dissociation, obtaining single-cell suspensions. Cells were washed with ACK lysing buffer and assessed for viability and count using trypan blue staining. Subsequently, utilizing the GEXSCOPER procedure, single-cell suspensions (1–3 × 105 cells/ml) were captured using the GEXSCOPER microfluidic chip. Each microwell contained a magnetic bead and a cell, labeled uniquely with multiple molecular labels (Unique Molecular Identifiers or UMIs). These UMIs enabled efficient RNA capture upon release from the captured cells, facilitating hybridization with the beads. Magnetic bead-bound cells were subsequently collected in 1.5 ml Eppendorf tubes for reverse transcription. The GEXSCOPER Single Cell RNA Library Kit was employed for cDNA synthesis, followed by PCR amplification. To ensure quality, cDNA fragment size was assessed using Qubit (Thermo, Waltham, MA, USA) for quantification and a fragmentation analyzer, maintaining a repetition rate below 20%. Library construction involved several sequential processes: fragmentation, adapter ligation, purification, PCR amplification, size selection, and rigorous quality control assessments. The indexed libraries were subjected to high-throughput sequencing on the Illumina HiSeq X platform, utilizing 150 bp paired-end reads.
The CellRanger software (version 3.1.0) was employed for read alignment, barcode filtering, and quantification of unique molecular identifiers (UMIs) using the mouse genome (mm10) as the reference for FASTQ inputs. Subsequently, the gene expression matrix was imported into the R software (version 4.0.0). For further downstream analysis, the Seurat package (version 4.0.0) was utilized. The gene expression matrix obtained was subjected to normalization and scaling procedures for each sample. Genes expressed in more than 10 cells were retained for further analysis. Cells with a high proportion (≥10%) of UMIs mapping to mitochondrial genes were excluded. Additionally, cells with detected genes below 200, or more than 5000 were removed. The DoubletFinder function was used for doublet detection and to filter cells with a doublet score exceeding 0.25. The top 2000 highly variable genes were identified using the “vst” method with the FindVariableFeatures function. After data integration, the expression levels of these highly variable genes were scaled and centered along each gene. Principal Component Analysis (PCA) was performed on the selected variable genes, and the top 20 principal components used for cell clustering (with a resolution of 0.5) and Uniform Manifold Approximation and Projection (UMAP) dimensional reduction. Clusters were identified by the FindCluster function in Seurat, employing a resolution parameter of 0.2. The choice of resolution parameter was adjusted based on the cell population being clustered. Differential expression analysis between the identified clusters was conducted using the FindAllMarkers function.
Cell type annotation
Marker genes were identified utilizing the FindAllMarkers function. Cell types were annotated based on top-ranked marker genes and well-established classical markers. Our comprehensive analysis revealed the presence of four major cell types within the studied population, encompassing an array of immune cell types, alongside a diverse range of non-immune cell types including epithelials, endothelials, and fibroblasts. The second round of clustering was conducted according to the same parameters to identify clusters within the major cell types previously mentioned.
Identification of malignant cells
To delineate malignant cells within epithelial populations, we employed the CopyKAT algorithm to estimate copy number variations (CNVs). Through a systematic approach, we selectively sampled a range of 10–100 malignant cells, incrementing by 10 cells at each step. Cells that were inferred as aneuploid by CopyKAT were focused for further analysis.
Scissor analysis to identify survival-associated subpopulations
The Scissor function was employed to identify subpopulations that were associated with bulk phenotypes. In our study, the Scissor pipeline utilized input data consisting of scRNA-seq data, the TCGA-HNSCC expression matrix, and matched clinical phenotype information from the TCGA dataset. A correlation matrix was then constructed to quantitatively assess the similarity between the scRNA-seq data and the clinical phenotype information from TCGA-HNSCC. By analyzing the signs of the estimated regression coefficients, the individual cells were categorized into two groups: Scissor-positive (Scissor+) cells and Scissor-negative (Scissor−) cells. These groups represented cell populations that exhibited positive and negative associations with patient survival, respectively.
Cell–cell communication analysis
To further investigate the intercellular communication within the tumor microenvironment, we employed the CellPhoneDB (https://www.cellphonedb.org) and CellChat (version 1.1.3) functions. Specifically, scRNA-seq data from distinct cell types, including tumor cells, CD4+ T cells, CD8+ T cells, γδ + T cells, neutrophils, and MoMΦDC cells, were utilized as input for CellPhoneDB analysis. MoMΦDC was further divided into PF4+ Macro, Thbs1+ Mono, Prdx1+ Macro, Lars2+ Macro, Ccl17+ Macro, Clec9a+ Macro. Cell–cell interaction heatmaps displaying the number of interactions were generated using the pheatmap R package. Additionally, the CellChat R package (version 1.1.3) was utilized to infer the intercellular communication network between clusters by creating a cellchat object with the normalized expression matrix and visualizing the data using functions such as netVisual_heatmap, netVisual_aggregate, and netAnalysis_contribution function.
Classification of CD8+ T-cell subclusters and trajectory analysis
Based on a comprehensive review of pertinent literature, three distinct gene signatures were delineated: the Cytotoxic Gene Array (comprising GZMB, PRF1, IFNG, GNLY), the Immune-Suppressive Gene Array (featuring PDCD1, TIGIT, HAVCR2, CTLA4, LAG3), and the Effector Memory T-cell (Tem) Gene Array (encompassing TCF7, CCR7, LEF1, SELL). A quantitative assessment of these gene sets was executed utilizing the AddModuleScore algorithm. Pertaining to the analysis of publicly available CD8+ T-cell datasets, we procured single-cell RNA-sequencing datasets specific to Head and Neck Squamous Cell Carcinoma (HNSCC) from three peer-reviewed publications, bearing the PubMed identifiers PMIDs 31924475, 32472114, and 34921143. Subsequent to the acquisition, we employed dimensionality reduction and cluster analysis techniques in accordance with previously established methodologies to facilitate a more nuanced categorization of the CD8+ T-cell populations. Trajectory analyses were conducted to prognosticate the differentiation pathways among distinct CD8+ T cells subtypes by Monocle3 (version 1.0.0).
Flow cytometry
As aforementioned, a single-cell suspension of mouse oral tumor cells was prepared. Prior to staining, the cells were washed in staining buffer (2% bovine growth serum PBS) and resuspended at a concentration of 1 × 107 cells/mL. The cells were stained with antibodies and then resuspended in the staining buffer for 2 h (4 °C) to accomplish extracellular staining. Antibodies and dyes used for the flow cytometer (CytoFLEX, Beckman Coulter) included CD45 PE-Cyanine7 (60-0451, Tonbo Biosciences), CD3 APC (20-0032, Tonbo Biosciences), F4/80 FITC (123107, Biolegend), CD8a VioletFluo 450 (ab242272, Abcam), GZMB PerCP/Cy5.5 (372211, Biolegend), CXCR6 PE (151103, Biolegend), PDCD1 PE-Cyanine5 (Tonbo, 55-5773-U100), PF4 violet FluorTM 450 (Abcam, ab303494). Data were detected using NovoExpress software, and samples were examined using a flow cytometer (CytoFLEX, Beckman Coulter).
CD8+ T cell isolation, activation, and expansion
Fresh oral tumor samples from mice were collected, chopped, and digested in RPMI1640 with type I collagenase (Sigma). The digested cells were filtered through a nylon net (70 micrometers) and rinsed with PBS. For FACS, single-cell suspensions of digested tumors were Incubated for 15 m with 500 ng of Fc blocker (anti-CD16/32, Elabscience), and then for 30 m at 4 °C with anti-CD8a VioletFluo 450 (75-1886, Tonbo Biosciences) (1:200). This procedure was done in order to sort intratumoral CD8+ T cells. Live CD8+ T cells were sorted on a flow cytometer (LSRFOrtessa, BD).
The isolated CD8+ T cells were activated at a 1:1 ratio with CD3/CD28 dynabeads (Thermo Scientific) and cultured in a complete culture detection medium (DMEM-GlutaMAX™-I, 10% FBS) supplemented with 1% penicillin-streptomycin and 100 U/ml IL-2 (Peprotech), at a cell density of 1 × 106 cells/ml, changing the medium every 2–3 days. On day 10, the CD8+ T cells were restimulated with the aforementioned CD3/CD28 dynabeads, with the medium changed every 2–3 days.
PF4+ Macrophage isolation and expansion
Fresh oral tumor samples from mice were collected, chopped, and digested in RPMI1640 with type I collagenase (Sigma). The digested cells were filtered through a nylon net (70 micrometers) and rinsed with PBS. For FACS, single-cell suspensions of digested tumor were Incubated for 15 m with 500 ng of Fc blocker (anti-CD16/32, Elabscience), and then for 30 m at 4 °C with F4/80 FITC (123107, Biolegend) and PF4 PerCP/Cy5.5 (ab303494, Abcam) (1:200). This procedure was done in order to sort intratumoral PF4+ Macrophage. Live PF4+ Macrophage were sorted on a flow cytometer (LSRFOrtessa, BD). The isolated PF4+ Macrophage cultured in complete culture detection medium (DMEM-GlutaMAX™-I, 10% FBS) supplemented with 1% penicillin-streptomycin and 100 U/ml IL-2 (Peprotech) with M-CSF (HY-P7085A, MedChemExpress) and IL4 (HY-P70750G, MedChemExpress), at a cell density of 1 × 106 cells/ml, changing the medium every 2–3 days.
Generation of anti-CD3 expressing Moc1 cells
For lentiviral transfection, 293T/17 packaging cells (ATCC) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 10% heat-inactivated/gamma-irradiated fetal bovine serum (FBS, Invitrogen) and 1 mM MEM non-essential amino acids (Gibco) at 37 °C and 5% CO2. At 70–90% confluence, 293T/17 cells were lentivirally transfected with pCDH-UbC-CD14-OKT3-EF1a-Hygro expression plasmid or a pCDH-UBC-EF1a-Hygro empty plasmid (for mock controls) and a master mix of packaging plasmids (pVSV-G, pPACK-HI-GAG, and Rev-pPACK-HI-REV) using Lipofectamine and Opti-MEM (Gibco). The supernatant containing lentivirus was collected 48 h later, filtered through a 0.2 µm filter (EMD Millipore), and aliquots were stored at −80 °C for transduction of tumor cells. A Lenti-X™ p24 Rapid Titer Kit (Clontech) was used to determine lentiviral titers of the collected supernatants. All tumor cells were maintained in RPMI Medium 1640 – GlutaMAX™-I (Gibco) supplemented with 10% FBS, at 37 °C and 5% CO2. Moc1 cells were grown to 80% confluence and were transduced using lentiviral supernatants at a multiplicity of infection (MOI) of 1 and 10, based on p24 ELISA results, in the presence of polybrene for 48 h. Positively expressing cells were selected using hygromycin B (Invitrogen) for 1 week. Cells were then maintained in hygromycin (30 µg/ml for PC-9 and NCI-H3255; 60 µg/ml for NCI-H1975), passaged, and expanded for all experiments. Anti-CD3 transgene expression was confirmed in cells transduced at an MOI of 1 and 10 by flow cytometry staining, and all future experiments were conducted with cells transduced at an MOI of 1.
Co-culture system
The sorted and activated CD8+ T cells and PF4+ Macrophages were co-cultured with Moc1 in a 48-well plate. In the co-culture system, the Moc1 cells were treated with different proportions of CD8+ T cells and PF4+ Macrophages, or with different concentrations of CXCL16 recombinant protein (HY-P700266, MCE). PF4+ Macrophages and CD8+ T cells were separately collected by flow cytometry. The proportion of GZMB+ CXCR6+ CD8+ T cells or PDCD1+ CXCR6+ CD8+ T cells was determined using the aforementioned flow cytometry method.
ChIP-qPCR
In the process of Chromatin Immunoprecipitation followed by quantitative PCR (ChIP-qPCR), cells are cross-linked with 1% formaldehyde at room temperature for 10 min to capture interacting proteins and DNA, subsequently quenched with 0.125 M glycine. The cells are then lysed to liberate cellular components and dissolve the cross-linked protein-DNA complexes. The soluble chromatin is precipitated with antibodies against Nkfb1 (3035, CST), or with control IgG (B900610-1MG, Proteintech). Real-time qPCR is performed using PerfectStart® Green qPCR SuperMix (AQ601, TransGen Biotech), executed by Bio-Rad CXF96 Real-Time System (Bio-Rad, USA). Data is reported in terms of relative enrichment multiples. The primers used for ChIP-qPCR are listed in the Key Resources Table. Primers used for ChIP-qPCR are listed in Supplementary Data 12.
Statistics
The experiments were replicated at least three times. Differences between two experimental groups were analyzed by the Student’s t-test, whereas comparisons among three or more groups were made by ANOVA followed, if significant, by the Bonferroni post hoc test. All mathematical analyses of the data were performed using GraphPad Prism 8.0, and p < 0.05 was considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The scRNA-seq data generated in this study have been deposited in the Genome Sequence Archive in the National Genomics Data Center under the accession number CRA011974. The publicly available data used in this study include the previously published HNSCC scRNA-seq data from the GEO database, specifically under accession codes GSE139324 and GSE164690. The TCGA HNSCC data is available in the Genomic Data Commons (GDC) Data Portal under accession code TCGA-HNSC. The TCGA Pan-Cancer data can be accessed under accession code TCGA [https://portal.gdc.cancer.gov]. The remaining data are available within the Article, Supplementary Information or Source Data file. Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author Demeng Chen (chendm29@mail.sysu.edu.cn). Source data are provided with this paper.
Code availability
All custom code and algorithms used in this study are available in a DOI-minting repository. The specific DOI for our code repository is https://zenodo.org/record/1306857554. Subsequent versions of the code will be managed in this repository under an open-source initiative-approved license.
References
Robert, C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 11, 3801 (2020).
Waldman, A. D., Fritz, J. M. & Lenardo, M. J. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 20, 651–668 (2020).
Kontos, F. et al. B7-H3: an attractive target for antibody-based immunotherapy. Clin. Cancer Res. 27, 1227–1235 (2021).
Chen, D. et al. Targeting BMI1(+) cancer stem cells overcomes chemoresistance and inhibits metastases in squamous cell carcinoma. Cell Stem Cell 20, 621–634.e6 (2017).
Wang, C. et al. CD276 expression enables squamous cell carcinoma stem cells to evade immune surveillance. Cell Stem Cell 28, 1597–1613.e7 (2021).
Aggarwal, C. et al. Dual checkpoint targeting of B7-H3 and PD-1 with enoblituzumab and pembrolizumab in advanced solid tumors: interim results from a multicenter phase I/II trial. J. Immunother Cancer 10, e004424 (2022).
Shenderov, E. et al. Neoadjuvant enoblituzumab in localized prostate cancer: a single-arm, phase 2 trial. Nat. Med. 29, 888–897 (2023).
Zhang, L. et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 181, 442–459.e429 (2020).
Picarda, E., Ohaegbulam, K. C. & Zang, X. Molecular pathways: targeting B7-H3 (CD276) for human cancer immunotherapy. Clin. Cancer Res. 22, 3425–3431 (2016).
Demaria, O. et al. Harnessing innate immunity in cancer therapy. Nature 574, 45–56 (2019).
Miyamoto, T. et al. B7-H3 suppresses antitumor immunity via the CCL2-CCR2-M2 macrophage axis and contributes to ovarian cancer progression. Cancer Immunol. Res. 10, 56–69 (2022).
Bikfalvi, A. & Billottet, C. The CC and CXC chemokines: major regulators of tumor progression and the tumor microenvironment. Am. J. Physiol. Cell Physiol. 318, C542–C554 (2020).
Hoeft, K. et al. Platelet-instructed SPP1(+) macrophages drive myofibroblast activation in fibrosis in a CXCL4-dependent manner. Cell Rep. 42, 112131 (2023).
Pucci, F. et al. PF4 promotes platelet production and lung cancer growth. Cell Rep. 17, 1764–1772 (2016).
Domschke, G. & Gleissner, C. A. CXCL4-induced macrophages in human atherosclerosis. Cytokine 122, 154141 (2019).
Sartori, M.T. et al. Platelet-Derived Microparticles Bearing PF4 and Anti-GAGS Immunoglobulins in Patients with Sepsis. Diagnostics (Basel) 10, 627 (2020).
Santos-Zas, I. et al. Cytotoxic CD8(+) T cells promote granzyme B-dependent adverse post-ischemic cardiac remodeling. Nat. Commun. 12, 1483 (2021).
Bassez, A. et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med. 27, 820–832 (2021).
Karaki, S. et al. CXCR6 deficiency impairs cancer vaccine efficacy and CD8(+) resident memory T-cell recruitment in head and neck and lung tumors. J. Immunother Cancer 9, e001948 (2021).
Zhang, J. et al. Osr2 functions as a biomechanical checkpoint to aggravate CD8(+) T cell exhaustion in tumor. Cell 187, 3409–3426.e24 (2024).
Mabrouk, N. et al. CXCR6 expressing T cells: functions and role in the control of tumors. Front. Immunol. 13, 1022136 (2022).
Wang, B. et al. CXCR6 is required for antitumor efficacy of intratumoral CD8(+) T cell. J. Immunother Cancer 9, e003100 (2021).
Dudek, M. et al. Auto-aggressive CXCR6(+) CD8 T cells cause liver immune pathology in NASH. Nature 592, 444–449 (2021).
Piehl, N. et al. Cerebrospinal fluid immune dysregulation during healthy brain aging and cognitive impairment. Cell 185, 5028–5039.e13 (2022).
Bagati, A. et al. Integrin alphavbeta6-TGFbeta-SOX4 pathway drives immune evasion in triple-negative breast cancer. Cancer Cell 39, 54–67.e9 (2021).
Vannini, A. et al. alphavbeta3-integrin regulates PD-L1 expression and is involved in cancer immune evasion. Proc. Natl Acad. Sci. USA 116, 20141–20150 (2019).
Busenhart, P. et al. Inhibition of integrin alphavbeta6 sparks T-cell antitumor response and enhances immune checkpoint blockade therapy in colorectal cancer. J. Immunother Cancer 10, e003465 (2022).
Gao, R. et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat. Biotechnol. 39, 599–608 (2021).
Chen, J. et al. Aberrant translation regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine modification drives head and neck squamous cell carcinoma progression. Cancer Commun. 42, 223–244 (2022).
Sun, D. et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat. Biotechnol. 40, 527–538 (2022).
Bakos, E. et al. CCR2 regulates the immune response by modulating the interconversion and function of effector and regulatory T cells. J. Immunol. 198, 4659–4671 (2017).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Li, M. et al. Integrins as attractive targets for cancer therapeutics. Acta Pharm. Sin. B 11, 2726–2737 (2021).
Cooper, J. & Giancotti, F. G. Integrin signaling in cancer: mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell 35, 347–367 (2019).
Liu, F., Wu, Q., Dong, Z. & Liu, K. Integrins in cancer: emerging mechanisms and therapeutic opportunities. Pharm. Ther. 247, 108458 (2023).
Slack, R. J., Macdonald, S. J. F., Roper, J. A., Jenkins, R. G. & Hatley, R. J. D. Emerging therapeutic opportunities for integrin inhibitors. Nat. Rev. Drug Discov. 21, 60–78 (2022).
Ribas, A. Adaptive immune resistance: how cancer protects from immune attack. Cancer Discov. 5, 915–919 (2015).
Vesely, M. D., Zhang, T. & Chen, L. Resistance mechanisms to anti-PD cancer immunotherapy. Annu. Rev. Immunol. 40, 45–74 (2022).
Wang, Z. et al. Syngeneic animal models of tobacco-associated oral cancer reveal the activity of in situ anti-CTLA-4. Nat. Commun. 10, 5546 (2019).
Breuss, J. M. et al. Expression of the beta 6 integrin subunit in development, neoplasia and tissue repair suggests a role in epithelial remodeling. J. Cell Sci. 108, 2241–2251 (1995).
DeNardo, D. G. & Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 19, 369–382 (2019).
Mantovani, A., Allavena, P., Marchesi, F. & Garlanda, C. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov. 21, 799–820 (2022).
Mariathasan, S. et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548 (2018).
Pan, Y., Yu, Y., Wang, X. & Zhang, T. Tumor-associated macrophages in tumor immunity. Front. Immunol. 11, 583084 (2020).
Nakamura, K. & Smyth, M. J. Myeloid immunosuppression and immune checkpoints in the tumor microenvironment. Cell Mol. Immunol. 17, 1–12 (2020).
Schmall, A. et al. Macrophage and cancer cell cross-talk via CCR2 and CX3CR1 is a fundamental mechanism driving lung cancer. Am. J. Respir. Crit. Care Med. 191, 437–447 (2015).
Zheng, J. et al. Chemokine receptor CX3CR1 contributes to macrophage survival in tumor metastasis. Mol. Cancer 12, 141 (2013).
Cascio, S. et al. Cancer-associated MSC drive tumor immune exclusion and resistance to immunotherapy, which can be overcome by Hedgehog inhibition. Sci. Adv. 7, eabi5790 (2021).
Zhang, W. et al. Crosstalk between IL-15Ralpha(+) tumor-associated macrophages and breast cancer cells reduces CD8(+) T cell recruitment. Cancer Commun. 42, 536–557 (2022).
Di Pilato, M. et al. CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell 184, 4512–4530.e22 (2021).
Blanchard, C. et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J. Clin. Invest. 116, 536–547 (2006).
Hua, C., Shukla, H. I., Merchant, T. E. & Krasin, M. J. Estimating differences in volumetric flat bone growth in pediatric patients by radiation treatment method. Int J. Radiat. Oncol. Biol. Phys. 67, 552–558 (2007).
Zhang, C. et al. ITGB6 modulates resistance to anti-CD276 therapy in head and neck cancer by promoting PF4+ macrophage infiltration. Code repository, https://zenodo.org/record/13068575 (2024).
Braun, D. A. et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat. Med. 26, 909–918 (2020).
Kurten, C. H. L. et al. Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing. Nat. Commun. 12, 7338 (2021).
Cillo, A. R. et al. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity 52, 183–199.e9 (2020).
Acknowledgements
We thank the members of the Peng and Lin labs for reagents, discussions and data-analysis support. This work is supported by the National Natural Science Foundation of China (82173362, 81872409, 82304069, and 823B2079), Guangzhou Municipal Science and Technology Bureau (2024B03J1384), and China Postdoctoral Science Foundation (2023M734003). The schematic diagrams in Figs. 1A, E, 4A, 5E, 9A, Supplementary Figs. 2, 3B, 5A, C, M, 8A, G, 11A, H, 12E were made with MedPeer.
Author information
Authors and Affiliations
Contributions
Conceptualization, C.Z., K.L., M.C., C.W., and D.C.; Methodology, C.Z., K.L., H.Z., M.C., S.C., and R.L.; Data Analysis and Curation, C.Z., K.L., M.C., S.C., R.L., and D.C.; Investigation and Validation, C.Z., K.L., M.C., S.C., R.L., and D.C.; Resources, H.Z., S.C., C.W., and D.C.; Writing-Original Draft, K.L., M.C., and D.C.; Writing-Review & Editing, K.L., M.C., and D.C.; Supervision and funding acquisition, S.C., K.L., and D.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jorrit De Waele and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, C., Li, K., Zhu, H. et al. ITGB6 modulates resistance to anti-CD276 therapy in head and neck cancer by promoting PF4+ macrophage infiltration. Nat Commun 15, 7077 (2024). https://doi.org/10.1038/s41467-024-51096-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51096-0
This article is cited by
-
Mechanisms for fibrate lipid-lowering drugs in enhancing bladder cancer immunotherapy by inhibiting CD276 expression
BMC Cancer (2025)
-
Yuanhuacine suppresses head and neck cancer growth by promoting ASCT2 degradation and inhibiting glutamine uptake
Acta Pharmacologica Sinica (2025)
-
Resistance to anti-LAG-3 plus anti-PD-1 therapy in head and neck cancer is mediated by Sox9+ tumor cells interaction with Fpr1+ neutrophils
Nature Communications (2025)
-
ITGB6 modulates resistance to anti-CD276 therapy in head and neck cancer by promoting PF4+ macrophage infiltration
Nature Communications (2024)