Abstract
Microeukaryotes are key contributors to marine carbon cycling. Their physiology, ecology, and interactions with the chemical environment are poorly understood in offshore ecosystems, and especially in the deep ocean. Using the Autonomous Underwater Vehicle Clio, microbial communities along a 1050 km transect in the western North Atlantic Ocean were surveyed at 10–200 m vertical depth increments to capture metabolic signatures spanning oligotrophic, continental margin, and productive coastal ecosystems. Microeukaryotes were examined using a paired metatranscriptomic and metaproteomic approach. Here we show a diverse surface assemblage consisting of stramenopiles, dinoflagellates and ciliates represented in both the transcript and protein fractions, with foraminifera, radiolaria, picozoa, and discoba proteins enriched at >200 m, and fungal proteins emerging in waters >3000 m. In the broad microeukaryote community, nitrogen stress biomarkers were found at coastal sites, with phosphorus stress biomarkers offshore. This multi-omics dataset broadens our understanding of how microeukaryotic taxa and their functional processes are structured along environmental gradients of temperature, light, and nutrients.
Similar content being viewed by others
Introduction
The western North Atlantic Ocean encompasses a diverse collection of ecosystems, including a subtropical gyre, a continental margin, and productive coastal waters, differing in temperature, nutrient availability, and organic carbon levels. The warm subtropical gyre waters are largely separated from cold, productive subpolar waters by the western boundary Gulf Stream current, with mixing occurring across the front1,2,3 (Fig. 1). These distinct oceanographic properties select for the type of microorganisms present and their functional capabilities, with distance from shore and temperature acting as key influences shaping microbial communities4. In oligotrophic gyre waters of the North Atlantic, nitrogen (N) limitation is a primary bottom-up control on phytoplankton abundance, with phosphorus (P) and iron (Fe) additionally limiting or co-limiting growth5,6,7. Little is understood about the metabolic status of microbial communities occupying transitionary waters between the subtropical gyre and the continental shelf, and how the differing nutrient supplies and physical dynamics influence phytoplankton physiology along this gradient.
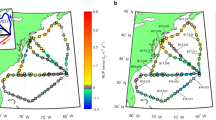
Biological sampling was performed using the AUV Clio and additional parameters were collected via CTD rosette at each station. The surface transect is shown overlaying sea surface temperature and satellite-derived chlorophyll a concentrations between June 19–28th 2019, obtained from NASA MODIS. Continuous beam transmission (turbidity), temperature, and dissolved oxygen measurements were obtained via the ship CTD, and dissolved nutrients were measured from discrete samples from the top 1500 m of the water column (see Supplementary Fig. 1 for full-depth temperature and oxygen sections). Note that surface nitrate+nitrite concentrations dropped below the detection limit of 0.06 μM in surface waters of St. 3, 6, and 7, and silicate concentrations were below the detection limit of 0.19 μM in the top 100 m throughout the section (Supplementary Data 1). Seawater samples collected by Clio for metaproteomics and metatranscriptomics were obtained from the depths shown in the bottom section plots.
The planktonic communities in the western North Atlantic experience strong seasonal forcings, subject to interannual and decadal-scale climate variability8. A major source of nutrients to the North Atlantic is seasonally variable aerosol dust from the Saharan Desert9,10,11,12, with most of the deposition in Bermuda occurring in the late summer13. These deposition events introduce dissolved Fe and N to surface waters, which are rapidly consumed to support phytoplankton growth and may drive the system towards P limitation14. Toward the subpolar-influenced northeast US continental margin, overall phytoplankton biomass increases, and the shelf/slope becomes a dominant source of Fe15,16,17. The offshore (subtropical) to coastal (subpolar) section thus represents a natural biogeochemical nutrient gradient (Fig. 1). The North Atlantic continental shelf ecosystem is also experiencing rapid warming, and the chemical and ecological ramifications for phytoplankton communities are unclear18.
Microeukaryotes, or protists, serve critical roles in the marine environment by performing primary production, transferring organic material to higher trophic levels, moving carbon to deeper layers of the ocean, and facilitating symbiotic relationships19. Microeukaryotes are capable of strict autotrophy, strict heterotrophy, or a combination of the two trophic strategies through mixotrophy20. Three decades of rich biogeochemical data collected at the Bermuda Atlantic Time-series Study (BATS) site located in the Sargasso Sea has established a baseline for phytoplankton production and community structure in this region21. Surface phytoplankton communities are generally dominated by small cells including cyanobacteria and haptophytes22, with cyanobacteria abundant in the winter and picoeukaryotes/nanoeukaryotes abundant in the spring, coinciding with a shoaling of the mixed layer and the annual North Atlantic spring bloom23,24. Deep sea microeukaryote communities in this region are distinct from surface communities, and include Acantharea and Polycystinea radiolaria and Euglenozoa discoba25,26. Although considerable progress has been made in cataloging microeukaryote diversity and ecology in the oligotrophic North Atlantic, we lack an understanding of how these oligotrophic communities differ from those in continental margin ecosystems, and which environmental factors are responsible for changes in community structure and physiology along such transition zones.
Obtaining an accurate understanding of microeukaryote metabolism and their relationships to nutrient availability over large spatial expanses of the open ocean requires specialized sampling approaches. In particular, vertical profiling autonomous underwater vehicles (AUVs) capable of in situ filtration are particularly useful for the collection of microbial biomass27,28. Clio is a specially designed AUV that actively transits vertically in the water column to a maximum depth of 6000 m, drifts in a Lagrangian manner, and is outfitted with sensors for measuring chlorophyll fluorescence, salinity, pressure, temperature, dissolved oxygen, and optical backscatter27. Clio can collect biomass by holding position at <5 cm vertical resolution27, and therefore enables an unparalleled opportunity to characterize microeukaryote metabolism using a highly efficient vertical sampling scheme.
Here we present microbial community structure and metabolic profiles obtained with Clio along a western North Atlantic Ocean offshore-coastal transect, with a focus on microeukaryotes. The ecosystems along this transect were characterized by physicochemical parameters, macronutrients, pigments, and dissolved trace metals. Physiology and ecological interactions were inferred through complementary data-dependent-acquisition (DDA) mass spectrometry metaproteomics and short-read metatranscriptomics. Our results demonstrate that the microeukaryote communities were primarily composed of stramenopiles, dinoflagellates, and ciliates, which were detected throughout the water column. We identify several incidents of concordance and discordance between the transcript and protein pools investigated, which together construct a view of the epipelagic, mesopelagic, and bathypelagic microbial communities and their functional roles. Along the environmental gradient, the nutrient physiology of major microeukaryotic groups shifted with evidence of nitrogen stress in coastal waters and phosphate stress in the oligotrophic gyre. We furthermore examined the vertical structure of microeukaryote metabolism through the water column, revealing functional processes coupled to depth zone and co-occurrences among members of different taxonomic groups.
Results and discussion
This study region encompasses several nutrient regimes: a subtropical oligotrophic ecosystem, a transitional continental margin region impacted by the Gulf Stream, and coastal productive waters receiving subpolar influence. Prominent oceanographic features captured include a persistent low oxygen zone, a sedimentary nepheloid layer along the continental margin, and deep chlorophyll maxima (Fig. 1, Supplementary Fig. 1). This transect is similar to the Western leg of the GEOTRACES GA03 North Atlantic Zonal Transect (NAZT), also known as Line W29.
Taxonomic composition of RNA and proteins
Transcripts were assembled using sequences derived from poly-A-selected mRNA, which enriches for eukaryotic members of the community (Supplementary Fig. 2A). Approximately 23% of the annotated open reading frames (ORFs) belonged to prokaryotes (Supplementary Fig. 2C), indicating that bacterial mRNA was not excluded in the library preparation process. Up to 50% of bacterial mRNA can undergo polyadenylation, and this process may be associated with a variety of functions including mRNA degradation, stabilization, or translation, in contrast to the eukaryotes, which polyadenylate most of their mRNA primarily to carry out translation30,31. The prokaryotic poly-A selected mRNA in this analysis may, therefore, not be reflective of the true prokaryotic community, and was instead used as a qualitative assessment of prokaryotes along the transect and depth zones. The poly-A-based metatranscriptome assembly enabled the identification of both eukaryotes and bacteria in the metaproteomes (Supplementary Fig. 2B). Eukaryotes were not enriched during protein processing as they were during mRNA processing, and the metaproteomes, therefore, more accurately reflect the natural proportion of prokaryotes relative to eukaryotes in the water column (Supplementary Fig. 2B).
A notable feature revealed by the metaproteomes is that of the prokaryotes detected, cyanobacteria consistently peaked in relative abundance at the deep chlorophyll maxima (DCMs) throughout the transect (Supplementary Fig. 2B), despite the relatively small number of cyanobacteria proteins predicted from the metatranscriptome assembly (Supplementary Fig. 2C). This is consistent with previous observations of elevated Prochlorococcus abundance at the DCM in the Sargasso Sea, where this organism thrives under warm, low macronutrient and low light conditions32. For the majority of this analysis, we focus our investigations on the microeukaryotic component of this diverse microbial community. Additional features of the pelagic prokaryotic community of the Sargasso Sea determined with Clio have been reported elsewhere27 and will be reported in a future study.
Of the total community detected on filters, approximately one-quarter of the transcript reads, and protein spectral counts were associated with predicted ORFs that could not be distinguished at the broadest classification level (“NA”; Supplementary Fig. 2). These are ORFs that were similar to multiple, distantly related references in our taxonomic database such that they were not possible to confidently assign using our sequence homology and LCA classification approach33.
The 10–200 m resolution sampling scheme of our metaproteomic approach allows for the visualization of strong vertical shifts in eukaryotic community composition at both the supergroup level and at finer taxonomic levels. Surface communities were comprised of dinoflagellates (Gymnodiniales, unidentified marine alveolates [MALVs]), haptophytes (Prymnesiales), mixed stramenopiles (dictyophytes, diatoms, pelagophytes), ciliates (Spirotrichea) and chlorophytes (Fig. 2; Supplementary Figs. 3–6). The diversity of surface phytoplankton communities is supported by pigments measured from the water column demonstrating the presence of diatoms (fucoxanthin), haptophytes (19’hex-fucoxanthin and fucoxanthin), chlorophytes (chlorophyll b), and cyanobacteria (zeaxanthin and divinyl chlorophyll a)34 (Supplementary Fig. 12). Dinoflagellates are also known to harbor many of the eukaryotic pigments, gained through secondary and tertiary endosymbiosis35. Although pigments have long been used to decipher marine phytoplankton community composition, biomarker pigments may be variable within and among taxonomic groups, complicating efforts to directly relate them to taxonomic assemblages36. The coherence among pigments, transcripts, and extracted protein pools is further discussed below (See Quantitative estimates of biomass).
Relative abundance of transcript reads assigned to open reading frame (ORF) predicted proteins (a), and exclusive proteomic spectral counts assigned to ORFs (b) within the eukaryotes. Each row represents a station and depth (“7_700” = St. 7700 m). Only microeukaryote transcripts having a EUKulele taxonomic annotation are shown. Reads associated with unannotated ORFs are not shown. Note that 30 m and 115 m surface samples at BATS (St. 1) were collected using McLane pumps instead of Clio, with separate 0.2–3 μm (“0.2”) and 3–51 μm (“3”) filter fractions. NA represent microbes with lineage-conflicted taxonomic assignments and are unable to be classified at the eukaryotic supergroup level. c The relative abundance of taxonomically annotated eukaryotic ORFs contained in the translated metatranscriptome database is displayed as a single stacked barplot.
Mesopelagic communities >200 m reflected a greater relative abundance of phaeophyte and pelagophyte stramenopiles, foraminifera (Rotaliidae), radiolaria (Symphyacanthida), discobans (Euglenozoa), and picozoa (Fig. 2, Supplementary Figs. 7–9). Many of these groups are similarly relatively abundant in the Sargasso Sea based on 18 S rRNA V4 amplicons, with communities also shifting toward heterotrophic taxa below the DCM26,37. Dinoflagellates, stramenopiles, fungi, and radiolaria are furthermore present in the deep ocean sediment and are hypothesized to be important taxa involved in carbon export38. These taxa contain autotrophic, heterotrophic, and mixotrophic members, and our deep omic signatures could reflect cells both passively sinking from the photic to aphotic zone, and resident communities adapted to the deep ocean. The vertical community shifts were more pronounced in the protein pool, whereas transcripts were more evenly distributed throughout the water column (Fig. 2a, b).
Although the proteomic assessment demonstrated heterotrophic microeukaryotes as key taxa in deep waters, foraminifera in the mesozooplankton size fraction (>500 μm) was in highest absolute concentration in surface waters along the continental margin, as determined via Underwater Vision Profiler (UVP) imagery (Supplementary Fig. 13). UVP imagery also supports radiolarian (Collodaria, Phaeodaria, and Acantharea) presence across the transect with highest concentrations in surface waters towards the coastline where plankton biomass accumulates (Supplementary Fig. 13). These cell counts differ from our protein profiles (Fig. 2b) and previously reported relative 18 S rDNA composition26, in which rhizaria (including radiolaria) of the Sargasso Sea notably increase with depth. These differences could be due to rhizaria being relatively dominant microeukaryotes in the mesopelagic where overall plankton biomass is low. It is important to note that the omics method captured microeukaryotes in the 0.2–51 μm size range while UVP captured mesozooplankton >500 μm, thus these methods are not directly comparable. Deep sea radiolaria, in particular, may be smaller than commonly observed surface taxa or have sufficiently different morphology, complicating direct comparisons between imaging and omic data sets39. In addition, the radiolarian transcripts and proteins detected could be derived from the <5 μm gamete-like swarmer reproductive stage40 captured on Clio filters, while images are biased towards the larger adult stage. Radiolaria are hypothesized to sink down below the mesopelagic zone to complete their reproduction cycle40, which is consistent with radiolarian proteins observed >200 m along the transect (Fig. 2b). It is also worth noting that we are less confident in fine-scale rhizaria taxonomic assignments given their poor representation in existing databases33.
Microeukaryote community composition continued to shift through the mesopelagic and into the bathypelagic. Ascomycota fungal proteins were enriched below 3000 m at St. 6, the only site where bathypelagic water was collected (Fig. 2b, Supplementary Fig. 10). Fungi are important microeukaryotes driving biogeochemical cycling and are poorly understood in offshore marine systems41,42. These results are consistent with 18 S rDNA data that show that this group comprises key members of the global bathypelagic zone, along with rhizaria, stramenopiles, and alveolates (including dinoflagellates and ciliates)43. These fungal proteins may represent small cells or structures (<51 μm), similar to small fungi detected in the 3–10 μm filter fraction in coastal ecosystems44. Large fungal structures (>51 μm) would have been missed in this omic analysis unless captured as cell debris during remineralization or broken apart during filtration through the 51 μm membrane. Approximately half of these annotated fungal proteins above the seafloor (4050–4100 m) were aldehyde dehydrogenases, involved in a number of cellular processes including detoxification and biosynthesis pathways45. A benthic nepheloid layer was observed at this site as determined with beam transmission (turbidity), appearing at ~4000 m (Supplementary Fig. 14). A nepheloid layer in this region has been previously characterized on the GEOTRACES NAZT expedition along the Line W transect, containing elevated suspended particulate matter above the continental margin, largely composed of lithogenic material with slight enrichment in particulate organics46. Particulate trace metal concentrations are additionally elevated in this nepheloid layer, potentially serving as a metal source to bathypelagic microbes47,48. Fungi are consumers of organic detrital material49,50, are hypothesized to contribute to methane and nitrogen cycling51, and are active degraders of carbohydrates and proteins throughout the water column and in the subseafloor52,53,54. It is unclear whether fungi benefit from enhanced particulate material in the nepheloid layer above the seafloor, or are generally abundant at these depths of the western North Atlantic. Additional deep ocean surveys are required to further understand the vertical spatial extent of pelagic fungi in the bathypelagic55 and their relationship to nepheloid layers and the seafloor41,56.
Concordance between transcript and protein pools
This analysis provided a valuable opportunity to evaluate protein and transcript concordance in the marine ecosystem, and to examine how community composition and functional assessments differ based on the molecular pool examined. Instances of protein and transcript concordance are insightful, as transcripts and proteins are not expected to uniformly agree given that the two pools have inherently different cellular concentrations, degradation times, and regulatory controls (summarized in Supplementary Data 2). Proteins can be 30,000 times more abundant in cells than transcripts57 and reflect cell activity, whereas transcripts are sensitive to rapid changes in the environment and subject to post-transcriptional regulation processes that may prevent transcripts from representing cell phenotype58. Importantly, there are key differences in the half-lives of these molecules. Transcripts are quickly produced and degraded in cells, and decrease once protein needs are met, resulting in large dynamic ranges. In contrast, proteins are generally more stable with larger inventories, though have variable turnover times59. Hence, proteins and transcripts are distinct molecular pools that are not universally expected to correlate. In environmental data sets, the comparison between these two pools is further complicated by the differences in functional and taxonomic coverage (Fig. 2a, b), potentially due to filtration time, extraction, amplification, or analytical biases (Supplementary Data 2). Nevertheless, environmentally responsive transporters and enzymes that accumulate in both transcript and protein fractions may indicate a cellular effort to maximize substrate uptake, and can result in an increase in inventory and resultant strong signal detected in both molecular fractions, as predicted in a recent modeling study using mRNA and protein turnover times60,61.
We investigated dataset-wide relationships between individual microeukaryote transcripts and proteins. To achieve this, a partial least squares (PLS) regression was performed on genes of interest (Fig. 3). Linear agreement was shown with nitrogen biomarkers, with a nitrate transporter (NRT) protein positively correlated with NRT, nitrate reductase, ferredoxin-nitrite reductase, and glutamate synthase transcripts. These transcripts were also positively correlated with cytochrome b6f complex proteins, consistent with nitrogen utilization and photosynthesis co-occurring in the upper water column. Transcripts and proteins of the respiratory protein NADH dehydrogenase furthermore positively correlated, specifically subunit 3. NADH dehydrogenase protein subunits 2 and 3 demonstrated some of the strongest positive relationships with transcripts also expressed in surface waters, including a zinc transporter (ZIP2), flavodoxin, carbonic anhydrase, and the inorganic phosphate transporter PHO84 (Fig. 3). Negative correlative relationships were also identified, and but were largely related to environmental distributions of where metabolic processes were occurring rather than intracellular biochemistry. For example, NRT proteins were detected in surface waters where nitrogen uptake to fuel photosynthetic growth occurs, and negatively correlated to ferredoxin and Cu/Zn SOD transcripts in deep water where they were presumably used in metal-dependent heterotrophic metabolism. Conversely, surface transcripts related to nitrogen assimilation and photosynthesis (flavodoxin, nitrate reductase) were negatively correlated with peptide/nickel transport system proteins that were primarily detected in the deep ocean.
Transcripts are labeled as rows (horizontal), proteins as columns (vertical), and clustered based on similarity. Annotations are color-coded by nutrient and trace metal functional categories133. Note: more KEGG Orthologs were detected in the metatranscriptome than in the metaproteome, resulting in more rows on the x axis.
Next, linear relationships were investigated among individual ORFs and aggregated on the Kyoto Encyclopedia of Genes and Genomes (KEGG) Ortholog functional level. There was no agreement between individual transcript and proteins on the ORF level (r2 = 0.006), or when averaged by KEGG IDs (r2 = 0.04; Supplementary Fig. 15A, B). Using Rubisco ORFs as an example functional group, there was also no agreement (r2 = 0.006; Supplementary Fig. 15C). However, there was linear agreement when counts associated with ORFs were aggregated (summed) to the KEGG functional level (r2 = 0.54) (Supplementary Fig. 15D). Some of the most abundant and positively correlated processes between the fractions were the carbon fixation enzyme Rubisco and the cytoskeletal protein tubulin (Supplementary Fig. 15D). We interpret this to indicate certain ORFs receive peptide spectral counts that transcripts do not, and vice versa, potentially due to redundant taxonomic features and the nature of assigning peptide-spectrum matches to representative protein groups. When summarized to higher functional classification, processes that were relatively abundant were reflected in both pools.
On the other hand, transcripts and proteins differed in functional and taxonomic agreement in many instances (summarized in Supplementary Data 3). In surface waters, haptophyte proteins were well-represented, reflecting 17% of eukaryotic proteins <200 m (Fig. 2b), but transcripts suggest they were minor members of the surface community, only comprising 7% of eukaryotic reads (Fig. 2a). Similarly, metazoan transcripts were relatively abundant (Fig. 2a), with most transcript reads belonging to a single sponge reference in the EukProt database (Vazella pourtalesii; Supplementary Fig. 11), while metazoa were comparatively minor constituents of the protein pool (Fig. 2b). Differences were additionally apparent at finer taxonomic levels. Silica skeleton-containing Polycystinea radiolaria transcripts were detected throughout the water column, yet proteins were mainly detected in the mesopelagic and largely represented by strontium sulfate-containing Symphyacanthida instead62 (Supplementary Fig. 9). Transcripts indicated a diverse ciliate population including Armophorea, Oligohymenophorea, and Spirotrichea, while proteins indicated that the ciliate population was nearly exclusively composed of Spirotrichea (Supplementary Fig. 6). On the metaproteomic side, these patterns could in part be due to shared peptides between closely related taxa masking diversity63, and/or not detecting rarer peptides that are sequenced in transcriptomes. In the transcript fraction, certain organisms may be relying on post-transcriptional modification for protein regulation64, resulting in a larger transcript pool (Supplementary Data 2).
In addition to these taxonomic distinctions between biochemical pools, we observed an unexpected difference in the proportion of the eukaryotic communities able to be confidently classified. On average, 21 ± 6% of annotated metatranscriptomic communities (n = 44) were associated with ORFs unable to be classified at the supergroup level, while an average of 38 ± 5% of metaproteomic communities (n = 86) were associated with these lineage-conflicted ORFs (two-tailed p = 1.01 × 10−34, Student’s t test, t = 17.03, degrees of freedom=128; Fig. 2: “Not Annotated (NA)”). These lineage-conflicted proteins were composed of a higher relative abundance of functional annotated33 cytoskeletal proteins (Supplementary Fig. 16). We attribute the large portion of lineage-conflicted eukaryotic ORF proteins to the collection of evolutionary conserved protein machinery with high sequence similarity across supergroups, such as exoskeleton components65, potentially with large inventories and slow degradation rates. These proteins may belong to a collection of genomically diverse eukaryotes sharing conserved proteins, or could instead belong to one or a few taxa dominant in the environment that are not included in the taxonomic database used here. It also is possible Clio captured these abundant and conserved proteins shed from organisms across trophic levels in the ecosystem as “eProtein”, similar to “eDNA”66, in addition to collecting biomolecules from intact cells. It is unlikely that the DDA mass spectrometry method was the reason behind this bias in conserved proteins. Although taxonomically conserved peptides may be abundant in extracted protein pools, a dynamic exclusion approach was used, which limits the continued fragmentation of a given parent ion.
The PLS correlation analysis showed most transcript/protein pairings lacked strong linear relationships, including ferritin, Cu/Zn superoxide dismutase, and the inorganic phosphate transporter (Fig. 3). Phosphate transporter transcripts and proteins have been shown to be correlated in the marine diatom Thalassiosira pseudonana, with both upregulated under phosphate stress67. The cause of the discordance here is likely related to differences in taxonomic coverage between the molecular pools, with only a few detected PHO84 and PiT proteins belonging to stramenopiles and haptophytes, while transcripts were detected from a greater assortment of protists (Supplementary Data 9, 10). We further investigated specific functional processes that were highly divergent between molecular pools (Supplementary Fig. 15D), being abundant in the protein fraction yet weakly expressed in transcripts and vice versa. In the proteins, some of the largest relative abundance counts were associated with a protein related to the eukaryotic cytoskeleton (neurofilament medium polypeptide, vimentin and/or desmin), and a cilia-associated protein (Bardet-Biedl syndrome 5 protein). It is possible cytoskeletal proteins are stabilized through post-translational modifications or by incorporation into a cytoskeletal interlinked filament lattice68, therefore leading to enrichment in the protein fraction. In transcripts, high counts were associated with ribosomal proteins (large subunits 6 and 22, small subunit 2), and a calcium/calmodulin-dependent serine/threonine protein kinase. A weak negative relationship between mitochondrial ribosomal proteins and transcripts has also been documented in human lymphocytes69, potentially related to ribosomal mRNA being more stable than ribosomal proteins69,70. Thus, differences in mRNA/protein stability and inventory may both contribute to the cause of the disconnect between transcripts and proteins detected here.
In addition to taxonomic coverage and molecule stability, other possible explanations for the observed discrepancies in our dataset include lineage-specific loss of proteins or RNA during the extraction, amplification, or analytical steps71,72,73, and/or a biological distinction in their biochemical inventories with differences in RNA/protein ratios among taxonomic groups74 (Supplementary Data 2). It is furthermore probable that as biomass accumulates on filters during in situ filtration, a portion of cells lyse under pressure75. This may lead to a loss in soluble RNA and proteins, but the retention of membrane-bound proteins along with particulate material on filters, thus contributing to observed differences between protein/RNA fractions. Large, fragile eukaryotes with gelatinous structures may be especially vulnerable to cell rupture during filtration. Both protein- and RNA-based community composition assessments may also be biased by inefficient zooplankton feeding and host-specific viral lysis, leading to the leakage of organic material from certain microeukaryotic groups76. Both fractions may also be influenced by growth rate, with faster-growing cells producing more ribosomal machinery and especially more RNA77,78,79, thereby reflecting higher relative abundance in the community. A multitude of analytical and biological factors, therefore, contribute to the taxonomic profiles observed here, which may differ from assessments based on bulk seawater cell densities. These results highlight that a combination of taxonomic metrics is more insightful than any one individual in reconstructing microbial community composition.
Quantitative estimates of biomass
Biomass was assessed using three metrics: total summed pigment concentrations (ng kg−1), protein abundance (spectral counts L−1), and semi-quantitative eukaryotic transcript estimates (copies L−1) (Supplementary Figs. 17 and 18). Note that internal standards were added after RNA extractions and prior to sequencing. As a result, this estimate assumes efficient yield of RNA during extractions and are likely high estimates (see methods). The highest levels of aggregated transcripts and proteins were measured in surface waters, but with transcripts being on average ~130-fold higher in surface waters than in the deep ocean, and proteins being on average 16-fold higher, reflecting a less dynamic range (Supplementary Fig. 17). Pigments consistently showed highest concentrations at the deep chlorophyll maxima (Supplementary Fig. 12), which may represent both increases in biomass and increases in chlorophyll a content per cell under low light conditions80. Transcripts (copies L−1) increased alongside pigments (ng kg−1) with a strong positive correlation evident in surface waters(r2 = 0.7) (Supplementary Fig. 18). Total proteins (spectral counts L−)1 showed a moderate positive correlation with pigment concentrations (r2 = 0.5). Transcripts and proteins showed similar moderate agreement along the transect (r2 = 0.48) (Supplementary Fig. 18). These results support that RNA and protein pools81,82 can be useful estimates of phytoplankton biomass (Supplementary Fig. 18). Observed disconnects among these measurements may be partially explained by protein pools containing abundant prokaryotes not reflected in the pigments and transcripts (Supplementary Fig. 2B, Supplementary Fig. 12), and with transcript data enriched for eukaryotes by use of poly-A transcript selection (Supplementary Fig. 2A).
At the St. 8 coastal site, pigments, and eukaryotic transcripts peaked in surface waters (40 m), where the thermocline shoaled and CTD chlorophyll fluorescence spiked (Supplementary Fig. 1), suggesting elevated biomass. A previous survey along a similar Sargasso Sea-coastal transect also showed highest chlorophyll a biomass north of the Gulf Stream where temperatures decreased, in which large plankton (>10 μm) were elevated in abundance32. In particular, we observed the highest contribution of fucoxanthin pigments at this site (Supplementary Fig. 12). There was a disconnect with omics data, with the relative abundance of stramenopile/diatom transcripts and proteins not similarly increasing at this location compared to other nearby sites (Fig. 2, Supplementary Fig. 3). This could suggest a limited ability of relative/omics-based community composition data to reflect bulk changes in community composition.
Metatranscriptomics has been demonstrated to generally reconstruct phytoplankton groups using microscopy-based observations83,84, but both molecular pools and pigment concentrations may be influenced by genetic factors and/or cell physiology, and therefore deviate from microscopy or image-based cell concentrations. The use of several RNA standards may produce more reliable quantitative transcript measures. Additional direct measurements of biomass (e.g. flow cytometry, microscopy) are recommended in future studies to further establish quantitative links between omics and cell densities, although these methods each have their own strengths and weaknesses. For example, flow cytometry excludes >100 μm eukaryotes, and traditional microscopy misses many picoplankton. Additionally, the size of organisms needs to be taken into account, as large organisms reach lower absolute cell concentrations given their larger volume, but still contribute to the biomolecular pool, perhaps even disproportionately. These results demonstrate that there is no perfect method for capturing eukaryotic community biomass, and multiple approaches are required to reconstruct community dynamics. Recommended methods will vary depending on the question and community members of interest.
Eukaryotic metabolism between depth zones
Nonmetric multidimensional scaling (NMDS) was performed on taxonomically and functionally annotated transcripts and proteins to examine the spatial structure of microeukaryotic communities. Depth overshadowed lateral effects in both molecular fractions (Supplementary Fig. 19), with surface (<100 m), upper water column (100–400 m), and deepest communities (400–4100 m) separating in ordination space.
Co-occurrence networks highlighted taxonomic associations and linked depth-dependent functional processes. The transcript network consisted of 21,347 nodes, or eukaryotic supergroups with KEGG functional annotations, and 21,346 edges, or interactions between nodes. These nodes clustered into 156 modules (Supplementary Fig. 20). The protein network contained a smaller set of annotated ORFs, and consisted of 2938 nodes and 2937 edges which clustered into 56 modules (Fig. 4). Whereas many modules were taxonomically diverse, some were dominated by one or few supergroups (Fig. 4a). A large number of modules represent distinct sets of linked metabolic processes, and may represent different taxonomic groups at a fine taxonomic level, or co-regulated functional processes. These co-occurrences may be a result of similar niche space occupation, endosymbioses, predator-prey dynamics, or animal microbiome communities85,86.
Taxonomic (a) and functional (b) composition of protein network modules. Normalized spectral abundance factor (NSAF) counts were summed to the supergroup and KEGG annotation level (e.g., Chlorophyta_ko:K00053). KEGG IDs were grouped into BRITE categories for a broader functional classification. KEGG IDs not included in BRITE categories are shown in dark gray. Only relatively abundant categories are shown; the full legend is in Supplementary Fig. 20. c Environmental context for the modules was determined by calculating eigengene values and correlating module eigengene values with environmental data (see methods). Insignificant Spearman rank correlations are denoted with an “x” (p > 0.05). Examples of surface ocean modules are highlighted with a yellow asterisk and deep water modules with a navy asterisk, and both groups are opaque for visibility in a, b.
The network modules showed strong representation of haptophytes, stramenopiles, and dinoflagellates, with several modules dominated by the heterotrophic taxa discoba, fungi, or metazoa. In contrast to other groups, metazoa were not relatively abundant in the protein pool (Fig. 2a–c), yet still comprised a large portion of certain modules, potentially reflecting consistent interactions with protists (Fig. 4a). The number of repeated KEGG Orthology genes (derived from different supergroups) contained in modules was low, ranging from 1–4, indicating that modules did not group by individual KEGG function. However, at a coarser functional level (KEGG BRITE category), differences in metabolic composition were observed. Several modules with an enrichment in photosynthesis were associated with higher relative abundances of photosynthetic taxa such as stramenopiles and haptophytes (Fig. 4b). These modules were positively correlated with oxygen, temperature and dMn, indicating elevated abundance in surface waters (Fig. 4c, Supplementary Fig. 20C).
One module in the protein network (Fig. 4, Supplementary Data 6) and eight modules in the transcript network (Supplementary Fig. 20, Supplementary Data 7) were significantly negatively correlated with temperature, and reflect deep ocean metabolism. These modules, along with several others showing negative (non-significant) correlations with surface parameters, were enriched in heterotrophic taxa such as discoba and foraminifera, with reduced photosynthetic functionality (Fig. 4). In particular, the deep ocean protein module 13 was mostly composed of discoba, metazoa, dinoflagellates, and picozoa, and contained a number of peptidases involved in amino acid catabolism (Supplementary Data 6). Other protein modules were positively correlated with temperature, yet negatively correlated with total dissolved Fe (dFe) (modules 27, 36), and proteins in these modules included chlorophyll components (chlorophytes, prasinodermophytes), and nitrate transporters (haptophytes). These modules therefore could reflect surface communities engaging in photosynthesis and assimilating macronutrients, thereby drawing down dFe concentrations to facilitate growth.
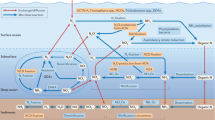
Nitrogen, phosphorus and iron metabolism along the lateral gradient
Transcripts and protein biomarkers of carbon fixation, nutrient status, and metal metabolism were surveyed across the offshore-coastal transect and aggregated (summed) across all eukaryotic groups to investigate bulk metabolic fingerprints. Their spatial distributions were evaluated in terms of relative abundance. Rubisco concentrations were higher closer to the coastline, and belonged to dinoflagellates, haptophytes, stramenopiles, cryptophytes, and unclassifiable eukaryotes (Fig. 5, Supplementary Data 9 and 10). However, they peaked at different coastal locations between the molecular pools, with Rubisco transcripts elevated in subsurface Gulf Stream waters (St. 7; Fig. 5a), whereas Rubisco proteins reached a maximum in coastal surface waters of St. 8 (Fig. 5b), which aligns with maximum pigment concentrations at St. 8 (Supplementary Fig. 12). This could represent differences in transcript/protein cellular ratios or turnover times. Growth rates of the phytoplankton populations between these two sites may also play a role.
Relative abundance of proteins (normalized spectral abundance factor [NSAF]) (a) and transcripts (transcripts per million [TPM]) (b) for functional processes of interest in the upper 1000 m. Eukaryotic open reading frames (ORFs) were summed to the supergroup level to examine a given KEGG functional gene. Ferritin (FTN) and flavodoxin were only detected in the transcript fraction. (See Supplementary Data 9 and 10 for individual open reading frame annotations). c Dissolved iron (Fe) and copper (Cu) concentrations in the top 1000 m of the water column. Station numbers are labeled in red. Note that biomass for omics was not collected from the coastal site St. 9.
Macronutrient stress biomarkers suggest N stress occurred towards the continental shelf while indications of P stress was apparent in offshore, oligotrophic gyre waters. The nitrate stress biomarker NRT is a nitrate transporter in phytoplankton, upregulated under nitrogen starvation87. NRT transcripts peaked at the surface at the most coastal site (St. 8) with a low subsurface signal elsewhere along the transect, while NRT proteins reached their highest levels in subsurface waters of the Gulf Stream (St. 7) with weaker surface signatures also at St. 8 and in subsurfaces throughout the transect (Fig. 5). This suggests that nitrogen may have been limiting phytoplankton growth in coastal communities. NRT also belonged to dinoflagellates, haptophytes, stramenopiles, cryptophytes, and unclassifiable eukaryotes (Supplementary Data 9 and 10).
Phosphate transporters showed a different distribution from NRT, with inorganic phosphate transporter PHO84, a major facilitator superfamily membrane transporter part of the Phosphate H+ Symporter family activated under phosphate stress88, having highest transcript concentration at the surface along the middle of the surface transect (Fig. 5). These transcripts and proteins belonged to dinoflagellates, haptophytes, stramenopiles, discoba, centroplasthelida and unclassifiable eukaryotes. A second inorganic phosphate transporter (PiT) was detected in transcripts in offshore surface waters at St. 1, 2, and 6, and in deep waters, reflecting heterotrophic metabolism or sinking euphotic zone cells. Protein concentrations for both the PiT and PHO84 phosphorus transporters reached maximum concentrations in subsurface oligotrophic waters (St. 2) and in the Gulf Stream (St. 7), but were undetected at other sites. PiT transcripts and proteins belonged to dinoflagellates, haptophytes, stramenopiles, cryptophytes, and ciliates. Alkaline phosphatase D, which is a metalloprotein that converts organic phosphorus to inorganic phosphorus, was only detected in the transcript fraction and was highly expressed in deep water at St. 1 & 2 (800 m), with a weak surface signal apparent in transitionary waters, matching PHO84 gene expression pattern. These transcripts were found in phototrophic taxa (dinoflagellates, stramenopiles, haptophytes) and heterotrophic taxa including choanoflagellates (Supplemental Fig. 21). Alkaline phosphatase has been demonstrated to be important in the subtropical Atlantic where inorganic phosphate concentrations are low, and enzyme activity increases with Saharan dust addition89. Although not detected in eukaryotic proteins with the functional database used here, additional investigations into the prokaryotic fraction could provide insights into broader microbial use. In agreement with these findings, the North Atlantic Ocean has been recently identified as a region experiencing P stress based on cyanobacterial biomarkers in metagenomes90.
These omic signatures are consistent with a previous study conducted along a similar transect in the western North Atlantic in early spring, in which the inorganic N:P ratio of surface seawater was above Redfield ratio (16:1) in the oligotrophic gyre, and dropped closer to and just below 16:1 north of the Gulf Stream, indicating the potential for phosphorus and nitrogen limitation in the oligotrophic gyre and coastal waters, respectively32. However, there are strong seasonal shifts in N:P at BATS, with summer ratios dropping below 1632, and nitrate concentrations below detection limit for much of the year apart from deep mixing spring periods. Indeed in our summer analysis, surface phosphate and nitrate + nitrite concentrations were consistently low (with nitrate + nitrite below the detection limit of 0.06 μM at several sites), with no clear transition in the N:P ratio towards the coastline (Fig. 1). As this survey was completed in early summer, it is possible that bioindications of nitrogen stress eventually appear in the oligotrophic gyre into the late summer and early fall, and a temporal analysis would be beneficial to further tease apart seasonal variations in nutrient physiology.
In addition to macronutrients, metalloproteins were also evaluated with respect to dissolved trace metal concentrations. The iron storage protein ferritin functions to maintain iron homeostasis91,92 and is commonly elevated in diatoms under Fe enrichment93,94. Two different ferritin genes were identified: “ferritin heavy chain” (FTH1: K00522) and “ferritin” (FTN: K02217). Although the only FTH1 protein signal (belonging to cnidarian metazoa) was detected in deep water, both FTN and FTH1 transcripts (comprised of metazoa, ciliates, foraminifera, telonemia, dinoflagellates, chlorophytes, streptophytes and unclassifiable eukaryotes) were found in the center of the surface transect as well as in deep water (Fig. 5a, b). Transcripts of flavodoxin, a Fe-independent photosynthetic electron acceptor typically diagnostic of Fe stress95, were abundant in surface transitionary waters alongside ferritin transcripts. These flavodoxin transcripts belonged to dinoflagellates, haptophytes, and cryptophytes. These spatial patterns of ferritin and flavodoxin coincide with 1.06 ± 0.38 nM dFe and 0.93 ± 0.10 nM dCu in surface waters (10 m) along the center of the transect (St. 2–3), which could be sourced from aerosol dust (Fig. 5c). Fe availability would presumably lead to Fe sufficiency in cells, and some phytoplankton continue to rely on flavodoxin even under Fe enrichment96 as an evolved strategy in oligotrophic ecosystems. The addition of eolian-sourced Fe and N may promote P stress14, which is consistent with high surface PHO84 and PIT transcript signatures alongside high FTH/FTN, FLDA, and lower NRT transcript signatures in oligotrophic surface waters. This spatial pattern supports P being an important driver of microeukaryotic growth and community diversity in the Sargasso Sea37,97,98.
Future directions
This analysis demonstrated that both metatranscriptomics and metaproteomics are advantageous in answering ecological questions, providing complementary views into microbial community dynamics. Metatranscriptomics enabled an in-depth examination with greater molecule coverage and clear view of nutrient status shifts along the transect (Fig. 5), but with metaproteomics offering insight into shifts into microbial communities with depth (Fig. 2). We recommend multiomic approaches be used in future oceanographic surveys to the extent possible, given the power of complementary approaches to account for biases specific to one method (Supplementary Data 2).
The data presented in this study also leveraged sampling efficiencies enabled by the coupled use of an autonomous underwater vehicle with shipboard sampling. Using AUV Clio, an average of 2320 ± 898 L of seawater was filtered per dive, concentrating biomass at 8–18 depths per station along a 1050 km transect in 11 days, providing mid- and full-depth metabolic profiles (Supplementary Data 8). The average dive duration was 12.7 ± 2.3 h and sampling range was 10–4100 m (although typically 10–1000 m, except for one deep dive); during this time, other deck operations using the ship’s J-frame could continue, including trace metal rosette, CTD, and complementary in situ McLane pump deployments. Typical in situ underwater pumps are attached to the ship’s winch line and pump water at specific depths, needing manual adjustments for subsequent biomass collection at additional depths, and requiring the ship to wait on station until filtration is complete. Autonomous collection instruments such as Clio are thus highly efficient for oceanographic surveys requiring large filtration volumes. As station time is a major constraining factor in the design of sectional sampling expeditions, the efficiencies demonstrated in this study have implications for future study design. Clio is particularly valuable in efforts to jointly sample chemical parameters and omics material, as is the vision of the international BioGeoSCAPES program99, and can provide insights into microbial metabolism within physical and chemical features, such as nepheloid layers, oxygen minimum zones, and transitions in water masses. Sensors attached to Clio reproduced physicochemical profiles obtained with a traditional CTD (Supplementary Fig. 22). Future expeditions will benefit from further integrating Clio sensor data, and testing bulk seawater collection by Clio for trace metal quantification. Such advances may obviate simultaneous CTD deployments, and Clio alone could be used for multi-parameter collection.
In conclusion, here we present vertical microeukaryote profiles that detail community composition and functionality across a coastal-offshore biogeochemical gradient, obtained using a recently designed autonomous underwater vehicle. The microeukaryote communities were predominantly represented by stramenopiles, dinoflagellates, and ciliates, and were detected throughout the water column, but with the emergence of heterotrophic taxa such as foraminifera, radiolaria, discoba, picozoa, and fungi in deep ocean protein pools (Fig. 6). The metabolic signatures of these surface and deep communities were functionally distinct and indicated viable cells regulating metabolism in response to environmental conditions. We identified several distinctions between transcripts and proteins in taxonomic and functional composition, underscoring the utility of paired metatranscriptomics-metaproteomics for comprehensively reconstructing microbial community dynamics. Positive correlations between protein and transcript fractions included Rubisco and a biomarker of nitrogen stress, which were both relatively abundant in coastal waters, and support a coupling between carbon and nitrogen-related assimilation processes. This multi-omics dataset thus captures shifts in microbial community composition across resource gradients, highlights potential regions of nutrient stress, and offers new insights into the complex relationship between transcripts and proteins in marine microeukaryotes.
Transcript and proteins indicated nitrate stress in surface coastal waters (green), with transcriptional signatures of elevated phosphate transport and iron use in offshore waters. Stramenopiles were detected throughout the transect with diatom and dictyophyte proteins in the surface (yellow), and diatom and pelagophyte proteins at depth (blue/gray). Diatom pigments were highest in concentration towards the coast. Haptophytes proteins were relatively more abundant in surface waters. Dinoflagellate and ciliate proteins and transcripts were detected throughout the water column. Foraminifera, discoba, picozoa and radiolarian proteins were relatively more abundant in the mesopelagic (blue). Radiolarian proteins reflected a vertical community shift with Polycystinea in the surface and Symphycanthida at depth. Fungi appeared in both transcripts and proteins in the bathypelagic (gray), though only one site above the continental shelf was surveyed.
Methods
Oceanographic sampling
Oceanographic samples were collected onboard the R/V Atlantic Explorer between June 16th - 28th 2019, along a transect beginning at the Bermuda Atlantic Time-series Study (BATS) site and terminating in northeast US continental shelf waters (Woods Hole Oceanographic Institution [WHOI]) (Fig. 1). Biomass was collected using the biogeochemical AUV Clio27. Clio payloads contained SUPR cartridges housing 142 mm, 0.2 µm porosity polyethersulfone filters (Pall Supor) positioned below 51 µm mesh nylon filters. Alternatively, for pigments, 142 mm combusted GFF filters were used. At Station 1 (BATS), SUPR cartridges with 0.2 µm polyethersulfone filters failed to pump, and biomass was instead collected from Clio GFF filters. To complement the vertical collection by Clio at this site only, in situ battery-operated McLane pumps were used for surface biomass collection (142 mm, 0.2–3, and 3–51 µm). Filters for the biomolecular analysis were sectioned and frozen at −80 °C analysis until RNA and protein extractions were performed in the laboratory. GFF filter fractions were additionally preserved for high-performance liquid chromatography pigment analyses throughout the surface transect. The R/V Atlantic Explorer CTD provided physicochemical contextualization (including temperature, salinity, dissolved oxygen, fluorescence, turbidity used here) and was deployed at all stations except the last one, St. 9, due to time restrictions. Once deployed, Clio filtrations happened simultaneously with ship wire operations.
After Clio was deployed, trace metal sampling was performed using a trace metal clean rosette equipped with 12 8 L X-Niskin bottles on an Amsteel winch line100. Upon retrieval, X-Niskins were brought into a class-100 clean van, and Niskins were pressurized with filtered nitrogen gas. Seawater was filtered through acid-rinsed 142 mm 0.2 µm Supor membranes to separate the particulate and dissolved fractions. Filtered seawater collected into 250 mL HCl-cleaned LDPE bottles was placed in the dark for dissolved metal analyses, and filters for the particulate metal analysis were kept frozen in acid-cleaned cryovials at −80 °C until analysis. Subsamples for dissolved nutrients were collected from 0.2 µm-filtered seawater and stored at −20 °C. Neither Clio nor the trace metal rosette was deployed at St. 4 due to unfavorable weather conditions, although physicochemical data was collected using the ship’s standard CTD.
An UVP 5 HD (UVP)101 was deployed with the CTD. The UVP records quantitative, in situ images of particles in the water column at a rate of ~15 Hz during CTD descent. Particle images > 0.5 mm equivalent spherical diameter (ESD) are recorded, processed using the Zooprocess software102, identified using the EcoTaxa website103, and validated by a trained taxonomist (full methodology in ref. 104). Microeukaryotes were identified to the lowest taxonomic category possible from the images (generally class or order) and abundances were binned by depth, with 40 m wide bins from 0–200 m, 100 m wide from 200–1400 m, and 200 m wide bins >1400 m.
Metatranscriptomic sequencing and bioinformatic workflow
Forty-four samples across the transect were selected for the metatranscriptomic analysis to achieve coverage along the lateral and vertical ocean gradient. One-sixteenth of 142 mm filters were removed from the −80 °C freezer and kept on ice in between handling steps. Total RNA was extracted using the Qiagen RNeasy Mini kit following the quick start protocol with the following modifications. Acid-rinsed 0.1 mm silica beads (Thomas Scientific) were added to each sample along with Qiagen RLT extraction buffer, which was pre-heated to 65 °C and contained 2-mercaptoethanol per manufacturer recommendation. Samples were vortexed and physically disrupted in a mini Bead-Beater homogenizer, centrifuged, and filters were removed using forceps that were sterilized with ethanol and RNaseZap (Invitrogen). Lysate was transferred to a Qiashredder (Qiagen) column to remove viscosity and improve yields for all but the first set of six randomly chosen samples. Molecular biology grade ethanol (Fisher Scientific) was used to prepare kit buffers for most samples, though the first round of randomly selected six samples were used with reagent grade ethanol and contained impurities. On-column digestion of DNA was performed using DNase I (Qiagen). Elution buffer was pre-heated to 70 °C, 30 µL elution volumes were used, and eluate was passed through the column a second time to concentrate RNA.
RNA yields and quality were assessed using a Nanodrop spectrophotometer and Agilent 2100 bioanalyzer. mRNA internal standards were added to each sample for later conversion of sequence read counts to transcript copies105 using poly-adenylated ArrayControl RNA spike #8 (Invitrogen)106 at a standard:sample ng RNA ratio of 0.22%. Since internal standards were not added prior to the extraction, our copies L−1 estimates likely overestimate true levels, as nucleic acid loss is expected during the extraction process107. As many samples contained less than 100 ng of total RNA, the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech) for cDNA synthesis, followed by the NexteraXT DNA library prep kit (Illumina). Libraries were sequenced on the Illumina NovaSeq 6000 platform (2 × 150 bp PE) with an average yield of 38 million PE reads per sample (Supplementary Data 4). Library preparation and sequencing was performed at the Columbia Genome Center.
Sequences were processed using the eukrhythmic eukaryotic metatranscriptomic workflow108. This workflow enabled assemblies, read alignments, and ORF predictions in an automated, scalable and reproducible Snakemake v5.16.0 framework. Poor quality bases were trimmed using trimmomatic v0.39109, and spiked mRNA internal standard reads were removed using bbmap v38.79 (sourceforge.net/projects/bbmap/). Four different transcriptome assemblers were used on each sample individually: Trinity v2.9.1110, MEGAHIT v1.2.9111, velvet v1.2.10112 and Transabyss v2.0.1113. Assembled contigs were clustered at 100% alignment coverage with cd-hit v4.8.1114 to remove redundant contigs. The assemblies created using the four assemblers were merged by sample, and de-replicated with cd-hit (100% alignment coverage). Next, all 44 samples were merged together using MMseqs2 v13.45111115 to produce a single dataset-wide final merged assembly. ORFs were predicted using TransDecoder v5.5.0 (github.com/TransDecoder), retaining nucleotide coding sequences with a minimum length of 300 base pairs. Reads were aligned to both the full-length assembly contigs and the ORF coding nucleotide sequences using Salmon v1.2.0116, which produced pseudocounts and community-wide normalized Transcripts Per Million (TPM) counts, and allowed for a relative comparison across samples. Since the ORFs only include coding regions and sequences longer than 100 amino acids (300 bp), the mapping percentage is approximately half of that obtained with the full (non-coding) contigs (Supplementary Data 4). In addition to relative analysis, pseudo-read counts were converted to copies L−1 following105, with the number of internal standard transcript copies added to samples being approximately 500,000, and accounting for seawater volume filtered. A factor was applied to correct for sample dilution (ng extracted RNA divided by ng RNA used in library)117, with 0.5 ng RNA submitted for sequence library preparation. Taxonomic assessments were performed on ORFs to be consistent with the metaproteomic analysis in which only ORFs (encoded proteins) are considered, as opposed to full-length contigs including non-coding sequences. Functional annotations were performed using eggNOG-mapper v2.1.1–1118 with default settings. For some ORFs, more than 1 KEGG ID was assigned (e.g., K11251 [histone H2A], K11252 [histone H2B], K11254 [histone H4]), and counts were distributed to all KEGG IDs.
The ORFs were taxonomically annotated by testing four sequence databases that differed in the reference organisms included: MMETSP + MarRef119,120,121, EukZoo (github.com/zxl124/EukZoo-database), EukProt122, and PhyloDB (allenlab.ucsd.edu/data). The taxonomic annotations were performed using EUKulele v2.0.0 with a last common ancestor approach (LCA)121. The LCA conservatively annotated ORFs only when bitscores associated within the top 3% of hits were in taxonomic agreement. If lineage conflicts were present, ORFs were labeled as “NA” (not annotated). In preliminary tests with taxonomic databases, it was determined the MMETSP + MarRef database was missing ecologically relevant protistan and bacterial sequences contained by PhyloDB, EukZoo, and/or EukProt, including Polycystinea radiolaria, known inhabitants of Sargasso Sea deep water25,26, and Spirotrichea ciliates, which were both relatively abundant in our dataset. We found that prokaryotic ORFs were being erroneously assigned to eukaryotes since they represented the closest available match in the database exceeding quality thresholds (E-value < 10−5), as determined using the NCBI non-redundant protein sequence (nr) database which includes a greater number of prokaryotic references. A custom protein database was therefore built using PhyloDB (25,996 taxonomic references, version 1.075), NCBI’s RefSeq complete translated genomes (1,114 references), Release 211, ftp.ncbi.nih.gov/refseq/release/complete), two key EukZoo references, and the recently released EukProt database (993 references, version 3) which contains transcriptomes and genomes of diverse eukaryotic lineages122. This combined protein database was not de-replicated, and manual efforts were made to best match coarse classifications belonging to major eukaryotic supergroups at the chosen taxonomic annotation levels analyzed here. Future annotation efforts may benefit from using the entire nr NCBI database which additionally includes incomplete genomes and marine environmental sequences. After top alignment hits within 3% of the highest bit score were identified, taxonomic cutoff levels were called following default EUKulele settings using 7 taxonomic levels. At least 15% sequence identity was needed to be met at the domain level, 20% at supergroup, 30% at division, 50% at class, 65% at order, 80% at family, and 95% at genus, with 75% consistency among hits needed to make a given taxonomic cutoff121.
Metaproteomic analysis
One half of the 142 mm filters (0.2–51 μm) were processed for metaproteomics. At St. 1 (BATS), Clio failed to recover biomass from the surface layer due to the SUPR cartridges being misaligned, and McLane pumps were used instead with separate 0.2–3 μm (“0.2”) and 3–51 μm (“3”) filter fractions. At this site, additional protein biomass from 20, 175, 250 and 350 m was obtained from Clio combusted GFF filters, which were successful in sample collection. Total seawater volumes filtered by Clio varied between 30–350 L during this expedition, with generally less volume in surface waters and more at depth due less biomass in deeper layers of the ocean (Supplementary Data 5). The full protein extraction procedure is described in123,100. Proteins were extracted in an 1% SDS-based detergent in 50 mM HEPES at pH 8.5, reduced with dithiothreitol, alkylated with iodoacetamide, and purified using a polyacrylamide electrophoresis tube gel method. Protein quantification was performed using a BSA assay (BioRad). Trypsin was added to the protein-bead mixture in a 1:20 trypsin:protein ratio. Peptides were purified using C18 tips and diluted to a concentration of 0.1 μg μL−1.
Approximately 2–5 µg of purified peptides were injected onto a Dionex UltiMate 3000 RSLCnano LC system with an additional RSLCnano pump, run in online 2D active modulation mode interfaced with a Thermo Fusion mass spectrometer124. The mass spectrometer acquired MS1 scans from 380 to 1,580 m/z at 240 K resolution in the orbitrap. MS2 were collected in data-dependent mode in the ion trap with a cycle time of 2 s between scans and acquisition of charge states 2 to 10. MS2 scans had 1.6 m/z isolation window, 50 ms maximum injection time and 5 s dynamic exclusion time.
The metatranscriptomic ORFs were used as the protein database, and peptide-spectrum matches were performed using the Sequest algorithm within IseNode Proteome Discoverer 2.2.0.388 (Thermo Fisher Scientific) with a parent ion tolerance of 10 ppm and fragment tolerance of 0.6 Da, and 0 max missed cleavage. Identification criteria consisted of a peptide threshold of 98% (false discovery rate [FDR] = 0.1%) and protein threshold of 99% (1 peptide minimum, FDR = 1.5%) in Scaffold 5.1.2 (Proteome Software) resulting in 77,438 proteins and 3,155,061 exclusive spectral counts.
Using the default Scaffold protein inference method (experiment-wide grouping with binary peptide-protein weights), proteins that shared all of their detected tryptic peptides were grouped together and considered the same protein reference (a “protein group”), with the ORF ID randomly chosen among the proteins. Approximately 26% of the ORFs with protein spectral counts were represented by a protein group. Incidents of shared tryptic peptides across distinct taxonomic lineages are generally rare based on investigations in marine prokaryotes63. In cases where a given peptide matched more than 1 protein or protein group, peptides were assigned to the protein with the highest spectral counts. Scaffold’s “exclusive spectral counts” were used for the proteomic analysis to avoid double counting of spectral counts, i.e. assigning counts to more than one protein/protein group125. Protein abundances in spectral counts per volume seawater (Supplementary Figs. 17 and 18) were calculated using Eq. (1), adapted from125:
Macronutrients and pigments
Dissolved nitrate + nitrite, nitrite and silicate were measured on an Alpkem Rapid Flow Analyzer, and ammonium and phosphate were measured on a Technicon AutoAnalyzer II at Oregon State University. Limits of detection were calculated as the standard deviation among 8–10 replicates (phosphate: 0.006 μM, nitrate + nitrite: 0.06 μM, silicate: 0.19 μM, nitrite: 0.007 μM, ammonium: 0.03 μM). For most stations, nutrients were collected and measured from trace metal rosette niskin bottles. However, St. 8 nutrients showed oceanographically inconsistent profiles, potentially due to samples thawing in transit. Nutrients collected from Clio at St. 8 were used instead, with general agreement between trace metal rosette and Clio-collected nutrients at other stations and depths.
Pigments were collected onto 142 mm GFF filters by Clio at the same depths that proteins were collected on Supor filters. GFF 25 mm punchouts were used for the pigment analysis, and the estimated volume filtered through 25 mm was used to quantify pigments per volume. Concentrations are reported as ng/kg assuming the standard density of seawater (1025 kg/m3). At stations 1 and 3, GFF fractions for pigments were not collected by Clio. Instead, pigments were analyzed from the CTD rosette at these stations whereby samples (4 L) were filtered directly onto 25 mm GFF glass fiber filters and immediately frozen in liquid nitrogen. Samples were analyzed on an Agilent 1100 series high performance liquid chromatography (HPLC) using the method of126 and instrument calibration was performed with certified pigment standards from the Danish Hydraulic Institute.
Dissolved trace metals
Filtered seawater was acidified to pH 1.8 using hydrochloric acid (Optima grade, Fisher Chemical), and samples intended for dissolved trace metal analyses were stored for 6 months at room temperature in the dark before metal quantification. Seawater preconcentration was performed using the seaFAST automated preconcentration system127 followed by quantification via inductively coupled plasma-mass spectrometry. Reagents consisted of a 4 M ammonium acetate pH 6.0 buffer prepared using high purity ammonium hydroxide and acetic acid (Optima grade, Fisher Chemical), a 1% nitric acid rinse solution (Optima grade, Fisher Chemical), 10% nitric acid elution acid, and a second internal standard 10% nitric elution acid solution containing 10 ppb indium (115In) (SPEX CertiPrep). All solutions were prepared with 18.2 Ω Milli-Q purified water (Millipore). Polypropylene conical tubes used with the autosampler were HCl acid-soaked and pH 2-rinsed prior to use. Process blanks consisted of Milli-Q HCl-acidified to pH 2 (Optima grade, Fisher Chemical), and were run alongside samples. A stable isotope cocktail, which consisted of 57Fe, 61Ni, 65Cu, 67Zn and 111Cd, was spiked (50 µL) into each 15 mL sample to account for recovery and matrix effects. Stable isotope spike standards were prepared by dissolving metal oxides into 50% nitric acid and diluted using volumetric flasks (Optima grade, Fisher Chemical). Spike concentrations were chosen based on the amount needed to generate an approximate 1:1 ratio of spiked:stable isotope in the 15 mL sample128,129.
Following offline seaFAST preconcentration to 500 μL, the samples were analyzed using an iCAP Q inductively coupled plasma-mass spectrometer (Thermo Scientific). An external standard curve was used with multi-element and indium (In) standards (SPEX CertiPrep), diluted to range from 1–10 ppb in 5% nitric acid. Dissolved metal concentrations were determined using the following isotope dilution Eq. (2):
where, using Fe for example, R = ratio of 56Fe/57Fe in sample, Rspike = 57Fecps / (56Fecps + 57Fecps) in the spike, Rnatural = 57Fecps / (56Fecps + 57Fecps) in 10 ppb standard, atm weight = average atomic weight between the two isotopes, fspike = 56Fecps / (56Fecps + 57Fecps) in spike, fnatural = 56Fecps / (56Fecps + 57Fecps) in 10 ppb standard, and ng spike = the metal concentration in the spike, determined empirically using 57Fecps of spike, and manually tuned to the value that allowed for the 10 ppb + spike standard to equal 5 ng. As there is no second stable isotope for manganese (Mn), concentrations were determined using the 57Fe isotope spike.
Nitric acid (5%) injection blanks were subtracted from sample metal cps values except for Zn, in which injection blanks were generally higher than process blanks. Accuracy was determined using the 2009 GEOTRACES surface coastal (GSC) seawater intercalibration standard (n = 5): dFe = 1.65 ± 0.19 nM [GSC = 1.56 ± 0.12 nM], dZn = 1.65 ± 0.17 nM [GSC = 1.45 ± 0.10 nM], dCu = 1.38 ± 0.16 nM [GSC = 1.12 ± 0.15 nM], dCd = 0.37 ± 0.03 nM [GSC = 0.37 ± 0.02 nM], dNi = 4.28 ± 0.17 nM [GSC = 4.5 ± 0.21 nM], and dMn = 2.42 ± 0.29 nM [GSC = 2.23 ± 0.08 nM].
The limit of detection was determined by calculating 3x the standard deviation of process blanks dataset-wide: dFe = 0.23 nM (n = 14), dZn = 0.35 nM (n = 16), dCu = 0.03 nM (n = 18), dCd =0.0103 nM (n = 16), dNi = 0.07 nM (n = 18), dMn = 0.03 nM (n = 18). Blanks that were overtly contaminated with Fe (4 of 18), Zn (2 of 18) or Cd (2 of 18) were not included in the LOD estimation. In the case of Zn, high deep water (>1000 m) concentrations altered the spiked:stable isotope ratio, and accurate concentrations were not able to be obtained. Measured concentrations below the LOD are reported as “0” in section plots and correlation matrices. The full dissolved metal dataset has been uploaded to BCO-DMO under project 765945.
Statistical approaches, normalization, and visualization
Normalizations were performed at the community-wide level for ordinations, network analyses, and PLS regression analyses. For these community-wide analyses, transcripts were normalized following the transcripts per million approach, which enables relative comparison of gene expression across samples by accounting for differences in library sizes and the length of ORFs. Proteomic exclusive spectral counts were normalized using the normalized spectral abundance factor (NSAF) calculation following the TPM approach, in which counts associated with a given ORF were normalized by the length of the ORF in amino acid space (length in base pairs divided by 3), multiplied by a scalar of 103, and divided by the sum of length-normalized ORF counts in that sample.
Nonmetric multidimensional scaling (NMDS) ordinations were performed using community-wide normalized transcripts and protein counts associated with ORFs having both a taxonomic and functional annotation (eukaryotic ORFs assigned by EUKulele and KEGG Orthology ID assigned by eggNOG-mapper130). Counts were summed to the supergroup level and log2 normalized, and Bray-Curtis was used to calculate distance using the metaMDS function in vegan v2.6-4.
Undirected networks were built to identify co-occurring eukaryotic transcripts or proteins. To reduce the size of the networks, only ORFs with a taxonomic and functional annotation were used. Counts were summed to the supergroup and KEGG annotation level (e.g., Chlorophyta_ko:K00053). Annotated ORFs with row sums of TPM > 10 or NSAF counts >0.01 were retained for the analysis, and counts were log2 transformed. A correlation matrix was calculated using Spearman rank correlation distances in igraph v1.2.11131. Correlations >|0.65| were considered. The graph was converted to a minimum spanning tree (MST) to identify the minimal number of edges needed for all nodes to be connected, thus decreasing the size of the network. Small, unconnected subnetworks were removed. Clusters of correlated genes (modules) were identified using fast greedy modularity optimization (“cluster fast greedy”) and the networks were visualized with the Kamada-Kawai algorithm. Modules were investigated through taxonomic and functional composition. KEGG IDs were grouped into BRITE categories for a broader functional classification, minus the “Enzyme” category as this grouping was considered too broad to be biologically meaningful. Environmental context for the modules was determined by calculating eigengene values, or the first principle component of gene/protein expression representing each module using tmod, and calculating Spearman rank correlations with environmental data. These correlations were calculated for stations/depths where corresponding trace metal data was collected.
PLS regression was performed to correlate protein abundance and gene expression. Only eukaryotic ORFs with a taxonomic and functional annotation were used. Counts were summed across all eukaryotic taxa to the KEGG functional level. Annotated ORFs with row sums of TPM > 5 or NSAF counts >0.01 were retained for the analysis. KEGG Orthology carbon and nutrient transporters largely compiled by ref. 132 were identified in the dataset, excluding the following proteins: an archaeal phosphate transport substrate-binding protein (pstS; K02040) and bacterial iron complex outer membrane receptor protein (TC.FEV.OM; K02014). ORF sequences annotated as these proteins showed high percent identity to eukaryotes in our early comprehensive taxonomic database (PhyloDB + RefSeq), but upon further examination, were closer matches to prokaryotes contained within the NCBI non-redundant protein database, which includes other relevant prokaryotic references as well as marine metagenome-assembled genomes. The PLS regression was performed using mixOmics v6.13.114 with default scaling and using two components and was evaluated using leave one out cross validation133.
Proportions of ORFs containing lineage-conflicted annotations between annotated metatranscriptomes and metaproteomes were compared using a Student’s t-test after passing a Shapiro-Wilk normality test and Brown-Forsythe equal variance test in Sigmaplot v14.5.
Contour section plots were created using Ocean Data View v5.3.0 with DIVA gridding.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The AE1913 expedition (CASHEW: Clio Atlantic Sectional Hoedown Ending at Woods Hole) CTD data, processed trace metal, pigment, and macronutrient concentrations, expedition logs, and other environmental data are publicly available on BCO-DMO. Raw sequences have been deposited to the NCBI SRA database under BioProject ID PRJNA903389. Metatranscriptomic assembly (the fasta database used for metaproteomics: merged.fasta.transdecoder.pep.zip), annotations, and count data are available through Zenodo. Metaproteomic mass spectrometry data is available on ProteomeXchange through PRIDE accession PXD045395.
Code availability
Analysis notebooks and compiled data files are available on GitHub (https://github.com/cnatalie/BATS).
References
Gaube, P., McGillicuddy, D. J. Jr., Chelton, D. B., Behrenfeld, M. J. & Strutton, P. G. Regional variations in the influence of mesoscale eddies on near-surface chlorophyll. J. Geophys. Res. C: Oceans 119, 8195–8220 (2014).
Wenegrat, J. O. et al. Enhanced mixing across the gyre boundary at the Gulf Stream front. Proc. Natl Acad. Sci. USA 117, 17607–17614 (2020).
Rypina, I. I., Pratt, L. J. & Lozier, M. S. Near-surface transport pathways in the North Atlantic ocean: looking for throughput from the subtropical to the subpolar gyre. J. Phys. Oceanogr. 41, 911–925 (2011).
Wang, Z. et al. Microbial communities across nearshore to offshore coastal transects are primarily shaped by distance and temperature. Environ. Microbiol. 21, 3862–3872 (2019).
Browning, T. J. et al. Iron limitation of microbial phosphorus acquisition in the tropical North Atlantic. Nat. Commun. 8, 15465 (2017).
Paerl, H. W. et al. Rainfall stimulation of primary production in western Atlantic Ocean waters:roles of different nitrogen sources and co-limiting nutrients. Mar. Ecol. Prog. Ser. 176, 205–214 (1999).
Sedwick, P. N. et al. Assessing phytoplankton nutritional status and potential impact of wet deposition in seasonally oligotrophic waters of the mid‐Atlantic bight. Geophys. Res. Lett. 45, 3203–3211 (2018).
Bates, N. R. Interannual variability of oceanic CO2 and biogeochemical properties in the Western North Atlantic subtropical gyre. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 48, 1507–1528 (2001).
Jickells, T. D., Baker, A. R. & Chance, R. Atmospheric transport of trace elements and nutrients to the oceans. Philos. Trans. A Math. Phys. Eng. Sci. 374, 20150286 (2016).
Prospero, J. M. Saharan dust transport over the north Atlantic ocean and Mediterranean: an overview. In: Environ. Sci. Technol., 133–151 (Springer Netherlands, Dordrecht, 1996).
Sedwick, P. N. et al. Iron in the Sargasso Sea (Bermuda Atlantic Time-series Study region) during summer: Eolian imprint, spatiotemporal variability, and ecological implications. Global Biogeochem Cycles 19, GB4006 (2005).
Wu, J. & Boyle, E. Iron in the Sargasso Sea: implications for the processes controlling dissolved Fe distribution in the ocean. Glob. Biogeochem. Cycles 16, 33–1–33–8 (2002).
Jickells, T. D. The inputs of dust derived elements to the Sargasso Sea; a synthesis. Mar. Chem. 68, 5–14 (1999).
Baker, A. R., Kelly, S. D., Biswas, K. F., Witt, M. & Jickells, T. D. Atmospheric deposition of nutrients to the Atlantic Ocean. Geophys. Res. Lett. 30, 2296 (2003).
Saba, V. S. et al. Physical associations to spring phytoplankton biomass interannual variability in the U.S. Northeast Continental Shelf. J. Geophys. Res. Biogeosci. 120, 205–220 (2015).
Hatta, M. et al. An overview of dissolved Fe and Mn distributions during the 2010–2011 U.S. GEOTRACES north Atlantic cruises: GEOTRACES GA03. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 116, 117–129 (2015).
Fitzsimmons, J. N. et al. Partitioning of dissolved iron and iron isotopes into soluble and colloidal phases along the GA03 GEOTRACES North Atlantic Transect. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 116, 130–151 (2015).
Thomas, A. C. et al. Seasonal trends and phenology shifts in sea surface temperature on the North American northeastern continental shelf. Elementa 5, 48 (2017).
Worden, A. Z. et al. Environmental science. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347, 1257594 (2015).
Stoecker, D. K., Hansen, P. J., Caron, D. A. & Mitra, A. Mixotrophy in the marine plankton. Ann. Rev. Mar. Sci. 9, 311–335 (2017).
Steinberg, D. K. et al. Overview of the US JGOFS Bermuda Atlantic Time-series Study (BATS): a decade-scale look at ocean biology and biogeochemistry. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 48, 1405–1447 (2001).
Lomas, M. W. & Bates, N. R. Potential controls on interannual partitioning of organic carbon during the winter/spring phytoplankton bloom at the Bermuda Atlantic time-series study (BATS) site. Deep Sea Res. Part I 51, 1619–1636 (2004).
DuRand, M. D., Olson, R. J. & Chisholm, S. W. Phytoplankton population dynamics at the Bermuda Atlantic time-series station in the Sargasso Sea. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 48, 1983–2003 (2001).
Bolaños, L. M. et al. Small phytoplankton dominate western North Atlantic biomass. ISME J. 14, 1663–1674 (2020).
Countway, P. D. et al. Distinct protistan assemblages characterize the euphotic zone and deep sea (2500 m) of the western North Atlantic (Sargasso Sea and Gulf Stream). Environ. Microbiol. 9, 1219–1232 (2007).
Blanco-Bercial, L. et al. The protist community traces seasonality and mesoscale hydrographic features in the oligotrophic Sargasso Sea. Front. Mar. Sci. 9, 897140 (2022).
Breier, J. A. et al. Revealing ocean-scale biochemical structure with a deep-diving vertical profiling autonomous vehicle. Sci. Robot 5, eabc7104 (2020).
Jakuba, M. V., Breier, J. A., Gómez-Ibáñez, D., Tradd, K. & Saito, M. A. Clio: an autonomous vertical sampling vehicle for global ocean biogeochemical mapping. In: 2018 IEEE/OES Autonomous Underwater Vehicle Workshop (AUV) 1–8 (2018).
Joyce, T. M., Dunworth-Baker, J., Pickart, R. S., Torres, D. & Waterman, S. On the Deep Western boundary current south of Cape Cod. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 52, 615–625 (2005).
Sarkar, N. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 66, 173–197 (1997).
Mohanty, B. K. & Kushner, S. R. Bacterial/archaeal/organellar polyadenylation. Wiley Interdiscip. Rev. RNA 2, 256–276 (2011).
Cavender-Bares, K. K., Karl, D. M. & Chisholm, S. W. Nutrient gradients in the western North Atlantic Ocean: relationship to microbial community structure and comparison to patterns in the Pacific Ocean. Deep Sea Res. Part I 48, 2373–2395 (2001).
Krinos, A. I. et al. Missing microbial eukaryotes and misleading meta-omic conclusions. bioRxiv https://doi.org/10.1101/2023.07.30.551153 (2023).
Wang, J.-X. et al. CHEMTAX analysis of phytoplankton assemblages revealed potential indicators for blooms of haptophyte Phaeocystis globosa. Ecol. Indic. 131, 108177 (2021).
Zapata, M., Fraga, S., Rodríguez, F. & Garrido, J. L. Pigment-based chloroplast types in dinoflagellates. Mar. Ecol. Prog. Ser. 465, 33–52 (2012).
Catlett, D. et al. Integrating phytoplankton pigment and DNA meta‐barcoding observations to determine phytoplankton composition in the coastal ocean. Limnol. Oceanogr. https://doi.org/10.1002/lno.12274 (2022).
Villiot, N., Maas, A. E., Poulton, A. J. & Blanco-Bercial, L. Organic and inorganic nutrients modulate taxonomic diversity and trophic strategies of small eukaryotes in oligotrophic oceans. FEMS Microbes 4, xtac029 (2022).
Cordier, T. et al. Patterns of eukaryotic diversity from the surface to the deep-ocean sediment. Sci. Adv. 8, eabj9309 (2022).
Mars Brisbin, M., Brunner, O. D., Grossmann, M. M. & Mitarai, S. Paired high‐throughput, in situ imaging and high‐throughput sequencing illuminate acantharian abundance and vertical distribution. Limnol. Oceanogr. 65, 2953–2965 (2020).
Decelle, J. and Not, F. Acantharia. In: eLS, John Wiley & Sons, Ltd (Ed.). https://doi.org/10.1002/9780470015902.a0002102.pub2 (2015).
Page, T. M. & Lawley, J. W. The next generation is here: a review of transcriptomic approaches in marine ecology. Front. Mar. Sci. 9, 1–7 (2022).
Peng, X. et al. Planktonic marine fungi: a review. J. Geophys. Res. Biogeosci. 129, e2023JG007887 (2024).
Pernice, M. C. et al. Large variability of bathypelagic microbial eukaryotic communities across the world’s oceans. ISME J. 10, 945–958 (2016).
Priest, T., Fuchs, B., Amann, R. & Reich, M. Diversity and biomass dynamics of unicellular marine fungi during a spring phytoplankton bloom. Environ. Microbiol. 23, 448–463 (2021).
Shortall, K., Djeghader, A., Magner, E. & Soulimane, T. Insights into aldehyde dehydrogenase enzymes: a structural perspective. Front. Mol. Biosci. 8, 659550 (2021).
Lam, P. J., Ohnemus, D. C. & Auro, M. E. Size-fractionated major particle composition and concentrations from the US GEOTRACES North Atlantic zonal transect. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 116, 303–320 (2015).
Noble, A. E., Ohnemus, D. C., Hawco, N. J., Lam, P. J. & Saito, M. A. Coastal sources, sinks and strong organic complexation of dissolved cobalt within the US North Atlantic GEOTRACES transect GA03. Biogeosciences. 14, 2715–2739 (2016).
Revels, B. N., Ohnemus, D. C., Lam, P. J., Conway, T. M. & John, S. G. The isotopic signature and distribution of particulate iron in the North Atlantic Ocean. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 116, 321–331 (2015).
Richards, T. A., Jones, M. D. M., Leonard, G. & Bass, D. Marine fungi: their ecology and molecular diversity. Ann. Rev. Mar. Sci. 4, 495–522 (2012).
Bachy, C. et al. Marine protists: a Hitchhiker’s guide to their role in the marine microbiome. In: Stal, L. J., Cretoiu, M. S. (eds) The Marine Microbiome. vol 3 https://doi.org/10.1007/978-3-030-90383-1_4 Springer, Cham. (2022).
Sen, K., Sen, B. & Wang, G. Diversity, abundance, and ecological roles of planktonic fungi in marine environments. J. Fungi (Basel) 8, 491 (2022).
Breyer, E., Zhao, Z., Herndl, G. J. & Baltar, F. Global contribution of pelagic fungi to protein degradation in the ocean. Microbiome 10, 143 (2022).
Baltar, F., Zhao, Z. & Herndl, G. J. Potential and expression of carbohydrate utilization by marine fungi in the global ocean. Microbiome 9, 106 (2021).
Orsi, W. D. et al. Carbon assimilating fungi from surface ocean to subseafloor revealed by coupled phylogenetic and stable isotope analysis. ISME J. 16, 1245–1261 (2022).
Pernice, M. C., Forn, I., Logares, R. & Massana, R. A fungi hotspot deep down the ocean: explaining the presence of Gjaerumia minor in Equatorial Pacific bathypelagic waters. bioRxiv https://doi.org/10.1101/2024.01.25.577184 (2024).
Hassett, B. T., Vonnahme, T. R. & Peng, X. Gareth Jones, E. B. & Heuzé, C. Global diversity and geography of planktonic marine fungi. Botanica Mar. 63, 121–139 (2020).
MacCoss, M. J. et al. Sampling the proteome by emerging single-molecule and mass spectrometry methods. Nat. Methods 20, 339–346 (2023).
Van Der Kelen, K., Beyaert, R., Inzé, D. & De Veylder, L. Translational control of eukaryotic gene expression. Crit. Rev. Biochem. Mol. Biol. 44, 143–168 (2009).
Maier, T., Güell, M. & Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 583, 3966–3973 (2009).
Walworth, N. G. et al. Why environmental biomarkers work: transcriptome–proteome correlations and modeling of multistressor experiments in the marine bacterium trichodesmium. J. Proteome Res. 21, 77–89 (2022).
Moran, M. A. et al. Sizing up metatranscriptomics. ISME J. 7, 237–243 (2013).
Zettler, L. A., Sogin, M. L. & Caron, D. A. Phylogenetic relationships between the Acantharea and the Polycystinea: a molecular perspective on Haeckel’s Radiolaria. Proc. Natl Acad. Sci. USA 94, 11411–11416 (1997).
Saunders, J. K. et al. METATRYP v 2.0: metaproteomic least common ancestor analysis for taxonomic inference using specialized sequence assemblies-standalone software and web servers for marine microorganisms and coronaviruses. J. Proteome Res. 19, 4718–4729 (2020).
Roy, S., Jagus, R. & Morse, D. Translation and translational control in dinoflagellates. Microorganisms 6, 30 (2018).
Erickson, H. P. Evolution of the cytoskeleton. Bioessays 29, 668–677 (2007).
Stat, M. et al. Ecosystem biomonitoring with eDNA: metabarcoding across the tree of life in a tropical marine environment. Sci. Rep. 7, 12240 (2017).
Dyhrman, S. T. et al. The transcriptome and proteome of the diatom thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS One 7, 1–10 (2012).
Safaei, R. & Fischer, I. Turnover of cytoskeletal proteins in vivo. Brain Res 533, 83–90 (1990).
Nicolet, B. P. & Wolkers, M. C. The relationship of mRNA with protein expression in CD8+ T cells associates with gene class and gene characteristics. PLoS One 17, e0276294 (2022).
Perry, R. P. Balanced production of ribosomal proteins. Gene 401, 1–3 (2007).
McCain, J. S. P. & Bertrand, E. M. Prediction and consequences of cofragmentation in metaproteomics. J. Proteome Res. 18, 3555–3566 (2019).
van Dijk, E. L., Jaszczyszyn, Y. & Thermes, C. Library preparation methods for next-generation sequencing: tone down the bias. Exp. Cell Res. 322, 12–20 (2014).
Saito, M. A. et al. Results from a multi-laboratory ocean metaproteomic intercomparison: effects of LC-MS acquisition and data analysis procedures. https://doi.org/10.5194/egusphere-2023-3148 (2024).
Delogu, F. et al. Integration of absolute multi-omics reveals dynamic protein-to-RNA ratios and metabolic interplay within mixed-domain microbiomes. Nat. Commun. 11, 4708 (2020).
Edgcomb, V. P. et al. Comparison of Niskin vs. in situ approaches for analysis of gene expression in deep Mediterranean Sea water samples. Deep Sea Res. Part 2 Top. Stud. Oceanogr. https://doi.org/10.1016/j.dsr2.2014.10.020 (2016).
Myklestad, S. M. Dissolved organic carbon from phytoplankton. In: The Handbook of Environmental Chemistry 111–148 (Springer-Verlag, Berlin/Heidelberg, 2005).
Rees, T. A. V. & Raven, J. A. The maximum growth rate hypothesis is correct for eukaryotic photosynthetic organisms, but not cyanobacteria. N. Phytol. 230, 601–611 (2021).
Isanta-Navarro, J. et al. Revisiting the growth rate hypothesis: towards a holistic stoichiometric understanding of growth. Ecol. Lett. 25, 2324–2339 (2022).
Lin, J. & Amir, A. Homeostasis of protein and mRNA concentrations in growing cells. Nat. Commun. 9, 4496 (2018).
Marañón, E. et al. Deep maxima of phytoplankton biomass, primary production and bacterial production in the Mediterranean Sea. Biogeosciences 18, 1749–1767 (2021).
Kleiner, M. et al. Assessing species biomass contributions in microbial communities via metaproteomics. Nat. Commun. 8, 1558 (2017).
Kleiner, M. Metaproteomics: much more than measuring gene expression in microbial communities. mSystems 4, e00115–e00119 (2019).
Campbell, L., Gaonkar, C. C. & Henrichs, D. W. Chapter 5 - integrating imaging and molecular approaches to assess phytoplankton diversity. In: Advances in Phytoplankton Ecology (eds. Clementson, L. A., Eriksen, R. S. & Willis, A.) 159–190 (Elsevier, 2022).
Gong, W., Hall, N., Paerl, H. & Marchetti, A. Phytoplankton composition in a eutrophic estuary: comparison of multiple taxonomic approaches and influence of environmental factors. Environ. Microbiol. 22, 4718–4731 (2020).
Apprill, A. Marine animal microbiomes: toward understanding host–microbiome interactions in a changing ocean. Front. Mar. Sci. 4, 222 (2017).
Sunagawa, S. et al. Tara Oceans: towards global ocean ecosystems biology. Nat. Rev. Microbiol. 18, 428–445 (2020).
Rogato, A. et al. The diatom molecular toolkit to handle nitrogen uptake. Mar. Genomics 24, Part 1, 95–108 (2015).
Lin, S., Litaker, R. W. & Sunda, W. G. Phosphorus physiological ecology and molecular mechanisms in marine phytoplankton. J. Phycol. 52, 10–36 (2016).
Mahaffey, C., Reynolds, S., Davis, C. E. & Lohan, M. C. Alkaline phosphatase activity in the subtropical ocean: insights from nutrient, dust and trace metal addition experiments. Front. Mar. Sci. 1, 00073 (2014).
Ustick, L. J. et al. Metagenomic analysis reveals global-scale patterns of ocean nutrient limitation. Science 372, 287–291 (2021).
Botebol, H. et al. Central role for ferritin in the day/night regulation of iron homeostasis in marine phytoplankton. Proc. Natl. Acad. Sci. 112, 1–6 (2015).
Cohen, N. R. et al. Iron storage capacities and associated ferritin gene expression among marine diatoms. Limnol. Oceanogr. 63, 1677–1691 (2018).
Marchetti, A. et al. Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature 457, 467–470 (2009).
Marchetti, A. et al. Development of a molecular-based index for assessing iron status in bloom-forming pennate diatoms. J. Phycol. 53, 820–832 (2017).
Whitney, L. P. et al. Characterization of putative iron responsive genes as species-specific indicators of iron stress in thalassiosiroid diatoms. Front. Microbiol. 2, 234 (2011).
Marchetti, A. et al. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc. Natl Acad. Sci. 109, E317–E325 (2012).
Lomas, M. W., Swain, A., Shelton, R. & Ammerman, J. W. Taxonomic variability of phosphorus stress in Sargasso Sea phytoplankton. Limnol. Oceanogr. 49, 2303–2309 (2004).
Van Mooy, B. A. S. et al. Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458, 69–72 (2009).
Saito, M. et al. The dawn of the BioGeoSCAPES program: ocean metabolism and nutrient cycles on a changing planet. Oceanography 37, 162–166 (2024).
Saito, M. A. et al. Multiple nutrient stresses at intersecting Pacific Ocean biomes detected by protein biomarkers. Science 345, 1173–1177 (2014).
Picheral, M. et al. The Underwater Vision Profiler 5: an advanced instrument for high spatial resolution studies of particle size spectra and zooplankton. Limnol. Oceanogr. Methods 8, 462–473 (2010).
Gorsky, G. et al. Digital zooplankton image analysis using the ZooScan integrated system. J. Plankton Res. 32, 285–303 (2010).
Picheral, M., Colin, S. & Irisson, J. O. EcoTaxa, a tool for the taxonomic classification of images. URL https://ecotaxa.obs-vlfr.fr (2017).
Barth, A. & Stone, J. Comparison of an in situ imaging device and net-based method to study mesozooplankton communities in an oligotrophic system. Front. Mar. Sci. 9, 898057 (2022).
Gifford, S., Satinsky, B. & Moran, M. A. Quantitative microbial metatranscriptomics. Methods Mol. Biol. 1096, 213–229 (2014).
Zeigler Allen, L. et al. The Baltic Sea virome: diversity and transcriptional activity of DNA and RNA viruses. mSystems 2, e00125–16 (2017).
Lever, M. A. et al. A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Front. Microbiol. 6, 476 (2015).
Krinos, A.I., Cohen, N. R., Follows, M. J. & Alexander, H. Reverse engineering environmental metatranscriptomes clarifies best practices for eukaryotic assembly. https://doi.org/10.1101/2022.04.25.489326 (2022).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Zerbino, D. R. Using the Velvet de novo assembler for short-read sequencing technologies. Curr. Protoc. Bioinformatics Chapter 11, Unit 11.5 (2010).
Robertson, G. et al. De novo assembly and analysis of RNA-seq data. Nat. Methods 7, 909–912 (2010).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Perneel, M., Lagaisse, R., Mortelmans, J., Maere, S. & Hablützel, P. I. Seasonal metabolic dynamics of microeukaryotic plankton: a year-long metatranscriptomic study in a temperate sea. MBio 15, e0038324 (2024).
Huerta-Cepas, J. et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122 (2017).
Klemetsen, T. et al. The MAR databases: development and implementation of databases specific for marine metagenomics. Nucleic Acids Res 46, D692–D699 (2018).
Johnson, L. K., Alexander, H. & Brown, C. T. Re-assembly, quality evaluation, and annotation of 678 microbial eukaryotic reference transcriptomes. Gigascience 8, giy158 (2019).
Krinos, A., Hu, S., Cohen, N. & Alexander, H. EUKulele: Taxonomic annotation of the unsung eukaryotic microbes. J. Open Source Softw. 6, 2817 (2021).
Richter, D. J. et al. EukProt: a database of genome-scale predicted proteins across the diversity of eukaryotes. Peer Community J. 2, e56 (2022).
Cohen, N. R. et al. Dinoflagellates alter their carbon and nutrient metabolic strategies across environmental gradients in the central Pacific Ocean. Nat. Microbiol. 6, 173–186 (2021).
McIlvin, M. R. & Saito, M. A. Online nanoflow two-dimension comprehensive active modulation reversed phase-reversed phase liquid chromatography high-resolution mass spectrometry for metaproteomics of environmental and microbiome samples. J. Proteome Res. 20, 4589–4597 (2021).
Saunders, J. K. et al. Microbial functional diversity across biogeochemical provinces in the central Pacific Ocean. Proc. Natl. Acad. Sci. USA 119, e2200014119 (2022).
Andersen, R. A. Algal Culturing Techniques. (Academic Press, 2005).
Cohen, N. R. et al. Hydrothermal trace metal release and microbial metabolism in the northeastern Lau Basin of the South Pacific Ocean. Biogeosciences 18, 5397–5422 (2021).
Tan, D. et al. Optimizing the ratio of the spike to sample for isotope dilution analysis: a case study with selenium isotopes. Acta Geochim. 39, 192–202 (2020).
Wu, J. & Boyle, E. A. Determination of iron in seawater by high-resolution isotope dilution inductively coupled plasma mass spectrometry after Mg(OH)2 coprecipitation. Anal. Chim. Acta 367, 183–191 (1998).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Csárdi, G. & Nepusz, T. The igraph software package for complex network research. Complex Syst. 1695 (2006).
Sun, Y., Debeljak, P. & Obernosterer, I. Microbial iron and carbon metabolism as revealed by taxonomy-specific functional diversity in the Southern Ocean. ISME J. 15, 2933–2946 (2021).
Rohart, F., Gautier, B., Singh, A. & Lê Cao, K.-A. mixOmics: an R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol. 13, e1005752 (2017).
Acknowledgements
We are grateful to the captain, crew, and science party of the AE1913 BATS cruise onboard the R/V Atlantic Explorer. The Clio deployments and recoveries were made possible by the expertise of the BIOS team. We thank members of the Seth John Lab (USC), especially Rachel Kelly and Xiaopeng Bian, for guidance and useful discussions regarding the isotope dilution calculations. We thank Maggi Mars Brisbin for valuable proteomic comments and discussion. The NIH/NCI Cancer Center Support Grant P30CA013696 supported Columbia’s Genomics and High Throughput Screening Shared Resource where metatranscriptomes were sequenced. Bioinformatic processing and computational analyses were performed on Woods Hole Oceanographic Institution’s high-performance computing cluster, Poseidon. NRC was supported by Simons Foundation #544236 and the University of Georgia Skidaway Institute of Oceanography. AIK was supported by the DOE Computational Science Graduate Fellowship (DE-SC0020347). H.A. was supported by the Simons Foundation Early Career Investigator in Aquatic Microbial Ecology and Evolution Award (award #931886). J.A.B. and M.V.J. were supported by NSF OCE-1333212, OCE-1658067, OCE-1924508, and NOAA #NA16SEC4810009. M.A.S., M.R.M., P.Z.L., and D.M.M. and metaproteome and environmental analyses were supported by Simons Foundation #1038971 and NSF OCE-1658030, OCE-2123055, and OCE-2125063.
Author information
Authors and Affiliations
Contributions
N.R.C. performed the trace metal and nutrient sampling on the R/V Atlantic Explorer, conducted the RNA extractions, performed the meta-omic analysis, and wrote the first draft of the manuscript. A.I.K. and H.A. designed and optimized the EUKulele and eukrhythmic software used in this study and oversaw its application. R.M.K., R.J.C., P.Z.L., B.A.A., E.W.C., J.A.B., M.V.J., and M.A.S. conducted sampling procedures with Clio. D.M.M. performed protein extractions, and M.R.M. led mass spectrometry efforts with the guidance of M.A.S. A.B. and J.S. contributed UVP imagery. R.J. led CTD deployments and the pigment analysis. J.A.B., M.V.J., and M.A.S. developed Clio and led the expedition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cohen, N.R., Krinos, A.I., Kell, R.M. et al. Microeukaryote metabolism across the western North Atlantic Ocean revealed through autonomous underwater profiling. Nat Commun 15, 7325 (2024). https://doi.org/10.1038/s41467-024-51583-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-51583-4
This article is cited by
-
Digital twin-driven swarm of autonomous underwater vehicles for marine exploration
Communications Engineering (2026)
-
Estimating variability of microeukaryotic community during summer at Potter Cove (Antarctica), using a multiprimer approach
Polar Biology (2025)
-
Missing microbial eukaryotes and misleading meta-omic conclusions
Nature Communications (2024)