Abstract
Strains of the Bacillus cereus (Bc) group are sporulating bacteria commonly associated with foodborne outbreaks. Spores are dormant cells highly resistant to extreme conditions. Nevertheless, the pathological processes associated with the ingestion of either vegetative cells or spores remain poorly understood. Here, we demonstrate that while ingestion of vegetative bacteria leads to their rapid elimination from the intestine of Drosophila melanogaster, a single ingestion of spores leads to the persistence of bacteria for at least 10 days. We show that spores do not germinate in the anterior part of the intestine which bears the innate immune defenses. Consequently, spores reach the posterior intestine where they germinate and activate both the Imd and Toll immune pathways. Unexpectedly, this leads to the induction of amidases, which are negative regulators of the immune response, but not to antimicrobial peptides. Thereby, the local germination of spores in the posterior intestine dampens the immune signaling that in turn fosters the persistence of Bc bacteria. This study provides evidence for how Bc spores hijack the intestinal immune defenses allowing the localized birth of vegetative bacteria responsible for the digestive symptoms associated with foodborne illness outbreaks.
Similar content being viewed by others
Introduction
Organisms are subjected to various environmental stresses including starvation, temperature variation, chemicals, and microbes. Healthy individuals overcome these assaults by engaging defense mechanisms that maintain their homeostasis. Among the stressors, opportunistic enteric bacteria become pathogenic when host defenses are diminished or inefficient.
The evolutionarily conserved innate immune system is the first line of defense against bacteria, and adult Drosophila has proven to be a powerful model for innate immunity studies1. In the midgut, the local innate immune system is mainly mounted in the anterior parts1,2. First, anterior enterocytes can rapidly sense the presence of allochthonous (i.e., non-commensal) bacteria and secrete reactive oxygen species (ROS) in a DUOX-dependent manner to block bacterial proliferation3,4. Concomitantly, luminal ROS are perceived by a subpopulation of anterior enteroendocrine cells that respond by releasing the DH31/CGRP neuropeptide, which promotes visceral muscle spasms to provoke the expulsion of bacteria from the midgut in less than 4 h post-ingestion4. Nevertheless, if the load of allochthonous bacteria is higher and/or if the immune ROS and visceral spasms are insufficient to eliminate them, the allochthonous bacteria can start to proliferate, releasing muropeptides from the peptidoglycan (PGN), a bacterial cell wall component, that bind to the transmembrane and intracellular immune receptors PGRP-LC and PGRP-LE, respectively5,6. Consequently, the Immune deficiency (Imd)/NF-κB pathway is activated, leading to the expression of anti-microbial peptides (AMPs) beginning 4–6 h post-ingestion7,8,9,10. Because a prolonged activation of the local innate immunity is responsible for chronical inflammation which is detrimental for the individual, a robust negative feedback is turned on to tune down the Imd pathway once the bacteria are cleared10. For instance, the PGRP-SC1 & 2 and PGRP-LB amidases have been described as negative regulators of the Imd pathway mainly acting by digesting the extracellular PGN fragments and thus blocking the recognition process by the Imd pathway receptors6,11,12,13. Altogether, these combined means of defense allow an efficient sensing and elimination of the allochthonous bacteria allowing the survival of the individual.
However, the detection of bacterial spores by the local innate immune system of the intestine and their elimination remains challenging for host organisms. Indeed, spores can resist many biological, chemical, or physical treatments14,15. Among opportunistic bacteria, the widespread environmental spore-forming bacteria belonging to the Bacillus cereus (Bc) group are well-known worldwide food poisoning pathogens that cause diarrheal and/or emetic-type illnesses16,17. Bc is the first cause of foodborne outbreaks (FBOs) in France18,19,20 and in Europe21. When nutrient-rich conditions are encountered, Bc spores can germinate, giving rise to vegetative cells which can even proliferate. Such favorable conditions are present in the small intestine of mammals, where it is assumed that spores are able to germinate and probably proliferate to ultimately trigger diarrhea due to the production of enterotoxins14,22. In vitro experiments have indicated that Bc vegetative cells can be destroyed by the acidic pH of the stomach and the biliary salt of the duodenum while spores can resist22,23,24,25,26,27. The effectiveness of the gut innate immune system to fight Bc vegetative cells has been demonstrated4. In contrast, nothing is known concerning the behavior and the fate of Bc spores in the intestine in vivo and the related immune response mounted by the host.
Here, thanks to in vivo studies in Drosophila melanogaster, we demonstrate that spores of Bc group can persist for at least ten days in the intestine and we could detected their germination only in the posterior part of the midgut. Next, we demonstrate that the spores do not trigger any detectable immune responses in the anterior parts of the Drosophila midgut. Once in the posterior midgut, germinated cells trigger the Imd immune pathway in a PGRP-LE-dependent manner. Strikingly, we found the amidases PGRP-SC1/2 and PGRP-LB, which are negative regulators of the Imd pathways, to be induced while the AMPs were repressed. In flies deficient in the PGRP-LE receptor, the cytosolic members of the Imd pathway or the PGRP-SC1/2 and PGRP-LB amidases, the persistence of the bacteria in midgut was reduced. Surprisingly, removing Relish, the NF-κB-like transcription factor, downstream of the Imd pathway has no impact on bacterial persistence. However, the depletion of Dif, another NF-κB-like transcription factor, together with Relish provided proof of a critical cooperation of both transcription factors in regulating amidase and AMP expressions in the posterior midgut and thus the bacterial clearance. Altogether, we provide evidence that spores belonging to the Bc group persist in the intestine when ingested as spores and escape the anterior immune response. Spores reach the posterior regions of the midgut where they germinate, and vegetative bacteria induce expression of amidases that act as negative regulators of the Imd pathway, dampening the production of AMPs, and consequently fostering the persistence of the bacteria.
Results
Spores of the Bc group persist in the Drosophila intestine
The Bc group is subdivided into at least eight phylogenetically very close genomospecies14,28,29. For this study, we selected two Bc sensu stricto strains: the Bc ATCC 14579 type strain30 and the probiotic strain, Bc Bactisubtil, whose ingestion of spores have been used to alleviate gastrointestinal disorders31. We also selected two Bacillus thuringiensis subspecies kurstaki (Btk) strains (Btk SA-11 and Btk ABTS-351) because of their broad use as microbial pesticides and the fact that Btk has been suspected to be responsible for FBOs19,32,33.
To determine whether, upon ingestion, Bc or Btk spores behaved the same as vegetative cells, we fed flies continuously with contaminated food (Fig. 1a) and assessed the amount of Bc or Btk bacteria in the Drosophila midgut at different times. We observed that regardless of the strain used, Bc/Btk were still present in the Drosophila midgut at least 10 days after the initial contact with spore-contaminated food (Fig. 1b). These data were very different from what we observed using Bc ATCC 14579 or Btk SA-11 vegetative cells, whose loads in the intestine rapidly decreased within 24 h (Fig. S1a). Unlike Bc, during sporulation, Btk produces Cry toxins embedded in a crystal, which displays specific entomopathogenic properties. Btk is widely used specifically to kill lepidopteran larvae that are broad crop pests14. Because the presence of Cry toxins might influence the behavior of Btk in the midgut, we engineered a Btk SA-11ΔCry strain cured of its plasmids and therefore devoid of Cry toxins (see Experimental Procedures). We also used a Btk ΔCry obtained from the Bacillus Genetic Stock Center (#4D22, https://bgsc.org/) also devoid of Cry toxins. Importantly, the SA-11ΔCry and the Btk ΔCry spores behaved like Btk SA-11 and Bc ATCC 14579 spores (Fig. 1b), refuting any possible role of the Cry toxins in Btk persistence. The commercially available preparation of the Btk ABTS-351 spores is known to contain 46% of additives (ec.europa.eu). To assess the potential involvement of those additives in the intestinal persistence of Btk ABTS-351, we extended our study to the commercial preparation, which we compared with a Btk ABTS-351 spore preparation made in our laboratory without any additives. We noticed that the commercial spores as well as the “homemade” Btk ABTS-351 spores behave similarly in the Drosophila midgut (Figs. 1b and S1b), suggesting that the additives present in the commercial preparation did not contribute to the persistence of Btk ABTS-351 spores.
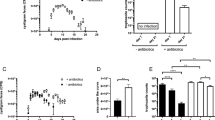
a Experimental setup to assess bacterial load after a continuous ingestion of spores. b Bacterial loads of dissected midguts after continuous ingestion of spores from Btk or Bc strains. The dot indicates the mean number of colony-forming units (CFUs) of at least three independent experiments per condition and time point. Each experiment corresponds to the mean of five midguts. CFUs correspond to spore and vegetative cell counts. Error bars correspond to the SEM. Source data are provided as a Source Data file. c Experimental setup to assess bacterial load after an acute ingestion of spores. Flies are in contact with the contaminated medium for 30 min and then transferred to fresh vial devoid of spores. d Bacterial loads of dissected midguts after acute ingestion of spores from Btk or Bc strains. The dot indicates the mean number of CFUs (spores + vegetative cells) of at least three independent experiments per condition and time point. Each experiment corresponds to the mean of five midguts. Error bars correspond to the SEM. * represent a statistically significant difference (p < 0.05) between Bc Bactisubtil and the other strains 10 days post-feeding using the two-sided Non-parametric Mann–Whitney’s test against each individual condition at 240 h. Source data are provided as a Source Data file. e Bacterial loads in split Drosophila midguts after acute intoxication with Btk (SA-11) or Bc ATCC 14579 spores. Dots correspond to independent experiments and are the mean of five pooled midgut domains. Error bars correspond to the SEM. The one-side Mann–Whitney tests were applied. Asterisks represent a statistically significant difference between bacterial loads in the anterior and the posterior midguts: **p < 0.01, *p < 0.05. Source data are provided as a Source Data file.
Because spores could germinate and proliferate on the fly medium, we checked this possibility by counting the number of Btk SA-11 bacteria on the fly medium in the absence of flies. We applied a heat treatment in order to kill all germinated vegetative cells. We noticed that 2 days after spore deposit on the fly medium, some of them started their germination and even proliferated 4 days after deposit (Fig. S1c). Hence, to remove this limitation in the persistence assessment, we performed acute feeding. Flies were fed with spores for 30 min before being transferred onto fresh food medium (i.e. without spores) (Fig. 1c). We first verified that upon acute ingestion of Bc ATCC 14579 or Btk SA-11 vegetative cells, they were readily cleared from Drosophila midguts4 (Fig. S1d). We then monitored the persistence of spores in the Drosophila midgut upon acute feeding (Fig. 1d). We used 30 min of spore feeding as food intake internal control. At 30 min of spore feeding, the bacterial load averaged 104 cells per midgut regardless of Bc/Btk strain. Interestingly, the bacteria could persist up to 10 days in the Drosophila midgut after acute spore ingestion (Figs. 1d and S1e). Noteworthy, we included the monitoring of the persistence of the probiotic Bc Bactisubtil strain and we found that Bc Bactisubtil load was significantly higher 10 days after acute feeding compared to all the other strains tested (Fig. 1d).
Because a recontamination of flies through their feces could occur during the 10 days of our experiments, we examined the amount of bacteria present in the feces and found only a small number of spores and vegetative cells (Fig. S1f). We also considered the potential for contamination by spores on the fly body during dissection. After examining fly bodies at 2, 24, and 48 h post-feeding, we found an average of 70 spores at 2 h, which decreased to 20 spores at 48 h (Fig. S1g). Our findings indicate that there is no evidence of contamination during the dissection or the reingestion of spores present in the feces that could account for this persistence. We also observed that the bacterial persistence in the midgut was not dependent on the genetic background of Canton S Drosophila since similar amounts of bacteria were detected over the 10 days of monitoring in either w1118 or w1118 isogenic flies (Fig. S1h). Finally, the persistence was also not affected in axenic flies suggesting that the commensal flora does not influence the Bc/Bt intestinal persistence (Fig. S1h).
The Drosophila midgut is subdivided into five major anatomical regions (R1 to R5) (Fig. S1i)34,35. To analyze in detail the localization of Bc/Btk along the midgut, we quantified the bacterial load in the anterior and posterior midgut after acute feeding. We did not focus on the acidic region due to its small size and the difficulty to dissect it accurately. During the first two hours after acute ingestion, we found that Bc/Btk bacteria were present in both regions of the midgut (anterior and posterior). However, from 4 h onward, the posterior midgut harbored a significantly higher load of Bc/Btk cells compared with the anterior midgut (Fig. 1e). Collectively, our results demonstrate that Bc/Btk persist for up to 10 days in the midgut and may accumulate preferentially in the posterior regions.
Spores of the Bc group germinate preferentially in the posterior midgut
Spores are metabolically dormant and resistant to extreme environmental conditions, allowing them to survive to extreme conditions15. However, the presence of nutrients can trigger the process of germination, in which spores emerge from dormancy, growing into vegetative cells. Since the intestine is a favorable environment for spore germination, we hypothesized that Bc/Btk spores might germinate in the Drosophila midgut. To address this question, we developed a robust fluorescent staining technique suitable for visualizing spores and differentiating them from their outgrowing vegetative cells. Dormant Bc/Btk spores harbored a red fluorophore cross-linked to the spore outer membrane. Once germinated, vegetative cells started to express the green fluorescent protein (GFP). The use of this novel tool endowed with dual red/green (R/G) labeling allowed us to follow the process of germination in real-time (Fig. 2a and supplementary movie 1).
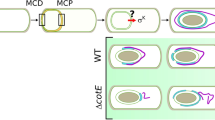
a Time-lapse images of SA-11R/G spores during germination. b Monitoring of SA-11R/G germination in vivo in Drosophila midgut 4 h post-spore-ingestion. Insets show enlargement of a single focal plan. Note that anteriorly GFP (i.e. germinating spores) are barely detectable. c Plots of the average fluorescence intensity (represented as mean gray value) of SA-11R/G germination measured along the Drosophila midgut presented in b. For all plot analyses of average fluorescence shown in this paper, the red line represents the average of the spore fluorescence (NHS-ester Alexa546) and the green line represents the average fluorescence of vegetative cells (GFP). d, e Bacterial load of Btk (SA-11) or Bc (ATCC 14579) in anterior (d) and posterior (e) Drosophila midguts. Green bars (non-heated samples) represent the whole Btk or Bc bacterial loads (spores and vegetative cells). Red bars (heated samples) represent the proportion of spore loads. Data represent the mean ± SEM of at least five independent experiments. Each experiment corresponds to five split midgut. Source data are provided as a Source Data file.
The use of the Btk SA-11R/G fluorescent strain in vivo first revealed that at 0.5- and 2-hours post-ingestion, Btk spores occupied the lumen of the whole midgut (Fig. S2a, b). Few vegetative cells were detectable in the posterior midgut 2 h post-ingestion (inset in Fig. S2b). Btk SA-11R/G spore germination was evident in the posterior midgut 4 h after ingestion (Fig. 2b, c). Interestingly, we found that regardless of the Bc group strain, excepted Bc Bactisubtil, spore germination occurred markedly in the Drosophila posterior midgut at 4 h after ingestion (Fig. S2d). Twenty-four hours post-ingestion, we detected mostly vegetative cells in the posterior midgut (Fig. S2c). Noteworthy, very few germinated cells were observed for Bc Bactisubtil 24 h post-acute feeding (Fig. S2d). To further confirm that spores mainly germinated in the posterior midgut, we performed measurements of colony-forming units (CFUs) in the anterior and posterior parts of the midgut by comparing heat-treated intestinal samples (to kill germinating spores and vegetative cells but not spores) to non-heat-treated samples (cumulating spores, germinating spores and vegetative cells). Interestingly, we did observe in the anterior midgut region the almost exclusive presence of Bc/Btk spores, even 3 days after acute ingestion (Fig. 2d). However, in the posterior midgut, we observed the appearance of the first Bc/Btk germinating spores as early as 30 min after ingestion (Fig. 2e). Together, these results demonstrate that the germination of Bc group spores begins 30 min after oral ingestion and occurs mainly in the posterior midgut of Drosophila melanogaster.
Spores do not trigger detectable Drosophila midgut innate immune response
The persistence of spores in the Drosophila midgut raises the question of how the local innate immune system can tolerate spores and/or vegetative cells. As mentioned previously, in response to enteric infection, the anterior Drosophila midgut (R1 and R2 regions) initiates immune responses via the luminal release of ROS and, if necessary, AMPs2,4,5,36,37,38. To test the release of local immune ROS (HOCl) in response to Btk vegetative cells or spores, we used the R19S probe, a HOCl sensitive fluorescent dye38,39. First, we confirmed that Btk vegetative cells induced ROS only in the anterior region 1-hour post-ingestion (Fig. 3a, middle panel). Surprisingly, Drosophila fed with Btk spores did not show ROS induction either in the anterior or in the posterior regions of the Drosophila midgut at that time (Fig. 3a, lower panel), though spores germinated in the posterior midgut (Fig. 2 and S2). We investigated the potential ROS production at later time points (i.e. 4, 8 and 24 h) in the posterior midgut and no HOCl production was detected (Fig. S3a). In parallel, we specifically knocked down the expression of Duox in Drosophila enterocytes by RNA interference and examined the resulting impact on the spore persistence. We first confirmed that, 4 h after ingestion of vegetative cells, the silencing of Duox in the enterocytes increased the load of Btk compared with control intestines4 (Fig. 3b). However, Duox silencing in Drosophila enterocytes did not impact the Btk persistence after spore ingestion (Fig. 3b). Based on these data, we inferred that ingestion of spores does not induce the production of Duox-dependent ROS.
a ROS monitoring one-hour post-acute feeding with SA-11 spores or vegetative cells. ROS production in the midgut is visualized by the HOCl-specific R19S probe (orange). DAPI (blue) marks the nuclei. b SA-11 loads in midguts knocked down for the expression of Duox in enterocytes 0.5 or 4 h after acute feeding with vegetative cells or spores. The horizontal axis indicates the mean number of CFUs. Dots correspond to independent experiments of five pooled midguts. c DptA-Cherry expression (red) in the anterior R1 midgut region (upper panel) and in the posterior R5 midgut region (bottom panel) of Drosophila fed for 30 min with H2O, Ecc15, SA-11 vegetative cells (Btvg) or SA-11 spores (Btsp) and observed 24 h later. In each panel anterior is to the left. Measured quantities are shown on the right graphs. The results are given as the relative expression compared with the control (H2O). At least three independent experiments were performed and each dot correspond to one midgut. d AttD-Gal4 UAS-Cherry expression (red, AttD>Cherry) in the anterior R1 midgut region (upper panel) and in posterior R4 midgut region (bottom panel) of Drosophila fed for 30 min with H2O, Ecc15, SA-11 vegetative cells (Btvg), or SA-11 spores (Btsp) and observed 24 h later. Measured quantities are shown on the right graphs. The results are given as the relative expression compared with the control (H2O). At least three independent experiments were performed and each dot correspond to one midgut. e qRT-PCR analyses of AMP expression in midgut upon acute feeding with SA-11 or Bc spores. UC corresponds to flies fed with water. For RT-qPCR results, mRNA levels in unchallenged wild-type flies were set to 100 and all other values were expressed as a percentage of this value. RT-qPCR results are shown as mean ± SEM. Dots correspond to independent experiments of 10 pooled female flies. f Bacterial load in the midguts of ∆AMP14 mutant flies 0.5 or 4 h after acute feeding with SA-11 spores. The horizontal axis indicates the mean number of CFUs per midgut. Each dot corresponds to an independent biological replicate where each replicate is the mean of five midguts. g Representative confocal images showing SA-11R/G spore germination in the anterior and posterior midgut of WT (Canton S) and ∆AMP14 mutant flies 2 h after acute feeding with spores. DAPI (blue) marks the nuclei. Spores are in red, vegetative cells in green. The yellow fluorescence corresponds to germinating spores. Error bars correspond to the SEM. The two-sided Mann–Whitney test was applied in b–d and f. A two-sided Student’s t-tests were used to analyze data in e. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ns non-significant (exact P values are provided in the source data). Source data are provided as a Source Data file.
We next investigated the induction of AMP genes in the Drosophila midgut following acute ingestion of vegetative cells vs. spores. Using the DiptericinA-Cherry (DptA-Cherry) and AttacinD-Gal4 UAS-Cherry (AttD>Cherry) reporters, two readouts for the activation of the Imd pathway in the midgut40, we first observed that the acute ingestion of vegetative cells of the Erwinia carotovora carotovora (Ecc15) opportunistic bacteria-induced DptA and AttD expression in the anterior midgut (Figs. 3c and S3b, c). Ecc15 was also capable of promoting the spreading of AttD expression in the posterior R4 region (Fig. S3c). However, Btk vegetative cells did not show significant changes in DptA and AttD expression in either the anterior or posterior midgut (Figs. 3c, d, and S3b, c). These data are consistent with the fact that early ROS induction followed by the visceral spasms are sufficient to rapidly eliminate Btk vegetative cells upon acute ingestion (Fig. 3a, b)4, at least before the Imd pathway can be induced. RT-qPCR analyses of the expression of DptA, Defensin, and AttD genes on dissected midguts confirmed the non-induction of AMP genes after acute ingestion of Btk (or Bc) vegetative cells (Fig. S3d).
Monitoring AMP expression upon acute spore feeding revealed that neither DptA-Cherry nor AttD>Cherry reporter genes were induced in vivo in the anterior midgut (Figs. 3c, d and S3b, c). Strikingly, repression of the DptA-Cherry reporter expression in the posterior midgut was observed (Figs. 3c and S3b). RT-qPCR analyses confirmed the repression of DptA expression as well as the repression of Defensin, AttD, and Drosomycin (Drs) expression (Figs. 3e and S3e). Importantly, ingestion of Bc Bactisubtil spores that did not germinate in posterior in midgut (Fig. S2d) did not led to the repression of AMP expression (Fig. 3e). This emphasizes the significance of localized spore germination in the posterior midgut, which is essential for promoting the down-regulation of AMP expression. Together these data suggest that Bc/Btk persistence upon spore ingestion and germination in the posterior midgut could be supported by decreased expression of AMPs.
To assess the involvement of the AMPs in Btk SA-11 persistence, we used a fly strain in which the 14 AMP genes were deleted (ΔAMP14)41,42. We quantified the Btk SA-11 load in the midgut of Wild Type (WT - Canton S) and ΔAMP14 flies. While both genotypes ingested a similar amount of spores during the 30 min of feeding (Fig. 3f), 120 h post-feeding, in ΔAMP14 mutant flies, we found a significantly higher Btk SA-11 load compared with wild-type flies (Fig. 3f). This result suggests that AMPs could kill germinating cells in the posterior midgut. To further challenge this hypothesis, we monitored the fate of the Btk SA-11R/G fluorescent strain in ΔAMP14 mutant flies after acute ingestion of spores. Interestingly, as soon as two hours post-ingestion, confocal imaging showed higher levels of germinating spores in ΔAMP14 mutant posterior midguts than in WT posterior midguts (Fig. 3g). We did not observe obvious changes in the anterior midguts where only spores were present (Fig. 3g). Collectively, our data suggest that, though repressed upon spore ingestion and their germination in the posterior midgut, the weak production of AMPs is necessary to limit the Bc/Btk bacterial load.
Amidases contribute to the intestinal persistence of spores
Since AMPs expression was downregulated after spore ingestion, we wondered whether the amidases, which exert negative feedback on the Imd pathway, could be induced by germinating spores11,12,40. First, we verified by RT-qPCR analyses that the ingestion of Bc/Btk vegetative cells could induce the expression of the three amidase encoding genes in the midgut (Fig. S4). Interestingly, after spore ingestion, we observed that PGRP-SC1 and -SC2 were consistently induced both 4 h and 24 h post-feeding while PGRP-LB was only induced at 4 h and was repressed at 24 h (Fig. 4a). Noteworthy, Bc Bactisubtil did not induce any amidases confirming that the germination of spores in the posterior compartment is required to modulate Imd target genes (Fig. 4a). We next assessed the Btk SA-11 intestinal load in mutant flies homozygous for either the PGRP-SC1/2 or PGRP-LB loss-of-function alleles (PGRP-SC1/2 Δ and PGRP-LB Δ respectively)12. In midguts from these mutant animals, no difference in bacterial load was observed compared with control flies 30 min after spore feeding (Fig. 4b). However, 120 h post-feeding, the loss of PGRP-SC1/2 or PGRP-LB was associated with a significant decrease in the number of Btk SA-11 in the midgut compared with the WT flies (Fig. 4b). Because spores accumulated and germinated in the posterior midgut of WT flies as early as 4 h post-ingestion (Fig. 2), we monitored the fate of the spores in PGRP-SC1/2 Δ and PGRP-LB Δ deficient flies. Six hours after acute ingestion, confocal imaging showed the presence of fewer red/green germinating spores in the posterior midguts of PGRP-SC1/2 ∆ or PGRP-LB Δ flies compared with WT flies (Fig. 4c), suggesting that in the absence of amidases, the production of AMPs was able to kill germinating spores.
a qRT-PCR analyses of amidase expressions in midguts upon SA-11 or Bc spore acute feeding. UC corresponds to flies fed with water. Results are shown as mean ± SEM. The dots correspond to independent experiments of 10 pooled female flies. b Bacterial load in midguts of PGRP-SC1/2 Δ double mutant or PGRP-LB ΔE mutant 0.5 or 120 h after SA-11 acute feeding with spores. The horizontal axis indicates the mean number of CFUs per midgut. Each dot corresponds to an independent biological replicate where each replicate is the mean of five midguts. c Representative confocal images showing SA-11R/G spore germination in the anterior and posterior midgut of WT (Canton S), PGRP-SC1/2 Δ double mutant or PGRP-LB ΔE mutant flies 6 h after spore acute feeding. DAPI (blue) marks the nuclei. Spores are in red, vegetative cells in green. The yellow fluorescence corresponds to germinating spores (Fig. 2a). d qRT-PCR analyses of AMP expressions in midguts of PGRP-SC1/2 Δ mutants following acute feeding with SA-11 spores. UC corresponds to PGRP-SC1/2 Δ flies fed with water. mRNA levels in unchallenged PGRP-SC1/2 Δ flies were set to 100 and all other values were expressed as a percentage of this value. Results are shown as mean ± SEM. Dots correspond to independent experiments of the mean of 10 pooled female midguts. e SA-11 load in midguts of flies silenced for PGRP-SC2 or PGRP-LB or overexpressing PGRP-SC2 specifically in enterocytes (using the myo1Ats driver) 0.5 or 120 h after acute feeding with spores. The horizontal axis indicates the mean number of CFUs per midgut. Each dot corresponds to an independent biological replicate where each replicate is the mean of five midguts. Error bars represent SEM. Two-sided Student’s t-tests were used to analyze data in a and d. Two-sided Mann–Whitney test was used to analyze data in b and e. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ns non-significant (P values are provided in the source data file). Source data are provided as a Source Data file.
We therefore investigated whether the repression of AMPs expression upon spore ingestion was indeed dependent of amidases. As expected, expression of AMPs was not repressed; indeed, it was even induced by Btk SA-11 spore ingestion in a PGRP-SC1/2 Δ or PGRP-LB Δ mutant background (Fig. 4d compared with 3e). Because amidases can be produced by the enterocytes or by the fat body (a systemic immune tissue) and can act at a distance from the site of production40, we specifically silenced PGRP-SC2 or PGRP-LB in enterocytes and, while food intake was not affected after the 30 min of feeding, we found a significant decrease in the load of Btk SA-11 in the midgut 120 h post-feeding (Fig. 4e). Conversely, the overexpression of PGRP-SC2 in enterocytes resulted in an increased bacterial load 120 h post-feeding without any effect on food intake measured after 30 min (Fig. 4e). Overall, our data suggest that spores are not detected by the anterior midgut immune response (i.e., no production of ROS or AMPs), and reach the posterior regions where they germinate and can activate the Imd signaling target genes PGRP-SC1, PGRP-SC2, and PGRP-LB. In turn, amidases promote a repression of the basal expression of AMP-encoding genes. Consequently, downregulation of AMP expression favors Bc/Btk persistence in the posterior Drosophila midgut.
The Imd pathway contributes to the intestinal persistence of bacteria
Because the expression of amidases is under the control of the Imd pathway in the intestine, we first tested the involvement of the two Imd pathway receptors, PGRP-LC and -LE, in Btk SA-11 persistence. In flies homozygous viable for the loss-of-function mutant for either receptor, similar amounts of Btk SA-11 were ingested compared with WT flies after 30 min of spore feeding (Fig. 5a). Nevertheless, 120 h after feeding, only PGRP-LE112 mutant flies showed a significant decrease in Btk SA-11 intestinal load (Fig. 5a) similar to that observed for flies lacking the amidases (Fig. 4b). We further tested mutants for intracellular components of the Imd pathway. Loss-of-function mutants for the cytoplasmic components Imd (imdShaddok) or Dredd (DreddF64) also displayed a decrease in Btk SA-11 persistence 120 h post-ingestion (Fig. 5a). Unexpectedly, the bacterial load in flies homozygous mutant for the downstream Imd pathway NF-κB-like transcription factor Relish (RelE20)43 was similar to control flies (Fig. 5b), although Rel has been found to be absolutely required in midgut epithelial cells to respond to enteropathogenic bacteria5,8,44. To further understand the apparent absence of Rel function in our model of spore infection, we inhibited Relish expression specifically in enterocytes. Silencing Relish in enterocytes did not change Btk bacterial abundance in the midgut (Fig. 5b). In addition, we monitored the fate of the Btk SA-11R/G fluorescent strain in RelE20 mutants, six hours after acute ingestion. No obvious differences between WT and RelE20 were observed in the Drosophila midgut (Fig. 5c). We also performed an epistasis experiment, removing both PGRP-SC1 and -SC2 amidases in the RelE20 loss of function background. Interestingly, while food intake measured after 30 min was not affected, the bacterial load was lower 120 h post-feeding compared with the control (Fig. 5b) to a similar extent to that of mutants for amidases alone (Fig. 4b). This observation suggests that amidases act downstream of Relish to control Btk persistence. Finally, we overexpressed in enterocytes an activated form of Relish known to strongly induced AMP expression45. As expected, we observed a reduced bacterial load 120 h after spore ingestion (Fig. 5b). Together, these data suggest that Relish has no significant role in the control of amidase expression in the posterior midgut and that Btk persistence depends on finely-tuned expression levels of amidases and AMPs.
a, b SA-11 bacterial load in midguts of different genotypes for components of the Imd pathway 0.5 or 120 h after acute feeding with spores. a Homozygous loss of function mutants for PGRP-LE112, PGRP-LC Δ, Imdshaddok or DreddF64. b Homozygous loss of function mutant for RelE20 or double homozygous loss of function mutant for PGRP-SC1/2 Δ; RelE20. Silenced (RelRNAi) or overexpressed (RelVP16) Relish in enterocytes (using the myo1Ats driver). The horizontal axis indicates the mean number of CFUs per midgut. Each dot corresponds to an independent biological replicate where each replicate is the mean of five midguts. c Representative confocal images showing SA-11R/G spore germination in the anterior and posterior midgut of WT flies (Canton S) and RelE20 mutant flies 6 h after acute feeding with spores. DAPI (blue) marks the nuclei. Spores are in red, vegetative cells in green. The yellow fluorescence corresponds to germinating spores (see Fig. 2a). d–g RT-qPCR analyses of the expression of AMPs (d and f) and amidases (e and g) in Rel E20 (d and f) and PGRP-LE 112 (e and g) mutant flies 4 h and 24 h after acute feeding with SA-11 spores. UC corresponds to flies fed with water. The dots correspond to independent experiments of 10 pooled female flies. RT-qPCR are represented as relative level of expression normalized to RP49 and Dp1 genes. Error bars represent SEM. Two-sided Mann–Whitney test was used to analyze data in a and b. Two-sided Student’s t tests were used to analyze data in d–g. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ns non-significant (P values are provided in the source data file). Source data are provided as a Source Data file.
Given these observations, we wanted to test the involvement of Relish in controlling the remaining AMP expression in WT flies fed with spores (Fig. 3e). In RelE20 unchallenged flies, while DptA and Defensin were drastically downregulated, AttD expression was not affected (Fig. S5a). In addition, still in unchallenged flies, the absence of Relish correlated with the downregulation of PGRP-SC1a/b and PGRP-LB expression but, unexpectedly, PGRP-SC2 was strongly upregulated (Fig. S5b). Hence, the above data suggest that Relish is involved in the repression of PGRP-SC2 expression and is not necessary for the basal AttD expression.
We further investigated the midgut expression of AMPs and amidases in a RelE20 mutant background upon spore ingestion. The expression of AMPs was neither induced nor repressed (as one would have expected since spore feeding induced AMP repression in a wild-type background, Fig. 3e) 4 h or 24 h post-feeding with spores (Fig. 5d). Even though PGRP-SC2 expression remained high, the expression of amidases was not induced in RelE20 mutants (Fig. 5e). Gathering these data suggests that Btk intestinal persistence is not affected upon spore ingestion by RelE20 mutant flies because, first, the upregulation of PGRP-SC2 likely compensates for the downregulation of PGRP-SC1 and -LB and second, the normal level of AttD expression might be sufficient to limit Btk persistence, knowing that AttD expression was downregulated upon spore ingestion by WT flies (Fig. 3e).
Because in the absence of PGRP-LE receptor Btk load was lower 120 h after spore feeding (Fig. 5a), we wondered whether in PGRP-LE112 loss of function mutant flies both the expressions of AMPs and amidases were impacted. In PGRP-LE112 unchallenged flies, the expressions of DptA, Defensin, PGRP-SC1 and -SC2 were lowered while AttD and PGRP-LB expressions were unaffected (Fig. S5c, d). Upon feeding PGRP-LE112 flies with spores, none of the amidases were induced (Fig. 5g) confirming that PGRP-LE is the primary receptor in the posterior midgut that regulates amidase expression5. Conversely, both Defensin and AttD were induced 4 h post-ingestion (Fig. 5f). Interestingly, though mild, this rise in AMP expression was correlated with the reduced bacterial load observed 120 h post-ingestion (Fig. 5a).
Taken together our data demonstrate that PGRP-LE is required to sense Bc/Btk geminating cells in the posterior midgut and consequently to activate the expression of the amidases. In turn, amidases provoke a reduction of AMPs expression by tuning down the Imd pathway but likely also by tuning down another pathway since Defensin and AttD are still induced by the ingestion of spores in absence of PGRP-LE. Consequently, the decrease in AMP levels favor the local Bc/Btk persistence in the posterior midgut.
Toll pathway participates to the regulation of amidases and AMPs in the posterior midgut in response to spore ingestion
The above data prompted us to investigate the possible involvement of the immune Toll signaling pathway in the Drosophila posterior midgut. The NF-κB-related transcription factor, the Dorsal-related immunity factor (Dif), acts downstream of the Toll pathway during the systemic immune response46,47,48,49. To understand whether Dif could also be involved in Bc/Btk persistence, we took advantage of flies homozygous viable for the Dif loss-of-function allele, Dif 1. While Dif 1 and WT flies ingested similar amounts of spores during the 30 min of feeding, 120 h later, we observed a significant decrease of Btk SA-11 loads in Dif 1 mutant flies (Fig. 6a). Confocal microscopy analysis confirmed the decrease of Btk SA-11R/G fluorescent cells, primarily in the posterior midguts of Dif 1 mutant flies, compared with WT (Fig. 6b). We confirmed the involvement of the Toll signaling pathway first by feeding Myd88 loss of function flies with spores. Myd88 is an adapter acting downstream of Toll receptor and upstream of Dif 50,51. Second, we specifically silenced Toll expression in enterocytes. In both mutants, the bacterial load was reduced 120 h after feeding with spores (Fig. 6a). To further analyze the role of the Toll pathway in the intestinal immune response, we monitored the midgut expression of AMPs and amidases in Dif 1 or Myd88 mutant flies by qRT-PCR. In uninfected flies, the expression of DptA and Defensin was significantly lowered in both mutants compared with WT, while AttD was not affected (Fig. S6a, c). The expression of PGRP-SC2 was also not affected in these mutants (Fig. S6b, d). Nevertheless, PGRP-SC1 was downregulated in Dif 1 mutant flies but upregulated in Myd88 mutant flies (Fig. S6b, d). Finally, PGRP-LB expression was unaffected in Dif 1 mutant flies but downregulated in Myd88 mutant flies (Fig. S6b, d).
a SA-11 bacterial load in midguts of Dif1 or Myd88 KG03447 homozygous loss of function mutant, or in midguts silenced for Toll (Toll RNAi) in enterocytes (using the myo1Ats driver) 0.5 or 120 h after acute feeding with spores. The horizontal axis indicates the mean number of CFUs per midgut. Each dot corresponds to an independent biological replicate where each replicate is the mean of five midguts. b Representative confocal images showing SA-11R/G spore germination in the anterior and posterior midgut of WT flies (Canton S) and Dif1 homozygous mutant flies 6 h after acute feeding with spores. DAPI (blue) marks the nuclei. Spores are in red, vegetative cells in green. The yellow fluorescence corresponds to germinating spores (Fig. 2a). c–f RT-qPCR analyses of the expression of AMPs (c and e) and amidases (d and f) in Dif1 (c and d) or Myd88KG0344 (e and f) homozygous mutant flies 4 and 24 h after acute feeding with SA-11 spores. Unchallenged flies (UC) corresponds to flies fed with water. The dots correspond to independent experiments of 10 pooled midguts. Error bars represent mean ± SEM of at least three independent experiments. Two-sided Mann–Whitney test was used to analyze data in A. Two-sided Student’s t tests were used to analyze data in c–f. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ns non-significant (P values are provided in the source data file). Source data are provided as a Source Data file.
Feeding Dif 1 mutant flies with spores only promoted the repression of Defensin 24 h post-feeding (Fig. 6c). Notably, the global relative levels of AMP expression upon spore ingestion were higher in Dif 1 than in RelE20 (compare Figs. 6c and 5d), likely explaining why the bacterial load was reduced in Dif 1 mutant (Fig. 6a) but not in RelE20 (Fig. 5b). While amidases were all induced in a WT background upon spore ingestion (Fig. 4a), in the absence of Dif, only PGRP-LB remained inducible 24 h post-feeding (Fig. 6d). In Myd88 mutant flies fed with spores, we observed an induction of the 3 AMPs 4 h post ingestion (Fig. 6e) in correlation with the reduced bacterial load reported in Fig. 6a. This result also pinpoints a role of Toll/Myd88 in repressing AMP expression in WT background (Fig. 3e). Interestingly, all amidases were induced 24 h post spore-feeding (Fig. 6f) suggesting that Toll/MyD88 signaling is not involved in amidases upregulation upon spore ingestion.
Dif cooperates with Relish to modulate the intestinal immune response to spore ingestion
To confirm the cooperation of both immune signaling and their downstream NF-κB transcription factors in regulating the expression of amidases and AMPs in the posterior midgut, we generated a Dif 1; RelE20 double mutant flies. We first monitored Btk persistence upon spore ingestion in this genetic background. Dif 1; RelE20 double mutant and WT flies ingested a similar amount of spores during the 30 min of feeding (Fig. 7a). However, 120 h later there was a significantly higher Btk SA-11 load in the double mutant flies compared with WT (Fig. 7a). Consistently, 6 h after ingestion of Btk SA-11R/G spores, more fluorescent bacteria were present in the posterior midguts of Dif 1; RelE20 double mutant flies compared with WT (Fig. 7b). In this double mutant background and in absence of spore feeding, the expressions of AMPs were lowered (Fig. S7a) suggesting that Dif and Relish act as cofactors to maintain basal levels of DptA and Defensin expressions while Dif and Relish act redundantly to maintain the basal level of AttD expression (Figs. S5a, S6a, and S7a). Our analyses of amidases expression in absence of spores revealed that while both factors act as cofactors to maintain the basal expression of PGRP-SC1, Relish was required to repress PGRP-SC2 but Dif appears necessary for PGRP-SC2 upregulation in absence of Relish (Figs. S5b, S6b, and S7b). Finally, PGRP-LB was only regulated by Relish since its expression in the Dif 1; RelE20 double mutant was similar to the RelE20 single mutant (Figs. S5b and S7b).
a SA-11 bacterial load in midguts of Dif1;RelE20 homozygous mutants 0.5 or 120 h after acute feeding with spores. The horizontal axis indicates the mean number of CFUs per midgut. Each dot corresponds to an independent biological replicate where each replicate is the mean of five midguts. b Representative confocal images showing SA-11R/G spore germination in the anterior and posterior midgut of WT flies (Canton S) and Dif 1;RelE20 homozygous mutant flies 6 h after acute feeding with spores. DAPI (blue) marks the nuclei. Spores are in red, vegetative cells in green. The yellow fluorescence corresponds to germinating spores. c, d RT-qPCR analyses of the expression of AMPs (c) and amidases (d) in Dif 1;RelE20 homozygous mutant flies 4 and 24 h after acute feeding with SA-11 spores. UC corresponds to flies fed with water. The dots correspond to independent experiments of 10 pooled female flies. Data represent mean ± SEM. Two-sided Mann–Whitney test was used to analyze data in a. Two-sided Student’s t tests were used to analyze data in c and d. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ns = non-significant (P values are provided in the source data file). Source data are provided as a Source Data file. e Bacterial persistence upon ingestion of spores. Upper part: ingestion of vegetative cells triggers the release of ROS in the lumen by the Duox enzyme located in anterior enterocytes. In addition to their bacteriostatic activity, ROS induce visceral spasms that accelerate bacterial clearance. Then, the binding of PGNs to the PGRP-LC transmembrane receptor activates the IMD pathway, leading to the release of AMPs which in turn kill the remaining bacteria. Lower part: ingested spores are not perceived by the anterior midgut. Spores reach the posterior midgut where they encounter favorable conditions for their germination. The release of PGNs by the germinating bacteria stimulates the cytoplasmic PGRP-LE receptor directly and the Toll receptor indirectly. The activated IMD and Toll pathways converge on the NF-kB factors Relish and Dif, which activate the genes encoding amidases. The secreted amidases, by digesting PGNs, exert a negative feedback on AMPs production in the posterior midgut, favoring bacterial persistence.
Upon spore ingestion, in the Dif 1; RelE20 double mutant, the overall levels of AMP expression were still low with no repression observed (Fig. 7c). These low levels of AMPs correlated with the increased bacterial load observed (Fig. 7a). The expressions of amidases were also not upregulated in the Dif 1; RelE20 double mutant (Figs. 7d and 4a) confirming that Rel and Dif were necessary together for PGRP-SC1 induction while only Rel was required for PGRP-LB induction and Dif for PGRPSC-2 induction (Figs. 5e, 6e, and 7d). Overall, these results demonstrate that Dif and Relish cooperate to tightly balance the expression of AMPs and amidases in the posterior midgut of unchallenged as well spore-fed flies.
Discussion
The majority of Bc-dependent FBOs, is due to the ingestion of Bc bacteria, which must grow in the gut and subsequently produce pore-forming enterotoxins responsible for the onset of diarrhea symptoms16. However, the mechanisms by which Bc bacteria colonize the gut and produce toxins remain poorly understood, and several questions unanswered. Is the disease due to the ingestion of vegetative bacteria or spores? What is the distribution and the fate of ingested spores along the gastrointestinal tract? How does the intestinal innate immune system detect and fight the infection? Here, we deciphered the behavior and fate of Bc cells in the intestine of Drosophila melanogaster and demonstrated that spores escape the innate immune system to reach the posterior part of the midgut/small intestine, where they can germinate and persist for days (Fig. 7e).
First of all, our findings confirm in vivo that after ingestion of vegetative cells, the Bc load in the Drosophila intestine remains low and that the bacteria are cleared in less than 24 h4,52,53. In Drosophila, it has also been shown that the presence of vegetative cells in the anterior midgut is rapidly detected, triggering the production of ROS and visceral spasms, both cooperating to quickly evict the undesired bacteria4. Therefore, the minimum infectious dose required to cause intestinal disorders (at least 105 CFU22,) is likely difficult to reach upon ingestion of vegetative cells. On the contrary, it has been suggested that the capacity of spores to withstand extreme conditions would allow them to overcome stomach acidity and bile salt attacks in the duodenum, favoring germination in the posterior small intestine. Consequently, the infectious dose could be more readily achievable, as illustrated by the 103 spores/g of food that can be associated with FBOs19,54. The spores of all Bc group strains we tested persisted up to 10 days post-ingestion. Consistent with our observations, studies have shown that Bc can persist at least 18 days in the intestine of rats transplanted with human-flora55 and 30 days in mouse intestine56. Moreover Bt could be detected in fecal samples in greenhouse workers five days after cessation of bioinsecticide use57. Interestingly, we also showed in vivo that spores accumulate and germinate (except for the Bc Bactisubtil strain) in the posterior part of the Drosophila midgut. Similarly, data suggest that spores derived from the probiotic B. subtilis germinate in the jejunum and eventually in the ileum of mice58,59. Our data also highlight the very rapid germination of spores in the posterior parts of the intestine (in less than 2 h). Indeed, while the proximal regions of the Drosophila midgut is quite acidic and produce digestive enzymes to break down food, the more distal parts of the intestine harbor a more basic and anaerobic environment with nutrient availability35,60,61. Interestingly, it has been shown, in vitro, that anaerobic conditions slow down the growth rate of Bc but favor the production of CytK, Nhe, and Hbl enterotoxins16,22,62,63,64,65,66. Hence, all the conditions for spore germination and enterotoxin production are encountered in the posterior Drosophila midgut, which accounts for the occurrence of diarrheic symptoms when a critical bacterial load is reached. However, contrary to what it has been observed for the B. subtilis probiotic strain that can proliferate in the intestine with an intestinal bacterial load either maintained or increasing over a short period58,59, we never observed such an event with the different Bc/Bt strains studied, even with the Bc Bactisubtil probiotic strain. Moreover, in all our microscopic confocal observations, we did not detect dividing bacteria. Hence, the capacity of probiotic strains to proliferate could be one important feature allowing their establishment in the intestine and the manifestation of their beneficial effects.
Importantly, our data also show that the local innate immune response is ineffective in eliminating vegetative cells in the posterior regions of the Drosophila midgut, which enables Bc/Bt persistence. Using Drosophila genetic tools, we first show that spores are not detected in the anterior midgut, unlike vegetative cells, which rapidly trigger immune ROS production4,36. Strikingly, although spores germinate in the posterior midgut, there is no release of immune ROS. Immune ROS are normally produced in a DUOX-dependent manner in response to uracil secretion by allochthonous bacteria36. Uracil is thought to serve as bacterial growth factor, promoting proliferation67. Hence, we can assume that either Bc/Bt vegetative cells in the posterior midgut do not produce uracil or the host receptor for uracil68 is absent from the posterior midgut. Moreover, the germination of spores in the posterior midgut, through the activation of the negative regulators, amidases, dampens the production of AMPs. Interestingly, repression of AMP expression was observed in lepidopteran larvae fed with commercial spores of Bt69 or with spores of Bt HD73 strain70 suggesting a conserved mechanism, at least in insects. Consequently, the combination of the absence of ROS and the reduced levels of AMPs favor Bc/Bt persistence.
Why does spore germination in the posterior midgut induce only genes encoding amidases and not those encoding AMPs? It has been shown that the Imd pathway cytosolic receptor PGRP-LE is required in the anterior midgut to activate AMP genes in response to pathogenic bacteria and to upregulate the amidases PGRP-SC1 and -LB in the posterior midgut in response to commensal bacteria. The transmembrane PGRP-LC receptor is also required in cooperation with PGRP-LE in the anterior midgut to activate the expression of AMPs in response to pathogenic bacteria, however, PGRP-LC is dispensable in the posterior midgut5,11,34,45,71,72,73. Consistent with this observation, we found that only PGRP-LE is involved in response to spore ingestion and local germination in the posterior midgut. Therefore, in the posterior midgut, the germination of spores allows the activation of the Imd pathway in a PGRP-LE-dependent manner, leading to the induction of three amidases (including PGRP-SC2) but not of AMPs. Noteworthy, many convergent data suggest that only amidase genes are inducible in a PGRP-LE-dependent manner in the posterior midgut while AMPs are poorly inducible5,11,12,40. Hence the germination of spores of Bc/Bt in the posterior compartment are perceived as if they were commensal bacteria, inducing a tolerance response5,74,75 through the induction of amidases that in turn dampen AMPs expression. Our spore ingestion paradigm also reveals that when the Imd pathway is only mobilized in the posterior midgut (spores are not detected in the anterior compartment) in a PGRP-LE-dependent manner, the only response elicited is the induction of amidases, even if the ingested bacteria are non-commensal ones. Consistently, the transcription repressor Caudal and the negative Imd regulator Pirk have been shown to be involved in the repression of AMP expression specifically in the posterior midgut5,12,76. Hence the midgut could be separated into two distinct immune domains: the anterior midgut is competent to fight pathogenic bacteria ingested along with the food, and the posterior midgut is immune-tolerant to sustain commensal flora. Bc/Bt spores have developed a strategy to hijack this physiological state for their own benefit, allowing them to escape the strong anterior immune response that would otherwise kill the germinated cells. Consistent with this model, it has been well demonstrated that the Drosophila posterior midgut is capable of increased cell turnover, when compared to the anterior midgut, in order to overcome the damages caused by pathogens35,77,78,79,80, likely to compensate for a weaker innate immune response.
Importantly, we cannot compare what we observe in this work using spore feeding for 30 min with all the previous data published using continuous feeding with more elevated doses of vegetative bacteria. In this latter case, most of the immune events occur in the anterior midgut2,81 while with spores that germinate in the posterior midgut, almost nothing happens in the anterior midgut. Our spore paradigm is similar to that of commensal vegetative bacteria, which do not trigger an immune response in the anterior midgut, but induce amidases in the posterior midgut to allow their tolerance by the host5,11,72,82.
Unexpectedly, our work in Drosophila also revealed that Imd is not the sole signaling pathway driving the innate immune response in the posterior midgut. Indeed it has been well demonstrated that Relish is absolutely required in the anterior midgut downstream of the Imd pathway to mount an efficient immune response against pathogens5,8,83,84. Interestingly, we observed that in the absence of PGRP-LE and consequently in absence of amidases induction, the AMPs were induced upon the germination of spores. This suggests the existence of an alternative pathway that can trigger the induction of AMPs in response to vegetative bacteria in the posterior midgut. We show that in the posterior midgut, Dif intervenes to control DptA and Defensin activation, probably in cooperation with Relish, since the absence of one of the NF-κB factors is sufficient to shut down their expression. In agreement, it has been shown that during the systemic immune response, both NF-κB factors were able to form hetero- and homodimers to differentially control AMP genes75,85,86,87. Similarly, Relish and Dif likely cooperate to activate PGRP-SC1, since its expression is reduced in either Dif or Rel loss-of-function mutants. Interestingly, Relish and Dif appears to act redundantly to control AttD expression since both have to be removed to observe its downregulation. Finally, the induction of PGRP-LB appears to be only under the control of Relish, and Dif and Relish exert opposite effects on PGRP-SC2 expression. While Rel represses its expression, Dif is required for its induction upon spore feeding. Interestingly, it has been shown that the IκB factor Pickle can bind to Relish homodimers, converting them into transcriptional repressors of AttD expression75. Therefore, a combination of NF-κB homo- and hetero-dimers, plus the presence of specific negative regulators, fine-tune the posterior immune response, limiting the level of expression of AMPs to enable commensal flora to become established, but also unfortunately allowing some opportunistic bacteria to persist. Interestingly, a role for Dif in shaping the intestinal commensal flora, downstream of the Toll pathway, has recently been uncovered88. Along with our results, this suggests that the Toll pathway is also active in the posterior midgut to contribute to the immune response against pathogens.
Together, our data shed light on the conserved behavior and strategy of Bc/Bt spores to escape the innate immune response in the proximal part of the intestine, allowing them to reach and germinate in the distal region. Our work also provides useful tools for further investigation to understand when and how enterotoxins are produced and trigger diarrheic symptoms. Our work also highlights that the persistence and load of Bc/Bt can be enhanced and could potentially lead to more severe symptoms in immunocompromised individuals.
Methods
Bacterial strains
The two bioinsecticide strains (SA-11 and ABTS-351) were used as commercial formulations. In parallel, the strain ABTS-351 was also used after bacterial isolation and “home-made” spore production as described below. The Btk ΔCry (#4D22) and Bc Bactisubtil (#6A8, also written Bactisubtyl) strains were collected from the Bacillus Genetics Stock Center (www.bgsc.org)31,89. The Bc (#ATCC 14579) was provided by ANSES Maisons-Alfort. B. toyonensis strain were selected in this work. Erwinia carotovora carotovora 15 (Ecc 15) was kindly provided by Bruno Lemaitre90. Bacterial strains were grown in LB medium at 30 °C for 16 h.
Construction of SA-11∆Cry
The mutant SA-11ΔCry was obtained from the WT strain SA-11, by a procedure of plasmid curing, as follows. After isolation on TSA-YE agar (Biomérieux, 18 h culture at 30 °C), the strain SA-11 was sub-cultured successively 3 times in 10 ml of brain-heart Infusion (BHI, Oxoid) broth at 42 °C with agitation, for 64, 48 and 36 h respectively. The first BHI culture was inoculated from an isolated colony, and the subsequent cultures were inoculated with 100 µl of the previous ones. Clones from the last culture were isolated on TSA-YE agar after serial dilution, then subcultured on the sporulating medium hydrolysate of casein tryptone (HCT) + 0.3% Glc, to select clones unable to produce crystals visible by phase contrast microscopy. The absence of plasmids carrying the cry genes was checked by sequencing. Briefly, the genomic DNA of SA-11ΔCry and SA-11 WT were extracted using the KingFisher cell and Tissue DNA kit (ThermoFisher) and sequenced with Illumina technology at the Institut du Cerveau et de la Moelle Epinière (ICM) platform, as previously described19, (NCBI accession number: SAMN23436137 and SAMN23455549, respectively). The absence of cry genes in SA-11ΔCry has been confirmed from raw reads, using KMA91. Consistently, a plasmid reconstruction made with Mob-Suite92 suggested the loss of 4 plasmids in SA-11ΔCry compared with SA-11 WT.
BtkΔCry-GFP, SA-11GFP, SA-11ΔCryGFP, BcGFP, Bc BactisubtilGFP, and B. toyonensisGFP strains
The GFP coding sequence was inserted into the pHT315 plasmid (bearing the erythromycin-resistant gene)93 (gift from Didier Lereclus). The pHT315-GFP recombinant plasmid was transfected and amplified into competent dam-/dcm- E. Coli (NEB#C2529H) which allowed it to be demethylated. pHT315-GFP was then extracted and purified using either the Pureyield plasmid miniprep kit (Promega #A1223) or the QIAGEN® Plasmid Mini Kit (QIAGEN). For the extraction using the QIAGEN® Plasmid mini Kit, the DNA solution was concentrated by isopropanol precipitation following the manufacturer’s recommendations and resuspended in PCR-grade water. The DNA concentrations were measured using the NanoDrop1000 spectrophotometer (Thermo Fisher Scientific).
The different strains from the Bc group were transfected with the pHT315-GFP plasmid as follows. Strains were plated on TSA-YE agar at room temperature for 48 h, then subcultured in 10 ml of BHI for 18 h at 30 °C, after inoculation from isolated colonies. The cultures were diluted 1/100 in 100 ml BHI and incubated at 37 °C under agitation until an OD600nm of about 0.3 was reached. Bacteria were washed in 10 ml of cold electroporation buffer (400 mM sucrose, 1 mM MgCl2, phosphate-buffered saline 1X, pH 6.8) and then resuspended in 850 µl of cold electroporation buffer. A hundred µl of each suspension was incubated with 250 ng of plasmid DNA in ice for 5 min, then submitted to electroporation using the MicroPulser Electroporator (Biorad, program Sta), and 2 mm electroporation cuvettes. After the addition of 0.9 ml of BHI, bacteria were incubated for 2 h at 37 °C and isolated on TSA-YE agar supplemented with 10 µg/ml of erythromycin. The selected clones were checked for the expression of GFP using fluorescence microscopy.
Spore production
Spores were produced as described in details in ref. 94. To summarize, strains were plated on LB-agar plates and grown overnight at 30 °C. Bacteria were grown at 30 °C in HCT-agar medium (pH 7.2) containing per 1 L: 5 g tryptone (Oxoid), 2 g casein hydrolysate (Oxoid), 15 g agar, 3 g glucose, and salts as previously reported in a sporulation-specific medium. After 10 days of incubation, spores were washed with 0.15% NaCl and heat-treated for 20 min at 70 °C. Then cells were centrifuged at 10,000×g, 8 °C for 20 min. Spores were washed with sterile deionized water and centrifuged at 10,000×g, 8 °C for 20 min. The supernatant was discarded, and the washing was repeated once. The last pellets were taken up in 10 ml and lyophilized (freeze-drying equipment model: RP2V). The numbers of spores produced were determined by estimating the CFUs on LB plates after serial dilution of lyophilized spores.
NHS-ester-547 spore labeling (sporeR/G)
Btk ΔCry-GFP/SA-11GFP/SA-11ΔCry-GFP/BcGFP/B. toyonensisGFP/Bc BactisubtilGFP spores (with known titer) were resuspended in 500 µl of sterile water and incubated 20 min at 70 °C (to remove any residual germinating or vegetative GFP cells). 10 µl of 1 mM NHS-ester-547 (Interchim #1H0880) were added and the sample was incubated for 1 h with gentle shaking at 4 °C. After centrifugation at 10,000×g, at 4 °C for 10 min, the supernatant was removed and the pellet of spores was washed with 1 ml of cold sterile water. This operation is performed twice. The final pellet was resuspended in the required volume of cold sterile water to get the desired concentration spores/100 µl.
Time-lapse fluorescence of SA-11R/G spore germination
SA-11R/G spores were placed on 1.5% agarose pads on a microscopy slide and covered with a cover glass. The use of agarose pad allowed for stabilizing spores to be achieved in the microscopy samples. The agarose pads were incubated at 37 °C for 60 min to accelerate spore germination process. The time lapse images were taken once every 5 min for 90 min to avoid bleaching. Images were acquired using the Zeiss LSM 880 microscope equipped with the AiryScan detector.
Drosophila rearing and stocks
Flies were reared on our standard laboratory medium95 in 12 h/12 h light/dark cycle-controlled incubators. We used the following stocks: WT Canton S (Bloomington #64349); w1118 (Bloomington #3605); w1118 isogenic (gift from Bruno Lemaître); RelE20 (Bloomington #55714); w; PGRP-LC ΔE (Bloomington #55713); yw; PGRP-LE112 (Bloomington #33055); w; PGRP-SC Δ (Bloomington #55724); w; PGRP-LB Δ (Bloomington #55715; gift from B. Charroux); Dredd F6496 (gift from B. Charroux); imd Shaddok (gift from B. Charroux); w; Dif ] (Bloomington #36559); Myd88 KG03447 (Bloomington #14091); ΔAMP14 (gift from B. Lemaitre)97; w; AttD-Gal4 UAS-cherry (gift from Leopold Kurz)98; w; DptA-cherry (gift from Leopold Kurz, Bloomington #55706); UAS-DUOX RNAI (Bloomington #38907); UAS-RELRNAI (Bloomington #33661); UAS-PGRP-LB RNAI (Bloomington #67236); UAS-PGRP-SC2 RNAI (Bloomington #56915); UAS-TollRNAi (Bloomington #35628); UAS-RelHA-VP16 (Bloomington #36547); UAS-PGRP-SC2 #8/CyO (gift from Heinrich Jasper)13; w; myo1A-Gal4; tubGal80ts UAS-GFP/TM6b (gift from Nicolas Tapon)99. The w; Dif 1; Rel E20 homozygous viable double mutant was obtained using classic mendelian genetic crosses. Axenic Canton S flies were obtained as described in ref. 95. It should be noted that we did not isogenized flies because we wanted to understand whether the persistence we observed was a general mechanism occurring independently of the genetic background, which is indeed the case.
Drosophila oral intoxication
Five-six-days old virgin females Drosophila were reared at 25 °C. For conditional expression of UAS-GAL80tslinked transgenes, flies were developed at room temperature, then shifted to 29 °C for 7 days to induce transgene expression. Before intoxication, Drosophila females were put into a new vial without medium for starvation for 2 h at 25 °C or at 29 °C for UAS-GAL80ts flies. This allows the synchronization of food intake once in contact with the contaminated medium. Ten females were transferred into a Drosophila narrow vial containing fly medium covered with a filter disk where the spore solution was deposited. The inoculum used for continuous and acute intoxication were respectively 106 CFU/5 cm2/fly and 108 CFU/5 cm2/fly respectively. For the acute intoxication, Drosophila were fed for 30 min with the spore inoculum, then transferred to a new sterile vial until dissection. For the continuous intoxication, Drosophila were let in contact with the spore inoculum until the dissection time. For control conditions, Drosophila females were fed with sterile deionized water in the same conditions.
Bacterial load quantification (CFU) in Drosophila midgut
Flies were washed first in ethanol 70% and then in PBS1X before guts dissection in PBS1X. Five whole midguts or 5 split parts (anterior and posterior regions) were crushed in 200 µL of LB at various times post-ingestion using a micro pestle and the homogenate was serially diluted in LB and incubated overnight at 30 °C on LB agar plates. Colony counting was performed the day after. Importantly the time point 30 min correspond to the end of the acute feeding and serve as food intake control. In dot graphs, each dot corresponds to 5 intestines. Each experiment (5 midguts/time point) was carried out at least in three independent replicates.
Bacterial load quantification (CFU) on the filter disk
The filter disk was washed and vortexed in 1 ml of sterile water. The homogenate was serially diluted in sterile water and incubated overnight at 30 °C on LB agar plates. Colony counting was performed the day after. Three independent replicates were performed.
Bacterial load quantification (CFU) in feces
After 2 h of starvation followed by 30 min of feeding with the spore inoculum, 10 flies were placed in a new vial containing 5 mL of sterile food and surrounded by filter paper. Then, flies were removed, the filter paper was washed and vortexed in 5 ml of sterile water. The homogenate was serially diluted in sterile water and incubated overnight at 30 °C on LB agar plates. The colony counting was performed the following day. All experiments were conducted at 25 °C. Three independent replicates were performed.
Bacterial load quantification (CFU) on fly body
After acute intoxication, 10 flies were transferred in a new sterile vial. Then 4, 24 or 48 h later, flies were removed from their vial and washed first 20 s in ethanol 70% and then 10 min in PBS1X. After, flies were immersed in 100 µl of LB and the LB further plated on LB agar plates over night at 30 °C. Three independent replicates were performed.730
Heat treatment
The intestinal samples or the filter disk samples were heated at 75 °C for 25 min to kill the germinating spores and the vegetative cells. Afterward, the spores were enumerated as described above.
In vivo monitoring of spore germination
Flies were fed with Btk SA-11R/G. Guts were dissected and fixed with 4% formaldehyde in PBS1X for 20 min and immediately mounted in Fluoroshield-DAPI medium. Images acquisition was performed at the microscopy platform of the Institut Sophia Agrobiotech (INRAE 1355-UCA-CNRS 7254-Sophia Antipolis) with the macroscope Zeiss AxioZoom V16 with an Apotome 2 and a Zeiss LSM 880 microscope equipped with the AiryScan detector. Images were analyzed using ZEN and Photoshop software and ImageJ.
RNA extraction and Real-time qPCR for Drosophila guts
At least three biological replicates were independently generated for each condition. Total RNA was extracted from 10 Drosophila midguts using Microelute Total RNA kit (Omega Bio Tek) and dissolved in 20 µl of RNase-free water. The quantity and quality of RNA were assessed using a Thermo Scientific™ NanoDrop 2000. 550 ng of extracted RNA was reverse transcribed to cDNA using Qscript™. Real-time PCR was performed on AriaMX Real-Time (Agilent) in a final volume of 20 µl, using the EvaGreen kit. Three technical replicates were done for each experiment. Relative expression data were normalized to RP49 and Dp1 genes and calculated according to the delta-delta Ct method100. All the results were analyzed with geNorm software. In brief, this software allows normalization of each gene expression level against a geometrical mean of the two reference genes, as well as the integration of the technical replicates and amplification efficiencies and associated errors (primer sequences are listed in Table S1).
HOCl staining with R19S
The protocol is described in ref. 38.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism v.7.00 or Microsoft Excel software (Anastats spreadsheet for Mann–Whitney’s test, http://www.anastats.fr/). Data are presented as mean and SEM. For all comparisons throughout our study, we performed two-sided unpaired Non-parametric Mann–Whitney’s test or Student’s t-tests, as specified on each figure legends. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ns = non-significant. For the t-tests (RT-qPCR), the exact P values are provided in the source data file. Diagrams and figures were produced using PowerPoint.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All source data needed to evaluate the conclusions are present in the paper as a Source Data file. Source data are provided with this paper. SAMN23436137 and SAMN23455549 sequences have been deposited at the NCBI and are freely accessible. Source data are provided with this paper.
References
Capo, F., Wilson, A. & Di Cara, F. The intestine of Drosophila melanogaster: an emerging versatile model system to study intestinal epithelial homeostasis and host-microbial interactions in humans. Microorganisms 7, microorganisms7090336 (2019).
Royet, J. & Charroux, B. Mechanisms and consequence of bacteria detection by the Drosophila gut epithelium. Gut 4, 259–263 (2013).
Kim, S. H. & Lee, W. J. Role of DUOX in gut inflammation: lessons from Drosophila model of gut-microbiota interactions. Front Infect. Microbiol 3, 116 (2014).
Benguettat, O. et al. The DH31/CGRP enteroendocrine peptide triggers intestinal contractions favoring the elimination of opportunistic bacteria. PLoS Pathog. 14, e1007279 (2018).
Bosco-Drayon, V. et al. Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe 12, 153–165 (2012).
Kaneko, T. et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 7, 715–723 (2006).
Bonfini, A., Liu, X. & Buchon, N. From pathogens to microbiota: how Drosophila intestinal stem cells react to gut microbes. Dev. Comp. Immunol. 64, 22–38 (2016).
Buchon, N., Broderick, N. A., Poidevin, M., Pradervand, S. & Lemaitre, B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5, 200–211 (2009).
Chakrabarti, S., Liehl, P., Buchon, N. & Lemaitre, B. Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe 12, 60–70 (2012).
Zhai, Z., Huang, X. & Yin, Y. Beyond immunity: the Imd pathway as a coordinator of host defense, organismal physiology and behavior. Dev. Comp. Immunol. 83, 51–59 (2018).
Costechareyre, D. et al. Tissue-specific regulation of Drosophila NF-x03BA;B pathway activation by peptidoglycan recognition protein SC. J. Innate Immun. 8, 67–80 (2016).
Paredes, J. C., Welchman, D. P., Poidevin, M. & Lemaitre, B. Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 35, 770–779 (2011).
Guo, L., Karpac, J., Tran, S. L. & Jasper, H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell 156, 109–122 (2014).
Ehling-Schulz, M., Lereclus, D. & Koehler, T. M. The Bacillus cereus group: Bacillus species with pathogenic potential. Microbiol Spectr. https://doi.org/10.1128/microbiolspec.GPP3-0032-2018. (2019).
Setlow, P. Spore resistance properties. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.TBS-0003-2012. (2014).
Jovanovic, J., Ornelis, V. F. M., Madder, A. & Rajkovic, A. Bacillus cereus food intoxication and toxicoinfection. Compr. Rev. Food Sci. Food Saf. 20, 3719–3761 (2021).
Dietrich, R., Jessberger, N., Ehling-Schulz, M., Märtlbauer, E. & Granum, P. E. The food poisoning toxins of Bacillus cereus. Toxins (Basel) 13, 98 (2021).
Santé publique France. https://www.santepubliquefrance.fr/. (2024).
Bonis, M. et al. Comparative phenotypic, genotypic and genomic analyses of Bacillus thuringiensis associated with foodborne outbreaks in France. PLoS ONE 16, e0246885 (2021).
Glasset, B. et al. Bacillus cereus-induced food-borne outbreaks in France, 2007 to 2014: epidemiology and genetic characterisation. Eur. Surveill. 21, 30413 (2016).
EFSA & ECDC. The European Union One Health 2022 Zoonoses Report. EFSA J. 21, e8442 (2023).
Berthold-Pluta, A., Pluta, A. & Garbowska, M. The effect of selected factors on the survival of Bacillus cereus in the human gastrointestinal tract. Microb. Pathog. 82, 7–14 (2015).
Clavel, T., Carlin, F., Lairon, D., Nguyen-The, C. & Schmitt, P. Survival of Bacillus cereus spores and vegetative cells in acid media simulating human stomach. J. Appl. Microbiol. 97, 214–219 (2004).
Barbosa, T. M., Serra, C. R., La Ragione, R. M., Woodward, M. J. & Henriques, A. O. Screening for bacillus isolates in the broiler gastrointestinal tract. Appl. Environ. Microbiol. 71, 968–978 (2005).
Ceuppens, S. et al. Survival and germination of Bacillus cereus spores without outgrowth or enterotoxin production during in vitro simulation of gastrointestinal transit. Appl. Environ. Microbiol. 78, 7698–7705 (2012).
Ceuppens, S. et al. Survival of Bacillus cereus vegetative cells and spores during in vitro simulation of gastric passage. J. Food Prot. 75, 690–694 (2012).
Ceuppens, S., Uyttendaele, M., Hamelink, S., Boon, N. & Van de Wiele, T. Inactivation of Bacillus cereus vegetative cells by gastric acid and bile during in vitro gastrointestinal transit. Gut Pathog. 4, 11 (2012).
Carroll, L. M., Cheng, R. A., Wiedmann, M. & Kovac, J. Keeping up with the Bacillus cereus group: taxonomy through the genomics era and beyond. Crit. Rev. Food Sci. Nutr. 62, 1–26 (2021).
Carroll, L. M., Wiedmann, M. & Kovac, J. Proposal of a taxonomic nomenclature for the bacillus cereus group which reconciles genomic definitions of bacterial species with clinical and industrial phenotypes. mBio 11, e00034–20 (2020).
Ivanova, N. et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423, 87–91 (2003).
Hoa, N. T. et al. Characterization of Bacillus species used for oral bacteriotherapy and bacterioprophylaxis of gastrointestinal disorders. Appl. Environ. Microbiol. 66, 5241–5247 (2000).
Johler, S. et al. Enterotoxin production of Bacillus thuringiensis isolates from biopesticides, foods, and outbreaks. Front. Microbiol. 9, 1915 (2018).
Biggel, M. et al. Whole genome sequencing reveals biopesticidal origin of Bacillus thuringiensis in foods. Front. Microbiol. 12, 775669 (2021).
Buchon, N. et al. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 3, 1725–1738 (2013).
Marianes, A. & Spradling, A. C. Physiological and stem cell compartmentalization within the Drosophila midgut. Elife 2, e00886 (2013).
Lee, K. A. et al. Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153, 797–811 (2013).
Tzou, P. et al. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13, 737–748 (2000).
Hachfi, S., Benguettat, O. & Gallet, A. Hypochlorous acid staining with R19-S in the Drosophila intestine upon ingestion of opportunistic bacteria. Bio-Protoc. 9, e3246 (2019).
Chen, X. et al. A specific and sensitive method for detection of hypochlorous acid for the imaging of microbe-induced HOCl production. Chem. Commun. (Camb.) 47, 4373–4375 (2011).
Charroux, B. et al. Cytosolic and secreted peptidoglycan-degrading enzymes in Drosophila respectively control local and systemic immune responses to microbiota. Cell Host Microbe 23, 215–228.e4 (2018).
Hanson, M. A. et al. Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. eLife 8, e44341 (2019).
Carboni, A. L., Hanson, M. A., Lindsay, S. A., Wasserman, S. A. & Lemaitre, B. Cecropins contribute to Drosophila host defense against a subset of fungal and Gram-negative bacterial infection. Genetics 220, iyab188 (2022).
Hedengren, M. et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell 4, 827–837 (1999).
Vodovar, N. et al. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl Acad. Sci. USA 102, 11414–11419 (2005).
Zhai, Z., Boquete, J. P. & Lemaitre, B. Cell-specific Imd-NF-kappaB responses enable simultaneous antibacterial immunity and intestinal epithelial cell shedding upon bacterial infection. Immunity 48, 897-910.e7 (2018).
Petersen, U. M., Björklund, G., Ip, Y. T. & Engström, Y. The dorsal-related immunity factor, Dif, is a sequence-specific trans-activator of Drosophila Cecropin gene expression. EMBO J. 14, 3146–3158 (1995).
Manfruelli, P., Reichhart, J. M., Steward, R., Hoffmann, J. A. & Lemaitre, B. A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. EMBO J. 18, 3380–3391 (1999).
Rosetto, M., Engström, Y., Baldari, C. T., Telford, J. L. & Hultmark, D. Signals from the IL-1 receptor homolog, Toll, can activate an immune response in a Drosophila hemocyte cell line. Biochem Biophys. Res. Commun. 209, 111–116 (1995).
Meng, X., Khanuja, B. S. & Ip, Y. T. Toll receptor-mediated Drosophila immune response requires Dif, an NF-kappaB factor. Genes Dev. 13, 792–797 (1999).
Tauszig-Delamasure, S., Bilak, H., Capovilla, M., Hoffmann, J. A. & Imler, J. L. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat. Immunol. 3, 91–97 (2002).
Horng, T. & Medzhitov, R. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc. Natl Acad. Sci. USA 98, 12654–12658 (2001).
Rolny, I. S., Minnaard, J., Racedo, S. M. & Pérez, P. F. Murine model of Bacillus cereus gastrointestinal infection. J. Med. Microbiol. 63, 1741–1749 (2014).
Loudhaief, R. et al. Apoptosis restores cellular density by eliminating a physiologically or genetically induced excess of enterocytes in the Drosophila midgut. Development 144, 808–819 (2017).
EFSA BIOHAZ. Risks for public health related to the presence of Bacillus cereus and other Bacillus spp. including Bacillus thuringiensis in foodstuffs. EFSA J. 14, e04524 (2016).
Wilcks, A. et al. Germination and conjugation of Bacillus thuringiensis subsp. israelensis in the intestine of gnotobiotic rats. J. Appl. Microbiol. 104, 1252–1259 (2008).
Oliveira-Filho, E. C. et al. Toxicity assessment and clearance of Brazilian microbial pest control agents in mice. Bull. Environ. Contam. Toxicol. 83, 570–574 (2009).
Jensen, G. B. et al. Bacillus thuringiensis in fecal samples from greenhouse workers after exposure to B. thuringiensis-based pesticides. Appl. Environ. Microbiol. 68, 4900–4905 (2002).
Casula, G. & Cutting, S. M. Bacillus probiotics: spore germination in the gastrointestinal tract. Appl. Environ. Microbiol. 68, 2344–2352 (2002).
Tam, N. K. et al. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 188, 2692–2700 (2006).
Osman, D. et al. Autocrine and paracrine unpaired signaling regulate intestinal stem cell maintenance and division. J. Cell Sci. 125, 5944–5949 (2012).
Shanbhag, S. & Tripathi, S. Epithelial ultrastructure and cellular mechanisms of acid and base transport in the Drosophila midgut. J. Exp. Biol. 212, 1731–1744 (2009).
Duport, C., Zigha, A., Rosenfeld, E. & Schmitt, P. Control of enterotoxin gene expression in Bacillus cereus F4430/73 involves the redox-sensitive ResDE signal transduction system. J. Bacteriol. 188, 6640–6651 (2006).
Duport, C., Thomassin, S., Bourel, G. & Schmitt, P. Anaerobiosis and low specific growth rates enhance hemolysin BL production by Bacillus cereus F4430/73. Arch. Microbiol. 182, 90–95 (2004).
Zigha, A., Rosenfeld, E., Schmitt, P. & Duport, C. Anaerobic cells of Bacillus cereus F4430/73 respond to low oxidoreduction potential by metabolic readjustments and activation of enterotoxin expression. Arch. Microbiol. 185, 222–233 (2006).
van der Voort, M. & Abee, T. Transcriptional regulation of metabolic pathways, alternative respiration and enterotoxin genes in anaerobic growth of Bacillus cereus ATCC 14579. J. Appl. Microbiol. 107, 795–804 (2009).
Miller, B. M., Liou, M. J., Lee, J. Y. & Bäumler, A. J. The longitudinal and cross-sectional heterogeneity of the intestinal microbiota. Curr. Opin. Microbiol. 63, 221–230 (2021).
Du, E. J. et al. TrpA1 regulates defecation of food-borne pathogens under the control of the duox pathway. PLoS Genet. 12, e1005773 (2016).
Lee, K. A. et al. Bacterial uracil modulates Drosophila DUOX-dependent gut immunity via hedgehog-induced signaling endosomes. Cell Host Microbe 17, 191–204 (2015).
Tamez-Guerra, P. et al. Detection of genes encoding antimicrobial peptides in Mexican strains of Trichoplusia ni (Hübner) exposed to Bacillus thuringiensis. J. Invertebr. Pathol. 98, 218–227 (2008).
Li, S. et al. Bacillus thuringiensis suppresses the humoral immune system to overcome defense mechanism of plutella xylostella. Front. Physiol. 9, 1478 (2018).
Neyen, C., Poidevin, M., Roussel, A. & Lemaitre, B. Tissue- and ligand-specific sensing of gram-negative infection in drosophila by PGRP-LC isoforms and PGRP-LE. J. Immunol. 189, 1886–1897 (2012).
Onuma, T. et al. Recognition of commensal bacterial peptidoglycans defines Drosophila gut homeostasis and lifespan. PLoS Genet. 19, e1010709 (2023).
Rodgers, F. H. et al. Functional analysis of the three major PGRPLC isoforms in the midgut of the malaria mosquito Anopheles coluzzii. Insect Biochem. Mol. Biol. 118, 103288 (2020).
Bonnay, F. et al. big bang gene modulates gut immune tolerance in Drosophila. Proc. Natl Acad. Sci. USA 110, 2957–2962 (2013).
Morris, O. et al. Signal integration by the IκB protein pickle shapes drosophila innate host defense. Cell Host Microbe 20, 283–295 (2016).
Ryu, J. H. et al. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782 (2008).
Apidianakis, Y., Pitsouli, C., Perrimon, N. & Rahme, L. Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc. Natl Acad. Sci. USA 106, 20883–20888 (2009).
Jiang, H. et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 (2009).
Zhou, F., Rasmussen, A., Lee, S. & Agaisse, H. The UPD3 cytokine couples environmental challenge and intestinal stem cell division through modulation of JAK/STAT signaling in the stem cell microenvironment. Dev. Biol. 373, 383–393 (2013).
Tamamouna, V. et al. Evidence of two types of balance between stem cell mitosis and enterocyte nucleus growth in the Drosophila midgut. Development 147, dev189472 (2020).
Ferguson, M. & Foley, E. Microbial recognition regulates intestinal epithelial growth in homeostasis and disease. FEBS J. 289, 3666–3691 (2022).
Broderick, N. A., Buchon, N. & Lemaitre, B. Microbiota-induced changes in drosophila melanogaster host gene expression and gut morphology. MBio 5, e01117–14 (2014).
Buchon, N., Broderick, N. A., Chakrabarti, S. & Lemaitre, B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23, 2333–2344 (2009).
Cronin, S. J. et al. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325, 340–343 (2009).
Han, Z. S. & Ip, Y. T. Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. J. Biol. Chem. 274, 21355–21361 (1999).
Tanji, T., Yun, E. Y. & Ip, Y. T. Heterodimers of NF-kappaB transcription factors DIF and Relish regulate antimicrobial peptide genes in Drosophila. Proc. Natl Acad. Sci. USA 107, 14715–14720 (2010).
Chowdhury, M. et al. An in vitro study of NF-κB factors cooperatively in regulation of Drosophila melanogaster antimicrobial peptide genes. Dev. Comp. Immunol. 95, 50–58 (2019).
Bahuguna, S. et al. Bacterial recognition by PGRP-SA and downstream signalling by Toll/DIF sustain commensal gut bacteria in Drosophila. PLoS Genet. 18, e1009992 (2022).
Gonzalez, J. M. Jr., Brown, B. J. & Carlton, B. C. Transfer of Bacillus thuringiensis plasmids coding for delta-endotoxin among strains of B. thuringiensis and B. cereus. Proc. Natl Acad. Sci. USA 79, 6951–6955 (1982).
Basset, A. et al. The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl Acad. Sci. USA 97, 3376–3381 (2000).
Clausen, P., Aarestrup, F. M. & Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 19, 307 (2018).
Robertson, J. & Nash, J. H. E. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 4, e000206 (2018).
Theoduloz, C., Vega, A., Salazar, M., González, E. & Meza-Basso, L. Expression of a Bacillus thuringiensisdelta-endotoxin cry1Ab gene in Bacillus subtilis and Bacillus licheniformis strains that naturally colonize the phylloplane of tomato plants (Lycopersicon esculentum, Mills). J. Appl. Microbiol. 94, 375–381 (2003).
Fichant, A. et al. New approach methods to assess the enteropathogenic potential of strains of the Bacillus cereus group, including Bacillus thuringiensis. Foods 13, 1140 (2024).
Nawrot-Esposito, M. P. et al. Bacillus thuringiensis bioinsecticides induce developmental defects in non-target Drosophila melanogaster larvae. Insects 11, 697 (2020).
Leulier, F., Rodriguez, A., Khush, R. S., Abrams, J. M. & Lemaitre, B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 1, 353–358 (2000).
Carboni, A. L., Hanson, M. A., Lindsay, S. A., Wasserman, S. A. & Lemaitre, B. Cecropins contribute to Drosophila host defense against a subset of fungal and Gram-negative bacterial infection. Genetics 220, iyab188 (2022).
Tavignot, R., Chaduli, D., Djitte, F., Charroux, B. & Royet, J. Inhibition of a NF-κB/Diap1 pathway by PGRP-LF is required for proper apoptosis during drosophila development. PLoS Genet. 13, e1006569 (2017).
Shaw, R. L. et al. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development 137, 4147–4158 (2010).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Acknowledgements
We would like to thank Bernard Charroux, François Leulier, Julien Royet, Heinrich Jasper, Leopold Kurz, Mark Hanson, Samuel Rommelaere, and Bruno Lemaitre for kindly providing fly stocks. We also thank Didier Lereclus for providing the pHT315 plasmid. We are grateful to Leanne Jones, Raphaël Rousset, Carmelo Luci, and Bernard Charroux for their advice on the project and the manuscript. We especially thank Bruno Lemaitre for the critical reading of the manuscript. We also thank Olivier Pierre and Anne Doye for their help with the microscopy, and Alexia Danné, Audrey Amate, and Olivia Benguettat for their technical help. Our thanks to the Université Côte d’Azur Office of International Scientific Visibility for English language editing of the manuscript. We also thank the Space, Environment, Risk and Resilience Academy of the Université Côte d’Azur for their financial support. This work has been supported by the French government, through the UCAJEDI Investments in the Future project managed by the National Research Agency (ANR) with the reference number ANR-15-IDEX-01 to A.G. and L.B., and through the ANR-13-CESA-0003-01 (ImBio) and the ANR-22-CE35-0006-01 (BaDAss) to A.G. This work has also been supported by the Plan ECOPHYTO II+ Axe 3 - Action 11 under the N°OFB.21.0450 to M.B., by the “Fondation ARC pour la recherche sur le cancer” (20171206145) and by the “Institut Olga Triballat” (AAP2021; institut-olgatriballat.org) to R.Ro., and by the Anses (N°ANSES-22-EST-203) to R.Ru.; A.F. was funded by a PhD grant from INRAE and Anses.
Author information
Authors and Affiliations
Contributions
Conceptualization: S.H., L.B. and A.G. Methodology: S.H., A.B.-B., A.F., P.M., M.-P.N.-E., G.M. and M.B. Validation: S.H., A.B.-B., P.M., M.B., L.B. and A.G. Formal Analysis: S.H., A.B.-B., P.M., L.B. and A.G. Investigation: S.H., A.B.-B., A.F., P.M., M.-P.N.-E. and M.B. Data Curation: S.H., A.B.-B., A.F., P.M., M.B., L.B. and A.G. Writing – Original Draft: S.H. and A.G. Writing – Review & Editing: S.H., A.B.-B., R.Ro., M.B., L.B. and A.G. Visualization: S.H., L.B. and A.G. Supervision: L.B and A.G. Project Administration: L.B. and A.G. Funding Acquisition: M.B., R.Ru., R.Ro., L.B. and A.G.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Fengliang Jin, Yoshitomo Kikuchi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hachfi, S., Brun-Barale, A., Fichant, A. et al. Ingestion of Bacillus cereus spores dampens the immune response to favor bacterial persistence. Nat Commun 15, 7733 (2024). https://doi.org/10.1038/s41467-024-51689-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-51689-9