Abstract
CD5 antigen-like (CD5L), also known as Spα or AIM (Apoptosis inhibitor of macrophage), emerges as an integral component of serum immunoglobulin M (IgM). However, the molecular mechanism underlying the interaction between IgM and CD5L has remained elusive. In this study, we present a cryo-electron microscopy structure of the human IgM pentamer core in complex with CD5L. Our findings reveal that CD5L binds to the joining chain (J chain) in a Ca2+-dependent manner and further links to IgM via a disulfide bond. We further corroborate recently published data that CD5L reduces IgM binding to the mucosal transport receptor pIgR, but does not impact the binding of the IgM-specific receptor FcμR. Additionally, CD5L does not interfere with IgM-mediated complement activation. These results offer a more comprehensive understanding of IgM and shed light on the function of the J chain in the immune system.
Similar content being viewed by others
Introduction
Immunoglobulin M (IgM) plays a critical role in both humoral and mucosal immunity. In serum, IgM is primarily found in its pentameric form, consisting of five IgM monomers linked together by the joining chain (J chain). The J chain is also essential for the transportation of the IgM pentamer to the mucosal surface, and achieves this by interacting with the polymeric immunoglobulin receptor (pIgR), the extracellular region of which becomes an integral subunit of secretory IgM known as the secretory component (SC).
The discovery of IgM can be traced back to 1937 when Heidelberger and Pedersen observed that horses immunized with pneumococcus polysaccharide produced antibodies with a remarkably large molecular weight1. Despite its long history, our understanding of IgM remains incomplete. The presence of an additional IgM-associated protein in human serum IgM, apart from the J chain, was initially detected by Tissot et al. using two-dimensional polyacrylamide gel electrophoresis in 1993–19942,3. Gebe et al. cloned and characterized human Spα in 1997 as a macrophage-secreted protein4, which belongs to the scavenger receptor cysteine-rich (SRCR) superfamily. Miyazaki et al. subsequently identified mouse AIM (Apoptosis inhibitor of macrophage) in 1999 and found that it can promote the survival of thymocytes against apoptosis5. It was soon realized that AIM is the mouse homolog of human Spα6. In 2002, it was confirmed that human Spα/AIM, also known as CD5 antigen-like or CD5L, corresponds to the additional protein found in IgM7. Recently, Oskam et al. performed state-of-the-art mass spectrometry analyses and demonstrated that CD5L/Spα/AIM is an integral subunit of human circulatory IgM, leading to the redefinition of the predominant form of circulatory IgM as a J-chain-linked IgM pentamer plus one CD5L [(IgM)5:(J)1:(CD5L)1]8. For consistency with this study and in compliance with UniProtKB recommendation, we will primarily refer to CD5L/Spα/AIM as CD5L in this study.
In the past two decades, a wide range of functions of CD5L have been documented9,10. CD5L is believed to act as a pattern recognition receptor (PRR) that recognizes a broad range of microbial pathogens, including both bacterial and fungal cells11,12,13,14, as well as endogenous harmful substances such as cell debris15,16,17. In addition to its role in inhibiting the apoptosis of thymocytes, CD5L has been shown to suppress the apoptosis of T cells and natural killer T cells in mice18, as well as hepatocytes after hepatic ischaemia-reperfusion (I/R) injury19. CD5L also regulates lipid biosynthesis and the pathogenicity of T helper 17 cells20. Furthermore, CD5L has been implicated in diseases such as obesity, atherosclerosis, and IgA nephropathy21,22,23. Recently, Oliveira et al. reported that intravenous administration of recombinant CD5L protein significantly reduces sepsis-related mortality in mice, suggesting that CD5L could serve as a potential biological therapeutic for treating sepsis24. Although the molecular mechanism of CD5L function remains elusive, it is clear that it plays an important role in immune homeostasis and diseases.
The IgM pentamer is believed to serve as a carrier for CD5L, shielding it from renal excretion. However, the molecular basis of the IgM–CD5L interaction remains unclear, and the regulatory role of CD5L in IgM function remains to be further investigated. In this study, we present a cryo-electron microscopy structure of the IgM-Fc pentamer core in complex with CD5L (Fcμ–J–CD5L), shedding light on the organizational principles of the IgM–CD5L complex. Furthermore, we demonstrate that CD5L diminishes IgM binding to pIgR while leaving the binding of the IgM-specific receptor FcμR unaffected. Additionally, we show that CD5L has no impact on IgM-mediated complement activation. These results support recent findings by Oskam et al.8 and offer a more comprehensive understanding of human IgM.
Results
CD5L binds to IgM in a Ca2+-dependent manner
The SRCR proteins are characterized by one or multiple repeats of highly conserved SRCR domains25,26. Structural analyses have shown that SRCR domains commonly feature a dual-cation-binding site that plays a crucial role in ligand binding27. To biochemically characterize the IgM–CD5L interaction and explore its dependence on divalent cations such as Ca2+, which is typically found in serum (at levels of 1.3–1.5 mM in healthy human blood), we generated CD5L, as well as Fcμ–J, the pentameric IgM core comprising an IgM-Fc pentamer and the J chain, using HEK293F cells (Supplementary Fig. 1a). We then conducted surface plasmon resonance (SPR) experiments to evaluate the binding affinity of CD5L with Fcμ–J. Fcμ–J was first immobilized on a CM5 sensor chip using the amine coupling technique, with CD5L serving as the analyte. The results revealed that CD5L binds to immobilized Fcμ–J in the presence of Ca2+, but not in its absence (Supplementary Fig. 2). To mitigate potential issues linked to amine coupling, we also employed the twin-strep tag present at the N-terminus of Fcμ and immobilized Fcμ–J on the streptavidin (SA) sensor chip. Subsequent SPR measurements showed that CD5L binds to the SA-coupled Fcμ–J with a Kd of 18.1 ± 3.8 nM (Fig. 1a), which is comparable to the results obtained from the CM5 immobilization method (Table 1). Bovine serum albumin (BSA) was used as a negative control, and no binding interaction was observed (Supplementary Fig. 2).
a Human CD5L binds to immobilized human Fcμ–J with a Kd of 18.1 ± 3.8 nM in the presence of Ca2+. Fcμ–J was immobilized on a SA chip the twin-strep tag at N-terminus of Fcμ–J, and serial dilutions of human CD5L (from 200 to 12.5 nM) were flowed over the chip. SPR data were fitted to a 1:1 binding model using Biacore Evaluation Software, with each experiment repeated at least twice. The colored curve represents experimental data, while the black curves depict the fitting results. b Cat CD5L binds to immobilized cat Fcμ–J in the presence of Ca2+. Serial dilutions of cat CD5L (200–12.5 nM) were flowed over immobilized cat Fcμ–J on an SA chip for the SPR measurement. c Mouse CD5L binds to immobilized mouse Fcμ–J in the presence of Ca2+. Immobilized mouse Fcμ–J on an SA chip and serial dilutions of mouse CD5L (200–12.5 nM) were utilized for the measurement.
CD5L is highly conserved in mammals (Supplementary Fig. 3a). Previous research has indicated that cat CD5L binds to cat IgM with a 1000-fold greater affinity compared to mouse CD5L and IgM28, and this strong IgM–CD5L binding in cats was suggested to contribute to their increased susceptibility to renal disease10. However, our measurements indicate that both cat and mouse CD5L exhibit similar affinities for their respective Fcμ–J in the presence of Ca2+. The Kd values on the SA sensor chip were 14.4 ± 8.4 nM for cat CD5L-Fcμ–J and 36.1 ± 10.3 nM for mouse CD5L-Fcμ–J (Fig. 1b, c; Table 1). Notably, measurements using the CM5 immobilization method produced similar results, and no binding occurred in the absence of Ca2+ (Supplementary Fig. 2). Collectively, these results suggest that CD5L binds to IgM in a Ca2+-dependent manner and that this interaction is evolutionarily conserved among mammals.
Cryo-EM structure of human Fcμ–J–CD5L
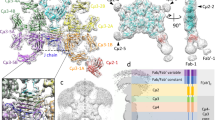
The core region of the IgM pentamer displays an asymmetrical arrangement that resembles a hexagon with a missing piece29,30,31,32,33. Negative-stain electron microscopy images indicate that CD5L binds to the gap of the IgM pentamer29. To reveal the molecular basis of IgM–CD5L interaction, we incubated human Fcμ–J complex and CD5L together to assemble the Fcμ–J–CD5L complex (Supplementary Fig. 1b), and then analyzed it using cryo-electron microscopy (cryo-EM). The overall structure was determined at a resolution of 3.4 Å (Fig. 2a, Supplementary Fig. 4, Supplementary Table 1), and revealed an asymmetrical arrangement of five Fcμ molecules with a gap occupied by the J chain and CD5L, in agreement with previous findings. The SRCR3 domain of CD5L binds to the center of Fcμ–J and interacts with several regions of the J chain. SRCR2 swings towards Fcμ5B, and presses the β5–β6 hairpin of the J chain onto Fcμ5B. When viewed from the side, the SRCR2–SRCR3 module was found to be non-uniformly distributed within the Fcμ–J plane, concentrating more on one side. This further accentuates the asymmetrical nature of the core region of IgM. Weak densities were observed for the SRCR1 domain of CD5L, as well as the Cμ2 domains of Fcμ, preventing clear visualization of these regions.
a The cryo-EM structure of the human Fcμ–J–CD5L complex is depicted in two views. Five Fcμ chains are shown in teal, and five are shown in pale cyan. The J chain, CD5L and Ca2+ are shown in light coral, gold and green, respectively. Consistent color schemes are applied across all figures unless specified otherwise. b Interactions between the SRCR2–SRCR3 junction of CD5L and the β5–β6 hairpin of the J chain. Polar interactions are denoted by dashed lines, whereas amino acids highlighted in blue boxes indicate CD5L sites mutated in this study. c Interactions between the SRCR3 domain of CD5L and the J chain. Ca2+-coordinating interactions and polar interactions involving D271, E339, and K107J are denoted with dashed lines. d Interactions between the SRCR2 domain of CD5L and the Cμ3 of Fcμ5B. A disulfide bond forms between C191S and C414Fcμ5B.
Local refinement was performed to enhance the EM densities surrounding CD5L and the Fcμ–J–CD5L interface (Supplementary Fig. 4, Supplementary Table 1). Both the SRCR2 and SRCR3 domains of CD5L were found to be crucial in binding to the J chain, particularly the β5–β6 hairpin of the J chain (Fig. 2b). It is worth noting that this hairpin structure, observable in IgA structures34,35,36, was disordered in all previous IgM structures without CD5L. In SRCR2, K173 and R183 form salt bridges with E77J and E75J (J chain residues are denoted by subscripts), respectively; whereas V185–T187 interact with N80J–I82J through hydrophobic and van der Waals interactions. In SRCR3, R328 forms a salt bridge with D72J, while F329 and F332 form hydrophobic interactions with P73J.
Further interactions have been observed between SRCR3 and the C-terminal region of the J chain. Sequence and structural analyses indicate that SRCR3 contains a complete dual-cation-binding site, while SRCR2 has a degenerate one27 (Supplementary Fig. 3). The cryo-EM density map is not inconsistent with the existence of two Ca2+ ions in SRCR3, coordinated by D270, D271, D311, D334, and E339 (Supplementary Fig. 4g, h). The presence of Ca2+ likely plays a role in stabilizing the negatively charged surface patch in SRCR3, as circular dichroism (CD) spectroscopy analyses indicate that Ca2+ does not induce a global conformational change in CD5L (Supplementary Fig. 5). Notably, a pocket between these two Ca2+-binding sites cradles Lys107J (Fig. 2c). In addition, F295 in SRCR3 interacts with P135J.
CD5L also directly interacts with Fcμ. H230 and D231 from SRCR2 were involved in making polar contacts with Fcμ5B. Furthermore, a disulfide bond was observed between Cys191 in SRCR2 and Cys414 in the Cμ3 domain of Fcμ5B (Fig. 2d, Supplementary Fig. 4k), consistent with previous analyses8,29. Indeed, the presence of a disulfide linkage between CD5L and Fcμ–J was confirmed by non-reducing SDS-PAGE (Supplementary Fig. 6a).
Interaction between CD5L mutants and Fcμ–J
In order to confirm the functional importance of these molecular interactions, we engineered CD5L mutants and conducted pull-down experiments. Consistent with the findings of the SPR analyses (Fig. 1, Supplementary Fig. 2), wild-type (WT) CD5L specifically binds to Fcμ–J in the presence of Ca2+, and this binding is disrupted in the presence of EDTA (Fig. 3a, b). Mutations of the putative Ca2+-binding sites in SRCR1 (D50A/D51A and E118A) did not affect the interaction between CD5L and Fcμ–J (Fig. 3b), supporting the cryo-EM structure that SRCR1 is not involved in the binding interface. On the other hand, mutations in the cation-binding sites of SRCR3 (D270A/D271A, E339A, and D270A/D271A/E339A) led to a complete loss of interaction with Fcμ–J. Furthermore, mutations (R183A/V185N and F329N/G331S) that introduced N-linked glycans at positions 185 (in SRCR2) and 329 (in SRCR3) in CD5L also significantly reduced the interaction with Fcμ–J (Fig. 3c); since the presence of these bulky glycans would interfere with the interaction between CD5L and the Jchain β5–β6 hairpin (Fig. 2b). SPR experiments further corroborated these results, demonstrating that both D270A/D271A/E339A and R183A/V185N mutants do not bind to Fcμ–J (Supplementary Fig. 6b, c).
a Lack of Ca2+ results in no binding between CD5L and Fcμ–J in a pull-down assay. This experiment has been repeated at least three times. b Mutations in putative Ca2+-binding sites in SRCR1 (D50A/D51A and E118A) do not influence CD5L-Fcμ–J interaction. However, mutations in SRCR3’s cation-binding sites (D270A/D271A, E339A, and D270A/D271A/E339A) abolish interaction with Fcμ–J in the presence of Ca2+. This experiment has been repeated at least three times. c The R183A/V185N and F329N/G331S mutations disrupt the interaction with Fcμ–J, whereas C191S does not. This experiment has been repeated at least three times. d C191S (green) is capable of forming a complex with Fcμ–J similar to the wild-type (orange) in size-exclusion chromatography. SDS–PAGE analysis of the peak fractions from size-exclusion chromatography experiments is presented on the right. This experiment has been repeated at least three times. e SPR analyses of the interaction between human Fcμ–J and the C191S mutant in the presence of Ca2+.
To our surprise, the mutation of C191 to Ser (C191S) in CD5L did not significantly impact the binding between CD5L and Fcμ–J (Fig. 3c). In fact, C191S is able to form a complex with Fcμ–J on size-exclusion chromatography, akin to the WT CD5L (Fig. 3d). Furthermore, SPR analyses indicated that C191S binds to Fcμ–J with a Kd of 13.2 ± 2.2 nM (Fig. 3e, Table 1), a value similar to that of the wild-type CD5L. These findings suggest that a noncovalent association plays a crucial role in the formation of the IgM–CD5L complex. The disulfide bond involving CD5L and Fcμ5B likely enhances the stability of the fully formed IgM–CD5L complex in circulation, akin to the role of the disulfide bond between pIgR-SC and IgA in secretory IgA.
CD5L reduces IgM binding to pIgR but not to FcμR
The integration of CD5L as a subunit of IgM prompts an inquiry into its impact on IgM function. To address this, we started by investigating the influence of CD5L on the interaction between the IgM pentamer and the specific IgM Fc receptor, FcμR37. Surface plasmon resonance (SPR) experiments revealed that Fcμ–J–CD5L and Fcμ–J exhibit similar high-affinity interactions with the D1 domain of FcμR (FcμR-D1), with apparent Kd values of 0.4 ± 0.04 and 0.2 ± 0.1 nM, respectively (Fig. 4a, b; Table 1). This suggests that the presence of CD5L does not significantly affect the interaction between IgM and FcμR. Structural analyses further support this conclusion, indicating that CD5L is unlikely to impede the recruitment of FcμR-D1 by IgM (Fig. 4c). Similar conclusions were drawn from SPR experiments conducted in the reverse manner, where Fcμ–J–CD5L and Fcμ–J were immobilized on the SPR chip, albeit with lower apparent affinities (Kd values of 8.8 ± 0.3 and 7.4 ± 0.4 nM, Supplementary Fig. 7a, Table 1). The observed affinity differences are likely due to avidity effects, as a single Fcμ–J can engage with up to four FcμR-D1 molecules on either side38. When FcμR-D1 is anchored to the SPR chip, Fcμ–J in the aqueous solution can participate in a multivalent interaction with FcμR, resulting in higher avidity and, higher apparent binding affinity. In contrast, FcμR-D1 in the mobile phase can only bind to the Fcμ–J in a monovalent manner, leading to an apparent binding affinity that reflects the average binding affinity of all the individual binding sites.
a and b SPR analyses of the interactions between immobilized FcμR-D1 and Fcμ–J or Fcμ–J–CD5L. Twofold serial dilutions of Fcμ–J–CD5L or Fcμ–J (both from 40 to 2.5 nM) were passed through immobilized FcμR-D1 on the CM5 chip. c Structural insights indicate that CD5L is unlikely to disrupt the IgM–FcμR interaction. Teal, light coral, and gold represent Fcμ chains, J chain, and CD5L of the Fcμ–J–CD5L complex, while the grey and purple illustrate Fcμ–J and FcμR-D1 from the human Fcμ–J–FcμR-D1 complex (PDB ID: 7YTD), respectively. d and e SPR analyses of the interactions between immobilized pIgR-SC and Fcμ–J or Fcμ–J–CD5L. Twofold serial dilutions of Fcμ–J–CD5L (from 80 to 5 nM) or Fcμ–J (from 320 to 20 nM) were passed through biotinylated pIgR-SC immobilized on a SA chip. f Structural analyses suggest that CD5L hinders the IgM–pIgR interaction. Fcμ–J and pIgR of the human Fcμ–J–pIgR complex (PDB ID: 6KXS) are shown in grey and blue, respectively.
We were unable to measure the binding between IgM and FcαμR, the Fc receptor for both IgM and IgA39, due to challenges in producing well-behaved FcαμR protein. We then evaluated how CD5L influences the binding between IgM and pIgR. Fcμ–J–CD5L associates with the C-terminally anchored pIgR-SC (the extracellular region of pIgR) with substantially decreased affinity when compared to the interaction between Fcμ–J and pIgR-SC (Fig. 4d, e; Table 1). Consistently, a reduced binding affinity between pIgR-SC and Fcμ–J in the presence of CD5L was also observed when pIgR-SC was used in the mobile phase (Supplementary Fig. 7b). A structural comparison between Fcμ–J–CD5L and Fcμ–J–pIgR-SC revealed that CD5L could substantially collide with the D3–D5 domains of pIgR-SC (Fig. 4f). It should be noted that only the D1 domain of pIgR-SC is involved in binding to Fcμ–J, and the five domains of pIgR-SC can undergo substantial conformational changes31. Therefore, despite the steric hindrance posed by CD5L on the D3–D5 domains of pIgR-SC, conformational changes in pIgR-SC may allow its D1 domain to be more exposed for interaction with Fcμ–J. This provides a rationale for the observed low-affinity binding between the Fcμ–J–CD5L complex and pIgR-SC. Nevertheless, the data clearly indicate that the presence of CD5L significantly impairs the binding of Fcμ–J to pIgR-SC.
CD5L does not interfere with IgM-mediated complement activation
IgM is capable of recruiting the C1q complex and initiating the classical complement pathway. The C1q-binding site is situated in the Cμ3 domain of Fcμ and includes residues 432–436 in the FG loop40, which does not participate in interacting with CD5L. To assess the effect of CD5L on IgM-mediated complement activation, we generated a recombinant anti-CD20 IgM molecule following previously established methods41. Subsequently, we conducted complement-dependent cytotoxicity experiments using OCI-Ly10 cells, which naturally express CD20. The results revealed that in the presence of human serum complement, the anti-CD20 IgM effectively induced the lysis of OCI-Ly10 cells (Fig. 5). Preincubation of this IgM with increasing amounts of CD5L did not show any significant effect on complement activation. In contrast, VAR2CSA, a malaria protein known for its potent inhibition of IgM-mediated complement cytotoxicity in this context41, effectively suppressed IgM-triggered cell killing. These findings suggest that CD5L does not exert a substantial influence on IgM-mediated complement activation.
IgM-mediated complement activation was assessed using OCI-Ly10 cells, which naturally express CD20. Serially diluted CD5L (ranging from 320 to 10 nM) or the malaria protein VAR2CSA was incubated with anti-CD20 IgM (at 6 nM) for 3 h. Subsequently, the protein mixtures were combined with normal human serum complement and applied to the OCI-Ly10 cultures. Cell viability was determined by measuring ATP levels using the CellTiter-Glo reagent and was shown by dots. For comparison, the statistical analysis was performed using an unpaired Student’s t-test with 95% confidence intervals and described statistical significance with two-sided p-values in GraphPad Prism. * Indicates statistical significance at p ≤ 0.0001. Plots indicate the mean (top line of box) and standard deviation (error bars).
Discussion
In this study, we have elucidated the molecular mechanism of the IgM–CD5L interaction. Our findings indicate that CD5L primarily engages with the J chain in a Ca2+-dependent manner, and the formation of a disulfide bond between CD5L and Fcμ further stabilizes the IgM–CD5L complex. Moreover, we have demonstrated that CD5L significantly reduces IgM binding to pIgR but does not affect the binding of FcμR or complement activation. By impeding the interaction between IgM and pIgR, CD5L likely hinders the mucosal transcytosis of IgM, thereby contributing to the retainment of IgM within the bloodstream. Collectively, these results corroborate recent findings by Oskam et al. and support the notion that CD5L is an integral component of circulatory IgM8.
Despite these advancements, the precise function of CD5L remains elusive. It is hypothesized that when bound to IgM, CD5L is shielded from renal elimination but appears to be functionally inactive10. Upon disruption of immune homeostasis, CD5L dissociates from IgM and plays a role in immune regulation9,15,16. Our results suggest that CD5L dissociation would require conditions that promote the disruption of the disulfide bond, and possibly a decrease in Ca2+ concentration. Deciphering the molecular events that trigger CD5L release from IgM remains a critical area for future research. Furthermore, the existence of specific receptors for CD5L is yet to be investigated. Regardless, the fact that CD5L serves as a subunit of circulatory IgM alters our understanding of IgM and warrants immediate attention in related fields.
Finally, we have shed light on the critical, yet previously underappreciated, role of the J chain. The significance of the J chain was first reported by Marian Koshland in 1970, and in her 1985 review42, she highlighted that “the existence of a third immunoglobulin polypeptide, the J chain, is often overlooked.” Recent structural insights from our research and others have revealed the molecular mechanism of the J chain in promoting IgM and IgA polymerization, as well as its role in the recognition of IgM and IgA by pIgR31,32,34,35,36. In this study and another study43, we further demonstrate that the J chain also plays a crucial role in mediating the interaction between the IgM pentamer and CD5L, as well as between systemic IgA and FcRL4. It is clear that the J chain imparts unique characteristics to polymeric IgM and IgA, enabling them to be specifically recognized by distinct molecular partners to fulfil their functions.
Methods
Protein expression and purification
The DNA fragment encoding the residues 20–347 of human CD5L (Uniprot O43866) was cloned into a modified pcDNA vector with an N-terminal IL-2 signal peptide followed by an 8-residue His-tag. The plasmid expressing CD5L was transfected into the HEK293F cells by polyethyleneimine (Polysciences), and the cells were cultured in SMM 293T-I medium (Sino Biological Inc.) at 37 °C, with 5% CO2 and 55% humidity. Four days following transfection, the conditioned media were collected by centrifugation, then concentrated via a Hydrosart Ultrafilter (Sartorius) and exchanged into the binding buffer (25 mM Tris–HCl, pH 7.4, 150 mM NaCl). The recombinant protein was isolated by Ni-NTA (Smart-Lifesciences) affinity chromatography and eluted with the binding buffer supplemented with 500 mM imidazole. The sample was further purified using a Superdex 200 increase column (GE Healthcare) and eluted using the binding buffer. Mouse CD5L (residues 22–352, Uniprot Q9QWK4) and cat CD5L (3-SRCR isoform, residues 20–355, Uniprot A0A1E1GEY0) were produced similarly. The Fcμ and the J chain of human Fcμ–J complex were co-expressed in the HEK293F cells, then purified using Ni-NTA affinity chromatography and Superose 6 increase column (GE Healthcare), as previously described31. Cat Fcμ (residues 94–441, NCBI BAA32231.1)–J (residues 1–158, Uniprot M3XBL5) and mouse Fcμ (residues 104–454, Uniprot P01872)–J (residues 1–159, Uniprot P01592) complexes were also expressed and purified in similar manners.
To obtain the human Fcμ–J–CD5L complex for cryo-EM analyses, Fcμ–J and CD5L were co-expressed in the HEK293F cells and purified first by Ni-NTA affinity chromatography, and subsequently subjected to size-exclusion chromatography using a Superose 6 increase column. The elution buffer comprises the binding buffer supplemented with 5 mM CaCl2.
Circular dichroism spectroscopy assay
The circular dichroism spectroscopy experiments were performed using the MOS-500 spectropolarimeter (Bio-Logic). Human CD5L was purified using a Superdex 200 increase column (GE Healthcare) and then concentrated to ~20 μM sample concentration. The measurement wavelength range was set to 190–250 nm, with a slit width of 2 nm, a scanning speed of 1 nm/s, and an acquisition time of 1 s per point. A 1-mm colorimetric cell was used for scanning. The experiments were repeated twice, and the data were analyzed using the GraphPad Prism software.
Surface plasmon resonance
The surface plasmon resonance experiments were performed on the Biacore T200 (GE Healthcare). Fcμ–J complex was captured by the SA chip (GE Healthcare) via the twin-strep tag at N-terminus of Fcμ–J. CD5L was injected over the flow cell at a range of five concentrations prepared by serial twofold dilutions at a flow rate of 30 μL/min using a single-cycle kinetics program. The SPR assays were performed at 25 °C. The running buffer comprises 1× HBS-EP Buffer (Cytiva) (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.05% Tween-20). Afterwards, the CD5L proteins were passed through using a single-cycle kinetics program. Bovine Serum Albumin (BSA, BBI Life Sciences) was used as a negative control.
The affinity of CD5L for Fcμ–J was also evaluated by immobilizing Fcμ–J on a CM5 chip (GE Healthcare) at pH 4.5 using the amine coupling procedure at a 10 μL/min flow rate. The twofold serial dilutions of CD5L flowed through the cell at five concentrations using a single-cycle kinetics program. Running buffer containing 1× HBS–EP buffer or 1× HBS–EP buffer with the addition of 8 mM of CaCl2 was used to test the effect of calcium ions on Fcμ–J and CD5L binding.
To investigate the effect of CD5L on the interaction between Fcμ–J and FcμR, FcμR-D1 was expressed and purified as previously described38 and was immobilized on the CM5 chip via amine coupling. Serial dilutions of Fcμ–J–CD5L and Fcμ–J were then passed through using a multiple-cycle kinetics program. To investigate the effect of CD5L on the interaction between Fcμ–J and pIgR, a pIgR-SC with a C-terminal Avi-tag (amino acid sequence: GLNDIFEAQKIEWHE) was first expressed in the HEK293F cells and purified by Ni-NTA affinity method. A GST (Glutathione S-transferases)-tagged BirA, a biotin ligase, was expressed in BL21 (DE3) E. coli and purified by the GST affinity method. Biotinylation was performed in a solution containing 100 μM Avi-tagged pIgR-SC in 1× phosphate-buffered saline (PBS) supplemented with 5 mM MgCl2, 2 mM ATP, 1 μM BirA, and 0.15 mM D-Biotin. After the reaction, BirA and excess D-Biotin were removed from the biotinylated pIgR-SC using a Superdex 200 increase column. The C-terminal biotinylated pIgR-SC was then immobilized on the SA chip for subsequent SPR measurements. The experiments were also conducted in a reverse manner, with Fcμ–J–CD5L and Fcμ–J immobilized on the CM5 chips via amine coupling. Binding studies were then performed by passing twofold serial dilutions of purified FcμR-D1 or pIgR-SC over these chips.
The interaction between immobilized pIgR-SC and Fcμ–J–CD5L was analyzed using both a two-state reaction model and a 1:1 binding model with the Biacore Evaluation Software v2.0, as shown in Table 1. The remaining SPR data were fitted to a 1:1 binding model. Each SPR experiment was repeated at least twice, as shown in Supplementary Fig. 8.
Cryo-EM sample preparation and data collection
Prior to cryo-EM sample preparation, the gel filtration peak fractions containing Fcμ–J–CD5L tripartite complex were collected and concentrated to 0.25 mg/mL, then cross-linked with 0.05% glutaraldehyde (Sigma) at 20 °C for 5 min. For grid preparation, 4 μL of Fcμ–J–CD5L was applied to glow-discharged holy-carbon gold grids (Quantifoil, R1.2/1.3, 300 mesh), blotted with filter paper in 100% humidity at 4 °C, and put into the liquid ethane with a Vitrobot Mark IV (FEI). A Talos Arctica microscope equipped with a Ceta camera (FEI) was used to screen grids. Data collection was performed on a Titan Krios electron microscope (FEI) operated at 300 kV with the Gatan Imaging Filter (GIF) energy-filtering slit width set at 20 eV. Movie stacks were recorded on a K3 Summit direct electron detector (Gatan) in super-resolution mode at a magnification of 105,000× using the EPU software (E Pluribus Unum, v2.1.2, Thermo Scientific), which corresponded to a pixel size of 0.83 Å per pixel. The dose rate was 16 e–/pixel/s. The defocus range was set from –1.0 to –1.5 μm. The micrographs were recorded for 2.56 s and subdivided into 32 frames with a total electron exposure of 59.5 electrons per Å2.
Imaging processing, model building and structure refinement
For 3D reconstruction of the Fcμ–J–CD5L complex, a total of 6962 movie stacks were recorded. All image processing was performed with cryoSPARC (v3.2.1)44. Raw movie frames were motion-corrected and dose-weighted using the Patch motion correction (multi) module. The contrast transfer function (CTF) parameters of each summed image were estimated by the Patch CTF estimation (multi) module. The images were screened manually to remove low-quality ones with the Manually Curate Exposures module. A set of 979,111 particles was picked without reference by the Blob picker module and subjected to reference-free 2D classification to generate templates for template particle picking. Subsequently, a total of 894,255 particles were template-picked, and extracted particles were subjected to several rounds of 2D classification to remove noise and fuzzy particles. 769,364 particles were kept for initial model generation and 3D classification, using Ab-Initio Reconstruction and Heterogeneous Refinement, respectively. The particles in qualified groups (381,470 particles) were combined and subjected to Homogeneous Refinement, resulting in a map with a 3.39 Å overall resolution, according to the Fourier shell correlation (FSC) = 0.143 criterion. Local Refinement was performed with a soft mask which encompasses the CD5L–Fcμ–J interface, yielding a map at the resolution of 3.41 Å with a clearer visualization of the interface. The mask was created using UCSF chimera (v1.16)45. The local resolution map was analyzed by the Local Resolution Estimation module of cryoSPARC and displayed using UCSF ChimeraX (v1.5rc202211120143).
The initial model of CD5L was predicted by Alphafold46, as well as the structures of Fcμ from the Fcμ–J–pIgR-SC complex (PDB ID: 6KXS) [https://doi.org/10.2210/pdb6KXS/pdb] and the J chain from the SIgA complex (PDB ID: 6LX3) [https://doi.org/10.2210/pdb6LX3/pdb], was docked into the cryo-EM map using UCSF chimeraX and then manually adjusted using Coot (v0.9.4.1)47. The SRCR1 domain of CD5L is absent in the structure because it displays poor densities, perhaps due to potential conformational flexibility. Refinement was performed using the real-space refinement in Phenix (v1.19)48. Figures were prepared with UCSF ChimeraX.
StrepTactin pull-down assay
CD5L mutations were introduced through PCR-based mutagenesis. Afterwards, wild-type and mutant CD5L proteins were purified with the Ni-NTA affinity method. For the pull-down assays, CD5L was first incubated with purified Fcμ–J complex on ice for 1 hr. The mixture was then incubated with the StrepTactin beads (Smart Lifesciences) and rotated at 4 °C, in the binding buffer containing 25 mM Tris–HCl, pH 7.4, 150 mM NaCl, 5 mM CaCl2 or 5 mM EDTA. A twin-strep tag is present on Fcμ. After 1 h of incubation, the beads were washed three times with the binding buffer, and the bound proteins were eluted with the binding buffer supplemented with 10 mM desthiobiotin (IBA Lifesciences). The samples were analyzed by SDS–PAGE and detected by Coomassie staining.
Complement-dependent cytotoxicity assay
Anti-CD20 IgM was produced as described previously41. OCI-Ly10 cells, which express CD20, were used for the complement-dependent cytotoxicity assay. Serially diluted CD5L were incubated with anti-CD20 IgM for 3 h in 50 μL of RPMI-1640. Then the proteins were mixed with equal amounts of normal human serum complement (1:12.5, Quidel) and OCI-Ly10 cultures (20,000 cells) before being put into a 96-microwell plate. 50 μL of CellTiter-Glo reagent (Promega, G7572) was added to each well and incubated for 10 min at room temperature after 6 h of incubation at 37 °C. A Cytation 5 cell imaging multi-mode reader (BioTek) was used to quantify luminescence. The experiment was repeated four times and the statistics were analyzed by two-tailed, unpaired Student’s t-test in GraphPad Prism v9.0.0.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Cryo-EM density maps of Fcμ–J–CD5L have been deposited in the Electron Microscopy Data Bank with accession codes EMD-37936 (overall) and EMD-37937 (local). Structural coordinates have been deposited in the Protein Data Bank with the accession codes 8WYR (overall) and 8WYS (local). A list of primers and codon-optimized DNAs is provided in the Source Data file. Source data are provided with this paper.
References
Heidelberger, M. & Pedersen, K. O. The molecular weight of antibodies. J. Exp. Med. 65, 393–414 (1937).
Tissot, J. D., Hochstrasser, D. F., Spertini, F., Schifferli, J. A. & Schneider, P. Pattern variations of polyclonal and monoclonal immunoglobulins of different isotypes analyzed by high‐resolution two‐dimensional electrophoresis. Electrophoresis 14, 227–234 (1993).
Tissot, J. D. et al. Two-dimensional polyacrylamide gel electrophoresis analysis of cryoglobulins and identification of an IgM-associated peptide. J. Immunol. Methods 173, 63–75 (1994).
Gebe, J. A. et al. Molecular cloning, mapping to human chromosome 1 q21-q23, and cell binding characteristics of Spα, a new member of the scavenger receptor cysteine-rich (SRCR) family of proteins. J. Biol. Chem. 272, 6151–6158 (1997).
Miyazaki, T., Hirokami, Y., Matsuhashi, N., Takatsuka, H. & Naito, M. Increased susceptibility of thymocytes to apoptosis in mice lacking AIM, a novel murine macrophage-derived soluble factor belonging to the scavenger receptor cysteine-rich domain superfamily. J. Exp. Med. 189, 413–422 (1999).
Gebe, J. A., Llewellyn, M., Hoggatt, H. & Aruffo, A. Molecular cloning, genomic organization and cell-binding characteristics of mouse Spalpha. Immunology 99, 78–86 (2000).
Tissot, J. D. et al. IgM are associated to Spα (CD5 antigen‐like). Electrophoresis 23, 1203–1206 (2002).
Oskam, N. et al. CD5L is a canonical component of circulatory IgM. Proc. Natl Acad. Sci. USA 120, e2311265120 (2023).
Sanjurjo, L., Aran, G., Roher, N., Valledor, A. F. & Sarrias, M. R. AIM/CD5L: a key protein in the control of immune homeostasis and inflammatory disease. J. Leukoc. Biol. 98, 173–184 (2015).
Miyazaki, T., Yamazaki, T., Sugisawa, R., Gershwin, M. E. & Arai, S. AIM associated with the IgM pentamer: attackers on stand-by at aircraft carrier. Cell. Mol. Immunol. 15, 563–574 (2018).
Joseph, S. B. et al. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell 119, 299–309 (2004).
Sarrias, M.-R. et al. A role for human Spα as a pattern recognition receptor. J. Biol. Chem. 280, 35391–35398 (2005).
Martinez, V. G. et al. The macrophage soluble receptor AIM/Api6/CD5L displays a broad pathogen recognition spectrum and is involved in early response to microbial aggression. Cell. Mol. Immunol. 11, 343–354 (2014).
Gao, X., Yan, X. X., Zhang, Q., Yin, Y. B. & Cao, J. CD5L contributes to the pathogenesis of methicillin-resistant Staphylococcus aureus-induced pneumonia. Int. Immunopharmacol. 72, 40–47 (2019).
Maehara, N. et al. AIM/CD5L attenuates DAMPs in the injured brain and thereby ameliorates ischemic stroke. Cell Rep. 36, 109693 (2021).
Arai, S. et al. Apoptosis inhibitor of macrophage protein enhances intraluminal debris clearance and ameliorates acute kidney injury in mice. Nat. Med. 22, 183–193 (2016).
Maehara, N. et al. Circulating AIM prevents hepatocellular carcinoma through complement activation. Cell Rep. 9, 61–74 (2014).
Kuwata, K. et al. AIM inhibits apoptosis of T cells and NKT cells in Corynebacterium-induced granuloma formation in mice. Am. J. Pathol. 162, 837–847 (2003).
Li, J. J., Lin, W. & Zhuang, L. CD5L-induced activation of autophagy is associated with hepatoprotection in ischemic reperfusion injury via the CD36/ATG7 axis. Exp. Ther. Med. 19, 2588–2596 (2020).
Wang, C. et al. CD5L/AIM regulates lipid biosynthesis and restrains Th17 cell pathogenicity. Cell 163, 1413–1427 (2015).
Kurokawa, J. et al. Macrophage-derived AIM is endocytosed into adipocytes and decreases lipid droplets via inhibition of fatty acid synthase activity. Cell Metab. 11, 479–492 (2010).
Arai, S. et al. A role for the apoptosis inhibitory factor AIM/Spα/Api6 in atherosclerosis development. Cell Metab. 1, 201–213 (2005).
Takahata, A. et al. Crucial role of AIM/CD5L in the development of glomerular inflammation in IgA nephropathy. J. Am. Soc. Nephrol. 31, 2013–2024 (2020).
Oliveira, L. et al. CD5L as a promising biological therapeutic for treating sepsis. Nat. Commun. 15, 4119 (2024).
Resnick, D., Pearson, A. & Krieger, M. The SRCR superfamily: a family reminiscent of the Ig superfamily. Trends Biochem. Sci. 19, 5–8 (1994).
Sarrias, M. R. et al. The scavenger receptor cysteine-rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit. Rev. Immunol. 24, 1–37 (2004).
Reichhardt, M. P., Loimaranta, V., Lea, S. M. & Johnson, S. Structures of SALSA/DMBT1 SRCR domains reveal the conserved ligand-binding mechanism of the ancient SRCR fold. Life Sci. Alliance 3, e201900502 (2020).
Sugisawa, R. et al. Impact of feline AIM on the susceptibility of cats to renal disease. Sci. Rep. 6, 35251 (2016).
Hiramoto, E. et al. The IgM pentamer is an asymmetric pentagon with an open groove that binds the AIM protein. Sci. Adv. 4, eaau1199 (2018).
Sharp, T. H. et al. Insights into IgM-mediated complement activation based on in situ structures of IgM-C1-C4b. Proc. Natl Acad. Sci. USA 116, 11900–11905 (2019).
Li, Y. X. et al. Structural insights into immunoglobulin M. Science 367, 1014–1017 (2020).
Kumar, N., Arthur, C. P., Ciferri, C. & Matsumoto, M. L. Structure of the human secretory immunoglobulin M core. Structure 29, 564–571 e3 (2021).
Chen, Q., Menon, R., Calder, L. J., Tolar, P. & Rosenthal, P. B. Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer. Nat. Commun. 13, 6314 (2022).
Kumar, N., Arthur, C. P., Ciferri, C. & Matsumoto, M. L. Structure of the secretory immunoglobulin A core. Science 367, 1008–1014 (2020).
Wang, Y. X. et al. Structural insights into secretory immunoglobulin A and its interaction with a pneumococcal adhesin. Cell Res. 30, 602–609 (2020).
Bharathkar, S. K. et al. The structures of secretory and dimeric immunoglobulin A. Elife 9, e56098 (2020).
Kubagawa, H. et al. Identity of the elusive IgM Fc receptor (FcmuR) in humans. J. Exp. Med. 206, 2779–2793 (2009).
Li, Y. et al. Immunoglobulin M perception by FcmuR. Nature 615, 907–912 (2023).
Shibuya, A. et al. Fc alpha/mu receptor mediates endocytosis of IgM-coated microbes. Nat. Immunol. 1, 441–446 (2000).
Arya, S. et al. Mapping of amino acid residues in the C mu 3 domain of mouse IgM important in macromolecular assembly and complement-dependent cytolysis. J. Immunol. 152, 1206–1212 (1994).
Ji, C. et al. Plasmodium falciparum has evolved multiple mechanisms to hijack human immunoglobulin M. Nat. Commun. 14, 2650 (2023).
Koshland, M. E. The coming of age of the immunoglobulin J chain. Annu Rev. Immunol. 3, 425–453 (1985).
Su, C., Wang, Y., Yang, X. & Xiao, J. J chain dictates the recognition of IgA by FcRL4. bioRxiv 12, 572144 (2023). 202318.
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D 66, 213–221 (2010).
Acknowledgements
We are grateful to the Core Facilities at the School of Life Sciences, Peking University, for their assistance with negative-staining EM; the Cryo-EM Platform of Peking University for their support with data collection; and the High-performance Computing Platform of Peking University for their aid with computation. We also acknowledge the National Center for Protein Sciences at Peking University for their help with Biacore, BioTek, and Bio-Logic facilities. This work received support from the National Natural Science Foundation of China (32325018) and the Qidong-SLS Innovation Fund to J.X.
Author information
Authors and Affiliations
Contributions
Y.W. and C.S. performed protein expression, purification, cryo-EM sample screening and data collection. Y.W. carried out the cryo-EM data processing, model building, structure refinement, and the circular dichroism spectroscopy experiment. C.S. performed the SPR and the complement-dependent cytotoxicity assay with assistance from C.J., Y.W., and C.S. conducted the pull-down experiments. J.X. conceived and supervised the project and wrote the manuscript, with inputs from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yigong Shi, Brian Sutton and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Y., Su, C., Ji, C. et al. CD5L associates with IgM via the J chain. Nat Commun 15, 8397 (2024). https://doi.org/10.1038/s41467-024-52175-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-52175-y