Abstract
Understanding the interface between nanomaterials and lipoproteins is crucial for gaining insights into their impact on lipoprotein structure and lipid metabolism. Here, we use graphene oxide (GOs) nanosheets as a controlled carbon nanomaterial model to study how surface properties influence lipoprotein corona formation and show that GOs have strong binding affinity with low-density lipoprotein (LDL). We use advanced techniques including X-ray reflectivity, circular dichroism, and molecular simulations to explore the interfacial interactions between GOs and LDL. Specifically, hydrophobic GOs preferentially associate with LDL’s lipid components, whereas hydrophilic GOs tend to bind with apolipoproteins. Furthermore, these GOs distinctly modulate a variety of lipid metabolism pathways, including LDL recognition, uptake, hydrolysis, efflux, and lipid droplet formation. This study underscores the importance of structure analysis at the nano-biomolecule interface, emphasizing how nanomaterials’ surface properties critically influence cellular lipid metabolism. These insights will inspire the design and application of future biocompatible nanomaterials and nanomedicines.
Similar content being viewed by others
Introduction
Recent research has indicated that nanomaterials, upon contact with body fluids, interact with biomolecules to form a biomolecular corona on their surface1,2,3,4. Lipoproteins, particularly low-density lipoprotein (LDL), high-density lipoprotein (HDL), and very low-density lipoprotein (VLDL), are pivotal components of this protein corona5,6,7,8. While studies have shown that lipoprotein adsorption enhances blood circulation9,10 and facilitates targeted delivery of nanomaterials to tissues11, the effects of this adsorption on the biological fate and physiological functions of lipoprotein coronas are still largely unknown. Lipoproteins function as carriers of cholesterol and lipids, and changes in their compositions and structures can significantly influence physiological processes, including lipid metabolism12 and inflammation13,14,15. However, the biological consequences of lipoprotein adsorption onto nanomaterials remain largely unexplored. Therefore, the investigations into the interactions between lipoproteins and nanomaterials, and the subsequent biological impacts, are crucial for establishing guidelines for the safe application of nanomaterials.
Among lipoproteins, LDL has garnered significant interest due to its association with the pathogenesis of atherosclerosis. LDL is a spherical nanoparticle composed of various lipids and apolipoprotein B-100 (ApoB-100), which surrounds the LDL particle and is responsible for receptor recognition and uptake16,17. The intricate structure of LDL features an outer monolayer of phospholipids surrounded by ApoB-100 protein and cholesterols, with the inner core comprising cholesterols, triglycerides, and unsaturated fatty acids18,19. Structural changes or chemical modifications to LDL can prompt its aggregation and fusion, leading to accelerated uptake by macrophages via scavenger receptors or phagocytosis. This, in turn, results in an imbalance in lipid metabolism and the accumulation of lipid droplets20,21,22, ultimately contributing to the formation of foam cells, a hallmark of atherosclerosis and posing a significant threat to human health23,24,25,26. LDL has been identified as a primary component in lipoprotein coronas, yet our understanding of how nanomaterials interact with lipoproteins and the potential modifications they induce in lipoprotein structure, composition, and the physiological functions of LDL remains limited. Previous studies have indicated that LDL adsorbed on the surface of SiO2 nanoparticles retains functional epitopes recognized by specific LDL receptors or scavenger receptors, thereby facilitating the internalization of SiO2 nanoparticles27,28. However, significant challenges persist in conducting structural analyses of nano-lipoprotein interfaces, leaving the interaction process and structural details of how lipoproteins attach and organize on nanomaterial surfaces, and the associated cellular behaviors, yet to be fully revealed.
As we know, the thickness of a phospholipid bilayer is approximately 7–8 nm29, thus the outer structure of LDL is a phospholipid monolayer with a quite thin thickness. Conventional techniques confront challenges in quantitatively characterizing the interface structure between phospholipid monolayer and the nanomaterial with high resolution and high sensitivity in liquid environments30,31,32,33. Comparably, X-ray reflectivity (XRR) is advantageous for providing quantitative information on nanomaterials and phospholipid interactions, such as electronic density of the surface and the thickness of lipids in the liquid phase which is powerful for resolving interfacial details. Additionally, synchrotron radiation-circular dichroism (SR-CD) is a powerful method for studying nanomaterial and protein interactions. At a far ultraviolet region (160–195 nm), SR-CD can capture more detailed information for protein secondary structures than conventional CD with higher signal-to-noise and signal intensity34 that enables structural measurement for apolipoprotein in the presence of lipoproteins and nanomaterials. To better understand the interaction of nanomaterials with lipoproteins, two-dimensional graphene oxide nanosheets (GOs) serve as a model carbon nanomaterial. GOs possess sp2- and sp3-hybrid carbons, demonstrating unique properties such as a high specific surface area, controllable oxidation degrees for tunable hydrophilicity or amphipathic features, a flat surface, and biocompatibility, enabling them promising candidates for biomedical applications35,36.
In this work, we aim to develop analytical methods for investigating the interfacial structure of nano-lipoproteins and correlate the structural, compositional, and chemical alterations in lipoproteins with cellular lipid metabolism. Firstly, we study the interaction of GOs with human serum and observe that GOs with varying degrees of hydrophilicity can adsorb and enrich LDL, resulting in diverse changes in its structure and morphology. To gain deeper insights, we subsequently use advanced techniques including SR-CD and XRR, complemented by molecular simulations, to elucidate the interfacial structures formed between GOs and the components of LDL, such as phospholipids and apolipoproteins. Finally, we establish connections between structural changes in LDL and various lipid metabolic processes in macrophages, encompassing LDL uptake, lipid trafficking, accumulation, and efflux in macrophages, at both the molecular and cellular levels. Our findings demonstrate the important role of nanomaterials’ physicochemical properties in modulating lipoprotein structure, physiological functions, and fate, particularly in the context of lipid metabolism in macrophages, thereby facilitating future safety evaluations and biomedical applications of nanomaterials.
Results and discussion
The hydrophilic/hydrophobic properties of GOs
GO nanomaterials with varying levels of hydrophilicity were synthesized using modified Hummer methods37. The schematic representation in Fig. 1a illustrates that hydrophilic GO possesses more hydrophilic functional groups, such as carboxyl and hydroxyl, with increasing oxidation degrees (M-GO and H-GO). In contrast, the most hydrophobic L-GO retains more sp2 structure compared to M-GO and H-GO. The atomic force microscopy (AFM) images in Fig. 1b and Supplementary Fig. 1a and 1b show that all three GO samples (L-GO: low oxidation degree, M-GO: mild oxidation degree, and H-GO: high oxidation degree) exhibit sheet-like structures with similar monolayer heights of 1 nm and lateral diameters ranging from 100 to 200 nm (Supplementary Fig. 2) based on the TEM scanning. Chemical analysis using X-ray photoelectron spectroscopy (XPS), X-ray absorption spectroscopy (XAS), and Fourier Transform Infrared (FTIR) measurements determined the carbon species and hydrophobicity of the three GOs. Figure 1c and Supplementary Figs. 1c, e, f demonstrate that GOs with higher oxidation levels (from L-GO to H-GO) contain more oxygen-containing functional groups, such as -C-O-C and O = C-O. Consequently, M-GO and H-GO with higher oxidation degrees exhibit more negative surface charges, as indicated by the Zeta potential in Fig. 1d. Due to their hydrophilicity, GO showed high stability in suspension of water or cellular medium and maintained the negative surface charges (Supplementary Fig. 3). UV-Vis absorption spectra of GO samples show a characteristic adsorption peak at 235 nm (Supplementary Fig. 1d). Moreover, contact angle measurements (θ) of the three GOs reveal values lower than 90°, with θ decreasing with increasing oxidative levels of GOs (Fig. 1e). This suggests that the surface of H-GO is more hydrophilic than that of M-GO and L-GO. Additionally, the intensity ratio of the D band and G band in GO samples (ID/IG) based on Raman spectra measurements also confirms the higher degree of oxidation in H-GO compared to M-GO and L-GO (Fig. 1f). In conclusion, the three GOs demonstrate tunable degrees of surface hydrophilicity based on the modulation of oxidation degrees.
a Schematic illustrating the ball-and-stick model of three types of GOs with varying degrees of oxidation: low (L-GO), medium (M-GO), and high (H-GO). Carbon, oxygen and hydrogen are presented in black, blue and green, respectively. b AFM images showing GOs with different oxidation degrees. Scale bar represents 1 μm. c Quantitative analysis of oxidation levels and functional groups (C-O-C, C = O, O-C = O, and C-C) determined by XPS. d Surface charges of GOs evaluated by zeta potential measurements. e Hydrophilicity of the three types of GOs measured by contact angle analysis. Data are presented as mean ± standard deviation for three replicates (n = 3). f Raman spectra for the three types of GOs. Source data are provided as a Source Data file.
High enrichment of apolipoproteins by GOs
Upon entry into the body, nanomaterials are enveloped by proteins in the physiological milieu, forming a protein corona1,2,3,4,38. In this study, we utilized LC-MS/MS-based proteomics to identify the major components of the protein corona on the surface of GOs following exposure to a 10% human serum solution (Supplementary Data 1). Classification of corona proteins by function revealed significant components, including apolipoproteins, complements, acute phase proteins, immunoglobulins, coagulation factors, and others (Fig. 2a, Supplementary Fig. 4). Additionally, top twenty abundant proteins of GOs upon interaction with human serum were identified (Supplementary Data 1). To assess the degree of protein enrichment on GOs, we calculated the protein enrichment factor by comparing the protein abundance on GOs with their abundance in serum. Among the significantly enriched protein types, lipoproteins, coagulation proteins, and complements stood out (Fig. 2b). Notably, lipoproteins exhibited the highest enrichment, indicating a strong affinity of GOs for lipoproteins compared to other proteins. For a more detailed analysis of lipoprotein composition, we examined the enrichment factor (Fig. 2c) and the relative abundance of apolipoproteins in the GOs’ corona (Supplementary Fig. 4b and Supplementary Data 1). Interestingly, ApoB-100 from LDL and Apo-A1 from HDL were particularly prominent, with their relative abundance above 5% (Supplementary Fig. 4b) and enrichment approximately five times higher (Fig. 2c). To further validate these findings, we conducted sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), which confirmed the presence of serum ApoB-100 protein or LDL and Apo-A1 protein or HDL on the GOs after incubation in 10% human serum under gentle stirring at 37 °C for 1 h (Supplementary Fig. 5). Notably, the hydrophobicity of GOs influenced the adsorption of LDL and HDL differently, with higher hydrophobicity favoring LDL binding (L-GO > M-GO > H-GO) and greater hydrophilicity favoring HDL binding (H-GO > M-GO » L-GO). Given the physiological and pathological significance of LDL, we focused on the interaction between GOs and LDL, as well as the subsequent effects of GOs/LDL complexes on lipid metabolism.
a Classification of components on three types of GOs (L-GO, M-GO and H-GO) by biological function. The pie chart indicates the relative abundance of coronal proteins in six categories: apolipoproteins, complements, acute phase, immunoglobulins, coagulation factors, and others. Albumin, due to its highest abundance in serum, is classified as an individual category. b Comparison of functional protein groups on the surface of graphene oxides. c Enrichment factor of apolipoproteins on different graphene oxide nanosheets. The enrichment factors (b, c) represent the comparison of protein or protein group abundance on GOs to that in the serum (coronal complex of GOs/serum). Protein compositions are identified through manual search using the Human Database via the UniProt website. Data are presented as mean ± standard deviation of triplicate biological samples (n = 3). Proteomics data are presented in Supplementary Data 1. Source data are provided as a Source Data file.
To determine the lipid composition of the corona on GOs, we utilized LC-MS/MS to quantify lipid components in both positive and negative ion modes (Supplementary Data 2). Our results revealed slight changes in the lipid compositions of phospholipids, sphingolipids, and derivatized lipids on the surface of GOs compared to the control (Supplementary Fig. 6a, b and Supplementary Data 2). Specifically, the percentage of phospholipids and neutral lipids in the enriched lipid corona slightly decreased with increased hydrophobicity of GOs, while the percentage of sphingolipids and derivatized lipids increased slightly with increased hydrophobicity of GOs (Supplementary Fig. 6a and Supplementary Data 2). Enriched lipids on the three GOs were classified into five and four types at the positive and negative modes, respectively. However, the abundance of lipid compositions on GOs did not significantly differ from the control or among the GO types based on this classification (Supplementary Fig. 6a, b). We then performed detailed analysis of most lipid compositions using principal component analysis (PCA) and volcano plot and heatmaps. PCA results showed significant differences in the compositions and the number of lipid corona adsorbed on the three types of GOs compared to the control with varying hydrophobicity (Supplementary Fig. 6c), with L-GOs potentially inducing a stronger impact on the adsorbed lipid composition than other GOs (Supplementary Data 2). Volcano plots and heatmaps corroborated the PCA results (Supplementary Fig. 6d, 7 and Supplementary Data 2). Furthermore, we compared the differences in lipid corona compositions among the three GOs with different surface hydrophobicity based on PCA (Supplementary Figs. 6e–g, 8). Consistent with the PCA results, volcano plots and heatmaps indicated that there were slight differences in both the composition and abundance of lipids on H-GO compared to M-GO (Supplementary Figs. 6e, f and 8a, b; Supplementary Data 2), but the composition and abundance of adsorbed lipids on H-GO and M-GO showed significant alterations compared to L-GO (Supplementary Fig. 8b, c). These findings provide valuable insights into the interaction between GOs and lipid molecules in the blood.
Cellular uptake of GO/LDL complexes
We investigated how the surface properties of GOs influence cellular uptake and subsequent lipid accumulation in macrophages. Initially, we assessed the cytotoxicity of GOs at various concentrations after exposing THP-1 cells to them for 24 h in 1640 RPMI medium with or without 10% FBS (Supplementary Fig. 9). The results showed that cell viability dropped below 80% at GO concentrations exceeding 20 μg/mL (Supplementary Fig. 9b). Therefore, for subsequent studies, we selected a concentration of 20 μg/mL. In our experiments, we incubated THP-1 cells with a combination of 20 μg/mL of GOs and 50 μg/mL of LDL in serum-free medium. Surprisingly, we did not observe any significant cytotoxicity caused by the GO/LDL complex (Supplementary Fig. 9c). However, GOs in serum-free medium did induce cytotoxicity (Supplementary Fig. 9b). Subsequently, we used Raman imaging (Fig. 3a, Supplementary Table 1) and the laser reflection technique (Fig. 3b, Supplementary Fig. 10) to investigate the cellular uptake of GO/LDL complexes. Interestingly, we found that macrophages internalized a significantly higher amount of the L-GO/LDL complex compared to H-GO/LDL and M-GO/LDL. This observation suggests a preference of macrophages for taking up more hydrophobic GOs in the presence of LDL.
a Cellular uptake of GO/LDL complexes assessed by Raman mapping of GOs, showing D-band (red color) and G-band (green color) signal intensity. Scale bar represents 10 μm. b Confocal images depicting the uptake of GO/LDL complexes using laser reflectivity microscopy. Fluorescent signals in red, green, and blue correspond to cytoplasmic membrane, GOs, and the nucleus, respectively. Scale bars represent 7.5 μm. c Subcellular localization of GOs and lipid droplets observed through TEM images. Red arrows indicate lipid droplets, while blue arrows indicate GOs. Scale bar represents 1 μm. Cells are treated with 50 μg/mL LDL and a mixture of 50 μg/mL LDL with 20 μg/mL GOs for 24 h. d Accumulation of lipid droplets in macrophages when treated with LDL and GO/LDL complexes, visualized by confocal imaging. Red color indicates lipid droplets, while blue indicates nucleus. Scale bar represents 10 μm. e Quantitative analysis of total lipid droplet mean fluorescent intensity (MFI) per cell using high-content analysis. Data are presented as mean ± standard deviation of n = 3 biological replicates. Statistical significance is calculated by one-way ANOVA with Tukey’s multiple comparisons test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; n.s., not significant (p > 0.05). f, g Soft X-ray cryo-tomographic images of lipid droplets in macrophages before and after exposure to LDL and GO/LDL, obtained through soft X-ray transmission microscopic imaging and 3D reconstruction (nano-CT). Cells are treated with 50 μg/mL LDL or a mixture of 50 μg/mL LDL with 20 μg/mL GOs for 24 h, and wet samples are frozen in liquid nitrogen for nano-CT imaging. f Soft X-ray transmission cryo-microscopic and 3D tomographic imaging of frozen cells and lipid droplets at 520 eV and −80 °C. Red stars indicate lipid droplets, while purple stars represent organelles. g Spatial distribution of lipid droplets (in yellow) and organelles (in red) based on 3D reconstruction of the macrophages. Scale bar represents 5 μm. Source data are provided as a Source Data file.
Less lipid accumulation in macrophages induced by hydrophilic GO/LDL
We further investigated the localization of GOs and resulting changes in subcellular structure after their uptake, using transmission electron microscopy (TEM). After 24 h of treatments with LDL, we observed the formation of lipid droplets (LDs) with relatively dense structures, which were significantly more abundant compared to LDL-untreated cells (control). This finding suggests that macrophages may engulf native LDL and transform into lipid-laden macrophages. Upon incubation of three types of GOs with LDL, we observed the formation of lipid droplets in the macrophages, while this phenomenon was not evident when cells were exposed to pure GOs (Supplementary Fig. 11). Notably, the number of LDs in cells treated with L-GO/LDL was significantly greater than in those treated with M-GO/LDL and H-GO/LDL (Fig. 3c). To quantitatively characterize the level of LDs and the degree of foam cell formation, we utilized a specific fluorescence dye (HCS LipidTOX Red neutral lipid stain) to stain the intracellular accumulation of neutral lipids. As shown in Fig. 3d and Supplementary Fig. 12, abundant LDs were stored in the macrophages after treatment with native LDL and L-GO/LDL. In comparison, far fewer LDs were observed in cells exposed to H-GO/LDL (Fig. 3d, Supplementary Fig. 12c), M-GO/LDL, and pure GOs (Supplementary Fig. 12a, b). The quantitative analysis of the fluorescent intensity of total lipid droplets per cell through high-content analysis (Fig. 3e) corroborated the TEM and confocal image results, indicating a significant increase in intracellular LDs with the increasing hydrophobicity of GOs during their interaction with macrophages. To further support our hypothesis and conclusions, we conducted another cellular experiment using mouse primary macrophages and mouse J774A.1 cell lines. Foam cell formation only occurred in the macrophage cells of LDL-treated group and L-GO/LDL-treated group (Supplementary Fig. 13a, b), which was similar to the findings in THP-1 macrophages.
Moreover, we employed a label-free synchrotron radiation soft X-ray transmission cryo-microscope (TXM) along with three-dimensional tomography (Nano-CT) to visualize LDs in macrophages with high spatial resolution (~30 nm) under aqueous conditions (Supplementary Movies 1-5). LDs, being dense lipid aggregates, exhibit a stronger adsorption coefficient for soft X-rays at an energy of 520 eV compared to the cytoplasm and organelles39. Nano-CT effectively distinguished LDs from subcellular structures and organelles. To distinguish between organelles and LDs, we used the linear absorption coefficient (LAC) to identify different types of subcellular structures from the reconstructed Nano-CT greyscale images. Due to the different molecular compositions and densities of organelles, they exhibit distinct capacities for soft X-ray absorption when soft X-rays transmit through the samples40. Following the incubation of macrophages with GO/LDL and LDL, we observed LDs inside the cells through TXM contrast 2D imaging (Fig. 3f) and spatial distribution of LDs (Fig. 3g) in single cells. Our findings reinforce the conclusion that the higher hydrophobicity of GOs leads to increased storage of LDs inside cells, consistent with the results obtained from TEM, confocal images, and high-content analysis. In contrast, pure GOs were taken up by macrophages, but they did not induce obvious LDs in the cells (Supplementary Figs. 11, 12), suggesting that the GO/LDL complex, rather than the nanosheets themselves, played a crucial role in regulating lipid metabolism. The higher uptake of the L-GO/LDL complex (Fig. 3a, b) likely explains why hydrophobic L-GO induced more storage of LDs.
Surface hydrophobicity of GOs mediating the change in LDL shape and lipid component structure
In our ongoing investigation, we concentrated on the interaction between GOs and LDL to determine if changes in LDL structure affected the uptake of the GO/LDL complex. LDL is an amphiphilic particle composed of phospholipid molecules with hydrophilic phosphate headgroups and hydrophobic aliphatic carbon tails. It also contains a single copy of ApoB-100 protein encircling the surface, along with other hydrophobic lipids such as triglycerides and cholesteryl ester in the core41. ApoB-100 protein plays a crucial role in maintaining the structural integrity of LDL particles and mediating their interactions with the LDL receptor. On the surface of ApoB-100, there is a receptor-binding domain (residue 3359-3367) containing positively charged sequences due to the presence of basic amino acids like L-Arg and L-Lys41,42. This enables LDL to bind to the negatively charged surface of hydrophilic GOs through electrostatic interactions. On the other hand, the hydrophobic tails of unsaturated phospholipids, such as 1-palmitoyl-2-lauroyl-sn-glycero-3-phosphocholine (PLPC) and sphingomyelin (SM), as well as hydrophobic lipids like triglycerides and cholesteryl esters, may interact with the carbocyclic ring of GOs through hydrophobic forces. Previous studies have shown that changes in the shape and structure of LDL, as well as modifications or oxidation of LDL components, can influence its functions, leading to increased cellular uptake of LDL by macrophages43,44,45,46,47. Based on these findings, we hypothesized that GOs with different hydrophilicity would selectively bind to the lipid and protein components of LDL, potentially altering its structure and functions.
To investigate the impact of GOs on the structure of LDL and its protein composition, specifically ApoB-100, we utilized circular dichroism spectroscopy. SR-CD is suited for measuring more detailed changes in protein secondary structures with high sensitivity and a higher signal-to-noise ratio compared to conventional CD34. According to the SR-CD results, the secondary structures of ApoB-100, such as α-helix and β-sheet, were significantly altered by more hydrophilic H-GO/M-GO nanosheets, but not by the relatively hydrophobic L-GO (Fig. 4a–d, Supplementary Table 2). The higher oxidation level of GOs results in more carboxyl groups at the edge, giving them more negative charges. Consequently, hydrophilic GOs readily interact with the positively charged residues of ApoB-100 proteins through electrostatic forces, leading to substantial alterations in secondary structures. Interestingly, in comparison with M-GO and H-GO, the CD spectra of the less hydrophilic L-GO showed negligible change at 208–240 nm, suggesting that ApoB-100 maintained its secondary structures after incubation with L-GO (Fig. 4a, d).
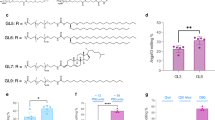
a–c Changes in the secondary structure of apolipoprotein, ApoB-100, after incubating LDL with three types of GOs, as determined by SR-CD. Time-dependent studies are conducted in a mixture of 20 μg/mL GOs with 50 μg/mL LDL, dispersed in phosphate buffer at pH 7.4. d Alterations in the relative content of LDL protein secondary structure treated with GOs at different time points. e Schematic diagram of the LDL receptor binding domain stained by RED-NHS protein labeling kit. Figure 4e Created in BioRender. Wu, J. (2024) BioRender.com/w06z982 released under a CC-BY-4.0 license. f Quantitative analysis of MFI of LDL labeling in the supernatant after incubation with 20 μg/mL GOs for 1 h. Data are presented as mean ± standard deviation of n = 3 biological replicates. Statistical significance is calculated by one-way ANOVA with Tukey’s multiple comparisons test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; n.s., not significant (p > 0.05). Source data are provided as a Source Data file.
To investigate the correlation between the interaction of GOs and ApoB-100 with cellular uptake, it is crucial to determine whether ApoB-100 adsorption on GOs influences its recognition by the LDL receptor and subsequent binding to LDL. To achieve this, we used the RED-NHS protein labeling Kit to label five positively charged residues (two L-lysine and three L-arginine) in the LDL receptor binding domain (RBD, containing 9 residues) (Fig. 4e). Our findings revealed that LDL exhibited the strongest fluorescent intensity, whereas the L-GO/LDL sample displayed a significantly stronger intensity compared to M-GO/LDL and H-GO/LDL. As the hydrophilicity of GOs increased, the labeling efficiency dramatically decreased, indicating that more residues located in the RBD of ApoB-100 were occupied and blocked by hydrophilic GOs due to their strong adsorption with ApoB-100 (Fig. 4f). Consequently, hydrophilic H-GO and M-GO exhibited a preference for binding to RBD residues of ApoB-100, interfering with the binding of LDL to LDLR during cellular uptake of LDL. This finding partially explains why H-GO/LDL and M-GO/LDL induced less formation of LDs from the uptake aspect.
Change in the shape of LDL and the structure of lipid components by GOs as modulated by surface hydrophobicity
The modifications of lipids, shape, and structure of LDL also contribute to increased uptake by macrophages43,44,45,46. Given the strong adsorption of LDL particles on GOs, we used AFM imaging to investigate LDL adsorption profiles on GOs. As shown in Fig. 5a, the average height of native LDL nanoparticles viewed in AFM was 20 nm. After incubating LDL with GOs, LDL particles adhered to the surface of all three types of GOs. Notably, L-GOs induced the aggregation and fusion of LDL, resulting in larger particles, making it difficult to observe LDL particles with their normal size and spherical morphology on L-GOs. This finding suggests that the adsorbed LDL particles significantly altered the shape and boundary of LDL, possibly due to a strong interaction between L-GO and phospholipids, the main component on the outer layer of LDL particles, which maintains the shape of LDL. In contrast, M-GO and H-GO did not induce the fusion of LDL, and the bound LDL particles retained their spherical shape, indicating a weaker interaction with the outer monolayer phospholipid structure of LDL.
a Morphological images of LDL and GO/LDL complexes observed by AFM. Scale bar represents 500 nm. b, c X-ray liquid interface scattering results of structures between phospholipid (SOPC) and GOs. b XRR data of GOs adsorption and the SOPC thickness. c Electron density profiles derived from the XRR data in b. POPC self-assembles to form a monolayer on the air-water surface and GOs at 20 μg/mL are incubated with SOPC monolayer under a surface pressure of 20 mN/m for XRR characterization. d–f Molecular interaction between graphene or GO nanosheets and a model lipid droplet (representing LDL particles) based on coarse-grained molecular dynamics. d Schematic diagrams showing the structure of a lipid droplet. The outer layer of the lipid droplet includes POPC monolayer (in cyan) and free cholesteryl (in orange). The inner components contain cholesteryl ester (in yellow) and triglyceride (in red). e Snapshots of the lipid droplet structure depositing on graphene and GOs. For the GO models, the hydrophilic carbon is assigned as carbonyl groups, while the hydrophobic carbon is assigned as sp2 and sp3 carbon. For graphene (GRA), GO1, and GO2, the percentage of carbonyl carbon among all carbons is assigned as 0, 30%, and 50% beads, correlating to the hydrophilicity of pristine graphene, L-GO, and H-GO, respectively. f Illustration of POPC lipids at the edge of lipid droplet bound to graphene/GOs. Each panel contains 40 POPC molecules. g–j Density profiles of the lipids along the vertical direction to graphene/GOs. k–n Interaction energy between the lipid components and the graphene/GOs. Each simulation is performed for at least 2 μs when the interaction energy between POPC and the graphene/GOs increases slowly, indicating that no more POPC molecules spread on the surface. Source data are provided as a Source Data file.
Subsequently, we employed X-ray liquid interface scattering techniques to verify the effect of GOs on LDL structure. X-ray reflectivity (XRR) measurement was used to study the interaction between GOs and a model SOPC monolayer phospholipid at the air-water interface. As shown in Fig. 5b, the Qz value, corresponding to the minimum of the reflectivity curve, under constant surface pressure (20 mN/m), shifted from 0.28 Å−1 to 0.35 Å−1 (Orange) after incubation with 20 μg/mL of L-GO in water, compared to the SOPC monolayer (grey), indicating that L-GO decreased the thickness of the SOPC monolayer (Supplementary Table 3). Moreover, the concentration-dependent change in the SOPC monolayer upon its interaction with three GOs was also observed (Supplementary Fig. 14 and Supplementary Table 3). However, with a higher level of hydrophilicity, the changes in QZ values for the systems of SOPC incubating with M-GO (blue) and H-GO (pink) were less pronounced than with L-GO (orange), suggesting that the more hydrophilic H-GO/M-GO did not significantly affect the structure of the SOPC monolayer. The electron density profile was consistent with the thickness variation (Fig. 5c). In summary, our results demonstrate that the physical adsorption of LDL on the GO surface can induce structural changes in LDL. Specifically, more hydrophilic H-GO/M-GO have a preference for binding and altering the secondary structures of the ApoB-100 protein, while hydrophobic L-GOs tend to adsorb the phospholipid structure. As a consequence, LDL may fuse with the disintegrated shape of LDL particles after binding to the L-GO surface. In contrast, LDL particles remain more intact on H-GO/M-GO, as observed in the AFM image (Fig. 5a), due to the different binding behaviors compared to L-GOs.
We employed coarse-grained molecular dynamics simulations to investigate the molecular and structural aspects of the interaction between LDL particles and GOs. Due to the extensive unknown regions within the ApoB-100 structure, we simplified the lipid droplet model to simulate how GOs interact with the lipid composition of LDL. For the ApoB-100 protein structure, we used SR-CD to determine the structural changes directly. To model LDL particles, we used a lipid droplet model (Fig. 5d) and chose hydrophobic graphene and relatively hydrophilic graphene oxides (GO1 and GO2) as surfaces. The simulation results showed that the lipid droplet adhered to and spread on the surfaces of all these materials, forming a hemispherical structure after deposition (Fig. 5e, and Supplementary Table 4; the dynamic processes are presented in Supplementary Movies 6–11). The major binding sites between GOs and the model lipid droplet were the planar carbon rather than the boundary carbon. Notably, as the surface hydrophobicity increased, the spreading area of lipid nanodroplets on the GO surface also increased, leading to more contact between neighboring lipid droplets and fusion between lipid nanodroplets on the graphene. In contrast, the limited spreading area hindered the fusion of neighboring lipid nanodroplets on GO2 (Fig. 5e). Consequently, lipid nanodroplets only formed numerous small hemispherical protrusions on the hydrophilic surface, consistent with observations from AFM images (Fig. 5a). The differences in the spreading area of lipid droplets were influenced by their interaction with the materials, mainly determined by the presence of POPC as the major component on the outer layer (Supplementary Table 4). The simulation provided representative structures indicating the positioning of POPC molecules on the three graphene material surfaces: nearly all the POPC tails were on the hydrophobic graphene surface; a few POPC tails adhered to the GO1 surface, with the remaining POPC tails perpendicular to the surface and their heads adhering to it; on the GO2 surface, only a few POPC tails were present, and most of the POPC molecules were perpendicular to the surface (Fig. 5f). This demonstrated that the lipid nanodroplet tended to interact with the hydrophobic surface through lipid extraction (graphene/GO1), promoting the spreading of POPC on the surface. Conversely, lipid nanodroplets interacted with the hydrophilic surface through adhesion with the polar heads of POPC (GO2). These simulation results were consistent with the XRR findings regarding the changes in the phospholipid monolayer structure (Fig. 5b).
Given the complexity of lipid droplet components, we conducted a thorough analysis of the interactions between graphene/GOs and different lipid components using simulation systems. We observed that most CHYO and TOG molecules were located within 3 nm of the nanomaterial surface, while the majority of the outer POPC and CHOL lipids adhered directly to the graphene/GO surfaces (within 1 nm above the surface, as shown in Fig. 5g–j). This suggested the formation of a monolayer at the interface of the lipid droplet and the graphene/GO, primarily consisting of POPC and CHOL molecules. We next quantified the molecules interacting with the graphene/GO surface and detailed the proportional composition of various lipid components on the nanomaterial’s surface (Supplementary Fig. 15). Our initial findings indicated a predominance of POPC molecules across all surfaces (Supplementary Fig. 15a), which exhibited much higher interaction energies than other molecules (Fig. 5k). Additionally, we noted an increase in the number of lipids in correlation with surface hydrophobicity (Supplementary Fig. 15a), suggesting that increased hydrophobicity facilitated LDL spreading on the nanosheets’ surface. Furthermore, we observed a tendency where more hydrophobic molecules (CHYO, TOG) showed a propensity to adsorb on the hydrophobic graphene surface, while more hydrophilic molecules (POPC, CHOL) were inclined to adsorb on the comparatively hydrophilic GO surface (Supplementary Fig. 15b). As a result, the interior CHYO and TOG lipids, being highly hydrophobic, showed a strong interaction only with the hydrophobic graphene surface and a much weaker interaction with the GO surface (Fig. 5l, n). The hydrophobicity of the materials significantly influenced their interactions with the lipids, thereby affecting the configuration of the lipids and the lipid redistribution at the lipid droplet-water interface. For example, upon the adsorption of the lipid droplet on the material surface, more C = C bonds of the interior CHYO molecules were exposed to the surrounding water (Supplementary Fig. 16), making them more susceptible to oxidation. The possibility of C = C bond oxidation increased with the increasing hydrophobicity of the material surface, correlating with the lipid peroxidation results (Fig. 6d).
a Detection of carbon radicals present in the GOs using Electron Spin Resonance (ESR). b, c Comparison of the pro-oxidant capacity of three GOs in promoting oxidation reactions. b Measurement of the MFI of ROS using DCF fluorescence spectra. c Quantitative analysis of DCF fluorescence intensity to assess ROS generation by three types of GO suspensions. Data are presented as mean ± standard deviation of n = 3 biological replicates. d Impact of three GOs on the lipid peroxidation of LDL under oxygenated or deoxidated conditions. Malondialdehyde (MDA). Data are presented as mean ± standard deviation of n = 4 biological replicates. e Oxygen consumption determined by ESR after incubating GOs and LDL for 0 min and 3 h. Statistical significance was calculated by one-way ANOVA with Tukey’s multiple comparisons test (c) and two-way ANOVA with Tukey’s multiple comparisons test (d). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; n.s., not significant (p > 0.05). Source data are provided as a Source Data file.
Based on our simulations and AFM images (Fig. 5), the differences in the uptake of the graphene-lipoprotein complex and the GO/lipoprotein complex may arise from two reasons. First, the curvature of the adsorbed lipoprotein on the GOs surface is much higher than that on the graphene surface, which could hinder the binding of ApoB-100 with cellular receptors. Second, the exposed surface region of apolipoprotein is hydrophilic and would prefer to interact directly with the hydrophilic surface rather than the hydrophobic surface of GOs. Therefore, when the lipoprotein adheres to a hydrophobic surface, the ligands of apolipoprotein are more likely to be exposed to the surrounding water, potentially enhancing cellular recognition.
Accelerated oxidation of LDL by GOs and chemical mechanism
The mechanisms underlying the interaction between GOs and lipid membranes have been suggested to involve the chemical oxidation of lipids following physical contact and disruption48,49. However, it remains unclear whether the physical interaction between LDL particles and GOs is responsible for LDL oxidation. Therefore, our primary objective was to determine whether GOs have the ability to oxidize lipids. Figure 6a illustrates that GOs possess innate carbon radicals, as measured by ESR. Furthermore, we observed that GOs with higher hydrophobicity had more carbon radicals, indicating their capacity for chemical oxidation. To ascertain whether GOs-mediated oxidation occurs via oxidative stress, we incubated GOs with H2DCFDA, an ROS indicator. The results showed that GOs were capable of oxidizing H2DCFDA, converting it into DCF with high-intensity fluorescence (Fig. 6b). Additionally, GOs with greater hydrophobicity facilitated the conversion of H2DCFDA into DCF (Fig. 6b, c), confirming that the chemical oxidation of GOs originated from oxidative stress. Considering the physical contact between GOs and LDL, we hypothesized that GOs oxidize LDL particles after interaction due to their oxidizability.
To assess the oxidative degree of LDL, we measured the malondialdehyde (MDA) content, an important parameter reflecting the potential peroxidant capacity of biomolecules. Initially, we incubated GOs with LDL under aerobic conditions for 24 h. As expected, GOs enabled the oxidation of LDL under aerobic conditions, while LDL alone did not exhibit oxidation. Additionally, GOs with higher hydrophobicity led to the production of higher MDA levels in LDL due to their increased carbon radicals. To investigate the impact of oxygen availability on LDL oxidation, we conducted experiments under anoxic conditions using nitrogen gas. Under anoxic conditions, lower levels of MDA were produced compared to aerobic conditions, regardless of the oxidized degree of GOs. These findings indicated that the oxidation of LDL was inhibited in the absence of oxygen (Fig. 6d). Subsequent ESR oxygen consumption experiments confirmed these results (Fig. 6e). Notably, GOs with higher hydrophobicity resulted in increased oxygen consumption after incubation with LDL for 3 h. In conclusion, our results demonstrate that GOs, particularly those with higher hydrophobicity, accelerate the oxidation of LDL due to their endogenous carbon radicals under aerobic conditions, leading to the generation of more ROS.
We also found the enrichment of HDL (Apolipoprotein AI, II, and IV) in the protein corona of GOs (Fig. 2c, Supplementary Figs. 17a-c). However, its affinity was 100 times lower than that of LDL with GOs (Supplementary Fig. 18), which is one of the impact factors on the oxidation of HDL by GOs. After incubation of HDL proteins with GOs for 3 h and 8 h, there was a slight change in the secondary structure of protein components of HDL based on SR-CD (Supplementary Fig. 17d, e and Supplementary Table 5). Moreover, we incubated HDL with GOs for 24 h and measured MDA of the mixture. The results revealed that HDL did not suffer from oxidation after incubation with GOs (Supplementary Fig. 17f). One reason may be the binding affinity of GOs with HDL was 100 folds lower than that of GOs with LDL. In addition, compared to the LDL and GOs interaction, the difference in the states of HDL may be ascribed to different sizes, molecular structures and compositions of proteins including multiple types of apolipoproteins, enzymes, lipid transfer proteins, acute-phase proteins, and others between LDL and HDL. Additionally, HDL possesses more apolipoproteins and less lipids and cholesterol than LDL, which endows HDL with high capability of anti-oxidation50.
Mechanism about lipid metabolism regulation after interaction with GO/LDL complexes in vitro and in vivo
As demonstrated, GOs with varying hydrophilicity can selectively alter the structure of LDL’s phospholipid monolayer and apolipoprotein ApoB-100, influencing their behavior in macrophages. Next, we explored the mechanism of how GO/LDL modulates lipid metabolism. LDL participates in macrophage lipid metabolism, and activated macrophages can be further stimulated by chemically or biologically modified LDL via ROS, RNS, or enzymatic modification24,25,41,43,45,51,52 within the vessel. Additionally, LDL particles undergo fusion and aggregation20,21,22,47,53,54,55 with physical changes in their structure and composition, leading to disturbed lipid metabolism and eventual development of inflammatory atherosclerosis24,51. In our study, the physical adsorption of LDL on GOs resulted in structural and property changes of LDL, while GOs themselves also promoted the lipid peroxidation of LDL chemically. Therefore, we hypothesized that the adsorbed LDL on GOs might induce dysregulation of lipid metabolism.
To understand how GOs modulate selective lipid metabolism fates, we examined the expression of genes and proteins responsible for LDL uptake, transport, efflux, and storage using Western Blotting and real-time PCR assays (Fig. 7a, b, Supplementary Figs. 19, 21). To determine the mechanism of internalization of GO/LDL complexes, we first used the Biolayer interferometry (BLI) technique to analyze the interaction between GO/LDL complexes and LDL receptor proteins or CD36 proteins (Supplementary Fig. 22). Results showed that GO/LDL complexes had higher KD values when interacting with CD36 protein than with LDL receptor (Fig. 7c, d). GO/LDL complexes could bind to more CD36 proteins than to the LDL receptor proteins, suggesting rapid internalization into THP-1 macrophages through scavenger receptor CD36, explaining why THP-1 macrophages internalized more GOs in the presence of LDL. To confirm this, we blocked the CD36 and LDLR receptor by their antibodies and found that macrophages internalized LDL and GO/LDL mainly via CD36, and LDLR (Supplementary Fig. 23), suggesting both CD36, and LDLR contributed to the recognition by macrophages and cellular uptake of LDL and GO/LDL.
a The change in mRNA level of lipid metabolism when macrophages are treated with LDL, L-GO/LDL, M-GO/LDL, and H-GO/LDL for 6 h according to the fatty acid metabolism PCR Array. b Effects of LDL and the GOs/LDL complexes on protein levels of cholesterol metabolism. All blots are performed with three independent replicates for biological experiments. Original and uncropped blots are shown in Supplementary Fig. 20. Binding affinity of GOs/LDL complexes with CD36 receptors (c) and with LDL receptors (d) in vitro by BLI. e Schematic illustration of the different pathways of cellular uptake and export of cholesterol when macrophages are treated with LDL and hydrophilic GO/LDL complexes. Figure 7e Created in BioRender. Wu, J. (2024) BioRender.com/j78q721 released under a CC-BY-4.0 license. Accumulation of lipid droplets in macrophages after knockdown of the expression of ABC transporter using ABCA1 siRNA (f) and ABCG1 siRNA (g) when treated with LDL and GO/LDL complexes, visualized by confocal imaging. Red color indicates lipid droplets, while blue indicates nucleus. Three biological experiments are repeated independently. The scale bar represents 35 μm. Source data are provided as a Source Data file.
After translocating into the cytoplasm, LDL was esterified into cholesterol ester and stored in lipid droplets in the presence of Acyl coenzyme A-cholesterol acyltransferase (ACAT)51,56. Simultaneously, Acetyl CoA acyltransferase 2 (ACAA2) facilitated fatty acid β-oxidation and fatty acid degradation crucial for lipid metabolism and adipocyte differentiation57. Comparing LDL and L-GO/LDL with H-GO/LDL and M-GO/LDL, we observed that H-GO/LDL and M-GO/LDL significantly decreased the expression of ACAA2 and ACAT1 (Fig. 7a, b). This suggests that hydrophilic GOs weakened the macrophages’ ability to esterify cholesterol into cholesterol esters, effectively inhibiting lipid droplet formation.
Furthermore, stored lipid droplets are lipolyzed by neural cholesterol ester hydrolase (nCEH) into free cholesterol, which can be exported from macrophages by ATP-binding cassette A1 (ABCA1) and ATP-binding cassette G1 (ABCG1)51. We also observed that LDL treatment downregulated the expression of nCEH and ABCA1 levels (Fig. 7a, b), indicating that neutral cholesterol ester from LDL was stored in lipid droplets and retained in the cytoplasm after LDL treatment (Fig. 3c–g). Compared to LDL and L-GO/LDL, H-GO/LDL upregulated the levels of ABCG1 and nCEH at the mRNA level. The higher the hydrophilicity of GOs, the more upregulation of these gene or protein expressions (Fig. 7a). Consequently, H-GO can significantly alleviate lipid accumulation in macrophages (Fig. 3d–g, Supplementary Fig. 12). In conclusion, our findings demonstrate that GOs can influence the fate of lipid metabolism in macrophages. Specifically, hydrophilic GOs may alleviate foam cell formation by upregulating the expression of lipid transport and hydrolysis-related proteins such as ABCA1, ABCG1, and nCEH, compared to cells treated with LDL and hydrophobic GOs (Fig. 7e).
Significantly, ABC transporters play crucial role in cholesterol efflux, and their expression is further regulated by PPARγ and LXRα58,59. To assess the functional role of ABC transporters, we initially examined the protein levels of ABC transporters, PPARγ and LXRα. As shown in Fig. 7b and Supplementary Fig. 21, only LDL treatment of THP-1 macrophages resulted in the downregulation of PPARγ and LXRα expression, leading to the suppression of ABCA1 or ABCG1 and the inhibition of cholesterol efflux. Conversely, treatment with GOs/LDL complexes upregulated the expression of PPARγ and LXRα. To further verify the role of ABC transporters in foam cell formation, we used siRNA to knock down the expression of ABCA1 and ABCG1 (Supplementary Fig. 24), followed by treatment with LDL or GOs/LDL to assess the lipid accumulation in THP-1 macrophages. The results indicated no significant difference in the fluorescent intensity of GOs determined by laser reflectivity imaging (Supplementary Figs. 10, 25), suggesting that neither ABCA1 nor ABCG1 predominantly mediates the uptake of GOs/LDL in macrophages. However, lipid droplet analysis indicated that ABCA1 and ABCG1 play major roles in lipid accumulation, leading to the formation of lipid droplets due to reduced cholesterol efflux after siRNA treatment for 48 h, followed by treatment with of LDL or GOs/LDL complexes (Fig. 7f, g).
As we know, the accumulation of cholesterols contributes to the differentiation of macrophages into pro-inflammatory M1-like type, leading to endoplasmic reticulum stress, inflammatory cytokine production, and apoptosis60. However, the activation of LXR-targeted gene expression will contribute to enhancing cholesterol efflux and acquisition of anti-inflammatory properties in M2-like type macrophages61. We thus studied the profiles of macrophage polarization after treatment with LDL or GOs/LDL complexes. The results indicated that macrophages tend to differentiate into anti-inflammatory M2-like types (CD206+ in F4/80+/CD11b+ subset) after treatment with GOs/LDL complexes, comparing to LDL treatment. Moreover, GOs with a higher hydrophilicity induced more polarization of macrophages in other words, more production of M2-like ones (Supplementary Figs. 26-28). This can be explained by the fact that GOs/LDL complexes upregulate the expression of LXR to enhance the anti-inflammatory properties in macrophages. As a result, the level of lipid metabolism modulated by GOs/LDL complexes affects the activation of macrophages and their functions.
We next wonder whether GO/LDL complexes could impact lipid metabolism in mice in vivo, taking into account the potential hemolytic effects induced by pure GOs in vivo. To mitigate this effect, we preincubated GOs and LDL. As shown in Fig. 8a, 12 weeks old mice received repeated injections of LDL or GOs/LDL through the tail vein, with doses of 100 μg of GOs and 200 μg of LDL, once every 3 days for a total of five injections in total. Seven days after the final injection, the mice were euthanized, and blood and livers were harvested for measurement of total cholesterol, triglycerides, LDL-cholesterol, and HDL-cholesterol to evaluate lipid metabolism. Injection of LDL five times exacerbated the higher levels of total cholesterol, triglycerides, and LDL-C in both blood serum and liver. In contrast, GO/LDL complexes appeared to reduce lipid accumulation in blood and liver (Fig. 8b–d, f–h). The oxidation degree of GO, correlated with the decrease in lipid storage, as evidenced by the higher level of HDL-C involved in lipid reverse transport (Fig. 8e, i). Thus, analysis of hepatic and blood-lipid levels indicated that GO/LDL complexes could regulate hyperlipidemia and disturbed lipid metabolism triggered by LDL in vivo.
a Overview of timeline for treatment with LDL or GOs/LDL in C57 BL/6 J mice. Figure 8a Created in BioRender. Wu, J. (2021) BioRender.com/n18b700 released under a CC-BY-4.0 license. Effects of LDL or GOs/LDL on serum total cholesterol (TC) levels (b), triglyceride (TG) levels (c), LDL-C levels (d), and HDL-C levels (e). Effects of LDL or GOs/LDL on liver TC levels (f), TG levels (g), LDL-C levels (h), and HDL-C levels (i). Data represent mean ± standard deviation of n = 7 biological replicates. Statistical significance was calculated by one-way ANOVA with Tukey’s multiple comparisons test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; n.s., not significant (p > 0.05). Source data are provided as a Source Data file.
In summary, our study underscores the crucial role of surface chemistry in modulating intracellular lipid metabolism during interactions between nanomaterials and macrophages. GOs serve as a valuable model for hydrophobic carbon nanomaterials, illustrating how surface chemistry influences the formation of a specific lipoprotein corona. This, in turn, leads to changes in the structure and functions of lipoproteins, ultimately regulating lipid metabolism at the molecular interface and cellular level. By precisely controlling the oxidation degrees and surface functional groups of GOs, such as carboxyl and hydroxyl groups, distinct hydrophobicity/hydrophilicity can be achieved. This enables hydrophilic or hydrophobic GOs to effectively enrich lipoproteins on their surface, resulting in different corona compositions of apolipoproteins in the presence of serum.
Hydrophobic GOs tend to adsorb more ApoB-100 (LDL), while hydrophilic nanosheets preferentially bind more ApoA-1 (HDL) proteins. Further investigations have revealed that both types of GOs can adsorb a large number of LDL, but they induce different behaviors in lipid metabolism within macrophages, including lipid accumulation and efflux. Through the use of SR-CD, XRR, and MD simulation, we have elucidated the underlying mechanisms. Hydrophobic GOs preferentially bind to the phospholipids on LDL’s outer monolayer, altering the structures and morphology of LDL and accelerating lipid peroxidation. Conversely, hydrophilic GOs tend to adsorb onto the protein component of LDL, ApoB-100, effectively blocking the sites where LDL binds to the LDL receptor (LDLR). These distinct nano-bio interactions ultimately determine the uptake fate of GO/LDL by activated macrophages and their ability to induce lipid storage and efflux within the macrophages (Fig. 9). Specifically, hydrophobic GOs with LDL are more readily internalized, leading to lipid storage and the formation of foam cells. Conversely, the complex formed by hydrophilic GOs and LDL is less internalized by macrophages, but it promotes the upregulation of lipid hydrolysis and efflux-related gene expression, facilitating the removal of lipids from macrophages. In conclusion, our findings highlight the pivotal role of nanomaterials surface chemistry in regulating the corona composition of lipoproteins and selectively altering the interfacial structures of low-density lipoprotein through preferential interactions with lipids or protein components. This, in turn, significantly affects the lipid metabolism of macrophages. Understanding the pathological effects of nanomaterials caused by surface chemistry provides valuable insights for biomedical applications, particularly in the modulation of lipid metabolism.
a Schematic illustration of how graphene oxide nanosheets determine lipid metabolism in macrophages. b Influence of surface properties of GOs on the structure and physiological function of bound LDL. GOs with distinct surface hydrophilicity/hydrophobicity exhibit preferred modes of binding the components of LDL such as lipoprotein ApoB-100 and phospholipids, which ultimately affect lipid metabolism in macrophages. Upon absorption of LDL, hydrophobic GOs preferentially bind lipid components, disrupting the integrity of LDL phospholipid monolayer, accelerating the fusion of LDL and lipid peroxidation, and inducing foam cell formation. In comparison, hydrophilic GOs prefers to bind ApoB-100, altering its secondary structures, which effectively blocks LDL receptor binding and cellular uptake, preventing the abnormal lipid metabolism in macrophages. Figure 9a Created in BioRender. Wu, J. (2024) BioRender.com/r16c552 released under a CC-BY-4.0 license. Figure 9b Created in BioRender. Wu, J. (2024) BioRender.com/o28z461 released under a CC-BY-4.0 license.
Methods
Preparation of GOs
Single-layer GO with varying oxidative degrees was prepared using modified Hummer methods37. Briefly, 0.5000 g of graphite was stirred in 60 mL of concentrated sulfuric acid (H2SO4) for 6 h at room temperature. Subsequently, the mixture was cooled to 0 °C, and KMnO4 powder was added, followed by another 2 h reaction at 37 °C. Afterwards, 240 mL of distilled water was added to the mixture at room temperature, and a 30% H2O2 solution was introduced to completely remove unreacted KMnO4. The resulting GO product underwent several washes with 5% HCl solution and was dialyzed in diluted water for a week to liminate any remaining saline and acid. Finally, the graphite oxide solution was ultrasonically exfoliated for 5 h (using a cycle of 5-s on and 5-s off, at a power of 400 W) and then centrifuged at 13,800 g to obtain the GO solution. The oxidative levels of the GOs were determined based on the amount of added KMnO4, and the final products were named L-GO, M-GO, and H-GO, corresponding to additions of 1 g, 2 g, and 3 g of KMnO4, respectively.
Preparation for serum proteomics of GOs
Human blood was collected from various healthy volunteers at the Shenzhen Blood Center with the donors’ consent. The collection of human blood samples was approved by Shenzhen Blood Center’s and Shenzhen Institute of Advanced Technology’s ethics committee (ethical approval number: SIAT-IRB-230415-H0648) and all relevant ethical regulations were followed. The proteomics samples were prepared following established procedures described in previous publications. The blood was drawn into serum collection tubes (BD Vacutainer™). After allowing the blood to clot at room temperature for 30 min, human serum was obtained by centrifuging the samples at 1100\(\times\)g for 10 min. The collected serum was then aliquoted and stored at −80 °C until further use in the preparation of GOs and serum proteomics samples. Prior to incubation with the GOs samples, the serum underwent a second centrifugation step at 10,600\(\times\)g for 10 min at 4 °C to separate protein aggregates from the serum. Subsequently, three different GOs samples (at 0.5 mg/mL) were incubated with 10% human serum in PBS for 1 h at 37 °C under continuous stirring. After the incubation, the GOs/protein complexes were separated from the serum by centrifugation at 21,600\(\times\)g for 15 min at 4 °C. The supernatant was discarded, and the pellet was washed with ice-cold PBS. This washing process was repeated three times, and the pellets at the bottom of the tubes were resuspended in ultra-pure water and stored at −20 °C for LC-MS/MS analysis. The protein concentration of the mixture was determined using the BCA assay. To elute corona proteins from the GOs samples, 100 µL of 5% SDS was added to the GOs/corona complexes, and the mixture was heated at 95 °C for 15 min. Finally, the samples were centrifuged at 10,600\(\times\)g for 15 min and digested for label-free LC-MS/MS analysis.
LC-MS/MS analysis
The digested products were separated using a Thermo Ultimate 3000 nano-UPLC system with a flow rate of 0.300 µL/min for a 120 min gradient elution, which was directly interfaced with a Thermo Fusion LUMOS mass spectrometer. The system employed an Acclaim PepMap RSLC column (75 µm ID, 250 mm length, C18), with mobile phase A (0.1% formic acid) and mobile phase B (80% acetonitrile and 0.1% formic acid). The Fusion LUMOS mass spectrometer operated in data-dependent acquisition mode using Xcalibur 4.1.50 software, starting with a single full-scan mass spectrum in the Orbitrap (300–1800 m/z, 60,000 resolution), followed by 20 data-dependent MS/MS scans. MS/MS spectra from each LC-MS/MS run were then searched against the selected database using Proteome Discoverer software (Thermo Scientific, ver. 2.2). Identified corona proteins on GOs samples were manually assigned by searching the human taxon in the UniProt database. Protein abundance was calculated based on label-free quantification intensities relative to the total sum of protein intensities for each sample (three biological replicates).
Observation of lipid accumulation in THP-1 macrophages by TXM imaging
Single-cell imaging was conducted to observe organelles and lipid droplets within THP-1 derived macrophages. The cryo-soft X-ray transmission microscope (TXM) at beamline BL07W of the National Synchrotron Radiation Laboratory (NSRL, Hefei, China) was utilized for sample observation. THP-1 cells were seeded and allowed to differentiate into macrophages for 16 h in the presence of 100 ng/mL PMA. The macrophages adhered to a nickel grid with a carbon-free supporting film (ZhongJingKeYi Technology Co., Ltd., China). Subsequently, the macrophages were treated with fresh medium containing 20 μg/mL GOs, 50 μg/mL LDL, and the GO/LDL complexes for 24 h, respectively. Following this, the cells were washed three times with PBS solution and fixed with a 4% paraformaldehyde solution for 20 min. Finally, the cells on the supported nickel grids were immersed in liquid ethane and inserted into a sample holder in liquid nitrogen, which was then transferred to the TXM chamber. Using a soft X-ray beam at 520 eV, the cells of interest were focused, and their images were magnified by a zone plate and an objective, then captured by a CCD camera with a 13-μm field of view and a 30 nm spatial resolution. The cells were rotated from -60° to +60°, and a series of projected images were sequentially collected for 121 times, with 1-degree intervals and an exposure time of 3 s. The tilt series of images were aligned using XMController, and 3D tomographic reconstruction was performed on these images using XMReconstruction and visualized. Because various organelles contain quite different molecular compositions with different matter density, lipid droplets and other organelles exhibit distinct capacity for X-ray absorption when soft X-ray transmit the samples40. This characteristic enables various organelles to exhibit distinct coefficients for X-ray absorption. To distinguish the organelles and lipid droplets, we thus used linear absorption coefficient (LAC) to identify the different types of subcellular structures from the reconstructed Nano-CT greyscale images. Segmented cellular structures, such as cytoplasm, organelles, and lipid droplets, were identified and labeled in red, cyan, and yellow, respectively, based on the difference in their LAC, via Amira (FEI Company).
Assessment of the interaction of GOs with the phospholipids
The X-ray reflectivity (XRR) experiment was conducted using a liquid surface reflectometer on the ChemMatCARS 15-ID-C beamline at the Advanced Photon Source, Argonne National Laboratory62,63,64,65. Incident X-rays with an energy of 10 keV and a wavelength of 1.23894 Å were utilized. For in situ XRR experiments, a 60 mL single-barrier Langmuir trough measuring 7.78 cm (W) × 17.8 cm (L) × 0.3 cm (H) was employed. To investigate the absorption of GOs with the outer phospholipid monolayer of LDL and the interfacial structure, a model phospholipid, 1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine (SOPC), was spread on the surface of water to form a SOPC phospholipid monolayer. Specifically, 1 mg/mL of SOPC phospholipid dissolved in chloroform was gently dropped onto the water’s surface using a microsyringe. Within 10 min, the SOPC phospholipids spread and formed a monolayer after the chloroform volatilized. A surface pressure of 20 mN/m was maintained using a barrier, mimicking the physiological state of biological membranes66. Next, 20 μg/mL of well-dispersed GO solutions were injected into the subphase (water) and mixed homogeneously to facilitate the adsorption of GO to SOPC through continuous stirring for 40 min at 120 rpm using a Teflon-coated magnetic bar65,67. The reflectivity signal was detected using a two-dimensional hybrid pixel array X-ray detector (PILATUS 100 K). After data collection, the XRR data were analyzed, and the data fitting was performed based on a model-dependent and iterative fitting procedure68,69.
Molecular dynamic simulation system for the interaction of graphene/GOs with a lipid nanodroplet model
All the coarse-grained molecular dynamics (CGMD) simulations were performed using the GROMACS package with the modified MARTINI 2.0 force field70,71. The CG model for graphene/GOs was created from the atomistic model using a 4-to-1 mapping scheme, preserving their hexagonal lattice geometry72. According to the MARTINI force field, the hydrophobic beads of GOs were represented as C4, while the hydrophilic beads were represented as P1. To prevent water freezing on the substrate surface during the simulations, the CSJ water model was utilized73,74. To mimic LDL, mainly composed of lipids and occupying a significant the surface area, a lipid droplet model was used with a similar lipid composition and surface properties75,76. Each lipid droplet consisted of 1,352 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC), 600 cholesterol (CHOL), 1,400 cholesteryl esters (cholesteryl oleate, CHYO), and 170 triglycerides (glyceryl trioleate, TOG)75,76. The lipid droplet was equilibrated in water and placed approximately 1 nm above the surface of the graphene or GOs as the initial setup. The total system size was 55 × 55 × 27 nm3, containing 883,715 CG beads. During the simulations, the system was maintained at a temperature of 310 K using the NVT ensemble. Temperature boundary conditions were set using the Nose-Hoover algorithm, with the water and other molecules independently coupled with relaxation times of 0.20 and 1.00 ps, respectively. The graphene/GOs substrates were considered fixed by freezing the position of the beads. The time step for all simulations was 10 fs, and the nonbonding pair list was updated every 5 steps with a neighboring search cutoff of 1.60 nm. Visulazation of MD simulation was used VMD software (version 1.9.3).
Animal experiments
Male C57BL/6 J mice (12 weeks old, 20–25 g) and male BALB/c mice (10–12 weeks old) were purchased from SPF Biotechnology Co., Ltd. (Beijing, China). All animal experiments were approved by Institute of High Energy Physics’ ethics committee (ethical approval number: IHEPLLSC) and all relevant ethical regulations were followed. The mice were kept in the specific pathogen-free (SPF) animal facility in a temperature-controlled (22 °C) and a strict 12-h light cycle environment, in which mice were fed with a standard diet and water with 60% relative humidity, with the cages changed every 2 days. After an adaptation period of 7 days, 7 mice in each group were repeatedly injected with LDL or GOs/LDL through the tail vein at 100 μg of GOs and 200 μg of LDL once every 3 days for five injections in total. Mice in control groups were injected with the same volume of saline intravenously. At days 7 after the final injection, the mice were sacrificed to harvest blood and livers for measurement of total cholesterol, triglyceride, LDL-cholesterol, and HDL-cholesterol.
Blood samples were collected at room temperature for 3 h and centrifugated at 1,200\(\times\)g for 15 min. Serum in supernatant were obtained and 10 μL of serum sample was mixed with 1 mL of working reagent and incubated at 37 °C for 10 min, following by measurement of OD value under 500 nm according to the manufacturer’s protocol. Positive control was conducted by mixing 10 μL of standard cholesterol solution (6.56 mM) or triglyceride solution (2.39 mM) with 1 mL of working reagent and incubated at 37 °C for 10 min. Negative control was conducted by mixing 10 μL of PBS solution with 1 mL of working reagent and incubated at 37 °C for 10 min.
For measurement the LDL-cholesterol (4.1 mM) and HDL-cholesterol (1.32 mM) in serum sample, 10 μL of serum samples were first mixed with 750 μL of reagent 1 and incubated at 37 °C for 5 min and corresponding OD values were recorded under 600 nm and set as A1. Then 250 μL of reagent 2 was added into above solution and continued to incubate for another 5 min, following by measurement of OD values under 600 nm and set as A2. The OD values of samples were calculated as \(\triangle\)A = A2-A1. Positive control was conducted by mixing 10 μL of standard LDL cholesterol solution (4.1 mM) or HDL cholesterol solution (1.32 mM) with 750 μL of reagent 1 and 250 μL of reagent 2. Negative control was conducted by using 10 μL of PBS solution. Liver samples were weighed and homogenized with icy PBS solution followed by sonication and centrifugation at 600\(\times\)g at 4 °C for 15 minutes to collect the supernatant. The procedure of OD values measurement was the same as blood samples.
Statistical analysis
The results are presented as mean values from replicate experiments or samples in one representative experiment, as indicated in the figure legends. Statistical significance was determined using the two-tailed Student’s t-test for comparisons between two groups, and one- or two-way ANOVA with Tukey’s or Sidak’s multiple comparisons test. Statistical analysis was performed using GraphPad Prism and p < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data generated in this study is available with the article, Supplementary Information, Supplementary Data files and Supplementary Movies. Source data is available for Figs. 1–8 and Supplementary Figs. 1-3, 9, 12, 14-24, and 28 in the associated source data file. The proteomics data generated by mass spectrometry, including raw data and search results have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier: PXD044475. The human proteomic dataset is available on the UniProt website (https://www.uniprot.org). The identified protein corona parameters, including relative abundance, accession code, enrichment factors (in Fig. 2, Supplementary Figs. 4, 5) generated in this study are presented in Supplementary Data 1. The identified coronal lipid components, including relative abundance, volcano plots, heatmaps, PCA analysis results (in Supplementary Figs. 6-8) generated in this study are presented in Supplementary Data 2. For clustering heatmaps, the data were normalized using z-scores of the intensity areas of differential metabolites and were plotted by Pheatmap package in R language (version 4.0.5). The secondary structure of Apo B-100 was analyzed using CDtool software (http://cdtools.cryst.bbk.ac.uk). The lipid droplets in macrophages visualized by TXM and Nano-CT are presented in Supplementary Movies 1-5. The simulation results are presented in Supplementary Movies 6–11. The primer sequences for cholesterol metabolism-related genes are listed in Supplementary Data 3. The schematic illustration of the ball-and-stick model of GOs (in Fig. 1a) were visualized by ChemBioDraw software (version 14.0.0.117). Other illustrations presented in the manuscript (in Figs. 4e, 7e, 8a and 9) were created in BioRender.com released under a CC-BY-4.0 license. Visualization of MD simulations (in Fig. 5d, e) is rendered by VMD software (version 1.9.3). Source data are provided with this paper.
References
Cedervall, T. et al. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl Acad. Sci. USA 104, 2050–2055 (2007).
Ke, P. C. et al. A decade of the protein corona. ACS Nano 11, 11773–11776 (2017).
Walkey, C. D. & Chan, W. C. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 41, 2780–2799 (2012).
Hadjidemetriou, M. & Kostarelos, K. Nanomedicine: evolution of the nanoparticle corona. Nat. Nanotechnol. 12, 288–290 (2017).
Ngo, W. et al. Identifying cell receptors for the nanoparticle protein corona using genome screens. Nat. Chem. Biol. 18, 1023–1031 (2022).
Cedervall, T. et al. Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew. Chem. Int. Ed. 46, 5754–5756 (2007).
Lundqvist, M. et al. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl Acad. Sci. USA 105, 14265–14270 (2008).
Sacchetti, C. et al. Surface polyethylene glycol conformation influences the protein corona of polyethylene glycol-modified single-walled carbon nanotubes: potential implications on biological performance. ACS Nano 7, 1974–1989 (2013).
Lu, X. et al. Tailoring the component of protein corona via simple chemistry. Nat. Commun. 10, 4520 (2019).
Li, M. et al. Nanoparticle elasticity affects systemic circulation lifetime by modulating adsorption of apolipoprotein A-I in corona formation. Nat. Commun. 13, 4137 (2022).
Liu, K. et al. Multiomics analysis of naturally efficacious lipid nanoparticle coronas reveals high-density lipoprotein is necessary for their function. Nat. Commun. 14, 4007 (2023).
Vaziri, N. D. HDL abnormalities in nephrotic syndrome and chronic kidney disease. Nat. Rev. Nephrol. 12, 37–47 (2016).
Gistera, A. & Hansson, G. K. The immunology of atherosclerosis. Nat. Rev. Nephrol. 13, 368–380 (2017).
Tall, A. R. & Yvan-Charvet, L. Cholesterol, inflammation and innate immunity. Nat. Rev. Immunol. 15, 104–116 (2015).
Rajamaki, K. et al. Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: a novel link between cholesterol metabolism and inflammation. PLoS One 5, e11765 (2010).
Ren, G. et al. Model of human low-density lipoprotein and bound receptor based on cryoEM. Proc. Natl Acad. Sci. USA 107, 1059–1064 (2010).
Kumar, V. et al. Three-dimensional cryoEM reconstruction of native LDL particles to 16A resolution at physiological body temperature. PLoS One 6, e18841 (2011).
Hevonoja, T. et al. Structure of low density lipoprotein (LDL) particles: basis for understanding molecular changes in modified LDL. Biochim. Biophys. Acta 1488, 189–210 (2000).
Griffin, B. A. Lipid metabolism. Surg. (Oxf.) 31, 267–272 (2013).
Hoff, H. F., Zyromski, N., Armstrong, D. & O’Neil, J. Aggregation as well as chemical modification of LDL during oxidation is responsible for poor processing in macrophages. J. Lipid Res. 34, 1919–1929 (1993).
Pentikiiinen, M. O., Lehtonen, E. M. P. & Kovanen, P. T. Aggregation and fusion of modified low density lipoprotein. J. Lipid Res. 37, 2638–2649 (1996).
Lu, M. & Gursky, O. Aggregation and fusion of low-density lipoproteins in vivo and in vitro. Biomol. Concepts 4, 501–518 (2013).
Moore, K. J. & Tabas, I. Macrophages in the pathogenesis of atherosclerosis. Cell 145, 341–355 (2011).
Weber, C. & Noels, H. Atherosclerosis: current pathogenesis and therapeutic options. Nat. Med. 17, 1410–1422 (2011).
Glass, C. K. & Witztum, J. L. Atherosclerosis: the road ahead. Cell 104, 503–516 (2001).
Lusis, A. J. Atherosclerosis. Nature 407, 233–241 (2000).
Lara, S. et al. Identification of receptor binding to the biomolecular corona of nanoparticles. ACS Nano 11, 1884–1893 (2017).
Lara, S. et al. Differential recognition of nanoparticle protein corona and modified low-density lipoprotein by macrophage receptor with collagenous structure. ACS Nano 12, 4930–4937 (2018).
Hochmuth, R. M., Evans, E. A., Wiles, H. C. & Mccown, J. T. Mechanical measurement of red-cell membrae thickness. Science 220, 101–102 (1983).
Wang, L. et al. Revealing the binding structure of the protein corona on gold nanorods using synchrotron radiation-based techniques: understanding the reduced damage in cell membranes. J. Am. Chem. Soc. 135, 17359–17368 (2013).
Leroueil, P. R. et al. Nanoparticle interaction with biological membranes: does nanotechnology present a Janus face? Acc. Chem. Res. 40, 335–342 (2007).
Xiao, M. et al. Monitoring antimicrobial mechanisms of surface-immobilized peptides in situ. Langmuir 34, 2057–2062 (2018).
Wu, L., Zeng, L. & Jiang, X. Revealing the nature of interaction between graphene oxide and lipid membrane by surface-enhanced infrared absorption spectroscopy. J. Am. Chem. Soc. 137, 10052–10055 (2015).
Miles, A. J. & Wallace, B. A. Synchrotron radiation circular dichroism spectroscopy of proteins and applications in structural and functional genomics. Chem. Soc. Rev. 35, 39–51 (2006).
Pattnaik, S., Swain, K. & Lin, Z. Graphene and graphene-based nanocomposites: biomedical applications and biosafety. J. Mater. Chem. B 4, 7813–7831 (2016).
Bitounis, D. et al. Prospects and challenges of graphene in biomedical applications. Adv. Mater. 25, 2258–2268 (2013).
William, S., Hummers, J. & Offeman, R. E. Preparation of graphitic oxide. J. Am. Chem. Soc. 80, 1339–1339 (1958).
Tenzer, S. et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 8, 772–781 (2013).
Varsano, N. et al. Development of correlative cryo-soft X-ray tomography and stochastic reconstruction microscopy. A study of cholesterol crystal early formation in cells. J. Am. Chem. Soc. 138, 14931–14940 (2016).
Liu, J. et al. Quantitative imaging of Candida utilis and its organelles by soft X-ray Nano-CT. J. Microsc. 270, 64–70 (2018).
Segrest, J. P., Jones, M. K., De Loof, H. & Dashti, N. Structure of apolipoprotein B-100 in low density lipoproteins. J. Lipid Res. 42, 1346–1367 (2001).
Law, A. & Scott, J. A cross-species comparison of the apolipoprotein B domain that binds to the LDL receptor. J. Lipid Res. 31, 1109–1120 (1990).
Berliner, J. A., Leitinger, N. & Tsimikas, S. The role of oxidized phospholipids in atherosclerosis. J. Lipid Res. 50, S207–S212 (2009).
Khoo, J. C., Miller, E., McLoughlin, P. & Steinberg, D. Enhanced macrophage uptake of low density lipoprotein after self-aggregation. Arteriosclerosis 8, 348–358 (1988).
Quinn, M. T., Parthasarthy, S., Fong, L. G. & Steinberg, D. Oxidatively modified low density lipoproteins: a potential role in recruitment and retention of monocyte/macrophages during atherogenesis. Proc. Natl Acad. Sci. USA 84, 2995–2998 (1987).
Stocker, R. & Keaney, JohnF. J. Role of oxidative modifications in atherosclerosis. Physiol. Rev. 84, 1381–1478 (2004).
Öörni, K., Pentikäinen, M. O., Ala-Korpela, M. & Kovanen, P. T. Aggregation, fusion, and vesicle formation of modified low density lipoprotein particles molecular mechanisms and effects on matrix interactions. J. Lipid Res. 41, 1703–1714 (2000).
Tu, Y. et al. Destructive extraction of phospholipids from Escherichia coli membranes by graphene nanosheets. Nat. Nanotechnol. 8, 594–601 (2013).
Lu, X. et al. Enhanced antibacterial activity through the controlled alignment of graphene oxide nanosheets. Proc. Natl Acad. Sci. USA 114, E9793–E9801 (2017).
Shah, A. S., Tan, L., Long, J. L. & Davidson, W. S. Proteomic diversity of high density lipoproteins: our emerging understanding of its importance in lipid transport and beyond. J. Lipid Res. 54, 2575–2585 (2013).
Moore, K. J., Sheedy, F. J. & Fisher, E. A. Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol. 13, 709–721 (2013).
Babaev, V. R. et al. Macrophage lipoprotein lipase promotes foam cell formation and atherosclerosis in low density lipoprotein receptor-deficient mice. J. Biol. Chem. 275, 26293–26299 (2000).
Griffin, E. E., Ullery, J. C., Cox, B. E. & Jerome, W. G. Aggregated LDL and lipid dispersions induce lysosomal cholesteryl ester accumulation in macrophage foam cells. J. Lipid Res. 46, 2052–2060 (2005).
Lu, M., Gantz, D. L., Herscovitz, H. & Gursky, O. Kinetic analysis of thermal stability of human low density lipoproteins: a model for LDL fusion in atherogenesis. J. Lipid Res. 53, 2175–2185 (2012).
Bancells, C. et al. Aggregated electronegative low density lipoprotein in human plasma shows a high tendency toward phospholipolysis and particle fusion. J. Biol. Chem. 285, 32425–32435 (2010).
Brown, M. S., Ho, Y. K. & Goldstein, J. L. The cholesteryl ester cycle in macrophage foam cells-continual hydrolysis and re-esterification of cytoplasmic cholesteryl esters. J. Biol. Chem. 255, 9344–9352 (1980).
Zhang, Y. et al. Acetyl-coenzyme A acyltransferase 2 promote the differentiation of sheep precursor adipocytes into adipocytes. J. Cell. Biochem. 120, 8021–8031 (2019).
Chawla, A. et al. A PPARγ-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell 7, 161–171 (2001).
He, X. W. et al. Anti-atherosclerotic potential of baicalin mediated by promoting cholesterol efflux from macrophages via the PPARγ-LXRα-ABCA1/ABCG1 pathway. Biomed. Pharmacother. 83, 257–264 (2016).
Yan, J. & Horng, T. Lipid metabolism in regulation of macrophage functions. Trends Cell Biol. 30, 979–989 (2020).
Hou, P. et al. Macrophage polarization and metabolism in atherosclerosis. Cell Death Dis. 14, 691 (2023).
Parratt, L. G. Surface studies of solids by total reflection of X-rays. Phys. Rev. 95, 359–369 (1954).
Pershan, P. S. Effects of thermal roughness on X-ray studies of liquid surfaces. Colloids Surf. A Physicochem. Eng. Asp. 171, 149–157 (2000).
Schlossman, M. L. et al. A synchrotron X-ray liquid surface spectrometer. Rev. Sci. Instrum. 68, 4372–4384 (1997).
Wang, L. et al. Stability of ligands on nanoparticles regulating the integrity of biological membranes at the nano-lipid interface. ACS Nano 13, 8680–8693 (2019).
Gidalevitz, D. et al. Interaction of antimicrobial peptide protegrin with biomembranes. Proc. Natl Acad. Sci. USA 100, 6302–6307 (2003).
Quan, P. et al. Structure of polymer-capped gold nanorods binding to model phospholipid monolayers. J. Phys. Mater. 4, 034004 (2021).
Meron, M. et al. Following transient phases at the air/water interface. Eur. Phys. J. Spec. Top. 167, 137–142 (2009).
Bu, W. et al. Observation of a rare earth ion-extractant complex arrested at the oil-water interface during solvent extraction. J. Phys. Chem. B 118, 10662–10674 (2014).
Marrink, S. J. et al. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B 111, 7812–7824 (2007).
Van Der Spoel, D. et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005).
Ruiz, L., Xia, W., Meng, Z. & Keten, S. A coarse-grained model for the mechanical behavior of multi-layer graphene. Carbon 82, 103–115 (2015).
Chiu, S.-W., Scott, H. L. & Jakobsson, E. A coarse-grained model based on morse potential for water and N-alkanes. J. Chem. Theory Comput. 6, 851–863 (2010).
Bai, X. et al. Adsorption of phospholipids at the air-water surface. Biophys. J. 117, 1224–1233 (2019).
Vuorela, T. et al. Role of lipids in spheroidal high density lipoproteins. PLoS Comput. Biol. 6, e1000964 (2010).
Koivuniemi, A., Sysi-Aho, M., Oresic, M. & Ollila, S. Interfacial properties of high-density lipoprotein-like lipid droplets with different lipid and apolipoprotein A-I compositions. Biophys. J. 104, 2193–2201 (2013).
Acknowledgements
This work was supported by the National Basic Research Program of China (2021YFA1200900 to C.C. and L.W. and 2020YFA0710702 to L.W.), the National Natural Science Foundation of China (32371469 to L.W., 31971322 to L.W., 32350410425 to D.B., and 22388101 to C.C.), Science and Technology Innovation Project in IHEP (E25459U210 to L.W.), Directional Institutionalized Scientific Research Platform relies on Beijing Synchrotron Radiation Facility of the Chinese Academy of Sciences (E12982U813 to L.W.), Major Instrument Project of the National Natural Science Foundation of China (22027810 to C.C.), Beijing Natural Science Foundation (IS23023 to D.B.), the New Cornerstone Science Foundation (NCI202318) and China Postdoctoral Science Foundation (2023M742048 to D.B.). We thank Scale Biomedicine Technology Co. LTD for help with the lipidomics analysis.
Author information
Authors and Affiliations
Contributions
The conception were proposed and experiments were designed by L.W., J.W., and C.C. and most experiments were performed by J.W. assisted by L.W. and D.B. Simulation was performed by X.B. and G.H. GO were prepared and characterized by L.Y. LDL-GO interaction and LC-MS/MS experiments for proteomics and lipidomics were performed and analyzed by J.W., D.B., S.L., S.Y., J.Wang, Y.Chong, R.C and Y.L. In vivo evaluation for lipid metabolism was done by J.W., and S.L. XRR experiment were performed and analyzed by P.Q., J.W., L.W., W.B., and B.L. Nano-CT imaging was performed and analyzed by Y.C., J.W., Y.G., and L.W. Manuscript was prepared by J.W. and L.W. and discussed with C.C., L.Y. and Y.Z. with input from other authors. All authors have proofread the paper and approved its publication in the present form.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yi Cao, Leandro Carreño, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, J., Bai, X., Yan, L. et al. Selective regulation of macrophage lipid metabolism via nanomaterials’ surface chemistry. Nat Commun 15, 8349 (2024). https://doi.org/10.1038/s41467-024-52609-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52609-7