Abstract
Cryptosporidium parvum and C. hominis are parasites that cause life-threatening diarrhea in children and immunocompromised people. There is only one approved treatment that is modestly effective for children and ineffective for AIDS patients. Here, screening 278 compounds from the Merck KGaA, Darmstadt, Germany collection and accelerated follow-up enabled by prior investigation of the compounds identifies a series of pyrazolopyrimidine human phosphodiesterase (PDE)-V (hPDE-V) inhibitors with potent anticryptosporidial activity and efficacy following oral administration in C. parvum-infected male mice. The lead compounds affect parasite host cell egress, inhibit both C. parvum and C. hominis, work rapidly, and have minimal off-target effects in a safety screening panel. Interestingly, the hPDE-V inhibitors sildenafil and the 4-aminoquinoline compound 7a do not affect Cryptosporidium. C. parvum expresses one PDE (CpPDE1) continuously during asexual growth, the inhibited life stage. According to homology modeling and docking, the lead compounds interact with CpPDE1. Bulkier amino acids (Val900 and His884) in the CpPDE1 active site replace alanines in hPDE-V and block sildenafil binding. Supporting this, sildenafil kills a CRISPR-engineered Cryptosporidium CpPDE1 V900A mutant. The CpPDE1 mutation also alters parasite susceptibility to pyrazolopyrimidines. CpPDE1 is therefore a validated pyrazolopyrimidine molecular target to exploit for target-based optimization for improved anticryptosporidial development.
Similar content being viewed by others
Introduction
Cryptosporidiosis, the diarrheal disease caused by Cryptosporidium parasites, is one of the most important causes of life-threatening diarrhea in infants worldwide, causes incurable diarrhea in AIDS and transplant patients, and is the most identified cause of waterborne diarrheal outbreaks in the United States and Europe1,2,3. Cryptosporidiosis is also strongly associated with child malnutrition, growth stunting, and delayed cognitive development4. Two species, Cryptosporidium hominis and Cryptosporidium parvum, account for almost all human infections. Unfortunately, there is no protective vaccine, and the only approved treatment, nitazoxanide, is only modestly effective in children ( ~ 56%) and equivalent to a placebo in AIDS patients5,6.
The need for improved anticryptosporidials has spurred research over the last decade, with various teams using phenotypic and/or target-based drug development approaches7,8,9,10,11,12,13,14,15,16,17,18,19,20. The current drug pipeline includes compounds targeting phosphatidyl-inositol-4-kinase (PI4K)18, calcium-dependent kinase 1 (CDPK1)11,12,13, cleavage and polyadenylation specific factor 3 (CPSF3)19, tRNA synthetase inhibitors (methionyl-, phenyl-, and lysyl-)8,10,21, and a partially optimized triazolopyridazine with unknown mechanism-of-action20,22. Two pre-clinical drug candidates are poised for human studies18,19. However, a recent dairy calf efficacy study of a methionine tRNA-synthetase inhibitor failed due to emergence of drug-resistant C. parvum with target gene mutations within days of beginning treatment23. Given the small number of validated targets and likelihood of drug resistance, there remains an urgent need to identify drug leads with novel mechanisms-of-action.
Since the heaviest burden of Cryptosporidium infection falls on young children in low- and middle-income countries, there is little financial incentive for anticryptosporidial drug development. The market for treatment of agricultural animals, especially cattle, is more favorable than for humans but still too small to stimulate de novo drug development. Realistic Cryptosporidium drug development efforts, as for any neglected disease, must therefore simultaneously respect the biological and pharmacological requirements of the intended use (i.e. for human cryptosporidiosis, treatment of small intestinal epithelial infection in young, often malnourished children and/or immunocompromised individuals) and the financial realities that impede target-based drug development. Drug repurposing has been widely touted as a solution to this mismatch between public health needs and financial reality24, and the approach, coupled with hypothesis, has successfully yielded treatments for several conditions (e.g. remdesivir for Covid-1925). However, as has recently been highlighted regarding efforts to identify existing drugs for treatment of Covid-19, hypothesis-free screening for drug repurposing has resulted in no approved treatments despite widespread efforts and substantial research funding over approximately the last 20 years26. In the case of Cryptosporidium, numerous existing drugs inhibit parasite growth in vitro but lack activity in animal models27, possibly in part because anticryptosporidial drug efficacy appears to require sustained drug levels in the intestine while most clinically available drugs are optimized for oral absorption28. And clofazimine, a drug repurposing candidate with efficacy in one of two Cryptosporidium mouse models assessed, appears to be parasitistatic and failed in an early-stage human trial of AIDS patients with cryptosporidiosis9,20,29.
We believe a more promising approach than straight-forward drug repurposing is identification of chemical starting points by screening well-characterized compounds for which there has been substantial study for other purposes but that are at a development stage that permits lead optimization. This approach applies the principle of Selective Optimization of Side Activities (SOSA) to high quality, advanced compounds and leverages prior medicinal chemistry, pharmacokinetic (PK) and pharmacodynamic (PD) data, and safety studies to accelerate screening hit prioritization and early-stage optimization, while still enabling optimization of potency and PK/PD characteristics for specific uses30. Towards this end, a growing number of pharmaceutical companies are providing researchers in academia and other non-profit organizations access to chemical libraries of well-characterized compounds.
Using this strategy, we now report whole-cell screening to identify Cryptosporidium growth inhibitors amongst well-characterized drug-like compounds made available through the Merck KGaA, Darmstadt, Germany Open Innovation Portal31,32. The included compounds possess a wide range of molecular targets encompassing enzymes (kinases, phosphatases, proteases, cyclic nucleotide phosphodiesterases, etc.), hormone or neurotransmitter receptors (serotonin, angiotensin II, endothelin, etc.), ion channels and several others. After confirmatory growth inhibition studies, anticryptosporidial screening hits were prioritized based on existing data and availability of analogs for early-stage optimization. The most promising compounds were then tested in vivo, which yielded a pyrazolopyrimidine phosphodiesterase-V (PDE-V) inhibitor series. The potent human PDE-V inhibitor sildenafil does not affect C. parvum growth, suggesting a target other than human PDE-V. Consistent with inhibition of a C. parvum PDE as the primary mechanism-of-action, the new lead series docks in silico with high confidence to an AlphaFold-based C. parvum PDE1 (CpPDE1; CryptoDB cgd3_2320) model, while sildenafil does not bind due to steric hindrance in the CpPDE1 active site from bulkier amino acids than those present in human PDE-V. Motivated by this model, we humanized the CpPDE1 binding site with a V900A mutation which conferred parasite susceptibility to sildenafil. Furthermore, the V900A mutation altered susceptibility to the PDEi lead compounds PDEi2 and PDEi5, confirming that CpPDE1 is a target of the anticryptosporidial series. This work collectively identifies the pyrazolopyrimidine PDE inhibitors as high-value drug leads and establishes CpPDE1 as a drug target.
Results
Compound screening and down-selection of identified C. parvum growth inhibitors
We used a previously optimized high-content microscopy assay of C. parvum (Iowa) growth in the colon cancer cell line HCT-827 to screen two compound libraries provided by the Merck KGaA, Darmstadt, Germany Open Innovation Portal31,32, the Open Global Health Library and the Mini-Library (together, 278 unique compounds)31,32. Twenty-three compounds inhibited Cryptosporidium growth by over 70% at 5 μM (eight at 1 μM) without affecting host cell numbers, yielding overall screening hit rates at 5 and 1 μM of 8.3% and 2.9%, respectively. Screening hits were selected for resupply and confirmatory dose-response testing based on availability and apparent potency, yielding thirteen confirmed inhibitors with 50% effective concentrations (EC50) ranging from 0.050 μM to 8.0 μM. Based on nuclear staining with Hoechst and quantifying host cell nuclei, none of the confirmed inhibitors affected host cell numbers at the maximum concentration tested (50 μM).
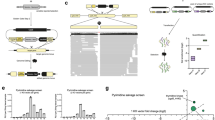
The confirmed C. parvum inhibitors were preliminarily prioritized based on a combination of potency and existing data regarding the mammalian target, PK characteristics, safety, and available chemical variants for follow-up testing. This yielded three chemical series of interest for further studies (Fig. 1): 1) PI3Ki1 (EC50 = 0.14 μM), a human phosphatidylinositol-3-kinase inhibitor; 2) PDEi1 (EC50 = 0.91 μM), a human phosphodiesterase V (PDE-V) inhibitor; and 3) IKK2i1 (EC50 = 4.1 μM), an inhibitor of the human inhibitor of NF-κB kinase 2 (IKK2). Both PDEi1 and IKK2i1 had previously been tested in human phase I/phase II and phase I studies respectively, making these particularly attractive series to investigate. Of note, although the thiazolidine-2,4-dione side chain of PI3Ki1 is associated with nuisance compounds that give false positive screening results (termed pan-assay interference compounds (PAINS))33, existing data indicated no false positives with PI3Ki1 in other assays and published reports on related compounds indicated that they are highly selective and form non-covalent binding interactions with PI3K in crystal structures34,35.
The shown phosphatidyl-inositol-3-kinase inhibitor (PI3Ki), phosphodiesterase-5 inhibitor (PDEi) and IkappaB kinase 2 inhibitor (IKK2i) compounds were prioritized for follow-up based on confirmatory anticryptosporidial EC50 data, existing safety and pharmacologic data, and the availability of chemical analogs for initial follow-up studies.
Nearest neighbor structure activity relationship development
We next conducted structure activity relationship (SAR) studies with analogs available in the chemical libraries provided by Merck KGaA, Darmstadt, Germany, both to judge the likelihood that significant progress might be made through a future medicinal chemistry program and to identify compounds worthy of testing for anticryptosporidial activity in animals. For this, the IKK2i, PI3Ki and PDEi chemical series were used for nearest neighbor chemical substructure searches36 and target inhibitor-based searches of the chemical libraries to identify compounds for dose-response testing. Of 26 IKK2i nearest neighbors and one other IKK2i series assayed, only one inhibited C. parvum at EC50 < 5 μM. The lone exception was IKK2i2, an unrelated pyrazole chemotype IKK2 inhibitor (Table S2) that was exceptionally potent with an EC50 of 0.020 µM. Only five total PI3Ki analogs were available for follow up testing that had C. parvum EC50s ranging from 0.070 to 0.14 μM.
On the other hand, 252 PDEi1 benzothienylpyrimidine analogs and an additional ten closely related chemotypes were available for follow up testing (Fig. 1). C. parvum EC50s for these series ranged from 0.60 to >25 μM. Most of the SAR was provided by the benzothienopyrimidines. Limited data was available for the N-benzyl position (green in Fig. 1) as only eight comparator analogs were available and all were N-benzylated. A much richer SAR was gleaned from the cyclohexylcarboxylic acid/benzoic acid position (magenta in Fig. 1) where benzoic acid (e.g., PDEi3) appeared to be the preferred substituent and COOH was preferred off the aryl ring but was not essential. Finally replacement of the benzothiophene was tolerated by other chemotypes (blue in Fig. 1) such as benzofurans, pyrazoles (e.g., PDEi2), indoles (e.g., PDEi4), thiophenes and pyrroles. From this analysis, 1-methyl-3-ethylpyrazolopyrimidines were a preferred chemotype relative to the others and comparable in potency to the benzothienopyrimidines while providing potential for improved pharmacokinetics due to reduced sp2 character. These three pyrazolopyrimidines had potency comparable to the original hit and differed only in substitution off the benzyl ring (N-benzyl, N-CH2(4-chlorophenyl) (PDEi2), and N-CH2(4-methoxy-3-chlorophenyl)). These three analogs all had C. parvum EC50s below 1 µM.
In an effort to see if the early SAR trend held, we designed a simple 4-fluorobenzyl analog of PDEi2, rationalizing that if 4-H and 4-Cl were potent, the 4-F would also be potent and make a useful comparator. This N-CH2(4-fluorophenyl) (PDEi5) analog was synthesized over six steps from a known aminocyanopyrazole intermediate I (Supplemental Fig. 1)37. Nitrile I was hydrolyzed, condensed with urea, and chlorinated with PCl3/PCl5 to give dichloropyrimidine intermediate IV. Regioselective nucleophilic displacement with 4-fluorobenzylamine followed by Suzuki coupling and ester hydrolysis gave compound PDEi5. PDEi5 was found to have similar potency with C. parvum EC50 of 1.8 µM.
In vivo efficacy and pharmacokinetics
Cryptosporidium parvum infected NOD SCID gamma (NSG) mice were used to assess efficacy of the PI3Ki, IKK2i, and PDEi series in vivo20. Using existing Merck KGaA, Darmstadt, Germany in vitro metabolic stability and mouse PK data on the chemical analogs studied, nine compounds (two IKK2i, three PI3Ki, and four PDEi) were selected and tested for in vivo efficacy (Table S1 shows characteristics of compounds tested in vivo, Table S2 provides compound structures, Supplemental Fig. 2 shows mouse efficacy data, Supplemental Fig. 3 shows mouse weights). Both IKK2is tested were ineffective, and all three PI3Kis resulted in weight loss at the dose tested, necessitating study cessation. The most promising PI3Ki (PI3Ki1) was retested at a dose of 10 mpk twice daily, below NOAEL of 30 mg/kg known from unpublished Merck KGaA, Darmstadt, Germany analyses, and proved to be ineffective. On the other hand, three of four PDEis tested in the proof-of-principle experiment appeared effective (p < 0.05) or potentially effective (p = 0.14) with a 4-day dosing and no adverse effects were noted (Supplemental Figs. 2 and 3).
The PDEis were studied in follow-up with 7-day dosing regimens in two independent experiments, using pyrazolopyrimidine PDEi2, newly synthesized analog PDEi5, and the indolopyrimidine PDEi4 (Fig. 2). PDEi4 had no significant effect. PDEi2 (Fig. 2A) and PDEi5 (Fig. 2B), in contrast, both reduced parasite shedding by ~99% after 7 days of treatment (p = 0.02), although infection relapsed within a week of stopping treatment. Supplemental Fig. 3D, E show mouse weights before and after PDEi dosing. The mean weight of mice receiving PDEi5 fell ~5% in the final days of treatment before rebounding after treatment cessation.
The indicated compounds were tested for efficacy in established cryptosporidiosis in male NSG mice. C. parvum infection was allowed to progress for either 7 days (A) or 14 days (B) prior to oral administration of 50 mpk twice daily of the indicated compounds or 1000 mpk twice daily of paromomycin (positive control) by oral gavage for 7 days. Parasite shedding in the feces was determined at the onset of treatment (D1), completion of treatment (D8), and one week after completion of treatment (D15). Data are the mean and SEM (n = 4 mice per experimental group) of parasite fecal shedding per mg of feces, and data points below the limit of qPCR detection are shown on the x-axis. p values vs. vehicle control determined by Kruskal–Wallis test are shown. Source data for (A) and (B) are provided as a Source Data file.
PDEi2 had kinetic solubility = 190 μM (2 h, PBS, pH 7.4) and other favorable properties for in vivo work such as high metabolic stability, modest in vivo half-life and oral bioavailability (Table S1). Single-dose IV and oral mouse PK studies were therefore performed with PDEi2 to further understand the determinants of in vivo efficacy for this series. PDEi2 was rapidly cleared from plasma (T1/2 ~ 1 h) following IV administration (Fig. 3A). In contrast, after a single 10 mg/kg oral dose, total drug concentrations in successive intestinal tissue segments persisted at high levels for at least 7 h and were still detectable after 24 h (Fig. 3B). Because tissue lysates mix interstitial and intracellular fluids, we cannot know how drug is partitioned between the extracellular and intracellular compartments38. We also cannot accurately determine the free (unbound) concentration in the lysate, since its protein composition is different from plasma. Thus, the precise unbound concentration at the site of action, presumably within the parasite which itself is located within the parasitophorous vacuole in the enterocyte, cannot be readily determined. Nevertheless, these data show that the total PDEi2 concentration in the intestinal tissue persisted for hours at levels that exceeded multiples of the EC90 for inhibition of parasite growth. Drug concentrations in plasma were also prolonged with oral versus IV administration, likely due to continued gastrointestinal absorption, but did not reach the same levels measured in intestinal tissues (Fig. 3A).
A Time courses of total drug concentration determined in plasma with single dose IV and oral administration at indicated doses. B Time courses of total drug concentration determined in washed intestinal tissue segments with single dose 10 mg/kg oral administration. Data for (A) and (B) are mean and SEM (n = 3 mice per experimental group where SEM shown; n = 2 mice per group where no SEM shown). Source data for (A) and (B) are provided as a Source Data file.
Activity against C. hominis, rate-of-action, and anticryptosporidial mechanism-of-action
Most human cryptosporidiosis is caused by either C. parvum or C. hominis, but most work (including our screening assays and confirmatory studies) is done using C. parvum because C. hominis oocysts can only be sourced from patients or using an expensive gnotobiotic piglet model. Nonetheless, prior to investing time and resources into a given drug lead series, confirmation of activity against both species is criticalto meet the desired target compound profile. We used the C. hominis Tu502 strain and the cell-based assay method used for C. parvum to test the activity of PDEi2, PDEi4, and PDEi5. Activity was comparable for both species (Table S1); for lead compounds PDEi2 and PDEi5, the C. hominis EC50s and 95% confidence intervals were 0.61 (undetermined-3.8) μM and 4.7 (2.4–9.1), respectively.
Given the desire to develop anticryptosporidials with efficacy for immunocompromised patient populations, we use rate-of-action curves at early stages of our developmental cascade to enable prioritizing compounds that rapidly eliminate C. parvum in tissue culture and are likely parasiticidal20. Figure 4 shows rate-of-action data in the HCT-8 cell culture system for nitazoxanide and PDEi5. Decay curves were fit after referencing data at each time point to the vehicle control to isolate the effect of compounds from the spontaneous decline in C. parvum numbers that begins after 48–60 h of in vitro culture. Like previously reported data suggesting parasitistatic activity19,20, no nitazoxanide-dependent parasite elimination occurred at the highest tolerated dose. PDEi5, on the other hand, drove progressive parasite elimination with an exponential decay half-life of ~ 10.7 h. Thus, ~107 h (<5 days) would be required for 99.9% parasite reduction by PDEi5 in the absence of host immune help. Consistent with a class effect which has been typical with this rate-of-action assay8,10,19,20,21, results were similar with PDEi2 (Supplemental Fig. 4). These data were also in keeping with the in vivo efficacy in the NSG mouse with twice daily administration, since the intestinal tissue concentration of PDEi2 following a single 10 mg/kg dose exceeded the EC90 for over 7 h.
Rate-of-action curves for (A) nitazoxanide (NTZ) and (B) PDEi5 showing elimination of C. parvum from infected HCT-8 cells. One-phase exponential decay curves were fit for NTZ and PDEi5. Data are the means and SD for 4 culture wells per condition, normalized for each time point to the mean of 28 vehicle (DMSO) control culture wells. Results from one of two experiments shown. Source data are provided as a Source Data file. A No exponential decay curve could be fit for NTZ at the EC90 which was the highest non-toxic concentration. Blue (DMSO); black (NTZ EC90). B Rapid parasite elimination by PDEi5. Curves are shown for 3×EC90 (gray) and 6×EC90 (cyan) (the highest concentration tested), or the DMSO control (blue).
PDEi2 potently inhibits the mammalian cGMP-specific phosphodiesterase PDE-V (IC50 = 20 nM) and less potently inhibits other cGMP-specific phosphodiesterases, including PDE-I, PDE-II, PDE-IV, and PDE-VI (Table S3). To determine if the anticryptosporidial activity of the PDEi series resulted from inhibition of host PDE-V or parasite phosphodiesterases, we compared the anticryptosporidial activity of two PDEi analogs (PDEi1 and PDEi5; structures given in Table S2) to the potent mammalian PDE-V inhibitors sildenafil and the 4-aminoquinoline compound 7a (Sigma-Aldrich, cat#508957) (IC50 for human PDE-V = 3.5 nM and 0.28 nM, respectively)39,40. The EC50 of sildenafil for C. parvum growth in HCT-8 cells exceeded 50 μM and that of compound 7a was 36 μM, far in excess of the concentrations needed to inhibit host PDE-V and suggesting that Cryptosporidium growth inhibition by the lead PDEi series was independent of host cell PDE-V and more likely resulted from inhibition of one or more Cryptosporidium phosphodiesterases or another off-target effect (Supplemental Fig. 5A).
BLAST of hPDE5A (https://www.ncbi.nlm.nih.gov/gene/8654) against the Cryptosporidium genome identifies three Cryptosporidium PDEs (CryptoDB: cgd3_2320 (E = 9e-18), cgd6_500 (E = 6e-16), and cgd6_4020 (E = 3e-08), previously reported as CpPDE1, CpPDE2 and CpPDE3, respectively41). None of these proteins have been characterized experimentally. Only CpPDE1 is predicted to be cGMP-specific like hPDE5A, with homology to hPDE5A limited to 260 of 997 total amino acids (29% identity, 46% similarity) that correspond to the predicted CpPDE1 active site (sequence alignment in Supplemental Fig. 6A). Publically available single-cell RNA-seq data and other published transcriptomics data indicate CpPDE1 is highly expressed in sporozoites and asexual forms42. CpPDE2 is expressed only at low levels that peak in HCT-8 cell cultures after ~36 h of asexual growth42. CpPDE3 expression, on the other hand, increases dramatically at 48 h in cell culture when gametocytes become predominant43 and expression is nearly restricted to male gametocytes42. Based on these expression patterns, we limited subsequent analyses to CpPDE1 which is expressed during the asexual growth phase reflected by the HCT-8 cell culture assay for anticryptosporidial activity.
We next used the available AlphaFold structure model for CpPDE144,45 and X-ray crystal structures of human PDE-V (hPDE5) to determine if the lead PDEi series compounds are likely to bind CpPDE1 and to understand the inability of sildenafil to block Cryptosporidium growth. The CpPDE1 model (AlphaFold: A3FQ29-F1-model_v4) provides a high confidence predicted structure starting near GLY590 that corresponds to the C-terminal catalytic domain but lacks active site metal ions known to interact with cGMP/cAMP that would potentially interact with the novel PDE inhibitors. Therefore, to enable addition of the active site Zn2+ and Mg2+ metal ions to the models, we overlayed the crystal structure of a Schistosoma mansoni PDE (SmPDE4 (pdb 6EZU46)) that is 36% identical and 53% similar to CpPDE1 in the active site. The AlphaFold model was truncated to remove the low-confidence and non-catalytic N-terminal region, and the active site metal ions and waters from the SmPDE structure, also observed in related PDEs, were incorporated by merging followed by the restrained minimization routine using the OPLS4 forcefield in Schrödinger Maestro and aligned to produce a structure suitable for molecular docking studies (Supplemental Fig. 6B). Using this model, PDEi5 was docked to CpPDE1 using the Glide SP docking routine in Schrödinger Maestro47. Docking scores for the top three poses ranged from -9.5 to -9.9 kcal/mol, predictive of strong binding interactions (comparable to sildenafil docked to human PDE-V crystal structure 1UDT with a Glide SP docking score of −9.2 kcal/mol). The CpPDE1 docking model was further refined using the Induced Fit docking routine in Schrödinger Maestro, giving docking scores as low as -11.9 kcal/mol against CpPDE1 and suggesting PDEi5 has a high likelihood of binding this target (Supplemental Fig. 7). All of the docked poses of PDEi5 involved a direct interaction of the carboxylic acid moiety with the two metal ions, mimicking the cGMP/cAMP phosphate interaction with the metals observed in SmPDE4. While positioning of the benzyl and pyrazolopyrimidine core varied, structurally conserved Phe935 formed π-π stacking with PDEi5. Both PDEi5 and sildenafil docked with similar docking scores in human PDE-V, but sildenafil docked poorly to the CpPDE1 model (best SP docking score -5.9 kcal/mol) due to its inability to adopt the same binding pose it has in the human structure, likely due to the bulky 2-ethoxy-5-sulfonamide-aryl ring (Fig. 5). Ala873 in hPDE-V is replaced by bulkier Val900 in CpPDE1, leading to a clash with the ethoxy group of sildenafil. Furthermore, Ala767 in hPDE-V is replaced by bulkier His884 in CpPDE1, leading to clashing with the N-Me of sildenafil. Of note, seven of the 18 residues within 4 Å of PDEi5 docked to CpPDE1 differ from hPDE-V, suggesting potential to design variants of PDEi5 with improved selectivity for CpPDE1 over hPDE-V.
Only the seven of the 18 total residues within 4 Å of the PDEi5 ligand that differ between the two proteins are shown, along with Val900 (5 Å). The ligand binding pocket of CpPDE1 is shown as a gray surface. A Induced fit docked pose of PDEi5. B Overlay of sildenafil from hPDE-V structure (pdb 1UDT). Arrows indicate steric clashing with Val900 and His884 preventing sildenafil from obtaining the same docking pose in CpPDE1 vs. hPDE-V.
We used CRISPR to make mutant parasite lines and test if the lead PDE inhbitors target CpPDE1. Parasites engineered to constitutively overexpress a second copy of CpPDE1 located in the disposable thymidine kinase locus were not viable. We therefore humanized the native CpPDE1 locus with attempted single and double amino acid mutations of the active site bulky Val900 and His885 to the corresponding Ala residues present in hPDE-V (Fig. 6A). CpPDE1 His885Ala and the double mutant parasite lines could not be recovered. However, a CpPDE1 Val900Ala mutant and its corresponding transgenic wildtype (wt) control parasite (i.e., with wildtype CpPDE1 amino acid sequence but otherwise recoded as for the Val900Ala mutant and with insertion of the downstream constitutively expressed nanoluciferase reporter and neomycin resistance cassette) were both viable and shed from infected mice in numbers similar to wt Iowa strain parasites. Modification of the CpPDE1 locus was confirmed by PCR using primers flanking the insertion site, and accuracy of the modifications was verified by Sanger sequencing (Fig. 6B, C). We first tested susceptibility of the CpPDE1 Val900Ala line to sildenafil using the HCT-8 cell growth assay. As predicted by the in silico docking model (Fig. 5B), replacing bulky Val900 with Alanine sensitized CpPDE1 Val900Ala to sildenafil, shifting the EC50 from >50 μM to 14 μM (Fig. 6C). The transgenic wt control line had susceptibility similar to that of the wt Iowa strain. The Val900Ala mutation also improved the potency of PDEi2 and PDEi5, thus demonstrating that CpPDE1 is a target of the PDEi drug leads (Fig. 6D, E).
A CRISPR strategy for humanizing CpPDE1. A Val900Ala mutation and nano-luciferase/Neo resistance cassette were incorporated in the CpPDE1 locus. Transgenic control parasites were also made. B PCR (primers P1 and P2 in (A)) confirming modification of CpPDE1 (representative of 2 experiments). C Sanger sequencing confirming the designed mutation. For (D–F), dose response assays were conducted with the indicated inhibitor and CpPDE1 Val900Ala, CpPDE1 transgenic wt, and C. parvum(Iowa). Graphs show the means of 4 culture wells per condition (one of two experiments per inhibitor). Orange = CpPDE1 Val900Ala; gray = transgenic wt; black = C. parvum(Iowa). D Sensitization of the CpPDE1 Val900Ala parasite to sildenafil. E Effect of Val900Ala mutation on growth inhibition by PDEi5. F Effect of Val900Ala mutation on growth inhibition by PDEi2. Source data for all graphs are provided as a Source Data file.
We next employed a suite of C. parvum life cycle-based assays48 to begin investigating the mode-of-action of the PDEi lead series and the likely function(s) of CpPDEs in Cryptosporidium biology. The PDEi inhibitors were inactive in a cell invasion assay (Supplemental Fig. 5B) but strongly active in an assay that measures a combination of Cryptosporidium egress and invasion of new host cells (Supplemental Fig. 5C and representative images in Supplemental Fig. 8), suggesting their primary activity is to block parasite egress. They also blocked sexual maturation, since they reduced expression of the macrogametocyte marker DMC1 (Supplemental Fig. 5D). Since anticryptosporidial pipeline diversity is critical and the mechanism of many compounds in development is unknown, we used a clustering algorithm previously reported with these phenotypic assays and publicly available assay data to generate a dendrogram and assess if the PDEi’s mode-of-action was distinct (Supplemental Fig. 9)48. Both PDEis clustered together, as expected, and separately from other compound series in development.
Safety assessment and identification of potential liabilities
Pyrazolopyrimidines PDEi2 and PDEi5 were both non-toxic to the HCT-8 cell line at 100 μM, the highest concentration tested (note that per our data, the kinetic solubility of PDEi2 is 190 μM and kinetic solubility of PDEi5 is 200 μM); selectivity of lead PDEi2 exceeded 125-fold and selectivity of lead PDEi5 exceeded 55-fold. The 50% toxic concentration (TC50) of the indolopyrimidine PDEi4 (kinetic solubility 21 μM) was 87 μM (selectivity index = 100). This in combination with superior activity in the mouse led us to focus further safety assessment on the pyrazolopyrimidine series.
We contracted Eurofins (St. Charles, MO) to conduct off-target safety profiling for PDEi5, the pyrazolopyrimidine PDEi2 analog that we had synthesized independently in our lab. PDEi5 was tested in dose-response up to 30 μM in the SafetyScan47 panel of cell-based assays that assesses effects on 47 human targets (including hERG; 78 assays total, as many of the targets are run in both agonist and antagonist modes)49. PDEi5 had EC50 values > 30 μM for 45 of 47 targets (see Table S4 for full list of targets and assay results). The only off-target enzymes inhibited were human PDE-IIIA and PDE-IVD2 with EC50 values of 10.2 and 8.2 μM, respectively. To assess human PDEs not included in the SafetyScan47 panel, we also contracted with Eurofins to measure PDEi5 inhibition of 13 mammalian PDEs (Table S5). PDEi5 most potently inhibited human PDE-V and PDE-VI, but PDE-IVB1 and PDE-XIA were also inhibited >50% at a test concentration of 10 μM. PDEi5 was also tested by Eurofins to demonstrate it has suitable aqueous solubility (200 µM, kinetic) and has low potential to inhibit the hERG channel (−14.7% at 10 µM in a [3H]-dofetilide competitive binding experiment). We also evaluated the potential for toxicity of PDEi5 using in silico methodology using ProTox-II, which indicated a low probability for toxicity (Supplemental Fig. 10)50. Finally, Eurofins tested in vitro inhibition of a panel of seven cytochrome p450s (CYPs), demonstrating modest inhibition of CYP2C8 and CYP2C9 (~31% and 18% respectively at 10 μM) (Table S6).
Discussion
This work establishes Cryptosporidium PDE1 (CpPDE1) as a target for anticryptosporidial development and the pyrazolopyrimidine phosphodiesterase inhibitors (i.e., PDEi2 and PDEi5) as a promising chemical series. Lead compounds PDEi2 and PDEi5 possess many desirable attributes as starting points to develop an improved Cryptosporidium treatment, since they rapidly eliminate parasites in vitro, have comparable activity against both C. parvum and C. hominis, are highly efficacious in a stringent mouse model of infection, have minimal off-target effects on a panel of human targets tested, and have only modest potential for drug-drug interactions due to CYP inhibition. Homology/docking studies done using high-quality AlphaFold structures predict that the pyrazolopyrimidine PDEis bind well to CpPDE1 and further predict steric clashing likely to reduce binding affinity of sildenafil and explain sildenafil’s inability to inhibit Cryptosporidium growth. Furthermore, humanizing CpPDE1 by CRISPR/Cas9 genome editing to incorporate homology/docking predicted amino acid mutations sensitizes C. parvum to sildenafil and alters the potency of drug leads PDEi2 and PDEi5. These genetic data validate the AlphaFold-based homology/docking model and indicate that CpPDE1 is a molecular target for the pyrazolopyrimidine drug lead series. Phenotypic assays with the compounds suggest a primary function of CpPDE1 in the HCT-8 cell culture system in parasite egress from host cells. Finally, the overall pattern of effects in phenotypic assays indicates that the pyrazolopyrimidine PDEi series works via a mechanism that is distinct amongst current compound series in the developmental pipeline, and thus contributes important drug pipeline diversity. This work collectively establishes the pyrazolopyrimidines as a high priority series for medicinal chemistry optimization and genetically validates CpPDE1 as an anticryptosporidial drug target.
Based on a combination of structural modeling/docking studies, CRISPR/Cas9 genome modification to incorporate a model-predicted mutation into CpPDE1, and phenotypic assays, the pyrazolopyrimidine PDEi series appears to impact Cryptosporidium growth at least in part by inhibiting CpPDE1 and affecting parasite egress from infected host cells, an essential step in progression of the asexual parasite life cycle. Our work focused on CpPDE1, since it is expressed during asexual parasite growth that is measured by the C. parvum HCT-8 cell infection assay. However, it is possible that CpPDE3, which is expressed in male gametocytes, is an additional target of the pyrazolopyrimidine lead series or could be otherwise targeted. The potential to similarly humanize CpPDE3 and confer susceptibility to sildenafil presents an intriguing chemical-genetic approach to selectively target C. parvum sexual reproduction and test whether sexual reproduction is truly obligatory (and thus, targetable for prevention or treatment), as predicted by a recent publication detailing C. parvum development that provided insights into the parasite life cycle51. Furthermore, evaluation of structural models of PDEi binding and presence of 39% variation in active site residues (CpPDE1 vs hPDE-V) suggests the potential to improve selectivity for CpPDE1 and possibly other CpPDEs over human PDE-V through medicinal chemistry optimization. Such information could be used in combination with in vitro assays using recombinant enzymes to guide drug optimization. In support of such optimization strategies, we are currently working to express functional recombinant CpPDEs and to make transgenic parasites with modifications to CpPDE3.
It is likely that nitazoxanide fails in malnourished children and immunocompromised people because it is parasitistatic rather than parasiticidal for Cryptosporidium species and requires assistance from the host immune system to clear cryptosporidiosis; unlike nitazoxanide, PDEi2 and PDEi5 rapidly eliminate C. parvum from in vitro culture. A growing body of published data indicate that in vivo efficacy in immunocompromised animals correlates with a compound’s rate-of-action10,19,20,21; furthermore, rapid vs. slow or static anticryptosporidial activity appears to be conserved amongst chemical analogs with a given mechanism of action. Since PDEi2 and PDEi5 rapidly eliminate C. parvum from tissue culture, we believe the pyrazolopyrimidines are a lead series for which optimization could yield a treatment effective for all patient groups impacted by cryptosporidiosis.
Consistent with this, PDEi2 and PDEi5 are efficacious in an NSG mouse model that tests the ability to impact established infection. Numerous mouse models have been used for anticryptosporidial efficacy studies, but most mice infected by C. parvum develop an acute infection that resolves spontaneously over ~17 days9,52,53,54. In the absence of effective drugs to benchmark different models, the utility of any animal model for predicting efficacy in children and immunocompromised people with cryptosporidiosis is, of course, unknown. We use the NSG mouse model, because NSG mice develop persistent infection localized to the ileum and cecum that enables delaying study drug administration until infection is fully established and monitoring for recrudescence after treatment cessation20. Relative to self-resolving infection models, the NSG mouse model of cryptosporidiosis is stringent, and compounds active in acute models of infection are frequently inactive in the NSG mouse model (e.g., clofazimine9). PDEi2 and PDEi5, on the other hand, both reduced C. parvum shedding by infected NSG mice by ~99% when dosed orally for 7 days, establishing the pyrazolopyrimidine PDE inhibitors as drug leads worthy of further optimization.
C. parvum infected NSG mice treated with both PDEi2 and PDEi5 relapsed with the dosing regimens tested; thus, improving in vivo efficacy is a key goal to optimize the pyrazolopyrimidine series. To do this, a medicinal chemistry program is underway to improve in vitro anticryptosporidial potency while simultaneously optimizing the PK/PD characteristics that drive in vivo efficacy. A growing body of data for other lead series indicates that the key determinant of in vivo efficacy of anticryptosporidials is achieving adequate compound presence in the intestinal lumen and tissue28. The same likely applies to the PDE inhibitors, since PDEi2 is rapidly cleared from the plasma but persists in intestinal tissue at levels above the EC90 for parasite growth inhibition. In the case of PDEi2 and PDEi5, the maximal rate of parasite elimination in vitro was achieved at a concentration of 3x EC90, and rate-of-action data similar to that for PDEi2 and PDEi5 can be used to provide target tissue compound concentrations for more potent analogs as we work towards curative compounds.
Safety is essential for drugs intended for outpatient administration to young children in whom drug toxicities are difficult to monitor. PDEi2 is non-toxic to HCT-8 cells at concentrations exceeding 100 μM, yielding a selectivity index for C. parvum greater than 125. Furthermore, testing close analog PDEi5 with the Eurofins SafetyScan47 assay panel to screen for off-target effects only identified modest potency for inhibition of several additional mammalian PDEs, and in silico safety evaluation of PDEi5 using ProTox-II also indicates a low probability of toxicity50. Finally, minimal CYP450 inhibition suggests a low likelihood of drug-drug interactions that might interfere with treating AIDS patients or organ transplantation patients, both of whom often chronically take CYP450-metabolized medications.
Despite the critical role of phosphodiesterases in controlling levels of cellular cAMP and cGMP and their proven druggability, there is limited precedence for targeting parasite phosphodiesterases for drug development. Trypanosoma brucei PDEB1 is a chemically validated therapeutic target for Human African Trypanosomiasis (HAT) for which the most advanced compounds are curative in a mouse model55,56,57,58,59. Benzoxaborole and other inhibitors of Schistosoma mansoni PDE4A (the enzyme used here to incorporate metal ions into the CpPDE1 AlphaFold docking model) have been identified60,61, but the best compounds only modestly reduce worm burden in S. mansoni infected mice, raising questions about suitability of the target62,63. For Apicomplexa like Cryptosporidium, potent inhibitors of Plasmodium falciparum and Toxoplasma phosphodiesterases have been identified and proven useful to demonstrate the function of cyclic nucleotides in Apicomplexa64. Consistent with the effects of PDEi1 and PDEi5 on Cryptosporidium cell egress and reinvasion, these inhibitors appear to block Plasmodium proliferation by disrupting protein kinase G-dependent cell egress64. A medicinal chemistry effort has produced more potent inhibitors of Plasmodium PDEs, but in vivo efficacy has not been reported65. Due to their different locations in the host, the drug pharmacodynamics required for treating malaria, babesiosis, toxoplasmosis, and cryptosporidiosis differ and make a pan-apicomplexan drug unlikely. In addition to anticryptosporidial lead optimization studies, however, pathogen-hopping studies are warranted to determine if the pyrazolopyrimidine anticryptosporidial drug leads described here can provide starting points to develop drugs for other Apicomplexa.
In conclusion, using a SOSA approach, we discovered a class of anti-Cryptosporidium compounds, pyrazolopyrimidines, which act at least in part through inhibition of CpPDE1. Lead compounds proved to have good in vitro and in vivo antiparasitic potency with appropriate selectivity profiles in order to achieve in vivo cure while minimizing potential toxicity. Further optimization can now be rationally supported by CpPDE1 and hPDE-V docking models. Furthermore, genetic validation studies reveal CpPDE1 as an attractive target for drug discovery.
Methods
Chemical compounds
The Merck KGaA, Darmstadt, Germany Mini-Library and Open Global Health Library were supplied through the Open Innovation Portal as 10 mM DMSO stock solutions in microtiter plates. All screening hits available for follow-up were reacquired for confirmatory dose-response assays. Nearest neighbor and chemical substructure searches were performed using available Merck KGaA, Darmstadt, Germany compound libraries to further prioritize the confirmed screening hits for follow-up and available compounds were sourced directly from Merck KGaA, Darmstadt, Germany. The PDEi analog PDEi5 was synthesized by the Meyers Lab (St. Louis University); compound structures and purity ≥95% were verified by 1H NMR and LC-MS. Sildenafil (Sigma-Aldrich, cat#SML3033) and compound 7a (Sigma-Aldrich, cat#508957) were purchased for use in phenotypic assays.
General synthesis procedure
Unless otherwise stated, all reagents and solvents purchased were used as received without further purifications. Normal and reverse-phase chromatography were performed using CombiFlash® Rf+ (Teledyne Isco) with SiliaFlash F60 40 − 63 μm (230 − 400 mesh) silica gel (SiliCycle Inc.) eluting with hexanes/ethyl acetate gradient and RediSep Rf Gold pre-packed C18 cartridge or prep-HPLC on a ACCQ-Prep HP150 (RediSep Prep C18, 100 Å, 5 µm, 250 mm × 20 mm or 30 mm column) eluting with acetonitrile/water gradient. Thin layer chromatography was carried out using TLC Silica gel 60 F254 Glass plates 2.5 × 7.5 cm (Merck KGaA, Darmstadt, Germany) and visualized with a UV lamp (254/365 nm UV/6-watt, BioGrow®). Liquid chromatography mass spectrometry (LCMS) was performed with an Agilent 1100/1946 HPLC/MSD electrospray mass spectrometer in positive ion mode with a scan range of 100 − 1000 Da. 1H NMR spectra of intermediates and final compound were recorded and acquired in CDCl3 or DMSO-d6 as solvents using Bruker 400 MHz spectrometer at ambient temperature (400 MHz for 1H). Chemical shifts for 1H NMR (400 MHz) spectra are reported in parts per million (ppm) from either CDCl3 (7.26 ppm) or DMSO-d6 (2.50 ppm) with multiplicity (s = singlet, bs = broad singlet, d = doublet, t = triplet, q = quartet, dd = doublet of a doublet, td = triplet of a doublet, and m = multiplet) and coupling constants (J) in Hz. High resolution mass spectrum (HRMS) was obtained with an ABSciex 5600+ instrument. The verified purity of final compound was ≥95% as determined by HPLC UV absorbance unless noted otherwise.
5-amino-3-ethyl-1-methyl-1H-pyrazole-4-carboxamide (II)
Concentrated sulfuric acid (H2SO4, 35 mL) was poured into a round-bottom flask and cooled in an ice water bath to 0 oC. 5-Amino-3-ethyl-1-methyl-1H-pyrazole-4-carbonitrile (I, 8.12 g, 54.1 mmol) was slowly added to the cooled concentrated H2SO4 at 0 °C37. The ice bath was removed, and the resulting solution allowed to warm to room temperature with stirring overnight. After this time, the reaction was poured on an ice and neutralized with concentrated sodium hydroxide solution to pH 8. The aqueous layer was transferred into a separatory funnel and extracted three times with ethyl acetate. The organics were combined, washed with brine, and dried over magnesium sulfate before filtration. The filtrate was concentrated on a rotary evaporator and the resulting light brown solid product (5.10 g, 30.3 mmol) was > 98% purity on LCMS with a yield of 56% and was used in the subsequent step without further purification. 1H NMR (400 MHz, DMSO-d6) δ 6.50 (br. s., 2H), 6.13 (s, 2H), 3.46 (s, 3H), 2.65 (q, J = 7.46 Hz, 2H), 1.13 (t, J = 7.46 Hz, 3H). LC-MS m/z (M + H)+ Calcd for C7H12N4O 169; found 169.
3-ethyl-1-methyl-1H-pyrazolo[3,4-d]pyrimidine-4,6(5H,7H)-dione (III)
Urea (6.35 g, 106 mmol) and 5-amino-3-ethyl-1-methyl-1H-pyrazole-4-carboxamide (II, 2.60 g, 15.5 mmol) were weighed into a round bottom flask. The flask was heated to 200 °C for 2 h. The reaction was cooled to room temperature and 2 M NaOH solution was added to the flask. The resulting solution was acidified with conc. HCl to pH 5 and the solids formed were filtered off. The solids were dried overnight and purified via column chromatography (0 to 20% MeOH/EtOAc) to afford the desired product in 74% yield (2.23 g, 11.5 mmol). 1H NMR (400 MHz, DMSO-d6): δ ppm 1.19 (t, J = 7.6 Hz, 3H), 2.69 (q, J = 7.6 Hz, 2H), 3.68 (s, 3H), 10.07 (s, 1H), 11.84 (s, 1H). LC-MS m/z (M + H)+ Calcd for C8H10N4O2 195; found 195.
4,6-dichloro-3-ethyl-1-methyl-1H-pyrazolo[3,4-d]pyrimidine (IV)
To a round bottom flask containing 3-ethyl-1-methyl-1H-pyrazolo[3,4-d]pyrimidine-4,6(5H,7H)-dione (III, 500 mg, 2.57 mmol) was added phosphorus (V) chloride (1.00 g, 5.14 mmol) and phosphoryl chloride (10 mL). The mixture was refluxed for 9 h and then allowed to cool room temperature. After POCl3 was removed by rotary evaporation, the solid was washed with water, and the yellow solid residue was purified by flash chromatography on silica gel (EtOAc/Hexane) to provide pure white solid intermediate compound (185 mg, 0.797 mmol, 31% yield). 1H NMR (400 MHz, DMSO-d6): δ ppm 1.34 (t, J = 7.2 Hz, 3H), 3.06 (q, J = 7.2 Hz, 2H), 3.96 (s, 3H). LC-MS m/z (M + H)+ Calcd for C8H8Cl2N4 231; found 231.
6-chloro-3-ethyl-N-(4-fluorobenzyl)-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine (V)
4,6-dichloro-3-ethyl-1-methyl-1H-pyrazolo[3,4-d]pyrimidine (IV, 1.00 g, 4.33 mmol) was dissolved in 10 mL acetonitrile before adding DIPEA (1.10 mL, 6.50 mmol) and 4-fluorobenzyl amine (0.800 mL, 6.50 mmol). The resulting solution was stirred at room temperature for 6 hrs and concentrated. The crude product was purified via silica gel chromatography (20% MeOH/EtOAc) to afford the desired product in 65% yield (897 mg, 2.81 mmol). 1H NMR (400 MHz, DMSO-d6) δ 8.09 (t, J = 5.87 Hz, 1H), 7.40 (dd, J = 5.62, 8.56 Hz, 2H), 7.15 (t, J = 8.80 Hz, 2H), 4.69 (d, J = 5.99 Hz, 2H), 3.78 (s, 3H), 2.98 (q, J = 7.46 Hz, 2H), 1.23 (t, J = 7.46 Hz, 3H). LC-MS m/z (M + H)+ Calcd for C15H15ClFN5 320; found 320.
Ethyl 4-(3-ethyl-4-((4-fluorobenzyl)amino)-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)benzoate
6-chloro-3-ethyl-N-(4-fluorobenzyl)-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine (V, 300 mg, 0.940 mmol), (4-(ethoxycarbonyl)phenyl)boronic acid (328 mg, 1.69 mmol), Pd(dppf)Cl2 (95.1 mg, 0.130 mmol), K2CO3 (299 mg, 2.16 mmol), 12 mL DMF and 3.0 mL H2O were weighed into a 20 mL microwave vial and capped. The vial was vacuumed and backfilled with Argon gas. This process was repeated three times and the vial kept under positive Argon pressure. Anhydrous DMF and degassed distilled water were added to the vial in a 4:1 ratio. The microwave vial was placed in a heating block and the block heated to 100 oC overnight with stirring. After this time, the reaction was cooled to room temperature and filtered through a short pad of Celite eluting with ethyl acetate. The organic was transferred into a separatory funnel containing water. The aqueous layer was extracted two times with ethyl acetate. All the organics were combined, washed with brine, dried over magnesium sulfate, and filtered. The filtrate was concentrated, and the crude product purified via silica gel chromatography eluting with hexanes and ethyl acetate. To ensure sufficiently pure compounds to submit for bioassay, a second reversed phase chromatography eluting with water and acetonitrile with no modifiers was performed. The product was isolated in 56% yield as an off-white solid (226 mg, 0.526 mmol). 1H NMR (400 MHz, DMSO-d6) δ 8.49 (d, J = 8.44 Hz, 2H), 8.05 (d, J = 8.44 Hz, 2H), 7.89 (s, 1H), 7.51 (dd, J = 5.69, 8.50 Hz, 2H), 7.15 (t, J = 8.93 Hz, 2H), 4.85 (d, J = 5.50 Hz, 2H), 4.35 (q, J = 7.09 Hz, 2H), 3.92 (s, 3H), 3.05 (q, J = 7.46 Hz, 2H), 1.35 (t, J = 7.09 Hz, 3H), 1.29 (t, J = 7.46 Hz, 3H). LC-MS m/z (M + H)+ Calcd for C24H24FN5O2 434; found 434.
4-(3-ethyl-4-((4-fluorobenzyl)amino)-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)benzoic acid (PDEi5)
Ethyl 4-(3-ethyl-4-((4-fluorobenzyl)amino)-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)benzoate (100 mg, 0.231 mmol) was weighed into a round bottom flask before adding KOH (23.3 mg, 0.347 mmol) and 10 mL ethanol. The flask was placed in a heating block and heated to 100 °C for 2 h with a condenser on top. After this time, the reaction was cooled to room temperature, acidified with 1 M HCl solution to pH 6 and concentrated. The resulting crude product was purified via reversed phase chromatography eluting with H2O:CH3CN. The product was isolated in 73% yield as a white solid (68.3 mg, 0.169 mmol). 1H NMR (400 MHz, DMSO-d6) δ 13.03 (br. s., 1H), 8.48 (d, J = 8.31 Hz, 2H), 8.03 (d, J = 8.31 Hz, 2H), 7.92 (t, J = 5.87 Hz, 1H), 7.51 (dd, J = 5.81, 8.25 Hz, 2H), 7.15 (t, J = 8.80 Hz, 2H), 4.86 (d, J = 5.50 Hz, 2H), 3.91 (s, 3H), 3.05 (q, J = 7.46 Hz, 2H), 1.28 (t, J = 7.46 Hz, 3H). LC-MS m/z (M + H)+ = 406. HRMS (ESI) m/z: [M + H]+ Calcd for C22H20FN5O2 406.1679; found 406.1670.
Cell culture and parasites
Human ileocecal adenocarcinoma (HCT-8) cells (ATCC; catalog # CCL-244) were cultured in RPMI 1640 medium (ThermoFisher; catalog # 11875093) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich; catalog # 12306 C), 120 U/mL penicillin, and 120 μg/mL streptomycin (ATCC; catalog # 30-2300) at 37 °C and 5% CO2. HCT-8 cells were used between passages 9 and 39 for all experiments. C. parvum Iowa strain oocysts were purchased from Bunch Grass Farm (Deary, ID), and stored in phosphate-buffered saline (PBS) with penicillin and streptomycin at 4 °C for up to 5 months prior to use. C. hominis TU502 strain oocysts were purchased from the Tzipori laboratory (Tufts University) and used within 1 month of acquisition.
Immunofluorescence assay for drug screening and follow-up growth inhibition assays
The same assay method was used to measure both C. parvum and C. hominis growth, except for adjustment in the inoculum size used to account for reduced C. hominis infectivity27. Oocysts were treated with 10 mM hydrochloric acid (10 min, 37 °C), followed by exposure to 2 mM sodium taurocholate (Sigma-Aldrich; catalog # A18346.03) in PBS (10 min, 16 °C) to initiate excystation. The mixture of treated oocysts and released sporozoites was then added to >95% confluent HCT-8 cell monolayers in 384-well plates, using material derived from 5500 C. parvum or C. hominis oocysts per well. Nitazoxanide was included on all assay plates as a positive control. Drug compounds were added 3 h post infection for both screening and follow-up assays and parasites were allowed to grow for 48 h post infection, at which time the plates were washed three times with PBS, fixed with 4% paraformaldehyde in PBS (15 min, room temperature), permeabilized with 0.25% Triton X-100 (10 min, 37 °C), washed three times with PBS with 0.1% Tween 20, and blocked with 4% bovine serum albumin (BSA) in PBS for 2 h at 37 °C or overnight at 4 °C. Parasitophorous vacuoles were then stained66 with 1.33 μg/ml fluorescein-labeled Vicia villosa lectin (Vector Laboratories) diluted in 1% BSA in PBS with 0.1% Tween 20 (1 h, 37 °C), followed by addition of Hoechst 33258 (AnaSpec) at a final concentration of 90 μM in water (15 min, 37 °C). Wells were then washed five times with PBS with 0.1% Tween 20. Images were acquired using a Nikon Eclipse TE2000 epifluorescence microscope (Nikon, USA) with an automated stage programmed to acquire a 3 × 3 tiled image of the center of each well using an Exi Blue fluorescence microscopy camera (QImaging, Canada) and a 20x objective (numerical aperture 0.45). The nucleus and parasite image channels were analyzed using NIH ImageJ (National Institutes of Health) and previously published macros27.
Cryptosporidium rate-of-action assays
C. parvum oocysts were treated to induce excystation and used to infect >90% confluent HCT-8 cells in 384-well plates as described above under growth assay. Compounds were added after 24 h of infection at each compound’s EC90 concentration and multiples thereof. At 24 h after infection (i.e. time of compound addition) and the shown time-intervals thereafter, the plates were washed, fixed, and prepared for parasite enumeration using epifluorescence microscopy as described for the growth inhibition assay described above. A separate 384-well plate was used for each time point. Parasite numbers were normalized to host cell nucleus numbers and dimethyl sulfoxide (DMSO) controls on each assay plate to determine the effect of compound inhibition at each time point.
Assays to determine the life-stage affected by drug compounds
Parasite stage-specific assays were used to assess the life-cycle stage affected by inhibitors used at two times the EC90 measured in the C. parvum growth assay48. Each of the assays used C. parvum oocysts induced to excyst and infection of HCT-8 cell monolayers as described above (see assay for drug screening and growth inhibition). Each life-cycle stage assay was performed as follows.
To assay host cell invasion, HCT-8 cell monolayers were pre-treated with two times the EC90 of each compound and incubated for 1 h prior to infection. Oocysts induced to excyst with HCl and Na+ taurocholate were added and invasion was allowed to proceed for 3 h at 37 °C before washing with PBS with 111 mM D-galactose to remove extracellular parasites. The monolayers were then fixed, permeabilized, and stained for microscopy as above to enumerate the parasitophorous vacuoles.
Because C. parvum infection in the HCT-8 system begins crudely synchronized, the number of parasitophorous vacuoles ratchets up with each round of asexual replication, cell egress, and invasion of new cells48. Therefore, to quantify C. parvum egress and re-invasion of host cells, HCT-8 monolayers grown in 384-well plates were infected and exposed to compounds as for the growth assay, and the number of parasite vacuoles was measured at 6 h and 19.5 h following infection, which are time points bracketing the time required for the first round of asexual replication. Data were expressed as the ratio of parasite vacuoles at 19.5 h and 6 h and viewed relative to the vehicle control and the known egress inhibitor MMV40367948.
Finally, development of female gametocytes was assayed by measuring the percent of C. parvum that stain for the female gametocyte marker DNA meiotic recombination protein (DMC1) which typically is expressed in female gametocytes at ~ 48 h in the HCT-8 culture system48. HCT-8 cells were infected and treated as for the growth assay, except that compounds were added at 30 h post-infection to exclude early effects on asexual replication. The percent of parasites expressing DMC1 at 48 h was then determined by staining all parasite vacuoles with V. villosa lectin and staining DMC1 with an anti-C. parvum DMC1 mouse monoclonal antibody (clone 1H10G7 (IgG2b, kappa) used as undiluted culture supernatant at 25 μL/well) and a secondary Alexa Fluor 568 goat anti-mouse IgG antibody (Invitrogen, catalog# A-11004) at 1:500 dilution (4 µg per mL).
Host cell cytotoxicity assays
Host cell toxicity was measured using CellTiter AQueous assay (Promega, USA) according to the manufacturer’s instructions. This method measures optical absorbance to measure reduction of the tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS] by NADH and NADPH produced by dehydrogenases in viable cells. Assays were conducted using >95% confluent HCT-8 cells grown in 384-well plates for 48 h (37 °C) with varying concentrations of compounds. The corner wells of each plate were trypsinized to remove cells and were used as blanks for measuring absorbance at 490 nm, and data were expressed as the percentage of cell viability for vehicle controls on each assay plate (DMSO). GraphPad PRISM software, version 9.4.1, was used to calculate the 50% toxic concentration (TC50) and selectivity indices were determined as the ratio of C. parvum EC50 to TC50.
NOD SCID gamma mouse model of cryptosporidiosis
All mouse experiments were performed in compliance with animal care guidelines and were approved by the University of Vermont Institutional Animal Care and Use Committee. Sex was not considered in study design because prior work indicates no sex-based difference in Cryptosporidium mouse infection. Male NOD SCID gamma mice (Mus musculus NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and housed for at least a week for acclimatization. Mice were housed by experimental group (4 mice per cage) in Super Mouse 750 Ventilated Cages (Micro-Isolator individually ventilated cages (Lab products, LLC) and fed Prolab IsoPro RMH 3000, 22% protein pellets (Lab Diet product # 5P75). The mouse facility was maintained with 12 h light/12 h dark cycle at a temperature range of 68-79 °F between 30-70% humidity. At the age of 4 to 5 weeks, mice were infected with 105 C. parvum oocysts and compounds were dosed by oral gavage beginning 7 days post-infection according to the regimen specified for each experiment (four mice per experimental group). Aliquoted DMSO stock solutions were stored for <10 days at −80 °C. Compound doses were prepared on the day of dosing by thawing and sonicating an aliquot of the DMSO stock, followed by dilution in 1% hydroxypropyl methylcellulose (HPMC) to a final concentration of 5% DMSO in 100 μL of 1% HPMC per dose. Oocyst shedding in feces was monitored at the specified time points qPCR using a previously described protocol67.
Pharmacokinetic studies
All procedures were performed in accordance with the Institutional Animal Care and Use Committee of Saint Louis University. Both intravenous and oral pharmacokinetic studies were conducted using 6–8-week-old male CD-1 mice (Mus musculus)(Charles River Labs). Prior to each study, they were housed in a temperature- and light-controlled environment for 5 days. All mice had free access to standard feed and water prior to and throughout the study. Intravenous dosing solution was prepared the day of the study in 60% Saline/40% Formal Glycerol to a final concentration of 0.24 mg/mL, which resulted in a clear solution. Drug was administered into the tail vein (3 mice/timepoint) at 1 mg/kg. At the given time point (0.08, 0.17, 0.5, 1, 2, 4, 6 h), the mouse was euthanized by CO2 exposure and samples collected. Oral administration of drug (2.4 mg/mL, 10 mg/kg) was performed with a solution of 0.5% Tween 80/0.5% Carboxymethyl cellulose in water. Drug was given by mouth using a stainless-steel feeding needle (3 in x2.9 mm ball). At the given time point (0.25, 0.5, 1, 2, 4, 7, 24 h), the mouse was euthanized by CO2 exposure and samples collected. In both studies, blood was obtained by cardiac puncture into sodium heparin tube (Vacutainer) and placed on ice. The blood was separated by centrifugation and the plasma stored at -80 °C until analysis. Plasma proteins were denatured by the addition of 3 volumes of ice-cold acetonitrile containing internal standard (5 ng/mL enalapril). Plasma samples were then vortexed and centrifuged at 12,000×g for 10 min. Supernatants were then transferred to a 96-well sample plates for analysis.
The intestinal tissue was obtained by dissection. Briefly, after exposing the abdominal cavity, the entire length of the intestine was removed and 4 cm segments excised from the beginning (duodenum), middle (jejunum), and end (ileum) of the small intestine, and from the proximal colon. The segments were sliced along the longitudinal axis, and the contents thoroughly rinsed away with sterile water. The tissues were stored at −80 °C until futher processed for analysis.
Intestinal tissue was macerated in a bead beater in PBS, pH 7.2. Tissue mascerates were centrifuged at 12,000 g for 10 min. The supernate was then treated with 3 volumes of ice-cold acetonitrile containing internal standard (5 ng/mL enalapril) followed by centrifugation. The final supernate was then transferred to a 96-well sample plate for analysis.
Both plasma and tissue samples were analyzed by liquid chromatography-tandem mass spectrometry (LC/MS/MS), using compound spiked into control plasma as a standard. Noncompartmental analysis was performed using Phoenix WinNonlin version 8.2 (Certara, Clayton, MO).
In silico sequence analysis and docking studies
Putative C. parvum PDEs were identified using CryptoDB and BLAST to search for genes encoding proteins homologous to human PDE5A (NCBI sp|076074)68,69. CryptoDB gene cgd3_2320 gene was designated as CpPDE1. The AlphaFold CpPDE1 model (AlphaFold: A3FQ29-F1-model_v4 (CpPDE1) was accessed on August 22, 2023 (last updated in AlphaFold DB version 2022-11-01)44,45. The crystal structure of a Schistosoma manosoni PDE4A (SmPDE4 (pdb 6EZU46)) was overlayed to enable addition of active site metal ions. Merged AlphaFold/metal ion structures were minimized using the Protein Preparation panel in Schrödinger Maestro to converge heavy atoms to RMSD 0.30 Å using the OPLS4 forcefield. Schrödinger Maestro and the Glide SP docking routine were used to produce structures for molecular docking studies47. The model was further refined using the Induced Fit docking routine in Schrödinger Maestro with and without active site waters.
Plasmid construction for CRISPR
To produce the Cas9/guide RNA plasmid, a 20-bp CpPDE1 guide sequence (5’TTTCTAGGTTTAGAGTGAGA3’) was cloned into the BbsI restriction site of the Aldo-Cas9-ribo plasmid (a gift from Boris Striepen, Univ. of Pennsylvania)70. Sanger sequencing was performed to verify integration of the CpPDE1 guide RNA into the Cas9/guide RNA plasmid. Gene fragment synthesis was employed to generate repair cassettes with or without the indicated single amino acid mutation and recoding the gene including and distal to the PAM site to prevent repeated gene cutting. Gene fragments for 50-bp homology arm to CpPDE1, a 432-bp fragment of the recodonized version of CpPDE1 with or without mutations (beginning from amino acid number 855), 3’UTR sequence of the enolase gene (cgd5_1960), the enolase promoter-nluc-neomycin resistance gene, and a 140-bp fragment of CpPDE1 3’UTR were purchased from Twist Bioscience (San Francisco, CA) (DNA sequences provided in Source Data File). Repair plasmids were constructed utilizing NEBuilder HiFi DNA assembly master mix (New England BioLabs; catalog number E5520S) and cloned into a pUC19 vector (New England BioLabs; catalog number N3041). Repair plasmids were sequence verified using whole plasmid sequencing performed by Plasmidsaurus (Oxford Nanopore Technology).
Generation of transgenic C. parvum by CRISPR/Cas9 genome editing
Transgenic C. parvum strains were generated using CRISPR/Cas9 genome editing and a method which involves in vitro electroporation of excysted sporozoites followed by infection of IFN-γ−/− mice and paromomycin selection70. All mouse handling and experiments for the creation of transgenic C. parvum strains were approved by the University of Vermont Institutional Animal Care and Use Committee. To transfect C. parvum sporozoites, the repair cassette for each mutant was PCR-amplified using the following homology end primers: 5’repair cassette homology_fwd (P1) 5’GAGACAGATATGCATAGGCATTTTG3’ and 3’repair cassette homology_rev (P2) 5’GGGATGATAGGTGCTTGAAATCAAC3’. 5× 107 C. parvum oocysts were bleached on ice, thoroughly washed with 1× PBS, and excysted by treatment with 10 mM hydrochloric acid followed by 0.2 mM sodium taurocholate in PBS. Transfection was performed using the SF Cell line 4D-Nucleofector X Large cuvette kit (Lonza; catalog number V4XC-2012) which includes a proprietary buffer and supplements. Excysted sporozoites were resuspended in 100 μl of complete SF transfection buffer (85.6 μl SF buffer + 14.4 μl Supplement 1) with 50 µg of the Cas9/gRNA plasmid and 50 µg of linearized repair template. Electroporation of the sporozoites with Cas9/guide plasmid and repair DNA template was performed using a Lonza Nucleofector 4D electroporator70.
Mouse laparotomy was performed to infect C57BL/6 ifn-γ−/− male mice (Jackson Laboratory) aged 4 to 6 weeks (n = 4) with transfected sporozoites. Transfected sporozoites were directly injected into an externalized loop of the ileum of the small intestine. Transgenic parasite selection was facilitated by the inclusion of paromomycin (GoldBio; 1263894, 16 g/liter) in the mouse drinking water. Fecal samples were collected from cages to detect presence of transgenic parasites using a nano-luciferase assay kit (Nano-Glo; Promega; catalog number N1120)70. For this, one or two pellets of mouse fecal matter and ~10 3 mm glass beads (Fisher Scientific; catalog number 11-312 A) were suspended in fecal lysis buffer (50 mM Tris-HCl, pH 7.4, 2 mM EDTA, 2Mm DTT, 10% glycerol, 1% Triton X-100) in a 1.5 mL centrifuge tube and vortexed thoroughly. The fecal pellets were incubated at 4 °C for 30 mins. Meanwhile, Nano-Glo substrate was diluted 1:50 in Nano-Glo buffer at room temp. After incubation, diluted Nano-Glo substrate was mixed 1:1 with fecal pellet homegenate in a 384-well white polystyrene plate (Corning; catalog number 3574) and read immediately using a Synergy H1 plate reader to measure luminescence (integration time: 0.3 seconds). Mouse feces containing transgenic parasites were then passaged by infection of male NOD SCID gamma mice (Jackson Laboratory) which become chronically infected with C. parvum20. To confirm the correct 5′ and 3′ integration events following homologous recombination, total fecal DNA was obtained using an E.Z.N.A. stool DNA kit (Omega Bio-Tek) according to the manufacturer’s protocol, barring the inclusion of six freeze-thaw cycles at the first step. Then, primers complementary to the 5′ 50-bp homologous CpPDE1 region and the CpPDE1 3′ UTR (primers P1 and P2 (see Fig. 6A and plasmid construction above)) were used to PCR-amplify the repair cassette from fecal DNA originating from infected NOD SCID gamma mice, and Sanger sequencing was used to validate accuracy of the incorporated mutations.
Purification of transgenic oocysts from mouse feces
Oocysts for subsequent experimentation were purified as follows71. Feces from NOD SCID gamma mice infected with the transgenic C. parvum strains were collected for 1–2 weeks into 50 mL conical centrifuge tubes. Cold water was added to about 35 mL fecal material in a 50 mL conical and blended using a Cole-Parmer LabGEN 125 Homogenizer (catalog number EW-04727-11) with autoclavable omni plastic tip. Two filter apparatuses with 850 µm and 250 µm mesh filters inserted into PVC fittings (Bel-Art mini sieve micro sieve set; catalog number F37845-1000) were assembled. The homogenized fecal slurry was first poured over the 850 µm utilizing a sterile spatula to guide the sample through the mesh filter into a conical below. The solid residues were resuspended in 35 mL cold water in 50 mL falcon tube and re-homogenized then filtered through the 850 µm. Homogenization and filtration were repeated an additional 5-6 times. The 850 µm mesh filter apparatus was replaced with a 250 µm mesh filter apparatus and the homogenized, filtered sample was passed though the 250 µm mesh filter into a sterile conical below. The 250 µm filtered sample was poured into a 500 mL conical. Solid residue on top the 250 µm mesh filter was vortexed in 35 mL cold water and re-filtered. Filtrate from each filtration step (about 3–4 steps) was transferred into a 500 mL conical, until about 400 mL filtered sample was attained.
The 500 mL conical with filtrate was centrifuged at 1000×g for 10 min at 4 °C. Supernatant was discarded into an autoclavable liquid waste container. The pellet was resuspended in 50 mL cold water and transferred into a 50 mL falcon tube. Sample was centrifuged at 1000×g for 10 min at 4 °C and the supernatant was discarded. Then, pellet was resuspended in 50 mL cold water and split into two conical tubes, 25 mL each. 25 mL sucrose floatation solution (1.33 specific gravity) was added to each tube and mixed gently by inversion. Samples were immediately centrifuged at 1000×g for 10 min at 4 °C. Supernatant from each tube was poured into a clean 500 mL conical and 300 mL cold water was added to sample to collect oocysts. The 500 mL conical was centrifuged at 1500×g for 15 min at 4 °C and supernatant was discarded in an autoclavable liquid waste container following centrifugation. 0.85% NaCl was added to the pellet (with oocysts) up to 5 mL volume and mixed by pipetting. To separate oocysts, 0.8 mL cold 1.25 M CsCl2 solution was added to ten clean 1.5 mL centrifuge tubes followed by the careful addition of 0.5 mL pelleted oocysts on top of the CsCl2 solution without allowing the solutions to mix. Tubes were centrifuged 16,000×g for 3 min at 4 °C. 1 mL of the supernatant (with floating oocysts) from each tube was transferred to new centrifuge tubes and mixed with 0.5 mL 0.85% NaCl. Oocysts were centrifuged 16,000×g for 3 min at 4 °C. The oocyst pellets from each tube were combined into one tube using 1 mL 0.85% NaCl immediately followed by centrifugation at 16,000×g for 3 min, 4 °C. Supernatant was discarded and pellet was resuspended in 1 mL PBS with 120 U/mL penicillin and 120 μg/mL streptomycin for storage at 4 °C.
SafetyScan47 assay panel, mammalian phosphodiesterase assay panel, CYP inhibition panel, hERG binding and aqueous solubility
Potential off target effects of PDEi5 were screened in 78 assays (47 targets) with the Eurofins SafetyScan47 panel49. Assay technologies used include: cAMP secondary messenger assays using cell lines expressing non-tagged GPCRs, a calcium secondary messenger assay to measure GPCR activity in live cells, a nuclear hormone receptor assay (NHR Pro) and Nuclear Translocation (NHR NT) assays, a proprietary kinase screening platform (KINOMEScan) that uses competition to quantify kinase binding, a neurotransmitter uptake assay, potassium and membrane potential assays, and specific enzymatic assays used to measure consumption of a substrate or production of a product. Mammalian PDE inhibition assays were also performed by Eurofins, using PDEs sourced as indicated in Table S5. The assay method for each enzyme was to measure production of tritiated product (either [3H]-5’GMP or [3H]-5’AMP (substrates indicated in Table S5)) by scintillation counting. Results were expressed as a percent inhibition of control specific activity in the presence of 10 μM PDEi1. CYP inhibition was measured by Eurofins using the indicated recombinant human enzymes. hERG affinity was measured by Eurofins at 10 µM as % inhibition of the binding of [3H]-dofetilide to the human potassium channel hERG. Aqueous solubility was measured by Eurofins as kinetic solubility in aqueous solution at pH 7.4.
Statistical information and figure preparation
GraphPad Prism software (version 9.5.1) was used to prepare all graphs, calculate compound dose-response curves, calculate decay curves for time-kill assays, and perform statistical analyses. The limit of detection of the qPCR assay performed on mouse feces is ~ 100 oocysts per gram of dried feces, and samples for which no signal was detercted were plotted at the limit-of-detection (LOD) for purposes of graphing and statistical analysis. Statistical significance was assessed as indicated in figure legends. Graphs were exported as.eps files, and final figures were prepared using Adobe Illustrator.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. Source data are provided with this paper. The following accession numbers were used for this study: hPDE5A [https://www.ncbi.nlm.nih.gov/gene/8654]; CpPDE1 [https://www.ncbi.nlm.nih.gov/search/all/?term=cgd3_2320]; CpPDE2 [https://www.ncbi.nlm.nih.gov/search/all/?term=cgd6_500]; CpPDE3 [https://www.ncbi.nlm.nih.gov/search/all/?term=cgd6_4020]; CpEnolase [https://www.ncbi.nlm.nih.gov/search/all/?term=cgd5_1960]; and SmPDE4A [https://www.rcsb.org/structure/6EZU]. Source data are provided with this paper.
References
Checkley, W. et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect. Dis. 15, 85–94 (2015).
Hlavsa, M. C. et al. Surveillance for waterborne disease outbreaks and other health events associated with recreational water–United States, 2007–2008. MMWR Surveill. Summ. 60, 1–32 (2011).
Liu, J. et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 388, 1291–1301 (2016).
Khalil, I. A. et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob. Health 6, e758–e768 (2018).
Amadi, B. et al. Effect of nitazoxanide on morbidity and mortality in Zambian children with cryptosporidiosis: a randomised controlled trial. Lancet 360, 1375–1380 (2002).
Amadi, B. et al. High dose prolonged treatment with nitazoxanide is not effective for cryptosporidiosis in HIV positive Zambian children: a randomised controlled trial. BMC Infect. Dis. 9, 195 (2009).
Bessoff, K. et al. Identification of Cryptosporidium parvum active chemical series by Repurposing the open access malaria box. Antimicrob. Agents Chemother. 58, 2731–2739 (2014).
Buckner, F. S. et al. Optimization of methionyl tRNA-synthetase inhibitors for treatment of cryptosporidium infection. Antimicrob. Agents Chemother. https://doi.org/10.1128/AAC.02061-18 (2019).
Love, M. S. et al. A high-throughput phenotypic screen identifies clofazimine as a potential treatment for cryptosporidiosis. PLoS Negl. Trop. Dis. 11, e0005373 (2017).
Baragana, B. et al. Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis. Proc. Natl Acad. Sci. USA 116, 7015–7020 (2019).
Castellanos-Gonzalez, A. et al. A novel calcium-dependent protein kinase inhibitor as a lead compound for treating cryptosporidiosis. J. Infect. Dis. 208, 1342–1348 (2013).
Murphy, R. C. et al. Discovery of potent and selective inhibitors of calcium-dependent protein kinase 1 (CDPK1) from C. parvum and T. gondii. ACS Med. Chem. Lett. 1, 331–335 (2010).
Schaefer, D. A. et al. Novel bumped kinase inhibitors are safe and effective therapeutics in the calf clinical model for cryptosporidiosis. J. Infect. Dis. 214, 1856–1864 (2016).
Guo, F. et al. Amelioration of Cryptosporidium parvum infection in vitro and in vivo by targeting parasite fatty acyl-coenzyme A synthetases. J. Infect. Dis. 209, 1279–1287 (2014).
Guo, F., Zhang, H., McNair, N. N., Mead, J. R. & Zhu, G. The existing drug vorinostat as a new lead against cryptosporidiosis by targeting the parasite histone deacetylases. J. Infect. Dis. 217, 1110–1117 (2018).
Jain, V. et al. Targeting prolyl-tRNA synthetase to accelerate drug discovery against malaria, leishmaniasis, toxoplasmosis, cryptosporidiosis, and coccidiosis. Structure 25, 1495–1505.e1496 (2017).
Huang, W. et al. 5-Aminopyrazole-4-carboxamide-based compounds prevent the growth of cryptosporidium parvum. Antimicrob. Agents Chemother. https://doi.org/10.1128/AAC.00020-17 (2017).
Manjunatha, U. H. et al. A Cryptosporidium PI(4)K inhibitor is a drug candidate for cryptosporidiosis. Nature 546, 376–380 (2017).
Lunde, C. S. et al. Identification of a potent benzoxaborole drug candidate for treating cryptosporidiosis. Nat. Commun. 10, 2816 (2019).
Jumani, R. S. et al. A novel piperazine-based drug lead for cryptosporidiosis from the medicines for malaria venture open-access malaria box. Antimicrob. Agents Chemother. 62, e01505–e01517 (2018).
Vinayak, S. et al. Bicyclic azetidines kill the diarrheal pathogen Cryptosporidium in mice by inhibiting parasite phenylalanyl-tRNA synthetase. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.aba8412 (2020).
Oboh, E. et al. Optimization of the urea linker of triazolopyridazine MMV665917 results in a new anticryptosporidial lead with improved potency and predicted hERG safety margin. J. Med. Chem. 64, 11729–11745 (2021).
Hasan, M. M. et al. Spontaneous selection of cryptosporidium drug resistance in a calf model of infection. Antimicrob. Agents Chemother. https://doi.org/10.1128/AAC.00023-21 (2021).
Heemskerk, J., Tobin, A. J. & Bain, L. J. Teaching old drugs new tricks. Meeting of the Neurodegeneration Drug Screening Consortium, 7-8 April 2002, Washington, DC, USA. Trends Neurosci. 25, 494–496 (2002).
De Pascale, G. et al. Remdesivir plus Dexamethasone in COVID-19: a cohort study of severe patients requiring high flow oxygen therapy or non-invasive ventilation. PLoS ONE 17, e0267038 (2022).
Edwards, A. What are the odds of finding a COVID-19 drug from a lab repurposing screen? J. Chem. Inf. Model. 60, 5727–5729 (2020).
Bessoff, K., Sateriale, A., Lee, K. K. & Huston, C. D. Drug repurposing screen reveals FDA-approved inhibitors of human HMG-CoA reductase and isoprenoid synthesis that block Cryptosporidium parvum growth. Antimicrob. Agents Chemother. 57, 1804–1814 (2013).
Arnold, S. L. M. et al. Necessity of bumped kinase inhibitor gastrointestinal exposure in treating Cryptosporidium infection. J. Infect. Dis. 216, 55–63 (2017).
Iroh Tam, P. et al. Clofazimine for treatment of cryptosporidiosis in human immunodeficiency virus infected adults: an experimental medicine, randomized, double-blind, placebo-controlled phase 2a trial. Clin. Infect. Dis. 73, 183–191 (2021).
Wermuth, C. G. Selective optimization of side activities: the SOSA approach. Drug Discov. Today 11, 160–164 (2006).
Merck-KGaA. Biopharma Mini Library, <https://www.emdgroup.com/en/research/open-innovation/biopharma-open-innovation-portal/biopharma-mini-library.html> (2023).
Merck-KGaA. Open Global Health Library, <https://www.emdgroup.com/en/research/open-innovation/biopharma-open-innovation-portal/open-global-health-library.html> (2023).
Baell, J. B. & Holloway, G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010).
Camps, M. et al. Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat. Med. 11, 936–943 (2005).
Pomel, V. et al. Furan-2-ylmethylene thiazolidinediones as novel, potent, and selective inhibitors of phosphoinositide 3-kinase gamma. J. Med. Chem. 49, 3857–3871 (2006).
Maggiora, G., Vogt, M., Stumpfe, D. & Bajorath, J. Molecular similarity in medicinal chemistry. J. Med. Chem. 57, 3186–3204 (2014).
Burch, H. A. Nitrofuryl heterocycles. VII. 4-Amino-6-(5-nitro-2-furyl)-1H-pyrazolo[3,4-d]pyrimidines. J. Med. Chem. 11, 79–83 (1968).
Mouton, J. W. et al. Tissue concentrations: do we ever learn? J. Antimicrob. Chemother. 61, 235–237 (2008).
Rawson, D. J. et al. The discovery of UK-369003, a novel PDE5 inhibitor with the potential for oral bioavailability and dose-proportional pharmacokinetics. Bioorg. Med. Chem. 20, 498–509 (2012).
Fiorito, J. et al. Synthesis of quinoline derivatives: discovery of a potent and selective phosphodiesterase 5 inhibitor for the treatment of Alzheimer’s disease. Eur. J. Med. Chem. 60, 285–294 (2013).
Moss, W. J., Brusini, L., Kuehnel, R., Brochet, M. & Brown, K. M. Apicomplexan phosphodiesterases in cyclic nucleotide turnover: conservation, function, and therapeutic potential. mBio 15, e0305623 (2024).
Walzer, K. A. et al. Transcriptional control of the Cryptosporidium life cycle. Nature 630, 174–180 (2024).
Tandel, J. et al. Life cycle progression and sexual development of the apicomplexan parasite Cryptosporidium parvum. Nat. Microbiol. 4, 2226–2236 (2019).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Brown, D. G. et al. The structure of Schistosoma mansoni Phospohodiesterase 4A in complex with cAMP. (RCSB Protein Data Bank, 2018).
Friesner, R. A. et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 (2004).
Jumani, R. S. et al. A suite of phenotypic assays to ensure pipeline diversity when prioritizing drug-like Cryptosporidium growth inhibitors. Nat. Commun. 10, 1862 (2019).
Eurofins. In Vitro Safety Pharmacology Profiling Panels. <https://www.eurofinsdiscovery.com/solution/safety-panels> (2023).
Banerjee, P., Eckert, A. O., Schrey, A. K. & Preissner, R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 46, W257–W263 (2018).
English, E. D., Guerin, A., Tandel, J. & Striepen, B. Live imaging of the Cryptosporidium parvum life cycle reveals direct development of male and female gametes from type I meronts. PLoS Biol. 20, e3001604 (2022).
Campbell, L. D., Stewart, J. N. & Mead, J. R. Susceptibility to Cryptosporidium parvum infections in cytokine- and chemokine-receptor knockout mice. J. Parasitol. 88, 1014–1016 (2002).
Smith, L. M., Bonafonte, M. T., Campbell, L. D. & Mead, J. R. Exogenous interleukin-12 (IL-12) exacerbates Cryptosporidium parvum infection in gamma interferon knockout mice. Exp. Parasitol. 98, 123–133 (2001).
Gorla, S. K. et al. Validation of IMP dehydrogenase inhibitors in a mouse model of cryptosporidiosis. Antimicrob. Agents Chemother. 58, 1603–1614 (2014).
Veerman, J. et al. Synthesis and evaluation of analogs of the phenylpyridazinone NPD-001 as potent trypanosomal TbrPDEB1 phosphodiesterase inhibitors and in vitro trypanocidals. Bioorg. Med. Chem. 24, 1573–1581 (2016).
Blaazer, A. R. et al. Targeting a subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) enables the structure-based discovery of selective inhibitors with trypanocidal activity. J. Med. Chem. 61, 3870–3888 (2018).
de Heuvel, E. et al. Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors. Bioorg. Med. Chem. 27, 3998–4012 (2019).
Amata, E., Bland, N. D., Campbell, R. K. & Pollastri, M. P. Evaluation of pyrrolidine and pyrazolone derivatives as inhibitors of trypanosomal phosphodiesterase B1 (TbrPDEB1). Tetrahedron Lett. 56, 2832–2835 (2015).
Zheng, Y. et al. Discovery of 5-phenylpyrazolopyrimidinone analogs as potent antitrypanosomal agents with in vivo efficacy. J. Med. Chem. 66, 10252–10264 (2023).
Long, T. et al. Phenotypic, chemical and functional characterization of cyclic nucleotide phosphodiesterase 4 (PDE4) as a potential anthelmintic drug target. PLoS Negl. Trop. Dis. 11, e0005680 (2017).
Sebastian-Perez, V. et al. Discovery of novel Schistosoma mansoni PDE4A inhibitors as potential agents against schistosomiasis. Future Med. Chem. 11, 1703–1720 (2019).
Zheng, Y. et al. To target or not to target schistosoma mansoni cyclic nucleotide phosphodiesterase 4A? Int. J. Mol. Sci. https://doi.org/10.3390/ijms24076817 (2023).
Botros, S. S. et al. The phosphodiesterase-4 inhibitor roflumilast impacts Schistosoma mansoni ovipositing in vitro but displays only modest antischistosomal activity in vivo. Exp. Parasitol. 208, 107793 (2020).
Howard, B. L. et al. Identification of potent phosphodiesterase inhibitors that demonstrate cyclic nucleotide-dependent functions in apicomplexan parasites. ACS Chem. Biol. 10, 1145–1154 (2015).
Zheng, Y. et al. Structural optimization of BIPPO analogs as potent antimalarials. Molecules https://doi.org/10.3390/molecules28134939 (2023).
Gut, J. & Nelson, R. G. Cryptosporidium parvum: synchronized excystation in vitro and evaluation of sporozoite infectivity with a new lectin-based assay. J. Eukaryot. Microbiol. 46, 56S–57S (1999).
Parr, J. B. et al. Detection and quantification of Cryptosporidium in HCT-8 cells and human fecal specimens using real-time polymerase chain reaction. Am. J. Trop. Med. Hyg. 76, 938–942 (2007).
Amos, B. et al. VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 50, D898–D911 (2022).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Vinayak, S. et al. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523, 477–480 (2015).
Pawlowic, M. C., Vinayak, S., Sateriale, A., Brooks, C. F. & Striepen, B. Generating and maintaining transgenic Cryptosporidium parvum parasites. Curr. Protoc. Microbiol. 46, 20B 22 21–20B 22 32 (2017).
Acknowledgements
We thank Merck KGaA, Darmstadt, Germany and the Open Innovation Portal for generous provision of compounds for screening and follow up studies, and for generous sharing of pre-existing data. CDH, MJM, and DWG were funded by NIAID R21R33 AI141184.
Author information
Authors and Affiliations
Contributions
C.D.H., D.W.G., and M.J.M. conceived and supervised the project. J.A., J.E.T., E.B.M., B.K.M., D.W.G., T.S., M.J.M., and C.D.H. jointly designed experiments. J.A., E.B.M., B.K.M., and P.M. generated CRISPR-modified parasites. JA conducted all life cycle assays and experiments with transgenic parasites. JET and ES conducted all compound screening, dose-response testing, and rate-of-action assays. JET conducted in vitro cytotoxicity assays. A.C.C. and P.M. conducted all animal compound efficacy testing. M.G. and M.J.M. planned and synthesized compound PDEi5. S.D.C. and D.W.G. conducted pharmacokinetic/pharmacodynamic studies. T.S. helped analyze compound screening results, conducted the nearest neighbor structure activity relationship analysis, and provided existing data where available. M.J.M. directed chemical synthesis and conduct of outsourced metabolism and safety assays, and conducted in silico docking analyses. C.D.H. and M.J.M. wrote the manuscript which was edited by J.A., D.W.G., and T.S.
Corresponding authors
Ethics declarations
Competing interests
C.D.H., M.J.M., and M.G. are inventors on a pending patent application jointly filed by the University of Vermont and St. Louis University that covers use of pyrazolopyrimidines as anti-infective agents (PCT Application No. 63/504,716 filed May 26, 2023). All other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks William Witola and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ajiboye, J., Teixeira, J.E., Gasonoo, M. et al. Identification of potent and orally efficacious phosphodiesterase inhibitors in Cryptosporidium parvum-infected immunocompromised male mice. Nat Commun 15, 8272 (2024). https://doi.org/10.1038/s41467-024-52658-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52658-y
This article is cited by
-
Targeted CRISPR screens reveal genes essential for Cryptosporidium survival in the host intestine
Nature Communications (2025)
-
Computational drug repurposing: approaches, evaluation of in silico resources and case studies
Nature Reviews Drug Discovery (2025)