Abstract
Rose is an important ornamental crop cultivated globally for perfume production. However, our understanding of the mechanisms underlying scent production and molecular breeding for fragrance is hindered by the lack of a reference genome for tea roses. We present the first complete telomere-to-telomere (T2T) genome of Rosa gigantea, with high quality (QV > 60), including detailed characterization of the structural features of repetitive regions. The expansion of genes associated with phenylpropanoid biosynthesis may account for the unique tea scent. We uncover the release rhythm of aromatic volatile organic compounds and their gene regulatory networks through comparative genomics and time-ordered gene co-expression networks. Analyzes of eugenol homologs demonstrate how plants attract pollinators using specialized phenylpropanoids in specific tissues. This study highlights the conservation and utilization of genetic diversity from wild endangered species through multi-omics approaches, providing a scientific foundation for enhancing rose fragrance via de novo domestication.
Similar content being viewed by others
Introduction
As the ancestor of tea roses, Rosa gigantea (RG) contributed to the tea scent, yellow color, large flowers, and vigorous growth of modern roses. Before the 17th century, roses were widely cultivated in the East and West1,2. After the establishment of sea lines of communication, Western botanists discovered Chinese roses, with a unique scent characterized by a subtle medicinal fragrance and gentle, refreshing sweetness, described as a tea scent by Westerners because it was reminiscent of another famous import from China, green tea3. The wild tea rose R. gigantea, with the typical tea scent and large yellow flowers, formed the early tea roses (Supplementary Fig. 1). Two famous Chinese garden roses, ‘Hume’s Blush Tea-scented China’ and ‘Parks’ Yellow Tea-scented China’, were introduced from China to England and then to France in the early 19th century4. Together with R. chinensis ‘Old Blush’ and R. chinensis var. semperflorens, gene exchange between these varieties and European roses started the first revolution in rose breeding5. Symbolized by ‘La France’ in 1867, Hybrid Tea Rose became the most popular owing to its large diameter, high-centered shape, single long stem, and diverse colors6.
Human intervention has generated roses with highly diverse colors, covering all shades except blue, but a limited fragrance, with a single characteristic scent. Hybrid Tea Rose has been continuously improved to create colorful hybrid roses7. Currently, most popular cut-flower roses are derived from Hybrid Tea Rose8. The aromatic trait has been neglected during the breeding process, and the tea scent has been lost6. In addition, to ensure a stable quality, oil roses with the classic scent have been propagated through strictly asexual reproduction, resulting in the loss of fragrance diversity. However, there has been a shift in consumer attitudes, whereby people are no longer satisfied with colored but odorless roses or single-scented rose essential oil products. There is demand for the ancestral tea scent and the development of new plant essential oil products with biological activities and multi-functionalities.
Multi-omics analyzes provide insights into the preservation of genetic resources and a basis for reviving the tea scent. Despite the importance of R. gigantea for the restoration of the tea scent, it has been over-exploited and population sizes have decreased sharply in the wild (Supplementary Fig. 2). Therefore, preservation and genetic research are urgently needed. Using traditional sequencing methods, it is difficult to assemble complex regions, such as telomeres and their surrounding long repetitive sequences, ribosomal DNA, and tandem repeat sequences. The three published rose genomes9,10,11 are incomplete, with gaps, and cannot serve as references for tea-scented or yellow roses. The combination of Nanopore ultra-long read sequencing (to span overlapping clusters across chromosomal arms and peri-centromeric regions) and PacBio HiFi sequencing (to fill in gaps) provides a strategy to overcome this complexity12,13,14. In this study, the first complete telomere-to-telomere (T2T) genome of the endangered ancestor of tea rose, R. gigantea, was generated. The sensory characteristics, release patterns, and regulatory networks15 related to the unique tea scent were elucidated using multi-omics technologies16.
Results
A complete T2T reference genome for R. gigantea
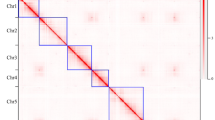
The first complete T2T gap-free genome for Rosa was generated, containing seven chromosomes with a total length of 549.76 Mb (Fig. 1a, b), similar to the estimated genome size (530.96 Mb). The result was consistent with a karyotype test (Supplementary Fig. 3) and k-mer (k = 19) analysis (Supplementary Fig. 4). Additionally, 159 bp candidate centromeric repeat sequences were found and designated as CEN159 satellites, representing the centromeric region of R. gigantea (RG). This region was the same length as the centromeric satellite OBC22617 identified in R. chinensis ‘Old Blush’ (RC). Oligo-FISH with CEN159 monomers as probes demonstrated clear and specific signals at most centromeres of RG (Fig. 1d, e).
a Circos plot of gene features at 500-kb intervals across the 7 chromosomes. TRF, Tandem Repeat Finder. SVs, Structural Variation. b Hi-C chromatin interaction map of the RG assembly. c Positions of centromeres and telomeres on 7 chromosomes. d Circos representation of the distribution of RgCEN159 (blue), the pericentromeric region (red), Ty3/Gypsy (orange), and Ty1/Copia repeat elements (yellow) along the seven chromosomes. e FISH with RgCEN159 oligo probes on R. gigantea chromosomes. f FISH with 45S rDNA oligo probes on R. gigantea chromosomes. Each experiment of (e and f) was repeated independently with similar results at least three times. Source data are provided as a Source Data file.
Thirteen telomeres and one 45S rDNA end were identified. While no telomere was found at the end of chromosome 3 in three assembly versions, 1198 45S rDNA sites were specifically detected in the region (Fig. 1c). This was similar to the discoveries of short-arm 45S rDNA clusters at the ends of chromosomes 2 and 4, instead of telomeres, in Arabidopsis18 and rDNA repeats on the short arms of human acrocentric chromosomes19,20. To verify the sequencing results, the telomere and 45S rDNA sequences were visualized by FISH, revealing 45S rDNA sites on two sister chromatids of chromosome 3 (Fig. 1f). In addition to new insights into high-copy repeats in non-coding regions (Supplementary Table 1), functionally important protein-coding genes were also discovered (Supplementary Table 2). All 33 gaps in the RC assembly were filled in the RG assembly. These gaps were distributed throughout the genome. There were six predicted genes in gap regions, suggesting that these genes would not be identified using RC as a reference genome. These genes included COX1 (cytochrome c oxidase subunit 1), a well-established mitochondrial gene involved in horizontal gene transfer among angiosperms21 (Supplementary Fig. 5).
The R. gigantea genome assembly had a higher completeness and accuracy than those of other rose genomes. First, the gap-free assembly showed a significant improvement in continuity over those of all previously released genome assemblies. The contig N50 lengths of RG were higher than those reported for RC, R. wichuraiana (RW), R. rugosa (RR), and R. persica (RP) (Supplementary Table 3). Second, ~99.0% of the core conserved plant genes (1597 out of 1614 BUSCOs) were complete in the RG genome assembly, indicating high genomic completeness (Supplementary Table 4). In particular, 7 centromeres, 13 telomeres, and one 45S rDNA site were assembled in the R. gigantea genome (Supplementary Table 5), while none of the telomeres were captured in the other rose genomes. Finally, the consensus quality values (QV) for the genome (60.53) and each chromosome (57.30–62.49) indicated a high accuracy (Supplementary Table 6).
T2T genome of a wild ancestral species contributes genetic diversity to the Rosa pan-genome
RG has not experienced whole-genome duplication (WGD) events22, as evidenced by the one-to-one syntenic depth ratio between RG and RC (Supplementary Fig. 6). This suggests that the RG genome only underwent the core eudicot-specific gamma whole-genome triplication. Phylogenetic trees with divergence time estimates were generated based on 271 single-copy orthologous genes from 13 species, including 5 species of Rosa, 6 species from other genera in the family Rosaceae, Vitis vinifera as a distant relative, and Apostasia shenzhenica as an outgroup. A total of 464,307 genes were identified and grouped into 79,059 orthologous gene families in all 13 species (Supplementary Table 7). In Rosaceae, Prunus, Malus, and Pyrus diverged early from Rubus, Rosa and Fragaria. In Rosa, the order of species emergence was RP, RR, RG, RC, and RW (Fig. 2a).
Within Rosa, there is variation in various phenotypic traits, such as plant type, inflorescence, color, and fragrance (Supplementary Table 8, Fig. 2b). There was 116–365 Mb of syntenic regions in comparisons between the R. gigantea genome and the genomes of the other four roses (Supplementary Table 9). The whole-genome alignment can serve as a reliable data set to predict levels of evolutionary constraint23. The gap-free genome provides an opportunity to characterize structural variants (SVs) that might be missed owing to limitations in the assembly technology24. Therefore, 4165–44,181 SVs were identified between the R. gigantea assembly and R. persica, R. rugosa, R. chinensis ‘Old Blush’, and R. wichuraiana genomes (Supplementary Fig. 7), including 622,584 presence/absence variants, 90–249 inversions, 2526–16,939 translocations, and 1540–29504 duplications. There were also 1,669,504–3,638,390 SNPs, 110,442–271,481 insertions, 91,077–233,107 deletions, 172–26,767 highly diverged regions, and 6227–58,596 unaligned regions. SVs were unevenly distributed along the chromosomes. The lengths of these regions are listed in Supplementary Table 10, showing substantial differentiation within Rosa (Fig. 2c). Therefore, the R. chinensis ‘Old Blush’ reference genome is not suitable for analyzes of R. gigantea, including promoter prediction or feature extraction. In addition, a collinearity analysis showed that the newly obtained T2T genome, with the most complete assembly, provided the best framework for constructing a high-quality Rosa pan-genome, combined with published genomes for Rosa. Furthermore, a 15 Mb specific inversion, including 1416 genes, was identified on the chr2 arm that was not detected in the other four roses. There was also a 22 Mb inversion on chr4 differentiating R. gigantea from R. rugosa, including 2000 genes.
Based on the same protein-coding gene set25, evolutionarily conserved genomic regions were characterized by identifying shared syntenic orthologous genes in Rosa species. Among 33042 pan-gene families, the five species contained 22713–27229 gene families. A total of 15703 (57.67–69.14%) were shared among all species, probably representing the core gene set of the Rosa species complex (Supplementary Fig. 8). In addition, there were 13981 dispensable gene families and 3358 species-specific gene families. Notably, R. gigantea possessed 1646 species-specific gene families, significantly more than the estimates for the other four Rosa species. This indicates that the genome of R. gigantea could contribute additional sequence diversity to the Rosa pan-genome. Increasing evidence suggests that alleles in wild species may play a key role in regulating adaptive traits in crops26. For example, in turbocharging introgression breeding of apple, allelic diversity in fruit germplasm provides a useful resource for the introduction of new exotic genes to meet consumer preferences and environmental challenges27. However, alleles in wild species were lost in cultivated varieties due to genetic bottlenecks during domestication. A similar situation may explain the evolution of rose scent, further emphasizing the importance of the studying genomes of wild roses for pan-genome construction and scent improvement.
Expansion of genes involved in phenylpropanoid biosynthesis contributes to tea scent evolution
Scent-related genes in phenylpropanoid/benzenoid, terpenoid, and fatty acid derivative biosynthetic pathways of R. gigantea were identified and characterized (Supplementary Data 1, Supplementary Fig. 9). Although only a few fatty acid derivatives contribute to rose floral scents, no related enzymes have been isolated or characterized from roses. More enzymatic genes were derived from tandem or proximal duplications within the phenylpropanoid/benzenoid metabolic pathway compared to in the terpenoid biosynthesis pathways28. A phylogenetic analysis of 13 species in Rosa showed that three odor-related pathways, including phenylpropanoid biosynthesis, terpenoid backbone biosynthesis, and sesquiterpenoid and triterpenoid biosynthesis pathways, were significantly enriched in the 854 gene family expansions in R. gigantea (Supplementary Fig. 10), potentially explaining the unique tea scent. Therefore, the major volatile organic compound (VOC) metabolic pathways were reconstructed, including phenylpropanoid/benzenoid (Supplementary Fig. 11) and terpenoid (Supplementary Fig. 12) biosynthesis. According to the main aroma components associated with the tea scent, two branching pathways involved in the synthesis of rose benzenoids are potentially linked to flower scent formation in R. gigantea.
Multiple O-methyltransferases (OMTs)29 involved in the production of TMB and DMT were identified (Supplementary Fig. 13). In R. chinensis, phloroglucinol O-methyltransferase (POMT) catalyzes the first methylation step of phloroglucinol (PLG) to 3,5-dihdroxyanisole (DHA). The expression level of a candidate POMT was found to be 30 times higher in R. chinensis than in R. gigantea, which may be related to high TMB synthesis in R. chinensis and low TMB in R. gigantea. DHA is converted to TMB by two orcinol O-methyltransferases in R. chinensis. Despite sharing 96.5% similarity at the amino acid level, RcOOMT1 and RcOOMT2 exhibit different substrate specificities in phenolic methyl ether biosynthesis. RcOOMT1 may have evolved from an OOMT2-like gene, and its emergence was a critical step in the evolution of scent production in Chinese roses. Both RcOOMT1 and RcOOMT2 efficiently methylate orcinol to DMT, contributing to the characteristic floral scent of R. gigantea. Four OOMT paralogs were identified in R. chinensis ‘Old Blush’; however, only two (RchiOBHm_Chr2g0119291 and RchiOBHm_Chr2g0128091) were highly expressed (~1000× higher than levels of other paralogs), suggesting that they corresponded to RcOOMT1 and RcOOMT2 and contributed to TMB synthesis. Five OOMT paralogs were identified in R. gigantea and three of these showed specific, high expression (Fig. 3 a–c). Functionally distinct OMTs are believed to have evolved from a common ancestral gene through gene duplication and mutation. RgOOMT1 (RgigChr2G00139230) and RgOOMT2 (RgigChr2G00139280) originated from a proximal duplication of another OOMT paralog (RgigChr2G00139210), resulting from a WGD. RgOOMT3 (RgigChr2G00153620) formed another OOMT cluster with RgigChr2G00153520 through the transposed duplication of RgigChr2G00139210. There were only one or two OOMT paralogs in RP, RR, and RW. RgigChr2G00139210 and RgigChr5G00009200 were paralogs resulting from the WGD event in Rosa, and subsequent proximal and transposed duplications of RgigChr2G00139210 in R. gigantea led to the high production of specific DMT. As the expression of RgOOMT1 was highly coincident with the production of DMT, it was the main gene involved in the synthesis of DMT in R. gigantea.
a Maximum likelihood tree based on POMT and OOMT homologs in Rosa. Scale bar represents 0.3 substitutions per site; b Localization and expression of OOMT on the genome of five roses; c Localization and expression of POMT on the genome of five roses; d Maximum likelihood tree based on EGS and IGS homologs. Scale bar represents 0.6 substitutions per site; e Localization and expression of IGS on the genome of five roses; f Localization and expression of EGS on the genome of five roses; RG, R. gigantea. RC, R. chinensis ‘Old Blush’. RW, R. wichuraiana. RR, R. rugosa. RP, R. persica. Scale bar represents 0.6 substitutions per site. Source data are provided as a Source Data file.
Five eugenol synthases (EGSs) were identified in all five roses. EGS1 was separated from a cluster containing EGS2–EGS5 (Fig. 3d–f), indicating that the duplication of the first ancestral orthologous EGS1 occurred before the species diverged. According to the high eugenol emission of R. gigantea while low eugenol emission of R. chinensis ‘Old Blush’, the accumulation of eugenol was not correlated with the number of EGS genes (Supplementary Fig. 14). All EGSs were located on chr6 with two EGS clusters. The first cluster was formed by a proximal repeat of RgEGS1 and a tandem repeat between RgEGS2 and RgEGS3. RgEGS1 (RgigChr6G00350620) reduces coniferyl ester to eugenol, as evidenced by the similar expression trends. The expression levels of RgEGS3 (Chr6G00350680) were much higher than those of other homologs, suggesting its important role in eugenol production. RgEGS2 (RgigChr6G00350670) was highly expressed in the gynoecium specifically, laying the substrate foundation for the production of methyleugenol. The other cluster contained RgEGS4 and RgEGS5, and the expression levels of these genes were not correlated with eugenol levels (Supplementary Figs. 14–15). There was an isoeugenol synthase (IGS) cluster on chr5 of three roses and only one singleton in the other two roses, indicating that the IGS duplication occurred between the R. rugosa and R. gigantea lineages30. RgIGS2 (RgigChr5G0056550) plays a major role in isoeugenol production and its expression is ten times higher than RgIGS1 (RgigChr5G0056530) expression. This gene was R. gigantea-specific.
Tissue-specific release rules and regulatory networks of tea scent in R. gigantea
Angiosperms have flowers with complex traits to advertise the flower and enhance foraging efficiency31,32,33,34. Tissue-specific and spatial emission patterns have been identified in the flowers of Petunia × hybrida35,36,37 and Silene latifolia38 according to the types or ratios of volatiles. An odor emission gradient is distinguished by bees as a directional road map. Therefore, the plant-pollinator mutualism mediated by scent might significantly influence crop yield39. For RG, a sensory evaluation showed that the petals, androecium, and gynoecium exhibited the highest aroma intensity among five flower tissue types (Supplementary Fig. 16). These flower parts also displayed an obvious neutral red color. In particular, an emission gradient towards reproductive organs was observed along the epidermis (Fig. 4a, b), with the area most proximal to the reproductive organs emitting the deepest red, indicating the most intense fragrances. This is similar to the spatial patterns found in Polianthes tuberosa and Ranunculus acris40,41. The center-oriented pattern attracts corresponding pollinator(s) to the reproductive organs. The petal base adjacent to reproductive organs emits significantly higher levels of volatile compounds than those of the distal petal rim because volatile compound levels were correlated with cell density. In RG, there were more cells in the region near the base than near the edge or middle sites (Fig. 4c). A similar gradient has been reported in Petunia × hybrida42 and Rosa hybrida43, where the cells of the upper petal part are wider than those of the basal part44.
a Five tissues of flower. b Neutral red solution staining. c Morphology of epidermal cells observed by scanning electron microscopy (SEM). d Potential regulatory networks involved in tissue-specific eugenols. The percentages represent the contribution of each tissue to the aroma of one flower. Source data are provided as a Source Data file.
The main site of VOC production in petals is the epidermis, which is often characterized by unique conical-shaped cells43,45,46,47,48,49. In addition to the horizontal layout of the release of fragrance compounds on the petals, there are also longitudinal differences. For example, the volatile emission from the adaxial side is ~20 times greater than that on the abaxial side in Polianthes tuberosa. Similar emission patterns have also been observed in Dianthus caryophyllus and Argyranthemum frutescens42. For RG, adaxial petal epidermal cells have a typical conical, papillate shape, whereas abaxial petal epidermal cells are flat, indicating a difference in the release environment on the epidermis (Fig. 4c). However, both epidermal layers are capable of producing and emitting scent volatiles43. A cytological study has also shown that there are no major differences in the anatomy of the petals of scent and scentless roses.
A total of 48 aroma VOCs were screened (Supplementary Table 11, Supplementary data 2). Comparison among tissues revealed that β-elemen was a leaf-specific volatile. In addition, 4-methylhexan-1-ol, benzyl acetate, (E)-isoeugenol, and γ-muurolene were not detectable in the androecium. Methyl octanoate and decanol were specifically detected in green odorless tissues such as the sepal, receptacle, and leaf. There was significant tissue specificity in the amounts of VOCs (Supplementary Fig. 17). Levels of DMT, borneol, caryophyllene, (E)-β-ionone, and the aspirant release were high in the petal. Androecium-specific VOCs included (E)-anethole and tetradecanal. Green leaf volatiles are emitted as a deceptive signal to attract predatory wasp pollinators50, and several fatty acid derivatives were released at high levels in the sepal.
Eugenol homologs, the major aroma components in RG (Supplementary Fig. 18), are known to attract moth, bee, and fruit fly pollinators of orchids. Interestingly, in RG, these homologs were not only detected but were also distributed in different tissues with different functions. Eugenol accumulation in the androecium was 20–100 times higher than that in other tissues. Isoeugenol and methyleugenol were highly released in the petal and gynoecium, respectively, and were not detected in the androecium. These results provide novel insight into how plants attract pollinators through highly specialized C6–C3 phenylpropanoids. Two co-expression modules were identified based on expression patterns similar to those of eugenols (Supplementary Fig. 19). In the androecium-specific module, RgEGS1 played a key role in eugenol accumulation via 13 transcription factors (TFs) based on analyzes of the transcription factor binding sites (TFBS) the potential binding affinity for gene promoters. In the gynoecium-specific module, RgEGS2 was identified as hub gene that interacts with a C4H gene and 12 TFs, forming a regulatory network involved in methyleugenol production in the gynoecium (Fig. 4d).
Time-order release rhythm and regulatory networks of tea scent in R. gigantea
To determine the circadian rhythm in tea scent release, six time points were observed from buds (0:00) to blooming (20:00) during a day (Fig. 5a). There were no significant differences in 46 aroma VOCs among time points. Decanol and p-hydroxyacetophenone were not detected at 00:00–08:00 but were volatilized at 12:00–20:00. The remaining 44 VOCs were detected at all time points, with variations in the levels (Fig. 5b). There were very few differentially expressed genes (DEGs) between 0:00 and 8:00 but a number of down-regulated genes from 8:00 to 12:00, consistent with the changes in VOC content and pollinator number. For plants, VOCs attract pollinators and defenders to ensure reproductive success51. In this study, the number of bees was much higher at 8:00 and 12:00 than at other time points (Fig. 5a). The results of a principal component analysis (PCA) of gene expression were consistent with VOC trends. PC2 separated the two points (08:00 and 12:00) in which flowers had a strong odor from the other four points when flowers were odorless. Flowers at 00:00–08:00 and 12:00–20:00 were clearly separated along PC1 in chronological order from right to left. These results are consistent with RG blooming from 8:00 to 12:00, the release of floral components, and the attraction of pollinators, which involved the down-regulation of various genes.
a Six flower sampling time points (T00–T20) for Rosa gigantea (RG) and pollinator records. b Pattern of major VOC changes in RG from T00 to T20. c Predicted gene co-expression networks and connections among VOCs (rhombus), enzymatic genes (hexagon), and TFs (circle) involved in pathways for the biosynthesis of phenylpropanoids/benzenoids (yellow nodes), terpenoids (pink nodes), and fatty acid derivatives (purple nodes). Heatmaps of average TPMs (z-score normalized) at each level of TO-GCNs at each sampling point in RG. Three stages in flower scent were identified, initial (T00-T08), transitional (T12), and terminal (T16-T20). L1 to L9 indicate the levels identified in the TO-GCN. Edges between enzymatic genes are not shown. d Subnetwork of the TO-GCN for isoeugenol biosynthesis. Larger points indicate greater support for the prediction based on TFBS analyzes. e Gene expression levels (TPM) and TF binding site (TFBS) detected in the 2 kb upstream sequences of RgIGS and potential regulators in RG. Source data are provided as a Source Data file.
The time lag between the expression levels of TFs, enzymatic genes, and VOCs was considered. Time-ordered gene co-expression networks (TO-GCN) were used to analyze the regulation of aroma compounds. The major GCN consisted of nine time-ordered levels (denoted L1 to L9 in Fig. 5c), matching the order of expression time of the TF genes over six time points, as revealed by the yellow squares (high expression levels) along the diagonal in the heat maps of mean normalized TPMs and VOCs (z-scores). A clear developmental transition during the scent-releasing process was divided into three periods: the initial (corresponding to T00-T08 and L1-4), transitional (corresponding to T12 and L5-6), and terminal periods (corresponding to T16-T20 and L7-9). In the TO-GCN, more than 80% of VOCs from phenylpropanoid/benzenoid and terpenoid pathways were released during the initial process, while VOCs from fatty acid derivative pathways were distributed throughout the entire process. The TO-GCN showed the regulatory relationship between volatiles and genes in the flowering process of RG, revealing the key genes and TFs that regulate the main volatiles (Fig. 5c). RgIGS2 was associated with isoeugenol in L2, with an important role in isoeugenol synthesis; however, its upstream regulators remain unknown. Thus, we chose RgIGS2 to identify up-stream regulators. The TO-GCN was used to predict candidate direct regulators of RgIGS2, which should be co-expressed with RgIGS2 at the same or previous period. Based on the final hierarchical network, bZIP and C2H2 were direct regulators and 29 TF genes, including MYB52, NAC, WRYK, and bHLH, were intermediate second regulators of RgIGS2 (Fig. 5d,e).
Species-specific VOCs and regulatory networks of tea scent in R. gigantea
To confirm the fragrance difference, RG (Fig. 6a) and RC (Fig. 6i) were compared with seven rose scent standards (Supplementary Fig. 20). RG exhibited a stronger tea scent and a higher accumulation of VOCs than those of odorless RC. Both the scent and VOCs were up-regulated from young buds to initial-open flowers and decreased from half-open to full-open flowers (Fig. 6c, g). The major aroma VOCs of RG included eugenol, DMT, methyleugenol, (E)-isoeugenol, 2-phenylethanol, linalool, and (E)-β-ionone (Fig. 6b). In contrast, the major components for RC were TMB, 2-methylbutan-1-ol, β-elemen, α-cadinol, calamenene, γ-cadinene, and 1S-β-pinene (Fig. 6h). The tea scent was characterized by VOCs from the benzenoid/phenylpropanoid pathway, as evidenced by the specific up-regulation in RG, including DMT, eugenol, methyleugenol, and (E)-isoeugenol, consistent with the gene family expansion specific to RG.
a Five flower developmental time points (S1–S5) of RG. b Major VOC changes in RG from S1 to S5. c Changes in aroma intensity during flowering in Rosa gigantea (RG). The darker shades correspond to greater intensity. d Heatmaps of average TPMs (z-score normalized) at each level of TO-GCNs at each flowering time point in RG. Three flower scent-related stages were identified based on expression profiles: initial (S1), transitional (S2-S3), and terminal (S4–S5). e Predicted gene co-expression networks and connections among VOCs (rhombus), enzymatic genes (hexagon), and TFs (circle) involved in phenylpropanoid/benzenoid (yellow nodes), terpenoid (pink nodes), and fatty acid derivative (purple nodes) biosynthetic pathways. L1 to L7 indicate the levels identified in three time-ordered gene co-expression networks (RG-specific, RC-specific, and the consensus TO-GCNs). Edges between enzymatic genes are not shown. f Changes in aroma intensity during flowering in R. chinensis ‘Old Blush’ (RC). g Heatmaps of average TPMs (z-score normalized) at each level of TO-GCNs at each flowering time point in RC. Three flower coloring stages were identified based on expression profiles: initial (S1), transitional (S2–S3), and terminal (S4–S5). h Major VOC changes in RC from S1 to S5. i Five flower developmental time points (S1–S5) of RC. Source data are provided as a Source Data file.
TO-GCNs were constructed separately for RG and RC to investigate regulatory genes associated with fragrance differences53. Two species-specific TO-GCNs (RG- and RC-specific) as well as a consensus TO-GCN were obtained (Fig. 6e, Supplementary Fig. 21). Based on expression patterns associated with flower scent, the time-ordered sub-networks could be assigned to three major processes: initial (S1, corresponding to L1–L3 for RG-GCN and L1–L2 for RC-GCN, when petals were not open and the scent was weak), transitional (S2-S3; corresponding to L4–L5 for RG-GCN and L3–L5 for RC-GCN, when the scent increased), and terminal (S4–S5; corresponding to L6–L7, when the scent decreased) (Fig. 6d, f). The release of RC double-flowers was later than that of RG single-flowers. Few VOCs but a number of genes were detected at the initial stage, indicating a time lag between VOC emission and gene expression. Many genes exhibited conserved co-expression between the two roses at the initial stage (Fig. 7a). In the phenylpropanoid/benzenoid-specific sub-network, 18, 21, and 11 enzymatic genes were identified in the RG-specific GCN, RC-specific GCN, and consensus GCN, respectively (Fig. 7b). At the other two stages, the limited similarity in TO-GCNs between the two roses facilitated the identification of species-specific scent regulatory mechanism. At the transitional stage, there were 26 enzymatic genes in the phenylpropanoid/benzenoid pathway in the RG-specific GCN, much more than the number in RC. The dominant TF families in RG were MYB, bZIP, ERF, bHLH, and NAC, while those in the RC-specific GCN were WRKY, NAC, bHLH, and GeBP.
a Numbers of transcription factors (TFs), volatile organic compounds (VOCs), and enzymatic genes involved in phenylpropanoid/benzenoid, terpenoid, and fatty acid derivative biosynthetic pathways at the initial, transitional, and terminal stages among three Time-ordered gene co-expression networks (TO-GCNs) (RG-specific, RC-specific, and consensus TO-GCNs). b Subnetwork of the TO-GCN for phenylpropanoid/benzenoid biosynthesis. c Subnetwork of the RG-specific TO-GCN for 3,5 dimethoxytoluene biosynthesis. Edges between enzymatic genes are not shown. Source data are provided as a Source Data file.
Key regulatory and enzymatic genes and their hierarchical regulation were comprehensively predicted by examining species-specific networks. The differences in enzyme genes and TFs elucidated the specific high release of DMT in RG compared to TMB in RC. RgOOMT3 and RgOOMT1 located in the RG-specific GCN played an important role in DMT production. By examining the predicted network, RgOOMT3 and RgOOMT1 exhibited hierarchical regulation, involving three TFs (MYB, NAC, and B3) as the third regulators, four TF genes (WRKY, Nin-like, NAC, and GeBP) as intermediate second regulators, and two TFs (C2H2, MYB) as direct regulators (Fig. 7c). Moreover, other TF families were identified as potential regulators, and these TFs were MYB-related (three), basic helix-loop-helix (bHLHs), bZIP, etc. These TFs might also regulate other enzyme genes in the pathway, such as RgEOMT, RgC4H, and RgCAD. To validate the network, structural genes RgOOMT3 and RgOOMT1 and the four MYBs involved in the network were studied as examples. The expression levels of these genes determined by qRT-PCR were consistent with the transcriptome assay results (Supplementary Fig. 22). RgPOMT and RgOOMT sequences were checked with high consistency with genomic sequences, corresponding to RhOOMT4 (Supplementary Fig. 23), with AdoMet_MTases Superfamily and dimerization domains. RgEOBII-like was clustered with PhEOBII54, which positively regulates ODO1, thereby affecting the formation of phenylcyclic and phenylpropane volatiles by modulating upstream precursors of the mangiferic acid and phenylalanine pathways. RgPH4-like was clustered with PhPH455, which also has established roles in floral color and flavor (Supplementary Fig. 24). In addition, RgMYB091-like1 regulates the RgOOMT1 promoter (Supplementary Fig. 25), which was further verified by a protoplast dual luciferase assay. Overall, the regulatory networks provided a reference for further analyzes of the molecular mechanism underlying tea scent in roses.
Discussion
Modern roses emerged through the combination of Chinese and European roses. During the peak period of global species exchange in the 18th century, Chinese garden roses altered the structure of rose breeding worldwide56. They not only exhibited bright colors and a long flowering period but also possessed the unique tea scent and robust vitality of R. gigantea, forming the genetic basis of modern roses57. Is this the end of the rose evolution? Certainly not. Previous research has provided insight into how certain important species were cultivated from wild into ancient roses and then into diverse modern roses over the past 3000 years. The history of mutations and recombination has created lineages with larger flowers, easier reproduction, better maintenance, and longer ornamental periods. However, many beneficial genes remain underutilized in efforts to obtain new roses58. For example, the tea scent characteristic of R. gigantea has been lost during the breeding process, leaving most modern roses lacking notable fragrance (Fig. 8). Technological advancements make de novo domestication from wild resources possible, providing a basis for precise crop improvement59. Combining genome editing with selective breeding is faster and more efficient than either technology alone60. The successful assembly of the T2T gap-free R. gigantea genome, with the highest continuity, completeness, consistency, and accuracy among rose genomes to date, represents an important milestone in research aimed at decoding all DNA sequences, structures, epigenetic signatures, and gene functions61. Ancestral R. gigantea will serve as a bridge for gene exchange between ancient and modern roses62, offering favorable genes to alter the fragrance of modern roses and igniting a second revolution in rose breeding23.
A single reference genome cannot fully represent the substantial genomic diversity within a species26. For example, a recently published super pan-genome of maize (about 6.71 Gb) includes about 4.57 Gb of sequences absent from the B73 reference genome. Since the publication of the first draft genome of R. multiflora in 201763, genomes of Rosa chinensis ‘Old Blush’11, R. rugosa9, and R. wichuraiana ‘Basye’s Thornless’10 have been reported, laying a foundation for analyzing traits and gene function. However, with over 200 species and 35000 varieties of Rosa exhibiting diverse plant shapes, flower colors, and fragrance types, a single reference genome is insufficient for research26. Our analysis of five rose genomes, including the newly sequenced R. gigantea genome, reveals a substantial number of species-specific gene families and SVs. Consequently, there is a critical need to construct a more comprehensive and complex pan-genomic database with genomic resources for diverse traits. The T2T gap-free genome of R. gigantea provides a new high-quality reference genome for roses with the tea scent or yellow color and will facilitate the construction of a pan-genome58. Telomeres and rDNA play pivotal roles in genomic stability, serving as hotspots for chromosomal fragmentation and gross chromosomal rearrangements63,64,65,66,67. In addition to R. gigantea, some wild Rosa species are diploid with two 45S rDNA sites68,69. Further studies of the evolutionary process and disappearance of telomeres on acrocentric chromosomes are needed70.
The conservation of genetic resources is crucial, as numerous species in various regions face the risk of extinction. Therefore, the comprehensive genetic database for R. gigantea with multiple dimensions, including data for different organs, time points, floral tissues, and flowering stages, establishes a fundamental basis for preserving precious information on endangered species71. In addition, aroma mapping through sensory evaluation and VOC characterization revealed the key aromatic compounds, release rhythm, and optimal harvesting and pollination times to facilitate the development of natural resources for new plant essential oils while promoting the application of wild germplasms.
VOCs specific to each tissue reflected the volatile-diversity strategy in plant tissues. Allelic natural variants also underscored the diversity of floral products72. High-throughput sequencing technology facilitates in-depth investigations into the molecular mechanisms underlying the transcriptional regulation of bioactive compounds73, thereby enhancing the efficiency of targeted improvement in rose fragrance. The specific distribution and regulatory networks of eugenol analogs, the major aroma components of tea scent, were revealed in different floral tissues. Further studies are needed to understand how plants attract pollinators through highly specialized C6-C3 phenylpropanoids73,74. In addition, DMT, the unique “tea element” from Chinese roses, was abundant in RG, while RC contained abundant TMB, a structurally similar compound. Olfactory threshold testing revealed that both DMT and TMB are odorless, suggesting that they act as fixatives in tea scent by promoting the stable long-lasting volatilization of aromatic components and inducing a calming and relaxing effect. Comparative genomics showed differences in the number of OOMT genes between the two Chinese roses. Therefore, new scent synthesis pathways like NUDIX found in R. chinensis ‘Old Blush’ were not found in all roses. RcOOMT1 and RcOOMT2 reported in previous studies were not applicable to all tea-scented roses. We discovered new OOMT genes and regulatory networks for DMT, requiring further functional validation.
Methods
Plant materials
For the whole-genome assembly, a 15-year-old adult R. gigantea individual was selected in 2022 (Supplementary Fig. 2). Young leaves were sampled for chromosome analyzes and genome sequencing. Five organs (root, stem, flower, fruit, and leaf) were sampled for Iso-seq to assist in assembling, examining, and correcting the spliced genome. To identify transcriptomic and chemical changes, samples were collected from five flower blooming stages, young bud stage (S1), bud stage (S2), initial-open stage (S3), half-open stage (S4), and full-open stage (S5)), six time points within a day (00:00 (T00), 04:00 (T04), 08:00 (T08), 12:00 (T12), 16:00 (T16), and 20:00 (T20)), and six tissue types (leaf, petal, androecium, gynoecium, receptacle, and sepal) for SPME-GC-MS and RNA sequencing (RNA-seq). In addition, five flower blooming stages of R. chinensis ‘Old Blush’ were sampled as a control variety to compare floral traits. All samples were harvested and immediately frozen in liquid nitrogen between 10:00 and 11:00 in the morning in 2021, except for the analysis of different time points. For sensory testing, fresh flowers of seven rose cultivars (Supplementary Table 12) were sampled with peduncles. All materials were cultivated in the South Tropical Garden located in Kunming, Yunnan, China (24.86°N, 102.98°E).
Sensory test
To prevent any natural odor interference, a clean, odor-free, noise-free sensory evaluation room, following the guidelines of international standard ISO 11136-2014 (2014), was carefully prepared. Flowers were harvested with stems of ~6 cm long from 7:30 to 8:00 am, weighed to obtain 5 g, and placed into glass containers with water for 1.5 h of air circulation. The room temperature was maintained at 20 °C and the relative humidity was controlled between 40% and 55%. At 9:30 am, samples were loaded into acrylic boxes to collect the fragrance for 30 min. Based on the fragrance observation and measurement, the intensity of the fragrance was the strongest before 12:00 pm. Therefore, the test was arranged between 10:00 am and 12:00 pm. The test steps were as follows: olfactory testing, fragrance comparison, sensory scoring, and description. More details on sensory survey are available in the Supplementary Note 1.
Genome assessment, generation, assessment and annotation
Genome assessments were performed by karyotype analysis (Supplementary Note 2) and K-mer frequency analysis (Supplementary Note 3).
Whole-genome sequences was generated using different platforms: Illumina HiSeq X Ten (HiFi), Oxford Nanopore Technologies (ONT), and HiC sequencing. More details on genome assembly are provided in the Supplementary Note 4 and Supplementary Note 5.
Telomeres and centromeres were identified by sequences serching (Supplementary Note 6) and verified by fluorescence in situ hybridization (Supplementary Note 7).
Quality assessment was performed considering continuity, accuracy and completeness, respectively, which was provided the details in the Supplementary Note 8.
Genome annotation was also included repeat sequence annotation (Supplementary Note 9), protein-coding gene structure annotation (Supplementary Note 10), and noncoding RNA prediction (Supplementary Note 11).
Comparative genomic analysis
Evolutionary analysis was performed in the genomes of 13 species, including Apostasia shenzhenica as an outgroup of monocotyledonous plants, Vitis vinifera as a representative species in Rosanae, six representative species in Rosaceae (F. vesca, M. × domestica, P. mume, P. persica, P. bretschneideri, and R. occidentalis), and five representative species in Rosa (R. persica, R. chinensis, R. gigantea, R. rugosa, and R. wichuraiana). The software and parameters were detailed in Supplementary Note 12. Synteny and structural variation analyzes was performed in five roses, including R. persica, R. rugosa, R. gigantea, R. chinensis, and R. wichuraiana. The software and parameters were detailed in Supplementary Note 13.
Gene family identification
To evaluate the evolution of major aroma VOCs in RG, a phylogenetic analysis of EGS/IGS genes in Rosa was conducted using the ML method, using the Petunia homologs sp|Q15GI3 | IGS1 and sp|B2WSN1 | EGS1 for reference, based on genomic sequences of RR, RC, and RW available in the Genome Database for Rosaceae (GDR, www.rosaceae.org) as well as recently sequenced RB and R. gigantea data. The ML tree showed five well-resolved clades, numbered EGS1 to EGS5, and a clade named IGS. Other sequences could not be assigned to a clade due to significant sequence divergence; these were associated with eugenol or isoeugenol synthesis. EGS and IGS were related to PLRs, IFRs, and PCBERs in the PIP family of NADPH-dependent reductases. EGS1 and IGS1 showed the highest similarity to two groups of sequences from various plants that currently lack a clear functional annotation (designated in databases as NAD_binding and cl21454, with a possible role in phenylpropene biosynthesis or mechanistically similar reductive pathways in plants).
For OMT gene family, Arabidopsis OMT and Petunia × hybrida EGS protein sequences were downloaded from the National Center for Biotechnology Information (NCBI) and used for a BLASTp search against R. gigantea, R. chinensis, R. wichurana, R. rugosa, and R. persica proteins. The protein sequences of the candidate OMT genes were analyzed to predict the conserved structural domains using CDD (https://www.ncbi.nlm.nih.gov/) and SMART to determine whether the OMT candidate proteins had pfam00891 (Methyltransf_2), pfam01596 (Methyltransf_3), or cl17173 (AdoMet_MTases superfamily) conserved domains75,76,77. To ensure the integrity of the gene sequence, the candidate OMT and EGS sequences were screened for an amino acid length greater than 20078.
In addition, other scent-related genes in fragrance related pathways were summarized and downloaded published protein/nucleic acid sequences from UniProt, NCBI, or the literature. Blastn (version 2.5.0+ ; parameters: -evalue 1e-5, -max_target_seqs 1) was used to align sequences. Genes whose identity exceeded 40% were obtained as candidate sequences. PlantRegMap79 was used to identify TFs (Supplementary data 3).
Phylogenies, conserved motif analyzes, and chromosomal distribution
OMT and EGS amino acid sequences were used for phylogenetic analyzes. An ML tree was constructed using Geneious Prime with the plugin PhyML80. Conserved structural domains were analyzed using MEME based on the amino acid sequences of OMT and EGS81. Duplicate OMT and EGS genes were removed (i.e., only one gene per locus was retained). The gene position on the chromosome was determined using gene start and end sites, and chromosome length files were obtained. Genes were mapped on chromosomes using TBtools82.
Metabolomic assessment
Samples from three individual plants of RG and RC were harvested, weighed, and immediately frozen in liquid nitrogen, followed by storage at –80 °C. Upon use, the samples were ground to a powder in liquid nitrogen. For semi-quantitative calculation of VOC content, 50 μg mL–1 3-hexanone-2,2,4,4-d4 (10 μL, chromatographically pure) was added as an internal standard. More details on VOCs detection, qualitative and quantitative analysis are provided in the Supplementary Note 14. The identified metabolites were subjected to an orthogonal partial least-squares discriminate analysis (OPLS-DA), and metabolites with |log2FC | ≥ 1, p < 0.05, and variable importance in projection (VIP) score ≥ 1 were considered differentially accumulated VOCs.
Transcriptomic analysis
Total RNA was isolated using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China). RNA-seq library construction was performed as described by Zhou et al.83. A total of 66 RNA-seq libraries were obtained, including those for five flowering stages of R. gigantea and R. chinensis ‘Old Blush’, six tissues of R. gigantea, and six time points within a day for R. gigantea. RNA-seq data were generated from three biological replicates with high repeatability, as determined by a sample correlation analysis. StringTie (version 2.1.4)84 with default parameters was employed to predict new genes, and RSEM85 was used to calculate the gene alignment and determine the transcripts per kilobase million (TPM) for each gene based on its length. DESeq286 was applied to identify DEGs using gene counts. DEGs were filtered based on fold change values ≥ 2 and a Benjamini–Yekutieli false discovery rate (FDR) of ≤ 0.01 using TBtools82.
Weighted gene co-expression network analysis
To study the distribution and expression of odor-related genes in different tissues of R. gigantea, the Weighted Gene Co-expression Network Analysis (WGCNA) package87 in R was used. To identify genes related to the major VOCs, 60979 genes detected in six tissues, including the leaf, petal, androecium, gynoecium, receptacle, and sepal, were analyzed using WGCNA Shiny. Then, 12715 genes were filtered out with a threshold TPM in 90% of samples of > 1 and the MAD method. Finally, 15 modules were analyzed. Hub genes were defined as nodes with a high degree in networks (gene significance > 0.90 and eigengene-based connectivity > 0.9).
Time-ordered gene co-expression network analysis
To investigate the regulatory mechanism underlying the circadian rhythm of compound release in R. gigantea, a TO-GCN was constructed following the method described by Chang (2019)53. The generation process was detailed in Supplementary Note 15.
In addition, TO-GCNs were also constructed for a rose with a strong scent (R. gigantea-specific TO-GCN), an odorless rose (R. chinensis ‘Old Blush’-specific TO-GCN), and a consensus TO-GCN between the two networks, using flower samples from five different developmental stages (S1 to S5). The generation and verifying process was detailed in Supplementary Note 16 and Supplementary Note 17.
Neutral red solution staining
Neutral red solution was used to initially locate the osmophere parts in the test material, and staining with the neutral red solution was performed following the methods of Kong et al.88. Complete flowers were selected and the pedicels were sealed with wax before immersion in 0.01% neutral red solution. The staining process was conducted in a dark environment for 24 h.
Scanning electron microscope observation
Round petal slices with a diameter of 5 mm were taken from three positions using a hole punch and fixed in 5 mL of 2.5% glutaraldehyde. The samples were rinsed with phosphate buffer (pH 7.2) three times (10 min each) to remove the residual fixative. Gradient dehydration was carried out using 30%, 50%, 70%, 90%, 95%, and 100% ethanol solutions for 15 min each, and then samples were immersed in 50%, 70%, and 100% tert-butanol-ethanol solutions for 15 min each. The prepared samples were put into Petri dishes and dried continuously for 8 h using a Labconco FreeZone 4.5 L vacuum freeze dryer, coated using a Hitachi Ion Sputter MC 1000, and placed under a Hitachi Cold Field Emission Scanning Electron Microscope SU 8010 for observation and obtaining images.
Gene cloning
Total RNA and DNA were extracted from blooming flowers of R. gigantea and R. chinensis ‘Old Blush’ using an OminiPlant RNA Kit (CWBIO, Beijing, China) and NuClean Plant Genomic DNA Kit (CWBIO), respectively, according to the manufacturer’s instructions. Standard procedures were followed to assess the quality of the extracted RNA and DNA. Genomic DNA was used for promoter cloning. RNA was reversely transcribed into cDNA using the Hieff CloneTM Plus One Step Cloning Kit (Yeasen, Shanghai, China), which was used for gene cloning and expression evaluation. Specific primers (Supplementary Table 18) were designed based on genome sequences.
Western blotting and phylogenetic analysis
The OOMT genes were subcloned into the pET-32a vector. Recombinant proteins were induced by 0.25–0.80 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 16 °C for 20 h. Afterwards, the cells were harvested by centrifugation and disrupted by sonication. The crude proteins were applied to a Ni-TED Sefinose column (Sangon Biotech Co. Ltd., Shanghai, China) for purification72. The precipitated protein extracts were solubilized in 1 x PBS and heated at 100 °C for 6 min. Proteins (50 μg/lane) were resolved by SDS-PAGE and electro-transferred to PVDF membranes. Membranes were probed at 4 °C overnight with Anti His-Tag Mouse Monoclonal Antibody (CWBIO) at 1:2000 dilution. Thereafter, this was incubated at room temperature for 1 h with Goat Anti-Mouse IgG, HRP Conjugated (CWBIO) at 1:5,000 dilution. Detection was performed by ECL using the Omni-ECL™ Pico Light Chemiluminescence Kit (Epizyme, Shanghai, China). RgOOMT proteins were submitted to Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) to perform multiple sequence alignment. For phylogenetic evaluation, the aligned sequences were subjected to MEGA-X to generate a maximum likelihood tree with bootstrap analysis.
RT-qPCR analysis
SYBR Green (Toyobo, Osaka, Japan)-based quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays were carried out using a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) in a total reaction volume of 10 µL containing 5 µL of 2× Master Mix (TOYOBO, Osaka, Japan), 0.5 µM of each primer (Supplementary Table 18), and 1 µL of template. The reaction was performed in triplicate under the following conditions: 95 °C for 60 s, followed by 40 cycles at 95 °C for 5 s and 60 °C for 60 s. RhTUB and RcUBI were used as internal controls. Relative expression was calculated using the 2−ΔΔCт formula89 (Livak and Schmittgen 2001)90. All measurements were performed in triplicate71,88.
Transient dual luciferase reporter assay
Promoter sequences were amplified from genomic DNA using PCR and inserted upstream of the 0800 Luciferase (LUC) CDS using the GoldenBraid 2.0 cloning strategy to yield promoter-LUC reporter vectors. For an internal control, the expression of the Renilla luciferase gene (REN) was driven by the CaMV35S promoter in a reporter vector. The empty vector with chloramphenicol acetyltransferase (CAT) was used as the negative control (CK). A. thaliana (Columbia-0) used in this study was grown in a greenhouse under a light/dark photoperiod of 16/8 h at 22 °C. Protoplasts for transfection were isolated from 4- to 5-week-old A. thaliana leaves. Co-transfection assays were performed using reporter plasmids and internal control vectors. The results were analyzed and quantified 16 h post-transfection. Gene expression levels were determined by calculating the ratio of LUC to REN activity91.
Statistics & reproducibility
For each sample type, three biological replicates from three individual plants were included. Sample sizes were chosen for satisfied the request of each experiments. WPS Office, Origin 2021, and IBM SPSS Statistics 24 were used for statistical analyzes and charting. Cytoscape was used to visualize the prediction results and co-expression networks92. Adobe Photoshop 2021 was used to splice the sub-figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequencing data for the PacBio HiFi reads, ONT long-reads, Illumina short reads, and Hi-C Illumina reads generated for R. gigantea in this study have been deposited in the Genome Sequence Archive at the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation under accession code CRA014350. The genomic data for R. persica was released at the BioProject database under the accession code PRJNA576752. The details of methods used in this study are provided in the Supplementary Notes. The detailed statistics data used in this study are available in the Supplementary Tables and Supplementary dataset. Source data are provided with this paper.
References
Qin, H. et al. Volatiles, polysaccharides and total polyphenols in Chinese rose tea infusions and their antioxidant activities. J. Food Process. Pres. 42, 1 (2018).
G. L. Wang. et al. Old roses in China (Science Press, 2015).
Buist, R. et al. American Flower-garden Directory: Containing Practical Directions For The Culture Of Plants In The Flower-garden, Hot-house, Green-house, Rooms, Or Parlor Windows, For Every Month In The Year. (A. Hart, late Carey and Hart, Philadelphia, 1851).
Parsons, S. B. et al. THe Rose: Its History, Poetry, Culture, And Classification. (Wiley & Putnam, New York, 1847).
Quest-Ritson, C. Tea roses through the ages. Plantsman 14, 214–221 (2015).
Bendahmane, M., Dubois, A., Raymond, O. & Bris, M. L. Genetics and genomics of flower initiation and development in roses. J. Exp. Bot. 64, 847–857 (2013).
Bombarely, A. Roses for Darwin. Nat. Plants 4, 406–407 (2018).
Cherri-Martin, M., Jullien, F., Heizmann, P. & Baudino, S. Fragrance heritability in hybrid tea roses. Sci. Hortic. 113, 177–181 (2007).
Chen, F. et al. A chromosome-level genome assembly of rugged rose (Rosa rugosa) provides insights into its evolution, ecology, and floral characteristics. Hort. Res. 8, 141 (2021).
Zhong, M. et al. Rose without prickle: genomic insights linked to moisture adaptation. Natl. Sci. Rev. 8, 12 (2021).
Raymond, O. et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 50, 772–777 (2018).
Deng, Y. et al. A telomere-to-telomere gap-free reference genome of watermelon and its mutation library provide important resources for gene discovery and breeding. Mol. Plant. 15, 1268–1284 (2022).
Song, J. et al. Two gap-free reference genomes and a global view of the centromere architecture in rice. Mol. Plant. 14, 1757–1767 (2021).
Chen, J. et al. A complete telomere-to-telomere assembly of the maize genome. Nat. Genet. 55, 1221–1231 (2023).
Zheng, H. et al. Transcriptional regulatory network of high-value active ingredients in medicinal plants. Trends Plant Sci. 28, 429–446 (2023).
Formenti, G. et al. The era of reference genomes in conservation genomics. Trends Ecol. Evol. 37, 197–202 (2022).
Saint-Oyant, L. H. et al. A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat. Plants 4, 473–484 (2018).
Naish, M. et al. The genetic and epigenetic landscape of the Arabidopsis centromeres. Science 374, eabi7489 (2021).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Guarracino, A. et al. Recombination between heterologous human acrocentric chromosomes. Nature 617, 335–343 (2023).
Zhang, C. et al. Horizontal gene transfer has impacted cox1 gene evolution in Cassytha filiformis. J. Mol. Evol. 88, 361–371 (2020).
Sun, P. et al. WGDI: A user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Mol. Plant. 15, 1841–1851 (2022).
Wu, Y. et al. Phylogenomic discovery of deleterious mutations facilitates hybrid potato breeding. Cell 186, 2313–2328 (2023).
Ruggieri, A. et al. A butterfly pan-genome reveals that a large amount of structural variation underlies the evolution of chromatin accessibility. Genome Res 32, 276122–276839 (2022).
Li, L. et al. Genome sequences of five Sitopsis species of Aegilops and the origin of polyploid wheat B subgenome. Mol. Plant. 15, 488–503 (2022).
Shi, J., Tian, Z., Lai, J. & Huang, X. Plant pan-genomics and its applications. Mol. Plant. 16, 168–186 (2023).
Satish, K., Elena, H., Cecilia, H. D. & Claire, M. Turbocharging introgression breeding of perennial fruit crops: A case study on apple. Hort. Res. 7, 1–7 (2020).
Nie, S. et al. Gapless genome assembly of azalea and multi-omics investigation into divergence between two species with distinct flower color. Hort. Res. 10, uhac241 (2023).
Yauk, Y. et al. The O-methyltransferase gene MdoOMT1 is required for biosynthesis of methylated phenylpropenes in ripe apple fruit. Plant J. 82, 937–950 (2015).
Van Belleghem, S. M. et al. High level of novelty under the hood of convergent evolution. Science 379, 1043–1049 (2023).
Fenster, C., Armbruster, W., Wilson, P., Dudash, M. & Thomson, J. Pollination syndromes and floral specialization. Annu. Rev. Ecol. Evol. Syst. 12, 375–403 (2004).
Morel, P. et al. Divergence of the floral a-function between an asterid and a rosid species. Plant. Cell. 29, 1605–1621 (2017).
Moyroud, E. & Glover, B. J. The physics of pollinator attraction. N. Phytol. 216, 350–354 (2017).
Muhlemann, J. K., Klempien, A. & Dudareva, N. Floral volatiles: from biosynthesis to function. Plant Cell Environ. 37, 1936–1949 (2014).
Underwood, G. et al. Patterns in microphytobenthic primary productivity: Species-specific variation in migratory rhythms and photosynthetic efficiency in mixed-species biofilms. Limnol. Oceanogr. 50, 755–767 (2005).
Verdonk, J. C. et al. Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry 62, 997–1008 (2003).
Boachon, B. et al. Natural fumigation as a mechanism for volatile transport between flower organs. Nat. Chem. Biol. 15, 583 (2019).
Dötterl, S. & Jürgens, A. Spatial fragrance patterns in flowers of Silene latifolia: lilac compounds as olfactory nectar guides? Plant Syst. Evol. 255, 99–109 (2005).
Cai, M., Xu, W., Xu, Y., Pan, H. & Zhang, Q. Analysis of Spatial-Temporal variation in floral volatiles emitted from Lagerstroemia caudata by headspace solid-phase microextraction and GC-MS. Molecules. 28, 2 (2023).
Bergström, G., Dobson, H. E. M. & Groth, I. Spatial fragrance patterns within the flowers of Ranunculus acris (Ranunculaceae). Plant Syst. Evol. 195, 221–242 (1995).
Maiti, S. & Mitra, A. Morphological, physiological and ultrastructural changes in flowers explain the Spatio-Temporal emission of scent volatiles in Polianthes tuberosa L. Plant Cell Physiol. 58, 2095–2111 (2017).
Skaliter, O. et al. Spatial patterning of scent in petunia corolla is discriminated by bees and involves the ABCG1 transporter. Plant J. 106, 1746–1758 (2021).
Bergougnoux, V. et al. Both the adaxial and abaxial epidermal layers of the rose petal emit volatile scent compounds. Planta 226, 853–866 (2007).
Yamada, K. et al. Cell division and expansion growth during rose petal development. J. Jpn. Soc. Hortic. 78, 356–362 (2009).
Dudareva, N., Cseke, L., Blanc, V. M. & Pichersky, E. Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant. Cell. 8, 1137–1148 (1996).
Effmert, U. et al. Volatile composition, emission pattern, and localization of floral scent emission in Mirabilis jalapa (Nyctaginaceae). Am. J. Bot. 92, 2–12 (2005).
Kolosova, N., Gorenstein, N., Kish, C. M. & Dudareva, N. Regulation of circadian methyl benzoate emission in diurnally and nocturnally emitting plants. Plant. Cell. 13, 2333–2347 (2001).
Raguso, R. A. & Pichersky, E. New perspectives in pollination biology: floral fragrances. a day in the life of a linalool molecule: chemical communication in a plant‐pollinator system. Part 1: linalool biosynthesis in flowering plants. Plant Species Biol. 14, 95–120 (1999).
Whitney, S. M., Houtz, R. L. & Alonso, H. Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiol. 155, 27–35 (2011).
Perkins, J., Hayashi, T., Peakall, R., Flematti, G. R. & Bohman, B. The volatile chemistry of orchid pollination. Nat. Prod. Rep. 40, 819–839 (2023).
Slavkovic, F. & Bendahmane, A. Floral phytochemistry: Impact of volatile organic compounds and nectar secondary metabolites on pollinator behavior and health. Chem. Biodivers. 20, 4 (2023).
He, G., Zhang, R., Jiang, S., Wang, H. & Ming, F. The MYB transcription factor RcMYB1 plays a central role in rose anthocyanin biosynthesis. Hort. Res. 10, uhad80 (2023).
Chang, Y. et al. Comparative transcriptomics method to infer gene coexpression networks and its applications to maize and rice leaf transcriptomes. Proc. Natl Acad. Sci. USa. 116, 3091–3099 (2019).
Spitzer-Rimon, B. et al. EOBII, a gene encoding a flower-specific regulator of phenylpropanoid volatiles’ biosynthesis in petunia. Plant Cell 22, 1961–1976 (2010).
Cna’Ani, A. et al. Two showy traits, scent emission and pigmentation, are finely coregulated by the MYB transcription factor PH4 in petunia flowers. N. Phytol. 208, 708–714 (2015).
Pierre-Joseph Redouté et al. The Bible Of Roses (Guangming Daily Publishing House, 2012).
Smulders, M. J. M. et al. In the name of the rose: a roadmap for rose research in the genome era. Hort. Res. 6, 65 (2019).
Yan, H. et al. Pangenomic analysis identifies structural variation associated with heat tolerance in pearl millet. Nat. Genet. 55, 507–518 (2023).
Fernie, A. R. & Yan, J. De novo domestication: an alternative route toward new crops for the future. Mol. Plant. 12, 615–631 (2019).
Bartlett, M. E., Moyers, B. T., Man, J., Subramaniam, B. & Makunga, N. P. The power and perils of de novo domestication using genome editing. Annu. Rev. Plant Biol. 74, 727–750 (2023).
Li, G. et al. A new phase of treasure hunting in plant genebanks. Mol. Plant. 16, 503–505 (2023).
Gahlaut, V., Kumari, P., Jaiswal, V. & Kumar, S. Genetics, genomics and breeding in Rosa species. J. Hortic. Sci. Biotechnol. 96, 1–15 (2021).
Nakamura, A. et al. High performance plasma amyloid-beta biomarkers for Alzheimer’s disease. Nature 554, 249–254 (2018).
Dvořáčková, M., Fojtová, M. & Fajkus, J. Chromatin dynamics of plant telomeres and ribosomal genes. Plant J. 83, 18–37 (2015).
Mondello, C., Pirzio, L. M., Azzalin, C. M. & Giulotto, E. Instability of interstitial telomeric sequences in the human genome. Genomics 68, 111–117 (2000).
Ruiz-Herrera, A., Nergadze, S., Santagostino, M. & Giulotto, E. Telomeric repeats far from the ends: mechanisms of origin and role in evolution. Cytogenet. Genome Res. 122, 219–228 (2008).
Stagno, D., Alcontres, M., Palacios, A., Mejias, D. & Blasco, M. A. TopoIIα prevents telomere fragility and formation of ultra thin DNA bridges during mitosis through TRF1-dependent binding to telomeres. Cell Cycle 13, 1463–1481 (2014).
Rocha, L. et al. Functional repetitive sequences and fragile sites in chromosomes of Lolium perenne L. Protoplasma. 252, 451–60(2015).
Min, T. et al. FISH analysis of 45S rDNA on the chromosomes of Chinese old garden roses (in Chinese). J. Yunnan Agric. Univ. 28, 380–385 (2013).
Ding, X. et al. Distribution of 45S rDNA in modern rose cultivars (Rosa hybrida), Rosa rugosa, and their interspecific hybrids revealed by fluorescence in situ hybridization. Cytogenet. Genome Res. 149, 226–235 (2016).
Jr, R. J. J. F. et al. Addressing the problem of scale that emerges with habitat fragmentation. Glob. Ecol. Biogeogr., (2023).
Bao, T. et al. Allelic variation of terpene synthases drives terpene diversity in the wild species of the Freesia genus. Plant Physiol. 192, 2419–2435 (2023).
Zheng, P. et al. Integrated spatial transcriptome and metabolism study reveals metabolic heterogeneity in human injured brain. Cell Rep. Med. 4, 101057 (2023).
Yan, H. et al. Functional characterization of the eugenol synthase gene (RcEGS1) in rose. Plant Physiol. Biochem. 129, 21–26 (2018).
Koeduka, T. et al. Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl Acad. Sci. USa. 103, 10128–10133 (2006).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res 42, D222–D230 (2014).
Letunic, I., Doerks, T. & Bork, P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Researchnucleic Acids Res. 43, D257–D260 (2014).
Marchler-Bauer, A. et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Researchnucleic Acids Res. 45, D200–D203 (2016).
Jin, J. et al. An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Mol. Biol. Evol. 32, 1767–1773 (2015).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biologysystematic Biol. 59, 307–321 (2010).
Bailey, T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Chen, C. et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202 (2020).
Zhou, L. et al. Volatile metabolome and transcriptome reveal fragrance release rhythm and molecular mechanisms of Rosa yangii. J. Integr. Agr. 22, 2111–2125 (2023).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Kong, Y., Sun, M., Pan, H. & Zhang, Q. Composition and emission rhythm of floral scent volatiles from eight lily cut flowers. J. Am. Soc. Horticultural Sci. J. Am. Soc. Hort. Sci. 137, 376–382 (2012).
Livak, K. & Schmittgen, T. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2-ΔΔCt method. Methods. 25, 402–408 (2000).
Yang, S. et al. Characterization of Terpene synthase variation in flowers of wild Aquilegia species from Northeastern Asia. Hortic. Res. 9, uhab020 (2022).
Li, Y. et al. MicroTom metabolic network: rewiring tomato metabolic regulatory network throughout the growth cycle. Mol. Plant. 13, 1203–1218 (2020).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003).
Acknowledgements
C.Y. thanks the Fundamental Research Funds for the Central Universities (QNTD202306), National Key R&D Program of China (2019YFD1000400) and National Natural Science Foundation of China (32071818). The authors express their sincere gratitude to Yuyong Yang (Kunming Yang Chinese Rose Gardening Co., Ltd.) for the pivotal role in the collection and sharing of Rosa germplasm resources. Appreciation is also extended to Xiaoling Tian, Lyu Jiang, and Peifeng Lyu (Beijing Forestry University, China) for their support with photographic documentation. Acknowledgment is due to Jiahe Sun (Southwest University, China) and Yongming Chen (China Agricultural University, China) for their expert guidance in the utilization of Orthoveen3 and Genetribe, respectively. The authors also wish to thank Quan Ji, Yaqi Dong, Deliang Lyu, and Wentong Chen for their assistance in sample collection. Furthermore, gratitude is extended to BenaGen (www.benagen.com) and MetWare (www.metware.cn) for providing essential testing platforms. Special thanks to all of Professor Gao Xiang’s team members (Northeast Normal University, China) for their guidance and assistance in the gene function identification assays. Finally, the authors are profoundly indebted to every member of the Fishpond family, whose collective efforts were instrumental in the fruition of this work.
Author information
Authors and Affiliations
Contributions
C.Y., X.G., L.L. and Q.X.Z. conceived and designed the project. C.Y., L.L., L.J.Z, R.H.H, and Y.Y.C. maintained the plant materials and coordinated sequencing data generation. K.Z. and L.J.Z. assembled the genomes and performed genome validations and annotations. L.J.Z., S.H.W., Y.Y.C., Q.Y.M, T.H.L., and Y.C.L. conducted bioinformatic analysis and prepared figures and tables. B.X.C. and H.T.P. assisted in the result interpretation and discussion. X.G., L.J.Z, and S.H.W completed the gene function verification experiment. L.J.Z. and S.H.W. wrote the manuscript. C.Y., X.G., L.L. and Q.X.Z. revised the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhou, L., Wu, S., Chen, Y. et al. Multi-omics analyzes of Rosa gigantea illuminate tea scent biosynthesis and release mechanisms. Nat Commun 15, 8469 (2024). https://doi.org/10.1038/s41467-024-52782-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-52782-9
This article is cited by
-
The telomere-to-telomere haplotype genome provides in-depth insights into the molecular mechanisms of the anthocyanin deficiency phenotype in Prunus mume
Molecular Horticulture (2026)
-
Salicylic acid regulates biosynthesis of floral fragrance (E)-β-farnesene via NPR3-WRKY1 module in chrysanthemum
Molecular Horticulture (2025)
-
A chromosomal-scale reference genome for Rosa hugonis
Scientific Data (2025)