Abstract
Membrane contact sites (MCSs) are hubs allowing various cell organelles to coordinate their activities. The dynamic nature of these sites and their small size hinder analysis by current imaging techniques. To overcome these limitations, we here design a series of reversible chemogenetic reporters incorporating improved, low-affinity variants of splitFAST, and study the dynamics of different MCSs at high spatiotemporal resolution, both in vitro and in vivo. We demonstrate that these versatile reporters suit different experimental setups well, allowing one to address challenging biological questions. Using these probes, we identify a pathway in which calcium (Ca2+) signalling dynamically regulates endoplasmic reticulum-mitochondria juxtaposition, characterizing the underlying mechanism. Finally, by integrating Ca2+-sensing capabilities into the splitFAST technology, we introduce PRINCESS (PRobe for INterorganelle Ca2+-Exchange Sites based on SplitFAST), a class of reporters to simultaneously detect MCSs and measure the associated Ca2+ dynamics using a single biosensor.
Similar content being viewed by others
Introduction
In eukaryotes, membrane-bound organelles allow the separation of sometimes incompatible biochemical reactions. Nevertheless, cell organelles do not work as isolated structures but coordinate their activities by forming specialized platforms, called membrane contact sites (MCSs), whereby their membranes are closely juxtaposed (typically in the 10–30 nm range)1, 2. At MCSs, key cell pathways take place, including the exchange of metabolites, signalling molecules and information1. The efficiency of these processes is indeed strongly favoured by the very close proximity between organelle membranes and is aided by protein-protein or protein-lipid interactions. Specific protein and lipid compositions characterize each type of MCS, ensuring the performance of their specialized functionalities2.
Remarkably, early disturbances in MCS structure and/or function have been described in different high-incidence disorders, such as neurodegenerative diseases and several cancers3, 4. This suggests the tantalizing hypothesis that such alterations might underlie pathogenesis, prompting an increasing interest in studying MCSs and the associated metabolic/signalling activities.
However, a comprehensive study of MCSs is hampered by the limitations of the currently available techniques. Indeed, because of the dynamic nature of MCSs1, 2, their study requires monitoring changes that occur in response to specific needs with high spatiotemporal resolution, something that current tools do not provide2, 5. For instance, optical microscopy allows to follow gross changes over time, but its diffraction-limited spatial resolution does not match the nanometric size of most MCSs. Conversely, electron microscopy (EM) and super-resolution microscopy can resolve membrane proximity in space, but need specialized equipment, are limited to fixed samples (e.g., EM) and/or require long acquisitions, which are poorly compatible with MCS dynamics. Recently, the introduction of self-complementing fluorescent probes based on split-GFP overcame the problem of spatial resolution and allowed the detection of MCSs6, 7. However, in most of these systems, the complementation reaction leading to fluorescence maturation is slow (taking tens of minutes to hours) and poorly reversible8, possibly stabilizing MCSs and/or limiting the detection of MCS changes occurring within seconds. On the other hand, Förster resonance energy transfer (FRET)-based probes have been developed to detect MCSs9. These rely on the expression of specific couples of donor and acceptor fluorescent proteins (relatively large in size, ~25 kDa each one) on the membranes of juxtaposed organelles. Of note, FRET efficiency is deeply affected by the distance between donor and acceptor, which must be closer than 10 nm and in a proper orientation. Moreover, FRET measurements need specialized equipment and the evaluation of the FRET signal requires careful calibration experiments, mostly relying on the rapamycin-induced dimerization of the reporters, which might force MCSs9. Finally, current methods allow to study either MCS morphology (i.e., the extent of inter-organelle apposition) or functionality (i.e., the efficiency of a specific activity associated with MCSs, such as inter-organelle Ca2+ transfer), but visualizing MCSs with high spatial resolution and simultaneously measuring their activities over time has been challenging.
To overcome these limitations, here we generate a series of reversible chemogenetic reporters, focusing on the well characterized juxtaposition between endoplasmic reticulum (ER), mitochondria and the plasma membrane (PM). Specifically, we engineer splitFAST (split Fluorescence-activating and Absorption Shifting Tag), a small, split fluorescent protein tag recently introduced for the monitoring of dynamic protein-protein interactions10. The splitFAST fragments show modest affinity; however, when in close proximity, they assemble into a reporter that binds and stabilizes the fluorescent state of exogenously applied multicolour fluorogenic dyes (fluorogens) (Fig. 1a), which are otherwise dark when unbound. SplitFAST complementation is rapid and promptly reversible10, making it an ideal system for studying MCS dynamics in physiological conditions.
a The cartoon represents the splitFAST system, formed by the NFAST and the CFAST portions, that are not fluorescent per se, unless assembled and bound to a fluorogen: green, red or far-red (this latter requiring frNFAST). Created in BioRender. Garcia Casas, P. (2023) BioRender.com/l67i901. b, c The cartoon (b) represents the tailoring of splitFAST to mark ER-mit MCSs. Created in BioRender. Garcia Casas, P. (2023) BioRender.com/e32h348. By targeting the NFAST portion to the ER membrane and CFAST10 to the OMM, both separated by unstructured linkers of different sizes as modelled by AlphaFold (c), two different probes have been generated to study ER-mit MCSs: the short ER-mit splitFAST, with a length of ~12 nm, and the long ER-mit splitFAST, of ~25 nm. The proposed AlphaFold structures provide static models of the probes in an extended conformation, but the unstructured linkers can bend and adopt multiple conformations (see Methods). d Representative confocal images of HeLa cells co-expressing either the short or the long ER-mit splitFAST (exhibiting a dotted pattern along mitochondria) with a mit-RFP. e Representative confocal image of a COS-7 cell co-expressing the short ER-mit splitFAST and an ER-GFP, stained by MitoTracker Deep Red. A dotted ER-mit splitFAST signal colocalizes with both the ER and mitochondria, marking sites of close contact between the two organelles. f Representative confocal images of stable HeLa cell clones expressing either the short (S1, S2, S3) or the long (L1, L2, L3) version of the ER-mit splitFAST probes. A dotted signal along mitochondria (labelled by MitoTracker Deep Red) is shown, allowing the calculation of the percentage of the mitochondrial surface (% mit) covered by either short or long contacts with the ER (box plots on the right, see “Methods”). The cell indicated with a white asterisk is shown in the corresponding binary image used for analysis. g The box plots represent the dynamic changes of ER-mit MCSs (calculated as in panel (f)) in HeLa cells expressing the short ER-mit splitFAST, upon treatments with Tunicamycin (5 μg/ml). Recovery of basal levels of ER-mit MCSs was observed after Tunicamycin removal. h, i Representative confocal images of (h) HeLa cells co-expressing OMM-RFP, OMM-Cepia3 and short ER-mit fr-splitFAST probe, or (i) of COS-7 cells co-expressing ER-GFP, short ER-mit splitFAST and in which mitochondria were marked with MitoTracker Deep Red. Scale bar: 10 μm (d–f, h, i).
We demonstrate that these reporters are endowed with high versatility, enabling researchers to: (a) detect MCSs in vitro and in vivo; (b) follow complex MCS fusion/fission events associated with deep organelle and cytoskeleton remodelling; (c) measure perturbations of organelle coupling in different physiological and pathological conditions; (d) track the transient recruitment of proteins at MCSs; (e) reveal MCS rearrangements upon specific cell stimulations and characterize the underlying mechanisms.
Moreover, by tailoring the splitFAST system and combining it with suitable Ca2+-sensing modules, we generate a series of single reporters capable of simultaneously detecting MCSs and measuring the associated Ca2+ signals, that we dub PRINCESS (PRobe for INterorganelle Ca2+-Exchange Sites based on SplitFAST). By integrating high spatiotemporal resolution, fluorescence imaging and Ca2+-sensing capabilities, we show that PRINCESS allows the study of MCS morphology and function using a single probe, opening avenues for designing biosensors which are able to address fundamental biological questions.
Results
Design of splitFAST-based probes to detect MCSs in vitro and in vivo
SplitFAST is a fluorescence complementation system formed by two parts, NFAST and CFAST, previously designed through bisection of the 14-kDa protein FAST for real-time visualization of transient protein-protein interactions10, 11 (Fig. 1a). CFAST fragments of respectively 10 (CFAST10) or 11 (CFAST11) amino acids have been reported, with the former displaying a lower self-complementation and thus a higher dynamic range10. We firstly confirmed the prompt reversibility of NFAST-CFAST10 interaction (Supplementary Fig. 1a, b and Supplementary Movie 1). Then, to test whether splitFAST can be tailored to detect MCSs, while preserving its dynamicity, we targeted NFAST and CFAST10 to the ER- and the outer mitochondrial membrane (OMM), respectively (Fig. 1b). Disordered linkers of different lengths were inserted between the OMM targeting sequence and CFAST10, generating two probes to detect either short (short ER-mit splitFAST) or long (long ER-mit splitFAST) ER-mitochondria contacts, potentially complementing at distances between ER and OMM of, respectively, less than 12 nm and 25 nm, as modelled by AlphaFold12 (Fig. 1c). The expression of these probes in HeLa (Fig. 1d) and COS-7 (Fig. 1e) cells revealed a punctate pattern overlapping with the mitochondrial network, in correspondence of regions of ER-mitochondria co-localization (Fig. 1e). The fluorescent dots appeared few seconds after the addition of the fluorogens (Supplementary Fig. 1c), confirming the high cell-permeability of these latter and the prompt splitFAST complementation. Different HeLa cell clones, expressing either the short or the long ER-mit splitFAST probe, displayed a larger fraction of mitochondrial surface engaged in the formation of long contacts with the ER, compared to the short ones (Fig. 1f and Supplementary Fig. 1d). Moreover, the treatment of cells expressing the short probe with the ER-stressor tunicamycin induced a transient increase, followed by a decrease, of ER-mitochondria contacts (ER-mit MCSs; Fig. 1g and Supplementary Fig. 1e), in line with previous data13 and confirming that the splitFAST-based probes can dynamically follow MCS changes. Consistently, either decreased or increased ER-mit MCSs were associated with different cell treatments/conditions (Supplementary Fig. 1f). SplitFAST complementation at ER-mit MCSs was visualized by three different hydroxybenzylidene rhodanine (HBR)-derived fluorogens: HMBR (green fluorescence; Fig. 1d), HPAR-3OM (far-red fluorescence; Fig. 1h) and HBR-3,5DOM (red fluorescence, Fig. 1i), with HPAR-3OM requiring the replacement of NFAST with frNFAST (far-red NFAST14; see Methods and Fig. 1a). Notably, the high spectral flexibility of the system enables the combined study of MCSs with multiple fluorescent reporters and represents an advantage compared with previous techniques.
Importantly, an ideal method to study MCSs should not interfere with their dynamics. The choice of CFAST10, rather than CFAST11, is instrumental to this purpose, as it is endowed with a very low affinity for NFAST10. To further minimize possible MCS conditioning, we introduced in our probes RspA-splitFAST, an improved, orthologous splitFAST version (derived from the photoactive yellow protein (PYP) of the bacteria Rheinheimera sp. A13L, RspA) endowed with even lower self-complementation, greater dynamic range and higher brightness compared with splitFAST15. RspA-NFAST and RspA-CFAST fragments were targeted, respectively, to the ER membrane and the OMM (hereafter referred as ER-mit RspA-splitFAST), giving rise to a bright, dotted fluorescent signal (Fig. 2a), precisely at regions corresponding to close ER-mitochondria juxtaposition, as revealed by correlative light electron microscopy (CLEM) experiments (Fig. 2b and Supplementary Movie 2). Importantly, by this approach, we verified that the fraction of mitochondrial perimeter in close contact with the ER was similar in control (untransfected) and ER-mit RspA-splitFAST-expressing cells (Fig. 2b), indicating that the probe does not force MCS formation. Moreover, the width of the contacts marked by ER-mit RspA-splitFAST was calculated by CLEM to be on average 13.8 ± 2.3 nm (n = 45 contacts from 16 different mitochondria), in line with the value estimated by AlphaFold modelling for short ER-mit splitFAST (Fig. 1c). ER-mit RspA-splitFAST, complemented with either HMBR or HBR-3,5DOM, allowed to monitor the dynamic changes of ER-mit MCSs triggered by tunicamycin treatment (Supplementary Fig. 1g), thus maintaining the dynamicity displayed by ER-mit splitFAST (Fig. 1g).
a Representative confocal images of HeLa cells expressing short ER-mit RspA-splitFAST and stained with MitoTracker Deep Red. The box plot shows the percentage of mitochondrial surface co-localized with ER-mit RspA-splitFAST. b Representative confocal image (top) of a region of a HeLa cell expressing short ER-mit RspA-splitFAST and stained with MitoTracker Red. The fluorescent splitFAST dots (two representative dots were marked “1”/“2”) correspond to sites of close ER-mit membrane juxtaposition, as revealed by the EM slice of the very same cell region (bottom; see also Supplementary Movie 2). The fraction of mitochondrial perimeter in close contact with ER, analysed by CLEM, is similar in untransfected and ER-mit RspA-splitFAST-expressing (transfected) cells (box plots). c Representative confocal images of HeLa cells expressing ER-PM RspA-splitFAST, in which PM was labelled by either WGA staining or MyrPalm-D1cpv expression. ER-PM MCSs were visualized by addition of respectively HMBR (left) or HBR-3,5DOM (right). d Representative confocal images of PM-mit RspA-splitFAST-expressing HeLa cells, stained with MitoTracker and WGA (PM). The box plots represent the percentage of mitochondrial surface in contact with either ER (same as in panel (a)) or PM. e Representative confocal images of HeLa cells co-expressing ER-PM fr-splitFAST and ER-mit RspA-splitFAST. The far-red signal is exclusively emitted by ER-PM fr-splitFAST (marking ER-PM MCSs); the green signal can be emitted by both ER-mit RspA-splitFAST and ER-PM fr-splitFAST (as fr-splitFAST can also incorporate the green and red fluorogens). As the lifetimes of the ER-mit RspA-splitFAST and ER-PM fr-splitFAST green signals are different (see FLIM Phasor Plot, Supplementary Fig. 1i and Methods), ER-mit and ER-PM MCSs were distinguished in the very same cells by FLIM. f Representative confocal images of a HeLa cell, co-expressing ER-long-RspA-NFAST, ER-short-frNFAST and OMM-short-RspA-CFAST to simultaneously visualize short and long ER-mit MCSs. The signals were separated by FLIM (see e). Arrows indicate few long MCSs that do not colocalize with short ones (see box plots on the right). g Tomographic 3D reconstruction of a portion of HeLa cell, showing mitochondrial surface (red) and the regions of juxtaposition with ER, either close (<12 nm, cyan areas, corresponding to short ER-mit MCSs) or wider (12–24 nm range, yellow areas, corresponding to long ER-mit MCSs). Most short ER-mit MCSs are continuous with long ones (white arrows, front view box). The box plot represents the surface of either the few long contacts not containing short ones (Long, white arrowheads in the top view) or those continuous with short MCSs. See also Supplementary Movie 3. h Representative confocal images of the Caenorhabditis elegans strain pHX6743 (expressing ER-mit RspA-splitFAST under the myo3 promoter) or N2 (not expressing RspA-splitFAST). The fluorescent signals of ER-mit RspA-splitFAST (complemented with HBR-3,5DOM) and of background were separated by FLIM (See Phasor Plot). Scale bar: 10 µm (a, c–f, h); 2 µm (b); 500 nm (g).
Co-expression of ER-RspA-NFAST and PM-targeted RspA-CFAST (ER-PM RspA-splitFAST; Supplementary Fig. 1h) produced a dotted pattern corresponding to ER-PM MCSs (Fig. 2c), suggesting that this system can be tailored to detect different types of MCSs. By expressing this probe, in few cells we additionally observed some dots corresponding to ER-intracellular vesicles (likely early endosomes - EE) MCSs (Fig. 2c). This result, possibly associated with a partial endocytosis of PM-RspA-CFAST when highly expressed, supports the existence of close ER-EE MCSs, as previously reported16, 17.
In addition, we co-expressed OMM-RspA-NFAST with PM-RspA-CFAST (PM-mit RspA-splitFAST) to detect PM-mit MCSs (Fig. 2d) and observed that a lower fraction of mitochondrial surface is in close contact with the PM (Fig. 2d), compared to that in contact with the ER (Fig. 2a). Moreover, by exploiting the spectral flexibility and versatility of splitFAST variants, the combined expression of PM-frNFAST, ER-RspA-CFAST and OMM-RspA-NFAST allowed us to simultaneously visualize ER-mit and ER-PM MCSs in the very same cells. Using fluorescence-lifetime imaging microscopy (FLIM), we could distinguish the signals of RspA-splitFAST (marking ER-mit MCSs) and fr-splitFAST (marking ER-PM MCSs) not only by their different spectral properties, but also by their different fluorescence lifetime (Fig. 2e and Supplementary Fig. 1i, see legend and “Methods”), providing an additional information of help to fully separate the different components. Similarly, to investigate the relationship between short and long ER-mit MCSs, we simultaneously visualized them by co-expressing short ER-mit fr-splitFAST and long ER-mit RspA-splitFAST. We found that almost all short ER-mit MCSs co-localize with, and are possibly wrapped within, the long ones, while the majority (~70%) of the long co-localizes with the short ones (Fig. 2f). However, since we inserted disordered linkers between the organelle-targeting sequences and the splitFAST fragments, we reasoned that the long probe could bend and mark the short contacts in addition to the long ones, potentially complicating the interpretation of the results. Therefore, to better clarify the mutual relationship between these types of contacts, we performed electron tomography (Fig. 2g and Supplementary Movie 3). In line with the data obtained by the combined expression of the long and short splitFAST variants, we observed that almost all the regions of close ER-mit juxtaposition (<12 nm, corresponding to short MCSs) are continuous with and/or enclosed within areas whereby the distance between the two organelle membranes is slightly wider (in the 12–24 nm range, corresponding to long MCSs). On the other hand, the majority (79.3%) of long ER-mit MCSs is continuous with short ones, whereas the remaining fraction (20.7%) is not and covers smaller surfaces (Fig. 2g). Overall, the close association between short and long ER-mit contacts suggests that they might represent different maturation stages of single contacts.
Finally, we generated a C. elegans strain expressing ER-mit RspA-splitFAST under the myo3 promoter, to specifically visualize sarcoplasmic reticulum (SR)-mitochondria MCSs in body wall muscle cells (see “Methods”). By a FLIM-based approach, we could clearly identify SR-mitochondria MCSs in living worms separating them from background signal (Fig. 2h), suggesting these probes are suitable for in vivo investigations.
The dynamics of ER-mit MCSs associate with ER, mitochondria and cytoskeleton remodelling
We followed organelle and MCS dynamics in live HeLa clones and COS-7 cells, expressing ER-mit RspA-splitFAST and stained for ER and/or mitochondrial markers. Imaging was performed by Airyscan-based super-resolution microscopy or lattice light-sheet microscopy (LLSM), allowing fast 4D acquisitions while minimizing photo-damage and preserving good spatial resolution. Imaris-based surface analysis (see “Methods”) revealed that the volume of most ER-mit RspA-splitFAST dots is <0.1 µm3, with the larger ones assuming an ellipsoidal shape (Fig. 3a). We observed that ER-mit MCSs are frequently associated with regions of mitochondrial branching and confirmed that they can be maintained during organelle movements18 (Fig. 3b,c and Supplementary Movies 4, 5). Moreover, in line with previous findings, we noticed that they mark sites of dynamin-related protein 1 (DRP1)-puncta formation19 and mitochondrial fission20 (Fig. 3d, Supplementary Fig. 2a and Supplementary Movie 6), but additionally observed frequent cases in which transient mitochondrial fusion (‘kiss-and-run’ mechanism)21 occurs at ER-mit MCSs (Fig. 3e and Supplementary Movie 7). Importantly, by Imaris-based tracking, we noticed that several MCSs undergo themselves frequent fission/fusion events (Fig. 3f and Supplementary Movie 8), tuning their continuous remodelling.
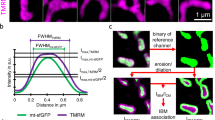
a Representative Airyscan 3D image of a COS-7 cell co-expressing ER-mit RspA-splitFAST (surface rendering, yellow), the ER-marker ER-StayGold and stained with MitoTracker Deep Red. Distribution graphs represent the Kernel density (orange) and the histogram (green) of volume, area and sphericity values of ER-mit RspA-splitFAST dots (see “Methods”). Dispersion plots show a negative correlation between volume and sphericity (left) and a positive correlation between volume and ellipticity (right). b Representative lattice light-sheet microscopy 3D image of a HeLa cell clone expressing ER-mit RspA-splitFAST (surface rendering, yellow) and stained with MitoTracker Deep Red. The time-lapse shows the maintenance of an ER-mit MCS during an extensive mitochondrial movement, followed by branching at the MCSs (arrows). See also the corresponding Supplementary Movie 4. c Representative Airyscan image of a COS-7 cell as in panel (a). The time-lapse shows co-sliding of mitochondria, ER and ER-mit RspA-splitFAST dots (arrow), as well as the disappearance of an ER-mit MCS (circle, first frame) upon mitochondria detachment from ER tubules. See also the corresponding Supplementary Movie 5. d Representative Airyscan 3D image (surface rendering) of a COS-7 cell co-expressing ER-mit RspA-splitFAST, mCherry-Drp1 and stained with MitoTracker Deep Red. The time-lapse shows mCherry-Drp1 localization at a site of mitochondria and ER-mit MCS fission (arrow). See also the corresponding Supplementary Movie 6. e Representative image as for panel (b). The time-lapse shows an ER-mit MCS movement (arrow) at a site of mitochondria “Kiss-and-run” fusion (40–70 s) followed by organelle fission (100–140 s). See also the corresponding Supplementary Movie 7. f Representative 3D image of a HeLa cell expressing ER-mit RspA-splitFAST (surface rendering). ER-mit RspA-splitFAST dots (tracked using IMARIS, see Methods) move in space and interact each other. Tracks (coloured continuous lines) report MCS movements and interactions. ER-mit MCSs undergoing fusion/fission are considered part of the same track. Enlarged regions show the time-lapse of a MCS undergoing fission (309–329 s), and two MCSs fusing together (448–488 s). Charts represent the percentage of interacting ER-mit MCS subgroups (tracks) for each range of fission/fusion events, as indicated. See also the corresponding Supplementary Movie 8. g Representative Airyscan 3D image of a COS-7 cell co-expressing ER-mit RspA-splitFAST, ER-StayGold and stained with MitoTracker Deep Red. Enlarged regions show the time-lapse of the appearance (38 s) of an ER-mit MCS (arrow), upon ER and mitochondria membrane docking, and its following disappearance (153 s), upon organelle distancing (arrow). See also the corresponding Supplementary Movie 9. h Representative 3D image of a COS-7 cell co-expressing ER-mit RspA-splitFAST, ER-StayGold and stained with MitoTracker Deep Red. ER-StayGold signal has been segmented (see “Methods”) to separate ER tubules from sheets. Manders’ co-localization M2 coefficient of ER-mit MCSs with ER sheets/tubules is shown (box plots). Scale bar: 5 µm (a–h).
Moreover, Airyscan-based analysis revealed that the transient docking of ER tubules to mitochondria is promptly matched with the appearance and vanishing of ER-mit RspA-splitFAST fluorescent dots (Fig. 3g and Supplementary Movie 9), confirming that the probe is dynamic and fully reversible. The tracking of ER-mit MCSs allowed to calculate an average half-life of ~270 s (Supplementary Fig. 2b). Interestingly, in COS-7 cells, we found that the majority of short ER-mit MCSs involves ER sheets, rather than tubules (Fig. 3h), though the remodelling of ER tubules frequently underlies ER-mit MCS fusion/fission (Supplementary Fig. 2c and Supplementary Movie 10). Of note, the half-life of ER-mit MCSs involving tubules is shorter than that of MCSs formed with ER sheets (Supplementary Fig. 2d). Finally, we observed that most ER-mit MCSs associate with tubulin- and filamentous actin (F-actin)-positive structures (Supplementary Fig. 2e and Supplementary Movie 11), in line with the key role of different cytoskeleton elements in organelle dynamics and MCS rearrangement18, 19, 21.
Detection of MCS perturbations in Alzheimer’s disease (AD) cell models by splitFAST-based probes
We and others have previously demonstrated that ER-mitochondria association is strengthened in different AD models3, 22,23,24. To test whether ER-mit RspA-splitFAST can efficiently detect these alterations, we firstly confirmed it can be used in AD-relevant primary cells, including microglia (Fig. 4a), astrocytes (Fig. 4b, c) and cortical neurons (Fig. 4d, e), in which we observed that ER-mit MCSs distribute throughout the soma, axons and dendrites (Fig. 4d and Supplementary Movie 12).
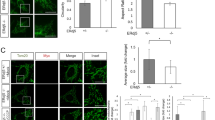
a–f Representative confocal images of different ER-mit RspA-splitFAST-expressing cells. Where indicated, cells were co-stained with MitoTracker Deep Red or mitoDsRed, used as mitochondrial marker. ER-mit MCSs are shown in mouse primary microglia (a), astrocytes (b, c), cortical neurons (d, e) and human fibroblasts (f). In (b, c, e, f) the box plots represent the percentage of mitochondrial surface co-localized with ER-mit RspA-splitFAST for the indicated cell types. In b, astrocytes from WT or AD mice were compared. In (c) astrocytes from WT mice were exposed (WT + Aβ) or not (WT) to a conditioned medium containing naturally generated Aβ peptides (see Methods). In (e) cortical neurons from WT or AD (AppNL-G-F, shortened as NLGF) mice were compared. In (f) primary human skin fibroblasts from either a healthy donor (WT) or a familial AD patient (PS2-N141I) were compared. *p < 0.05; **p < 0.01; ***p < 0.001. d Confocal microscopy 3D projection of the ER-mit RspA-splitFAST fluorescent signal in a cortical neuron from WT mice. The colour bar on the left represents the depth (in µm) along the z-axis. See also the corresponding Supplementary Movie 12. Scale bar: 10 µm (a–c, e, f); 50 µm (d).
Astrocytes from an AD transgenic mouse model (B6.152H)25 displayed a significantly higher fraction of mitochondrial surface covered by the ER-mit RspA-splitFAST signal, compared to wild type (WT) cells (Fig. 4b). A similar increase in ER-mit MCSs was detected in astrocytes from WT mice upon acute exposure to naturally generated amyloid β (Aβ) peptides (Fig. 4c; see Methods), as well as in cortical neurons from the AppNL-G-F knock-in AD mouse model26, 27 compared to WT (Fig. 4e), suggesting that multiple pathways involved in AD might converge on alterations of ER-mitochondria juxtaposition. Similarly, ER-mit RspA-splitFAST efficiently detected a higher ER-mitochondria tethering in human fibroblasts from a patient harboring a familial AD mutation in Presenilin 2, compared to those from an age-matched healthy donor (Fig. 4f), as previously reported in the same cell models by different techniques24, 28, 29. Overall, these data demonstrate that the splitFAST-based probes can track MCS alterations in disease models.
PRINCESS design and characterization
Several cell pathways are regulated by Ca2+ signalling events occurring at MCSs. For instance, during inositol 1,4,5-trisphosphate (IP3)-linked cell stimulations, transient microdomains of high Ca2+ concentration are formed at the ER-mitochondria interface, sustaining mitochondrial Ca2+ uptake30, 31. Similarly, upon depletion of ER Ca2+ content, the activation of store-operated Ca2+ entry (SOCE) at ER-PM MCSs is instrumental in recovering ER Ca2+ content32.
However, the detection of these rapid and spatially restricted Ca2+ hot spots has been challenging. Indeed, the limitations of current techniques, mostly relying on slow super-resolution microscopy or plagued by limited spatial resolution and complex pixel-by-pixel analyses9, 33, hinder the possibility of unambiguously locating the signal of interest to a specific MCS.
We tackled this problem using two different approaches. First, we co-expressed either short or long ER-mit RspA-splitFAST (to identify ER-mit MCSs) with a reported Ca2+ probe (Cepia3)34, that we targeted to the whole OMM (Fig. 5a). Importantly, Cepia3 was chosen for this experiment because its Ca2+ affinity (Kd ~6–7 µM; Fig. 5b) well-suits the reported cation concentration reached at ER-mitochondria interface (~10–15 µM)9, 33. IP3-linked cell stimulations induced an increase of OMM-Cepia3 ratio (Fig. 5a; see Methods), associated with ER Ca2+ release and cytosolic Ca2+ rise experienced by the OMM. However, by restricting the analysis to the sub-regions of OMM-Cepia3 co-localized with either short or long ER-mit RspA-splitFAST signal, larger increases in Cepia3 ratio were observed in both cases (Fig. 5a), suggesting that ER-mit MCSs are sites of high Ca2+-microdomain generation and privileged inter-organelle Ca2+ transfer. Of note, we found that approximately half of ER-mit MCSs experiences local high Ca2+-levels upon histamine-evoked cell stimulation (Fig. 5c), possibly suggesting that not all IP3R clusters respond to IP3-generation and/or that not all ER-mit MCSs associate with active IP3R clusters. Moreover, we observed few transient, short-lasting Ca2+ spikes at some MCSs prior to stimulus exposure (Fig. 5c). To better investigate this point, we co-expressed ER-mit RspA-splitFAST with OMM-targeted GCaMP6f, a higher affinity Ca2+ probe. We indeed observed that Ca2+ levels at ER-mit MCSs are on average higher than in the bulk mitochondrial surface also in basal conditions (Fig. 5d), possibly hinting at a constitutive, low-level ER-mitochondria Ca2+-transfer35.
a Representative images of the 475/390 nm ratio (R, see Methods) of OMM-Cepia3, co-expressed in HeLa cells with either short or long ER-mit RspA-splitFAST to mark short or long ER-mit MCSs. The ratio images of OMM-Cepia3 and of the portion of OMM-Cepia3 co-localized with ER-mit RspA-splitFAST (ER-mit MCSs Cepia3 Ratio) are shown, before (basal) and upon histamine (100 µM) stimulation. On the right, traces represent the ratios of OMM-Cepia3 (OMM) or of the portion of OMM-Cepia3 co-localized with short ER-mit RspA-splitFAST (ER-mit) upon histamine stimulation (arrows), for the three cells on the left. The box plots represent the ΔR of OMM-Cepia3 upon histamine stimulation either in the bulk OMM or in the regions co-localized with ER-mit MCSs, in cells expressing either short or long ER-mit RspA-splitFAST, as indicated. b OMM-Cepia3, expressed in HeLa cells, was calibrated by permeabilizing cells with digitonin (25 µM) in intracellular-like buffer and adding fixed Ca2+ concentrations. Ratiometric 475/390 nm measurements were performed (see Methods). Mean ± SEM. c The heat map represents the OMM-Cepia3 Ratio over time in the sub-regions co-localized with short ER-mit MCSs (identified as single objects, y axis) of a HeLa cell (as in a). Upon histamine stimulation, ~50% of ER-mit MCSs experiences high Ca2+ concentrations (box plot on the right). d The box plot represents the 475/390 nm ratio (R, see Methods) of OMM-GCaMP6f in HeLa cells in basal conditions, either in the bulk OMM or at short ER-mit MCSs (identified by co-expression and co-localization with ER-mit RspA-splitFAST). e The cartoon represents the rationale behind ER-mit PRINCESS design. Mutated Calmodulin (CaM*) from Cepia3 and the M13 peptide were incorporated within ER-mit splitFAST, to provide Ca2+-sensing capabilities. Created in BioRender. Garcia Casas, P. (2023) BioRender.com/f06c444. f Representative traces of ER-mit PRINCESS or OMM-PRINCESS fluorescence in HeLa cells, upon histamine (100 µM) stimulation in Ca2+-free mKRB (see Methods), or CaCl2 (2 mM) addition (SOCE) after 6 min depletion of ER Ca2+ content (obtained by histamine and thapsigargin (100 nM) stimulation in Ca2+-free mKRB). On the right, box plots represent the peaks of OMM- or ER-mit PRINCESS fluorescence (expressed as F/F0) upon the indicated treatments. g Representative confocal images of HeLa cells co-expressing OMM-RFP (used to normalize the fluorescent signal) with either ER-mit PRINCESS or OMM-PRINCESS, before (basal) or upon histamine (100 µM) stimulation in Ca2+-free mKRB. The PRINCESS/OMM-RFP ratio images are also shown. See also the corresponding Supplementary Movies 13 and 14. On the right, the corresponding traces (mean ± SEM) of the ratio signals for the indicated conditions are shown. Scale bar: 10 µm (a, g).
In these experiments, the co-expression of ER-mit RspA-splitFAST is helpful to increase the specificity of the analysis, allowing to simultaneously visualize ER-mit MCSs in a different colour. However, though it represents an improvement, this approach does not completely overcome the limitations of previous techniques, because it requires the expression of two different sensors (i.e., ER-mit RspA-splitFAST and the OMM-Cepia3 Ca2+ probe) and the co-localization of their different fluorescent signals, inevitably retaining some resolution constraints.
An ideal method would be that of unambiguously marking MCSs and specifically measuring the associated Ca2+ dynamics by a single sensor, endowed with sufficient spatiotemporal resolution to report local, fast changes of Ca2+ concentration. To design such a probe, that we dubbed PRINCESS, we integrated into a single reporter both splitFAST (tailored to detect MCSs) and a couple of known Ca2+-sensing protein-domains (i.e., Calmodulin -CaM- and the M13 peptide), thus endowing it with Ca2+-detection capabilities (Fig. 5e). Importantly, to match the range of Ca2+ concentrations potentially reached at MCSs, we selected the low Ca2+ affinity CaM mutant (CaM*) of Cepia334 (Fig. 5b), which should restrict the increase in splitFAST complementation only where Ca2+ hotspots are generated. We hypothesized that, in resting conditions, PRINCESS should maintain a low-level, spontaneous NFAST-CFAST complementation at MCSs, allowing to mark organelle contacts. However, upon sustained Ca2+ peaks, the transient CaM*-M13 binding could dynamically change the equilibrium of the reaction, favouring a Ca2+-dependent complementation of a larger fraction of NFAST-CFAST complexes (Fig. 5e and Supplementary Fig. 3a).
We generated different versions of this probe, to specifically investigate Ca2+ dynamics at ER-mit (ER-mit PRINCESS; Fig. 5e and Supplementary Fig. 3a) or ER-PM (ER-PM PRINCESS; Supplementary Fig. 3b) MCSs. HeLa cells expressing ER-mit PRINCESS displayed a dotted pattern along the mitochondrial network (Supplementary Fig. 3c), like that observed with the ER-mit RspA-splitFAST (Fig. 2a) and corresponding to ER-mit MCSs. Notably, IP3-linked cell stimulations triggered a fast, transient increase of ER-mit PRINCESS fluorescence (Fig. 5f, g). Conversely, the activation of Ca2+ entry through the PM by SOCE, which does not generate microdomains of high Ca2+ concentration at ER-mit MCS33, led to a slower and lower fluorescence rise (Fig. 5f). These data suggest that the probe can promptly detect Ca2+ peaks generated at ER-mitochondria interface upon specific cell stimulations. To further verify this point, we compared the response of ER-mit PRINCESS with that of a similar, control probe, obtained by targeting both the M13-NFAST and CaM*-CFAST fragments to the whole OMM (OMM PRINCESS; Fig. 5f, g). The prediction is that OMM PRINCESS should detect Ca2+ rises occurring along the entire OMM, and not specifically at ER-mit MCSs. Upon histamine-induced, IP3-dependent Ca2+ rises, the response of ER-mit PRINCESS was faster and higher than that of OMM PRINCESS, whereas the two probes displayed a similar behaviour upon SOCE activation (Fig. 5f). Importantly, the signal of PRINCESS can be intrinsically affected by the expression levels of the probe, possibly complicating a quantitative comparison of changes in fluorescence intensity between different cells. However, by co-expressing ER-mit or OMM PRINCESS together with an OMM-RFP, we performed ratiometric measurements that confirmed a higher response of ER-mit PRINCESS upon histamine-induced cell stimulations (Fig. 5g and Supplementary Movies 13, 14).
Interestingly, when similar experiments were performed with ER-PM PRINCESS, we observed high and fast fluorescence increases upon Ca2+ rises triggered either by histamine or SOCE (Supplementary Fig. 3d). Therefore, ER-PM MCSs experience high Ca2+ concentrations upon both types of cell stimulation. This is in line with the reported intense IP3-receptor activity36, as well as the known activation of Orai channels during SOCE32, at ER-PM junctions.
ER-PM and ER-mitochondria MCSs are remodelled by Ca2+ signals
The data above confirm previous findings indicating that MCSs host specific Ca2+ signalling events. By taking advantage of the dynamic complementation of splitFAST-based probes, we wondered whether the opposite is also true, i.e., whether Ca2+ signals in turn modulate organelle juxtaposition. Total internal reflection fluorescence (TIRF)-based experiments have corroborated a Ca2+-mediated regulation of ER-PM MCSs37; however, studying the possible existence of similar mechanisms for MCSs involving organelles located far away from the PM has been challenging.
First, we checked whether ER-PM RspA-splitFAST is sensitive enough to detect the reported strengthening of ER-PM junction upon release of ER Ca2+ content, associated with the recruitment of the ER transmembrane proteins Stromal Interaction Molecule 1/2 (STIM1/STIM2) at ER-PM MCSs and activation of SOCE32, 38. To better follow the process, we also tailored RspA-splitFAST to specifically investigate the migration of STIM1 close to PM and tested that this system is promptly reversible (Supplementary Fig. 4a). Upon depletion of ER Ca2+, we indeed observed a fast, transient recruitment of STIM1 underneath PM and found that this process precedes a progressive increase of ER-PM RspA-splitFAST signal (which reaches a plateau after ~10 min) (Supplementary Fig. 4b), possibly suggesting that STIM1 recruitment is critical for triggering the formation/expansion of ER-PM MCSs, but that additional proteins contribute to their stabilization.
As to ER-mit MCSs, their architecture is critical for the formation of Ca2+ microdomains facilitating inter-organelle Ca2+ transfer, as confirmed by ER-mit PRINCESS. However, whether and how these signals might, in turn, modulate ER-mitochondria juxtaposition has been poorly investigated. In HeLa cells, we did not observe rapid changes of the ER-mit RspA splitFAST signal upon histamine-elicited elevations of cytosolic Ca2+ (Supplementary Fig. 4c and Supplementary Movie 15), implying that transient Ca2+ peaks do not directly affect ER-mit MCSs. However, we noted a tendency of the fluorescent signal to slightly and progressively increase after few minutes following drug addition (Fig. 6a, black trace; see also Supplementary Fig. 4d), an effect that was enhanced by sustaining histamine-induced Ca2+ peaks via cell co-stimulation with thapsigargin (Tg, a SERCA pump inhibitor) (Fig. 6a, red trace), and was also observed by eliciting IP3-dependent Ca2+ stimulation through direct photolysis of caged-IP3 (Supplementary Fig. 4e). Consistently, IP3-linked cell stimulations triggered a sustained rise of ER-mit MCSs in HEK WT, but not in HEK IP3Rs-3KO cells (Supplementary Fig. 4f). To verify whether long-lasting cytosolic Ca2+ elevations could underlie ER-mit MCS potentiation, we pre-incubated cells with BAPTA-AM, a known intracellular Ca2+ buffer. Surprisingly, we observed an even higher increase in ER-mit RspA-splitFAST signal (i.e., in ER-mitochondria juxtaposition) upon histamine-Tg exposure (Fig. 6a, green trace), suggesting that cytosolic Ca2+ is not involved. Moreover, in this condition, we observed that the signal of the probe was slightly increasing ahead of cell stimulation (Fig. 6a, green trace). We reasoned that this effect could be linked to a partial depletion of ER Ca2+ content during cell pre-incubation with BAPTA-AM, as previously reported39 and here confirmed (Supplementary Fig. 4g). Furthermore, the presence of BAPTA-AM remarkably accelerates the discharging of ER Ca2+ upon histamine-Tg stimulation (Supplementary Fig. 4g), possibly explaining the observed faster increase in ER-mitochondria juxtaposition (Fig. 6a). To assess whether ER Ca2+ depletion potentiates ER-mit MCSs, we directly and acutely buffered Ca2+ in the lumen of the ER by TPEN, a low-affinity Ca2+ chelator that specifically sequesters Ca2+ within the ER (Supplementary Fig. 4h) whilst not interfering with cytosolic Ca2+ rises40. Acute TPEN addition progressively increases the ER-mit RspA-splitFAST signal (Fig. 6a, blue trace); along the same line, different treatments partially reducing ER Ca2+ content (i.e., downregulation of SERCA pump, TPEN treatment or incubation with BAPTA-AM; Supplementary Fig. 4g–i) enhance the fraction of mitochondrial surface engaged in the formation of MCSs with the ER (Supplementary Fig. 4j). Remarkably, we observed that this effect is reversible because the refilling of ER Ca2+ content restored basal levels of ER-mit MCSs, though the recovery of organelle tethering was delayed of several minutes (Fig. 6b). Overall, these data support a hitherto unknown mechanism, in which the content of Ca2+ in the lumen of the ER modulates ER-mitochondria coupling. This mechanism is also relevant in vivo, as Tg-induced depletion of ER Ca2+ content in the C. elegans strain expressing ER-mit RspA-splitFAST increased ER-mit MCSs (Fig. 6c).
a Representative traces of ER-mit RspA-splitFAST fluorescence in HeLa cells upon ER Ca2+ depletion by different treatments (arrow): 100 μM histamine (HIST); 100 μM histamine plus 100 nM thapsigargin (HIST Tg); pre-treatment (30 min) with 10 µM BAPTA-AM followed by 100 μM histamine plus 100 nM thapsigargin (BAPTA-AM); 500 μM TPEN. The box plot represents the change in ER-mit RspA-splitFAST fluorescence (ΔF/F0) 15 min after the indicated cell stimulations. b Representative confocal images of HeLa cells, expressing ER-mit RspA-splitFAST and labelled with MitoTracker, in different conditions: at basal, (CTRL); 15 min after stimulation with 100 μM histamine plus 20 μM cyclopiazonic acid (Hist CPA) and 15, 30, or 40 min after washing of Hist+CPA and re-addition of 2 mM CaCl2 (to refill ER Ca2+). The box plots represent the corresponding percentages of mitochondrial surface in contact with ER. c Representative confocal images (max projection) of ER-mit RspA-splitFAST signal in pHX6743 worms, either treated with thapsigargin (Tg, see “Methods”) or not (CTRL). The box plot represents the percentage of cell area covered by ER-mit MCSs. d Representative traces of ER-mit RspA-splitFAST fluorescence upon ER Ca2+ depletion (Hist Tg) in HeLa cells transfected with control (siCTRL) or STIM1-, STIM2- or TMEM110-specific siRNAs. The box plot represents the change in ER-mit RspA-splitFAST fluorescence (ΔF/F0) 15 min after cell stimulation. e Traces (mean ± SEM) of either ER-mit RspA-spiltFAST or STIM1-mit RspA-splitFAST fluorescence upon Hist Tg treatment in HeLa cells. Right, representative images of HeLa cells expressing STIM1-mit RspA-splitFAST, before (t = 0 s) or 180 s after exposure to Hist Tg. f Traces (mean ± SEM) of STIM1-mit RspA-splitFAST fluorescence (F/F0) upon ER Ca2+ depletion (Hist Tg) in HeLa cells, co-transfected with either control or TMEM110-specific siRNA. g Representative STED images of HeLa cells untreated (NT) or stimulated for 15 min with Hist Tg, immunolabeled with αSTIM1 + αTOM20 antibodies. The box plot represents the Pearson’s co-localization coefficient between STIM1 and TOM20 signals, after 15 min in the indicated conditions (500 μM TPEN was used as an alternative treatment to deplete ER Ca2+). h The box plots represent mitochondrial Ca2+ peaks upon histamine-induced ER Ca2+ release in Hela cells, expressing mitochondrial Aequorin (see Methods) and transfected with control (siCTRL) or STIM1-specific siRNAs. Before the experiments, cells were transiently (15 min) treated (ERdep) or not with 500 μM TPEN. i Representative images of HeLa cells expressing STIM1-mit PRINCESS, before (t = 0 s) and 2 s after stimulation with histamine (Hist). Representative traces of OMM-PRINCESS or STIM1-mit PRINCESS fluorescence in HeLa cells, treated with histamine (Hist) in Ca2+-free mKRB, are shown. On the right, the box plots represent the peaks of OMM- or STIM1-mit PRINCESS fluorescence (expressed as F/F0) upon histamine stimulation (OMM-PRINCESS data as in Fig. 5f). Scale bar: 10 µm (b, c, e, g, i).
STIM1 is involved in the Ca2+-mediated modulation of ER-mit MCSs
By distinguishing the signals of co-expressed ER-mit RspA-splitFAST and ER-PM fr-splitFAST through their different lifetime (see also Fig. 2e), we verified in the very same cells that both ER-mit and ER-PM MCSs are increased by the release of ER Ca2+ (Supplementary Fig. 5a). We then explored the molecular mechanism of the Ca2+-mediated modulation of ER-mit MCSs by a small, siRNA-based screening of some possible protein effectors, chosen because previously reported to be Ca2+-sensitive and/or involved in the modulation of different MCSs (Supplementary Table 1). Among these proteins, we excluded an involvement of PERK (Protein Kinase RNA-Like ER Kinase), previously shown to tether ER-mitochondria in basal conditions41 and strengthen ER-PM juxtaposition by a Ca2+-dependent mechanism42. Indeed, although we confirmed that in resting conditions PERK downregulation reduces ER-mit MCSs (Supplementary Fig. 5b), following ER Ca2+ depletion, neither the silencing (Supplementary Fig. 5c), nor the knockout of PERK (PERK-KO) (Supplementary Fig. 5d) significantly affected their increase.
Conversely, we observed that the downregulation of STIM1, but not of STIM2, dampens the increase of ER-mitochondria juxtapositions upon release of ER Ca2+ (Fig. 6d). Intriguingly, a similar effect was detected by downregulating the STIM1 partner TMEM110 (Transmembrane protein 110, also called STIMATE; Fig. 6d)43, 44, strongly supporting a possible involvement of this pathway in ER-mit MCS remodelling.
Experiments performed in SH-SY5Y cells double KO for both STIM1 and STIM2 (STIM1/2-dKO)45 confirmed a significant reduction of the effect of ER Ca2+-depletion on ER-mit MCSs, completely rescued by the re-expression of STIM1 WT, but not of the STIM1-D78G mutant46, which displays a reduced affinity for ER luminal Ca2+ and only partially recovers this parameter (Supplementary Fig. 5e). Importantly, in this latter condition, we observed more ER-mit MCSs in basal conditions (i.e., independently of ER Ca2+-release) (Supplementary Fig. 5f), in line with the constitutive activity of the STIM1-D78G mutant46, providing further evidence that ER Ca2+ regulates ER-mit MCSs by modulating STIM1 activity. Next, we tailored RspA-splitFAST to test whether STIM1, upon sensing the depletion of ER Ca2+ content by its luminal EF-hand domains, migrates at ER-mit MCSs (Supplementary Fig. 5g), as it does towards ER-PM MCSs (Supplementary Fig. 4a, b)32. We indeed observed such a recruitment, preceding the rise of ER-mitochondria juxtaposition (Fig. 6e). Moreover, we found that TMEM110 is important to stabilize STIM1 close to mitochondria, as its downregulation substantially reduces the Ca2+-induced recruitment of STIM1 (Fig. 6f). Conversely, by fusing RspA-NFAST to TMEM110, we found that this protein is present at ER-mit MCSs in basal conditions (as suggested by the presence of fluorescent dots), but is not further recruited upon ER Ca2+ release, nor the presence of STIM1 affects this parameter (Supplementary Fig. 5h). Importantly, using STED microscopy to effectively improve the diffraction-limited spatial resolution of confocal microscopy (Supplementary Fig. 5i), we confirmed that a fraction of endogenous STIM1 is recruited, and forms puncta, close to mitochondria following ER Ca2+ depletion (Fig. 6g), excluding possible artifacts linked to its overexpression. Next, we wondered whether STIM1 can also act as a regulator of ER-mitochondria tethering in resting conditions (i.e., without reducing ER Ca2+ content). Our results showed that the fraction of mitochondrial surface in contact with ER is not affected by STIM1 downregulation (Supplementary Fig. 5j), implying a specific role of this protein in boosting ER-mit MCSs only upon ER Ca2+-depletion.
We speculated that, upon reduction of ER Ca2+ concentration, the STIM1-mediated strengthening of organelle connectivity might be instrumental for sustaining physiological, constitutive levels of ER-to-mitochondria Ca2+ transfer, in turn critical to stimulating mitochondrial activity and cell bioenergetics35, 47. In line with this hypothesis, after a transient TPEN-mediated partial depletion of ER Ca2+ content (see Methods), we measured a higher efficiency of ER-mitochondria Ca2+ shuttling in controls, but not in cells in which STIM1 was downregulated (Fig. 6h). This suggests that STIM1 might be specifically involved in the remodelling of those ER-mit MCSs hosting Ca2+ transfer activity, i.e., where Ca2+ hot spots are generated. Indeed, by designing a PRINCESS probe to monitor Ca2+ dynamics where STIM1 is in contact with mitochondria (STIM1-mit PRINCESS; Supplementary Fig. 5k), we observed that these regions experience microdomains of high Ca2+ concentration upon IP3-linked cell stimulations (Fig. 6i). Overall, these data support a role of STIM1 in the modulation of ER-mitochondria Ca2+ shuttling at MCSs.
Discussion
By tailoring specific variants of splitFAST endowed with low self-complementation, we here designed several chemogenetic reporters to dynamically investigate different MCSs and associated signalling. As in the case of previously reported bimolecular fluorescent complementation (BiFC) approaches, these reporters ensure high and tunable spatial resolution, thanks to an interaction-dependent generation of the fluorescent signal. However, compared with other BiFC systems, the incorporation of improved splitFAST versions guarantees a rapid and fully reversible complementation of the split fragments. This key property endows these reporters with high spatiotemporal resolution, enabling real-time monitoring of dynamic MCS changes, whilst minimizing the possible impact on physiological MCS rearrangements. Moreover, their wide spectral flexibility allows the combination with different probes for performing multi-colour imaging in the very same cell, while the small size of the FAST fragments reduces the risk of dysfunctional protein fusions.
The modest affinity of NFAST and CFAST fragments leads to a low-level, promptly reversible FAST self-complementation, which involves only a minor fraction of the molecules10, 15. This implies that, depending on the sensitivity of the detection method, relatively high expression levels of the probe might be necessary to guarantee a sufficiently bright fluorescent signal at the MCSs under investigation. The fluorogens also contribute to increasing NFAST/CFAST affinity15; thus, the optimal fluorogen concentration should be tested in each experimental setup. We recommend using low fluorogen concentration (<3–5 µM), avoiding excessive amounts to minimize the possible impact on MCS dynamics and reduce background. Similar conclusions were suggested by a very recent report, describing the use of splitFAST-based probes to investigate different MCSs48. By generating a C. elegans strain and substantially improving the signal-to-noise ratio by FLIM, we demonstrated that these reporters can efficiently detect MCSs in vivo. We envisage that in vivo experimental setups will benefit from the availability of the far-red emitting fr-splitFAST (Fig. 1h), enabling a deeper tissue imaging and reducing phototoxicity.
By taking advantage of the dynamic and reversible complementation of our probes, we interrogated the relationship between Ca2+ signalling and MCSs, identifying a hitherto unknown pathway in which ER Ca2+ content modulates ER-mitochondria juxtaposition. Intriguingly, by an siRNA-based, small-scale screening of candidate proteins selected from literature, we identified STIM1 and TMEM110 as key molecules involved in this regulation. While the precise definition of the underlying molecular mechanism will require additional investigations, we further benefited from the versatility of these chemogenetic reporters, generating specific probes that allowed us to demonstrate the prompt recruitment of STIM1 at ER-mit (as well as ER-PM) MCSs upon ER Ca2+ discharge.
The intrinsic dynamicity of MCSs makes cell imaging the ideal application for our reporters; however, the built-in fluorogenic properties provided by splitFAST might enable to carry out larger screening by techniques such as cytometry. As a hint of this possible approach, we used fluorescence-activated cell sorting (FACS) to select the cell clones expressing ER-mit splitFAST (Supplementary Fig. 1d).
Last, by integrating Ca2+-sensing capabilities with both the high spatiotemporal resolution and the fluorescence activation provided by splitFAST, we introduced PRINCESS. In this series of reporters, the spontaneous, low-level NFAST/CFAST self-complementation can be transiently boosted by the Ca2+-mediated binding of CaM* to M13, which favours a fully reversible interaction of the two fragments. The recent development of MAM-Calflux49, a ratiometric probe combining the application of BiFC strategy (to detect ER-mit MCS structural features) with a Bioluminescence Resonance Energy Transfer (BRET)-based Ca2+ indicator (to confer Ca2+-sensitivity), pioneered the generation of reporters with a dual functionality. Similarly, we demonstrated that the properties of PRINCESS allow to simultaneously detect MCSs and investigate the associated Ca2+ dynamics using a single probe. While not being per se a ratiometric sensor (though we could perform ratiometric measurements by co-expressing it with a RFP as a normalizer signal, Fig. 5g), PRINCESS is promptly reversible (thus minimizing the possible interference with ER-mit MCS structure linked to the self-assembly of Venus fragments)49 and allows fast acquisitions (overcoming the need of long frame-to-frame exposures as consequence of the relatively low BRET signal of MAM-Calflux)49, thereby suiting the analysis of short-lasting Ca2+ dynamics well (see also Supplementary Table 2). We envisage that this approach will pave the way to develop an innovative class of biosensors, enabling multiple parameters to be monitored at MCSs and addressing a large diversity of unanswered biological questions.
Methods
Ethical statement
Experiments were performed under European and Italian legislation on animal and human research. All procedures were approved by the Italian Ministry of Health (ref. no. D2784.N.D6J) or by the Stockholm Animal Ethical Committee (ref. no. 15758-2019). Human fibroblasts were purchased from the Coriell Institute for Medical Research; informed consent was obtained based on the guidelines in place at the time of biospecimen collection and approved by the Coriell IRB.
Cell culture and transfection
HeLa (ATCC CCL-2) and COS-7 (ATCC CRL-1651) cells were maintained in DMEM (Sigma-Aldrich, D5671) supplemented with 10% FCS, 2 mM L-glutamine, 100 U/ml penicillin and 0.1 mg/ml streptomycin. Cells were growth in a humidified Heraeus incubator at 37 °C, with 5% CO2.
Tetracyclin-Regulated Expression (T-Rex) HeLa cells were maintained as previously described50 in DMEM (Sigma-Aldrich, D5671), supplemented with 10% FCS, 2 mM L-glutamine, 100 U/ml penicillin and 0.1 mg/ml streptomycin. The expression of ER-mit splitFAST in the different clones was induced by addition of tetracyclin (2 µg/ml) for 16 h.
SH-SY5Y WT or STIM1/STIM2-dKO cells were previously described45 and maintained in DMEM (Gibco 41966-029) supplemented with 10% FCS and 1% MEM NEAA (Gibco 11140-035).
HeLa cells PERK-KD (shP) and relative controls (EV), as well as MEF WT and PERK-KO, were maintained as previously described42 in DMEM (Sigma-Aldrich, D5671) supplemented with 0.11 g/L sodium pyruvate, 2 mM L-glutamine, 100 U/ml penicillin, 0.1 mg/ml streptomycin and 10% FCS. HEK293 WT and IP3Rs-1/2/3 triple KO (HEKs IP3Rs-3KO, obtained from Kerafast, #EUR030) were maintained as described51 in DMEM (Sigma-Aldrich, D5671) supplemented with 0.11 g/L sodium pyruvate, 2 mM L-glutamine, 100 U/ml penicillin, 0.1 mg/ml streptomycin and 10% FCS.
To express all the splitFAST-based probes, cells were either transfected or transduced.
Transfection was performed at 50–60% confluence, 24–48 h after seeding, with either TransIT-LT1 (Mirus Bio, HeLa cells) or LipofectamineTM 2000 (Thermo Fisher, COS-7, SH-SY5Y and HEK cells). Imaging was performed 24 h after transfection. Co-transfection of siRNA was performed with TransIT-X2 (Mirus Bio) and experiments performed 48 h later.
Transduction was performed with lentiviruses (Vector Builder, see plasmids) in medium containing polybrene (1 µg/ml). 24 h later, the medium was replaced with fresh medium. Imaging (or antibiotic selection) was performed at least 72–96 h after transduction.
Primary cell cultures
Primary human skin fibroblasts from a FAD-PS2-N141I female patient (81 year old, AG09908) and a healthy female donor (82 year old, AG08269) were obtained from the Coriell Institute for medical research and grown in DMEM (Sigma-Aldrich, D5671) supplemented with 15% FCS, 2 mM L-glutamine, 100 U/ml penicillin and 0.1 mg/ml streptomycin. Fibroblasts were transduced with lentiviral particles for expression of ER-mit RspA-splitFAST (Vector Builder, see plasmids).
Cryopreserved embryonic cortical neurons isolated from day-17 mouse embryos were purchased from ThermoFisher Scientific (A15586). Cells were revived in Neurobasal Plus Medium (ThermoFisher Scientific, A3582901), complemented with 0.5 mM GlutaMAX I Supplement (ThermoFisher Scientific, 35050) and 2% B-27 Plus Supplement (ThermoFisher Scientific, A3582801), following manufacturer’s instructions. Neurons were then plated on Poly-D-Lysine (Sigma-Aldrich, P6407)-coated 18 mm coverslips at the density of 300,000 neurons/coverslip. Half conditioned medium was changed with fresh medium every third day to feed neurons and prevent excessive evaporation. At 6-days-in-vitro (6 DIV), neurons were transduced with lentiviral particles for the expression of ER-mit RspA-splitFAST (Vector Builder, see plasmids) in conditioned medium supplemented with polybrene (1 µg/ml). 24 h later, medium was replaced with 0.5 ml of conditioned medium (collected before transduction) and 0.5 ml of fresh medium. Imaging was performed 7 days after transduction.
Primary cortico-hippocampal neurons from WT and AppNL-G-F (AD) mice were isolated at embryonic day 17, generated from inbred C57B6/J parents and homozygous AppNL-G-F parents with same genetic background, housed at Karolinska Institutet animal facility under conditions of controlled temperature (22–23 °C), a 12-h light/12-h dark cycle and food and water available ad libitum. All experimental procedures were approved by Stockholm Animal Ethical Committee (ref. no. 15758-2019). Sex was not considered as embryos were used. Brain tissue was digested in 0.08% Trypsin-EDTA (ThermoFisher Scientific, 25200056) diluted in HBSS (ThermoFisher Scientific, 14025092) supplemented with 0.3% fatty acid free BSA (FAF-BSA) for 12 min. Trypsinization was stopped by adding 10% FBS. The tissue suspension was centrifuged at 300 × g for 5 min, followed by mechanical dissociation with HBSS/0.3% FAF-BSA using a flame-polished Pasteur pipette. The cell suspension was filtered using a 40 μm cell strainer (Corning, 352340) and centrifuged again at 300 × g for 5 min. Neurons were seeded in 10 mm bottom-glass MatTek dishes (Mattek, P35G-1.5-10-C) pre-coated with 0.1 mg/mL poly-D-lysine (Sigma-Aldrich, A3890401) at the density of 18,000 neurons/dish, in Neurobasal Medium (ThermoFisher Scientific, 21103049) supplemented with 100 mM GlutaMAX (ThermoFisher Scientific, 35050038) and 2% B-27 Supplement (ThermoFisher Scientific, 17504044). At day 6, half of the medium was changed to freshly supplemented neurobasal medium. At day 8, neurons were transfected using lipofectamine 2000 reagent (ThermoFisher, 11668019) following manufacture recommendations. Per dish, 0.5 µL of lipofectamine and 0.15 µg of each plasmid was used and diluted in OptiMEM reduced serum media (ThermoFisher Scientific, 31985062), and the transfection was performed for 75 min. Neurons were imaged at day 10.
For astrocyte and microglia primary cultures, we used a previously published protocol52, with few modifications. The brains from P0-P2 wild-type (WT) and B6.152H (AD) mice were extracted using forceps and submerged in 30 ml cold dissection medium (DMEM high glucose/pyruvate (Thermo Fisher Scientific, #41966-029) and 0.5% Penicillin-Streptomycin (10,000 U/ml; Gibco). Thereafter, the meningeal layers were completely removed, cortexes were isolated and washed in dissection medium for 2 min to remove blood cells. The cortex was minced and dissociated using a P1000 pipette. The suspension was centrifuged (200 × g, 10 min, RT) and the pellet resuspended in pre-heated 2 ml DMEM high glucose/pyruvate supplemented with 10% heat inactivated FCS, and 0.5% Penicillin-Streptomycin, referred to as supplemented DMEM. Cell clumps were eliminated from the mixed glial culture by passing the suspension through a 70 µm-cell strainer (Corning, #431751), and the suspension was seeded into a Poly-D-lysine (PDL; Sigma-Aldrich, A-003-E)-pre-coated 75 cm2-cell culture flask containing 10 ml of the supplemented DMEM. On day 2, mixed cultures were washed twice with pre-heated D-PBS (pH 7.5; Sigma-Aldrich, D8537) and supplemented DMEM was added. The medium was replaced every 3–4 days.
After 10 days, fresh medium supplemented with 100 ng/mL recombinant murine M-CSF (Peprotech, #315-02), which promotes development and proliferation of microglia53, was added. After 4–5 days, microglia was harvested by tapping and seeded to flasks/well plates for further experiments in supplemented DMEM. Microglia harvest was performed up to 4 times in intervals of 2 to 5 days. After final microglia isolation, the remaining attached cells, expected to be enriched in astrocytes, were harvested by 2.5% trypsin/0.5 mM EDTA for 7–10 min at 37 °C and seeded in flasks/well plates for further experiments in supplemented DMEM. Both microglia and astrocytes were transduced with lentiviral particles for expression of ER-mit RspA-splitFAST (Vector Builder, see plasmids) in medium containing polybrene (1 µg/ml). 24 h later, the medium was replaced with fresh medium. Imaging was performed 72 h after transduction. Where indicated, astrocytes were exposed for 16 h to a conditioned medium obtained from either WT CHO cells or 7PA2-CHO cells (containing high levels of naturally generated Aβ peptides), as described22.
Animals were bred in-house in individually ventilated cages (IVCs) under specific-pathogen-free (SPF) conditions. Water and food were provided ad libitum. All animals were maintained in compliance with the Italian animal protection law and the local animal welfare committee. Organ harvesting was approved by the institutional review.
Cloning, plasmids, siRNA
Unless differently specified, the plasmids generated in this study have been obtained either by PCR amplification or by gene synthesis (Life Technologies), followed by digestion with specific restriction enzymes and insertion into pcDNA3, allowing mammalian expression under the control of the CMV promoter. All the plasmids and the detailed sequences will be available on request from the corresponding author R.F. Requests will be processed in 1–2 weeks.
ER-NFAST
The ER-membrane targeting sequence from ER-ABKAR (Addgene #61508) was amplified by PCR. The NFAST sequence was amplified by PCR from the Addgene plasmid pAG148 FRB-NFAST (Addgene #130812) and ligated at the 3’ of ER-targeting sequence into pcDNA3.
OMM-short-CFAST10
The OMM targeting sequence encoding the N-terminal 72 aminoacids of human TOM70 was amplified by PCR from the previously described TOM70-YFP plasmid54, including the CFAST10-encoding sequence in the reverse primer. The fragment was ligated into pcDNA3.
OMM-long-CFAST10
The OMM targeting sequence coding the N-terminal 111 aminoacids of human TOM70 was amplified by PCR from the previously described TOM70-YFP plasmid54, including the CFAST10-encoding sequence in the reverse primer. The fragment was ligated into pcDNA3. The cytosolic region of TOM70 from the end of the N-terminal transmembrane domain till aminoacid 111 is flexible and works as a linker, as predicted by AlphaFold.
ER-NFAST in pcDNA™FRT⁄TO vector (used to generate inducible HeLa T-Rex clones)
The sequence encoding ER-NFAST was obtained by enzymatic digestion (HindIII + XhoI) of the above described ER-NFAST plasmid and ligated into pcDNA™FRT⁄TO vector (ThermoFisher Scientific).
ER-frNFAST
The sequence encoding frNFAST was amplified by PCR from the plasmid pAG499-FRB-N-frFAST14 and ligated in place of the NFAST sequence in the above described ER-NFAST plasmid. Please note that in Fig. 2f this plasmid is called ER-short-frNFAST for an easier comprehension of that specific experiment.
ER-RspA-NFAST
The sequence encoding RspA-NFAST was amplified by PCR from the plasmid pAG573 FRB-RspA(N)-IRES-mTurquoise215 and ligated in place of the NFAST sequence in the above described ER-NFAST plasmid.
ER-long-RspA-NFAST
The sequence encoding a flexible linker of 55 aminoacids was generated by gene synthesis (Life Technologies) and ligated between the ER-targeting and the RspA-NFAST encoding sequences of the ER-RspA-NFAST plasmid described above, previously digested with the KpnI and BamHI restriction enzymes. The sequence of the linker is: GGTACCGACTACAAAGACGATGACGACAAGGGCAGCGGAGGAGGCTCCGGAGGAGGCGGCAGCGGCGGAGGGAAGAAAAGCGGAGGCGGAGGCTCCGGAGGAGGCGGCAGCGGCGGAGGGAAGAAATCCGGAGGAGGCGGCAGCGGCGGAGGGTCTGGTGGATCC
OMM-short-RspA-CFAST
The OMM targeting sequence coding the N-terminal 72 aminoacids of human TOM70 was amplified by PCR from the previously described TOM70-YFP plasmid54, including the RspA-CFAST-encoding sequence15 in the reverse primer. The fragment was ligated into pcDNA3.
PM-RspA-CFAST
PM-RspA-CFAST was generated by gene synthesis (Life technologies), fusing the RspA-CFAST encoding sequence15 at the 3’ of the Lyn11 PM-targeting sequence. The coding fragment was ligated into pcDNA3.
MyrPalm-D1cpv
The PM-targeting sequence MyrPalm was amplified by PCR and ligated at the 5’ of the cameleon D1cpv-encoding sequence (Addgene #37479) into pcDNA3.
PM-frNFAST
The sequence encoding frNFAST was amplified by PCR from the plasmid pAG499-FRB-N-frFAST14 and ligated in place of the RspA-CFAST sequence in the above described PM-RspA-CFAST plasmid. This construct was used to simultaneously detect ER-PM and ER-mit MCSs in the very same cells. For this approach, we used ER-RspA-CFAST (see below) to compliment both PM-frNFAST and OMM-short-RspA-NFAST (see below).
ER-RspA-CFAST
ER-RspA-CFAST was generated by gene synthesis (Life Technologies), fusing the sequence encoding RspA-CFAST15 at the 3’ of the ER-membrane targeting sequence of ER-NFAST. The insert was then ligated into pcDNA3.
OMM-short-RspA-NFAST
The sequence encoding RspA-NFAST was extracted by enzymatic digestion (BamHI + XhoI) from the plasmid ER-RspA-NFAST and ligated in place of the RspA-CFAST-encoding fragment into the OMM-short-RspA-CFAST plasmid described above.
OMM-Cepia3
The sequence encoding Cepia3 was amplified by PCR from the plasmid pCMV CEPIA3mt (Addgene #58219) and ligated at the 3’ of the OMM-targeting sequence encoding the N-terminal 72 aminoacids of human TOM70.
OMM-GCaMP6f
The OMM targeting sequence of OMM-RFP (a gift of G. Hajnoczky)55 was amplified by PCR, eliminating with synonymous mutations the BamHI and EcoRI restriction sites inside the targeting sequence with the following primers:
Fw:ACGAAGCTTGCCACCATGGCAATCCAGTT
Rv:CGCGAATTCGGATCCTTTAGATAGGATAGCACCAGCACCAGCACCAGCACCTGGATTCGCAGATCTGGTGAGATCC
GCaMP6f sequence was amplified by PCR from the Addgene plasmid #40755 with the following primers:
Fw:CGCGGATCCATGGGTATGGCTAGCATGACTG
Rv:GCGTCTAGATTACTTCGCTGTCATCATTTG
The two fragments were then ligated into pcDNA3 between HindIII and XbaI restriction sites.
ER-mit PRINCESS
ER-mit PRINCESS is based on the co-expression of ER-M13-NFAST and OMM-CaM*-CFAST10. To generate ER-M13-NFAST, the M13-encoding sequence was amplified by PCR from the plasmid pCMV CEPIA3mt (Addgene #58219) and ligated between the ER-membrane targeting sequence and the NFAST-encoding sequence in the above described ER-NFAST plasmid. OMM-CaM*-CFAST10 was generated by amplifying by PCR the sequence encoding mutated Calmodulin (CaM*) from the plasmid pCMV CEPIA3mt (Addgene #58219). The CaM*-encoding fragment was then ligated between the OMM-targeting sequence and the CFAST10-encoding sequence in the OMM-short-CFAST10 plasmid described above.
ER-PM PRINCESS
ER-PM PRINCESS is based on the co-expression of ER-M13-NFAST (see above ER-mit PRINCESS) and PM-CaM*-CFAST10. To generate PM-CaM*-CFAST10, the CaM*-CFAST10-encoding fragment was extracted by enzymatic digestion from the OMM-CaM*-CFAST10 plasmid (see above ER-mit PRINCESS) and ligated in place of the RspA-CFAST sequence in the above described PM-RspA-CFAST plasmid.
OMM PRINCESS
OMM PRINCESS is based on the co-expression of OMM-M13-NFAST and OMM-CaM*-CFAST10 (see above ER-mit PRINCESS). OMM-M13-NFAST was generated by extracting the M13-NFAST-encoding sequence from the plasmid ER-M13-NFAST (see above ER-mit PRINCESS) and ligating it in place of the CFAST10-encoding sequence in the above described OMM-long-CFAST10 plasmid.
STIM1-RspA-NFAST
The sequences encoding STIM1 and RspA-NFAST were amplified by PCR from respectively the human STIM1-YFP plasmid (Addgene #19754, a gift from A. Rao) and the pAG573 FRB-RspA(N)-IRES-mTurquoise215 plasmid. The RspA-NFAST-encoding fragment was then ligated at the 3’ of the STIM1-encoding fragment into pcDNA3.
TMEM110-RspA-NFAST
The sequence encoding TMEM110 was generated by gene synthesis (Life Technologies), isolated by enzymatic digestion with HindIII and BamHI and then ligated at the 5’ of the RspA-NFAST-encoding sequence (isolated from the above described STIM1-RspA-NFAST plasmid by eliminating the STIM1-encoding sequence) into pcDNA3.
STIM1-mit PRINCESS
STIM1-mit PRINCESS is based on the co-expression of STIM1-M13-NFAST and OMM-CaM*-CFAST10 (see above ER-mit PRINCESS). STIM1-M13-NFAST was generated by extracting the M13-NFAST-encoding sequence from the plasmid ER-M13-NFAST (see above ER-mit PRINCESS) and ligating it in place of the RspA-NFAST-encoding fragment in the STIM1-RspA-NFAST plasmid described above.
Lentiviral plasmid VB220320-1120bts for the expression of ER-mit RspA-splitFAST
The lentiviral vector VB220320-1120bts was generated by gene synthesis (VectorBuilder) and used to generate lentiviral particles for the expression and/or stable integration of inserts allowing the expression of ER-mit RspA-splitFAST. In this vector, the above described ER-RspA-NFAST and OMM-RspA-CFAST sequences were inserted under the control of respectively CMV and SV40 promoters, allowing the simultaneous expression of the two complimentary fragments. A puromycin-resistance gene was included under the control of mPGK promoter, allowing the selection of cell clones with stable integration of the lentiviral vector.
C. elegans plasmid for ER-mit RspA-splitFAST
Plasmid p L4665 (a gift from Andrew Fire, Addgene #1667) carrying the C. elegans myo 3 promoter was used as backbone to clone an insert allowing expression of the ER-mit RspA-splitFAST. The sequence TOMM70::RspACFAST::SL2::EMB8::RspANFAST was generated by gene synthesis and cloned into pL4665 using BamHI and NcoI restriction enzymes. We used the targeting sequences of C. elegans TOMM70 and EMB8 to respectively target RspA-CFAST and RspA-NFAST to the OMM and the ER-membrane. The C. elegans-specific SL2 sequence was inserted between TOMM70-RspA-CFAST and EMB8-RspA-NFAST to guarantee co-expression of the two fragments by a single plasmid. The resulting plasmid [Pmyo-3::TOMM70::RspACFAST::SL2::EMB8:: RspANFAST::unc-54 3’UTR, pRF4] allows the expression of ER-mit RspA-splitFAST on body wall muscle cells. Plasmid pRF4 (rol-6; su1006) was used as co-injection marker for pHX6743 C. elegans strain generation (see below).
The plasmids encoding mit-RFP and H2B-GCaMP6f were previously described54. The plasmid encoding OMM-RFP was a gift of G. Hajnoczky. The plasmid encoding ER-GFP (GFP-Sec61β) was a gift of G. Voeltz. The following plasmids were from Addgene: ER-StayGold (Addgene #186296), mCherry-Drp1 (Addgene #49152), Tubuline-GFP (Addgene #56450), LifeActGFP (Addgene #58470), mCherry-ER-3 (Addgene #55041), pLV-mitoDsRed (#44386).
For RNAi experiments, we used the following siRNA (all from Merck):
MISSION® siRNA Universal Negative Control #1 (SIC001); STIM1 (SASI_Hs01_00107803); STIM2 (SASI_Hs02_00354130); TMEM110 (SASI_Hs01_00204728); SERCA-2A (SASI_Hs01_00047711); ESYT1 (SASI_Hs01_00188940); VPS13A (SASI_Hs02_00306926); VPS13D (SASI_Hs01_00155099).
Reagents and fluorogens
HMBR ((Z)-5-(4-hydroxy-3-methylbenzylidene)-2-thioxo-1,3-thiazolidin-4-one; TFLime, 480541-250), HBR-3,5DOM ((Z)-5-(4-Hydroxy-3,5-dimethoxybenzylidene)-2-thioxothiazolidin-4-one; TFCoral 516600-250) and HPAR-3OM (4-hydroxy-3-methoxy-phenyl)allylidene rhodanine; TFPoppy 555670-250) were from Twinkle Factory. All the other reagents were from Merck, unless differently specified.
Fluorescence microscopy
Imaging of the different splitFAST-based probes for MCSs by fluorescence microscopy was performed on a Thunder Imager 3D Cell Culture (Leica), equipped with a LED8 illumination system, both 40×/1.30 oil immersion (HC PL Fluotar 340) and 63×/1.40 oil immersion objective and with a cool camera (Hamamatsu Flash 4.0 V3). Cells seeded and transfected on 18 mm coverslips, were mounted just before the imaging experiments on a chamber and bathed in mKRB (in mM: 140 NaCl, 2.8 KCl, 2 MgCl2, 1 CaCl2, 10 HEPES, 10 glucose, pH 7.4 at 37 °C). Where indicated, CaCl2 concentration was 100 µM, or replaced with EGTA 0.5 mM. The probes were complemented with either Lime (4 µM, Twinkle Factory 480541-250), Coral (4 µM, Twinkle Factory 516600-250) or Poppy (5 µM, Twinkle Factory 555670-250) and imaged using respectively the 475 nm (15%), 555 nm (20%) or 575 nm (20%) LED8 excitation lines. Excitation and emission light was filtered by GFP-, TRITC- or far-red dedicated sets of filters (Chroma for GFP and TRITC, Leica CYR71010 for far-red), respectively. In most experiments, a 60 ms exposure time and a frame rate of 15 s/frame were set, unless differently specified (see figure legends). For fast acquisitions, frame rates of 100 ms/frame and 250 ms/frame were used for OMM-Cepia3 or PRINCESS- -based experiments, respectively. Where indicated, different cell stimuli/treatments were acutely added to the experimental medium, including histamine (Hist, 100 µM), Thapsigargin (Tg, 100 nM), N,N,N′,N′-tetrakis(2-pyridinylmethyl)-1,2-ethanediamine (TPEN, 500 µM), bradykinin (BK, 100 nM), ATP (100 µM). In some experiments, a 30 min pre-incubation with BAPTA-AM (10 µM), added to mKRB supplemented with CaCl2 1 mM, was performed, as indicated. For the calculation of the genuine change of fluorescence of the different MCS splitFAST-based probes, we performed parallel experiments for each experimental condition/stimulus in cells expressing either cytosolic RspA-NFAST (plasmid pAG573 FRB-RspA(N)-IRES-mTurquoise2)15 and RspA-CFAST (plasmid pAG580 FKBP-RspA(C)-IRES-iRFP670)15, or NFAST (pAG148 FRB-NFAST, Addgene #130812) and CFAST10 (pAG241 FKBP-CFAST10, Addgene #130814), keeping the same illumination/exposure parameters. The average fluorescent signals (expressed as ΔF/F0) of the cells expressing cytosolic RspA-splitFAST (or splitFAST) were subtracted from those of the cells expressing the corresponding MCS probes, to normalize for bleaching and other possible fluorescence changes not linked to MCS remodelling (see Supplementary Fig. 4d as an example). All the data shown in this manuscript were normalized for the corresponding fluorescence changes of cytosolic RspA-splitFAST (or splitFAST).
Confocal, STED and FLIM microscopy
Confocal analysis was performed on a Leica TCS-II SP5 STED CW microscope, equipped with a 100×/1.4 N.A. Plan Apochromat objective, a tunable WLL laser, both PMT and HyD (Leica) detectors for signal collection, and a suitable resonant scanner (8000 Hz) for fast acquisitions. Alternatively, a Leica Stellaris 8 microscope, equipped with a 63×/1.4 N.A. Plan Apochromat objective, a WLL and HyD detectors was used. The WLL was set at 488, 555 and 600 nm to respectively excite the different splitFAST-based probes complemented with either Lime (3 µM, Twinkle Factory 480541-250), Coral (5 µM, Twinkle Factory 516600-250) or Poppy (3 µM, Twinkle Factory 555670-250, this latter to complement fr-splitFAST). When a quantitative comparison between different experimental conditions was performed, images were collected by keeping the same excitation/emission, zoom, and image size parameters. Where indicated, 3D-stacks were collected with z-steps of 0.4 µm. Images were background-subtracted and analyzed with specific ImageJ (NIH) plugins. In particular, the calculation of the percentage of mitochondrial surface in contact with either ER or the PM was performed by staining mitochondria for 15 min in mKRB (or medium) supplemented with MitoTracker Deep Red (300 nM, Thermo Fisher M22426) or MitoTracker Green (300 nM, Thermo Fisher M7514). PM staining was performed incubating the cells in 2 µg/ml WGA (Thermo Fisher W32464) for 25 min in mKRB. After washing, cells were imaged in mKRB, supplemented with Lime (3 µM). Images for both channels were background subtracted and preprocessed using a custom-written Python script56.
The following dedicated Python libraries have been employed: opencv and scikit-image.
Images were min-max normalized and filtered with a Gaussian filter with a standard deviation of 1 using the “filters.gaussian” function from scikit-image. Images were binarized using a threshold set at half of the mean value of the pixel intensity values exceeding 20% of the pixel intensity distribution.
To locate spatial overlaps between ER-mit or ER-PM RspA-splitFAST and MitoTracker, we merged the binarized images using pixel multiplication. The fraction of overlap has been calculated on the MitoTracker image and the result saved using the ‘to_excel’ function from pandas.
To improve the signal-to-noise ratio, after background subtraction some of the displayed images were further processed with the automatized ImageJ plugin enhance image, but all analyses were performed before this modification. For fast acquisitions (such as those for the experiments with PRINCESS), resonant scanner was used, with the following parameters: image size 1024 × 512 pixels, bidirectional acquisition, line accumulation 3×, frame average 3×, 293 ms/frame.
STED and FLIM analyses were performed on a Leica Stellaris 8 TauSTED microscope, equipped with a 100×/1.4 N.A. Plan Apochromat objective (for STED imaging) or a 63×/1.4 N.A. Plan Apochromat objective (for FLIM analysis), a WLL, HyD detectors, a continuous 660 nm STED laser and a pulsed 775 nm STED laser.
For STED analysis of endogenous STIM1 recruitment close to mitochondria, after the indicated treatments to deplete ER Ca2+ content (see Fig. 6e legend), HeLa cells were washed with PBS, fixed in 4% formaldehyde (10 min), washed 3× with PBS and permeabilized for 2 h in blocking solution (0.3% Triton X-100, 1% BSA, 5% goat serum in PBS). Cells were incubated overnight at 4 °C with primary antibodies (α-TOM20 Santa Cruz # sc-11415; α-STIM1 Merck WH0006786M1) diluted (1:200) in antibody dilution buffer (0.3% Triton X-100, 1% BSA in PBS), washed 3 × 5 min with PBS and incubated for 1 h at RT with secondary antibodies (α-mouse STAR 580 and α-rabbit STAR 635 P, Abberior) diluted 1:300 in antibody dilution buffer. Coverslips were washed 4 × 5 min in PBS and mounted with Aqua-Poly/mount (Polysciences, 18606). STED images were acquired by sequential acquisition of each channel, setting the WLL at respectively 570 nm and 635 nm, by activating (30%) the 775 nm pulsed depletion-laser. Excitation/emission parameters were kept constant, avoiding signal saturation. Pixel size was set at 20 nm/pixel. Images were background-subtracted, and the Manders’ and Pearson’s co-localization coefficients calculated by the ImageJ Co-localization Analysis plugin.
For FLIM analysis, we used the Phasor Method57, available within the FALCON > FLIM application module of the Leica Application Suite X (4.6.0) software. Briefly, each pixel of the FLIM image was transformed into a unique position in the phase diagram by Fourier transformation of the decay curve, with the x-axis representing the real component and the y-axis the imaginary component. The different lifetime components were visualized as clustering of pixels in defined regions of the phasor plot, that were isolated by adjusting the size and position of specific cursors. The different lifetime components, identified by each of these cursors, were then broken down into separate channels by the “Separate” function provided by the FALCON module. To ascertain the molecular identity of each specific component, control experiments were performed by preparing samples in which single splitFAST-based probes were expressed separately.
Lattice light-sheet and airyscan microscopy
HeLa cells stable clones were seeded on µ-Dish 35 mm (Ibidi) and 24–48 h later incubated for 10 min in culture media (37 °C, 5% CO2) with MitoTracker Deep Red (200 nM, Thermo Fisher M22426). Imaging was performed at room temperature in mKRB using ZeissLattice Lightsheet 7, to get gentle volumetric live imaging. ER-mit RspA-splitFAST (marked with 3 µM TFLime) and MitoTracker Deep Red signals were simultaneously acquired in two channels using a dual camera configuration with bandpass 505–545 nm, and 656–750 nm detection. Excitation was combined 488 nm and 640 nm with a Sinc3 30 × 1000 lightsheet. Excitation intensities were balanced to minimize photobleaching and maintain a good signal to noise ratio. Integration time was set to 15 ms. The imaged volumes were recorded in 500 slices with step 0.2 µm and pixel size 0.145 µm, Time lapse recordings were made with 6 volumes per minute for 10 min. Data were postprocessed by iterative deconvolution followed by deskewing, resulting in a coverslip transformed image volume of 297 × 100 × 38 µm for each time point. Surface rendering was performed by IMARIS software after median (3 × 3 × 3) filtering of the ER-Mit RspA-splitFAST signal to reduce background noise. Surface tracking was calculated using the “Connected Components” algorithm and data collected from IMARIS “Statistics” module.
COS-7 cells were seeded on µ-Dish 35 mm (Ibidi), co-transfected 24–48 h later with plasmids encoding ER-Mit RspA-splitFAST and either ER-StayGold, mCherry-Drp1, Tubulin-GFP or LifeActGFP. The day after, cells were incubated for 10 min in culture media (37 °C, 5% CO2) with MitoTracker Deep Red and then imaged at room temperature in mKRB (supplemented with either 3 µM Lime or 3 µM Coral) using a Plan-Apochromat 63×/1.4 NA oil objective on an inverted Zeiss 980 LSM Airyscan2 confocal microscope. Multi-color timelapse imaging was performed by sequentially scanning three channels configured for 639 nm excitation with longpass 660 nm detection, 561 nm excitation with shortpass 615 nm detection, and 488 nm excitation with bandpass 495–555 nm detection. For 3D timelapse imaging, a full z-stack of 5–15 slices was acquired per laser line before switching. Zoom factor was set at 5× yielding maximum pixel-dwell time of ~0.69 μs per pixel and a frame time 325 ms at Nyquist sampling (~40 × 40 × 150 nm per voxel). High resolution images were obtained by Airyscan processing in Zen Blue (3.8) using standard 3D Auto defined strength. Surface rendering was performed by using IMARIS software.
To segment ER structures of COS-7 cells into “sheets” (rough ER) and “tubules” (smooth ER), we have used a morphology-based approach. On average, the width of tubules measure between 10-12 pixels in our recordings (Pixelsize: 42.3 nm), while the sheet dimensions are larger. The thickness range of tubules span 5 to 21 pixels (10.5 ± 2.5 pixels, mean ± SEM, n = 5 cells). We used the morphological operators erode and dilate in ImageJ and created a mask by eroding 6 times (eliminating structures <12 pixels) that isolates the sheets. The sheets were then subtracted from the original ER channel leaving the tubules. In this manner we separated the different structures from each other to enable co-localization analysis with the splitFAST signal in the other channel. Structures <5 pixels were regarded as unspecific noise/background and removed.
Morphological segmentation requires high resolution, which was achieved by Airyscan processing (see above) or deconvolution of Leica Stellaris 8 data acquired with Resonant Scanner at 8000 Hz through 8 slices (step size 0.4 µm) and pixel size 42.95 nm, in time lapse recordings of 10 min. COS-7 cells were co-transfected with ER-mit RspA-splitFAST (marked with 3 µM Lime) and mCherry-ER-3 (see plasmids), and imaged at 488 and 575 nm, respectively. For data preprocessing in Fiji, we used the “Denoise” function prior to deconvolution with “DeconvolutionLab2” and the “Richardson-Lucy” algorithm. Theoretical point spread functions were generated using the Fiji plugin “PSF Generator”.
For the ER morphological segmentation, we developed an ImageJ macro, which was applied in all image sequences. The macro used for image analysis can be accessed at Github58.
MCS lifetimes were calculated from Lattice Light-Sheet data (see above) or ER-segmented movies from Resonant Scanner acquisitions. In this latter case, lifetimes were calculated for contacts involving only ER sheets or tubules after tracking of colocalized surfaces of the MCSs (calculated with IMARIS “Surface-Surface Contact Area” XTension) with those of segmented ER sheets or tubules. Tracking was calculated using the IMARIS “Brownian Motion” algorithm, allowing a maximal gap of 3 frames, and “track duration” data were collected from IMARIS “Statistics” module.
After quantification, performed as detailed above, representative images and Movies were obtained calibrating the signal through the IMARIS “Display Adjustment” console.
The Kernel Density and the correlation Spearman index were calculated by Excel XLSTAT (Microsoft).
Flow cytometry
14 days after selection with either Hygromycin (300 µg/ml, HeLa T-Rex clones expressing ER-mit splitFAST; induction of ER-mit splitFAST expression by overnight treatment with tetracyclin 2 µg/ml) or puromycin (3 µg/ml, for stable HeLa clones expressing ER-mit RspA-splitFAST after transduction with lentiviral particles, see plasmids), cells were bathed in PBS supplemented with Lime (5 µM). The cells expressing the probe of interest were separated by Fluorescence-activated cell sorting (FACS), using a S3e TM Cell Sorter (BIORAD) endowed with a 488 nm laser and a 525/30 nm emission filter. Analysis was performed with ProSort 1.6 (BIORAD). After sorting, single cell clones were isolated from the positive cell population by limiting dilution cloning in 96 well-plates, followed by a screening for optimal probe-expression performed by fluorescence microscopy.
CLEM
HeLa cells were seeded on a 35 mm Dish with gridded glass coverslip (MatTek In Vitro Life Science Laboratories) and transfected with plasmids to express ER-mit RspA-splitFAST. 24 h after transfection, cells were incubated for 20 min with MitoTracker™ Red CMXRos (200 nM, Thermo Fisher M7512) to stain mitochondria, washed in PBS and bathed in PBS containing Lime (5 µM). Confocal images of cells expressing ER-mit RspA-splitFAST were acquired by a FluoVIEW 3000 RS system (Evident Scientific, Waltham, MA) equipped with an UPLSAPO 60X/1.3 Silicon objective. The entire thickness of the cell was acquired with optical section depth of 0.21 μm and the images were deconvolved with Huygens Software before alignment with the matching EM images (SVI, Hilvertum, Netherlands). After the acquisition of the full z-stack, fixative was added (final concentration 4% PFA in 0.1 M Hepes) and the field was acquired for further 10 min. Dishes were removed from the microscope stage and postfixed with 2,5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 1 h at RT. Samples were postfixed with reduced osmium (1% OsO4, 1.5% potassium ferrocyanide in 0.1 M cacodylate buffer, pH 7.4) for 2 h on ice. After several washes in milli-Q water, sections were incubated in 0.5% uranyl acetate overnight at 4 °C. Samples were then dehydrated with increasing concentration of ethanol, embedded in epoxy resin and polymerized for 48 h at 60 °C. Ultrathin serial sections were obtained using an ultramicrotome (UC7, Leica microsystem), collected on formvar-carbon coated copper slot grids, stained with uranyl acetate and Sato’s lead solutions and observed in a Transmission Electron Microscope Talos L120C (FEI, Thermo Fisher Scientific) operating at 120 kV. Images were acquired with a Ceta CCD camera (FEI, Thermo Fisher Scientific) and aligned with confocal images using ec-CLEM plugin of ICY software. To evaluate the fraction of mitochondrial surface in contact with ER, analysis was performed with Microscope Image Browser (MIB) software59.
Serial section electron tomography
HeLa cells, cultured in Minimum Essential Medium (MEM, Sigma) supplemented with 10% FBS, 2 mM glutamine, 1 mM sodium pyruvate and 0.1 mM non-essential amino acids, were transfected with HRP-KDEL construct60, 61 to better visualize ER; 24 h after transfection, cells were fixed in 1% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 30 min at room temperature, followed by incubation with 0.3 mg/ml 3,3’-diaminobenzidine tetrahydrochloride (DAB) and 0.03% hydrogen peroxide in 0.1 M sodium cacodylate buffer pH 7.4 for 20 min at room temperature. Samples were rinsed in sodium cacodylate buffer and post-fixed with 1.5% potassium ferrocyanide, 1% osmium tetroxide in 0.1 M sodium cacodylate buffer (pH 7.4) for 1 h on ice. After enbloc staining with 0.5% uranyl acetate in dH2O overnight at 4 °C in the dark, samples were dehydrated with increasing concentrations of ethanol, embedded in Epon and cured in an oven at 60 °C for 48 h. Ultrathin sections (70–90 nm) were obtained using an ultramicrotome (UC7, Leica microsystem, Vienna, Austria), collected, stained with uranyl acetate and Sato’s lead solutions, and observed in a Transmission Electron Microscope Talos L120C (FEI, Thermo Fisher Scientific) operating at 120 kV. Images were acquired with a Ceta CCD camera ((FEI, Thermo Fisher Scientific).
Serial thick sections (130–150 nm) were collected on formvar-coated copper slot grids. Gold fiducials (10 nm) were applied on both surfaces and the grids were stained with 2% methanolic uranyl acetate and Sato’s lead citrate. The samples were imaged using a 120 kV Talos L120C (Thermo Fisher Scientific). Tilted images (+65/−65 according to a Saxton scheme) were acquired with Tomography 4.0 acquisition software (Thermo Fisher Scientific) using a 4k × 4k Ceta16M camera (Thermo Fisher Scientific). Tilted series alignment, tomography reconstruction, and serial tomograms joining were performed using the IMOD software package62. Mitochondria and ER were segmented with a deep learning approach using Empanada Napari plugin63. Labeled ER and mitochondria layers were imported in Microscope Image Browser (MIB)59 and manually inspected for errors in the segmentation. Distance from ER and mitochondria were calculated using MCalc plugin in MIB setting a contact cutoff of approximately 12 nm (3 pixels) and 24 nm (6 pixels) respectively for short and long ER-mit MCSs.
Segmented model generated in MIB was imported in IMOD and cloudcompare (https://www.danielgm.net/cc/) for 3D and ER-mitochondria distance visualization.
Generation and maintenance of C. elegans strains
C. elegans strains were cultured and maintained following standard methods as previously described64. Specifically, worms were maintained in Nematode Growth Medium (NGM) agar plates seeded with E. Coli OP50 as food source. Worms were transferred to new plates before bacteria were fully consumed to avoid starvation. Worms were always kept in a thermostatic incubator at 20 °C. The pmyo3::ER-mit RspA-splitFAST allele sybIs6743 (see in plasmids) was generated at SunyBiotech (http://www.sunybiotech.com) by plasmid microinjection together with a rol6 co-injection marker, followed by X-ray irradiation to randomly integrate the sequence in the worms genome and generate a stable C. elegans strain. All experiments were performed on synchronized worms generated by L4 larvae isolation, and maintained at 20 °C.
C. elegans worms (N2, control not injected; pHX6743, sybIs6743) were evaluated in a 3% agarose pad slide, pre-treated with Levamisole 1 mM, and surrounded by a thin silicone line. When levamisole was completely absorbed by the agarose pad, 3 μL of the fluorogen solution (TFCoral 40 μM in M9 Buffer) were placed in the center of the pad and the worms transferred into the solution. The worms were covered with a cover slip and incubated O/N on the fluorogen solution. Imaging was performed 16 h later.
Synchronized pHX6743 worms at D3 or D4 of adulthood were incubated in the fluorogen solution O/N in agitation at RT. 4 h before confocal imaging, Thapsigargin 10 μM or DMSO were added to the solution and incubated at room temperature. Then worms were mounted for imaging as described above and confocal analysis was performed on a Leica TCS SP5X confocal microscope equipped with a 63×/1.4 N.A Plan Apochromat objective, a tunable WLL laser, and PMT detectors for signal collection. 3D-stacks were collected with z-steps of 0.8 μm. Images were background-substracted, filtered with a median filter (standard deviation of 0.8), binarized and analyzed by the “analyze particles” plugin on ImageJ, calculating the % of cell area covered by MCSs.
Ca2+ measurements
For Aequorin Ca2+ measurements, HeLa cells (0.6 × 105 cells/well) were plated on coverslips (13 mm diameter) and transfected with plasmids for the expression of either cytosolic Aequorin or mitochondrial mutated Aequorin, together with the indicated siRNA (see above).
48 h after transfection, cells were incubated for 1 h at 37 °C in mKRB supplemented with native coelenterazine (5 µM, BIOTIUM), and then incubated for 15 min in mKRB supplemented with either TPEN (500 µM, to induce partial depletion of ER Ca2+ content) or EtOH (as control). After incubation with EtOH, control cells were additionally incubated for 30 s in mKRB supplemented with TPEN (500 µM). This brief treatment was performed to match the possible contribution of residual TPEN in buffering Ca2+ peaks, while not being long enough to induce an appreciable increase of ER-mit juxtaposition, which was observed to only occur at least 4–5 min after depletion of ER Ca2+. The coverslips were then transferred to the perfusion chamber and perfused with mKRB. Histamine (100 µM) was added in Ca2+-free, EGTA-containing mKRB. The experiments ended by permeabilizing cells with digitonin (100 µM) in a hypotonic Ca2+-rich solution (10 mM CaCl2 in H2O) to discharge the remaining aequorin pool and calibrate the signal. Luminescence was collected by a photon counter (Package Photon Counting 9125, Sens-Tech) and analysis performed by the associated software (Sens-Tech). The light signal was analysed and converted into Ca2+ concentrations as described65.
ER Ca2+ content was measured by ratiometric GEM-CEPIA1er34. HeLa cells, expressing GEM-CEPIA1er and treated as indicated, were mounted into an open-topped chamber in mKRB and imaged by a Thunder Imager 3D Cell Culture (Leica), equipped with a LED8 illumination system, a 40×/1.30 oil immersion (HC PL Fluotar 340) objective and a cool camera (Hamamatsu Flash 4.0 V3). Excitation was performed by the 390 nm LED8 line, and emissions at either 530 or 480 nm were simultaneously collected by a beam splitter (502 nm dichroic mirror, 535/30 nm and 480/40 nm emission filters, Chroma Technologies). Exposure time was 60 ms; images were acquired every 2.5 s. After background subtraction, the ratio between the blue and the green emission (proportional to ER Ca2+ concentration) was calculated by the LASX software (Leica).
For ratiometric OMM-Cepia3 and OMM-GCaMP6f measurements, HeLa cells co-expressing OMM-Cepia3 and ER-mit RspA-splitFAST (either short or long) or OMM-GCaMP6f and short ER-mit RspA-splitFAST were mounted into an open-topped chamber in mKRB supplemented with TFCoral (5 µM) and imaged by a Thunder Imager 3D Cell Culture (Leica), equipped with a LED8 illumination system, a 63×/1.40 oil immersion (HC PL Fluotar 340) objective and a cool camera (Hamamatsu Flash 4.0 V3). ER-mit RspA-splitFAST was excited with the 555 nm led line and emission collected by a TRITC filter (Chroma Technologies), whereas OMM-Cepia3 or OMM-GCaMP6f were sequentially excited with the 475 nm (10 ms exposure) and 390 nm (50 ms exposure) led lines (in both cases, we used a 495 nm dichroic mirror and a 525/50 nm emission filter). Indeed, we exploited the isosbestic point of Cepia3 and GCaMP6f (~395 nm) to perform ratiometric Ca2+ measurements, sequentially exciting at 475 nm (Ca2+-sensitive wavelenght) and at 390 nm (Ca2+-insensitive wavelenght). The overall frame rate was 100 ms/frame. A 2 × 2 binning was set. Images were background substracted and the ratio between the two wavelenghts performed by the LASX software. To specifically evaluate the OMM-Cepia3 (or OMM-GCaMP6f) ratio at the level of ER-mit MCSs, we used the “AND” function of ImageJ between the OMM-Cepia3 (or OMM-GCaMP6f) ratio image and the corresponding ER-mit RspA-splitFAST image. To calculate the % of ER-mit MCSs experiencing high Ca2+ concentrations upon Histamine stimulation, images of the regions of OMM-Cepia3 ratio co-localized with ER-mit RspA-splitFAST (obtained as described above) were processed using a custom-written Python script56. To track the signal intensity over a time series, we initially identified contours of each ER-mit MCS region in the first frame of the image where pixel values were greater than 0.1 of the maximal value. Then, for each following frame, the mean intensity of the pixels within the contour of each single ER-mit MCS was calculated. Those MCSs displaying a peak of Cepia3 ratio higher than that of the mean + 1 SD of the pixels correponding to the bulk OMM-Cepia3 were calculated as “highly” responding.
For photoactivation of caged IP3, just before the experiments cells were loaded for 40 min at 37 °C with ci-IP3/PM (Tocris, 6210) at the final concentration of 1 µM in mKRB, in accordance with manufacturer instructions. To photoactivate ci-IP3 and generate active IP3 during the imaging experiments, where indicated cells were exposed for 300 ms at UV light, generated by the simultaneous activation (100% power intensity) of the 340 nm and 380 nm lines of a pE-340fura led (coolLED), installed on a Thunder Imager 3D Cell Culture microscope (Leica). A 40×/1.30 oil immersion objective (HC PL Fluotar 340) was used.
Molecular modelling
First, the disorder propensity of ER-mit splitFAST and ER-mit PRINCESS sequences was predicted by flDPnn tool66 based on machine learning (http://biomine.cs.vcu.edu/servers/flDPnn) to assess the putative flexibility of the linkers. Then we proceeded with the generation of the 3D-models by generating initial starting coordinates using AlphaFold; the linker portions, predicted to have not a secondary structure and be disordered by flDPnn, were subjected to a cycle of manual stretching and energy minimization steps to evaluate the lenght of the complexes. In detail, the initial 3D-models of short and long ER-mit splitFAST were generated by AlphaFold-multimer67 through ColabFold68. ER-mit PRINCESS model was generated with AlphaFold369 using AlphaFold Server. The structures were stretched and minimized (AMBER force field) with the molecular modelling suite MOE by the Chemical Computing Group (CCG).
Statistics and reproducibility
All data are representative of at least 3 independent experiments (biological replicates). The sample size (n) for each experimental condition is indicated in the specific figure panels (and the associated Source Data files) and represents the number of cells, unless differently specified. When a pairwise comparison with the control group was performed, significance was calculated by unpaired Student’s t-test for normally distributed and Wilcoxon Mann–Whitney U test for not normally distributed data. For paired data, significance was calculated by paired Student’s t-test for normally distributed and Wilcoxon signed-rank test for not normally distributed data. When more than two groups were compared, statistical significance was calculated using one-way analysis of variance (ANOVA) with post-hoc comparisons (Tukey) for normally distributed data or Kruskall-Wallis test for not normally distributed data, setting p < 0.05 as a level of significance. Statistical analysis was performed by GraphPad Prism software v9.2. *p < 0.05, **p < 0.01, ***p < 0.001. For each experimental condition, the statistical test used and the exact p values are reported in the Source Data files. In all the displayed box plots, the bounds of the box represent the 25th percentile (first quartile) and the 75th percentile (third quartile), the centre represents the median, the two whiskers extend from the first quartile to the smallest value and from the third quartile to the largest value; where present, the cross represents mean. No statistical method was used to predetermine sample size; no data were excluded from the analyses. Randomization was not applied because all samples were handled in the same way and experiments were always carried out in parallel for control and treated groups. Data collection and data analysis were not blinded, because they are strictly quantitative.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All the plasmids generated in this study and the detailed sequences will be available on request from the corresponding author R.F. Requests will be processed in 1–2 weeks. Source data are provided with this paper.
References
Prinz, W. A., Toulmay, A. & Balla, T. The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 21, 7–24 (2020).
Scorrano, L. et al. Coming together to define membrane contact sites.Nat. Commun. 10, 1287 (2019).
Filadi, R., Theurey, P. & Pizzo, P. The endoplasmic reticulum-mitochondria coupling in health and disease: molecules, functions and significance. Cell Calcium 62, 1–15 (2017).
Markovinovic, A., Greig, J., Martín-Guerrero, S. M., Salam, S. & Paillusson, S. Endoplasmic reticulum-mitochondria signaling in neurons and neurodegenerative diseases.J. Cell Sci. 135, jcs248534 (2022).
Rossini, M., Pizzo, P. & Filadi, R. Better to keep in touch: investigating inter-organelle cross-talk. FEBS J. 288, 740–755 (2021).
Cieri, D. et al. SPLICS: a split green fluorescent protein-based contact site sensor for narrow and wide heterotypic organelle juxtaposition. Cell Death Differ 25, 1131–1145 (2018).
Vallese, F. et al. An expanded palette of improved SPLICS reporters detects multiple organelle contacts in vitro and in vivo. Nat. Commun. 11, 6069 (2020).
Romei, M. G. & Boxer, S. G. Split Green Fluorescent Proteins: Scope, Limitations, and Outlook. Ann. Rev. Biophys. 48, 19–44 (2019).
Csordas, G. et al. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell 39, 121–132 (2010).
Tebo, A. G. & Gautier, A. A split fluorescent reporter with rapid and reversible complementation. Nat. Commun. 10, 2822 (2019).
Plamont, M. A. et al. Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo. Proc. Natl. Acad. Sci. USA 113, 497–502 (2016).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Bravo, R. et al. Increased ER-mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. J. Cell Sci. 124, 2143–2152 (2011).
Li, C. et al. A far-red emitting fluorescent chemogenetic reporter for In Vivo molecular imaging. Angew. Chemie - Int. Ed. 59, 17917–17923 (2020).
Rakotoarison, L. M. et al. Improving split reporters of protein-protein interactions through orthology-based protein engineering. ACS Chem. Biol. 19, 428–441 (2024).
Rowland, A. A., Chitwood, P. J., Phillips, M. J. & Voeltz, G. K. ER contact sites define the position and timing of endosome fission. Cell 159, 1027–1041 (2014).
Raiborg, C., Wenzel, E. M. & Stenmark, H. E. R. endosome contact sites: molecular compositions and functions. EMBO J. 34, 1848–1858 (2015).
Friedman, J. R., Webster, B. M., Mastronarde, D. N., Verhey, K. J. & Voeltz, G. K. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 190, 363–375 (2010).
Korobova, F., Ramabhadran, V. & Higgs, H. N. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467 (2013).
Friedman, J. R. et al. ER tubules mark sites of mitochondrial division. Science (80-.) 334, 358–362 (2011).
Liu, X., Weaver, D., Shirihai, O. & Hajnóczky, G. Mitochondrial ‘kiss-and-run”: interplay between mitochondrial motility and fusion–fission dynamics’. EMBO J. 28, 3074 (2009).
Hedskog, L. et al. Modulation of the endoplasmic reticulum-mitochondria interface in Alzheimer’s disease and related models. Proc. Natl. Acad. Sci. USA 110, 7916–7921 (2013).
Zampese, E. et al. Presenilin 2 modulates endoplasmic reticulum (ER)-mitochondria interactions and Ca2+ cross-talk. Proc. Natl. Acad. Sci. USA 108, 2777–2782 (2011).
Area-Gomez, E. et al. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 31, 4106–4123 (2012).
Ozmen, L., Albientz, A., Czech, C. & Jacobsen, H. Expression of transgenic APP mRNA is the key determinant for beta-amyloid deposition in PS2APP transgenic mice. Neurodegener. Dis. 6, 29–36 (2009).
Saito, T. et al. Single App knock-in mouse models of Alzheimer’s disease. Nat. Neurosci. 17, 661–663 (2014).
Naia, L. et al. Mitochondrial hypermetabolism precedes impaired autophagy and synaptic disorganization in App knock-in Alzheimer mouse models. Mol. Psychiatry 28, 3966–3981 (2023).
Filadi, R. et al. Presenilin 2 modulates endoplasmic reticulum-mitochondria coupling by tuning the antagonistic effect of mitofusin 2. Cell Rep. 15, 2226–2238 (2016).
Rossini, M., García-Casas, P., Filadi, R. & Pizzo, P. Loosening er—mitochondria coupling by the expression of the presenilin 2 loop domain. Cells 10, 1968 (2021).
Rizzuto, R. et al. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science (80-.) 280, 1763–1766 (1998).
Csordás, G., Thomas, A. P. & Hajnoczky, G. Quasi-synaptic calcium signal transmission between endoplasmic reticulum and mitochondria. EMBO J. 18, 96–108 (1999).
Luik, R. M., Wang, B., Prakriya, M., Wu, M. M. & Lewis, R. S. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature 454, 538–542 (2008).
Giacomello, M. et al. Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels. Mol. Cell 38, 280–290 (2010).
Suzuki, J. et al. Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nat. Commun. 5, 4153 (2014).
Cardenas, C. et al. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 142, 270–283 (2010).
Thillaiappan, N. B., Chavda, A. P., Tovey, S. C., Prole, D. L. & Taylor, C. W. Ca2+ signals initiate at immobile IP3 receptors adjacent to ER-plasma membrane junctions. Nat. Commun. 8, 1–16 (2017). 2017 81.
Giordano, F. et al. XPI(4,5)P2-Dependent and Ca2+-Regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153, 1494 (2013).
Soboloff, J., Rothberg, B. S., Madesh, M. & Gill, D. L. STIM proteins: dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 13, 549–565 (2012).
Fedeli, C., Filadi, R., Rossi, A., Mammucari, C. & Pizzo, P. PSEN2 (presenilin 2) mutants linked to familial Alzheimer disease impair autophagy by altering Ca(2+) homeostasis. Autophagy. 15, 2044–2062 (2019).
Hofer, A. M., Fasolato, C. & Pozzan, T. Capacitative Ca2+ entry is closely linked to the filling state of internal Ca2+ stores: a study using simultaneous measurements of ICRAC and intraluminal [Ca2+]. J. Cell Biol. 140, 325–334 (1998).
Sassano, M. L. et al. PERK recruits E-Syt1 at ER–mitochondria contacts for mitochondrial lipid transport and respiration. J. Cell Biol. 222, e202206008 (2023).
van Vliet, A. R. et al. The ER stress sensor PERK coordinates ER-plasma membrane contact site formation through interaction with Filamin-A and F-Actin remodeling. Mol. Cell 65, 885–899.e6 (2017).
Jing, J. et al. Proteomic mapping of ER-PM junctions identifies STIMATE as regulator of Ca2+ influx. Nat. Cell Biol. 17, 1339 (2015).
Quintana, A. et al. TMEM110 regulates the maintenance and remodeling of mammalian ER-plasma membrane junctions competent for STIM-Orai signaling. Proc. Natl. Acad. Sci. USA 112, E7083–E7092 (2015).
Ramesh, G. et al. A short isoform of STIM1 confers frequency-dependent synaptic enhancement. Cell Rep. 34, 108844 (2021).
Schober, R. et al. Sequential activation of STIM1 links Ca2+ with luminal domain unfolding. Sci. Signal. 12, eaax3194 (2019).
Denton, R. M. Regulation of mitochondrial dehydrogenases by calcium ions. Biochim. Biophys. Acta 1787, 1309–1316 (2009).
Li, X. et al. A fluorogenic complementation tool kit for interrogating lipid droplet-organelle interaction. bioRxiv Prepr. Serv. Biol. https://doi.org/10.1101/2023.11.29.569289 (2023).
Cho, E. et al. Ratiometric measurement of MAM Ca2+ dynamics using a modified CalfluxVTN. Nat. Commun. 14, 3586 (2023).
Tighe, A., Staples, O. & Taylor, S. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J. Cell Biol. 181, 893–901 (2008).
Alzayady, K. J. et al. Defining the stoichiometry of inositol 1,4,5-trisphosphate binding required to initiate Ca2+ release. Sci. Signal. 9, ra35 (2016).
Zelenka, L. et al. Novel protocol for the isolation of highly purified neonatal murine microglia and astrocytes. J. Neurosci. Methods 366, 109420 (2022).
Kierdorf, K. & Prinz, M. Microglia: Same same, but different. J. Exp. Med. 216, 2223–2225 (2019).
Filadi, R. et al. TOM70 sustains cell bioenergetics by promoting IP3R3-Mediated ER to Mitochondria Ca(2+) Transfer. Curr. Biol. 28, 369–382 e6 (2018).
Csordas, G. et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174, 915–921 (2006).
García Casas, P. et al. Simultaneous detection of membrane contact dynamics and associated Ca2+ signals by reversible chemogenetic reporters. Github https://doi.org/10.5281/ZENODO.13381018. (2024).
Digman, M. A., Caiolfa, V. R., Zamai, M. & Gratton, E. The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 94, L14–L16 (2008).
García Casas, P. et al. Simultaneous detection of membrane contact dynamics and associated Ca2+ signals by reversible chemogenetic reporters. Github https://doi.org/10.5281/ZENODO.13380661. (2024).
Belevich, I., Joensuu, M., Kumar, D., Vihinen, H. & Jokitalo, E. Microscopy image browser: a platform for segmentation and analysis of multidimensional datasets. PLoS Biol. 14, e1002340 (2016).
Mesa, D. et al. A tripartite organelle platform links growth factor receptor signaling to mitochondrial metabolism. Nat. Commun. 15, 5119 (2024).
Schikorski, T., Young, S. M. & Hu, Y. Horseradish peroxidase cDNA as a marker for electron microscopy in neurons. J. Neurosci. Methods 165, 210–215 (2007).
Mastronarde, D. N. Dual-axis tomography: an approach with alignment methods that preserve resolution. in Journal of Structural Biology 120 343–352 (Academic Press Inc., 1997).
Conrad, R. & Narayan, K. Instance segmentation of mitochondria in electron microscopy images with a generalist deep learning model trained on a diverse dataset. Cell Syst. 14, 58–71.e5 (2023).
Stiernagle, T. Maintenance of C. elegans. WormBook 1–11 https://doi.org/10.1895/WORMBOOK.1.101.1 (2006).
Brini, M. et al. Transfected aequorin in the measurement of cytosolic Ca2+ concentration ([Ca2+]c): a critical evaluation. J. Biol. Chem. 270, 9896–9903 (1995).
Hu, G. et al. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 12, (2021).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021.10.04.463034 https://doi.org/10.1101/2021.10.04.463034 (2021).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Acknowledgements
We owe Tullio Pozzan, who provided us with precious advices and was a mentor and a friend for many of us. The authors gratefully acknowledge L. Ozmen and F. Hoffmann-La Roche Ltd (Basel, Switzerland) for kindly donating the AD (B6.152H) mouse model used in this study (MTA 02-01-2022-UniPD), E. Trevisson, V. Morbidoni (University of Padua) and A. Miranda Vizuete for helping with the setting of the C. elegans strains, P. Romani for helping with the setting of confocal microscopy, D. Sandonà for suggestions on cloning strategies, I. Costa and M. Gintoli for training and suggestions on FLIM acquisitions and P. Magalhães for proofreading of the manuscript. Part of the schemes presented in this work were created with BioRender.com. P.G.C fellowship was supported by Ayudas de recualificación del Sistema Universitario Margarita Salas (2021–2023), University of Valladolid-Spanish Ministry of Education, financed by the European Union through EU Next Generation. T.F. fellowship was supported by PRIN 2022943TH9. S.S. was supported by grants from the European Research Council (ERC-CoG2020 101002280) and Associazione Italiana per la Ricerca sul Cancro (AIRC IG 24415). This work was supported by grants from the Italian Ministry of University and Scientific Research (PRIN2017XA5J5N and PRIN2022943TH9, financed by the EU, NextGenerationEU); the University of Padova (SID2019); Cure Alzheimer’s Fund (USA) to P.P.; the Dynamic Imaging program of the Chan-Zuckerberg Initiative DAF (grant number 2023-321185), an advised fund of Silicon Valley Community Foundation, to A.G. and R.F; the Italian Ministry of University and Scientific Research grant (PRIN P20225R4Y5, financed by the European Union, NextGenerationEU) to P.P. and R.F. The authors acknowledge Euro-BioImaging [https://www.eurobioimaging.eu/] for providing access to imaging technologies and services via the Advanced Light Microscopy Italian Node (Laboratory of Ca2+ and cAMP signaling in physiology and pathology, Padua, Italy; and ALEMBIC, Milan, Italy); the Swedish National Microscopy Infrastructure, NMI (VR-RFI 2019-00217); EuroBioImaging-ERIC and the PNRR infrastructure, SEELIFE n. IR00023 (financed by the European Union, NextGenerationEU, Missione 4, Componente 2, CUP B53C22001810006).
Author information
Authors and Affiliations
Contributions
P.G.C., M.R., P.P., and R.F. were responsible for experimental design and data interpretation. P.G.C., M.R., L.P., M.SA., T.F., I.D., L.N., H.B. and R.F. performed and analyzed experiments. N.A. and S.SO. prepared primary glial and neuronal cell cultures. M.B. conceived the scripts for analyzing images. V.B. and A.R. performed CLEM experiments and image analysis. E.B., S.SI. and A.R. designed and supervised electron tomography experiments. M.S.T. performed AlphaFold analysis. M.L.S., P.A., B.A.N. provided reagents, suggestions and discussed experiments. L.N. and M.A. provided suggestions and discussed experiments. A.G. provided all the original splitFAST-based tools, reagents, expertise on splitFAST and suggestions. P.P. and R.F. conceived the research and secured funds. R.F. wrote the manuscript with the help of the other authors. All the authors have agreed to publish the current version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.G. is co-founder and holds equity in Twinkle Bioscience/The Twinkle Factory, a company commercializing the FAST and split-FAST technologies. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Antonella De Matteis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
García Casas, P., Rossini, M., Påvénius, L. et al. Simultaneous detection of membrane contact dynamics and associated Ca2+ signals by reversible chemogenetic reporters. Nat Commun 15, 9775 (2024). https://doi.org/10.1038/s41467-024-52985-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52985-0
This article is cited by
-
Design and evaluation of a tripartite chemogenetic fluorescent reporter for visualizing ternary protein complexes
Nature Communications (2025)
-
A tunable and versatile chemogenetic near-infrared fluorescent reporter
Nature Communications (2025)
-
The complex web of membrane contact sites in brain aging and neurodegeneration
Cellular and Molecular Life Sciences (2025)