Abstract
Multi-modal image analysis using deep learning (DL) lays the foundation for neoadjuvant treatment (NAT) response monitoring. However, existing methods prioritize extracting multi-modal features to enhance predictive performance, with limited consideration on real-world clinical applicability, particularly in longitudinal NAT scenarios with multi-modal data. Here, we propose the Multi-modal Response Prediction (MRP) system, designed to mimic real-world physician assessments of NAT responses in breast cancer. To enhance feasibility, MRP integrates cross-modal knowledge mining and temporal information embedding strategy to handle missing modalities and remain less affected by different NAT settings. We validated MRP through multi-center studies and multinational reader studies. MRP exhibited comparable robustness to breast radiologists, outperforming humans in predicting pathological complete response in the Pre-NAT phase (ΔAUROC 14% and 10% on in-house and external datasets, respectively). Furthermore, we assessed MRP’s clinical utility impact on treatment decision-making. MRP may have profound implications for enrolment into NAT trials and determining surgery extensiveness.
Similar content being viewed by others
Introduction
Breast cancer (BC) is the most common cancer worldwide, being responsible for of 31% female cancers1. Neoadjuvant therapy (NAT) followed by surgery is the standard therapeutic regimen for patients with inoperable or extensive but resectable BC2. Pathological complete response (pCR), defined as the absence of residual invasive cancer cells in histopathological specimens of breast and axillary lymph nodes after NAT (ypT0/is, ypN0), is associated with favorable survival outcomes3,4,5. However, it is challenging to accurately assess treatment response at different treatment timepoints, which is a multidisciplinary task that requires integrating various information streams from cancer staging of Pre-NAT to Post-NAT, as shown in Fig. 16,7. According to the National Comprehensive Cancer Network guidelines8, there is currently very little variation in the choice of NAT for breast cancer patients. However, some patients may not respond well to standard therapy and experience toxicity without clear benefits, which could be avoided if response could accurately be predicted. In the Post-NAT phase, all patients are required surgery, even if they have already achieved a pCR. Therefore, there is a clinical need to develop a system that could cope with both personalized patient and clinician preferences while handling longitudinal multi-modal information. For example, it could help to decide whether to consider neoadjuvant therapy trials in the phase of Pre-/Mid-NAT phase9 or whether to consider surgery-reducing trials potential in the phase of Post-NAT10,11.
a Pre-NAT phase. Breast cancer (BC) is diagnosed following a tumor screening/diagnosis (mammography and/or ultrasound) and biopsy, subsequent histopathology analysis, and a staging breast MRI. These measures help derive demographic, radiological, clinical and histopathological variables describing the patient’s state at diagnosis. cTNM refers to tumor node metastasis. b Mid-NAT phase. The mid-NAT MRI is performed to assess the response and see if therapy adjustments for unresponsive patients. c Post-NAT phase. Breast MRI is used to assess if patients have achieved pathological complete response (pCR). Subsequently, patients undergo surgery, and a histological examination is performed, assessing the post-NAT pathological tumor(ypT) and lymph node staging (ypN) scores (together ypTN)68, which is the international standard for post-neoadjuvant therapy pathology reporting.
Previous efforts12,13,14 to predict pCR have explored serological or genetic biomarkers, but these have not been widely applied in clinical practice due to their cost and limited validation on large-scale populations. Recent advances15,16,17,18,19 in machine learning (ML) have shown promise in predicting response to NAT. Some studies20,21,22,23 have utilized radiomics (i.e., extraction of differentiating quantitative features from medical images) to successfully predict pCR in patients with breast cancer. However, the clinical applicability of these radiomics studies are limited by potential risks of overfitting due to small sample sizes20. On the other hand, several studies15,17,24,25,26 have shown that breast radiological and clinical fusion features based on deep learning (DL) could predict pCR in BC patients. Additionally, integrating histopathological information has also been shown to complement tumor heterogeneity assessment and the predictive capacity of DL models for patient prognosis16,19,27,28. However, current artificial intelligence (AI) studies often focus only on exploring multi-modal characteristics extraction, ignoring whether their model design and rigid input structures fit applicably with the clinical pathway context. Their practical feasibility in different (multi-center) hospital settings and overall generalizability are not often explored. Moreover, the clinical utility and potential impact of existing AI systems for predicting responses in different NAT scenarios are often left unexplored. For example, addressing clinical concerns at different phases of treatment trajectories demands different attention and emphasis.
In this study, we propose a multi-modal response prediction (MRP) system tailored to response prediction of neoadjuvant therapy in breast cancer patients. The MRP system capitalizes on the utilization of longitudinal data throughout the NAT process within real clinical contexts, as illustrated in Fig. 2. The system integrates multi-modal data sources from 3,352 eligible breast cancer participants, encompassing 4,802 Pre-NAT mammogram exams, 3,719 longitudinal MRI exams (specifically, subtracted contrast-enhanced T1-weighted imaging), along with associated radiological findings. Additionally, MRP incorporates essential histopathological information such as molecular subtype, tumor histology, tumor type, and tumor differentiation. Personal factors including weight, gender, age, the age of menarche, menopausal status, genetic mutations, as well as clinical data such as clinical tumor node metastasis (cTNM) Pre-NAT and therapy details, are also considered within the MRP system. It can, therefore, validate the potential benefits of integrating multi-modal (including radiological, histopathological, clinical, and personal) information throughout the entire neoadjuvant therapy care to enhance the performance of AI models for predicting pathological complete response in breast cancer. We have made the code of our model public at https://github.com/yawwG/MRP/. On the other hand, existing methodologies17,29,30, which are primarily developed for response prediction based either solely on a single time point imaging (Pre-NAT) or completely on longitudinal data (Pre- and Post-NAT). These approaches are, therefore, constrained for clinical use due to their rigid input requirements. In contrast, the MRP system is purposefully designed to accommodate missing data inputs and embeds temporal information into longitudinal inputs. Additionally, to ensure adaptability to diverse therapeutic protocols used in different hospital centers, and given the frequent availability of imaging data, we prioritize improving the model’s ability to extract image features and enhance the contribution of imaging modalities. To achieve this, we implement a cross-modal knowledge mining strategy on top of the extracted imaging features, designed to enhance visual representation learning.
a Model development. We developed and evaluated a deep learning system to predict the treatment response of breast cancer patients across neoadjuvant therapy (NAT). The system incorporates deep neural networks trained on Pre-NAT mammogram images and longitudinal MRI scans, along with rhpc information (radiological assessments, histopathological assessments, personal patient records, and clinical data). After data retrieval, iMGrhpc and iMRrhpc were modeled independently, where iMGrhpc is based on Pre-NAT mammogram and rhpc data, while iMRrhpc is based on longitudinal MRIs embedding temporal information and rhpc data. Both models include two modules: one module is for cross-modal knowledge learning that predicts rhpc information using only imaging features, and another module is for response prediction using integrated features of rhpc-based and imaging features. These models were further combined into the Multi-modal Response Prediction (MRP) system. MLP refers to a two-layer multi-layer perceptron with an output dimension of 256. b Datasets. The internal dataset was collected from the Netherlands Cancer Institute and was randomly partitioned into training, validation, and test subsets. For evaluating our system on unseen data, we collected three external datasets from different centers: Duke University (United States; n = 288), Fuzhou Province Hospital (China; n = 85), and I-SPY2 (United States; n = 508). c NAT response assessment of AI model and reader study. We assessed MRP's ability to predict pathological response (pCR vs. non-pCR) at different stages-Pre-NAT (before administration of NAT), Mid-NAT (during therapy), and Post-NAT (prior to surgery)-using standard metrics: AUROC (Area Under Receiver Operating Characteristic Curve) and AUPRC (Area Under Precision-Recall Curve). To compare the performance of MRP with human experts, we conducted a reader study involving six international breast radiologists. The average performance of the readers is indicated with a red “+R" in the plot. d Personalizing management in clinical practice. We simulated two scenarios to assess the system’s ability to personalize treatment: identifying non-pCR patients before NAT in whom toxic treatments may be timely adapted, and identifying pCR patients before surgery for the potential reduction of surgical procedures. Circled C indicates current clinical practice; Circled AI indicates our MRP system suggested strategy.
We evaluate the system’s performance on internal and external international datasets using traditional decision accuracy metrics (area under the receiver operating characteristic (AUROC), area under the precision-recall curve (AUPRC), sensitivity, specificity, positive predictive value (PPV), and negative predictive value (FPV)). We compare the predictive performance of MRP to that of radiologists in a retrospective reader study under different clinical scenarios. Additionally, we analyze the system’s performance in various patient subgroups and investigate cases where MRP fails to produce correct diagnoses. We also conduct feature importance analysis to understand how structured and image-based features/modalities contribute to pCR prediction. To gain insights into the contributions of implemented modules, we perform ablation experiments involving the cross-modal knowledge predictor and the temporal information embedding component. Finally, we assess the clinical impact of the MRP model on treatment decision-making (decision curve analysis31), which is explored in two specific clinical scenarios: personalizing Pre-/Mid-NAT management of non-pCR patients for consideration of enrollment into neoadjuvant clinical trials of alternative therapies9 and optimizing Post-NAT management of pCR patients to consider the potential of surgery reducing trials10,11.
Results
MRP overview
Following clinical practice pathway32, MRP combines the longitudinal multi-modal data to evaluate the therapy response in BC patients. As illustrated in Fig. 2. a, MRP comprises two independently trained models: iMGrhpc and iMRrhpc. The iMGrhpc model takes inputs including Pre-NAT mammogram as well as rhpc (r:radiological, h:histopathological, p:personal, c:clinical variables). On the other hand, the iMRrhpc model leverages rhpc data and visual inputs of longitudinal MRI sequences that embed specified temporal information. This aims to mimic the evaluation process employed by radiologists, utilizing longitudinal images that encompass both baseline (Pre-NAT image) and follow-up (Mid-NAT or Post-NAT image) data to evaluate treatment response. Ultimately, the MRP combines the predicted probability of pCR generated by iMGrhpc and iMRrhpc. For a more comprehensive understanding of the model, further details can be found in the “Methods” section.
Patient data
The study included a total of 3,352 patients with breast cancer who were treated with NAT in the Netherlands Cancer Institute between January 1, 2004, and December 31, 2020. Within the training cohort (80%), 1,134 patients underwent MRI examinations for staging and during NAT while pre-treatment mammograms were available for 1,556 patients (see Supplementary Fig. 1a). Additionally, 120 patients from the same institute were randomly selected to comprise test cohort A. External test cohort B (see Supplementary Fig. 1b), consisting of 288 patients who underwent Pre-NAT MRI examinations, was obtained from Duke University. For the external test cohort C (see Supplementary Fig. 1c), we collected data from 85 patients who underwent pre- and post-NAT MRIs as well as pre-NAT mammogram examinations at Fujian Provincial Hospital in China. For external test cohort D, as depicted in Supplementary Fig. 1d, we included data from 508 patients who underwent MRIs at multiple time points (pre-, mid-, and post-NAT), derived from the I-SPY2 study33,34. For detailed cohorts information, please refer to “Inclusion and ethics statement”.
Primary results
MRP demonstrates promising potential in predicting pCR across various NAT scenarios
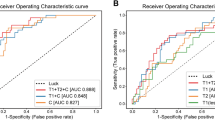
In the Pre-NAT phase (Fig. 3a), the MRP achieves an AUROC of 0.883 (95% confidence interval (CI): 0.821-0.941) and an AUPRC of 0.772 (95% CI: 0.630-0.900) for predicting breast cancer treatment response. Compared to the performance of baseline models, the MRP demonstrates a significant improvement in response prediction. When compared to the uni-modal (i.e., without radiological images integrated) rhpc model, MRP exhibited a ΔAUROC of 10.4% with a corresponding p-value of 0.003. Additionally, when compared to the iMGrhpc model, the MRP also outperforms it with a ΔAUROC of 4.9% and a p-value of 0.048. In the Mid-NAT phase (Fig. 3b), the MRP’s performance moderately enhances that of the Pre-NAT period, although the improvement is not statistically significant (p = 0.96). It achieves an AUROC of 0.889 (95% CI: 0.827-0.948) and an AUPRC of 0.784 (95% CI: 0.642-0.906). Compared to baseline models, the MRP remains superior. It significantly outperforms the uni-modal rhpc model with a ΔAUROC of 11% and a p-value of 0.009, the iMGrhpc model with a ΔAUROC of 5.5% and a p-value of 0.045, and the iMRrhpc model with a ΔAUROC of 4.1% and a p-value of 0.049. Interestingly, when comparing the performance of Mid-NAT with the Post-NAT phase (Fig. 3c), the MRP maintains a similarly competitive level of performance (p = 0.71). In the Post-NAT phase, it achieves an AUROC of 0.889 (95% CI: 0.826-0.948) and an AUPRC of 0.783 (95% CI: 0.644-0.903). Besides, MRP remains the top-performing model, surpassing the uni-modal rhpc model with a ΔAUROC of 11% and a p-value of 0.008, the iMGrhpc model with a ΔAUROC of 5.5% and a p-value of 0.044, and the iMRrhpc model with a ΔAUROC of 4.3% and a p-value of 0.041. For more detailed results regarding breast cancer molecular subtypes, please refer to Supplementary Fig. 2. For comparisons of models in predicting responses across all NAT scenarios, see Supplementary Fig. 3 and Tab. 1. Please be aware that the performance metrics on the in-house cohort were derived from the independent test set.
(Top) ROC curves with 95% CIs in bracket calculated with boot-strapping. (Bottom) PRCs with 95% CIs. From left to right: Pre-NAT(Staging), Mid-NAT, Post-NAT(Pre-surgical). rhpc refers to the model trained by radiological assessments (r), histopathological assessments (h), personal patient records (p), and clinical data (c), detailed definitions can be found in Methods and Fig. 1. iMGrhpc is based on Pre-NAT mammogram and rhpc data, while iMRrhpc is based on single/longitudinal MRI(s) embedding with temporal information and rhpc data. MRP aggregates and optimizes the outputs of iMGrhpc model and iMRrhpc model.
In cohort B, where only Pre-NAT MRI data is available, the MRP model cannot be applied due to the absence of MG information. Nevertheless, as shown in Supplementary Fig. 4, the iMRrhpc achieves an AUROC of 0.797 (95% CI: 0.736-0.850) and an AUPRC of 0.587 (95% CI: 0.468-0.717), showing an improvement of 12% in AUROC and 10.8% in AUPRC compared to the uni-modal rhpc model. Comprehensive statistical analysis confirms the significant superiority of iMRrhpc over rhpc, with a notable difference in AUC (p <0.001). For results on breast cancer molecular subtypes, please see Supplementary Fig. 5.
In cohort C, as illustrated in Supplementary Fig. 6, the model’s performance consistently improves with the addition of successive levels of multi-modal imaging and longitudinal MRI. At the pre-NAT stage, compared to the uni-modal rhpc model, MRP increases the AUROC to 0.821, representing an 8.4% improvement over the rhpc model, and AUPRC to 0.726, reflecting a 17.2% increase. At the post-NAT stage for the longitudinal scenarios setting, MRP achieves an AUROC of 0.842 and AUPRC of 0.758. The differences in AUC between the rhpc and MRP models are statistically significant at each NAT stage (pre-NAT: p = 0.009, post-NAT: p = 0.006). For results on breast cancer molecular subtypes, please refer to Supplementary Fig. 7.
In cohort D, as illustrated in Supplementary Fig. 8, the model achieves superior performance with the integration of multi-modal data and longitudinal MRIs. The iMRrhpc model, using pre-NAT data, shows a 12.3% increase in AUROC (0.799) and a 16.3% increase in AUPRC (0.701) compared to the uni-modal rhpc model. Further integration of pre- and mid-NAT data results in AUROC improvements of up to 15% (0.826) and AUPRC improvements of up to 20.3% (0.741). Additionally, pre- and post-NAT data integration leads to AUPRC improvements of up to 20.8% (0.746). Compared to the rhpc model, the multi-modal model that integrated imaging demonstrates a significant performance improvement at each NAT scenario (p < 0.001). For details on the results of breast cancer molecular subtypes, please see Supplementary Fig. 9. These results (i.e., Supplementary Figs. 4–9 and detailed metrics in Table 1) also highlight the need for increased access to comprehensive clinical information and imaging modalities in public datasets to foster multi-modal AI research.
MRP demonstrates favorable performance to breast radiologists in pCR prediction on both the in-house and external datasets
Six international radiologists interpreted all cases from the cohort A set, and two radiologists furthermore evaluated another 100 cases sampled from the cohort B set. All radiologists were board-certified and had between 2 and 12 years of experience interpreting breast MRI exams. In cohort A, at the Pre-NAT phase shown in Fig. 3a, readers achieve an average performance of 0.740 of AUROC (95% CI: 0.643-0.828) and 0.506 of AUPRC (95% CI: 0.368-0.705). During the Mid-NAT phase shown in Fig. 3b, readers achieve an average performance of 0.852 of AUROC (95% CI: 0.779-0.914) and 0.750 of AUPRC (95% CI: 0.606-0.863); at the Post-NAT period, readers achieve an average performance of 0.840 AUROC of (95% CI: 0.759–0.907) and 0.710 of AUPRC (95% CI: 0.547-0.841). The difference between MRP and radiologists is statistically significant in the Pre-NAT phase shown in Fig. 3c. In a comparison between the MRP and six international radiologists’ AUROC, significant differences were found in the Pre-NAT phase where the MRP outperformed all six readers (ΔAUROC ranging from 0.159 to 0.373, DeLong’s test: p-values < 0.001). In the Mid-NAT and Post-NAT phases, the MRP continued to significantly outperform four readers (Mid-NAT: ΔAUROC ranging from 0.106 to 0.166, with p-values between 1.798e-02 and 3.0823e-03; Post-NAT: ΔAUROC ranging from 0.093 to 0.140, with p-values between 2.437e-02 and 4.175e-03). Further detailed results for each reader’s evaluation can be found in Supplementary Fig. 3 and Supplementary Tab. 1.
For cohort B shown in Supplementary Fig. 4b, iMRrhpc performance on the reader study subset is 0.828 (0.733–0.917) for AUROC and 0.520 (0.298–0.755) for AUPRC while the best performance from two readers is 0.775 (0.663–0.872) for AUROC and 0.480 (0.267–0.682) for AUPRC. The difference between iMRrhpc and one of the radiologists demonstrated statistical significance (p = 0.041).
Modality contribution analysis
To conduct a systematic evaluation of the influential features driving the breast cancer therapy response predictions, we analyzed the feature coefficients presented in Fig. 4 using the Integrated Gradients method35. Consistently across different multi-modal based models (rhpc, iMGrhpc, iMRrhpc, MRP), the top five significant features positively associated with the prediction of pCR were related to histopathological and radiological factors, and image-wise inputs. Among the histopathological features, molecular subtype, in situ status (presence of an in situ component next to the invasive component), and tumor histology exhibited strong positive associations. Not unexpectedly, our findings indicate that the type of therapy (a clinical marker including neoadjuvant chemotherapy, neoadjuvant endocrine therapy, and neoadjuvant immunotherapy), which is closely related to molecular subtype, also demonstrated significant positive correlations. Regarding general radiological contributions, mammography, MRI, and factors such as multifocality and mammographic density were found to be useful for accurate pCR prediction. In contrast, the consistently identified unrelated markers (that are not significantly associated with pCR prediction) were primarily related to tumor location and laterality which is understandable from a clinical perspective. Interestingly, also the presence of mutations does not seem to influence pCR prediction. Our analysis thus demonstrates the vital features influencing the prediction of pCR in a systematic manner.
Comparison of coefficient importance among four deep learning models : rhpc (n = 120), iMGrhpc (n = 120), iMRrhpc (n = 120), and MRP (n = 120), organized by coefficients in descending order. The horizontal bar plot displays normalized coefficients, derived from averaged contributing values among cases and normalized across the included attributes, highlighting comparable trends among these attributes. Positive values stand for favorable attributes for response prediction, and vise versa for negative values. The horizontal line (error bar) represents the standard deviation centered on the corresponding coefficients. Each attribute associated different group (radiological(r), histopathological(h), personal(p), clinical(c)) is viewed in the colors legend in the upper right. iMGrhpc is based on Pre-NAT mammogram and rhpc data, while iMRrhpc is based on pre-NAT MRI and rhpc data. MRP aggregates and optimizes the outputs of iMGrhpc model and iMRrhpc model.
In addition, we evaluated the association between pCR and the top five contributing factors identified in the MRP model. As indicated in Table 2 and Supplementary Tab. 2, the pCR varied by molecular subtype. Compared to ER+/HER2- cancers (3.8% pCR rate, odds ratio (OR): 1, as reference), the highest pCR OR was observed for ER-/HER2+ (54.9%, OR: 9.18, p = 0.021) and TNBC (50.1%, OR: 7.02, p < 0.001) subtypes. Many studies have reported that in situ components show lower proliferation rates, and their non-invasive nature contributes to chemoresistance36,37. Our study confirmed that a known in situ component is predictive of poorer response to therapy (24.8%, OR: 0.51, p < 0.001). Regarding tumor histology characteristics, patients with ductal carcinoma (36.2%, OR: 6.19, p < 0.001) have a higher likelihood of achieving pCR compared to those with lobular carcinoma (6.2%, OR: 1.00, as reference). This observation aligns with previous reports38,39 and underscores the need for histology-specific treatment approaches in breast cancer management. Consistent with previously published study40, multi-focal tumors (24.1%, OR: 0.53, p < 0.001) exhibited a lower rate of pCR compared to uni-focal tumors (41%, OR: 1.00, as reference). Prior studies41,42 suggest considering mammographic density in planning and optimizing treatment strategies for breast cancer patients. However, while our study demonstrates a slightly higher likelihood of pCR in patients with dense breasts (20.5%, OR: 1.05, p = 0.159) compared to patients with less dense breasts (18.8%, OR: 1.00, as reference), this was not statistically significant.
Subgroup analysis
We conduct detailed analyses on a wide range of patient subgroups in the internal test set. This experiment aims to determine if the model is underperforming in any assessed categories and to explore which groups may derive greater benefits from the model at different time points. We divided them with respect to the above-mentioned top positive favorable elements of response prediction, which are histopathological characteristics (in situ component, tumor differentiation, tumor histology, molecular subtype), clinical characteristics (different therapy, cTN stage); radiological characteristics (uni-/multi-focal); and personal characteristics (gene mutation, age). The evaluated metric values for each subgroup are available in supplemental tables across each NAT phase (Pre-NAT Supplementary Tab. 3, Mid-NAT Supplementary Tab. 4, and Post-NAT Supplementary Tab. 5). Despite frequent concerns in radiological DL systems that often show relatively high performance in less common groups, our analysis revealed that the MRP performed similarly across all groups.
Personalized management considering both non-pCR and pCR patients
To enhance decision-making in treatment planning, we investigate using MRP predictions in specific scenarios. We explore the specificity of predictions in the Pre-/Mid-NAT phase to enable timely change of NAT regimen for non-pCR cases to consider the enrollment into neoadjuvant clinical trials of alternative therapies9. Simultaneously, we seek to improve prediction sensitivity in the Post-NAT stage to identify pCR cases, which would allow patients to consider the potential chance of breast surgery reducing trials10,11.
We conducted the analysis in two ways. First, we compared the trade-offs between correctly avoiding unnecessary therapy and missed pCR patients at Pre-NAT, as well as surgery and missed non-pCR patients at Post-NAT. We utilized various decision thresholds to convert probabilities of pCR into binary outcomes. This comparison involved equally weighting the number of successfully opted-out and missed predictions. Our findings indicated that two operating points yielded promising results based on Fig. 3. In the Pre-NAT phase, MRP correctly determined 35.8% of non-pCR cases, enabling a potential selection of better-suited therapies trials, without any missed pCR cases. In the Post-NAT stage, MRP identified 16.7% of pCR cases to consider surgery-reducing trials, as well as 37% of non-pCR patients for considering neoadjuvant treatment trials, all without increasing false predictions. Further increasing the decision threshold allowed for the identification of more therapy response lesions, resulting in more patients opting out of breast surgery, but it also led to a higher rate of missed non-pCR cancers.
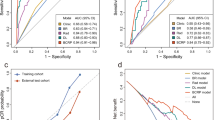
For the second approach, we employed the Decision Curve Analysis (DCA) methodology43 to incorporate patients’ and clinicians’ preferences into decision-making to explore whether using the model could be clinically beneficial or harmful. This approach evaluated the clinical usefulness of the MRP at various decision thresholds. Unlike the trade-off technique described earlier, DCA considers the missed predictions’ number value versus the unnecessary therapy or surgery’ number value. Specifically, we assessed whether the MRP’s predictions could select non-pCR patients in the Pre-NAT phase, enabling early modification of therapy, and whether it could select pCR patients in the Post-NAT stage, thereby potentially enrolling in surgery-reducing trials. Our DCA results demonstrated that using the MRP’s predictions not only allowed for the benefits of non-pCR patients at the pre/mid-NAT stage (Fig. 5a, b) but also pCR patients at the post-NAT stage (Fig. 5c). When comparing our model with human readers, although there is a benefit offered by adding radiologists’ findings to the standard clinical approach (black curve, i.e., y = 0), the MRP (red curve) still outperformed human readers at each time point. Our results indicated that incorporating the MRP into patient care led to a net benefit, even at low decision thresholds. For instance, at a decision threshold of 10% of non-pCR (Fig. 5a), the MRP resulted in a net reduction of 220 ineffective therapies per 1000 patients. In the Post-NAT scenario (Fig. 5c), with a decision threshold of 10% of pCR, our approach might yield a net benefit of 132 treatment-sensitive breast tumors for pCR findings per 1000 patients. As decision thresholds increased, the benefits became more substantial, with ~430 per 1000 pCR patients at a threshold of 21%. We did not evaluate higher decision thresholds, as they are rarely clinically relevant and may not represent most patients’ or clinicians’ preferences reliably. Decision thresholds below 6% did not provide a convincing net benefit in using our system, but these patients were not the focus of this analysis and should always undergo standard therapy or surgery.
Left and Middle: The percentage of net interventions avoided per 1000 patients with non-pCR findings in Pre-/Mid-NAT (y-axis). The black curve (y = 0) of Pre-/Mid-NAT is a standard therapy-all approach (current clinical choice). Right: The percentage of net interventions avoided per 1000 patients with pCR (ypT0) findings in the Post-NAT (Pre-surgical) phase (y-axis). The black curve (y = 0) of Post-NAT is surgery-all approach (current clinical choice).
The ablation studies for model
w/o cross-modal knowledge predictor
As illustrated in Supplementary Figs. 10–14, it is evident that the models (i.e. iMGrhpc, iMRrhpc, MRP) without the cross-modal knowledge predictor exhibit notably decreased performance in both in-house and external cohorts. Especially, for the external validation results, this technique leads to an average 7.6% increase in AUC, with cohorts B, C, and D showing improvements of 8.6%, 6.2%, 8.0%, respectively, compared to the same model without cross-modal learning. This signifies the clear advantage brought by the cross-modal knowledge predictor in enhancing the model’s generalization capabilities. Likewise, as demonstrated in Supplementary Fig. 11, there is a noticeable 15.3% and 12.1% increase in AUROC for pCR prediction in ER+/HER2- and ER+/HER2+ subtypes, respectively. This outcome is not surprising given that the cross-modal knowledge mining module incorporates the task of molecular subtype prediction from the images. We believe these specific tasks mutually benefit each other during the learning process44. When we included the predictor, the modal contribution analysis depicted in Fig. 4 compared to Supplementary Fig. 15 revealed enhanced image-wise contributions from MRI and mammogram. This finding suggests that not only do the features derived from the image itself play a crucial role in improving prediction performance, but our strategy of employing a cross-modal knowledge predictor also helps to extract valuable clinically relevant information to enhance the model performance and its generalizability.
w/o time-embedding of longitudinal information learning
As illustrated in Supplementary Figs. 16–18, the models (i.e. iMRrhpc, MRP) without time-embedding exhibit inferior performance in comparison to the final model, with decreases in evaluated metric values in longitudinal-based scenarios. This outcome is expected, as the embedded temporal information helps the model understand the relationship between inputs before and after NAT. This is crucial because the ability to predict response to NAT can vary when utilizing information before NAT versus information after NAT, as supported by previous research19,45. Most importantly, in the time-embedding setting, the model’s input becomes more adaptable, making it applicable in various real-world scenarios.
Reader study error analysis
Last, we performed an error analysis of predictions made by MRP using the reader study. The primary objective was to compare MRP’s predictions with radiologists’ predictions and investigate cases with significant discrepancies. Upon qualitative assessment, we observed that in most instances, MRP’s predictions closely matched the radiologists’ predictions. Notably, MRP exhibited high accuracy when predicting cases without pathological complete response (non-pCR), consistently assigning them a low probability of achieving pCR (Supplementary Fig. 19). This was evident in Supplementary Figs. 20 and 21. Moreover, we noticed MRP’s predictions were notably lower than those of radiologists, indicating its potential in identifying non-pCR patients who may not benefit from therapy adjustments. Similarly, MRP effectively identified pCR cases by assigning them a high probability, as depicted in Supplementary Figs. 22 and 23. However, we did encounter a few cases where our model assigned a high probability to non-pCR cases (Supplementary Figs. 24 and 25), or conversely, a low probability to positive cases of pCR (Supplementary Figs. 26 and 27). These situations were similar to those encountered by human radiologists, who also faced challenges in interpreting such cases. For example, for certain cases, we found that most of the missed pCR cases were ductal carcinomas in situ (ypTis), which would anyway not qualify for skipping surgery. In a small number of cases (Supplementary Figs. 28 and 29), where MRP’s predictions diverged from radiologists’ assessments, the underlying reasons for these discrepancies remained unclear. This underscores the importance of future research in the field of deep learning uncertainty.
Discussion
In this study, we developed a multi-modal deep learning model, MRP, for predicting treatment response in breast cancer patients across clinical scenarios of NAT. The model was designed to closely resemble physician evaluation in real clinical settings, taking into account data from different time points in the treatment process and considering the dependence between modalities and the phase of treatment. Due to the use of histopathological and radiological data for predictions, the model increases the applicability compared to that of a uni-modal model. Our study is based on >3000 patients and covers all breast cancer molecular subtypes, across the full process of NAT care courses. It represents the biggest BC cohort in the literature for these types of analyses. The MRP model demonstrates a superior ability to accurately predict pathological complete response compared to models based on clinicopathological features alone, achieving an AUROC of 0.889 with the integration of multi-modal radiological imaging and longitudinal MRIs.
MRP model demonstrates a superior ability to accurately predict pathological complete response
Initial studies have shown that deep learning has the potential to identify patients who will achieve pCR, and the addition of breast imaging data improves the accuracy of treatment response assessment beyond clinical factors alone15,26. In our study, compared to the non-imaging-based model (rhpc), we observed the AUROCs constantly improved with our developed multi-modal models: iMGrhpc (incorporating mammography) improved by 5.5% in cohort A and 4.2% in cohort C; iMRrhpc (incorporating MRI) improved by 9.5% in cohort A, 12% in cohort B, 9.6% in cohort C, and 15.3% in cohort D, and the MRP system (which combines iMGrhpc and iMRrhpc) improved by 11% in cohort A and 10.5% in cohort C. These findings indicate that the integration of image-derived data with auxiliary features increases the predictive value. Implementing a clinical baseline model using our uni-modal rhpc framework demonstrated similar performance to a previously published clinical predictor27 using the same clinical features. In contrast, our NAT routine-used imaging (pre-NAT only) combined model showed a significant improvement over the baseline clinical model (cohort B: iMRrhpc: p = 2.358e-03; cohort C: iMGrhpc: p = 1.640e-02; iMRrhpc: p = 7.584e-03; MRP: p = 4.956e-04). Although the differences between iMGrhpc/iMRrhpc and rhpc were statistically significant, it is important to note that our rhpc model differs from conventional clinico-pathologic combined models. The rhpc model incorporates four attributes from radiological findings, five histopathological attributes from pathology examinations, nine personal attributes from patient records, and four clinical attributes determined by the clinician. Consequently, rhpc captures multidisciplinary features using a deep learning algorithm that encompasses clinical abnormalities as well as underlying molecular characteristics. Therefore, it could also validate the potential benefits of integrating radiological, histopathological, clinical, and personal information throughout neoadjuvant therapy care to enhance the performance of artificial intelligence models for predicting pathological complete response in breast cancer. Moreover, our final integrated response prediction model, referred to as MRP, which combines iMGrhpc and iMRrhpc, demonstrated even better performance in predicting pathological complete response when compared to rhpc (cohort A: p = 0.003; cohort C: p = 0.009). This superior predictive performance of MRP can be attributed to the integration of heterogeneous radiological and histopathological features, enabling a comprehensive assessment of both macro-structural and micro-structural characteristics of the tumor. Histopathological features had the largest contribution to the prediction of response (Fig. 4), with tumor-specific features like histology type and molecular subtype being the most important. Although radiological data, which capture tumor-phenotype features, contributed less overall, the heterogeneity of breast cancer response related to mammographic density41,42 and tumor distribution (uni-focal or multi-focal)40 also demonstrated additional predictive power in our model.
MRP presented comparable robustness and generalizability to breast radiologists, significantly outperforming humans in pCR prediction on Pre-NAT phase
To address the challenges of transparency and reproducibility in the application of artificial intelligence in medicine46, we followed established guidelines47,48, which provide best practices for study protocols and reporting specific to machine learning. According to these guidelines, we conducted external validation studies using international datasets. The results demonstrated robust accuracy for the prediction of pCR, with AUROC of 0.797 (95% CI 0.736-0.850) in cohort B. In longitudinal clinical settings for cohort C and cohort D, the MRP demonstrated a consistently increasing AUROC from the Pre-NAT stage (cohort C: 0.821, 95% CI 0.721-0.908; cohort D: 0.799, 95% CI 0.761-0.836) to Post-NAT stage (cohort C: 0.842, 95% CI 0.753-0.918; cohort D: 0.829, 95% CI 0.794-0.864). This finding suggests that MRP has the potential to be applicable in real-world clinical settings. In order to facilitate the adoption of MRP in other settings, we have made our methods and algorithm code publicly available. Regarding the applicability of multi-modal imaging, breast MRI is currently the preferred imaging modality for assessing tumor response to neoadjuvant therapy49. Mammography remains the standard imaging technique for breast cancer diagnosis50 and can be used as an independent predictor in the pre-NAT scenario. While we have chosen not to include ultrasound in this study due to its operator dependency51, we aim to work towards incorporating it into our model in the future, considering its effectiveness in predicting tumor size49 and assessing lymph node status51. Although this will require retraining the deep learning architecture described in this manuscript and further validation, the benefit of our model is that features from any data modality can be added or removed to generate predictions. In addition to external validation, we also conducted a reader study using both internal and external cohorts. In cohort A, the findings of our study demonstrate that MRP achieved remarkably superior pCR prediction after NAT when compared to human readers. We identified several cases where MRP successfully predicted pCR that the readers had missed. One common scenario involved the presence of residual ductal carcinomas in situ, where Post-NAT imaging still exhibited enhancement, albeit not indicative of actual invasive tumor presence (see Supplementary Fig. 23). Additionally, we observed that radiologists, due to their training, focus primarily on image characteristics, and occasionally overlook important tumor characteristics, such as the molecular subtype, in their decision-making process. It is worth noting that MRP also exhibited statistically significant better predictive performance in the Pre-NAT phase in cohort A, surpassing the results of all six readers (all p < 1e-04). To further investigate the performance of our system in the Pre-NAT phase, we conducted an additional reader study in cohort B, involving 100 patients and two breast radiologists. The results showed that MRP significantly outperformed one radiologist, with a margin of 15% AUROC (p = 0.041), and exhibited comparable performance to the second radiologist, with a non-statistically significant margin of 4.5% AUROC. It is important to note that this analysis had some limitations, such as the small number of readers (two) and their different countries of origin compared to the data source. Nevertheless, the study demonstrated that MRP performs at least comparable to board-certified radiologists when predicting pCR on pre-treatment MRI examinations and clinicopathological data, indicating its robustness in handling distribution shifts. The superior performance of the MRP model compared to human readers shows that it is difficult to include all relevant factors in human reasoning, and consequently highlights the importance of using models in clinical reasoning.
MRP shows its interpretability in design for training/inference
It is essential to ensure that the model exhibits good interpretability and predictable performance. In the case of our multi-modal based models (i.e., iMGrhpc, iMRrhpc, MRP), we not only consider their clinical application during the design phase but also explicitly demonstrate the contribution of the various input modalities in the model’s training. Their design mimics the process in clinical practice, integrating longitudinal multi-modal data to evaluate therapy response in breast cancer patients during the therapy. However, it is important to acknowledge that in the context of multi-modal and longitudinal data, there may be missing data in the model’s input settings. This issue becomes even more pertinent when the model is applied to external datasets or deployed in clinical practice, as the input factors may not always align due to varying clinical treatment regimens across hospitals. To maintain the model’s stable generalization ability and given the frequent availability of imaging data, we incorporate a cross-modal knowledge predictor that solely utilizes the imaging modality to predict other modal inputs for extracting the clinically relevant features. This approach prioritizes improving the efficiency of imaging data and increases the contribution of the imaging modality. Ablation studies with cross-modal knowledge mining reveal enhanced image-wise data contribution (Fig. 4, Supplementary Fig. 15) and consistently increased AUCs for both iMGrhpc and iMRrhpc models, as demonstrated in Supplementary Figs. 12, 13, and 14 (for results descriptions, please see “w/o time-embedding of longitudinal information learning”). Consequently, the unified model MRP remains compatible with missing data input while minimizing the impact on prediction performance. Regarding the longitudinal inputs, their contribution to treatment response prediction varies depending on the different time points of neoadjuvant therapy. The ability to predict response to NAT differs when using Pre-NAT information versus Post-NAT information, as evidenced by previous studies19,45. We train the longitudinal model using both single-time point images and multi-time point inputs in the meantime embedding with temporal information, effectively capturing the heterogeneity of clinical outcomes throughout the NAT timeline. In contrast to most existing studies, which typically employ fixed inputs consisting of either single or multiple time points for predicting treatment responses, our multi-modal based models are designed to accommodate flexible input data. Furthermore, in the context of multi-modal breast data, combining all paired MRI and mammogram images for patients poses significant challenges. Therefore, our iMRrhpc and iMGrhpc models also support such independent usage, providing a practical solution in situations where combining both modalities is not feasible.
MRP may provide a potential clinical tool in breast cancer personalized therapy management across NAT care
Several studies29,30 have successfully developed deep learning models that utilize Pre-NAT MRI and clinical information to predict therapy response in breast cancer patients. However, the clinical applicability of these models remains unclear due to common limitations. Firstly, many of the studies had small sample sizes and lacked external validation, which increases the risk of overfitting and limits the generalizability of the findings. Secondly, their approach may not fully address the evolving clinical needs throughout the course of care. Specifically, different time points in the treatment trajectory require different attention and emphasis. For instance, in the Pre-/Mid-treatment phase, a higher specificity is desired to identify only the non-responsive patients who may benefit from stopping of ineffective therapies and adjusting their treatment regimen accordingly. In the Post-NAT or end-treatment phase, a fair sensitivity of identifying pCR patients is crucial to consider the safe surgery reducing trials for patients who have already responded well to treatment, while securing the surgery opportunity for all women who have not achieved pCR. In our study, we evaluate the clinical utility of MRP by employing a decision curve analysis (DCA) methodology31. DCA indirectly incorporates the preferences of both patients and doctors by calculating the net benefit or reduction associated with alternative strategies, such as selectively performing surgeries based on high DL-based pCR probabilities, compared to the default strategy of performing surgeries for all patients. Despite being a relatively recent methodology, DCA has gained recognition in medical journals and has been adopted in numerous studies15,52,53,54. The benefits of utilizing the MRP were consistent across various threshold probability values that are clinically relevant. By employing this approach, we hope to provide valuable insights into the potential clinical implications and associated benefits of integrating DL models based on MPR into breast cancer therapy management. MRP may potentially also provide a clinical guide for personalized breast cancer NAT, as it can predict the effect of different therapies as well.
Despite encouraging findings, our study possesses several limitations that need to be acknowledged. Firstly, the design of our reader study and analysis may not fully capture the impact of MRP on radiologists’ decision-making processes when it would be implemented in real-world hospital systems. To gain further insights into these effects, it would be valuable to conduct a multireader, multicase study, where the MRP is used as a concurrent or second read tool. Further, careful interpretation and integration of the MRP model into clinical practice by clinicians are necessary to ensure its safe and effective use, considering multiple factors such as clinical judgment and patient preferences. The safety of adapting regular clinical processes should be tested in randomized clinical trials. Secondly, while our DCA analysis provides valuable information about the clinical utility of the MRP, a more comprehensive analysis should be conducted in the future to estimate the potential benefits and costs associated with implementing such an AI system. Thirdly, the validation of our model was based on retrospective data. Although this study represents the biggest breast cancer cohort in the literature to our knowledge that integrates longitudinal multi-modal breast data with NAT outcomes, further prospective clinical trials are required for enhancing the model’s clinical evidence. Fourthly, survival analysis is necessary to further assess the prognostic potential of MRP. This aspect should be addressed in future investigations. Fifthly, this study demonstrates the designed MRP system’s efficiency in integrating rhpc data with breast radiological imaging modalities for NAT response prediction. However, our current models were developed solely using two radiological modalities (mammography and MRI). Recent studies16,27, known to utilize digital pathology data with whole-slide images, have advanced predictions of response to NAT of breast cancer. Future work will explore incorporating additional imaging modalities, such as pathological slides and ultrasound, which may enhance the performance of our models and contribute to their overall improvement.
In conclusion, the proposed multi-modal AI system MRP enables predicting the probability of breast cancer response during the whole NAT period and the model exhibits superior performance compared to breast imaging experts in predicting the probability of breast cancer patients achieving pCR. The MRP design, including a cross-modal knowledge predictor and temporal information embedding, ensures its capability in handling longitudinal multidisciplinary information across different clinical realities, whilst being compatible with missing data. Additionally, the generalizability of our model across different populations by evaluating its performance was shown on three international external datasets. These findings may have significant clinical implications, due to the potential to personalize breast cancer patient therapy management. In the clinical workflow, the MRP could be applied to patients across neoadjuvant therapy stages. In the Pre- and Mid-NAT setting, patients predicted to have treatment-resistant tumors could be promptly considered for enrollment into neoadjuvant clinical trials of alternative therapies9. In the Post-NAT setting, patients predicted to have achieved pCR may be considered for clinical trials of reducing surgery10,11, contingent upon further validation.
Methods
Study design
The purpose of this study is to develop and evaluate a longitudinal multi-modal model known as MRP. It is designed to predict the response of breast cancer to neoadjuvant therapy while effectively addressing the various clinical scenarios encountered across the world in neoadjuvant breast cancer care. According to the standard of NAT care addressed in Fig. 1, our data collection includes essential patient information, as shown in Table 2, such as age, sex, the age of menarche, menopausal status, and genetic mutations related to TP53, BRCA1, BRCA2, and CHEK2. Additionally, we have gathered a Pre-NAT mammogram dataset comprising 4,802 examinations, which includes radiological findings like tumor location, laterality, density, and multifocality. We also acquire histopathological data regarding tumor cell characteristics, which includes molecular subtype, tumor histological type, tumor differentiation degree, and in situ component findings. Furthermore, our dataset encompasses longitudinal sets of DCE-MRI data from 3,719 patients, completed with radiological observations. Lastly, we include clinical information, such as cTNM staging and treatment details. All collected training data are sourced from the Netherlands Cancer Institute. In addition, we use independent, international datasets for external validation of our model. The MRP system is built upon two multi-modal models, namely, iMRrhpc and iMGrhpc. The final MRP model is an enhanced ensemble that integrates optimized weightings from both the iMRrhpc and iMGrhpc models. Beyond analyzing the multi-modal based model performance with the uni-modal based rhpc model, we compare their performance with that of an international group of radiologists. We also explore how these models/modalities’ features are weighted to predict and explain pCR outcomes by feature importance analysis. Last, we used DCA methodology to demonstrate that MRP’s predictions can identify non-pCR at Pre-/Mid-NAT stage to help in adjusting therapy promptly, and pCR patients at Post-NAT stage to help in avoiding unnecessary surgeries.
Cohorts construction
Patient pathway
This study follows the clinical practice pathway32 in breast cancer neoadjuvant therapy response evaluation illustrated in Fig. 1 that we now detail. In the Pre-NAT phase – i.e. during the initial staging of breast cancer, patient information is collected. This includes weight, gender, age, the age of menarche, menopausal status, and genetic mutations related to TP53, BRCA1, BRCA2, and CHEK2. Primary screening is conducted through mammograms and/or ultrasound examinations, concluding with critical radiological findings that have given rise to the indication to biopsy a lesion. Breast MRI and CT scans (if available) are utilized to establish the clinical tumor node metastasis classification (cTNM) of the cancer, yielding the intial breast cancer stage. The diagnostic biopsies are performed to extract suspected tissue samples that are subjected to histopathological examinations to gather essential pathological variables. After diagnosing the patient with a specific type and stage of breast cancer, the standard therapeutic regimen is initiated, which consists of neoadjuvant therapy in most larger ( >2 cm) and/or node positive cancers. Mid-NAT stage involves the collection of a second round of MRI studies. These studies serve as follow-ups and are used to compare with the baseline Pre-NAT MRI for evaluating if the patient needs to adjust the treatment plan. In the Post-NAT phase – after completion of NAT and prior to surgery – a third MRI study is conducted, known as the Post-NAT MRI, to evaluate if the patient achieves the pCR. During surgery, both the breast tissue and axillary lymph nodes are examined to assess the treatment’s effectiveness. This response assessment is based on the pathological tumor and lymph node staging after NAT (ypTN).
Inclusion and ethics statement
In-house dataset. The protocol was approved by the medical ethics committee of The Netherlands Cancer Institute (registration number: IRBd21-059). We collected histopathological attributes from pathology examinations, personal attributes from patient records, clinical attributes determined by the clinician, breast imaging (including DCE-MRI and mammogram), and related radiological findings for participants with breast cancer between August 2004 and December 2020. All patients treated at our institution undergo pre- and post-NAT breast MRI. Interim scans were standard between 2010 and 2015. Before 2010, and after 2015, this practice was performed for trial participants or when treatment efficacy was in doubt. As Supplementary Fig. 1. a shows, the inclusion criteria were: (1) pathologically confirmed diagnosis of breast cancer and treatment with neoadjuvant NAT (chemotherapy, hormone therapy, immune therapy); (2) at least one time point scans available from either of the NAT timepoints (Pre-NAT scans available within 1 year prior to the initiation of NAT; Mid-NAT scans available between the initiation and end of NAT; Post-NAT scans available after NAT but before surgery). The exclusion criteria were: (1) patients had other concurrent malignancies or were receiving another oncologic treatment; (2) patients that did not complete the entire course of therapy or did not undergo surgery. Patients were also excluded if the MRI/MG quality was insufficient.
In total, 3352 patients were included in this multi-modality study, where pre-NAT MG and multi-timepoint MRI datasets were collected. We first randomly divided the MG dataset at the patient level, allocating 80% for training and the remaining 20% split equally for validation and testing. In the test subset, 120 patients were further selected to form the test cohort A. These patients had both available pre-NAT MG and longitudinal MRI examinations at three different time points (pre-, mid-, and post-NAT), allowing for model evaluation across these stages. The remaining cases from the test subset were added to the validation set. Consequently, for the MRI dataset division, we ensured a consistent test set of 120 patients with the MG dataset for comparative purposes, while still maintaining a 20% allocation for validation and test. The variables of collected patient characteristics are detailed in Table 2.
To demonstrate the generalizability of our model across different populations, we collected datasets from international institutions in the Netherlands, the United States, and China. These external datasets were preprocessed using the same pipeline as our in-house data, including resampling, reorientation to the left-posterior-superior (LPS) orientation.
Duke University. The Duke Breast Cancer MRI dataset55, publicly accessible through The Cancer Imaging Archive (TCIA), includes data from patients recruited at Duke University School of Medicine (Durham, USA) between January 2000 and March 2014. The dataset comprises 922 examinations of invasive breast cancer, with 312 eligible patients having received NAC. To meet the requirements of our model, we identified pre- and post-contrast sequences to generate subtracted images and performed inference to produce predictions.
For this study, as Supplementary Fig. 1. b shows, we first excluded those with missing ground truth on pCR (i.e. 12 patients treatment response assessment unavailable). We also excluded 12 examinations for size mismatch between pre- and postcontrast sequences or lacking information. Ultimately, 288 imaging exams were included from the Duke dataset, for further details please refer to Supplementary Tab. 6. Of note, these only comprise the MRI scans obtained in the pre-NAT phase, as mammograms and examinations obtained during and after NAT are not available.
Fujian Provincial Hospital Dataset. The cohort consists of 220 breast cancer patients who received neoadjuvant therapy between January 1st, 2019, and December 31st, 2023, collected from Fujian Provincial Hospital (FJPH) in China. The dataset comprises pre-NAT mammograms, longitudinal dynamic contrast-enhanced MRI data before and after NAT, clinical information, and histopathologic outcome data. To ensure comparable validation between the iMGrhpc and iMRrhpc models across the NAT period, we excluded 135 patients due to either missing multi-modal imaging or longitudinal MRI, or unqualified imaging, as shown in Supplementary Fig. 1. c. The remaining 85 cases were included. For further patient details, please refer to Supplementary Tab. 7.
I-SPY2 Dataset. In the I-SPY2 (Investigation of Serial studies to Predict Your Therapeutic Response with Imaging And moLecular analysis 2)33,34 collection, from 2010 to 2016, 719 breast cancer patients were enrolled across multiple clinical sites in the United States, including dynamic contrast-enhanced MRI and clinicopathologic data. In this study, we excluded 211 patients who were either size mismatched between pre- and post-contrast sequences or missing multi-time points (pre-, mid-, and post-NAT) MRIs, as shown in Supplementary Fig. 1. d. Ultimately, 508 cases and corresponding 2,032 imaging exams were included in the I-SPY2 dataset. For further patient details, please refer to Supplementary Tab. 8.
Ground Truth: Definition of pathological complete response
pCR is defined as the absence of invasive disease in breast and axillary nodes (ypT0/is, ypN0) at time of surgery, which allows for the presence of residual DCIS (ypTis) in the pathological specimen. Patients in whom residual invasive breast cancer was found and patients who progressed before surgery are labeled as non-pCR. Note that for multi-center cohorts, a unified pCR standard2 was used.
Model development
We first trained the uni-modal rhpc-based model as the baseline model, which extracts features from rhpc characteristics (r: radiological, h: histopathological, p: personal, c: clinical) as presented in Table 2. Age of patients was modeled as a continuous variable, while all other characteristics were encoded as one-hot categorical variables. These variables were then embedded through a linear layer followed by an Exponential Linear Unit (ELU) activation function56. Subsequently, the features from all variables were concatenated and fed into a two-layer multi-layer perceptron (MLP) with an output dimension of 256. Finally, the MLP output is passed through a linear classifier to estimate the treatment response of breast cancer patients.
For multi-modal models development, as indicated by Fig. 2, the proposed iMGrhpc/iMRrhpc takes the imaging (Mammogram/MRI) and rhpc-based signatures as input, and outputs including rhpc and response prediction. Each model network architecture consists of two modules: one is a cross-modal knowledge mining that predicts rhpc information through extracted imaging features only, and another is for response prediction through integrated features of rhpc-based signatures and imaging features. The design of cross-modal knowledge mining is to enhance the model capacity of imaging feature extraction and increase the contribution of the imaging modality.
It is understood that there easily exists incomplete multi-modality data due to different therapy workflows in clinical reality. Thus our model is designed to maintain the contribution from imaging data, which is frequently available across different medical centers. Thus minimizing the model dependence on data from other modalities. Specifically, we insert a branch topped on the extracted imaging features where it is trained to predict all structured input and therapy response. For the MG and MRI imaging features extractors, which are respectively based on 2D-ResNet18 and 3D-ResNet1857 architecture with pre-trained backbones. Particularly, the visual encoder of iMRrhpc consists of two identical 3D-ResNet18 with shared parameters. Subsequently, the extracted image features are embedded with respective stage-specific temporal information. This includes three temporal signals for Pre-, Mid- and Post-NAT, which we converted into one-hot categorical variables. The one-hot vectors are then constructed using a linear layer followed by the Exponential Linear Unit (ELU) activation function, a method commonly used in previous studies58,59, allowing for the integration of temporal representations. The rhpc extractor is a sequential layer that passes through a Multi-layer Perceptron (MLP) utilizing fully connected layers activated by the ELU activation function. Eventually, the imaging and rhpc features flow to the therapy response prediction modules.
For the sake of both explainability and flexibility during training and inference, the ensemble method, MRP, is structured to involve the separate training of iMGrhpc and iMRrhpc models, followed by the integration of optimized weightings from both models. For a given inference sample x, if F1 denotes the model trained locally on iMGrhpc and F2 is the model trained locally on iMRrhpc, then the prediction y for the ensemble strategy is given as y = W1 × F1 + W2 × F2, where W1 and W2 are the weights for aggregating each model’s prediction to provide a best overall prediction performance during validation, which use the same weights for testing.
Training details
The training was performed with two separated customized data loaders (MRI-based, mammogram-based), which, for each patient, picked random combinations of available data per phase of NAT, from all present imaging, temporal information, and corresponding structured data to be fed into each modal model. For training of the MRI-based model, we loaded weights from the model Pre-trained on the medical dataset60. For validation and testing, patient-level predictions were calculated by averaging over the probability predictions produced by iMGrhpc and iMRrhpc. The MRP is based on the optimized weights to select the best multi-modal fusion performance.
Imaging acquisition and processing
The full field digital mammography images were acquired using a HOLOGIC Selenia Dimensions mammography system. Before being input into the model, the mammograms with standard DICOM format undergo preprocessing steps. Firstly, the images are converted into 16-bit PNG format. Then, the whole breast region is segmented to exclude the background. To ensure a consistent size for all images, zero-padding and resizing are applied, resulting in images with dimensions of 512 by 1024 pixels while preserving the relative scale and aspect ratio. Lastly, the images are normalized using the min-max method. Additionally, standard data augmentation techniques are employed during training to enhance model robustness and prevent overfitting. These augmentation techniques include random flipping, brightness adjustment, and contrast enhancement, as described in61.
The MRIs were acquired using Philips Ingenia 3.0-T scanners. Specifically, subtraction images were collected following the radiologists’ preferences. Subtraction images are generated by subtracting the precontrast volume from each postcontrast volume, following a simple matrix subtraction method: X_subtraction = X_post - X_pre, where X_post represents one of the postcontrast volumes, and X_pre represents the precontrast volume. To ensure consistency in spatial resolution, all MRI images are resampled to a voxel size of 1 mm × 1 mm × 1 mm. After resampling, the images are cropped to retain the bilateral breast area, resulting in a final size of 176 × 176 × 352. Various augmentation techniques are also applied to the MRI images, including geometric transformations, intensity-based modifications, and MRI-specific augmentations62.
Ablation analysis
In order to examine the necessity of different components within the proposed multi-modal network for accurately predicting responses in multi-center data, we conducted a comparative analysis by simplifying the network. Firstly, we eliminated the cross-modal knowledge predictor, which serves as a functional module that topped the extracted image representations. Consequently, the network focused solely on one module of response prediction without incorporating any branch functions. This modification allowed us to assess the significance of cross-modal knowledge mining in enhancing the model’s generalization capability, as well as its influence on the contribution of the imaging modality. Secondly, we removed the time-embedding component responsible for encoding stage-specific temporal information within the network. This alteration provided us with the opportunity to evaluate the importance of time embedding across various NAT phases, considering flexible data inputs of both longitudinal and single time points.
Reader study
We conducted a retrospective reader study to evaluate the performance of our model compared to radiologists in predicting therapy response. The study employed a standalone assessment approach, in which six board-certified breast radiologist attendings from five countries interpreted the imaging exams. For the reader study set, we selected imaging exams from two test cohorts. To ensure the dataset reflected the clinical reality, we enriched it with a regular distribution of pCR and non-pCR cases. We also considered different molecular breast cancer subtypes. During the study, radiologists were provided with MRI images, as well as clinical and pathological information such as Pre-therapy clinical stage, molecular subtype, and treatment received. They could also access subtraction MRI images. All the imaging exams used in the study were pseudonymized and stored in a grand-challenge platform dedicated to reader studies, which can be found at https://grand-challenge.org/reader-studies. Radiologists used this platform to report their predictions guided by RECIST criteria32. Before participating in the study, the recruited radiologists were required to familiarize themselves with the study instructions and the DICOM viewer, specifically the LesionTracker system described by Urban et al.63. Once ready, radiologists were asked to provide the prediction for each imaging exam. A probability estimation for the patient achieving pCR, ranging from zero (indicating non-pCR) to one (indicating pCR). Breast tumors were evaluated at each phase of NAT, with subsequent images being blinded to the radiologists. The assessments were performed sequentially, starting with the Pre-NAT MRI, followed by the addition of Mid-NAT MRI, and finally the Post-NAT MRI inclusion. This approach ensured that the radiologists made their predictions without knowledge of the subsequent images, mimicking the real clinical scenario.
Statistical analysis
In our study, we employed several methods to assess the performance of the models and to evaluate inter-reader agreement. To evaluate model performance, we utilized ROC and PR curves. We calculated the AUC for both ROC and PR curves using a nonparametric (trapezoidal) method. Additionally, we reported sensitivity and specificity. To obtain confidence intervals (CIs) for the results, we used the bootstrap method outlined by Carpenter and Bithell64, to derive 95% CIs. For comparisons between two models or a model and a specific reader, we performed DeLong’s test65 on the AUROCs. We considered p-values <0.05 as statistically significant. To assess the clinical utility of the models, we employed decision curve analysis (DCA). We used the dcurves Python package (available at https://github.com/MSKCC-Epi-Bio/dcurves) to generate DCA curves and calculate the Net Reduction of Intervention (NRI). In DCA, we established a range of threshold probabilities for the test set and reported a wide range of threshold probabilities for the full population. The NRI is defined in the following equation:
Here, TP refers to true positives, FP refers to false positives, FN refers to false negatives, TN refers to true negatives, and N represents the total number of cases or observations. To prevent overestimation of the net benefit66, we performed bootstrapping with 1000 replicates and reported decision curves with 95% CIs. We performed a disproportionality analysis67 to evaluate the association between pCR and the top significant contributing factors identified in the MRP model by calculating the Odds Ratio (OR), using the most common group as the comparator.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw in-house data are protected and are not available due to data privacy laws, while supporting the findings including the imaging data can be available under restricted access for non-commercial and academic purposes only. Access can be obtained by request to the corresponding author, and all requests will be promptly reviewed within 15 working days. The Duke Breast Cancer MRI55 data are available at TCIA (https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=70226903). The I-SPY2 Breast Cancer MRI datasets33,34 are available at TCIA (https://www.cancerimagingarchive.net/collection/ispy2/). Because those external datasets originally do not provide information on pre- and post-contrast sequences as well as labels formatted for deep learning research, we publish files and scripts to enable processing of them for deep learning purposes. Source data are provided with this paper.
Code availability
Custom codes related to the deep learning models are available on https://github.com/yawwG/MRP/.
References
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA. Cancer J. Clin. 73, 17–48 (2023).
Cortazar, P. & Geyer, C. E. Pathological complete response in neoadjuvant treatment of breast cancer. Ann. Surg. Oncol. 22, 1441–1446 (2015).
van der Valk, M. J. et al. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the international watch & wait database (iwwd): an international multicentre registry study. Lancet 391, 2537–2545 (2018).
Smith, J. J. et al. Assessment of a watch-and-wait strategy for rectal cancer in patients with a complete response after neoadjuvant therapy. JAMA Oncol. 5, e185896–e185896 (2019).
Dattani, M. et al. Oncological and survival outcomes in watch and wait patients with a clinical complete response after neoadjuvant chemoradiotherapy for rectal cancer: a systematic review and pooled analysis. Ann. Surg. 268, 955–967 (2018).
Von Minckwitz, G. et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 30, 1796–1804 (2012).
Spring, L. M. et al. Pathologic complete response after neoadjuvant chemotherapy and impact on breast cancer recurrence and survival: a comprehensive meta-analysispcr and association with clinical outcomes in breast cancer. Clin. Cancer Res. 26, 2838–2848 (2020).
Gradishar, W. J. et al. Breast cancer, version 3.2020, NCCN clinical practice guidelines in oncology. J. Natl Compr. Cancer Netw. 18, 452–478 (2020).
Symmans, W. F. et al. Long-term prognostic risk after neoadjuvant chemotherapy associated with residual cancer burden and breast cancer subtype. J. Clin. Onco. 35, 1049 (2017).
Kuerer, H. M. et al. Eliminating breast surgery for invasive breast cancer in exceptional responders to neoadjuvant systemic therapy: a multicentre, single-arm, phase 2 trial. Lancet Oncol. 23, 1517–1524 (2022).
Johnson, H. M. et al. Eliminating breast surgery for invasive cancer with exceptional response to neoadjuvant systemic therapy: prospective multicenter clinical trial planned initial feasibility endpoint. J. Am. College Surg. 237, 101–108 (2023).
Li, G., Hu, J. & Hu, G. Biomarker studies in early detection and prognosis of breast cancer. Adv. Exp. Med. Biol. 1026, 27–39 (2017).
Arriaga-Canon, C. et al. The clinical utility of lncRNAs and their application as molecular biomarkers in breast cancer. Int. J. Mol. Sci. 24, 7426 (2023).
Jafari, S. H. et al. Breast cancer diagnosis: Imaging techniques and biochemical markers. J. Cell. Physiol. 233, 5200–5213 (2018).
Huang, Y. et al. Longitudinal MRI-based fusion novel model predicts pathological complete response in breast cancer treated with neoadjuvant chemotherapy: a multicenter, retrospective study. EClinicalMedicine 58, 101899 (2023).
Ogier du Terrail, J. et al. Federated learning for predicting histological response to neoadjuvant chemotherapy in triple-negative breast cancer. Nat. Med. 29, 135–146 (2023).
Jin, C. et al. Predicting treatment response from longitudinal images using multi-task deep learning. Nat. Commun. 12, 1851 (2021).
Skarping, I., Larsson, M. & Förnvik, D. Analysis of mammograms using artificial intelligence to predict response to neoadjuvant chemotherapy in breast cancer patients: proof of concept. Eur. Radiol. 32, 3131–3141 (2022).
Huang, Z. et al. Artificial intelligence reveals features associated with breast cancer neoadjuvant chemotherapy responses from multi-stain histopathologic images. NPJ Precision Oncol. 7, 14 (2023).
Zhang, T. et al. Radiomics and artificial intelligence in breast imaging: a survey. Artif. Intell. Rev. 56, 857–892 (2023).
Caballo, M. et al. Four-dimensional machine learning radiomics for the pretreatment assessment of breast cancer pathologic complete response to neoadjuvant chemotherapy in dynamic contrast-enhanced MRI. J. Magn. Reson. Imaging. 57, 97–110 (2023).
Zeng, Q. et al. Radiomics based on DCE-MRI for predicting response to neoadjuvant therapy in breast cancer. Acad. Radiol. Suppl 2, S38–S49 (2023).
Liu, S. et al. A delta-radiomic lymph node model using dynamic contrast enhanced MRI for the early prediction of axillary response after neoadjuvant chemotherapy in breast cancer patients. BMC Cancer 23, 15 (2023).
El Adoui, M., Drisis, S. & Benjelloun, M. Multi-input deep learning architecture for predicting breast tumor response to chemotherapy using quantitative mr images. Int. J. Comp. Assist. Radiol. Surg. 15, 1491–1500 (2020).
Duanmu, H. et al. Prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer using deep learning with integrative imaging, molecular and demographic data. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part II 23, 242–252 (Springer, 2020).
Wang, X. et al. Artificial intelligence in breast imaging. Clin. Radiol. 74, 357–366 (2022).
Sammut, S.-J. et al. Multi-omic machine learning predictor of breast cancer therapy response. Nature 601, 623–629 (2022).
Duanmu, H. et al. A spatial attention guided deep learning system for prediction of pathological complete response using breast cancer histopathology images. Bioinformatics 38, 4605–4612 (2022).
Comes, M. C. et al. Early prediction of neoadjuvant chemotherapy response by exploiting a transfer learning approach on breast DCE-MRIs. Sci. Rep.11, 14123 (2021).
Joo, S. et al. Multimodal deep learning models for the prediction of pathologic response to neoadjuvant chemotherapy in breast cancer. Sci. Rep. 11, 18800 (2021).
Fitzgerald, M., Saville, B. R. & Lewis, R. J. Decision curve analysis. JAMA 313, 409–410 (2015).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised recist guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009).
Rugo, H. S. et al. Adaptive randomization of veliparib–carboplatin treatment in breast cancer. N. Engl. J. Med. 375, 23–34 (2016).
Park, J. W. et al. Adaptive randomization of neratinib in early breast cancer. N. Engl. J. Med. 375, 11–22 (2016).
Sundararajan, M., Taly, A. & Yan, Q. Axiomatic attribution for deep networks. In International Conference on Machine Learning, 3319–3328 (PMLR, 2017).
Burstein, H. J., Polyak, K., Wong, J. S., Lester, S. C. & Kaelin, C. M. Ductal carcinoma in situ of the breast. N. Engl. J. Med. 350, 1430–1441 (2004).
Ernster, V. L., Barclay, J., Kerlikowske, K., Grady, D. & Henderson, I. C. Incidence of and treatment for ductal carcinoma in situ of the breast. Jama 275, 913–918 (1996).
Petrelli, F. & Barni, S. Response to neoadjuvant chemotherapy in ductal compared to lobular carcinoma of the breast: a meta-analysis of published trials including 1,764 lobular breast cancer. Breast Cancer Res. Treat. 142, 227–235 (2013).
Tubiana-Hulin, M. et al. Response to neoadjuvant chemotherapy in lobular and ductal breast carcinomas: a retrospective study on 860 patients from one institution. Ann. Oncol. 17, 1228–1233 (2006).
Ataseven, B. et al. Impact of multifocal or multicentric disease on surgery and locoregional, distant and overall survival of 6,134 breast cancer patients treated with neoadjuvant chemotherapy. Ann. Surg. Oncol. 22, 1118–1127 (2015).
Di Cosimo, S. et al. Mammographic density to predict response to neoadjuvant systemic breast cancer therapy. J. Cancer Res. Clin. Oncol. 148, 775–781 (2022).
Skarping, I. et al. Mammographic density is a potential predictive marker of pathological response after neoadjuvant chemotherapy in breast cancer. BMC Cancer 19, 1–11 (2019).
Vickers, A. J. & Elkin, E. B. Decision curve analysis: a novel method for evaluating prediction models. Med. Decision Making 26, 565–574 (2006).
Crawshaw, M. Multi-task learning with deep neural networks: a survey. Preprint at https://doi.org/10.48550/arXiv.2009.09796 (2020).
Qu, Y.-H. et al. Prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer using a deep learning (dl) method. Thoracic Cancer 11, 651–658 (2020).
Haibe-Kains, B. et al. Transparency and reproducibility in artificial intelligence. Nature 586, E14–E16 (2020).
Rivera, S. C. et al. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the spirit-ai extension. Lancet Digital Health 2, e549–e560 (2020).
Liu, X. et al. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the consort-ai extension. Lancet Digital Health 2, e537–e548 (2020).
Fowler, A. M., Mankoff, D. A. & Joe, B. N. Imaging neoadjuvant therapy response in breast cancer. Radiology 285, 358–375 (2017).
Bleyer, A. & Welch, H. G. Effect of three decades of screening mammography on breast-cancer incidence. N. Engl. J. Med. 367, 1998–2005 (2012).
Croshaw, R., Shapiro-Wright, H., Svensson, E., Erb, K. & Julian, T. Accuracy of clinical examination, digital mammogram, ultrasound, and MRI in determining postneoadjuvant pathologic tumor response in operable breast cancer patients. Ann. Surg. Oncol. 18, 3160–3163 (2011).
Kerr, K. F., Brown, M. D., Zhu, K. & Janes, H. Assessing the clinical impact of risk prediction models with decision curves: guidance for correct interpretation and appropriate use. J. Clin. Oncol.34, 2534 (2016).
Deniffel, D. et al. Using decision curve analysis to benchmark performance of a magnetic resonance imaging–based deep learning model for prostate cancer risk assessment. Eur. Radiol. 30, 6867–6876 (2020).
Witowski, J. et al. Improving breast cancer diagnostics with deep learning for mri. Sci. Transl. Med. 14, eabo4802 (2022).
Saha, A. et al. A machine learning approach to radiogenomics of breast cancer: a study of 922 subjects and 529 DCE-MRI features. Br. J. Cancer 119, 508–516 (2018).
Clevert, D.-A., Unterthiner, T. & Hochreiter, S. Fast and accurate deep network learning by exponential linear units (elus). Preprint at https://doi.org/10.48550/arXiv.1511.07289 (2015).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proc. IEEE Conference on Computer Vision and Pattern Recognition, 770–778 (Las Vegas, NV, 2016).
Gorishniy, Y., Rubachev, I. & Babenko, A. On embeddings for numerical features in tabular deep learning. Adv. Neural Inform. Proc. Syst. 35, 24991–25004 (2022).
Ching, T. et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 15, 20170387 (2018).
Chen, S., Ma, K. & Zheng, Y. Med3d: Transfer learning for 3d medical image analysis. Preprint at https://doi.org/10.48550/arXiv.1904.00625 (2019).
Wang, X. et al. Disasymnet: Disentanglement of asymmetrical abnormality on bilateral mammograms using self-adversarial learning. Preprint at https://doi.org/10.48550/arXiv.2307.02935 (2023).
Pérez-García, F., Sparks, R. & Ourselin, S. Torchio: a python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning. Comput. Methods Progr. Biomed. 208, 106236 (2021).
Urban, T. et al. Lesiontracker: extensible open-source zero-footprint web viewer for cancer imaging research and clinical trials. Cancer Res. 77, e119–e122 (2017).
Carpenter, J. & Bithell, J. Bootstrap confidence intervals: when, which, what? a practical guide for medical statisticians. Stat. Med. 19, 1141–1164 (2000).
DeLong, E. R., DeLong, D. M. & Clarke-Pearson, D. L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44, 837–45 (1988).
Capogrosso, P. & Vickers, A. J. A systematic review of the literature demonstrates some errors in the use of decision curve analysis but generally correct interpretation of findings. Med. Decision Making 39, 493–498 (2019).
Bate, A. & Evans, S. Quantitative signal detection using spontaneous adr reporting. Pharmacoepidemiol. Drug Safety 18, 427–436 (2009).
Provenzano, E. et al. Standardization of pathologic evaluation and reporting of postneoadjuvant specimens in clinical trials of breast cancer: recommendations from an international working group. Modern Pathol. 28, 1185–1201 (2015).
Acknowledgements
We acknowledge and are grateful to the six participating breast radiologists for involving in the reader study of NAT response prediction. Our thanks also to Sofia Ventura for designing Fig. 1. Laura Estacio, George Agrotis, Luuk Balkenende for fruitful discussions, ideas, supporting analysis, and engineering that either directly or indirectly made this work possible. The authors thank Grand Challenge for offering the reader study platform. The authors thank the support of the China Scholarship Council (202107720016, 202006930001, and 202006240065), Guangzhou Elite Project (TZ-JY201948), Macao Polytechnic University Grant (RP/FCA-15/2022) and Science and Technology Development Fund of Macao (0105/2022/A).
Author information
Authors and Affiliations
Contributions
Y.G., R.M., T.T., and X.W. conceived the conception and design of the study; Y.G. developed the deep learning model; J.T. contributed to device support for model training and inference; Y.G., R.M., R.T., S.V., X.W., T.Z, L.H., and M.H. contributed to acquisition and annotation of the data; Y.G., R.M., T.T., X.W., F.D., M.K., H.H., H.Z., and Z.L. contributed to analysis and interpretation of the data; S.V, Z.X, A.W., X.L., A.D., and V.L. conducted the reader study. Y.G. wrote the initial manuscript; T.T. and R.M. supervised the work. All authors contributed to the revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Aleix Prat, Rami Vanguri and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, Y., Ventura-Diaz, S., Wang, X. et al. An explainable longitudinal multi-modal fusion model for predicting neoadjuvant therapy response in women with breast cancer. Nat Commun 15, 9613 (2024). https://doi.org/10.1038/s41467-024-53450-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53450-8
This article is cited by
-
Multicenter study provides radiomic and biological insights into neoadjuvant chemotherapy response and prognosis in luminal breast cancer
Cancer Imaging (2026)
-
Prediction of pathological complete response to neoadjuvant therapy in breast cancer using deep learning with multi-modal radiological image and biopsy whole slide images: a two-center study
Cancer Imaging (2026)
-
Diagnostic accuracy of artificial intelligence-assisted 18f-fdg pet/ct for predicting pathological complete response to neoadjuvant chemotherapy in breast cancer: a systematic review and meta-analysis
Annals of Nuclear Medicine (2026)
-
Circulating tumor DNA dynamics predict pathological response and guide therapy personalization in the neoadjuvant setting
Discover Oncology (2026)
-
Artificial intelligence-assisted PET imaging for predicting neoadjuvant chemotherapy response in breast cancer: a systematic review and meta-analysis
European Journal of Nuclear Medicine and Molecular Imaging (2026)