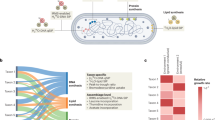
Abstract
Soil microbial growth, a vital biogeochemical process, governs both the accrual and loss of soil carbon. Here, we investigate the biogeography of soil microbiome potential growth rates and show that microbiomes in resource-rich (high organic matter and nutrients) and acid-neutral soils from cold and humid regions exhibit high potential growth. Conversely, in resource-poor, dry, hot, and hypersaline soils, soil microbiomes display lower potential growth rates, suggesting trade-offs between growth and resource acquisition or stress tolerance. In addition, the potential growth rates of soil microbiomes positively correlates with genome size and the number of ribosomal RNA operons but negatively correlates with optimum temperature, biomass carbon-to-phosphorus and nitrogen-to-phosphorus ratios. The spatial variation of microbial potential growth rates aligns with several macroecological theories. These findings not only enhance our understanding of microbial adaptation to diverse environments but also aid in realistically parameterizing microbial physiology in soil carbon cycling models.
Similar content being viewed by others
Introduction
The feedback between soil carbon (C) and climate change is fraught with uncertainty in Earth System Models due to the vast variation in model structure, parameter values, and initial conditions1,2. The physiology of the soil microbiome mediates the biogeochemical cycling of C, nitrogen (N), and phosphorus (P)3,4. Over seventy microbial models have been developed to realistically model C cycling in soils5. However, the current understanding of microbial feedback to temperature and the corresponding representation in soil C cycling models are incomplete. Soil microbial growth, a critical microbial physiological trait, governs both the accrual, contributing to soil non-living organic C through biomass turnover, and the loss, through respiration metabolism, of soil C6,7. Therefore, understanding the response of microbial growth to climatic factors could help better explain the global patterns of soil C under climate change.
Growth of organisms, which integrates morphology with physiological processes, is a fundamental ecological and evolutionary trait8,9. The latitudinal/temperature compensation hypothesis suggests that cold-adapted organisms from high latitudes have a growth advantage to compensate for the low temperature and short growing season10,11,12. However, trade-offs between growth and adaptation to stress significantly influence the physiology and distribution of both macro-species13 and micro-species14. Specifically, organisms with high growth rates are generally more vulnerable to environmental stress, such as cold or arid climates15. Therefore, organisms in high-latitude regions enduring cold stress may have low growth rates. These two contrasting mechanisms lead to different patterns of growth rates along latitudinal/temperature gradients among different plant species16,17,18,19,20,21. The biogeography of soil microbiome potential growth may also be associated with microbial biomass stoichiometry. The growth rate hypothesis suggests that rapid growth results in a high demand for P used in ribosomes for the synthesis of mRNA relative to the demand for N, leading to a low N:P ratio in rapidly growing organisms22. Although spatial variabilities in microbial stoichiometric ratios have been noted23,24, and a previous global analysis found a decreasing trend in microbial N:P with increasing latitude25, whether microbial growth covaries with biomass N:P at a biogeographic scale is unclear. In addition, microbial activity is known to be reduced with increasing aridity; consequently, microbial strategies depend on the ability to resist desiccation in more arid environments to continue the production of C-rich organelles and molecules (increasing biomass C:N and N:P)24. Therefore, trade-offs between microbial growth and stress tolerance may relate to microbial biomass stoichiometry. However, measurements of the growth rates of soil microbiome at geographical scale are limited, lagging substantially behind those for macro-organisms, despite their crucial importance to terrestrial ecosystems26. A recent substrate-independent approach using 18O-H2O incorporation into DNA now allows to investigate the large-scale variation of microbial growth rate under relative realistic condition.
Community aggregated microbial traits, like community-weighted means of traits proposed for plants27, represent the average functional profile of thousands of microbial species co-existing within a local habitat, which controls microbial metabolic potential and their response to environmental changes28. Several microbial traits, such as genome size (an expansion of metabolic capacities), the number of ribosomal RNA operons (rrn copy number, a proxy of maximum growth rate), and guanine plus cytosine content, were used to construct the life history strategy scheme28,29. However, one of the current challenges is the association between microbial traits and their realistic adaptation to the environment in nature28. For example, the strongest expectation is that oligotrophic microbes characterized by smaller genome size are adapted to resource-poor soil30,31, however, Wang et al.32 suggested that oligotrophic microbes tend to have larger genomes. Experimental evidence linking microbial traits from omics to ecological processes in nature is lacking. Therefore, exploring the association between microbial growth and critical microbial traits would help advance our understanding of the life history strategies of soil microbiomes.
Here, we investigated the potential growth rate of the soil microbiome using an 18O-labeled water approach on a national scale, encompassing 112 sample sites (49 grasslands and 63 forests). This survey spanned a broad spectrum of geoclimatic patterns. Specifically, the latitude ranged from 18.4° to 53.3°N, the longitude ranged from 81.2° to 129.0°E, the mean annual temperature ranged from −4.8 °C to 24.4 °C, and the De Martonne aridity index (a greater aridity index indicates a more humid climate) ranged from 7.5 to 93.5. As potential drivers of microbial growth, we evaluated climate (mean annual temperature and aridity index), edaphic properties (soil texture and pH), microbial resources (soil organic C, total N, total P, dissolved organic C, and available N), microbial community structure (dominant fungal and bacterial phyla), and microbial traits (genome size, rrn copy number, optimum temperature, and biomass stoichiometry). Our aims were to determine how microbial biomass-specific potential growth rate (microbial growth rate per unit microbial biomass C, Gmass) changes across geoclimatic gradients, identify the potential drivers of this change, and investigate the implications for soil C cycling.
In this work, microbiomes in resource-rich (high organic matter and nutrients) and acid-neutral soils from cold and humid regions exhibit high potential growth rates. Conversely, microbiomes in hypersaline soil from dry and hot regions display lower potential growth rates. The potential growth rate of soil microbiomes positively correlates with genome size and the number of ribosomal RNA operons but negatively correlates with optimum temperature, biomass carbon-to-phosphorus and nitrogen-to-phosphorus ratios. Overall, the spatial variation of microbial growth rates, driven by climate, edaphic properties, microbial resources, community structure, and genome traits, aligns with macroecological theories of the latitudinal/temperature compensation hypothesis, the trade-off between growth and stress survival, and the growth rate hypothesis. Our study also offers a robust benchmark for microbial-explicit soil C models, because microbial growth rate is a key parameter in these models.
Results and discussion
Biogeographical pattern of microbial growth rates
The potential Gmass of the soil microbiome exhibited significant spatial variation on a national scale (Fig. 1a). Alpine and boreal forests had the highest potential Gmass, whereas desert grasslands had the lowest (Fig. 1b). The biome-specific potential Gmass was predominantly regulated by aridity rather than mean annual temperature. First, the correlation coefficient between potential Gmass and the aridity index was greater than that between potential Gmass and mean annual temperature (Figs. 1b, c and 2a). Second, variance decomposition analysis further showed that the aridity index explained more of the variance in potential Gmass than mean annual temperature (Supplementary Fig. 1). The soil microbiome in dry soils exhibited lower potential Gmass compared to that in humid soils (Fig. 1b), likely supporting the trade-offs between growth and adaptation to drought stress33,34. Desiccation-tolerant microorganisms produce and secrete mixtures of compatible solutes to protect against drought, safeguarding sensitive molecules and cells from desiccation damage35,36. Drought resistance, achieved by active osmoregulation, requires significant C investment due to the high energetic cost of osmolyte production37,38. Therefore, the soil microbiome exposed to water stress likely possesses traits associated with drought tolerance at the cost of growth29.
a One-way analysis of variance tests the differences in microbial biomass-specific potential growth rates (Gmass) among ecosystems of desert grassland (n = 33), temperate grassland (n = 45), alpine grassland (n = 69), tropical forest (n = 24), subtropical forest (n = 57), temperate forest (n = 54), boreal forest (n = 30), and alpine forest (n = 24) (One-way analysis of variance with two-sided test: F7,328 = 52.61, P < 0.001). Lowercase letters above the box indicate a significant difference among ecosystems (Duncan multiple comparisons with two-sided test). Center line is the median. Box limits are the upper and lower quartiles. Whiskers are 1.5 times interquartile range. b Relationship between Gmass and aridity index (Two-sided test, y = 0.76ln(x) – 2.49). MBC is microbial biomass carbon. c Relationship between Gmass and mean annual temperature within grasslands (Two-sided test, y = −0.12x + 0.10) and forests (Two-sided test, y = −0.02x + 0.71). ln is natural logarithm. Source data are provided as a Source Data file.
a Relationship between microbial biomass-specific potential growth rate (Gmass) and climate, edaphic factors, microbial resources, and microbial community structure (Two-sided test). b Partial correlation showing the effect of community aggregated traits on potential Gmass after controlling for climate, edaphic factors, microbial resources, and microbial community structure (Two-sided test). C is carbon, N is nitrogen, and P is phosphorus. * is 0.01 < P < 0.05, ** is 0.001 < P < 0.011, *** is P < 0.001. Source data are provided as a Source Data file.
Despite the weak correlation between potential Gmass and mean annual temperature (Fig. 2a), the negative effect of mean annual temperature on potential Gmass was observed in both forest and grassland ecosystems (Fig. 1c), corroborating the latitudinal/temperature compensation hypothesis10,11,12. Adaptation to the short growing seasons of cold ecosystems is another potential explanation for this negative relationship between potential Gmass and mean annual temperature. The soil microbiome in frozen soils is predominantly limited by the absence of liquid water rather than low temperature39. Thousands of psychrophilic microbial taxa adapt to cold environments through metabolic and structural adjustments40, as indicated by high microbial diversity, biomass, and metabolic activity during winter in cold ecosystems23,41,42. Additionally, dormancy is a prevalent life history strategy among microbes43,44. Soil microbial species may achieve cold resistance through dormancy, while stress avoidance is typically linked with high growth rates45,46. We found that the optimum temperature of the microbial community in warm regions is greater than that in cold regions, and that the optimum temperature also had a significant negative effect on potential Gmass. Consequently, the soil microbiome of cold ecosystems has adapted to low temperature and short growing seasons with a high potential growth rate.
Microbial C:P and N:P adversely affected the potential Gmass (Fig. 2b), supporting the growth rate hypothesis47. These patterns were consistent with a previous study that microbial growth of isolated strains was negatively correlated with their biomass C:P and N:P from leaf litter in a semi-arid Mediterranean grassland ecosystem48. High-latitude regions had greater soil P than that in low-latitude regions (Supplementary Fig. 2). We also found a negative effect of soil P on microbial C:P and N:P (Supplementary Fig. 2). Therefore, P limitation may also explain the low potential Gmass in low-latitude regions with warm climates. It is noteworthy that the biomass N:P is not necessarily equal to the N:P ratio derived from RNA and protein contents49, because the storage of elements in excess, their reuse, and other processes may cause significant variation in actual biomass N:P ratios49,50. These mechanisms probably result in small correlation coefficients between potential microbial growth and biomass C:P or N:P (Fig. 2), consistent with a previous study48.
The potential Gmass of the soil microbiome peaked in neutral pH-balanced soils (Supplementary Fig. 3), indicating that extreme pH directly impacts microbial physiology, reducing growth or selecting for microbes investing in pH stress-tolerant traits at the expense of fast growth potential. At local, regional, and global scales, soil pH was found to be the dominant factor regulating the diversity, community structure, and community aggregated traits of the soil microbiome28,32,51,52,53. Soil pH influences membrane-bound proton pumps and protein stability; extreme pH levels can impose physiological stress on microorganisms, reducing the net growth of specific taxa when the soil pH deviates from a certain range51,54.
There are expected positive correlations between potential Gmass and soil resources (soil organic C, dissolved organic C, total N, total P, and available N; Fig. 2a) because the growth of almost all organisms is resource limited evolutionarily. Potential Gmass was more closely related to C than to nutrients (Fig. 2a), supporting the idea that microbial growth is primarily limited by C and only secondarily by nutrients55. We also found a positive correlation between potential Gmass and clay plus silt content, probably because clay- and silt-size minerals increase the capacity of soils to hold water, C, and nutrients.
Our measurements of microbial growth aggregate responses from thousands of individual microbial species, each growing at different rates. Understanding individual taxonomic groups may provide insights into how these microorganisms will impact soil C cycling under future climate change scenarios6,56. In this study, both dominant phyla of fungi and bacteria were found to influence microbial potential Gmass. Specifically, potential Gmass was positively correlated with Basidiomycota, Acidobacteriota, and Proteobacteria, but negatively correlated with Ascomycota, Actinobacteriota, and Gemmatimonadota (Fig. 2a). The former phyla predominantly occur in humid, resource-rich soils (high organic matter and nutrients), whereas the latter phyla are prevalent in regions with dry, resource-poor, hypersaline, and alkaline soils (Fig. 2a).
Partial least squares path modeling showed that microbial community structure (dominant phyla of fungi and bacteria) influenced potential Gmass indirectly by altering microbial traits (genome size, rrn copy number, optimum temperature, and biomass stoichiometry) rather than through a direct pathway (Fig. 3; Supplementary Table 1; the insignificant pathway between microbial community structure and potential Gmass was removed from the model). Genome size is a crucial life-history trait impacting the abundance, growth rate, metabolic activity, and dispersal capability of soil microbiome28,30,32. A recent global study28 and a recent national-scale survey across the US found a negative correlation between genome size and soil pH57. Consistent with these two studies, we found that humid regions with acidic soils had greater bacterial genome sizes than arid regions (Supplementary Figs. 3 and 4). However, fungal genome size peaked in neutral soils with moderate humidity (Supplementary Figs. 3 and 4). Therefore, the universal pattern of fungal genome size along environmental gradient requires further study. The consistent positive effects of fungal and bacterial genome size on potential Gmass align with a recent proposal highlighting the dominant role of genome size in shaping the first dimension of life history strategies, ranging from streamlined genomes with simple metabolisms to larger genomes with expanded metabolic capacities28. However, theoretical and laboratory studies suggest that streamlined genomes are associated with rapid growth, as smaller genomes reduce the nutrient demand for genome replication58,59,60. Together with findings from Piton et al.28, we suggest that the growth rate of the soil microbiome in nature may be less constrained by genome size than by environmental stress and community interactions.
The partial least squares path model depicts factors influencing microbial potential growth rates through direct and indirect pathways. Blue and red arrows indicate positive and negative effects, respectively, while the indicated values on the arrows are the path coefficients for the inner model. The path coefficients for outer models of the partial least squares path modeling are shown in Supplementary Table 1. C is carbon, N is nitrogen, and P is phosphorus. The soil microbiome in acid-neutral soils with high organic matter and nutrients (resource-rich) in humid regions, dominated by Basidiomycota, Acidobacteriota, and Proteobacteria, exhibits a large genome size and low biomass C:P and N:P ratios, indicating a high potential growth rate. Conversely, in resource-poor, dry, and hypersaline soils, the microbiome, dominated by Ascomycota, Actinobacteriota, and Gemmatimonadota, displays a lower potential growth rate, suggesting that resource acquisition and stress tolerance tradeoff with growth rate. Source data are provided as a Source Data file.
Spatial isolation is another important factor limiting microbial growth in soil matrices61. A larger genome size would also increase the metabolic pathways in such limited and variable space. Furthermore, the soil microbiome produces extracellular enzymes to break down complex resources, and the subsequent uptake of dissolved organic C depends on various membrane transporters29. Metabolic complexity may favor soil microbiomes in producing diverse enzymes and in uptake of diverse substrates, thus promoting microbial growth. Additionally, rapid microbial growth requires a substantial increase in cellular ribosomes compared to slow growth, as multiple rrn copies in genomes help to effectively amplify rRNA gene dosage during rapid growth62. Therefore, the rrn copy number has been suggested as a predictor of microbial growth rate62, and a positive correlation between potential Gmass and rrn copy number is shown here (Fig. 2b).
Our national-scale study had several limitations. First, our calculation of genome size was based on species-specific traits from NCBI and GTDB datasets (see Methods). However, the genome sizes of many microbial species were not reported in current datasets. Second, our measurements of microbial growth were conducted under optimal conditions, which may differ from patterns observed in the field. Although the 18O-labeled water approach has been used to study microbial growth in the field56, this method is challenging to implement on a national scale.
Implications for microbial strategies and soil C cycling
The spatial variation of microbial growth, driven by climate, edaphic properties, microbial resources, community structure, and genome traits, aligns with several macroecological theories. These include the latitudinal/temperature compensation hypothesis, the trade-off between growth and stress survival, and the growth rate hypothesis. While omics datasets have been extensively utilized to quantify traits associated with microbial life history strategies, such as the competitor-stress tolerant-ruderal28,63 and high yield-resource acquisition-stress tolerance triangle29, growth remains a challenging property to extract from these datasets due to an incomplete understanding of its genetic determinants29. Additionally, maximum potential growth rates estimated through genomic analyses or culture-based experiments64,65 may not accurately reflect actual growth rates in situ soils26,29,60. Our large-scale study shows that low resource availability and high stress decrease potential growth rates, partly consistent with the idea of trade-offs between growth and resource acquisition or stress tolerance29.
Microbial growth rate is a key parameter in biogeochemical models of C flux26. Our study offers a robust benchmark for microbial-explicit soil C models. Furthermore, accelerated microbial growth is likely to promote microbial necromass formation66, which has been suggested to be more physically protected in organo-mineral associations than plant-derived C, due to its closer proximity and interactions with the soil mineral matrix67,68. Recent evidence indicates a positive correlation between microbial C use efficiency and soil C storage69, suggesting that accelerated microbial growth could enhance soil organic C storage. This is expected through ongoing iterative cycles of microbial proliferation, growth, and death, particularly leading to the incorporation of higher amounts of microbial-derived C into stable formations, such as mineral-associated organic C66,70.
Methods
Sites and sampling
Complying with all regional laws and regulations for research sampling, we sampled a total of 112 sites, comprising 49 grasslands and 63 forests, across China from June to August 2022, including 11 desert grasslands, 15 temperate grasslands, 23 alpine grasslands, 8 tropical forests, 19 subtropical forests, 18 temperate forests, 10 boreal forests, and 8 alpine forests. At each site, we established three sample plots within well-protected areas to minimize the effects of anthropogenic disturbance. The plots in grasslands measured 5 × 5 m and were spaced more than 10 m apart, while the plots in forests measured 20 × 20 m and were spaced more than 30 m apart. We collected topsoil samples from the upper 0–20 cm layer at ten random locations within each plot. These samples were then combined to form a composite sample for each plot, yielding a total of 336 soil samples across China (Fig. 1a). We sieved the fresh soil to 2 mm, homogenized it, and removed fine roots and other plant debris in the field. We stored 1 kg of soil from each sample on ice and transported it to the laboratory within 48 h of collection. Once the soil samples were transported to the laboratory, each soil sample was homogenized and then divided into two subsamples. One subsample was air-dried, while the other subsample was stored at -20°C for microbial analyses. We also recorded the latitude, longitude, and elevation of the 112 studied sites. The mean annual temperature and precipitation were extracted from WorldClim 2.0. The De Martonne aridity index was calculated as the ratio of mean annual precipitation to mean annual temperature plus 10 °C.
Edaphic factor analyses
Soil pH was determined in a 1:2.5 soil-to-deionized water mixture and analyzed using a pH electrode (FE28-Standard, Mettler, Switzerland). Soil texture was measured with a Malvern laser particle size analyzer (MS2000; Malvern Instruments, Malvern, UK). Soil organic C was measured using a Multi N/C 2100 s analyzer (Analytik Jena AG, Germany) after the removal of inorganic C by hydrochloric acid. Soil available N (ammonium and nitrate) was extracted with 2 M potassium chloride. Soil total N was extracted by digesting the soil sample with sulfuric acid and adding copper sulfate and potassium sulfate, while total P was extracted by digesting the soil sample with sulfuric acid and perchloric acid. All extracted solutions were analyzed for N and P concentrations with an Auto Analyzer (BRAN + LUEBBE-AA3, Germany).
The funnel-filter paper-drainage method was used to measure the maximum water holding capacity71,72. Specifically, deionized water was first used to pre-saturate the filter paper in the funnel. Then 10 g of air-dried soil was placed into the wet filter paper funnel. The soil was saturated by wetting the outer rim of the exposed filter paper. After the soil was glistening and a small amount of water was showing on the surface, the funnel was covered with cling film with small holes to minimize evaporation. After 6 h, the drained soil was weighed and then oven-dried to obtain the dry weight.
Microbial biomass
Microbial biomass C, N, and P were measured using the chloroform fumigation extraction method73. The frozen soil (stored at −20 °C) was thawed on ice. The thawed soil was placed in a brown bottle for a 7-day preincubation at 25 °C in the dark with 60% water holding capacity. The bottle was covered with cling film with several holes, which allowed gas exchange and slowed down water evaporation. We adjusted the water-holding capacity on the fourth and seventh days during preincubation. Our measurements were conducted in winter with outdoor air temperature ranging from −10 to −30 °C, therefore, the indoor high temperature (keeping at ~25 °C) environment had very dry air. Therefore, we must adjust the water holding capacity every three or four days (a lot of previous studies adjust the water holding capacity every 1 week in carbon mineralization incubation experiment). We found that the water content of most soil samples had dropped lower than 50% water holding capacity before adding water. After that, the soil was fumigated or unfumigated for 24 h at 25 °C. The fumigated and unfumigated soil samples were used to extract C and N with 0.5 M potassium sulfate and P with 0.5 M sodium bicarbonate. The C concentration in the extracts was determined by a Multi N/C 2100 analyzer (Analytik Jena AG, Germany), while the N and P concentrations were analyzed with an Auto Analyzer (BRAN + LUEBBE-AA3, Germany). Microbial biomass C, N, and P were calculated as the difference between fumigated and nonfumigated subsamples multiplied by the respective conversion factors, i.e., 0.45, 0.45, and 0.40 for C, N, and P, respectively.
Microbial growth
We determined microbial growth using the substrate-independent method, which involves the incorporation of 18O from water into microbial genomic DNA (dsDNA74). Ten grams of soil was placed in a 50 ml tube for a 7 days preincubation at 25 °C in the dark with 60% water-holding capacity (as in the preincubation described above). We adjusted the water-holding capacity on the fourth day during preincubation. After 7 days preincubation, we placed duplicate aliquots of 500 mg of each preincubated soil into 2 mL brown chromatographic vials. One vial served as the natural 18O abundance control, and the other as the labeled sample. For one replicate, we adjusted the 18O content of soil water to 20.0 at% 18O by adding H218O, while the other replicate received the same volume of non-labeled water. Additional Millipore ultrapure water was added to maintain 60% water holding capacity. We then placed the vials in 20 ml headspace bottles, setting three blank bottles without soil for each batch of test samples as controls.
After 24 h of incubation at 25 °C in the dark with 60% water-holding capacity, we extracted DNA from both labeled and unlabeled soils using a DNA extraction kit (FastDNA™ SPIN Kit for Soil; MP Biomedicals). We determined the DNA concentration via the picogreen fluorescence assay. The remaining DNA extracts were pipetted into silver cups, dried at 45 °C for 5 h to remove all water, and folded into silver capsules for analysis. We measured the abundance of 18O and total O content using an IRMS-TC/EA (Thermo Scientific) at the Laboratory of Ecological Indicators Analysis (Institute of Geographic Sciences and Natural Resources Research, Chinese Academy of Sciences, Beijing, China).
Microbial DNA production was estimated by multiplying sample O content by the 18O excess of DNA relative to the natural abundance of 18O in DNA measured in unlabeled samples. A DNA‐oxygen content of 31.21% was applied to estimate the dsDNA produced (\({{\mbox{DNA}}}_{{\mbox{produced}}}\)) by microbial growth during the incubation period:
where \({{\mbox{O}}}_{{\mbox{Total}}}\) is the total O content of the dried DNA extract, \({{\mbox{at}}\%}_{{\mbox{excess}}}\) is the at% excess 18O of the labeled sample compared to the mean at% 18O of unlabeled samples, and \({{\mbox{at}}\%}_{{\mbox{final}}}\) is the 18O at% of soil water at the beginning of incubation (20.0% in our study). A conversion factor (\({f}_{{\mbox{DNA}}}\)) was calculated as the ratio of soil microbial biomass C to \({{\mbox{DNA}}}_{{\mbox{produced}}}\). Microbial growth was calculated based on the \({{\mbox{DNA}}}_{{\mbox{produced}}}\) and \({f}_{{\mbox{DNA}}}\):
where DW is the dry weight of soil and t is the incubation time.
Illumina sequencing and bioinformatic analysis
For the bacterial community, we amplified the 16S rRNA genes using primers 338 F (ACTCCTACGGGAGG CAGCAG) and 806 R (GGACTACHVGGGTWTCTAAT). For the fungal community, we amplified the ITS1 regions using primers ITS1F (CTTGGTCATTTAGAGGAAGTAA) and ITS2R (GCTGCGTTCTTCATCGATGC). We conducted PCR using TransStart Fastpfu DNA Polymerase (TransGen Biotech, Beijing, China) in a GeneAmp 9700 thermal cycler (Applied Biosystems, Foster City, CA, United States). The thermal cycling protocol included an initial denaturation step at 95 °C for 3 min, followed by 35 cycles for fungi and 27 cycles for bacteria of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s, with a final extension at 72 °C for 10 min. We extracted the PCR products from 2% agarose gels and concentrated them using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA). We sequenced the purified amplicons on an Illumina Novaseq PE250 platform (Illumina, San Diego, CA, USA) at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). We demultiplexed, quality-filtered, and processed reads using QIIME2. We determined the taxonomic assignment of the 16S rRNA and ITS sequences based on the bacterial SILVA reference database and fungal UNITE reference database using RDP Classifier.
Data analysis
We downloaded the datasets associated with genome size at the species level from NCBI and Madin et al.75. We integrated these two datasets to calculate the community aggregated bacterial genome size (weighted average by the abundance of individual species), while the dataset from NCBI was used to calculate fungal genome size. We also calculated the bacterial traits of rrn copy number and optimum temperature from Madin et al.75. We focused on the relative abundances of the dominant microbial populations at the phylum level. Spearman’s correlation was used to explore the relationships among variables (R package of Hmisc). Partial correlation was used to explore the effect of community aggregated traits on potential Gmass after controlling for climate, edaphic properties, microbial resources, and microbial community structure (R package of ppcor). Finally, partial least squares path modeling was used to explore the direct and indirect effects of climate, edaphic properties, microbial resources, microbial community structure, and community aggregated traits on potential Gmass (R package of plspm). Insignificant pathways were removed from the partial least squares path model. All the statistical analyses were conducted using R 4.3.276.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that supports the findings of this study is available in Supplementary Data 1. Source data are provided with this paper.
Code availability
The code that supports the findings of this study is available in Supplementary Code 1.
References
Shi, Z., Crowell, S., Luo, Y. & Moore, B. III Model structures amplify uncertainty in predicted soil carbon responses to climate change. Nat. Commun. 9, 2171 (2018).
Todd-Brown, K. et al. Changes in soil organic carbon storage predicted by Earth system models during the 21st century. Biogeosciences 11, 2341–2356 (2014).
Singh, B. K., Bardgett, R. D., Smith, P. & Reay, D. S. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 8, 779–790 (2010).
Ren, C. et al. Thermal sensitivity of soil microbial carbon use efficiency across forest biomes. Nat. Commun. 15, 6269 (2024).
Chandel, A. K., Jiang, L. & Luo, Y. Microbial models for simulating soil carbon dynamics: a review. J. Geophys. Res. Biogeosci. 128, e2023JG007436 (2023).
Purcell, A. M. et al. Decreased growth of wild soil microbes after 15 years of transplant‐induced warming in a montane meadow. Glob. Change Biol. 28, 128–139 (2022).
Calabrese, S., Chakrawal, A., Manzoni, S. & Van Cappellen, P. Energetic scaling in microbial growth. Proc. Natl. Acad. Sci. USA 118, e2107668118 (2021).
Li, B., Suzuki, J.-I. & Hara, T. Latitudinal variation in plant size and relative growth rate in Arabidopsis thaliana. Oecologia 115, 293–301 (1998).
Kempes, C. P., Dutkiewicz, S. & Follows, M. J. Growth, metabolic partitioning, and the size of microorganisms. Proc. Natl. Acad. Sci. USA 109, 495–500 (2012).
Levinton, J. S. The latitudinal compensation hypothesis: growth data and a model of latitudinal growth differentiation based upon energy budgets. I. Interspecific comparison of Ophryotrocha (Polychaeta: Dorvilleidae). Biol. Bull. 165, 686–698 (1983).
Yuan, Z. Y., Chen, H. Y. & Reich, P. B. Global-scale latitudinal patterns of plant fine-root nitrogen and phosphorus. Nat. Commun. 2, 344 (2011).
Conover, D. O. & Present, T. M. Countergradient variation in growth rate: compensation for length of the growing season among Atlantic silversides from different latitudes. Oecologia 83, 316–324 (1990).
Fletcher, L. R. et al. Testing the association of relative growth rate and adaptation to climate across natural ecotypes of Arabidopsis. N. Phytol. 236, 413–432 (2022).
Garcia, M. O. et al. Soil microbes trade-off biogeochemical cycling for stress tolerance traits in response to year-round climate change. Front. Microbiol. 11, 507526 (2020).
Grime, J. P. Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am. Nat. 111, 1169–1194 (1977).
Vile, D. et al. Leaf economics and slow-fast adaptation across the geographic range of Arabidopsis thaliana. Sci. Rep. 9, 10758 (2019).
Ramírez‐Valiente, J. A., López, R., Hipp, A. L. & Aranda, I. Correlated evolution of morphology, gas exchange, growth rates and hydraulics as a response to precipitation and temperature regimes in oaks (Quercus). N. Phytol. 227, 794–809 (2020).
Leites, L. P., Rehfeldt, G. E. & Steiner, K. C. Adaptation to climate in five eastern North America broadleaf deciduous species: Growth clines and evidence of the growth-cold tolerance trade-off. Perspect. Plant Ecol., Evol. Syst. 37, 64–72 (2019).
Vasseur, F. et al. Adaptive diversification of growth allometry in the plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 115, 3416–3421 (2018).
Jung, E.-Y., Gaviria, J., Sun, S. & Engelbrecht, B. M. J. Comparative drought resistance of temperate grassland species: testing performance trade-offs and the relation to distribution. Oecologia 192, 1023–1036 (2020).
Sorensen, J. G., White, C. R., Duffy, G. A. & Chown, S. L. A widespread thermodynamic effect, but maintenance of biological rates through space across life’s major domains. Proc. R. Soc. B-Biol. Sci. 285, 1775 (2018).
Elser, J. et al. Biological stoichiometry from genes to ecosystems. Ecol. Lett. 3, 540–550 (2000).
Xu, X., Thornton, P. E. & Post, W. M. A global analysis of soil microbial biomass carbon, nitrogen and phosphorus in terrestrial ecosystems. Glob. Ecol. Biogeogr. 22, 737–749 (2013).
Delgado-Baquerizo, M. et al. Aridity decouples C: N: P stoichiometry across multiple trophic levels in terrestrial ecosystems. Ecosystems 21, 459–468 (2018).
Li, P., Yang, Y., Han, W. & Fang, J. Global patterns of soil microbial nitrogen and phosphorus stoichiometry in forest ecosystems. Glob. Ecol. Biogeogr. 23, 979–987 (2014).
Caro, T. A., McFarlin, J., Jech, S., Fierer, N. & Kopf, S. Hydrogen stable isotope probing of lipids demonstrates slow rates of microbial growth in soil. Proc. Natl. Acad. Sci. USA 120, e2211625120 (2023).
Violle, C. et al. Let the concept of trait be functional! Oikos 116, 882–892 (2007).
Piton, G. et al. Life history strategies of soil bacterial communities across global terrestrial biomes. Nat. Microbiol. 8, 2093–2102 (2023).
Malik, A. A. et al. Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. ISME J. 14, 1–9 (2020).
Westoby, M. et al. Cell size, genome size, and maximum growth rate are near-independent dimensions of ecological variation across bacteria and archaea. Ecol. Evol. 11, 3956–3976 (2021).
Chen, Y., Neilson, J. W., Kushwaha, P., Maier, R. M. & Barberán, A. Life-history strategies of soil microbial communities in an arid ecosystem. ISME J. 15, 649–657 (2021).
Wang, C. et al. Bacterial genome size and gene functional diversity negatively correlate with taxonomic diversity along a pH gradient. Nat. Commun. 14, 7437 (2023).
Skirycz, A. & Inzé, D. More from less: plant growth under limited water. Curr. Opin. Biotechnol. 21, 197–203 (2010).
Schimel, J. P. Life in dry soils: effects of drought on soil microbial communities and processes. Annu. Rev. Ecol. Evol. Syst. 49, 409–432 (2018).
Narvaez-Reinaldo, J. J., Barba, I., Gonzalez-Lopez, J., Tunnacliffe, A. & Manzanera, M. Rapid method for isolation of desiccation-tolerant strains and xeroprotectants. Appl. Environ. Microbiol. 76, 5254–5262 (2010).
Manzanera, M. Dealing with water stress and microbial preservation. Environ. Microbiol. 23, 3351–3359 (2021).
Manzoni, S., Schaeffer, S. M., Katul, G., Porporato, A. & Schimel, J. P. A theoretical analysis of microbial eco-physiological and diffusion limitations to carbon cycling in drying soils. Soil Biol. Biochem. 73, 69–83 (2014).
Schimel, J., Balser, T. C. & Wallenstein, M. Microbial stress-response physiology and its implications for ecosystem function. Ecology 88, 1386–1394 (2007).
Clarke, A. The thermal limits to life on Earth. Int. J. Astrobiol. 13, 141–154 (2014).
Margesin, R. & Collins, T. Microbial ecology of the cryosphere (glacial and permafrost habitats): current knowledge. Appl. Microbiol. Biotechnol. 103, 2537–2549 (2019).
Yin, S., Liang, G., Wang, C. & Zhou, Z. Asynchronous seasonal patterns of soil microorganisms and plants across biomes: a global synthesis. Soil Biol. Biochem. 175, 108859 (2022).
Zifcakova, L., Vetrovsky, T., Howe, A. & Baldrian, P. Microbial activity in forest soil reflects the changes in ecosystem properties between summer and winter. Environ. Microbiol. 18, 288–301 (2016).
Roszak, D. B. & Colwell, R. R. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 51, 365–379 (1987).
Jones, S. E. & Lennon, J. T. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. USA 107, 5881–5886 (2010).
Berger, J., Palta, J. & Vadez, V. Review: an integrated framework for crop adaptation to dry environments: Responses to transient and terminal drought. Plant Sci. 253, 58–67 (2016).
Salazar, A., Sulman, B. N. & Dukes, J. S. Microbial dormancy promotes microbial biomass and respiration across pulses of drying-wetting stress. Soil Biol. Biochem. 116, 237–244 (2018).
Michaels, A. F. Ecological stoichiometry-The biology of elements from molecules to the biosphere. Science 300, 906–907 (2003).
Mouginot, C. et al. Elemental stoichiometry of Fungi and Bacteria strains from grassland leaf litter. Soil Biol. Biochem. 76, 278–285 (2014).
Karpinets, T. V., Greenwood, D. J., Sams, C. E. & Ammons, J. T. RNA: protein ratio of the unicellular organism as a characteristic of phosphorous and nitrogen stoichiometry and of the cellular requirement of ribosomes for protein synthesis. BMC Biol. 4, 1–10 (2006).
Mooshammer, M., Wanek, W., Zechmeister-Boltenstern, S. & Richter, A. Stoichiometric imbalances between terrestrial decomposer communities and their resources: mechanisms and implications of microbial adaptations to their resources. Front. Microbiol. 5, 22 (2014).
Zhou, Z., Wang, C. & Luo, Y. Meta-analysis of the impacts of global change factors on soil microbial diversity and functionality. Nat. Commun. 11, 3072 (2020).
Rousk, J. et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351 (2010).
Fierer, N. & Jackson, R. B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 103, 626–631 (2006).
Luan, L. et al. Integrating pH into the metabolic theory of ecology to predict bacterial diversity in soil. Proc. Natl. Acad. Sci. USA 120, e2207832120 (2023).
Soong, J. L. et al. Microbial carbon limitation: the need for integrating microorganisms into our understanding of ecosystem carbon cycling. Glob. Change Biol. 26, 1953–1961 (2020).
Purcell, A. M. et al. Rapid growth rate responses of terrestrial bacteria to field warming on the Antarctic Peninsula. ISME J. 17, 2290–2302 (2023).
Chuckran, P. F. et al. Edaphic controls on genome size and GC content of bacteria in soil microbial communities. Soil Biol. Biochem. 178, 108935 (2023).
Lynch, M. Streamlining and simplification of microbial genome architecture. Annu. Rev. Microbiol. 60, 327–349 (2006).
Hessen, D. O., Jeyasingh, P. D., Neiman, M. & Weider, L. J. Genome streamlining and the elemental costs of growth. Trends Ecol. Evol. 25, 75–80 (2010).
Li, J. et al. Predictive genomic traits for bacterial growth in culture versus actual growth in soil. ISME J. 13, 2162–2172 (2019).
Zhou, J. et al. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68, 326–334 (2002).
Roller, B. R., Stoddard, S. F. & Schmidt, T. M. Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat. Microbiol. 1, 1–7 (2016).
Krause, S. et al. Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front. Microbiol. 5, 251 (2014).
Long, A. M., Hou, S., Ignacio-Espinoza, J. C. & Fuhrman, J. A. Benchmarking microbial growth rate predictions from metagenomes. ISME J. 15, 183–195 (2021).
Weissman, J. L., Hou, S. & Fuhrman, J. A. Estimating maximal microbial growth rates from cultures, metagenomes, and single cells via codon usage patterns. Proc. Natl. Acad. Sci. USA 118, e2016810118 (2021).
Prommer, J. et al. Increased microbial growth, biomass, and turnover drive soil organic carbon accumulation at higher plant diversity. Glob. Change Biol. 26, 669–681 (2020).
Sokol, N. W., Sanderman, J. & Bradford, M. A. Pathways of mineral-associated soil organic matter formation: Integrating the role of plant carbon source, chemistry, and point of entry. Glob. Change Biol. 25, 12–24 (2019).
Liang, C., Schimel, J. P. & Jastrow, J. D. The importance of anabolism in microbial control over soil carbon storage. Nat. Microbiol. 2, 17105 (2017).
Tao, F. et al. Microbial carbon use efficiency promotes global soil carbon storage. Nature 618, 981–985 (2023).
Zhou, Z. et al. Global turnover of soil mineral-associated and particulate organic carbon. Nat. Commun. 15, 5329 (2024).
Robertson, G. P. Standard soil methods for long-term ecological research vol. 2 (Oxford University Press, 1999).
Nelson, J. T. et al. A simple, affordable, do-it-yourself method for measuring soil maximum water holding capacity. Communications in Soil Science and Plant Analysis, 1–15 (2024).
Brookes, P., Landman, A., Pruden, G. & Jenkinson, D. Chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 17, 837–842 (1985).
Spohn, M., Klaus, K., Wanek, W. & Richter, A. Microbial carbon use efficiency and biomass turnover times depending on soil depth-Implications for carbon cycling. Soil Biol. Biochem. 96, 74–81 (2016).
Madin, J. S. et al. A synthesis of bacterial and archaeal phenotypic trait data. Sci. Data 7, 170 (2020).
Team, R. C. R: A language and environment for statistical computing, R Foundation for Statistical, Vienna, Austria. Computing (2023).
Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (2021YFD2200401) to Chuankuan Wang, the National Natural Science Foundation of China (42377345 to Chengjie Ren and 31901293 to Zhenghu Zhou), and the Fundamental Research Funds for the Central Universities (2572021CG07) to Zhenghu Zhou.
Author information
Authors and Affiliations
Contributions
Z.Z., C. W., and C.R. conceived and designed the project; C.R. collected the data; Z.Z. conducted the data analyses and wrote the manuscript; Z.Z., C.W., X.C., T.Z., X.P., F.Z., X.H., G.Y., G.W., and C.R. contributed to discussing the results, writing, and editing the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Gabin Piton, Wolfgang Wanek and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhou, Z., Wang, C., Cha, X. et al. The biogeography of soil microbiome potential growth rates. Nat Commun 15, 9472 (2024). https://doi.org/10.1038/s41467-024-53753-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53753-w
This article is cited by
-
The rhizosphere exhibits higher microbial carbon use efficiency compared to bulk soil
Biology and Fertility of Soils (2026)
-
Experimental mapping of bacterial fitness landscapes reveals eco-evolutionary fingerprints
Scientific Reports (2025)