Abstract
Zoonotic infections (swine-human) caused by influenza A viruses (IAVs) have been reported and linked to close contact between these species. Here, we describe eight human IAV variant infections (6 mild and 2 severe cases, including 1 death) detected in Paraná, Brazil, during 2020–2023. Genomes recovered were closely related to Brazilian swIAVs of three major lineages (1 A.3.3.2/pdm09, 1B/human-like, and H3.1990.5), including three H1N1v, two H1N2v, two H3N2v and one H1v. Five H1v were closely related to pdm09 lineage, one H1v (H1N2v) grouped within 1B.2.3 clade, and the two H3v grouped within a clade composed exclusively of Brazilian H3 swIAV (clade H3.1990.5.1). Internal gene segments were closely related to H1N1pdm09 isolated from pigs. IAV variant rarely result in sustained transmission between people, however the potential to develop such ability is of concern and must not be underestimated. This study brings into focus the need for continuous influenza surveillance and timely risk assessment.
Similar content being viewed by others
Introduction
Continued influenza surveillance remains important especially since the emergence of a novel Influenza A virus (IAV) with pandemic potential continues to be a worldwide threat. The first pandemic event of this century was caused by a triple reassortant virus, named 2009 pandemic influenza A H1N1 virus (H1N1pdm09), which was simultaneously reported in Mexico and USA and spread rapidly across the globe1,2,3. IAV is an enveloped virus which belongs to the Orthomyxoviridae family and has an octa-segmented genome composed of single strand negative sense RNA4. The eight gene segments are composed of hemagglutinin (HA) and neuraminidase (NA), which encode the surface glycoproteins; and the internal genes polymerase basic 1 and 2 (PB1 and PB2), polymerase acidic (PA), and nucleoprotein (NP), which encode the ribonucleoprotein (RNP) complex, matrix gene (M) and the non-structural protein (NS)4. Currently, 18 HA (H1–H18) and 11 NA subtypes (N1–N11) are known, including H17N10 and H18N11 described in fruit bats in Central and South America5,6. During the replication cycle of IAV, two main evolutionary processes can occur: antigenic drift, characterized by point mutations due to the absence of polymerase proofreading activity that causes antigenic changes commonly observed in influenza seasonal viruses; and antigenic shift, when two or more distinct viruses infect the same cell and generate a new virus with a novel gene constellation, which can be potentially pandemic4.
The IAVs are continuously evolving, eventually resulting in a new strain causing epidemics and requiring annual revision of influenza vaccine composition for the Northern and Southern Hemispheres. Despite these efforts, the major concern regarding a potential influenza pandemic event is associated with animal-human transmission7. Better understanding the dynamics of this interface is crucial to identify and mitigate influenza cross-species transmission. Swine are susceptible to avian and human influenza viruses since they have compatible sialic acid (SA) receptor binding sites for both avian (alpha ɑ−2.3) and human (ɑ−2.6) viruses. This facilitates reassortment events among avian/swine/human strains, generating new viruses that can be transmitted to humans8,9. During 2009, the emergence of the IAV H1N1pdm09 as a pandemic strain heightened public concern about the bidirectional transmission of IAV between humans and swine, recognized as a major, hard to control, global threat. During 2012, over 300 human cases of swine-origin IAV (H3N2)v were reported in the United States, predominantly acquired through close contact with pigs at agricultural fairs, leading to 11 hospitalizations10,11,12.
The IAV H1N1 and H3N2 are both established subtypes in humans, with a seasonal and well-described circulation worldwide. However, swine IAVs (swIAVs) are occasionally transmitted to humans, especially due to close contact with these animals. In this case, a “v” from “variant” is added to the virus subtype identifier, indicating its swine origin11. The subtypes H1N1, H1N2, and H3N2 are enzootic in swine populations, causing contagious respiratory disease in pigs in many regions of the world13. These three subtypes are reservoirs of a vast genetic diversity that is mainly the result of reverse-zoonosis events, i.e., when human seasonal viruses are transmitted to pigs, followed by sustained transmission among these animals, and establishing novel and distinct lineages of human-origin IAVs in swine14,15. The genetic classification of swIAVs follows a phylogenetic nomenclature system that classifies swIAV H1 into three major lineages: 1 A or classical swine lineage, 1B or human seasonal lineage, and 1 C or Eurasian avian lineage. Swine H3N2 viruses are classified by decade of introduction of human seasonal influenza viruses16. Although swIAVs are genetically related to human IAV, they are mainly limited to their reservoir host and only sporadically cross the host range barrier and infect humans17. It has occurred with IAV H1N1pdm09, a virus bearing gene segments from classical swine lineage (H1, NP, and NS), human seasonal viruses (PB1), North American avian-like swine viruses (PA and PB2), and Eurasian avian-like swine viruses (M and NA)18,19. Viruses with a similar genomic constellation circulated in the North American swine population from 2005 to 2009, causing sporadic zoonotic transmission events1,20,21.
As recommended by the Brazilian Ministry of Health (MoH), influenza A or B not subtyped by the influenza panel that is available to the LACEN are immediately sent to a World Health Organization (WHO) National Influenza Centre (NIC), such as the one located at Oswaldo Cruz Institute, Fiocruz, Rio de Janeiro for complementary molecular and phenotypic analysis22,23. Whenever a new variant virus is identified, the MoH, Pan American Health Organization (PAHO)/WHO and the International Health Regulation (IHR) must be notified. These organizations immediately team up with local authorities to conduct a thorough epidemiological investigation that aims at providing actionable results to support public health control measures. The Brazilian Agricultural Research Corporation (EMBRAPA) Swine and Poultry monitors influenza in the swine population since 2009, when outbreaks of acute respiratory infections in pigs became more frequent, and were associated with the IAV H1N1pdm0924,25. In the following years, H1N2 and H3N2 IAVs began to be identified26,27,28, and subsequent studies revealed a comprehensive genetic diversity of Brazilian swIAVs that was mostly shaped by many incursions of human seasonal influenza viruses. Four genetic lineages of H1 viruses circulate in pigs in Brazil: 1 A.3.3.2 (H1pdm09), 1B.2.3, 1B.2.4 and 1B.2.626,28. H1N1pdm09 viruses were repeatedly introduced into swine in Brazil since the beginning of the 2009 pandemic but only a few of these introductions resulted in onward transmission in swine26. In contrast, it was estimated that Brazilian swIAV of 1B lineage emerged from three distinct human-to-swine transmission events that occurred in the mid-1980s (1B.2.6), early 2000s (1B.2.4), and mid-2000s (1B.2.3). Similarly, swIAV H3 emerged in the late-1990s after a human seasonal IAV H3N2 spillover to swine27,28. Regarding the NA, an even bigger genetic diversity was observed, including distinct N2 (six) and N1 (one) incursions, all of them derived from pre-2009 human seasonal H1N1, H1N2 and H3N2 viruses27. Viral diversity increased after several introductions of H1N1pdm09 virus into swine, followed by reassortment with endemic swIAVs and replacement of all the internal genes27,28. These surveillance efforts in Brazil were fundamental to describe a case of influenza zoonotic transmission that occurred in Castro, a municipality in Southern Brazil, in 2015: an H1N2v that was detected in a 16-year-old girl, only three months after she started working in a pig farm29. Events like this reinforce the need for integrated surveillance under the One Health approach. Since 2005, WHO has reported 38 cases of influenza A(H1N2)v, 18 IAV (H1N1)v and 439 IAV (H3N2)v30. In this study, we describe in detail eight cases of zoonotic infections of swIAV in humans that occurred in Brazil from 2020 to 2023.
Results
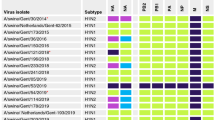
Between April 2020 and December 2023, eight cases of human infections with variant IAVs were detected in four municipalities in Paraná State, Southern Brazil: Ibiporã, Irati, Toledo, and Santa Helena (Fig. 1). These cities have intensive swine production and, except for case 8, most cases were reported to have direct or indirect contact with swine. One H1N2v was identified in Ibiporã (case 1), one H1N2v in Irati (case 2), three H1N1v (cases 3, 6 and 8) and one H1v (case 7) in Toledo, and two H3N2v (cases 4 and 5) in Santa Helena. All variant viruses were found closely related to Brazilian swIAVs of three major lineages (H1-1A.3.3.2 or pdm09, H1-1B or human-like, and H3 1990.5) (Figs. 2 to 6). In Table 1, we summarized the main epidemiological features of the eight cases. Detailed epidemiological data for each case is described in Supplementary Material and RT-PCR details for Influenza are in Supplementary Table 1. No respiratory virus co-detections were found in the analyzed samples.
a Locations in the State of Paraná, Brazil, where cases were detected. Concentric circles depict the proportion of cases identified in each location. b Genetic composition of viruses identified in the swine-to-human transmissions. The vertical bars correspond to the colors of the locations in (a). Horizontal bars identify the eight genomic segments of IAVs: (i) PB2, (ii) PB1, (iii) PA, (iv) HA, (v) NP, (vi) NA, (vii) MP, and (viii) NS. The figure key shows the subtype of each segment.
Swine-to-human transmission event in Ibiporã: H1N2
Case 1 (#3625/2020/H1N2) was detected in 2020, in a pig slaughterhouse worker that lived in Ibiporã. The HA from this variant virus clustered with swIAVs that belong to the genetic lineage 1B.2.3 (SH-aLRT 100% and UFBoot 100%) (Fig. 2). This lineage emerged in swine after a human-to-swine spillover event that occurred between 2004 and 200927,28. Phylogenetically, the IAVs from clade 1B.2.3 are most closely related to human seasonal H1N1 influenza viruses that circulated in humans during 2006 and were later replaced by the H1N1pdm09. Curiously, the first IAV variant reported in Brazil, A/Parana/720/2015 (H1N2)v, was related to another Brazilian clade (1B.2.4) which arose from a human-to-swine transmission event that occurred between 2001 and 200326,28. Both clades are well established in swine herds in Brazil. On the N2 phylogeny, the variant #3625/2020/H1N2 clustered within the largest Brazilian N2 clade (previously characterized as Clade N2-#4)28, and close to the variant #28600/2020/H1N2 (case 2) (Fig. 3B). The internal gene segments of Case 1 virus were closely related to swIAVs of the H1N1pdm09 lineage and displayed a distant evolutionary relationship with other cases (Table 2, Supplementary Figs. 1–6).
Swine sequences isolated in Brazil contains the state of isolation within its names (Brazil_MG = Minas Gerais, Brazil_MS = Mato Grosso do Sul, Brazil_PR = Paraná, Brazil_RS = Rio Grande do Sul, Brazil_SC = Santa Catarina, Brazil_SP = São Paulo). Branches are colored by host as shown in the figure key. The tree is midpoint rooted for clarity, and all branch lengths are drawn to scale representing nucleotide substitutions per site. Black-circles at the internal nodes identify clades supported by SH-aLRT > 80% and/or UFB > 95%. A Phylogenetic tree showing the three genetic clades of H1 viruses of the 1B lineage that circulate in swine in Brazil (1B.2.3, 1B.2.4, and 1B.2.6). The swine-to-human introduction is highlighted by a gray-shaded form. B Topology details for Clade H1 1B.2.3 that comprises the HA of A/Parana/3625/2020(H1N2)v.
Swine sequences isolated in Brazil contains the state of isolation within its names (Brazil_PR = Paraná, Brazil_RS = Rio Grande do Sul, Brazil_SC = Santa Catarina). The tree is midpoint rooted for clarity. Branches are colored by host and cases are color-coded according to the figure key. Branch lengths are drawn to scale representing nucleotide substitutions per site. Black-circles at the internal nodes identify clades supported by SH-aLRT > 80% and/or UFB > 95%. A Phylogenetic tree indicating the swine-to-human introductions occurred in Brazil by gray-shaded forms. B Topology details for N2 #4 Brazilian clade. C Topology details for N2 #2 Brazilian clade.
Case 1 patient sought medical assistance and was fully recovered after treatment. Notably, one co-worker presented similar symptoms at the same week, and 218 employees of the slaughterhouse presented ILI symptoms within a window of one month before and two months after Case 1 diagnostic (Supplementary material). Despite the lack of sampling, the possibility that other individuals have been infected by this H1N2v, or the occurrence of human-to-human transmission, should not be dismissed.
Swine-to-human transmission event in Irati: H1pdm N2
Case 2 (#28600/2020/H1pdmN2) was detected in a girl living in a farm with livestock production (cattle, sheep, poultry, and swine) in the Irati municipality. The HA segment of this variant clustered within a well-characterized clade of H1N1pdm09 viruses circulating among swine in Brazil26, but it’s not directly related to any of the other cases described here (Fig. 4). On the N2 phylogeny, Case 2 was directly linked to another variant sample already described previously (A/Parana/720/2015/H1N2)29 isolated in 2015 in the municipality of Castro (Fig. 3), 127 km from Irati. These two samples clustered within the largest Brazilian swine N2 clade (Fig. 3B)27. All internal segments of Case 2 belong to the H1N1pdm09 clade, and similarly to the HA segment, all of them show a distant evolutionary relationship to the other cases identified in this study (Supplementary Figs. 1–6, Table 2). This result indicates that different lineages of H1N1pdm09 are being transmitted from swine to humans in Paraná.
Swine sequences isolated in Brazil contains the state of isolation within its names (Brazil_PR = Paraná, Brazil_RS = Rio Grande do Sul, Brazil_SC = Santa Catarina,). Branches are colored by host and cases are color-coded according to the figure key. Branch lengths are drawn to scale representing nucleotide substitutions per site. Black-circles at the internal nodes identify clades supported by SH-aLRT > 80%. A Phylogenetic tree indicating swine-to-human introductions occurred in Brazil by gray-shaded forms. B Topology details for Clades H1 pdm #1 and #2. c Topology details for Clade H1 pdm #3.
Swine-to-human transmission event in Toledo: H1pdm N1pdm
Cases 3 (#10835/2021/H1N1), 6 (#20675/2022/H1N1) and 8 (#138697/2023/H1N1) were all identified in Toledo. Together with Case 7, for which only the HA segment could be sequenced, all samples were classified within the H1N1pdm09 lineage, including its internal segments (Figs. 4, 5 and S1–S6, Table 2).
The HA segment of Case 3 falls within a clade composed of human and swine sequences but presents no evolutionary relationship with any of the other variant samples described in this study (Fig. 4C). On the other hand, the NA segment grouped in a well-supported clade with Case 6 and Case 8 (Fig. 5B) inside of a well-characterized H1N1pdm09 swine cluster26. Corroborating this result, all the phylogenetic reconstructions for the internal segments suggest a close evolutionary relationship of Cases 3, 6 and 8. The uneven phylogenetic relationship between HA and all the other segments could be explained by reassortment, most likely prior to the swine-to-human transmission26.
Swine sequences isolated in Brazil contains the state of isolation within its names (Brazil_MG = Minas Gerais, Brazil_PR = Paraná, Brazil_RS = Rio Grande do Sul, Brazil_SC = Santa Catarina). Branches are colored by host and cases are color-coded according to the figure key. Branch lengths are drawn to scale representing nucleotide substitutions per site. Black-circles at the internal nodes identify clades supported by SH-aLRT > 80%. A Phylogenetic tree indicating swine-to-human introductions occurred in Brazil by gray-shaded forms. B Topology details for Clade N1 pdm #1.
The HA segments of Case 6, Case 7 and Case 8 grouped together in a well-supported clade that shares a common ancestor with swine sequences (Fig. 4). This swine cluster was previously characterized and represents one of the major H1N1pdm09 lineage to be circulating among swine in Brazil26. The close relationship among these three cases is corroborated by all phylogenetic reconstructions of the available internal segments (Supplementary Figs. 1–6). The accordant clustering and by assuming that these are not related human infections indicate that this lineage is circulating among pigs without any reassortment. The recognition that this lineage is being maintained in the swine population and managed to be transmitted from swine to humans, on at least three different occasions, is alarming and suggests that this combination of genes may facilitate the jump of the virus between the two species.
Swine-to-human transmission event in Santa Helena: H3N2
In July 2021, two human cases involving an H3N2v virus were detected in individuals from the same family, a mother (Case 4, #440706/2021) and her son (Case 5, #51528/2021), residents of Santa Helena (Fig. 1). The two H3N2v strains contain identical gene segments, although the N2 NA of the variant in Case 4 could not be successfully sequenced. On the H3 HA phylogenetic tree, these variants were most closely related to an H3N2 virus isolated from swine in 2019, in Nova Santa Rosa (Paraná), 76 km from Santa Helena (SH-aLRT 100% and UFBoot 100%) (Fig. 6). Both H3v belong to the swine lineage H3 1990.5 and clustered within the genetic clade H3 1990.5.1. It was previously estimated that viruses from this lineage derived from a human seasonal H3N2 virus introduced in the mid-late 1990s in pigs in Brazil, that subsequently evolved in pigs to form three clades (1990.5.1, 1990.5.2 and 1990.5.3)27. Viruses of genetic clade 1990.5.1 have been detected in several Brazilian states and are most closely related to human viruses that circulated around 199624,25. The N2 of Case 5’ variant virus (#51528/2021) clustered with four swIAVs collected in 2019, also in Paraná, that together compose the Brazilian swine genetic clade N2-#2 (Fig. 3C). It was estimated previously that this genetic clade emerged in pigs between 2011 and 2017 after a human seasonal H3N2 virus spillover into swine27,28. The most closely related human seasonal virus circulated around 2010-2011 (e.g., A/Idaho/03/2011 (H3N2)). All the internal gene segments of Case 4 and Case 5 were of swine lineage and classified within the H1N1pdm09 lineage (Supplementary Figs. 1–6, Table 2). Although these H3N2v presented a single genomic composition, some internal genes (PB2, PB1, PA, PA, NP) showed a close relationship with other variants (Cases 3, 6, 7 and 8) (Table 2), indicating additional reassortment events that likely occurred in swine prior to the zoonotic transmission. The epidemiological investigation revealed that four close contacts of Cases 4 and 5 developed ILI symptoms (no samples collected) and only one of them (Case 4’s brother) reported direct contact with pigs. Taken together, these findings suggest the H3N2v likely was transmitted to other individuals of the family that lived in the same house. No additional cases of influenza A in neighboring municipalities were detected within one month before and one month after the case (description of cases in the Supplementary material).
Swine sequences isolated in Brazil contains the state of isolation within its names (Brazil_MG = Minas Gerais, Brazil_MS = Mato Grosso do Sul, Brazil_PR = Paraná, Brazil_RS = Rio Grande do Sul, Brazil_SC = Santa Catarina, Brazil_SP = São Paulo). Branches are colored by host and cases are color-coded according to the figure key. The tree is midpoint rooted for clarity, and all branch lengths are drawn to scale representing nucleotide substitutions per site. Black-circles at the internal nodes identify clades supported by SH-aLRT > 80% and/or UFB > 95%. A Phylogenetic tree showing the major clade of H3 viruses that circulate in swine in Brazil (H3 1990.5). The swine-to-human introduction is highlighted by a gray-shaded form. B Topology details of the H3 Brazilian clade and subclades (1990.5.1, 1990.5.2, and 1990.5.3), with emphasis on the variant viruses.
The evaluation of point mutations revealed a high number of mutations in the detected viruses compared to the components of the current Southern Hemisphere vaccine strains and vaccine candidate viruses (CVV) are represented in Supplementary Data 1 and 231. No available vaccine strain or CVV is in the genetic group correspondent to the clades of variants found in Paraná, Brazil.
In relation to the antiviral resistance markers (Supplementary Data 3), we evaluated each of the M1, NA and PA relevant sites for antiviral susceptibility previously investigated, according to WHO32,33. Therefore, we observed the NA K249I/R substitution in A/Parana/3625/2020 and A/Parana/28600/2020 A(H1N2) viruses. However, this mutation has never been related to antiviral resistance, but another alteration in the same site (NA K249E) has previously been seen in viruses displaying a borderline reduced inhibition by Oseltamivir phosphate34,35,36. Additionally, we detected the S331R mutation in the A/Parana/51528/2021 A(H3N2), which has been formerly related to borderline reduced inhibition by Oseltamivir phosphate and Zanamivir37.
Regarding the genetic markers associated with high pathogenicity or increase of virulence, only HA mutation in positions D204S and D239N38,39,40 were found in IAV variant detected in Brazil; and NS1 mutation F103L41,42 (Supplementary Data 3). Some genetic markers associated with host adaptation or increase of transmissibility (host specificity shift) in humans were described in Supplementary Data 3. However, due to the inability to isolate the IAV variants for functional testing, we could not investigate potential new genetic markers, and the real effect of the reported genetic markers found in IAV variant detected in Brazil.
Discussion
Here, we described eight cases of human infection with swine-origin IAVs that occurred in Brazil between 2020 and 2023. The three subtypes of IAV that are enzootic in pigs (H1N1, H1N2 and H3N2) were found in individuals who had direct or indirect contact with these animals, and lived in the state of Paraná, Southern Brazil. Pig farming is a well-established livestock activity in Brazil, which is the world’s fourth largest pork producer and exporter43. About 72% of the pork production in the country is located in the Southern region, represented by three states, Santa Catarina (32.33%), Rio Grande do Sul (20.40%), and Paraná (19.21%)43. Paraná is Brazil’s third largest pork producer, therefore one of the major hubs for pig farming. For human influenza surveillance is recommended collecting 5 samples from ILI cases per sentinel unit each week44. However, the distribution of sentinel units in each Brazilian State is heterogeneous when compared to the number of inhabitants per State. Unlike other Brazilian states, Paraná, by state decision, has the highest number of sentinel units in the country, totalizing 34 units for monitoring ILI cases. Some of these sites are located near municipalities with a high density of pigs, which could have facilitated the detection of the variant cases described here. Recently, in an effort to improve the surveillance and sampling in other states, the Brazilian MoH decided to increase the number of samples collected to 20 per week45,46. One of the requirements established by the MoH for a city to have a sentinel unit was a population of over 300,000 inhabitants44. This criterion excludes many rural areas where variant influenza likely circulates initially. Strategically placing units in rural regions of swine and poultry production or advising the population and workers in these areas to seek care at existing sentinel units, can help enhance the capacity to detect influenza variants.
Influenza A viruses can be regularly transmitted from humans to swine15. The viruses can adapt to the swine host, evolve over time, and might then be reintroduced to humans as novel reassortant viruses. The eight influenza variant viruses described here were first detected by LACEN and sent for sequencing at the NIC, that is responsible for characterizing strains for which regional laboratories detect either influenza A or B viruses but are not able to identify the viruses to the subtype or lineage level. With few exceptions, the gene segments of all variant viruses grouped within swine clades, exhibiting a full-genome of swine-origin. In a few cases, some gene segments were closely related to human viruses (for instance, the HA of case 3); however, the intensive surveillance of human influenza along with the long branch observed in the phylogenetic tree suggest the absence of swine sequences rather than unsampled human viruses.
The IAV (H1N2)v of Case 1, detected in the municipality of Ibiporã in 2020, had apparently no onward transmission in humans and presented a swine-origin genomic composition not seen in the other cases. This H1N2v carries an HA derived from a pre-2009 H1N1 human seasonal virus that disappeared from the human population after 2009 pandemic, but continued to evolve in pigs, ultimately giving rise to a well-established and antigenically distinct lineage of H1 viruses in Brazilian pig population. Phylogenetic analysis of sequences sampled from Case 6, Case 7, and Case 8 show a close phylogenetic relationship among these samples for all available segments. These sequences were isolated in the municipality of Toledo and were found clustering together within a well-characterized H1pdm09 swine cluster circulating in Brazil since 201026. The HA segment isolated from Case 2 was also found within this well-characterized swine H1pdm cluster, despite being identified in a different geographic area (Irati), and all the other segments were distantly related to Case 6, Case 7, or Case 8. On the other hand, Case 3 has an HA that clusters outside the big swine H1pdm clade, but all the remaining segments clustered together with Case 6, Case 7 and Case 8. In total, four of the eight variants described in this study harbor HA segments derived from the same swine clade. This result is noteworthy and suggests that the major H1N1pdm09 lineage circulating in swine, already detected in different states in Brazil26, has ‘genetic potential’ to be transmitted from pigs back to humans. The same clustering pattern across different phylogenetic trees for some of the cases (especially Cases 6, 7 and 8) also brings into the discussion the possibility of a sustained transmission of this lineage within the human population instead of separate swine-to-human transmissions. Also, the genomic pattern observed in the H3N2v in Case 4 and Case 5, together with the epidemiological data, suggest the occurrence of limited transmission among individuals sharing the same residence. These findings highlight the remarkable ability of swIAVs in crossing inter-species barriers and to quickly be transmitted between close contacts. The phylogenetic clades defined for the influenza A virus variants described in this study also revealed differences in the genome constellations. This demonstrates the high diversity of strains that can cross the animal-human barrier, reinforcing the need to improve surveillance both throughout the country and in the region.
A constellation of genes in a particular influenza strain within a specific host determine the pathogenicity or virulence of an influenza virus. Few described genetic markers associated with pathogenicity, increased transmissibility, antiviral resistance, or virulence were observed in the analyzed genomes. However, these viral genomes, with this set of mutations, are new to the human population, and no phenotypic studies have been conducted on them. Despite causing limited infections, we cannot exclude the possibility that these viruses possess as-yet unidentified markers that may confer an advantage in adapting to the human species. We emphasize the need to preserve the collected samples to ensure viral integrity and allow the virus isolation for functional and phenotypic testing.
The comparison of the current seasonal influenza vaccine strains, H1N1 and H3N2 components, recommended for the Southern Hemisphere31 with the variants reported in this study revealed a significant genetic distance from the current vaccine, consistent with the phylogenetic analysis. The genetic data observed may indicate antigenic mismatch and a potential lack of immune protection against the IAV variants.
Increased surveillance of swine and human viruses in this region of Paraná could potentially contribute to the solution of this issue and even help control the spread of these virus strains. Besides, vaccination of pigs in Brazil may be perhaps the easiest way to control and block new infections in humans, since cartography data indicates that no vaccine recommended for controlling the influenza virus in humans would offer protection against the H1 and H3 viruses currently circulating in swine47. The worker’s swine industry should be included in the preferential group for influenza vaccination of the Brazilian MoH, once the transmission of human seasonal influenza viruses to swine has heavily influenced the extensive genetic diversity observed in swIAVs (15).
Continuous influenza surveillance and monitoring are decisive to timely assess the associated risks of seasonal, zoonotic, and pandemic outbreaks. The strengthening of One Health approaches would allow the monitoring of cross-species transmission events and the identification of potential pandemic situations48. These new Brazilian IAV variant cases were considered as sporadic cases and had good clinical outcomes that did not spread throughout the local human population. However, a worst-case scenario could have occurred. Maintaining sensitive, articulated, and actionable surveillance systems remains the key to early detection of the threat posed by new influenza variants.
Methods
Ethical aspects
This study was conducted under the terms established by the Ethical Certificate presented for appreciation (CAAE) to the Brazilian Platform (http://plataformabrasil.saude.gov.br/) and approved by the number 68118417.6.0000.5248. The SISGEN (National System for Management of Genetic Heritage and Associated Traditional Knowledge) number is A15FAF3.
The samples were collected within the National Surveillance System for Influenza viruses, and individual consent is not required. In this report, individual identities have been safeguarded by ensuring that personal data, such as age or other numeric information, is presented in aggregate or averaged for relevant identifier categories. Information on sex and/or gender has been maintained in compliance with the study’s sex and gender reporting policies.
Molecular detection of Influenza A viruses
As part of the National Influenza Surveillance established by the Brazilian Ministry of Health (MoH), Influenza-like Illness (ILI) and Severe Acute Respiratory Infections (SARI) are continuously monitored. In the state of Paraná, sentinel sites collect respiratory samples from suspect cases and the Central Public Health Laboratory of Paraná State (LACEN-PR) routinely performs real time RT-PCR tests for influenza A and B, SARS-CoV-2 and other respiratory viruses (Respiratory Syncytial Viruses, Parainfluenzavirus 1, 2 and 3, Metapneumovirus, Adenovirus, Rhinovirus, Bocavirus, human Coronavirus (hCoV) hCoV-HKU, hCoV-NL63, hCoV-OC43, hCoV-229E)49,50,51,52,53. The real time RT-PCR influenza assay performed by LACEN-PR covers all influenza seasonal viruses, with IAV being recognized by the target InfA (M gene), and the subtypes H1N1 and H3N2 by the targets H1pdmInfa (NP gene), H1 or H3 (HA gene) and N1 or N2 (NA gene)54, Supplementary Table 1. Additionally, Influenza B, Victoria and Yamagata lineages are detected by the targets InfB (NP gene), HA-Yam and HA-Vic (HA gene for each lineage)55.
Influenza A virus molecular characterization
As part of the routine surveillance, samples containing unsubtyped IAVs were sent to NIC Fiocruz for further characterization. The viral RNA was extracted from 140 µL of clinical samples (nasopharyngeal swab) using the RNA Viral mini kit (Qiagen) in a final 80 µL AVE elution, following the manufacturer’s instructions. The NIC repeated the molecular analysis performed at LACEN Paraná and was also not able to identify a seasonal IAV subtypes. Thus, the samples were submitted to additional testing for exotics IAV HA and NA genes, such as H5a, H5b, H7, HA seasonal Eurasian, H9, H1 and seasonal N1. All reactions were performed using the SuperScript ™ III One-Step RT-PCR kit (Invitrogen) and following RT-PCR protocols established by the NIC54.
Influenza A virus isolation
Each clinical sample was inoculated in Madin-Darby Canine Kidney (MDCK) cell culture in the Biosafety Level 3 (BSL3) laboratory. In summary, MDCK cells were seeded at a density of 2.0E + 5 per well in a 24-well culture plate and incubated for 24 h in Dulbecco’s Modified Eagle’s - Medium (DME-M) supplemented with pen-strep and 10% Fetal Bovine Serum (FBS). The viral inoculum was prepared by mixing 200 µL of the original clinical sample with 100 µL of an inoculation medium constituted of DMEM supplemented with 2% bovine serum albumin (BSA), antibiotic-antimycotic (A/A solution 1x), and 0,1% TPCK (N-tosyl-L-phenylalanine chloromethyl ketone) trypsin. Prior to inoculation, MDCK monolayers were washed once with Phosphate Buffered Saline (PBS) and once with inoculation medium. Inoculum was then added to cells and incubated for one hour at 37 °C. After that, the inoculum was removed, and cells were cultured in the inoculation medium described above for up to 72 h or until cultures displayed evidence of cytopathic effect.
Influenza A whole-genome sequencing
To recover the whole genome, 8 µL of the viral RNA extracted from the clinical samples was submitted to a multisegmented reverse transcription PCR (M-RTPCR) protocol56, which uses the IAV universal primers for the amplification of the eight gene segments of all influenza subtypes in a multiplex reaction. Then, the library was constructed using the Nextera XT DNA Library Preparation Kit (Illumina) and submitted to sequencing by the Illumina MiSeq System using MiSeq Reagent Kit v2 Micro (300 cycles, Illumina) to achieve an estimative of at least 3000x depth of genome coverage. Conventional Sanger sequencing was also used in some cases due to the low coverage observed in some genes. The first round of PCR was performed using the universal primers and a second round of PCR using HA gene specific primers was performed according to the CDC protocol (available by CDC under request). Subsequently, the DNA fragments were purified using the ExoSAP-IT™ PCR Product Cleanup Reagent (Invitrogen) and quantified by Qubit™ dsDNA HS Assay Kit (Thermo Fisher Scientific). The products were then sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific), 3.2 µM of the internal primers, and the DNA products were read by a 96-capillary 3730xl DNA Analyzer (Thermo Fisher Scientific).
Influenza A genomic assembling
The bioinformatics pipeline for assembling the reads and obtaining the consensus was carried out by the CLC Genomics platform (Qiagen). First, reads were mapped against a database containing all IAV available references (a panel including around 120 influenza genomes from different subtypes). Then, the genes with the best score of reads were selected to be mapped, at this point just genes with the highest number of reads were mapped, and then the consensus was retrieved for further phylogenetic analysis.
Data set construction
In addition to the eight viral genomes sequenced for this study, all available South American human, and swine H1N1, H1N2 and H3N2 IAV genomes were downloaded from GISAID Epiflu, BV-BRC, and NCBI Influenza Virus databases. Additionally, the consensus of each gene segment was submitted to BLASTN integrated in GISAID and NCBI and the 100 sequences with the closest identity with the target viral gene were recovered. WHO recommended human seasonal HA/NA vaccine and Reference sequence data were also included. Each dataset was submitted to the octoFLU classifier pipeline to filter out unrelated sequences of distinct lineages57. These data were manually curated, and sequences were removed when: (i) presented a mixed serotype, (ii) data with “lab” or “laboratory” in the host record, (iii) sequences with more than 5 ambiguous bases, and (iv) sequences with less than 75% of the total alignment size. Duplicate sequences were identified, and a single representative was retained. This process resulted in 11 discrete datasets: H1pdm (1 A.3.3.2 lineage; 2851 viruses), H1hu (pre-2009 human seasonal virus-like or 1B lineage; 374 viruses), H3 (3634 viruses), N1pdm (2158 viruses), N2 (3209 viruses), and the 6 internal gene segments: PB2 (2378 viruses), PB1 (2485 viruses), PA (2406 viruses), NP (632 viruses), MP (601 viruses) and NS (501 viruses). Each data set included swine and human IAV sequences isolated within the period from 1969 to 2023.
Phylogenetic and evolutionary analysis
Sequence alignments were generated separately for each data set using MAFFT v7.453 with default options58 followed by manual correction using AliView59. Maximum likelihood (ML) phylogenetic trees were inferred with IQ-TREE v.2.1.360, following the standard automatic best-fit model selection for each alignment (Supplementary Table 2). Statistical support for branches within the inferred trees was assessed using SH-like approximate Likelihood-Ratio Test (SH-aLRT)61 and Ultrafast Bootstrap (UFBoot) approximation with 1000 pseudoreplicates60,62. The visualization and annotation of phylogenetic trees were performed using FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/) and edited using Inkscape v.1.2 (https://inkscape.org/). For amino acid comparisons and analysis of identity, the Flusurver (https://flusurver.bii.a-star.edu.sg/) was used.
Epidemiological and additional actions to elucidate the zoonotic cases
The objectives of the epidemiological investigation were to describe the events in terms of person, time, and place; to identify potential causal factors contributing to the case, and to provide evidence-based recommendations for prevention and control measures. The strategies used for the investigation included videoconferencing with local surveillance to discuss the case, provide guidance, and establish the necessary following steps. Furthermore, field investigations were conducted, which involved interviewing the infected individual and/or their family members, and, when appropriate, collecting new samples. A search for the contacts of the index case, relatives, patients, and healthcare professionals who remained in the same ward and attended to the case was conducted. Additionally, at the reference health unit for the case, all records of ILI treated during the investigation period (1 month before and 1 month after the onset of symptoms) were surveyed to identify potential cases that meet the case definition, whether or not they have undergone sample collection for detailed investigation. Also, SARI positive cases for IAV were used to survey the circulating respiratory viruses among the residents of the municipality or region where the case occurred.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The sequencing data generated in this study have been deposited in the NCBI GenBank under the accession numbers PQ451850-PQ451906, additionally the sequences were also submitted to the EpiFlu database in GISAID (Global Initiative on Sharing Avian Flu Data) platform under the accession numbers listed in Table 1. Additional metadata associated with the sequences, including information on sample collection and some clinical characteristics are available in the metadata of both platforms. Previously published sequences used in this study are listed in Supplementary Data Set 1. All other data supporting the findings of this study are included within the manuscript and its supplementary information files.
References
Shinde, V. et al. Triple-Reassortant Swine Influenza A (H1) in Humans in the United States, 2005–2009. N. Engl. J. Med. 360, 2616–2625 (2009).
Trifonov, V., Khiabanian, H. & Rabadan, R. Geographic Dependence, Surveillance, and Origins of the 2009 Influenza A (H1N1) Virus. N. Engl. J. Med. 361, 115–119 (2009).
Trifonov, V., Khiabanian, H., Greenbaum, B. & Rabadan, R. The origin of the recent swine influenza A(H1N1) virus infecting humans. Eur. Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 14, 19193 (2009).
Webster, R. G., Bean, W. J., Gorman, O. T., Chambers, T. M. & Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56, 152–179 (1992).
Tong, S. et al. New World Bats Harbor Diverse Influenza A Viruses. PLoS Pathog. 9, e1003657 (2013).
Tong, S. et al. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. 109, 4269–4274 (2012).
Dobson, A. P. et al. Ecology and economics for pandemic prevention. Science 369, 379–381 (2020).
Scholtissek, C., Bürger, H., Kistner, O. & Shortridge, K. F. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 147, 287–294 (1985).
Ito, T. et al. Molecular Basis for the Generation in Pigs of Influenza A Viruses with Pandemic Potential. J. Virol. 72, 7367–7373 (1998).
Bowman, A. S. et al. Influenza A(H3N2) Virus in Swine at Agricultural Fairs and Transmission to Humans, Michigan and Ohio, USA, 2016. Emerg. Infect. Dis. 23, 1551–1555 (2017).
Centers for Disease Control and Prevention, C. D. C. Update: Influenza A (H3N2)v transmission and guidelines - five states, 2011. MMWR Morb. Mortal. Wkly. Rep. 60, 1741–1744 (2012).
Centers for Disease Control and Prevention, C. D. C. Influenza A (H3N2) variant virus-related hospitalizations: Ohio, 2012. MMWR Morb. Mortal. Wkly. Rep. 61, 764–767 (2012).
Brown, I. H. History and Epidemiology of Swine Influenza in Europe. in Swine Influenza (eds. Richt, J. A. & Webby, R. J.) vol. 370 133–146 (Springer Berlin Heidelberg, Berlin, Heidelberg, 2011).
Nelson, M. I. & Vincent, A. L. Reverse zoonosis of influenza to swine: new perspectives on the human–animal interface. Trends Microbiol. 23, 142–153 (2015).
Anderson, T. K. et al. Swine Influenza A Viruses and the Tangled Relationship with Humans. Cold Spring Harb. Perspect. Med. 11, a038737 (2021).
Anderson, T. K. et al. A Phylogeny-Based Global Nomenclature System and Automated Annotation Tool for H1 Hemagglutinin Genes from Swine Influenza A Viruses. mSphere 1, e00275–16 (2016).
Brockwell‐Staats, C., Webster, R. G. & Webby, R. J. Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1). Influenza Other Respir. Viruses 3, 207–213 (2009).
Garten, R. J. et al. Antigenic and Genetic Characteristics of Swine-Origin 2009 A(H1N1) Influenza Viruses Circulating in Humans. Science 325, 197–201 (2009).
Smith, G. J. D. et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459, 1122–1125 (2009).
Newman, A. P. et al. Human Case of Swine Influenza A (H1N1) Triple Reassortant Virus Infection. Wis. Emerg. Infect. Dis. 14, 1470–1472 (2008).
Shu, B. et al. Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990–2010. Virology 422, 151–160 (2012).
Ministério da Saúde. Guia para a Rede Laboratorial de Vigilância de Influenza no Brasil, (2016).
World Health Organization. Manual for the laboratory diagnosis and virological surveillance of influenza. (2011).
Schaefer, R. et al. Isolation and characterization of a pandemic H1N1 influenza virus in pigs in Brazil. Isol. Charact. Pandemic H1N1 Influenza Virus Pigs Braz. 31, 761–767 (2011).
Rajão, D. S. et al. Genetic characterization of influenza virus circulating in Brazilian pigs during 2009 and 2010 reveals a high prevalence of the pandemic H1N1 subtype. Influenza Other Respir. Viruses 7, 783–790 (2013).
Junqueira, D. M. et al. Human-to-swine introductions and onward transmission of 2009 H1N1 pandemic influenza viruses in Brazil. Front. Microbiol. 14, 1243567 (2023).
Tochetto, C. et al. Introductions of Human-Origin Seasonal H3N2, H1N2 and Pre-2009 H1N1 Influenza Viruses to Swine in Brazil. Viruses 15, 576 (2023).
Nelson, M. I., Schaefer, R., Gava, D., Cantão, M. E. & Ciacci-Zanella, J. R. Influenza A Viruses of Human Origin in Swine, Brazil. Emerg. Infect. Dis. 21, 1339–1347 (2015).
Resende, P. C. et al. Whole-Genome Characterization of a Novel Human Influenza A(H1N2) Virus Variant, Brazil. Emerg. Infect. Dis. 23, 152–154 (2017).
CDC. Fluview interactive. Novel Influenza A virus Infection. (2024).
World Health Organization. Recommended composition of influenza virus vaccines for use in the 2024 southern hemisphere influenza season. (2023).
World Health Organization. Summary of polymerase acidic (PA) protein amino acid substitutions analysed for their effects on baloxavir susceptibility. (2024).
World Health Organization. Summary of neuraminidase (NA) amino acid substitutions associated with reduced inhibition by neuraminidase inhibitors (NAIs). (2023).
Sheu, T. G. et al. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob. Agents Chemother. 52, 3284–3292 (2008).
Gubareva, L. V. et al. Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors, 2015-2016. Antivir. Res. 146, 12–20 (2017).
Zhong, J. et al. Genetic mutations in influenza H3N2 viruses from a 2012 epidemic in Southern China. Virol. J. 10, 345 (2013).
Takashita, E. et al. Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors, 2013-2014. Antivir. Res. 117, 27–38 (2015).
Tscherne, D. M. & García-Sastre, A. Virulence determinants of pandemic influenza viruses. J. Clin. Invest. 121, 6–13 (2011).
Chutinimitkul, S. et al. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J. Virol. 84, 11802–11813 (2010).
Liu, Y. et al. Altered receptor specificity and cell tropism of D222G hemagglutinin mutants isolated from fatal cases of pandemic A(H1N1) 2009 influenza virus. J. Virol. 84, 12069–12074 (2010).
Dankar, S. K. et al. Influenza A virus NS1 gene mutations F103L and M106I increase replication and virulence. Virol. J. 8, 13 (2011).
Wang, B. X., Brown, E. G. & Fish, E. N. Residues F103 and M106 within the influenza A virus NS1 CPSF4-binding region regulate interferon-stimulated gene translation initiation. Virology 508, 170–179 (2017).
Brazilian Association of Animal Protein (ABPA). Annual Report 2023. (2023).
Brasil. Ministério da Saúde. Nota Técnica no 31/2022 - CGPNI/DEIDT/SVS/MS. Informações técnicas e recomendações sobre a vigilância epidemiológica da influenza no Brasil. (2022).
Brasil. Ministério da Saúde. Nota Técnica No 13/2023-CGVDI/DIMU/SVSA/MS. Orientações sobre a estratégia e operacionalização da coleta de amostras de aspirado de nasofaringe (ANF) ou swab combinado (nasal/oral) para diagnóstico laboratorial dos vírus respiratórios, no contexto da vigilância sentinela de Síndrome Gripal (SG) e da vigilância de Síndrome Respiratória Aguda Grave (SRAG).
Brasil. Ministério da Saúde. Guia de Vigilância Em Saúde. vol. 1 (Ministério da Saúde, Brasília, 2023).
Lopes, S. et al. Antigenic and Genetic Diversity of H1 and H3 Influenza A Viruses in Swine in Brazil. http://biorxiv.org/lookup/doi/10.1101/2023.12.01.569635 (2023).
Karesh, W. B. et al. Ecology of zoonoses: natural and unnatural histories. Lancet 380, 1936–1945 (2012).
Dare, R. K. et al. Human Coronavirus Infections in Rural Thailand: A Comprehensive Study Using Real‐Time Reverse‐Transcription Polymerase Chain Reaction Assays. J. Infect. Dis. 196, 1321–1328 (2007).
Kodani, M. et al. Application of TaqMan Low-Density Arrays for Simultaneous Detection of Multiple Respiratory Pathogens. J. Clin. Microbiol. 49, 2175–2182 (2011).
Lu, X. et al. Real-Time Reverse Transcription-PCR Assay for Comprehensive Detection of Human Rhinoviruses. J. Clin. Microbiol. 46, 533–539 (2008).
Lu, X. et al. Real-Time PCR Assays for Detection of Bocavirus in Human Specimens. J. Clin. Microbiol. 44, 3231–3235 (2006).
Kim, C. et al. Comparison of Nasopharyngeal and Oropharyngeal Swabs for the Diagnosis of Eight Respiratory Viruses by Real-Time Reverse Transcription-PCR Assays. PLoS ONE 6, e21610 (2011).
Shu, B. et al. Design and Performance of the CDC Real-Time Reverse Transcriptase PCR Swine Flu Panel for Detection of 2009 A (H1N1) Pandemic Influenza Virus. J. Clin. Microbiol. 49, 2614–2619 (2011).
Biere, B., Bauer, B. & Schweiger, B. Differentiation of influenza B Virus Lineages Yamagata and Victoria by Real-Time PCR. J. Clin. Microbiol. 48, 1425–1427 (2010).
Zhou, B. et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza A Viruses. J. Virol. 83, 10309–10313 (2009).
Chang, J., Anderson, T. K., Zeller, M. A., Gauger, P. C. & Vincent, A. L. octoFLU: automated classification for the evolutionary origin of influenza A virus gene sequences detected in U.S. Swine. Microbiol. Resour. Announc. 8, e00673–19 (2019).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Larsson, A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278 (2014).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Acknowledgements
We acknowledge the WHO Global Influenza Surveillance and Response System (GISRS) which provides the mechanism for detection and monitoring of emerging zoonotic influenza viruses. The EMBRAPA team for sharing some current genomic influenza, all teams in LACEN, the team SESA-PR to perform the local surveillance with the support of the Ministry of Health surveillance team. We are thankful for the viability to use some equipments (3130XL and Bioanalyzer) at Sequencing Platform and perform some experiments at the Biosafety Level 3 facility both located at Oswaldo Cruz Institute. We also acknowledge the Global Initiative on Sharing All Influenza Data (GISAID) for the EpiFlu database, and other sequence databases which were used to share gene sequences and associated information (Supplementary Data 4). We would like to thank the financial support of the Laboratories General Coordination of the Brazilian Ministry of Health (CGLab/MoH) and Coordination of Health Surveillance and Reference laboratories from Oswaldo Cruz Foundation (CVSLR/FIOCRUZ). Caroline Tochetto is a postdoc student (FUNARBE/ARS/USDA) number 13856. Elisa Pereira is a postdoc supported by Global Immunology and Immune Sequencing for Epidemic Response (GIISER) funding project and Leticia Lima is supported by INOVA project. Financial support from the State of Rio Grande do Sul, through FAPERGS (23/2551-0000879-3, RS). CNPq CABBIO (Grant number 423857/2021-5, PCR); CNPq productivity research fellowship (311759/2022-0, PCR). CNPq productivity research fellowship (313403/2018-0, MMS).
Author information
Authors and Affiliations
Contributions
P.C.R., D.M.J., and C.T. wrote the manuscript and performed the molecular and phylogenetic analyses. L.A. and L.M. performed the whole genome sequencing. P.C.R., E.C.P., and L.F.L. carried out the assembly of Influenza A variant genomes. B.C. and A.M. performed the attempt of virus isolation. P.C.R., D.M.J., C.T., E.C.P., and L.F.L. carried out the assembly of genomes and phylogenetic analysis. I.R., M.C.D., G.N.B., A.A., M.O., F.C.M., and J.L. performed molecular detection at LACEN-PR and NIC-Fiocruz. T.S., A.M.L.F.N., R.A.P., A.C.D.V., and W.A. conducted epidemiological investigation of cases. D.B., R.S., and M.M.S. supervised the work. All authors contributed to the manuscript review.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Resende, P.C., Junqueira, D.M., Tochetto, C. et al. Zoonotic transmission of novel Influenza A variant viruses detected in Brazil during 2020 to 2023. Nat Commun 15, 10748 (2024). https://doi.org/10.1038/s41467-024-53815-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53815-z
This article is cited by
-
Whole-genome analysis of influenza A(H1N1)pdm09 viruses in Cameroon (2019–2024) using nanopore sequencing
BMC Infectious Diseases (2025)
-
Passive surveillance for Influenza A virus among swine, Brazil, 2009–2023
Brazilian Journal of Microbiology (2025)