Abstract
The epigenetic landscape plays a critical role in cancer progression, yet its therapeutic potential remains underexplored. Glucocorticoids are essential components of treatments for lymphoid cancers, but resistance, driven in part by epigenetic changes at glucocorticoid-response elements, poses a major challenge to effective therapies. Here we show that glucocorticoid treatment induces distinct patterns of chromosomal organization in glucocorticoid-sensitive and resistant acute lymphoblastic leukemia xenograft models. These glucocorticoid-response elements are primed by the pioneer transcription factor PU.1, which interacts with the glucocorticoid receptor. Eviction of PU.1 promotes receptor binding, increasing the expression of genes involved in apoptosis and facilitating a stronger therapeutic response. Treatment with a PU.1 inhibitor enhances glucocorticoid sensitivity, demonstrating the clinical potential of targeting this pathway. This study uncovers a mechanism involving PU.1 and the glucocorticoid receptor, linking transcription factor activity with drug response, and suggesting potential therapeutic strategies for overcoming resistance.
Similar content being viewed by others
Introduction
Acute lymphoblastic leukemia (ALL) is the most common cancer among children. Glucocorticoids, such as prednisone and dexamethasone, are critical components of most multi-agent chemotherapy regimens used to treat lymphoid malignancies1,2,3,4. However, the emergence of glucocorticoid resistance remains a significant barrier to cure3,4,5, glucocorticoid-based therapies rarely successfully treat myeloid malignancies6, and the molecular basis for these observations remains elusive7,8.
Glucocorticoids function via interactions with the glucocorticoid receptor (GR) in the cytoplasm9, after which the GR translocates to the nucleus to act as a transcription factor (TF) by binding to accessible glucocorticoid-response elements (GREs)10,11,12. The accessibility of GREs can be characterized by histone H3 modifications (e.g., H3K4Me3 and H3K27Ac) at cis-regulatory elements13,14. The GR binding further induces chromatin conformation changes to form transcriptionally active DNA loops, also known as topologically associated domains (TADs)15,16,17. We have previously identified a series of cis-regulatory conditions (i.e., low accessibility and hypermethylation at GREs) associated with glucocorticoid resistance in ALL18,19,20,21. However, upstream mechanisms responsible for regulating the epigenetic landscape at GR binding sites remain elusive.
Whole exome and transcriptome sequencing have identified many genetic and epigenetic mutations in relapsed/refractory ALL18,22,23,24,25,26,27,28,29,30. Although various mechanisms for glucocorticoid resistance have been proposed, they do not converge into a single common cause for all patients. For instance, loss of BTG1 or PTEN can cause glucocorticoid resistance by inhibiting GR expression or translocation to the nucleus31,32, but primary ALL cells rarely show blocked GR function26. Similarly, even though some ALLs exhibit mutations in epigenetic regulators, such as KMT2D, CREBBP, and HDAC7, no specific mutations account for abnormal chromatin accessibility across all patients. Nonetheless, the significance of BIM as a key gene in glucocorticoid-induced apoptosis in ALL is well recognized. Mutations in various genes deregulating different signaling pathways33,34,35,36 have been associated with the dysregulation of BIM and glucocorticoid resistance. We have demonstrated that epigenetic abnormalities at lymphocyte-specific genes, including an enhancer of BIM18 and mutation in methyltransferase gene NSD221, occur in ALLs with normal and mutant karyotypes, and contribute to the development of glucocorticoid resistance. Importantly, while the high diversity of genetic mutations may hinder the identification of a single genetic cause for drug resistance, epigenetic variation, including chromatin accessibility and methylation, are much more common characteristics of relapsed/refractory ALL.
Lineage-specific pioneer TFs, such as PU.1, EBF1, and TCF3, can initiate nucleosome remodeling during lymphocyte development, regulate the transcription of lineage-specific genes37,38, and recruit TET2 and DNMT3b to modulate DNA methylation of target genes, which in turn regulates gene expression during hematopoietic differentiation39. Chromatin conformation provides a structural framework for TF binding and thus gene regulation40,41. From 0.8 million open chromatin domains identified by DNase I-hypersensitive site (DHS) sequencing (derived from ENCODE), we have previously defined over 11,000 lymphocyte-specific open chromatin domains (LSOs)18. By analyzing glucocorticoid-induced epigenetic alterations in vivo using ALL patient-derived xenografts (PDXs), we identified around 2000 LSOs with GR binding (GR-LSOs), including 42 GR-activated and 61 GR-repressed LSOs with abnormal hypermethylation in glucocorticoid-resistant ALL.
To date, the precise mechanism by which lineage-specific pioneer factors contribute to the epigenetic aberrations seen at GR-LSOs in ALL remains unknown. We hypothesize that during tumor evolution, these factors prime regulatory domains which in turn leads to abnormal chromatin remodeling and the development of glucocorticoid resistance in ALL. In this work, we aim to explore common upstream events that contribute to the aberrant chromatin accessibility patterns that we previously described in ALL, and identify targets and strategies for reversing glucocorticoid resistance.
Results
Distinct regions of higher-order chromatin are associated with dexamethasone response in ALL
To explore the relationship between chromatin conformation and glucocorticoid response, we conducted chromosome conformation capture sequencing (HiC) in two pediatric ALL PDXs following dexamethasone (DEX) treatment in vivo. Both PDXs have normal cytogenetics and comparative basal gene expression profiles but ALL-54S is sensitive and ALL-50R is resistant to DEX in vivo and in vitro18,28.
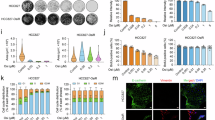
We first integrated the HiC measures with our database of epigenetic profiles from a previous study of the same PDXs18. The genome was divided into 60,000 bins, each with a size of 50 kb, and we compared their principal components (PCA, representing contact intensities) across 0.8 million DHS domains in ALL-54S and ALL-50R. Analysis of these data showed that regions with an increasing signal of open chromatin (Low to High DHS) also had a significant increase in the number of HiC interactions (PCA Value) in both PDXs, validating the principles of nuclear organization from other publications42 (Supplementary Data 1 and Fig. 1A, first panel). Representative genomic regions of DHS-Low, -Mid, and -High bins are shown in Supplementary Fig. 1. Focusing on LSOs and LSCs (i.e., DHS domains open and closed in lymphocytes, respectively), HiC contact intensities were significantly higher in LSOs (Fig. 1A) with a trend for significantly increasing contact intensities for genome regions (50 kb bins) that contained higher frequencies of LSOs (Fig. 1B). LSOs that were also bound by the GR18, showed significantly higher contact intensities compared to those without GR binding (non-GR-LSOs) (Fig. 1A). Finally, limiting our analyses to the subset of the 42 activated and 61 repressed GR-LSOs (i.e., associated with changes in key gene expressions18), we found that the ALL-54S PDX had higher basal contact intensity that significantly increased upon in vivo DEX treatment for the 42 activated GR-LSOs but remained the same for ALL-50R, while no significant difference was observed for the 61 repressed GR-LSOs (Fig. 1A, panel four; Supplementary Data 1). Furthermore, genes within DHS-High regions showed higher RNA transcriptional activity compared to DHS-Low regions in both PDXs, and similarly, genes within LSOs demonstrated higher RNA transcriptional activity compared to LSCs (Fig. 1C). Notably, Dex treatment made no global changes in RNA transcriptional activity compared to untreated control. Taken together, these data support the concept that LSOs have increased accessibility facilitating higher genomic contacts and increased transcriptional activity compared to LSCs.
Violin plots of HiC contact intensities quantified using principal component analysis (PCA) in genomic regions for A (i) 0–9 DHS domains (DHS-Low), (ii) 10-19 DHS domains (intermediate, DHS-Mid), (iii) ≧20 DHS domains (DHS-High), (iv) lymphocyte-specific open regions (LSOs), (v) lymphocyte-specific closed regions (LSCs), (vi) LSOs with GR binding (GR-LSOs), (vii) LSOs without GR binding (Non-GR-LSOs), (viii) 42 activated GR-LSOs and (ix) 61 repressed GR-LSOs; for B genomic regions with different abundance of LSOs. Statistics by Wilcoxon matched-pairs signed rank test, ***p < 0.001; ****p < 0.0001; NS, not significant. See Supplementary Data 1 for exact p values. C Gene expression levels measured by RNA-seq at the regions defined in A. Data are presented as normalized transcript counts across transcription start sites (TSS) to transcription end sites (TES) of all genes located in each bin category of genomic regions. D DEX-induced HiC contact intensity changes in ALL-50R and ALL-54S in vivo. Dynamic bins are genome regions that changed in HiC intensity. IN, increase from negative to positive PCA values or >1.5-fold increase; DE, decrease from positive to negative PCA values or >1.5-fold decrease). Stable bins (ST) are genome regions with no change of HiC contact intensities, including ST-Low Contacts with negative PCA values and ST-High-Contacts with positive PCA values. E Volcano plots depicting differential expressed bins of HiC contact intensities between control and dex-treated groups in ALLs. A generalized linear model “estimateGLMCommonDisp” in R package EdgeR (version 4.2.1) was applied to calculate the statistical significance for the exponential transformed PCA values of each HiC bin (n = 56380). F Heatmap of HiC contact intensities in ALLs. The top row indicates the 103 (i.e., 42 GR-activated or 61 GR-repressed) LSOs as well as their adjacent 50 kb regions (42 GR-LSOs +/− nearby bin) and the bottom row highlights genes identified by a ΔΔHiC analysis (ΔΔHiC = ΔHiC of ALL-54S (Dex – Ctrl) - ΔHiC of ALL-50R (Dex – Ctrl)). See also Supplementary Data 1–3 and Supplementary Fig. 1. Source data are provided as a Source Data file.
Next, we investigated DEX-induced chromatin conformation changes on a genome-wide scale (Supplementary Data 2). Most contact intensities remained unchanged (i.e., stable) but we found a ~2-fold increase in dynamic changes (i.e., bins with increasing or decreasing contact intensities) in ALL-54S compared to ALL-50R (Fig. 1D). Therefore, we set out to identify genomic regions that showed significantly different contact intensities between the sensitive and resistant cells post-treatment with DEX (i.e., ALL-54S and ALL-50R). In total, 4645 bins were significantly different, including 135 dynamic bins containing GR-LSOs and their associated genes including the previously identified BIM (BCL2L11)18,28. (Supplementary Data 3, Fig. 1E). Combining the dynamic changes and overlaying with the 42 GR-activated and 61 GR-repressed LSOs (Fig. 1F), we further identified a number of regions containing genes of interest, including ZBTB16, BIM (BCL2L11), SUPT4H1, and others linked to aberrant chromatin accessibility and glucocorticoid resistance18,22,23,24,25,26,27,28,29,30. These data suggest that the ability to dynamically change higher-order chromatin interactions is key to GR sensitivity, warranting further interrogation of regulatory factors of chromatin conformation.
PU.1 is a transcriptional regulator of higher order chromatin regions associated with DEX response in ALL
To better understand the cis-regulatory mechanism of these chromatin conformation dynamics and their associated response to DEX we performed an enrichment analysis using i-cisTarget to identify potential upstream regulators43,44. Our in silico analysis focussed on GR-LSOs enriched for H3K27Ac (enhancer mark) in ALL-54S but not ALL-50R post-DEX treatment (Supplementary Data 4). The DNA binding motifs for 77 TFs were identified with the highest enrichment scores found for steroid receptor family members including GR, androgen receptor (AR), and mineralocorticoid receptor (MR), as well as key TFs involved in lymphocyte development, such as PU.1, EBF1, and TCF3 (Fig. 2A, Supplementary Data 5). While TFs may share binding motifs, PU.1 (SPI1) had the highest enrichment score among the ETS family proteins comparable to GR (median NES of 3.01 vs. 3.25). EBF1 had the highest score among non-ETS family proteins (median NES of 0.88). The structural protein CTCF, which is considered to play a critical role in stabilizing GR-induced TADs despite no interaction with the GR15,16,18, exhibited no enrichment (median NES of −0.19; Fig. 2B).
A Motif enrichment analysis using i-cisTarget at genomic regions associated with HiC contact intensity changes in ALL-54S and ALL-50R pre- and post-treatment with DEX. The normalized enrichment scores (NES) for all TF motifs are shown in blue and the top 10 TF motifs are highlighted in red and annotated. B Violin plots of the NES for the top 10 ranked TF motifs, two lineage-specific TFs EBF1 and TCF3, and structural protein CTCF. C GR-interacting proteins identified using Rapid Immunoprecipitation Mass Spectrometry (RIME). Interacting affinities were calculated as a differential abundance of the GR-bound proteins in DEX-treated vs. control samples in ALL-50R and ALL-54S. GR co-regulators, previously reported GR-interacting proteins46,47,48,49,50 to mediate GR actions. TF, transcription factor; FC, fold change. Statistics were performed using limma through the FragPipe-Analyst (http://fragpipe-analyst.nesvilab.org/). N = 3, the data was generated from PDX cells in 3 separate mice of each group. D Relative changes of TF-GR interacting affinities in ALL-54S compared with ALL-50R upon DEX treatment. Δlog2FC = log2FC in ALL-54S – log2FC in ALL-50R based on the data from C. N = 3. E Violin plots of PU.1 and CTCF ChIP-seq enrichment in ALL-50R and ALL-54S pre- and post-treatment with DEX at genomic regions described in Fig. 1D. ST = genomic bins with stable HiC-contact intensities after DEX treatment, IN = bins with increased HiC-contact intensities after DEX treatment; DE = bins with decreased HiC-contact intensities after DEX treatment. ALL-50R: IN, n = 3956; DE, n = 3299; ST, n = 49125. ALL-54S: IN, n = 7036; DE, n = 6578; ST, n = 42766. See also Supplementary Data 4-5. Source data are provided as a Source Data file.
To complement this in silico analysis and directly identify GR co-factors that mediate tissue-specific glucocorticoid response, we performed rapid immunoprecipitation mass spectrometry (RIME) analysis45 of GR-binding proteins in ALL-54S and ALL-50R post-DEX treatment (Fig. 2C, D). The interacting affinity between the GR and all previously reported GR co-regulators46,47,48,49,50 was analyzed, including the GR itself (as a GR dimer) and proteins involved in structural and enzyme-interacting, chromatin-remodeling, methyltransferases, histone acetyltransferase/deacetylase, and TFs. Previous work has identified long-range GR interactions depend on interaction with members of the cohesin complex46,47,48. In this study, we did see that the sister chromatid cohesion proteins PDS5A and PDS5B were enriched in RIME samples, but their interacting affinity with the GR was similar in ALL-54S and ALL-50R, showing |Δlog2FC| < 1. Instead, PAXIP1, which is required for stabilizing cohesin on chromatin48, revealed enhanced GR interactions in ALL-54S with Δlog2FC = 1.25 (Fig. 2D). Furthermore, clusters of SMARC proteins manipulating chromatin structure and Mediators of RNA polymerase II were highly interacted with the GR in ALL-54S compared to ALL-50R, showing highly positive Δlog2FC (Fig. 2D), which is consistent with the more active glucocorticoid-induced signaling cascade in ALL-54S. Furthermore, notably, PU.1 was one of the highest enriched GR-interacting proteins in DEX-treated ALL-54S compared to control, but not in ALL-50R (Fig. 2C). This DEX-induced change in PU.1-GR interaction in ALL-54S vs. ALL-50R (Δlog2FC) is the highest among ETS-related proteins and all GR co-regulators (Fig. 2D). There was no significant enrichment for ELF1, EBF1, or CTCF proteins in either PDX. Together, the in-silico prediction and RIME experimentation provide compelling evidence for PU.1 serving as a key co-regulator of GR in glucocorticoid response of ALL.
This finding led to a global analysis of PU.1 binding in ALL-54S and ALL-50R by ChIP-seq with CTCF as control. Comparing binding profiles in dynamic bins pre- and post-treatment with DEX, we found that CTCF binding remained unchanged, while PU.1 binding was globally decreased after DEX treatment in ALL-54S but not ALL-50R (Fig. 2E). These data suggest that the interaction between GR and PU.1 may lead to the eviction of PU.1 following DEX treatment and warranted further studies of PU.1 and GR co-binding at these cis-regulatory elements.
PU.1 eviction mediates DEX response via recruitment of the GR in DEX sensitive but not resistant ALLs
Next, binding profiles of GR, PU.1, and CTCF at key clusters of glucocorticoid-response genomic regions, i.e., LSCs, LSOs, non-GR-LSOs, and GR-LSOs, were analyzed. We found that GR and PU.1 were bound more frequently to LSOs than LSCs (green curves vs. orange curves) while CTCF binding was generally low and did not exhibit any group preference. Furthermore, while PU.1 was enriched at both GR-LSOs and non-GR-LSOs, its enrichment was much stronger at GR-LSOs, providing further evidence for the cooperation of PU.1 and GR at LSOs (Fig. 3A). Moreover, we have previously shown that there was no difference in chromatin accessibility or CTCF binding at either GR-LSOs or non-GR-LSOs in ALL-54S and ALL-50R18. However, we found a marked difference in PU.1 binding at GR-LSOs and non-GR-LSOs between ALL-54S and ALL-50R. In response to DEX treatment, the binding of PU.1 increased in ALL-50R but decreased in ALL-54S at GR-LSOs and non-GR-LSOs (Fig. 3B).
A GR, PU.1, and CTCF ChIP-seq binding profile plots at GR-LSOs, non-GR-LSOs, LSOs, and LSCs in DEX-treated ALL-54S. BS, binding sites. B PU.1 and CTCF ChIP-seq binding profile plots at GR-LSOs and non-GR-LSOs in ALL-54S and ALL-50R pre- and post-DEX treatment. C Pie chart showing the binding and co-occurrence of PU.1, CTCF, and EBF1 in ALL-54S pre- and post-treatment with DEX. D Summary of binding dynamics of PU.1, CTCF, and EBF1 at GR-LSO in ALL-54S pre- and post-treatment with DEX. See also Supplementary Data 6 and Supplementary Fig. 2. Source data are provided as a Source Data file.
To further explore these patterns, we interrogated PU.1 binding at each individual GR-LSO upon DEX treatment and used CTCF and the lineage-specific transcription factor (TF) EBF1 as controls. We found that sites exclusively bound by PU.1 decreased by 7% and those bound by both PU.1 & EBF1 decreased by 8% in ALL-54S (Fig. 3C; Supplementary Data 6). These losses were accompanied by an 8% and 7% increase in sites lacking TF binding or sites bound exclusively by EBF1, respectively. In line with these data, among the 1773 GR-LSOs, 1009 did not show any changes in the binding patterns of PU.1, EBF1, and CTCF in ALL-54S (depicted by black bars in Fig. 3D). However, we identified three clusters of GR-LSOs in ALL-54S, consisting of 127, 97, and 39 sites, which exclusively exhibited loss of PU.1 binding while maintaining an unchanged pattern of EBF1 and CTCF binding (depicted by blue bars in Fig. 3D). As a control, the top two clusters of GR-LSOs with altered EBF1 binding demonstrated binding loss at 48 regions and binding gain at 41 regions exclusively in ALL-54S (depicted by green bars in Fig. 3D). In contrast, the PU.1 eviction upon DEX treatment was not observed in ALL-50R (Supplementary Fig. 2A and B), and PU.1 binding in ALL-50R revealed a similar pattern to EBF1 (positive z-score) in a bootstrapping analysis compared to a distinct PU.1 action in ALL-54S (negative z-score; Supplementary Fig. 2C).
Taken together, these data suggest that PU.1 primes chromatin in treatment-naive cells, and that PU.1 eviction is linked to GR binding post-treatment with DEX, in DEX-sensitive but not resistant leukemia cells.
PU.1 priming and eviction lead to epigenetic changes at GREs and the formation of a densely connected gene regulatory network centered around GR
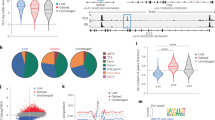
Next, we integrated the above data with regions of open chromatin (ATAC-seq) in gene regulatory elements defined by the presence of histone modifications H3K4Me3 (promoter region), H3K4Me1 (enhancer region), H3K27Ac (active region) and H3K27Me3 (repressed region). Using these histone marks, the genome was partitioned into regulatory regions including promoters, enhancers, and dual promotor-enhancer regions. These were then further divided into silent (S), active (A), repressed (R) and bivalent (Bi) regions (Fig. 4A). Reconciling these regions with the ChIP-seq data, CTCF and PU.1 were more frequently bound at repressed regions when compared to the GR across all conditions, while PU.1 but not CTCF was more frequently bound to enhancers than promoters. However, we also found differences between conditions, for example, PU.1 showed a different binding frequency at active enhancers in ALL-54S compared to ALL-50R (Fig. 4A). Similarly, at GR-LSOs, the binding patterns of GR and PU.1 were constrained to the distribution of the GR-LSOs (Supplementary Fig. 3A). Taken together, while these data show that high GR enrichment was critical for the activation of promoters and enhancers, a low enrichment of PU.1 was associated with enhancer activation in ALL-54S.
A Epigenome categorization in ALL-54S and ALL-50R pre- and post-DEX treatment. Top panel: Combinatorial code of histone modifications for epigenome categorization. Pro, promoter with H3K4Me3; Enh, enhancer with H3K4Me1; four sub-categories under Pro or Enh: S, silent; A, active; R, repressed; Bi, bivalent. RO, repressed mark H3K27Me3 only; AO, active mark H3K27Ac only; NS, no signal (regions lacking ChIP enrichment). Lower panel: GR, PU.1, and CTCF binding at defined epigenome categories in ALL-54S and ALL-50R pre- and post-treatment with DEX. B RNA-seq signal of GR-LSO-associated genes and H3K27Ac/H3K27Me3 ChIP-seq signal at GR-LSOs in ALL-54S and ALL-50R pre- and post-DEX treatment. HiC data was used to associate LSOs with genes and the five genes with the highest scores in each LSO were used for the analysis. H3K27Ac and H3K27Me3 ChIP-seq profiles are +/− 5 kb surrounding GR-LSOs. Red box, clusters 2 showing relatively higher H3K27Ac enrichment after DEX treatment in ALL-54S; yellow box, genomic regions showing lower H3K27Me3 enrichment in ALL-54S compared to ALL-50R. C Bar-graph of -log (p-values) calculated using the Fisher exact test in Ingenuity Pathway Analysis to determine the molecular pathways enriched between ALL-54S and ALL-50R upon DEX treatment in vivo based on genes expression levels (RNA-Seq; N = 87), protein interactions (RIME; N = 125), and gene regulatory elements (ChIP-seq; N = 247). D Cystoscope model integrating i-cisTarget motifs, TF ChIP-seq, and histone marks ChIP-seq at LSOs. See also Supplementary Data 6-7 and Supplementary Fig. 3. Source data are provided as a Source Data file.
Furthermore, we associated GR-LSOs with their interacting gene promoters using HiC (Supplementary Data 7) and then integrated with gene expression measured by RNA-seq. Unsupervised hierarchical clustering analysis of differentially expressed genes identified four clusters that variably correlated with the identity of gene regulatory elements (left panel in Fig. 4B). For example, in ALL-54S, we found that H3K27Ac signal in clusters 2 (red box, indicating active enhancers and promoters) was increased and in line with up-regulated gene expression but remained unchanged in clusters 3 and 4 despite the downregulation of target genes, suggesting that additional factors might impact the down-regulated gene expression (right and lower panel in Fig. 4B and Supplementary Fig. 3B). Interestingly, we found that the genomic regions in cluster 2 had a significant decrease in PU.1 binding in ALL-54S but not ALL-50R (Supplementary Fig. 3C and D). In ALL-50R, however, H3K27Me3 levels were elevated at promoters of selected target genes (yellow boxes), including BIM and ZBTB16, in line with their stable gene expression pre/post-DEX treatment (left panel in Fig. 4B). These data suggest that genes differentially regulated in ALL-50R and ALL-54S, including BIM and ZBTB16, are associated with differential H3K27Ac and H3K27Me3 signals at genomic regions bound by PU.1.
To link potential signaling pathways regulated by the GR-interacting protein networks, we performed an ingenuity pathway analysis (IPA, Fig. 4C) of differentially expressed genes identified from RNA-seq, proteins identified from RIME, and gene regulatory elements with DEX-induced PU.1 loss identified in Fig. 3D (first three blue bars, n = 263 regions). The GR interactome via RIME analyses demonstrated that the “gene expression pathway” was the most enriched upon DEX treatment, validating the critical role of GR-interacting proteins in triggering gene transcription. RNA-seq, RIME, and ChIP-seq analyses demonstrated that both up- or down-regulated genes, GR protein-protein interactions, and PU.1 eviction-affected genes were significantly enriched in “cellular development, proliferation, death & survival pathways” (involving both pro- and anti-apoptotic genes), and “cell cycle pathways”. This supports GR and its interacting proteins in regulating proliferation arrest and cell death of ALL cells following DEX treatment in vivo. Particularly, as the most significant one on the list, enrichment of the “cellular development pathway” is consistent with the interaction of GR with the lineage-specific transcriptional factor PU.1 in response to glucocorticoid treatment.
Next, we used Cytoscape51 to construct a regulatory network model centered around GR-LSOs by integrating data from i-cisTarget43,44 motif analyses of GR and PU.1 compared to other lineage-specific TFs such as EBF1 and TCF3 (Fig. 4D). This analysis revealed a core set of genes that are co-regulated by GR and PU.1, compared to rare connections between the GR and two other TFs (EBF1 and TCF3). ZBTB16, a regulator of lymphoid development52, had multiple connections with both GR and PU.1. ZBTB16 was also ranked highly in two independent analyses of DEX-induced HiC contact intensities (Fig. 1E, F) and an integrative transcriptome and epigenome analysis (RNA-seq, H3K27Ac/me3 and H3K4me1/me3 ChIP-seq; Fig. 4B and Supplementary Fig. 3B), which suggests that ZBTB16 may play a role in the GR - PU.1 pathway and might thus be a mediator of DEX response in ALL.
Multi-omics annotation of DEX-induced De Novo TADs confirms the eviction of PU.1 as a recurrent epigenetic pattern in DEX-sensitive but not resistant ALLs
We next compared changes in TAD formation pre- and post-treatment with DEX in the sensitive and resistant PDXs, and found that the formation of DEX-induced de novo TADs is common at genomic regions surrounding GR-LSOs. First, our analysis identified the formation of a de novo TAD spanning a 300 kb region around the BIM LSO in ALL-54S but not ALL-50R (Fig. 5A top). This TAD had two contact-enriched sub-regions, including a breakpoint at the BIM enhancer18, and ΔHiC analysis confirmed an increase in contact frequency in ALL-54S but a decrease in frequency in ALL-50R post-treatment with DEX (Fig. 5A bottom). In ALL-50R, the decrease was in-line with a lack of conclusive protein binding at the TAD borders (i.e., CTCF marked only one of the two TAD borders) pre- and post-treatment. In ALL-54S, however, we found an overall increase in contact frequencies which was particularly high in regions adjacent to TAD borders post-treatment (Fig. 5B, top). Additionally, while CTCF marked TAD borders pre- and post-treatment, PU.1 binding adjacent to TAD borders was reduced post-treatment. Similarly, at LSOs, we found CTCF binding pre- and post-treatment, while a decrease in PU.1 signal and an increase in GR and H3K27Ac signal were observed post-treatment (Fig. 5B, bottom). Notably, BIM expression was also upregulated post-treatment, while ACOXL expression, a gene located upstream to the TAD, remained unchanged (Fig. 5F).
HiC interaction map and ΔHiC analysis at BIM TAD (A) and ZBTB16 TAD (C) of ALL-50R and ALL-54S pre- and post-treatment with DEX (top) and directionality index (DI; bottom). Triangles indicate TADs. DI shows a preference to interact with either an up or downstream region. (B, D) UCSC Genome Browser tracks at BIM locus (B) and ZBTB16 locus (D) showing LSO-interacting genomic regions by HiC and CTCF, PU.1, GR, H3K27Ac, H3K4Me1, and H3K27Me3 ChIP-seq. Blue and green highlights are the up and downstream TAD borders, yellow highlight is LSO, pink highlight is super-enhancer (SE). E UCSC Genome Browser tracks of HiC interaction at ZBTB16 SE in ALL-50R and ALL-54S pre- and post-DEX treatment. F Heatmap of RNA-seq data showing gene expression within the BCL2L11 (BIM) TAD and ZBTB16 TAD in ALL-50R and ALL-54S pre- and post-DEX administration in vivo. G UCSC Genome Browser tracks of ATAC-seq data showing accessible genomic regions at the ZBTB16 locus in a panel of DEX-sensitive and resistant ALL PDXs pre- and post-treatment with DEX. T-ALL 8I has intermediate resistance to glucocorticoids. See also Supplementary Fig. 4.
Similarly, we observed de novo TAD formation surrounding GR-LSOs at ZBTB16 and RASA1 loci (Fig. 5C and Supplementary Fig. 4A), which followed the same epigenetic blueprint pre- and post-treatment with DEX (Fig. 5D and Supplementary Fig. 4B). The TAD at ZBTB16 locus was again divided into three sub-sections by the LSO and its two TAD borders (Fig. 5C). Interestingly, we found a relatively larger increase in contact intensities in the sub-section downstream of the LSO insulated by the two TAD borders. In ALL-54S, but not ALL-50R, DEX treatment led to a particularly high increase in H3K27Ac signal, and this region was identified as a super-enhancer (SE) by ROSE53 spanning 32 kb (ZBTB16 SE, Fig. 5D). As in ALL-54S and ALL-50R, both TAD borders were insulated by CTCF, we further shifted our focus to the interaction between LSO, ZBTB16 SE, and other gene regulatory elements. In ALL-54S, but not ALL-50R, PU.1 binding was partly lost, and the signal of GR and H3K27Ac was partly gained at both LSO and ZBTB16 SE after DEX treatment (Fig. 5D). Particularly, interactions between ZBTB16 SE and genes within the TAD were enhanced post-treatment (Fig. 5E). This was consistent with expression activation of ZBTB16 and other genes within the TAD, including ZW10, USP28 and H3TR3A post-treatment (Fig. 5F and Supplementary Fig. 4C). Of note, genes such as TMPRSS5 and H3TR3B, which located at the border of TAD or had no interaction with the SE, revealed no response to DEX treatment.
Interactions between ZBTB16 LSO, promoter and SE were more frequent in ALL-54S than ALL-50R and these interactions were further augmented after DEX treatment in ALL-54S (Supplementary Fig. 4D and E). Additionally, the LSO interacted more frequently with the SE, compared to the promoter, suggesting that the LSO regulated ZBTB16 expression via the SE in ALL (Supplementary Fig. 4E). Furthermore, a genome-wide HiC study revealed upregulated SE activities at IN HiC dynamic bins in DEX-treated ALL-54, indicating SE may play an important role in mediating chromatin conformation dynamics in response to glucocorticoid (Supplementary Fig. 4F).
To assess these observations across a larger panel of sensitive vs. resistance ALL samples upon DEX treatment in vivo, ATAC-seq studies were performed to determine chromatin accessibility at the ZBTB16 and BIM locus in a panel of DEX-sensitive and -resistant PDXs (Fig. 5G and Supplementary Fig. 4G). While the ZBTB16 SE was accessible in sensitive and resistant ALL, the ZBTB16 and BIM LSOs were only accessible in glucocorticoid-sensitive PDXs but not resistant PDXs. These data indicate a more cell-type-specific role of the LSO rather than the SE in mediating glucocorticoid activities.
The multi-omics annotation of two TADs, at the BIM and ZBTB16 loci, illustrate how TFs drive the cooperation of gene regulatory elements and SE to remodel chromatin, and identified a recurrent pattern of selective PU.1 eviction followed by GR recruitment and H3K27Ac engagement in DEX-sensitive ALL. This suggests PU.1 eviction as an upstream regulatory mechanism underlying DEX response in ALL.
PU.1 inhibition enhances glucocorticoid sensitivity of ALL
To validate the function of ZBTB16 LSO and SE, we identified six H3K27Ac-enriched regions within the ZBTB16 SE (Supplementary Fig. 5A) and performed luciferase reporter assays (Supplementary Fig. 5B). Interestingly, while the full-length ZBTB16 LSO did not significantly enhance luminescence we found that the insertion of SE peaks #2 and #6 significantly increased luciferase expression upon DEX treatment in Nalm6 cells (Supplementary Fig. 5B). Next, we performed luciferase reporter assays with the PU.1 inhibitor DB2313. When compared to the treatment with DEX alone, combination treatment with DB2313 significantly increased the luminescence intensities driven by BIM, RASA1 and ZBTB16 LSO-insert in Nalm6 cells (Supplementary Fig. 5C).
We have recently demonstrated that the displacement of PU.1 by DB2313 is reliant upon the enrichment of A/T nucleotides surrounding the core PU.1 ETS motif54 (Fig. 6A, left panel). We found that GR-LSOs had a higher log-odds score for the A/T-rich motifs than non-GR-LSOs (Fig. 6A, right panel) which suggested a potential synergy between DB2313 and glucocorticoids. To test this hypothesis, we performed PU.1 ChIP-seq in a Dex-resistant B-ALL cell line KOPN8 treated with Dex alone or with a combination of DEX and DB2313. The combination treatment reversed Dex resistance in KOPN8 cells (Supplementary Fig. 6A) and, when compared to the treatment with Dex treatment alone, led to a further loss of PU.1 binding sites at LSOs and GR-LSOs (Fig. 6B; Supplementary Fig. 6B, C). We further annotated the PU.1 binding sites that were lost in GR LSOs with coding genes and performed IPA pathway analysis (Supplementary Fig. 6D, E). We found that these genes were significantly enriched for pathways associated with “Cellular Growth and Proliferation” (rank 2) and “Cellular Death and Survival” (rank 4;) supporting the hypothesis that PU.1 removal, and potential GR recruitment to these sites, might induce apoptosis. Similarly, we performed ATAC-Seq and found that chromatin accessibility at LSOs (Fig. 6C and GR-LSOs (Supplementary Fig. 7A) in NALM6 cells and PDXs were enhanced by the DEX & DB combinational treatment compared to DEX alone after 48 h in vitro. Genes linked to the LSOs with enhanced chromatin accessibility are enriched in a published glucocorticoid-induced gene set55 and signaling pathways including PI3-AKT, MAPK, and B-cell receptor signaling pathways that have been reported to be involved in glucocorticoid actions56 (Fig. 6C). Furthermore, RNA-seq analysis confirmed that glucocorticoid-induced signaling cascades were enhanced by the combinational treatment compared to DEX alone (Fig. 6D and Supplementary Fig. 7B). Particularly, the GR-LSOs with PU.1 Loss demonstrated higher transcriptional activities following combination treatment compared to GR-LSOs and LSOs (upper panel in Fig. 6D), indicating critical cooperation between GR binding and PU.1 eviction in enhancing glucocorticoid sensitivity in ALL.
A Left: Schematic of PU.1 motif that is most responsive to DB2313 treatment (DB-sensitive) and showed PU.1 binding loss after DB2313 treatment54. Right: log-odds ratio for enrichments of DB-sensitive PU.1 motif in GR-LSOs (n = 1510) and non-GR-LSOs (n = 10707). Statistics were performed using receiver operating characteristics (ROC) analysis. B PU.1 ChIP-seq profiles and heatmap plots (DeepTools) at LSOs in a B-ALL cell line (kopn8) exposed to DEX (Dex, 1 μM) alone or a combination treatment of 1 μM Dex and 1 μM of a PU.1 inhibitor DB2313 (DB) for 16 h in vitro. C ATAC-seq analysis of Nalm6 and ALL-54S cells treated with DEX ± DB2313 for 48 h in vitro. Upper left panel normalized ATAC-seq intensity at LSOs. Gene-set enrichment analysis was performed using the “RHEIN ALL GLUCOCORTICOID THERAPY UP55” gene set (RAGTU, lower left panel) and the top activated signaling pathways (right panel) comparing Dex + DB with Dex-treated Nalm6 cells. See the Methods section for statistics. D RNA transcriptional activities in Nalm6 treated with DEX ± DB2313 for 48 h in vitro. The data are presented as normalized transcript counts across TSS to TES of genes associated with LSOs (LSO-related Genes), GR-LSOs (GR-LSO Genes), GR-LSOs with PU.1 Loss (PU.1 Loss at GR-LSOs), RAGTU gene set, and differentially expressed genes comparing Dex treatment and control Nalm6 cells (DEG). E Time course study of DEX ± DB2313 induced gene expression in Nalm6 cells. Cells were exposed to DEX (1 μM) ± DB2313 (1 μM). GILZ is a direct target of GR and was used as a positive control. F Cytotoxicity assays of DEX (1 μM) ± DB2313 (1 μM) in two ALL cell lines (Nalm6 and 697) and two ALL PDXs (RJ-1 and RJ-10) treated for 72 h in vitro. G Cytotoxicity assay to determine synergistic effects of DEX and DB in inducing cell death of Nalm6 Cells. Interactive analysis is performed using SynergyFinder. A synergy score >10 is considered significant. Data in E, F, and G are presented as the mean ± SEM of three biological replicates. P value was calculated using two-sided student t-test. See also Supplementary Fig. 5–7. Source data are provided as a Source Data file.
We then validated the findings from multi-omics analysis. Firstly, a time-course analysis showed an increased expression for BIM, RASA1 and ZBTB16 after combination treatment compared to DEX alone (Fig. 6E). Secondly, combination treatment significantly promoted cell death in ALL cell lines and PDXs compared to DEX treatment alone (Fig. 6F) and synergy profiling in Nalm-6 cells confirmed a synergistic cytotoxic effect (SynergyFinder score: 11.95; Fig. 6G). These data suggest that inhibition of PU.1 enhanced the sensitivity of ALL cells to DEX.
To further test these findings, we developed an inducible CRISPR knockout system and performed BIM and PU.1 (SPI1) knockout (KO) in vivo in ALL-54S. The doxycycline-induced BIM KO led to DEX resistance and shorter event-free survival (EFS), while the PU.1 KO significantly delayed leukemic growth and led to an increase in EFS (Fig. 7A, B). To better understand the impact of gene editing at both BIM and PU.1 target sites, we analyzed the insertions and deletions (INDELs) generated by CRISPR/Cas9 in the BIM KO and PU.1 KO clones. INDELs in BIM KO clones displayed varying lengths in all frames while INDELs in PU.1 exclusively generated in-frame mutations with lengths being multiples of 3 bp particularly for DEX treated mice (Fig. 7C, D). Protein structures of two representative PU.1 mutants with 3 or 6 bp insertions were predicted using Aphafold257 (Supplementary Fig. 7C). The insertions are located 140 amino acids upstream of the DNA binding domain of the PU.1 protein and do not alter its core structure. This suggests that frame-shift mutations were lethal and the extended EFS post-DEX treatment was due to loss of all frameshift mutated PU.1 KO clones and therefore evicted from GR binding sites whereas clones that retained PU.1 were selected for during DEX treatment due to lack of eviction and conferring glucocorticoid resistance.
Engraftment (A) and Event-free Survival (EFS) curves (B) of ALL-54S cells with BIM KO, PU.1 KO or Non-Targeting Control (NTC) guide RNA in mouse peripheral blood (PB). Gene KO was induced by doxycycline (Dox) feeding. Leukemia event is defined as 25% human cells in mouse PB. Gene KO was induced by doxycycline (Dox) feeding. Leukemia event is defined as 25% human cells in mouse PB. Statistics were performed using log-rank (Mantel-Cox) test, n = 6/group except for the BIM KO DEX-treated group has n = 5 due to mouse loss from non-leukemia related disease. Insertion and deletion profiles of relapsed BIM KO (C) and PU.1 KO (D) ALL-54S clones following saline (Ctrl)/DEX treatment in vivo. Genomic DNA of PDX cells from two independent mice were sequenced and representative profiles from each group were shown. The TIDE webtool (http://tide.nki.nl) was used for sequencing data analysis. See also Supplementary Fig. 7. Source data are provided as a Source Data file.
Discussion
Epigenomic landscapes play a crucial role in the evolution of leukemia, but their therapeutic utility is yet to be fully realized. Previous studies have defined the genomic and epigenomic landscapes that mediate glucocorticoid resistance in ALL18,21 and this study identified the developmental pioneer TF PU.1 as a critical upstream regulator. Furthermore, glucocorticoids have been in clinical use for decades, and their tissue- and cell-specific activity is a recurring clinical observation7,58. To this end, this study revealed the molecular basis of glucocorticoid response, and this could further be exploited to guide the clinical application of glucocorticoids and form a foundation for developing strategies to reverse resistance in ALL.
We have published a series of studies on epigenetic inhibition of the pro-apoptotic BIM gene associated with glucocorticoid resistance in ALL18,26,28,59,60. Dysregulation of an intronic GR-bound enhancer at the BIM locus that triggers BIM transcription and apoptosis of ALL upon glucocorticoid treatment was attributed to glucocorticoid resistance in ALL for the first time28. It was further demonstrated that the BIM enhancer was lymphocyte-specific and could be activated in normal and malignant lymphoid cells. Furthermore, glucocorticoid resistance driven by different mutations harbored a similar mechanism: aberrant chromatin accessibility20,21.
While cancer stem cells have been identified as the initiators of chemo-resistance and relapse in various tumor types61,62,63 including acute myeloid leukemia, lymphoid cells at various developmental stages have been implicated in key aspects of ALL, such as oncogenesis, chemo-resistance, and relapse64,65,66,67,68,69. Thus, the critical role of developmental factors in ALL biology is becoming increasingly evident. In this study, we provide evidence through both bioinformatic and experimental work that PU.1, a well-studied lineage-specific pioneer protein, is highly interactive with the GR and associated with TAD dynamics in ALL PDXs upon treatment with DEX. Even though various transcription levels and mutations were identified at gene loci coding for PU.1 and other TFs that are potentially associated with TAD alterations (RNA-seq and whole exome sequencing assays in Supplementary Fig. 8 and Supplementary Data 8), no specific gene dysregulation or loss-of-function mutation has been identified in the PDXs used in this study. This suggests that TF mutations may not be a cause for the abnormal chromatin accessibility in glucocorticoid-resistant ALL.
Alterations in tumor-associated TADs are often oncogenic, and DNA structural variation, together with enhancer-hijacking, play critical roles in the oncogenesis of various cancer types70,71. Several recent studies identified recurrent changes of TADs in leukemia subtypes and suggested their application as genomic signatures to stratify leukemias71,72,73. In this study, we identified a similar pattern, and reported TADs associated with distinct patterns of PU.1 binding in DEX-sensitive vs. resistant ALL, and PU.1 priming of lineage-specific cis-regulatory elements, rendering them as accessible before GR binding, in DEX-sensitive but not resistant ALLs. Our findings suggest that the PU.1 priming and eviction are critical events leading to the formation of DEX-induced de novo TADs in ALL, which cannot be achieved by either the structural protein CTCF that stabilizes TADs or other developmental TFs such as EBF1 or TCF3.
Furthermore, the ZBTB family comprises a diverse group of TFs, with several members considered to be critical regulators of lymphoid cell development74. For example, MIZ-1 (encoded by ZBTB17) regulates B-cell differentiation at early progenitor stages75, while PLZF (encoded by ZBTB16) directs differentiation of the NKT cell lineage52 and ZBTB16 translocations have been reported critical in driving leukemogenesis, such as ZBTB16-RARA in acute promyelocytic leukemia (APL)76 and ZBTB16-ABL1 in T-ALL77,78. Our data suggest that ZBTB16 was combinatorically regulated by GR and PU.1 at its LSO and SE, leading to the formation of a de novo TAD upon DEX treatment.
Overall, while epigenetic changes underlying DEX resistance may differ across different ALLs, this multi-omics analyses provides insights into a pathway of developmental factors modulating cancer epigenomes and sheds light on the unified upstream mechanism driven by developmental pioneer factor PU.1. These findings provide a link between the lineage-specific transcription factors, response to glucocorticoids and the activity of epigenetic modulators and provide a path to translate fundamental epigenetic research into the clinic.
Methods
ALL xenograft model and primary patient samples
The process of establishing continuous xenografts from ALL biopsies in immunodeficient NOD/SCID interleukin (IL)-2 receptor gamma chain null (NSG) mice has been previously described18,28. ALL biopsies were obtained for xenografting with informed, written consent approved by the Human Research Ethics Committees of the South Eastern Sydney Illawarra Area Health Service and UNSW Sydney. All animal studies had approval from the Animal Care and Ethics Committee of UNSW Sydney. The fidelity of all PDXs is routinely validated by high-density SNP (single nucleotide polymorphism) arrays as previously described24.
There is no restriction from the Ethics Committees on the maximal leukemia burden in mice models.
In vivo treatments
ALL xenograft cells were inoculated by tail-vein injection into NSG mice and engraftment was monitored weekly as previously described18,28. Mice were housed under a 12-hour light/dark cycle, with a temperature range of 22–26 °C and a relative humidity range of 40%-70% (optimum humidity: 55%-60%). Mice were randomized and treated with either DEX (15 mg/kg) or saline vehicle control by intraperitoneal injection when the %huCD45+ cells in the peripheral blood reached >50% and euthanized 8 hours thereafter. Cell suspensions of spleens were prepared, and mononuclear cells were enriched to >97% human cells by density gradient centrifugation. After harvesting, cells were immediately resuspended in fetal calf serum (FCS) containing 10% DMSO, frozen, and stored in liquid nitrogen for further use.
Animal euthanasia: The CO2 used for euthanasia is sourced from the compressed CO2 system, which is available in all holding and procedure rooms within the animal facility. The outlets are equipped with gas pressure and flow regulators to ensure controlled CO2 flow into the euthanasia chamber. Mice are euthanized in their home cages to minimize stress from unfamiliar environments. The transparent cages allow visual monitoring of the mice, ensuring the confirmation of loss of consciousness followed by death. A maximum of six mice can be euthanized at any given time to ensure proper oversight during the process.
Assessment of DEX sensitivity
In vitro DEX sensitivity was assessed by mitochondrial activity using Alamar Blue assay as described previously18,28. The half-maximal inhibitory concentration (IC50) was calculated from the dose-response curves. In vivo, DEX sensitivity was determined by the leukemia growth delay (LGD, treated-control). The sensitivities of ALL cells to DEX in vitro and in vivo have been described in our previous publications18,28. The PDXs were stratified into glucocorticoid-sensitive or resistant groups based on their in vitro IC50 and in vivo LGD.
Rapid Immunoprecipitation Mass Spectrometry (RIME)
RIME was done with some minor modifications to the original protocol79. Briefly, 1 × 108 cells were collected from ALL-54S or ALL-50R upon acute treatment with DEX or vehicle control (8 hours), pelleted, and initially cross-linked using 2 mM disuccinimidyl glutarate (DSG) in PBS for 20 min with shaking at RT. After pelleting the cells, the cells were resuspended in 1% formaldehyde in PBS and incubated at RT with shaking for 10 min to double-cross-linked the samples45. Crosslinking was quenched by the addition of glycine to a final concentration of 0.125 M and an incubation at RT for 5 min. The cells were washed with ice-cold PBS, and then resuspended in PBS containing protease inhibitors (PI). The nuclear fraction was extracted by resuspending the pellet in 1 mL LB1 (50 mM HEPES-KOH pH7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Igepal CA-630, 0.25% Triton X-100) containing PI, followed by rotating the samples at 4 °C for 10 min. After pelleting the cells, cells were resuspended in 1 mL LB2 (10 mM Tris-HCl pH8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) containing PI, mixed by rotation at 4 °C for 5 min, then pelleted. The cells were then resuspended in 300 µl LB3 (10 mM Tris-HCl pH8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-Deoxycholate, 0.5% N-laurosylsarcosine) containing PI, and sonicated for 7 cycles (30 seconds on, 30 seconds off). To the sonicated samples, 10% Triton X-100 in LB3 was added to a final concentration of 1%, followed by centrifugation at 20,000xg for 10 mins at 4 °C to remove the debris. The supernatant, which is the nuclear fraction, was then collected for immunoprecipitation. Protein A/G magnetic beads (Thermo Fisher Scientific) were firstly blocked with Pierce Protein-Free PBS blocking buffer (Thermo Fisher Scientific) and resuspended in LB3 containing 1% Triton X-100. The blocked beads were incubated with the supernatant containing the nuclear fraction for 1 hour rotating at RT to preclear the samples. The pre-cleared samples were incubated with 10 µg anti-NR3C1 antibody (Atlas Antibodies) overnight at 4 °C with rotation. The samples/antibody mixtures were then incubated with blocked beads for 1 hour at RT with rotation. The beads were washed 10 times with LiCl RIPA buffer (50 mM HEPES pH7.6, 1 mM EDTA, 0.7% Na-Deoxycholate, 1% Igepal CA-630, 0.5 M LiCl), followed by washing twice in 100 mM ammonium hydrogen carbonate (AMBIC) solution. The beads were further washed three times with 25 mM AMBIC solution before tryptic on-bead digestion was performed by adding 10 µl of sequencing grade trypsin (0.02 µg/µl in 25 mM AMBIC solution) and incubating overnight at 37 °C. An additional 10 µl of sequencing grade trypsin (0.02 µg/µl in 25 mM AMBIC solution) was added to the beads, and on-bead digestion was performed for another 1 hour at 37 °C. The supernatant containing peptides was collected and combined with TFA to give a final concentration 0.1%. Peptides were loaded onto EvoTip Pure tips for desalting and as a disposable trap column for nanoUPLC using an EvoSep One system. A pre-set EvoSep 100 SPD gradient was used with an 8 cm EvoSep C18 Performance column (150 mm×1.5 mm). The nanoUPLC system was interfaced to a timsTOF HT mass spectrometer (Bruker) with a CaptiveSpray ionization source (Source). Positive PASEF-DIA, ESI-MS, and MS2 spectra were acquired using Compass HyStar software (version 6.2, Thermo). Instrument source settings were: capillary voltage, 1500 V; dry gas, 3 l/min; dry temperature; 180 °C. Spectra were acquired between m/z 100-1,700. Custom TIMS settings were applied as 1/K0 0.−1.60 V.s/cm2; Ramp time, 100 ms; Ramp rate 9.42 Hz. PASEF-DIA acquisition specified DIA-windows of 25 Th width between 400–1201 Th and a mobility range of 0.6–1.43 1/K0. The total cycle time was 1.8 s. Collision energy was interpolated between 20 eV at 0.6 V.s/cm2 to 59 eV at 1.6 V.s/cm2. Peak lists in.d format were imported into DIA-NN software (ver. 1.8)80 for identification and relative quantification. An in-silico spectral library was created from the human subset of SwissProt appended with common proteomic contaminants. The two-pass method was used in DIA-NN, first searching against the in-silico library and then iterating against the DIA data from these samples for a second search. The search was run at 1% FDR. DIA-NN results in peptide-centric.tsv format were filtered to peptide q-values < 0.01, then pivoted on protein accessions before further filtering to require a minimum of 2 unique peptides per protein match and protein q-values < 0.01. For statistical comparison between groups, missing values were imputed with minimum values, and differential abundance was tested using limma through the FragPipe-Analyst (http://fragpipe-analyst.nesvilab.org/), run as a local installation in R-shiny. Three biological replicates were used for each group and the Hochberg and Benjamini correction was used for multiple test correction.
HiC library preparation and sequencing
HiC was performed using a modified version of a previously described protocol81. Briefly, 10 million cells were collected and cross-linked in 10 ml of PBS containing 1% formaldehyde for 10 minutes at room temperature. The reaction was stopped by 0.125 M glycine solution. A cross-linked cell pellet was used to prepare nuclei, which were subjected to digestion with restriction enzyme MboI followed by end-filling to create biotin-labeled blunt ends. Blunt ends were ligated by T4 DNA ligase at room temperature for 4 hours with rotation. Nuclei were treated with proteinase K and 10% sodium dodecyl sulfate (SDS) at 55 °C for 30 minutes. Following sodium acetate and ethanol precipitation, DNA was isolated and dissolved in Tris buffer. DNA was sonicated using a Bioruptor sonicator (Diagenode SA, Belgium) to obtain 150-300 bp fragments, followed by double-sided size selection using AMPure XP beads at 0.55X and 0.3X volume for HiC. Biotinylated HiC material was then purified using Dynabeads MyOne Streptavidin T1 beads (Life technologies). While on beads, DNA ends were blunt-ended and dA-tailed, followed by ligation of the Illumina sequencing adapter with T4 DNA ligase. On-bead PCR of 8 cycles (95 °C for 2 min, followed by 94 °C for 80 s, 65 °C for 30 s, 72 °C for 30 s) was performed and the final library was size selected using 1.5% agarose gel to obtain 400–700 bp fragments. Sequencing was performed on the Illumina HiSeq2500 to obtain 150 bp paired-end reads at a sequencing depth of 100 million reads per sample.
ChIP-seq
ChIP and ChIP-seq were carried out as previously described18,28. Briefly, in vivo treated PDX cells were revived from cryostorage and fixed with 1% formaldehyde for 10 min at room temperature. Nuclei were then extracted, lysed, and chromatin was fragmented to 200-500 bp using a Bioruptor sonicator (Diagenode SA, Belgium) at 4 °C with 30 sec on/off for 10 cycles. Separate immunoprecipitates were produced using immunoglobulin raised against GR (Cell Signaling, Danvers, MA, USA, Cat. #3660, Rabbit, 1:100), PU.1 (Cell signaling, Cat. #2258, Rabbit, 1:50), CTCF (Cell Signaling, Cat. #3418, Rabbit, 1:50), histone marks (Cell Signaling, H3K4Me3, Cat. #9727, Rabbit, 1:100; H3K4Me1, Cat. #5326, Rabbit, 1:50; H3K27Ac, Cat. #8173, Rabbit, 1:100; and H3K27Me3, Cat. #9733, Rabbit, 1:50). DNA from protein-associated complexes were reverse crosslinked and extracted using minElute columns (QIAGEN) in 20 µl water. ChIP DNA was then amplified and sequenced on Hiseq2500 platform for 50 bp single-end reads at 20 million reads per sample.
RNA-seq
For gene expression studies, RNA was extracted immediately after cell harvesting using the RNeasy Mini Kit (QIAGEN, Valencia, CA, USA) following manufacturer’s instructions. RNA samples with integrity number (RIN) > 8.0 were sent to NovogeneAIT Genomics (Beijing, China) for library preparation and sequencing using Hiseq4000 platform for 150 bp paired-end reads at 40 million reads per sample.
ATAC-seq
ATAC-seq was carried out as previously described18,82. Briefly, frozen PDX cells that were harvested from mice after dexamethasone or vehicle control treatment in vivo were revived from cryostorage, and 50000 cells per sample were lysed in lysis buffer [10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2 and 0.1% (v/v) Igepal (Sigma, St Louis, MO, USA)] on ice. Nuclei were extracted by centrifugation at 500 × g for 10 min at 4 °C, and then incubated with the Nextera Tn5 Transposase (Nextera kit, Illumina) at 37 °C for 30 min. After the tagmentation, qPCR was performed to determine the optimal number of cycles to amplify the library to reduce artifacts associated with saturation PCR of complex libraries. PCR was then performed for the optimum number of cycles using the following PCR conditions: 72 °C for 3 min; 98 °C for 30 s; and thermocycling at 98 °C for 10 s, 63 °C for 30 s and 72 °C for 1 min; following by 72 °C 1 min. After the PCR reaction, libraries were purified with the Qiagen MinElute PCR Purification Kit. Libraries were using Hiseq4000X10 (150PE) platform with 100 M reads per sample.
HiC data analysis
HiC data were first trimmed and then mapped against hg19 human reference genome using runHi-C pipeline based on the 4DN consortium. Specifically, Burrows-Wheeler Aligner (BWA) was used for the FASTQ file alignment, and low-quality aligned reads and PCR duplicates were removed. Aligned reads were then paired and filtered for fragments containing ligations of at least two different restriction fragments. These reads were then binned at 5-kb resolution. To generate the contact matrix at multiple resolutions (5, 10, 25, 40, 50, 100, 250, 500 kb, 1, 2.5, 5, and 10 Mb), we used the run-cool2multirescool script from 4DN consortium, which performed the ICE normalization at the same time. We used Coolbox to visualize ICE-normalized genomic HiC data. The juicer tool was also used to generate multiresolution.hic files, which can be visualized using Juicebox.
In Fig. 1, we divided the whole genome into 60,000 bins at 50 kb resolution and grouped them based on the enrichment of DHS domains. We have previously analyzed DHS-seq datasets from 78 different cell types in the ENCODE database18, which identified 0.8 million open chromatin domains (DHS domains) and defined a series of categories of DHS domains based on their chromatin accessibility and histone modification data. Here, in this study, we aligned the PCA values derived from HiC contact intensities with the domain categories.
To study DEX-induced chromatin conformation changes on a genome-wide scale, we compared HiC contact intensities before and after DEX treatment in ALL-54S and ALL-50R, and defined dynamic bins and stable bins as described in the legend of Fig. 1D. To further interrogate the subset of stable bins, we divided these regions into those with high (−0.2 < PCA < 0.2 and values of each group > 0) and low (−0.2 < PCA < 0.2 and values of each group <0) contact intensities. Next, ΔΔHiC was calculated as ΔHiC of ALL-54S (DEX – Control) - ΔHiC of ALL-50R (DEX – Control), which depicts differential bin activity between control and DEX-treated group in ALL-50R and -54S following DEX treatment in vivo. ΔΔHiC Fold change < −0.24 or > 0.24 and -log10 (adj.p-Value) > 0.2 were considered as significant changed bins.
To generate a volcano plot between Dex vs Ctrl of ALL-50R and ALL-54S groups, a generalized linear model were applied to calculate the statistical significance for the exponential transformed PCA values of each HiC bin. The PCA values of each bin were 10-based exponential transformed and R package edgeR (version 4.2.1) was used to estimate statistical significance of the transformed values. Dispersion parameters were estimated and implemented by “estimateGLMCommonDisp” of EdgeR, setting estimating method of “deviance”.
ChIP-seq, ATAC-seq and RNA-seq data analysis
ChIP-seq, ATAC-seq, and RNA-seq analyses were performed as previously described83. In brief, ChIP-seq reads were aligned to human reference genome hg19 using the Burrows-Wheeler Aligner (BWA, default parameters)84. Peak calling was performed using three algorithms, HOMER (FDR 1e-3 and 1e-2)38, MACS85 (p < 1e-5 and p < 1e−9) and SPP86 (FDR 1e-2 and FDR 1e-5). Peaks identified by ≥2 algorithms were kept in the final peak list. These sites were assigned as regulatory regions to specific genes using the genomic regions enrichment of annotations tool (GREAT) analysis package87. Paired-end RNA-seq reads were aligned to the human reference genome hg19 using the software STAR (v2.5.0b)88 with standard parameters. Gene expression levels were quantified using htseq (v0.9)89 and TMM normalized using the software package DESeq (v3.6) in the R statistical analysis software (v3.3.3)90,91. To visualize read intensities, aligned reads were further processed using the DeepTools function bamCoverage which normalized these datasets to reads per kilobase per million mapped reads and then visualized the results using the DeepTools functions plotProfile and plotHeatmap, or on the UCSC Genome Browser. Furthermore, signal intensities of regulatory regions were defined by normalizing the absolute reads of each peak to the total reads of the dataset and region size. Heatmaps of gene expression profiles and epigenetic profiles at related regulatory regions were generated using Genepattern (Broad Institute, USA). Hierarchical clustering was performed using the commercially available package Partek Genomics Suite (version 6.6).
Pathway analysis from ATAC-seq datasets: Trimmed reads were aligned with Bowtie2 (v.2.4.1) to hg19, with a maximum insert size of 2000 bases. Aligned bam files were sorted (Samtools v.1.9), and duplicates were marked (Picard v.2.22.8). Genrich (v. 0.6.1) was used to perform peak calling, and R (v.4.2.0) and package DiffBind (v.3.8.4) were used for differential binding analysis. Gene-to-peak associations were determined using the GREAT online service (v.4.0.4; http://great.stanford.edu/public/html/) with default settings. GREAT was also used to determine the gene set enrichment, with default settings. Then package clusterProfiler (v.4.6.2) was used for pathway analysis.
GSEA analysis: Trimmed reads were aligned with bwa (v.0.7.10) to hg19. Gene counts were generated using featureCounts (v.2.0.1) and differential expression analysis was performed using edgeR (v.4.2.1). GR glucocorticoid-associated gene sets were retrieved from the databases of MSigDB. GSEA analysis was carried out using the R package clusterProfiler (v.4.6.2) and the enrichplot (v.1.24.0) was used for the visualization of enrichment results. P value was calculated using an empirical phenotype-based permutation test with one-tailed tests, and adjustments were made for multiple comparisons.
Link enhancer with promoter using HiC data
To map enhancer with promoter using HiC data, interaction normalization, matrix creation, and significant interaction identification at 10 kb resolution were performed using HOMER tools (v4.11.1)92. Gene annotations (version GeneCode V42lift37) were retrieved from UCSC93,94. Gene TSS regions 1.5 kb upstream or downstream of the interactions of GR-LSOs were extracted based on the significant interactions of the HiC data. Interacting pairs were deduplicated and sorted by HiC scores.
Co-binding Z-score calculation
Bootstrapping was used to assess the statistical significance of combinatorial binding events involving CTCF, EBF1, PU.1, and GR across all 15 potential binding patterns as previously described40. In brief, we used a conservative estimate of 80,000 binding sites per protein95,96 to establish a background distribution of combinatorial binding events and then calculated a standardized z-score to measure the deviation between the number of combinatorial binding events (i.e., determined by ChIP-seq) from the expected mean of the background distribution.
Motif analysis
Motif analyses were performed using ChIP-seq data of H3K4Me1, H3K27Ac, H3K4Me3, H3K27Me3, and GR in ALL-50R and -54S before and post in vivo DEX treatment. Next, GR-bound regions matching the following criteria were selected (Supplementary Data 4)
-
a.
defined as Pro-S or Pr-Enh-S or RO or NS in ALL-50R control, AND
-
b.
defined as Pro-S or Pr-Enh-S or RO or NS in DEX-treated ALL-50R, AND
-
c.
defined as Pro-S or Pr-Enh-S or RO or NS or Enh-S in ALL-54S control, AND
-
d.
defined as Pro-A or Enh-A or Pro-Enh-A or AO in DEX-treated ALL-54S
The selected GR-bound regions were analyzed using an integrative genomics method for the prediction of regulatory features and cis-regulatory modules from i-cisTarget43,44. The regulatory network model was built based on analysis of connectivity between GR, PU.1, EBF1, and TCF3 with their regulated genes by i-cisTarget and was visualized by Cytoscape51.
Motif analysis of DB2313-induced PU.1 Loss
The extended “DB-sensitive” PU.1 motif (derived from examination of DB2115 displaced PU.1 binding in MOLM13 cells from Taylor et al.54) was used to calculate a log-odds score for GR-LSOs and non-GR-LSOs using the annotated peaks function from the Homer package with the -m and -mscore options enabled38.
Cell line authentication and mycoplasma testing
DNA was extracted by a commercial kit from Corning Incorporated (catalog number: AP-EMN-BL-GDNA-250G). The twenty STRs including Acerogenin locus were amplified by six multiplex PCR and separated on ABI 3730XL Genetic Analyzer. The signals were then analyzed by the software GeneMapper.
The Mycoplasma test was performed using MycoBlue Mycoplasma Detector kit from Nanjing Vazyme Biotech Co., Ltd (catalog number: D101-01). Briefly, after three days of cell culture, samples were centrifuged at 2300 rpm (500 × g) for 5 minutes and the supernatant was collected. The experiment was performed following the manufactory instructions. After incubation of 24 μl MycoBlue Buffer, 1 μl MycoBlue Enzyme, and 1 μl supernatant for 1 hour at 60 oC, observe the color change. If the reaction solution remains purple, the sample is negative for Mycoplasma. If the solution turns sky blue, the sample is positive for Mycoplasma.
Luciferase reporter assay
Sequences of target regulatory elements were synthesized as dsDNA by Integrated DNA Technologies (Coralville, IA, USA) and inserted into pGL3-promoter (pGL3p) vector (Promega, Madison, Wisconsin, USA) between SalI and BamHI cutting sites downstream of the firefly luciferase gene. The cloned vector was co-transfected with pRL-TK renilla luciferase control reporter vectors (Promega) into Nalm6 cells. The firefly and renilla luminescence were detected in the transfected cells treated with DB2313 (MCE, NJ, USA) in the presence or absence of DEX (Sigma) using the Dual-Glo Luciferase Assay System (Promega). The firefly luminescence was normalized to renilla luminescence for each condition. Fold inductions were then calculated by normalizing to the luminescence signal of the pGL3p control sample.
Cytotoxicity assay
Nalm6 cell line (ATCC, Manassas, USA, Cat. #CRL-3273) was cultured in RPMI 1640 media supplemented with 10% FCS, 1 mmol/L pyruvate, nonessential amino acids, 10 mmol/L 2-mercaptoethanol, and 2 mmol/L L-glutamine. Nalm6 cells were seeded in 96-well U-bottomed plates at 20,000 cells per 100 μL medium per well. DB2313 or vehicle control was added to a final concentration of 1 μM in triplicate wells. After 48 hr, 10 μM DEX (Sigma) or vehicle control was added to the cells pre-treated with either DB2313 or vehicle control. The cells were incubated for a series of time points. Following the incubation, Cell Counting Kit-8 (Vazyme) was added. After an additional 4 hr incubation, fluorescence was measured (450 nm) and cell viability was expressed relative to vehicle-treated cells. The presumed additive effect of combination treatment (DB2313 and DEX) was shown by viability% of DB2313 × viability% of DEX-treated conditions. Synergy score was calculated by SynergyFinder (https://synergyfinder.fimm.fi). The p values were calculated as the combination vs. the presumed additive effect at each concentration point.
qRT-PCR
Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out as previously described18,28. Briefly, total RNA was isolated using TRIzol (life technologies) and cDNA was synthesized using HiScriptIII All-in-one RT SuperMix kit (Vazyme). Primers and probes for targeted genes were purchased from Life Technologies and qRT-PCR was carried out in triplicate under cycling conditions according to the manufacturer’s instructions. qRT-PCR was conducted using the ChamQ Universal SYBR qPCR Master Mix (Vazyme) on a ViiA 7 qRT-PCR system (Applied Biosystems, Waltham, MA). Relative expression of target genes was calculated using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression as control. The sequences of primers used are as follows:
BIM forward primer, ‘5-TAAGTTCTGAGTGTGACCGAGA-3’
reverse primer, ‘5-GCTCTGTCTGTAGGGAGGTAGG-3’;
RASA1 forward primer, ‘5-ACTTGACAGAACGATAGCAGAAG-3’
reverse primer ‘5-GCCTCCGATCACTCTCTCTTA-3’;
ZBTB16 forward primer, ‘5-GAGATCCTCTTCCACCGCAAT-3’
reverse primer ‘5-CCGCATACAGCAGGTCATC-3’;
GILZ forward primer, ‘5-AACACCGAAATGTATCAGACCC-3’
reverse primer, ‘5-TGTCCAGCTTAACGGAAACCA-3’;
GAPDH forward primer, ‘5-GGAGCGAGATCCCTCCAAAAT-3’
reverse primer, ‘5-GGCTGTTGTCATACTTCTCATGG-3’.
CRISPR/Cas9-Mediated In Vivo Gene Knockout (KO)
Stable Cas9-expressing PDX cells were generated using the FU-Cas9-mCherry plasmid (Addgene #70182). In brief, HEK293FT cells (Invitrogen) were transfected using Lipofectamine™ 2000 (Thermo Fisher Scientific 11668019) with FU-CAS9-mCherry and the pMD2.G envelope (Addgene #12259) and psPAX2 packaging plasmids (Addgene #12260) to generate lentiviral particles. PDX cells were revived and transduced ex vivo with lentivirus for 24 hours, washed in PBS, and injected into NSG mice (Australian Bioresources) for in vivo expansion (1 million cells per mouse). Mice were sacrificed at high leukemia burden (>50% huCD45+% in mouse peripheral blood) and PDX cells were harvested from the spleen. mCherry+ cells were sorted by fluorescence-activated cell sorting (FACS) to generate a purified Cas9-mCherry population and reinjected into naïve NSG mice for in vivo expansion.
For gene KO, single guide RNA (sgRNA) sequences against human SPI1 (PU.1), BCL2L11 (BIM), or non-targeting sgRNA sequences were cloned into the FgH1tUTG (Addgene #70138) GFP vector and lentiviral particles made as described above. Cas9-mCherry+ PDX cells were transduced, in vivo expanded, and sorted by FACS to generate a purified mCherry+ GFP+ population and reinjected into naïve NSG mice for in vivo expansion. sgRNA sequences are as follows:
Targeted Genes | sgRNA sequence |
SPI1 | AATACTCGTGCGTTTGGCGT |
BCL2L11 | GCCCAAGAGTTGCGGCGTAT |
Human non-targeting sequence | ACGGAGGCTAAGCGTCGCAA |
For in vivo gene KO and drug efficacy testing, Cas9-mCherry+ sgRNA-GFP + PDX cells were injected into NSG mice (6 mice/treatment group) and mice switched to Doxycycline-impregnated food (600 mg/kg Doxycycline) 7 days post-injection to activate sgRNA expression. Leukemia level was tracked weekly by tail vein bleeding and quantifying huCD45+ cells in mouse peripheral blood. At 1% huCD45+, DEX treatment was administered via intraperitoneal injection at 15 mg/kg daily, 5 days on/2 days off for 4 weeks. A leukemia event is defined as 25% huCD45+, or when the animal is euthanized due to leukemia-related morbidity, and event-free survival (EFS) was used to assess therapeutic enhancement with statistical significance defined as p < 0.01. At the leukemia event, mice were sacrificed and PDX cells were harvested from the spleen for quantification of CRISPR efficiency.
Evaluation of Indel mutagenesis
The CRISPR/Cas9-induced indel frequencies were quantified by Tracking of Indels by Decomposition (TIDE)97. Genomic DNA was extracted using an Isolate II Genomic DNA Kit (Bioline BIO-52066) following the manufacturer’s instructions. 100 ng genomic DNA was PCR amplified using Q5 polymerase (NEB M0494X) and primers flanking the sgRNA target region. The PCR products are next subjected to standard Sanger sequencing and the sequence data files of the edited samples and the wild-type samples were analyzed by the TIDE webtool (http://tide.nki.nl). It requires one pair of standard capillary (Sanger) sequencing reactions of the wild-type (wt) genome and the CRISPR/Cas9-edited genome (Cas9-targeted locus amplified by standard PCR). The sequence traces are then analyzed by a specially developed decomposition algorithm that identifies the major induced mutations in the projected editing site and determines their frequency in a cell population. Specifically, the TIDE software decomposes the composite sequence trace into its individual components by means of multivariate non-negative linear modeling, with the control sequence trace (wt) serving as a template to model the individual INDEL components. This decomposition results in an estimate of the relative abundance of every possible INDEL. The software provides the R2 value as a goodness-of-fit measure and calculates the statistical significance for each INDEL.
Primer sequences used for INDEL quantification by TIDE assay are as follows:
Genes | Forward primer (5’-3’) | Reverse primer (5’-3’) |
SPI1 | GGAAGAAATGAACCCCGCAC | GAGGGCTGTAGGTCCAACG |
BCL2L11 | GGTTGGAATGTTTTCAGTTCTTGC | TCCTTTGCTGCCTCCTACTG |
Protein structure analysis
Mutated PU.1 proteins structures were predicted using Aphafold257 based on the following amino acid sequences and the results were viewed with PyMOL98.
Mutant #1: Serine insertion
MLQACKMEGFPLVPPQPSEDLVPYDTDLYQRSQTHEYYPYLSSDGESHSDHYWDFHPHHVHSEFESFAENNFTELQSVQPPQLQQLYRHMELEQMHVLDTPMVPPHPSLGHQVSYLPRMCLQYPSLSPAQPSSDEEEGERQSPPLEVSDGEADGLEPGPGLLPGETGSKKKIRLYQFLLDLLRSGDMKDSIWWVDKDKGTFQFSSKHKEALAHRWGIQKGNRKKMTYQKMARALRNYGKTGEVKKVKKKLTYQFSGEVLGRGGLAERRHPPH
Mutant #2: Glycine and Valine insertion
MLQACKMEGFPLVPPQPSEDLVPYDTDLYQRGVQTHEYYPYLSSDGESHSDHYWDFHPHHVHSEFESFAENNFTELQSVQPPQLQQLYRHMELEQMHVLDTPMVPPHPSLGHQVSYLPRMCLQYPSLSPAQPSSDEEEGERQSPPLEVSDGEADGLEPGPGLLPGETGSKKKIRLYQFLLDLLRSGDMKDSIWWVDKDKGTFQFSSKHKEALAHRWGIQKGNRKKMTYQKMARALRNYGKTGEVKKVKKKLTYQFSGEVLGRGGLAERRHPPH
Statistics & reproducibility
HiC-seq, RNA-seq, and ChIP-seq studies were performed using PDX cells from three randomized ALL-engrafted mice at each condition of treatments. Gene expression and cytotoxicity studies were performed with three independent experiments. Quantitative variables of normally distributed data were compared by the two-sided student t-test and non-normally distributed data were compared by the Mann-Whitney U test. All statistical tests were two-sided and p values < 0.05 were considered statistically significant.
No statistical method was used to predetermine the sample size. No data were excluded from the analyses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The ChIP-seq publicly available data used in this study are available in the NCBI’s Gene Expression Omnibus repository under accession code GSE10994918. The ChIP-seq and HiC data generated in this study have been deposited in the NCBI’s Gene Expression Omnibus repository under accession code GSE236085. The dataset is publicly available. The RNA-seq and ATAC-seq data generated in this study have been deposited in the Genome Sequence Archive99 in National Genomics Data Center100, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences under accession code HRA008240 and HRA008250. The datasets are accessible through a request process. The data do not include genetic information on individuals of Chinese people, and therefore are not subject to restrictions from the Chinese government. However, according to the GSA genetic data sharing policy, users must clarify how they intend to use the data in their research, including the research title, chosen research period, research purpose, methods, and techniques. Both data producers and users must sign a ‘Data Access Agreement of GSA-Human’. Once this agreement is approved by both parties, the datasets will be available for download. Upon granting access, the data will be available for a period of two years. For any questions, please contact Duohui Jing, the corresponding author of this paper; we will respond within two weeks. Please refer to the guidance of GSA, https://ngdc.cncb.ac.cn/gsa-human/document/GSA-Human_Request_Guide_for_Users_us.pdf. The RIME data generated in this study has been deposited in the ProteomeXchange under accession code PXD056483 and available to download from MassIVE (doi:10.25345/C52N4ZW0F; ftp://massive.ucsd.edu/v08/MSV000096019/). The dataset is publicly available. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided in this paper.
References
Kim, I. K. et al. Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells. Nat. Med. 21, 1010–1017 (2015).
Palumbo, A. et al. Daratumumab, bortezomib, and dexamethasone for multiple myeloma. N. Engl. J. Med. 375, 754–766 (2016).
Bhojwani, D. & Pui, C. H. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 14, e205–e217 (2013).
Pui, C. H. & Evans, W. E. A 50-year journey to cure childhood acute lymphoblastic leukemia. Semin. Hematol. 50, 185–196 (2013).
Hunger, S. P. & Mullighan, C. G. Acute lymphoblastic leukemia in children. N. Engl. J. Med. 373, 1541–1552 (2015).
Klein, K. et al. Glucocorticoid-induced proliferation in untreated pediatric acute myeloid leukemic blasts. Pediatr. Blood Cancer 63, 1457–1460 (2016).
Gruver-Yates, A. L. & Cidlowski, J. A. Tissue-specific actions of glucocorticoids on apoptosis: a double-edged sword. Cells 2, 202–223 (2013).
Cain, D. W. & Cidlowski, J. A. Immune regulation by glucocorticoids. Nat. Rev. Immunol. 17, 233–247 (2017).
Watson, L. C. et al. The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals. Nat. Struct. Mol. Biol. 20, 876–883 (2013).
Vockley, C. M. et al. Direct GR binding sites potentiate clusters of TF binding across the human genome. Cell 166, 1269–1281 e1219 (2016).
Guo, B., Huang, X., Cooper, S. & Broxmeyer, H. E. Glucocorticoid hormone-induced chromatin remodeling enhances human hematopoietic stem cell homing and engraftment. Nat. Med 23, 424–428 (2017).
Swinstead, E. E. et al. Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 165, 593–605 (2016).
Aranda, S., Mas, G. & Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 1, e1500737 (2015).
Hnisz, D., Day, D. S. & Young, R. A. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell 167, 1188–1200 (2016).
McDowell, I. C. et al. Glucocorticoid receptor recruits to enhancers and drives activation by motif-directed binding. Genome Res. 28, 1272–1284 (2018).
D’Ippolito, A. M. et al. Pre-established Chromatin interactions mediate the genomic response to Glucocorticoids. Cell Syst. 7, 146–160.e147 (2018).
Johnson, G. D. et al. Human genome-wide measurement of drug-responsive regulatory activity. Nat. Commun. 9, 5317 (2018).
Jing, D. et al. Lymphocyte-specific chromatin accessibility pre-determines glucocorticoid resistance in acute lymphoblastic leukemia. Cancer Cell 34, 906–921.e908 (2018).
Bergeron, B. P. et al. Epigenomic profiling of glucocorticoid responses identifies CIS-regulatory disruptions impacting steroid resistance in childhood acute lymphoblastic leukemia. Leukemia 36, 2374–2383 (2022).
Krivtsov, A. V. et al. A Menin-MLL inhibitor induces specific chromatin changes and eradicates disease in models of MLL-rearranged leukemia. Cancer Cell 36, 660–673.e611 (2019).
Li, J. et al. PRC2 inhibitors overcome glucocorticoid resistance driven by NSD2 mutation in pediatric acute lymphoblastic leukemia. Cancer Discov. 12, 186–203 (2022).
Liu, Y. F. et al. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine 8, 173–183 (2016).
Li, J. F. et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an International study of 1223 cases. Proc. Natl Acad. Sci. USA 115, E11711–E11720 (2018).
El-Hoss, J. et al. A single nucleotide polymorphism genotyping platform for the authentication of patient derived xenografts. Oncotarget 7, 60475–60490 (2016).
Liem, N. L. et al. Characterization of childhood acute lymphoblastic leukemia xenograft models for the preclinical evaluation of new therapies. Blood 103, 3905–3914 (2004).
Bachmann, P. S. et al. Divergent mechanisms of glucocorticoid resistance in experimental models of pediatric acute lymphoblastic leukemia. Cancer Res. 67, 4482–4490 (2007).
Lee, E. M. et al. Efficacy of an Fc-modified anti-CD123 antibody (CSL362) combined with chemotherapy in xenograft models of acute myelogenous leukemia in immunodeficient mice. Haematologica 100, 914–926 (2015).
Jing, D. et al. Opposing regulation of BIM and BCL2 controls glucocorticoid-induced apoptosis of pediatric acute lymphoblastic leukemia cells. Blood 125, 273–283 (2015).
Jones, L. et al. A review of new agents evaluated against pediatric acute lymphoblastic leukemia by the Pediatric Preclinical Testing Program. Leukemia 30, 2133–2141 (2016).
Jin, L. et al. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell 5, 31–42 (2009).
Piovan, E. et al. Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia. Cancer Cell 24, 766–776 (2013).
van Galen, J. C. et al. BTG1 regulates glucocorticoid receptor autoinduction in acute lymphoblastic leukemia. Blood 115, 4810–4819 (2010).
Jones, C. L. et al. MAPK signaling cascades mediate distinct glucocorticoid resistance mechanisms in pediatric leukemia. Blood 126, 2202–2212 (2015).
Serafin, V. et al. Glucocorticoid resistance is reverted by LCK inhibition in pediatric T-cell acute lymphoblastic leukemia. Blood 130, 2750–2761 (2017).
Nagao, K., Iwai, Y. & Miyashita, T. RCAN1 is an important mediator of glucocorticoid-induced apoptosis in human leukemic cells. PLoS One 7, e49926 (2012).
Cialfi, S. et al. Glucocorticoid sensitivity of T-cell lymphoblastic leukemia/lymphoma is associated with glucocorticoid receptor-mediated inhibition of Notch1 expression. Leukemia 27, 485–488 (2013).
Boller, S., Li, R. & Grosschedl, R. Defining B cell chromatin: lessons from EBF1. Trends Genet. 34, 257–269 (2018).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
de la Rica, L. et al. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 14, R99 (2013).
Beck, D. et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood 122, e12–e22 (2013).
Corces, M. R. et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016).
Dekker, J. et al. The 4D nucleome project. Nature 549, 219–226 (2017).
Imrichova, H., Hulselmans, G., Atak, Z. K., Potier, D. & Aerts, S. i-cisTarget 2015 update: generalized CIS-regulatory enrichment analysis in human, mouse and fly. Nucleic Acids Res. 43, W57–W64 (2015).
Herrmann, C., Van de Sande, B., Potier, D. & Aerts, S. i-cisTarget: an integrative genomics method for the prediction of regulatory features and cis-regulatory modules. Nucleic Acids Res. 40, e114 (2012).
Papachristou, E. K. et al. A quantitative mass spectrometry-based approach to monitor the dynamics of endogenous chromatin-associated protein complexes. Nat. Commun. 9, 2311 (2018).
Rinaldi, L. et al. The glucocorticoid receptor associates with the cohesin loader NIPBL to promote long-range gene regulation. Sci. Adv. 8, eabj8360 (2022).
Jann, J.-C. & Tothova, Z. Cohesin mutations in myeloid malignancies. Blood 138, 649–661 (2021).
Mayayo-Peralta, I. et al. PAXIP1 and STAG2 converge to maintain 3D genome architecture and facilitate promoter/enhancer contacts to enable stress hormone-dependent transcription. Nucleic Acids Res. 51, 9576–9593 (2023).
Subramanian, S. et al. Genome-wide transcription factor-binding maps reveal cell-specific changes in the regulatory architecture of human HSPCs. Blood 142, 1448–1462 (2023).
Weikum, E. R., Knuesel, M. T., Ortlund, E. A. & Yamamoto, K. R. Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat. Rev. Mol. Cell Biol. 18, 159–174 (2017).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Savage, A. K. et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Taylor, S. J. et al. Pharmacological restriction of genomic binding sites redirects PU.1 pioneer transcription factor activity. Nat. Genet. 56, 2213–2227 (2024).
Rhein, P. et al. Gene expression shift towards normal B cells, decreased proliferative capacity and distinct surface receptors characterize leukemic blasts persisting during induction therapy in childhood acute lymphoblastic leukemia. Leukemia 21, 897–905 (2007).
Choi, J. et al. Molecular targets of glucocorticoids that elucidate their therapeutic efficacy in aggressive lymphomas. Cancer Cell 42, 833–849.e812 (2024).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Quatrini, L. & Ugolini, S. New insights into the cell- and tissue-specificity of glucocorticoid actions. Cell Mol. Immunol. 18, 269–278 (2021).
Bachmann, P. S., Gorman, R., Mackenzie, K. L., Lutze-Mann, L. & Lock, R. B. Dexamethasone resistance in B-cell precursor childhood acute lymphoblastic leukemia occurs downstream of ligand-induced nuclear translocation of the glucocorticoid receptor. Blood 105, 2519–2526 (2005).
Bachmann, P. S. et al. Epigenetic silencing of BIM in glucocorticoid poor-responsive pediatric acute lymphoblastic leukemia, and its reversal by histone deacetylase inhibition. Blood 116, 3013–3022 (2010).
Bonnet, D. & Dick, J. E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3, 730–737 (1997).
Visvader, J. E. & Lindeman, G. J. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat. Rev. Cancer 8, 755–768 (2008).
Essers, M. A. & Trumpp, A. Targeting leukemic stem cells by breaking their dormancy. Mol. Oncol. 4, 443–450 (2010).
Kong, Y. et al. CD34+CD38+CD19+ as well as CD34+CD38-CD19+ cells are leukemia-initiating cells with self-renewal capacity in human B-precursor ALL. Leukemia 22, 1207–1213 (2008).
le Viseur, C. et al. In childhood acute lymphoblastic leukemia, blasts at different stages of immunophenotypic maturation have stem cell properties. Cancer Cell 14, 47–58 (2008).
Rehe, K. et al. Acute B lymphoblastic leukaemia-propagating cells are present at high frequency in diverse lymphoblast populations. EMBO Mol. Med. 5, 38–51 (2013).
Spinella, J. F. et al. Mutational dynamics of early and late relapsed childhood ALL: rapid clonal expansion and long-term dormancy. Blood Adv. 2, 177–188 (2018).
Ma, X. et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat. Commun. 6, 6604 (2015).
Kim, J. C. et al. Transcriptomic classes of BCR-ABL1 lymphoblastic leukemia. Nat. Genet. 55, 1186–1197 (2023).
Johnstone, S. E. et al. Large-scale topological changes restrain malignant progression in colorectal cancer. Cell 182, 1474–1489 e1423 (2020).
Kloetgen, A. et al. Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia. Nat. Genet. 52, 388–400 (2020).
Xu, J. et al. Subtype-specific 3D genome alteration in acute myeloid leukaemia. Nature 611, 387–398 (2022).
Schuetzmann, D. et al. Temporal autoregulation during human PU.1 locus SubTAD formation. Blood 132, 2643–2655 (2018).
Constantinides, M. G. & Belkaid, Y. Early-life imprinting of unconventional T cells and tissue homeostasis. Science 374, eabf0095 (2021).
Kosan, C. et al. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity 33, 917–928 (2010).
Chen, Z. et al. Fusion between a novel Krüppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 12, 1161–1167 (1993).
Chen, B. et al. Identification of fusion genes and characterization of transcriptome features in T-cell acute lymphoblastic leukemia. Proc. Natl Acad. Sci. USA 115, 373–378 (2018).
Dai, Y. T. et al. Transcriptome-wide subtyping of pediatric and adult T cell acute lymphoblastic leukemia in an international study of 707 cases. Proc. Natl Acad. Sci. USA 119, e2120787119 (2022).
Mohammed, H. et al. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 11, 316–326 (2016).
Demichev, V., Messner, C. B., Vernardis, S. I., Lilley, K. S. & Ralser, M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020).
Belaghzal, H., Dekker, J. & Gibcus, J. H. Hi-C 2.0: An optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods 123, 56–65 (2017).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21–29 (2015). 21–29.
Chacon, D., Beck, D., Perera, D., Wong, J. W. & Pimanda, J. E. BloodChIP: a database of comparative genome-wide transcription factor binding profiles in human blood cells. Nucleic Acids Res. 42, D172–D177 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Kharchenko, P. V., Tolstorukov, M. Y. & Park, P. J. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 26, 1351–1359 (2008).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Lin, Y. C. et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat. Immunol. 13, 1196–1204 (2012).
Nassar, L. R. et al. The UCSC Genome Browser database: 2023 update. Nucleic Acids Res. 51, D1188–D1195 (2023).
Rosenbloom, K. R. et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 41, D56–D63 (2013).
Wilson, N. K. et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7, 532–544 (2010).
Tijssen, M. R. et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev. Cell 20, 597–609 (2011).
Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Schrodinger, L. L. C. The PyMOL Molecular Graphics System, Version 1.8. (2015).
Chen, T. et al. The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteom. Bioinforma. 19, 578–583 (2021).
Members, C.-N. Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 50, D27–D38 (2022).
Acknowledgements
This research was funded by grants from the National Key R&D Program of China (2022YFE0200100, 2023YFC2508901), the National Natural Science Foundation of China (NSFC 82070144, 82270155, 82070227, 82270187, 82050410359, 32271165), the Innovation Program of Shanghai Science and Technology Committee (23141903000, 21430711800), Ideas Grant funding from the National Health and Medical Research Council of Australia (APP1181666), Kids Cancer Alliance (KCA) New Project Grant (KCA Project Code: 114), Perpetual’s 2019 IMPACT Philanthropy Fund (IPAP2019/0902), UNSW-SJTU Collaborative Research Seed Grant (RG200056), the Mobility Program of the Joint Committee of the Sino-German Center for Research Promotion by the NSFC and the Deutsche Forschungsgemeinschaft (M-0337), Innovative research team of high-level local universities in Shanghai, Hebei Province Major Science and Technology Support Project (242W7712Z). RBL was supported by a fellowship from The National Health and Medical Research Council of Australia (NHMRC Fellowship APP1157871). BBSRC-Australia Partnering Award to ANH, KSB, CdB, and RL: BB/X018288/1, DJK, and KSB are funded by the Kay Kendall Leukemia Fund, Intermediate Fellowship KKL1377 to KSB. WZ and ANH are funded by the BBSRC Responsive Mode Grant Award to ANH: BB/V000071/1. The York Center of Excellence in Mass Spectrometry was created thanks to a major capital investment through Science City York, supported by Yorkshire Forward with funds from the Northern Way Initiative, and subsequent support from EPSRC (EP/K039660/1; EP/M028127/1). The authors acknowledge the ENCODE Consortium for generating DNaseI HS datasets and ChIP-seq datasets. The authors acknowledge the Translational Cancer Research Network of the Cancer Institute of NSW and NSW Pathology. Children’s Cancer Institute Australia is affiliated with UNSW Sydney and The Sydney Children’s Hospitals Network.
Author information
Authors and Affiliations
Contributions
D.J., D.B., R.B.L., and C.E.B. designed the study. D.J., H.C., Y.H., M.J., X.T., H.J.K., D.J.K., K.S.B., W.Z., A.N.H., and A.A.D. performed experiments and analyzed the data. D.B., F.T., and T.A.C. performed bioinformatics analysis. D.J., D.B., H.C., C.E.B., and R.B.L. interpreted the data and wrote the manuscript. H.Z., W.X., G.S., Y.W., and M.W. assisted in performing experiments and data analysis. D.C.F., Y.W., and S.J.T. contributed to bioinformatics analysis. C.Z., J.W., and J.Q.M. contributed to the data interpretation and revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jason Carroll, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Beck, D., Cao, H., Tian, F. et al. PU.1 eviction at lymphocyte-specific chromatin domains mediates glucocorticoid response in acute lymphoblastic leukemia. Nat Commun 15, 9697 (2024). https://doi.org/10.1038/s41467-024-54096-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54096-2