Abstract
Immune checkpoint inhibitors (ICI) represent new anticancer agents and have been used worldwide. However, ICI can potentially induce life-threatening severe cutaneous adverse reaction (SCAR), such as Stevens-Johnson syndrome/toxic epidermal necrolysis (SJS/TEN), hindering continuous ICI therapy. We examine 6 cohorts including 25 ICI-induced SJS/TEN patients and conduct single-cell RNA sequencing (scRNA-seq) analysis, which shows overexpression of macrophage-derived CXCL10 that recruits CXCR3+ cytotoxic T lymphocytes (CTL) in blister cells from ICI-SJS/TEN skin lesions. ScRNA expression profiles and ex vivo blocking studies further identify TNF signaling as a pathway responsible for macrophage-derived CXCL10 and CTL activation. Based on the trajectory analysis, ICI-activated T cells from whole blood are proposed to serve as the initial cells involved in inflammation, that lead to monocytes differentiating into macrophages and increasing their susceptibility to migrate to the lesion sites. Compared with systemic corticosteroids treatment, ICI-induced SJS/TEN patients treated with biologic TNF blockade showed a significantly rapid recovery and no recurrence of SCAR with continuous ICI therapy. Our findings identify that macrophage-eliciting CTL contribute to the pathogenesis of ICI-induced epidermal necrolysis and provide potential therapeutic targets for the management and prevention of SCAR induced by ICI therapy.
Similar content being viewed by others
Introduction
Immune checkpoint inhibitors (ICI), such as antibodies against cytotoxic T lymphocyte-associated molecule 4 (CTLA-4), programmed cell death protein 1 (PD-1), and programmed death-ligand 1 (PD-L1), represent a new class of anticancer agents and have been approved to treat various advanced cancers1,2,3. Although ICI are effective across several cancer types, they can still induce autoimmune-like toxicities referred as immune-related adverse events (irAEs)4,5,6 classified by different clinical features, including cutaneous reactions, pneumonitis, colitis, and myocarditis7,8. ICI-induced cutaneous adverse drug reactions (cADR) can range from mild maculopapular exanthema and lichenoid dermatitis to life-threatening severe cutaneous adverse reaction (SCAR), such as Stevens-Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN)9,10,11,12. Most of the cADR induced by ICI are relatively benign, but life-threatening SJS/TEN are worth to be noticed. According to U.S. Food and Drug Administration database, there has been reported more than 100 cases of ICI-induced SJS/TEN13, which hinder the further ICI therapy.
SJS and TEN are typically induced by drugs, and cause high morbidity and mortality (10–50%)14,15. Traditional small chemical drug(s)-induced SJS/TEN is known to be associated with the activation of effector CD8+cytotoxic T lymphocytes (CTL), which recognize culprit drugs presented by human leukocyte antigen (HLA) molecules or their altered peptide repertoires on antigen-presenting cells16,17,18,19,20,21,22. We previously identified granulysin (GNLY) as the key mediator that triggers disseminated keratinocyte apoptosis in small molecular drug(s)-induced SJS/TEN23,24,25. ICI, such as anti-CTLA-4 and anti-PD-1/PD-L1 antibodies, can directly restore T cell activity through immune checkpoint molecules26, and the immune mechanisms underlying ICI-induced SJS/TEN remain unclear. To date, there is no consensus for the management of ICI-induced SCAR. Although systemic corticosteroids remain the recommended first-line treatment for irAEs27, the use of systemic corticosteroids to treat SJS/TEN remains controversial28. Moreover, patients who suffer from ICI-induced SJS/TEN are often forced to interrupt ICI therapy29. Currently, no strategy has been developed to prevent SCAR induced by ICI.
In this study, we investigate the immune mechanism of patients with ICI-induced severe epidermal necrolysis. Our results lead to the discovery of potential therapeutic targets and method as well as prevention strategies for this important adverse event associated with promising cancer immunotherapy.
Results
Study design and patient enrollment
In this study, we performed scRNA-seq for comprehensive analysis of immune cell populations obtained from patients undergoing ICI therapy who experienced irAEs, with a specific focus on SJS/TEN. The scRNA-seq results were confirmed by flow cytometry, cytokine array/enzyme-linked immunosorbent assay (ELISA), immunofluorescence, and ex vivo lymphocyte activation studies (Fig. 1a). A total of 148 participants were enrolled in this study, including 25 patients with ICI-induced SJS/TEN, characterized as grade III to V irAEs; 41 patients with mild ICI-induced cADR, characterized as grade I to II irAEs; 46 patients who were classified as ICI-tolerant; 15 burn patients, 18 healthy donors (HD), and 3 patients with small molecule drug-induced SJS/TEN. Patient characteristics are displayed in Fig. 1b and Supplementary Table 1. Representative images of patients with SJS/TEN and mild cADR phenotypes induced by ICI are presented in Fig. 1c and Supplementary Fig. 1a–l, respectively. The detailed characteristics and diagnostic scores of ICI-SJS/TEN are displayed in Supplementary Table 2.
a Workflow showing sample collection and processing for 10X Genomics single-cell RNA sequencing (scRNA-seq) and the confirmation of results by flow cytometry, ex vivo lymphocyte activation test (LAT), BD Rhapsody scRNA-seq, ELISA/cytokine array, NanoString RNA-seq, and immunofluorescence assay performed in this study. Lesional blister cells (lesional BC) or peripheral blood mononuclear cells (PBMC) were obtained from enrolled patients with immune-related adverse events (irAEs) or control participants. b Characteristics of the participants enrolled in this study, such as groups with “immune checkpoint inhibitor (ICI)-cADR” (including ICI-induced Stevens-Johnson syndrome/toxic epidermal necrolysis [SJS/TEN], ICI-induced mild cADR) and “Control participants” (including small molecule drug-induced SJS/TEN, ICI-tolerant patients treated with ICI for at least 6 months with no evidence of drug-induced hypersensitivity reactions, burn patients, and healthy donors [HD] with no cADR history). Detailed clinical information can be found in Supplementary Table 1. c Representative clinical pictures of skin detachment for patients with ICI-induced SJS/TEN are shown. Other pictures of ICI-induced SJS/TEN (e.g., oral mucosal involvement and ocular injury) and ICI-induced mild cADR (lichenoid dermatitis) can be found in Supplementary Fig. 1a–l. d The identification of all cell clusters following 10X Genomics scRNA-seq of lesional BC and PBMC samples from patients with ICI-induced SJS/TEN, mild cADR and control participants (including lesional BC from 5 patients with ICI-induced SJS/TEN, PBMC from 5 the same patients with ICI-induced SJS/TEN, PBMC from 1 patient with an ICI-induced mild cADR [lichenoid dermatitis], and 5 each of sex-, age-matched ICI-tolerant patients and 6 of sex-, age-matched HD). A total of 115,327 cells are analyzed. e Violin plots show the expression of canonical marker genes across different clusters; the y-axis represents normalized values of marker gene expression (detailed gene expression profiles for different clusters can be found in Supplementary Fig. 1m). f Frequencies of cells in each cluster for each enrolled patient with ICI-cADR and control participants. ISB indicates ICI-induced SJS/TEN lesional BC; ISP indicates ICI-induced SJS/TEN PBMC; IMP indicates ICI-induced mild cADR (lichenoid dermatitis) PBMC; ITP indicates ICI-tolerant PBMC; HD indicates HD PBMC. g Distributions of all cell clusters colored for different groups of enrolled ICI-cADR patients and control participants. ISB: 18,010 cells; ISP: 22,156 cells; IMP: 6,354 cells; ITP: 34,477; HD: 34,330 cells. Figure 1a created with BioRender.com, released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Global analysis of immune cell populations in ICI-induced SJS/TEN and mild cADR
We first performed scRNA-seq using 10X Genomics 5’ barcoding technology as the discovery cohort in five sample groups, including (1) blister cells from the skin lesions (lesional BC) of five patients with ICI-induced SJS/TEN; (2) peripheral blood mononuclear cells (PBMC) from five patients with ICI-induced SJS/TEN (five of whom were the same patients examined in the blister cell group); (3) PBMC from 1 patient with mild ICI-induced cADR; (4) PBMC from five age-, sex-, and underlying diseases-matched ICI-tolerant cancer patients; and (5) PBMC from six age- and sex-matched HD (Patient characteristics in the scRNA-seq cohort are shown in Supplementary Table 3). High-quality transcriptomes were obtained for 115,327 total cells.
After performing dimensional reduction and unsupervised clustering, the integrated data revealed 20 distinct clusters, comprising all expected cell populations, including T cells, B cells, macrophage/monocyte/myeloid cells, NK/NKT cells, platelets, and erythrocytes (Fig. 1d–f, and Supplementary Fig. 1m). The distribution of the uniform manifold approximation and projection (UMAP) plot for all cells from ICI-cADR patients (including ICI-SJS/TEN and ICI-mild cADR) and control participants (including ICI-tolerant patients and HD) is shown in Fig. 1g. Compared with control participants, lesional BC from patients with ICI-induced SJS/TEN showed predominant macrophage/monocyte/myeloid (cluster 11) and CD8+ T cell (clusters 6) lineages (Fig. 1e–g and Supplementary Fig. 1n–p).
scRNA expression signatures in macrophage/monocyte/myeloid lineages
We next analyzed the scRNA expression profiles associated with ICI-induced SJS/TEN in macrophage/monocyte/myeloid lineages (from clusters 9–14 in Fig. 1d; defined by SingleR30,31 and annotated based on canonical marker genes32,33,34), sub-clustered into myeloid/monocytes (clusters 1–8), macrophages (clusters 9–10), and dendritic cells (cluster 11) (Fig. 2a–c and Supplementary Fig. 2a). The distribution of the UMAP plot for macrophage/monocyte/ myeloid lineages for these five sample groups is shown in Fig. 2d. We identified that lesional BC from patients with ICI-induced SJS/TEN were predominantly comprised of macrophages (clusters 9–10; Fig. 2b–d). Cluster 9 represents macrophages with highly co-expressed CD8+CTL-related genes, such as CD3D, GNLY, and GZMB (shown as a low doublet score in Supplementary Fig. 2b and similar to findings described for ICI-induced colitis33). We found that the same significant differentially expressed genes (DEG) in macrophage/monocytes/ myeloid cells from patients of ICI-SJS/TEN lesional BC compared to HD or ICI-tolerant (tolerant) PBMC were C1QB, APOE, APOC1, IFI27, CXCL9, CXCL10, GNLY, CD163, CD86, TNFRSF1A, etc. (Fig. 2e, f). Pathway enrichment analysis with MSigDB Hallmark gene sets showed the top upregulated gene sets in ICI-SJS/TEN lesional BC compared with HD PBMC or ICI-tolerant PBMC were those associated with tumor necrosis factor (TNF) signaling via nuclear factor kappa B (NF-κB), the interferon-gamma (IFN-γ) response, and the interferon-alpha (IFN-α) response in macrophage/monocyte/myeloid lineage clusters (Fig. 2g, h). Further comparisons were performed between different sample groups, including ICI-SJS/TEN lesional BC against ICI-SJS/TEN PBMC (Supplementary Fig. 2c), “ICI-cADR” PBMC against “tolerant & HD” PBMC (Supplementary Fig. 2d), and ICI-SJS/TEN PBMC against ICI-tolerant (tolerant) PBMC (Supplementary Fig. 2e). The results showed similar significant DEG with upregulated TNF, complement, and IFN-γ-related genes in ICI-induced SJS/TEN and mild cADR.
a Sub-clustering of macrophage/monocyte/myeloid cells selected from patients with ICI-induced SJS/TEN, ICI-induced mild cADR, and control participants. A total of 31,046 cells were defined as macrophage/monocyte/myeloid cells by SingleR annotation and well-studied marker genes. b Violin plots display the marker genes of macrophage/monocyte/myeloid cell clusters. For each cluster, the selected top 10 genes of the highest expression levels with cell type–specificity score, and a highly expressed well-defined subset marker are shown. The y-axis represents normalized values for different gene expression levels (detailed gene expression profiles for different clusters are presented in Supplementary Fig. 2a). c Frequencies of macrophage/monocyte/myeloid cells in each cluster for each enrolled patient with ICI-cADR and control participants. ISB: ICI-induced SJS/TEN lesional BC; ISP: ICI-induced SJS/TEN PBMC; IMP: ICI-induced mild cADR PBMC; ITP: ICI-tolerant PBMC; HD: healthy donors PBMC. d Distributions of macrophage/monocyte/myeloid cells across clusters among patients with ICI-cADR patients and control participants. Numbers of cells defined as macrophage/monocyte/myeloid cells by group were ISB: 5579 cells; ISP: 7558 cells; IMP: 984 cells; ITP: 3247 cells; HD: 13,678. e Ranking of the significant and relevant differentially expressed genes (DEG) in macrophage/monocyte/myeloid cells comparing between ICI-SJS/TEN lesional BC and HD PBMC. f Ranking of the significant and relevant DEG in macrophage/monocyte/myeloid cells comparing between ICI-SJS/TEN lesional BC and ICI-tolerant (tolerant) PBMC. The significance of DEG was defined a using a two-sided non-parametric Wilcoxon rank-sum test and Bonferroni correction. g Functional enrichment analysis of significant and relevant hallmark gene sets identified as differentially expressed genes in macrophage/monocyte/myeloid cells comparing between ICI-SJS/TEN lesional BC and HD PMBC. NES, normalized enrichment score. h Functional enrichment analysis of significant and relevant hallmark gene sets identified as differentially expressed in macrophage/monocyte/myeloid cells comparing between ICI-SJS/TEN lesional BC and ICI-tolerant (tolerant) PMBC. All significant the significant and relevant differentially expressed genes and hallmark gene sets are shown in the source data.
scRNA expression signatures in CD8+ T cells
We further evaluated the CD8+ T cell lineages from the clusters showing CD8A expression (clusters 3–7 in Fig. 1d, e; these cells were further defined by SingleR30,31 and annotated based on published signatures and cell markers32,33,34), which were sub-clustered into tissue-resident memory T cells (Trm)-like, cytotoxic effector T cells, central memory (CM)/Naïve, mucosal-associated invariant T cells (MAIT), and cycling T cells (Fig. 3a–c and Supplementary Fig. 3a). The major cells in lesional BC from ICI-induced SJS/TEN patients were cycling T cells (cluster 7) and Trm-like cells (cluster 1) (Fig. 3a–d). By comparing ICI-SJS/TEN lesional BC with HD PBMC or ICI-tolerant (tolerant) PBMC, we identified the same significant DEG in CD8+ T cell lineages, including GZMB, GNLY, HMGB2, LAG3, CTLA4, PDCD1, TIGIT, HAVCR2, TNFRSF18, CXCR3, etc. (Fig. 3e, f), some which were also identified as differentially expressed in other between-group comparisons (Supplementary Fig. 3b–d). The overexpression of these genes was also noticed in cycling T cell and Trm clusters in lesional BC from patients with ICI-induced SJS/TEN (Fig. 3g). With the Hallmark gene sets, we further demonstrated that the top upregulated gene sets in CD8+ T cell lineage clusters in lesional BC from patients with ICI-induced SJS/TEN compared with PBMC from HD or ICI-tolerant participants were IFN-γ response, IFN-α response, TNF signaling via NF-ĸB, and IL-2_STAT5 signaling (Fig. 3h, i).
a Sub-clustering of CD8+ T cells selected from patients with ICI-induced SJS/TEN, ICI-induced mild cADR, and control participants. A total of 29,134 cells were defined as CD8+ T cells by SingleR annotation and well-studied marker genes. CM: central memory T cells; MAIT: mucosa-associated invariant T cells; Trm: tissue-resident memory T cells. b Violin plots display marker genes for CD8+ T cell clusters. For each cluster, the selected top 10 genes of the highest expression levels with cell type–specificity score, and a highly expressed well-defined subset marker are shown. The y-axis represents the normalized values for gene expression (detailed gene expression profiles for different clusters can be found in Supplementary Fig. 3a). c Frequencies of CD8+ T cells in each cluster for each enrolled patient with ICI-cADR and control participant. ISB: ICI-induced SJS/TEN lesional BC; ISP: ICI-induced SJS/TEN PBMC; IMP: ICI-induced mild cADR (lichenoid dermatitis) PBMC; ITP: ICI-tolerant PBMC; HD: healthy donors PBMC. d Distributions of CD8+ T cells across clusters from ICI-cADR patients and control participants. CD8+ T cell numbers according to group were ISB: 6388 cells; ISP: 4462 cells; IMP: 1570 cells; ITP: 10,319 cells; HD: 6395 cells. e Ranking of the significant and relevant differentially expressed genes (DEG) in CD8+ T cells comparing between ICI-SJS/TEN lesional BC and HD PBMC. f Ranking of the significant and relevant DEG in CD8+ T cells comparing between ICI-SJS/TEN lesional BC and ICI-tolerant (tolerant) PBMC. The significance of DEG was defined a using a two-sided non-parametric Wilcoxon rank-sum test and Bonferroni correction. g The expression patterns of cytotoxic proteins, co-inhibitory receptors, and tumor necrosis factor receptor (TNFR) in UMAP of CD8+ T cell sub-clusters. h Functional enrichment analysis of significant and relevant hallmark gene sets identified as differentially expressed in CD8+ T cells comparing between ICI-SJS/TEN lesional BC and HD PMBC. NES, normalized enrichment score. i Functional enrichment analysis of significant and relevant hallmark gene sets identified as differentially expressed in CD8+ T cells comparing between ICI-SJS/TEN lesional BC and ICI-tolerant (tolerant) PBMC. All significant the significant and relevant differentially expressed genes and hallmark gene sets are shown in the source data.
Dominant TCRs in patients with ICI-induced SJS/TEN
We further investigated T cell receptor (TCR) V(D)J gene expression and clonal relationships among CD8+ T cells across different groups (Supplementary Fig. 4a, b). The higher proportions of clonal expanded CD8+ T cells (TCR clonotypes ≥0.01, defined by the fraction of cells with the certain clonotype) were found in ICI-induced SJS/TEN lesional BC and PBMC as well as in ICI-tolerant PBMC (Supplementary Fig. 4b). Importantly, the cycling T cell- and Trm-associated markers (e.g., ITGAE, SELL, and CD27), cytotoxic markers (e.g., GNLY and GZMB), immune checkpoint regulatory markers (e.g., HAVCR2 and LAG3), and TNF receptor (TNFRSF18) genes were highly expressed in clonally expanded CD8+ T cells (TCR clonotypes ≥0.01) in lesional BC from patients with ICI-induced SJS/TEN compared with those without clonal expansion (TCR clonotypes <0.01) or with other sample groups (Supplementary Fig. 4c). The shared TCR clonotypes were found in PBMC and lesional BC of individual ICI-SJS/TEN patients (Supplementary Fig. 4d–f). These data suggested that specific populations of clonally expanded CD8+CTL participate in the pathogenesis of ICI-induced SJS/TEN.
Interactions between CD8+ T cells and macrophage/monocyte/myeloid lineages
To determine the interactions among immune cell populations, we first analyzed the ligand-receptor network in the group of “ICI-SJS/TEN” patients compared with “ICI-tolerant & HD” controls across all cell populations and found the interactions between C-X-C motif chemokine ligand 9 (CXCL9), CXCL10, and C-X-C motif chemokine receptor 3 (CXCR3) were exclusively identified between the macrophage/monocyte/myeloid and CD8+ T cells lineages in patients with ICI-SJS/TEN (cluster 11 and 14 interacted with clusters 1, 5, and 6) (Fig. 4a, upper; and Supplementary Fig. 5a, b) but not observed in “ICI-tolerant & HD” group (Fig. 4a, lower). CXCL9 and CXCL10 were elevated in macrophages (Fig. 4b, top), whereas their receptor, CXCR3, was highly expressed in CD8+ T cells (Fig. 4c, top). Violin and heatmap plots further display that CXCL9, CXCL10, and CXCR3 were dominantly expressed in lesional BC from patients with ICI-induced SJS/TEN compared with the other sample groups (Fig. 4b, c, red in bottom). Furthermore, we analyzed the skin-homing genes35 in the clusters predominantly concentrated in the lesional blister cells (clusters 9 and 11 from the total cell populations) which were classified as macrophage/monocyte/myeloid lineages (Supplementary Fig. 5c). The level of CCL20 expression was found higher in ICI-SJS/TEN lesional BC than in ICI-SJS/TEN PBMC, ICI-tolerant PBMC, and HD PBMC.
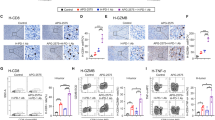
a Analysis of chemokine receptor–ligand pairs (CXCR3–CXCL9 and CXCR3–CXCL10) across clusters in all cell datasets shown in Fig. 1d. The “ICI-SJS/TEN” group includes lesional BC and PBMC from 5 patients with ICI-induced SJS/TEN patients, and the “ICI-tolerant & HD” group includes PBMC from 5 ICI-tolerant patients and 6 healthy donors (HD). b The scaled and normalized gene expression levels for CXCL9 and CXCL10 in all cell datasets (related to Fig. 1d) was shown, whereas the violin plot and heatmap beneath was drawn from the subset of macrophage/monocyte/myeloid clusters. c The scaled and normalized expression levels of CXCR3 in all cell datasets (related to Fig. 1d) were shown, whereas the violin plot and heatmap beneath were drawn from the subset of CD8+ T cell clusters. ISB: ICI-SJS/TEN lesional BC; ISP: ICI-SJS/TEN PBMC; IMP: ICI-mild cADR PBMC; ITP: ICI-tolerant PBMC; HD: healthy donors PBMC. d Heatmap showing Z score–normalized averaged expression levels of the indicated cytokines/cytotoxic proteins/inflammatory proteins and its related receptor genes compared among ICI-SJS/TEN, ICI-mild cADR, and control groups. The normalized differential gene expression (DEG) levels in CD8+ T cell clusters (clusters 3–7 in Fig. 1d; defined by SingleR annotation) and macrophage/monocyte/myeloid clusters (clusters 9–14 in Fig. 1d; defined by SingleR annotation) were shown. The indicated relevant genes list is based on the significant P values for ICI-SJS/TEN lesional BC group comparing to “ICI-tolerant & HD” group, which were calculated by Wilcoxon rank-sum test. The genes associated with the TNF signaling pathway were denoted in a red color. e Scatterplot showing DEG, confirming RNA expression levels in formalin-fixed paraffin-embedded (FFPE) skin tissue samples from patients with ICI-induced SJS–TEN (ICI-SJS/TEN; n = 5) or ICI-induced lichenoid dermatitis (ICI-mild cADR; n = 7) compared with those from HD (HD; n = 8). Each dot denotes an individual gene with a Benjamini–Hochberg-adjusted P value (two-sided unpaired Mann–Whitney U test) <0.05 and average log2 fold change (FC) > 2 in ICI-SJS/TEN samples compared with HD samples (ICI-SJS/TEN vs. HD) and in ICI-mild cADR samples compared with HD samples (ICI-mild cADR vs. HD). The blue-labeled genes represent genes that were significantly elevated in both the ICI-SJS/TEN vs. HD and ICI-mild cADR vs. HD comparisons, whereas the red-labeled genes were only significantly elevated in the ICI-SJS/TEN vs. HD comparison. f Immunofluorescence staining with anti-CD8 (green) and anti-CXCR3 (red) antibodies, and 4’,6-diamidino-2-phenylindole (DAPI; nuclear stain; blue) in skin tissues from 7 patients with ICI-SJS/TEN and 4 ICI-mild cADR and from 4 HD control participants (sample list shown in source data). The upper figures represent 200×, whereas the lower figures represent 400× magnification. g Immunofluorescence staining with anti-CXCL10 (green) and anti-CD163 (red) antibodies and DAPI (blue). The upper figures represent 200×, whereas the lower figures represent 400× magnification.
We further investigated the expression patterns of cytokines/cytotoxic proteins/inflammatory proteins and their receptors in CD8+ T cells and macrophage/monocyte/myeloid cell populations (Fig. 4d). A number of ligand-receptor pairs were upregulated in patients with ICI-induced SJS/TEN. In CD8+ T cells, highly expressed receptor genes for cytokines/cytotoxic proteins included TNFRSF18, IL12RB2, HAVCR2, IL6ST, IL18R1, and IL1R1 within lesional BC from patients with ICI-induced SJS/TEN and TNFRSF1B, and IL1R2, and TRAF2 within both lesional BC and PBMC from patients with ICI-induced SJS/TEN. Highly expressed ligand genes included HMGB1, GNLY, GZMB, CSF2, IL21, and IFITM1 within lesional BC from patients with ICI-induced SJS/TEN. In macrophage/monocyte/myeloid cells, several receptor genes for cytokines/inflammatory proteins, including TNFRSF18, CD81, IL1R2, C1QB, HAVCR2, and TRAF2; whereas ligand genes, including TNFSF18, IFITM1, IFNG, APOE, IFI27, and IL18, were upregulated in lesional BC from patients with ICI-induced SJS/TEN. These results provide evidence for an upregulated TNF-related gene expression signature in both CD8+ T cells and macrophage/monocyte/myeloid cells in patients with ICI-induced SJS/TEN.
To analyze the cell states in ICI-cADR (including ICI-SJS/TEN and ICI-mild cADR) patients, we performed monocle trajectory analysis for T cells, macrophage/monocyte/myeloid cells, and NK/NKT cells in lesional BC and PBMC from patients with ICI-cADR (Supplementary Fig. 6a–e). The trajectory analysis revealed that T cells in PBMC from ICI-cADR patients were distributed to the initial state of the pseudotime (Supplementary Fig. 6c–g), whereas macrophages in lesional BC from ICI-induced SJS/TEN patients were distributed to the second stage of the pseudotime. Finally, the CD8+ T cells in lesional BC from patients with ICI-induced SJS/TEN patients were regarded as the last stage of the pseudotime (Supplementary Fig. 6g).
We further enrolled another separate samples of ICI-induced cADR patients and control participants to confirm the scRNA-seq results. We first utilized bulk NanoString RNA-seq in formalin-fixed paraffin-embedded (FFPE) skin tissues from 20 ICI-cADR patients and controls (including 5 patients with ICI-SJS/TEN, 7 with ICI-mild cADR, and 8 HD; Fig. 4e), and found that CD86, TNFSF13, and IL32 were only significantly increased in ICI-induced SJS/TEN comparing to HD; while CXCL10, CXCL11, CXCL9, GZMB, S100A8, LAG3, STAT1, C1QB, CD163, TNFRSF1B, CXCR3, etc., were significantly highly expressed in both ICI-SJS/TEN and ICI-mild cADR comparing to HD (Fig. 4e). The scoring of different cell types showed that CD8+ T cells, macrophages, Th1 cells, and cytotoxic cells were significantly upregulated in samples from patients with ICI-SJS/TEN compared with samples from patients with ICI-mild cADR or HD (Ctrl) (Supplementary Fig. 7a). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for the NanoString results revealed the upregulation of several genes in the TNF signaling pathway, such as TNFR2, TNFR1, and CXCL10, in lesional samples from patients with ICI-induced SJS/TEN compared with samples from HD (Supplementary Fig. 7b)36.
We then performed immunofluorescence assays to confirm the expression of CXCR3 and CXCL10 proteins in CD8+ T cells and macrophage, which showed that CXCR3+ CD8+ T cells (Fig. 4f) and CXCL10+ CD163+ macrophages (Fig. 4g) were predominantly localized in the skin lesions of patients with ICI-SJS/TEN, with fewer of these cells identified in ICI-mild cADR (lichenoid dermatitis) patients and none found in samples from HD. We also measured the cytokine/chemokine/cytotoxic protein levels in plasma samples from ICI-cADR patients and control participants. Our results showed immune mediators identified in the scRNA-seq analysis, such as CXCL10, TNF, GNLY, and GZMB, were significantly upregulated in the blister fluids from patients with ICI-induced SJS/TEN compared with those obtained from patients with burn-related injuries (Fig. 5a). In addition, the plasma levels of chemokines (CXCL10 and CXCL9), cytokines/inflammatory proteins (TNF, IFN-γ, IL-6, and IL-8), and cytotoxic proteins (GNLY and GZMB) were significantly increased in patients with ICI-SJS/TEN compared with patients with ICI-mild cADR and control participants (Fig. 5b). Higher levels of CXCL10, CXCL9, and TNF were identified both in patients’ plasma and blister fluid of ICI-induced SJS/TEN. The expressions of these different immune mediators in the individual patient were shown in Supplementary Fig. 8a. Some cases showed extremely high plasma levels of IL-6 and IFN-γ, and we subsequently analyzed the correlation of plasma level of CXCL10 with these two cytokines; however, there was no significant correlation between CXCL10 with these two cytokines (Supplementary Fig. 8b, c). We further measured the expression level of CXCL10 for other inflammatory skin diseases, including psoriasis and atopic dermatitis. Our data demonstrated the plasma CXCL10 level was markedly elevated in ICI-induced SJS/TEN compared with psoriasis and atopic dermatitis (Supplementary Fig. 8d).
a The secretion of CXCL10, TNF, GNLY, and GZMB were measured by ELISA in the blister fluids of 6 patients with ICI-SJS/TEN and 15 burn patients (Burns). b Plasma expression levels of CXCL10, CXCL9, TNF, IFN-γ, IL-6, IL-8, GNLY, and GZMB were measured by cytokine array or ELISA in 8 patients with ICI-induced SJS/TEN (ICI-SJS/TEN), 32 patients with ICI-induced mild cADR (ICI-mild cADR), and 9 control participants (Ctrl; including 4 participants with ICI-tolerant and 5 with HD). Plasma from patients with ICI-induced cADR were obtained during the acute stage of cADR phenotypes. Data in (a) and (b) are presented as the mean ± SD. P values were calculated by a two-sided unpaired Mann–Whitney U test. *P < 0.05, **P < 0.01, ***P < 0.001.
Different expressions in ICI-induced SJS/TEN and drug-induced SJS/TEN
To distinguish the differences between ICI-induced SJS/TEN and drug(s)-induced SJS/TEN, we further performed scRNA-seq for lesional BC and PBMC from three patients with small molecular drug-induced SJS/TEN. The clinical characteristics and culprit drug(s) information of drug-induced SJS/TEN patients are shown in Supplementary Tables 1 and 3, respectively. The distinct cell type clusters and distribution of the UMAP plot for all immune cells from ICI-induced SJS/TEN, drug(s)-induced SJS/TEN, and HD groups is shown in Supplementary Fig. 9a–d. Compared to drug(s)-induced SJS/TEN, we found the significant DEG in macrophage/monocytes/myeloid cells of patients with ICI-induced SJS/TEN lesional BC were CCL4, CXCL2, C1QB, CCL3, IL1B, GNLY, CD163, CD86, TNFRSF1B, TNFAIP3, etc. (Fig. 6a); while the significant DEG in CD8+ T cells of patients with ICI-induced SJS/TEN lesional BC were ITGAE, TTN, IRF1, IL12RB2, ITGAL, PDCD1, CD38, LAG3, CTLA4, TIGIT, etc. (Fig. 6c). The cell clusters across these sample groups are shown in Supplementary Fig. 9c, d. Other comparisons between different sample groups were shown in Fig. 6b, d, and Supplementary Fig. 9e–h.
a scRNA-seq were performed for lesional BC and PBMC samples from 5 patients with ICI-induced SJS/TEN and 3 patients with drug-induced SJS/TEN. Ranking of the significant and relevant differentially expressed genes (DEG) in macrophage/monocyte/myeloid cells comparing between ICI-SJS/TEN lesional BC and drug-induced SJS/TEN lesional BC. b Ranking of the significant and relevant DEG in macrophage/monocyte/myeloid cells comparing between ICI-SJS/TEN PBMC and drug-SJS/TEN PBMC. c Ranking of the significant and relevant DEG in CD8+ T cells comparing between ICI-SJS/TEN lesional BC and drug-SJS/TEN lesional BC. d Ranking of the significant and relevant DEG in CD8+ T cells comparing between ICI-SJS/TEN PBMC and drug-SJS/TEN PBMC. e scRNA-seq analysis of PBMC collected during the recovery stage of a patient with ICI-induced SJS/TEN (ICI-SJS/TEN-1) after ex vivo culture with phosphate-buffered saline (PBS, solvent control), ICI stimulation (ICI), and ICI plus anti-TNF (ICI+anti-TNF). f Identification of cell clusters across sample groups. g Frequencies of different immune cells in each cluster across groups treated with PBS, ICI, and ICI+anti-TNF. The Violin plots display canonical marker genes for these immune cell clusters are shown in Supplementary Fig. 11. h Violin plots display the expression of selected ICI-induced SJS/TEN-associated genes, including GZMB, CD163, TNF, CXCR3, and CXCL10, in CD8+ T cell or macrophage/monocyte cell clusters after ex vivo culture. i The macrophage/monocyte phenotype after ex vivo culture was measured by flow cytometric assay. The percentage of CD163+CXCL10+ cells was determined in CD14+ monocyte cells (detailed gating information for flow cytometric assay is shown in Supplementary Fig. 12). i-1 The populations of CD163+CXCL10+ cells in CD14+ monocytes after ex vivo treatment. j Cytotoxic CD8+ T cell phenotypes after ex vivo culture were measured. GranzymeB (GZMB)+ cells were determined in CD8+ T cells (detailed gating information for the flow cytometric assay is shown in Supplementary Fig. 12). j-1 The populations of GZMB+ cells in CD8+ T cells identified in PBMC after ex vivo treatment are shown. k Expression levels of CXCL10 in ex vivo cultured supernatant were detected. The significance of DEG was defined a using a two-sided non-parametric Wilcoxon rank-sum test and Bonferroni correction. Data in (i–j) are presented with boxplot showing individual data points, the first quartile, the median, and the third quartile. A black * indicates that P values were calculated by two-sided unpaired Mann–Whitney U test; a blue * indicates that P values were calculated by two-sided paired Wilcoxon matched-pairs signed-rank test.
We then analyzed the TNF and CXCL10 expressions in the scRNA-seq data, and identified that macrophage/monocytes/myeloid cells of all SJS/TEN lesional BC both expressed high levels of TNF and CXCL10 (Supplementary Fig. 10a), but there was weak or even no expressions of TNF and CXCL10 in the PBMC of SJS/TEN patients and HD. Moreover, the expressions of TNF and CXCL10 in macrophage/monocytes/myeloid cells were higher in the ICI-SJS/TEN lesional BC (clusters 9 and 15) than in the drug-induced SJS/TEN lesional BC (clusters 9 and 11; Supplementary Fig. 10a). We also analyzed the TNF expression in CD8+ T cells (Supplementary Fig. 10b), and found that the TNF level in CD8+ CTL (cluster 7) was higher in drug-induced SJS/TEN than in ICI-induced SJS/TEN (clusters 2, 3, 5, and 7; Supplementary Fig. 10b).
Ex vivo assay to determine the role of CXCL10-CXCR3 axis and anti-TNF effects on ICI-induced cADR
We then performed ex vivo assays and utilized BD Rhapsody scRNA-seq to analyze the PBMC cells from one patient with ICI-induced SJS/TEN both with and without treatment with ICI and concurrent TNF blockade (etanercept) after 6 days of culture. High-quality transcriptomes were obtained from 3730 total cells (Fig. 6e). The scRNA-seq results revealed that the percentages of macrophages and macrophage-monocyte-derived cells increased after ICI stimulation (see clusters 5 and 6 in Fig. 6f, g and Supplementary Fig. 11). The gene signatures for GZMB and CXCR3 in CD8+ T cells, those for CD163 and CXCL10 in macrophages, and those for TNF and CXCL10 in macrophage-monocyte-derived cells significantly increased after ICI stimulation (Fig. 6h). Furthermore, the percentages of macrophages/macrophage-monocyte-derived cells and their associated genes signatures were largely reduced when ICI was combined with TNF blockade (Fig. 6f–h).
We collected more PBMC from patients with ICI-cADR (including 5 patients with ICI-SJS/TEN and 6 with mild cADR) and 8 ICI-tolerant patients for evaluation using the ex vivo TNF blockade assay and the determination of cell populations and CXCL10 secretion using flow cytometry (Supplementary Fig. 12a) and ELISA, respectively, to confirm scRNA-seq findings. Our result showed that the percentages of CD163+CXCL10+ (Fig. 6i and i-1) and CD86+CXCL10+cells (Supplementary Fig. 12b, c), in addition to GZMB+CD8+ T cells (Fig. 6j and j-1) and CXCR3+CD8+ T cells (Supplementary Fig. 12d, e), significantly increased after ICI treatment, and were significantly restored after treatment with TNF blockade. We also measured the protein levels of CXCL10 in cultured supernatants, and the result was consistent with the changes observed in patients and ex vivo experiments (Fig. 6k).
Successful treatment for patients with progressive ICI-induced SJS/TEN by targeting TNF
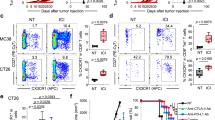
We retrospectively analyzed the clinical outcome of these 25 patients with ICI-induced SJS/TEN. Among these patients, there were 8 progressive ICI-SJS/TEN patients received TNF blockade therapy (Supplementary Table 4). One patient of ICI-SJS/TEN (Case no. 1), who was resistant to high-dose systemic corticosteroids, was dramatically improved after administration of TNF blockade therapy (Etanercept, 50 mg, bi-weekly for 3 doses). After TNF blockade therapy, patient’s skin lesions showed to stop the progression of blister formation with rapid skin healing within 10 days (the clinical presentation and treatment course were shown in Supplementary Fig. 13). We performed 10X Genomic scRNA-seq in PMBC derived from three patients before and after TNF blockade treatment (Fig. 7a–c, and Supplementary Fig. 14a) and found a significant reduction in the proportions of myeloid/monocyte cells (from 39.86% to 12.25%) (Fig. 7c). In myeloid/monocyte cell clusters, the gene signatures for IL1R2, HMGB2, CD163, and IFNGR1 were significantly upregulated before TNF blockade therapy (Supplementary Fig. 14b), and the gene expression of IL7R, IFNGR1, TNFAIP3, GNLY, HMGB2, and TNFRSF1B were significantly upregulated in CD8+ T cell clusters (Supplementary Fig. 14c) before TNF blockade therapy. Violin and UMAP plots showed that CD163 and TNFRSF1B expressions decreased in myeloid/monocyte cell clusters 10 and 12, whereas TNFRSF1B, IFNGR1, and GNLY expressions decreased in CD8+ T cell cluster 4 (Supplementary Fig. 14d–i). Computed tomography imaging further showed a durable tumor treatment response with complete remission after this ICI-SJS/TEN patient recovered from her SJS/TEN episode (Fig. 7d, e). An additional 7 patients with ICI-induced SJS/TEN were treated with TNF blockade therapy (Supplementary Table 4; the best cancer treatment outcome within TNF blockade therapy are also shown in Table) and demonstrated great improvement in skin lesions (skin complete healing time: 9.9 ± 2.1 days) comparing to systemic corticosteroids treatment (P = 0.002) (Supplementary Table 5). TNF blockade therapy decreased the SCORTEN-based predicted mortality rate for these ICI-SJS/TEN patients and no mortality was found (predicted and observed rates: 36.31 ± 27.07% and 0%, respectively; Supplementary Table 5). Further analysis of plasma levels after treatment with TNF blockade showed that CXCL10 was significantly decreased (P < 0.05) (Fig. 7f and Supplementary Fig. 14j).
a 10X Genomic single-cell RNA sequencing (scRNA-seq) analysis in PBMC from three patients with ICI-induced SJS-TEN. UMAP plot generated from a merged dataset of the three patients with ICI-SJS/TEN pre- and post-TNF blockade treatment (total n = 31,904 cells). b UMAP plots segregated according to cellular origin. Dark red indicates the cell clusters before TNF blockade, whereas green indicates those after TNF blockade. Dotted regions highlight cytotoxic cells (including CD8+ T cells and NK/NKT cells) and myeloid/monocyte clusters both before and after TNF blockade. c Pie charts of three ICI-SJS/TEN patients showing relative cluster abundances before and after TNF blockade. The populations of total myeloid/monocyte cells before and after TNF blockade are shown in red. d, e Tumor images in the ICI-induced SJS-TEN overlapping case (ICI-SJS/TEN-1) with metaplastic breast carcinoma were evaluated by computed tomography (CT), revealing a durable complete response after recovery from the SJS/TEN episode. The red arrow indicates the tumor location. f Serial plasma CXCL10, TNF, and GZMB levels in 6 available patients with ICI-induced SJS/TEN during the clinical course from acute to late recovery stages. Day 1 on the horizontal axis is defined as the first day of hospitalization. The arrow in each line chart indicates TNF blockade (etanercept) administration. * of CXCL10 level represent P value calculated by two-sided paired Wilcoxon matched-pairs signed-rank test. g Clinical course, skin presentation, main tumor evaluation by CT, and serial plasma CXCL10 and TNF levels for one patient with ICI-induced SJS/TEN (ICI-SJS/TEN-15) who tolerated to continuous the same type of ICI immunotherapy combined with concurrent TNF blockade. Day 1 on the horizontal axis is defined as the first day of hospitalization. The blue arrow indicates TNF blockade (etanercept) administration time points, and the red arrow indicates the ICI (atezolizumab) treatment and re-challenge time points. The dotted line, arrow range, and values in the CT image indicate the tumor size.
Prevention of ICI-induced SCAR by administering TNF blockade during ICI re-challenge
Most (6 in 8 patients) of ICI-SJS/TEN patients treated with TNF blockade received another targeted therapy after their skin recovery for cancer treatment, and none of them had further relapsed SCAR for at least 3-month follow-up. Two ICI-induced SJS/TEN patients were resumed with the same type of ICI after their recovery ICI-induced SCAR. One hepatocellular carcinoma (HCC) patient who suffered from ICI (atezolizumab)-induced SJS/TEN (Case no. 15) recovered following TNF blockade (3 doses of etanercept; Fig. 7g, left). After the patient recovered from his SJS episode, he was resumed atezolizumab therapy with concurrent etanercept. The concurrent use of TNF blockade successfully prevented this patient from suffering from the recurrence of SJS/TEN or other systemic irAEs for 2 years following ICI use (Fig. 7g, right). The measurement of plasma CXCL10, TNF, and GZMB showed the maintenance of normal levels throughout the duration of the re-challenge course (Fig. 7g and Supplementary Fig. 15). The tumor also showed a sustainable response to continuous ICI treatment, with a largely decrease in size (the longest diameter decreased from 11.2 cm to 4.9 cm; Fig. 7g), and the levels of HCC tumor marker alpha-fetoprotein (AFP) remained within the normal range (Supplementary Fig. 15). Another patient with ICI-induced SJS/TEN (Case no. 9) underwent a re-challenge with the same ICI combined with concurrent TNF blockade and also showed a partial remission of his cancer outcome to continued ICI treatment over six months (Supplementary Fig. 16). These are cases of ICI-induced severe epidermal necrolysis which have been successfully re-challenged with ICI immunotherapy.
Discussion
ICI can still cause severe, life-threatening irAEs, such as SJS/TEN, pneumonitis, and myocarditis. In this study, we first identified the accumulation of macrophage-derived CXCL10-recruiting CXCR3+ CD8+CTL in lesional BC from patients with ICI-induced SJS/TEN. ScRNA gene expression profiles further identified TNF signaling via NF-κB as the key pathway responsible for macrophage-derived CXCL10 and CD8+CTL activation, which was confirmed by an ex vivo TNF blockade assay.
Our clinical and histological analyses, scRNA expression profiles, monocle trajectory analysis, and immunofluorescence staining experiments provided evidence suggesting that macrophage/monocyte/myeloid lineages serve as the crucial immune cells in ICI-induced cADR, especially in severe epidermal necrolysis. Macrophages in tissues are known to release chemokines and cytokines for localization, differentiation, survival, and recruitment of effector T cells, Trm and Trm precursors37,38. Furthermore, our results revealed that cycling T and Trm-like cells were the most predominant T cells among lesional BC from patients with ICI-induced SJS/TEN. A large population of Trm cells have been reported and are known to be involved in the pathomechanism of delayed-type hypersensitivity reactions39,40,41,42, but their roles in SCAR induced by ICI have not yet been identified. Previous studies reported a subset of Trm cells were likely the primary immune cells responsible for ICI-induced vitiligo43 and circulating activated memory T cells were associated with ICI-induced autoimmune-like toxicity44. A large population of cycling T cells, which involved in the rapid onset of adverse events, was identified in ICI-induced colitis33. Cycling T cells might also participate in the rapid progression of ICI-induced SJS/TEN.
Inflammatory monocyte/macrophage cells are key produces of the TNF superfamily of costimulatory signals, which promote T cell activation38,45. In this study, ICI-induced SJS/TEN-associated macrophage/monocyte/myeloid lineage cell populations expressed high levels of CXCL9 and CXCL10, especially in the macrophages from lesional BC. Based on the trajectory analysis in this study, ICI-activated T cells from PBMC are proposed to serve as the initial cells involved in the pathogenesis of irAE, leading to the differentiation of monocytes into macrophages and increasing their susceptibility to migrate to skin lesion sites. Macrophages of ICI-SJS/TEN patients can release chemokines (e.g., CXCL10 and CXCL9) to recruit CD8+CTL (e.g., CXCR3+ cells) from blood into skin lesions, resulting in epidermal necrosis and detachment. Previous studies also proposed a role for CXCL9 and CXCL10 in patients with ICI-induced colitis and nephritis46,47,48,49. Our ex vivo study revealed that CXCL10 released by monocyte/macrophage cells was increased by ICI stimulation in PBMC from ICI-cADR patients but not in PBMC from ICI-tolerant participants. Increased CXCL10 secretion could be further attenuated by TNF blockade, suggesting that the CXCL9/CXCL10-dependent recruitment of memory CD8+CTL could be disrupted by biologic anti-TNF agent.
CD8+CTL activations are known to be involved in massive epidermal necrosis for traditional small-molecule drug(s)-induced SJS/TEN patients23,24,50,51. We found that CD8+CTL are also abundant in lesional BC of ICI-induced SJS/TEN patients. The expression levels of immune checkpoint receptors (e.g., CTLA-4, TIGIT, LAG3, and PDCD1) on CD8+CTL and macrophage-derived chemokines (e.g., CCL4 and CXCL2) were found to be significantly higher in ICI-induced SJS/TEN than in small-molecule drug(s)-induced SJS/TEN patients. Our results revealed the significant increase of TNF-mediated macrophage/monocyte activation and immune checkpoint receptors-mediated T-cell reactions in ICI-induced SJS/TEN comparing with drug(s)-induced SJS/TEN. Previous studies have shown that neutrophils also play a potential cellular role in the inflammation of traditional small chemical drugs-induced SJS/TEN52. However, we did not find a significant neutrophil signature in our scRNA-seq results. This may be explained by the different pathomechanisms between traditional small chemical drugs-induced SJS/TEN and ICI-induced SJS/TEN.
TNF and IFN-α2 expression levels have previously been correlated with high-grade irAEs49,53. However, IFN-γ blockade may have negative effects on anti-tumor immunity54. TNF blockade can attenuate macrophage-derived CXCL10 release and a prophylactic TNF blockade has previously shown to prevent immunotoxicity of ICI-induced colitis in a murine model55,56. A randomized controlled trial exploring the safety and tolerability of treating metastatic melanoma using combination ICI and TNF blockade (infliximab or certolizumab) therapy is currently ongoing57. According to the current guidance of the National Comprehensive Cancer Network, and Society for Immunotherapy of Cancer, a further re-challenge of ICI is not recommended for patients who ever developed ICI-induced SJS/TEN29,58. One recent review paper showed a potential treatment for this recalcitrant ICI-SJS/TEN including a combination treatment of corticosteroids, intravenous immunoglobulin and TNF inhibitor59. Our study provides a useful strategy to prevent the recurrence of ICI-induced severe cutaneous reactions with concurrent TNF blockade therapy. Of note, TNF blockade therapy is contraindicated in patients with severe sepsis, which should be noticed in clinical practice.
This study still has several limitations: first, all ICI-cADR patients are metastatic cancer patients, and some of these patients refused to provide skin lesion samples. For these ICI-cADR patients who refused to provide skin samples, we utilized blister fluids and blood samples as well as performed ex vivo assay to investigate the immune mechanism. Second, the cell viability of several blister cells is difficult to meet the standard experimental criteria for scRNA-seq, we confirmed the cytokine/chemokine levels in blister fluids instead of blister cells from ICI-SJS/TEN and burn patients. Third, some ICI-tolerant individuals still had fever, fatigue, or non-immune mediated gastrointestinal side effects. Finally, the sample size of ICI-induced SJS/TEN patients is still limited.
In summary, our results showed the overexpression of macrophage-derived CXCL10, contributing to the recruitment of CXCR3+ CTL to skin lesional in patients with ICI-induced SJS/TEN, and TNF signaling via NF-κB as the key pathway responsible for macrophage-derived CXCL10 and CTL activation. Most importantly, we provide an effective strategy for the management and prevention of fatal cutaneous irAEs using TNF blockade therapy. Further clinical trial examining larger cohorts of patients remains necessary to validate the finding.
Methods
We enrolled the ICI-cADR patients and control participants from the Taiwan Severe Cutaneous Adverse Reaction consortium (e.g., Taipei, Linkou, Keelung, and Taoyuan Chang Gung Memorial Hospitals, Taipei and Taichung Veterans General Hospital, and National Cheng Kung University Hospital) in Taiwan, the Xiamen Chang Gung Memorial Hospital, China, and the University of Michigan Hospital, USA. Each participant enrolled in this study were with written informed consent, which had been approved by the institutional review board (IRB) and ethics committee of each hospital (IRB. NO. 105-3600C, 201800463B0, 201802027B0, 201902171A3, 202300921A3, and 202300997A3). Informed consent was obtained from all participants.
ICI-induced cADR phenotypic classification
SJS/TEN phenotypes were classified using the consensus RegiSCAR definitions51,60,61,62. SJS/TEN was characterized by the rapid development of blistering exanthema involving purpuric macules and target-like lesions, accompanied by mucosal involvement and skin detachment. Skin detachment was assessed according to total body surface area (TBSA), with epidermal detachment <10% of TBSA classified as SJS, >30% of TBSA classified as TEN, and 10–30% of TBSA classified as SJS–TEN overlapping. ICI-induced mild cADR mainly includes ICI induced lichenoid dermatitis and maculopapular exanthema. Lichenoid dermatitis was characterized by a generalized, pruritic, papulosquamous eruption that spared the face, palms, soles, and mucous membranes. Maculopapular exanthema was defined as self-limited, erythematous macules, diffuse, and papules without any systemic involvement.
The irAE grade was based on the Common Terminology Criteria for Adverse Events (CTCAE) version 5.063,64. From 2015 to 2023, we enrolled a total of 25 patients with ICI-induced SJS/TEN, ranging from grade III to V irAEs (including 16 patients with SJS, 6 with SJS-TEN overlapping, and 3 with TEN), and 41 patients with ICI-induced mild cADR, ranging from grade I to II irAEs (including 26 patients with lichenoid dermatitis, and 15 with maculopapular exanthema). All of the ICI-induced cADR patients were assessed by at least two dermatologists. Furthermore, 46 patients who had received ICI for more than 4 times and the cumulative duration was for more than 6 months without any evidence of hypersensitivity reactions were enrolled as ICI-tolerant participants.
For patients with ICI-induced severe SJS/TEN, we discontinued the ICI therapy immediately when the SJS/TEN symptoms occurred (index day or onset day), such as skin detachment, mucosal involvement or blister formation. For patients with ICI-induced mild cADR, most of patients discontinued the ICI therapy immediately when skin symptoms occurred, and a small number of patients continued ICI therapy because their symptoms were mild during their initial illness. Clinically, the patients could suffer from the ICI-induced SJS/TEN and cADR with a diverse period after ICI use. We’ve collected the biological samples within the acute stage (before immunosuppressants treatment) or maximum stage of skin detachment after the onset of symptoms and signs of ICI-SJS/TEN and cADR. The average sampling time was 0.7 ± 3.3 and 1.4 ± 5.6 days after onset for ICI-induced SJS/TEN and ICI-induced cADR patients, respectively. The maximal stage was defined as the day with maximal extent of total body surface area.
Patient care was conducted in compliance with the study guideline based on our approved IRB and ethics committee. Several therapeutic methods for small drug(s)-induced SJS/TEN patients (non-ICI induced) have been reported, including corticosteroid, intravenous immunoglobulin (IVIg), cyclosporine, TNF blockade, etc. However, there is still no consensus for the therapeutics for patients with ICI-induced SJS/TEN. ICI recently was also found to potentially induce life-threatening SJS/TEN, but the exact pathomechanism remains unknown, and there is also no consensus for the treatment. We retrospectively analyzed the clinical outcome of skin healing and cancer treatment for the enrolled 25 patients with ICI-induced SJS/TEN.
Among these ICI-induced SJS/TEN patients, there was three patients complicated with acute kidney injury. In these patients with ICI-SJS/TEN and acute kidney injury, the renal function of all these three patients was improved at the recovery stage of ICI-SJS/TEN with the use of concurrent TNF blockade therapy.
Drug causality assessment
To determine drug causality, the culprit drug inducing SJS/TEN was determined using the algorithm of drug causality for epidermal necrolysis (ALDEN)65,66 and Naranjo algorithm, whereas the Naranjo algorithm was used to assess the causal drugs for milder ADR phenotypes67. Patients with an ICI (such as nivolumab, pembrolizumab, ipilimumab, or atezolizumab) identified as the probable or definite cause of ADR (ALDEN score ≥4 or Naranjo algorithm ≥5) were recruited. The majority of ICI-SJS/TEN cases (23/25) in this study were identified as “probable and definite” cases. 2 (2/25) of ICP-SJS/TEN cases were determined as “possible” cases by ALDEN (due to the onset time was longer than 60 days), and no other drug causality was identified. To further ensure no confounding medications, the medication profiles for all enrolled patients were cross-checked to identify concomitantly used drugs listed in the ALDEN drug notoriety list, and showed no high-risk medication and “probable and definite” drug used. The clinical course, ICI dosage, ICI duration, systemic involvement, and mortality were also recorded.
Cell isolation and live-cell enrichment
Peripheral blood mononuclear cells (PBMC) were freshly isolated using density gradient centrifugation from ethylenediaminetetraacetic acid (EDTA)-anticoagulated blood samples. Lesional blister cells (lesional BC) from ICI-induced SJS/TEN were isolated by centrifugation at 1000 × g for 5 min. PBMC or lesional BC were either processed immediately for scRNA-seq and ex vivo testing or stored at −80 °C in freezing medium for later use. For scRNA-seq, live cells were enriched by fluorescence-activated cell sorting (FACS). The cells were first stained using an APC Annexin V Apoptosis Detection Kit (BioLegend, Cat.#640932), then sorted on an S3e cell sorter (Bio-Rad Laboratories Taiwan Ltd). Dead cells were excluded by using a conventional forward versus side scatter gating strategy, and Annexin V/propidium iodide double-negative live cells were collected in a new tube containing 0.5 ml 1× phosphate-buffered saline (PBS) supplemented with 2% fetal bovine serum (FBS).
10X Genomics scRNA-seq
Sorted live cells were centrifuged and washed with PBS containing 0.04% bovine serum albumin (Sigma-Aldrich). Cell numbers for each sample were counted by using an automated cell counter (The Countess II FL, ThermoFisher). Single-cell capture and downstream library construction were conducted using the Chromium Single Cell 5′ v2 library preparation kit, based on the manufacturer’s protocol (10X Genomics). Chips were loaded into the 10X Genomics Chromium Controller for the single-cell partitioning. All steps, including emulsions, cDNA isolation, and library preparation, were completed according to the manufacturer’s protocol. After that, cDNA was amplified for 14 cycles and allocated for T-cell receptor (TCR) enrichment/library preparation with the Chromium Single Cell V(D)J TCR kit or for the preparation of the mRNA expression library. The library quality was determined using a 2200 TapeStation system (Agilent) and libraries were sequenced on the Illumina NovaSeq 6000 platform with a 150-bp. We obtained more than 30,000 reads per cell for all samples, which was particularly important to ensure sufficient sampling for gene expression and TCR V(D)J libraries. Detailed information on sample list and sequencing results of scRNA-seq was shown in Supplementary Table 3 and Supplementary Table 6, respectively.
scRNA-seq data analysis
scRNA-seq reads were demultiplexed and aligned to the ENSEMBL GRCh38 v.3.0.0 human transcriptome to generate gene expression matrices using 10X Genomics Cell Ranger (v7.0.1). Unique molecular identifiers (UMIs) were also counted (by using the “cellranger count” function). Each sample obtained from “ICI-cADR” patients and “control participants” was aggregated using the “cellranger aggr” function without depth normalization. We further analyzed these matrices using Seurat (v4.1.1R package)68,69 with default parameters, unless otherwise indicated. We filtered the matrices to exclude low-quality cells using a standard panel with the following criteria: (1) number of detected transcripts (nFeature_RNA) >upper quantile (the value under which 75% of data points are found when arranged in increasing order) + 1.5 interquartile range (IQR); (2) number of detected transcripts (nFeature_RNA) < 200; and (3) percent of reads mapped to mitochondrial genes >20%. Seurat filtration criteria and the final cell number of each sample were shown in Supplementary Table 7.
Cell clusters identification and batch effect assessment
For downstream analysis of scRNA-seq data, the whole gene expression matrix for all cells was performed using Seurat (v4.1.1R package)68,69 for downstream analyses. Firstly, only genes expressed in more than 1 cells were retained, and the UMI count matrix was normalized using the ‘NormalizeData’ function, in which the number of UMIs for each gene is divided by the total number of UMIs in each cell and multiplied by a factor of 10,000, followed by the addition of a pseudocount of 1 for each gene and a natural-log transformation. Based on the normalized gene expression matrix, 2000 highly variable genes were identified using the ‘FindVariableFeatures’ function with the ‘vst’ method for each dataset. The ‘ScaleData’ function was used to scale and center the gene expression matrix after regressing out the heterogeneity associated with mitochondrial contamination. We clustered cells using the FindNeighbors and FindClusters functions and performed nonlinear dimensional reduction with the “RunPCA” (npcs = 30) and ‘RunUMAP’ (dims = 1:20).
To evaluate whether cells were clustered together driven by batch effects, we used the entropy plot for assessment70,71. However, the results showed that the cell clusters of the combined “ICI-cADR” patients and “control participants” might drive by batch effect. We further applied with mutual nearest neighbors (MNN) batch effect correction for the scRNA-seq analysis.
Annotation of cell clusters and data visualization
We used two complementary approaches to annotate the identities of different cell clusters: (1) we applied an unbiased cell type recognition method, named SingleR (v1.8.1)30,31 which leverages reference transcriptomic datasets of known cell types for annotation; (2) we also checked whether well-studied marker genes for different immune cell types32,33,34 were among the top-ranked differentially expressed genes (DEG) of queried clusters and assigned the most likely identity for each cell cluster. We first applied SingleR to determine whether the predicted annotations for a queried cluster were consistent with various reference datasets and assigned this cluster to the predicted cell type annotation; alternatively, we used the “FindAllMarkers” function in Seurat to confirm the identified markers for each cluster and also verified the top-ranked DEG for this cluster. The uniform manifold approximation and projection (UMAP) was applied to visualize the single-cell transcriptional profile72. Furthermore, the clusters expressing two or more canonical cell-type markers of cluster 9 for Macrophage/Monocyte/Myeloid sub-clustering (Supplementary Fig. 2b) were classified as doublet cells evaluated by DoubletFinder73.
Differential expression testing was performed using the “FindMarkers” function in Seurat with parameter “test.use=wilcox” by default. The DEG between each group for gene-set enrichment analysis (GSEA) analysis were determined by “FindMarkers” functions, where “min.pct = 0.25” was set to avoid genes that were infrequently expressed. We used scaled log-normalized expression levels in UMAP plot projections to create violin plots or heatmaps. The GSEA of DEG was performed using clusterProfiler v4.2.274,75. We downloaded hallmark gene sets from the Molecular Signatures Database (MsigDB v7.4)76 and fed each DEG list, together with averaged log (fold change) of each gene and the hallmark gene sets, to the ‘fgsea’ function. The hallmark gene sets with adjusted P value < 0.05 were considered to be functionally enriched biological states or processes.
TCR V(D)J analysis
Demultiplexing, gene quantification, and TCR clonotype assignment were performed using CellRanger v7.0.1 (10X Genomics). FASTQ reads were aligned to the human GRCh38 v7.0.1V(D)J reference genome (10X Genomics) using the “cellranger vdj” function, resulting in the assembly of V(D)J sequences and clonotypes. A TCR diversity metric was obtained, containing clonotype frequency and barcode information. Using barcode information, T cells with prevalent TCR clonotypes were projected on a UMAP plot.
Trajectory and cell–cell communication analysis
To analyze cell states in patients with ICI-cADR, we performed trajectory analysis on T cell, macrophage/monocyte/myeloid cell, and NK/NKT cell subsets using the merged dataset from lesional BC and PBMC obtained from patients with ICI-cADR (including ICI-induced SJS/TEN and mild cADR) using Monocle v2.22.077,78. We included a total of 42,857 cells and reduced dimensionality using the DDRTree algoithm77. We estimated visualized trajectories using pseudo-time plots. To analyze cell–cell communications and perform ligand–receptor analysis at the molecular level, three groups were generated, including the “ICI-cADR”, “ICI-SJS/TEN”, and “ICI-tolerant & HD” groups. We analyzed all cell clusters by using the CellChat v1.1.379 tool.
Immunofluorescence staining
Formalin-fixed paraffin-embedded (FFPE) skin tissue samples from patients with ICI-induced SJS/TEN, ICI-induced mild cADR (lichenoid dermatitis) and non-cADR HD participants were soaked in xylene, followed by sequential treatment with 100%, 95%, and 70% ethanol for paraffin wax removal and rehydration. Antigen unmasking was performed by heating the slides in retrieval buffer, after which they were allowed to cool to room temperature. Protein blockade treatment was applied to the tissues to prevent non-specific protein binding. The following primary antibodies were used: rabbit monoclonal anti-human CXCL10-FITC antibody (1:100 dilution, LS-C123903, LSBio), mouse monoclonal anti-human CD163 antibody (1:100 dilution, ab156769, Abcam), mouse monoclonal anti-human CD8 antibody (1:100 dilution, ab199016, Abcam), and rabbit polyclonal anti-human CXCR3 antibody (1:200 dilution, ab71864, Abcam). All antibodies and 4’,6-diamidino-2-phenylindole (DAPI; nuclear stain; D1306, ThermoFisher) were applied to the tissue sections according to the manufacturer’s instructions for 30 min at room temperature. The sections were washed three times for 5 min each with PBS, followed by incubation with Alexa Fluor 488-conjugated goat anti-mouse IgG (1:400 dilution, ab150113, Abcam), Texas Red-conjugated goat anti-rabbit IgG (1:400 dilution; ab6719, Abcam) or Texas Red-conjugated goat anti-mouse IgG (1:400 dilution; ab6787, Abcam) for 30 min at room temperature. Slides were washed three times for 5 min each in PBS and mounted with ProLong Gold (Molecular Probes) antifade reagent. Fluorescence images were captured using a Zeiss Axio. For each sample, 2–3 randomly chosen high-powered fields were evaluated at 400× or 200× magnification by at least two experienced researchers in a blinded manner.
Cytokine array and ELISA
Concentrations of 24 cytokines/chemokines present in patients’ blister fluids or plasma were determined using the Luminex MAGPIX system (ThermoFisher Scientific), according to the manufacturer’s instructions. ELISA was used to measure the levels of granulysin (DY3138, R&D), granzyme B (3486-1H-6S, Mabtech), TNF (DY210-05, R&D), and CXCL10 (DY266, R&D). The fold change in each sample was normalized to the solvent control.
Ex vivo lymphocyte activation test (LAT) and anti-TNF blocking assay
PBMC from the recovery stage of patients with ICI-cADR were isolated from EDTA-containing whole-blood samples using Ficoll-Paque (Pharmacia Fine Chemicals, Uppsala, Sweden) density gradient centrifugation. Isolated PBMC (2.0 × 105 cells/well) were cultured in 96-well microplates in RPMI-1640 medium (GIBCO, Cat.#A16491-01, Invitrogen, Life Technologies, Carlsbad, CA), supplemented with 10% human AB serum (Sigma-Aldrich, Darmstadt, Germany), 5 U/mL human IL-2 (Croyez, Cat.#C01004), and ICI (including ipilimumab, pembrolizumab, nivolumab, and atezolizumab) at 37 °C in 5% CO2. The three test treatments included PBS, ICI, and ICI plus anti-TNF (Etanercept 5 µg/mL) and were applied in a 200-µL volume of culture medium. Drugs were diluted in medium to obtain concentrations similar to 10-fold physiological therapeutic levels (pembrolizumab: 250 µg/ml; nivolumab: 80 µg/ml; atezolizumab: 488 µg/ml; ipilimumab: 50 µg/ml). In addition, dimethyl sulfoxide (DMSO) was added to the medium as the solvent control. After 6 days of culture, supernatants were harvested by enzyme-linked immunosorbent assay (ELISA), and cells were subjected to BD Rhapsody single-cell RNA sequencing (scRNA-seq) or flow cytometry analyses.
BD Rhapsody scRNA-seq
For determining the single-cell RNA transcriptome of PBMC from patients with ICI-cADR following ex vivo treatment with ICI or biologic anti-TNF, we utilized the BD Rhapsody scRNA-seq system (BD Biosciences). After treatment with ICI or ICI plus biologic anti-TNF for 6 days, single cells were sequentially labeled with the BD Human Single-Cell Multiplexing Kit (BD Biosciences, #633781). Briefly, cells were labeled with sample tags. Each sample was washed three times with PBS before pooling all samples together, and the pooled cells were then stained with Calcein AM (Thermo Fisher Scientific, Cat.#C1430) and Draq7 (Thermo Fisher Scientific, Cat.#564904) to measure cell viability using a BD Rhapsody Scanner (BD Biosciences). Only pooled single cells with a >75% survival rate were loaded onto a BD Rhapsody Cartridge for single-cell capture using the BD Rhapsody Single-Cell Analysis system (BD Biosciences, Doc ID:210966). cDNA libraries were prepared using the BD Rhapsody System mRNA Whole Transcriptome Analysis (WTA) and Sample Tag Library Preparation Protocol (Part Number: 23-21712-00), according to the manufacturer’s instructions.
The quality of the final libraries was assessed using an Agilent 2100 bioanalyzer and quantified using a Qubit Fluorometer using the Qubit dsDNA HS kit (ThermoFisher, Cat.#Q32854). Final libraries were diluted to 2 nM and multiplexed for paired-end (150-bp) sequencing on an Illumina NovaSeq 6000 (Illumina).
Analyzing (WTA) data from BD Rhapsody scRNA-seq via Seven Bridges
We uploaded and analyzed the Fastq sequencing files on the Seven Bridges website (Seven Bridges Genomics) using the “BD Rhapsody whole transcriptome analysis (WTA) Analysis Pipeline” (BD Biosciences, San Jose, CA), according to the manufacturer’s protocol. After performing alignment, filtering, and sample tag detection, we downloaded and used the pipeline’s final outputs, including sample tag calls and molecule count information, for further analysis in R (v4.1.0) using Seurat (v4.1.1 R package)68,69. Using the standard processing workflow in Seurat, we acquired clustering and gene expression data. Detailed information on sequencing results of BD Rhapsody scRNA-seq was shown in Supplementary Table 8.
Flow cytometry analysis
To identify the cell populations, the cultured 5 × 105 of patients’ PBMC were incubated with Horizon V450-labeled anti-human CD8 (1:40 dilution, Cat#560347; BD Biosciences), Alexa 647-labeled anti-human CD163 (1:40 dilution, Cat#562669; BD Biosciences), BB700-labeled anti-human CD86 (1:40 dilution, Cat#747524; BD Biosciences), and APC-Cy7-labeled anti-human CD14 (1:40 dilution, Cat#557831; BD Biosciences) at 4 °C for 30 min. After the cell surface staining, the cells of each sample were fixed/permeabilized according to the instructions of BD fixation/permeabilization kit (Cat#554714; BD Bioscience). Then the cells were stained with PE-labeled anti-human CXCL10 (IP-10) (1:40 dilution, Cat# 555049; BD Bioscience), Horizon BV711-labeled anti-human CXCR3 (CD183) (1:40 dilution, Cat#563156; BD Bioscience) and PE-Cy7-labeled anti-human granzyme B (1:40 dilution, Cat#372214; Biolegend). After washing, the stained cells were acquired with FACSDiva v.8.0 (BD Biosciences), and data were analyzed using FlowJo v.10 software (LLC).
RNA extraction for FFPE samples
Three 10-µm sections of FFPE skin tissues from patients with ICI-induced SJS/TEN, ICI-induced mild cADR (lichenoid dermatitis) and non-cADR HD were collected for RNA extraction, which was performed using the Roche HighPure FFPET RNA Isolation spin-column kit (Catalog #06650775001), according to manufacturer’s specifications. RNA was quantified using the NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Oxfordshire, UK), and any samples with concentrations were below 20 ng/µL were concentrated using the RNA Clean and Concentrator Columns (Zymo Research, Cat.#11-325), according to the manufacturer’s specifications.
NanoString nCounter System Processing
A total of 100–500 ng purified RNA was hybridized with the nCounter PanCancer human immune profiling panel code set (NanoString Technologies, Seattle, WA, USA) at 65 °C overnight. Further purification and binding of the hybridized probes to the optical cartridge were performed using nCounter Elements technology (NanoString Technologies), following the manufacturer’s protocol, and the cartridge was scanned on the nCounter Digital Analyzer. Raw counts from each gene were imported into the nSolver Analysis Software v4.0 (NanoString Technologies; https://www.nanostring.com/products/analysis-software/nsolver), normalized against background and housekeeping genes, and overall assay performance was assessed using built-in positive controls. Data were also analyzed using the R-based nSolver software v4.0.
Statistical analysis
The statistical analyses for scRNA-seq and NanoString RNA were performed using R (v4.1.0) and nSolver 4.0, respectively. All analyses are described in the figure legends or source data, and all analyses were corrected for multiple comparisons when appropriate. The two-sided non-parametric Wilcoxon rank-sum test was used to compare between groups, and adjusted P values were based on Bonferroni correction in the dataset.
Fisher exact tests, Chi-square tests, and odds ratios (ORs) with their 95% confidence intervals (CIs) calculation for patient characteristics of two treatment groups were performed by RStudio-1.2.1335 (Northern Ave, Boston, MA). ORs were calculated by using a Haldane modification, and added 0.5 to accommodate possible zero counts.
Statistical analyses of flow cytometry data and cytokine/chemokine/cytotoxic protein levels were performed using Prism v.8.4.1 (GraphPad). Within-group comparisons were performed using the Wilcoxon signed-rank test, whereas between-group comparisons were performed using the Mann–Whitney U test. The correlations of plasma level of CXCL10 with IL-6 and IFN-γ were analyzed by using Spearman’s rank correlation coefficient.
Serial plasma cytokines/chemokine/cytotoxic protein levels in 6 available patients with ICI-induced SJS/TEN during the clinical course from the maximal stage and 3 weeks after initiation of TNF inhibitor treatment were calculated by two-sided paired Wilcoxon matched-pairs signed-rank test. These data represent the mean ± standard deviation (SD); ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data for Figs. 1–7 and Supplementary Figs. 1–16 are provided with the paper. The matrix and raw data for scRNA-seq reported in this paper have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number (GSE273720). These data are freely available without any restrictions or time limits. All other data are available in the article and its Supplementary files or from the corresponding author upon request. Source data are provided with this paper.
Code availability
Representative code is available on GitHub (https://github.com/phoebee-h/MS_NatureComm2024). This code is openly available with no restriction or time limit. Any queries or further requests can be addressed to the corresponding authors.
References
Reck, M. et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 375, 1823–1833 (2016).
Ribas, A. et al. Pembrolizumab versus investigator-choice chemotherapy for ipilimumab-refractory melanoma (KEYNOTE-002): a randomised, controlled, phase 2 trial. Lancet Oncol. 16, 908–918 (2015).
Weber, J. S. et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 16, 375–384 (2015).
Davar, D. & Kirkwood, J. M. PD-1 immune checkpoint inhibitors and immune-related adverse events: understanding the upside of the downside of checkpoint blockade. JAMA Oncol. 5, 942–943 (2019).
Ramos-Casals, M. et al. Immune-related adverse events of checkpoint inhibitors. Nat. Rev. Dis. Prim. 6, 38 (2020).
Taylor, C. A. et al. IL7 genetic variation and toxicity to immune checkpoint blockade in patients with melanoma. Nat. Med. 28, 2592–2600 (2022).
Kumar, V. et al. Current diagnosis and management of immune related adverse events (irAEs) induced by immune checkpoint inhibitor therapy. Front. Pharmacol. 8, 49 (2017).
Chen, C. B. et al. Severe cutaneous adverse reactions induced by targeted anticancer therapies and immunotherapies. Cancer Manag. Res. 10, 1259–1273 (2018).
Curry, J. L. et al. Diverse types of dermatologic toxicities from immune checkpoint blockade therapy. J. Cutan. Pathol. 44, 158–176 (2017).
Phillips, G. S. et al. Treatment outcomes of immune-related cutaneous adverse events. J. Clin. Oncol. 37, 2746–2758 (2019).
Roujeau, J. C. & Stern, R. S. Severe adverse cutaneous reactions to drugs. N. Engl. J. Med. 331, 1272–1285 (1994).
Hung, S. I. et al. Severe cutaneous adverse reactions. Nat. Rev. Dis. Prim. 10, 30 (2024).
Raschi, E. et al. Serious cutaneous toxicities with immune checkpoint inhibitors in the U.S. Food and Drug Administration adverse event reporting system. Oncologist 24, e1228–e1231 (2019).
Roujeau, J. C. et al. Medication use and the risk of Stevens-Johnson syndrome or toxic epidermal necrolysis. N. Engl. J. Med. 333, 1600–1607 (1995).
Wang, Y. H. et al. The medication risk of Stevens-Johnson syndrome and toxic epidermal necrolysis in Asians: the major drug causality and comparison with the US FDA label. Clin. Pharm. Ther. 105, 112–120 (2019).
Illing, P. T. et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 486, 554–558 (2012).
Pan, R. Y., Dao, R. L., Hung, S. I. & Chung, W. H. Pharmacogenomic advances in the prediction and prevention of cutaneous idiosyncratic drug reactions. Clin. Pharm. Ther. 102, 86–97 (2017).
Chung, W. H. et al. Medical genetics: a marker for Stevens-Johnson syndrome. Nature 428, 486 (2004).
Gibson, A. et al. Updates on the immunopathology and genomics of severe cutaneous adverse drug reactions. J. Allergy Clin. Immunol. 151, 289–300.e284 (2023).
Chung, W. H. et al. Genetic variants associated with phenytoin-related severe cutaneous adverse reactions. JAMA 312, 525–534 (2014).
Wang, C. W. et al. Whole genome sequencing identifies genetic variants associated with co-trimoxazole hypersensitivity in Asians. J. Allergy Clin. Immunol. 147, 1402–1412 (2021).
Jiang, H. et al. Functional and structural characteristics of HLA-B*13:01-mediated specific T cells reaction in dapsone-induced drug hypersensitivity. J. Biomed. Sci. 29, 58 (2022).
Chung, W. H. et al. Granulysin is a key mediator for disseminated keratinocyte death in Stevens-Johnson syndrome and toxic epidermal necrolysis. Nat. Med. 14, 1343–1350 (2008).
Chen, C. B. et al. Detecting lesional granulysin levels for rapid diagnosis of cytotoxic T lymphocyte-mediated bullous skin disorders. J. Allergy Clin. Immunol. Pr. 9, 1327–1337.e1323 (2021).
Wang, C. W. et al. A new nucleic acid-based agent inhibits cytotoxic T lymphocyte-mediated immune disorders. J. Allergy Clin. Immunol. 132, 713–722.e711 (2013).
Pardoll, D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012).
Brahmer, J. R. et al. Management of immune-related adverse events in patients treated with immune checkpoint inhibitor therapy: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 36, 1714–1768 (2018).
Yamane, Y. et al. Retrospective analysis of Stevens-Johnson syndrome and toxic epidermal necrolysis in 87 Japanese patients—treatment and outcome. Allergol. Int. 65, 74–81 (2016).
Thompson, J. A. et al. NCCN guidelines insights: management of immunotherapy-related toxicities, version 1.2020. J. Natl Compr. Cancer Netw. 18, 230–241 (2020).
Abdelaal, T. et al. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 20, 194 (2019).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Martin, J. C. et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178, 1493–1508.e1420 (2019).
Luoma, A. M. et al. Molecular pathways of colon inflammation induced by cancer immunotherapy. Cell 182, 655–671.e622 (2020).
Smillie, C. S. et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e722 (2019).
Matsushima, H. & Takashima, A. Bidirectional homing of Tregs between the skin and lymph nodes. J. Clin. Invest. 120, 653–656 (2010).
Luo, W. & Brouwer, C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831 (2013).
Dufour, J. H. et al. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J. Immunol. 168, 3195–3204 (2002).
Chu, K. L., Batista, N. V., Girard, M. & Watts, T. H. Monocyte-derived cells in tissue-resident memory T cell formation. J. Immunol. 204, 477–485 (2020).
Trubiano, J. A. et al. Analysis of skin-resident memory T cells following drug hypersensitivity reactions. J. Invest. Dermatol. 140, 1442–1445.e1444 (2020).
Villani, A. P. et al. Massive clonal expansion of polycytotoxic skin and blood CD8(+) T cells in patients with toxic epidermal necrolysis. Sci. Adv. https://doi.org/10.1126/sciadv.abe0013 (2021).
Schunkert, E. M., Shah, P. N. & Divito, S. J. Skin resident memory T cells may play critical role in delayed-type drug hypersensitivity reactions. Front Immunol. 12, 654190 (2021).
Wang, C. W., Divito, S. J., Chung, W. H. & Hung, S. I. Advances in the pathomechanisms of delayed drug hypersensitivity. Immunol. Allergy Clin. North Am. 42, 357–373 (2022).
Han, J. et al. Resident and circulating memory T cells persist for years in melanoma patients with durable responses to immunotherapy. Nat. Cancer 2, 300–311 (2021).
Lozano, A. X. et al. T cell characteristics associated with toxicity to immune checkpoint blockade in patients with melanoma. Nat. Med. 28, 353–362 (2022).
Croft, M. The role of TNF superfamily members in T-cell function and diseases. Nat. Rev. Immunol. 9, 271–285 (2009).
Perazella, M. A. & Shirali, A. C. Nephrotoxicity of cancer immunotherapies: past, present and future. J. Am. Soc. Nephrol. 29, 2039–2052 (2018).
Abdallah, M. et al. CXCL-10 and Interleukin-6 are reliable serum markers for vitiligo activity: a multicenter cross-sectional study. Pigment Cell Melanoma Res. 31, 330–336 (2018).
Khan, S. et al. Immune dysregulation in cancer patients developing immune-related adverse events. Br. J. Cancer 120, 63–68 (2019).
von Itzstein, M. S., Khan, S. & Gerber, D. E. Investigational biomarkers for checkpoint inhibitor immune-related adverse event prediction and diagnosis. Clin. Chem. 66, 779–793 (2020).
Pan, R. Y. et al. Identification of drug-specific public TCR driving severe cutaneous adverse reactions. Nat. Commun. 10, 3569 (2019).
Wang, C. W. et al. Randomized, controlled trial of TNF-alpha antagonist in CTL-mediated severe cutaneous adverse reactions. J. Clin. Invest 128, 985–996 (2018).
Kinoshita, M. et al. Neutrophils initiate and exacerbate Stevens-Johnson syndrome and toxic epidermal necrolysis. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.aax2398 (2021).
Husain, B. et al. Inflammatory markers in autoimmunity induced by checkpoint inhibitors. J. Cancer Res. Clin. Oncol. 147, 1623–1630 (2021).
Mandai, M. et al. Dual faces of IFNγ in cancer progression: a role of PD-L1 induction in the determination of pro- and antitumor immunity. Clin. Cancer Res. 22, 2329–2334 (2016).
Perez-Ruiz, E. et al. Prophylactic TNF blockade uncouples efficacy and toxicity in dual CTLA-4 and PD-1 immunotherapy. Nature 569, 428–432 (2019).
Montfort, A. et al. The TNF paradox in cancer progression and immunotherapy. Front Immunol. 10, 1818 (2019).
Montfort, A. et al. Combining nivolumab and ipilimumab with infliximab or certolizumab in patients with advanced melanoma: first results of a phase Ib clinical trial. Clin. Cancer Res. 27, 1037–1047 (2021).
Brahmer, J. R. et al. Society for Immunotherapy of Cancer (SITC) clinical practice guideline on immune checkpoint inhibitor-related adverse events. J. Immunother. Cancer https://doi.org/10.1136/jitc-2021-002435 (2021).
Satoh, T. K., Neulinger, M. M., Stadler, P. C., Aoki, R. & French, L. E. Immune checkpoint inhibitor-induced epidermal necrolysis: a narrative review evaluating demographics, clinical features, and culprit medications. J. Dermatol. 51, 3–11 (2024).
Roujeau, J. C. Clinical heterogeneity of drug hypersensitivity. Toxicology 209, 123–129 (2005).
Auquier-Dunant, A. et al. Correlations between clinical patterns and causes of erythema multiforme majus, Stevens-Johnson syndrome, and toxic epidermal necrolysis: results of an international prospective study. Arch. Dermatol. 138, 1019–1024 (2002).
Mockenhaupt, M. Epidemiology of cutaneous adverse drug reactions. Allergol. Sel. 1, 96–108 (2017).
Khoja, L., Day, D., Wei-Wu Chen, T., Siu, L. L. & Hansen, A. R. Tumour- and class-specific patterns of immune-related adverse events of immune checkpoint inhibitors: a systematic review. Ann. Oncol. 28, 2377–2385 (2017).
Hsiehchen, D., Watters, M. K., Lu, R., Xie, Y. & Gerber, D. E. Variation in the assessment of immune-related adverse event occurrence, grade, and timing in patients receiving immune checkpoint inhibitors. JAMA Netw. Open 2, e1911519 (2019).
Rzany, B. et al. Epidemiology of erythema exsudativum multiforme majus, Stevens-Johnson syndrome, and toxic epidermal necrolysis in Germany (1990-1992): structure and results of a population-based registry. J. Clin. Epidemiol. 49, 769–773 (1996).
Sassolas, B. et al. ALDEN, an algorithm for assessment of drug causality in Stevens-Johnson Syndrome and toxic epidermal necrolysis: comparison with case-control analysis. Clin. Pharm. Ther. 88, 60–68 (2010).
Naranjo, C. A. et al. A method for estimating the probability of adverse drug reactions. Clin. Pharmacol. Ther. 30, 239–245 (1981).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Haghverdi, L., Lun, A. T. L., Morgan, M. D. & Marioni, J. C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018).
Kinchen, J. et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell 175, 372–386.e317 (2018).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. https://doi.org/10.1038/nbt.4314 (2018).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e324 (2019).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Acknowledgements
We thank Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung; and Cancer Vaccine and Immune Cell Therapy Core Laboratory, Chang Gung Memorial Hospital, Linkou, Taiwan, for supporting the scRNA-seq analysis in this study. This study was supported by research grants from the National Science and Technology Council, Taiwan (grant no. NSTC 108-2314-B-182A-006-MY3 to C.B.C., 110-2320-B-182A-014-MY3 to C.W.W., 110-2326-B-182A-003- to W.H.C., 111-2314-B-182A-111-MY3 to W.H.C., 111-2314-B-182A-111-MY3 to C.B.C., 112-2326-B-182A-003-MY3 to C.B.C., 112-2326-B-182A-001- to W.H.C., 113-2320-B-182A-003-MY3 to C.W.W.) and Chang Gung Memorial Hospital, Taiwan (grant no. CMRPG2J0221-2 to C.B.C., CMRPG3L0491 to C.B.C., CMRPG3I0381-2 to W.H.C., CIRPG3I0024-5 to W.H.C., CIRPG3I0044-5 to S.I.H., CIRPG2I0014-5 to C.B.C., CMRPG3P0471 to C.W.W., OMRPG3N0011 to W.H.C.). We would also like to thank the organizing committee and instructors of the Formosa Immunology Spring School & Symposium for their inspiration and feedback on this research.
Author information
Authors and Affiliations
Contributions
Drs. C.B.C., C.W.W., C.L.K. and W.H.C. had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Drs. C.L.K., C.W.W. and W.H.C. contributed equally to this article as corresponding authors. Study concept and design: Drs. C.B.C., S.I.H., C.W.W. and W.H.C. Acquisition, analysis, or interpretation of data: All authors. Drafting of the manuscript: Drs. C.B.C., C.W.W. and W.H.C. Critical revision of the manuscript for important intellectual content: All authors. Performing scRNA-seq., immunofluorescence staining, NanoString RNA-seq, ex vivo lymphocyte activation test, cytokine array/ELISA, and flow cytometry analysis: H.Y.Y., C.C.T., Y.Y.W.L., Chih-Jung Chang and C.W.W. scRNA-seq analysis: Mr/Mrs. Y.C.T. and F.M.H. Statistical analysis: Drs. C.B.C., C.W.W., Y.C., H.C.L., Chee Jen Chang and Y.J.L. Patients enrollment: Drs. C.B.C., J.W.C.C., C.K.Y., D.H.K.M., C.W.L., W.T.C., C.L.W., P.H.C., J.W., Y.W.T., L.Y.L., R.C.Y.H., C.K.H., C.N.L., Y.J.C., C.C.C., H.C.H., Y.C.C. and W.H.C. Obtained funding: Drs. C.B.C., J.W.C.C., C.W.W., Y.C. and W.H.C. Administrative, technical, or material support: All authors. Study supervision: Drs. S.I.H., C.L.K., C.W.W. and W.H.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Tetsuo Shiohara and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, CB., Hung, SI., Chang, J.WC. et al. Immune checkpoint inhibitor-induced severe epidermal necrolysis mediated by macrophage-derived CXCL10 and abated by TNF blockade. Nat Commun 15, 10733 (2024). https://doi.org/10.1038/s41467-024-54180-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-54180-7