Abstract
Salicylic acid (SA) production in Brassicaceae plants is uniquely accelerated from isochorismate by EPS1, a newly identified enzyme in the BAHD acyltransferase family. We present crystal structures of EPS1 from Arabidopsis thaliana in both its apo and substrate-analog-bound forms. Integrating microsecond-scale molecular dynamics simulations with quantum mechanical cluster modeling, we propose a pericyclic rearrangement lyase mechanism for EPS1. We further reconstitute the isochorismate-derived SA biosynthesis pathway in Saccharomyces cerevisiae, establishing an in vivo platform to examine the impact of active-site residues on EPS1 functionality. Moreover, stable transgenic expression of EPS1 in soybean increases basal SA levels, highlighting the enzyme’s potential to enhance defense mechanisms in non-Brassicaceae plants lacking an EPS1 ortholog. Our findings illustrate the evolutionary adaptation of an ancestral enzyme’s active site to enable a novel catalytic mechanism that boosts SA production in Brassicaceae plants.
Similar content being viewed by others
Introduction
Salicylic acid (SA), also known as 2-hydroxybenzoic acid, is a simple phenolic acid essential in higher plants for both local and long-distance defense responses after pathogen exposure1. Beyond its defense functions, SA serves auxiliary roles in photosynthesis, ion uptake and transport, and growth regulation2. Despite its chemical simplicity and significance in defense signaling, the full scope of various SA biosynthetic pathways across the plant kingdom remains incompletely explored. Two major plant SA biosynthetic pathways have been identified downstream of chorismate to date: the phenylpropanoid-derived pathway3 and the isochorismate-derived pathway4. In the model plant Arabidopsis thaliana, a minor amount of SA is generated through phenylalanine ammonia-lyase (PAL), converting cinnamic acid to benzoic acid via side-chain shortening and 2‐hydroxylation5,6,7,8. However, during pathogen attacks, the primary SA biosynthesis route relies on isochorismate synthase, an enzyme in plastids that converts chorismate to isochorismate, encoded by the SALICYLIC ACID INDUCTION DEFICIENT 2 (SID2) gene9 (Fig. 1a). The A. thaliana gene SALICYLIC ACID INDUCTION DEFICIENT 1 (SID1) encodes a multidrug and toxin extrusion (MATE) transporter, which is localized in the chloroplast envelope and transports isochorismate, synthesized by SID2, from the plastid to the cytosol10,11,12. Unlike the bacterial pathway in which SA is produced directly from isochorismate via an isochorismate pyruvate lyase (IPL)13, plants employ a cytosolic GH3 acyl adenylase family enzyme PBS3 that catalyzes regiospecific conjugation of isochorismate with L-glutamate to form isochorismoyl-glutamate A (IGA) (Fig. 1a)12,14. IGA is unstable and decays spontaneously into the byproduct N-pyruvoyl-L-glutamate (NPG) and the phytohormone SA12,14.
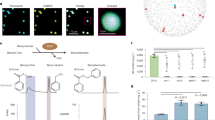
a The SA biosynthetic pathway in Brassicaceae plants. b Simplified maximum likelihood tree of the EPS1 clade (purple), the EPS1 sister clade (blue) and its most closely related HXXXD-type acyl-transferase family protein ancestral clade (green). Note that the blue EPS1 sister clade is seemingly lost in the majority of Brassicaceae plants. The purple EPS1 clade enzymes and not the neighboring clades display strict conservation for the serine substitution for the characteristic histidine in the (HXXXD) motif. c Simplified taxonomy of Brassicaceae plants displaying the presence or absence of the green ancestor clade, the blue EPS1 sister clade or the purple true EPS1 clade enzymes. d In A. thaliana, the EPS1 gene (purple arrow and shading) is adjacent to its closest homolog AT5G67150.1 (green arrow and shading). In the related species A. lyrata, EPS1 is sandwiched between the AT5G67150.1 ortholog AL8G44210.t1 (green arrow and shading) and the even more closely related A. lyrata AL8G44190.t1 (blue arrow and shading). Our analyses suggest that in the common ancestor of Brassicaceae plants, the HXXXD-clade progenitor duplicated in situ to yield two derived copies that founded the EPS1 clade and the EPS1 sister clade, respectively. Whereas the EPS1 clade acquired IPGL activity, the EPS1 sister clade genes were then lost in A. thaliana and the majority of other profiled extant Brassicaceae species.
Unique to the Brassicaceae plants, the spontaneous decay of IGA is accelerated by a lineage-specific isochorismoyl-glutamate pyruvoyl-glutamate lyase (IPGL) encoded by the ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 (EPS1) gene14 (Fig. 1a). The A. thaliana eps1 mutant is hypersusceptible to the bacterial pathogen Pseudomonas syringae and is defective in de novo SA production upon P. syringae infection15. EPS1 is a member of the plant BAHD acyltransferase family, which is named after the first four enzymes characterized within the family16. The BAHD family has undergone extensive radiation during land plant evolution, giving rise to functionally diverse acyltransferases widely distributed in many plant natural product biosynthetic pathways17. Canonical BAHD enzymes catalyze the transfer of the acyl group from an acyl-CoA thioester substrate to an alcohol or amine-containing acceptor molecule to form the corresponding ester or amide product. These proteins are also annotated as HXXXD family enzymes due to a highly conserved HXXXD motif. While most BAHD enzymes contain the conserved catalytic histidine as part of the HXXXD motif, which serves to coordinate the -OH or -NH2 group of the acyl acceptor substrate to initiate the catalytic cycle18, A. thaliana EPS1 (AtEPS1) and other Brassicaceae orthologs contain a serine substitution at this conserved histidine residue. In addition, AtEPS1 does not contain an acyl-CoA thioester-binding motif. Both of these observations suggest the emergence of a new catalytic function in AtEPS114. Indeed, our recent work on the coumarin synthase (COSY) enzyme is an example of neofunctionalization within the BAHD enzyme family, showcasing an unconventional active site that catalyzes the intramolecular isomerization and lactonization of substrates to produce coumarins19.
In this study, we present in-depth characterization of crystal structures of AtEPS1 in both the apo form and holo form bound to an inert substrate analog. Our approach combines computational methods, site-directed mutagenesis, and reconstitution of the SA biosynthesis pathway in Saccharomyces cerevisiae to pinpoint crucial active-site residues affecting AtEPS1’s lyase activity and stability. To examine this Brassicaceae plant trait in an orthologous plant species, we proceeded to introduce AtEPS1 transgenically into soybeans, which do not naturally possess an EPS1 ortholog. This step allowed us to delve deeper into whether gaining IPGL activity in a non-Brassicaceae plant could boost SA production efficiency. These findings not only illuminate the adaptable nature of BAHD-family enzymes through evolution but also propose a novel approach to enhancing defense responses in agricultural crops.
Results and discussion
The evolutionary history of EPS1
To trace the evolutionary history of EPS1 and uncover its role, we conducted a targeted phylogenetic analysis of 62 annotated BAHDs encoded by the A. thaliana genome, placing EPS1 within the Ib clade. While some enzymes in clade I were linked to flavonoid biosynthesis, the exact functions of these clade Ib enzymes remain largely unknown (Supplementary Fig. 1)16,20. Expanded phylogenetic analysis of additional EPS1-like homologs from multiple reference plant genomes revealed that EPS1 falls into a subgroup of BAHDs within the Ib clade that are restricted to Brassicaceae species (Fig. 1b, c). Within this subgroup, a canonical HXXXD-type clade (green, represented by AT5g67150 from A. thaliana, which has an unknown function) is present in all Brassicaceae species examined and is likely the immediate ancestor of the closely related EPS1 clade (purple) and EPS1 sister clade (blue). (Fig. 1b). Synteny analysis of the EPS1 genomic loci across several Brassicaceae plant genomes provided further details regarding the evolutionary birth of EPS1 (Fig. 1d). In the A. thaliana genome, AtEPS1 is adjacent to its most closely related HXXXD-type clade homolog AT5G67150, which is also the case in the syntenic regions for 7 of the 8 Brassicaceae species examined (Fig. 1d). The syntenic region in the A. lyrata genome harbors a third homologous gene AL8G44190, belonging to the EPS1 sister clade, on the other side of AlEPS1 (Fig. 1d). This tandem gene duplication event likely gave rise to EPS1, as well as the EPS1 sister clade gene which was subsequently lost in A. thaliana and the majority of other profiled Brassicaceae species (Fig. 1c).
The overall structure of AtEPS1
To understand how IPGL activity could arise from a progenitor BAHD acyltransferase, we solved the crystal structures of apo AtEPS1 and the substrate analog (2-(3-carboxyphenoxy)acetyl)-L-glutamic acid (CAG) (Supplementary Figs. 2-8) bound AtEPS1, at 1.9 Å and 1.7 Å resolution, respectively (Fig. 2a, b and Supplementary Table 1). Similar to several previously reported BAHD structures, AtEPS1 displays pseudo-symmetric N-terminal (residues 1-181) and C-terminal (residues 227-434) domains, connected by a large crossover loop (residues 182-226) which runs around ¾ of the circumference of the protein18,21,22,23,24,25,26,27 (Fig. 2a). In the AtEPS1 active site, located at the interface between the two domains, the aryl carboxylate of the substrate analog CAG is coordinated through the backbone carbonyl of Pro390, in addition to the positively charged guanidino groups of Arg44 and Arg395. The aromatic ring of CAG is stabilized by the side chains of Leu42, Val357, Val363, and Leu392, while Phe39 likely functions as part of this hydrophobic pocket to hold the acetyl portion of CAG or the corresponding acrylate group of the true IGA substrate. The ɑ-carboxyl group of the glutamate portion of CAG is highly coordinated by the guanidino groups of Arg36 and Arg282 as well as the hydroxyl group of Thr306, while the γ-carboxyl group of the glutamate portion forms a H-bond with the hydroxyl group of Thr365.
a The CAG-bound AtEPS1 structure. The N-terminal domain is displayed in salmon, the crossover loop is displayed in slate blue, the dynamic portion of the crossover loop is colored in cyan, the C-terminal domain is displayed in maroon, and the lid loop is colored in purple. |2Fo – Fc| electron density map for the CAG ligand is contoured at 1.5 σ. b Structure of the EPS1 ligand (CAG) and the native substrate (IGA). c The CAG-bound AtEPS1 and the gray superimposed p-coumaroylshikimate-bound AtHCT (PDB: 5KJU). AtEPS1 residues corresponding to the AtHCT catalytic triad and the bulky residues filling the acetyl donor site are individually labeled and colored in salmon. The two catalytic residues of AtHCT are labeled and colored in gray. d Superimposition of the AtEPS1 CAG-bound and apo structures. The apo structure is displayed in bluewhite with the dynamic loops rendered in black. The dynamic loops from both structures are displayed in cartoon putty where the radius of the loops represents relative b-factor. Val355, Ser356, and Val357, annotated in the CAG-bound model, are the only dynamic-loop residues in both structures, within 6 Å distance from CAG.
Interestingly, when super-positioning CAG-bound AtEPS1 with the product-bound structure of A. thaliana hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase (AtHCT) (PDB ID: 5KJU)18, it is apparent that the CAG-binding site in AtEPS1 corresponds to only the acyl acceptor portion of the AtHCT active site. Several bulky residues in AtEPS1, notably Leu42, Phe156, Tyr158, Ile168, and Trp169, fill the space that otherwise would host the acyl donor in a canonical BAHD acyltransferase (Fig. 2c). In addition to the substitution of the conserved catalytic histidine to serine in AtEPS1, Trp371 (AtHCT numbering), a residue that serves as an oxyanion hole to stabilize the tetrahedral intermediate of the acylation reaction, conserved in a large fraction of BAHD acyltransferases18, is substituted to serine (Ser367) in AtEPS1 (Fig. 2c).
Two dynamic loops gate the active site entrance
Amongst previously reported plant BAHD structures, the size and arrangement of the crossover loop on the acyl-donor side of the enzyme vary little between structures, while portions of the crossover loop on the acyl-acceptor side of the enzyme vary substantially corresponding to individual enzymes’ acyl-acceptor substrates. Despite its overall high sequence and structural similarity with AtHCT, AtEPS1 adopts a crossover-loop conformation more similar to that observed in the vinorine synthase and anthocyanin malonyltransferase structures27,28, indicating a possible preferred configuration for larger substrates, such as IGA, vinorine, or anthocyanin, compared to the smaller HCT acyl acceptor substrate shikimate (Supplementary Fig. 9, Supplementary Table 2, and Supplementary Table 3).
Comparison of the apo- and CAG-bound AtEPS1 structures reveals conformational changes of a dynamic portion of the crossover loop (Pro212-Asn226), a phenomenon which was not observed in other BAHD structures (Fig. 2d). In the CAG-bound AtEPS1 structure, crossover-loop residues Pro212-Asn226 are rotated away from the central ligand-binding cavity, and an adjacent dynamic loop (Ile351-Lys360), recently described as the lid loop29, is positioned directly above the substrate analog. Conversely, in the apo AtEPS1 structure, the lid loop (Ile351-Lys360) and the crossover-loop residues Pro212-Asn226 are displaced to open up a larger entrance to the enzyme active site (Fig. 2d). Moreover, 28.04% volume constriction is observed at the active-site upon binding of CAG ligand in AtEPS1 (Supplementary Fig. 10), possibly indicating an induced fit mechanism in EPS1. Molecular dynamics (MD) simulations were next employed (Supplementary Table 4, and Supplementary Table 5) to investigate the apparent coordinated movement of these active-site loops with ligand binding. Clustering analysis of altogether 16-μs trajectories revealed markedly reduced flexibility of both loops upon EPS1’s transition from its apo to the holo state (Supplementary Fig. 11a). Between the two dynamic loops, the crossover loop appears to be more tightly locked upon such transitions, while the lid loop retains greater flexibility in the holo state (Supplementary Fig. 11b). These changes in loop dynamics may contribute to selective substrate binding of EPS1.
Structural basis for substrate selectivity and catalytic mechanism of AtEPS1
Guided by the CAG positioning in the AtEPS1 active site, we docked the native substrate IGA into the active site, refined the five highest-affinity poses with simulated annealing, and corroborated the refined binding pose of IGA using microsecond-long MD simulations (Supplementary Fig. 12 and Supplementary Fig. 13). Simulated CAG and IGA binding to AtEPS1 both conform to a nearly identical binding position compared to the substrate analog in the CAG-bound AtEPS1 crystal structure, where their differences derive from the non-planar isochorismate ring and the presence of an additional alkene double bond in the native substrate IGA (Fig. 3a, Supplementary Fig. 14, and Supplementary Fig. 15). The refined binding pose of the simulated IGA is well retained between plain MD as well as simulated annealing calculations (Supplementary Fig. 14). In contrast, simulations of chorismoyl-glutamate A (CGA), an isomer of IGA, illustrate a different orientation of the ring from the native substrate, corroborating previous findings that CGA serves as a poor substrate for EPS1 (Supplementary Fig. 15)14.
a Binding pose of IGA (yellow) in the EPS1 active site revealed by docking, simulated annealing and MD simulations. The pose of CAG (green) is superposed. b Structural difference of IGA in water (dashed line) and when bound to EPS1 (solid line) measured by three representative distances: d1:H2-C9 (blue), d2: ether O-amide H (orange), and d3: H2-N (green). c IPGL process of IGA in EPS1. Rearrangement results in intramolecular strain release and separation of products through a 57° flipping of SA adjusted by two residues (Arg395 and Arg44) and anchoring of NPG with Thr306, leading to an irreversible IPL process. d Proposed pericyclic reaction where the hydrogen atom at C2 is transferred to C9 of the side chain simultaneously with C-O cleavage. e The structure of the geometry optimized QM cluster model starting from the MD snapshots. The secondary structure is depicted with an orange cartoon representation. Carbons of key amino acids are shown in gray, and the substrate carbons are shown in light blue. All other atoms are colored as follows: nitrogen in blue, oxygen in red, and hydrogen in white. f Relative ΔG energy profiles in kcal/mol of the enzymatic reaction (black) and non-enzymatic reaction (red) calculated at the B3LYP/def2-TZVP level of theory.
Interestingly, comparison of simulated IGA in the AtEPS1 holo complex and in water reveals that the molecules adopt similar conformations in both with only the d2 distance between the ether oxygen and the amide hydrogen showing a recognizable decrease in interatomic distance in the enzyme context (Fig. 3b). The decrease in the d2 distance could support a previously proposed IGA nonenzymatic E1 elimination mechanism12, however, the long d3 distance ( > 4 Å) would preclude the required deprotonation of the ring hydrogen by the amide to complete the proposed C-O bond cleavage. To explore the catalytic mechanism of IPGL, we next simulated AtEPS1 bound to its products SA and NPG. These simulations indicate that EPS1-mediated IGA decomposition to form SA and NPG results in a 1.4 Å translation and 57° rotation of the SA ring in addition to a 2.8 Å shift of the oxygen atom in the pyruvoyl moiety of NPG compared to the modeled IGA substrate (Fig. 3c). These apparent changes in the products’ positions and orientations relative to the substrate suggest that the enzyme likely imposes an unfavorable constraint on the native substrate IGA. Here, aromatization alters the pseudodiaxial conformation, elongating the C3-O distance, placing strain on the IGA substrate to break the C-O bond, and steering the separation of the products through a pericyclic mechanism (Fig. 3c, d)30.
To investigate the energetics for the proposed mechanism of AtEPS1, we constructed large (415 atoms) and medium (207 atoms) sized quantum mechanical (QM) cluster models, starting from the minimized centroid of the MD simulations (Supplementary Fig. 16 and Supplementary Data 1)31. We first performed density functional theory (DFT) relaxed potential energy scans at the B3LYP/6-31G* level of theory for the large QM cluster model and identified a putative transition state (TS) with a ΔE‡ of 13.9 kcal/mol (Fig. 3e and Supplementary Fig. 17a)32,33. To further investigate the TS, we ran a nudged elastic band (NEB) calculation for the medium-size QM cluster model and confirmed a TS with a ΔE‡ of 13.8 kcal/mol and a single imaginary frequency (i326.6 cm−1) using partitioned rational-function optimization (P-RFO) (Supplementary Fig. 17b, c). The bond lengths of the confirmed TS were then applied to the large QM cluster model and geometry optimized, holding the TS bond lengths fixed at the same level of theory (Supplementary Fig. 18). Additional free energy calculations were performed on the reactant, product, and confirmed transition state with B3LYP/6-31G* with single points performed with a larger def2-TZVP basis set (Supplementary Table 6, and Supplementary Table 7)34,35. The resulting free energies reveal that the proposed mechanism is thermodynamically favored, with a ΔG of −56.5 kcal/mol for the products relative to the reactants (Fig. 3f, Supplementary Fig. 16, and Supplementary Fig. 17). The identified putative TS for the large QM cluster model had a moderate free energy barrier, ΔG‡, of 9.5 kcal/mol relative to the reactant. However, other steps in the reaction such as conformational changes or binding may influence the observed reaction rate.
To better understand the catalytic role of the enzyme active site, we next investigated the non-enzymatic reaction. We first identified the lowest energy conformation of IGA using CREST as a conformer search tool. We then performed NEB calculations of the proposed reaction, without AtEPS1, in implicit solvent with a dielectric constant of 80 chosen to mimic an aqueous environment (Supplementary Fig. 19 and Supplementary Table 8). The maximum energy structure from the NEB calculation was then optimized with a P-RFO calculation and a single imaginary frequency (i301.7 cm−1) was identified (Supplementary Fig. 20). Free energy calculations were then performed on all intermediates and transition states at the same level of theory and single points were performed with a larger def2-TZVP basis set revealing a free energy barrier, ΔG‡, of 18.0 kcal/mol. We observed that without the stabilizing effect of the binding pocket, the free energy barrier was 8.5 kcal/mol higher (Fig. 3f and Supplementary Table 8). This observation supports AtEPS1’s role catalyzing the proposed mechanism by potentially stabilizing a more favorable conformation.
Structure-function roles of AtEPS1 active-site residues
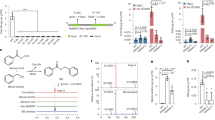
Previously, we characterized the biochemical function of EPS1 in vitro using a pre-assay that enzymatically synthesizes the EPS1 substrate IGA from chorismate using recombinant SID2 and PBS314. However, the unstable nature of several intermediates, including chorismate, isochorismate, and IGA, complicate the utility of this setup to quantitatively assess the relative activities of site-directed EPS1 mutants. Thus, we sought to reconstitute the isochorismate-derived SA biosynthetic pathway in transgenic Saccharomyces cerevisiae, which would allow us to measure the relative activities of EPS1 mutants in vivo. When experimenting with various combinations of A. thaliana genes that enable SA production in S. cerevisiae, we discovered that transgenic co-expression of SID2 and PBS3 alone is sufficient to elicit SA production in yeast, while the addition of AtEPS1 further led to seven- and six-fold increase in SA and NPG production, respectively, with a concomitant depletion of IGA levels by more than ten fold (Fig. 4a–c). Although the isochorismate-derived SA biosynthetic pathway is naturally compartmentalized in plastid and cytoplasm in plants, it can be reconstituted in the yeast cytosol without the need for the MATE transporter, SID1.
Error bars indicate standard error of the mean (SEM) based on biological triplicates. Statistical analysis was conducted by two-tailed unpaired t-test. *P < 0.05, **P < 0.01, and ***P < 0.001. a IGA. b SA. c NPG. d Relative depletion of IGA in AtEPS1 alanine mutants alongside rescuing mimetic mutations (blue bars) in the heterologous yeast system which additionally express SID2 and PBS3.
We utilized our newly developed heterologous yeast screening system to investigate the effects of various active-site residues of AtEPS1 using site-directed mutagenesis. Subsequently, we measured in vivo IPGL activity by assessing the relative depletion of IGA, acknowledging the limitation that mutational outcomes in yeast may not perfectly mirror their effects in plants. Creating a panel of AtEPS1 mutants with single alanine substitutions in 19 active-site residues, we introduced them into an S. cerevisiae background strain co-expressing SID2 and PBS3 where the reduction in IGA levels relative to wild-type AtEPS1 served as a measure of the mutants’ IPGL activities (Fig. 4a–c). Notably, mutants Arg44Ala, Asp164Ala, Thr306Ala, and Arg395Ala exhibited significant increases in IGA levels, indicating inactive or improperly folded EPS1 variants, whereas mimetic mutations restored IPGL activities comparable to wild type. Substituting alanine, but not lysine, for Arg44 or Arg395 significantly reduces IPGL activity, supporting their roles in coordinating the aryl carboxylate group of IGA via salt bridges and facilitating the leaving of SA product during pericyclic reactions. The Thr306Ala mutation renders AtEPS1 inactive in the yeast system (Fig. 4d), while the Thr306Cys mutant maintains wild-type activity. The Asp164Ala mutant exhibits minimal activity, but is restorable by an asparagine substitution. Since substituting polar residues with alanine can induce protein instability and misfolding, we conducted a western blot analysis to assess the expression and solubility of our alanine mutants in the yeast system. All four key-residue mutants were extracted from yeast, with the Arg44Ala and Arg395Ala mutants being soluble. In contrast, the Asp164Ala and Thr306Ala mutants were insoluble, suggesting a destabilizing effect on EPS1 folding when expressed in yeast (Supplementary Fig. 21). It should be noted that while we confirm soluble expression of the Arg395Ala mutant by western blot, the band intensity is lower than WT and Arg44Ala. Consequently, due to the nature of our experimental design, we cannot determine whether the lower IPGL activity of Arg395Ala is due to a reduced protein amount, decreased activity, or effects on protein stability when expressed in the yeast system. Sequence conservation analysis of the active-site residues among EPS1-clade enzymes and EPS1-sister-clade enzymes demonstrates that while Arg44 and Asp164 are required for IPGL activity, these residues are also conserved in the EPS1 sister clade and therefore may not indicate adaption specific for EPS1 neofunctionalization (Supplementary Fig. 22). Thr306 and Arg395, however, are conserved among the EPS1-clade sequences, but are substituted to other amino acids in the EPS1 sister clade, suggesting these two essential residues are selected to support IPGL activity among EPS1 orthologs.
To further investigate the role of the aforementioned residues, we conducted additional MD simulations on the site-directed mutants in the holo state with IGA or IPGL products (i.e., SA and NPG). Given that our in silico mutants are created using the WT protein as a template, these microsecond simulations primarily sample the local, rather than global, structural impact of the mutation. By comparing them with the WT simulations, we can probe how the in situ replacement of a given residue by alanine may impact ligand coordination rather than the folding and stability of the entire protein. Although Asp164Ala and Thr306Ala are not soluble, we propose that simulating them could still provide valuable insights into how other enzymes drive pericyclic reactions and could offer further mechanistic insights into EPS1.
As shown in Supplementary Fig. 23, the ligand RMSD reveals that IGA and/or IPGL products either precariously bind to or do not bind at all to alanine-substituted mutants. Due to the missing coordination of its aryl carboxylate, Arg44Ala and Arg395Ala mutants fail to retain the active position of IGA in the binding pocket (Supplementary Fig. 24). Such missing coordination impacts the binding of IPGL products even more severely, with SA detached from the active pocket almost from the beginning of Arg44Ala and Arg395Ala simulations (Supplementary Fig. 23). For the Asp164Ala mutant, simulations capture the local disruption of protein structure, which primarily arises from an unsettled small loop (Asn286-Val303) due to the mutation (Supplementary Fig. 25a). The alanine substitution of Asp164 cuts off its original binding to Arg288 and frees this small loop, resulting in its significantly elevated B factor as well as altered ligand coordination (Supplementary Fig. 25).
Interestingly, although Thr306Ala significantly reduces the solubility and IPGL activity of the EPS1 enzyme, the binding between Thr306 and IGA as revealed by the WT simulations (Supplementary Fig. 26) is dynamic, hinting at a more complex and subtle function of Thr306 than tight coordination of the substrate as offered by Arg44 and Arg395. We notice that a nearby Arg282 transiently flips towards the substrate during simulations, where its guanidino group interacts with the α-carboxylate of IGA, thereby distorting the original, active pose of the substrate (Fig. 5a). Upon such transient distortion, the occasional hydrogen bond offered by Thr306 stops IGA from further displacement and severs its interaction with Arg282, restoring the substrate to its active pose (Fig. 5a and Supplementary Movie 1). A similar guarding effect is provided by the hydrophilic Cys306, with its increased direct contact to Arg282 further hindering the latter’s binding to IGA (Supplementary Fig. 27 and Supplementary Movie 2). The distorted, inactive pose of IGA is retained when the guarding residue at position 306 is substituted with an alanine (Supplementary Movie 3). This distinction is also captured by the simulations with IPGL products, where the original binding pose of NPG is distorted by Arg282 in the Thr306Ala but not the WT and Thr306Cys simulations (Supplementary Fig. 27).
a Transiently altered binding pose of IGA due to flipped Arg282 only restored in WT and T306C. Populated structures of IGA and Arg282 in the WT simulations are displayed transparently. The red and green arrows indicate the variation upon Arg282 flipping and after restoration, respectively. Hydrogen bonds are illustrated by the yellow dashed lines. b Probability distribution of the minimum distance between the α-carboxylate of IGA and the guanidine group of Arg282 (d282). c Probability distribution of the distance between the C4 atom of IGA from a given simulation and the corresponding C4’ atom of product SA in the top cluster centroid structure from the WT simulations (ΔdC4).
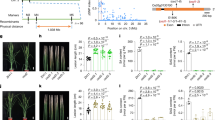
Transgenic expression of AtEPS1 in soybean leads to SA overproduction
In Arabidopsis, the primary flux of pathogen-induced SA is derived from isochorismate, with a minor contribution coming from the phenylalanine pathway4. Conversely, both of these pathways have been shown to be equally important to SA accumulation in soybean, as suppression of either pathway results in reduced SA levels36. Previous work has shown that overexpression of isochorismate synthase and IPL genes from bacteria in Nicotiana plants resulted in increased SA levels and triggered systemic acquired resistance37. We generated stable transgenic soybean plants overexpressing AtEPS1 to examine if SA levels were affected. Eighteen independent transgenic lines were generated and grown under greenhouse conditions for phenotypic characterization. Among these, we observed varying levels of severity in the stunted growth phenotype, accompanied by leaf yellowing, which is known to be associated with SA overaccumulation14. Compared to the wild type, these growth phenotypes were categorized as wild-type-like (1), moderate (2), and severe (3) (Fig. 6a). Metabolic profiling of leaf extracts from three representative lines within these categories of transgenic plants revealed that SA levels in the wild-type-like and moderate phenotypes fell within the error range of the wild type. In contrast, the severe phenotype exhibited nearly a five-fold increase in SA levels (Fig. 6b). To determine if the severity of the stunted growth phenotype and SA levels correlated with the levels of AtEPS1 transgenic expression, we employed quantitative reverse transcription-polymerase chain reaction (qRT-PCR). As expected, no AtEPS1 transcript was detected in the wild-type soybean plants. Plants exhibiting wild-type-like and moderate phenotypes showed moderate expression levels, while the plant with the severe phenotype exhibited significantly higher transgene expression. Our data suggest that ectopic overexpression of AtEPS1 in soybean plants, which lack an EPS1 ortholog, leads to SA overaccumulation and a stunted growth phenotype. This indicates the functional and evolutionary significance of EPS1 in enhancing SA production efficiency in planta.
a Four-week-old, greenhouse-grown wild-type soybean plant and three representative independent lines of transgenic soybean overexpressing AtEPS1. Phenotypes were categorized as wild-type-like (1), moderate (2), and severe (3). b SA levels in the individual plants as pictured in a. Results are presented as mean peak area values, based on three biological replicates obtained from distinct leaf samples of the same plant, with SEM indicated as error bars. Statistical analysis was conducted using a two-tailed, unpaired t-test. ***P < 0.001. c Relative expression levels of the AtEPS1 transgene in the individual plants as pictured in a. The relative expression levels were quantified using the relative quantification (ΔΔCt) method based on three technical replicates.
The ability to mount a vigorous defense against invading pathogens at local tissue while sending warning signals to the unaffected distal tissue is critical for plants to survive the challenging biotic environments they live in. Although the defense hormone SA could be produced through spontaneous decay of IGA in plants that contain PBS3, natural selection has propelled the common ancestors of the Brassicaceae plants to recruit a gene-duplication-derived BAHD progenitor enzyme, refurbish its active site, and ultimately gain specific IPGL activity that accelerates this last step of SA biosynthesis. In this case of neofunctionalized catalytic machinery, EPS1 developed a highly coordinated substrate-binding site that forces an energetically unfavorable conformation for its substrate IGA, thereby facilitating the breakdown of IGA to yield SA and NPG through a pericyclic reaction (Fig. 3d). While pericyclic reactions are regularly used in organic chemistry, enzymes known to catalyze pericyclic reactions have only been sparsely characterized in cellular metabolism, including IPL and chorismate mutase38,39. Both enzymes employ a pericyclic dissociative mechanism to cleave a C−O bond, which points to a broader mechanism of pericyclic elimination from shikimate-derived metabolites. Interestingly, despite IPL and EPS1 employing similar mechanisms, their overall folds are entirely different (Supplementary Fig. 28). Our observation that SA production can be enhanced in soybean plants through transgenic overexpression of AtEPS1 illuminates the evolutionary advantage that could be gained from the neofunctionalization of a BAHD-type enzyme to acquire new IPGL activity within the Brassicaceae family. Furthermore, our findings not only underscore the remarkable adaptability and specificity of plant metabolic pathways but also open avenues for using EPS1 as a novel plant trait to improve pathogen resistance in non-Brassicaceae plants.
Methods
Sequence alignment, phylogenetic analysis, and homology modeling
Sequence alignments were built using ClustalW2 with default settings40. Maximum likelihood tree shown in Supplementary Fig. 1b and Supplementary Fig. 1 were determined using MEGAX41. The bootstrap consensus trees were inferred from 500 replicates to represent the local BAHD phylogeny of AtEPS1 which encompassed homologous sequences from the phytozome SMB clade with an e-value threshold of 1e-40. The local EPS1 tree was then generated as described above from only the EPS1 clade sequences and a probable sister and ancestor clade sequences. Conservation of the active-site residues between AtEPS1 and probable sister clade was displayed using WebLogo42.
Molecular cloning, heterologous expression, and recombinant protein production
Total RNA from A. thaliana was extracted from six-week-old plants grown in long-day greenhouse conditions using the RNeasy Mini Kit (Qiagen). First-strand cDNAs were synthesized using the Invitrogen SuperScript III kit (Invitrogen) with the oligo(dT)20 primer. The coding sequences (CDS) of candidate genes were amplified from cDNAs by PCR using gene-specific primers. Point mutations were produced with Gibson assembly mutagenesis using the initial cloning primers and the mutation primers. The open reading frame (ORF) of target SA biosynthetic genes were cloned into p423TEF, p425TEF, and p426TEF 2μ plasmids for constitutive expression in S. cerevisiae or into pHis8-4, a bacterial expression vector containing an N-terminal 8xHis tag followed by a tobacco etch virus (TEV) cleavage site for recombinant protein production in E. coli. Note, recombinant AtEPS1 was produced and purified in a previous study14. 15 mL cultures of transgenic S. cerevisiae BY4743 strains were grown in 50 mL mini bioreactor tubes for 24 h with shaking at 30 °C. The cultured cells were subsequently pelleted at 2500 x g, washed, disrupted, and clarified for LC-HRAM-MS metabolic profiling43. The raw data were processed using MZmine 244 and further analyzed using metaboanalyst45. Statistical analysis was conducted using Prism 8 (GraphPad Prism version 8.0.0 for Mac, GraphPad Software, San Diego, California USA, www.graphpad.com).
Metabolomic profiling by LC-HRAM-MS
Metabolite profiling was performed using a Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled with a Dionex UltiMate 3000 UPLC system. Samples (2 µL) were injected onto a C18 column (Kinetex 2.6 μm, 100 Å, 150 × 3 mm) with a gradient of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) at a flow rate of 0.8 mL/min. The gradient was 5% B for 2 min, 5%–80% B over 40 min, 95% B for 4 min, and 5% B for 5 min. The mass spectrometer operated in polarity-switching mode (70–1000 m/z), with key settings including a spray voltage of 3.0 kV, capillary temperature of 275 °C, and HESI probe temperature of 350 °C. Data were acquired at 70,000 resolution with a maximum injection time of 20 ms.
Chemical synthesis general methods
All reactions were performed under nitrogen unless otherwise noted. All reagents and solvents were used as supplied without further purification unless otherwise noted. Column chromatography was conducted using Silicycle SiliaFlash P60 SiO2 (40–63 μm). Analytical TLC was conducted using Millipore SiO2 60 F254 TLC (0.250 mm) plates. HPLC was conducted using a pair of Shimadzu LC-20AP pumps, a Shimadzu CBM-20A communications module, a Shimadzu SPD-20A UV-Vis detector, and a Phenomenex Kinetex 5 u C18 100 Å Axia column (150 ×21.2 mm). Melting points were obtained using a Mel-Temp II apparatus and are uncorrected. 1H and 13C NMR spectra were obtained using a Bruker Avance Neo 400 MHz spectrometer equipped with a 5 mm Bruker SmartProbe. IR spectra were obtained using a Bruker Alpha 2 with a Platinum ATR accessory. Mass spectrometric analysis was performed on a JEOL AccuTOF-DART. Syntheses were inspired by previously described approaches towards polymer‐supported IBX46.
Protein crystallization and structural determination
Crystals for AtEPS1 and (2-(3-carboxyphenoxy)acetyl)-L-glutamic acid bound AtEPS1 were grown at 4 °C by hanging-drop vapor diffusion method with the drop containing 0.9 µL of protein sample and 0.9 µL of reservoir solution at a reservoir solution volume of 500 µL. The crystallization buffer for the both AtEPS1 structures were composed of 0.2 M Potassium Sodium Tartrate 20 % (w/v) PEG 3350. The ligand-bound AtEPS1 drop also contained 500 µM (2-(3-carboxyphenoxy)acetyl)-L-glutamic acid. Crystals were cryogenized with an additional 15% weight/volume ethylene glycol. The structures were determined first by molecular replacement using the native HCT structure from Coffea canephora (PDB:4G0B)25 as the search model in Molrep47. The resulting model was iteratively refined using Refmac 5.248 and then manually refined in Coot 0.7.149.
Transgenic soybean
AtEPS1 under the control of the CaMV 35S promoter was introduced into soybean plants as a transgene using Agrobacterium-mediated transformation. The transformation was performed by the Wisconsin Crop Innovation Center following the protocol described in U.S. patent US 11,266,086 B2. 18 Independent T1 transformants were obtained. Transgenic plants were grown at 22 °C in a greenhouse under long-day conditions (16-h light/8-h dark) before metabolic profiling and qRT-PCR analysis.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from soybean leaves using RNeasy Plant Mini Kit (Qiagen), per the manufacturer’s instructions. Reverse transcription (RT) and first-strand cDNA synthesis were carried out using the SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative real-time PCR was performed using PowerUp SYBR Green Master Mix (Applied Biosystems) on a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) with the default cycling program50. Primers were designed to amplify gene-specific PCR products of <200 bp in size. GmACT11 was used as an internal control to normalize the cDNA. The primers used to amplify AtEPS1 were 5′-CTCAAGGACATCACTGAGCTC-3′ and 5′-CCTTTAGCTGGTCGTCTTGTG-3′. The primers used to amplify GmACT11 were 5′-ATCTTGACTGAGCGTGGTTATTCC-3′ and 5′-GCTGGTCCTGGCTGTCTCC-3′. Gene expression was quantified using the relative quantification (ΔΔCt) method based on three technical replicates.
Immunoblot
100 mg of wet cell pellets from yeast harvested at mid-log phase were resuspended in 500 µL lysis buffer (50 mM sodium phosphate pH 6.2, 500 mM NaCl, 0.01% Triton X-100) supplemented with protease inhibitors (Roche, Catalog # 11836170001), and lysed with 0.5 mm Zirconia/Silica beads (BioSpec) for 30 min using a TissueLyser II (Qiagen) at a frequency of 30 per second. Following lysis, samples were centrifuged at 21,000 x g to separate the soluble lysate from the insoluble pellet fraction. Soluble and insoluble fraction samples were mixed with Laemmli buffer and proteins from each sample were separated by SDS-PAGE and immunoblotted with Anti-AtEPS1 polyclonal antibody (custom-produced by Genscript). The anti-AtEPS1 polyclonal antibody was 1:1000 diluted in Tris-buffered saline with 0.1% Tween® 20 detergent (TBST) buffer containing 5% milk. The secondary antibody used was Goat anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, horseradish peroxidase (HRP) (Thermo Fisher Scientific, Catalog # G-21040, RRID AB_2536527) used at 1:10000 dilution in TBST containing 5% milk.
MD simulations
Molecular dynamics simulations of apo- and CAG-bound AtEPS1 were constructed using the corresponding crystal structures. For holo-AtEPS1 in complex with the native substrate IGA, molecular docking using Autodock vina 1.1.251 was employed to generate initial, candidate binding poses of IGA, which were then refined by simulated annealing using GROMACS 2020.252. Specifically, five docking poses with the highest affinity produced by Autodock vina were solvated and ionized with 0.05 M NaCl and then adopted as the starting structures for simulated annealing (Supplementary Fig. 12). Following a 1000-step energy minimization and a 1-ns NVT equilibration at 300 K, the temperature was increased to 500 K at a speed of 0.1 K/ps; after a 50-ns NPT equilibration, the temperature was reduced to 400 K at a speed of 0.05 K/ps, followed by a 5-ns NPT equilibration. The temperature was then brought to 300 K at the same speed, followed by another 5-ns NPT equilibration, and, finally, a 1000-step energy minimization. During the above simulations, protein backbones of α-helices and β-sheets were restrained with the exception of residues within 3 Å of IGA, the latter of which, along with the substrate, were allowed to move freely. The simulated annealing simulations were conducted in six replicas per docking pose, producing altogether 30 refined structures. The centroid structure of the top cluster obtained from clustering analysis over these 30 refined structures (Supplementary Fig. 12) was taken as the input structure for the subsequent, altogether 8-μs MD simulations of holo-AtEPS1 in complex with IGA. Clustering analysis of these simulation trajectories yields the binding pose of IGA shown in Fig. 3a, which is similar to the aforementioned centroid structure revealed by simulated annealing calculations (Supplementary Fig. 13). Simulations of AtEPS1 in complex with the yielded products (SA and NPG) and CGA were initialized by superimposing these ligands to the aforementioned refined structure of AtEPS1 in complex with IGA. Single mutations of AtEPS1, including R44A, R395A, D164A, T306A and T306C, were simulated in complex with IGA or IPGL products based on our in-silico models. All the structures of AtEPS1 mutants were initially prepared using Modeller 10.053, where the wild-type models of IGA-bound or IPGL-product-superimposed AtEPS1 were employed as templates.
After initialization, 2-μs MD simulations in four replicas were performed using GROMACS 2020.252 for each system (Supplementary Table 4). These systems were placed in a dodecahedron box with a margin of at least 12 Å from any protein atoms, solvated by explicit water molecules described by the TIP3P model54, and then neutralized by 0.05 M NaCl. The initial atomic velocity was assigned according to the Maxwell-Boltzmann distribution at 300 K. All systems were subjected to energy minimization, followed by a 1-ns NVT equilibration and a 1-ns NPT equilibration, with heavy atoms of the protein backbone positionally restrained. The resulting structures then were used to launch the microsecond production runs listed in Supplementary Table 4.
All simulations were performed using the CHARMM36 force field55 with the ligands parameterized using CHARMM General Force Field (CGenFF)56. Initial parameters of the ligands were obtained from the CGenFF interface57,58 and then optimized using force field toolkit (FFTK)59 in VMD 1.9.460 as well as Gaussian0961. In all simulations, van der Waals interactions were smoothly switched off from 8 Å to 9 Å, while electrostatic interactions were calculated using the particle mesh Ewald (PME) method62 with a cutoff of 9 Å. All systems were regulated with velocity-rescaling temperature coupling (300 K)63 and Berendsen’s pressure coupling (1 bar)64. All bonds with H atoms were constrained using the LINCS algorithm65. Clustering and other analyses were performed on all four replica trajectories of a given system. The representative binding pose in a given set of MD simulations is taken to be the centroid structure of the top cluster derived from clustering analysis of the trajectories using the GROMOS method66. The hydrogen bonds are defined and measured with a cutoff heavy-atom distance of 3.6 Å and a cutoff angle of 63°. PyMOL 2.4.067 was used for visualization while atomic distances were measured by VMD 1.9.460. Final parameters of ligands along with their corresponding structures as well as the input and output files from the MD simulations of each system are available at GitHub repository: https://github.com/TianjieLi-Jason/EPS1.git.
QM cluster model preparation
A large QM cluster model was constructed from the minimized centroid of the IGA-bound MD simulations31. The QM cluster model included all amino acids in contact with the substrate and adjacent residues to preserve the active site’s structure. Backbone atoms were fixed to maintain the conformation of the active site. The QM cluster model included the following residues: R36, L37, R38, F39, G40, Y41, L42, N43, R44, Y158, N159, S160, N302, V303, S304, Q305, T306, V363, V364, T365, P390, Y391, L392, N393, G394, and R395. We aimed to include continuous chains of amino acids, where residues with side chains oriented away from the substrate were substituted with alanine to minimize the size of the cluster model without introducing covalent cuts. The residues that were substituted for alanine include: L37, R38, Y41, N43, N159, V303, V364, Y391, and N393. The final QM cluster model contained 415 atoms and maintained charge neutrality. The medium sized QM cluster model included the following residues, which were selected to maintain the same overall charge and the residues most likely to impact substrate positioning: R36, R44, Q302, Q305, V363, T365, L392, and R395. The coordinates of the QM cluster model are included in the Source Data zip file.
QM calculations
Nudged elastic band (NEB) calculations and geometry optimizations were calculated with ORCA version 5.0.3. The constrained geometry scan for the large QM cluster model was calculated with developer version 1.9 of the GPU-accelerated quantum chemistry package TeraChem68,69,70. The geometry optimizations were initially performed at the B3LYP/6-31 G* level of theory to balance cost accuracy trade-offs32. Reactants, products, and transition state (TS) energies were also computed with B3LYP using the larger def2-TZVP basis71 in ORCA, with no qualitative effect on the results (Supplementary Tables 6–8). Semi-empirical DFT-D3 with default Becke-Johnson damping was also applied to account for long-range interactions within the QM cluster model69. The large QM cluster model calculations were carried out with an implicit conductor-like polarizable continuum model (C-PCM) to approximate the protein’s internal dielectric72,73, whereas the QM calculations of the substrate in solution used to approximate the dielectric of water34. To conserve the active site structure, backbone atoms were frozen, and all other atoms were allowed to move freely. For the QM calculations of the substrate in solution, all atoms were allowed to move freely. For the large QM cluster model, an initial putative transition state was identified using the TeraChem TRIC implementation of the constrained geometry scan74. The distance between C9 and the C2 hydrogen was used as a reaction coordinate and was fixed incrementally while optimizing all other degrees of freedom. ΔG energies were computed using frequency calculations to obtain zero-point vibrational energy corrections at the B3LYP/6-31 G* level of theory. The computed zero-point vibrational energy corrections were also applied to the B3LYP/def2-TZVP calculations (Supplementary Tables 6–8). The coordinates of the scans are included in the Source Data zip file.
Transition state calculations
While the large QM cluster model of the AtEPS1 active site is prohibitively large to confirm the TS (415 atoms), we confirmed the TS with a single imaginary frequency for the medium-size QM cluster model and for the non-enzymatic reaction in solution (Supplementary Fig. 17, and Supplementary Fig. 20). For TS searching, we employed ORCA 5.0.3 as TeraChem version 1.9 does not support analytical Hessians. Optimization of the TS was performed by first running an NEB calculation at the B3LYP/6-31 G* level of theory. The maximum from the NEB calculation was then used as an initial guess for a partitioned rational-function optimization (P-RFO), and a frequency calculation was performed for the reactants, products, and TS to ensure that an imaginary frequency corresponding to the TS was observed (Supplementary Fig. 17) To approximate the TS of the large AtEPS1 cluster model more accurately, the geometry of the optimized TS from medium-size QM cluster model was used to model the putative TS of the large AtEPS1 cluster model by restraining the following distances and performing a constrained geometry optimization: C2···H = 1.17 Å, C9···H = 1.85 Å, C9···C8 = 1.37 Å, C8···O = 1.30 Å, C3···O = 2.00 Å, and C2···C3 = 1.48 Å (Supplementary Fig. 17, and Supplementary Fig. 18). Single points were run for all intermediates and transition states using the larger def2-TZVP basis in ORVA with a free energy correction calculated at the B3LYP/6-31 G* level of theory.
Volume calculations
Volume calculation was carried out using KVFinder75 by detecting cavities around active-site (residues 36, 39, 42, 44, 160, 282, 284, 306, 257, 363, 365, 390, and 395 in Chain A) with a “probe in” size of 1.4 Å, a “probe out” size of 5.0 Å, padding size of 3.5 Å, removal distance of 2.4 Å, and volume cutoff of 150 Å3. 6WAO (CAG-bound) results in volume of 1537.49 Å3, whereas 6WCS (apo) results in volume of 2136.67 Å3, which indicates 28.04% of active-site volume constriction.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All atomic coordinates and structure factors generated in this study have been deposited in the Protein Data Bank database under accession codes 6WCS (AtEPS1 Apo), and 6WAO (AtEPS1+CAG). The final parameters of ligands along with their corresponding structures as well as the input and output files from the MD simulations of each system presented in this work are available at GitHub repository: https://github.com/TianjieLi-Jason/EPS1.git. Atomic coordinate files used in computational analyses are included in Supplementary Data 1. Source data are provided with this paper.
References
Ding, P. & Ding, Y. Stories of salicylic acid: a plant defense hormone. Trends Plant Sci. 25, 549–565 (2020).
Raskin, I. Role of salicylic acid in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 439–463 (1992).
León, J., Shulaev, V., Yalpani, N., Lawton, M. A. & Raskin, I. Benzoic acid 2-hydroxylase, a soluble oxygenase from tobacco, catalyzes salicylic acid biosynthesis. Proc. Natl Acad. Sci. USA 92, 10413–10417 (1995).
Wildermuth, M. C., Dewdney, J., Wu, G. & Ausubel, F. M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565 (2001).
Silverman, P. et al. Salicylic acid in rice (biosynthesis, conjugation, and possible role). Plant Physiol. 108, 633–639 (1995).
Chadha, K. C. & Brown, S. A. Biosynthesis of phenolic acids in tomato plants infected with Agrobacterium tumefaciens. Can. J. Bot. 52, 2041–2047 (1974).
Yalpani, N., Leon, J., Lawton, M. A. & Raskin, I. Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiol. 103, 315–321 (1993).
Meuwly, P., Molders, W., Buchala, A. & Metraux, J. P. Local and systemic biosynthesis of salicylic acid in infected cucumber plants. Plant Physiology 109, 1107–1114 (1995).
Strawn, M. A. et al. Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 282, 5919–5933 (2007).
Rogers, E. E. & Ausubel, F. M. Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9, 305–316 (1997).
Nawrath, C., Heck, S., Parinthawong, N. & Métraux, J.-P. EDS5, an essential component of salicylic acid–dependent signaling for disease resistance in arabidopsis, is a member of the MATE transporter family. Plant Cell 14, 275–286 (2002).
Rekhter, D. et al. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 365, 498–502 (2019).
Mercado-Blanco, J. et al. Analysis of the pmsCEAB gene cluster involved in biosynthesis of salicylic acid and the siderophore pseudomonine in the biocontrol strain Pseudomonas fluorescensWCS374. J. Bacteriol. 183, 1909–1920 (2001).
Torrens-Spence, M. P. et al. PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. Mol. Plant 12, 1577–1586 (2019).
Zheng, Z., Qualley, A., Fan, B., Dudareva, N. & Chen, Z. An important role of a BAHD acyl transferase-like protein in plant innate immunity. Plant J. 57, 1040–1053 (2009).
D’Auria, J. C. Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 9, 331–340 (2006).
Weng, J.-K. & Noel, J. P. The remarkable pliability and promiscuity of specialized metabolism. Cold Spring Harb. Symp. Quant. Biol. 77, 309–320 (2012).
Levsh, O. et al. Dynamic conformational states dictate selectivity toward the native substrate in a substrate-permissive acyltransferase. Biochemistry 55, 6314–6326 (2016).
Kim, C. Y. et al. Emergence of a proton exchange-based isomerization and lactonization mechanism in the plant coumarin synthase COSY. Nat. Commun. 14, 597 (2023).
Tuominen, L. K., Johnson, V. E. & Tsai, C.-J. Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues. BMC Genomics 12, 236 (2011).
Levsh, O., Pluskal, T., Carballo, V., Mitchell, A. J. & Weng, J.-K. Independent evolution of rosmarinic acid biosynthesis in two sister families under the Lamiids clade of flowering plants. J. Biol. Chem. 294, 15193–15205 (2019).
Chiang, Y.-C., Levsh, O., Lam, C. K., Weng, J.-K. & Wang, Y. Structural and dynamic basis of substrate permissiveness in hydroxycinnamoyltransferase (HCT). PLoS Comput. Biol. 14, e1006511 (2018).
Eudes, A. et al. Exploiting the substrate promiscuity of hydroxycinnamoyl-coa: shikimate hydroxycinnamoyl transferase to reduce lignin. Plant Cell Physiol. 57, 568–579 (2016).
Walker, A. M. et al. Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme A-dependent transferases and synthases. Plant Physiol. 162, 640–651 (2013).
Lallemand, L. A. et al. A structural basis for the biosynthesis of the major chlorogenic acids found in coffee. Plant Physiol. 160, 249–260 (2012).
Manjasetty, B. A. et al. Structural basis for modification of flavonol and naphthol glucoconjugates by Nicotiana tabacum malonyltransferase (NtMaT1). Planta. 236, 781–93 (2012).
Unno, H. et al. Structural and mutational studies of anthocyanin malonyltransferases establish the features of BAHD enzyme catalysis. J. Biol. Chem. 282, 15812–15822 (2007).
Ma, X., Koepke, J., Panjikar, S., Fritzsch, G. & Stöckigt, J. Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J. Biol. Chem. 280, 13576–13583 (2005).
Wang, C. et al. Structural and biochemical insights into two BAHD acyltransferases (SHT and SDT) involved in phenolamide biosynthesis. Front. Plant Sci. 11, 610118 (2020).
Ohashi, M. et al. An enzymatic Alder-ene reaction. Nature 586, 64–69 (2020).
Himo, F. & de Visser, S. P. Status report on the quantum chemical cluster approach for modeling enzyme reactions. Commun. Chem. 5, 29 (2022).
Lee, C., Yang, W. & Parr, R. G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter 37, 785–789 (1988).
Stephens, P. J., Devlin, F. J., Chabalowski, C. F. & Frisch, M. J. Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem. 98, 11623–11627 (1994).
Kendall, R. A., Dunning, T. H. & Harrison, R. J. Electron affinities of the first-row atoms revisited. Systematic basis sets and wave functions. J. Chem. Phys. 96, 6796–6806 (1992).
Woon, D. E. & Dunning, T. H. Jr. Gaussian basis sets for use in correlated molecular calculations. III. The atoms aluminum through argon. J. Chem. Phys. 98, 1358–1371 (1993).
Shine, M. B. et al. Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 212, 627–636 (2016).
Verberne, M. C., Verpoorte, R., Bol, J. F., Mercado-Blanco, J. & Linthorst, H. J. Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nat. Biotechnol. 18, 779–783 (2000).
DeClue, M. S., Baldridge, K. K., Künzler, D. E., Kast, P. & Hilvert, D. Isochorismate pyruvate lyase: a pericyclic reaction mechanism? J. Am. Chem. Soc. 127, 15002–15003 (2005).
Lamb, A. L. Pericyclic reactions catalyzed by chorismate-utilizing enzymes. Biochemistry 50, 7476–7483 (2011).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Torrens-Spence, M. P., Pluskal, T., Li, F.-S., Carballo, V. & Weng, J.-K. Complete pathway elucidation and heterologous reconstitution of rhodiola salidroside biosynthesis. Mol. Plant 11, 205–217 (2018).
Pluskal, T., Castillo, S., Villar-Briones, A. & Oresic, M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 11, 395 (2010).
Xia, J. & Wishart, D. S. Using metaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinform. 55, 14.10.1–14.10.91 (2016).
Mülbaier, M. & Giannis, A. The synthesis and oxidative properties of polymer-supported IBX. Angew. Chem. Int. Ed. Engl. 40, 4393–4394 (2001).
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015).
Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 54, 5.6.1–5.6.37 (2016).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Vanommeslaeghe, K. et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010).
Vanommeslaeghe, K. & MacKerell, A. D. Jr. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 52, 3144–3154 (2012).
Vanommeslaeghe, K., Raman, E. P. & MacKerell, A. D. Jr. Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 52, 3155–3168 (2012).
Mayne, C. G., Saam, J., Schulten, K., Tajkhorshid, E. & Gumbart, J. C. Rapid parameterization of small molecules using the Force Field Toolkit. J. Comput. Chem. 34, 2757–2770 (2013).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graphics 14, 33–38 (1996).
Frisch, M. et al. Gaussian 09 Revision E.01. Gaussian, Inc. (2009).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: AnN⋅log(N) method for Ewald sums in large systems. The J. Chem. Phys. 98, 10089–10092 (1993).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Levy, R. S., Tendler, C., VanDevanter, N. & Cleary, P. D. A group intervention model for individuals testing positive for HIV antibody. Am. J. Orthopsychiatry 60, 452–459 (1990).
Hess, B., Bekker, H., Berendsen, H. J. C. & Johannes G. E. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Lindahl, Abraham, Hess & van der Spoel. GROMACS 2020 Manual. Downloadable at https://doi.org/10.5281/ZENODO.3562512 (2020).
Lilkova, E., Petkov, P., Ilieva, N. & Litov, L. The PyMOL molecular graphics system, version 2.0. Schrodinger, LLC (2015).
Seritan, S. et al. TeraChem: accelerating electronic structure and ab initio molecular dynamics with graphical processing units. J. Chem. Phys. 152, 224110 (2020).
Grimme, S., Antony, J., Ehrlich, S. & Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 132, 154104 (2010).
Kulik, H. J., Luehr, N., Ufimtsev, I. S. & Martinez, T. J. Ab initio quantum chemistry for protein structures. J. Phys. Chem. B 116, 12501–12509 (2012).
Weigend, F. & Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: design and assessment of accuracy. Phys. Chem. Chem. Phys. 7, 3297–3305 (2005).
Lange, A. W. & Herbert, J. M. A smooth, nonsingular, and faithful discretization scheme for polarizable continuum models: the switching/Gaussian approach. J. Chem. Phys. 133, 244111 (2010).
Darrin, M. & Karplus, M. A Smooth Solvation Potential Based on the Conductor-Like Screening Model. J. Phys. Chem. A 103, 11060–11079 (1999).
Wang, L.-P. & Song, C. Geometry optimization made simple with translation and rotation coordinates. J. Chem. Phys. 144, 214108 (2016).
Oliveira, S. H. P. et al. KVFinder: steered identification of protein cavities as a PyMOL plugin. BMC Bioinforma. 15, 197 (2014).
Acknowledgements
We thank the Metabolite Profiling Core Facilities and the Bioinformatics and Research Computing of the Whitehead Institute for help with the metabolomics experiments and data analysis. This work was supported by the Pew Scholar Program in the Biomedical Sciences (J.K.W.), the Searle Scholars Program (J.K.W.), Keck Foundation (J.K.W.), the National Science Foundation (CHE-1709616, J.K.W.), and the Hong Kong research grants council (project 14306819, Y.W.). We thank Pingtao Ding for constructive comments on this manuscript.
Author information
Authors and Affiliations
Contributions
M.P.T.S. and J.K.W. designed the research. M.P.T.S., C.M.G., J.O.M., and J.S. performed the cloning and metabolomics analyses. C.M.G. performed chemical synthesis. J.O.M. and J.S. characterized the transgenic soybean, and performed western blot analyses of AtEPS1 expressed in yeast. M.P.T.S. and J.K.W. generated and processed the crystallographic data. T.L., Z.W., and Y.W. conducted and analyzed the molecular dynamics simulations. D.W.K., C.Y.K., and H.J.K. conducted and analyzed the QM simulations. All authors are involved in the interpretation of the results and writing of the paper.
Corresponding author
Ethics declarations
Competing interests
J.K.W. is a member of the Scientific Advisory Board and a shareholder of DoubleRainbow Biosciences, Galixir, and Inari Agriculture, which develop biotechnologies related to natural products, drug discovery, and agriculture. All other authors have no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Torrens-Spence, M.P., Matos, J.O., Li, T. et al. Mechanistic basis for the emergence of EPS1 as a catalyst in salicylic acid biosynthesis of Brassicaceae. Nat Commun 15, 10356 (2024). https://doi.org/10.1038/s41467-024-54437-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54437-1