Abstract
Mitogen-activated protein kinase (MAPK) cascades are evolutionarily conserved in both plants and animals and play critical roles in activating innate immunity to defend against various pathogens. However, the role of MAPK cascades in positively regulating or enhancing viral infections in plants is unclear. In this study, we investigate the involvement of MAPK cascades in infection by the positive-strand RNA virus tomato chlorosis virus (ToCV). Our findings reveal that ToCV infection activates MAPK cascades, promoting virus spread within plants. Specifically, ToCV P7, a pathogenicity determinant protein, localizes to the plasma membrane and recruits NbMPK3/6 from the nucleus. Subsequently, P7 is directly phosphorylated on serine 59 by NbMPK3/6. Phosphorylated P7 interacts with NbREM1.1 and inhibits its ability to induce callose deposition at plasmodesmata. These results demonstrate that NbMPK3/6 directly phosphorylate ToCV P7, modulating antiviral defence mechanisms by downregulating callose deposition at plasmodesmata and thereby enhancing ToCV transmission in N. benthamiana. This study sheds light on the intricate arms race between host defence and viral counter-defence strategies.
Similar content being viewed by others
Introduction
Unlike animals, plants cannot escape the inevitable threat posed by diverse pathogens, including viruses, fungi and bacteria. In the process of coevolution with pathogens, plants have evolved sophisticated multilayered defences against pathogen infection, such as calcium flux, activation of several mitogen-activated protein kinases (MAPKs), production of reactive oxygen species (ROS), and induction of phytohormone biosynthesis1,2,3. A typical MAPK cascade is composed of at least one MAPK (MPK), one MAPK kinase (MAPKK, MKK, or MEK), and one MAPKK kinase (MAPKKK, MKKK, or MEKK)3. The MAPK pathway is divided into two branches according to the substrates combined with kinases in Arabidopsis: MEKK1-MKK1/MKK2-MPK4 and MEKKs-MKK4/MKK5-MPK3/MPK64,5. In Solanaceae plants, there are orthologues to the three tiers of kinases in Arabidopsis; for example, two MKKs, MEK1 and MEK2, and two MPKs, NTF6 and wounding-induced protein kinase (WIPK), are involved in stress signal transduction in tomato6. Both branches of the MAPK cascade transduce stress signals to downstream targets and induce defence mediated by the hypersensitive-like response (HR), reactive oxygen species (ROS), extensive transcriptional reprogramming and phytohormones7. The upstream effectors that activate MAPK cascades and downstream targets collectively constitute a sophisticated and intricate defence network in plants. Although the molecular basis of the MAPK cascade is relatively clear, the downstream targets of the MAPK cascade and its molecular mechanisms mediating plant defence are largely unknown.
Plant defence responses against bacterial and fungal pathogens involve activation of the MAPK cascade through various mechanisms. Extensive research has focused on upregulation of defence responses mediated by the MEKKs-MKK4/MKK5-MPK3/MPK6 branch of the MAPK cascade2,8. For instance, the flagellin peptide flg22 triggers phosphorylation of the SUPPRESSOR OF NPR1-1 CONSTITUTIVE 1 (SNC1) protein by activating MPK3/MPK6, leading to enhanced salicylic acid (SA)-mediated resistance9. In Arabidopsis and Oryza sativa, certain bacterial pathogens, such as Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas campestris pv. campestris (Xcc), activate the MAPK phosphatase 1 (MKP1)-mediated MAPK cascade. This activation downregulates transcription factor MYB4-mediated vascular-specific lignification immunity by inhibiting lignin biosynthesis10. Moreover, MPK3 and/or MPK6 directly phosphorylate two 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE (ACS) isoforms, ACS2 and ACS6, preventing their degradation in the 26S proteasome and thereby enhancing ethylene biosynthesis11,12. In response to the fungal pathogen Botrytis cinerea, MPK3 and MPK6 phosphorylate the transcription factor WRKY33, which upregulates pathogen-induced camalexin biosynthesis in Arabidopsis13. Furthermore, the transcription factors ETHYLENE RESPONSE FACTOR1 (ERF1) and WRKY33 integrate the ethylene and JA pathways to induce camalexin biosynthesis by directly upregulating camalexin biosynthetic genes14. In addition, the MEKK1-MKK1/MKK2-MPK4 branch in the MAPK cascade has also been found to downregulate plant defence responses. For instance, the flagellin peptide flg22 activates MPK4, inhibiting SA biosynthesis and ROS accumulation but having no effect on callose biosynthesis, thus negatively regulating plant defence responses15,16. Recent reports have also revealed that MPK3 downregulate plant defence responses, as the rice blast pathogen Magnaporthe oryzae subverts the rice Blast Disease Resistance 1 (OsBDR1)-MPK3 cascade to negatively regulate the JA-related defence response17.
While extensive research has been conducted on the role of MAPK cascades in defence responses against bacterial and fungal pathogens, few studies have documented their effects on virus-host interactions. However, it is known that the plant DNA virus geminivirus βC1 protein suppresses MAPK cascade-mediated defence by targeting MKK2 and MPK4 to promote viral infection18. Moreover, the interaction of beet black scorch virus (BBSV) coat protein with 14-3-3a protein results in degradation of MAPKKKα, suppressing MAPKKKα-activated MAPK cascade antiviral defence19. As a strategy to defend against viruses, plants defend against viral infections by utilizing MAPK cascades to phosphorylate viral proteins. For example, in barley (Hordeum vulgare), the HvMPK3 kinase phosphorylates nucleoprotein of barley yellow striate mosaic virus (BYSMV), limiting the spread of the virus within the plant20. Furthermore, some studies have shown that human or animal viruses can activate MAPK cascades to enhance their own infection in the host. The MAPK pathway positively regulates virus replication in diverse groups of viruses, such as encephalomyocarditis virus, influenza A viruses and herpes simplex virus21. MAPK pathway activation is also required for multiplication of viruses, such as Junín virus and Vaccinia virus22. Hepatitis C virus (HCV) infection triggers TAB1-dependent p38 MAPK activation to phosphorylate the HCV core protein, promoting HCV replication23. Compared with studies of human or animal viruses, there is little evidence to show that MAPK cascades can be utilized by plant viruses to enhance viral infection. Nonetheless, the soybean (Glycine max) homologues of MPK4 and GmMPK4s negatively regulate SA accumulation and the defence response against downy mildew and soybean mosaic virus (SMV)24. Overall, the detailed molecular mechanisms by which viruses manipulate MAPK cascades to promote their infection require further investigation.
Tomato chlorosis virus (ToCV) is an important disease-causing virus that was first identified in tomatoes and has been found to infect a wide range of economically important vegetable crops and a large number of wild plants25. ToCV was first identified in the mid-1990s from greenhouse-grown tomato plants in north-central Florida, USA26,27. The typical symptoms of ToCV-infected tomato plants include interveinal bright yellowing beginning on the lower leaves and gradually progressing to the upper part of the plant, reduced fruit growth and delayed ripening; severe growth reduction occurs if ToCV infects at the seedling stage25. The ToCV genome has the typical organization of bipartite rhinoviruses, with two molecules of linear, positive-sense, single-stranded RNA, namely, RNA1 (8593-8596 nt) and RNA2 (8242-8247 nt), separately encapsulated in long, flexuous, rod-like virions28,29. The genome map of ToCV is shown in Supplementary Fig. 3. RNA1 contains four open reading frames (ORFs) encoding proteins associated with virus replication, suppression of gene silencing, and auxin signalling-mediated defence; RNA2 contains nine ORFs (ORF4 to ORF12) possibly encoding proteins putatively involved in virus encapsulation, cell-to-cell movement, membrane association, whitefly transmission and suppression of gene silencing25,30. The presence of the unique P7 protein gene at the 3’-end of ToCV RNA2 distinguishes it from all members of the family Closteroviridae29. The ToCV P7 was a transmembrane protein, but its function remains unknown31.
In this work, we confirm that plant MPK3/6 pathway can positively regulate ToCV infection. Our research reveals that activating MAPK cascades in Nicotiana benthamiana enhances ToCV infection. The small protein P7 encoded by ToCV acts as a pathogenicity determinant, recruiting NbMPK3/6, which translocates from the nucleus to the plasma membrane or cytoplasm and phosphorylates the P7 protein at serine 59. The phosphorylated P7 inhibits the ability of NbREM1.1 on callose-dependent antiviral defence, leading to promote viral spread in cells. These findings provide novel insights into the role of MAPK cascades in mediating plant-virus interactions and highlight the significance of P7 as a key player in ToCV infection and pathogenicity.
Results
The MAPK cascade positively regulates ToCV infection
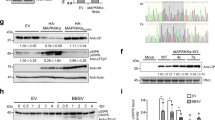
To investigate whether ToCV infection can activate the MAPK cascade, we infiltrated N. benthamiana plants at 4-leaf stage with Agrobacterium containing a ToCV infectious cDNA clone, and we analysed phosphorylation of MAPK pathway components in the systemic leaves at 7 to 30 days post infiltration (dpi). As the mock treatment, N. benthamiana plants were infiltrated with Agrobacterium containing empty vector (EV). Western blot analysis revealed that the phosphorylation level of MPK3/6 and accumulation of ToCV CP gradually increased in systemic leaves of infected plants from 7 to 30 dpi (Fig. 1a). To further assess the impact of the MAPK pathway on ToCV infection, we utilized tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) to silence key components of the MAPK cascade, including NbMAPKKKα, NbMKK2, NbMPK3 and NbMPK6. The silencing efficiency was confirmed by quantitative RT-PCR (qRT-PCR) and western blot analysis at 7 dpi (Supplementary Fig. 1), in which the protein level of NbMAPKKKα was not assessed, because there was no specific antibody for NbMAPKKKα. The silenced plants were infiltrated with Agrobacterium containing the ToCV infectious cDNA clone for 30 days. Compared with control TRV-00-inoculated plants, silencing of NbMAPKKKα, NbMKK2, NbMPK3, and NbMPK6 resulted in reduced ToCV CP and RNA accumulation and reduced phosphorylation of two MAPKs: MPK3 and MPK6 (Fig. 1b, c).
a MAPK activation in ToCV-infected N. benthamiana. Four-leaf-stage N. benthamiana plants were infiltrated with Agrobacterium containing empty vector (EV) (Mock) or a ToCV infectious cDNA clone. Systemic leaves were collected at the indicated time points. The accumulation levels of ToCV and that of MAPK phosphorylation were detected by western blot analysis with an anti-CP or anti-phospho-p44/42 MAPKs (anti-pTEpY) antibody. Actin served as the loading control. Each lane represents a sample from an individual experiment. Each treatment included three biological replicates. The molecular weights of pMPK6, pMPK3, CP and Actin are 44, 42, 30 and 43 kDa, respectively. b Western blot analysis of ToCV CP accumulation and MAPK phosphorylation in N. benthamiana plants inoculated with TRV-00 control, TRV-MPK3, TRV-MPK6, TRV-MKK2 and TRV-MAPKKKα. Actin served as the loading control. Each lane represents a sample from an individual experiment. Each treatment included three biological replicates. c Accumulation of ToCV RNA1 and RNA2 in N. benthamiana plants inoculated with TRV-00 control, TRV-MPK3, TRV-MPK6, TRV-MKK2 and TRV-MAPKKKα, as determined by qRT-PCR using P22 and CP gene-specific primers. Data are presented as means ± SDs and were analysed using Student’s t-test (one-sided, P = 0.0022, 0.0023, 0.0004, 0.0021, 0.0038, 0.0016, 0.0015, 0.0024, respectively). Each treatment included three biological replicates. **P < 0.01, ***P < 0.001. d Overexpression of MAPKKKα enhances ToCV infection. Agrobacterium harbouring ToCV infectious cDNA was infiltrated into different N. benthamiana leaves for 27 days. Then, Agrobacterium containing MAPKKKα-HA, MAPKKKαK236M-HA or empty vector (EV) was infiltrated into ToCV-infected leaves. After 3 days, accumulated levels of ToCV or MAPK phosphorylation were detected by western blot analysis with an anti-CP or anti-pTEpY antibody. MAPKKKα-HA and MAPKKKαK236M-HA protein levels were determined by western blot analysis with an anti-HA antibody. Actin served as the loading control. Each lane represents a sample from an individual experiment. Each treatment included three biological replicates. The molecular weights of MAPKKKα-HA and MAPKKKαK236M-HA are 75 kDa. e Accumulation of ToCV RNAs in N. benthamiana plants coinfected with ToCV and EV, ToCV and MAPKKKαK236M-HA or ToCV and MAPKKKα-HA determined by qRT-PCR using P22 and CP gene-specific primers. Data are presented as means ± SDs and were analysed using Student’s t-test (one-sided, P = 0.1472, 0.0036, 0.4097, 0.0001, respectively). Each treatment included three biological replicates. **P < 0.01, ***P < 0.001, ns, not significant. Source data are provided as a Source Data file.
To further determine the role of MPK3/6 pathway in ToCV infection, Agrobacterium harbouring ToCV infectious cDNA was infiltrated into different N. benthamiana leaves for 27 days. Then, we transiently overexpressed MAPKKKα with an HA-tag and the kinase-inactive mutant MAPKKKαK236M with an HA-tag19,32 in the ToCV-infected plants. At 3 dpi, phosphorylation levels of MPK3 and MPK6, which act downstream of MAPKKKα19,32, were higher in leaves overexpressing MAPKKKα than in those overexpressing MAPKKKαK236M or the EV (empty vector) control. Furthermore, accumulation of ToCV CP and RNA was greater in leaves overexpressing MAPKKKα than in those overexpressing EV. However, overexpression of MAPKKKαK236M had no significant effect on ToCV CP or RNA accumulation (Fig. 1d, e). Expression of MAPKKKα-HA and MAPKKKαK236M-HA was confirmed by western blot analysis (Fig. 1d, e). These results collectively indicate that the MPK3/6 pathway positively regulates ToCV infection.
P7 promotes ToCV infection by activating the MAPK pathway and is associated with membrane localization of P7
To investigate the role of P7 during ToCV infection in plants, we obtained P7-overexpressing (OE-P7) plants (Supplementary Fig. 2). Subsequently, RNA-seq analysis was conducted on wild-type (WT) and OE-P7 plants. Our analysis revealed 508 deferentially expressed genes (DEGs) induced in OE-P7 plants, with 300 genes upregulated and 208 genes downregulated according to a log2 FC change threshold of >1 and <−1 and a one-sided t-test p-value < 0.05 (Supplementary Data 1). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed significant enrichment of DEGs involved in plant-pathogen interaction and the MAPK signalling pathway (Fig. 2a). These results suggest that the P7 protein likely activates the plant MPK3/6 pathway.
a Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of all differentially expressed genes in OE-P7 N. benthamiana plants. b The key genes in the MAPK signalling pathway verified by qRT-PCR. Nbactin was used as the internal reference gene to normalize relative expression. Each treatment included three biological replicates. Values are means ± SDs of three biological replicates (one-sided t-test, P = 0.0095, 0.0308, 0.0494, 0.0326, respectively). *P < 0.05, **P < 0.01. c The phosphorylation level of the MAPK pathway was detected by western blot using an anti-pTEpY antibody. Expression of P7 was detected by western blot analysis with an anti-P7 antibody. Actin served as the loading control. Each lane represents a sample from an individual experiment. Each treatment included three biological replicates. The molecular weight of P7 is approximately 10 kDa. d Symptoms of P7-enhanced PVX-infected N. benthamiana plants (scale bar = 1 cm). e Accumulation of PVX CP in the assayed N. benthamiana samples was determined through western blot analysis using a PVX CP-specific antibody. Coomassie blue (CBB) staining was used to assess protein loading. The molecular weight of the PVX CP is 30 kDa. f The CP expression level was quantified by qRT-PCR using PVX CP gene-specific primers. Data were presented as means ± SDs and were analysed using Student’s t-test (one-sided, P = 0.0037, 0.0096, 0.0010, respectively). Each treatment had three biological replicates. **P < 0.01. g The subcellular location of P7 tagged with GFP in mesophyll cells was determined by laser scanning confocal microscopy (scale bar = 50 μm). The first and second columns show the subcellular localization of P7 and AtPIP2A determined by the GFP and mCherry channels, respectively. The third column shows the subcellular localization of P7 and AtPIP2A determined by merging the GFP and mCherry channels, with a magnified view inserted. The fourth column shows overlapping fluorescence spectra analysis of P7-GFP and AtPIP2A-mCherry signals marked with white arrows. h Analysis of the subcellular location of P7 tagged with GFP by western blot using proteins extracted from cell fractions. T, total protein; M, membrane protein; C, cytoplasmic protein; N, nuclear protein. H+-ATP served as a plasma membrane marker. The molecular weights of P7-GFP and H+-ATP are 37 and 100 kDa, respectively. i The subcellular location of mP7 tagged with GFP in mesophyll cells (scale bar = 50 μm). mP7-GFP and AtPIP2A-mCherry were transiently coexpressed in leaves of N. benthamiana. The first and second columns show the subcellular localization of mP7 and AtPIP2A determined by the GFP and mCherry channels, respectively. The third column shows the subcellular localization of mP7 and AtPIP2A determined by merging the GFP and mCherry channels, with a magnified view inserted. The fourth column shows overlapping fluorescence spectra analysis of mP7-GFP and AtPIP2A-mCherry signals marked with white arrows. j Subcellular location analysis of mP7 tagged with GFP by western blot using proteins extracted from cell fractions. T total protein; M membrane protein; C cytoplasmic protein; N nuclear protein. Actin served as a cytoplasmic marker. H3 served as a nuclear marker. The molecular weights of mP7-GFP and H3 are 37 and 17 kDa, respectively. k Symptoms of N. benthamiana plants infiltrated with ToCV and ToCVmP7 at 30 dpi (scale bar = 3 cm); a magnified view is shown. N. benthamiana plants were infiltrated with Agrobacterium containing empty vector (EV) as a mock control. l The ToCV CP mRNA level quantified by qRT-PCR. Data are presented as means ± SDs and were analysed using Student’s t-test (one-sided, P = 0.0008). Each treatment had three biological replicates. ***P < 0.001. m Accumulated levels of ToCV or MAPK phosphorylation were detected by western blot analysis with an anti-CP or anti-pTEpY antibody. Actin served as the loading control. Each lane represents a sample from an individual experiment. Each treatment included three biological replicates. For (g–j) the experiments were repeated three times with similar results. Source data are provided as a Source Data file.
To further explore the MAPK signalling pathway affected by the P7 protein, we conducted qRT-PCR analysis on the enriched DEGs. Among the 11 DEGs enriched in the plant MAPK signalling pathway, 10 were upregulated (Supplementary Fig. 4). Notably, expression of key genes in the MAPK pathway, such as MPK3, MKKK17, MKKK3, and MPKKK18, was significantly upregulated, consistent with the RNA-seq data (Fig. 2b). These findings suggest that P7 may activate the MAPK pathway in plants. To corroborate these findings, we assessed the phosphorylation status of proteins in the MAPK pathway in OE-P7 plants using an anti-pTEpY antibody at 5 weeks after planting. The results indicated a notable increase in phosphorylation levels of proteins related to the MAPK pathway (Fig. 2c).
To investigate the role of P7 during ToCV infection in plants, we constructed a PVX expression vector carrying the P7 (PVX:P7) or P7 fused with GFP at the C-terminal end (PVX:P7-GFP) and then inoculated it into N. benthamiana plants, respectively. PVX expression vector carrying GFP (PVX:GFP) was used as a control. At 4 dpi, the phenotype of plants infected with PVX:P7 or PVX:P7-GFP was similar to that of plants infected with PVX:GFP. At 8 dpi and 12 dpi, more severe mosaic disease, yellowing, and leaf deformities were observed in the PVX:P7- or PVX:P7-GFP-inoculated plants compare to that in the PVX:GFP-inoculated plants (Fig. 2d, Supplementary Fig. 5a). Western blot and qRT-PCR analysis revealed both protein and mRNA accumulation of PVX CP in the systemic leaves of PVX:P7-inoculated plants was significantly increased compared to that in PVX:GFP-inoculated plants at 4-12 dpi (Fig. 2e, f). However, the PVX CP accumulation in the systemic leaves of PVX:P7- and PVX:P7-GFP -inoculated plants was equal at 4-12 dpi (Supplementary Fig. 5b). These results strongly indicate that P7 functions as a pathogenic factor that enhances viral infection in N. benthamiana.
Several studies have highlighted the importance of pathogenic factors and their subcellular localization18,33. In our study, we analysed the subcellular localization signal of P7 using TMHMM software (http://www.cbs.dtu.dk/services/TMHMM-2.0/). The prediction revealed a transmembrane region spanning amino acid residues 5 to 27 of P7, suggesting that P7 might localize to the plasma membrane (Supplementary Fig. 6a). To confirm this prediction, we constructed P7 fused with GFP at the C-terminal end (P7-GFP) and coexpressed it with Arabidopsis thaliana plasma membrane intrinsic protein 2 A (AtPIP2A)-mCherry in N. benthamiana. AtPIP2A-mCherry serves as a marker for plasma membrane localization34. At 48 h post infiltration (hpi), laser scanning confocal microscopy (LSCM) confirmed that P7-GFP colocalized with AtPIP2A-mCherry at the plasma membrane (Fig. 2g). Western blot analysis with subcellular fractionation also verified the presence of P7-GFP at the plasma membrane (Fig. 2h). Meanwhile, we have fused GFP to the N-terminus of P7 (GFP-P7) and transiently expressed in N.benthamiana leaves. The empty GFP was used as control. The results shown that both P7-GFP and GFP-P7 were localized in plasma membrane while the empty GFP was localized in cytoplasm and nucleus (Supplementary Fig. 7). These results provided evidence that P7 is a plasma membrane-localized protein.
To further investigate the subcellular localization and significance of the predicted membrane region of P7, we constructed a mutant of P7 with mutations of valine (V) to aspartic acid (D) at positions 15, 16, 20 and 26 (designated mP7) that cannot localize to the plasma membrane. The expression vector was designed to destroy the transmembrane domain of mP7 through TMHMM software analysis (Supplementary Fig. 6b). Coexpression of mP7-GFP with AtPIP2A-mCherry in N. benthamiana revealed that mP7-GFP did not colocalize with AtPIP2A-mCherry at 48 hpi (Fig. 2i). Western blot analysis with subcellular fractionation confirmed that mP7 was present in the cytoplasm and nucleus (Fig. 2j). These data demonstrate that P7 is a plasma membrane-localized protein.
To assess the correlation of the membrane-associated region of P7 with its pathogenicity, we constructed a mutant ToCV infectious cDNA clone (ToCVmP7) in which the P7 is mutated to mP7. Compared with plants infected with ToCV, alleviated symptoms at 30 dpi were observed for the plants infected with ToCVmP7 (Fig. 2k). Furthermore, both viral RNA and CP accumulation were reduced in the plants infected with ToCVmP7 (Fig. 2l, Fig. 2m upper panel). The phosphorylation level of MPK3/6 pathway in ToCVmP7-infected plants was similar to that in ToCV-infected plants (Fig. 2m, middle panel), suggesting that mP7 expressed from virus activates MPK3/6 like the virus-expressed wild type P7 but has lost the capacity to enhance viral infection. Taken together, these findings indicate that P7 promotes ToCV infection and that this function is closely associated with membrane localization of P7.
P7 interacts with NbMPK3/6 in vivo and in vitro
To elucidate the detailed mechanism by which P7 regulates the MAPK pathway, we conducted liquid chromatography-mass spectrometry (LC-MS/MS) to screen for potential host factors that interact with P7. We transiently expressed P7-GFP in N. benthamiana plants for 48 h and conducted LC-MS/MS analysis to screen for P7-interacting proteins among the P7-GFP immunoprecipitation (IP) products. The identified peptides were annotated at NCBI protein database (https://www.ncbi.nlm.nih.gov/gene). Numerous interacting proteins, including NbMPK3, were co-immunoprecipitated with P7-GFP (Supplementary Table 1). Because the MPK3/6 in the MAPK cascade has been well documented13,35 and P7 could increase phosphorylation level of MPK3/6, we focused on verifying interactions between P7 and NbMPK3/6.
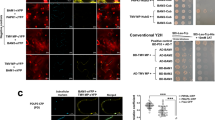
We used pull-down assays and a luciferase complementation imaging (LCI) assay to confirm the interaction between P7 and NbMPK3/6. These experiments demonstrated that P7 interacted with NbMPK3/6 both in vitro and in vivo (Fig. 3a, b). To investigate where P7 interacts with NbMPK3/6 in cells, we conducted a bimolecular fluorescence complementation (BiFC) assay using the leaves of N. benthamiana plants. At 48 hpi, YFP fluorescence was detected in leaf cells coexpressing P7-nYFP and NbMPK3/6-cYFP (Fig. 3c). In contrast, no YFP fluorescence was detected in leaf cells coexpressing AtPIP2A-nYFP and NbMPK3/6-cYFP, P7-nYFP and AtPIP2A-cYFP, P7-nYFP and cYFP, or nYFP and NbMPK3/6-cYFP (control) (Fig. 3c). Western blot analysis with subcellular fractionation also showed NbMPK3 in both the plasma membrane and cytoplasm in cells coexpressing P7-nYFP+NbMPK3-cYFP (Fig. 3d, left panel); NbMPK6 was located in the cytoplasm in cells coexpressing P7-nYFP+NbMPK6-cYFP (Fig. 3d, right panel). In addition, to investigate the subcellular localization of NbMPK3/6 in the absence of P7, we examined the subcellular localization of NbMPK3/6 fused with GFP at the C-terminus (NbMPK3-GFP, NbMPK6-GFP) at 48 hpi in N. benthamiana plants using LSCM. We observed that NbMPK3-GFP and NbMPK6-GFP were present in both the cytoplasm and nucleus (Supplementary Fig. 8a). Western blot analysis confirmed the presence of NbMPK3/6-GFP in protein extracts from both the cytoplasm and nucleus (Supplementary Fig. 8b). These results collectively indicate that P7 inhibits the entry of MPK3 and MPK6 into the nucleus by interacting with them. To further investigate whether the mP7 is interacted with NbMPK3/6, we used BIFC assay and LCI assay to confirm the interaction between mP7 and NbMPK3/6. These results show that the mP7 was unable to interact with NbMPK3 and NbMPK6 (Supplementary Fig. 9a–f).
a GST pull-down analysis of the interaction between NbMPK3 and P7. P7 was incubated with NbMPK3/6-GST or GST. Input and pull-down products were analysed by western blot with anti-His and anti-GST antibodies. The molecular weights of GST, NbMPK3-GST, NbMPK6-GST and P7-His are 28, 70, 72 and 27 kDa, respectively. b LCI assay was used to detect interaction between P7 and NbMPK3/6. Luminescence signals were recorded in N. benthamiana leaves at 48 hpi. nLUC-SGT1 and cLUC-RAR1 served as positive controls. nLUC and cLUC served as negative controls. c BiFC assay to detect interaction of P7 with NbMPK3/6. P7-nYFP was coexpressed with NbMPK3-cYFP, NbMPK6-cYFP, AtPIP2A-cYFP or cYFP. NbMPK3-cYFP was coexpressed with AtPIP2A-nYFP or nYFP. NbMPK6-cYFP was coexpressed with AtPIP2A-nYFP or nYFP. YFP signals in N. benthamiana leaves were recorded at 48 hpi (bar = 50 µm). AtPIP2A-cYFP, AtPIP2A-nYFP, nYFP and cYFP served as negative controls. d Subcellular location of P7 interacting with NbMPK3/6 detected by western blot using a polyclonal YFP antibody. T total protein; M membrane protein; C cytoplasmic protein; N nuclear protein. H+-ATP served as a plasma membrane marker. Actin served as a cytoplasmic marker. H3 served as a nuclear marker. The molecular weights of P7-nYFP, NbMPK3-cYFP and NbMPK6-cYFP are 27, 55 and 57 kDa, respectively. For (a, c, d) the experiments were repeated three times with similar results. Source data are provided as a Source Data file.
NbMPK3/6 phosphorylates the Ser59 residue of P7 and enhances viral infection
Given MPK3/6 acts as the protein kinase3 and P7 interacts with NbMPK3/6, we hypothesized that P7 might serve as a substrate of NbMPK3/6, leading to its phosphorylation. To test this hypothesis, we generated NbMPK3-overexpressing and NbMPK6-overexpressing transgenic lines (OE-NbMPK3 and OE-NbMPK6, respectively) and a mpk3 mutant in which MPK6 is silenced by RNA interference (mpk3−/−mpk6RNAi) (Supplementary Fig. 10). To confirm the phosphorylation level of P7, we transiently expressed P7 containing Flag tag and cYFP36 in wild type (WT) N. benthamiana plants for 48 h. We conducted immunoprecipitation of P7-Flag and subsequently detected phosphoproteins using the pIMAGO reagent37. This analysis revealed that phosphorylated P7-Flag was present in WT plants (Fig. 4a, first panel). To elucidate the importance of NbMPK3/6 in P7 phosphorylation, P7-Flag was transiently over-expressed in the OE-NbMPK3, OE-NbMPK6 and mpk3−/−mpk6RNAi plants. N. benthamiana plants were infiltrated with Agrobacterium containing EV as a mock control. At 48 hpi, P7-Flag fusion was isolated through immunoprecipitation and then subjected to phosphoproteins analysis using the pIMAGO reagent37. The results showed that the phosphorylation levels of P7 and the activation of the MPK3/6 pathway were higher in OE-NbMPK3 and OE-NbMPK6 plants than those in WT plants (Fig. 4a). In contrast, these levels were lower in mpk3−/−mpk6RNAi plants (Fig. 4a).
a In vivo phosphorylation of P7-Flag. P7-Flag was immunoprecipitated with anti-Flag from the total soluble proteins extracted from the leaves of WT, OE-NbMPK3, OE-NbMPK6 and mpk3−/−mpk6RNAi N. benthamiana plants transiently overexpressing P7-Flag and harvested at 48 hpi. N. benthamiana plants were infiltrated with Agrobacterium containing empty vector (EV) as the mock control. Phosphorylation levels of P7 were analysed by western blot with pIMAGO analysis. Accumulation of P7 was analysed by western blot with an anti-Flag antibody. The level of MAPK phosphorylation was detected by western blot analysis with an anti-pTEpY antibody. The molecular weight of P7-Flag is approximately 25 kDa. Each lane represents a sample from an individual experiment. Each treatment included three biological replicates. b In vitro phosphorylation assays showing phosphorylation of P7 by NbMPK3/6. P7-GST protein was incubated with preactivated NbMPK3/6-GST or NbMPK3m/NbMPK6m-GST in 1× kinase reaction buffer. The NbMPK3m-GST and NbMPK6m-GST in the TEY mutant (T218A/Y220A) served as negative control. The phosphorylation level of P7 was analysed by western blot with pIMAGO analysis. Coomassie blue (CBB) staining was used to assess protein loading. The molecular weight of P7-GST is 35 kDa. c Phosphorylation assays of P7S59A-Flag in vivo. P7S59A-Flag was immunoprecipitated with an anti-Flag antibody from the total soluble proteins extracted from the leaves of transiently over-expressing P7S59A-Flag in WT, OE-NbMPK3, OE-NbMPK6 and mpk3−/−mpk6RNAi N. benthamiana harvested at 48 hpi. N. benthamiana plants were infiltrated with Agrobacterium containing empty vector (EV) as the mock control. Phosphorylation levels of P7 S59A were analysed by western blot with pIMAGO analysis. Accumulation of P7S59A was analysed by western blot with an anti-Flag antibody. The level of MAPK phosphorylation was detected by western blot analysis with an anti-pTEpY antibody. The molecular weight of P7S59A-Flag is approximately 25 kDa. Each lane represents a sample from an individual experiment. Each treatment included three biological replicates. d Phosphorylation assays of P7S59A by NbMPK3/6 in vitro. The P7S59A-GST protein was incubated with preactivated NbMPK3/6-GST or NbMPK3m/NbMPK6m-GST in 1× kinase reaction buffer. The NbMPK3m-GST and NbMPK6m-GST served as negative control. The phosphorylation level of P7S59A was analysed by western blot with pIMAGO analysis. CBB staining was used to assess protein loading. The molecular weight of P7 S59A-GST is 35 kDa. e Symptoms of N. benthamiana plants infiltrated with ToCV, ToCVS59D and ToCVS59A at 30 dpi (scale bar = 3 cm); a magnified view is shown. N. benthamiana plants were infiltrated with Agrobacterium containing empty vector (EV) as the mock control. f Western blot analysis of ToCV CP accumulation in ToCV-, ToCVS59D- and ToCVS59A-inoculated N. benthamiana plants. Actin served as the loading control. Each lane represents a sample from an individual experiment. Each treatment included three biological replicates. g Symptoms of WT or mpk3−/−mpk6RNAi plants infiltrated with Agrobacteria containing ToCV, ToCVS59D or ToCVS59A at 30 dpi; a magnified view is shown (scale bar = 3 cm). N. benthamiana plants were infiltrated with Agrobacterium containing empty vector (EV) as the mock control. h Western blot analysis of ToCV CP accumulation in ToCV-, ToCVS59D- and ToCVS59A-inoculated WT or mpk3−/−mpk6RNAi plants. Actin served as the loading control. Each lane represents a sample from an individual experiment. Each treatment included three biological replicates. i Symptoms of WT, OE-NbMPK3 and OE-NbMPK6 plants infiltrated with Agrobacteria containing ToCV, ToCVS59D and ToCVS59A at 30 dpi; a magnified view is shown (scale bar = 3 cm). N. benthamiana plants were infiltrated with Agrobacterium containing empty vector (EV) as the mock control. j Western blot analysis of ToCV CP accumulation in ToCV-, ToCVS59D- and ToCVS59A-inoculated WT OE-NbMPK3 or OE-NbMPK6 plants. Actin served as the loading control. Each lane represents a sample from an individual experiment. Each treatment included three biological replicates. For (b, d) the experiments were repeated three times with similar results. Source data are provided as a Source Data file.
To confirm P7 phosphorylation by NbMPK3 or NbMPK6, we purified recombinant NbMPK3-GST, NbMPK6-GST, NbMPKK5DD-His and P7-GST proteins from Escherichia coli (E. coli). NbMPK3-GST or NbMPK6-GST was activated through incubation with NbMPKK5DD-His in reaction buffer at 30 °C for 30 min. Then, NbMPKK5DD-His was removed from the reaction buffer using His-agarose beads. The results of in vitro phosphorylation assays showed no phosphorylation signal for P7-GST alone, but when P7-GST was incubated with preactivated NbMPK3-GST or preactivated NbMPK6-GST, a clear phosphorylation signal was detected (Fig. 4b). The NbMPK3m-GST (the kinase-inactive version of NbMPK3) harbouring T202A and Y204A mutants on the TEY motif was defective in phosphorylating P7 (Fig. 4b, left panels). Similar with NbMPK3m, NbMPK6m-GST (the kinase-inactive version of NbMPK6) harbouring T218A and Y220A mutants on the TEY motif was defective in phosphorylating P7 (Fig. 4b, right panels). To identify the precise amino acid residues of P7 phosphorylated by NbMPK3/6, LC‒MS/MS analysis was performed on phosphorylated P7. Four potential sites in P7 phosphorylated by NbMPK3 were identified at serine residues 39, 54, and 59 (designated S39, S54, and S59, respectively) and at tyrosine residue 45 (designated Y45). NbMPK6 was responsible for phosphorylation of P7 at threonine residues 4 and 7 (designated T4 and T7, respectively) and S59 (Supplementary Table 2). Among them, S59 was consistently identified in four independent assays and commonly phosphorylated by NbMPK3 and NbMPK6 (Supplementary Table 2, Supplementary Fig. 11a, b). S59 was selected for further study. Further, we overexpressed P7-Flag in TRV:00-, TRV: NbMPK3- or TRV: NbMPK6-inoculated plants and phosphorylation of P7 at S59 was monitored using a parallel reaction monitoring (PRM) system, which is a targeted method of quantification performed using high-resolution hybrid mass spectrometers38. The results showed that phosphorylation of P7 at S59 was significantly lower in TRV: NbMPK3- or TRV: NbMPK6-inoculated plants than in TRV:00-inoculated plants (Supplementary Fig. 11c). Moreover, phosphorylation of P7 at S59 was almost undetectable in the PRM system when P7-Flag was overexpressed in mpk3−/−mpk6RNAi plants (Supplementary Fig. 11d). Interestingly, the alignment amino acid sequences of ToCV P7 reveals that S59 and Gln60 (Q60) is conserved among 43 ToCV isolates in Supplementary Fig. 12. To determine whether the SQ motif of P7 is the phosphorylation target sites of NbMPK3/NbMPK6, we generated two P7 mutants: the P7Q60P by substituting Q60 with Proline (P) and the P7Q60A by substituting Q60 with alanine (A, a nonfunctional residue) then performed in vivo phosphorylation assays. The results indicate that the phosphorylation level of P7Q60P was equal to that of P7 in WT, OE-NbMPK3, OE-NbMPK6 and mpk3−/−mpk6RNAi plants. However, the phosphorylation level of P7Q60A was lower than that of P7 and P7Q60P in WT, OE-NbMPK3, OE-NbMPK6 and mpk3−/−mpk6RNAi plants (Supplementary Fig. 13). The results reveal that the function of Gln60 was equal to Pro60, the SQ motif is the phosphorylation target sites of NbMPK3/NbMPK6 same as the SP/TP motif39.
To further evaluate phosphorylation of P7 by NbMPK3 or NbMPK6 at S59, we generated a P7 mutant (P7S59A) by substituting S59 with alanine (A, a non-phosphorylatable residue) and performed in vivo and in vitro phosphorylation assays. The results indicate that S59 is the critical residue phosphorylated by NbMPK3 or NbMPK6 (Fig. 4c, d). To study the importance of S59 in P7 during ToCV infection, we generated two mutant ToCV infectious clones (ToCVS59A and ToCVS59D) in which S59 is substituted with A or with aspartic acid (D, a mimic the phosphorylated residue). The mutations at S59 have no effect on the expression or stability of P7. Expression of P7/P7S59D/P7S59A-Flag was detected by western blot analysis with an anti-Flag antibody (Supplementary Fig. 14a). After 30 days, ToCVS59A-inoculated plants showed weaker mosaic symptoms, while ToCVS59D-inoculated plants showed stronger mosaic symptoms than ToCV-inoculated plants (Fig. 4e). Western blot assays showed that accumulation of ToCV CP in the systemic leaves of ToCVS59D-inoculated plants increased but that accumulation of ToCV CP in the systemic leaves of ToCVS59A-inoculated plants decreased (Fig. 4f). These findings collectively demonstrate that P7 is phosphorylated at S59 to enhance viral infection. To further investigate whether the phosphorylation status of P7 is related to its interaction with NbMPK3/6, we used BIFC assay and LCI assay to confirm the interaction between P7S59A and NbMPK3/6. These results show that the P7S59A interacts with NbMPK3/6 (Supplementary Fig. 9g–j). These results indicate that the phosphorylation status of P7 have no effect on the interaction between P7 and NbMPK3/6.
To investigate the role of NbMPK3/6 in ToCV infection, ToCV, ToCVS59A and ToCVS59D were inoculated into WT and mpk3−/−mpk6RNAi plants. At 30 dpi, the mosaic symptoms in ToCVS59A- or ToCVS59D-inoculated mpk3−/−mpk6RNAi plants were similar to that in WT, though ToCV-inoculated mpk3−/−mpk6RNAi plants exhibited weaker mosaic symptoms compared to WT. Moreover, ToCVS59A- and ToCVS59D-inoculated plants displayed weaker and stronger mosaic symptoms compared to ToCV-inoculated plants, respectively (Fig. 4g). Moreover, at 30 dpi, ToCV-inoculated OE-NbMPK3 or OE-NbMPK6 plants exhibited markedly stronger mosaic symptoms than did ToCV-inoculated WT plants, ToCVS59D-inoculated OE-NbMPK3 or OE-NbMPK6 plants showed severe mosaic symptoms similar to those of ToCVS59D-inoculated WT plants, and ToCVS59A-inoculated OE-NbMPK3 or OE-NbMPK6 plants exhibited weak mosaic symptoms similar to those of ToCVS59A-inoculated WT plants (Fig. 4i). These results were confirmed through western blot analysis using a ToCV CP-specific antibody (Fig. 4h, j). Taken together, our results demonstrate that NbMPK3/6 phosphorylates S59 of P7, enhancing viral infection.
Phosphorylated P7 interacts with NbREM1.1 to reduce callose deposition
Considering that P7 is phosphorylated at the plasma membrane and enhances viral infection, we sought to identify host factors involved in P7 functions at the plasma membrane. Among the candidates obtained from the LC‒MS/MS analysis (Supplementary Table 1), we focused on NbREM1.1, which has been implicated in callose deposition processes40,41. Through LCI and pull-down assays, we confirmed interaction between P7 and NbREM1.1 both in vivo and in vitro (Fig. 5a, b). Interestingly, we observed that WT P7 and P7S59D were capable of interacting with NbREM1.1 but not P7S59A according to LCI and pull-down analyses (Fig. 5a, b). This finding suggests that the phosphorylation status of P7 at S59 might regulate callose deposition and, consequently, enhance viral transmission.
a LCI assay to detect interactions between P7 and mutants with NbREM1.1. Luminescence signals were recorded in N. benthamiana leaves at 48 hpi. nLUC-SGT1 and cLUC-RAR1 served as positive controls. b Pull-down analysis of the interaction between P7 and its mutants with NbREM1.1. P7/P7S59A/P7S59D-His or His was incubated with NbREM1.1-GST. Input and pull-down products were analysed by western blot with anti-His and anti-GST antibodies. The molecular weights of NbREM1.1-GST, His, P7/P7S59A/P7S59D-His are 60, 20 and 27 kDa, respectively. The experiments were repeated three times with similar results. c Callose fluorescence at PD upon aniline blue staining. (scale bar = 20 μm). PDLP5-RFP served as a PD marker. d Quantification of the callose staining intensity. The relative callose staining intensity was set as 1 for untreated WT plants. Value is the mean ± SD (one-way ANOVA with Tukey’s test, n = 3 biologically independent experiments, P-values are shown in the Source Data file). Different lowercase letters indicate significant differences (P < 0.05). e Fluorescence images of N. benthamiana leaf tissue coexpressing PVX-GFP with EV, P7-Flag, P7S59D-Flag and P7S59A-Flag. GFP fluorescence signals were recorded by an inverted fluorescence microscope at 3 dpi (scale bar = 100 μm). f Statistical analysis of PVX-GFP movement efficiency shown in (e). Value is the mean ± SD (one-way ANOVA with Tukey’s test, n = 5 biologically independent experiments, P-values are shown in the Source Data file). Different lowercase letters indicate significant differences (P < 0.01). For box plot, the horizontal lines from top to bottom represent the maximum, first quartile, median, third quartile, and minimum of the total data, respectively. Source data are provided as a Source Data file.
To investigate the effect of different P7 phosphorylation states on callose deposition, we individually inoculated ToCV, ToCVS59D and ToCVS59A into WT, OE-NbMPK3, OE-NbMPK6 and mpk3−/−mpk6RNAi plants through Agrobacterium infiltration. These plants were infiltrated with Agrobacterium containing EV as a mock control. Additionally, an untreated control was used to visualize and measure basal callose levels. Plasmodesmata-located protein 5 (PDLP5)-RFP is a plasmodesmata marker42. At 30 dpi, the quantity of PD-associated callose was measured by in vivo aniline blue staining. Callose deposition in ToCV-inoculated WT plants was lower than that in the untreated or the mock treatment. This finding suggests that the reduction of callose levels upon infection occurs during normal ToCV infection. Callose deposition in ToCVS59D-inoculated plants was lower than that in the ToCV-inoculated plants. Callose deposition in ToCVS59A-inoculated plants was higher than that in the ToCV-inoculated plants. These findings collectively demonstrate that phosphorylated P7 inhibits the callose deposition to enhance ToCV infection. In OE-NbMPK3 and OE-NbMPK6 plants that were inoculated with ToCV, the level of callose deposition was lower than in WT plants, but higher than that in mpk3−/−mpk6RNAi plants (Fig. 5c, d). These results show that phosphorylation at S59 of P7 by NbMPK3 and NbMPK6 is pivotal for callose deposition.
To investigate the effect of P7 phosphorylated by NbMPK3/6 on viral spread, we coexpressed PVX-GFP and P7/P7S59D/P7S59A-Flag in WT or mpk3−/−mpk6RNAi leaves. Because NbMPK3 and NbMPK6 were tagged with GFP in the OE-NbMPK3 and OE-NbMPK6 transgenic plants, we transiently overexpressed NbMPK3 with an HA-tag (NbMPK3-HA) and NbMPK6 with an HA-tag (NbMPK6-HA) to substitute for these overexpressing plants. Expression of P7/P7S59D/P7S59A-Flag, NbMPK3-HA and NbMPK6-HA was confirmed by western blot analysis (Supplementary Fig. 14c). Before infiltration, Agrobacterium harbouring PVX-GFP was diluted 10000-fold (OD600 = 0.0001)43, mixed with Agrobacterium harbouring P7/P7S59D/P7S59A-Flag (OD600 = 0.2), and coinfiltrated into N. benthamiana leaves. At 5 dpi, the areas of green fluorescent infection foci in P7S59D-Flag- or P7S59A-Flag-inoculated NbMPK3-HA, NbMPK6-HA and mpk3−/−mpk6RNAi plants were similar to those in WT plants, while the areas of infection foci in P7-Flag-inoculated NbMPK3-HA and NbMPK6-HA plants were larger than those in WT and mpk3−/−mpk6RNAi plants. Moreover, the area of infection foci in P7S59D-Flag-inoculated plants was greater than that in P7S59A-Flag-inoculated plants (Fig. 5e, f). Overall, these findings demonstrate the importance of P7 phosphorylated by NbMPK3/6 at S59 in regulating callose deposition and facilitating virus transmission during the interaction of ToCV with the host plant.
NbREM1.1 negatively regulates ToCV infection
To investigate the role of NbREM1.1 in ToCV infection, we obtained transgenic plants overexpressing NbREM1.1 (OE-REM1) and a dual NbREM1.1/NbREM1.2 (NbREM1) knockout mutant plant lines (ΔNbrem1)44. After 30 days of ToCV infection, we observed that accumulation levels of ToCV CP and RNA were significantly lower in OE-REM1 plants than in WT plants, as demonstrated by western blot and qRT-PCR analyses (Fig. 6a, b). Conversely, in ΔNbrem1 plants, accumulation of ToCV RNA and CP was significantly greater than that in WT plants (Fig. 6a, b). Additionally, to assess callose deposition in these plants, we employed aniline blue staining to stain callose and PDLP5-RFP as a PD marker42. Compared with that in WT plants, callose deposition in OE-REM1 plants was significantly increased, whereas callose deposition in ΔNbrem1 plants was decreased (Fig. 6c, d). These results show that NbREM1.1 positively regulates callose deposition and inhibits ToCV infection.
a Accumulation of ToCV CP was analysed by western blot with an anti-CP antibody. Ponceau staining (Ponceau S) revealed equal protein loading in each lane. N. benthamiana plants were infiltrated with Agrobacterium containing empty vector (EV) as the mock control. b The ToCV RNA expression level was quantified by qRT-PCR using ToCV CP gene-specific primers. Data are presented as means ± SDs and were analysed using Student’s t-test (one-sided, P = 0.0224, 0.0033, respectively). Each treatment had three biological replicates. *P < 0.05, **P < 0.01. c Callose staining with aniline blue in WT, OE-REM1 and ΔNbrem1 plants (scale bar = 20 μm). PDLP5-RFP served as a PD marker. d The relative intensity of callose staining in (c). The relative callose staining intensity was set as 1 in wild-type. Value is the mean ± SD (one-way ANOVA with Tukey’s test, n = 3 biologically independent experiments, P-values are shown in the Source Data file). Different lowercase letters indicate significant differences (P < 0.05). e. Fluorescence images of PVX-GFP in WT, OE-REM1 and ΔNbrem1 plants coexpressing EV, P7-Flag, P7S59D-Flag or P7S59A-Flag. GFP fluorescent signals were recorded by confocal microscopy at 3 dpi (scale bar = 100 μm). f Statistical analysis of PVX-GFP movement efficiency shown in (e). Value is the mean ± SD (one-way ANOVA with Tukey’s test, n = 5 biologically independent experiments, P-values are shown in the Source Data file). Different lowercase letters indicate significant differences (P < 0.05). For box plot, the horizontal lines from top to bottom represent the maximum, first quartile, median, third quartile, and minimum of the total data, respectively. Source data are provided as a Source Data file.
To further investigate the effect of the interaction between P7 and NbREM1.1 on virus transmission, we coexpressed PVX-GFP and P7/P7S59D/P7S59A-Flag in WT, OE-REM1 or ΔNbrem1 leaves. Expression of P7/P7S59D/P7S59A-Flag was confirmed by western blot analysis (Supplementary Fig. 14d). Before infiltration, Agrobacterium harbouring PVX-GFP was diluted 10000-fold (OD600 = 0.0001)43, mixed with Agrobacterium harbouring P7/P7S59D/P7S59A-Flag (OD600 = 0.2), and coinfiltrated into N. benthamiana leaves. At 5 dpi, the area of infection foci in OE-REM1 plants was smaller than in WT plants while the infection foci were larger in ΔNbrem1 plants (Fig. 6e, f). Furthermore, P7-Flag and P7S59D-Flag increased the area of infection foci area compared to P7S59A-Flag. These findings indicate that phosphorylated P7 inhibits the ability of NbREM1.1 to suppress viral spread (Fig. 6e, f).
In summary, our results suggest that NbREM1.1 plays a negative regulatory role in ToCV infection, as demonstrated by reduced viral accumulation and increased callose deposition in OE-REM1 plants and increased viral accumulation and reduced callose deposition in ΔNbrem1 plants. Additionally, phosphorylated P7 disrupts the ability of NbREM1.1 to inhibit viral spread, further highlighting the interplay between P7, NbMPK3/6, and NbREM1.1 in the context of ToCV infection.
Discussion
During the process of pathogen coevolution, plants have developed sophisticated pathways to finely regulate their defence network45. MAPK cascades, which are evolutionarily conserved in both plants and animals, play a central role in activating defence responses against pathogens2. However, in the context of plant‒virus interactions, research investigating plant MAPK cascades is limited, and most studies have shown that these cascades can downregulate infections caused by DNA and RNA viruses, such as TYLCCNV, TMV, and BBSV18,19,46. Here, our results showed that ToCV also activated the MPK3/6 pathway in N. benthamiana (Fig. 1a). Interestingly, ToCV accumulation was reduced upon inhibition of the MPK3/6 pathway but increased upon activation of the MPK3/6 pathway in N. benthamiana (Fig. 1b–e). This finding represents a significant contribution, as it is likely that plant MPK3/6 can promote ToCV infection. Our results suggest that the MPK3/6 pathway may play a novel role during ToCV infection, at least in N. benthamiana plants. Moreover, our results showed that ToCV P7 activates the MPK3/6 pathway (Fig. 2a–c) and that P7 is critical for its pathogenicity (Fig. 2d–f). Although mP7 also activated the plant MPK3/6 pathway, it did not localize to the plasma membrane and lacked the capacity to enhance viral infection (Fig. 2g–m). These results suggest that amino acids at position 15, 16, 20 and 26 are critical for P7 localization and pathogenicity. MAPK cascades play essential roles in transferring extracellular stimulus signals from the plasma membrane to the nucleus to activate defence responses47. We observed that the expression levels of NbMAPKKKα and NbMKK2 were increased in ToCV- and ToCVmP7- infiltrated plants (Supplementary Fig. 15). These results indicate that P7/mP7 likely mediates the activation of upstream components of the MAPK cascade to activate the MPK3/6 pathway. The P7 protein likely functions as a stimulus signal to initiate activation of N. benthamiana MPK3/6 pathway in the plasma membrane and cytoplasm, while enhancing ToCV infection depends on plasma membrane localization.
The viral protein disrupts the function of the host protein by altering its localization48,49. In our study, P7 interacted with NbMPK3/6 both in vivo and in vitro (Fig. 3a, b). BiFC assays revealed that the P7 and NbMPK3/6 interacted in the plasma membrane and cytoplasm, respectively (Fig. 3c, d). However, NbMPK3/6 was localized in the cytoplasm and nucleus (Supplementary Fig. 8). These results indicate that P7 inhibits the entry of NbMPK3/6 into the nucleus by interacting with NbMPK3 and NbMPK6 on the plasma membrane and in the cytoplasm, respectively. We also found that NbMPK6 interacted with P71–33 (containing the transmembrane region) (Supplementary Fig. 9k-l). The interaction between NbMPK6 and P71–33 may disrupt the membrane localization of P7, resulting to P7 interacting with NbMPK6 in the cytoplasm. However, NbMPK3 interacted with P734–65 but not P71–33, resulting to P7 recruited NbMPK3 to the plasma membrane (Supplementary Fig. 9m). The TLCYnV C4 could direct NbSKη to cytoplasm membrane and reduces the accumulation of nucleus-localized NbSKη, then facilitated viral replication48. HCV initiates the activation of the MAPK pathway, which leads to the phosphorylation of the HCV core protein and subsequently enhances the replication of the virus23. Some studies have shown that MPK3/6 phosphorylates transcription factors in the nucleus to elicit an immune response. WRKY33 is phosphorylated by MPK3/6 in the nucleus and responds to pathogen infection50. We also observed that the upregulation or blockade of MPK3/6 promoted or inhibited the replication of ToCV in protoplasts (Supplementary Fig. 16). We can assume that P7 inhibits the immune response mediated by NbMPK3/6 through inhibiting the entry of NbMPK3/6 into the nucleus, thereby benefiting ToCV replication. We also demonstrated that ToCV P7 is phosphorylated at S59 in the SQ motif by NbMPK3/6 both in vivo and in vitro (Fig. 4a–d, Supplementary Fig. 11). This finding is different from those of previous studies in which the conserved (S/T) P motif was shown to be a substrate of the MAPKs phosphorylation3,39. Nevertheless, there are a few exceptions, such as the SD motif51 or TD motif52. In vivo phosphorylation assays revealed that the SQ motif of P7, which is the same as the SP motif, is the phosphorylation target sites of NbMPK3/NbMPK6 (Supplementary Fig. 13), which might expand our knowledge regarding substrates phosphorylated by MAPKs. We also found that substitution of S59 with A weakened viral mosaic symptoms, whereas substitution S59 with D enhanced viral mosaic symptoms (Fig. 4e, f). Moreover, ToCV accumulation was significantly lower in ToCV-infected mpk3−/−mpk6RNAi plants (Fig. 4g, h) and greater in ToCV-infected OE-NbMPK3/6 plants (Fig. 4I, j) than in ToCV-infected WT plants. These results showed that P7 phosphorylation by NbMPK3/6 promoted ToCV infection. It has been reported that MPK3 is phosphorylated by BDR1 in the plasma membrane to promote the infection of Magnaporthe oryzae through the inhibition of JA signalling and terpenoid biosynthesis17. In our study, NbMPK3 and phosphorylated P7 were localized in the plasma membrane to facilitate ToCV infection (Figs. 3c, 4e–i). However, the mechanism by which NbMPK3 functions in the plasma membrane requires further investigation. Based on these results, we speculate P7 was phosphorylated by NbMPK3/6 in the cytoplasm in cells and then phosphorylated P7 recruits NbMPK3 to plasma membrane. In addition, the mP7 lost the capacity to enhance viral infection because it cannot interact with NbMPK3/6. Taken together, we putative phosphorylated P7 recruits NbMPK3 to plasma membrane may be beneficial to viral infection but the function should be further investigated. Furthermore, we also found that the NbMPK6 was detained in the cytoplasm through interacting with P7. Previous studies have shown that the Arabidopsis MPK3 and MPK6 are required in different ways for the activation of response to B. cinerea53. Thus, we speculated that the function of NbMPK6 in cytoplasm recruited by P7 was different to that of NbMPK3 and the function of NbMPK6 localizing to cytoplasm should be further investigated.
The remorin family is among the best-characterized plasma membrane nanodomain-associated proteins in plants54,55. Remorins are markers of membrane rafts that are enriched at plasmodesmata56. Remorins are well known to be involved in plant‒pathogen interactions56,57,58. LCI and pull-down assays revealed that P7 or P7S59D, but not P7S59A, interacts with NbREM1.1 both in vivo and in vitro (Fig. 5a, b). The remorin protein-callose module plays a pivotal role in the interaction between plants and viruses44. Some studies have reported that remorin can restrict viral infection59. The accumulation of ToCV CP and RNA was reduced in OE-REM1 plants but elevated in ΔNbrem1 plants (Fig. 6a, b). These results suggest that NbREM1.1 negatively regulates ToCV infection. Callose is localized in the cell wall, and its deposition near the neck zone of plasmodesmata regulates their size exclusion limit (SEL), which controls the cell-to-cell transport of molecules60. Callose deposition at plasmodesmata may be involved in early defence responses against virus attack43,61. The plant immune signal restricts viral movement by inducing callose deposition at plasmodesmata, whereas viruses counteract this defence mechanism by suppressing callose deposition at plasmadesmata43. In a recent study, rice stripe virus (RSV) was found to interfere with the S-acylation of remorin and induce its autophagic degradation to reduce callose deposition and facilitate viral infection44. Callose deposition was lower in ToCV-infected WT plants than in untreated and mock-treated plants (Fig. 5c, d). Moreover, the phosphorylation status of P7 regulated callose deposition. Compared with that in ToCV-infected WT plants, callose deposition was lower in ToCVS59D-infected WT plants and greater in ToCVS59A-infected plants. The phosphorylation status of P7 was correlated with NbMPK3/6. Compared with that in ToCV-infected WT plants, callose deposition was reduced in ToCV-infected OE-NbMPK3/6 plants and increased in ToCV-infected mpk3−/−mpk6RNAi plants (Fig. 5c, d). These results suggest that the phosphorylation of P7 regulates its ability to inhibit callose deposition via interaction with NbREM1.1. In line with some studies that have reported remorin-mediated inhibition of viral movement44,62,63, P7 and P7S59D promoted PVX transmission but not P7S59A compared with mock treatment in WT plants (Fig. 5e, f). PVX transmission increased when P7 was coincubated with NbMPK3/6 and was lower in mpk3−/−mpk6RNAi plants than that in WT plants. These results show that P7 is phosphorylated by NbMPK3/6 through their interaction and that phosphorylated P7 then interacts with NbREM1.1 in the plasma membrane to inhibit callose deposition and promote viral movement. In summary, P7 enhanced ToCV infection in a manner dependent on viral replication and movement. The phosphorylation status of a viral protein may regulate its the function64. We speculated that nonphosphorylated and phosphorylated P7 increased ToCV replication in the cytoplasm, while phosphorylated P7 was further targeted to NbREM1.1 for inhibiting its callose-inducing activity at plasmodesmata, thereby increasing viral cell-to-cell movement and allowing viral transmission.
In summary, in this study, we propose a model in which P7 was phosphorylated by MPK3/6 and inhibits callose-dependent antiviral defence, thereby promoting ToCV infection (Fig. 7). Under normal conditions, the MPK3/6 was inactive, and the callose was localized in the cell wall. Upon ToCV infection, the MPK3/6 pathway was activated. P7 interacts with NbMPK3/6 and inhibits the entry of NbMPK3/6 into the nucleus. P7 might inhibit the immune response mediated by NbMPK3/6 and facilitate ToCV replication. P7 was phosphorylated by NbMPK3/6 then targets NbREM1.1 to inhibit its callose-inducing activity to promote viral movement in N. benthamiana plants. Although our results revealed a novel role of MPK3/6 in plant viral infection, this study has some limitations. For example, 1) the currently available ToCV infectious clones are unable to infect natural host but can efficiently infect N. benthamiana plants via agroinfiltration65,66, and therefore, all of our experiments were performed in N. benthamiana plants; 2) only one viral protein of one virus (ToCV) was investigated here, and the results might be specific to the MPK3/6 pathway. Nevertheless, our results, together with previous data, suggest the existence of a complex relationship in the ongoing arms race between plants and viruses.
Under normal condition (left column), the MPK3/6 was inactive, and the callose was localized in the cell wall. Upon ToCV infection (right column), the MPK3/6 was activated. ToCV P7 interacts with NbMPK3/6 and inhibits the entry of NbMPK3/6 into the nucleus. P7 then inhibits the immune response mediated by NbMPK3/6 and promotes ToCV replication. Subsequently, NbMPK3/6 phosphorylates P7 at S59. The phosphorylated P7 and NbMPK3 then localized to the plasma membrane. The phosphorylated P7 targets NbREM1.1 at the plasma membrane to inhibit its callose-inducing activity to promote ToCV movement in N. benthamiana plants.
Methods
Plant growth and virus inoculation
Nicotiana benthamiana and transgenic plants were grown in a greenhouse at 23 ± 1 °C with a 16 h/8 h (light/dark) cycle. The tomato chlorosis virus (ToCV) full genomic cDNA infectious clones pCa-ToCR1 and pCa-ToCR2 (containing the full-length RNA1 or RNA2 sequence of the ToCV Beijing isolate) were a kind gift from Tao Zhou (China Agricultural University) and reported previously65; potato virus X (PVX)-based infectious clones were obtained from a previously reported source67. The pCa-ToCR1 and pCa-ToCR2 plasmids were individually transformed into Agrobacterium tumefaciens strain GV3101. Agrobacterium cultures were grown overnight, pelleted through centrifugation, and incubated for 2 h in infiltration buffer (100 mM MES, pH 5.2, 10 mM MgCl2, and 200 mM acetosyringone). Agrobacterium cultures carrying pCa-ToCR1 (OD600 = 0.6–0.8) were mixed with an equal volume of Agrobacterium cultures carrying pCa-ToCR2 or one of its mutants. The mixed cultures were infiltrated individually into leaves of N. benthamiana plants using needleless syringes, as reported previously65. The Agrobacterium cultures carrying PVX-based infectious clones or with inserted genes were infiltrated as reported previously67.
Plasmid construction
Three mutant ToCV infectious clones with S59D, S59A and mP7 were generated by overlapping PCR using the plasmid pCa-ToCR2 as the template. The ToCV P7 gene sequence was inserted into the pGWB505-GFP, pET-32a-His (containing the Trx-tag), pGEX-4T-2-GST, pCV-nYFP68 and pCambia1300-cLUC and pEarleyGate-202-Flag (containing the cYFP)36 vectors. The coding sequence of NbMPK3 (GenBank accession no. AB098729.1) was cloned and inserted into the pGWB505, pGEX-4T-2, pCV-cYFP68 and pCambia1300-nLUC vectors. The coding sequence of NbMPK6 (GenBank accession no. AB373025.1) was cloned and inserted into the pGWB505, pGEX-4T-2, pCV-cYFP68 and pCambia1300-nLUC vectors. The coding sequence of NbREM1.1 (GenBank accession no. ON392760.1) was cloned and inserted into the pGEX-4T-2 and pCambia1300-nLUC vectors. The primers used in this study are listed in Supplementary Table 3. The above clones were confirmed by Sanger DNA sequencing.
Generation of transgenic plants
Constitutive expression of the NbMPK3 gene with a GFP tag under the control of its native promoter in N. benthamiana was used to construct OE-NbMPK3 plants. Constitutive expression of the NbMPK6 gene with a GFP tag under the control of its native promoter in N. benthamiana was used to construct OE-NbMPK6 plants. Additionally, we conducted the mpk3−/−mpk6RNAi lines employing CRISPR/Cas9 technology to knockout NbMPK3 in N. benthamiana. Meanwhile, MPK6 transcripts are targeted by an RNA interference (RNAi) in the mpk3−/−mpk6RNAi lines. Sanger sequencing revealed the 1 bp insertion at the NbMPK3 target site in mpk3−/−mpk6RNAi plants (Supplementary Fig. 10a, b). NbMPK6 mRNA level was significantly decreased in mpk3−/−mpk6RNAi plants compared with WT plants, as evidenced by qRT-PCR (Supplementary Fig. 10c). The overexpression of NbMPK3 and NbMPK6 protein in the transgenic plants were confirmed using western blot with an anti-GFP antibody (Supplementary Fig. 10d). ToCV P7 gene (OE-P7) N. benthamiana transgenic plants were obtained via Agrobacterium-mediated transformation. Western blots with anti-GFP antibody (1:5000, TransGen Biotech, China, Cat. No. HT801) were performed to screen the positive OE-NbMPK3 and OE-NbMPK6 transgenic plants. Western blot with anti-P7 antibody (1:1000, HUABIO, China, customer order) was performed to screen the positive OE-P7 transgenic plants. To screen the positive mpk3−/−mpk6RNAi plants, DNA fragment corresponding to the gRNA-targeted DNA sequence regions was amplified from genomic DNA of transgenic plants using corresponding primers listed in Supplementary Table 3, and DNA sequencing was carried out to examine the insertion or deletion at the target site.
RNA-seq
The leaf samples from WT and OE-P7 plants were collected. Three leaves from different plants were collected as one biological replicate, and three biological replicates were used for each group. Total RNA was extracted using the TRIzol reagent (Invitrogen, CA, USA). After quantification and qualification, total RNAs were pooled for cDNA library construction. cDNA library preparation and sequencing were carried out on an Illumina NovaSeq 6000 platform with 150-bp pair-end reads in OE Biotech Co., Ltd. (Shanghai, China). Total reads were mapped to the Nicotiana benthamiana transcriptome using Hisat2 (v2.0.5). The expression level of each gene was calculated based on expected number of Fragments Per Kilobase of transcript sequence per Million’s base pairs sequenced (FPKM). Differential expression analysis of two groups was performed using the DESeq2 R package (1.20.0). P-value < 0.05 and |log2(foldchange)| > 1 and <−1 were set as the threshold for significantly differential expression.
RNA extraction and qRT-PCR
Total RNA was extracted from N. benthamiana samples using HiPure Plant RNA Mini Kit (Magen, China, R4165-02). qRT-PCR was performed as previously reported, with minor modifications64. First-strand cDNAs were synthesized using random primers, 1 μg of total RNA per 20 μL of reaction, and a First Strand cDNA Synthesis Kit (Vazyme, China, Cat. No. R312-02). The actin gene (GenBank accession no. XM_016647918) served as the internal control. Relative expression levels of the assayed genes were calculated using the 2−ΔΔCt method. Each treatment had three biological replicates with three technical replicates each. The primers used for qRT-PCR analysis are listed in Supplementary Table 3.
BiFC and LCI assays
For BIFC assays, Agrobacterium mixtures containing expression plasmids different BiFC vectors68 were diluted to an OD600 = 0.6 with infiltration medium and coinfiltrated into N. benthamiana leaves. At 48 hpi, a Leica TCS SP8 confocal microscope (Leica Microsystems, Heidelberg, Germany) was used to observe the reconstituted YFP signal. The YFP signal was confirmed by western blot using a polyclonal YFP antibody (1:5000, HUABIO, China, Cat. No. R1312-2). LCI assays were performed as described previously, with minor modifications64. For LCI assays, Agrobacterium harbouring cLUC-P7 and nLUC-NbMPK3/6 was diluted to an OD600 = 0.6 with infiltration medium and coinfiltrated into N. benthamiana leaves. At 3 dpi, images of Luc signals were captured with Lumazone Sophia. The cLUC-RAR1 (required for Mla12 resistance) and nLUC-SGT1 (suppressor of the G2 allele of skp1) served as a positive control69.
Identification of the phosphorylation site of P7 through LC‒MS/MS
The P7-Flag fusion was transiently expressed in N. benthamiana leaves and then enriched through immunoprecipitation with anti-Flag M2 magnetic beads (Sigma-Aldrich, USA, Cat. No. M8823). The enriched P7-Flag fusion protein was detected through western blot using an anti-Flag antibody (1:5000, TransGen Biotech, China, Cat. No. HT201) or through separation by SDS‒PAGE followed by Coomassie Brilliant Blue staining. The P7-Flag band was cut from the gel and digested overnight at 37 °C with trypsin, followed by LC‒MS/MS.
MAPK activation assay
The MAPK activation assay was performed as previously described, with minor modifications19. Total protein was extracted in protein extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 1× protease inhibitor cocktail, 1 mM Na3VO4, 1 mM NaF, and 20 mM β-glycerophosphate) and detected by western blot with an anti-phospho-p44/42 MAPK antibody (pTEpY) (1:2000, Cell Signaling Technology, USA, Cat. No. #4370).
In vitro phosphorylation assay
In vitro phosphorylation assays were performed as described previously20. P7-pGEX-4T-2, P7S59A-pGEX-4T-2, NbMPK3-pGEX-4T-2 and NbMPK6-pGEX-4T-2 plasmids were transformed into E. coli (Rosetta), and P7-GST, P7S59A-GST, NbMPK3-GST and NbMPK6-GST proteins were purified by Mag-Beads GST Fusion Protein Purification (Sangon Biotech, China, Cat. No. C650031). 2 μg NbMPK3-GST and 2 μg NbMPK6-GST were incubated with 2 μg His-MPKK5DD20 in a reaction mixture (50 mM Tris-HCl, pH 7.5; 10 mM MgCl2; 1 mM DTT; 0.2 mM ATP) for 30 min at 30 °C. Then, His-MPKK5DD was removed using Ni-NTA 6FF Sefinose (TM) (BBI, China, Cat. No. C600332) and preactivated NbMPK3/6 was purified using Mag-Beads GST Fusion Protein Purification (Sangon Biotech, China, Cat. No. C650031). For in vitro phosphorylation assays, preactivated NbMPK3/6-GST or GST was incubated with 2 μg P7-His or P7S59A-His in 30 μL of 1× kinase reaction buffer (25 mM Tris-HCl, pH 7.5; 10 mM MgCl2; 10 mM CaCl2; 10 mM DTT; 0.025 mM ATP) at 30 °C for 30 min. Next, 5 μL of 5× SDS sample buffer was added to terminate the reactions. Proteins were separated by SDS-PAGE, and phosphorylated proteins were detected by pIMAGO (Tymora Analytical, USA, Cat. No. 800-10)37.
In vivo phosphorylation assay
P7-Flag fusion was transiently expressed in WT, OE-NbMPK3, OE-NbMPK6 or mpk3−/−mpk6RNAi N. benthamiana leaves and then enriched through immunoprecipitation (IP) with anti-Flag M2 magnetic beads (Sigma-Aldrich, USA, Cat. No. M8823) for 3 h. After washing 6 times with 1× PBS buffer containing 1× protease inhibitor cocktail (Roche, Cat. No. 11836153001), the products were detected by pIMAGO (Tymora Analytical, USA, Cat. No. 800-10)37.
Pull-down assay
Pull-down assays were used to confirm interaction between P7 and NbMPK3/6 or between P7 and NbREM1.1. Recombinant P7-His was expressed and purified from E. coli (Supplementary Fig. 17). First, 6 μg of GST- or His-tagged protein was incubated with 20 μL of Mag-Beads GST Fusion Protein Purification (Sangon Biotech, China, Cat. No. C650031) or Ni-IDA-Sefinose Resin 6FF (Sangon Biotech, China, Cat. No. C600029) in 600 μL of 1× PBS buffer for 3 h at 4 °C. The beads were washed six times with 1× PBS buffer and analysed by western blot with anti-GST (1:5000, TransGen Biotech, China, Cat. No. HT601) or anti-His (1:5000, TransGen Biotech, China, Cat. No. HT501) antibodies.
Virus-induced gene silencing (VIGS)
To silence NbMPK3, NbMPK6, NbMKK2 or NbMAPKKKα expression in N. benthamiana, 300 bp fragments representing partial sequences of NbMPK3, NbMPK6, NbMKK2 or NbMAPKKKα were inserted into the TRV2 vector70, respectively. Agrobacterium cultures carrying pTRV1 and pTRV2 (referred to as TRV:00) or pTRV1 and pTRV2:NbMPK3, NbMPK6, NbMKK2 or NbMAPKKKα (TRV:NbMPK3, TRV:NbMPK6, TRV:NbMKK2 or TRV:NbMAPKKKα) were infiltrated into leaves of N. benthamiana plants, and the infiltrated plants were grown inside a climate growth chamber at 25 °C. After 7 days, the plants were inoculated again with ToCV. Plants inoculated with TRV:00 were used as controls. The silencing efficiency of NbMPK3, NbMPK6, NbMKK2 and NbMAPKKKαwas confirmed using qRT-PCR or western blot analysis. Specifically, the efficiency of NbMPK3 and NbMPK6 silencing was verified with an anti-MPK3 antibody (1:10000, Sigma-Aldrich, USA, Cat. No. M8318) and an anti-MPK6 antibody (1:10000, Sigma-Aldrich, USA, Cat. No. A7104), respectively. The silencing efficiency of NbMKK2 was confirmed with an anti-MKK2 antibody (1:10000, Beyotime, China, Cat. No. AG2961).
Isolation of cytoplasmic, membrane and nuclear proteins
Subcellular fractionation was performed as previously reported, with minor modifications33,71. To obtain cytoplasmic and membrane-rich fractions, 0.5 g of Agrobacterium-infiltrated leaves at 2 dpi were gently ground with 1 mL of homogenization buffer [50 mM Tris-HCl, pH 8.0; 10 mM KCl; 3 mM MgCl2; 1 mM EDTA; 1 mM DTT; 0.1% BSA; 0.3% dextran; and 13% (w/v) sucrose]. The mixture was centrifuged at 3000 × g for 20 min at 4 °C to remove nuclei and large cellular debris, and the resulting supernatant was ultracentrifuged at 30,000 × g for 1 h at 4 °C to produce supernatant (cytoplasm) and microsomal (membrane-rich) fractions. The membrane-rich fraction was resuspended in homogenization buffer. All fractions were subjected to western blot analysis. To obtain the nuclear fraction, 0.5 g Agrobacterium-infiltrated leaves at 2 dpi were harvested, ground to a fine powder in liquid nitrogen and mixed with 5 mL of lysis buffer (20 mM Tris-HCl, pH 7.5; 20 mM KCl; 2 mM EDTA; 2.5 mM MgCl2; 25% glycerol; 250 mM sucrose; and 5 mM DTT) supplemented with protease inhibitor cocktail (Roche, 11836153001). The homogenate was filtered through a double layer of Miracloth. The flow-through was centrifuged at 1500 × g for 10 min. The pellet was washed with 5 mL of nuclear resuspension buffer (20 mM Tris-HCl, pH 7.4, 25% glycerol, 2.5 mM MgCl2, and 0.2% Triton X-100). The samples were subsequently centrifuged at 16,000 × g for 10 min at 4 °C. The final nuclear pellet was resuspended in 400 μL of lysis buffer. As a quality control for fractionation, H+-ATP was used as a plasma membrane marker, actin was used as a cytoplasmic marker, and histone H3 was used as a nuclear marker. These components were confirmed by western blot using anti-H+-ATP antibody (1:5000, Amyjet Scientific, China, Cat. No. AS07-260), anti-actin (1:5000, Abbkine, China, Cat. No. ABM40122), anti-H3 antibody (1:5000, Cell Signaling Technology, USA, Cat. No. #4499).
Callose staining
Callose staining was performed as previously reported, with minor modifications43,44. As a plasmodesmata marker, Agrobacterium harbouring the PDLP5-RFP was diluted 1000-fold (OD600 = 0.001) and infiltrated into N. benthamiana leaves for 3 days. Callose was stained with 0.025 mg/mL aniline blue fluorochrome. Callose quantification was performed by measuring the aniline blue fluorescence focus number and focus intensity using ImageJ image processing software (http://rsbweb.nih.gov/ij/) with the callose Quant plug-in, which, after setting parameters, localizes fluorescent callose foci and quantifies the callose fluorescence intensity of each focus automatically43. P-values for significant differences between mean values were determined by one-way ANOVA with Tukey’s test. The statistical tests were performed with GraphPad Prism 8 software.
PVX transmission experiments
PVX transmission experiments were performed as previously reported, with minor modifications43,44. For PVX transmission experiments, Agrobacterium harbouring PVX-GFP was diluted 10000-fold (OD600 = 0.0001), mixed with Agrobacterium harbouring P7/P7S59D/P7S59A-Flag (OD600 = 0.2), and coinfiltrated into N. benthamiana leaves for 5 days. Green fluorescence was distributed among multiple cell lines, as assessed by confocal microscopy. The extent of PVX-GFP movement was quantified by measuring the area of green fluorescence using confocal microscopy, followed by statistical calculations using one-way ANOVA with Tukey’s test.
N. benthamiana protoplast isolation and transfection
N. benthamiana protoplast isolation and transfection were performed as previously reported, with minor modifications72. Nicotiana benthamiana protoplasts were isolated from 21-day-old plants. In brief, young leaves were chopped and immersed in enzyme solution (0.5 M mannitol, 1.5% cellulose RS (Yakult Honsha, Tokyo, Japan), 0.75% macerozyme R10 (Yakult Honsha), 1 mM CaCl2, and 0.1% BSA). The mixture was incubated in a shaking incubator (60 rpm) in the dark at room temperature for 4–5 h and then filtered through Miracloth. The protoplasts were pelleted by centrifugation at 200 × g for 5 min and then resuspended in an equal volume of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl and 1.5 mM MES, adjusted to pH 5.7), followed by centrifugation and resuspension in MMG solution (0.4 M mannitol, 15 mM MgCl2 and 4.7 mM MES, adjusted to pH 5.7). Plasmid DNA (10 or 20 µg) was added to the protoplast solution and transfected with 40% polyethylene glycol (PEG) solution [40% PEG4000 (Sigma, Cat. No. 102078930), 0.4 M mannitol, and 100 mM Ca(NO3)2] at room temperature for 20 min. W5 solution was gradually added to dilute the PEG solution, which was subsequently discarded. The transfected protoplasts were incubated overnight at room temperature, after which RNA was extracted.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data supporting the findings of this study are available within the paper and its Supplementary Information files. RNA-seq raw data have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA:CRA020063) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. LC-MS/MS raw data have been successfully submitted to ProtenmeXchange Via the PRIDE database. Project accession: PXD057403. Source data are provided with this paper.
References
Cristina, M., Petersen, M. & Mundy, J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649 (2010).
Meng, X. & Zhang, S. MAPK cascades in plant disease resistance signaling. Annu Rev. Phytopathol. 51, 245–266 (2013).
Zhang, M. & Zhang, S. Mitogen‐activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 64, 301–341 (2022).
Kong, Q. et al. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24, 2225–2236 (2012).
Meng, X. et al. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25, 1126–1142 (2013).
Ekengren, S. K., Liu, Y., Schiff, M., Dinesh-Kumar, S. P. & Martin, G. B. Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 36, 905–917 (2003).
Thulasi Devendrakumar, K., Li, X. & Zhang, Y. MAP kinase signalling: interplays between plant PAMP-and effector-triggered immunity. Cell. Mol. Life Sci. 75, 2981–2989 (2018).
Zhang, M., Su, J., Zhang, Y., Xu, J. & Zhang, S. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 45, 1–10 (2018).
Bartels, S. et al. MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21, 2884–2897 (2009).
Lin, H. et al. An MKP-MAPK protein phosphorylation cascade controls vascular immunity in plants. Sci. Adv. 8, eabg8723 (2022).
Liu, Y. & Zhang, S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16, 3386–3399 (2004).
Joo, S., Liu, Y., Lueth, A. & Zhang, S. MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54, 129–140 (2008).
Zhou, J. et al. Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in Arabidopsis. Plant Cell 32, 2621–2638 (2020).
Zhou, J. et al. Multilayered synergistic regulation of phytoalexin biosynthesis by ethylene, jasmonate, and MAPK signaling pathways in Arabidopsis. Plant Cell 34, 3066–3087 (2022).
Petersen, M. et al. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120 (2000).
Berriri, S. et al. Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell 24, 4281–4293 (2012).
Wang, L. et al. The OsBDR1-MPK3 module negatively regulates blast resistance by suppressing the jasmonate signaling and terpenoid biosynthesis pathway. Proc. Natl Acad. Sci. USA 120, e2211102120 (2023).
Hu, T. et al. βC1 protein encoded in geminivirus satellite concertedly targets MKK2 and MPK4 to counter host defense. PLoS Pathog. 15, e1007728 (2019).
Gao, Z. et al. Coat proteins of necroviruses target 14-3-3a to subvert MAPKKKalpha-mediated antiviral immunity in plants. Nat. Commun. 13, 716 (2022).
Ding, Z. H. et al. MAPKs trigger antiviral immunity by directly phosphorylating a rhabdovirus nucleoprotein in plants and insect vectors. Plant Cell 34, 3110–3127 (2022).
Kumar, R. et al. Role of MAPK/MNK1 signaling in virus replication. Virus Res 253, 48–61 (2018).
Mohanta, T. K. et al. Molecular Insights into the MAPK Cascade during Viral Infection: Potential Crosstalk between HCQ and HCQ Analogues. BioMed. Res. Int. 2020, 1–9 (2020).
Cheng, Y. et al. Virus-induced p38 MAPK activation facilitates viral infection. Theranostics 10, 12223–12240 (2020).
Liu, J.-Z. et al. Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol. 157, 1363–1378 (2011).
Fiallo-Olive, E. & Navas-Castillo, J. Tomato chlorosis virus, an emergent plant virus still expanding its geographical and host ranges. Mol. Plant Pathol. 20, 1307–1320 (2019).
Wisler, G. C., Duffus, J. E., Liu, H. Y. & Li, R. H. Ecology and epidemiology of whitefly-transmitted closteroviruses. Plant Dis. 82, 270–280 (1998).
Wisler, G. C., Li, R. H., Liu, H. Y., Lowry, D. S. & Duffus, J. E. Tomato chlorosis virus: a new whitefly-transmitted, Phloem-limited, bipartite closterovirus of tomato. Phytopathology 88, 402–409 (1998).
Wintermantel, W. M. et al. The complete nucleotide sequence and genome organization of tomato chlorosis virus. Arch. Virol. 150, 2287–2298 (2005).
Lozano, G., Moriones, E. & Navas-Castillo, J. Complete nucleotide sequence of the RNA2 of the crinivirus tomato chlorosis virus. Arch. Virol. 151, 581–587 (2006).
Liu, S. et al. Tomato chlorosis virus-encoded p22 suppresses auxin signalling to promote infection via interference with SKP1-Cullin-F-box(TIR1) complex assembly. Plant Cell Environ. 44, 3155–3172 (2021).
Zhao, R. et al. Characterization and full genome sequence analysis of a Chinese isolate of tomato chlorosis virus. Acta Virol. 58, 92–94 (2014).
del Pozo, O., Pedley, K. F. & Martin, G. B. MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23, 3072–3082 (2004).
Mei, Y., Yang, X., Huang, C., Zhang, X. & Zhou, X. Tomato leaf curl Yunnan virus-encoded C4 induces cell division through enhancing stability of Cyclin D 1.1 via impairing NbSKeta -mediated phosphorylation in Nicotiana benthamiana. PLoS Pathog. 14, e1006789 (2018).
Marmagne, A. et al. Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell Proteom. 3, 675–691 (2004).
Zhao, J., Sun, Y., Li, X. & Li, Y. Cysteine-rich receptor-like kinase5 (CRK5) and CRK22 regulate the response to Verticillium Dahliae toxins. Plant Physiol. 190, 714–731 (2022).
Lu, Q. et al. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61, 259–270 (2010).
Bredow, M. et al. Phosphorylation-dependent subfunctionalization of the calcium-dependent protein kinase CPK28. Proc. Natl Acad. Sci. USA 118, e2024272118 (2021).
Lei, L., Stevens, D. M. & Coaker, G. Phosphorylation of the pseudomonas effector AvrPtoB by Arabidopsis SnRK2.8 is required for bacterial virulence. Mol. Plant 13, 1513–1522 (2020).
Pitzschke, A. Modes of MAPK substrate recognition and control. Trends Plant Sci. 20, 49–55 (2015).
Perraki, A. et al. Plasma membrane localization of Solanum tuberosum remorin from group 1, homolog 3 is mediated by conformational changes in a novel C-terminal anchor and required for the restriction of potato virus X movement. Plant Physiol. 160, 624–637 (2012).
Perraki, A. et al. REM1.3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog. 14, e1007378 (2018).
Tee, E. E., Johnston, M. G., Papp, D. & Faulkner, C. A PDLP-NHL3 complex integrates plasmodesmal immune signaling cascades. Proc. Natl Acad. Sci. USA 120, e2216397120 (2023).
Huang, C. et al. dsRNA-induced immunity targets plasmodesmata and is suppressed by viral movement proteins. Plant Cell 35, 3845–3869 (2023).
Fu, S. et al. Rice stripe virus interferes with s-acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol. Plant 11, 269–287 (2018).
Dangl, J. L. & Jones, J. D. Plant pathogens and integrated defence responses to infection. Nature 411, 826–833 (2001).
Zhang, D. et al. The MAPK-Alfin-like 7 module negatively regulates ROS scavenging genes to promote NLR-mediated immunity. Proc. Natl Acad. Sci. USA 120, e2214750120 (2023).
Bi, G. & Zhou, J.-M. MAP kinase signaling pathways: a hub of plant-microbe interactions. Cell Host Microbe 21, 270–273 (2017).
Mei, Y., Yang, X., Huang, C., Zhang, X. & Zhou, X. Tomato leaf curl Yunnan virus-encoded C4 induces cell division through enhancing stability of Cyclin D 1.1 via impairing NbSKη -mediated phosphorylation in Nicotiana benthamiana. PLOS Pathog. 14, e1006789 (2018).
Mei, Y. et al. Nucleocytoplasmic shuttling of geminivirus C4 protein mediated by phosphorylation and myristoylation is critical for viral pathogenicity. Mol. Plant 11, 1466–1481 (2018).
Mao, G. et al. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653 (2011).
Xing, Y. et al. MPK6‐mediated HY5 phosphorylation regulates light‐induced anthocyanin accumulation in apple fruit. Plant Biotechnol. J. 21, 283–301 (2022).
Xiang, Y. et al. ZmMPK5 phosphorylates ZmNAC49 to enhance oxidative stress tolerance in maize. N. Phytologist 232, 2400–2417 (2021).
Galletti, R., Ferrari, S. & De Lorenzo, G. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 157, 804–814 (2011).
Gouguet, P. et al. Connecting the dots: from nanodomains to physiological functions of REMORINs. Plant Physiol. 185, 632–649 (2021).
German, L., Yeshvekar, R. & Benitez-Alfonso, Y. Callose metabolism and the regulation of cell walls and plasmodesmata during plant mutualistic and pathogenic interactions. Plant Cell Environ. 46, 391–404 (2023).
Raffaele, S. et al. Remorin, a Solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21, 1541–1555 (2009).
Perraki, A. et al. StRemorin1.3 hampers Potato virus X TGBp1 ability to increase plasmodesmata permeability, but does not interfere with its silencing suppressor activity. FEBS Lett. 588, 1699–1705 (2014).
Son, S., Oh, C. J. & An, C. S. Arabidopsis thaliana remorins interact with SnRK1 and play a role in susceptibility to beet curly top virus and beet severe curly top virus. Plant Pathol. J. 30, 269–278 (2014).
Yue, N. et al. Palmitoylation of γb protein directs a dynamic switch between Barley stripe mosaic virus replication and movement. EMBO J. 41, e110060 (2022).
Wu, S. W., Kumar, R., Iswanto, A. B. B. & Kim, J. Y. Callose balancing at plasmodesmata. J. Exp. Bot. 69, 5325–5339 (2018).
Beffa, R. S., Hofer, R. M., Thomas, M. & Meins, F. Jr. Decreased susceptibility to viral disease of [beta]−1,3-glucanase-deficient plants generated by antisense transformation. Plant Cell 8, 1001–1011 (1996).
Wang, A. et al. REM1.3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLOS Pathog. 14, e1007378 (2018).
Wang, A. et al. The Barley stripe mosaic virus γb protein promotes viral cell-to-cell movement by enhancing ATPase-mediated assembly of ribonucleoprotein movement complexes. PLOS Pathog. 16, e1008709 (2020).
Li, J. et al. Phosphorylated viral protein evades plant immunity through interfering the function of RNA-binding protein. PLoS Pathog. 18, e1010412 (2022).
Zhao, R. et al. P22 of tomato chlorosis virus, an RNA silencing suppressor, is naturally expressed in the infected plant. Acta Virol. 60, 423–425 (2016).
Orílio, A. F., Fortes, I. M. & Navas-Castillo, J. Infectious cDNA clones of the crinivirus tomato chlorosis virus are competent for systemic plant infection and whitefly-transmission. Virology 464-465, 365–374 (2014).
Chapman, S., Kavanagh, T. & Baulcombe, D. Potato virus X as a vector for gene expression in plants. Plant J. 2, 549–557 (1992).
LU, Y. et al. Garlic virus X 11-kDa protein granules move within the cytoplasm and traffic a host protein normally found in the nucleolus. Mol. Plant Pathol. 12, 666–676 (2011).
Chen, H. et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376 (2008).
Liu, Y., Schiff, M. & Dinesh-Kumar, S. P. Virus-induced gene silencing in tomato. Plant J. 31, 777–786 (2002).
Wang, W. et al. An importin beta protein negatively regulates MicroRNA activity in Arabidopsis. Plant Cell 23, 3565–3576 (2011).
Jiang, Y. et al. NbWRKY40 positively regulates the response of Nicotiana benthamiana to tomato mosaic virus via salicylic acid signaling. Front Plant Sci. 11, 603518 (2020).
Acknowledgements
We are grateful to Dr. Yi Xu at the Nanjing Agricultural University for gifting the seeds of transgenic lines of OE-REM1 and ΔNbrem1 Nicotiana benthamiana, and Dr. Tao Zhou at the China Agricultural University for providing the ToCV full cDNA infectious clone, and Dr. Tong Zhou (Institute of Plant Protection, Jiangsu Academy of Agricultural Science) for providing valuable assistance with writing the manuscript. The study was supported by the National Natural Science Foundation of China (32030088 and 31972242 to Y.L), the China Agricultural Research System (CARS-23-D-02 to Y. L), the Innovation Project of Agricultural Science of Hunan Province (2022CX08 to Y.L) and the Science and Technology Projects of Hunan Province (2020TJ-Y05 to Y.L).
Author information
Authors and Affiliations
Contributions
S.Z., J.Y., and Y.L. conceived and directed the project. S.Z. and J.Y. designed the study. L.F. performed the experiments. X.L., L.H., Y.Z., S.L., Z.Z., X.Y., X.W., X.O., X.S. and D.Z. performed and analysis the the RNA-seq. F.L., X.T., J.C., J.Y., S.Z. and Y.L. analysed the data. L.F., J.Y., and S.Z. wrote the manuscript. All authors have read and approved the manuscript for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Feng, L., Luo, X., Huang, L. et al. A viral protein activates the MAPK pathway to promote viral infection by downregulating callose deposition in plants. Nat Commun 15, 10548 (2024). https://doi.org/10.1038/s41467-024-54467-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-54467-9
This article is cited by
-
Global phosphoproteome analysis identifies PeVYV P4-induced host protein phosphorylation changes that facilitate viral proliferation
BMC Plant Biology (2025)
-
From perception to action: MAPKs in plant–virus interactions
Physiology and Molecular Biology of Plants (2025)