Abstract
Pluripotent stem cells possess a unique nuclear architecture characterized by a larger nucleus and more open chromatin, which underpins their ability to self-renew and differentiate. Here, we show that the nucleolus-specific RNA helicase DDX18 is essential for maintaining the pluripotency of human embryonic stem cells. Using techniques such as Hi-C, DNA/RNA-FISH, and biomolecular condensate analysis, we demonstrate that DDX18 regulates nucleolus phase separation and nuclear organization by interacting with NPM1 in the granular nucleolar component, driven by specific nucleolar RNAs. Loss of DDX18 disrupts nucleolar substructures, impairing centromere clustering and perinucleolar heterochromatin (PNH) formation. To probe this further, we develop NoCasDrop, a tool enabling precise nucleolar targeting and controlled liquid condensation, which restores centromere clustering and PNH integrity while modulating developmental gene expression. This study reveals how nucleolar phase separation dynamics govern chromatin organization and cell fate, offering fresh insights into the molecular regulation of stem cell pluripotency.
Similar content being viewed by others
Introduction
In eukaryotes, the linear genome is folded and packaged into three-dimensional chromatin structures in the nucleus, a highly organized and dynamic organelle1,2,3. The organization and reconfiguration of chromatin into different compact versus open states allow fine-tuning gene regulation during cell differentiation and embryonic development. Pluripotent embryonic stem cells (ESCs) are ideal for studying gene expression programs and developmental control due to their unlimited self-renewal capacity and multilineage differentiation properties that often co-occur with the acquisition of the heterochromatin compartment. In particular, perinucleolar heterochromatin (PNH) formation is required for both mouse4,5 and human6 ESC differentiation and early embryo development7,8.
Transcription factors, structural RNAs, and RNA binding proteins (RBPs) have been identified as important regulators for chromatin organization through multivalent interactions. The self-assembly of these macromolecules and the immiscibility of each, known as phase separation9,10, give rise to a series of membraneless sub-compartments (or nuclear bodies), including nucleoli11, paraspeckles12, P granules13, and nuclear speckles14. The mammalian nucleolus is the largest nuclear body assembled around the tandem repeated rDNA clusters. Each nucleolus is composed of three morphologically and functionally distinct sub-regions named the fibrillar center (FC), dense fibrillar center (DFC), and granular component (GC). Aside from the classical function in ribosome biogenesis (RiBi)15,16, nucleoli are also involved in phase-separated protein quality control17 and stem cell differentiation, the latter of which co-occurs with the decrease in RiBi4,18,19,20. Although widely recognized for their roles in providing an inter-chromosomal hub for 3D chromatin organization21,22, nucleoli functions and the underlying mechanisms in chromatin organization and reconfiguration during cell differentiation or development remain poorly understood.
DEAD-box RNA helicases are evolutionarily conserved regulators of RNA-containing phase-separated organelles23, including nucleoli24. DDX18 was one of the 86 proteins whose depletion in the siRNA screen impacted nucleolar structure maintenance in HeLa cells25. By studying DDX18 in human ESCs (hESCs), we discovered that DDX18 plays a critical role in controlling human pluripotency by maintaining high RiBi and highly organized membraneless nucleolar architecture. Mechanistically, DDX18 modulates liquid-liquid phase separation (LLPS) properties of the nucleoli through its intrinsically disordered regions (IDRs) and the dynamic multivalent interactions with nucleolar RNAs and proteins. Specifically, heterotypic interactions of DDX18 with nucleophosmin (NPM1) in nucleolar GC and with snoRNAs and rRNAs in the DFC/FC interface modulate the biophysical properties of the nucleolus, limiting the heterochromatin formation and preventing the initiation of differentiation program in hESCs. Finally, we developed a DDX18-mediated nucleolus-specific CasDrop26, dubbed NoCasDrop. We successfully manipulated centromere clustering around the nucleolus, demonstrating that the redirection of constitutive heterochromatin organization towards a different compartment alone is sufficient to drive the derepression of development-related genes, initiating an early event associated with the exit of pluripotency and cell differentiation.
Results
DDX18 is required for hESC maintenance through the regulation of RiBi and protein synthesis
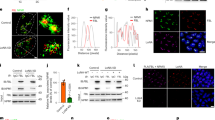
DDX18 is highly enriched in pluripotent hESCs relative to differentiated27 or somatic28 cells (Fig. S1a) and abruptly downregulated upon embryoid body (EB) differentiation at kinetics even faster than NANOG and OCT4 (Fig. S1b), suggestive of its specific function in hESC maintenance. shRNA-mediated DDX18 knockdown (KD) (shDDX18) at both protein (Fig. 1a) and RNA (Fig. 1b) levels, relative to control KD (shLuc), functionally validated its critical roles in maintaining hESCs, manifested by downregulation of the core pluripotency and stem cell surface marker genes and upregulation of lineage-specific and developmental genes (Fig. 1a–c), as well as reduced alkaline phosphatase staining (Fig. 1d). RNA sequencing (RNA-seq) in control and DDX18 KD cells (Fig. 1e) revealed 789 significantly downregulated genes for “stem cell population maintenance”, “rRNA processing”, and “translational initiation”, and 1903 significantly upregulated genes for multilineage differentiation and development based on both the gene ontology (GO) analysis (Fig. 1f) and gene set enrichment analysis (GSEA) (Figs. 1g, S1c, d). Of note, the “ectoderm differentiation” is not enriched for the upregulated genes (Fig. S1c, d), indicating lineage skewing towards mesendoderm differentiation of DDX18 KD hESCs (Fig. S1e). When compared to the transcriptomes of the three germ layers29, the transcriptome of DDX18 KD cells clustered with those of endoderm and mesoderm but not ectoderm cells, further highlighting the skewed mesendoderm differentiation of DDX18 KD cells (Fig. S1f). Among the upregulated genes enriched with the “embryonic organ development” (Fig. 1g) are HOXA-D clusters (e.g., HOXA in Fig. 1h), key developmental regulators that are not expressed but held in bivalent chromatin domains in pluripotent stem cells to be resolved and activated upon differentiation30,31,32.
a Western blot analysis of pluripotency and lineage-specific transcription factors in control (shLuc) and DDX18 knockdown (KD) cells with two independent shRNAs (shDDX18#1 and shDDX18#3). β-Actin was used as the loading control. The experiment was repeated independently 3 times with similar results. b Quantitative RT–PCR analysis of pluripotency genes and differentiation genes. Bars represent the average of three independent experiments. T is also known as Brachyury. c Flow cytometric analysis of the pluripotency-related surface markers SSEA-4 and TRA-1-60. Data represent three replicates of two independent shRNAs. d Alkaline phosphatase (AP) staining of control and DDX18 KD cells. Biological replicates were performed using two shRNAs targeting DDX18. e Volcano plot showing dysregulated genes in DDX18 KD cells by RNA-seq. The green dots denote downregulated genes in DDX18 KD samples (log2 [fold change] <−1, p < 0.05); red dots denote upregulated genes in DDX18 KD samples (log2 [fold change] >1, p < 0.05). Biological replicates from two hESC lines were analyzed. The p-value was calculated with edgeR. f Gene Ontology analysis of the downregulated and upregulated genes in DDX18 KD cells relative to control cells. The most significantly enriched biological processes GO terms with p values are plotted. The p values were calculated using the DAVID bioinformatics resource system. g Geneset enrichment analysis (GSEA) for the terms “Cytoplasmic translation”, “Ribosome biogenesis”, and “Embryonic organ development” in DDX18 KD hESCs. P value was calculated using the GSEA web tool. h IGV track showing HOXA cluster genes are upregulated upon DDX18 KD. i SUnSET assay measuring relative puromycin levels incorporated into proteins during translation in DDX18 KD and controls human ESCs, indicated by the anti-puromycin western blot. The Coomassie blue staining gel and the Vinculin blot are the loading controls. WT and mutant DDX18 protein levels under KD and rescue conditions are also indicated. The experiment was repeated independently twice with similar results. Source data are provided as a Source Data file.
The prevalent association of translation-related GO and biological processes with downregulated genes upon DDX18 KD prompted us to investigate the potential translational defects of DDX18 KD hESCs. First, a SUnSET assay to measure the newly synthesized peptides33 revealed a marked decrease of total protein synthesis in DDX18 KD relative to control cells, a global translational defect that can be rescued with wild-type DDX18 (WT) but not the RNA helicase inactive DQAD mutant34,35 (Fig. 1i). Next, SILAC-MS (stable isotope labeling by amino acids coupled with mass spectrometry) quantitative proteomics (Supplementary Data 1) identified 879 downregulated and 319 upregulated proteins with the enrichment of translation-related and differentiation-related GO terms, respectively, in DDX18 KD relative to control KD hESCs (Fig. S1g, h). By integrating the RNA-seq and SILAC data (Fig. S1i), we found that, while many genes/proteins are positively regulated at both transcriptional and translational levels (Groups I and VI), there are more genes/proteins regulated at translational levels without significant transcriptional changes (Groups II and V). Only a small number of genes/proteins exhibit anti-correlation between transcription and translation (Groups III and IV) (Fig. S1i; Supplementary Data 2). Furthermore, consistent with our previous findings in mouse ESCs (mESCs)19, our DDX18 ChIP-seq experiment in hESCs revealed that no significant DDX18 binding signals could be detected in other genomic regions beyond the rDNA loci (Fig. S5f), excluding the direct transcriptional effects of DDX18 on the pluripotency and development gene expression programs except for the rDNA gene expression.
Together, these results establish the roles of DDX18 and its RNA helicase activity in maintaining hESCs through its nucleolar functions in RiBi and translational control, a finding in line with emerging evidence linking translational control and protein synthesis to ESC maintenance and embryonic development20,36,37.
DDX18 undergoes phase separation in vitro and in vivo
To understand how DDX18 may exert its nucleolar functions in regulating RiBi and translation for hESC pluripotency, we first performed 3D structured illumination microscopy (3D-SIM) and confirmed its sub-nucleolar localization in the NPM1-containing GC layer in hESCs (Fig. 2a, b). Nucleolus assembly is driven by liquid-liquid phase separation (LLPS), facilitated by nucleolar proteins harboring intrinsically disordered regions (IDRs) that mediate multivalent interactions to assist macromolecular assembly24,38,39. Employing PONDER VSL2 webtool40 for protein disorder predictions we identified two IDRs at DDX18’s N-terminal and C-terminal ends, respectively (Fig. S2a). Phase separation properties of DDX18 under or near physiological salt concentration (150 mM NaCl) were first verified in vitro using DDX18-GFP recombinant protein (Fig. 2c, conditions A’, B’, and C’), which does not require the molecular crowding reagent polyethylene glycol (PEG). DDX18 droplets can be disrupted by higher salt concentrations (Fig. 2c, d, condition series from D’ to F’); however, this disruption could be overcome by increasing the protein concentration (Fig. 2d). The LLPS nature of DDX18 in vivo was also confirmed in hESCs expressing DDX18-GFP at the endogenous DDX18 locus (Fig. 2e) by time-lapse microscopy, revealing that DDX18-GFP undergoes fusion and fission within minutes (Fig. 2f) comparable with the nucleolus fusion speed in vivo11,38, and by fluorescence recovery after photobleaching (FRAP) with the average time for DDX18 to fully recover after the photobleaching around 104 ± 8 s (Figs. 2g, S2b), slightly slower than GC protein NPM138.
a Schematic depiction of a eukaryotic cell with representative nuclear and nucleolus structures. FC (fibrillar center), DFC (dense fibrillar component), GC (granular component). (Created in BioRender. Malik, V. (2025) https://BioRender.com/v27l155). b Representative hESC SIM images showing the localization of DDX18 relative to DFC (FBL) and GC (NPM1). The experiment was repeated independently twice with similar results. c Representative images of liquid-like droplets formed with various NaCl concentrations (A’-F’), visualized by GFP emission of DDX18-GFP. DDX18-GFP is 5 µM. The experiment was repeated independently twice with similar results. d Phase diagrams for LLPS of DDX18 under different concentrations of protein and NaCl determined by turbidity assays. DDX18 concentrations spans from 0.5 µM to 20 µM, and NaCl from 50 mM to 300 mM. Representative images from series A’ to F’ are shown in (c). e Schematic diagram for the DDX18 C-terminal GFP fusion reporter knock-in allele. f Time-lapse images showing the fusion and fission of DDX18-GFP within the nucleolus. g Quantification of FRAP data for DDX18-GFP signal. The bleaching event occurs at t = 0 s. DDX18-GFP exhibits fast dynamics with nearly complete recovery (>90%) on a timescale of τ = 104 \(\pm\) 8 s. Data represent three independent experiments and are presented as mean values \(\pm\) SD. h Representative images of liquid-like droplets formed by fusion proteins containing 10 µM DDX18 full-length (FL), N-terminal IDR deletion (∆NIDR), C-terminal IDR deletion (∆CIDR), and combined N-terminal/C-terminal IDR deletion (∆IDR) detected by GFP (top, in vitro) in 150 mM NaCl, 25 mM Tris-HCl, pH 8.0; anti-Flag immunostaining (middle, in vivo), and the merged images (bottom, in vivo) showing 3xFlag-DDX18 (red), FBL(green) and DAPI (blue) in human ESCs. The experiment was repeated independently twice with similar results. i Phase diagrams for LLPS of DDX18 full-length and truncated mutants (∆NIDR, ∆CIDR, ∆IDR) under various concentrations of proteins (0.5 µM to 20 µM) in the presence of 150 mM NaCl, 25 mM Tris-HCl, pH 8.0 by turbidity assays. Source data are provided as a Source Data file.
DEAD-box helicases are conserved from bacteria to mammals containing the N-terminal domain, helicase core domain, and C-terminal domain41. The interactions implicated in the phase separation include long-range electrostatic effects, prion-like property-mediated interactions, and aromatic amino acid-mediated π-π interactions42. To explore which domains and which types of molecular interactions confer DDX18 functions in phase separation, we analyzed the fraction of charged residues and net charge per residues (NCPR) (Fig. S2c, top track), presence of prion-like domain (PLD) (Fig. S2c, middle track), and propensity for π-π contacts (Fig. S2c, bottom track). We found neither PLD nor π-π contacts were involved in DDX18 phase separation. Instead, NCPR analysis identified positively charged basic tracts (blue) highly enriched within NIDR and CIDR of DDX18 (Fig. S2c). Furthermore, we found that both NIDR and CIDR are required for DDX18 phase separation: lacking either domain (ΔNIDR or ΔCIDR) or both (ΔIDR) greatly reduced the droplet formation in vitro (Fig. 2h, i) and in vivo (Figs. 2h, S2d). When fused with a far-red fluorescent protein (faRFP), NIDR alone, but not CIDR alone, was enough to drive the fusion protein into the nucleolus in vivo (Fig. S2d). Interestingly, we found NIDR harbors a predominant positively charged basic tract consisting of an evolutionarily conserved K-stretch (8 Lysines and 1 Arginine) (Fig. S2e, f), previously reported to be enriched in disordered regions and interact with RNA to drive and regulate cellular condensations43. Accordingly, we found that the K stretch mutant (9 K/R to all A mutation; 9 A) was compromised in its nucleolar localization in vivo (Fig. S2g) and RNA-facilitated formation of the “core-shell” like structure44 compared with WT (Fig. S2h). Of note, the DQAD helicase mutant of DDX18 failed to form any core-shell-like structure (Fig. S2h).
To further corroborate the above findings, we conducted a rescue experiment using wild-type (WT), DQAD, and IDR-deleted versions of DDX18 in DDX18 KD cells, followed by qPCR analysis. Our data demonstrated that only the wild-type DDX18 could fully restore hESC maintenance (Fig. S2i) and pluripotency/lineage gene expression programs (Fig. S2j) during differentiation, thus confirming the essential role of DDX18’s RNA helicase activity and phase separation property in its functionality. Together, these results establish the LLPS nature of DDX18 that is promoted by its RNA helicase activity and the RNA-interacting NIDR, suggesting that DDX18 may contribute to the LLPS of nucleoli in regulating RiBi and protein synthesis for pluripotency of hESCs.
DDX18 binds nucleolar localized RBPs and rRNAs/snoRNAs
To investigate how DDX18 exerts such nucleolar functions through its phase separation properties, we identified DDX18-interacting proteins and RNA targets in hESCs. First, we established a transgenic hESC line expressing 3xFlag-DDX18 for affinity purification of DDX18 and its interacting partners in the hESC nuclear extracts using anti-Flag beads (Fig. S3a). Then, with a stringent cut-off (number of unique peptides ≥4 and ratio of 3xFlag-DDX18/IgG ≥ 4), we identified 140 DDX18-interacting proteins (Supplementary Data 3). GO analysis of these proteins linked them to rRNA processing, translational initiation, and RNA secondary structure unwinding (Fig. 3a). As an important hub in maintaining genome stability, the nucleolus is highly enriched for DNA repair proteins, some of which were identified as DDX18-binding proteins with validated nucleolar localization and functional involvement in RiBi, including PARP145, SMC346, and TRIM2847. However, we cannot exclude the possibility that DDX18 may also play a role in DNA damage repair, a topic worthy of future investigation.
a Summary of the DDX18 interactome showing representative GO terms and corresponding proteins. Nucleolar GC marker proteins NPM1 and NCL are also shown under rRNA processing. b RNA-dependent interactions between DDX18 and NPM1. Western blotting detection of DDX18 and NPM1 protein levels after Flag-IP with and without RNase A treatment. IgG serves as the negative control IP. The experiment was repeated independently twice with similar results. c Representative live imaging of reconstituted BiFC fluorescence. Green fluorescence shows the reconstitution of YFP as an indicator of the protein-protein interaction between DDX18 and NPM1. The experiment was repeated independently twice with similar results. d Relative enrichment of different RNA species identified from DDX18 iCLIP-seq. e HOMER motif analysis of DDX18 iCLIP-seq showing the top consensus sequences. The conserved motif of the C/D box snoRNAs is shown at the top. P value was calculated by HOMER. f GO analysis of biological processes for DDX18 bound mRNAs. Source data are provided as a Source Data file.
Notably, the two well-known nucleolar GC marker proteins, NCL and NPM1, were also detected in the DDX18 interactome. We further confirmed the DDX18-NPM1 interaction by co-immunoprecipitation (co-IP) (Fig. 3b) and bimolecular fluorescence complementation (BiFC)48 (Figs. 3c, S3b), in line with their GC co-localization (Fig. 2b) and supporting a GC-specific function of DDX18. Importantly, the DDX18-NPM1 interaction was attenuated by RNase A treatment in co-IP (Fig. 3b) and BiFC (Fig. 3c) assays, supporting an RNA-facilitated interaction between DDX18 and NPM1.
To identify the RNA targets of DDX18 in hESCs, we performed iCLIP (individual-nucleotide-resolution crosslinking and immunoprecipitation) with the established 3xFlag-DDX18 hESC line (Fig. S3c). DDX18 binds to a diverse set of RNAs, among which rRNAs were most highly represented (Fig. 3d; Supplementary Data 4), while some snoRNAs were also bound by DDX18, consistent with its nucleolus-specific localization (Fig. 2a, b). Upon evaluating our results in relation to iCLIP-seq data for ADAR149, a well-studied nuclear-localized RBP in hESCs, we noted an elevated degree of rRNA and snoRNA binding to DDX18 over ADAR1 (compare Fig. 3d with Fig. S3d). Interestingly, the C/D box snoRNA motif RUGAUGA was ranked among the top three in motif analysis (Fig. 3e), and DDX18 binding to C/D box snoRNAs (SNORDs) was confirmed by CLIP-qPCR, exemplified by a few SNORD members (Figs. S3e, S3f, top panel). We also noticed that DDX18 could bind protein-coding mRNAs (Figs. 3d, S3f, bottom panel; Supplementary Data 5). Of note, mRNA of genes residing in DNA nucleolus-associated domains (NADs) are highly enriched in the nucleolus50, and we found that the DDX18-interacting mRNAs were also enriched for GO terms in RiBi and translation (Fig. 3f). Furthermore, unlike the DFC component FBL51, DDX18 does not bind to 1∼414 nt (5′ ETS-1) region of 47S pre-rRNAs localized in DFC (Fig. S3g), further supporting the nucleolar GC-specific functions of DDX18 in hESCs. These data establish physical and functional interactions between DDX18 and nucleolar RBPs and RNA species that underlie their roles in maintaining the pluripotency of hESCs.
DDX18 maintains nucleolar structural integrity through multiphase liquid miscibility and immiscibility with GC and DFC proteins, respectively
We next investigated how DDX18 may contribute to the structural integrity of the nucleolus, particularly in the context of DFC and GC. GC phase is known for enriching negatively charged proteins such as NPM1 and NCL, and phase separation of NPM1 is enhanced by positively charged peptides52,53. As DDX18 binds to NPM1 both in vitro and in vivo (Figs. 3a–c, S3a, b), we compared the biophysical properties of DDX18 and NPM1 by testing their droplet miscibility. We found that NPM1-RFP cannot form droplets in vitro without the PEG crowding reagent (Fig. 4a), as reported54. Interestingly, adding NPM1-RFP solution without droplets to the preformed DDX18 droplets, we observed a profound “core-shell” like structure containing homogeneous DDX18-GFP and NPM1-RFP mixture in the same compartment, phase-separated from the empty core structures, mimicking the three-layered structure of the nucleolus (Fig. 4b). The hollow-like condensates also underwent shell fusion and cavity fusion (see “Supplementary Movie”). In contrast, neither homogenous droplets nor “core-shell” structures were observed upon mixing preformed DDX18-GFP and RFP-FBL droplets together (Fig. S4a, b), verifying the immiscibility of DDX18 and FBL droplets. The observed differential miscibility of DDX18 with NPM1 and FBL droplets was consistent with NPM1 and FBL being the most representative components of the phase-separated GC and DFC regions within the nucleolus, respectively38. To further probe the potential contribution of DDX18 to nucleolus architecture through its multiphase liquid (im)miscibility, we simultaneously mixed DDX18-GFP, NPM1-CFP, and RFP-FBL recombinant proteins. We observed a co-existing multiphase structure mimicking the regular nucleolus structure (Fig. 4c, top panel). As RNA helicase activity is critical for DDX18 functions (Figs. 1i, S2h–j), we tested how DDX18-bound RNAs (rRNA and snoRNA) may contribute to forming this particular multiphase structure. Intriguingly, the introduction of snoRNA or rRNA leads to a significant increase in the proportion of structures resembling the nucleolus (Fig. 4c, middle and bottom panels). This observation suggests a dynamic interplay between these RNA components and the formation of nucleolar-like structures.
a, b Biophysical properties of the mixtures of purified DDX18-GFP (20 μM) and NPM1-RFP (10 μM) recombinant proteins in vitro in a buffer comprised of 150 mM NaCl, 25 mM Tris-HCl, pH 8.0. Adding NPM1-RFP solution without PEG (i.e., no droplets formed) to the preformed DDX18 (a) established droplets with “core-shell” like structure, imaged with RFP, GFP, and merging RFP/GFP (b). The experiment was repeated independently twice with similar results. c Biophysical properties of the mixtures of purified DDX18-GFP (20 μM), NPM1-CFP (10 μM), and RFP-FBL (20 μM) with or without rRNA/snoRNA in a buffer comprised of 150 mM NaCl, 25 mM Tris-HCl, pH 8.0, detected by GFP, CFP, and RFP, respectively. Merged images and quantification of the formation of indicated core-shell structures are shown, where the percentages reflect the proportion of fields of view exhibiting the core-shell structures. The experiment was repeated independently twice with similar results. d Images of normal (shLuc) and disrupted (shDDX18#1) nucleolar condensate organization. FBL labels the dense fibrillar component (DFC); NPM1 labels the granular component (GC). The experiment was repeated independently twice with similar results. e Quantification of immunostaining images from control and KD cells (shDDX18#1 and shDDX18#3) showing the percentage of inclusive DFC localized cells (represented by the top part of panel d) or exclusive DFC localized cells (represented by the bottom part of (d). Quantifications were performed using 152 cells from shLuc, 211 cells from shDDX18#1, and 202 cells from shDDX18#3, based on three technical replicate cultures. Data are presented as mean values \(\pm\) SD. f A proposed model showing that DDX18 coordinates with NPM1 to safeguard nucleolus organization. (Created in BioRender. Malik, V. (2025) https://BioRender.com/v27l155). Source data are provided as a Source Data file.
To further understand its nucleolar functions in maintaining hESC pluripotency in vivo, we addressed how DDX18 depletion may modulate nucleolar architecture by immunostaining and 3D-SIM imaging in control and DDX18 KD hESCs. In hESCs with control KD (shLuc), the DFC marker FBL is completely encompassed (or included) in the NPM1-marked GC as immiscible multiphase droplets (Fig. 4d, top). In contrast, upon DDX18 depletion (shDDX18), the DFC region starts “leaking/escaping” (or excluded) from the GC and forming “nucleolar cap” or “nucleolar segregation” like structures (Fig. 4d, bottom) without significant changes of the core nucleolar protein levels including the GC markers NPM1 and NCL, and the DFC marker FBL (Fig. S4c, d). Quantification of the representative images on such “inclusive/exclusive” structures indicated that more than 40% of the DDX18 KD hESCs, compared to less than 10% of the control KD hESCs, showed mislocalization of FBL (Fig. 4e). Importantly, mislocalization of FBL, coincident with nucleolar enlargement, was also observed more prominently in the mesendoderm lineage than in the ectoderm lineage upon the directed trilineage differentiation of hESCs (Fig. S4e, f).
Our data thus establish a critical role of DDX18 in maintaining hESCs by preserving the structural integrity of the nucleolus through its multiphase liquid miscibility and immiscibility with the key GC and DFC proteins NPM1 and FBL, respectively (Fig. 4f, left). DDX18 depletion in hESCs, under the KD or differentiation condition, results in the mislocalization of DFC as “nucleolar caps” protruding from the GC (Fig. 4f, right) and lineage-specific nucleolar enlargement (Fig. S4e), causing the defects in RiBi and translation and, consequently, the loss of hESC pluripotency with the skewed differentiation towards mesendoderm lineage.
DDX18 restricts perinucleolar heterochromatin organization via its liquid miscibility and immiscibility with NPM1 and HP1 droplets, respectively
Nucleolar structure alteration is reported to cause chromatin reorganization55,56,57. During ESC differentiation, the initially dispersed H3K9me3-marked and HP1-containing heterochromatin foci (i.e., PNH) start accumulating around nucleoli5,20,58,59. Tethering heterochromatin at nucleoli promotes transcriptional activation of differentiation genes, leading to the exit of pluripotency in mESCs20. We thus asked whether the PNH formation could be facilitated by DDX18 KD, leading to differentiation of hESCs as we observed (Fig. 1). Indeed, we found that DDX18 depletion led to the redistribution of dispersed HP1α/β foci, characteristic features of heterochromatin, from nucleoplasm in control cells (shLuc) closer to nucleolus in DDX18 KD cells (shDDX18) (Fig. 5a, b), colocalizing with heterochromatin-associated H3K9me3 (Fig. S5a), indicative of the PNH accumulation. These findings establish the requirement of nucleolus structural integrity for restricting PNH formation and maintaining the pluripotent dispersed heterochromatin state.
a, b DDX18 depletion leads to perinucleolar heterochromatin organization in hESCs. Representative images of immunostaining for nucleolar NPM1, nucleolus-associated heterochromatin markers HP1α and HP1β, nuclear DAPI, and their merge are shown (a). The yellow arrows, labeled from I to II, indicate the direction of ImageJ immunofluorescence signal quantification shown as line charts (b). Black arrows indicate that HP1α and HP1β can dock at or enter the nucleolus when DDX18 is depleted. The experiment was repeated independently twice with similar results. c Images of the mixtures of purified recombinant proteins DDX18-GFP (10 μM) and RFP-HP1α (40 μM) (top) and NPM1-CFP (10 μM) and RFP-HP1α (40 μM) (bottom) in a buffer comprised of 150 mM NaCl, 25 mM Tris-HCl, pH 8.0 and 5% PEG8000, detected by respective reporters. The experiment was repeated independently twice with similar results. d, e Images of the mixtures of purified DDX18-GFP (10 μM), NPM1-CFP (10 μM), and RFP-HP1α (40 μM) in a buffer comprised of 150 mM NaCl, 25 mM Tris-HCl, pH 8.0 and 5% PEG8000, detected by respective reporters under immunofluorescence microscope (d) and the line chart showing the ImageJ quantified intensity of NPM1-CFP, RFP-HP1α and DDX18-GFP fluorescence signals (e) along the direction indicated in (d). The experiment was repeated independently twice with similar results. f, g DDX18 KD results in centromere clustering around the nucleolus. Representative images for nucleolus (NPM1, red), centromeres (CENPA, green), and DNA (DAPI, blue) (f) quantification of the number of nucleolus-localized centromeres under control and DDX18 KD conditions (g) are shown. Each dot in g represents one cell. ***P < 0.001 (The P-values were calculated using a two-sided t-test and indicated). h ChIP-qPCR analysis of NPM1 genomic occupancy at the centromere locus in control (shLuc) and DDX18 KD (shDDX18#1, shDDX18#3) hESCs. Data are mean±SD of three independent experiments. Chromosome Y serves as a negative control for female H9 hESCs. IgG serves as the ChIP negative control. i A proposed model showing that DDX18 restricts centromere from clustering heterochromatin organization around the nucleolus. (Created in BioRender. Malik, V. (2025) https://BioRender.com/v27l155). Source data are provided as a Source Data file.
To explore the mechanisms by which DDX18 restricts PNH formation, we first sought to analyze the (im)miscibility of the droplets formed by GC phase proteins and the heterochromatin protein HP1α, given that HP1α phase separation regulates the liquid droplet-like properties of heterochromatin60,61,62,63. By mixing RFP-HP1α with DDX18-GFP and NPM1-CFP, respectively, we observed that HP1α droplets were miscible with both DDX18 and NPM1 in droplet formation (Fig. 5c). NPM1 contains two evolutionarily conserved HP1α binding motifs (PXXVXL and PXVXL) (Fig. S5b) with reported roles in regulating HP1α oligomerization and promoting the formation of phase-separated droplets of HP1α60, explaining the miscibility between HP1α and NPM1 droplets. The fact that PNH is not accumulated in undifferentiated hESCs despite normal expression levels of heterochromatin HP1 proteins suggests additional factors may act upon HP1α and NPM1 to restrict their liquid miscibility. We thus tested DDX18 action in this context by simultaneously mixing DDX18-GFP, NPM1-CFP, and RFP-HP1α recombinant proteins to explore their multiphase organization behaviors. We found that NPM1 preserves the homogeneous droplet formation with DDX18 (merged image of the cyan droplets in Fig. 5d), consistent with our prior in vitro observation (Fig. 4c). However, NPM1 suddenly lost its liquid miscibility with HP1α when DDX18 was present with HP1α being pushed outside of the DDX18/NPM1 condensates, resulting in the formation of a shell-like structure (Fig. 5d, e). These data indicate that, while maintaining its liquid miscibility with NPM1 droplets for GC integrity, DDX18 prevents the HP1α-NPM1 droplet miscibility, hindering the PNH accumulation at GC (see Discussion).
Next, we addressed why and how the heterochromatin prefers to dock/accumulate at the nucleolus region during ESC differentiation after DDX18 depletion. As multiphase organization could be regulated by interconnected complexes (nodes) of multivalent proteins63,64, we sought to find the connecting node between PNH and the nucleolus. In mESCs, it has been reported that transcriptionally repressive regions near centromeres are often closer to the nucleolus21. Within the human genome, 45S rDNA tandem repeats are localized as rDNA gene arrays at the short arms of five chromosomes (Chr13, 14, 15, 21, and 22) flanked by the centromere and telomere (Fig. S5c). We detected centromere positioning by staining with the centromere markers CENPA and CENPT and observed a significant increase of centromere clustering around the nucleolus upon DDX18 depletion (Figs. 5f, g, S5d, e). Genome-wide occupancy of DDX18 by ChIP-seq revealed that DDX18 mainly binds to the rDNA transcribed regions (Fig. S5f), which is consistent with our previous study in mESCs19. In contrast, it is well established that the heterochromatin proteins HP1α/β bind to centromeres/pericentromeres65 and that the GC protein NPM1 binds to the centromere66,67,68,69. Thus, we hypothesized that centromere and centromere-binding proteins from the nucleolus might provide the connecting node for PNH to dock around the nucleolus. Indeed, we found that DDX18 depletion resulted in increased centromere binding of NPM1 within hESCs (Fig. 5h).
Together, these data support a model whereby DDX18 coordinates with NPM1 to safeguard nucleolar GC integrity and restrict centromere clustering and PNH formation (Fig. 5i, left). When DDX18 is absent or depleted, leading to the nucleolar cap formation (Fig. 4d, e), NPM1 droplets become miscible with HP1 droplets with increased centromere binding, promoting heterochromatinization (Fig. 5i, right), an early event required for ESC differentiation20.
DDX18 depletion results in nuclear reorganization
To address how chromatin may be reorganized at a genome-wide scale contributing to changes in gene expression and, consequently, the differentiation of DDX18 KD hESCs, we applied the Hi-C seq technology to generate chromatin interaction data in control and DDX18 KD cells. Hi-C data allow the partition of the genome into two compartments called A/B compartments, containing relatively active (A) and inactive (B) regions, respectively64. The saddle plots (Fig. S6a) revealed that DDX18 depletion leads to moderate changes in A/A (an increase from 1.369 to 1.419) and B/B (a decrease from 1.346 to 1.307) homotypic interactions, indicating a decrease in chromatin interaction within heterochromatin. In contrast, more minor changes in A/B or B/A heterotypic interaction were observed (Fig. S6a). Further using principal component analysis (PCA) at 100-kb resolution to classify the genome into A and B compartments, we identified regions that changed compartments upon DDX18 KD. We found 3.2% of regions switched from A to B and 3.3% from B to A (Fig. S6b). However, integrating these results with RNA-seq data showed that over 90% of significantly up- or down-regulated genes were not in compartment-switching regions (Fig. S6c). We then analyzed gene expression changes in 100-kb bins based on their PC1 value changes, indicating shifts towards active or inactive states. We found that a small proportion of bins with extreme PC1 changes correlated with significant gene regulation, with only 195 and 17 genes (out of >2000 differentially expressed) in bins showing PC1 changes greater than −2 and +2, respectively (Fig. S6d). Further analysis of upregulated lineage-associated genes showed most were in regions with moderate PC1 changes (−0.5 to +0.5) (Fig. S6e). This weak correlation between PC1 changes and transcription was also observed globally (Fig. S6f). Together, our results suggest that, while locus-specific transcriptional changes resulting from the compartment switch do occur (Fig. S6g), a vast majority of transcriptional changes observed upon DDX18 KD arise from mechanisms independent of compartment organization. This observation is consistent with the fact that DDX18 is an RBP rather than a typical transcription factor or chromatin remodeler and that DDX18 mainly binds to the rDNA transcribed regions in the nucleolus (Fig. S5f). Hence, it is more likely to influence gene expression via its unique nucleolar functions.
A/B compartments are primarily defined by intrachromosomal interactions within each chromosome70. By analyzing the intrachromosomal interaction for all five rDNA-containing chromosomes (Chr13, 14, 15, 21, and 22), we only found two interacting regions (R1 and R2) of Chr21 showing significantly increased contact frequencies after DDX18 KD (Figs. 6a, S6h). Both interacting regions are close to the pericentromere. R1 is mediated by rDNA closer locus (Site I, Chr21: 9.4 MB-10.1 MB) and chromosome arm locus (Site III, Chr21: 24.9 MB-26.5 MB), while R2 is mediated by pericentromeric locus (Site II, Chr21: 14.7 MB-15.1 MB) and chromosome arm locus (Site III, Chr21: 24.9 MB-26.5 MB) (Fig. 6b). Interestingly, we found that Site III is localized in the lamina-associated domain (LAD)71 (Fig. S6i). The increased interactions of R1 and R2 in DDX18 KD relative to control KD samples suggest that DDX18 depletion could affect nucleolus-associated domain (NAD) and LAD exchange (Fig. 6c). Together with our findings that DDX18 depletion impairs nucleolus integrity (Fig. 4) and PNH organization (Fig. 5), we concluded that DDX18 could play a pivotal role in regulating centromere-telomere proximal rDNA-containing chromosome (i.e., Chr21) organization, i.e., heterochromatin formation.
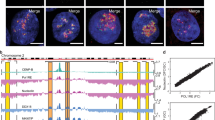
a, b Hi-C interaction heatmaps (a) and schematic diagram (b) depicting increased chromatin interactions at Chr21 region R1 (I↔III) and R2 (II↔III) in DDX18 KD vs. control KD hESCs. c A proposed model. See the main text for a detailed explanation. d Boxplots showing the change in the strength of significant interactions (the numbers of interactions are indicated) upon DDX18 KD categorized by chromosome arms. The edges of the box represent the 25th and 75th percentile, while the line in the middle of the box is the median. The whiskers extend from the box edges to 1.5 times the interquartile range. e Heatmap showing log10(ratio) of mean normalized interchromosomal contacts per 250-kb bin in DDX18 KD vs. control KD hESCs. Acrocentric chromosomes are highlighted in black boxes. f Illustration of the contact domains and chromatin loops for a 3 Mbp region of chromosome 7p containing HOXA cluster genes from Hi-C data with the epigenetic modifications of the HOXA cluster from public datasets shown. Circles on the matrix denote increased interaction frequency of the HOXA cluster genes with nearby genes in DDX18 KD vs. control KD hESCs. g, h Quantification (g) and representative DNA FISH images (h) of the control and DDX18 KD cells for HOXA9-13 alleles (red) compared with nucleolar (NPM1, green) and nucleoplasm (DAPI, blue). The total numbers (n) of nuclei from two independent experiments were scored. i, j Representative confocal microscopy merged images of RNA-FISH detecting HOXA9 (i) and HOXA11 (j) mRNAs in control and DDX18 KD H9 hESCs. Dotted circles indicate nucleoli detected by Nucleolus Bright Green Dye reaction to predominantly nucleolar and cytoplasmic RNAs. k, l Quantification of HOXA9 (k) and HOXA11 (l) mRNAs RNA-FISH signals at nucleolar (NAD) and nuclear periphery (LAD) proximities in control and DDX18 KD hESCs (representative cells are shown in i, j). RNA-FISH signals outside the nuclear periphery were not counted. Equal numbers of shLuc and shDDX18 cells (N = 39 for HOXA9 and N = 30 for HOXA11), randomly selected from three independent experiments, were analyzed for signal quantifications (mean ± SD). All cells were counterstained with DAPI. The red and green arrows indicate the increase and decrease trends, respectively. Source data are provided as a Source Data file.
Since the nucleolus is a hub for interchromosomal interactions, we next examined the contacts between ribosomal DNAs (rDNAs) from nucleolar organizer region (NOR)-bearing chromosomes (Chr13, 14, 15, 21, and 22). The analysis of chromatin interactions in the short arms of NOR-bearing acrocentric chromosomes is challenging due to their repetitive nature and gaps in the human reference genome GRCh38. Utilizing the mHi-C pipeline72, which retains multimapping reads, enabled us to analyze rDNA interactions, revealing improved detection in the repetitive short arms of acrocentric chromosomes 15, 21, and 22 (Fig. S6j). However, gaps in the GRCh38 reference genome (Fig. S6k top) still limited the analysis. Remapping Hi-C data to the updated T2T-CHM13 reference genome73 allowed us to fill many gaps, enhancing the resolution of contact maps in acrocentric chromosomes (Fig. S6k bottom). Focusing on the impact of DDX18 KD, we detected significant changes in chromatin organization, with 31% of interactions shared between control KD and DDX18 KD conditions (Fig. S6l). Next, to specifically study the impact of DDX18 KD on rDNA-containing chromosomal interactions, we initially calculated the change in interaction strength (observed/expected counts) upon DDX18 KD of all shared interactions across the genome and categorized our results by chromosome arm. Interestingly, this analysis showed a preferential weakening of Hi-C interactions, specifically in the short arms of acrocentric chromosomes containing rDNA repeats. In contrast, long arms and non-acrocentric chromosomes showed no significant change (Fig. 6d). Importantly, an increase in interchromosomal contacts among acrocentric chromosomes was observed (Fig. 6e and Fig. 6c model), suggesting that DDX18 plays a crucial role in organizing NORs.
Nuclear reorganization upon DDX18 depletion results in developmental gene expression changes
To understand how nuclear reorganization involving LAD-NAD exchange and NORs may lead to gene expression changes in DDX18-depleted hESCs, we studied the impact of DDX18 KD on the HOXA gene cluster for the following reasons. First, the HOXA gene cluster is in a gene-poor region of Chr7 that, in human cancer cells, is sequestered by the Nup93 sub-complex for HOXA repression at the nuclear periphery (LAD). The derepression of HOXA genes upon Nup93 depletion results in the LAD to nuclear interior (closer to NAD) relocation74,75. Second, the HOXA gene cluster represents the CTCF-mediated chromatin loops and chromatin status changes during embryonic development and disease progression76,77,78. Intriguingly, a published study reported the interchromosomal interaction between the NOR-bearing Chr21 and HOXA gene-residing Chr7 and its resultant t(7;21) translocation in a human leukemia patient79. Third, the HOXA cluster genes were upregulated in our DDX18 KD hESCs (Fig. 1h) coincident with weakened interchromosomal interactions of HOXA-residing Chr7 with Sites II and III in Chr21 (Fig. S6m), suggesting that the DDX18 KD effect on the 3D conformation of those NOR-bearing chromosomes (e.g., Chr21) could indirectly affect chromatin organization (e.g., Chr7) and developmental gene expression (e.g., HOXA genes) (see Fig. 6c model).
To substantiate our model whereby DDX18 depletion could cause the NAD and LAD exchange and HOXA gene derepression (Fig. 6c), we performed the following studies. First, we performed additional Hi-C data analysis on chromatin interactions involving the HOXA locus and revealed that, despite the lack of a major compartment switch in the DDX18 KD hESCs (Fig. S6a–c), loops involving the HOXA locus within Chr7 appeared strengthened in the DDX18 KD hESCs (Fig. 6f). In addition, DDX18 KD hESCs showed reduced interaction frequencies within the HOXA gene cluster (Fig. S6n, o, left panels) and increased the external interactions between the HOXA cluster and the nearby genes (e.g., HOTTIP) (Fig. S6n, o, right panels). Furthermore, DNA FISH analysis revealed that HOXA9-13 loci are relocated away from the nucleolus (NAD) (i.e., reduced FISH signals) towards the nuclear periphery (LAD) (i.e., increased FISH signals) in DDX18 KD relative to control KD hESCs (Fig. 6g, h). Importantly, RNA FISH analysis also detected the corresponding upregulation of HOXA transcripts, exemplified by increased RNA FISH signals per nucleus of HOXA9 (Fig. 6i, k) and HOXA11 (Fig. 6j, l) at the nuclear periphery (LAD), while their NAD signals were slightly and appreciably reduced. These findings demonstrate that DDX18 depletion modulates the heterochromatin via the LAD and NAD exchanges associated with the NOR-bearing chromosomes, resulting in nuclear reorganization and derepression of developmental genes (e.g., Chr7 reorganization at the HOXA locus; Fig. 6c model).
Nucleolus-specific NoCasDrop restructures the genome and controls gene expression
We have thus far established the roles of DDX18 in maintaining the intrinsic nucleolar architecture (Fig. 4f) and extrinsic heterochromatin organization (Fig. 5i) for pluripotency and lineage commitment/differentiation of hESCs. However, it remains an open question whether the extrinsic mechanism, i.e., centromere clustering and PNH accumulation, is the cause or consequence of hESC differentiation. Therefore, inspired by CasDrop, a recently developed method successfully applied in mammalian cells to activate gene expression80, we created NoCasDrop (Nucleolus specific CasDrop) to address whether tethering heterochromatin towards nucleolus would impair hESC pluripotency.
To this end, we took advantage of the nucleolus-specific phase separation by fusing full-length (FL) DDX18 and its individual domains (NIDR, HD, and CIDR) with the dCas9-mCherry (Figs. 7a, S7a). We found that both FL and NIDR of DDX18 fusion, but not the other domains, could recruit dCas9-mCherry protein to the nucleolus (Fig. S7a). However, FL, but not NIDR, could activate the pre-rRNA transcription (Fig. S7b). Therefore, to avoid any indirect effects resulting from rRNA overexpression, we used NIDR NoCasDrop for our further study (Fig. 7a). To overcome the challenge of manipulating a whole trunk of chromosome fragment by a single dCas9, we targeted tandem repeats in centromere and satellite DNA by infecting the NIDR NoCasDrop stable cell line with a human centromere-specific alpha-satellite sgRNA (sgAlpha) and a non-specific control sgRNA (sgNS), both of which have been functionally validated in human cell lines previously81. This allows multiple copies of dCas9 to coordinately target the same gene locus by the same sgRNA leading to a combined/concerted action in controlling the chromosome localization (Fig. 7a). Then, we performed immunostaining after 48 h of sgRNA infection and found that the centromere (Fig. 7b, c) and heterochromatin maker HP1α (Fig. 7d, e) could be tethered to the perinucleolar region by NoCasDrop and α-satellite specific sgRNA. More importantly, we found the derepression of developmental genes (e.g., HOXA cluster genes) by this NoCasDrop-mediated perinucleolar centromere clustering (Fig. 7f). In contrast, pluripotency genes and RiBi genes were minimally affected within the tested time window (Fig. S7c), indicating the specificity of NoCasDrop and corroborating the Hi-C data. These results also suggest that the derepression of HOXA cluster genes is an early event leading to the loss of pluripotency in DDX18 KD hESCs.
a Schematic diagram of NoCasDrop (Nucleolus CasDrop). Repetitive loci such as centromeres can be tethered by α-satellite specific sgRNA and dCas9-mCherry-NIDR (DDX18) to nucleolus hub in vivo. (Created in BioRender. Malik, V. (2025) https://BioRender.com/v27l155). b, c Alpha (α)-satellite sgRNAs (sgAlpha) and NoCasDrop recruit centromere to the nucleolus. Representative images for nucleolus (NPM1, red), centromeres (CENPA, green), and nucleus (DAPI, blue), as well as merged images (b) and quantification of the number of centromere foci around nucleolus from more than 50 cells (c) are shown. Each dot in c represents one cell. ***P < 0.001 (The P-value was calculated using a two-sided t-test). sgNS acts as a non-specific sgRNA control. d, e NoCasDrop with sgAlpha recruits perinucleolar heterochromatin (HP1α, magenta) at the nucleolus (NPM1, green) in hESCs. Representative images of immunostaining for nucleolar NPM1, nucleolus-associated heterochromatin markers HP1α and HP1β, nuclear DAPI, and their merge are shown (d). The yellow arrows, labeled from I to II, indicate the direction of ImageJ immunofluorescence signal quantification, as shown in line charts (e). The experiment was repeated independently twice with similar results. f qRT-PCR analysis of HOXA cluster genes upon sgAlpha and NoCasDrop treatment. sgNS acts as a non-specific control. g, h A summary model. Under the pluripotent state, DDX18 is highly expressed. It maintains the nucleolar structural integrity while restricting perinucleolar heterochromatin (PNH) formation through its strong liquid miscibility with the core GC factor NPM1 and relatively weaker miscibility with heterochromatin protein HP1 and immiscibility with the core DFC factor FBL (g). When DDX18 is depleted (e.g., through genetic manipulation) or downregulated (e.g., during hESC differentiation), NPM1 increases its binding to the centromere and acquires the liquid miscibility with HP1, resulting in the escape of DFC from the center of the nucleolus and the formation of “nucleolar caps” with accumulated PNH and altered nuclear organization. Consequently, intrachromosomal and interchromosomal reorganization leads to the derepression of developmental genes (e.g., HOXA) (h). (Created in BioRender. Malik, V. (2025) https://BioRender.com/v27l155). Source data are provided as a Source Data file.
These data reinforce a major function of DDX18 in restricting centromere clustering around the nucleolus for the repression of developmental genes (e.g., HOXA cluster genes) in undifferentiated hESCs and provide a molecular tool to tether centromere clustering at the perinucleolar region for cell fate manipulation.
Discussion
Accumulating evidence indicates that the nucleolus functions as the ribosome biogenesis (RiBi) site for protein synthesis and plays a role in chromatin organization and transcriptional regulation in mESCs47,82. Using a human pluripotent stem cell paradigm, we demonstrate that, as a nucleolus-specific RNA helicase, DDX18 controls RiBi and global translation while simultaneously preventing PNH formation through the modulation of nucleolus phase separation in maintaining human pluripotency. Specifically, under the pluripotent state, DDX18 maintains the nucleolar structural integrity and prevents PNH formation through its liquid miscibility with the core GC factor NPM1 and its immiscibility with the core DFC factor FBL (Fig. 7g). In contrast, when DDX18 is depleted (e.g., through genetic manipulation) or downregulated (e.g., during development or hESC differentiation), NPM1 increases its binding to the centromere and acquires the liquid miscibility with HP1, resulting in the escape of DFC from the center of the nucleolus and the formation of “nucleolar caps” with accumulated PNH (Fig. 7h). While specific nucleolar caps were reported to be located adjacent to the distal junction sequences nearby the short arm of the acrocentric chromosome anchored in PNH83, it remains to be determined whether the “nucleolar caps” from those escaped DFCs in DDX18 KD hESCs (Fig. 4f) are the source of or overlapped with PNH. Notably, the mesendoderm lineage-specific increase of FBL mislocalization and enlargement of nucleoli in directed trilineage differentiation of hESCs (Fig. S4e, f) are reminiscent of enhanced nucleolar caps (Fig. 4d, e) and skewed mesendoderm differentiation (Figs. 1 and S1) of DDX18 KD cells. In addition, surface tension is crucial in biological processes, affecting how liquid droplets behave within cells. The phase separation of the initially miscible NPM1 and HP1 droplets in the presence of DDX18 (compare Fig. 5d with Fig. 5c) suggests that DDX18 may prevent the NPM1-HP1 droplet formation in pluripotent cells by its stronger interaction/miscibility with NPM1 than with HP1, evidently supported by DDX18-NPM1 droplets are being more homogeneous (Fig. 4a–c) than DDX18-HP1 droplets (Fig. 5c top; note the merged unpure yellow droplets with tainted red and green dots). Thus, our study uncovers a previously unappreciated role of the nucleolar RNA helicase DDX18 in coordinating nucleolus phase separation and chromatin organization to control the pluripotency of hESCs.
We previously reported that Ddx18 maintains mESC pluripotency by antagonizing PRC2 to protect rDNA from epigenetic silencing, ensuring high RiBi in mESCs19. However, this epigenetic mechanism is likely not active in hESCs. First, Ezh2/EZH2 depletion affects mESC differentiation but causes severe self-renewal and growth defects in hESCs84,85. Second, the differentiation phenotype of DDX18 loss can be partly rescued by PRC2 depletion in mouse19 but not human (Fig. S7d) ESCs. Third, the DDX18-PRC2 partnership is detectable in mouse19 but not human (Fig. 3a; Supplementary Data 3) ESCs. Fourth, mouse and human ESCs represent distinct pluripotency states at functional86,87 and molecular88 levels, indicating that human pluripotency cannot be inferred from mouse studies. Our findings provide molecular insights into nucleoli’s role in chromatin organization, beyond being a RiBi compartment, in controlling stem cell pluripotency and lineage commitment.
The nucleolar GC protein NPM1 maintains mESCs, with its loss resulting in differentiation89. NPM1 forms liquid-like droplets in the presence of RNA52 and is required for centromere clustering and anchoring to the nucleolus90. An outstanding question is how such an essential and ubiquitous nucleolar function can meet the specific needs of different cell types and/or during various developmental stages. DEAD-box (DDX) or DEAH-box (DHX) RNA helicase proteins are evolutionarily conserved with cell/tissue-specific expression profiles and prominent roles in LLPS23. In contrast to DDX18, which maintains the undifferentiated state in hESCs, DHX9 is dispensable for ESC maintenance but required for differentiation, processing IGS-rRNA into mature rDNA promoter RNA (pRNA)91 to promote TIP5 interaction and rRNA gene silencing, leading to low RiBi in differentiated cells92. This pRNA maturation initiates heterochromatin formation in the nucleolus, maintaining some undifferentiated ESC features while promoting the exit from pluripotency20. We demonstrate that DDX18 binds to both rRNAs and snoRNAs, the latter assisting the 2’-O-methylation of the former93,94. While the interaction of rRNA52 or nucleolar localized lncRNA95,96 with nucleolar proteins was reported to contribute to their phase separation, our study also implicates snoRNAs and likely rRNA modifications in DDX18’s coordination of nucleolar multiphase separation. Furthermore, the requirement of the RNA helicase activity of DDX18 to rescue the DDX18 KD defects also suggests the involvement of rRNA/snoRNAs secondary structures in nucleolar LLPS properties. Future investigations are needed to unravel these details further.
It is noteworthy that HOXA clusters can change their nuclear location under physiological or leukemic conditions97,98. Intriguingly, supporting the inter-chromosomal interaction between the NOR-bearing Chr21 and HOXA gene-residing Chr7 in hESCs (Figs. S6m, 6c), the interaction between Chr21-Chr7 and its resultant t(7;21) translocation were reported in a case of acute myeloid leukemia79. In addition, while pluripotent cells were known to be relatively depleted for H3K9me3-marked constitutive heterochromatin99,100, differentiation of hESCs is indeed associated with H3K9me3 accumulation at the HOXA loci for their activation, as reported30, a finding that is itself counterintuitive but nonetheless supports our hypothesis. More importantly, published studies have established that HOXA genes, especially HOXA9, play an important role in the differentiation of hESCs towards mesodermal and hematopoietic fates101. In addition, mouse HoxA genes were also reported to play an essential role in specifying mesendoderm lineage during mESC differentiation and embryonic development102. Overall, HOXA genes’ upregulation appears to be an important step in the exit from pluripotency and commitment to the lineages derived from mesendoderm (reviewed by31,32).
Our findings contribute to an increased understanding of the nucleolus in the nuclear (i.e., genome/chromatin) organization, which is currently under-investigated compared with the well-recognized role of the nuclear periphery in genome organization. Heterochromatic domains are always associated with the nuclear lamina (LADs) and nucleoli (NADs). Therefore, manipulating the chromatin organization within the nucleus to control gene transcription is desirable for studying cell biology. However, the membrane-less compartment and LLPS nature of the nucleolus24 render it technically more difficult to identify NADs than LADs71,103, a major challenge in studying NADs. Here, we developed a nucleolus-specific NoCasDrop technique to dissect how phase separation might target genomic domains in the nucleolus. By using NoCasDrop, we successfully tethered the centromere to the nucleolus and reprogrammed the developmental gene expression, providing direct support for DDX18 functions in controlling human pluripotency by maintaining nucleolus phase separation and restricting centromere clustering around nucleolus in hESCs. The molecular features of our NoCasDrop-treated hESCs are analogous to those of mESCs with nucleolar pRNA-tethered heterochromatin at rDNA loci, initiating structural remodeling toward a highly condensed nuclear heterochromatin, resulting in the upregulation of genes involved in cell differentiation while maintaining the undifferentiated state with normal ESC proliferation, expression of pluripotency genes, morphology, and AP staining20. Our findings and the NoCasDrop tool should be applicable for further studies of nucleolar functions in development and disease and for cell fate manipulation.
Methods
Cell lines
H9 and HES2 hESCs were initially obtained from WiCell Research Institute and maintained in the Wang lab.
DDX18 C-terminal GFP knock-in cell line (H9-DDX18GFP) was established by the CRISPR-Cas9 method and maintained in the Wang lab. First, H9 hESCs were transfected with a plasmid expressing Cas9/sgRNA (5’-GCAAGAAATCATCTGACAGC-3’) in the presence of a recombination GFP donor DNA flanked by right and left homology arms (HA) of DDX18. Then, three days after transfection, GFP-positive cells were selected by FACS, expanded, and validated by immunoblotting.
The 3xFlag-DDX18 transgenic cell line was established by PiggyBac transposon-mediated reversible genetic modification strategy and maintained in Wang lab. We co-transfected PB-3xFlag-DDX18 with PiggyBac transposase (PBase) to create the stable hESC lines (H9 and HES2) with the selection for hygromycin resistance. DDX18 gene and protein expression levels were confirmed by qPCR and western blotting analyses.
Plasmid construction
shRNA cloning
shRNAs were designed using the RNAi Consortium (TRC) siRNA selection program from MIT Whitehead Institute (http://sirna.wi.mit.edu/). pLKO.1-GFP is a replication-incompetent lentiviral vector chosen by the TRC for the expression of shRNAs. Oligos were first annealed and then cloned into pLKO.1-GFP vector following the protocol described by Addgene (www.addgene.org). Target sequences: DDX18 shRNA1 (shDDX18#1), CCAAGGTTCCATTAAGTGAAT; DDX18 shRNA3 (shDDX18#3), CGTGCATACCTATGGCTTGAT; shRNA targeting the firefly luciferase was used as a control (shLuc).
cDNA cloning
Human DDX18 cDNA was purchased from OriGene and was amplified by PCR using primers designed with 5’ NheI and 3’ PacI restriction sites (Supplementary Data 6) and cloned into the PiggyBac (PB) vector with 3xFlag tag in it.
NoCasDrop plasmid construction
Full-length (FL) DDX18 and its domains (NIDR, HD, and CIDR) were amplified by PCR with the primers containing 5’ NotI and 3’ XhoI restriction sites (Supplementary Data 6). PCR products and the dCas9-mCherry vector70 were digested with restriction enzymes, purified, and ligated with T4 DNA ligase. All plasmids were confirmed by Sanger sequencing.
Construction of plasmids for recombinant proteins
Full-length human NPM1-RFP, NPM1-CFP, RFP-FBL, and RFP-HP1α were PCR amplified by G5 High Fidelity DNA Polymerases (EnzyArtisan, China) and then sub-cloned into a modified pET28b vector with a SUMO protein fused at the N terminus after the 6xHis tag by One-Tube Cloning Kit (EnzyArtisan, China). In addition, full-length human DDX18 and its truncations or mutants were PCR amplified and subcloned into a modified pET vector with an 8xHis tag followed by a GFP protein fused at the C-terminus. The plasmids were then extracted using the SPARKeasy Superpure Mini Plasmid Kit (Sparkjade Science Co., Ltd., China), and all inserted sequences of plasmids were validated by Sanger sequencing.
Human ESC culture
All hESC lines were cultured at 37 °C in a controlled humidified atmosphere of 5% CO2, as described previously88. The hESC lines used were H9 and HES2 (WiCell), authenticated by the provider, and routinely tested negative for mycoplasma. hESCs were maintained on irradiated inactivated MEFs in hESC medium composed of DMEM/F12 (Corning) containing 20% Knockout serum (Invitrogen), penicillin/streptomycin, L-glutamine, non-essential amino acids, β-mercaptoethanol (β-ME, Sigma), and bFGF (R&D Systems). For feeder-free culture, hESCs were cultured in mTeSR Plus (STEMCELL Technologies) on Matrigel hESC-Qualified Matrix (Corning) coated wells and passaged using ReLeSR (STEMCELL Technologies) every 3 to 4 days.
EB differentiation
For non-lineage-specific differentiation, we used the previously described EB formation protocol104. H9 hESCs were cultured in a feeder-free mTeSR medium. The cells were treated with collagenase B (Roche) for 15–30 min and collected by scraping. After centrifuging, cell pellets were resuspended in differentiation media, StemPro 34 (Invitrogen) supplemented with 2 mM L-Gln, 4 × 10−4 M monothioglycerol (MTG), 50 μg/ml ascorbic acid, and 1 mM transferrin (Sigma). EBs were grown in ultra-low adherent plates (Costar), and the medium was changed every three days. After eight days of differentiation, EBs were collected, resuspended in DMEM 10%, and transferred to gelatin-coated dishes to allow them to attach and differentiate for 4–7 additional days. Whole-cell lysates were collected at specific time points during the differentiation.
Direct differentiation of human embryonic stem cells into trilineage
Human ESCs (H9) were differentiated into three germ layers using STEMdiff™ Trilineage Differentiation Kit (STEMCELL Technologies, Cat#05230). HESCs were cultured in mTeSR™1 medium and differentiated following the trilineage differentiation protocol. Cell differentiation markers for each lineage were detected by immunostaining. PAX6 antibody (Santa Cruz, sc-81649), Brachyury antibody (R&D, AF2085), and CXCR-4 antibody (Santa Cruz, sc-53534) were used for ectoderm, mesoderm, and endoderm, respectively.
Production of viral supernatants
Calcium phosphate transfection of 293 T cells protocol was used to produce viruses. 293 T cells were cultured in a DMEM complete medium (DMEM supplemented with 10% FBS, Glutamine, and Penicillin/Streptomycin). 293 T cells were seeded at a density of 12 × 106 cells per 10 cm plate in a DMEM complete medium without antibiotics 24 h before transfection. On the day of transfection, the culture was changed to 10 ml DMEM medium containing 25 nM chloroquine one hour before the transfection. The transfection mix was prepared by adding 14 µg target plasmid DNA, 7 µg pVSVg, 14 µg pCMVΔR8.2, and ddH2O to 438 µl in a 15-ml sterile tube, followed by adding 62 µl 2 M CaCl2 to a final volume of 500 µl. Next, 500 µl of 2 × HBS were added to the above transfection mix dropwise with gentle mixing. The whole mixture was incubated for 15 min at RT and then added directly to the 293 T cells dropwise through the medium. After 24 h incubation, the medium was changed without chloroquine for cell culture. Supernatants were collected 48- and 72-hours post-transfection. Viruses were concentrated using an Amicon Ultra-15 centrifugal filter unit (Millipore) at 3000 g at 4 °C for 20 minutes. Concentrated virus particles were aliquoted in sterile 1.5-ml centrifuge tubes and stored at –80 °C.
Virus infection
For virus infection, 30–50 μl of the concentrated virus was used to infect 500,000 hESCs in the presence of 6 μg/ml polybrene. A total of 1 ml of cells (~500,000) were plated per well of a 6-well plate coated with Matrigel. Cells were mixed with the virus gently and incubated overnight at 37 °C. After 20–24 h post-infection, the viruses were removed by changing to a fresh medium. After 48 h post-infection, shRNA-transduced cells showed GFP positivity under a fluorescent microscope, and puromycin (1 μg/ml for hESCs) was added to kill non-transduced cells. RNA and protein were isolated after four days of transduction.
Protein synthesis measurements using puromycin incorporation (SUnSET)
Puromycin incorporation was performed as previously described31. H9 hESCs with or without ectopic expression of shRNA-resistant DDX18 WT and the DQAD mutant were infected with DDX18 shRNA viruses. Four days later, control KD and WT or mutant rescue cells were incubated with puromycin (1 μM final concentration) for 30 min, followed by protein extraction and western blotting using an anti-puromycin antibody (MABE343, Millipore). Coomassie blue staining was used as the loading control.
Real-time quantitative PCR
Total RNA was extracted from cells using TRIzol Reagent (Invitrogen) and subsequently column-purified with the RNeasy Kit (Qiagen) and treated with RNase-free DNase (Qiagen). One microgram of total RNA was reverse-transcribed into cDNA using the qScript cDNA SuperMix (Quantabio). For ChIP-qPCR analysis, 1 ng ChIP-DNA was used for each PCR. All qPCR analysis was performed on the QuantStudio 5 PCR system with Fast SYBR Green Master Mix (Applied Biosystems). The results were analyzed and normalized to β-ACTIN gene expression.
Western blotting
Total cellular protein was extracted using RIPA lysis buffer (20 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1 mM EDTA; 1 mM EGTA; 1% NP-40; 1% sodium deoxycholate; and protease inhibitor cocktail) and then quantified using the Bradford method (Thermo Fisher). The protein lysates were separated by SDS–PAGE, electro-blotted onto polyvinylidene difluoride (PVDF) membranes (Millipore), probed with the indicated primary antibody followed by the HRP-conjugated secondary antibody, and then detected by enhanced chemiluminescence.
SILAC and mass spectrometry (SILAC-MS)
To perform the quantitative mass spectrometry-based whole-proteome comparison of control KD and DDX18 KD hESCs, we cultured control KD and DDX18 KD hESCs in SILAC heavy medium and SILAC light medium, respectively. For the SILAC heavy condition, hESCs were grown for three passages in DMEM/F12 with corresponding complete supplements but deficient in both L-lysine and L-arginine and supplemented with heavy 13C615N4 L-arginine and 13C615N2 L-lysine (Cambridge Isotope Laboratories). The medium was supplemented with 10% dialyzed FBS for SILAC (Thermo Fisher Scientific), and all other gradients followed the normal condition for conventional primed hESC culture (with regular L-lysine and L-arginine as SILAC light condition).
Complete labeling (>95%) was confirmed by MS before initiating the experiment. To quantitatively identify changes in protein levels upon DDX18 depletion, cells were infected with shRNA-expressing viruses and selected with puromycin for four days. After washing with ice-cold PBS, the control and DDX18-knockdown cells were harvested and counted. An equivalent number of SILAC-labeled cells (5 × 106) were lysed and sonicated in RIPA buffer on ice. After centrifugation at 14,000 rpm for 10 min at 4 °C, proteins of an equal number of cells were combined, and the concentrations were estimated using a BCA protein assay kit (Thermo Fisher). The proteins were boiled with SDS loading buffer and subjected to SDS-PAGE followed by colloidal blue staining. Gel fragments were excised, washed, and sent for liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) to identify proteins.
Affinity purification coupled with mass spectrometry (AP-MS)
To isolate endogenous DDX18-associated protein complexes, affinity purification coupled with mass spectrometry (AP-MS) was performed as described105 with some modifications. First, nuclear extraction was prepared from 10 × 15 cm dishes of 3xFlag-DDX18 hESCs with the nuclear extraction buffer [20 mM HEPES (pH 7.9), 25% glycerol (v/v), 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT (add fresh), 0.2 mM PMSF (add fresh), 1x protease inhibitor cocktail (add fresh)], then, nuclear extraction was pre-cleared with protein G agarose beads (Roche), using 100 μl beads (200 μl slurry) per 10 mg total protein. 25 μg of Flag antibody (Sigma, F1804) and 25 μg IgG (Millipore) per IP were incubated with the pre-cleared nuclear extraction in the cold room with rotation overnight without Benzonase nuclease to maintain the nucleic acid-mediated protein interactions. The next day, equilibrate protein G agarose beads (200 μl beads) with the nuclear extraction buffer, transfer the beads to the lysate/Ab mixture, and rotate in the cold room for 3 h. The immunoprecipitated samples were washed with wash buffer [20 mM HEPES (pH 7.9), 20% glycerol (v/v), 100 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT (add fresh), 0.02% NP40, 0.2 mM PMSF (add fresh), 1x protease inhibitor cocktail (add fresh)] five times, dry the beads by aspirating all the washing buffer using 20 μl tip at the last wash, beads were boiled with an equal volume of 2× SDS loading buffer for 5 min, and separated by SDS-PAGE. Finally, whole lanes were excised from the gel for LC-MS/MS to sequence and identify DDX18-associated proteins.
Immunoprecipitation (IP) and co-immunoprecipitation (Co-IP)
For immunoprecipitation, 2 μg Flag antibody was coupled to Dynabeads at 4 °C for 2 h in 200 μl lysis buffer, and nuclear extracts from 3xFlag-DDX18 hESCs were prepared and incubated with anti-Flag antibody overnight. For RNase A treatment, the immunoprecipitant was incubated in a lysis buffer containing 200 μg/ml RNase A (Sigma) at 37 °C for 10 min. The beads were washed four times with lysis buffer, resuspended in 1xSDS loading buffer, and boiled for analysis by SDS-PAGE. 3xFlag-DDX18 and NPM1 were identified by western blotting using anti-Flag M2 (Sigma) and anti-NPM1 (Santa Cruz, sc-47725) antibodies, respectively.
RNA-seq library preparation, sequencing, and data analysis
RNA was isolated with TRIzol (Invitrogen). RNA integrity was evaluated using a Eukaryotic RNA 6000 Nanochip on an Agilent 2100 Bioanalyzer (Agilent Technologies). Up to 1 µg of total RNA from each sample was used for library preparation with the TruSeq RNA Sample Preparation Kit (Illumina). A common adapter was used for all samples, and barcode sequences present in the reverse primers were introduced by 12–16 cycles of amplification. Each library was assessed for quality and size distribution using an Agilent High Sensitivity Assay bioanalyzer chip and quantified by PCR. Equimolar amounts of each barcoded library were mixed, and single-end sequenced on an Illumina HiSeq Sequencing System. For each sample, 14–21.7 M 50-nt reads were obtained, pre-processed with the FASTX-toolkit suite (http://hannonlab.cshl.edu/fastx_toolkit/), and aligned to the human genome (hg19 assembly) using TopHat mapper.
ChIP-sequencing and data analysis
ChIP-seq was performed as described previously106. Chromatin was sheared by sonication to 100-500 base pair fragments. ChIP DNA libraries were made following the Illumina ChIP-Seq Library preparation kit (New England BioLabs). Samples were subjected to Illumina Solexa Cluster Station sequencing. Sequencing was performed on Illumina HiSeq2500 with a single read of 100 nt. Images acquired were processed through the image extraction pipeline and were aligned to the human genome hg19 using Bowtie. The reads that mapped to multiple locations in the genome were discarded. To map DDX18 ChIP-seq data to the rDNA, we used the consensus DNA sequence of the 43-kb ribosomal locus from NCBI (GeneBank ID: U13369.1). Employing this specific 43-kb region, we utilized the Bowtie algorithm with standard parameters for mapping to the Hg19 human genome build. The coverage was then visualized using IGV.
Hi-C sequencing and data analysis
Human ESCs were infected with shLuc and shDDX18 viruses. Samples were collected on day 4. ESCs were washed with PBS and fixed with 2% formaldehyde for 10 min at RT. Hi-C samples were prepared with the Arima Hi-C Kit, and libraries were prepared with the KAPA HyperPrep Kit following the manufacturer’s instructions. Paired-end reads were combined and pre-processed by Trim Galore (v.0.6.3) and Cutadapt programs. Hi-C library reads were processed using HiC-Pro (v3.0.0) and aligned to the hg19 reference genome using Bowtie2. All valid pairs of contacts were generated after filtering and removing duplicates in HiC-Pro. Valid contact read pairs of samples were applied to obtain the correlation coefficient matrices using Homer (v4.11), and PC1 values were exported in bedgraph.
Fluorescence recovery after photobleaching (FRAP) assay
H9-DDX18GFP cells were cultured on a Matrigel (Corning) pretreated 35 mm 60 μ-dish (ibidi). 60% confluent cells were used for the FRAP experiment. All FRAP assays were performed on a ZEISS microscope with a 63× oil immersion objective and operated with the software. Spots of approximately 1 μm in diameter were photobleached with 20% laser power for 2 s using 488 nm lasers. Time-lapse images were acquired over 2 min after bleaching. Intensity recovery curves were normalized and corrected for photobleaching. The recovery curves were plotted by GraphPad Prism 9.
Measurement of centromere numbers and perinucleolar heterochromatin size
H9 hESCs were infected with DDX18 and control shRNAs as described previously. Three days after infection, cells were fixed and permeabilized for immunostaining. A primary antibody against NPM1 (Santa Cruz, sc-47725) was used for nucleolus staining. In addition, the primary antibody against HP1α (Abcam, ab202533) was used for perinucleolar heterochromatin marker, and antibodies against CENPA (GeneTex, GTX13939) and CENPT (Invitrogen, PA583540) were utilized for centromere staining. The images acquired with a confocal microscope (Leica SP8) were then measured by Image J software using particle analysis function. The average number of CENPA/CENPT foci and volume of HP1α foci in each cell were calculated and plotted.
Live cell imaging
All live-cell imaging was carried out on a Leica SP8 imaging system, equipped with a 60× /1.42 NA Plan Apo oil-immersion objective, 100×/1.40 NA Plan Apo oil-immersion objective (Olympus), and live-cell imaging environment control system (Live Cell Instrument). H9-DDX18GFP cells were cultured on 29 mm No.1.5 glass-bottom (ibidi). In addition, DDX18-GFP knockin hESCs were cultured in mTeSR Plus medium. For imaging of the single time point, six z sections with 0.2 μm spacing were acquired, and for imaging of time, cells were imaged in a single layer per 15 s for a single channel or per 30 s for dual channels. For presentation in figures, raw data were processed by Fiji/ImageJ.
Structured illumination microscopy (SIM) procedure
For all SIM experiments, H9 hESCs were seeded on High-Performance No.1.5 18 × 18 mm glass coverslips (ibidi), fixed and permeabilized. To visualize DDX18, NPM1, and FBL, cells were blocked with 1% BSA for 1 h. Then, antibodies were 1:200 diluted in blocking buffer and incubated for 1 h at RT. After washing with 1xPBS 3 times, fluorescent secondary antibodies were 1:2,000 diluted in blocking buffer and incubated for 1 h at RT. Samples were mounted in ProLong Diamond Antifade Mountant medium (Thermo Fisher). Images were acquired with a 100×/1.4 Apo-TIRF objective lens on an Eclipse Ti microscope with 3D N-SIM (Nikon Instruments, Melville, NY). Imaging and SIM reconstruction parameters were set empirically for each field of view to achieve optimal signal intensity and reconstruction. Focus stabilization (Perfect Focus System, Nikon) was used to maintain focus during image acquisition. Z-stacks were collected with a step size of 0.2 μm and a total depth of 2.5–3 μm.
DNA fluorescence in situ hybridization with immunofluorescence (immuno-DNA FISH)
FISH probes were procured from Empire Genomics, and the BAC clone RP11-1025G19, including a part of HOXA9-13, was utilized to label the HOXA locus. The FISH procedure involved modifications to a previously employed protocol107. Cells were firstly fixed in 3% paraformaldehyde in PBS for 10 min at room temperature and washed in PBST for 5 min 3 times. Cells were then permeabilized with 0.1% Triton-X at room temperature for 10 mins, then washed in PBST for 5 min 3 times. Cells were then incubated with rabbit anti-NPM1 antibodies diluted 1:1000 in blocking buffer overnight at 4 °C, washed with PBST 5 min for three times at room temperature, incubated for 1 hour at RT with Alexa488-labeled secondary goat anti-rabbit IgG (Thermo Fisher). Cells were then subjected to the FISH protocol. Coverslips were treated with PBST washing and ice incubation with 0.1 M HCl/0.7% Triton X-100/2× SSC for 10 min. Afterwards, coverslips were washed with 2× SSC, and 2 μl FISH probes were added, followed by mounting and sealing with rubber cement. Samples were denatured on a heat block at 76 °C for 3 min and then hybridized for 18–72 h at 37 °C on a heat block. Coverslips underwent washing at 37 °C in 2× SSC and 60 °C in 0.1× SSC, and at room temperature in 4× SSC. Finally, coverslips were mounted in DAPI-containing antifade mounting media, and images were captured using a Nikon A1plus-RSI confocal microscope.
Hybridization chain reaction RNA fluorescence in situ hybridization (HCR RNA-FISH)
HCR RNA-FISH probes for each target mRNA were designed and ordered from Molecular Instruments (www.molecularinstruments.com). Human ESCs (H9) were cultured in mTeSR1 Plus medium (StemCell Technologies) on Matrigel-coated 6-well plates. Lentiviral shDDX18 and shLuc were added to H9 cells with polybrene. After 24 h post-infection, viruses were removed, and puromycin (1 µg/ml) was added to the fresh medium to kill non-transduced hESCs for 24 h. On Day 3, the cells were trypsinized, counted, and seeded into 8-well chambered glass bottom #1.5 µ-slides (ibidi). Cells were grown to the desired confluency (~70%) for 24–48 h with puromycin in the medium. Growth media were aspirated, and each chamber was washed with DPBS. Cells were fixed with 4% formaldehyde and then permeabilized overnight with 70% ethanol at −20 °C. The Multiplexed HCR RNA-FISH (V3.0) encompassing the detection and amplification stages was performed following the protocol provided by the Molecular Instruments. DAPI was used to counterstain the nucleus. Nucleolus Bright Green Dye (Dojindo Cat#N511), which reacts to RNAs present in both nuclear (predominantly nucleolar) and cytoplasmic RNAs, was used to detect nucleolus when co-stained with DAPI. The cells were examined under a Stellaris confocal microscope.
Individual nucleotide-resolution UV crosslinking and immunoprecipitation (iCLIP) and analysis
The iCLIP protocol was performed as described previously108 with several modifications. First, 3xFlag-DDX18 H9 cells were irradiated by UV once with 160 mJ/cm2 using Stratalinker 1800 at 254 nm. Next, 3xFlag-DDX18 was immunoprecipitated with protein A Dynabeads (10002D, Invitrogen) conjugated to mouse anti-Flag (Sigma-Aldrich, F1804-1MG). The region corresponding to 75–100 kDa complexes was excised from the membrane to isolate the RNA and sequenced using Illumina HiSeq, generating 50-nt single-end reads. The iCLIP analysis is based on the instructions of the CLIP Tool Kit (CTK, https://zhanglab.c2b2.columbia.edu/index.php/CTK_Documentation). Analysis of the reproducibility of crosslinked sites, identification of the significant iCLIP crosslinked clusters, and z-score analysis of enriched pentamers were performed as described. Data were processed by the iCount software (https://github.com/tomazc/iCount). Duplicates were collapsed before alignment. After collapsing duplicates, barcode sequences were trimmed. Adapters were cleaned after adapter removal. Alignment was done according to CTK instructions. Sequences were aligned to hg19 using Burrows-Wheeler Aligner (BWA) and reads shorter than 15 bp were removed. Peak calling was done using tag2Peak.pl script with statistical assessment.
Protein expression and purification
For protein expression, Escherichia coli Transetta (DE3) cells (TransGen Biotech, China) bearing expression plasmids were grown to OD 0.6–0.8 at 37 °C in LB media containing kanamycin and chloramphenicol and then induced at 18 °C with 0.1 mM Isopropyl β-D-1-thiogalactopyranoside for an additional 16–18 h. Cell pellets were harvested and resuspended in lysis buffer [50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 mM 2-mercaptoethanol, and protease inhibitor cocktail]. Cells were then broken by sonication and cleared by ultracentrifugation at 18,000 rpm for 50 min at 4 °C. Next, the supernatant was subjected to Ni-NTA agarose beads (QIAGEN, USA) for affinity purification. The resin was then washed with lysis buffer, and the bound protein was eluted in elution buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, and 300 mM imidazole). For NPM1-RFP, NPM1-CFP, RFP-FBL, and RFP-HP1α, the His-SUMO tag was removed by on-column digestion with Ulp1 protease at 4 °C for 12 h. Proteins were further purified by size-exclusion chromatography on a Hiload Superdex 75 or Hiload Superdex 200 column (GE Healthcare, USA), equilibrated with 25 mM Tris-HCl, pH 8.0, and 500 mM NaCl.
Furthermore, full-length DDX18-GFP proteins and their truncations or mutants were injected on a Mono S 5/50 GL cation-exchange column (GE Healthcare, USA) and further purified by size-exclusion chromatography on a Superdex 200 column. NPM1-RFP and NPM1-CFP were injected on a Mono Q 5/50 GL anion-exchange column (GE Healthcare, USA) and further purified by size-exclusion chromatography on a Superdex 200 column. The purified proteins were concentrated and stored in the high salt buffer (25 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 2 mM DTT) at −80 °C to inhibit droplet formation. Concentrations were measured by UV absorbance at 280 nm.
Phase separation assays in vitro
Purified human DDX18-GFP, NPM1-RFP, NPM1-CFP, RFP-FBL, and RFP-HP1α proteins were mixed with varying volumes of salt buffer (25 mM Tris-HCl, pH 8.0, and varying NaCl concentrations) to obtain desired protein/salt concentrations. To visualize the droplet formation, 3–6 μl of the protein solution was immediately loaded onto a homemade chamber comprising a glass slide with a coverslip fixed by two parallel strips of double-sided tape. For HP1α droplet formation, we used nonphosphorylated HP1α and added 5% PEG8000 to facilitate its phase separation. Fluorescent images were taken by a Zeiss LSM 710 microscope.
DDX18 protein disorder plots
Intrinsic disorder regions (IDR) for DDX18 (NCBI Reference Sequence: NP_006764.3) were performed using the PONDR VSL2 webtool (http://www.pondr.com/). Prion-like domains (PLD) prediction was performed on PLAAC. Pi-Pi (π-π) contacts predictor was used online on Forman-Kay’s laboratory website (http://pound.med.utoronto.ca/~JFKlab/). Finally, CIDER was used to analyze the net charge per residue (NCPR) distribution.
Quantification and statistical analysis
Statistical analyses were carried out using Excel or R (version 3.3.0). Data are presented as mean ± SD. The statistical tests are analyzed using the student’s t-test if it is not stated in the relevant figure legends. All the statistical details can be found in the figure legends or results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The DDX18 affinity purification and SILAC mass spectrometry (MS) data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD057813. The RNA-seq, ChIP-seq, iCLIP-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE182804 and Hi-C sequencing GSE182855. Source data are provided with this paper.
References
Jerkovic, I. & Cavalli, G. Understanding 3D genome organization by multidisciplinary methods. Nat. Rev. Mol. Cell Biol. 22, 511–528 (2021).
Hug, C. B. & Vaquerizas, J. M. The birth of the 3D genome during early embryonic development. Trends Genet. 34, 903–914 (2018).
Zheng, H. & Xie, W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 20, 535–550 (2019).
Watanabe-Susaki, K. et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells 32, 3099–3111 (2014).
Gupta, S. & Santoro, R. Regulation and roles of the nucleolus in embryonic stem cells: from ribosome biogenesis to genome organization. Stem Cell Rep. 15, 1206–1219 (2020).
Bartova, E., Krejci, J., Harnicarova, A. & Kozubek, S. Differentiation of human embryonic stem cells induces condensation of chromosome territories and formation of heterochromatin protein 1 foci. Differ. Res. Biol. Divers. 76, 24–32 (2008).
Jachowicz, J. W., Santenard, A., Bender, A., Muller, J. & Torres-Padilla, M. E. Heterochromatin establishment at pericentromeres depends on nuclear position. Gene Dev. 27, 2427–2432 (2013).
Burton, A. et al. Heterochromatin establishment during early mammalian development is regulated by pericentromeric RNA and characterized by non-repressive H3K9me3. Nat. Cell Biol. 22, 767–778 (2020).
Gibson, B. A. et al. Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e421 (2019).
Hyman, A. A., Weber, C. A. & Julicher, F. Liquid-liquid phase separation in biology. Annu Rev. Cell Dev. Biol. 30, 39–58 (2014).
Brangwynne, C. P., Mitchison, T. J. & Hyman, A. A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl Acad. Sci. USA 108, 4334–4339 (2011).
West, J. A. et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 214, 817–830 (2016).
Putnam, A., Cassani, M., Smith, J. & Seydoux, G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 26, 220–226 (2019).
Fei, J. et al. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J. Cell Sci. 130, 4180–4192 (2017).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Abraham, K. J. et al. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature 585, 298–302 (2020).
Frottin, F. et al. The nucleolus functions as a phase-separated protein quality control compartment. Science 365, 342–347 (2019).
Woolnough, J. L., Atwood, B. L., Liu, Z., Zhao, R. & Giles, K. E. The regulation of rRNA gene transcription during directed differentiation of human embryonic stem cells. PloS ONE 11, e0157276 (2016).
Zhang, H. et al. DEAD-box helicase 18 counteracts PRC2 to safeguard ribosomal DNA in pluripotency regulation. Cell Rep. 30, 81–97.e87 (2020).
Savic, N. et al. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 15, 720–734 (2014).
Quinodoz, S. A. et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174, 744–757.e724 (2018).
Stevens, T. J. et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017).
Hondele, M. et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148 (2019).
Lafontaine, D. L. J., Riback, J. A., Bascetin, R. & Brangwynne, C. P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 22, 165–182 (2021).
Stamatopoulou, V., Parisot, P., De Vleeschouwer, C. & Lafontaine, D. L. J. Use of the iNo score to discriminate normal from altered nucleolar morphology, with applications in basic cell biology and potential in human disease diagnostics. Nat. Protoc. 13, 2387–2406 (2018).
Shin, Y. et al. Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175, 1481–1491.e1413 (2018).
Gifford, C. A. et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153, 1149–1163 (2013).
Tanaka, Y. et al. Transcriptome signature and regulation in human somatic cell reprogramming. Stem Cell Rep. 4, 1125–1139 (2015).
Tsankov, A. M. et al. Transcription factor binding dynamics during human ES cell differentiation. Nature 518, 344–349 (2015).
Atkinson, S. P. et al. Epigenetic marking prepares the human HOXA cluster for activation during differentiation of pluripotent cells. Stem Cells 26, 1174–1185 (2008).
Bhatlekar, S., Fields, J. Z. & Boman, B. M. Role of HOX genes in stem cell differentiation and cancer. Stem Cells Int. 2018, 3569493 (2018).
Seifert, A., Werheid, D. F., Knapp, S. M. & Tobiasch, E. Role of Hox genes in stem cell differentiation. World J. Stem Cells 7, 583–595 (2015).
Schmidt, E. K., Clavarino, G., Ceppi, M. & Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009).
Cordin, O., Banroques, J., Tanner, N. K. & Linder, P. The DEAD-box protein family of RNA helicases. Gene 367, 17–37 (2006).
Rocak, S., Emery, B., Tanner, N. K. & Linder, P. Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res. 33, 999–1009 (2005).
Sampath, P. et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2, 448–460 (2008).
Corsini, N. S. et al. Coordinated control of mRNA and rRNA processing controls embryonic stem cell pluripotency and differentiation. Cell Stem Cell 22, 543–558.e512 (2018).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Riback, J. A. et al. Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214 (2020).
Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018).
Linder, P. & Jankowsky, E. From unwinding to clamping—the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516 (2011).
Guthmann, M., Burton, A. & Torres-Padilla, M. E. Expression and phase separation potential of heterochromatin proteins during early mouse development. EMBO Rep. 20, e47952 (2019).
Ukmar-Godec, T. et al. Lysine/RNA-interactions drive and regulate biomolecular condensation. Nat. Commun. 10, 2909 (2019).
Alshareedah, I., Moosa, M. M., Raju, M., Potoyan, D. A. & Banerjee, P. R. Phase transition of RNA-protein complexes into ordered hollow condensates. Proc. Natl Acad. Sci. USA 117, 15650–15658 (2020).
Boamah, E. K., Kotova, E., Garabedian, M., Jarnik, M. & Tulin, A. V. Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet. 8, e1002442 (2012).
Harris, B. et al. Cohesion promotes nucleolar structure and function. Mol. Biol. Cell 25, 337–346 (2014).
Percharde, M. et al. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174, 391–405.e319 (2018).
Hu, C.-D., Chinenov, Y. & Kerppola, T. K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 (2002).
Chen, T. et al. ADAR1 is required for differentiation and neural induction by regulating microRNA processing in a catalytically independent manner. Cell Res. 25, 459–476 (2015).
Fazal, F. M. et al. Atlas of subcellular RNA localization revealed by APEX-seq. Cell 178, 473–490.e426 (2019).
Yao, R. W. et al. Nascent pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol. Cell 76, 767–783.e711 (2019).
Mitrea, D. M. et al. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 5, e13571 (2016).
Mitrea, D. M. et al. Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat. Commun. 9, 842 (2018).
Latonen, L. Phase-to-phase with nucleoli—stress responses, protein aggregation and novel roles of RNA. Front Cell Neurosci. 13, 151 (2019).
Harding, S. M., Boiarsky, J. A. & Greenberg, R. A. ATM dependent silencing links nucleolar chromatin reorganization to DNA damage recognition. Cell Rep. 13, 251–259 (2015).
Potapova, T. A. & Gerton, J. L. Ribosomal DNA and the nucleolus in the context of genome organization. Chromosome Res.: Int. J. Mol. Supramol. Evolut. Asp. Chromosome Biol. 27, 109–127 (2019).
Pontvianne, F. et al. Identification of nucleolus-associated chromatin domains reveals a role for the nucleolus in 3D organization of the A. thaliana genome. Cell Rep. 16, 1574–1587 (2016).
Schlesinger, S. & Meshorer, E. Open chromatin, epigenetic plasticity, and nuclear organization in pluripotency. Dev. Cell 48, 135–150 (2019).
Mattout, A. et al. Heterochromatin protein 1beta (HP1beta) has distinct functions and distinct nuclear distribution in pluripotent versus differentiated cells. Genome Biol. 16, 213 (2015).
Larson, A. G. et al. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017).
Keenen, M. M. et al. HP1 proteins compact DNA into mechanically and positionally stable phase separated domains. Elife 10, e64563 (2021).
Sanulli, S. et al. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575, 390–394 (2019).
Strom, A. R. et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017).
Sanders, D. W. et al. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324 e328 (2020).
LeRoy, G. et al. Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol. 13, R68 (2012).
Foltz, D. R. et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484 (2009).
Dunleavy, E. M. et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 (2009).
Chun, Y. et al. New centromeric component CENP-W is an RNA-associated nuclear matrix protein that interacts with nucleophosmin/B23 protein. J. Biol. Chem. 286, 42758–42769 (2011).
Thakur, J., Fang, H., Llagas, T., Disteche, C. M. & Henikoff, S. Architectural RNA is required for heterochromatin organization. bioRxiv, https://doi.org/10.1101/784835 (2019).
Harris, H. L. et al. Chromatin alternates between A and B compartments at kilobase scale for subgenic organization. Nat. Commun. 14, 3303 (2023).
Meuleman, W. et al. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 23, 270–280 (2013).
Zheng, Y., Ay, F. & Keles, S. Generative modeling of multi-mapping reads with mHi-C advances analysis of Hi-C studies. eLife 8, e38070 (2019).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Labade, A. S., Karmodiya, K. & Sengupta, K. HOXA repression is mediated by nucleoporin Nup93 assisted by its interactors Nup188 and Nup205. Epigenet. Chromatin 9, 54 (2016).
Labade, A. S., Salvi, A., Kar, S., Karmodiya, K. & Sengupta, K. Nup93 and CTCF modulate spatiotemporal dynamics and function of the HOXA gene locus during differentiation. J. Cell Sci. 134, jcs259307. (2021).
Yang, L. et al. 3D genome alterations associated with dysregulated HOXA13 expression in high-risk T-lineage acute lymphoblastic leukemia. Nat. Commun. 12, 3708 (2021).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Zheng, H. et al. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell 63, 1066–1079 (2016).
Moon, Y., Horsman, D. E., Humphries, R. K. & Park, G. A novel translocation involving RUNX1 and HOXA gene clusters in a case of acute myeloid leukemia with t(7;21)(p15;q22). Immune Netw. 13, 222–226 (2013).
Allshire, R. C. & Madhani, H. D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19, 229–244 (2018).
Gao, X. D. et al. C-BERST: defining subnuclear proteomic landscapes at genomic elements with dCas9-APEX2. Nat. Methods 15, 433–436 (2018).
Sun, Z. et al. LIN28 coordinately promotes nucleolar/ribosomal functions and represses the 2C-like transcriptional program in pluripotent stem cells. Protein Cell. 13, 490–512 (2021).
van Sluis, M. & McStay, B. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev. 29, 1151–1163 (2015).
Collinson, A. et al. Deletion of the polycomb-group protein EZH2 leads to compromised self-renewal and differentiation defects in human embryonic stem cells. Cell Rep. 17, 2700–2714 (2016).
Shan, Y. et al. PRC2 specifies ectoderm lineages and maintains pluripotency in primed but not naive ESCs. Nat. Commun. 8, 672 (2017).
Theunissen, T. W. & Jaenisch, R. Mechanisms of gene regulation in human embryos and pluripotent stem cells. Development 144, 4496–4509 (2017).
Weinberger, L., Ayyash, M., Novershtern, N. & Hanna, J. H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 17, 155–169 (2016).
Huang, X. et al. OCT4 cooperates with distinct ATP-dependent chromatin remodelers in naïve and primed pluripotent states in human. Nat. Commun. 12, 5123 (2021).
Abujarour, R., Efe, J. & Ding, S. Genome-wide gain-of-function screen identifies novel regulators of pluripotency. Stem Cells 28, 1487–1497 (2010).
Padeken, J. & Heun, P. Nucleolus and nuclear periphery: velcro for heterochromatin. Curr. Opin. Cell Biol. 28, 54–60 (2014).
Santoro, R., Schmitz, K. M., Sandoval, J. & Grummt, I. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep. 11, 52–58 (2010).
Leone, S., Bar, D., Slabber, C. F., Dalcher, D. & Santoro, R. The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation. EMBO Rep. 18, 1248–1262 (2017).
Lafontaine, D. L. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 22, 11–19 (2015).
Kiss, T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109, 145–148 (2002).
Xing, Y. H. et al. SLERT regulates DDX21 rings associated with Pol I transcription. Cell 169, 664–678.e616 (2017).
Wu, M. et al. lncRNA SLERT controls phase separation of FC/DFCs to facilitate Pol I transcription. Science 373, 547–555 (2021).
David Wang, X. Q. et al. Three-dimensional regulation of HOXA cluster genes by a cis-element in hematopoietic stem cell and leukemia. bioRxiv https://doi.org/10.1101/2020.04.16.017533 (2020).
Oka, M. et al. Chromatin-bound CRM1 recruits SET-Nup214 and NPM1c onto HOX clusters causing aberrant HOX expression in leukemia cells. eLife 8, e46667 (2019).
Wen, B., Wu, H., Shinkai, Y., Irizarry, R. A. & Feinberg, A. P. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 41, 246–250 (2009).
Loh, Y. H., Zhang, W., Chen, X., George, J. & Ng, H. H. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 21, 2545–2557 (2007).
Ramos-Mejia, V. et al. HOXA9 promotes hematopoietic commitment of human embryonic stem cells. Blood 124, 3065–3075 (2014).
Iacovino, M. et al. HoxA3 is an apical regulator of haemogenic endothelium. Nat. Cell Biol. 13, 72–78 (2011).
Peric-Hupkes, D. et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 38, 603–613 (2010).
Carvajal-Vergara, X. et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature 465, 808–812 (2010).
Fidalgo, M. et al. Zfp281 coordinates opposing functions of Tet1 and Tet2 in pluripotent states. Cell Stem Cell 19, 355–369 (2016).
Ding, J. et al. Tex10 coordinates epigenetic control of super-enhancer activity in pluripotency and reprogramming. Cell Stem Cell 16, 653–668 (2015).
Chen, Y. et al. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 217, 4025–4048 (2018).
Modic, M. et al. Cross-regulation between TDP-43 and paraspeckles promotes pluripotency-differentiation transition. Mol. Cell 74, 951–965.e913 (2019).
Acknowledgements
We thank Drs. Ling-Ling Chen for providing NPM1-RFP and RFP-FBL bacterial expressing plasmids, Dan Li at the Wang lab for eCLIP-seq expertise, and Tajda Klobucar at the Modic lab for technical advice on HCR-FISH. We also thank Columbia Confocal and Specialized Microscopy Shared Resource for 3D-SIM imaging, Columbia Medicine Microscopy Core for Stellaris confocal microscope services (funded by NIH-1S10OD032447), the CCTI Flow Cytometry Core for cell sorting services. Laboratories of Drs. Chen and Li were supported by grants (2023YFA1800403 to Y.C., NSFC32071195, 31900934, and 23QC1400600 to Y.L.). Research in the Wang laboratory was supported by NIH (GM129157, HD095938, HD097268, HD114122, and R21HD116446).
Author information
Authors and Affiliations
Contributions
X.S., Y.L., H.Z., and X. Hou designed and conducted the experiments and wrote the manuscript with contributions from all other authors. V.M., F.F., J.D., and L.C. performed the experiments. J.Y., M.M., X. Huang, X.B., and W.Z. performed the computational analysis. S.R.M. and E.A. performed Hi-C data analyses presented in Figs. 6d, e and S6b-f,l. F.-C.Y., M.X., and W.X. provided technical advice and helpful discussions. J.W. conceived the project and, together with Y.C., designed the experiments, interpreted the results, and prepared and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Fulai Jin, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shi, X., Li, Y., Zhou, H. et al. DDX18 coordinates nucleolus phase separation and nuclear organization to control the pluripotency of human embryonic stem cells. Nat Commun 15, 10803 (2024). https://doi.org/10.1038/s41467-024-55054-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-55054-8
This article is cited by
-
Unveiling the dynamic drivers: phase separation’s pivotal role in stem cell biology and therapeutic potential
Stem Cell Research & Therapy (2025)