Abstract
Recent advancements in biological technologies have enabled the measurement of spatially resolved multi-omics data, yet computational algorithms for this purpose are scarce. Existing tools target either single omics or lack spatial integration. We generate a graph neural network algorithm named COSMOS to address this gap and demonstrated the superior performance of COSMOS in domain segmentation, visualization, and spatiotemporal map for spatially resolved multi-omics data integration tasks.
Similar content being viewed by others
Introduction
With the explosion of spatially resolved single-cell omics development, the measurements of spatially resolved multi-omics on the same tissue sections have recently been achieved1,2,3,4,5,6,7,8,9. However, the computational algorithms that are designed to analyze such spatially resolved multi-omics are lacking. Existing tools like SpaGCN10 and SpaceFlow11 focus on spatially resolved single-omics data, while multi-omics integration tools such as GLUE12, WNN13, MIRA14, totalVI15 perform integration without using spatial information. Algorithms like Moscot16 handle co-embedding for spatially resolved multi-omics data that do not share the same spatial locations. To our knowledge, the only algorithms designed specifically to integrate paired spatially resolved multi-omics data are CellCharter17, SpaMultiVAE18 and SpatialGlue19. However, CellCharter and SpaMultiVAE concatenate two omics into a single matrix for integrated embedding, failing to extract and utilize the complementary features and unique contributions of each omics. SpatialGlue attempts to learn the contribution of each modality, but it does not perform well on the predictive accuracy and computational efficiency of segmentation analysis when applied to simulated and actual experimental datasets. Here we design a graph neural network algorithm to achieve COoperative integration of Spatially resolved Multi-OmicS data (COSMOS) that extracts complementary features from different modalities and gives an integrated embedding that outperforms analysis on each modality and other competing integration methods. We apply COSMOS to two simulated and three real-world spatially resolved multi-omics datasets from different platforms and diverse tissue types, and demonstrate its performance in domain segmentation, low-dimensional visualization, and spatiotemporal map generation.
Results
Introduction to COSMOS
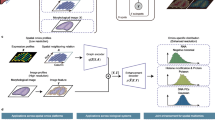
The input of COSMOS is a spatially resolved multi-omics dataset consisting of two sets of molecular omics data (e.g., transcriptomics and epigenomics, or transcriptomics and proteomics) and a set of shared spatial positions between two omics. The input multi-omics data were encoded using two separate graph convolutional networks (GCN). These representations were then integrated into a unified format with modality weights determined by the Weighted Nearest Neighbor (WNN) algorithm13. Following the strategy in the Deep Graph Infomax (DGI) model20, we permuted the integrated layer to form a negative representation and built a contrastive discriminator that contrasts mutual information between local node representation hi and global summary vector s in the original graph (\({{{\bf{H}}}}_{{{\bf{int}}}},{{\bf{A}}}\)) and permuted graph (\({\widetilde{{{\bf{H}}}}}_{{{\bf{int}}}},{{\bf{A}}}\)). A spatial regularization was added to the discriminator for the training of the model (Fig. 1a). The output is an integrated embedding that can be applied to downstream analyses including domain segmentation, spatially consistent low-dimensional visualization, pseudo-spatiotemporal map (pSM) generation and pSM gene extraction (Fig. 1b).
a COSMOS takes spatially resolved multi-omics data with shared spatial positions as input. Each omics dataset is encoded using a graph convolutional network (GCN), with edges defined by spatial distances. b COSMOS produces an integrated embedding of the multi-omics data, which can then be used for downstream analyses.
Results on two simulated datasets
Due to the infancy stage of spatially resolved multi-omics technologies, experimental datasets based on these methods are rare. Therefore, we first simulated a spatially resolved multi-omics dataset from mouse visual cortex STARmap spatial transcriptomics data21, which consists of 1207 cells annotated by six layer labels: L1, L2/3, L4, L5, L6, and HPC/CC (Fig. 2a). We simulated two datasets (termed as “Omics-1” and “Omics-2” in the following content) by shuffling cells in L4/L5 and L5/L6, respectively, and then added Gaussian noise to the expression values (see Methods). In this design, cells in L4/L5 cannot be separated by Omics-1 alone, and cells in L5/L6 cannot be separated by Omics-2 alone. We applied five multi-omics integration algorithms (WNN, SpatialGlue, CellCharter, SpaMultiVAE, COSMOS) and one single-omics algorithm (SpaceFlow) to the simulated data to assess whether cells in L4/L5/L6 could be separated by integrating Omics-1 and Omics-2. We evaluated the performance of these algorithms using the Adjusted Rand Index (ARI), a common metric for validating clustering results. A higher ARI value indicates greater alignment with curated annotations. Our analysis revealed that COSMOS achieved the highest ARI (0.84) in domain segmentation, outperforming all other algorithms (with ARI values ≤ 0.66), as shown in Fig. 2a–b. In addition, the modality weights distributions for each layer type in COSMOS displayed that L4 cells in Omics-1 and L6 cells in Omics-2 have smaller modality weights (Fig. 2c). These distributions of weights were expected because L4 cells cannot be separated from L5 in Omics-1 and contributed less to the integrated embeddings, so are the L6 cells in Omics-2. We further used UMAP to visualize the low-dimensional representation of cells by SpatialGlue, SpaMultiVAE, and COSMOS, and found that L1-L6 layers were better separated in COSMOS than in SpaMultiVAE (Fig. 2d). After plotting pseudo-Spatiotemporal map (pSM) by running the diffusion pseudo-time11,22 using the low dimensional embedding by SpatialGlue, SpaMultiVAE, and COSMOS, we observed that the pSM of COSMOS showed better consistency with the anatomical structure of mouse visual cortex sample than SpatialGlue and SpaMultiVAE (Fig. 2e).
a The first simulated dataset was created by shuffling L5/L4 and L5/L6 regions of a spatially resolved STARmap mouse visual cortex transcriptomics dataset, with Gaussian noise added to the expression values. We compared the best domain segmentation results from the six algorithms, noting that SpaceFlow, which operates on single omics, was contrasted with algorithms capable of integrating two omics. b We evaluated the Adjusted Rand Index (ARI) of domain segmentations produced by the six algorithms at different clustering resolutions using “leiden” method. c The distribution of modality weights for the two omics in COSMOS is shown. d UMAP visualizations of integrated embeddings from SpatialGlue, SpaMultiVAE, and COSMOS are presented. e The pseudo-spatiotemporal maps (pSM) generated by these three algorithms are illustrated. f–j Similar analyses were conducted on a second simulated dataset, created by shuffling EPL/MCL and EPL/GL regions of a spatially resolved Stereo-Seq mouse olfactory bulb dataset, with Gaussian noise added to the expression values. SpaMultiVAE was excluded from this comparison due to its instability in analyzing the sparse Stereo-Seq dataset. The full names of the abbreviations of brain regions are: hippocampus (HPC), corpus callosum (CC), layer 1 (L1), layer 2/3 (L2/3), layer 4 (L4), layer 5 (L5), layer 6 (L6), rostral migratory stream (RMS), granule cell layer (GCL), internal plexiform layer (IPL), mitral cell layer (MCL), external plexiform layer (EPL), glomerular layer (GL) and olfactory nerve layer (ONL). Source data are provided as a Source Data file.
To demonstrate the general applicability of COSMOS on different tissue types and experimental platforms, we used another mouse olfactory bulb dataset generated by Stereo-Seq technology23 to create the second simulated dataset, which consists of 18196 cells annotated by six layer types: RMS, GCL/IPL, MCL, EPL, GL, and ONL (Fig. 2f). Similarly, we shuffled the cells in EPL/MCL and EPL/GL separately and added Gaussian noise to simulate two omics datasets (termed as Omics-3 and Omics-4 below). We applied SpaceFlow, WNN, SpatialGlue, CellCharter, and COSMOS to integrate these two datasets and observed that COSMOS (ARI = 0.63) outperformed all other algorithms (ARI ≤ 0.56, Fig. 2f–g). The MCL cells in Omics-3 and GL cells in Omics-4 have smaller modality weights (Fig. 2h), which is consistent with the simulation design. COSMOS helped to generate better clustering results than SpatialGlue in UMAP visualization. For example, EPL cells (blue) and GL cells (orange) were more separatable in COSMOS than in SpatialGlue (Fig. 2i). COSMOS also generated more consistent pSM results with the anatomical structure of mouse olfactory bulb sample (Fig. 2j). The analyses on the two simulated spatially resolved multi-omics data demonstrated that COSMOS outperforms other competing algorithms in domain segmentation, cell type clustering, and pseudo-spatiotemporal map generation.
Results on three real-world datasets
Next, we applied COSMOS to analyze a spatially resolved multi-omics dataset based on real-world experiments instead of simulated data. This experimental dataset contains P22 mouse brain coronal sections with a joint profiling of spatial chromatin accessibility (ATAC) and spatial transcriptome (RNA)7. By aligning the sample with the P56 mouse brain coronal section from Allen Mouse Brain Atlas (Fig. 3a), we made a manual annotation for the sample and highlighted several regions of interest for analysis: ACB, CP, VL, L1-3, L4, L5, L6a/b, and ccg/aco (Fig. 3b). We first applied SpaceFlow to each modality individually and then used WNN, SpatialGlue, CellCharter, and COSMOS to integrate the datasets with two omics. As shown in Fig. 3c, COSMOS achieved the highest domain segmentation with an ARI of 0.63, outperforming SpatialGlue (ARI = 0.43), CellCharter (ARI = 0.50), SpaceFlow on ATAC (ARI = 0.58), and SpaceFlow on RNA (ARI = 0.45). Comparing the segmentation results of SpaceFlow to COSMOS, we observed that ATAC modality contributed dominantly to the separation of L1-L6 layers, while RNA modality played an auxiliary role in separating VL, CP and ccg/aco layers (Fig. 3c). We plotted the modality weight distributions of COSMOS and found that the cells from ATAC modality had larger overall weights than the cells from RNA modality, but the distributions of weights differed across various layers. The RNA modality has the smallest weights in L1-L6 layers but the largest weights in VL, CP, ACB, and ccg/aco (Fig. 3d), which reflected distinct contributions of both modalities to the clustering of different regions. Next, we generated UMAP visualization and pSM from the low-dimensional representation by SpatialGlue and COSMOS (Fig. 3e–f). COSMOS presented the clusters in a more compact way (Fig. 3e) and organized the clusters in the pSM that is consistent with the anatomical structure of the mouse brain (Fig. 3f, Fig. S1). Based on the pseudo-time inferred from COSMOS representation, we selected 577 pseudo-time associated genes and found that these genes are enriched in biological processes related to brain development (e.g., Neurogenesis, Fig. 3g). The pseudo-time associated genes were differentially expressed in different stages of the pseudo-time (Fig. 3h). From these we identified six marker genes that well characterized the major anatomical regions of the brain: Nexn (CP), Bcl11b (CP and ACB), Mbp (ccg and aco), Nfix (L1-L6), Mef2c (L1-L4), and Cux2 (L1-L3) (Fig. 3i). These results demonstrated that COSMOS integration of spatially resolved multi-omics performs domain segmentation with a higher accuracy than that obtained from single modality analysis or other competing integration methods, and generates a pseudo-time that aligns well with anatomical structure and marker genes expression patterns in the tissue.
a Annotation of P56 mouse brain coronal sections from the Allen Brain Atlas. b Manual annotation of tissue from P22 mouse brain coronal sections in Zhang et al., overlaid on the image from a. c Comparison of domain segmentation results from five algorithms. Note that SpaMultiVAE was excluded as it is not designed for ATAC-RNA multi-omics data analysis. d Distribution of modality weights for RNA and ATAC in COSMOS analysis. e UMAP visualizations of integrated embeddings produced by SpatialGlue and COSMOS. f Pseudo-spatiotemporal maps (pSM) generated by SpatialGlue and COSMOS. (g) Gene Ontology Biological Process (GOBP) analysis of genes associated with pseudo-time. h Gene expression profiles for the top 15 positive and 15 negative pSM-associated genes. i Spatial mapping of gene expressions for brain development genes negatively (Nexn, Bcl11b, Mbp) and positively (Nfix, Mef2c, Cux2) correlated with pseudo-time, as identified by COSMOS. The full names of the abbreviations of brain regions are: nucleus accumbens (ACB), caudoputamen (CP), lateral ventricle (VL), genu of corpus callosum (ccg) and anterior commissure olfactory limb (aco). Source data are provided as a Source Data file.
To demonstrate the applicability of COSMOS on different data types, we applied COSMOS to a spatially resolved RNA-Protein multi-omics dataset. This dataset contains 1789 spatially resolved cells from E10 mouse embryo brain regions with a joint profiling of 22 proteins and 254 genes with DBiT-seq1 (Fig. 4a). We applied six algorithms to perform domain segmentation, identifying ten domains with each method. We found that COSMOS produced a smoother mapping compared to WNN, SpatialGlue, CellCharter, and SpaMultiVAE (Fig. 4d–h). We also noticed that SpaceFlow on RNA exhibited different clustering patterns from SpaceFlow on Protein. For example, C2 (green) cluster is more clearly identified in SpaceFlow on RNA (Fig. 4b), while C1 (red) cluster is more narrowly defined in SpaceFlow on Protein (Fig. 4c). The distinct clustering patterns in both modalities were well preserved in COSMOS mapping (Fig. 4h). The narrower C1 cluster in SpaceFlow on Protein corresponds to the brain region with high expression of MAdCAM-11. This region was accurately captured by COSMOS, but not by WNN, SpatialGlue, CellCharter and SpaMultiVAE, which mixed this region with the neighboring regions (Fig. 4d–h). In the modality weights distributions of COSMOS, we observed that cells in C1 and C6 have larger weights on Protein modality, while cells in C2 and C8 have larger weights on RNA modality (Fig. 4i). This reflects distinct contributions of both modalities to the clustering of different regions. The regions identified by COSMOS correspond to differential expression patterns from both modalities: C1, C6, and C5 correspond to regions with high-expression of proteins MAdCAM-1, CD55, and ESAM, respectively (Fig. 4j); C2, C8 and C10 correspond to regions with high-expression of genes Myh7, Msx1 and Hba-x, respectively (Fig. 4k). These results demonstrate that COSMOS can extract and preserve informative features from both modalities and produces an integrated embedding that achieves smoother and more accurate domain segmentation than the embedding on single modality and the embedding by other integration methods. In addition to DBiT-seq data, we also applied COSMOS to RNA-Protein multi-omics data generated by spatial-CITE-seq from human tonsil cells4. We observed that COSMOS provided smoother domain segmentation (Fig. S2a) and more closely matched protein and gene expression profiles compared to other algorithms (Fig. S2b–c).
a H&E-stained image of a mouse embryo brain region. b–h Domain segmentation results from six algorithms, with ten domains identified by each method. In the COSMOS mapping, six regions are highlighted with dashed curves: C1, C2, C5, C6, C8, and C10. i Distribution of modality weights for RNA and Protein omics in the COSMOS analysis. j Proteomic profiles of three proteins—MAdCAM-1, CD55, and ESAM—with dashed curves indicating the boundaries of regions C1, C6, and C5 identified by COSMOS. k Transcriptomic profiles of three genes—Myh7, Msx1, and Hba-x—with dashed curves indicating the boundaries of regions C2, C8, and C10 identified by COSMOS. Source data are provided as a Source Data file.
Discussion
We applied COSMOS to two simulated and three experimental spatially resolved multi-omics datasets, demonstrating that COSMOS outperformed other integration algorithms in domain segmentation, low-dimensional visualization, spatiotemporal map generation, and differentially expressed gene extraction. We also want to highlight that COSMOS can be extended to analyze three or more spatially resolved omics data when these data are available in the near future.
We applied COSMOS to analyze single-cell level omics data throughout the manuscript, as most available spatially resolved multi-omics datasets are at this resolution. Consequently, we did not apply COSMOS to spot-level multi-omics data. However, we believe that COSMOS could be adapted to process spot-level data when combined with appropriate deconvolution methods.
COSMOS was designed to integrate spatially resolved multi-omics datasets by extracting informative features from both omics. It performs best when these two omics contain distinct features that are complementary to each other, as showcased in the non-spatial single-cell multi-omics integration by the WNN algorithm13. As a GCN-based method, COSMOS constructs a graph based on the spatial positions of cells and assumes local spatial dependency among cells of the same type. This assumption is valid for the two simulated datasets and three real datasets discussed in the manuscript. However, in cases where cells of different types are spatially mixed, such as cells in the Slide-Tag human melanoma RNA-ATAC multi-omics data9, this spatial dependency assumption may not hold. Consequently, COSMOS may not outperform non-spatial integration algorithms like WNN in distinguishing cell types in such scenarios (Fig. S3). Therefore, COSMOS is more advantageous when applied to spatial profiling datasets with complementary omic features and clear local spatial dependencies among cells. With the rapid development of spatially resolved profiling technologies, we anticipate that COSMOS will demonstrate its capabilities to their fullest extent as increasingly diverse and informative multi-omics datasets are generated in the future.
Methods
Data preprocessing
We used two simulated and four real-world experimental datasets based on spatially resolved multi-omics technologies. For the mouse visual cortex STARmap data21, we used the “scanpy” Python package to normalize the unique molecular identifier (UMI) counts so that each cell or spot has a total count equal to the median of total counts per cell, then we transformed them to a natural log scale. For mouse olfactory bulb Stereo-Seq data23, we first did the filtering by selecting genes that are expressed in more than three cells and selecting cells that contain more than 100 genes with non-zero expression, then we did the normalization and log-transformation as we did for mouse visual cortex data. For the ATAC-RNA-seq mouse brain multi-omics data7 and slide-tags human melanoma RNA-ATAC multi-omics data9, we followed the preprocessing protocols used by the original authors. For the DBiT-seq mouse embryo brain RNA-Protein multi-omics data1 and spatial-CITE-seq human tonsil RNA-Protein multi-omics data4, we followed the preprocessing in Tian et al.18.
Data annotations
The annotations for the first three datasets are generated by manually marking the anatomical regions. The anatomical references of mouse visual cortex STARmap data and mouse olfactory bulb Stereo-Seq data are obtained from original papers. The reference of mouse brain ATAC-RNA data is obtained from a P56 mouse brain coronal section from Allen Mouse Brain Atlas (Allen Institute for Brain Science: https://mouse.brain-map.org/static/atlas). The annotations of these datasets were provided in Supplementary Data 1-3. The full names of the abbreviations of brain regions are: hippocampus (HPC), corpus callosum (CC), layer 1 (L1), layer 2/3 (L2/3), layer 4 (L4), layer 5 (L5), and layer 6 (L6); nucleus accumbens (ACB), caudoputamen (CP), lateral ventricle (VL), genu of corpus callosum (ccg), anterior commissure olfactory limb (aco); rostral migratory stream (RMS), granule cell layer (GCL), internal plexiform layer (IPL); mitral cell layer (MCL), external plexiform layer (EPL), glomerular layer (GL), olfactory nerve layer (ONL).
Simulated spatially resolved multi-omics data
Current simulation algorithms for spatially resolved data, such as scDesign324 serve as powerful tools to simulate different kinds of single-cell omics datasets that well preserve the characteristics of the original test datasets. However, we found two challenges in applying scDesign3 to achieve the goal of our simulation. First, scDesign3 has been shown to effectively simulate both single-cell multi-omics and spatially resolved transcriptomics datasets. However, when we applied scDesign3 to simulate spatially resolved ATAC datasets and paired spatial single-cell multi-omics datasets, such as spatial RNA and spatial ATAC, which are the focus of our analysis in this manuscript, we found that scDesign3 is not compatible with these types of datasets. Second, scDesign3 was designed to preserve the gene and cell-specific characteristics. However, the aim of our simulation is to generate a paired multi-omics dataset in which the two omics have non-overlapping features that are complementary to each other, allowing us to demonstrate the unique strengths of COSMOS in ‘cooperative’ integration. We found that scDesign3 was not designed to simulate data with such a specific feature. Thus, we generated simulated spatially resolved multi-omics data from single transcriptomics. The simulation contains two steps. First, we selected two different sets of regions with some overlapping and shuffled the gene expressions of the cells in each set of regions separately. In mouse visual cortex STARmap data, we shuffled the gene expressions of cells in L4/L5 and L5/L6 separately. In mouse olfactory bulb Stereo-Seq data, we shuffled the cells in MCL/EPL and EPL/GL. This step is to simulate two complementary sets of Omics that cannot separate certain regions on their own, but can separate all these regions in the integration of the two omics by extracting informative features from each of them. Second, we added Gaussian noise to the expressions of each of the shuffled data. This step is to simulate the noise in the two omics. The new expression values \({{{\rm{X}}}}_{{{\rm{ij}}}}^{{\prime} }\) were generated by:
Where Xij is the gene expression of cell i and gene j in each omics after shuffling, ε is a value denoting noise taken from the Gaussian distribution of N(0,0.0025).
There is a limitation in our simulation method as it relies solely on gene expression profiles. Nonetheless, this approach effectively tests COSMOS’s ability to perform cooperative integration by evaluating how well it extracts and utilizes complementary features from two omics datasets, thereby achieving better clustering results compared to analyzing each omics dataset individually.
COSMOS model
The general framework is a two-channel Deep Graph Infomax (DGI) with a spatial regularization. The DGI model was first proposed to perform unsupervised node classification for graph-structured data20. When applied to spatially resolved transcriptomics data, DGI has the advantages of capturing both the expression patterns and the neighborhood micro-environment of cells, as well as global patterns such as the pseudo-time. Specifically, the input data is an expression matrix of cells \({{\bf{X}}}=({{{\boldsymbol{x}}}}_{{{\boldsymbol{1}}}},\,{{{\boldsymbol{x}}}}_{{{\boldsymbol{2}}}},\ldots,\,{{{\boldsymbol{x}}}}_{{{\boldsymbol{N}}}})\) and a spatial adjacency matrix \({{\bf{A}}}\in {R}^{N\times N}\), where N is the number of cells, xi is the expression vector of cell i, \({{{\bf{A}}}}_{{{\rm{ij}}}}\) is equal to 1 if there is an edge between cell i and j and otherwise 0. The adjacency matrix is calculated by k-nearest neighbor algorithm. A graph can be constructed by treating the expression profiles of the cells as nodes and the adjacency matrix as edges.
The DGI generates an encoder \({{\rm{\varepsilon }}}\left({{\bf{X}}},{{\bf{A}}}\right)={{\bf{H}}}=\left({{{\boldsymbol{h}}}}_{{{\boldsymbol{1}}}},{{{\boldsymbol{h}}}}_{{{\boldsymbol{2}}}},{{\boldsymbol{\ldots }}},{{{\boldsymbol{h}}}}_{{{\boldsymbol{N}}}}\right)\) which represents node embeddings for the cells. The encoder reduces the high dimensional input data to be a representation with fixed low dimension (D = 50), enabling the integration of multi-omics data with significant dimensional differences. A node embedding hi summarizes a patch of graph that centers around a node which captures the neighborhood information of the node. The encoder is learned by maximizing the local mutual information, i.e. generating local node representations that best maintain the global information of the entire graph represented by a summary vector s. A discriminator is defined to quantify the local mutual information:
Where M denotes learnable weights, s is a summary of all the node embeddings obtained by a readout function \({{\boldsymbol{s}}}={{\mathcal{R}}} {({{\bf{H}}})}\). We simply average all the node features to get s. A negative discriminator is defined by paring the summary vector s with a corrupted graph \((\widetilde{{{\bf{X}}}},\widetilde{{{\bf{A}}}})\) obtained by randomly permuting the nodes. In permutation, X is shuffled while the adjacent matrix A remains the same: \(\widetilde{{{\bf{A}}}}={{\bf{A}}}\). Direct shuffling for negative pairs generation offers simplicity and preserves overall data characteristics, but it may be less effective for highly structured data with clearly separated cell types. In our analysis, all five spatially resolved datasets displayed cell types in a continuous manner with minimal separation (e.g., Figs. 2d, i; 3e), making the current shuffling strategy effective for the DGI model. In future work, we plan to explore more sophisticated sampling strategies to enhance COSMOS’s adaptability to diverse datasets, such as adversarial approaches or cluster-based methods to generate negative pairs.
DGI employs a contrastive strategy by maximizing the following objective function:
Where hi is the embedding node of the graph of input data, \(\tilde{{{{\boldsymbol{h}}}}_{{{\boldsymbol{J}}}}}\) is the embedding node of the corrupted graph. This contrastive strategy does not calculate the contrastive loss directly from pairs of positive and negative samples. Instead, it pairs the embeddings of positive and negative samples with a single global summary vector s. The summary vector s is computed as sigmoid of the mean of all positive sample embeddings. Consequently, the contrastive learning in DGI is much less computationally intensive with a computational complexity of O(N) rather than O(N2).
While GCNs in many other works primarily focus on local neighborhood aggregation within the spatial graph, DGI goes a step further by capturing global information using local node representations. This approach enables the GCN encoder to integrate globally relevant data across the entire tissue, resulting in richer and more comprehensive node embeddings that reflect both local and global graph structures. Consequently, DGI enhances the overall performance and robustness of our model compared to traditional GCN methods.
Because the real-world spatially resolved multi-omics experimental data we studied have been focusing on two paired omics that could extract two sets of omics information from the same cells, we adapted the traditional DGI model to a two-channel model, which takes the input of two paired omics data with shared spatial positions through two encoders \({{{\rm{\varepsilon }}}}_{1}\left({{{\bf{X}}}}_{{{\boldsymbol{1}}}},{{\bf{A}}}\right)\) and \({{{\rm{\varepsilon }}}}_{2}\left({{{\bf{X}}}}_{{{\boldsymbol{2}}}},{{\bf{A}}}\right)\), where \({{{\bf{X}}}}_{{{\boldsymbol{1}}}},{{{\bf{X}}}}_{{{\boldsymbol{2}}}}\) are the expression profiles of two omics, \({{\bf{A}}}\) is the shared spatial adjacency matrix. The node embeddings of the two graphs \({{{\boldsymbol{h}}}}_{{{\bf{i}}}}^{{{\bf{1}}}},{{{\boldsymbol{h}}}}_{{{\bf{i}}}}^{{{\bf{2}}}}\) are integrated by the Weighted Nearest Neighbor (WNN) algorithm13:
Where \({w}_{i}^{1}\), \({w}_{i}^{2}\) represent WNN weights for each cell \(i\) in the two omics. The WNN weights are calculated by applying WNN algorithm to node embeddings of the two omics:
Where \({r}_{1,2}(i)\) is pairwise affinity ratio between omics 1 and 2 on cell \(i\). The pairwise affinity ratio is calculated by defining the within and cross-modality affinities between within and cross-modality predicated and actual embedding values for each node. See the details of WNN weights calculation in Hao et al.13. The approach that uses a weighted sum to achieve integration may exacerbate the noise effects that exist in each modality. However, the modality weights used here are determined by the WNN algorithm, which calculates the contributions of cells in each modality to the predictability of expression profiles of the cells. Thus, the weights are related to the noise in the modality. For example, if a group of cells has a high level of noise in RNA modality, the RNA modality weights of these cells would be small, and the RNA profiles of these cells would have limited contributions to the integrated embedding. Therefore, the calculation of weights by WNN helps to mitigate modality-specific noise during the integration process. We plan to explore the incorporation of noise-robust fusion methods, such as the Kalman Filter, to further enhance the integration process of COSMOS in future work.
The integrated embedding nodes from corrupted graphs are calculated with the same WNN weights:
Where \(\widetilde{{{{\boldsymbol{h}}}}_{i}^{1}},\widetilde{{{{\boldsymbol{h}}}}_{i}^{2}}\) represent node embeddings of corrupted graphs for the two omics. The same contrastive strategy in Eq. (3) is applied to the integrated node embeddings which gives the objective function of the two-channel DGI model:
Where \({{{\boldsymbol{h}}}}_{{{\rm{i}}}}^{{{\rm{int}}}}\) is the integrated embedding node of the two graphs calculated by Eq. (4), \(\tilde{{{{\boldsymbol{h}}}}_{j}^{int}}\) is the integrated embedding node of the two corrupted graphs calculated by Eq. (6). To enforce the spatial consistency of the embeddings, we added a spatial regularization term to the objective function to make the closeness of embedding points similar as the spatial proximity of cells. The final objective function can be expressed as:
Where \({D}_{i,j}^{s}\) is the spatial distance between cell \(i\) and \(j\), \({D}_{i,j}^{e}\) is the embedding distance between cell \(i\) and \(j\) in the embedding space. \(\alpha\) is the parameter to control the spatial regularization strength.
COSMOS shares a similar structure with SpatialGlue, but there are two key differences in how the GCN model and omics integration are implemented in these methods. First, when learning each omic, SpatialGlue implemented a GCN-encoder for spatial adjacency graph and feature graph separately to generate two node representations. The feature graph was constructed by simply applying KNN (K = 20) on the PCA embeddings which only involves local information. In contrast, COSMOS used DGI to obtain node representations that capture the global patterns in the whole graph. Thus, COSMOS is more effective in integrating global spatial information and better preserves the global spatiotemporal patterns of the spatial omics data (Figs. 2e,j; 3f). Second, SpatialGlue uses attention coefficients to weigh the two graph-specific node representations before integrating them into a final node representation. This approach introduces additional trainable weights, increasing the complexity of the weight space and making the model more susceptible to noise, as reflected in the noisy domain segmentation results of SpatialGlue in Figs. 3c and 4e. In contrast, COSMOS employs the widely used Weighted Nearest Neighbor (WNN) method to calculate the weights of each omics. The WNN weight calculation is independent of the DGI training, which simplifies the model and enhances its stability.
Training of COSMOS
The WNN weights are computationally slow to calculate and are independent of the parameters of the two-channel DGI model. For stability and efficiency of the training, we did not calculate WNN weights at every iteration, but fixed the WNN weights first and updated them one time after a given number of iterations (“wnn_epoch”) or the maximal number of patience (“max_patience_bef”) is reached. The patience is defined as the accumulated number of iterations that have larger loss than the most recent minimal loss. After the calculation of WNN weights, we reset the patience to 0 and fixed WNN weights until the number of iterations reaches “total_epoch” or the number of new patience reaches “max_patience_aft”. The training procedures of COSMOS are summarized below:
-
1.
Setting all the WNN weights of both omics to be 0.5.
-
2.
Training the model by maximizing the objective function in Eq. (8) with fixed WNN weights which were set in step 1.
-
3.
Calculating WNN weights by Eq. (5) when the number of iterations reaches “wnn_epoch” or the number of patience reaches “max_patience_bef”, then resetting the patience to be 0.
-
4.
Training the model by maximizing the objective function in Eq. (8) with fixed WNN weights which were updated in step 3, until the number of iterations reaches “total_epoch” or the number of new patience reaches “max_patience_aft”.
Parameters of COSMOS
One of the key parameters in COSMOS is the spatial regularization strength \(\alpha\), which controls the alignment between embedding distances and spatial distances. This regularization helps preserve the fine spatial structure of certain cell types. For instance, some cell types cannot be effectively separated when \(\alpha\) = 0, such as RMS cells in the simulated Stereo-Seq mouse olfactory bulb data (Fig. S4b), the C2 cluster in the DBiT-seq mouse embryo brain RNA-Protein data (Fig. S5a), and the C6 cluster in the spatial-CITE-seq human tonsil RNA-Protein data (Fig. S5b). However, excessive regularization can reduce the effectiveness of omics expression profiles. For example, the Adjusted Rand Index (ARI) significantly decreases when \(\alpha\) exceeds 0.1 in the STARMap mouse visual cortex data (Fig. S4a), the simulated Stereo-Seq mouse olfactory bulb data (Fig. S4b), and the ATAC-RNA-seq mouse brain multi-omics data (Fig. S4c). Based on our analysis of these five datasets, we recommend setting \(\alpha\) within the range of [0, 0.1]. The specific \(\alpha\) values used for generating the figures in these datasets are listed in Supplementary Data 4.
Another parameter influencing the embedding result is ‘max_patience_bef’ (denoted as ‘P’ in the figures), which controls the timing of performing WNN. If this parameter is too small, it may result in insufficient training of the DGI model before performing WNN. Conversely, if it is too large, it can lead to model overfitting and sub-optimal results. As the number of iterations increases, the modality weights for clusters from the two modalities converge to stable values (Fig. S6d-f, j-l). We found that an intermediate value for ‘max_patience_bef’ (P = 10, Fig. S6b, h) provides better results compared to both smaller (P = 1, Fig. S6a, g) and larger values (P = 50, Fig. S6c, i). For example, with P = 50, more sub-clusters appear in the CP and ACB regions in the ATAC-RNA-seq mouse brain multi-omics data (Fig. S6c), and the C1 cluster (defined in Fig. 4h) cannot be distinguished from neighboring regions in the DBiT-seq mouse embryo brain RNA-Protein data (Fig. S6i). Based on the analysis of five datasets, we recommend setting ‘max_patience_bef’ within the range of [5, 30]. When we observed a slow decrease in loss during initial iterations, as seen in the spatial-CITE-seq human tonsil RNA-Protein and slide-tags human melanoma RNA-ATAC multi-omics data, we set ‘max_patience_bef’ to be 20. The parameters used for generating the figures in these datasets are listed in Supplementary Data 4.
Some other parameters were set to be the same throughout the analyses: the optimizer for training the DGI is “Adam” with a default learning rate set as lr = 0.001, the maximal number of iterations before running WNN is set as wnn_epoch = 500, the maximal number of total epochs is set as total_epoch = 1000. The early stopping strategy was used to avoid overfitting. The minimal number of epochs before early stopping is set as min_stop = 200, and the patience for the loss after WNN is set as max_patience_aft = 30. We fixed the random seed to be 20 for reproducibility in the COSMOS results.
Computational cost of the model
The computational cost of the model consists of two parts: the two-channel DGI training and the WNN computation. For the two-channel DGI training, the contrastive strategy in DGI (Eq. (3)) has a computational complexity of \({\rm O}(N)\), and the cost largely depends on the calculation of spatial regularization loss in Eq. (8). The pairwise embedding distances calculation has a computational complexity of \({\rm O}({N}^{2})\), where \(N\) is the number of cells. However, we only computed the distances of a random fixed number of cell pairs when \(N\) is larger than 5000, which reduces the quadratic cost to constant. We fixed the number of cell pairs at 1,000,000, which is significantly smaller than the full set of edges for over 5000 cells. However, this number is sufficiently large to approximate the results obtained with the full set, as demonstrated in the ATAC-RNA-seq mouse brain multi-omics data with 9215 cells (Fig. S7). For the WNN computation, the computational complexity is \({\rm O}({N}^{2})\), but it’s a one-time calculation and thus feasible for large datasets. Thus, the total computational complexity of COSMOS for a large dataset is Ο(N). The COSMOS training function, executed on the Tesla V100-SXM2-32GB GPU equipped with 5120 CUDA cores and 32 GB of memory, took 24 seconds for the mouse visual cortex dataset containing 1207 cells and 230 seconds for the mouse olfactory bulb dataset containing 18,196 cells.
Domain segmentation
After integration of the paired omics in each dataset, we performed clustering for the integrated embeddings of ATAC-RNA-seq mouse brain multi-omics data by “louvain” and all other data by “leiden” with “scanpy” python package.
UMAP analysis
We performed umap analysis by using “umap-learn” python package. The metric is set as default, the min_dist is set as 0.3, the n_neighbor is set as 30.
Pseudo-spatiotemporal map generation
The pseudo-spatiotemporal map (pSM) is generated by running diffusion pseudotime (DPT) algorithm by “scanpy” python package with default parameters. We manually set the root cell to be the first cell of a given cell type. The cell type selected to define root is “HPC” in STARmap Stereo-Seq mouse visual cortex data, “RMS” in mouse olfactory bulb data, “CP2” in ATAC-RNA-seq P22 mouse brain data.
Pseudo-time associated genes
For each gene, we calculated the Pearson correlation coefficient between the gene expressions across cells and the pSM values (by COSMOS) of the cells. The significantly correlated genes with P < 1e-10 were selected as pseudo-time-associated genes.
GOBP analysis for pseudo-time-associated genes
We used Gene Set Enrichment Analysis (GSEA) (https://www.gsea-msigdb.org/gsea/index.jsp) to perform the Gene Oncology Biological Process (GOBP) analysis for the pseudo-time associated genes. The positively and negatively associated genes were analyzed separately.
Statistics & reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Algorithms availability
We used the default parameters in SpatialGlue, CellCharter and SpaMultiVAE from the original scripts. The parameters in SpaceFlow and WNN can be found in Supplementary Data 4.
SpaceFlow: https://github.com/hongleir/SpaceFlow
WNN: https://github.com/dylkot/pyWNN
SpatialGlue: https://github.com/JinmiaoChenLab/SpatialGlue
CellCharter: https://github.com/CSOgroup/cellcharter
SpaMultiVAE: https://github.com/ttgump/spaVAE
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this study can be obtained in raw from the original publications. STARmap mouse visual cortex transcriptomics data is available at the Dropbox (https://www.dropbox.com/sh/f7ebheru1lbz91s/AADm6D54GSEFXB1feRy6OSASa/visual_1020/20180505_BY3_1kgenes?dl=0&subfolder_nav_tracking=1). Stereo-Seq mouse olfactory bulb transcriptomics is available at the github page (https://github.com/JinmiaoChenLab/SEDR_analyses/tree/master/data). The ATAC-RNA-seq mouse brain multi-omics data is available at UCSC Cell Browser (https://brain-spatial-omics.cells.ucsc.edu). The DBiT-seq mouse embryo brain RNA-Protein multi-omics data is available at figshare database (https://figshare.com/articles/dataset/Spatial_genomics_datasets/21623148/5). The Spatial-CITE-seq human tonsil RNA-Protein multi-omics data is available at figshare database (https://figshare.com/articles/dataset/Spatial_genomics_datasets/21623148/5). The Slide-tags human melanoma RNA-ATAC multi-omics data is available at Broad Institute Database (https://singlecell.broadinstitute.org/single_cell/study/SCP2176/slide-tags-multiomic-snrna-seq-snatac-seq-on-human-melanoma#/). The processed data is available at Zenodo (https://zenodo.org/records/13932144). Source data are provided with this paper.
Code availability
The tutorial for implementing COSMOS to analyze spatially resolved paired multi-omics data is available at: https://github.com/Lin-Xu-lab/COSMOS.git and https://cosmos-tutorials.readthedocs.io/en/latest/index.html. It is also deposited at Zenodo dataset “14114770”. The GitHub repository was linked to Zenodo with the https://doi.org/10.5281/zenodo.1411477025.
References
Liu, Y. et al. High-Spatial-Resolution Multi-Omics Sequencing via Deterministic Barcoding in Tissue. Cell 183, 1665–1681 e1618 (2020).
Vickovic, S. et al. SM-Omics is an automated platform for high-throughput spatial multi-omics. Nat. Commun. 13, 795 (2022).
Deng, Y. et al. Spatial-CUT&Tag: Spatially resolved chromatin modification profiling at the cellular level. Science 375, 681–686 (2022).
Liu, Y. et al. High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq. Nat. Biotechnol. 41, 1405–1409 (2023).
Jiang, F. et al. Simultaneous profiling of spatial gene expression and chromatin accessibility during mouse brain development. Nat. Methods 20, 1048–1057 (2023).
Ben-Chetrit, N. et al. Integration of whole transcriptome spatial profiling with protein markers. Nat. Biotechnol. 41, 788–793 (2023).
Zhang, D. et al. Spatial epigenome-transcriptome co-profiling of mammalian tissues. Nature 616, 113–122 (2023).
Chen, J. et al. Spatially resolved multi-omics unravels region-specific responses, microenvironment remodeling and metabolic reprogramming in aristolochic acid nephropathy. Innov. Med. 100066 (2024).
Russell, A. J. C. et al. Slide-tags enables single-nucleus barcoding for multimodal spatial genomics. Nature 625, 101–109 (2024).
Hu, J. et al. SpaGCN: Integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat. Methods 18, 1342–1351 (2021).
Ren, H., Walker, B. L., Cang, Z. & Nie, Q. Identifying multicellular spatiotemporal organization of cells with SpaceFlow. Nat. Commun. 13, 4076 (2022).
Cao, Z. J. & Gao, G. Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nat. Biotechnol. 40, 1458–1466 (2022).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529 (2021).
Lynch, A. W. et al. MIRA: joint regulatory modeling of multimodal expression and chromatin accessibility in single cells. Nat. Methods 19, 1097–1108 (2022).
Gayoso, A. et al. Joint probabilistic modeling of single-cell multi-omic data with totalVI. Nat. Methods 18, 272–282 (2021).
Klein, D. et al. Mapping cells through time and space with moscot. bioRxiv, 2023.2005. 2011.540374 (2023).
Varrone, M., Tavernari, D., Santamaria-Martinez, A., Walsh, L. A. & Ciriello, G. CellCharter reveals spatial cell niches associated with tissue remodeling and cell plasticity. Nat. Genet 56, 74–84 (2024).
Tian, T., Zhang, J., Lin, X., Wei, Z. & Hakonarson, H. Dependency-aware deep generative models for multitasking analysis of spatial omics data. Nat. Methods (2024).
Long, Y. et al. Deciphering spatial domains from spatial multi-omics with SpatialGlue. Nat Methods (2024).
Veličković, P. et al. Deep graph infomax. arXiv preprint arXiv:1809.10341 (2018).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Haghverdi, L., Buttner, M., Wolf, F. A., Buettner, F. & Theis, F. J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792 e1721 (2022).
Song, D. et al. scDesign3 generates realistic in silico data for multimodal single-cell and spatial omics. Nat. Biotechnol. 42, 247–252 (2024).
Zhou, Y. et al. Cooperative Integration of Spatially Resolved Multi-Omics Data with COSMOS. COSMOS, https://doi.org/10.5281/zenodo.14114770 (2024).
Acknowledgements
The resources of the high-performance computing environment from Quantitative Biomedical Research Center (QBRC) and BioHPC at UT Southwestern Medical Center, as well as the Texas Advanced Computing Center (TACC) at The University of Texas at Austin, are gratefully acknowledged. We also thank Ms. Jessie Norris for proofreading this manuscript. This work was supported by the following funding: the Rally Foundation, Children’s Cancer Fund (Dallas), the Cancer Prevention and Research Institute of Texas (RP180319, RP200103, RP220032, RP170152 and RP180805), and the National Institutes of Health funds (R21CA259771, P30CA142543, HG011996, and R01HL144969) (to L.X.); the National Institutes of Health (1R01GM115473, 1R01GM140012, 5R01CA152301, P30CA142543, P50CA70907, R35GM136375); and the Cancer Prevention and Research Institute of Texas (RP180805, RP190107) (to G. X.).
Author information
Authors and Affiliations
Contributions
Y.Z. and L.X. conceived and designed the study. Y.Z. and X.X. developed the COSMOS algorithm and performed the data analysis. L.D. performed the bioinformatics analyses. C.T. generated GitHub page for COSMOS software. L.X. and G.X. acquired the funding. Y.Z., G.X. and L.X. wrote and revised the manuscript. All authors have read, revised, and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhou, Y., Xiao, X., Dong, L. et al. Cooperative integration of spatially resolved multi-omics data with COSMOS. Nat Commun 16, 27 (2025). https://doi.org/10.1038/s41467-024-55204-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-55204-y