Abstract
Cell type deconvolution methods can impute cell proportions from bulk transcriptomics data, revealing changes in disease progression or organ development. But benchmarking studies often use simulated bulk data from the same source as the reference, which limits its application scenarios. This study examines batch effects in deconvolution and introduces SCCAF-D, a computational workflow that ensures a Pearson Correlation Coefficient above 0.75 across simulated and real bulk data for various tissue types. Applied to non-alcoholic fatty liver disease, SCCAF-D unveils meaningful insights into changes in cell proportions during disease progression.
Similar content being viewed by others
Introduction
Understanding the cell-type composition change during disease progression is critical in studying disease mechanisms1. To date, some large-scale databases, including the Gene Expression Omnibus (GEO)2, ArrayExpress3, Genotype-Tissue Expression (GTEx)4, The Cancer Genome Atlas (TCGA)5, Therapeutically Applicable Research To Generate Effective Treatments (TARGET) (https://ocg.cancer.gov/programmes/target), International Cancer Genome Consortium (ICGC)6, and etc, have amassed extensive repositories of bulk RNA sequencing (RNA-seq) data resources crucial for both biological and clinical investigations. Of note, bulk RNA-seq fails to discern gene expression differences among individual cells, thereby masking cellular heterogeneity. For instance, a significant risk factor for the progression from non-alcoholic fatty liver disease (NAFLD) to hepatocellular carcinoma (HCC) is the degree of liver fibrosis7. The existing databases encompass vast quantities of bulk RNA-seq data pertaining to liver fibrosis staging, yet they lack key information on cellular proportion changes during disease progression. Deconvolving bulk RNA-seq samples using reference data provides an alternative, which may make full reuse of the existing bulk RNA-seq data by generating the cell composition of these samples8,9,10,11,12,13,14,15,16,17, indicating the biological variation during disease progression.
Tens of computational deconvolution methods have been proposed, while reference-based methods are more accurate when reliable reference data is available11,18. Before applying deconvolution algorithms, it is necessary to benchmark their capabilities and limitations with “gold standard” datasets of known cell proportions. For the establishment of “gold standard” benchmark data, one approach is to determine the cell proportion in bulk data using alternative experiments, e.g., flow cytometry and immunohistochemistry, when identical samples are available. Another approach takes single-cell RNA sequencing19 (scRNA-seq) data as ground truth and simulates bulk RNA-seq data, as ‘pseudobulk’18,20, by aggregating the expression values in scRNA-seq, where the cell composition is known. Based on this ‘pseudobulk’ simulation, some previous benchmark studies18 used a ‘self-reference’ setting, in which both the simulated ‘pseudobulk’ and the reference data originate from the same dataset. However, this “self-reference” setting may not accurately represent a realistic scenario where batch effects exist. In realistic applications of deconvolution, the bulk data and the reference data normally come from different experimental batches, studies or sources, termed as a ‘cross-reference’ setting. Considering batch effects can arise when adding replicates from different sources, such as laboratories, donors, or sequencing platforms, using reference data in cell type deconvolution from a different source may lead to unexpected variations. Therefore, we term this situation as batch effects in deconvolution, which should be benchmarked with a ‘cross-reference’ setting. To date, benchmarking of cell type deconvolution has exclusively been conducted under the ‘self-reference’ setting, leaving the performance of deconvolution in the ‘cross-reference’ setting unclear.
This work demonstrates that batch effects in the ‘cross-reference’ setting affect the deconvolution accuracy to a large extent, while the existing deconvolution approaches show limited capabilities in dealing with such batch effects. Therefore, we present ‘SCCAF-D’ (Single-Cell Clustering Assessment Framework optimised reference for Deconvolution), a framework that achieves stable accuracies about 0.75 Pearson Correlation Coefficient (PCC) in a ‘cross-reference’ setting via optimally prepared reference data. In particular, we delineate the capabilities and robustness of SCCAF-D through the benchmarking of both simulated datasets across five different tissues and eight real bulk datasets from both healthy and diseased samples. Applying SCCAF-D to bulk NAFLD data, we discover the continuous cell proportion changes of cholangiocytes, hepatic stellate cells (HSCs), and hepatocytes during disease progression.
Results
Batch effects hinder cell type deconvolution
We have observed inconsistencies in cell proportion statistics across 15 datasets due to donor, study, and sequencing technology effects (Fig. 1 and Supplementary Figs. 1, 2). According to the human pancreas single-cell datasets from Baron et al.21, distinct cell types are clearly discriminated in the Uniform Manifold Approximation and Projection (UMAP) through our analysis (Fig. 1a). Donor effects manifest in both variations in the gene expression profile (Fig. 1b) and the cell proportion (Fig. 1c and Supplementary Data 1). For instance, acinar cells were exclusively observed in donors 1 and 3 but not in donors 2 and 4. Notably, such batch effects in the transcriptional profiles and in the cell proportions also exist between different single-cell studies (Fig. 1d–f). And the cell proportions also differ among samples in bulk RNA-seq (Fig. 1g) or scRNA-seq (Fig. 1h), resulting in unforeseen biases in deconvolution.
The UMAP plots show the Baron et al. dataset according to a cell type and b donor. c The stacked bar plot shows the cell proportions of each donor (Y axis) in the Baron et al. dataset. The colour bar indicates the cell types. The UMAP shows the Baron et al. dataset and Segerstolpe et al. dataset according to cell type d and study e. f The stacked bar plot shows the cell proportion of each study (Y axis) for the Segerstolpe et al. dataset and Baron et al. dataset. The colour bar is the same with d. g The stacked bar plot shows the real cell proportion for each sample within the PBMC bulk RNA-seq dataset, with cell type determinations based on flow cytometry from Finotello et al.31 dataset. The plot displays the distribution of cell types as determined by flow cytometry across all samples. h The stacked bar plot shows the cellular proportions of individual healthy samples within a 10X single-cell dataset from Lee et al. Each bar represents the relative abundance of different cell types within each sample. The scatter plots show the results of DWLS using the different donors as reference or pseudobulk. Specifically, data from donor 1 is used to simulate pseudobulk, while data from donor 2 i and donor 4 j are used as references. The scatter plots show the results of DWLS using different studies as reference or pseudobulk. The data from Marquina-Sanchez et al.26 is used as pseudobulk, the data from Muraro et al.27 is used as reference in panel k and it is the opposite in panel l. X axis is the predicted cell proportion, Y axis is the true cell proportion. Panels a–c show within dataset batch effects, panels d–f show between dataset batch effects, panels g, h show between technologies comparison, while panels i–l show deconvolution results from cross-donor i, j and cross-study k, l scenarios. Each group of panels are coloured in the same colour scheme. Two-sided t test is used to calculate the p values in panels i–l. The p value is nearing zero in i–l. MHC class ll: major histocompatibility complex ll; PSC: pancreatic stellate cell; T: T cell; CD4 + T: CD4 positive T cell; CD8 + T: CD8 positive T cell; Treg regulatory T cell, B B cell, NK natural killer cell, Mono monocyte cell, DC dendritic cell, HSPC hematopoietic stem/progenitor cell, RBC red blood cell, AT1 alveolar epithelial type I, AT2 alveolar epithelial type ll. Source data are provided as a Source Data file.
To understand the deconvolution capability in a ‘cross-reference’ setting, we first identified candidate algorithms from a ‘self-reference’ benchmarking of 25 deconvolution algorithms on 44 datasets (Supplementary Data 2). Here, we define ‘self-reference’ as the situation when the simulated pseudobulk data for testing comes from the same dataset as the reference data, while ‘self-reference’ is assumed to be a simpler case than ‘cross-reference’. Six top-ranking algorithms (DWLS22, FARDEEP14, MuSiC23, NNLS24, RLR (see Methods), and EpiDISH12) (Supplementary Fig. 3 and Supplementary Data 3) according to the ‘self-reference’ results, were used for ‘cross-reference’ studies. Several ‘cross-reference’ studies were designed as below. In a ‘cross-donor’ setting, single-cell data (Barga et al.25) from one donor is used to simulate ‘pseudobulk’, while data from another donor is used as reference. The deconvolution approach using the DWLS algorithm yields varying accuracies depending on simulated bulk and reference data utilised (Fig. 1i, j). Subsequently, in a ‘cross-study’ setting, data from one study is used to simulate ‘pseudobulk’, while another study of the same experiment is used as reference. When the dataset from Marquina-Sanchez et al.26 was treated as pseudobulk and the dataset from Muraro et al.27 was used as the reference, the predicted proportions of the seven cell types (including alpha, beta, and duct cells) showed a PCC of 0.59 with the ground truth (Fig. 1k). Conversely, when the Muraro et al.27 dataset was used as pseudobulk and the Marquina-Sanchez et al.26 dataset served as the reference, the predicted cell type proportions exhibited a higher PCC of 0.85 with the ground truth (Fig. 1l). It shows similar results as in the ‘cross-donor’ setting, while certain cell types are either overestimated or under-estimated, indicating the batch effects. Importantly, all algorithms are affected by such batch effects to a certain extent (Supplementary Fig. 4), suggesting the necessity of alleviating batch effects in deconvolution.
SCCAF-D: Alleviating batch effects in deconvolution using optimised references
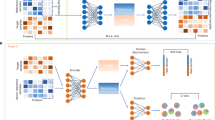
The deconvolution accuracy highly depends on the reference data, whose quality is not detectable a priori. To achieve stable deconvolution accuracy, we introduce SCCAF-D, a computational framework designed to alleviate batch effects through optimised single-cell references, coupled with the DWLS algorithm to infer cell-type composition from bulk-profiled samples. It first integrates multiple datasets and selects the ‘self-consistent’ part of the data as reference. Given that a single dataset can be biased on certain batch effects, SCCAF-D integrates multiple datasets to optimise the cell type annotation for the reference data. Specifically, SCCAF-D integrates datasets by using Harmony28 and then re-annotates the cell type according to the Leiden clustering and cell type labels obtained from the original publications, resulting in a more coherent cell type annotation across datasets.
Although multiple datasets suffer from batch effects, they share the same biology, which should be ‘self-consistent’. Therefore, if a machine learning model can effectively extract the biology from part of the dataset, it should successfully recover the cell type labels in the rest of the data. SCCAF-D selects a ‘self-consistent’ part of the data as an optimised reference based on a ‘self-projection’ approach29. It first trains a machine learning (Logistic Regression) model based on part of the data together with the cell type labels, and predicts the cell types labels on the rest of the data, known as ‘self-projection’29 (Fig. 2, see Methods). A cell whose original cell type label is the same as the machine learning assigned label is ‘self-consistent’, meaning that its expression profile is discriminative in machine learning. We hypothesise that each cell type has a set of differentially expressed genes that allow it to distinguish from other cell types. If the labels are self-consistent, the differentially expressed genes can be encoded both by the machine learning model and its original gene expression profile. These self-consistent cells, who are discriminative to each other, can better represent their biology and be a good reference for deconvolution.
SCCAF-D first combines different single-cell datasets from the same tissue to an integrated dataset and optimises the cell annotation. It then identifies representative cells, which are self-consistent, using a ‘self-projection’ approach from SCCAF (the block boxed by orange dashed line). The integrated dataset is split into training set and testing set, whereas training set is used to train a machine learning model of logistic regression. The machine learning model is applied to the test set to give a predicted cell type score matrix. This predicted score matrix is compared with the original cell type labels from the optimised cell annotation. For each cell type, the top 100 cells with the highest predicted scores are selected as reference data. Subsequently, this optimised reference is used to perform cell type deconvolution with DWLS, yielding cell proportions. The UMAP visualisation of single-cell reference data before and after SCCAF-D optimisation, with grey shape indicating cells excluded by SCCAF-D and other cells of other colours as selected representative cells.
SCCAF-D achieves a high accuracy on simulated bulk datasets
The performance of SCCAF-D was tested on simulated ‘pseudobulk’ data of a ‘cross-reference’ setting, where the ‘pseudobulk’ data and the reference data come from two single-cell datasets of known cell proportions, see Methods. SCCAF-D was compared with other five top-ranking deconvolution algorithms (FARDEEP, MuSiC, RLR, EpiDISH, and NNLS) on 20 scRNA-seq datasets of five tissue types (4 datasets each), including pancreas (n = 34 specimens), lung (n = 61 specimens), peripheral blood mononuclear cells (PBMCs) (n = 90 specimens), brain (n = 43 specimens) and spleen (n = 21 specimens). For each tissue, one dataset was used to simulate the pseudobulk, while each of the other three datasets is used as the reference in the ‘cross-reference’ setting. In comparison, SCCAF-D integrates these three datasets as an optimised reference. We then quantitatively evaluate the deconvolution performance by computing PCC, Root-mean-square error (RMSE) (Supplementary Data 4), and Jensen-Shannon Divergence (JSD) values (Supplementary Fig. 5a–c, and Supplementary Fig. 6a, b) between the estimated cell-type composition and ground truth.
When deconvolving the pancreas pseudobulk of Baron et al.21 using the DWLS algorithm (Fig. 3a and Supplementary Fig. 7a), reference Marquina et al.26 yielded PCCs of 0.37, while reference Segerstolpe et al.30 resulted in PCC of 0.60. Similarly, when using reference Muraro et al.27 to deconvolve the pseudobulk of Baron et al.21, PCCs were 0.15, 0.15 and 0.40 for EpiDISH, RLR and MuSiC, respectively. Although DWLS demonstrates top-ranking accuracies in many cases, PCC can decrease to 0.37 in certain cases (e.g., when Baron et al.21 dataset is used for pseudobulk, Marquina-Sanchez et al.26 dataset as reference), while other algorithms also exhibit low PCCs in some cases (EpiDISH: 0.15; FARDEEP: −0.63; MuSiC: 0.40; NNLS: 0.11; RLR: 0.15) indicating potential failures.
The heatmaps show the deconvolution results of PCCs on simulated bulk data from a pancreas, b lung, c PBMC, d brain and e spleen tissue, respectively. Each panel is a row of heatmaps. X axis shows the references, and Y axis shows simulated pseudobulk. ‘Optimised Ref’ labels the optimised reference data from SCCAF-D. Two-sided t test is used to calculate the p values. R: pearson correlation coefficient. Source data are provided as a Source Data file.
As for lung (Fig. 3b and Supplementary Fig. 7b) and PBMC datasets (Fig. 3c and Supplementary Fig. 7c), SCCAF-D also demonstrates PCCs above 0.82 and 0.89, respectively, suggesting a reliable performance. Specifically, DWLS performs better than others when using one dataset as reference, yet its accuracy depends on the choice of reference data. In lung datasets, the lowest PCCs for DWLS, FARDEEP, MuSiC, NNLS, RLR, EpiDISH are 0.66, 0.42, 0.62, 0.11, 0.42, 0.42, respectively. The same PCCs are as low as 0.60, 0.09, 0.04, 0.16, 0.16, 0.16 in PBMC datasets.
We also performed simulations on the human brain (Fig. 3d and Supplementary Fig. 8a) and spleen (Fig. 3e and Supplementary Fig. 8b) datasets to evaluate the performance of SCCAF-D, which achieved PCCs above 0.93 and 0.80, respectively. Of note, when using DWLS with a single reference, the PCCs vary from −0.24 to 0.99 and RMSEs change from 0.02 to 0.22. By contrast, using the SCCAF-D optimised reference, the PCCs improved to the range of 0.93 to 0.99. Furthermore, using any spleen dataset as a reference, DWLS consistently shows a positive correlation between predicted and true cell proportions, outperforming the other five algorithms. Similarly, the accuracy of DWLS on the spleen is influenced by the choice of the reference (PCCs from 0.41 to 0.99), while the accuracy for the SCCAF-D optimised reference increased to a range of 0.80 to 0.99.
The deconvolution accuracy of the DWLS algorithm depends on the choice of the reference dataset, with PCCs ranging from 0.37 to 0.99, 0.66 to 0.98, 0.60 to 0.98, −0.24 to 1, and 0.41 to 0.99 for pancreas, lung, PBMC, brain, and spleen pseudobulk datasets, respectively. In comparison, SCCAF-D improves PCCs to ranges of 0.86 to 0.99, 0.82 to 0.98, 0.89 to 0.98, 0.93 to 0.99, and 0.80 to 0.99, respectively, guaranteeing reliable deconvolution performance (>0.80 PCC).
Deconvolving real bulk datasets using SCCAF-D
Further benchmarks of these six algorithms were performed on real bulk PBMC datasets of RNA-seq or microarray (Finotello et al.31, Newman et al.32 and Monaco et al.33, see Methods), whose cell proportions were known a priori according to the flow cytometry count data. The single-cell reference datasets were sourced from Arunachalam et al.34 (n = 12 specimens), Lee et al.35 (n = 15 specimens), Schulte et al.36 (n = 49 specimens) and Wilk et al.37. (n = 14 specimens). To evaluate both the accuracy and robustness of SCCAF-D in predicting cell type proportion changes in disease states, SCCAF-D was applied to type 2 diabetes (T2D) data from Fadista et al.38 (n = 89 specimens) and idiopathic pulmonary fibrosis (IPF) data from McDonough et al.39 (n = 84 specimens), Sivakumar et al.40 (n = 72 specimens), and Furusawa et al.41 (n = 206 specimens).
In cross-reference benchmarking with a single reference dataset, DWLS shows higher accuracies than others (Fig. 4a, b). When deconvolving the Finotello et al.31 dataset, DWLS demonstrated the highest PCC of 0.86 and lowest Root-mean-square error (RMSE) of 0.06 with Wilk et al.37 dataset as reference, being higher than the second PCC of 0.64 (RMSE of 0.10) from MuSiC. Similarly, when using reference datasets from Arunachalam et al.34, Lee et al.35, and Schulte et al.36, DWLS consistently outperformed other methods, exhibiting PCCs of 0.82, 0.64, and 0.78 (RMSE: 0.09, 0.12 and 0.08), respectively. For Monaco et al.33 (microarray) dataset, when utilising the reference dataset from Schulte et al.36, the DWLS achieves the highest PCC of 0.67 and lowest RMSE of 0.10, while NNLS ranked second with a PCC of 0.51 and a RMSE of 0.26 using Arunachalam et al.34 as reference. When deconvolving the RNA-seq dataset from Monaco et al.33, DWLS maintained its superior performance over other algorithms (EpiDISH: 0.47, RLR: 0.47, FARDEEP: 0.45, MuSiC: 0.35, NNLS: 0.44), achieving the highest PCC of 0.87 and the lowest RMSE of 0.07 using Arunachalam et al.34 as reference. Using Lee et al.35 dataset as reference, DWLS achieved the highest PCC of 0.94 and lowest RMSE of 0.07 when deconvolving the Newman et al.32 dataset, while MuSiC obtained a PCC of 0.92 and an RMSE of 0.10. In spite of the top performance of DWLS in both simulated and real bulk data, its accuracies highly depend on the choice of the reference, showing a wide range of accuracies. In the datasets of Finotello et al.31, Monaco et al.33 (RNA-seq and microarray), and Newman et al.32, its PCCs ranged from 0.64 to 0.86, 0.47 to 0.67, 0.14 to 0.87 and −0.16 to 0.94, respectively (Supplementary Fig. 9). Consequently, without a clear understanding of which reference data to utilise, the deconvolution performance is not promising.
The histograms show a PCC and b RMSE for each real bulk dataset. Each bar represents the PCC or RMSE between estimated cell proportions and cell proportions quantified by flow cytometry, with colours indicating different datasets. * represents p value < 0.05. c The scatter plots show the PCC of each cell type for each reference using DWLS in the Newman et al.32 dataset. Each point represents an individual donor within each dataset. ‘Optimised Ref’ is the optimised reference from SCCAF-D, which is shown as the yellow bars in a, b and the yellow dots in c. Two-sided t test is used to calculate the p values. Error bands mean the 95% confidence region. CD4T: CD4 positive cell; CD8T: CD8 positive cell. Source data are provided as a Source Data file.
Fortunately, SCCAF-D shows top-ranking performance. This approach consistently achieves top-ranking accuracies across microarray dataset from Monaco et al.33, while also demonstrating exceptional performance on the other RNA-seq datasets. In comparison with DWLS with a single reference dataset (of lowest PCC −0.16 and highest RMSE of 0.32) (Fig. 4a, b), SCCAF-D achieves a lowest PCC of 0.75 and a highest RMSE of 0.10, suggesting a reliable accuracy (Supplementary Data 4). The other five algorithms (EpiDISH, FARDEEP, MuSiC, NNLS, and RLR) show improved deconvolution accuracy across four real bulk datasets by using optimised references, though none outperforms SCCAF-D. Following deconvolution with optimised references, the algorithms of EpiDISH, FARDEEP, MuSiC, NNLS, and RLR show higher PCCs on the Finotello et al.31 dataset compared to the lowest values (0.58, 0.24, 0.24, 0.19, 0.58, for these five algorithms, respectively.). Furthermore, SCCAF-D achieves stably high accuracies on other three datasets (microarray and RNA-seq in Monaco et al.33, and Newman et al.32). But the combinations between optimised reference and other five deconvolution algorithms do not always give improved accuracies as SCCAF-D, which is the combination between optimised reference and DWLS. Additionally, for each cell type across different bulk samples, relative cell proportion predicted from SCCAF-D exhibits a reliable positive correlation with the experiment determined proportion (Fig. 4c, Supplementary Figs. 10–12 and Supplementary Data 5), except for some minor cell populations (e.g., cDC), indicating the potential application in cross sample comparison.
Using the optimised reference derived from Baron et al.21, Marquina et al.26, Muraro et al.27 and Segerstolpe et al.30, 89 human pancreatic islet samples reported by Fadista et al.38 were deconvolved by SCCAF-D (Supplementary Fig. 13a). We identified a reduced proportion of β cells in diabetes samples, with a negative correlation observed between these proportions and haemoglobin A1C (HbA1c) levels (Supplementary Fig. 13b, c and Supplementary Data 5). This distribution aligns with existing knowledge42,43, suggesting the potential of SCCAF-D in inferring cell type composition under disease conditions.
In addition, SCCAF-D is able to recover the cell type proportion changes in IPF disease, which often leads to respiratory failure and death, with pathological features including alveolar epithelial cell injury, and excessive fibroblast proliferation44,45. SCCAF-D was applied to three human bulk RNA-seq datasets from both IPF (n = 198 specimens) and healthy lung tissues (n = 164 specimens), using two optimised references. One generated from the Adams44 (n = 107 specimens), Reyfman46 (n = 17 specimens), and Tsukui47 (n = 15 specimens) datasets (Fig. 5a), while another generated from the Habermann48 (n = 29 specimens), Valenzi49 (n = 11 specimens), and Morse50 (n = 17 specimens) datasets (Fig. 5b and Supplementary Data 5). In the IPF group, the proportions of alveolar epithelial type I (AT1) and type II (AT2) cells decrease compared to the control group, while the proportions of fibroblasts and basal cells increase, in consistency with known pathological features.
a The box plot shows the cell type proportions deconvolved using optimised reference 1. Optimised reference 1 was derived from Adams et al.44 (n = 107 specimens), Reyfman et al.46 (n = 17 specimens), and Tsukui et al.47 (n = 15 specimens) using SCCAF-D. Real bulk data were sourced from McDonough et al.39 (Nomal: 35 specimens; IPF: 49 specimens), Sivakumar et al.40 (Nomal: 26 specimens; IPF: 46 specimens), and Furusawa et al.41 (Nomal: 103 specimens; IPF: 103 specimens). Each column represents one bulk dataset. Blue represents samples from normal conditions, while red denotes samples from IPF. b The box plot shows the cell type proportions deconvolved using optimised reference 2. Optimised reference 2 was sourced from Habermann et al.48 (n = 29 specimens), Valenzi et al.49 (n = 11 specimens), and Morse et al.50 (n = 17 specimens) using SCCAF-D. Real bulk datasets and box colour are consistent with those in a. All box plots were plotted based on quartile values. The horizontal line within each box represents the median, while the lower and upper hinges correspond to the first and third quartiles, respectively. The upper whisker extends from the upper hinge to the largest value within 1.5 times the interquartile range (IQR) from the hinge. SMC: smooth muscle cell; EC endothelial cell. Source data are provided as a Source Data file.
SCCAF-D captures changes in cell proportions during NAFLD progression
Given that SCCAF-D achieves stable accuracies (PCCs above 0.75) on both simulated and real bulk data, it may reveal actual cell proportion changes during disease progression. Taking the Non-alcoholic fatty liver disease (NAFLD) as a test case, single-cell liver datasets, including MacParland et al. 51, Guilliams et al.52, Wang et al.53 and Tabula Sapiens-Liver et al.54, were used as references to deconvolve the bulk samples at liver fibrosis stages from 0 to 4, where 0 means no fibrosis and 4 means severe fibrosis. NAFLD bulk RNA-seq data with clinical information from Powell et al.55 (87 specimens) and Govaere et al.56 (206 specimens) were collected and the major 14 cell types were considered in SCCAF-D deconvolution (Fig. 6).
a The UMAP shows the cell type of the integrated dataset prior to SCCAF-D selection for the optimised reference. b The dot plot shows the expression of the differentially expressed genes, which are identified by SCCAF-D, across liver cell types. c The UMAP shows optimised reference selected by SCCAF-D. d The stacked bar plot shows the predicted cell proportion in different fibrosis stages of NAFLD bulk dataset. Different cell types are shown with different colours. e The violin plots show the estimated proportions of each cell type across fibrosis stages for all 87 specimens. Each point represents a different patient in the bulk datasets from Powell et al. 55 **** represents p value ≤ 0.0001, *** represents p value ≤ 0.001, ** represents p value ≤ 0.01, * represents p value ≤ 0.05, and ns represents p value ˃0.05. Two-sided t test is used to calculate the p values. HSC: hepatic stellate cells. Source data are provided as a Source Data file.
SCCAF-D first integrated the four reference datasets and optimised the cell type labels according to leiden clusters, Fig. 6a. Based on this integrated data with optimised labels, SCCAF-D trains a logistic regression model, in which the cell type related marker genes are encoded, as shown in Fig. 6b. Subsequently, SCCAF-D selects the ‘self-consistent’ cells, whose original cell type label is the same as assigned by the logistic regression model. These cells show clear separation between each other on the UMAP plot (Fig. 6c), indicating that they are representing more discriminative features for each cell type.
Proportions of main liver cell types, including hepatocytes, cholangiocytes, endothelial cells, HSC, plasma cells and T cells, change significantly across the fibrosis stages (Fig. 6d and Supplementary Data 6). The relative cell proportion of cholangiocytes increases gradually during fibrosis progression, while HSCs show significant increase in stage 4 compared with stage 0. In contrast, the relative cell proportion of hepatocytes decreases over fibrosis stages (Fig. 6e). Given the limited sample size in one dataset (87 specimens in Powell et al.55), data from Govaere et al.56 with 206 NAFLD specimens were utilised for validation. SCCAF-D with the same optimised reference was applied to Govaere et al.56, revealing similar patterns of cell type proportion changes across fibrosis stages (Supplementary Fig. 14).
Discussion
Batch effects between bulk and reference data have not yet been considered in cell type deconvolution. Due to the limited availability of reference datasets, previous studies and benchmarks always depend on a single reference dataset and a ‘self-reference’ setting which is over optimistic. In a more realistic ‘cross-reference’ setting, where the bulk (or simulated bulk) data from different sources than the reference, many deconvolution algorithms show low accuracies in certain datasets, despite demonstrating top-ranking accuracy in ‘self-reference’ tests (Figs. 3 and 4).
As the single-cell datasets accumulate over the past decade, more than one reference dataset are available for each tissue type. However, it is difficult to know which dataset to use as reference to achieve a good deconvolution result. But using a single reference dataset may result in a failure in certain cases that can lead to false conclusions when applying to real datasets57. Our benchmark results on both simulated and real bulk data highlight that the success of deconvolution depends on the choice of the reference data. Furthermore, the PCC evaluation metric demonstrates a partial bias of high values when multiple cell types are predicted around zero, which may hinder the deconvolution of heterogeneous datasets. For instance, with Lee et al.35 as the reference dataset, the PCC for CD8+ T cells in Newman et al.32 dataset reaches 0.66, yet the predicted cell proportion approaches zero (Fig. 4c). The bias is evidently present due to the prevalence of zero values, a phenomenon also observed by Jin et al.58.
To mitigate the effects from the reference data, SCCAF-D takes the advantage of multiple available reference datasets to obtain an optimised reference, which can achieve reliably high accuracies in combination with DWLS. It first optimises the cell type annotation through data integration, and then selects the discriminative (or self-consistent) cells as reference using a self-projection approach. In both simulated bulk and real bulk tests, it illustrated PCCs above 0.75 on all the datasets, guaranteeing a general success of deconvolution, which cannot be achieved by other single reference approaches. Furthermore, we applied SCCAF-D to real bulk data from T2D and IPF, inferring the cell-type composition of disease-relevant tissues. Given the established knowledge that beta cells are gradually lost during T2D progression42,43, estimates of cell type abundances from various computational deconvolution methods in some studies23,59 further corroborate the cell type proportion changes revealed by SCCAF-D. For real bulk applications in IPF, Adams et al.44 used scRNA-seq to reveal an increased proportion of basal cells and substantial decline in alveolar epithelial cells (AT1 and AT2) in IPF lungs compared to non-diseased tissue, which supports our findings. Similarly, Mayr et al.45 deconvoluted Visium and scRNA-seq data, observing an increase in fibroblast and basal cell frequencies and a decrease in AT1 and AT2 cell frequencies in IPF compared to control patient lung tissue. These validation cases demonstrate that the correct inference of cell type proportion changes by SCCAF-D exerts potentials in identifying cellular targets for treatment. Application on two distinct real NAFLD datasets, SCCAF-D consistently demonstrated gradual changes in cell proportion of cholangiocytes, HSCs and hepatocytes during disease progression. In consistency with our results, several NAFLD studies60,61 have provided additional clues to our discovery of an increased cell proportion of cholangiocytes and HSCs in the late fibrotic stages.
However, obtaining ‘gold standard’ datasets of known cell proportions for evaluating deconvolution methods remains a key challenge. While most bulk samples with cell type proportions are from flow cytometry, the accuracy of cell counting can be compromised for rare cell types due to challenges in sample isolation, enrichment, and potential retention in the flow cytometer62. We cannot rule out the possibility that this can be one explanation for the observed negative correlation between the estimated and flow cytometry defined proportions of cDCs in Finotello et al.31. As single-cell technology demonstrates good capability in quantifying rare cell types, analysing the same sample with bulk RNA-seq together with scRNA-seq may generate alternative benchmark data for understanding the cell proportions. On the other hand, since the deconvolution accuracy depends on the choice of the reference data, deconvolving disease samples with healthy reference data from atlases need to consider the possible existence of disease-specific cell types.
With the stable accuracy of SCCAF-D, it may exert additional potential in deconvolving large-scale databases of different tissue types. To achieve this, further efforts could be made in providing a best reference dataset for each tissue type. With the accumulation of human large-scale single-cell atlas, such as the Human Cell Atlas63, HuBMAP64, and Human Tumor Atlas Network (HTAN)65, the issue of selecting optimal reference datasets for tissue-specific deconvolution using SCCAF-D is poised to be further addressed.
Methods
Data collection
All bulk RNA-seq and scRNA-seq utilised in this study were collected from published publications, Gene Expression Omnibus (GEO)2 (https://www.ncbi.nlm.nih.gov/geo/), ArrayExpress3 (https://www.ebi.ac.uk/arrayexpress/), UCSC Cell Browser66 (https://cells.ucsc.edu/), COVID-19 cell atlas (https://www.covid19cellatlas.org/), Neuroblastoma Cell Atlas67 (https://www.neuroblastomacellatlas.org/) and CELLxGENE68 (https://cellxgene.cziscience.com/). Detailed description of all the dataset can be found in Supplementary Data 2 and 7. The bulk datasets were processed as counts or Transcripts Per Million (TPM), as indicated in the table. ID conversion of bulk data, including the conversion of ‘Probe ID’, ‘entrez’ or ‘ensembl’ to ‘gene symbols’, was performed using the AnnoProbe69(v.0.1.7) or AnnotationDbi70 (v.1.60.2) R packages.
Single-cell data processing
We downloaded the raw count matrix and cell type annotations for each dataset from the corresponding websites (Supplementary Data 2 and 7). All single-cell datasets were analysed by SCANPY71 (v. 1.9.1) and Seurat72 (v. 4.4.0) packages. Cells with fewer than 200 detected genes and genes expressed in fewer than 3 cells were excluded. For datasets from Hrvatin et al.73, Liao et al.74, Aizarani et al.75, and Segerstolpe et al.30, we removed cells annotated as ‘not applicable’ or ‘unclassified’. ERCC genes were removed from the data for Muraro et al.27. For data from Moncada et al.76, the cell type labels were reannotated as follows: ‘Cancer clone A’ and ‘Cancer clone B’ were combined into ‘Cancer’; ‘Ductal - APOL1 high/hypoxic,’ ‘Ductal - CRISP3 high/centroacinar like,’ ‘Ductal - MHC Class II,’ and ‘Ductal - terminal ductal like’ were consolidated into ‘Ductal’; ‘Macrophages A’ and ‘Macrophages B’ were merged into ‘Macrophages’; and ‘mDCs A’ and ‘mDCs B’ were combined into ‘mDCs.’ For data from Vieira et al.25, the cell labels ‘Type 1’ and ‘Type 2’ were consolidated into ‘Alveolar’ (Supplementary Fig. 3).
For human lung48,49,77,78 and PBMC datasets (Arunachalam et al.34, Lee et al.35, Wilk et al.37 and Schulte-Schrepping et al.36), cells with total counts below 200 or exceeding 6000 were removed. Additionally, cells with a percentage of mitochondrial contents exceeding 15% of the total counts were discarded. Minor cell types of fewer than 40 cells from per dataset were removed. Doublets were predicted by ‘Scrublet’ (v. 0.2.3) and cells with doublet scores higher than 0.25 were excluded. For human pancreas datasets, cells with more than 20% mitochondrial content were excluded, while for human brain datasets, the threshold was set at 10%. For spleen datasets and IPF datasets from Adams et al.44, Reyfman et al.46, and Tsukui et al.47, cells with more than 6000 detected genes were excluded, as were those with mitochondrial content exceeding 20%. Additionally, cells with doublet scores above 0.3, as predicted by Scrublet (v.0.2.3), were removed. For spleen datasets and IPF datasets from Habermann et al.48, Valenzi et al.49, and Morse et al.50, cells with fewer than 400 or more than 8,000 detected genes were excluded, along with those exhibiting mitochondrial content greater than 12%.
For MacParland et al.51, Guilliams et al.52, and Tabula Sapiens-Liver54 datasets from human liver tissue, we applied the same thresholds for total counts, genes and doublet scores. Considering the liver tissue is metabolically more active the threshold for percentage of mitochondrial contents is set to 50%. We removed cells expressing more than 100,000 genes in the Wang et al.53 dataset.
After quality control, we performed standard SCANPY single-cell data analysis workflow, including normalisation, highly variable gene selection (2000 genes), principal component analysis (PCA, 50 components) and UMAP visualisation. Datasets from different studies were integrated by Harmony28 with the batch key set as the ‘sampleID’. FindNeighbors and FindClusters functions were used on the Harmony derived latent space. For cell type annotation, we retained the original annotations from each study and manually curated them to achieve a consistent level of granularity (Supplementary Fig. 15).
SCCAF-D workflow for generating optimised references
SCCAF-D first integrates multiple datasets to optimise cell type annotation for reference data. Considering batch effects from different studies normally do not overlap each other, integrating multiple datasets may mitigate batch effects. While it is still difficult to recognise which dataset is a better reference when only two datasets are available, we propose to use at least three datasets for integration, copying the common number of biological replicates in experiments79. Specifically, SCCAF-D makes cell type annotations more consistent across datasets by integrating datasets using Harmony28 and then re-annotating cell types based on Leiden clustering and cell type labels obtained from the original publication.
Single-cell data information can be divided into biological and batch parts. Different data sources will be affected by batch, resulting in differences between different data, but the biological part of single-cell data from different sources should be the same, which is called ‘self-consistent’. Based on this assumption, we use SCCAF-D to select the optimised reference.
When preparing an optimised reference from single-cell data, we follow the Single-Cell Clustering Assessment Framework (SCCAF)29 workflow, which is a method for the automated identification of putative cell types from scRNA-seq according to the self-projection. We first divide the data into two parts: a training set and a test set. Our splitting strategy is based on the number of cell types. If the number of cells for a particular cell type exceeds 500, we randomly select 500 cells from this type as the training set. However, if the number of cells for a cell type is less than 500, we split it in half, randomly selecting one half of the cells as the training set and the other half as the test set. Next, we use a logistic regression model for training, as it has already demonstrated advantages in well-established algorithms (such as SCCAF29, CellHint80 and CellTypist81) in identifying cell types. This model learns to predict cell types based on the data in the training set and assigns a prediction score to each cell within each cell type, generating a score matrix with cells as rows and cell types as columns. We assume that the training set contains sufficient information for cells to distinguish themselves from cells of other cell types, and this is known as self-projection.
Once the model is trained, we apply it to the test set and compare the predicted cell types of the test set with the true cell types in the original data, this process called ‘self-consistent’. We select those cells whose predictions match the true cell types and sort them based on their prediction scores. Finally, we select the top 100 cells with the highest prediction scores from each cell type to generate the final single-cell reference dataset.
User defined reference and deconvolution in SCCAF-D
The SCCAF-D computational framework allows users to customise the preparation of optimised reference data and select from 25 deconvolution methods, including DWLS22, FARDEEP14, MuSiC23, NNLS24, RLR82, EpiDISH12, OLS83, EPIC84, ElasticNet85, Lasso85, ProportionsInAdmixture86, Ridge, CIBERSORT8, SCDC59, BisqueRNA87, CDSeq88, CPM89, DCQ90, DSA17, DeconRNASeq91, TIMER92, Deconf93, Dtangle11, ssFrobenius16, and ssKL94 (Supplementary Data 3). All these deconvolution algorithms have been implemented as a package, Critical Assessment of Transcriptomic Deconvolution (CATD)20. For optimised reference preparation, users have the flexibility to choose their preferred methods for building reference from single-cell datasets, including data processing, integration, clustering, and cell type annotation, or they can choose the default SCCAF-D settings. By default, we use the standard procedure in SCCAF-D to prepare optimised references and perform cell type deconvolution on the input data using DWLS.
To achieve an unbiased comparison, we incorporated 25 available deconvolution algorithms methods (Supplementary Data 3) into SCCAF-D, allowing users to estimate cell-type proportions from their own bulk RNA-seq data. When conducting deconvolution with SCCAF-D, users can customise their approach based on data type, deconvolution methods, and parameter settings: (a) reference data in Seurat format can be user-provided or generated by SCCAF-D as optimised references; (b) the bulk matrix can be derived from counts or normalised data; (c) four data input transformations are available (none, log, sqrt, vst); (d) 18 normalisation methods are supported (column, row, mean, column z-score, global z-score, column min-max, global min-max, LogNormalize, none, Quantile Normalisation (QN), Trimmed Mean of M-values (TMM), Upper Quartile (UQ), median ratios, TPM, SCTransform, scran, scater, Linnorm); (e) all other parameters follow the default settings of specific deconvolution algorithms. Besides, we advise users to keep consistent normalisation methods for both reference and bulk data to ensure reliable results.
Benchmarking workflow
In the ‘self-reference’ setting, we benchmarked all 25 deconvolution methods using data from 44 datasets and selected six methods with better performance: DWLS22, EpiDISH12, FARDEEP14, MuSiC23, NNLS24, and RLR82 for comparative analysis (Supplementary Fig. 3). To simulate pseudobulk, we generated a total of 1000 synthetic simulated bulk profiles as samples of simulated bulk data. For each simulated sample, we randomly selected 10,000 cells with replacement to ensure that the number of each cell type is greater than one and that the sum of all cell type proportions is 1. The selection was based on successful deconvolution across all datasets, with PCCs above 0.3. According to the type of reference provided, these methods can be classified into bulk and single-cell methods. EpiDISH, FARDEEP, NNLS and RLR are common bulk deconvolution methods, while DWLS and MuSiC take single-cell expression profiles as input to get the specific features in order to deconvolve. We allocated 50% of the single-cell data to generate pseudobulk samples, utilising the remaining 50% as a reference dataset to deconvolute the simulated bulk data.
Under the ‘cross-reference’ setting, we evaluated each single-cell reference dataset and selected the best combination of method and reference. The same bulk data (simulated or real bulk data) was used to assess the performance of each reference and algorithm. Twenty scRNA-seq datasets, with four datasets from each tissue type (pancreas, lung, PBMC, brain, and spleen), were used for the deconvolution of simulated ‘pseudobulk’ data. For each tissue type, one dataset was used to simulate pseudobulk (following the methods described in the ‘self-reference’ benchmarking), while the remaining three datasets were processed using the SCCAF-D standard workflow to generate optimised references.
For the deconvolution of real bulk datasets, four PBMC datasets from Arunachalam et al.34, Lee et al.35, Wilk et al.37, and Schulte et al.36 were used to generate the optimised reference. Deconvolution with flow cytometry data as ground truth was performed utilising four bulk datasets with known cell type proportions, which were derived from the studies of Finotello et al.31, Newman et al.32 and Monaco et al.33. Considering that the flow cytometry count data included some finer-granularity cell types not present in our reference data, we summarised the proportions of these finer cell types with their corresponding broader cell types. In the analysis of cell type proportions determined by flow cytometry as reported in Finotello et al.31, we have added the proportions of regulatory T cells (Tregs) into the counts for CD4 positive T cells. For the flow cytometry count data from Monaco et al.33, we have attributed the proportions of the following CD4 positive T cell subsets to the overall CD4 positive T cell count: ‘T CD4 Naive’, ‘Tregs’, ‘Tfh’, ‘Th1’, ‘Th1/Th17’, ‘Th17, and ‘Th2’. Similarly, the proportions of the following CD8 positive T cell subsets have been added to the total CD8 positive T cell count: ‘T CD8 Naive’, ‘T CD8 CM’, ‘T CD8 EM’, and ‘T CD8 TE’.
Deconvolution analysis of bulk-profiled disease samples
We obtained one T2D bulk dataset associated with HbA1c levels from Fadista et al.38, three IPF datasets from McDonough et al.39, Sivakumar et al.40, and Furusawa et al.41, and two NAFLD datasets linked to liver fibrosis stages (0–4) from Powell et al.55 and Govaere et al.56. The corresponding reference data were obtained for T2D from Baron et al.21, Marquina-Sanchez et al.26, Segerstolpe et al.30 and Muraro et al.27; for IPF, datasets from Adams et al.44, Reyfman et al.46, and Tsukui et al.47 were used to generate the first set of optimised reference, while those from Habermann et al.48, Valenzi et al.49 and Morse et al.50 were used for the second; and for NAFLD, references were drawn from MacParland et al.51, Guilliams et al.52, Wang et al.53 and Tabula Sapiens-Liver et al.54. First, optimised references for T2D, IPF, and NAFLD were generated from single-cell data using the standard SCCAF-D workflow. No transformations were applied to the bulk data. Both the optimised references and bulk data were then subjected to TMM normalisation. Finally, deconvolution was performed using the optimised reference in combination with the DWLS algorithm following the default settings.
Graphics visualisation
Heatmaps of PCCs between estimated cell type proportions and ground truth were generated using the ComplexHeatmap95 (v.2.15.4) package. All bar plots, scatter plots, violin plots, and box plots were visualised with the ggpubr96 (v.0.6.0) and ggplot297 (v.3.4.2) packages. Sankey plots comparing original labels to manually curated cell type labels were visualised using Matplotlib98 (v.3.8.1).
Metrics for deconvolution performance evaluation
Pearson correlation coefficient (PCC, or Pearson’s R) and Root mean square error (RMSE) were used to estimate the performance of cell type deconvolution. These two metrics were implemented as in CATD programme and calculated using the ‘dplyr’ (v. 1.1.3) R package. Details of these two metrics as well as the Jensen-Shannon divergence are introduced as below.
For the calculation of Pearson correlation coefficient between the true cell type proportions X and the deconvolution derived cell type proportions Y (equation 1), N is the number of observations, while X and Y are vectors. Therefore, \({x}_{i}\) means the proportion of each cell type for sample in X, while \({y}_{i}\) corresponds to that in a sample from Y. \(\bar{x}\) and \(\bar{y}\) are mean values for X and Y, respectively.
For the calculation of RMSE, \({x}_{i}\), \({y}_{i}\) and N are the same as in PCC.
The Jensen-Shannon divergence (JSD) is a symmetric and smoothed variant of the Kullback-Leibler (KL) divergence, used to measure the similarity between two probability distributions. JSD is implemented using the philentropy (v.0.8.0) R package. The KL divergence is defined as follows:
In the calculation, \({P}\) and \({Q}\) are the predicted and true cell type probability distributions over cell type space T, whereas log is base 2.
For the calculation of JSD, it is defined as:
in which,
Statistics and reproducibility
No statistical methods were used to pre-determine sample sizes. No data were excluded from the analyses. No randomisation was used in our study. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The publicly available single-cell datasets used in this study can be accessed using the following accession codes or URLs: GSE84133, GSE142465, GSE85241, E-MTAB-5061 for human pancreas; syn21041850, GSE135893, GSE128169, and PRJEB31843 for human lung; GSE155673, GSE149689, GSE150728, and EGAS00001004571 for PBMCs; GSE115469, GSE192742, GSE189539, and Tabula Sapiens-Liver (https://cellxgene.cziscience.com/e/6d41668c-168c-4500-b06a-4674ccf3e19d.cxg/) for Liver; GSE186538, GSE157827, GSE160189, and GSE179590 for brain; E-MTAB-11536, PRJEB31843, GSE159929 for spleen; GSE136831, GSE122960, GSE132771, GSE135893, GSE128169, and GSE128033 for IPF. The bulk data used in this article is publicly available and can be downloaded via Gene Expression Omnibus (GEO) with accession number GSE107572, GSE127813, GSE107019, GSE225740, GSE50244, GSE124685, GSE134692, GSE150910, and GSE135251. Source data are provided with this paper.
Code availability
An open source implementation of SCCAF-D is available at GitHub github.com/rnacentre/SCCAF-D/, and Zenodo (https://doi.org/10.5281/zenodo.14211888)99 while codes for reproducing the paper is available at: github.com/rnacentre/SCCAF-D_reproducibility/. Example data for code execution is available at: https://doi.org/10.6084/m9.figshare.27232896.
References
Kuhn, A., Thu, D., Waldvogel, H. J., Faull, R. L. M. & Luthi-Carter, R. Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain. Nat. Methods 8, 945–947 (2011).
Edgar, R., Domrachev, M. & Lash, A. E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Parkinson, H. et al. Array express-a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 35, D747–D750 (2007).
GTEx Consortium et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
International Cancer Genome Consortium et al. International network of cancer genome projects. Nature 464, 993–998 (2010).
Zhang, J. et al. International cancer genome consortium data portal-a one-stop shop for cancer genomics data. Database 2011, bar026 (2011).
Taylor, R. S. et al. Association between fibrosis stage and outcomes of patients with nonalcoholic fatty liver disease: a systematic review and meta-analysis. Gastroenterology 158, 1611–1625.e12 (2020).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Du, R., Carey, V. & Weiss, S. T. deconvSeq: deconvolution of cell mixture distribution in sequencing data. Bioinformatics 35, 5095–5102 (2019).
Frishberg, A. et al. Cell composition analysis of bulk genomics using single-cell data. Nat. Methods 16, 327–332 (2019).
Hunt, G. J., Freytag, S., Bahlo, M. & Gagnon-Bartsch, J. A. dtangle: accurate and robust cell type deconvolution. Bioinformatics 35, 2093–2099 (2018).
Teschendorff, A. E., Breeze, C. E., Zheng, S. C. & Beck, S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in Epigenome-Wide Association Studies. BMC Bioinform. 18, 105 (2017).
Aliee, H. & Theis, F. J. AutoGeneS: Automatic gene selection using multi-objective optimization for RNA-seq deconvolution. Cell Syst. 12, 706–715.e4 (2021).
Hao, Y., Yan, M., Heath, B. R., Lei, Y. L. & Xie, Y. Fast and robust deconvolution of tumor infiltrating lymphocyte from expression profiles using least trimmed squares. PLoS Comput. Biol. 15, e1006976 (2019).
Li, Z. & Wu, H. TOAST: improving reference-free cell composition estimation by cross-cell type differential analysis. Genome Biol. 20, 190 (2019).
Gaujoux, R. & Seoighe, C. CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics 29, 2211–2212 (2013).
Zhong, Y., Wan, Y.-W., Pang, K., Chow, L. M. L. & Liu, Z. Digital sorting of complex tissues for cell type-specific gene expression profiles. BMC Bioinform. 14, 89 (2013).
Avila Cobos, F., Alquicira-Hernandez, J., Powell, J. E., Mestdagh, P. & De Preter, K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat. Commun. 11, 5650 (2020).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
Pournara, A. V. et al. CATD: a reproducible pipeline for selecting cell-type deconvolution methods across tissues. Bioinform. Adv. 4, vbae048 (2024).
Baron, M. et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346–360.e4 (2016).
Tsoucas, D. et al. Accurate estimation of cell-type composition from gene expression data. Nat. Commun. 10, 2975 (2019).
Wang, X., Park, J., Susztak, K., Zhang, N. R. & Li, M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 10, 380 (2019).
NNLS: The Lawson-Hanson Algorithm for Non-Negative Least Squares (NNLS). Comprehensive R Archive Network (CRAN) https://CRAN.R-project.org/package=nnls (2024).
Vieira Braga, F. A. et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 25, 1153–1163 (2019).
Marquina-Sanchez, B. et al. Single-cell RNA-seq with spike-in cells enables accurate quantification of cell-specific drug effects in pancreatic islets. Genome Biol. 21, 106 (2020).
Muraro, M. J. et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394.e3 (2016).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Miao, Z. et al. Putative cell type discovery from single-cell gene expression data. Nat. Methods 17, 621–628 (2020).
Segerstolpe, Å. et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607 (2016).
Finotello, F. et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 11, 34 (2019).
Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782 (2019).
Monaco, G. et al. RNA-Seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 26, 1627–1640.e7 (2019).
Arunachalam, P. S. et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 369, 1210–1220 (2020).
Lee, J. S. et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 5, eabd1554 (2020).
Schulte-Schrepping, J. et al. Severe COVID-19 Is marked by a dysregulated myeloid cell compartment. Cell 182, 1419–1440.e23 (2020).
Wilk, A. J. et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 26, 1070–1076 (2020).
Fadista, J. et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Natl. Acad. Sci. USA. 111, 13924–13929 (2014).
McDonough, J. E. et al. Transcriptional regulatory model of fibrosis progression in the human lung. JCI Insight 4, e131597 (2019).
Sivakumar, P. et al. RNA sequencing of transplant-stage idiopathic pulmonary fibrosis lung reveals unique pathway regulation. ERJ Open Res. 5, 00117–2019 (2019).
Furusawa, H. et al. Chronic hypersensitivity pneumonitis, an interstitial lung disease with distinct molecular signatures. Am. J. Respir. Crit. Care Med. 202, 1430–1444 (2020).
Sayyed Kassem, L., Rajpal, A., Barreiro, M. V. & Ismail-Beigi, F. Beta-cell function in type 2 diabetes (T2DM): Can it be preserved or enhanced? J. Diabetes 15, 817–837 (2023).
Hara, M., Fowler, J. L., Bell, G. I. & Philipson, L. H. Resting beta-cells - A functional reserve? Diabetes Metab 42, 157–161 (2016).
Adams, T. S. et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 6, eaba1983 (2020).
Mayr, C. H. et al. Spatial transcriptomic characterization of pathologic niches in IPF. Sci. Adv. 10, eadl5473 (2024).
Reyfman, P. A. et al. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 199, 1517–1536 (2019).
Tsukui, T. et al. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat. Commun. 11, 1920 (2020).
Habermann, A. C. et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6, eaba1972 (2020).
Valenzi, E. et al. Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease. Ann. Rheum. Dis. 78, 1379–1387 (2019).
Morse, C. et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. 54, 1802441 (2019).
MacParland, S. A. et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018).
Guilliams, M. et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379–396.e38 (2022).
Wang, Z. et al. Single-cell analysis reveals a pathogenic cellular module associated with early allograft dysfunction after liver transplantation. bioRxiv https://doi.org/10.1101/2022.02.09.479667 (2022).
Tabula Sapiens Consortium*. et al. The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022).
Powell, N. R. et al. Clinically important alterations in pharmacogene expression in histologically severe nonalcoholic fatty liver disease. Nat. Commun. 14, 1474 (2023).
Govaere, O. et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci. Transl. Med. 12, eaba4448 (2020).
Garmire, L. X. et al. Challenges and perspectives in computational deconvolution of genomics data. Nat. Methods 21, 391–400 (2024).
Jin, H. & Liu, Z. A benchmark for RNA-seq deconvolution analysis under dynamic testing environments. Genome Biol. 22, 102 (2021).
Dong, M. et al. SCDC: bulk gene expression deconvolution by multiple single-cell RNA sequencing references. Brief. Bioinform. 22, 416–427 (2021).
Richardson, M. M. et al. Progressive fibrosis in nonalcoholic steatohepatitis: association with altered regeneration and a ductular reaction. Gastroenterology 133, 80–90 (2007).
Kisseleva, T. & Brenner, D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat. Rev. Gastroenterol. Hepatol. 18, 151–166 (2021).
Nguyen, H., Nguyen, H., Tran, D., Draghici, S. & Nguyen, T. Fourteen years of cellular deconvolution: methodology, applications, technical evaluation and outstanding challenges. Nucleic Acids Res. 52, 4761–4783 (2024).
Regev, A. et al. The human cell atlas. Elife 6, e27041 (2017).
HuBMAP Consortium. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
Rozenblatt-Rosen, O. et al. The human tumor atlas network: charting tumor transitions across space and time at single-cell resolution. Cell 181, 236–249 (2020).
Speir, M. L. et al. UCSC Cell Browser: visualize your single-cell data. Bioinformatics 37, 4578–4580 (2021).
Kildisiute, G. et al. Tumor to normal single-cell mRNA comparisons reveal a pan-neuroblastoma cancer cell. Sci. Adv. 7, eabd3311 (2021).
Megill, C. et al. Cellxgene: A performant, scalable exploration platform for high dimensional sparse matrices. bioRxiv https://doi.org/10.1101/2021.04.05.438318 (2021).
Annotate the Gene Symbols for Probes in Expression Array [R package AnnoProbe version 07]. (2022).
AnnotationDbi. Bioconductor http://bioconductor.org/packages/AnnotationDbi/ (2024).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15 (2018).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Hrvatin, S. et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci. 21, 120–129 (2018).
Liao, M. et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020).
Aizarani, N. et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204 (2019).
Moncada, R. et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 38, 333–342 (2020).
Travaglini, K. J. et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020).
Madissoon, E. et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol 21, 1 (2019).
Blainey, P., Krzywinski, M. & Altman, N. Points of significance: replication. Nat. Methods 11, 879–880 (2014).
Xu, C. et al. Automatic cell-type harmonization and integration across Human Cell Atlas datasets. Cell 186, 5876–5891.e20 (2023).
Domínguez Conde, C. et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science 376, eabl5197 (2022).
Support Functions and Datasets for Venables and Ripley’s MASS [R package MASS version 7.3–61]. (2024).
Chambers, J., Hastie, T. & Pregibon, D. Statistical Models in S. Compstat. 317–321 (1990).
Racle, J., de Jonge, K., Baumgaertner, P., Speiser, D. E. & Gfeller, D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife 6, e26476 (2017).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010).
Applied Research Applied Research Press. WGCNA: An R Package for Weighted Correlation Network Analysis. (2015).
Jew, B. et al. Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat. Commun. 11, 1971 (2020).
Kang, K. et al. CDSeq: A novel complete deconvolution method for dissecting heterogeneous samples using gene expression data. PLoS Comput. Biol. 15, e1007510 (2019).
Song, L., Sun, X., Qi, T. & Yang, J. Mixed model-based deconvolution of cell-state abundances (MeDuSA) along a one-dimensional trajectory. Nat. Comput. Sci. 3, 630–643 (2023).
Altboum, Z. et al. Digital cell quantification identifies global immune cell dynamics during influenza infection. Mol. Syst. Biol. 10, 720 (2014).
Gong, T. & Szustakowski, J. D. DeconRNASeq: a statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics 29, 1083–1085 (2013).
Li, T. et al. TIMER: A Web Server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77, e108–e110 (2017).
Repsilber, D. et al. Biomarker discovery in heterogeneous tissue samples -taking the in-silico deconfounding approach. https://doi.org/10.1186/1471-2105-11-27 (2010).
Gaujoux, R. & Seoighe, C. Semi-supervised Nonnegative Matrix Factorization for gene expression deconvolution: a case study. Infect. Genet. Evol. 12, 913–921 (2012).
Gu, Z. Complex heatmap visualization. Imeta 1, e43 (2022).
Kassambara, A. ‘ggplot2’ Based Publication Ready Plots [R package ggpubr version 0.6.0]. (2023).
Create Elegant Data Visualisations Using the Grammar of Graphics [R package ggplot2 version 3.5.1]. (2024).
Hunter, J. D. Matplotlib: A 2D graphics environment. https://ieeexplore.ieee.org/document/4160265 (2007).
Feng, S. & Miao, Z. Alleviating batch effects in cell type deconvolution with SCCAF-D. Zenodo. https://doi.org/10.5281/ZENODO.14211888. (2024).
Acknowledgements
Shuo Feng would like to thank Prof. Haiyan Liu for supervision and Dr. Yin Huang for discussion. This work is funded by the National Key R&D Programmes of China (2023YFF1204700, 2023YFF1204701), the Natural Science Foundation of China (32270707), the R&D Programmes of Guangzhou Laboratory, Grant No. GZNL2024A01002, GZNL2023A01006, SRPG22-003, SRPG22-006, SRPG22-007, HWYQ23-003, YW-YFYJ0102, the Postdoctoral Research Project Funding of Guangzhou (BSHF23-087).
Author information
Authors and Affiliations
Contributions
S.F. designed the algorithm, conducted the analyses and wrote the manuscript. Z.M. conceived the project, designed the algorithm, did the analyses and wrote the manuscript. A.V.P. contributed the analyses. L.H. interpreted the results and wrote the manuscript. Z.H. designed the algorithm. X.Y., Y.Z. and M.S. contributed to the collection of scRNA-seq data. A.B., M.S., I.P. and Z.M. Supervised the project. All authors reviewed, edited, and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Mingon Kang, Yang Xiao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Feng, S., Huang, L., Pournara, A.V. et al. Alleviating batch effects in cell type deconvolution with SCCAF-D. Nat Commun 15, 10867 (2024). https://doi.org/10.1038/s41467-024-55213-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55213-x