Abstract
Lilies are economically important monocots known for their ornamental flowers, bulbs, and large genomes. The absence of their genomic information has impeded evolutionary studies and genome-based breeding efforts. Here, we present reference genomes for Lilium sargentiae (lily, 35.66 Gb) and Gloriosa superba (flame lily, 5.09 Gb). The giant lily genome is shaped by recent long terminal repeat retroelements. Phylogenetic analysis reveals diverse, independent origins of lily cultivars. Gene families involved in sucrose and starch metabolism are significantly expanded in the lily genome. Key homologs of XTH22, SOC1, and AP1/FUL-like genes regulate the development, bud growth transition, and floral bud growth transition of lily bulbs. Colchicine biosynthetic gene clusters are identified in G. superba but are absent in L. sargentiae, highlighting independent colchicine evolution in Colchicaceae. These genomic insights enhance understanding of Liliales evolution, providing a foundation for future breeding and molecular research.
Similar content being viewed by others
Introduction
Lilies (the Lilium genus) are perennial bulbous plants of Liliaceae within the Liliales order, with significant economic, ecological, and ornamental value. However, they harbor enormous genomes1,2, which currently pose challenges for genomic accessibility. With a vast genetic diversity comprising over 100 wild lily species, including 55 ones originating from China, Lilium’s center of origin, lilies hold a prominent place in horticulture worldwide3,4. The Lilium species are mainly distributed in the cold and temperate regions of the Northern Hemisphere, such as Eastern Asia, Europe, and North America5. Lilium has been divided into seven taxonomic sections according to their morphological characteristics, including bulb forms: Lilium, Martagon, Pseudolirium, Archelirion, Sinomartagon, Leucolirion, and Oxypetalum4,6. The rich genetic reservoir of lilies has been pivotal in breeding efforts, yielding numerous lily cultivars with desirable traits. Through hybridization, domestication, natural mutation, etc., breeders have developed a multitude of lily cultivars with diverse attributes (e.g., pathogen resistance), such as the widely cultivated Trumpet (T) and Oriental × Trumpet (OT) hybrids4,7. Lilium sargentiae E. H. Wilson serves as the primary progenitor for the T and OT lily hybrids, and was introduced to American and European gardens by the famous plant hunter E. H. Wilson8. Globally, there are approximately 10,000 registered lily cultivars, with over 300 novel cultivars added annually. Lilies belong to the most important bulbous crops generating considerable economic returns. The commercial significance of lilies is reflected by the millions of cutting and potted flowers sold each season in flower stores and auctions. The total global area of lily cut flower cultivation is approximately 90,000 hectares, with a total trade value of around 20 billion US dollars (including bulb trade) (International Statistics Flowers and Plants 2022, https://aiph.org/giic/international-statistical-yearbook/).
Lily species exhibit distinctive flower structures characterized by six petaloid tepals (three outer tepals and three inner tepals), the constituent parts of the undifferentiated perianth. These gorgeous flowers are renowned for their vibrant hues and enchanting fragrance, endowing lilies with significant ornamental and economic value. Another important feature of lilies is their underground bulbs, which are modified stems made up of overlapping scales and play a crucial role in the plant’s growth. They store essential nutrients, primarily carbohydrates, and serve as water reservoirs4,9. The bulbs of various Lilium species have long been utilized in traditional medicine and cuisines in different countries3. Furthermore, in commercial lily cultivation, bulbs are extensively employed in asexual reproduction7. All parts of a lily plant, including its flowers, originate from the basal plate of the bulb. The apical meristems of bulbs give rise to the inflorescence, while the axillary meristems produce side bulblets (small bulbs); however, due to these bulbs have prolonged reproductive cycle, they can be a limiting factor in lily development4. Moreover, successful lily flowering necessitates bulb expansion to a specific size, a process facilitated by cooling, which triggers the growth transition, and by vernalization, which is essential for flowering10,11. Consequently, expediting the development and growth transition of lily bulbs, along with understanding the associated mechanisms, are pivotal for enhancing the quality and efficiency of lily production industries and resource conservation. Nonetheless, the evolutionary and genetic mechanisms governing lily bulb development and growth transition remain largely unknown10,12,13. Gloriosa superba, also known as flame lily, a member of the Colchicaceae family (Liliales order), holds significance not only as a popular ornamental crop renowned for its striking flame-like flowers but also as the primary natural source of colchicine. Colchicine is used as a drug in treating gout, arthritis, and inflammation and has a prominent role in inducing polyploidy in plants14,15.
Lily and flame lily are amongst the most popular ornamental plants and cutting flowers in the Liliales order. They share similar floral structures but differ in their stem modifications: lily has bulbs while flame lily has rhizomes, ideal for the comparative study of organ development in Liliales. While only the Colchicaceae family can synthesize colchicine16, a comparison of the genomes of lily and flame lily offers insights into the origin and evolution of colchicine. Additionally, their contrasting genome sizes will enable the exploration of the underlying evolutionary mechanisms of the giant lily genome.
In this study, we employ a combination of PacBio high-fidelity (HiFi) and Hi-C technologies to elucidate the expansive genomes of L. sargentiae and G. superba and provide important genomic resources for the evolutionary, comparative genomics, and molecular study of the plant species in Liliales.
Results
Genome sequencing, assembly, and annotation of L. sargentiae and G. superba
For de novo whole-genome sequencing, we generated 602.05 Gb and 82.50 Gb of PacBio HiFi long-read data for L. sargentiae and G. superba, respectively, and 1.07 terabytes (Tb) and 594.24 Gb Hi-C short-read data for L. sargentiae and G. superba, respectively. We also separately generated short-read data for L. sargentiae (2.12 Tb) and G. superba (362.28 Gb) to estimate their genome size and heterozygosity. By k-mer analysis, the genome size of L. sargentiae and G. superba were estimated to be ~35.66 Gb and ~5.09 Gb, respectively, with heterozygosity calculated as 3.35% and 0.70%, respectively (Supplementary Fig. 1a, c). The genome sizes of L. sargentiae and G. superba were estimated to be ~31.8 Gb and 5.7 Gb, respectively, by flow cytometry analysis (Supplementary Fig. 1b, d). Additionally, we obtained transcriptome data for a total of 291 samples of Liliales species, with an average of 8.63 Gb per sample, resulting in a total of 2.63 Tb of short-read data (Supplementary Data 1 and 2). Therefore, a total of approximately 7.45 Tb of sequencing data was obtained. The final assembly of the L. sargentiae genome sequence was 47.09 Gb in length (contig N50 = 0.93 Mb; scaffold N50 = 2.85 Gb) with 35 scaffolds (longest scaffold length = 5.31 Gb and average scaffold length = 1.35 Gb) (Supplementary Table 1 and 2), while the final assembly of the G. superba genome sequence was 5.15 Gb in length (contig N50 size = 0.48 Mb; scaffold N50 size = 0.56 Gb) with 11 chromosomes (longest length = 0.93 Gb and average length = 0.47 Gb) (Supplementary Tables 3 and 4). BUSCO17 analysis of the L. sargentiae and G. superba genome revealed the complete gene information at 90.8% and 91.8%, respectively; BUSCO analysis of the L. sargentiae and G. superba protein sets revealed the complete gene information at 88.3% and 85.9%, respectively (Supplementary Tables 5 and 6). Based on ab initio and evidence-based predictions, 47,139 (L. sargentiae) and 54,617 (G. superba) protein-coding genes were annotated and the average lengths were 42,804 and 9,984 bp, respectively, both are longer than those in other published angiosperm genomes (Supplementary Tables 7, 8, and 9).
The evolution and lineage-specific WGD events in lily and flame lily
Although the genomes of 8 monocot plant orders have been sequenced, the order Liliales, which includes economically significant crops such as lilies and tulips, remains unsequenced. As the sequenced two genomes from Liliales, our assemblies of lily and flame lily provide an invaluable opportunity to explore the phylogenetic position of the Liliales order within the monocots. We conducted a phylogenomic analysis of 26 flowering plant species from 18 orders, including 9 of the 12 monocot orders. The resulting phylogenetic tree revealed consistent evolutionary positions for the 8 monocot orders examined (Fig. 1a)18,19,20. We found that L. sargentiae and G. superba form a monophyletic clade, which is sister to the Asparagales species, consistent with the previous findings (Fig. 1a)18,21. We also confirmed that the Liliales and Asparagales species constitute a distinct clade, which is sister to the clade comprising Arecales, Zingiberales, and Poales species, consistent with previous reports (Fig. 1a)21,22.
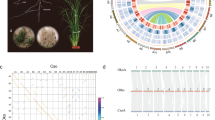
a Phylogenetic tree of Lilium sargentiae and Gloriosa superba along with 24 other plants. The whole genome duplication (WGD) and triplication events were labeled on the tree. b Ks distributions of paralogous gene pairs within species. The WGD events and their associated time were labeled accordingly. Ks, synonymous substitutions per site; Mya, million years ago; Gsu, G. superba; Lsa, L. sargentiae; Osa, Oryza sativa. c Long terminal repeat (LTR) insertion analysis in the genomes of L. sargentiae and G. superba. d Statistics of the ratio of the solo-LTR number to the intact-LTR number is plotted against their genome sizes. The ratios for other species were obtained from previous study24.
Genome comparisons within and between species showed clear evidence that both L. sargentiae and G. superba have experienced two rounds of whole genome duplication (WGD) events (Fig. 1a, b); the ratio of syntenic blocks was 3:1 in self-comparison of L. sargentiae or G. superba (Supplementary Fig. 2), while the ratio of syntenic blocks was 4:1 in both G. superba vs. L. sargentiae and Acorus calamus vs. G. superba comparisons (Supplementary Fig. 3). Analysis of synonymous substitutions per site (Ks) distributions in the genomes of L. sargentiae, G. superba, and O. sativa all showed a peak at 0.92, corresponding to the monocot-shared WGD event (τ) at 120–130 million years ago (Mya) (Fig. 1a, b). The distributions for L. sargentiae and G. superba showed an additional peak at Ks = 0.73 and Ks = 0.55, respectively, indicating two separate WGD events occurred at 95–103 Mya and 72–78 Mya, respectively (Fig. 1b and Supplementary Fig. 4). These results demonstrate that L. sargentiae and G. superba underwent lineage-specific WGD events in Liliaceae and Colchicaceae, respectively (Fig. 1a, b).
LTR-RTs contribute to the two huge lily genomes
A total of 38.77 Gb and 4.45 Gb of the L. sargentiae genome and G. superba genome sequences were repetitive elements with 47.4% and 57.9% belonging to long terminal repeat retroelements (LTR-RTs), respectively (Supplementary Tables 10 and 11). Ty3/Gypsy and Ty1/Copia are the main two classes of LTR-RTs in higher plants; they comprise 32.2% and 14.6% of the LTR-RTs in L. sargentiae, as well as 42.5% and 15.1% of the LTR-RTs in G. superba, respectively (Supplementary Tables 10 and 11). We found continuous proliferation of LTR-RTs during recent evolution (0-3 Mya) in both L. sargentiae and G. superba (Fig. 1c). In particular, there is a more pronounced activity of LTR-RTs (mainly the Ty1/Copia type) in L. sargentiae within 0–3 Mya (Fig. 1c). Moreover, L. sargentiae accumulated significantly more LTR-RTs than G. superba in the genome, with 22.32 Gb and 2.98 Gb LTR-RTs, respectively (Supplementary Tables 10 and 11). These results indicated that the expansion of LTR-RTs has shaped the giant lily genome. The most abundant Copia LTR-RTs were Tork (43.2%) and Ale (20.6%) in lily and flame lily, respectively, while the most abundant Gypsy LTR-RTs were Tekay (17.1%) and Tekay (5.7%) in lily and flame lily, respectively (Supplementary Fig. 5). Through GO enrichment analysis of genes located within 2 kb of Tork and Tekay LTR-RTs, or containing these LTR-RTs within their introns in the lily genome, we discovered a significant enrichment (P < 0.05) of GO terms associated with biological processes involved in asexual organ propagation (e.g., bulblet initiation and development), such as ‘vascular transport’ (GO:0010232), ‘callose localization’ (GO:0052545), and ‘multicellular organism reproduction’ (GO:0022412), or sensing changes in temperature and light in underground organs, which are closely linked to the growth transition of lilies, e.g., ‘response to red light’ (GO:0010114), ‘cellular response to external stimulus’ (GO:0071496), and ‘cellular response to water deprivation’ (GO:0071496) (Supplementary Fig. 6). This implies that the expansion of LTR-RTs, besides genome size expansion, might also have influenced the evolution and structure of genes underlying crucial bulb-related traits, including bulb formation as well as growth transition.
The greater-than-usual average gene lengths in L. sargentiae and G. superba can be largely attributed to intron lengths. Both L. sargentiae and G. superba featured an abundance of lengthy introns exceeding 10 kb, which are rare in rice with a compact genome (Supplementary Fig. 7a). However, it is worth noting that the L. sargentiae genome presented a higher prevalence of long introns in comparison to the G. superba genome (Supplementary Fig. 7a). The average intron lengths in L. sargentiae and G. superba reached 9.1 kb and 5.0 kb, respectively, both are substantially larger than that (0.5 kb) in the other 55 genome-sequenced angiosperms which have smaller genomes (genome size <5 Gb)23 (Supplementary Tables 7a and 8a). Moreover, the ratios of total intron length to total exon length in L. sargentiae and G. superba were higher than those in other angiosperms but similar to the ones in representative gymnosperms with large genomes, and there is a positive correlation (R2 = 0.62, P < 0.01) between the ratio of total intron length to total exon length and genome size in plants (Supplementary Fig. 8a), suggesting that large genomes in different lineages tend to contain enlarged introns. Within the introns of L. sargentiae and G. superba, 73.2% and 15.5% of all genes contain LTR-RTs in their introns, respectively (Supplementary Table 12). The insertion patterns of intronic LTR-RTs in both L. sargentiae and G. superba exhibited a bimodal distribution with a pronounced bias towards the long end, exhibiting longer LTR-RTs within introns when compared to rice (Supplementary Fig. 7b). Both L. sargentiae and G. superba genomes showed a clear peak around 100 bp in LTR length (Supplementary Fig. 7b). Additionally, L. sargentiae had a noticeable peak near 10 kb, while G. superba exhibited another peak around 3.2 kb in LTR length (Supplementary Fig. 7b). Moreover, the genome of L. sargentiae contained a greater number of longer LTR insertions when compared to G. superba’s genome (Supplementary Fig. 7b). These indicated that the large intron size of L. sargentiae is also related to the LTR-RT expansion in intronic regions.
In L. sargentiae, the intergenic space was expanded with an average length of one megabase, which is 3.1- and 9.3-fold longer than the average intergenic length in Vicia faba (327.9 kb) and G. superba (107.7 kb), respectively (Supplementary Fig. 8b). A total of 97.6% and 95.0% of LTR-RTs were found in the intergenic regions in the L. sargentiae and G. superba genomes, respectively (Supplementary Table 12), indicating that the large genome size of lily is mainly due to the huge intergenic regions with high LTR-RT contents. Along with their proliferation, LTR-RTs are also actively removed through ectopic homologous recombination, which generates solo-LTRs24. Interestingly, the ratios of the number of solo-LTRs to that of intact-LTRs in L. sargentiae (2.6) and G. superba (3.9) were both higher than those in other angiosperms, but similar to the ratios found in the three gymnosperms studied (Torreya grandis, Ginkgo biloba, and Welwitschia mirabilis), which have all undergone expansions of ancient LTR-RT bursts (10–30 Mya)24. This suggests that LTR removal rates are higher in these above-mentioned large genomes driven by LTR-RT expansion (Fig. 1d).
Phylogeny of Lilium and origins of lily cultivars
Lilium is a diverse genus with ~100 wild species4, and numerous cultivars have been developed by methods such as hybridization. We took advantage of the lily genome and transcriptome datasets of a wide range of lilies to conduct the most comprehensive to-date phylogenomic analysis of lily (Fig. 2a). Based on 360 nuclear genes, the lily phylogeny of 51 wild Lilium species and 34 lily breeding cultivars was overall well-resolved and supported (Fig. 2a). The lilies were separated into two distinct monophyletic groups (Clade 1 and Clade 2) (Fig. 2a). The wild lily species have been classified into seven sections based on morphological characteristics; while the sections Martagon and Archelirion were monophyletic, the sections Leucolirion and Sinomartagon were both polyphyletic (Fig. 2a). For instance, Leucolirion 6a and 6b were distributed in Clade 1-1 and Clade 9, representing two different parental lineages with trumpet-shaped flowers (Fig. 2a). The phylogeny revealed that lily cultivars have multiple independent origins, including the T hybrids in Clade 1-1, OT hybrids in Clade 1, and Asiatic (A) hybrids in Clade 10 (Fig. 2a). Interestingly, we found that all but one species (L. eupetes, a newly identified species with no specified classification) of the traditional Sinomartagon 5c section and the embedded Nomocharis genus were clustered in Clade 2-1 without any lily cultivar (Fig. 2a). The results indicate that these wild lilies in Clade 2-1 might not have been widely used in the past lily breeding programs, highlighting a previously untapped genetic reservoir, which may offer immense potential for future breeding endeavors.
a The phylogenetic tree of the wild Lilium species and cultivated lilies was inferred from combined RNA-seq data of wild Lilium species and Lilium cultivars. The blue numbers labeled on the tree represent the classified clades. Bootstrap values for important branches were labeled above branches. b Heatmap showing shared genomic haplotype patterns among different lily species. Each cell represents the ratio of the number of transcripts with the highest gene similarity (identity >95%) between two species to the total number of transcripts within the corresponding species in that row (excluding self-aligning transcripts). The intensity of the box color reflects the proportion of gene similarity (best match ratio, BMR), with redder shades indicating higher similarity. The yellow rectangle highlights shared genomic haplotypes between clades, while boxes of species within the same clade are delineated by black rectangles. The order of species represented vertically (from top to bottom) and horizontally (from left to right) in the heatmap corresponds to the order of species in the phylogenetic tree represented in (a). c The ratios of the number of gene clusters obtained through Orthofinder to the number of genes within a cluster were depicted for different lilies within Clade 1-1 (upper panel) and Clade 1-2 (lower panel). Source data are provided as a Source Data file.
Our analysis of core-gene sequence similarities revealed a significantly higher proportion of shared genomic haplotypes among lilies within clades, compared to those among lilies from different clades in the phylogenetic tree (Fig. 2b). In Clade 4, however, we did not detect significant shared genomic haplotype ratios between the wild lilies of Clade 9 (Leucolirion 6b) and Clade 10 (Sinomartagon), nor between the Leucolirion 6b wild lilies and cultivated lilies (Fig. 2b). Notably, L. browni, originating from China and widely distributed across 17 provinces4, showed distinct shared genomic haplotypes with nearly all examined lilies (Fig. 2b). It thus may have been extensively involved in the formation of wild and cultivated lilies. These findings imply that the wild lilies of Leucolirion 6a likely served as original parents of T and OT hybrids, while wild species of Archelirion were likely the parent lines for O and OT hybrids as well. Wild lilies of Sinomartagon 5b and 5a lilies are potential parents for all the three types (A, OA, and L) of lily hybrids (Fig. 2b). In addition, we discovered shared genomic haplotypes between Clade 1-1 and Clade 1-2, where four OT hybrids (‘Black Beauty; ‘Palazzo’, ‘Conca d’Or’, and ‘Zelmira’) from Clade 1-2 exhibited shared genomic haplotypes with lilies from Clade 1-1, suggesting that these OT hybrids have parental lines from both Clade 1-1 and 1-2, whereas other OT hybrids have parental lines exclusively from Clade 1-1 (Fig. 2b).
For lilies in Clade 1, we used transcriptome assemblies from different lilies and the longest transcripts of the L. sargentiae genome assembly to calculate the number of gene clusters for each species (Fig. 2c). Among the lilies from Clades 1-1 and 1-2, three cultivated lilies (‘Regale Album’, ‘Red Morning’, and ‘Judith Saffigna’) from Clade 1-1, and four (‘Black Beauty; ‘Palazzo’, ‘Conca d’Or’, and ‘Zelmira’) from Clade 1-2 exhibited a higher number of clusters with multi-copy transcripts (two or three copies) compared to the number of clusters with single-copy transcripts, indicating the presence of genetic materials from more than one parental line (Fig. 2c). For instance, ‘Red Morning’ showed the highest number of gene clusters with two-copy transcripts (Fig. 2c). In contrast, wild lilies, and some other cultivated lilies (‘Beijing Moon’, ‘Siberia’, ‘Marlon’, ‘Sorbonne’, ‘Isabella’, and ‘Brasilia’) predominantly showed single-copy transcripts (Fig. 2c). This implies that the seven cultivated lilies with a higher number of clusters with multi-copy transcripts likely underwent hybridization, as also evidenced by their positions at the outermost branches of clades (Fig. 2a, c). While the other cultivated lilies with the highest number of clusters with single-copy genes are likely bred through alternative methods, such as domestication and mutation breeding (Fig. 2c).
Evolution of genes involved in the formation of lily bulb and flame lily rhizome
Bulbs of lily and rhizomes of flame lily are modified stem-originated rootstocks serving as both storage and propagation organs. They enable plants to survive adverse conditions. These two types of modified stems exhibit distinct different structures: lily bulbs consist of imbricating scales with a central bud being surrounded inside, while rhizomes lack the distinct layering structure like lily bulbs and have short internodes instead. To investigate the evolution and development of lily bulbs and flame lily rhizomes, we performed comparative genomic and transcriptomic analysis using the huge amount of high-resolving genomic and transcriptomic data of the two species. Our phylogenetic analyses identified significant expansions of gene families involved in starch and sucrose metabolism (Fig. 3a, b and Supplementary Figs. 9–14), which play important roles in the initiation and expansion of storage organs13,25,26,27,28,29. L. sargentiae has an average eight more copies of Sucrose Synthase 1/4 (SUS1/4), ADP Glucose Pyrophosphorylase 2/3/4 (APL2/3/4), Starch Synthase 2 (SS2), Beta-Amylase 1 (BAM1), BAM3, and Cell Wall Invertase (CWINV) homologs, mainly generated through tandem duplication (TD) (Fig. 3a, b and Supplementary Figs. 9–14). For instance, we identified a total of 15 SUS1/4 homologs in L. sargentiae, all of which were tandem duplicates, while only five SUS1/4 homologs were identified in G. superba (Fig. 3a, b and Supplementary Fig. 9). The extra copies in L. sargentiae can be mainly attributed to two tandem arrays on Scaffold 4 (coordinates: 4.65–4.66 Gb, and 4.68–4.69 Gb) which contained six and seven SUS1/4 homologs, respectively (Fig. 3b and Supplementary Fig. 9). Moreover, G. superba has 12 Sucrose Transporter 2 (SUT2) homologs (including seven TD-generated ones), a notable expansion relative to only two copies in L. sargentiae (Fig. 3b and Supplementary Fig. 13). Interestingly, the two lily and flame lily might have followed independent trajectories along the same direction; the gene families expanded in L. sargentiae were related to biosynthesis or degradation of sucrose and starch, while only the SUT gene family related to sucrose transport was significantly expanded in G. superba (Fig. 3a).
a Statistics on genes involved in starch and sucrose metabolism across 10 monocot plant species. Stars represent significant increase of gene copy numbers by tandem duplication. The bubble size represents the number of genes. Genes involved in biosynthesis, degradation, and transport are in red, blue, and black fonts, respectively. SUS, Sucrose Synthase; Starch Synthase, SS2; ADP Glucose Pyrophosphorylase, APL; Beta-Amylase, BAM; Cell Wall Invertase, CWINV; Sucrose Transporter, SUT2. b Microsynteny analysis of the duplicated SUS1/4 homologs between Lilium sargentiae and Gloriosa superba. c, d The representative images of the base of bulb scales without and with newly generated bulblets. During the lily bulblet initiation process, a cluster of undifferentiated cells is generated; during the bulblet expansion process, the shoot apical meristem (SAM) is generated, which represents the formation of new bulblets. e Expression of LaXTH in TRV2 control lines and TRV2-LaXTH silenced lines as well as EV control lines and OE-LaXTH overexpressed lines, respectively. Error bars are shown as mean ± s.e.m. of three technical repeats (two-sided Student’s t-test; **: P < 0.01). **P = 0.0004 (TRV2-LaXTH-1), **P = 0.0002 (TRV2-LaXTH-2), **P = 0.0009 (TRV2-LaXTH-3), **P < 0.0001 (OE-LaXTH-1), **P = 0.0005 (OE-LaXTH-2), **P < 0.0001 (OE-LaXTH-3). f The statistics of bulb induction rate (bulblet initiation rate plus bulblet expansion rate) of the TRV2 (n = 5) and TRV2-LaXTH (n = 7) lines as well as EV (n = 5) and OE-LaXTH (n = 7) lines, respectively. Error bars stand for mean ± s.e.m. (two-sided Student’s t-test; **: P < 0.01). **P = 0.0076 (TRV2-LaXTH), **P = 0.001 (OE-LaXTH). g A proposed model of the evolutionary and genetic mechanisms of bulblet initiation and bulblet expansion in L. sargentiae. JA jasmonic acid, BR brassinolide. The detailed functions of the involved genes could be found in Supplementary Data 3. More candidate genes were listed in Supplementary Fig. 15. Source data are provided as a Source Data file.
We collected samples from the scale bases and the callus/bulblets on them across four development stages from bulblet initiation to bulblet expansion (abbreviated as S0–S4) of lily plants for RNA-seq (Fig. 3c, d) (see detailed characteristics in Supplementary Fig. 15) and conducted RNA-seq. During the development stages S0 to S4, we found that 9 of 15 SUS1/4 homologs exhibited higher expression levels at S4, four of which were exclusive to bulbs (Supplementary Fig. 16). At S0, we found that eight out of ten BAM1s exhibited higher expression levels (Supplementary Fig. 16).
To identify more potential genes related to bulblet development in lily, we performed weighted gene co-expression network analysis (WGCNA). We identified a gene co-expression module (the turquoise module) correlated with bulblet expansion as well as the green module correlated with both bulblet initiation and bulblet expansion (Supplementary Fig. 15), both of which contained multiple members of the xyloglucan endotransglucosylase/hydrolase (XTH) family. The XTH genes play a crucial role in cell wall loosening for plant cell expansion, an important process in bulb development22,30. Interestingly, we found that the XTH gene family was also expanded in L. sargentiae (81 copies) compared with G. superba (47 copies), and most of the extra copies in L. sargentiae were produced through TD (Supplementary Fig. 17a, b, c). Additionally, five LsaXTHs were only expressed in bulbs (Supplementary Fig. 17d). To evaluate the importance of these LsaXTHs in bulb formation, we produced transient Virus-Induced Gene Silencing (VIGS) (TRV-LaXTH) and transient over-expression lily lines of LaXTH (OE-LaXTH). We found that the expression levels of LaXTH in TRV-LaXTH and OE-LaXTH lines were significantly lower and higher than their controls, respectively (Fig. 3e). Moreover, the bulblet induction rate (bulblet initiation and bulblet expansion rate) in TRV-LaXTH lines and OE-LaXTH lines are significantly lower and higher, respectively, than observed in their respective controls (Fig. 3f). The positive correlation between bulb induction and LaXTH expression suggests that LsaXTHs relate to the bulblet initiation and bulblet expansion processes, and the expansion of XTHs may have contributed to the formation of bulbs in L. sargentiae (Fig. 3g). Moreover, we identified other candidate genes (e.g., homologs of JAZ1, SOC1, and PMEI41) of lily bulblet initiation and bulblet expansion (Fig. 3g and Supplementary Fig. 15). Several of these genes showed high expression patterns exclusively during bulblet initiation and bulblet expansion in bulbs. Given their established functional roles and involvement in biological processes in other plants, this suggests they might participate in lily bulb development22,25,28,29,30,31,32,33,34 (Fig. 3g and Supplementary Fig. 18).
In flame lily, we also identified several genes that might be related to rhizome expansion, including the rhizome-specific ones, e.g., the Granule Bound Starch Synthase 1 homolog (GsuGBSS1, Gs05G074250) (Supplementary Figs. 19, 20b, and 21). During rhizome expansion, three SUS1/4 homologs were highly expressed and six SUT2 homologs showed the highest expression levels in rhizomes among all the examined tissues (Supplementary Figs. 19 and 20a). Among them, GsuSUT2 (Gs05G154810) was only expressed in rhizomes and displayed significantly higher expression level at 60 days after sprouting (DAS) in G. superba (Supplementary Figs. 19 and 20). Additionally, we found that genes encoding enzymes which catalyze starch biosynthesis, including GsuSS1 (Gs05G116640), GsuSS2 (Gs10G066010), GsuGBSS1 (Gs05G074250), and GsuAPL2/3/4 (Gs03G099340), all showed the highest expression levels at 40 DAS, followed by down-regulation at 60 DAS and subsequent up-regulation at 80 DAS (Supplementary Fig. 20b). GsuGBSS1 was specifically expressed in rhizomes, suggesting the coordinated regulation of starch biosynthesis and rhizome development (Supplementary Fig. 20b). In summary, these results indicate that the expansion and expression of SUS and SUT genes monitor enhanced metabolism of sucrose and starch during bulb and rhizome development, the predominant natural constituents of bulbs and rhizomes25.
Floral organ identity genes in lily and flame lily
The lilies and flame lilies feature flower structures with petaloid organs called tepals, which contribute significantly to their ornamental values. Here we elucidate the molecular basis of the floral morphology of L. sargentiae and G. superba by identifying the floral organ identity genes (including class A, B, C, and E MADS-box genes) and analyzing their expression patterns (Supplementary Figs. 22, 23, and 24). We identified a total of four class A, three class B, one class C, and three class E MADS genes in L. sargentiae, and two class A, three class B, two class C, and two class E MADS genes in G. superba (Supplementary Fig. 22). Regarding the class B PI-like genes, both PI-like genes in L. sargentiae and G. superba underwent independent duplication events after their divergence (Supplementary Fig. 22). Additionally, both PI-like genes (GsuPIa and GsuPIb) in G. superba contain the PI-motif. In contrast, only one PI-like gene (LsaPIa) in L. sargentiae possesses the PI-motif, while the other (LsaPIb) does not (Supplementary Fig. 24a). This finding is in agreement with the previous reports that there are two types of PI-like genes in lilies with one containing the PI motif35. As for the AGL2 homologs in the class E MADS genes, the two identified AGL2 homologs (LsaAGL9 and LsaAGL6) in lily are distributed across two separate clades: one clustering with AtAGL9 and another grouping more closely with AtAGL2/3/4 (Supplementary Fig. 22)36.
For L. sargentiae, the expression profiles of various floral tissues, including tepals, stamens, filaments, ovaries, and stigmas, were analyzed across five consecutive stages (S1–S5) from the bud stage to full maturity. For G. superba, tepals, stamens, and carpels were analyzed at the bud stage (Supplementary Fig. 23). We found that the class A and B MADS genes in L. sargentiae and G. superba exhibited broader expression in floral organs than their counterparts in eudicot model systems (Supplementary Fig. 23). Compared to class A MADS genes, both LsaAGL6 and GsuAGL6 exhibited significantly higher expression levels in tepals, indicating their A-class function, similar to the roles of AGL6 homologs reported in other plants, e.g., rice37,38 (Supplementary Fig. 23). LsaAG in lily and two AG genes in flame lily almost exhibited no expressions in tepals, but those showed high levels of expression in the reproductive organs (Supplementary Fig. 23). For class E genes, GsuAGL2 and GsuAGL6 showed high expression levels in all examined flower tissues at the bud stage (Supplementary Fig. 23). While in lily, class E LsaAGL2 showed lower expressions than the other two class E genes, LsaAGL9 and LsaAGL6. LsaAGL9 was expressed in all tissues, while LsaAGL6 was not expressed in stamens (Supplementary Fig. 23).
Genes related to growth transition of bulbs in lily
Lily bulbs need to undergo low-temperature conditions to facilitate growth transition and vernalization required for flowering. Incomplete growth transition inhibits the later-stage vegetative and reproductive growth of lilies, ultimately affecting the quality and yield of lily flowers. In L. sargentiae, we found a huge number (107) of the Suppressor of overexpression of CO 1 (SOC1) homologs, which is substantially (35.7-fold) more than the three SOC1 homologs in G. superba (Supplementary Fig. 25). LoSOC1, a vernalization-related candidate gene was recently identified in the RNA-seq study of dormancy bulbs and dormancy released bulbs10. We compared the ratios of bud length to bulb length in TRV and TRV-LoSOC1-infiltrated bulbs under 8 weeks of long-term cold treatment (4 °C). We observed that the central bud of TRV-LoSOC1-infiltrated bulbs exhibited significantly slower growth compared to that of the control bulbs, that is, the rate of bud length to bulb length of the control bulbs was about 1.45 times of that of the TRV-LoSOC1-infiltrated bulbs (Fig. 4a, c), indicating that the expansion of SOC1 homologs might influence the evolution of the bud transition growth trait of lilies. Additionally, we identified a dormancy-related module (the turquoise module) using WGCNA, which contains an AP1/FUL-like homolog (lili00G414190) with higher expressions in dormancy released bulbs, suggesting that this gene might also be involved in growth transition in lily (Supplementary Fig. 26). We observed that LoFUL expression is significantly induced in the shoot apical meristems with 12 °C cold treatment (bud-forcing treatment) to accelerate germination in lily cultivar ‘Siberia’ (Fig. 4d). After bud-forcing treatment for 15 days, the number of flower buds and the flower bud transition rate of the TRV-LoFUL-infiltrated bulbs were both significantly lower than those in the TRV-infiltrated bulbs (Fig. 4b, e, f), demonstrating that the AP1/FUL-like homolog is associated with the floral bud growth transition in lily.
a Representative images of bulbs in TRV control lines and TRV-LoSOC1 silenced lines after infiltration under 4 °C long-temperature cold treatment for 8 weeks. b Representative images of bulbs in TRV control lines and TRV-LoFUL silenced lines under 12 °C bud-forcing treatment for 15 days. TP tepal primordium, FAM flower apical meristem, SAM, shoot apical meristem. c Statistics of the ratios of central bud length to bulb length of TRV (n = 15 per replicate) and TRV-LoSOC1 (n = 15 per replicate) lines after infiltration under 4 °C long-temperature cold treatment for 8 weeks. Data are presented as mean ± s.d. of three technical replicates (two-sided Student’s t-test; **: P < 0.01). **P = 0.0047. d The expression pattern of LoFUL in SAMs under 4 °C from 0 week to 8 weeks and SAMs under 12 °C bud-forcing treatment from 3 days to 15 days. W, week; d, day. Error bars are shown as the mean ± s.d. of three biological replicates. ANOVA with Turkey’s HSD tests for pairwise comparisons is used for data analysis and the letters represent significant differences (P = 0.0001). **P = 0.0001. e Statistics of the number of flower buds of TRV and TRV-LoFUL lines (n = 15) with 12 °C bud-forcing treatment for 15 days. Error bars stand for mean ± s.d. (two-sided Student’s t-test; **: P < 0.01). **P < 0.0001. f Statistics of flower bud transition rate of TRV (n = 15) and TRV-LoFUL (n = 15) lines with 12 °C bud-forcing treatment for 15 days. Error bars stand for mean ± s.d. of three technical replicates (two-sided Student’s t-test; **: P < 0.01). **P < 0.0001. Source data are provided as a Source Data file.
The origin, evolution, and function of colchicine in flame lily
In flame lily, we detected distinct structural types of alkaloids, including high abundance of colchicine in leaves, flowers, and rhizomes (Fig. 5a, b and Supplementary Fig. 35a). We identified multiple homologs of the previously identified colchicine biosynthesis genes39,40 (Fig. 5c and Supplementary Figs. 27–30). Specifically, a gene cluster (named CYP75A109-OMTs gene cluster) spanning a 1.12 Mb region on Chromosome 5 contained homologs of all reported colchicine-related O-methyltransferase genes (GsOMT1, GsOMT2, GsOMT3, and GsOMT4), as well as the homolog (GsuCYP75A109, Gs05G056020) of the reported cytochrome P450 (P450) gene (GsCYP79A109)40. Additionally, this cluster includes genes encoding retrotransposons or RNA-dependent DNA polymerases (Fig. 5d and Supplementary Fig. 32). These are homologs to key genes encoding enzymes that are involved in the transformation of the (S)-1-phenethylisoquinoline scaffold to O-methylandrocymbine40. GsuOMT1 (Gs05G056070), GsuOMT2 (Gs05G055960), GsuOMT3a (Gs05G056090), GsuOMT3b (Gs05G056290), and GsuCYP75A109 were all highly expressed at 40 DAS in rhizomes (Fig. 5c), among which GsuOMT1, GsuOMT3a, and GsuCYP75A109 belong to the same co-expression module (the magenta module) identified by WGCNA (Supplementary Fig. 31), suggesting coordinated expression of genes in a cluster41. Synteny analyses found that in the genomic region corresponding to the CYP75A109-OMTs gene cluster, only the flame lily genome had the GsCYP75A109 homologous gene, while other examined monocots had different number of uncharacterized OMT genes (for example, the lily genome only has one OMT gene in this region) (Fig. 5d and Supplementary Fig. 33a). While plant OMTs constitute a large family of enzymes that methylate the oxygen atom of a variety of secondary metabolites, only GsOMT1, GsOMT2, GsOMT3, and GsOMT4 in flame lily are reported to function in colchicine biosynthesis40,42. Thus, the roles of other OMT members in colchicine biosynthesis remain to be explored. In addition, we found two tandem arrays of P450 encoding genes in G. superba, including ten paralogs of GsCYP71DA12 (encoding enzymes that catalyze the oxidation of N-formyldemecolcine) and eleven paralogs of GsCYP71FB1 (encoding enzymes that catalyze the transformation of O-methylandrocymbine to N-formyldemecolcine by the unusual oxidative ring expansion reaction) (Supplementary Fig. 32). Similarly, the CYP71FB1 tandem array in G. superba were not found in the respective syntenic genomic regions in other examined monocots (Supplementary Figs. 33b and 34b). In the syntenic genomic region of the CYP71DA12 tandem array, we identified a corresponding CYP71DA12 tandem array in the lily genome (Supplementary Fig. 34a). However, only two lily CYP71DA12 genes exhibit synteny with the CYP71DA12 genes from the flame lily (Supplementary Figs. 33c and 34). Liquid Chromatograph-Mass Spectrometer (LC-MS) analysis of L. sargentiae did not detect any traces of colchicine in bulbs and buds (Fig. 5a and Supplementary Fig. 35b), consistent with the absence of these colchicine-related gene clusters in L. sargentiae. Based on these results, the absence of these colchicine-related clusters in the other examined monocots including L. sargentiae, is suggestive of a scenario where colchicine biosynthesis has originated independently within the Colchicaceae.
a The extracted ion chromatogram (EIC) of the theoretical m/z value (400.1755, [M + H]+) of colchicine in the total ion chromatograms (TICs) of the colchicine standard, the rhizome of Gloriosa superba, and the bulb of Lilium sargentiae. b Contents of the detected metabolites of the colchicine biosynthesis pathway in G. superba. Sampling time points of 20, 40, 60, and 80 days after sprouting (DAS) in G. superba were named S1, S2, S3, and S4, respectively. Flowers on the bud stage (Bud), initial opening stage (Ini), turning stage (Tur), and mature stage (Mat) were sampled from the G. superba plants at the time of 60 DAS. L leaf, T Tepal, R rhizome. c Heatmap showing expression patterns of potential colchicine-related genes in flame lily. Genes from the CYP75A109-OMTs gene cluster are in red fonts (refer to Supplementary Fig. 32). d Microsynteny analysis of the colchicine-related O-methyltransferase genes and CYP75A109 between G. superba and L. sargentiae. Source data are provided as a Source Data file.
Discussion
In this study, we unveil the reference genome assemblies for both L. sargentiae (47.09 Gb) and G. superba (5.15 Gb) of Liliales. Notably, L. sargentiae represents the largest sequenced eukaryotic genome published to date, surpassing even that of the Chinese pine genome (Pinus tabuliformis, 25 Gb), as well as the lungfish genome (Neoceratodus forsteri, 37 Gb)43. The extraordinary size of L. sargentiae is mainly attributed to the substantial activity of LTR-RTs, which are recurrent factors in the evolution of large plant genomes23,44,45. While the genomes of most monocot orders have been sequenced, genomic information for the order Liliales remains sparse. The large genome sizes in Liliales, with an exceptionally high mean 1 C value of 39.26 pg (38.39 Gb) for monocots46, have evidently increased the challenge of resolving their genomes. The genome size diversity among Liliaceae, Melanthiaceae, and Colchicaceae has been evaluated1,47,48. For example, Lilium species exhibit a wide range of estimated genome sizes, from 44.88 pg (43.89 Gb) in L. souliei to 167.58 pg (163.93 Gb) in L. grayi2, which offers a valuable opportunity to compare the underlying evolutionary mechanisms driving genome size diversity in plants. We will gain more detailed insights into genome size and diversity of Liliales as more genomes within Liliales are sequenced in the future.
Several phylogenetic studies have utilized plastome (the chloroplast genome) sequences to infer relationships among species in Lilium49,50,51,52,53,54,55. However, relying solely in plastome sequences has limitations that could compromise the reliability of the resulting phylogeny. A main limitation is that plastid gene sequences only reflect maternal lineages, making them insufficient for detecting hybridization events55. Furthermore, the limited taxon in these previous studies may have further reduced the accuracy of the evolutionary conclusions from phylogenetic analyses49. In our study, nuclear sequences from 85 lilies were analyzed, resulting in the most comprehensive and up-to-date phylogenetic study of the Lilium genus. Unlike previous reports, our findings revealed that Lilium was divided into two major clades: one consisting solely of Asian species (the Leucolirion 6a + Archelirion species) and the other comprising species from Asia, Europe, and North America49,50,51,55. Notably, we found that Sinomartagon is polyphyletic, with species distributed across three different clades, and that Leucolirion 6a and 6b are independent sections rather than subsections, consistent with earlier findings49,50,51,54. Additionally, we further demonstrated that L. henryi and L. rosthornii belonged to Leucolirion 6a, while L. brownii was part of Leucolirion 6b, respectively49,50,56. However, the composition of sections within the three Sinomartagon clades differs from the previous reports49,50. Our results also confirmed that the Nomocharis genus is closely related to Sinomartagon 5c49,54. Our results confirmed that Martagon was monophyletic and sister to Sinomartagon 5a, rather than derived from Sinomartagon49,50. We also validated that numerous wild lilies belonging to Sinomartagon 5c and Nomocharis have not undergone hybridization with the dominant cultivated lily species. This highlights the existence of untapped genetic reservoirs and offers immense potential for future lily breeding endeavors. We detected shared genome haplotypes among wild and cultivated lily species. While significant advancements in lily breeding have been made in the past 50 years, particularly through interspecific hybridization57, we found that some modern lilies may have been developed through methods other than hybridization, such as domestication and breeding. Our classification of the Lilium genus, the origin of cultivated lilies, and hybridization patterns at the nuclear gene level provide extensive insights into the relationships among major wild and cultivated lily species and offer directions for future breeding efforts.
To date, while several genomes of bulbous and rhizomatous plants have been previously documented21,22,58,59, our understanding of these unique plant structures remains limited. In our proposed module for lily bulb development, we identified biological pathways similar to those found during bulb formation in onion, which are known to play crucial roles in storage organ formation22. Notably, we discovered that multiple gene families involved in sucrose and starch metabolism, key processes in the initiation and expansion of storage organs13,25,26,27,28,29, have undergone significantly TD in lily. Furthermore, members of the XTH gene family in lily are also significantly expanded due to TD; these genes have been previously reported to exhibit differential expressions during storage organ development33. Importantly, we provide the experimental evidence of the role of LsaXTH in bulb formation in plants. The research on rhizome development in flame lilies is more fragmentary compared to that of lily bulb formation60. In this study, we proposed a model for rhizome formation in flame lily, highlighting the potential importance of sucrose metabolism/transport and starch biosynthesis pathways in this process. On the basis of the crucial roles of StSUT1 and StGBSS genes in potato tuber development61,62,63. We propose that the rhizome specific GsuSUT2 and GsuGBSS1 genes may be vital for flame lily rhizome formation. Specifically, in the flame lily rhizome, GsuSUT2 likely facilitates the transport of sucrose for subsequent starch biosynthesis in amyloid, while GsuGBSS1 is involved in the following amylose synthesis process. Additionally, the two significantly expanded gene families in lily and flame lily are different, providing a valuable opportunity to understand the formation of distinct modified stem structures in these two species.
Although a near-complete colchicine biosynthesis pathway in G. superba has recently been elucidated, the genomic foundation of this pathway remains largely unexplored39,40. We identified gene clusters responsible for colchicine biosynthesis that are unique to flame lily. Notably, the critical gene clusters associated with colchicine biosynthesis and colchicine-related metabolites, are absent in L. sargentiae, suggesting that this species lacks the complete pathway to synthesize colchicine. This finding is consistent with the previous report that colchicine production is restricted to the Colchicaceae family16, supporting the hypothesis of an independent origin of colchicine biosynthesis within this family. Within the syntenic genomic region of the colchicine-related CYP71DA12 tandem array in flame lily, the lily genome also contains a CYP71DA12 tandem array. However, only two of the lily CYP71DA12 genes are syntenic homologs of the flame lily CYP71DA12 genes. Interestingly, each of these two lily CYP71DA12 genes forms multiple syntenic gene pairs with the flame lily CYP71DA12s, particularly LsaCYP71DA12a (lili00G285310). This suggests that after the divergence of the lily and flame lily, their ancestral CYP71DA12 genes underwent TD in flame lily, leading to functional divergence of these genes. Subsequently, the CYP71DA12 genes in flame lily have evolved to encode enzymes for colchicine biosynthesis. However, it remains to be verified whether the CYP71DA12 genes in lily exhibit similar function to the CYP71DA12s of flame lily. Furthermore, in the phylogenetic analysis of CYP75A genes, we identified one GsCYP75A109 homolog, and multiple GsCYP75A110 and GsCYP75A109/110-like homologs in flame lily. In contrast, we identified only one CYP75A gene in both the lily genome and the transcriptome assemblies of various lilies, which was found to occupy the most basal branches relative to the homologs of GsCYP75A109 and GsCYP75A110 (Supplementary Fig. 29c). Considering that GsCYP75A109 and GsCYP75A110 play distinct roles within colchicine biosynthesis, specifically in the phenethylisoquinoline scaffold hydroxylation and phenol ring coupling, respectively39,40, we propose that the CYP75A109 and CYP75A110 genes in flame lily may have undergone functional divergence following the formation of multiple copies and have acquired novel functions. In contrast, the single CYP75A gene in lily is not likely to contain these specialized functionalities.
In summary, our work provides insights into the evolution of expansive plant genomes, which offers valuable genomic resources for the future study of Liliales plants and acceleration of the selection and breeding of lilies and flame lilies.
Methods
Plant materials
The 51 Lilium wild species as well as 34 lily cultivars used in RNA-seq were cultivated at the Yanqing Base in Beijing, China under natural conditions. The L. sargentiae plants used in genome sequencing and NGS were cultivated in Kunming, Yunnan Province, China (25.04°N, 102.72°E) under natural conditions. The G. superba (the cultivated variety named ‘Passion flame’) plants used in genome sequencing, RNA-seq, and NGS were cultivated in Yuxi, Yunnan Province, China (25.04°N, 102.72°E) under natural conditions. Lily cultivars ‘Tresor’ and ‘Siberia’ used in gene cloning were planted in the Science Research Garden at China Agricultural University, Beijing, China (39.90°N, 116.40°E) under natural conditions. The leaves of L. sargentiae and G. superba plants were used to isolate DNA for genome sequencing and NGS. The detailed list of plant samples used in RNA-seq in this study can be found in Supplementary Data 1 and 2.
Genome sequencing and assembly
PacBio library construction and sequencing. According to PacBio’s standard protocol for third-generation sequencing, the PacBio DNA library was built and then sequenced on the Sequel platform using the CCS mode of the single-molecule real-time sequencing technology. To obtain sub-reads, long sequencing reads were initially corrected. The size of fragments selected for sequencing was approximately 20 kb. HiFi reads were generated by merging sub-reads.
Hi-C library construction and sequencing were conducted on the Illumina HiSeq X Ten system using the 150-bp pair-end reads. The Hi-C libraries were constructed to anchor scaffolds onto chromosomes64. DNA isolated from L. sargentiae and G. superba leaves were digested with HindIII overnight, respectively. Sticky ends were biotinylated and proximity-ligated, and then physically sheared to a size of 500–700 bp and enriched to make chimeric junctions. The cross-linked long-distance physical connections were next processed into chimeric fragments, followed by reverse cross-linking, purification, and PCR amplification, and were subsequently used to create paired-end sequencing libraries. For the PCR amplification reaction, the cycling conditions are set as follows: (1) initial denaturation: 95 °C for 3 min for (1 cycle); (2) denaturation: 98 °C for 20 s (10 cycles); (3) annealing: 60 °C for 15 s (10 cycles); (3) extension: 72 °C for 30 s(10 cycles); (4) final extension: 72 °C for 5 min (1 cycle); (5) hold: 4 °C. To evaluate the Hi-C sequencing, the Hi-C-Pro program (v3.1.0) was then used65.
Short-read library construction and sequencing. The 150-bp paired-end libraries were constructed from the high-quality DNA for NGS and were subsequently sequenced using the Illumina X-ten platform (Illumina, CA, USA).
For transcriptome sequencing, cDNA libraries were constructed from the integrated and high-quality RNA of the mixture of bulbs, stems, leaves, and flowers from lilies as well as rhizomes, leaves, and flowers from flame lily. Before the Agilent Bioanalyzer 2100 system was used to evaluate the cDNA library, the PCR products were purified. The libraries were sequenced using Illumine HiSeq X Ten system according to the manufacturer’s instructions. Trimommatic (v 0.39) was used to clean up the raw RNA-seq readings by removing low-quality bases, adaptors, duplications, and potential contaminants66.
Genome size and heterozygosity estimation
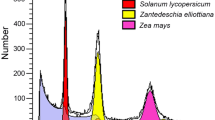
Genome size and heterozygosity were estimated by K-mer frequency distribution analysis. Initially, the short reads were filtered using fastp (v0.19.4) with default settings. K-mers were then counted using Jellyfish (v2.2.10) with the parameters “-C -m 17 -s 1 G -t 56”67,68. The resulting output file was subsequently used as input for GenomeScope (v2.0)69, which estimated the genome size and heterozygosity using default parameters. The 17-mer frequency distribution was analyzed based on genome characteristics, considering the pattern of Poisson distribution. The genome sizes of L. sargentiae and G. superba were also estimated by flow cytometry using DNA from fresh leaves. Zea mays (2 C = 4.7 pg) and Triticum aestivum (2 C = 30.9 pg) were used as an internal standard for the evaluation of the genome sizes of G. superba and L. sargentiae, respectively.
Genome assembly and evaluation
HiFi-asm (v 0.9) (https://github.com/chhylp123/hifiasm) was used to assemble PacBio HiFi reads to obtain assembled contigs using the HiFi read sequencing data70. Purge-dup (https://github.com/dfguan/purge_dups) was used for sequence dehybridization to obtain the purged assembled contigs. Juicer (v 1.5.6)71 was employed for the exclusive mapping of purified Hi-C reads obtained from data quality control to the cleansed contig assemblies. Additionally, 3D-DNA (v 180922)72 and JuiceBox (v 2.0)73 were used to construct the scaffolds. The completeness of the genome annotation was evaluated using BUSCO (v5.4.0)17 with the embryophyta_odb10, which includes 1,664 single-copy orthologs.
Genome annotation
Gene structure annotation was performed through the method of combining evidence-based prediction and de novo prediction (De novo). For evidence-based annotation, we used Maker (https://gmod.org/wiki/MAKER_Tutorial) to utilize RNA-seq data as evidence files. Using RNA-seq data and genome alignment information, the Augustus training model was constructed through Program to Assemble Spliced Alignments (v2.4.1)74, and then De novo was performed by Augustus (http://bioinf.uni-greifswald.de/augustus/). Then the Maker and De novo results were integrated based on the principle that the evidence results were prior to the predicted results. Finally, the genes encoding proteins with a repeat ratio of less than 50% and with a length greater than 50 amino acids, and with FPKM (fragments per kilobase of exon per million mapped reads) greater than 0 in at least one transcriptome sequencing sample were retained.
The repeat sequences of the L. sargentiae and G. superba genomes were annotated by methods including homologous sequence alignment (Homolog) and De novo. RepeatModeler (http://www.repeatmasker.org/RepeatModeler/), which can automatically perform RECON (v 1.08)75 and RepeatScout (v1.0.5)76, and LTR_FINDER(v1.06) (http://tlife.fudan.edu.cn/ltr_finder/) were used to build de novo repeat libraries of the L. sargentiae and G. superba genomes, respectively. By the combination of the RepBase v21.12 database (http://www.girinst.org/repbase), the repeat sequences were predicted using RepeatMasker (v 4.0.7)77.
Gene functional annotation
To perform functional annotation, orthology assignments, and domain prediction, the identified proteins was used in eggNOG-Mapper v278. The annotation results were counted in Supplementary Tables 13 and 14. The GO annotation information on genes were obtained from the results of eggNOG-Mapper. TBtools (v 1.098696) was used to conduct GO enrichment analysis of genes in proximity (within 2 kb) to Tork and Tekay LTR-RTs or containing these LTR-RTs in their introns in the lily genome79. The P-value cutoff for measuring whether the enrichment of the GO term is significant is 0.05.
Phylogenetic tree reconstruction
The species phylogenetic tree of 26 plant species including L. sargentiae and G. superba was constructed by 893 single-copy orthologous genes obtained by BUSCO (Benchmarking Universal Single-Copy Orthologs)17. Protein sequences from these single-copy homologous genes were aligned using MAFFT (v 7.467) with default settings, followed by trimming with default parameters of trimAL (v 1.4.1)80,81. The maximum likelihood tree was then constructed using FastTree (v2.1)82 default settings and visualized with MEGA (v 7.0.26)83.
Synteny analysis
The syntenic dot plots of syntenic gene pairs were drawn by WGDI (v0.4.7)84. Ks values were calculated for each gene pair in the aligned blocks obtained by WGDI (v0.4.7)84. The distributions of all Ks values were plotted to infer WGD events that occurred during the evolutionary history. To correlate the Ks peak representing the monocot-shared τ event, we applied a Ks correction method as follows: if species A and B experienced the same duplication event, it should have occurred simultaneously in both, meaning their Ks peaks should be equal (i.e., KsA = KsB). However, due to different evolutionary rates among species, the actual KsA and KsB are not identical. Assuming that after the duplication event, species A and B evolved at rates VA and VB, respectively, and that the divergence point between them, O, reflects an ancestral evolutionary rate of V, the correction factor for species A is
and for species B is
Thus, the corrected Ks value between species A and B is
The correction for key evolutionary events within species A and B are
LTR insertion time estimation
Careful classification of repeat sequences (solo-LTRs, intact-LTRs, and the subfamilies of LTRs) was performed using EDTA (v1.9.9)85. The time calculation module within LTR_retriever (v2.9.5)86 was used to determine the insertion times of LTR elements. Annotation of solo-LTRs was performed using the solo_finder.pl script integrated into the above software.
The origin of cultivated lilies
A total of 85 transcriptomes comprising 34 cultivated and 51 wild lilies were selected for phylogenetic tree analysis (Supplementary Data 1). Trinity (v2.1.1)87 was used for transcriptome assembly, yielding the longest transcript. TransDecoder (v5.5) (https://www.msi.umn.edu/sw/transdecoder) was then used for opening reading frame prediction, resulting in the extraction of the protein sequences from the longest transcripts. Orthofinder (v2.3.1)88 was then used to identify single-copy genes, resulting in the detection of 360 single-copy orthologous genes. IQ-TREE(v1.6.2)89 was used for model selection and 1000 bootstrap tests. The JTT + F + I + G4 model was selected for phylogenetic tree construction. To access the similarity of lily transcripts (the shared genome haplotypes) across different species, an all-to-all blast calculation was performed among lily species. For each lily species, the proportion of transcripts exhibiting the highest gene similarities (identities > 95%) with those of other species was calculated, excluding self-alignments within species. The resulting data were visualized using the pheatmap package in R (1.0.12) to generate a heatmap. For lilies within Clade 1, transcript assembly was performed using Trinity (v2.1.1)87. Orthofinder (v2.3.1)88 was employed to identify gene clusters using these transcripts, along with the longest transcripts of the L. sargentiae genome. Gene clusters containing transcripts from more than 80% of these species were preserved. For each species, the number of gene clusters of one to ten genes were calculated.
Gene family identification
Gene family members were identified by HMMER (v3.0)90 and BLASTP (v2.2.26)91. Multiple sequence alignment was performed by MAFFT (v 7.467)80, and the maximum likelihood tree was built using FastTree (v2.1)82. For the identification of MADS genes, we used IQ-TREE (v1.6.12)89 to construct the phylogenetic trees using the most appropriate evolutionary model selected based on the data. To further investigate the presence or absence of colchicine-related P450 and OMT genes in Liliales, transcripts from a total of 71 transcriptome assemblies of wild Liliales species were analyzed. This included data from species including Colchicum autumnale, G. superba, Helonias bullata, Smilax bona-nox, Xerophyllum asphodeloides, and L. sargentiae, which were downloaded from the 1KP database (https://db.cngb.org/onekp/) (Supplementary Data 4). The gene IDs corresponding to the gene symbols mentioned in this study can be found in Supplementary Data 5.
RNA-seq analysis
The RNA-seq results and samples of L. sargentiae and G. superba can be found Supplementary Data 1 and 2, respectively. Flame lily rhizomes were sampled for RNA-seq at 20, 40, 60, and 80 DAS, along with leaves at 60 DAS, and flowers at various stages (bud, initial opening, turning, and mature) at 60 DAS. Three biological replicates were collected for each sample. For L. sargentiae, flower tissues, including tepals, stamens, filaments, ovaries, and stigmas, were sampled at five consecutive stages from the bud stage to the mature stage. Additionally, whole flowers at the mature stage were also collected for RNA-seq analysis (at least two biological replicates were used). Using hisat2 (v 2.2.0) with the default parameters, clean reads were mapped into the assembled L. sargentiae and G. superba genomes92. Cutfflinks (v2.1.1) were used to obtain the expression levels (FPKM)93. WGCNA (v3.2.5)94 and Cytoscape (v3.6.1)95 were used to analyze and show gene co-expression networks, respectively. For WGCNA, the weighted network was unsigned. Genes with a mean gene expression level (FPKM) less than 1 were filtered. The used top 10,000 genes were selected by median absolute deviation. The minimum module size was 30. The public RNA-seq data from ‘Tresor’ and ‘Siberia’ were aligned to the L. sargentiae genome and subsequently used in WGCNA analysis to study bulb development and growth transition, respectively (Supplementary Data 1). TBtools (v 1.098696) was used to display gene expression patterns79.
Silencing of LaXTH, LoSOC1, and LoFUL in lily scales by VIGS
LaXTH was chosen by BLASTP91 using the protein sequence of lili00G412220, the homolog of AtXTH22. LaXTH was cloned from lily cultivar ‘Tresor’. LoSOC1 and LoFUL (the homolog of lili00G414190) were cloned from lily cultivar ‘Siberia’. CDS sequences of these three genes could be found in Supplementary Data 6. The pTRV2, pTRV2-LaXTH, pTRV2-LoSOC1, and pTRV2-LoFUL lines were obtained using the VIGS method. The gene fragments of LaXTH, LoSOC1, and LoFUL were used to generate the constructs pTRV2-LaXTH, pTRV2-LoSOC1, and pTRV2-LoFUL, respectively. These constructs, along with pTRV1, pTRV2, and the modified pTRV2 vectors, were separately introduced into Agrobacterium tumefaciens strain EHA105. The transformed A. tumefaciens was cultured in liquid Luria-Bertani medium supplemented with 50 mg/L kanamycin and 25 mg/L rifampicin at 28 °C, then harvested by centrifugation. The bacterial pellets were resuspended in an infiltration buffer (10 mM MgCl2, 200 μM acetosyringone, and 10 mM MES at pH 5.8) to a final optical density at 600 nm (OD600) of 1.0. For agroinfiltration, a 1:1 (v/v) mixture of A. tumefaciens cultures carrying pTRV1 and pTRV2-LaXTH (or pTRV2-LoSOC1 or pTRV2-LoFUL) was prepared, while a mixture of pTRV1 and pTRV2 served as the control. For the pTRV2 and pTRV2-LaXTH lines, the infiltrated scales were dried on moist absorbent cotton in glass culture bottles and were incubated in the dark for 14 days at room temperature (22 °C) for statistics of bulb induction rate. Bulblet initiation rate and bulblet expansion rate are the rates of callus formation and the rates of bulblet formation, respectively. In the pTRV2 lines, the bulb induction rate was counted at S0 when it was a merely lily scale, while in the pTRV2-LaXTH lines, the bulb induction rate was measured at S1 when callus formation happened. The experiments in pTRV2 lines and pTRV2-LaXTH lines were repeated five and seven times using 10 scales in each repetition, respectively. For the pTRV2 and pTRV2-LoSOC1 lines, the bud length and bulb length were calculated using ImageJ/FIJI. Student’s t-test (*: P < 0.05; **: P < 0.01) was used in statistical analyses.
Transient over-expression of LaXTH in lily scales
LaXTH was inserted into the pCAMBIA2300 vector, and the empty pCAMBIA2300 vector was used as the control. The EV (35S:eGFP/pCAMBIA2300) and OE-LaXTH (35S:LaXTH-eGFPpCAMBIA2300) vectors were transformed into Agrobacterium tumefaciens GV3101, respectively to get EV and OE-LaXTH lines10. The bulb induction rate was counted as described in the section of ‘Silencing of LaXTH, LoSOC1, and LoFUL in lily scales by VIGS”. Student’s t-test (*: P < 0.05, **: P < 0.01) was used in statistical analyses.
SEM observation
Scanning electron microscopy (SEM) was performed using central buds of lily bulbs. The central buds were collected and fixed in FAA solution (a mixture of 50% ethanol, 5% acetic acid, and 3.7% formaldehyde) for 8 h, followed by a 30-min vacuum treatment. The fixed buds were gradually dehydrated in an ethanol series, undergoing submersion with gentle agitation at concentrations of 50%, 70%, and 90% ethanol for 1 h each, and twice in 100% ethanol for 1 h each at room temperature. The sample was placed on the Hitachi TM-4000 scanning electron microscopy and their tiny scales were removed under the stereo microscope.
qRT-PCR analysis
Total RNA was extracted from SAMs of lily cultivars ‘Siberia’ under two conditions: 4 °C from 0 to 8 weeks, and 12 °C bud-forcing treatment from 3 to 15 days. Additionally, RNA was extracted from tissue located 2 mm above the base of the lily scales in pTRV2, pTRV2-LaXTH, EV, and OE-LaXTH lines at the S1 stage. The extraction was performed using the FastPure Plant Total RNA Isolation Kit (Vazyme, Nanjing, China), to analyze the expression profiles of LoFUL and LaXTH, respectively. The qRT-qPCR experiment was performed on the Applied Biosystems StepOnePlus real-time PCR system using the ChamQ SYBR qPCR kit (Vazyme, Nanjing, China). FP was used as the internal control gene. All experiments were run in three replicates and the expression data were calculated by the method of the 2−ΔΔCt. Primer sequences were listed in Supplementary Table 15.
Detection of colchicine in lily and flame lily
A total of 50 mg of plant materials were ground in liquid nitrogen, and then 1 ml of 80% methanol-water solution was added. The mixture was sonicated for 15 min, and the supernatant was collected after centrifugation. The sample was then added to a sample bottle for detection. Thermo U3000-QEplus liquid chromatography-mass spectrometry system (Waters C18 T3, 2.1 × 150 mm, and 3.5 µm chromatography column) was used in detection. Mobile phase A was 0.1% formic acid in water, while mobile phase B was 0.1% formic acid in acetonitrile. The flow rate was set at 0.2 ml/min with the following gradient: 0–7 min, 5–90% B; 7–8 min, 90% B; 8.5–10 min, 5% B. The colchicine standard purchased from Shanghai Yuan Ye was optimized with a molecular weight of 400.1755 and the colchicine retention time is 6.36 min.
Untargeted metabolomics
Flame lily rhizomes were sampled for metabolite sequencing at 20, 40, 60, and 80 DAS, along with leaves at 60 DAS, and flowers at various stages (bud, initial opening, turning, and mature) at 60 DAS. Three biological replicates were collected for each sample. Biological samples were processed using a Scientz-100F vacuum freeze-dryer for freeze-drying. The resulting material was pulverized with a zirconia bead in a mixer mill (MM 400, Retsch) for 1.5 min at a frequency of 30 Hz. A portion of 50 mg of the powdered sample was dissolved in 1.2 mL of 70% methanol solution and subjected to vertexing for 30 s every 30 min, repeated six times in total. After centrifuging at 12,000 rpm for 3 min, the supernatant was filtered through a 0.22 µm pore size membrane (SCAA-104, ANPEL, Shanghai, China; http://www.anpel.com.cn/) and prepared for UPLC-MS/MS analysis.
The analysis was carried out on a UPLC-ESI-MS/MS system, comprising a UPLC unit (ExionLCTM AD, https://sciex.com.cn/) and a mass spectrometer (Applied Biosystems 6500 Q TRAP, https://sciex.com.cn/). The chromatographic separation was performed on an Agilent SB-C18 column (1.8 µm, 2.1 mm × 100 mm). The mobile phase consisted of solvent A (water containing 0.1% formic acid) and solvent B (acetonitrile containing 0.1% formic acid). A gradient program was applied, starting with 95% A and 5% B, transitioning to 5% A and 95% B within 9 min, maintaining this ratio for 1 min, then reverting to 95% A and 5% B over 1.1 min and holding this condition for 2.9 min. The flow rate was 0.35 mL/min, with the column oven set at 40 °C and an injection volume of 2 µL. The effluent was connected to an ESI-triple quadrupole-linear ion trap mass spectrometer. The ESI source was operated at a source temperature of 500 °C with an ion spray voltage of 5500 V in positive mode and −4500 V in negative mode. Source gas I, source gas II, and curtain gas were set at 50, 60, and 25 psi, respectively, with a high collision-activated dissociation setting. Multiple reaction monitoring (MRM) transitions were acquired with nitrogen as the collision gas set to medium. Declustering potential and collision energy were optimized for each MRM transition, and specific MRM transitions were monitored for metabolites based on their elution times.
Data acquisition was performed using the MRM mode, with instrument control and raw data acquisition carried out via Analyst (v 1.6.3)96 software. After data collection, metabolite identification was conducted using Analyst (v 1.6.3)96 in conjunction with the self-built Metware Database from Wuhan Metware Biotechnology Co., Ltd. The identification process involved comparing the MS/MS spectra of detected metabolites with database entries, using secondary mass spectral information to achieve high-confidence qualitative analysis. During data processing, isotopic signals, duplicate signals containing K+, Na+, and NH4+ ions, and fragment ion signals that were duplicates of larger molecular weight substances were removed to ensure data accuracy. For quantitative analysis, chromatographic peaks of all detected substances were integrated based on peak areas by MultiQuant (v 3.0)97. Quantification was achieved by normalizing and correcting the peak areas of the same metabolite across different samples, ensuring result consistency and accuracy98.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequencing data generated in this study have been deposited in the Sequence Read Archive of the NCBI database under the accession PRJNA1037021. The genome assemblies with annotations and metabolomics raw data are available at FigShare [https://figshare.com/articles/dataset/_b_The_giant_genome_of_lily_provides_insights_into_the_hybridization_of_cultivated_lilies_b_/27933375]99. The accession numbers in the NCBI GenBank database of the previously reported functional genes used in this study are available in Supplementary Data 7. Source data are provided with this paper.
References
Leitch, I. J. et al. Punctuated genome size evolution in Liliaceae. J. Evol. Biol. 20, 2296–2308 (2007).
Du, Y. P. et al. Genome size diversity in Lilium (Liliaceae) is correlated with karyotype and environmental traits. Front. Plant Sci. 8, 1303 (2017).
Zhou, J., An, R. & Huang, X. Genus Lilium: a review on traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 270, 113852 (2021).
Dhiman M. R., Sharma P. & Bhargava B. Lilium: conservation, characterization, and evaluation. In: Floriculture and Ornamental Plants (eds Datta, S. K. & Gupta, Y. C.) 1–36 (Springer, Singapore, 2021) https://www.bexis.uni-jena.de/ddm/data/Showdata/27087.
Du, Y. P. et al. Lilium spp. pollen in China (Liliaceae): taxonomic and phylogenetic implications and pollen evolution related to environmental conditions. PloS one 9, e87841 (2014).
Comber, H. F. A new classification of the genus Lilium. In: Lily Year Book of RHS (ed. Chittenden, F. J.) 13, 86–105 (Royal Horticultural Society, London, 1949) https://www.scirp.org/reference/referencespapers?referenceid=3215680.
Bakhshaie, M., Khosravi, S., Azadi, P., Bagheri, H. & van Tuyl, J. M. Biotechnological advances in Lilium. Plant Cell Rep. 35, 1799–1826 (2016).
Pride, G. H. Lilies and the arnold arboretum. Arnoldia 34, 125–132 (1974).
Du, F. et al. Identification of differentially expressed genes in flower, leaf and bulb scale of Lilium oriental hybrid ‘Sorbonne’ and putative control network for scent genes. BMC Genomics 18, 899 (2017).
Pan, W. et al. Epigenetic silencing of callose synthase by VIL1 promotes bud-growth transition in lily bulbs. Nat. Plants 9, 1451–1467 (2023).
Lazare, S. & Zaccai, M. Flowering pathway is regulated by bulb size in Lilium longiflorum (Easter lily). Plant Biol. 18, 577–584 (2016).
Li, X. et al. Transcriptome analysis of carbohydrate metabolism during bulblet formation and development in Lilium davidii var. unicolor. BMC Plant Biol. 14, 358 (2014).
Xin, Y. et al. Auxin regulates bulbil initiation by mediating sucrose metabolism in Lilium lancifolium. Hortic. Res 11, uhae054 (2024).
Larsson, S. & Rønsted, N. Reviewing Colchicaceae alkaloids - perspectives of evolution on medicinal chemistry. Curr. Top. Med. Chem. 14, 274–289 (2014).
Stander, E. A., Papon, N. & Courdavault, V. Puzzling out the colchicine biosynthetic pathway. ChemMedChem 16, 621–623 (2020).
Vinnersten, A. & Larsson, S. Colchicine is still a chemical marker for the expanded Colchicaceae. Biochem. Syst. Ecol. 38, 1193–1198 (2010).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Timilsena, P. R. et al. Phylogenomic resolution of order- and family-level monocot relationships using 602 single-copy nuclear genes and 1375 BUSCO genes. Front. Plant Sci. 13, 876779 (2022).
Zhang, G. Q. et al. The Apostasia genome and the evolution of orchids. Nature 549, 379–383 (2017).
Shi, T. et al. The slow-evolving Acorus tatarinowii genome sheds light on ancestral monocot evolution. Nat. Plants 8, 764–777 (2022).
Sun, X. et al. A chromosome-level genome assembly of garlic (Allium sativum) provides insights into genome evolution and allicin biosynthesis. Mol. Plant 13, 1328–1339 (2020).
Hao, F. et al. Chromosome-level genomes of three key Allium crops and their trait evolution. Nat. Genet 55, 1976–1986 (2023).
Niu, S. et al. The Chinese pine genome and methylome unveil key features of conifer evolution. Cell 185, 204–217.e214 (2022).
Cossu, R. M. et al. LTR retrotransposons show low levels of unequal recombination and high rates of intraelement gene conversion in large plant genomes. Genome Biol. Evol. 9, 3449–3462 (2017).
Zierer, W., Ruscher, D., Sonnewald, U. & Sonnewald, S. Tuber and tuberous root development. Annu Rev. Plant Biol. 72, 551–580 (2021).
Hao, C. et al. Sucrose function on the bulbil formation of Lilium lancifolium. Sci. Hortic. 323, 112538 (2024).
Liang, J. et al. Cytokinins influence bulblet formation by modulating sugar metabolism and endogenous hormones in Asiatic hybrid lily. Ornament Plant Res. 3, 19 (2023).
Seng, S. et al. ADP-glucose pyrophosphorylase gene plays a key role in the quality of corm and yield of cormels in gladiolus. Biochem. Bioph. Res. Co. 474, 206–212 (2016).
Seng, S. et al. Silencing GhAGPL1 reduces the quality and quantity of corms and cormels in gladiolus. J. Am. Soc. Hortic. Sci. 142, 119–125 (2017).
Ishida, K. & Yokoyama, R. Reconsidering the function of the xyloglucan endotransglucosylase/hydrolase family. J. Plant Res. 135, 145–156 (2022).
Guo, L., Plunkert, M., Luo, X. & Liu, Z. Developmental regulation of stolon and rhizome. Curr. Opin. Plant Biol. 59, 101970 (2021).
Atif, M. J. et al. Mechanism of Allium crops bulb enlargement in response to photoperiod: A review. Int J. Mol. Sci. 21, 1325 (2020).
Li, M. et al. Expression analysis of XTH in stem swelling of stem mustard and selection of reference genes. Genes 11, 113 (2020).
Huang, S. et al. RNA interference knockdown of the brassinosteroid receptor BRI1 in potato (Solanum tuberosum L.) reveals novel functions for brassinosteroid signaling in controlling tuberization. Sci. Hortic. 290, 110516 (2021).
Chen, M. K., Hsieh, W. P. & Yang, C. H. Functional analysis reveals the possible role of the C-terminal sequences and PI motif in the function of lily (Lilium longiflorum) PISTILLATA (PI) orthologs. J. Exp. Bot. 63, 941–961 (2011).
Tzeng, T. Y., Hsiao, C. C., Chi, P. J. & Yang, C. H. Two lily SEPALLATA-like genes cause different effects on floral formation and floral transition in Arabidopsis. Plant Physiol. 133, 1091–1101 (2003).
Dreni, L. The ABC of flower development in monocots: the model of rice spikelet. Methods Mol. Biol. 2686, 59–82 (2023).
Hu, J. et al. The pineapple MADS-box gene family and the evolution of early monocot flower. Sci. Rep. 11, 849 (2021).
Nett, R. S. & Sattely, E. S. Total biosynthesis of the tubulin-binding alkaloid colchicine. J. Am. Chem. Soc. 143, 19454–19465 (2021).
Nett, R. S., Lau, W. & Sattely, E. S. Discovery and engineering of colchicine alkaloid biosynthesis. Nature 584, 148–153 (2020).
Nutzmann, H. W., Scazzocchio, C. & Osbourn, A. Metabolic gene clusters in eukaryotes. Annu Rev. Genet 52, 159–183 (2018).
Lam, K. C., Ibrahim, R. K., Behdad, B. & Dayanandan, S. Structure, function, and evolution of plant O-methyltransferases. Genome 50, 1001–1013 (2007).
Meyer, A. et al. Giant lungfish genome elucidates the conquest of land by vertebrates. Nature 590, 284–289 (2021).
Lou, H. et al. The Torreya grandis genome illuminates the origin and evolution of gymnosperm-specific sciadonic acid biosynthesis. Nat. Commun. 14, 1315 (2023).
Jayakodi, M. et al. The giant diploid faba genome unlocks variation in a global protein crop. Nature 615, 652–659 (2023).
Leitch, I. J., Beaulieu, J. M., Chase, M. W., Leitch, A. R. & Fay, M. F. Genome size dynamics and evolution in monocots. J. Exp. Bot. 2010, 1–18 (2010).
Pellicer, J., Kelly, L. J., Leitch, I. J., Zomlefer, W. B. & Fay, M. F. A universe of dwarfs and giants: genome size and chromosome evolution in the monocot family Melanthiaceae. N. Phytol. 201, 1484–1497 (2013).
Fridlender, A., Brown, S., Verlaque, R., Crosnier, M. & Pech, N. Cytometric determination of genome size in Colchicum species (Liliales, Colchicaceae) of the western Mediterranean area. Plant Cell Rep. 21, 347–352 (2002).
Duan, Q. et al. Phylogenetic analysis of wild species and the maternal origin of cultivars in the genus Lilium using 114 plastid genomes. Front. Plant Sci. 13, 865606 (2022).
Kim, H. T., Lim, K. B. & Kim, J. S. New insights on Lilium phylogeny based on a comparative phylogenomic study using complete plastome sequences. Plants 8, 547 (2019).
Zhou, N. et al. Historical biogeography and evolutionary diversification of Lilium (Liliaceae): New insights from plastome phylogenomics. Plant Divers 46, 219–228 (2024).
Du, Y. P. et al. Complete chloroplast genome sequences of Lilium: insights into evolutionary dynamics and phylogenetic analyses. Sci. Rep. 7, 5751 (2017).
Kim, J. S. & Kim, J. Updated molecular phylogenetic analysis, dating and biogeographical history of the lily family (Liliaceae: Liliales). Bot. J. Linn. Soc. 187, 579–593 (2018).
Li, J. et al. Phylogeny, age, and evolution of tribe Lilieae (Liliaceae) based on whole plastid genomes. Front. Plant Sci. 12, 699226 (2021).
Givnish, T. et al. Evolution, geographic spread and floral diversification of the genus Lilium with special reference to the lilies of North America. Evolution 74, 26–44 (2020).
Du, Y. P. et al. Molecular phylogeny and genetic variation in the genus Lilium native to China based on the internal transcribed spacer sequences of nuclear ribosomal DNA. J. Plant Res. 127, 249–263 (2014).
van Tuyl, J. M. & Arens, P. Lilium: Breeding history of the modern cultivar assortment. Acta Hortic. 900, 223–230 (2011).
Cheng, S. P. et al. Haplotype-resolved genome assembly and allele-specific gene expression in cultivated ginger. Hortic. Res. 8, 188 (2022).
Li, H. L. et al. Haplotype-resolved genome of diploid ginger (Zingiber officinale) and its unique gingerol biosynthetic pathway. Hortic. Res. 8, 189 (2022).
Bass, J. S., Gang, D. R., Kutchan, T. M. & Sivakumar, G. Gloriosa superba and Colchicum autumnale multi-tissue transcriptome analysis for colchicine pathway and rhizome development candidate gene identification. Research Square. https://arxiv.org/abs/1234.56789 (2019).
Kuhn, C. et al. The sucrose transporter StSUT1 localizes to sieve elements in potato tuber phloem and influences tuber physiology and development. Plant Physiol. 131, 102–113 (2003).
van der Steege, G., Nieboer, M., Swaving, J. & Tempelaar, M. J. Potato granule-bound starch synthase promoter-controlled GUS expression: regulation of expression after transient and stable transformation. Plant Mol. Biol. 20, 19–30 (1992).
Andersson, M. et al. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 36, 117–128 (2017).
Burton, J. N. et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 31, 1119–1125 (2013).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Vurture, G. W. et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33, 2202–2204 (2017).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017).
Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016).
Haas, B. J. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003).
Bao, Z. & Eddy, S. R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12, 1269–1276 (2002).
Price, A. L., Jones, N. C. & Pevzner, P. A. De novo identification of repeat families in large genomes. Bioinformatics 21, i351–i358 (2005).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinforma. 25, 4.10.1–4.10.14 (2009).
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P. & Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829 (2021).
Chen, C. et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202 (2020).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Sun, P. et al. WGDI: a user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Mol. Plant 15, 1841–1851 (2022).
Ou, S. et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20, 275 (2019).
Ou, S. & Jiang, N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176, 1410–1422 (2018).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Nguyen, L., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Finn, R. D., Clements, J. & Eddy, S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Wen, G. A simple process of RNA-sequence analyses by Hisat2, Htseq and DESeq2. In: Proceedings of the 2017 International Conference on Biomedical Engineering and Bioinformatics 11–15 (Association for Computing Machinery, New York, 2017)https://dl.acm.org/doi/10.1145/3143344.3143354.
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 9, 559 (2008).
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. & Ideker, T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011).
Analyst Software (Version 1.6.3) [Computer software]. Sciex, Retrieved from Reference Guide-1.6 Version.Sciex. https://sciex.com/products/software/analyst-software.
MultiQuant Software (2013 Version) [Computer software]. Sciex. Retrieved from https://sciex.com/products/software/multiquant-software.
Fraga, C. G., Clowers, B. H., Moore, R. G. & Zink, E. M. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Anal. Chem. 82, 4165–4173 (2010).
Liang, Y. W. The giant genome of lily provides insights into the hybridization of cultivated lilies. figshare. Dataset. https://doi.org/10.6084/m9.figshare.27933375 (2024).
Acknowledgements
This research is founded by Scientific and Technological Talents and Platform Program of Yunnan Province (Academician Expert Workstation) (202305AF150165 to L.Zhang), Xingdian Talent Support Project (CYRC2020004 to F.L.), “Pioneer” and “Leading Goose” R&D Program of Zhejiang (2023C02028 to L.Zhang), the National Natural Science Foundation of China (32272750 to L.Zhang), the Australian Research Council (FT210100366 to Z.C.), Horticulture Innovation Australia (LP18000 to Z.C.), Future Food Systems Corporate Research Centre (P2-016 and P2-018 to Z.C), the National Natural Science Foundation of China (32372740 to J.Wu). We thank for Dr. Manuel Spannagl from the Helmholtz Center Munich for advice on this manuscript. Y.Liang thanks for the support from the China Scholarship Council.
Author information
Authors and Affiliations
Contributions
L.Zhang designed the project. L.Zhang, J.Wang, M.K.F.X., and X.Zhou conceived the study. Y.Liang wrote the original manuscript. Y.Liang, M.K.F.X., Z.C., X.Zhou, and L.Zhang reviewed and edited the manuscript. Q.G., Y.Liang, and X.Zhou assembled and annotated the genomes. Y.Liang, Q.G., F.L., Y.D., J.Wu, X.S., Y.Li, D.L., and Z.W. analyzed the data. J.Wu, W.P, and S.W. verified gene functions. L.Zhong and F.Z. detected colchicine contents. X.Zhang, M.Z., Y.D., Q.D., S.L., C.J., P.Z., and Y.G. provided the plant materials.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yuepeng Han, Akira Kanno and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liang, Y., Gao, Q., Li, F. et al. The giant genome of lily provides insights into the hybridization of cultivated lilies. Nat Commun 16, 45 (2025). https://doi.org/10.1038/s41467-024-55545-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-55545-8
This article is cited by
-
Expression profiling and functional characterization of LoNCED gene in the pollen abortion of Lilium spp.
BMC Plant Biology (2025)
-
Biomod2 modeling for predicting the potential ecological distribution of Lilium Davidii var. Willmottiae (E. H. Wilson) Raffill
Scientific Reports (2025)
-
Genomic and epigenomic insight into giga-chromosome architecture and adaptive evolution of royal lily (Lilium regale)
Nature Communications (2025)