Abstract
Current assays fail to address breast cancer’s complex biology and accurately predict treatment response. On a retrospective cohort of 1082 female breast tissues, we develop and validate mFISHseq, which integrates multiplexed RNA fluorescent in situ hybridization with RNA-sequencing, guided by laser capture microdissection. This technique ensures tumor purity, unbiased whole transcriptome profiling, and explicitly quantifies intratumoral heterogeneity. Here we show mFISHseq has 93% accuracy compared to immunohistochemistry. Our consensus subtyping and risk groups mitigate single sample discordance, provide early and late prognostic information, and identify high risk patients with enriched immune signatures, which predict response to neoadjuvant immunotherapy in the multicenter, phase II, prospective I-SPY2 trial. We identify putative antibody-drug conjugate (ADC)-responsive patients, as evidenced by a 19-feature T-DM1 classifier, validated on I-SPY2. Deploying mFISHseq as a research-use only test on 48 patients demonstrates clinical feasibility, revealing insights into the efficacy of targeted therapies, like CDK4/6 inhibitors, immunotherapies, and ADCs.
Similar content being viewed by others
Introduction
Breast cancer (BCa) is a heterogeneous disease with distinct biology leading to differences in response to treatment modalities and clinical outcomes1. The discovery of molecularly distinct subgroups of BCa (luminal A (LumA), luminal B (LumB), HER2-overexpressing (HER2-OE), basal- and normal-like)2,3,4 has fundamentally changed our understanding of BCa biology and paved the way for a union between genomic and clinical classification of BCa subtypes. Various multigene signatures have emerged that provide important diagnostic, predictive, and prognostic insights to inform appropriate treatment5,6. However, assignment of subtypes/risk groups show only moderate reproducibility at the individual tumor level depending on the array platform, tumor composition, and gene list and associated thresholds7,8,9,10. The underlying clinical and molecular factors driving discordance remain obscure, fostering uncertainty about which multigene prognostic signature to use and whether combining signatures improves performance.
With a rapidly expanding therapeutic arsenal, existing BCa biomarkers fail to accurately predict novel treatment response. Neoadjuvant immunotherapy is the new standard of care for high clinical risk TNBC regardless of PD-L1 status, which failed to predict response, leaving only clinical risk to stratify patients11. Ongoing trials (KEYNOTE-765, CheckMate 7FL) aim to expand immunotherapies to hormone receptor positive (HR+)/HER2− patients in the neoadjuvant setting but patients are selected based on clinical risk and PD-L1 is only a secondary outcome measure. Identifying effective predictive biomarkers is critical in these early settings, especially in HR+/HER2− patients who have relatively immune cold tumors12.
Novel therapeutics such as antibody-drug conjugates (ADCs) are effective, but the optimal patient subgroups who benefit most from ADCs remain elusive. The pivotal trials for trastuzumab deruxtecan and sacituzumab govitecan demonstrated that responses cannot be explained by the mere presence of the ADC’s antigen target13,14,15, revealing efficacy in patients expressing even low/negative levels of their antigen targets when assessed using immunohistochemistry (IHC). This shows new tools are needed that go beyond the ADC target and consider other ADC-related markers such as the cytotoxin target and cellular pathways involved in ADC processing (e.g., endocytosis, lysosome).
A key limitation of current biomarkers is that they ignore spatial information. All currently used multigene assays are based on bulk processing/crude macrodissection, thus losing spatial context, which results in limited info about the tumor microenvironment (TME), and may introduce erroneous gene expression from non-tumor elements16. Indeed, tumor heterogeneity, whether it is histological, genetic, or proteomic, occurs both spatially and temporally and contributes to diagnostic inconsistencies and inappropriate treatment17. Laser capture microdissection (LCM) is a powerful tool that circumvents these limitations, ensuring unbiased interrogation of tumor heterogeneity, tumor purity, and scalability for clinical diagnostics.
Here, we address these issues by developing and validating mFISHseq, a diagnostic tool that uses a multiplexed RNA fluorescent in situ hybridization (RNA-FISH) panel consisting of estrogen (ESR1), progesterone (PGR), and HER2 (ERBB2) receptors and Ki67 (MKI67) to assess tumor heterogeneity and identify regions of interest for guiding LCM. This facilitates the collection of spatially resolved tumor-enriched samples from a single specimen that can be used for downstream total RNA sequencing (Fig. 1a). On a retrospective cohort (MDX-BRCA) of 1082 FFPE BCa samples with detailed clinicopathological information (Supplementary Data 1), we demonstrate that mFISHseq 1) resolves single sample discordance of multigene subtyping and prognostic classifiers by combining information from multiple classifiers, yielding improved relapse and survival prediction, a finding that was validated in two external datasets, METABRIC and TCGA cohorts; and 2) leverages transcriptome profiling of ADC processing-related genes and gene signatures to provide clinical insights into ADC treatment response. Furthermore, we demonstrate mFISHseq’s clinical feasibility in predicting the efficacy of diverse therapeutics, including chemotherapies and targeted therapies, in 48 patients that underwent a research-use only (RUO) version of the test (Supplementary Data 2).
a A retrospective cohort of 1,082 formalin-fixed paraffin-embedded (FFPE) breast cancer samples underwent multiplexed RNA-FISH-guided laser capture microdissection (LCM) coupled with RNA-sequencing. Annotation of the tumor on a hematoxylin and eosin (H&E) section and the biomarker expression derived from multiplexed RNA-FISH were used to select regions of interest (ROIs) for LCM from cresyl violet sections. These tumor-enriched samples were then sequenced to characterize gene expression signatures to provide diagnostic, prognostic, and predictive inferences from the cohort clinical data. DEGs, Differential expressed genes; IHC, immunohistochemistry. Created with BioRender.com. b Analytical validity of mFISHseq compared to immunohistochemistry assessed by receiver operating characteristic (ROC) curves in 1013 breast tumors stratified into 70:30 training (n = 701) and test (n = 312) datasets. AUC, area under the curve. The table shows biomarker thresholds defined in the training set by maximizing concordance (Cohen’s κ) between RNA-SEQ transcripts per million (TPM) expression values and immunohistochemistry results for each biomarker. These thresholds were then applied to the test set. Source data are provided as a Source Data file.
Results
mFISHseq identifies discordance between molecular subtyping classifiers
To assess the analytical validity of mFISHseq, we split our dataset into training and test cohorts (70:30) and constructed receiver operating characteristic (ROC) and precision-recall curves to compare mFISHseq to the known IHC results for PR, ER, HER2, and Ki67. Based on biomarker thresholds prespecified in the training set (Fig. 1b, table), we observed excellent concordance between mFISHseq and IHC with all biomarkers having ROC and precision-recall AUCs > 0.90, except for the precision-recall curves for ERBB2, which showed AUCs > 0.85 (Fig. 1b, Supplementary Fig. 1, Supplementary Table 1). This is consistent with prior reports comparing RNA-SEQ with IHC18,19 and demonstrates the analytical validity of mFISHseq in assessing IHC BCa biomarkers.
To classify BCa specimens into molecular subtypes, we identified 293 genes relevant for subtyping through differential gene expression (Supplementary Table 2, Supplementary Data 3, and Supplementary Methods). Semi-supervised consensus clustering of these 293 genes was benchmarked against IHC-surrogate, PAM50, and Absolute Intrinsic Molecular Subtyping (AIMS) classifications (Supplementary Fig. 2; Supplementary Fig. 3a shows the top 10 DE genes for each subtype comparison using the 293-gene mFISHseq list). While samples clustered into 5 groups representing the intrinsic subtypes, considerable discordance was observed between the four subtyping methods. Multigene subtyping methods classified fewer LumA samples and more HER2-OE and basal-like samples relative to the IHC surrogate subtypes (Supplementary Fig. 3b). While all subtyping methods showed similar overall survival curves (Supplementary Fig. 3c), there was only moderate single sample concordance between each multigene classifier and the IHC surrogate subtype (Supplementary Fig. 3d). In contrast to prior reports7,8, we found substantial concordance between multigene approaches, potentially due to the use of LCM.
Overall, 45% (459/1013) of IHC surrogate subtype samples showed discordance in ≥1 multigene classifier leading to clinically relevant differences in survival (Supplementary Fig. 3e). IHC surrogate LumA patients that had discordant results showed poorer survival than patients classified as LumA by all multigene classifiers (Supplementary Fig. 3e; top panel). In contrast, IHC surrogate LumB patients that had discordant results by two classifiers survived longer than patients classified as LumB by all multigene classifiers (Supplementary Fig. 3e; middle-top panel). While discordant HER2-OE patients showed comparable survival to concordant HER2-OE patients (Supplementary Fig. 3e; middle-bottom panel), discordant TNBC samples interestingly showed poorer survival than concordant TNBC samples (Supplementary Fig. 3e; bottom panel). Discordant samples, relative to concordant samples, showed evidence of instability as demonstrated by lower PAM50 centroid correlations (Supplementary Fig. 3f).
mFISHseq consensus subtyping mitigates discordance between molecular subtyping classifiers
To improve sample level agreement, we implemented a consensus intrinsic subtype by using a simple voting scheme for the three subtyping approaches (mFISHseq, PAM50, and AIMS) and found there were considerable differences between IHC surrogate and gene-expression based consensus subtypes that had prognostic implications. For example, 24% (102/432) of IHC surrogate LumA patients were reclassified as LumB, showing poorer overall survival for node negative patients relative to patients unanimously classified as LumA by all subtyping methods (Fig. 2a). The IHC surrogate LumB subtypes showed high discordance with 62% (194/313) disagreeing with one or more classifiers. Around 15% (46/313) of patients were reclassified as basal-like and 21% (65/313) as LumA by consensus subtyping and these patients had poorer and better survival, respectively, compared to patients unanimously classified as LumB (Fig. 2b). We observed the least discordance in the HER2+ IHC surrogate patients where 27% (20/74) displayed disagreement among classifiers, resulting in reclassification of 19% (14/74) of patients as basal-like, whom had equivalent survival compared to consensus HER2-OE patients (Fig. 2c). Disagreement among classifiers occurred in 29% (53/181) of TNBC IHC surrogate patients, resulting in reclassification of 4% (7/181) of patients as LumA/B and normal-like subtypes and 13% (23/181) of samples into HER2-OE subtype. HER2-OE reclassified patients, both node +/−, showed poorer survival when compared to consensus basal-like samples (Fig. 2d).
Overall survival (OS) according to consensus subtyping with respect to the IHC (immunohistochemistry) surrogate subtype according to IHC results, including Luminal A (a, n = 432), Luminal B (b, n = 313), HER2+ (c, n = 87), and TNBC (d, n = 181) stratified by nodal status (left and right panels depict node negative and positive, respectively). (e) Sankey diagram shows the IHC surrogate subtypes and the proportion (%) of samples reclassified by consensus subtyping. (f) Overall survival of consensus molecular subtypes (n = 1013). Survival curves were analyzed using the log-rank test to assess statistical significance. (g) Forest plots showing univariate and multivariate Cox proportional hazards models comparing prognostic utility of IHC surrogate vs Consensus molecular subtypes (n = 1013). Multivariate models included both tumor size (pT1 vs pT2-pT4) and node status (pN0 vs pN1-pN3). Hazard ratios show the overall survival estimates with 95% CIs, where the center of the interval represents the point estimate. P-values were obtained from the Wald test. Source data are provided as a Source Data file.
We ascertained discordance at the single sample level by exploring the 41% (415/1013) of samples uniquely classified into a specific molecular subtype by a single classifier. Each approach uniquely classified some patients as LumA that no other classifier agreed upon, and these patients showed poorer survival and higher genomic/clinical risk relative to patients classified as LumA by all tools (LumA All; Supplementary Fig. 4a, b). These misclassified samples, however, were rescued by taking the consensus of the three multigene classifiers with 83% (n = 91/109) being reclassified as LumB (Supplementary Fig. 4b, bottom right panel). In contrast, mFISHseq and PAM50 uniquely classified LumB patients showed better overall survival and enriched intermediate-/low-risk scores relative to samples classified as LumB by all tools (LumB All; Supplementary Fig. 4c, d). Consensus subtyping reclassified 97% (n = 69/71) of these patients as LumA thus assigning a subtype that better matched their survival and risk (Supplementary Fig. 4d, bottom right panel). Although samples uniquely classified as LumB by the IHC surrogate approach had similar survival to LumB All, consensus subtyping stratified these patients into LumA (n = 29)/normal-like (n = 6) subtypes and HER2-OE (n = 24)/basal-like (n = 46; Supplementary Fig. 4d, bottom right panel), which showed either more favorable or poorer survival, respectively (Supplementary Fig. 4c, inset).
Samples uniquely classified as HER2-OE showed disparate findings depending on the classifier (Supplementary Fig. 4e,f). Relative to HER2 All samples, IHC surrogate HER2+ patients had equivalent survival, high genomic/clinical risk, and were reclassified as basal-like by consensus subtyping. AIMS HER2-OE samples had poorer survival relative to HER2 All samples, high genomic/clinical risk, and most samples were reclassified as basal-like (n = 15/24) or LumB (n = 8/24). The mFISHseq and PAM50 HER2-OE patients had better survival and a subset of these patients (n = 9) with lower GENE70 and/or clinical risk experienced no deaths and were predominantly reclassified into LumA (n = 7/9).
TNBC/basal-like samples showed the least uniquely identified samples (n = 34). While IHC surrogate only TNBC patients showed poorer overall survival compared to those classified as TNBC/basal-like by all methods (Basal-like All), patients classified as basal-like by multigene classifiers had better prognosis (Supplementary Fig. 4g, h). IHC surrogate only TNBC samples encompassed 13 patients with low GENE70 or clinical risk and favorable survival, and 15 patients with high risk and poor survival (Supplementary Fig. 4g, inset, 4 h). Consensus subtyping stratified the samples into clinically meaningful subgroups with deaths occurring in 10/22 patients reclassified as HER2-OE/basal-like and only 1/5 patients reclassified as LumA/B (Supplementary Fig. 4f). AIMS and PAM50 independently classified normal-like patients were reclassified by consensus subtyping to basal-like (n = 18), LumA (n = 14), and HER2-OE (n = 3), providing more clinically relevant subtyping (Supplementary Fig. 5a, b).
Consensus subtyping reclassified 214 patients uniquely classified by IHC surrogate subtyping into intrinsic subtypes that better fit survival data. Similarly, patients that were uniquely classified by mFISHseq (n = 55), AIMS (n = 83), and PAM50 (n = 63) were reclassified by a consensus of the other two multigene classifiers, which yielded more reproducible single sample assignment to the appropriate intrinsic subtype that matched the survival and genomic/clinical risk.
Overall, 30% (305/1013) of samples were reclassified from their IHC surrogate subtype into a different consensus subtype (Fig. 2e) showing prognostic utility in stratifying the lower-risk LumA patients from other subtypes (Fig. 2f). This improved concordance between the consensus subtypes and other multigene classifiers to near perfect agreement, even for challenging luminal samples (Supplementary Fig. 3d). Moreover, consensus subtyping showed superior prognostic utility compared to IHC surrogate subtypes in both univariate and multivariate Cox models (Fig. 2g).
Molecular drivers of molecular subtyping discordance
While survival may be a suitable ground truth to indicate a better classification when comparing LumA patients to other more aggressive subtypes (and vice-versa), it is challenging to interpret for other subtype comparisons that have similar outcomes. Therefore, we used another ground truth metric to benchmark reclassified samples that included a panel of genes and gene signatures that are important for molecular subtyping (luminal, proliferation, HER2, and basal-like markers). Samples reclassified from IHC surrogate LumA to LumB showed increased expression of proliferation markers (MKI67, AURKA, PCNA, TOP2A, Module11_Prolif) compared to both ground truth LumA (consensus LumA) and LumB samples (consensus LumB without the reclassified LumB samples) but samples reclassified as LumB were statistically more like consensus LumB (Supplementary Fig. 6a). Samples reclassified as LumB also had reduced expression of luminal markers (PGR and the GSEA Estrogen Response Early signature) but only relative to consensus LumA (Supplementary Fig. 6b) and showed intermediate ERBB2 expression (Supplementary Fig. 6c). A similar finding was observed for samples reclassified from LumB to LumA, albeit with the opposite changes in gene expression (Supplementary Fig. 6d–f).
IHC surrogate LumB samples reclassified as HER2-OE had a molecular phenotype that resembled consensus HER-OE samples as demonstrated by elevated expression of HER2 amplicon genes and gene signatures (ERBB2, STARD3, GRB7, Supplementary Fig. 7a) and reduced expression of estrogen markers (XBP1, FOXA1, GATA3, Supplementary Fig. 7b). IHC surrogate LumB samples reclassified as TNBC/Basal-like had a molecular phenotype similar to consensus TNBC/Basal-like sample, which was driven by increased basal markers (KRT14, ID4, MYC) and immune/DNA damage repair/hypoxia signatures (Chemokine 12, Dendritic cells, and VC_pred) as well as reduced estrogen markers (Supplementary Fig. 7c, d).
While IHC surrogate HER2+ samples reclassified as TNBC/Basal-like did not show any changes in survival (Fig. 2c), their tumor biology was more revealing. Unexpectedly, these samples had a HER2 phenotype closer to consensus HER2-OE samples but elevated expression of TNBC/Basal-like markers (Supplementary Fig. 8a, b), which indicates an appropriate reclassification. Samples reclassified from IHC surrogate TNBC to consensus HER2-OE had intermediate expression of HER2 amplicon markers (ERBB2, Module7_ERBB2) and reduced expression of TNBC/Basal-like markers (Supplementary Fig. 8c, d). Relative to consensus HER2-OE, samples that were reclassified as TNBC/Basal-like and consensus TNBC/Basal-like had higher levels of ABC transporters (Supplementary Fig. 8b, d) suggesting TNBC/Basal-like have higher de novo resistance to treatments.
mFISHseq improves prognostic risk classification
We also investigated the performance and concordance of several multigene prognostic risk assays since they provide information on the effectiveness of adjuvant chemotherapy in ER+/HER2− patients with 0-3 positive lymph nodes. We compared clinical risk (assessed using MINDACT criteria), to research-based versions of OncotypeDX, the PAM50 Risk of Recurrence by Sample (ROR-S), GENE70 (i.e., MammaPrint), and the Genomic Grade Index (GGI). Relative to clinical risk, all multigene assays classified fewer patients as high risk (Supplementary Fig. 9). While all risk classifiers had comparable prognostic utility (Supplementary Fig. 9b–f), agreement between multigene classifiers and clinical risk was only fair and agreement among multigene classifiers was moderate to substantial (Supplementary Fig. 9g).
Only 39.2% of patients (n = 222/567) were unanimously classified by all five approaches as either high (n = 162) or low (n = 60) risk leaving 61.8% of patients (n = 345) with a discordant result in at least one classifier, mirroring the results of the OPTIMA Prelim Trial9. Within these discordant patients, another 27.5% of patients (n = 156/567) were categorized as either high (n = 52) or low (n = 104) by four classifiers and 23.5% of patients (n = 133/567) were categorized as either high (n = 33) or low (n = 100) by three classifiers. Thus, the five prognostic classifiers reached a majority (i.e., at least 3 classifiers in agreement) to stratify 43.6% (n = 247/567) and 47.1% (n = 267/567) of patients into high and low risk, respectively. Notably, patients differed markedly in terms of outcome depending on the number of concordant classifiers for a particular risk category (Fig. 3a). Patients with at least four classifiers in agreement for either high or low risk showed the poorest and best progression free survival (PFS) at 10 years (59.2% for 4/5 classifiers - 65.1% for 5/5 classifiers in agreement for high risk vs 89.0% for 4/5 classifiers - 88.2% for 5/5 classifiers in agreement for low risk; Fig. 3a). When separating patients based on whether a majority (≥3 classifiers) or minority (1-2 classifiers) of the classifiers predicted the same risk category, we observed intermediate PFS in the minority group compared to the majority as well as patients classified unanimously as high or low risk (Fig. 3b). Patients classified as high risk by the majority of classifiers had lower expression of PGR and higher MKI67 relative to patients classified as high risk by the minority of classifiers (Fig. 3c).
a Concordance of each classifier for high- and low-risk samples as illustrated by the number of concordant classifiers and (b) after consolidating into majority (agreement in ≥3 prognostic classifiers) and minority (agreement in 1-2 prognostic classifiers) categories. c Distribution of mRNA expression for estrogen receptor (ESR1, yellow, left panel), progesterone receptor (PGR, green, left/middle panel), HER2 receptor (ERBB2, red, right/middle panel), and Ki67 marker of proliferation (MKI67, orange, right panel) in patients that were classified as high risk by a particular number of concordant classifiers (i.e., 0-5 concordant classifiers). Statistical comparisons were performed using the Kruskal-Wallis test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Data are presented as scatter dot plots with the dotted line as the median. d Exploratory decision tree describing criteria for consensus classification of patients into high-, low-, and ultra-low-risk categories. e Kaplan Meier plots show progression free survival (PFS) and overall survival (OS) for each consensus prognostic risk category (high, low, and ultra-low). All analyses contain 567 patient samples that would be eligible for prognostic multigene tests in a real-world clinical setting (ER or PR+, HER2−, and 0-3 positive lymph nodes). Survival curves were analyzed using the log-rank test to assess statistical significance. Source data are provided as a Source Data file.
Each classifier independently categorized some patients into high, intermediate, and low risk while no other classifier agreed (Supplementary Fig. 10a) and this discordance remained after combining intermediate and high risk into a single risk category (Supplementary Fig. 10b, Supplementary Table 3). OncotypeDX and ROR-S had more intermediate-/high-risk uniquely classified samples, whereas GENE70 and GGI had more low-risk samples. Clinical risk uniquely classified samples in both risk categories. The number of uniquely classified samples influenced the frequency in which a prognostic classifier was present in the majority or minority (Supplementary Table 4). GENE70, which had the fewest uniquely classified samples (n = 12) appeared in the majority in 90% of classifications, while clinical risk (n = 57) and OncotypeDX (n = 48) had the most uniquely classified sample and consequently appeared in the majority only in 77% and 78% of classifications, respectively (Supplementary Fig. 10c). When stratified by intermediate/high vs low risk, Clinical risk, OncotypeDX, and ROR-S had higher proportions of majority intermediate-/high-risk classifications, suggesting these are the classifiers that drive intermediate-/high-risk classifications. GGI had a higher proportion of majority low-risk classifications and GENE70 had an equivalent proportion of majority intermediate/high and low classifications, suggesting that GGI and GENE70 primarily drive low risk. Uniquely classified intermediate-/high-risk patients had better PFS relative to patients classified as intermediate/high risk by all classifiers (Supplementary Fig. 10d, e). Patients independently categorized as low risk by GGI or GENE70 had poorer survival and patients uniquely classified as low risk by Clinical risk were similar relative to patients classified as low risk by all classifiers (Supplementary Fig. 10d, e).
Given the discordance observed at the single-patient level, we constructed consensus prognostic risk categories by combining the results for all five classifiers into 3-risk categories: high risk (If ≥3 prognostic classifiers agree on intermediate/high risk), low risk (If ≥3 prognostic classifiers agree on low risk), and ultra-low risk (If all 5 prognostic classifiers agree on low risk and the patient is node negative and PR+; Fig. 3d). Consensus high-risk patients (n = 300) showed poor outcomes with 49 relapses and 36 deaths within 5-years and another 24 relapses and 17 deaths from 5-10 years. Consensus low-risk patients (n = 214) had better outcomes with 13 relapses and 10 deaths within 5-years and another 10 relapses and 7 deaths from 5-10 years. Consensus ultra-low-risk patients (n = 53) showed the best outcomes with 1 relapse and 0 deaths within 5-years and another 2 relapses and 1 death from 5-10 years (Fig. 3e). When stratifying consensus risk groups by treatment (Supplementary Fig. 10f), we observed that high-risk patients benefited most from chemoendocrine therapy, while low/ultra-low patients did not benefit from chemoendocrine therapy, highlighting the clinical validity of our consensus prognostic risk categories in de-escalating overtreatment in low-/ultra-low-risk patients.
Clinical and molecular markers associated with discordant risk classification
To better understand the drivers of concordance/discordance among the four multigene prognostic signatures and clinical risk, we first compared their similarities. The multigene prognostic signatures had minimal overlap with no genes being present in all classifiers (Fig. 4a, Supplementary Table 5). GGI and ROR-S shared the most genes (n = 14), followed by OncotypeDX and ROR-S (n = 11), GENE70 and GGI (n = 8), OncotypeDX and GGI (n = 5), GENE70 and ROR-S (n = 3), and OncotypeDX and GENE70 with the least (n = 1, SCUBE2), which partially explained the concordance between signatures (Supplementary Fig. 9g). ROR-S shared the most genes with all three other classifiers (n = 28) followed by GGI (n = 27), OncotypeDX (n = 17), and GENE70 (n = 12). Several proliferation-related genes (BIRC5, CCNB1, KNTC2, MELK, MKI67, MYBL2) occurred in three of the gene lists, highlighting that proliferation and cell-cycle related genes form a core component of these multigene prognostic signatures.
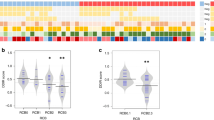
a Venn diagram depicting the number of overlapping genes among each of the four multigene prognostic classifiers. b Dot plot of clinical parameters significantly associated with high and low risk. Statistical analysis was performed using a Chi-square test. Stacked bar graphs illustrate the proportion of selected clinical parameters associated with discordance in either high or low risk, including tumor size (n = 575) (c), node status (n = 575) (d), tumor grade (n = 573) (e), histological subtypes (n = 509) (f), clinical risk as described in the MINDACT trial (n = 568) (g), and treatment with chemotherapy (n = 556) (h). The percentages at the top of each bar denote the proportion of patients with pT2-pT3 tumors (c), node positive status (d), grade 1 (G1) tumors (e), invasive lobular carcinoma (f), low clinical risk (g), and adjuvant chemotherapy treatment (h). Missing or ambiguous clinical data resulted in some clinical parameters having <575 patients. The heatmaps (i) show the significant genes/gene signatures associated with discordance in 1 or 2 prognostic classifiers relative to patients with unanimous agreement (group labeled as 0) for either high (left heatmap) or low (right heatmap) risk. The legends refer to the row metadata for gene/gene signature group, −log10 (adjusted p-value) for all significant Kruskal-Wallis tests (adjusted p-value < 0.05), results from Dunn’s multiple comparison tests for each pairwise comparison (gray box, adjusted p-value < 0.05; white box, adjusted p-value > 0.05), and the z-score normalized expression. Note that the data in the high- and low-risk heatmaps are the same as presented in Supplementary Fig. 11, but the labels for low risk have been changed from 5, 4, and 3 to 0, 1, and 2. Source data are provided as a Source Data file.
We observed several clinical parameters that were associated with discordance between high and low risk, including tumor grade, clinical risk (MINDACT criteria and AJCC stage), chemotherapy, tumor size, and node status (Fig. 4b). When comparing discordance within a single risk category (i.e., high risk with all classifiers in agreement or only 1 or 2 in disagreement), we also observed changes in the same clinical parameters in patients with one or two discordant classifiers compared with those with unanimous concordance. High-risk patients that had 1 or 2 discordant classifiers, compared to those with unanimous concordance for high risk, had greater proportions of T1 tumors, node negative cases, grade 1-2 tumors, low-risk scores, and patients treated less frequently with adjuvant chemotherapy (Fig. 4c–h). Low-risk patients that had 1 or 2 discordant classifiers, compared to those with unanimous concordance for low risk, displayed the opposite pattern, with greater proportions of T2-3 tumors, node positive cases, grade 2-3 tumors, high-risk scores, and patients treated more frequently with adjuvant chemotherapy (Fig. 4c–h). Interestingly, both high- and low-risk patients that had discordance showed enrichment in the proportion of invasive lobular carcinomas (Fig. 4f), suggesting histology is a factor that drives discordance.
Next, we interrogated a list of 92 genes and 110 gene signatures that span cancer hallmark pathways and treatment response (Supplementary Data 4 shows a list of these signatures categorized into groups) to ascertain the tumor biology of high- and low-risk discordance. When comparing samples that were classified as high or low risk by the consensus of all five prognostic classifiers (i.e., those where the majority (≥3) of classifiers agreed), we found 121 genes/gene signatures significantly different between risk groups, when high risk was used as the reference group for discordance (Supplementary Fig. 11). Expectedly, these were largely dominated by proliferation (FOXM1, AURKA, MKI67, CCNB1, Module11_Prolif), cell cycle (RB/PTEN loss, E2F4), DNA damage repair (RAD51, PARP1, TYMS, HALLMARK_DNA_REPAIR), luminal/estrogen pathways (PGR, Correlation_to_centroid_PAM50_LumA), and multigene prognostic signature scores. Many signatures involved in immune function (STAT1, Mast cells, Module3_IFN), metabolism/angiogenesis/hypoxia (Metabolic_sig_Trop2_TNBC_Survival), and ADC mechanisms of action were also altered between high- and low-risk patients, an interesting finding considering many multigene prognostic signatures do not capture the biology of these pathways. Regarding ADCs, these spanned antigen targets for ADCs (F3, ROR1/2, TPBG, FOLH1, ERBB2, TACSTD2), endocytosis (CAV1, Endocytic_uncoat), and lysosome function (LAMP2, CTSB). As the number of discordant classifiers increased in reference to high risk (Supplementary Fig. 11), the expression patterns of genes and gene signatures steadily increased (or decreased), highlighting the continuum of gene expression that separates high- and low-risk patients.
When comparing discordance within a single risk category, where either all multigene prognostic classifiers were unanimous or had 1 or 2 discordant classifiers (but retained the assigned consensus risk category), the significantly altered genes/gene signatures markedly differed for discordant high- and low-risk samples (Fig. 4i). Discordant low-risk patients were distinguished by 23 genes/gene signatures composed of proliferation, cell cycle, and estrogen response markers (Fig. 4i, right heatmap) In contrast, discordant high-risk patients differed in 70 genes/gene signatures that spanned the same pathways observed in discordant low-risk samples as well as pathways such as DNA damage repair and PARP sensitivity, metabolism/hypoxia/angiogenesis, and immune activation (Fig. 4i, left heatmap), suggesting high-risk tumor biology is more diverse than low risk.
Cellular states associated with risk discordance and immunotherapy response
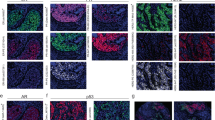
The differences in immune markers led us to use EcoTyper/CIBERSORTx digital cytometry20,21 to elucidate the abundances of 11 cell types stratified into 71 cell states across high- and low-risk patients. High- versus low-risk patients showed marked differences in cellular composition with 50 out of 71 cell states reaching statistical significance (Supplementary Fig. 12a). This revealed unique patterns of immune states with high-risk patients displaying markers of active adaptive and innate immune response, while low-risk patients were enriched in immunoregulatory and resting (normal) immune cells. Interestingly, when investigating cell type and state abundances in the context of discordance within a single risk category, discordant high-risk samples had significantly altered TMEs relative to discordant low-risk samples, with the former and latter differing in 39% (28/71) versus 11% (8/71) of cell states, respectively (Fig. 5a, b). High-risk patients, relative to low risk, had active immune TMEs comprised of exhausted/effector memory CD8/CD4 T cells, mature immunogenic dendritic cells, and M2-like proliferative macrophages (Fig. 5c–f, top panels) as well as reduced composition of resting or normal immune cells (Supplementary Fig. 12b–e, top panels). Non-immune cells like epithelial, endothelial, and fibroblasts were also differentially expressed in high- and low-risk patients, with high-risk patients having enriched pro-inflammatory epithelial cells and pro-migratory-like fibroblasts (Fig. 5f–i, top panels). Interestingly, high-risk patients with 1 or 2 discordant multigene prognostic signatures showed reduced active immune responses (Fig. 5c–f, bottom panels), increased resting/normal immune cells (Supplementary Fig. 12b–e bottom panels), and epithelial/endothelial/fibroblast cells (Fig. 5g-h, bottom panels; Supplementary Fig. 12f–i, bottom panels) that resembled the immune cold TMEs of low-risk patients. Altogether, high- and low-risk patients (and patients with discordance in high- and low-risk assignment) have unique immune ecosystems, which could be a source of discordance since multigene prognostic signatures have minimal overlap on immune genes.
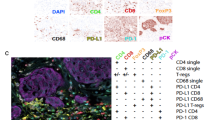
The heatmaps show the significantly different cell types/states in patients that have one or two (groups 1-2) prognostic classifiers that are discordant with patients that are unanimously classified (group 0) as either high (a) or low (b) risk as the reference group. The legends refer to the row metadata for group (cell type), −log10 (FDR) with red values denoting significant results from a Kruskal-Wallis test (FDR < 0.05), results from Dunn’s multiple comparison tests for each pairwise comparison (gray box, FDR < 0.05; white box, FDR > 0.05; note that post hoc comparisons are depicted even if the Kruskal-Wallis test was not significant), and the cube root transformed cell type/state abundances. The data in the high (a) and low (b) risk heatmaps are the same as presented in Supplementary Fig. 12, but the labels for low risk have been changed from 5, 4, and 3 to 0, 1, and 2. Box and whisker plots illustrate the exemplary cell types/states that are differentially expressed when comparing high- and low-risk samples (top panel graphs) as well as patients that have 1 or 2 discordant classifiers for high risk relative to those who have unanimous agreement (bottom panels). Cell types/states include CD8 T cells – Exhausted/effector memory (SO3) (c), CD4 T cells – Exhausted / effector memory / Treg (SO1) (d), Dendritic cells – Mature immunogenic (S03) (e), Monocytes/Macrophages – M2-like proliferative (S07) (f), Epithelial cells – Pro-inflammatory (SO4) (g), Fibroblasts – Pro-migratory-like (SO8) (h). Statistical comparisons are only shown for bottom panels and were performed using the Kruskal-Wallis test (n = 575). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Boxes denote the interquartile range with the line plotted as the median, whiskers show the 5 and 95 percentiles, and dots are individual samples outside this range. (i) Bar graphs show the percentage of patients (n = 40) who achieved pathological complete response following neoadjuvant treatment with paclitaxel combined with pembrolizumab in the I-SPY2 trial stratified by biomarker subgroups. Source data are provided as a Source Data file.
Given the promising results of neoadjuvant immunotherapy in hormone receptor positive breast cancer as shown in the prospective KEYNOTE-756 and I-SPY2 trials, we sought to determine whether these cell state signatures that are enriched in high-risk patients could serve as biomarkers for patient stratification. To test this hypothesis, we utilized the publicly available clinical and microarray data from the 40 HR+/HER2− patients in the paclitaxel combined with pembrolizumab (PEMBRO) arm of the multicenter, phase 2 prospective I-SPY2 trial, where 30% (12/40) of patients achieved the primary endpoint of pathological complete response (pCR). Using the online EcoTyper/CIBERSORTx tool, we deconvolved bulk microarray gene expression data into cell type/state signatures and dichotomized them into high and low subgroups based on median expression in the PEMBRO arm. Strikingly, PEMBRO arm patients with high cutoff signatures for Exhausted/effector memory CD8 T cells, mature immunogenic dendritic cells, and classical M1 monocytes/macrophages all experienced a higher proportion of pCRs (55%, n = 11/20) relative to those with low signatures (5%, n = 1/20), an absolute pCR gain of 25% compared to all PEMBRO arm patients (Fig. 5i). Combining all three signatures yielded a subgroup with 65% (n = 11/17) of patients achieving pCR (Fig. 5i). These cell type/state signatures were among the top 20 differentially expressed signatures between high- and low-risk patients (Supplementary Fig. 12a, b) and the top 10 signatures between high-risk discordant samples (Fig. 5a), highlighting the notion that high consensus prognostic risk patients may be ideal candidates for neoadjuvant immunotherapy. Indeed, these signatures performed similarly to the MammaPrint High 2 (55%, n = 6/11 patients with pCR) and response-predictive subtyping-5 (RPS-5, HER2−/Immune+; 69%, n = 11/16 patients with pCR) biomarker groups characterized in I-SPY2 (Fig. 5i), while covering a slightly larger eligible patient population.
Validation of consensus subtyping on METABRIC and TCGA cohorts
We used both The Cancer Genome Atlas breast cancer (TCGA-BRCA)22,23 and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC)24,25 cohorts to externally validate our consensus molecular subtyping and prognostic risk. For benchmarking subtype classifications, we obtained ground truth PAM50 classifications from the flagship METABRIC paper24 and Perou and colleagues’ molecular analysis of TCGA breast cancer histologic types26 (see Supplementary Methods). Individual classifiers showed more variability in the proportions of luminal subtypes with AIMS classifying the least number of luminal subtypes in all datasets (Supplementary Fig. 13a, 14a). Normal-like classifications were 1.5-3x higher in METABRIC and TCGA datasets, relative to our retrospective MDX-BRCA cohort that underwent LCM, suggesting the greater presence of non-tumor elements, which influence the proportion of normal-like subtype calls. There were also 82 METABRIC and 24 TCGA samples with indeterminate subtypes (i.e., each molecular classifier assigned a different subtype), considerably higher than the 8 indeterminate samples found in our MDX-BRCA cohort.
While all subtype classifiers showed prognostic utility in METABRIC (Supplementary Fig. 13b), only mFISHseq and PAM50 ground truth did in TCGA (Supplementary Fig. 14b. Concordance between IHC surrogate and molecular subtypes was only moderate in both datasets; while the concordance between molecular classifiers was moderate/substantial (Supplementary Figs. 13c, 14c), having slightly lower concordance than our MDX-BRCA cohort (Supplementary Figs. 13c, 14c), supporting our observation that LCM may improve concordance.
In METABRIC and TCGA, 57% (1109/1929) and 52% (490/951) of samples showed discordance in ≥1 classifier, respectively, and similar to our retrospective findings, some discordant samples had altered survival, especially in the case of LumB (Supplementary Figs. 13d, 14d). As the number of discordant samples for a given subtype increased, the correlation with the PAM50 centroid decreased suggesting changes in gene expression underlie the discordance (Supplementary Figs. 13e, 14e). Inspection of samples that were uniquely classified by a single approach revealed altered survival, especially in LumA patients who had poorer outcomes as well as changes in biomarker expression (Supplementary Figs. 15–18), relative to those unanimously classified as LumA. For example, uniquely classified LumA samples had higher MKI67 and lower PGR expression relative to samples classified as LumA by all multigene classifiers. Conversely, uniquely classified LumB samples had lower MKI67 and higher PGR expression relative to samples classified as LumB by all multigene classifiers.
Like our MDX-BRCA cohort, taking the consensus subtype call from the three multigene classifiers mitigated the discordance, ultimately assigning 27% of METABRIC and 32% of TCGA patients to subtypes that better fit their biology and outcome (Supplementary Figs. 19 and 20). In the case of METABRIC, these consensus reclassifications had clinically relevant differences in outcome when compared with their original IHC surrogate classifications, with Consensus LumA and Consensus LumB having better and poorer overall survival, respectively (Supplementary Figs. 19a, b). While the differences in clinical outcome were less evident in the TCGA cohort, in both TCGA and METABRIC, consensus subtyping had better prognostic utility in both univariate and multivariate Cox models when compared with IHC surrogates (Supplementary Figs. 19g and 20g). Our investigation of the correctness of the reclassified samples using a panel of genes and gene signatures that are important for molecular subtyping revealed nearly identical comparisons in TCGA, METABRIC, and MDX-BRCA cohorts (Supplementary Figs. 21 and 22). Samples reclassified by consensus subtyping displayed tumor markers that closely aligned with their new subtype rather than the original IHC surrogate or discordant multigene classification.
Validation of consensus risk groups on METABRIC and TCGA cohorts
Compared to our MDX-BRCA cohort, in the METABRIC and TCGA cohorts, each prognostic classifier assigned risk groups in similar patterns (e.g., GENE70/GGI had the most low-risk patients and Clinical risk/OncotypeDX had the most high-risk patients, Supplementary Figs. 23a and 24a) and fewer classifiers showed prognostic utility (Supplementary Figs. 23b–f and 24b–f). Concordance among prognostic classifiers was also dramatically lower than our MDX-BRCA cohort, both between each multigene classifier and clinical risk and among multigene classifiers (Supplementary Figs. 23g and 24g). Only 22.8% (n = 188 high risk, n = 11 low risk/872 eligible patients) of METABRIC and 26.2% (n = 97 high risk, n = 25 low risk/465 eligible patients) of TCGA patients were unanimously assigned to the same risk group by all five classifiers, highlighting the substantial single sample discordance. As the number of discordant classifiers increased for either high or low risk, patients differed markedly in terms of outcome (Supplementary Fig. 25a,b). Stratification based on whether a majority (≥3 classifiers) or minority (1-2 classifiers) of the classifiers predicted the same risk category revealed similar differences in outcome, with the largest differences observed in the 5–10-year range (Supplementary Fig. 25c, d).
We further explored single sample discordance by investigating the outcomes of patients classified into a risk group by a single classifier, which no other classifier agreed upon. All three cohorts (MDX, METABRIC, and TCGA) showed similar patterns of uniquely classified samples with clinical risk and OncotypeDX having the most unique high-risk samples and clinical risk and GGI having the most unique low-risk samples (Supplementary Figs. 26a, b and 27a, b). These classifiers were less frequently present in the majority consensus risk category than ROR-S and GENE70 (Supplementary Figs. 26c and 27c). Uniquely classified samples also showed clinically relevant differences in PFS when benchmarked against samples classified into a particular risk group by all classifiers: uniquely classified high-risk samples had better PFS whereas uniquely classified low-risk samples had poorer PFS relative to unanimously classified samples (Supplementary Figs. 26d, e and 28d, e). Similar to our MDX cohort, taking the consensus of the five risk classifiers yielded high- and low-risk groups that had differences in PFS/RFS past 10 years in both METABRIC and TCGA and in OS past 20 years in METABRIC. The ultralow-risk group, although limited in patients due to the low concordance for low risk, displayed excellent PFS/RFS, with only a single relapse across both cohorts Supplementary Fig. 25e, f). Overall, these results highlight specific biases for each prognostic classifier that spanned over three independent cohorts, thus underlining potential misclassifications that may arise by using a single risk classification scheme and providing a useful consensus approach to mitigate these limitations.
Laser capture microdissection (LCM) enables tumor-specific gene expression and accurate multigene profiling
Breast tumors display considerable intra-tumoral heterogeneity spanning histological, morphological/cellular, genetic, and molecular features, which has important implications for patient diagnosis, treatment, and prognosis. Particularly important for multigene signatures is the tissue and cellular composition of the tumor because specimens with low tumor content (or tumor cellularity) can lead to spurious gene expression results and may be an important source of discordance. We found approximately 12.2% of tissues (n = 132/1082) contained either histological (n = 94/1082 = 8.7% for mixed histology, which includes mixed histological subtypes (e.g., invasive ductal carcinoma mixed with mucinous carcinoma) and mixed invasive-DCIS/LCIS) or biomarker heterogeneity (n = 16/1082 = 1.5% for all or none spatial expression and n = 22/1082 = 2.0% high/low spatial expression; see Methods). This intratumoral heterogeneity is likely underestimated considering we are only assaying single tissue sections for H&E and RNA-FISH that come from a single FFPE block. To investigate the effects of tumor content and heterogeneity on gene expression, we selected a panel of 41 samples with low (<10%), intermediate (≥10 to <30%), and high (≥30%) tumor content that were representative of each molecular subtype and compared the transcriptome profiles of LCM with adjacent sections that did not undergo LCM (Fig. 6a). LCM samples showed enrichment for each marker (PGR, ESR1, ERBB2, and MKI67), but only for samples that were classified as IHC-positive (Fig. 6b). IHC-negative genes showed reduced expression when comparing LCM samples to matched undissected sections, except for ERBB2, which was either unaltered or enriched (Fig. 6c), an observation maybe related to ERBB2/Her2-low (see27,28,29). LCM, compared to no LCM, resulted in a broader dynamic range for all markers (Fig. 6d), presumably because more sequencing reads are distributed to transcripts derived from cancerous tissues rather than normal, healthy epithelial, connective, and adipose tissues. In support of this, LCM enriched Cadherin-1 (CDH1) expression, a cell-type marker of breast glandular epithelial cells (i.e., tumor cell marker) and reduced the expression of Vimentin (VIM) and Platelet and Endothelial Cell Adhesion Molecule 1 (PECAM1), markers for fibroblasts and endothelial cells, respectively (Fig. 6e).
a Photomicrographs depict examples of hematoxylin and eosin-stained resected tumor specimens with low, intermediate, or high tumor content represented in shaded annotations. Scale bars represent 2 mm length. Bar plots with individual data points show change in gene expression of PGR, ESR1, ERBB2, and MKI67 in specimens that were classified as IHC (immunohistochemistry) positive (b) or IHC negative (c). Error bars represent mean ± SEM. d Dot plots show the dynamic range of gene expression for each biomarker in LCM vs no LCM matched samples. Dotted lines represent the median. e Bar plots with individual data points showing expression of cell-type specific markers in LCM vs no LCM samples containing either low, intermediate, or high tumor content. Error bars represent mean ± SEM. Sankey diagrams illustrate change in mFISHseq consensus subtypes (f) and PAM50 risk of recurrence by subtype (ROR-S) classification (g) for LCM vs no LCM samples. All analyses were performed with a sample size of n = 41, except for panel (f), which was performed with n = 40 due to an indeterminate consensus subtype classification. Source data are provided as a Source Data file.
We further explored the impact of LCM on molecular subtyping and prognostic risk classifiers as many of these procedures rely on bulk processing or tumor content thresholds to ensure reliable results. Molecular subtyping was particularly susceptible to the presence of non-tumor tissue with 61% (25/41) of AIMS and 32% (13/41) of both mFISHseq and PAM50 samples switching molecular subtype when comparing paired LCM and no LCM samples. This also resulted in 41% (17/41) samples switching their consensus subtype (Fig. 6f). Expectedly, the most common consensus subtype change was to the normal-like subtype (15/41) and then two samples changed from LumB to LumA and one sample from LumB to HER2 (Fig. 6f). Bulk processing also influenced the prognostic classifiers to a lesser extent with 41% (17/41) of ROR-S, 22% (9/41) of GGI, 15% (6/41) of GENE70, and 10% (4/41) of OncotypeDX samples switching prognostic risk groups. All samples that switched prognostic risk groups were classified as a lower-risk group (e.g., high to intermediate or high to low), which could have profound implications for treatment since low-risk individuals may forego receiving potentially beneficial chemotherapy. For ROR-S, 12 patients (6 high and 6 intermediate risk by LCM) switched to the low prognostic risk group (Fig. 6g) and these individuals would be incorrectly recommended to not receive chemotherapy. Overall, this highlights the importance of LCM in enriching tumor specific gene expression to provide accurate assessment of the four main breast cancer biomarkers and classification by multigene signatures. Methodologies that fail to adequately eliminate non-tumor elements may lead to erroneous gene expression, misclassification of patients, and inappropriate treatment regimens.
Single-cell enrichment and specificity of multigene expression classifiers
Our observation that sample classification is dependent on the amount of tumor content led us to determine if particular gene expression assays may be more susceptible to non-tumor tissue due to non-specific gene expression. We analyzed two single-cell transcriptomic datasets: a healthy breast tissue dataset30 from the Human Protein Atlas (HPA)31,32 and a breast cancer dataset33 from the Broad Institute’s Single Cell Portal34. A comprehensive overview of single cell expression data from both healthy and cancer single cell atlases is shown for each classifier in Supplementary Figs. 28–33.
When comparing molecular subtyping classifiers (Fig. 7a–c), mFISHseq contained the highest proportion of Cell Enhanced genes (65%) in breast glandular epithelial cells followed by PAM50 (50%) and AIMS (43%). AIMS contained the highest proportion of Cell Enhanced genes in non-breast glandular cells (24%), including adipocytes (6%), mesenchymal cells (7%), and blood and immune cells (8%), whereas mFISHseq contained the least proportion of Cell Enhanced genes in non-breast glandular cells (12%), including adipocytes (<1%), mesenchymal cells (4%), and blood and immune cells (6%). PAM50 had low proportions of Cell Enhanced genes in non-breast glandular cells (14%), which was dominated by mesenchymal cells (2%), and blood and immune cells (12%). Group Enriched and Group Enhanced genes displayed similar patterns of expression among the three classifiers. PAM50 contained the highest proportion of Low specificity genes (20%) followed by AIMS (14%) and mFISHseq (8%). Notably, seven (out of 17) of the low specificity PAM50 genes comprise the subset of 18 genes used to calculate the proliferation score used in the ROR algorithm, including CCNE1, CDC20, CDC6, CENPF, ORC6, TYMS, and UBET2. Many of these genes showed the highest expression in immune cells in healthy breast tissue (Supplementary Fig. 30), highlighting how non-tumor elements may influence subtype and risk classifications. These results may also explain why AIMS, which had the highest proportion of enhanced/enriched genes in non-glandular cell types (e.g., adipose and stromal cells) showed the highest susceptibility to non-tumor tissue when comparing samples that did or did not undergo LCM (Fig. 6f).
Stacked bar graphs show the number of genes classified as Cell Enriched, Cell Enhanced, Cell Group Enriched, Cell Group Enhanced, Low Specificity, and Not detected using slightly modified criteria from The Human Protein Atlas (see Supplementary Methods). The distribution and specificity overview of the metadata from Supplementary Figs. 28–33 is provided for each classifier, including (a) mFISHseq, (b) AIMS, (c) PAM50 and PAM50 ROR-S, (d) OncotypeDX, (e) GENE70, and (f) GGI. Bars are colored based on the major cell type defined by The Human Protein Atlas portal. Source data are provided as a Source Data file.
Compared to molecular subtyping, the prognostic risk classifiers showed lower proportions of gene enrichment/enhancement in breast glandular epithelial cells, greater enrichment/enhancement in immune cells, and more low specificity genes (Fig. 7d–f). OncotypeDX contained the highest proportion of Cell Enhanced genes (48%) in breast glandular epithelial cells followed by GENE70 (24%) and GGI (16%). GENE70 contained the highest proportion of Cell Enhanced genes in non-breast glandular cells (32%), including blood and immune cells (16%), endothelial cells (6%), and mesenchymal cells (6%), whereas OncotypeDX contained the least proportion of Cell Enhanced genes in non-breast glandular cells (14%) with all being enhanced in blood and immune cells. GGI also contained a high proportion of Cell Enhanced genes in non-breast glandular cells (31%) with most enhanced in blood and immune cells (23%). Strikingly, over half of the genes in GGI were Low specificity (51%), which is likely due to this signature being developed by a differential gene expression analysis comparing low versus high histological grade tumors apparently without respect to tumor content/cellularity. Thus, many non-tumor related genes would be represented in this analysis. GENE70 and OncotypeDX had 44% and 38% of Low specificity genes, respectively, overall revealing considerable susceptibility to non-tumor elements in determining risk scores. Out of all classifiers, mFISHseq had the most genes that were Not detected (14%), which could be due to the differences in RNAseq and scRNAseq library preparation and whether skin is present in the breast sample, as some of these genes are expressed in bulk RNAseq datasets from the HPA in either breast or skin tissue (e.g., AKR1B15, A2ML1, CASP14, GREP1, IGHV1-69D, IVL, KRT83, KIAA0319, SULT1C3).
When comparing cancer to healthy tissue, most genes were elevated in cancer epithelial cells, however, some genes showed higher expression in healthy tissue. BAG1, an important regulator of the oncogene BCL2, is a shared gene in OncotypeDX and PAM50, one of the most highly expressed in both multigene panels, showing the highest expression in normal breast epithelial cells (Supplementary Figs. 30, 31). In PAM50 and AIMS, the basal cytokeratins (KRT5, 14, and 17) that are commonly associated with the basal-like molecular subtype all showed higher expression and in a higher proportion of normal cells relative to cancer cells (Supplementary Figs. 29, 30). Although there are more examples, the last to highlight is GRB7, an important gene that is often co-amplified in HER2+ breast cancers as part of the HER2 amplicon. This gene, which is included in mFISHseq, AIMS, PAM50, and OncotypeDX, had slightly higher expression in normal vs cancer epithelial cells (Supplementary Figs. 28-31). Taken together, many genes show substantial expression in normal tissue cells (epithelial, immune, etc.) and could have profound influences on the final subtype or risk group call depending on tumor content/cellularity and the methodology used to enrich (e.g., LCM vs macrodissection) or not enrich for exclusively tumor cells.
ADC markers and association with survival
Intrigued by the finding that ADC-related markers showed altered expression in high- and low-risk patients, we curated a list of 70 genes and 32 gene signatures related to endocytosis, lysosomal function, payload targets, and resistance pathways (Supplementary Data 4) and sought to characterize their expression across BCa subtypes and their association with survival. These ADC processing genes/gene signatures displayed marked variability and semi-supervised consensus clustering showed stratification along molecular subtypes (Supplementary Fig. 34). Indeed, gene expression analysis of 20 ADC targets (FDA/EMA-approved or undergoing clinical trials) revealed high variability, differences between healthy and invasive tumor, and subtype-specific enrichment (Supplementary Figs. 35). Many ADC targets were associated with survival and in some instances associations with survival depended on subtype (Supplementary Fig. 36a, b), which could be prognostic in ADC treated populations. For example, TACSTD2 (TROP2), targeted by sacituzumab govitecan (and datopotamab deruxtecan), was associated with favorable survival in LumA but poor survival in TNBC; ERBB2, ERBB3, and TPBG predicted favorable survival in LumB but poor survival in TNBC.
Development of a classifier for sensitivity to T-DM1
Because the sensitivity and resistance to ADCs likely encompasses broad cellular targets/pathways involved in ADC processing, we posited that combining markers may elucidate ADC-responsive subgroups that can be exploited for effective patient selection. Given that our retrospective cohort did not include ADC-treated patients, we re-analyzed the trastuzumab emtansine (T-DM1) arm and control arms of the multicenter, adaptive-randomized, phase 2 prospective I-SPY2 (Investigation of Serial studies to Predict Your Therapeutic Response with Imaging and Molecular AnaLysis 2, NCT01042379) clinical trial to determine whether combinations of ADC-processing markers would predict response to T-DM1. The T-DM1 arm contains 52 HER2+ patients (35 ER+ and 17 ER− treated with T-DM1 in the neoadjuvant setting with pathological complete response (pCR) as the primary endpoint (see Methods for control arm details). Univariate logistic regression on 71 pre-specified ADC-relevant markers revealed associations with pCR (Fig. 8a). Elastic net multivariate logistic regression (10-fold cross validation) yielded a 19-feature classifier (Fig. 8b) that displayed superior predictive utility than ERBB2 mRNA alone (ROC AUC of 0.99 vs 0.87, Fig. 8c). Importantly, the T-DM1 predictor only had moderate/low predictive utility in the trastuzumab/pertuzumab and chemotherapy control arms (ROC AUC of 0.78 and 0.62; Fig. 8d), underlining its specificity to T-DM1 rather than anti-HER2 targeted therapies. The dominant features in the signature predicting T-DM1 efficacy were related to the antigen, endocytosis (FLOT1/FLOT2 ratio, RAB5A), lysosome function (GLB1, HTT), maytansine payload (TUBA1B), and resistance markers (e.g., the multidrug resistance transporter, ABCC3; Fig. 8e). By demonstrating that ADC-processing markers can be integrated into predictive models for effective patient selection, this evidence provides a foundation for future prospective clinical validation.
a Univariate logistic regression analysis of 71 prespecified ADC-related genes/gene signatures and their association with pathologic complete response (pCR) in the T-DM1 arm of the I-SPY2 trial (n = 52). Significance was assessed using the likelihood ratio test. b The 19 genes/gene signatures selected in the multivariate logistic regression with elastic net modeling using 10-fold cross validation and their association with pCR. Green bars denote signatures associated with pCR; red bars indicate signatures associated with no pCR; black bars depict signatures not associated with either pCR or no pCR. c Receiver operating characteristic (ROC) curves showing performance of two T-DM1_pred classifiers in the test set relative to ERBB2 mRNA alone in the T-DM1 arm (n = 52). Univariate T-DM1_pred is a single score derived from all 19 features, while multivariate T-DM1_pred includes all 19 features in a multivariate regression model. d ROC curves showing performance of the T-DM1_pred classifier in the test set relative to ERBB2 mRNA alone in both the pertuzumab (n = 44) and taxane/anthracycline (n = 31) control arms. AUC, area under the curve. e Scatter plots showing the distribution of selected genes/gene signature scores of the T-DM1_pred classifier in patient samples (n = 52) according to pCR. Red lines denote the median. Statistical comparisons were performed using the Mann-Whitney test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Source data are provided as a Source Data file.
Real-world implementation of mFISHseq
To demonstrate the feasibility of implementing mFISHseq in a real-world setting, we conducted an RUO version of the test on 48 patients, which included assigning consensus subtyping/prognostic risk groups and assessing 40 genes and 28 gene signatures spanning cancer pathways relevant for treatment and prognosis (Fig. 9a). These patients comprised all clinical settings and molecular subtypes (Fig. 9b). The most frequently recommended therapies were novel, targeted therapies such as ADCs, PARP inhibitors, and immunotherapies (Fig. 9c and Supplementary Methods for details on how therapies are recommended). Supplementary Figs. 37-41 show the RUO testing results in several patient vignettes that describe clinicopathological information, relevant mFISHseq results, and information about patient follow up data to determine potential efficacy of the predictive signatures. A detailed report of each patient can be found at https://multiplex8.com/medical-professional (Note that the patient ID numbers do not reveal any identifying information). The RUO version of mFISHseq (called Multiplex8+) provided unique insights for each patient that could identify potential treatments (including subsequent lines of therapy) and helped to explain why prior treatments performed poorly. Like our retrospective study, there was exceptional concordance between IHC results and mFISHseq with agreement in 91% (n = 164/181) of cases for all four biomarkers, with HER2 IHC and ERBB2 mFISHseq showing 96% agreement. Most discordant results were observed for ESR1 (n = 7) followed by MKI67 (n = 5) and PGR (n = 3). In two patients (#7 and #12), both ER and PR were in gray zone areas for IHC (i.e., ER/PR low ≤10%) and mFISHseq classified them as negative. Notably, both patients were classified as basal-like by consensus subtyping, mesenchymal (M) TNBC subtype, and had high expression of proliferation, immune, and/or angiogenesis suggesting the mFISHseq results reflected the underlying tumor biology better than IHC.
a Table outlining the number of genes and gene signatures and their relevant drug targets/pathways that were used for RUO testing. b Proportion of 48 patients according to treatment setting (left bar), consensus molecular subtype (middle bar, includes 58 samples total to account for 10 patients that had 2 subtypes collected by LCM), and TNBC subtype (right bar, contains 36 samples total comprised of 31 patients with 4 patients who had two subtypes collected by LCM and one patient that had both a primary and metastatic tumor analyzed). c Frequency of therapies that were recommended as the top three according to expression of genes and gene signatures tailored to each patient. Expression of 20 ADC antigen targets d as well as targets relevant to payloads for topoisomerase and microtubule inhibitors (e), endocytosis (f), lysosome activity (g), and resistance (h). The center line of the box and whisker plots represents the median, the box denotes the interquartile range, and the whiskers extend to the minimum and maximum values of the dataset. Dots show individual data points. i Examples of patients belonging to putative ADC treatment-responsive groups based on expression of ADC relevant biomarkers (shown as a percentile score). Patients 1, 18, 15, and 23 are predicted to respond to sacituzumab govitecan (SG), trastuzumab deruxtecan (T-DXd), both SG and T-DXd, and neither SG nor T-DXd, respectively. Note that these are hypothetical ADC treatment-responsive groups and no patients in the RUO cohort were recommended ADCs based on this framework. Source data are provided as a Source Data file.
To further illustrate a hypothetical framework that could be used in a future prospective validation for identifying patients responsive to ADCs, we interrogated the expression of targets related to ADC antigens, cytotoxic payloads, endocytosis, lysosome function, and resistance (Fig. 9d–h) in these RUO patients. Expression of ADC markers was compared with either all patient samples from our retrospective cohort or samples restricted to the relevant IHC-surrogate subtype to assign a percentile ranking score and then categorized into tertiles (high, intermediate, low). Based on the expression of these ADC relevant genes and gene signatures, patients could be stratified into putative groups that may be responsive or unresponsive to ADCs (Fig. 9i), which could then serve as prespecified biomarker groups in future prospective studies.
Discussion
The mFISHseq test utilizes two orthogonal methods, RNA-FISH and RNA-SEQ, to characterize breast tumor biology. Simultaneously visualizing gene expression of the four main breast cancer biomarkers in a multiplexed and multicolor RNA-FISH reaction allows users to identify ROIs based on cellular phenotypes and tumor heterogeneity and then precisely capture these ROIs using LCM for downstream spatially resolved transcriptomics. Total RNA-SEQ facilitates transcriptome-wide expression profiling to classify the molecular subtype of breast cancer and quantify predictive and prognostic gene signatures.
Although multigene subtyping and prognostic risk classifiers provide clinically relevant information, discordance at the single-sample level remains unresolved7,8. This limitation is often attributed to RUO versions and differences in data processing; however, in the OPTIMA Prelim Trial, multigene tests showed marked disagreement in 40.7% of patients for intrinsic subtyping and 60.6% of patients for prognostic risk assessment, resulting in only moderate concordance between each test, despite each test being conducted by the respective vendors9. Our consensus subtyping alleviated this discordance by reclassifying 30% of patients into subtypes that better corresponded to their overall survival, prognostic risk, and subtype-specific molecular markers thus providing a more holistic approach to patient stratification using multigene subtyping.
Like consensus subtyping, we found that combining information from 5 different prognostic classifiers into a consensus prognostic classification system of high-, low-, and ultra-low-risk groups provided more accurate results for cases with discordance in at least one classifier. Moreover, the ultra-low-risk patients had excellent PFS and OS up to 15 years. With this consensus prognostic classification system, high-risk individuals showed the poorest OS and more relapses within 5 and 10 years and benefitted the most from chemoendocrine therapy. Low-risk patients had better OS, more relapses after 10 years, and showed no benefit from chemotherapy in addition to endocrine therapy. Notably, many of these patients received only 5 years of endocrine therapy and may constitute a group of patients that could benefit from extended endocrine therapy for up to 10 years. The ultra-low-risk group contained about 10% of patients that had low probability of relapse and excellent survival. This group may overlap with ultra-low-risk categories identified by other studies35, which constitutes 10-25% of all patients depending on screening and may represent patients with indolent tumors that need only surgery and no adjuvant systemic therapy at all.
Importantly, the key components of our consensus subtyping approach were validated in two external cohorts, METABRIC and TCGA. Although molecular subtyping and prognostic classifiers showed prognostic utility in both cohorts, there was considerable discordance among classifiers, with discordant samples showing altered outcomes and gene expression patterns. Moreover, our consensus subtyping and prognostic risk approach was able to mitigate this single sample discordance by reclassifying patients into subtypes/risk groups that better matched their outcome and tumor biology. Notably, when compared to our retrospective MDX cohort, both external cohorts yielded higher proportions of normal-like subtypes, indeterminate samples in the consensus, greater discordance among classifiers, and more subtle improvements in risk classification. These differences may be attributed in part to spurious changes in gene expression due to a greater presence of non-tumor tissue in METABRIC and TCGA, which is known to increase normal-like subtype calls and confound gene expression signatures such as PAM50 and AIMS36,37,38.
We made strides in defining the clinical and molecular features associated with discordance among multigene prognostic classification of low and high risk. Similar to other reports39,40, classification as high or low risk was primarily driven by a continuum of gene expression programs for proliferation/cell cycle and estrogen (luminal) pathways. This continuum highlights the challenges in classifying patients residing in the middle ranges of gene expression. Patients with discordant results in one or more prognostic signatures had intermediary phenotypes for proliferation/cell cycle genes making them more susceptible to discordant classifications based on the disparate gene lists, algorithms, and cutoff thresholds used by a particular signature to assign risk. Other pathways were also implicated in the biology of high vs low risk, including DNA damage repair, metabolism / hypoxia / angiogenesis, and immune states, an interesting finding, since many multigene prognostic signatures do not adequately cover these pathways. Prognostic signatures that better capture the nuanced biology in these pathways may help to resolve discordance at the individual tumor level. The finding that high-risk patients, especially those with all five prognostic classifiers in complete agreement, have immune hot TMEs contributes to a growing body of evidence that high-risk HR+ (luminal) breast cancer patients may be candidates for targeted immunotherapy. By deconvolving our bulk RNA expression data, we identified a variety of immune cell types and states differentially expressed in high- and low-risk patients as well as high-risk patients with discordance in risk assignment. Using data from patients treated with neoadjuvant pembrolizumab in the I-SPY2 trial, we showed that several of these signatures effectively doubled the predicted pCR rates compared to classical patient stratification based on risk and receptor status. Moreover, they performed equivalent to transcriptomic signatures shown to predict pCR in the I-SPY2 trial such as MammaPrint High-2 (MP2) and the HER2−/Immune+ RPS-5 groups41,42, underlining the potential of high transcriptomic risk patients, especially those with complete agreement in all prognostic signatures, to benefit from neoadjuvant immunotherapy.
The substantial inter-predictor concordance we observed without consensus subtyping/risk assessment was interestingly higher than prior reports7,8,9,10, potentially due to using LCM. Most other studies and assays use bulk/macrodissected specimens, which sacrifice the spatial information about biomarker expression and may introduce erroneous gene expression from non-tumor tissue, leading to misclassification of BCa samples8,36,37,43,44,45. For example, the normal-like subtype is simply a byproduct of insufficient tumor content/cellularity and the six TNBC subtypes were refined to four when LCM was used46,47. We found that bulk processing, compared to LCM, resulted in substantial gene expression changes and molecular subtype/prognostic risk group assignment, resulting in downscaling to less aggressive subtypes and risk groups. This has important implications for treatment as some patients, especially those in gray zones (e.g., bordering high-/low-risk thresholds) may be misclassified and consequently inappropriately treated. This stresses the importance of incorporating LCM into the mFISHseq workflow to minimize contamination from non-tumor elements and obtain spatially defined, tumor-enriched cell populations, ultimately facilitating precise and accurate biomarker quantification and multiparameter testing. However, the evidence supporting LCM relies on indirect comparisons (e.g., scRNAseq and independent datasets), evidence from the literature, and the limited pilot experiment on samples that either did or did not undergo LCM. Definitive proof of the benefits of LCM in the mFISHseq workflow should be an area of future investigation.
Our scRNAseq analysis of healthy and cancerous breast tissue revealed insights into the cell type distribution and specificity of multigene subtyping and risk panels. Surprisingly, each multigene classifier had genes that were either enriched in non-breast glandular epithelial cells or displayed low specificity, providing unequivocal evidence that gene expression from non-tumor cells could contribute and even confound the algorithms used for subtyping and risk classification. This analysis raises some important questions for future research: (1) to what extent does tumor-intrinsic and tumor-extrinsic gene expression influence classifications and are there circumstances when non-tumoral gene expression is necessary to derive an accurate classification? (2) Are particular methods (e.g., RT-PCR, RNA-SEQ, microarray) more or less susceptible to the presence of non-tumor elements? (3) Will gene signatures developed on LCM specimens provide the same information when applied to gene expression data derived from non-microdissected tissues (and vice-versa)? By focusing on tumor-intrinsic gene expression, our approach may be limited in understanding how other cells in the TME (e.g., stromal and immune cells) define clinical features, tumor biology, and treatment response. However, multi-region sampling of different cellular compartments (stromal, immune, tumor) by LCM may help to dissociate the relative contributions of TME cell types, while precisely controlling for the cell type proportions, an important factor that is challenging for bulk processing methods. This comprehensive analysis of single cell expression of multigene panels is an important prerequisite to test these questions.
A limitation of the study is that we compared research-based multigene classifiers rather than the genomic tests performed by the manufacturers. Research-based signatures may use different methodologies, gene lists, and approaches to normalization (see Supplementary Methods), which may lead to scaling effects that do not exactly recapitulate the commercial test48. Altogether these differences suggest caution when extrapolating the results from research-based signatures to their commercial counterparts. However, several lines of evidence suggest these research-based signatures generated from RNA-SEQ data are valid for assessing discordance and showing that combining multigene signatures mitigates discordance and improves prognostic performance. First, multigene signatures show discordance at the individual tumor level, this is true for both research-based signatures performed in silico7,8,49 and comparisons of multigene assays ran by the manufacturers on identical patient samples in clinical trial cohorts like transATAC and OPTIMA Prelim9,50,51,52,53. Second, combining information from more than one multigene signature may enhance prognostic performance50,51,52,54. Lastly, RNA-SEQ research-based classifiers are strongly correlated with the vendor-based tests at the gene level and show even better agreement when stratifying patients into risk groups55,56,57,58. Of note, the concordance between a research-based RNA-SEQ prognostic classifier and the respective vendor test on risk classification was higher than the concordance between two vendor tests performed on the same sample9. What remains unknown though, is the clinical utility of mFISHseq and whether the consensus of multiple subtyping and/or prognostic signatures can predict response to adjuvant chemotherapy, which would need to be demonstrated in a prospective trial.
The clinical trials for the recently approved ADCs, trastuzumab deruxtecan13 and sacituzumab govitecan14,15, suggest that protein expression of the antigen target is insufficient to predict treatment response, highlighting an unmet need to identify novel biomarkers that can select treatment-sensitive patients. We conducted one of the most comprehensive analyses of ADC processing-related markers in BCa, expanding on the findings of Bosi and colleagues59 by characterizing subtype-specific expression of substantially more ADC markers and their association with survival. The results portray unique subtype-specific expression patterns and at-risk patient groups that may be relevant for stratifying patients into ADC-responsive subgroups. Using data from the T-DM1 arm of I-SPY2, we developed a 19-feature classifier containing features related to the ADC target (ERBB2), receptor endocytosis, lysosome function, and microtubule targets of the maytansine payload. Moreover, this signature outperformed ERBB2 expression alone in a multivariate logistic regression, thus supporting the notion that combining ADC processing features into a predictive model has clinical potential for patient selection. Notably, both ERBB2 mRNA and our T-DM1_pred classifier had substantially more predictive utility than HER2 IHC, since 42.3% of patients (n = 22/52) did not respond despite being HER2 IHC positive. An important next step will be to validate this signature as a prespecified, qualified biomarker in a larger external cohort. These data create a foundation and roadmap for ADC patient selection by tailoring gene signatures to key features of the ADC: antigen and payload target (topoisomerase, microtubule, or DNA), cleavable (enzyme or acid labile) or noncleavable (lysosome processing) linker, and mechanisms of resistance (ABC transporters, glucuronidation enzymes).
Although we demonstrated the feasibility of implementing mFISHseq in a real-world setting, our RUO testing results are limited by the lack of clinical outcome and treatment response data, making it challenging to assess the clinical potential and utility. However, the insights gained provide a strong foundation for future prospective validation.
In summary, we developed and validated mFISHseq on a cohort of 1082 breast tumors, demonstrating excellent analytical validity and improved molecular subtyping and prognostic classification, which was validated on two external cohorts). We characterized the immune microenvironment of high-risk breast cancer and the expression patterns and association with survival of ADC-relevant markers, thus highlighting approaches to predict treatment response to neoadjuvant immunotherapy and ADCs, respectively. We further demonstrated the clinical feasibility and implementation of mFISHseq in a real-world setting on 48 patients who received an RUO version of the test, named Multiplex8+ (patient reports are located at www.multiplex8.com in the medical professional section).
Methods
Study Design
We developed and validated our mFISHseq BCa diagnostic test using 1082 archived FFPE BCa samples collected from two European biobanks, Biobank Graz of the Medical University of Graz, Austria and PATH Biobank (Munich, Germany), one hospital (Malaga, Spain), and two commercial companies (AMS Bio and Precision for Medicine). Details about the sample size/power analysis are provided in the Supplementary Methods. Informed consent to use these FFPE specimens and associated clinicopathological data was obtained from the source of the tissue. No tissues were processed without informed consent. Clinicopathological information associated with each sample (age, receptor / histological status, tumor grade, therapy history, survival data, etc.) was accrued in collaboration with the biobanks and follows the Biospecimen Reporting for Improved Study Quality (BRISQ) criteria and used to perform association analyses with molecular data. Inclusion criteria for the study consisted of females with histologically confirmed invasive BCa, availability of anonymized data regarding pathological diagnosis (IHC status, TNM staging), therapy (hormone/targeted/chemo- or radiotherapy), and survival (progression-free survival, overall survival), as well as signed and dated informed consent. The only exclusion criteria were pre-existing conditions or concurrent diagnosis of a cancer other than breast cancer or other disease that may influence the interpretation of the study results.
Out of a starting cohort of 1082 breast samples, we excluded one sample for revoked informed consent, four samples for damaged FFPE blocks or sections rendering them unable to be processed, 63 samples because pathology review revealed benign/healthy tissue or DCIS/LCIS, and one sample had missing clinical data. This left a cohort of 1013 breast tumors available for later analyses. Depending on the analyses, some data points may have been excluded due to missing data (e.g., missing survival data, IHC receptor status, etc.) or eligibility (e.g., patients with positive HER2 expression and/or more than three positive lymph nodes were excluded from analyses involving prognostic signatures). Details of missing data are described in Supplementary Data 1. Supplementary Figs. 2 and 34 show gene expression in 1254 breast cancer samples comprised of 1014 patients with invasive breast cancer (1 sample has no clinical data, yielding the 1013 breast tumors used in most analyses), 99 subtype samples from patients who had an extra ROI collected by LCM, 25 patients with in situ carcinoma (24 DCIS/1 LCIS), 24 no tumor tissues (i.e., tissues dissected from tumor specimens that contained only healthy, atypical ductal hyperplasia, or other benign cells upon pathological review), 12 true healthy samples, 41 scroll samples used for the LCM vs. no LCM experiment, and 39 positive control samples.
Patient specimens were processed in batches (see Supplementary Methods) using a stratified randomization approach to ensure that each batch contained a representative sampling of the IHC surrogate subtypes (i.e., LumA, LumB, HER2+, TNBC). Researchers who processed batches and conducted the data processing and analysis were blind to IHC biomarker status (e.g., ER, PR, HER2, and KI67) and other clinical information.
To assess the analytical validity of mFISHseq, the dataset was divided into a training and test set (70:30 split) using a stratified randomization approach to ensure similar proportions of positive and negative biomarkers (as defined by IHC) and sufficient patient outcomes. Other analyses like consensus subtyping and characterization of genes/gene signatures utilized the full dataset.
For the research-use only (RUO) cohort of 48 patients, tissues were obtained from several participating hospitals and clinics in Slovakia (Supplementary Data 2). Informed consent was obtained from the patient using a standard form and approval from their primary oncologist was mandatory prior to processing the specimen. Participants were not compensated. The Ethics Committee of the Bratislava Self-Governing Region also gave ethical approval for this work (Ref. No. 05320/2020/HF).
Tissue processing and H&E staining
We obtained at least eight 5 μm sections from FFPE BCa specimens using a Leica Histocore Multicut. Two sets of adjacent sections were collected in the following order: 1 section on a glass slide for H&E, 1 section on a functionalized PEN membrane slide for LCM, 1 section on a glass slide for RNA-FISH, and 1 section taken as a scroll and frozen at –20 °C for later RNA extraction and RNA sequencing (see Supplementary Methods). To ensure proper identification of invasive breast cancer, we stained one section using H&E, cover slipped, and then obtained a whole-slide scan. The resulting image was annotated by a trained researcher to identify the invasive breast cancer component for later microdissection. If necessary, a board-certified pathologist either annotated or reviewed challenging cases. Importantly, the H&E-stained section was adjacent to the PEN membrane slide used for LCM to ensure comparable anatomical morphology between the slide that was annotated and the slide that was microdissected.
Multiplexed RNA-FISH
For RNA-FISH we used the Advanced Cell Diagnostics RNAscope™ Multiplex Fluorescent V2 Assay to detect PGR, ESR1, ERBB2, and MKI67 according to the manufacturer’s instructions. These markers were detected using Akoya Opal 690, 620, 520, and 570 fluorophores, respectively. Visualization of these markers allowed us to capture the heterogeneity of the tumor tissue and isolate key regions of interest using LCM to obtain tumor-specific regions of interest, while eliminating otherwise healthy tissue, stroma, and adipose cells that may mask true gene expression differences.
Whole-slide imaging, image annotation, and image analysis
Following H&E staining and RNA-FISH, we used the Akoya Vectra Polaris™ imaging system to obtain brightfield and fluorescent whole slide scans (20x objective lens, 0.5 µm/pixel resolution, standard Akoya MOTiF™ multispectral imaging filters) that could be further analyzed and annotated for microdissection. The H&E whole slide scans were annotated by a trained researcher using the open-source program QuPath v0.4.3 (https://qupath.github.io/). Annotations were color-coded to identify invasive breast cancer (segregated by histological subtype if more than one is present in a specimen), ductal carcinoma in situ (DCIS), and healthy/normal tissue. If necessary, a board-certified pathologist either annotated or reviewed challenging cases. The RNA-FISH whole slide scans were annotated by a trained researcher using Akoya’s Phenochart™ software v1.1.0. The RNA-FISH annotation consisted of a qualitative overview of the intensity and distribution of fluorescent signals from ESR1, PGR, ERBB2, and MKI67. Based on the annotated H&E and RNA-FISH images, specific regions of interest were selected for LCM with an emphasis on regions that displayed the expression of biomarkers of interest (e.g., hotspots), areas identified as invasive by a trained researcher/pathologist, and the margins of invasive tumors. Moreover, in the case of specimens that displayed histologic or biomarker expression heterogeneity in the form of different molecular expression patterns (e.g., ER/PR+ and HER2− regions versus ER/PR− and HER2+ regions) or histological subtypes (e.g., invasive ductal vs invasive lobular carcinoma) distinct regions of interest were annotated and separately subjected to LCM and downstream analyses (Supplementary Methods).
For RNA-FISH image analysis, regions of interest that were dissected by LCM were stamped on the digital whole slide scans for further processing of biomarker signals using Akoya’s InForm® software v2.6.0. At least 1-3 stamped regions, depending on the size of the area dissected by LCM, were analyzed. The analysis consisted of the following steps: (1) spectral unmixing and autofluorescence isolation using a synthetic spectral library; (2) using machine learning algorithms to segment the tissue into different regions (tumor versus stroma) as well as to segment individual cells into nuclear and cytoplasmic components; and (3) scoring the expression of each biomarker. The average fluorescence intensity for each marker was assessed specifically in the tumor segment of the image and the researcher conducting the analysis was blinded to the known IHC results and the clinicopathological data.
Laser capture microdissection
We followed established protocols from Leica for conducting LCM in a manner that maintained RNA integrity. This included conducting a rapid, cresyl violet stain, limiting dissection times to under 1 hour per sample, and taking precautionary measures to ensure RNA integrity. Regions selected for dissection were identified by comparing the annotated H&E and RNA-FISH images with the adjacent cresyl violet stained section. We aimed to dissect approximately 10-20 mm2 of tissue per sample to ensure an adequate amount of material for RNA extraction. For samples with less tumor area, we conducted LCM on multiple PEN membrane slides to obtain sufficient tissue.
RNA isolation and quality control
The Macherey Nagel NucleoSpin totalRNA FFPE XS kit was used for RNA isolation (see Supplementary Methods). After RNA isolation, RNA quantity was measured using the Qubit RNA HS (High Sensitivity) Assay Kit with a Qubit 4 Fluorometer and RNA quality using the Agilent High Sensitivity RNA ScreenTape with an Agilent 4150 TapeStation. The DV200 value of the sample (i.e., the percentage of fragments ≥ 200 bases in length) was calculated as recommended by Illumina. Samples with DV200 values > 15% were considered as viable samples for library preparation.
RNA library preparation and sequencing
We used the Takara SMARTer Stranded Total RNA-Seq Kit v3 - Pico Input Mammalian kit to prepare total RNA-SEQ libraries following the manufacturer’s instructions. To control for batch library preparation effects, we included a single natural positive control sample in each library preparation batch and a synthetic spike-in control in each sample (see Supplementary Methods). Following library preparation, the quantity and fragment size range of the library were assessed using both the Qubit dsDNA HS kit (Qubit 4 Fluorometer) and the Agilent High Sensitivity DNA ScreenTape kit (Agilent 4150 TapeStation). Successfully prepared libraries contained sufficient library (≥4 ng/μl) to pool on an Illumina NovaSeq 6000 sequencing instrument and fragment range spanning 200–1000 bp, with a local maximum ~250–350 bp. Individual sequencing libraries were pooled and sequenced on an Illumina NovaSeq 6000 using SP, S1, S2, or S4 flow cells depending on pool size. Pooled libraries were spiked with 10% PhiX as recommended by both Illumina and Takara for low-complexity libraries sequenced on patterned flow cells. Paired-end sequencing (2 × 100 bp) was conducted with the aim of obtaining approximately 100 million reads per sample. The bioinformatics pipeline for RNA sequencing is described in Supplementary Methods. The detailed list of reagents used in this study is provided in Supplementary Data 5.
Statistics & Reproducibility
We performed a power analysis and sample size estimate using the RnaSeqSampleSize software (version 3.18, https://bioconductor.org/packages/release/bioc/html/RnaSeqSampleSize.html, Supplementary Methods). All statistical tests were conducted using GraphPad Prism 9, R packages described in the text, and the following python packages: pandas 2.1.3, numpy 1.26.2, scipy 1.11.4, statsmodels 0.14.2, and scikit-posthoc 0.9.0. Unless otherwise stated, the level of significance was set at p < 0.05 for both adjusted and unadjusted p-values, with all statistical tests being two-sided unless otherwise specified. For determining the significance of continuous variables, unpaired Mann-Whitney U tests (independent samples) were conducted for comparisons of two groups with non-normal data (nonparametric), while non-normal data for three or more groups were analyzed using Kruskal-Wallis tests. Appropriate corrections for multiple comparisons were conducted using Dunn’s multiple comparison test (nonparametric) followed by Bonferroni or Benjamini-Hochberg to adjust the p-values or control the false discovery rate (FDR), respectively. ROC and precision-recall curves were constructed using either GraphPad Prism 9 or the R package pROC 1.3.1. Diagnostic performance metrics were calculated using the R package caret 6.0-94. All measurements were obtained from biological replicates. If a biological specimen was measured more than once due to technical issues (e.g., poor sequencing metrics), only one value was reported based on the most reliable measurement.
Cohort descriptive statistics
Standard descriptive statistics presented as either median or mean with the percentage of samples represented in parenthesis were used to summarize sample characteristics (e.g., data for biospecimen information, demographics, pathological, therapy type, survival) for all specimens in the study and were segregated by source and IHC surrogate subtype.
Survival/Outcome analyses
Kaplan-Meier (KM) analysis and/or Cox Proportional Hazards models were used to quantify associations made between specific dependent variables and/or genes and gene signature predictors with known clinical outcome data (overall survival, progression-free survival). For KM analysis, we used the log-rank (Mantel-Cox) test to determine if one or more curves were significantly different. Both univariate and multivariate Cox analyses with clinical parameters (tumor size, pT1 vs pT2-pT4 and node status, pN0 vs pN1-pN3) were conducted using the R package survival (v3.5-7) or GraphPad Prism 9. For each parameter estimate in the Cox model, two-sided p-values were obtained by testing the null hypothesis that the true parameter estimate (beta) is equal to zero using the maximum log partial likelihood estimate (Wald test). Results were adjusted for multiple comparisons using FDR. Log-rank comparisons were not performed when certain groups had <10 patients or <3 events and no formal comparisons were conducted on the normal-like subtype group, since this subtype has been shown to be an artifact due to contamination from the presence of normal/healthy tissue.
Benchmarking consensus subtypes and prognostic risk groups
To determine if reclassified samples by consensus subtyping resulted in a better classification, we used a panel of genes and gene signatures for molecular subtyping as the ground truth. Nonparametric Kruskal-Wallis tests using Bonferroni p-value correction for multiple hypothesis testing were applied to compare the group of reclassified samples with the consensus subtype from which the samples were originally classified by IHC surrogate subtyping to the consensus subtype where the samples were reclassified. This was followed by pairwise comparisons using Dunn’s multiple comparisons test.
To assess the clinical and molecular parameters associated with high- and low- risk assignments, we stratified patients into groups based on the number of discordant classifiers (0-5) for high (intermediate and high risk combined) risk. Thus, a single dataset was used for this analysis with high risk as the reference for discordance. Group 0 is defined as unanimous classification as high risk by all five classifiers, while Group 5 is defined as unanimous classification as low risk by all five classifiers. Groups 1-4 are defined by the number of discordant classifiers for high risk. Groups 0-2 are classified as high risk and groups 3-5 are classified as low risk by the consensus. To assess discordance within an assigned risk group (low or high), we stratified patients into groups based on the number of discordant classifiers (0-2) for either low or high (intermediate and high risk combined). Since more than 2 discordant classifiers would result in assignment to the other risk group, this approach allowed us to identify factors influencing discordance in an exclusive risk group. For the analyses of continuous values (transcripts per million (TPM) or variance stabilizing transformation (vst) values for genes and vst values for gene signatures), we used nonparametric Kruskal-Wallis tests using Bonferroni p-value correction for multiple hypothesis testing followed by Dunn’s multiple comparisons tests. Two separate analyses were performed using all groups (0-5) to explore expression changes between high and low risk (Supplementary Fig. 11) and groups split into high (0-2) and low (3-5) risk categories to assess expression changes within a risk group (Fig. 4i). Note that the two high- and low-risk heatmaps presented in Fig. 4i are the same data presented in the heatmap in Supplementary Fig. 11 but have been separated into two heatmaps and the low-risk discordant labels were changed from 5, 4, and 3 (Supplementary Fig. 11) to 0, 1, and 2 (Fig. 4i) for clarity. The data illustrated in the heatmaps was obtained by first calculating individual sample z-scores for each gene and gene signature across all groups (e.g., 0-5 discordant classifiers for high risk). Then, these individual sample z-scores were averaged within each group (i.e., individual z-scores from all samples in group 0 were averaged) to obtain a single z-score value for each gene/gene signature and each group.
For the analyses of clinical parameters that were categorical, we generated contingency tables and used Chi-square tests for significance. Chi-square tests were unadjusted for multiple comparisons because they were independent null hypotheses.
The deconvolved bulk RNA-SEQ data from Ecotyper yielded cell type/state and Cellular Ecotype abundance values and was analyzed using the same approach outlined above for genes and gene signatures, except for the approaches for multiple correction testing and normalization. The data were analyzed using nonparametric Kruskal-Wallis tests (Benjamini-Hochberg FDR corrections) followed by Dunn’s multiple comparisons tests. To illustrate this data in heatmaps, the mean abundances for each group (as outlined above) were calculated for each cell type/state and then cube root transformed using the formula: each abundance i in array x is: yi=xi1/3. Cube root transformation was selected because of the large differences in abundances and below zero values. Note that the data in the high (Fig. 5a) and low (Fig. 5b) risk heatmaps are the same as presented in Supplementary Fig. 12, but the labels for low risk have been changed from 5, 4, and 3 to 0, 1, and 2.
Univariate and multivariate logistic regression using I-SPY2 trial data
We downloaded data from the T–DM1, control, and pertuzumab arms of the I-SPY2 trial from NCBI’s Gene Expression Omnibus (GEO) under accession code GSE181574. The T-DM1 arm contains 52 patients (ER+/HER2+: n = 35, ER−/HER2+: n = 17) treated with T-DM1 and pertuzumab with 30 patients achieving pathological complete response (pCR). The control arm contains 31 patients (ER+/HER2+: n = 19, ER−/HER2+: n = 12) treated with paclitaxel and trastuzumab with 8 patients achieving pCR. The pertuzumab arm (used as a control arm for pertuzumab in the T-DM1 arm) contains 44 patients (ER+/HER2 + : n = 29, ER−/HER2+: n = 15) treated with paclitaxel, pertuzumab, and trastuzumab with 26 patients achieving pCR. The T-DM1 dataset was stratified into training and test (50:50) data based on pCR and hormonal status. To associate the gene/gene signature with pCR, we performed univariate logistic regression in R through lmtest (v0.9.4) with the likelihood ratio test assessing significance. We further combined genes and gene signatures into the multivariate logistic model with elastic net regularization using tidymodels (v 1.1.1) with hyperparameter tuning by 10-fold cross-validation. The final 19-feature classifier (called multivariate T-DM1_pred) was then applied to the T-DM1 test set and compared with ERBB2 alone. We also constructed a single score (called univariate T-DM1_pred) by taking the individual features and multiplying them by a weighted coefficient, which was determined by multiplying the beta coefficient from the univariate analysis by the log of the p-value (e.g., univariate T-DM1_pred score = \({\sum }_{i=1}^{19}{sign}\left({\beta }_{i}\right)\times \log \left({p}_{i}\right)\times {X}_{i}\) where \({X}_{i}\) is normalized expression value of feature i, \({\beta }_{i}\) is \(\beta\) coefficient of feature i and \(\log \left({p}_{i}\right)\) is log of p-value of feature i). The sum of the weighted value of each feature was then averaged and transformed into z-scores.
Validation using METABRIC and TCGA cohorts
We used both TCGA-BRCA22,23 and METABRIC24,25 cohorts to externally validate the consensus molecular subtyping and prognostic risk approaches. For benchmarking subtype classifications, we obtained several ground truth PAM50 classifications from the flagship METABRIC paper24 and Perou and colleagues’ molecular analysis of TCGA breast cancer histologic types26 and compared with several implementations of the GeneFu PAM50 ROR-S using different sample compositions and scaling approaches (see Supplementary Methods and Supplementary Fig. 42 for additional details).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RNA-sequencing data and a de-identified clinicopathological data table from the retrospective MDX-BRCA cohort have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE283522. This comprises 1,254 breast tissue samples: 1,014 patients with invasive breast cancer (1 sample has no clinical data), 99 subtype samples from patients who had an extra ROI collected by LCM, 25 patients with in situ carcinoma (24 DCIS/1 LCIS), 24 no tumor tissues (i.e., tissues dissected from tumor specimens that contained only healthy, atypical ductal hyperplasia, or other benign cells upon pathological review), 12 true healthy samples, 41 scroll samples used for the LCM vs. no LCM experiment, and 39 positive control samples.
For both the retrospective (n = 1,082) and RUO (n = 48) cohorts, derived prognostic/gene signatures, digitized whole slide images of H&E (with and without pathology annotations), digitized whole slide images and analyzed region of interest from multiplexed RNA-FISH cannot be publicly shared due to existing material and data transfer agreements between MultiplexDX and participating biobanks and commercial companies. Qualified researchers may apply for access to these data through the MultiplexDX Data Access Committee (DAC) by sending an initial request to the lead corresponding author (P.Č., pavol@multiplexdx.com) or the following email address: info@multiplexdx.com. Then the qualified researcher would submit a brief research proposal and a standard form describing the project, data/materials requested, applicable ethics, and purpose. Requests will be reviewed and discussed by the DAC based on scientific merit, existing collaborations, and commercial agreements. The time frame of response to an initial request is about 1-2 months. After approval, the parties will agree on the conditions of a data access/sharing agreement and restrictions of use, which may increase the total time frame to around 6 months. Alternatively, qualified researchers may contact Biobank Graz of the Medical University of Graz, Austria and PATH Biobank (Munich, Germany) to request access to clinicopathological data and patient tumor specimens. Reports from the 48 patients that underwent a research use only version of the diagnostic test (called Multiplex8 + ) can be found at https://www.multiplex8.com/medical-professional.
Regarding external datasets, data from the paclitaxel + pembrolizumab (PEMBRO), T–DM1, control, and pertuzumab arms of the I-SPY2 trial can be downloaded from NCBI’s Gene Expression Omnibus (GEO) under accession code GSE181574 (T-DM1 only) or GSE194040 (all 988 patients in 10 arms). METABRIC microarray and clinical data for 2,509 patients was downloaded from the cBioPortal for Cancer Genomics (https://www.cbioportal.org/study/summary?id=brca_metabric). TCGA-BRCA data was downloaded from several different sources: 1. TCGA RNA-sequencing data was downloaded from the NIH GDC Data Portal database (https://portal.gdc.cancer.gov/), filtering for TCGA-BRCA, transcriptomic profiling, RNA-seq and Gene Expression Quantification to get 1231 samples; 2. TCGA clinical data was downloaded from cBioPortal for Cancer Genomics from both the TCGA, GDC dataset (n = 1,103) and the TCGA, PanCancer Atlas (n = 1,084); 3. TCGA PAM50 Ground truth subtypes were downloaded from cBioPortal for Cancer Genomics from the flagship dataset (TCGA, Nature 2012, n = 825 with 521 samples having PAM50 subtype calls) and a more updated analysis from Perou and colleagues molecular analysis of TCGA breast cancer histologic types (PMID: 35465400). Source data are provided with this paper.
Code availability
All software used for data collection, analysis, and bioinformatics was either open source or commercially available as documented in the Methods, Supplementary Information, and Reporting Summary; therefore, no new source code was generated in this paper.
References
Yersal, O. & Barutca, S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J. Clin. Oncol. 5, 412–424 (2014).
Sørlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. PNAS 98, 10869–10874 (2001).
Sørlie, T. et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. PNAS 100, 8418–8423 (2003).
Perou, C. M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000).
Litton, J. K., Burstein, H. J. & Turner, N. C. Molecular testing in breast cancer. Am. Soc. Clin. Oncol. Educ. Book 39, e1–e7 (2019).
Kittaneh, M., Montero, A. J. & Glück, S. Molecular profiling for breast cancer: a comprehensive review. Biomark. Cancer 5, 61–70 (2013).
Mackay, A. et al. Microarray-based class discovery for molecular classification of breast cancer: analysis of interobserver agreement. JNCI: J. Natl Cancer Inst. 103, 662–673 (2011).
Weigelt, B. et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol. 11, 339–349 (2010).
Bartlett, J. M. S. et al. Comparing breast cancer multiparameter tests in the OPTIMA prelim trial: no test is more equal than the others. J. Natl Cancer Inst. 108, djw050 (2016).
Haibe-Kains, B. et al. A three-gene model to robustly identify breast cancer molecular subtypes. J. Natl Cancer Inst. 104, 311–325 (2012).
Schmid, P. et al. Pembrolizumab for early triple-negative breast cancer. N. Engl. J. Med. 382, 810–821 (2020).
Jacob, S. L., Huppert, L. A. & Rugo, H. S. Role of immunotherapy in breast cancer. JCO Oncol. Pr. 19, 167–179 (2023).
Modi, S. et al. Trastuzumab Deruxtecan in previously treated HER2-low advanced breast cancer. N. Engl. J. Med 387, 9–20 (2022).
Bardia, A. et al. Sacituzumab Govitecan in metastatic triple-negative breast cancer. N. Engl. J. Med 384, 1529–1541 (2021).
Bardia, A. et al. Biomarker analyses in the phase III ASCENT study of sacituzumab govitecan versus chemotherapy in patients with metastatic triple-negative breast cancer. Ann. Oncol. 32, 1148–1156 (2021).
Liao, J., Lu, X., Shao, X., Zhu, L. & Fan, X. Uncovering an organ’s molecular architecture at single-cell resolution by spatially resolved Transcriptomics. Trends Biotechnol. https://doi.org/10.1016/j.tibtech.2020.05.006 (2020).
Turashvili, G. & Brogi, E. Tumor Heterogeneity in Breast Cancer. Front Med (Lausanne) 4, 227 (2017).
Brueffer, C. et al. Clinical value of RNA sequencing–based classifiers for prediction of the five conventional breast cancer biomarkers: a report from the population-based multicenter Sweden Cancerome Analysis Network—Breast Initiative. JCO Precis. Oncol. 1–18 (2018) https://doi.org/10.1200/PO.17.00135.
Atout, S., Shurrab, S. & Loveridge, C. Evaluation of the suitability of RNAscope as a technique to measure gene expression in clinical diagnostics: a systematic review. Mol. Diagn. Ther. 26, 19–37 (2022).
Luca, B. A. et al. Atlas of clinically distinct cell states and ecosystems across human solid tumors. Cell 184, 5482–5496.e28 (2021).
Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782 (2019).
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Ciriello, G. et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 163, 506–519 (2015).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Pereira, B. et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479 (2016).
Thennavan, A. et al. Molecular analysis of TCGA breast cancer histologic types. Cell Genom. 1, 100067 (2021).
Corti, C. et al. HER2-Low Breast Cancer: a New Subtype? Curr. Treat Options Oncol. https://doi.org/10.1007/s11864-023-01068-1 (2023).
Nicolò, E., Boscolo Bielo, L., Curigliano, G. & Tarantino, P. The HER2-low revolution in breast oncology: steps forward and emerging challenges. Ther. Adv. Med Oncol. 15, 17588359231152842 (2023).
Baez-Navarro, X. et al. Selecting patients with HER2-low breast cancer: Getting out of the tangle. Eur. J. Cancer 175, 187–192 (2022).
Bhat Nakshatri, P., et al. A single-cell atlas of the healthy breast tissues reveals clinically relevant clusters of breast epithelial cells. Cell Rep. Med. 2, (2021).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Karlsson, M. et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 7, eabh2169 (2021).
Wu, S. Z. et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet 53, 1334–1347 (2021).
Tarhan, L. et al. Single Cell Portal: an interactive home for single-cell genomics data. 2023.07.13.548886 Preprint at https://doi.org/10.1101/2023.07.13.548886 (2023).
Lopes Cardozo, J. M. N. et al. Outcome of patients with an ultralow-risk 70-Gene signature in the MINDACT Trial. J. Clin. Oncol. 40, 1335–1345 (2022).
Bastien, R. R. et al. PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC Med. Genomics 5, 44 (2012).
Elloumi, F. et al. Systematic bias in genomic classification due to contaminating non-neoplastic tissue in breast tumor samples. BMC Med. Genomics 4, 54 (2011).
Paquet, E. R. & Hallett, M. T. Absolute assignment of breast cancer intrinsic molecular subtype. J. Natl Cancer Inst. 107, 357 (2015).
Vieira, A. F. & Schmitt, F. An Update on Breast Cancer Multigene Prognostic Tests—Emergent Clinical Biomarkers. Front. Med. 5, 248 (2018).
Buus, R. et al. Molecular Drivers of Oncotype DX, Prosigna, EndoPredict, and the Breast Cancer Index: A TransATAC Study. J. Clin. Oncol. 39, 126–135 (2021).
Ríos-Hoyo, A. et al. Neoadjuvant Chemotherapy and Immunotherapy for Estrogen Receptor–Positive Human Epidermal Growth Factor 2–Negative Breast Cancer. JCO JCO.23.02614 https://doi.org/10.1200/JCO.23.02614 (2024).
Wolf, D. M. et al. Redefining breast cancer subtypes to guide treatment prioritization and maximize response: Predictive biomarkers across 10 cancer therapies. Cancer Cell 40, 609–623.e6 (2022).
Lien, T. G. et al. Sample preparation approach influences PAM50 risk of recurrence score in early breast cancer. Cancers 13, 6118 (2021).
Nielsen, T. O. et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor positive breast cancer. Clin. Cancer Res 16, 5222–5232 (2010).
Nielsen, T. et al. Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer 14, 177 (2014).
Lehmann, B. D. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest 121, 2750–2767 (2011).
Lehmann, B. D. et al. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS One 11, e0157368 (2016).
Bartlett, J. M. S. et al. Computational approaches to support comparative analysis of multiparametric tests: Modelling versus Training. PLOS ONE 15, e0238593 (2020).
Fan, Cheng et al. Concordance among Gene-Expression–based predictors for breast cancer. N. Engl. J. Med. 355, 560–569 (2006).
Kelly, C. M. et al. Agreement in risk prediction between the 21-Gene Recurrence Score Assay (Oncotype DX®) and the PAM50 Breast Cancer Intrinsic ClassifierTM in Early-Stage Estrogen Receptor–Positive Breast Cancer. Oncologist 17, 492–498 (2012).
Dowsett, M. et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. JCO 31, 2783–2790 (2013).
Sgroi, D. C. et al. Prediction of late distant recurrence in patients with oestrogen-receptor-positive breast cancer: a prospective comparison of the breast-cancer index (BCI) assay, 21-gene recurrence score, and IHC4 in the TransATAC study population. Lancet Oncol. 14, 1067–1076 (2013).
Sestak, I. et al. Comparison of the performance of 6 prognostic signatures for estrogen receptor–positive breast cancer: a secondary analysis of a randomized clinical trial. JAMA Oncol. 4, 545–553 (2018).
Bartlett, J. M. S. et al. Comparative survival analysis of multiparametric tests-when molecular tests disagree-A TEAM Pathology study. NPJ Breast Cancer 7, 90 (2021).
Sinicropi, D. et al. Whole Transcriptome RNA-Seq analysis of breast cancer recurrence risk using formalin-fixed paraffin-embedded tumor tissue. PLOS ONE 7, e40092 (2012).
Staaf, J. et al. RNA sequencing-based single sample predictors of molecular subtype and risk of recurrence for clinical assessment of early-stage breast cancer. npj Breast Cancer 8, 1–17 (2022).
Mittempergher, L. et al. MammaPrint and BluePrint molecular diagnostics using targeted RNA next-generation sequencing technology. J. Mol. Diagn. 21, 808–823 (2019).
Picornell, A. C. et al. Breast cancer PAM50 signature: correlation and concordance between RNA-Seq and digital multiplexed gene expression technologies in a triple negative breast cancer series. BMC Genomics 20, 452 (2019).
Bosi, C. et al. Pan-cancer analysis of antibody-drug conjugate targets and putative predictors of treatment response. Eur. J. Cancer 195, 113379 (2023).
Acknowledgements
We gratefully acknowledge Biobank Graz of the Medical University of Graz, Austria and PATH Biobank (Munich, Germany) for providing the FFPE breast cancer specimens and associated clinicopathological data; Jeff Palatini, Dorota Adamska, Krzysztof Goryca, and other staff at the Centre for New Technologies, University of Warsaw, Genomics Core Facility for providing first-class sequencing and bioinformatic services; Dr. Karol Kajo, a board-certified pathologist, at the St. Elizabeth Cancer Institute Hospital in Bratislava for assistance in annotating digital whole slide images of H&E-stained tissues; MUDr. Tomáš Sieber, MPH, MUDr. Andrea Cipková, M.D. Mária Višňovská from the East Slovak Oncology Institute in Košice (Východoslovenský onkologický ústav a.s.) for their participation and feedback on patient follow-up data for the RUO cohort study. RNA-sequencing data was obtained using the computational resources of the Computing Center of the Slovak Academy of Sciences procured in the national project: National competence centre for high-performance computing (project code: 311070AKF2) funded by European Regional Development Fund, EU Structural Funds Informatization of society, Operational Program Integrated Infrastructure. This project was supported by the European Union’s Horizon 2020 research and innovation program under an EIC Accelerator grant (agreement No 946693) awarded to MultiplexDX s.r.o. (Pavol Cekan as PI). FP is funded in part by an NIH/NCI P50 CA24779 01 grant. Research reported in this publication was supported in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (Grant No. P30CA008748 funding for FP). The funding sources had no role in the design and conduct of the study, interpretation of the data, writing of the manuscript, and decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
E.D.P, B.H., N.Val., N.M., S.B., V.Man., T.T., F.P., J.N.K., and P.Č. conceptualized and planned the study. E.D.P. and P.Č. jointly supervised the project. B.H., D.G., S.G., H.I., T.O., J.B., K.B., Z.K., and N.Voj., sectioned FFPE tissues, conducted histological staining and RNA-FISH, annotated H&E images, quantified RNA-FISH images, and performed laser capture microdissection. N.Val., N.M., L.B., S.B., D.D., M.K., D.L., V.Mam., and V.Man., extracted RNA, prepared RNA-SEQ libraries, conducted RNA/cDNA quality control, and pooled libraries for sequencing. N.Val., S.G., H.I., M.G., M.R., and P.M. conducted bioinformatic analyses. E.D.P., B.H., N.Val., N.M., D.G., S.G., H.I., T.O., M.G., L.B., S.B., J.B., K.B., D.D., Z.K., M.K., D.L., V.Mam., V.Man., N.Voj., M.R., I.C.M, I.A., P.M., T.T., F.P., J.N.K., and P.Č. verified the underlying data, analyzed, and interpreted the data, prepared figures, and wrote the manuscript. All authors had full access to the data, provided critical comments and feedback on the manuscript, and accepted responsibility to submit the manuscript for publication.
Corresponding authors
Ethics declarations
Competing interests
E.D.P., B.H., N.Val., N.M., D.G., S.G., H.I., T.O., M.G., L.B., S.B., J.B., K.B., D.D., Z.K., M.K., D.L., V.Mam., V.Man., N.Voj., M.R. and P.Č. are current, or former employees of MultiplexDX, a biotechnology company that is developing a lab developed diagnostic test called Multiplex8+ (https://www.multiplexdx.com/products/multiplex-eight-plus), which is based on the research presented in the manuscript. P.Č. and E.D.P. are inventors and MultiplexDX, s.r.o is the assignee on patent applications that were filed in relation to the technology and research outlined in the manuscript. These include a family of patents entitled “METHOD FOR DIAGNOSING DISEASES USING MULTIPLEX FLUORESCENCE AND SEQUENCING” (WO/2020/070325, EP3775277, CA3114689, AU2019354863, SG11202103466T, KR1020210071003, CN113366118, BR112021006454, US20230037279, JP2022513333, IL282067, and NZ774986) as well as submitted EPO and PCT patents that are not published. T.T., F.P., and J.N.K. are members of the Scientific Advisory board at MultiplexDX. F.P. reports consulting services and serving on the advisory board for AstraZeneca. J.N.K. declares consulting services for Owkin, France, DoMore Diagnostics, Norway, Panakeia, UK, Scailyte, Switzerland, Cancilico, Germany, Mindpeak, Germany, and Histofy, UK; furthermore, he holds shares in StratifAI GmbH, Germany, has received a research grant by GSK, and has received honoraria by AstraZeneca, Bayer, Eisai, Janssen, MSD, BMS, Roche, Pfizer and Fresenius. All other authors declare no competing interests.
Ethics & Inclusions
This study complies with all relevant ethical regulations for human research participants in accordance with the Declaration of Helsinki. The collection and use of human tissue in the retrospective clinical validation study on 1082 patients and the RUO testing cohort of 48 patients were approved by the Ethics Committee of the Bratislava Self-Governing Region (Ref. No. 05320/2020/HF). Additionally, the retrospective cohort received approval from the Ethics Commission of the Medical University of Graz on behalf of Biobank Graz (No. 34-354 ex 21/21, 1158−2022). Informed consent was obtained for tissues in the retrospective study from tissue sources including biobanks (PATH Biobank and Biobank Graz), a hospital (Malaga), and commercial companies (AMSBio and Precision for Medicine). For the RUO cohort, all 48 patients signed informed consent forms, with their oncologists approving them prior to tissue processing. The RUO testing occurred through routine testing with collaborating hospitals in Slovakia and included consultations and participation of local oncologists and researchers for patient selection, collecting clinicopathological data, obtaining informed consent, and explaining results. No discrimination occurred in the selection of patients for RUO testing and both clinical partners and MultiplexDX ensured sensitive patient information was anonymized when appropriate and secured both physically and electronically. A local pathologist (Dr. Karol Kajo) was also consulted with in both the retrospective and RUO cohorts to identify invasive breast cancer or unique histology in more challenging cases.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Paul, E.D., Huraiová, B., Valková, N. et al. The spatially informed mFISHseq assay resolves biomarker discordance and predicts treatment response in breast cancer. Nat Commun 16, 226 (2025). https://doi.org/10.1038/s41467-024-55583-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55583-2
This article is cited by
-
Antibody–drug conjugates in cancer therapy: current landscape, challenges, and future directions
Molecular Cancer (2025)
-
Multimodal histopathologic models stratify hormone receptor-positive early breast cancer
Nature Communications (2025)