Abstract
African swine fever virus is highly contagious and causes a fatal infectious disease in pigs, resulting in a significant global impact on pork supply. The African swine fever virus RNA polymerase serves as a crucial multifunctional protein complex responsible for genome transcription and regulation. Therefore, it is essential to investigate its structural and functional characteristics for the prevention and control of African swine fever. Here, we determine the structures of endogenous African swine fever virus RNA polymerase in both nucleic acid-free and elongation states. The African swine fever virus RNA polymerase shares similarities with the core of typical RNA polymerases, but possesses a distinct subunit M1249L. Notably, the dynamic binding mode of M1249L with RNA polymerase, along with the C-terminal tail insertion of M1249L in the active center of DNA-RNA scaffold binding, suggests the potential of M1249L to regulate RNA polymerase activity within cells. These results are important for understanding the transcription cycle of the African swine fever virus and for developing antiviral strategies.
Similar content being viewed by others
Introduction
African swine fever (ASF) is an acute, highly contagious, and fatal infectious disease caused by the African swine fever virus (ASFV). The presence of highly virulent strains of ASFV can lead to mortality rates of nearly 100%1,2. ASFV is the sole member of the Asfarviridae family and belongs to the nucleocytoplasmic large DNA virus (NCLDV) family3. ASFV is an icosahedral DNA virus 200 nm in diameter that possesses an envelope, capsid, inner membrane, core shell, and nucleoid4. The viral genome is a linear double-stranded DNA molecule, is approximately 170~190 kb in length and encodes 150~200 viral proteins, including 68 structural proteins and over 100 nonstructural proteins5. Variations in the genome, such as the loss or repetition of specific sequences, play a crucial role in distinguishing ASFV strains across generations and identical strains from different sources3. The major capsid protein P72, known for its high degree of conservation, is utilized for genotyping ASFV strains6. Although ASFV primarily infects monocyte-macrophage pairs, our understanding of the molecular mechanisms underlying infection and replication within host cells remains limited. Regrettably, effective vaccines or antiviral drugs for preventing or treating ASF are currently unavailable, leading to significant economic losses in animal husbandry each year7.
RNA polymerases (RNAPs) play a crucial role in transcribing RNA from the genome and are indispensable across all domains of life. In recent years, extensive research has been conducted using cryo-electron microscopy (cryo-EM) to investigate the structural basis of RNAPs in mammals8,9,10, yeasts11,12,13, archaea14,15 and DNA viruses16,17,18 using cryo-electron microscopy (cryo-EM). Notably, members of the NCLDV family exhibit cytoplasmic transcriptional activity necessitating a virus-encoded RNAP for generating mature mRNA from the viral genome. The structurally well-characterized NCLDV RNAP is Vaccinia virus (VACV) RNAP, which provided initial insights into the structural basis of NCLDV RNAP16,17,18. The VACV core RNAP consists of eight subunits (Rpo147, Rpo132, Rpo35, Rpo22, Rpo19, Rpo18, Rpo7 and Rpo30), which are conserved with their counterparts (RPB1, RPB2, RPB3-11 fusion, RPB5, RPB6, RPB7, RPB10 and TFIIS, respectively) in classical RNA polymerase II (Pol II).
Like other NCLDVs, ASFV encodes a transcription machinery similar to eukaryotic RNA polymerase, including core subunits and general transcription factors involved in initiation, elongation, and termination processes19,20,21. Approximately twenty proteins encoded by ASFV actively participate in the transcription and modification of mRNAs. Recently, the structure of ASFV RNAP has been solved through recombinant expression22, which is composed of eight core subunits (vRPB1, vRPB2, vRPB3-11, vRPB5, vRPB6, vRPB7, vRPB9, and vRPB10). However, it is still only rudimentarily investigated and it remains uncertain if this structure is consistent with the natural RNAP of ASFV. Here, we isolated endogenous ASFV RNAP from infected cells and determined the structures of RNAP in multiple conformations. These structural findings not only validate the conserved molecular mechanism of multi-subunit RNAPs but also shed light on the characteristic features exhibited by ASFV RNAP. Our investigations hold significant implications for establishing a mechanistic understanding of the transcription cycle of ASFV and facilitating the design of antiviral drugs, thus underscoring their paramount importance in advancing our knowledge in this field.
Results
Purification of the ASFV RNAP complex
The eight core components of RNAP have counterparts in the ASFV genome and were named vRPB1~vRPB10 in this study (Supplementary Table 1). Based on evolutionary tree analysis, it has been determined that ASFV RNAP belongs to a distinct evolutionary branch from other structurally known RNAPs. Additionally, the sequences of the two large subunits of RNAP are highly conserved across various types of ASFV strains (Fig. 1a). To obtain endogenous ASFV RNAP, we generated a recombinant strain called ASFV CN/GS/2018-strep derived from the ASFV CN/GS/2018-GFP strain and expressing the C-terminal Twin-Strep-tagged subunit vRPB2 (Supplementary Fig. 1a). The replication rate of ASFV CN/GS/2018-strep was comparable to that of the untagged parental ASFV CN/GS/2018-GFP strain for the infection of porcine alveolar macrophages (PAMs) (Supplementary Fig. 1b). This observation suggests that the presence of a tag on vRPB2 does not impede the transcriptional activity or replication capacity of the virus.
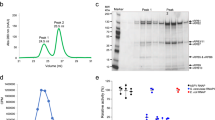
a Phylogenetic analysis of the two largest subunits of RNAP to determine evolutionary relationships. b ASFV RNAP was purified from cell extracts infected with ASFV CN/GS/2018-strep and fractionated on a sucrose gradient ranging from 10% to 30%. The respective subunit bands were visualized using silver staining on SDS-PAGE. This is a representative group of gels, which were repeated three times under the same condition with similar results. Source data are provided in a Source Data file. c The RNA polymerization activities using different template were assessed by measuring the incorporation of [α−32P]-ATP and normalized against the negative controls lacking RNA polymerase. The data represents the mean values ± standard deviation (error bars) from three independent experiments, with source data provided in a Source Data file. Statistical significance was determined using an unpaired two-tailed t-test, resulting in a P-value of 0.0003 with a 95% confidence interval (−8.491,−5.182) and degrees of freedom of 4. All calculations were conducted using Graphpad Prism 8 software.
After propagation of ASFV CN/GS/2018-strep with PAMs, purification of ASFV RNAP was carried out from infected cells using strep beads and sucrose gradient centrifugation (Supplementary Fig. 1c). Subsequently, the fractions were resolved via SDS-PAGE and visualized through silver staining (Fig. 1b). Mass spectrometry analysis was performed on all components (Supplementary Data 1), revealing that the elution containing the known core subunits of ASFV RNAP (Supplementary Table 1). Notably, M1249L emerged as a factor associated with ASFV RNAP upon elution. To evaluate the catalytic activity of ASFV RNAP, we performed a test for RNA polymerization activity by measuring the incorporation of [α-32P]-ATP into RNA using dsDNA-RNA as template, followed by precipitation with cold trichloroacetic acid (TCA) and quantification through scintillation counting22. Our biochemical results revealed that the sample exhibits in vitro elongation capability and functions as an active catalytic RNA polymerase. When using dsDNA as a template, we observed reduced activity, indicating that dsDNA template can direct RNA synthesis in a non-specific manner and highlighting the potential utility of an RNA primer in elongation (Fig. 1c).
Structure of the ASFV RNAP core complex and M1249L C-tail occupied complex
Fractions containing the most complete RNAP core subunits were subjected to cryo-EM single-particle analysis (Fig. 1b). Through unbiased 3D classification during data processing, we obtained a total of five distinct conformations of RNAP at a nominal resolution of 2.4~3.0 Å. Based on the overall size of the complex and protein density in the active center, these five states were designated as follows: the core complex (CC), M1249L C-tail occupied complexes 1 to 4 (MCOC1 to MCOC4) respectively (Supplementary Fig. 2). In all five structures, clear identification was made for the eight core subunits of ASFV RNAP, and their corresponding AlphaFold223-predicted models were confidently fitted into the map. Following manual adjustment and auto-refinement, most side chains within these eight core subunits exhibited excellent agreement with the density maps.
However, in the MCOC1 to 4 states of ASFV RNAP, we observed additional density on the outer surface and within the active center of the structure. We employed DeepTracer24 for de novo modeling based on density and further refined it manually using Coot (v0.9.8)25. The results confirmed that this increase in density corresponds to the ASFV M1249L protein. Amongst these four structures, M1249L is predominantly found in MCOC1, with most of its domains clearly identifiable in the cryo-EM map. In contrast, varying amounts of M1249L domains dissociate from MCOC1 in the MCOC2 to 4 states. The composition of the MCOC1 structure includes all eight core subunits, one M1249L subunit, seven structural zinc ions, and a catalytic magnesium ion A (Fig. 2a).
a Overall structure of the ASFV MCOC1, shown in cartoon depiction with helices depicted as cylinders. Subunits are colored by chains. The active site magnesium and zinc ions are shown as red and cyan spheres, respectively. b Comparison of subunit compositions among ASFV RNAP, H. sapiens Pol II (PDB: 5IYD8), and VACV RNAP (PDB: 6RIC16). These structures are depicted in surface representation. Homologous subunits are indicated in the table and colored accordingly. c The specific regions of ASFV RNAP are highlighted on the structure, while the remaining conserved regions are depicted as transparent gray surfaces.
The overall architecture of ASFV RNAP exhibits a crab claw-like structure, which is evolutionarily conserved across all domains of life (Fig. 2a). While the eight core subunits share structural homology with their counterparts in Pol II and VACV RNAP, they possess distinct features (Supplementary Figs. 3–8). The two major subunits, vRPB1 and vRPB2, constitute the central body of RNAP and encompass all conserved domains essential for the active center (Supplementary Figs. 3–5). Subunits vRPB3-11 and vRPB10 form an assembly platform at the posterior region of the RNAP body, akin to Rpo35 and Rpo7 in VACV RNAP. Similar to Rpo35, vRPB3-11 represents an RPB3/11 fusion polypeptide; however, compared to Rpo35, vRPB3-11 bears greater resemblance to eukaryotic Pol II (Supplementary Figs. 3 and 8). Subunits vRPB5, vRPB6 and vRPB7 assemble into a small crab claw-like shape and form the periphery of the RNAP. This configuration bears resemblance to Rpo22, Rpo19, and Rpo18 of VACV RNAP. In contrast to VACV RNAP, ASFV RNAP includes vRPB9 which comprises two zinc-ribbon domains and exhibits close similarity to RPB9 found in typical Pol II (Supplementary Figs. 3 and 8).
Conserved core and specific periphery of the ASFV RNAP
To investigate the functional roles of individual subunits within ASFV RNAP, we conducted a comparative analysis with H. sapiens Pol II and VACV RNAP, employing both structural comparison (Fig. 2b and Supplementary Fig. 8) and structure-based sequence alignment (Supplementary Figs. 4–7). While the core of RNAP is highly conserved across multi-subunit RNAPs, there are notable differences in the periphery and active center of ASFV RNAP, including the presence of an additional subunit M1249L and several extended regions within core subunits (Fig. 2c). The two major subunits, vRPB1 and vRPB2, exhibit significant similarity to their respective counterparts RPB1/Rpo147 and RPB2/Rpo132 (Supplementary Fig. 3). However, it is noteworthy that the jaw and clamp domains of vRPB1 undergo a rotation of 28° and 19°, respectively, resulting in a slightly narrower active center (Supplementary Fig. 8). Additionally, an intriguing feature of vRPB2 is the presence of a characteristic motif within its external1 domain comprising residues 678~710. This motif encompasses an alpha helix that interacts with the zinc-ribbon2 domain of vRPB9. Similar to vRPB1, the lobe domain of vRPB2 exhibits movement towards the protrusion region leading to a narrowing cleft (Supplementary Fig. 8).
As corresponding to a fusion protein of Pol II RPB3/11, the vRPB3-11 subunit is similar to the Rpo35 subunit of VACV RNAP. Positioned at the posterior region of RNAP, this subunit forms an assembly platform in conjunction with subunit vRPB10, thereby anchoring the two large subunits similar to other multi-subunit RNAPs26,27,28,29,30. In contrast to Rpo35 of VACV RNAP, both the overall structure and location of vRPB3-11 closely resemble those observed for its counterparts RPB3 and RPB11 (Supplementary Fig. 8). However, it is worth noting that vRPB3-11 lacks a zinc finger domain but instead possesses an additional short helix motif spanning 28 amino acids (residues 13~40) located on the periphery of the complex. The subunit vRPB10 occupies a similar location as Pol II RPB10 and the VACV RNAP Rpo7. However, within the zinc-bundle domain, there exists a distinct loop (residues 29-49) that extends further and establishes additional interactions with vRPB2 and M1249L (Supplementary Fig. 8). Notably, ASFV and VACV RNAP lack the RPB12. The assembly of vRPB3-11/vRPB10 serves as a viral RNAP platform analogous to the RPB3/10/11/12 subassembly observed in Pol II.
vRPB9 is positioned similarly and exhibits a comparable structure to Pol II RPB9, with the exception of the orientation of the zinc-ribbon2 domain, which is absent in VACV RNAP (Supplementary Fig. 8). Structurally resembling RPB5 and Rpo22, vRPB5 occupies a similar spatial location. The jaw domain of vRPB5 is shorter than that of RPB5, and together with the vRPB1 jaw domain, it forms the DNA entry path to the cleft (Supplementary Figs. 3b and 8). As for vRPB6, it serves as a structural and functional homolog of RPB6 by interacting with vRPB1 to facilitate RNAP assembly and stability. Notably, the elongated N-terminal tail of vRPB6 deeply inserts into the complex’s cleft region and potentially engages in interactions with nucleic acids—a characteristic distinct from other known structures (Fig. 2c and Supplementary Fig. 8). The tip and OB-fold domain of vRPB7 form a protruding stalk from the RNAP body, resembling the RPB7 domain of typical Pol II. However, unlike any known RNAP including VACV, which is also an NCLDV, there is no sequence or structural homology observed in the CTD of vRPB7. Consequently, the CTD represents a characteristic specific to ASFV RNAP and occupies a position analogous to RPB4 for maintaining complex stability (Supplementary Fig. 8).
These comparisons have revealed that ASFV RNAP exhibits a conserved core. However, the extension regions of vRPB2, vRPB3-11, vRPB10, and vRPB7 along with M1249L are prominently exposed on the outermost surface of the complex and are specific to ASFV. These regions potentially play a crucial role in binding to ASFV-specific transcription factors. Consequently, it can be inferred that ASFV RNAP has evolved features tailored towards its specific transcriptional environment and requirements.
Subunit M1249L stabilizes RNAP as a cage
In the MCOC1 to 4 of ASFV RNAP, an additional subunit known as M1249L tightly associates with the eight core subunits (Fig. 3a). Although not traditionally considered a component of RNAP, M1249L exhibits high abundance in the isolated RNAP fractions we obtained (Fig. 1b and Supplementary Data 1), indicating its significant role in RNAP function. Cryo-EM map clearly reveals most regions of M1249L, including D1~D5 and the C-tail domains, within MCOC1 (Supplementary Fig. 9). Furthermore, our constructed structure differs from the predicted structure of M1249L generated by AlphaFold2 (Supplementary Fig. 10a), suggesting that M1249L integrates into the RNAP core complex in a specific manner to fulfill specific functions. It has been previously reported that M1249L may serve as a structural protein within the capsid of ASFV4; however, our findings indicated that M1249L may have the potential role in regulating the RNAP function.
a Distribution of the functional domains of M1249L and its structure in ASFV MCOC1. The core subunits are shown as transparent surfaces and colored as indicated in Fig. 2. b Depiction of interfaces between M1249L domains and core subunits. The protein subunits are shown using surface presentation, using the same color scheme as depicted in Fig. 2. The interface area on each protein surface is colored to match its corresponding binding protein.
The M1249L domains are distributed ubiquitously throughout the complex and interact with nearly all core subunits of the RNAP, thereby increasing the stability of the complex by forming a cage structure around it (Fig. 3a and Supplementary Fig. 10). The D1 domain (residues 67~235) binds to the upper region of the RNAP clamp and engages in interactions with vRPB1. Furthermore, D1 integrates into the vRPB5-vRPB6-vRPB7 module while interacting with both the N-tail domain of vRPB6 and CTD domain of vRPB7, thus fortifying the periphery of RNAP (Fig. 3b and Supplementary Fig. 11). The D2 domain (residues 250~517) interacts with vRPB1 and vRPB2 while serving as a foundation for binding to vRPB6. The D3 domain, spanning residues 673~817 and located at the protrusion of vRPB2, plays a crucial role in stabilizing the assembly platform of RNAP by binding to vRPB3-11 and vRPB10. A linker composed of residues 817 ~ 845 connects the D3 with D4 (residues 845~987). The D4 domain engages in diverse interactions as it binds to the funnel of vRPB1, extending towards vRPB9 (Fig. 3b and Supplementary Fig. 11). Comprising residues 1011~1193, the D5 domain interact with both the lobe of vRPB2 and zinc ribbon of vRPB9. Additionally, the C-tail (residues 1194~1249) extends from the D5 and inserts into the active center where it interacts with both vRPB1 and vRPB2, occupying a nucleic acid binding position (Fig. 3b and Supplementary Fig. 11). Consequently, M1249L exhibits high modularity serving as an essential scaffold for stabilizing RNAP akin to a cage structure.
Structures of the ASFV RNAP elongation complex
To gain deeper insights into the molecular mechanism underlying the recognition and transcription of substrates by RNAP, we devised a DNA-RNA scaffold. This scaffold was constructed using double-stranded DNA containing a mismatch bubble, following a previously established strategy16. The template strand was derived from an early gene found within the ASFV genome, encoding the vRPB1 subunit of RNAP. To mimic nucleic acid present in actively transcribing complex, an RNA molecule was designed to be complementary to the template strand. Subsequently, native RNAP sample was incubated with a DNA-RNA scaffold and subjected to cryo-EM single-particle analysis (Supplementary Fig. 12). Using unbiased 3D classification, we solved four RNAP conformations in the absence of nucleic acids at a nominal resolution (2.5~2.8 Å). These conformations resembled the M1249L C-tail occupied complexes and core complex. Additionally, we solved a conformation with nucleic acids bound at a resolution of 2.6 Å, representing elongation complex (EC) composed of eight core subunits and nucleic acids in the active center cleft (Fig. 4a).
a The overall structure of the ASFV EC is depicted in cartoon representation. The subunit colors are identical to those in Fig. 2. Helices are represented as cylinders. Nucleic acids are shown in blue (template strand DNA), sky blue (non-template strand DNA) and purple (RNA). The active site magnesium and zinc ions are shown as red and cyan spheres, respectively. b A close-up view of the active center of the ASFV EC. The protein and nucleic acid are represented as sticks. The cryo-EM density is shown as a gray mesh. c Comparison of the active center in ASFV EC with that of the VACV EC (PDB: 6RID16). Residues interacting with nucleic acids are generated by PDBsum46 and are indicated. Residues interacting with nucleic acids are shown as sticks in ASFV EC (green) and VACV EC (gray). Residues specific to ASFV EC are highlighted in red. d A schematic representation of the nucleic acid scaffold used in this study. Individual bases are shown as squares, and the bases are abbreviated using one-letter codes. The bases visible in the EC structure are represented as solid squares, while the invisible bases are shown as hollow squares. The active-site Mg2+ is depicted as a red circle. Residues specific to ASFV EC are highlighted in red.
The EC structure revealed the active state of ASFV RNAP and contained eight core subunits of vRPB1~vRPB10. However, the unusual CTD of vRPB7 is partially obscured by its dynamics. Overall, the ASFV EC closely resembled the MCOC1 except for the absence of the M1249L subunit. A 9 bp DNA-RNA hybrid is bound to the active center cleft, which is consistent with the findings of other RNAPs, which all bind to a hybrid of 8~9 bp31,32. Within the RNAP cleft, no nucleoside triphosphate substrate is present at its binding site, whereas the +1 template base adopts a conformation along the bridge helix suitable for base pairing (Fig. 4b). Therefore, ASFV RNAP EC occurs in an active, post-translocated state. The location and structure of the DNA-RNA hybrid resembled to those of the VACV RNAP EC structure (Fig. 4c). Most residues that participate in nucleic acid interactions are conserved, indicating the conserved fundamental catalytic mechanism of DNA-dependent RNA polymerase. Furthermore, ASFV-specific residues were found to be responsible for nucleic acid binding. The active center cleft primarily comprises charged residues, particularly basic arginine and lysine residues. Residues from vRPB2 are implicated in binding the DNA-RNA hybrid, while those from vRPB1 coordinate the DNA double strands. Notably, at downstream of the template strand DNA, a lysine residue (K100) from vRPB5 interacts with nucleotides (Fig. 4c, d).
The M1249L C-tail occupies the nucleic acid binding site in RNAP
In the EC structure, the DNA-RNA hybrid binds to the active center (Fig. 5a). Notably, the vRPB6 N-tail (residues 9–27), which inserts into the RNAP active center in the MCOC1 structure, was not visible in the EC structure. Upon comparing these two structures, we found that when occupying the active site, M1249L C-tail (residues 1230~1249) significantly clashes with DNA-RNA hybrid. Similarly, the vRPB6 N-tail (D12, D13, L14, and E16) were also found to clash with RNA strand, including U23 and A24 (Fig. 5b). The discovered C-tail of M1249L and N-tail of vRPB6 deeply penetrate into the active center, possibly regulating nucleic acid binding and transcription processes. It suggested that displacement of M1249L C-tail may be necessary for transcriptional activity. Furthermore, the high conservation of M1249L sequences across different ASFV strains, particularly in its C-tail region, suggests its crucial role in regulating viral gene expression during the cellular replication phase or the transition from a packaged state to an actively transcribing state (Supplementary Fig. 13).
a The overall structures of the ASFV RNAP EC and MCOC1 are depicted in cartoon representation. The subunits colors are identical to those in Fig. 2. Helices are represented as cylinders. The nucleic acids are shown in blue (template strand DNA), sky blue (non-template strand DNA), and purple (RNA). Proteins, except for the M1249L C-tail and vRPB6 N-tail in the MCOC1 structure, are shown transparently. b The active center in the EC structure is occupied by the M1249L C-tail. The ASFV RNAP EC and MCOC1 structures are superposed. The nucleic acids in the EC structure, the M1249L C-tail and vRPB6 N-tail in the MCOC1 structure (transparent) are depicted as sticks. c The RNA polymerization activities before and after anti-M1249L antibody purification were assessed by measuring the incorporation of [α−32P]-ATP and normalized against the negative controls lacking RNA polymerase. Each data point represents the mean value of three independent experiments, with error bars indicating standard deviation. Source data are provided as a Source Data file. All calculations were performed using Graphpad Prism 8 software.
To investigate whether M1249L has an inhibitory effect on RNAP activity, we removed RNAP-M1249L from the sample using a home-made mouse anti-M1249L antibody, resulting in a significant reduction of M1249L within the sample (Supplementary Fig. 14). Subsequently, RNA polymerization activity was assessed by measuring the incorporation of [α-32P]-ATP into RNA utilizing dsDNA-RNA as template, followed by precipitation with cold TCA and quantification via scintillation counting22. The comparable activity observed before and after antibody purification implied that M1249L does not inhibit RNAP activity (Fig. 5c). Therefore, M1249L may regulate RNAP activity through a more complicated mechanism, which requires further investigation.
Discussion
ASFV poses a significant threat to global pig production and pork supply due to its high mortality rates and the absence of antiviral drugs and vaccines. Similar to other NCLDVs, ASFV encodes a transcription apparatus for gene expression completion. However, there is limited understanding of the molecular mechanism and temporal regulation of ASFV transcription, which hampers effective prevention and control measures against ASF disease. In this study, we successfully isolated endogenous ASFV RNAP from infected cells and determined its structures in multiple conformations. By comparing these structures with known RNAPs, we not only confirmed the conserved molecular mechanism of multi-subunit RNAPs but also elucidated the features specific to ASFV RNAP.
RNAP represents a promising target for the development of chemotherapeutic antiviral drugs against DNA viruses. The human Monkeypox virus (MPXV) has been reported in 99 countries and territories, with approximately 50,000 confirmed cases33. To identify effective MPXV inhibitors, researchers modeled MPXV RNAP using its homolog VACV RNAP and proposed multiple inhibitors; these candidates were subsequently employed as a starting point for the development of an innovative treatment strategy targeting MPXV34. ASFV RNAP is resistant to the bacterial RNAP and eukaryotic Pol II inhibitors rifampicin and α-amanitin, respectively, suggesting that ASFV RNAP-specific inhibitors may exist35,36. Our structural analysis lays a crucial foundation for studying RNAP-targeted inhibitors, while investigating how inhibitors interact with RNAP in the presence of the M1249L protein may be the next research priority.
The transcription machinery is composed of core subunits and numerous additional factors involved in the initiation, elongation, and termination phases of transcription. These factors are responsible for synthesizing mRNA with 5′-terminal cap and 3′-terminal polyadenylation. Extensive studies on VACV transcription have demonstrated its dependence on multiple transcription factors, including the Vaccinia virus-specific factor Rap94, the termination factor/capping enzyme VTF/CE, the initiation factor VETF, and the helicase NPH-I16,17,18. As a member of NCLDVs, ASFV encodes eight homologs of core subunits as well as homologs of key transcription factors such as VETF, VTF/CE, NPH-I, TFIIB, and TFIIS. These enable precise temporal regulation of ASFV gene expression during different stages of ASFV infection20,21,36,37. In this study, we purified endogenous ASFV RNAP from infected cells with Twin-Strep-tagged vRPB2 and resolved the structure of ASFV RNAP, which included eight core subunits and M1249L. No known transcription factors have been identified, but our purification strategy closely resembles the method employed for isolating the VACV RNAP complete complex16,17,18. This could be attributed to a very low proportion of transcription factors or unstable binding, resulting in our inability to resolve the structure of the transcription factor-bound complex. By comparing MCOC1 with the VACV RNAP complete complex, we observed conservation in the core subunits, suggesting that the core of ASFV RNAP may potentially interact with transcription factors in vivo to carry out its function (Supplementary Fig. 15a, b). Furthermore, it is worth noting that there are some clashes between transcription factors present in VACV RNAP and ASFV RNAP. The transcription factors VETF and CE in VACV RNAP exhibit clashes with vRPB7 and M1249L of ASFV RNAP (Supplementary Fig. 15c). The VACV RNAP subunit Rpo30 resembles to TFIIS and partially overlaps with vRPB1 and M1249L (Supplementary Fig. 15d). M1249L occupies the sites designated for the Rap94 (Supplementary Fig. 15e). These clashes primarily arise from M1249L, suggesting that it may potentially regulate the binding of transcription factors through partial or complete dissociation. A more comprehensive understanding of this regulatory mechanism requires further elucidation through subsequent structural analysis involving a more complete complex configuration.
In this study, we resolved multiple structures of ASFV RNAP that represent various stages of the RNAP transcription process. The dynamic binding mode observed between M1249L and RNAP suggests a potential regulatory mechanism for M1249L in modulating RNAP activity. In the absence of nucleic acids, M1249L can fully attach to RNAP (MCOC1), attach without D2 (MCOC2), without D1-D2 (MCOC3), or without D1-D2-D3-D4 (MCOC4). In cells, these states should be interchangeable with CC state, but the regulatory interactions among them are unclear (Supplementary Fig. 16a and Supplementary Movie 1). Notably, in all M1249L binding states we resolved, the C-terminus of M1249L consistently occupies the nucleic acid binding site on RNAP, suggesting that this occupation may serve as a major functional role for M1249L. Furthermore, upon addition of nucleic acids, we obtained an EC structure which provides evidence for the existence of active RNAP within our sample (Supplementary Fig. 16a). However, it remains challenging to ascertain whether RNAP in the CC state directly binds nucleic acids, or if the M1249L-bound state underwent conformational changes enabling binding. Comparing the conformations of MCOC1 and EC, it can be observed that the active center cleft widens from 26.2 Å to 30.6 Å (Supplementary Fig. 16b and Supplementary Movie 2). Superimposing CC-EC structures reveals minimal changes, except for the absence of vRPB2 protrusion domain, which serves as an entrance to the active cleft. This suggests that the occupancy of active center may contribute to stabilizing this domain. Additionally, dissociation of M1249L leads to complex instability around vRPB7 CTD (Supplementary Fig. 16c). Therefore, it is likely that M1249L plays a significant role in maintaining complex stability and its C-tail should be released for nucleic acid binding. However, further investigation is required to elucidate the molecular mechanism underlying assembly/dissociation of M1249L from ASFV RNAP.
During the course of our manuscript preparation, we observed that a recombinant ASFV RNAP lacking the M1249L subunit has been published22. Upon comparing our CC structure with this recombinant structure, it shows that the overall architecture of the ASFV RNAP remains consistent across different preparation methods. This suggests a high stability in the assembly patterns among the core subunits of RNAP (Supplementary Fig. 17). In our study, we have identified an intriguing viral factor known as M1249L which associates with RNAP and exhibits interactions with nearly all core subunits. Remarkably modular, M1249L envelops the complex and acts as a scaffold to enhance its stability (Fig. 3). Comprising D1 ~ D5 and C-tail domains, M1249L integrates into the RNAP core complex through a specific mechanism. Firstly, the M1249L enhances the stability of the complex by interacting with core subunits and dispersing throughout the entire complex. It forms a cage that potentially hinders the binding of other transcription factors (Supplementary Fig. 15). Additionally, its C-tail occupies the nucleic acid binding site and must be released for transcription (Fig. 5). Furthermore, different states of ASFV RNAP can interconvert through dynamic interactions between multiple domains of M1249L and core subunits (Supplementary Fig. 16a). Considering that M1249L promotes complex stability and occupies the nucleic acid binding site, it is plausible to propose that its multistage binding might regulate ASFV RNAP activity and viral gene expression during cellular replication.
In conclusion, our study reveals the various conformations of ASFV RNAP, confirming the conserved molecular mechanism of multi-subunit RNAPs and elucidating specific features of ASFV RNAP. The peripheral domains of vRPB2, vRPB3-11, vRPB7, and vRPB10 are exclusive to ASFV and may be responsible for binding to virus-specific factors. The dynamic binding mode of M1249L with RNAP suggests an intricate regulation mechanism for M1249L in modulating RNAP activity. These states should be interchangeable with the multiple domains of M1249L binding the core complex dynamically and nucleic acids binding (Supplementary Movie 1). Importantly, these results provide a foundation for mechanistic analysis of the transcription cycle of ASFV and antiviral drug design that is essential in discovering strategies to prevent ASF disease spreading.
Methods
Cell culture and virus propagation
Primary porcine alveolar macrophages (PAMs) were collected from six, 60~70-day-old Duroc-Large White-Landrace cross specific pathogen-free (SPF) piglets by washing porcine lungs twice with PBS. After centrifugation and cell counting, the cells were seeded in T225 cell culture flasks and maintained in 10% fetal bovine serum (FBS) in RPMI 1640 medium (BasalMedia, China) at 37 °C with 5% CO2. The genotype II ASFV CN/GS/2018-GFP was provided by the Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences. The virus titer was measured using the tissue culture infectious dose 50% (TCID50) of the PAMs, calculated by the Reed and Muench method.
Generation of recombinant ASFV CN/GS/2018-strep
ASFV CN/GS/2018-strep was derived from the ASFV CN/GS/2018-GFP strain with a tandem Strep tag (Twin-Strep tag) inserted at the C-terminus of the EP1242L gene (encoding vRPB2). For insertion of the Twin-Strep tag, a pUC57-1242 transfer vector was constructed. An mCherry fragment with the ASFV p72 (B646L gene) promoter (p72mCherry) was inserted at the middle of the intron located between the K421R gene and the EP1242L gene. DNA fragments (termed A and B), flanking approximately 1000 bp of each side of the insertion site of p72mCherry, were amplified via PCR with the primers 1242-L-F/1242-L-R (A fragment) and 1242-R-1-F/1242-R-2-F/1242-R-3-F/1242-R-4-F/1242-R-1-R (B fragment) synthesized by Tsingke Biotech, as listed in Supplementary Table 2. A second round of PCR linked the A fragment and p72mCherry to product C with the primers 1242-L-F/p72mCherry-R. A third round of PCR linked the C and B fragments to D with the primers 1242-L-F/1242-R-1-R. The PCR product D was cloned and inserted into the pUC57 plasmid (MiaoLingBio, China) using the ClonExpress II One Step Cloning Kit (Vazyme, China). The sequence of the resulting construct pUC57-1242 was confirmed. ASFV CN/GS/2018-strep, which can express red fluorescent protein and green fluorescent protein, was constructed by transfecting pUC57-1242 into PAMs using Lipo3000 transfection reagent (Invitrogen, USA) and infecting them with the ASFV CN/GS/2018-GFP strain. The ASFV CN/GS/2018-strep strain was purified by several rounds of limited dilution.
Viral replication analysis
Replications of the recombinant strain ASFV CN/GS/2018-strep and the parental strain ASFV CN/GS/2018-GFP were performed using a TCID50 assay. PAMs were grown in 6-well cell culture plates and infected with virus at a multiplicity of infection (MOI) of 0.01. After incubating for 1 h at 37 °C, the medium was replaced with fresh RPMI 1640 medium, and the samples were collected at 2, 24, 48, and 72 hours post infection (hpi). After the freeze‒thaw cycle, the supernatants and cell lysates were titrated by a TCID50 assay on PAMs. The assay was performed in triplicate.
ASFV RNAP purification
To purify ASFV RNAP from infected cells, PAMs were grown in T225 cell culture flasks. The cells were infected with purified ASFV CN/GS/2018-strep at an MOI of 0.5. After 60 h, the cells were pelleted and resuspended in lysis buffer (50 mM HEPES (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 0.5% [v/v] NP-40, 1 mM DTT, and complete EDTA-free protease inhibitor cocktail). For ASFV RNAP purification, the extract was incubated for 6 h at 4 °C with 60 μL of Strep-Tactin®XT 4 Flow® high capacity (IBA Life Sciences, Germany). The beads were washed five times with wash buffer (50 mM HEPES, pH 7.5; 150 mM NaCl; 1.5 mM MgCl2; 0.1% [v/v] NP-40; and 1 mM DTT) and equilibrated with elution buffer (50 mM HEPES, pH 7.5; 150 mM NaCl; 1.5 mM MgCl2; and 1 mM DTT). The bead-bound proteins were eluted with 50 mM biotin (Sigma Aldrich, USA), resolved via 12.5% SDS‒PAGE and visualized via silver staining. To purify native RNAP, the eluate was concentrated to 1 mg/mL, layered on top of a 10~30% sucrose gradient, and centrifuged for 16 h at 35,000 rpm and 4 °C. The gradient fractions were fractioned manually and separated via SDS‒PAGE, after which the proteins were visualized via silver staining.
To prepare the elongation complex, oligonucleotides (template-stranded DNA, non-template-stranded DNA and RNA) were synthesized by Sangon Biotech (Supplementary Table 2). Native RNAP was purified as described above; then, 4 mM of template strand-RNA scaffold was added, and the sample was incubated at room temperature for 20 min. Next, 8.45 mM non-template strand DNA (corresponding to a scaffold:RNAP molar ratio of approx. 20:1) was added and incubated at room temperature for 20 min.
Separation of RNAP from RNAP-M1249L complexes
To separate RNAP from RNAP-M1249L complexes, we performed purification using mouse anti-M1249L antibody (Wuhan GeneCreate Biological Engineering, China) and Pierce® Crosslink Immunoprecipitation Kit (ThermoFisher Scientific, USA) following the manufacturer’s instructions. For preparation of anti-M1249L antibody, firstly, six 8-week-old SPF Balb/c mice were immunized with the purified M1249L (673-1249aa) protein followed by a serum titer test. Then, the B cells of the mouse with the highest potency (1:128,000) were fused with myeloma cells to obtain hybridoma cells. After western blot validation and subtype identification, hybridoma cell line 1D9-1E7 was selected to prepare ascites. Finally, high-purity monoclonal antibodies were purified from ascites. The titer, sensitivity and specificity of the antibody were verified by Enzyme-Linked ImmunoSorbent Assay (ELISA) and western blot. For purification of RNAP-M1249L complexes, briefly, resin slurry was washed three times with PBS and centrifuged at 1000 g for 1 min. The anti-M1249L antibody was incubated with protein A/G agarose resin for 2 h at room temperature on a flip shaker. After washing three times with elution buffer, the RNAP-M1249L mixture was incubated with protein A/G agarose resin coupled with anti-M1249L antibody for 1.5 h at room temperature and flow-through was collected. The flow-through underwent three rounds of repetitive incubation using fresh resin. Samples before and after antibody purification were analyzed by western blot and silver staining. The mouse anti-M1249L antibody (1:5000 diluted) mentioned above was utilized for the detection of M1249L. The anti-Strep-Tag II monoclonal antibody (mouse, clone 8C12, cat. #ABT2230, lot #ATXD18021, 1:2000 diluted) was procured from Abbkine and utilized for the detection of vRPB2.
In vitro transcription assay
To evaluate the transcription activity of ASFV RNAP, an in vitro transcription assay was conducted. ASFV RNAP was incubated with 500 µM UTP/GTP/CTP, 6 µM ATP, 0.2 µL 3000 Ci/mmol [α-32P]-ATP, and 60 µM dsDNA-RNA/dsDNA template (Sangon Biotech) in 50 µL transcription buffer (25 mM HEPES pH 8.0, 5 mM MgCl2, 1.5 mM DTT, 0.1 mg/ml recombinant albumin (NEB)). Components were mixed on ice and then incubated for 10 min at 37 °C. Reactions were halted by placing on ice and addition of 60 mM EDTA-NaOH pH 8.0 before transferring to 0.8 mL cold 5% Trichloroacetic acid (TCA) and incubated for another 15 min on ice. Precipitated nucleic acids were collected on circular 25 mm Glass Microfiber GF-F Filters (GE Healthcare), washed with ice-cold 5% TCA, and the filter was transferred into 2 mL of Ecoscint A Scintillation Cocktail (National Diagnostics) in a scintillation tube. The signal was measured as counts per minute (CPM) using a Hidex 300 SL instrument (HIDEX) with 1 min signal count time.
Mass spectrometry analysis
The protein sample was subjected to 12.5% SDS‒PAGE and analyzed as described below. The gel bands containing the protein sample were destained and dehydrated in 100% acetonitrile Reduction (10 mM, DTT in 25 mM NH4HCO3 for 45 min at 56 °C) and alkylation (40 mM iodoacetamide in 25 mM NH4HCO3 for 45 min at room temperature in the dark) were performed, and the gel plugs were washed with 50% acetonitrile in 25 mM ammonium bicarbonate twice. The gel plugs were then dried using a SpeedVac and digested with sequence-grade modified trypsin (40 ng for each band) in 25 mM NH4HCO3 overnight at 37 °C. The enzymatic reaction was stopped by adding formic acid to a 1% final concentration. The tryptic peptide mixtures obtained from the digestion were then analyzed by LC‒MS/MS. The nanoLC-MS/MS experiments were performed on a Q Exactive (Thermo Scientific) instrument equipped with an Easy-nLC 1000 HPLC system (Thermo Scientific). The peptides bound on the column were eluted with a 100-min linear gradient. Solvent A consisted of 0.1% formic acid in water, and solvent B consisted of 0.1% formic acid in acetonitrile.
MS analysis was performed with a Q Exactive mass spectrometer (Thermo Scientific). With the data-dependent acquisition mode, the MS data were acquired at a high resolution of 70,000 (m/z 200) across the mass range of 300 ~ 1600 m/z. The raw data from Q Exactive were analyzed with Proteome Discoverer (version 1.4.0.288) using the Sequest HT search engine against the Uniprot_proteome_ASFV database (updated on 11.2019) for protein identification. The Main searching parameters were set as follows: trypsin as the enzyme with up to two missed cleavages allowed; the mass tolerance of precursor was 10 ppm and the product ions tolerance was 0.02 Da; carbamidomethylation of cysteine was selected as a fixed modification amd oxidation of methionine was set as variable modifications. FDR analysis was performed with Percolator, and an FDR < 1% was used for protein identification. The peptide confidence was set as high for the peptide filter.
Structure determination
Following sucrose gradient purification, the fractions containing RNAP were merged and concentrated in a Vivaspin concentrator to a concentration of approximately 1 mg/mL to remove sucrose. For cryo-EM analysis, the purified sample (3 μL) was applied to an H2/O2 glow-discharged, 300-mesh-R1.2/1.3 amorphous nickel-titanium alloy (ANTA) film grid38. The grid was blotted for 9 s at 4 °C and 100% humidity and frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). Cryo-EM images were collected using a 300 kV Titan Krios G3i electron microscope (Thermo Fisher Scientific) equipped with a K3 Summit detector (Gatan). The micrographs were automatically collected using EPU in super-resolution mode, with a nominal magnification of 105,000× and a total accumulated dose of 60 electrons/Å2, resulting in a final pixel size of 0.82 Å. A total of 8576 micrographs for ASFV RNAP and 8849 micrographs for elongation complexes were collected with a defocus range between -1.0 and -2.0 μm.
All image processing and reconstruction were performed in cryoSPARC (v4.1)39 and Relion (v4.0)40. Movies were preprocessed through patch motion correction and patch CTF estimation. Particle picking was performed using a Blob Picker, followed by particle extraction with a box size of 256 pixels (bin2). The particles were then subjected to multiple rounds of 2D classification to remove “junk” particles and to sort different orientations of RNAP. The selected particles were subjected to template picking or topaz picking followed by 2D classification. After three rounds of 2D classification, ~1013 K particles with good 2D averages were chosen and subjected to Relion 3D classification, generating five classes. A map of each class was generated in Chimera (v1.17.1)41. The particles from the good class were selected and subjected to nonuniform refinement42 and subsequent local refinement to yield maps at 2.4~3.0 Å resolution with no symmetry imposed. The local resolution map was calculated using Local Resolution Estimation in cryoSPARC and displayed in Chimera.
For the elongation complex, ~2479 K particles with good 2D averages were chosen and subjected to Relion 3D classification, generating five classes. Then, the particles were subjected to nonuniform refinement42 and subsequent local refinement to yield maps at 2.5~2.8 Å resolution with no symmetry imposed.
The full cryo-EM data processing workflows are described in Supplementary Figs. 2 and 12.
Model building and structure refinement
For the M1249L C-tail occupied complex 1 (MCOC1) structure, the AlphaFold models of the eight core subunits were docked into the map in Chimera and manually adjusted in Coot to acquire the atomic model of ASFV RNAP. The M1249L model was constructed de novo according to density by using DeepTracer and manually adjusted in Coot. Model refinement was performed on the main chain of the atomic model using the real-space refinement module of PHENIX (v1.20.1-4487)43 with secondary structure and geometry restraints to prevent overfitting. After manual adjustment was performed in Coot, the MCOC1 model was subjected to real-space refinement in PHENIX. The models of MCOC2, MCOC3, MCOC4 and the core complex were derived from the MCOC1 model with minor adjustments.
The structure of the elongation complex (EC) was modeled by placing the core complex structure at density, followed by rigid body fitting and adjustment in Coot. The quality of the nucleic acid density is good, and de novo modeling can be performed. The EC structure was real-space refined using PHENIX.
The data validation and model refinement statistics are summarized in Supplementary Table 3.
Figures were created with UCSF Chimera41 and UCSF ChimeraX (v1.7.1)44.
Quantification and statistical analysis
No statistical methods were used to predetermine the sample size. The experiments were not randomized, and the investigators were not blinded to the allocation of the participants during the experiments or outcome assessments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The atomic coordinates of the ASFV RNAP reported in this paper have been deposited in the Worldwide Protein Data Bank (PDB) (http://www.rcsb.org) and are available under accession codes 8XX5 (MCOC1), 8XXT (MCOC2), 8XY6 (MCOC3), 8Y0E (MCOC4), 8XXP (CC) and 8XX4 (EC), respectively. The corresponding maps have been deposited in the Electron Microscope Data Bank (EMDB) (http://emdatabank.org) and are available under accession codes EMD-38746 (MCOC1), EMD-38760 (MCOC2), EMD-38767 (MCOC3), EMD-38802 (MCOC4), EMD-38757 (CC) and EMD-38745 (EC), respectively. The structures used for this study are: 5IYD (H. sapiens Pol II), 6RIC (VACV core RNAP), 6RID (VACV EC), 1WCM (S. cerevisiae Pol II), 7NVS (S. scrofa Pol II), 6RFL (VACV complete RNAP), 8Q3B (ASFV recombinant RNAP). The mass spectrometry data reported in this paper was deposited to the ProteomeXchange Consortium via the PRIDE45 partner repository with the dataset identifier PXD058133. Other source data are also provided with this paper. Source data are provided with this paper.
References
Portugal, R. et al. Related strains of African swine fever virus with different virulence: genome comparison and analysis. J. Gen. Virol. 96, 408–419 (2015).
Gallardo, C. et al. Dynamics of African swine fever virus (ASFV) infection in domestic pigs infected with virulent, moderate virulent and attenuated genotype II ASFV European isolates. Transbound. Emerg. Dis. 68, 2826–2841 (2021).
Dixon, L. K., Chapman, D. A., Netherton, C. L. & Upton, C. African swine fever virus replication and genomics. Virus Res, 173, 3–14 (2013).
Wang, N. et al. Architecture of African swine fever virus and implications for viral assembly. Science 366, 640–644 (2019).
Yang, S. et al. Structure and function of African swine fever virus proteins: current understanding. Front. Microbiol. 14, 1043129 (2023).
Gaudreault, N. N., Madden, D. W., Wilson, W. C., Trujillo, J. D. & Richt, J. A. African swine fever virus: an emerging DNA arbovirus. Front Vet. Sci. 7, 215 (2020).
Zhang, H. et al. Vaccines for African swine fever: an update. Front. Microbiol. 14, 1139494 (2023).
He, Y. et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533, 359–365 (2016).
Abdella, R. et al. Structure of the human Mediator-bound transcription preinitiation complex. Science 372, 52–56 (2021).
Chen, X. et al. Structural insights into preinitiation complex assembly on core promoters. Science 372, eaba8490 (2021).
Plaschka, C. et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 518, 376–380 (2015).
Plaschka, C. et al. Transcription initiation complex structures elucidate DNA opening. Nature 533, 353–358 (2016).
Schilbach, S., Aibara, S., Dienemann, C., Grabbe, F. & Cramer, P. Structure of RNA polymerase II pre-initiation complex at 2.9 A defines initial DNA opening. Cell 184, 4064–4072.e4028 (2021).
Pilotto, S. et al. Structural basis of RNA polymerase inhibition by viral and host factors. Nat. Commun. 12, 5523 (2021).
Jun, S. H. et al. Direct binding of TFEalpha opens DNA binding cleft of RNA polymerase. Nat. Commun. 11, 6123 (2020).
Hillen, H. S. et al. Structural basis of Poxvirus transcription: transcribing and capping Vaccinia complexes. Cell 179, 1525–1536 e1512 (2019).
Grimm, C. et al. Structural basis of Poxvirus transcription: Vaccinia RNA polymerase complexes. Cell 179, 1537–1550.e1519 (2019).
Grimm, C., Bartuli, J., Boettcher, B., Szalay, A. A. & Fischer, U. Structural basis of the complete poxvirus transcription initiation process. Nat. Struct. Mol. Biol. 28, 779–788 (2021).
Zhang, Y. et al. ASFV transcription reporter screening system identifies ailanthone as a broad antiviral compound. Virol. Sin. 38, 459–469 (2023).
Cackett, G. et al. The African swine fever virus transcriptome. J. Virol. 94, e00119–e00120 (2020).
Cackett, G., Sykora, M. & Werner, F. Transcriptome view of a killer: African swine fever virus. Biochem Soc. Trans. 48, 1569–1581 (2020).
Pilotto, S., Sykora, M., Cackett, G., Dulson, C. & Werner, F. Structure of the recombinant RNA polymerase from African Swine Fever Virus. Nat. Commun. 15, 1606 (2024).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Pfab, J., Phan, N. M. & Si, D. DeepTracer for fast de novo cryo-EM protein structure modeling and special studies on CoV-related complexes. Proc. Natl Acad. Sci. USA 118, e2017525118 (2021).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. Biol. Crystallogr 60, 2126–2132 (2004).
Cramer, P. et al. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288, 640–649 (2000).
Cramer, P., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292, 1863–1876 (2001).
Hoffmann, N. A. et al. Molecular structures of unbound and transcribing RNA polymerase III. Nature 528, 231–236 (2015).
Engel, C., Sainsbury, S., Cheung, A. C., Kostrewa, D. & Cramer, P. RNA polymerase I structure and transcription regulation. Nature 502, 650–655 (2013).
Hirata, A., Klein, B. J. & Murakami, K. S. The X-ray crystal structure of RNA polymerase from Archaea. Nature 451, 851–854 (2008).
Cramer, P. Common structural features of nucleic acid polymerases. Bioessays 24, 724–729 (2002).
Martinez-Rucobo, F. W. & Cramer, P. Structural basis of transcription elongation. Biochim Biophys. Acta 1829, 9–19 (2013).
Salcedo, R. M. & Madariaga, M. G. Monkeypox (hMPXV infection): a practical review. Am. J. Med 136, 234–243 (2023).
Abduljalil, J. M. & Elfiky, A. A. Repurposing antiviral drugs against the human monkeypox virus DNA-dependent RNA polymerase; in silico perspective. J. Infect. 85, 702–769 (2022).
Caeiro, F. & Costa, J. V. In vitro transcription by cytoplasmic extracts from cells infected with African swine fever virus. Virology 173, 728–732 (1989).
Kuznar, J., Salas, M. L. & Vinuela, E. DNA-dependent RNA polymerase in African swine fever virus. Virology 101, 169–175 (1980).
Zheng, Y. et al. Transcriptome profiling in swine macrophages infected with African swine fever virus at single-cell resolution. Proc. Natl Acad. Sci. USA 119, e2201288119 (2022).
Huang, X. et al. Reprint of “Amorphous nickel titanium alloy film: a new choice for cryo electron microscopy sample preparation. Prog. Biophys. Mol. Biol. 160, 5–15 (2021).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Kimanius, D., Dong, L., Sharov, G., Nakane, T. & Scheres, S. H. W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem J. 478, 4169–4185 (2021).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput Chem. 25, 1605–1612 (2004).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 (2002).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2021).
Laskowski, R. A., Jablonska, J., Pravda, L., Varekova, R. S. & Thornton, J. M. PDBsum: structural summaries of PDB entries. Protein Sci. 27, 129–134 (2018).
Acknowledgements
We wish to thank the Electron Microscopy Centre (EMC) of Lanzhou University for the microscopy and microanalysis of our specimens. We would also like to thank Yong Peng, Dongsheng Lei, and Xia Deng from the EMC for their help in the data collection. We thank Xiang Ding and Zhensheng Xie from the Institute of Biophysics, Chinese Academy of Science (IBP, CAS), for their assistance with the LC‒MS analysis. Special thanks are due to Dr. Hong-Jie Zhang (Core Facility for Protein Research, IBP, CAS) and Bin Yuan (State Key Laboratory of Respiratory Disease, the First Affiliated Hospital of Guangzhou Medical University) for their technical expertise in radioactivity detection and Dr. Yujia Zhai (IBP, CAS) for her insightful recommendations on experimental design. This work was supported by grants from the National Key R&D Program of China (2021YFD1800100, awarded to Z.-X. Zhu), the Joint Research Foundation of Gansu Province (24JRRA813, awarded to G.-L. Zhu), the Fundamental Research Funds for the Central Universities (awarded to H.-X. Zheng and F. Yang), the Open Competition Program of Top Ten Critical Priorities of Agricultural Science and Technology Innovation for the 14th Five-Year Plan of Guangdong Province (2024KJ14, awarded to H.-X. Zheng), the China Agriculture Research System of Ministry of Finance and Ministry of Agriculture and Rural Affairs (CARS-35, awarded to H.-X. Zheng), the Lanzhou Veterinary Research Institute (CAAS-ASTIP-JBGS-20210101, awarded to Z.-X. Zhu), the Project of National Center of Technology Innovation for Pigs (NCTIP-XD/C03, awarded to H.-X. Zheng), the Innovation Group of Gansu Province (23JRRA1515, awarded to J.-J. He; 23JRRA546, awarded to Z.-X. Zhu), the Major Science and Technology Project of Gansu Province (22ZD6NA001 and 22ZD6NA012, awarded to H.-X. Zheng), and the Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-CSLPDCP-202302 and CAAS-ASTIP-2024-LVRI, awarded to H.-X. Zheng).
Author information
Authors and Affiliations
Contributions
G.Z., F.X., W.Z., Y.Zhao., W.C., C.L., F.Y., Y.R., S.X., S.Z., H.L., H.T., F.Y., B.L., S.S., H.S, B.S., X.Z., L.T., K.L., J.H., J.G., Y.Zhu., and Z.Z. performed experiments; H.Z., F.S., Z.Z., Y.Zhu., and G.Z. designed the study; all authors analyzed data; G.Z., Y.Zhu., and Z.Z. wrote the paper with the substantial input from F.S., and H.Z. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics and facility statements
All animal-related experiments complied with the Animal Welfare Act, Guide for the Care and Use of Laboratory Animals, and were approved by the Gansu Animal Experiments Inspectorate and the Gansu Ethical Review Committee (License no. SYXK [GAN] 2020-0010). All the experiments involving live virus were conducted in biosafety level 3 (BSL-3) facilities at the Lanzhou Veterinary Research Institute (LVRI) of the Chinese Academy of Agricultural Science (CAAS), and approved by the Ministry of Agriculture and Rural Affairs.
Peer review
Peer review information
Nature Communications thanks Clemens Grimm and Linda Dixon for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, G., Xi, F., Zeng, W. et al. Structural basis of RNA polymerase complexes in African swine fever virus. Nat Commun 16, 501 (2025). https://doi.org/10.1038/s41467-024-55683-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55683-z