Abstract
Hypoplastic left heart syndrome (HLHS) is a severe congenital heart disease associated with microcephaly and poor neurodevelopmental outcomes. Here we show that the Ohia HLHS mouse model, with mutations in Sap130, a chromatin modifier, and Pcdha9, a cell adhesion protein, also exhibits microcephaly associated with mitotic block and increased apoptosis leading to impaired cortical neurogenesis. Transcriptome profiling, DNA methylation, and Sap130 ChIPseq analyses all demonstrate dysregulation of genes associated with autism and cognitive impairment. This includes perturbation of REST transcriptional regulation of neurogenesis, disruption of CREB signaling regulating synaptic plasticity, and defects in neurovascular coupling mediating cerebral blood flow. Adult mice harboring either the Pcdha9 mutation, which show normal brain anatomy, or forebrain-specific Sap130 deletion via Emx1-Cre, which show microcephaly, both demonstrate learning and memory deficits and autism-like behavior. These findings provide mechanistic insights indicating the adverse neurodevelopment in HLHS may involve cell autonomous/nonautonomous defects and epigenetic dysregulation.
Similar content being viewed by others
Introduction
Congenital heart disease (CHD) affects up to 1% of live births1. With recent surgical advances allowing infants born with CHD to survive to adulthood, there are now more adults living with CHD than infants born annually with CHD. Unfortunately, people with CHD often suffer developmental delay with high risk of neurodevelopmental impairment that can significantly degrade quality of life2. Thus, a “neurobehavioral signature” has been described that includes learning disabilities, impaired social/communication skills, autism spectrum disorders, and other cognitive, behavioral, and neuropsychiatric deficits2,3. Lack of mechanistic insights into the causes for the neurodevelopmental impairment has precluded the development of targeted therapy for early intervention2.
The major contributing factors for the poor neurodevelopment were previously thought to arise from hypoxic injury and complications related to multiple cardiac surgeries. However, outcome studies have pointed to patient intrinsic factors4,5,6. This is consistent with the findings of small head circumference, microcephaly, and brain abnormalities identified in utero or at birth before cardiac surgery7,8,9,10. That these intrinsic factors may encompass genetic factors is suggested by the finding of de novo pathogenic variants significantly associated with CHD with neurodevelopmental anomalies, such as in chromatin modifiers11. Interestingly, chromatin modifiers are also reported to cause autism, intellectual disability, cognitive impairment, and neuropsychiatric disorders outside of the CHD population12. In addition, genetic disorders and other patient-specific factors such as socioeconomic status have been reported to increase the risk for adverse neurodevelopmental outcome in CHD6,13.
Among people with CHD, the highest morbidity and mortality is observed with hypoplastic left heart syndrome (HLHS), a severe CHD now survivable with a three-stage surgical palliation. However, HLHS is associated with high risk of neurodevelopmental delay and neurobehavioral and cognitive impairment2. This is associated with reductions in functional status and quality of life, increase in behavioral symptoms, poor adaptive skills required for navigating school and daily living, and intellectual disability with deficits in full scale IQ14,15. HLHS is also associated with impaired brain development. Microcephaly, holoprosencephaly, and agenesis of the corpus callosum are observed in 25% of HLHS fetuses2,10. Microcephaly was shown to predict early adverse neurologic outcomes16. While the brain volume reductions were correlated with lower cerebral substrate delivery, neurodevelopmental outcome was not improved after in utero fetal intervention with aortic valvuloplasty, suggesting intrinsic brain deficits17.
To investigate the potential causes of poor neurodevelopment in HLHS, here we leverage the Ohia mouse model of HLHS18. This mouse model was recovered from a large scale forward genetic screen for CHD. It was shown to harbor two recessive mutations that in combination cause HLHS, Sin3A-associated chromatin modifier Sap130, and protocadherin cell adhesion protein Pcdha9 from the α-protocadherin gene cluster.19 Both the Sin3A complex and the clustered protocadherins have essential roles in development of the nervous system. Of note, we previously identified mutations in Sap130 and Pcdha13 in a person with HLHS, highlighting the relevance of these genes to human disease19.
The Sin3A protein is part of the histone deacetylase (HDAC) repressor complex and is essential for brain development. Thus, SIN3A deficiency causes Witteveen-Kolk syndrome, a neurodevelopmental disorder associated with intellectual disability, autism, microcephaly, and facial dysmorphism20. Of note, cardiac abnormalities including thickened aortic valve, atrial septal defect, patent ductus arteriosus, and patent foramen ovale have also been identified in subjects with Witteveen-Kolk syndrome21,22. Also found in the Sin3A complex is MECP2, an X-linked methyl CpG binding protein associated with Rett syndrome, the most common cause of cognitive impairment in females23. Further, the Sin3A complex is known to regulate DNA methylation (DNAm) via interactions with the Tet family of methylcytosine dioxygenases. We note DNAm signatures have been shown to be diagnostic of neurodevelopmental/neuropsychiatric disorders24,25.
The clustered protocadherins also play critical roles in development of the nervous system. They are cell adhesion proteins encoding cell surface diversity that specify neuronal identity patterning synaptic connectivity in the brain26. Mice with deletion of the entire Pcdha gene cluster exhibit deficits in synaptic connectivity and behavioral impairment27. In the human population, the PCDHA gene cluster has been clinically implicated in both autism and CHD. Thus, in a study of 841 families, which included 1,411 children with autism, 5 single nucleotide polymorphisms in the PCDHA gene cluster were found to show significant genetic association with autism28. Further, we previously identified a common deletion in the PCDHA cluster to be significantly associated with HLHS, and bicuspid aortic valve, a CHD phenotype linked to HLHS29.
Leveraging the Ohia mouse model, we examined the impact of Sap130/Pcdha9 mutations on development of the brain, examining structure and differentiation of the brain with histopathological analyses, immunoconfocal microscopy, and molecular profiling with RNA sequencing (RNAseq), Sap130 chromatin immunoprecipitation sequencing (ChIPseq), and genome wide DNA methylation analyses. To investigate the role of Sap130 in the microcephaly phenotype, we generated mice with forebrain-specific deletion of Sap130. As these mice and mice harboring only the Ohia Pcdha9 mutation are adult viable, functional effects of the Sap130 and Pcdha9 mutations were examined with neurobehavioral assessments. Together these studies yielded insights into the cellular and molecular mechanisms contributing to the poor neurodevelopmental outcome clinically associated with HLHS.
Results
Ohia Mutant Mouse Model of HLHS Exhibit Brain Abnormalities
Ohia mutant mice double homozygous for the Sap130/Pcdha9 mutations (Ohiam/m) were previously shown to exhibit HLHS and other CHD with incomplete penetrance19. Analysis of double homozygous mutants (n = 83) showed 56.7% displayed craniofacial/head defects. Of these, 48.2% exhibited both CHD and craniofacial/head defects, 8.5% showed only craniofacial/head defects, 24% showed only CHD, and 19.3% showed neither CHD nor craniofacial/head defects. Severity of the craniofacial/head defects was variable, ranging from mild micrognathia to severe agnathia often accompanied by low set ears, dome shaped head, and eye defects (Figure S1). Examination of gross brain anatomy in Ohia mice (n = 40) from E14.5–E18.5 revealed 81% have forebrain hypoplasia, with 59% exhibiting severe and 22% moderate microcephaly (Fig. 1A–D). In half of the severe cases (49.6%), holoprosencephaly was also observed. Analysis of severe mutants using episcopic confocal microscopy (ECM) for serial section histological 3D reconstruction revealed hypoplastic/aplastic olfactory bulbs, thin cerebral cortex, hypoplasia of the cerebellum, reduction of cerebellar fissures, dilation of the lateral ventricles, and hypoplasia of the corpus callosum (Fig. 1E–J). Quantification confirmed significant cortical thinning, indicating defects in cortical plate development (Fig. 1K–N).
A–D E16.5 wildtype A and Ohiam/m mutant mouse brains exhibiting moderate B and severe C forebrain hypoplasia, with the severe mutant also showing holoprosencephaly. The distribution of brain phenotype severity in Ohiam/m mice is shown in the pie chart D. E–J Episcopic confocal imaging of the head shown in the sagittal and coronal plane of a wildtype (+/+) E, H and two Ohiam/m mice, one with mild F, I and the other with severe G, J forebrain hypoplasia. Note hypoplastic F or absent G olfactory bulb, thin cortex F, G, hypoplastic corpus callosum I, and holoprosencephaly J. K–N. Cresyl violet stained sections from control K, and two Ohia mutants with mild L vs. severe M forebrain hypoplasia. Quantification of 5 Ohia mutants and 5 littermate controls showed reduced cortical thickness N. Graph represents mean ± SD, p = 0.003 with 2-tailed unpaired t-test. Cx cortex, OB olfactory bulb, Cb cerebellum, CC corpus callosum, Lat V lateral ventricle, Holo holoprosencephaly. Scale bars = 0.5 mm. Source data are provided as a Source Data file.
Impaired cortical neurogenesis with reduction of intermediate progenitors
Cortical plate formation is orchestrated via the expansion of neural progenitors in the ventricular and subventricular zone of the developing brain. These progenitors give rise to cells that migrate upwards to form layers II through VI of the cortical plate, with the innermost layer, layer VI, being the first to form, and Layer II, the outermost layer being the last to form. Immunostaining using antibodies for different progenitor lineages and different layers of the cortical plate showed no change in the Pax6+ apical progenitors (radial glial cells) in the ventricular zone, but Tbr2+ intermediate progenitors in the subventricular zone were markedly reduced (Fig. 2A–L). Satb2, a marker for postmitotic neurons in the upper layers of the developing cortex was also markedly reduced, indicating failure to form the later born neurons in layers II-IV (Fig. 2Q–T vs. M-P, quantified in Fig. 2U–Y). In contrast, the Tbr1+ earlier born neurons of layers VI, and Ctip2+ neurons in layer V-VI were present, but abnormally distributed in overlapping domains as compared to their normal pattern of deployment in wildtype mice (Fig. 2Q–T vs. M-P). These observations indicate a cortical neurogenesis defect in the Ohiam/m mutant brain involving failure of the intermediate progenitors to expand, thereby causing deficiency in the later born neurons in layers II-IV.
A–L Sections of the brain of E16.5 control A–D and Ohiam/m mutant mice with mild E–H or severe I–L forebrain hypoplasia were stained with cresyl violet A, E, I to delineate the brain tissue architecture. Antibody staining was conducted for markers of apical progenitor cells (Pax6 in B, F, J), intermediate progenitor cells (Tbr2 in C, G, K), and merged image showing both markers D, H, L. Boxed regions in panels A,E,I are shown in magnified view under darkfield illumination in B–D, F–H, J–L respectively. M–T Section of the cortical plate of E16.5 wildtype (M–P) and mutant (Q–T) mouse brain was stained with antibodies to Satb2, marker for cortical layers II–IV (M, Q), Ctip2 for cortical layers V–VI (O, S), and Tbr1 (N, R) for cortical layer VI, and merged image showing all three markers (P, T). U–Y Quantification of number of cells stained by Pax6 (U, P = 0.9639), Tbr2 (V, P < 0.0001), Ctip2 (W, P = 0.2598), Tbr1 (X, P = 0.3494), and Satb2 (Y, P < 0.0001) in the brain of 5 Ohiam/m mutant and 5 littermate controls. Graphs represent mean ± SD. P-values were calculated by 2-tailed, unpaired t-test. Scale bars in A–L indicate 500 µm. Scale bars in M–T indicate 100 µm. CP cortical plate, SVZ subventricular zone, VZ ventricular zone. Source data are provided as a Source Data file.
Loss of neural progenitors associated with mitotic block and centrosome defects
To elucidate the pathogenic mechanism underlying the cortical neurogenesis defect in the Ohiam/m brain, we quantified cell proliferation and apoptosis. Analysis of E14.5 Ohiam/m brain tissue showed a significant increase in pH3 positive cells in the subventricular zone, and also increased TUNEL throughout the ventricular and subventricular zones (Fig. 3A–F, quantified in G), findings reminiscent of those seen in the Ohiam/m HLHS heart tissue19. The fraction of mitotic cells in anaphase/telophase decreased, indicating metaphase block. This likely accounts for the apparent increase in cell proliferation (Fig. 3H). Similar findings have been reported in mouse models of primary microcephaly and are associated with mitotic spindle defects, such as with mutations in ASPM, Assembly Factor for Spindle Microtubules, and WDR62, another centrosome/spindle protein, two genes that account for over 50% of primary microcephaly30,31.
A–H Comparison of the brain cortex from E14.5 Ohiam/m mutant (D–F) vs. wildtype littermate embryos (A–C) showed more pH3 and TUNEL staining (G) in the ventricular (VZ) and subventricular (SVC) zones in the mutants. A decrease was also noted in mitotic cells in anaphase-telophase (H, n = 3, Ohiam/m mutants, n = 3 littermate controls). I–K Centrosomes were visualized by confocal microscopy in E14.5 brain tissue sections from 4 Ohiam/m mutant (n = 766) and 3 wildtype control (n = 1394) embryos using γ-tubulin (green) antibody staining. Coimmunostaining with Tbr2 was conducted to track cells in the intermediate zone that will give rise to cortical layers II–IV. 3D serial confocal imaging stacks were collected to investigate centrosome distribution. Commonly observed were paired centrosomes of similar size, predicting normal bipolar spindles I. Also observed were abnormal juxtapositioning of three or more centrosomes of varying sizes that would predict multipolar spindle defects J. The insets show magnified views of the same regions indicated by the arrowheads. Quantification showed a significant increase of the abnormal centrosomes in the mutant brain tissue (J, K). L–R. Confocal imaging was conducted using MEFs from wildtype (n = 3) (L-N) and Ohiam/m mutant embryos (n = 3) (O–Q) with α-tubulin and γ-tubulin used to visualized the mitotic spindles. >500 cells analyzed per embryo. Mutant MEFs showed increase in multipolar spindles (R). S–U. Ohiam/m MEFs showed reduction in the proportion of mitotic cells in anaphase/telophase (S) and an increase in lagging chromosomes during anaphase (T, U) Q, R. CP cortical plate, IZ intermediate zone, SVZ subventricular zone, VZ ventricular zone, P/M prophase/metaphase, A/T anaphase/telophase. Scale bars = 100 µm. All graphs represent mean ± SD. p-values were obtained using 2-way, unpaired t-test. Source data are provided as a Source Data file.
To investigate further the role of spindle defects in the Ohia micrcephaly phenotype, we conducted confocal microscopy of E14.5 brain tissue sections to investigate for centrosome abnormalities as a proxy for spindle defects. The limited resolution of histological sections precluded direct visualization of the spindle apparatus. Centrosomes visualized with a γ-tubulin antibody showed mostly paired centrosomes of similar size representing mother and daughter centrioles, an arrangement consistent with normal bipolar spindles (Fig. 3I). Also observed, but much less frequently, were occasional abnormal clusters of three or more centrosomes of varying sizes that would predict mitotic spindle defects (Fig. 3J). Quantification showed such centrosome abnormalities are significantly increased in the Ohia mutant brain tissue (Fig. 3K; p = 0.058 × e−3), supporting the involvement of spindle defects in the microcephaly. This was associated with the Satb2 expressing cells that will give rise to the later formed cortical layers II-IV, which are largely absent in the Ohia mutant brain (Fig. 2I–K).
To confirm the presence of mitotic spindle defects, we also generated mouse embryonic fibroblasts (MEFs) from Ohiam/m mutant embryos and littermate controls and conducted high resolution confocal microscopy to visualize the spindle apparatus in the MEFs plated as monolayers. The MEFs were immunostained with α-tubulin to visualize the spindle apparatus and γ-tubulin for the centrosome. This revealed a marked increase in multi-polar spindles in the Ohiam/m MEFs, which is seldom seen in wildtype MEFs (Fig. 3L–N vs. 3I–K, quantified in Fig. 3). This was associated with an increase of mitotic cells at prophase/metaphase (P/M), while mitotic cells at anaphase/telophase (A/T) decreased, indicating a mitotic block (Fig. 3P). A mitotic spindle defect was further supported by the observation of a significant increase in lagging chromosomes in mitotic cells at anaphase (Fig. 3Q,R). Together these findings provide further evidence supporting a spindle defect contributing to the loss of later born neurons in cortical layers II-IV in the emergence of microcephaly in Ohiam/m mutant mice.
Dysregulated gene expression and defects in neurodevelopment
Transcriptome profiling was conducted with RNAseq analysis of E13.5-E14.5 brain tissue from Ohiam/m mutants with severe brain phenotype (n = 3) and wildtype littermate controls (n = 5). This yielded 1549 differentially expressed genes (DEGs), 1081 downregulated and 468 upregulated (FDR 0.05; Supplementary Data 1). Metascape analysis yielded many nervous system related terms, including forebrain development, synapse assembly, glutamatergic synapses, and learning and memory (Fig. 4A). Ingenuity Pathway Analysis (IPA) recovered three upstream regulators: REST - repressor element 1 silencing transcription factor, which was upregulated, and DTNBP1: dystrobrevin binding protein 1, and SGK1: serum glucocorticoid regulated kinase 1, both downregulated (Fig. 4B). The recovery of REST is particularly notable in the context of microcephaly, as REST controls expansion of neural progenitors via recruitment of HDACs to the Sin3A complex32. DTNBP1 regulates neurotransmitter release and receptor signaling, while SGK1 regulates glutamate receptor expression and long-term potentiation33,34. These upstream regulators and their downstream genes are predicted to perturb biological processes that are neurobehavioral related (Fig. 4B).
Differentially expressed genes (DEGs) recovered from RNASeq analysis of E14.5 Ohia mutant brain were analyzed for pathway enrichment using Metascape (A), Ingenuity Pathway Analysis (IPA) for upstream regulators of downstream neurodevelopmental outcomes (B, red for upregulated, green for downregulated), IPA canonical pathway (C), IPA curated CREB signaling pathway (D, purple outline indicates genes found to be differentially expressed, green fill indicates downregulated and red fill indicates upregulated DEGs), as well as ToppGene Disease analysis (E).
Further IPA canonical pathway analysis recovered cyclic AMP response element binding protein (CREB) signaling, synaptic depression/potentiation, synaptogenesis signaling, GABA receptor signaling, and calcium signaling (Fig. 4C; Supplementary Data 1). Associated with the CREB signaling pathway is reduction in glutamate receptor signaling and downregulation of PKC, PLC, PKA, calmodulin, and CAMKII/CAMK4 (Fig. 4D). This pathway has critical roles in synaptic plasticity mediating long term memory. Interestingly, also recovered was circadian rhythm, likely a reflection of the known role of CREB in circadian rhythm regulation (Fig. 4C)35. ToppGene disease pathway enrichment identified autism, mental depression, intellectual disability, seizures, neurodegenerative diseases, and neuropsychiatric disorders (Fig. 4E, Supplementary Data 1).
NO signaling defect and the perturbation of neurovascular coupling
The top IPA pathway recovered from the Ohia brain RNAseq analysis was neurovascular coupling, an autoregulatory mechanism by which the brain’s energy demands are closely coordinated with cerebral blood flow (Fig. 4C, Fig. 5, Supplementary Figs. 2–4)36. Such cerebral autoregulation is mediated by neuronal activity dependent production of nitric oxide (NO), which promotes vascular smooth muscle relaxation and dilation of cerebral arteries to increase blood flow to the brain (Supplementary Fig. 2, 3). This cerebral autoregulation, which has been described in the context of CHD, causes enhanced cerebral perfusion providing a brain sparing effect37. In the Ohia mutant brain, we observed marked downregulation of NO synthase 1 (NOS1) (Fig. 5; FDR = 8.03E-14, Supplementary Data 1) and guanylate cyclase, a NO receptor and sensor generating cGMP, a second messenger mediating NO signaling with downstream repression of protein kinase G (PKG) to modulate vasodilation and neuronal activity (Fig. 5). In contrast to NOS and guanylate cyclase, PKG expression was upregulated, perhaps a compensatory response to the reduction in NO and cGMP (Fig. 5). However, as cGMP is required to activate the enzymatic function of PKG, PKG overexpression without elevation of cGMP would not recover PKG enzymatic activity. Interestingly, the IPA analysis also returned “Effect of sildenafil”, a drug commonly used to promote vasodilation by enhancing NO levels via inhibition of phosphodiesterase which breaks down cGMP (Fig. 4C). As NO signaling also has additional roles in CREB regulated neuronal gene expression important in learning/memory, NO deficiency may also cause learning/memory deficits38.
Correlation of DNA methylation with transcriptomic profiles and brain phenotypes
We investigated for possible involvement of epigenetic regulation in the brain defects of the Ohia mutant mice, as the Sin3A-HDAC repressor complex to which Sap130 is bound is known to regulate DNAm by recruitment of Tet DNA demethylases39. This was investigated with methylome analysis of DNA from the forebrain tissue of E15.5 Ohiam/m mutant mice (n = 3) with severe microcephaly phenotype vs. C57BL/6 J wildtype mice (n = 3) using the Illumina Infinium mouse methylome array containing probes for over 285,000 CpG sites genome wide39. This analysis recovered 5,117 differentially methylated regions (DMRs) associated with 4,179 genes (FDR < 0.05; Supplementary Data 3). By way of example, we show CpG methylation associated with a DMR in a highly conserved genomic region at the terminal exon of Klf13, a known negative regulator of CREB signaling essential for axonogenesis and neurite outgrowth (Fig. 6A)40.
A Increased methylation of CpG sites in a DMR found in Klf17 in the Ohiam/m brain tissue. B Venn diagrams show two-way intersection of downregulated DEGs with genes associated with DMR and three-way intersection of down regulated DEGs, genes associated with DMR, and genes recovered from Sap130 ChiPSeq. C, D ToppGene enrichment analysis for Mouse Phenotypes (C) and Diseases (D) recovered from the two-way intersection of the downregulated DEGs with genes associated with DMRs.
Intersection of DEGs that are downregulated with DMR associated genes identified 297 genes (Fig. 6B, Supplementary Data 3). ToppGene pathway enrichment analysis of these 297 intersecting genes for Mouse Phenotypes recovered terms related to brain, behavior, and neurological function (Fig. 6C, Supplementary Data 3), and for Disease, we observed autism, intellectual disability, epilepsy, and other neurological diseases known to be associated with HLHS (Fig. 6D; Supplementary Data 3). In contrast, only 61 genes were recovered from interrogating the DEG/DMR intersecting genes comprising the upregulated DEGs, with pathway enrichment analysis recovering mostly metabolic pathways (Supplementary Fig. 7, Supplementary Data 3).
These observations suggest DNAm changes associated with the downregulated genes may drive the adverse neurodevelopmental outcome associated with HLHS. To investigate this possibility further, we analyzed DNAm in Ohia mutants with mild phenotype comprising those with little or no forebrain hypoplasia (Supplementary Fig. 5). DMR analysis yielded only 1921 genes as compared to 4179 genes for DNAm analysis of the severe brain phenotype mutants (Figure S6A). In the severe mutants, 2539 (60%) of the 4179 genes were exclusive to the severe phenotype, but for the mild brain phenotype, only 343 (17.9%) of 1921 genes were exclusive to the mild brain phenotype (Supplementary Fig. 6A). Analysis of all the DMR associated genes in the mild phenotype mutants using ToppGene DisGeNET focused analysis yielded only 6 of 10 top neurological diseases obtain from all DMR associated genes of the severe phenotype, all showing much lower p-values (Supplementary Data 3). For example, the p-value for schizophrenia is e-23 for severe and e-9 for mild, or for neurodevelopmental disorder, e-07 for severe and e-02 for mild (Supplementary Data 3). Similar interrogation of DMRs exclusive to the severe or mild mutants yielded no significant disease terms for mild, while the same disease terms with lower p values were observed for the severe phenotype (Supplementary Fig. 6B; Supplementary Data 3). Additional analysis of DMR associated genes shared between the mild and severe mutants yielded only two disease terms, schizophrenia and intellectual disability, both with low p-values (Supplementary Fig. 6B; Supplementary Data 3). Overall, these observations support DNAm playing a role in the transcriptional changes associated with Ohia mutant brain phenotype severity.
Sap130 epigenetic regulation of neurogenesis and neurological disease pathways
We further investigated whether Sap130 may contribute to the epigenetic regulation by conducting Sap130 chromatin immunoprecipitation sequencing (ChIPseq) analysis to identify potential targets of Sap130 regulation. Sap130 ChIPseq analysis of E13.5 wildtype mouse brain tissue yielded 16,231 Sap130 binding sites encompassing 10,753 genes (Supplementary Data 2). Among the genes recovered were transcription regulators of neurogenesis such as Rest, Sin3A, Ctcf, Satb2, Klf13, and also microcephaly-related genes such as Aspm and Wdr62 (Fig. 7A). Transcription factor binding site analysis identified enrichment for SP, KLF, IRF, ELF, and RFX motifs (Fig. 7B). SP and KLF are closely related zinc finger proteins that bind GC rich promoters, and are commonly associated with pluripotency and cell cycle genes41. KLFs, Kruppel like transcription factors, also have important roles in brain and heart development, with KLF2 regulating vascular tone in response to blood flow40,42,43.
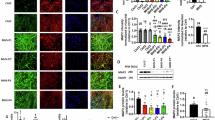
A Sap130 occupancy in the 5’ upstream promoter region of selected genes. B Motif enrichment analysis of the Sap130 ChIPseq target genes identified putative Sap130 DNA binding sequences. C–F Pathways in GO Biological Process (C) and Cellular Components (D), as well as Human Phenotype (E), and Disease (F) terms recovered from ToppGene gene enrichment analysis of Sap130 ChIPseq target genes. G–I Cilia visualization in sections of wildtype (G) and Ohiam/m mutant brain tissue (H) immunostained with Arl13b (cilia, green), γ-tub (centrosome, red) and DAPI. Quantification showed reduction of cilia in the mutant brain (I, n = 3 Ohia mutant and 4 littermate controls). The graph represents the mean ± SD, with P = 0.0274 determined by 2-tailed, unpaired t-test. Magnification in panels G and H are the same, with scale bar = 200 µm. Source data are provided as a Source Data file.
Pathway enrichment analysis yielded GO biological processes comprising chromosome organization, cell cycle progression, and neurogenesis (Fig. 7C). GO cellular components identified mitochondrion, centrosome, glutamatergic synapse and transcriptional regulation (Fig. 7D). Human Phenotype and Disease Pathway enrichment analyzes identified many of the same terms seen with the down regulated DEGs, such as microcephaly and intellectual disability (Fig. 7E). Among Human Phenotype recovered were aplasia/hypoplasia of the cerebrum, decreased head circumference, hypotonia, and for Disease terms, neurodevelopmental delay, neurodegenerative disorders, epilepsy, autism, and ciliopathies (Fig. 7E, F, Supplementary Data 4). As ciliopathies are often associated with neurodevelopmental defects, we investigated ciliogenesis in the Ohia mutant brain and showed a ciliation defect (Fig. 7G–I). Overall, these findings point to a role for Sap130 in the epigenetic regulation of gene expression changes underlying the brain defects in the Ohia mutant mice.
To further assess impact of the Sap130 mutation on DNAm and transcriptional changes in the Ohia mutant brain, we examined three-way intersection of the DEGs, the DMR associated genes, and genes recovered from the Sap130 ChIPseq analysis. Such three-way intersection of the downregulated DEGs yielded 161 genes (Fig. 6B), while only 43 genes were recovered with three-way intersection of the upregulated DEGs (Supplementary Fig. 7). Pathway enrichment for the downregulated DEGs for Mouse Phenotype and Disease terms yielded similar results to that seen in analysis of the DEG and DEG/DMR intersecting genes (Fig. 8A, B, Supplementary Data 3). In contrast, no significant Mouse Phenotype or Disease terms were recovered from analysis of the upregulated DEGs. This suggests the genes contributing to the neurological diseases and phenotypes are targets of Sap130 mediated epigenetic repression.
A Circle plots showing Disease and Mouse Phenotype association identified by ToppGene enrichment analysis of the down regulated DEGs (first column), genes that are both down regulated DEGs and DMR associated (second column) and genes that are down regulated DEGs, DMR associated, and ChIPseq target genes (third column). All three analyzes yielded the same Disease and Mouse Phenotype terms, but with differing p-values. B Circle plots showing Disease and Mouse Phenotype associations identified via ToppGene analysis of the genes with direct physical overlap of the DMRs and the ChIPseq binding sites.
To investsigate this possibility further, the DMRs and Sap130 ChIPseq peaks with direct physical overlap were recovered and genes localized in the DMR/ChipSeq peaks were identified. This yielded 1366 genes (Supplementary Data 3), of which 968 had DMRs at the transcriptional start site (Supplementary Data 3). Focusing on the latter genes, we conducted ToppGene analysis and recovered many of the same or similar Disease and Mouse Phenotype pathways (Fig. 8B) as observed with analysis of the intersecting genes independently recovered from the RNAseq and DMR analyses (Fig. 8A; Supplementary Data 4). Together these findings suggest Sap130 mediated epigenetic repression may underlie the transcriptomic changes observed in the Ohia mutant mice.
Protein–protein interactome network analysis
To further functionalize the downregulated/differentially methylated genes in the Ohia brain, STRING-db was used to construct a protein-protein interaction (PPI) network (Supplementary Data 5). The PPI network assembled comprised 132 genes, 82 from the 127 hypomethylated/downregulated genes (FDR = 0.05) (Supplementary Fig. 8A) and 50 additional genes recovered by STRING-db (Supplementary Fig. 8A; Supplementary Data 5). ToppGene analysis yielded significant terms for Mouse Phenotype, Human Phenotype, Disease, and Pathway. In Human Phenotype, we recovered polymicrogyria (Supplementary Fig. 8B), a defect involving abnormal small cortical folds in the forebrain, a phenotype that has previously been reported among human fetuses with HLHS and in primary microcephaly44,45. Also relevant to clinical findings in people with HLHS, we observed abnormal hippocampal morphology and multiple seizure related terms (Supplementary Fig. 8B). For Mouse Phenotype, terms such as cerebral hemisphere morphology, abnormal forebrain morphology, abnormal neuron morphology, and abnormal inhibitory postsynaptic currents were recovered (Supplementary Fig. 8C). Disease terms recovered included several also seen with the DMR/DEG analysis, such as neurodevelopmental disorder, autism, impaired cognition, and epilepsy (Supplementary Fig. 8D). Interestingly, Pathway enrichment yielded multiple cilia related terms, with the top pathway being “cargo targeting to cilium”, and also “cargo trafficking to periciliary membrane” and “cilium assembly” (Supplementary Fig. 8E). These findings further point to the disturbance of cilia, supporting the recovery of ciliopathies in the Sap130 ChIPseq analysis, and the ciliation defect observed in the Ohia mutant brain.
Forebrain specific deletion of Sap130 causes microcephaly
To investigate cell autonomy of the microcephaly phenotype in the Ohiam/m mutant mice, we conducted Cre deletion using the forebrain specific Emx1-Cre driver together with a Sap130 floxed allele. This Cre driver is expressed in excitatory neurons and glia in the developing cerebral cortex from E9.5 onwards46. The Emx1-Cre:Sap130f/− mice are adult viable without CHD and yet they had microcephaly with smaller forebrain (Fig. 9A). This was associated with a reduction in the brain to body weight ratio (Fig. 9B), suggesting the microcephaly is cell autonomous and unrelated to altered cardiovascular hemodynamics. Unlike the Ohia mutant, this phenotype showed complete (100%) penetrance in the Emx1-Cre:Sap130f/− mice. Real time PCR analysis showed marked reduction in Sap130 transcripts in the forebrain (Fig. 9C). Among the 28 genes known to cause primary microcephaly, many of which are centrosome related, four were up regulated in the Emx1-Cre:Sap130f/− forebrain (Wdr62, Cit, Ncapd2, Pcnt; Supplementary Data 6), and confirmed by real time PCR analysis (Wdr62 shown in Fig. 9D). We note centrosome amplification can cause spindle defects, which is one of the pathogenic mechanisms associated with primary microcephaly47. These findings indicate a cell autonomous requirement for Sap130 in expansion of neural progenitors, a deficiency that likely underlies the microcephaly phenotype in the Ohia mutant mice.
A Wildtype newborn control (left) and Emx1-cre Sap130f/− mutant (right) brains. Scale bar = 500 µm. B Brain/body weight ratio for newborn control (n = 18) and Emx1-cre Sap130f/− (n = 6) mice confirm brain hypoplasia in the Emx1-cre Sap130f/− mice. C qPCR of Sap130 transcripts in E14.5 forebrain tissue from 4 control and 4 Emx1-cre Sap130f/− mice showed decrease in Sap130 transcripts, demonstrating efficacy of the Emx1-Cre mediated Sap130 deletion. The graph represents the mean ± SD, with P = 0.0108 determined by 2-tailed, unpaired t-test. D qPCR of Wdr62 transcripts in E14.5 forebrain tissue from 4 control and 4 Emx1-cre Sap130f/− mice showed increase transcript expression, indicating possible centrosome amplification. The graph represents the mean ± SD, with P = 0.0307 determined by 2-tailed, unpaired t-test. E–H IPA and ToppGene analysis of DEGs from RNAseq of E14.5 forebrain tissue from Emx1-cre Sap130f/− vs. control mice. Shown are the results obtained with IPA Canonical Pathway analysis (E), Gene Set analysis (F) for hallmark centrosome, hypoxia, and mitotic spindle genes, and ToppGene analysis for Mouse Phenotype (G) and Disease (H). Source data are provided as a Source Data file.
RNAseq analysis of the E14.5 forebrain of Emx1-cre:Sap130f/− mice (n = 3) and littermate controls (n = 3) recovered 369 DEGs, 202 down and 167 upregulated (FDR < 0.1; Supplementary Data 6). IPA pathway enrichment analysis of the DEGs identified axonal guidance as the top pathway, followed by cell cycle, Wnt signaling, and semaphorin signaling in neurons (Fig. 9E). Also observed were GADD45 signaling, a stress activated pathway induced by DNA damage and cell cycle checkpoint control, DNA break repair, and HOTAIR regulation - a lncRNA regulating cell cycle and apoptosis via epigenetic gene silencing by providing a scaffold for recruitment of proteins in the Sin3A complex such as REST/Co-REST and lysine demethylase LSD1 (Fig. 9E). Gene set enrichment also recovered “Centrosome” and “Mitotic Spindle”, further supporting mitotic spindle defects, and “Hypoxia” was also observed, despite the normal cardiovascular system in the Emx1-cre:Sap130f/− mice (Fig. 9F).
Mouse Phenotype terms recovered included abnormal neuronal differentiation, abnormal forebrain morphology, abnormal limbic system, small olfactory bulb, abnormal neural precursor, abnormal social interaction and abnormal neocortex (Fig. 9G). For “Disease” enrichment, intellectual disability was recovered as the top pathway, and additionally many Disease terms were recovered that were also seen in the Ohia mutant mice such as impaired cognition, neurodevelopmental disorders, microcephaly, and autism (Fig. 9H). Cerebrovascular accidents and hypoxic-ischemic encephalopathy were also identified, suggesting a role for hypoxia in the neurological deficits independent of any cardiac defects (Fig. 9H). Together these findings suggest microcephaly in Ohia mutant mice is not secondary to CHD, but may reflect a cell autonomous requirement for Sap130 in normal forebrain neurodevelopment.
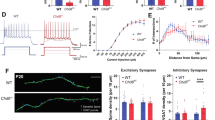
Adult Pcdha9 m/m and Emx1-Cre deleted Sap130 mice exhibit behavioral deficits
The molecular profiling of the Ohiam/m fetal brain predicts impaired cognitive function with possible learning/memory defects and autism spectrum disorder. To assess for these neurobehavorial deficits, we generated viable adult mice homozygous for the Pcdha9 mutation, and mice with forebrain deletion of Sap130 (Emx1-Cre:Sap130f/−). Brain anatomy of the Emx1-cre:Sap130f/− and Pcdha9m/m adult mice were analyzed using anatomical brain MRI. The Emx1-cre:Sap130f/− mice showed significant reduction in total forebrain volume, and volume of the corpus callosum, cortex, and hippocampus (Fig. 10A–F). In the Pcdha9 mutant mice, only mild hippocampal dysplasia was observed but no other appreciable changes in brain structure or size (Fig. 10G, H). Neurobehavioral phenotypes were further assessed using three tests to evaluate spatial learning and memory, fear associative learning, and sociability. These tests were administered on 18 female and 6 male Pcdha9m/m mice and 6 male and 3 female Emx1-cre:Sap130f/− mice. For the Pcdha9m/m mice, equal number of sex and age matched C57BL/6 J wildtype mice were assessed as controls. For the Emx1-cre:Sap130f/− mice, littermate controls were used comprising Emx1-Cre:Sap130f/+ mice and Sap130f/+ mice without Emx1-cre.
A–F MRI of the brain of adult wildtype (A), and Emx1-cre;Sap130f/− mice (B) showed significant reduction in brain volumes associated with the corpus callosum (C, n = 4 control, n = 4 mutant), the cerebral cortex (D, n = 8 control, n = 8 mutant), the hippocampus (E, n = 8 control, n = 8 mutant), and forebrain (F, n = 4 control, n = 8 mutant). The values shown represent mean ± SD. P-values were obtained using 2-way unpaired t-test. G, H MRI of the wildtype (G) and adult Pcdha9m/m mutant (H) mouse brain showed no detectable change in anatomical structure. Scale bars in panels A, B, G, H = 500 µm. I–N. Neurobehavioral testing was conducted on Pcdha9m/m mice (I, K, M) and Emx1-cre:Sap130f/− mice (J, L, N) using three tests: Morris water maze (I, n = 18 control, n = 18 mutant; J, n = 11 control, n = 9 mutant), cued fear conditioning (K, n = 10 control, n = 19 mutant; L, n = 9 control, n = 9 mutant), and three chamber sociability test (M, n = 8 control, n = 8 mutant; N, n = 10 control, n = 7 mutant). In Pcdha9m/m female mice and male/female Emx1-cre:Sap130f/− mice, significant deficits were observed for fear conditioning (K, L) and sociability (M, N), but not in the Morris water maze (I, J). Similar analysis of Pcdha9m/m male mice showed no change relative to wildtype male mice (Supplementary Fig. 10). Morris water maze analyzed by 2-way repeated measures ANOVA, fear conditioning analyzed by 2-way repeated measures ANOVA, and sociability tests analyzed by two-way and three-way ANOVA. All graphs represent mean ± SD. Source data are provided as a Source Data file.
Spatial learning and memory were interrogated using the Morris water maze whereby mice were trained to find a hidden platform in a pool of water. After training, no significant difference in the time (latency) required to find the hidden platform was observed for either the Pcdha9m/m or Emx1-Cre:Sap130f/− mice vs. controls (Fig. 10I, J, Supplementary Figs. 9,10). In the fear conditioning test, mice learn to associate a cue (a tone) with an adverse event (foot shock) administered after the cue. After training, mice typically freeze upon hearing the cue, but return to normal activity post cue. Emx1-Cre:Sap130f/− mice and Pcdha9m/m female, but not male mice, showed increased post-cue freezing, indicating impaired associative learning (Fig. 10K, L; Supplementary Figs. 9, 10). We also conducted sociability testing in which mice are presented with a novel object vs. a novel mouse. While wildtype and littermate control mice showed preference for the novel mouse (Fig. 10M, N), Emx1-Cre:Sap130f/− mice and Pcdha9m/m female mice, but not male mice, showed equal time spent exploring the novel object and novel mouse (Fig. 10M, N, Supplementary Figs. 9, 10), indicating social interaction deficits. It was not possible to assess gender effects in the Emx1-Cre:Sap130f/− mice given the smaller cohort size.
Discussion
CHD poses increased risk for adverse neurodevelopmental outcome that can significantly degrade the quality of life. While clinical studies have shown this is associated with brain abnormalities and neurobehavioral impairment, the underlying mechanisms remain unclear. Here, our studies of the Ohia mouse model of HLHS show the Pcdha9 and Sap130 mutations causing HLHS also can cause brain and neurodevelopmental abnormalities. This is congruent with the fact that many genes highly expressed in the heart are also highly expressed in the brain11. Moreover, de novo mutations linked to CHD are enriched for genes known to cause neurodevelopmental defects11. In Ohia HLHS mouse model, Pcdha9 is part of the Pcdha gene cluster known to provide unique cell surface barcodes required for patterning neural circuit assembly, while Sap130 is part of the Sin3A-HDAC repressor complex known to regulate the expansion of neural progenitors. Mutations in both genes have been clinically associated with various adverse neurological/neurobehavioral outcomes.
We showed Ohia fetal mice exhibit brain defects comprising forebrain hypoplasia with a spectrum of phenotype severity that spans overt microcephaly to milder phenotype with modest to no obvious forebrain hypoplasia. We also observed a cortical neurogenesis defect with deficiency of later born neurons in cortical layers II-IV. This cortical neurogenesis defect was observed regardless of whether the brain is microcephalic. The cortical neurogenesis defect was associated with the loss of intermediate neural progenitors in the subventricular zone. We showed this is likely from a centrosome defect causing multipolar spindle formation with mitotic block at metaphase. A cortical neurogenesis defect has also been reported clinically with MRI interrogation of HLHS fetuses; volume reduction was noted in the cortical subplate and intermediate and ventricular zones48. Transcriptome profiling of the Ohia mutant brain indicated the disturbance of REST, a master transcriptional regulator that recruits HDACs to the Sin3A complex to epigenetically regulate neural progenitor expansion and neural stem cell maintenance32,49,50,51.
Microcephaly with loss of intermediate progenitors has also been observed with disturbance of REST via HDAC deficiency and with mutations in ZNF335, another component of the Sin3A complex52,53. Together these findings suggest the Sap130 mutation via the Sin3A complex may be the main driver of the microcephaly phenotype in the Ohia mutant mice. Consistent with this, microcephaly was also observed with forebrain targeted deletion of Sap130 with Emx1-Cre. With Emx1-Cre, Sap130 deletion is expected in excitatory neurons and glia in the forebrain. While microcephaly showed complete penetrance in the Emx1-Cre:Sap130f/− mice, in the Ohia mutant penetrance was incomplete with the forebrain hypoplasia spanning a spectrum. This is likely due to the hypomorphic nature of the Ohia Sap130 mutation vs. creation of a Sap130 null allele by Cre recombinase.
The recovery of “centrosome” in the Sap130 ChIPseq analysis, and “centrosome” and “mitotic spindle” in the RNAseq analysis of the Emx1-Cre:Sap130flx/− mutant would suggest centrosome defect may also contribute to the microcephaly phenotype. This is consistent with the well described association of centrosome mutations and multipolar spindle defects in primary microcephaly. Wdr62, one of the major genetic causes of primary microcephaly54, has been shown to cause multipolar spindle defects, and CHD subjects with rare variants in Wdr62 have been shown to have multipolar spindles and impaired cardiomyocyte proliferation, suggesting the tantalizing possibility that centrosome defects may contribute to hypoplasia of both the brain and the heart55. In Ohia mutant brain, we observed elevated expression of Wdr62 and several other microcephaly associated centrosome genes, suggesting centrosome amplification, a pathogenic mechanism also seen in primary microcephaly47,56. Importantly, Wdr62 mutations have been implicated in autism, and neuronal deletion of Wdr62 in mice caused impaired learning/memory and social interaction deficits57.
Unexpectedly, the Sap130 ChIPseq analysis yielded “ciliopathy”. Multiple cilia-related pathways were also recovered in our PPI network analysis. As cilia assembly is templated on a basal body derived from the centrosome, centrosome defects can contribute to defects in ciliogenesis. Consistent with this, we documented a ciliation defect in the Ohia mutant brain. An increased burden for ciliopathy related variants have been reported in subjects with HLHS. Conversely subjects with various ciliopathies have been reported to have HLHS58. As HLHS and ciliopathies are both rare birth defects, their cooccurrence would suggest a mechanistic link. The finding of holoprosencephaly among Ohia mutants and people with HLHS would suggest a defect in cilia transduced hedgehog signaling59.
Disease pathway enrichment in the molecular profiling yielded mostly neurological diseases such as autism, intellectual disability, and impaired cognition, neurobehavioral deficits also seen clinically in HLHS. This is supported by our mouse neurobehavioral assessments which showed autism-like social interaction deficits and impaired learning/memory. This was observed in both the Emx1-Cre:Sap130f/− mice with microcephaly and the Pcdha9 mutant mice with normal brain anatomy, indicating these two genes may act on convergent pathways. Our molecular profiling would suggest this may involve the disturbance of CREB signaling, which can cause cognitive impairment, and perturbation of SGK1, an upstream regulator of glutamate receptor expression with important roles in learning and hippocampal long term potentiation34,35. As CREB signaling is known to regulate dopaminergic neurons implicated in autism, this may also contribute to the social interaction deficits60. CREB disturbance also has been linked to neurodegenerative memory related diseases such as Alzhemier’s61, and relevant to this, HLHS and other single ventricle CHD subjects have been reported to have elevated risk for early onset dementia61,62.
It is notable that all the neurological phenotypes and diseases recovered from molecular profiling of the Ohia and Emx1-Cre:Sap130f/− mutant mice were associated with downregulated DEGs. As these DEGs are also differentially methylated and are targets of Sap130 binding, this would suggest neurodevelopment is epigenetically regulated by Sap130. As expression of Pchda9 and other genes with the CTCF motif requires promoter demethylation mediated by TET demethylases recruited via the Sin3A complex63,64, this would suggest transcription from the Pcdha gene cluster is subject to epigenetic regulation by Sin3A-Sap13065. This convergence could explain the similar neurobehavioral presentations of the Emx1-Cre:Sap130f/− and Pcdha9 mutant mice. Disturbance of DNAm in the clustered protocadherins has been associated with various neurodevelopmental disorders, including William syndrome with aortic valve defects66. In William syndrome, there is increase in DNA methylation with hypermethylation of Pcdha9 and other genes in the clustered protocadherins reported67. Supporting the important role of DNAm in neurodevelopmental disorders22, DNAm signatures have been identified for over 40 neurodevelopmental disorders, including Witteveen-Kolk syndrome with Sin3A mutations.
We also observed brain volume reduction in the Ohia mutant mice, consistent with in utero imaging of human fetuses with HLHS68,69. This was correlated with reduced brain oxygenation, a finding indictive of a disruption of neurovascular coupling - a cerebral autoregulatory mechanism that normally shunts blood to the brain during hypoxia37. Clinical studies with in utero ultrasound imaging showed impairment of such cerebral autoregulation with HLHS44. The recovery of hypoxemic encephalopathy in transcriptome profiling of the Emx1-Cre:Sap130f/− mouse brain would suggest neurovascular coupling disturbance may occur independent of altered cardiovascular hemodynamics. Impaired neurovascular coupling in CHD patients has been reported in a study using blood oxygen level dependent (BOLD) brain MRI70. This was associated with low NO, consistent with the reduction in NOS seen in the Ohia mutant mice. Importantly, the CHD patients with low NO also showed impaired cognitive function, supporting the importance of neurovascular coupling in maintaining brain health70.
Overall, our findings in the Ohia mutant mice point to a developmental etiology for the brain abnormalities associated with HLHS. One limitation of our study is the lack of single cell RNAseq data for defining the transcriptome profile on the single cell level. Nevertheless, we expect gene expression changes observed are likely to reflect the underlying cell/tissue composition alterations seen with thinning of the cortex and varying degrees of microcephaly, phenotypes seen both in the Ohia mutant mice and in patients with HLHS. Supporting congruence of the mouse and human HLHS findings, some of the same disease pathways seen with HLHS such as intellectual disability, impaired cognition, and autism spectrum disorder are observed both in the RNAseq and the Sap130 ChIPseq analysis.
Regarding clinical relevance of the Ohia mutations in human HLHS, we observed pathways identified as significantly perturbed in the Ohia mutant brain are shared with HLHS patients, thus providing insights into pathomechanisms underlying the neurodevelopmental deficits associated with HLHS. However, whether Sap130/Pcdha9 mutations may cause HLHS in the human population cannot be addressed currently, given any genomic sequencing analysis of HLHS patients will likely be underpowered given the known genetic heterogeneity19,71,72 and multigenic etiology19,73 of HLHS. Nevetheless, it is interesting to note that we previously identified a subject with HLHS harboring mutations in Sap130 and Pcdha13, the latter being another gene in the Pcdha gene cluster19 that is closely related to Pcdha9.
In summary, our study uncovered cell autonomous and nonautonomous defects together with epigenetic dysregulation contributing to the neurodevelopmental defects in the Ohia HLHS mouse model. We also uncovered a corticoneurogenesis defect associated with centrosome abnormalities causing the loss of intermediate progenitors. Microcephaly was observed, but with variable penetrance that may also involve centrosome defects, but likely affecting an earlier forebrain progenitor population. Our findings pointing to a prominent role for DNAm changes in the adverse neurodevelopment in HLHS would suggest altenrative avenues for therapy to improve neurodevelopmental outcome with manipulation of the epigenome, such as via diet or pharmacological interventions74. We expect these findings will have broad clinical relevance beyond HLHS, as adverse neurodevelopmental outcome is observed among CHD of a broad spectrum.
Methods
Animal studies
Mouse studies were conducted under an approved University of Pittsburgh Institutional Animal Care and Use Committee animal study protocol (21120410). Mice were housed at the Rangos Research Center Animal Facility, a pathogen free facility, with temperatures maintained at 20–26 °C with 30–70% humidity under a 12 h dark/light cycle. The adult mice used in this study do not have any overt disease phenotype, and are checked twice a day to ensure they have adequate food and water and are in good health. Euthanasia was performed by exposure to a highly concentrated environment of C02 gas as recommended by the American Veterinary Medical Association.
Mouse strain and husbandry
CRISPR gene edited Ohia and Pcdha9 (c.2389_2399del; [p.Asp796Phefs∗]) mice, and mice with the floxed and knockout Sap130 allele were generated in our previous study19 and maintained in the C57BL/6 J background. The B6.129S2-Emx1tm1(cre)Krj/J mice with the Emx1-cre driver were purchased from Jackson Laboratory (Strain #005628) and intercrossed with Sap130f/f and Sap130+/− mice to generate Emx1-cre positive Sap130f/− animals for analysis19. Oligonucleotide primer sequences used for genotyping these various different mutant and CRISPR gene edited mice are provided in Supplementary Data 7. Mice 2-6 months old were bred to obtain fetal mice for the various histological, and cell and molecular profiling studies of the fetal mouse brain. The neurobehavioral assessments were initiated with adult mice at 6-8 weeks of age and spanned a period of three weeks, beginning with the sociability testing, followed by the Morris water maze, and finally the fear conditioning test.
Magnetic resonance imaging
Following behavioral testing, mice underwent in vivo brain MRI carried out using a Bruker BioSpec 70/30 USR spectrometer (Bruker BioSpin MRI, Billerica, MA, USA) operating at 7-Tesla with quadrature radio-frequency volume coil with inner-diameter of 35 mm75. For in vivo MRI, mice were anesthesized using inhaled isoflurane and then maintained with 1–2% iLeisoflurane in oxygen and transferred to the animal bed for imaging. Different brain areas, including the hippocampus, cerebral cortex, cerebellum, corpus callosum and forebrain, were manually defined by 2 blinded independent operators in ITK-SNAP, and volume for each brain region was calculated.
Immunostaining and confocal microscopy
Ohiam/m fetuses were obtained at E14.5, E155, and E16.5, the head was removed and drop fixed in 4% PFA overnight, and then processed for cryoembedding. Frozen sections were collected for immunostaining. Cryosections were stained with antibodies to Pax6 (BioLegend; 901301), Tbr1 (Abcam; ab31940), Tbr2 (EB Bioscience; 14-4875-80), Satb2 (Abcam; ab51502), Ctip2 (Abcam; ab18465), pH3 (Abcam; ab197502), Arl13b (Neuromab, N295B/66). TUNEL labeling was also performed with TUNEL assay kit from Roche (12156792910). Sample size for each experiment is listed in associated figure legends. Confocal imaging was conducted with the Leica SP8 confocal microscope and quantitatively analyzed with ImageJ. Figures were prepared with Adobe photoshop.
Mouse embryonic fibroblast analysis
Mouse embryonic fibroblasts (MEFs) were isolated from E14.5–E15.5 embryos, with three independent MEF lines analyzed. Cells were plated on glass coverslips, fixed in 4% PFA, stained with antibodies to α-tubulin (Abcam; ab15246), and γ-tubulin (Sigma; T6557) and counterstained with DAPI (Thermo Fisher Scientific; D1306).
Histological reconstructions using episcopic confocal microscopy
E14.5 or newborn mice were euthanized, the heads were removed, fixed in 4% paraformaldehyde, followed by embedding in paraffin for espicopic confocal microscopy (ECM)19. This entailed sectioning the paraffin blocks using a Leica sledge microtome mounted with a Leica LSI confocal macroscope that captures images of the block face at the microtome’s fixed photoposition. The 2D serial image stacks thus collected were digitally resliced in different imaging plane and also 3D reconstructed using the OsiriX Dicom viewer (https://www.osirix-viewer.com) to assess for changes in brain anatomical structures.
RNA sequencing
Total RNA was isolated from whole brain tissue samples collected from 3 Ohiam/m animals with severe brain defects and 5 littermate control animals at E13.5–E14.5. Of the Ohiam/m animals used for this analysis, two showed HLHS heart defects while the third showed aortic defects. Additional analysis was conducted on 3 Emx1-cre:Sap130f/− and 3 littermate control Sap130f/− animals without Emx1-cre using the RNeasy plus mini kit (Qiagen). Libraries were constructed with a TruSeq RNA Sample Preparation Kit v2 (Illumina) and sequenced with an Illumina HiSeq 2000 platform (BGI Americas) with 100-bp paired-end reads. Reads were aligned to mm10 (NCBI build 38) with TopHat2 (v2.0.9)76, and gene-level counts were calculated with HTSeq-count (v0.5.4p5)77. Differential expression analyses were performed with edgeR78, and mouse sex and litter were included as covariates in the analysis. Differentially expressed genes were recovered for Ohiam/m mutant tissue with false discovery rate (FDR) ≤ 0.05 (Benjamini–Hochberg) and no fold change cut off, and from Emx1-cre:Sap130f/− with FDR < 0.1 and no fold change cut off. In total, 1,549 DEGs in the Ohiam/m brain and 369 DEGs in the Emx1-cre:Sap130f/− forebrain were identified and used to perform pathway analysis with Ingenuity (www.ingenuity.com), ToppFun from the ToppGene Suite79, and Metascape80. For the protein-protein interaction analysis, genes differentially expressed were recovered using FDR cut-off of 0.05. Genes with logFC>0 were considered up-regulated while log FC < 0 were considered down-regulated.
Chromatin immunoprecipitation sequencing
Sap130 chromatin immunoprecipitation was conducted with E13.5 embryonic brain tissue from wildtype C57BL/6 J mice performed using iDeal ChIP–seq Kit for Transcription Factors (Diagenode) with rabbit anti-Sap130 antibody (A302-491A, Bethyl laboratories). Cross-linking was performed with 1% formaldehyde and stopped with glycine, and samples were sonicated to 100–300-bp fragments with a Covaris S2 instrument. Sonicated lysates were incubated with Sap130 antibody-bound Protein A magnetic beads (2 μg antibody per 20 μl beads). ChIP–seq libraries were then generated with an NEBNext Ultra DNA Library Prep Kit Illumina (NEB), and sequencing was carried out on an Illumina HiSeq 4000 platform (BGI Americas). Reads were aligned to the mouse reference genome (mm10) with Bowtie1 (version 1.1.2)81, and potential Sap130 target regions were identified with MACS1.4.2 software82, and pathway analysis was carried out using ToppFun from the ToppGene Suite. Motif enrichment analysis was performed using MEME suite version 5.4.183.
DNA methylation analysis
For methylation analysis, forebrain tissue samples were collected from 3 Ohiam/m mice with severe head phenotype, 3 Ohiam/m mice with mild head phenotype, and 3 wildtype C57BL/6 J control mice, all at E15.5 and flash frozen in liquid nitrogen. DNA was isolated using the QIAamp DNA mini kit (Quiagen) and analyzed using the Illumina Infinium mouse methylation beadchip to assess DNA methylation (Illumina; 20041558). Bisulphite conversion of 250 ng DNA was carried out using the EZ DNA Methylation™ Kit (Zymo Research Corp., CA), and bisulfite DNA samples were applied to the Infinium arrays and hybridized 16–24 hours at 48°C. After further posthybridization processing, the beadchip was scanned using an Illumina iSCAN and the data analyzed using Genome Studio. Raw intensities were read into R using the R package minfi (v 1.36.0)84, and R methylation manifest and annotation packages to use with minfi were created using the Manifest and Annotation files provided by Illumina.
Samples with >1% of sites with detection p-value > 0.01 were removed. Normal-exponential out-of-band (NOOB) normalization was performed. Poor quality probes with ≥20% of samples with detection p-value > 0.01 were filtered, as well as probes on sex chromosomes, and those that overlap SNPs were removed. Samples were pooled using the shared set of probes and final beta values were calculated. Probe-level differential methylation analyzes was performed using the limma package85, including sex as covariate in the design matrix. Probes with an FDR ≤ 0.1 were considered significantly differentially methylated. Further, differentially methylated regions (DMRs) were identified using the R package DMRcate (v 2.4.1)86, with sex as a covariate, with an FDR of 0.05. DMR with maxdiff > 0 were considered hypermethylated regions and those with maxdiff <0 were considered hypomethylated regions. Pathway analysis was carried out using ToppFun from the ToppGene Suite.
Protein–protein interaction analysis of downregulated genes using STRING-db
Gene expression with logFC>0 were considered up-regulated, while logFC <0 were considered down-regulated, and those overlapping with hypermethylated regions were considered hypermethylated and those overlapping with hypomethylated regions were considered hypomethylated genes. Genes were categorized based on differential DNA methylation and RNA expression, with the analysis focusing on the downregulated genes, yielding hypermethylated and hypomethylated down regulated genes. These two gene sets were then used to retrieve interacting genes using the String-db database (STRING, https://string-db.org) to predict functional protein-protein interactions, and the top 50 interactors retrieved were then incorporated into an expanded network which was used for ToppGene pathway enrichment analysis.
Mouse behavioral phenotyping
Mouse behavior was assessed using three training paradigms: the Morris water maze, fear conditioning, and sociability. Mice were group housed by sex during testing. For mice that underwent all three of these tests, testing was completed in order of least stressful to most stressful with sociability being conducted first followed by the Morris water maze, and then fear conditioning. On the day of testing, mice were transferred to the testing facility and allowed to habituate for at least one hour prior to test start. Testing apparatuses were cleaned with 70% ethanol between each animal tested. Testing was completed by operator blinded to genotype. For all tests, mice were weighed on the day of testing, and mice with loss of >10% of baseline weight between test days were excluded from analysis. Additionally, if mice appeared ill including little to no movement, piloerection, abnormal posture, or trouble breathing, they were excluded from analysis. This resulted in exclusion of 3 Emx1cre Sap130f/− mice total.
Morris water maze
The Morris water maze87,88 set-up consisted of a pool of 90 cm in diameter and 60 cm high filled with water (depth 28 cm, 20–22 °C) located in a 2.5 × 2.5 meter room with multiple extra-maze cues (black shapes on white wall) that remained constant throughout the experimental studies. A hidden platform 10 cm in diameter and 27 cm high (i.e.,1 cm below the water’s surface) was used to assess the mouse’s ability to learn spatial relations between distal cues and escape via this platform. Mice were given 120 sec per trial to find the hidden platform, and if the mouse failed to find the platform after 120 sec it was placed on the platform. Following each trial, mice remained on the platform for 30 sec before being moved to a heated incubator between trials. Mice were given 4 trials per day, starting at each of the 4 quadrants of the pool, and were tested over a period of 5 days. On Day 6 a probe test was completed, and testing was also done with a visible platform. The water maze pool and incubator were cleaned and disinfected daily. A video tracking system (AnyMaze) in conjunction with a digital camera mounted over the pool was used to record and quantitate the swimming behavior of the animals.
Fear conditioning
The fear conditioning assessment89 was conducted using an apparatus consisting of a box (30.5 × 24.1 × 21 cm, Medical Associates) with an electric grid floor (1.6 cm spacing, 4.8 mm diameter rods). Animals initially underwent a training protocol in which they were placed in the box and after 120 sec, a 10 sec tone (75 dB, 32.8 kHz) was given, and during the last second of this tone, a 1 mA 1 sec long footshock was delivered through the grid floor. The tone and footshock co-terminated. The tone and foot shock were given sequentially for a total training period of 9 min, with a 60 sec intertrial interval. 24 hours after training, contextual fear conditioning was assessed by placing the animals in the box, and freezing was measured for 3 min. One hour after the assessment of contextual fear conditioning, cued fear conditioning was assessed by placing the animals in an altered chamber (the box was cleaned and scented with a novel odorant, a white noise of 75 dB is given, and a paper roll lined with print is placed within the box to hide the walls). The animal’s freezing behavior was measured for 3 min during which the tone (75 dB, 32.8 kHz) is delivered for the last minute, yielding the freezing behavior measurement with “cue”. No footshocks were delivered during this assessment. Following the cue the animal is observed for an additional minute to measure “post-cue” freezing behavior. Freezing behavior was quantified using a video-based analysis system (FreezeFrame, Coulburn Instruments) and verified by manual scoring.
Sociability
Mouse social behavior was examined using a three-chambered social approach task90,91. We used a video camera and EthovisionXT video tracking software (Noldus) to follow subject mice as they freely explored a three-chamber sociability cage. A test mouse was placed into the center compartment of the sociability cage, which is a box consisting of three compartments. In the other two compartments, we placed either a mouse with which the test mouse was unfamiliar, or an empty cage. At the beginning of the experiment the test mouse was placed into the middle chamber and habituated for 5 min. Following habituation, the novel mouse and empty cage were placed into either side of the three-chamber apparatus and the test mouse was able to freely explore each compartment for 10 min. The amount of time the test mouse spent with the empty cage versus with the unfamiliar mouse was quantified.
Statistics and Reproducibility
Data were analyzed using GraphPad Prism 9 (GraphPad Software). No statistical methods were used to pre-determine sample size. Sample sizes for immunostaining, RNA sequencing, and DNA methylation experiments were chosen to ensure an n of at least 3 for each condition analyzed and no data was excluded from these analysis. For cell quantification independent samples T-test was used. Statistics for the RNAseq, ChIPseq, and DNAm analyzes are described in the respective Methods section above detailing these specific experiments. Sample size for behavioral testing was based on the number of mice of each genotype that could be generated with full capacity breeding, with 3 mice excluded due to poor health. The testing was conducted in a blinded fashion with investigators blinded to the animal genotype. For mouse behavioral analysis, data were analyzed by two-way, two-way repeated measures, or three-way analysis of variance (ANOVA), with a p-value < 0.05 considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNAseq, ChIPseq, and DNA Methylation raw and processed data are publicly available via the Gene Expression Omnibus (GEO) database under accession codes GSE283523 and GSE283526 for the RNAseq, GSE283524 for the ChIPseq, and GSE283525 for the DNA methylation analyzes). Source data are provided with this paper.
References
Hoffman, J. I. & Kaplan, S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39, 1890–1900 (2002).
Marino, B. S. et al. Neurodevelopmental outcomes in children with congenital heart disease: evaluation and management: a scientific statement from the American Heart Association. Circulation 126, 1143–1172 (2012).
Marelli, A., Miller, S. P., Marino, B. S., Jefferson, A. L. & Newburger, J. W. Brain in congenital heart disease across the lifespan. Circulation 133, 1951–1962 (2016).
Marelli, A., Miller, S. P., Marino, B. S., Jefferson, A. L. & Newburger, J. W. Brain in congenital heart disease across the lifespan: the cumulative burden of injury. Circulation 133, 1951–1962 (2016).
Gaynor, J. W. et al. Patient characteristics are important determinants of neurodevelopmental outcome at one year of age after neonatal and infant cardiac surgery. J. Thorac. cardiovascular Surg. 133, 1344–1353. e3 (2007).
Newburger, J. W. et al. Early developmental outcome in children with hypoplastic left heart syndrome and related anomalies: the single ventricle reconstruction trial. Circulation 125, 2081–2091 (2012).
Hinton, R. B. et al. Prenatal head growth and white matter injury in hypoplastic left heart syndrome. Pediatr. Res. 64, 364–369 (2008).
Miller, S. P. et al. Abnormal brain development in newborns with congenital heart disease. N. Engl. J. Med. 357, 1928–1938 (2007).
Owen, M., Shevell, M., Majnemer, A. & Limperopoulos, C. Abnormal brain structure and function in newborns with complex congenital heart defects before open heart surgery: a review of the evidence. J. Child Neurol. 26, 743–755 (2011).
Glauser, T. A., Rorke, L. B., Weinberg, P. M. & Clancy, R. R. Congenital brain anomalies associated with the hypoplastic left heart syndrome. Pediatrics 85, 984–990 (1990).
Homsy, J. et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350, 1262–1266 (2015).
Schanen, N. C. Epigenetics of autism spectrum disorders. Hum. Mol. Genet. 15, R138–R150 (2006).
Bucholz, E. M. et al. Socioeconomic status and long-term outcomes in single ventricle heart disease. Pediatrics 146, e20201240 (2020).
Goldberg, C. S. et al. Behavior and Quality of Life at 6 Years for Children With Hypoplastic Left Heart Syndrome. Pediatrics 144 (2019).
Siciliano, R. E. et al. Cognitive function in pediatric hypoplastic left heart syndrome: systematic review and meta-analysis. J. Pediatr. Psychol. 44, 937–947 (2019).
Hangge, P. T. et al. Microcephaly is associated with early adverse neurologic outcomes in hypoplastic left heart syndrome. Pediatr. Res. 74, 61–67 (2013).
Laraja, K. et al. Neurodevelopmental outcome in children after fetal cardiac intervention for aortic stenosis with evolving hypoplastic left heart syndrome. J. Pediatr. 184, 130–136.e4 (2017).
Li, Y. et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 521, 520–524 (2015).
Liu, X. et al. The complex genetics of hypoplastic left heart syndrome. Nat. Genet. 49, 1152–1159 (2017).
Witteveen, J. S. et al. Haploinsufficiency of MeCP2-interacting transcriptional co-repressor SIN3A causes mild intellectual disability by affecting the development of cortical integrity. Nat. Genet. 48, 877–887 (2016).
Uctepe, E. et al. A Witteveen-Kolk Syndrome Patient with Reflux Disease and a de novo Deletion of the SIN3A Gene. Molecular Syndromology, 1–6 (2024).
Coenen-van der Spek, J. et al. DNA methylation episignature for Witteveen-Kolk syndrome due to SIN3A haploinsufficiency. Genet Med. 25, 63–75 (2023).
Amir, R. E. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188 (1999).
Shirvani-Farsani, Z., Maloum, Z., Bagheri-Hosseinabadi, Z., Vilor-Tejedor, N. & Sadeghi, I. DNA methylation signature as a biomarker of major neuropsychiatric disorders. J. Psychiatr. Res. 141, 34–49 (2021).
Aref-Eshghi, E. et al. Evaluation of DNA methylation episignatures for diagnosis and phenotype correlations in 42 mendelian neurodevelopmental disorders. Am. J. Hum. Genet. 106, 356–370 (2020).
Thu, C. A. et al. Single-cell identity generated by combinatorial homophilic interactions between α, β, and γ protocadherins. Cell 158, 1045–1059 (2014).
Mountoufaris, G. et al. Multicluster Pcdh diversity is required for mouse olfactory neural circuit assembly. Science 356, 411–414 (2017).
Anitha, A. et al. Protocadherin α (PCDHA) as a novel susceptibility gene for autism. J. Psychiatry Neurosci. 38, 192–198 (2013).
Teekakirikul, P. et al. Common deletion variants causing protocadherin-alpha deficiency contribute to the complex genetics of BAV and left-sided congenital heart disease. HGG Adv. 2, 100037 (2021).
Nicholas, A. K. et al. The molecular landscape of ASPM mutations in primary microcephaly. J. Med. Genet. 46, 249–253 (2009).
Nicholas, A. K. et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 42, 1010–1014 (2010).
Gao, Z. et al. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 31, 9772–9786 (2011).
Bray, N. J. et al. Haplotypes at the dystrobrevin binding protein 1 (DTNBP1) gene locus mediate risk for schizophrenia through reduced DTNBP1 expression. Hum. Mol. Genet. 14, 1947–1954 (2005).
Lang, F., Strutz‐Seebohm, N., Seebohm, G. & Lang, U. E. Significance of SGK1 in the regulation of neuronal function. J. Physiol. 588, 3349–3354 (2010).
Gau, D. et al. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron 34, 245–253 (2002).
Stackhouse, T. L. & Mishra, A. Neurovascular coupling in development and disease: focus on astrocytes. Front Cell Dev. Biol. 9, 702832 (2021).
Donofrio, M. et al. Autoregulation of cerebral blood flow in fetuses with congenital heart disease: the brain sparing effect. Pediatr. Cardiol. 24, 436–443 (2003).
Riccio, A. et al. A nitric oxide signaling pathway controls CREB-mediated gene expression in neurons. Mol. Cell 21, 283–294 (2006).
Williams, K. et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011).
Ávila-Mendoza, J., Subramani, A. & Denver, R. J. Krüppel-like factors 9 and 13 block axon growth by transcriptional repression of key components of the cAMP signaling pathway. Frontiers in Molecular Neuroscience, 217 (2020).
Jiang, J. et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. cell Biol. 10, 353–360 (2008).
Lavallée, G. et al. The Kruppel‐like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 25, 5201–5213 (2006).
Dekker, R. J. et al. Endothelial KLF2 links local arterial shear stress levels to the expression of vascular tone-regulating genes. Am. J. Pathol. 167, 609–618 (2005).
Kinnear, C. et al. Abnormal fetal cerebral and vascular development in hypoplastic left heart syndrome. Prenat. Diagn. 39, 38–44 (2019).
Bhat, V. et al. Mutations in WDR62, encoding a centrosomal and nuclear protein, in Indian primary microcephaly families with cortical malformations. Clin. Genet. 80, 532–540 (2011).
Gorski, J. A. et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314 (2002).
Zhang, Y. et al. Overexpression of WDR62 is associated with centrosome amplification in human ovarian cancer. J. ovarian Res. 6, 1–6 (2013).
Rollins, C. K. et al. Regional brain growth trajectories in fetuses with congenital heart disease. Ann. Neurol. 89, 143–157 (2021).
Qureshi, I. A., Gokhan, S. & Mehler, M. F. REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle 9, 4477–4486 (2010).
Ballas, N., Grunseich, C., Lu, D. D., Speh, J. C. & Mandel, G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657 (2005).
Huang, Y., Myers, S. J. & Dingledine, R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 2, 867–872 (1999).
Tang, T. et al. HDAC1 and HDAC2 regulate intermediate progenitor positioning to safeguard neocortical development. Neuron 101, 1117–1133.e5 (2019).
Yang, Y. J. et al. Microcephaly gene links trithorax and REST/NRSF to control neural stem cell proliferation and differentiation. Cell 151, 1097–1112 (2012).
Chen, J.-F. et al. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat. Commun. 5, 1–13 (2014).
Hao, L. et al. WDR62 variants contribute to congenital heart disease by inhibiting cardiomyocyte proliferation. Clin. Transl. Med. 12, e941 (2022).
Marthiens, V. et al. Centrosome amplification causes microcephaly. Nat. cell Biol. 15, 731–740 (2013).
Xu, D. et al. WDR62-deficiency Causes Autism-like Behaviors Independent of Microcephaly in Mice. Neurosci. Bull. 39, 1333–1347 (2023).
Geddes, G. C., Stamm, K., Mitchell, M., Mussatto, K. A. & Tomita-Mitchell, A. Ciliopathy variant burden and developmental delay in children with hypoplastic left heart syndrome. Genet. Med. 19, 711–714 (2017).
Nanni, L. et al. The mutational spectrum of the sonic hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Hum. Mol. Genet. 8, 2479–2488 (1999).
Servili, E., Trus, M., Sajman, J., Sherman, E. & Atlas, D. Elevated basal transcription can underlie timothy channel association with autism related disorders. Prog. Neurobiol. 191, 101820 (2020).
Saura, C. A. & Valero, J. The role of CREB signaling in Alzheimer’s disease and other cognitive disorders. Rev. Neurosci. 22, 153–169 (2011).
Bagge, C. N. et al. Risk of dementia in adults with congenital heart disease: population-based cohort study. Circulation 137, 1912–1920 (2018).
Salas-Armenteros, I. et al. Human THO-Sin3A interaction reveals new mechanisms to prevent R-loops that cause genome instability. EMBO J. 36, 3532–3547 (2017).
Grunseich, C. et al. Senataxin mutation reveals how r-loops promote transcription by blocking DNA methylation at gene promoters. Mol. Cell 69, 426–437 e7 (2018).
Canzio, D. et al. Antisense lncRNA transcription mediates DNA demethylation to drive stochastic protocadherin α promoter choice. Cell 177, 639–653.e15 (2019).
El Hajj, N., Dittrich, M. & Haaf, T. Epigenetic dysregulation of protocadherins in human disease. Semin Cell Dev. Biol. 69, 172–182 (2017).
Strong, E. et al. Symmetrical dose-dependent DNA-methylation profiles in children with deletion or duplication of 7q11. 23. Am. J. Hum. Genet. 97, 216–227 (2015).
Sadhwani, A. et al. Fetal brain volume predicts neurodevelopment in congenital heart disease. Circulation 145, 1108–1119 (2022).
Limperopoulos, C. et al. Brain volume and metabolism in fetuses with congenital heart disease: evaluation with quantitative magnetic resonance imaging and spectroscopy. Circulation 121, 26–33 (2010).
Schmithorst, V. J. et al. Impaired neurovascular function underlies poor neurocognitive outcomes and is associated with nitric oxide bioavailability in congenital heart disease. Metabolites 12, 882 (2022).
Hinton, R. B. Jr. et al. Hypoplastic left heart syndrome is heritable. J. Am. Coll. Cardiol. 50, 1590–1595 (2007).
McBride, K. L. et al. Linkage analysis of left ventricular outflow tract malformations (aortic valve stenosis, coarctation of the aorta, and hypoplastic left heart syndrome). Eur. J. Hum. Genet. 17, 811–819 (2009).
McBride, K. L. et al. Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: Segregation, multiplex relative risk, and heritability. Am. J. Med. Genet. A 134A, 180–186 (2005).
Rasmi, Y. et al. The role of DNA methylation in progression of neurological disorders and neurodegenerative diseases as well as the prospect of using DNA methylation inhibitors as therapeutic agents for such disorders. IBRO Neurosci. Rep. 14, 28–37 (2023).
Shang, P. et al. Paired immunoglobulin-like receptor B is an entry receptor for mammalian orthoreovirus. Nat. Commun. 14, 2615 (2023).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Chen, J., Bardes, E. E., Aronow, B. J. & Jegga, A. G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic acids Res. 37, W305–W311 (2009).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1–10 (2019).
Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinforma. Chapter 11, Unit 11 7 (2010).
Feng, J., Liu, T., Qin, B., Zhang, Y. & Liu, X. S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012).
Bailey, T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Aryee, M. J. et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Peters, T. J. et al. Calling differentially methylated regions from whole genome bisulphite sequencing with DMRcate. Nucleic Acids Res. 49, e109 (2021).
Barnhart, C. D., Yang, D. & Lein, P. J. Using the Morris water maze to assess spatial learning and memory in weanling mice. PLoS One 10, e0124521 (2015).
Vorhees, C. V. & Williams, M. T. Assessing spatial learning and memory in rodents. ILAR J. 55, 310–332 (2014).
Phillips, R. G. & LeDoux, J. E. Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behav. Neurosci. 106, 274–285 (1992).
Moy, S. S. et al. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 3, 287–302 (2004).
Nadler, J. J. et al. Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain Behav. 3, 303–314 (2004).
Acknowledgements
This research was supported by NIH grants HD097967 (GCG), HL14278 (CWL), AHA CDA847524 (HY). YLW was supported by funding from AHA (18CDA34140024), NIH (EB023507, NS121706), and DoD (W81XWH1810070).
Author information
Authors and Affiliations
Contributions
GCG, BJG, TT, MGS performed mouse husbandry, pregnancy monitoring, and genotyping, GCG and BJG performed mouse brain immunostaining and confocal imaging, and mouse fibroblast cell culture work; GCG, HY performed RNAseq, ChIPseq, and DNA methylation experiments and LH and AB performed bioinformatic analysis; MG assisted with PPI visualization, GCG and DS performed mouse behavioral testing and data analysis; GCG, WTR performed episcopic confocal microscopy analysis of mouse brains; MCS and YLW performed brain MRI imaging and imaging data analysis, GCG and CWL developed the experimental design, and conducted analysis, integration and interpretation of results; GCG and CWL wrote the manuscript; GCG, BJG, AP, CWL performed editing of the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jian Zhou, Yutaka Suzuki and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gabriel, G.C., Yagi, H., Tan, T. et al. Mitotic block and epigenetic repression underlie neurodevelopmental defects and neurobehavioral deficits in congenital heart disease. Nat Commun 16, 469 (2025). https://doi.org/10.1038/s41467-024-55741-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-55741-6