Abstract
Dissecting the mechanisms underlying heat tolerance is important for understanding how plants acclimate to heat stress. Here, we identify a heat-responsive gene in Arabidopsis thaliana, RNA-DIRECTED DNA METHYLATION 16 (RDM16), which encodes a pre-mRNA splicing factor. Knockout mutants of RDM16 are hypersensitive to heat stress, which is associated with impaired splicing of the mRNAs of 18 out of 20 HEAT SHOCK TRANSCRIPTION FACTOR (HSF) genes. RDM16 forms condensates upon exposure to heat. The arginine residues in intrinsically disordered region 1 (IDR1) of RDM16 are responsible for RDM16 condensation and its function in heat stress tolerance. Notably, RDM16 produces two alternatively spliced transcripts designated RDM16-LONG (RDL) and RDM16-SHORT (RDS). RDS also forms condensates and can promote RDL condensation to improve heat tolerance. Our findings provide insight into the cooperative condensation of the two RDM16 isoforms encoded by RDM16 splice variants in enhancing heat tolerance in Arabidopsis.
Similar content being viewed by others
Introduction
Plants are exposed to a variety of abiotic stresses during their growth and development, one of which is high temperature. In agriculture, excessive temperature (heat stress, HS) negatively affects plant growth and leads to significant drops in crop yield and quality1. Therefore, dissecting the complex mechanisms by which plants respond to HS provides crucial information for breeding heat-tolerant crops.
Heat shock transcription factors (HSFs) and heat shock proteins (HSPs) play key roles in establishing basal and/or acquired HS tolerance2,3,4,5,6. The Arabidopsis (Arabidopsis thaliana) genome encodes 21 HSFs, which are classified into three groups (A, B, and C) based on their structural domains7,8. The transcription of the HSFA3 gene is induced by heat, and its encoded protein functions as an important regulator of acquired HS tolerance, as evidenced by the defects in HS memory displayed by the Arabidopsis hsfa3-1 knockout mutant. Moreover, HSFA3 maintains high expression of the HS memory genes HSA32, HSP18.2, HSP22, and HSP21 following exposure to HS9.
Alternative splicing (AS) involves the splicing of one pre-mRNA to produce multiple distinct mRNA splice isoforms, some of which are then translated into different proteins with similar, different, or no functions, and many of them harbor premature translation termination codons (PTC) and are degraded by nonsense mediated mRNA decay (NMD) pathway. RNA splicing relies on the pre-mRNA that is being spliced and the RNA splicing complex (spliceosome) that performs the splicing10. The core spliceosome comprises five small nuclear ribonucleoproteins (snRNPs), U1, U2, U4/U6, and U5, each containing a specific small nuclear RNA (snRNA) of the same name11. Various types of splicing factors also participate in the splicing of pre-mRNAs into mature mRNAs12,13,14,15. AS is involved in many aspects of plant biology, especially stress responses16,17,18. For instance, HSF primary transcripts undergo AS to produce different variants that function in HS tolerance. Under high-temperature stress, HSFA2 produces the splicing variant HSFA2-III, which encodes a short protein that promotes HSFA2 expression by binding to a heat shock element (HSE) in its promoter3. Splicing factors also play important roles in responses to stress. PRP31 is a conserved pre-mRNA splicing factor that regulates the formation of the U4/U6.U5 snRNP complex in fungi and animals; in plants, PRP31 contributes to pre-mRNA splicing associated with cold-responsive regulation of gene expression and cold tolerance19.
RDM16 is a splicing factor in the RNA-directed DNA methylation (RdDM) pathway that is homologous to the yeast (Saccharomyces cerevisiae) pre-mRNA splicing factor Pre-mRNA Processing 3 (Prp3). Prp3 was first identified in yeast as one of the components of the U4/U6.U5 tri-snRNP complex and is required for pre-mRNA splicing20. Transcriptome deep sequencing (RNA-seq) analysis in Arabidopsis revealed that pre-mRNA splicing events are significantly altered in the knockout mutant rdm16-4 compared to wild-type (WT) plants, confirming an essential role for RDM16 in pre-mRNA splicing in plants21. The rdm16 mutant shows morphological defects and is hypersensitive to abscisic acid (ABA) treatment and salinity stress22, but whether RDM16 participates in plant responses to other stresses is unknown.
Nucleoli and stress granules (SGs) define a class of membrane-less compartments that are involved in regulating cellular activities via liquid–liquid phase separation (LLPS)23,24. LLPS events form droplets that typically involve intracellular protein–protein, protein–RNA, or RNA–RNA interactions, with protein–protein interactions often being driven by intrinsically disordered regions (IDRs) and/or low-complexity sequences within the protein component of these droplets25,26. The formation of LLPS-based droplets occurs during adaptive and innate immune signaling, autophagy, the regulation of transcription, and the assembly of SGs, heterochromatin, and the miRNA-induced silencing complex27,28,29,30,31,32,33. Moreover, LLPS has emerged as a mechanism to sense changes in the environment, including during several adverse biotic and abiotic stress conditions27,34,35,36,37,38,39.
In this study, we describe a previously unknown role for RDM16 in HS tolerance and identify naturally occurring AS variants of RDM16. In addition to the full-length mRNA RDM16-LONG (RDL), RDM16 also produces RDM16-SHORT (RDS), encoding a shorter protein. RDL shows heat-responsive condensation, and the arginine residues within its IDR are responsible for driving condensate formation and heat tolerance. Notably, RDS promotes the condensation of RDL, supporting its role in HS tolerance.
Results
RDM16 plays an important role in HS tolerance in Arabidopsis
To analyze the molecular mechanisms behind heat tolerance in plants, we explored an mRNA-sequencing (RNA-seq) dataset comprising gene expression profiles of seedlings from the Arabidopsis accession C24 grown at 22 °C or exposed to different durations of HS treatment at 37 °C (Supplementary Data 1). Trend analysis of the transcriptome data, using Short Time-series Expression Miner (STEM)40 software on the OmicShare tools platform (www.omicshare.com/tools), revealed 2626 genes that were continuously upregulated under heat treatments (Supplementary Fig. 1a and Data 1). Some of them were enriched in ‘spliceosome’ pathway through Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis (Supplementary Fig. 1b and Data 2). Since snRNPs are core components of the spliceosome and play important roles in plant stress tolerance41,42,43, we focused on them and identified 19 genes whose expression was induced by high temperature from the ‘spliceosome’ pathway (Supplementary Fig. 1b and Data 3).
Given the emerging key roles of biomolecular condensates in stress perception and responses44, we screened these snRNPs for the ability to form biomolecular condensates in response to HS. We cloned the coding sequences of the candidates and fused them in-frame with the coding sequence of green fluorescent protein (GFP). We transfected Col-0 protoplasts with each construct for imaging under control conditions (22 °C) and following a challenge by HS. Among the list of candidates snRNPs, At1g28060 (encoding RDM16) fusion protein appeared as bright round speckles in response to HS treatment at 42 °C (Supplementary Fig. 2). RDM16 participates in the RdDM pathway and shares 30.2% amino acid sequence similarity with yeast Prp322.
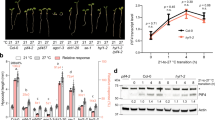
To investigate the role of RDM16 in HS responses, we obtained two independent Arabidopsis mutant lines in different genetic backgrounds, rdm16-1 in the C24 background and rdm16-4 in the Col-0 background21,22. Reverse transcription–quantitative PCR (RT-qPCR) analysis indicated that relative RDM16 transcript levels in rdm16-1 were 44% of those in the corresponding WT (Supplementary Fig. 3a). There is a premature stop codon in the sixth exon of the RDM16 sequence in rdm16-4 (Supplementary Fig. 3b–d) as previously reported21. We subjected these mutants and their respective WT plants to two types of heat treatments (Fig. 1a and Supplementary Fig. 4a). First, we conducted one-step basal heat tolerance assays with 8-day-old seedlings grown at 22 °C and exposed to 45 °C for 35 or 40 min before returning them to 22 °C for 6 days of recovery45. We observed no significant differences in the survival rates and fresh weight but stronger electrolyte leakage of the two mutants after HS treatment compared to the WT (Supplementary Fig. 4b–i). Second, we performed two-step acquired heat treatment (AHT) assays by growing Arabidopsis seedlings for 8 days at 22 °C before exposing them to 37 °C for 30 min, followed by a short recovery period at 22 °C for 30 min and an additional heat treatment at 45 °C for 80 min (Fig. 1a). After 6 days of recovery at 22 °C, both mutants exhibited significantly lower survival rates and higher electrolyte leakage than the WT (Fig. 1b–g); we therefore used AHT to explore the role of RDM16 in heat tolerance.
a Diagram of the heat treatment used in this study. CK, control treatment at 22 °C; AHT, acquired heat treatment. b, e Representative photographs of rdm16 mutant seedlings and wild type under CK or AHT conditions. c, g Relative electrolyte leakage of wild type and rdm16 mutant seedlings after a 6-day recovery from heat treatment. Interactive P-value between genotype and treatment is <0.001, suggesting that the rdm16 mutants are hypersensitive to heat treatment. d, f Survival rates of wild type and rdm16 mutant seedlings after a 6-day recovery from heat treatment. h Relative RDM16 transcript levels in Col-0, rdm16-4, and rdm16-4 RDM16pro:RDM16-GFP (RDM16:RDM16-GFP) transgenic lines, as determined by RT-qPCR. i Representative photographs of Col-0, rdm16-4, and RDM16:RDM16-GFP seedlings under CK or AHT conditions. j Survival rates of Col-0, rdm16-4, and RDM16:RDM16-GFP seedlings after heat treatment. k Relative electrolyte leakage of Col-0, rdm16-4, and RDM16:RDM16-GFP seedlings. l Splicing efficiency of transcripts for the indicated heat stress–responsive genes in 8-day-old Col-0, rdm16-4, and RDM16:RDM16-GFP seedlings subjected to heat stress as indicated (TR1, 37 °C for 30 min; TR2, 30 min 37 °C, 30 min 22 °C, and 80 min 45 °C). The PCR products either spanned exon–exon junctions to detect spliced transcripts or exon–intron junctions to detect unspliced transcripts. Splicing = spliced/(spliced + unspliced). Data in (c, d, f, g, h, j, k and l) are means ± standard deviation (SD), n = 3. Different letters in (c, g, k and l) indicate a significant difference by two-way ANOVA with post hoc Holm–Šídák test (P < 0.05). In (d, f), significant differences were determined using two-sided Student’s t-test (* P < 0.05; *** P < 0.001). Different letters in (h, j) indicate a significant difference by one-way ANOVA with post hoc Tukey HSD test (P < 0.05). Experiments in (b–k) were performed at least three times with similar results. Experiments in (l) were performed at least twice with similar results.
We further generated complementation lines by introducing a transgene consisting of the RDM16 promoter driving RDM16-GFP (RDM16:RDM16-GFP) in the rdm16-4 background. RT-qPCR analysis showed that relative RDM16 expression levels were higher in the resulting RDM16:RDM16-GFP transgenic plants than in Col-0 and rdm16-4 (Fig. 1h). The lower survival rate and greater electrolyte leakage observed in rdm16-4 upon heat treatment were restored to WT values by introducing the RDM16:RDM16-GFP transgene (Fig. 1i–k). In addition, the seedlings overexpressing RDM16 (RDM16 OE) showed significantly higher survival rates and lower electrolyte leakage values than Col-0 transformed with the empty vector (EV) after HS treatment (Supplementary Fig. 5). These results indicate that RDM16 is important for acquired HS tolerance in Arabidopsis.
RDM16 is a homolog of Prp3 from yeast, a component of the U4/U6 snRNP and it was previously shown to be involved in pre-mRNA splicing in planta21,22. To investigate how RDM16 functions in HS tolerance, we examined the splicing efficiency of transcripts from 20 HSF genes, as many of them have been proved important for the HS response and acclimation in plants5,6,9,46,47. We collected seedlings grown at 22 °C (CK: control condition), after an initial exposure to 37 °C for 30 min (TR1: heat treatment 1), and after a second exposure to 45 °C for 80 min (TR2) for splicing analysis of these HSF mRNA (Fig. 1l). RT-PCR results showed that most of the HSFs except for HSFA1b and HSFA9 undergo intron retention events (Supplementary Fig. 6). Further RT-qPCR analysis using gene specific PCR primers indicated that, upon TR1, the splicing efficiency of HSFA1a, HSFA1b, HSFA1d, HSFA1e, HSFA2, HSFA4a, HSFA4c, HSFA5, HSFA6a, HSFA7a, HSFA7b, HSFA8, HSFB1, HSFB2a, HSFB2b, HSFB4, and HSFC1 was significantly reduced in the rdm16-4 mutant, and recovered in RDM16:RDM16-GFP lines, compared to Col-0. After TR2, the splicing efficiency of most transcripts showed no significant changes in Col-0, except for HSFA3, which exhibited a marked increase in splicing in Col-0 but very low levels of splicing in rdm16-4 (Fig. 1l). Taken together, these results suggest that the function of RDM16 in heat tolerance may associate with the splicing of the HSFs.
To investigate whether RDM16 also affects gene expression, we performed transcriptomic profiling of Col-0 and rdm16-4 seedlings before and after heat treatments. The differentially expressed genes (DEGs) were determined with the criteria | Log2 (fold changes) | ≥ 1 and q-value < 0.05. In total, 3138 DEGs (1386 upregulated and 1752 down-regulated) were identified in rdm16-4 versus Col-0 comparison after TR1, and 5871 DEGs (1999 upregulated and 3872 down-regulated) were identified in rdm16-4 versus Col-0 after TR2 (Supplementary Fig. 7a and Data 4). Of these, 5582 DEGs were both regulated by RDM16 and heat stress (Supplementary Fig. 7b and Data 4). Among these DEGs, the expression of nine HSFs and their known target genes involved in cellular defense (Hop3, DAG6, HSA32, APX2, At3g03270, PR4, and COR15A), cellular transport (LTP4 and At5g52760), metabolism (GolS2, MIPS2 and PEPC1), transcription (WRKY30), and protein fate (UBP21)9,48,49, changed significantly upon heat treatment (Supplementary Fig. 7c and Data 5), which were verified by RT-qPCR analysis (Supplementary Fig. 7d). In addition, we performed differential AS analysis on the transcriptomes of Col-0 and rdm16-4. The number of significant AS events increased from 2981 to 3601 at TR1, and decreased to 2827 at TR2. Specifically, retained intron (RI) events were predominant before and after HS treatment in the comparison between Col-0 and rdm16-4, and the proportion of RI in total significant AS events decreased from 61.7% to 51.4% and 51.3%, after HS treatments of TR1 and TR2, respectively (Supplementary Table 1). These results suggest that RDM16 plays an important role in regulating gene expression and mRNA splicing.
RDM16 undergoes condensation in response to HS
RDM16 contains the N-terminal structural domain CC1, a low-complexity domain (LCD), and four disordered regions, a structure potentially associated with protein condensation, as predicted by the Predictor of Natural Disordered Regions (PONDR) online tool (Fig. 2a). Considering the role of RDM16 in heat tolerance (Fig. 1), we examined the subcellular pattern of RDM16-GFP before and after heat treatment in the root cells of transgenic Arabidopsis RDM16:RDM16-GFP seedlings (Fig. 2b). Under normal conditions, most RDM16 signals appeared as even distribution with some occasional granules close to the peripheral ring of the nucleus. After 30 min of heat treatment at 37 °C, more granular structures formed in the nucleus, together with irregularly shaped structures in the nucleus (Fig. 2b, c). When RDM16-GFP seedlings roots were treated at 45 °C for one min, the similar level of granules formed rapidly (Fig. 2d, e). Moreover, when the heat-treated seedlings were returned to normal growth conditions for ~150 min, the granules in the cells disappeared, similar to the untreated state (Fig. 2b–e), indicating that the heat-induced granules are dynamically regulated.
a Diagram of RDM16. Top, functional domains of RDM16. CC1, splicing factor domain present in the CC1-like family. Prp3, pre-mRNA processing factor 3. DUF, domain of unknown function. Bottom, potential intrinsically disordered regions (IDRs) of RDM16, as analyzed by the Predictor of Natural Disordered Regions online tool (PONDR; http://www.pondr.com/). The black lines with asterisk and the cyan numbers underneath indicate the IDR and its average score. b, d and h Confocal microscopy of root cells of RDM16:RDM16-GFP (b, d) and overexpressing RDM16-GFP (RDM16-GFP OE) (h) transgenic seedlings that were treated as indicated. Scale bars, 10 μm (b, d) or 5 μm (h). c, e Quantitative analysis of the RDM16-GFP fluorescent spots in (b, d). n = 239, 247, and 221 for 22 °C, 37 °C and recovery (c); n = 234, 248, and 212 for 22 °C, 45 °C and recovery (e). The number on the right represents the number of granules per cell. f, g and i Representative micrographs and quantification of fluorescence recovery after photobleaching (FRAP) of RDM16-GFP granules after treatment as indicated. The arrowheads indicate the condensate targeted for photobleaching. Scale bars, 5 μm. n = 5 (f), n = 6 (g) or n = 7 (i). j, k Representative micrographs and quantification of RDM16-GFP in protoplasts before and during heat treatment at 37 °C. Scale bars, 5 μm. n = 90, 122, and 88 for 0-, 15-, and 30-min treatments at 37 °C, respectively. The boxes represent the interquartile range, with the middle line defining the median. The lines extending from the quartiles of the box are called ‘whiskers’ and show the maximum and minimum values. In (c, e and k), different letters indicate significant differences by non-parametric version of one-way ANOVA: Kruskal–Wallis test followed by Dunn’s multiple comparisons test (P < 0.05). In (f, g and i), data are means ± SD. The maximum recovery fraction (Rmax) and half-time of recovery (t1/2) was calculated from the logarithmic curve and the best-fit values were generated by GraphPad. All experiments were performed at least three times with similar results.
We further assessed the dynamics of RDM16 granules in the nuclei of Arabidopsis root cells after heat treatments at 37 °C and 45 °C using fluorescence recovery after photobleaching (FRAP). In the experiments performed at 37 °C, we observed a fast recovery in the fluorescence of the bleached RDM16-GFP region after 5 to 25 s (Fig. 2f). By contrast, after heat treatment at 45 °C, the fluorescence in the bleached RDM16-GFP region was much less recoverable (Fig. 2g). As biomolecular condensation can be promoted by increased protein abundance29,50, we examined RDM16 level in response to heat by analyzing RDM16-GFP abundance in RDM16:RDM16-GFP seedlings grown at 22 °C or exposed to heat treatment at 37 °C. After heat treatment at 37 °C for 30 min, RDM16-GFP level was significantly reduced, while the levels of RDM16 mRNA showed no change within 1 h heat treatment (Supplementary Fig. 8a, b), suggesting that the formation of RDM16 granules we observed was not due to protein accumulation. In addition to the RDM16:RDM16-GFP complementation lines, we generated transgenic lines overexpressing RDM16-GFP (RDM16-GFP OE). These plants contained punctate granules and foci in the nuclei of root cells under normal conditions and became more abundant after heat treatment (Fig. 2h). A FRAP assay of these punctate granules demonstrated their highly dynamic nature (Fig. 2i). Like the complementation lines, the overexpression lines also showed reduced RDM16-GFP abundance in response to heat treatment (Supplementary Fig. 8c).
In addition, we exposed Col-0 protoplasts transfected with RDM16-GFP plasmids to heat treatment at 37 °C for 15 or 30 min. The number of RDM16-GFP fluorescent granules increased in the nucleus in response to heat treatment (Fig. 2j, k). Together, these results indicate that RDM16 undergoes condensation in response to HS treatment in vivo.
RDM16 undergoes condensation in vitro
We purified recombinant GFP-RDM16 fused to a His tag at its N-terminus. The GFP-RDM16 fusion protein could condensate upon the addition of crowding reagents (polyethylene glycol 4000 [PEG4000] or Ficoll 400)51 (Fig. 3a, b). Similarly, increasing the concentration of GFP-RDM16 also promoted the formation of more and larger droplets (Fig. 3c), whereas the addition of 1,6-hexanediol, a reagent that affects hydrophobic interactions, disrupted the formation of these RDM16 droplets52 (Fig. 3d). Moreover, the number of GFP-RDM16 droplets increased along with the increase of GFP-RDM16 concentration, but reduced with the increase of NaCl (Fig. 3e), and small GFP-RDM16 droplets could fuse to form larger ones (Fig. 3f).
a In vitro phase separation assays of 10 μM GFP or GFP-RDM16 in the presence of 10% (w/v) PEG4000. Scale bars, 5 μm. b In vitro phase separation assay of 10 μM GFP or GFP-RDM16 in the presence of 15% (w/v) Ficoll 400. Scale bars, 5 μm. c Phase separation assay of purified GFP-RDM16 at different concentrations in the presence of 10% (w/v) PEG4000. Scale bars, 5 μm. d In vitro phase separation assay of 10 μM GFP or GFP-RDM16 in the presence of 10% (w/v) PEG4000. The addition of 15% (w/v) 1,6-hexanediol solution disrupted RDM16 phase separation. Scale bars, 5 μm. e GFP-RDM16 phase separation assay with different concentrations of protein and NaCl. Scale bars, 5 μm. f Fusion of GFP-RDM16 liquid droplets. Scale bars, 2 μm. g FRAP of GFP-RDM16 droplets (10 μM concentration) in the presence of 10% (w/v) PEG4000. Time 0 s indicates the time of the photobleaching pulse. Scale bars, 1 μm. h Recovery of GFP-RDM16 fluorescence after photobleaching. Data are means ± SD, n = 6. i Effect of temperature treatment on 10 μM GFP-RDM16 droplets in HEPES buffer with and without 10% (w/v) PEG4000. GFP-RDM16 droplets increase as the temperature rises from 22 °C to 37 °C or 45 °C. Scale bars, 10 μm. j, k Droplet size (j) and fluorescence intensity (k) of GFP-RDM16 shown in (i). n = 199, 240, 219, 216, and 217 for 22 °C, 37 °C, recovery (37 °C–22 °C), 45 °C, and recovery (45 °C–22 °C). l Images and quantification of FRAP of GFP-RDM16 droplets after photobleaching under the condition of HS at 37 °C. Scale bars, 2 μm. Data are means ± SD, n = 6. The maximum recovery fraction (Rmax) and half-time of recovery (t1/2) in (h, l) was calculated from the logarithmic curve and the best-fit values were generated by GraphPad. In (j, k), different letters indicate significant differences by non-parametric version of one-way ANOVA: Kruskal–Wallis test followed by Dunn’s multiple comparisons test (P < 0.05). All experiments were performed at least twice with similar results.
We performed FRAP assays to probe the dynamics of RDM16 droplets. The intensity of GFP-RDM16 fluorescence recovered rapidly (within 30 s) after photobleaching (Fig. 3g, h). Consistent with the observation that RDM16 formed granule-like structures under HS in root cells (Fig. 2), raising the temperature from 22 °C to 37 °C or 45 °C enhanced the phase separation of RDM16 in solution (Fig. 3i–k). Moreover, GFP-RDM16 droplets restored to their initial state when they were placed at 22 °C for 1 h (Fig. 3i–k). We further performed FRAP experiments to probe the effect of temperature treatment at 37 °C on the mobility of GFP-RDM16 droplets, the result showed that the intensity of RDM16 fluorescence signal recovered partially after photobleaching (Fig. 3l). Together, these results demonstrate that RDM16 undergoes condensation, which can be enhanced by HS.
Arginine (Arg) residues in IDR1 mediate RDM16 condensation in vitro and in vivo
To delineate the regions within RDM16 responsible for its condensation, we tested the ability of truncated variants of RDM16 to undergo condensation. The online tool PONDR identified four IDRs in RDM16 (Fig. 2a), and two IDRs (IDR1 and IDR2) in the N-terminus were focused first, as they spanned long protein regions (Fig. 4a). We produced and purified recombinant GFP fused to RDM16 lacking IDR1 (ΔIDR1) or IDR2 (ΔIDR2) and examined their capability to undergo condensation. The deletion of IDR2 had little influence on RDM16 droplet formation, whereas the deletion of IDR1 completely abolished the formation of liquid droplets (Fig. 4b).
a Diagram of RDM16 with functional domains and truncations. IDR1 and IDR2 are two predicted disordered regions by PONDR. b Phase separation assay of purified GFP-RDM16 and its truncated variants. Phase separation condition, 10% (w/v) PEG4000. Scale bars, 5 μm. c Pie chart showing the relative proportion of each amino acid in the 30–70 amino acid (aa) region of RDM16. d Diagram of RDM16mu, harboring Ala (A) residues in place of all Arg (R) residues within aa 30–70 of RDM16. e In vitro phase separation assays of GFP, GFP-RDM16, and GFP-RDM16mu in the presence of 10% (w/v) PEG4000. Scale bars, 5 μm. f Subcellular localization of RDM16-GFP and RDM16mu-GFP in protoplasts at 22 °C (CK) or during heat exposure at 37 °C for 30 min. Enlarged images of the nuclei are shown in the upper right corner of each figure panel. Scale bars, 5 μm. g RDM16mu-GFP (n = 103, 99 for CK and 37 °C) forms fewer condensates per nucleus than RDM16-GFP (n = 95, 140 for CK and 37 °C) in protoplasts after heat treatment. Different letters indicate significant differences by non-parametric version of one-way ANOVA: Kruskal–Wallis test followed by Dunn’s multiple comparisons test (P < 0.05). The boxes represent the interquartile range, with the middle line defining the median. The lines extending from the quartiles of the box are called ‘whiskers’ and show the maximum and minimum values. h Confocal microscopy images of root tip cells from Arabidopsis seedlings RDM16:RDM16-GFP or RDM16:RDM16mu-GFP. The seedlings were exposed to 37 °C for 30 min. Enlarged images of the nuclei are shown in the upper left corner of each figure panel. Scale bars, 5 μm. All experiments were performed at least three times with similar results.
To narrow down the region governing RDM16 condensation within IDR1, we generated additional truncations within IDR1 and tested their ability to form condensates in vitro (Supplementary Fig. 9a). Purified GFP-T2 (lacking amino acids [aa] 71–113 of RDM16) and GFP-T3 (lacking aa 114–184) retained the ability to form droplets, whereas the deletion of the N-terminal 70 aa of IDR1 (GFP-T1) had a strong effect on the formation of GFP-RDM16 condensates (Supplementary Fig. 9b). In the Arabidopsis protoplasts transformed with 35S:T1-GFP plasmids, we detected strong fluorescence in a single large structure, but no multiple fluorescent droplets, within the nucleus at 22 °C and following exposure to 37 °C (Supplementary Fig. 9c), suggesting that the T1 region is important for the formation and response of RDM16 condensates to temperature. We divided the T1 region into the T4 and T5 regions, lacking aa 1–29 or aa 30–70 from RDM16, respectively (Supplementary Fig. 9a). Recombinant purified GFP-T4 formed droplets, but GFP-T5 did not, showing nearly no phase separation in solution (Supplementary Fig. 9b). To test this finding in vivo, we transfected protoplasts with the 35S:T5-GFP plasmid, which revealed no condensate foci under normal conditions or upon high-temperature treatment (Supplementary Fig. 9d).
We sought to pinpoint the key residues within the IDR1 of RDM16 that are responsible for its condensation. We noticed that the 30–70 aa region of RDM16 contains 15 (or 37%) Arg residues (Fig. 4c). Thus, we suspected that the Arg residues in IDR1 of RDM16 may supply adequate intermolecular interactions to drive its condensation. We replaced all 15 Arg (R) residues with Ala (A), yielding the mutated variant RDM16mu (Fig. 4d). In protein solutions, recombinant purified GFP-RDM16mu exhibited no capacity to form liquid droplets, even with the addition of 10% PEG4000 (Fig. 4e). To explore the role of the R-to-A mutations on the condensation of RDM16-GFP in response to HS, we transfected Arabidopsis protoplasts with the RDM16mu-GFP construct. RDM16mu-GFP accumulated in the nuclei of the protoplasts, but we observed little condensate formation, even upon HS treatment (Fig. 4f, g). We also generated transgenic RDM16mu-GFP lines driven by RDM16 promoter harboring the R-to-A mutations in rdm16-4. Similar to the results obtained in protoplasts, the R-to-A mutations of RDM16 significantly impaired its ability to form granules under HS (Fig. 4h). To explore whether the Arg mutations affect RDM16 structure, Circular Dichroism (CD) spectra technique was utilized, which indicate that RDM16 and RDM16mu are both composed of helix, β-sheet, β-turn, and random coil structures, but no clear spectral changes was observed between these two versions (Supplementary Fig. 10a, b). Therefore, although these Arg mutations do not alter the secondary structure of RDM16, they are able to affect RDM16 condensation.
Condensation of RDM16 is essential for its function in HS
We investigated whether the condensation of RDM16 contributes to its function in protecting plants from HS by testing the effect of RDM16mu on plant heat tolerance. We examined the HS tolerance of transgenic Arabidopsis plants RDM16 promoter driving RDM16mu or full-length RDM16 by exposing them to 37 °C for 30 min, followed by a short recovery period at 22 °C for 30 min and an additional heat treatment at 45 °C for 80 min (Fig. 1a). The RDM16:RDM16mu-GFP seedlings showed significantly lower survival rates and higher electrolyte leakage values compared to Col-0 and RDM16:RDM16-GFP after HS treatment (Fig. 5a–c), suggesting that Arg residues of IDR1 are essential for HS tolerance. Moreover, the splicing efficiency of HSFA3 in the RDM16:RDM16mu-GFP lines was significantly lower than that of Col-0 and RDM16:RDM16-GFP lines specifically after HS treatment at 37 °C (Fig. 5d).
a Representative photographs of Col-0, rdm16-4, RDM16:RDM16-GFP, and RDM16:RDM16mu-GFP seedlings under control (CK) or heat treatment (AHT, acquired heat treatment) conditions. b Survival rates of Col-0, rdm16-4, RDM16:RDM16-GFP, and RDM16:RDM16mu-GFP seedlings after heat treatment. c Relative electrolyte leakage of Col-0, rdm16-4, RDM16:RDM16-GFP, and RDM16:RDM16mu-GFP under CK and AHT conditions. d Relative splicing of HSFA3 transcripts in Col-0, rdm16-4, RDM16:RDM16-GFP, and RDM16:RDM16mu-GFP seedlings under CK or after 37 °C for 30 min. e RNA immunoprecipitation (RIP) RT-qPCR analysis of RDM16 binding to snRNAs and HSFA3 transcripts. Total RNA from Col-0, RDM16:RDM16-GFP, and RDM16:RDM16mu-GFP were immunoprecipitated (IP) with anti-GFP antibody. Relative enrichment in IP samples was calculated against input samples. At5g51880 (also reported by OGIO) served as a negative control. Data in (b–e) are means ± SD, n = 3. Different letters in (b, e) indicate a significant difference by one-way ANOVA with post hoc Tukey HSD test (P < 0.05). Different letters in (c, d) indicate a significant difference by two-way ANOVA with post hoc Holm–Šídák test (P < 0.05). Experiments in (a–c) were performed at least three times with similar results. Experiments in (d, e) were performed at least twice with similar results.
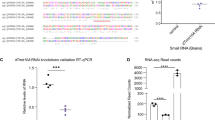
Human Prp3, a homolog of RDM16, binds to U4/U6 di-snRNA fragments to facilitate the stepwise assembly of a mature spliceosome53. We therefore tested whether RDM16 binds directly to snRNAs in vivo by performing an RNA immunoprecipitation (RIP) assay using an anti-GFP antibody on Col-0, RDM16:RDM16-GFP, and RDM16:RDM16mu-GFP seedlings, followed by RT-qPCR analysis of the immunoprecipitated RNAs with primers specific to the U1, U2, U3, U4, U5, or U6 snRNAs. The anti-GFP antibody immunoprecipitated significant amounts of U4 and U6 snRNAs and relatively low amounts of U1, U2, U3, and U5 snRNAs from RDM16:RDM16-GFP and RDM16:RDM16mu-GFP seedlings compared to Col-0 seedlings under HS; the enrichment of RDM16 at the same snRNAs following RIP using RDM16:RDM16mu-GFP seedlings was significantly lower than that in RDM16:RDM16-GFP seedlings (Fig. 5e). We reasoned that RDM16 might associate with its target mRNAs to regulate their splicing. To test this hypothesis, we selected to check HSFA3, which showed differences in pre-mRNA splicing efficiency in the rdm16-4 mutant (Fig. 1l), in RDM16:RDM16-GFP, and RDM16:RDM16mu-GFP seedlings compared to Col-0, using the 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily gene At5g51880 (also reported as OGIO) as a control54. HSFA3 mRNAs were enriched in RDM16:RDM16-GFP, and mutating the Arg residues of RDM16 affected its association to HSFA3 mRNAs (Fig. 5e). These results indicate that RDM16 associates with these snRNAs and HSFA3 mRNAs, and that the condensation of RDM16 plays an important role in this process.
To further demonstrate that RDM16 condensation regulated by the Arg residues in IDR1 is important for its biological function, we fused the LCD of fused in sarcoma (FUS), a phase-separated disease-related RNA-binding protein55, to the N-terminus of RDM16mu. Confocal imaging showed that GFP-FUS-RDM16mu in solution formed liquid droplets, similar to GFP-RDM16 (Supplementary Fig. 11a). When RDM16-GFP, RDM16mu-GFP, and FUS-RDM16mu-GFP plasmids were transformed into protoplasts, FUS-RDM16mu could restore the ability of RDM16mu to form condensation under heat stress conditions (Supplementary Fig. 11b, c). To analyze if FUS-RDM16mu has function in gene splicing, we cloned the genomic DNA of HSFA3 (containing two exons and one intron) and linked it to a vector containing luciferase (LUC) to generate a 35S:HSFA3-LUC construct. If the HSFA3 intron is transcribed and spliced normally, LUC activity can be detected. If intron is retained and a PTC will be produced, and LUC cannot be formed (Supplementary Fig. 11d). RDM16-GFP, RDM16mu-GFP, and FUS-RDM16mu-GFP plasmids were individually transformed together with 35S:HSFA3-LUC into Col-0 protoplasts for LUC imaging analysis. The combination of RDM16mu-GFP and 35S:HSFA3-LUC gave lower LUC activity than that of RDM16-GFP and 35S:HSFA3-LUC, whereas the LUC activity was enhanced in FUS-RDM16mu-GFP and 35S:HSFA3-LUC combination (Supplementary Fig. 11e). These results suggest that RDM16 condensation is important for its function on gene splicing.
RDM16 locus produces two mRNA splicing variants
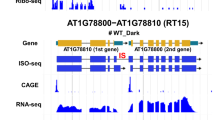
While amplifying the full-length coding sequence of RDM16 from Col-0 cDNA by RT-PCR, we noticed two PCR bands produced by a pair of RDM16 primers, as shown on an agarose gel, suggesting that RDM16 undergoes AS in Col-0 (Fig. 6a). Sanger sequencing indicated that in addition to the coding sequence supported by the annotation in TAIR (https://www.arabidopsis.org/), the RDM16 locus produces another splicing product with an additional 79-bp intron within the second exon of the typical RDM16 pre-mRNA, which may lead to the introduction of a PTC. We designated this new splice variant as RDS (shorter open reading frame of RDM16 or RDM16-SHORT) and the longer typical RDM16 transcript as RDL (longer open reading frame of RDM16 or RDM16-LONG) (Fig. 6b, c). As RDS mRNA harbors a PTC that may cause RDS degradation by the NMD pathway56, we therefore checked its stability relative to RDL but did not detect a significant difference between them (Fig. 6d). To determine if RDS encodes a protein, we performed immunoblot analysis and detected RDL and RDS proteins in Col-0 protoplast cells transformed with GFP-RDM16gDNA plasmids driven by the RDM16 promoter (Fig. 6e). RT-qPCR analysis indicated that RDL and RDS were upregulated with increasing temperatures (from 22 °C to 37 °C) or increasing treatment times at 37 °C (Fig. 6f, g) and that both transcripts are expressed in different tissues of Arabidopsis seedlings or plants (Fig. 6h). Moreover, like RDL-GFP (identical to RDM16-GFP used earlier), RDS-GFP fusion protein, also accumulated in the nuclei of Arabidopsis protoplasts; RDL-GFP and RDS-GFP both co-localized with the nuclear localization marker SV40-DsRed1 (Supplementary Fig. 12a).
a Identification of alternatively spliced RDM16 transcripts by RT-PCR. The cDNA and genomic DNA samples were analyzed using the primers (F and R) indicated in (c). No RT and DNase/RNase-free water were used as negative controls. b Nucleotide sequence alignment of the isoforms RDM16-LONG (RDL) and RDM16-SHORT (RDS). c Diagram of the RDM16 locus and the RDM16 splice variants RDL and RDS. The horizontal lines indicate introns and untranslated regions; orange rectangles indicate exons. The blue rectangle indicates an additional 79-bp intron within the second exon of the typical RDM16 pre-mRNA. The red arrow indicates a premature termination codon (PTC) in exon 2 of RDS. The dashed arrow shows the PCR primer spanning the 79-bp junction to specifically detect RDS. d The RDM16 mRNA stability shows no significant difference between RDL and RDS. Two-week-old Col-0 seedlings were treated with Cordycepin for different times (0.5, 1, 2, 4, 6, 12 h). Different letters indicate significant differences by two-way ANOVA with post hoc Holm–Šídák test (P < 0.05). e Immunoblot analysis of RDS- and RDL-GFP using an anti-GFP antibody. “1” and “2” indicate Col-0 protoplasts transformed with RDM16: RDS-GFP and RDM16: GFP-RDM16gDNA plasmids. f Relative expression levels of RDL and RDS in Col-0 seedlings grown at 22 °C or exposed to 28 °C or 37 °C for 12 h. g Relative expression levels of RDL and RDS following transfer from 22 °C to 37 °C for the indicated duration. h Relative expression levels of RDL and RDS in different tissues of Col-0 seedlings. In (d, f, g and h), data are means ± SD, n = 3. Different letters in (f, g) indicate significant differences by one-way ANOVA with post hoc Tukey HSD test (P < 0.05). Experiments in (a, e) were performed at least three times with similar results. Experiments in (d, f, g and h) were performed at least twice with similar results.
RDS contains only the splicing factor (SF)-CC1 domain within IDR1 that is important for condensation of RDM16 (Fig. 4a, b). We therefore investigated whether RDS undergoes condensation following the same methods used for RDM16. We purified recombinant GFP-RDS, which formed liquid droplets at a protein concentration at 1.25–10 μM upon the addition of 10% (w/v) PEG4000 (Supplementary Fig. 12b). Like GFP-RDM16, GFP-RDS droplets could be dissolved by adding 1.6-hexanediol (Supplementary Fig. 12c). GFP-RDS also formed liquid droplets upon the addition of 15% (w/v) Ficoll 400 (Supplementary Fig. 12d). Consistent with condensates of GFP-RDL, the intensity of GFP-RDS fluorescence recovered quickly after photobleaching in a FRAP assay (Supplementary Fig. 12e, f). Taken together, these results indicate that RDS also has the ability to undergo condensation in vitro.
RDS interacts with RDL and promotes its condensation
We wondered if RDS interacts with RDL and affect its function, as they both contain IDR1, which is required for condensate formation. We thus explored their possible interaction using multiple approaches. In a bimolecular fluorescence complementation (BiFC) assay using Arabidopsis protoplasts, we observed the reconstitution of yellow fluorescent protein (YFP), suggestive of a positive interaction, when protoplasts were co-transfected with YNE-RDL (encoding a fusion of RDL to the N-terminal half of YFP) and YCE-RDS (encoding a fusion of RDS to the C-terminal half of YFP), YNE-RDL and YCE-RDL, and YNE-RDS and YCE-RDS (Fig. 7a). We confirmed the interaction of RDS and RDL by protein pull-down assays using purified recombinant GST-RDS and His-MBP-RDL (Fig. 7b). We validated the interaction by conducting a co-immunoprecipitation (Co-IP) assay with an anti-GFP antibody using protein extracts from protoplasts prepared from RDM16:RDM16-GFP transgenic plants transfected with the RDS-FLAG construct, but not with the FLAG control construct (Fig. 7c).
a Bimolecular fluorescence complementation (BiFC) assays showing that RDS interacts with RDL in Arabidopsis protoplasts. YNE, N-terminal half of yellow fluorescent protein (YFP); YCE, C-terminal half of YFP. Scale bars, 10 μm. b MBP pull-down assay showing that RDS interacts with RDL in vitro. MBP fused to RDL (MBP-RDL) was tested for binding to GST fused to RDS (GST-RDS). MBP alone was used as a negative control in the pull-down assays. M, molecular weight of the protein marker. c In vivo co-immunoprecipitation (Co-IP) assay validating the association between RDS-FLAG and RDM16-GFP. RDS-FLAG or FLAG was co-transfected into protoplasts prepared from RDM16:RDM16-GFP plants. M, molecular weights of the protein markers. d RDS promotes condensation of GFP-RDL in vitro. MBP was used as a negative control. Scale bars, 5 μm. e, f Quantification of GFP-RDL droplet size (e) and fluorescence intensity (f) in the samples with the addition of the control protein (MBP) (n = 810) or MBP-RDS (n = 438). g–i Overexpression of RDS promotes of RDL-GFP condensation in Arabidopsis root cells. Representative micrographs of root cells from Arabidopsis seedlings (g) and quantitative analysis of the RDL-GFP fluorescent spots (h) and intensity (i) are shown. In (g), scale bars, 10 μm. n = 154 (h, i). j RDS overexpression does not influence RDL protein abundance. In (e, f, h and i), significant differences were determined using two-sided non-parametric Mann–Whitney U test (*** P < 0.001). All experiments were performed at least three times with similar results.
We investigated the relationship between of RDL and RDS condensation by mixing purified recombinant GFP-RDL with MBP (as a control) or MBP-RDS at a 1:1 ratio (equal protein concentrations). Notably, the addition of MBP-RDS significantly increased the droplet size of GFP-RDL, and the fluorescent signal was significantly enhanced compared to the addition of MBP alone (Fig. 7d–f). To obtain in vivo support for these results, we generated the stable transgenic Arabidopsis line overexpressing RDL-GFP and crossed it to a line overexpressing RDS; confocal microscopy imaging showed that the RDL-GFP + RDS plants contained more fluorescent spots with stronger fluorescence intensity than RDL-GFP plants (Fig. 7g–i), although the difference in fluorescence intensity was not due to different protein levels (Fig. 7j). By contrast, recombinant purified MBP-RDL did not significantly influence the droplet size or fluorescence intensity of GFP-RDS droplets in solution (Supplementary Fig. 13a–c). Overall, these results suggest that RDS promotes RDL condensation.
The splice isoforms and condensation of RDM16 are essential for its role in HS tolerance
To better understand the function of RDS, we generated transgenic lines in the rdm16-4 background harboring a construct consisting of the RDM16 promoter driving RDS. The expression of RDS in the resulting rdm16-4 RDM16pro:RDS (rdm16-4 RDS) seedlings was significantly higher than that in Col-0 (Supplementary Fig. 14a). We treated 8-day-old Col-0, rdm16-4, and rdm16-4 RDS seedlings with HS (30 min at 37 °C, 30 min at 22 °C and 80 min at 45 °C). The survival rates of rdm16-4 RDS seedlings were not significantly different from those of rdm16-4, indicating that the rdm16-4 RDS failed to rescue the defects of rdm16-4 (Fig. 8a–c). However, overexpressing RDS in Col-0 promoted heat tolerance (Fig. 8d–f), suggesting that RDS may contribute to heat tolerance in the presence of RDL.
a Representative photographs of rdm16-4 RDM16pro:RDS (rdm16-4 RDS) seedlings grown under control conditions (CK) or subjected to heat treatment (AHT, acquired heat treatment) as indicated in Fig. 1a. b Survival rates of Col-0, rdm16-4, and rdm16-4 RDS seedlings after heat treatment. c Relative electrolyte leakage of Col-0, rdm16-4, and rdm16-4 RDS seedlings under CK and AHT conditions. d Representative photographs of RDS overexpression (Col-0 RDS) and Col-0 seedlings under CK or subjected to AHT as indicated in Supplementary Fig. 5a. e Survival rates of Col-0 and Col-0 RDS seedlings after heat treatment. f Relative electrolyte leakage of Col-0 and Col-0 RDS lines. g Representative photographs of RDL and RDL + RDS overexpression seedlings under CK and AHT conditions as indicated in Supplementary Fig. 5a. h, i Survival rates (h) and relative electrolyte leakage (i) of RDL and RDL + RDS seedlings under CK and AHT conditions. j Relative RDL transcript levels in RDL and RDL + RDS seedlings. k Relative splicing of HSFA3 in RDL and RDL + RDS seedlings under CK or after as indicated in Fig. 1l (TR1, 37 °C for 30 min; TR2, 30 min 37 °C, 30 min 22 °C, and 80 min 45 °C). In (b, c, e, f, h, i, j and k), data are means ± SD, n = 3. Different letters in (b, e) indicate a significant difference by one-way ANOVA with post hoc Tukey HSD test (P < 0.05). Different letters in (c, f, i and k) indicate a significant difference by two-way ANOVA with post hoc Holm–Šídák test (P < 0.05). In (h, j), significant differences were determined using two-sided Student’s t-test (*** P < 0.001; ns no significant difference). Experiments in (a–i) were performed at least three times with similar results. Experiments in (j, k) were performed at least twice with similar results.
To specifically test this hypothesis, we generated transgenic Arabidopsis overexpressing RDL or RDS and crossed them to obtain the plants overexpressing both RDL and RDS (RDL + RDS). In these seedlings, the presence of RDS promoted heat tolerance, as evidenced by the higher survival rates and lower electrolyte leakage after heat treatment compared to the parental lines (Fig. 8g–i). The enhanced heat tolerance in the presence of Col-0 RDS OE was not due to altered RDL transcript levels or RDL protein abundance (Figs. 8j and 7j). Finally, we examined the splicing efficiency of HSFA3 and determined that RDS improved the pre-mRNA splicing of HSFA3 compared to the RDL lines (Fig. 8k). Taken together, these results suggest that RDS increases heat tolerance by cooperating with RDL in Arabidopsis.
Discussion
RDM16 is a component of the U4/U6 snRNP similar to yeast Prp3. Although knockdown of RDM16 was shown to increase sensitivity to salinity stress and ABA treatment during seed germination in Arabidopsis22, whether RDM16 plays a role in HS tolerance has not been reported. Recent studies have shown that the splicing machinery is important to plant HS responses. For instance, a mutation of STABILIZED1 (STA1), encoding a U5 snRNP-associated pre-mRNA splicing factor, leads to the lack of HS tolerance45. The observation that RDM16 was upregulated by HS treatment prompted us to investigate the role of RDM16 in HS responses (Supplementary Fig. 1). The rdm16-1 and rdm16-4 mutants were hypersensitive to HS, which could be rescued to WT levels by introducing of an intact copy of RDM16 in complementation lines (Fig. 1a–k). In agreement with this finding, transgenic plants overexpressing RDM16 were more tolerant to HS than the WT (Supplementary Fig. 5). The reduced heat tolerance of rdm16-4 seedlings was correlated with lower splicing efficiency of the pre-mRNAs from a list of HSFs (Fig. 1l), whose roles in heat tolerance were reported previously6,7,9,47. Notably, HSFA3 and HSFA2 are specifically required for acquired thermotolerance9, which may explain the heat-sensitive phenotype of the rdm16-4 mutant following AHT (Fig. 1l). In support of this idea, RDM16 is able to associate with HSFA3 mRNA in our RIP assays (Fig. 5e). Interestingly, RDM16 also associates with U4, U6, and U3 snRNAs, suggesting that it may play a role in processing pre-ribosomal RNA (Fig. 5e).
RDM16 contains an SF-CC1 domain (Fig. 2a), which is present in a subfamily of RNA splicing factors. This domain is characterized by an N-terminal arginine-rich region and an LCD57,58,59. LCDs, which are widespread in proteins, participate in the formation of various membrane-less organelles or biomolecular condensates through their roles in phase separation or phase transitions35. RDM16 possesses the capacity for phase separation in vitro and in vivo, which is enhanced under HS conditions (Figs. 2 and 3). RDM16 no longer formed droplets in vitro after the deletion of IDR1 (Fig. 4b). The Arg residues within IDR1 mediate this ability for heat-sensitive phase separation, as evidenced by the lack of condensate formation in the RDM16mu variant (Fig. 4c–h). Similarly, the Arg-rich C-terminal domain of RNase E is required for the assembly of the core bacterial ribonucleoprotein body (BR-body)60,61, and the 207th arginine in the C-terminal domain is a key site mediating phase separation of the SGF29 protein62. Although the Arg mutations prevent RDM16 from condensation but they show no influence on its secondary protein structure (Supplementary Fig. 10), possibly the intermolecular interaction forces (such as electrostatic and cation-π interactions, etc.) are altered between RDM16mu and the typical version, which leads to differential protein condensation23,63,64. For example, the IDR in DEAD-box helicase 4 (DDX4) contains a number of phenylalanine (Phe) - glycine (Gly) repeats, and its aromatic ring appears to facilitate phase separation by forming cation-pi interactions with Arg residues both intramolecularly and intermolecularly65. Phase separation of the RNA-binding protein FUS is mediated by cation-π interactions between C-terminal Arg and N-terminal tyrosine (Tyr) that are regulated by arginine methylation66. Importantly, the condensation character of RDM16 is essential for its function in heat tolerance, as the seedlings containing RDM16 promoter driving RDM16mu (with Arg residues replaced with Ala) were sensitive to HS (Fig. 5a–c) and could not properly regulate the splicing of the downstream regulatory transcript HSFA3 (Fig. 5d). In support of this conclusion, FUS-RDM16mu is able to rescue the ability of RDM16mu to condensate and implement the function on HSFA3 intron splicing (Supplementary Fig. 11).
AS, an important post-transcriptional regulatory mechanism, generates multiple transcripts from the pre-mRNA produced by a single locus, thereby increasing transcriptome plasticity and protein diversity67. Growing evidence suggests that the contribution of AS to environmental stress responses is not negligible68. Here, we present several lines of evidence for the importance of RDM16 AS in regulating heat tolerance in Arabidopsis. RDM16 produces the two splice variants: RDL and RDS. Due to an additional intron in the second exon, RDS alone does not function in heat tolerance, but it can enhance plant heat tolerance conferred by RDL (Fig. 8a–f). Mechanically, RDS can interact with RDL and promote its condensation to achieve a stronger heat tolerance capacity in Arabidopsis seedlings (Figs. 7 and 8, Supplementary Fig. 12), although RDL does not influence the condensation ability of RDS (Supplementary Fig. 13). This RDL–RDS intra-regulatory module underlying splice variants appears to represent a never reported mechanism, as most proteins encoded by splice variants from the same locus in plants function by differentially or competitively binding to their regulated genes or interacting proteins. For instance, TaHSFA6e-II and TaHSFA6e-III in wheat (Triticum aestivum) both bind to the HSE in the promoters of three HSP70 candidate targets, with TaHSFA6e-III enhancing the transcriptional activity of these target genes to a greater extent than TaHSFA6e-II69. Two major FLM isoforms encoded by the two splice variants, FLM-β and FLM-δ, regulate flowering by competing for interactions with the floral repressor SHORT VEGETATIVE PHASE (SVP)70.
In summary, we identified a distinct role for RDM16 isoforms encoded by two splice variants and their cooperative condensation characteristics in HS tolerance, adding an additional regulatory layer to the HS tolerance pathway. Our findings provide valuable clues for engineering heat-tolerant crops in the future.
Methods
Plant materials and growth conditions
The Arabidopsis (Arabidopsis thaliana) rdm16-4 mutant in the Columbia-0 (Col-0) background was provided by Dr. Zhaojun Ding from Shandong University, China. WT C24 and the rdm16-1 mutant are gifts from Dr. Jian Kang Zhu from Southern University of Science and Technology, Shenzhen, China. The seeds were sown on half-strength Murashige and Skoog (1/2 MS, Solarbio, China) medium with 1% agarose (w/v) without sucrose, incubated in the dark at 4 °C for 3 days, and transferred to the growth chamber under long-day conditions (22 °C, 16 h light/8 h dark) with illumination provided by LED lights (Philips, F25T8/TL841) with a fluence rate of 153 mmol m−2 s−1 and 70% relative humidity.
RNA-seq analysis
Transcriptome analysis was conducted by LC-Bio Technologies in Hangzhou Co., Ltd. For trend analysis of gene expression, 12-day-old C24 seedlings after different durations of HS treatment at 37 °C were collected for RNA isolation. The quantity and purity of the total RNA were analyzed using a Bioanalyzer 2100 and RNA 1000 Nano LabChip Kit (Agilent, CA, USA) with RIN > 7.0. StringTie was used to analyze the expression levels of the mRNAs by calculating Reads Per Kilobase Million (RPKM) values71. Trend analysis results were visualized using Short Time-series Expression Miner (STEM) software (Ernst and Bar-Joseph, 2006) on the OmicShare tools platform (www.omicshare.com/tools). Gene expression was normalized with log2 (Fold-change) and gene expression trends were set to 20 profiles using the Trend Analysis tool with a P-value < 0.05 and the minimum fold change of the trend > 2. Pathway analysis of the identified snRNPs proteins was performed based on the online Omicshare software using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database.
For DEG analysis, 8-day-old Col-0 and rdm16-4 seedlings before and after HS treatment were prepared for transcriptome sequencing as described above. HS treatment conditions were set as: CK, 22 °C; TR1, 37 °C for 30 min; TR2, 37 °C for 30 min followed by 22 °C recovery for 30 min and then 45 °C for 80 min. DEG analysis between sample groups was performed using DESeq2 to obtain DEGs sets between the different samples. The DEGs were identified based on | log2 (Fold-change) | ≥ 1 and q-value < 0.05. Heatmaps of gene expression were generated using the OmicShare tools.
For differential AS analysis, replicate Multivariate Analysis of Transcript Splicing (rMATS) tool was utilized72. False discovery rate (FDR) < 0.05 and | IncLevelDifference | > 0.1 were used to screen for the differential AS event between two groups. The P-value was calculated using the likelihood-ratio test to indicate the difference between the two sample groups at the IncLevel (Inclusion Level) level, and the P-value was corrected to obtain the FDR value using the Benjamini-Hochberg algorithm. IncLevelDifference is the difference in IncLevel between the two samples.
Vector construction and transformation
For the proRDM16:RDL-GFP and proRDM16:RDS constructs, the genomic sequence of RDM16 encompassing a 1.7-kb promoter fragment and 1.0 kb of 3′ untranslated region was amplified from Col-0 genomic DNA by PCR using PrimerSTAR Max DNA polymerase (Takara Bio, Dalian, China, cat: R045Q). The coding regions of RDL and RDS were amplified by PCR from cDNA prepared from RNA extracted from Col-0 tissues. The GFP coding sequence was amplified by PCR using the plasmid pCAMBIA1305-GFP as a template. All DNA fragments were cloned and ligated into the pCAMBIA1300 vector by restriction digestion and ligation with T4 DNA ligase (Takara Bio, Dalian, China, cat: 2011A).
RNA extraction and RT-qPCR analysis
Arabidopsis seedlings at the four-leaf stage were harvested, frozen in liquid nitrogen, and stored at −80 °C. Total RNA was extracted from the samples using the hot phenol method73 and reverse transcribed to first-strand cDNA using a Perfect Real Time Reverse Transcription kit (TaKaRa, RR047A) according to the manufacturer’s instructions. qPCR analysis was performed using diluted cDNA as template in a Roche LightCycler 480 PCR machine. Primers used for RT-qPCR are listed in Supplementary Data 6; the UBC21 (UBC) gene was used as an internal reference gene. The 2−ΔCt method was used to calculate the normalized expression levels of the target genes.
Heat stress treatment and plant survival assay
For the acquired HS tolerance assays, 8-day-old seedlings grown on 1/2 MS plates were subjected to a stepwise heat treatment of 37 °C for 30 min followed by 22 °C for 30 min and then 45 °C for 80 or 90 min; the seedlings were then returned to normal growth conditions for further growth. For the basal HS tolerance assays, the seedlings were subjected to 45 °C for 35 or 40 min and transferred to normal growth conditions for further growth.
Relative electrolyte leakage assay
Eight-day-old Arabidopsis seedlings grown on 1/2 MS plates were subjected to basal or acquired HS tolerance assays as described above. Immediately after treatment, the seedlings (0.2 g) were transferred into a 50 mL tube and washed three times with double distilled H2O (ddH2O). After adding 20 mL H2O to each tube, the samples were subjected to vacuum treatment at −0.8 psi for 30 min. The initial electrolyte leakage (Ei) was measured using a conductivity meter (IS228-Basic, InsMark, China). The conductivity of H2O (Ei0) was measured to establish the background. All tubes, including one containing only H2O, were boiled for 30 min, cooled to room temperature, and the final electrolyte leakage (Ef), including that for H2O alone (Ef0), measured. The relative electrolyte leakage (%) was calculated as (Ei–Ei0) * 100/(Ef–Ef0).
Immunoblot analysis
Total proteins were extracted from 8-day-old seedlings with protein extraction buffer (50 mM Tris-HCl pH 7.5, 2.5 mM EDTA, 150 mM NaCl, 0.1% [v/v] IGEPAL, 10% [v/v] glycerol, 1% [v/v] Triton X-100, 0.1% [w/v] SDS, 10 mM β-mercaptoethanol, 1 mM PMSF, and 1× Roche protease inhibitor cocktail). The proteins were separated by SDS-PAGE, and the target proteins were probed by immunoblotting with anti-GFP (Roche, cat.:11814460001, lot: 42903200, dilution: 1:1000) or anti-Actin (Abcam, cat.: ab197345, lot: 1037553-5, dilution: 1:2000) antibodies.
In vitro LLPS assay
The purified recombinant GFP fusion proteins maltose-binding protein (MBP)-GFP-RDL and MBP-GFP-RDS were stored in stock solution (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 2 mM DTT) and subjected to an in vitro phase separation29. MBP-GFP-RDL and MBP-GFP-RDS were diluted to the indicated concentrations and mixed gently with polyethylene glycol 4000 (PEG4000, Sigma-Aldrich, Darmstadt, Germany, cat: 1.09727) or Ficoll 400 (Solarbio, Beijing, China, cat: F8150) to a final concentration of 10% or 15%, respectively (both w/v). Five microliters of the solution were transferred to a glass slide and imaged by confocal microscopy (Zeiss LSM800, Jena, Germany; laser excitation, 488 nm; collection bandwidth, 500–530 nm; digital gain, 1.0). To check the influence of salt and protein concentration on RDM16 condensation, the same procedure was applied, using different amount of protein (5 to 15 μM) and NaCl (125 to 1000 mM) followed by gently mixing the protein solution before microscopy imaging.
Fluorescence recovery after photobleaching (FRAP) assay
For the in vitro FRAP assay, the purified recombinant GFP fusion proteins GFP-RDL and GFP-RDS were subjected to fluorescence imaging under a confocal microscope. After the target regions were focused and imaged, a small portion of GFP signal was bleached with 50 pulses of the laser beam set to 100% intensity. The fluorescence of each image was collected (one image every 2–3 s) and quantified to generate a recovery curve. For in vivo tests, the roots of transgenic Arabidopsis seedlings expressing RDM16-GFP driven by the native RDM16 or cauliflower mosaic virus (CaMV) 35S promoter were subjected to fluorescence imaging under a confocal microscope. After the target regions were focused and imaged, a small portion of GFP signal was bleached with five pulses of the laser beam at 100% intensity.
RNA immunoprecipitation (RIP)
Procedures for crosslinking, nuclear protein extraction, and immunoprecipitation were performed as described previously with minor modifications74. Briefly, 3 g of 2-week-old seedlings was crosslinked with 0.5% (w/v) formaldehyde, and the nuclear fraction was isolated and sonicated. After the chromatin was prewashed with protein A/G magnetic beads, and the samples were precipitated with anti-GFP (Abcam, Cambridge, UK, cat.: ab290, lot: GR3251545-1, dilution: 1:1000) antibodies. The obtained protein-RNA complexes were treated with 20 μg proteinase K and incubated for 1 h at 50 °C. To reverse the crosslinking, each sample was treated with 16 μL 5 M NaCl and incubated at 65 °C for 1 h. The RNA samples were purified in phenol:chloroform:isoamyl-alcohol (25:24:1, pH 4.3, v/v/v), and the aqueous phase was transferred to ethanol in the presence of 0.15 M sodium acetate (pH 5.3) and 0.3 mg GlycoBlue (Invitrogen). Following precipitation, the RNA pellet was dissolved in ddH2O, and reverse transcribed into first-strand cDNA using a Perfect Real Time Reverse Transcription kit (TaKaRa, RR047A) according to the manufacturer’s instructions. RT-qPCR was performed with the primers listed in Supplementary Data 6.
mRNA decay analysis
mRNA decay analysis was performed as previously described29. Briefly, 12-day-old Col-0 seedlings were incubated in 5 mL of incubation buffer with rotation at 75 rpm in 6-well plates for 30 min. 1 mM cordycepin (Solarbio, Beijing, China, cat: 70-03-0) was added to treat the seedlings for a series of time. Then the plant samples were frozen in liquid nitrogen, and subjected to RNA extraction for RT-qPCR analysis as described above.
Bimolecular fluorescence complementation (BiFC) assays
For BiFC, the RDL coding sequence was cloned into the pUC-SPYNE vector and the RDS coding sequence was cloned into the pUC-SPYCE vector. The resulting plasmids were co-transfected into Arabidopsis Col-0 protoplasts via PEG-mediated transfection75. After 24 h of incubation in the dark, the protoplasts were imaged using a Zeiss confocal imaging system.
Pull-down assay
The coding sequence of RDL was cloned into the vector pET28a-His-MBP to produce the MBP-RDL fusion protein. The coding sequence of RDS was cloned into the vector pGEX-GST to produce the glutathione S-transferase (GST)-RDS fusion protein. The purified MBP or MBP-RDL was incubated overnight at 4 °C in equal volumes with GST-RDS. His beads (Sigma-Aldrich, H9914) were washed with 1× phosphate-buffered saline (PBS, pH 7.2) and added to the protein mixture for incubation at 4 °C for 4 h. Magnetic bead-protein complexes were collected using a magnetic holder. After three washes with washing buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8.0) and protein separation by SDS-PAGE, the target proteins were identified by immunoblotting with anti-His (Abcam, cat.: ab184607, lot: GR3224572-4, dilution: 1:2000) or anti-GST (Abmart, cat.: M20007, lot: 324184, dilution: 1:5000) antibodies.
Co-immunoprecipitation (Co-IP) assay
The pLGNL-FLAG or pLGNL-RDS-FLAG vectors were constructed and cotransformed into transgenic rdm16-4 RDM16pro:RDL-GFP protoplasts and incubated in the dark for 24 h. The protoplasts were collected by brief centrifugation, and the proteins were extracted by incubation in protein extraction buffer. The target protein was identified by immunoblotting with anti-GFP (Roche, cat.: 11814460001, lot: 42903200, dilution: 1:1000) or anti-FLAG (MBL, Sapporo, Hokkaido, Japan, cat.: M185, lot: 012, dilution: 1:10,000) antibodies.
Circular dichroism (CD) spectroscopy
Far-UV CD spectra were recorded for purified MBP-RDM16 and MBP-RDM16mu to determine their secondary structures. 300 μL protein sample with the final concentration of 0.1 μg/μL (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, and 2 mM DTT) were added into 1 mm quartz cuvette (Starna Scientific, 1/Q/1/CD). Far-UV tests using a CD spectrometer (Bio-Logic Science Instruments, MOS-500, France) were performed with the parameter setting: begin wavelength 180 nm, end wavelength 260 nm, step 1 nm, repeat 1 time, acquisition period 1 s/point, cuvette width 1 cm. Three scans were conducted for each sample. Finally, Circular Dichroism Neural Network (CDNN) software was used to fit and calculate the secondary structure of the proteins (200 to 260 nm).
Dual-luciferase assay
The HSFA3 genomic sequence (containing two exons and one intron as shown in Supplementary Fig. 11d) was cloned into the pCAMBIA0800-35S-LUC vector to generate 35S:HSFA3-LUC. The RDM16, RDM16mu, and FUS-RDM16mu coding region was cloned into the pCAMBIA1300-GFP vector to generate RDM16-GFP, RDM16mu-GFP, and FUS-RDM16mu-GFP. To test the influence of RDM16, RDM16mu, and FUS-RDM16mu on HSFA3 splicing, the RDM16-GFP, RDM16mu-GFP, and FUS-RDM16mu-GFP constructs were co-transfected together with 35S:HSFA3-LUC respectively into Col-0 protoplasts. After 24 h of cultivation, the protoplasts were collected for the quantitative analysis of LUC activity. Fluc (Firefly luciferase) activity and the Rluc (Renilla luciferase) activity were measured using a Dualucif Firefly & Renilla Assay Kit (cat.: F6075M; BioScience, Shanghai, China) with an Ensight™ Multimode Plate Reader (Perkin Elmer, America) according to the manufacturer’s instructions.
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism (Version 10.1.2) and SPSS 18.0. Quantification of protein abundance was performed in ImageJ. Sample size, P values and statistical methods employed are described in the respective figure legends or in the Source data. Differences were considered significant at P < 0.05. All experiments were repeated at least twice, and multiple biological replicates were used in each experiment. No data were excluded from the analyses. The investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All sequencing data generated in this study have been deposited in the NCBI database under accession code PRJNA1162775 and PRJNA1165901. The sequence data used in this study are available in the Arabidopsis Information Resource database under the following accession numbers: RDM16 (At1g28060), UBC (At1g50490), and HSFA3 (At5g03720). All other data generated in this study are provided in the Supplementary Information/Source Data file or from the corresponding author upon request. Source data are provided with this paper.
References
Ohama, N., Sato, H., Shinozaki, K. & Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65 (2017).
McLoughlin, F. et al. Class I and II small heat-shock proteins protect protein translation factors during heat stress. Plant Physiol. 172, 1221–1236 (2016).
Liu, J. et al. An autoregulatory loop controlling arabidopsis HsfA2 expression: role of heat shock-induced alternative splicing. Plant Physiol. 162, 512–521 (2013).
Zhong, L. et al. Chloroplast small Heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell 25, 2925–2943 (2013).
Charng, Y.-Y. et al. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in arabidopsis. Plant Physiol. 143, 251–262 (2007).
Ikeda, M., Mitsuda, N. & Ohme-Takagi, M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 157, 1243–1254 (2011).
Scharf, K. D., Berberich, T., Ebersberger, I. & Nover, L. The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta 1819, 104–119 (2012).
Baniwal, S. K. et al. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 29, 471–487 (2004).
Friedrich, T. et al. Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat. Commun. 12, 3426 (2021).
Will, C. L. & Lührmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3, a003707 (2011).
Jurica, M. S. & Moore, M. J. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12, 5–14 (2003).
Jang, Y. H. et al. A homolog of splicing factor SF1 is essential for development and is involved in the alternative splicing of pre-mRNA in Arabidopsis thaliana. Plant J. 78, 591–603 (2014).
Lee, K. C. et al. RRM domain of Arabidopsis splicing factor SF1 is important for pre-mRNA splicing of a specific set of genes. Plant Cell Rep. 36, 1083–1095 (2017).
Zhang, W., Du, B., Liu, D. & Qi, X. Splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Biochem. Biophys. Res. Commun. 455, 312–317 (2014).
Liu, M. et al. GAMETOPHYTIC FACTOR 1, involved in pre-mRNA splicing, is essential for megagametogenesis and embryogenesis in Arabidopsis. J. Integr. Plant Biol. 51, 261–271 (2009).
Staiger, D. & Brown, J. W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25, 3640–3656 (2013).
Keller, M. et al. Alternative splicing in tomato pollen in response to heat stress. DNA Res. 24, 205–217 (2017).
Rosenkranz, R. R. E., Ullrich, S., Löchli, K., Simm, S. & Fragkostefanakis, S. Relevance and regulation of alternative splicing in plant heat stress response: current understanding and future directions. Front Plant Sci. 13, 911277 (2022).
Du, J. L. et al. The splicing factor PRP31 is involved in transcriptional gene silencing and stress response in Arabidopsis. Mol. Plant 8, 1053–1068 (2015).
Anthony, J. G., Weidenhammer, E. M. & Woolford, J. L. Jr The yeast Prp3 protein is a U4/U6 snRNP protein necessary for integrity of the U4/U6 snRNP and the U4/U6.U5 tri-snRNP. RNA 3, 1143–1152 (1997).
Lv, B. et al. The pre-mRNA splicing factor RDM16 regulates root stem cell maintenance in Arabidopsis. J. Integr. Plant Biol. 63, 662–678 (2020).
Huang, C. F. et al. A Pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. PLoS Genet. 9, e1003779 (2013).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Field, S., Jang, G.-J., Dean, C., Strader, L. C. & Rhee, S. Y. Plants use molecular mechanisms mediated by biomolecular condensates to integrate environmental cues with development. Plant Cell 35, 3173–3186 (2023).
Alberti, S., Gladfelter, A. & Mittag, T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019).
Zavaliev, R., Mohan, R., Chen, T. & Dong, X. Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell 182, 1093–1108.e1018 (2020).
Huang, S., Zhu, S., Kumar, P. & MacMicking, J. D. A phase-separated nuclear GBPL circuit controls immunity in plants. Nature 594, 424–429 (2021).
Wang, W. et al. The P-body component DECAPPING5 and the floral repressor SISTER OF FCA regulate FLOWERING LOCUS C transcription in Arabidopsis. Plant Cell 35, 3303–3324 (2023).
Li, Y. R., King, O. D., Shorter, J. & Gitler, A. D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 (2013).
Zhang, H., Qin, W., Romero, H., Leonhardt, H. & Cardoso, M. C. Heterochromatin organization and phase separation. Nucleus 14, 2159142 (2023).
Sheu-Gruttadauria, J. & MacRae, I. J. Phase transitions in the assembly and function of human miRISC. Cell 173, 946–957.e916 (2018).
Noda, N. N., Wang, Z. & Zhang, H. Liquid-liquid phase separation in autophagy. J. Cell Biol. 219, e202004062 (2020).
Jung, J. H. et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585, 256–260 (2020).
Zhu, S. et al. Liquid-liquid phase separation of RBGD2/4 is required for heat stress resistance in Arabidopsis. Dev. Cell 57, 583–597.e586 (2022).
Zhu, P., Lister, C. & Dean, C. Cold-induced arabidopsis FRIGIDA nuclear condensates for FLC repression. Nature 599, 657–661 (2021).
Huang, X. et al. ROS regulated reversible protein phase separation synchronizes plant flowering. Nat. Chem. Biol. 17, 549–557 (2021).
Dorone, Y. et al. A prion-like protein regulator of seed germination undergoes hydration-dependent phase separation. Cell 184, 4284–4298.e4227 (2021).
Wang, B. et al. Condensation of SEUSS promotes hyperosmotic stress tolerance in Arabidopsis. Nat. Chem. Biol. 18, 1361–1369 (2022).
Ernst, J. & Bar-Joseph, Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinforma. 7, 191 (2006).
Gao, C. et al. Phylogenetic analysis and stress response of the plant U2 small nuclear ribonucleoprotein B” gene family. BMC Genomics 23, 744 (2022).
dFA, M. et al. The U1 snRNP Subunit LUC7 modulates plant development and stress responses via regulation of alternative splicing. Plant Cell 30, 2838–2854 (2018).
Ohtani, M. Plant snRNP biogenesis: a perspective from the nucleolus and cajal bodies. Front. Plant Sci. 8, 2184 (2018).
Liu, Q., Liu, W., Niu, Y., Wang, T. & Dong, J. Liquid-liquid phase separation in plants: advances and perspectives from model species to crops. Plant Commun. 5, 100663 (2024).
Kim, G. D., Cho, Y. H., Lee, B. H. & Yoo, S. D. STABILIZED1 modulates pre-mRNA splicing for thermotolerance. Plant Physiol. 173, 2370–2382 (2017).
Nishizawa-Yokoi, A. et al. HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 52, 933–945 (2011).
Lin, K.-F., Tsai, M.-Y., Lu, C.-A., Wu, S.-J. & Yeh, C.-H. The roles of Arabidopsis HSFA2, HSFA4a, and HSFA7a in the heat shock response and cytosolic protein response. Bot. Stud. 59, 15 (2018).
Nishizawa, A. et al. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547 (2006).
Andrási, N. et al. The mitogen-activated protein kinase 4-phosphorylated heat shock factor A4A regulates responses to combined salt and heat stresses. J. Exp. Bot. 70, 4903–4918 (2019).
Fang, X. et al. Arabidopsis FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. Nature 569, 265–269 (2019).
Wegmann, S. et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049 (2018).
Krainer, G. et al. Reentrant liquid condensate phase of proteins is stabilized by hydrophobic and non-ionic interactions. Nat. Commun. 12, 1085 (2021).
Liu, S. et al. A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing. elife 4, e07320 (2015).
Škiljaica, A. et al. Evaluation of reference genes for RT-qPCR gene expression analysis in Arabidopsis thaliana exposed to elevated temperatures. Plant Biol. 24, 367–379 (2022).
Ji, Y., Li, F. & Qiao, Y. Modulating liquid–liquid phase separation of FUS: mechanisms and strategies. J. Mater. Chem. B 10, 8616–8628 (2022).
Kalyna, M. et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40, 2454–2469 (2012).
Kuldau, G. A. & Raju, N. B. NL G. Repeat-induced point mutations in Pad-1, a putative RNA splicing factor from Neurospora crassa, confer dominant lethal effects on ascus development. Fungal Genet. Biol. 23, 169–180 (1998).
Imai, H., Chan, E. K., Kiyosawa, K. & Fu, X. D. EM T. Novel nuclear autoantigen with splicing factor motifs identified with antibody from hepatocellular carcinoma. J. Clin. Invest. 92, 2419–2426 (1993).
Jung, D. J., Na, S. Y. & Na, D. S. JW L. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J. Biol. Chem. 277, 1229–1234 (2002).
Al-Husini, N., Tomares, D. T., Bitar, O., Childers, W. S. & Schrader, J. M. α-Proteobacterial RNA degradosomes assemble liquid-liquid phase-separated RNP bodies. Mol. Cell 71, 1027–1039.e1014 (2018).
Al-Husini, N. et al. BR-bodies provide selectively permeable condensates that stimulate mRNA decay and prevent release of decay Intermediates. Mol. Cell 78, 670–682.e678 (2020).
Yan, K. et al. SGF29 nuclear condensates reinforce cellular aging. Cell Discov. 9, 110 (2023).
Poudyal, M. et al. Intermolecular interactions underlie protein/peptide phase separation irrespective of sequence and structure at crowded milieu. Nat. Commun. 14, 6199 (2023).
Borcherds, W., Bremer, A., Borgia, M. B. & Mittag, T. How do intrinsically disordered protein regions encode a driving force for liquid–liquid phase separation? Curr. Opin. Struct. Biol. 67, 41–50 (2021).
Nott, T. J. et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015).
Qamar, S. et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine Cation-π interactions. Cell 173, 720–734.e715 (2018).
Lee, Y. & Rio, D. C. Mechanisms and regulation of alternative Pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323 (2015).
Laloum, T., Martín, G. & Duque, P. Alternative splicing control of abiotic stress responses. Trends Plant Sci. 23, 140–150 (2018).
Wen, J. et al. Alternative splicing of TaHSFA6e modulates heat shock protein-mediated translational regulation in response to heat stress in wheat. N. Phytol. 239, 2235–2247 (2023).
Pose, D. et al. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417 (2013).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Shen, S. et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl Acad. Sci. USA 111, E5593–E5601 (2014).
Li, P., Tao, Z. & Dean, C. Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev. 29, 696–701 (2015).
Kim, D.-H., Xi, Y. & Sung, S. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 13, e1006939 (2017).
Wang Y, et al. Molecular variation in a functionally divergent homolog of FCA regulates flowering time in Arabidopsis thaliana. Nat. Commun. 11, 5830 (2020).
Acknowledgements
We thank Dr. Jian Kang Zhu from the Frontier Biotechnology Research Institute of Southern University of Science and Technology for the gift of rdm16-1 seeds and Dr. Zhaojun Ding from the School of Life Sciences of Shandong University for rdm16-4 seeds. This work was supported by the National Key Research and Development Program of China (2022YFD1201803) to P.J.L, and the Anhui Provincial Major Science and Technology Project (202203a06020005) to P.J.L.
Author information
Authors and Affiliations
Contributions
P.J.L., J.M., S.L., and C.H.W. designed the experiments and analyzed the data. J.M., S.L., C.H.W., T.Y.W., Z.T., S.J.H., N.L., and Y.B.Z. performed the experiments. P.J.L. and J.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Cesar Cuevas-Velazquez, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ma, J., Li, S., Wang, T. et al. Cooperative condensation of RNA-DIRECTED DNA METHYLATION 16 splicing isoforms enhances heat tolerance in Arabidopsis. Nat Commun 16, 433 (2025). https://doi.org/10.1038/s41467-025-55850-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-55850-w