Abstract
Bacterial transcription activator-like effectors (TALEs) promote pathogenicity by activating host susceptibility (S) genes. To understand the pathogenicity and host adaptation of Xanthomonas citri pv. malvacearum (Xcm), we assemble the genome and the TALE repertoire of three recent Xcm Texas isolates. A newly evolved TALE, Tal7b, activates GhSWEET14a and GhSWEET14b, different from GhSWEET10 targeted by a TALE in an early Xcm isolate. Activation of GhSWEET14a and GhSWEET14b results in water-soaked lesions. Transcriptome profiling coupled with TALE-binding element prediction identify a pectin lyase gene as an additional Tal7b target, quantitatively contributing to Xcm virulence alongside GhSWEET14a/b. CRISPR-Cas9 gene editing supports the function of GhSWEETs in cotton bacterial blight and the promise of disrupting the TALE-binding site in S genes for disease management. Collectively, our findings elucidate the rapid evolution of TALEs in Xanthomonas field isolates and highlight the virulence mechanism wherein TALEs induce multiple S genes to promote pathogenicity.
Similar content being viewed by others
Introduction
Cotton (Gossypium spp.) stands as a valuable crop, cultivated worldwide for its versatility in providing fiber, livestock feed, and oil1. The allotetraploid species G. hirsutum, commonly known as upland cotton, is predominantly grown for commercial cotton production. Despite its economic importance, cotton yields are constrained by environmental stresses, including pathogens. Bacterial blight of cotton (BBC), caused by Xanthomonas citri pv. malvacearum (Xcm), is a destructive foliar disease2. The disease can affect any aerial part of the plant, and typically manifests as water-soaked lesions. These lesions can progress to become necrotic, causing premature defoliation and boll loss2,3. Historically, BBC control relied on measures such as acid-delinted seeds and the cultivation of BBC-resistant cotton varieties4. Around 2011, a resurgence of BBC occurred across the southern United States, impacting previously resistant cotton varieties3,5,6. The factors contributing to this resurgence, such as pathogen population composition, genomic organization, and virulence dynamics, remain elusive.
The key virulence factors linked to the pathogenicity of Xanthomonas spp. are transcription activator-like effectors (TALEs), which emulate the function of eukaryotic transcription factors7,8,9. These TALEs exhibit a conserved structure comprising a type III secretion signal at the amino (N)-terminus, followed by a central repeat region (CRR) containing 1.5 to 33.5 tandem repeats, each consisting of nearly identical 33-34 amino acids. The amino acids between different repeats primarily differ at the 12th and 13th residues, designated as the repeat variable di-residue (RVD). Following the CRR are nuclear localization signals and an acidic activation domain at the carboxyl (C)-terminus7,8,9. Each RVD within the CRR has a selectivity for a specific nucleotide, and the arrangement of RVDs within the CRR corresponds to a binding site within the promoter region of a host target gene, referred to as the effector-binding element (EBE)4,7. The first identified TALE, AvrBs3, in Xanthomonas campestris pv. vesicatoria (Xcv), the causal agent of bacterial leaf spot on pepper10, induces the expression of UPA20, encoding a basic helix-loop-helix (bHLH) transcription factor that regulates plant cell hypertrophy11.
The plant SWEET gene family, crucial for sugar transport and accumulation in plants12,13, encodes proteins primarily located in the cell membrane. These proteins play a role in the uptake, efflux, and intercellular transport of sugars, with the family divided into four clades based on the different sugar they transport: clade I and II for hexose, clade III for sucrose, and clade IV for fructose13,14. Clade III SWEET genes in different plants are targeted by various Xanthomonas spp. for pathogenicity, termed susceptibility (S) genes. For example, rice OsSWEET1115, OsSWEET1316, and OsSWEET1417,18,19 are S genes targeted by TALEs from X. oryzae pv. oryzae (Xoo). Pepper UPA16 and cassava MeSWEET10a are targeted by TALEs from Xcv and X. axonopodis pv. manihotis (Xam), respectively11,20. Cotton GhSWEET10 is targeted by TALE Avrb6 in Xcm strain H1005 to induce water-soaked lesions21. The upregulation of clade III SWEET genes, such as GhSWEET10 by Avrb6, leads to the efflux of sucrose from the host cell into the apoplast21,22. This released sucrose may potentially serve as a carbon and energy source for bacterial infection or alter the apoplast water potential for water-soaked lesion development in plant leaves16,19,20,23,24,25,26.
Here, we show reemerging Xcm isolates from BBC-infected cotton fields in Texas do not induce GhSWEET10. Instead, they induce the expression of GhSWEET14a and GhSWEET14b, two additional members of the clade III GhSWEET gene family21, highlighting the dynamic nature of Xcm-cotton interactions. The shift in the induction of specific SWEET genes implies an ongoing evolutionary adaptation, suggesting the potential evolution of new TALEs in Xcm to induce alternative S genes in cotton. To gain insight into the evolutionary adaptation of Xcm and its interaction with cotton, we sequence and assemble the whole genomes of three recent Xcm Texas field isolates and examine their TALE repertoires (TALomes). By generating a series of genetic knockout mutants of individual TALEs and screening them in cotton, we identify a newly evolved TALE, Tal7b, contributing to water-soaked lesion development and activating the expression of GhSWEET14a and GhSWEET14b. While the activation of GhSWEET14a or GhSWEET14b alone by gene-specific designer TALEs (dTALEs) weakly induced water-soaked lesions, the simultaneous activation of both genes by dTALEs more strongly induces water-soaked lesions, suggesting partial redundancy of function. CRISPR-Cas9-mediated gene editing at the EBE or coding regions of GhSWEET10 gene results in a loss of susceptibility to Xcm H1005, providing additional support for the role of SWEET genes in mediating lesion development. To further understand the virulence mechanism of Tal7b, we deploy whole-transcriptome analysis and in silico TALE EBE predictions, identifying a pectin lyase gene, GhPL1, as an additional Tal7b target. Importantly, GhPL1 quantitatively contributes to water-soaked lesion development, and during simultaneous activation alongside GhSWEET14a and GhSWEET14b using dTALEs, acts synergistically, restoring water-soaking to the level of induction by Tal7b. Collectively, our findings exemplify the rapid evolution of TALEs in Xanthomonas spp. and elucidate a mechanism that the cotton pathogen uses to promote disease by simultaneously activating, through a single TALE, members of two functionally distinct gene families that contribute synergistically to water-soaked lesion development.
Results
Virulence diversity and whole-genome sequence analysis of recent field Xcm isolates
To examine the dynamics of BBC disease, a series of Xcm isolates were collected from infected cotton fields across high cotton-yielding regions in Texas between 2010 and 2015, and nine isolates designated Xcm TX1 to Xcm TX9 were further characterized21. Among them, isolates TX4 and TX7 exhibited the most significant upregulation of the GhSWEET14a and GhSWEET14b genes, whereas isolate TX9 displayed the least or no upregulation of GhSWEET14a and GhSWEET14b21. Given their distinct effects on GhSWEET gene expression, these three Xcm isolates, TX4, TX7, and TX9, were selected for further investigation in this study (Fig. 1A). Upon inoculation into the cotton germplasm Acala 44E (Ac44E), Xcm isolates TX4 and TX7 elicited virulence, characterized by the development of water-soaked lesions (Fig. 1B, C). This pattern resembled that of previously characterized virulent Xcm isolates Xcm H1005 and Xcm MSCT1. In contrast, Xcm TX9 showed very minor or no water-soaked lesions, more similar to Xcm HM2.2, a multiple TALE deletion mutant of Xcm H100527, which showed nearly no water-soaked lesions upon infection (Fig. 1B, C). Therefore, Xcm TX4 and TX7 trigger water-soaked lesion development, whereas Xcm TX9 shows almost no detectable water-soaked symptoms in cotton Ac44E.
A Geographical locations and collection years of Xcm isolates. Xcm TX4, TX7, and TX9 were from infected cotton leaves in Texas. Xcm H1005 is a derivative of Xcm H from Oklahoma30 and Xcm MSCT1 is from Mississippi29. Green circle sizes indicate the relative acreage of cotton production in the region. Created in BioRender. Mormile, B. (2025) https://BioRender.com/g97n131/. B Xcm isolates induce variable degrees of water-soaked lesions in cotton. Xcm isolates were syringe-inoculated at OD600 = 0.1 into ten-day-old cotton cotyledons with photos shown at five days post-inoculation (dpi). C Xcm TX4 and TX7 elicit more severe water-soaked lesions than Xcm TX9. Water-soaked lesions in (B) were analyzed using ImageJ FIJI81. Data are shown as the negative grayscale means ± s.e. of water-soaked lesions for Xcm isolates relative to Xcm HM2.2 from 9 independent repeats (n = 9) The asterisks indicate a significant difference (***p < 0.001, ns = no significance) compared to Xcm HM2.2 control using a two-tailed Student’s t-test. D Genome comparisons of Xcm isolates. Genome alignment was conducted using progressiveMAUVE67 with default parameters. Colored rectangles represent locally collinear blocks of homology without any rearrangement across the aligned sequences. Xcm TX4, TX7, and TX9 each harbor a circular 5-megabase pair (Mb) chromosome, two 52-kilobase plasmids (pXCM52.4 kb and pXCM52kb), and a 15-kb plasmid (pXCM15kb), with overall sequence identities exceeding 99%. Xcm TX4 contains an inverted region in the chromosome and an additional tandem 12 bp (*) sequence in pXCM15kb compared to Xcm TX7 and TX9. Xcm MSCT1 harbors a circular 5-Mb chromosome, one 15 kb plasmid (pMSCT15kb), one 60 kb plasmid (pMSCT60kb), and one 44 kb plasmid (pMSCT44kb). Xcm H1005 contains a 5 Mb chromosome and one 88 kb plasmid. The genome sequences of Xcm H1005 and MSCT1 were from the National Center of Biotechnology Information (NCBI) database. The experiments in B, C were repeated four times with a similar result. Source data are provided as a Source Data file.
To explore potential genome variations contributing to virulence, we employed whole-genome sequencing with the Pacific Biosciences (PacBio) single-molecule real-time (SMRT) sequencing platform, generating long reads capable of spanning the repeat-rich TALE genes for efficient de novo genome assembly. The resulting assemblies revealed a common genomic architecture among Texas Xcm isolates, each harboring a 5-megabase (Mb) circular chromosome, along with one 15 kb28 plasmid (pXCM15kb) and two 52 kb plasmids (pXCM52.4 kb and pXCM52kb) (Fig. 1D, Supplementary Fig. 1, and Supplementary Tables 1 and 2). Genome alignment demonstrated remarkable similarities in overall genomic structures among Xcm TX4, TX7, and TX9 (Fig. 1D, Supplementary Figs. 1 and 2). Noteworthy differences emerge as an inverted region within the chromosome and an additional tandem 12 bp segment in plasmid pXCM15kb of Xcm TX4 compared to TX7 and TX9 (Fig. 1D). Despite variations in virulence, Xcm TX4, TX7, and TX9 share an almost identical overall genome sequence.
Xcm MSCT1 was isolated in Mississippi during the 2011 outbreak29. To facilitate a comparative analysis of Xcm isolates associated with the BBC resurgence in different states, genome alignment was performed to compare Mississippi isolate MSCT1 with Texas isolates. Given the high genomic similarity among Texas Xcm isolates, TX4 was chosen as a representative for the pairwise comparison. MSCT1 harbors a 5 Mb circular chromosome, one 15 kb plasmid (pMSCT15kb), one 60 kb plasmid (pMSCT60kb), and one 44 kb plasmid (pMSCT44kb) (Fig. 1D). In line with different Texas isolates, the chromosome and plasmid pMSCT15kb of Xcm MSCT1 are similar to those of Xcm TX4. Notably, dissimilarities emerge in the two large plasmids, with relatively few locally collinear blocks (Fig. 1D).
However, Xcm H1005, an isolate collected in 1968 from Oklahoma30, exhibited divergence from Xcm TX4 and MSCT1 in both the chromosome and plasmids. Genomic alternations, including chromosome translocations, inversions, and deletions, were observed in the chromosome between Xcm TX4 and H1005. The divergence is even more pronounced in the plasmids, where Xcm H1005 carries only one plasmid, which has limited collinear blocks with the 52 kb plasmids of Xcm TX4. Both Mississippi isolate MSCT1 and Texas Xcm isolates are divergent from the early Oklahoma isolate (Fig. 1D).
Diversity in the TALE repertoire among different Xcm isolates
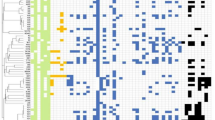
TALEs are key determinants for Xanthomas pathogenicity7,8,9. To gain an understanding of the TALE diversity and evolution, we analyzed the TALE repertoires of Texas Xcm isolates using the AnnoTALE tool31. Each Texas Xcm isolate has eight intact TALE genes: one on the chromosome (Tal1), five on plasmid pXCM52.4 kb (Tal2 to Tal6), and two on plasmid pXCM52kb (Tal7b and Tal8) (Fig. 2A). Of note, we named the TALEs in Texas isolates based on their order in the genome from the origin of replication and by replicon (chromosome first, then plasmids in order)32,33. The nomenclature of Tal7b is attributed to the presence of a partial TALE sequence immediately upstream, with an incomplete 5’ region and only one repeat. Xcm TX4 and MSCT1 exhibited a similar TALE composition and identity across their genomes (Fig. 2A–D). Note that the TALEs of MSCT1 were previously named numerically as well29, but not in order of position in the genome, so the numbers do not correspond to those of the Texas isolate TALEs. Immunoblot analysis with α-TALE antibodies revealed that the shared number and relative sizes of TALEs in Xcm MSCT1 and the Texas isolates are not conserved in strain H1005 and strain N1003, a derivative of Xcm N collected from Burkina Faso in 196830 (Fig. 2B).
A TALE distribution in the chromosome and plasmids of Xcm TX4 and MSCT1. TALEs in Xcm TX4 were designated numerically from the replication origin of the chromosome and plasmids. TALEs in Xcm MSCT1 were designated previously using a different scheme29. The same-colored boxes indicate TALEs with identical RVD sequences. B Xcm TX4 and MSCT1 share similar TALome distinct from those of Xcm H1005 and N1003. Xcm strains were subjected to immunoblotting using α-TALE antiserum72. Molecular weight (kDa) is shown on the left of the gel. Asterisks (*) indicate annotated TALEs at their predicted sizes. The experiment was repeated twice with a similar result. C The repeat variable di-residues (RVD) and predicted effector binding elements (EBE) of the TALEs from Xcm MSCT1 and TX4, TX7, and TX9. RVDs are identical within the TALome of Xcm TX4 and MSCT1. Xcm TX9 exhibits three RVD mismatches in Tal2, one mismatch in Tal4, and two mismatches (green highlight) and a deletion (red highlight) in Tal7b relative to Xcm TX4 and MSCT1. Xcm TX7 has two RVD mismatches (yellow highlight) within Tal2 relative to Xcm TX4 and MSCT1. TALEs from Xcm MSCT1 are in parentheses. D Relationships of TALE from Xcm isolates based on their predicted DNA targets using FuncTAL35. Thirty-one unique RVD sequences from eighty TALEs from nine Xcm isolates. FuncTAL clusters TALEs based on functional relatedness by calculating similarities in predicted EBE probabilities according to the RVD-DNA code. Highlighted boxes with colors show the different classes (Tal followed by a two-letter code) assigned by AnnoTALE. Individual TALEs are designated as RVD followed by the number of repeats within their central repeat region (CRR) and a lowercase letter to distinguish among TALEs with the same number of repeats. Numbers in parentheses indicate TALEs from different isolates with the same RVD sequence. The genome sequences of Xcm HD-1, AR81009, and MS14003 are from the NCBI database. E Phylogenetic analysis of TALEs from Xcm isolates. Eighty TALEs in (D) were analyzed using DisTAL35 which clusters based on CRR nucleotide sequence similarities. Only TALEs from Xcm isolates with available whole-genome sequences were used in this analysis. Source data are provided as a Source Data file.
The CRRs harboring RVDs in TALEs of Texas Xcm isolates and Xcm MSCT1 range in length from 14 to 28 copies of the tandem amino acid sequence. Given the importance of RVD composition in the recognition and binding of TALEs to their target DNA sequence, we compared TALE RVDs among recent Xcm isolates: MSCT1, TX4, TX7, and TX9. Notably, water-soaked lesion-inducing Xcm isolates MSCT1, TX4, and TX7 share nearly identical RVD sequences within all eight TALEs, except Tal2TX7, which contains two RVD differences relative to its orthologs in Xcm MSCT1 and TX4 (Fig. 2C). In contrast, the virulence-attenuated Xcm TX9 exhibits several RVD dissimilarities in Tal2, Tal4, and Tal7b compared to the counterparts in Xcm MSCT1 and TX4 (Fig. 2C). Considering the role of RVDs in directing TALE to its host target gene and inducing susceptibility, the divergence of these sequences in Tal2, Tal4, and Tal7b of the weakly virulent strain suggests that one or more of these TALEs may contribute to bacterial pathogenicity and water-soaked lesion development.
Next, we conducted an analysis to assess the functional relatedness of TALEs derived from the nine Xcm strains collected across diverse geographic locations and spanning different isolation dates (Supplementary Table 3). In addition to Xcm TX4, TX7, TX9, MSCT1, N1003, and H1005, we included three other Xcm strains with available full genome sequencing data at the time of our analysis: MS14003 isolated from Mississippi in 2014, AR81009 isolated from Argentina in 198134, and HD-1 isolated from China in 2017 (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_009671025.1/, GenBank ID: GCA_009671025.1). In total, 80 TALEs are present across nine Xcm strains. Among the 80 TALEs, thirty-one unique RVD sequences are represented, with the number of repeats in individual proteins ranging from 12 to 28 (Supplementary Fig. 3). These 31 RVD sequences were grouped into 21 classes based on their predicted corresponding DNA target sequences (EBEs), using FuncTALE35 and AnnoTALE31 (Fig. 2D). Since TALEs can tolerate a certain number of mismatches between their RVDs and corresponding EBEs, the relatively high number of EBE classes compared to unique RVDs suggests a greater degree of sequence diversity within the CRRs. This plasticity is possibly driven by frequent recombination events within the CRRs, allowing for adaptation to different host genotypes36. Further analysis, using DisTAL35 revealed that the groups based on predicted EBEs almost exclusively consist of phylogenetically related TALEs (Fig. 2E). For example, the FuncTAL class TalGY contains Tal1 from three Texas isolates and the corresponding TALE in two Mississippi isolates. Moreover, phylogenetically related reemerging Xcm isolates exhibited similar TALomes, indicating a common ancestral origin. Notably, despite the RVD polymorphisms observed in Tal2 and Tal4 among Texas isolates (Fig. 2C), Tal2 and Tal4 from TX9 belong to the same FuncTAL class as Tal2 and Tal4, respectively, from Xcm TX4 and TX7 (Fig. 2E). This indicates that differences in TAL2 and TAL4 RVD composition in different Texas isolates do not result in gross alterations to their EBE target sites. In contrast, Tal7b from Xcm TX9 belongs to a distinct FuncTAL class compared to Tal7b from Xcm TX4 and TX7, implying a different EBE target site for Tal7b from Xcm TX9 in comparison to Xcm TX4 and TX7 (Fig. 2E). This also hints at the role of Tal7b in mediating bacterial virulence.
Tal7b mediates GhSWEET14a/b induction and water-soaked lesion development
Texas Xcm isolates TX4 and TX7 induced the expression of GhSWEET14a and GhSWEET14b in cotton21. To identify the TALE responsible for this induction, we constructed a broad-host-range vector pKEB1 expressing individual TALEs from Xcm TX4 and transformed them into Xcm HM2.2, a multiple TALE deletion mutant of Xcm H1005. Quantitative reverse transcription-polymerase chain reaction (RT-qPCR) was used to analyze transcriptional changes of GhSWEET14a and GhSWEET14b following inoculation of Xcm HM2.2 expressing individual TALEs from Xcm TX4. Xcm HM2.2 carrying Tal7b, but not other TALEs, significantly upregulated GhSWEET14a, comparable to the expression level induced by Xcm TX4 (Fig. 3A). Similarly, Xcm HM2.2 carrying Tal7b, but not others, induced the expression of GhSWEET14b (Fig. 3B). Importantly, Xcm HM2.2 carrying Tal7b, but not other TALEs, strongly induced water-soaked lesions (Fig. 3C and Supplementary Fig. 4A). These results indicate that Tal7b upregulates the expression of GhSWEET14a and GhSWEET14b and mediates water-soaked lesion development.
A, B Tal7b upregulates the expression of GhSWEET14a/b in cotton. Significant differences are in comparison to Xcm HM2.2 (-). C Tal7b restores water-soaked symptom development to Xcm HM2.2. Photos taken 9 dpi. D, E Xcm TX4-induced GhSWEET14a/b expression depends on Tal7b. Significant differences are in comparison to wild-type (WT) Xcm TX4. Xcm TX4-triggered water-soaked lesion development depends on Tal7b. Photos taken 6 dpi (F). Water-soaked lesions were counted on the 3rd true leaves of vacuum infiltrated cotton 9 dpi (G). Significant differences are in comparison to WT Xcm TX4. Tal7b activates pGhSWEET14a::LUC (H) and pGhSWEET14b::LUC (I) in N. benthamiana. Luciferase activity was normalized to the mock control (set as 1). (J) Tal7b immunoprecipitates the endogenous EBE-containing sequences of GhSWEET14a (GhSWEET14aEBE) and GhSWEET14b (GhSWEET14bEBE) in cotton protoplasts. The relative enrichment was normalized to the mock control (set as 1). K GhSWEET14aEBE and GhSWEET14bEBE are critical for Tal7b immunoprecipitation in Arabidopsis protoplasts. The relative enrichment was normalized to an empty vector control (-) set as 1. L Biotin-tagged GhSWEET14a or GhSWEET14b EBE sequences bind to Tal7b, but not mutant (m) EBE sequences. Tal7b or Avrb6 expressed in Arabidopsis protoplasts were subjected to immunoblotting (IB) using α-TALE antibodies. Top panel: Tal7b or Avrb6 expression after pull-down (PD). Middle panel: Tal7b or Avrb6 inputs before PD. Bottom panel: Coomassie brilliant blue (CBB) staining of rubisco (RBC). Molecular weight (kDa) is shown on the left. Inoculations in A, B, C, D, E, F were done as in Fig. 1B. Experiments in H, I, J, K, L were repeated three times with a similar result. Data are shown as mean ± s.d. from independent repeats (n = 3) in A, B, G; (n = 4) in D, E, H, I; mean ± s.e. from four independent repeats (n = 4) in J, K. Asterisks indicate a significant difference (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001) using a two-tailed Student’s t-test. Source data are provided as a Source Data file.
Next, we investigated whether Xcm mutants with Tal2, Tal4, or Tal7b missing are less virulent in cotton by generating a series of TX4 TALE mutant strains (Supplementary Figs. 4B and 4C). RT-qPCR analysis showed that Xcm TX4Δtal7b, but not TX4Δtal2 or TX4Δtal4, induced GhSWEET14a and GhSWEET14b less than wild-type (WT) TX4 (Fig. 3D, E). The capacity to induce GhSWEET14a and GhSWEET14b was restored upon introducing pKEB1 carrying Tal7b into Xcm TX4Δtal7b (Fig. 3D, E). Notably, the induction of GhSWEET14b remained lower in the complemented strain compared with the WT, implying partial complementation effects. Furthermore, unlike Xcm TX4, TX4Δtal7b could not induce the water-soaked phenotype in cotton when infiltrated into the leaf, and water-soaking was restored by complementation with pKEB1-Tal7b (Fig. 3F). Deletion of Tal2 or Tal4, TX4Δtal2 or TX4Δtal4, did not affect water-soaked lesion development compared to Xcm TX4 (Fig. 3F). The strains were also evaluated by counting the number of lesions that developed following vacuum-inoculation. The number of water-soaked lesions was reduced upon inoculation with Xcm TX4Δtal7b compared to TX4, TX4Δtal2, and TX4Δtal4 (Fig. 3G) and pKEB1-Tal7b complemented this phenotype (Fig. 3G). Notably, in planta bacterial growth assay showed no claimable difference between Xcm TX4 and TX4Δtal7b populations over the course of seven or fourteen days (Supplementary Figs. 4D, E). Thus, Tal7b triggers the elevated expression of GhSWEET14a and GhSWEET14b and elicits water-soaked lesions but does not affect bacterial multiplication in planta. This resembles the function of Avrb6 in Xcm H1005, which enhances water-soaked lesion development by inducing GhSWEET10 expression but does not contribute to bacterial growth in cotton leaves21,27. Interestingly, TX4Δtal7b shows reduced bacterial populations on the leaf surface compared to TX4 (Supplementary Fig. 4F), suggesting that Tal7b may play a role in facilitating bacterial release to the leaf surface. Together, gain-of-function and loss-of-function assays of individual TALEs indicate that Tal7b from Xcm TX4 plays a role in the induction of GhSWEET14a and GhSWEET14b genes, as well as water-soaked lesion development in cotton.
The induction of GhSWEET14a and GhSWEET14b by Tal7b prompted us to test whether their promoters, fused with the firefly luciferase (LUC) reporter gene, could be activated by Tal7b in a heterologous system. Individually, we fused the 1 kb regions upstream of the translational start site of GhSWEET14a and GhSWEET14b with LUC and assayed these constructs by Agrobacterium-mediated transient transformation in Nicotiana benthamiana followed 24 h later by infiltration of Xcm TX4 or HM2.2 carrying Tal7b. Each Xanthomonas strain activated both pGhSWEET14a::LUC and pGhSWEET14b::LUC compared to mock treatment or Xcm HM2.2 alone (Fig. 3H, I). The Xcm TX4Δtal7b inoculation resulted in a lower induction of pGhSWEET14a::LUC and pGhSWEET14b::LUC promoters than WT TX4 (Fig. 3H, I). Screening additional Texas field isolates collected spanning 2002 and 2024 for the pGhSWEET10::LUC and pGhSWEET14a::LUC promoter activation indicated that most of these isolates strongly activated GhSWEET14a while most isolates either did not or only moderately activated GhSWEET10 (Supplementary Table 4 and Supplementary Figs. 5A and 5B), implying complex and dynamic potential acquisition TAL effectors in the field contributing to the virulence shift of the host target genes.
In addition, chromatin immunoprecipitation (ChIP)-qPCR assays using Tal7b expressed in cotton protoplasts demonstrated that Tal7b, but not the control vector (Ctrl.), enriched the endogenous EBE sequences of GhSWEET14a and GhSWEET14b promoters (Fig. 3J). To determine the requirement of the EBE sequence in mediating such binding, we further co-expressed Tal7b with GhSWEET14a under the control of its WT EBE promoter (pGhSWEET14a::GhSWEET14a-HA) or the promoter carrying EBE mutation (pGhSWEET14amEBE::GhSWEET14a-HA), and GhSWEET14b under the control of its WT EBE promoter (pGhSWEET14b::GhSWEET14b-HA) or the promoter carrying EBE mutation (pGhSWEET14bmEBE::GhSWEET14b-HA) in Arabidopsis protoplasts. As shown in Fig. 3K, Tal7b enriched the WT pGhSWEET14a::GhSWEET14a-HA or pGhSWEET14b::GhSWEET14b-HA, but not the EBE mutant pGhSWEET14amEBE::GhSWEET14a-HA or pGhSWEET14bmEBE::GhSWEET14b-HA, implying the importance of the EBE sites of pGhSWEET14a and pGhSWEET14b in binding to Tal7b. Furthermore, we also synthesized biotin-tagged EBE sequences of GhSWEET14a and 14b as well as their cognate EBE mutants and conjugated them to streptavidin agarose beads. As shown in Fig. 3L, biotin-tagged EBE sequences, but not the cognate EBE mutant sequences from the GhSWEET14a or GhSWEET14b promoters bind to Tal7b. In contrast, the biotin-tagged pGhSWEET10-EBE sequence specifically binds to Avrb6, but not to Tal7b (Fig. 3L). Taken together, data suggest that Tal7b binds to EBE sequences in the GhSWEET14a and GhSWEET14b promoters.
GhSWEET14a/b mediate Tal7b-triggered disease susceptibility
To determine the role of GhSWEET14a and GhSWEET14b in Tal7b-triggered water-soaked lesion development, we silenced GhSWEET14a and GhSWEET14b in cotton by Agrobacterium-mediated virus-induced gene silencing (VIGS) followed by inoculations of Xcm TX4 or HM2.2 carrying Tal7b (Fig. 4A). The tetraploid upland cotton contains A and D subgenomes37. GhSWEET14a has two homoeologous genes in both A and D subgenomes, referred to as GhSWEET14aA and GhSWEET14aD, respectively, and they share 99.1% sequence identity at the nucleotide level (Supplemental Fig. 6A). GhSWEET14b is only present in the D subgenome, and thus referred to as GhSWEET14bD (Supplemental Fig. 6A). GhSWEET14bD is 80.5% and 80.6% identical at the nucleotide level to GhSWEET14aA and GhSWEET14aD, respectively. We constructed a VIGS vector, VIGS-GhSWEET14a/b, that reduced the expression of GhSWEET14a (including both GhSWEET14aA and GhSWEET14aD) and GhSWEET14b, in comparison to the vector control (Supplemental Fig. 6B). Once gene silencing was established, Xcm TX4 or HM2.2 carrying Tal7b was syringe-inoculated into cotton leaves. Compared to those inoculated with the VIGS vector control, the GhSWEET14a/b-silenced cotton true leaves exhibited a noticeable reduction in water-soaked symptoms caused by Xcm TX4, confirming the role of GhSWEET14a/b in water-soaked symptom development (Fig. 4A, top panel). Furthermore, the water-soaked symptom development triggered by Xcm HM2.2 carrying Tal7b was also reduced in the GhSWEET14a/b-silenced cotton true leaves compared to VIGS vector-control leaves (Fig. 4A, bottom panel). Moreover, GhSWEET14a/b-silenced cotton true leaves vacuum-infiltrated with Xcm TX4 or HM2.2 carrying Tal7b showed fewer water-soaked lesions than VIGS control leaves inoculated with those strains (Fig. 4B, C).
A Silencing GhSWEET14a/b reduces the severity of Xcm TX4 and HM2.2 (Tal7b)-induced water-soaked symptoms. The 2nd true leaves were syringe-inoculated with Xcm TX4 or HM2.2 (Tal7b) at OD600 = 0.1. Photos taken 5 dpi. B, C Silencing GhSWEET14a/b reduces the lesion number following Xcm TX4 or HM2.2 (Tal7b) infection. Water-soaked lesions were counted on the 3rd true leaves of vacuum infiltrated cotton 10 dpi. D Xcm HM2.2 (Tal7b) enhances GhSWEET14a/b protein expression in a EBE sequence-dependent manner. Protein expression in Nicotiana benthamiana was assessed by immunoblotting (IB) with α-HA (top) or α-TALE antibodies (middle). Coomassie brilliant blue (CBB) staining of rubisco (RBC) was used as a loading control (bottom). Molecular weight (kDa) is shown on the left. E Xcm HM2.2 (Tal7b) promotes more severe water-soaked lesions under the WT EBE GhSWEET14a/b promoters compared with their cognate EBE mutants in N. benthamiana. F RVDs of designer TALEs (dTALE) and EBEs of GhSWEET14a/b, with RVDs in shaded boxes above their corresponding EBE. The numbers below indicate EBE positions relative to the translational start codon. G, H dTALEs induce the expression of target genes. Significant differences are in comparison to the vector control. Upregulation of GhSWEET14a/b by dTALEs induces water-soaked symptoms, in an incompletely additive, partially redundant manner. Photos were taken at 5 dpi (I). Lesions were analyzed in (J) as in Fig. 1C. Data are shown as the negative grayscale means ± s.e. of lesions relative to Xcm HM2.2 from 6 independent repeats (n = 6). Significant differences are comparisons between treatments. Inoculations in G, H, I, and J were done as in Fig. 1B. Experiments in I, J were repeated three times with a similar result. Data are shown as mean ± s.e. from five independent repeats (n = 5) in C. Data are shown as mean ± s.d. from three independent repeats (n = 3) in G, H. Asterisks indicate a significant difference (*p < 0.05, ***p < 0.001) using a two-tailed Student’s t-test. Source data are provided as a Source Data file.
When co-expressing Tal7b together with GhSWEET14a or GhSWEET14b tagged with an HA epitope in N. benthamiana, Tal7b enhanced GhSWEET14a-HA or GhSWEET14b-HA protein levels under the control of the WT EBE promoters (pGhSWEET14a::GhSWEET14a-HA or pGhSWEET14b::GhSWEET14b-HA), but not the EBE mutant promoters (pGhSWEET14amEBE::GhSWEET14a-HA or pGhSWEET14bmEBE::GhSWEET14b-HA) (Fig. 4D), corroborating the importance of Tal7b in binding to EBE sites to activate GhSWEET14a and GhSWEET14b expression, thus by enhancing protein expression levels. In line with this, co-inoculation with Xcm HM2.2 (Tal7b) also promoted much more severe and faster water-soaked lesion development when pGhSWEET14a::GhSWEET14a-HA or pGhSWEET14b::GhSWEET14b-HA was expressed compared to pGhSWEET14amEBE::GhSWEET14a-HA or pGhSWEET14bmEBE::GhSWEET14b-HA (Fig. 4E). Notably, inoculation of Agrobacterium carrying pGhSWEET14a::GhSWEET14a-HA or pGhSWEET14b::GhSWEET14b-HA, Xcm HM2.2 or Xcm HM2.2 (Tal7b) did not trigger water-soaked lesion development (Supplementary Fig. 7). The more severe water-soaked lesion development by pGhSWEET14a::GhSWEET14a-HA or pGhSWEET14b::GhSWEET14b-HA compared to pGhSWEET14amEBE::GhSWEET14a-HA or pGhSWEET14bmEBE::GhSWEET14b-HA upon Xcm HM2.2 (Tal7b) inoculation could be observed from both the abaxial and adaxial sides of multiple leaves (Supplementary Fig. 7). Together, these data further support the importance of Tal7b targeting GhSWEET14a/b via EBEs for water-soaked lesion development in promoting bacterial virulence.
Gene-specific dTALEs were engineered to individually induce the expression of GhSWEET14a or GhSWEET14b. The best DNA sequence match to the RVD sequence of Tal7b in each gene was predicted using the Target Finder tool of TALE-NT 2.038, and the dTALEs were designed to target sequences distinct from that candidate EBE in each gene promoter (Fig. 4F). RT-qPCR confirmed the specific induction of GhSWEET14a by dTAL-SWEET14a and GhSWEET14b by dTAL-SWEET14b when expressed in Xcm HM2.2 (Fig. 4G, H). Notably, HM2.2 harboring either dTALE alone caused a water-soaked phenotype, but to a lesser extent than HM2.2 carrying Tal7b (Fig. 4I, J). Co-inoculating Xcm HM2.2 carrying dTAL-SWEET14a and dTAL-SWEET14b induced a stronger water-soaked phenotype than the strains individually, yet still at a lower level than Tal7b, suggesting partially redundant function with respect to susceptibility (Fig. 4I, J). The data indicate that GhSWEET14a and GhSWEET14b contribute quantitatively and partially redundantly to the development of water-soaked symptoms, but they also hint at one or more additional S gene(s) targeted by Tal7b.
GhSWEET14a/b play dual roles in Xcm virulence and cotton boll development
To further investigate the roles of GhSWEET14a and GhSWEET14b, we targeted GhSWEET14a and GhSWEET14b for mutagenesis through multiplexed CRISPR-Cas9-mediated gene editing. The gRNA-GhSWEET14a/b construct was designed to simultaneously target GhSWEET14a and GhSWEET14b in both the A and D subgenomes. Given the sequence conservation within upland cotton A and D subgenomes, gRNA-GhSWEET14a targeted both GhSWEET14aA and GhSWEET14aD. As GhSWEET14b is only present in the D subgenome, gRNA-GhSWEET14b targeted GhSWEET14bD. These gRNAs were arranged in a tandem array of tRNA-gRNA motifs expressed as a single polycistronic tRNA-gRNA gene39. Cleavage of primary transcripts by the endogenous tRNA-processing system releases mature gRNA molecules40.
gRNA-GhSWEET14a/b, along with the Cas9 gene in the pRGEB32-GhU6.9 vector39, was transformed into the cotton variety Coker 312 through standard tissue culture regeneration to generate GhSWEET14a/b-edited lines. Among five lines we screened by targeted sequencing of GhSWEET14a/b, line 5-2 contained a 2-nucleotide (nt) deletion in the first exon of both GhSWEET14aA and GhSWEET14aD, as well as a 1-nt deletion in the first exon of GhSWEET14bD, each resulting in a disruptive frameshift (Fig. 5A). Following Xcm TX4 inoculation, water-soaked lesion development was reduced in the CRISPR-Cas9-GhSWEET14a/b edited cotton line compared to the control line transformed with GFP (Fig. 5B). The result substantiates the role of GhSWEET14a and GhSWEET14b in Xcm TX4-triggered water-soaked lesion development and suggests a strategy for conferring bacterial blight resistance in allotetraploid cotton by targeting multiple S genes.
A The CRISPR-GhSWEET14a/b gene-edited cotton line harbors early frameshift deletions in both GhSWEET14a and GhSWEET14b. The target site was amplified by PCR from genomic DNAs of the edited line for amplicon sequencing. Nucleotide deletions are denoted by red dashes with amino acid sequences displayed above and below, frameshift mutations indicated by red letters and shaded boxes for gRNA target sequences and PAM sites with numbers relative to the translational start codon as 1. B Water-soaked symptom development is reduced in CRISPR-GhSWEET14a/b cotton upon Xcm TX4 infection. Cotton plants were syringe-inoculated with Xcm TX4 or HM2.2 at OD600 = 0.05 with photos shown at 5 dpi. C The CRISPR-pGhSWEET10D line contains a 7-nucleotide deletion in the GhSWEET10D promoter at the Avrb6 EBE, determined by PCR amplificon sequencing. Nucleotide deletions are denoted by red dashes with sequencing chromatogram shown below, shaded boxes for the gRNA target sequence and PAM site and the underlined sequence for Avrb6 EBE. D Xcm H1005-mediated upregulation of GhSWEET10D is reduced in the CRISPR-pGhSWEET10D line. RT-qPCR analysis was done as in Fig. 3A, B. Data are shown as mean ± s.d. (n = 3) from three independent repeats. Asterisks indicate a significant difference (*p < 0.05) compared to the control line using a two-tailed Student’s t-test. E Lesion development induced by Xcm H1005 is reduced in the CRISPR-pGhSWEET10D line. Twenty-one-day-old cotton plants were vacuum-infiltrated with Xcm H1005 at OD600 = 0.0001 followed by counting water-soaked lesions on the 3rd true leaf at 10 dpi. Data are shown as mean ± s.e. (n = 6) from six independent repeats. Asterisks indicate a significant difference (*p < 0.05) compared to the control line using a two-tailed Student’s t-test. F The CRISPR-pGhSWEET10D line exhibits reduced water-soaked symptoms following Xcm H1005 infection. Inoculations were done as in Fig. 1C. Photos shown at 4 dpi. The experiments in E, F were repeated three times with a similar result. Source data are provided as a Source Data file.
However, it’s noteworthy that the CRISPR-GhSWEET14a/b plants did not produce bolls. We observed a contrast in reproductive development between the CRISPR-GhSWEET14a/b plants and control plants. While the control plants exhibited typical flower withering and subsequent boll formation a few days after bloom, the flowers of the CRISPR-GhSWEET14a/b plants persisted for over 20 days without any observable boll development (Supplementary Fig. 8A). This phenomenon corroborates the significance of sugar transporters, including SWEET genes, in development and reproduction stages of plants13. The absence of boll development in the CRISPR-GhSWEET14a/b plants suggests the essential nature of GhSWEET14a and GhSWEET14b in cotton development and reproduction.
Gene editing of the GhSWEET10 promoter EBE for Avrb6 confers resistance to Xcm H1005
As illustrated above and observed by others, knockout of SWEET genes can be detrimental (Supplementary Fig. 8A)25,41. Mutagenesis of TALE EBE sites in the S genes could offer a strategy for achieving resistance without compromising growth or reproduction42,43. As a proof-of-concept in cotton, we designed a gRNA-pGhSWEET10 construct to target the EBE of GhSWEET10, the S gene induced by Avrb6 in Xcm H100521. Notably, the induction of GhSWEET10D by Avrb6 is much higher than its A subgenome homoeolog GhSWEET10A, indicating GhSWEET10D as a primary S gene in Xcm H1005 infection21.
Through CRISPR-Cas9-mediated gene editing, we introduced a 7-nucleotide deletion in the Avrb6 EBE within the GhSWEET10D promoter in a cotton line designated CRISPR-pGhSWEET10D (Fig. 5C). RT-PCR analysis revealed that the CRISPR-pGhSWEET10D line maintained a comparable basal level of GhSWEET10D transcripts to the cotton line transformed with GFP as controls (Supplementary Fig. 8B), indicating undisturbed endogenous expression of GhSWEET10D in the CRISPR-pGhSWEET10D line. Importantly, Xcm H1005-mediated induction of GhSWEET10D was abrogated in the CRISPR-pGhSWEET10D line compared to the control line (Fig. 5D). The induction of GhSWEET10D by Xcm H1005 in the CRISPR-pGhSWEET10D line was similar to that by Xcm HM2.2, which lacks Avrb6 (Fig. 5D), suggesting that Avrb6 delivered by Xcm H1005 was ineffective at inducing GhSWEET10D in this line.
Furthermore, the CRISPR-pGhSWEET10D line displayed a reduction in water-soaked lesion development and lesion count following either hand- or vacuum-inoculation with Xcm H1005, in comparison to the control line (Fig. 5E, F). Notably, bacterial counting assays following Xcm H1005 infection showed no detectable difference in bacterial populations between the CRISPR-pGhSWEET10D line and the control line over a seven-day period (Supplementary Fig. 8C). This lack of effect on bacterial multiplication aligns with our prior results obtained by knockdown of GhSWEET10 using VIGS21. The CRISPR-pGhSWEET10D line displayed no growth defects, with normal boll development. Cases in other crops, such as rice and cassava, where EBE edits provided resistance without adverse developmental effects, support the potential of this approach. For example, EBE edits in OsSWEET11, OsSWEET13, and OsSWEET14 disrupted TAL effector binding without impairing endogenous gene function or causing developmental defects43,44. Conversely, knockout of OsSWEET15 in rice impairs fertility45. Similar results were observed in cassava, where EBE editing of MeSWEET10a conferred resistance to bacterial blight while maintaining fertility. In contrast, a knockout of MeSWEET10a led to non-viable seed production in female flowers46.
Integration of transcriptome analysis and EBE site prediction reveals GhSWEET14a/b as Tal7b targets
To uncover possible additional targets of Tal7b, we performed whole-genome RNA-sequencing (RNA-seq) analysis using the Illumina HiSeq 2000 platform to identify Tal7b-induced G. hirsutum genes. Cotyledons from the BBC susceptible cotton variety FiberMax FM 2322GL (FM 2322GL) were inoculated with either Xcm HM2.2 carrying Tal7b or an empty vector before being harvested 24 h post-inoculation (hpi). Our analysis identified 707 genes induced by Xcm HM2.2 (Tal7b) compared to the Xcm HM2.2 control using cutoffs of log2 fold change >1 and p-value < 0.05 (Fig. 6A). Next, we screened the G. hirsutum promoterome for potential Tal7b EBEs using the TALE-NT 2.0 Target Finder tool38 and further manually checked these candidates with knowledge of the codes, including the context of the EBE in the promoter. Since the score ratios for the Tal7b candidate EBEs in the GhSWEET14a/b promoters are greater than 3.0 (Supplementary Table 5), typically considered indicative of a weak candidate EBE, we relaxed the score ratio cutoff in this screen to 4.0. This analysis identified 397 genes containing predicted Tal7b EBEs within their promoters (Fig. 6A). By cross-referencing 707 Tal7b-induced genes with the 397 genes containing predicted Tal7b EBEs within their promoters, we uncovered 26 candidate gene targets of Tal7b (Fig. 6A and Supplementary Table 5). Among the top ten candidates, GhSWEET14aA and GhSWEET14bD exhibit the EBE score ratios with EBE positions around 100 bp upstream of the start codon and were induced approximately 20-fold by Xcm HM2.2 (Tal7b) compared to Xcm HM2.2 (Fig. 6B, Supplementary Table 5, and Supplementary Fig. 9). Given the molecular and physiological evidence that we obtained for GhSWEET14a and GhSWEET14b as targets of Tal7b, these findings underscore the feasibility of RNA-seq analysis coupled with EBE site prediction in identifying TALE target genes. However, they also emphasize the fact that EBE score ratios for valid targets may exceed the commonly used cutoff for the prediction of candidates. In addition to the Tal7b EBEs in the GhSWEET14a/b gene promoters described here, this is the case for the EBE of the X. oryzae pv. oryzae TALE AvrXa27 in the promoter of the rice Xa27 gene47.
A Venn diagram of Tal7b-induced genes by RNA-sequencing (RNA-seq) analysis and Tal7b-candidate target genes by TALE-NT 2.0-based EBE predication with a score ratio cutoff 4.0. B Tal7b RVDs and predicted EBEs. Colored boxes depict the RVDs of each repeat of Tal7b, with the number above indicating the RVD position within the CRR. Tal7b EBEs predicted in the promoters using TALE-NT 2.038 are listed below RVDs. C Silencing GhPL1 reduces water-soaked symptoms caused by Xcm TX4 or HM2.2 carrying Tal7b. Assay was done the same as in Fig. 4A. Photos taken at 5 dpi. D Silencing GhPL1 reduces the water-soaked lesion development caused by Xcm TX4 or HM2.2 carrying Tal7b. Assay was done the same as in Fig. 4B. Photos taken at 9 dpi. E GhPL1 is induced by dTAL-PL. Assay was done the same as in Fig. 4G. Data are shown as mean ± s.d. (n = 3) from three independent repeats. Asterisks indicate a significant difference (*p < 0.05) compared to the vector control using a two-tailed Student’s t-test. F, G GhPL1 quantitatively contributes to the water-soaked symptom development, synergistically with GhSWEET14a/b. Xcm HM2.2 carrying dTAL-PL, dTAL-SWEET14a or dTAL-SWEET14b was syringe-inoculated into seven-day-old cotton cotyledons at OD600 = 0.1. Equal proportions of each bacterial inoculum were combined with an empty vector co-inoculum. Xcm HM2.2 carrying Tal7b, co-inoculated with HM2.2 carrying empty vector to maintain the same relative OD600 as each of the others, was included as a control. Photos (F) and water-soaked lesions were analyzed using ImageJ FIJI (G) the same as in (Fig. 1C). Data are shown as the negative grayscale means ± s.e. of lesions relative to Xcm HM2.2 from 9 independent repeats (n = 9) four dpi. Asterisks indicate a significant difference (*p < 0.05, ***p < 0.001, ns = no significance) between treatments using a two-tailed Student’s t-test. The experiments in C, D, F, & G were repeated three times with a similar result. Source data are provided as a Source Data file.
A pectin lyase functions synergistically with GhSWEET14a/b to induce water-soaked lesions
Three of the top ten candidate genes with reasonable EBE positions and Tal7b score ratios were more strongly induced than GhSWEET14aA (A04G0861) and GhSWEET14bD (D02G1767), with expression levels reaching up to ~200-fold higher. Two of these, GhD08G1143 and GhA08G0935, encode proteins of unknown function. Their predicted EBE sites are positioned more than 400 bp upstream from the start codon, a location typically deemed less likely as a target gene48. The third, GhD02G1231, with an over 100-fold change in expression, encodes a pectin lyase-like protein, which was termed GhPL1. To investigate whether the expression of GhPL1 contributes to BBC disease symptom development, we silenced it using Agrobacterium-mediated VIGS, followed by Xcm inoculations. Notably, following syringe-inoculation with Xcm TX4 or HM2.2 carrying Tal7b, GhPL1-silenced cotton exhibited reduced water-soaked symptoms compared to a non-silenced vector-only control (Fig. 6C). Similarly, GhPL1-silenced cotton produced fewer water-soaked lesions than the vector-only control when vacuum-infiltrated with Xcm TX4 or HM2.2 carrying Tal7b (Fig. 6D). These results suggest that the induction of GhPL1 also contributes to the development of water-soaked lesions.
To further test this conclusion, dTAL-PL was constructed to induce the expression of GhPL1 through an EBE distinct from that of Tal7b. RT-qPCR analysis confirmed the effective induction of GhPL1 by HM2.2 expressing dTAL-PL (Fig. 6E). The upregulation of GhPL1 by dTAL-PL was associated with weak water-soaked lesions, intermediate to the barely discernible water-soaked lesions induced by HM2.2 with an empty vector and the more obvious water-soaked lesions induced by co-inoculated transformants of HM2.2 expressing dTAL-SWEET14a and dTAL-SWEET14b (Fig. 6F, G). To assess the relationship of GhPL1 and GhSWEET14a/b in the development of BBC disease symptoms, we compared the severity of the water-soaked lesions caused by dTAL-PL, dTAL-SWEET14a and dTAL-SWEET14b together, and all three in combination, relative to that caused by Tal7b. Visually and when quantified by image analysis using ImageJ, water-soaked lesions were strongest when all three dTALEs were tested in combination, reaching a level comparable to that caused by Tal7b (Fig. 6F, G). Thus, it is likely that the pectin lyase plays an integral role in the development of BBC disease symptoms synergistically with GhSWEET14a and GhSWEET14b.
Discussion
Identifying resistance (R) or susceptibility (S) genes in tetraploid cotton remains a challenge. Initially, GhSWEET10 was identified as an S gene targeted by Avrb6 in the early Xcm H1005 isolate from BBC-infected cotton fields, but its current relevance to protection against BBC is uncertain, as GhSWEET14a and GhSWEET14b rather than GhSWEET10 are induced by reemerging Xcm isolates21. In this study, we identified Tal7b in a collection of Xcm isolates from Texas, USA, targeting GhSWEET14a and GhSWEET14b to promote disease, specifically water-soaked symptoms. Despite the overall similarity in genome sequence and organization between Xcm H1005 and reemerging Texas isolates, their TALomes exhibit distinct RVD compositions, suggesting rapid evolution of TALEs associated with virulence shifts over time. However, the TALomes of three Texas Xcm isolates, TX4, TX7, and TX9, exhibit a high degree of overall sequence identity and share similar genome/plasmid locations among them, as well as with those of an isolate from Mississippi, MSCT1. Similar to H1005 collected in 1968, three Xcm isolates collected in Africa during the 1950s and 1980s also harbor Avrb649. Although the genome sequence is unavailable, the TALEs in Xcm Xss-V2-18, a virulent strain in China50, resemble those in Xcm TX4 and MSCT151. Among six TALEs in Xcm Xss-V2-18, four of them, Tal4, Tal1, Tal5, and Tal3, share identical RVD sequences with Tal3, Tal5, Tal6, and Tal8, respectively, in Xcm TX451. Notably, Xcm Xss-V2-18 lacks Tal7b, crucial for Xcm TX4-triggered water-soaked lesion development. Conversely, Tal2 in Xcm Xss-V2-18, responsible for its virulence, exhibits two mismatches compared to the corresponding Tal2 in Xcm TX4. This suggests that these reemerging Xcm isolates likely originated from a recent common ancestral but have developed distinct virulence strategies to infect cotton in different regions.
SWEET genes are common S gene targets of TALEs across several Xanthomonas spp. Multiple TALEs from different X. oryzae pv. oryzae strains, including AvrXa7, PthXo3, TalC, and Tal5, target OsSWEET14 to promote infection19,26. In some cases, different TALEs within the same X. oryzae pv. oryzae strain target functionally redundant SWEET genes, as seen with AvrXa7 and PthXo2 from strain MAFF311018 targeting OsSWEET14 and OsSWEET13, respectively16,52. In cassava bacterial blight, Tal20 of X. axonopodis pv. manihotis promotes disease by activating MeSWEET10a20. And, we previously demonstrated in cotton bacterial blight that Xcm strain H1005 depends on Avrb6 targeting GhSWEET10D to cause water-soaked lesions21. In the present study, we demonstrate that Tal7b found in Xcm strain TX4 and other reemerging isolates induces GhSWEET14a and GhSWEET14b, leading to the partially additive promotion of water-soaked lesions in cotton. This observation illustrates an alternative model relative to the ones mentioned above, in which a single effector induces the expression of multiple SWEET genes to achieve a high level of SWEET expression, crucial for SWEET-mediated susceptibility. The targeting and induction of both GhSWEET14a and GhSWEET14b in cotton by Tal7b suggests an adaptive mechanism whereby the pathogen increases its evolutionary advantage. By simultaneously targeting two functionally similar S genes, the pathogen ensures an additive effect, thereby enhancing virulence while safeguarding against host evolution of a single TALE-insensitive S gene allele.
It has been suggested that TALE-mediated sucrose efflux in the apoplast serves as a carbon source for bacteria, aiding in their survival and proliferation16,19,20,23,24,25,26. However, the Tal7b-mediated induction of GhSWEET14a and GhSWEET14b does not appear to influence bacterial multiplication in planta. Similar results were observed with Avrb6-mediated induction of GhSWEET10D, which does not affect in planta multiplication of Xcm H100521,27. One explanation for this observation is that sucrose efflux might affect the osmotic potential of the apoplast, contributing to water-soaked lesions and benefiting bacterial virulence by regulating stomatal movement. Stomatal pores serve as entry points for many pathogens to invade leaf apoplasts53. Once inside, the aqueous environment within the leaf apoplast promotes pathogen colonization54,55. Furthermore, a high level of sucrose could induce stomatal closure56, and thereby increase water potential in the apoplast. Notably, the TALE AvrHah1 from X. gardneri also contributes to water-soaked lesion development but does not affect bacterial multiplication in planta57. It was proposed that activating bHLH transcription factor genes that in turn activate a pectate lyase, AvrHah1 modifies the plant cell wall to promote water uptake that facilitates bacterial egress from the apoplast to the leaf surface57. Finally, Tal2g of the rice bacterial leaf streak pathogen X. oryzae pv. oryzicola contributes to lesion expansion and bacterial egress to the leaf surface, without impacting total bacterial populations, by activating OsSULTR3;6, encoding a putative sulfate transporter58,59. These findings collectively suggest that TALEs may contribute to pathogen virulence by modulating apoplast water dynamics and osmotic potential to create a favorable environment for colonization and dissemination.
The integration of transcriptome analysis and EBE site prediction58 serves as an alternative strategy for identifying S genes targeted by Xanthomonas spp. TALEs21,57,60,61,62,63. Through this approach, we identified the pectin lyase gene GhPL1 as an additional target of Tal7b, quantitively contributing to water-soaked symptom development, synergistically with GhSWEET14a and GhSWEET14b. Pectin is a major component of the plant cell wall. Pectolytic enzymes in plants function in diverse processes, from cell wall remodeling to fruit ripening, and others. Cotton young bolls appear to have higher pectin levels than cotyledons and true leaves (Supplementary Fig. 10). Degradation of pectin by secreted enzymes of soft rot bacteria in the genera Pectobacterium and Dickeya leads to plant tissue maceration, electrolyte loss, and cell death64. How GhPL1 contributes to water-soaked lesions development is not clear, but as illustrated by AvrHah1 of X. gardneri, discussed above, TALE-mediated induction of pectolytic enzymes may be a virulence strategy common to several Xanthomonas species. In addition to the pectate lyase gene induced by X. gardneri, a TALE-targeted pectate lyase gene was identified as a candidate S gene in rice bacterial blight of rice62. Also, X. axonopodis pv. manihotis deploys a TALE, Tal14, that induces two pectate lyases63. Our findings, validated through gain-of-function (dTALEs) and loss-of-function (VIGS) approaches, demonstrate that GhPL1 functions as a bona fide S gene in cotton, contributing to water-soaked lesion development.
Our study adds to the growing body of literature showing the effectiveness of CRISPR-Cas9-mediated gene editing of S genes for resistance to diseases caused by Xanthomonas spp. Despite the complexities posed by the tetraploid genome of G. hirsutum, we targeted GhSWEET14a in both subgenomes and the sole copy of GhSWEET14b, resulting in knockouts of all three and reduction of Xcm-mediated water-soaked symptoms. However, the edits also resulted in a defect in reproductive development, the failure to produce bolls. This observation aligns with findings in rice, where knockout of OsSWEET11 led to defective grain filling and abnormal pollen development41, underscoring the roles of at least some SWEET genes in plant reproduction and development. To guard against potential negative consequences of disrupting endogenous S gene functions, in the case of TALE targets, mutagenesis of EBEs rather than coding regions can be carried out, as has been demonstrated in rice and citrus42,44. We demonstrated the effectiveness of this approach in BBC through targeted mutagenesis of the Avrb6 EBE in the GhSWEET10D promoter, resulting in resistance to strain H1005. Since we have shown that the virulence contribution of Tal7b in water-soaked lesion development involves simultaneous activation of GhSWEET14a, GhSWEET14b, and GhPL1, multiplexed editing of the Tal7b EBEs in each of these genes in each subgenome in which they occur, could be an effective strategy for resistance to the currently prevalent Xcm genotypes that depend on Tal7b for virulence. This is especially true given our finding also that blocking the activation of either the GhSWEETs or the GhPL gene reduces water-soaked lesion development in leaves without impacting bacterial population growth, alleviating selection imposed on the pathogen population that would be imposed by other forms of resistance that restrict pathogen multiplication.
Methods
Plant materials, bacterial strains, and growth conditions
Gossypium hirsutum germplasm Acala44E (Ac44E) and FiberMax FM 2322GL (FM 2322GL) were cultivated in 3.5-inch square pots containing Metro Mix 900 soil (SunGro Horticulture, Agawam, MA, USA) in a growth chamber at 28 °C, 65% relative humidity, and 100 μE m-2s-1 light with a 12 h light/12-hr dark photoperiod. Nicotiana benthamiana were cultivated in 3.5-inch square pots containing Metro Mix 366 soil (SunGro Horticulture, Agawam, MA, USA) in a growth chamber at 22 °C, 30% humidity and 100 μE m-2s-1 light with a 12 h light/12 h dark photoperiod. Cotton plants inoculated with Xanthomonas citri pv. malvacearum (Xcm) were transferred into a growth chamber at 28 °C, 65% humidity, and 100 μE m-2s-1 light with a 12 h light/12 h dark photoperiod. Escherichia coli DH5α and Agrobacterium tumefaciens GV3101 were incubated in Luria-Bertani (LB) media overnight with appropriate antibiotics (ampicillin 100 µg/mL, gentamicin 20 µg/mL, kanamycin 50 µg/mL, spectinomycin 50 µg/mL, or tetracycline 10 µg/mL) on a rotary shaker at 37 °C or 28 °C, respectively. Xcm strains were incubated in Nutrient Broth (NB; 0.5% peptone, 0.3% yeast extract, 0.5% NaCl) media with appropriate antibiotics (gentamicin 20 µg/mL, kanamycin 50 µg/mL, or rifampicin 50 µg/mL) on a rotary shaker at 28 °C. Xcm field isolates were collected from BBC-infected cotton fields in Texas by excising water-soaked lesions from infected cotton leaves with a sterile scalpel. Cut lesions were macerated in sterile water, serial diluted, and plated on Nutrient Agar (NA; 0.5% peptone, 0.3% yeast extract, 0.5% NaCl, and 1.5% agar). Individual colonies were re-streaked and inoculated into cotton to confirm pathogenicity.
Xcm DNA isolation for whole-genome sequencing
Texas Xcm field isolates were incubated overnight in 30 mL of glucose yeast extract media (2% w/v glucose, 1% w/v yeast extract) for total DNA extraction. The bacterial suspension was centrifuged at 3000 g at 4 °C, and the pellet was washed twice with sterile NE buffer (150 mM NaCl and 50 mM EDTA). The bacterial pellet was resuspended in 2.5 mL TE buffer (50 mM Tris-HCl, pH 8.0, and 50 mM EDTA) and mixed with 10 µL Ready-Lyse lysozyme (Epicentre Biotechnologies, Madison, WI, USA) and 50 µL RNase (10 mg/mL). The bacterial mix was stored at 4 °C for 45 min before adding 1 mL STEP solution (0.5% SDS, 50 mM Tris-HCl, pH 7.5, 40 mM EDTA, and 2 mg/mL protease K) and incubating for 1 h at 37 °C. The cell lysate was neutralized with 1.8 mL 7.5 M ammonium acetate before adding 10 mL phenol:chloroform:IAA (25:24:1) and phase separated by centrifuging at 7000 g for 10 min. The supernatant was collected, and 10 mL chloroform:IAA was added before centrifuging at 7000 g for 10 min. The supernatant was collected, and 2x volume of 95% ethanol was added. The precipitated DNA was transferred to a 2 mL tube using a plastic pipette tip and centrifuged at 2000 g for 5 min. Excess liquid was removed, and the DNA pellet was washed with 70% ethanol. The DNA pellet was briefly air-dried and dissolved in TE buffer overnight at 4 °C. The quantity and quality of DNA was checked using a NanoDrop One Microvolume UV-Vis Spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA).
PacBio whole-genome sequencing and de novo genome assembly of Xcm isolates
DNA library preparation and sequencing were conducted following the manufacturer’s instructions65 and performed at the Genomics Core Facility of the Icahn School of Medicine at Mount Sinai (New York, NY, USA). Electrophoresis was conducted using standard methods to detect small plasmids in each strain, which could potentially be lost during library preparation18. Sequencing was conducted to >120 coverage with two Single-molecule real-time (SMRT) cells per isolate. Analysis of the read-length distribution revealed a fat tail, with approximately 20% of the coverage post-adaptor removal contained in subreads ≥15,000 bp. The whole genomes of Xcm TX4, TX7, and TX9 were assembled using HGAP366 and validated by performing local assemblies of reads containing TALE genes. These local assemblies were then compared to the whole-genome assembly65.
Chromosomes and plasmids were aligned using progressiveMAUVE67 with default parameters, employing a progressive alignment approach. ProgressiveMAUVE was chosen due to its effectiveness in handling large-scale rearrangements, inversions, and other genomic rearrangements commonly observed in microbial genomes. Additionally, it produces alignments that uncover both conserved regions and genomic rearrangements across multiple genomes, facilitating comparative genomic and evolutionary analysis. The D-GENIES web tool68 with alignment by Minimap2 v2.2669 was used to map pair-wise comparisons of chromosomes and plasmids to generate percent identity scores. Proteins for Xcm TX4, TX7, TX9 were predicted using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP)70. Phylogenetic analysis was conducted using OrthoFinder v.2.3.1171, which uses the STAG and STRIDE algorithms for rooting. Xcm H1005, an isolate collected in 1968 from Oklahoma, was later sub-cultured through three laboratories (D. Gabriel, University of Florida; A. Bogdanove, Cornell University; and L. Shan, Texas A&M University) before being sequenced by Cox et al.21.
Immunoblot analysis of Xcm TALomes
Xcm strains were incubated overnight in NB media on a rotary shaker at 28 °C. Bacterial pellets were harvested by centrifuge and washed twice with sterile double-distilled66 H2O to remove exopolysaccharides. Bacterial cells were diluted to OD600 = 0.5, and 120 µL was added to 40 µL of 4x SDS buffer (250 mM Tris-HCl pH 6.8, 4% SDS, 0.1% bromophenol blue, 40% glycerol, and 4% β-mercaptoethanol) and heated to 95 °C for 10 min. Proteins were separated in a 7.5% SDS-PAGE gel before being transferred to a PVDF membrane. PBST solution (phosphate-buffered saline solution with 0.1% Tween-20) with 5% nonfat milk was used to block the membrane for 3 h at room temperature. TALEs were detected by immunoblotting with primary α-TALE antibodies72 (1:2000) in 5% milk PBST solution overnight at 4 °C, followed by secondary goat-α-rabbit-IgG-HRP antibody (ThermoFisher Scientific, Waltham, MA, USA) in 5% milk PBST solution for 2 h at room temperature. Protein bands were analyzed using a ChemiDoc Gel Imaging System (Bio-Rad, Hercules, CA, USA).
Construction of TALE expression vectors
TALEs were individually cloned into an in-house custom Gateway entry vector pBADZ2 using the Gibson assembly approach to TALE gene cloning first developed by Li et al.73. pBADZ2 was digested with restriction enzymes SpeI and AvrII, while 30 µg gDNA of Xcm was digested with SphI at 37 °C for 4 h. Linearized pBADZ2 was purified by 1.5% agarose gel extraction. gDNA was placed in an 80 °C water bath for 15 min to stop the digestion. DNA was then precipitated by adding 10% of 3 M potassium acetate pH 5.2 and 2x volume of 100% ethanol before centrifuging at 15,000 g for 2 min. The pellet was washed in 70% ethanol and centrifuged at 15,000 g for 5 min. The supernatant was discarded, and the DNA pellet was resuspended in sterile water. DNA quantity and quality were checked using a NanoDrop One Microvolume UV-Vis Spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA). Gibson assembly was performed according to the manufacturer’s instructions using NEBuilder® HiFi DNA Assembly (New England BioLabs Inc., Ipswich, MA, USA). E. coli DH5a was transformed with a 5 µL reaction mix, and transformants were selected on LB plates with appropriate antibiotics (gentamicin 20 µg/mL). Thirty colonies were selected for plasmid miniprep and screened to confirm TALE inserts by digestion with restriction enzymes StuI, targeting the plasmid backbone, and AatII, targeting the insert. Gateway cloning was used to shuttle TALEs into the destination vector pKEB1 using the manufacturer’s instructions from Gateway LR Clonase II Enzyme mix (ThermoFisher Scientific, Waltham, MA, USA). Colonies were selected for plasmid miniprep and screened by restriction enzyme digestion using SphI to confirm TALE inserts.
Reporter construct pGhSWEET10::LUC was reported previously21. pHBT-pGhSWEET14a::LUC and pHBT-pGhSWEET14b::LUC were constructed by PCR amplifying the GhSWEET14aA or GhSWEET14bD promoter from gDNA of Ac44E using primers containing BamHI and NcoI restriction sites. Promoter sequences were designated as 1000 bp upstream from the translational start site. PCR amplicons were cloned into plant expression vector pHBT immediately upstream of a luciferase gene. EBE mutations were introduced by site-directed mutagenesis with primers substituting three nucleotides ‘CCC’ to ‘GGG’ in the EBE to pHBT-pGhSWEET14a::LUC and pHBT-pGhSWEET14b::LUC to create pHBT-pGhSWEET14amEBE::LUC and pHBT-pGhSWEET14bmEBE::LUC vectors. The wild-type EBE or mEBE-containing promoter was shuttled into the pHBT-p35S::SWEE14a-HA or pHBT-p35S::SWEE14b-HA vector by XhoI and BamHI to generate pHBT-pGhSWEET14a::SWEET14a-HA, pHBT-pGhSWEET14amEBE::SWEET14a-HA, pHBT-pGhSWEET14b::SWEET14b-HA, or pHBT-pGhSWEET14bmEBE::SWEET14b-HA. The pGhSWEET14a/b::SWEET14a/b-HA and pGhSWEET14a/bmEBE::SWEET14a/b-HA were shuttled into binary vector pCAMBIA1300 by XhoI and EcoRI. All constructs for the promoter- or gene-of-interest were confirmed by Sanger sequencing.
Construction of Xcm TALE mutants
Suicide vector pTeM5 was transformed into Xcm TX4 by electroporation74. pTeM5 contains a kanamycin resistance gene flanked by the central repeat region of AvrXa5 from Xoo strain PXO99A and the N- and C- termini from Xoc strain BLS256. Mutants were generated by single or double crossover from plasmid integration into the Xcm genome. The genomic DNA extracted from potential mutants was subjected to digestion with the restriction enzymes XmnI and EcoRI. These enzymes cleave DNAs at sites outside the TALE open reading frame, generating unique DNA fragments for each effector. Following digestion, the resulting DNA fragments were separated by agarose gel electrophoresis until bands were adequately separated. Southern blotting was conducted to transfer the DNA fragments from the gel onto a membrane, which was subsequently hybridized with a TALE-specific probe. The resulting Southern blot images were examined to determine the presence or absence of DNA fragments corresponding to the TALE loci in the mutant samples compared to wild-type controls. To validate TALE mutations in Xcm, genomic DNA digested with SphI was ligated into the pBADZ2 vector, which had been previously digested with SpeI and AvrII, by Gibson assembly using the NEBuilder® HiFi DNA Assembly kit and following the manufacturer’s instructions (New England BioLabs Inc., Ipswich, MA, USA). The resulting Gibson assembly products were transformed into 50 µL of DH5α competent E. coli cells. Bacterial colonies were selected based on antibiotic marker resistance, and PCR amplicons derived from the insertion regions were subjected to Sanger sequencing. To characterize the TALE repertoire in each Xcm mutant, the cloned DNA sequences were translated into amino acids using the Expasy DNA translation tool (https://web.expasy.org/translate/). This translation facilitated the identification of the RVDs associated with different TALEs. Individual TALE mutants were identified by the presence of a kanamycin resistance gene flanked by RVDs corresponding to a specific TALE. Xcm mutants lacking specific TALEs were used for the assays.
Bacterial tri-parental mating
Broad-host-range vector pKEB1, a gateway-compatible pUFR047 derivative21,75, carrying TALEs, designer TALEs, or empty vector controls was transformed into Xcm HM2.2 by bacterial tri-parental mating. Helper plasmid pRK2073 and modifier plasmid pUFR054 were included to facilitate plasmid transfer from E. coli to Xcm75. Overnight liquid cultures of E. coli and Xcm were centrifuged at 2000 g for 5 min. Bacterial pellets were washed with Nutrient broth (NB) and centrifuged again at 2,000 g for 5 min. Bacterial cells were individually resuspended in NB, with Xcm HM2.2 having a higher density compared to the other strains, in a ratio of 5:1:1:1 (recipient:donor:helper:modifier). Subsequently, 5 µL of each suspension was spot inoculated on top of each other on Nutrient agar (NA) plates. NA plates were incubated overnight at 28 °C. Conjugated bacteria were suspended in NB, spread onto NA plates with antibiotics, and incubated for 2 days at 28 °C. Single colonies were re-streaked on NA plates with antibiotics to purify the bacteria. Conjugants were confirmed by colony PCR.
Protoplast-based chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) assay
Cotton protoplasts were isolated from 2-week-old cotton cotyledons. The edges and middle veins of the cotyledons were removed before cutting them into 0.5 mm strips using a razor blade. These strips were digested in an enzyme solution (1.5% cellulase R10 [Yakult Pharmaceutical Industry], 0.4% macerozyme R10 [Yakult Pharmaceutical Industry], 0.4 M mannitol, 20 mM KCl, 20 mM MES, pH 5.7, 2% sucrose). Arabidopsis protoplasts were isolated from fully expanded leaves of 4-week-old Arabidopsis leaves. The leaves were cut into 0.5 mm strips using a razor blade and digested in an enzyme solution (1% cellulase R10, 0.2% macerozyme R10, 0.4 M mannitol, 20 mM KCl, 20 mM MES pH 5.7). The leaf strips, submerged in their respective enzyme solutions, were placed in Petri dishes, covered with foil, and vacuum infiltrated using a desiccator for 30 min. Digestion was then continued for an additional 2-3 h without further vacuuming. The released protoplast were washed with W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 5.7) and subsequently transformed using PEG-Ca2+ mediated transfection21,76. ChIP-qPCR assay to examine Tal7b binding to the endogenous GhSWEET14a/b promoters in cotton was performed using cotton protoplasts transformed with pUCSupGW-MSCT1-Tal7b. Tal7b binding to the wild-type EBE, but reduced binding to EBE mutants of the GhSWEET14a/b promoters was performed using Arabidopsis protoplasts transformed with pUCSupGW-MSCT1-Tal7b together with pHBT-pGhSWEET14a/b::SWEET14a/b-HA or pHBT-pGhSWEET14a/bmEBE::SWEET14a/b-HA, followed by ChIP-PCR assays.
Nuclei were freshly extracted from transformed protoplast. Briefly, 5 mL of transformed protoplast were crosslinked with 1% formaldehyde for 20 min and then quenched with glycine for 5 min. Nuclei were isolated77,78, and a small aliquot was used for immunoblotting as input controls using α-TALE72 antibodies. Subsequently, nuclei lysates aliquoted into two tubes with one for ChIP and the other one as the control were sheared into about 200 bp using Covaris M220 Focused Ultra-sonicator (Peak Incident Power: 175 W, Duty Factor: 20%, Treatment Time: 180 s). The lysed and sheared nuclei were centrifuged at 10,000 g for 5 min at 4 °C to remove debris, and the supernatants were collected and diluted with ice-cold ChIP dilution buffer (1% v/v Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 × protease inhibitor cocktail [Roche, Cat # 11873580001]). The diluted supernatants were aliquoted into four tubes and co-incubated with or without α-TALE antibodies in chromatin solution mixtures overnight at 4 °C with gentle agitation, followed by co-incubation with pre-washed protein G agarose beads (IgG, Sigma, Cat # 16-201) for 3 h at 4 °C with gentle agitation. Beads were collected by centrifugation at 1500 g for 3 min at 4 °C and washed one time with low salt washing buffer (0.1% w/v SDS, 1% v/v TritonX-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.0), 150 mM NaCl), one time with high salt washing buffer (0.1% w/v SDS, 1% v/v Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.0), 500 mM NaCl), one time with LiCl washing buffer (0.25 M LiCl, 1% v/v NP-40, 1 mM EDTA, 10 mM Tris-HCl [pH 8.0], 1% w/v sodium deoxycholate), and three times with TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA), followed by elution using freshly prepared elution buffer (1% w/v SDS, 0.1 M NaHCO3) at 65 °C for 15 min. Supernatants after centrifugation at 5000 g for 3 min at RT were collected with three-time elution. Combined eluted supernatants were added with 5 M NaCl and protein-DNA eluate was reverse crosslinked at 65 °C overnight79. The elution was further incubated with proteinase K 0.5 M EDTA (pH 8.0) and 1 M Tris-HCl (pH 6.5) at 37 °C for 1 h followed by phenol/chloroform/isoamyl alcohol (25:24:1) extraction to remove proteins. DNA was precipitated with 100% ethanol for qPCR analyses with primers listed in Supplemental Table S9. The relative enrichment fold changes were calculated by normalizing the IgG to 1.
Biotinylated EBE promoter oligo pull-down TAL effector assays
Biotinylated DNA oligos corresponding to the EBE or mEBE of GhSWEET14a or GhSWEET14b promoter (Sequences shown in Supplemental Table S11) were synthesized by Sigma-Aldrich, St. Louis, MO, USA. Pre-washed streptavidin agarose resins (15942-050, Invitrogen) were incubated in binding buffer (20 mM HEPES, pH 7.4, 100 mM KCl, 0.5 mM EDTA, 1.5 mM MgCl2, 20% glycerol, 1 mM DTT, and protease inhibitor cocktail [Roche, Cat#11873580001]) with or without biotinylated DNA oligos for 1 h at 23 °C. Biotinylated DNA oligo-linked resins were washed twice using binding buffer and resuspended in 100 µL binding buffer. Avrb6 or Tal7b was expressed in Arabidopsis protoplasts by transfection of pHBT-p35S::Avrb6-HA or pUCSupGW-MSCT1-Tal7b76. Five mL of transformed protoplasts expressing Tal7b (5 aliquots) or 1 mL protoplasts expressing Avrb6 were harvested by centrifuging at 100 g for 2 min, and lysed in 200 ul lysis buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP40, and 5% glycerol, protease inhibitor). Ten ul lysate was saved for immuno-blotting using α-TALE antibodies as input controls. After centrifugation at 3000 g for 30 s, the protoplast lysate supernatant was incubated with 20 µL biotinylated DNA oligo-linked streptavidin agarose resins in 400 µL binding buffer at 4 °C for 2 hrs with gentle agitation. Resins were collected by centrifugation at 3000 g for 30 s and washed three times using washing buffer (25 mM Tris, pH 7.4, 15 mM NaCl, 1% NP40, and 0.5% sodium deoxycholate) followed by elution three times using 500 mM NaCl elution buffer for 5 min. Combined elution supernatants were added with an equal volume of 2 x SDS-PAGE sample buffer (63 mM Tris-HCl, pH 6.8, 25% Glycerol, 2% SDS, 0.01% bromophenol blue, and 5% β-mercaptoethanol) and heated at 95 °C for 10 min. The samples were separated by 7.5% SDS-PAGE for immuno-blotting using α-TALE antibodies.
Xcm inoculations and water-soaked lesion quantification
Xcm strains were incubated in NB liquid media with appropriate antibiotics (gentamicin 20 µg/mL, kanamycin 50 µg/mL, or rifampicin 50 µg/mL) overnight on a rotary shaker at 28 °C. For syringe inoculations, bacterial suspensions were adjusted to OD600 = 0.05 in sterile ddH2O with 10 mM MgCl2. Small punctures were made to facilitate infiltration on the underside of expanded cotton cotyledons using a 10 µL pipette tip. Bacterial solutions were injected into plant tissue using a needleless syringe. The severity of the water-soaked lesion was quantified using ImageJ version FIJI80,81. The grayscale mean was determined by averaging the grayscale values in the RGB channels from independent repeats. For vacuum infiltration, bacterial suspensions were adjusted to OD600 = 0.0001 in ddH2O with 10 mM MgCl2 and 0.04% Silwet L-77 surfactant solution. Four-week-old cotton plants were submerged in the bacterial solution inside a desiccator connected to a vacuum pump. Plants were vacuum infiltrated for 5 min at 76 mmHg. Water-soaked lesions were manually quantified using ImageJ. The mean was calculated from three or more biological replicates.
TAL effector-induced water-soaked lesion assays in Nicotiana benthamiana
Agrobacterium tumefaciens strain GV3101 harboring pGhSWEET14a::GhSWEET14a-HA, pGhSWEET14amEBE::GhSWEET14a-HA, pGhSWEET14b::GhSWEET14b-HA, or pGhSWEET14bmEBE::GhSWEET14b-HA was cultured in liquid Luria-Bertani (LB; 1% tryptone, 0.5% yeast extract, 1% NaCl) medium containing 50 mg/mL kanamycin, 50 mg/mL gentamycin, 10 mM MES, and 20 mM acetosyringone on a rotary shaker at 28 °C overnight. Agrobacterial cells were pelleted by 1000 g centrifugation at 23 °C for 10 min, resuspended in infiltration solution (10 mM MES, 10 mM MgCl2, and 200 mM acetosyringone) and incubated at 23 °C for 3 h. Xcm HM2.2 carrying Tal7b (Xcm HM2.2 (Tal7b)) or Xcm HM2.2 strains were incubated in liquid Nutrient broth (NB; 0.5% peptone, 0.3% yeast extract, 0.5% NaCl) media containing 20 µg/mL gentamicin and 50 µg/mL kanamycin overnight on a rotary shaker at 28 °C. A. tumefaciens bacterial suspensions were adjusted to OD600 = 1.0, and Xcm HM2.2 (Tal7b) or Xcm HM2.2 for OD600 = 0.1 in infiltration solution. Bacterial suspension was inoculated using a needless syringe into N. benthamiana leaves. Inoculated plants were covered with a plastic dome overnight, and after removing the dome, plants were kept at 22 °C, 30% humidity, 100 μE m-2s-1 light with a 12 h light/12 h dark photoperiod for five days. Detached infiltrated leaves were sprayed with water and wrapped with a wet paper towel for 20 min. The excess water was removed by gently wiping the leaf surface to further observe the water-soaked lesion development.
Cotton RNA isolation for RT-PCR and RT-qPCR analysis
Total RNA was extracted from cotton leaves using a Spectrum Plant Total RNA Kit (Sigma-Aldrich, Burlington, MA, USA) according to the manufacturer’s protocol. RNA was extracted at 24 h post-inoculation (hpi) for analyzing treatment-induced gene expression. Extracted RNA was treated with DNase to remove residual genomic DNA, and 1 µg of purified RNA was used for reverse transcription by first-strand synthesis of cDNA with oligo(dT). Conditions for PCR amplification were 30 cycles of 15 s at 98 °C, 30 s at 55 °C, and 45 s at 72 °C. GhACTIN was used as an internal control for RT-PCR. RT-qPCR analysis was performed using iTaq SYBR green supermix (Bio-Rad Hercules, CA, USA) with a Bio-Rad CFX384 Real-Time PCR System. Gene expression was measured in relation to GhUBQ21,82.
Xcm bacterial counting
Xcm strains were washed and resuspended in sterile ddH2O with 10 mM MgCl2. For in planta counting, bacterial solutions at OD600 = 0.0001 were syringe inoculated into cotyledons. Leaf discs from three biological repeats were collected at 0, 1-, 3-, 5-, and 7- or 1-, 7-, 10-, and 14 days post-inoculation and macerated in 100 µL of sterile ddH2O. Bacterial surface counting was performed as previously described83. Briefly, bacterial solutions at OD600 = 0.1 were syringe inoculated into cotyledons. Five days post-inoculation, 100 µL of sterile ddH2O was applied to the leaf surface of three biological repeats covering an area of approximately 1 cm2 and collected with a pipette. This process was repeated ten times across the leaf surface, resulting in the collection of a total volume of 1 mL. Serial dilutions from in planta or leaf surface assays were plated on NA plates with appropriate antibiotics (gentamicin 20 µg/mL or kanamycin 50 µg/mL) and incubated at 28 °C. Bacterial colony-forming units were counted after two days.
Pectin measurement
For gravimetric analysis, pectin content was measured following a previously described protocol with modifications84. Briefly, one gram of cotton tissues was collected and dried in a 75 °C incubator before grinding in liquid nitrogen. The tissues were ground into fine powder and resuspended in 10 mL of 0.1 M sodium carbonate to extract pectin. The mixture was then incubated at 80 °C for 3 h. The resulting mixture was filtered through filter paper (Cat. 28320-020VWR International, Radnor, PA, USA) to remove debris, and pectin was precipitated by gradually adding an equal volume of 0.5% hydrogen chloride in ethanol. After allowing the pectin to precipitate for 30 min at 23 °C, the solution was centrifuged at 2000 g for 20 min to collect the pectin pellet. The pellet was resuspended in 1 mL of 0.1 M sodium hydroxide and precipitated using 1 mL of 0.1 M copper sulfate to form a copper-pectate complex. The copper-pectate complex was collected on pre-weighed filter paper and dried at 75 °C. The dried copper pectate was weighed, and the pectin content was calculated based on the initial tissue mass and moisture content of the sample. The following formula was used for calculating pectin content, where the percentage of pectin (X) is calculated based on the weight of copper pectate obtained from the sample. m is the weight of the dry filter paper (grams); m1is the total weight of the dried copper pectate and filter paper (grams); m0 is the weight of the initial plant tissue sample (grams); w is the moisture content of the sample (%).
For histochemical analysis, cotyledons and true leaf sections were prepared by first carefully removing the abaxial epidermal tissues using a dual tape system. The adaxial leaf surface was first attached to a clear tape strip. A second tape strip was applied to the abaxial epidermis and gently peeled off, to successfully remove the abaxial epidermis while leaving the underlying tissue intact and adhered to the first tape strip. Thin sections of cotton bolls were prepared by slicing the tissue using a razor blade. Chlorophylls were removed by incubating the tissue sections in 75% ethanol at 37 °C for 12 h. Tissue sections were stained with 0.02% ruthenium red for 10 min. The samples were analyzed with a Leica Stereo Microscope MZFLIII.
Luciferase reporter assay
Reporter constructs pGhSWEET14a::LUC and pGhSWEET14b::LUC were constructed by PCR amplifying the GhSWEET14aA and GhSWEET14bD promoters from gDNA of Ac44E using primers containing BamHI and NcoI restriction sites. Promoter sequences were designated as 1000 bp upstream from the translational start site. PCR amplicons were cloned into binary vector pCB302 immediately upstream on a luciferase gene85. Reporter constructs were transformed into A. tumefaciens GV3101 using electroporation. The leaves of five-week-old N. benthamiana were syringe-infiltrated with Agrobacterium containing reporter constructs and resuspended in infiltration buffer (10 mM MES, pH 5.7, 10 mM MgCl2, and 200 µL acetosyringone) at OD600 = 1.5. Inoculated N. benthamiana were covered with a plastic dome and placed under ambient light for 24 h before co-infiltrated with Xcm strains resuspended in 10 mM MgCl2 at OD600 = 0.5. Leaf discs from co-infiltrated tissue were harvested 24 h post-inoculation and evenly sprayed with 0.2 mM luciferin with 0.2% Silwet L-77. Luciferase activity was detected by Glomax Multi-Detection System (PromegaTM Madison, WI, USA).
Virus-induced gene silencing
A conserved region between GhSWEET14aA, GhSWEET14aD, and GhSWEET14bD, or GhPL1 was PCR amplified using primers containing EcoRI and KpnI restriction sites and cDNA from G. hirsutum. PCR amplicons were cloned into modified Tobacco rattle virus vector pYL156 (pTRV-RNA2). A. tumefaciens containing pYL156-GhSWEET14a/b, pYL156-GhPL1, pYL156-empty vector, or pYL192 (pTRV-RNA1) were resuspended in infiltration buffer (10 mM MES, pH 5.7, 10 mM MgCl2, and 200 µL acetosyringone) at OD600 = 1.5. Bacterial suspensions of pYL192 were mixed with pYL156-GhSWEET14a/b, pYL156-GhPL1, or pYL156-empty vector at a 1:1 ratio before syringe inoculating into seven-day-old cotton cotyledons. Inoculated cotton plants were covered with a plastic dome and placed under ambient light overnight, then moved to a growth chamber at 22 °C, 30% humidity, and 100 μE m-2s-1 light with a 12 h light/12 h dark photoperiod86. Disease assays were performed on plants 3-weeks post-inoculation, when silencing was established.
Construction of designer TALEs
Designer TALEs were constructed using the Golden Gate TALEN and TALE Kit 2.0 (Addgene, Watertown, MA, USA) according to the manufacturer’s protocol87. EBE target sequences were evaluated for dTALE binding efficiency using the TAL Target Finder tool38. The central repeat region of each dTALE consisted of repeats containing RVD sequences corresponding with the respective nucleotide. dTALEs were shuttled into broad-host-range vector pKEB1 using the Gateway LR Clonase II Enzyme mix (ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
Construction of CRISPR-Cas9 vectors
Single-stranded oligonucleotides were synthesized comprising of the guide RNA (gRNA) target sequence for pGhSWEET10D (5’-AGAACGGGCTGGGGATG-3’) with a 5’-CGGA motif on the forward oligonucleotide and a 5’-AAAC motif on the reverse oligonucleotide. The oligonucleotides were annealed together and cloned into the BsaI-linearized pTX171 vector using the Golden Gate method88.
For mutating GhSWEET14a and GhSWEET14b, single-stranded oligonucleotides were designed according to established protocols40. The pGTR plasmid, containing a fused tRNA-gRNA scaffold fragment, served as the DNA template for PCR to construct a polycistronic tRNA-gRNA (PTG) gene encompassing the gRNA target sequences for GhSWEET14a (5’-ATATACCAAAAACAACAGCCA-3’) and GhSWEET14b (5’-AGACACCAAATGCAACTGCCA-3’). Subsequently, the PTG gRNA-GhSWEET14a/b product was cloned into the pRGEB32-GhU6.9 vector using the Golden Gate method.
All gRNA target sequences were generated using the gRNA design tool CRISPR-P (http://cbi.hzau.edu.cn/crispr). The resulting Golden Gate ligation products were transformed into 50 μl of competent E. coli DH5α cells. Bacterial colonies were selected based on antibiotic marker resistance, and vectors were validated by Sanger sequencing of PCR amplicons derived from the insertion regions.
Generation of transgenic cotton
Agrobacterium-mediated cotton variety Coker 312 transformation was performed at Texas A&M Multicrop Transformation Facility following a previously described protocol with modifications89. Briefly, cotton seedlings were cultivated using Murashige and Skoog (MS) medium. Hypocotyls were excised and infected with A. tumefaciens LBA4404 containing the CRISPR-Cas9 construct or a GFP control. Explants and Agrobacterium were co-cultivated on P1 [4.31 g/L MS salts, 100 mg/L myoinositol, 0.4 mg/L thiamine HCl, 5 mg/L N6-(2-isopentenyl)adenine, 0.1 mg/L α-naphthaleneacetic acid (NAA), 3% glucose, 1 g/L magnesium chloride hexahydrate, pH 5.8, 0.2% Phytagel medium supplemented with 50 µM acetosyringone for 72 h under light at 25 °C before being transferred to P1 media containing 15 mg/L hygromycin. Explants were cultivated in a growth chamber at 28 °C for 42 days until callus tissue developed at the cut surface of explants. Callus tissue was excised under a microscope, transferred to P1 media containing 15 mg/L hygromycin, and incubated under light at 28 °C. Somatic embryos were transferred to EG3 (2.16 g/L MS salts, 0.5% glucose, 100 mg/L myoinositol, 0.4 mg/L thiamine HCL, 0.01 mg/L NAA, pH 6.5, 0.2% Phytagel) media for regeneration and subsequent plantlets were transferred to MS3 (2.16 g/L MS salt, 0.5% glucose, 0.14 mg/L thiamine HCl, 0.1 mg/L pyridoxine HCl, 0.1 mg/L nicotinic acid, pH 5.8, 0.08% Phytagel, 0.6% Bacto agar) media for further growth and development before being transferred to soil. Genomic DNA was extracted from the edited line, and the target site was amplified by PCR, and the amplicon sequenced to identify homozygous lines. GFP lines were further confirmed by confocal microscopy observation.
RNA sequencing and transcriptomic profiling
Two-week-old cotyledons of cotton variety FM 2322GL were syringe-inoculated with Xcm HM2.2 expressing Tal7b or a vector control. Total RNA was extracted 24 h post-inoculation (hpi) from three independent biological repeats. RNA-seq libraries were prepared using Illumina TruSeq Stranded mRNA Sample Preparation Kit following the manufacturer’s protocol and sequenced using an Illumina HiSeq 2000 platform with 2 × 150-nucleotide pair-end reads at the Texas A&M Institute for Genome Sciences and Society (College Station, TX, USA). RNA-seq analysis was performed as previously described90. Briefly, the Trimmomatic tool91 was used to preprocess raw RNA-seq reads by performing quality filtering and trimming of low-quality bases from sequencing reads to ensure that only high-quality reads were used for alignment and assembly. The alignment of RNA-seq reads to the G. hirsutum (AD1) TM-1 genome28 was executed using HISAT292, employing an indexing scheme based on the Burrows-Wheeler transform and the Ferragina-Manzini index. Transcriptional assembly was conducted using StringTie93, which reconstructed full-length transcripts from aligned reads while estimating their abundance and quantifying gene expression levels. Differential gene expression was analyzed using Cuffdiff94, a component of the Cufflinks suite, which facilitated the comparison of transcript abundances between different treatments. Genes were considered differentially expressed if they demonstrated a log2 fold expression change >1 and an adjusted p-value < 0.05. These cutoffs were employed to ensure that only genes displaying statistically significant changes in expression between treatments were included in the analysis.
EBE predictions
TALE-NT 2.0 Target Finder was used to predict TALE EBE sites within the promoterome of G. hirsutum38. Promoter regions were defined as 1000 bp upstream of the transcriptional start site. Promoter sequences were identified from the G. hirsutum (AD1) TM-1 genome assembled using SOAPdenovo1228. Binding sites passing the Target Finder score ratio cutoff of 4.0 were ranked based on Target Finder output and genomic context using a machine learning classifier58,95. The primers used in this study are in Supplementary Tables 6–11.
Quantification and statistical analysis
Data for quantification analyses are presented as mean ± standard error (s.e.) or standard deviation (s.d.) as indicated in the figure legends. The statistical analyses were performed by unpaired two-tailed Student’s t-test. The number of biologically independent replicates is shown in the figure legends. The p-values are provided in the graphs.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The whole-genome sequencing data generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) database under BioProject PRJNA1117961 [https://www.ncbi.nlm.nih.gov/bioproject/1117961]. The previously published whole-genome sequencing data used in this study can be found in the NCBI database under BioProject accession PRJNA298765 [https://www.ncbi.nlm.nih.gov/bioproject/298765], PRJNA298770 [https://www.ncbi.nlm.nih.gov/bioproject/298770], PRJNA299817 [https://www.ncbi.nlm.nih.gov/bioproject/299817], PRJNA396899 [https://www.ncbi.nlm.nih.gov/bioproject/396899] and PRJNA587534 [https://www.ncbi.nlm.nih.gov/bioproject/587534]. RNA-sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE270600. Source data are provided in this paper. Source data are provided with this paper.
References
Ahmad, S. & Hasanuzzaman, M. Cotton production and uses. Agronomy, Crop Protection, and Postharvest Technologies (Springer Nature Singapore Pte Ltd.) (2020).
Jalloul, A., Sayegh, M., Champion, A. & Nicole, M. Bacterial blight of cotton. Phytopathologia Mediterranea, 1, 3–20 (2015).
Cox, K. L. Jr, Babilonia, K., Wheeler, T., He, P. & Shan, L. Return of old foes-recurrence of bacterial blight and Fusarium wilt of cotton. Curr. Opin. Plant Biol. 50, 95–103 (2019).
An, S. et al. Mechanistic insignts into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol. Rev. 44, 1–32 (2020).
Kemerait, B. et al. Identification and management of bacterial blight of cotton. Cotton Incorporated Bulletin, Cotton Incorporated, Cary, NC (2017).
Wheeler, T. A. in Beltwide cotton conference, San Antonio, TX. 11–18.
Timilsina, S. et al. Xanthomonas diversity, virulence and plant-pathogen interactions. Nat. Rev. Microbiol. 18, 415–427 (2020).
Perez-Quintero, A. L. & Szurek, B. A Decade Decoded: Spies and Hackers in the History of TAL Effectors Research. Annu. Rev. Phytopathol. 57, 459–481 (2019).
Boch, J., Bonas, U. & Lahaye, T. TAL effectors - pathogen strategies and plant resistance engineering. N. Phytologist 204, 823–832 (2014).
Bonas, U., Stall, R. E. & Staskawicz, B. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet. MGG 218, 127–136 (1989).
Kay, S., Hahn, S., Marois, E., Hause, G. & Bonas, U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318, 648–651 (2007).
Jeena, G. S., Kumar, S. & Shukla, R. K. Structure, evolution and diverse physiological roles of SWEET sugar transporters in plants. Plant Mol. Biol. 100, 351–365 (2019).
Xue, X. Y., Wang, J., Shukla, D., Cheung, L. S. & Chen, L. Q. When SWEETs Turn Tweens: Updates and Perspectives. Annu. Rev. Plant Biol. 73, 379–403 (2022).
Ji, J. et al. Plant SWEET family of sugar transporters: Structure, evolution and biological functions. Biomolecules 12, 205 (2022).
Chu, Z. et al. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20, 1250–1255 (2006).
Zhou, J. et al. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643 (2015).
Hutin, M., Sabot, F., Ghesquière, A., Koebnik, R. & Szurek, B. A knowledge‐based molecular screen uncovers a broad‐spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 84, 694–703 (2015).
Yu, Y. et al. Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin-3 Os11N3 gene. Mol. Plant-Microbe Interact. 24, 1102–1113 (2011).
Antony, G. et al. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22, 3864–3876 (2010).
Cohn, M. et al. Xanthomonas axonopodis Virulence Is Promoted by a Transcription Activator-Like Effector-Mediated Induction of a SWEET Sugar Transporter in Cassava. Mol. Plant-Microbe Interact. 27, 1186–1198 (2014).
Cox, K. L. et al. TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nat. Commun. 8, 15588 (2017).
Chen, L.-Q. et al. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211 (2012).
Huang, S. et al. The broadly effective recessive resistance gene xa5 of rice is a virulence effector-dependent quantitative trait for bacterial blight. Plant J. 86, 186–194 (2016).
Romer, P. et al. Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. N. Phytologist 187, 1048–1057 (2010).
Yang, B., Sugio, A. & White, F. F. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl Acad. Sci. 103, 10503–10508 (2006).
Streubel, J. et al. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. N. Phytologist 200, 808–819 (2013).
Yang, Y., Yuan, Q. & Gabriel, D. W. Watersoaking function (s) of XcmH1005 are redundantly encoded by members of the Xanthomonas avr/pth gene family. Mol. Plant Microbe Interact. 9, 105–113 (1996).
Zhang, T. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537 (2015).
Showmaker, K. C. et al. The genome of the cotton bacterial blight pathogen Xanthomonas citri pv. malvacearum strain MSCT1. Stand. Genom. Sci. 12, 1–12 (2017).
Hunter, R., Brinkerhoff, L. & Bird, L. The development of a set of Upland cotton lines for differentiating races of Xanthomonas malvacearum. Phytopathology 58, 830–832 (1968).
Grau, J. et al. AnnoTALE: bioinformatics tools for identification, annotation and nomenclature of TALEs from Xanthomonas genomic sequences. Sci. Rep. 6, 21077 (2016).
Salzberg, S. L. et al. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9, 1–16 (2008).
Bogdanove, A. J. et al. Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J. Bacteriol. 193, 5450–5464 (2011).
Phillips, A. Z. et al. Genomics-enabled analysis of the emergent disease cotton bacterial blight. PLoS Genet. 13, e1007003 (2017).
Pérez-Quintero, A. L. et al. QueTAL: a suite of tools to classify and compare TAL effectors functionally and phylogenetically. Front. Plant Sci. 6, 545 (2015).
Denance, N. et al. Two ancestral genes shaped the Xanthomonas campestris TAL effector gene repertoire. N. Phytologist 219, 391–407 (2018).
Chen, Z. J. et al. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 145, 1303–1310 (2007).
Doyle, E. L. et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40, W117–W122 (2012).
Wang, P. et al. High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 16, 137–150 (2018).
Xie, K., Minkenberg, B. & Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. 112, 3570–3575 (2015).
Ma, L. et al. Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 58, 863–873 (2017).
Peng, A. et al. Engineering canker‐resistant plants through CRISPR/Cas9‐targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519 (2017).
Xu, Z. et al. Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 12, 1434–1446 (2019).
Oliva, R. et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350 (2019).
Hu, Z. et al. Knockout of OsSWEET15 impairs rice embryo formation and seed-setting. Plant Cell Physiol. 64, 258–268 (2023).
Elliott, K. et al. CRISPR/Cas9-generated mutations in a sugar transporter gene reduce cassava susceptibility to bacterial blight. Plant Physiol. 195, 2566 (2024).
Moscou, M. J. & Bogdanove, A. J. A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501–1501 (2009).
Grau, J. et al. Computational predictions provide insights into the biology of TAL effector target sites. PLoS Comput. Biol. 9, e1002962 (2013).
Pérez-Quintero, A. L. et al. Comparative genomics identifies conserved and variable TAL effectors in African strains of the cotton pathogen Xanthomonas citri pv. malvacearum. Phytopathology 113, 1387–1393 (2023).
Shah, S. M. A. et al. Two TAL effectors of Xanthomonas citri pv. malvacearum target GhSWEET15 as the susceptibility genes for bacterial blight of cotton. bioRxiv, 2024.2008. 2006.606744 (2024).
Haq, F. et al. Identification of a virulence tal gene in the cotton pathogen, Xanthomonas citri pv. malvacearum strain Xss-V2-18. BMC Microbiol. 20, 91 (2020).
Pérez-Quintero, A. L. et al. An improved method for TAL effectors DNA-binding sites prediction reveals functional convergence in TAL repertoires of Xanthomonas oryzae strains. PloS One 8, e68464 (2013).
Hou, S., Rodrigues, O., Liu, Z., Shan, L. & He, P. Small holes, big impact: Stomata in plant-pathogen-climate epic trifecta. Mol. Plant 17, 26–49 (2024).
Xin, X. F. et al. Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539, 524–529 (2016).
Liu, Z. et al. Phytocytokine signalling reopens stomata in plant immunity and water loss. Nature 605, 332–339 (2022).
Lima, V. F. et al. Toward multifaceted roles of sucrose in the regulation of stomatal movement. Plant Signal. Behav. 13, e1494468 (2018).
Schwartz, A. R., Morbitzer, R., Lahaye, T. & Staskawicz, B. J. TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proc. Natl. Acad. Sci. 114, E897–E903 (2017).
Cernadas, R. A. et al. Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLoS Pathog. 10, e1003972 (2014).
Scinto-Madonich, N. J. et al. Initial characterization of a bacterial leaf streak susceptibility gene suggests it encodes a membrane transporter that influences seed nutrition and germination. Physiological Mol. Plant Pathol. 126, 102031 (2023).
Sivaraman, S. et al. TAL effectors and the predicted host targets of pomegranate bacterial blight pathogen Xanthomonas citri pv. punicae. Curr. Genet. 68, 361–373 (2022).
Zárate-Chaves, C. et al. TAL effector repertoires of strains of Xanthomonas phaseoli pv. manihotis in commercial cassava crops reveal high diversity at the country scale. Microorganisms 315. https://doi.org/10.3390/microorganisms9020315, (2021)
Mücke, S. et al. Transcriptional reprogramming of rice cells by Xanthomonas oryzae TALEs. Front. plant Sci. 10, 429072 (2019).
Cohn, M., Morbitzer, R., Lahaye, T. & Staskawicz, B. J. Comparison of gene activation by two TAL effectors from Xanthomonas axonopodis pv. manihotis reveals candidate host susceptibility genes in cassava. Mol. Plant Pathol. 17, 875–889 (2016).
Collmer, A. & Keen, N. T. The role of pectic enzymes in plant pathogenesis. Annu. Rev. Phytopathol. 24, 383–409 (1986).
Booher, N. J. et al. Single molecule real-time sequencing of Xanthomonas oryzae genomes reveals a dynamic structure and complex TAL (transcription activator-like) effector gene relationships. Microbial. Genom. 1, e000032 (2015).
Chin, C.-S. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013).
Darling, A. E., Mau, B. & Perna, N. T. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS One 5, e11147 (2010).
Cabanettes, F. & Klopp, C. D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ 6, e4958 (2018).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Tatusova, T. et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624 (2016).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14 (2019).
Read, A. C., Hutin, M., Moscou, M. J., Rinaldi, F. C. & Bogdanove, A. J. Cloning of the rice Xo1 resistance gene and interaction of the Xo1 protein with the defense-suppressing Xanthomonas effector Tal2h. Mol. Plant-Microbe Interact. 33, 1189–1195 (2020).
Li, C. et al. An efficient method to clone TAL effector genes from Xanthomonas oryzae using Gibson assembly. Mol. Plant Pathol. 20, 1453–1462 (2019).
Falahi Charkhabi, N. et al. Complete genome sequencing and targeted mutagenesis reveal virulence contributions of Tal2 and Tal4b of Xanthomonas translucens pv. undulosa ICMP11055 in bacterial leaf streak of wheat. Front. Microbiol. 8, 1488 (2017).
De Feyter, R., Yang, Y. & Gabriel, D. Gene-for-genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol. Plant-Microbe Interact. 6, 225–237 (1993).
He, P., Shan, L. & Sheen, J. The use of protoplasts to study innate immune responses. Methods Mol. Biol. 354, 1–9 (2007).
Li, B. et al. Phosphorylation of trihelix transcriptional repressor ASR3 by MAP KINASE4 negatively regulates Arabidopsis immunity. Plant Cell 27, 839–856 (2015).
Gao, X. et al. Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 9, e1003127 (2013).
Lee, J. H., Jin, S., Kim, S. Y., Kim, W. & Ahn, J. H. A fast, efficient chromatin immunoprecipitation method for studying protein-DNA binding in Arabidopsis mesophyll protoplasts. Plant Methods 13, 1–12 (2017).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Elliott, K., Berry, J. C., Kim, H. & Bart, R. S. A comparison of ImageJ and machine learning based image analysis methods to measure cassava bacterial blight disease severity. Plant Methods 18, 1–12 (2022).
Gao, X., Britt Jr, R. C., Shan, L. & He, P. Agrobacterium-mediated virus-induced gene silencing assay in cotton. J Vis. Exp. 54, e2938 (2011).
Yang, Y., Feyter, R. D. & Gabriel, D. W. Host-specific symptoms and increased release of Xanthomonas citri and X. campestris pv. malvacearum from leaves are determined by the 102-bp tandem repeats of pthA and avrb6, respectively. Mol. Plant-Microbe Interact. 7, 345–355 (1994).
Wang, F., Du, C., Chen, J., Shi, L. & Li, H. A new method for determination of pectin content using spectrophotometry. Polymers 13, 2847 (2021).
Li, F. et al. Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity. Cell Host Microbe 16, 748–758 (2014).
Gao, X. et al. Cotton GhBAK 1 Mediates Verticillium Wilt Resistance and Cell Death. J. Integr. Plant Biol. 55, 586–596 (2013).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82–e82 (2011).
Tang, X., Zhong, Z., Zheng, X. & Zhang, Y. Construction of a single transcriptional unit for expression of Cas9 and single-guide RNAs for genome editing in plants. Bio-Protoc. 7, e2546–e2546 (2017).
Rathore, K. S., Campbell, L. M., Sherwood, S. & Nunes, E. Cotton (Gossypium hirsutum L.). Agrobacterium Protoc. ume 2, 11–23 (2015).
Babilonia, K. et al. A nonproteinaceous Fusarium cell wall extract triggers receptor-like protein-dependent immune responses in Arabidopsis and cotton. N. Phytologist 230, 275–289 (2021).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Ghosh, S. & Chan, C.-K. K. Analysis of RNA-Seq data using TopHat and Cufflinks. Methods Mol. Biol. 1374, 339–361 (2016).
Wilkins, K. E., Booher, N. J., Wang, L. & Bogdanove, A. J. TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Front. Plant Sci. 6, 536 (2015).
Acknowledgements
We thank Baden Aniline and Soda Factory (BASF) for providing FiberMax cotton seeds; Drs. Shi-En Lu (Mississippi State University) and Thomas Isakeit (Texas A&M University) for Xcm strains; Leslie Wells (Texas A&M AgriLife Research) for cotton seed propagation; and members of the laboratories of L.S. and P.H. for discussions and comments on the experiments. The study was supported by the United States Department of Agriculture-National Institute of Food and Agriculture (USDA-NIFA) (2018-67013-28513) to L.S and A.J.B., and National Science Foundation (NSF) (IOS-2421016), USDA-NIFA (2023-67013-41927 and 2020-67013-41537) to P.H. and Cotton Incorporated (18-123TX) to L.S. B.W.M. and Y.Y. contributed equally to the work.
Author information
Authors and Affiliations
Contributions
B.W.M., P.H., A.B., and L.S. conceived the project, designed experiments, and analyzed data. B.W.M. performed most of the bacterial inoculation, disease phenotyping, gene expression, and RNA-seq sample preparation. Y.Y. generated vectors and performed TALE-EBE expression and binding assays, and created vectors and performed water-soaked lesion assays in planta. T.B. and L.W. generated the Xcm TX4 mutants and designer TALEs. R.C.R. and S.C.D.C. performed genome assembly and analysis of Texas Xcm isolates. C.D.C. performed transcriptome analysis of RNA-seq data. K.C. initially performed RT-PCR analysis, isolated Xcm genomic DNA, and constructed VIGS-SWEET14a/b vector. L.Z. constructed the CRISPR vectors. X.M. performed TALomes immunoblot analysis. T.A.W. and J.K.D. collected Xcm isolates from Texas cotton fields. B.W.M., Y.Y., P.H., A.J.B., and L.S. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Alvaro Pérez-Quintero and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mormile, B.W., Yan, Y., Bauer, T. et al. Activation of three targets by a TAL effector confers susceptibility to bacterial blight of cotton. Nat Commun 16, 644 (2025). https://doi.org/10.1038/s41467-025-55926-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-55926-7