Abstract
A critical goal in functional genomics is evaluating which non-coding elements contribute to gene expression, cellular function, and disease. Functional characterization remains a challenge due to the abundance and complexity of candidate elements. Here, we develop a CRISPRi-based approach for multi-locus screening of putative transcription factor binding sites with a single truncated guide. A truncated guide with hundreds of sequence match sites can reliably disrupt enhancer activity, which expands the targeting scope of CRISPRi while maintaining repressive efficacy. We screen over 13,000 possible CTCF binding sites with 24 guides at 10 nucleotides in spacer length. These truncated guides direct CRISPRi-mediated deposition of repressive H3K9me3 marks and disrupt transcription factor binding at most sequence match target sites. This approach can be a valuable screening step for testing transcription factor binding motifs or other repeated genomic sequences and is easily implemented with existing tools.
Similar content being viewed by others
Introduction
Over 1 million human cis-regulatory elements (CREs) have been cataloged across various cell and tissue types1,2,3,4. CREs include the promoters, enhancers, insulators, and silencers that direct gene expression, sometimes in dynamic interplay or synergy. CRE function is further influenced by cell state and multiple transcription factor (TF) binding sites. TFs recruit proteins and complexes to orchestrate gene expression. TFs bind with various strengths, often dictated by cell state and genomic contexts such as motif combinations and orientations5,6,7. However, the determinants for TF binding to one motif over another and the effect of that binding are not well understood. Connecting CREs and TF binding with functional outputs is important for interpreting disease associated genetic variation3,8,9 and may help nominate regions for clinical interventions. Together, TFs and CREs direct the intricate regulatory networks that govern cell function and disease.
CRISPR interference (CRISPRi) consists of a catalytically dead Cas9 (dCas9) that can be fused to a zinc-finger repressive protein (KRAB) for transcriptional silencing. Several studies have relied on CRISPRi-directed targeting of CREs followed by RNA measurement or flow cytometry to detect gene expression changes10,11,12,13,14,15,16,17,18,19. However, efforts to characterize CREs at scale have been complicated by the large number of putative elements and mild effect sizes. High multiplicity of infection (MOI) delivery of guides paired with single-cell RNA-seq provided a multiplexed testing approach16, though at the cost of many viral integration events. As such, while CRISPRi-based approaches can effectively assess significant CREs, there is a critical need for improving their scalability.
The Cas9 nuclease is guided by a spacer sequence that determines targeting specificity. Typically, spacers are 20 nucleotides (nt) in length and target a single genomic site based on sequence complementarity. Early studies posited that spacers with minor 5’ truncations or mismatches retain Cas9-mediated, on-target cleavage20,21,22. The 3’ end of the spacer sequence, also termed the seed sequence, is necessary though not alone sufficient for on-target cleavage. Activity was observed with truncated spacers of 17nt while 15nt or shorter spacers failed to demonstrate cleavage activity20,23,24,25. However, an important distinction exists in the requirements for Cas9 binding and cleavage that is illuminated with dCas9 protein. Indeed, spacers as short as 10nt sufficed for dCas9-VPR (CRISPRa) activity at a single target site26. We postulated that KRAB-dCas9 (CRISPRi) would perform similarly and, further, target multiple intended sites simultaneously.
Here, we explored the ability of truncated guides to direct CRISPRi to multiple sites simultaneously in the genome for multi-locus repression. Truncated guides resulted in reliable on-target efficacy down to spacer lengths of 9nt. TF motifs, which are often less than 14nt, presented ideal genomic loci for multiplexed repression. We target TF motifs in a CRE of the EPB41 gene and observe comparable on-target efficiencies with full-length and truncated guides. We screened a truncated guide library targeting thousands of CTCF motif sites and discovered significant CTCF disruption. This approach offers a distinct opportunity to simultaneously perturb CREs at scale and effectively prioritize genomic loci for further study.
Results
Truncated guides direct CRISPRi to a sequence match site
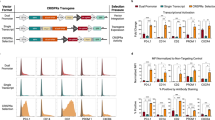
We first set out to characterize the minimum guide length required for CRISPRi-mediated repression. All experiments termed “CRISPRi” utilized a KRAB-dCas9 lentivrial construct. CD81, a stably expressed, non-essential cell-surface protein, served as a reporter of on-target efficiency by flow cytometry (Fig. 1a). We selected a high performing 20nt S. pyogenes spacer (sgCD81i-1)27, directed to the CD81 transcriptional start site (TSS) and tested successive truncations. By convention, guides are cloned with a guanine in the 5’ position to improve Pol III transcription levels28, sometimes resulting in the guanine complementing the target sequence (see Methods and Supplementary Fig. 1a). Therefore, here we use brackets to denote the length of guide sequence that complements a single target site. For example, sgCD81i-1 g[12nt] consists of a 5’ mismatched guanine and 12 complementary bases to the CD81 TSS. Successive 5’ truncations of sgCD81i-1 resulted in repression with each guide down to a 9nt target match in Jurkat (T lymphocyte) cells (Fig. 1b), with sgCD81i-1 g[9nt] active and sgCD81i-1 g[8nt] exhibiting a complete loss of on-target activity. We next tested additional CD81 TSS 20nt guides with less effective on-target efficiency (sgCD81i-2 and sgCD81i-3). These truncated guides resulted in similar and sometimes better CD81 repression relative to the respective 20nt guide (Supplementary Fig. 1b). We expected that CRISPR knockout would be ineffective with sizeable truncations based on prior studies23,24,25 and designed 2 guides that target exon 1 of CD81 (sgCD81-KO-1 and -2). CD81 knockout was effective at lengths down to a 17nt target match, consistent with prior findings (Fig. 1c). Indeed, guide length requirements for Cas9 cleavage and CRISPRi diverge at <17nt guide lengths, highlighting opportunities for CRISPRi targeting with truncated guides that are not possible with Cas9 cleavage.
a Schematic of truncated guide experiments. b CRISPRi in Jurkat cells with truncated CD81 promoter targeting guide and analyzed for CD81 cell surface expression. c Cas9 cleavage in Jurkat cells treated with two guides targeting CD81 and truncated versions. d CRISPRi in A375 cells with truncated CD81 promoter targeting guide and analyzed for CD81 cell surface expression. e Gene expression in A375 cells in 20nt and g[9nt] sgCD81i-1 CRISPRi populations. p-value cutoff at >2 and log fold-change >2 and <−2 using a linear mixed model and two-sided test that adjusts for multiple hypotheses. Significantly upregulated genes in 20nt and g[9nt] are 2 and 34, respectively. For b–d, cells were stained with CD81-FITC antibody analyzed by flow cytometry 7 days post lentiviral transduction for CD81 targeting guide, safe harbor control (Safe), or untransduced (UT). Flow gating strategy found in Supplementary Fig. 1f. Data are mean +/− SD from biological triplicate. Source data are provided as a Source Data file.
Next, we investigated the specificity of truncated guide repression. Unpaired bases at the 5’ end of 20nt guides can impact their activity. We lengthened the 5’ end of sgCD81i-1 10nt with 1–3 additional bases. Either 1 or 2 unpaired bases on the 5’ end resulted in effective repression, while 3 unpaired bases (gcc) completely abrogated repression (Supplementary Fig. 1c, d). Additionally, we tested dCas9 alone (no KRAB) with truncations of sgCD81i-1 and observed modest repression and more variability compared to the 20nt guides (Supplementary Fig. 1e). It is noteworthy that steric hindrance by dCas9 binding at the sgCD81i-1 target site is contributing to CD81 repression. We next tested sgCD81i-1 full-length and truncated constructs in A375 (melanoma) cells with CRISPRi and demonstrated similar CD81 repression as observed in the Jurkat experiments (Fig. 1d), providing evidence that truncated guides are active in an additional cellular context. RNA sequencing in A375 showed similar levels of CD81 repression at 20nt and g[9nt] lengths along with 32 additional downregulat ed targets in the g[9nt] treatment (Fig. 1e and Supplementary Data 1). In sum, 5’ truncated guides can direct dCas9 components to induce repression at target promoters.
Enhancer disruption with truncated guides
We next asked whether an active enhancer is targetable with truncated guides directed toward multiple TF motif sequences. We selected a 570 bp locus with several putative TF binding sites, 2.8 kb upstream of the EPB41 gene (Fig. 2a, chr1:28,883,749-28,884,318). This locus was previously identified as a possible regulator of EPB41 in a K562 CRISPRi screen16. We selected 4 TF motifs in this enhancer (PU.1, SP1, YY1 and NR2), each containing an ideally positioned NGG sequence for the Cas9 protospacer adjacent motif (PAM) (Fig. 2b). TF motifs were positioned at the 3’ end of the guide including the PAM and two truncated versions (g[13nt] or 14nt and 11nt). The full-length guides match only the EPB41 enhancer locus while the 11nt guides matched hundreds of additional genomic sites (Fig. 2c). We transduced K562 cells and measured on-target efficiency for EPB41 knockdown by real-time quantitative PCR of EPB41 and compared to 3 guides identified from the prior screen16 as well as a promoter targeting guide (Fig. 2d). EPB41 expression was reduced to levels comparable to the respective 20nt guide in 3 out of 4 11nt guides (PU.1, YY1, NR2) (Fig. 2d and Supplementary Fig 2a). In aggregate, the full-length and truncated guides tested significantly decreased EPB41 expression as compared to safe harbor control (Fig. 2e and Supplementary Fig. 2b, one-way ANOVA P < 0.0001). CRISPRi-directed truncated guides can effectively disrupt an enhancer.
a Schematic of EPB41 genomic regulatory region and full-length/truncated guides targeting each of 4 TF motifs. b TF motifs (JASPAR) targeted by the guides with protospacer adjacent motif (PAM) orientation denoted. c Number of sequence match target sites in the genome (hg38) for the indicated motif directed guide. d Quantitative real time PCR analysis of EPB41 expression in K562 cells with respective guide CRISPRi treatment conditions. Safe (Safe Harbor-targeting) guide = g[20nt], promoter guide = 21nt, enh1 = g[20nt], enh2 = 21nt, enh3 = g[20nt] (enhancer guides from Gasperini et al. 2019). Data are mean +/− SEM. e Violin plot summary of EPB41 expression data in panel d organized by guide length or targeting region. TF-targeting guides (n = 16; 4 guides per spacer length), sgEnh (n = 12; 3 guides), and Promoter (n = 4; 1 guide, all points shown) are the product of 2 biological replicates per guide run in duplicate. Safe is the sum of 3 biological replicates run 2-3 times each (n = 8). Data are normalized to the mean CT value of Safe Harbor actin-b control probe. Results are analyzed by one-way ANOVA. ****P < 0.0001 for all guide lengths compared with Safe Harbor guide. P = 8.24 × 10−7, 9.95 × 10−7, 5.62 × 10−4, 3.98 × 10−5, and 0.01 for Safe vs. 21nt,g[20nt]; 14nt,g[13nt]; 11nt; sgEnh; and Promoter respectively. Source data are provided as a Source Data file.
A CTCF-directed truncated guide library
To test the utility of truncated guides for multi-locus TF perturbation, we selected CCCTC-binding factor (CTCF) sites to screen. CTCF is a ubiquitously expressed TF whose role in genomic insulation is dependent on convergently oriented consensus sequences29. Leveraging the 3’ NGG PAM sequence in the CTCF motif (Fig. 3a), we designed a library of 24 10nt guides targeting a total of 13,352 sequence match CTCF binding sites (Fig. 3a–c). Based on CTCF ChIP-seq in Jurkat cells, approximately half of these sites are CTCF-bound (6228) and represent 10.8% of all CTCF peaks (Fig. 3c and Supplementary Fig. 3a). This library allowed us to test CTCF binding sites, partitioned by guide, ranging from a minimum of 182 sites (sg1) to maximum of 1123 sites (sg24). As a control we targeted the CTCF locus itself with full-length guides for gene repression (Fig. 3d and Supplementary Fig. 3c). We packaged guides into a lentiviral library, transduced Jurkat cells near an MOI of 0.5, and collected cells over 21 days. We measured guide enrichment and depletion as a proxy for fitness. We quantified the scale of this effect with a z-score (see Methods) relative to 15 full-length safe harbor guides. Our results indicated that most truncated guides were not lethal, as we observed moderate shifts in guide representation relative to safe harbor guides (Fig. 3d and Supplementary Datas 2 and 3). A subset of guides resulted in enrichment, suggesting changes that may promote proliferation. We also identified guides sg4 and sg20 as broadly depleted, though not to the degree of CTCF knockdown. This initial screen provided evidence that certain truncated guides can induce fitness changes in Jurkat cells.
a Guide selection strategy based on top Homer generated motif from CTCF ChIP in Jurkat cells. b CTCF targeting schematic of a single 10nt guide directing CRISPRi to many motifs simultaneously (CTCF-bound and unbound). c Target site distribution (black) and CTCF occupancy based on Jurkat ChIP (blue) for each of the 10nt guide sequences. d, e Guide enrichment and depletion in pooled screen for the indicated cell lines. Pooled screens were run in triplicate, while K562 was in duplicate. Data are mean +/− SD. CTCF promoter targeting full length guide knockdown (KD) used as a control. Source data are provided as a Source Data file.
We next screened the CTCF library in additional cell lines to compare with Jurkat results. We processed A375 cells for CTCF ChIP-seq and found 6140 library target sites bound, representing 14% of all CTCF peaks (Supplementary Fig. 3a, b). In comparison, 6228 of CTCF library sites are bound in Jurkat. We further include K562 (T-lymphocytes), MV4-11 (AML) and HEK293 as additional models representing diverse cellular contexts for screening. Cells were transduced with the CTCF library and assessed for guide representation after 21 days. Fitness effects in these additional cell models largely recapitulated trends observed in Jurkat cells (Fig. 3e and Supplementary Data 3). This could be attributed to invariance of CTCF binding sites across tissues30,31. However, we observed some instances of cell-specific fitness effects, particularly with sg2, sg22, and sg23. While sg2 and sg23 impacted more than one cell line, sg22 was strongly depleted in A375 only.
As an additional test of guide-sequence specificity, we screened 11nt guides by adding every base to each 10nt guide in the CTCF library, totaling 96 guides. The additional 5’ base had modest changes on fitness outcomes when compared to the respective 10nt guide outcome (pearson correlations ranged from 0.86 to 0.94 in Jurkat, 0.64 to 0.85 in K562, 0.79 to 0.94 in MV4-11, 0.86 to 0.96 in HEK293, and 0.73 to 0.87 in A375, Supplementary Fig. 4). This reinforced our prior finding that a single base mismatch was not detrimental to targeting, whereas ≥2 mismatches can disrupt activity (Supplementary Fig. 1c, d). Testing in 5 cell lines, we find that addition of a single 5’ base to a 10nt guide did not often alter guide effects.
Simultaneous targeting of CTCF binding sites
We selected sg4 for further exploration due to its effects on fitness. Guide sg4 targets 357 sites in the genome with 10nt and NGG PAM complementarity, termed “perfect match” sites. Perfect match sites were determined regardless of the 5’ guanine present on all guides. We transduced Jurkat cells with sg4 and CRISPRi for 6 or 7 days (in contrast to 21 days for the fitness screens above) and performed CTCF and H3K9me3 ChIP-seq. The analysis revealed a significant drop in CTCF occupancy at perfect match sites (t-test, P < 10−5) (Fig. 4a, c and Supplementary Fig. 5a, c). We found it promising that CTCF, a strong binder to chromatin, was displaced at many sites simultaneously. Concurrent with CTCF loss was a significant increase in H3K9me3 signal at perfect match sites (t-test, P < 10−5) (Fig. 4b, d and Supplementary Fig. 5b). H3K9me3 is a histone mark that indicates KRAB-dCas9 binding and recruitment of repressive proteins11. An example track of a perfect match site depicts decreased CTCF binding and concurrent increased H3K9me3 signal (Fig. 4e). We next performed dCas9 ChIP-seq and found strong binding at perfect match sites (Fig. 4f and Supplementary Fig. 6). These initial findings presented compelling on-target CTCF disruption at multiple sequence match genomic loci with truncated guides.
a CTCF ChIP of the CTCF bound sites (134) targeted by sg4 in Jurkat cells. Significance with t-test ***P = 5.6 × 10−13. b H3K9me3 ChIP of sg4 targeted perfect match sites (357) with genomic bins plotted (416). Two bins were included in cases where an sg4 site targeted a bin boundary. Significance with t-test ***P = 1.3 × 10−38. Data are aggregated from two independent replicates and presented as median values +/− 1 SD (box), with whiskers extending 1.5x below the first quartile and 1.5x above the third quartile in a and b. c Scatter plot CTCF binding at JASPAR motifs (880k) with 357 perfect match sites (black) and partial match sites (gray). d H3K9me3 density plot for Safe Harbor (S.H., light purple) and sg4 (dark purple) treated Jurkat cells. e Track view of a targeted region depicting CTCF and H3K9me3 signal from 2 replicate experiments (rep). Bars on the bottom depict CTCF perfect match (black) and partial match sites (gray). f dCas9 density plot for Safe Harbor (S.H., light purple) and sg4 (dark purple) treated Jurkat cells. g Volcano plot of CTCF bound sites (400k) with significant 10nt match CTCF sites (black), significant partial match sites (79), and not significant sites. Significance was determined at cutoffs (abs [LFC] > 0.5 and -log10 P > 5) using a linear mixed model and two-sided test that adjusts for multiple hypotheses. h Histogram depicting significant CTCF disrupted sites as a fraction of CTCF bound sites with 0 mismatches (black) up to 4 mismatches (blue). Raw values depicted on each bar. i Logogram of significant perfect match sites (top, 79 sites) and significant partial match sites (bottom, 79 sites) from Jurkat sg4 experiments. Source data are provided as a Source Data file.
We next evaluated multi-locus targeting specificity with a slightly longer spacer sequence. We designed a 13nt guide based on our prior findings that 3 mismatched bases disrupt targeting of CD81 TSS (Supplementary Fig. 1c, d). We appended 3 bases (aag) to the 5’ end of sg4, termed aag[sg4], resulting in a guide with 77 expected perfect match sites. We transduced Jurkat cells with the guide and CRISPRi and processed cells for CTCF ChIP-seq. Examining the 26 CTCF-bound genomic regions targeted by both guides, we observed significantly lower CTCF signal in the aag[sg4] and sg4 samples compared to safe harbor guide (Supplementary Fig. 7a, b). Targeting with aag[sg4] led to a greater CTCF loss (mean binding ratio of 0.34 versus safe) than with sg4 (mean binding ratio of 0.54 versus safe) at these sites (Supplementary Figs. 5a and 7c). Sg4 and aag[sg4] were not significantly different from one another; however, we observed a lower mean CTCF signal in aag[sg4].
An additional truncated guide from the CTCF library was investigated further for its efficiency. We selected sg8 (10nt) which had 465 sequence match sites and little fitness impact on Jurkat cells. We transduced Jurkat cells with sg8 and CRISPRi and processed the cells for ChIP-seq after 7 days. CTCF binding was severely impacted at sequence match bound sites (t-test, P < 10−5, 411 sites) along with an increase in H3K9me3 signal (t-test, P < 10−5) (Supplementary Fig. 8a–c). This complemented the CTCF loss and H3K9me3 gain results observed with sg4. Therefore, truncated guide CRISPRi can deplete CTCF binding events in a bulk population and at a few hundred genomic loci.
We next explored gene expression effects with CTCF-directed truncated guides. Samples of sg4 and sg8 were collected 7 days post lentiviral transduction and processed for RNA sequencing. Significant genes were determined with dream differential expression analysis32, which aggregates data across replicates for confidence. Gene expression for sg4 indicated no differential genes (Supplementary Fig. 9). However, targeting with sg8 resulted in upregulation of 67 genes (Supplementary Fig. 9) which cluster in keratin filament and intermediate filament gene programs (Supplementary Data 5). Interestingly, the sg8 motif is bound by CTCF at 88.4% of sequence match sites compared to only 46.6% for all tested 10nt guides (Fig. 3c). Further investigation is needed to characterize the changes in enhancer-promoter interactions due to CTCF insulator loss33,34,35,36. CTCF disruption with one truncated guide led to no differential gene expression while another guide induced gene upregulation.
Characterization of disrupted CTCF sites
We next pursued a comprehensive approach to identify significantly impacted CTCF sites. We generated a list of over 880k JASPAR CTCF binding sites in the genome (see Methods). We plotted CTCF ChIP-seq read counts mapping to these regions for sg4 or sg8 relative to Safe Harbor controls (Fig. 4c, Supplementary Figs. 5c and 8c and Supplementary Data 4). This allowed for proper visualization of both targeted and non-targeted CTCF sites. We then applied dream differential analysis to determine significant sites. This analysis determined that 64% of Jurkat sg4 sites were CTCF depleted (79 out of 123 perfect match bound sites, P < 10−5, LFC < 0.5) (Fig. 4g, h). Similarly, dream analysis of sg8 Jurkat samples showed significant loss at 55.7% of CTCF sites (Supplementary Fig. 8d, 229 of 411 perfect match bound sites, P < 10−5, LFC < 0.5). We concluded that hundreds of CTCF sites can be depleted with truncated guides and over half of targeted CTCF peaks are lost at this scale.
We next investigated CTCF loss at sites other than perfect match loci. We filtered the 880k JASPAR annotated motifs to find putative CTCF binding sites with ≤9nt complementarity to the 10nt guide that we collectively termed “partial match” sites. We analyzed partial match sites in sg4 containing 1 to 5 mismatches and observed 78 out of 29,908 sites had significant CTCF loss (P < 10−5, LFC < 0.5) (Fig. 4g, h). Most partial match sites with CTCF loss had a 1nt mismatch. Motif analysis further illuminated mismatch tolerance positions in the guide at A-T rich bases and the 5’ end for sg4 (Fig. 4i and Supplementary Fig. 10a) and sg8 (Supplementary Figs. 8e). Additional analysis on CTCF bound 1 mismatch sites revealed the importance of PAM-proximal bases (Supplementary Fig. 10b). It is noteworthy that most partial match sites remained unaffected by the CRISPRi truncated guides, indicating strong sequence specificity for targeted CTCF sites.
TFs besides CTCF may be explored with this approach. We screened JASPAR motifs for ideally positioned NGG PAMs and highlight 88 putative vertebrate proteins for targeting with truncated guides (Supplementary Data 6 and Supplementary Fig. 11). We provide ENCODE accessions of ChIP-seq data available for these proteins. In sum, truncated guides can enhance TF binding site evaluation with a motif-centered approach.
Discussion
Here we show effective CRISPRi targeting at multiple genomic loci with truncated guides as short as 9nt. This is a unique property of dCas9 moieties, as catalytically active Cas9 is incapable of on-target cleavage with guides shorter than 17nt. A single truncated guide can target CRISPRi to hundreds of TF binding sites, prospectively targeting functional regulatory elements. A library of 24 10nt guides enabled screening of over 13,000 CTCF sites, representing 10.8% and 14% of bound CTCF sites in Jurkat and A375 cells respectively, demonstrating scalable utility. Chromatin binding analysis revealed simultaneous disruption of multiple CTCF binding events with a single truncated guide at most sequence match sites.
The activity of a guide depends on seed sequence, which impacts best outcomes of both 20nt and 10nt guides. When designing a truncated guide library, it is important to consider exact seed sequence as some TF motifs will be better targeted than others. PAM distal mismatches of a few bases are tolerated with full-length guide-directed dCas937 and surprisingly we observed the same 5’ flexibility with 10-13nt guides. PAM proximal deviations on the 3’ end of the guide are not tolerated. Additionally, there are considerations beyond the guide sequence that determine dCas9 targeting efficiency. Not all CREs or TF motifs may be amenable to CRISPRi perturbation. Furthermore, it is important to consider how much H3K9me3 is for disruption of TF binding. A recent study demonstrated better enhancer and promoter targeting with a KRAB-dCas9-MeCP2 system than with KRAB-dCas9 alone38, suggesting the importance of the repressive marks. We tested truncated guides with dCas9 alone and observed repressive activity. Ultimately, without repressive H3K9me3, we expect guide performance and proximity to the TF binding site will dictate the degree of steric repressive activity. In this work we targeted TF binding motifs directly, making it plausible that steric hindrance also contributed to TF displacement from chromatin.
Selection of targeted TF motifs and guide length are critical when planning truncated guide screens. We selected motifs containing an NGG PAM to maximize the likelihood of dCas9 binding. dCas protein variants with expanded PAM requirements will widen the available target sequence space. The shortest amenable truncated guide lengths will need to be determined in these variant systems. In our experiments, 10nt and 13nt guides both significantly disrupted target CTCF binding, however the 13nt guide generated a greater effect size at its targets. Explanations for this are either that longer guides form a more stable R-loop structure with dCas9 resulting in a stronger binding affinity39,40 or longer guides benefit from a more favorable ratio of KRAB-dCas9 protein units to fewer target sites. The maximum number of sites simultaneously targetable with a single guide is yet to be determined. Here we investigate guides with hundreds of sequence match sites (sg4 and sg8), but the exact multi-locus limit will depend on specific guide sequences and expression levels of the KRAB-dCas9 construct. It stands to reason the more truncated the guide, the greater the likelihood for false negatives. It may be beneficial to screen libraries with guides of various sizes (such as both 10nt and 13nt) to accommodate combinatorial effects and improve confidence, prior to secondary screening or in tandem with full length guides. Lastly, we suggest calculating the fraction of sequence match sites bound by the target TF (via ATAC-seq, ChIP-seq, or CUT&Tag data, including any of the 88 factors provided based on motif) to assess screen efficiency. In Jurkat cells, 46.6% of sequence match CTCF sites for all guides in the library were bound by the protein. The bound fraction may be lower for other TFs and DNA-binding proteins.
The truncated guide method is a first pass discovery tool for targeting repeated genomic loci. This approach is particularly useful when TF knockout results in negative fitness outcomes or lethality, such as with CTCF. TFs can have alternate cellular functions, such as RNA binding41, that can confound TF knockout studies. This is avoided with truncated guide experiments since binding sites themselves are reliably assayed. Another advantage to this approach is its applicability to cell models with low lentiviral efficiencies or rare cell populations, such as primary cells. A single truncated guide provides a rich landscape of tested outcomes with few transduction events. Finally, we anticipate that truncated guide perturbation will provide a rich readout of gene and TF regulatory networks in single cell assays.
Methods
All experiments and methodology in this research study are compliant with and have been approved by the Institutional Review Board of the Broad Institute. No human participants were used in this study.
Cell culture
Jurkat, K562, and MV4-11 cells were cultured in RPMI (Gibco) + 10% FBS (Sigma) supplemented with 1% penicillin/streptomycin (Gibco). HEK293FT and A375 cells were cultured in GlutaMAX High Glucose DMEM (Gibco) + 10% FBS supplemented with 1% penicillin/streptomycin. Cells were tested monthly (negative) for mycoplasma contamination and maintained in a 37 °C humidity-controlled incubator with 5% CO2. Jurkat, K562, MV4-11 cell lines were obtained from the Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle/home), HEK293FT cells from Invitrogen, and A375 cells from ATCC. STR profiling was used to confirm cell line identities upon arrival.
Flow cytometry
Cells were incubated for 30 min at room temperature in 0.5% BSA PBS with 1:50 CD81-FITC antibody (Biolegend, 349504) or mouse IgG1 FITC isotype control antibody (Biolegend, 400107). Cells were washed twice prior to analysis on a Cytoflex (BD) cell analyzer. The gating strategy can be found in Supplementary Fig. 1f.
Guide selection
All guide sequences in this study can be found in Supplementary Data 2. This includes the CTCF motif-directed library for the 24 guides (10nt, selected based on TF motif) and 96 (11nt, by each base to the 5’ end of each 10nt guide). To assess the effect of CTCF knockdown on cell fitness, we used CRISPick to select 20 sgRNAs targeting the CTCF promoter. After screening, one guide was selected based on lethality across all cell lines and was included as the knockdown data found in Fig. 2c. Additionally, 15 safe harbor sgRNAs42 were included as negative controls.
Vectors and virus production
Annealed oligos were cloned into an all-in-one KRAB-dCas9-puro vector (pXPR_066, Broad GPP) using the BsmBI restriction enzyme for backbone linearization and T7 ligase for CD81 promoter targeting experiments, the EPB41 enhancer locus experiments, and the CTCF pooled library and follow up. sgCD81i-1 g[9nt] and g[8nt] guides required golden gate assembly (NEB) due to the short length of the oligonucleotides. The CTCF library with varying guide lengths involved the production of 3 separate pooled libraries, one for each of 10nt, 11nt, and 20nt guide lengths. These libraries were then mixed at a balanced (equimolar) ratio to produce the final library.
Single plasmids were chemically transformed into One Shot STBL3 chemically competent coli (Invitrogen C737303). Bacterial cultures were shaken at 225 rpm for one hour at 37 °C and then plated on an ampicillin agar dish. After overnight growth at 37 °C, single colonies were picked into LB and shaken overnight at 225 rpm and 37 °C. Plasmids were isolated the next day using a Plasmid Miniprep Kit (Qiagen) and quantified using a Qubit fluorometer.
Pooled plasmid libraries were electroporated into ElectroMAX™ Stbl4™ electrocompetent cells (Invitrogen 11635018) and spread on to bioassay plates. After overnight incubation at 30 °C, bacterial colonies were collected and isolated using the Plasmid Plus Midi Kit (Qiagen 12941). 20nt, 11nt, and 10nt sequences of the CTCF pooled library were cloned as individual pools and then combined to reduce drift. Guide sequences can be found in Supplementary Data 2. All guides were cloned with a guanine base at the 5’ end to improve transcription from the U6 promoter28,43.
For single plasmids, 1 × 106 HEK293FT cells were seeded in each 6-well in 2 ml of DMEM + 10% FBS 24 h prior to transfection. A DNA mixture was prepared consisting of 250 µl Opti-MEM, 0.25 µg pCMV_VSVG (Addgene 8454), 1.25 µg psPAX2 (Addgene 12260), 1 µg of the all-in-one CRISPR vector (pXPR_066), and 7.5ul TransIT-LT1 (Mirus) transfection reagent. After a 20-min incubation, the solution was added dropwise to the 6-well and incubated for 6–8 h. Fresh media was added to the cells and collected 36 h later and either snap frozen or added to cells.
For CTCF pooled library, 8 × 106 HEK293FT cells were seeded in each of 2 T75 flasks in 12 ml of DMEM + 10% FBS 24 h prior to transfection. Next, pCMV_VSVG (Addgene, 8454, 1.5 µg), psPAX2 (Addgene 12260, 9 µg), the guide containing vector (pXPR_066, 7.5 µg), and 66 µl TransIT-LT1 (Mirus MIR 2306) were combined with 2.1 ml of Opti-MEM to produce TransIT-LT1:DNA complexes. After a 20-min incubation, the solution was added dropwise to the 6-well and incubated for 6–8 h, then the media was changed. After 36 h, the lentivirus was collected, filtered, and either snap frozen or used for cell transduction.
Viral transduction
CTCF pooled library frozen viral supernatant (300 µl) was thawed and added to 700 µl target cells in 12-wells with a final volume of 10 µg/mL polybrene, resulting in a 30–50% transduction efficiency, corresponding to an MOI of ~0.35–0.70. Cells with viral supernatant were centrifuged at 2000 × g for 20 min at 22 °C and incubated overnight. After 18–24 h, cells were fed fresh media and maintained at 2 × 105 cells/mL for suspension cells (Jurkat, K562, and MV4-11) and 1-2 × 105 cells/cm2 for adherent cells (A375 and HEK293). Cells were passaged into media supplemented with 1 µg/mL puromycin 3 days after transduction. Seven days after transduction, cells were passaged into 0.5 µg/mL puromycin (1/2 dose) and cultured continuously for the duration of the screen. At day 21, pellets of 1 × 106 cells were snap frozen on dry ice and stored at −80 °C in preparation for gDNA isolation.
Single transductions were performed identically to the pooled production, with the exception that viral supernatant varied based on viral titer.
Genomic DNA preparation and sequencing
Genomic DNA was isolated using DNeasy Blood and Tissue Kit (Qiagen 69504). PCR, sequence adaptor barcoding, cleanup, sequencing, and data deconvolution were carried out as previously described44. PCR primers were Argon and Beaker (Broad Institute GPP). At the PCR stage, CTCF pooled library plasmid DNA (pDNA) was diluted to 10 ng for amplification. All PCR reactions were carried out for 28 cycles. Libraries were prepared using TruSeq amplicon construction and single end sequenced on a MiSeq50. Fastq files were deconvolved using PoolQ (https://portals.broadinstitute.org/gpp/public/software/poolq). Apron (Broad Institute GPP) was used to analyze the distribution of each guide relative to the plasmid DNA, enabling enrichment/depletion measurements.
ChIP-seq sample preparation
Frozen crosslinked cell pellets (1 × 107 cells) were suspended in cell lysis buffer (20 mM Tris pH 8.0, 85 mM KCl, 0.5% NP40) with protease inhibitors (cOmplete EDTA-free Protease Inhibitor Tablets, Sigma Aldrich), incubated on ice for 10 min, then centrifuged at 1000 × g for 5 min. Cell pellets were resuspended for a second time in cell lysis buffer with protease inhibitors, incubated on ice for 5 min and centrifuged for 5 min at 1000 × g. The pellets were resuspended in nuclear lysis buffer (10 mM Tris-HCl pH7.5, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS) with protease inhibitors for 10 min and subsequently sheared in a sonifier (Branson).
The chromatin was quantified after sonication to determine the cell number in each sample. H3K9me3 ChIP-seq samples were prepared with 1.5 × 106 cells and 0.4 µg H3K9me3 antibody (Abcam ab176916). CTCF ChIP-seq samples were prepared with 3 × 106 cells and 1 µg CTCF antibody (Diagenode C15410210). dCas9 ChIP-seq samples were prepared with 4 × 106 cells and 4 µl Cas9 antibody (Diagenode C15310258). ChIP Dilution Buffer (16.7 mM Tris-HCl pH 8.1, 167 mM NaCl, 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA) with protease inhibitors was added to bring the ChIP volume to 0.5 mL. ChIP-seq samples were rotated overnight at 4 °C. The following day, Protein A Dynabeads (Invitrogen) were added for 1 hour to enrich fragments of interest. The ChIP-seq samples were removed from rotation, spun briefly and placed on a magnet to isolate the beads. The beads were washed with a series of buffers, low salt RIPA buffer, high salt RIPA buffer, LiCl buffer (250 mM LiCl, 0.5% NP40, 0.5% sodium deoxycholate, 1 mM EDTA,10 mM Tris-HCl pH 8.1) and finally Low TE. The Protein A beads were then suspended in 50 µl direct ChIP elution buffer (10 mM Tris-Cl pH 8.0, 5 mM EDTA, 300 mM NaCl, 0.1% SDS and 5 mM DTT directly before use) and 8 µl of reverse crosslinking mix (250 mM Tris-HCl pH 6.5, 1.25 M NaCl, 62.5 mM EDTA, 5 mg/ml Proteinase K, and 62.5 µg/ml RNAse A). The suspended beads were incubated at 65 °C for a minimum of 3 h. After incubation, the supernatants were transferred to a clean tube. The DNA was SPRI purified, eluted, and quantified by Qubit. Libraries with 6 µg of input were prepared using the KAPA Hyper Prep Kit.
Quantitative real time PCR
Real time PCR was performed as described previously45. In brief, RNA extraction was performed with the RNeasy Plus Micro Kit (Qiagen) and cDNA was synthesized using the Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen). Probes for EPB41 and actin beta transcripts (EPB41_1_For AACTTCCCAGTTACCGAGCA, EPB41_1_Rev CTTGAGTCCGGCCACTGTAT, EPB41_2_For CTGCTCTAGTGGCCTTCTGG, EPB41_2_Rev CTGCTCGGTAACTGGGAAGT, actin-b_For CATCGAGCACGGCATCGTCA, and actin-b_Rev TAGCACAGCCTGGATAGCAAC) were paired with Power SYBR Green PCR Master Mix (Applied Biosystems) for quantification. Samples were analyzed on a BioRad CFX Opus 384 Real-Time PCR System. All samples were normalized to the average Ct across all replicates of actin beta safe harbor.
RNA sequencing sample preparation and analysis
Jurkat and A375 cells were transduced with all-in-one KRAB-dCas9-puro (pXPR_066) vector carrying safe harbor, CD81 CRISPRi (20nt or g[9nt]), CTCF-sg4 (10nt), or CTCF-sg8 (10nt) guides (Supplementary Data 2) were pelleted and stored in −80 °C. RNA was isolated with RNeasy Plus Micro kit (Qiagen) according to the manufacturer’s protocol and ensuring RIN values greater than 7. Libraries were prepared first with Poly-A enrichment using magnetic oligo(dT)-beads (Invitrogen), then ligated to RNA adaptors for sequencing. Paired end sequencing (2 × 150 bp) was carried out on an Illumina Nextseq or Novaseq (Illumina).
Statistics and reproducibility
No statistical method was used to determine sample size. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Data analysis
Pooled screening
Log fold change calculations were calculated with the starting plasmid DNA pool as reference. Initial quality control of pooled screening data included running pairwise comparisons on replicates to assess replicate consistency. Based on these tests, one replicate of the screen in K562 cells was excluded, as multiple comparisons test of these replicates identified a significant difference between replicate LFC values (Repeated measures one-way ANOVA, P < 0.0001) and a post-hoc multiple comparisons test identified significant differences between Rep A and Rep B as well as between Rep B and Rep C (Tukey’s, P < 0.0001 for both tests) while there was no significant difference between Rep A and Rep C (Tukey’s, p = 0.794). Based on these findings, Rep B was excluded from further analysis while the 2 other replicates from this screen were retained. All other cell line replicates were not significantly different.
Z-scores for pooled screen analysis were calculated using the following equation:
RNA-seq data processing
RNA-seq data for A375 sgCD81, Jurkat sg4 and sg8 were processed using the Kallisto v0.46.1 alignment and quantification tool (kallisto quant -i {transcriptome_index} -o output -b 50 ~{read1} ~{read2} -t 4 -g {gtf}). Transcriptome index and gtf for human was taken from https://github.com/pachterlab/kallisto-transcriptome-indices/releases. The data, composed of paired-end reads in fastq format and included multiple replicates per condition. Output files of interest to this analysis were the.h5 count matrices and alignment logs.
RNA-seq quantification and analysis
The.h5 count matrices were imported into R using tximport::tximport. Raw counts were aggregated to the gene level. Genes with zero counts across all samples were subsequently removed from the matrix for differential expression analysis. Differential expression followed the workflow outlined in the vignette provided within the dream32 statistical package. To visualize the impact of the guide in comparison to the Safe Harbor, we generated volcano plots (Fig. 1e and Supplementary Fig. 9a, b). We set the significance criteria to have an absolute log fold-change (abs(logFC)) >2 and an adjusted P < 0.001. The top differentially expressed genes were visualized through heatmaps using pheatmap::pheatmap, with hierarchical clustering revealing that replicates clustered together. For gene set enrichment analysis, upregulated genes identified in Jurkat sg8 cell gene expression (normalized to Jurkat safe harbor) were entered into Enrichr46,47,48 and GO Cellular Component 2023 results exported (found in Supplementary Data 5).
Comparison of CTCF guide target sites and CTCF binding events
Perfect match sites were identified using Cas-OFFinder v2.449 (http://www.rgenome.net/cas-offinder/) with hg38 2 bit as reference. Alt chromosomal matches were excluded from the analysis. “N” bases were added to guide sequences such that they met the minimum threshold of 15 nt.
Putative CTCF binding site determination
CTCF sites were selected using the JASPAR MA0139 matrix profile (https://jaspar.genereg.net/matrix/MA0139.1/) and filtered down to the 880k sites using the R library and steps previously detailed50,51. These were exported to a.bed file and a.saf file.
ChIP-seq data processing
For Fig. 3c and Supplementary Fig. 3a, b, Jurkat and A375 CTCF ChIP-seq datasets were processed using the ENCODE ChIP-seq pipeline v2.1.5 (https://github.com/ENCODE-DCC/chip-seq-pipeline2). Both replicates were processed using the default “tf” options for the pipeline with the MACS2 peak-caller. To obtain a final peak-set of CTCF binding events, we utilized the IDR-optimal output peak calls at an IDR threshold of <0.05 for each cell line and merged overlapping peaks. We then extended the CTCF peaks symmetrically by +/−50 bp, corresponding to a stringent perturbation radius, and used bedtools to obtain overlapping sites between each CTCF guide target site and CTCF binding peaks.
For all other figures, ChIP-seq data for CTCF and H3K9me3 were processed using the ENCODE ChIP-seq pipeline v2.2.0 with default parameters. The pipeline_type parameter for CTCF and H3K9me3 was set to “tf” and “histone”, respectively. The data, composed of single-end reads in fastq format, included two replicates per condition. Output files of interest to this analysis were the bam files and QC html reports.
ChIP-seq quantification and analysis
CTCF bigwig files were created using bamCoverage (bamCoverage -b $1 -o “$2.bw” -bs 50 -p 4 --effectiveGenomeSize 2913022398 --normalizeUsing bpm). We used deeptools to calculate normalized signal using counts within a +/−3Kb window centered at perfect match sites. To observe the effect of the guide in the H3K9me3 and dCas9 landscapes, we extracted the windows overlapping perfect match sites and plotted the aggregate histone signal and dCas9 signal in Safe Harbor and sg4 samples (Fig. 4d, f). We also generated CTCF profile heatmaps (Supplementary Fig. 5a) and H3K9me3 and dCas9 (Supplementary Fig 6a, b). The bigwigs were used with the karyoploteR R package to generate genome tracks (Fig. 4e). A minimum sequencing depth of 20 million reads were collected for each CTCF and H3K9me3 ChIP-seq samples; a minimum of 500,000 reads for each Cas9 ChIP-seq sample.
For visualization and differential binding analysis (Fig. 4 and Supplementary Figs. 5c, 7, and 8), we created a CTCF count matrix using featureCounts (featureCounts(files, allowMultiOverlap = T, largestOverlap = T, annot.ext = “jaspar_motifs.saf”, readExtension3 = 200, ignoreDup = T), where jaspar_motifs.saf contains the putative CTCF binding sites with window size 500 bp in a.saf format. The same was done for a H3K9me3 count matrix, except the.saf file contains genome-wide non-overlapping windows of size 5Kb. Raw counts were stored in a.tsv file.
To observe the effect of the guide in CTCF binding at perfect match sites, we first loaded the count matrix in R (rows: ~880k putative binding sites from JASPAR, columns: samples) and used edgeR::cpm to normalize the data. We extracted the bins that overlapped a perfect match site and filtered out bins if they had <5 CPM in the Safe Harbor samples. CTCF binding was averaged between sample replicates. We observed a significant difference in mean CTCF binding between Safe Harbor and sg4 samples, using stats::t.test (Fig. 4a and Supplementary Fig. 5c). A similar procedure was used for H3K9me3 (Fig. 4b and Supplementary Fig. 5b).
To observe the genome-wide effect of the guide in CTCF binding, we took the above normalized CTCF count matrix and filtered out bins if they had <5 CPM in the Safe Harbor samples. We plotted the counts in the remaining bins and colored points if they overlap a sg4 or aag[sg4] perfect match site (Supplementary Fig. 7a, b).
For the differential analysis, we utilized the dream statistical package in R, as outlined by Hoffman and Roussos, 2021. We established significance criteria with a requirement for absolute log fold change (abs(logFC)) >0.5 and an adjusted P < 10−5. (Fig. 4g, Supplementary Fig. 8d). While many significant sites overlapped perfect match sites, we also observed some sites that showed significant CTCF loss where the sequence did not perfectly match the guide target sequence. We extracted the sequence at these sites from JASPAR MA0139, and used Biostrings::consensusMatrix and ggseqlogo:: ggseqlogo to look at the logogram of the sequences (Fig. 4i, Supplementary Fig 8e). We observed that the sequences matched closely with the guide target sequence. To investigate the significance of nucleotide positions for efficient truncated guide binding, we extracted all CTCF sequences that have 0–4 mismatches to the target sg4 sequence using Cas-OFFinder49. These sites were intersected with our list of bound CTCF sites in the Jurkat Safe Harbor, and we calculated the ratio of CTCF sites with significant loss to the total CTCF sites for each level of partial sequence match (from 0 to 4 mismatches) (Fig. 4h). Additionally, we assessed the role of each nucleotide position in guide binding by examining 1-mismatch sequences that resulted in significant CTCF loss. The analysis revealed that most mismatches occurred at the 5’ end (Supplementary Fig. 10a). Conversely, sequences that did not show significant loss had mismatches primarily in the PAM-proximal bases, underscoring the critical role of these bases in proper guide binding (Supplementary Fig. 10b).
Additional transcription factor motif search
Beginning with the JASPAR vertebrates motif database48 we parsed individual Position Frequency Matrices (PFMs) in MEME format and filtered them to match the guide’s design. First, all motifs were selected that match the PAM sequence of the guides, which correspond to PFM matrices that contain a strong CCN (>0.8 probability on each nt) in the first half of the motif, or a strong NGG in the second half of the MEME PFM. In addition to PAM, a robust nucleotide profile was required. For each motif, we extracted the substring delimited by the PAM sequence and required at least 5 nucleotides with 0.5 or more probability in the PFM. We further filtered the motifs and selected those that have an Information Content Matrix (ICM) score higher than 0.9 in at least 5 nucleotides from the PAM location to the end of the motif. We used the “ggseqlogo” R library to plot the motifs52. Two scripts are provided.
R (version 4.1.2), Python (version 3.7), and Graphpad Prism (version 10) were used for visualization.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The ChIP-seq and RNA-seq data generated in this study have been deposited and are unrestricted under GEO accession GSE247143. Source data are provided with this paper.
Code availability
The code used in this study can be found at GitHub [https://github.com/broadinstitute/gro-crispri-ctcf] and Zenodo [https://doi.org/10.5281/zenodo.14606569].
References
Gerstein, M. B. et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Roadmap Epigenomics Consortium. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
ENCODE Project Consortium. et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020).
Avsec, Ž. et al. Base-resolution models of transcription-factor binding reveal soft motif syntax. Nat. Genet. 53, 354–366 (2021).
Zeitlinger, J. Seven myths of how transcription factors read the cis-regulatory code. Curr. Opin. Syst. Biol. 23, 22–31 (2020).
Amit, I. et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326, 257–263 (2009).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Gusev, A. et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 95, 535–552 (2014).
Fulco, C. P. et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773 (2016).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Canver, M. C. et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527, 192–197 (2015).
Korkmaz, G. et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 34, 192–198 (2016).
Sanjana, N. E. et al. High-resolution interrogation of functional elements in the noncoding genome. Science 353, 1545–1549 (2016).
Reilly, S. K. et al. Direct characterization of cis-regulatory elements and functional dissection of complex genetic associations using HCR-FlowFISH. Nat. Genet. 53, 1166–1176 (2021).
Gasperini, M. et al. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell 176, 1516 (2019).
Thakore, P. I. et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015).
Nasser, J. et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 593, 238–243 (2021).
Chen, Z. et al. Integrative dissection of gene regulatory elements at base resolution. Cell Genom 3, 100318 (2023).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 (2013).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Zhang, J.-P. et al. Different effects of sgRNA length on CRISPR-mediated gene knockout efficiency. Sci. Rep. 6, 28566 (2016).
Fu, Y., Sander, J. D., Reyon, D., Cascio, V. M. & Joung, J. K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284 (2014).
Cencic, R. et al. Protospacer adjacent motif (PAM)-distal sequences engage CRISPR Cas9 DNA target cleavage. PLoS One 9, e109213 (2014).
Kiani, S. et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Methods 12, 1051–1054 (2015).
Sanson, K. R. et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 9, 5416 (2018).
Ma, H. et al. Pol III Promoters to Express Small RNAs: Delineation of Transcription Initiation. Mol. Ther. Nucleic Acids 3, e161 (2014).
de Wit, E. et al. CTCF binding polarity determines chromatin looping. Mol. Cell 60, 676–684 (2015).
Kim, T. H. et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231–1245 (2007).
Vietri Rudan, M. et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 10, 1297–1309 (2015).
Hoffman, G. E. & Roussos, P. Dream: powerful differential expression analysis for repeated measures designs. Bioinformatics 37, 192–201 (2021).
Flavahan, W. A. et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016).
Liu, X. S. et al. Editing DNA methylation in the mammalian genome. Cell 167, 233–247.e17 (2016).
Hnisz, D. et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458 (2016).
Nora, E. P. et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017).
Boyle, E. A. et al. High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc. Natl. Acad. Sci. USA. 114, 5461–5466 (2017).
Morris, J. A. et al. Discovery of target genes and pathways at GWAS loci by pooled single-cell CRISPR screens. Science 380, eadh7699 (2023).
Kocak, D. D. et al. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 37, 657–666 (2019).
Josephs, E. A. et al. Structure and specificity of the RNA-guided endonuclease Cas9 during DNA interrogation, target binding and cleavage. Nucleic Acids Res. 43, 8924–8941 (2015).
Oksuz, O. et al. Transcription factors interact with RNA to regulate genes. Mol. Cell 83, 2449–2463.e13 (2023).
Hess, G. T. et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat. Methods 13, 1036–1042 (2016).
Gao, Z., Harwig, A., Berkhout, B. & Herrera-Carrillo, E. Mutation of nucleotides around the +1 position of type 3 polymerase III promoters: The effect on transcriptional activity and start site usage. Transcription 8, 275–287 (2017).
Najm, F. J. et al. Orthologous CRISPR–Cas9 enzymes for combinatorial genetic screens. Nat. Biotechnol. 36, 179–189 (2017).
Najm, F. J. et al. Chromatin complex dependencies reveal targeting opportunities in leukemia. Nat. Commun. 14, 448 (2023).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Xie, Z. et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 1, e90 (2021).
Bae, S., Park, J. & Kim, J.-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Dozmorov, M. G. et al. CTCF: an R/bioconductor data package of human and mouse CTCF binding sites. Bioinform. Adv. 2, vbac097 (2022).
Fornes, O. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020).
Wagih, O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics 33, 3645–3647 (2017).
Acknowledgements
We thank E. Gaskell, N. Durand, A. Hall, G. Griffin, A.G. Reyna, and C. White for helpful discussions, and E. Donnard and E. Roberts for reagents. The Broad Institute Flow Core and Genomics Platform provided experimental support. This project was supported by funds from the Gene Regulation Observatory at the Broad Institute. B.E.B. is the Richard and Nancy Lubin Family Endowed Chair at the Dana Farber Cancer Institute and an American Cancer Society Research Professor. Figures 1a and 3b were created with BioRender.com and publication licenses granted to F.J.N. F.J.N. is supported by funding from the Gene Regulation Observatory, BroadIgnite, and the Broad NextGen Fund.
Author information
Authors and Affiliations
Contributions
M.M.M., R.I., A.C., A.J.C., and Y.L. conducted the experiments. S.W., N.J., S.C.G., and E.M. processed the data. J.D.B., C.B.E., J.G.D., B.E.B., N.S. and F.J.N. provided guidance and direction. M.M.M. and F.J.N. conceived the study and wrote the paper with help from all co-authors. This work was conducted in line with Nature’s inclusion and ethics policies. Roles and responsibilities were agreed amongst collaborators ahead of the research and all authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.G.D. consults for Microsoft Research, Abata Therapeutics, Servier, Maze Therapeutics, BioNTech, Sangamo, and Pfizer. J.G.D. consults for and has equity in Tango Therapeutics. J.G.D. serves as a paid scientific advisor to the Laboratory for Genomics Research, funded in part by GlaxoSmithKline. J.G.D. receives funding support from the Functional Genomics Consortium: Abbvie, Bristol Myers Squibb, Janssen, Merck, and Vir Biotechnology. J.G.D.’s interests were reviewed and are managed by the Broad Institute in accordance with its conflict of interest policies. J.D.B. is on the scientific advisory board for Camp4 and seqWell and is a consultant at the Treehouse Family Foundation. B.E.B. declares outside interests in Fulcrum Therapeutics, Arsenal Biosciences, HiFiBio, Cell Signaling Technologies, Design Pharmaceuticals, and Chroma Medicine. A provisional patent has been filed on this work (M.M.M. and F.J.N.). The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Qianxin Wu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Moore, M.M., Wekhande, S., Issner, R. et al. Multi-locus CRISPRi targeting with a single truncated guide RNA. Nat Commun 16, 1357 (2025). https://doi.org/10.1038/s41467-025-56144-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56144-x