Abstract
CRISPR activation (CRISPRa) is a powerful tool for endogenous gene activation, yet the mechanisms underlying its optimal transcriptional activation remain unclear. By monitoring real-time transcriptional bursts, we find that CRISPRa modulates both burst duration and amplitude. Our quantitative imaging reveals that CRISPR-SunTag activators, with three tandem VP64-p65-Rta (VPR), form liquid-like transcriptional condensates and exhibit high activation potency. Although visible CRISPRa condensates are associated with some RNA bursts, the overall levels of phase separation do not correlate with transcriptional bursting or activation strength in individual cells. When the number of SunTag scaffolds is increased to 10 or more, solid-like condensates form, sequestering co-activators such as p300 and MED1. These condensates display low dynamicity and liquidity, resulting in ineffective gene activation. Overall, our studies characterize various phase-separated CRISPRa systems for gene activation, highlighting the foundational principles for engineering CRISPR-based programmable synthetic condensates with appropriate properties to effectively modulate gene expression.
Similar content being viewed by others
Introduction
Transcription factors (TFs) play a central role in eukaryotic gene regulation1,2,3. They consist of a DNA-binding domain (DBD) for sequence recognition and an activation domain (AD) that interacts with other transcriptional regulators to facilitate transcription4,5,6. To activate gene expression, nuclease-dead Cas9 (dCas9) variants have been fused to ADs, creating CRISPR activation (CRISPRa) systems7,8,9. Initially, dCas9-VP64 was developed as the first-generation activator, but it had limited efficiency10,11,12,13. To improve CRISPRa, second-generation activators have been developed, including VPR (dCas9-VP64-p65-Rta)14, SAM (dCas9-VP64/sgRNA-p65-HSF1)15, SunTag10xVP64 (or SunTag24xVP64)16, SunTag10x(p65-HSF1) (also known as SPH)17. CRISPR-SunTag activators utilize dCas9 or RNA bacteriophage coat proteins (MCP or PCP) fused with a tandem array of GCN4 peptides to recruit multiple ADs by fusing them with the single-chain antibody fragment (scFV) against GCN418,19. This enables ADs to be directed to specific target sites via dCas9 or single-guide RNA (sgRNA) scaffolds harboring MS2 or PP7 hairpins, thereby increasing TF concentration near the transcription start site (TTS) of target genes. TF clustering could enhance transcription potentially in many ways, including accelerating TF target search20,21,22,23,24,25.
The capacity of TFs to form condensates, known as TF condensates, has garnered significant attention26,27,28,29,30. The term ‘TF condensates’ generally refers to any nuclear compartment that concentrates biomolecules involved in transcription26. The latest development in CRISPRa technology is based on the principle of condensates formed by liquid-liquid phase separation (LLPS), which theoretically concentrates TFs to enhance gene activation31,32. However, the role of transcriptional condensates in gene regulation remains controversial. Numerous studies have pointed towards a positive functional role of condensates in transcriptional activity20,23,25,33,34,35,36,37,38,39,40,41. Alternative perspectives from certain groups propose that transcriptional condensates may have neutral or inhibitory effects on transcription42,43,44,45. Much of the controversy derives from the challenges to prove that transcriptional condensates form at precise genomic locations and directly regulate dynamic transcriptional output in living cells. Therefore, it is imperative to continuously develop methodologies and technologies to evaluate the significance of phase separation in transcriptional regulation.
Due to the ongoing debate regarding the impact of transcriptional condensates on transcriptional activation, it is crucial to explore the superiority of phase-separated CRISPRa methods compared to existing CRISPRa systems and determine strategies for controlling TF clustering to achieve optimal activation. Several studies have compared the efficiency of different CRISPRa systems46,47,48. However, the evaluation of gene activation often relies on two methods: quantifying RNA abundance in a population of cells using quantitative PCR and assessing protein expression through fluorescent labeling. Neither strategy can accurately capture dynamic transcriptional changes. Therefore, more precise evaluation and further optimization of CRISPRa tools require novel approaches. For instance, labeling nascent RNAs produced by the target gene in living cells enables direct observation of transcriptional dynamics49,50,51,52, offering better insights into molecular mechanisms underlying CRISPRa-mediated gene activation and facilitating the development of CRISPRa tools.
To address these technical challenges, we have established an experimental framework that enables real-time visualization of the transcriptional dynamics induced by CRISPRa systems. Additionally, we have engineered CRISPR-SunTag activators to control the formation of transcriptional condensates with distinct properties and monitored their dynamics simultaneously with the transcriptional output of target genes. Our findings suggest that the dynamicity and liquidity of transcriptional condensates are crucial for achieving high transcriptional activity. Our results collectively unveil crucial principles for developing gene activation tools based on TF clustering.
Results
Visualization of CRISPRa-mediated gene activation at the single-cell level
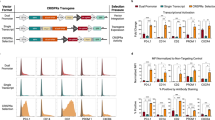
To better understand CRISPRa systems, we utilized our previously developed TriTag system49 to simultaneously image nascent RNA production and protein expression levels in live cells. We applied a miniCMV promoter to facilitate TriTagmTagBFP expression and a sgRNA targeting this promoter for CRISPRa. Using a lentiviral vector, we established a stable cell line by integrating miniCMV-TriTagmTagBFP into the HeLa cell genome. In this system, newly transcribed RNAs would be detected by stdMCP-tdTomato, while mTagBFP expression can be quantified through fluorescent imaging (Supplementary Fig. 1a). To conduct a comparative analysis of various CRISPRa systems, we used the Dox-inducible TRE3G promoter for dCas9 expression and the CMV promoter for co-factors (Supplementary Fig. 1b). The dCas9-VP64, VPR, SAM and SunTag10xPH (10xPH) systems were built and tested (Supplementary Fig. 2). We first isolated a clonal cell line of miniCMV-TriTagmTagBFP in which BFP expression was relatively weak but exhibited low heterogeneity (Supplementary Fig. 3a, b). CRISPR-mediated DNA labeling and quantitative PCR (qPCR) assays indicated that this cell line had incorporated only a single active miniCMV site, enabling accurate quantification of transcriptional activity (Supplementary Fig. 3c–g). Upon CRISPR activation, we observed the presence of a single transcription site of miniCMV in the reporter cell line. However, in the absence of activation, we rarely detected nascent transcripts in the negative control, possibly due to the low transcriptional activity of the miniCMV promoter. Our imaging analysis revealed that the 10xPH system demonstrated the highest activation potency, resulting in the highest levels of nascent RNA production (Supplementary Fig. 4a, b). We conducted quantitative microscopy imaging of BFP expression to measure the relative BFP intensity in single cells, thereby assessing the activation strength of individual CRISPRa systems. The results indicated that the activation strength of 10xPH was the highest (Supplementary Fig. 4c, d). Subsequently, we performed flow cytometry (FCM) analysis to quantify the activation ratio, which is defined as the percentage of cells with fluorescence intensity exceeding that of all cells in the negative control group. Surprisingly, FCM analysis indicated that the 10xPH system only activated miniCMV expression in 8.4% of successfully transfected cells, lower than SAM (13.0%) and VPR (12.4%) (Supplementary Fig. 4e). Collectively, our findings suggest that the 10xPH system, which recruited the largest number of activation domains (10 copies of p65-HSF1 in theory), exhibits the highest activation potency among the established CRISPRa systems. However, its activation ratio is relatively low, possibly due to associated cellular toxicity.
CRISPRa tools modulate transcriptional bursting kinetics
Numerous studies have shown that increasing the number of activation domains (ADs) recruited by each dCas9 is an effective strategy for optimizing CRISPRa7,8,9. The VPR system, which concatenates three ADs, can recruit more ADs when combined with the SunTag design than the corresponding PH system. Therefore, we utilized the SunTag system to construct SunTag3xVPR, SunTag5xVPR, and SunTag10xVPR systems and compared their performance with the VP64, SAM, VPR and SunTag10xPH systems (Fig. 1a). By tracing nascent transcripts produced at the sites of miniCMV-TriTagmTagBFP, we observed transcriptional bursts upon CRISPR activation, alternating between active (on) and inactive (off) states (Fig. 1b, c). Using this system, we examined how different CRISPRa systems regulate transcriptional bursting by analyzing burst duration, pause duration, and burst amplitude as indicators of transcriptional activity. Intriguingly, we observed significant variations in burst durations among all systems, while the duration of transcriptional silencing remained relatively consistent at around 13 min. The dCas9-VP64 system displayed the shortest burst duration (14 min), while the VPR and SAM systems exhibited burst durations of around 25 min. The 10xPH system showed significantly extended burst durations (70 min). Notably, the 3xVPR system demonstrated robust RNA production from miniCMV, outperforming the 10xPH system, with an average burst duration of approximately 95 min. However, the 5xVPR and 10xVPR systems had shorter average burst durations compared to the 3xVPR system, at 37 and 50 min, respectively (Fig. 1d). Furthermore, the burst amplitude of the 3xVPR system is comparable to that of the 10xPH system, yet significantly higher than other systems (Fig. 1e). These findings collectively illustrate the capacity of CRISPRa tools to enhance transcriptional activation by prolonging burst durations and increasing burst amplitude. Additionally, our results uncover an unexpected phenomenon indicating a potential decline in the efficiency of CRISPR-SunTag activators as the number of SunTag-AD arrays increases.
a Schematics illustrating the design of different CRISPRa systems. b Time-lapse images depicting transcriptional bursts of the miniCMV-TriTagmTagBFP reporter induced by various CRISPRa systems. Arrows highlight nascent RNAs labeled with stdMCP-tdTomato at transcription sites. Each cell was imaged for 2 h with 2-min intervals. Only images taken between 30 to 84 min are displayed. Scale bars, 5 μm. c Representative traces (red) demonstrating the kinetics of transcriptional bursting at miniCMV loci induced by various CRISPRa systems. Gray traces indicate the intensity of the background signal. d Bar graph presenting the average durations of bursts and pauses induced by CRISPRa. From left to right, the burst numbers are 117, 150, 217, 103, 140, 119, and 106, while the pause numbers are 123, 93, 237, 49, 134, 99 and 95, respectively. Data are shown as mean ± SEM. e Burst amplitude determined by measuring the maximum total intensity of stdMCP-tdTomato spots within each burst. From left to right, cell numbers are 54, 89, 128, 135, 115, 111 and 115. Data are shown as mean ± SEM. P values were calculated by one-way ANOVA test, comparing to the VP64 group. f Fluorescent imaging-based quantification of BFP expression levels driven by miniCMV following CRISPR activation for 48 h. n = 100 cells. Data are shown as mean ± SEM. P values were calculated by one-way ANOVA test, with the negative control (NC) as the reference group. g Flow cytometry analysis of BFP expression levels to determine the activation ratio of individual CRISPRa systems. Transfection-positive cells were gated based on an internal fluorescent reporter. Data are shown as mean ± SEM. n = 3 independent experiments. P values were calculated by one-way ANOVA test, comparing to the NC group. In (f) and (g), negative controls (NC) were conducted by transfecting dCas9 lacking AD fusions and sgRNA targeting miniCMV. Source data are provided as a Source Data file.
SunTag3xVPR outperforms SunTag10xVPR for gene activation
To further validate the efficacy of CRISPRa tools, we assessed the expression levels of miniCMV-driven BFP at the single-cell level. We quantified the fluorescence intensity of protein expression in each cell using flow cytometry to measure the activation ratios of the CRISPRa system. At 48 h post-transfection, the 3xVPR system displayed significantly higher activation than VP64, 5xVPR, 10xVPR, and 10xPH systems (Fig. 1f). FCM analysis revealed a 48.6% activation ratio for the 3xVPR system, surpassing other systems. VP64, SAM, VPR, 5xVPR, 10xVPR, and 10xPH exhibited activation ratios of 13.2%, 35.8%, 18.8%, 23.6%, 5.1% and 34.3%, respectively (Fig. 1g). We then expanded our analysis to include endogenous genes in different cell types. BFP reporters for endogenous HSPB1, HSPB8, HPDL, LMNA, and BAG3 were established in HeLa or HEK 293T cells. These reporters were generated by integrating BFP tags at the N- or C-terminus of the target genes using Cas9-mediated homology-directed repair, enabling the quantification of gene activation (Supplementary Fig. 5). Through single-cell imaging and FCM analysis, the 3xVPR system consistently demonstrated stronger activation and larger activation ratios than VPR, 5xVPR, 10xVPR and 10xPH for all targets. Protein imaging analysis revealed that 3xVPR displayed higher activation strength than 3xPH for certain targets while maintaining similar activation ratios as determined by FCM analysis. When comparing 3xPH with 10xPH, the activation strength was similar, but the activation ratio of 3xPH was significantly higher (Supplementary Fig. 6). We also validated these findings in HEK 293T cells for endogenous LMNA and HSPB1 by quantifying protein expression levels (Supplementary Fig. 7a, b). Furthermore, qPCR analysis of protein-coding genes (ASCL1, TTN, MIAT, RHOXF2, Neurog2, MYOD1, and HBG1) and non-coding RNAs (HOTTIP and LINC00925) consistently showed higher activation efficacy for 3xVPR compared to VPR, 5xVPR, and 10xVPR. Of note, we observed that 3xPH did not perform effectively in 293T cells, whereas 3xVPR demonstrated superior performance compared to 10xPH for most genes (Supplementary Fig. 7c). These analyses further validate that increasing the number of ADs in the CRISPR-SunTag system may not yield superior outcomes.
Activation features of CRISPR-SunTag activators
To determine the optimal assembly of activation domains for robust gene activation, we engineered different CRISPRa-SunTag systems with three or ten arrays of ADs. Each array comprised one, two, or three distinct ADs (Fig. 2a and Supplementary Fig. 8a). The common components in each comparison group (dCas9-3xADs vs. dCas9-10xADs) are scFV-AD and sgRNA. To ensure that the differences in gene activation are not due to varying expression levels of dCas9 fused with three or ten GCN4 peptide arrays, we performed Western blot and flow cytometry analyses. Using GFP as the indicator, both methods indicated consistent expression levels of dCas9 fused with either 3 or 10 GCN4 peptide arrays (Supplementary Fig. 8b). We then evaluated the activation capacity using exogenous (miniCMV) and endogenous (HSPB1, HPDL and LMNA) reporter systems by fluorescent imaging and FCM analysis. When fusing scFv with a single AD, including VP64, p65, Rta and HSF1, the highest activation efficiency was observed with 3xVP64 and 10xp65. Of note, VP64 consists of four VP16 copies (Supplementary Fig. 8c–f). We further compared 3x/10x/24xVP64 and 3x/10x/24xp65 systems and found that increasing the number of AD copies to 24 significantly reduced the activation strength. The activation ratio of 3xVP64 was higher than that of 10xVP64, but the activation strength was slightly weaker than that of 10xVP64 (Supplementary Fig. 9). Notably, 10xVP64 is the first developed CRISPR-SunTag systems16. When scFv was fused with two or three ADs, SunTag activators containing three copies of GCN4 repeats exhibited much higher activation efficiency than those with ten copies (Fig. 2b–e). This consistent conclusion was observed for both single sgRNA and three sgRNAs used for CRISPRa. These results suggest that, in the SunTag system, harboring more copies of ADs does not necessarily lead to higher activation. Thus, it is crucial to determine the optimal number of AD copies to achieve the highest activation ratio and maximal activation level.
a Schematic presenting various designs of CRISPR-SunTag activators. b Flow cytometry assessing the expression levels of the exogenous reporter (miniCMV-BFP) and showing the activation ratios of individual CRISPRa systems. c Fluorescent imaging analysis of miniCMV-BFP expression levels in single cells. n = 80 cells for each group, except for 10xVP (n = 29) and 3xVP (n = 81). Data are shown as mean ± SEM. d Schematic diagram using HSPB1 as an example to show the target locations of three sgRNAs upstream of the transcription start site (TTS). e, f Flow cytometry analysis evaluating the CRISPR activation ratios using either a mixture of three sgRNAs as shown in (e) or a single sgRNA (f). All data in (b, e), and (f) were analyzed from cells expressing transfection markers. Three biological replicates are displayed as mean ± SEM. Negative controls (NC) were performed by transfecting dCas9 lacking AD fusions and on-target sgRNAs. P values were calculated by one-way ANOVA test. Source data are provided as a Source Data file.
Transcriptional condensates, formed partly through ADs with intrinsically disordered regions (IDRs), are proposed as regulators of transcription28,53,54. Recent studies have harnessed the N-terminal IDR sequence of human oncogene Fused in Sarcoma (FUSn), which undergoes LLPS in the nucleus and in vitro55,56,57,58, to develop phase-separation assisted CRISPRa tools31,32. We therefore constructed dCas9-VPR-FUSn (VPRF), dCas9-SunTag3x,6x,10xVPRF (referred to as NxVPRF) and PCP-SunTag3x,6x,10xVPRF (Fig. 3a). In the PCP-SunTagNxVPRF systems, scFV-VPRF was recruited to sgRNA-2xPP7 via PCP-SunTag. Each sgRNA-2xPP7 can bind four copies of PCP, thus PCP-SunTag3x, 6x or 10XVPRFare equivalent to recruiting 12x, 24x and 40xVPRF, respectively. These three systems are denoted as PCP-12x, 24x, and 40xVPRF for simplicity. We compared the efficiency of SunTag3xVPR to these FUSn-assisted CRISPRa systems using the miniCMV exogenous reporter system and three endogenous gene reporter systems in HeLa cells. First, we confirmed that none of the tested CRISPRa systems activated these reporters when targeting Gal4 (Supplementary Fig. 10a). Therefore, we included only 3xVPR and sgGal4 as the negative control for each reporter. Our imaging quantifications indicate that VPRF, 3xVPR, and 3xVPRF are comparable and exhibit superior efficiency to other systems, including VPR (Fig. 3b, c). Intriguingly, activation strength significantly decreased when an excessive number of VPRF were recruited via dCas9 or PCP. We also conducted flow cytometry analysis to validate the protein expression quantified by fluorescence microscopy. The two methods were largely consistent in demonstrating the activation efficiency of the various CRISPRa systems (Fig. 3d). We tried increasing the transient transfection amounts of CRISPRa plasmids and optimizing the ratio of dCas9-3/10xGCN4 to scFV-ADs. However, under all conditions, the activation efficiency of 10xVPR and 10xVPRF consistently remained between 0% and 0.2% (Supplementary Fig. 10b). These findings reveal the importance of precise control over phase separation for achieving robust transcriptional activation. An alternative explanation could be that excessive assembly of AD impedes the entry of CRISPR activators into the cell nucleus, thereby limiting their effectiveness.
a Schematic of FUSn-assisted CRISPRa systems. N-terminal IDR of FUS (FUSn) was fused to AD arrays. b Representative images illustrating the activation of the endogenous reporter BFP-LMNA by CRISPRa systems. BFP-LMNA expression is indicated by cyan. mCherry indicates the successful transfection of CRISPRa plasmids. Scale bars, 5 μm. c Quantification of gene activation efficiency by measuring the protein expression level in single cells based on fluorescent imaging. Target genes include three endogenous reporters (LMNA, HSPB1 and HPDL) and one exogenous reporter (miniCMV-BFP). All results are shown as mean ± SEM. Each dot represents a single cell. n = 60 in LMNA and HPDL groups except NC, VPR, VPRF, 3xVPR and 3xVPRF from the HPDL group (n = 100), n = 100 in HSPB1 and miniCMV groups. d Flow cytometry analysis of BFP expression levels to determine the activation efficiency of individual CRISPRa systems. Transfection-positive cells were gated based on an internal fluorescent reporter. Data are shown as mean ± SEM. Each dot represents the average BFP intensity of all transfection-positive cells from a single experiment. n = 3 independent experiments. P values in (c) and (d) were calculated by one-way ANOVA test, comparing to the negative control (NC) group (3xVPR + sgGal4). Source data are provided as a Source Data file.
CRISPR-SunTag activators form phase-separated condensates with distinct properties
In order to investigate the significance of the number of activation domains (ADs) in CRISPRa design for efficient transcriptional activation, we established a GFP fluorescence reporter system. This system allowed us to visualize the subcellular localization of eight dCas9 activators, including VPR, VPRF, 3xVPR, 10xVPR, 3xVPRF, 10xVPRF, PCP-12xVPRF, and PCP-40xVPRF. By directly fusing GFP to dCas9 in all systems, we could observe the molecular complex associated with dCas9 (referred to as dCas9-GFP-ADs) (Fig. 4a and Supplementary Fig. 11). To ensure consistent dCas9 expression levels across different CRISPRa systems, we used GFP as an indicator due to its direct fusion with dCas9. Flow cytometry analysis revealed comparable dCas9 expression levels among these systems (Supplementary Fig. 12a). At 48 h post-transfection, cells were imaged to monitor dCas9 behaviors. Most GFP signals entered the cell nucleus, except for a fraction in the PCP-12xVPRF and PCP-40xVPRF systems that remained in the cytoplasm, likely due to the large size of the complexes hindering nuclear entry. The VPR system showed limited, irregular dCas9-GFP-ADs clusters. Although FUSn is expected to promote phase separation, the distribution of VPRF was similar to that of VPR. In contrast, the other CRISPRa systems displayed brighter and round condensates, indicating a higher degree of dCas9 clustering. The number of bright condensates significantly increased in the 10xVPR system compared to 3xVPR. Furthermore, the addition of FUSn further enhanced condensate formation in both 3xVPR and 10xVPR (Fig. 4b). Thus, in this study, we use the term ‘condensate’ to describe microscopically detectable clusters that concentrate CRISPRa components.
a Schematic illustrating the fusion of GFP to dCas9 for imaging its subcellular localization in different CRISPRa systems. b Representative images depicting dCas9-GFP puncta formed in each CRISPRa system before or after 1.5% 1,6-hexanediol (1,6-HD) for 2 min. Scale bars, 5 μm. GFP punta number and total signal intensity of puncta per cell were quantified for individual conditions. VPR: n = 41, VPRF: n = 48, 3xVPR: n = 50, 10xVPR: n = 54, 3xVPRF: n = 48, 10xVPRF: n = 42, PCP-12xVPRF: n = 52, PCP-40xVPRF: n = 50. All plots represent mean ± SEM; P values were calculated by unpaired two-tailed t-test. c Representative images of the FRAP experiment. Bleaching was performed at the indicated time points. A representative puncta was indicated with a white box. Images were recorded with 10 s intervals. Scale bars, 5 μm. d FRAP recovery curves showing the GFP intensity overtime in the bleach region in (c). The mobile fraction (Fm) and recovery half time (1/2 t) were calculated for the recovery curves. Data are shown as mean ± SEM. The number of cells quantified in (d), from left to right, is 10, 20, 12, 12, 10, 15, 12 and 13, respectively. Source data are provided as a Source Data file.
We then conducted short-term (1-h) and long-term (6-h) real-time imaging to monitor the dynamics of dCas9-GFP-ADs. For short-term imaging, which started 48 h post-transfection, we observed no fusion dynamics between bright spots in both VPR and VPRF systems. However, droplet-like behaviors and fusion were observed in condensates formed in CRISPR-SunTag systems (Supplementary Fig. 12b). For long-term imaging, we initiated time-lapse imaging 12 h after transient transfection of plasmids when the expression level of dCas9 was still very low. In all CRISPR-SunTag systems, we observed the formation of small condensates, which were more abundant in the 10xVPRF and PCP-12x/40xVPRF systems. Throughout the imaging, we frequently observed the fusion of smaller condensates into larger ones in these three systems (Supplementary Fig. 13a, b).
To gain further insights into the properties of condensates formed in these CRISPRa systems at 48 h post-transfection, we employed 1,6-hexanediol (1,6-HD) treatment. 1,6-HD is known to disrupt hydrophobic protein-protein/protein-RNA interactions required for droplet formation30,39,40,59,60,61. After 1,6-HD treatment, minimal changes were observed in VPR and VPRF systems, while 3xVPR and 3xVPRF showed significant decreases in GFP puncta number and fluorescence intensity. Some disruption was observed in 10xVPR, but no significant changes were observed in 10xVPRF, PCP-12xVPRF, and 40xVPRF, indicating stronger inter-molecular interactions within these condensates (Fig. 4b). We then conducted fluorescence recovery after photobleaching (FRAP) experiments to assess whether CRISPRa-SunTag condensates can continually exchange molecules with surrounding partners. The results indicated restricted mobility of dCas9-GFP-ADs clusters in VPR and VPRF. Conversely, the condensates of 3xVPR and 3xVPRF displayed high mobility and were susceptible to disruption by 1,6-HD, indicating their liquid-like behavior. FRAP analysis demonstrated impaired mobility in condensates formed by 10xVPR, 10xVPRF, PCP-12xVPRF, and PCP-40xVPRF (Fig. 4c, d). We then used 3xVPRF and 10xVPRF as representative systems to quantify reporter protein expression and conduct FRAP analysis on the condensates for measuring the half-time (1/2t) and mobile fraction (Fm) in the same cell (Supplementary Fig. 13c, d). Plotting reporter expression against 1/2t or Fm revealed distinct clusters for 3xVPRF and 10xVPRF. 3xVPRF exhibited higher condensate mobility and gene activation, whereas 10xVPRF condensates showed much lower mobility and activation activity (Supplementary Fig. 13e). This suggests that condensate properties are related to gene activation.
By examining the hallmark behaviors62 of phase separation (Supplementary Table 1), we conclude that 3xVPR and 3xVPRF form liquid-like transcriptional condensates through phase separation. On the other hand, the higher presence of scaffold sequences in the 10xVPR, 10xVPRF, PCP-12xVPRF, and PCP-40xVPRF systems facilitates the formation of more extensive molecular networks, resulting in solid-like transcriptional condensates with reduced dynamicity and liquidity. We validated the localization of CRISPR activators and their activation capacities in HCT116 cells, and the results were consistent with those obtained in HeLa cells (Supplementary Fig. 14). To assess the specificity of CRISPR-SunTag activators forming TF condensates, we performed RNA-seq analysis on samples where individual CRISPRa systems (VPR, VPRF, 3xVPR and 3xVPRF) targeted HSPB8. The correlation in gene expression between each activator and the control sample was highly similar (R > 0.98), indicating that none of the activators broadly influenced gene expression. Notably, HSPB8 was consistently the most upregulated gene across all samples, demonstrating the activators’ high specificity (Supplementary Fig. 15a). Additionally, single-cell quantitative analysis via fluorescence imaging showed that only the target-specific sgRNA could activate the target gene (Supplementary Fig. 15b). Thus, despite forming numerous TF condensates, 3xVPR and 3xVPRF systems maintain high specificity in gene activation.
Overall levels of visible CRISPRa condensates do not correlate with gene activation potency
Our previous experiments have established that CRISPR-SunTag activators form phase-separated transcriptional condensates with either liquid- or solid-like states. To assess the relationship between visible condensate formation and transcriptional activation, we quantified the cumulative signals of all dCas9-GFP-ADs condensates within a cell. Concurrently, we measured the protein expression level of target genes within the same cell using exogenous and endogenous reporters (miniCMV-BFP and HSPB1-BFP) (Supplementary Fig. 16a). Subsequently, we calculated the correlation between the total intensity of condensates and transcriptional activation in individual cells. We found that the correlation coefficients were consistently below 0.1 for all individual CRISPRa systems, with most values closely approaching 0 (Supplementary Fig. 16b, c). These results suggest that high levels of phase separation are not required for efficient transcriptional activation. Notably, VPRF, 3xVPR and 3xVPRF consistently showed high gene activation potency in this set of experiments. We propose that achieving optimal levels of phase separation, indicative of the appropriate extent of multivalent interactions within the CRISPRa platform, establishes a microenvironment surrounding the transcription start site that facilitates efficient transcriptional activation. Taken together, our findings suggest that the liquidity and dynamicity of synthetic transcriptional condensates, rather than the overall level of phase separation, are critical factors in facilitating transcriptional activation.
Dynamic imaging of CRISPRa condensates and transcriptional bursts at target sites
Since 3xVPR and 3xVPRF consistently show high activation efficiency in all the tested genes and cell lines, we sought to investigate whether visible condensates formed at the target sites in these systems. We employed live-cell imaging to monitor the formation, localization, and dynamics of CRISPRa condensates while simultaneously quantifying the transcriptional levels at the single-cell level with the miniCMV-TriTagmTagBFP reporter system. Live-cell imaging started 12 h after transient transfection of CRISPRa plasmids and continued for 6 h. Over time, dCas9-GFP-ADs concentration increased within the cell nucleus. The miniCMV locus displayed transcriptional bursting, which only occurred upon CRISPR activation. This observation further highlights the remarkable activation potency of 3xVPR and 3xVPRF systems. Initially, no visible GFP puncta were detected in the 3xVPR system, but they appeared later. In contrast, 3xVPRF immediately formed numerous small condensates, which eventually merged into larger ones (Supplementary Fig. 17a). We then examined whether CRISPRa condensates localized at the transcription sites. Exceptionally bright condensates did not co-localize with nascent RNA. Instead, small condensates were found near or co-localized with nascent RNA (Supplementary Fig. 17b, c). Given the increase in condensate number and size over time, we analyzed their influence on transcriptional activation by quantifying burst duration and amplitude during the initial and final 3 h of imaging. In the 3xVPR system, both burst duration and amplitude slightly decreased, while in 3xVPRF cells, transcriptional parameters remained minimal changes (Supplementary Fig. 17d, e). Moreover, we examined the presence of closely adjacent or colocalized condensates with nascent RNAs during active miniCMV transcription. Our results showed that the total amount of nascent RNA was significantly higher in the presence of condensates compared to their absence (Supplementary Fig. 17f, g).
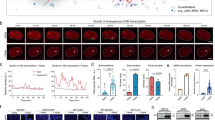
To validate the results observed at the miniCMV-TriTag site, we traced activation at the endogenous LMNA locus using a clonal cell line harboring one endogenous LMNA locus labeled with TriTag49. This locus shows low basal transcription, with nascent RNA being rarely detectable. 3xVPR and 3xVPRF systems significantly activated transcription of this locus, displaying long burst durations and high amplitudes (Fig. 5a–c). Similar to the miniCMV locus, longer transfection time led to more and larger CRISPRa condensates without further enhancing transcription activity (Fig. 5d, e). Real-time imaging revealed that CRISPRa condensates were often enriched at transcription sites. Our quantifications indicated that the nascent RNA levels were significantly higher when condensates were present compared to when they were absent, suggesting that condensates may promote transcription (Fig. 5f, g). We then analyzed over 84 RNA bursts in around 40 cells for both 3xVPR and 3xVPRF, tracing condensate dynamics at LMNA transcription sites before, during, and after each burst. Whether examining individual bursts or averaging all bursts, we found no significant relationship between condensate dynamics and RNA burst occurrence (Fig. 5h). Intriguingly, VRPF did not form visible condensates, but showed similar transcriptional bursting to both 3xVPR and 3xVPRF at both miniCMV and LMNA loci (Supplementary Fig. 18). Notably, small, undetectable condensates may influence our analysis, potentially complicating data interpretation. Thus, advanced imaging and quantitative tools are needed to fully understand the role of TF condensates in gene activation.
a Time-lapse images showing nascent RNA (stdMCP-tdTomato) at TriTag-LMNA loci and dCas9-HaloTag-ADs condensates (3xVPR or 3xVPRF) over 6 h with 2-min intervals. White arrows point to nascent RNAs at active transcription sites, while yellow arrows highlight the association of nascent RNA and CRISPRa condensates. Scale bars, 5 μm. b, c Real-time traces of nascent RNA (red) and dCas9-HaloTag-ADs cluster (green) at transcription sites. The light green and light red lines represent the background signal changes of RNA and HaloTag, respectively. Enlarged snapshots highlight the association of RNA bursts and CRISPRa condensates. Scale bars, 1 μm. d, e Bar graphs displaying the total duration and average amplitude of RNA bursts occurred in the first and second halves of the 360-min imaging period. The sample sizes in (d), from left to right, are 59, 59, 106 and 135, while for (e), they are 55, 55, 145 and 156. Data of burst durations are mean ± SEM. Dotted lines in burst amplitude plots represent the median values with 25th and 75th percentiles. f, g Bar graphs of total RNA intensity at LMNA transcription sites with (+) and without (−) CRISPRa condensates in 3xVPR (f) and 3xVPRF (g) groups. Data are mean ± SEM. Each dot represents an RNA spot labeled by stdMCP-tdTomato that appears during a transcriptional burst. 3xVPR, n = 1342 (+ condensate) and 1735 (− condensate); 3xVPRF, n = 1491 (+ condensate) and 2256 (− condensate). P values in (d–g) were calculated by unpaired two-tailed t tests. h Time-course plots of RNA bursts and condensate dynamics during LMNA activation by 3xVPR (left) or 3xVPRF (right). The graphs are aligned as three types: burst initiation, burst peak and burst decline. Condensate dynamics analyzed 30 min before and after time zero. Magenta lines show burst period, light magenta lines show RNA background, green lines show condensate intensity, light green lines show GFP background. Data are mean ± SEM at each time points. Number of bursts analyzed is highlighted in the figure. 3xVPR, n = 39 cells; 3xVPRF, n = 44 cells. P values from one-way ANOVA show no significant differences in condensate brightness between any two time points. Source data are provided as a Source Data file.
Dynamic properties of CRISPRa condensates impact gene activation
The higher activation efficiency of 3xVPR and 3xVPRF than VPR might be due to increased VPR copy number rather than phase-separated condensates. To investigate this, we fixed the VPR copy number while varying FUSn copies by constructing dCas9-VPR-FUSn, dCas9-VPR-2xFUSn, dCas9-VPR-3xFUSn, dCas9-VPR-SunTag-3xFUSn, and dCas9-VPR-SunTag-10xFUSn. Nucleoporin 98 (NUP98) and survival of motor neuron 1 (SMN1) have been reported as phase separation-associated proteins, which we confirmed using a web server designed to predict phase-separating proteins63. Previous studies have shown that incorporating NUP98 IDR (NUP98n) or SMN1 to dCas9-VPR enhanced its gene activation capability32. Therefore, we also replaced FUSn with NUP98n and SMN1, creating dCas9-VPR-SunTag-3xNUP98n, dCas9-VPR-SunTag-10xNUP98n, dCas9-VPR-SunTag-3xSMN1, and dCas9-VPR-SunTag-10xSMN1 (Supplementary Fig. 19a). Testing the activation of miniCMV-BFP, HSPB1-BFP, and BFP-LMNA revealed that increasing FUSn copies with a single VPR copy boosts gene activation efficiency. However, replacing FUSn with NUP98n or SMN1 significantly decreased activation efficiency, falling below that of VPR alone (Supplementary Fig. 19b). Labeling these CRISPRa systems with GFP showed that fusing 3x or 10x NUP98n to VPR significantly promoted the formation of dCas9-VPR condensates. These condensates were not significantly disrupted by 1,6-HD, indicating a lack of liquid-like properties. In contrast, VPR-2x/3xFUSn formed fewer visible condensates (~5 per cell), significantly disrupted by 1,6-HD. VPR-10xFUSn formed more varied-sized condensates (~11 per cell), with only smaller condensates effectively disrupted by 1,6-HD. Notably, its gene activation potency was lower than VPR-2x/3xFUSn (Supplementary Fig. 19c, d). These findings suggest that the liquidity of CRISPRa condensates affects their gene activation capability. Optimal multivalency between transcription factors via IDR or other scaffolds in the CRISPRa platform likely plays a crucial role in efficient transcriptional activation.
Next, we aimed to understand why decreased liquidity of CRISPRa condensates significantly reduces gene activation capability. We hypothesized that these less dynamic condensates reduce the availability of CRISPRa molecules at target sites and sequester other transcriptional co-activators, thereby limiting their involvement in transcriptional activation. To test this, we examined the components of co-activator complexes, including Mediator and p300, which are recruited through transcription factors to regulate the target gene expression64,65,66,67. We performed antibody staining for endogenous Mediator complex subunit 1 (MED1) and directly imaged HaloTag-labeled endogenous p300 in living cells generated through CRISPR knockin. We found that both co-activators were enriched in condensates formed by 3xVPR, 10xVPR, 3xVPRF, and 10xVPRF, with co-localization ratios ranging from 81.8% to 99.5% (Fig. 6a–d). Further quantitative analysis indicated that the total fluorescence intensity of MED1 and p300 within the condensates was significantly greater in the 10xVPR system than in the 3xVPR system. Correspondingly, the average nucleoplasmic signal of both co-activators outside the condensates was reduced in the 10xVPR system compared to the 3xVPR system. This trend was also observed in the 10xVPRF and 3xVPRF systems (Fig. 6e, f). These observations suggest that fewer MED1 and p300 molecules freely diffuse outside the condensates in the 10xVPR and 10xVPRF systems. Through 1,6-HD treatment and FRAP analysis, we found that the liquidity of p300 was consistent with that of CRISPRa condensates. Specifically, p300 exhibited high liquidity in 3xVPR and 3xVPRF, whereas its liquidity was significantly reduced in 10xVPR and 10xVPRF (Fig. 6g, h). These results revealed that low liquidity in CRISPRa condensates can trap co-activators, resulting in the loss of gene activation capability. Our findings suggest that the dynamicity and liquidity of transcriptional condensates are crucial for effective transcriptional activity.
a–d Representative images and line scan of fluorescent intensity showing the colocalization of MED1 (a, b) or p300 (c, d) and CRISPRa condensates (dCas9-GFP-ADs). Colocalization ratios (CR) on the corresponding plots indicate the percentage of CRISPRa condensates enriched with MED1 or p300 signals (3xVPR: n = 314, 10xVPR: n = 436, 3xVPRF: n = 438, 10xVPRF: n = 1109). Scale bars, 5 μm. e Quantifications of MED1 or p300 signals within CRISPRa condensates, as shown in (a) and (c). Each dot represents the total fluorescence intensity of all MED1 (or p300) condensates formed within the same cell (n = 50). Data are shown as mean ± SEM. f Quantifications of MED1 or p300 signals outside CRISPRa condensates, as shown in (a) and (c). Each dot represents the mean fluorescence intensity of the MED1 (or p300) signal from the nucleoplasm, excluding CRISPRa condensates, within each cell (n = 50). P values in (e) and (f) were calculated using unpaired two-tailed t test. g Representative images depicting p300 (magenta) and dCas9-GFP-ADs (green) puncta formed in each CRISPRa system before and after 1.5% 1,6-hexanediol treatment for 2 min. Scale bars, 10 μm. Correlation analysis of the change in total intensity ratios of p300 and dCas9-GFP-ADs in individual cells after 1,6-hexanediol (1,6-HD) treatment. Black dashed lines divide cells into two groups: less sensitive to 1,6-HD on the left and more sensitive on the right. R2 represents the coefficient of determination. Each dot represents a single cell (n = 40). h Representative images of the FRAP experiment. Bleaching was performed at the indicated time points. A representative punctum was highlighted with a white box. Images were recorded with 10-second intervals. Scale bars, 5 μm. i FRAP recovery curves showing the p300 intensity overtime in the bleach region in (h). Data are shown as mean ± SEM. The number of cells quantified in (i), from left to right, is 14, 12, 18 and 16, respectively. The mobile fraction (Fm) and recovery half time (1/2 t) were calculated for the recovery curves. Source data are provided as a Source Data file.
Design and comparative analysis of FUSn-assisted CRISPRa systems
Given that VPRF and 3xVPRF are two efficient FUSn-assisted CRISPRa systems, we aimed to investigate the possibility of developing even more effective systems based on similar design principles. Both systems involved the fusion of FUS IDR (FUSn), enabling its interaction with transcription factors and co-activators, thereby enhancing transcriptional activation. Additionally, FUSn can undergo phase separation, leading to the formation of membraneless compartments55,58,68. However, the VPRF system displayed limited phase separation, whereas the 3xVPRF system formed substantial condensates. The single-component VPRF system demonstrated advantages in practical applications. Therefore, we attempted to replace VPR in the VPRF system with various individual activation domains (ADs) such as VP64, p65, Rta, and HSF1, as well as combinations of dual ADs including p65-HSF1, VP64-HSF1, p65-Rta, VP64-p65, and VP64-Rta. To visualize the subcellular localization of dCas9, we fused it with GFP (Supplementary Fig. 20a). Through single-cell imaging and analysis of exogenous (miniCMV) and endogenous (HSPB1 and LMNA) gene activation, we consistently observed that VPRF demonstrated the highest activation capability across these three targets. Of note, only dCas9-GFP-FUSn-p65 displayed condensate formation 48 h post-transfection (Supplementary Fig. 20b).
Next, we constructed and evaluated additional CRISPRa systems resembling 3xVPRF by substituting VPR with various combinations of ADs. These designs included 3xPF, 3xVF, 3xPHF, 3xPRF, 3xVPF and 3xVRF (Supplementary Fig. 21a). Quantitative imaging analysis of endogenous genes (HSPB1 and LMNA) and exogenous miniCMV activation consistently showed that 3xPRF and 3xVRF achieved activation levels comparable to 3xVPRF. Furthermore, we increased the numbers of AD and FUSn in the CRISPRa system by modifying the SunTag design, resulting in constructs such as 10xPRF, PCP-12xPRF and PCP-40xPRF. Interestingly, our results revealed a dramatic decrease in activation efficiency when increasing the number of tandem AD-FUSn repeats in the CRISPR-SunTag system, which is consistent with our previous findings. Throughout this series of experiments, we included 3xVPR as a control and consistently observed its performance to be comparable to that of 3xVPRF (Supplementary Fig. 21b). Given the consistent activation efficiency observed across genes of the VPRF, 3xVPR and 3xVPRF components, we conducted further evaluations to assess their effectiveness in activating specific genes within stem cells.
SunTag3xVPR triggers robust cell differentiation by activating a single transcription factor
To examine CRISPRa efficiency in stem cells, we first utilized a GFP knockin reporter in mouse embryonic stem cells (mESCs). In this reporter system, the open reading frame of the endogenous Sox1 gene was replaced with the coding sequence of GFP69. Using two sgRNAs that have been used for activating Sox1 expression70, we quantified GFP expression to assess CRISPRa efficiencies (Fig. 7a). The quantitative results indicated that the activation efficiency of 3xVPR was superior to other CRISPRa systems, including VPR, VPRF, 10xVPR, and 10xPH (Fig. 7b). The capability to selectively enhance the expression of genes offers a powerful approach for reshaping cellular identity. It has been shown that overexpressing a single transcription factor, neurogenic differentiation factor 1 (NEUROD1), is sufficient to promote human pluripotent stem cells differentiated into induced neurons (iNeurons) either by ectopically expression or CRISPRa-mediated gene activation14,71,72. Therefore, we attempted to test whether 3xVPR could induce sufficient expression of NEUROD1 to trigger differentiation using the same set of sgRNAs as reported before14. H1 hESC cells were transfected with CRISPRa plasmids and were monitored for phenotypic changes. Two days following the electroporation of CRISPRa plasmids, we performed immunostaining of cells using the neuronal marker neurofilament 200 and quantified the proportion of cells exhibiting a neuronal-like phenotype. Our results showed that the VPRF, 3xVPR and 3xVPRF systems demonstrated similar levels of differentiation efficiency, which were significantly more potent than VPR, 10xPH, 10xVPR and 10xp65. Notably, when using a single sgRNA, only 3xVPR and 3xVPRF among these CRISPRa systems were able to induce high levels of stem cell differentiation (Fig. 7c–e). These results suggest that in gene activation assays, particularly in genome-wide CRISPRa screenings, 3xVPR and 3xVPRF may be the most effective options when using a single sgRNA. Taken together, this study consistently demonstrated that 3xVPR outperformed well-known CRISPRa systems by evaluating multiple target genes and various cell lines. However, the effectiveness of these activators varied depending on the target genes analyzed. The observations made in this study, along with previous reports31,46,73,74,75, strongly indicate that the performance of CRISPRa tools can be influenced by factors such as the targeting site of the sgRNA and the cell type being utilized.
a Schematic illustrating an endogenous Sox-1 reporter in mouse embryonic stem cells for testing the efficiency of CRISPR activation. GFP expression is only detectable upon efficient CRISPR activation. b Quantification of Sox-1 promoter driven GFP expression levels based on fluorescent imaging in single cells. Negative controls (NC) were performed by transfecting VPR without the on-target sgRNA. Each dot represents a single cell (n = 60). All values are mean ± SEM. P values were determined by one-way ANOVA, comparing to the negative control group. c Representative immunofluorescence images of Hoest33342 (blue, nuclear marker) and anti-neurofilament 200 staining (magenta, neuronal marker) in human embryonic stem cells at various conditions. Cells exhibiting neuronal phenotype are highlighted in the white box. Scale bars, 200 μm (original view) and 50 μm (enlarged view). d Quantification of neuronal cells generated by various CRISPRa systems following the use of single or mixed sgRNAs. Data are shown as the mean ± SEM (n = 3 biological replicates). Each dot represents a biological replicate. Dots of the same color indicate that they are from the same batch of experiments. P values were determined by one-way ANOVA, comparing to the negative control (NC, dCas9 + sgNEUROD1) group. e A diagram summarizing the findings of this study. Increasing the number of SunTag-AD arrays (e.g., 3xVPR and 3xVPRF) results in liquid-like condensates and boosts the transcriptional activation of the CRISPRa-SunTag platform. However, when the number of SunTag scaffolds is increased to 10 or more, solid-like condensates form, sequestering co-activators such as p300 and MED1, which exhibit low dynamicity and liquidity, significantly reducing activation strength. We propose that optimal phase separation, reflecting appropriate multivalent interactions within the CRISPRa platform, creates a conducive microenvironment around the transcription start site for efficient activation. Source data are provided as a Source Data file.
Discussion
The continuous improvement of CRISPRa technology is guided by a common principle: maximizing the recruitment of transcriptional activators near the transcription start site using the dCas9 system9,76,77,78,79. This strategy is analogous to the mechanism of super-enhancer activation, where TFs are highly enriched at transcription sites20. To achieve this goal, many CRISPR activators utilized SunTag to generate a repeating peptide array fused with multiple copies of activator domains (ADs)16,17,18. However, there is still much to explore and understand regarding the basic features of CRISPRa-mediated gene activation. In this study, we comprehensively compared different CRISPRa systems via quantitatively assessing transcriptional dynamics within the same cellular context. We discovered that the transcriptional activation capacity can be enhanced by increasing the number of SunTag-AD arrays in CRISPR activators within a certain range. However, exceeding this range, such as having ten copies of VPR, results in loosing activation capability. This observed pattern appears to be consistent across a wide range of SunTag activators by testing genes with varying expression levels in different cell types.
Incorporating multivalent molecules into the CRISPRa platform is likely an effective strategy to enhance gene activation80. IDR-rich proteins often have increased effective valence through nucleic acid binding or direct oligomerization81,82,83,84. FUS IDR (FUSn) has been proposed to function as a transcriptional activation domain that can recruit TFs and Pol II and mediate transcription68,85,86,87. Gene activation in the CRISPRa system increased with additional FUS IDRs, but excessive FUSn (e.g., ≥10) resulted in condensate resistance to 1,6-HD and reduced gene activation. Substituting the FUS IDR with NUP98 IDR or SMN1 resulted in condensates that were highly resistant to 1,6-HD and incapable of activating genes. Similar behavior was observed in the 10xVPR(F) and PCP-12/40xVPRF systems. We observed that CRISPRa condensates sequester coactivators such as p300 and MED1, with their fluidity and dynamics resembling those of CRISPRa molecules. This sequestration likely explains why excessive multivalent interactions within CRISPR-SunTag systems diminish activation capacity. Therefore, optimal phase separation, which reflects appropriate multivalent interactions within the CRISPRa platform, creates a conducive microenvironment around the transcription start site, therefore promoting efficient activation (Fig. 7e). Thus, our studies lend support to the feasibility of utilizing CRISPR-based synthetic condensates as a programmable platform, wherein their composition and functionalities can be precisely designed to meet specific requirements88,89,90.
The term “transcriptional condensate” refers to nuclear compartments that concentrate biomolecules involved in transcription without implying that the formation of these structures is exclusively driven by phase separation26. Transcriptional condensates have been hypothesized to concentrate high levels of TFs and co-activators, which super-enhancers can trigger to regulate key cell identity genes20,24,91. The controversy surrounding the role of transcriptional condensates in gene activation could be explained by various factors, including the compositions or properties of condensates, as well as cell-type and gene-specific effects26,81,92. The material properties of heterotypic condensates depend on the relative abundance and interaction strengths of individual factors93,94,95,96. By manipulating the compositions and properties of CRISPRa-based transcriptional condensates, we found that gene activation strength correlates positively with the dynamicity and liquidity of condensates rather than the overall condensate formation. Additionally, our real-time imaging demonstrated that transcriptional condensates formed by 3xVPR and 3xVPRF were closely associated with transcription sites at certain time points. Under these conditions, transcription levels were significantly higher than when no condensates were present at the transcription sites. Our findings imply that these dCas9-guided transcriptional condensates may play a regulatory role in facilitating RNA bursts. However, the active transcription may also occur outside of condensates25,97. Based on our studies, we propose that an optimal phase-separated CRISPRa system could generate transcriptional condensates, which may not be visible but effectively concentrate transcription factors and coactivators near transcription sites through dynamic interactions with surrounding molecules (Fig. 7e). While larger, detectable condensates may not directly drive transcriptional activation, but must retain fluidity and dynamicity to facilitate the free diffusion of molecules. Collectively, our findings emphasize the significance of maintaining balanced homotypic and heterotypic interactions to ensure the proper functionality of condensates89,98.
CRISPRa provides precise, versatile, and scalable methods for gene activation, enabling the study of gene function and regulation, as well as potential disease treatments resulting from transcriptional misregulation99,100,101,102,103. Therefore, extensive efforts are dedicated to optimizing its efficiency. The phase-separated CRISPRa platform, formerly known as DropCRISPRa31, represents a new generation of CRISPRa tools. Our analysis revealed that 3xVPR and 3xVPRF form phase-separated condensates and consistently exhibit higher activation potency compared to other systems. We have previously developed a method termed Narta, which utilizes multiple binding sites within nascent RNA introns to recruit abundant TFs to transcription sites, thereby achieving high transcriptional levels104. Therefore, the substantial enrichment of transcription factors at the transcription start site is indeed a promising strategy for achieving highly potent gene activation. By incorporating the design principles uncovered in our study, future efforts can focus on selecting optimal scaffolds and modulating multivalent interactions within synthetic transcriptional condensates to maximize gene activation.
Methods
Cell culture
HeLa cells (ATCC, CCL-2) and HEK 293T cells (ATCC, CRL-3216) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with high glucose (HAKATA), 10% fetal bovine serum (HAKATA), and 1% penicillin/streptomycin (Gibco). HCT116 cells (National collection of authenticated cell culture, TCHu 99) were cultured in McCoy’s 5A medium (Procell) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. The primed hESC line H1 (WiCell, WA01-PCBC) was cultured in mTeSR medium (Stem cell technologies) on matrigel-coated plates at 37 °C and 5% CO2 in a humidified incubator. Mouse mESCs (46C, gift from Austin Smith’s lab) were maintained in a complete ESC culture medium under standard ESC culture conditions as previously described105. All cells were cultured at 37 °C and 5% CO2 and regularly tested for mycoplasma.
Plasmids construction
All plasmids generated in this study were constructed using either the Gibson Assembly method (Beyotime, D7010M) or restriction cloning (New England Biolabs) followed by ligation using T4 DNA ligase (Abclonal, RK21501). Oligonucleotides were synthesized by Tsingke Biotech. DNA fragments were directly synthesized by BGI or amplified from corresponding DNA templates by PCR. Backbones and the required fragments were digested using restriction endonucleases (New England Biolabs), following the manufacturer’s instructions. All cloned plasmids were confirmed by Sanger sequencing (Tsingke). All sgRNAs used for CRISPRa are listed in Supplementary Table 2 and all plasmids used in this study are shown in Supplementary Data 1.
Generation of reporter cell lines
To analyze CRISPRa efficiency at the single-cell level, this study utilized a series of reporter cell lines, including both exogenous and endogenous reporters. These cell lines have been previously reported in our previous studies104. The exogenous reporter, miniCMV-TriTagmTagBFP, was established by lentiviral infection followed by clonal cell isolation. Clonal cell lines that harbored an active miniCMV transcription site upon CRISPRa activation were selected for CRISPRa testing. To achieve DNA labeling of miniCMV-TriTagmTagBFP locus, the cells were co-infected with dCas9-GFP1114X and GFP1–10106 lentiviruses. To enhance the infection efficiency of each lentivirus, polybrene was added at a concentration of 5 µg/ml. The endogenous reporters, including HSPB1-BFP, HSPB8-BFP, BFP-LMNA, HPDL-BFP, BAG3-BFP and HaloTag-p300, were generated through Cas9-mediated knockin. HeLa or 293T cells were cultured in 24-well plates and transiently transfected with 400 ng of donor plasmids, 100 ng of Cas9 protein-expressing vectors, and 250 ng of sgRNA expression vectors. Three to four days after transfection, fluorescence-activated cell sorting (FACS) was conducted to isolate knock-in positive cells. The LMNA-TriTagBFP single-cell clone cell line was established in our previous study49. This cell line contains a single endogenous LMNA locus labeled with TriTag and stably expresses stdMCP-tdTomato and dCas9-GFP14x, enabling the visualization of both the LMNA-TriTagBFP DNA locus and the nascent RNA produced during transcription. sgRNAs used for CRISPR knockin are listed in Supplementary Table 3.
Transfection
Transient transfections of 293T or Hela cells were performed in 8-well or 24-well plates. To generate CRISPR knockin cell lines, cells were transfected with plasmids using FuGENE transfection reagent (Promega). For CRISPRa testing, all transfections were performed using PEI (Polysciences) following the manufacturer’s recommended protocol. To achieve inducible CRISPRa, the quantity ratio of Tet-On 3G and TRE3G plasmids was maintained at 2:1. At 6 h post-transfection, the cell culture medium was replaced with non-penicillin/streptomycin medium supplemented with doxycycline (1 μg/ml). Following 12-h incubation with doxycycline, cells were subjected to imaging analysis to examine the production of nascent RNAs in Fig. 1b and Supplementary Fig. 4a. Protein expression levels and relative mRNA abundance were quantified 48 h after adding doxycycline in Fig. 2 and Supplementary Figs. 4b–d, 6, 7, 8c–f, 9. The transfections for Figs. 1b–f, 2c, 3b, c, 4, 5, 6, 7b and Supplementary Figs. 3a–d, 4a–d, 5, 6a, b, 7a, 8d, 9, 12b, 13, 14a–c, 16, 17, 18, 19c, d, 20, 21 were conducted in 8-well chambered coverglass, and imaging was performed 12, 24 or 48 h after transfection. For FACS or qPCR analysis, cells were seeded in 24-well plates and analyzed 48 h after transfection or doxycycline induction. To achieve CRISPRa, a total of 600 ng sgRNAs were used for individual transfections. To improve the adhesion of 293T cells, 100 μl of Gelatin (40 mg/ml) was added to each well before seeding cells. To evaluate the efficiency of CRISPRa in mouse embryonic stem cells, transient transfection of CRISPRa plasmids was carried out using Lipofectamine 2000. The usage of each plasmid for each group of transfection is listed in Supplementary Table 4.
Flow cytometry
To assess the activation ratio of CRISPRa, the protein expression level was analyzed by flow cytometry using a BD Fortessa instrument (BD Biosciences). Transfection-positive cells were specifically analyzed by incorporating GFP, mCherry or HaloTag into one of the CRISPRa plasmids. If HaloTag was used, cells were incubated with 250 nM Jenelia Fluor HaloTag ligands (Promega) for 20 min before detaching them from the culture plate using trypsin digestion. Cells were first gated for the intact cell population based on forward scatter versus side scatter plots, followed by gating for single cells using forward scatter W versus forward scatter H. Using the basal expression level of the CRISPRa target gene as the control, transfection-positive cells expressing the target gene at a higher level than the basal expression were identified as CRISPR activation positive cells.
RNA extraction and qPCR analysis
Cells were grown in the 24-well plate and collected 48 h after transfection. RNA extraction was performed using the MolPure® Cell/Tissue Total RNA Kit (YEASEN), and the RNA concentration was determined by measuring the absorbance at A260/280. cDNA synthesis was carried out using oligo-dT primers (HiScript II Q RT SuperMix for qPCR, Vazyme) using 800 ng of RNA per cDNA reaction. For lincRNA, a 5-min pre-denaturation was included before reverse transcription. PCR reactions were prepared using the 2x Universal SYBR Green Fast qPCR Mix (ABclonal) and performed on the CFX96 Real-Time PCR system (Bio-Rad). A 2 μl aliquot of a 1:10 cDNA dilution was used per reaction in a total reaction volume of 10 μl. All reactions were performed in triplicate or more. The RNA abundance was normalized to the endogenous reference gene UBC or ACTB and calculated using the delta-delta threshold cycle (ΔΔCt) method. The expression levels of reporter RNAs were normalized to the levels of beta-actin mRNA (DCT) and presented as the fold-change compared to the mock control. The primers used for qRT-PCR are listed in Supplementary Table 5.
CRISPR-dCas9-mediated DNA labeling
Both miniCMV-TriTag and TriTag-LMNA clonal cell lines stably express dCas9-14xGFP11 and GFP1-10, forming dCas9-14xGFP, which enhances the signal-to-noise ratio of DNA labeling106. To label the genomic loci of miniCMV-TriTag or TriTag-LMNA, cells were plated in an 8-well plate and transfected 12 h later with a single sgRNA plasmid (600 ng) targeting the repetitive TS1 sequence (12 copies) in TriTag49. 24 h post-transfection, DNA labeling can be visualized using a fluorescence microscope with a 63x or 100x oil immersion objective. The specific sgRNA used for DNA imaging is listed in Supplementary Table 6.
Copy number of TriTag cassette
To determine the copy number of miniCMV-TriTagBFP integrated into the genome of the clonal cell line, the CRISPR imaging method we had previously developed was applied to label miniCMV genomic locus. To confirm the result, quantitative PCR was used to calculate the number of copies of miniCMV-TriTag. TriTag-LMNA, a single-copy gene validated in our previous studies49, was used as a reference locus. TriTag fragments were cloned into a vector. Following an established protocol107, standard curves were generated with dilutions of pPF75-TriTag plasmid, ranging from 103 to 107 copies per 5 µl. A standard curve was generated by plotting the log of plasmid copy number (load) against Ct value, with the regression line slope (m) and intercept (b) used to calculate the absolute copy number. The curve was created using GraphPad. Genomic DNA was diluted to 100 ng/µl as the template, and qPCR was performed in triplicate to determine the Ct value. The number of TriTag cassette copies was estimated by calculating the absolute copy numbers for both the target (miniCMV-TriTag) and the reference (TriTag-LMNA) using the measured Ct values and the equation: total copies = 10([Ct - b]/m), where b and m represent the intercept and slope of the respective plasmid standard curve. The ratio of miniCMV-TriTag to TriTag-LMNA copy numbers was then calculated, and the copies of miniCMV-TriTag per genome were determined. qPCR was carried out using the ChamQ Universal SYBR qPCR Master Mix (Vazyme) on the Bio-Rad CFX96 Real-Time PCR system. qPCR primers are listed in Supplementary Table 7.
Immunostaining
To detect endogenous MED1, samples were first washed once with PBS and then fixed with 4% paraformaldehyde for 20 min. Permeabilization was carried out using a solution of 0.5% Triton X-100, 3% BSA, and PBS for 15 min. The samples were then incubated with anti-MED1 (Bethyl, A300-793A) in a blocking buffer at room temperature for 3 h. Following three washes with PBS, the samples were incubated with Alexa647-conjugated secondary antibody at room temperature for 1 h, followed by three additional PBS washes. The secondary antibody was Alexa Fluor 647–labeled Donkey Anti-Rabbit IgG H&L (Abcam, ab150075). All staining steps were performed at room temperature.
Immunostaining of CRISPRa-induced iNeurons
The primed hESC line H1 was seeded in a 12-well plate. To introduce CRISPRa plasmids into H1 cells, the electroporation method was employed. First, cells were detached from the culture plate one day after plating using TrypLETM Express enzyme (Gibco). Then, the desired amount of plasmid DNA was mixed with the cells in an appropriate electroporation buffer. The cell-plasmid mixture was transferred to an electroporation cuvette or chamber and subjected to an electric pulse using an electroporation device. Two days after electroporation, the cells were subjected to immunostaining. Immunofluorescence staining was performed to detect neurofilament 200, serving as a marker for CRISPRa-induced iNeurons. The samples were washed once with PBS, fixed with 4% paraformaldehyde for 20 min, and permeabilized with a solution of 0.5% Triton X-100, 3% BSA, and PBS for 15 min. Subsequently, the samples were incubated with anti-neurofilament 200 (Sigma, N4142) in a blocking buffer at room temperature for 3 h. After three washes, the samples were incubated with Alexa647-conjugated secondary antibody at room temperature for 1 h and washed three times again. The secondary antibody used was Alexa Fluor 647–labeled Donkey Anti-Rabbit IgG H&L (Abcam, ab150075). Hoechst 33342 dye was used as a nuclear stain during the immunofluorescence process. All staining steps were performed at room temperature.
1,6-Hexanediol treatment
Cells were cultured in an 8-well chambered coverglass and transfected with the respective CRISPRa reporters. After 48 h of transfection, the cells were treated with 1,6-Hexanediol (Sigma). The 1,6-HD was dissolved in H2O to create a storage solution with a concentration of 15%. Prior to use, a working solution was prepared by diluting the stock solution with DMEM medium containing 10% FBS to a final concentration of 1.5%. The cells were then treated with this working solution for 2 min and subsequently imaged using confocal microscopy.
Western blot
To quantify the protein expression levels of dCas9-GFP-3xGCN4 and dCas9-GFP-10xGCN4, the relevant plasmids were transiently transfected into 293T cells. 48 hours post-transfection, GFP-positive cells were sorted as successfully transfected cells using flow cytometry (BD, FACSAria II) and subsequently used for Western blot analysis. Protein samples were separated on SDS-PAGE gels and transferred onto cellulose acetate membranes. The membranes were blocked with 5% non-fat milk and incubated with primary antibodies against GFP (Roche, 11814460001) and the loading control, Tubulin (Earthox, E021030). Protein bands were visualized using the ECL Enhanced Plus kit (ABclone, RM00021P) and detected with the Bio-Rad Chemidoc XRS+ imaging system.
RNA-seq analysis
To evaluate the specificity of CRISPRa systems, 293T cells were seeded into 12-well plates. The following day, using the same transient transfection conditions as described in Fig. 3, plasmids for the VPR, VPRF, 3XVPR, and 3XVPRF systems were individually transfected into each well. RNA extraction was performed 48 h post-transfection. Samples were collected for RNA-seq library construction and subsequent sequencing, with single-end 50-bp reads. The analysis procedures were carried out as previously described in our work104. The sequencing reads were aligned to the human GRCh38 genome by STAR. Reads that mapped to multiple genomic locations were eliminated. Gene expression counts for each sample were calculated using featureCounts108. The normalized expression counts for each gene were obtained using the count function from the DESeq2 package (https://github.com/mikelove/DESeq2). For differential expression analysis, the gene expression counts were first normalized and log2 transformed by the edgeR package (https://bioconductor.org/packages/release/bioc/html/edgeR.html) with default parameters. Genes with low expression levels were filtered out if their maximum Counts Per Million (CPM) value across all samples was less than 1. The differential up-regulated genes between the treatment group (targeting HSBP8) and the control group (targeting Gal4) were identified using the limma package (https://github.com/cran/limma). The criteria for significance included an adjusted p-value (t-test q-value < 0.05 with FDR correction) and a fold change greater than 2.0.
Fluorescence recovery after photobleaching (FRAP) assays
FRAP assays were used to evaluate the mobility of fluorescently labeled proteins within cells. Cells were seeded on 8-well chambered coverglass and imaged 48 h after transfection with the desired CRISPRa reporters. FRAP experiments were conducted on a Nikon A1 confocal microscope with a ×60 oil objective. A square region of interest (ROI) was selected for photobleaching using a 488 nm line at 80% laser power. Prior to photobleaching, three pre-bleaching frames were recorded, followed by the recording of fluorescence recovery within the bleached area over 18 frames. The time interval between capturing each frame was precisely set to 10 seconds. The recovery curve was then analyzed using the Fiji plugin FRAP Profiler V2 (https://worms.zoology.wisc.edu/research/4d/4d.html)109. The half-life values (1/2 t) and mobile fraction (Fm) were determined by selecting single exponential recovery as curve fitting method.
Microscopy
Fluorescent images, except for Figs. 4c, 6h, 7c and Supplementary Figs. 3a, 5, 13c, 13d, were obtained using an Olympus IX83 fluorescence microscope equipped with spinning-disk confocal scanner (Yokogawa CSU-W1), a 60× NA 1.49 oil Apochromat objective, an sCMOS camera (Prime 95B), 405/488/561/640 nm lasers (OBIS), and a PEIZO stage (ASI) with stage incubator (Tokai Hit). Cells for live-cell imaging were maintained at 37°C and 5% CO2 in a humidified chamber. For confocal imaging, cells were plated into 8-well chambered coverglass. Real-time imaging in Fig. 1b was performed 18 h post-transfection (with Dox added for 12 h) and lasted for 2 h. Multiple z-planes, spaced by 1 μm at intervals of 2 min, were captured during this period. Using the same imaging settings, the consecutive images in Fig. 5a and Supplementary Fig. 13a, 17a, 18a, 18d were acquired 12 h after transfection and recorded for 6 h. However, real-time imaging in Supplementary Fig. 12 was captured 48 h post-transfection, with multiple z-planes spaced 1 μm apart at intervals of 30 seconds. The images in Figs. 1b, 3b, 4b, 5a, b, 6a, c, g, 7c and Supplementary Figs. 3b, c, 4a, 4c, 12b, 13a, 14a, 16a, 17a–c, 18a, 18d, 19c, 20a, 21a are presented as maximum-intensity projections derived from z stacks, while Figs. 4c, 6h and Supplementary Figs. 5, 6a, 13c, d were obtained through single-plane imaging. FRAP images in Figs. 4c, 6h and Supplementary Fig. 13d and the images in Supplementary Figs. 3a, 5 and 13c were acquired using a Nikon A1 confocal microscope with a 60× oil objective. Images in Fig. 7c were obtained using a Nikon Ti2-E fluorescence microscope equipped with a 10×/0.3 Plan Fluor objective, an LED source (SPECTRA 4), an sCMOS camera (ZYLA 4.2MP Plus), a Perfect Focus unit (Nikon), and a motorized stage (Nikon).
Statistics and reproducibility
Fluorescence imaging data were analyzed by ImageJ to calculate the mean intensity of fluorescent reporter proteins and the total intensity or the area of fluorescent spots (representing nascent RNA and CRISPRa condensates). The maximum intensity observed during a burst was measured to define the burst amplitude. Burst durations were calculated by measuring the time interval from the start of the burst (when the signal of nascent RNAs exceeds the background) to the end of the burst (when the nascent RNA signal returns to baseline). Pause durations were determined by the time gap between the end of a burst and the initiation of the next burst. To generate the curves depicting transcriptional bursting or condensate formation around transcription sites over time in Figs. 1b and 5a, the time-lapse imaging data was imported into the Olympus cellSens imaging software. Specific regions corresponding to the transcription sites and a background region for control were selected. The dynamic ROI (Region of Interest) was adjusted over time to accommodate any movement of the transcription sites or cells. The “Intensity profile” function was applied, specifically selecting “maximum intensity” to extract the relevant values. These values were exported for plotting the curves using GraphPad Prism. To elucidate the relationship between condensate dynamics and transcriptional bursts in Fig. 5h, each individual RNA burst and the total condensate intensity at each time point were quantified. The burst analysis was categorized into three types: (1) ‘burst initiation’, using the burst initiation point as time zero and quantifying the burst from initiation up to 20 min; (2) ‘burst peak’, using the burst peak as time zero and covering 20 min before and after the burst peak; (3) ‘burst decline’, using the burst termination point as time zero and quantifying the 20 min preceding termination. For each type, condensate intensity was measured every 2 min within a 30-min window around time zero. All statistical analyses were performed using GraphPad Prism.
To quantify the overall level of CRISPRa condensates in a cell for Supplementary Fig. 16, Fiji software was used to identify all visible condensates in each cell with signals above the background and measured the total fluorescence intensity of individual condensates. The sum of the fluorescence intensities of all condensates within a cell was then calculated to indicate the level of phase separation in that cell. To quantitatively assess the efficiency of stem cell differentiation in Fig. 7c, z-s tack images were converted into maximum projection images and opened using Fiji software. The total number of cells was determined by applying the “Find Maximum” function in Fiji, with appropriate parameter settings to detect Hoechst33323 stained nuclei. Similarly, the count of neural-like cells positive for neurofilament 2000 staining was performed using the same method. By comparing these two counts, the ratio of differentiated cells can be calculated, providing a measure of the differentiation efficiency. GraphPad Prism software version 8 (GraphPad Software, La Jolla, CA, USA; https://www.graphpad.com) was utilized for calculating mean values, P-values, correlation coefficients (R2), and standard error of the mean (SEM) for statistical analysis. Statistical significance between two groups was determined using the student t-test, while significance among three or more groups was assessed using one-way ANOVA. Flow cytometry data were analyzed using FlowJo v10 software (FlowJo LLC). No statistical method was used to predetermine the sample size. No data were excluded from the analyses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw-data of RNA-Seq has been deposited in the Gene Expression Omnibus (GEO) database with the accession number GSE278565. All data supporting the findings of this study are available within the article and its Supplementary Information file. Other potential type of data used to interpret the findings can be provided upon request to the corresponding author. Source data are provided with this paper.
References
Lambert, S. A. et al. The human transcription factors. Cell 172, 650–665 (2018).
Lu, F. & Lionnet, T. Transcription factor dynamics. Cold Spring Harb. Perspect. Biol. 13, a040949 (2021).
Wagh, K., Stavreva, D. A., Upadhyaya, A. & Hager, G. L. Transcription factor dynamics: one molecule at a time. Annu. Rev. Cell Dev. Biol. 39, 277–305 (2023).
Soto, L. F. et al. Compendium of human transcription factor effector domains. Mol. Cell 82, 514–526 (2022).
Keegan, L., Gill, G. & Ptashne, M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science 231, 699–704 (1986).
Weirauch, M. T. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014).
Chen, M. & Qi, L. S. Repurposing CRISPR system for transcriptional activation. Adv. Exp. Med. Biol. 983, 147–157 (2017).
Dominguez, A. A., Lim, W. A. & Qi, L. S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 17, 5–15 (2016).
Casas-Mollano, J. A., Zinselmeier, M. H., Erickson, S. E. & Smanski, M. J. CRISPR-Cas activators for engineering gene expression in higher eukaryotes. CRISPR J. 3, 350–364 (2020).
Maeder, M. L. et al. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977–979 (2013).
Perez-Pinera, P. et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 10, 973–976 (2013).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015).
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S. & Vale, R. D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014).
Zhou, H. et al. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat. Neurosci. 21, 440–446 (2018).
Kunii, A. et al. Three-component repurposed technology for enhanced expression: highly accumulable transcriptional activators via branched tag arrays. CRISPR J. 1, 337–347 (2018).
Pan, C. et al. CRISPR-Act3.0 for highly efficient multiplexed gene activation in plants. Nat. Plants 7, 942–953 (2021).
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K. & Sharp, P. A. A phase separation model for transcriptional control. Cell 169, 13–23 (2017).
Mazzocca, M., Fillot, T., Loffreda, A., Gnani, D. & Mazza, D. The needle and the haystack: single molecule tracking to probe the transcription factor search in eukaryotes. Biochem. Soc. Trans. 49, 1121–1132 (2021).
Sabari, B. R. Biomolecular condensates and gene activation in development and disease. Dev. Cell 55, 84–96 (2020).
Schneider, N. et al. Liquid-liquid phase separation of light-inducible transcription factors increases transcription activation in mammalian cells and mice. Sci. Adv. 7, eabd3568 (2021).
Shrinivas, K. et al. Enhancer features that drive formation of transcriptional condensates. Mol. Cell 75, 549–561.e547 (2019).
Wei, M. T. et al. Nucleated transcriptional condensates amplify gene expression. Nat. Cell Biol. 22, 1187–1196 (2020).
Stortz, M., Presman, D. M. & Levi, V. Transcriptional condensates: a blessing or a curse for gene regulation? Commun. Biol. 7, 187 (2024).
Ferrie, J. J., Karr, J. P., Tjian, R. & Darzacq, X. Structure”-function relationships in eukaryotic transcription factors: the role of intrinsically disordered regions in gene regulation. Mol. Cell 82, 3970–3984 (2022).
Boija, A. et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e1816 (2018).
Wagh, K., Garcia, D. A. & Upadhyaya, A. Phase separation in transcription factor dynamics and chromatin organization. Curr. Opin. Struct. Biol. 71, 148–155 (2021).
Chong, S. et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 (2018).
Ma, S. et al. Phase-separated DropCRISPRa platform for efficient gene activation in mammalian cells and mice. Nucleic Acids Res. 51, 5271–5284 (2023).
Liu, J. et al. CRISPR-assisted transcription activation by phase-separation proteins. Protein Cell 14, 874–887 (2023).
Lu, Y. et al. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat. Cell Biol. 22, 453–464 (2020).
Zhang, H. et al. Reversible phase separation of HSF1 is required for an acute transcriptional response during heat shock. Nat. Cell Biol. 24, 340–352 (2022).
Kim, Y. J. et al. Light-activated macromolecular phase separation modulates transcription by reconfiguring chromatin interactions. Sci. Adv. 9, eadg1123 (2023).
Ma, L. et al. Co-condensation between transcription factor and coactivator p300 modulates transcriptional bursting kinetics. Mol. Cell 81, 1682–1697.e1687 (2021).
Kawasaki, K. & Fukaya, T. Functional coordination between transcription factor clustering and gene activity. Mol. Cell 83, 1605–1622.e1609 (2023).
Wu, J. et al. Modulating gene regulation function by chemically controlled transcription factor clustering. Nat. Commun. 13, 2663 (2022).
Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018).
Cho, W. K. et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018).
Du, M. et al. Direct observation of a condensate effect on super-enhancer controlled gene bursting. Cell 187, 331–344.e317 (2024).
Li, C., Li, Z., Wu, Z. & Lu, H. Phase separation in gene transcription control. Acta Biochim. Biophys. Sin. (Shanghai) 55, 1052–1063 (2023).
Chong, S. et al. Tuning levels of low-complexity domain interactions to modulate endogenous oncogenic transcription. Mol. Cell 82, 2084–2097.e2085 (2022).
Trojanowski, J. et al. Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol. Cell 82, 1878–1893.e1810 (2022).
Meeussen, J. V. W. et al. Transcription factor clusters enable target search but do not contribute to target gene activation. Nucleic Acids Res. 51, 5449–5468 (2023).
Chavez, A. et al. Comparison of Cas9 activators in multiple species. Nat. Methods 13, 563–567 (2016).
Martella, A. et al. Systematic evaluation of CRISPRa and CRISPRi modalities enables development of a multiplexed, orthogonal gene activation and repression system. ACS Synth. Biol. 8, 1998–2006 (2019).
Omachi, K. & Miner, J. H. Comparative analysis of dCas9-VP64 variants and multiplexed guide RNAs mediating CRISPR activation. PLoS ONE 17, e0270008 (2022).
Xu, H. et al. TriTag: an integrative tool to correlate chromatin dynamics and gene expression in living cells. Nucleic Acids Res. 48, e127 (2020).
Wan, Y. et al. Dynamic imaging of nascent RNA reveals general principles of transcription dynamics and stochastic splice site selection. Cell 184, 2878–2895.e2820 (2021).
Muramoto, T. et al. Live imaging of nascent RNA dynamics reveals distinct types of transcriptional pulse regulation. Proc. Natl Acad. Sci. USA 109, 7350–7355 (2012).
Liu, X. et al. MONITTR allows real-time imaging of transcription and endogenous proteins in C. elegans. J. Cell Biol. 224, iyac068 (2025).
Brodsky, S. et al. Intrinsically disordered regions direct transcription factor in vivo binding specificity. Mol. Cell 79, 459–471.e454 (2020).
Gaglia, G. et al. HSF1 phase transition mediates stress adaptation and cell fate decisions. Nat. Cell Biol. 22, 151–158 (2020).
Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Crozat, A., Aman, P., Mandahl, N. & Ron, D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 363, 640–644 (1993).
Rabbitts, T. H., Forster, A., Larson, R. & Nathan, P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat. Genet. 4, 175–180 (1993).
Owen, I. et al. The oncogenic transcription factor FUS-CHOP can undergo nuclear liquid-liquid phase separation. J. Cell Sci. 134, jcs258578 (2021).
Lin, Y. et al. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789–802.e712 (2016).
Lu, H. et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323 (2018).
Strom, A. R. et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017).
Gao, Y., Li, X., Li, P. & Lin, Y. A brief guideline for studies of phase-separated biomolecular condensates. Nat. Chem. Biol. 18, 1307–1318 (2022).
Sun, J. et al. Precise prediction of phase-separation key residues by machine learning. Nat. Commun. 15, 2662 (2024).
Roeder, R. G. 50+ years of eukaryotic transcription: an expanding universe of factors and mechanisms. Nat. Struct. Mol. Biol. 26, 783–791 (2019).
Korzus, E. et al. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279, 703–707 (1998).
Reiter, F., Wienerroither, S. & Stark, A. Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 43, 73–81 (2017).
Allen, B. L. & Taatjes, D. J. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166 (2015).
Kwon, I. et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 (2013).
Ying, Q. L., Stavridis, M., Griffiths, D., Li, M. & Smith, A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183–186 (2003).
Baumann, V. et al. Targeted removal of epigenetic barriers during transcriptional reprogramming. Nat. Commun. 10, 2119 (2019).
Busskamp, V. et al. Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol. 10, 760 (2014).
Zhang, Y. et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798 (2013).
Zhang, X. et al. Gene activation in human cells using CRISPR/Cpf1-p300 and CRISPR/Cpf1-SunTag systems. Protein Cell 9, 380–383 (2018).
Tak, Y. E. et al. Augmenting and directing long-range CRISPR-mediated activation in human cells. Nat. Methods 18, 1075–1081 (2021).
Lebar, T., Lainšček, D., Merljak, E., Aupič, J. & Jerala, R. A tunable orthogonal coiled-coil interaction toolbox for engineering mammalian cells. Nat. Chem. Biol. 16, 513–519 (2020).
Du, D. & Qi, L. S. An introduction to CRISPR technology for genome activation and repression in mammalian cells. Cold Spring Harb. Protoc. 2016, pdb.top086835 (2016).
La Russa, M. F. & Qi, L. S. The new state of the Art: Cas9 for gene activation and repression. Mol. Cell Biol. 35, 3800–3809 (2015).
Becirovic, E. Maybe you can turn me on: CRISPRa-based strategies for therapeutic applications. Cell Mol. Life Sci. 79, 130 (2022).
Dominguez, A. A. et al. CRISPR-mediated synergistic epigenetic and transcriptional control. CRISPR J. 5, 264–275 (2022).
Chen, R. et al. Specific multivalent molecules boost CRISPR-mediated transcriptional activation. Nat. Commun. 15, 7222 (2024).
Sharp, P. A., Chakraborty, A. K., Henninger, J. E. & Young, R. A. RNA in formation and regulation of transcriptional condensates. RNA 28, 52–57 (2022).
Smith, J. et al. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. Elife 5, e21337 (2016).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Mitrea, D. M. et al. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife 5, e13571 (2016).
Yang, L., Gal, J., Chen, J. & Zhu, H. Self-assembled FUS binds active chromatin and regulates gene transcription. Proc. Natl Acad. Sci. USA 111, 17809–17814 (2014).
Raczynska, K. D. et al. FUS/TLS contributes to replication-dependent histone gene expression by interaction with U7 snRNPs and histone-specific transcription factors. Nucleic Acids Res. 43, 9711–9728 (2015).
Shao, Y. et al. A chaperone-like function of FUS ensures TAZ condensate dynamics and transcriptional activation. Nat. Cell Biol. 26, 86–99 (2024).
Chang, H. Y. & Qi, L. S. Reversing the Central Dogma: RNA-guided control of DNA in epigenetics and genome editing. Mol. Cell 83, 442–451 (2023).
Mitrea, D. M., Mittasch, M., Gomes, B. F., Klein, I. A. & Murcko, M. A. Modulating biomolecular condensates: a novel approach to drug discovery. Nat. Rev. Drug Discov. 21, 841–862 (2022).
Dai, Y., You, L. & Chilkoti, A. Engineering synthetic biomolecular condensates. Nat. Rev. Bioeng. 1, 466–480 (2023).
Blobel, G. A., Higgs, D. R., Mitchell, J. A., Notani, D. & Young, R. A. Testing the super-enhancer concept. Nat. Rev. Genet. 22, 749–755 (2021).
Donovan, B. T. & Larson, D. R. Regulating gene expression through control of transcription factor multivalent interactions. Mol. Cell 82, 1974–1975 (2022).
Li, W. & Jiang, H. Nuclear protein condensates and their properties in regulation of gene expression. J. Mol. Biol. 434, 167151 (2022).
Espinosa, J. R. et al. Liquid network connectivity regulates the stability and composition of biomolecular condensates with many components. Proc. Natl Acad. Sci. USA 117, 13238–13247 (2020).
Sundaravadivelu Devarajan, D. et al. Sequence-dependent material properties of biomolecular condensates and their relation to dilute phase conformations. Nat. Commun. 15, 1912 (2024).
Schuster, B. S. et al. Biomolecular condensates: sequence determinants of phase separation, microstructural organization, enzymatic activity, and material properties. J. Phys. Chem. B 125, 3441–3451 (2021).
Cai, D. et al. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol. 21, 1578–1589 (2019).
Do, S., Lee, C., Lee, T., Kim, D. N. & Shin, Y. Engineering DNA-based synthetic condensates with programmable material properties, compositions, and functionalities. Sci. Adv. 8, eabj1771 (2022).
Bendixen, L., Jensen, T. I. & Bak, R. O. CRISPR-Cas-mediated transcriptional modulation: The therapeutic promises of CRISPRa and CRISPRi. Mol. Ther. 31, 1920–1937 (2023).
Matharu, N. et al. CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science 363, eaau0629 (2019).
Kemaladewi, D. U. et al. A mutation-independent approach for muscular dystrophy via upregulation of a modifier gene. Nature 572, 125–130 (2019).
Nuñez, J. K. et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell 184, 2503–2519.e2517 (2021).
Thakore, P. I., Black, J. B., Hilton, I. B. & Gersbach, C. A. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat. Methods 13, 127–137 (2016).
Liang, Y. et al. Gene activation guided by nascent RNA-bound transcription factors. Nat. Commun. 13, 7329 (2022).
Shen, X. et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502 (2008).
Chen, B., Zou, W., Xu, H., Liang, Y. & Huang, B. Efficient labeling and imaging of protein-coding genes in living cells using CRISPR-Tag. Nat. Commun. 9, 5065 (2018).
Johnson, S. M., Carlson, E. L. & Pappagianis, D. Determination of ribosomal DNA copy number and comparison among strains of Coccidioides. Mycopathologia 179, 45–51 (2015).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Serre, J. M. et al. C. elegans srGAP is an α-catenin M domain-binding protein that strengthens cadherin-dependent adhesion during morphogenesis. Development 149, dev200775 (2022).
Acknowledgements
The authors thank Hui Yang for sharing the CRISPRa (SPH) plasmids; Peiguo Yang, Heng Xu and Huasong Lu for fruitful discussions; Yanwei Li and Shuangshuang Liu from the core facilities at Zhejiang University School of Medicine for their technical support with imaging and flow cytometry analysis. This work was supported by grants from the National Natural Science Foundation of China (32371507 and 32171444 to B.C., 32370822 to W.Z., and 32350023 and 8237012 to N.L.) and the National Key Research and Development Program of China (2023ZD0500503 and 2021YFC2700904 to B.C.).
Author information
Authors and Affiliations
Contributions
Conceptualization, B.C. and W.Z.; study supervision, B.C., W.Z., and N. L.; methodology, B.C., H.L., Y.Y., P.X., Y.F., X.Y., and S.L.; acquisition of data, Y.F., X.Y., S.L., C.M., Y.A., Y.L., and S.S.; analysis of data, Y.F., X.Y., S.L., Tao C., Tianyi C., Y.Z., and J.W.; Resources, X.W. and N.L.; drafting of manuscript, B.C., W.Z., Y.F., and X.Y.; manuscript reviewing, all authors; coordination, B.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fu, Y., Yang, X., Li, S. et al. Dynamic properties of transcriptional condensates modulate CRISPRa-mediated gene activation. Nat Commun 16, 1640 (2025). https://doi.org/10.1038/s41467-025-56735-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56735-8
This article is cited by
-
The rheology and interfacial properties of biomolecular condensates
Biophysical Reviews (2025)