Abstract
C4 photosynthesis exemplifies convergent evolution of complex traits. Herein, we construct chromosome-scale genome assemblies and perform multi-omics analysis for five Flaveria species, which represent evolutionary stages from C3 to C4 photosynthesis. Chromosome-scale genome sequence analyses reveal a gradual increase in genome size during the evolution of C4 photosynthesis attributed to the expansion of transposable elements. Systematic annotation of genes encoding C4 enzymes and transporters identify additional copies of three C4 enzyme genes through retrotranspositions in C4 species. C4 genes exhibit elevated mRNA and protein abundances, reduced protein-to-RNA ratios, and comparable translation efficiencies in C4 species, highlighting a critical role of transcriptional regulation in C4 evolution. Furthermore, we observe an increased abundance of ethylene response factor (ERF) transcription factors and cognate cis-regulatory elements associated with C4 genes regulation. Altogether, our study provides valuable genomic resources for the Flaveria genus and sheds lights on evolutionary and regulatory mechanisms underlying C4 photosynthesis.
Similar content being viewed by others
Introduction
C4 photosynthesis is a complex trait that evolved from ancestral C3 types approximately 35 million years ago1,2. Due to its high efficiencies in light, water, and nitrogen use3,4, C4 photosynthesis has been proposed for integration into C3 crops to enhance crop yield5,6,7. In contrast to C3 photosynthesis, C4 photosynthesis allocates more enzymes to carbon fixation, with these enzymes compartmentalized in mesophyll cells (MCs) or bundle sheath cells (BSCs)8,9. All known genes that involved in C4 photosynthesis have orthologs in C3 species10,11,12,13. The same C4 orthologous genes, which exhibit relatively high transcript abundances, are employed to support C4 metabolism across different C4 lineages in parallel12,14. However, it remains largely unclear how C4 genes evolved to acquire increased transcript abundances necessary for C4 photosynthesis.
Among dicotyledonous model systems for C4 photosynthesis, the genus Flaveria is notable for encompassing C3, C4, and many intermediate species15. Flaveria intermediate species are classified into C3–C4 and C4-like types, with the latter performing a full C4 metabolic pathway alongside C3 metabolic pathway. C3–C4 species are featured with decreased CO2 compensation points as a result of performing photorespiratory glycine shuttle, which was acquired by localizing glycine decarboxylase activity into BSCs and thus resulting in high CO2 concentration in BSCs2. C3–C4 species are further classified into type I C3–C4 species (with no or minimal C4 metabolism) and type II C3–C4 (with moderate C4 metabolism)16. Note that the term intermediate species does not necessarily refer to transitional forms, but may instead represent alternative evolutionary outcomes within the spectrum of photosynthetic strategies17. The Flaveria genus serves as an ideal model for investigating how C4 genes evolved from non-photosynthetic genes and adapted to function in C4 photosynthesis.
Decades of research on the genus of Flaveria have significantly advanced our understanding of the evolution of C4 photosynthesis15,18,19,20. Recently published short-read assembly-based reference genomes of four Flaveria specie provide valuable resources for protein-coding gene annotation in Flaveria genus21. Analyses of gene regulatory networks (GRNs) using long-read transcriptomic sequencing have provided critical insights into the evolution of C4 photosynthesis in the genus Flaveria20, emphasizing the pivotal role of transcriptional regulatory mechanisms in shaping the C4 pathway across different Flaveria species. However, due to a lack of high-quality reference genomes, our understanding of the regulation of C4 photosynthetic genes in the Flaveria genus is still incomplete. Existing short-read assembly-based Flaveria reference genomes are fragmented, potentially limiting their utility in identifying cis-regulatory elements (CREs) crucial for C4 photosynthesis. In this study, we present chromosome-scale reference genomes of five Flaveria species, generated using long-read genome sequencing technology. Based on high-quality Flaveria genomes generated, we further conducted an integrated multi-omics study focusing on gene duplications and transcriptional and translational regulations during the evolution of C4 photosynthesis, aiming to uncover the mechanisms underlying the elevated mRNA and protein levels of C4 genes in C4 photosynthesis.
Results
Chromosome-scale genome assemblies of five Flaveria species
The genome sequences of five Flaveria species, i.e., F. robusta (Frob, C3), F. sonorensis (Fson, type I C3–C4), F. linearis (Flin, type I C3–C4), F. ramosissima (Fram, type II C3–C4), and F. trinervia (Ftri, C4)22,23 were constructed using PacBio RSII single-molecule real-time (SMRT) sequencing technology (Fig. 1a). The assembled genome size gradually increased during the evolution of C4 photosynthesis in this genus, ranging from 0.55 Gb in the C3 species Frob to 1.26–1.66 Gb in the C3–C4 species, and reaching 1.8 Gb in the C4 species Ftri (Supplementary Fig. 1a). These findings were corroborated by flow cytometry analysis (Supplementary Data 1). Chromatin conformation capture (Hi-C sequencing) analysis revealed that 98% to 99% of the assembled genome sequences were anchored to 18 pseudo-chromosomes (Fig. 1b, Supplementary Fig. 1b and Supplementary Data 2). To verify the chromosome number of sequenced species, we examined the chromosome number of Frob, Flin, and Ftri using fluorescence in situ hybridization (FISH). All three analyzed species exhibited a chromosome number of 2 × 18 (Fig. 1c), consistent with the reported diploid chromosome number of 36 (2n) for all five Flaveria species15.
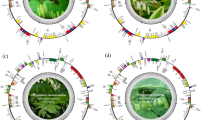
a Summary of phylogeny and timescale of the five Flaveria species and the three indicated outgroup species, i.e., Arabidopsis thaliana (Atha), Helianthus annuus (Hann, sunflower), Lactuca sativa (Lsat, lettuce). Bars represent 95% confidence intervals of the estimated divergence time. Whole genome duplications are shown at the corresponding node/branch. The plant image of Atha, Hann and Lsat were created in BioRender. b The circular representation of pseudochromosomes. From outer to inner side: blue: long terminal repeats density per million base pair (Mb), purple: exon density per Mb, pink: transcript abundance per gene in log10 TPM (transcript per kilobase per million mapped reads). Lines in the inner circle represent links between synteny-selected paralogs. c Fluorescence in situ hybridization images to assess the chromosome numbers in Ftri, Flin, and Frob. This experiment was conducted in three biological replicates and one representative result (c) was shown. d Ks between different species and within species. Ks of Frob vs. Hann and Ftri vs. Hann are shown to represent a speciation between Flaveria and Hann. Ks of Lsat vs. Hann, Frob vs. Lsat are shown to represent the speciation of Asteraceae and Lsat, and Ks of Hann vs. Atha, Lsat vs. Atha are shown to represent the speciation of Asterids II with Atha. Ks of paralogs in Frob and Hann are shown representing a whole genome duplication event (WGD). Frob and Hann shared the WGD2 indicated as the second peak in orange “WGD2”. The first peak for Ks curves for either Frob or Hann represents tandem duplication found in each species. (Frob F. robusta, Fson F. sonorensis, Flin F. linearis, Fram F. ramosissima, Ftri F. trinervia).
The genome completeness of the Flaveria species sequenced in this study was assessed using Benchmarking Universal Single-Copy Orthologues (BUSCO) genes and showed coverage ranging from 92.5% to 99.2%. In line with the high genome completeness, the average genome mapping rate of RNA-seq reads across the five Flaveria species was 94.8%, ranging from 86.7% to 97.6% (Supplementary Data 3). A relatively lower RNA-seq genome mapping rate (86%) was observed in Flin compared to the other four species, likely due to the use of different accessions of Flin for RNA-seq (Sugarloaf Key population) and genome assembly (Yucatan population)22.
A recent whole genome duplication event (WGD), referred to as WGD2, occurred in Asteraceae species, including in Helianthus annuus (sunflower) approximately 29 million years ago (mya)24. Flaveria and sunflower were estimated to have diverged approximately 31.7 mya based on our calibrations (Fig. 1a). To determine whether Flaveria shared the WGD2 with sunflower, we analyzed the Ks distribution of paralogs in both Ftri and sunflower. Two distinct peaks were identified in sunflower, with the higher Ks peak corresponding to WGD2. Similarly, Flaveria species exhibited a peak within the same Ks range (Fig. 1d and Supplementary Fig. 2), suggesting that Flaveria shared the WGD2 with sunflower.
Although the genome size of the C4 species Ftri was tripled compared to the C3 species Frob, the number of protein-coding genes remained comparable, with 35,875 protein-coding genes in Frob (C3), 32,915 in Ftri (C4), and 37,028 to 38,652 predicted in the C3–C4 species (Supplementary Fig. 1a). The synteny of 18 chromosomes was conserved across the five Flaveria species, with 50% to 75% of protein-coding genes being colinear between Frob and the other species (Supplementary Data 2). We compared the predicted protein-coding genes from our assembly with those from Taniguchi’s assembly for the shared species F. robusta (Frob). The results showed that approximately 93.1% of the genes with protein-coding regions of at least 100 amino acids from Taniguchi’s assembly21 were readily covered by our assembly (Blastp, E-value < 0.001, coverage ≥80%) (Supplementary Data 4). Subsequently, we cross-referenced the annotated C4 genes in both assemblies and identified several crucial C4 genes, including CA1, PEPC1, and NADP-ME4, in our assemblies that were absent in earlier assemblies (Supplementary Data 4). This finding underscores the importance of our high-quality chromosome-scale genome assembly in improving the annotation of protein-coding genes in Flaveria species.
Annotating the genes encoding C4 enzymes and transporters
The chromosome-scale assembly of genome sequences and improved gene annotations of five Flaveria species enabled the identification of functional C4 gene copies and provided insights into the evolutionary trajectory by which non-photosynthetic genes evolved into photosynthetic genes. We identified eight enzymes and seven transporters as functional copies of C4 genes by integrating both phylogenetic analysis and transcript abundance data (Fig. 2a and Supplementary Data 5, also see “Methods”), These enzymes include carbonic anhydrase 1 (CA1), phosphoenolpyruvate carboxylase 1 (PEPC1), PEPC kinase 1 (PEPC-k1), NADP-dependent malate dehydrogenase (NADP-MDH), Aspartate aminotransferase (AspAT), alanine aminotransferase (AlaAT), NADP-dependent malic enzyme 4 (NADP-ME4), pyruvate orthophosphate dikinase (PPDK), and PPDK regulatory protein (PPDK-RP). The transporters identified were dicarboxylate transporter 2.1 (DiT2.1 or DCT)25,26, bile acid sodium symporter 2 (BASS2)27, sodium: hydrogen antiporter 1 (NHD1)27, oxaloacetate/malate transporter (OMT or DiT1)28, and phosphate/phosphoenolpyruvate translocator 1 (PPT1)29,30,31. In addition to these verified transporters, we included BASS4, which has been proposed as a pyruvate transporter in bundle sheath chloroplast32,33. Higher transcript abundance of the gene encoding BASS4 was observed in the C4 species compared to the C3 and C3-C4 species (Supplementary Data 5). Our data showed that PEPC-k1 and was absent in the Fram plant sequenced in this study (Supplementary Data 6). Notably, the chromosomal locations of all 15 C4 versions of C4 genes were conserved throughout evolution (Fig. 2b).
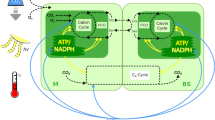
a The diagram of the core C4 pathway in Flaveria C4 species, C4 enzymes, and transports are labeled in red. b Collinearity of chromosomes among Flaveria species. Genes encoding C4 enzymes and transporters are drawn in red line. Dashed lines represent either failure in anchoring to chromosome (NADP-ME4 in Flin) or deletion from the genome (PEPC-k1 in Fram). c Gene tree of PEPC orthologs, PEPCs from Arabidopsis thaliana (Atha) are used as outgroups. PEPC1 (indicated with red circles) is the functional version according to the highest expression levels among all PEPCs. Bars on right of tree show gene expression in transcript per kilobase per million mapped reads (TPM). The bars show mean values ± SD. (n = 3 biological replicates). d Comparative of PEPC1 in Frob (C3) and Ftri (C4). Inverted repeats (blue background) were observed adjacent to the conserved region of FtriPEPC1.3. A 4-bp motif (purple background) flanks the inverted repeats, resembling a target site duplication (TSD) in a transposition event mediated by retrotransposons. (MC mesophyll cell, BSC bundle sheath cell, ALA Alanine, ASP aspartate, CBB Calvin-Benson-Bassham cycle, MAL malate, OAA oxaloacetate, PEP phosphoenolpyruvate, PRY pyruvate, AspAT aspartate aminotransferase, PEPC1 phosphoenolpyruvate carboxylase 1, NHD1 sodium: hydrogen antiporter 1, PEPC-k1 PEPC kinase 1, DCT dicarboxylate transport 2.1 (or DiT2.1), AlaAT alanine aminotransferase, BASS2 bile acid sodium symporter 2, NADP-MDH NADP-dependent malate dehydrogenase, BASS4 bile acid sodium symporter 4, NADP-ME4 NADP-dependent malic enzyme 4, OMT oxaloacetate/malate transporter or dicarboxylate transporter 1 (DiT1), PPDK pyruvate orthophosphate dikinase, PPDK-RP PPDK regulatory protein, PPT1 phosphate/phosphoenolpyruvate translocator 1, CA1 carbonic anhydrase 1.). Source data are provided as a Source Data file.
Since functional copies of C4 enzymes, but not their orthologs in C3 species, have been reported to be induced by light in C4 species34, we verified the predicted functional copies of seven C4 enzymes by examining their responsiveness to light. PEPC and PPDK showed light induction in both C3 and C4 species, but the light induction occurred more rapidly in C4 species than in C3 species, i.e., 2 h upon illumination in the C4 species Ftri compared to 4 h upon illumination in the C3 species Frob. Light induction of CA was observed in C4 (Ftri) and type II C3–C4 species (Fram and Flin) but not in C3 species (Frob) or type I C3–C4 species (Fson). Light induction of PEPC-k and NADP-ME was observed in Fram (type II C3–C4) and Ftri (C4), whereas induction of NADP-MDH and PPDK-RP was restricted in Ftri (C4). Therefore, the light induction of C4 enzymes was most pronounced in C4 species and largely intermediate in C3–C4 species (Supplementary Fig. 3a). These findings align with previous reports30,35, indicating that C4 genes have evolved to become light-responsive over time. Given that orthologs of these C4 genes play roles in primary metabolism within C3 species36, the acquisition of light responsiveness during the evolution of C4 photosynthesis enables these genes to better synchronize their activities with those of other photosynthetic genes, which predominantly exhibit light responsiveness. This supports the accuracy of identifying functional copies of C4 genes and highlights the gradual acquisition of light responsiveness during C4 evolution. Our results also indicate that light responsiveness may serve as a potential criterion for identifying novel C4-related genes.
We also measured enzyme activity for PEPC, NADP-MDH, NADP-ME, and PPDK. The C4 species showed significantly higher enzyme activities compared to all four enzymes than the C3 and C3–C4 species (two-tailed Wilcoxon rank sum tests, BH-adjusted p < 0.05, the exact adjusted p values were provided in the Supplementary Fig. 3b). Specifically, the C4 species displayed approximately 10-fold higher enzyme activities for PEPC and NADP-MDH compared to the C3 species. For NADP-ME, the increase was even more pronounced, aligning with recent observations of enzyme activity in Flaveria species22. Remarkably, Fram (C3–C4) exhibited enzyme activities that were comparable to those of C3 species (PEPC and NADP-MDH) or intermediate between C3 and C4 species (NADP-ME and PPDK) (Supplementary Fig. 3b). Western blot experiment further demonstrated that NADP-ME exhibited intermediate levels between C3 and C4 species in Fram. The other two C3–C4 species, Fson and Flin, showed enzyme activities comparable to those of the C3 species Frob (Supplementary Fig. 3c).
CA, PEPC, and PEPC-k showed additional copies in the C4 species Ftri (Fig. 2c and Supplementary Fig. 4). For example, the C4 version of PEPC, identified as PEPC1 due to its highest transcript abundance among paralogs in C4 species, contained three copies in the C4 species Ftri but only one copy in the other four Flaveria species. The three paralogs of PEPC1 in Ftri, designated as FtriPEPC1.1, FtriPEPC1.2, and FtriPEPC1.3, were located on the same chromosome (Chr3). The presence of the three FtriPEPC1s paralogs on the chromosome was verified by PCR (Supplementary Data 7). Such duplication events of CA, PEPC, or PEPC-k were not observed in other C4 species, including Zea mays (corn; herein Zmay), Setaria italica (foxtail millet), or Sorghum bicolor (sorghum) (Supplementary Data 8), suggesting that these C4-specific gene duplications were not universal.
Retrotransposons are important mediators for gene duplications through retroposition37,38,39,40,41,42. To determine whether the observed C4 gene duplications were associated with retrotransposons in Ftri, we closely examined the evolution and sequences of FtriPEPC1s. Among the three FtriPEPC1 paralogs, FtriPEPC1.1 was predicted to be the ancestral copy, as its mesophyll expression module 1 (MEM1) in the promoter of FtriPEPC1.1 was conserved with that of PEPC1 from the other four Flaveria species (Frob, Fson, Fram, and Flin). In contrast, the MEM1 of FtriPEPC1.2 and FtriPEPC1.3 contained a 109-bp deletion (Supplementary Data 9). Beyond the coding region, sequences approximately 2500 bp upstream and 2000 bp downstream of the coding sequences were also conserved among three FtriPEPC1 paralogs (Supplementary Data 9). Closely examining the sequences near the conserved region showed 9-bp inverted repeat sequences, i.e., 5’-AAAATAAAG-3’. Besides, a 4-bp motif, i.e., 5’-TTTT-3’ (Fig. 2d), immediately flanked the invert repeats, resembling a target site duplication (TSD) characteristic of retrotransposon-mediated transposition events. In line with the observation that all three FtriPEPC1 paralogs shared conserved gene flanking sequences, particularly the MEM1 motif43 (Supplementary Data 9), their transcript abundances were similar and higher than those of PEPC1s in the other four species (Fig. 2c).
A major role of transcriptional regulation in elevated protein levels of C4 genes in Ftri
The increased transcript abundance of C4 genes in C4 species is well-documented44,45. Here, we investigated how protein abundances were modified during evolution using proteomics. We performed proteomics measurements for the five species with six biological replicates for each (Supplementary Fig. 5). To compare protein and transcript levels of paralogous genes across different Flaveria species, we incorporated RNA-seq data of five species from our previous study, which included six replicates for each species20. We found that correlations between samples from the same species were higher than those between different species based on either transcript abundances of detected 27,684 genes or protein abundances of 4908 detected proteins (Supplementary Fig. 6a, b), implying the reliability of RNA and protein quantifications.
Transcript and protein abundances of C4 genes were generally higher in C4 species compared to C3 and C3-C4 species (Fig. 3a). To investigate whether transcriptional or translational regulation is primarily responsible for the observed differences in protein abundance among species with different photosynthetic types, we compared the protein-to-mRNA ratios (PTR) between genes in five Flaveria species. Low PTR genes were defined as those with PTR values less than the mean PTR minus one standard deviation (SD), while high PTR genes had PTR values exceeding the mean PTR plus one SD. The remaining genes were classified as moderate PTR genes (Fig. 3b). An average, 181 low PTR genes (ranging from 138 to 238) and 418 high PTR genes (ranging from 372 to 469) were obtained across the five species (Supplementary Fig. 6c–e and Supplementary Data 10–12). In general, a positive correlation was observed between mRNA and protein levels, with Pearson correlations ranging from 0.36 to 0.53, and most genes exhibited moderate PTRs (Fig. 3c). In C4 species, seven C4 genes were identified as low PTR genes, whereas three or fewer C4 orthologous genes were classified as low PTR genes in the C3 and C3–C4 species (Fig. 3c).
a Heatmaps show relative transcript and protein abundances of C4 genes in the five Flaveria species. Transcript abundance in TPM and protein abundance in iBAQ were normalized using Z-score normalization. PEPC-k1 was excluded as the protein level of C4 version of PEPC-k1 could not be detected in any of these species. b The protein-to-mRNA ratio (PTR) distribution of genes across the five Flaveria species. High PTR and low PTR genes are defined as genes with PTR higher than the mean plus one standard deviation (SD) and with PTR values lower than the mean minus one SD respectively. Enriched function of conserved high PTR and low PTR genes across the five Flaveria species and their enriched function were shown. c Scatter plots of protein vs. transcript abundance of the five Flaveria species. Low PTR and high PRT C4 genes were indicated with arrows. Pearson correlation (PC) between protein abundance and transcript abundance is shown in the parentheses on top of each panel. d PTR values for the C4 gene set in the five Flaveria species, showing that C4 genes have significantly lower PTRs in C4 species Ftri than in the non-C4 species. Data are presented as mean values ± SD. Note that no such decrease is shown for photorespiratory genes, photosynthesis genes, or 100 times of resampling dataset (randomly choosing 14 genes from each species for each resampling). The statistical significance was determined by a one-way ANOVA procedure followed by a two-tailed Wilcoxon rank sum test, p values were adjusted with “BH” (*p < 0.05, **p < 0.01, ***p < 0.001). (Abbreviations for the C4 gene are the same as Fig. 2). Source data are provided as a Source Data file.
The low PTR genes were enriched in gene ontology (GO) categories related to photosynthesis, including chloroplast, light harvesting, and PSII (Supplementary Fig. 6f). This result aligns with a previous study in Arabidopsis, which reported that photosynthesis-related genes exhibited significantly lower PTRs than other genes in photosynthetic leaf tissues46 (Supplementary Data 12). C4 genes showed significantly lower PTRs in C4 species compared to their orthologs in C3 and C3–C4 species (two-tailed Wilcoxon rank sum test, BH-adjusted p < 0.05, the exact adjusted p values were provided in Fig. 3d). In contrast, photorespiratory genes and photosynthesis genes (excluding C4 genes) showed comparable PTRs between C3 and C4 Flaveria species (Fig. 3d). These results suggest that the elevated protein levels of C4 genes in C4 species during the evolution of C4 photosynthesis might be primarily attributed to increased transcriptional abundances.
Translation efficiency of C4 genes between C3 and C4 Flaveria species
In addition to transcriptional regulation, factors such as RNA stability, translation efficiency, and protein stability contribute to increased protein abundance. One noteworthy aspect influencing both transcription and translation is the frequencies of G + C at the third positions of codons (GC3)47,48. We compared the GC3 values of C4 orthologous genes in the five Flaveria species and found no significant differences in GC3 among them (Fig. 4a).
a GC3 comparisons of C4 genes across five Flaveria species. As FramPEPC-k1 is missing, its paralog (FramPEPC-k2, FramNA03444, see Supplementary Fig. 4) was utilized as a comparison for Fram in this context. b Abundances of ribosome protected fragment (RPF) of C4 genes. Data are presented as mean values ± SD. (n = 2 biological replicates). c, d Distribution of RPF-to-RNA ratio and protein-to-RPF ratio. e Scatter plots of RPF vs. RNA of Frob and Ftri. Low/high/moderate RPF-to-RNA genes are labeled in pink/light blue/gray. f Scatter plots of protein vs. RPF of Frob and Ftri. Low/high/moderate protein-to-RPF genes were labeled in pink/light blue/gray. Low protein-to-RPF C4 genes were indicated with red arrows. C4 and photorespiratory genes are labeled in red and blue, and Pearson correlation (PC) between RPF vs. RNA, protein vs. RPF is shown in the parentheses on top of each panel in (e) and (f). g, h RPF-to-RNA ratio and protein-RPF ratio for C4 genes and photorespiratory genes. Data are presented as mean values ± SD. Statistical significance was determined by two-tailed Wilcoxon rank sum test (*p < 0.05, **p < 0.01, ***p < 0.001). Source data are provided as a Source Data file.
To further examine whether significant differences in translation efficiency arose during evolution, we performed ribosome profiling (Ribo-seq) on two representative species: the C3 species Frob and the C4 species Ftri, with two biological replicates for each. As Ribo-seq captures the positions of ribosomes on mRNAs, it provides a direct measure of translational activity. In parallel, RNA-seq was conducted on the same samples for Frob and Ftri to measure transcript abundances for further translational efficiency estimation (Supplementary Fig. 7a). After filtering out rRNA sequences, approximately 35% and 25% of the reads mapped to the genomes of Frob and Ftri, respectively (Supplementary Fig. 7a), which was comparable to those reported in other species, such as 12% in maize49 and 16% in Saccharomyces cerevisiae50. The Ribo-seq data demonstrated clear triplet periodicity on codons in the reference transcriptome (Supplementary Fig. 7b). The read length distribution peaked at 27 to 32 nucleotides, with 94% of fragments mapping to the gene region (UTR exons, coding exons, and introns) originating from the coding exons (Supplementary Fig. 7c, d).
Principal component analysis (PCA) based on transcript per kilobase per million mapped reads (TPM) of either RNA-seq or Ribo-seq data showed that samples from Frob were well separated from those of Ftri, with the first principal component explaining 63% and 65% of the total variance, respectively (Supplementary Fig. 7e). Consistent with RNA-seq results, C4 genes from Ftri exhibited higher transcript abundances compared to their counterparts in Frob based on Ribo-seq (Fig. 4b). We then estimated the translation efficiency of each gene as a ratio of ribosome-protected fragment (RPF) abundance to RNA abundance and further compared the transcriptional efficiency between species. To ensure comparability, the translation efficiency was normalized by the mean translation efficiency of all photosynthesis genes, excluding C4 genes. The distribution of translation efficiencies was comparable between Frob and Ftri after normalization (Fig. 4c). Similarly, the distribution of protein-to-RPF abundance ratios was also comparable between Frob and Ftri (Fig. 4d).
A relatively high correlation between RPF and RNA abundances was observed, with Pearson correlation coefficients of 0.81 and 0.88 in Frob and Ftri, respectively (Fig. 4e). The correlation between protein and RPF abundances was intermediate to the RPF vs. RNA and protein vs. RNA correlations, with Pearson correlation coefficients of 0.57 in Frob and 0.58 in Ftri (Fig. 4f). Given that translation efficiency varied over a wide range due to the detection of a relatively large number of genes with both RNA and RPF (~17,000 genes), we defined high and low translation efficiency genes as the top 5% and bottom 5% of genes ranked by translation efficiency, resulting in 447 and 507 high-translation-efficiency genes and 447 and 508 low-translation-efficiency genes in Frob and Ftri, respectively. Notably, all C4 genes, except PEPC-k in Ftri, and a large proportion of photorespiratory genes were classified as having intermediate translation efficiency in both species (Fig. 4e). Consistently, translation efficiency did not differ significantly between Frob and Ftri for either C4 genes or photorespiratory genes (Fig. 4g).
We also defined high and low protein-RPF-ratio genes the same way as high and low translation-efficiency genes, 254 and 250 high-ratio genes and 250 and 253 low-ratio genes in Frob and Ftri, respectively (Fig. 4f). In line with the protein and RNA comparisons, more C4 genes fell into the category of low protein-RPF-ratio genes, and C4 proteins showed significantly lower protein-RPF ratios in Ftri compared to Frob (two-tailed Wilcoxon rank sum test, p = 0.019, Fig. 4h). In contrast, photorespiratory proteins exhibited comparable protein-RPF ratios between these two species. These findings suggest that the observed decreased protein-RPF ratios for C4 genes may primarily attributable to the increased transcriptional abundances rather than changes in translation efficiency.
Predicted cis-regulatory elements and transcription factors associated with the regulation of C4 genes in Ftri
Having established a major role of the transcriptional regulation in C4 genes during evolution, we then explored how cis-regulatory elements (CREs) were modified along the evolution of C4 genes. We first investigated the enriched CREs within the promoter regions of C4 genes in the C4 species Ftri. The results revealed that C4 genes in Ftri were enriched with nine known CREs, three of which were identified as ethylene response factor (ERF) CREs. In contrast, the other four species showed at most one enriched CRE of their C4 orthologous genes in ERF (Supplementary Fig. 8). To ascertain whether the ERF CREs were localized within accessible chromatin regions (ACRs) of C4 genes in the C4 species Ftri, we analyzed the enriched CREs within ACRs (ACR-CREs) using data from two biological replicates of transposase-accessible chromatin sequencing (ATAC-seq) experiments (Supplementary Fig. 9).
During ATAC-seq experiments, the Tn5 transposase enzyme shows strong preferential binding to nucleosome-free DNA regions, generating sequencing tags that correspond to open chromatin. Consequently, Tn5 transposase-sensitive sites often exhibit peaks at gene transcription start sites. Our data also showed that Tn5 transposase-sensitive sites in the ATAC-seq reads showed a prominent peak upstream of gene transcription start sites (Supplementary Fig. 9a, b). We obtained 14,443 conserved peaks from the two replicates after applying Irreducible Discovery Rate (IDR) less than 0.05 (Supplementary Data 13), with 48% of Tn5 peaks mapping to the gene promoter region (3k bp upstream of the start codon) (Supplementary Fig. 9b). Tn5 peaks were evident in the promoter regions of photosynthetic genes and C4 genes (Supplementary Fig. 9c), including Rubisco small subunit 1b (RUBSC1b), Light-harvesting complex a 1b (Lhca1b), and proton gradient regulation 5-like (PGR5-like). Due to the complete sequence identity in the upstream regions of the three FtriPEPC1 paralogs, chromatin accessibility showed consistent patterns across these genes (Fig. 5a).
a Integrated Genome Viewer (IGV) of RNA-seq reads and two-biological replicates of ATAC-seq reads of three PEPC1s in Ftri (C4). b Word cloud shows the log2-transformed frequency of enriched cis-regulatory elements (CREs) identified through Monte Carlo permutation testing (FDR < 0.05) in accessible chromatin regions (ACR)-CREs associated with C4 genes based on ATAC-seq in Ftri. c, d Bar plots show the top five most abundant TF families of all annotated TFs and TFs that are from the C4 gene-TF regulatory network, respectively, with the latter were termed as C4GRN TFs. e C4 gene-TF regulatory network of the C4 species Ftri. Lines represent predicted regulatory interactions between TFs and C4 genes. ERFs are highlighted in pink. f Electrophoretic mobility shift assay (EMSA) was performed with ERF TFs and cy5-labled partial DNA sequence (probe) of their regulated C4 genes’ promoter from Ftri. Labeled probes were incubated with GST-TF-10xHis protein. GST represents GST−10xHis protein without TF. For competition analysis, the binding reaction was performed with addition of 200-fold of corresponding unlabeled probe. Bands corresponding to DNA-protein complexes (shift) or free probes are indicated by arrows. Predicted ERF motif was shown on the bottom of each panel. The EMSA experiment was conducted in three biological replicates and one representative result (f) was shown. g Structure of reporter and effector plasmids for transient transcription assay. For the reporter constructs, 35S promoter, C4 gene (CA1.1 or PEPC1.1) promoter, firefly luciferase (LUC) and Renilla luciferase (REN) are indicated. For the effector construct, ERF TF (ERF12 or ERF61) was driven by the 35S promoter. h, i ERF12 and ERF61 activates the promoters of CA1.1 and PEPC1.1 (Significance was calculated with two tailed T-test, n = 3 biological replicates). Data are presented as mean values ± SD. Leaf epidermal cells of Nicotiana benthamiana were transfected with infiltration buffer (Mock), reporter DNA (pCA1.1 or pPEPC1.1) with Flag, reporter DNA with ERF-Flag (ERF12 or ERF61). Source data are provided as a Source Data file.
We categorized gene-associated ACRs-CREs into three types according to their distance from the nearest gene: genic (gACR-CREs; overlapping a gene), upstream (upACR-CREs; within 3 kb upstream of a gene’s start codon), and downstream (downACR-CREs; within 3 kb downstream of a gene’s stop codon). We then calculated enriched CREs in ACR-CREs (“Methods”). Among all three types of ACR-CREs, ERF CREs were the most abundant enriched CREs (Supplementary Fig. 9d). Moreover, ERF CREs dominated the enriched ACR-CREs of C4 genes (Fig. 5b) and were also prevalent in photosynthetic and photorespiratory genes (Supplementary Fig. 9e). Notably, ERF CREs were abundant in photosynthesis-related genes of other C4 species, including maize, foxtail millet, and sorghum (Supplementary Data 14).
We have constructed gene regulatory networks (GRNs) for Frob, Fson, Fram, and Ftri based on at least 22 RNA-seq datasets previously20. In this study, the GRNs for these four species were further refined by incorporating species-specific gene annotations. Additionally, we developed a GRN for Flin, leveraging data from 18 RNA-seq datasets explicitly generated for this study (Supplementary Data 15). Besides, transcription factors (TFs) without predicted cognate CRE families within 3 kb upstream of the start codon were filtered out. Sub-GRNs comprising C4 genes and their regulating TFs were constructed for each species and were termed C4GRNs. The C4GRNs of Frob, Fson, Fram, and Ftri reconstructed in this study were largely consistent with previously constructed GRNs annotated using transcriptomic data of Fram20. However, the number of regulated TFs was increased due to improved TF annotations based on our high-quality genome assemblies (Supplementary Data 15).
Regarding the total annotated TFs, the number of TFs within each TF family remained comparable across all five Flaveria species. However, notable distinctions were observed for TFs within the C4GRN (Fig. 5c, d and Supplementary Fig. 10). The ERF, bHLH, MYB, NAC, and C2H2 emerged as the top five most abundant TF families in the C4GRN of the five Flaveria species (Fig. 5d). In the C4 species Ftri, 323 TFs were predicted to regulate C4 genes (Fig. 5e). Notably, ERF TFs were much more prevalent in the C4GRN of the C4 species compared to other species, either considering the total number of ERF TFs in C4GRN or the number of ERF TFs per C4 genes (Supplementary Data 15). In contrast, the number of predicted ERF TFs were comparable across all five Flaveria species (Fig. 5c). These findings suggest a preferential recruitment of ERF TFs for regulating C4 genes during the evolution of the genus Flaveria.
We performed an electrophoretic mobility-shift assay (EMSA) to verify the predicted regulation of C4 genes by ERF TFs in the C4 species Ftri. Seven ERF TFs predicted to regulate FtriCA1, PEPC1.1 and PEPC-k1.1 were selected for the EMSA experiment. Cognate ERF TF binding site (TFBS) located within 2 kb upstream of their (CA1, PEPC1.1, or PEPC-k1.1) start codon were selected to perform EMSA. EMSA experiments verified the binding of ERF12 to the promoter of Ftri PEPC1.1 and PEPC-k1.1, as well as the binding of ERF61, ERF51, and ERF1 to the promoter of CA1.1 (Fig. 5f). We then performed transient transcription assay to further verify the regulation of ERF12 and ERF61 on C4 genes. We used dual-luciferase reporter plasmids, containing the firefly luciferase (LUC) gene driven by CA1.1 promoter (200 bp from the start codon) and PEPC1.1 promoter (250 bp from the start codon) and the Renilla luciferase (REN) gene driven by the constitutive 35S promoter, in the analysis (Fig. 5g). The results showed that ERF12 and ERF61 displayed significantly higher LUC/REN ratio compared with Flag tag (Fig. 5h, i), suggesting the activation role of ERF12 and ERF61 on CA1.1 and PEPC1.1.
Taken together, these results indicate that ERF CREs and ERF TFs are involved in the regulation of C4 genes in the C4 species Ftri.
Discussion
The high-quality chromosome-level genome assemblies and multi-omics data from five Flaveria species provide valuable resources for investigating the evolutionary and regulatory mechanisms of C4 photosynthesis. Our comprehensive study revealed that tandem duplication, rather than whole-genome duplication (WGD), likely contributed to the increased transcript abundance of C4 genes. Mechanisms controlling the up-regulation of C4 genes’ protein abundances during evolution were also explored, revealing that increased RNA abundances may predominantly drive the observed increased ribosome-protected fragments (RPFs) and protein abundances of C4 genes. Additionally, ethylene response factor (ERF) transcription factors (TFs) and their cognate cis-regulatory elements (CREs) were identified as being associated with the regulation of C4 genes in the C4 species Ftri.
Transposable elements have contributed to the expansion of genome size in Flaveria species during evolution (Supplementary Fig. 1). In this study, we found that three C4 genes (CA1, PEPC1, and PEPC-k1) gained additional copies through retrotransposons, contributing to the elevated transcript abundances of these genes in the C4 species Ftri (Fig. 2c and Supplementary Fig. 4). While the duplication is intriguing, its specific role in C4 evolution remains speculative. We propose that it might be a beneficial event facilitating the progression from the C3–C4 intermediate to a full C4 state, without implying an inevitable progression towards a full C4 photosynthetic pathway. Given that the chromosome number remains consistently at 2 × 18 for known Flaveria species, with the exception of two F. pringlei collections, the only known polyploids in this genus15, WGD events appear to be rare in the Flaveria genus. This contrasts with Gynandropsis gynandra51,52, where WGD has been implicated as a major factor contributing to the increased gene copy number during the evolution of C4 photosynthesis. The discovery of retrotransposon-mediated gene duplications offers critical insights into the genomic complexity and adaptability that facilitated the evolution of C4 photosynthesis, highlighting the diverse evolutionary strategies underlying C4 photosynthesis.
It has been known that RNA and protein levels of C4 genes are elevated in C4 species compared to C3 species32,44,45, but the dominant mechanism underlying these changes remains unclear. Transcriptional regulation has been identified as a key mechanism in the evolution of C4 photosynthesis53,54. In Flaveria, notably in the C4 species Ftri, C4 genes exhibited elevated RNA, RPFs, protein levels compared to non-C4 species (Figs. 3a, 4b and Supplementary Fig. 3). Despite overall comparable protein-to-RNA (PTR) values across the five Flaveria species, C4 genes exhibited lower PTR ratios in C4 species compared to non-C4 species (Fig. 3c, d). Moreover, the translation efficiency (defined as the ratio of RPF to RNA) of C4 genes in C4 species and their counterparts in non-C4 species is comparable (Fig. 4e, g), further suggesting that increased RNA abundances predominantly drive the increased RPF and protein abundances of C4 genes. This is consistent with our finding that GC3, which is widely recognized for its impact on translation efficiency47,48, is comparable among these C4 orthologs across the five Flaveria species (Fig. 4a). Nevertheless, it is important to acknowledge that other factors, such as epigenetic regulation and post-transcriptional/post-translational processes, could also influence protein abundance, warranting further exploration.
The acquisition of new cis-regulatory elements and new transcription factors has been crucial in enhancing the transcript levels of C4 genes53,54. Our research revealed a pronounced association between ERF CREs and ERF TFs with C4 genes in C4 species compared to non-C4 species (Fig. 5). Notably, ERF CREs were highly abundant in genes related to photosynthesis in C4 species, such as maize, foxtail millet, and sorghum (Supplementary Data 14), suggesting the widespread presence of ERF cis-regulatory elements in photosynthesis-related genes across various plants. ERF transcription factors are widely recognized for their roles in plant stress responses55,56,57, and their heightened association with C4 genes suggests a contributions to the evolution of C4 photosynthesis, possibly as an adaptation to environmental stressors such as low CO2, drought, high light, and high-temperature conditions58,59. Transposable elements have also been linked to plant stress responses60,61 and are capable to drive rapid phenotypic changes62. The observed associations between transposable elements, ERF transcription factors, and the evolution of C4 photosynthesis highlight C4 photosynthesis as a strategic adaptation of plants to environmental stresses.
ATAC-seq is an important genomic approach for facilitating the genome-wide identification of cis-regulatory elements63,64,65. However, obtaining high-quality ATAC-seq data remains challenging, especially for non-model species, including those in Flaveria genus. In this study, we obtained ATAC-seq data only from C4 species (Ftri). Although considerable effort has been devoted to ATAC-seq experiments in other Flaveria species, we were unable to obtain ATAC-seq data of comparable quality to that of the C4 species (Ftri). Based on ATAC-seq data from C4 species (Ftri), we provided evidence that ERF CREs were enriched in the open chromatin regions associated with C4 genes (Fig. 5b). Importantly, the electrophoretic mobility-shift assay (EMSA) and transient transcription assay further verified the regulation of ERF TFs on the expression of C4 genes. Nevertheless, high-quality ATAC-seq data from species of Flaveria genus other than C4 species (Ftri) are critical for further deepening our understanding of the regulatory and evolutionary mechanisms underlying the formation of C4-specific photosynthesis, which requires further exploration.
The Flaveria genus has historically been used as a model system to study the evolution of C4 photosynthesis, leading a substantial body of knowledge on this topic. With the high-quality chromosome-level reference genomes and comprehensive genomic resources provided in this study, we anticipate that the Flaveria genus will accelerate the investigation into the genetic basis of C4 photosynthesis evolution. Initial analyses supported by these data highlight the critical role of transcriptional control, particularly the ERF TFs, in the regulation of C4 gene expression. By leveraging the comprehensive genomic data generated in this study, researchers can further explore the genetic and regulatory elements that drive the development of C4 photosynthesis in Flaveria and other plant species. Moreover, this study provides a wealth of data that can serve as a foundation to explore the genomic features and evolutionary stages of different intermediate species within the Flaveria genus. For instance, our comprehensive dataset allows detailed comparisons between C3–C4 species from clade A (Fson and Fram) and clade B (Flin) of this genus. Such analyses may uncover the mechanisms underlying the absence of true C4 photosynthesis in clade B, thereby providing deeper insights into the evolutionary dynamics and genetic factors that influence photosynthetic pathway development.
Methods
Plant materials and fluorescence in situ hybridization assay
F. robusta (Frob, C3) and F. ramosissima (Fram, C3–C4) were provided by Professor Peter Westhoff from Heinrich Heine University, Germany. F. sonorensis (Fson, C3–C4), F. linearis (Flin, C3–C4), and F. trinervia (Ftri, C4) were obtained from Professor Rowan F. Sage at the University of Toronto, Canada. The plants were grown in soil in a greenhouse as depicted in ref. 30.
The chromosome numbers of Frob, Flin, and Ftri were determined using fluorescence in situ hybridization assay (FISH). Mitotic metaphase spreads of meristem root tip cells were prepared following66. FISH was performed following67 with slight modifications, which were depicted in Supplementary Data 2.
Genome sequencing
Genomic DNA was extracted from young leaves. PacBio sequencing libraries were constructed following the guidelines of Pacific Biosciences (USA). DNA fragments of 0.5–18 kb were selected using BluePippin electrophoresis (Sage Science, USA). Libraries were then sequenced on the PacBio Sequel platform (PacBio, USA). The N50 of PacBio reads ranged from 16.4 to 21.9 kbp. Approximately 120 GB of data were produced for each species on average. Genome coverage ranged from 66.9-fold (Ftri) to 232.2-fold (Frob). Besides, short reads were sequenced using the Illumina X Ten platform in paired-end 150 bp mode. Approximately 200 million short reads were obtained for each species and used for genome assembly polishing and completeness estimation. Hi-C libraries were constructed following68. Two Hi-C libraries were constructed for each species with an insert size of ~350 bp, and sequenced on the Illumina X Ten platform. Between 291 Gb and 325 Gb of 150-bp paired-ended clean data were generated for each species.
De novo assembly
Flaveria nuclear genome sequences were assembled into 18 pseudochromosomes in a step-wise way. Sequencing adapters were removed, and low-quality or short reads were filtered using PacBio SMRT Analysis package with the following parameters: readScore = 0.75; minSubReadLength = 50. The remaining high-quality PacBio subreads were then corrected and contigs were assembled using Canu (v1.8)69 with the following parameters: useGrid = true, minThreads = 4, genomeSize = 1200 m, minOverlapLength = 500, minReadLength = 1000. For contig polishing, the Illumina paired-end reads were mapped to assembled contigs applying bwa mem (bwa v0.7.17)70, low qualified mapped reads were filtered off applying samtools (v1.11)71 with q30 setting. Pilon (v1.22)72 was used for polishing with the following parameters: –mindepth 10 –changes –fix bases.
For Fram specifically, the BioNano next-generation mapping system was used to facilitate high-quality genome assembly. DNA was labeled at Nt.BspQI sites using the IrysPrep kit (BioNano Genomics, USA). Molecules collected from BioNano chips (BioNano Genomics, USA) were de novo assembled using RefAligne and Assembler from the BioNano73 using the following parameters: -U -d -T 20 -j 4 -N 10 -i 5, resulting in the optical genome maps. Next, genome assembly generated from Pilon (v1.22)72 mentioned above was then evaluated and corrected by aligning with the optical genome maps. Corrected contigs and optical genome maps were aligned and merged using hybridScaffold.pl73, resulting in hybrid scaffolds. Next, HERA74 was used to fill gaps in the obtained hybrid scaffold in the following parameters: InterIncluded_Side = 30000, InterIncluded_Identity = 99, InterIncluded_Coverage = 99, MinIdentity = 97, MinCoverage = 90, MinLength = 5000, MinIdentity_Overlap = 97, MinOverlap_Overlap = 1000, MaxOverhang_Overlap = 100, MinExtend_Overlap = 500. Obtained hybrid scaffolds were then used for the following assembly.
Subsequently, assembled genome sequences were improved using Hi-C data in two steps. First, contigs were corrected using Hi-C data. Briefly, low-quality Hi-C data (over 10% N base pairs or Q10 < 50%) were removed, and remaining reads were mapped to assembled contigs applying bwa (v0.7.17)70 with “aln” settings and other parameters were in default (https://bio-bwa.sourceforge.net/bwa.shtml). Only uniquely mapped reads were used for re-assembly. Invalid mapping was filtered using HiC-Pro (v2.11.1)75 with the following settings: mapped_2hic_fragments.py -v -S -s 100 -l 1000 -a -f -r -o. Next, corrected contigs were re-assembled into a scaffold using LACHESIS76 with the following parameters: CLUSTER MIN RE SITES = 770, CLUSTER MAX LINK DENSITY = 2, CLUSTER NONINFORMATIVE RATIO = 2, ORDER MIN N RES IN TRUNK = 578, ORDER MIN N RES IN SHREDS = 593.
Annotation of transposable elements
To predict transposable elements (TEs), whole genome sequences of the five Flaveria species were searched for repetitive sequences individually. A de novo repeat sequence library was constructed using RepeatModeler (RepeatModeler-Open-1.0.5) with the following parameters: RepeatModeler -database database_name -engine ncbi -pa [int]. RepeatMasker (RepeatMasker-Open-4.1.0) was then used to search for similar TEs against the de novo library with the following parameters: RepeatMasker genome. fa -lib de_novo_library -nolow -no_is -q -engine rmblast -pa [int] –norna. Intact long terminal repeat retrotransposons (LTR-RTs) were identified using LTR_FINDER (v1.07)77 and LTRharvest (v1.5.10)78. Then LTR_Retriever (v2.9.0)79 was used to merge the above results with the parameters: LTR_retriever -genome genome.fa -inharvest species.harvest.scn -infinder species.finder.scn –nonTGCA species.harvest.nonTGCA.scn. The insertion time of intact LTR-RT was extracted from LTR-Retriever analysis.
Annotation of protein-coding genes
Gene models were predicted using a combination of de novo prediction, homology-based, and transcriptome-based strategies. Briefly, Augustus (v2.4)80, GlimmerHMM (v3.0.4)81, GeneID (v1.4)82, and Genscan (http://genes.mit.edu/GENSCAN.html) were used in combination for de novo prediction. GeMoMa (v1.3.1)83 was used for homology-based prediction. To facilitate gene annotation, 18 to 32 Illumina RNA-seq datasets were generated either in this study (for Flin, as depicted below) or generated in our previous work20. Clean RNA-seq reads were mapped to the genome using Hisat2 (v2.0.4)84 with “ -k 5”, and genome-based transcript assembly was performed applying StringTie (v1.2.3)85 with “-T 0 -F 0”. Additionally, de novo transcript assembly was conducted using PASA (v2.0.2)86 in default parameters based on RNA-seq data. All predicted gene structures were integrated into consensus gene models using EVidenceModeler (v1.1.1)87, and pseudogenes were predicted applying GeneWise (v2.4.1)88. Coding sequence (CDS) failed to be translated either lacking an open reading frame (ORF) or having premature stop codons were removed.
The completeness of protein repertoire was estimated based on: (1) using BUSCO (v3.0.2)89 against viridiplantae reference, (2) RNA-seq reads mapping to genome applying STAR (v2.7.3a)90, and (3) 150-bp paired-ended DNA sequencing reads mapping to genome apply bowtie2 (v2.3.4.3)91 (Supplementary Data 3).
Putative gene functions were assigned using the best matches to GO, KEGG, Swiss-Prot, TrEMBL, and a non-redundant protein database (NR) using BLASTP (v2.2.31+)92 with the E value threshold of 1e-5.
Transcription factors were predicted using the online website PlantTFDB (v5.0)93,94 (http://planttfdb.gao-lab.org/prediction.php). Cis-regulatory elements (CREs) in promoter regions (3 kb upstream of the start codon) were predicted using Plantpan (v3.0)95 with a score threshold of 0.85.
Orthologous genes prediction and gene evolution
To predict orthologous groups, protein-coding genes from the five Flaveria species, Arabidopsis thaliana (Atha), Helianthus annuus (Hann, sunflower), and Lactuca sativa (Lsat, lettuce) were processed using Orthofinder (v2.3.11)96 “diamond” was used for sequence search, and “fasttree” was used for tree inference. The protein sequences of Atha (TAIR10), Hann (v1.0), and Lsat (v7) were downloaded from Phytozome (v13) (https://phytozome.jgi.doe.gov/pz/portal.html). For genes with multiple alternative transcripts, the longest one was kept to represent the protein-coding gene.
Phylogeny and divergence time analysis
To construct the phylogenetic tree, CDS sequences of 1:1 orthologous genes were aligned using MUSCLE (v3.8.31)97 with the options “-stable -quiet”. Alignments of all the CDS were concatenated to create a supermatrix, and then RAxML (v7.9.3)98 was applied to infer phylogenetic tree using the following model: GTR (General Time Reversible nucleotide substitution model) + GAMMA (variations in sites follow GAMMA distribution) + I (a portion of Invariant sites in a sequence). To calibrate the evolutionary time, CDS were aligned codon-wisely guided by protein alignment using pal2nal (v14)99. The evolutionary time was calibrated using mcmctree in PAML package (v4.9)100 using the following parameters: seqtype = 0 (nucleotides), clock = 2 (independent), model = 0 (JC69). The reported fossil divergence time between Hann and Lsat (34–40 million years), as inferred from timetree (http://timetree.org/), was used for calibration. The phylogenetic tree and calibrated evolutionary time were visualized using FigTree (http://tree.bio.ed.ac.uk/software/Figuretree/).
Synteny analysis between Flaveria species
To identify syntenic gene blocks in each species and between Frob and other four species, all-against-all BLASTP (E value < 1e − 10, top five matches) (v2.2.31+)92 was performed for protein coding genes for each genome pair. Syntenic blocks were determined according to the presence of at least five synteny gene pairs using MCScanX (-e 1e-10) (v0.8)101. The colinearity of the five species were visualized with JCVI (https://github.com/tanghaibao/jcvi). The circular graphic was plotted using Circos (v0.69-5).
Estimation of genome duplications and speciation
To estimate the duration of whole genome duplication events and speciation events, pair-wise paralogs and orthologs were aligned in protein sequences, and CDS alignment codon-wise was generated based on protein alignment using pal2nal software (v14)99. Synonymous substitution (Ks) values were then calculated using the codeml program in the PAML package (v4.9)100. The following parameters were used: runmode = −2, seqtype = 1(codon sequences), codonFreq = 2 (F2X4) and alpha fixed to 0.
Verification of functional copy of C4 genes using qRT-PCR
As most C4 genes belong to multiple-gene families12. The functional copy of C4 genes was determined with the following criteria: (1) the highest transcript abundances within its paralogous group in C4 species and (2) higher transcript abundance in C4 species compared to its counterparts in C3 species. Since the functional copy of C4 genes exhibits faster light responsiveness in C4 species but not in C3 species30,35, we verified the identified C4 version of C4 genes by investigating the changes of gene expression in response to light induction using quantitative real-time PCR (qRT-PCR). Flaveria species were placed in a dark room at 6:00 p.m. The dark-adapted plants were illuminated at 9:00 a.m. the next day. Fully expanded leaves, typically the 2nd or 3rd leaf pair counted from the top, were collected after illumination for 0, 2, or 4 h, and immediately flash-frozen in liquid nitrogen. Samples were stored at −80 °C before processing. RNA isolation and qRT-PCR were performed as previously described20. Relative transcript abundances were calculated using ACTIN7 as the reference gene, and the primers used were as described in our previous study20.
C4 enzyme western blot and enzyme activity measurements
Western blots for PEPC, NADP-ME, and PPDK were performed using 0.6 g of fresh, fully expanded leaf tissue. Actin was used as a loading control. The antibody of PEPC and NADP-ME were custom-developed by Orizymes Biotechnologies Company (Shanghai). The antibody of PPDK was from Orizymes Biotechnologies Company (Shanghai) (catalog number: PAB07103). The antibody of Actin was from Yamei (Shanghai) (catalog number: LF208S). For all experiments, these antibodies were diluted to a working concentration of 1:5000.
PEPC activity was assayed following the method described in reference102. NADP-ME and NADP-MDH activities were determined following the method described in reference103. The PPDK activity was assayed following the method described in reference104. A FlexA-200HT UV Spectrophotometer (VARIAN Co. Ltd., USA) was used to monitor the consumption or generation of NAD(P)H at 340 nm.
RNA-seq and transcriptional quantification for Flaveria species
RNA-seq data of Flin were obtained from plants grown under low CO2 (100 ppm) and normal CO2 (380 ppm) for 2 weeks and 4 weeks, respectively. Additionally, plants grown under high light (with PPFD of 1400 μmol m−2 s−1) and control light condition (500 μmol m−2 s−1) were sequenced independently. Growth conditions were as described in ref. 20. For RNA extraction, the young fully expanded leaf typically situated on the 2nd or 3rd pair of leaves counting started from the top was used. The selected leaves were cut and immediately frozen into liquid nitrogen and stored thereafter at −80 °C until further processing. Total RNA was then isolated following the protocol of the PureLInkTM RNA kit (Thermo Fisher Scientific, USA). The RNA sequencing was performed on the Illumina platform in paired-end mode with a read length of 150 bp. RNA-seq data of the other four species were obtained from our previous study20.
RNA-seq data for Frob and Ftri were obtained from mature leaves from plants grown in the phytotron with a PPFD of 500 μmol m−2 s−1, a temperature of 25 °C ± 2 °C, 70% relative humidity, and a 16-h light/8-h dark photoperiod. Two biological replicates were used for each species. Following RNA extraction, mRNA was enriched using mRNA Capture Beads. The RNA sequencing was performed on the Illumina NovaSeq X Plus by Gene Denovo Biotechnology Co., Ltd (Guangzhou, China).
To quantify the expression level of Flaveria genes, raw reads were trimmed using fastp (v0.20.0)105 in default parameters, filtering reads if 40% of bases were unqualified (phred quality < 15). Transcript abundance of genes were calculated by mapping RNA-seq reads to the assembly genome sequence of corresponding species using RSEM (v1.3.3)106, with STAR (v2.7.3a)90 as the mapping tool.
Proteomics of five Flaveria species
Approximately 0.1 g of mature leaves were collected from 1-month-old plants and immediately frozen in liquid nitrogen. The plants were grown in the phytotron under the same conditions as those mentioned for the RNA-seq samples of Frob and Ftri. Six biological replicates were prepared for each species. Frozen leaf samples were finely ground and then incubated in 0.6 ml lysis buffer (100 mM Tris-Base, 100 mM EDTA, 50 mM Borax, 50 mM Ascorbic Acid, 30% (m/v) Sucrose, Triton X-100 (final concentration 1%), 10 mM TCEP, 1 mM PMSF, complete EDTA-free protease inhibitor cocktail (PIC) (Roche)). The lysis buffer was freshly prepared, and its pH was adjusted to 8.2 using ammonium hydroxide (NH4OH). After adding TCEP, the pH was readjusted to 8.0. The buffer was stored at −80 °C until needed and thawed at room temperature before use. Samples were centrifuged at 14,000 × g for 10 min at 4 °C. The supernatant was retained for total protein extraction. Total protein concentration was determined with a Bradford assay107.
Details of protein digestion, HPLC fractionation, and LC-MS/MS analysis are provided in Supplementary Data 11. Briefly, peptides were pre-fractionated to generate data data-dependent acquisition (DDA) library. Fractionated peptides were mixed from all the 30 samples (a total of 200 μg). The mixture was separated using a linear gradient and 30 fractions were combined into 15 components. Raw data from each species were utilized to construct libraries based on their respective protein sequences. As a result, five peptide libraries were obtained, one for each species. Finally, data-independent acquisition (DIA) was performed using Spectronaut (version 14.7, Biognosys, Zurich, Switzerland). Default settings for MS1-level quantification were applied. The mass spectrometry proteomics data have been deposited in the PRteomics IDEntifications Database (PRIDE).
For the inter-species comparison among Flaveria species, orthologous gene pairs between the remaining four Flaveria species and Frob were predicted using blast (v2.2.31+)92. The top hits were identified with an E-value threshold of 1e−5 and a sequence identity requirement of at least 60%. A K-means clustering analysis was performed separately on the transcript abundance and protein abundance data, using the unified Frob annotation.
Ribosome profiling of Frob and Ftri
For ribosome profiling (Ribo-seq), mature leaves of Frob and Ftri were collected from same plants used for RNA-seq, as described above. Two biological replicates were prepared for each species. The leaves were immediately frozen in liquid nitrogen and ground into fine powder.
The ribosome profiling was performed with slight modifications to a previously reported protocol108. Specifically, the powder was resuspended in 400 μL of lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 100 μg/mL cycloheximide, and 1% [v/v] Triton X-100). The mixture was incubated on ice for 10 min and then centrifuged at 20,000 × g for 10 min at 4 °C. The supernatant was collected.
Ribosome footprints (RFs) were prepared by adding 10 μL of RNase I and 6 μL of DNase I (NEB, Ipswich, MA, USA) to 400 μL of the collected supernatant and incubated at room temperature for 45 min. The nuclease digestion was terminated by adding 10 μL of SUPERase-In RNase inhibitor (Ambion, Austin, TX, USA). Next, 100 μL of the digested RFs was loaded onto a pre-equilibrated size exclusion column (Illustra MicroSpin S-400 HR Columns; GE Healthcare) and eluted by centrifugation at 600 × g for 2 min. RFs longer than 17 nucleotides (nt) were isolated using an RNA Clean and Concentrator-25 kit (Zymo Research). Antisense DNA probes complementary to ribosomal RNA (rRNA) sequences were used to remove rRNA, and the RFs were further purified using magnetic beads (Vazyme, China). Ribo-seq libraries were prepared using the NEBNext Multiple Small RNA Library Prep Set for Illumina® (NEB, E7300S and E7300L). Sequencing was performed on the Illumina NovaSeq X Plus by Gene Denovo Biotechnology Co., Ltd. (Guangzhou, China).
For Ribo-seq data analysis, low-quality reads and adapter sequences were filtered and trimmed using fastp (v0.20.0)105. Reads with lengths ranging between 20 and 40 bp were retained for subsequent analysis. Remaining reads were mapped to the rRNA database, GenBank, and Rfam database using bowtie2 (v2.3.4.3)91. Reads aligned to rRNA, transfer RNAs, small nuclear RNAs, small nucleolar RNAs, and microRNAs were excluded. The remaining reads were aligned to the respective genome using STAR (v2.7.3a)90 with 2-pass setting enabled. Gene expression levels were quantified with RSEM (v1.3.3)106. RFs were assigned to different genomic features (5’UTR, CDS, 3’UTR and intron) according to the position of the 5’ end of the alignments. Three-nucleotide periodicity was visualized using the riboWaltz package (v2.0)109.
ATAC-seq for the C4 species Ftri
To isolate nuclei from the C4 species Ftri, fully expanded mature leaves were harvested at 1:00 pm. Approximately 3 g of fresh leaves from five plants were used for each of the two biological replicates. Leaf material was ground in ice in 10 ml 4xNE buffer (40 mM MES -KOH, PH5.4, 40 mM NaCl, 40 mM KCl, 10 mM EDTA, 1 M Sucrose, 0.1 mM spermidine, 0.5 mM spermine and 1 mM DTT). Next, the debris was removed by sieving the mixture through two layers of 70 μm nylon cell strainer into precooled flasks. The filtrate was then centrifuged at 200 × g at 4 °C for 3 min to further remove debris. The supernatant was centrifuged at 2000 × g at 4 °C for 5 min to spin down Nuclei. Nuclei were lysed by adding 1X NE buffer containing 0.1% (v/v) NP40, and 0.1 (v/v) Tween-20, followed by incubation on ice for 3 min. Nuclei were pelleted by centrifugation at 2000 × g at 4 °C for 5 min. Pellets were incubated in RS buffer (Tn5 mix, 10 mM Tris-HCL, PH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.01% digitonin, 0.1% OM and 0.1% Tween-20) at 37 °C for 30 min. The Tn5 tagmentation was then terminated under 95 °C for 2 min. DNA was purified using a spin column (Qiagen, Germany) and amplified using index primers matching the Illumina Nextra adapter. The above protocol was provided by Orizymes Biotechnologies (Shanghai) Co., Ltd.
ATAC-seq libraries containing DNA inserts of 50–150 bp were gel-purified and sequenced in Illumina X Ten platform in paired-end 150 bp mode. Raw reads were trimmed using fastp (v0.20.0)105 in default parameters. Sequencing reads were mapped to the genome sequence of Ftri (C4) using bowtie2 (v2.3.4.3)91 with the parameter “-k 10”. Mapping results were sorted using the “sort” function in samtools (v1.11)71, and low-quality was filtered off using “view” function in samtools with -q = 10. We then used “samtools collate” to group reads with the same names, and “samtools fixmate -m” to fill in mate coordinates and add mate score tags, and “samtools markdup -r” to remove duplicate reads. The start position of each read based on strand information was adjusted using alignmentSieve in deepTools (v3.5.0)110 with –ATACshift. Peaks were identified using MACS2 (v2.2.7.1)111 using the following parameters: -f BAMPE -g 1.7e9 -q 0.05 –nomodel –keep-dup all –nolambda –shift -100 –extsize 200. The consistency of the two biological replicates was assessed using the irreproducibility discovery rate (IDR) analysis with the IDR package (v2.0.4), following ENCODE guidelines. An IDR threshold of 0.05 was applied to filter irreproducible peaks.
Genes associated with peaks were identified using the “closest” function in bedtools (v2.29.2)112 with the parameter “-k 2,” considering the two nearest genes (upstream and downstream). The distribution of ATAC-seq reads relative to genome features were assessed using the “computeMatrix” function in deepTools (v3.5.0)110 with the following parameters: –skipZeros –reference Point TSS -a 3000 -b 3000. The results were visualized using the “plotHeatmap” in the same tool. peaks was visualized with IGV (v2.16.0)113.
Based on accessible chromatin regions (ACRs) from ATAC-seq data of C4 species (Ftri), we employed both a permutation-based method and a Fisher’s exact test-based method to predict the enriched CREs associated with C4 genes (including the regions 3 kb upstream of start codons, 3 kb downstream of stop codons, and the gene bodies). FIMO of MEME suite (v5.0.2)114 was used to identify the occurrences of known CREs of plants within the entire set of ACRs, applying a q-value threshold of 0.05. The CRE annotations were sourced from PlantPAN 3.095 (https://plantpan.itps.ncku.edu.tw/plantpan3/download/home.php). This analysis identified 1,471,751 occurrences of 277 distinct CREs, with 1858 occurrences of 117 CREs were associated with C4 genes. To assess whether specific CREs were overrepresented near C4 genes beyond random chance, we conducted a Monte Carlo permutation test. For each of the 117 CREs, observed occurrences were compared against a distribution of expected occurrences estimated from 1000 permutations. In each permutation, 1858 CRE occurrences were randomly selected from the total pool, and the frequency of each CRE was recorded. Following the completion of all permutations, the p value for each CRE was calculated as the proportion of permutations where CRE occurrences surpassed the observed value. To control for multiple testing, we applied the Benjamini–Hochberg procedure to adjust for the false discovery rate (FDR). For the Fisher’s exact test-based method, we evaluated the enrichment of each CRE associated with C4 genes against the background of 1,471,751 total CRE occurrences. The CRE enrichment results were largely consistent between these two methods (Supplementary Data 13).
Furthermore, to predict enriched CREs in ACRs from various genomic contexts, including within gene bodies, upstream and downstream of genes, as well as those associated with photosynthetic and photorespiratory genes, we employed the Monte Carlo permutation test as described above.
Analysis of enriched CREs in the promoters of C4 genes in five Flaveria species
We employed the HOMER package115 to identify enriched motifs within the promoters (3 kb upstream of the start codons) of C4 genes and their orthologous counterparts in each Flaveria species. For each species, the promoter sequences (3 kb upstream of the start codon) of non-C4 genes were used as the background to account for potential genomic distribution bias. Regarding sequence composition bias, the HOMER package automatically selects background regions from the promoter sequences of non-C4 genes that match the GC-content distribution of the promoter sequences of C4 genes (in 5% increments), as detailed in the HOMER manual (http://homer.ucsd.edu/homer/ngs/peakMotifs.html). Specifically, if the promoter sequences of C4 genes (input) are highly GC-rich, HOMER selects random regions from GC-rich regions of the promoter sequences of non-C4 genes (background) as a control. In addition to accounting for GC-content bias, HOMER package also applies “autonormalization of sequence bias” to eliminate bias introduced by lower-order oligo sequences associated with the promoter sequences of C4 genes. The HOMER package operates under the assumption that the promoter sequences of C4 genes (input) and non-C4 genes (background) should not exhibit imbalances in 1-mers, 2-mers, 3-mers, etc. After calculating these imbalances for each oligonucleotide, HOMER adjusts the weights of background sequences slightly to normalize the imbalances. This analytical procedure ensured that the enrichment analysis accounted for potential biases in sequence composition and genomic distribution between the promoters of C4 and non-C4 genes.
Electrophoretic mobility shift assay
To construct plasmids for recombinant protein production, the coding sequences of target ERF proteins were PCR-amplified from cDNA and inserted into pGST (His-ERF) vector to create fusions with 3×Flag, 10×His and GST tags, respectively. The recombinant proteins were expressed in E. coli strain Rosetta (DE3) and induced with 0.5 mM isopropyl β-D-thiogalactoside (IPTG) for 2 h at 37 °C for ERF12 (Ftri15G25371, ortholog in Arabidopsis is AT1G28360), ERF11 (Ftri17G17198, ortholog in Arabidopsis is AT3G23240) and ERF57 (Ftri1G11197, ortholog in Arabidopsis is AT5G65130), 0.1 mM IPTG for 1.5 h at 37 °C for ERF61 (Ftri13G23465, ortholog in Arabidopsis is AT1G64380). Bacterial cells were collected and lysed in lysis buffer (20 mM Tris-HCl, 300 mM NaCl, 0.5 mM DTT, and 1× protease inhibitor cocktail). The proteins were released from the collected cells by sonication (100 V, 20 m) and purified with Ni column. DNA fragments were end-labeled with Cy5. The fluorescence-labeled DNA (20 nM) was incubated with purified protein in 5x EMSA/Gel-Shift bidding buffer according to manufactory’s instructions (Beyotime, GS005, China) for 30 min at 25 °C. For competition assays, 400 nM unlabeled competitor DNA was also added in the reaction. For the empty-TF control, Glutathione-S-transferase (GST)-3×Flag-10×His without TF proteins was produced in E. coli Rosetta (DE3) as described above. The reaction mixture was electrophoresed at 4 °C on a 6% native polyacrylamide gel in 0.5×TBE for 50 min at 100 V. Fluorescence-labeled DNA on the gel was then detected with Typhoon (TyphoonTM, Cytiva). All PCR primers are listed in Supplementary Data 15.
Transient transcription assay
To construct an effector plasmid, the full-length CDS of ERF12 and ERF61 was cloned into pCambia1300, driven by the cauliflower mosaic virus (CaMV) 35S promoter to generate Pro35S::ERF12-Flag and Pro35S::ERF61-Flag. To construct a reporter plasmid, the CA promoter (200 bps upstream of the start codon) and PEPC promoter (250 bps upstream of the start codon) were cloned into pGreenII-0800 to generate pCA::LUC and pPEPC:LUC.
Transient transcription dual-LUC assays were performed using Nicotiana Benthamiana. The effector and report plasmids were transformed into Agrobacterium strain GV3101 carrying the helper plasmid pSOUP1P19. Agrobacterium cultures were cultured overnight and collected by centrifugation at 5000 × g for 3 min and resuspended in MES buffer (10 mM MgCl2, 10 mM MES, 100 μm acetosyringone, PH = 6.0) to 1.5 OD600. Mixed Agrobacterium with effector and reporter were incubated at room temperature for 2 h. The Agrobacterium suspension was then gently press-infiltrated into healthy leaves of 3-week-old N. benthamiana plants with a 1-mL needleless syringe. The plants were grown under 25 °C with photoperiod of 16/8 h day/night for 2–3 days. Luciferase activity was imaged with a CCD camera or quantified with a luminometer (Promega 20/20) using LUC reaction reagents according to the manufacturer’s instructions (Yeasen, China).
Statistics and reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyses except for those failing quality control checks. All statistical tests were performed in R (version 4.2.1) with a Benjamini–Hochberg correction applied where applicable. The statistical analysis for each experiment has been described in the main text and figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The genome assemblies, gene annotations, proteomics data, and raw reads of transcriptome data, Ribo-seq data and ATAC-seq data are available at the China National GeneBank (CNGB) [https://db.cngb.org/codeplot/datasets/flaveria] with project ID CPN0003058. The genome assemblies, gene annotations, transcriptome data, and proteomics data are also available at figshare [https://doi.org/10.6084/m9.figshare.19918876.v4]. The genome assemblies are also available at the National Center for Biotechnology Information (NCBI) with accession numbers: SAMN14943594 for Frob, SAMN14943595 for Fson, SAMN14943597 for Flin, SAMN14943596 for Fram, and SAMN14943598 for Ftri. The mass spectrometry proteomics data were submitted to the PRoteomics IDEntifications Database (PRIDE)116 with accession number PXD024720. RNA-seq data of Flin were also submitted to Gene Expression Omnibus (GEO) in the NCBI database under accession number PRJNA827625. RNA-seq data of Frob, Fson, Fram, and Ftri were obtained from published data under project accession PRJNA600545. Source data are provided with this paper.
References
Sage, R. F. The evolution of C4 photosynthesis. New Phytol. 161, 341–370 (2004).
Sage, R. F., Sage, T. L. & Kocacinar, F. Photorespiration and evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 63, 19–47 (2012).
Zhu, X. G., Long, S. P. & Ort, D. R. What is the maximum efficiency with which photosynthesis can convert solar energy into biomass? Curr. Opin. Biotechnol. 19, 153–159 (2008).
Vogan, P. J. & Sage, R. F. Water-use efficiency and nitrogen-use efficiency of C3-C4 intermediate species of Flaveria Juss. (Asteraceae). Plant Cell Environ. 34, 1415–1430 (2011).
Maurino, V. G. & Weber, A. P. Engineering photosynthesis in plants and synthetic microorganisms. J. Exp. Bot. 64, 743–751 (2013).
Zhu, X.-G., Shan, L., Wang, Y. & Quick, W. P. C4 rice—an ideal arena for systems biology research. J. Integr. Plant Biol. 52, 762–770 (2010).
Long, S. P., Marshall-Colon, A. & Zhu, X. G. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66 (2015).
Hatch, M. D. C4 photosynthesis—a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta 895, 81–106 (1987).
Slack, C. R. & Hatch, M. D. Comparative studies on the activity of carboxylases and other enzymes in relation to the new pathway of photosynthetic carbon dioxide fixation in tropical grasses. Biochem. J. 103, 660–665 (1967).
Christin, P. A., Petitpierre, B., Salamin, N., Buchi, L. & Besnard, G. Evolution of C4 phosphoenolpyruvate carboxykinase in Grasses, from genotype to phenotype. Mol. Biol. Evol. 26, 357–365 (2009).
Christin, P. A. et al. Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biol. Evol. 5, 2174–2187 (2013).
Moreno-Villena, J. J., Dunning, L. T., Osborne, C. P. & Christin, P. A. Highly expressed genes are preferentially co-opted for C4 photosynthesis. Mol. Biol. Evol. 35, 94–106 (2018).
Williams, B. P., Aubry, S. & Hibberd, J. M. Molecular evolution of genes recruited into C4 photosynthesis. Trends Plant Sci. 17, 213–220 (2012).
Emms, D. M., Covshoff, S., Hibberd, J. M. & Kelly, S. Independent and parallel evolution of new genes by gene duplication in two origins of C4 photosynthesis provides new insight into the mechanism of phloem loading in C4 species. Mol. Biol. Evol. 33, 1796–1806 (2016).
Powell, A. M. Systematics of Flaveria (Flaveriinae Asteraceae). Ann. Missouri Bot. Gard. 65, 590–636 (1978).
Edwards, G. E. & Ku, M. S. B. Biochemistry of C3-C4 intermediates. in The Biochemistry of Plants (eds Hatch, M. D. & Boardman, N. K.) 275–325 (Academic Press, 1987).
Lyu, M. J. et al. Evolution of gene regulatory network of C4 photosynthesis in the genus Flaveria reveals the evolutionary status of C3-C4 intermediate species. Plant Commun. 4, 100426 (2023).
Sage, T. L. et al. Initial events during the evolution of C4 photosynthesis in C3 species of Flaveria. Plant Physiol. 163, 1266–1276 (2013).
Gowik, U. & Westhoff, P. The path from C3 to C4 photosynthesis. Plant Physiol. 155, 56–63 (2011).
Lyu, M. J. et al. Evolution of gene regulatory network of C4 photosynthesis in the genus Flaveria reveals the evolutionary status of C3-C4 intermediate species. Plant Commun. https://doi.org/10.1016/j.xplc.2022.100426 (2022).
Taniguchi, Y. Y. et al. Dynamic changes of genome sizes and gradual gain of cell-specific distribution of C4 enzymes during C4 evolution in genus Flaveria. Plant Genome e20095. https://doi.org/10.1002/tpg2.20095 (2021).
Adachi, S. et al. The evolution of C4 photosynthesis in Flaveria (Asteraceae): insights from the Flaveria linearis complex. Plant Physiol. 191, 233–251 (2023).
Monson, R. K., Moore, B. D., Ku, M. S. & Edwards, G. E. Co-function of C3-and C4-photosynthetic pathways in C3, C4 and C3-C4 intermediate Flaveria species. Planta 168, 493–502 (1986).
Badouin, H. et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546, 148 (2017).
Renne, P. et al. The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J. 35, 316–331 (2003).
Weissmann, S. et al. Interactions of C4 subtype metabolic activities and transport in maize are revealed through the characterization of DCT2 mutants. Plant Cell 28, 466–484 (2016).
Furumoto, T. et al. A plastidial sodium-dependent pyruvate transporter. Nature 476, 472–475 (2011).
Kinoshita, H. et al. The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism. Plant J. 65, 15–26 (2011).
Knappe, S. et al. Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis–AtPPT1 may be involved in the provision of signals for correct mesophyll development. Plant J. 36, 411–420 (2003).
Lyu, M. J. et al. What matters for C4 transporters: evolutionary changes of phosphoenolpyruvate transporter for C4 photosynthesis. Front. Plant Sci. 11, 935 (2020).
Schluter, U., Denton, A. K. & Brautigam, A. Understanding metabolite transport and metabolism in C4 plants through RNA-seq. Curr. Opin. Plant Biol. 31, 83–90 (2016).
Gowik, U., Brautigam, A., Weber, K. L., Weber, A. P. & Westhoff, P. Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? Plant Cell 23, 2087–2105 (2011).
Furumoto, T. Pyruvate transport systems in organelles: future directions in C4 biology research. Curr. Opin. Plant Biol. 31, 143–148 (2016).
Xu, J., Brautigam, A., Weber, A. P. & Zhu, X. G. Systems analysis of cis-regulatory motifs in C4 photosynthesis genes using maize and rice leaf transcriptomic data during a process of de-etiolation. J. Exp. Bot. 67, 5105–5117 (2016).
Burgess, S. J. et al. Ancestral light and chloroplast regulation form the foundations for C4 gene expression. Nat. Plants 2, 16161 (2016).
Aubry, S., Brown, N. J. & Hibberd, J. M. The role of proteins in C3 plants prior to their recruitment into the C4 pathway. J. Exp. Bot. 62, 3049–3059 (2011).
Kaessmann, H., Vinckenbosch, N. & Long, M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat. Rev. Genet. 10, 19–31 (2009).
Tan, S. et al. DNA transposons mediate duplications via transposition-independent and -dependent mechanisms in metazoans. Nat. Commun. 12, 4280 (2021).
Xiao, H., Jiang, N., Schaffner, E., Stockinger, E. J. & van der Knaap, E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319, 1527–1530 (2008).
Tan, S. J. et al. LTR-mediated retroposition as a mechanism of RNA-based duplication in metazoans. Genome Res. 26, 1663–1675 (2016).
Zhu, Z., Tan, S., Zhang, Y. & Zhang, Y. E. LINE-1-like retrotransposons contribute to RNA-based gene duplication in dicots. Sci. Rep. 6, 24755 (2016).
Hu, Y. et al. Rapid genome evolution and adaptation of Thlaspi arvense mediated by recurrent RNA-based and tandem gene duplications. Front. Plant Sci. 12, 772655 (2021).
Akyildiz, M. et al. Evolution and function of a cis-regulatory module for mesophyll-specific gene expression in the C4 dicot Flaveria trinervia. Plant Cell 19, 3391–3402 (2007).
Billakurthi, K. et al. Transcriptome dynamics in developing leaves from C3 and C4 Flaveria species reveal determinants of Kranz anatomy. BioRxiv. https://doi.org/10.1101/473181 (2020).
Mallmann, J. et al. The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. Elife 3, e02478 (2014).
Mergner, J. et al. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 579, 409–414 (2020).
Zhou, Z. P. et al. Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc. Natl Acad. Sci. USA 113, E6117–E6125 (2016).
Chiapello, H., Lisacek, F., Caboche, M. & Hénaut, A. Codon usage and gene function are related in sequences of Arabidopsis thaliana. Gene 209, Gc1–Gc38 (1998).
Lei, L. et al. Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 84, 1206–1218 (2015).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Huang, C. F. et al. Whole-genome duplication facilitated the evolution of C4 photosynthesis in Gynandropsis gynandra. Mol. Biol. Evol. 38, 4715–4731 (2021).
Hoang, N. V. et al. The Gynandropsis gynandra genome provides insights into whole-genome duplications and the evolution of C4 photosynthesis in Cleomaceae. Plant Cell. https://doi.org/10.1093/plcell/koad018 (2023).
Hibberd, J. M. & Covshoff, S. The regulation of gene expression required for C4 photosynthesis. Annu. Rev. Plant Biol. 61, 181–207 (2010).
Schluter, U. & Weber, A. P. M. Regulation and evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 71, 183–215 (2020).
Xie, Z., Nolan, T. M., Jiang, H. & Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 10, 228 (2019).
Merchante, C., Alonso, J. M. & Stepanova, A. N. Ethylene signaling: simple ligand, complex regulation. Curr. Opin. Plant Biol. 16, 554–560 (2013).
Wu, Y. et al. ERF subfamily transcription factors and their function in plant responses to abiotic stresses. Front. Plant Sci. 13, 1042084 (2022).
Sage, R. F., Christin, P. A. & Edwards, E. J. The C4 plant lineages of planet Earth. J. Exp. Bot. 62, 3155–3169 (2011).
Sage, R. F., Sage, T. L. & Kocacinar, F. Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 63, 19–47 (2012).
Ito, H. Environmental stress and transposons in plants. Genes Genet. Syst. 97, 169–175 (2022).
Wang, D. et al. Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants. Plant J. 90, 133–146 (2017).
Niu, X. M. et al. Transposable elements drive rapid phenotypic variation in Capsella rubella. Proc. Natl Acad. Sci. USA 116, 6908–6913 (2019).
Maher, K. A. et al. Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Plant Cell 30, 15–36 (2018).
Dai, X. et al. Chromatin and regulatory differentiation between bundle sheath and mesophyll cells in maize. Plant J. 109, 675–692 (2022).
Swift, J. et al. Exaptation of ancestral cell-identity networks enables C(4) photosynthesis. Nature 636, 143–150 (2024).
Deng, C. L. et al. Identification of sex chromosome of spinach by physical mapping of 45 s rDNAs by FISH. Caryologia 65, 322–327 (2012).
Li, S. F. et al. The landscape of transposable elements and satellite DNAs in the genome of a dioecious plant spinach (Spinacia oleracea L.). Mob. DNA 10, 3 (2019).
Mascher, M. et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433 (2017).
Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014).
Pendleton, M. et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods 12, 780–786 (2015).
Du, H. & Liang, C. Assembly of chromosome-scale contigs by efficiently resolving repetitive sequences with long reads. Nat. Commun. 10, 5360 (2019).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015).
Burton, J. N. et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 31, 1119–1125 (2013).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268 (2007).
Ellinghaus, D., Kurtz, S. & Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinforma. 9, 18 (2008).
Ou, S. & Jiang, N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176, 1410–1422 (2018).
Stanke, M. & Morgenstern, B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33, W465–W467 (2005).
Majoros, W. H., Pertea, M. & Salzberg, S. L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20, 2878–2879 (2004).
Parra, G., Blanco, E. & Guigo, R. GeneID in Drosophila. Genome Res. 10, 511–515 (2000).
Keilwagen, J., Hartung, F. & Grau, J. GeMoMa: homology-based gene prediction utilizing intron position conservation and RNA-seq data. Methods Mol. Biol. 1962, 161–177 (2019).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9, R7 (2008).
Birney, E., Clamp, M. & Durbin, R. GeneWise and genomewise. Genome Res. 14, 988–995 (2004).
Seppey, M., Manni, M. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962, 227–245 (2019).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–U354 (2012).
Camacho, C. et al. BLAST plus: architecture and applications. BMC Bioinformatics 10, https://doi.org/10.1186/1471-2105-10-421 (2009).
Jin, J. et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045 (2017).
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J. & Gao, G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113 (2020).
Chow, C. N. et al. PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 47, D1155–D1163 (2019).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Suyama, M., Torrents, D. & Bork, P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Wang, Y. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012).
Fukayama, H. et al. Activity regulation and physiological impacts of maize C4-specific phosphopyruvate carboxylase overproduced in transgenic rice plants. Photosynth. Res. 77, 227–239 (2003).
Tsuchida, H. et al. High level expression of C4-specific NADP-malic enzyme in leaves and impairment of photoautotrophic growth in a C3 plant, rice. Plant Cell Physiol. 42, 138–145 (2001).
Wang, D., Portis, A. R. Jr., Moose, S. P. & Long, S. P. Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold tolerance of carbon assimilation in Miscanthus x giganteus. Plant Physiol. 148, 557–567 (2008).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 12, 323 (2011).
Bradford, M. M. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M. & Weissman, J. S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012).
Lauria, F. et al. riboWaltz: optimization of ribosome P-site positioning in ribosome profiling data. PLoS Comput. Biol. 14, e1006169 (2018).
Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Thorvaldsdottir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Acknowledgements
We appreciate Prof. Rowan F. Sage, Prof. Peter Westhoff, Dr. Udo Gowik, and Dr. Matt Stata for sharing Flaveria materials, and Ms. Lin Huang (State Key Laboratory of Genetic Engineering, Fudan University) for help on proteomic analysis and Zhen Cao (Zhe Jiang Agricultural and Forestal University) for help on plastid construction. We also thank suggestions from Prof. Haiyang Hu. The work is funded by the National Key Research and Development Program of China (2020YFA0907600, X.G.Z.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB0630101, M.J.A.L.; XDB0630301, X.G.Z.), the general program of the National Science Foundation of China (31870214, X.G.Z.).
Author information
Authors and Affiliations
Contributions
X.G.Z., T.L., C.L., and M.J.A.L. conceived and designed the study. H.D., Y.H., Z.Z., Y.Z., H.L., L.F., Q.G., Y.Q., and M.J.A.L. performed genome assembly and annotation. M.J.A.L., Y.H., and X.N. performed genome comparison analysis, qRT-PCR, and ATAC-seq analysis. M.J.A.L., H.Y., Z.Z., F.M., and Y.W. conducted RNA-seq and proteomics analysis. M.J.A.L. and F.C. performed gene regulatory network construction. Y.Y.Z. performed PCR verification of the three paralogs of PEPC1s in Ftri. Q.T. performed Ka/Ks analysis. X.C. and M.J.A.L. performed transcription factor prediction. Q.Z. and J.Z. performed syntenic analysis. T.Y. constructed Flaveria workspace in China National GeneBank (CNGB). M.J.A.L., X.G.Z., T.L., C.L., and G.C. drafted the manuscript. M.J.A.L. and X.G.Z. revised the manuscript. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks David Wickell, Dijun Chen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lyu, MJ.A., Du, H., Yao, H. et al. A dominant role of transcriptional regulation during the evolution of C4 photosynthesis in Flaveria species. Nat Commun 16, 1643 (2025). https://doi.org/10.1038/s41467-025-56901-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56901-y