Abstract
Despite the high prevalence of gastric diseases like gastric cancer and peptic ulcer disease attributed to Helicobacter pylori infections, there is still only a limited understanding of the underlying mechanisms. Existing in vitro models are either two-dimensional systems lacking the structural complexity of the gastric architecture, or complex three-dimensional systems that pose challenges for experimental access. In this study, we introduce a patterned homeostatic human gastric organoid-on-a-chip system with bilateral access that is capable of modeling H. pylori niche establishment and persistent colonization of the gastric epithelium. We show that in physiological apical acidic conditions, our organ-on-a-chip can generate pit cells of higher maturity in contrast to traditionally grown organoids. Upon infection with H. pylori for up to 6 days, these mature pit cells exhibit a distinctive response from other cell types, which was previously uncharacterized. Beyond its application in studying H. pylori infection, the increased structural and functional relevance of our model offers broader significance as a versatile platform for advancing our understanding of gastric epithelial cell interactions, gastric mucosal immunity, and host-pathogen interactions.
Similar content being viewed by others
Introduction
Various gastric pathologies, including gastric cancer, are primarily driven by persistent Helicobacter pylori infection and remain a global health concern due to the high prevalence of this bacteria1,2. Upon ingestion, the bacteria colonize the gastric epithelium by attaching to the cell junctions, creating a niche where they remain protected from the harsh acidic stomach environment3,4,5. In proximity to cells, H. pylori can persist throughout the lifetime of an infected individual, causing local epithelial damage and inducing inflammation and immune responses. Patients often remain asymptomatic for decades, but infections can eventually progress to peptic ulcers or gastric adenocarcinomas6. However, many mechanisms underlying these diseases remain inadequately understood, such as the early interplay between individual bacteria and host cells and the transition from chronic inflammation to malignancy.
These mechanisms elude understanding primarily due to limitations apparent in widely used model systems, as traditional in vitro models fail to replicate the physiological conditions present in the stomach, such as variations in pH levels, mucus production, and/or the diverse differentiation states of gastric epithelial cells7. The glandular epithelium of the stomach antrum, the primary site of H. pylori infections6, is composed of stem-and early progenitor cells, MUC6 mucin-expressing neck cells, prostaglandin C (PGC)-expressing chief cells and MUC5AC mucin-producing pit cells8. Moreover, the antral glands also contain acid-producing parietal cells, however to a much lesser extent than the glands of the corpus, and specialized cell types such as diverse hormone producing enteroendocrine cells (chromogranin A (CHGA)-positive) and Tuft cells. The tissue is organized in a stereotypical manner with the stem- and progenitor cells residing at the bottom of the glands together with neck and chief cells, and the pit cells present towards the surface of the epithelium.
Recently, the improved physiological relevance of human gastric organoid cultures, as introduced by Bartfeld and colleagues9, has provided a tool to generate many of those cell types and thus a potential avenue for exploring mechanisms underlying H. pylori-induced diseases. For instance, organoids grown in expansion conditions are populated by progenitor and neck cells, and upon H. pylori infection, these cells highlighted an IL–8-dominant cytokine response9,10. Conversely, reduced responsiveness to H. pylori was seen in organoids cultured in differentiation conditions (without Wnt signaling), which are mainly composed of pit cells. Showing a similar increase in cytokine production in Wnt-active cultures compared to those without, investigations employing a transwell system accentuated this impact of the level of cellular differentiation on H. pylori-induced inflammatory responses11,12. However, in investigations into the infection responses within gastric organoids and organoid–cell-derived 2D cultures, H. pylori showed a preference for attaching to mature pit cells, as determined by single-cell RNA sequencing (scRNAseq) on sorted infected and uninfected cells13. Regardless, the clarity of these findings remains obscured by disparities in experimental conditions, such as resultant variations in mucus production, which affect the infection efficiency and thus hinder a direct comparison between cell types. Furthermore, the technical challenges faced using traditional 2D monolayer and organoid models to sustain long-term co-cultures of host cells with H. pylori restricts the comprehensive understanding of chronic infection dynamics and disease progression over extended periods. While the gastric mucosoid cultures described by Boccellato and colleagues11 support long-term co-cultures (up to 4 weeks), the model does not recapitulate the spatial organization of cell types within the gastric glands and fails to maintain a constant bacterial burden over time. Consequently, current models are limited in their ability to investigate chronic H. pylori infection in a relevant 3D setting.
To investigate H. pylori infection responses and resolve inherent inconsistencies between prior in vitro studies, we adapted here our previous observable murine gastric organoid model system14 to generate an analogous human system. We show the successful generation of a bilaterally accessible, homeostatic, and geometrically patterned model system that simultaneously encompasses progenitor and fully differentiated pit cells. Compared to traditional 3D organoid cultures, scRNAseq showed that our model was able to generate pit cells that reach a higher level of maturity upon introduction of physiologically relevant apically acidic conditions. We then used this model to systematically test the acute (2 days) and prolonged (6 days) cellular responses upon H. pylori infections under physiological settings. Our findings reveal that all cell types responded to H. pylori infections, albeit in a distinct manner. While progenitor and immature pit cells showed a cytokine-dominant response in line with previous observations, mature pit cells responded to the infection by upregulating genes involved in cellular junctions or antibacterial responses. Overall, this model enhanced our understanding of H. pylori infections and promises to bolster research into gastric health and disease progression.
Results
Generation of patterned and homeostatic human gastric organoids-on-a chip
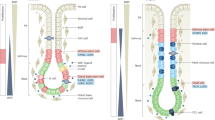
Building upon our previous success in constructing highly observable models of murine gastrointestinal epithelia with tissue-mimicking geometries14, we developed an analogous model system for the human gastric epithelium. We leveraged our previous setup, termed Transgels, for culturing epithelial cells on a geometrically pre-patterned hydrogel surface within a poly(dimethylsiloxane) (PDMS) device granting experimental access to both sides of the epithelium. To form an initial epithelial monolayer, we seeded organoid-derived human gastric cells on the glandular surface and incubated them for three days in expansion medium rich in growth factors conducive to stem cell maintenance (Wnt3a, R-Spondin 3, Noggin) (Fig. 1a). We found that, irrespective of cell density, the resultant monolayer did not perfectly conform to the geometry of the hydrogel (Supplementary Fig. 1a). Speculating that manipulating the medium prior to seeding could optimize epithelial attachment, we tried i) pre-treating the cells by removing the Alk inhibitor from the standard medium and ii) modulating Wnt signaling and thus the level of cellular differentiation by either adding the Wnt activator CHIR or removing Wnt, R-Spondin, and Noggin. While the latter did not improve cell attachment, removing the Alk inhibitor resulted in the robust formation of an epithelial monolayer that conformed to the pre-defined glandular geometry of the hydrogel within 3 days (Supplementary Fig. 1b).
a A Polydimethylsiloxane (PDMS) device is employed to hold a patterned hydrogel as growth substrate for epithelial cells as published previously14. Gastric epithelial cells obtained from organoid cultures are seeded onto the hydrogel and let to grow until confluency. Apical and basal side media can be controlled independently. b Brightfield overview and detailed (extended depth of field, inset) images of fully established model 7 days after seeding. Representative images from > 20 independent experiments. c 3D reconstruction (left) and single planes (right) of confocal images of fixed sample stained for Nuclei (DAPI, blue), F-Actin (pink) and Mucus (UEA-FITC, green). Scale bar, 100 µm. d Confocal image of fluorescently stained cryosection of a 7-day old sample illustrating SOX9 and KI67-positive cells in the gland regions, including single-channel split-ups below. Representative images from three independent experiments. e Frequency of SOX9+ and KI67+ cells within the gland and surface regions, respectively. Each dot represents the quantification of the fraction of positive cells within one image with 1–3 glands, originating from three independent experiments. Bars represent mean and error bars SD values. Statistics were performed using a paired two-sided t-test. ****, p < 0.0001. Source data are provided as a Source Data file. f Confocal images of fluorescently stained cryosections of 7-day old samples illustrating cells positive for differentiation markers. Representative images from three independent experiments. Scale bars, 500 µm (b), 100 µm (c, d, f).
To next simulate the physiological context wherein stem cell-inducing growth factors originate from the underlying mesenchyme, on the third day of culture, we transitioned to a medium with reduced growth factors on the apical side while maintaining the maximal growth factor concentration on the basal side (see “methods” for details). Despite this transition, we could maintain the epithelium stably for extended periods (up to 13 days) while preserving its geometry (Fig. 1b, c, Supplementary Fig. 1c). After that, local disruptions of the epithelium can be observed occasionally. During medium changes, apically shed dead cells are removed (Supplementary Fig. 1d). To subsequently assess the differentiation level of the epithelium after four days in these conditions, we conducted an immunofluorescence analysis of cryosections. We found stem and progenitor cells marked by the expression of the gland-marker15 SRY-box transcription factor 9 (SOX9) and marker of proliferation Kiel 67 (KI67) predominantly localized in the gland regions of the model (Fig. 1d, e) coexisting alongside differentiated pit cells expressing MUC5AC as well as occasionally MUC6- and PGC-expressing neck/chief cells and CHGA-expressing enteroendocrine cells (Fig. 1f). The functionality of pit cells was evidenced by the presence of a viscous mucus layer on the apical side (Supplementary Fig. 1e, Supplementary Movie 1). Collectively, we here introduce a homeostatic human gastric epithelial organoid model that faithfully recapitulates architectural features of native tissue, including cell-type localization, and, while predominantly composed of pit cells, manifests cell type diversity similar to the human antrum.
Pit cells show higher level of maturity in transgels
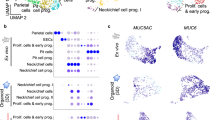
Our next objective was to benchmark the Transgel cultures against traditionally grown organoids using single-cell RNA sequencing (scRNAseq). To achieve this, we cultured 3–day-old organoids for another four days in either expansion medium or differentiation medium (lower growth factor concentrations). This has been shown to yield two distinct types of organoids in conventional 3D organoid cultures: the first with a cellular composition similar to the gland region of the stomach and the second comprising mainly pit cells9,13. We also extended our investigation by examining the effects of acidifying the apical medium to simulate physiological conditions, using Transgel organoids cultivated as before. Additionally, we explored the induction of air-liquid interface (ALI) conditions based on reports indicating their potential to enhance cellular maturation11 (Fig. 2a). The Transgel epithelia accommodated these alterations well, as we did not detect any discernible changes in the morphology or viability of the cells when exposed to apical acidic or ALI conditions (Fig. 2b). Employing a multiplexing strategy for scRNAseq, we profiled a total of 3311 cells. To annotate cell types, we followed standard multidimensionality reduction and unsupervised clustering approaches and identified a cluster of proliferating and early progenitor cells marked by the proliferation marker MKI67 and the isthmus stem cell marker STMN116 (Fig. 2c, Supplementary Fig. 2a). Additionally, we identified three distinct clusters of pit cells expressing MUC5AC and GKN1 along with one cluster of neck cells expressing MUC6 and PGC. We then determined the maturity level of the pit cells using a maturity score based on a published dataset of freshly extracted cells13 (detailed in methods). From this, we labeled one of the clusters as mature pit cells while the two other clusters had lower levels of maturity, labeled as immature pit cells I and II (Supplementary Fig. 2b). Consistent with previous findings, we show that organoids in expansion medium are predominantly composed of progenitor and neck cells, while upon differentiation, they contain mostly immature pit cells9,13 (Fig. 2d, e). Transgel organoids are primarily composed of pit-type cells, with a presence of progenitor cells and a notable reduction in neck cells. In contrast to organoids, however, the proportion of mature pit cells increased, especially in cultures exposed to acidic and ALI conditions. Overall, our investigation highlights the unique cellular composition of Transgel organoids, characterized by a pronounced abundance of pit-type cells of higher maturity than organoids, which is further enhanced under physiologically relevant acidic conditions that are not easily achieved in other 3D systems.
a Experimental overview. Organoids or Transgel organoids were grown as indicated for a total of 7 days and single-cell RNA sequencing was performed using a multiplexing strategy (see “methods” for details). b Brightfield images (Extended field of depth) of organoids and Transgels in the conditions used. Representative images from three independent experiments. Scale bars, 100 µm (c) UMAP plot of all cells with annotations and relative gene expression values of marker genes (right). Marker genes used for annotations are provided in Source Data file. d UMAP plots with subset of cells corresponding to indicated conditions highlighted. e Cellular composition of the different model systems. Source data are provided as a Source Data file. c–e Colors represent annotated cell types and are consistent throughout.
Live observation of Helicobacter pylori infection shows niche formation and microcolony growth in physiological conditions
Unlike acidophilic bacteria, H. pylori struggles to survive in a highly acidic environment, such the gastric lumen. To endure these harsh conditions and to persist in the stomach, these bacteria strategically establish a microenvironment at the surface of gastric epithelial cells via adhesion molecules, which help them adhere to the epithelium, and secreted factors such as urease, which aids in neutralizing acidity5. To explore the suitability of our system in understanding these host-pathogen interactions, we introduced bacteria into either acidic or neutral medium on the apical side of fully differentiated Transgel epithelia. Aiming to remove unattached bacteria, we washed the epithelium twenty-four hours post-infection. Following this procedure, no freely swimming bacteria were observed anymore (Supplementary Fig. 3a, b). We then cultured these models for six days with daily medium changes mimicking gastric emptying, which maintained consistent bacterial presence and continuous epithelium integrity (Fig. 3a, Supplementary Fig. 3c).
a Timeline brightfield images (gray, extended field of depth) and confocal images of bacteria (red, maximum intensity projection). Bacteria were introduced apically in either neutral or acidic medium at 7-day-old samples. Apical medium was exchanged every 24 h thereafter. Scale bars, 100 µm. b Confocal images (maximum intensity projection) of fixed and stained samples 6 days p.i. Overview image (left) and high-magnification image with split channel images (right). Scale bars, 100 µm (overview), 10 µm (zoom). Representative images from three independent experiments. c Assessment of colony forming units (CFUs) per sample 2 days and 6 days p.i. Each dot represents one of four independent samples. Indicated p-values result from a two-way ANOVA. Source data are provided as a Source Data file.
Fluorescent staining of fixed samples at six days post-infection confirmed bacterial attachment to cell-cell junctions and formation of bacterial clusters (Fig. 3b, Supplementary Movie 2). Using two differently colored bacterial strains, we confirmed the clonality of those clusters (Supplementary Fig. 3d). Moreover, the viability of the bacteria was confirmed by the presence of high colony-forming units (CFUs) throughout the experiment, even in acidic conditions (Fig. 3c). Importantly, the consistent CFU counts from day 2 to day 6 post-infection in both conditions suggest the bacteria’s ability to establish a microenvironment that protects against acidity. This shows the successful construction of a model for homeostatic H. pylori infection, presenting opportunities to explore both immediate and subsequent secondary ( > 48 h) effects of the infection.
Cell–type-specific responses to acute and persistent helicobacter pylori infection
To then examine longer-term cellular responses to a persistent infection, we performed single-cell RNA sequencing on H. pylori-infected and uninfected Transgel organoids, in both apically neutral and acidic conditions. Using two samples of different patient cell lines per condition, we obtained a total of 6986 and 4517 cells at two- and six-days post infection, respectively. Unsupervised clustering of the individual datasets per patient line and timepoint reveals separate clusters containing cells from infected samples (Supplementary Fig. 4a). In order to gain robustness and to remove technical and patient-specific differences, we performed dataset alignment and annotated the cell types as before, though now also distinguished between MUC5AChigh and GKNhigh cells within the mature pit cell cluster (Fig. 4a, Supplementary Fig. 4b). To match experimental conditions of previous in vitro studies11,12,13, we first focused on the samples infected in neutral apical conditions. In line with previous reports9,11,12, infected progenitor cells, neck cells, and immature pit cells responded to infection by producing lipocalin (LCN2), the polymeric immunoglobulin receptor (PIGR), and inflammatory cytokines, such as CCL20, CXCL8 (IL-8), CXCL1, CXCL2, and CXCL3 (Fig. 4b, Supplementary Fig. 4c). As reported previously9,10,11, pit cells showed limited capacity to upregulate inflammatory cytokines upon infection. However, we found another transcriptional response of mature pit cells: These cells also upregulated the expression of LCN2, CCL20, and PIGR, similar to immature and progenitor cells, but in addition they upregulate the expression of IgGFc-binding protein (FCGBP) and the dual oxidase 2 (DUOX2) and dual oxidase maturation factor 2 (DUOXA2) that form a hydrogen–peroxidase-producing heterodimeric NADPH oxidase complex at the apical membrane. KEGG pathway analysis corroborated these findings and the specific response to this bacterium, revealing terms related to cytokine production among the most enriched pathways, which occurred predominantly in the progenitor, neck, and immature pit populations, together with the pathway “Epithelial Cell Signaling in Helicobacter pylori infection” (Supplementary Fig. 5a). Conversely, enrichment in mature cell populations occurred in pathways related to metabolism and epithelial integrity maintenance.
a Dimensionality reduction (UMAP) plot with annotation of clusters of single cell RNA sequencing results of infected and uninfected samples. b Highly upregulated genes (Fold change > 2 & adjusted p-value < 10-4 in at least one cell type, using a likelihood-ratio test for single cell gene expression with the Seurat FindMarkers (bimod) approach) upon infection per cell type cluster in apically neutral (left) and acidic (right) smples. c Scatter plots of log-fold-changes upon infection in neutral versus acidic conditions with annotation of selected genes. Complete figure is presented as Supplementary Fig. 4d. b–c) Data are provided in Source Data file. d Immunofluorescent staining images for DUOX2 indicating predominant upregulation upon H. pylori (Hp) infection in apically acidic conditions, 6 d p.i. Scale bar, 100 µm. e Live images of infected samples with addition of compound 521 (C521, right) and DMSO control (left) with fluorescent images for bacteria (Hp, red) and dead cells (DRAQ7, magenta). Scale bars, 200 µm. d, e Representative images from three independent experiments.
We then examined the data from samples infected under acidic conditions, comparing the responses to the previously observed patterns. The earlier response was largely preserved in this more physiologically relevant model, notably the upregulation of LCN2, CCL20, and other cytokines (Fig. 4b). However, we found an additional response by progenitors and immature pit cells, which displayed upregulated metallothioneins, including MT1E, MT1G, MT1X, and MT2A at two days post-infection. Intriguingly, mature pit cells in acidic conditions displayed significantly intensified upregulation of both DUOX2 and DUOXA2 six days post-infection, together with the upregulation of genes producing proteins involved in cell-cell junctions, such as occludin (OCLN), claudin 7 (CLDN7) and the LIM domain 7 (LMO7). (Fig. 4c, Supplementary Fig. 4d). The KEGG analysis again confirmed this data, showing enrichment in terms related to tight junctions, adherence junctions, and cell adhesion molecules (Supplementary Fig. 5b). We further confirmed the upregulation of DUOX2 on a protein level by immunofluorescent analysis (Fig. 4d) and showed the direct antimicrobial importance of the DUOX2/DUOXA2 complex by an inhibition experiment using the small molecule inhibitor Compound 521 (ref. 17). The bacterial load as well as the number of dead cells notably increased during infections with DUOX2 inhibition (Fig. 4e, Supplementary Figs. 4e, f). This suggests that in acidic conditions—presumably due to a tighter interaction between the bacteria and the cells—mature pit cells respond more strongly to H. pylori, characterized by the upregulation of cell junction proteins as well as a direct antibacterial response spearheaded by the DUOX2/DUOXA2 complex.
In summary, our findings reveal intricate cell-specific responses to H. pylori infection, similar to previous reports9,11 and highlight our model’s ability to dissect cellular responses in a longer-term infection. Specifically, under physiologically relevant acidic apical conditions, we identified additional responses specific to mature pit cells that had not been observed before.
Discussion
Here we introduce a homeostatic model system of the human antral epithelium that mirrors native tissue architecture due to its spatial organization of coexisting progenitors and mature pit cells. Unlike conventional 3D organoids, our model offers bilateral accessibility, facilitating long-term infection experiments and simulations of an acidic apical environment. Apically introduced bacteria preferentially attached to the epithelial cells near cell-cell junctions and survived for 6 days in acidic conditions without notably damaging the epithelial monolayer, reminiscent of persistent infections. Importantly, the bacterial burden remained stable over time, a key feature for modeling chronic infection.
Gene expression profiling of infected cells demonstrated substantial responses in all major antral gastric epithelial cell types. Progenitor, neck, and immature pit cells exhibited an anticipated inflammatory response and the upregulation of specific genes like LCN2 and CCL20. Surprisingly, mature pit cells, previously considered less responsive, displayed a unique response, particularly under acidic conditions. Here, they not only upregulated genes involved in cellular junctions, suggesting potential damage thereof, but also expressed antimicrobial systems, such as the DUOX2/DUOXA2 complex. Intriguingly, DUOX2/DUOXA2 has been shown to be important in preventing H. felis infection in mice18, but to date, this was not verified experimentally in human stomach cells. Considering H. pylori-induced IL-8 upregulation is reported to be independent of the cag pathogenicity island and bacterial viability12, we hypothesize that this response might not depend on direct physical interactions, whereas such close interactions, solely recapitulated in apically acidic conditions, are crucial for the DUOX2 response in mature pit cells. Our system enables future investigations with differently virulent strains, particularly those with different cag/vac genotypes, to elucidate their effects on host-pathogen interactions and immune responses. In summary, we describe a comprehensive platform that models the architecture of the human gastric epithelium and can be leveraged to study long-term host-pathogen dynamics in persistent H. pylori infections under physiologically relevant conditions. This model provides a versatile system for further studies of gastric epithelial cell type-specific interactions in physiological and pathological settings.
Methods
Transgel hydrogel production
Polydimethylsiloxane (PDMS)-holder fabrication, hydrogel loading and surface patterning has been done as previously described14. Briefly, molds for the PDMS holder device components were fabricated using conventional photolithography and the components assembled using oxygen plasma bonding. For hydrogel loading, a PDMS stamp with 300 μm long and 50 μm wide pillars was added on top of the hydrogel chamber. 5 mg/ml Collagen type I (Koken, KKN-IAC-50) was neutralized (1:4 addition of 5X DMEM supplemented with 50 mM NaHCO3 (Thermo Scientific, J63025AK)) and mixed with Matrigel (Corning, growth factor reduced and phenol red-free formulation) at a ratio 4:1, added to the hydrogel chamber and let to gelate at 37 °C for 30 min. Hydrogels were washed and stored in 1X phosphate-buffered saline pH 7.4 (PBS, Gibco, 16210064) at 4 °C until use. Hydrogels were equilibrized for > 4 h with growth medium.
L-WRN-conditioned medium
Triple Wnt/R-Spondin/Noggin (WRN)-conditioned medium was produced using L-WRN cells (ATCC, CRL-3276) similar to a previously published protocol19. L-WRN cells were grown as described until confluency. Advanced DMEM/F-12 (Gibco, 12634010) supplemented with 10 mM HEPES (Gibco, 15630080), 1X GlutaMAX (Gibco, 35050061), 20% fetal bovine serum (FBS, Gibco, 10270106) and 50 U/ml Penicillin-Streptomycin (Gibco, 15070063) was then used for continuous culture of the cells and the medium was collected every 24 h for 9 days. Collections of three consecutive days were combined and diluted 1:1 in medium without FBS (final FBS concentration: 10%), sterile filtered, and stored at -20 °C.
Ethics statement
Consenting patients donating gastric biopsies were recruited at least two days before the planned panendoscopy. Full informed written consent was obtained from all participants. All procedures were in accordance with internal and national ethical laws and standards and the project was approved by the Commission cantonale d'éthique de la recherche sur l'être humain (CER-VD, Project ID 2019-01144).
Organoid extraction and culture
Gastric forceps biopsies of healthy tissues were obtained from consenting patients undergoing routine panendoscopy for surveillance. Biopsies were kept in DMEM/F-12 on ice for maximal 2 h before processing. Epithelial cell extraction was performed following a previously published protocol20. Briefly, obtained epithelial cell clusters were embedded in Matrigel and cultured in L-WRN medium supplemented with 50 ng/ml EGF (Peprotech, 315-09), 200 ng/ml FGF-10 (Peprotech 100-18B), 10 nM [Leu15]-Gastrin (Sigma-Aldrich, G9145), 0.5 μM A83-01 (Expansion medium). For the first two days in culture and for the first two days after passaging, 10 mM Nicotinamide (Millipore, 481907) and 10 μM Y-27632 (Selleckchem, S1049) was added. Additionally, the medium was supplemented with 100 μg/ml Primocin (Invivogen, ant-pm-2) during the initial 2–4 passages.
Passaging was performed every 5–8 days, by collecting the organoids in ice-cold adv. DMEM/F-12 and mechanical disruption using a glass Pasteur pipette (witeg Labortechnik, 4100230). Fragments were pelleted by centrifugation (200 g, 3 min) and diluted in fresh Matrigel as before.
For differentiating organoids, the medium was replaced 3 days after passaging with differentiation medium containing reduced concentration of WRN-conditioned medium (10%) and EGF (5 ng/ml) and replenished after two days. Organoids were analyzed after 4 days of differentiation together with organoids kept in expansion medium for the same total time of 7 days.
Cell seeding and culture on transgel devices
Organoids grown in expansion medium were cultured for one day in expansion medium lacking A83-01, unless otherwise stated and then collected in ice-cold adv. DMEM/F-12 and dissociated using TrypLE Express (Gibco, 12605028) for 12 min at 37 °C. Alternative pre-treatments during one day before seeding were the addition of 3 μM CHIR99021 (STEMCELL, 100-1042) or the removal of L-WRN medium. Medium for equilibration was removed from the Transgel devices and 8 μl of a 12.5 mio cells/ml single cell solution was added on top of the hydrogel, avoiding the cells to spread on the PDMS. Cells were let to sediment (30 min, 37 °C) before adding expansion medium without A83-01 but with Nicotinamide and Y-27632. On day 3, the apical side expansion medium was replaced with differentiation medium, supplemented with 10 mM HCl where indicated (pH ~ 3), and replenished every two days. Alternatively, the medium on the apical side was completely removed to induce air-liquid interface (ALI) conditions. The basal side medium was replaced with expansion medium every two days. For infection experiments, WRN-conditioned medium was replaced with recombinant factors at the time of infection, to obtain serum-free medium: Basal: Wnt3a 50 ng/ml (Time Bioscience, rmW3aL-010), R-spondin 0.5 μg/ml, Noggin 0.1 μg/ml (both EPFL Protein Expression Core Facility); Apical: 20% thereof.
Immunofluorescence and histological staining
Immunofluorescent analyzes were perfomed as previously described14. Briefly, collagen gels were harvested and fixed in 4% paraformaldehyde (ABCR, AB351601) for 30 min at room temperature (RT) and then washed with PBS three times for 1 h at RT and incubated in 50% Cryomatrix embedding resin (Epredia, 6769006) in PBS at 4 °C overnight. Samples were then embedded in 100% Cryomatrix and frozen on dry ice. 10 μm-thin sections were obtained using a Leica CM3050 S cryostat, permeabilized with 0.2% Triton X-100 (Sigma-Aldrich, X100) in PBS (15 min, RT) and blocked in 10% goat serum in PBS containing 0.01% Triton X-100 (30 min, RT). Sections were incubated with primary antibodies (Supplementary Table 1) diluted in blocking buffer (overnight, 4 °C) and washed with blocking buffer three times for 30 min. Samples were then incubated with secondary antibodies, DAPI, and fluorophore-conjugated phalloidin (Supplementary Table 1) for 2 h at RT and washed three times with PBS. Whole mount stainings were performed similarly but with longer incubation times: Permeabilization, 1 h; Blocking, 3 h; Primary and secondary antibodies, overnight; Washes, 3x3h plus overnight.
Microscopy and image processing
Brightfield and widefield fluorescent imaging of living samples was performed using a Nikon Eclipse Ti2 inverted microscope system using a DS-Qi2 camera and 4x/0.20NA and 10x/0.30NA objectives. Dead cells were stained with 1 μM DRAQ7 (Biolegend, 424001) dye for 10 min before imaging. Live confocal imaging was performed with a Leica SP8 inverted microscope system equipped with a supercontinuum laser (range 470 nm-670 nm) and hybrid photon counting detectors (HyD), using a 2.4 mm WD HC FLUOTAR 25x/0.95NA water-immersion objective.
Fixed and stained samples were imaged with a Leica SP8 microscope system equipped with 405 nm, 488 nm, 552 nm and 638 nm solid state lasers and HyD detectors using HC PL APO 20x/0.75NA and HC PL APO 63x/1.40NA objectives.
Image processing was performed using FIJI ImageJ (v1.54 d) using standard contrast and intensity level adjustments, for noise filtering, as well as for generating z-maximum intensity projections. Extended depth of field (EDF) images were generated with LAS X v3.7.4.23462 (Leica) and NIS-Elements AR 5.11.02 (Nikon Corporation) softwares.
Single-cell RNA sequencing
Single cell suspensions of samples have been obtained by digesting the collagen hydrogel with 10’000 U/ml Collagenase Type 1 (Gibco, 17100017) for 12 min at 37 °C and by subsequently dissociating the cells with TrypLE Express for 10 min at 37 °C. For multiplexing, cells were immunolabelled with TotalSeq-C anti-human Hashtag antibodies (BioLegend, 394661, 394663, 394665, 394667, 394669, 394671, 394673, 394675, 394677, 394679, 394683, 394685) at a dilution of 1:500 in staining buffer (PBS + 0.1% Bovine Serum Albumin (BSA, Gibco, 15260037)) for 30 min on ice and washed 2 times in 4 ml staining buffer with intermittent centrifugation (300 g, 3 min). Cells were strained using tubes with 35 μm cell strainer caps (Corning, 100–087) and counted and assessed for cell viability using an automated cell counter Countless II (Invitrogen). Cell viability was > 97% for all pooled cell suspensions. Single-cell RNA sequencing was then performed following the 10X Genomics pipeline using a Chromium Controller and the Chromium Next GEM Single Cell 5’ v2 (Dual Index) chemistry for the cDNA and HTO library preparation, following the protocol CG000330 Rev F. Library quantity and quality were assessed with a Qubit Fluorometer (Invitrogen) and DNF-474 HS NGS Fragment Analyzer (Aligent). Sequencing was performed on a NovaSeq 6000 Instrument (Illumina).
The reads were aligned to GRCh38 using Cell Ranger v.6.1.2. Raw count matrices were imported in RStudio 2023.09.0 + 463 and analysis was performed using the Seurat pipeline. Briefly, single, live cells were selected and demultiplexing of cells was performed using HTODemux function. The data was normalized and log-transformed. The datasets of the infection experiment were aligned using harmony package. Standard dimensionality reduction and unsupervised clustering was performed and clusters were annotated to cell types manually inspecting the expression patterns of typical marker genes. Pit cell maturity scoring was performed by calculating the sum of the gene expression values of pit cell markers minus the sum of the gene expression values of immature pit cell markers. The marker lists were obtained from analyzed freshly extracted gastric cells13. Gene set enrichment analysis were performed with the fgsea function and the msigdbr database. Data visualizations were created utilizing internal functions within Seurat or with ggplot2, and esthetic refinements were applied using Adobe Illustrator.
Helicobacter pylori strains and culture
The previously published tdTomato- or GFP- expressing H. pylori PMMS1 strains21 were grown on Columbia blood agar plates for 2–5 days at 37 °C in an humified 10% CO2 environment. Bacteria were then subcultured overnight in brucella broth (BD, 211088) supplemented with 10% FBS on a shaker at 37 °C in microaerophilic environment generated with the GasPak system (BD, 260680).
Helicobacter infections
Overnight liquid cultures were diluted in apical side medium (neutral or acidic pH) to an OD600 of 0.015. 300 μl of solution was added to 7-day old epithelial models on the apical side. Every 24 h the medium was removed and replaced with fresh medium whereby unattached bacteria were removed. Where specified, 5 μM of Compound C521 DUOX2-inhibitor17 or the DMSO equivalent as control was added to the media (apical and basal) during the infection.
CFUs
Samples were digested in 500 μl of 1% saponin (Sigma-Aldrich, 47036) in adv. DMEM/F-12 for 5 min at 37 °C while shaking, followed by vortexing for 30 sec. 5 μl of 1:2, 1:4 and 1:10 dilutions in brucella broth were plated on a Columbia blood agar plate and let dry for 10–15 min. The plates were incubated as described above and colonies were manually counted 5 days later.
Statistics & reproducibility
No statistical methods were employed to predetermine sample size. The experiments were not randomized, but samples were randomly assigned to different experimental groups. The Investigators were not blinded to allocation during experiments and outcome assessment. All experiments with qualitative results were independently repeated at least three times, yielding consistent results. Statistical analyzes, where applicable, are indicated in the relevant sections of the manuscript.
Manuscript writing
Manuscript writing was assisted by a Large Language Model (ChatGPT v3.5, OpenAI Inc, California, USA).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the results in this study are available within the paper and its Supplementary Information. Source data are provided with this paper. Source scRNA-seq data has been deposited to the Gene Expression Omnibus (GEO) public repository with the accession code GSE255276 and processed data is available in souce data file. Source data are provided with this paper.
Code availability
Custom analysis code is available on request.
References
Hooi, J. K. Y. et al. Global prevalence of helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 153, 420–429 (2017).
Malfertheiner, P. et al. Helicobacter pylori infection. Nat. Rev. Dis. Prim. 9, 19 (2023).
Noach, L. A., Rolf, T. M. & Tytgat, G. N. J. Electron microscopic study of association between Helicobacter pyloni and gastric and duodenal Mucosa. J. Clin. Pathol. 47, 699–704 (1994).
Mahdavi, J. et al. Helicobacter pylori sabA adhesin in persistent infection and chronic inflammation. Sci. 297, 573–578 (2002).
Salama, N. R., Hartung, M. L. & Müller, A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat. Rev. Microbiol. 11, 385–399 (2013).
Carrasco, G. & Corvalan, A. H. Helicobacter pylori -induced chronic gastritis and assessing risks for gastric cancer. Gastroenterol. Res. Pract. 2013, 529–541 (2013).
Pompaiah, M. & Bartfeld, S. Gastric organoids: an emerging model system to study helicobacter pylori pathogenesis. Curr. Top. Microbiol Immunol. 400, 149–168 (2017).
Choi, E. et al. Cell lineage distribution atlas of the human stomach reveals heterogeneous gland populations in the gastric antrum. Gut 63, 1711–1720 (2014).
Bartfeld, S. et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148, 126–136.e6 (2015).
Sebrell, T. A. et al. A novel gastric spheroid co-culture model reveals to the gastric epithelium. Cell. Mol. Gastroenterol. Hepatol. 8, 157–171.e3 (2019).
Boccellato, F. et al. Polarised epithelial monolayers of the gastric mucosa reveal insights into mucosal homeostasis and defence against infection. Gut 68, 400–413 (2019).
Uotani, T. et al. Changes of tight junction and interleukin-8 expression using a human gastroid monolayer model of Helicobacter pylori infection. Helicobacter 24, 1–12 (2019).
Aguilar, C. et al. Helicobacter pylori shows tropism to gastric differentiated pit cells dependent on urea chemotaxis. Nat. Commun. 13, 5878 (2022).
Hofer, M., Duque-Correa, M. A. & Lutolf, M. P. Patterned gastrointestinal monolayers with bilateral access as observable models of parasite gut infection. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-024-01313-4 (2024).
Sashikawa Kimura, M., Mutoh, H. & Sugano, K. SOX9 is expressed in normal stomach, intestinal metaplasia, and gastric carcinoma in humans. J. Gastroenterol. 46, 1292–1299 (2011).
Han, S. et al. Defining the identity and dynamics of adult gastric isthmus stem cells. Cell Stem Cell 25, 342–356.e7 (2019).
Lu, J. et al. Characterization of potent and selective iodonium-class inhibitors of NADPH oxidases. Biochem. Pharmacol. 143, 25–38 (2017).
Grasberger, H., El-Zaatari, M., Dang, D. T. & Merchant, J. L. Dual oxidases control release of hydrogen peroxide by the gastric epithelium to prevent helicobacter felis infection and inflammation in mice. Gastroenterology 145, 1045–1054 (2013).
Miyoshi, H. & Stappenbeck, T. S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8, 2471–2482 (2013).
Bartfeld, S. & Clevers, H. Organoids as model for infectious diseases: culture of human and murine stomach organoids and microinjection of helicobacter pylori. J. Vis. Exp. 1–9 (2015).
Fung, C. et al. High-resolution mapping reveals that microniches in the gastric glands control Helicobacter pylori colonization of the stomach. PLoS Biol. 17, 1–28 (2019).
Acknowledgements
We thank Dr. Mikhail Nikolaev for help producing the microfabricated mold for stamps, Dr James H. Doroshow (NIH) for the DUOX2 inhibitor and Dr. Tania Hübscher, Dr. Antonius Chrisnandy, Bilge Sen Elci and Dr. L. Francisco Lorenzo-Martin for valuable discussions. We thank Julia Prébandier, Lucie Tillard, Valentin Borgeat and Rachel Cooper for administrative and technical support. We acknowledge support from CMi, BIOP, GECF and HCF EPFL core facilities. This work was funded by the National Center of Competence in Research Bio-Inspired Materials, the EU Horizon 2020 research program INTENS (www.intens.info; no. 668294-2) and the Swiss National Science Foundation research grant no. 310030_179447 (M.P.L., M.H.). This work was further supported by the Novo Nordisk Foundation grant NNF19OC0056411 and National Institutes of Health grant 5U19AI116484-10 (M.R.A., Y.K.).
Author information
Authors and Affiliations
Contributions
M.H. and M.P.L. conceived the study, designed experiments, interpreted data and wrote the manuscript. M.H. conducted experiments and analysis. Y.K., J.A.K. and M.R.A. contributed to the conceptualization, setting up and interpretation of H. pylori infection experiments. N.B. pre-processed scRNAseq data. F.G. conducted and organized biopsy harvesting.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Gregor Gorkiewicz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hofer, M., Kim, Y., Broguiere, N. et al. Accessible homeostatic gastric organoids reveal secondary cell type-specific host-pathogen interactions in Helicobacter pylori infections. Nat Commun 16, 2767 (2025). https://doi.org/10.1038/s41467-025-57131-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-57131-y