Abstract
Dysregulation of mitochondrial function has been implicated in Parkinson’s disease (PD), but the role of mitochondrial metabolism in disease pathogenesis remains to be elucidated. Using an unbiased metabolomic analysis of purified mitochondria, we identified alterations in α-ketoglutarate dehydrogenase (KGDH) pathway upon loss of PD-linked CHCHD2 protein. KGDH, a rate-limiting enzyme complex in the tricarboxylic acid cycle, was decreased in CHCHD2-deficient male mouse brains and human dopaminergic neurons. This deficiency of KGDH led to elevated α-ketoglutarate and increased lipid peroxidation. Treatment of CHCHD2-deficient dopaminergic neurons with lipoic acid, a KGDH cofactor and antioxidant agent, resulted in decreased levels of lipid peroxidation and phosphorylated α-synuclein. CHCHD10, a close homolog of CHCHD2 that is primarily linked to amyotrophic lateral sclerosis/frontotemporal dementia, did not affect the KGDH pathway or lipid peroxidation. Together, these results identify KGDH metabolic pathway as a targetable mitochondrial mechanism for correction of increased lipid peroxidation and α-synuclein in Parkinson’s disease.
Similar content being viewed by others
Introduction
Parkinson’s disease is a common neurodegenerative disorder that is pathologically characterized by the preferential loss of dopaminergic neurons in the substantia nigra. These neurons are energetically active and, consequently, highly susceptible to mitochondrial dysfunction and oxidative stress1. Indeed, mitochondrial dysfunction has been extensively implicated in the pathogenesis of both familial and sporadic forms of PD2. Over the past two decades, genetic studies have identified several PD-linked genes that are primarily linked to mitochondrial dysfunction, such as PINK1/parkin-mediated mitochondrial quality control and dopamine oxidation pathways3,4,5,6,7,8. Recent metabolomic studies revealed alterations in mitochondrial metabolites in PD brain tissues and patient iPSC-derived cells9,10,11. However, the molecular mechanisms through which mitochondrial metabolic pathways contribute to the pathogenesis of PD have been largely unknown.
Mutations in CHCHD2 have been identified as a cause of familial PD12. CHCHD2 protein is localized in the intermembrane space (IMS) of mitochondria and plays a role in stabilizing cytochrome C and maintaining cristae structure12,13,14, as well as regulating the transcription of mitochondrial genes under stress conditions14,15. CHCHD2 and its close homolog CHCHD10 share an evolutionary origin from the same ancestral gene and are both implicated in the maintenance of mitochondrial cristae structure16,17,18,19,20,21,22,23,24. Mutations in CHCHD10 cause amyotrophic lateral sclerosis/frontotemporal dementia (ALS/FTD)23,25,26, characterized by degeneration of upper and lower motor neurons. Coding variants of CHCH10 have also been linked to Charcot-Marie-Tooth disease type 2 (CMT2), Jokela type spinal muscular atrophy (SMAJ) and mitochondrial myopathy27,28,29,30. Notably, both CHCHD2 and CHCHD10 are expressed in dopaminergic and motor neurons31,32. This implies the presence of distinct mitochondrial signaling pathways regulated by CHCHD2 and CHCHD10 that may be implicated in PD and ALS/FTD, respectively.
In this study, we examined mitochondrial metabolism in CHCHD2- and CHCHD10-deficient cells and found that tricarboxylic acid (TCA) cycle rate-limiting enzyme complex KGDH was preferentially impaired by deficiency of CHCHD2 but not CHCHD10. This deficiency of KGDH resulted in disrupted conversion of α-KG to downstream succinate, accompanied by an increase in lipid peroxidation and phosphorylated α-synuclein. Treatment of CHCHD2-deficient dopaminergic neurons with lipoic acid, a KGDH cofactor and antioxidant agent, was able to reduce lipid peroxidation and prevent the accumulation of phosphorylated α-synuclein. Together, these results highlight an important role of KGDH-related metabolic dysfunction in the pathogenesis of Parkinson’s disease.
Results
CHCHD2-deficient cells exhibit reduced KGDH enzyme complex and increased mitochondrial α-ketoglutarate
We performed a comprehensive analysis of hydrophilic metabolites of purified mitochondria by liquid chromatography-mass spectrometry. We labeled CHCHD2 knockout and control HEK293T cells with HA-EGFP-OMP25 and immunopurified mitochondria with high purity and enrichment using anti-HA magnetic beads (Fig. S1A–S1E). We found that TCA cycle metabolite α-KG was increased in CHCHD2-deficient cells compared to controls (Fig. 1A–E and Supplementary Data 1). In addition, CHCHD2-deficient cells exhibited a decrease in GSH/GSSG ratio in mitochondria (Fig. 1K). Mitochondrial α-KG is generated from both citrate/isocitrate and glutamate and converted to succinyl CoA and then succinate utilizing NAD+ as a cofactor (Fig. 1D). We found that the levels of citrate/isocitrate and glutamate were not significantly changed (Fig. 1F,G), whereas the α-KG/succinate ratio and NAD+ level were increased in CHCHD2-deficient cells (Fig. 1H–J), suggesting that the conversion of α-KG to succinate may be disrupted.
A Schematic illustration of purification of mitochondria and metabolomic analysis. D2 KO: CHCHD2 knockout. Mito IP: mitochondria immunopurification. Created in BioRender. Gao, G. (2025) https://BioRender.com/a45x634. B Heatmap representing metabolites abundance of CHCHD2-deficient (KO) and control (ctrl) HEK293T cells. Red and blue reflect high and low abundance, respectively. N = 4 and 3 for control and CHCHD2 knockout cells, respectively. C Volcano plot showing the changes of mitochondrial metabolites in CHCHD2-deficient HEK293T cells compared to controls. N = 3 and 4 biological replicates for knockout and control cells, respectively. Significant metabolites were defined as those with a fold change >2 or <0.5, and a P < 0.05 (two-sided t test). Metabolites that are significantly increased and decreased in CHCHD2-deficient cells are highlighted in red and blue, respectively. D Schematic of TCA cycle and glutaminolysis metabolites. The levels of mitochondrial α-KG (E), citrate/iso citrate (F), glutamate (G) and succinate (H) of control and CHCHD2-deficient HEK293T cells. I The ratio of mitochondrial α-KG to succinate of control and CHCHD2-deficient cells. J The level of mitochondrial NAD+ of control and CHCHD2-deficient cells. K The ratio of mitochondrial GSH to GSSG of control and CHCHD2-deficient cells. For (E–K), all values were relative to the average levels of control conditions, and each data point represents a biological replicate (N = 4 and 3 for control and CHCHD2 knockout cells, respectively). All data shown are mean ± SEM. ns, not significant. Two-sided t test. Source data are provided as a Source Data file.
We thus examined the glucose flux in the TCA cycle by labeling CHCHD2 knockout and control HEK293T cells with U-13C6 glucose. After the first turn of TCA cycle, pyruvate incorporates three carbons (M + 3), while citrate, α-KG, succinate, fumarate, and malate each incorporate two carbons (M + 2) derived from the initial U-13C6-labeled glucose (Fig. 2A). After 6 hours of labeling, the levels of M + 3-labeled pyruvate, M + 2-labeled citrate/isocitrate, α-KG and glutamate were not different between control and CHCHD2-deficient HEK293T cells (Fig. 2B–D, H and Supplementary Data 2). This indicates that the glucose entry into the TCA cycle and the exchange between α-KG and glutamate were not affected by the loss of CHCHD2. However, CHCHD2-deficient cells exhibited reduced levels of M + 2-labeled succinate, fumarate and malate compared to control cells (Fig. 2E–G and Supplementary Data 2), suggesting that glucose flux was blocked at the conversion of α-KG to succinate.
A Schematic of U-13C6 glucose flux into the TCA cycle. After the first turn of TCA cycle, pyruvate incorporates three carbons (M + 3), while citrate, α-KG, succinate, fumarate and malate each incorporate two carbons (M + 2) derived from the initial U-13C6-labeled glucose. B–H The levels of unlabeled (M + 0) and 13C labeled (M + N) pyruvate, citrate/iso citrate, α-KG, succinate, fumarate, malate and glutamate in CHCHD2-deficient and control HEK293T cells after 6 h of labeling with U-13C6 glucose. The levels of metabolites were normalized by the levels of 13C6 glucose (Glc) for each sample. N = 3 biological replicates for CHCHD2 knockout and control cells. All data shown are mean ± SEM. ns, not significant. Two-sided t test. I Schematic of the conversion of α-KG to Succinyl-CoA catalyzed by KGDH enzyme complex. TPP: thiamine pyrophosphate, LA: lipoic acid, DHLA: dihydrolipoic acid, E1: oxoglutarate dehydrogenase (OGDH), E2: dihydrolipoyl succinyltransferase (DLST), E3: dihydrolipoyl dehydrogenase (DLD). Created in BioRender. Gao, G. (2025) https://BioRender.com/j39s594. J–M Protein levels of OGDH, DLST and DLD in control and two independent clones of CHCHD2 knockout HEK293T cells. N = 12, 11, and 6 biological replicates for control, D2 KO #1 and D2 KO #2, respectively. All data shown are mean ± SEM. One-way ANOVA with Dunnett’s test. N, O KGDH enzyme activities of control and CHCHD2-deficient cells. N = 6 biological replicates. Data shown are mean ± SEM. One-way ANOVA with Dunnett’s test. P–S Immunoblots and quantifications of OGDH, DLST and DLD protein levels of control and CHCHD10-deficient (D10 KO) HEK293T cells. N = 4 biological replicates. Data shown are mean ± SEM. ns, not significant. Two-sided t test. T, U KGDH enzyme activities of control and CHCHD10-deficient cells. N = 5 biological replicates. Data shown are mean ± SEM. ns, not significant. Two-sided t test. Source data are provided as a Source Data file.
Alpha-ketoglutarate dehydrogenase (KGDH), a rate-limiting enzyme complex of the TCA cycle, catalyzes the conversion of α-KG to succinyl CoA (Fig. 2I). The enzyme complex comprises multiple copies of three subunits: oxoglutarate dehydrogenase (OGDH, E1), dihydrolipoyl succinyltransferase (DLST, E2) and dihydrolipoyl dehydrogenase (DLD, E3) (Fig. 2I). Downregulation or inhibition of KGDH enzyme complex results in accumulation of α-KG33,34. We found that the protein levels of the three KGDH subunits and the activity of the enzyme complex were decreased in CHCHD2-deficient cells compared to controls (Fig. 2J–O). Mitochondrial content remained unaffected as indicated by unchanged levels of citrate synthase (CS, mitochondrial matrix marker) and COXIV (inner mitochondrial membrane marker) (Fig. S2A–S2C). In addition, the protein levels of IDH2 and IDH3A, which catalyze the conversion of citrate/isocitrate to α-KG, were not affected by the loss of CHCHD2 (Fig. S2D and S2E). These findings demonstrate that the deficiency of CHCHD2 leads to reduction in KGDH enzyme complex, disrupting the conversion of α-KG to downstream succinate.
Inhibition of KGDH enzyme complex limits NADH production and consequently impairs mitochondrial respiration35,36,37. Indeed, CHCHD2-deficient cells exhibited decreased oxygen consumption rate compared to control cells (Fig. S2F–S2H). The assembly of mitochondrial complex I was not affected by CHCHD2 deficiency (Fig. S2I and S2J). The deficiency of ALS/FTD-linked CHCHD10, a close homolog of CHCHD2, did not affect the protein levels or enzyme activity of KGDH complex in HEK293T cells (Fig. 2P–U, S3A–S3D, S3F and S3G). The levels of mitochondrial α-KG were not significantly affected by the loss of CHCHD10 (Fig. S3O). Thus, KGDH enzyme complex is impaired by the deficiency of CHCHD2 but not CHCHD10.
CHCHD2 deletion or T61I mutation result in KGDH deficiency in human dopaminergic neurons and mouse brain
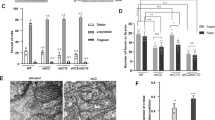
We next investigated whether CHCHD2 regulates KGDH enzyme complex in human induced pluripotent stem cells (iPSC)-derived dopaminergic (DA) neurons and mouse brains. DA neurons derived from CHCHD2-deficient iPSCs exhibited decreased levels of OGDHL, OGDH, DLST and DLD subunits of the KGDH complex (Fig. 3A–E, S4A–S4E), accompanied by a reduction in KGDH enzyme activity (Fig. 3F, G). We then generated a knock-in mouse model with PD-causing T61I mutation of CHCHD2. The levels of Chchd2 protein were reduced in T61I knock-in mice compared to controls (Fig. 3H, I). In addition, the levels of Ogdh, Dlst and Dld subunits of the KGDH enzyme complex were reduced in the striatum of T61I knock-in mice (Fig. 3H, J–L), whereas the mitochondrial content marker citrate synthase (Cs) was not affected (Figs. 3H, S2K). We also generated mutation-corrected isogenic controls for a PD patient-derived iPSC line with CHCHD2 T61I/+ mutation (Fig. S4A–S4C). DA neurons with T61I mutation exhibited decreased protein levels of CHCHD2 and KGDH subunits OGDHL, DLST and DLD compared to isogenic controls (Fig. 3M–R, S4F and S4G). On the other hand, DA neurons derived from CHCHD10 knockout or ALS/FTD-linked S59L knock-in iPSC lines exhibited unchanged levels of OGDHL, OGDH, DLST and DLD subunits compared to controls (Fig. S3H–S3L and S3N). Together, these data demonstrated that CHCHD2 deletion or PD-causing T61I mutation resulted in decreased KGDH enzyme complex in human DA neurons and mouse brains.
A–E Immunoblots and quantifications of OGDHL, OGDH, DLST and DLD protein levels in control and CHCHD2-deficient human dopaminergic (DA) neurons. N = 4 biological replicates. Data shown are mean ± SEM. Two-sided t test. F, G KGDH enzyme activities in control and CHCHD2-deficient dopaminergic neurons. N = 6 biological replicates. Data shown are mean ± SEM. Two-sided t test. H–L Immunoblots and quantifications of Chchd2, Ogdh, Dlst and Dld protein levels in striatum of wildtype (WT) and Chchd2 T61I/+ knock-in (KI) mice. n = 3 mice. Data shown are mean ± SEM. Two-sided t test. M–R Immunoblots and quantifications of CHCHD2, OGDHL, OGDH, DLST, and DLD protein levels in DA neurons derived from patient iPSC line with PD-linked CHCHD2 T61I/+ mutation (T61I/ + PD) and mutation-corrected isogenic controls (iso ctrl). N = 3 biological replicates for 3 N and 3 P, and 6 biological replicates for 3 O, 3Q, and 3 R. Data shown are mean ± SEM. Two-sided t test. S–X The levels of unlabeled (M + 0) and 13C labeled (M + N) pyruvate, citrate/iso citrate, α-KG, succinate, fumarate and malate in T61I/ + PD and isogenic control DA neurons after 6 h of labeling with U-13C6 glucose. The levels of metabolites were normalized by the levels of 13C6 glucose for each sample. N = 4 biological replicates. All data shown are mean ± SEM. ns, not significant. Two-sided t test. Source data are provided as a Source Data file.
To investigate whether KGDH-linked mitochondrial metabolism was disrupted by CHCHD2 T61I PD mutation, we performed U-13C6 glucose fluxomic analysis in T61I mutant and isogenic control DA neurons. T61I mutant DA neurons exhibited decreased levels of M + 2-labeled succinate, fumarate and malate, while the levels of M + 3-labeled pyruvate and M + 2 labeled α-KG remained unaffected after 6 hours of labeling with U-13C6 glucose (Fig. 3S–X and Supplementary Data 3). These results suggest that the glucose flux was blocked at the conversion of α-KG to succinate while the glucose entry into the TCA cycle was unaffected, further supporting that CHCHD2 T61I mutation led to the deficiency of KGDH.
Deficiency of CHCHD2 but not CHCHD10 leads to lipid peroxidation
KGDH deficiency and the resulting α-KG accumulation have been associated with a decline in NADH-linked respiration and generation of reactive oxygen species (ROS)38,39,40. Deletion of DLST subunit of KGDH results in increased malondialdehyde, a product of lipid peroxidation41. Moreover, glutathione (GSH), which was decreased in CHCHD2-deficient cells (Fig. 1J), is required for glutathione peroxidase 4 (GPX4)-mediated protection from excessive lipid peroxidation42. We thus investigated whether the deficiency of CHCHD2 affects the capacity of cells to defend against lipid peroxidation (Fig. 4A). Using C11-BODIPY as lipid peroxidation sensor, we found that CHCHD2-deficient cells showed higher lipid peroxidation levels at baseline compared to control cells (Figs. 4B, C and S5A). Under stressed conditions where cells were challenged by RSL3- or ML162-induced GPX4 inhibition, and Erastin-induced cystine deprivation (Fig. 4A), CHCHD2-deficient cells also exhibited higher sensitivity to lipid peroxidation compared to controls (Figs. 4B, D, F, S5A, S5H–S5K). Since iron is required for the initiation of lipid peroxidation and associated ferroptosis, we examined the effects of iron chelators on lipid peroxidation in CHCHD2 knockout cells. Treatment of CHCHD2-deficient cells with iron chelators Deferoxamine (DFO) and Ciclopirox (CPX) prevented the induction of lipid peroxidation under RSL3- and ML162-stressed conditions (Figs. 4E, F and S5B–S5G).
A Alterations of metabolites of CHCHD2-deficient cells are highlighted by red arrows. Created in BioRender. Gao, G. (2025) https://BioRender.com/t66n226. B Histograms of the intensities of oxidized and reduced C11-BODIPY signals of control and CHCHD2-deficient HEK293T cells under DMSO- and RSL3-induced conditions. The ratio of oxidized to reduced C11-BODIPY fluorescence intensities of control and two CHCHD2 knockout clones of HEK293T cells under DMSO- (C) and RSL3-treated (D) conditions. N = 12, 10 and 6 biological replicates for control, D2 KO #1 and D2 KO #2, respectively. All data shown are mean ± SEM. One-way ANOVA with Dunnett’s test. E, F The ratio of oxidized to reduced C11-BODIPY fluorescence intensities of control and CHCHD2 KO #1 HEK293T cells under RSL3- or ML162-stressed conditions treated with or without iron chelator deferoxamine (DFO). N = 5 and 4 biological replicates for (E, F), respectively. All data shown are mean ± SEM. One-way ANOVA with Dunnett’s test. G Representative images of oxidized and reduced C11-BODIPY signals of control and CHCHD2-deficient human dopaminergic neurons under DMSO- and RSL3-treated conditions. Scale bar, 20 μm. N = 3 independent experiments. H The ratio of oxidized to reduced C11-BODIPY fluorescence intensities of control and CHCHD2-deficient dopaminergic neurons under basal (left) and RSL3-treated (right) conditions. Images were collected and analyzed from N = 3 independent experiments. Data shown are mean ± SEM. Two-sided t test. I Representative images of oxidized and reduced C11-BODIPY signals of DA neurons with CHCHD2 T61I mutation and isogenic controls under basal condition. Scale bar, 20 μm. N = 3 independent experiments. J The ratio of oxidized to reduced C11-BODIPY fluorescence intensities of T61I/ + PD and isogenic control DA neurons. Images were collected and analyzed from N = 3 independent experiments. Data shown are mean ± SEM. Two-sided t test. K Histograms of the intensities of oxidized and reduced C11-BODIPY signals of control and CHCHD10-deficent HEK293T cells under basal and RSL3-induced conditions. The ratio of oxidized to reduced C11-BODIPY fluorescence intensities of control and CHCHD10-deficient cells under basal (L) and RSL3-treated (M) conditions. N = 3 independent experiments. Data shown are mean ± SEM. ns, not significant. Two-sided t test. Source data are provided as a Source Data file.
Using live cell imaging, we confirmed increased lipid peroxidation under basal conditions in iPSC-derived DA neurons with CHCHD2 deletion (Fig. 4G, H left). Upon RSL3 treatment, CHCHD2-deficient DA neurons also showed higher sensitivity to lipid peroxidation than control neurons (Fig. 4G, H right). DA neurons with PD-linked T61I mutation also exhibited increased levels of lipid peroxidation compared to isogenic controls (Fig. 4I,J). Notably, the deficiency of CHCHD10 does not affect the levels of lipid peroxidation, either at baseline, or upon RSL3- or erastin-induced stress (Figs. 4K–M, and S6A–S6E).
We next examined whether specific components of the KGDH enzyme complex contributed to the increased levels of lipid peroxidation observed in CHCHD2-deficient cells. Using lentiviral short hairpin RNA (shRNA)-mediated RNA interference (RNAi), we achieved reduced expression levels of DLST, the E2 catalytic subunit of KGDH (Fig. 5A, B). The protein levels of the E1 subunit OGDH exhibited a significant decrease in cells expressing shRNA constructs specifically targeting DLST (Fig. 5A,C), whereas the protein level of E3 subunit DLD remained unaffected (Fig. 5A, D), indicating that maintaining the proper copy number of the E2 subunit is crucial for the stability of the E1 subunit within the enzyme complex. We analyzed the KGDH-linked mitochondrial metabolism in DLST knockdown HEK293T cells by performing the U-13C6 glucose flux assay. The levels of M + 2-labeled α-KG were increased whereas the levels of M + 3-labeled pyruvate and M + 2-labeled citrate/isocitrate were unaffected in DLST knockdown cells compared to non-targeting controls (Fig. 5E–G and Supplementary Data 4). Importantly, disruption of KGDH led to decreased levels of M + 2-labeled succinate, fumarate and malate (Fig. 5H–J and Supplementary Data 4), as observed in cells with CHCHD2 deletion or T61I mutation.
A–D Immunoblots and quantifications of DLST, OGDH and DLD protein levels of non-targeting control (NT ctrl) and two independent lines of DLST knockdown (KD #1 and KD #2) HEK293T cells. N = 8 biological replicates. All data shown are mean ± SEM. ns, not significant. One-way ANOVA with Dunnett’s test. E–J The levels of unlabeled (M + 0) and 13C labeled (M + N) pyruvate, citrate/iso citrate, α-KG, succinate, fumarate and malate in non-targeting control and DLST knockdown HEK293T cells after 6 h of labeling with U-13C6 glucose. The levels of metabolites were normalized by the levels of 13C6 glucose for each sample. N = 5 biological replicates. All data shown are mean ± SEM. ns, not significant. Two-sided t test. K Histograms of the intensities of oxidized and reduced C11-BODIPY signals of control and DLST knockdown HEK293T cells under basal and RSL3-induced conditions. The ratio of oxidized to reduced C11-BODIPY fluorescence intensities of control and DLST knockdown cells under basal (L) and RSL3-treated (M) conditions. N = 4 biological replicates. All data shown are mean ± SEM. One-way ANOVA with Dunnett’s test. Source data are provided as a Source Data file.
We found that, like CHCHD2 deficiency, disruption of the E2 and E1 subunits within the KGDH complex led to increased levels of lipid peroxidation under basal condition (Fig. 5K,L). DLST knockdown cells also exhibited higher sensitivity to RSL3-induced lipid peroxidation (Fig. 5K, M). Thus, deficiency of KGDH results in the disruption in the conversion of α-KG to succinate, leading to increased levels of lipid peroxidation.
Alpha-lipoic acid decreases lipid peroxidation and prevents the accumulation of phosphorylated α-synuclein
We next investigated whether enhancing KGDH enzyme activity could mitigate increased lipid peroxidation observed in CHCHD2-deficient cells. Lipoic acid is an essential cofactor for mitochondrial enzyme complexes including KGDH and pyruvate dehydrogenase (PDH), and it displays antioxidant and metal chelating properties43,44. Lipoic acid treatment has been shown to enhance the activity of KGDH enzyme complex and protect it from oxidative damage during aging45,46,47,48. We found that treatment of CHCHD2-deficient HEK293T cells with α-lipoic acid (α-LA) reduced the levels of mitochondrial α-KG and decreased lipid peroxidation in a dose-dependent manner (Figs. 6A, S7A–S7C and Supplementary Data 5). We further demonstrated that treatment with α-LA rescued the sensitivity of CHCHD2-deficient DA neurons to RSL3-induced lipid peroxidation (Fig. 6B, C). Treatment with α-LA inhibited the RSL3-induced lipid peroxidation in control but not DLST knockdown cells (Figs. S7D and S7E), suggesting that the beneficial effects of α-LA treatment were at least partly mediated through the activation of KGDH enzyme.
A The levels of mitochondrial α-KG of CHCHD2 knockout HEK293T cells (KO #1) treated with vehicle or 50 μM α-lipoic acid (α-LA). N = 4 biological replicates. Data shown are mean ± SEM. Two-sided t test. B Representative images of oxidized and reduced C11-BODIPY signals of CHCHD2-deficient dopaminergic neurons at baseline (DMSO condition) and RSL3-induced conditions pretreated with serial concentrations of α-LA. Scale bar, 20 μm. N = 4 independent experiments. C The ratio of oxidized to reduced C11-BODIPY fluorescence intensities of CHCHD2-deficient dopaminergic neurons at baseline (DMSO condition) and RSL3-induced conditions pretreated with serial concentrations of α-LA. Images were collected and analyzed from N = 4 independent experiments. Data shown are mean ± SEM. One-way ANOVA with Dunnett’s test. D Immunoblots and quantifications of pS129 α-synuclein (α-Syn) levels in striatum of 24-month-old wildtype and Chchd2 T61I knock-in mice. n = 3 mice. Data shown are mean ± SEM. Two-sided t test. E Immunoblots and quantifications of pS129 α-synuclein levels in DA neurons derived from T61I/ + PD patient and isogenic control iPSCs. N = 3 biological replicates. Data shown are mean ± SEM. Two-sided t test. F, G Immunoblots and quantifications of pS129 α-synuclein levels of control and CHCHD2-deficient human DA neurons. N = 6 biological replicates. Data shown are mean ± SEM. Two-sided t test. Immunoblots and quantifications of pS129 α-synuclein levels in RIPA- soluble (H) and insoluble (I) fractions of CHCHD2-deficient human DA neurons treated with vehicle, 50 μM or 100 μM α-LA. CBB, Coomassie brilliant blue. N = 3 biological replicates. Data shown are mean ± SEM. One-way ANOVA with Dunnett’s test. Source data are provided as a Source Data file.
Lipid peroxidation-induced oxidative modifications are implicated in the oligomerization of α-synuclein49,50,51. We found that the levels of phosphorylated α-synuclein at Ser129 (pS129 α-synuclein) were elevated in the striatum of two-year-old CHCHD2 T61I knock-in mice and human DA neurons with CHCHD2 deletion or T61I mutation (Fig. 6D–G). The levels of pS129 α-synuclein were unchanged in DA neurons with CHCHD10 deletion or ALS/FTD-linked S59L mutation (Fig. S3H, S3M). Treatment of CHCHD2-deficient DA neurons with α-LA was able to reduce the levels of pS129 α-synuclein (Fig. 6H, I). Thus, the KGDH cofactor and antioxidant agent α-lipoic acid protects neurons from lipid peroxidation and prevents the accumulation of phosphorylated α-synuclein.
Discussion
In this study, we found that Parkinson’s disease-linked CHCHD2 resulted in reduction in KGDH enzyme complex in human dopaminergic neurons and mouse brains, and that dysregulated KGDH led to increased lipid peroxidation. Treatment of CHCHD2-deficient neurons with α-lipoic acid, a cofactor of KGDH, was able to reduce lipid peroxidation and prevent the accumulation of phosphorylated α-synuclein. This metabolic pathway was not affected by the deficiency of CHCHD10, a close homolog of CHCHD2 that is linked to ALS/FTD. KGDH enzyme complex resides in the mitochondrial matrix facing the inner mitochondrial membrane, whereas CHCHD2 is localized in the intermembrane space of mitochondria, suggesting that the regulation of KGDH subunits by CHCHD2 may not occur through direct protein-protein interaction. It has been reported that CHCHD2 can localize to the nucleus and promote transcription of a subset of mitochondrial genes including COX subunit 4 isoform 2 (COX4I2) under hypoxic conditions14,15. Another study in Drosophila reported that CHCHD2 prevents the YME1L-mediated degradation of OPA1 by competitive binding with chaperone-like protein P32, potentially displacing YME1L17,52. These suggest that CHCHD2 may regulate the protein levels of KGDH subunits through transcriptional regulation, and/or by affecting the stability/degradation of the subunits. Given that disruption of one subunit can destabilize the complex, it is also possible that CHCHD2 may primarily impact one specific subunit of KGDH.
Lipid peroxidation is a process of oxidative modification on phospholipids with polyunsaturated fatty acid chains, which damages mitochondrial and plasma membrane and may trigger ferroptosis in the context of iron dyshomeostasis53,54,55. Reactive adducts resulting from lipid peroxidation additionally induce oxidative modifications on enzymes participating in mitochondrial metabolism and α-synuclein oligomerization49,50,51,56, potentially contributing to Parkinson’s disease pathology. Of note, 4-hydroxy-2-nonenal (4-HNE), a highly reactive toxic product of lipid peroxidation, irreversibly modifies the lipoic acid covalently bound to the E2 DLST subunit of KGDH, leading to inhibition of KGDH activity56. Fruit flies lacking CG5010, a homologous gene of CHCHD2, showed elevated levels of 4-HNE in TH-positive neurons in the brain13. Our work suggests that deficiency in KGDH enzyme complex sensitizes cells to lipid peroxidation, potentially due to increased reactive oxygen species, which in turn depletes reducing agent GSH. Therefore, a toxic feedback loop may exist between dysregulated KGDH and increased lipid peroxidation, leading to the disruption of mitochondrial function and accumulation of phosphorylated α-synuclein in CHCHD2-deficient dopaminergic neurons. Increased levels of pS129 α-synuclein in CHCHD2 mutant DA neurons and T61I knock-in mouse brains is consistent with the presence of Lewy pathology with pS129 α-synuclein staining in brain tissues of a PD patient carrying T61I mutation57,58,59.
CHCHD2 and CHCHD10 are highly homologous genes that share an evolutionary origin, stemming from the same ancestral gene21,22. Both CHCHD2 and CHCHD10 are localized in the mitochondrial intermembrane space and their physical interaction has been observed19,31,32,60, indicating a potential involvement in overlapping mitochondrial pathways. Indeed, both CHCHD2 and CHCHD10 have been functionally associated with the stability of OPA1 and the MICOS complex, which regulate mitochondrial fusion and cristae structure16,17,18,19,20,24. In mice with the knock-in of CHCHD10 S55L mutation (the equivalent of human ALS/FTD-linked S59L mutation), CHCHD10 co-aggregates with CHCHD2, leading to mitochondrial integrated stress response61. Studies in mammalian cell lines and mouse models revealed compensatory responses between CHCHD2 and CHCHD10 in the regulation of cristae morphology, and the activation of OMA1 and mitochondrial integrated stress response62,63, suggesting potentially shared mechanisms through which CHCHD2 and CHCHD10 contribute to the maintenance of mitochondrial ultrastructure and homeostasis.
On the other hand, mutations in CHCHD2 cause PD, while mutations in CHCHD10 are linked to ALS/FTD, CMT2, SMAJ and mitochondrial myopathy, suggesting that the functional disparities between these two proteins may potentially contribute to preferential neurodegeneration in these disorders. Our work demonstrated that CHCHD2, but not CHCH10, plays an important role in maintaining the function of TCA cycle rate limiting enzyme complex KGDH, protecting cells from excessive lipid peroxidation. We did not observe an obvious compensation between these two proteins, and the reduction of KGDH subunits was not exacerbated by CHCHD2/CHCHD10 double knockout compared to CHCHD2 single knockout (Fig. S3A, S3E, S3P–S3S). CHCHD10 S55L knock-in mice showed induction of mitochondrial integrated stress response, leading to depletion of mitochondrial DNA and taurine64. Fibroblasts of ALS patients carrying R15L mutation in CHCHD10 exhibited an increase in the NADH/NAD+ ratio65. Therefore, CHCHD2 and CHCHD10 may exert distinct effects on mitochondrial metabolism that lead to preferential degeneration of dopaminergic and motor neurons. Further studies dissecting the molecular details of CHCHD10-linked metabolic dysfunction will be required to elucidate a distinct role of CHCHD10 in ALS/FTD.
We found that CHCHD2 was increased in DA neurons derived from PD patient iPSC lines carrying LRRK2 G2019S/+ or GBA1 84GG/+ mutation compared to mutation-corrected isogenic controls (Fig. S8), suggesting a potential compensatory response by CHCHD2 in these LRRK2 and GBA1 mutant DA neurons. Other recent studies revealed decreased levels of succinate in the cortex and putamen regions of PD patients9, and decreased levels of succinate, fumarate and malate, as well as OGDHL, the brain-specific E1 isoform of KGDH complex, in neural precursor cells derived from sporadic PD iPSC lines10. Together, these findings suggest that KGDH deficiency may play a role in other forms of PD.
In sum, our data highlight an important role of dysregulated mitochondrial metabolism in the pathogenesis of CHCHD2-linked PD and suggest a therapeutic approach through targeting of α-ketoglutarate dehydrogenase.
Methods
Ethics statement
Skin fibroblasts from healthy control donor (See also Fig. S4A) were obtained from the Northwestern University Movement Disorders Center (MDC) Biorepository. The generation and use of the healthy control line followed the guidelines approved by the Institutional Review Board of Northwestern University (STU00091585). All mouse work followed the guidelines approved by the Institutional Animal Care and Use Committee of Northwestern University for this project (protocol number: IS00003894).
Cells and animals
HEK293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, 11995065) supplemented with 10% (vol/vol) heat inactivated fetal bovine serum (Hi-FBS) (GeminiBio). Skin fibroblasts from healthy control donor were reprogrammed into induced pluripotent stem cells (iPSCs) by Sendai particles-mediated expression of Yamanaka factors Oct3/4, Sox2, Klf4 and c-Myc (Sendai Reprogramming Kit, Invitrogen)66. The iPSC line derived from a PD patient carrying CHCHD2 T61I mutation was a gift from Nobutaka Hattori lab12,59. The LRRK2 G2019S PD iPSC line was obtained from the Parkinson’s Progression Markers Initiative (PPMI). GBA1 84GG PD patient and mutation-corrected isogenic control iPSC lines were generated and characterized previously67,68. In this study, human iPSC lines with CHCHD2 or CHCHD10 deletion were generated in a healthy control background, isogenic control lines with corrections of CHCHD2 T61I or LRRK2 G2019S mutations were generated from PD patient iPSC lines, and CHCHD10 S59L mutant lines were created by introducing the mutation into a healthy control background, all using CRISPR/Cas9-mediated genome editing. All iPSC lines generated using CRISPR/Cas9-mediated genome editing exhibited a normal karyotype. Pluripotency marker expression was verified through immunocytochemistry. Sequencing of genomic regions with predicted off-target sites located in exons confirmed the absence of off-target edits (Supplementary Table 1). Human iPS cells were cultured in complete mTeSR Plus medium (STEMCELL Technologies, 100-0276) and passaged manually every 6–7 days. All cells were maintained at 37 °C in incubator with 5% CO2 and tested negative for mycoplasma contamination.
The mice used in this study were maintained on a C57BL/6 J background and were housed in the facility of Northwestern University Center for Comparative Medicine with a 12-h light/dark cycle and fed a regular diet ad libitum. All experiments were carried out according to regulatory standards. Wildtype and Chchd2 T61I knock-in mice aged six weeks and two years were utilized for immunoblotting experiments of KGDH subunits and pS129 α-synuclein, respectively, with 3 male mice per condition.
Differentiation of dopaminergic neurons
Human iPSCs were differentiated into midbrain dopaminergic neurons as previously described69,70. On day 13 of differentiation, cells were manually passaged in small chunks with size of 1–2 mm and plated on 10 cm dishes pre-coated with poly-D-lysine (PDL) and laminin. On day 25, cells were dissociated into single cells using Accutase (Sigma) and plated on PDL and laminin-coated plates for downstream assessments. Differentiated neurons were cultured in Neurobasal medium (Life Technologies) supplemented with Neurocult SM1 (Stemcell technologies), BDNF, ascorbic acid, GDNF, TGF-β3, Dibutyryl-cAMP and DAPT. By day 50–60, neurons were used for immunocytochemistry, live cell imaging and KGDH activity assays. Neurons at day 90 and day 120 were used for immunoblotting experiments of KGDH subunits and pS129 α-synuclein, respectively.
CRISPR/Cas9-mediated genome editing
Deletion of CHCHD2 or CHCHD10 gene in HEK293T cells were generated by transfection of Px459 plasmid containing sgRNA targeting sequence and S.p.Cas9 coding sequence. Briefly, sgRNA targeting sequences were cloned into Px459 backbone (a gift from Feng Zhang)71 using BbsI sites through T4 DNA Ligase. DNA constructs were transfected in HEK293T cells using X-tremeGENE HP DNA Transfection Reagent (Roche). 48 h post transfection, cells were selected using 2 µg/ml of Puromycin for 2 continuous days. Single cells were sorted into 96-well plates with one cell per well and cultured in complete DMEM medium. Knockout clones were screened and validated by Sanger sequencing and immunoblotting. For generation of CHCHD2 and CHCHD10 knockout lines in HEK293T cells, sgRNAs targeting exon 2 were designed using CRISPOR to maximize on-target efficiency while minimizing off-target effects. CHCHD2 sgRNA targeting sequence: 5’- GCTCTTCTGCTGCTGCGCCC-3’, and CHCHD10 sgRNA targeting sequence: 5’- AGATGGCGACCACGGCCGCA-3’.
Deletion of CHCHD2 gene in human iPSC line was performed by electroporation of CRISPR-Cas9 ribonucleoprotein (RNP) complex. For each 10 μl electroporation reaction, 2.2 μM Alt-R CRISPR-Cas9 crRNA and tracrRNA (Integrated DNA Technologies, IDT) were annealed to form crRNA:tracrRNA duplex, and then assembled with 1.8 μM Alt-R Cas9 nuclease (IDT). The assembled Cas9 RNP complex was transfected in iPS cells using Neon Transfection System (Invitrogen). Electroporated iPSCs were cultured overnight in complete mTeSR Plus medium supplemented with the ROCK inhibitor, then maintained in regular mTeSR Plus medium for 2 additional days to allow expansion. Cells were dissociated using Accutase into single cells and replated at a serial dilution to reach a low density for single cells to grow into separate colonies. Cells were cultured in complete mTeSR Plus medium supplemented with 10% CloneR (STEMCELL Technologies, # 05888) for 3 days to enhance survival, then maintained in regular mTeSR Plus medium until colonies expanded. Single clones were manually picked under a dissection microscope and cultured in 96-well plates, with one clone per well. Knockout clones were screened and validated by Sanger sequencing and immunoblotting. The sgRNA targeting sequence used for the deletion of CHCHD2 in healthy control iPSCs was designed using IDT, which is as follows: 5’- CATTGGGTCACGCCATTACT-3’.
Correction of CHCHD2 T61I or LRRK2 G2019S mutation in PD patient-derived iPSC lines was performed by electroporation of Px459 plasmid containing sgRNA targeting sequence and S.p.Cas9 coding sequence, together with single-stranded oligodeoxynucleotide (ssODN) as repair template. Briefly, sgRNA targeting sequences were cloned into Px459 backbone using BbsI sites through T4 DNA Ligase, and ssODNs were obtained from IDT. The sgRNA/Cas9-expressing plasmid and ssODN repair template were transfected in iPS cells using Neon Transfection System (Invitrogen). Electroporated iPSCs were cultured overnight in complete mTeSR Plus medium supplemented with the ROCK inhibitor, then selected using 0.25 μg/ml Puromycin for 2 days. After selection, cells were dissociated using Accutase into single cells and replated at a serial dilution to reach a low density for single cells to grow into separate colonies. Cells were maintained and single clones were screened as described above. The sgRNAs were designed within 10 bp of the mutation site using CRISPOR to maximize on-target efficiency while minimizing off-target effects. CHCHD2 T61I correction sgRNA targeting sequence: 5’-AGCTGCAGTGATTGCCATCT-3’, and ssODN: 5’-AGCACCAGTCGCTCAGCCACCAGCAGCGGCACCCCCATCTGCAGTTGGCTCTTCTGCTGCTGCGCCCCGGCAGCCAGGTCTGATGGCCCAGATGGCAACCACTGCAGCTGGCGTGGCTGTGGGCTCTGCTGTGGGGCACACATTGGGTCACGCCATTACTGGGGGCTTCAGTGGAGGAAGTAATGCTGAGCCT-3’. The T61I mutation of CHCHD2 was incorporated into the sgRNA to specifically target the genome of the patient line. LRRK2 G2019S correction sgRNA targeting sequence: 5’-GTCAGCAATCTTTGCAATGA-3’, and ssODN: 5’- ACCGAGACCTGAAACCCCACAATGTGCTGCTTTTCACACTGTATCCCAATGCTGCgATCATTGCAAAGATTGCTGACTACGGCATTGCTCAGTACTGCTGTAGAATGGGGATAAAAACATCAGAGGGCACACCAGGTAGGTGATCAGGTCTGTCTCATAATTCTATCTTCAGGATGGAT-3’. A synonymous mutation was incorporated into the ssODN for G2019S correction to prevent recutting and enhance knock-in efficiency.
Deletion of CHCHD10 in healthy control iPSC line was performed by electroporation of CRISPR-Cas9 RNP complex as described above. Knock-in of S59L mutation in healthy control iPSC line was performed by electroporation of CRISPR-Cas9 RNP complex, together with ssODN donor template. Electroporated iPSCs were maintained and single clones were screened as described above. The sgRNA for CHCHD10 editing was designed within 10 bp of the mutation site using IDT to maximize on-target efficiency while minimizing off-target effects, which is as follows: 5’- TCGGCTGTGGGACACGTCAT-3’. The ssODN donor template for knock-in of CHCHD10 S59L mutation was designed alongside the sgRNA as a set using IDT, which is as follows: 5’- GCTCCCCCCGCTGAAGGCTCCGGTCAGGGCGCTGCCCATCACATGTCCCACAGCCAAGCCCACGGCTACCCCTGCGGCCGTGGTCGCCATCTGAGC-3’. Two synonymous mutations were incorporated into the ssODN for S59L knock-in to prevent recutting and enhance knock-in efficiency.
For the generation of Chchd2 T61I knock-in mice, we chose the target sequence [5’-GGCCCAGATGGCTACCACCG (CGG, PAM)] in exon 2 of the mouse Chchd2 gene. A single-stranded oligonucleotides (ssODN) of 198nt containing a C > T change (ACC > ATC) was synthesized. This ssODN was designed for introduction of the T61I mutation into the mouse Chchd2 gene by homologous recombination after Cas9 cleavage at the targeting site. Microinjection of the SpCas9 mRNAs (from 100 ng/μl), sgRNA (50 ng/μl) and the ssODN (100 ng/μl) into the cytoplasm of fertilized eggs of the C57BL/6 J strain and embryo transplantation were performed at the Northwestern University Transgenic and Targeted Mutagenesis Facility. The founder mice were genotyped using PCR-sequencing with a pair of primers in introns 1 and 2 of the mouse Chchd2 (forward: 5’-CATTTGTCTGGGACCTGAGGA; reverse: CATCTCAGAAAGGAGAAGCAG). Six potential off-target sites were also sequenced. Founders were crossbred with C57BL/6 J mice, and the mouse lines were subsequently maintained on the C57BL/6 J genetic background.
Purification of mitochondria and extraction of metabolites
Rapid immunopurification of mitochondria was performed following an established protocol with minor modifications72,73. Briefly, 3xHA-EGFP-OMP25 or 3xMyc-EGFP-OMP25 tagged HEK293T cells were cultured on 15 cm dishes for 2 days (~80% confluency). Media were removed and cells were washed twice with ice-cold PBS, and then collected in 1 ml of ice-cold KPBS buffer (136 mM KCl and 10 mM KH2PO4, pH 7.25, in LC/MS-grade water) followed by centrifugation at 1000 × g for 2 min at 4 °C. KPBS buffer was removed, and cells were resuspended gently by pipetting in 1 ml cold KPBS buffer. Cells were homogenized using a syringe with 23 G x 1 in. needle (BD, 305271) for 15 strokes on ice. 50 μl cell homogenate was boiled in 2x Laemmli sample buffer (BioRad) as input fraction. The rest cell homogenate was centrifuged at 1000 × g for 2 min at 4 °C, and 800 μl mitochondria-containing supernatant lysate was incubated with prewashed anti-HA magnetic beads (100 μl beads slurry per reaction) (Thermo Fisher Scientific, 88837) for 5 min with gentle rotation at 4 °C. Beads were pelleted using a magnetic stand and gently washed four times with 1 ml cold KPBS buffer. After the final washing, beads/KPBS solution was transferred to a new tube. 20% of the final beads in KPBS solution were pelleted using a magnetic stand and boiled in 100 μl Laemmli sample buffer for immunoblotting analysis. The rest 80% of the final beads in KPBS solution were pelleted, and total hydrophilic metabolites were extracted in 50 μl pre-chilled 80% methanol for 5 min on ice. Samples were briefly centrifuged, and beads were pelleted on a magnetic stand. Metabolites-containing supernatant was transferred to a new tube followed by centrifugation at 17,000 × g for 15 min at 4 °C. The final supernatant was transferred to a new tube and dried using a SpeedVac Concentrator. Dried samples were stored at −80 °C until analysis. The cell equivalents of each immunoprecipitation reaction were calculated following the previous protocol73. Immunoprecipitation reactions with similar number of cell equivalents were used for downstream metabolomics.
U-13C6 glucose flux assay
HEK293T cells were plated and cultured in standard DMEM supplemented with 10% (vol/vol) Hi-FBS for 2 days to reach ~70% confluency. Six hours prior to cell harvesting, the medium was replaced with glucose- and sodium pyruvate-free DMEM (Gibco, 11966025) supplemented with 10 mM 13C6 D-glucose (Cambridge Isotope Lab, CLM-1396-1), 1 mM sodium pyruvate and 10% (vol/vol) Hi-FBS. Human iPSC-derived mature DA neurons were maintained in standard Neurobasal medium supplemented with Neurocult SM1. Six hours prior to cell harvesting, the medium was replaced with glucose- and sodium pyruvate-free Neurobasal-A Medium (Gibco, A2477501) supplemented with 20 mM 13C6 D-glucose (Cambridge Isotope Lab, CLM-1396-1), 0.227 mM sodium pyruvate, 2 mM L-Glutamine and Neurocult SM1. Cells were washed twice with normal saline solution (0.9% NaCl), and cell pellets were collected from N ≥ 3 independent biological replicates. Total hydrophilic metabolites were extracted in 1 mL of pre-chilled 80% (vol/vol) methanol by homogenizing the samples with a 23 G x 1 in. syringe needle (BD, 305271) for 15 strokes on ice. The samples were stored at −80 °C overnight to precipitate proteins. The following day, samples were centrifuged at 20,000 × g for 15 min at 4 °C, and the metabolite-containing supernatant was collected for HPLC-MS/MS analysis. The levels of metabolites were normalized by the levels of 13C6 glucose for each sample.
HPLC-MS/MS analysis of metabolites
Dried samples were reconstituted in 60% acetonitrile by vortexing for 30 sec, followed by centrifugation at 20,000 × g for 15 min at 4 °C. The supernatant was collected for HPLC-MS/MS analysis. The system comprised a Thermo Q-Exactive coupled with an electrospray source and an Ultimate3000 series HPLC system (Thermo), consisting of a binary pump, degasser, and autosampler equipped with a Xbridge Amide column (Waters; dimensions: 3.0 mm × 100 mm; particle size: 3.5 µm). The mobile phase A consisted of 95% (vol/vol) water, 5% (vol/vol) acetonitrile, 10 mM ammonium hydroxide, and 10 mM ammonium acetate (pH = 9.0); mobile phase B was 100% acetonitrile. The gradient profile was as follows: 0 min, 15% A; 2.5 min, 30% A; 7 min, 43% A; 16 min, 62% A; 16.1–18 min, 75% A; 18–25 min, 15% A, with a flow rate of 150 μL/min. The capillary temperature of the ESI source was set to 275 °C, with sheath gas at 35 arbitrary units, auxiliary gas at 5 arbitrary units, and the spray voltage at 4.0 kV. For positive/negative polarity switching mode, an m/z scan range from 60 to 900 was selected, with MS1 data collected at a resolution of 70,000. The automatic gain control (AGC) target was set at 1 × 106, and the maximum injection time was 200 ms. The top 5 precursor ions were subsequently fragmented in a data-dependent manner using the higher-energy collisional dissociation (HCD) cell set to 30% normalized collision energy in MS2 at a resolution power of 17,500. In addition to matching m/z, metabolites were identified by aligning retention times with analytical standards and/or MS2 fragmentation patterns. Data acquisition and analysis were performed using Xcalibur 4.1 (Thermo Fisher Scientific), Tracefinder 4.1 (Thermo Fisher Scientific) and MetaboAnalyst 5.0 (https://metaboanalyst.ca).
Immunoblotting
HEK293T cells or iPSC-derived dopaminergic neurons were washed with ice-cold PBS and homogenized in RIPA lysis buffer with protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, 78440). Striatum tissues of mice were homogenized and sonicated in RIPA lysis buffer with protease and phosphatase inhibitor cocktail. Samples were lysed on ice for 30 min followed by centrifugation at 16,000 × g for 15 min at 4 °C. The supernatant lysate was collected. RIPA-insoluble pellets were further boiled and sonicated in 2% SDS/50 mM Tris buffer (pH 7.4). The samples were centrifuged at 20,000 × g for 15 min at 4 °C, and the supernatant lysate was collected. Protein concentration was measured using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23225). Proteins were denatured by boiling in Laemmli sample buffer (BioRad), and then separated using 4–20% or 4–12% Tris-Glycine protein gel (Invitrogen), followed by transferring onto nitrocellulose membrane using BIO-RAD Trans-Blot Turbo Transfer system. Membranes were blocked with 5% nonfat milk in 1x TBST buffer and then incubated with primary antibodies overnight on an orbital shaker at 4 °C. Membranes were washed 3 times with 1x TBST buffer and incubated with anti-mouse or anti-rabbit secondary antibodies conjugated with horseradish peroxidase (HRP) for 1 h at room temperature. The signal was developed using Clarity Western ECL Substrate (BioRad) or West Femto Substrate (ThermoFisher) and imaged on a ChemiDoc imaging station equipped with a 16-bit CCD camera (BioRad). Signal intensities were quantified using Image Studio (LI-COR). Protein levels were normalized to α-tubulin (Sigma-Aldrich, T5168) or GAPDH (Sigma-Aldrich, MAB374) for HEK293T cells and β-III-tubulin (BioLegend, 801202) for dopaminergic neurons. Rabbit anti-CHCHD2 (Sigma-Aldrich, HPA027407), Rabbit anti-OGDHL (Proteintech, 17110-1-AP), Rabbit anti-OGDH (Proteintech, 15212-1-AP), Rabbit anti-DLST (Cell Signaling Technology, 11954 and ABclonal, A13297), Rabbit anti-DLD (Proteintech, 16431-1-AP), Rabbit anti-CHCHD10 (Proteintech, 25671-1-AP), Rabbit anti-pS129 α-synuclein (Cell Signaling Technology, 23706), Rabbit anti-Tyrosine hydroxylase (Sigma, AB152), mouse anti-Vimentin (BD Pharmingen, 550513), Rabbit anti-CS (Cell Signaling Technology, 14309), Rabbit anti-COXIV (Cell Signaling Technology, 4850), Rabbit anti-VDAC3 (Proteintech, 14451-1-AP), Mouse anti-LAMP1 (DSHB, H4A3), Rabbit anti-PEX5 (Cell Signaling Technology, 83020), Rabbit anti-Calreticulin (CALR) (Cell Signaling Technology, 12238), Mouse anti-NDUFB8 (Abcam, ab110242), Rabbit anti-ATP5A1 (Proteintech, 14676-1-AP), Rabbit anti-IDH2 (Proteintech, 15932-1-AP) and Rabbit anti-IDH3A (Proteintech, 15909-1-AP) antibodies were used in this study.
Blue native PAGE
HEK293T cells cultured on 10 cm dish (with ~70% confluency) were washed with ice-cold PBS, and then collected in 1 ml cold PBS, followed by centrifugation at 1000 × g for 2 min at 4 °C. PBS was removed and cells were homogenized in 1% n-dodecyl-β-D-maltoside (DDM) buffer (1% DDM, 1x NativePAGE Sample Buffer (Invitrogen, BN2008), and 1x protease and phosphotase inhibitor cocktail (Thermo Fisher Scientific, 78440)) using a syringe with 23 G x 1 in. needle (BD, 305271) for 10 strokes on ice. Samples were lysed for 20 min on ice, and protein lysate was collected after centrifugation at 18,000 x g for 30 minutes at 4 °C. Lysate was stored in small aliquots to avoid freeze-thaw cycles at −80 °C. Protein concentration was measured using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23225). Proteins were diluted in NativePAGE Sample Buffer (Invitrogen, BN2008) following manufacturer’s instructions. 20 μg proteins were loaded per lane onto NativePAGE 3–12% Bis-Tris protein gel (Invitrogen, BN1003BOX). After separation, proteins were wet transferred onto PVDF membrane on ice. Membranes were blocked with 5% nonfat milk in 1x TBST buffer and then used for immunoblotting of mitochondrial complex I (anti- NDUFB8) and complex V (anti-ATP5A1).
KGDH activity assay
KGDH activity assay was conducted using the a-Ketoglutarate Dehydrogenase Activity Assay Kit following manufacturer’s instructions (Abcam, ab185440). Briefly, 5 million HEK293T cells or iPSC-derived dopaminergic neurons were collected in ice-cold PBS and washed with PBS. Cells were pelleted by centrifugation at 1000 × g for 2 min at 4 °C. Cells were homogenized in 300 μl of assay buffer with protease inhibitor cocktail (Thermo Fisher Scientific, 87785) using a syringe with 23 G x 1 in. needle (BD, 305271) for 10 strokes on ice. Samples were incubated for 5 min on ice, and protein lysate was collected after centrifugation at 10,000 × g for 5 min at 4 °C. Protein concentration was measured to ensure equal amount of proteins were used across conditions. 50 μg and 80 μg of proteins were used per reaction well for HEK293T and dopaminergic neurons, respectively. Background control wells were set up for each cell line or treatment condition on the same plate in each experiment. For background control wells, substrate was replaced by assay buffer. The absorbance at 450 nm (OD 450 nm) was measured in kinetic mode every minute for 60 min at 37 °C using a plate reader. The OD 450 nm value of background well was subtracted from the value of corresponding sample well for each time point. The rate of NADH production by KGDH enzyme complex was interpolated from NADH standard curve, and the KGDH activity was calculated as the NADH production (nmol) per minute per μg protein.
Seahorse assay
Oxygen consumption rate was measured by a Seahorse assay. HEK293T cells cultured on XF24 cell culture plate (≥90% confluency) were washed and equilibrated in warmed assay medium (XF Base DMEM Medium with 10 mM glucose, 2 mM L-glutamine and 1 mM pyruvate, pH 7.4) for 1 h without CO2 at 37 °C. Cells were sequentially incubated with 1 μM Oligomycin, CCCP, and antimycin A. The change in oxygen concentration per minute was measured using a Seahorse XF24 Analyzer (Agilent). Cells were lysed using RIPA lysis buffer, and protein concentration was measured. The oxygen consumption rate was calculated as the change in oxygen concentration per minute per μg protein.
Chemicals and treatments
For lipid peroxidation assay in HEK293T cells, cells were treated with 0.5 μM RSL3 (Cayman chemicals) or 0.5 μM ML162 (SelleckChem) for 4 h, or 15 μM Erastin (Selleckchem) for 18 h before staining with C11-BODIPY (Invitrogen, D3861). To induce lipid peroxidation in iPSC-derived dopaminergic neurons, cells were treated with 2 μM RSL3 for 4 h before incubation with C11-BODIPY. For assays involving iron chelators, cells were incubated overnight (~16 h) with 10 μM Deferoxamine (DFO) or 10 μM Ciclopirox (CPX) before the addition of RSL3 or ML162. For α-lipoic acid treatment assays, HEK293T cells were preincubated with 5 μM, 10 μM or 50 μM α-lipoic acid (α-LA, Sigma-Aldrich) for 24 h before adding RSL3. Human iPSC-derived DA neurons were preincubated with 50 μM or 100 μM α-LA every other day for 7 days before adding RSL3. For quantification of mitochondrial α-KG levels, CHCHD2-deficient HEK293T cells were treated with vehicle or 50 μM α-LA overnight and again 4 h before mitochondrial purification and subsequent HPLC-MS/MS analysis. For cell fractionation and pS129 α-synuclein immunoblotting assay, CHCHD2-deficient iPSC-derived dopaminergic neurons were treated with 50 μM or 100 μM α-LA every other day for two weeks before harvesting. DMSO and ethanol were used as vehicle controls for RSL3/ML162/erastin- and α-LA- treated conditions, respectively.
Flow cytometry
Lipid peroxidation was quantified by flow cytometry in HEK293T cells. Cells were incubated with 1 μM C11-BODIPY for 30 min (protected from light) at 37 °C in incubator with 5% CO2. Cells were dissociated using TrypLE (Gibco, 12605) and pelleted by centrifugation at 500 × g for 1 min. Cells were washed in PBS by gentle pipetting followed by centrifugation. PBS was removed, and cells were resuspended in HBSS solution (Gibco, 14025092) and kept on ice until analysis. Fluorescence of oxidized and reduced signal were quantified using FITC/Alexa Fluor 488 (green) and Cy3 (red) filters on a LSRFortessa Cell Analyzer (BD). Lipid peroxidation level was quantified by calculating the ratio of fluorescent intensity obtained from FITC/Alexa Fluor 488 channel to that from Cy3 channel. Representative gating strategies were shown in Supplementary Fig. S9.
Generation of lentivirus
DNA fragments of 3xHA-EGFP-OMP25 and 3xMyc-EGFP-OMP25 from pMXs-3xHA-EGFP-OMP25 (Addgene, 83356) and pMXs-3xMyc-EGFP-OMP25 (Addgene, 83355) plasmids72, respectively, were cloned into pER4 lentiviral expression vector11. Lentiviral shRNA constructs (MISSION pLKO.1-puro) for non-target control and DLST were obtained from Millipore Sigma. For lentiviral packaging, HEK293FT cells were transfected with lentiviral expression constructs together with psPAX2 (Addgene, 12260) and pLP3 (Invitrogen) using X-tremeGENE HP DNA Transfection Reagent (Roche). A media change was performed the day after transfection. 48 h after transfection, virus-containing supernatant was collected and cleared by filtering through a 0.45μm PES Filter (CELLTREAT Scientific Products). Virus particles were concentrated using Lenti-X Concentrator (Takara Bio, 631232) following manufacturer’s instructions. The concentration of virus particles was determined by ELISA using RETRO-TEK HIV-1 p24 Antigen ELISA Kit (ZeptoMetrix, 0801111).
Labeling of mitochondria with 3xHA-EGFP or 3xMyc-EGFP tags
HEK293T cells were infected with lentivirus expressing 3xHA-EGFP-OMP25 or 3xMyc-EGFP-OMP25 at MOI of 1. Two days later, GFP positive cells were sorted into 96-well plate with one cell per well and cultured in complete DMEM medium. GFP fluorescence were inspected under a fluorescent scope, and clones with moderate fluorescence intensity were kept. HA and EGFP expression levels of 3xHA-EGFP-OMP25-labeled wildtype control and CHCHD2-deficient cells were quantified by western blotting, and clones with similar transgene expression levels were used for downstream mitochondria purification.
Lentiviral short hairpin RNA (shRNA)-mediated RNA interference
To generate non-target control and DLST knockdown cell lines, HEK293T cells were infected with non-target control or DLST shRNA lentiviruses at MOI of 2. Two days later, infected cells were selected using 2 μg/ml of Puromycin for 3 continuous days. Knockdown efficiency was determined by western blotting. Two independent DLST-targeting shRNA constructs were used.
Immunocytochemistry
Human iPSCs and differentiated DA neurons cultured on coverslips were washed with PBS and fixed in 4% paraformaldehyde (PFA) for 20 min at room temperature. After fixation, cells were washed three times with PBS, and then permeabilized with 0.1% saponin (with 10% FBS in PBS) for 1 h at room temperature. Permeabilized cells were incubated with primary antibodies overnight at 4 °C. After incubation, cells were washed three times with PBS and incubated with anti-mouse or anti-rabbit secondary antibodies conjugated with AlexaFluor for 1 h at room temperature protected from light. Samples were washed and mounted with DAPI+ mounting media. Images were acquired on a Leica DMI4000B confocal microscope. Rabbit anti-OCT4 (Abcam, ab19857, 1:500), Mouse anti-SSEA-4 (Sigma, MAB4304, 1:500), Rabbit anti-NANOG (Abcam, ab21624, 1:500) and Mouse anti-TRA-1-81 (Sigma, MAB4381, 1:200) antibodies were used for detecting the expression of pluripotency markers. Mouse anti- β-III-tubulin (BioLegend, 801202, 1:200) and Rabbit anti-Tyrosine hydroxylase (Sigma, AB152, 1:200) antibodies were used for characterization of differentiated dopaminergic neurons.
Live cell imaging
On day 25 of differentiation, human iPSC-derived dopaminergic neurons were plated on 35 mm 4-chamber glass bottom live cell dishes (Cellvis, D35C4-20-1.5-N) at density of 180,000 cells per quarter. Live cell imaging assays were conducted on day 50–60. A media change was performed the day before the assay. For examination of lipid peroxidation, neurons were incubated with 1 μM C11-BODIPY for 30 min (protected from light) at 37 °C in incubator with 5% CO2. Neurons were gently washed twice with warmed Neurobasal medium followed by two additional washes using warmed HBSS solution. After washing, 500 μl warmed HBSS solution was added to each quarter. Fluorescence of oxidized and reduced signals were imaged using GFP and RFP channels on a Nikon Ti2 Widefield fluorescence scope with a 60x objective and a 5% CO2 chamber at 37 °C. Lipid peroxidation level was quantified by calculating the ratio of fluorescent intensity obtained from GFP channel to that from RFP channel using Fiji (ImageJ).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 9. Two-sided t tests were used for comparisons between two groups and one-way ANOVA with Dunnett’s correction tests were used for comparisons across three or more groups. All data were collected from N ≥ 3 independent experiments and presented as mean ± SEM in histogram graphs. Statistical significance was determined by P < 0.05. Exact P values were indicated in Figures and provide in Source Data file. Detailed information regarding sample sizes, number of experiments and statistical tests was described in Figure Legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are included in the paper and its Supplementary Information. The metabolomics data generated in this study are provided in Supplementary Data 1–5. Source data are provided with this paper.
References
Surmeier, D. J., Obeso, J. A. & Halliday, G. M. Selective neuronal vulnerability in Parkinson disease. Nat. Rev. Neurosci. 18, 101–113 (2017).
Ye, H., Robak, L. A., Yu, M., Cykowski, M. & Shulman, J. M. Genetics and Pathogenesis of Parkinson’s Syndrome. Annu Rev. Pathol. 18, 95–121 (2023).
Park, J. et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 (2006).
Clark, I. E. et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 (2006).
Narendra, D. P. et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. Plos Biol. 8. https://doi.org/10.1371/journal.pbio.1000298 (2010).
Burbulla, L. F. et al. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science 357, 1255–1261 (2017).
Nguyen, M., Wong, Y. C., Ysselstein, D., Severino, A. & Krainc, D. Synaptic, mitochondrial, and lysosomal dysfunction in Parkinson’s Disease. Trends Neurosci. 42, 140–149 (2019).
Song, P. et al. Parkinson’s disease-linked parkin mutation disrupts recycling of synaptic vesicles in human dopaminergic neurons. Neuron 111, 3775–3788 e3777 (2023).
Kalecky, K. & Bottiglieri, T. Targeted metabolomic analysis in Parkinson’s disease brain frontal cortex and putamen with relation to cognitive impairment. NPJ Parkinsons Dis. 9, 84 (2023).
Schmidt, S. et al. A reversible state of hypometabolism in a human cellular model of sporadic Parkinson’s disease. Nat. Commun. 14, 7674 (2023).
Peng, W., Schroeder, L. F., Song, P. P., Wong, Y. C. & Krainc, D. Parkin regulates amino acid homeostasis at mitochondria-lysosome (M/L) contact sites in Parkinson’s disease. Sci. Adv. 9. https://doi.org/10.1126/sciadv.adh3347 (2023).
Funayama, M. et al. CHCHD2 mutations in autosomal dominant late-onset Parkinson’s disease: a genome-wide linkage and sequencing study. Lancet Neurol. 14, 274–282 (2015).
Meng, H. et al. Loss of Parkinson’s disease-associated protein CHCHD2 affects mitochondrial crista structure and destabilizes cytochrome c. Nat. Commun. 8, 15500 (2017).
Aras, S. et al. MNRR1 (formerly CHCHD2) is a bi-organellar regulator of mitochondrial metabolism. Mitochondrion 20, 43–51 (2015).
Aras, S. et al. Oxygen-dependent expression of cytochrome c oxidase subunit 4-2 gene expression is mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic Acids Res. 41, 2255–2266 (2013).
Genin, E. C. et al. CHCHD10 mutations promote loss of mitochondrial cristae junctions with impaired mitochondrial genome maintenance and inhibition of apoptosis. EMBO Mol. Med. 8, 58–72 (2016).
Liu, W. et al. Chchd2 regulates mitochondrial morphology by modulating the levels of Opa1. Cell Death Differ. 27, 2014–2029 (2020).
Sato, S. et al. Homeostatic p62 levels and inclusion body formation in CHCHD2 knockout mice. Hum. Mol. Genet 30, 443–453 (2021).
Zhou, W. et al. PD-linked CHCHD2 mutations impair CHCHD10 and MICOS complex leading to mitochondria dysfunction. Hum. Mol. Genet. 28, 1100–1116 (2019).
Liu, T. et al. CHCHD10-regulated OPA1-mitofilin complex mediates TDP-43-induced mitochondrial phenotypes associated with frontotemporal dementia. FASEB J. 34, 8493–8509 (2020).
Cavallaro, G. Genome-wide analysis of eukaryotic twin CX9C proteins. Mol. Biosyst. 6, 2459–2470 (2010).
Imai, Y., Meng, H., Shiba-Fukushima, K. & Hattori, N. Twin CHCH proteins, CHCHD2, and CHCHD10: key molecules of parkinson’s disease, amyotrophic lateral sclerosis, and frontotemporal dementia. Int. J. Mol. Sci. 20. https://doi.org/10.3390/ijms20040908 (2019).
Bannwarth, S. et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain 137, 2329–2345 (2014).
Genin, E. C. et al. CHCHD10 and SLP2 control the stability of the PHB complex: a key factor for motor neuron viability. Brain 145, 3415–3430 (2022).
Johnson, J. O. et al. Mutations in the CHCHD10 gene are a common cause of familial amyotrophic lateral sclerosis. Brain 137. https://doi.org/10.1093/brain/awu265 (2014).
Muller, K. et al. Two novel mutations in conserved codons indicate that CHCHD10 is a gene associated with motor neuron disease. Brain 137, e309 (2014).
Auranen, M. et al. CHCHD10 variant p.(Gly66Val) causes axonal Charcot-Marie-Tooth disease. Neurol. Genet. 1, e1 (2015).
Pasanen, P. et al. Intrafamilial clinical variability in individuals carrying the CHCHD10 mutation Gly66Val. Acta Neurol. Scand. 133, 361–366 (2016).
Penttila, S. et al. CHCHD10 mutations and motor neuron disease: the distribution in Finnish patients. J. Neurol. Neurosurg. Psychiatry 88, 272–277 (2017).
Ajroud-Driss, S. et al. Mutation in the novel nuclear-encoded mitochondrial protein CHCHD10 in a family with autosomal dominant mitochondrial myopathy. Neurogenetics 16, 1–9 (2015).
Burstein, S. R. et al. In vitro and in vivo studies of the ALS-FTLD protein CHCHD10 reveal novel mitochondrial topology and protein interactions. Hum. Mol. Genet. 27, 160–177 (2018).
Huang, X. et al. CHCHD2 accumulates in distressed mitochondria and facilitates oligomerization of CHCHD10. Hum. Mol. Genet. 27, 3881–3900 (2018).
Atlante, S. et al. alpha-ketoglutarate dehydrogenase inhibition counteracts breast cancer-associated lung metastasis. Cell Death Dis. 9, 756 (2018).
Bailey, P. S. J. et al. ABHD11 maintains 2-oxoglutarate metabolism by preserving functional lipoylation of the 2-oxoglutarate dehydrogenase complex. Nat. Commun. 11, 4046 (2020).
Tretter, L. & Adam-Vizi, V. Inhibition of Krebs cycle enzymes by hydrogen peroxide: a key role of [alpha]-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. J. Neurosci. 20, 8972–8979 (2000).
Chinopoulos, C., Tretter, L. & Adam-Vizi, V. Depolarization of in situ mitochondria due to hydrogen peroxide-induced oxidative stress in nerve terminals:: Inhibition of α-ketoglutarate dehydrogenase. J. Neurochem. 73, 220–228 (1999).
Lucas, D. T. & Szweda, L. I. Declines in mitochondrial respiration during cardiac reperfusion: age-dependent inactivation of alpha-ketoglutarate dehydrogenase. Proc. Natl Acad. Sci. USA 96, 6689–6693 (1999).
Vatrinet, R. et al. The alpha-ketoglutarate dehydrogenase complex in cancer metabolic plasticity. Cancer Metab. 5, 3 (2017).
Shin, D., Lee, J., You, J. H., Kim, D. & Roh, J. L. Dihydrolipoamide dehydrogenase regulates cystine deprivation-induced ferroptosis in head and neck cancer. Redox Biol. 30, 101418 (2020).
Quinlan, C. L. et al. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J. Biol. Chem. 289, 8312–8325 (2014).
Calingasan, N. Y. et al. Influence of mitochondrial enzyme deficiency on adult neurogenesis in mouse models of neurodegenerative diseases. Neuroscience 153, 986–996 (2008).
Seibt, T. M., Proneth, B. & Conrad, M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic. Biol. Med. 133, 144–152 (2019).
Packer, L., Witt, E. H. & Tritschler, H. J. Alpha-Lipoic acid as a biological antioxidant. Free Radic. Bio Med. 19, 227–250 (1995).
Solmonson, A. & DeBerardinis, R. J. Lipoic acid metabolism and mitochondrial redox regulation. J. Biol. Chem. 293, 7522–7530 (2018).
Arivazhagan, P., Ramanathan, K. & Panneerselvam, C. Effect of DL-alpha-lipoic acid on mitochondrial enzymes in aged rats. Chem. Biol. Interact. 138, 189–198 (2001).
Savitha, S., Sivarajan, K., Haripriya, D., Kokilavani, V. & Panneerselvam, C. Efficacy of levo carnitine and alpha lipoic acid in ameliorating the decline in mitochondrial enzymes during aging. Clin. Nutr. 24, 794–800 (2005).
Ambrus, A., Tretter, L. & Adam-Vizi, V. Inhibition of the alpha-ketoglutarate dehydrogenase-mediated reactive oxygen species generation by lipoic acid. J. Neurochem. 109, 222–229 (2009).
Hiller, S. et al. alpha-Lipoic acid protects mitochondrial enzymes and attenuates lipopolysaccharide-induced hypothermia in mice. Free Radic. Biol. Med. 71, 362–367 (2014).
Qin, Z. et al. Effect of 4-hydroxy-2-nonenal modification on alpha-synuclein aggregation. J. Biol. Chem. 282, 5862–5870 (2007).
Nasstrom, T. et al. The lipid peroxidation products 4-oxo-2-nonenal and 4-hydroxy-2-nonenal promote the formation of alpha-synuclein oligomers with distinct biochemical, morphological, and functional properties. Free Radic. Biol. Med. 50, 428–437 (2011).
Bae, E. J. et al. Lipid peroxidation product 4-hydroxy-2-nonenal promotes seeding-capable oligomer formation and cell-to-cell transfer of alpha-synuclein. Antioxid. Redox Signal. 18, 770–783 (2013).
Song, Z. Y., Chen, H. C., Fiket, M., Alexander, C. & Chan, D. C. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178, 749–755 (2007).
Rochette, L. et al. Lipid peroxidation and iron metabolism: two corner stones in the homeostasis control of ferroptosis. Int. J. Mol. Sci. 24. https://doi.org/10.3390/ijms24010449 (2022).
Zheng, J. & Conrad, M. The metabolic underpinnings of ferroptosis. Cell Metab. 32, 920–937 (2020).
Li, J. et al. Ferroptosis: past, present and future. Cell Death Dis. 11, 88 (2020).
Humphries, K. M. & Szweda, L. I. Selective inactivation of α-ketoglutarate dehydrogenase and pyruvate dehydrogenase: reaction of lipoic acid with 4-hydroxy-2-nonenal. Biochemistry 37, 15835–15841 (1998).
Fan, L. et al. CHCHD2 p.Thr61Ile knock-in mice exhibit motor defects and neuropathological features of Parkinson’s disease. Brain Pathol. 33, e13124 (2023).
Torii, S. et al. Involvement of casein kinase 1 epsilon/delta (Csnk1e/d) in the pathogenesis of familial Parkinson’s disease caused by CHCHD2. EMBO Mol. Med. 15, e17451 (2023).
Ikeda, A. et al. Mutations in CHCHD2 cause alpha-synuclein aggregation. Hum. Mol. Genet. 28, 3895–3911 (2019).
Straub, I. R. et al. Loss of CHCHD10-CHCHD2 complexes required for respiration underlies the pathogenicity of a CHCHD10 mutation in ALS. Hum. Mol. Genet. 27, 178–189 (2018).
Anderson, C. J. et al. ALS/FTD mutant CHCHD10 mice reveal a tissue-specific toxic gain-of-function and mitochondrial stress response. Acta Neuropathol. 138, 103–121 (2019).
Liu, Y. T. et al. Loss of CHCHD2 and CHCHD10 activates OMA1 peptidase to disrupt mitochondrial cristae phenocopying patient mutations. Hum. Mol. Genet. 29, 1547–1567 (2020).
Ruan, Y. et al. CHCHD2 and CHCHD10 regulate mitochondrial dynamics and integrated stress response. Cell Death Dis. 13, 156 (2022).
Sayles, N. M. et al. Mutant CHCHD10 causes an extensive metabolic rewiring that precedes OXPHOS dysfunction in a murine model of mitochondrial cardiomyopathy. Cell Rep. 38. https://doi.org/10.1016/j.celrep.2022.110475 (2022).
Straub, I. R., Weraarpachai, W. & Shoubridge, E. A. Multi-OMICS study of a CHCHD10 variant causing ALS demonstrates metabolic rewiring and activation of endoplasmic reticulum and mitochondrial unfolded protein responses. Hum. Mol. Genet. 30, 687–705 (2021).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Burbulla, L. F. et al. A modulator of wild-type glucocerebrosidase improves pathogenic phenotypes in dopaminergic neuronal models of Parkinson’s disease. Sci. Transl. Med. 11. https://doi.org/10.1126/scitranslmed.aau6870 (2019).
Kim, S., Wong, Y. C., Gao, F. & Krainc, D. Dysregulation of mitochondria-lysosome contacts by GBA1 dysfunction in dopaminergic neuronal models of Parkinson’s disease. Nat. Commun. 12, 1807 (2021).
Kriks, S. et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 480, 547–551 (2011).
Schröder, L. F. et al. VPS13C regulates phospho-Rab10-mediated lysosomal function in human dopaminergic neurons. J. Cell Biol. 223. https://doi.org/10.1083/jcb.202304042 (2024).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Chen, W. W., Freinkman, E., Wang, T., Birsoy, K. & Sabatini, D. M. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell 166, 1324–1337.e1311 (2016).
Chen, W. W., Freinkman, E. & Sabatini, D. M. Rapid immunopurification of mitochondria for metabolite profiling and absolute quantification of matrix metabolites. Nat. Protoc. 12, 2215–2231 (2017).
Acknowledgements
We thank Jianhua Yan for helping with mouse culture and genotyping, Wesley Peng for subcloning of 3xHA-EGFP-OMP25 and 3xMyc-EGFP-OMP25 into pER4 vector11, Dr. Nobutaka Hattori for providing the CHCHD2 T61I PD iPSC line12,59, PPMI for providing the LRRK2 G2019S PD iPSC line, Pingping Song for providing the protein lysate of GBA1 84GG mutant and isogenic control DA neurons, Ezekiel Bello for assistance with cell culture, and Peng Gao for assistance with HPLC-MS/MS analysis of metabolites. We thank Yvette Wong, Leonie F. Schröder, Diptaman Chatterjee and fellow members of Krainc Laboratory for advice. Skin fibroblasts from healthy control donor were obtained from the Northwestern University Movement Disorders Center (MDC) Biorepository. We thank MDC Biorepository investigators Rizwan Akhtar MD, PhD; Tanya Simuni MD; Puneet Opal MD, PhD; Niccolo Mencacci MD, PhD; Joanna Blackburn MD; Monika Szela MHA; and Lisa Kinsley MS, CGC. Metabolomics services were performed by the Metabolomics Core Facility at Robert H. Lurie Comprehensive Cancer Center of Northwestern University. Flow cytometry assays were conducted at Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Core Facility. Imaging experiments were performed at the Center for Advanced Microscopy/Nikon Imaging Center. Schematic graphs were generated in https://BioRender.com and Adobe Illustrator. This work was supported by R35 NS122257 to D.K. and R01 NS099623, R21 NS114765 to H.-X. D.
Author information
Authors and Affiliations
Contributions
G. Gao and D. Krainc designed the study. G. Gao performed the experiments and analyzed data. Y. Shi and H.-X. Deng generated knock-in mice and helped with mouse experiments. G. Gao and D. Krainc wrote the manuscript with input from H.-X. Deng.
Corresponding author
Ethics declarations
Competing interests
D.K. is the founder and scientific advisory board chair of Vanqua Bio, serves on the scientific advisory boards of The Silverstein Foundation, Intellia Therapeutics, AcureX, and is a venture partner at OrbiMed. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Derek Narendra, Alessandro Prigione, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, G., Shi, Y., Deng, HX. et al. Dysregulation of mitochondrial α-ketoglutarate dehydrogenase leads to elevated lipid peroxidation in CHCHD2-linked Parkinson’s disease models. Nat Commun 16, 1982 (2025). https://doi.org/10.1038/s41467-025-57142-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57142-9
This article is cited by
-
Glycerol 3-phosphate acyltransferase exacerbates α-synuclein-induced toxicity by increasing lipid peroxidation
Nature Communications (2026)
-
Bridging pathway dysfunction and therapy: novel compounds for neuroprotection
3 Biotech (2026)