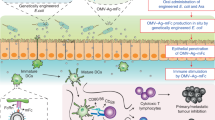
Abstract
Genetically engineered commensal bacteria are promising living drugs, however, their therapeutic molecules are frequently confined to their colonization sites. Herein, we report an oral protein delivery technology utilizing an engineered bacterial type zero secretion system (T0SS) via outer membrane vesicles (OMVs). We find that OMVs produced in situ by Escherichia coli Nissle 1917 (EcN) can penetrate the intact gut epithelial barrier to enter the circulation and that epithelial transcytosis involves pinocytosis and dynamin-dependent pathways. EcN is engineered to endogenously load various enzymes into OMVs, and the secreted enzyme-loaded OMVs are able to stably catalyze diverse detoxification reactions against digestive fluid and even enter the circulation. Using hyperuricemic mice and uricase delivery as a demonstration, we demonstrate that the therapeutic efficacy of our engineered EcN with a modified T0SS outperforms that with a direct protein secretion apparatus. The enzyme-loaded OMVs also effectively detoxify human serum samples, highlighting the potential for the clinical treatment of metabolic disorders.
Similar content being viewed by others
Introduction
Oral protein delivery is superior in terms of noninvasiveness, patient compliance and convenience among various methods of drug administration1,2. Nevertheless, few therapeutic proteins have been approved for oral administration in clinical practice, because of their easy degradation, fragile tertiary structure and poor epithelial permeability in the digestive tract3. The development of oral protein delivery system is in urgent need, which should overcome multiple physiological barriers of the gastrointestinal tract, including harsh acid and proteases in the stomach, the viscous mucus layer, and the epithelial cell barrier connected by tight junctions in the intestine3. To overcome these absorption barriers, nanoparticle (NP)-based drug delivery systems have been developed for oral protein delivery4,5, while suffering from complicated fabrication procedures, inefficient cargo loading and delivery. Therefore, the development of facile and efficient oral protein delivery systems is highly important for the clinical use of therapeutic proteins.
Genetically engineered commensal bacteria are emerging as live biotherapeutic products, which have shown substantial efficacy in treating numerous diseases, including inflammatory bowel disease (IBD), tumors and neurological disorder, etc6,7,8,9,10. Commensal bacteria are able to tolerate harsh gastric environment and colonize the colon, and their synthetic gene circuits enable improved catalytic activities of the cells or programable release of therapeutic molecules in the gut11. Several strategies have been developed to harness synthetic commensal bacteria to secrete metabolites12,13, peptides14,15 or proteins16,17 in the colon. Through metabolic engineering, recombinant bacteria with heterologous synthesis pathways were able to overproduce and secrete chemicals after intestinal colonization18. Gram-negative commensal bacteria outfitted with a modified type III secretion system (T3SS) obtained the capacity to secrete nanobodies in the gut19. Moreover, efforts have been made to program bacteria to lyse and release therapeutic proteins around their colonization site20. Synthetic commensal bacteria are promising living delivery platforms21,22, however, the delivery distance of their therapeutic payloads is frequently limited to their surroundings, and the phase 3 clinical failure of the engineered probiotic, SYNB193423,24,25, in the treatment of phenylketonuria might be attributed to its restricted efficacy in lowering toxic blood metabolites merely by degrading the metabolites in the gut. How to deliver stable proteins in the digestive tract and further penetrate through the gut barrier into the circulation is the main challenge to realizing oral protein delivery by orally administered commensal bacteria.
Outer membrane vesicles (OMVs) are considered bacterial type zero secretion system (T0SS), which mediate microbe‒host or microbe‒microbe interactions in the gut26. OMVs are formed by the budding of the outer membrane and subsequently detach from the cell body, and the OMV content of gram-negative bacteria is isolated from the cytoplasm by the inner membrane. As a nanoscale derivative of bacterial cells, intestinal microbial-derived OMVs may have the ability to enter the circulatory system27. Escherichia coli Nissle 1917 (EcN) is a classic gram-negative probiotic chassis for synthetic biology, which has been approved by the FDA as GRAS (generally recognized as safe)28. Here, we report an oral protein delivery technology enabled by genetically engineered EcN outfitted with a modified T0SS (Fig. 1). Notably, we find that OMVs from EcN can penetrate the intact gut epithelial barrier and enter the circulation. Pinocytosis and dynamin-dependent pathways are involved in this transcellular transport. Genetically engineered EcN equipped with a modified T0SS and endogenous protein loading system is able to secrete protein payloads encapsulated in OMVs with high protein stability against proteolysis and circulation entry capability. This T0SS-based protein secretion system is compatible with various protein payloads, and can arrange multiple distinct protein cargos into individual OMV, making protein-loaded OMVs promising biocatalysts for diverse detoxifying reactions. Using a murine model of hyperuricemia and uricase delivery as a demonstration, we demonstrate that our engineered EcN outfitted with modified T0SS is superior to recombinant EcN equipped with direct protein secretion apparatus in terms of therapeutic efficacy. Moreover, the secreted uricase-loaded or lactate oxidase-loaded OMVs effectively detoxify circulating urate or lactate in sixteen human serum samples, proving that our protein delivery system based on synthetic EcN is promising for the clinical treatment of a wide range of metabolic disorders.
EcN was engineered to increase OMV biosynthesis by genomic deletion of an outer membrane lipoprotein encoding gene nlpI, heterologously overexpress protein of interest (POI) on a stable plasmid in the absence of antibiotic selection (through deleting an essential gene thyA in the genome and compensate it on the plasmid), and endogenously load POI into OMVs guided by a periplasm-targeted secretion signal peptide. After oral administration of our synthetic EcN outfitted with modified T0SS, the protein-loaded OMVs produced in situ by engineered EcN could resist proteolysis, penetrate the intact gut epithelial barrier and enter the circulation. Pinocytosis and dynamin-dependent pathways are involved in this transcellular transport. Synthetic EcN outfitted with modified T0SS can deliver therapeutic proteins to the circulation and detoxify circulating metabolites, presenting an oral protein delivery strategy. Created in BioRender. Gong, X. (2025) https://BioRender.com/o53n846.
Results
Development of a modified T0SS for protein secretion
To establish protein secretion via the T0SS in EcN, genetically engineered EcN was explored for programmed assembly of specific proteins into OMV as well as enhanced OMV synthesis. First, nlpI, which encodes an outer membrane lipoprotein involved in modulating peptidoglycan dynamics, was deleted in EcN to increase OMV production29. The OMV yield of EcNΔnlpI in M9 medium was 2.83 ± 0.24 times greater than that of wild-type EcN, with conserved OMV morphology, zeta potential and size distribution (Supplementary Fig. 1). In the case of the gram-negative bacteria EcN, its native secretion systems serve to deliver the cytoplasmic protein with a signal peptide into the periplasmic space, including the general secretion (Sec), twin-arginine translocation (Tat) and signal-recognition particle (Srp)-dependent pathways, and the signal peptide is cleaved from the secretory protein by a signal peptidase located on the inner membrane30. Next, we investigated whether, as in other E. coli strains31,32, heterologously overexpressed proteins labeled with signal peptides could be channeled into the periplasm and further encapsulated into OMVs of EcN. The fluorescent protein superfolder GFP labeled with a Sec signal peptide33 at its N-terminus was overexpressed in EcNΔnlpI, resulting in the recombinant strain EcNΔnlpI-OG. The OMV of EcNΔnlpI-OG was isolated by ultracentrifugation, and the presence of GFP in the OMV sample was confirmed by both western blot analysis using an anti-GFP antibody and fluorescence images, demonstrating that heterologously overexpressed protein labeled with a periplasm-targeted signal peptide in engineered EcN could be successfully packaged into the spherical buds of the outer membrane (Fig. 2a and Supplementary Fig. 2).
a Western blot analysis of GFP abundance in OMV samples from EcNΔnlpI-OG (abbreviated as “OG”) and EcNΔnlpI-GR (abbreviated as “OGR”) and EcNΔnlpI (abbreviated as “NC”) using an anti-GFP antibody. b The synthetic EcN outfitted with modified T0SS and endogenous loading system was able to secrete protein-loaded OMVs with an encapsulation ratio of 97.9%. c The OMV encapsulation ratio of GFP was tested by Nano-flow cytometry, which was calculated as the ratio of GFP-loaded OMVs to total OMVs derived from EcNΔnlpI-OG, using EVMembrane Red to stain the membrane of OMVs. d Western blot analysis of RFP abundance in OMV samples from EcNΔnlpI-OR (abbreviated as “OR”), EcNΔnlpI-GR (abbreviated as “OGR”) and EcNΔnlpI (abbreviated as “NC”) using an anti-RFP antibody. e The synthetic OMVs of engineered EcN could encapsulate multiple protein payloads simultaneously in individual OMV vehicles, making the synthetic OMV a promising cascade biocatalyst. f Engineered OMVs were able to simultaneously encapsulate GFP and RFP in individual OMVs, as visualized via Polar-SIM super-resolution microscope. Scale bar=2 μm. a, d, f Each experiment was repeated three times independently with similar results. b, e Created in BioRender. Gong, X. (2025) https://BioRender.com/i12g295.
The OMV encapsulation ratio of the protein payload was studied by Nano-flow cytometry, which was calculated as the ratio of GFP-loaded OMVs to total OMVs derived from EcNΔnlpI-OG, using GFP as the protein payload and EVMembrane Red to stain the membrane of OMVs (Fig. 2b). In comparison to OMVs derived from EcNΔnlpI, GFP-loaded OMVs from EcNΔnlpI-OG presented strongly increased GFP fluorescence signals (Fig. 2c). The OMV encapsulation ratio of GFP cargos was as high as 97.9%, which was calculated as the ratio of GFP-loaded OMVs to EVMembrane Red-stained OMVs. The fluorescence images of GFP-loaded OMVs from EcNΔnlpI-OG stained with EVMembrane Red demonstrated that most OMVs from EcNΔnlpI-OG stained with EVMembrane Red simultaneously presented GFP signals (Supplementary Fig. 3), verifying the homogeneity of the engineered OMVs in this endogenous loading system. The conventional exogenous protein loading approaches include electroporation, coincubation, extrusion, freeze‒thawing, sonication and surfactant treatment, etc, whose encapsulation efficiencies are approximately 20%-50%34. Compared with the exogenous loading strategy, our endogenous loading system is superior in terms of the encapsulation ratio and self-programmed assembly.
Furthermore, we investigated whether the endogenous loading system could arrange multiple distinct protein payloads into individual OMVs. In addition to GFP, RFP fused with a Srp-signal peptide was overexpressed in EcNΔnlpI, and recombinant EcN could secrete RFP within OMV vehicles, as verified by both western blot analysis of the extracted OMV sample using an anti-RFP antibody and fluorescence images (Fig. 2d and Supplementary Fig. 2). EcNΔnlpI was subsequently engineered to co-overexpress two heterologous proteins, i.e., GFP labeled with an N-terminal Sec signal peptide and RFP fused with an N-terminal Srp signal peptide. OMVs from the resulting strain EcNΔnlpI-GR were isolated and analyzed. Both GFP and RFP were detected in the OMV sample via western blot assay (Fig. 2a, d). The fluorescence images of OMVs from EcNΔnlpI-GR revealed that a single OMV emitted both green and red fluorescence signals (Fig. 2e, f and Supplementary Fig. 4), showing that the individual OMVs encapsulated both the GFP and RFP payloads. These results demonstrate that the modified T0SS-based protein secretion system in engineered EcN could assemble multiple distinct cargos in individual OMV vehicles, making synthetic OMVs promising cascade biocatalysts.
The T0SS-based protein secretion system is compatible with various enzymes and enables diverse detoxifying reactions
An array of enzymes were tested for the compatibility of the T0SS-based protein secretion system. The enzyme payloads were labeled with an N-terminal Tat signal peptide and a protein tag (His or HA), and overexpressed in EcNΔnlpI, including the phenylalanine deaminase StlA (from Photorhabdus luminescens), catalase KatE (from EcN), lactate oxidase Lox (from Aerococcus viridans), and uricase Uox (from Candida utilis). Following the isolation of OMV samples from the engineered EcN strains, the enzyme payloads in OMVs were detected through western blot assay using their respective anti-tag antibodies (Fig. 3a, b). In comparison to the OMVs of EcNΔnlpI, the synthetic OMVs with enzyme payloads obtained the catalytic activities of their corresponding enzyme cargos, i.e., synthetic OMVs containing StlA degraded phenylalanine at a rate of 0.38 ± 0.04 μM/h/mg, OMVs encapsulating KatE degraded hydrogen peroxide at a rate of 2.92 ± 0.07 μM/h/mg, OMVs containing Lox degraded lactate at a rate of 0.36 ± 0.02 μM/h/mg, and OMVs containing Uox degraded uric acid (UA) at a rate of 0.34 ± 0.03 μM/h/mg (Fig. 3c-g). These results demonstrated that engineered EcN outfitted with modified T0SS could self-assemble and secrete functional enzyme payloads within OMV vehicles, and such T0SS-based protein secretion system is compatible with a variety of proteins.
a Western blot analysis of StlA (abbreviated as “S”) abundance in OMVs secreted by EcNΔnlpI-OS (abbreviated as “OS”), Lox (abbreviated as “L”) abundance in OMVs secreted by EcNΔnlpI-OL (abbreviated as “OL”) and Uox (abbreviated as “U”) abundance in OMVs secreted by EcNΔnlpI-OU (abbreviated as “OU”) using an anti-His antibody. OMVs secreted by EcNΔnlpI are indicated as “NC”. b Western blot analysis of KatE (abbreviated as “K”) abundance in OMVs secreted by EcNΔnlpI-OK (abbreviated as “OK”) and EcNΔnlpI (abbreviated as “NC”) using an anti-HA antibody. c–f OMVs from EcNΔnlpI-OS (c), EcNΔnlpI-OK (d), EcNΔnlpI-OL (e) and EcNΔnlpI-OU (f) were able to degrade phenylalanine, H2O2, lactate and UA in vitro, respectively (n = 3 biologically independent samples). OMVs secreted by EcNΔnlpI are indicated as “NC”. The data are presented as the mean ± SD. g Enzyme-loaded OMVs effectively catalyzed the detoxification reaction. The reactions were classified into two groups: decomposition reactions and NADH-independent oxidation reactions. StlA-loaded OMVs and KatE-loaded OMVs could catalyze the decomposition reactions. The oxidases (Uox and Lox) couple with indigenous catalase in the OMV to catalyze oxidation reactions dependent on H2O2/O2 cycling instead of NAD+/NADH. Source data are provided as a Source Data file.
Synthetic OMVs loaded with various enzymes are promising for catalyzing diverse detoxification reactions (Fig. 3g). The performance of StlA-loaded and KatE-loaded OMVs indicated that synthetic OMVs are able to catalyze decomposition reactions. Additionally, it was found that the OMVs from EcNΔnlpI could substantially degrade hydrogen peroxide at a rate of 1.61 ± 0.05 μM/h/mg. The presence of indigenous catalase-peroxidase KatG in EcN OMVs was revealed by proteomic analysis of OMV samples (Supplementary Table 1). These results confirmed that the OMV vehicles from EcN contain indigenous catalase and can degrade hydrogen peroxide to oxygen and water. It was noticed that both Uox and Lox catalyze the oxidation reaction and produce hydrogen peroxide (Fig. 3g), whereas negligible amount of hydrogen peroxide was detected when Uox-loaded or Lox-loaded OMVs were used to catalyze the degradation of UA or lactate, respectively (data not shown), indicating that the oxidases (Uox and Lox) couple with indigenous catalase in the OMV to catalyze oxidation reactions dependent on H2O2/O2 cycling instead of NAD+/NADH (Fig. 3g). Collectively, these results demonstrated that the synthetic OMVs from our engineered EcN equipped with modified T0SS can catalyze diverse detoxification reactions, including decomposition and oxidation reactions.
Proteins encapsulated in OMVs exhibit enhanced stability in simulated intestinal fluid
Next, we evaluated the performance of the protein payload from our modified T0SS-based secretion system in a simulated intestinal environment, through comparing its protein stability and catalytic activity with that of the common secretion system, using the Uox payload as an example (Fig. 4a). First, to achieve stable expression of the heterologous gene in engineered EcN in the gut environment, a stable plasmid system in the absence of antibiotic selection was established, through deleting an essential gene thyA in the genome of EcNΔnlpI and compensating for it in the expression vector. The growth of EcNΔnlpI∆thyA was dependent on exogenous thymidine (Supplementary Fig. 5a), while EcNΔnlpI∆thyA carried the stable plasmid grew well as that of EcNΔnlpI in the absence of exogenous thymidine (Supplementary Fig. 5a), demonstrating the successful retention of the stable plasmid in antibiotic-free microenvironment (Supplementary Fig. 5b). The recombinant EcN strain EcN∆nlpI∆thyA-SU overexpressed Uox labeled with a Tat-signal peptide and a His tag on the stable plasmid, and was also proved to secrete Uox as encapsulated by OMV (Fig. 4b). A genetically engineered EcN strain, EcN∆thyA-SUT, with a heterologous type I secretion system (T1SS)35 was established to represent the direct protein secretion strategy, and Uox fused with the HlyA signal peptide and a His tag was observed in the cell culture supernatant, as tested by western blot analysis using an anti-His antibody (Supplementary Fig. 6a, b).
a Schematic of the Uox secretion approach involving the direct protein secretion apparatus T1SS or modified T0SS-based protein secretion system. Created in BioRender. Gong, X. (2025) https://BioRender.com/r83g400. b Western blot analysis of Uox abundance in the intracellular fraction, supernatant and OMVs of EcN∆nlpI∆thyA-SU and EcN∆thyA-SUT using an anti-His antibody. The stable plasmid system did not affect the function of the recombinant EcN strains outfitted with T0SS or T1SS. c EcN∆nlpI∆thyA-SU with T0SS and EcN∆thyA-SUT with T1SS presented similar UA degradation rates in M9 medium supplemented with 0.5 mM UA under microaerobic culture condition (n = 3 biologically independent samples). d EcN∆nlpI∆thyA-SU with T0SS exhibited greater UA degradation efficiency than that of EcN∆thyA-SUT with T1SS in simulated intestinal fluid supplemented with 0.5 mM UA under microaerobic culture condition (n = 3 biologically independent samples). e Comparison of the enzyme activities of purified protein (T1SS-Uox) or protein-loaded OMVs (OMV-Uox) from EcN∆thyA-SUT or EcN∆nlpI∆thyA-SU, respectively, tested in PBS at various time points at 37 °C (n = 3 biologically independent samples). f, g The stabilities of T1SS-Uox and OMV-Uox were assessed by monitoring the enzyme activities (f) and Uox protein levels (g) in the simulated intestinal fluid at various time points at 37 °C (n = 3 biologically independent samples). The data are presented as the mean ± SD. The P value was determined by two-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test. Source data are provided as a Source Data file.
The recombinant EcN strains outfitted with T0SS or T1SS (Fig. 4a), i.e., EcN∆nlpI∆thyA-SU and EcN∆thyA-SUT, were incubated in M9 media supplemented with 0.5 mM UA, and presented similar UA degradation rates (Fig. 4c), indicating that there is no significant difference in protein secretion efficiency between the T0SS and T1SS in these engineered EcN strains. However, in simulated intestinal fluid containing trypsin and 0.5 mM UA, EcN∆nlpI∆thyA-SU with T0SS exhibited significantly greater UA degradation efficiency than EcN∆thyA-SUT with T1SS (Fig. 4d), indicating that the Uox secreted by T0SS is more resistant to proteolysis than that secreted by T1SS. Furthermore, the protein stability and catalytic activity of Uox secreted by T0SS and T1SS were analyzed and compared in simulated intestinal fluid. The Uox encapsulating OMVs from EcN∆nlpI∆thyA-SU (abbreviated as OMV-Uox) and Uox fused with the HlyA secretion signal sequence and His tag for purification (abbreviated as T1SS-Uox) were extracted and quantified via western blotting (Supplementary Fig. 7a, b). OMV-Uox and T1SS-Uox containing similar amounts of Uox were incubated in PBS or simulated intestinal fluid, and their enzyme activities at different time points were assayed (Fig. 4e, f). It was observed that OMV-Uox and T1SS-Uox had similar enzyme activities and stabilities in PBS within 2 h (Fig. 4e). However, the enzyme activity of T1SS-Uox decayed rapidly in the simulated intestinal fluid, remaining 37.39 ± 4.91% of its original enzyme activity after 1 h (Fig. 4f). In contrast, the decay rate of the enzyme activity of OMV-Uox was substantially lower than that of T1SS-Uox, conserving over 79.81 ± 2.51% of its original activity after 1 h of incubation with trypsin (Fig. 4f, g). These results demonstrated that the OMV vehicles protect the protein cargos from proteolysis, indicating that the protein payloads delivered via our modified T0SS-based protein secretion strategy are more stable and have greater catalytic activity than those delivered via the common direct secretion route in the digestive tract.
Protein loaded OMVs of EcN can penetrate the gut barrier and enter the circulation
Recent studies have reported the observation of blood OMVs in healthy individuals36, therefore, we speculated that some intestinal OMVs derived from the gut microbiota may enter the circulation. To investigate this hypothesis, we explored whether OMVs of EcN could penetrate the intact gut barrier. Healthy mice with ligated colons were injected with Cy5.5-labeled OMVs from EcN∆nlpI∆thyA into the colon lumen. Eight hours after colonic incubation, fluorescent OMVs were observed inside the villi on the basolateral side of the colon as well as the liver and kidney from healthy mice treated with OMVs rather than PBS (Fig. 5a and Supplementary Fig. 8), demonstrating that the OMVs of EcN are capable of penetrating the intact gut epithelial barrier and entering the circulation. Furthermore, we explored whether protein-loaded OMVs produced in situ by intestinal synthetic EcN could cross the gut barrier and enter the circulation in healthy mice without intestinal permeability defects. Healthy mice were orally administered EcN∆nlpI∆thyA-SU or EcN∆nlpI∆thyA-OK, and the in vivo distributions of Uox-loaded or KatE-loaded OMVs were tracked at various time points after gavage via anti-His or anti-HA immunofluorescence staining (pink), respectively. Four hours after gavage of EcN∆nlpI∆thyA-SU or EcN∆nlpI∆thyA-OK, OMVs encapsulating Uox or KatE were detected in the colon of mice using an anti-His or anti-HA antibody (pink), respectively (Fig. 5b and Supplementary Fig. 9). At 12 and 24 hours postgavage of EcN∆nlpI∆thyA-SU or EcN∆nlpI∆thyA-OK, OMVs were observed in the liver and kidney in addition to the colon (Fig. 5b and Supplementary Fig. 9), demonstrating that protein-loaded OMVs produced in situ by engineered EcN could penetrate the gut barrier and enter the circulation in healthy mice. At 36 hours after the gavage of EcN∆nlpI∆thyA-SU or EcN∆nlpI∆thyA-OK, few OMVs were detected in vivo (Fig. 5b and Supplementary Fig. 9), indicating that the circulating OMVs derived from intestinal EcN could persist for up to 36 hours after one dose of engineered EcN. In comparison, no circulating OMVs were detected in healthy mice treated with the vehicle (phosphate-buffered saline, PBS). Moreover, the GFP-expressing strain, EcNΔnlpIΔthyA-OK-G, was utilized to trace the location of the engineered EcN. It was observed that the strain was present only in the gut lumen but not in the liver or kidney after 12 h of administration to healthy mice (Supplementary Fig. 10). This finding indicates that the signal observed above through immunofluorescence stems from the OMV itself rather than the bacteria. These results collectively demonstrated that protein-loaded OMVs derived from engineered EcN in the gut are capable of entering the circulation.
a Representative image of colonic sections after the injection of Cy5.5-labeled OMVs (abbreviated as “Cy5.5-OMV”) or PBS (abbreviated as control) into the colon lumen of healthy mice. Nuclei were counterstained with DAPI (blue). Scale bar = 50 μm. b Representative images of different tissues (colon, liver and kidney) from healthy mice orally administered EcNΔnlpIΔthyA-SK or PBS. OMVs were detected using an anti-HA antibody (pink). Nuclei were counterstained with DAPI (blue). Scale bar = 50 μm. a, b Each experiment was repeated three times independently with similar results.
It has been reported that some bacterial OMVs contain degradative enzymes that are likely responsible for accelerating their rates to pass through mucus pores within the mucus layer to reach gut epithelial cells37. We compared the protein content of natural OMVs and GFP-loaded OMVs from wild-type EcN and EcNΔnlpIΔthyA-SG, respectively, through SDS-PAGE and proteomics analysis of the OMV samples. The results revealed that the protein profiles of OMVs from wild-type or engineered EcN were similar (Supplementary Fig. 11), and there was no significant difference in the protein abundance of enzymes, e.g., proteases, hydrolases, glycosidases, hexosaminidase and sulfatase, between OMVs from wild-type or engineered EcN (Supplementary Table 1). These results demonstrated that the heterologous and secreted proteins via the modified T0SS system of engineered EcN do not interfere with the indigenous proteins in OMVs, hence, the protein-loaded OMVs of engineered EcN conserve the ability to pass through the mucus layer.
Further, we investigated the mechanism underlying the ability of protein-loaded OMVs to penetrate the intestinal epithelial cell barrier, using an in vitro intestinal epithelium model of Caco-2 monolayers grown in Transwell cultures (Fig. 6a and Supplementary Fig. 12). To determine whether the OMVs are transported across the intestinal epithelium through an active or passive mechanism, RFP-loaded OMVs were incubated with the intestinal epithelium in the apical compartment of Transwell chambers at either 37 °C or 4 °C for 4 hours. The fluorescence signals detected in the basolateral chamber were significantly greater when they were incubated at 37 °C than that at 4 °C, indicating that OMV transfer across the intestinal epithelial monolayer involves an active mechanism under physiological conditions (Fig. 6b). Furthermore, we explored the mechanism underlying the epithelial transcytosis of OMVs by focusing on the first endocytosis step of transcytosis, using various chemical inhibitors38 targeting different endocytosis pathways. The addition of chlorpromazine (inhibits clathrin-dependent endocytosis), cytochalasin D (inhibits membrane fusion), wortmannin (inhibits phagocytosis), or nystatin (inhibits caveolin-mediated endocytosis and lipid raft formation) did not significantly affect the transcytosis efficiency of RFP-loaded OMVs across the intestinal epithelium (Fig. 6c). In contrast, dynasore (an inhibitor of dynamin-dependent endocytosis) and amiloride (an inhibitor of pinocytosis) significantly reduced OMV transcytosis in a dose-dependent manner, in comparison to the control group treated with the solvent (dimethyl sulfoxide, DMSO) (Fig. 6c). Additionally, incubation with endocytosis inhibitors along with OMVs did not alter the permeability of the intestinal epithelium to Lucifer Yellow (Fig. 6d). These results suggest the involvement of the dynamin-dependent pathway and pinocytosis in the transcytosis of protein-loaded OMVs from engineered EcN across the intestinal epithelial cell layer.
a The mechanism underlying the ability of EcN OMVs to penetrate the gut epithelial cell barrier was studied using an in vitro intestinal epithelium model of Caco-2 monolayers grown in Transwell cultures. Various inhibitors targeting different endocytosis pathways and RFP-loaded OMVs were sequentially added to the upper chamber. Created in BioRender. Gong, X. (2025) https://BioRender.com/d95j675. b To determine whether epithelial transcytosis of OMVs occurs through an active or passive mechanism, RFP-loaded OMVs were incubated in the apical compartment of Transwell chambers at 37 °C or 4 °C for 4 hours. Fluorescence signals were detected in the basolateral chamber, and further normalized to that of the 37 °C group (n = 3 biologically independent samples). c The mechanism underlying epithelial transcytosis of OMV was examined by focusing on the first endocytosis step of transcytosis, using various chemical inhibitors targeting different endocytosis pathways at various concentrations (low, medium and high concentrations), including the vehicle (dimethyl sulfoxide, DMSO), chlorpromazine (inhibition of clathrin-dependent endocytosis), cytochalasin D (inhibition of membrane fusion), wortmannin (inhibition of phagocytosis), amiloride (inhibition of pinocytosis), nystatin (inhibition of caveolin-mediated endocytosis and lipid raft formation), and dynasore (inhibitors of dynamin-dependent endocytosis). Low concentration: 0.05 μM wortmannin; 0.25 μM cytochalasin D; 10 μM chlorpromazine; 20 μM amiloride; 0.1 μM nystatin and 20 μM dynasore; Medium concentration: 0.1 μM wortmannin; 0.5 μM cytochalasin D; 20 μM chlorpromazine; 100 μM amiloride; 0.2 μM nystatin and 100 μM dynasore; High concentration: 1 μM wortmannin; 2 μM cytochalasin D; 50 μM chlorpromazine; 200 μM amiloride; 1 μM nystatin and 200 μM dynasore. Fluorescence signals were detected in the basolateral chamber and were further normalized to that of the DMSO group (n = 3 biologically independent samples). d The permeability of the epithelial monolayers after incubation with OMVs along with DMSO or inhibitors was assayed with Lucifer Yellow (n = 3 biologically independent samples). The data are presented as the mean ± SD. The P value was determined by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test. Source data are provided as a Source Data file.
In vivo safety evaluation of engineered EcN equipped with modified T0SS
To test whether oral administration of synthetic EcN equipped with modified T0SS is a safe therapeutic approach, potential toxicity was evaluated in healthy mice orally administered saline, EcN, or EcN∆nlpI∆thyA-SU for one week, followed by histological analyzes of major organs as well as hematological and blood biochemical analyzes. Compared with those in saline-treated mice, no significant changes in white blood cell (WBC) counts, red blood cell (RBC) counts, platelet (PLT) counts, or hemoglobin (HGB) levels were detected in EcN- or EcN∆nlpI∆thyA-SU-treated mice (Supplementary Figs. 13a–d). These results demonstrated that neither the EcN chassis nor the circulating OMVs derived from the synthetic EcN equipped with modified T0SS have adverse effects on the hematopoietic system. Furthermore, biochemical analysis of plasma samples revealed that treatment with EcN or EcN∆nlpI∆thyA-SU did not increase the levels of alanine aminotransferase (ALT), creatinine (CRE) or blood urea nitrogen (BUN), demonstrating that our synthetic EcN does not induce liver or kidney toxicity in vivo (Supplementary Fig. 13e–g). Moreover, hematoxylin and eosin (H&E) staining analysis of major organs (including the heart, liver, spleen, lung, and kidney) revealed no signs of organ damage in any group, demonstrating that the administration of our synthetic EcN does not produce significant histopathological abnormalities (Supplementary Fig. 14). Collectively, the in vivo safety assessment proved that our synthetic EcN outfitted with modified T0SS is biocompatible and safe, and has potential for clinical application.
Oral delivery of Uox by engineered EcN equipped with modified T0SS showed superior therapeutic efficacy in treating hyperuricemic mice
The therapeutic efficacy and superiority of our engineered EcN outfitted with modified T0SS were evaluated for its ability to treat metabolic diseases, in comparison with that of engineered EcN equipped with direct protein secretion apparatus (T1SS), using hyperuricemia as a demonstration. A murine model of hyperuricemia was induced via a UA-supplemented diet and daily intraperitoneal injection of potassium oxonate (PO) for 28 days, where PO serves as a murine Uox inhibitor39. Healthy mice gavaged with saline and intraperitoneally administered with the vehicle served as the control group. Hyperuricemic mice were gavaged with saline, wild-type EcN, or recombinant EcN strains outfitted with T0SS or T1SS, i.e., EcN∆nlpI∆thyA-SU or EcN∆thyA-SUT, respectively, every two days during the 28-day UA- and PO-challenged period (Fig. 7a). A murine model of hyperuricemia treated with saline showed a substantial increase in the serum UA concentration to ~599.7 µM, which was ~2.18 times greater than that of the normal control (serum UA concentration ~274.7 µM). Among the hyperuricemic mice dosed with wild-type EcN, EcN∆nlpI∆thyA-SU or EcN∆thyA-SUT, respectively, treatment with wild-type EcN or recombinant EcN equipped with T1SS mildly lowered the serum UA to ~514.3 µM or ~464.8 µM, respectively. Notably, treatment with our engineered EcN outfitted with T0SS exhibited therapeutic performance in reducing the concentration of circulating UA, which significantly decreased the serum UA concentration from ~599.7 µM to ~279.3 µM, approaching the level of the healthy control (serum UA concentration of ~274.7 µM) (Fig. 7b). These results proved that our engineered EcN outfitted with T0SS is superior to the recombinant EcN equipped with T1SS in alleviating hyperuricemia, demonstrating that OMV encapsulated Uox secreted by T0SS is prominent in degrading circulating UA due to its superior protein stability and wide delivery range beyond the gut.
a Experimental design of UA- and PO-induced murine model of hyperuricemia as well as treatment procedures. Mice fed a normal diet and intraperitoneally injected with vehicle are indicated as “Con”. Hyperuricemic mice were induced via a UA-supplemented diet (2% UA in the diet) and daily intraperitoneal injection of 250 mg/kg PO for 4 weeks. At the onset of hyperuricemia induction, hyperuricemic mice were gavaged with saline (abbreviated as “UA”), EcN (abbreviated as “UA + E”), EcN∆thyA-SUT (abbreviated as “UA + T”) or EcN∆nlpI∆thyA-SU (abbreviated as “UA + O”) (n = 6). b–e Serum UA (b), urine UA (c), serum CRE (d) and IL-1β (e) were measured. (n = 6 mice) (f) Representative histopathological images of colon slices obtained from five groups of mice. The slices were stained with hematoxylin and eosin (H&E). The black arrow indicates infiltration of immune cells, and the red arrow indicates sloughed-off epithelial cells. Scale bar = 50 μm. g, h The UA (g) and glucose (h) degradation efficacy of Uox-loaded OMVs derived from EcN∆nlpI∆thyA-SU on serum samples from hyperuricemic patients, in response to OMV treatment for 30 min (n = 9 biologically independent samples). i, j Lactic acid (i) and glucose (j) degradation efficacy of Lox-loaded OMVs derived from EcNΔnlpI-OL on serum samples from lung cancer patients (n = 7 biologically independent samples), in response to OMV treatment for 30 min. Data are presented as the mean ± SD. The P value was determined by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test (b–e) or a two-tailed paired t test (g–j). Source data are provided as a Source Data file.
The performance of our engineered EcN outfitted with modified T0SS to ameliorate hyperuricemia symptoms was further assessed. Hyperuricemia is usually accompanied by increased urine UA content40, which was also observed in hyperuricemic mice models (Fig. 7c). Compared with that in healthy controls, the level of urine UA was increased ~2.25 times in hyperuricemic mice treated with saline (Fig. 7c). Treatment with EcN, EcN∆thyA-SUT or EcN∆nlpI∆thyA-SU significantly reduced the urine UA of hyperuricemic mice by ~21.59%, 41.40% and 46.70%, respectively (Fig. 7c). UA is widely recognized as a damage-associated molecular pattern (DAMP) that can activate the NLRP3 inflammasome and increase the release of inflammatory cytokines, e.g., IL-1β41. Hence, the serum level of the inflammatory cytokine IL-1β was examined to assess systemic inflammation, and the level of serum creatinine (CRE) was assayed to assess renal function as well as the biosafety of drug treatment. Compared with those in healthy controls, the levels of serum IL-1β and serum creatinine (CRE) were significantly increased in hyperuricemic mice treated with saline (Fig. 7d, e). Compared with hyperuricemic mice dosed with saline, administration of EcN∆nlpI∆thyA-SU, rather than EcN∆thyA-SUT or wild-type EcN, significantly decreased the serum CRE and serum IL-1β levels by ~15.94% and ~32.91%, respectively (Fig. 7d, e). In addition, histological analysis of the colon, liver, and kidney tissues was performed through H&E staining (Fig. 7f). In hyperuricemic mice dosed with saline or wild-type EcN, significant immune cell infiltration was observed in the liver and kidneys, whereas the groups treated with EcN∆nlpI∆thyA-SU presented a significant reduction in immune cell infiltration (Fig. 7f). These results showed that our synthetic EcN outfitted with modified T0SS was superior for the oral delivery of Uox and the degradation of serum UA, and consequently exhibited outstanding efficacy in alleviating the pathological symptoms of hyperuricemia as well as good biosafety. Using hyperuricemia as a demonstration, we proved that, in comparison to conventional recombinant commensals that secrete therapeutic proteins into their surroundings within the gut, our engineered EcN outfitted with modified T0SS is superior for treating metabolic disorders, probably benefitted from superior stability of the therapeutic proteins encapsulated in OMVs, high endogenous encapsulation ratio and wide delivery range beyond the gut.
Engineered OMVs effectively detoxify clinical samples from patients
To further investigate the clinical potential of our engineered EcN outfitted with T0SS for treating metabolic diseases, the efficacy of protein-loaded OMVs derived from synthetic EcN for treating human serum samples was evaluated. Serum samples from nine hyperuricemic patients were collected and incubated with Uox-loaded OMVs derived from EcNΔnlpIΔthyA-SU. After incubation for 30 min, the levels of serum UA were all substantially reduced by OMV treatment, with an average reduction of 42.6% from 496.9 μM to 285.3 μM after OMV incubation (Fig. 7g), whereas the levels of serum glucose in these nine samples were not influenced by OMV incubation (Fig. 7h). These results demonstrated that the synthetic Uox-loaded OMVs derived from our engineered EcN are effective and specific for degrading UA in the blood of hyperuricemic patients. Moreover, the serum samples of seven lung cancer patients with elevated lactate levels were collected and incubated with Lox-loaded OMVs derived from EcNΔnlpI-OL. The incubation of Lox-loaded OMVs for 30 min led to a reduction in the serum lactate levels of all seven human samples, which decreased from ~5.67 mM to ~5.50 mM on average (Fig. 7i). Similarly, Lox-loaded OMVs significantly reduced the serum levels of lactate but did not influence the serum levels of glucose (Fig. 7j). Overall, we demonstrated that the therapeutic proteins delivered by our engineered EcN outfitted with modified T0SS could effectively detoxify circulating human metabolites, and our protein delivery system based on synthetic EcN is promising for the clinical treatment of a wide range of metabolic disorders.
Discussion
Oral protein delivery represents a convenient and highly compliant route of drug administration for patients. However, oral delivery efficiency is tremendously limited by gastrointestinal physiological barriers, including harsh acid and proteases in the stomach, the intestinal mucus layer, and the gut epithelial barrier. To overcome these physiological barriers, NP-based drug delivery systems have made significant progress to improve the efficiency of oral protein delivery, through material engineering, surface modification and so on42,43. Despite these findings, NP-based drug delivery systems suffer from complicated fabrication procedures, inefficient cargo loading and delivery. In our work, we established an oral protein delivery technology, which could be facilely implemented via the oral administration of genetically engineered EcN outfitted with modified T0SS. The EcN chassis is tolerant of the digestive tract, and in situ-secreted OMVs were found to penetrate the intact gut barrier and enter the circulation, consequently the protein payloads were escorted sequentially by engineered EcN and secreted OMV vehicles for efficient and programmed oral protein delivery. The protein payloads were encapsulated in OMVs in vivo by an endogenous protein loading strategy, whose encapsulation ratio was 97.9%, which is much greater than that of exogenous protein loading methods in vitro for EVs and NPs. A filtration and ultracentrifugation-based method was employed here for crude OMV isolation from bacterial culture medium, these OMVs exhibit high heterogeneity in size. While the engineered OMVs show homogeneity in loading the protein cargo of interest, other non-target molecular cargos could vary significantly among OMV populations. Thus, our engineered EcN outfitted with modified T0SS provides a self-programmed and facile oral protein delivery technology.
The presence of blood bacterial extracellular vesicles (EVs) has been reported primarily in patients with gut leakage, intestinal dysbiosis and bacterial translocation36. Few studies have reported the presence of bacterial OMVs in the circulation of healthy individuals, and it has been speculated that the translocation of gut bacteria may lead to the production of circulating OMVs27. Some researchers have reported that immune cells are hijacked for the delivery of engineered bacterial EVs via surface modification with targeting ligands44,45. Our work revealed that EcN OMVs without surface modification could enter the circulation through the intact gut barrier via bacterial translocation, indicating the physiological role of bacterial EVs in host‒microbe interactions beyond the gut without intestinal permeability defects or bacterial translocation. On the basis of this important finding, EcN was engineered to improve OMV synthesis and endogenously load proteins of interest into OMVs. OMV vehicles confer the advantages of high protein stability against proteolysis and circulation entry capability. The in vitro assay revealed that the epithelial transcytosis efficiency of EcN OMVs was approximately 3%, similar to that of receptor ligand-free engineered nanoparticles in traversing an in vitro blood-brain barrier model in a previous study46. The T0SS-based protein secretion system was compatible with various protein payloads, and was able to self-package multiple distinct protein cargos in individual OMV vehicles for cascade reactions. Moreover, EcN OMVs contain indigenous redox enzymes, including peroxide reductase, making OMVs ideal biocatalysts for detoxification, including decomposition, cascade and NADH-independent oxidation reactions. Our engineered EcN outfitted with modified T0SS was superior in detoxifying circulating metabolites, ascribe to its high stability of protein payloads and wide delivery range beyond the gut, presenting a promising oral protein delivery system based on synthetic bacteria for the clinical treatment of a wide range of metabolic disorders.
Methods
Ethics statement regarding animals, cells and clinical serum samples
This research complies with all relevant ethical regulations for both animal and human studies. All animal experiments were approved by the Institutional Animal Use and the Animal Experimentation Ethics Committee at Tsinghua University. The acquisition and processing of patient serum samples were approved by the China-Japan Friendship Hospital Clinical Research Ethics Committee, with approval number 2023-KY-007. The written informed consent was obtained from all study participants and the participants were compensated with free blood biochemical test. This study was conducted in accordance with the ethical standards outlined in the 2013 Declaration of Helsinki. The samples were obtained deidentified and no demographics were collected.
Male Kunming mice (4-5 weeks old) were obtained from Beijing Vital River Laboratory Animal China and used for all animal studies in this work. The mice were housed under specific pathogen-free (SPF) conditions with a 12 h light/dark cycle, enriched water and ad libitum feeding, with temperature kept at 22-24 °C, and humidity at 40-70%.
Caco-2 cells were obtained from the China Center for Type Culture Collection (Serial: TCHu146), and cultivated in DMEM supplemented with 20% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin in a humidified incubator with 5% CO2 at 37 °C.
Materials
DAPI (BL739B) was obtained from Biosharp Life Science. The cell dishes, 96-well culture plates, kanamycin (GC301008-5 g), IPTG (GC205011-1 g), chloramphenicol (GC301018-5 g), penicillin‒streptomycin (G4003-100ML), Dulbecco’s modified Eagle’s medium (DMEM; G4525-500 mL), anti-HA-Tag antibody (GB151252-100, for western blot), anti-GFP antibody (GB15603-100), anti-RFP antibody (GB125053-100) and L-phenylalanine (GC304010-10 g) were purchased from Servicebio Biotechnology. Cy5.5-NHS ester (A8103) was purchased from APExBIO Technology. The anti-HA-tag antibody (26183 for immunofluorescence), anti-His-tag antibody (MA121315), goat anti-IgG (H + L) highly cross-adsorbed secondary antibody conjugated with Alexa Fluor 647 (A32728TR) and fetal bovine serum (FBS; A5669401) were purchased from Thermo Fisher Scientific. Mouse IL-1β ELISA Kit (KE10003), goat anti-mouse IgG (H&L)-HRP (PR30012) and goat anti-rabbit IgG (H&L)-HRP (PR30011) for western blotting were purchased from Proteintech. EVMembrane Red Stain (NEPU-638-50T) was purchased from NanoFCM. Sodium carboxymethyl cellulose (IS9000), Uric Acid Content Assay Kit (BC1365), Creatinine Content Assay Kit (BC4915) and Urea Nitrogen Content Assay Kit (BC1535) were obtained from Solarbio Science & Technology. Cytochalasin D (gc13440), chlorpromazine HCl (gc14216), dynasore (gc10395), nystatin (gc10090), amiloride HCl (gc17853) and wortmannin (gc12338) were purchased from Glpbio Technology. Uric acid (U0018) and potassium oxonate (O0164) were purchased from Tokyo Chemical Industry. Sodium lactate (S108838) was purchased from Aladdin Scientific.
Bacterial strains and culture conditions
A list of the bacterial strains used in this investigation is provided in Supplementary Table 2. The E. coli S17-1 strain was used for plasmid construction, and the E. coli WM3064 strain served as the donor strain for conjugation. The katE gene was amplified from E. coli Nissle 1917 genomic DNA (GenBank CP007799.1). Heterologous genes, namely, uox from C. utilis (UniProt Entry P78609), hlyBD-hlyA from E. coli J9635, stlA from P. luminescens TT0147, and lox from A. viridans (UniProt Entry Q44467), were codon optimized, synthesized and cloned and inserted into the pYYD plasmid48 under the anaerobic promoter by GenScript (Nanjing, China). The plasmids were subsequently transformed into the E. coli WM3064 strain, which was subsequently conjugated with EcN.
The suicide plasmid pRE11249 was used for the in-frame genomic deletion of the nlpI and thyA genes in EcN. Plasmids were constructed via Gibson assembly in E. coli S17-1, which were subsequently transformed into E. coli WM3064 for subsequent conjugation with EcN. The single-crossover mutant was selected on LB agar medium supplemented with chloramphenicol and verified via colony PCR. The resulting double-crossover mutant was selected on LB agar medium supplemented with 10% sucrose and 3 mM thymidine (for the ∆thyA mutants), and further confirmed by colony PCR and sequencing. EcN and E. coli S17-1 were cultivated in LB broth (10 g tryptone, 10 g NaCl, 5 g yeast extract per liter) at 37 °C. E. coli WM3064 was cultured in LB medium supplemented with 50 µg/mL 2,6-diaminopimelic acid at 37 °C. Antibiotic selection involved the use of 50 µg/mL kanamycin (for pYYD-derived plasmids) and 25 μg/mL chloramphenicol (for pRE112-derived plasmids). The strains were cryopreserved at −80 °C in 15% glycerol.
In vitro UA consumption assay
To assess the ability of bacteria to degrade UA, logarithmically growing bacteria in LB medium were resuspended in modified M9 minimal medium (6.78 g Na2HPO4, 3 g KH2PO4, 1 g NH4Cl, 0.5 g NaCl, 5 g glucose, 1 g yeast extract per liter) supplemented with 0.5 mM UA for microaerobic culture, resulting in a concentration of 109 cells/mL. Sampling was conducted at various time intervals, and a Uric Acid Content Assay Kit was used to quantify the UA concentration.
Outer membrane vesicle isolation and characterization
To isolate and purify OMVs, the supernatant from the E. coli culture was 0.22-μm syringe-filtered after centrifugation at 4 °C to remove large contaminants and bacteria. The resulting mixture was concentrated with a 100 kDa cutoff centrifugal filter (Millipore, USA) and further ultracentrifuged at 120000 × g for 2 hours at 4 °C. The OMV pellet was washed with PBS, subjected again to ultracentrifugation, 0.22-μm syringe-filtered, and suspended in PBS. The morphology of the OMVs was characterized via a Hitachi transmission electron microscopy (TEM) system. Zeta potentials were measured with a Zetasizer Nano ZS (Malvern, UK) at 25°C under standard settings (refractive index, 1.330; viscosity, 0.89 cP). The hydrodynamic diameters of the OMVs were measured via ZetaVIEW (Particle Metrix, Germany) at 25 °C under a sensitivity of 80, a frame rate of 30 frames per second and a shutter value of 100.
Supernatant purification
The supernatant from bacterial culture was 0.22-μm syringe-filtered after centrifugation at 4 °C to remove large contaminants and bacteria. Then, the supernatant was concentrated with a 3 kDa cutoff centrifugal filter (Millipore, USA) to obtain concentrated solution for subsequent SDS-PAGE and Western Blot analysis.
Nano-flow cytometry
The isolated OMV samples were analyzed using the Flow NanoAnalyzer (nanoFCM) instrument equipped with 488 nm and 638 nm lasers. OMVs derived from EcNΔnlpI and EcNΔnlpI-OG were labeled with EVMembrane Red Stain at room temperature for 30 minutes. The fluorescently labeled samples were washed twice with PBS to remove excess dye via the isolation method described above. The particles were then resuspended in PBS and measured via calibrated instruments with 50 mW laser power, 10% SS decay, and a sampling pressure of 1 kPa. Data analysis was conducted via NF professional 2.0 software. Through analyzing the green and red fluorescence signals of the PBS control and OMVs derived from EcNΔnlpI, the threshold lines of green and red fluorescence were determined to reduce the reads of nanoscale impurities and background signals ( < 5%). The cutoff lines were drawn that the ratio of reads from P1 + P2 or P1 + P4 in PBS sample were <5%.
Measurement of metabolites
UA, creatinine and urea nitrogen were quantified via urea acid, creatinine and urea nitrogen content assay kits, respectively. Other metabolites (e.g., phenylalanine and lactate) in culture media or serum were measured via high-performance liquid chromatography (HPLC). Phenylalanine quantification was conducted via an HPLC system equipped with a UV detector and a ZORBAX SB-C18 chromatographic column. The mobile phase for gradient elution consisted of acetonitrile and 1.5% acetic acid. Detection occurred at 260 nm, and the column temperature was maintained at 40 °C. Lactate quantification was conducted via an HPLC system equipped with an RID-10A refractive index detector and an Aminex HPX-87H column. The samples were run with 5 mM sulfuric acid at 50 °C. Metabolite identification and quantification were accomplished by comparing the retention time and chromatographic peak area with those of established standards.
Animal experiments
The induction of hyperuricemia in male Kunming mice was performed with 2% UA in the diet (Jiangsu Xietong Pharmaceutical Bioengineering Co., Ltd.) and daily intraperitoneal injections of 250 mg/kg PO (dissolved in 0.5% CMC-Na solution) for 4 weeks. At the onset of hyperuricemia induction, four groups of mice were gavaged with EcN (5 × 109 CFU), EcN∆thyA-SUT (5 × 109 CFU), EcN∆nlpI∆thyA-SU (5 × 109 CFU), or an equal volume of saline, respectively, every two days during the entire 28 days of hyperuricemia induction. On the last day of the study, the mice were euthanized via CO2 inhalation. Blood samples were collected via cardiac puncture, and the liver, kidney and colon were harvested for histological analysis.
For in vivo safety evaluation of engineered EcN equipped with modified T0SS, three groups of healthy male Kunming mice were gavaged with EcN (5 × 109 CFU), EcN∆nlpI∆thyA-SU (5 × 109 CFU), or an equal volume of saline, respectively, every two days for one week. The mice were euthanized on the last day of the study. Blood samples were collected via cardiac puncture for routine blood examination and serum biochemistry assays, and the major organs (heart, liver, spleen, lung and kidney) were harvested for histological analysis.
Western blot and immunofluorescence
The quantification of total protein content in the samples was conducted via the Bradford assay. The purified OMVs or proteins were heated at 95 °C for 5 min, followed by SDS‒PAGE and subsequent transfer to a polyvinylidene difluoride (PVDF) membrane. After transfer, the membranes were blocked with 5% bovine serum albumin (BSA) for 1 h and incubated overnight at 4 °C with gentle shaking with specific primary antibodies. Afterward, the membranes were subjected to three 10-minute washes with TBST and further incubated with goat anti-mouse IgG (H&L)-HRP or goat anti-rabbit IgG (H&L)-HRP for an additional hour, and the bands were visualized by enhanced chemiluminescence (ECL).
For immunofluorescence, 7-μm sections of OCT-embedded frozen colon, liver, or kidney tissue were mounted on slides. After incubation in 0.5% Triton X-100 for 1 h, the slides were blocked for 2 h in 10% sheep serum and incubated with primary antibodies overnight at 4 °C. The slides were then incubated with the appropriate secondary antibody for 2 h at room temperature. Images were acquired via the Andor DragonFly confocal imaging system.
In vitro transcytosis studies
Caco-2 cells were seeded onto Transwell inserts and cultured for 3 weeks. The integrity of the epithelial monolayer was assessed through transepithelial electrical resistance (TEER) assays employing an EVOM TEER meter, and only monolayers with TEER values of at least 300 Ω·cm2 were used for transcytosis studies. To investigate the transport of OMVs via this model, OMVs containing RFP were introduced into the apical chamber ( ~ 1010 particles in 100 μL of media). The influence of temperature was examined by incubating this model at either 4 or 37 °C. To evaluate the mechanism of transcytosis, cells were pretreated with commercially available inhibitors (0.05, 0.1, or 1 μM wortmannin; 0.25, 0.5 or 2 μM cytochalasin D; 10, 20 or 50 μM chlorpromazine; 20, 100 or 200 μM amiloride; 0.1, 0.2 or 1 μM nystatin; 20, 100 or 200 μM dynasore) for 1 h at 37 °C before OMV addition, as described previously50. After 8 h, media from the basolateral chamber were collected, and fluorescent signals were quantified via a fluorescence microplate reader. To evaluate the impact of OMVs on epithelial barrier integrity, Lucifer Yellow (LY) was added to the Transwell insert, and the fluorescence intensity in both the apical and basolateral chamber media was measured after 1 h with a fluorescence microplate reader. The percentage of LY rejection was calculated via the following formula: % LY rejection = 100 (1 – RFUbasolateral/RFUapical).
Detection of metabolites in clinical serum samples
To detect the efficacy of OMVs in degrading metabolites in human blood, 50 μL of human serum sample was incubated with 100 μg of purified OMVs for 30 minutes. Then, the concentrations of UA, glucose, or lactate in the samples were detected as described above. Sex was not considered in the study design.
Statistics & reproducibility
The data are expressed as the mean ± SD or mean ± SEM. Statistical differences between the control and experimental groups were analyzed by one-way or two-way ANOVA with Tukey’s post-hoc test. Paired samples were compared via a paired t test. No data were excluded from the analyzes.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All the data are available in the main text or the supplementary materials. The OMV mass spectrometry proteomics data generated in this study have been deposited in the ProteomeXchange Consortium via the iProX partner repository with the dataset identifier PXD053377 [https://www.iprox.cn/page/project.html?id=IPX0008897000]. Source data are provided with this paper.
References
Spain, C. V., Wright, J. J., Hahn, R. M., Wivel, A. & Martin, A. A. Self-reported barriers to adherence and persistence to treatment with injectable medications for type 2 diabetes. Clin. Ther. 38, 1653–1664.e1651 (2016).
Crawford, A., Jewell, S., Mara, H., McCatty, L. & Pelfrey, R. Managing treatment fatigue in patients with multiple sclerosis on long-term therapy: the role of multiple sclerosis nurses. Patient Prefer Adherence 8, 1093–1099 (2014).
Brown, T. D., Whitehead, K. A. & Mitragotri, S. Materials for oral delivery of proteins and peptides. Nat. Rev. Mater. 5, 127–148 (2020).
Hong, S. et al. Protein-based nanoparticles as drug delivery systems. Pharmaceutics 12, 604 (2020).
Liu, J., Zhou, Y., Lyu, Q., Yao, X. & Wang, W. Targeted protein delivery based on stimuli-triggered nanomedicine. Exploration 4, 20230025 (2024).
Inda, M. E., Broset, E., Lu, T. K. & de la Fuente-Nunez, C. Emerging frontiers in microbiome engineering. Trends Immunol 40, 952–973 (2019).
Gurbatri, C. R. et al. Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci. Transl. Med. 12, eaax0876 (2020).
Chowdhury, S. et al. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat. Med. 25, 1057–1063 (2019).
Praveschotinunt, P. et al. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat. Commun. 10, 5580 (2019).
Wang, S. et al. Gut-to-brain neuromodulation by synthetic butyrate-producing commensal bacteria. Innovation Life 2, 100082 (2024).
Shen, H. et al. Engineered microbial systems for advanced drug delivery. Adv. Drug Deliv. Rev. 187, 114364 (2022).
Sanmarco, L. M. et al. Lactate limits CNS autoimmunity by stabilizing HIF-1α in dendritic cells. Nature 620, 881–889 (2023).
Yan, X. et al. Construction of a sustainable 3-hydroxybutyrate-producing probiotic Escherichia coli for treatment of colitis. Cell. Mol. Immunol. 18, 2344–2357 (2021).
Xie, W. et al. A bovine lactoferricin-lactoferrampin-encoding Lactobacillus reuteri CO21 regulates the intestinal mucosal immunity and enhances the protection of piglets against enterotoxigenic Escherichia coli K88 challenge. Gut Microbes 13, 1956281 (2021).
Duan, F. F., Liu, J. H. & March, J. C. Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes 64, 1794–1803 (2015).
Chen, K. et al. A probiotic yeast-based immunotherapy against Clostridioides difficile infection. Sci. Transl. Med. 12, eaax4905 (2020).
Hendrikx, T. et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 68, 1504–1515 (2019).
Gong, X. et al. Metabolic engineering of commensal bacteria for gut butyrate delivery and dissection of host-microbe interaction. Metab. Eng. 80, 94–106 (2023).
Lynch, J. P. et al. Engineered Escherichia coli for the in situ secretion of therapeutic nanobodies in the gut. Cell Host Microbe 31, 634–649.e638 (2023).
Zhang, X. et al. A red light-controlled probiotic bio-system for in-situ gut-brain axis regulation. Biomaterials 294, 122005 (2023).
Kuang, Z. et al. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science 365, 1428–1434 (2019).
Wang, Y. et al. The gut microbiota reprograms intestinal lipid metabolism through long noncoding RNA Snhg9. Science 381, 851–857 (2023).
Adolfsen, K. J. et al. Improvement of a synthetic live bacterial therapeutic for phenylketonuria with biosensor-enabled enzyme engineering. Nat. Commun. 12, 6215 (2021).
Puurunen, M. K. et al. Safety and pharmacodynamics of an engineered E. coli Nissle for the treatment of phenylketonuria: a first-in-human phase 1/2a study. Nat. Metab. 3, 1125–1132 (2021).
Vockley, J. et al. Efficacy and safety of a synthetic biotic for treatment of phenylketonuria: a phase 2 clinical trial. Nat. Metab. 5, 1685–1690 (2023).
Guerrero-Mandujano, A., Hernández-Cortez, C., Ibarra, J. A. & Castro-Escarpulli, G. The outer membrane vesicles: Secretion system type zero. Traffic 18, 425–432 (2017).
Schaack, B. et al. Microbiota-derived extracellular vesicles detected in human blood from healthy donors. Int. J. Mol. Sci. 23, 13787 (2022).
Sonnenborn, U. & Schulze, J. The non-pathogenic Escherichia coli strain Nissle 1917 – features of a versatile probiotic. Microbial Ecology Health Disease 21, 122–158 (2009).
McBroom, A. J., Johnson, A. P., Vemulapalli, S. & Kuehn, M. J. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188, 5385–5392 (2006).
Green, E. R. & Mecsas, J. Bacterial secretion systems: an overview. Microbiol Spectr 4, VMBF–0012-2015 (2016).
Kesty, N. C. & Kuehn, M. J. Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J. Biol. Chem. 279, 2069–2076 (2004).
Bartolini, E. et al. Recombinant outer membrane vesicles carrying Chlamydia muridarum HtrA induce antibodies that neutralize chlamydial infection in vitro. J. Extracell. Vesicles 2, 20181 (2013).
Aronson, D. E., Costantini, L. M. & Snapp, E. L. Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon. Traffic 12, 543–548 (2011).
Chen, C. et al. Single-particle assessment of six different drug-loading strategies for incorporating doxorubicin into small extracellular vesicles. Anal. BioAnal. Chem. 415, 1287–1298 (2023).
Tzschaschel, B. D., Guzmán, C. A., Timmis, K. N. & de Lorenzo, V. An Escherichia coli hemolysin transport system-based vector for the export of polypeptides: export of Shiga-like toxin IIeB subunit by Salmonella typhimurium aroA. Nat. Biotechnol. 14, 765–769 (1996).
Jones, E. et al. The origin of plasma-derived bacterial extracellular vesicles in healthy individuals and patients with inflammatory bowel disease: a pilot study. Genes (Basel) 12, 1636 (2021).
Schwechheimer, C. & Kuehn, M. J. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 13, 605–619 (2015).
Mulcahy, L. A., Pink, R. C. & Carter, D. R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 3, 24641 (2014).
Lu, J. et al. Mouse models for human hyperuricaemia: a critical review. Nat. Rev. Rheumatol. 15, 413–426 (2019).
Wu, Y. et al. Limosilactobacillus fermentum JL-3 isolated from “Jiangshui” ameliorates hyperuricemia by degrading uric acid. Gut Microbes 13, 1–18 (2021).
Cabău, G., Crișan, T. O., Klück, V., Popp, R. A. & Joosten, L. A. B. Urate-induced immune programming: Consequences for gouty arthritis and hyperuricemia. Immunol. Rev. 294, 92–105 (2020).
Mitchell, M. J. et al. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 20, 101–124 (2021).
Zhang, F. et al. Lipid-based intelligent vehicle capabilitized with physical and physiological activation. Research (Wash D C) 2022, 9808429 (2022).
Yue, Y. et al. Antigen-bearing outer membrane vesicles as tumour vaccines produced in situ by ingested genetically engineered bacteria. Nat. Biomed. Eng. 6, 898–909 (2022).
Li, Y. et al. Rapid surface display of mRNA antigens by bacteria-derived outer membrane vesicles for a personalized tumor vaccine. Adv. Mater. 34, e2109984 (2022).
Chen, Z. A. et al. Receptor ligand-free mesoporous silica nanoparticles: a streamlined strategy for targeted drug delivery across the blood-brain barrier. ACS Nano 18, 12716–12736 (2024).
Williams, J. S., Thomas, M. & Clarke, D. J. The gene stlA encodes a phenylalanine ammonia-lyase that is involved in the production of a stilbene antibiotic in Photorhabdus luminescens TT01. Microbiology (Read.) 151, 2543–2550 (2005).
Yang, Y. et al. Enhancing bidirectional electron transfer of Shewanella oneidensis by a synthetic flavin pathway. ACS Synth. Biol. 4, 815–823 (2015).
Edwards, R. A., Keller, L. H. & Schifferli, D. M. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157 (1998).
Zingl, F. G. et al. Outer membrane vesicles of vibrio cholerae protect and deliver active cholera toxin to host cells via porin-dependent uptake. mBio 12, e0053421 (2021).
Acknowledgements
The authors would like to thank Dr. Yunzi Luo (from Tianjin University, China) for sharing the E. coli Nissle 1917 strain. We would like to express our gratitude to Fanyu Zeng (Analytical Testing Center, Chinese National Vaccine and Serum Institute) for her assistance in collecting the nano-flow cytometric data, Gang Wang, and Xichuan Gai (Airy-tech Company) for their assistance with Polar-SIM super-resolution microscopy of OMVs. The authors acknowledge BioRender (www.biorender.com) for assisting in creating the illustrative diagrams. This research was funded by the National High Level Hospital Clinical Research Funding (2023-NHLHCRF-LXYZ-02 and 2022-NHLHCRF-LX-03-0303, F.Y.) and the National Natural Science Foundation of China (No. 82202336, J.C. and No. 31900069, Y.Y.).
Author information
Authors and Affiliations
Contributions
Y.Y., X.-J.L., and F.X. supervised the project. X.G., S.L., B.Z., Y.W., and Z.W. acquired the data. X.G., S.L., B.X., and Y.Y. contributed to the data analyzes and to the interpretation of the results., S.Z., J.C., and F.X. provided advice and support. X.G. wrote the manuscript. F.X., Y.Y., and X.-J.L. edited the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Tappei Takada and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gong, X., Liu, S., Xia, B. et al. Oral delivery of therapeutic proteins by engineered bacterial type zero secretion system. Nat Commun 16, 1862 (2025). https://doi.org/10.1038/s41467-025-57153-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57153-6