Abstract
Senescence-associated secretory phenotype (SASP) mediates the biological effects of senescent cells on the tissue microenvironment and contributes to ageing-associated disease progression. ACSS2 produces acetyl-CoA from acetate and epigenetically controls gene expression through histone acetylation under various circumstances. However, whether and how ACSS2 regulates cellular senescence remains unclear. Here, we show that pharmacological inhibition and deletion of Acss2 in mice blunts SASP and abrogates the pro-tumorigenic and immune surveillance functions of senescent cells. Mechanistically, ACSS2 directly interacts with and promotes the acetylation of PAICS, a key enzyme for purine biosynthesis. The acetylation of PAICS promotes autophagy-mediated degradation of PAICS to limit purine metabolism and reduces dNTP pools for DNA repair, exacerbating cytoplasmic chromatin fragment accumulation and SASP. Altogether, our work links ACSS2-mediated local acetyl-CoA generation to purine metabolism through PAICS acetylation that dictates the functionality of SASP, and identifies ACSS2 as a potential senomorphic target to prevent senescence-associated diseases.
Similar content being viewed by others
Introduction
Cellular senescence promotes tissue and organismal ageing, as well as ageing-associated pathologies1,2. Cellular senescence is characterized by cell cycle arrest and the secretion of pro-inflammatory and growth factors by senescent cells, known as senescence-associated secretory phenotype (SASP)3. Various stresses, including oncogene activation, telomere shortening, mitochondria dysfunction, and others, can trigger senescence4. SASP mediates the biological functions of the senescent cells and exhibits context-dependent effects on the tissue microenvironment5,6. While critical for wound healing and immune surveillance, SASP adversely contributes to the chronic progression of diseases such as cancer7,8, tissue fibrosis9, and metabolic disorders10. By virtue of this, selective elimination of senescent cells by “senolytics” or suppression of SASP generation by “senomorphics” have been considered potential strategies to promote healthy ageing or prevent ageing-associated diseases3,11,12,13.
Persistent DNA damage response (DDR) and compromised DNA repair result in genomic instability of senescence and trigger SASP14,15,16. Upon DNA damage, senescent cells release γH2AX-expressing nuclear chromatin fragments into the cytoplasm, known as cytoplasmic chromatin fragments (CCFs)17,18. In this context, cytosolic DNA sensor cGAS recognizes CCFs and produces second messenger cyclic GMP-AMP (cGAMP), which binds and activates STING, activating NFκB signaling to trigger SASP gene transcription19,20,21. Additionally, apoptotic stress also enables the release of mitochondrial DNA (mtDNA) into the cytosol, where mtDNA, in turn, activates the cGAS-STING pathway22.
Metabolic deregulation is one hallmark of senescence1,23,24. Senescent cells show increased glycolysis and mitochondrial dysfunction, among other metabolic abnormalities23,25,26,27,28. Recent studies demonstrate that deregulated metabolism also promotes CCF formation. For example, mitochondria-derived oxidative stress facilitates senescence induction and SASP through DNA damage and CCF formation29,30. Notably, suppressed nucleotide metabolism results in deoxyribonucleotide triphosphate (dNTP) deficiency, leading to aberrant DNA replication, DNA repair blockade, and genomic instability during oncogene-induced senescence (OIS)31. In line with this, exogenous dNTPs supplement or increasing nucleotide, especially purine biosynthesis, facilitates DNA repair, senescence bypass, and cancer development31,32,33,34. However, the regulation of nucleotide metabolism in cellular senescence and subsequent effects on SASP remain poorly understood.
Acetyl-CoA, the lipogenic precursor and acetylation substrate, plays compartment-specific roles in physiological and pathological processes, including cellular senescence35. Intracellular acetyl-CoA is mainly catalyzed by citrate-dependent ATP-citrate lyase (ACLY) and acetate-dependent acetyl-CoA synthetase 2 (ACSS2). ACLY synthesizes the major pool of acetyl-CoA, and promotes DNA repair through histone acetylation in proliferating cancer cells36. In non-cancerous cells, ACLY deficiency has been shown to induce senescence through p53 activation37. The emerging evidence demonstrates that ACSS2 undergoes cytosol-to-nuclear translocation and regulates gene transcription during neural differentiation or under glucose starvation, through on-site generation of acetyl-CoA and histone acetylation38,39,40,41. A recent study also showed that ACSS2 homolog Acs2 is required for histone H4K16 acetylation to maintain the telomere silencing and regulate the senescence in budding yeast42. While these studies uncover important roles of nuclear ACSS2 in gene regulation, whether and how ACSS2 regulates mammalian cellular senescence is unknown.
Here, we show that ACSS2 drives SASP by inhibiting PAICS-dependent de novo purine biosynthesis (DNPB) and promoting CCF formation. Our work links ACSS2-mediated local acetyl-CoA generation to non-histone acetylation of a metabolic enzyme that dictates the functionality of SASP. Our results also suggest that ACSS2 can be a potential senomorphic target to treat senescence-associated diseases.
Results
ACSS2 drives SASP
We used human primary embryonic lung fibroblast (IMR90) to establish a 4-Hydroxy-tamoxifen-activatable oncogenic RAS-induced senescence model (OIS). Starting on day eight post-initiation, most cells enter senescence accompanied by strong SASP43. To explore the role of ACSS2 in OIS, we employed shRNAs to knock down ACSS2 at day 8 (Fig. 1a). ACSS2 deficiency resulted in the accumulation of its substrate acetate, but did not affect senescence-associated growth arrest or expression of senescence markers, including senescence-associated beta-galactosidase (SA-β-gal) activity and upregulation of p16 and p21 (Supplementary Fig. 1a and Fig. 1b–d). However, the global transcriptomic profiling among groups of proliferating (Pro), senescent (Sen) cells with or without ACSS2 knockdown (shACSS2) showed a significant enrichment of SASP-related pathways, such as cytokine-cytokine interaction, chemokine signaling and NFκB pathways (Fig. 1e, f). The SASP genes were indeed significantly upregulated in OIS cells while reduced upon ACSS2 knockdown (Fig. 1g and Supplementary Fig. 1b). Furthermore, consistent results were obtained by treating OIS cells with ACSS2 inhibitor (ACSS2i) (Supplementary Fig. 1c, d). Similar SASP decline by ACSS2 inhibition was also observed in other forms of senescence, including chemotherapy drug etoposide treatment-induced senescence (TIS) (Supplementary Fig. 1e, f) or irradiation-induced senescence (Supplementary Fig. 1g-j). Notably, ACSS2 expression positively correlated with the expression of several SASP genes in human pancreatic intra-epithelial neoplasia harboring oncogenic RAS-induced senescent cells44 (Supplementary Fig. 1k). We then compared the secreted SASP components from proliferating and OIS cell groups using antibody-based cytokine array. Consistently, the secreted protein levels significantly decreased in ACSS2-knockdown senescent cells (Fig. 1h).
a ER:RAS-expressing IMR90 cells were induced to senesce by adding 100 nM 4-OHT, expressing non-targeting shRNA (shCtrl) or ACSS2 targeted shRNA (shACSS2). The puromycin-selected cells were harvested and analysed for expression of the indicated proteins by immunoblots. b–d The indicated cells were examined by SA-β-gal staining and colony formation (b). SA-β-gal-positive cells were quantified (c), the integrated intensity of the colonies was quantified using NIH Image J software (d). e, f Heat map of the RNA-seq data of the genes for which expression was significantly changed among proliferating and senescent cells with or without ACSS2 knockdown (e), KEGG analysis of genes altered by ACSS2 knockdown is shown (fold change, FC ≥ 2, P < 0.05) (f). g Heat map of SASP genes in senescent cells with ACSS2 knockdown vs. control knockdown determined by RNA-seq analysis. h The secretion of SASP components under the indicated conditions was detected by antibody arrays. The relative FC in comparison to the senescent cell with control shRNA was shown (n = 2 independent repeats). i Average signal from ChIP-seq analysis of H3K27ac occupancy in proliferating and senescent cells with or without knockdown of ACSS2. j Overlap between genes with expression upregulated in senescence while decreased by ACSS2 knockdown and genes with increased H3K27ac signal while decreased by ACSS2 knockdown (FC ≥ 2). RNA-seq and ChIP-seq analysis, n = 3 biologically independent repeats. k Gene ontology analysis of the overlap genes from (j). l Tracks of H3K27ac distribution on the representative SASP genes in groups of proliferating and senescent cells with or without ACSS2 knockdown. Data represent the mean ± s.d. of three biologically independent experiments. The P values were calculated using One-way ANOVA (Tukey’s multiple-comparison test) (c, d) or two-tailed Fisher Exact test (j) or one-sided Fisher exact test with Benjamini-Hochberg adjustment (f, k). Representative blot of n = 3 independent experiments was shown (a). Source data are provided as a Source Data file.
To determine if the effect of ACSS2 deficiency is specific to the senescence condition, we performed ACSS2 knockdown or ACSS2i treatment in proliferating cells and found that neither deficiency nor inhibition of ACSS2 significantly impacts the proliferative state, as shown by the expression of senescence-associated cell cycle arrest markers, SA-β-gal activity, and SASP gene expression (Supplementary Fig. 2).
ACSS2 can drive gene transcription through histone acetylation-dependent mechanisms39,41. Histone acetylation on SASP gene super-enhancers and enhancers featured by lysine 27 acetylated histone H3 (H3K27ac), and the interaction with transcription factor NFκB, play pivotal roles in the activation of SASP gene transcription45,46. We next sought to determine whether ACSS2 could regulate SASP gene expression directly via epigenetic mechanisms. The immunoblots of H3ac and H3K27ac showed that global histone acetylation did not change by ACSS2 knockdown compared to the control knockdown cells (Supplementary Fig. 1l). Furthermore, ChIP-sequencing results showed that the global distribution of H3K27ac did not change significantly in ACSS2 knockdown compared to control knockdown OIS cells (Fig. 1i). Through integrated analysis of the H3K27ac ChIP-seq with RNA-seq datasets comparing control and ACSS2 knockdown OIS cells, 1260 genes were upregulated in senescence while downregulated by ACSS2 knockdown, and 296 out of 1260 genes exhibited reduced H3K27ac (more than twofold) deposition in ACSS2 knockdown OIS cells, indicating that ACSS2 regulates these genes potentially through histone acetylation. Gene ontology analysis of the 296 genes showed significant enrichment (P = 2.2 × 10−61, Fisher’s exact test) of signaling transduction and cell adhesion (Fig. 1j, k). Notably, although the SASP gene expression was increased in senescence while significantly decreased by ACSS2 knockdown or inhibition, and H3K27ac deposition on the SASP genomic loci indeed dramatically elevated in senescent cells compared to proliferating cells, ACSS2 knockdown did not change the H3K27ac occupancy on these SASP gene loci, indicating that downregulated SASP gene expression by ACSS2 loss is not regulated by H3K27ac (Fig. 1l). Similar results were confirmed in TIS, where ACSS2 knockdown did not change H3K27ac occupancy on SASP gene loci, as revealed by H3K27ac ChIP-qPCR analysis (Supplementary Fig. 1m). Altogether, these data suggest that ACSS2 controls SASP independently of H3K27ac-marked epigenetic regulation.
ACSS2 promotes CCF formation in senescent cells
CCF formation is an upstream molecular event of SASP gene transcription, and CCFs are recognized by the cytosolic DNA sensor cGAS that produces cyclic GMP-AMP (cGAMP) to activate the adaptor protein STING which leads to activation of NFκB signaling20,21. We next sought to determine whether ACSS2 knockdown alters SASP gene expression through CCF formation. By co-staining with CCF marker γH2AX and cGAS in proliferating and senescent cells, we observed that ACSS2 but not ACLY deficiency dramatically suppressed CCF formation in OIS, TIS, and irradiation-induced senescence (Fig. 2 and Supplementary Fig. 3a, b). However, ACSS2 deficiency did not impact proliferating cells in which CCFs were not obvious (Supplementary Fig. 3c, d). Given that ACSS2 nucleus localization coincides with its nuclear function, we wondered if the cellular localization of ACSS2 changed during senescence. The immunostaining results showed that ACSS2 displayed a similarly diffused pattern in the cytoplasm of both proliferating and senescent cells, indicating a potential role of cytosolic ACSS2 in controlling SASP through CCF formation (Supplementary Fig. 3e–j).
a, b IMR90 cells were induced to senesce by oncogene RAS (a) or Etoposide (b). The proliferating cells and senescent cells expressing non-targeting shRNA, ACSS2, or ACLY-targeted shRNAs were infected with cGAS-FLAG encoding lentivirus. All groups of cells were subjected to γH2AX and cGAS-FLAG immunostaining. CCFs were indicated by arrows. Scale bar = 20 μm. c, d the CCF-positive cells were quantified among groups as described in (a, b). Randomly imaged fields with over 100 cells were analysed. Data represent the mean values of six different fields ± s.d. The P values were calculated using One-way ANOVA (Tukey’s multiple-comparison test). Representative of n = 3 independent experiments were shown (c, d). Source data are provided as a Source Data file.
Aberrant purine and pyrimidine metabolism in ACSS2-deficient senescent cells
The cytosolic ACSS2 location led us to explore potential metabolic mechanisms for ACSS2 to regulate CCF formation. To this, we performed targeted metabolomics analysis of proliferating cells, senescent cells with or without ACSS2, and included ACLY knockdown senescent cells for comparison (Fig. 3a). Higher NADP+/NADPH ratios and lower GSH/GSSG ratios were detected in OIS compared to proliferating cells, indicating redox imbalance in senescence as previously reported26,28 (Supplementary Fig. 4a, b). Consistent with previous studies, ACLY-knockdown senescent cells had much lower acetyl-CoA levels than ACSS2-knockdown senescent cells (Supplementary Fig. 4c). However, ACSS2 deficiency elicited a distinct metabolic profile compared to control or ACLY-knockdown senescent cells (Fig. 3a). Moreover, metabolic pathway analysis uncovered pyrimidine and purine metabolism as two top-ranked pathways enriched in ACSS2 deficient senescent cells (Fig. 3b). Indeed, knockdown of ACSS2 significantly increased the intermediate metabolites of pyrimidine metabolism, including dCTP and dUMP (Fig. 3c), as well as metabolites of purine metabolism, including ATP and dATP (Fig. 3d). However, these metabolites were not consistently accumulated in ACLY-knockdown senescent cells. We also measured the intracellular concentration of dNTPs in OIS with or without ACSS2 knockdown using a fluorescence-based method47, and confirmed that ACSS2 deficiency increased the dNTPs levels in senescent cells (Fig. 3e). As previously reported, enhanced purine biosynthesis facilitates DNA repair in tumor cells by increasing dNTPs pools, and exogenous dNTPs delay senescence-associated cell cycle arrest32,33. We thus reasoned that ACSS2 deficiency potentially enhances nucleotide metabolism to promote DNA repair that reduces CCF formation and functionally dampens SASP generation. Indeed, exogenous nucleosides supplement remarkably reduced CCF formation, decreased SASP mRNA levels, as well as p16 and p21 protein levels in OIS cells, whereas supplement of purine nucleosides failed to do so (Fig. 3f–h and Supplementary Fig. 4d–g). Collectively, these results connect ACSS2 to nucleotide metabolism that functionally controls SASP.
a ER:RAS-expressing cells were induced to senesce by adding 100 nM 4-OHT. At day 8 post-induction, cells were infected with lentivirus encoding non-targeting shRNA, ACSS2, or ACLY-targeted shRNAs. The puromycin-selected cells were harvested to perform targeted metabolomics analysis. Hierarchical clustering of significantly altered metabolites among groups was shown (n = 3 each group). b Top-ranked metabolic pathways in senescent cells with ACSS2 knockdown compared to control shRNA knockdown. P values were calculated using hypergeometric distribution (MetaboAnalyst v6.0). c, d Heat map analysis of the metabolites involved in the pyrimidine pathway (c) and the purine pathway (d). e ER:RAS-expressing cells were induced to senesce by adding 100 nM 4-OHT. On day 8 post-induction, cells were infected with lentivirus-encoding non-targeting shRNA or ACSS2-targeted shRNA. The puromycin-selected cells were harvested to measure cellular dNTP levels. f–h ER:RAS-expressing cells were induced to senesce by adding 100 nM 4-OHT. At day 8 post-induction, cells were supplemented with or without nucleosides or purine-only nucleosides for 14 days. Cells were stained for γH2AX (f). Three randomly imaged fields were analysed (g). The SASP mRNA expression was examined by qRT-PCR (h). Scale bar = 20 μm. Data represent mean ± s.d. of three biologically independent experiments. The P values were calculated using an unpaired two-tailed Student’s t-test (e) or One-way ANOVA (Tukey’s multiple-comparison test) (g, h). Pro, proliferating cells; Sen, RAS induced senescence; NS, nucleosides; A + G, Adenosine+Guanosine. Source data are provided as a Source Data file.
ACSS2 interacts with purine biosynthetic enzymes in senescent cells
As ACSS2 functions differently from ACLY to impact CCF-SASP, presumably through nucleotide metabolism, we postulated that the protein, rather than the common metabolite acetyl-CoA, mediated this specific role of ACSS2 in senescence. To elucidate how ACSS2 specifically modulates nucleotide metabolism, we conducted a TurboID-based proximity labeling assay to map ACSS2 interactome. ACSS2-V5 fused with biotin ligase was expressed in senescent cells, the biotinylated proteins were captured by streptavidin beads followed by LC-tandem MS (LC-MS/MS) (Supplementary Fig. 5a). This proteomic analysis identified 221 proteins significantly enriched (p < 0.05, log2fold change >1) in ACSS2-V5-TurboID over TurboID control group, including ACSS2 itself as a positive control. Among the 221 proteins, we detected significant enrichment of PAICS and AK1, two enzymes of de novo purine biosynthesis (DNPB) pathway (Fig. 4a and Supplementary Fig. 5b). The DNPB consists of a series of metabolic reactions that convert phosphoribosyl pyrophosphate (PRPP) into inosine 5′-monophosphate (IMP), then to dATP and dGTP, respectively. Of note, purine biosynthetic enzymes compartmentalize into a dynamic complex called the purinosome to achieve a greater DNPB efficiency, and PAICS plays a key role in purinosome organization48,49,50.
a Volcano plot showing Log2 fold changes in protein intensities on the X-axis and −Log10 P values on the Y-axis. Significantly enriched proteins were in blue (P < 0.05) and non-significant in gray. P values were calculated using a two-tailed Student’s t-test. b–e ER:RAS-expressing cells were induced to senesce by adding 100 nM 4-OHT, and the proliferating cells and senescent cells expressing non-targeting shRNA, ACSS2 targeting shRNA together with PAICS or AK1 specific shRNAs were subjected to immunoblots for the indicated proteins (b), or CCF detection (c). Scale bar = 10 μm. d The CCF-positive cells were quantified among groups. Four Randomly imaged fields with over 100 cells were analysed. Bar graphs represent mean values of different fields ± s.d. e Proliferating and senescent cells, as described in (b), were subjected to qRT-PCR analysis for expression of the indicated SASP genes. f Proliferating and OIS cells with shCtrl or shACSS2 were harvested and the labeled 15N-Glutamine incorporation into purine intermediates were analysed by LC-MS/MS. Data represent mean ± s.d. of three biologically independent experiments. The P values were calculated using One-way ANOVA (Tukey’s multiple-comparison test) (d–f). Representative of n = 3 independent experiments were shown (b, d). Source data are provided as a Source Data file.
Focusing on PAICS, we validated the interaction between ACSS2 and PAICS by co-immunoprecipitation (co-IP) assays in 293 T cells (Supplementary Fig. 5c). To determine whether the identified PAICS and AK1 functionally regulate SASP, we performed PAICS and AK1 knockdown respectively in senescent cells, and examined the effects on SASP gene expression and CCF formation (Supplementary Fig. 5d). Compared to the control group, loss of either gene significantly increased CCF formation and further upregulated SASP gene expression (Supplementary Fig. 5e, f). Overall, these results suggest that DNPB enzymes negatively regulate CCF formation and SASP generation.
We next examined whether inhibition of purine metabolism could rescue the effect of ACSS2 deficiency on SASP. To this, we further knocked down PAICS or AK1 in ACSS2-knockdown senescent cells, and compared CCF formation and SASP gene expression (Fig. 4b). While ACSS2 knockdown cells harbored fewer CCFs and lower SASP mRNA levels, knockdown of PAICS or AK1 in these cells significantly recovered CCF formation and SASP gene expression (Fig. 4c–e). To further confirm that intervention of purine or dNTPs synthesis can recover CCF formation and SASP gene expression elicited by ACSS2 inactivation, we treated senescent cells with methotrexate (MTX), the purine biosynthesis inhibitor51. Alternatively, we knocked down PRPS1, the rate-limiting enzyme of DNPB that produces phosphoribosyl pyrophosphate (PRPP), the common precursor for de novo purine and pyrimidine synthesis52. Indeed, either MTX treatment or PRPS1 knockdown significantly rescued the CCF formation and SASP expression induced by ACSS2 loss (Supplementary Fig. 5g–l). These data have demonstrated that elevating nucleotide biosynthesis contributes to impaired SASP production upon ACSS2 inactivation.
To further examine whether the metabolic activity of DNPB is regulated by ACSS2, we measured relative flux of stable isotope-labeled glutamine (amide-15N) in proliferating and senescent cells with or without ACSS2 knockdown (Fig. 4f). Compared to proliferating cells, the labeling of purine intermediates e.g. AMP and GMP were significantly decreased in senescent cells, which were increased by ACSS2 deficiency (Fig. 4f). Consistently, ACSS2 deficiency elevated the level of SAICAR, the PAICS product (Supplementary Fig. 5m). These data suggest that DNPB were blocked in senescence while can be reactivated by ACSS2 deficiency.
Collectively, we conclude that ACSS2 interacts with DNPB enzymes including PAICS in senescent cells, and ACSS2 deficiency enhances DNPB to alleviate DNA damage and then reduce CCF formation and SASP generation.
ACSS2 promotes acetylation and degradation of PAICS
We next defined how ACSS2 regulates PAICS. We first examined the interaction by co-IP assays using V5-tagged ACSS2 and HA-tagged PAICS in proliferating and senescent cells (Supplementary Fig. 6a). The endogenous association between ACSS2 and PAICS in both proliferating and senescent cells with or without ACSS2 knockdown were further confirmed (Fig. 5a). Moreover, ACSS2-PAICS interaction was confirmed in pull-down assays using recombinant proteins, where bacterially expressed and purified ACSS2 and PAICS directly binds to each other (Fig. 5b and Supplementary Fig. 6b, c).
a Proliferating and OIS cells with or without ACSS2 knockdown were harvested. Co-immunoprecipitation analysis of endogenous PAICS and ACSS2 in extracts from the indicated IMR90 cells was performed and subjected to immunoblots of the indicated proteins. b Purified human protein of ACSS2 and PAICS was subjected to in vitro pull-down assay. Immunoblot of the indicated proteins was shown. c Proliferating and OIS cells were infected with HA-PAICS encoding lentivirus, and then treated with or without ACSS2i for 48 h. PAICS protein was immunoprecipitated, and the acetylation was analysed by immunoblot with the indicated antibody. d HEK293T cells were transfected with HA-tagged wildtype (WT) or K36R, K47R, and K36RK47R mutants of PAICS. HA-PAICS protein was immunoprecipitated, and the acetylation was analyzed by immunoblot with the indicated antibody. e, f ER:RAS-expressing cells were induced to senesce by adding 100 nM 4-OHT. On day 8 after the induction, the senescent cell was infected with lentivirus encoding non-targeting shRNA or ACSS2 targeted shRNA (e) or treated with ACSS2i for 48 h (f). The cells were harvested and analysed by immunoblot for expression of the indicated proteins. g, h Proliferating and senescent cells described in (e) were treated with CHX (100 μg/mL) for 0, 6, or 12 h, and the expression of the indicated proteins was analysed by immunoblot (g) and quantified (h). i OIS cells were subjected to co-immunoprecipitation analysis of endogenous PAICS and PCAF, immunoblots of the indicated proteins were shown. j, k OIS cells expressing non-targeting shRNA or PCAF-targeted shRNA were infected with HA-PAICS encoding lentivirus. PAICS protein was immunoprecipitated, and the acetylation was analyzed by immunoblot with the indicated antibody (j) and quantified (k). l Proliferating and OIS cells expressing non-targeting shRNA or PCAF-targeted shRNA were subjected to immunoblots of the indicated proteins. Data represent mean ± s.d. of three biologically independent experiments. The P values were calculated using an unpaired two-tailed Student’s t-test (k). Representative of n = 3 independent experiments were shown (a–g, i, j, l). Source data are provided as a Source Data file.
Since ACSS2 facilitates the direct transferring of the acetyl group onto histone, the interaction between ACSS2 and PAICS prompted us to explore the potential acetylation of PAICS regulated by ACSS2. Indeed, immunoblot analysis using anti-acetylated lysine (ac-K) antibody revealed obvious PAICS acetylation in proliferating and senescent cells (Fig. 5c). Importantly, enzymatic inhibition by ACSS2i dramatically suppressed PAICS acetylation in senescent cells (Fig. 5c), indicating that ACSS2 associates with PAICS and promotes its acetylation through acetyl-CoA production.
To identify the acetylated residues in PAICS, HA-tagged PAICS was expressed in senescent cells, immunoprecipitated, and analysed by LC-MS/MS. Two lysine residues (K36 and K47) were identified for subsequent analysis (Supplementary Fig. 6d). Interestingly, single mutation of either lysine residue to arginine (K36R or K47R) substantially reduced acetylation of PAICS, while double mutation (K36RK47R) displayed further reduction of the acetylation (Fig. 5d), indicating that both K36 and K47 residues contribute to PAICS acetylation.
Given acetylation can affect protein stability53, to assess whether acetylation affects PAICS stability, we performed cycloheximide (CHX) chase assay to compare the stability of wildtype (WT), single- and double-mutant PAICS. Time course-dependent degradation of WT PAICS was observed within 12 h, while single- and double-mutation dramatically protected PAICS from degradation, supporting that acetylation promotes PAICS degradation (Supplementary Fig. 6e, f). To further determine whether ACSS2 controls PAICS stability, we first compared PAICS protein levels among proliferating and senescent cells with or without ACSS2 inactivation. Compared to proliferating cells, PAICS protein levels were significantly reduced in OIS, TIS, and irradiation-induced senescent cells, which were rescued by ACSS2 knockdown or enzymatic inhibition (Fig. 5e, f and Supplementary Fig. 6g–i). Of note, PAICS mRNA levels did not change accordingly among proliferating and senescent cells with or without ACSS2 knockdown (Supplementary Fig. 6j). Conversely, overexpression of ACSS2 decreased PAICS protein but not mRNA levels in senescent cells (Supplementary Fig. 6k, l). Notably, ACLY knockdown did not recover the decline of PAICS protein in senescence (Supplementary Fig. 6m–o). Moreover, the CHX assay showed that ACSS2 deficiency counteracted the progressive degradation of PAICS in senescent cells (Fig. 5g, h). Collectively, these results consistently support that ACSS2 enhances PAICS degradation through acetylation during senescence.
We next performed proximal labeling for PAICS integrated with LC-MS/MS in senescent cells, and identified PCAF as the potential acetyltransferase for PAICS protein (Supplementary Fig. 6p, q). While typically functions as nuclear histone acetyltransferase, PCAF can shuttle among extranuclear compartments, recognize non-histone proteins and transfer the acetyl group onto the lysine of target proteins including p53, IDH2, PKM2, which affects their activity or stability54,55,56. The subcellular fractionation showed that PCAF localized in both cytosol and nucleus in proliferating and senescent cells (Supplementary Fig. 6r). We next validated the endogenous protein-protein interaction between PCAF and PAICS (Fig. 5i). Importantly, PCAF knockdown leads to markedly reduced acetylation but increased protein of PAICS without affecting the mRNA level, indicating that PCAF is responsible for the acetylation of PAICS and governs its stability in senescence (Fig. 5j–l and Supplementary Fig. 6s).
Acetylation promotes autophagy-mediated PAICS degradation in senescence
We next investigated how PAICS acetylation promotes its degradation. To test the involvement of the ubiquitin-proteasome pathway, we treated senescent cells with the proteasome inhibitor MG132, but failed to restore PAICS protein levels (Fig. 6a and Supplementary Fig. 7a). However, treatment with Bafilomycin A1 (BafA1), an inhibitor of both macroautophagy and chaperone-mediated autophagy (CMA), markedly restored PAICS protein levels (Fig. 6b and Supplementary Fig. 7b, c), indicating that PAICS is degraded through autophagy. Because macroautophagy is highly activated in senescence and mediates the massive degradation of senescence regulators57,58,59, we further ascertained the type of autophagy for PAICS degradation. By knocking down macroautophagy protein ATG7 or CMA receptor LAMP2, we observed that ATG7 but not LAMP2 knockdown restored PAICS protein levels (Fig. 6c and Supplementary Fig. 7d), suggesting that PAICS is degraded by macroautophagy. To further verify this, we investigated the interaction between PAICS and microtubule-associated protein 1 A/1B-light chain 3 (LC3), an essential step for macroautophagy. Although input PAICS protein levels from senescent cells were much lower than those of proliferating cells, LC3-immunoprecipitated PAICS levels were comparable between the two groups, suggesting the enhanced LC3-PAICS interaction in senescent cells (Fig. 6d-f). Notably, the PAICS acetylation mutant (K36RK47R) exhibited much weaker interaction with LC3 than the WT counterpart, consistent with the increased protein stability of these mutants (Fig. 6g–i and Supplementary Fig. 6e).
a, b ER:RAS-expressing cells were induced to senesce by adding 100 nM 4-OHT. At day 8, senescent cells were treated with or without MG132 (0.1 μM) (a) or BafA1 (30 nM) for 48 h (b); the cells were harvested and analysed for expression of the indicated proteins by immunoblot. c OIS cells expressing non-targeting shRNA or ATG7 targeted shRNA were harvested for expression of the indicated proteins by immunoblot. d–f Co-immunoprecipitation analysis of endogenous PAICS and LC3 was performed in proliferating and OIS cells (d), and the PAICS or LC3 IP bands were normalized to the LC3 or PAICS IP and the input bands, respectively (e, f). g–i HA-tagged WT or mutant PAICS and FLAG-tagged LC3 were co-transfected into HEK293T cells and analysed by co-immunoprecipitation (g), the HA or FLAG IP bands were normalized to the FLAG or HA IP and the input bands respectively (h, i). j–l HA-tagged WT PAICS and FLAG-tagged WT or mutant LC3 were co-transfected into HEK293T cells, and analysed by co-immunoprecipitation (j), the HA or FLAG IP bands were normalized to the FLAG or HA IP and the input bands respectively (k, l). m Schematic of the three LIR motifs in PAICS protein. n–p HA-tagged WT or LIR-motif-mutant PAICS and FLAG-tagged LC3 were co-transfected into HEK293T and analysed by co-immunoprecipitation (n), the HA or FLAG IP bands were normalized to the FLAG or HA IP and the input bands respectively (o, p). q, r HA-tagged WT or LIR-motif-mutant PAICS was transfected into HEK293T. At 48 h post-transfection, cells were treated with CHX (100 μg/mL) for 0, 6, or 12 h, and the expression of the indicated proteins was analysed by immunoblot (q) and quantified (r). Data represent mean ± s.d. of three biologically independent experiments. The P values were calculated using an unpaired two-tailed Student’s t-test (e, f, h, i) or One-way ANOVA (Tukey’s multiple-comparison test) (k, l, o, p). Representative of n = 3 independent experiments were shown (a–d, g, j, n, q). Source data are provided as a Source Data file.
Thus, we further dissected the amino acid residues responsible for the LC3–PAICS interaction. As reported, F52 and L53, as well as R70, residues on the hydrophobic surface of LC3, are required for the binding of LC3 to its interacting proteins60. We disrupted these residues on LC3 and found that both F52AL53A and R70A substitution exhibited impaired interaction with PAICS, suggesting that they are essential residues for LC3 to recognize PAICS (Fig. 6j–l).
We subsequently mapped the LC3-interacting domain in PAICS. The autophagy substrates bind to LC3 through a core consensus sequence known as the LC3-interacting region (LIR) motif, which typically comprises acidic residues (E/D) followed by aromatic and hydrophobic residues (W/F/Y)XX(L/I/V)61. We identified three LIR-like motifs in different regions of PAICS (Fig. 6m). We then substituted individual aromatic and hydrophobic residues of each LIR-like motif and performed co-IP assays to compare the interactions between LC3 and PAICS substitutions (Fig. 6n–p). All three PAICS double substitutions (Y22AL25A, Y231AL234A, or W363AL366A) displayed impaired interaction with LC3. Consistently, PAICS double substitutions exhibited higher protein stability than WT protein in the CHX chase assay (Fig. 6q, r). These results collectively support that PAICS-LC3 interaction contributes to macroautophagy-mediated PAICS degradation during senescence.
To further dissect whether these sites, which we have identified to be essential for the protein stability of PAICS, have any effect on the enzyme activity of PAICS and DNPB pathway, we knocked down endogenous PAICS, re-introduced HA-tagged and shRNA-resistant wildtype or mutant PAICS into the senescent cells. Immunoblot results revealed that mutant PAICS display higher protein levels than the WT counterpart, probably due to the relatively higher protein stability of these mutants (Supplementary Fig. 7e, f). 15N-Glutamine tracing data revealed that the flux into purine intermediates is significantly abolished by PAICS knockdown, but can be recovered by WT and mutant PAICS (Supplementary Fig. 7g). Notably, senescent cells rescued with mutant PAICS displayed even higher activity of DNPB compared to the WT counterpart (Supplementary Fig. 7g). In addition, both WT and mutant PAICS dramatically rescued the CCF formation and SASP gene expression (Supplementary Fig. 7h, i). To further distinguish whether the higher activity of DNPB of these cells expressing mutant PAICS is due to higher protein levels or the enzyme activity of PAICS, we compared the enzymatic activity of WT and the series of mutant PAICS. The in vitro enzyme assay by using an equal amount of PAICS displayed no significant difference in the enzymatic activity among WT and mutant PAICS, indicating that these amino acid residues are dispensable for the catalytic activity of PAICS (Supplementary Fig. 7j). Altogether, these results confirm that these K36, Y22, Y231, W363 amino acids are essential for the protein stability and thus the functionality of PAICS.
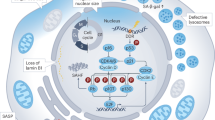
Based on these, we propose a model in which ACSS2 directly binds to PAICS and facilitates acetylation-dependent PAICS-LC3 interaction required for macroautophagy-mediated PAICS degradation in senescent cells. Consequently, PAICS deficiency limits purine biosynthesis for DNA repair, which in turn aggravates DNA damage and CCF formation, functionally triggering SASP (Supplementary Fig. 7k).
Targeting ACSS2 blunts SASP and immune surveillance in vivo
SASP promotes the growth of tumor cells both in vitro and in an immunocompromised mouse model4. To validate the role of ACSS2 in SASP regulation and the biological impacts, we treated an ovarian cancer line with conditioned media (CM) collected from OIS cells with or without ACSS2 knockdown. While control senescent cell-derived CM dramatically promoted cancer cell growth, such effect was significantly diminished in CM from ACSS2-deficient senescent cells (Supplementary Fig. 8a). Consistent with this, the tumor growth-stimulating effects of co-injected senescent cells were markedly compromised by ACSS2 knockdown in the xenograft model (Fig. 7a, b). Collectively, these results suggest that ACSS2 deficiency dampens the pro-tumorigenic function of senescent cells.
a, b TOV21G cells were co-injected with proliferating or senescent IMR90 cells into NCG mice. The tumor volume was measured (a) and the tumor weight was measured, (b). n = 5 mice per group. c A scheme of the experimental procedure. d–f One week after ionizing irradiation, Livers from vehicle or ACSS2i treated mice were analysed by immunohistochemistry for γH2AX (d), the mean values of γH2AX positive cells from nine randomly imaged fields were quantified per mouse (e), and SASP mRNA expression was analysed (f). Scale bar = 20 μm. n = 5 for non-IR group, n = 7 per IR group. g–j 18-month-old mice were administered with vehicle or ACSS2i for one week (g). Livers were harvested and analyzed by immunohistochemistry for γH2AX (h), and the mean values of γH2AX positive cells from ten randomly imaged fields were quantified per mouse (i) and SASP mRNA expression was analysed (j). Scale bar = 20 μm. n = 5 for the young or vehicle-treated old mice group, n = 7 for the treated old mice group. k Schematic model of the constructs and the experimental design. l–n Representative images of SA-β-gal staining of the liver tissues at day 7 and 14 post-injection (l), and quantified (m, n). n = 6 each group. o, p Representative images of CD45 immunofluorescence staining and NRas-dsRed at day 7 post-injection (o), and the mean value of the double positive clusters from five randomly imaged fields per mouse were calculated (p). Scale bar = 100 μm. q, r Representative images of CD45 IHC staining of the liver tissues at day 7 and 14 post-injection (q). Arrows point to examples of immune clusters. CD45+ cells in the indicated groups at day 7 post-injection were quantified (r). Scale bar = 100 μm. n = 6 each group. Data represent the mean ± s.d. The P values were calculated using One-way ANOVA (Tukey’s multiple-comparison test) (a, b, e, f, i, j) or an unpaired two-tailed Student’s t-test (m, n, p, r). Source data are provided as a Source Data file.
It has been shown that sublethal ionizing irradiation (IR) induces senescence, CCF formation, and the SASP in mouse liver21,29 (Supplementary Fig. 8b, c). We then assessed whether ACSS2-controlled SASP could be targeted through a pharmacologic strategy in IR-induced senescence in vivo. We applied a small molecule inhibitor (ACSS2i) that shows antitumour efficacy and safety in preclinical breast cancer models62. C57BL/6 mice were administered vehicle or ACSS2i for one week post-IR, liver samples were harvested and analysed by immunohistochemical (IHC) staining and RT-qPCR. IR resulted in the accumulation of DNA damage marker γH2AX and upregulated SASP components in vehicle hepatocytes (Fig. 7d–f). However, IR-exposed and ACSS2i-treated liver sections exhibited fewer cells bearing γH2AX and SASP gene expression (Fig. 7d–f).
We further confirmed that ACSS2 maintains functional SASP in vivo by exposing wild-type (WT) and Acss2-null (null) mice to IR (Supplementary Fig. 8d, e). While IR promoted γH2AX and IL1α accumulation, CCF formation, and SASP gene expression in WT mouse livers, these phenotypes were remarkably abrogated in the null mice (Supplementary Fig. 8f–k). The colocalization of IL1α and hepatocyte marker Albumin proved that the expression of IL1α is caused by the senescence of hepatocytes rather than other cell types (Supplementary Fig. 8l, m). Interestingly, Paics protein levels increased in Acss2-null compared to WT livers post IR, while the mRNA levels remained comparable among groups (Supplementary Fig. 8n–p). Together, these results provide in vivo evidence for the negative regulation of Paics protein by Acss2, and Acss2 drives DNA damage-induced SASP in tissue ageing.
To explore whether ACSS2i can restrain natural ageing-related inflammation, we treated 18-month-old mice with vehicle or ACSS2i. We found that both senescence-associated γH2AX and the expression of inflammatory factors increased in the aged liver, while these features were significantly alleviated by ACSS2 inhibition (Fig. 7g–j).
Given that SASP plays a vital role in the immune surveillance of pre-malignant oncogene-induced senescent cells, we finally investigated the role of Acss2 in regulating SASP-mediated immune surveillance. Hydrodynamic tail-vein injection (HDTI) of a vector expressing transposase and transposon vector expressing DsRed-NRasG12V was performed and then treated with vehicle or ACSS2i (Fig. 7k). In this model, oncogenic NRasG12V acutely triggers senescence and SASP in hepatocytes at early time point, which activates immune surveillance for subsequent clearance of senescent hepatocytes at a later time point46,63. At day 7 post-injection, NRasG12V-induced hepatocyte senescence was detected comparably in both vehicle and ACSS2i treated livers, as revealed by SA-β-gal staining (Fig. 7l, m). As a control, the NRasG12VD38A mutant, which is incapable of inducing senescence, failed to trigger senescence or induce the immune cell clusters (Supplementary Fig. 9a, b). However, day 7 ACSS2i administrated livers exhibited less immune infiltration marked by diminished CD45+ immune cell clusters near NRas-expressing (DsRed+) cells compared to vehicle-treated mice (Fig. 7o–r). At day 14 post-injection, while most senescent cells had been eliminated in the vehicle-treated group, more SA-β-gal+ cells were still retained in ACSS2i-treated livers, indicating inefficient immune surveillance (Fig. 7n). Similar results were obtained in WT and Acss2-null mice post HDTI, in which the livers from Acss2-null mice exhibited less immune cell infiltration at day 7 and subsequently retained more senescent cells in the livers at day 14 (Supplementary Fig. 9c–i). Together, these findings support that Acss2 maintains the SASP-mediated immune surveillance in vivo, and targeting Acss2 might be a therapeutic option to counteract senescence-related inflammation.
Discussion
Although metabolic deregulation has been established as a notable feature of cellular senescence, the metabolic regulators and underlying mechanisms for SASP control remain largely unknown. Here, we show that the metabolic enzyme ACSS2 maintains SASP gene expression in multiple forms of senescence and dictates the tumor-promoting tissue inflammation as well as immune surveillance functions of senescent cells. ACSS2 is best known to regulate cell growth and gene expression through cell-intrinsic mechanisms primarily involving histone acetylation38,41. However, we have revealed ACSS2 functions by limiting nucleotide biosynthesis to maintain the DNA damage-induced CCF formation and SASP independently of histone acetylation. Specifically, ACSS2 interacts with purine biosynthetic enzyme PAICS and promotes autophagy-mediated PAICS degradation to limit purine biosynthesis in senescence. While ACSS2-PAICS interaction is also observed in proliferating cells, PAICS protein stability is significantly decreased in senescent cells, primarily due to highly active autophagy. Interestingly, ACSS2 promotes PAICS acetylation that facilitates PAICS-LC3 interaction, a prerequisite for macroautophagy, and ACSS2 enzyme activity is required for PAICS acetylation. We further defined PCAF as the lysine acetyltransferase for PAICS, but how exactly ACSS2 and PCAF collaborate to achieve the specific acetylation of PAICS needs to be addressed in future studies. Nonetheless, our results suggest that acetylation is one critical post-translational modification regulating PAICS protein stability. Notably, we observed that supplement of nucleosides but not purine-only could recover SASP, and ACSS2 inhibition also increases pyrimidine metabolites during cellular senescence. Whether and how ACSS2 may regulate pyrimidine metabolism warrants further investigation. A recent study has demonstrated that GOT1 promotes de novo pyrimidine biosynthesis by facilitating the formation of a multi-enzyme complex, the ‘pyrimidinosome’64. We assume an indirect regulation on GOT1 or other target genes by ACSS2 may mediate the upregulated pyrimidine synthesis by ACSS2 loss, but the underlying mechanisms remain to be dissected in detail.
It has been reported that ACSS2 inhibition using the small molecule inhibitor (ACSS2i) bolsters T cell activity and potentiates antitumour immunity, underscoring its role in shaping the immune microenvironment62. Using multiple in vitro and in vivo senescence and ageing models, we provide compelling evidence to show that genetic or pharmacological ACSS2 inhibition abrogates the impact of senescent cells on the tissue microenvironment. Notably, Acss2 deletion in mice does not cause significant defect, and these mice are viable and fertile, suggesting that targeting ACSS2 should be well-tolerated, and ACSS2 might be a potential senomorphic target.
In summary, we have identified ACSS2 as an indispensable metabolic regulator of SASP through a non-histone acetylation-dependent mechanism. We further provide a proof-of-principle for potential therapeutic intervention of ACSS2 to prevent ageing and senescence-related inflammatory disorders.
Methods
Cells and culture conditions
IMR90 primary human diploid lung embryonic fibroblasts were were purchased from ATCC (CCL-186) and cultured under low oxygen tension (2%) in DMEM medium supplemented with 10% fetal bovine serum (Sigma), L-glutamine, sodium pyruvate, non-essential amino acids, and sodium bicarbonate (all from Gibco). Human IMR90 ER:RAS cells were established and cultured as described previously43,65. The ovarian cancer TOV21G cell line was cultured in RPMI 1640 medium with 10% FBS and 1% penicillin–streptomycin under 5% CO2. 293T (ATCC, CRL-3216) were cultured in DMEM medium with 10% FBS and 1% penicillin–streptomycin under 5% CO2.
Reagents, plasmids, and antibodies
4-hydroxytamoxifen (4-OHT), EmbryoMax nucleosides (50x, ES-008-D), Etoposide (E1383) and MG132 (M8699) were purchased from Sigma. Adenosine (HY-B0228), Guanosine (HY-N0097), Methotrexate (HY-14519) and CAIR (HY-133154) were purchased from MedChemExpress. ACSS2 inhibitor (S8588), Cycloheximide (S7418), Bafilomycin A1 (S1413) and were purchased from Selleckchem. ACSS2 inhibitor VY-3-135 was synthesized from TopSCIENCE (Taoshu) and dissolved in 10% DMSO, 10% Solutol, and 80% PBS.
The scramble control shRNA (#1864), the viral packaging plasmids pMD2.G (#12259), and psPAX2 (#12260) were obtained from Addgene. FLAG-LC3 plasmid (P36332) and pLV3-CMV-cGAS(human)-3×FLAG-Puro plasmid (P64508) were obtained from MiaoLing Biology. The following shRNAs were used: ACSS2 (#TRCN0000045566, #TRCN0000045567), ACLY (#TRCN0000078283, # TRCN0000078284), PAICS (#TRCN0000045774, # TRCN0000045775), AK1 (#TRCN0000006018, #TRCN0000006021), ATG7 (#TRCN0000007586, #TRCN0000007587), LAMP2 (#TRCN0000029260, #TRCN0000029262), PCAF (#TRCN0000018528, #TRCN0000018529), PRPS1 (#TRCN0000010123, # TRCN0000010125).
HA-tagged PAICS and V5-tagged ACSS2 were subcloned into the pCDH vector. Site-specific mutation of HA-tagged PAICS and FLAG-tagged LC3 was generated following ClonExpress II One Step Cloning Kit (Vazyme), all the primers corresponding to the mutation sites were listed in the Supplementary Data 1. Turbo biotin ligase was amplified from pcDNA3-3xHA-TurboID (Addgene, #107171) and subcloned into the pCDH vector, then ACSS2 or PAICS were subcloned into the pCDH-TurboID. All primers are listed in the Supplementary Data 1.
The following antibodies were purchased from the indicated suppliers. For immunoprecipitation and western blots: anti-ACSS2 (Boster, cat. no. BM4830; 2 μg per immunoprecipitation, Cell Signaling, cat. no. 3658; 1:1000 for western blots), anti-ACLY (Cell Signaling, cat. no. 4332; 1:1000 for western blots), anti-LC3 (Cell Signaling, cat. no. 3868; 2 μg per immunoprecipitation, 1:1000 for western blots), anti-PAICS (Bethyl, cat. no. A304-546A; 2 μg per immunoprecipitation, Proteintech, cat. no. 12967-1-AP; 1:1000 for western blots), anti-ATG7 (Abmart, cat. no. T57051S; 1:1000 for western blots), anti-LAMP2 (Abcam, cat. no. ab125068; 1:10,000 for western blots), anti-p16 (Santa Cruz Biotechnology, cat. no. sc-56330; 1:1000 for western blots), anti-RAS (Becton Dickinson, cat. no. 610001; 1:1000 for western blots), anti-p21 (Abcam, cat. no. 7960; 1:1000 for western blots), anti-acetylated-Lysine (Cell Signaling, cat. no. 9441; 1:1000 for western blots), anti-PCAF (Abmart, cat. no. T58354S; 2 μg per immunoprecipitation, 1:1000 for western blots). For immunohistochemistry (IHC) and immunofluorescence (IF): anti-IL1α (R&D Systems, cat. no. AF-400; 1:150 for IHC), anti-γH2AX (Cell Signaling, cat. no. 9718; 1:500 for IHC, 1:200 for IF), anti-CD45 (Cell Signaling, cat. no. 70257; 1:100 for IHC and IF), anti-FLAG (Proteintech, cat. no. 66008; 1:500 for IF).
Immunohistochemistry and immunofluorescence
Fresh mouse liver tissues were fixed overnight in 4% PFA in PBS at 4 °C or embedded in OCT for cryosection. For immunohistochemistry staining, the slides were deparaffinized, rehydrated, quenched in 0.6% hydrogen peroxide in methanol for 15 minutes, and boiled for 20 minutes in 10 mM citrate (pH 6.0) buffer for antigen retrieval. The slides were incubated with blocking buffer (5% serum, 1% BSA, and 0.5% Tween 20 in PBS) for 1 h at room temperature and then incubated overnight with primary antibodies diluted in blocking buffer at 4 °C. The slides were further incubated with biotinylated secondary antibodies, followed by ABC solution, and developed with 3,3′-diaminobenzidine (Vector Laboratories). The slides were finally counterstained with hematoxylin, dehydrated, and mounted with Permount (Thermo Fisher). For quantification, all slides were scanned using Evident Olympus VS200, and nine or ten different fields per mouse were counted. IMR90 cells or liver tissues were fixed with 4% paraformaldehyde (PFA) for 30 minutes at room temperature, followed by permeabilization with 0.2% Triton X-100 in PBS for 5 minutes. After blocking with 3% BSA in PBS, the cells were incubated overnight with primary antibody at 4 °C and Alexa Fluor-conjugated secondary antibody (Life Technologies) for 1 h at RT, followed by DAPI staining. The slides were visualized on an OLYMPUS IX83 microscope.
Immunoprecipitation and immunoblot
For immunoprecipitation, cells were lysed with buffer (150 mM NaCl, 1% NP40, 20% Glycerol, 1 mM EDTA, 50 mM Tris-HCl pH 8.0, 1 mM PMSF, EDTA-free protease inhibitor cocktail). Incubate 1-2 mg of cell lysates with anti-LC3, anti-PAICS, anti-ACSS2, anti-PCAF, anti-HA, anti-V5 or the corresponding IgG controls overnight at 4 °C. Protein A/G beads (Thermo Fisher) were incubated with proteins for 1.5 h at 4 °C. After three washes (150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, 50 mM Tris-HCl pH 7.5, 1 mM PMSF, EDTA-free protease inhibitor cocktail), beads were boiled in 1X sample buffer (10% glycerol, 2% SDS, 0.01% bromophenol blue, 0.1 M dithiothreitol and 62.5 mM Tris-HCl pH 6.8) and analysed by immunoblot. Protein was isolated and separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane (Millipore). Following blocking and incubation with the primary and secondary antibodies, membranes were imaged by Tanon Imager 4200. To detect immunoprecipitated PAICS, use the secondary antibody (M21008, Abmart) to avoid heavy/light chain interference or regular secondary antibody (Cell Signaling Technology).
Senescence induction, SA-β-gal staining, and colony formation
For ER:RAS senescence induction, IMR90 cells were infected with retrovirus encoding a 4-hydroxytamoxifen-inducible ER:RAS construct (pLNC-ER:Ras), following two weeks of selection with G418, cells were treated with 4-hydroxytamoxifen at a final concentration of 100 nM. For etoposide-induced senescence, IMR90 cells at 60–70% confluency were treated with 100 μM etoposide for 48 h, washed with PBS, and then cultured in normal medium. For irradiation-induced senescence, IMR90 cells at 40–50% confluency were irradiated with 10 Gy and recovered overnight29. Cells were fixed in 2% formaldehyde and 0.2% glutaraldehyde and then incubated overnight in solution (150 mM NaCl, 40 mM Na2HPO4 (pH 6.0), 2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6 and 1 mg mL−1 X-gal) at 37 °C in a non-CO2 incubator. For mouse liver tissue, frozen sections were fixed and stained at pH 5.5 for 5–8 h, as previously described66. Cells were seeded in six-well plates at a density of 3,000 cells per well and cultured for ten days before staining with 0.05% crystal violet for visualization. Analysis was performed based on integrated density using the NIH ImageJ software (v1.48).
Reverse-transcriptase qPCR
Total RNA was harvested using TRIzol (Invitrogen) and isolated using an RNeasy mini kit (Qiagen, 74106). The extracted RNA was used for reverse-transcriptase PCR using a High-capacity cDNA reverse transcription kit (Thermo Fisher). Quantitative PCR was performed using a QuantStudio 5 real-time PCR system. The primers used for RT-qPCR are listed in Supplementary Data 1.
Antibody array analysis
The antibody array for secreted factors was performed using a Human cytokine antibody array C3 (RayBiotech, cat. no. AAHCYT34). As previously described46, conditioned medium was generated from cells incubating in serum-free DMEM for 48 h. The conditioned medium was filtered (0.2 μm) and incubated overnight on the array at 4 °C. Following the manufacturer’s instructions, the signals were measured and quantified using the NIH ImageJ software (v1.48), followed by normalization to the cell number from which the conditioned medium was generated.
Expression and purification of proteins from E. coli
Human ACSS2 and PAICS were cloned into pET15b, and transformed into BL21 (DE3) competent cells. Protein expression was induced by 0.2 mM IPTG (0.2 mM, OD600 = 0.8) for 16 h at 16 °C. The bacteria were harvested in the buffer containing 25 mM Tris (pH 8.0) and 150 mM NaCl and lysed using sonication. After centrifugation at 12000 g for 60 minutes, the supernatant was collected and loaded onto Ni-NTA affinity resin (Qiagen) and rinsed with the buffer containing 25 mM Tris (pH 8.0), 150 mM NaCl, and 25 mM imidazole. The protein was eluted with a buffer containing 25 mM HEPES Tris (pH 8.0), 150 mM NaCl, and 250 mM imidazole. Purified protein was confirmed with Coomassie blue staining.
Measurement of dNTP concentration
The intracellular dNTP was extracted as described47. Briefly, 1 × 106 cells were centrifuged for 3 minutes at 3,000 × g at 4 °C. Cell pellets were resuspended in 500 μL of ice-cold 60% methanol, vortexed, and boiled at 97 °C for 3 minutes. After sonication, the extracts were centrifuged, and the supernatants were passed through an Amicon ultra-0.5 mL filter (3 kDa). The filtrate was dried with RapidVap (Labconco) and dissolved in nuclease-free water. The extracts were measured by fluorescence-based assay as previously described47.
Cycloheximide (CHX) chase assay
Cells were treated with 100 µg/mL CHX for 0, 6, and 12 h, and harvested for immunoblots.
PAICS acetylation and LC-MS/MS analysis
ER:RAS IMR90 was induced to senesce by 4-OHT. On day 10, senescent cells were infected with HA-PAICS encoding lentivirus and collected for lysis with RIPA buffer after 48 h of infection. A sample of 5 mg of lysate was incubated with 100 µL of HA-coated beads at 4 °C overnight. The beads were washed with RIPA buffer five times and eluted. Samples were submitted to the proteomics facility (School of Life Sciences) at Fudan University for LC-MS analysis. LC-MS analysis was performed using a nanoflow EASYnLC 1200 system (Thermo Fisher Scientific, Odense, Denmark) coupled to an Orbitrap Exploris480 mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). A one-column system was adopted for all analyses. Samples were analysed on a home-made C18 analytical column (75 µm i.d. × 25 cm, ReproSil-Pur 120 C18-AQ, 1.9 µm (Dr. Maisch GmbH, Germany)67. The mobile phases consisted of Solution A (0.1% formic acid) and Solution B (0.1% formic acid in 80% ACN). The peptides for LFQ analysis were eluted using the following gradients: 5-8% B in 3 minutes, 8-44% B in 100 minutes, 44-70% B in 5 minutes 70-100% B in 2 minutes, 100% B for 10 minutes, at a flow rate of 200 nL/min. High-field asymmetric-waveform ion mobility spectrometry (FAIMS) was enabled during data acquisition with compensation voltages set as −40 and −60 V. MS1 data were collected in the Orbitrap (60,000 resolution). Charge states between 2 and 7 were required for MS2 analysis, and a 45 s dynamic exclusion window was used. Cycle time was set at 1.5 s. MS2 scans were performed in the Orbitrap with HCD fragmentation (isolation window 1.6; 15,000 resolution; NCE 30%, max injection time 30 ms). The data were processed with UniProt human protein database (75,004 entries, download in 20200701) and Protein Discoverer (v2.4, Thermo Fisher Scientific) with Mascot (v2.7.0, Matrix Science). The mass tolerances were 10 ppm for precursor and fragment Mass Tolerance 0.05 Da. Up to two missed cleavages were allowed. The carbamidomethylation on cysteine is a fixed modification, and acetylation on the protein N-terminal and oxidation on methionine are variable modifications.
Proximity labeling by TurboID
TurboID-based proximity labeling assay was performed as described68. Specifically, OIS cells were infected with ACSS2-Turbo or Turbo-only lentivirus for 48 h, and then incubated with 500 μM biotin (Sigma B4501) at 37 °C for 30 minutes. The cells were collected and lysed with RIPA buffer. A sample of 1-2 mg of lysate was incubated with 100 μL of streptavidin-coated beads (Thermo Fisher, cat. no. 88816) at 4 °C overnight. Streptavidin beads were washed twice with 1% SDS and BC500 (50 mM Tris-HCl pH 7.6, 2 mM EDTA, 500 mM KCl), and once with BC100, BC100 containing 2 M urea. Biotinylated proteins were eluted and sent to the proteomics facility (School of Life Sciences) at Fudan University for LC-MS analysis. Significant hits were defined as proteins detected by Log2FC (TurboID-ACSS2 or PAICS/control TurboID group) > 1, P ≤ 0.05. Differential analysis was performed in R (v4.2.2) using the packages ggplot2 package (v3.4.4) (https://ggplot2.tidyverse.org/).
Targeted metabolomics
To extract metabolites, proliferating and different groups of senescent cells were directly chilled into 2 mL of ice-cold 80% HPLC grade methanol containing 0.1% formic acid, followed by centrifugation at 10,000 g for 20 minutes at 4 °C. The supernatant was collected and dried with RapidVap (Labconco). The extract was dissolved in 20% acetonitrile, and the insoluble material was removed by centrifugation. The samples were sent to the metabolomics facility (Institute of Metabolism and Integrative Biology) of Fudan University for LC-MS/MS analysis. The analysis was performed using AB SCIEX ExionLC AD interfaced with a QTRAP 6500+ system (AB SCIEX). Metabolites were separated through iHILIC-(P) Classic HPLC HILIC column (150 × 2.1 mm with 5 μm particle size). Mobile phases consisted of 5% acetonitrile,20 mM ammonium acetate in 10 mM ammonia water (mobile phase A), and acetonitrile (mobile phase B) and were run at a flow rate of 0.2 mL/min. The gradient was as follows: 0 minute, 85% B; 2 minutes, 85% B; 7 minutes, 60% B; 12 minutes, 35% B; 12.1 minutes, 20% B; 15.9 minutes, 20% B; 16 minutes, 85% B; 23 minutes, 85% B. The mass spectrometer was run in multiple reactions monitoring (MRM) mode. The ESI source parameters: source temperature of 500 °C, the ion source gas 1 and 2 at 60 psi, the curtain gas at 35 psi, and the ion spray voltage at 5500 V or -4500 V for positive or negative modes. Data were processed using SCIEX OS software and were normalized with packed cell volume (relative to proliferating cells). Significantly changed metabolites (FC ≥ 1.5) were determined and visualized in R v4.2.2 using the pheatmap package (v1.0.12, https://cran.r-project.org/web/packages/pheatmap/index.html).
15N-Glutamine labeling and flux analysis
Cells were incubated in DMEM medium containing 10% dialyzed FBS and 15N-Glutamine for 24 h. Cells were extracted using buffer (acetonitrile, methanol, and water in a ratio of 40:40:20). The extracts were processed and measured by LC-MS. For data analysis, the area of 14N and various 15N isotopologue peaks were integrated using El-MAVEN software (v0.12.1). Correction for natural isotope abundance was performed using the R package AccuCor69. The labeling fraction of a specific isotopologue was calculated as the area of that isotopologue divided by the sum of all isotopologues’ area. The spectral peaks of the metabolites are included in Supplementary Data 2.
ChIP-sequencing and analysis
Briefly, cells were fixed with 1% formaldehyde (Sigma, cat. no. F8775) for 10 minutes at RT, quenched with 2.5 M glycine, washed twice with cold PBS, and then lysed and sheared using Bioruptor (Diagenode). The lysate was incubated overnight with 8 μg H3K27ac antibody, 1 μg Spike-in Antibody (Active Motif, Cat. No. 61686) and 1 μg Spike-in Chromatin (Active Motif, Cat. No. 53083) or IgG at 4 °C. After incubation with protein A/G Dynabeads (Thermo Fisher) for 2 h at 4 °C, the ChIP samples were washed and the DNA was eluted and incubated overnight at 65 °C to reverse the cross-linking. The next day, the samples were digested with proteinase K and RNase A. Quantitative PCR was performed on the immunoprecipitated DNA using the primers listed in Supplementary Data 1. For ChIP-seq, 10 ng ChIP or input DNA was used for the library construction using an NEBNext ultra DNA library prep kit (NEB, E7645). The qualified libraries were pooled and sequenced on Illumina platforms with PE150 in Novogene Bioinformatics Technology Co., Ltd (Beijing, China). For ChIP-seq analysis, FastQC (v0.12.1) was used for quality control operations on raw sequencing data. The clean reads mapping of the reference genome of humans (HG38) and Drosophila (BDGP6) was achieved through the use of default parameters in bowtie2 (v2.4.4). Mapping reads that were uniquely paired at least once were retained. Sambamba (v1.0.1) was used to remove duplicate reads and obtain the final number of tags without duplicate reads. The Drosophila sample with the lowest tag count was compared with the tag counts of other Drosophila samples to obtain the normalization factor. Using the normalization factor of each sample obtained by the aforementioned calculation method for downsampling, the final BAM files were obtained. The bamCoverage tool in DeepTools (v3.5.5) converts BAM files into bigWig files. Use the BigWigCompare (v3.5.5) tool to calculate the ratio between each group of samples and their input samples. Use macs2 (v2.2.7.1) to call the peaks, with the significance q value ≤ 0.05. The merge command within bedtools (v2.30.0) was used to merge the overlapping peaks of the merged sample. According to the bigWig files, the programs “computeMatrix,” “plotProfile,” and “plotHeatmap” within DeepTools (v3.5.5) were used to compute ChIP-seq signals and visualize them. The ChIPseeker package (v1.28.3) was used for annotating and visualizing peaks of interest.
Enzyme activity assay
PAICS proteins were immunoprecipitated from senescent cells expressing HA-tagged WT or mutant PAICS using HA antibody. As previously reported70, equal amounts of PAICS protein were incubated with 20 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT, 50 mM sodium bicarbonate, 0.30 mM CAIR, 1 mM L-aspartate, 0.25 mM ATP. ADP generated from the reaction was quantified by ADP Assay Kit (ab83359, Abcam) according to the manufacturer’s instructions. The luminescence was measured, and ADP concentrations were calculated using an ADP standard curve.
In vivo mouse models
All animal protocols were approved by the Institutional Animal Care and Use Committee at Fudan University. The maximal tumor size permitted by the ethics committee is 2000 mm3 and the maximal tumor size in this study was not exceeded. Acss2+/+ and Acss2−/− mice in C57BL/6 background were purchased from GemPharmatech Co., Ltd (Nanjing, China), and 18-month-old mice were purchased from Wukong Biotech Co., Ltd (Shandong, China). All mice were maintained at 22–23 °C with 40–60% humidity and a 12-hour light–12-hour dark cycle in an SPF animal facility at Fudan University.
For xenograft mouse models, 1 × 106 TOV21G cells with 2 × 106 IMR90 cells mixed with Matrigel at a 1:1 volume ratio were co-injected subcutaneously into 6-8 week NCG mice. Tumor size was measured with a caliper and calculated using the formula: tumor size (mm3) = (d2 × D) / 2, where d and D are the shortest and the longest diameters, respectively. For ionizing radiation, mice were subjected to a sub-lethal dose of 4 Gy irradiation, and were harvested seven days post irradiation. For hydrodynamic tail-veil injection, 8-9 week mice were used. Endotoxin-free transposon-based construct expressing NRas (25 μg) together with transposase plasmid (5 μg) in saline at a volume of 10% of the mouse body weight were mixed and delivered into the mice within 5–8 s as previously described46. ACSS2i VY-3-135 was administered i.p. at 100 mg/kg daily as previously published62.
Statistical analysis
Experiments were repeated at least three times unless otherwise stated. All bar graphs show mean values with error bars (s.d. as defined in figure legends). Generally, an unpaired two-tailed Student’s t-test was used to compare two groups, and an ANOVA analysis was used to compare more than two groups.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data described in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the identifiers PXD048937, PXD048965 and PXD54616. H3K27ac ChIP-seq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE275173. The RNA-seq data are accessible GSE274295. The raw metabolomics data is available in MetaboLights under the identifier MTBLS12215, MTBLS12216 and MTBLS12217. Source data are provided with this paper.
References
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: An expanding universe. Cell 186, 243–278 (2023).
Di Micco, R., Krizhanovsky, V., Baker, D. & d’Adda di Fagagna, F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 22, 75–95 (2021).
Birch, J. & Gil, J. Senescence and the SASP: many therapeutic avenues. Genes Dev. 34, 1565–1576 (2020).
Coppe, J. P., Desprez, P. Y., Krtolica, A. & Campisi, J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev. Pathol. 5, 99–118 (2010).
McHugh, D. & Gil, J. Senescence and aging: Causes, consequences, and therapeutic avenues. J. Cell Biol. 217, 65–77 (2018).
Hernandez-Segura, A., Nehme, J. & Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 28, 436–453 (2018).
Faget, D. V., Ren, Q. & Stewart, S. A. Unmasking senescence: context-dependent effects of SASP in cancer. Nat. Rev. Cancer 19, 439–453 (2019).
Li, F. et al. FBP1 loss disrupts liver metabolism and promotes tumorigenesis through a hepatic stellate cell senescence secretome. Nat. Cell Biol. 22, 728–739 (2020).
Schafer, M. J. et al. Cellular senescence mediates fibrotic pulmonary disease. Nat. Commun. 8, 14532 (2017).
Spinelli, R. et al. Increased cell senescence in human metabolic disorders. J. Clin. Invest. 133, e169922 (2023).
Niedernhofer, L. J. & Robbins, P. D. Senotherapeutics for healthy ageing. Nat. Rev. Drug Discov. 17, 377 (2018).
Amor, C. et al. Senolytic CAR T cells reverse senescence-associated pathologies. Nature 583, 127–132 (2020).
Chaib, S., Tchkonia, T. & Kirkland, J. L. Cellular senescence and senolytics: the path to the clinic. Nat. Med 28, 1556–1568 (2022).
Rodier, F. et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11, 973–979 (2009).
Seluanov, A., Mittelman, D., Pereira-Smith, O. M., Wilson, J. H. & Gorbunova, V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc. Natl Acad. Sci. USA 101, 7624–7629 (2004).
Mao, Z. et al. Sirtuin 6 (SIRT6) rescues the decline of homologous recombination repair during replicative senescence. Proc. Natl Acad. Sci. USA 109, 11800–11805 (2012).
Ivanov, A. et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 202, 129–143 (2013).
Miller, K. N. et al. Cytoplasmic DNA: sources, sensing, and role in aging and disease. Cell 184, 5506–5526 (2021).
Yang, H., Wang, H., Ren, J., Chen, Q. & Chen, Z. J. cGAS is essential for cellular senescence. Proc. Natl Acad. Sci. USA 114, E4612–E4620 (2017).
Gluck, S. et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19, 1061–1070 (2017).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017).
Victorelli, S. et al. Apoptotic stress causes mtDNA release during senescence and drives the SASP. Nature 622, 627–636 (2023).
Miwa, S., Kashyap, S., Chini, E. & von Zglinicki, T. Mitochondrial dysfunction in cell senescence and aging. J. Clin. Invest. 132, e158447 (2022).
Wiley, C. D. & Campisi, J. The metabolic roots of senescence: mechanisms and opportunities for intervention. Nat. Metab. 3, 1290–1301 (2021).
Moiseeva, O., Bourdeau, V., Roux, A., Deschenes-Simard, X. & Ferbeyre, G. Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol. Cell Biol. 29, 4495–4507 (2009).
Nacarelli, T. et al. NAD(+) metabolism governs the proinflammatory senescence-associated secretome. Nat. Cell Biol. 21, 397–407 (2019).
Kaplon, J. et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 498, 109–112 (2013).
Dou, X. et al. PDK4-dependent hypercatabolism and lactate production of senescent cells promotes cancer malignancy. Nat. Metab. 5, 1887–1910 (2023).
Vizioli, M. G. et al. Mitochondria-to-nucleus retrograde signaling drives formation of cytoplasmic chromatin and inflammation in senescence. Genes Dev. 34, 428–445 (2020).
Correia-Melo, C. et al. Mitochondria are required for pro-ageing features of the senescent phenotype. EMBO J. 35, 724–742 (2016).
Aird, K. M. et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 3, 1252–1265 (2013).
Zhou, W. et al. Purine metabolism regulates DNA repair and therapy resistance in glioblastoma. Nat. Commun. 11, 3811 (2020).
Liu, R. et al. Innate immune response orchestrates phosphoribosyl pyrophosphate synthetases to support DNA repair. Cell Metab. 33, 2076–2089.e2079 (2021).
Bester, A. C. et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145, 435–446 (2011).
Guertin, D. A. & Wellen, K. E. Acetyl-CoA metabolism in cancer. Nat. Rev. Cancer 23, 156–172 (2023).
Sivanand, S. et al. Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination. Mol. Cell 67, 252–265.e256 (2017).
Lee, J. H. et al. ATP-citrate lyase regulates cellular senescence via an AMPK- and p53-dependent pathway. FEBS J. 282, 361–371 (2015).
Gao, X. et al. Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat. Commun. 7, 11960 (2016).
Mews, P. et al. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546, 381–386 (2017).
Mews, P. et al. Alcohol metabolism contributes to brain histone acetylation. Nature 574, 717–721 (2019).
Li, X. et al. Nucleus-Translocated ACSS2 Promotes Gene Transcription for Lysosomal Biogenesis and Autophagy. Mol. Cell 66, 684–697.e689 (2017).
Chen, W. et al. The SESAME complex regulates cell senescence through the generation of acetyl-CoA. Nat. Metab. 3, 983–1000 (2021).
Innes, A. J. & Gil, J. IMR90 ER:RAS: A Cell Model of Oncogene-Induced Senescence. Methods Mol. Biol. 1896, 83–92 (2019).
Guerra, C. et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell 19, 728–739 (2011).
Tasdemir, N. et al. BRD4 Connects Enhancer Remodeling to Senescence Immune Surveillance. Cancer Discov. 6, 612–629 (2016).
Liu, P. et al. m(6)A-independent genome-wide METTL3 and METTL14 redistribution drives the senescence-associated secretory phenotype. Nat. Cell Biol. 23, 355–365 (2021).
Wilson, P. M. et al. A novel fluorescence-based assay for the rapid detection and quantification of cellular deoxyribonucleoside triphosphates. Nucleic Acids Res 39, e112 (2011).
He, J., Zou, L. N., Pareek, V. & Benkovic, S. J. Multienzyme interactions of the de novo purine biosynthetic protein PAICS facilitate purinosome formation and metabolic channeling. J. Biol. Chem. 298, 101853 (2022).
Pedley, A. M., Pareek, V. & Benkovic, S. J. The Purinosome: A Case Study for a Mammalian Metabolon. Annu Rev. Biochem 91, 89–106 (2022).
Chou, M. C. et al. PAICS ubiquitination recruits UBAP2 to trigger phase separation for purinosome assembly. Mol. Cell 83, 4123–4140.e4112 (2023).
Fairbanks, L. D. et al. Methotrexate inhibits the first committed step of purine biosynthesis in mitogen-stimulated human T-lymphocytes: a metabolic basis for efficacy in rheumatoid arthritis? Biochem J. 342, 143–152 (1999).
Moffatt, B. A. & Ashihara, H. Purine and pyrimidine nucleotide synthesis and metabolism. Arabidopsis Book 1, e0018 (2002).
Narita, T., Weinert, B. T. & Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20, 156–174 (2019).
Liu, L. et al. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell Biol. 19, 1202–1209 (1999).
Savoia, M. et al. P300/CBP-associated factor regulates transcription and function of isocitrate dehydrogenase 2 during muscle differentiation. FASEB J. 33, 4107–4123 (2019).
Lv, L. et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol. Cell 42, 719–730 (2011).
Dou, Z. et al. Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 (2015).
Narita, M. et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332, 966–970 (2011).
Hao, X. et al. ADAR1 downregulation by autophagy drives senescence independently of RNA editing by enhancing p16(INK4a) levels. Nat. Cell Biol. 24, 1202–1210 (2022).
Kraft, L. J., Nguyen, T. A., Vogel, S. S. & Kenworthy, A. K. Size, stoichiometry, and organization of soluble LC3-associated complexes. Autophagy 10, 861–877 (2014).
Birgisdottir, A. B., Lamark, T. & Johansen, T. The LIR motif - crucial for selective autophagy. J. Cell Sci. 126, 3237–3247 (2013).
Miller, K. D. et al. Acetate acts as a metabolic immunomodulator by bolstering T-cell effector function and potentiating antitumor immunity in breast cancer. Nat. Cancer 4, 1491–1507 (2023).
Kang, T. W. et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479, 547–551 (2011).
Yang, C. et al. De novo pyrimidine biosynthetic complexes support cancer cell proliferation and ferroptosis defence. Nat. Cell Biol. 25, 836–847 (2023).
Young, A. R. et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 23, 798–803 (2009).
Krizhanovsky, V. et al. Senescence of activated stellate cells limits liver fibrosis. Cell 134, 657–667 (2008).
Kovalchuk, S. I., Jensen, O. N. & Rogowska-Wrzesinska, A. FlashPack: Fast and Simple Preparation of Ultrahigh-performance Capillary Columns for LC-MS. Mol. Cell Proteom. 18, 383–390 (2019).
Cho, K. F. et al. Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat. Protoc. 15, 3971–3999 (2020).
Su, X., Lu, W. & Rabinowitz, J. D. Metabolite Spectral Accuracy on Orbitraps. Anal. Chem. 89, 5940–5948 (2017).
Shon, H. et al. Evidence Supporting Substrate Channeling between Domains of Human PAICS: A Time-Course Analysis of (13)C-Bicarbonate Incorporation. Biochemistry 61, 575–582 (2022).
Acknowledgements
We thank the members of Dr. Fuming Li’s laboratory for their helpful discussions and insights on the manuscript. We thank Dr. Qingqing Yan for discussing the TurboID proximity labeling experiment. We are grateful to the core facilities of proteomics (School of Life Sciences) and metabolomics (Institute of Metabolism and Integrative Biology) of Fudan University for technical assistance. This work was supported by the National Natural Science Foundation of China (32270798 to P. Liu), the National Key R&D Program of China (2023YFC3603300 to P. Liu), and the Shanghai Municipal Science and Technology Commission original exploration program (23ZR1479800 to P. Liu).
Author information
Authors and Affiliations
Contributions
L.Y., X.Y., W.M., J.X., and L.Z. performed the experiments and analysed data. P.L. and Y.L. designed the experiments. J.Y. and L.C. helped with metabolomics and data analysis. R.J. and Y.Y. analysed the ChIP-seq data. Y.Z. and R.R. purified the human PAICS protein. M.T. and D.M. contributed to the enzyme activity assay. F.L. contributed to the study design. P.L. and Y.L. wrote the manuscript. P.L. supervised and conceived the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jesus Gil and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, L., You, J., Yang, X. et al. ACSS2 drives senescence-associated secretory phenotype by limiting purine biosynthesis through PAICS acetylation. Nat Commun 16, 2071 (2025). https://doi.org/10.1038/s41467-025-57334-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57334-3
This article is cited by
-
SLC6A6 imports taurine into mitochondria to sustain mitochondrial translation and tumour growth
Nature Metabolism (2026)
-
Cellular senescence in cancer: from mechanism paradoxes to precision therapeutics
Molecular Cancer (2025)
-
α-Ketoglutarate dictates AMPK protein synthesis for energy sensing in human cancers
Nature Chemical Biology (2025)