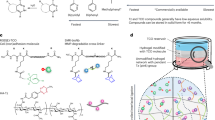
Abstract
Expansion microscopy enhances the microscopy resolution by physically expanding biological specimens and improves the visualization of structural and molecular details. Numerous expansion microscopy techniques and labeling methods have been developed over the past decade to cater to specific research needs. Nonetheless, a shared limitation among current protocols is the extensive sample processing time, particularly for challenging-to-expand biological specimens (e.g., formalin-fixed paraffin-embedded (FFPE) sections and large three-dimensional specimens). Here we present BOOST, a rapid and robust expansion microscopy workflow that leverages a series of microwave-accelerated expansion microscopy chemistry. Specifically, BOOST enables a single-step 10-fold expansion of cultured cells, tissue sections, and even the challenging-to-expand FFPE sections under 90 minutes. Notably, BOOST pioneers a 10-fold expansion of large millimeter-sized three-dimensional specimens, previously unattainable to the best of our knowledge. The workflow is also easily adaptable based on stable and common reagents, thus boosting the potential adoption of expansion microscopy for applications.
Similar content being viewed by others
Introduction
The need for better imaging methods is nearly universal for biological research. As the cornerstone of understanding biological processes, microscopy techniques have played a pivotal role in numerous scientific breakthroughs and discoveries. A recent addition is expansion microscopy (ExM) introduced by Boyden’s lab in 20151, enabling high-resolution imaging of biological samples by physically expanding the specimens, thereby overcoming the diffraction-limited resolution barrier of conventional light microscopy. Over the decade, a myriad of ExM techniques have been developed to cater to specific research aims. Recent advancements included achieving ~ 20 nm resolution with iterative ultrastructure expansion microscopy2, ~ 25 nm resolution with a single linear expansion3,4,5, < 20 nm resolution achieved by a recent single linear 20-fold expansion protocol6, and revelation of morphological details utilizing DNA and proteomic labeling dyes7,8. When coupled with fluorescence fluctuation analysis, ExM revealed ultrastructural details of biological samples with higher-resolution4,9 and even achieved the visualization of single proteins with an astonishing 1 nm resolution by recent ONE microscopy10. The utilization of ExM techniques also revealed previously unknown assembly and organization of biological nanostructures11,12.
Nevertheless, the integration of ExM on a wider scale has been constrained by the time-consuming nature of the sample preparation process. This is particularly true for challenging-to-expand biological specimens such as formalin-fixed paraffin-embedded (FFPE) sections, with the latest protease-free protocol taking around three days to expand the FFPE samples into tenfold for imaging4, which precludes potential time-sensitive applications such as nanoscale pathology requiring same day diagnosis. In addition, despite progress achieved in expanding large biological samples up to fourfold their original size over extended periods (often spanning days)13,14, the absence of protocols enabling higher fold expansion for large specimens remains a critical limitation, rendering unlocking hidden dimensions within large biological specimens an elusive goal. Therefore, in order to facilitate the widespread implementation of ExM, the development of a method that is rapid, robust, and compatible with a wide range of specimens is of paramount importance.
In the context of developing microscopic methods, microwave (MW) irradiation has emerged as a valuable technique for expediting sample processing. This approach speeds up both electron microscopy and optical microscopy sample preparation15,16,17,18,19,20. Notably, Mayers pioneered the use of microwaves in histopathology during the late 1970s, successfully fixing tissues21. Subsequently, researchers explored microwave irradiation for various biological sample processing procedures, including fixation, histochemical staining, and immunohistology20. Particularly, microwaves have gained recognition for antigen retrieval in paraffin-embedded tissue sections20. Brief exposure to microwave irradiation considerably expedited these procedures, yielding more consistent results with enhanced histochemical and immunohistological staining15. The underlying mechanisms of microwave action have been suggested to involve both generated heat and primary molecular kinetics, which contribute to the acceleration of chemical diffusion and subsequent reactions22.
We reasoned that the primary processes of ExM, i.e., monomer infiltration, gelation, anchoring, and denaturation, could be compatible with microwave irradiation if properly designed chemistry is used as shown in other sample processing methods20,23. Also, gelation of the hydrogel monomers (e.g., acrylamide) is essentially a free radical reaction known to be favored by microwave irradiation24. Hence, it was deemed prudent to explore the potential of integrating microwave irradiation and developing microwave-compatible chemistry to expedite the traditionally time-consuming ExM. In this study, we opted for a microwave tissue processor with features of automated temperature control and programable parameters to avoid potential unevenness in processing25.
Here, we developed a rapid and robust expansion microscopy workflow, BOOST, which aims to boost capability and broaden implementation in various applications. In brief, we developed microwave-compatible ExM chemistry, including anchoring chemistry, gel formulation, denaturation solutions, and labeling, together with optimized microwave irradiation protocols to expedite entire ExM workflows from gelation to final expansion. BOOST substantially reduces the processing time from several days3,4,5,26 to less than 2 h to 4.5 h, depending on the sample type and staining required. In addition to the rapid processing facilitated by microwave-compatible chemistry, we demonstrated the robustness of the BOOST technique. Notably, BOOST exhibited improved gel uniformity and mechanical properties, achieved through a simplified gel formulation that eliminated sodium acrylate (SA), and avoided protease-based digestion for better preservation of epitopes for high-quality post-expansion staining27. Our experiments successfully demonstrated expansion and imaging across a diverse range of biological samples, spanning sizes from a few micrometers to millimeters. These samples included cultured cells, tissue sections, FFPE sections, organoids, and whole lymph nodes, achieving an approximate tenfold expansion.
Results
BOOST chemistry and workflow
We developed a series of microwave-assisted chemistry and protocols for the whole workflow of ExM, from anchoring, gelation, denaturation, wash and expansion. The running time for various sample types varies while shortening the expansion process for all sample types to under two hours (Fig. 1A). When compared to other protocols to date3,4,5,26,28, there has been a significant reduction in the amount of time used, making it about 6–40 times faster (Fig. 1B), while achieving ~ 10-fold single step linear expansion factor. Hence, we termed this ExM protocol as BOOST.
A Optimized BOOST microwave-assisted protocol for three sample types. Reagents and processing times are listed for each step. B Comparison of processing times for selected ExM techniques for three types of samples. C Microwave-assisted BOOST hydrogel chemistry uses free-radical initiated polymerization of acrylamide (AA) and N,N’-Methylenebis(acrylamide) (bis), followed by alkaline hydrolysis to convert amide groups into charged carboxyl groups, transforming the gel into a swellable polyelectrolyte hydrogel. D Microwave-assisted anchoring with the reactions between the amine groups of the biomolecules and aldehyde of PFA to produce methylol groups, followed by reactions with AA to be anchored to the hydrogel. E A schematic diagram of biomolecules undergoing microwave-assisted gelation, hydrolysis, denaturation, and expansion; green dots and purple sticks depict the anchorable gel-reactive groups on biomolecules. F Optimization of concentrations of AA and bis for maximizing the expansion factor (EF) under 20-minute microwave-assisted hydrolysis (mean ± SD, n = 3 from three independent experiments). G Gel mechanical performance measurement showing the deformation indexes of BOOST gel formulations and the TREx gel. Squares in the background indicate 5 mm × 5 mm areas. Additional note: graphical icons in (1A, 1C–E) are created using Biorender; source data are provided as a Source Data file.
At the early stage of the development, we selected the hydrogel chemistry without SA addition due to many reported problems, such as quality inconsistency of different sources and SA-induced tissue shrinkage5,28,29. An acrylamide-only gel recipe introduced by ZOOM28 based on 30% (w/v) acrylamide (AA) and a cross-linking agent 0.01% (w/v) N,N’-methylenebisacrylamide (bis) was initially employed here to develop the early microwave-compatible gelation and denaturation processes. For the gelation process, like conventional polyacrylamide gels, bis crosslink polymer chains to the network under free radical polymerization30, which can be accelerated by low-energy microwave irradiation24 and we subsequently confirmed this by carrying out the gelation process under microwave using the acrylamide-only gel recipe (Fig. 1C). For the denaturation process, as SA is absent in the crosslinked hydrogel, simultaneous hydrogel hydrolysis is required to transform the neutral polyacrylamide gel into a swellable polyelectrolyte hydrogel, facilitating the expansion process (Fig. 1C), which can also be theoretically assisted with microwave irradiation under alkaline conditions31. To validate, we tested hydrolysis of the crosslinked blank hydrogel (30% w/v AA + 0.01% w/v bis) with an alkaline buffer with pH ranging from 9 to 12 under microwave irradiation, a maximal pH of 12 was chosen here for 12 is the highest pH reported for non-destructive treatment for proteins32. We observed an increase in expansion factor (EF) with the increasing pH, reaching a maximum of ~ 5 × EF (Supplementary Fig. 1A). Our findings also indicate that a 20 min duration is sufficient to achieve the maximum EF for this specific gel formulation (Supplementary Fig. 1B). Simultaneous to hydrogel hydrolysis, anchored proteins need to be digested to ensure uniform expansion. We optimized a denaturation method based on heat and chaotropic reagent, sodium dodecyl sulfate (SDS), which is a recognized method from SDS-PAGE33 and microwave compatible23, and is being suggested to facilitate better epitope preservation for better immunostaining29,34. We, therefore, also employed 10% w/v SDS in the hydrolysis buffer. Toward intact morphology and protein preservation, we then evaluated the impact of hydrolysis buffer pH on the preservation of morphology and proteins with samples embedded. We performed N-Hydroxysuccinimide (NHS) ester staining on ZOOM-processed mouse liver sections and observed no significant differences in protein intensity or morphology across the tested pH range under microwave hydrolysis (pH 10 to pH 12) (Supplementary Fig. 1C).
In addition to the microwave-adapted gelation and hydrolysis step, we set out to seek a microwave-compatible anchoring strategy to maximize biomolecular tethering to the hydrogel within a limited time. To start, we questioned whether the widely used anchoring strategy by Acroylol-X (AcX)29 would be compatible with microwave irradiation. The Ten-fold Robust Expansion Microscopy (TREx) protocol3 was used in this case with only the anchoring step being modified into several testing conditions (Supplementary Fig. 2A). Notably, microwaved samples exhibited inferior preservation compared to the standard AcX-incubated samples. Instead, the microwaved samples resembled the one without any anchoring, suggesting that the acceleration of AcX and amine reactions was ineffective under microwave conditions (Supplementary Fig. 2A). Next, we tested different anchoring strategies (Supplementary Fig. 2B) and finalized with a microwave-assisted anchoring strategy with 30% AA + 4% PFA, similarly as reported previously27. In this anchoring strategy, amine groups on biomolecules are initially modified by PFA to form a reactive methylol group (Fig. 1D), which further reacts with surrounding excess acrylamide monomers to form a protein with a gel-reactive group that is later anchored to the gel matrix during polymerization (Fig. 1E)27,35. The acceleration of anchoring methods using PFA and AA is consistent with previous uses of PFA for protein fixation with improved effectiveness and uniformity under microwave irradiation36, ensuring that reactive methylols are rapidly available for further acrylamide reactions.
To achieve higher-resolution, we further developed the gel formulation to increase EFs while preserving adequate gel mechanical properties. We tested gel formulations with various AA and bis concentrations (Fig. 1F). The mechanical property measured by deformation index (DI) and corresponding expansion factor of blank gels of the selected formulas were measured and calculated3 (Fig. 1G), and the lowest DI of 0.05 was achieved by 17.5% w/v AA/0.0075% w/v bis recipe with an EF of 9.6 ×, and the largest EF of 10.5 × was achieved by 15% w/v AA/0.005% w/v bis formula with DI of 0.13. Both DIs indicate an overall stronger gel compared with the TREx formula, which obtained a DI of above 0.20 at a 10 × expanded state (Fig. 1G). The results obtained from the DI values of the selected formulas (0.05 and 0.13) demonstrate that they exhibit satisfactory mechanical strength3. The highest EF formula is potentially ideal for cultured cell samples, whereas the high mechanical strength formula could be used for larger tissue samples, which would need firmer gel for handling and transfer.
BOOST reveals ultrastructural details of cultured cells, tissue sections, and FFPE samples
To visualize the overall high-quality morphology with less time spent on staining, we optimized microwave-compatible proteomic covalent staining with NHS esters and nucleic acid staining with SYTOX Green for a wide range of biological samples (e.g., cultured cells, tissue sections, and mouse and human FFPE samples) (Fig. 2A). We achieved 4–12 times faster morphological staining than reported methods (Fig. 2B)2,3,4,7,8.
A Schematic of ultrastructural staining after BOOST denaturation. B Comparison of the microwave-assisted NHS ester staining time with ExM techniques using post-processing staining. C 3D visualization of a mitotic U2-OS cell, EF = 10.2 ×. D Left, stained U2-OS cell, EF = 10.2 ×. Blue arrow, granular component of nucleolus. Line profile along the dashed line indicating a partially complementary staining pattern of NHS ester and SYTOX Green in the nucleolus. E BOOST preserves ultrastructural details in vibratome-cut mouse tissue sections. (i) Synaptic structures in mouse brain section, EF = 9.5 ×. (ii) Mouse sperm annulus (green arrowhead), midpiece (below the annulus), and principal piece (above the annulus) in the testicle, EF = 9.4 ×. (iii) Mouse hepatocytes showing endoplasmic reticulum (purple arrowhead), mitochondria (blue arrowhead), and nuclear pores (green arrowhead) in the liver, EF = 9.1 ×. (iv) Mouse cardiomyocyte showing densely labeled A-band with intercalated M-line (green arrowheads) in heart, EF = 8.9 ×. (v) Mouse podocyte foot process (green arrow) and continuous globular basement membrane (blue arrow) in kidney with zoomed-in view (right), EF = 9.0 ×. F NHS ester stained mouse tumor-bearing lung FFPE section before and after BOOST treatment. Post-BOOST images were taken at ~ 2.6 × shrunken state in PBS. Magnified views showing the tumor before and after expansion. G NHS ester stained human prostate cancer FFPE section before and after BOOST processing. Magnified views showing resolved tumor cells after expansion, EF = 8.7 ×. H Visualization of mitochondria (green arrowheads) in the FFPE human prostate cancer section, EF = 8.7 ×. Additional note: graphical icons in 2A are created using Biorender. Scale bars (biological scale): 2 μm (C); 2 μm (D, left); 1 μm (D, right); 2 μm (E, left and top right); 0.2 μm (E(i), all four panels); 0.2 μm (E(ii)); 1 μm (E(iii), top and bottom); 0.5 μm (E(iv)); 2 μm (E(v), left); 0.5 μm (E(v), right); 1 mm (F, top left and top middle); 400 μm (F, bottom left and bottom middle); 500 μm (G, left and right normal views), 20 μm (G, left and right boxed zoomed-in views); 1 μm (H).
For cultured cells, we stained U2-OS cells with ATTO 488 NHS ester under two staining conditions (i.e., 1 h at room temperature as previous protocols suggested3,4,7 and 10 min under microwave irradiation). We found no significant differences in the staining (Supplementary Fig. 3A), both revealed ultrastructural details of organelles such as mitochondria by their morphological characteristics. We then pondered upon the choice of NHS ester dyes with different hydrophobicity and potential differential compartment labeling37. We tested three NHS esters that are conjugated with ATTO dyes (ATTO 488, ATTO 565, ATTO 647 N) owning different hydrophobicity on expanded U2-OS cells. We observed variations in the intensity of the nucleus labeling amongst the three, but all NHS esters were compatible with microwave irradiation and showed no major difference in the mitochondria structure visualization (Supplementary Fig. 3B). We then established the post-BOOST proteomic staining by NHS ester dyes with simultaneous SYTOX Green staining under the microwave. The staining time is 15 minutes for all three types of samples (Fig. 2A), which is considerably shorter than other protocols for post-processing proteomic staining (Fig. 2B). Using BOOST with the proteomic staining strategy, we visualized a mitosis U2-OS cell with down to 30 nm resolution (Fig. 2C and Supplementary Movie 1). We also visualized that SYTOX Green and NHS ester staining showed complementary structures of nucleolus, outlined by the differential compartment labeling in the nucleolar granular components (Fig. 2D).
For tissue sections, we demonstrated rapid and homogenous NHS ester staining on various types of 100 μm thick vibratome-cut mouse tissues. In the mouse brain, we observed well-resolved postsynaptic densities and presynaptic dense projections (Fig. 2E(i)). In the mouse testicle, BOOST resolved the annulus dividing the midpiece and principal piece of sperm (Fig. 2E(ii)). Nuclear envelopes with densely stained nuclear pore complexes as well as the endoplasmic reticulum and mitochondria in hepatocytes, were clearly discernable (Fig. 2E(iii)). In mouse cardiomyocytes, we observed the densely labeled A-band with the intercalated M-line (Fig. 2E(iv)). The podocyte foot processes and continuous glomerular basement membrane were clearly distinguishable in the mouse kidney (Fig. 2E(v)). Besides these nanoscopic details, we also validated the preservation of integrity on a larger scale across a wide range of types of tissues (Supplementary Fig. 4A).
Unlike the high-quality lateral visualization, the uniform staining of thick tissue throughout the thickness always poses an issue, even staining at the expanded state8. Notably, we observed a greatly increased staining uniformity of a 100 μm thick section under 15 min microwave processing compared to 1 h incubation under standard conditions (Supplementary Fig. 4B), likely due to the microwave enables increased diffusion and simultaneously, the NHS ester and amine reaction rate38. With the uniform staining in thick tissues, we demonstrated the preservation of nanoscale structural integrity in 3D (Supplementary Fig. 5 and Supplementary Movie2). We demonstrated the use of BOOST to visualize tangling sperms within the mouse testicle, the continuous glomerular basement membrane, and the ultrastructure in cardiomyocytes (Supplementary Fig. 5). In addition to the single NHS strategy, the dual NHS ester labeling approach was found to label complementary ultrastructures37 and potentially enable better visualization of morphological contrasts. We tested dual NHS ester labeling for expanded tissue sections such as brain (Supplementary Fig. 6A) and kidney (Supplementary Fig. 6B). We observed different NHS ester stained different structures (Supplementary Fig. 6A, B), in particular, ATTO 565 NHS ester showed strong labeling signals on the nucleus (Supplementary Fig. 6A).
Importantly, we optimized BOOST for FFPE samples. As FFPE samples have a highly cross-linked nature39, we integrated a step of 30 min microwave-assisted antigen retrieval step in Tris-EDTA buffer (pH 9) before anchoring to reverse heavy chemical crosslinks. The role of microwave treatment for antigen retrieval has been established17,40. We verified the importance of the antigen retrieval step for the BOOST; absence or insufficient antigen retrieval would lead to insufficient anchoring and, therefore, fractures at an expanded state (Supplementary Fig. 7). We tested the mouse lung tumor FFPE sample and found the whole section was well expanded by BOOST, with the ultrastructural details of the tumor region being clearly discerned (Fig. 2F and Supplementary Movie 3). We then tested BOOST on the human prostate tumor FFPE samples; after processing the sample, the morphology (NHS ester staining) of the tumor region was revealed with drastically improved details using the same microscope setup (Fig. 2G), with mitochondria in the prostate cancer cells also became discernable (Fig. 2H and Supplementary Movie 4). To evaluate the distortion over a large scale of FFPE samples, we analyzed the sample distortion after BOOST processing on a millimeter macroscale using mouse lung and human prostate FFPE samples stained with NHS ester. Low distortion was confirmed with under 2% over a millimeter (Supplementary Fig. 8).
BOOST is compatible and accelerates immunostaining
We optimized microwave-accelerated immunostaining for three types of gel-embedded biological samples (i.e., cultured cells, tissue sections, and FFPE sections) (Fig. 3A). Microwave-accelerated immunohistostaining, immunocytochemistry, and immunofluorescence have long been tested16,19,25,41 and benefits with microwave irradiation during antigen-antibody incubation were also verified18. We modified the previously reported microwave immunostaining protocol41 and achieved the microwave-assisted immunostaining of gel-embedded samples within an hour (Fig. 3B), which was ~ 10–80 times faster than other protocols for immunostaining of gel-embedded samples2,4,26,28.
A Schematic of microwave-assisted immunostaining after BOOST denaturation. B Comparison of immunostaining duration with selected ExM techniques using post-denaturation immunostaining. C Pre-BOOST U2-OS cells stained for α-tubulin (Sparse deconvolution applied, single Z plane) and expanded view (maximum projection, Z = 11), EF = 10.2 ×. D Measurement error versus measurement distance for pre-expansion (Sparse deconvolution applied) versus post-expansion U2-OS α-tubulin images. Solid line, mean; dashed line, SD; n = 4 biological replicates; average EF = 10.0 ×. E Post-BOOST immunolabeling of NPCs in U2-OS cells, EF = 9.6 ×. F NPC diameter quantified from dashed circles with SD (n = 120 NPCs from four biological replicates, average EF = 9.7 ×. G Pre-BOOST human FFPE prostate cancer images (top) labeled against keratin with SYTOX Green (Sparse deconvolution applied, single Z plane), and expanded view (maximum projection, Z = 10), EF = 8.7×. H, I Measurement error versus measurement distance for pre-expansion versus post-expansion FFPE prostate cancer with keratin labeling (H) and SYTOX Green (I). Solid line, mean; dashed line, SD; n = 3 biological replicates; average EF = 8.6 ×. J Post-expansion immunolabeling with NHS ester stain for vibratome-cut mouse tissue sections with magnified views. (i) Cardiac vessel stained against caveolin-1 (CAV1), EF = 9.0 ×. (ii) Podocytes stained against vimentin, EF = 9.1 ×. (iii) Synaptic structures stained against synaptophysin, EF = 9.5 ×. K Post-expansion immunolabeling with SYTOX Green for FFPE human prostate cancer specimens. (i) Keratin (maximum projection, Z = 30), EF = 8.6 ×; (ii) CAV1 (maximum projection, Z = 34), EF = 8.7 ×; (iii) Vimentin (maximum projection, Z = 32), EF = 8.6 ×; (iv) α-tubulin (maximum projection, Z = 30), EF = 8.6 ×; (v) ATPIF1 (maximum projection, Z = 5), EF = 8.7 ×. Additional note: graphical icons (3 A) are created using Biorender; source data are provided as a Source Data file. Scale bar (biological scale): 5 μm (C, left and right); 2 μm (E, left); 0.5 μm (E, right); 0.1 μm (F, top); 20 μm (G, top); 2 μm (G, bottom); 10 μm (J(i) and J(ii), top); 2 μm (J(i) and J(ii), bottom); 5 μm (J(iii), top); 0.2 μm (J(iii), bottom); 5 μm (K(i), (ii), (iii), (iv) and (v)).
To ensure the structures were uniformly expanded on the nanoscale, we quantified the error introduced by expansion by comparing microtubules immunostained and imaged before and after BOOST (Fig. 3C). The pre-BOOST images were processed with Sparse deconvolution42 to enhance the resolution for more accurate distortion assessments. The distortion measured was under 3% across the 10 μm distance measured, indicating low distortions (Fig. 3D).
We verified that BOOST is compatible with immunostaining both before and after the expansion procedure, with immunostaining after the expansion procedure producing strong signals (Supplementary Fig. 9A, B, and Supplementary Movie 5). This is consistent with works of molecular decrowding with ExM to improve epitope accessibility and to get better immunostaining results11,34. We reasoned immunostaining after BOOST would be a preferred method to achieve improved staining results.
We also validated BOOST by staining ultrastructural details in cells that were not distinguishable using conventional optical microscopes at a non-expanded state. We demonstrated the visualization of nuclear pores using immunostaining with antibodies for nuclear pore complex (NPC) (anti-NUP 153 and anti-NUP 96) (Fig. 3E). We quantified 120 NPCs measured from four replicates (Fig. 3F) and obtained NPC sizes of 964 nm ± 52 nm (99.5 nm ± 5.5 nm when translated into original size using average EF = 9.7 ×). This further translates the local EF of 9.0 × calculated from NPC sizes of 107 nm, with a 7.7% deviation from the average EF determined with the macroscopic method, which is more than acceptable as suggested by TREx3. We further evaluated the microscopic distortion of BOOST when applied to more complex FFPE samples. We compared pre-BOOST and post-BOOST images stained with Keratin antibody and nuclear stain (Fig. 3G) and found the distortion was in the range of 1-2% (Fig. 3H, I) across 100 μm assessed, showing more than acceptable level of distortions.
Next, we tested the microwave-accelerated post-BOOST immunostaining procedure for other types of samples. For 100 μm mouse tissue sections, post-BOOST epitope preservation and immunolabeling were verified using caveolin-1 to visualize cardiac blood vessels (Fig. 3J(i)), vimentin to stain kidney podocytes (Fig. 3J(ii)), and synaptophysin to label the neuron synapses (Fig. 3J(iii)). For human FFPE samples, post-BOOST immunolabeling was used to visualize five protein markers (i.e., keratin, caveolin-1, vimentin, α-tubulin, and ATPIF1) in prostate tumors (Fig. 3K(i), (ii), (iii), (iv) and (v)). We validated antibody specificity under microwave acceleration with post-BOOST staining with secondary antibody-only negative controls for the above specimens (Supplementary Fig. 10). We also compared microwave-accelerated immunostaining and standard 2-day immunostaining using post-BOOST FFPE human prostate cancer at a ~ 2-fold expansion state. We observed identical staining patterns against α-tubulin and prostate-specific membrane antigen (PSMA) (Supplementary Fig. 9C).
BOOST is capable of rapidly expanding large samples to ~ 10-fold
We further adjusted the anchoring, gelation, hydrolysis, and wash and expand steps of BOOST to expand larger biological samples with original sizes ranging from hundreds of micrometers to a few millimeters and achieved ~ 10 × expansion and obtained millimeter-to-centimeter-size expanded samples, which has not been reported by any current protocols. Here, we demonstrated the homogenous expansion of organoids and whole lymph nodes, which only took ~ 2 h and ~ 3 h, respectively (Fig. 4A).
A Schematic of BOOST workflow for large biological specimens. B 3D visualization of an expanded whole airway organoid stained with NHS ester and SYTOX Green, EF = 10.1 ×. C 3D visualization (left) of an airway organoid stained with NHS ester, SYTOX Green, and antibody against β-tubulin IV, with additional views (middle and right) to reveal the ultrastructural details of cilia, EF = 10.0 ×. D Processing workflow of an intact lymph node to achieve a 9-fold expansion. Each square in the background represents a 5 mm × 5 mm area. E 2D image of the middle section of intact lymph node stained with ATTO 565 NHS ester, and (F) a magnified 3D view of the boxed region, EF = 9 ×. G 3D and 2D view of selected regions in the intact lymph node. Additional note: graphical icons in 4 A are created using Biorender. Scale bar (biological scale): 100 μm (B); 10 μm (C, left); 2 μm (C, middle); 0.2 μm (C, right); 100 μm (E); 10 μm (F); 2 μm (G).
Specifically, we demonstrated BOOST to characterize the cellular composition and organization in three dimensions and morphology and ultrastructure of ~ 500 µm airway organoids with an EF of 10.1 × when employing 15% w/v AA / 0.005% w/v bis gel formulation. With the staining of SYTOX Green and NHS ester, we reconstructed the 3D morphology of the whole organoid in detail (Fig. 4B and Supplementary Movies 6 and 7). Combining microwave-assisted immunostaining and NHS staining, we visualized the distribution of β-tubulin IV on cilia of the airway organoid (Fig. 4C and Supplementary Movies 8 and 9). Nine bundles of microtubules in cilia axoneme could be occasionally resolved, yet sometimes challenging with cilia in certain orientations. Coupling with additional algorithms4 and larger expansion factor ExM methods2 may be required for fully visualizing the nine-fold symmetry.
We further demonstrated the capability of BOOST to expand even larger samples, using lymph nodes in the range of several millimeters. The robustness of BOOST enabled millimeter-sized samples to achieve an EF beyond just 4 × by existing techniques involving millimeter-sized sample13,14. We adjusted the BOOST protocol in the processing time for sufficient anchoring, gelation, hydrolysis, and staining to achieve whole lymph node visualization at ~ 9 × fold with 17.5% w/v AA / 0.0075% w/v bis gel formulation in ~ 4.5 h (Fig. 4A). We demonstrated the embedding of a lymph node with a measured vertical size of 1.95 mm; the lymph node was later expanded to 2.8-fold in PBS after denaturation/hydrolysis; and a further expansion in water led to a final vertical size of ~ 1.75 cm, indicating a 9× macroscopic EF with almost transparent appearance under normal lighting (Fig. 4D). We then combined proteomic staining to image the 3D morphology of the whole lymph node in detail; we imaged across the middle region of the lymph node to confirm the uniformity of the NHS ester labeling (Fig. 4E). A further magnified region was imaged to visualize morphological details of the expanded lymph node (Fig. 4F, G). Hence, BOOST achieved a ~ 10-fold expansion of millimeter-sized three-dimensional samples that was not feasible before, to the best of our knowledge.
Discussion
We developed BOOST—an hour-scale and robust workflow for expanding diverse biological specimens to boost ExM’s throughput and adoption in biological investigations (Supplementary Fig. 11). BOOST has made significant advances in the following aspects: (1) BOOST has dramatically improved the throughput of ExM by innovatively using a series of microwave-assisted chemistries for all major ExM procedures (i.e., monomer infiltration, anchoring, gelation, denaturation, staining, and expansion). (2) BOOST is highly robust and versatile. It uses common reagents and instruments, and it works effectively on various sample types, including cultured cells, tissues, organoids, and FFPE samples, and is also compatible with morphological staining and immunostaining. (3) BOOST has pioneered the single-step ~ 10-fold linear expansion of millimeter-sized large 3D biological specimens.
The improved throughput of BOOST was achieved largely due to the innovative implementation of microwave-compatible ExM chemistries. Microwave irradiation accelerates mainly monomer infiltration, anchoring, gelation, denaturation/hydrolysis, and staining chemistries, with the most significant acceleration for the denaturation step. The improvement in the throughput was particularly evident for expanding FFPE samples, as previously a homogenous linear expansion of 10-fold for FFPE samples was achieved by Magnify. BOOST reduced the whole workflow from days in Magnify4 to less than 4 h, inclusive of staining for ultrastructure and proteins of interest. The significant enhancement in throughput may facilitate time-sensitive applications, such as nanoscale pathology. Researchers have previously utilized ExM techniques to obtain ultrastructural details in renal pathological samples yet only with 4-fold expansion9,43. This reduction in processing times, when coupled with 10-fold expansion, could provide a crucial advantage for clinical applications.
The robustness and versatility of BOOST were demonstrated by gel rigidity, non-proteolytic digestion, and compatibility with various samples and different staining protocols. Importantly, BOOST utilized a simple gel formulation with only AA and bis and avoided SA, a gel monomer present in most ExM gel formulations to date29. The absence of SA helps to avoid potential issues, such as variable impurities from vendors, potential tissue shrinkage, and uneven distribution of ionic residues, as reported by other studies4,28,29. The use of heat and chaotropic reagents for denaturation improved epitope preservation and enabled high-quality post-expansion staining facilitated by protein decrowding27,29,34. All the reagents used in the BOOST workflow are reliable and easily accessible, which could potentially boost the adoption of the technique for biological investigations.
It is noteworthy that BOOST successfully accomplished a single-step ~ 10-fold expansion of large 3D biological specimens, a task previously unattainable to the best of our knowledge. This improvement is likely attributable to the enhanced monomer infiltration, anchoring, homogenous gelation, and denaturing within large specimens facilitated by microwave irradiation. In contrast to conventional 2D imaging of tissue sections, 3D imaging excels in capturing the intricacy of biological samples. The advanced capabilities of 3D imaging are crucial for comprehending cellular composition, and interactions between cells in intact biological samples. Microscopy imaging of large 3D volumes remains challenging. To attain large volumes of 3D information, BOOST could be coupled with a light sheet microscope equipped with an objective that is designed to achieve a large working distance with good NA for fast and detailed acquisition. We could also adjust the expansion factor using a salinity gradient to achieve the mapping of desired volumes and resolution for specific applications. For instance, we could perform a macroscale overview at a shrunken state in high salinity conditions and then locate the targeted region of interest for higher-resolution imaging at a fully expanded state. We foresee future developments in this area in both hardware and software for imaging and data processing.
In developing the current BOOST protocol, our primary focus was on enhancing the speed of major ExM steps. BOOST is advantageous for its rapid processing across various tissue types within hours, but the ~ 10-fold expansion limits its resolution for resolving ultrafine structures. There are minor variations in the expansion factor of different sample types and preparations. To achieve higher resolving power would require a higher expansion factor, such as iterative methods2,11 or the single-shot 20-fold ExM6 method to visualize features requiring < 20 nm resolving power. In addition to ExM methods to achieve a larger expansion factor, imaging with super-resolution imaging platforms3,44 and integrating computational methods9,10 can enhance the resolution of BOOST to resolve finer structures. These integrations will allow for more accurate assessments of nanoscale distortions than the current assessments.
To foster the future adoption of BOOST in both research and clinical settings, several potential advancements could be explored. For instance, we have demonstrated compatibility with immunostaining and preservation of proteins. Validation of the preservation of other biomolecules, such as RNA, should be considered. We have demonstrated the processing of a broad range of sample types and developed detailed protocols in this study, but specific types of tissues may require further optimization. The application of microwave-assisted processing may also be extended to other steps of the workflow, including long-verified sample fixation and washing20. In addition, the integration of computational methods with BOOST could potentially enhance resolution to a further scale, which may prove advantageous for specific applications.
Methods
Ethical statement
The protocol of the mouse experiment was approved by The Committee on the Use of Live Animals on Teaching and Research at the University of Hong Kong. All the housing conditions for the mice were strictly complied with the regulations of the Center for Comparative Medicine Research at The University of Hong Kong. For the intravenous tumor model, no specific maximum tumor size was imposed, animals were closely monitored and euthanized as soon as they reached a predetermined humane endpoint that the mice lose 10% of their body weight, to prioritize their welfare. All procedures adhered to the institutional guidelines of the Committee on the Use of Live Animals in Teaching and Research at the University of Hong Kong. No animal in this study exceeded that limit. All prostate cancer paraffin tissues were obtained from the Third Affiliated Hospital of Southern Medical University, all human tissue samples were collected with informed consents from patients and approved by the ethics committee at the Third Affiliated Hospital of Southern Medical University.
Chemicals
acrylamide (A9099, Sigma-Aldrich), N,N’-Methylenebis(acrylamide) (146072, Sigma-Aldrich), ammonium persulfate (A7460, Sigma-Aldrich), TEMED (1610801, Bio-Rad), sodium chloride (S3014, Sigma-Aldrich), paraformaldehyde (15700, EMS), glutaraldehyde (16501, EMS), sodium dodecyl sulfate (L3771, Sigma-Aldrich), sodium hydroxide (S5881, Sigma-Aldrich), boric acid (A01298, 3 A), ATTO 488, 565, 647 N NHS esters (ATTO-TEC), SYTOX Green (S7020, Thermo Fisher), tris base (T6066, Sigma-Aldrich), ethylenediaminetetraacetic acid disodium salt (E5134, Sigma-Aldrich), poly-L-lysine solution (P8920, Sigma-Aldrich), Sigmacote (SL2, Sigma-Aldrich), DAPI (62248, Thermo Fisher), xylene (UN1307, DUKSAN), ethanol absolute (UN1170, Scharlau), Phosphate-buffered saline (PBS) (Gibco 10010023), Bovine serum albumin (BSA) (9998S, ENERGY), Triton X-100 (X100, Sigma-Aldrich), Tween20 (P9416, Sigma-Aldrich), sodium acrylate (408220, Sigma-Aldrich), 4-hydroxy TEMPO (E020145, Energy).
Antibodies
rabbit polyclonal anti-α-tubulin (11224-1-AP, Proteintech), rabbit polyclonal anti-NUP98-NUP96 (12329-1-AP, Proteintech), rabbit polyclonal anti-NUP153 (14189-1-AP, Proteintech), rabbit polyclonal anti-PSMA/GCPII (13163-1-AP, Proteintech), rabbit monoclonal anti-beta IV tubulin antibody (EPR16776, Abcam), rabbit polyclonal synaptophysin antibody (17785-1-AP, Proteintech), rabbit polyclonal ATPIF1 antibody (12067-1-AP, Proteintech), rabbit polyclonal vimentin antibody (10366-1-AP, Proteintech), rabbit polyclonal pan-keratin antibody (26411-1-AP, Proteintech), rabbit monoclonal caveolin-1 (D46G3) XP antibody (3267S, Cell Signaling Technology), Alexa Fluor 647-conjugated donkey anti-rabbit IgG (ab150063, Abcam), Alexa Fluor 568-conjugated donkey anti-rabbit IgG (ab175470, Abcam).
Cultured cell samples
Homo sapiens bone osteosarcoma U2-OS HTB-96 (ATCC) cells were cultured in DMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin at 37 °C in the presence of 5% CO2. To prepare cells for fixation, imaging, and expansion, cells were seeded on 12 mm × 12 mm coverslips and cultured until they reached 50–60% confluency for fixation. Cells on coverslips were normally fixed using 4% PFA and 0.1% GA in PBS buffer for 10 minutes. For NUP 98-96 & NUP153 immunostaining, a different fixation strategy, as reported previously, was used to better preserve nuclear pores2,45. Briefly, cells were firstly prefixed using 2.4% PFA in PBS for 30 s, followed by incubation with 0.4% Triton X-100 in PBS for 3 min and two additional 5-minute washes in PBS on a rocking platform. Subsequently, the cells were fixed with 2.4% PFA in PBS for 20 min at room temperature and then washed twice for 5 min with PBS on a rocking platform. A second permeabilization step was carried out using a 0.2% Triton X-100 in PBS for 10 min, followed by two additional 5-minute washes in PBS on a rocking platform.
Mouse tissue section and lymph node samples
C57BL/6 J adult mice (12 weeks) were anesthetized using 2% Xylazine: 10% Ketamine: PBS mixed in 1:2:9, and perfused transcardially with 20 mL of PBS and 20 mL of cold fixative solution (4% PFA in PBS). Tissues (i.e., brain, liver, kidney, intact lymph nodes) were then harvested and fixed for 48–72 h with the same fixative solution at 4 °C. Fixed tissues were then sectioned to 100 µm-thick slices using a vibratome (easiSlicer, TED PELLA).
Mouse FFPE samples
The lung tumor was established by intravenously injecting 5 × 105 - 1 × 106 mouse LLC cells into C57BL/6 J mice. The lung tissue was collected after 4 weeks of injection. Animals were closely monitored on a routine basis, if the mice lose 10% of their body weight, to prioritize their welfare we will terminate the experiment and euthanize the mice. All procedures adhered to the institutional guidelines of the Committee on the Use of Live Animals in Teaching and Research of the University of Hong Kong. Tissues were then fixed in 4% PFA in PBS for at least 24 h. Fixed tissues were then dehydrated and processed as follows: 80% ethanol for 15 min, 95% ethanol for 30 min for 3 times, 100% ethanol for 45 minutes for 3 times, toluene for 15 min for twice, and pre-warmed paraffin for 45 minutes for 3 times. Then samples were embedded in fresh paraffin.
Human FFPE samples
All prostate cancer paraffin tissues were obtained from the Third Affiliated Hospital of Southern Medical University with informed consent from patients and approved by the ethics committee at the institution. The pathological diagnosis of the patients was confirmed by two experienced pathologists. Briefly, after surgical resection of the tumor, the samples were fixed with formalin for dehydration immediately and then embedded with paraffin. 5 μm thickness sections were prepared and mounted on glass slides for subsequent analysis.
Lung airway organoid samples
The lung organoids were cultured in an expansion medium and passaged every 2 to 3 weeks with a ratio between 1:3 to 1:10, as described previously46. The lung organoids were cultured in the expansion medium for 7–10 days after passaging. To generate apical-out airway organoids, lung organoids were recovered from matrigel and then suspension-cultured in proximal differentiation medium (PneumaCult™-ALI medium + 10 μM Y-27632 + 10 μM DAPT + 100 μg/mL primocin) in Nunclon Sphera culture plate (Thermo Scientific, Waltham, MA, USA) for 14 days. Differentiated airway organoids were then fixed with 4% PFA in PBS for 1 h at room temperature before processing.
Microwave set-up
A PELCO BioWave® Pro+ Microwave Processing System (36700, TED PELLA) was used for all the microwave processing in BOOST, with ColdSpot Plus and EM Pro Vacuum Chamber used for specified steps. All the detailed microwave parameters for each step are included in Supplementary Table 1 and Supplementary Table 2.
Immunostaining for cultured cells
Cells were washed twice with PBS at room temperature, 5 min each, on a rocking platform. Then, samples were permeabilized with PBS containing 0.2% Triton X-100 for 10 min at room temperature and washed three times with PBS, 5 min each, either on a rocking platform or under microwave irradiation (Supplementary Table 1). Then, samples were blocked using 3% BSA in PBS for 5 min under programmed microwave irradiation (Supplementary Table 1), followed by incubation with primary antibodies diluted (1:200 dilution) with 1% BSA in PBS for 5 min under programmed microwave irradiation (Supplementary Table 1), then washed three times with PBS, 5 min each. Finally, samples were incubated with fluorophore-conjugated secondary antibodies (1:250 dilution) diluted with 1% BSA in PBS for 5 min under programmed microwave irradiation (Supplementary Table 1) and washed in PBS for three times, 5 min each. Samples will be used for BOOST.
FFPE sample preparation
FFPE sections mounted on glass slides were deparaffined. Briefly, slides were baked in the oven for 1 h at 60 °C, incubated in fresh xylene for 10 min twice, and then processed at room temperature in the following flow, 3 min for each step: xylene × 3, 100% ethanol × 3, 95% ethanol × 1, 70% ethanol × 1, ddH2O × 1, PBS × 3. Antigen retrieval was performed with the slide placed in a microwavable container with the antigen retrieval buffer (Tris/EDTA pH 9) and irradiated with microwave for 30 min using the pre-set program (Supplementary Table 1). The samples were cooled to room temperature and then washed with PBS three times.
Gelation chamber design
For sample with thickness < 300 μm (i.e., cells, tissue sections, FFPE, organoids)
Two strips of 300 μm-thick silicone sheet (pre-treated with ethanol to remove impurities) were placed on the parafilm-wrapped or Sigmacote-treated glass slides with appropriate distance left in between for the sample inclusion. The gelation chamber will produce the final gel with a thickness of 300 μm.
For thick samples (i.e., organoids, whole lymph nodes)
A silicone sheet of appropriate thickness was selected, or several 300 μm-thick silicone sheets could be stacked for a deeper chamber. Then the sheet or stacked sheets were punched with a hole of appropriate size sufficient to cover the entire sample volume.
BOOST monomer infiltration, anchoring, and gelation
Cultured cells
Cultured cells on the coverslip was placed in a 12-well plate, incubated with anchoring solution (15% w/v AA, 0.005% w/v bis, 0.65 M sodium chloride (NaCl), and 4% PFA in PBS) for 5 min under programmed microwave irradiation (Supplementary Table 1). Then, the coverslip was placed on the gelation chamber on ice, with the gelation solution (15% w/v AA, 0.005% w/v bis, 0.65 M NaCl, 4% PFA, 0.1% w/v TEMED and 0.1% w/v APS in PBS) being mixed well and injected into the space between. The chamber was transferred to the microwave processor for 10 min under programmed microwave irradiation (Supplementary Table 1) for gelation. The coverslip with the gel could then be lifted off the chamber, and the gel was cut into a desired size.
Tissue sections
The 100 μm vibratome-cut section was first washed in PBS supplemented with 0.2% Triton X-100 three times, 5 min each. Then, it was placed in an anchoring solution (17.5% w/v AA, 0.0075% w/v bis, 0.65 M NaCl, 4% PFA in PBS) and incubated for 10 min under programmed microwave irradiation in a vacuum environment (Supplementary Table 1). Then, the tissue was transferred to the gelation chamber on ice. A coverslip was placed on the silicone strips to cover the tissue. The gelation solution (17.5% w/v AA, 0.0075% w/v bis, 0.65 M NaCl, 4% PFA, 0.1% w/v TEMED, and 0.1% w/v APS in PBS) was mixed well and injected into the space between and transferred to the microwave processor for 10 min under programmed microwave irradiation in a vacuum environment (Supplementary Table 1) for gelation. The coverslip with the gel could then be lifted off the chamber, and the gel was cut into a desired size.
FFPE sections
After FFPE pre-processing step, the slide with the FFPE section was cut to the desired size, placed in infiltration/anchoring solution (17.5% w/v AA, 0.0075% w/v bis, 0.65 M NaCl, and 4% PFA in PBS) and incubated for 15 min under programmed microwave irradiation in a vacuum environment (Supplementary Table 1). Then, the slide was placed on ice, and two 300 μm silicone strips were placed near the tissue section to construct a chamber on the same slide. A Sigmacote-treated coverslip was placed over the tissue section, and the gelation solution (17.5% w/v AA, 0.0075% w/v bis, 0.65 M NaCl, 4% PFA, 0.1% w/v TEMED, and 0.1% w/v APS in PBS) was mixed well and injected into the space between. The chamber was then transferred to the microwave processor for 15 min under programmed microwave irradiation (Supplementary Table 1) for gelation. The coverslip with the gel could then be lifted off the chamber, and the gel was cut into a desired size.
Organoids
The fixed organoids were permeabilized for 5 minutes using PBS + 0.5% Triton X-100. Monomer anchoring was then performed in an anchoring solution (15% w/v AA, 0.005% w/v bis, 0.65 M NaCl, and 4% PFA in PBS) for 20 min under programmed microwave irradiation in a vacuum environment (Supplementary Table 1). Then, the tissue was transferred to the gelation chamber on ice. The gelation solution (17.5% w/v AA, 0.0075% w/v bis, 0.65 M NaCl, 4% PFA, 0.1% w/v TEMED, and 0.1% w/v APS in PBS) was mixed well and injected into the chamber space to cover the sample, and a coverslip was placed on the silicone sheet to cover the chamber. The chamber was transferred to the microwave processor for 15 min under programmed microwave irradiation in a vacuum environment (Supplementary Table 1) for gelation. The coverslip with the gel could then be lifted off the chamber, and the gel was cut into a desired size.
Whole lymph node
The fixed lymph node was permeabilized for 10 min using PBS + 0.5% Triton X-100. Monomer anchoring was then performed in anchoring solution (17.5% w/v AA, 0.0075% w/v bis, 0.65 M NaCl, and 4% PFA in PBS) for 30 min under programmed microwave irradiation in a vacuum environment (Supplementary Table 1). Then, the tissue was transferred to the gelation chamber on ice. The gelation solution (17.5% w/v AA, 0.0075% w/v bis, 0.65 M NaCl, 4% PFA, 0.1% w/v TEMED, and 0.1% w/v APS in PBS) was mixed well and injected into the space to cover the sample, and a coverslip was placed on the silicone sheet to cover the chamber. The chamber was transferred to the microwave processor for 30 min under programmed microwave irradiation in a vacuum environment (Supplementary Table 1) for gelation. The coverslip with the gel could then be lifted off the chamber, and the gel was cut into a desired size.
BOOST denaturation and hydrolysis conversion
Culture cells and tissue sections
The gel was washed with PBS, then transferred with a paintbrush into a microwavable vessel containing denaturation buffer (10% w/v SDS, 0.05 M boric acid in ddH2O, pH 12) and processed immediately under microwave irradiation for 20 min (Supplementary Table 1). Then, the gel was washed in large volumes of PBS supplemented with 0.1% Tween 20 (PBST) for at least twice, either on a rocking platform or under microwave irradiation (Supplementary Table 1), 5 min each.
FFPE sections
The slide with the gel was washed with PBS, then placed into a microwavable vessel containing denaturation buffer (10% w/v SDS, 0.05 M boric acid in ddH2O, pH 12) and processed with programmed microwave irradiation for 30 minutes (Supplementary Table 1). The gel would normally detach from the slide in the first 5 min, the slide could be removed, and the gel was hydrolyzed for the rest of the 25 min. Then, the gel was washed in large volumes of PBST for at least twice, 5 min each.
Organoids/ Whole lymph nodes
The gel with the organoid/lymph node embedded was washed with PBS, then placed into a microwavable vessel containing denaturation buffer (10% w/v SDS, 0.05 M boric acid in ddH2O, pH 12) and processed with programmed microwave irradiation for 30/60 min.
BOOST post-denaturation NHS ester staining
Cultured cells, tissue sections, and FFPE sections
The denatured gel, after thorough washing, was incubated with ATTO NHS esters diluted in 1:200 in PBS and/or SYTOX Green dyes diluted in 1:500 for 15 min under microwave irradiation (Supplementary Table 1). Then, the gel was washed in large volumes of PBST for at least twice either on a rocking platform or under microwave irradiation (Supplementary Table 1), 5 min each.
Organoids/ Whole lymph nodes
The denatured gel after through washing, was incubated with ATTO NHS esters diluted in 1:200 in PBS and/or SYTOX Green dyes diluted in 1:500 for 25–50 min under microwave irradiation (Supplementary Table 1). Then, the gel was washed in large volumes of PBST for at least twice, 10 min each.
BOOST immunostaining after hydrolysis and denaturation
Cultured cells, tissue sections, FFPE sections and organoids
The denatured gel, after thorough washing, was incubated in the blocking buffer (3% BSA with 0.2% Triton X-100 in PBS) under microwave irradiation for 10–15 min (Supplementary Table 1). Then, the gel was incubated with primary antibodies diluted (1:100 dilution) with 1% BSA + 0.1% Triton X-100 in PBS for 10 min under programmed microwave irradiation (Supplementary Table 1). The gel was then washed three times with excessive PBST, 5 minutes each, either on a rocking platform or under microwave irradiation (Supplementary Table 1). The gel was finally incubated with fluorophore-conjugated secondary antibodies (1:150 dilution) in 1% BSA + 0.1% Triton X-100 in PBS for 10 min under programmed microwave irradiation (Supplementary Table 1). The gel was then washed for at least three times with excessive PBST, 5 min each.
BOOST expansion
For cultured cells, tissue sections, FFPE sections, organoids and whole lymph nodes, large volume of ddH2O was exchanged every 5–10 min, either on a rocking platform or under microwave irradiation (Supplementary Table 1) until fully expanded, usually four exchanges would be sufficient.
Gel mounting, imaging & analysis
The bottom of the plate/dish used for imaging was coated with poly-L-lysine to prevent gel drifting during imaging. Four microscope systems were employed. Epifluorescence images were captured on Keyence X810 (BZ-X800 Viewer software), using 4 × /0.13 NA air (Fig. 2F and Supplementary Fig. 7), 10 × /0.45 NA air (Fig. 2G and Supplementary Fig. 8), 20 × /0.45 NA air (Supplementary Fig. 9C). Confocal microscopy and structured illumination microscopy images were captured either using a ZEISS LSM900 upright microscope (Zen 3.5 blue edition software) equipped with Zeiss Plan-Neofluar 2.5 × /0.085 NA air (Figs. 4B, E), Zeiss N-Achroplan 10 × /0.3 NA water immersion (Fig. 4C, F and Supplementary Figs. 4A, 10F), Zeiss Plan-Apochromat 40×/1.0 NA water immersion (Figs. 2E, 2H, 3G, 3J, 3K, 4C, 4G and Supplementary Figs. 1C, 2A, 2B, 3A, 4B, 5, 6A, 10B, 10C, 10D, 10E) objectives or a Vt-iSIM inverted microscope (Micromanager version 2.0.0 software) equipped with Olympus UPLSAPO60XW 60 × /1.20 NA water immersion objective (Figs. 2C, 2D, 3C, 3E, 3F and Supplementary Figs. 3B, 9A, 9B, 10A). In addition, tested tile images were captured using a LiTone LBS Light-sheet Microscope equipped (LitScan version 3.3.0 software) with a Nikon 16 × /0.80 NA water dipping objective (Supplementary Fig. 6B).
Assessing hydrolysis conditions in protein and morphology preservation
The workflows were adapted as previously reported28 except for the hydrolysis step modified into three conditions (i.e., pH 10, pH 11, and pH 12) for the evaluation of morphological difference and protein retention under a 20-min microwave. Fixed liver slices were first incubated in a monomer solution (30% w/v AA, 0.01% w/v bis, 0.65 M NaCl, 4% w/v PFA, and 0.1% w/v TEMED in PBS) at room temperature for 3 h. Then, the tissue was transferred to the gelation chamber (monomer solution with 0.1% w/v APS). A coverslip was placed on the silicone strips to cover the tissue, and the fresh gelation solution was injected into the space between. The chamber was then placed at room temperature for 40 min to complete gelation, and the gel was cut into appropriate sizes and recovered in PBS. For the modified hydrolysis, hydrolysis buffer (0.2 M SDS, 0.05 M boric acid in DI water) for three pH conditions: pH 10, pH 11, and pH 12, was prepared separately. The recovered gel was then transferred with a paintbrush into a microwavable vessel containing a denaturation buffer of specified pH value and processed immediately under microwave irradiation for 20 min (Supplementary Table 1). The denatured gel was then washed in PBS four times, 1 h each. The washed gel was stained with ATTO 565 NHS ester diluted 1:200 in PBS for 1 h at room temperature, then washed in ddH2O three times, 15 min each, followed by an exchange of ddH2O every hour to a fully expanded state for imaging.
Optimization of microwave-assisted anchoring
The workflows were adapted from previously published3 except for the anchoring step modified into four conditions (i.e., no anchoring, 10 min microwave, 20 min microwave, and the standard TREx protocol of 1 h incubation) for the evaluation of AcX effectiveness under microwave. For standard TREx anchoring, liver sections were incubated in 100 μg/mL AcX diluted in PBS for 1 h at room temperature. For modified microwave-assisted anchoring, liver sections were incubated in 100 μg/mL AcX diluted in PBS and processed in a microwave processor for 10 min and 20 min (Supplementary Table 1), respectively. After the anchoring step, the sections were rinsed in PBS. Later incubated in TREx gelation solution (10.34% w/v SA, 14.22% w/v AA, 0.005% w/v bis, 0.15% w/v APS, 0.15% w/v TEMED, 0.0015% w/v TEMPO in PBS) for 20 min on ice. Then, the tissue section was transferred on the gelation chamber on ice. A coverslip was placed on the silicone strips to cover the tissue. The fresh gelation solution was then injected into the space between. The chamber was placed in a 37 °C incubator for 1 h to complete gelation, and the gel was cut into appropriate sizes and recovered in PBS. Gel was then placed into TREx disruption buffer (5% w/v SDS, 0.2 M NaCl, 0.05 M Tris in ddH2O, pH 7.5) in a tube, and the tube was transferred to an incubator set at 80 °C for 3 h. The denatured gel was then recovered in 0.4 M NaCl followed by two washes in PBS, 30 min each. The washed gel was stained with ATTO 565 NHS ester diluted 1:200 in PBS, then washed in ddH2O three times, 15 min each, followed by an exchange of ddH2O every hour to a fully expanded state for imaging.
Standard post-denaturation immunostaining for FFPE sample
The denatured FFPE gel after thorough washing, was incubated in the blocking buffer (3% BSA with 0.2% Triton X-100 in PBS) at room temperature for 3 h. Then, the gel was incubated with primary antibodies diluted (1:100 dilution) with 1% BSA + 0.1% Triton X-100 in PBS overnight at 4 °C. The gel was then washed three times with excessive PBST, 30 min each, on a rocking platform. The gel was finally incubated with fluorophore-conjugated secondary antibodies (1:150 dilution) in 1% BSA + 0.1% Triton X-100 in PBS for 3 h at room temperature. The gel was then washed for at least three times with excessive PBST, 30 min each.
Expansion factor determination
We used three strategies to determine expansion factors (Supplementary Fig. 12). First being the linear measurement of the longest distance of the unexpanded gel and expanded gel, second being the square root of the area ratio of the gel size in two states and lastly being the cell nucleus size quantification based on pre-BOOST SYTOX Green, NHS ester staining or NUP immunostaining of unexpanded and expanded images.
Sparse deconvolution
The pre-BOOST images used for distortion analysis (Fig. 3C and G) were processed using Sparse deconvolution to improve the resolution for more accurate distortion analysis42. For Fig. 3C, Sparse iteration was set to 100, image fidelity was set to 1000, and sparsity was set to 100 with 10 deconvolution times. For Fig. 3G, Sparse iteration was set to 100, image fidelity was set to 500, and sparsity was set to 50 with 10 deconvolution times.
Distortion and RMSE analysis
The unexpanded and expanded image distortion was evaluated using B-spline transformation in Elastix, and RMSE was analyzed according to protocols and script offered by Vaughan’s group47
Deformation index (DI) analysis
The blank gel was formed in a gelation chamber with a thickness of 800 μm, a 4 mm diameter region was punched to produce a circular gel, then hydrolysis was performed (only for BOOST formulations), and the gel was expanded to a fully expanded state. To calculate the DI, the gel was cut in half and stood against the ruler wall (with the curved side facing downwards to allow the flat surface to deform), images were taken and later processed in ImageJ for measurement of the best-fit deformed circle. Two BOOST formulations were used (17.5% w/v AA / 0.0075% w/v bis; 15% w/v AA /0.005% w/v bis), and the original TREx formulation was also evaluated (10.34% w/v SA, 14.22% w/v AA, 0.005% w/v bis, 0.15% w/v APS, 0.15% w/v TEMED in PBS).
Statistics and reproducibility
All experiments were carried out independently at least 3 times, besides those already stated in the figure legends. All data were recorded in the form of mean ± SD (standard deviation). Micrographs related experiments (Figs. 2E, F, 2G, H, 3E, J, K, 4C) were performed for at least three biological replicates with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets from the current study are available on Zenodo: https://doi.org/10.5281/zenodo.14878218. All other relevant data are provided in this paper. Source data are provided in this paper.
References
Chen, F., Tillberg, P. W. & Boyden, E. S. Expansion microscopy. Science 347, 543–548 (2015).
Louvel, V. et al. iU-ExM: nanoscopy of organelles and tissues with iterative ultrastructure expansion microscopy. Nat. Commun. 14, 7893 (2023).
Damstra, H. G. et al. Visualizing cellular and tissue ultrastructure using Ten-fold robust expansion microscopy (TREx). ELife 11, e73775 (2022).
Klimas, A. et al. Magnify is a universal molecular anchoring strategy for expansion microscopy. Nat. Biotechnol. 41, 858–869 (2023).
Truckenbrodt, S. et al. X10 expansion microscopy enables 25‐nm resolution on conventional microscopes. EMBO Rep. 19, e45836 (2018).
Wang, S. et al. Single-shot 20-fold expansion microscopy. Nat. Methods 21, 2128–2134 (2024).
M’Saad, O. & Bewersdorf, J. Light microscopy of proteins in their ultrastructural context. Nat. Commun/11, 3850 (2020).
Mao, C. et al. Feature-rich covalent stains for super-resolution and cleared tissue fluorescence microscopy. Sci. Adv. 6, eaba4542 (2020).
Kylies, D. et al. Expansion-enhanced super-resolution radial fluctuations enable nanoscale molecular profiling of pathology specimens. Nat.Nanotechnol. 18, 336–342 (2023).
Shaib, A. H. et al. One-step nanoscale expansion microscopy reveals individual protein shapes. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02431-9 (2024).
Sarkar, D. et al. Revealing nanostructures in brain tissue via protein decrowding by iterative expansion microscopy. Nat. Biomed. Eng. 6, 1057–1073 (2022).
Pownall, M. E. et al. Chromatin expansion microscopy reveals nanoscale organization of transcription and chromatin. Science 381, 92–100 (2023).
Sim, J. et al. Nanoscale Resolution Imaging of Whole Mouse Embryos Using Expansion Microscopy. ACS Nano https://doi.org/10.1021/acsnano.4c14791 (2025).
Steib, E. et al. TissUExM enables quantitative ultrastructural analysis in whole vertebrate embryos by expansion microscopy. Cell Rep. Methods 2, 100311 (2022).
Amrutha, N. Microwaves: A revolution in histoprocessing. J. Contemp. Dental Pract. 15, 149–152 (2014).
Boon, M. E. & Kok, L. P. Microwaves for immunohistochemistry. Micron 25, 151–170 (1994).
Cuevas, E. C. et al. Microwave antigen retrieval in immunocytochemistry: a study of 80 antibodies. J. Clin. Pathol.47, 448–452 (1994).
Jackson, P., Lalani, E. N. & Boutsen, J. Microwave-stimulated immunogold silver staining. Histochem. J. 20-20, 353–358 (1988).
Hougaard, D. M., Larsson, L. I. & Tornehave, D. Microwaving for double indirect immunofluorescence with primary antibodies from the same species and for staining of mouse tissues with mouse monoclonal antibodies. Histochem. Cell Biol. 113, 0019–0023 (2000).
Leong, A. S. Y. & Sormunen, R. T. Microwave procedures for electron microscopy and resin-embedded sections. Micron 29, 397–409 (1998).
Mayers, C. P. Histological fixation by microwave heating. J. Clin. Pathol. 23, 273–275 (1970).
Leong, A. S. Y. Microwaves and turnaround times in histoprocessing. Am. J. Clin. Pathol. 121, 460–462 (2004).
Horscroft, N. J. & Roy, P. Thermal denaturation of proteins for SDS-PAGE analysis by microwave irradiation. Biotechniques 22, 224–226 (1997).
Cook, J. P., Goodall, G. W., Khutoryanskaya, O. V. & Khutoryanskiy, V. V. Microwave‐assisted hydrogel synthesis: A new method for crosslinking polymers in aqueous solutions. Macromol. Rapid Commun. 33, 332–336 (2012).
Muñoz, T. E., Giberson, R. T., Demaree, R. & Day, J. R. Microwave-assisted immunostaining: a new approach yields fast and consistent results. J. Neurosci. Methods 137, 133–139 (2004).
Tillberg, P. W. et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34, 987–992 (2016).
Ku, T. et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 34, 973–981 (2016).
Park, H. E. et al. Scalable and isotropic expansion of tissues with simply tunable expansion ratio. Adv. Sci. 6, 1901673 (2019).
Truckenbrodt, S. Expansion microscopy: Super-resolution imaging with hydrogels. Anal. Chem. 95, 3–32 (2023).
Zhou, Y. & Jin, L. Hydrolysis-induced large swelling of polyacrylamide hydrogels. Soft Matter 16, 5740–5749 (2020).
Hussam-Aldeen Kalaleh, M. T., Yomen Atassi Preparation of poly(sodium acrylate-co-acrylamide) superabsorbent copolymer via alkaline hydrolysis of acrylamide using microwave irradiation. Preprint at https://doi.org/10.48550/arXiv.1502.03639 (2015).
Xiao, N., Chen, Y., Chen, A. & Feng, L. Enhanced bio-hydrogen production from protein wastewater by altering protein structure and amino acids acidification Type. Sci. Rep. 4, 3992 (2014).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Valdes, P. A. et al. Improved immunostaining of nanostructures and cells in human brain specimens through expansion-mediated protein decrowding. Sci. Transl. Med. 16, eabo0049 (2024).
Lai, H. M. et al. Rationalisation and validation of an acrylamide-free procedure in three-dimensional histological imaging. PLOS ONE 11, e0158628 (2016).
Katoh, K. Microwave-assisted tissue preparation for rapid fixation, decalcification, antigen retrieval, cryosectioning, and immunostaining. Int. J. Cell Biol. 2016, 1–9 (2016).
Sheard, T. M. D. et al. Differential labelling of human sub-cellular compartments with fluorescent dye esters and expansion microscopy. Nanoscale 15, 18489–18499 (2023).
Lee, J. H. et al. Rapid and facile microwave‐assisted surface chemistry forfunctionalized microarray slides. Adv. Funct. Mater. 22, 872–878 (2012).
Hoffman, E. A., Frey, B. L., Smith, L. M. & Auble, D. T. Formaldehyde crosslinking: A tool for the study of chromatin complexes. J. Biol. Chem. 290, 26404–26411 (2015).
Leong, A. S. Y. Microwaves in diagnostic immunohistochemistry. Eur. J. Morphol. 34, 381–384 (1996).
Katoh, K. Rapid fixation and immunofluorescent staining of cultured cells using microwave irradiation. J. Histotechnol. 34, 29–34 (2011).
Zhao, W. et al. Sparse deconvolution improves the resolution of live-cell super-resolution fluorescence microscopy. Nat. Biotechnol. 40, 606–617 (2022).
Bucur, O. & Zhao, Y. Nanoscale Imaging of Kidney Glomeruli Using Expansion Pathology. Front. Med. 5, 00322 (2018).
Zwettler, F. U. et al. Molecular resolution imaging by post-labeling expansion single-molecule localization microscopy (Ex-SMLM). Nat. Commun. 11, 3388 (2020).
Thevathasan, J. V. et al. Nuclear pores as versatile reference standards for quantitative superresolution microscopy. Nat. Methods 16, 1045–1053 (2019).
Chiu, M. C. et al. A bipotential organoid model of respiratory epithelium recapitulates high infectivity of SARS-CoV-2 Omicron variant. Cell Discov. 8, 57 (2022).
Chozinski, T. J. et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 13, 485–488 (2016).
Acknowledgements
This work is supported by the Hong Kong Research Grant Council General Research Fund (17102722, 17300523), and National Natural Science Foundation of China (32271445) to H.J. We thank the Imaging and Flow Cytometry Core at the Center for PanorOmic Sciences (CPOS) at HKU for the technical support in imaging acquisition. The work was conducted in the JC STEM Lab of Molecular Imaging, funded by The Hong Kong Jockey Club Charities Trust.
Author information
Authors and Affiliations
Contributions
H.J. and J.G. designed the experiments and wrote the paper. H.J., J.G., H.Y., C.L. (Chixiang Lu), D.C., M.Z., W.C., Q.Y., Z.L., M.C., S.Z., and J.H. performed cell and tissue-related experiments. C.L. (Cun Li) and J.Z. prepared organoid samples. All analyzed and assembled the results. HJ secured funding. All have commented on and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interest(s): Authors H.J. and J.G. are inventors of two pending patent applications covering the expansion method. Other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Guo, J., Yang, H., Lu, C. et al. BOOST: a robust ten-fold expansion method on hour-scale. Nat Commun 16, 2107 (2025). https://doi.org/10.1038/s41467-025-57350-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-57350-3
This article is cited by
-
Comparative analysis of algorithmic approaches in ensemble learning: bagging vs. boosting
Scientific Reports (2025)