Abstract
Putting pancreatic adenocarcinoma (PAAD) screening into perspective for high-risk individuals could significantly reduce cancer morbidity and mortality. Previous studies have profiled somatic mutations in PAAD. In contrast, the prevalence of mutations in PAAD predisposition genes has not been defined, especially in the Asian population. Using a multi-tier cohort design and whole genome/exome sequencing, we create a comprehensive germline mutation map of PAAD in 1,123 Chinese cancer patients in comparison with 11 pan-ethnic studies. For well-known pathogenic/likely pathogenic germline variants, Chinese patients exhibit overlapping but distinct germline mutation patterns comparing with Western cohorts, highlighted by lower mutation rates in known PAAD genes including BRCA1, BRCA2, ATM, CDKN2A, and CHEK2, and distinct mutations in CFTR, RAD51D, FANCA, ERCC2, and GNAS exclusive to Chinese patients. CFTR emerges as a top candidate gene following loss of heterozygosity analysis. Using an integrative multi-omics and functional validation paradigm, we discover that deleterious variants of uncertain significance may compromise CFTR’s tumor suppressor function, and demonstrate the clinical relevance by using patient derived organoids for drug screen. Our multifaceted approach not only deepens the knowledge of population differences in PAAD germline mutations but also unveils potential avenues for targeted therapeutic interventions.
Similar content being viewed by others
Introduction
Pancreatic adenocarcinoma (PAAD) is a highly fatal cancer with increasing morbidity worldwide1. Several studies have characterized the landscape of genomic abnormalities in PAAD2,3,4,5 and identified KRAS mutations as the most dominant somatic mutations, accounting for at least 78% of cases. Unfortunately, current clinical molecularly stratified KRAS-targeted therapy mostly targets the KRAS G12C mutation, which make up a mere 1.3% of all PAAD patients. Excitingly, discovery of PAAD susceptibility genes has yielded substantial therapeutic improvements. For example, PAAD patients with specific BRAC1 or BRAC2 variants have benefited from poly adenosinediphosphate-ribose polymerase (PARP) inhibitors6,7, and anti-PD-1 antibodies have been found to be effective in patients with Lynch syndrome8,9. In a metastatic PAAD screening clinical trial10 involving 3,315 patients, 2.7% individuals harbored germline BRCA1/2 mutation, 33% of which can be targetable. Thus, identification of previously uncharacterized PAAD germline genes might lead to both mechanistic insights and potential therapeutic strategies for this devastating cancer.
Hereditary factors play a crucial role in the pathogenesis of PAAD11,12,13,14. There are indications of ancestral differences in frequencies and distribution of germline mutations15. It is worth noting that the majority of PAAD research to identify germline mutations has been performed in cohorts of European descent16: in Queensland Centre for Medical Genomics (QCMG) cohorts3 and TCGA cohorts17, Asian patients represent only 4% and 6%, respectively. Thus, it is critical to extend germline studies to individuals from diverse ancestries. Additionally, previous studies have mostly focused on the known pathogenic genes, including BRCA1, BRCA2, ATM, PALB2, CDKN2A, MLH1, MSH2, STK11 and TP5318,19,20,21. However, much less effort has been devoted to the investigation of variants of uncertain significance (VUS), which constitute a large fraction of PAAD associated genetic variations. Systematic analyses of VUS in a large cohort using multi-omics approaches can reveal relevant function of specific genes in PAAD and their potential clinical significance. For example, variant allele frequencies (VAFs) obtained by sequencing matched tumor and normal samples can verify whether the germline variation of tumor suppressor gene is prone to loss of heterozygosity (LOH)17, and gene expression analysis can confirm whether germline truncation mutations lead to reduced expression. In addition, identifying PAAD susceptibility genes has substantial clinical implications6,7,8,9. However, most susceptibility genes identified so far do not yet have specific targeted therapies available.
Here, we define a paradigm to discover, validate, and functionally characterize pathogenic germline variations in PAAD. We aimed to assess the prevalence of deleterious germline variants in PAAD patients in Chinese population. Our study defined the landscape of pathogenic, likely pathogenic and VUS variants from 1,123 PAAD patients including discovery and validation cohorts, followed by functional genomics characterization and validation using tumor samples, cell lines, and patient derived organoids (Supplementary Data 1). The study design is presented in Supplementary Fig. 1. We further characterized one of our top candidate genes, CFTR, and revealed that identified germline mutations associated PAAD may impair CFTR’s role as a tumor suppressor gene. Overall, our study provided a valuable resource for studying PAAD in Chinese patients.
Results
Germline P/LP/VUS_D mutation landscape in Chinese PAAD patients
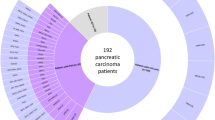
We applied three types of next-generation sequencing (NGS) of DNA to identify genomic mutations: (1) whole-genome sequencing (WGS) of 108 paired tumor-normal bulk samples; and (2) whole-exome sequencing (WES) of 281 paired samples. A total of 389 patients were incorporated into discovery cohort (denoted as the CH cohort); (3) targeted sequencing of 665 tumor-normal samples as a validation cohort; (4) WGS of 69 organoids with paired blood samples as functional validation cohort. We identified genome-wide germline mutations by conducting comparative analyses with a control group of Chinese reference cohort (n = 853) (Supplementary Data 2). A total of 6,423,173 SNVs and 145,652 indels were detected in CH cohort. Of these variants, 1,616 were considered pathogenic (P) or likely pathogenic (LP) according to ClinVar22 and InterVar23 based in-house pipeline. Out of 200,229 VUS variants, 6,791 were predicted to be damaging VUS (VUS_D) using our pipeline (Supplementary Data 3, Supplementary Fig. 2). Overall, 98% (383/389) of patients had at least one germline mutation classified as deleterious (P, LP, VUS_D) (Supplementary Data 3). In 150-curated cancer susceptibility genes (CSGs) (Supplementary Data 4), forty-eight mutated genes (identified in at least two patients) linked to hereditary pancreatic cancer risk, including genes involved in hereditary pancreatitis (CFTR, CASR, CPB1 and CTRC), homologous recombination repair (BRCA1, BRCA2, PALB2, NBN, BARD1 and RAD51D) and DNA mismatch repair (MSH2, MSH3, MSH6, MLH1, MSH4, MLH3 and PMS1) pathways, were detected among 123 patients (Fig. 1). The most frequently mutated genes in the CH cohort were CFTR (3.6%), BRCA1(1.8%), MSH2 (1.8%), BRCA2 (1.5%), MSH3 (1.5%), MSH6 (1.3%), PALB2 (1.3%), TSC2 (1.3%), EPCAM (1.0%), FANCA (1.0%) and GNAS (1.0%). About 86.3% of the deleterious variants in BRCA1, BRCA2 and ATM in cases were singleton, and the most frequent variant was p.P68L in BRCA1 (n = 3 in discovery cohort, n = 2 in validation cohort, and undetected in Chinese control cohort) (Supplementary Fig. 3, Supplementary Data 3 and 5). Sanger sequencing validated 100% (7/7) of the germline mutations (Supplementary Fig. 4a), confirming the accuracy of genotyping. Clinically, the proportion of PAAD patients with at least one first- or second-degree relative with a history of cancer was significantly higher in carriers of deleterious variants than in the non-carriers (27.6% vs. 15.7%, P = 0.002) (Supplementary Table 1). This suggests that deleterious cancer-predisposing variants might predispose individuals to PAAD and contribute to familiar clustering24. Moreover, the proportion of PAAD patients with a history of other cancers (history of prior malignancy) was higher in carriers of deleterious variants than in the non-carriers (10.5% vs. 4.6%, P = 0.015) (Supplementary Table 1), suggesting that the presence of deleterious germline variations might also increase the incidence of other cancers (most frequent in lung cancer, liver cancer, gastroesophageal cancer, and colorectal cancer).
Four cohorts were included in this study. The discovery cohort was consisted of 108 Whole Genome Sequencing (WGS) samples and 281 Whole Exome Sequencing (WES) samples. The validation cohort included two panels of a total of 665 tumor samples in which targeted sequencing was performed. A functional validation cohort was comprised of 69 patient-derived organoids. A germline reference cohort contained 853 healthy volunteers. A total of 867 million germline variants were called from WGS bam files. From WES bam files, 1.07 million germline variants were called. After in-house bioinformatics analysis, 210 pathogenic, 1,406 likely pathogenic, and 6,791 VUS_D variants were used for downstream analyses. Each column in the Oncoprint represents an individual tumor (n = 122) that harbors germline mutations. The top panel displays clinical features including patients’ sex, age, smoking history, alcohol use, histology, grading, and tumor stage, with a color key for each feature. The middle panel showcases the pathogenic, likely pathogenic, and VUS_D germline mutations in the 48 cancer predisposing genes within the discovery cohort. The bottom panel displays the somatic mutations in 4 important PAAD-related genes. The right panel displays the frequency of germline mutations in each cohort, including the discovery cohort, validation cohort 1 (PANEL 1), validation cohort 2 (PANEL 2), and functional validation cohort. For somatic mutation, sequences with an average depth of <50× were labeled as the “X” symbol. P, pathogenic; LP, likely pathogenic; VUS_D, variants of uncertain significance_damaging; ASC, adenosquamous carcinoma; IPMN, intraductal papillary mucinous neoplasms; PDAC, pancreatic ductal adenocarcinoma; WT, wide type; HRR, homologous recombination repair; MMR, mismatch repair; DDR, DNA damage repair.
Well-known germline P/LP landscape between Asian and Western PAADs
To investigate whether germline mutational landscape differs between Western and Asian pancreatic cancer population, we compared P/LP germline mutations in CSGs between seven Western cohorts17,24,25,26,27,28,29 and four Asian studies29,30,31. BRCA1 (75/6,540 vs. 16/3,353, P = 7.52e-4, Fisher’s exact test), BRCA2 (198/6,540 vs. 49/3,353, P = 1.11e-06, Fisher’s exact test), ATM (87/6,540 vs. 28/3,353, P = 0.029, Fisher’s exact test), CDKN2A (13/6,540 vs. 1/3,353, P = 0.044, Fisher’s exact test) and CHEK2 (48/6,540 vs.1/3,353, P = 5.49e-08, Fisher’s exact test) germline mutations were significantly more common in Western cohorts than in Asian cohorts (Fig. 2a). Based on data from Polo III studies29, Asians living in Western countries exhibited higher germline mutation frequencies of BRCA1 and BRCA2 comparing with other Asians. For BRCA2, we identified a total of 159 mutations in CH cohort; 113 of which were also found in Western cohorts and 52 in Asian cohorts (Fig. 2b). Interestingly, only 3.8% (6/159) of the mutations we identified in the CH cohort were previously reported, including p.I605fs in 2018 JAMA cohort, 2020 POLO III cohort, 2020 Japanese cohort; p.K437fs in 2020 POLO III cohort, 2022 Nanjing Cohort; p.L1908fs in 2020 POLO III cohort, 2022 Nanjing Cohort; p.N2135fs in 2020 POLO III cohort, 2022 Nanjing Cohort; p.S1882X in 2018 JAMA cohort, 2020 POLO III cohort, 2022 Nanjing Cohort and p.V1283fs in 2018 JAMA cohort, 2020 POLO III cohort, 2022 Nanjing Cohort (Fig. 2b, Supplementary Data 6). Importantly, mutations in five genes, i.e. CFTR, RAD51D, FANCA, ERCC2 and GNAS were exclusively identified in Asian patients (0/6,540 vs. 15/3,353, P = 8.76e-08, Fisher’s exact test; 0/6,540 vs. 6/3,353, P = 0.002, Fisher’s exact test; 1/6,540 vs. 4/3,353, P = 0.048, Fisher’s exact test; 0/6,540 vs. 4/3,353, P = 0.013, Fisher’s exact test; 0/6,540 vs. 3/3,353, P = 0.039, Fisher’s exact test; 0/65,40 vs. 21/3,353, P = 1.30e-10, Fisher’s exact test) (Fig. 2a).
a Bubble plot displays frequency of rare germline P/LP mutation as a percentage of cases in each cohort. The genes labeled in red are those with statistically significant differences in mutation frequency between Western and Asian cohorts. b Mirror distribution plot of BRCA2 germline P/LP mutations between Western and Asian cohorts. HRR, homologous recombination repair; MMR, mismatch repair; DDR, DNA damage repair.
Germline mutation and LOH events
To investigate the interplay between germline and somatic mutation profiles and to discover potential genes associated with PAAD, we conducted an analysis of LOH in paired normal and tumor samples from 458 PAAD patients (Supplementary Data 1). A total of 26 cancer-associated genes demonstrated LOH of the wild-type allele in 7.6% (35/458) patients (Fig. 3a–c, Supplementary Fig. 4b). Consistent with expectations, LOH was present in 2 of 7 (29%) and 1 of 7 (14%) of the of BRCA1 and BRCA2 genes, respectively. Other genes exhibiting variants with LOH include: MSH2, MSH3 and MSH6, all mismatch repair initiation factors that recognize DNA lesions (4 of 21, 19%); PALB2, ERCC2 and NBN, genes known to be activated by double-strand breaks (4 of 11, 36%); FAT1 and FH, transcriptional repressors involved in BRCA1- mediated cell growth suppression (2 of 8, 25%). WRN is also a highly likely LOH candidate. In total, 34 variants showed significant LOH. It is worth noting that CFTR emerges as a top candidate, as we detected five LOH events (5 of 18, 28%) of CFTR involving p.P140S, p.R279Q, p.E681V, p.C1355Y and p.S1456R. These five variants were located in transmembrane domain 1 (TMD1), regulatory domain (R), nucleotide-binding domain 2 (NBD2) and carboxy-terminal cytoplasmic tail of the protein (Fig. 3d). All LOH events were manually confirmed using the IGV Browser (Fig. 3e, Supplementary Fig. 4c–g).
a Identification of LOH in tumor suppressor genes through comparison of VAFs in tumor and normal samples. The diagonal line denotes neutral selection of the germline variant where the normal and tumor VAFs are identical. b Somatic copy number changes of genes detected for the tumors based on LOH analysis classification. Significant (n = 3), suggestive (n = 18), and no evidence (n = 165) of LOH are shown in red, green, and blue, respectively. Statistical significance was computed by two-sided Wilcoxon test. Boxplot shows the median (central line). c Counts of germline variants showing the various types of classified LOH. d A schematic representation of the CFTR protein integrated into the cell membrane, highlighting key features. Germline mutations identified in this study are depicted as stars. The LOH of CFTR variants is highlighted in bold. e IGV view of CFTR p.E681V mutation. The depths of mutation reads and wild type reads are displayed in red and green, respectively. The left panel illustrates a VAF of 0.44 in the germline, indicating a heterozygous mutation. The middle panel reveals LOH in the tumor, evidenced by an increased VAF of 0.72. In the right panel, RNA sequencing of the same patient reveals the presence of only the mutant transcript. f The mirror distribution plot illustrates the distribution of CFTR germline mutations. g Percentage of patients with at least one of the 18 germline mutations in a cohort of 9,253 pan-cancer patients. h Heatmap showing the percentage of specific CFTR mutations in pan-cancer and in individual cancer types. Values of mutation frequencies are included in Supplementary Data 9. TSG, tumor suppressor gene; LOH, loss of heterozygosity; VAF, variant allele frequency; MSD, membrane-spanning domain; NBD, nucleotide-binding domain; RD, regulatory domain; IGV, Integrative Genomics Viewer; VUS_D, variants of uncertain significance_damaging; PAAD, pancreatic adenocarcinoma; UCC, uterine cervical cancer; BRCA, breast invasive carcinoma; ESCA, esophageal carcinoma; OV, ovarian serous cystadenocarcinoma; LIHC, liver hepatocellular carcinoma; SCLC, small-cell lung cancer; STAD, stomach adenocarcinoma; COAD, colon adenocarcinoma; ICC, intrahepatic cholangiocarcinoma; NSCLC, non-small-cell lung cancer; EHCC, extrahepatic cholangiocarcinoma; UC, urothelial carcinoma.
Functional validation of CFTR VUS_D variants
VUS poses a daunting challenge, especially in under-represented populations where the prevalence of VUS is higher due to their lack of representation in existing public databases. In-house pipeline classified a total of 6791 variants as VUS_D. CFTR variants were present at the highest frequency in our cohorts, around 4.2% (35/842, in discovery, validation 2 and functional validation cohorts), consisting of 15 VUS_D variants (15 out of 18) (Fig. 3f, Supplementary Data 7). We compared these CFTR VUS_D variants with those in the CFTR2 database (http://cftr2.org), which collects mutations in the CFTR gene for the international cystic fibrosis (CF) genetics research community. Among the aforementioned mutations, p.L88X, p.G970D, p.R74W, and p.V201M have been previously reported in relation to CF32. Additionally, p.G451R, and p.G970D33 have been previously associated with pancreatitis. Notably, compared to CF, 72% (13 out of 18) of the mutations identified in our study were not linked to PAAD until now (Supplementary Data 7). CFTR p.F508del, the most frequent mutation in Western populations34, has been associated with increased risk of developing young-onset pancreatic cancer35. However, this mutation was not detected in our study or in another Asian pancreatic cancer cohort36. In Nanjing cohort30, four CFTR mutations, p.R74W, p.G622D, p.G970D, p.R1070Q, and one splice mutation, were reported. Out of the four mutations, p.R74W and p.G970D also existed in our cohort. The CFTR mutations (p.R74W, p.P140S, p.G451R, p.E681V, p.G970D, p.R1097C) were found in 2.7% (23/853) of individuals from the PAAD case-control study in Asians (Supplementary Data 2). Overall, deleterious variants rate of CFTR was higher in PAAD patients compared with control cohort (3.9% vs. 1.4%, P = 0.001, Supplementary Data 8). We further analyzed the differences in clinical characteristics between deleterious CFTR carriers and non-carriers among the 842 pancreatic cancer patients. We found that patients with deleterious CFTR mutations had a higher proportion of family cancer history, history of prior malignancy and history of chronic pancreatitis (42.4% vs. 24.3%, P = 0.019; 15.2% vs. 5.8%, P = 0.030; 18.2% vs. 3.0%, P < 0.001; Supplementary Table 1). Furthermore, the proportion of female patients with CFTR deleterious mutations was higher than that of patients without CFTR deleterious mutations (57.6% vs. 38.4%, P = 0.027, Supplementary Table 1). To investigate whether germline CFTR mutations are more prevalent in Chinese PAAD or pan-cancer patients, we further collected 9,253 tumor samples across 13 cancer types. Notably, CFTR germline mutations were associated with a higher frequency in uterine cervical cancer (UCC), uterine corpus endometrial carcinoma (UCEC) and breast invasive carcinoma (BRCA), which is consistent with their association with female-specific cancers (Fig. 3g). CFTR p.E681V, p.R74W, p.R1453W and p.G970D are hotspots in pan-cancer cohort. In particular, CFTR p.E681V was found in all cancers except bladder urothelial cancers (Fig. 3h, Supplementary Data 9).
CFTR as a converging genetic and epigenetic driver of PAAD
CFTR exhibited lower expression in both discovery cohort tumor samples and public tumor samples37 comparing to matched normal samples (Fig. 4a). Significantly lower CFTR expression in tumor was independently confirmed in organoid validation cohort as well as in a publicly available laser capture microdissection (LCM) dataset38 (Fig. 4b). Moreover, in organoids, the CFTR carriers displayed even lower expression compared with CFTR wild type (WT) samples (Fig. 4c). To evaluate the expression of the CFTR protein, immunohistochemical (IHC) staining was performed on an additional 200 specimens of PAAD. We found that the expression of CFTR in normal ductal cells was significantly higher than that in tumor cells (Fig. 4d). Interestingly, CFTR expression in ductal cells of chronic pancreatitis was also significantly higher than in tumor cells (Fig. 4d). The difference in the expression of CFTR between pancreatic cancer and para-cancerous tissues was consistent with the difference at the transcriptional level, and the difference was more statistically significant in protein, possibly due to the better delineation of the tumor and normal glands (Supplementary Fig. 5). Although no statistically significant difference in clinical outcomes was found in the CH cohort based on CFTR RNA expression, the analysis of bulk samples based on CFTR protein expression, the organoid cohort and the public LCM cohort (GSE17891)39 indicated an association between low CFTR expression and poor prognosis (Fig. 4e).
a CFTR expression between matched normal and tumor samples in the discovery cohort and a public cohort. b CFTR expression in the organoid and a public LCM cohort. c Lollipop graph and box plot were used to visualize trimmed mean of M-values of CFTR in individual samples. d CFTR opacity density in normal pancreatic tissue, chronic pancreatitis tissues, and PAAD. Representative immunohistochemistry slides are shown at 20 × magnification (scale bar = 50 μm). e Kaplan-Meier curves depicting overall survival were stratified based on CFTR expression. Significance was calculated using log-rank Mantel-Cox test. f CFTR expression in individual organoids were displayed in the barchat. DNA methylation status of the CFTR promoter in individual organoids was illustrated by the DNA methylation specific PCR gel. The names of organoid with high, low and intermediate expression of CFTR were highlighted in red, blue and green, respectively. CFPAC-1, a PAAD cell line carrying the CFTR p.F508del mutation, shows unmethylated of CFTR promoter. ddH2O is control. Three times each experiment was repeated independently with similar results. g A schematic graph illustrating the genetic and epigenetic mechanisms contributing to the silencing of CFTR. The red box represents the heterozygous germline mutation present in normal cells. In cancer cells, due to copy number variation loss of the wild-type CFTR allele, only one mutant CFTR allele remains. Epigenetically, cancer cells exhibit DNA methylation of the CFTR gene, resulting in gene silencing. The downstream RNA and protein expression effects, coupled with the emerging prognostic findings, indicate CFTR’s role as a tumor suppressor gene. Statistical significance was computed by two-sided Wilcoxon test in (a–d). Bar represents the SD with maxima (upper bar) indicated. Box indicates the 25th to 75th percentile values, the line indicates the median, with the lower whiskers representing the minimum 25% of data values and the whiskers above the box representing the 25% maximum values. Source data are available as a Source Data file. LCM, laser capture microdissection; NPT, normal pancreatic tissue; CPT, chronic pancreatitis tissues; PAAD, pancreatic adenocarcinoma; IHC, immunohistochemistry.
Surprisingly, we noticed that CFTR was downregulated in a subset of tumors without germline mutations, suggesting that other mechanisms may also contribute to the reduced expression of CFTR in PAAD, underscoring the functional relevance of CFTR expression. To investigate whether the epigenetic changes play a role in the changes of CFTR gene expression, we performed an integrative analysis of ATAC-seq and RNA-seq. We discovered a significant positive correlation between differential gene expression and differential chromatin accessibility signal in the CFTR gene promoter and enhancers40 (R = 0.46; R = 0.51; R = 0.65; Supplementary Fig. 6). When samples were classified based on expression levels of CFTR, we observed that the chromatin accessibility of CFTR promoter and two upstream enhancers was higher in the CFTR-high group than in the CFTR-low group. Furthermore, using DNA methylation-specific PCR, we demonstrated that the promoter region of the CFTR gene was methylated in most of the low-expression samples (Fig. 4f). Taken together, the convergence of genetic and epigenetic data strongly supports that CFTR has the characteristics of a tumor suppressor gene (Fig. 4g).
CFTR is a putative tumor suppressor gene in PAAD
CFTR gene encodes a membrane scaffold protein that forms the anion channel in vertebrates41. Recent studies have suggested CFTR germline mutation is associated with pancreatitis and pancreatic cancers33,42. However, CFTR alterations have conflicting interpretations of pathogenicity and the biological functions of CFTR alterations are still largely unknown in PAAD42,43,44,45,46. The result of Supplementary Fig. 7a showed that the majority of PAAD cell lines express extremely low level of CFTR in CCLE datasets47,48. We selected two PAAD cell lines, PANC-1 and AsPC-1, both lack CFTR expression, to investigate whether up-regulating CFTR expression would influence classical cellular function of PAAD cells. In both cell lines, CFTR over expression (Fig. 5a, b, Supplementary Fig. 7b–d) significantly decreased cell viability (Fig. 5c, d, Supplementary Fig. 7e, f), proliferation (Fig. 5e, Supplementary Fig. 7g–i), migration and invasion rates in vitro (Fig. 5f, Supplementary Fig. 7j), and enhanced apoptosis (Fig. 5g, Supplementary Fig. 7k, 8a and b) in vitro. By contrast, we knocked down CFTR levels in two other cell lines, Capan-1 and SU.86.86, by using two different shRNAs (Supplementary Fig. 9a). Downregulation of CFTR significantly promoted the capability of colony formation and cell viability for both cell lines (Supplementary Fig. 9b–d), as well as increased proliferation (Supplementary Fig. 9e–h) and decreased apoptosis (Supplementary Fig. 9i, j).
a, b PANC-1 cells transduced with lentivirus carrying wild-type CFTR or CFTR mutants were cultured for 2 weeks. CFTR RNA expression levels (a) were examined using qPCR (n = 3 technical replicates/group). CFTR protein and FLAG tags levels (b) were examined using Western blot. c–e The proliferation of PANC-1 cells were assessed using CCK-8 method (c, n = 5 technical replicates/group), colony formation assay (d, n = 3 technical replicates/group) and EdU incorporation assays (e, n = 6 technical replicates/group). f The migration and invasion of PANC-1 cells were measured following CFTR overexpression using the Transwell assay (Magnification: ×200, scale bar = 200 μm) (n = 9 technical replicates/group). g The apoptosis of PANC-1 cells using Caspase 3/7 assay (n = 3 technical replicates/group). h Tumor growth was measured at day 22 of inoculation (n = 5 biological replicates/group). In the accompanying photograph of mice bearing xenograft tumors, the tumors’ locations are indicated by arrows. Statistical significance was assessed by two-sided t-test. i The immunofluorescence staining of CFTR in PANC-1. Magnification: ×630, scale bar = 20 μm. Five times each experiment was repeated independently with similar results. j Western blot of the cell fractionation experiments. CFTR were detected in membrane fractions in PANC-1 CFTR OE, p.R31C, p.E681V and p.C1355Y. Three times each experiment was repeated independently with similar results. k Gene ontology analysis by comparing the EV and CFTR OE conditions. l Heatmap visualizations of pathway based on ssGSEA analysis for EV, p.L88X, p.C1355Y, and OE, showing comparisons between each mutant and the CFTR OE. Statistical analysis was performed using one-way ANOVA other than (h); multiple comparisons were performed according to Tukey’s method. Data were presented as mean ± standard deviation (SD). Bars represent the SD with minima (lower bar) and maxima (upper bar) indicated. P-values for comparisons with EV are indicated by asterisks, with OE are indicated by pound signs. ns: not significant; */#P < 0.05; **/##P < 0.01; ***/###P < 0.001; ****/####P < 0.0001. Statistical significance among PANC-1 cells comparisons is tabulated in Supplementary Table 2. Source data are available as a Source Data file.
To further investigate the impact of germline mutation of CFTR in pancreatic cell lines, we expressed 9 CFTR mutants in PANC-1 cell line and AsPC-1, including 3 VUS_ND (p.R31C, p.E217G and p.Q1352H), 1 pathogenic mutation (p.L88X) and 5 VUS_D mutation (p.L69F, p.E681V, p.R1097C, p.C1355Y, p.S1456R) (Fig. 5a, b, Supplementary Fig. 7b–d, Supplementary Fig. 10a). These specific mutations were chosen because they span across different domains of the CFTR protein. More than 2000 mutations in CFTR have been described (http://cftr2.org). They often interfere with protein biosynthetic and folding, leading to the rapid degradation of CFTR molecules that have not matured beyond the endoplasmic reticulum49. Six out of nine mutants showed reduced protein levels, with p.L88X representing a stop-codon gain mutation leading to missing expression of the protein (Fig. 5b, Supplementary Fig. 7b and d). Comparing with wild type CFTR (OE), four mutants (p.L69F, p.L88X, p.R1097C and p.C1355Y) were much less effective in decreasing cell viability in both PANC-1 and AsPC-1, shown by CCK-8 assay (Fig. 5c, Supplementary Fig. 7e, and 10b, c). Two mutants (p.L88X and p.C1355Y) were significantly less effective in reducing tumor colony formation in both cell lines (Fig. 5d, Supplementary Fig. 7f). Six mutants (p.L88X, p.E681V, p.R1097C, p.Q1352H, p.C1355Y, and p.S1456R) demonstrated significantly reduced efficacy in decreasing the proliferation of PANC-1 and AsPC-1 cells, as evidenced by the EdU incorporation assay (Fig. 5e, Supplementary Fig. 7g–i). Four mutants (p.L88X, p.E217G, p.E681V, and p.C1355Y) exhibited decreased apoptotic cell proportions compared to wild-type CFTR (OE) in both cell lines (Fig. 5g, Supplementary Fig. 7k, 8a, b). Next, we established a xenograft model to explore CFTR-mediated tumorigenesis in vivo. We found that tumor weight was dramatically decreased in the CFTR-expressing tumor-bearing mice in comparison with the EV control group (Fig. 5h). In addition, CFTR mutants (p.L88X and p.C1355Y) almost completely abolished the tumor suppressor effects compared with overexpression of wild type CFTR (Fig. 5h).
The wild type CFTR is normally localized to the cell membrane41. However, for certain CFTR mutants (p.C1355Y), although protein expression is observed, the protein fails to properly localize to the cell membrane (Fig. 5i, Supplementary Fig. 11). Compared to wild type CFTR (OE), less membrane CFTR expression was detected in CFTR p.C1355Y PANC-1 cell, using cell fractionation followed by western blot analysis (Fig. 5j). This aberrant localization is likely associated with downstream functional impairments of CFTR.
We utilized our WGS or WES data to interrogate the mutational signatures of the CFTR mutant cohort to determine if there is any difference in mutational signature. We used two methods to analyze mutation signatures: refitting of well-known mutational signatures and de novo extraction. These analyses did not reveal any statistically significant differences in the prevalence or patterns of mutational signatures (Supplementary Fig. 12). RNA-seq analysis in CFTR OE, EV, p.L88X, and p.C1355Y of PANC-1 and AsPC-1 reveals potential different mechanisms regulating cancer cellular behavior (Fig. 5k, l, Supplementary Fig. 13a–d). From three previous studies50,51,52, we collected differentially expressed genes (DEGs) from Cftr-deficient endothelial cells, colon, and small intestine of murine models. We compared these genes with DEGs we identified using our organoid system by comparing CFTR mutant and wild-type. We identified 8 upregulated and 10 downregulated genes shared between these studies which are associated with oncogenic and inflammatory pathways (Supplementary Fig. 14). It is worth noting that the overlap was small. We speculate that this may be due to the differences between the systems (mouse models vs. human tumor organoid), but it might also reflect the complex interplay between CFTR, different CFTR mutants, and immune regulation. Although it is clear that CFTR mutations have an impact on immune regulation, the nature and precise mechanism of this impact will need additional experimentation to elucidate.
Extending our cell line based analysis, we examined pathways associated with CFTR expression using both discovery cohort of 108 patients (Supplementary Fig. 15a) as well as TCGA data (Supplementary Fig. 15b) by performing ssGSEA analysis. We found a positive correlation between CFTR expression and pathways such as pancreatic beta-cell secretion (Supplementary Fig. 15c). We also found a negative correlation between CFTR expression and certain oncogenic pathways, including the Tumor Proliferation Rate pathway (Supplementary Fig. 15d). Analysis of TCGA data revealed a similar pattern of pathway enrichment associated with CFTR expression (Supplementary Fig. 15e, f).
To explore CFTR variants’ role in known CFTR downstream effects, we conducted chloride channel function and lipidomics analysis. The relative chloride flux was significantly reduced in the PANC-1 cells expressing CFTR variants compared to the wild-type cells. These findings suggest that these nine CFTR mutations impair chloride channel function in the PANC-1 cell line (Supplementary Fig. 16). Fatty acid metabolism plays a pivotal role in tumor development and progression53. Alpha-linolenic acid (ALA, an omega-3 fatty acid) and linoleic acid (an omega-6 fatty acid) are essential polyunsaturated fatty acids (PUFAs) that function as direct signaling molecules or precursors for other omega-3 and omega-6 PUFAs. An elevated omega-6 to omega-3 PUFA ratio serves as an inflammatory biomarker in tumor tissues of metastatic colorectal cancer patients54. Three PANC-1 cells carrying CFTR mutations (p.L88X, p.E681V, p.C1355Y) showed a significantly higher ratio of FA 20:4 (omega-6 PUFA) and FA 22:6 (omega-3 PUFA) compared to the CFTR WT PANC-1 cells (Supplementary Fig. 17a, Supplementary Data 12), which suggests that CFTR mutations may influence fatty acid metabolism or composition in cells. We have identified a subset of arachidonoyl species and docosahexaenoyl species. Among the arachidonoyl species, the compounds PC16:0_20:4 and PC18:0_20:4 showed higher normalized intensities in CFTR-mutated PANC-1 cells compared to CFTR OE. The compound PS18:0_20:4 exhibited a difference only between CFTR p.L88X PANC-1 cells and CFTR OE. The compounds PI16:0_20:4 and PI18:0_20:4 showed differences comparing CFTR p.L88X and p.C1355Y cells with CFTR OE. In the docosahexaenoyl species, the compounds PC18:0_22:6 and PI18:0_22:6 displayed higher normalized intensities in CFTR-mutated PANC-1 cells compared to CFTR OE. The compound PC16:0_22:6 exhibited differences comparing CFTR p.L88X and p.C1355Y cells with CFTR OE (Supplementary Fig. 17b, Supplementary Data 12). These findings aligned well with previous studies in CF mice, suggesting that CFTR mutations may affect specific phospholipid species and their concentrations55. In addition to confirming some of the reported differences, we also made several additional discoveries. We identified 2 OxPE species (PE 18:1_18:0;O, PE 18:0_20:4;O), 1 OxPC species (PC 16:0_18:1;O), and 1 OxPG species (PG 18:0_18:1;3 O). PE 18:1_18:0;O, PC 16:0_18:1;O, and PG 18:0_18:1;3 O all exhibited lower normalized intensity in CFTR-mutant PANC-1 cells compared to CFTR overexpressing cells. PE 18:0_20:4;O also displayed lower normalized intensity in CFTR p.L88X and p.C1355Y mutant PANC-1 cells compared to CFTR overexpressing cells (Supplementary Fig. 17c, Supplementary Data 12). OxPLs are ubiquitous and are formed in many inflammatory tissues, where they frequently mediate proinflammatory changes. Since OxPLs are mostly products of non-enzymatic lipid peroxidation56, it has been identified that the oxidized phospholipid OxPLcan induce lung injury and cytokine production by lung macrophages via the TLR4-TRIF pathway57.
Drug screening for pathogenic germline mutations
To determine whether germline mutations in PAAD are associated with altered drug response, we conducted a drug sensitivity screen using patient-derived 3D-organoid (PDO) culture58 and estimated correlation between drug sensitivity and genomic features of the organoids (Fig. 6a). A bespoke 63 compound library was assembled for screening, including chemotherapy (n = 8) and targeted drugs, which were selected based on treatment regimens targeting cell cycle (n = 4), DNA damage (n = 5), classical cellular signaling cascades (n = 34), and other cellular aspects (n = 12) (Supplementary Data 10). The library included AMG510, which is used clinically for KRAS G12C PAAD, as well as Fluorouracil and Gemcitabine, the first line chemotherapeutics for PAAD treatment. We were able to successfully conduct drug screen for 46 of the 69 tumor organoids in replicates, generating >7000 measurements of organoid-drug interactions (Supplementary Data 10). As positive controls, organoid DAC102, DAC58, DAC81, DAC85 and DAC53 harboring BRCA2 somatic mutations, as well as organoid DAC4, bearing a BRCA2 germline mutation, displayed the most pronounced response to the PARP inhibitor (Olaparib) (Fig. 6b). Interestingly, two other organoids (DAC76, and DAC99) also demonstrated efficacy with Olaparib despite lacking the BRCA1/2 mutation, although DAC99 harbored an ATM somatic mutation. One organoid bearing the KRAS p.G12C mutation exhibited sensitivity to AMG510 and MRTX849 (Fig. 6b), both of which demonstrated promising anticancer effects in patients with advanced solid tumors harboring the KRAS p.G12C mutation59,60. These results underscore the utility of personalized therapeutic assessment through PDO drug screening. The clustering of compounds based on their AUC demonstrated a diverse range of sensitivities across the organoids and identified three major sub-groups (Fig. 6b). One group was associated with sensitivity to many compounds (organoids DAC76, DAC58, DAC78, DAC2, DAC16, DAC57, DAC37, DAC82, DAC61 and DAC4), in contrast to the cluster (DAC80, DAC67, DAC75, DAC25, DAC73, DAC13, DAC83, DAC20, DAC56, DAC54, DAC110, DAC32, DAC30, DAC31, DAC22, DAC35, DAC88 and DAC5) exhibiting broad drug resistance. The remaining organoids had intermediate sensitivity.
a Assay description for individual organoids. Each column represents a PAAD organoid, and blue indicates that sequencing experiment or drug sensitivity data are available whereas gray indicates the absence of data. b The treatment response of 46 pancreatic adenocarcinoma (PAAD) organoids to 63 different drugs. The colors from red to blue represent Z-scores based on AUC (Methods). A low z-score or blue indicates high sensitivity and red indicates low sensitivity. The germline mutation and somatic mutation information of the PAAD organoid are shown in the top panel. c The treatment response of 15 PAAD organoids to 15 CFTR-related drugs, including CFTR potentiators, inhibitors, and correctors. d Dose-response curves generated from 15 PAAD organoids treated with CFTR correctors. Curve for CFTR p.R297Q PODs is labeled in blue. Curves for CFTR p.E681V PODs are labeled in yellow. Curve for CFTR p.H949P PODs is labeled in purple. PODs with wild type CFTR are labeled gray. e Comparing sensitivity to CFTR correctors between organoids with wild type CFTR (n = 11) and with CFTR mutations (n = 4). Statistical significance was computed by two-sided Wilcoxon test. Box indicates the 25th to 75th percentile values, the line indicates the median, with the lower whiskers representing the minimum 25% of data values and the whiskers above the box representing the 25% maximum values. Source data are available as a Source Data file.
A notable portion of organoids, 39%, exhibited resistance to chemotherapy. Among them, DAC13, DAC73, DAC110, DAC32 and DAC30 displayed a complete lack of sensitivity to all chemotherapy agents, with an IC50 of 0.5 mM. Intriguingly, these organoids bear CFTR VUS_D germline mutations. Extensive research in CF has produced two types of CFTR modulators: potentiators and correctors61. Potentiators keep CFTR ion channel open, allowing chloride to flow through the cell membrane. Correctors, on the other hand, assist CFTR in adopting correct 3D conformation, thus enabling the protein to traffic to cell membrane. To evaluate the effectiveness of commercial modulators on PAAD with CFTR mutations, fifteen PDOs (4 with CFTR mutations: p.R297Q, p.E681V, p.E681V, p.H949P, and 11 CFTR WT) were screened against a panel of 15 compounds. This panel included four potentiators, six inhibitors and five correctors. A dose–response analysis with 7 or 8 data points per compound was conducted to determine the therapeutic response. DNA and RNA sequencing confirmed the mutation status and CFTR expression level of these organoids. Excitingly, five CFTR correctors exhibited greater activity against CFTR mutants compared with CFTR wide-type PDOs (Fig. 6c, d). Tezacaftor, Lumacaftor, FDL169, Galicaftor and Elexacaftor exhibited a significant difference in AUC between CFTR mutant and wild-type PDOs (P-values were 0.006, 0.005, 0.006, 0.016 and 0.002, respectively, Wilcoxon test). Our results demonstrated the ability of the organoid platform to recapitulate known drug sensitivities associated with specific genetic alterations and to explore candidate gene-targeted potential drugs of interest.
Discussion
In this study, we present a comprehensive germline mutation landscape of PAAD in Chinese patients as a resource. In contrast to using conventional gene panels30, by combining WGS, WES and gene panels across several independent discovery and validation cohorts, we identify 8,407 P, LP and VUS_D. By comparing across multiple Asian and Western cohorts, we substantiated the large race and ethnic associated variability previously described in germline mutation screening17,24,25,26,27,28,29,30,31 into the pancreatic cancer setting. The highest rate of germline BRCA mutation was observed in the 2020 POLO III African American (13.8%). More importantly, our integrated germline and somatic study identified CFTR, BRCA1, BRCA2, RAD51C, RAD51D, FANCM and MSH6 germline mutations harbor LOH events.
We functionally explored our top candidate gene, CFTR, which harbors well known pathogenic germline mutations related to CF. CFTR mutations have been implicated in pancreatitis and PAAD35,45,46,62. However, most previous studies used gene panels to test known CFTR mutations which were not characterized in pancreatic cancer. Profoundly, our study discovered previously unreported pathogenic CFTR germline mutations in Asian patients associated with PAAD. CFTR is a pleiotropic factor. Thus, different mutations might impair different aspects of CFTR’s biological functions, some of which may be tumor suppressing in nature. By integrating genetic data, gene expression, chromatin accessibility and DNA methylation data, we confirm that both genetic factors, particularly germline mutations, and epigenetic factors such as DNA methylation, collectively contribute to the downregulation of CFTR RNA and protein expression in PAAD. Importantly, reduced CFTR expression serves as a prognostic marker for PAAD. We further confirm CFTR’s tumor suppressor function by manipulating CFTR expression in PAAD cell lines. Increased CFTR expression results in inhibition of cell growth and proliferation, whereas depleting CFTR results in opposite phenotypes. Finally, we demonstrate the utility of patient derived organoid in testing and screening therapeutic regimens. For example, Olaparib has demonstrated promising efficacy in targeting HRR germline mutations6, and our organoid model confirmed these findings. Using our organoid model, we identified candidate drugs, specifically, CFTR correctors, that hold promise in treating PAAD with CFTR germline mutations. In summary, the integration of high throughput discovery, experimental validation and drug screening offers the promise to identify the most functionally and clinically relevant variants for cancer risk assessment.
Methods
Ethics statement
The study was conducted in accordance with the national guidelines and was approved by the Ethics Committee of Changhai Hospital (Approval number: CHEC2017-147). All patients provided written informed consent for the use of their clinical data and surgical specimens, and consent to publish potentially identifiable clinical information was obtained.
Study design (Patient cohorts and samples)
Patients in all cohorts were included or excluded based on following criteria. Inclusion criteria: a. Patients with pancreatic cancer who underwent surgical resection or endoscopic ultrasound-guided fine-needle aspiration/biopsy (EUS-FNA/B). b. Patients who provided informed consent for the collection of biological samples and sequencing analysis. Exclusion criteria: c. Patients with confirmed non-pancreatic ductal adenocarcinoma based on histopathology. d. Patients with corrupted or low quality NGS sequencing data. The discovery cohort encompassed 389 pancreatic cancer patients, who were subjected to WGS (n = 108) and WES (n = 281) using their tumor and paired blood/normal samples. The validation cohort contained 665 pancreatic cancer patients and underwent two independent targeted sequencing (281 patients with PANEL 1, and 384 patients with PANEL 2). A total of 69 PDOs were included as a functional validation cohort. Tumor samples were collected from pancreatic cancer patients who received surgical resection at Shanghai Changhai Hospital. Tissue parts distant from the tumor in surgical specimens were used as normal samples. Samples were pathologically confirmed as tumor or normal tissue. Clinical variables of patients were collected from the prospective database of Shanghai Changhai Hospital. Sex information was self-reported by all patients.
The control cohort comprised of blood WES sequencing data from 853 healthy adults, obtained from the State Key Laboratory of Computer Architecture, Institute of Computing Technology, Chinese Academy of Sciences, Beijing, China. Among these, data from 287 female participants have been previously disclosed63. Pan-cancer CFTR germline variants data were obtained from OrigiMed Co. Ltd (Shanghai, China).
Cancer susceptible genes (CSGs) collection and curated
We have expanded the count of CSGs, originally outlined by Rahman et al.64 through a comprehensive literature review, from an initial 114 to a final total of 150 (Supplementary Data 4).
DNA and RNA isolation, quantification, and qualification
Genomic DNA was extracted from fresh frozen tissue and whole blood samples using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. In addition, the DNA concentration was measured by Qubit® DNA Assay Kit with a Qubit 3.0 fluorometer (Invitrogen, Carlsbad, CA, USA).
Total RNA was isolated from tumor tissues and adjacent normal tissues using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and purified using the RNAeasy Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s protocols. RNA degradation and contamination were assessed through agarose gel electrophoresis. The purity of RNA was measured using NanoPhotometer® spectrophotometer (IMPLEN, CA, USA) and the integrity of RNA was confirmed using the RNA Nano 6000 Assay Kit I Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
WGS library preparation
A total of 0.2 μg of DNA was used as input material for DNA library preparation per sample. The library was prepared using the NEB Next® Ultra™ DNA Library Prep Kit for Illumina (NEB, USA), following the manufacturer’s guidelines. Briefly, genomic DNA was randomly fragmented using the Covaris E220 Focused-ultrasonicator (Covaris, Brighton, UK) to an average size of 350 bp. The DNA fragments were then subjected to end-repairing, A-tailing, as well as ligation with index sequences incorporated within the paired-end adapters (Illumina, San Diego, California, USA). Subsequently, PCR amplification was performed.
WES library preparation
A total of 0.4 μg of genomic DNA was used for WES library preparation. The Agilent liquid capture system (Agilent Technologies, Santa Clara, USA) with the SureSelect All Exon v6 kit was employed following the manufacturer’s instructions. The genomic DNA was sheared using the Covaris E220 Focused-ultrasonicator (Covaris, Brighton, UK) to generate fragments with a size range of 180–280 bp. Subsequently, the DNA fragments underwent end-repairing, A-tailing, as well as ligation with the paired-end adapters (Illumina, San Diego, California, USA). The DNA libraries were then hybridized to biotin-labeled probes in a liquid phase, and subsequently captured using streptavidin-coupled magnetic beads. Captured libraries were subjected to PCR amplification to add index tags in preparation for sequencing.
Quality check and WGS/WES sequencing
After PCR, the products were purified using AMPure XP beads (Beckman Coulter, Beverly, USA). The concentration of the DNA libraries was quantified using the Qubit®3.0 Fluorometer (Invitrogen, Carlsbad, CA, USA), and the insert size distribution was assessed using NGS3K/Caliper. The index-coded samples were processed on a cBot Cluster Generation System using Illumina PE Cluster Kit (Illumina, San Diego, California, USA) for cluster generation. Finally, the DNA libraries were sequenced on an Illumina Hiseq XTEN platform to generate 150 bp paired-end reads (Illumina, San Diego, California, USA).
Targeted next-generation sequencing preparation and sequencing
We designed two targeted sequencing panels. The targeted sequencing library was generated using the Agilent SureSelect XT HS (Agilent Technologies) (PANEL 1) and NGS Panel (YouSuTM, OrigiMed) (PANEL 2) according to the manufacturer’s instructions. A total of 500 ng of DNA per sample was used as input material for DNA sample preparation. First, DNA was fragmented on an E220 focused ultrasonicator Covaris (Covaris, Woburn, MA, USA) to a size of 150–220 bp. Then, the DNA fragments were end-polished, A-tailed, and ligated with adapters. The exons of 436 cancer-related genes (PANEL 1, Supplementary Data 5) and the exons of 242 cancer-related genes and the introns of 43 genes (PANEL 2, Supplementary Data 5) were captured and amplified via PCR. After quality control and quantification using an Agilent 2100 Bioanalyzer (Agilent Technologies) and Qubit® 3.0 Fluorometer (Invitrogen), the DNA libraries were sequenced on an Illumina Next CN500 platform (Berry Genomics) by paired-end 150-bp to a mean unique read depth of 800X coverage.
Transcriptome library preparation and sequencing
At least 1 μg of total RNA per sample was used as input material for RNA library preparation. Stranded RNA-seq libraries were constructed using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s instructions. Briefly, mRNA was enriched from total RNA by oligo dT magnetic beads. Then, synthesis of first-strand cDNA was performed by using random hexamer primer, and followed by synthesis of second-strand cDNA using DNA Polymerase I and RNase H. Fragments size selection, adapter ligation as well as PCR amplification were performed subsequently. The cDNA library quality was evaluated on the Agilent Bioanalyzer 2100 system. The index-coded samples were carried out on a cBot Cluster Generation System using Illumina PE Cluster Kit (Illumina, San Diego, California, USA) for cluster generation, and cDNA libraries were sequenced on an Illumina Hiseq XTEN platform to produce 150 bp paired-end reads (Illumina, San Diego, California, USA).
ATAC-seq library preparation and sequencing
For each PDO, we prepared two sequencing libraries (technical replicates). PDOs were collected and washed with precooled PBS. Cells were washed once with 200 μL of cold PBS buffer and were then centrifuged for 5 min at 500 × g and 4 °C. The supernatant was removed and discarded. The cell pellet was resuspended in 50 μL of cold lysis buffer (10 mM Tris-HCl, pH 7.4; 10 mM NaCl, 3 mM MgCl2; 0.5% NP- 40) by gently pipetting up and down. The cells were immediately centrifuged at 500 × g for 5 min at 4 °C to collect nuclei, and the transposition reaction was immediately continued. ATAC-seq libraries were generated using a TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme, TD501) according to the manufacturer’s instructions. After cluster generation, the libraries were sequenced on the Illumina NovaSeq platform, and 150 bp paired-end reads were generated. Libraries containing <15 million aligned, deduplicated reads were sequenced again, and the additional reads were pooled prior to deduplication.
DNA sequence processing
First, we aligned raw sequence reads to the Human Genome Reference Consortium Build38 (GRCh38) using BWA v.0.7.1765 (BWA-mem). Next, we performed duplicate-read removal and base quality score recalibration using the GATK v.4.1.966.
Germline variant calling and filtering
The GATK, using its haplotype caller67 in single-sample mode and VarScan2 v.2.3.968 (with default parameters, except where --min-avg-qual 20 --min-normal-coverage 10 --min-tumor-coverage 10 --min-var-freq 0.04 --somatic-p-value 0.05,--strand-filter 0 were applied) were used to perform germline variant calling. All resulting variants were limited to coding regions, while resulting variants from targeted sequencing outside of these regions were filtered out. The final sets of variants were based on the intersection of GATK and VarScan2 calls. Germline variants that contained a minimum of five variant-containing reads and a variant allele frequency (VAF) of ≥ 0.1 were included.
Variants that passed quality control and variant filters were evaluated for pathogenicity. They were classified as “Pathogenic” (P), “Likely Pathogenic” (LP), “Variant of Uncertain Significance” (VUS), “Likely Benign” (LB), or “Benign” as follows:
Step 1: Variants annotated by a badged laboratory in the ClinVar69 database (20210123) as P, LP, VUS, LB, or B.
Step 2: Variants not listed in ClinVar69 were assessed using the InterVar23 database (20180118) and classified according to its designated pathogenicity category (P, LP, VUS, LB, or B).
Step 3: VUS from both Step 1 and 2 that were a high-impact variants (frameshift indels, stop gain/loss, or known splice sites) were categorized as LP.
Step 4: P and LP variants from Step 1, 2 and 3 were further filtered by population frequency (gnomAD_genome_EAS70 and our control frequency). For autosomal recessive (AR) genes, the variant allele frequency had to be ≤0.005, and non-AR genes, it had to be ≤ 0.001; otherwise, the variants were downgraded to VUS. The VUS category was further divided into in silico predicted damaging (‘Variant of Uncertain Significance damage’, VUS_D) or not-damaging (‘Variant of Uncertain Significance Non-Damage’, VUS_ND). If SIFT71 (predicted Damaging), MetaSVM72 (predicted Damaging), and CADD73 (score ≥ 20) predicted a variant as deleterious, it was categorized as ‘VUS_D’; otherwise, it was categorized as “VUS_ND”.
Step 5: A manual review of all P, LP and VUS_D variants was conducted, including a review of literature for confirmation of pathogenicity, a gene-specific database review, confirmation of the phenotype within the spectrum of the associated syndrome or cancer, and an evaluation of the mutation’s impact on gene function and the mechanism of action associated with that gene.
Somatic mutation calling
Somatic single-nucleotide variants (SNVs) and insertions/deletions (INDELs) in tumor DNA were identified using GATK (MuTect2)74. A minimum of five variant-containing reads and a variant allele frequency (VAF) ≥ 0.04 in the tumor were required for mutation calling. For hotspot mutations (such as KRAS), sequences with an average depth of <50× were labeled as low quality. Additionally, we used 389 matched normal samples from this study to build the panel of normals (PoNs) and removed any variants appearing in >1 PoN samples. ANNOVAR was used to annotate VCF (variant call format) files.
Mutational signature analysis
De novo mutational signature analysis using WGS data, and one case was removed for a small number of mutations (<50). We used “MutationalPatterns” R package and SigProfilerExtractor from SigProfiler bioinformatics tool suite (v1.1.23) to extract SBS signatures. For SigProfiler signature extraction, 500 iterations were performed (nmf_replicates = 500). Factorized signatures were compared with COSMIC mutational signatures v3.4 (https://cancer.sanger.ac.uk/cosmic/signatures) by computing all pairwise cosine similarities. Clustering by contribution of mutational signatures was performed using unsupervised hierarchical clustering with cosine distance and Ward.D linkage. Decomposition of mutational signatures was performed using deconstructSigs R package based on the set of COSMIC mutational signatures v3.4 in WGS samples (one case was removed for less 50 somatic mutations). To compare the differences in signatures among samples, we combined signatures with similar proposed etiology, and signatures with a distribution of <0.05 were classified as “Others”.
Somatic copy number variation and LOH analysis
Control-FREEC v.11.675 was used to detect genomic segments with somatic CNAs and LOH from matched normal and tumor WGS and WES mapped data. LOH and biallelic event analysis were conducted as previously described17,76. In summary, the following steps were performed: (1) VAF comparison between tumor and non-tumor samples: The VAFs of mutations in cancer susceptibility genes (CSGs) were obtained from bam files. Fisher’s exact test was used to compare the VAFs between tumor and normal samples. This involved constructing a 2 × 2 table with allele type (reference and variant) and sample type (tumor and normal). (2) VAF comparison between CSGs and non-CSGs: The analysis focused on determining whether the VAFs of variants in CSGs were higher than those in non-CSGs. To establish a null distribution, permutations were performed on the reference counts and variant counts of somatic events in all other genes. Tailed p-values were computed using this null distribution. The combined p-value was calculated by integrating the results from the above two steps. Significant and suggestive events were defined using p-value thresholds of 0.05 and 0.15, respectively. Additionally, suggestive LOH events were identified based on the criteria of tumor VAF > 0.6 and normal VAF < 0.6. The LOH events, both significant and suggestive, were further classified as follows: (1) Wild-type allele copy number deletion: The GISTIC77 CNV results indicated that the ploidy of the gene region was below the threshold. (2) Alternative allele copy number amplification: The GISTIC CNV results indicated that the ploidy of the gene region was equal to or higher than the threshold. In cases where it was equal to the threshold, it could be a LOH event with neutral copy number.
RNA-seq processing
Sequencing reads were mapped to GRCh38 using Hisat2 v.2.1.078. Subsequently, gene expression levels were quantified using FeatureCounts v2.0.279. To normalize the gene expression data, two methods were applied: FPKM (Fragments Per Kilobase of transcript per Million mapped reads) and TMM (weighted trimmed mean of M-values). These normalization techniques were implemented using the R package edgeR80. Differentially expressed genes (DEGs) were selected based on rigorous criteria, including an absolute log2 fold change >2 and a false discovery rate (FDR) < 0.05 when compared to the corresponding samples. The shared DEGs between the NATs subtypes and their associated paired tumors are presented in Fig. 5E. To gain insights into the biological functional implications of these DEGs, we performed Gene Ontology (GO) and KEGG pathway analyses using the R package clusterProfiler (v4.8.3). Significance was determined using a threshold of FDR < 0.05 for both GO and KEGG analyses. To investigated the relationship between CFTR expression and pancreatic cancer tumorigenesis, we utilized RNA data from a discovery cohort of 108 patients and 50 HALLMARK pathways along with the Tumor Proliferation Rate pathway81. Using the ssGSEA method, we calculated enrichment scores for each pathway in each sample. We then assessed the Pearson correlation between the ssGSEA scores of each pathway and CFTR expression.
ATAC-seq processing
Adapter and transposase sequences specific to the ATAC-seq protocol were removed from the paired-end reads using a customized script. The trimmed reads were aligned to the GRCh38 reference genome using BWA version 0.7.17 with the BWA-mem algorithm. Duplicate reads and reads mapped to the mitochondrial genome were removed using SAMtools version 1.1182. The remaining uniquely aligned reads were used for peak calling using macs2 version 2.2.7.183(default parameters, except for -B -q 0.05 --cutoff-analysis --call-summits).
Sanger sequencing
A set of 7 samples harboring CFTR variants underwent validation using Sanger sequencing performed on a 3730xl DNA Analyzer from Applied Biosystems. The Sanger sequencing primers utilized for this validation can be found in Supplementary Data 11.
Organoid culture
Tumor tissues obtained through resection were minced and then incubated in enzymatic digestion agent collagenase II (2.5 mg/mL with 10 mM Y-27632) at 37 °C for ~60 min. The digested cells were washed twice with RPMI-1640 medium containing 10 mM Y-27632 and 100 µg/mL primocin and centrifuged for 5 min at 1500 rpm at room temperature. The harvested cells were then re-suspended in a previously defined complete medium and embedded in Matrigel58. Upon the solidification of the Matrigel and complete medium mixture, an overlay of appropriate complete media was applied. The components of the complete medium are detailed in the supplementary materials. PDAC biopsy samples were cultured without undergoing enzymatic digestion, facilitating the spontaneous formation of organoids. The organoids were cultured in a 5% CO2 and 20% O2 environment, with media being replaced every 4 days. All established PDAC organoids are passaged either when 80% of them reach a diameter of 100-200 μm or when they occupy 60% of the field of view in matrigel under examination. This standard was uniformly applied across all samples. Organoids were preserved as cryo-stocks using a medium composed of the complete medium (90%) and 10% DMSO.
High-throughput screening of chemical and chemotherapeutic drugs
White, clear bottom 384-well plates were coated with 10 μL of collagen and allowed to incubate for at least 1 h at room temperature. Patient-derived organoid suspensions were prepared by dissociating PDOs with Tryp-LE and resuspending them in medium. The PDOs were then added to the 384-well plates at a density of 3000 cells per well. The following day, 63 + 15 compounds obtained from Selleck, along with DMSO controls, were added in duplicate to the wells using Bravo robotic workstation. Seven concentrations were used for each compound to measure sensitivity. Detailed compound information can be found in Supplementary Data 10. After 5 days of incubation with the compounds, 25 μL of CellTiter-Glo Reagent was added to each well to assess cell viability. The plates were gently shaken for 15 minutes at room temperature, and luminescence was measured using an Envision plate reader. Average inhibition rates were calculated from two independent experiments using Excel and visualized with GraphPad Prism 9. Viability values higher than the baseline were set to 100. Each drug concentration (nmol/L) was log10 transformed. The area under the curve (AUC) was calculated using GraphPad Prism 9, and the normalized AUC was obtained by dividing each AUC by the maximum AUC for that drug. An AUC heatmap for the secondary screening was generated using GraphPad Prism 9. The assay, library, screen and post-screen analysis are summarized in Supplementary Table 4.
Survival analysis
The institutional follow-up for this study was conducted by follow-up specialists, and third-party professional data were provided by LinkDoc Technology Co. Ltd. (Beijing, China). The follow-up methods employed in this study were previously described84. For patients who were still alive at the designated cut-off date for follow-ups, their follow-up data were censored at the last confirmed date of being alive. Patients who did not return for clinical follow-ups or could not be contacted were classified as lost to follow-up. Overall Survival (OS) was defined as the time from the surgical operation to death. Survival curves depicting OS were generated using the Kaplan-Meier method, and statistical analysis was performed using the log-rank test. The results were presented as median survival in months, accompanied by a 95% confidence interval (CI), and the number of patients at risk.
Immunohistochemistry
The specimens were fixed in 4% formalin and then processed for paraffin embedding. Immunohistochemistry assays were performed by incubating the tissue sections with a mixture of primary antibodies (Ab219337, Abcam) overnight at 4 °C. For detection, the HRP polymer DAB Detection System (K5007, DAKO) was used, followed by incubation in DAB for 5 minutes. Hematoxylin was used for counterstaining. Images were captured using an inverted fluorescence microscope (DMI4000B, Leica). ImageJ/FIJI software (National Institutes of Health, Bethesda, MD) was used to segment nuclei and quantify the area stained with DAB.
Validation of CFTR expression and survival analysis in public databases
To validate the different levels of CFTR expression in tumor and normal tissues, two public datasets were included (GSE28735, E-MEXP-1121). A third LCM sample datasets (GSE17891) was used to validate the association between CFTR expression and prognosis. RNA-seq and survival data were collected from R2 platform (http://r2.amc.nl).
Methylation-specific PCR
Cell lines and PDOs DNA extraction was performed using the QIAamp DNA Mini Kit (Qiagen, USA). Genomic DNA was then subjected to bisulfite conversion using the EpiTect Bisulfite Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocols. The primer sequences used were as follows: for methylated CFTR forward primer 5ʹ-GGAGGAGGAAGGTAGGTTTC-3ʹ and reverse primer 5ʹ-GACCTCTCTTTAAATCCAATTAACAAC-3ʹ; for unmethylated CFTR forward primer 5ʹ-GGAGGAGGAAGGTAGGTTTT-3ʹ and reverse primer 5ʹ-AACCTCTCTTTAAATCCAATTAACAAC-3ʹ. These primer sequences were designed using MethPrimer (http://www.urogene.org/methprimer/). The PCR protocol involved an initial denaturation step at 95 °C for 5 minutes, followed by 35 cycles of denaturation at 94 °C for 20 s, annealing at 60 °C for 30 s, and extension at 72 °C for 20 s. A final extension step was performed at 72 °C for 5 min. The PCR products (10 µL) were then loaded onto a 2% agarose gel, and electrophoresis was carried out at 120 V for 45 minutes to visualize and capture images.
Cell line and cell culture
The human pancreatic cancer cell lines CFPAC-1, PANC-1, AsPC-1, Capan-1, and SU.86.86 were obtained from the National Collection of Authenticated Cell Cultures. PANC-1 cells were cultured in DMEM supplemented with 10% fetal bovine serum. CFPAC-1 and Capan-1cells were cultured in IMDM with 10% or 20% fetal bovine serum. AsPC-1 and SU.86.86 cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum. All cells were maintained in a humidified atmosphere containing 5% CO2 at 37 °C.
Construction of CFTR-overexpressing and CFTR mutation cell lines
We used lentiviral vectors (OBiO Technology, Shanghai, China) to construct various recombinant vectors for overexpressing CFTR and its mutant variants. The vectors included CFTR overexpressing vector (CFTR OE), CFTR p.R31C overexpressing vector (CFTR p.R31C mt), CFTR p.L69F overexpressing vector (CFTR p.L69F mt), CFTR p.L88X overexpressing vector (CFTR p.L88X mt), CFTR p.E217V overexpressing vector (CFTR p.E217V mt), CFTR p.E681V overexpressing vector (CFTR p.E681V mt), CFTR p.R1097C overexpressing vector (CFTR p.R1097C mt), CFTR p.Q1352H overexpressing vector (CFTR p.Q1352H mt), CFTR p.C1355Y overexpressing vector (CFTR p.C1355Y mt), CFTR p.S1456R overexpressing vector (CFTR p.S1456R mt), and an empty plasmid serving as a negative control (EV).To introduce these vectors into PANC-1 and AsPC-1 cells, we infected cells at 40%‐50% confluence with lentiviral vectors at an appropriate multiplicity of infection (MOI). Following infection, stably transfected cells were selected using puromycin according to standard protocols.
shRNA-mediated inhibition of CFTR expression
To knockdown CFTR, we purchased short hairpin RNA (shRNA) specifically targeting the CFTR gene from Qiachang Biotechnology Co., Ltd (Shanghai, China). The shRNA was transfected into cells using an shRNA transfection reagent from Invitrogen, following the provided protocols. As a negative control, we used a non-targeting control shRNA. The sequences used were shRNA for CFTR:
shRNA-1 (F: GATCCGCGTTGAAAGCAGGTGGGATTCTTAATTCAAGAGATTAAGAATCCCACCTGCTTTCAACGTTTTTTG, R: AATTCAAAAAACGTTGAAAGCAGGTGGGATTCTTAATCTCTTGAATTAAGAATCCCACCTGCTTTCAACGGG) and shRNA-2 (F: GATCCGCACAAGCAGTTGATGTGCTTGGCTATTCAAGAGATAGCCAAGCACATCAACTGCTTGTGTTTTTTG, R: AATTCAAAAAACACAAGCAGTTGATGTGCTTGGCTATCTCTTGAATAGCCAAGCACATCAACTGCTTGTGCG).
qRT-PCR
Total RNA from cell cultures was extracted using the MolPure Cell/Tissue Total RNA kit (19221ES50, YEASEN) following the manufacturer’s instructions. The concentration and purity of the RNA were measured spectrophotometrically at a 260:280 ratio. Subsequently, 1 μg of total RNA was reverse transcribed into cDNA using the HifairII 1st Strand cDNA Synthesis Super Mix for qPCR (11123ES60, YEASEN) in a 10 μL reaction. Real-time PCR was performed using the Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (11184ES08, YEASEN) to assess mRNA expression levels. The GAPDH gene was amplified as an internal control. The primers used for quantitative RT-PCR analysis of CFTR and GAPDH are as follows (CFTR: F-AAAAGGCCAGCGTTGTCTCC, R-AAACATCGCCGAAGGGCATTA. GAPDH: FGGAGCGAGATCCCTCCAAAAT, R-GGCTGTTGTCATACTTCTCATGG). The reactions were run on an Applied Biosystems QuantStudio™ 7 Flexible (4485701, Thermo) with a thermal cycling program consisting of a 10-minute hot start at 95 °C, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 10 s, and extension at 70 °C for 10 s. After each run, a melting curve analysis was performed to confirm the amplification of a single PCR product with the expected melting temperature. Each treatment group was analyzed with three biological replicates and three technical replicates. The data were analyzed using the comparative Ct (ΔΔCt) method.
Western blot
Cells were lysed using Cell lysis reagent (C2978, Sigma-Aldrich) supplemented with protease inhibitor cocktail (#B14002, Bimake), using sonication. Protein concentrations were determined using the Pierce BCA Protein Assay (23225, ThermoFisher). A total of 20 μg of protein per sample was loaded onto an SDS-polyacrylamide gel, transferred to a nitrocellulose membrane (Millipore, Bedford, MA, USA), and incubated overnight at 4 °C with primary antibodies against CFTR (#78335, CST, USA, 1:1000) and β‐actin (66009-1-Ig, Proteintech). The following day, the membrane was incubated with HRP-conjugated secondary antibodies goat anti-rabbit IgG (H + L) (31460, ThermoFisher) or goat anti-mouse IgG (H + L) (31430, ThermoFisher) for 1 h at room temperature. The protein bands were detected using densitometric measurements (ImageJ), and the optical densities were normalized to internal controls. The data were expressed as fold changes relative to the control values.
Cell viability assay
Cells were seeded in 96-well plates at a density of 2000 cells per well. Cell viability was assessed at 0, 24, 48, 72 and 96 h using the CCK-8 kit (Beyotime Biotechnology). Absorbance measurements were taken at a wavelength of 450 nm using an Epoch microplate reader (Biotek US). Each experiment was performed in triplicate.
Colony formation assay and statistical analysis
Cells were seeded at a density of 1000 cells per well in 6-well plates and cultured in triplicate. The growth medium was refreshed every 48 h. After 2 weeks, the cells were fixed with 4% paraformaldehyde for 30 min and stained with 0.2% crystal violet for 10 min. Cell counting was performed, and each experiment was conducted in triplicate.
Transwell assay for migration
The effect of CFTR on cell migration was assessed using the Transwell Assay System (Corning, High Wycombe, UK). Cells were suspended in serum-free medium and seeded into the upper chamber at a concentration of 104 cells/200 µL. The lower chamber was filled with complete medium containing 10% FBS (600 µL). After 48 h of incubation, the cells adhered to the upper surface were gently scraped and washed with PBS. The migrated cells on the lower surface of the membrane were stained with 0.2% crystal violet and quantified. Each experiment was performed in triplicate.
Transwell assay for invasion
Cells were suspended at a concentration of 104 cells/200 μL in serum-free medium and seeded onto the upper compartment of an invasion chamber coated with extracellular matrix (ECM; MatrigelTM, BD Biosciences, Franklin Lakes, NJ). After 48 h of incubation, the cells migrated through the ECM layer towards the complete medium in the lower well. The migrated cells were then stained with 0.2% crystal violet for 10 minutes and quantified. Each experiment was performed three times.
Cell proliferation assays
Cells were seeded in 24-well plates. After 24 h of culture, cells were incubated with EdU (E607204, Sangon Biotech) for 2 h before fixation and permeabilization, and EdU stainings were performed according to the manufacturer’s protocol. The cell nuclei were stained with Hoechst 33342 (E607204, Sangon Biotech) at a concentration of 1 µg/mL for 10 min. The proportion of cells that incorporated EdU was determined through fluorescence microscopy.
Cell death/apoptosis analysis
To assess cell apoptosis, cells were seeded in 6-well plates. After 24 h of culture, cells were incubated with the Magic Red staining solution using Caspase 3/7 Assay Kit (ab270771, Abcam) for 1 h. Staurosporine (40334ES60, YEASEN Biotech) was used as a positive control for apoptosis induction85. Fluorescence from a population of 1 × 105 cells were detected using the BD FACSCantoTM flow cytometer (Becton Dickenson, Mountain View, CA, USA) and CellQuest Pro (IVD) software (Becton Dickenson, Mountain View, CA, USA). Assays were performed in duplicates and repeated in three independent experiments.
Cell fractionation western blot analysis
Briefly, 5 × 107 PANC-1 cell line were lysed in extraction buffer A for 10 minutes at 4 °C and immediately transferred to the filter cartridge. After centrifugation at 16000 g for 30 s, the filter cartridge was discarded and the pellet was resuspended. After centrifugation at 700 g for 60 s, the remaining pellet was collected as the “nuclear fraction”. The supernatant was centrifugation at 16000 g for 30 minutes, the remaining supernatant was collected as the “cytosolic fraction”. The pellet was resuspended in extraction buffer B and was then centrifugated at 7800 g for 5 minutes. The remaining supernatant was collected and centrifugated at 16000 g for 30 minutes. The remaining pellet was collected as the “membrane fraction”. The protein concentration of each fraction was measured by the Pierce BCA Protein Assay (23225, ThermoFisher) and the samples were analyzed by Western blotting. Fractions were normalized by volume and resolved on 12% SDS-PAGE, blotted onto NC membrane and probed with antibodies for the indicated proteins. ATP1A1 (14418-1-AP, Proteintech), β-Actin (66009-1-Ig, Proteintech), and Histone-H3 (30005ES50, YEASEN Biotech) were used as membrane marker, cytosolic marker, and nuclear marker, respectively.
Immunofluorescence staining
PANC-1 cells were cultured on glass coverslips (genecome, WHB-12-CS-LC) to facilitate adherence and growth. Subsequently, they were fixed in 4% paraformaldehyde for 10 minutes at room temperature, permeabilized with 0.2% Triton X-100 (Sangon Biotech) for 10 min, and then blocked with 1% BSA (Sigma-Aldrich) for 1 h. Following the blocking step, the samples were incubated overnight at 4 °C with primary antibodies against CFTR (#78335, CST), which were diluted 1:200 in the blocking buffer. A subsequent 1-h incubation was performed using the secondary goat-anti-rabbit Alexa-Fluor 488 antibody (ab150141, Abcam), also diluted 1:200 in the blocking buffer. Additionally, DAPI staining (Thermo Fisher Scientific) was applied to PANC-1 cells. Captured images were obtained using a Zeiss LSM 900 confocal microscope, with uniform settings maintained across all acquired images.
Chloride channel runction detection
The cultivation of PANC-1 cell lines is described in the manuscript. The cells were digested with TrypLE™ Expres (Gibco, 12604021) and the cell suspension was diluted to a final concentration of about 105 cells/ml. 100 μL cell suspension was added into each 96-well plate with 6 replicates per group of cells and cultured at 37 °C for 16 h. Prepare a 130 mM chloride buffer solution, including the following components: 10 mM HEPES (Gibco, 156-300080), 10 mM Glucose (Sigma-Aldrich, G5-767), 1 mM MgSO4 (BBI, A6-0250), 3 mM K2HPO4 (Sangon Biotech, A5-01212-0), 0.6 mM KH2PO4 (Sangon Biotech, A5-01211-0), 1 mM CaSO4 (BBI, A6-0266), 130 mM NaCl (BBI, A6-0476-0001). Prepare a 130 mM chloride-free buffer solution, including the following components: 10 mM HEPES, 10 mM Glucose, 1 mM MgSO4, 3 mM K2HPO4, 0.6 mM KH2PO4, 1 mM CaSO4, and 130 mM sodium gluconate (Sangon Biotech, A500852-0250). Remove DMEM from the 96-well plate, and add 60 μL of 20 mM MQAE (Beyotime, S1082) to each well at 37 °C for 2 h. Remove the MQAE solution, and wash the plate twice with 130 mM chloride buffer. For the third wash, add 130 mM chloride buffer to retain. The fluorescence value of each well was measured using a Multifunctional Enzyme Marker (TECAN, Spark) with an excitation wavelength of 340 nm and an emission wavelength of 465 nm, and recorded as F0. The 130 mM chloride buffer solution was removed, and 130 mM chloride-free buffer solution was added. The fluorescence value of each well was again measured, and recorded as Ft. The 130 mM chloride-free buffer solution was removed, and 130 mM buffer solution containing DID (Invitrogen, V22887) was added, and the cells were incubated at 37 °C for 15 minutes. The fluorescence value of each well was measured using an excitation wavelength of 560 nm and an emission wavelength of 665 nm, and recorded as Fc. The actual chloride ion permeability of each well was calculated as (Ft-F0)/Fc.
Lipid metabolism assays
The PANC-1 cells (CFTR EV, OE, p.L88X, p.E681V and p.C1355Y, n = 4 technical replicates in each group) were thawed on ice, and lipids were extracted with MTBE (Methyl Tertiary Butyl Ether) Buffer. Briefly, 50 mg of sample was added with 0.2 ml of precooled 50% methanol and homogenated with TissueLyser. The lipids were extracted with 0.6 ml MTBE and stored in 30 min at -20°C. After centrifugation at 3,000 rcf for 15 min, the supernatants were transferred into new tube to and vacuum dried. The lipid extractions were dried in a lyophilizer and then the lipids were resuspended in 100 μL isopropanol and methanal (volume 1:1). In addition, pooled QC samples (n = 6) were also prepared by combining 10 μL of each extraction mixture. Waters ACQUITY UPLC coupled to Obritrap-LTQ-Elite (Thermo Fisher, USA) was used for lipidomic analyses. A Waters ACQUITY UPLC BEH C18 column (2.1 mm × 100 mm, 1.7 μm) was used for lipid separation. The mobile phases consisted of 10 mM ammonium formate and 0.1% formic acid dissolved in 6:4 (v/v) ACN/H2O (phase A) and 1:1 (v/v) isopropanol/ACN (phase B). The flow rate was set at 0.3 mL/min, the injection volume was 4 μL, and the column temperature was at 45 °C. The elution gradient: 0 ~ 2 minutes, 70% ~ 57% A; 2 ~ 2.1 minutes, 57% ~ 45% A; 2.1 ~ 12 minutes, 45% ~ 35% A; 12 ~ 18 minutes, 35% ~ 15% A; 18 ~ 20 minutes, 15% ~ 0% A; 20 ~ 25 minutes, 0% ~ 70% A.
The MS parameters conducted for lipid detection were summarized as follows: the ion spray voltage of MS 3.5 kV; interface heater temperature 300 °C; source temperature 100 °C; sheath gas flow rate 35 arb; aux gas flow rate 10 arb. Full scan MS spectra were acquired at a mass range of m/z 200-1500.
ProteoWizard (version 3.0.18178) was used to convert raw data to mzXML format. Ion features were detected and extracted according to retention time (RT) and m/z using the XCMS package (version 3.9.3). A three dimensional matrix containing randomly assigned peak indices (retention time-m/z pairs), sample names (observations), and ion area was generated. Lipids identification was searched against a LipidBlast database (version 68). Mass tolerance was configured as 0.01 for MS1 and 0.05 for MS2 for both positive and negative modes. The minimum cutoff score for identification was set at 0.5. The MS2 identification entries from all files were integrated with the features quantified by MS1 m/z and RT using an in-house script. The intensity of peak data was further preprocessed by metaX. Those features that were detected in <50% of QC samples or 80% of biological samples were removed, the remaining peaks with missing values were imputed with the k-nearest neighbor algorithm to further improve the data quality. The metabolome data was normalized with Probabilistic Quotient Normalization method86. Quality control-based robust LOESS signal correction87 was fitted to the QC data with respect to the order of injection to minimize signal intensity drift over time. The online KEGG, HMDB database was used to annotate the metabolites by matching the exact molecular mass data (m/z) of samples with those from database. If a mass difference between observed and the database value was <10 ppm, the metabolite would be annotated and the molecular formula of metabolites would further be identified and validated by the isotopic distribution measurements. We also used a combination fragment database (Lipidblast Version 37, Massbank, HMDB Version 4.0 and in-house fragment spectrum library) to validate the metabolite identification.
Animal models
Four-week-old male BALB/c nude mice (n = 25, average weight 16 ± 2 g) were purchased from Shanghai Lab. Animal Research center (Shanghai, China). After 1 week of acclimatization, each group of five mice is housed in separated individual standard cleaned cages under automatically controlled air condition system with controlled temperature (22 ± 2 °C), humidity (about 60%), and lighting (12:12 h light–dark cycle). Diet and sterilized water are provided ad libitum throughout the experiments. Mice were inoculated subcutaneously into the right armpit with human pancreatic cancer cell line PANC-1 expressing CFTR EV, OE, p.L88X, p.E681V and p.C1355Y (5 × 106 cells in 200 μL of 20% Matrigel and serum-free culture medium, 1:4 v/v%) to develop a xenograft model of human pancreatic cancer. We observed the model until the tumor volume expanded to up to 1000 mm3. Implanted tumor size was assessed three times per week and calculated by tumor volume = 1/2(length × width2). Tumorigenicity was measured in the third week after injecting the cells (n = 5 per group). All animal experiments were undertaken in accord with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of Second Military Medical University (Shanghai, China). The maximal tumor size/burden permitted by our ethics committee review board is 20 mm in diameter (or 4000 mg) for subcutaneous tumors. The maximal tumor size/burden permitted by our institutional review board was not exceeded.
Statistical analysis
Statistical analysis is reported as appropriate in each Figure, Figure Legend or Supplementary Table. All statistical analyses were conducted using R (version 4.1.0; https://cran.r-project.org/). Statistical significance was defined as a two-sided P-value of <0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequencing data of all WGS, WES and RNA-Seq of Discovery cohort, and WGS of functional validation cohort created in this study have been uploaded to the Genome Sequence Archive [https://ngdc.cncb.ac.cn/gsa-human] with the accession number HRA005195. RNA-Seq and ATAC-seq data of organoids (functional validation cohort) and RNA-seq data of cell lines are available with the accession number HRA010277. Data and materials that can be shared will be released via a material transfer agreement to protect patient privacy. Single-nucleotide polymsorphism or deleterious germline mutation data of Control and Validation cohorts were provided in Supplementary data 2 and 3. The TCGA PAAD cohort is available at cBioPortal [https://www.cbioportal.org/]. Expression profilings by array used in this study are available in the Gene Expression Omnibus under accession code GSE28735 and GSE17891, and in ArrayExpress database under accession code E-MEXP-1121. The lipidomics raw data and processed data matrices have been deposited to Metabolomics Workbench (https://www.metabolomicsworkbench.org/) under the accession number ST003734. Source Data are provided with this study. All other data could be found within the main manuscript or Supplementary information or source data. Source data are provided with this paper.
Code availability
All original code has been deposited in Code Ocean88 (capsule number 1432248) under an MIT License at: https://doi.org/10.24433/CO.1432248.v1.
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Biankin, A. V. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012).
Bailey, P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016).
Witkiewicz, A. K. et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 6, 6744 (2015).
Cao, L. et al. Proteogenomic characterization of pancreatic ductal adenocarcinoma. Cell 184, 5031–5052 e5026 (2021).
Javle, M. et al. Olaparib monotherapy for previously treated pancreatic cancer with DNA damage repair genetic alterations other than germline BRCA variants: findings from 2 phase 2 nonrandomized clinical trials. JAMA Oncol. 7, 693–699, (2021).
Pokataev, I. et al. Efficacy of platinum-based chemotherapy and prognosis of patients with pancreatic cancer with homologous recombination deficiency: comparative analysis of published clinical studies. ESMO Open 5, e000578 (2020).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Eng. J. Med. 372, 2509–2520 (2015).
Golan, T. et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 381, 317–327 (2019).
Raimondi, S., Maisonneuve, P. & Lowenfels, A. B. Epidemiology of pancreatic cancer: an overview. Nat. Rev. Gastroenterol. Hepatol. 6, 699–708 (2009).
Hruban, R. H., Canto, M. I., Goggins, M., Schulick, R. & Klein, A. P. Update on familial pancreatic cancer. Adv. Surg. 44, 293–311 (2010).
Wang, W. et al. PancPRO: risk assessment for individuals with a family history of pancreatic cancer. J. Clin. Oncol. 25, 1417–1422 (2007).
Klein, A. P. et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 64, 2634–2638 (2004).
Salo-Mullen, E. E. et al. Identification of germline genetic mutations in patients with pancreatic cancer. Cancer 121, 4382–4388 (2015).
Zhan, W., Shelton, C. A., Greer, P. J., Brand, R. E. & Whitcomb, D. C. Germline variants and risk for pancreatic cancer: a systematic review and emerging concepts. Pancreas 47, 924–936 (2018).
Huang, K. L. et al. Pathogenic germline variants in 10,389 adult cancers. Cell 173, 355–370 e314 (2018).
Kastrinos, F. et al. Risk of pancreatic cancer in families with lynch syndrome. Jama 302, 1790–1795 (2009).
Roberts, N. J. et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2, 41–46 (2012).
Zhen, D. B. et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: a PACGENE study. Genet. Med. 17, 569–577 (2015).
Roberts, N. J. et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov. 6, 166–175 (2016).
Landrum, M. J. et al. ClinVar: improvements to accessing data. Nucleic Acids Res. 48, D835–D844 (2020).
Li, Q. & Wang, K. InterVar: Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am. J. Hum. Genet. 100, 267–280 (2017).
Hu, C. et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA 319, 2401–2409 (2018).
Holter, S. et al. Germline BRCA mutations in a large clinic-based cohort of patients with pancreatic adenocarcinoma. J. Clin. Oncol. 33, 3124–3129 (2015).
Mandelker, D. et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA 318, 825–835 (2017).
Shindo, K. et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J. Clin. Oncol. 35, 3382–3390 (2017).
Scarpa, A. et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 543, 65–71 (2017).
Golan, T. et al. Geographic and ethnic heterogeneity of germline BRCA1 or BRCA2 mutation prevalence among patients with metastatic pancreatic cancer screened for entry into the POLO trial. J. Clin. Oncol. 38, 1442–1454 (2020).
Yin, L. et al. Prevalence of germline sequence variations among patients with pancreatic cancer in China. JAMA Netw. Open 5, e2148721 (2022).
Mizukami, K. et al. Genetic characterization of pancreatic cancer patients and prediction of carrier status of germline pathogenic variants in cancer-predisposing genes. EBioMedicine 60, 103033 (2020).
Ni, Q. et al. Systematic estimation of cystic fibrosis prevalence in Chinese and genetic spectrum comparison to Caucasians. Orphanet J. Rare Dis. 17, 129 (2022).
Zou, W. B. et al. SPINK1, PRSS1, CTRC, and CFTR genotypes influence disease onset and clinical outcomes in chronic pancreatitis. Clin. Transl. Gastroenterol. 9, 204 (2018).
Organization, W. H. Report of a joint meeting of WHO/ECFTN/ICF (M) A/ECFS. https://iris.who.int/handle/10665/68702 (2002).
McWilliams, R. et al. Cystic fibrosis transmembrane regulator gene carrier status is a risk factor for young onset pancreatic adenocarcinoma. Gut 54, 1661–1662 (2005).
Matsubayashi, H. et al. Polymorphisms of SPINK1 N34S and CFTR in patients with sporadic and familial pancreatic cancer. Cancer Biol. Ther. 2, 652–655 (2003).
Zhang, G. et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin. Cancer Res. 19, 4983–4993 (2013).
Pilarsky, C. et al. Activation of Wnt signalling in stroma from pancreatic cancer identified by gene expression profiling. J. Cell Mol. Med. 12, 2823–2835 (2008).
Collisson, E. A. et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17, 500–503 (2011).
NandyMazumdar, M. et al. Looping of upstream cis-regulatory elements is required for CFTR expression in human airway epithelial cells. Nucleic acids Res. 48, 3513–3524 (2020).
Saint-Criq, V. & Gray, M. A. Role of CFTR in epithelial physiology. Cell Mol. Life Sci. 74, 93–115 (2017).
Cazacu, I. M. et al. Pancreatitis-associated genes and pancreatic cancer risk: a systematic review and meta-analysis. Pancreas 47, 1078–1086 (2018).
Schubert, S. et al. CFTR, SPINK1, PRSS1, and CTRC mutations are not associated with pancreatic cancer in German patients. Pancreas 43, 1078–1082 (2014).
Piepoli, A. et al. Lack of association between UGT1A7, UGT1A9, ARP, SPINK1 and CFTR gene polymorphisms and pancreatic cancer in Italian patients. World J. Gastroenterol. 12, 6343–6348 (2006).
Hamoir, C. et al. Clinical and morphological characteristics of sporadic genetically determined pancreatitis as compared to idiopathic pancreatitis: higher risk of pancreatic cancer in CFTR variants. Digestion 87, 229–239 (2013).
McWilliams, R. R. et al. Cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations and risk for pancreatic adenocarcinoma. Cancer 116, 203–209 (2010).
Barretina, J. et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Chambers, J. A. & Harris, A. Expression of the cystic fibrosis gene and the major pancreatic mucin gene, MUC1, in human ductal epithelial cells. J. Cell Sci. 105, 417–422 (1993).
Ward, C. L., Omura, S. & Kopito, R. R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121–127 (1995).
Than, B. L. N. et al. CFTR is a tumor suppressor gene in murine and human intestinal cancer. Oncogene 36, 3504 (2017).
Ikpa, P. T. et al. Transcriptome analysis of the distal small intestine of Cftr null mice. Genomics 112, 1139–1150 (2020).
Declercq, M. et al. Transcriptomic analysis of CFTR-impaired endothelial cells reveals a pro-inflammatory phenotype. Eur. Respir. J. 57, 2000261 (2021).
Martin-Perez, M., Urdiroz-Urricelqui, U., Bigas, C. & Benitah, S. A. The role of lipids in cancer progression and metastasis. Cell Metab.34, 1675–1699 (2022).
Tutino, V. et al. Elevated AA/EPA ratio represents an inflammatory biomarker in tumor tissue of metastatic colorectal cancer patients. Int. J. Mol. Sci. 20, 2050 (2019).
Dombrowsky, H., Clark, G. T., Rau, G. A., Bernhard, W. & Postle, A. D. Molecular species compositions of lung and pancreas phospholipids in the cftr(tm1HGU/tm1HGU) cystic fibrosis mouse. Pediatric Res. 53, 447–454 (2003).
Que, X. et al. Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice. Nature 558, 301–306 (2018).
Imai, Y. et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 133, 235–249 (2008).
Shi, X. et al. Integrated profiling of human pancreatic cancer organoids reveals chromatin accessibility features associated with drug sensitivity. Nat. Commun. 13, 2169 (2022).
Hong, D. S. et al. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N. Engl. J. Med. 383, 1207–1217 (2020).
Janne, P. A. et al. Adagrasib in non-small-cell lung cancer harboring a KRAS(G12C) mutation. N. Engl. J. Med. 387, 120–131 (2022).
Beekman, J. M. Individualized medicine using intestinal responses to CFTR potentiators and correctors. Pediatr. Pulmonol. 51, S23–S34 (2016).
Earl, J. et al. A comprehensive analysis of candidate genes in familial pancreatic cancer families reveals a high frequency of potentially pathogenic germline variants. EBioMedicine 53, 102675 (2020).
Yang, M. et al. DAGM: A novel modelling framework to assess the risk of HER2-negative breast cancer based on germline rare coding mutations. EBioMedicine 69, 103446 (2021).
Rahman, N. Realizing the promise of cancer predisposition genes. Nature 505, 302–308 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 43, 11 10 11–11 10 33 (2013).
Poplin, R. et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv https://doi.org/10.1101/201178 (2018).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Landrum, M. J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018).
Chen, S. et al. A genome-wide mutational constraint map quantified from variation in 76,156 human genomes. bioRxiv https://doi.org/10.1101/2022.03.20.485034 (2022).
Ng, P. C. & Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Kim, S., Jhong, J. H., Lee, J. & Koo, J. Y. Meta-analytic support vector machine for integrating multiple omics data. BioData Min. 10, 2 (2017).
Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J. & Kircher, M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic acids Res. 47, D886–d894 (2019).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 31, 213–219 (2013).
Boeva, V. et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28, 423–425, (2012).
Lu, C. et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nat. Commun. 6, 10086 (2015).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol.12, R41 (2011).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Bagaev, A. et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 39, 845–865.e847 (2021).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Guo, S. W. et al. A preoperative risk model for early recurrence after radical resection may facilitate initial treatment decisions concerning the use of neoadjuvant therapy for patients with pancreatic ductal adenocarcinoma. Surgery 168, 1003–1014 (2020).
Malsy, M., Bitzinger, D., Graf, B. & Bundscherer, A. Staurosporine induces apoptosis in pancreatic carcinoma cells PaTu 8988t and Panc-1 via the intrinsic signaling pathway. Eur. J. Med. Res. 24, 5 (2019).
Dieterle, F., Ross, A., Schlotterbeck, G. & Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 78, 4281–4290 (2006).
Dunn, W. B. et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6, 1060–1083 (2011).
Yin, X. et al. Pathogenic germline variants in Chinese pancreatic adenocarcinoma patients. code ocean. JAMA Netw. Open 5, e2148721 (2024).
Acknowledgements
We sincerely thank our patients and their families. This study was supported by the Shanghai Science and Technology Innovation Action Plan (23Y41900200, G.J.), “National Key R&D Program of China” fund (2022YFA1305700, J.L.), the Chinese Ministry of Science and Technology, National Natural Science Foundation of China (81602467, J.L.), the National Natural Science Foundation of China (82172712, G.J.), the Clinical Medicine Research Special Project for Major Disease Multi-center Clinical Research of Naval Medical University First Affiliated Hospital (2024LYA04, G.J.), the “Dawn” Program of Shanghai Education Commission, China (21SG33, J.L.), and the China Postdoctoral Science Foundation (2023M744284, X.Y.Y.). We would like to thank Jinghang Yuan for experimental assistance. We thank Shanghai Clinbrain Information Technology Co., Ltd. for establishing and maintaining the prospective database of Shanghai Changhai Hospital.
Author information
Authors and Affiliations
Contributions
Gang Jin, Jing Li and Xiaoyi Yin conceived and implemented the study. Haojie Jin and Hongbo Yang supervised the in vitro experiments and in vivo experiments, respectively. Xiaoyi Yin performed data analyses. Hui Shen performed the majority of wet lab experiments. Qingchen Wang, Shan Zhang and Qi Jia contributed to the in vivo experiments and in vitro cell line experiments, respectively. Huan Wang, Xiaohan Shi and Yunguang Li was responsible for the organoid culture and drug screening. Dong Gao supervised the analysis of organoid experiment results. Chunming Zhang, Gang Niu and Qiangzu Zhang collected and analyzed data from the Asian control cohort. Hui Shen, Shiwei Guo, Xiongfei Xu, Wenhui Zhang, Meilong Shi, Xuenan Zhao, Sheng Wang and Qingchen Wang provided support for basic statistics, proofreading, data storage, and experiments preparation. Suizhi Gao, Jiawei Han, Guoxiao Zhang, Yikai Li, Penghao Li, Wei Jing, Bin Song, Kailian Zheng, Gang Li and Yijie Zhang contributed to sample collection as surgeons. Bo Li and Hui Jiang performed pathology review of all samples. Cong Wu was responsible for biobanking. Zhijian Song analyzed the CFTR mutation in the pan-cancer cohort. Fengxian Han, Jianglong Guo and Zhen Sun provided assistance in revising the manuscripts and figures. Gang Jin, Jing Li and Xiaoyi Yin prepared the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Kai Kretzschmar, Nobuyuki Kakiuchi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yin, X., Shen, H., Wang, H. et al. Pathogenic germline variants in Chinese pancreatic adenocarcinoma patients. Nat Commun 16, 2214 (2025). https://doi.org/10.1038/s41467-025-57520-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57520-3
This article is cited by
-
Deficiencies in germline DNA repair are associated with early-onset gastrointestinal cancers and inform precision prevention strategies
Journal of Translational Medicine (2025)
-
Pathogenic CDKN2A germline variants are rare in a cohort of unselected pancreatic cancer patients from Pakistan
Familial Cancer (2025)