Abstract
Metabolites are small molecules that are useful for estimating disease risk and elucidating disease biology. Here, we perform two-sample Mendelian randomization to systematically infer the potential causal effects of 1099 plasma metabolites measured in 6136 Finnish men from the METSIM study on risk of 2099 binary disease endpoints measured in 309,154 Finnish individuals from FinnGen. We find evidence for 282 putative causal effects of 70 metabolites on 183 disease endpoints. We also identify 25 metabolites with potential causal effects across multiple disease domains, including ascorbic acid 2-sulfate affecting 26 disease endpoints in 12 disease domains. Our study suggests that N-acetyl-2-aminooctanoate and glycocholenate sulfate affect risk of atrial fibrillation through two distinct metabolic pathways and that N-methylpipecolate may mediate the putative causal effect of N6,N6-dimethyllysine on anxious personality disorder.
Similar content being viewed by others
Introduction
Metabolites are intermediate or end products of cellular metabolism with a wide range of functions1. Compared to gene transcripts and proteins, metabolites are more proximal to diseases, making them ideal biomarkers for estimating disease risk and understanding disease biology. Metabolite levels have shown associations with many human diseases, including type 2 diabetes, chronic kidney disease, and cardiovascular diseases2,3,4,5. Some metabolites have demonstrated potential for predicting future disease6,7. However, the causal effects of metabolites on human diseases have not been evaluated comprehensively.
Metabolite levels reflect both environmental and genetic influences1. With the advent of high-throughput metabolic profiling technology, measuring levels of thousands of metabolites for participants in population studies has become possible. Recent genome-wide association studies (GWAS) that combine high-throughput metabolic profiling and genotyping/sequencing in large samples have identified thousands of genetic associations for thousands of metabolites and metabolic features8,9,10,11,12,13,14,15. These studies usually measure metabolite levels in blood, which are widely considered to reflect metabolite aggregate concentrations across tissues16. Recently, we profiled plasma levels for 1391 metabolites using Metabolon non-targeted mass spectrometry technology in 6136 Finnish individuals of the Metabolic Syndrome in Men (METSIM) study17. GWAS identified 2030 genetic associations for 803 of the 1391 metabolites17. Integrating these metabolite GWAS with expression quantitative trait loci (eQTL) in 49 human tissues established associations of expression levels of 397 genes with levels of 521 plasma metabolites18. These GWAS deepen our understanding of genetic regulation of metabolic individuality, open an avenue to evaluate the putative causal effects of blood metabolites on human diseases using Mendelian randomization (MR), and have the potential to provide actionable disease interventions.
MR is an instrumental variable (IV) method to interrogate causal effects of heritable risk factors on diseases of interest using genetic variants as IVs19. MR tests whether IVs that affect the exposure have a proportional effect on the outcome. If MR assumptions about relevance, independence, and exclusion restriction are fulfilled20, the proportionality constant is an estimate for the causal effect of the exposure on the outcome. Recent method development allows increased robustness to violations of these assumptions. For example, MR using the robust adjusted profile score (MR-RAPS) can account for bias of weak and outlier genetic IVs21, and multivariable MR (GRAPPLE) enables testing causal effects of multiple potentially related exposures on the same outcome22,23.
MR is commonly used to test causal hypotheses that may be motivated by epidemiological studies24. Recently, MR has been used to comprehensively screen risk factors with potential causal effects on outcomes25,26. Latest studies have applied MR to search for causal blood metabolites for a wide range of diseases and traits, including type 2 diabetes27, neuroticism28, Alzheimer’s disease29, and rheumatoid arthritis30. These studies demonstrate the utility of MR to identify potential causal metabolites and metabolic pathways for human diseases. However, the existing studies are restricted to one or a few disease outcomes and a relatively limited set of metabolites8,9.
Here, we comprehensively evaluated potential causal effects of 1099 plasma metabolites on 2099 binary disease endpoints (hereafter disease traits) using a MR analysis in GWAS of METSIM plasma metabolites17 and FinnGen disease traits (release 7)31. We identified evidence for 282 putative causal effects of 70 plasma metabolites on 183 disease traits. Our study uncovered potential causal effects of plasma metabolites for a broad spectrum of human diseases. We also identified metabolites with broad potential causal effects across multiple disease types.
Results
Summary of MR results
We previously conducted GWAS for 1099 named plasma metabolites with annotated chemical identities in up to 6136 Finnish men aged 45–74 at enrollment from the METSIM study17. These 1099 metabolites included nine biochemical classes of small molecules related to the metabolisms of lipids (n = 548, 49.9%), amino acids (n = 215, 19.6%), xenobiotics (n = 163, 14.8%), peptides (n = 42, 3.8%), nucleotides (n = 42, 3.8%), cofactors and vitamins (n = 38, 3.5%), carbohydrates (n = 25, 2.3%), partially-characterized molecules (n = 16, 1.5%), and energy (n = 10, 0.9%) (Supplementary Data 1).
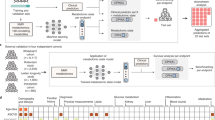
To identify potential causal plasma metabolites for human diseases, we carried out univariable MR analysis using MR-RAPS31 to evaluate causal effects of the 1099 metabolites on 2099 binary disease traits from the FinnGen study (release 7; Fig. 1a). In GWAS, we inverse normalized the metabolite measurements17 and measured disease trait associations by mixed-model logistic regression31. Our estimated causal effects can, therefore, be interpreted as the change in log odds of disease risk caused by an increase of one standard deviation of the normalized metabolite level. To identify independent IVs for the MR analysis, we performed linkage disequilibrium (LD) clumping in the GWAS summary statistics for each of the 1099 metabolites to ensure resulting variants achieve association P < 10−5 and each pair of variants within 1 megabase (Mb) distance satisfy LD r2 < 0.01. For the 1099 metabolites, we identified from 12 to 173 likely independent variants (mean = 42.3; median = 40.0) and used these as IVs (Supplementary Fig. 1).
a the overall design of univariable MR to test causal effects of 1099 metabolites on 2099 disease traits; b distribution of metabolites by the number of disease traits that they showed significant putative causal effects on; c distribution of metabolites by the number of disease categories that they showed significant putative causal effects on; d distribution of disease traits by the number of their associated putative causal metabolites. Source data are provided as a Source Data file.
We identified evidence for 282 potential causal effects of 70 plasma metabolites on 183 disease traits at a false discovery rate (FDR) threshold < 1% (Fig. 2 and Supplementary Data 2), highlighting the relevance of plasma metabolite levels to human health. These 282 metabolite-disease trait pairs showed strong robustness to IV selection and choice of MR method (Supplementary Figs. 2–5, Supplementary Methods). As a sensitivity analysis, we repeated our MR analysis after removing all IVs associated with another metabolite at P < 5 × 10−8 in METSIM metabolite GWAS17. The resulting estimates exhibited a strong correlation with the original ones (Pearson r = 0.92; Supplementary Fig. 6). Of the 282 putative causal effects originally identified, 70 (24.8%) between 16 metabolites and 68 disease traits remained significant and consistent at FDR < 5% (Supplementary Data 2). We note that this sensitivity analysis exhibited substantially reduced statistical power to detect causal effects and may be overly conservative. Multivariate MR suggested that the 282 putative causal relationships were likely independent of common potential lifestyle confounders alcohol drinking, cigarette smoking, and sleep duration (Supplementary Fig. 7 and Supplementary Data 3).
The x-axis denotes the 183 disease traits of 20 colored categories (from left to right). The y-axis denotes the 70 metabolites of eight colored biochemical classes (from bottom to top). The bar plots show the number of FinnGen disease traits that each metabolite confers potential causal effects on (on the left) and the number of putative causal metabolites for each disease trait (on the top). The color of cells denotes the direction of potential causal effects (red for positive and blue for negative effects) of metabolites on disease traits. Source data are provided as a Source Data file.
The 70 putative causal metabolites comprised lipids (n = 31, 44.3%), amino acids (n = 29, 41.4%), xenobiotics (n = 4, 5.7%), cofactors and vitamins (n = 2, 2.9%), and nucleotides, carbohydrate, peptide, and partially-characterized molecule (n = 1, 1.4% for each). Compared to the total set of 1099 metabolites evaluated, the 70 metabolites with putative causal effects were enriched in amino acids (odds ratio (OR) = 3.20, Chi-square test P = 4.0 × 10-6) and depleted in xenobiotics (OR = 0.33, Chi-square test P = 0.041). We found that amino acids had more IVs on average than xenobiotics (Student’s t-test P = 1.2 × 10−12), so the observed enrichment of amino acids may be a result of better power to detect effects. The enrichment could also be a consequence of which xenobiotics and amino acids are represented on the Metabolon platform or could indicate a more central role of amino acids in disease risk. The 70 plasma metabolites conferred significant putative causal effects on 1–26 disease traits (mean = 4.0; median = 1.0), with 32 (46%) showing significant putative causal effects on more than one disease trait (Fig. 1b, c). The 183 disease traits covered a broad spectrum of diseases. The FinnGen consortium grouped these disease traits into 20 categories, including cancers (e.g., colon cancers), cardiometabolic (e.g., type 2 diabetes), infectious (e.g., tularemia), neurological (e.g., Parkinson’s disease), and mental and behavioral diseases (e.g., anxiety personality disorder) (Supplementary Data 2). Each of the 183 disease traits had 1–6 potential causal metabolites (mean = 1.5; median = 1.0); 53 (29%) had ≥2 potential causal metabolites (Fig. 1d).
Potential causal metabolites for diseases
Among the 282 putative causal effects, we reproduced several known relationships. For example, we identified a potential causal effect of low plasma lipid glycosyl-N-stearoyl-sphingosine levels on increasing risk of coronary artery disease (β = −0.11, P = 1.0 × 10−6), reinforcing the important role of sphingolipid metabolism in coronary artery disease32. Studies have reported high levels of valine, a branched-chain amino acid, associated with increased risk of type 2 diabetes6,33. We validated, with nominal significance, the putative causal effect of plasma valine levels on risk of type 2 diabetes (β = 0.041, P = 5.0 × 10−3). In addition, we found that elevated plasma N-acetylvaline levels decreased risk of type 2 diabetes (β = −0.085, P = 1.1 × 10−8). N-acetylvaline is a derivative of valine and belongs to a class of N-acyl-alpha amino acids. Multivariable MR23 including both valine and N-acetylvaline, suggested that both metabolites have direct effects on type 2 diabetes (N-acetylvaline: β = −0.096, P = 2.7 × 10−12; valine: β = 0.087, P = 1.8 × 10−5), indicating a potentially important and complex role of valine metabolism in risk of type 2 diabetes. Interestingly, we found that high levels of two additional plasma N-acyl-alpha amino acids N-acetylglutamate (β = −0.11, P = 1.0 × 10−7) and N-acetylmethionine (β = −0.072, P = 5.5 × 10−7) potentially causally decreased the risk of type 2 diabetes. The three N-acyl-alpha amino acids N-acetylvaline, N-acetylglutamate, and N-acetylmethionine show substantial phenotypic correlation and share many IVs (Fig. 3, Supplementary Fig. 8). For these three N-acyl-alpha amino acids, our GWAS previously identified genome-wide significant associations at the ACY1 gene17, which encodes enzyme aminoacylase 1 that catalyzes the hydrolysis of acylated L-amino acids to L-amino acids. MR analysis using a single IV for plasma aminoacylase 1 levels identified by the deCODE project34 suggested that increased aminoacylase 1 levels may decrease levels of the three N-acyl-alpha amino acids (β < −1.20, P < 4.2 × 10−21) and increase risk of type 2 diabetes (β = 0.16, P = 2.6 × 10−4), directionally consistent with the known function of aminoacylase 1 and a recently reported putative risk effect of increasing aminoacylase 1 on type 2 diabetes35. These findings suggest a possible role of synthesis or degradation of N-acetylated proteins in type 2 diabetes. However, due to substantial sharing of IVs across the three N-acetyl amino acids, MR cannot identify whether this effect is due to one specific or multiple N-acetyl amino acids.
The color bar on the x-axis and y-axis denotes the biochemical classes of metabolites. In the upper left triangular heat map, each cell denotes the proportion of IVs with metabolite association at P ≤ 10−5 shared between the pair of metabolites. In the lower right triangular heat map, each cell denotes the IV correlation between the pair of metabolites. The diagonal cells are colored in dark gray to distinguish the upper and lower triangular heat maps. Source data are provided as a Source Data file.
Our study also identified potential causal metabolites for human diseases. MR recently suggested causal effects of plasma metabolites on the risk of dementia29,36,37. Among them, previous studies only reported 2-methoxyacetaminophen sulfate38 with a putative causal effect specifically on frontotemporal dementia, a type of dementia characterized by progressive loss of neurons in the brain’s frontal or temporal lobes. We identified a significant potential protective effect of high plasma lipid 2-arachidonoyl-GPC (20:4) levels on the risk of frontotemporal dementia (β = −0.89, P = 1.2 × 10−6). 2-arachidonoyl-GPC (20:4) is a lysophosphatidylcholine widely considered as a potent pro-inflammatory mediator39. Emerging evidence has demonstrated that neuroinflammation plays an important role in dementia40. Studies have identified a negative association of lysophosphatidylcholine with Alzheimer’s disease41. Consistent with these results, we found a potentially protective causal effect of increased 2-arachidonoyl-GPC (20:4) levels on risk of frontotemporal dementia. We previously identified genome-wide associations for 2-arachidonoyl-GPC (20:4) around the FADS1/FADS2, two fatty acid desaturase genes17. Interestingly, we found that low expression of FADS1/FADS2 in the whole blood but high expression in the brain significantly increased plasma 2-arachidonoyl-GPC (20:4) level18. FADS1 variants could regulate erythrocyte arachidonic acid biosynthesis that subsequently induces inflammation in Alzheimer’s disease42.
Chronic kidney disease affects >10% of the general population worldwide43, and its risk factors are still poorly understood. We found evidence that elevated plasma xenobiotic sulfate levels increased risk of chronic kidney disease (β = 0.080, P = 1.9 × 10−7). High sulfate levels have been previously found to be associated with disease progression and increased mortality in individuals with kidney disease44. Our previous GWAS identified a genome-wide significant association with plasma sulfate levels at the SLC13A1 gene17, which encodes a sulfate transmembrane transporter and mediates the first step of sulfate absorption. SLC13A1 is primarily expressed in the proximal renal tubules. We previously found that high expression of SLC13A1 decreased plasma sulfate abundance18. These results together suggest that SLC13A1 could serve as a potential drug target for chronic kidney disease through the regulation of plasma sulfate levels.
Potential causal metabolites shared across diseases
We identified evidence for 32 metabolites with putative causal effects on more than one disease trait (Figs. 1b and 2; see Summary of MR results). Of these 32 metabolites, 25 (78%) showed significant potential causal effects on two or more disease categories (Fig. 1c and Supplementary Data 2). The sharing of putative causal metabolites between diseases may partially explain observed phenotypic correlations and disease comorbidities. For example, we identified potential causal effects of plasma amino acid N-acetylvaline levels on optic atrophy (β = 0.53, P = 4.7 × 10−7) and myasthenia gravis (β = 0.53, P = 7.9 × 10−8), diseases with substantial comorbidity45. These results suggest that valine metabolism might play a role in both the cell cycle of retinal ganglion cell axons and communication between nerves and muscle. We found potential causal effects of plasma amino acid N-acetyl-aspartyl-glutamate (NAAG) levels on increased risk of both Parkinson’s disease (β = 0.11, P = 3.2 × 10−7) and autoimmune hypothyroidism (β = 0.039, P = 3.9 × 10−9), which also have substantial comorbidity46.
The metabolite linked to the largest number of disease traits was ascorbic acid 2-sulfate, with evidence of potential causal effects on 26 disease traits in 12 categories, including cardiomyopathy (disease of the circulatory system), arthropathy (disease of the musculoskeletal system and connective tissue), and acne (disease of the skin and subcutaneous tissue) (Supplementary Data 2). We found that elevated levels of ascorbic acid 2-sulfate may decrease risk of 12 disease traits including colon adenocarcinoma (β = −0.13, P = 9.3 × 10−8) and endometriosis of the fallopian tube (β = −0.48, P = 1.6 × 10−7) but increase risk of 14 others including conjunctiva cancer (β = 0.36, P = 2.8 × 10−14) and arthropathy (β = 0.028, P = 1.1 × 10−7).
Notably, the suggested putative causal effects of plasma ascorbic acid 2-sulfate showed heterogeneity across disease traits, even in the same category. For example, we found that elevated ascorbic acid 2-sulfate levels were potentially protective for acne (β = −0.18, P = 3.9 × 10−10) and lichen sclerosus (β = −0.15, P = 7.1 × 10−7) but putatively increase risk of dyshidrosis, a kind of eczema (β = 0.42, P = 4.2 × 10−10). These three conditions all affect skin but usually in different anatomical locations: the face, upper part of the chest, and back; the genital area; and the palms and fingers, respectively. Ascorbic acid 2-sulfate arises from the action of a liver-derived sulfotransferase on vitamin C, so it is possible that plasma levels of ascorbic acid 2-sulfate are a proxy for the action of liver-derived sulfotransferases or for vitamin C levels, or a combination of these. For example, we estimated that elevated ascorbic acid 2-sulfate levels are protective for colon adenocarcinoma (β = −0.13, P = 3.2 × 10−7), which is consistent with a report that high-dose vitamin C kills human colorectal cancer cells with KRAS or BRAF mutations47. Vitamin C is an essential nutrient for humans, acting as an antioxidant by protecting the body against oxidative stress, as a cofactor in enzymatic reactions including collagen synthesis, and as a structural component for blood vessels, cartilage, and muscle48. Vitamin C supplementation has been broadly recommended to help protect cells against the effects of free radicals and has generally been found to be safe. Further investigation is needed to understand whether the effects we identified are effects of vitamin C itself or other biological processes.
Potential independent causal metabolic pathways for the same disease
Our univariable MR identified 53 disease traits with more than one putative causal metabolite, which comprised 152 potential causal associations with 41 metabolites (see Summary of MR results). This could occur due to direct causal effects of multiple metabolites, mediation of effects of one metabolite by another, or it could result from heritable confounding of one metabolite by another. To gain a better understanding of these results and to reduce the risk of false positives due to heritable confounding, we used multivariable MR23 to jointly estimate the direct effects of all metabolites implicated for a single disease in the univariable MR analysis. Multivariable MR identified 20 significant putative causal effects of 17 metabolites on 23 disease traits at P < 0.05 (Supplementary Data 4). To provide additional insight, we computed both phenotypic correlation and correlation of IV effects (rIV) for each pair of the 70 significant metabolites (Fig. 3 and Supplementary Figs. 9–10; see “Methods”). We found strong correlations between some pairs of potential causal metabolites for the same disease traits (absolute rIV median = 0.84, mean = 0.64, range = 0.00033–0.99; Supplementary Fig. 11).
For atrial fibrillation, we identified a putative risk effect of plasma lipid N-acetyl-2-amino-octanoate (β = 0.068, P = 2.3 × 10−7) and potential protective effects of plasma amino acid N-delta-acetylornithine (β = −0.047, P = 5.1 × 10−7) and lipid glycocholate sulfate (β = −0.061, P = 2.9 × 10−8). N-acetyl-2-aminooctanoate and N-delta-acetylornithine have highly correlated IVs (rIV = 0.74), but neither has correlated IVs with glycocholenate sulfate (|rIV| < 0.08). Multivariable MR analysis identified direct potential causal effects on atrial fibrillation of lipids N-acetyl-2-amino-octanoate (β = 0.054, P = 7.2 × 10−3) and glycocholenate sulfate (β = −0.058, P = 2.6 × 10−7), but no causal effect of N-delta-acetylornithine, conditional on the other two metabolites (β = −0.020, P = 0.17; Supplementary Data 4). In the METSIM study, we identified 816 individuals with atrial fibrillation (see “Methods”). Logistic regression identified a significant association between plasma N-acetyl-2-amino-octanoate level and risk of atrial fibrillation (β = 0.080, P = 0.045), directionally consistent with the putative causal effect estimated in MR. We observed no significant associations with N-delta-acetylornithine (β = 0.057, P = 0.148) or glycocholenate sulfate levels (β = 0.072, P = 0.064), however, observational associations may be biased by unmeasured confounding variables.
For anxious personality disorder, we identified putative risk effects of plasma xenobiotic N-methylpipecolate (β = 0.28, P = 2.8 × 10−7) and amino acid N6,N6-dimethyllysine (β = 0.24, P = 8.6 × 10−8) and a potential protective effect of plasma lipid androsterone sulfate (β = −0.27, P = 1.5 × 10−7). N6,N6-dimethyllysine, and N-methylpipecolate have high IV correlation (rIV = 0.98) and share 42.4% of their IVs at a threshold of metabolite association P ≤ 1 × 10−5, but neither has correlated IVs with androsterone sulfate (|rIV| < 0.03). Because of the high IV correlation between N6,N6-dimethyllysine, and N-methylpipecolate, there is insufficient independent genetic signal to tease apart their putative causal effects on anxious personality disorder using multivariable MR. We performed two multivariable MR analyzes, including androsterone sulfate and either N-methylpipecolate or N6, N6-dimethyllysine. In both cases, the data were consistent with direct effects of both included metabolites N-methylpipecolate (β = 0.29, P = 6.2 × 10−8) and androsterone sulfate (β = −0.27, P = 7.6 × 10−8) or at N6, N6-dimethyllysine (β = 0.24, P = 5.0 × 10−7) and androsterone sulfate (β = −0.27, P = 2.5 × 10−7). N6, N6-dimethyllysine, and N-methylpipecolate are likely derived from lysine and pipecolate, respectively. Previous studies have suggested that pipecolate is an intermediate product of lysine metabolism by the cyclodeaminases RapL/FkbL49. The ratio of N6,N6-dimethyllysine, and N-methylpipecolate may indirectly reflect the relative levels of lysine and pipecolate. To further investigate the putative causal role of the relative levels of N6,N6-dimethyllysine and N-methylpipecolate on anxious personality disorder, we created a metabolite ratio between N6,N6-dimethyllysine and N-methylpipecolate and carried out a GWAS on the ratio, identifying six independent association signals in the AKR1C1/AKR1C2/AKR1C3/AKR1C4/AKR1C8, NAT8, PYROXD2, SLC6A20, and SLC7A9 regions (P < 5.0 × 10−8) (Supplementary Data 5 and Supplementary Fig. 12). MR identified evidence for a potential causal effect of increased N6,N6-dimethyllysine:N-methylpipecolate ratio on risk of anxious personality disorder (β = −0.34; P = 0.047; Supplementary Fig. 13; see “Methods”). The pattern we observe in which N6,N6-dimethyllysine and N-methylpipecolate both increase risk of anxious personality disorder, but an increase in their ratio confers a putative protective effect supports a hypothesis that N-methylpipecolate acts as a mediator in the potential causal pathway of N6,N6-dimethyllysine on anxious personality disorder (Fig. 4). This is consistent with previous reports that pipecolate is an intermediate product of lysine metabolism49.
Discussion
In this study, we systematically screened for potential causal effects of 1099 plasma metabolites on 2099 disease endpoints using two-sample univariable and multivariable MR analysis. We identified evidence for 282 putative causal effects of 70 plasma metabolites on 183 disease endpoints. We characterized the sharing of metabolite putative causal effects across 53 human diseases and showed the heterogeneity of causal metabolic pathways in disease pathophysiology. This study uncovers modifiable risk metabolites for disease intervention and underscores a potential causal role of plasma metabolites in human health.
We identified evidence for putative causal effects of 70 plasma metabolites on 183 human diseases. The relationships of many plasma metabolites with diseases have not been studied previously. These findings have several implications. First, they provide potential targets for disease intervention. Many plasma metabolites levels can be modified by diet and lifestyle changes. For example, we identified that high plasma sulfate levels increased the risk of chronic kidney disease. A wide range of food and beverages has been suggested as sources of dietary sulfate. We can, in principle, reduce plasma sulfate levels by reducing the consumption of these foods and beverages.
Second, these findings help elucidate disease biology and prioritize therapeutic targets for human diseases. For example, the risk of high plasma sulfate in chronic kidney disease suggested SLC13A1 as a potential drug target for chronic kidney disease. The potentially protective effect of high 2-arachidonoyl-GPC (20:4) level on frontotemporal dementia bolsters the hypothesis that neuroinflammation contributes to the pathophysiology of dementia40,42. We characterize the sharing of potential causal metabolites and their heterogeneity effects across human diseases. The sharing may help explain some disease comorbidity and reveal previously unappreciated connections between diseases. For example, we identified evidence for 126 heterogeneous putative causal effects of 15 N-acyl-alpha amino acids on 67 disease traits of 14 categories, highlighting an impact of synthesis or degradation of N-acetylated proteins on human health.
Our study showed that metabolites with significant univariable putative causal effects on the same disease traits might act in disease pathogenesis through separate metabolic pathways or through a metabolic cascade. We identified two independent metabolic pathways among three tested metabolites for atrial fibrillation and for anxious personality disorder, highlighting the heterogeneity of potential causal metabolic pathways in human diseases. We suggested that a putative causal effect of N-delta-acetylornithine on atrial fibrillation might be induced by IVs shared with N-acetyl-2-amino-octanoate. In contrast, we suggested that N-methylpipecolate might act as a downstream mediator in the causal pathway of N6,N6-dimethyllysine on anxious personality disorder, which could partially explain the strong IV correlation between N-methylpipecolate and N6,N6-dimethyllysine. Previous survival analyzes detected a significant positive association of glycocholenate sulfate levels with atrial fibrillation incidence50, while our analysis identified a negative association of plasma glycocholenate sulfate with atrial fibrillation. The effect of plasma glycocholenate sulfate on atrial fibrillation warrants further investigation.
MR is an advantageous method for screening causal hypotheses about relatively un-studied but heritable exposures because it relies on less domain-specific knowledge than traditional causal inference methods based on observational data. However, there are also limitations to the conclusions that can be drawn from MR estimates. MR effects may reflect causal effects of a related exposure that shares genetic regulation with the measured exposure51. In our case, MR estimates may reflect the causal effects of metabolite levels in non-plasma tissues or effects of related but unmeasured molecules in the same biochemical pathway. Despite this limitation, metabolome-wide MR remains a powerful tool for identifying pathways and molecules that influence disease susceptibility.
The MR estimates in this study differ from RCT estimates in several important ways24. MR estimates do not correspond to the effect of a specific intervention, and interventions on the plasma levels of implicated metabolites could have different effects than the ones estimated by MR. This could occur because (a) MR measures lifetime exposure effects, (b) plasma metabolite levels act as proxies for the activity of specific biological pathways, or (c) plasma metabolites act as proxies for metabolite levels in other tissues. The biochemical pathway regulating the metabolite may be the true causal factor, which warrants further investigation. For example, we identified a negative association of plasma 2-arachidonoyl-GPC (20:4) level with the risk of frontotemporal dementia, which might suggest a role of 2-arachidonoyl-GPC (20:4)-mediated neuroinflammation in the brain. However, further evidence is required to understand whether the modulation of plasma levels of 2-arachidonoyl-GPC (20:4) through intervention could modify dementia risk.
In addition, the complexity of metabolites and metabolic regulation presents another challenge for interpreting the metabolite-disease trait associations. We applied multivariable MR to estimate direct potential causal effects of multiple metabolites on the same disease. However, multivariable MR can only distinguish the effects of metabolites that have a sufficient number of distinct IVs. For example, we were unable to disentangle the effects of N-methylpipecolate and N6,N6-dimethyllysine on the risk of anxious personality disorder using multivariable MR because they share nearly all of their IVs. We were, therefore, only able to identify a potential causal effect related to the process that co-regulates the levels of these two metabolites. A metabolite ratio reflects the relative levels of two metabolites at a given time point. Our MR analysis identified a significant putative causal effect of the ratio between N6,N6-dimethyllysine, and N-methylpipecolate on anxious personality disorder. This causal estimate is consistent with the previous report that pipecolate is an intermediate product of lysine metabolism49. Though the metabolite ratio does not recapitulate the metabolic flux, we suggest that the relative levels of N-methylpipecolate and N6,N6-dimethyllysine may play a causal role in anxious personality disorder.
A limitation of our study is that METSIM and FinnGen study populations differ in some features. Our METSIM metabolite GWAS contained only non-diabetic males17, while FinnGen is not ascertained on sex or any disease status. Our putative causal effect estimates rely on the assumption that genetic regulation of metabolites is similar across sex and diabetes status. If these assumptions are violated, our estimates will be inaccurate. This issue is most likely to affect sexually differentiated metabolites such as androsterone sulfate.
In our MR analysis, we assumed that there was no sample overlap between the METSIM and FinnGen samples. METSIM is not directly part of the FinnGen study. However, since FinnGen is a nationwide biobank, there could be a small amount of sample overlap.
In conclusion, we systematically evaluated the potential causal effects of 1099 plasma metabolites on the risk of 2099 disease endpoints. We identified evidence for 282 putative causal effects of 70 plasma metabolites on 183 disease traits. Our study uncovered potential causal effects of plasma metabolites on a broad spectrum of human diseases. These findings highlight heterogeneous and shared potential causal effects of plasma metabolites on human diseases.
Methods
Ethics
In the present study, we used publicly available datasets from previous analyzes of the METSIM and FinnGen studies. All METSIM participants provided written informed consent. The Ethics Committee at the University of Eastern Finland and the Institutional Review Board at the University of Michigan approved the METSIM metabolomics study. FinnGen obtained participants' informed consent for biobank research based on the Finnish Biobank Act. Research cohorts collected prior to the Finnish Biobank Act coming into effect (September 2013) and the start of FinnGen (August 2017) obtained study-specific consents and later transferred the consents to the Finnish Biobank after the National Supervisory Authority for Welfare and Health (Fimea) approved the recruitment protocols. This study was approved by the Ethics Committee at the University of Eastern Finland and the Institutional Review Board at the University of Michigan. All the study procedures were in compliance with the Declaration of Helsinki.
Metabolic syndrome in men (METSIM) metabolomics study
METSIM is a single-site cohort study designed to investigate risk factors for type 2 diabetes and cardiovascular diseases52. It includes 10,197 Finnish men from Kuopio aged 45–74 years at baseline. We performed non-targeted metabolomics profiling in 6136 randomly selected non-diabetic participants using the Metabolon DiscoveryHD4 mass spectrometry platform (Durham, North Carolina, USA) on EDTA-plasma samples obtained after ≥10-h overnight fast during baseline visits from 2005 to 201017. We completed single-variant GWAS for 1391 metabolites, which identified 2030 independent metabolite associations17. For this study, we used GWAS summary statistics at 16.2 M genotyped or imputed genetic variants for the 1099 named metabolites with annotated biochemical identities17.
FinnGen study
FinnGen is designed to collect and analyze genome and healthcare data to identify diagnostic and therapeutic targets for human diseases31. FinnGen identified 3095 disease endpoints in release 7 using healthcare data from Finnish national registries: Drug Purchase and Drug Reimbursement and Digital and Population Data Services Agency; Digital and Population Data Services Agency; Statistics Finland; Register of Primary Health Care Visits (AVOHILMO); Care Register for Health Care (HILMO); and Finnish Cancer Registry. These registries recorded disease-relevant codes of the International Classification of Diseases (ICD) revisions 8, 9, and 10, cancer-specific ICD-O-3, Nordic Medico-Statistical Committee (NOMESCO) procedure, Finnish-specific Social Insurance Institute (KELA) drug reimbursement, and Anatomical Therapeutic Chemical (ATC)17. Each FinnGen participant was genotyped with an Illumina or Affymetrix array. Genotype imputation followed using the Finnish-specific Sequencing Initiative Suomi (SISu) v3 reference panel53. FinnGen carried out single-variant GWAS for each disease endpoint using mixed-model logistic regression in SAIGE54. For this study, we used GWAS summary statistics at 16.7 M genotyped or imputed genetic variants for all 3095 disease traits in up to 309,154 individuals from FinnGen release 7. After we finished the MR analysis, FinnGen made the release 8 publicly available, which includes GWAS summary statistics for 2202 disease traits. In comparison to FinnGen release 7, release 8 reduced the number of disease traits primarily by dropping redundant disease traits. To improve efficiency and reduce redundancy, we restricted our MR analysis results to 2099 of the 3095 disease traits that are included in FinnGen release 8.
Selection of IVs
We identified 16.2 M genetic variants shared between GWAS summary files across all the 1099 metabolites in METSIM and the 2099 disease traits in FinnGen release 7. To identify independent genetic variants as IVs for MR, we performed LD clumping in the GWAS results for each of the 1,099 metabolites in Plink to ensure resulting variants achieved association P < 10−5 and each pair of variants within 1 Mb distance has LD r2 < 0.0155. For LD calculation, we used genotypes in 8433 METSIM individuals without close relatives defined as pairwise kinship coefficients < 0.125.
Primary univariable MR analysis
To identify potential causal metabolites for human diseases, we performed two-sample univariable MR to test the putative causal effect of each of the 1099 plasma metabolites on each of the 2099 disease traits using MR–robust adjusted profile scoring (MR-RAPS)21. MR-RAPS allows for horizontal pleiotropy and enables the inclusion of IVs with weak effects by accounting for the precision of IV exposure and IV outcome associations21. We used over-dispersion and Tukey robust loss function parameters in MR-RAPS. We used the IVs for each metabolite as individual covariates in MR-RAPS. We conducted the MR-RAPS analysis using the mr.raps R package. To identify significant potential causal effects, we applied an FDR < 1% to account for multiple tests.
To test the potential causal effects of protein aminoacylase 1 on plasma levels of three N-acyl-alpha amino acids, N-acetylvaline, N-acetylglutamate, and N-acetylmethionine, and the risk of type 2 diabetes, we performed two-sample univariable MR. deCODE measured plasma aminoacylase 1 level using SomaScan version 4 in 35,559 Icelanders, followed by protein quantitative trait loci (pQTL) analysis, which identified three independent cis-pQTLs for aminoacylase 134. Among the three cis-pQTLs, the top pQTL site, rs121912698 was available in both METSIM and FinnGen. We used this variant as single IV and performed a Wald ratio test to evaluate causal effects of protein aminoacylase 1 on plasma levels of the three N-acyl-alpha amino acids and risk of type 2 diabetes in the two sample R packages.
Sensitivity analysis
For each of the 282 metabolite-disease trait pairs that we detected in MR-RAPS, we evaluated its potential causal association using four alternative MR methods and performed sensitivity tests using MR-Egger intercept, MR-PRESSO global, and Steiger filtering tests (see Supplementary Methods). The MR-RAPS method provides some robustness to false positives due to horizontal pleiotropy through the use of a robust loss function and allowance for over-dispersion. However, false positives may still occur if a metabolite and disease share a common heritable cause (heritable confounding or correlated horizontal pleiotropy). We performed an additional sensitivity analysis to evaluate the potential influence of heritable confounding mediated by other metabolites. We removed all IVs that were associated with another metabolite at P < 5 × 10−8 in the METSIM metabolite GWAS17 and repeated all the univariable MR-RAPS analyzes. This method is very conservative because not all shared variants result in bias in the MR estimate, and the removal of shared variants substantially reduces power in many cases.
Multivariable MR
To detect direct potential causal effects among metabolites with significant univariable putative causal effects on the same disease trait, we performed multivariable MR using genome-wide MR Analysis under Pervasive PLEiotropy (GRAPPLE)23. For each disease with multiple implicated metabolites, we combined IVs for all implicated metabolites and performed LD clumping as in the Selection of IVs to ensure that all IVs were nearly independent and the IVs with the lowest association p values across metabolites were prioritized. We used default parameters in GRAPPLE and applied a nominal P < 0.05 as the significance threshold.
To evaluate whether the metabolite-disease trait associations that we identified in the univariable MR were independent of common potential lifestyle confounders alcohol drinking, cigarette smoking, and sleep duration, we identified (a) GWAS for alcohol drinking status (GWAS ID: ukb-d-20117_2), ever smoked (GWAS ID: ukb-b-20261), and sleep duration (GWAS ID: ukb-b-4424) from the IEU OpenGWAS database (https://gwas.mrcieu.ac.uk) and (b) IVs for each of these three phenotypes (see Selection of IVs). We performed multivariable MR through including one of the three phenotypes at a time using GRAPPLE. We used FDR < 5% as the significance threshold.
Estimation of IV correlation between metabolites
To estimate the degree to which each pair of metabolites shares genetic IVs, we computed the proportion of overlapping IVs and the IV correlation. For each metabolite pair, we took the union of IVs for both metabolites. We then performed LD clumping using LD r2 < 0.01 in 1 Mb distance in Plink55 to remove correlated IVs. Finally, we extracted association statistics for the resulting set of IVs for both metabolites. For LD calculation, we used genotypes in 8433 METSIM individuals with pairwise kinship coefficients < 0.125. We calculated the proportion of IVs shared as the proportion of the LD clumped union set of IVs with association P ≤ 10−5 for both metabolites. We calculated the IV correlation, rIV, as the correlation of association statistics of the LD clumped union set of IVs with the two metabolites.
Associations of N-acetyl-2-aminooctanoate, N-delta-acetylornithine, and glycocholenate sulfate with atrial fibrillation in METSIM
Among the 6102 METSIM participants with measured plasma N-acetyl-2-aminooctanoate, N-delta-acetylornithine, and glycocholenate sulfate levels at baseline, we identified 816 with atrial fibrillation in METSIM as of June 2022. To test for associations between plasma metabolite levels and presence of atrial fibrillation, we used logistic regression with covariates baseline study age, body mass index (BMI), binary cigarette smoking status (ever smoker versus never smoker), alcohol drinking amount, baseline systolic and diastolic blood pressure, and lipid and hypertension medication use.
GWAS for metabolite ratio of N6,N6-dimethyllysine, and N-methylpipecolate and causal effect of the ratio on anxious personality disorder
In the 6136 METSIM participants17, we computed the ratio of N6,N6-dimethyllysine to N-methylpipecolate by dividing the level of N6,N6-dimethyllysine by the level of N-methylpipecolate. We regressed out covariates study age, Metabolon batches, and lipid-lowering medication status, and inverse normalized the residuals. We performed single-variant GWAS for the resulting residuals in Regenie v3.2.256. For the chromosomes on which we identified genome-wide significant associations (P < 5.0 × 10−8), we performed recursively a stepwise conditional test to identify near-independent association signals until no variant attained P < 5.0 × 10−817. To test the potential causal effect of the metabolite ratio on the risk of anxious personality disorder, we performed a univariable MR test using MR-RAPS21. We used the near-independent association signals for the metabolite ratio that are also available in the GWAS for anxious personality disorder as IVs. We conducted the MR-RAPS analysis with over dispersion and Tukey robust loss function parameters using the mr.raps R package.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
FinnGen genome-wide summary statistics are available at https://r7.finngen.fi. Full summary statistics from the genome-wide association studies of the 1099 plasma metabolites are available at https://pheweb.org/metsim-metab/. The MR results are available in Supplementary Data 2, 3, and 4. Source data are provided with this paper.
References
Wishart, D. S. Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 99, 1819–1875 (2019).
Ahola-Olli, A. V. et al. Circulating metabolites and the risk of type 2 diabetes: a prospective study of 11,896 young adults from four Finnish cohorts. Diabetologia 62, 2298–2309 (2019).
Lind, L., Fall, T., Ärnlöv, J., Elmståhl, S. & Sundström, J. Large-scale metabolomics and the incidence of cardiovascular disease. J. Am. Heart Assoc. 12, e026885 (2023).
Wen, D. et al. Metabolite profiling of CKD progression in the chronic renal insufficiency cohort study. JCI Insight 7, e161696 (2022).
Peng, L. et al. Increased soluble epoxide hydrolase activity positively correlates with mortality in heart failure patients with preserved ejection fraction: evidence from metabolomics. Phenomics 3, 34–49 (2023).
Wang, T. J. et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 17, 448–453 (2011).
Wu, Q. et al. Prediction of metabolic disorders using NMR-based metabolomics: the Shanghai Changfeng study. Phenomics 1, 186–198 (2021).
Surendran, P. et al. Rare and common genetic determinants of metabolic individuality and their effects on human health. Nat. Med. 28, 2321–2332 (2022).
Chen, Y. et al. Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat. Genet. 55, 44–53 (2023).
Schlosser, P. et al. Genetic studies of paired metabolomes reveal enzymatic and transport processes at the interface of plasma and urine. Nat. Genet. 55, 995–1008 (2023).
Shin, S. Y. et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 46, 543–550 (2014).
Chen, L. et al. Influence of the microbiome, diet, and genetics on inter-individual variation in the human plasma metabolome. Nat. Med. 28, 2333–2343 (2022).
Bar, N. et al. A reference map of potential determinants for the human serum metabolome. Nature 588, 135–140 (2020).
Long, T. et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 49, 568–578 (2017).
Illig, T. et al. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 42, 137–141 (2010).
Bartel, J. et al. The human blood metabolome-transcriptome interface. PLoS Genet. 11, e1005274 (2015).
Yin, X. et al. Genome-wide association studies of metabolites in Finnish men identify disease-relevant loci. Nat. Commun. 13, 1644 (2022).
Yin, X. et al. Integrating transcriptomics, metabolomics, and GWAS helps reveal molecular mechanisms for metabolite levels and disease risk. Am. J. Hum. Genet. 109, 1727–1741 (2022).
Richmond, R. C. & Davey Smith, G. Mendelian randomization: concepts and scope. Cold Spring Harb. Perspect. Med. 12, a040501 (2022).
Davies, N. M., Holmes, M. V. & Davey Smith, G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362, k601 (2018).
Zhao, Q., Wang, J., Hemani, G., Bowden, J. & Small, D. S. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann. Stat. 48, 1742–1769 (2020).
Burgess, S. & Thompson, S. G. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am. J. Epidemiol. 181, 251–260 (2015).
Wang, J. et al. Causal inference for heritable phenotypic risk factors using heterogeneous genetic instruments. PLoS Genet. 17, e1009575 (2021).
Sanderson, E. et al. Mendelian randomization. Nat. Rev. Method Prim. 2, 6 (2022).
Zheng, J. et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat. Genet. 52, 1122–1131 (2020).
Zhao, H. et al. Proteome-wide Mendelian randomization in global biobank meta-analysis reveals multi-ancestry drug targets for common diseases. Cell Genom. 2, None (2022).
Sun, Y., Lu, Y. K., Gao, H. Y. & Yan, Y. X. Effect of metabolite levels on type 2 diabetes mellitus and glycemic traits: a mendelian randomization study. J. Clin. Endocrinol. Metab. 106, 3439–3447 (2021).
Qian, L. et al. Genetically determined levels of serum metabolites and risk of neuroticism: a mendelian randomization study. Int J. Neuropsychopharmacol. 24, 32–39 (2021).
Lord, J. et al. Mendelian randomization identifies blood metabolites previously linked to midlife cognition as causal candidates in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 118, e2009808118 (2021).
Qin, Y. et al. Genome-wide association and Mendelian randomization analysis prioritizes bioactive metabolites with putative causal effects on common diseases. medRxiv https://doi.org/10.1101/2020.08.01.20166413 (2020).
Kurki, M. I. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518 (2023).
Shu, H. et al. Emerging roles of ceramide in cardiovascular diseases. Aging Dis. 13, 232–245 (2022).
Lotta, L. A. et al. Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: a Mendelian randomisation analysis. PLoS Med. 13, e1002179 (2016).
Ferkingstad, E. et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 53, 1712–1721 (2021).
Ngo, D. et al. Proteomic profiling reveals biomarkers and pathways in type 2 diabetes risk. JCI Insight 6, e144392 (2021).
Zhuang, Z. et al. Causal relationships between gut metabolites and Alzheimer’s disease: a bidirectional Mendelian randomization study. Neurobiol. Aging 100, 119.e115–119.e118 (2021).
Huang, S. Y. et al. Investigating causal relations between circulating metabolites and Alzheimer’s disease: a Mendelian randomization study. J. Alzheimers Dis. 87, 463–477 (2022).
Chen, H. et al. Assessing causal relationship between human blood metabolites and five neurodegenerative diseases with GWAS summary statistics. Front. Neurosci. 15, 680104 (2021).
Knuplez, E. & Marsche, G. An updated review of pro- and anti-inflammatory properties of plasma lysophosphatidylcholines in the vascular system. Int J. Mol. Sci. 21, 4501 (2020).
Kinney, J. W. et al. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimers Dement. 4, 575–590 (2018).
Semba, R. D. Perspective: the potential role of circulating lysophosphatidylcholine in neuroprotection against Alzheimer disease. Adv. Nutr. 11, 760–772 (2020).
Hammouda, S. et al. Genetic variants in FADS1 and ELOVL2 increase level of arachidonic acid and the risk of Alzheimer’s disease in the Tunisian population. Prostaglandins Leukot. Ess. Fat. Acids 160, 102159 (2020).
Kovesdy, C. P. Epidemiology of chronic kidney disease: an update 2022. Kidney Int. Suppl. 12, 7–11 (2022).
Niwa, T. Role of indoxyl sulfate in the progression of chronic kidney disease and cardiovascular disease: experimental and clinical effects of oral sorbent AST-120. Ther. Apher. Dial. 15, 120–124 (2011).
Leite, M. I. et al. Myasthenia gravis and neuromyelitis optica spectrum disorder: a multicenter study of 16 patients. Neurology 78, 1601–1607 (2012).
Li, X., Sundquist, J. & Sundquist, K. Subsequent risks of Parkinson disease in patients with autoimmune and related disorders: a nationwide epidemiological study from Sweden. Neurodegener. Dis. 10, 277–284 (2012).
Yun, J. et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science 350, 1391–1396 (2015).
Li, Y. & Schellhorn, H. E. New developments and novel therapeutic perspectives for vitamin C. J. Nutr. 137, 2171–2184 (2007).
Gatto, G. J. Jr, Boyne, M. T. 2nd, Kelleher, N. L. & Walsh, C. T. Biosynthesis of pipecolic acid by RapL, a lysine cyclodeaminase encoded in the rapamycin gene cluster. J. Am. Chem. Soc. 128, 3838–3847 (2006).
Alonso, A. et al. Serum metabolomics and incidence of atrial fibrillation (from the atherosclerosis risk in communities study). Am. J. Cardiol. 123, 1955–1961 (2019).
Burgess, S. et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res. 4, 186 (2019).
Laakso, M. et al. The metabolic syndrome in men study: a resource for studies of metabolic and cardiovascular diseases. J. Lipid Res. 58, 481–493 (2017).
Lim, E. T. et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 10, e1004494 (2014).
Zhou, W. et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 50, 1335–1341 (2018).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53, 1097–1103 (2021).
Acknowledgements
We thank all the participants and investigators in the METSIM and FinnGen studies. This work was supported by the National Institutes of Health (NIH) under awards U01 DK062370 (M.B.), R35 GM138121 (X.Q.W.), R01 DK119380 (X.Q.W.), the American Diabetes Association Postdoctoral Fellowship (1-19-PDF-061, X.Y.), the University of Michigan Precision Health Scholarship (X.Y.), the Academy of Finland under grant no. 321428 (M.L.), the Sigrid Juselius Foundation (M.L.), the Academy of Finland Center of Excellence in Complex Disease Genetics under grant no. 312062 and 336820 (S.R.), grant no. 312074 and 336824 (A.P.), the Finnish Foundation for Cardiovascular Research (S.R.), University of Helsinki HiLIFE Fellow and Grand Challenge grants, and Horizon 2020 Research and Innovation Program (grant no. 101016775 “INTERVENE”) (S.R.), National Natural Science Foundation of China (X.Y.), Jiangsu Professorship (X.Y.), and Nanjing Medical University under award NMUR20230003 (X.Y.).
Author information
Authors and Affiliations
Contributions
X.Y. and J.M. designed the study. M.B. and M.L. supervised the study. X.Y. performed the analysis and wrote the manuscript. X.Y., M.B., M.L., and J.M. revised the manuscript. L.F.S., A.O., M.L., S.R., M.D., and A.P. collected the data. J.L., D.B., J.O., A.K., A.U.J., X.M.C., H.M.S., L.L., R.Y.P., and Z.J.X. contributed to the data analysis and manuscript revision. L.J.S., C.F.B., E.B.F., and X.W. interpreted the results.
Corresponding authors
Ethics declarations
Competing interests
E.B.F. is an employee and stockholder of Pfizer. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Zhengbao Zhu, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yin, X., Li, J., Bose, D. et al. Assessing the potential causal effects of 1099 plasma metabolites on 2099 binary disease endpoints. Nat Commun 16, 3039 (2025). https://doi.org/10.1038/s41467-025-58129-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58129-2