Abstract
Frontotemporal lobar degeneration with neuronal inclusions of the TAR DNA-binding protein 43 (FTLD-TDP) is a fatal neurodegenerative disorder with only a limited number of risk loci identified. We report our comprehensive genome-wide association study as part of the International FTLD-TDP Whole-Genome Sequencing Consortium, including 985 patients and 3,153 controls compiled from 26 institutions/brain banks in North America, Europe and Australia, and meta-analysis with the Dementia-seq cohort. We confirm UNC13A as the strongest overall FTLD-TDP risk factor and identify TNIP1 as a novel FTLD-TDP risk factor. In subgroup analyzes, we further identify genome-wide significant loci specific to each of the three main FTLD-TDP pathological subtypes (A, B and C), as well as enrichment of risk loci in distinct tissues, brain regions, and neuronal subtypes, suggesting distinct disease aetiologies in each of the subtypes. Rare variant analysis confirmed TBK1 and identified C3AR1, SMG8, VIPR1, RBPJL, L3MBTL1 and ANO9, as novel subtype-specific FTLD-TDP risk genes, further highlighting the role of innate and adaptive immunity and notch signaling pathway in FTLD-TDP, with potential diagnostic and novel therapeutic implications.
Similar content being viewed by others
Introduction
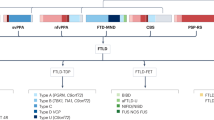
Frontotemporal lobar degeneration (FTLD) is one of the leading causes of dementia in individuals younger than 65 years, but can also affect individuals later in life. The predominant clinical presentations of FTLD are behavioral and language dysfunction resulting in behavioral variant frontotemporal dementia (bvFTD)1, semantic variant primary progressive aphasia (svPPA), or nonfluent variant primary progressive aphasia (nfvPPA)2. The diagnosis of FTLD can be established with certainty only with neuropathologic postmortem examination and is characterized neuropathologically by significant atrophy of the frontal and temporal lobes and accumulation of abnormal neuronal and/or glial inclusions upon immunohistochemical analysis. FTLD-TDP, characterized by neuronal and cytoplasmic aggregates of the DNA and RNA-binding protein TDP-43, is one of the two main pathological subtypes (the other being FTLD-Tau) and can be further classified into five FTLD-TDP subtypes (FTLD-TDP A-E) based on the distribution of the neuronal cytoplasmic TDP-43-positive inclusions and dystrophic neurites in the cortical layers3,4. The main subtypes are types A, B, and C. Type A includes patients with moderate to numerous TDP-43-immunoreactive neuronal cytoplasmic inclusions (NCIs) and short dystrophic neurites (DNs) concentrated mainly in the upper cortical layers II/III. Type B encompasses patients with moderate to numerous TDP-43-immunoreactive NCIs and sparse DNs distributed across all cortical layers. Type C is designated for patients where long dystrophic neurites are prevalent primarily in the upper cortices, and NCIs are infrequent. Extensive research is focused on the accurate prediction of the underlying FTLD neuropathology in clinical FTLD patients; however, despite some recent reports on the possible use of Tau and TDP43 levels in plasma extracellular vesicles, this remains a major diagnostic challenge5. Importantly, among FTLD-TDP patients, a few clinicopathological correlations exist. Specifically, FTLD-TDP C pathology is strongly overrepresented in svPPA patients6, whereas patients with bvFTD with concomitant amyotrophic lateral sclerosis (ALS) almost invariably present as FTLD-TDP B at autopsy2,4,7.
A small number of autosomal dominant genes and risk factors associated with FTLD-TDP have been reported8,9,10,11,12,13,14. A quarter of FTLD-TDP patients are associated with repeat expansions in C9orf72, which are mainly enriched in patients with FTLD-TDP B, while autosomal dominant loss-of-function (LOF) mutations in GRN explain 10–15% of familial patients and invariably lead to FTLD-TDP A15,16. The first FTLD-TDP genome-wide association study (GWAS) identified the TMEM106B locus (rs1990622); however, this signal was strongly driven by the FTLD-TDP A patients with GRN mutations included in that study17. Three additional FTLD-TDP loci, UNC13A, DPP6, and HLA-DQA2, were identified in phase I of the International FTLD-TDP whole-genome sequencing (WGS) consortium and require replication in larger datasets12. Importantly, most FTLD-TDP patients are not yet genetically explained, and the relatively small sample size precluded rare variant analyzes in phase I.
To validate and identify new genetic risk factors, we doubled the original sample size of the FTLD-TDP WGS consortium by not only sequencing more pathologically confirmed FTLD-TDP patients but also including clinically defined FTLD subtypes enriched for specific FTLD-TDP pathological subtypes at autopsy. In this work, we performed association study analyzes of both common (minor allele frequency, MAF, >1%) and rare (MAF < 1%) variants, followed by comprehensive gene-prioritization, enrichment analyzes, and co-localization studies. We identify novel FTLD-TDP risk loci, including novel risk genes and loci specific to FTLD-TDP pathological subtypes. Our study highlights similarities and differences between FTLD-TDP and other neurodegenerative diseases, while unique biological processes in specific tissues, brain regions, and cell types were found to characterize individual FTLD-TDP pathological subtypes.
Results
Common variant genome-wide association analysis (GWAS)
To identify novel common FTLD-TDP genetic risk factors, we performed single variant GWAS using an additive disease risk model for 6,568,099 common variants in 985 patients and 3153 controls free of neurodegenerative disorder that passed quality control (QC). Variants with MAF > 1% in patients or controls were considered. Combining all patients (FTLD-TDP All), we identified one genome-wide significant signal at the UNC13A locus (rs8111424, OR = 1.37, P = 1.17x10-8). We also performed separate GWAS within the FTLD-TDP A, FTLD-TDP B, and FTLD-TDP C subtypes (Fig. 1, Tables 1–2, Supplementary Data 1, Supplementary Fig. 1). The most significant locus identified in FTLD-TDP A was GRN (rs5848; OR = 1.89, P = 5.57 × 10−9). In phase I, this locus only reached genome-wide significance under an exploratory recessive model12, and also now, the recessive model provided an even stronger association (OR = 4.12, P = 8.28 × 10−15). We further detected 3 additional new genome-wide significant loci in FTLD-TDP A: TINAG (rs138698596), MZT1 (rs138959102), and FARP2 (rs886815). In FTLD-TDP B, we detected a genome-wide significant association at the UNC13A locus (rs12973192). The lead variant rs12973192 is in linkage disequilibrium (LD) with rs8111424 (D′ = 1; r2 = 0.43) identified in the FTLD-TDP All analysis. We further detected 3 new genome-wide significant risk loci in FTLD-TDP B: TNIP1 (rs871269), RCL1 (rs76742217), PDS5B (rs527749954), and one in FTLD-TDP C C19orf52 (also known as TIMM29, rs576561313). Given the large number of svPPA patients included in the FTLD-TDP C group and the evolving literature on the exact clinicopathological correlation of this clinical subtype with FTLD-TDP C, we performed an additional association study including only pathologically confirmed FTLD-TDP C (referred to as FTLD-TDP C*, Supplementary Fig. 2). This analysis showed 4 genome-wide significant loci in LRP1B (rs35902922), COL22A1 (rs146589681), TMEM135 (rs117642163), and TRPC4 (rs540663062). These loci were only nominally significant in the analysis including the svPPA patients (LRP1B: P = 1.72 × 10−4; OR = 4.66, COL22A1: P = 0.03; OR = 5.32; TMEM135: P = 1.68 × 10−3; OR = 2.19, and TRPC4 P = 1.25 × 10−4; OR = 2.91).
Association studies were performed using logistic regression with allele dosage as the predictor assuming log-additive allele effects adjusted for relevant covariates. Raw two-sided P are reported as −log10(P). A Manhattan plot of the FTLD-TDP All patients versus controls association study (inflation, λ = 1.05). B Manhattan plot of the FTLD-TDP A patients versus controls association study (λ = 1.07). C Manhattan plot of the FTLD-TDP B patients versus controls association study (λ = 1.06). D Manhattan plot of the FTLD-TDP C patients versus controls association study (λ = 1.05). The red-dotted line represents the genome-wide significance level (p = 5 × 10−8).
We also specifically assessed evidence for association at previously reported FTLD risk loci12,14,17,18. Besides the UNC13A locus, which we previously identified in our Phase I study, none of the other reported risk loci were replicated at the genome-wide significance level (Supplementary Data 2). Notably, TMEM106B rs1990622 almost reached the genome-wide significance threshold in the FTLD-TDP A subgroup (P = 2.7 × 10−7, OR = 0.80).
Gene prioritization
In order to prioritize risk genes and identify possible biological mechanisms, we applied a range of variant annotation and molecular quantitative trait loci (QTL)-GWAS integration analyzes as previously described19 on all variants (Supplementary Data 3–14). Genes included in the analyzes are listed in Supplementary Data 4. We integrated different levels of evidence using a weighting scheme and obtained a weighted sum of the hits in different subcategories for each gene. We grouped candidate risk genes in genome-wide significant loci and in subthreshold loci and prioritized them at two levels of confidence for being a likely risk gene as tier 1 (higher confidence) and tier 2 (lower confidence).
The gene prioritization analyzes nominated a total of 70 tier 1 and 286 tier 2 genes in 351 different loci for the 5 different GWAS analyzes (All, A, B, C, and C*; Fig. 2, Supplementary Data 4 and Supplementary Fig. 3–7). Our results showed that the nearest protein-coding genes were prioritized as tier 1 (n = 10) and tier 2 (n = 3) risk genes in the genome-wide significant loci for the distinct FTLD-TDP subtypes. Of the 10 tier 1 prioritized genes, 3 were found in common variant loci where molecular QTL-GWAS analyzes aided their prioritization (Fig. 2). First, in locus A4, GRN was prioritized through consistent expression QTL (eQTL) domain hits in bulk brain regions (eQTL PROSMAP DLPFC = 6.32 × 10−38 and betaROSMAP DLPFC = −0.25, eQTL colocalization (coloc) PPH4s of 81.8%-99.7%, and fine-mapped [posterior inclusion probability being 100%] expression transcriptome-wide association study [eTWAS] associations with P from 1.74 × 10−8 to 5.15 × 10−9 and Z-scores from −5.63 to −5.84) and in oligodendrocytes (cell type specific eQTL (ct-eQTL) coloc PPH4 = 90%) where genetic downregulation of GRN gene expression was associated with the FTLD-TDP A risk signal (Supplementary Data 6, 7, and 12), which was also observed in brain proteome-wide association study (PWAS) with the same effect direction for the FTLD-TDP A risk (PROSMAP DLPFC = 3.32 × 10−6, ZROSMAP DLPFC = −4.65, Supplementary Data 14). Second, in locus B1, TNIP1 was prioritized because the minor allele was associated with decreased TNIP1 expression (PROSMAP DLPFC = 2.40 × 10−4, betaROSMAP DLPFC = −0.10), the GWAS signal colocalized with a microglia splicing QTL (sQTL) associated with TNIP1 chr5:151032383-151035002 known splice junction (coloc PPH4 = 82.2%), and because the mQTL variants for cg03340667, a CpG ~3.7 kb upstream of the transcription start site (TSS) of the canonical transcript of TNIP1, colocalized with the GWAS variants (coloc PPH4 = 70%) in dorsolateral prefrontal cortex (DLPFC) (Supplementary Data 6, 8, and 9). Third, in locus B4, UNC13A was prioritized through an eQTL-GWAS colocalization in temporal cortex (coloc PPH4 = 81.82%, Supplementary Data 7). Furthermore, beyond genome-wide significant loci, we identified additional candidate prioritized risk genes in subthreshold regions through molecular QTL-GWAS coloc and TWAS analyzes, one important example being TMEM106B as the prioritized risk gene in locus A_S14. The FTLD-TDP A GWAS signal near TMEM106B colocalized with eQTL variants regulating TMEM106B gene expression in bulk brain regions (eQTL coloc PPH4s = 81.40% in MayoRNASeq temporal cortex and 89.66% in Genotype–Tissue Expression (GTEx) brain cortex, Supplementary Data 7). We also observed a significant eTWAS association in GTEx cortex (P = 4.13 × 10−7, Z = −5.06), together with a significant PWAS hit (P = 2.01 × 10−8, Zscore = 5.61) (Supplementary Data 12, 13). Finally, a significant hit in splicing TWAS in cortex (sTWAS, P = 6.66 × 10−7, Z = −4.97) predicted a decreased preference for the TMEM106B splice junction chr7:12224385-12229679 with the increased FTLD-TDP A GWAS risk, while we also observed methylation QTL (mQTL) coloc hits for two CpGs for TMEM106B (~500 bp upstream cg23422036 coloc PPH4 = 94.25% and intronic cg09613507 coloc PPH4 = 94.09%) (Supplementary Data 13, 9). We summarized our gene prioritization results in Fig. 2 for the genome-wide significant loci, and full results for all performed genome-wide association studies are presented in Supplementary Data 4–14 and Supplementary Fig. 3–7.
A visual summary of weighted evidence category scores for the prioritized genes within genome-wide significant loci in related FTLD subtype-specific GWAS summary statistics. Using the gene prioritization strategy in these selected loci, we prioritized a total of 13 genes in 12 genome-wide significant loci at two different confidence levels (10 tier 1 and 3 tier 2 prioritized genes). The leftmost squares, which are colored in red for FTLD-TDP A, in blue for FTLD-TDP B, in lighter green for FTLD-TDP C, and in darker green for FTLD-TDP C* specific analyzes, indicate the locus index numbers for the genome-wide loci. The types of evidence for each category are colored according to the six different domains to which they belonged. Weighted scores for each evidence category are rescaled to a 0–100 scale based on the maximum score a candidate gene can obtain from a category (see Supplementary Data 3). The darker colors represent higher scores in categories, while tier 1 prioritized genes are displayed in dark green, and tier 2 prioritized genes are displayed in light green. Only tier 1 and tier 2 genes are shown for each locus, whereas all candidate genes considered and scored can be found in Supplementary Data 4. MAFs (based on gnomAD v4 non-Finnish European samples) and CADD (v1.7) PHRED scores for rare and/or protein-altering rare variants are labeled in white within the respective squares. eQTL expression QTL, sQTL splicing QTL, mQTL methylation QTL, pQTL protein-expression QTL, haQTL histone acetylation QTL, coloc colocalization, eTWAS expression transcriptome-wide association study, sTWAS splicing transcriptome-wide association study, PWAS proteome-wide association study, Mon Mac monocytes and macrophages, LCL lymphoblastoid cell line, QTLCat the eQTL catalog.
Pathway analyzes
Next, we performed gene ontology (GO) analyzes on tier 1 prioritized genes, separately for each GWAS. The most significant term in the nominated genes in FTLD-TDP All was positive regulation of defense response to bacterium (P = 3.98 × 10−5). Lysosomal function appeared to be strongly affected in FTLD-TDP A with several genes such as GRN and TMEM106B (lysosomal organization GO term, P = 4.12 × 10−4) as well as cathepsin B (CTSB). We further detected enriched terms for retrograde transport in FTLD-TDP B (P = 2.21 × 10−3) driven by DENND2A and VPS53 genes, and for excitatory postsynaptic potential in FTLD-TDP C (P = 1.48 × 10−3) driven by DMPK and P2RX5 genes (Fig. 3 and Supplementary Data 15). Note that some genes belong to multiple significant GO terms; for instance, GRN is present in terms associated with FTLD-TDP A and B (Supplementary Data 15 and Supplementary Fig. 8). Finally, in the FTLD-TDP C* analysis (excluding svPPA patients), an enrichment for genes implicated in cellular homeostasis terms was observed (p = 8.77 × 10−03), driven by DMPK, CYBA, and CTRC (Supplementary Data 15 and Supplementary Fig. 2). Importantly, except for lysosomal transport, no terms overlapped between subtypes of FTLD-TDP, suggesting mostly distinct genetic etiologies in the different FTLD-TDP groups.
To further characterize genetic factors associated with FTLD-TDP, we performed gene-based analyzes on common variants with P < 10−5 using MAGMA. Analyzes of FTLD-TDP All did not yield exome-wide significant loci; however, FTLD-TDP A showed exome-wide significant signals for the two genes located at the GRN locus (FAM171A2, ITGA2B) and for TMEM106B (P = 4.74 × 10−7). The TMEM106B signal was driven by the rs10281425 variant (OR = 0.54, P = 2.12 × 10−7). No exome-wide significant signal was detected for the other FTLD-TDP pathological subtypes (FTLD-TDP B, C, and C*).
Cell type and brain region expression pattern of GWAS hits
To find tissues and cell types for which gene expression profiles were enriched for genes within FTLD-risk loci, we combined gene-based association statistics calculated using MAGMA with gene expression patterns from the GTEx project in a gene set enrichment analysis. Only loci with P < 10−5 for their respective traits were included. We observed an enrichment in genes expressed in brain tissue (cerebellum, frontal cortex, and cortex) in FTLD-TDP A and B (Fig. 4 and Supplementary Data 16). This was strikingly different from the signature observed in FTLD-TDP C, for which significant enrichment was only detected in non-central nervous system tissue, in particular small intestine terminal ileum (Fig. 4 and Supplementary Data 16). When removing svPPA patients (FTLD-TDP C* analysis), we again observed an enrichment in genes expressed in brain tissue, albeit without specificity for the frontal cortex (Supplementary Fig. 2). We also compared expression profiles of the genes at FTLD loci with expression of genes located at associated loci in Alzheimer’s disease and related disorders (ADRD)19 and ALS GWAS20 using available summary statistics and restricting our query to loci with P < 10−5 for their respective traits. FTLD-TDP subtypes presented with a distinct genetic signature as compared to these related disorders, highlighting the importance of regional specificity in FTLD-TDP.
A Enrichment of genes in multiple tissues, including 13 brain regions, and based on GTEX data in FTLD subgroups, ADRD, and ALS. Color represents the enrichment coefficient, and size indicates two-sided −log10 (FDR-adjusted Ps) of enrichment obtained by the linear regression model in the MAGMA gene property analysis. B Central nervous system cell type enrichment analyzes in FTLD subgroups, ADRD, and ALS. Color represents the enrichment coefficient, and size indicates two-sided −log10 (FDR-adjusted Ps) of enrichment obtained by the linear regression model in the MAGMA gene property analysis. Excitatory neurons and glial cells are highlighted in blue. Excitatory and inhibitory neurons from the PsychENCODE dataset were labeled based on their transcriptional profile from 1 to 8. Asterisks denote brain regions or cell types enriched with FDR P < 0.05. Cx cortex, Ex* Excitatory neuron, In* inhibitory neurons, Oligo oligodendrocytes, OPCs oligodendrocyte progenitor cells, Astro astrocytes, Endo endothelial cells, Per pericytes.
We subsequently queried PsychENCODE frontal-cortex single-cell RNA-seq datasets of human-derived brain samples to specify further which brain-specific enriched cell types express the genetic loci associated with FTLD-TDP risk (Fig. 4 and Supplementary Data 17). We observed a significant enrichment in genes expressed in excitatory neurons for FTLD-TDP A loci (Ex4 P = 3.55 × 10−2, Ex5b P = 2.72 × 10−2), and FTLD-TDP B loci (Ex8 P = 1.27 × 10−4), while no other cell type reached significance. While FTLD-TDP C loci were also significantly enriched in genes expressed in excitatory neurons (Ex3e P = 2.10 × 10−2), they were additionally enriched in genes expressed in astrocytes and oligodendrocyte progenitor cells (P = 2.53 × 10−2, P = 4.69 × 10−2). No significant enrichment was detected for FTLD-TDP C* (without svPPA) (Supplementary Fig. 2). Genes expressed in microglia were enriched only in ADRD gene loci (P = 1.90 × 10−2). Overall, loci comprising genes expressed in excitatory neurons were enriched in the three FTLD-TDP subtypes, with stronger specificity for specific neuron types in each FTLD-TDP subtype as compared to what was observed for ALS gene loci.
GWAS meta-analysis
To provide further support for the identified FTLD-TDP risk loci, we performed a meta-analysis of our FTLD-TDP cohort with the Dementia-seq study (phs001963.v1.p1), which includes 2102 clinical FTLD patients and 1748 controls. Given that this cohort lacks details on the FTLD pathology underlying each patient, pathological subgroup analyzes could not be performed. Meta-analysis confirmed UNC13A and identified the new TNIP1 locus as genome-wide significantly associated with FTLD (Prs12973192 = 8.85 × 10−10; Prs871269 = 3.42 × 10−8, respectively). Note that the most significant single-nucleotide variant (SNV) at the UNC13A locus was rs12608932 (P = 9.13 × 10−11), in strong LD with rs12973192 (r2 = 0.96, D′ = 0.99). Interestingly, while the TNIP1 signal in FTLD-TDP All was not genome-wide significant, it became significant when running the meta-analysis. The change in significance likely results from the inclusion of participants with bvFTD/ALS (with likely FTLD-TDP B pathology) as part of the DementiaSeq cohort.
Overlap with other neurodegenerative diseases
Both UNC13A and TNIP1 were previously associated with other neurodegenerative diseases19,21. Colocalization analyzes showed that our UNC13A signal was shared with ALS (coloc PPH4 = 95.71%) (Fig. 5A). On the contrary, for TNIP1, we found strong colocalization with the ADRD association signal19,21 (coloc PPH4 = 99.2%) and a weaker colocalization with ALS (71.5%), which was confirmed in a sensitivity analysis (coloc PPH4 = 20.1%, for π12 = 1.00 × 10−6), possibly reflecting multiple independent association signals in FTLD in this locus (Fig. 5B, C). Prompted by these findings, we performed global genetic correlations between FTLD-TDP (using FTLD-TDP All) and ALS and ADRD GWAS. A strong overall genetic correlation was observed between FTLD-TDP and ALS (P = 1.88 × 10−4, r = 0.88, standard error = 0.23), whereas no significant correlation was seen between FTLD-TDP and ADRD (P = 3.1 × 10−1, r = 0.22, standard error = 0.31).
A Genetic colocalization between the UNC13A locus in FTLD (meta-analysis) and ALS signal. B Genetic colocalization between the TNIP1 locus in FTLD (meta-analysis) and ALS. C Genetic colocalization between the TNIP1 locus in FTLD (meta-analysis) and ADRD. For A, B, C, chromosome position is located on the x-axis, and −log10 transformed raw two-sided P is represented on the y-axis. Each dot represents a SNV tested in the dataset for its association with disease status. Purple diamonds are the index SNVs reported. Linkage disequilibrium with index SNV is indicated by r2.
Rare variant analysis
To identify genes carrying rare variants contributing to FTLD-TDP, we performed a burden test in genes with rare variants (MAF < 1% in patients or controls; Supplementary Fig. 9) likely to affect protein function. Only variants that are frameshift (insertion/deletion/block substitution), stopgain, stoploss, and splicing SNVs (jointly defined as LOF variants), and non-synonymous SNVs with a REVEL pathogenicity prediction score above 0.7522 were included. In the FTLD-TDP All cohort, no exome-wide significant gene was detected except the known TBK1 risk gene (Table 3 and Supplementary Data 18). Even though we detected some inflation in our association tests, likely due to small cohort size, we did detect nine exome-wide significant signals within FTLD-TDP pathological subtypes (Table 3 and Supplementary Data 19–22). TBK1 was associated with disease status in FTLD-TDP A and B (P = 1.27 × 10−11, inflation = 2.18; P = 3.17 × 10−12, inflation = 1.49, respectively, Supplementary fig. 9). The signal was driven by 3 carriers in FTLD-TDP A patients (3/193 = 1.5%) and 5 carriers in FTLD-TDP B patients (5/288 = 1.7%) with no carriers in controls. We further identified 2 new genes in FTLD-TDP A (Supplementary Data 19) that were exome-wide significant: C3AR1 (P = 6.51 × 10−7, 3/193 FTLD-TDP A and 4/3153 controls) and SMG8 (P = 9.11 × 10−7, 3/193 FTLD-TDP A and 1/3153 controls). We also detected an enrichment in rare variants in VIPR1 in FTLD-TDP B (P = 4.65 × 10−7, 3/288 FTLD-TDP B and 1/3153 control; Fig. 6 and Supplementary Data 20) and 3 exome wide significant signals in FTLD-TDP C (inflation = 1.51, Supplementary Data 21 and Supplementary fig. 9): L3MBTL1 (P = 2.87 × 10−7, 8/467 FTLD-TDP C and 3/3153 controls), RBPJL (P = 6.39 × 10−7, 5/467 FTLD-TDP C and 3/3153 controls) and ANO9 (P = 1.39 × 10−6, 8/467 FTLD-TDP C and 6/3153 controls). For FTLD-TDP C* (without svPPA) only ANO9 reached exome-wide significance (P = 1.5 × 10−6, 4/199 FTLD-TDP C and 6/3153 controls; Supplementary Data 22). Rare variants in significantly associated genes can be found in Supplementary Data 23. Weighted gene coexpression network analysis using the ROSMAP dataset and the BrainExp database23 revealed that L3MBTL1 and RBPJL belonged to the same module (yellow, PL3MBTL1 = 1.32 × 10−45, PRBPJL = 1.00 × 10−79; Supplementary Fig. 10) that is enriched in neuroactive ligand-receptor interaction and the cytokine-cytokine receptor interaction GO terms (PFDR = 3.7 × 10−12, PFDR = 5.8 × 10−12, respectively). While expression of L3MBTL1 was throughout the central nervous system cells, RBPJL expression was restricted to inhibitory neurons and in particular, to Parvalbumin neurons (Supplementary Fig. 10). None of the rare variant carriers had a mutation in a known FTLD gene.
Schematic representation of C3AR1, SMG8, VIPR1, L3MBTL1, and RBPL protein structure (source Uniprot) showing a map of nonsense, splicing, frameshift, and missense rare variants with a REVEL score > 0.75 in patients and controls. Variants identified in patients are colored in orange, and variants identified in controls are colored in blue. n = number of carriers. When no number is indicated, the variant was observed in a single individual. Total number of subjects included in the analyzes was FTLD-TDP A (n = 193), FTLD-TDP B (n = 288), FTLD-TDP C (n = 467), and FTLD-TDP C* (n = 199), and controls (n = 3153).
Discussion
In this work, we report 12 new genome-wide significant FTLD-TDP risk loci and 6 new genes harboring rare variants contributing to FTLD-TDP risk by performing the largest FTLD-TDP WGS study to date, including 985 patients and 3153 controls. A comprehensive analysis of our data highlights the genetic overlap between FTLD-TDP, ADRD, and ALS while also defining tissue and cell type enrichment unique to FTLD-TDP. Most importantly, we highlight distinct genetic aetiologies for each of the three main FTLD-TDP pathological subtypes (A, B, and C), suggesting that multiple distinct pathomechanisms underlie the TDP-43 dysfunction and deposition in FTLD-TDP. A graphical summary of the significant findings across FTLD-TDP subtypes can be found in Fig. 7.
Genome-wide significant single variant loci, exome-wide significant genes, enriched gene ontology pathways, and tissues- and cell-types enriched for genome-wide significant risk loci are shown for each FTLD-TDP pathological subtype in rings moving from the center (genome-wide significant single variant loci in FTLD-TDP All) to the outer rings. Orange background shades correspond to FTLD-TDP A findings, green background shades to FTLD-TDP B findings, and blue background shades to FTLD-TDP C findings. Gene names in green font were exome-wide significant using a gene-based approach with common variants, while gene names in red font were exome-wide significant using a gene-based approach with rare variants. In addition to unique associations, some overlap between FTLD-TDP A and B exists (TBK1, lysosomal function, and inflammatory response), whereas FTLD-TDP C showed a unique and non-overlapping genetic profile. Note that genetic associations with LRP1B, COL22A1, TRPC4, and TMEM135 (identified in the FTLD-TDP C* GWAS focused solely on pathologically confirmed FTLD-TDP C patients) are not shown.
We replicate in an independent cohort for the first time, our previously reported GWAS signal at the UNC13A locus in FTLD-TDP patients12. This locus was also reported in a recent large GWAS focused on clinical FTLD patients and controls without reaching genome-wide significance (P = 1.07 × 10−3), possibly due to the heterogeneity of the underlying pathologies in this cohort, with only a subset having TDP-43 pathology18. The same UNC13A locus was previously linked to ALS24 and is known to modify the phenotype in ALS patients by increasing the risk of frontotemporal cortical atrophy and diminished cognitive performance, which are reminiscent of an FTLD clinical presentation25. From a biological perspective, the UNC13A risk haplotype tagged by rs12973192 and rs12608932 was previously shown to increase cryptic splicing of UNC13A in brain tissue by modulating TDP-43 binding26,27. This cryptic splicing leads to transcripts with premature stop codons and the subsequent loss of UNC13A protein, significantly impacting the release of vesicles in glutamatergic synapses28. UNC13A variants thus enhance the effect of TDP-43 dysfunction, yet both ALS and FTLD-TDP are characterized by TDP-43 pathology, suggesting that additional genetic or environmental mechanisms must exist to explain the tissue specificity of the pathology and associated clinical phenotypes in individual patients.
We further establish and replicate in an independent cohort a novel genetic association between the TNIP1 locus and FTLD-TDP. Recently, Restuadi et al. deeply characterized the GPX3/TNIP1 locus associated with ALS and suggested that GPX3 should be prioritized for deeper exploration into disease mechanisms related to this region29. GPX3, encoding for glutathione peroxidase 3, is a secreted enzyme involved in the regulation of oxidative damage, and its levels were found to be reduced in ALS sera30. Interestingly, however, the risk variant associated with FTLD-TDP (rs871269) is an expression quantitative trait locus for TNIP1 in the dorsolateral prefrontal cortex, and along with the fact that we only observed a weak colocalization signal with the ALS locus, we highlight TNIP1, and not GPX3, as the most likely gene candidate for FTLD-TDP. In fact, we observed a shared signal at this locus between our FTLD-TDP GWAS and the recent large ADRD GWAS19, whereas independent TNIP1 association signals were reported for ALS and ADRD31. The genetic overlap between FTLD-TDP and AD at this locus is of interest and suggests that TNIP1, an important ubiquitin-binding adaptor protein regulating cell death and innate immune responses through NF-kb activation32,33,34, modifies a disease process shared by AD and FTLD-TDP. The idea of a continuum of neurodegenerative disorders in which common pathological mechanisms are involved is further supported by recent GWAS19,35. Interestingly, TNIP1 undergoes phosphorylation by TBK1 and interacts with OPTN36, two proteins associated with FTLD-TDP etiology13,37. While this functional connection further supports TNIP1 as an FTLD-TDP risk gene, more work is needed to understand the mechanisms underlying disease onset. Overall, we substantiate the genetic overlap between ALS, ADRD, and FTLD-TDP and emphasize the need for deeper exploration into pathways underlying disease-specific risk.
One of the most striking conclusions from this phase II FTLD-TDP GWAS is the distinct association signals among FTLD-TDP pathological subtypes. Even the UNC13A and TNIP1 risk loci, which reach genome-wide significance in the meta-analysis stage, show stronger association in FTLD-TDP B alone, and for the first time, genome-wide significant common risk loci are reported for each of the individual pathological FTLD-TDP subtypes.
In FTLD-TDP A, in addition to individual genome-wide significant common variants assigned to GRN, TINAG, MZT1, and FARP2 risk loci, we identified exome-wide significant association with the burden of common variants in GRN and TMEM106B, in addition to multiple QTL-based analyzes prioritizing TMEM106B as a tier 1 risk gene, reinforcing the specific connection of these genes with FTLD-TDP A, even in patients without LOF GRN mutations12,15. While GRN and TMEM106B are also reported as AD risk genes19, an even stronger connection exists between these genes and limbic-predominant age-related TDP-43 encephalopathy (LATE)38,39, which has a more restricted neuroanatomical distribution of TDP-43 pathology as compared to FTLD-TDP but with some characteristics of FTLD-TDP A40,41. The TMEM106B signal is primarily influenced by rs10281425, a variant located in the 3′UTR of TMEM106B, which tags the previously reported TMEM106B risk haplotype17 associated with an increase in TMEM106B mRNA expression17 and a higher burden of insoluble disease-associated TMEM106B C-terminal fragments42. The GRN association is driven by rs5848, a variant located in the 3′UTR of GRN, which was shown to partially reduce plasma and CSF progranulin levels, independent of the presence of GRN LOF mutations43,44. More broadly, also including prioritized genes from the subthreshold regions, GO analysis in FTLD-TDP A revealed enrichment in genes implicated in lysosomal function driven by GRN, TMEM106B, but also CSTB, three genes which also had the highest individual gene scores in the prioritization analysis in FTLD-TDP A. CSTB encodes one of the most abundant lysosomal proteases in the brain45, and has been reported as a progranulin protease46,47. Genes involved in lysosomal dysfunction were also overrepresented in FTLD-TDP B, including GRN and PPT1. PPT1 is a lysosomal enzyme that facilitates the degradation of fatty-acylated proteins by lysosomal hydrolases. Mutations in PPT1 cause neuronal ceroid lipofuscinosis 148,49, and Ppt1 knock-out mice displayed fewer lipid droplets (LD) than wild type, indicating impairment of lipophagy, previously associated with FTLD/ALS50,51,52,53. Overall, our genetic data provide compelling evidence that lysosomal dysfunction contributes to the pathobiology of FTLD-TDP A, and, to a lesser extent, FTLD-TDP B.
For FTLD-TDP B, additionally, we identified individual genome-wide significant associations with variants in the RCL1 and PDS5B loci, and we observed enrichment for GO terms related to retrograde transport resulting from the VPS53 and DENND2A loci. VPS53 is part of the Golgi-associated retrograde protein (GARP) complex54,55 involved in intracellular cholesterol transport by targeting NPC2 to lysosomes56. Recently, laser capture microdissection and single-cell mass spectrometry-based proteomics in motor neurons of ALS patients revealed a strong reduction in endolysosomal trafficking complexes, such as the GARP complexes57. Limited information about DENN2A function is currently available, but structural and functional analysis indicate it may be involved in intracellular vesicle trafficking to the lysosome and to the Golgi through its guanine nucleotide exchange factor activity and regulation of RAB family GTPases58. However, retrograde transport has been previously implicated in ALS with, for instance, mutations in DCTN159,60 and KIF5A61,62, highlighting functional connections of prioritized genes from the subthreshold loci with TDP-43 dysfunction and ALS. Future GWAS with larger sample sizes, potentially combining FTLD-TDP B and ALS, are required to firmly establish a genetic contribution of this pathway to disease.
Focusing on rare variants, exome-wide significant association with TBK1 was observed in both FTLD-TDP A and B (but not FTLD-TDP C), confirming TBK1 mutations as the most common cause of FTLD-TDP after GRN and C9orf7213. Novel genes with a significantly increased cumulative frequency of rare variants in specific FTLD-TDP subtypes as compared to controls were also identified, and require confirmation in future studies. We acknowledge that limited sample sizes in these studies may have led to inflation and false positive findings; yet, this limitation is inherently linked to the unique and well-characterized study groups included in this study, the largest in the field of FTLD. We identified a significant association between rare variants in C3AR1 and FTLD-TDP A. C3AR1 plays a key role in the regulation of innate immunity and is involved in neuroinflammation63,64. Interestingly, Grn−/− mice present with upregulation of complement genes before onset of neurodegenerative features. Since GRN mutation carriers always present with FTLD-TDP A at autopsy, we suggest that aberrant activation of the complement pathway may play a major role in FTLD-TDP A pathology65. We also identified a significant association between rare variants in SMG8 and FTLD-TDP A. Homozygous pathogenic variants in the SMG8 gene have been identified as a novel cause of autosomal recessive neurodevelopmental disorder66. We further unveiled rare predicted pathogenic variants associated with FTLD-TDP B within VIPR1, which encodes for the vasoactive intestinal peptide (VIP) receptor 1. The variants are predicted to lead to an alteration of VIPR1 function, impairing the VIP biological pathway. Indeed, VIPR1 is activated upon binding by VIP, which exerts a neuroprotective effect mainly through glial cells67,68 even though neurons also express VIPRs69,70. Notably, VIP is also a key regulator of innate and adaptive immunity71, making it an important therapeutic target for multiple neurodegenerative diseases. Altogether, our studies suggest that lysosome dysfunction and/or alterations in the innate and adaptive immune system are important contributors to both FTLD-TDP A and B risk, yet to varying degrees in each pathological subtype and with likely important variability in the contribution from each pathway among individual patients.
FTLD-TDP C was previously recognized as a clinicopathological entity distinct from FTLD-TDP A and B72, and our genetic studies support this notion, showing no overlap in common or rare risk genes with the other FTLD-TDP types. Importantly, however, while often considered a sporadic FTLD subtype14,73,74, we implicate several genes and risk loci in FTLD-TDP C and uncover a potential role for mitochondrial membrane dysfunction and the notch signaling pathway. C19orf52 (TIMM29), which mediates the import and insertion of multi-pass transmembrane proteins into the mitochondrial inner membrane, was identified as the first genome-wide significant risk locus for FTLD-TDP C. This locus was identified when svPPA patients were included in the GWAS, and both pathologically confirmed FTLD-TDP C and svPPA patients were found to contribute to the association (MAF FTLD-TDP C: 0.028, svPPA: 0.019, controls: 0.001; Supplementary Data 24). This finding illustrates the power we gained by including the clinically diagnosed patients, notwithstanding the fact that the pathological diagnosis will not be FTLD-TDP C in all. The inclusion of svPPA patients in our study is somewhat complicated by the evolving clinical definitions of patients affected by focal anterior temporal lobe (ATL) atrophy in recent years. Those with left-predominant ATL atrophy show severe anomia and verbal semantic deficits leading to diagnoses of svPPA and (previously) semantic dementia2,75. However, patients with right ATL atrophy have been more challenging to fit into current diagnostic criteria and may have received diagnoses such as right-sided svPPA, right temporal variant of FTD, or, most recently, semantic behavioral variant of FTD76,77,78,79,80,81. At the start of our study in 2019, this field was just starting to evolve, and we allowed inclusion of left- and right-sided svPPA patients as both were thought to have predominant FTLD-TDP C pathology82. More recently, through large cohort studies and the establishment of an international Working Group83, systematic reassessment of clinically diagnosed patients with svPPA and bvFTD is ongoing with an emphasis on recognizing and diagnosing patients with right-temporal atrophy. As criteria are still in development and our clinical patients were not yet systematically assessed in this new framework, we also performed an additional GWAS including only pathologically confirmed FTLD-TDP C patients (FTLD-TDP C*). This analysis led to the identification of 4 novel genome-wide significant loci (LRP1B, COL22A1, TRPC4, and TMEM135). While these loci remained nominally significant in the combined analysis, inspection of the results showed no contribution from the svPPA patients to the association (Supplementary Data 24). Even with a heterogeneous population of clinical patients, one would have expected an increased frequency of risk alleles in patients as compared to controls, raising concern that these additional loci may represent type I errors. In fact, the rare-variant burden analyzes in FTLD-TDP C also showed more hits when including the svPPA patients. First of all, we observed rare predicted pathogenic variants in RBPJL, which encodes for the recombination signal binding protein for immunoglobulin kappa J region like transcription factor. RBPJL can repress Notch target gene expression (Hey1, Hey2, HeyL, and Notch3)84. As such, our findings align with a previous analysis of sub-genome-wide significant genes in clinical svPPA patients, which highlighted an overrepresentation of the Notch pathway85. Interestingly, RBPJL and L3MBTL1, the second gene carrying rare predicted pathogenic variants in FTLD-TDP C, are part of the same co-expression module, suggesting that they are functionally related. Moreover, L3MBTL1, a histone methyl-lysine binding protein, is a key regulator of proteotoxicity associated with C9orf72 dipeptide repeats and mutant SOD186 and was found to be increased in spinal cord of ALS patients. Furthermore, reduction of L3MBTL1 expression in Drosophila models with the C9orf72-associated dipeptides poly(PR) or poly(GR) ameliorated the rough-eye phenotype86, suggesting that loss of L3MBTL1 expression is beneficial. While no RNA samples were accessible from rare variant carriers, nonsense-mediated decay escape has been reported in other genes linked to ALS87. It is thus possible that the L3MBTL1 variants lead to the generation of truncated proteins with toxic gain-of-function, but additional work is necessary to understand the disease etiology fully. Finally, rare variants in ANO9 were also associated with FTLD-TDP C disease status. ANO9 encodes for anoctamin 9, for which the biological function is currently unclear. ANO9 was the only gene with exome-wide significance when svPPA patients were removed from the FTLD-TDP C subgroup.
When analyzed in sum, common variants associated with the different FTLD-TDP pathological subtypes appeared to be located in genes expressed in excitatory neurons, in contrast to AD risk variants, which are enriched in microglia. Interestingly, glutamatergic transmission impairment has been reported in FTLD88,89,90,91,92, and voxel-based brain changes have been significantly associated with spatial distribution of mGluR5 in symptomatic C9orf72 and GRN carriers93. Therefore, and in line with previously reported studies, our data suggest that neurons are the major players in disease etiology, as compared to what has been observed in ADRD. Whether specific neuronal subpopulations differentially express associated genes could be the focus of future studies once single-nuclei transcriptomic datasets from FTLD-TDP patients and controls are available. Interestingly, the distribution of risk loci was specific to the cerebellar hemisphere and the frontal cortex for FTLD-TDP A, B, and C*, as opposed to FTLD-TDP C (with participants with svPPA), where genes expressed in small intestine were enriched in risk loci. While the link between gut microbiome and FTLD remains limited94, our data suggest that the gut-brain axis might be of interest for future studies, especially in svPPA patients. In fact, emerging evidence also supports a role for the gut-brain axis in autoimmune diseases95, a group of disorders that were found to be enriched in svPPA patients96.
In prior studies, besides UNC13A, common variants in the HLA and DPP6 loci, in TMEM106B, and most recently in MAPT, APOE, and MOBP, were reported to be associated with FTLD12,14,18. The latter three loci and HLA-DR5 locus were identified as associated with clinical FTLD and likely do not represent risk factors specific to TDP-43 dysfunction. HLA-DQA2 and DPP6 loci were reported as overall FTLD-TDP risk loci in phase I12 but were not replicated in the current study. The relative composition of patients with FTLD-TDP pathological subtypes in phase I and II (e.g., less FTLD-TDP A in phase II) and inclusion of clinically diagnosed individuals in phase II may have contributed to this; however, it is also possible that the increase in sample size reduced type I errors from phase I. Importantly, we identified and replicated in two independent cohorts the UNC13A and TNIP1 loci associated with FTLD-TDP. Replication of the newly identified risk loci, each specific to distinct neuropathological FTLD-TDP subtypes, will require additional GWAS studies in the future. Obtaining sufficient samples will, however, be challenging, especially for FTLD-TDP A, which lacks a clear clinical correlate of the pathological phenotype. For FTLD-TDP C, the relative contribution of patients with left- and right-predominant ATL atrophy to the observed associations should be assessed once clinical criteria for patients with right temporal atrophy are finalized. Finally, functional characterization of the newly identified genes and loci may also provide mechanistic insight.
In conclusion, we confirmed UNC13A and identified 12 new genetic loci, i.e., TNIP1, GRN, TINAG, MZT1, FARP2, RCL1, PDS5B, C19orf52, LRP1B, COL22A1, TMEM135, and TRPC4, and 6 new genes with rare variants associated with FTLD-TDP risk, i.e., C3AR1, SMG8, VIPR1, RBPJL, L3MBTL1, and ANO9. Importantly, by enriching in neuropathologically confirmed patients and substantially increasing our cohort size, we uncovered distinct genetic aetiologies for each of the three main FTLD-TDP pathological subtypes. Our findings align with recent data obtained from cryo-electron microscopy, which identified distinct homomeric TDP-43 filaments in FTLD-TDP A and B and heteromeric amyloid filaments of ANXA11 and TDP-43 in FTLD-TDP C97,98,99. The recognition of individual FTLD-TDP subtypes as potentially distinct diseases with unique pathomechanism may have important implications for the design of clinical trials and therapeutic interventions.
Methods
Ethics approval and consent to participate
This study was approved by the Mayo Clinic Institutional Review Board. All participants or their guardians provided written informed consent.
Samples
Our current dataset includes previously generated data through the International FTLD-TDP WGS consortium phase I12 with 554 persons with clinicopathologically defined FTLD-TDP and newly generated phase II sequencing data from 32 FTLD-TDP A, 43 FTLD-TDP B, 66 FTLD-TDP C, 4 FTLD-TDP E, and 9 with unclassifiable FTLD-TDP pathology (abbreviated as FTLD-TDP U). To increase statistical power, we also sequenced 70 persons with clinical diagnosis of bvFTD/ALS, a clinical subtype associated with FTLD-TDP B, and 283 persons with svPPA, a clinical subtype associated with FTLD-TDP C. Overall, the total cohort pre-quality control was a combined FTLD-TDP cohort of 202 FTLD-TDP A, 237 FTLD-TDP B, 225 FTLD-TDP C, 4 FTLD-TDP D, 11 FTLD-TDP E, 29 FTLD-TDP U persons, 70 persons with bvFTD/ALS and 283 persons with svPPA (Table 1). After QC, 985 patients from 26 sites were included in the analysis (Supplementary Data 1 and Supplementary Data 25). Patients were diagnosed according to established diagnostic criteria1,2,75,100; however, in the subgroup of patients with svPPA, in addition to including svPPA patients diagnosed according to Gorno-Tempini et al.2, patients previously diagnosed with semantic dementia according to Neary et al75., and a few patients suspected to have the right temporal variant of FTD were included. For the latter group of patients, diagnostic criteria are only now being developed76,83, we thus relied on the expert knowledge of the specialized dementia centers to identify these patients, recognizing the limitations of this approach, especially since this was not performed systematically in all contributing centers. All persons clinically or pathologically diagnosed with FTLD are referred to as patients throughout the manuscript. We further used WGS data from 982 participants from the Mayo Clinic Biobank (from phase I), 12,101, 322 new controls free of neurodegenerative disorder from Mayo Clinic with WGS available, and 2,037 controls derived from the ADSP. C9orf72 repeat expansions were assessed in all patients using our previously reported two-step protocol, and Sanger sequencing was used to perform mutation analyzes of GRN8,10. Patients carrying repeat expansions in C9orf72 or LOF mutations in GRN, both associated with autosomal dominant forms of FTLD-TDP, were removed prior to WGS. Study protocols were reviewed and approved by the appropriate institutional review boards.
Whole genome sequencing
In phase I of the International FTD-TDP WGS consortium, WGS was generated on 554 patients with FTLD-TDP (512 passed QC in that study)12. Briefly, whole blood- or brain-derived DNA from 499 unrelated FTLD-TDP patients and 982 participants from the Mayo Clinic Biobank Study were sequenced at HudsonAlpha using the standard library preparation protocol using the NEBNext® DNA Library Prep Master Mix Set for Illumina® (New England BioLabs Inc., Ipswich, MA, USA). Concentration of the libraries was assessed by Qubit® 2.0 Fluorometer, and the quality of the libraries was estimated by a DNA 5 K chip on a Caliper GX. Accurate quantification was determined using the qPCR-based KAPA Biosystems Library Quantification kit (Kapa Biosystems, Inc., Woburn, MA, USA). Each sample was sequenced on one lane of Illumina’s HiSeq X instrument using v2 flow cells and reagents to target 30× genomic coverage. Fastq files previously generated on an Illumina HiSeq X for 55 FTLD-TDP patients were obtained from 3 sites: UCSF (n = 36), DZNE (n = 14), and NSW (n = 5).
In phase II, additional WGS of 507 patients with FTLD-TDP, svPPA, bvFTD/ALS, and 322 controls free of neurodegenerative disorders was performed at USUHS sequencing center or Mayo Clinic Rochester using the TruSeq DNA PCR-Free Library preparation Kit (Illumina), followed by Whole Genome Sequencing by synthesis (SBS) chemistry on HiSeq X Illumina platform using the HiSeq X Ten Reag. kit v2.5. Fastq files for all patients and controls were transferred to Mayo Clinic and processed through the Mayo Genome GPS v4.0 pipeline in batches of up to 75 samples using the Burrows–Wheeler Aligner to map reads to the human reference sequence (GRCh38 build). Local realignment around indels and variant calling were performed using Genome Analysis Toolkit (GATK) HaplotypeCaller, followed by variant recalibration (VQSR) according to GATK best practice recommendations. Participants from the Mayo Clinic Biobank with a possible clinical diagnosis or family history of a neurodegenerative disorder were removed during analysis.
To enhance our study, we incorporated genomic variant call format (gVCF) files from 2037 controls obtained from the Alzheimer’s Disease Sequencing Project (ADSP). gVCF files provide a comprehensive record of variant calls and reference positions, which are essential for accurate joint-genotyping. The gVCF files from ADSP controls were merged with our cohort’s gVCF files using the joint-genotyping approach implemented with the GATK. By merging these gVCFs, we ensured all our patients and controls were analyzed together, allowing for a more robust comparison and reducing batch effects. Overall, we obtained genomic data on a total of 1061 patients and 3341 controls.
Sample-level quality control and definition of subgroups
Samples with less than 30× coverage in more than 50% of the genome, call rate below 85%, sex error, and contamination defined by a FREEMIX score above 0.04 (4% or more of non-reference bases are observed in reference sites) were removed. In more detail, estimates of sample contamination were calculated using data on all autosomes with 1000 Genomes European array allele frequencies as reference. The estimated contamination value, FREEMIX 1.3, provides a sequence-only estimate of contamination on a 0–1 scale using excessive heterozygosity. In addition, we identified 31 non-European White participants (Admixture EUR Probability <0.7), which we removed from analysis because of the small sample size and to increase genetic background homogeneity. Information from chromosomes X and Y was used to estimate sex. All SNVs having MAF > 0.05, variant call-rate > 0.80, Hardy Weinberg Equilibrium p value > 1e-8, and LD r2 < 0.8 were used for these calculations. Samples whose reported sex did not match the estimated sex based on genomic data using the—check-sex PLINK command were removed. At this step, joint genotyping on all samples was performed. A final relatedness measurement was calculated using PREST102, and duplicates were removed, while only one individual per family was kept. In total, 985 pathologically confirmed FTLD-TDP or presumed FTLD-TDP patients clinically presenting with svPPA or bvFTD/ALS, as well as 3153 neurologically normal controls passed all QC measures (Supplementary Data 25). Age at onset of svPPA and bvFTD/ALS did not differ from the age at onset of FTLD-TDP C (P = 1) and FTLD-TDP B patients (P = 1), respectively. Based on these findings and the previously established associations between the svPPA and bvFTD/ALS clinical diagnoses with specific FTLD-TDP pathological subtypes, we combined svPPA with FTLD-TDP C and bvFTD/ALS with FTLD-TDP B patients in all analyzes (except where explicitly specified, such as in FTLD-TDP C* analyzes). Within our overall cohort of 193 FTLD-TDP A, 288 FTLD-TDP B (defined as FTLD-TDP B and bvFTD/ALS) and 467 FTLD-TDP C (defined as FTLD-TDP C and svPPA), the ages at onset and death differed significantly between the pathological FTLD-TDP subtypes (Table 1 and Supplementary Fig. 11). FTLD-TDP A patients had a later age at onset than FTLD-TDP B and FTLD-TDP C groups (P = 4.73 × 10−8, P = 1.37 × 10−13, respectively), and a later age at death (P = 4.00 × 10−15, P = 3.00 × 10−6, respectively). FTLD-TDP B had an earlier age at death as compared to FTLD-TDP C (P = 5.60 × 10−8). Differences in age distribution between patient groups were assessed using the Kruskal–Wallis test followed by Wilcoxon test, correcting for multiple testing. Corrected Bonferroni P values are provided.
Variant level quality control
Genotype calls with genotype quality < 20 and/or depth (DP) < 10 were set to missing, and variants with edit-distance > 4 and call rate <80% were removed from all subsequent analyzes, leading to a total of 85,345,466 variants. For all analyzes, only variants that pass VQSR (127,658 variants removed) and with a call rate > 95% in patients and controls were considered (591,431 variants removed). Functional annotation of variants was performed using ANNOVAR (version 2016Feb01). Rare loss-of-function variants frameshift insertion/deletion/block substitution, stopgain, stoploss, and splicing single-nucleotide variants (SNVs) and missense with REVEL score > 0.7522 identified in exome-wide significantly associated genes (Supplementary Data 18–23) were confirmed in patients by Sanger sequencing (primers available upon request). For the known neurodegenerative disease genes (GRN, MAPT, TBK1, OPTN, VCP, TARDBP, CHCHD10, SQSTM1, UBQLN2, hnRNPA1, hnRNPA2B1, CSF1R, FUS, CHMP2B, and LRRK2), potentially pathogenic rare variants were also identified and confirmed by Sanger sequencing (n = 25 and Supplementary Data 26).
Generation of principal components
Prior to running genetic association analyzes, principal component (PC) analysis was performed using a subset of variants meeting the following criteria: minor allele frequency (MAF) > 5% and full sample Hardy-Weinberg Equilibrium (HWE) P > 1 × 10−5. Influential regions such as the HLA region were removed, and variants were pruned by LD with r2 threshold of 0.1 prior to PC analysis. This analysis identified that the 13 top PCs were significantly associated with patient/control status.
Variant-level analysis of common variants
For the common variant GWAS, SNV with MAF > 0.01 in patients or controls (n = 7,178,250 variants), and HWE P > 1.00 × 10−6 in controls were analyzed (17,450 variants removed). In addition, since whole genome sequencing of FTLD-TDP patients and controls was performed at multiple sites, a test was performed to identify variants with significant differences in genotype distributions between sequencing batches, and 592,701 SNVs showing evidence of batch effects (p < 0.05) were removed, leading to a total of 6,568,099 variants analyzed.
For all remaining variants, association of genotypes with the patient/control status was assessed using logistic regression with allele dosage as the predictor, assuming log-additive allele effects. Genome-wide significance was defined as P < 5 × 10−8. Sex and the first 13 PCs were included as covariates in the models. The SNV-level analyzes were performed using PLINKv.00a23LM2, combining all FTLD-TDP patients (FTLD-TDP All) and in FTLD-TDP pathological subtypes. MAGMA (v1.6) was used to perform gene-based analysis of common variants using summary statistics. MAGMA is a tool designed to analyze associations at the gene level by aggregating SNV p values while accounting for LD. Dementia-seq vcf was processed the exact same way as our data, except that 10 PCs were included in the model to perform common variant association analysis. Identification of duplicate samples between our discovery dataset and the Dementia-seq data was performed, and all duplicates were removed. Meta-analyzes of FTLD-TDP phase II with publicly available datasets from the Dementia-seq project (phs001963.v2.p1) were performed under a fixed-effects model comparing our data with 2102 FTLD patients and 1748 controls from the Dementia-seq project using Metal103.
Colocalization analyzes
We performed colocalization analysis for UNC13A and TNIP1 loci (top SNVs ±100 kb) with ALS (GCST90027164) and ADRD (GCST90027158) using the “coloc” package version 4.0.4 in R using our meta-analyzes data. When the summary statistics of the other trait were expressed on another build than GRCh38, the variant alleles and positions were converted. We set the prior probabilities to π1 = 1 × 10−4, π2 = 1 × 10−4, and π12 = 1 × 10−5 for a causal variant in trait 1 or trait 2 and a shared causal variant between traits 1 and 2, respectively (default parameters). Sensitivity analysis was performed at π12 = 1 × 10−6. P < 0.05 was considered statistically significant.
Tissue and cell type enrichment analysis
Tissue and cell type enrichment analyzes were performed using the summary statistics (variants with P < 10−5) and FUMA104. Briefly, FUMA aggregates summary statistics per gene to calculate gene-wise association signals using MAGMA version 1.6 and subsequently tests whether tissues and cell types are enriched for expression of these genes. For tissue enrichment analysis, we used the GTEx version 8 reference set. P < 0.05 across all tissues (n = 54) was considered statistically significant. For cell type enrichment analyzes, we used human-derived single-cell RNA-seq data from major brain cell types (PsychENCODE). Excitatory and inhibitory neurons from the PsychENCODE dataset were labeled based on their transcriptional profile from 1 to 8105. P < 0.05 was considered statistically significant.
Gene prioritization and functional interpretation of GWAS
We performed the gene prioritization and functional interpretation analyzes for FTLD-TDP All and each FTLD-TDP pathological subtype separately by using the subtype-specific GWAS summary statistics on all variants. We adapted a systematic gene prioritization and functional interpretation strategy (as previously described in Bellenguez et al.19) to prioritize GWAS-implicated candidate risk genes and nominate possible downstream biological mechanisms. Briefly, six distinct domains, that are related to lead variant annotation and molecular QTL-GWAS integration analyzes (e.g., colocalization and TWAS) in FTLD-relevant tissues and cell types were systematically assessed: (1) variant annotation, (2) eQTL-GWAS integration, (3) sQTL-GWAS integration, (4) protein expression QTL (pQTL)-GWAS integration, (5) mQTL-GWAS integration, and (6) histone acetylation QTL (haQTL)-GWAS integration; for which detailed information on categories and subcategories is provided in Supplementary Data 3.
In the variant annotation domain, for each lead variant at each locus, we queried which candidate risk genes were the nearest protein-coding genes with respect to the genomic position of the lead variants, and/or whether the lead variant was a rare (MAF < 1% in gnomAD v4 non-Finnish European samples) and/or protein-altering (missense or predicted LOF) variant for the same nearest protein-coding genes. In the molecular QTL-GWAS integration domains, we leveraged molecular cis-QTL catalogs for different molecular phenotypes (i.e., gene expression, splicing, protein expression, methylation, and histone acetylation) in FTLD-relevant tissues and cell types, we performed genetic colocalization analyzes between molecular cis-QTL and GWAS signals, TWAS, and proteome-wide association studies (PWAS). For these analyzes, we processed and used publicly available molecular QTL catalogs; namely, FTLD-relevant bulk brain regions from AMP-AD106,107,108,109 (as reanalyzed in Bellenguez et al.19) and GTEx v8110 cohorts for the bulk brain eQTLs and sQTLs, eight major brain cell types (excitatory neurons, inhibitory neurons, astrocytes, oligodendrocytes, microglia, oligodendrocyte precursor cells/committed oligodendrocyte precursors [OPCs/COPs], pericytes, and endothelial cells) from Bryois et al.111 and primary microglia from Young et al.112 and the MiGA study113 for the brain cell-type-specific eQTLs (ct-eQTL) and for microglia sQTLs (from the MiGA study), dorsolateral prefrontal cortex (DLPFC) pQTLs from Wingo et al.114 (v2), and DLPFC mQTLs and haQTLs from Brain xQTL serve (June 2021 release)114,115. Finally, we also included naïve state monocyte and macrophage eQTL catalogs116,117,118,119,120,121 reanalyzed by eQTL Catalog (Release 6)122 and lymphoblastoid cell line (LCL) eQTLs from GTEx v8110 and the European Alzheimer & Dementia Biobank (EADB) Belgian LCL cohorts19. Using each of these molecular QTL catalogs, we first investigated whether the reported lead variants in this study were significant molecular QTLs for the quantified levels of molecular phenotypes in tissues and cell types of interest. Moreover, for each quantified molecular phenotype in these catalogs, we performed molecular QTL-GWAS coloc (v5.2.2) analyzes to determine if specific molecular QTL signals are colocalized (at coloc PP4 ≥ 70%) with FTLD subtype GWAS signals. Of note, for mQTL-GWAS integrative analyzes, the CpGs were assigned to genes using BECon annotations123 whenever available, and if not, the CpGs were mapped to the nearest genes based on GENCODE v24; while for haQTL-GWAS integrative analyzes, the histone acetylation peaks were assigned to the nearest genes based on GENCODE v24 as well. Finally, we conducted TWAS (using FUSION and S-PrediXcan [implemented in MetaXcan] tools) for each heritable feature modeled in gene expression (eTWAS; followed by eTWAS fine mapping with FOCUS124 [v0.803] within 1 Mb extended genome-wide significant lead variant genetic regions in each FTLD-TDP subtype GWAS), splicing (sTWAS), and PWAS reference panels derived from AMP-AD bulk brain106,107,108,109, GTEx bulk brain and LCL110, EADB Belgian LCL19, and Wingo et al. DLPFC data114, to identify the significant associations (after Bonferroni correction) between predicted levels of gene expression, splicing, and protein expression with each FTLD subtype-specific genetic risk. Detailed description and details (e.g., number of samples, significance criteria, references, and sources) of these molecular QTL catalogs used in this study for the systematic gene prioritization strategy and functional interpretation of FTLD-TDP GWAS results can be found in Supplementary Data 5.
Using a predetermined weighting scheme for each type of evidence (see Supplementary Data 3), we computed a gene prioritization score (between 0 and 87) for each gene, which was constructed by the weighted sum of the hits in different subcategories within six distinct domains described above. As described in Bellenguez et al.19 in detail, we gave higher weights for the hits obtained through the brain QTLs rather than other tissue QTLs, for the replicated hits across multiple catalogs or reference panels, and for the fine-mapped eTWAS hits. After obtaining weighted gene prioritization scores in each FTLD-TDP subtype-specific gene prioritization analysis, we first assigned each candidate risk gene (with gene prioritization score > 0) to the genome-wide significant loci if their gene coordinates (based on GENCODE v24) are positioned within a ± 1 Mb window of the identified lead variants (Table 2). The rest of the candidate risk genes in subthreshold regions (nominated by Coloc and TWAS analyzes only) were grouped together if they were positioned together (<1 Mb), and these subthreshold regions were indexed and named as subthreshold loci. The candidate risk genes in genome-wide significant and subthreshold loci were also annotated by the evidence of minimum P observed within 1 Mb of the gene coordinates in related FTLD-TDP subtype-specific GWAS summary statistics. We then ranked the protein-coding genes per locus in each FTLD subtype-specific analysis based on their total weighted scores, and investigated the relative score differences between the highest-ranked protein-coding gene and the other candidate risk genes in each locus, together with the overall total weighted score of the top-ranked gene. We then classified candidate risk genes in each locus as tier 1 and tier 2 prioritized risk genes, respectively having a higher and lower level of confidence for being a true risk gene in a given locus (see Bellenguez et al.19 for detailed description). As also described in Bellenguez et al.19, the gene prioritization pipeline determines a single tier 1 prioritized risk gene in each locus if there is adequate evidence, meanwhile additional tier 2 prioritized risk genes in the same loci or multiple tier 2 prioritized risk genes in a locus can also be assigned based on the score distribution of candidate genes in the investigated loci.
Gene ontology analyzes
GO on tier 1 genes identified in FTLD-TDP All or in individual FTLD-TDP subtype analyzes were performed using anRichment R package, which aggregates summary statistics and assesses GO term enrichment. GO terms were collapsed using the RVizgo R package. Only terms with two or more genes were considered in the analyzes. P < 0.05 was considered statistically significant.
Genetic correlation analyzes
We assessed genetic similarity between FTLD-TDP All and ALS and ADRD using the LD score regression (LDSC, https://github.com/bulik/ldsc). We estimated genetic correlations attributable to genome-wide SNPs (rg) between FTLD-TDP All and ALS, and FTLD-TDP All and ADRD using the default settings in the LDSC software and pre-calculated LD scores from the 1000 Genomes European reference population, supplied with the LDSC software.
Gene-level analysis of rare variants
Association of rare variants with the patient/control status was assessed using an unweighted burden test implemented using the SKAT_1.2.1R package. Only VQSR-pass variants with call rate > 90%, ED ≤ 4, and MAF < 0.01 in either patients or controls were included. We included only frameshift (insertion/deletion/block substitution), stopgain, stoploss, and splicing SNVs (jointly defined as LOF variants), and non-synonymous SNVs with REVEL score above 0.7522. Only genes with at least 3 patients carrying rare variants were retained. Sex and the first thirteen PCs were used as covariates. Exome-wide significance with p value < 2.5 × 10−6 (Bonferroni correction for 20,000 genes) was used. The rare variants in the top genes were confirmed by Sanger sequencing in all patient samples and visually inspected to eliminate sequencing errors. One gene enriched in rare variants in FTLD-TDP A (TDRD5) as compared to controls failed at the inspection stage due to the inclusion of a multi-allelic variant around the repetitive C-terminal end of the gene, which could not be definitively confirmed. Rare variants enriched in two genes in FTLD-TDP C (C4orf47 and TYRO3) failed confirmation via Sanger sequencing.
RBPJL and L3MBTL1 RNA expression
Assessment of module membership of RBPJL and L3MBTL1 was performed using the gene co-expression analysis from the BrainEXP-NPD23 website using default parameters. Single-nuclei RNA expression was assessed using the transcriptomic comparative viewer of the Seattle Alzheimer’s Disease Brain cell Atlas from middle temporal gyrus of 84 aged donors (42 cognitively normal and 42 with dementia).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The processed (summary statistics) data generated in this study have been deposited in the GWAS catalog database under accession codes GCP001210 (GCST90558311, GCST90558312, GCST90558313, and GCST90558314) [https://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/]. Whole genome sequencing data from the 507 FTLD-TDP patients and 322 controls generated in Phase II are available as follows: raw sequencing data and VCF for 435 FTLD-TDP patients and 19 control individuals have been deposited in the dbGAP platform as part of the dataset with accession code phs003309 [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003309.v1.p1]. For the 435 FTLD-TDP patients, access is restricted: 271 can be for General Research Use, 1 is for Health/Medical/Biomedical research only, and 163 are for ‘Disease-Specific (Neurodegenerative Disorders)’ research only. The 19 controls can also be used for Disease-Specific (Neurodegenerative Disorders) research only. Access can be obtained by applying for dbGaP Authorized Access via https://view.ncbi.nlm.nih.gov/dbgap-controlled. The gVCF genetic data from ADSP used in Phase II are available through restricted to not-for-profit organizations, access can be obtained by applying at https://dss.niagads.org/. The genetic data for Phase I participants12 and the remaining 72 FTLD-TDP patients and 303 controls from Phase II are not part of dbGAP accession phs003309 and are not available due to data sharing constraints related to the participants’ consent form. From the Phase II post quality control dataset, 303 controls from Mayo Clinic, USA, and 51 FTLD-TDP patients could not be shared (48 from Erasmus Medical Center, The Netherlands, 1 from Mayo Clinic, USA, 1 from Indiana University, USA, and 1 from University of California, San Francisco). The PsychENCODE data used in this study are publicly available through the FUMA platform [https://fuma.ctglab.nl/]. Dataset and molecular QTLs used in the gene prioritization are publicly available (see also Supplementary Data 5): The eQTLs and eTWAS reference panels in AD-relevant bulk brain regions from AMP-AD cohorts and in LCLs from the EADB Belgian cohort, as analyzed by Bellenguez et al.19 are publicly available in the Zenodo database under accession code 5745927. The sQTLs and sTWAS reference panels used in this study from AD-relevant bulk brain regions from AMP-AD cohorts and from LCLs from the EADB Belgian cohort, as analyzed by Bellenguez et al.19 are publicly available in the Zenodo database under accession code 5745929. The Bryois et al.111. ct-eQTL catalogs used in this study are publicly available in the Zenodo database under accession code 5543734. The eQTL data used in this study are publicly available in the eQTL catalog database [https://www.ebi.ac.uk/eqtl/]. The mQTL and haQTL catalogs used in this study are publicly available in the Brain xQTL Serve database [https://mostafavilab.stat.ubc.ca/xqtl/] issued from Ng et al.115. The eQTL and sQTL catalogs used in this study are available in the GTEX v8 database [https://www.gtexportal.org/]. The GTEx v8 expression and splicing prediction models for eTWAS/sTWAS used in this study are available at https://predictdb.org/post/2021/07/21/gtex-v8-models-on-eqtl-and-sqtl/#mashr-based-models. The microglial eQTL data used in this study are available in the Zenodo database under accession code 4118605. The microglial sQTL data used in this study are available in the Zenodo database under accession code 4118403. The microglial meta-analysis data used in this study are available in the Zenodo databse under accession code 4118676. The pQTL v2 data used in this study is available in the Synapse database under accession code syn2580853 [https://www.synapse.org/#!Synapse:syn23627957], from the publication Wingo et al.114. The BECon annotations used in this study are available at https://redgar598.shinyapps.io/BECon/.
References
Rascovsky, K. et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain 134, 2456–2477 (2011).
Gorno-Tempini, M. L. et al. Classification of primary progressive aphasia and its variants. Neurology 76, 1006–1014 (2011).
Lee, E. B. et al. Expansion of the classification of FTLD-TDP: distinct pathology associated with rapidly progressive frontotemporal degeneration. Acta Neuropathol. 134, 65–78 (2017).
Mackenzie, I. R. et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 122, 111–113 (2011).
Chatterjee, M. et al. Plasma extracellular vesicle tau and TDP-43 as diagnostic biomarkers in FTD and ALS. Nat. Med. 30, 1771–1783 (2024).
Mackenzie, I. R. et al. Heterogeneity of ubiquitin pathology in frontotemporal lobar degeneration: classification and relation to clinical phenotype. Acta Neuropathol. 112, 539–549 (2006).
Mackenzie, I. R. & Neumann, M. Reappraisal of TDP-43 pathology in FTLD-U subtypes. Acta Neuropathol. 134, 79–96 (2017).
Baker, M. et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442, 916–919 (2006).
Cruts, M. et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442, 920–924 (2006).
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011).
Renton, A. E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011).
Pottier, C. et al. Genome-wide analyses as part of the international FTLD-TDP whole-genome sequencing consortium reveals novel disease risk factors and increases support for immune dysfunction in FTLD. Acta Neuropathol. 137, 879–899 (2019).
Pottier, C. et al. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol. 130, 77–92 (2015).
Ferrari, R. et al. Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol. 13, 686–699 (2014).
Mackenzie, I. R. et al. The neuropathology of frontotemporal lobar degeneration caused by mutations in the progranulin gene. Brain 129, 3081–3090 (2006).
Mackenzie, I. R. & Neumann, M. Subcortical TDP-43 pathology patterns validate cortical FTLD-TDP subtypes and demonstrate unique aspects of C9orf72 mutation cases. Acta Neuropathol. 139, 83–98 (2020).
Van Deerlin, V. M. et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat. Genet. 42, 234–239 (2010).
Manzoni, C. et al. Genome-wide analyses reveal a potential role for the MAPT, MOBP, and APOE loci in sporadic frontotemporal dementia. Am. J. Hum. Genet. 111, 1316–1329 (2024).
Bellenguez, C. et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022).
Project Min, E.A.L.S.S.C. Project MinE: study design and pilot analyses of a large-scale whole-genome sequencing study in amyotrophic lateral sclerosis. Eur. J. Hum. Genet. 26, 1537–1546 (2018).
Benyamin, B. et al. Cross-ethnic meta-analysis identifies association of the GPX3-TNIP1 locus with amyotrophic lateral sclerosis. Nat. Commun. 8, 611 (2017).
Ioannidis, N. M. et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet 99, 877–885 (2016).
Jiao, C. et al. BrainEXP: a database featuring with spatiotemporal expression variations and co-expression organizations in human brains. Bioinformatics 35, 172–174 (2019).
van Es, M. A. et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat. Genet. 41, 1083–1087 (2009).
Placek, K. et al. UNC13A polymorphism contributes to frontotemporal disease in sporadic amyotrophic lateral sclerosis. Neurobiol. Aging 73, 190–199 (2019).
Brown, A. L. et al. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature 603, 131–137 (2022).
Ma, X. R. et al. TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A. Nature 603, 124–130 (2022).
Augustin, I., Rosenmund, C., Sudhof, T. C. & Brose, N. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature 400, 457–461 (1999).
Restuadi, R. et al. Functional characterisation of the amyotrophic lateral sclerosis risk locus GPX3/TNIP1. Genome Med. 14, 7 (2022).
Tanaka, H. et al. ITIH4 and Gpx3 are potential biomarkers for amyotrophic lateral sclerosis. J. Neurol. 260, 1782–1797 (2013).
Wainberg, M., Andrews, S. J. & Tripathy, S. J. Shared genetic risk loci between Alzheimer’s disease and related dementias, Parkinson’s disease, and amyotrophic lateral sclerosis. Alzheimers Res. Ther. 15, 113 (2023).
Oshima, S. et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature 457, 906–909 (2009).
Su, Z. et al. ABIN-1 heterozygosity sensitizes to innate immune response in both RIPK1-dependent and RIPK1-independent manner. Cell Death Differ. 26, 1077–1088 (2019).
Zhou, J. et al. A20-binding inhibitor of NF-kappaB (ABIN1) controls Toll-like receptor-mediated CCAAT/enhancer-binding protein beta activation and protects from inflammatory disease. Proc. Natl. Acad. Sci. USA 108, E998–E1006 (2011).
Shade, L. M. P. et al. GWAS of multiple neuropathology endophenotypes identifies new risk loci and provides insights into the genetic risk of dementia. Nat. Genet. 56, 2407–2421 (2024).
Lee, Y. et al. Coordinate regulation of the senescent state by selective autophagy. Dev. Cell 56, 1512–1525 e7 (2021).
Freischmidt, A. et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 18, 631–636 (2015).
Katsumata, Y. et al. Multiple gene variants linked to Alzheimer’s-type clinical dementia via GWAS are also associated with non-Alzheimer’s neuropathologic entities. Neurobiol. Dis. 174, 105880 (2022).
Katsumata, Y., Nelson, P. T., Ellingson, S. R. & Fardo, D. W. Gene-based association study of genes linked to hippocampal sclerosis of aging neuropathology: GRN, TMEM106B, ABCC9, and KCNMB2. Neurobiol. Aging 53, 193 e17–193 e25 (2017).
Nelson, P. T. et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain 142, 1503–1527 (2019).
Josephs, K. A. et al. Pathological, imaging and genetic characteristics support the existence of distinct TDP-43 types in non-FTLD brains. Acta Neuropathol. 137, 227–238 (2019).
Vicente, T. C. et al. C-terminal TMEM106B fragments in human brain correlate with disease-associated TMEM106B haplotypes. Brain 146, 4055–4064 (2023).
Swift, I. J. et al. A systematic review of progranulin concentrations in biofluids in over 7000 people-assessing the pathogenicity of GRN mutations and other influencing factors. Alzheimers Res. Ther. 16, 66 (2024).
Nicholson, A. M. et al. Progranulin protein levels are differently regulated in plasma and CSF. Neurology 82, 1871–1878 (2014).
Hook, V. et al. Cathepsin B in neurodegeneration of Alzheimer’s disease, traumatic brain injury, and related brain disorders. Biochim. Biophys. Acta Proteins Proteomics 1868, 140428 (2020).
Lee, C. W. et al. The lysosomal protein cathepsin L is a progranulin protease. Mol. Neurodegener. 12, 55 (2017).
Zhou, X. et al. Lysosomal processing of progranulin. Mol. Neurodegener. 12, 62 (2017).
Vesa, J. et al. Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature 376, 584–587 (1995).
Camp, L. A., Verkruyse, L. A., Afendis, S. J., Slaughter, C. A. & Hofmann, S. L. Molecular cloning and expression of palmitoyl-protein thioesterase. J. Biol. Chem. 269, 23212–23219 (1994).
Branchu, J. et al. Loss of spatacsin function alters lysosomal lipid clearance leading to upper and lower motor neuron degeneration. Neurobiol. Dis. 102, 21–37 (2017).
Han, S. M. et al. VAPB/ALS8 MSP ligands regulate striated muscle energy metabolism critical for adult survival in caenorhabditis elegans. PLoS Genet. 9, e1003738 (2013).
Cingolani, F. & Czaja, M. J. Regulation and functions of autophagic lipolysis. Trends Endocrinol. Metab. 27, 696–705 (2016).
Liu, Y. et al. A C9orf72-CARM1 axis regulates lipid metabolism under glucose starvation-induced nutrient stress. Genes Dev. 32, 1380–1397 (2018).
Conibear, E. & Stevens, T. H. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol. Biol. Cell 11, 305–323 (2000).
Liewen, H. et al. Characterization of the human GARP (Golgi associated retrograde protein) complex. Exp. Cell Res. 306, 24–34 (2005).
Wei, J. et al. The GARP complex is involved in intracellular cholesterol transport via targeting NPC2 to lysosomes. Cell Rep. 19, 2823–2835 (2017).
Guise, A.J. et al. TDP-43-stratified single-cell proteomics of postmortem human spinal motor neurons reveals protein dynamics in amyotrophic lateral sclerosis. Cell Rep. 43, 113636 (2024).
Yoshimura, S., Gerondopoulos, A., Linford, A., Rigden, D. J. & Barr, F. A. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191, 367–381 (2010).
Deshimaru, M. et al. DCTN1 binds to TDP-43 and regulates TDP-43 aggregation. Int. J. Mol. Sci. 22, 3985 (2021).
He, J., Yu, W., Liu, X. & Fan, D. An identical DCTN1 mutation in two Chinese siblings manifest as dHMN and ALS respectively: a case report. Amyotroph. Lateral Scler. Frontotemporal Degener. 23, 149–153 (2022).
Nicolas, A. et al. Genome-wide analyses identify KIF5A as a Novel ALS gene. Neuron 97, 1267–1288 (2018).
Xia, C. H. et al. Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A. J. Cell Biol. 161, 55–66 (2003).
Litvinchuk, A. et al. Complement C3aR inactivation attenuates tau pathology and reverses an immune network deregulated in tauopathy models and Alzheimer’s disease. Neuron 100, 1337–1353 e5 (2018).
Veerhuis, R., Nielsen, H. M. & Tenner, A. J. Complement in the brain. Mol. Immunol. 48, 1592–1603 (2011).
Kao, A. W., McKay, A., Singh, P. P., Brunet, A. & Huang, E. J. Progranulin, lysosomal regulation and neurodegenerative disease. Nat. Rev. Neurosci. 18, 325–333 (2017).
Alzahrani, F. et al. Recessive, deleterious variants in SMG8 expand the role of nonsense-mediated decay in developmental disorders in humans. Am. J. Hum. Genet 107, 1178–1185 (2020).
Brenneman, D. E., Hauser, J., Spong, C. Y. & Phillips, T. M. Chemokines released from astroglia by vasoactive intestinal peptide. Mechanism of neuroprotection from HIV envelope protein toxicity. Ann. NY Acad. Sci. 921, 109–114 (2000).
Brenneman, D. E. et al. Vasoactive intestinal peptide. Link between electrical activity and glia-mediated neurotrophism. Ann. NY Acad. Sci. 897, 17–26 (1999).
Chneiweiss, H., Glowinski, J. & Premont, J. Vasoactive intestinal polypeptide receptors linked to an adenylate cyclase, and their relationship with biogenic amine- and somatostatin-sensitive adenylate cyclases on central neuronal and glial cells in primary cultures. J. Neurochem. 44, 779–786 (1985).
Hosli, E. & Hosli, L. Autoradiographic localization of binding sites for vasoactive intestinal peptide and angiotensin II on neurons and astrocytes of cultured rat central nervous system. Neuroscience 31, 463–470 (1989).
Delgado, M. & Ganea, D. Vasoactive intestinal peptide: a neuropeptide with pleiotropic immune functions. Amino Acids 45, 25–39 (2013).
Baer, G. M. et al. Sporadic FTLD TDP type C is a distinct clinicopathological entity from FTLD types A/B (S7.007). Neurology 88, S7.007 (2017).
Rohrer, J. D. et al. The heritability and genetics of frontotemporal lobar degeneration. Neurology 73, 1451–1456 (2009).
Snowden, J., Neary, D. & Mann, D. Frontotemporal lobar degeneration: clinical and pathological relationships. Acta Neuropathol. 114, 31–38 (2007).
Neary, D. et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology 51, 1546–1554 (1998).
Younes, K. et al. Right temporal degeneration and socioemotional semantics: semantic behavioural variant frontotemporal dementia. Brain 145, 4080–4096 (2022).
Acocella, G. & Billing, B. H. The effect of rifamycin SV on pigment excretion in rats. Gastroenterology 49, 526–530 (1965).
Snowden, J. S. et al. Semantic dementia and the left and right temporal lobes. Cortex 107, 188–203 (2018).
Josephs, K. A. et al. Two distinct subtypes of right temporal variant frontotemporal dementia. Neurology 73, 1443–1450 (2009).
Kumfor, F. et al. On the right side? A longitudinal study of left-versus right-lateralized semantic dementia. Brain 139, 986–998 (2016).
Chan, D. et al. The clinical profile of right temporal lobe atrophy. Brain 132, 1287–1298 (2009).
Borghesani, V. et al. Regional and hemispheric susceptibility of the temporal lobe to FTLD-TDP type C pathology. Neuroimage Clin. 28, 102369 (2020).
Ulugut, H. et al. Clinical recognition of frontotemporal dementia with right anterior temporal predominance: a multicenter retrospective cohort study. Alzheimers Dement. 20, 5647–5661 (2024).
Pan, L. et al. Transcription factor RBPJL is able to repress notch target gene expression but is non-responsive to notch activation. Cancers 13, 5027 (2021).
Bonham, L. W. et al. Genetic variation across RNA metabolism and cell death gene networks is implicated in the semantic variant of primary progressive aphasia. Sci. Rep. 9, 10854 (2019).
Lu, J. et al. L3MBTL1 regulates ALS/FTD-associated proteotoxicity and quality control. Nat. Neurosci. 22, 875–886 (2019).
Guissart, C. et al. Premature termination codons in SOD1, causing amyotrophic lateral sclerosis, are predicted to escape the nonsense-mediated mRNA decay. Sci. Rep. 10, 20738 (2020).
Murley, A. G. & Rowe, J. B. Neurotransmitter deficits from frontotemporal lobar degeneration. Brain 141, 1263–1285 (2018).
Ferrer, I. Neurons and their dendrites in frontotemporal dementia. Dement. Geriatr. Cogn. Disord. 10, 55–60 (1999).
Premi, E. et al. Unravelling neurotransmitters impairment in primary progressive aphasias. Hum. Brain Mapp. 44, 2245–2253 (2023).
Bowen, D. M. et al. Imbalance of a serotonergic system in frontotemporal dementia: implication for pharmacotherapy. Psychopharmacology 196, 603–610 (2008).
Procter, A. W., Qurne, M. & Francis, P. T. Neurochemical features of frontotemporal dementia. Dement. Geriatr. Cogn. Disord. 10, 80–84 (1999).
Premi, E. et al. Early neurotransmitters changes in prodromal frontotemporal dementia: a GENFI study. Neurobiol. Dis. 179, 106068 (2023).
Ghezzi, L., Cantoni, C., Rotondo, E. & Galimberti, D. The gut microbiome-brain crosstalk in neurodegenerative diseases. Biomed. 10, 1486 (2022).
Khawar, M. M. Sr. et al. The gut-brain axis in autoimmune diseases: emerging insights and therapeutic implications. Cureus 15, e48655 (2023).
Miller, Z. A. et al. TDP-43 frontotemporal lobar degeneration and autoimmune disease. J. Neurol. Neurosurg. Psychiatry 84, 956–962 (2013).
Arseni, D. et al. Heteromeric amyloid filaments of ANXA11 and TDP-43 in FTLD-TDP type C. Nature 634, 662–668 (2024).
Arseni, D. et al. Structure of pathological TDP-43 filaments from ALS with FTLD. Nature 601, 139–143 (2022).
Arseni, D. et al. TDP-43 forms amyloid filaments with a distinct fold in type A FTLD-TDP. Nature 620, 898–903 (2023).
Brooks, B. R. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on motor neuron diseases/amyotrophic lateral sclerosis of the World Federation of Neurology research group on neuromuscular diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J. Neurol. Sci. 124, 96–107 (1994).
Olson, J. E. et al. The Mayo Clinic Biobank: a building block for individualized medicine. Mayo Clin. Proc. 88, 952–962 (2013).
McPeek, M. S. & Sun, L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am. J. Hum. Genet. 66, 1076–1094 (2000).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Lake, B. B. et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 36, 70–80 (2018).
Allen, M. et al. Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci. Data 3, 160089 (2016).
Mostafavi, S. et al. A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat. Neurosci. 21, 811–819 (2018).
Bennett, D. A. et al. Religious orders study and rush memory and aging project. J. Alzheimers Dis. 64, S161–S189 (2018).
Wang, M. et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci. Data 5, 180185 (2018).
Consortium, G. T. The GTEx consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Bryois, J. et al. Cell-type-specific cis-eQTLs in eight human brain cell types identify novel risk genes for psychiatric and neurological disorders. Nat. Neurosci. 25, 1104–1112 (2022).
Young, A. M. H. et al. A map of transcriptional heterogeneity and regulatory variation in human microglia. Nat. Genet. 53, 861–868 (2021).
Lopes, K. P. et al. Genetic analysis of the human microglial transcriptome across brain regions, aging and disease pathologies. Nat. Genet. 54, 4–17 (2022).
Wingo, A. P. et al. Integrating human brain proteomes with genome-wide association data implicates new proteins in Alzheimer’s disease pathogenesis. Nat. Genet. 53, 143–146 (2021).
Ng, B. et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 20, 1418–1426 (2017).
Alasoo, K. et al. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 50, 424–431 (2018).
Nedelec, Y. et al. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell 167, 657–669 e21 (2016).
Chen, L. et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell 167, 1398–1414 e24 (2016).
Momozawa, Y. et al. IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nat. Commun. 9, 2427 (2018).
Quach, H. et al. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell 167, 643–656 e17 (2016).
Fairfax, B. P. et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 (2014).
Kerimov, N. et al. A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nat. Genet. 53, 1290–1299 (2021).
Edgar, R. D., Jones, M. J., Meaney, M. J., Turecki, G. & Kobor, M. S. BECon: a tool for interpreting DNA methylation findings from blood in the context of brain. Transl. Psychiatry 7, e1187 (2017).
Mancuso, N. et al. Probabilistic fine-mapping of transcriptome-wide association studies. Nat. Genet. 51, 675–682 (2019).
Acknowledgements
We thank the Mayo Clinic Center of Individualized Medicine for collection and sequencing of the Mayo Clinic Biobank samples. CG would also like to acknowledge Dr Inger Nennesmo for the neuropathological characterization. Acknowledgment for the ADSP dataset can be found in Supplementary Notes. This work was made possible by the support of funding from the UG3/UH3 NS103870 (RR), P30 AG062677 (BFB), P01 AG003949 (DWD), P01 NS084974 (DWD), R01 AG037491 (KAJ), R01 DC012519 (JLW), R21 NS94684 (KAJ), R01 DC010367 (KAJ), P30 AG062422 (JSY, WWS, BLM, HJR), P01 AG019724 (DHG, BLM, ALB, WWS), U01 AG057195 (BCD), P30 AG077444 (MMM), P30 AG013854 (MMM, SW), P30 AG07297, P30 AG066468 (OLL, JK), P30 AG012300 (CLW), P30 AG072972 (JL-W), P30 AG010129 (JL-W), P50 AG005136 (DCK), P30 AG066509 (DCK), P30 AG066511 (JG), P01 AG066597 (MG), P30 AG072979 (EBL, VVD), R01 NS085770 (CG), R01 AG091388 (TG, CG, MMM), and R01 DC008552 (MMM). It was further supported by the ALLFTD Consortium (U19: AG063911, funded by the NIA and NINDS, to BFB, DWD, ALB) and the former ARTFL & LEFFTDS Consortia (ARTFL: U54 NS092089, to ALB, funded by the NINDS and NCATS; LEFFTDS: U01 AG045390, funded by the NIA and NINDS, to BFB). It was also supported by the Mayo ADRC (P30 AG62677, funded by the NIA, to RCP). A subset of samples was obtained through the National Centralized Repository for Alzheimer Disease and Related Dementias (NCRAD), which receives government support under a cooperative agreement grant (U24 AG021886) awarded by the National Institute on Aging (NIA), and was used in this study. ZKW was supported by Mayo Clinic Center for Regenerative Medicine, the gifts from the Donald G. and Jodi P. Heeringa Family, the Haworth Family Professorship in Neurodegenerative Diseases fund, the Albertson Parkinson’s Research Foundation, and PPND Family Foundation. This work was further supported by the Rainwater Charitable Foundation (DWD, BFB, WWS, DHG, JSY) and the Bluefield Project to Cure FTD (RR and JDR), the DEC Brain and BioBank (ECF), CIHR grant 74580 (IRM), the Canadian Consortium on Neurodegeneration in Aging, of the Canadian Institutes of Health Research (IRM), the University of Pittsburgh Brain Institute (JK). JDR was further supported by the NIHR UCL/H Biomedical Research Centre, the UK MRC (MR/M008525/1; MR/M023664/1), and the JPND (2019-02248). CLW was supported, in part, by the Winspear Family Center for Research on the Neuropathology of Alzheimer disease, the McCune Foundation, and the Nancy and Buster Alvord endowment. CG was supported by Grants provided by Swedish FTD Initiative- Schörling foundation, the Swedish Research Council (Dnr 521-2010-3134, 529-2014-7504, 2015-02926), Alzheimer Foundation Sweden, Brain Foundation Sweden, Swedish Brain Power, Gamla Tjänarinnor, Stohnes Foundation, Dementia Foundation Sweden, and the Stockholm County Council (ALF-project). The brain pathology was provided through in part by the Brain Bank at Karolinska Institutet, which was financially supported by Karolinska Institutet StratNeuro, Swedish Brain Power, Stockholm County Council core facility funding (CG), and Schörling Foundation. KS was supported in part by SAO-FRA 2021/0032. FK is supported by a postdoctoral fellowship (BOF 49758) from the University of Antwerp Research Fund and a postdoctoral fellowship from Brein Instituut. RR was supported by a grant from the Fund for Scientific Research, Flanders (G064223N).
Author information
Authors and Affiliations
Contributions
C.P., and R.R.: Study concept or design, acquisition of data, analysis or interpretation of data, statistical analysis or interpretation of data, drafting the manuscript for content, revising the manuscript for content, study supervision or coordination. M.B., F.K., A.B., G.D.J., J.M.B., C.L.D.: Sequencing, data analysis or interpretation of data, and drafting the manuscript for content, including writing of content. M.V.B., C.T.V., W.D.C., S.W., P.V.W., O.A.R., M.E.M., J.F., S.J.H., J.V.R., M.O.M., G.Y.R.H., C.G., L.Ö., M.N., Y.A., S.K.M., S.B., K.A.J., J.L.W.W., K.F.B., L.F., H.H., A.L.L., E.G.G., J.S.Y., A.P.O., M.F., Q.M., J.R.H., J.B.K., K.D.R., M.S., C.W., C.O., B.C.D., B.M.E., B.N.D., D.G.M., J.K., L.Z., E.R., E.S., T.G., C.G., S.W., J.D.S., M.R.F, D.E., B.K.W., R.J.C., L.L.K., E.D.H., E.M.R., S.M., A.K., S.R., A.L.N., N.E.T., D.S.K., R.C.P., L.P., R.J.U., Z.K.W., E.M.R., L.T.G., M.G.T., H.J.R., S.S., O.P., M.G., J.Q.T., D.C.K., J.L., J.P., D.H.G., R.A.R., C.C., B.G., G.M.H., T.G.B., G.E.S., T.A., J.H., A.L.B., L.S.H., J.P.V., O.L.L., J.K., C.L.W., M.G., J.G.l., J.D.R., D.J.I., E.B.L., V.V.D., R.C., M.M., M.C.T., E.C.F., C.T., S.A.F.S., B.L.M., H.S., N.G.R., B.F.B., I.R.A.M.K., J.C.V., W.W.S., K.S., and D.W.D.: Revised the manuscript for content, provided intellectual input and most shared samples. All authors: Read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.J.H. serves on the scientific advisory board (SAB) of Proximity Therapeutics, Psy Therapeutics, Frequency Therapeutics, Souvien Therapeutics, Sensorium Therapeutics, 4 M Therapeutics, Ilios Therapeutics, Entheos Labs, Alzheimer’s Drug Discovery Foundation, and the Kissick Family Foundation FTD Grant Program, none of whom were involved in the present study. S.J.H. has also received speaking or consulting fees from Amgen, AstraZeneca, Biogen, Merck, Regenacy Pharmaceuticals, Syros Pharmaceuticals, Juvenescence Life, as well as sponsored research or gift funding from AstraZeneca, JW Pharmaceuticals, Lexicon Pharmaceuticals, Vesigen Therapeutics, Compass Pathways, Atai Life Sciences, and Stealth Biotherapeutics. The funders had no role in the design or content of this article. ZKW serves as PI or Co-PI on Biohaven Pharmaceuticals, Inc. (BHV4157-206), Vigil Neuroscience, Inc. (VGL101-01.002, VGL101-01.201, PET tracer development protocol, Csf1r biomarker and repository project, and ultra-high field MRI in the diagnosis and management of CSF1R-related adult-onset leukoencephalopathy with axonal spheroids and pigmented glia), and ONO-2808-03 projects/grants. He serves as Co-PI of the Mayo Clinic APDA Center for Advanced Research and as an external advisory board member for the Vigil Neuroscience, Inc., and as a consultant for Eli Lilly & Company and for NovoGlia, Inc. R.R. and I.R.M. receive royalties from progranulin-related patent. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Oriol Dols-Icardo, Celeste Sassi, Emilia Vitale, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pottier, C., Küçükali, F., Baker, M. et al. Deciphering distinct genetic risk factors for FTLD-TDP pathological subtypes via whole-genome sequencing. Nat Commun 16, 3914 (2025). https://doi.org/10.1038/s41467-025-59216-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59216-0
This article is cited by
-
Divergent TDP43-regulated and TDP43-independent cryptic splicing in the cortex and spinal cord
Molecular Neurodegeneration Advances (2026)
-
Methylome analysis of FTLD patients with TDP-43 pathology identifies epigenetic signatures specific to pathological subtypes
Molecular Neurodegeneration (2025)